Patent application title: ENGINEERED MICROORGANISMS CAPABLE OF PRODUCING TARGET COMPOUNDS UNDER ANAEROBIC CONDITIONS
Inventors:
Thomas Buelter (Denver, CO, US)
Thomas Buelter (Denver, CO, US)
Peter Meinhold (Denver, CO, US)
Peter Meinhold (Denver, CO, US)
Reid M. Renny Feldman (Denver, CO, US)
Eva Eckl (Rohrbach, DE)
Andrew Hawkins (Parker, CO, US)
Andrew Hawkins (Parker, CO, US)
Aristos Aristidou (Highland Ranch, CO, US)
Catherine Asleson Dundon (Englewood, CO, US)
Catherine Asleson Dundon (Englewood, CO, US)
Doug Lies (Parker, CO, US)
Doug Lies (Parker, CO, US)
Sabine Bastian (Pasadena, CA, US)
Sabine Bastian (Pasadena, CA, US)
Frances Arnold (La Canada, CA, US)
Jun Urano (Aurora, CO, US)
Jun Urano (Aurora, CO, US)
IPC8 Class: AC12P716FI
USPC Class:
435160
Class name: Containing hydroxy group acyclic butanol
Publication date: 2010-06-10
Patent application number: 20100143997
Claims:
1. A recombinant microorganism comprising an engineered metabolic pathway
for producing isobutanol under aerobic and anaerobic conditions, wherein
said recombinant microorganism produces isobutanol under anaerobic
conditions at a rate higher than a parental microorganism comprising a
native or unmodified metabolic pathway.
2. The recombinant microorganism of claim 1, wherein said engineered metabolic pathway comprises an overexpressed transhydrogenase that converts NADH to NADPH.
3. The recombinant microorganism of claim 2, wherein said transhydrogenase is a membrane-bound transhydrogenase.
4. The recombinant microorganism of claim 3, wherein said membrane-bound transhydrogenase is encoded by the Escherichia coli pntAB genes.
5. The recombinant microorganism of claim 1, wherein said engineered metabolic pathway comprises an NADPH-dependent glyceraldehyde-3-phosphate dehydrogenase.
6. The recombinant microorganism of claim 5, wherein said NADPH-dependent glyceraldehyde-3-phosphate dehydrogenase is encoded by the Clostridium acetobutylicum gapC gene or the Kluyveromyces lactis GDP1 gene.
7. The recombinant microorganism of claim 1, wherein said engineered metabolic pathway comprises one or more enzymes catalyzing conversions in said engineered metabolic pathway that are not catalyzed by glyceraldehyde-3-phosphate dehydrogenase, and wherein said one or more enzymes have increased activity using NADH as a cofactor.
8. The recombinant microorganism of claim 7, wherein said engineered metabolic pathway comprises genes encoding an NADH-dependent ketol-acid reductoisomerase (KARI) and an NADH-dependent alcohol dehydrogenase (ADH).
9. The recombinant microorganism of claim 8, wherein said KARI and/or said ADH are identified in nature with increased activity using NADH as a cofactor as compared to the wild-type E. coli KARI llvC and a native E. coli ADH YqhD, respectively.
10. The recombinant microorganism of claim 9, wherein said KARI and/or said ADH show at least a 10-fold higher catalytic efficiency using NADH as the cofactor as compared to the wild-type E. coli KARI llvC and a native E. coli ADH YqhD, respectively.
11. The recombinant microorganism of claim 8, wherein said KARI and/or said ADH have been modified or mutated to have increased activity using NADH as a cofactor as compared to the wild-type E. coli KARI llvC and a native E. coli ADH YqhD, respectively.
12. The recombinant microorganism of claim 11, wherein said KARI and/or said ADH show at least a 10-fold higher catalytic efficiency using NADH as the cofactor as compared to the wild-type E. coli KARI llvC and a native E. coli ADH YqhD, respectively.
13. The recombinant microorganism of claim 11, wherein said KARI and/or said ADH have been modified or mutated to be NADH-dependent.
14. The recombinant microorganism of claim 8, wherein said KARI enhances the recombinant microorganism's ability to convert acetolactate to 2,3-dihydroxyisovalerate under anaerobic conditions.
15. The recombinant microorganism of claim 8, wherein said KARI enhances the recombinant microorganism's ability to utilize NADH for the conversion of acetolactate to 2,3-dihydroxyisovalerate.
16. The recombinant microorganism of claim 11, wherein said KARI comprises two or more mutations or modifications at positions corresponding to amino acids selected from the group consisting of: (a) alanine 71 of the wild-type E. coli llvC (SEQ ID NO 13); (b) arginine 76 of the wild-type E. coli llvC; (c) serine 78 of the wild-type E. coli llvC; and (d) glutamine 110 of the wild-type E. coli llvC.
17. The recombinant microorganism of claim 16, wherein said alanine 71 residue of said KARI is replaced with a serine residue, said arginine 76 residue is replaced with an aspartic acid residue, said serine 78 residue is replaced with an aspartic acid residue, and said glutamine 110 residue is replaced with a valine residue.
18. The recombinant microorganism of claim 16, wherein said KARI has at least about a 25% increased catalytic efficiency with NADH as compared to the wild-type KAR1.
19. The recombinant microorganism of claim 16, wherein the catalytic efficiency of the KARI with NADH is at least about 25% of the catalytic efficiency with NADPH of the wild-type KAR1.
20. The recombinant microorganism of claim 16, wherein the KARI preferentially utilizes NADH rather than NADPH.
21. The recombinant microorganism of claim 16, wherein the KARI demonstrates a switch in cofactor preference from NADPH to NADH as compared to a corresponding wild-type KAR1.
22. The recombinant microorganism of claim 16, wherein the KARI exhibits at least about a 1:1 ratio of catalytic efficiency (kcat/KM) with NADH over catalytic efficiency with NADPH.
23. The recombinant microorganism of claim 16, wherein the KARI exhibits at least about a 1:10 ratio of KM for NADH over KM for NADPH.
24. The recombinant microorganism of claim 16, wherein the KARI is selected from the group consisting of Escherichia coli (GenBank No: NP--418222, SEQ ID NO 13), Saccharomyces cerevisiae (GenBank No: NP--013459, SEQ ID NO: 70), Methanococcus maripaludis (GenBank No: YP--001097443, SEQ ID NO: 71), Bacillus subtilis (GenBank Nos: CAB14789, SEQ ID NO: 72), Piromyces sp (GenBank No: CAA76356, SEQ ID NO: 73), Buchnera aphidicola (GenBank No: AAF13807, SEQ ID NO: 74), Spinacia oleracea (GenBank Nos: Q01292 and CAA40356, SEQ ID NO: 75), Oryza sativa (GenBank No: NP--001056384, SEQ ID NO: 76) Chlamydomonas reinhardtii (GenBank No: XP--001702649, SEQ ID NO: 77), Neurospora crassa (GenBank No: XP--961335, SEQ ID NO: 78), Schizosaccharomyces pombe (GenBank No: NP--001018845, SEQ ID NO: 79), Laccaria bicolor (GenBank No: XP--001880867, SEQ ID NO: 80), Ignicoccus hospitalis (GenBank No: YP--001435197, SEQ ID NO: 81), Picrophilus torridus (GenBank No: YP--023851, SEQ ID NO: 82), Acidiphilium cryptum (GenBank No: YP--001235669, SEQ ID NO: 83), Cyanobacteria/Synechococcus sp. (GenBank No: YP--473733, SEQ ID NO: 84), Zymomonas mobilis (GenBank No: YP--162876, SEQ ID NO: 85), Bacteroides thetaiotaomicron (GenBank No: NP--810987, SEQ ID NO: 86), Vibrio fischeri (GenBank No: YP--205911, SEQ ID NO: 87), Shewanella sp (GenBank No: YP--732498, SEQ ID NO: 88), Gramella forsetti (GenBank No: YP--862142, SEQ ID NO: 89), Psychromonas ingrhamaii (GenBank No: YP--942294, SEQ ID NO: 90), and Cytophaga hutchinsonii (GenBank No: YP--677763, SEQ ID NO: 91).
25. The recombinant microorganism of claim 16, wherein the KARI is derived from a genus selected from the group consisting of Escherichia, Zymomonas, Staphylococcus, Corynebacterium, Clostridium, Salmonella, Pseudomonas, Bacillus, Lactobacillus, Lactococcus, Enterobactor, Enterococcus, Klebsiella, Saccharomyces, Kluyveromyces, Pichia, Hansenula, Candida, Trichosporon, Yamadazyma, Schizosaccharomyces, Cryptococcus, Aspergillus, Neurospora, Piromyces, Orpinomyces, and Neocallimastix, Piromyces, Buchnera, Spinacia, Oryza, Chlamydomonas, Neurospora, Schizosaccharomyces, Laccaria, Ignicoccus, Picrophilus, Acidiphilium, Cyanobacteria/Synechococcus, Zymomonas, Bacteroides, Methanococcus, Vibrio, Shewanella, Gramella, Psychromonas, and Cytophaga.
26. The recombinant microorganism of claim 16, wherein the KARI has further been codon optimized for expression in a host cell, and wherein said host cell is yeast.
27. The recombinant microorganism of claim 16, wherein the KARI is selected from the group consisting of SEQ ID NO: 17, SEQ ID NO: 19, SEQ ID NO: 23, SEQ ID NO: 25, SEQ ID NO: 34, SEQ ID NO: 36, SEQ ID NO: 38, SEQ ID NO: 40, SEQ ID NO: 42 and SEQ ID NO: 44.
28. The recombinant microorganism of claim 1, wherein said engineered metabolic pathway comprises a first dehydrogenase and a second dehydrogenase that catalyze the same reaction, and wherein the first dehydrogenase is NADH-dependent and wherein the second dehydrogenase is NADPH dependent.
29. The recombinant microorganism of claim 28, wherein said first dehydrogenase is encoded by the E. coli gene maeA and the second dehydrogenase is encoded by the E. coli gene maeB or wherein said first dehydrogenase is encoded by the E. coli gene maeA and the second dehydrogenase is encoded by the S. cerevisiae gene MAE1.
30. The recombinant microorganism of claim 1, wherein said engineered metabolic pathway comprises a replacement of a gene encoding for pyk or homologs thereof with a gene encoding for ppc or pck or homologs thereof.
31. The recombinant microorganism of claim 30, wherein said engineered metabolic pathway further comprises the overexpression of the genes mdh and maeB or wherein said engineered metabolic pathway further comprises the overexpression of the S. cerevisiae genes MDH1 and MAE1.
32. A recombinant microorganism according to claim 1, wherein said recombinant microorganism is selected from GEVO1846, GEVO1886, GEVO1993, GEVO2158, GEVO2302, GEVO1803, GEVO2107, GEVO2710, GEVO2711, GEVO2712, GEVO2799, GEVO2847, GEVO2848, GEVO2849, GEVO2851, GEVO2852, GEVO2854, GEVO2855 and GEVO2856.
33. The recombinant microorganism of claim 1, wherein said recombinant microorganism produces said isobutanol under anaerobic conditions at a yield which is at least about the same yield as under aerobic conditions.
34. The recombinant microorganism of claim 1, wherein said recombinant microorganism produces isobutanol at substantially the same rate under anaerobic conditions as the parental microorganism produces under aerobic conditions.
35. The recombinant microorganism of claim 1, wherein said engineered metabolic pathway is balanced with respect to NADH and NADPH as compared to a native or unmodified metabolic pathway from a corresponding parental microorganism, and wherein said native or unmodified metabolic pathway is not balanced with respect to NADH and NADPH.
36. A method of producing isobutanol under anaerobic conditions, comprising:(a) providing a recombinant microorganism according to claim 1;(b) cultivating the recombinant microorganism under anaerobic conditions in a culture medium containing a feedstock providing the carbon source, until a recoverable quantity of isobutanol is produced; and(c) recovering isobutanol.
37. The method according to claim 36, wherein the recombinant microorganism is selected from:(i) E. coli that produces isobutanol at a yield of greater than 80% theoretical; and(ii) Yeast that produces isobutanol at a yield of greater than 30% theoretical.
38. The method according to claim 36, wherein isobutanol is produced under anaerobic conditions at a yield which is at least about the same yield as under aerobic conditions.
39. A mutant ketol-acid reductoisomerase (KARI) comprising two or more mutations or modifications at positions corresponding to amino acids selected from the group consisting of: (a) alanine 71 of the wild-type E. coli llvC (SEQ ID NO: 13); (b) arginine 76 of the wild-type E. coli llvC; (c) serine 78 of the wild-type E. coli llvC; and (d) glutamine 110 of the wild-type E. coli llvC.
Description:
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001]This application claims priority to U.S. Provisional Application Ser. No. 61/110,543, filed Oct. 31, 2008; U.S. Provisional Application Ser. No. 61/121,830, filed Dec. 11, 2008; U.S. Provisional Application Ser. No. 61/184,580, filed Jun. 5, 2009; U.S. Provisional Application Ser. No. 61/184,605, filed Jun. 5, 2009; and U.S. Provisional Application Ser. No. 61/239,618, filed Sep. 3, 2009. This application is related to U.S. patent application Ser. No. 12/263,442, entitled "Methods for the Economical Production of Biofuel Precursors that is also a Biofuel from Biomass," filed Oct. 31, 2008. This application is also related to the U.S. patent application Ser. No. 12/263,436, entitled "Methods for the Economical Production of Biofuel from Biomass," filed Oct. 31, 2008. Accordingly, this application incorporates by reference in its entirety all subject matter of the above-referenced applications to the extent such subject matter is not inconsistent herewith.
FIELD OF THE INVENTION
[0003]The present invention is generally related to genetically engineered microorganisms, methods of producing such organisms, and methods of using such organisms for the production of beneficial metabolites, including C3-C5 alcohols such as isobutanol.
BACKGROUND
[0004]Biofuels have a long history ranging back to the beginning of the 20th century. As early as 1900, Rudolf Diesel demonstrated at the World Exhibition in Paris, France, an engine running on peanut oil. Soon thereafter, Henry Ford demonstrated his Model T running on ethanol derived from corn. Petroleum-derived fuels displaced biofuels in the 1930s and 1940s due to increased supply, and efficiency at a lower cost.
[0005]Market fluctuations in the 1970s coupled to the decrease in US oil production led to an increase in crude oil prices and a renewed interest in biofuels. Today, many interest groups, including policy makers, industry planners, aware citizens, and the financial community, are interested in substituting petroleum-derived fuels with biomass-derived biofuels. The leading motivations for developing biofuels are of economical, political, and environmental nature.
[0006]One is the threat of `peak oil`, the point at which the consumption rate of crude oil exceeds the supply rate, thus leading to significantly increased fuel cost results in an increased demand for alternative fuels. In addition, instability in the Middle East and other oil-rich regions has increased the demand for domestically produced biofuels. Also, environmental concerns relating to the possibility of carbon dioxide related climate change is an important social and ethical driving force which is starting to result in government regulations and policies such as caps on carbon dioxide emissions from automobiles, taxes on carbon dioxide emissions, and tax incentives for the use of biofuels.
[0007]Ethanol is the most abundant biofuel today but has several drawbacks when compared to gasoline. Butanol, in comparison, has several advantages over ethanol as a fuel: it can be made from the same feedstocks as ethanol but, unlike ethanol, it is compatible with gasoline at any ratio and can also be used as a pure fuel in existing combustion engines without modifications. Unlike ethanol, butanol does not absorb water and can thus be stored and distributed in the existing petrochemical infrastructure. Due to its higher energy content which is close to that of gasoline, the fuel economy (miles per gallon) is better than that of ethanol. Also, butanol-gasoline blends have lower vapor pressure than ethanol-gasoline blends, which is important in reducing evaporative hydrocarbon emissions.
[0008]Isobutanol has the same advantages as butanol with the additional advantage of having a higher octane number due to its branched carbon chain. Isobutanol is also useful as a commodity chemical. For example, it is used as the starting material in the manufacture of isobutyl acetate, which is primarily used for the production of lacquer and similar coatings. In addition, isobutanol finds utility in the industrial synthesis of derivative esters. Isobutyl esters such as diisobutyl phthalate (DIBP) are used as plasticizer agents in plastics, rubbers, and other dispersions.
[0009]A number of recent publications have described methods for the production of industrial chemicals such as isobutanol using engineered microorganisms. See, e.g., WO/2007/050671 to Donaldson et al., and WO/2008/098227 to Liao et al., which are herein incorporated by reference in their entireties. These publications disclose recombinant microorganisms that utilize a series of heterologously expressed enzymes to convert sugars into isobutanol. However, the production of isobutanol using these microorganisms is feasible only under aerobic conditions and the maximum yield that can be achieved is limited.
[0010]There is a need, therefore, to provide modified microorganisms capable of producing isobutanol under anaerobic conditions and at close to theoretical yield. The present invention addresses this need by providing modified microorganisms capable of producing isobutanol under anaerobic conditions and at high yields.
SUMMARY OF THE INVENTION
[0011]The present invention provides recombinant microorganisms comprising an engineered metabolic pathway capable of producing one or more C3-C5 alcohols under aerobic and anaerobic conditions. In a preferred embodiment, the recombinant microorganism produces the C3-C5 alcohol under anaerobic conditions at a rate higher than a parental microorganism comprising a native or unmodified metabolic pathway. In another preferred embodiment, the recombinant microorganism produces the C3-C5 alcohol under anaerobic conditions at a rate of at least about 2-fold higher than a parental microorganism comprising a native or unmodified metabolic pathway. In another preferred embodiment, the recombinant microorganism produces the C3-C5 alcohol under anaerobic conditions at a rate of at least about 10-fold, of at least about 50-fold, or of at least about 100-fold higher than a parental microorganism comprising a native or unmodified metabolic pathway.
[0012]In various embodiments described herein, the C3-C5 alcohol may be selected from 1-propanol, 2-propanol, 1-butanol, 2-butanol, isobutanol, 2-methyl-1-butanol, 3-methyl-1-butanol, and 1-pentanol. In a preferred embodiment, the C3-C5 alcohol is isobutanol. In another preferred embodiment, isobutanol is produced at a specific productivity of at least about 0.025 gl-1 h-1 OD-1.
[0013]In one aspect, there are provided recombinant microorganisms comprising an engineered metabolic pathway for producing one or more C3-C5 alcohols under anaerobic and aerobic conditions that comprises an overexpressed transhydrogenase that converts NADH to NADPH. In one embodiment, the transhydrogenase is a membrane-bound transhydrogenase. In a specific embodiment, the membrane-bound transhydrogenase is encoded by the E. coli pntAB genes or homologues thereof.
[0014]In another aspect, there are provided recombinant microorganisms comprising an engineered metabolic pathway for producing one or more C3-C5 alcohols under anaerobic and aerobic conditions that comprises an NADPH-dependent glyceraldehyde-3-phosphate dehydrogenase. In one embodiment, the NADPH-dependent glyceraldehyde-3-phosphate dehydrogenase is encoded by the Clostridium acetobutylicum gapC gene. In another embodiment, the NADPH-dependent glyceraldehyde-3-phosphate dehydrogenase is encoded by the Kluyveromyces lactis GDP1 gene.
[0015]In yet another aspect, there are provided recombinant microorganisms comprising an engineered metabolic pathway for producing one or more C3-C5 alcohols under anaerobic and aerobic conditions that comprises one or more enzymes catalyzing conversions in said engineered metabolic pathway that are not catalyzed by glyceraldehyde-3-phosphate dehydrogenase, and wherein said one or more enzymes have increased activity using NADH as a cofactor. In one embodiment, said one or more enzymes are selected from an NADH-dependent ketol-acid reductoisomerase (KARI) and an NADH-dependent alcohol dehydrogenase (ADH). In various embodiments described herein, the KARI and/or ADH enzymes may be engineered to have increased activity with NADH as the cofactor as compared to the wild-type E. coli KARI llvC and a native E. coli ADH YqhD, respectively. In some embodiments, the KARI and/or the ADH are modified or mutated to be NADH-dependent. In other embodiments, the KARI and/or ADH enzymes are identified in nature with increased activity with NADH as the cofactor as compared to the wild-type E. coli KARI llvC and a native E. coli ADH YqhD, respectively.
[0016]In various embodiments described herein, the KARI and/or ADH may show at least a 10-fold higher catalytic efficiency using NADH as a cofactor as compared to the wild-type E. coli KARI llvC and the native ADH YqhD, respectively. In a preferred embodiment, the KARI enhances the recombinant microorganism's ability to convert acetolactate to 2,3-dihydroxyisovalerate under anaerobic conditions. In another embodiment, the KARI enhances the recombinant microorganism's ability to utilize NADH from the conversion of acetolactate to 2,3-dihydroxyisovalerate.
[0017]The present invention also provides modified or mutated KARI enzymes that preferentially utilize NADH rather than NADPH, and recombinant microorganisms comprising said modified or mutated KARI enzymes. In general, these modified or mutated KARI enzymes may enhance the cell's ability to produce beneficial metabolites such as isobutanol and enable the production of beneficial metabolites such as isobutanol under anaerobic conditions.
[0018]In certain aspects, the invention includes KARIs which have been modified or mutated to increase the ability to utilize NADH. Examples of such KARIs include enzymes having one or more modifications or mutations at positions corresponding to amino acids selected from the group consisting of: (a) alanine 71 of the wild-type E. coli llvC (SEQ ID NO: 13); (b) arginine 76 of the wild-type E. coli NC; (c) serine 78 of the wild-type E. coli llvC; and (d) glutamine 110 of the wild-type E. coli llvC, wherein llvC (SEQ ID NO: 13) is encoded by codon optimized E. coli ketol-acid reductoisomerase (KARI) genes Ec_ilvC_coEc (SEQ ID NO: 11) or Ec_ilvC_coSc (SEQ ID NO: 12).
[0019]In one embodiment, the KARI enzyme contains a modification or mutation at the amino acid corresponding to position 71 of the wild-type E. coli llvC (SEQ ID NO: 13). In another embodiment, the KARI enzyme contains a modification or mutation at the amino acid corresponding to position 76 of the wild-type E. coli llvC (SEQ ID NO: 13). In yet another embodiment, the KARI enzyme contains a modification or mutation at the amino acid corresponding to position 78 of the wild-type E. coli llvC (SEQ ID NO: 13). In yet another embodiment, the KARI enzyme contains a modification or mutation at the amino acid corresponding to position 110 of the wild-type E. coli llvC (SEQ ID NO: 13).
[0020]In one embodiment, the KARI enzyme contains two or more modifications or mutations at the amino acids corresponding to the positions described above. In another embodiment, the KARI enzyme contains three or more modifications or mutations at the amino acids corresponding to the positions described above. In yet another embodiment, the KARI enzyme contains four modifications or mutations at the amino acids corresponding to the positions described above.
[0021]In one specific embodiment, the invention is directed to KARI enzymes wherein the alanine at position 71 is replaced with serine. In another specific embodiment, the invention is directed to KARI enzymes wherein the arginine at position 76 is replaced with aspartic acid. In yet another specific embodiment, the invention is directed to KARI enzymes wherein the serine at position 78 is replaced with aspartic acid. In yet another specific embodiment, the invention is directed to KARI enzymes wherein the glutamine at position 110 is replaced with valine. In yet another specific embodiment, the invention is directed to KARI enzymes wherein the glutamine at position 110 is replaced with alanine. In certain embodiments, the KARI enzyme contains two or more modifications or mutations at the amino acids corresponding to the positions described in these specific embodiments. In certain other embodiments, the KARI enzyme contains three or more modifications or mutations at the amino acids corresponding to the positions described in these specific embodiments. In an exemplary embodiment, the KARI enzyme contains four modifications or mutations at the amino acids corresponding to the positions described in these specific embodiments. In additional embodiments described herein, the KARI may further comprise an amino acid substitution at position 68 of the wild-type E. coli llvC (SEQ ID NO: 13).
[0022]In one embodiment, the modified or mutated KARI is selected from group consisting of SEQ ID NO: 17, SEQ ID NO: 19, SEQ ID NO: 23, SEQ ID NO: 25, SEQ ID NO: 34, SEQ ID NO: 36, SEQ ID NO: 38, SEQ ID NO: 40, SEQ ID NO: 42 and SEQ ID NO: 44.
[0023]Further included within the scope of the invention are KARI enzymes, other than the E. coli llvC (SEQ ID NO: 13), which contain alterations corresponding to those set out above. Such KARI enzymes may include, but are not limited to, the KARI enzymes encoded by the S. cerevisiae ILV5 gene, the KARI enzyme encoded by the E. coli ilvC gene and the KARI enzymes from Piromyces sp., Buchnera aphidicola, Spinacia oleracea, Oryza sativa, Chlamydomonas reinhardtii, Neurospora crassa, Schizosaccharomyces pombe, Laccaria bicolor, Ignicoccus hospitalis, Picrophilus torridus, Acidiphilium cryptum, Cyanobacteria/Synechococcus sp., Zymomonas mobilis, Bacteroides thetaiotaomicron, Methanococcus maripaludis, Vibrio fischeri, Shewanella sp, Gramella forsetti, Psychromonas ingrhamaii, and Cytophaga hutchinsonii.
[0024]In certain exemplary embodiments, the KARI to be modified or mutated is a KARI selected from the group consisting of Escherichia coli (GenBank No.: NP--418222, SEQ ID NO 13), Saccharomyces cerevisiae (GenBank No: NP--013459, SEQ ID NO: 70), Methanococcus maripaludis (GenBank No: YP--001097443, SEQ ID NO: 71), Bacillus subtilis (GenBank Nos: CAB14789, SEQ ID NO: 72), Piromyces sp (GenBank No: CAA76356, SEQ ID NO: 73), Buchnera aphidicola (GenBank No: AAF13807, SEQ ID NO: 74), Spinacia oleracea (GenBank Nos: Q1292 and CAA40356, SEQ ID NO: 75), Otyza sativa (GenBank No: NP--001056384, SEQ ID NO: 76) Chlamydomonas reinhardtii (GenBank No: XP--001702649, SEQ ID NO: 77), Neurospora crassa (GenBank No: XP--961335, SEQ ID NO: 78), Schizosaccharomyces pombe (GenBank No: NP--001018845, SEQ ID NO: 79), Laccaria bicolor (GenBank No: XP--001880867, SEQ ID NO: 80), Ignicoccus hospitalis (GenBank No: YP--001435197, SEQ ID NO: 81), Picrophilus torridus (GenBank No: YP--023851, SEQ ID NO: 82), Acidiphilium cryptum (GenBank No: YP--001235669, SEQ ID NO: 83), Cyanobacteria/Synechococcus sp. (GenBank No: YP--473733, SEQ ID NO: 84), Zymomonas mobilis (GenBank No: YP--162876, SEQ ID NO: 85), Bacteroides thetaiotaomicron (GenBank No: NP--810987, SEQ ID NO: 86), Vibrio fischeri (GenBank No: YP--205911, SEQ ID NO: 87), Shewanella sp (GenBank No: YP--732498, SEQ ID NO: 88), Gramella forsetti (GenBank No: YP--862142, SEQ ID NO: 89), Psychromonas ingrhamaii (GenBank No: YP--942294, SEQ ID NO: 90), and Cytophaga hutchinsonii (GenBank No: YP--677763, SEQ ID NO: 91).
[0025]In various embodiments described herein, the modified or mutated KARI may exhibit an increased catalytic efficiency with NADH as compared to the wild-type KARI. In one embodiment, the KARI has at least about a 5% increased catalytic efficiency with NADH as compared to the wild-type KARI. In another embodiment, the KARI has at least about a 25%, at least about a 50%, at least about a 75%, or at least about a 100% increased catalytic efficiency with NADH as compared to the wild-type KARI.
[0026]In some embodiments described herein, the catalytic efficiency of the modified or mutated KARI with NADH is increased with respect to the catalytic efficiency with NADPH of the wild-type KARI. In one embodiment, the catalytic efficiency of said KARI with NADH is at least about 10% of the catalytic efficiency with NADPH of the wild-type KARI. In another embodiment, the catalytic efficiency of said KARI with NADH is at least about 25%, at least about 50%, or at least about 75% of the catalytic efficiency with NADPH of the wild-type KARI. In some embodiments, the modified or mutated KARI preferentially utilizes NADH rather than NADPH.
[0027]In one embodiments, the invention is directed to modified or mutated KARI enzymes that demonstrate a switch in cofactor preference from NADPH to NADH. In one embodiment, the modified or mutated KARI has at least about a 2:1 ratio of kcat with NADH over kcat with NADPH. In an exemplary embodiment, the modified or mutated KARI has at least about a 10:1 ratio of kcat with NADH over kcat with NADPH.
[0028]In one embodiments, the invention is directed to a modified or mutated KARI enzyme that exhibits at least about a 1:10 ratio of catalytic efficiency (kcat/KM) with NADH over catalytic efficiency with NADPH. In another embodiment, the modified or mutated KARI enzyme exhibits at least about a 1:1 ratio of catalytic efficiency (kcat/KM) with NADH over catalytic efficiency with NADPH. In yet another embodiment, the modified or mutated KARI enzyme exhibits at least about a ratio of catalytic efficiency (kcat/KM) with NADH over catalytic efficiency with NADPH. In an exemplary embodiment, the modified or mutated KARI enzyme exhibits at least about a 100:1 ratio of catalytic efficiency (kcat/KM) with NADH over catalytic efficiency with NADPH.
[0029]In some embodiments, the modified or mutated KARI has been modified to be NADH-dependent. In one embodiment, the KARI exhibits at least about a 1:10 ratio of KM for NADH over KM for NADPH.
[0030]In additional embodiments, the invention is directed to modified or mutated KARI enzymes that have been codon optimized for expression in certain desirable host organisms, such as yeast and E. coli. In other aspects, the present invention is directed to recombinant host cells (e.g. recombinant microorganisms) comprising a modified or mutated KARI enzyme of the invention. According to this aspect, the present invention is also directed to methods of using the modified or mutated KARI enzymes in any fermentation process where the conversion of acetolactate to 2,3-dihydroxyisovalerate is desired. In one embodiment according to this aspect, the modified or mutated KARI enzymes may be suitable for enhancing a host cell's ability to produce isobutanol and enable the production of isobutanol under anaerobic conditions. In another embodiment according to this aspect, the modified or mutated KARI enzymes may be suitable for enhancing a host cell's ability to produce 3-methyl-1-butanol.
[0031]According to this aspect, the present invention is also directed to methods of using the modified or mutated KARI enzymes in any fermentation process where the conversion of 2-aceto-2-hydroxy-butyrate to 2,3-dihydroxy-3-methylvalerate is desired. In one embodiment according to this aspect, the modified or mutated KARI enzymes may be suitable for enhancing a host cell's ability to produce 2-methyl-1-butanol and enable the production of 2-methyl-1-butanol under anaerobic conditions.
[0032]In another aspect, there are provided recombinant microorganisms comprising an engineered metabolic pathway for producing one or more C3-C5 alcohols under anaerobic conditions, wherein said engineered metabolic pathway comprises a first dehydrogenase and a second dehydrogenase that catalyze the same reaction, and wherein the first dehydrogenase is NADH-dependent and wherein the second dehydrogenase is NADPH dependent. In an exemplary embodiment, the first dehydrogenase is encoded by the E. coli gene maeA and the second dehydrogenase is encoded by the E. coli gene maeB.
[0033]In another aspect, there are provided recombinant microorganisms comprising an engineered metabolic pathway for producing one or more C3-C5 alcohols under anaerobic conditions, wherein said engineered metabolic pathway comprises a replacement of a gene encoding for pyk or homologs thereof with a gene encoding for ppc or pck or homologs thereof. In another embodiment, the engineered metabolic pathway may further comprise the overexpression of the genes mdh and maeB.
[0034]In various embodiments described herein, the recombinant microorganisms may further be engineered to express an isobutanol producing metabolic pathway comprising at least one exogenous gene that catalyzes a step in the conversion of pyruvate to isobutanol. In one embodiment, the recombinant microorganism may be engineered to express an isobutanol producing metabolic pathway comprising at least two exogenous genes. In another embodiment, the recombinant microorganism may be engineered to express an isobutanol producing metabolic pathway comprising at least three exogenous genes. In another embodiment, the recombinant microorganism may be engineered to express an isobutanol producing metabolic pathway comprising at least four exogenous genes. In another embodiment, the recombinant microorganism may be engineered to express an isobutanol producing metabolic pathway comprising five exogenous genes.
[0035]In various embodiments described herein, the isobutanol pathway enzyme(s) may be selected from the group consisting of acetolactate synthase (ALS), ketol-acid reductoisomerase (KARI), dihydroxyacid dehydratase (DHAD), 2-keto-acid decarboxylase (KIVD), and alcohol dehydrogenase (ADH).
[0036]In another embodiment, the recombinant microorganism further comprises a pathway for the fermentation of isobutanol from a pentose sugar. In one embodiment, the pentose sugar is xylose. In one embodiment, the recombinant microorganism is engineered to express a functional xylose isomerase (XI). In another embodiment, the recombinant microorganism further comprises a deletion or disruption of a native gene encoding for an enzyme that catalyzes the conversion of xylose to xylitol. In one embodiment, the native gene is xylose reductase (XR). In another embodiment, the native gene is xylitol dehydrogenase (XDH). In yet another embodiment, both native genes are deleted or disrupted. In yet another embodiment, the recombinant microorganism is engineered to express a xylulose kinase enzyme.
[0037]In another embodiment, the recombinant microorganisms of the present invention may further be engineered to include reduced pyruvate decarboxylase (PDC) activity as compared to a parental microorganism. In one embodiment, PDC activity is eliminated. PDC catalyzes the decarboxylation of pyruvate to acetaldehyde, which is reduced to ethanol by alcohol dehydrogenases via the oxidation of NADH to NAD+. In one embodiment, the recombinant microorganism includes a mutation in at least one PDC gene resulting in a reduction of PDC activity of a polypeptide encoded by said gene. In another embodiment, the recombinant microorganism includes a partial deletion of a PDC gene resulting in a reduction of PDC activity of a polypeptide encoded by said gene. In another embodiment, the recombinant microorganism comprises a complete deletion of a PDC gene resulting in a reduction of PDC activity of a polypeptide encoded by said gene. In yet another embodiment, the recombinant microorganism includes a modification of the regulatory region associated with at least one PDC gene resulting in a reduction of PDC activity of a polypeptide encoded by said gene. In yet another embodiment, the recombinant microorganism comprises a modification of the transcriptional regulator resulting in a reduction of PDC gene transcription. In yet another embodiment, the recombinant microorganism comprises mutations in all PDC genes resulting in a reduction of PDC activity of the polypeptides encoded by said genes.
[0038]In another embodiment, the recombinant microorganisms of the present invention may further be engineered to include reduced glycerol-3-phosphate dehydrogenase (GPD) activity as compared to a parental microorganism. In one embodiment, GPD activity is eliminated. GPD catalyzes the reduction of dihydroxyacetone phosphate (DHAP) to glycerol-3-phosphate (G3P) via the oxidation of NADH to NAD.sup.+. Glycerol is produced from G3P by Glycerol-3-phosphatase (GPP). In one embodiment, the recombinant microorganism includes a mutation in at least one GPD gene resulting in a reduction of GPD activity of a polypeptide encoded by said gene. In another embodiment, the recombinant microorganism includes a partial deletion of a GPD gene resulting in a reduction of GPD activity of a polypeptide encoded by the gene. In another embodiment, the recombinant microorganism comprises a complete deletion of a GPD gene resulting in a reduction of GPD activity of a polypeptide encoded by the gene. In yet another embodiment, the recombinant microorganism includes a modification of the regulatory region associated with at least one GPD gene resulting in a reduction of GPD activity of a polypeptide encoded by said gene. In yet another embodiment, the recombinant microorganism comprises a modification of the transcriptional regulator resulting in a reduction of GPD gene transcription. In yet another embodiment, the recombinant microorganism comprises mutations in all GPD genes resulting in a reduction of GPD activity of a polypeptide encoded by the gene.
[0039]In various embodiments described herein, the recombinant microorganisms of the invention may produce one or more C3-C5 alcohols under anaerobic conditions at a yield which is at least about the same yield as under aerobic conditions. In additional embodiments described herein, the recombinant microorganisms of the invention may produce one or more C3-C5 alcohols at substantially the same rate under anaerobic conditions as the parental microorganism produces under aerobic conditions. In the various embodiments described herein, the engineered metabolic pathway may be balanced with respect to NADH and NADPH as compared to a native or unmodified metabolic pathway from a corresponding parental microorganism, wherein the native or unmodified metabolic pathway is not balanced with respect to NADH and NADPH.
[0040]In another aspect, the present invention provides a method of producing a C3-C5 alcohol, comprising (a) providing a recombinant microorganism comprising an engineered metabolic pathway capable of producing one or more C3-C5 alcohols under aerobic and anaerobic conditions; (b) cultivating the recombinant microorganism in a culture medium containing a feedstock providing the carbon source, until a recoverable quantity of the C3-C5 alcohol is produced; and (c) recovering the C3-C5 alcohol. In one embodiment, the recombinant microorganism is cultured under anaerobic conditions. In a preferred embodiment, the C3-C5 alcohol is produced under anaerobic conditions at a yield which is at least about the same yield as under aerobic conditions.
[0041]In various embodiments described herein, a preferred C3-C5 alcohol is isobutanol. In one embodiment, the microorganism produces isobutanol from a carbon source at a yield of at least about 5 percent theoretical. In another embodiment, the microorganism is selected to produce isobutanol at a yield of at least about 10 percent, at least about 15 percent, about least about 20 percent, at least about 25 percent, at least about 30 percent, at least about 35 percent, at least about 40 percent, at least about 45 percent, at least about 50 percent, at least about 55 percent, at least about 60 percent, at least about 65 percent, at least about 70 percent, at least about 75 percent, at least about 80 percent theoretical, at least about 85 percent theoretical, at least about 90 percent theoretical, or at least about 95 percent theoretical. In one embodiment, the C3-C5 alcohol, such as isobutanol, is produced under anaerobic conditions at about the same yield as under aerobic conditions.
[0042]In another aspect, the present invention provides a recombinant microorganism comprising a metabolic pathway for producing a C3-C5 alcohol from a carbon source, wherein said recombinant microorganism comprises a modification that leads to the regeneration of redox co-factors within said recombinant microorganism. In one embodiment according to this aspect, the modification increases the production of a C3-C5 alcohol under anaerobic conditions as compared to the parental or wild-type microorganism. In a preferred embodiment, the fermentation product is isobutanol. In one embodiment, the re-oxidation or re-reduction of said redox co-factors does not require the pentose phosphate pathway, the TCA cycle, or the generation of additional fermentation products. In another embodiment, the re-oxidation or re-reduction of said redox co-factors does not require the production of byproducts or co-products. In yet another embodiment, additional fermentation products are not required for the regeneration of said redox co-factors.
[0043]In another aspect, the present invention provides a method of producing a C3-C5 alcohol, comprising providing a recombinant microorganism comprising a metabolic pathway for producing a C3-C5 alcohol, wherein said recombinant microorganism comprises a modification that leads to the regeneration of redox co-factors within said recombinant microorganism; cultivating the microorganism in a culture medium containing a feedstock providing the carbon source, until a recoverable quantity of said C3-C5 alcohol is produced; and optionally, recovering the C3-C5 alcohol. In one embodiment, said microorganism is cultivated under anaerobic conditions. In another embodiment, the C3-C5 alcohol is produced under anaerobic conditions at about the same yield as under aerobic conditions. In a preferred embodiment, the C3-C5 alcohol is isobutanol.
[0044]In various embodiments described herein, the recombinant microorganisms may be microorganisms of the Saccharomyces clade, Saccharomyces sensu stricto microorganisms, Crabtree-negative yeast microorganisms, Crabtree-positive yeast microorganisms, post-WGD (whole genome duplication) yeast microorganisms, pre-WGD (whole genome duplication) yeast microorganisms, and non-fermenting yeast microorganisms.
[0045]In some embodiments, the recombinant microorganisms may be yeast recombinant microorganisms of the Saccharomyces clade.
[0046]In some embodiments, the recombinant microorganisms may be Saccharomyces sensu stricto microorganisms. In one embodiment, the Saccharomyces sensu stricto is selected from the group consisting of S. cerevisiae, S. kudriavzevii, S. mikatae, S. bayanus, S. uvarum. S. carocanis and hybrids thereof.
[0047]In some embodiments, the recombinant microorganisms may be Crabtree-negative recombinant yeast microorganisms. In one embodiment, the Crabtree-negative yeast microorganism is classified into a genera selected from the group consisting of Kluyveromyces, Pichia, Hansenula, or Candida. In additional embodiments, the Crabtree-negative yeast microorganism is selected from Kluyveromyces lactis, Kluyveromyces marxianus, Pichia anomala, Pichia stipitis, Hansenula anomala, Candida utilis, Issatchenkia orientalis and Kluyveromyces waltii.
[0048]In some embodiments, the recombinant microorganisms may be Crabtree-positive recombinant yeast microorganisms. In one embodiment, the Crabtree-positive yeast microorganism is classified into a genera selected from the group consisting of Saccharomyces, Kluyveromyces, Zygosaccharomyces, Debaryomyces, Candida, Pichia and Schizosaccharomyces. In additional embodiments, the Crabtree-positive yeast microorganism is selected from the group consisting of Saccharomyces cerevisiae, Saccharomyces uvarum, Saccharomyces bayanus, Saccharomyces paradoxus, Saccharomyces castelli, Saccharomyces kluyveri, Kluyveromyces thermotolerans, Candida glabrata, Z. bailli, Z. rouxii, Debaryomyces hansenii, Pichia pastorius, Schizosaccharomyces pombe, and Saccharomyces uvarum.
[0049]In some embodiments, the recombinant microorganisms may be post-WGD (whole genome duplication) yeast recombinant microorganisms. In one embodiment, the post-WGD yeast recombinant microorganism is classified into a genera selected from the group consisting of Saccharomyces or Candida. In additional embodiments, the post-WGD yeast is selected from the group consisting of Saccharomyces cerevisiae, Saccharomyces uvarum, Saccharomyces bayanus, Saccharomyces paradoxus, Saccharomyces castelli, and Candida glabrata.
[0050]In some embodiments, the recombinant microorganisms may be pre-WGD (whole genome duplication) yeast recombinant microorganisms. In one embodiment, the pre-WGD yeast recombinant microorganism is classified into a genera selected from the group consisting of Saccharomyces, Kluyveromyces, Candida, Pichia, Debaryomyces, Hansenula, Pachysolen, Issatchenkia, Yarrowia and Schizosaccharomyces. In additional embodiments, the pre-WGD yeast is selected from the group consisting of Saccharomyces kluyveri, Kluyveromyces thermotolerans, Kluyveromyces marxianus, Kluyveromyces waltii, Kluyveromyces lactis, Candida tropicalis, Pichia pasto'ris, Pichia anomala, Pichia stipitis, Debaryomyces hansenii, Hansenula anomala, Pachysolen tannophilis, Yarrowia lipolytica, Issatchenkia orientalis, and Schizosaccharomyces pombe.
[0051]In some embodiments, the recombinant microorganisms may be microorganisms that are non-fermenting yeast microorganisms, including, but not limited to those, classified into a genera selected from the group consisting of Tricosporon, Rhodotorula, or Myxozyma.
[0052]In certain specific embodiments, there are provided recombinant microorganisms comprising an engineered metabolic pathway for producing one or more C3-C5 alcohols under anaerobic conditions, wherein the recombinant microorganism is selected from GEVO1846, GEVO1886, GEVO1993, GEVO2158, GEVO2302, GEVO1803, GEVO2107, GEVO2710, GEVO2711, GEVO2712, GEVO2799, GEVO2847, GEVO2848, GEVO2849, GEVO2851, GEVO2852, GEVO2854, GEVO2855 and GEVO2856. In another specific embodiment, the present invention provides a plasmid is selected from the group consisting of pGV1698 (SEQ ID NO: 112), pGV1720 (SEQ ID NO: 115), pGV1745 (SEQ ID NO: 117), pGV1655 (SEQ ID NO: 109), pGV1609 (SEQ ID NO: 108), pGV1685 (SEQ ID NO: 111), and pGV1490 (SEQ ID NO: 104).
[0053]In yet another aspect, the present invention provides methods for the conversion of an aldehyde with three to five carbon atoms to the corresponding alcohol is provided. The method includes providing a microorganism comprising a heterologous polynucleotide encoding a polypeptide having NADH-dependent aldehyde reduction activity that is greater than its NADPH-dependent aldehyde reduction activity and having NADH-dependent aldehyde reduction activity that is greater than the endogenous NADPH-dependent aldehyde reduction activity of the microorganism; and contacting the microorganism with the aldehyde.
[0054]In another embodiment, a method for the conversion of an aldehyde derived from the conversion of a 2-ketoacid by a 2-ketoacid decarboxylase is provided. The method includes providing a microorganism comprising a heterologous polynucleotide encoding a polypeptide having NADH-dependent aldehyde reduction activity that is greater than its NADPH-dependent aldehyde reduction activity and having NADH-dependent aldehyde reduction activity that is greater than the endogenous NADPH-dependent aldehyde reduction activity of the microorganism; and contacting the microorganism with the aldehyde. In various embodiments described herein, the aldehyde may be selected from 1-propanal, 1-butanal, isobutyraldehyde, 2-methyl-1-butanal, or 3-methyl-1-butanal. In a preferred embodiment, the aldehyde is isobutyraldehyde.
[0055]In another embodiment, an microorganism include a heterologous polynucleotide encoding a polypeptide having NADH-dependent aldehyde reduction activity that is greater than its NADPH-dependent aldehyde reduction activity and having NADH-dependent aldehyde reduction activity that is greater than the endogenous NADPH-dependent aldehyde reduction activity of the microorganism is provided. The microorganism converts an aldehyde comprising three to five carbon atoms to the corresponding alcohol.
[0056]In another embodiment, an isolated microorganism is provided. The microorganism includes a heterologous polynucleotide encoding a polypeptide having NADH-dependent aldehyde reduction activity that is greater than its NADPH-dependent aldehyde reduction activity and having NADH-dependent aldehyde reduction activity that is greater than the endogenous NADPH-dependent aldehyde reduction activity of the microorganism. The microorganism converts an aldehyde derived from a 2-ketoacid by a 2-ketoacid decarboxylase. In one embodiment, the polypeptide is encoded by the Drosophila melanogaster ADH gene or homologs thereof. In a preferred embodiment, the Drosophila melanogaster ADH gene is set forth in SEQ ID NO: 60. In an alternative embodiment, the Drosophila melanogaster alcohol dehydrogenase is set forth in SEQ ID NO: 61. In another embodiment, the polypeptide possesses 1,2 propanediol dehydrogenase activity and is encoded by a 1,2 propanediol dehydrogenase gene. In a preferred embodiment, the 1,2-propanediol dehydrogenase gene is the Klebsiella pneumoniae dhaT gene as set forth in SEQ ID NO: 62. In an alternative embodiment, the 1,2-propanediol dehydrogenase is set forth in SEQ ID NO: 63. In another embodiment, the polypeptide possesses is encoded by a 1,3-propanediol dehydrogenase gene. In a preferred embodiment, the 1,3-propanediol dehydrogenase gene is the Escherichia coli fucO gene as set forth in SEQ ID NO: 64. In an alternative embodiment, the 1,3-propanediol dehydrogenase is set forth in SEQ ID NO: 65.
[0057]In yet another aspect, the present invention provides a recombinant microorganism producing isobutanol, wherein said recombinant microorganism i) does not overexpress an alcohol dehydrogenase; and ii) produces isobutanol at a higher rate, titer, and productivity as compared to recombinant microorganism expressing the S. cerevisiae alcohol dehydrogenase ADH2.
BRIEF DESCRIPTION OF THE DRAWINGS
[0058]Illustrative embodiments of the invention are illustrated in the drawings, in which:
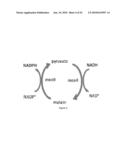
[0059]FIG. 1 illustrates an exemplary metabolic pathway for the conversion of glucose to isobutanol via pyruvate.
[0060]FIG. 2 illustrates a metabolic pathway for the conversion of glucose to isobutanol via pyruvate in which a transhydrogenase converts NADH from glycolysis to NADPH
[0061]FIG. 3 illustrates a metabolic pathway for the conversion of glucose to isobutanol via pyruvate in which an NADPH-dependent glyceraldehyde-3-phosphate dehydrogenase converts generates NADPH during glycolysis.
[0062]FIG. 4 illustrates a Transhydrogenase cycle converting NADH to NADPH
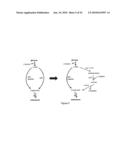
[0063]FIG. 5 illustrates an exemplary isobutanol pathway; on the left native conversion of PEP to pyruvate; on the right bypass of pyruvate kinase.
[0064]FIG. 6 illustrates an amino acid sequence alignment among various members of the KARI enzyme family.
[0065]FIG. 7 illustrates the structure alignment of E. coli KARI with rice KAR1.
[0066]FIG. 8 illustrates growth of GEVO1859 under anaerobic shift conditions over the course of the fermentation.
[0067]FIG. 9 illustrates isobutanol production of GEVO1859 under anaerobic shift conditions over the course of the fermentation.
[0068]FIG. 10 illustrates that microorganisms featuring an overexpressed E. coli pntAB operon (pGV1745) increased in OD600 from 6 h to 24 h by 0.2-1.1 under anaerobic conditions, while microorganisms lacking E. coli pntAB (pGV1720) decreased in OD600 by 0.5-1.2.
[0069]FIG. 11 illustrates that microorganisms featuring an overexpressed E. coli pntAB operon (pGV1745) continued isobutanol production under anaerobic conditions until the fermentation was stopped at 48 hours while microorganisms lacking E. coli pntAB (pGV1720) did not produce isobutanol between 24 and 48 hours
[0070]FIG. 12 illustrates that for strains GEVO1886, GEVO1859 and GEVO1846 stable OD values can be observed under anaerobic shift conditions over the course of the fermentation
[0071]FIG. 13 illustrates that over-expression of E. coli pntAB in either strain GEVO1846 or GEVO1886 leads to an improvement in isobutanol production over the course of the fermentation compared to the control strain GEVO1859 which does not over-express E. coli pntAB.
[0072]FIG. 14 illustrates that a strain lacking zwf without E. coli pntAB (Δzwf) grew to an OD of about 3, whereas the samples featuring E. coli pntAB (Δzwf+pntAB) reached OD values of about 5-6.
[0073]FIG. 15 illustrates an SDS PAGE of crude extracts of E. coli BL21(DE3) and GEVO1777 containing overexpressed KARI from plasmids pGV1777 and pET22[ilvC_co], respectively. The arrow highlights the KARI band. The protein marker (M) was an unstained 200 kDa ladder from Fermentas.
[0074]FIG. 16 illustrates an SDS PAGE of crude extract (C), purified KARI over a linear gradient (1), purified KARI over a step gradient (2), and PageRuler®unstained protein ladder (M, Fermentas). KARI was enriched to high purity with just one purification step.
[0075]FIG. 17 illustrates the structure alignment of E. coli KARI with spinach KAR1.
[0076]FIG. 18 illustrates the characterization of E. coli llvC and three variants resulting from the site saturation libraries: from top to bottom: Specific activities in U/mg, kcat in 1/s, and catalytic efficiencies in M-1*s-1. All proteins were purified over a nickel sepharose histrap column.
[0077]FIG. 19 illustrates the characterization of Ec_llvC.sup.B8-his6 and Ec_IlVC.sup.B8A71S-his6 compared to Ec_llvC.sup.his6, Ec_llvC.sup.Q110V-his6, Ec_llvC.sup.Q110A-his6, and Ec_llvC.sup.S78D-his6.
[0078]FIG. 20 illustrates a protein gel of cell lysates from the production strain GEVO1780 harboring the plasmids pGV1490, or pGV1661.
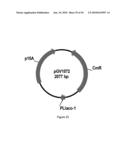
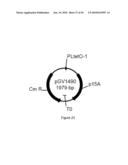
[0079]FIG. 21 illustrates plasmid pGV1102 (SEQ ID NO: 101).
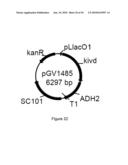
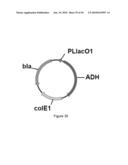
[0080]FIG. 22 illustrates plasmid pGV1485 (SEQ ID NO: 103).
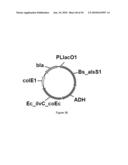
[0081]FIG. 23 illustrates plasmid pGV1490 (SEQ ID NO: 104).
[0082]FIG. 24 illustrates plasmid pGV1527.
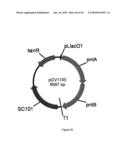
[0083]FIG. 25 illustrates plasmid pGV1572 (SEQ ID NO: 105).
[0084]FIG. 26 illustrates plasmid pGV1573 (SEQ ID NO: 106).
[0085]FIG. 27 illustrates plasmid pGV1575 (SEQ ID NO: 107).
[0086]FIG. 28 illustrates plasmid pGV1609 (SEQ ID NO: 108).
[0087]FIG. 29 illustrates plasmid pGV1631.
[0088]FIG. 30 illustrates plasmid pGV1655 (SEQ ID NO: 109).
[0089]FIG. 31 illustrates plasmid pGV1661 (SEQ ID NO: 110).
[0090]FIG. 32 illustrates plasmid pGV1685 (SEQ ID NO: 111).
[0091]FIG. 33 illustrates plasmid pGV1698 (SEQ ID NO: 112).
[0092]FIG. 34 illustrates plasmid pGV1711 (SEQ ID NO: 113).
[0093]FIG. 35 illustrates plasmids pGV1705-A, pGV1748-A, pGV1749-A, and pGV1778-A carrying the ADH genes Ec_yqhD, Ec_fucO, Dm_ADH, and Kp_dhaT, respectively.
[0094]FIG. 36 illustrates plasmids pGV1748, pGV1749, and pGV1778 carrying the ADH genes Ec_fucO, Dm_ADH, and Kp_dhaT, respectively.
[0095]FIG. 37 illustrates plasmid pGV1716 (SEQ ID NO: 114).
[0096]FIG. 38 illustrates plasmid pGV1720 (SEQ ID NO: 115).
[0097]FIG. 39 illustrates plasmid pGV1730 (SEQ ID NO: 116) and linearization for integration by NruI digest (SEQ ID NO: 116).
[0098]FIG. 40 illustrates plasmid pGV1745 (SEQ ID NO: 117).
[0099]FIG. 41 illustrates plasmid pGV1772.
[0100]FIG. 42 illustrates plasmid pGV1777 (SEQ ID NO: 118).
[0101]FIG. 43 illustrates plasmids pGV1824, pGV1994, pGV2193, pGV2238, and pGV2241 carrying the KARI genes Ec_ilvC_coSc, Ec_ilvC_coSc6E6, Ec_ilvC_coSc.sup.P2D1-his6, Ec_ilvC_coSc.sup.P2D1-A1-his6, and Ec_ilvC_coSc6E6-his6, respectively.
[0102]FIG. 44 illustrates plasmid pGV1914 (SEQ ID NO: 119).
[0103]FIG. 45 illustrates plasmids pGV1925, pGV1927, pGV1975 and pGV1776 carrying the Ec_fucO in combination with KARI genes Ec_ilvC_coEc, Ec_ilvC_coEc.sup.S78D, Ec_ilvC_coEc6E6 and Ec_ilvC_coEc2H10, respectively.
[0104]FIG. 46 illustrates plasmid pGV1936 (SEQ ID NO: 120).
[0105]FIG. 47 illustrates plasmid pGV1938.
[0106]FIG. 48 illustrates plasmid pGV2020 (SEQ ID NO: 121).
[0107]FIG. 49 illustrates plasmid pGV2082 (SEQ ID NO: 122).
[0108]FIG. 50 illustrates plasmids pGV2227 (SEQ ID NO: 123), pGV2242 (SEQ ID NO: 125) carrying the KARI genes Ec_ilvC_coScQ110V and Ec_ilvC_coSc.sup.P2D1, respectively.
DETAILED DESCRIPTION
Definitions
[0109]As used herein and in the appended claims, the singular forms "a," "an," and "the" include plural referents unless the context clearly dictates otherwise. Thus, for example, reference to "a polynucleotide" includes a plurality of such polynucleotides and reference to "the microorganism" includes reference to one or more microorganisms, and so forth.
[0110]Unless defined otherwise, all technical and scientific terms used herein have the same meaning as commonly understood to one of ordinary skill in the art to which this disclosure belongs. Although methods and materials similar or equivalent to those described herein can be used in the practice of the disclosed methods and compositions, the exemplary methods, devices and materials are described herein.
[0111]Any publications discussed above and throughout the text are provided solely for their disclosure prior to the filing date of the present application. Nothing herein is to be construed as an admission that the inventors are not entitled to antedate such disclosure by virtue of prior disclosure.
[0112]The term "microorganism" includes prokaryotic and eukaryotic microbial species from the Domains Archaea, Bacteria and Eukarya, the latter including yeast and filamentous fungi, protozoa, algae, or higher Protista. The terms "microbial cells" and "microbes" are used interchangeably with the term microorganism.
[0113]The term "prokaryotes" is art recognized and refers to cells which contain no nucleus or other cell organelles. The prokaryotes are generally classified in one of two domains, the Bacteria and the Archaea. The definitive difference between organisms of the Archaea and Bacteria domains is based on fundamental differences in the nucleotide base sequence in the 16S ribosomal RNA.
[0114]The term "Archaea" refers to a categorization of organisms of the division Mendosicutes, typically found in unusual environments and distinguished from the rest of the prokaryotes by several criteria, including the number of ribosomal proteins and the lack of muramic acid in cell walls. On the basis of ssrRNA analysis, the Archaea consist of two phylogenetically-distinct groups: Crenarchaeota and Euryarchaeota. On the basis of their physiology, the Archaea can be organized into three types: methanogens (prokaryotes that produce methane); extreme halophiles (prokaryotes that live at very high concentrations of salt (NaCl); and extreme (hyper) thermophiles (prokaryotes that live at very high temperatures). Besides the unifying archaeal features that distinguish them from Bacteria (i.e., no murein in cell wall, ester-linked membrane lipids, etc.), these prokaryotes exhibit unique structural or biochemical attributes which adapt them to their particular habitats. The Crenarchaeota consist mainly of hyperthermophilic sulfur-dependent prokaryotes and the Euryarchaeota contain the methanogens and extreme halophiles.
[0115]"Bacteria", or "eubacteria", refers to a domain of prokaryotic organisms. Bacteria include at least 11 distinct groups as follows: (1) Gram-positive (gram+) bacteria, of which there are two major subdivisions: (1) high G+C group (Actinomycetes, Mycobacteria, Micrococcus, others) (2) low G+C group (Bacillus, Clostridia, Lactobacillus, Staphylococci, Streptococci, Mycoplasmas); (2) Proteobacteria, e.g., Purple photosynthetic+non-photosynthetic Gram-negative bacteria (includes most "common" Gram-negative bacteria); (3) Cyanobacteria, e.g., oxygenic phototrophs; (4) Spirochetes and related species; (5) Planctomyces; (6) Bacteroides, Flavobacteria; (7) Chlamydia; (8) Green sulfur bacteria; (9) Green non-sulfur bacteria (also anaerobic phototrophs); (10) Radioresistant micrococci and relatives; (11) Thermotoga and Thermosipho thermophiles.
[0116]"Gram-negative bacteria" include cocci, nonenteric rods, and enteric rods. The genera of Gram-negative bacteria include, for example, Neisseria, Spirillum, Pasteurella, Brucella, Yersinia, Francisella, Haemophilus, Bordetella, Escherichia, Salmonella, Shigella, Klebsiella, Proteus, Vibrio, Pseudomonas, Bacteroides, Acetobacter, Aerobacter, Agrobacterium, Azotobacter, Spirilla, Serratia, Vibrio, Rhizobium, Chlamydia, Rickettsia, Treponema, and Fusobacterium.
[0117]"Gram positive bacteria" include cocci, nonsporulating rods, and sporulating rods. The genera of gram positive bacteria include, for example, Actinomyces, Bacillus, Clostridium, Corynebacterium, Erysipelothrix, Lactobacillus, Listeria, Mycobacterium, Myxococcus, Nocardia, Staphylococcus, Streptococcus, and Streptomyces.
[0118]The term "genus" is defined as a taxonomic group of related species according to the Taxonomic Outline of Bacteria and Archaea (Garrity, G. M., Lilburn, T. G., Cole, J. R., Harrison, S. H., Euzeby, J., and Tindall, B. J. (2007) The Taxonomic Outline of Bacteria and Archaea. TOBA Release 7.7, March 2007. Michigan State University Board of Trustees. [http://www.taxonomicoutline.org/]).
[0119]The term "species" is defined as a collection of closely related organisms with greater than 97% 16S ribosomal RNA sequence homology and greater than 70% genomic hybridization and sufficiently different from all other organisms so as to be recognized as a distinct unit.
[0120]The terms "modified microorganism," "recombinant microorganism" and "recombinant host cell" are used by inserting, expressing or overexpressing endogenous polynucleotides, by expressing or overexpressing heterologous polynucleotides, such as those included in a vector, by introducing a mutations into the microorganism or by altering the expression of an endogenous gene. The polynucleotide generally encodes a target enzyme involved in a metabolic pathway for producing a desired metabolite. It is understood that the terms "recombinant microorganism" and "recombinant host cell" refer not only to the particular recombinant microorganism but to the progeny or potential progeny of such a microorganism. Because certain modifications may occur in succeeding generations due to either mutation or environmental influences, such progeny may not, in fact, be identical to the parent cell, but are still included within the scope of the term as used herein.
[0121]The term "wild-type microorganism" describes a cell that occurs in nature, i.e. a cell that has not been genetically modified. A wild-type microorganism can be genetically modified to express or overexpress a first target enzyme. This microorganism can act as a parental microorganism in the generation of a microorganism modified to express or overexpress a second target enzyme. In turn, the microorganism modified to express or overexpress a first and a second target enzyme can be modified to express or overexpress a third target enzyme.
[0122]Accordingly, a "parental microorganism" functions as a reference cell for successive genetic modification events. Each modification event can be accomplished by introducing a nucleic acid molecule into the reference cell. The introduction facilitates the expression or overexpression of a target enzyme. It is understood that the term "facilitates" encompasses the activation of endogenous polynucleotides encoding a target enzyme through genetic modification of e.g., a promoter sequence in a parental microorganism. It is further understood that the term "facilitates" encompasses the introduction of heterologous polynucleotides encoding a target enzyme in to a parental microorganism.
[0123]The term "mutation" as used herein indicates any modification of a nucleic acid and/or polypeptide which results in an altered nucleic acid or polypeptide. Mutations include, for example, point mutations, deletions, or insertions of single or multiple residues in a polynucleotide, which includes alterations arising within a protein-encoding region of a gene as well as alterations in regions outside of a protein-encoding sequence, such as, but not limited to, regulatory or promoter sequences. A genetic alteration may be a mutation of any type. For instance, the mutation may constitute a point mutation, a frame-shift mutation, an insertion, or a deletion of part or all of a gene. In addition, in some embodiments of the modified microorganism, a portion of the microorganism genome has been replaced with a heterologous polynucleotide. In some embodiments, the mutations are naturally-occurring. In other embodiments, the mutations are the results of artificial mutation pressure. In still other embodiments, the mutations in the microorganism genome are the result of genetic engineering.
[0124]The term "biosynthetic pathway", also referred to as "metabolic pathway", refers to a set of anabolic or catabolic biochemical reactions for converting one chemical species into another. Gene products belong to the same "metabolic pathway" if they, in parallel or in series, act on the same substrate, produce the same product, or act on or produce a metabolic intermediate (i.e., metabolite) between the same substrate and metabolite end product.
[0125]The term "heterologous" as used herein with reference to molecules and in particular enzymes and polynucleotides, indicates molecules that are expressed in an organism other than the organism from which they originated or are found in nature, independently on the level of expression that can be lower, equal or higher than the level of expression of the molecule in the native microorganism.
[0126]On the other hand, the term "native" or "endogenous" as used herein with reference to molecules, and in particular enzymes and polynucleotides, indicates molecules that are expressed in the organism in which they originated or are found in nature, independently on the level of expression that can be lower equal or higher than the level of expression of the molecule in the native microorganism. It is understood that expression of native enzymes or polynucleotides may be modified in recombinant microorganisms.
[0127]The term "carbon source" generally refers to a substance suitable to be used as a source of carbon for prokaryotic or eukaryotic cell growth. Carbon sources include, but are not limited to, biomass hydrolysates, starch, sucrose, cellulose, hemicellulose, xylose, and lignin, as well as monomeric components of these substrates. Carbon sources can comprise various organic compounds in various forms, including, but not limited to polymers, carbohydrates, acids, alcohols, aldehydes, ketones, amino acids, peptides, etc. These include, for example, various monosaccharides such as glucose, dextrose (D-glucose), maltose, oligosaccharides, polysaccharides, saturated or unsaturated fatty acids, succinate, lactate, acetate, ethanol, etc., or mixtures thereof. Photosynthetic organisms can additionally produce a carbon source as a product of photosynthesis. In some embodiments, carbon sources may be selected from biomass hydrolysates and glucose. The term "substrate" or "suitable substrate" refers to any substance or compound that is converted or meant to be converted into another compound by the action of an enzyme. The term includes not only a single compound, but also combinations of compounds, such as solutions, mixtures and other materials which contain at least one substrate, or derivatives thereof. Further, the term "substrate" encompasses not only compounds that provide a carbon source suitable for use as a starting material, such as any biomass derived sugar, but also intermediate and end product metabolites used in a pathway associated with a modified microorganism as described herein.
[0128]The term "volumetric productivity" or "production rate" is defined as the amount of product formed per volume of medium per unit of time. Volumetric productivity is reported in gram per liter per hour (g/L/h).
[0129]The term "specific productivity" is defined as the rate of formation of the product. To describe productivity as an inherent parameter of the microorganism or microorganism and not of the fermentation process, productivity is herein further defined as the specific productivity in gram product per unit of cells, typically measured spectroscopically as absorbance units at 600 nm (OD600 or OD) per hour (g/L/h/OD).
[0130]The term "yield" is defined as the amount of product obtained per unit weight of raw material and may be expressed as g product per g substrate (g/g). Yield may be expressed as a percentage of the theoretical yield. "Theoretical yield" is defined as the maximum amount of product that can be generated per a given amount of substrate as dictated by the stoichiometry of the metabolic pathway used to make the product. For example, the theoretical yield for one typical conversion of glucose to isobutanol is 0.41 g/g. As such, a yield of butanol from glucose of 0.39 g/g would be expressed as 95% of theoretical or 95% theoretical yield.
[0131]The term "titre" or "titer" is defined as the strength of a solution or the concentration of a substance in solution. For example, the titre of a biofuel in a fermentation broth is described as g of biofuel in solution per liter of fermentation broth (g/L).
[0132]The term "total titer" is defined as the sum of all biofuel produced in a process, including but not limited to the biofuel in solution, the biofuel in gas phase, and any biofuel removed from the process and recovered relative to the initial volume in the process or the operating volume in the process.
[0133]A "facultative anaerobic organism" or a "facultative anaerobic microorganism" is defined as an organism that can grow in either the presence or in the absence of oxygen.
[0134]A "strictly anaerobic organism" or a "strictly anaerobic microorganism" is defined as an organism that cannot grow in the presence of oxygen and which does not survive exposure to any concentration of oxygen.
[0135]An "anaerobic organism" or an "anaerobic microorganism" is defined as an organism that cannot grow in the presence of oxygen.
[0136]"Aerobic conditions" are defined as conditions under which the oxygen concentration in the fermentation medium is sufficiently high for an aerobic or facultative anaerobic microorganism to use as a terminal electron acceptor.
[0137]In contrast, "Anaerobic conditions" are defined as conditions under which the oxygen concentration in the fermentation medium is too low for the microorganism to use as a terminal electron acceptor. Anaerobic conditions may be achieved by sparging a fermentation medium with an inert gas such as nitrogen until oxygen is no longer available to the microorganism as a terminal electron acceptor. Alternatively, anaerobic conditions may be achieved by the microorganism consuming the available oxygen of the fermentation until oxygen is unavailable to the microorganism as a terminal electron acceptor. "Anaerobic conditions" are further defined as conditions under which no or small amounts of oxygen are added to the medium at rates of <3 mmol/L/h, preferably <2.5 mmol/L/h, more preferably <2 mmol/L/h and most preferably <1.5 mmol/L/h. "Anaerobic conditions" means in particular completely oxygen-free (=0 mmol/L/h oxygen) or with small amounts of oxygen added to the medium at rates of e.g. <0.5 to <1 mmol/L/h.
[0138]"Dissolved oxygen," abbreviated as "DO" is expressed throughout as the percentage of saturating concentration of oxygen in water.
[0139]"Aerobic metabolism" refers to a biochemical process in which oxygen is used as a terminal electron acceptor to make energy, typically in the form of ATP, from carbohydrates. Aerobic metabolism occurs e.g. via glycolysis and the TCA cycle, wherein a single glucose molecule is metabolized completely into carbon dioxide in the presence of oxygen.
[0140]In contrast, "anaerobic metabolism" refers to a biochemical process in which oxygen is not the final acceptor of electrons contained in NADH. Anaerobic metabolism can be divided into anaerobic respiration, in which compounds other than oxygen serve as the terminal electron acceptor, and substrate level phosphorylation, in which the electrons from NADH are utilized to generate a reduced product via a "fermentative pathway."
[0141]In "fermentative pathways," NAD(P)H donates its electrons to a molecule produced by the same metabolic pathway that produced the electrons carried in NAD(P)H. For example, in one of the fermentative pathways of certain yeast strains, NAD(P)H generated through glycolysis transfers its electrons to pyruvate, yielding lactate. Fermentative pathways are usually active under anaerobic conditions but may also occur under aerobic conditions, under conditions where NADH is not fully oxidized via the respiratory chain. For example, above certain glucose concentrations, crabtree positive yeasts produce large amounts of ethanol under aerobic conditions.
[0142]The term "fermentation product" means any main product plus its coupled product. A "coupled product" is produced as part of the stoichiometric conversion of the carbon source to the main fermentation product. An example for a coupled product is the two molecules of CO2 that are produced with every molecule of isobutanol during production of isobutanol from glucose according the biosynthetic pathway described herein.
[0143]The term "byproduct" means an undesired product related to the production of a biofuel. Byproducts are generally disposed as waste, adding cost to a biofuel process.
[0144]The term "co-product" means a secondary or incidental product related to the production of biofuel. Co-products have potential commercial value that increases the overall value of biofuel production, and may be the deciding factor as to the viability of a particular biofuel production process.
[0145]The term "non-fermenting yeast" is a yeast species that fails to demonstrate an anaerobic metabolism in which the electrons from NADH are utilized to generate a reduced product via a fermentative pathway such as the production of ethanol and CO2 from glucose. Non-fermentative yeast can be identified by the "Durham Tube Test" (J. A. Barnett, R. W. Payne, and D. Yarrow. 2000. Yeasts Characteristics and Identification. 3rd edition. p. 28-29. Cambridge University Press, Cambridge, UK.) or by monitoring the production of fermentation productions such as ethanol and CO2.
[0146]The term "polynucleotide" is used herein interchangeably with the term "nucleic acid" and refers to an organic polymer composed of two or more monomers including nucleotides, nucleosides or analogs thereof, including but not limited to single stranded or double stranded, sense or antisense deoxyribonucleic acid (DNA) of any length and, where appropriate, single stranded or double stranded, sense or antisense ribonucleic acid (RNA) of any length, including siRNA. The term "nucleotide" refers to any of several compounds that consist of a ribose or deoxyribose sugar joined to a purine or a pyrimidine base and to a phosphate group, and that are the basic structural units of nucleic acids. The term "nucleoside" refers to a compound (as guanosine or adenosine) that consists of a purine or pyrimidine base combined with deoxyribose or ribose and is found especially in nucleic acids. The term "nucleotide analog" or "nucleoside analog" refers, respectively, to a nucleotide or nucleoside in which one or more individual atoms have been replaced with a different atom or with a different functional group. Accordingly, the term polynucleotide includes nucleic acids of any length, DNA, RNA, analogs and fragments thereof. A polynucleotide of three or more nucleotides is also called nucleotidic oligomer or oligonucleotide.
[0147]It is understood that the polynucleotides described herein include "genes" and that the nucleic acid molecules described herein include "vectors" or "plasmids." Accordingly, the term "gene", also called a "structural gene" refers to a polynucleotide that codes for a particular sequence of amino acids, which comprise all or part of one or more proteins or enzymes, and may include regulatory (non-transcribed) DNA sequences, such as promoter sequences, which determine for example the conditions under which the gene is expressed. The transcribed region of the gene may include untranslated regions, including introns, 5'-untranslated region (UTR), and 3'-UTR, as well as the coding sequence.
[0148]The term "expression" with respect to a gene sequence refers to transcription of the gene and, as appropriate, translation of the resulting mRNA transcript to a protein. Thus, as will be clear from the context, expression of a protein results from transcription and translation of the open reading frame sequence.
[0149]The term "operon" refers two or more genes which are transcribed as a single transcriptional unit from a common promoter. In some embodiments, the genes comprising the operon are contiguous genes. It is understood that transcription of an entire operon can be modified (i.e., increased, decreased, or eliminated) by modifying the common promoter. Alternatively, any gene or combination of genes in an operon can be modified to alter the function or activity of the encoded polypeptide. The modification can result in an increase in the activity of the encoded polypeptide. Further, the modification can impart new activities on the encoded polypeptide. Exemplary new activities include the use of alternative substrates and/or the ability to function in alternative environmental conditions.
[0150]A "vector" is any means by which a nucleic acid can be propagated and/or transferred between organisms, cells, or cellular components. Vectors include viruses, bacteriophage, pro-viruses, plasmids, phagemids, transposons, and artificial chromosomes such as YACs (yeast artificial chromosomes), BACs (bacterial artificial chromosomes), and PLACs (plant artificial chromosomes), and the like, that are "episomes," that is, that replicate autonomously or can integrate into a chromosome of a host cell. A vector can also be a naked RNA polynucleotide, a naked DNA polynucleotide, a polynucleotide composed of both DNA and RNA within the same strand, a poly-lysine-conjugated DNA or RNA, a peptide-conjugated DNA or RNA, a liposome-conjugated DNA, or the like, that are not episomal in nature, or it can be an organism which comprises one or more of the above polynucleotide constructs such as an agrobacterium or a bacterium.
[0151]"Transformation" refers to the process by which a vector is introduced into a host cell. Transformation (or transduction, or transfection), can be achieved by any one of a number of means including electroporation, microinjection, biolistics (or particle bombardment-mediated delivery), or agrobacterium mediated transformation.
[0152]The term "enzyme" as used herein refers to any substance that catalyzes or promotes one or more chemical or biochemical reactions, which usually includes enzymes totally or partially composed of a polypeptide, but can include enzymes composed of a different molecule including polynucleotides.
[0153]The term "protein" or "polypeptide" as used herein indicates an organic polymer composed of two or more amino acidic monomers and/or analogs thereof. As used herein, the term "amino acid" or "amino acidic monomer" refers to any natural and/or synthetic amino acids including glycine and both D or L optical isomers. The term "amino acid analog" refers to an amino acid in which one or more individual atoms have been replaced, either with a different atom, or with a different functional group. Accordingly, the term polypeptide includes amino acidic polymer of any length including full length proteins, and peptides as well as analogs and fragments thereof. A polypeptide of three or more amino acids is also called a protein oligomer or oligopeptide
[0154]The term "homologs" used with respect to an original enzyme or gene of a first family or species refers to distinct enzymes or genes of a second family or species which are determined by functional, structural or genomic analyses to be an enzyme or gene of the second family or species which corresponds to the original enzyme or gene of the first family or species. Most often, homologs will have functional, structural or genomic similarities. Techniques are known by which homologs of an enzyme or gene can readily be cloned using genetic probes and PCR. Identity of cloned sequences as homolog can be confirmed using functional assays and/or by genomic mapping of the genes.
[0155]A protein has "homology" or is "homologous" to a second protein if the nucleic acid sequence that encodes the protein has a similar sequence to the nucleic acid sequence that encodes the second protein. Alternatively, a protein has homology to a second protein if the two proteins have "similar" amino acid sequences. (Thus, the term "homologous proteins" is defined to mean that the two proteins have similar amino acid sequences).
[0156]The term "analog" or "analogous" refers to nucleic acid or protein sequences or protein structures that are related to one another in function only and are not from common descent or do not share a common ancestral sequence. Analogs may differ in sequence but may share a similar structure, due to convergent evolution. For example, two enzymes are analogs or analogous if the enzymes catalyze the same reaction of conversion of a substrate to a product, are unrelated in sequence, and irrespective of whether the two enzymes are related in structure.
The Microorganism in General
[0157]Microorganism Characterized by Producing C3-C5 Alcohols from Pyruvate Via an Overexpressed Metabolic Pathway
[0158]Native producers of butanol, and more specifically 1-butaanol, such as Clostridium acetobutylicum, are known, but these organisms generate byproducts such as acetone, ethanol, and butyrate during fermentations. Furthermore, these microorganisms are relatively difficult to manipulate, with significantly fewer tools available than in more commonly used production hosts such as E. coli. Additionally, the physiology and metabolic regulation of these native producers are much less well understood, impeding rapid progress towards high-efficiency production. Furthermore, no native microorganisms have been identified that can metabolize glucose into isobutanol in industrially relevant quantities or yields.
[0159]The production of isobutanol and other fusel alcohols by various yeast species, including Saccharomyces cerevisiae is of special interest to the distillers of alcoholic beverages, for whom fusel alcohols constitute often undesirable off-notes. Production of isobutanol in wild-type yeasts has been documented on various growth media, ranging from grape must from winemaking (Romano, et al., Metabolic diversity of Saccharomyces cerevisiae strains from spontaneously fermented grape musts, 19:311-315, 2003), in which 12-219 mg/L isobutanol were produced, supplemented to minimal media (Oliviera, et al. (2005) World Journal of Microbiology and Biotechnology 21:1569-1576), producing 16-34 mg/L isobutanol. Work from Dickinson, et al. (J Biol. Chem. 272(43):26871-8, 1997) has identified the enzymatic steps utilized in an endogenous S. cerevisiae pathway converting branch-chain amino acids (e.g., valine or leucine) to isobutanol.
[0160]A number of recent publications have described methods for the production of industrial chemicals such as C3-C5 alcohols such as isobutanol using engineered microorganisms. See, e.g., WO/2007/050671 to Donaldson et al., and WO/2008/098227 to Liao et al., which are herein incorporated by reference in their entireties. These publications disclose recombinant microorganisms that utilize a series of heterologously expressed enzymes to convert sugars into isobutanol. However, the production of isobutanol using these microorganisms is feasible only under aerobic conditions and the maximum yield that can be achieved is limited.
[0161]Recombinant microorganisms provided herein can express a plurality of target enzymes involved in pathways for the production isobutanol from a suitable carbon source under anaerobic conditions.
[0162]Accordingly, "engineered" or "modified" microorganisms are produced via the introduction of genetic material into a host or parental microorganism of choice thereby modifying or altering the cellular physiology and biochemistry of the microorganism. Through the introduction of genetic material the parental microorganism acquires new properties, e.g. the ability to produce a new, or greater quantities of, an intracellular metabolite under anaerobic conditions. As described herein, the introduction of genetic material into a parental microorganism results in a new or modified ability to produce isobutanol under anaerobic conditions. The genetic material introduced into the parental microorganism contains gene(s), or parts of genes, coding for one or more of the enzymes involved in a biosynthetic pathway for the production of isobutanol under anaerobic conditions and may also include additional elements for the expression and/or regulation of expression of these genes, e.g. promoter sequences.
[0163]An engineered or modified microorganism can also include in the alternative or in addition to the introduction of a genetic material into a host or parental microorganism, the disruption, deletion or knocking out of a gene or polynucleotide to alter the cellular physiology and biochemistry of the microorganism. Through the reduction, disruption or knocking out of a gene or polynucleotide the microorganism acquires new or improved properties (e.g., the ability to produce a new metabolite or greater quantities of an intracellular metabolite, improve the flux of a metabolite down a desired pathway, and/or reduce the production of undesirable by-products).
[0164]Microorganisms provided herein are modified to produce under anaerobic conditions metabolites in quantities not available in the parental microorganism. A "metabolite" refers to any substance produced by metabolism or a substance necessary for or taking part in a particular metabolic process. A metabolite can be an organic compound that is a starting material (e.g., glucose or pyruvate), an intermediate (e.g., 2-ketoisovalerate), or an end product (e.g., isobutanol) of metabolism. Metabolites can be used to construct more complex molecules, or they can be broken down into simpler ones. Intermediate metabolites may be synthesized from other metabolites, perhaps used to make more complex substances, or broken down into simpler compounds, often with the release of chemical energy.
[0165]Exemplary metabolites include glucose, pyruvate, and C3-C5 alcohols, including isobutanol. The metabolite isobutanol can be produced by a recombinant microorganism engineered to express or over-express metabolic pathway that converts pyruvate to isobutanol. An exemplary metabolic pathway that converts pyruvate to isobutanol may be comprised of a acetohydroxy acid synthase (ALS) enzyme encoded by, for example, alsS from B. subtilis, a ketolacid reductoisomerase (KARI) encoded by, for example ilvC from E. coli, a dihyroxy-acid dehydratase (DHAD), encoded by, for example ilvD from E. coli, a 2-keto-acid decarboxylase (KIVD) encoded by, for example kivd from L. lactis, and an alcohol dehydrogenase (ADH), encoded by, for example, by a native E. coli alcohol dehydrogenase gene, like Ec_yqhD.
[0166]Accordingly, provided herein are recombinant microorganisms that produce isobutanol and in some aspects may include the elevated expression of target enzymes such as ALS (encoded e.g. by the ilvIH operon from E. coli or by alsS from Bacillus subtilis), KARI (encoded e.g. by ilvC from E. coli), DHAD (encoded, e.g. by ilvD from E. coli, or by ILV3 from S. cerevisiae, and KIVD (encoded, e.g. by, ARO10 from S. cerevisiae, THI3 from S. cerevisiae, kivd from L. lactis).
[0167]The recombinant microorganism may further include the deletion or reduction of the activity of enzymes that (a) directly consume a precursor of the product, e.g. an isobutanol precursor, (b) indirectly consume a precursor of the product, e.g. of isobutanol, or (c) repress the expression or function of a pathway that supplies a precursor of the product, e.g. of isobutanol. These enzymes include pyruvate decarboxylase (encoded, e.g. by PDC1, PDC2, PDC3, PDC5, or PDC6 of S. cerevisiae), glycerol-3-phosphate dehydrogenase (encoded, e.g. by GPD1 or GPD2 of S. cerevisiae) an alcohol dehydrogenase (encoded, e.g., by adhE of E. coli or ADH1, ADH2, ADH3, ADH4, ADH5, ADH6, or ADH7 of S. cerevisiae), lacate dehydrogenase (encoded, e.g., by IdhA of E. coli), fumarate reductase (encoded, e.g., by frdB, frdC or frdBC of E. coli), FNR (encoded, e.g. by fnr of E. coli), 2-isopropylmalate synthase (encoded, e.g. by leuA of E. coli or by LEU4 or LEU9 of S. cerevisiae), valine transaminase (encoded, e.g. by ilvE of E. coli or by BAT1 or BAT2 of S. cerevisiae), pyruvate oxidase (e.g. encoded by poxB of E. coli), Threonine deaminase (encoded, e.g. by ilvA of E. coli or CHA1 or ILV1 of S. cerevisiae), pyruvate-formate-lyase (encoded, e.g. by pflB of E. coli), or phosphate acetyltransferase (encoded, e.g. by pta of E. coli), or any combination thereof, to increase the availability of pyruvate or reduce enzymes that compete for a metabolite in a desired biosynthetic pathway.
[0168]In yeast microorganisms, pyruvate decarboxylase (PDC) is a major competitor for pyruvate. During anaerobic fermentation, the main pathway to oxidize the NADH from glycolysis is through the production of ethanol. Ethanol is produced by alcohol dehydrogenase (ADH) via the reduction of acetaldehyde, which is generated from pyruvate by pyruvate decarboxylase (PDC). Thus, most of the pyruvate produced by glycolysis is consumed by PDC and is not available for the isobutanol pathway. Another pathway for NADH oxidation is through the production of glycerol. Dihydroxyacetone-phospate, an intermediate of glycolysis is reduced to glycerol 3-phosphate by glycerol 3-phosphate dehydrogenase (GPD). Glycerol 3-phosphatase (GPP) converts glycerol 3-phosphate to glycerol. This pathway consumes carbon from glucose as well as reducing equivalents (NADH) resulting in less pyruvate and reducing equivalents available for the isobutanol pathway. These pathways contribute to low yield and low productivity of C3-C5 alcohols, including isobutanol. Accordingly, deletion or reduction of the activity of PDC and GPD may increase yield and productivity of C3-C5 alcohols, including isobutanol.
[0169]Reduction of PDC activity can be accomplished by 1) mutation or deletion of a positive transcriptional regulator for the structural genes encoding for PDC or 2) mutation or deletion of all PDC genes in a given organism. The term "transcriptional regulator" can specify a protein or nucleic acid that works in trans to increase or to decrease the transcription of a different locus in the genome. For example, in S. cerevisiae, the PDC2 gene, which encodes for a positive transcriptional regulator of PDC1,5,6 genes can be deleted; a S. cerevisiae in which the PDC2 gene is deleted is reported to have only ˜10% of wildtype PDC activity (Hohmann, Mol Gen Genet, 241:657-666 (1993)). Alternatively, for example, all structural genes for PDC (e.g. in S. cerevisiae, PDC1, PDC5, and PDC6, or in K. lactis, PDC1) are deleted.
[0170]Crabtree-positive yeast strains such as Saccharomyces cerevisiae strain that contains disruptions in all three of the PDC alleles no longer produce ethanol by fermentation. However, a downstream product of the reaction catalyzed by PDC, acetyl-CoA, is needed for anabolic production of necessary molecules. Therefore, the Pdc-mutant is unable to grow solely on glucose, and requires a two-carbon carbon source, either ethanol or acetate, to synthesize acetyl-CoA. (Flikweert M T, de Swaaf M, van Dijken J P, Pronk J T. FEMS Microbiol Lett. 1999 May 1; 174(1):73-9. PMID:10234824 and van Maris A J, Geertman J M, Vermeulen A, Groothuizen M K, Winkler A A, Piper M D, van Dijken J P, Pronk J T. Appl Environ Microbiol. 2004 January; 70(1):159-66. PMID: 14711638).
[0171]Thus, in an embodiment, such a Crabtree-positive yeast strain may be evolved to generate variants of the PDC mutant yeast that do not have the requirement for a two-carbon molecule and has a growth rate similar to wild type on glucose. Any method, including chemostat evolution or serial dilution may be utilized to generate variants of strains with deletion of three PDC alleles that can grow on glucose as the sole carbon source at a rate similar to wild type (van Maris et al., Directed Evolution of Pyruvate Decarboxylase-Negative Saccharomyces cerevisiae, Yielding a C2-Independent, Glucose-Tolerant, and Pyruvate-Hyperproducing Yeast, Applied and Environmental Microbiology, 2004, 70(1), 159-166).
[0172]Another byproduct that would decrease yield of isobutanol is glycerol. Glycerol is produced by 1) the reduction of the glycolysis intermediate, dihydroxyacetone phosphate (DHAP), to glycerol-3-phosphate (G3P) via the oxidation of NADH to NAD.sup.+ by Glycerol-3-phosphate dehydrogenase (GPD) followed by 2) the dephosphorylation of glycerol-3-phophate to glycerol by glycerol-3-phosphatase (GPP). Production of glycerol results in loss of carbons as well as reducing equivalents. Reduction of GPD activity would increase yield of isobutanol. Reduction of GPD activity in addition to PDC activity would further increase yield of isobutanol. Reduction of glycerol production has been reported to increase yield of ethanol production (Nissen et al., Anaerobic and aerobic batch cultivation of Saccharomyces cerevisiae mutants impaired in glycerol synthesis, Yeast, 2000, 16, 463-474; Nevoigt et al., Method of modifying a yeast cell for the production of ethanol, WO 2009/056984). Disruption of this pathway has also been reported to increase yield of lactate in a yeast engineered to produce lactate instead of ethanol (Dundon et al., Yeast cells having disrupted pathway from dihydroxyacetone phosphate to glycerol, US 2009/0053782).
[0173]In one embodiment, the microorganism is a crab-tree positive yeast with reduced or no GPD activity. In another embodiment, the microorganism is a crab-tree positive yeast with reduced or no GPD activity, and expresses an isobutanol biosynthetic pathway and produces isobutanol. In yet another embodiment, the microorganism is a crab-tree positive yeast with reduced or no GPD activity and with reduced or no PDC activity. In another embodiment, the microorganism is a crab-tree positive yeast with reduced or no GPD activity, with reduced or no PDC activity, and expresses an isobutanol biosynthetic pathway and produces isobutanol.
[0174]In another embodiment, the microorganism is a crab-tree negative yeast with reduced or no GPD activity. In another embodiment, the microorganism is a crab-tree negative yeast with reduced or no GPD activity, expresses the isobutanol biosynthetic pathway and produces isobutanol. In yet another embodiment, the microorganism is a crab-tree negative yeast with reduced or no GPD activity and with reduced or no PDC activity. In another embodiment, the microorganism is a crab-tree negative yeast with reduced or no GPD activity, with reduced or no PDC activity, expresses an an isobutanol biosynthetic pathway and produces isobutanol.
[0175]Any method can be used to identify genes that encode for enzymes with pyruvate decarboxylase (PDC) activity. PDC catalyzes the decarboxylation of pyruvate to form acetaldehyde. Generally, homologous or similar PDC genes and/or homologous or similar PDC enzymes can be identified by functional, structural, and/or genetic analysis. In most cases, homologous or similar PDC genes and/or homologous or similar PDC enzymes will have functional, structural, or genetic similarities. Techniques known to those skilled in the art may be suitable to identify homologous genes and homologous enzymes. Generally, analogous genes and/or analogous enzymes can be identified by functional analysis and will have functional similarities. Techniques known to those skilled in the art may be suitable to identify analogous genes and analogous enzymes. For example, to identify homologous or analogous genes, proteins, or enzymes, techniques may include, but not limited to, cloning a PDC gene by PCR using primers based on a published sequence of a gene/enzyme or by degenerate PCR using degenerate primers designed to amplify a conserved region among PDC genes. Further, one skilled in the art can use techniques to identify homologous or analogous genes, proteins, or enzymes with functional homology or similarity. Techniques include examining a cell or cell culture for the catalytic activity of an enzyme through in vitro enzyme assays for said activity, then isolating the enzyme with said activity through purification, determining the protein sequence of the enzyme through techniques such as Edman degradation, design of PCR primers to the likely nucleic acid sequence, amplification of said DNA sequence through PCR, and cloning of said nucleic acid sequence. To identify homologous or similar genes and/or homologous or similar enzymes, analogous genes and/or analogous enzymes or proteins, techniques also include comparison of data concerning a candidate gene or enzyme with databases such as BRENDA, KEGG, or MetaCYC. The candidate gene or enzyme may be identified within the above mentioned databases in accordance with the teachings herein. Furthermore, PDC activity can be determined phenotypically. For example, ethanol production under fermentative conditions can be assessed. A lack of ethanol production may be indicative of a yeast microorganism with no PDC activity.
[0176]Any method can be used to identify genes that encode for enzymes with glycerol-3-phosphate dehydrogenase (GPD) activity. GPD catalyzes the reduction of dihydroxyacetone phosphate (DHAP) to glycerol-3-phosphate (G3P) with the corresponding oxidation of NADH to NAD+. Generally, homologous or similar GPD genes and/or homologous or similar GPD enzymes can be identified by functional, structural, and/or genetic analysis. In most cases, homologous or similar GPD genes and/or homologous or similar GPD enzymes will have functional, structural, or genetic similarities. Techniques known to those skilled in the art may be suitable to identify homologous genes and homologous enzymes. Generally, analogous genes and/or analogous enzymes can be identified by functional analysis and will have functional similarities. Techniques known to those skilled in the art may be suitable to identify analogous genes and analogous enzymes. For example, to identify homologous or analogous genes, proteins, or enzymes, techniques may include, but not limited to, cloning a GPD gene by PCR using primers based on a published sequence of a gene/enzyme or by degenerate PCR using degenerate primers designed to amplify a conserved region among GPD genes. Further, one skilled in the art can use techniques to identify homologous or analogous genes, proteins, or enzymes with functional homology or similarity. Techniques include examining a cell or cell culture for the catalytic activity of an enzyme through in vitro enzyme assays for said activity, then isolating the enzyme with said activity through purification, determining the protein sequence of the enzyme through techniques such as Edman degradation, design of PCR primers to the likely nucleic acid sequence, amplification of said DNA sequence through PCR, and cloning of said nucleic acid sequence. To identify homologous or similar genes and/or homologous or similar enzymes, analogous genes and/or analogous enzymes or proteins, techniques also include comparison of data concerning a candidate gene or enzyme with databases such as BRENDA, KEGG, or MetaCYC. The candidate gene or enzyme may be identified within the above mentioned databases in accordance with the teachings herein. Furthermore, GPD activity can be determined phenotypically. For example, glycerol production under fermentative conditions can be assessed. A lack of glycerol production may be indicative of a yeast microorganism with no GPD activity.
[0177]The recombinant microorganism may further include metabolic pathways for the fermentation of a C3-C5 alcohols from five-carbon (pentose) sugars including xylose. Most yeast species metabolize xylose via a complex route, in which xylose is first reduced to xylitol via a xylose reductase (XR) enzyme. The xylitol is then oxidized to xylulose via a xylitol dehydrogenase (XDH) enzyme. The xylulose is then phosphorylated via an xylulokinase (XK) enzyme. This pathway operates inefficiently in yeast species because it introduces a redox imbalance in the cell. The xylose-to-xylitol step uses NADH as a cofactor, whereas the xylitol-to-xylulose step uses NADPH as a cofactor. Other processes must operate to restore the redox imbalance within the cell. This often means that the organism cannot grow anaerobically on xylose or other pentose sugar. Accordingly, a yeast species that can efficiently ferment xylose and other pentose sugars into a desired fermentation product is therefore very desirable.
[0178]Thus, in one aspect, the recombinant microorganism is engineered to express a functional exogenous xylose isomerase. Exogenous xylose isomerases functional in yeast are known in the art. See, e.g., Rajgarhia et al, US20060234364, which is herein incorporated by reference in its entirety. In an embodiment according to this aspect, the exogenous xylose isomerase gene is operatively linked to promoter and terminator sequences that are functional in the yeast cell. In a preferred embodiment, the recombinant microorganism further has a deletion or disruption of a native gene that encodes for an enzyme (e.g. XR and/or XDH) that catalyzes the conversion of xylose to xylitol. In a further preferred embodiment, the recombinant microorganism also contains a functional, exogenous xylulokinase (XK) gene operatively linked to promoter and terminator sequences that are functional in the yeast cell. In one embodiment, the xylulokinase (XK) gene is overexpressed.
[0179]The disclosure identifies specific genes useful in the methods, compositions and organisms of the disclosure; however it will be recognized that absolute identity to such genes is not necessary. For example, changes in a particular gene or polynucleotide comprising a sequence encoding a polypeptide or enzyme can be performed and screened for activity. Typically such changes comprise conservative mutation and silent mutations. Such modified or mutated polynucleotides and polypeptides can be screened for expression of a functional enzyme using methods known in the art.
[0180]Due to the inherent degeneracy of the genetic code, other polynucleotides which encode substantially the same or a functionally equivalent polypeptide can also be used to clone and express the polynucleotides encoding such enzymes.
[0181]As will be understood by those of skill in the art, it can be advantageous to modify a coding sequence to enhance its expression in a particular host. The genetic code is redundant with 64 possible codons, but most organisms typically use a subset of these codons. The codons that are utilized most often in a species are called optimal codons, and those not utilized very often are classified as rare or low-usage codons. Codons can be substituted to reflect the preferred codon usage of the host, a process sometimes called "codon optimization" or "controlling for species codon bias."
[0182]Optimized coding sequences containing codons preferred by a particular prokaryotic or eukaryotic host (see also, Murray et al. (1989) Nucl. Acids Res. 17:477-508) can be prepared, for example, to increase the rate of translation or to produce recombinant RNA transcripts having desirable properties, such as a longer half-life, as compared with transcripts produced from a non-optimize sequence. Translation stop codons can also be modified to reflect host preference. For example, typical stop codons for S. cerevisiae and mammals are UAA and UGA, respectively. The typical stop codon for monocotyledonous plants is UGA, whereas insects and E. coli commonly use UAA as the stop codon (Dalphin et al. (1996) Nucl. Acids Res. 24: 216-218). Methodology for optimizing a nucleotide sequence for expression in a plant is provided, for example, in U.S. Pat. No. 6,015,891, and the references cited therein.
[0183]Those of skill in the art will recognize that, due to the degenerate nature of the genetic code, a variety of DNA compounds differing in their nucleotide sequences can be used to encode a given enzyme of the disclosure. The native DNA sequence encoding the biosynthetic enzymes described above are referenced herein merely to illustrate an embodiment of the disclosure, and the disclosure includes DNA compounds of any sequence that encode the amino acid sequences of the polypeptides and proteins of the enzymes utilized in the methods of the disclosure. In similar fashion, a polypeptide can typically tolerate one or more amino acid substitutions, deletions, and insertions in its amino acid sequence without loss or significant loss of a desired activity. The disclosure includes such polypeptides with different amino acid sequences than the specific proteins described herein so long as they modified or variant polypeptides have the enzymatic anabolic or catabolic activity of the reference polypeptide. Furthermore, the amino acid sequences encoded by the DNA sequences shown herein merely illustrate embodiments of the disclosure.
[0184]In addition, homologs of enzymes useful for generating metabolites are encompassed by the microorganisms and methods provided herein.
[0185]As used herein, two proteins (or a region of the proteins) are substantially homologous when the amino acid sequences have at least about 30%, 40%, 50% 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity. To determine the percent identity of two amino acid sequences, or of two nucleic acid sequences, the sequences are aligned for optimal comparison purposes (e.g., gaps can be introduced in one or both of a first and a second amino acid or nucleic acid sequence for optimal alignment and non-homologous sequences can be disregarded for comparison purposes). In one embodiment, the length of a reference sequence aligned for comparison purposes is at least 30%, typically at least 40%, more typically at least 50%, even more typically at least 60%, and even more typically at least 70%, 80%, 90%, 100% of the length of the reference sequence. The amino acid residues or nucleotides at corresponding amino acid positions or nucleotide positions are then compared. When a position in the first sequence is occupied by the same amino acid residue or nucleotide as the corresponding position in the second sequence, then the molecules are identical at that position (as used herein amino acid or nucleic acid "identity" is equivalent to amino acid or nucleic acid "homology"). The percent identity between the two sequences is a function of the number of identical positions shared by the sequences, taking into account the number of gaps, and the length of each gap, which need to be introduced for optimal alignment of the two sequences.
[0186]When "homologous" is used in reference to proteins or peptides, it is recognized that residue positions that are not identical often differ by conservative amino acid substitutions. A "conservative amino acid substitution" is one in which an amino acid residue is substituted by another amino acid residue having a side chain (R group) with similar chemical properties (e.g., charge or hydrophobicity). In general, a conservative amino acid substitution will not substantially change the functional properties of a protein. In cases where two or more amino acid sequences differ from each other by conservative substitutions, the percent sequence identity or degree of homology may be adjusted upwards to correct for the conservative nature of the substitution. Means for making this adjustment are well known to those of skill in the art (see, e.g., Pearson et al., 1994, hereby incorporated herein by reference).
[0187]The following six groups each contain amino acids that are conservative substitutions for one another: 1) Serine (S), Threonine (T); 2) Aspartic Acid (D), Glutamic Acid (E); 3) Asparagine (N), Glutamine (Q); 4) Arginine (R), Lysine (K); 5) Isoleucine (I), Leucine (L), Methionine (M), Alanine (A), Valine (V), and 6) Phenylalanine (F), Tyrosine (Y), Tryptophan (W).
[0188]Sequence homology for polypeptides, which is also referred to as percent sequence identity, is typically measured using sequence analysis software. See, e.g., the Sequence Analysis Software Package of the Genetics Computer Group (GCG), University of Wisconsin Biotechnology Center, 910 University Avenue, Madison, Wis. 53705. Protein analysis software matches similar sequences using measure of homology assigned to various substitutions, deletions and other modifications, including conservative amino acid substitutions. For instance, GCG contains programs such as "Gap" and "Bestfit" which can be used with default parameters to determine sequence homology or sequence identity between closely related polypeptides, such as homologous polypeptides from different species of organisms or between a wild type protein and a mutein thereof. See, e.g., GCG Version 6.1.
[0189]A typical algorithm used comparing a molecule sequence to a database containing a large number of sequences from different organisms is the computer program BLAST (Altschul, S. F., et al. (1990) "Basic local alignment search tool." J. Mol. Biol. 215:403-410; Gish, W. and States, D. J. (1993) "Identification of protein coding regions by database similarity search." Nature Genet. 3:266-272; Madden, T. L., et al. (1996) "Applications of network BLAST server" Meth. Enzymol. 266:131-141; Altschul, S. F., et al. (1997) "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs." Nucleic Acids Res. 25:3389-3402; Zhang, J. and Madden, T. L. (1997) "PowerBLAST: A new network BLAST application for interactive or automated sequence analysis and annotation." Genome Res. 7:649-656), especially blastp or tblastn (Altschul, S. F., et al. (1997) "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs." Nucleic Acids Res. 25:3389-3402). Typical parameters for BLASTp are: Expectation value: 10 (default); Filter: seg (default); Cost to open a gap: 11 (default); Cost to extend a gap: 1 (default); Max. alignments: 100 (default); Word size: 11 (default); No. of descriptions: 100 (default); Penalty Matrix: BLOWSUM62.
[0190]When searching a database containing sequences from a large number of different organisms, it is typical to compare amino acid sequences. Database searching using amino acid sequences can be measured by algorithms other than blastp known in the art. For instance, polypeptide sequences can be compared using FASTA, a program in GCG Version 6.1. FASTA provides alignments and percent sequence identity of the regions of the best overlap between the query and search sequences (Pearson, W. R. (1990) "Rapid and Sensitive Sequence Comparison with FASTP and FASTA" Meth. Enzymol. 183:63-98). For example, percent sequence identity between amino acid sequences can be determined using FASTA with its default parameters (a word size of 2 and the PAM250 scoring matrix), as provided in GCG Version 6.1, hereby incorporated herein by reference.
[0191]It is understood that a range of microorganisms can be modified to include recombinant metabolic pathways suitable for the production of C3-C5 alcohols, including isobutanol. In various embodiments, microorganisms may be selected from bacterial or yeast microorganisms. Microorganisms for the production of C3-C5 alcohols, including isobutanol may be selected based on certain characteristics:
[0192]One characteristic may include the ability to metabolize a carbon source in the presence of a C3-C5 alcohol, including isobutanol. A microorganism capable of metabolizing a carbon source at a high isobutanol concentration is more suitable as a production microorganism compared to a microorganism capable of metabolizing a carbon source at a low isobutanol concentration. Another characteristic may include the property that the microorganism is selected to convert various carbon sources into C3-C5 alcohols, including isobutanol. Accordingly, in one embodiment, the recombinant microorganism herein disclosed can convert a variety of carbon sources to products, including but not limited to glucose, galactose, mannose, xylose, arabinose, lactose, sucrose, and mixtures thereof.
[0193]Another characteristic specific to a yeast microorganism may include the property that the microorganism is able to metabolize a carbon source in the absence of pyruvate decarboxylase (PDC). In an embodiment, it is preferable that the yeast microorganism is able to metabolize 5- and 6-carbon sugar in the absence of PDC. In one embodiment, it is even more preferred that a yeast microorganism is able to grow on 5- and 6-carbon sugars in the absence of PDC.
[0194]Another characteristic may include the property that the wild-type or parental microorganism is non-fermenting. In other words, it cannot metabolize a carbon source anaerobically while the yeast is able to metabolize a carbon source in the presence of oxygen. Non-fermenting yeast refers to both naturally occurring yeasts as well as genetically modified yeast. During anaerobic fermentation with fermentative yeast, the main pathway to oxidize the NADH from glycolysis is through the production of ethanol. Ethanol is produced by alcohol dehydrogenase (ADH) via the reduction of acetaldehyde, which is generated from pyruvate by pyruvate decarboxylase (PDC).
[0195]Thus, in one embodiment, a fermentative yeast can be engineered to be non-fermentative by the reduction or elimination of the native PDC activity. Thus, most of the pyruvate produced by glycolysis is not consumed by PDC and is available for the isobutanol pathway. Deletion of this pathway increases the pyruvate and the reducing equivalents available for the isobutanol pathway. Fermentative pathways contribute to low yield and low productivity of isobutanol. Accordingly, deletion of PDC may increase yield and productivity of isobutanol. In one embodiment, the yeast microorganisms may be selected from the "Saccharomyces Yeast Clade", defined as an ascomycetous yeast taxonomic class by Kurtzman and Robnett in 1998 ("Identification and phylogeny of ascomycetous yeast from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences." Antonie van Leeuwenhoek 73: 331-371, see FIG. 2 of Leeuwenhook reference). They were able to determine the relatedness of yeast of approximately 500 yeast species by comparing the nucleotide sequence of the D1/D2 domain at the 5' end of the gene encoding the large ribosomal subunit 26S. In pair-wise comparisons of the D1/D2 nucleotide sequence of S. cerevisiae and the two most distant yeast in the Saccharomyces clade: K. lactic and K. marxianus, yeast from this clade share greater than 80% identity.
[0196]An ancient whole genome duplication (WGD) event occurred during the evolution of hemiascomycete yeast was discovered using comparative genomics tools (Kellis et al 2004 "Proof and evolutionary analysis of ancient genome duplication in the yeast S. cerevisiae." Nature 428:617-624. Dujon et al 2004 "Genome evolution in yeasts." Nature 430:35-44. Langkjaer et al 2003 "Yeast genome duplication was followed by asynchronous differentiation of duplicated genes." Nature 428:848-852. Wolfe and Shields 1997 "Molecular evidence for an ancient duplication of the entire yeast genome." Nature 387:708-713.) Using this major evolutionary event, yeast can be divided into species that diverged from a common ancestor following the WGD event (termed "post-WGD yeast" herein) and species that diverged from the yeast lineage prior to the WGD event (termed "pre-WGD yeast" herein).
[0197]Accordingly, in one embodiment, the yeast microorganism may be selected from a post-WGD yeast genus, including but not limited to Saccharomyces and Candida. The favored post-WGD yeast species include: S. cerevisiae, S. uvarum, S. bayanus, S. paradoxus, S. castelli, and C. glabrata.
[0198]In another embodiment, a method provided herein includes a recombinant organism that is a Saccharomyces sensu stricto yeast microorganism. In one aspect, a Saccharomyces sensu stricto yeast microorganism is selected from one of the species: S. cerevisiae, S. cerevisiae, S. kudriavzevii, S. mikatae, S. bayanus, S. uvarum, S. carocanis or hybrids thereof.
[0199]In another embodiment, the yeast microorganism may be selected from a pre-whole genome duplication (pre-WBD) yeast genus including but not limited to Saccharomyces, Kluyveromyces, Issatchenkia, Candida, Pichia, Debaryomyces, Hansenula, Pachysolen, Yarrowia and, Schizosaccharomyces. Representative pre-WGD yeast species include: S. kluyveri, K. thermotolerans, K. marxianus, K. waltii, K. lactis, C. tropicalis, P. pastoris, P. anomala, P. stipitis, D. hansenii, H. anomala, P. tannophilis, I. orientalis, Y. lipolytica, and S. pombe.
[0200]A yeast microorganism may be either Crabtree-negative or Crabtree-positive. A yeast cell having a Crabtree-negative phenotype is any yeast cell that does not exhibit the Crabtree effect. The term "Crabtree-negative" refers to both naturally occurring and genetically modified organisms. Briefly, the Crabtree effect is defined as the inhibition of oxygen consumption by a microorganism when cultured under aerobic conditions due to the presence of a high concentration of glucose (e.g., 50 g-glucose L-1). In other words, a yeast cell having a Crabtree-positive phenotype continues to ferment irrespective of oxygen availability due to the presence of glucose, while a yeast cell having a Crabtree-negative phenotype does not exhibit glucose mediated inhibition of oxygen consumption.
[0201]Accordingly, in one embodiment the yeast microorganism may be selected from a yeast with a Crabtree-negative phenotype including but not limited to the following genera: Kluyveromyces, Pichia, Issatchenkia, Hansenula, and Candida. Crabtree-negative species include but are not limited to: K. lactis, K. marxianus, P. anomala, P. stipitis, H. anomala, I. orientalis, and C. utilis.
[0202]In another embodiment, the yeast microorganism may be selected from a yeast with a Crabtree-positive phenotype, including but not limited to Saccharomyces, Kluyveromyces, Zygosaccharomyces, Debaryomyces, Pichia and Schizosaccharomyces. Crabtree-positive yeast species include but are not limited to: S. cerevisiae, S. uvarum, S. bayanus, S. paradoxus, S. castelli, S. kluyveri, K. thermotolerans, C. glabrata, Z. bailli, Z. rouxii, D. hansenii, P. pastorius, and S. pombe.
[0203]Bacterial Microorganisms may be selected from a number of genera, including but not limited to Arthrobacter, Bacillus, Brevibacterium, Clostridium, Corynebacterium, Cyanobacterium, Escherichia, Gluconobacter, Lactobacillus, Nocardia, Pseudomonas, Rhodococcus, Saccharomyces, Shewanella, Streptomyces, Xanthomonas, and Zymomonas. In another embodiment, such hosts are Corynebacterium, Cyanobacterium, E. coli or Pseudomonas. In another embodiment, such hosts are E. coli W3110, E. coli B, Pseudomonas oleovorans, Pseudomonas fluorescens, or Pseudomonas putida.
[0204]One exemplary metabolic pathway for the conversion of a carbon source to a C3-C5 alcohol via pyruvate begins with the conversion of glucose to pyruvate via glycolysis. Glycolysis also produces 2 moles of NADH and 2 moles of ATP. Two moles of pyruvate are then used to produce one mole of isobutanol (PCT/US2006/041602, PCT/US2008/053514). Alternative isobutanol pathways have been described in International Patent Application No PCT/US2006/041602 and in Dickinson et al., Journal of Biological Chemistry 273:25751-15756 (1998).
[0205]Accordingly, the engineered isobutanol pathway to convert pyruvate to isobutanol can be, but is not limited to, the following reactions:
1. 2 pyruvate→acetolactate+CO2 2. acetolactate+NADPH→2,3-dihydroxyisovalerate+NADP+3. 2,3-dihydroxyisovalerate→alpha-ketoisovalerate4. alpha-ketoisovalerate→isobutyraldehyde+CO2 5. isobutyraldehyde+NADPH→isobutanol+NADP.sup.+
[0206]These reactions are carried out by the enzymes 1) Acetolactate Synthase (ALS), 2) Ketol-acid Reducto-Isomerase (KARI), 3) Dihydroxy-acid dehydratase (DHAD), 4) Keto-isovalerate decarboxylase (KIVD), and 5) an Alcohol Dehydrogenase (ADH).
[0207]In another embodiment, the microorganism is engineered to overexpress these enzymes. For example, ALS can be encoded by the alsS gene of B. subtilis, alsS of L. lactis, or the ilvK gene of K. pneumonia. For example, KARI can be encoded by the ilvC genes of E. coli, C. glutamicum, M. maripaludis, or Piromyces sp E2. For example, DHAD can be encoded by the ilvD genes of E. coli, L. lactis, or C. glutamicum, or by the ILV3 gene from S. cerevisiae. KIVD can be encoded by the kivd gene of L. lactis. ADH can be encoded by ADH2, ADH6, or ADH7 of S. cerevisiae, by the adhA gene product of L. lactis, or by an ADH from D. melanogaster.
[0208]The microorganism of the invention may be engineered to have increased ability to convert pyruvate to a C3-C5 alcohol, including isobutanol. In one embodiment, the microorganism may be engineered to have increased ability to convert pyruvate to isobutyraldehyde. In another embodiment, the microorganism may be engineered to have increased ability to convert pyruvate to keto-isovalerate. In another embodiment, the microorganism may be engineered to have increased ability to convert pyruvate to 2,3-dihydroxyisovalerate. In another embodiment, the microorganism may be engineered to have increased ability to convert pyruvate to acetolactate.
[0209]Furthermore, any of the genes encoding the foregoing enzymes (or any others mentioned herein (or any of the regulatory elements that control or modulate expression thereof)) may be optimized by genetic/protein engineering techniques, such as directed evolution or rational mutagenesis.
[0210]It is understood that various microorganisms can act as "sources" for genetic material encoding target enzymes suitable for use in a recombinant microorganism provided herein. For example, In addition, genes encoding these enzymes can be identified from other fungal and bacterial species and can be expressed for the modulation of this pathway. A variety of eukaryotic organisms could serve as sources for these enzymes, including, but not limited to, Drosophila spp., including D. melanogaster, Saccharomyces spp., including S. cerevisiae and S. uvarum, Kluyveromyces spp., including K. thermotolerans, K. lactis, and K. marxianus, Pichia spp., Hansenula spp., including H. polymorpha, Candida spp., Trichosporon spp., Yamadazyma spp., including Y stipitis, Torulaspora pretoriensis, Schizosaccharomyces spp., including S. pombe, Cryptococcus spp., Aspergillus spp., Neurospora spp., or Ustilago spp. Sources of genes from anaerobic fungi include, but not limited to, Piromyces spp., Orpinomyces spp., or Neocallimastix spp. Sources of prokaryotic enzymes that are useful include, but not limited to, Escherichia coli, Klebsiella spp., including K. pneumoniae , Zymomonas mobilis, Staphylococcus aureus, Bacillus spp., Clostridium spp., Corynebacterium spp., Pseudomonas spp., Lactococcus spp., Enterobacter spp., and Salmonella spp.
Methods in General
Gene Expression
[0211]In another embodiment a method of producing a recombinant microorganism that converts a suitable carbon substrate to C3-C5 alcohols such as isobutanol is provided. The method includes transforming a microorganism with one or more recombinant polynucleotides encoding polypeptides that include but are not limited to, for example, ALS, KARI, DHAD, KIVD, ADH and a transhydrogenase. Polynucleotides that encode enzymes useful for generating metabolites including homologs, variants, fragments, related fusion proteins, or functional equivalents thereof, are used in recombinant nucleic acid molecules that direct the expression of such polypeptides in appropriate host cells, such as bacterial or yeast cells. It is understood that the addition of sequences which do not alter the encoded activity of a polynucleotide, such as the addition of a non-functional or non-coding sequence, is a conservative variation of the basic nucleic acid. The "activity" of an enzyme is a measure of its ability to catalyze a reaction resulting in a metabolite, i.e., to "function", and may be expressed as the rate at which the metabolite of the reaction is produced. For example, enzyme activity can be represented as the amount of metabolite produced per unit of time or per unit of enzyme (e.g., concentration or weight), or in terms of affinity or dissociation constants.
[0212]Those of skill in the art will recognize that, due to the degenerate nature of the genetic code, a variety of DNA compounds differing in their nucleotide sequences can be used to encode a given amino acid sequence of the disclosure. The native DNA sequence encoding the biosynthetic enzymes described herein are referenced herein merely to illustrate an embodiment of the disclosure, and the disclosure includes DNA compounds of any sequence that encode the amino acid sequences of the polypeptides and proteins of the enzymes utilized in the methods of the disclosure. In similar fashion, a polypeptide can typically tolerate one or more amino acid substitutions, deletions, and insertions in its amino acid sequence without loss or significant loss of a desired activity. The disclosure includes such polypeptides with alternate amino acid sequences, and the amino acid sequences encoded by the DNA sequences shown herein merely illustrate embodiments of the disclosure.
[0213]The disclosure provides nucleic acid molecules in the form of recombinant DNA expression vectors or plasmids, as described in more detail below, that encode one or more target enzymes. Generally, such vectors can either replicate in the cytoplasm of the host microorganism or integrate into the chromosomal DNA of the host microorganism. In either case, the vector can be a stable vector (i.e., the vector remains present over many cell divisions, even if only with selective pressure) or a transient vector (i.e., the vector is gradually lost by host microorganisms with increasing numbers of cell divisions). The disclosure provides DNA molecules in isolated (i.e., not pure, but existing in a preparation in an abundance and/or concentration not found in nature) and purified (i.e., substantially free of contaminating materials or substantially free of materials with which the corresponding DNA would be found in nature) forms.
[0214]Provided herein are methods for the expression of one or more of the genes involved in the production of beneficial metabolites and recombinant DNA expression vectors useful in the method. Thus, included within the scope of the disclosure are recombinant expression vectors that include such nucleic acids. The term expression vector refers to a nucleic acid that can be introduced into a host microorganism or cell-free transcription and translation system. An expression vector can be maintained permanently or transiently in a microorganism, whether as part of the chromosomal or other DNA in the microorganism or in any cellular compartment, such as a replicating vector in the cytoplasm. An expression vector also comprises a promoter that drives expression of an RNA, which typically is translated into a polypeptide in the microorganism or cell extract. For efficient translation of RNA into protein, the expression vector also typically contains a ribosome-binding site sequence positioned upstream of the start codon of the coding sequence of the gene to be expressed. Other elements, such as enhancers, secretion signal sequences, transcription termination sequences, and one or more marker genes by which host microorganisms containing the vector can be identified and/or selected, may also be present in an expression vector. Selectable markers, i.e., genes that confer antibiotic resistance or sensitivity, are used and confer a selectable phenotype on transformed cells when the cells are grown in an appropriate selective medium.
[0215]The various components of an expression vector can vary widely, depending on the intended use of the vector and the host cell(s) in which the vector is intended to replicate or drive expression. Expression vector components suitable for the expression of genes and maintenance of vectors in E. coli, yeast, Streptomyces, and other commonly used cells are widely known and commercially available. For example, suitable promoters for inclusion in the expression vectors of the disclosure include those that function in eukaryotic or prokaryotic host microorganisms. Promoters can comprise regulatory sequences that allow for regulation of expression relative to the growth of the host microorganism or that cause the expression of a gene to be turned on or off in response to a chemical or physical stimulus. For E. coli and certain other bacterial host cells, promoters derived from genes for biosynthetic enzymes, antibiotic-resistance conferring enzymes, and phage proteins can be used and include, for example, the galactose, lactose (lac), maltose, tryptophan (trp), beta-lactamase (bla), bacteriophage lambda PL, and T5 promoters. In addition, synthetic promoters, such as the tac promoter (U.S. Pat. No. 4,551,433), can also be used. For E. coli expression vectors, it is useful to include an E. coli origin of replication, such as from pUC, p1P, p1, and pBR.
[0216]Thus, recombinant expression vectors contain at least one expression system, which, in turn, is composed of at least a portion of PKS and/or other biosynthetic gene coding sequences operably linked to a promoter and optionally termination sequences that operate to effect expression of the coding sequence in compatible host cells. The host cells are modified by transformation with the recombinant DNA expression vectors of the disclosure to contain the expression system sequences either as extrachromosomal elements or integrated into the chromosome.
[0217]Moreover, methods for expressing a polypeptide from a nucleic acid molecule that are specific to yeast microorganisms are well known. For example, nucleic acid constructs that are used for the expression of heterologous polypeptides within Kluyveromyces and Saccharomyces are well known (see, e.g., U.S. Pat. Nos. 4,859,596 and 4,943,529, each of which is incorporated by reference herein in its entirety for Kluyveromyces and, e.g., Gellissen et al., Gene 190(1):87-97 (1997) for Saccharomyces. Yeast plasmids have a selectable marker and an origin of replication, also known as Autonomously Replicating Sequences (ARS). In addition certain plasmids may also contain a centromeric sequence. These centromeric plasmids are generally a single or low copy plasmid. Plasmids without a centromeric sequence and utilizing either a 2 micron (S. cerevisiae) or 1.6 micron (K. lactis) replication origin are high copy plasmids. The selectable marker can be either prototrophic, such as HIS3, TRP1, LEU2, URA3 or ADE2, or antibiotic resistance, such as, bar, ble, hph, or kan.
[0218]A nucleic acid of the disclosure can be amplified using cDNA, mRNA or alternatively, genomic DNA, as a template and appropriate oligonucleotide primers according to standard PCR amplification techniques and those procedures described in the Examples section below. The nucleic acid so amplified can be cloned into an appropriate vector and characterized by DNA sequence analysis. Furthermore, oligonucleotides corresponding to nucleotide sequences can be prepared by standard synthetic techniques, e.g., using an automated DNA synthesizer.
[0219]It is also understood that an isolated nucleic acid molecule encoding a polypeptide homologous to the enzymes described herein can be created by introducing one or more nucleotide substitutions, additions or deletions into the nucleotide sequence encoding the particular polypeptide, such that one or more amino acid substitutions, additions or deletions are introduced into the encoded protein. Mutations can be introduced into the polynucleotide by standard techniques, such as site-directed mutagenesis and PCR-mediated mutagenesis. In contrast to those positions where it may be desirable to make a non-conservative amino acid substitutions (see above), in some positions it is preferable to make conservative amino acid substitutions. A "conservative amino acid substitution" is one in which the amino acid residue is replaced with an amino acid residue having a similar side chain. Families of amino acid residues having similar side chains have been defined in the art. These families include amino acids with basic side chains (e.g., lysine, arginine, histidine), acidic side chains (e.g., aspartic acid, glutamic acid), uncharged polar side chains (e.g., glycine, asparagine, glutamine, serine, threonine, tyrosine, cysteine), nonpolar side chains (e.g., alanine, valine, leucine, isoleucine, proline, phenylalanine, methionine, tryptophan); beta-branched side chains (e.g., threonine, valine, isoleucine) and aromatic side chains (e.g., tyrosine, phenylalanine, tryptophan, histidine).
[0220]Although the effect of an amino acid change varies depending upon factors such as phosphorylation, glycosylation, intra-chain linkages, tertiary structure, and the role of the amino acid in the active site or a possible allosteric site, it is generally preferred that the substituted amino acid is from the same group as the amino acid being replaced. To some extent the following groups contain amino acids which are interchangeable: the basic amino acids lysine, arginine, and histidine; the acidic amino acids aspartic and glutamic acids; the neutral polar amino acids serine, threonine, cysteine, glutamine, asparagine and, to a lesser extent, methionine; the nonpolar aliphatic amino acids glycine, alanine, valine, isoleucine, and leucine (however, because of size, glycine and alanine are more closely related and valine, isoleucine and leucine are more closely related); and the aromatic amino acids phenylalanine, tryptophan, and tyrosine. In addition, although classified in different categories, alanine, glycine, and serine seem to be interchangeable to some extent, and cysteine additionally fits into this group, or may be classified with the polar neutral amino acids.
Overexpression of Heterologous Genes
[0221]Methods for overexpressing a polypeptide from a native or heterologous nucleic acid molecule are well known. Such methods include, without limitation, constructing a nucleic acid sequence such that a regulatory element promotes the expression of a nucleic acid sequence that encodes the desired polypeptide. Typically, regulatory elements are DNA sequences that regulate the expression of other DNA sequences at the level of transcription. Thus, regulatory elements include, without limitation, promoters, enhancers, and the like. For example, the exogenous genes can be under the control of an inducible promoter or a constitutive promoter. Moreover, methods for expressing a polypeptide from an exogenous nucleic acid molecule in yeast are well known. For example, nucleic acid constructs that are used for the expression of exogenous polypeptides within Kluyveromyces and Saccharomyces are well known (see, e.g., U.S. Pat. Nos. 4,859,596 and 4,943,529, for Kluyveromyces and, e.g., Gellissen et al., Gene 190(1):87-97 (1997) for Saccharomyces). Yeast plasmids have a selectable marker and an origin of replication. In addition certain plasmids may also contain a centromeric sequence. These centromeric plasmids are generally a single or low copy plasmid. Plasmids without a centromeric sequence and utilizing either a 2 micron (S. cerevisiae) or 1.6 micron (K. lactis) replication origin are high copy plasmids. The selectable marker can be either prototrophic, such as HIS3, TRP1, LEU2, URA3 or ADE2, or antibiotic resistance, such as, bar, ble, hph, or kan.
[0222]In another embodiment, heterologous control elements can be used to activate or repress expression of endogenous genes. Additionally, when expression is to be repressed or eliminated, the gene for the relevant enzyme, protein or RNA can be eliminated by known deletion techniques.
[0223]As described herein, any microorganism within the scope of the disclosure can be identified by selection techniques specific to the particular enzyme being expressed, over-expressed or repressed. Methods of identifying the strains with the desired phenotype are well known to those skilled in the art. Such methods include, without limitation, PCR, RT-PCR, and nucleic acid hybridization techniques such as Northern and Southern analysis, altered growth capabilities on a particular substrate or in the presence of a particular substrate, a chemical compound, a selection agent and the like. In some cases, immunohistochemistry and biochemical techniques can be used to determine if a cell contains a particular nucleic acid by detecting the expression of the encoded polypeptide. For example, an antibody having specificity for an encoded enzyme can be used to determine whether or not a particular microorganism contains that encoded enzyme. Further, biochemical techniques can be used to determine if a cell contains a particular nucleic acid molecule encoding an enzymatic polypeptide by detecting a product produced as a result of the expression of the enzymatic polypeptide. For example, transforming a cell with a vector encoding acetolactate synthase and detecting increased cytosolic acetolactate concentrations compared to a cell without the vector indicates that the vector is both present and that the gene product is active. Methods for detecting specific enzymatic activities or the presence of particular products are well known to those skilled in the art. For example, the presence of acetolactate can be determined as described by Hugenholtz and Starrenburg, Appl. Microbiol. Biotechnol. 38:17-22 (1992).
Identification of Genes in a Host Microorganism
[0224]Any method can be used to identify genes that encode for enzymes with a specific activity. Generally, homologous or analogous genes with similar activity can be identified by functional, structural, and/or genetic analysis. In most cases, homologous or analogous genes with similar activity will have functional, structural, or genetic similarities. Techniques known to those skilled in the art may be suitable to identify homologous genes and homologous enzymes. Generally, analogous genes and/or analogous enzymes can be identified by functional analysis and will have functional similarities. Techniques known to those skilled in the art may be suitable to identify analogous genes and analogous enzymes. For example, to identify homologous or analogous genes, proteins, or enzymes, techniques may include, but not limited to, cloning a gene by PCR using primers based on a published sequence of a gene/enzyme or by degenerate PCR using degenerate primers designed to amplify a conserved region among a gene. Further, one skilled in the art can use techniques to identify homologous or analogous genes, proteins, or enzymes with functional homology or similarity. Techniques include examining a cell or cell culture for the catalytic activity of an enzyme through in vitro enzyme assays for said activity, then isolating the enzyme with said activity through purification, determining the protein sequence of the enzyme through techniques such as Edman degradation, design of PCR primers to the likely nucleic acid sequence, amplification of said DNA sequence through PCR, and cloning of said nucleic acid sequence. To identify homologous or analogous genes with similar activity, techniques also include comparison of data concerning a candidate gene or enzyme with databases such as BRENDA, KEGG, or MetaCYC. The candidate gene or enzyme may be identified within the above mentioned databases in accordance with the teachings herein. Furthermore, enzymatic activity can be determined phenotypically. For example, ethanol production under fermentative conditions can be assessed. A lack of ethanol production may be indicative of a microorganism lacking an alcohol dehydrogenase capable of reducing acetaldehyde to ethanol.
Genetic Insertions and Deletions
[0225]Any method can be used to introduce a nucleic acid molecule into the chromosomal DNA of a microorganism and many such methods are well known. For example, lithium acetate transformation and electroporation are common methods for introducing nucleic acid into yeast microorganisms. See, e.g., Gietz et al., Nucleic Acids Res. 27:69-74 (1992); Ito et al., J. Bacterol. 153:163-168 (1983); and Becker and Guarente, Methods in Enzymology 194:182-187 (1991).
[0226]In an embodiment, the deletion of a gene of interest in a bacterial microorganism, including an E. coli microorganism occurs according to the principle of homologous recombination. According to this embodiment, an integration cassette containing a module comprising at least one marker gene is flanked on either side by DNA fragments homologous to those of the ends of the targeted integration site. After transforming the host microorganism with the cassette by appropriate methods, homologous recombination between the flanking sequences may result in the marker replacing the chromosomal region in between the two sites of the genome corresponding to flanking sequences of the integration cassette. The homologous recombination event may be facilitated by a recombinase enzyme that may be native to the host microorganism or may be heterologous and transiently overexpressed (Datsenko and Wanner, Proc. Natl. Acad. Sci. USA 97, 6640-6645, 2000).
[0227]In an embodiment, the integration of a gene of interest into a DNA fragment or target gene of a yeast microorganism occurs according to the principle of homologous recombination. According to this embodiment, an integration cassette containing a module comprising at least one yeast marker gene and/or the gene to be integrated (internal module) is flanked on either side by DNA fragments homologous to those of the ends of the targeted integration site (recombinogenic sequences). After transforming the yeast with the cassette by appropriate methods, a homologous recombination between the recombinogenic sequences may result in the internal module replacing the chromosomal region in between the two sites of the genome corresponding to the recombinogenic sequences of the integration cassette. (Orr-Weaver et al., Proc Natl Acad Sci USA 78:6354-6358 (1981))
[0228]In an embodiment, the integration cassette for integration of a gene of interest into a yeast microorganism includes the heterologous gene under the control of an appropriate promoter and terminator together with the selectable marker flanked by recombinogenic sequences for integration of a heterologous gene into the yeast chromosome. In an embodiment, the heterologous gene includes an appropriate native gene desired to increase the copy number of a native gene(s). The selectable marker gene can be any marker gene used in yeast, including but not limited to, HIS3, TRP1, LEU2, URA3, bar, ble, hph, and kan. The recombinogenic sequences can be chosen at will, depending on the desired integration site suitable for the desired application.
[0229]Additionally, in an embodiment pertaining to yeast microorganisms, certain introduced marker genes are removed from the genome using techniques well known to those skilled in the art. For example, URA3 marker loss can be obtained by plating URA3 containing cells in FOA (5-fluoro-orotic acid) containing medium and selecting for FOA resistant colonies (Boeke, J. et al, 1984, Mol. Gen. Genet, 197, 345-47).
[0230]Integration of all the genes of a metabolic pathway that lead to a product into the genome of the production strain eliminates the need of a plasmid expression system, as the enzymes are produced from the chromosome. The integration of pathway genes avoids loss of productivity over time due to plasmid loss. This is important for long fermentation times and for fermentations in large scale where the seed train is long and the production strain has to go through many doublings from the first inoculation to the end of the large scale fermentation.
[0231]Integrated genes are maintained in the strain without selection. This allows the construction of production strains that are free of marker genes which are commonly used for maintenance of plasmids. Production strains with integrated pathway genes can contain minimal amounts of foreign DNA since there are no origins of replication and other non coding DNA necessary that have to be in plasmid based systems. The biocatalyst with integrated pathway genes improves the performance of a production process because it avoids energy and carbon requiring processes. These processes are the replication of many copies of plasmids and the production of non-pathway active proteins like marker proteins in the production strain.
[0232]The expression of pathway genes on multi-copy plasmids can lead to overexpression phenotypes for certain genes. These phenotypes can be growth retardation, inclusion bodies, and cell death. Therefore the expression levels of genes on multi copy plasmids has to be controlled effectively by using inducible expression systems, optimizing the time of induction of said expression system, and optimizing the amount of inducer provided. The time of induction has to be correlated to the growth phase of the biocatalyst, which can be followed by measuring of optical density in the fermentation broth.
[0233]A biocatalyst that has all pathway genes integrated on its chromosome is far more likely to allow constitutive expression since the lower number of gene copies may avoid overexpression phenotypes.
[0234]Plasmids disclosed herein were generally based upon parental plasmids described previously (Lutz, R. & Bujard, H. (1997) Nucleic Acids Research 25(6):1203-1210). Plasmids pGV1698 (SEQ ID NO: 112) and pGV1655 (SEQ ID NO: 109) produce optimized levels of isobutanol pathway enzymes in a production host when compared to other expression systems in the art. Compared to the expression of the isobutanol pathway from pSA55 and pSA69 as described in (WO 2008/098227) BIOFUEL PRODUCTION BY RECOMBINANT MICROORGANISMS, pGV1698 and pGV1655 lead to higher expression of E. coli llvC and Bacillus subtilis AlsS and lower expression levels for Lactococcus lactis Kivd and E. coli ilvD. These changes are the result of differences in plasmid copy numbers. Also the genes coding for E. coli llvD and E. coli llvC were codon optimized for E. coli. This leads to optimized expression of the genes and it also avoids recombination of these genes with their native copies on the E. coli chromosome, thus stabilizing the production strain. The combination of two plasmids with the pSC101 and the ColE1 origin of replication in one cell as realized in a production strain carrying pGV1698 and pGV1655 is known to be more stable than the combination of two plasmids with p15A and ColE1 origins respectively as was used in the prior art (WO 2008/098227--BIOFUEL PRODUCTION BY RECOMBINANT MICROORGANISMS).
Reduction of Enzymatic Activity
[0235]Host microorganisms within the scope of the invention may have reduced enzymatic activity such as reduced alcohol dehydrogenase activity. The term "reduced" as used herein with respect to a particular enzymatic activity refers to a lower level of enzymatic activity than that measured in a comparable host cell of the same species. Thus, host cells lacking alcohol dehydrogenase activity are considered to have reduced alcohol dehydrogenase activity since most, if not all, comparable host cells of the same species have at least some alcohol dehydrogenase activity. Such reduced enzymatic activities can be the result of lower enzyme expression level, lower specific activity of an enzyme, or a combination thereof. Many different methods can be used to make host cells having reduced enzymatic activity. For example, a host cell can be engineered to have a disrupted enzyme-encoding locus using common mutagenesis or knock-out technology. See, e.g., Methods in Yeast Genetics (1997 edition), Adams, Gottschling, Kaiser, and Stems, Cold Spring Harbor Press (1998), Datsenko and Wanner, Proc. Natl. Acad. Sci. USA 97, 6640-6645, 2000.
[0236]In addition, certain point-mutation(s) can be introduced which results in an enzyme with reduced activity.
[0237]Alternatively, antisense technology can be used to reduce enzymatic activity. For example, host cells can be engineered to contain a cDNA that encodes an antisense molecule that prevents an enzyme from being made. The term "antisense molecule" as used herein encompasses any nucleic acid molecule that contains sequences that correspond to the coding strand of an endogenous polypeptide. An antisense molecule also can have flanking sequences (e.g., regulatory sequences). Thus antisense molecules can be ribozymes or antisense oligonucleotides. A ribozyme can have any general structure including, without limitation, hairpin, hammerhead, or axhead structures, provided the molecule cleaves RNA.
[0238]Host cells having a reduced enzymatic activity can be identified using many methods. For example, host cells having reduced alcohol dehydrogenase activity can be easily identified using common methods, which may include, for example, measuring ethanol formation via gas chromatography.
Increase of Enzymatic Activity
[0239]Host microorganisms of the invention may be further engineered to have increased activity of enzymes. The term "increased" as used herein with respect to a particular enzymatic activity refers to a higher level of enzymatic activity than that measured in a comparable yeast cell of the same species. For example, overexpression of a specific enzyme can lead to an increased level of activity in the cells for that enzyme. Increased activities for enzymes involved in glycolysis or the isobutanol pathway would result in increased productivity and yield of isobutanol.
[0240]Methods to increase enzymatic activity are known to those skilled in the art. Such techniques may include increasing the expression of the enzyme by increasing plasmid copy number and/or use of a stronger promoter and/or use of activating riboswitches, introduction of mutations to relieve negative regulation of the enzyme, introduction of specific mutations to increase specific activity and/or decrease the KM for the substrate, or by directed evolution. See, e.g., Methods in Molecular Biology (vol. 231), ed. Arnold and Georgiou, Humana Press (2003).
Microorganism in Detail
Microorganism Characterized by the Ability to Produce Isobutanol Under Anaerobic Conditions
[0241]Economic studies indicate that the aeration of a fermentation process leads to increased operating and capital expenses and thus makes such a fermentation process less desirable compared to a fermentation process that operates under anaerobic conditions. In addition, yield and aeration conditions are closely related. For example, oxygen used as the terminal electron acceptor in respiration leads to undesired loss of carbon in the form of carbon dioxide, resulting in a reduced yield of the target compound.
[0242]As exemplified in the examples below, the present inventors have overcome the problem of an oxygen requirement for the production of a fermentation product. For example isobutanol was produced anaerobically at rates, titers and yields comparable to those achieved under micro-aerobic conditions.
[0243]Thus, in one embodiment, a modified microorganism may produce said fermentation product under anaerobic conditions, conditions at higher rates, and yields, as compared to a the wild-type or parental microorganism.
[0244]In one embodiment, said modified microorganism may be engineered to balance cofactor usage during the production of said fermentation product under anaerobic conditions.
[0245]In a specific aspect, a modified microorganism in which cofactor usage is balanced during the production of isobutanol may allow the microorganism to produce said isobutanol under anaerobic conditions at higher rates and yields as compared to a modified microorganism in which the cofactor usage in not balanced during production of isobutanol. One compound to be produced by the recombinant microorganism according to the present invention is isobutanol. However, the present invention is not limited to isobutanol. The invention may be applicable to any metabolic pathway that is imbalanced with respect to cofactor usage. One of skill in the art is able identify pathways that are imbalanced with respect to cofactor usage and apply this invention to provide recombinant microorganisms in which the same pathway is balanced with respect to cofactor usage.
[0246]Any method, including the methods described herein may be used to provide a modified microorganism with a metabolic pathway for the production of a target compound in which the cofactor usage is balanced; i.e. said metabolic pathway utilizes the same cofactor that is produced during glycolysis.
[0247]In one embodiment, the microorganism may converts glucose, which can be derived from biomass into a target compound under anaerobic conditions with a yield of greater than 75% of theoretical. In another embodiment, the yield is greater than 80% of theoretical. In another embodiment the yield is greater than 85% of theoretical. In another embodiment, the yield is greater than 90% of theoretical. In another embodiment, the yield is greater than 95% of theoretical. In another embodiment, the yield is greater than 97% of theoretical. In another embodiment the yield is greater than 98% of theoretical. In yet another embodiment, the yield is greater than 99% of theoretical. In still another embodiment, the yield is approximately 100% of theoretical
[0248]In one aspect, the microorganism may convert glucose, which can be derived from biomass into isobutanol under anaerobic conditions with a yield of greater than 50% of theoretical. In one embodiment, the yield is greater than 60% theoretical. In another embodiment, the yield is greater than 70% of theoretical. In yet another embodiment the yield is greater than 80% of theoretical. In yet another embodiment, the yield is greater than 85% of theoretical. In another embodiment, the yield is greater than 90% of theoretical. In yet another embodiment, the yield is greater than 95% of theoretical. In yet another embodiment, the yield is greater than 97% of theoretical. In yet another embodiment the yield is greater than 98% of theoretical. In yet another embodiment, the yield is greater than 99% of theoretical. In still another embodiment, the yield is approximately 100% of theoretical.
[0249]It is understood that while in the present disclosure the yield is exemplified for glucose as a carbon source, the invention can be applied to other carbon sources and the yield may vary depending on the carbon source used. One skilled in the art can calculate yields on various carbon sources. Other carbon sources, such as including but not limited to galactose, mannose, xylose, arabinose, sucrose, lactose, may be used. Further, oligomers or polymers of these and other sugars may be used as a carbon source.
Microorganism Characterized by an Increased Product Yield
[0250]Economic studies indicate that the predominant factor accounting for the production cost for commodity chemicals and fuels from fermentation processes is attributed to the feedstock cost. In fact, as much as 60% of the variable cash operating costs or more may be attributable to feedstock costs. An important measure of the process economics is therefore the product yield. For a biocatalyst to produce a biofuel most economically, a single product is desired. Extra products reduce primary product yield increasing capital and operating costs, particularly if those extra, undesired products, or byproducts have little or no value. Extra products or byproducts also require additional capital and operating costs to separate these products from the product or biofuel of interest or may require additional cost for disposal.
[0251]As exemplified in the examples below, the present inventors have shown that, achieving cofactor balance increases the yield of fermentation products as compared to wild-type or parental organisms.
[0252]In an embodiment, a microorganism is provided in which cofactor usage is balanced during the production of a fermentation product and the microorganism produces the fermentation product at a higher yield compared to a modified microorganism in which the cofactor usage in not balanced.
[0253]In a specific aspect of the present invention, a microorganism is provided in which cofactor usage is balanced during the production of isobutanol and the microorganism produces isobutanol at a higher yield compared to a modified microorganism in which the cofactor usage in not balanced.
[0254]One compound to be produced by the recombinant microorganism according to the present invention is isobutanol. However, the present invention is not limited to isobutanol. The invention may be applicable to any microorganism comprising a metabolic pathway that leads to an imbalance with respect to cofactor usage. One of skill in the art is able to identify microorganisms comprising metabolic pathways that lead to an imbalance with respect to cofactor usage and apply this invention to provide recombinant microorganisms in which the microorganism comprising the same metabolic pathway is balanced with respect to cofactor usage.
[0255]Any method, including the methods described herein may be used to provide a modified microorganism with a metabolic pathway for the production of a target compound in which the cofactor usage is balanced; i.e. said metabolic pathway utilizes the same cofactor that is produced during glycolysis.
[0256]In one embodiment, the microorganism may convert glucose, which can be derived from biomass into a target compound with a yield of greater than 75% of theoretical. In another embodiment, the yield is greater than 80% of theoretical. In another embodiment the yield is greater than 85% of theoretical. In another embodiment, the yield is greater than 90% of theoretical. In another embodiment, the yield is greater than 95% of theoretical. In another embodiment, the yield is greater than 97% of theoretical. In another embodiment the yield is greater than 98% of theoretical. In yet another embodiment, the yield is greater than 99% of theoretical. In still another embodiment, the yield is approximately 100% of theoretical
[0257]In one aspect, the microorganism may convert glucose, which can be derived from biomass into isobutanol with a yield of greater than 75% of theoretical. In one embodiment, the yield is greater than 80% of theoretical. In one embodiment the yield is greater than 85% of theoretical. In another embodiment, the yield is greater than 90% of theoretical. In yet another embodiment, the yield is greater than 95% of theoretical. In yet another embodiment, the yield is greater than 97% of theoretical. In yet another embodiment the yield is greater than 98% of theoretical. In yet another embodiment, the yield is greater than 99% of theoretical. In still another embodiment, the yield is approximately 100% of theoretical.
[0258]It is understood that while in the present disclosure the yield is exemplified for glucose as a carbon source, the invention can be applied to other carbon sources and the yield may vary depending on the carbon source used. One skilled in the art can calculate yields on various carbon sources. Other carbon sources, such as including but not limited to galactose, mannose, xylose, arabinose, sucrose, lactose, may be used. Further, oligomers or polymers of these and other sugars may be used as a carbon source.
Microorganism Characterized by Balancing Cofactor Usage
[0259]The ideal production microorganism produces a desirable product at close to theoretical yield. For example the ideal isobutanol producing organism produces isobutanol according to the following equation:
1 glucose→isobutanol+2 CO2+H2O
[0260]Accordingly, 66% of the glucose carbon results in isobutanol, while 33% is lost as CO2. In exemplary metabolic pathways for the conversion of pyruvate to isobutanol described by Atsumi et al. (Atsumi et al., Nature, 2008 Jan. 3; 451(7174):86-9, which is herein incorporated by reference; International Patent Application No PCT/US2008/053514, which is herein incorporated by reference) two of the five enzymes used to convert pyruvate into isobutanol according to the metabolic pathway outlined in FIG. 1 require the reduced cofactor nicotinamide adenine dinucleotide phosphate (NADPH). NADPH is produced only sparingly by the cell--the reduced cofactor nicotinamide adenine dinucleotide (NADH) is the preferred equivalent. Respiration is required to produce NADPH in the large quantities required to support high-level production of isobutanol.
[0261]Even If competing pathways can be eliminated or reduced in activity by metabolic engineering, yield is limited to about 83% of theoretical. Carbon loss to carbon dioxide (CO2) remains the main limitation on yield in the aforementioned metabolic pathway for the production of isobutanol. Reducing the oxygen uptake rate (OUR) of the cells should decrease the loss of carbon to CO2 because it decreases the metabolic flux through the CO2-generating tricarboxylic acid (TCA) cycle and/or pentose phosphate pathway (PPP). However, a modified microorganism utilizing the aforementioned metabolic pathway for the production of isobutanol exhibits drastically decreased specific productivity under conditions where the OUR is decreased and isobutanol production under anaerobic conditions may not be possible.
[0262]The decreased yield and the loss of productivity upon O2 limitation indicate that the strain uses one or more metabolic pathways to generate the NADPH needed to support isobutanol production. In a modified cell utilizing the aforementioned metabolic pathway the production of isobutanol from glucose results in an imbalance between the cofactors reduced during glycolysis and the cofactors oxidized during the conversion of pyruvate to isobutanol. While glycolysis produces two moles of NADH, the isobutanol pathway consumes two moles of NADPH. This leads to a deficit of two moles of NADPH and overproduction of two moles of NADH per isobutanol molecule produced, a state described henceforth as cofactor imbalance.
[0263]The terms "cofactor balance" or "balanced with respect to cofactor usage" refer to a recombinant microorganism comprising a metabolic pathway converting a carbon source to a fermentation product and a modification that leads to the regeneration of all redox cofactors within the recombinant microorganism producing said fermentation product from a carbon source and wherein the re-oxidation or re-reduction of said redox cofactors does not require the pentose phosphate pathway, the TCA cycle or the generation of additional fermentation products.
[0264]Stated another way, the terms "cofactor balance" or "balanced with respect to cofactor usage" can refer to an advantageous modification that leads to the regeneration of all redox cofactors within the recombinant microorganism producing a fermentation product from a carbon source and wherein said re-oxidation or re-reduction of all redox cofactors does not require the production of byproducts or co-products.
[0265]Stated another way, the terms "cofactor balance" or "balanced with respect to cofactor usage" can refer to an advantageous modification that leads to the regeneration of all redox cofactors within the recombinant microorganism producing a fermentation product from a carbon source under anaerobic conditions and wherein the production of additional fermentation products is not required for re-oxidation or re-reduction of redox cofactors.
[0266]Stated another way, the terms "cofactor balance" or "balanced with respect to cofactor usage" can refer to an advantageous modification that leads to the regeneration of all redox cofactors within the recombinant microorganism producing a fermentation product from a carbon source and wherein said modification increases production of said fermentation product under anaerobic conditions compared to the parental or wild type microorganism and wherein additional fermentation products are not required for the regeneration of said redox cofactors.
[0267]The cell has several options for resolving a cofactor imbalance. One is to change the relative fluxes going from glucose through glycolysis and through the pentose phosphate pathway (PPP). For each glucose molecule metabolized through the PPP, two moles of NADPH are generated in addition to the two moles of NADH that are generated through glycolysis (a total of 4 reducing equivalents). Therefore, use of the PPP results in the generation of excess reducing equivalents since only two moles are consumed during the production of isobutanol. Under anaerobic conditions, and without an alternate electron acceptor, the cell has no way to reoxidize or regenerate these extra cofactors to NADP.sup.+ and metabolism thus stops. The excess reducing equivalents must instead be utilized for energy production through aerobic respiration which is only possible under aerobic conditions or for the production of byproducts. Another result of the flux through the PPP is that one additional molecule of CO2 is lost per molecule of glucose consumed, which limits the yield of isobutanol that can be achieved under aerobic conditions.
[0268]Another way the cell can generate NADPH is via the TCA cycle. Flux through the TCA cycle results in carbon loss through CO2 and in production of NADH in addition to the NADPH required for the isobutanol pathway. The NADH would have to be utilized for energy production through respiration under aerobic conditions (and without an alternate electron acceptor) or for the production of byproducts. In addition, the TCA cycle likely is not functional under anaerobic conditions and is therefore unsuitable for the production of stoichiometric amounts of NADPH in an anaerobic isobutanol process.
[0269]An economically competitive isobutanol process requires a high yield from a carbon source. Lower yield means that more feedstock is required to produce the same amount of isobutanol. Feedstock cost is the major component of the overall operating cost, regardless of the nature of the feedstock and its current market price. From an economical perspective, this is important because the cost of isobutanol is dependent on the cost of the biomass-derived sugars. An increase in feedstock cost results in an increase in isobutanol cost. Thus, it is desirable to utilize NADH-dependent enzymes for the conversion of pyruvate to isobutanol.
[0270]An enzyme is "NADH-dependent" if it catalyzes the reduction of a substrate coupled to the oxidation of NADH with a catalytic efficiency that is greater than the reduction of the same substrate coupled to the oxidation of NADPH at equal substrate and cofactor concentrations.
[0271]Thus, in one embodiment of the invention, a microorganism is provided in which cofactor usage is balanced during the production of a fermentation product.
[0272]In a specific aspect, a microorganism is provided in which cofactor usage is balanced during the production of isobutanol, in this case, production of isobutanol from pyruvate utilizes the same cofactor that is produced during glycolysis.
[0273]In another embodiment, a microorganism is provided in which cofactor usage is balanced during the production of a fermentation product and the microorganism produces the fermentation product at a higher yield compared to a modified microorganism in which the cofactor usage in not balanced.
[0274]In a specific aspect, a microorganism is provided in which cofactor usage is balanced during the production of isobutanol and the microorganism produces isobutanol at a higher yield compared to a modified microorganism in which the cofactor usage in not balanced.
[0275]In yet another embodiment, a modified microorganism in which cofactor usage is balanced during the production of a fermentation product may allow the microorganism to produce said fermentation product under anaerobic conditions at higher rates, and yields as compared to a modified microorganism in which the cofactor usage in not balanced during production of a fermentation product.
[0276]In a specific aspect, a modified microorganism in which cofactor usage is balanced during the production of isobutanol may allow the microorganism to produce isobutanol under anaerobic conditions at higher rates, and yields as compared to a modified microorganism in which the cofactor usage is not balanced during production of isobutanol.
[0277]One compound to be produced by the recombinant microorganism according to the present invention is isobutanol. However, the present invention is not limited to isobutanol. The invention may be applicable to any metabolic pathway that is imbalanced with respect to cofactor usage. One skilled in the art is able to identify pathways that are imbalanced with respect to cofactor usage and apply this invention to provide recombinant microorganisms in which the same pathway is balanced with respect to cofactor usage. One skilled in the art will recognize that the identified pathways may be of longer or shorter length, contain more or fewer genes or proteins, and require more or fewer cofactors than the exemplary isobutanol pathway. Further, one skilled in the art will recognize that in certain embodiments, such as a recombinant microbial host that produces an excess of NADPH, certain embodiments of the present invention may be adapted to convert NADPH to NADH.
Microorganism Characterized by Providing Cofactor Balance Via Overexpression of a Transhydrogenase
[0278]Conversion of glucose to pyruvate via glycolysis in E. coli leads to the production of two moles of NADH. A metabolic pathway that converts pyruvate to a target product that consumes either two moles of NADPH or one mole of NADH and one mole of NADPH leads to cofactor imbalance. For example, the isobutanol metabolic pathway that converts glucose to two moles of pyruvate via glycolysis to 1 mole of isobutanol generates two moles of NADH and consumes two moles of NADPH and thus is imbalanced with respect to cofactor usage.
[0279]The different ways in which the cell can provide NADPH to the isobutanol pathway show that utilization of the TCA cycle as well as the PPP has to be avoided to maximize the yield of the isobutanol process. Loss of CO2 as a byproduct in isobutanol producing microorganism described in the prior art (Atsumi et al., Nature, 2008 Jan. 3; 451(7174):86-9; International Patent Application No PCT/US2008/053514; International Patent Application No PCT/US2006/041602) indicates that either or both of these two yield-limiting pathways are currently active.
[0280]A Nicotinamide dinucleotide transhydrogenase (hereinafter may be referred to simply as "transhydrogenase") that catalyzes the interconversion of NADH and NADPH as disclosed herein may be used to provide cofactor balance in a metabolic pathway for the production of a target compound that is otherwise imbalanced with respect to cofactor usage and thus decrease the yield loss to CO2 in such a pathway (FIG. 2)
[0281]A preferred transhydrogenase under conditions in which the reduced cofactor NADPH is limiting is one that preferentially catalyzes the conversion of NADH to NADPH. For example, membrane-bound transhydrogenases have been described in bacteria that catalyze this reaction. Membrane bound transhydrogenases require energy in form of proton translocation to catalyze the reaction. As long as there is enough energy available to maintain the proton gradient across the cell membrane a transhydrogenase may thus be used to balance an otherwise imbalanced metabolic pathway. However, in some circumstances, a transhydrogenase that catalyzes the conversion of NADPH to NADH may be preferred. However, a preferred transhydrogenase under conditions in which the reduced cofactor NADH is limiting is one that preferentially catalyzes the conversion of NADPH to NADH.
[0282]The expression and specific activity of an endogenously expressed membrane-bound transhydrogenase might not be sufficient to maintain the high metabolic flux through the metabolic pathway for the production of a fermentation product (e.g. for isobutanol) that is required in a commercial process.
[0283]Thus, in one embodiment, the insufficient activity of the membrane-bound transhydrogenase may be compensated by overexpression of the coding genes of a membrane bound transhydrogenase.
[0284]In a preferred embodiment, the E. coli pntA (SEQ ID NO: 1) and pntB genes (SEQ ID NO: 3), encoding for the PntA (SEQ ID NO: 2) and PntB (SEQ ID NO: 4) enzymes respectively or homologs thereof may be overexpressed. These genes have been overexpressed in E. coli before for characterization of the enzyme (Clarke, D. M. and P. D. Bragg, Journal of Bacteriology, 1985. 162(1): p. 367-373) and have been used to regenerate NADPH cofactor in the production of chiral alcohols from ketones using a whole cell biocatalyst (Weckbecker, A. and W. Hummel, Biotechnology Letters, 2004. 26(22): p. 1739-1744) or to increase production of biosynthesized products that rely on NADPH-dependent biosynthetic pathways (U.S. Pat. No. 5,830,716).
[0285]In one embodiment, the E. coli pntAB operon (SEQ ID NO: 1 and SEQ ID NO: 3) is expressed in the presence of the isobutanol pathway. The E. coli pntAB operon may be cloned on a medium copy plasmid (p15A origin of replication) under the control of the LtetOl promoter, for example pGV1685 (SEQ ID NO: 111). The high level expression of membrane proteins can lead to the buildup of toxic intermediates and to inclusion bodies. Thus, in another embodiment, different copy numbers of the E. coli pntAB operons may be tested to find the optimum expression level of this membrane transhydrogenase.
[0286]In another embodiment, the E. coli pntAB operon may be integrated into the chromosome of the microorganism. For example, E. coli pntAB may be integrated into the E. coli genome.
[0287]In one aspect of the present invention, the pntAB operon may be integrated into the sthA locus of E. coli or the corresponding locus in another microorganism. The sthA gene codes for the soluble transhydrogenase of E. coli and has previously been shown to be utilized by the cell for the conversion of NADPH to NADH. To avoid the generation of a futile cycle E. coli pntAB may be integrated at the sthA site, thus removing the sthA gene and eliminating this reverse reaction.
[0288]The E. coli pntAB operon may be integrated into a wild-type E. coli W3110 and then transduced into a recombinant microorganism that produces a product via a metabolic pathway that is imbalanced with respect to cofactor usage. For example, the E. coli pntAB operon may be integrated into an isobutanol producing strain in which the isobutanol pathway is integrated into the chromosome.
[0289]For example the E. coli pntAB operon may be integrated into the isobutanol pathway strain GEVO1859 which has the pathway genes Bs_alsS1 and Ec_ilvC_coEc integrated into the pflB site and has Ll_kivd1 and Ec_ilvD_coEc genes integrated into the adhE site. All genes may be under the control of the LlacOl promoter.
[0290]The soluble E. coli transhydrogenase coded by sthA has been shown to be utilized by the cell for the conversion of NADPH to NADH. However overexpression of sthA was demonstrated to increase the yield of poly(3-hydroxybutyrate) production in E. coli. These results indicate that if a pathway is present in E. coli that consumes NADPH effectively, the soluble transhydrogenase can function in the direction of NADPH production. The advantages of using SthA as opposed to E. coli PntAB are that the soluble protein might be easier to overexpress and that this enzyme is energy independent. The sthA gene may be cloned into pGV1685, replacing E. coli pntAB. Decisive for the success of this approach is the affinity of E. coli llvC (KARI enzyme) for its cofactor and the steady state concentrations of NADH and NADPH in the cell that allow SthA to run "backwards" or in the direction of converting NADH to NADPH. It is to be expected that the concentration of the reduced cofactor NADPH has to be low in order for SthA to supply this cofactor. If this concentration is low enough to limit the activity of E. coli llvC and therefore the flux through the isobutanol pathway then this approach is not suitable for the isobutanol production strain without further modifications. These modifications could be identification of a KARI with a lower KM for NADPH, or mutagenesis and directed evolution to increase the affinity of E. coli llvC for its cofactor.
[0291]This approach may be used to provide cofactor balance in a metabolic pathway otherwise imbalanced with respect to cofactor usage if the steady state concentrations of NADH and NADPH in the cell are appropriate to allow SthA to run "backwards" or in the direction of converting NADH to NADPH. It is to be expected that the concentration of the reduced cofactor NADPH has to be low in order for SthA to supply this cofactor.
[0292]This embodiment may enable higher yields of a fermentation product in a microorganism. Further, this embodiment may enable economical anaerobic production of a fermentation product, which was not possible without the teachings of this embodiment. Further, this embodiment may enable aerobic production of a fermentation product at higher yield, which was not possible without the teachings of this embodiment.
Microorganism Characterized by Providing Cofactor Balance Via Overexpression of an NADPH-Dependent GAPDH
[0293]Conversion of glucose to pyruvate via glycolysis in E. coli leads to the production of two moles of NADH. A metabolic pathway that converts pyruvate to a target product that consumes either two moles of NADPH or one mole of NADH and one mole of NADPH leads to cofactor imbalance. For example, the isobutanol metabolic pathway that converts glucose to two moles of pyruvate via glycolysis to 1 mole of isobutanol generates two moles of NADH and consumes two moles of NADPH and thus is imbalanced with respect to cofactor usage.
[0294]GAPDH catalyzes the conversion of glyceraldehyde 3-phosphate (GAP) to 1,3-diphosphate glycerate as part of glycolysis. For example, in E. coli GAPDH is encoded by gapA which is NADH-dependent and is active in glycolysis as well as in gluconeogenesis [DellaSeta, F., et al., Characterization of Escherichia coli strains with gapA and gapB genes deleted. Journal of Bacteriology, 1997. 179(16): p. 5218-5221.]. GAPDH proteins from other organisms vary in their cofactor requirements.
[0295]Thus in an embodiment, a recombinant microorganism that produces a compound may express a GAPDH is that uses the same cofactor as the fermentative pathway for the production of said compound. For example, in case of an isobutanol biosynthetic pathway that consumes two moles of NADPH per mole of pyruvate an NADPH-dependent GAPDH may be utilized to provide a metabolic pathway that is balanced with respect to cofactor usage (FIG. 3). In such an embodiment, it may also be desirable to increase the concentration of NADPH in the cell by overexpression of other enzymes for the metabolic synthesis of NADPH cofactor. In other embodiments, it may also be desirable to increase the concentration of NADPH in the cell by overexpression of other enzymes for the metabolic synthesis of NADPH cofactor.
[0296]Thus, such an NADPH-dependent GAPDH may be expressed in a recombinant microorganism. NADPH-dependent GAPDH enzymes may be identified by analysis with an in vitro enzyme assay. Further, some NADPH-dependent GAPDH enzymes may be identified by analysis of protein identity, similarity, or homology. Further, genes that encode NADPH-dependent GAPDH enzymes may be identified by analysis of gene identity, similarity, or homology.
[0297]One NADPH-dependent GAPDH according to the present invention with reported high activity with NADPH is Gdp1 from Kluyveromyces lactis [Verho, R., et al., Identification of the first fungal NADP-GAPDH from Kluyveromyces lactis. Biochemistry, 2002. 41(46): p. 13833-13838.]. Gdp1 has been expressed in Saccharomyces cerevisiae to improve ethanol fermentations on xylose as a substrate [Verho, R., et al., Engineering redox cofactor regeneration for improved pentose fermentation in Saccharomyces cerevisiae. Applied and Environmental Microbiology, 2003. 69(10): p. 5892-5897.] Expression of Gdp1 improved the yield of the fermentation from 18 to 23% and from 24 to 41% when it was coupled to a zwf1 deletion which forces more flux through glycolysis. Purified Gdp1 was shown in the literature to be as active with NAD+ as it is with NADP+. Thus, the intracellular concentrations and more importantly the redox ratio of the cofactors in a recombinant microorganism according to the present invention will dictate which cofactor is used in glycolysis.
[0298]Another NADPH accepting GAPDH is found in Clostridium acetobutylicum and is coded by the gene gapC. Additional homologs of NADPH-dependent GAPDH enzymes may be found in thermotolerant bacteria. Other alternatives of such GAPDH enzymes are those found in cyanobacteria.
[0299]A different class of enzymes that can be used to generate NADPH from glucose during glycolysis is comprised of the NADP+-dependent GAPDH (non-phosphorylating). Such enzymes are designated as GapN. However, use of this enzyme results in a loss of one ATP per pyruvate produced. Thus, the production of one NADPH is coupled to a reduction of ATP yield by 1 ATP.
[0300]To provide cofactor balance in a recombinant microorganism via an NADPH-dependent GAPDH, it may be necessary to deactivate the native NADH-dependent GAPDH. For example, in the host strain E. coli the gapA gene may be deleted.
[0301]Another way to force the cell to produce NADPH with GDP1 is the elimination of flux through the PPP. This can be accomplished by deletion of the gene that encodes 6-Phosphogluconate dehydrogenase or decreasing the activity of 6-Phosphogluconate dehydrogenase. For example, in E. coli 6-Phosphogluconate dehydrogenase is encoded by zwf. The mutation of zwf eliminates flux through the PPP and may force the microorganism to utilize glycolysis in which the heterologously expressed GAPDH will utilize the cofactor NADP+ instead of NADH.
[0302]Alternatively, cofactor imbalance in a recombinant microorganism Alternatively, cofactor imbalance in a recombinant microorganism that produces a fermentation product may be alleviated by engineering the native GAPDH to accept NADPH as cofactor. A crystal structure is available from the Palinurus versicolor GAPDH which can be used to model the structures of GDP1, GapA (E. coli) and other GAPDH enzymes with different cofactor specificities. It is known that an aspartate residue in the NAD binding site is conserved among the NAD dependent GAPDHs. This residue is replaced by asparagine in GDP1.
[0303]Additional target amino acids may be found using sequence alignments and structure modeling for site directed mutagenesis. The gapA gene can be mutated using saturation mutagenesis or random mutagenesis according to protein engineering methods known to those skilled in the art. The library of mutant genes may be transformed into microorganisms carrying a zwf deletion and expressing a metabolic pathway that is imbalanced with respect to cofactor usage pathway genes. Mutant enzymes that are NADPH-dependent may be identified in those microorganism that grow on a growth medium. In certain embodiments, it may not be necessary to delete the zwf gene. Alternate genes known to one skilled in the art may be deleted from the organism that in effect inhibits flux through the pentose phosphate pathway.
[0304]This embodiment may enable higher yields of a fermentation product in a microorganism. Further, this embodiment may enable anaerobic production of a fermentation product, which was not possible without the teachings of this embodiment. Further, this embodiment may enable anaerobic production of a fermentation product at higher yield, which was not possible without the teachings of this embodiment.
Microorganism Characterized by Providing Cofactor Balance Via a Transhydrogenase Cycle
[0305]Conversion of glucose to pyruvate via glycolysis in E. coli leads to the production of two moles of NADH. A metabolic pathway that converts pyruvate to a target product that consumes either two moles of NADPH or one mole of NADH and one mole of NADPH leads to cofactor imbalance. For example, the isobutanol metabolic pathway that converts glucose to two moles of pyruvate via glycolysis to 1 mole of isobutanol generates two moles of NADH and consumes two moles of NADPH and thus is imbalanced with respect to cofactor usage.
[0306]This cofactor imbalance may be resolved using two dehydrogenase enzymes that catalyze the same reaction but use different cofactors. One example for such a pair of enzymes are the malic enzymes MaeA and MaeB. MaeA is NADH-dependent and MaeB is NADPH-dependent and both catalyze the conversion of malate to pyruvate [Bologna, F. P., C. S. Andreo, and M. F. Drincovich, Escherichia coli malic enzymes: Two isoforms with substantial differences in kinetic properties, metabolic regulation, and structure. Journal of Bacteriology, 2007. 189(16): p. 5937-5946.]. The reaction catalyzed by each of these two enzymes is reversible. The kinetics of the two malic enzymes and the different concentrations and redox ratios of the cofactors they use might allow the NADH-dependent enzyme to run in the oxidative direction while the NADPH-dependent enzyme catalyses the reductive direction of the same conversion. In effect the enzymes would catalyze the interconversion of pyruvate and malate coupled to the consumption of NADH and the generation of NADPH (FIG. 4).
[0307]Thus the two malic enzymes may function like a transhydrogenase. This cofactor conversion cycle is dependent on the redox ratios of the cofactors which depends on the kinetics of the enzymes in an metabolic pathway that is imbalanced with respect to cofactor, for example the isobutanol pathway enzyme E. coli llvc as well as GapA and the malic enzymes. Homologs of malic enzymes can be identified by those skilled in the art. Those enzymes may be used which show kinetic properties favoring the oxidative conversion with NAD+ as cofactor and the reductive conversion with NADPH. The E. coli enzymes may to perform these reactions but enzymes with more favorable kinetics may increase the performance of the cofactor conversion.
[0308]This embodiment may enable higher yields of a fermentation product in a microorganism. Further, this embodiment may enable anaerobic production of a fermentation product, which was not possible without the teachings of this embodiment. Further, this embodiment may enable anaerobic production of a fermentation product at higher yield, which was not possible without the teachings of this embodiment.
Microorganism Characterized by Providing Cofactor Balance Via Metabolic Transhydrogenation Via Ppc or Pyc
[0309]Conversion of glucose to pyruvate via glycolysis in E. coli leads to the production of two moles of NADH. A metabolic pathway that converts pyruvate to a target product that consumes either two moles of NADPH or one mole of NADH and one mole of NADPH leads to cofactor imbalance. For example, the isobutanol metabolic pathway that converts glucose to two moles of pyruvate via glycolysis to 1 mole of isobutanol generates two moles of NADH and consumes two moles of NADPH and thus is imbalanced with respect to cofactor usage.
[0310]To resolve this cofactor imbalance the metabolic flux may be diverted to allow the conversion of at least one mole of NADH into NADPH. Looking at the stoichiometric network in E. coli points to a pathway that allows such a conversion of cofactors (FIG. 5).
[0311]Flux from PEP to pyruvate can be replaced by flux from PEP to oxaloacetate, to malate, to pyruvate. To redirect the flux in such a way the native conversion from PEP to pyruvate has to be removed from the network by deletion of the genes coding for pyruvate kinase (pykA, pykF). The other enzymes required are phosphoenolpyruvate carboxylase (Ppc) or phosphoenolpyruvate carboxykinase (Pck) for the conversion of PEP to oxaloacetate, malate dehydrogenase (mdh) for the conversion of oxaloacetate to malate and MaeB for the conversion of malate to pyruvate. The choice whether to use ppc or pck for the conversion of PEP to oxaloacetate depends on the energy load of the isobutanol production strain. With the deletion of Pyk the ATP yield of the strain is reduced if Ppc is used. If Pck is used instead the ATP yield is the same as when the flux goes from PEP to pyruvate using Pyk. Under production condition the strain will only need limited amounts of ATP for cell maintenance. This energy requirement might be lower than the two ATP per glucose generated by glycolysis. By overexpressing ppc, pck or both enzymes the energy yield of the conversion of PEP to pyruvate can be varied between one and two moles of ATP.
[0312]The native expression levels of some or all of the enzymes used in the above described conversion from PEP to pyruvate is expected to be insufficient to sustain the high glycolytic flux necessary in the isobutanol production strain. As an example the expression level of mdh is reduced in the presence of glucose and it is further reduced two-fold under anaerobic conditions. Therefore these enzymes may be overexpressed. To allow conversion of 50% of the NADH generated through glycolysis to NADPH the NADH-dependent malic enzyme MaeA may be deleted. Further the enzyme Mqo was reported to catalyze the conversion of malate to oxaloacetate and may be deleted to allow maximum flux in the opposite direction. The thermodynamic equilibrium of the conversion of malate to oxaloacetate lies on the malate side and Mdh catalyzes the reduction of oxaloacetate under anaerobic respiration and under fermentative conditions.
[0313]Flux through the PPP may be avoided by adding the deletion of zwf to the strain which eliminates glucose 6-phosphate 1-dehydrogenase the first committed step of the oxidative PPP.
[0314]This embodiment may enable higher yields of a fermentation product in a microorganism. Further, this embodiment may enable anaerobic production of a fermentation product, which was not possible without the teachings of this embodiment. Further, this embodiment may enable anaerobic production of a fermentation product at higher yield, which was not possible without the teachings of this embodiment.
Yeast Microorganism Characterized by Providing Cofactor Balance
[0315]The aforementioned methods to provide cofactor balance are generally applicable to many microorganisms, including yeast microorganisms. Specifically, however, in yeast, metabolic transhydrogenation may accomplished by introduction of NADPH dependent malic enzyme into yeast. If the conversion of phosphoenol pyruvate to pyruvate by pyruvate kinase is disrupted then the carbon flux can go through a pyruvate kinase bypass that goes from PEP to oxaloacetate to malate and from there to pyruvate. The conversion of oxaloacetate to malate by Mdh consumes one NADH and the conversion of malate to pyruvate by the heterologous malic enzyme produces one NADPH. NADPH dependent malic enzymes are common in bacteria and one example is E. coli MaeB. If the NADPH cofactor is needed in the mitochondria the malic enzyme expression can be directed into this organelle instead of the cytoplasm by addition of mitochondrial targeting sequence to the N-terminus or C-terminus of the gene. Also, the yeast enzyme Mae1, which is physiologically localized in the mitochondria can be overexpressed. Malate as well as pyruvate is shuttled across the mitochondrial membranes enabling the pyruvate bypass to effectively convert one cytoplasmic NADH into a mitochondrial NADPH. In yeast the complete carbon flux can be diverted in this way since there is no phosphotransferase (pts) system for glucose import and all PEP generated by glycolysis is available. However, one ATP is lost per NADPH produced through the yeast pyruvate kinase bypass.
[0316]Yeast do not have transhydrogenases. The heterologous expression of bacterial, plant or other eukaryotic transhydrogenases in yeast can be used to provide cofactor balance. The transhydrogenases that natively convert NADH to NADPH are generally membrane proteins that use the proton motive force to drive the reaction they are catalyzing. Bacterial transhydrogenases are in the cell membrane while plant and mammalian transhydrogenases are located in the inner mitochondrial membrane. For the heterologous transhydrogenase expression these enzymes can be targeted either to the cytoplasmic membrane or to the mitochondrial membrane in yeast. To achieve this leader sequences have to be added to the heterologous proteins. The mechanisms of membrane targeting are well understood and the direction of normally cytosolic proteins to the mitochondrium has been demonstrated. These targeting mechanisms are well conserved throughout the eukaryotes, which was demonstrated by the use of plant mitochondrial targeting sequences in yeast. Eukaryotic transhydrogenases are expressed in yeast with their native targeting and sorting sequences. Bacterial transhydrogenases are fused to mitochondrial targeting and membrane sorting sequences that have been characterized in yeast membrane proteins.
[0317]An alternative approach for the production of NADPH is the use of biosynthetic pathway enzymes. An NADH kinase could phosphorylate NADH to NADPH. Then the NADP+ needs to be dephosphorylated to NAD+ to maintain NAD+ pool. This can be carried out by an NADP phosphatase.
Microorganisms Characterized by Providing Cofactor Balance Via Engineered Enzymes
[0318]Conversion of one mole of glucose to two moles of pyruvate via glycolysis leads to the production of two moles of NADH. A metabolic pathway that converts pyruvate to a target product that consumes either two moles of NADPH or one mole of NADH and one mole of NADPH leads to cofactor imbalance. One example of such a metabolic pathway is the isobutanol metabolic pathway described by Atsumi et al., (Atsumi et al., 2008, Nature 451(7174): 86-9) which converts two moles of pyruvate to one mole of isobutanol. In this five enzyme pathway, two enzymes are dependent upon NADPH: (1) KARI and (2) ADH, encoded by the E. coli ilvC and E. coli yqhD, respectively.
[0319]To resolve this cofactor imbalance, the present invention provides a recombinant microorganism in which the NADPH-dependent enzymes KARI and ADH are replaced with enzymes that preferentially depend on NADH (i.e. KARI and ADH enzymes that are NADH-dependent).
[0320]To further resolve this cofactor imbalance, the present invention in another embodiment provides recombinant microorganisms wherein the NADH-dependent KARI and ADH enzymes are overexpressed.
[0321]In one aspect, such enzymes may be identified in nature. In an alternative aspect, such enzymes may be generated by protein engineering techniques including but not limited to directed evolution or site-directed mutagenesis.
[0322]In one embodiment, the two NADPH-dependent enzymes within an isobutanol biosynthetic pathway that converts pyruvate to isobutanol may be replaced with ones that utilize NADH. These two enzymes may be KARI and an alcohol dehydrogenase (ADH).
[0323]In another embodiment, two NADH-dependent enzymes that catalyze the same reaction as the NADH-dependent enzymes are overexpressed. These two enzymes may be KARI and an alcohol dehydrogenase.
[0324]In one aspect, NADH-dependent KARI and ADH enzymes are identified in nature. In another aspect, the NADPH-dependent KARI and ADH enzymes may be engineered using protein engineering techniques including but not limited to directed evolution and site-directed mutagenesis.
[0325]There exist two basic options for engineering NADH-dependent isobutyraldehyde dehydrogenases or ketol-acid reductoisomerases: (1) increase the NADH-dependent activity of an NADPH-dependent enzyme that is active towards the substrate of interest and/or (2) increase the activity of an NADH-dependent enzyme that is not sufficiently active towards the substrate of interest.
NADH-Dependent KARI Enzymes
[0326]As shown in FIG. 1, the ketol-acid reductoisomerase (KARI) enzyme of the isobutanol biosynthetic pathway as disclosed by Atsumi et al (Atsumi et al., 2008, Nature 451(7174): 86-9, herein incorporated by reference in its entirety), requires the cofactor nicotinamide dinucleotide phosphate (NADPH) to convert acetolactate to 2,3-dihydroxyisovalerate. However, under anaerobic conditions, NADPH is produced only sparingly by the cell--nicotinamide adenine dinucleotide (NADH) is the preferred equivalent. Therefore, oxygen is required to produce NADPH in the large quantities to support high-level production of isobutanol. Thus, the production of isobutanol is feasible only under aerobic conditions and the maximum yield that can be achieved with this pathway is limited. Accordingly, KARI enzymes that preferentially utilize NADH rather than NADPH are desirable.
[0327]Other biosynthetic pathways utilize KARI enzymes for the conversion of acetolactate to 2-3-dihydroxyisovalerate. For example, KARI enzymes convert acetolactate to 2-3-dihydroxyisovalerate as part of the biosynthetic pathway for the production of 3-methyl-1-butanol (Atsumi et al., 2008, Nature 451(7174): 86-9, herein incorporated by reference in its entirety).
[0328]Yet other biosynthetic pathways utilize KARI to convert 2-aceto-2-hydroxy-butyrate to 2,3-dihydroxy-3-methylvalerate. This reaction is part of the biosynthetic pathway for the production of 2-methyl-1-butanol. (Atsumi et al., 2008, Nature 451(7174): 86-9, herein incorporated by reference in its entirety).
[0329]As used herein, the term "KARI" or "KARI enzyme" or "ketol-acid reductoisomerase" are used interchangeably herein to refer to an enzyme that catalyzes the conversion of acetolactate to 2,3-dihydroxyisovalerate and/or the conversion of 2-aceto-2-hydroxy-butyrate to 2,3-dihydroxy-3-methylvalerate. Moreover, these terms can be used interchangeably herein with the terms "acetohydroxy acid isomeroreductase" and "acetohydroxy acid reductoisomerase."
[0330]Enzymes for use in the compositions and methods of the invention include any enzyme having the ability to convert acetolactate to 2,3-dihydroxyisovalerate and/or the ability to convert 2-aceto-2-hydroxy-butyrate to 2,3-dihydroxy-3-methylvalerate. Such enzymes include, but are not limited to, the E. coli ilvC gene product and the S. cerevisiae ilv5 gene product, and the KARI enzyme from Piromyces sp, Buchnera aphidicola, Spinacia oleracea, Oryza sativa, Chlamydomonas reinhardtii, Neurospora crassa, Schizosaccharomyces pombe, Laccaria bicolor, Ignicoccus hospitalis, Picrophilus torridus, Acidiphilium cryptum, Cyanobacteria/Synechococcus sp., Zymomonas mobilis, Bacteroides thetaiotaomicron, Methanococcus maripaludis, Vibrio fischeri, Shewanella sp, Gramella forsetti, Psychromonas ingrhamaii, and Cytophaga hutchinsonii.
[0331]Preferred KARI enzymes are known by the EC number 1.1.1.86 and sequences are available from a vast array of microorganisms, including, but not limited to, Escherichia coli (GenBank Nos: NP--418222 and NC--000913, Saccharomyces cerevisiae (GenBank Nos: NP--013459 and NC--001144, Methanococcus maripaludis (GenBank Nos: CAF30210 and BX957220, and Bacillus subtilis (GenBank Nos: CAB14789 and Z99118) and the KARI enzymes from Piromyces sp (GenBank No: CAA76356), Buchnera aphidicola (GenBank No: AAF13807), Spinacia oleracea (GenBank Nos: Q01292 and CAA40356), Oryza sativa (GenBank No: NP--001056384) Chlamydomonas reinhardtii (GenBank No: XP--001702649), Neurospora crassa (GenBank No: XP--961335), Schizosaccharomyces pombe (GenBank No: NP--001018845), Laccaria bicolor (GenBank No: XP--001880867), Ignicoccus hospitalis (GenBank No: YP--001435197), Picrophilus torridus (GenBank No: YP--023851), Acidiphilium cryptutm (GenBank No: YP--001235669), Cyanobacteria/Synechococcus sp. (GenBank No: YP--473733), Zymomonas mobilis (GenBank No: YP--162876), Bacteroides thetaiotaomicron (GenBank No: NP--810987), Methanococcus maripaludis (GenBank No: YP--001097443), Vibrio fischeri (GenBank No: YP--205911), Shewanella sp (GenBank No: YP--732498), Gramella forsetti (GenBank No: YP--862142), Psychromonas ingrhamaii (GenBank No: YP--942294), and Cytophaga hutchinsonii (GenBank No: YP--677763).
[0332]As will be understood by one of ordinary skill in the art, modified KARI enzymes may be obtained by recombinant or genetic engineering techniques that are routine and well-known in the art. Mutant KARI enzymes can, for example, be obtained by mutating the gene or genes encoding the KARI enzyme of interest by site-directed or random mutagenesis. Such mutations may include point mutations, deletion mutations and insertional mutations. For example, one or more point mutations (e.g., substitution of one or more amino acids with one or more different amino acids) may be used to construct mutant KARI enzymes of the invention.
[0333]Ketol-acid reductoisomerase (KARI; EC 1.1.1.86) catalyzes the reduction of acetolactate to 2,3-dihydroxyisovalerate. The two-step reaction involves an alkyl migration and a ketone reduction that occurs at a single active site on the enzyme without dissociation of any reaction intermediates. The enzyme is NADPH-dependent. The cofactor specificity may be expanded or switched so that it will utilize both cofactors and preferentially NADH during the production of isobutanol. A study published in 1997 (Rane, M. J. and K. C. Calvo, Archives of Biochemistry and Biophysics, 1997. 338(1): p. 83-89) describes a supposed cofactor-switched KARI quadruplet variant of the E. coli ilvC gene product with mutations R68D, K69L, K75V and R76D). However, in-house studies indicate that although the ratio NADH/NADPH was 2.5, the specific activity of this variant on NADH was actually worse than wild-type (Table 25), rendering this enzyme not suited for the purpose of this disclosure.
Modified or Mutated KARI Enzymes
[0334]In accordance with the invention, any number of mutations can be made to the KARI enzymes, and in a preferred aspect, multiple mutations can be made to result in an increased ability to utilize NADH for the conversion of acetolactate to 2,3-dihydroxyisovalerate. Such mutations include point mutations, frame shift mutations, deletions, and insertions, with one or more (e.g., one, two, three, or four, etc.) point mutations preferred.
[0335]Mutations may be introduced into the KARI enzymes of the present invention using any methodology known to those skilled in the art. Mutations may be introduced randomly by, for example, conducting a PCR reaction in the presence of manganese as a divalent metal ion cofactor. Alternatively, oligonucleotide directed mutagenesis may be used to create the mutant KARI enzymes which allows for all possible classes of base pair changes at any determined site along the encoding DNA molecule. In general, this technique involves annealing an oligonucleotide complementary (except for one or more mismatches) to a single stranded nucleotide sequence coding for the KARI enzyme of interest. The mismatched oligonucleotide is then extended by DNA polymerase, generating a double-stranded DNA molecule which contains the desired change in sequence in one strand. The changes in sequence can, for example, result in the deletion, substitution, or insertion of an amino acid. The double-stranded polynucleotide can then be inserted into an appropriate expression vector, and a mutant or modified polypeptide can thus be produced. The above-described oligonucleotide directed mutagenesis can, for example, be carried out via PCR.
[0336]The invention further includes homologous KARI enzymes which are 5%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, or 99% identical at the amino acid level to a wild-type KARI enzyme (e.g., encoded by the Ec_ilvC gene or S. cerevisiae llv5 gene) and exhibit an increased ability to utilize NADH for the conversion of acetolactate to 2,3-dihydroxyisovalerate. Also included within the invention are KARI enzymes which are 50%, 60%, 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, or 99% identical at the amino acid level to a KARI enzyme comprising the amino acid sequence set out in SEQ ID NO: 13 and exhibit an increased ability to utilize NADH for the conversion of acetolactate to 2,3-dihydroxyisovalerate. The invention also includes nucleic acid molecules which encode the above described KARI enzymes.
[0337]The invention also includes fragments of KARI enzymes which comprise at least 50, 100, 150, 200, 250, 300, 350, 400, 450, 500, 550, or 600 amino acid residues and retain one or more activities associated with KARI enzymes. Such fragments may be obtained by deletion mutation, by recombinant techniques that are routine and well-known in the art, or by enzymatic digestion of the KARI enzyme(s) of interest using any of a number of well-known proteolytic enzymes. The invention further includes nucleic acid molecules which encode the above described mutant KARI enzymes and KARI enzyme fragments.
[0338]By a protein or protein fragment having an amino acid sequence at least, for example, 50% "identical" to a reference amino acid sequence it is intended that the amino acid sequence of the protein is identical to the reference sequence except that the protein sequence may include up to 50 amino acid alterations per each 100 amino acids of the amino acid sequence of the reference protein. In other words, to obtain a protein having an amino acid sequence at least 50% identical to a reference amino acid sequence, up to 50% of the amino acid residues in the reference sequence may be deleted or substituted with another amino acid, or a number of amino acids up to 50% of the total amino acid residues in the reference sequence may be inserted into the reference sequence. These alterations of the reference sequence may occur at the amino (N-) and/or carboxy (C-) terminal positions of the reference amino acid sequence and/or anywhere between those terminal positions, interspersed either individually among residues in the reference sequence and/or in one or more contiguous groups within the reference sequence. As a practical matter, whether a given amino acid sequence is, for example, at least 50% identical to the amino acid sequence of a reference protein can be determined conventionally using known computer programs such as those described above for nucleic acid sequence identity determinations, or using the CLUSTAL W program (Thompson, J. D., et al., Nucleic Acids Res. 22:4673 4680 (1994)).
[0339]In one aspect, amino acid substitutions are made at one or more of the above identified positions (i.e., amino acid positions equivalent or corresponding to A71, R76, S78, or Q110 of E. coli llvC). Thus, the amino acids at these positions may be substituted with any other amino acid including Ala, Asn, Arg, Asp, Cys, Gln, Glu, Gly, His, Ile, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, and Val. A specific example of a KARI enzyme which exhibits an increased ability to utilize NADH includes an E. coli llvC KARI enzyme in which (1) the alanine at position 71 has been replaced with a serine, (2) the arginine at position 76 has been replaced with an aspartic acid, (3) the serine at position 78 has been replaced with an aspartic acid, and/or (4) the glutamine at position 110 has been replaced with valine.
[0340]Polypeptides having the ability to convert acetolactate to 2,3-dihydroxyisovalerate and/or 2-aceto-2-hydroxy-butyrate to 2,3-dihydroxy-3-methylvalerate for use in the invention may be isolated from their natural prokaryotic or eukaryotic sources according to standard procedures for isolating and purifying natural proteins that are well-known to one of ordinary skill in the art (see, e.g., Houts, G. E., et al., J. Virol. 29:517 (1979)). In addition, polypeptides having the ability to convert acetolactate to 2,3-dihydroxyisovalerate and/or 2-aceto-2-hydroxy-butyrate to 2,3-dihydroxy-3-methylvalerate may be prepared by recombinant DNA techniques that are familiar to one of ordinary skill in the art (see, e.g., Kotewicz, M. L., et al., Nucl. Acids Res. 16:265 (1988); Soltis, D. A., and Skalka, A. M., Proc. Natl. Acad. Sci. USA 85:3372 3376 (1988)).
[0341]In accordance with the invention, one or more mutations may be made in any KARI enzyme of interest in order to increase the ability of the enzyme to utilize NADH, or confer other properties described herein upon the enzyme, in accordance with the invention. Such mutations include point mutations, frame shift mutations, deletions and insertions. Preferably, one or more point mutations, resulting in one or more amino acid substitutions, are used to produce KARI enzymes having an enhanced or increased ability to utilize NADH, particularly to facilitate the conversion of acetolactate to 2,3-dihydroxyisovalerate and/or the conversion of 2-aceto-2-hydroxy-butyrate to 2,3-dihydroxy-3-methylvalerate. In a preferred aspect of the invention, one or more mutations at positions equivalent or corresponding to position A71 (e.g., A71S), R76 (e.g., R76D), S78 (e.g. S78D), and/or Q110 (e.g. Q110V) and/or D146 (e.g. D146G), and/or G185 (e.g. G185R) and/or K433 (e.g. K433E) of the E. coli llvC KARI enzyme may be made to produce the desired result in other KARI enzymes of interest.
[0342]The corresponding positions of the KARI enzymes identified herein (e.g. E. coli llvC may be readily identified for other KARI enzymes by one of skill in the art. Thus, given the defined region and the assays described in the present application, one with skill in the art can make one or a number of modifications which would result in an increased ability to utilize NADH, particularly for the conversion of acetolactate to 2,3-dihydroxyisovalerate, in any KARI enzyme of interest. Residues to be modified in accordance with the present invention may include those described in Examples 14, 15, and 16.
[0343]In a preferred embodiment, the modified or mutated KARI enzymes have from 1 to 4 amino acid substitutions in amino acid regions involved in cofactor specificity as compared to the wild-type KARI enzyme proteins. In other embodiments, the modified or mutated KARI enzymes have additional amino acid substitutions at other positions as compared to the respective wild-type KARI enzymes. Thus, modified or mutated KARI enzymes may have at least about 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40 different residues in other positions as compared to the respective wild-type KARI enzymes. As will be appreciated by those of skill in the art, the number of additional positions that may have amino acid substitutions will depend on the wild-type KARI enzyme used to generate the variants. Thus, in some instances, up to 50 different positions may have amino acid substitutions.
[0344]The nucleotide sequences for several KARI enzymes are known. For instance, the sequences of KARI enzymes are available from a vast array of microorganisms, including, but not limited to, Escherichia coli (GenBank No: NP--418222), Saccharomyces cerevisiae (GenBank Nos: NP--013459, Methanococcus maripaludis (GenBank No: YP--001097443), Bacillus subtilis (GenBank Nos: CAB14789), and the KARI enzymes from Piromyces sp (GenBank No: CAA76356), Buchnera aphidicola (GenBank No: AAF13807), Spinacia oleracea (GenBank Nos: Q01292 and CAA40356), Oryza sativa (GenBank No: NP--001056384) Chlamydomonas reinhardtii (GenBank No: XP--001702649), Neurospora crassa (GenBank No: XP--961335), Schizosaccharomyces pombe (GenBank No: NP--001018845), Laccaria bicolor (GenBank No: XP--001880867), Ignicoccus hospitalis (GenBank No: YP--001435197), Picrophilus torridus (GenBank No: YP--023851), Acidiphilium cryptum (GenBank No: YP--001235669), Cyanobacteria/Synechococcus sp. (GenBank No: YP--473733), Zymomonas mobilis (GenBank No: YP--162876), Bacteroides thetaiotaomicron (GenBank No: NP--810987), Methanococcus maripaludis (GenBank No: YP--001097443), Vibrio fischeri (GenBank No: YP--205911), Shewanella sp (GenBank No: YP--732498), Gramella forsetti (GenBank No: YP--862142), Psychromonas ingrhamaii (GenBank No: YP--942294), and Cytophaga hutchinsonii (GenBank No: YP--677763).
Improved NADH-Dependent Activity
[0345]In one aspect, the NADH-dependent activity of the modified or mutated KARI enzyme is increased.
[0346]In a preferred embodiment, the catalytic efficiency of the modified or mutated KARI enzyme is improved for the cofactor NADH. Preferably, the catalytic efficiency of the modified or mutated KARI enzyme is improved by at least about 5% as compared to the wild-type or parental KARI for NADH. More preferably the catalytic efficiency of the modified or mutated KARI enzyme is improved by at least about 15% as compared to the wild-type or parental KARI for NADH. More preferably, the catalytic efficiency of the modified or mutated KARI enzyme is improved by at least about 25% as compared to the wild-type or parental KARI for NADH. More preferably, the catalytic efficiency of the modified or mutated KARI enzyme is improved by at least about 50% as compared to the wild-type or parental KARI for NADH. More preferably, the catalytic efficiency of the modified or mutated KARI, enzyme is improved by at least about 75% as compared to the wild-type or parental KARI for NADH. More preferably, the catalytic efficiency of the modified or mutated KARI enzyme is improved by at least about 100% as compared to the wild-type or parental KARI for NADH. More preferably, the catalytic efficiency of the modified or mutated KARI enzyme is improved by at least about 300% as compared to the wild-type or parental KARI for NADH. More preferably, the catalytic efficiency of the modified or mutated KARI enzyme is improved by at least about 500% as compared to the wild-type or parental KARI for NADH. More preferably, the catalytic efficiency of the modified or mutated KARI enzyme is improved by at least about 1000% as compared to the wild-type or parental KARI for NADH. More preferably, the catalytic efficiency of the modified or mutated KARI enzyme is improved by at least about 5000% as compared to the wild-type or parental KARI for NADH.
[0347]In a preferred embodiment, the catalytic efficiency of the modified or mutated KARI enzyme with NADH is increased with respect to the catalytic efficiency of the wild-type or parental enzyme with NADPH. Preferably, the catalytic efficiency of the modified or mutated KARI enzyme is at least about 10% of the catalytic efficiency of the wild-type or parental KARI enzyme for NADPH. More preferably, the catalytic efficiency of the modified or mutated KARI enzyme is at least about 25% of the catalytic efficiency of the wild-type or parental KARI enzyme for NADPH. More preferably, the catalytic efficiency of the modified or mutated KARI enzyme is at least about 50% of the catalytic efficiency of the wild-type or parental KARI enzyme for NADPH. More preferably, the catalytic efficiency of the modified or mutated KARI enzyme is at least about 75%, 85%, 95% of the catalytic efficiency of the wild-type or parental KARI enzyme for NADPH.
[0348]In a preferred embodiment, the KM of the KARI enzyme for NADH is decreased relative to the wild-type or parental enzyme. A change in KM is evidenced by at least a 5% or greater increase or decrease in KM compared to the wild-type KARI enzyme. In certain embodiments, modified or mutated KARI enzymes of the present invention may show greater than 10 times decreased KM for NADH compared to the wild-type or parental KARI enzyme. In certain embodiments, modified or mutated KARI enzymes of the present invention may show greater than 30 times decreased KM for NADH compared to the wild-type or parental KARI enzyme.
[0349]In a preferred embodiment, the kcat of the KARI enzyme with NADH is increased relative to the wild-type or parental enzyme. A change in kcat is evidenced by at least a 5% or greater increase or decrease in KM compared to the wild-type KARI enzyme. In certain embodiments, modified or mutated KARI enzymes of the present invention may show greater than 50% increased kM for NADH compared to the wild-type or parental KARI enzyme. In certain embodiments, modified or mutated KARI enzymes of the present invention may show greater than 100% increased kcat for NADH compared to the wild-type or parental KARI enzyme. In certain embodiments, modified or mutated KARI enzymes of the present invention may show greater than 200% increased kcat for NADH compared to the wild-type or parental KARI enzyme.
Cofactor Switch
[0350]In preferred embodiments, the cofactor specificity of the modified or mutated KARI enzyme is altered such that there is a cofactor switch from NADPH to NADH. In other words, these modified or mutated KARI enzymes will have an increase in NADH-dependent activity and a substantially simultaneous decrease in NADPH dependent activity. Thus, the methods of the present invention can be used to change the cofactor preference from NADPH to NADH.
[0351]"Cofactor specificity" is a measure of the specificity of an enzyme for one cofactor over another. Thus, the methods of the present invention may be used to alter the cofactor preference of the target enzyme, such that the preference for the less favored cofactor is increased by 20%, 50%, 100%, 300%, 500%, 1000%, up to 2000%. For example, a number of reductase enzymes have been described that favor NADPH over NADH (see WO 02/22526; WO 02/29019; Mittl, P R., et al., (1994) Protein Sci., 3: 1504 14; Banta, S., et al., (2002) Protein Eng., 15:131 140; all of which are hereby incorporated by reference in their entirety). As the availability of NADPH is often limiting, both in vivo and in vitro, the overall activity of the target protein is often limited. For target proteins that prefer NADPH as a cofactor, it would be desirable to alter the cofactor specificity of the target protein (e.g. a KARI enzyme) to a cofactor that is more readily available, such as NADH.
[0352]In a preferred embodiment, the cofactor specificity of the KARI enzyme is switched. By "switched" herein is meant, that the cofactor preference (in terms of catalytic efficiency (kcat/KM) of the KARI enzyme is changed to another cofactor Preferably, in one embodiment, by switching cofactor specificity, activity in terms of catalytic efficiency (kcat/KM) with the cofactor preferred by the wild-type KARI enzyme is reduced, while the activity with the less preferred cofactor is increased. This can be achieved, for example by increasing the kcat for less preferred cofactor over the preferred cofactor or by decreasing KM for the less preferred cofactor over the preferred cofactor or both.
[0353]In a preferred embodiment, the KARI enzyme is modified or a mutated to become NADH-dependent. The term "NADH-dependent" refers to the property of an enzyme to preferentially use NADH as the redox cofactor. An NADH-dependent enzyme has a higher catalytic efficiency (kcat/KM) with the cofactor NADH than with the cofactor NADPH as determined by in vitro enzyme activity assays. Accordingly, the term "NADPH-dependent" refers to the property of an enzyme to preferentially use NADPH as the redox cofactor. An NADPH dependent enzyme has a higher catalytic efficiency (kcat/KM) with the cofactor NADPH than with the cofactor NADH as determined by in vitro enzyme activity assays.
[0354]In a preferred embodiment, the catalytic efficiency of the KARI enzyme for NADH is enhanced relative to the catalytic efficiency with NADPH. The term "catalytic efficiency" describes the ratio of the rate constant kcat over the Michaelis-Menten constant KM. In one embodiment, the invention is directed to a modified or mutated KARI enzyme that exhibits at least about a 1:10 ratio of catalytic efficiency (kcat/KM) with NADH over catalytic efficiency with NADPH. In another embodiment, the modified or mutated KARI enzyme exhibits at least about a 1:1 ratio of catalytic efficiency (kcat/KM) with NADH over catalytic efficiency with NADPH. In yet another embodiment, the modified or mutated KARI enzyme exhibits at least about a 10:1 ratio of catalytic efficiency (kcat/KM) with NADH over catalytic efficiency with NADPH. In yet another embodiment, the modified or mutated KARI enzyme exhibits at least about a 100:1 ratio of catalytic efficiency (kcat/KM) with NADH over catalytic efficiency with NADPH. In an exemplary embodiment, the modified or mutated KARI enzyme exhibits at least about a 100:1 ratio of catalytic efficiency (kcat/KM) with NADH over catalytic efficiency with NADPH.
[0355]In a preferred embodiment, the KM of the KARI enzyme for NADH is decreased relative to the KM of the KARI enzyme for NADPH. In one embodiment, the invention is directed to a modified or mutated KARI enzyme that exhibits at least about a 10:1 ratio of KM for NADH over KM for NADPH. In one embodiment, the invention is directed to a modified or mutated KARI enzyme that exhibits at least about a 1:1 ratio of KM for NADH over KM for NADPH. In a preferred embodiment, the invention is directed to a modified or mutated KARI enzyme that exhibits at least about a 1:10 ratio of KM for NADH over KM for NADPH. In yet another embodiment, the invention is directed to a modified or mutated KARI enzyme that exhibits at least about a 1:20, 1:100, 1:1000 ratio of KM for NADH over KM for NADPH.
[0356]In another preferred embodiment, the kcat of the KARI enzyme with NADH is increased relative to kcat with NADPH. In certain embodiments, modified or mutated KARI enzymes of the present invention may show greater than 0.8:1 ratio of kcat with NADH over kcat with NADPH. In certain embodiments, modified or mutated KARI enzymes of the present invention may show greater than 1:1 ratio of kcat with NADH over kcat with NADPH. In a preferred embodiments, modified or mutated KARI enzymes of the present invention may show greater than 10:1 ratio of kcat with NADH over kcat with NADPH. In certain embodiments, modified or mutated KARI enzymes of the present invention may show greater than 100:1 ratio of kcat with NADH over kcat with NADPH
Identification of Corresponding Amino Acid Substitutions in Homologous Enzymes
[0357]An amino acid sequence alignment of 22 KARIs (including E. coli llvC, spinach KARI and rice KARI) was performed (FIG. 6). Some KARIs aligned with the E. coli KARI sequence at amino acid positions 71, 76, 78, and 110 and this allows to conclude that the beneficial mutations found for E. coli KARI confer the same effects in these KARI enzymes. Other sequences show deletions at about these positions and the sequence alignment is not sufficient to make any predictions.
[0358]A structure alignment of E. coli KARI (PDB ID NO. 1YRL) with rice KARI (PDB ID NO. 3FR8) as a representative of the shorter loop group was performed (FIG. 7). The sites of useful mutations in the E. coli context corresponded reasonably well with specific residues in the context of the shorter loop: Ser165, Lys166, and Ser167. Ser165 of (corresponding to A71 in E. coli) therefore may be substituted with aspartate. A charge reversal at position K166 (corresponding to position R76D) may yield the same result. Ser167 may correspond to Ser78 and a mutation to aspartate may be beneficial Mutations at 0110 may be transferable in all 22 KARIs aligned.
[0359]In the case of D146 (e.g. D146G), G185 (e.g. G185R), and K433 (e.g. K433E), surface charge changes took place. Glycine at position 185 and Lysine at position 433 are highly conserved among other KARIs. These mutations may therefore be transferable to other KARIs with a similar effect. Aspartate at position 146 is not as highly conserved.
NADH-Dependent ADH Enzymes
[0360]Several alcohol dehydrogenases may be suitable candidates for conversion into an NADH-dependent isobutyraldehyde dehydrogenase. Among the preferred enzymes for conversion are S. cerevisiae ADH1, Zymomonas mobilis ADHII, E. coli YqhD, herein referred to as Ec_YqhD, and S. cerevisiae ADH7.
[0361]As described in the prior art in PCT/US2008/053514, the S. cerevisiae ADH2 gene is expected to be functionally expressed from pSA55 and required for catalyzing the final step of the isobutanol biosynthetic pathway, namely the conversion of isobutyraldehyde to isobutanol. Thus, no isobutanol should be produced with the plasmid combination lacking ADH2 as adhE is deleted in JCL260. However, as exemplified in Example 10, the results of a fermentation using a strain without overexpression of any gene encoding an enzyme with ADH activity for the conversion of isobutyraldehyde to isobutanol showed that overexpression of an ADH enzyme is not required for isobutanol production in E. coli. In fact, isobutanol production for the system lacking ADH2 was higher than for the system with ADH2 expression. Volumetric productivity and titer showed 42% increase, specific productivity showed 18% increase and yield 12% increase. This suggests strongly that a native E. coli dehydrogenase is responsible for the conversion of isobutyraldehyde to isobutanol.
[0362]Surprisingly, this last step of the isobutanol biosynthetic pathway was found to be carried out by a native E. coli dehydrogenase in the aforementioned strains, as exemplified in Example 11: Approximately ˜80% of the isobutyraldehyde reduction activity is due to Ec_YqhD under certain culture conditions. Available literature on Ec_YqhD suggests that while it does prefer long-chain alcohols, it also utilizes NADPH (versus NADH) (Perez, J. M., et al., Journal of Biological Chemistry, 2008. 283(12): p. 7346-7353).
[0363]Switching the cofactor specificity of an NADPH-dependent alcohol dehydrogenase may be complicated by the fact that cofactor binding induces a conformational change, resulting in an anhydrous binding pocket that facilitates hydride transfer from the reduced cofactor to the aldehyde (Leskovac, V., S. Trivic, and D. Pricin, Fems Yeast Research, 2002. 2: p. 481-494; Reid, M. F. and C. A. Fewson, Critical Reviews in Microbiology, 1994. 20(1): p. 13-56). Mutations that are beneficial for binding NADH may have deleterious effects with respect to this conformational change.
[0364]Alternatively, isobutyraldehyde reduction activity of an NADH-dependent enzyme with little native activity towards this substrate may be increased. This approach has the advantages that (1) several specialized enzymes exist in nature that are highly active under fermentative conditions, (2) the binding sites of several of these enzymes are known, (3) mutational studies indicate that substrate specificity can easily be altered to achieve high activity on a new substrate.
[0365]Several alcohol dehydrogenase enzymes may be suitable candidates for conversion into an NADH-dependent isobutyraldehyde dehydrogenase: S. cerevisiae ADH1 and Zymomonas mobilis ADHII are NADH-dependent enzymes responsible for the conversion of acetaldehyde to ethanol under anaerobic conditions. These enzymes are highly active. The substrate specificity for these enzymes has been analyzed (Leskovac, V., S. Trivic, and D. Pricin, Ferns Yeast Research, 2002. 2: p. 481-494; Rellos, P., J. Ma, and R. K. Scopes, Protein Expression and Purification, 1997. 9: p. 89-90), the amino acid residues comprising the substrate binding pocket are known (Leskovac, V., S. Trivic, and D. Pricin, Ferns Yeast Research, 2002. 2: p. 481-494; Rellos, P., J. Ma, and R. K. Scopes, Protein Expression and Purification, 1997. 9: p. 89-90), and attempts to alter the substrate specificity by mutation have revealed that the substrate specificity can be altered (Rellos, P., J. Ma, and R. K. Scopes, Protein Expression and Purification, 1997. 9: p. 89-90; Green, D. W., H. Suns, and B. V. Plapp, Journal of Biological Chemistry, 1993. 268(11): p. 7792-7798). Ec_YqhD and S. cerevisiae ADH7 are NADPH-dependent enzymes whose physiological functions are not as well understood. Ec_YqhD has been implicated in the protection of the cell from peroxide-derived aldehydes (Perez, J. M., et al., Journal of Biological Chemistry, 2008. 283(12): p. 7346-7353). The substrate specificity of both enzymes is understood, and amino acids lining the substrate binding pocket are known (Perez, J. M., et al., Journal of Biological Chemistry, 2008. 283(12): p. 7346-7353). Based on the known amino acid residues implicated in substrate binding (S. cerevisiae ADH1, Z. mobills ADHII) or the cofactor binding site (Ec_yqhD), sites with the highest likelihood of affecting desired enzyme features such as substrate specificity or cofactor specificity may be mutated to generate the desired function.
[0366]One approach to increase activity of enzymes with NADH as the cofactor may be saturation mutagenesis with NNK libraries at each of the residues that interact with the cofactor. These libraries may be screened for activity in the presence of NADPH and NADH in order to identify which single mutations contribute to increased activity on NADH and altered specificity for NADH over NADPH. Combinations of mutations at aforementioned residues may be investigated by any method. For example, a combinatorial library of mutants may be designed based on the results of the saturation mutagenesis studies. For example, a combinatorial library of mutants may be designed including only those mutations that do not lead to decrease in NADH-dependent activity.
[0367]Another approach to increase the NADH-dependent activity of the enzyme is to perform saturation mutagenesis of a first amino acid that interacts with the cofactor, then isolate the mutant with the highest activity using NADH as the cofactor, then perform saturation mutagenesis of a second amino acid that interacts with the cofactor, and so on. Similarly, a limited number of amino acids that interact with the cofactor may be targeted for randomization simultaneously and then be screened for improved activity with NADH as the cofactor. The selected, best mutant can then be subjected to the same procedure again and this approach may be repeated iteratively until the desired result is achieved.
[0368]Another approach is to use random oligonucleotide mutagenesis to generate diversity by incorporating random mutations, encoded on a synthetic oligonucleotide, into the cofactor binding region of the enzyme. The number of mutations in individual enzymes within the population may be controlled by varying the length of the target sequence and the degree of randomization during synthesis of the oligonucleotides. The advantages of this more defined approach are that all possible amino acid mutations and also coupled mutations can be found.
[0369]If the best variants from the experiments described above are not sufficiently active with NADH as the cofactor, directed evolution via error-prone PCR may be used to obtain further improvements. Error-prone PCR mutagenesis of the first domain containing the cofactor binding pocket may be performed followed by screening for ADH activity with NADH and/or increased specificity for NADH over NADPH as the cofactor.
[0370]Surprisingly, alcohol dehydrogenase enzymes that are not known to catalyze the reduction of isobutyraldehyde to isobutanol were identified that catalyze this reaction. Thus, in another aspect, such an alcohol dehydrogenase may be encoded by an NADH-dependent 1,3-propanediol dehydrogenase. In yet another aspect, such an alcohol dehydrogenase may be encoded by an NADH-dependent 1,2-propanediol dehydrogenase. Preferred enzymes of this disclosure include enzymes listed in Table 1. These enzymes exhibit NADH-dependent isobutyraldehyde reduction activity, measured as Unit per minute per mg of crude cell lysate (U min-1 mg-1) that is approximately six-fold to seven-fold greater than the corresponding NADPH-dependent isobutyraldehyde reduction activity (Tables 2 and 23).
[0371]In addition to exhibiting increased activity with NADH as the cofactor as compared to the NADPH, alcohol dehydrogenases of the present invention may further be more active as compared to the native E. coli alcohol dehydrogenase Ec_YqhD. In particular, alcohol dehydrogenases of the present invention may exhibit increased activity and/or decreased KM values with NADH as the cofactor as compared to Ec_YqhD with NADPH as the cofactor. Exemplary enzymes that exhibit greater NADH-dependent alcohol dehydrogenase activity than the NADPH-dependent alcohol dehydrogenase activity are listed in Table 1; activity values are listed in Table 2 and Table 23.
TABLE-US-00001 TABLE 1 ADH genes tested in the following fermentations, and rationale for inclusion of each GENE Accession NAME SEQ ID NO Number Rationale for inclusion Drosophila 60 (nucleotide NT_033779, NADH-dependent, broad melanogaster sequence) REGION: substrate specificity, well- ADH 61 (amino acid 14615555 . . . 14618902 expressed in bacterial sequence) expression systems. Different class of enzyme versus others tested (short- chain, non-metal binding) Lactococcus 66 (nucleotide NADH-dependent alcohol lactis adhA sequence) dehydrogenase with activity 67 (amino acid using isobutyraldehyde as sequence) the substrate (Atsumi et al., Appl. Microbiol. Biotechnol., 2009, DOI 10.1007/s00253- 009-2085-6) Klebsiella 62 (nucleotide NC_011283 NADH-utilizing 1,2- pneumoniae sequence) propanediol dehydrogenase dhaT 63 (amino acid sequence) Escherichia 64 (nucleotide NC_000913.2 Homolog of K. pneumoniae coli fucO sequence) (2929887 . . . 2931038, dhaT, NADH-dependent 1,3- 65 (amino acid complement) propanediol dehydrogenase sequence)
TABLE-US-00002 TABLE 2 Kinetic parameters for the conversion of isobutyraldehyde to isobutanol by Ec_YqhD, Ec_FucO, Dm_Adh, and Kp_DhaT NADH NADPH Activity Activity (U/min-1 mg-1 (U/min-1 mg-1 KM crude KM crude Plasmid Adh (mM) lysate) (mM) lysate) pGV1705-A Ec_YqhD n.d. n.d. 0.25 0.09 pGV1748-A Ec_FucO 0.8 0.23 0.2 0.04 pGV1749-A Dm_Adh 0.9 6.60 2.7 1.70 pGV1778-A Kp_DhaT 1.3 0.56 0.6 0.08
[0372]Alcohol dehydrogenases of the present disclosure may also be utilized in metabolically-modified microorganisms that include recombinant biochemical pathways useful for producing additional alcohols such as 2-methyl-1-butanol, 3-methyl-1-butanol, 2-phenylethanol, 1-propanol, or 1-butanol via conversion of a suitable substrate by a modified microorganism.
[0373]Microorganisms producing such compounds have been described (PCT/US2008/053514). For example, these alcohols can be 1-propanol, 1-butanol, 2-methyl-1-butanol, 3-methyl-1-butanol or 2-phenylethanol and are generally produced from a metabolite comprising a 2-keto acid. In some aspects, the 2-keto acid includes 2-ketobutyrate, 2-ketovalerate, 2-keto-3-methylvalerate, 2-keto-4-methyl-pentanoate, or phenylpyruvate. The 2-ketoacid is converted to the respective aldehyde by a 2-ketoacid decarboxylase. For example, 2-ketobutyrate is converted to 1-propanal, 2-ketovalerate is converted to 1-butanal, 2-keto-3-methylvalerate is converted to 2-methyl-1-butanol, 2-keto-4-methyl-pentanoate is converted to 3-methyl-1-butanal, and phenylpyruvate is converted to phenylethanal by a 2-ketoacid decarboxylase. Thus, the recombinant microorganism includes elevated expression or activity of a 2-keto-acid decarboxylase, as compared to a parental microorganism. The 2-keto-acid decarboxylase may be encoded by kivd from Lactococcus lactis, or homologs thereof. The 2-keto-acid decarboxylase can be encoded by a polynucleotide derived from a gene selected from kivd from L. lactis, or homologs thereof.
[0374]In earlier publications (PCT/US2008/053514, Atsumi et al., Nature, 2008 Jan. 3; 451(7174):86-9), only NADPH-dependent alcohol dehydrogenases are described that convert the aforementioned aldehyde to an alcohol. In particular, S. cerevisiae Adh2p is described that converts the aldehyde to the respective aldehyde.
[0375]Thus, in one embodiment of this disclosure, a microorganism is provided in which the cofactor dependent final step for the conversion of the aldehyde to the respective alcohol is catalyzed by an NADH-dependent alcohol dehydrogenase. In particular, NADH-dependent alcohol dehydrogenases are disclosed that catalyze the reduction aldehydes to alcohols, for example, of 1-propanal to 1propanol, 1-butanal to 1-butanol, 2-methyl-1-butanal to 2-methyl-1-butanol, 3-methyl-1-butanal to 3-methyl-1-butanol, or phenylethanal to phenylethanol.
[0376]In a specific aspect, such an alcohol dehydrogenase may be encoded by the Drosophila melanogaster alcohol dehydrogenase Dm_Adh or homologs thereof. In another specific aspect, such an alcohol dehydrogenase may be encoded by the Lactococcus lactis alcohol dehydrogenase Ll_AdhA (SEQ ID NO: 67), as described by Atsumi et al. (Atsumi et al., Appl. Microbiol. Biotechnol., 2009, DOI 10.1007/s00253-009-2085-6) or homologs thereof.
[0377]Surprisingly, alcohol dehydrogenase enzymes that are not known to catalyze the reduction of isobutyraldehyde to isobutanol were identified that catalyze this reaction. Thus, in another aspect, such an alcohol dehydrogenase may be encoded by an NADH-dependent 1,3-propanediol dehydrogenase. In yet another aspect, such an alcohol dehydrogenase may be encoded by an NADH-dependent 1,2-propanediol dehydrogenase. Preferred enzymes of this disclosure include enzymes listed in Table 1.
[0378]In another embodiment, a method of producing an alcohol is provided. The method includes providing a recombinant microorganism provided herein; culturing the microorganism of in the presence of a suitable substrate or metabolic intermediate and under conditions suitable for the conversion of the substrate to an alcohol; and detecting the production of the alcohol. In various aspects, the alcohol is selected from 1-propanol, 1-butanol, 2-methyl 1-butanol, 3-methyl 1-butanol, and 2-phenylethanol. In another aspect, the substrate or metabolic intermediate includes a 2-keto acid-derived aldehyde, such as 1-propanal, 1-butanal, 2-methyl-1-butanal, 3-methyl-1-butanal, or phenylethanal.
Recombinant Host Cells Comprising a NADH-Dependent KARI and/or ADH Enzymes
[0379]In an additional aspect, the present invention is directed to recombinant host cells (i.e. metabolically "engineered" or "modified" microorganisms) comprising NADH-dependent KARI and/or ADH enzymes of the invention. Recombinant microorganisms provided herein can express a plurality of additional heterologous and/or native target enzymes involved in pathways for the production of beneficial metabolites such as isobutanol from a suitable carbon source.
[0380]Accordingly, metabolically "engineered" or "modified" microorganisms are produced via the introduction of genetic material (i.e. a NADH-dependent KARI and/or ADH enzymes) into a host or parental microorganism of choice, thereby modifying or altering the cellular physiology and biochemistry of the microorganism. Through the introduction of genetic material and/or the modification of the expression of native genes the parental microorganism acquires new properties, e.g. the ability to produce a new, or greater quantities of, an intracellular metabolite. As described herein, the introduction of genetic material and/or the modification of the expression of native genes into a parental microorganism results in a new or modified ability to produce beneficial metabolites such as isobutanol. The genetic material introduced into and/or the genes modified for expression in the parental microorganism contains gene(s), or parts of genes, coding for one or more of the enzymes involved in a biosynthetic pathway for the production of isobutanol and may also include additional elements for the expression and/or regulation of expression of these genes, e.g. promoter sequences.
[0381]Recombinant microorganisms provided herein may also produce metabolites in quantities not available in the parental microorganism. A "metabolite" refers to any substance produced by metabolism or a substance necessary for or taking part in a particular metabolic process. A metabolite can be an organic compound that is a starting material (e.g., glucose or pyruvate), an intermediate (e.g., 2-ketoisovalerate), or an end product (e.g., 1-propanol, 1-butanol, isobutanol, 2-methyl-1-butanol, 3-methyl-1-butanol) of metabolism. Metabolites can be used to construct more complex molecules, or they can be broken down into simpler ones. Intermediate metabolites may be synthesized from other metabolites, perhaps used to make more complex substances, or broken down into simpler compounds, often with the release of chemical energy.
[0382]Exemplary metabolites include glucose, pyruvate, 1-propanol, 1-butanol, isobutanol, 2-methyl-1-butanol, and 3-methyl-1-butanol.
[0383]The metabolite 1-propanol can be produced by a recombinant microorganism engineered to express or over-express a metabolic pathway that converts pyruvate to 1-propanol. An exemplary metabolic pathway that converts pyruvate to 1-propanol has been described in WO/2008/098227 and by Atsumi et al. (Atsumi et al., 2008, Nature 451(7174): 86-9), the disclosures of which are herein incorporated by reference in their entireties. In a preferred embodiment, metabolic pathway comprises a KARI and/or an ADH enzyme of the present invention.
[0384]The metabolite 1-butanol can be produced by a recombinant microorganism engineered to express or over-express a metabolic pathway that converts pyruvate to 3-methyl-1-butanol. An exemplary metabolic pathway that converts pyruvate to 3-methyl-1-butanol has been described in WO/2008/098227 and by Atsumi et al. (Atsumi et al., 2008, Nature 451(7174): 86-9), the disclosures of which are herein incorporated by reference in their entireties. In a preferred embodiment, metabolic pathway comprises a KARI and/or an ADH enzyme of the present invention.
[0385]The metabolite isobutanol can be produced by a recombinant microorganism engineered to express or over-express a metabolic pathway that converts pyruvate to isobutanol. An exemplary metabolic pathway that converts pyruvate to isobutanol may be comprised of a acetohydroxy acid synthase (ALS) enzyme encoded by, for example, alsS from B. subtilis, a ketolacid reductoisomerase (KARI) of the present invention, a dihydroxy-acid dehydratase (DHAD), encoded by, for example ilvD from E. coli, a 2-keto-acid decarboxylase (KIVD) encoded by, for example kivd from L. lactis, and an alcohol dehydrogenase (ADH) of the present invention.
[0386]The metabolite 3-methyl-1-butanol can be produced by a recombinant microorganism engineered to express or over-express a metabolic pathway that converts pyruvate to 3-methyl-1-butanol. An exemplary metabolic pathway that converts pyruvate to 3-methyl-1-butanol has been described in WO/2008/098227 and by Atsumi et al. (Atsumi et al., 2008, Nature 451(7174): 86-9), the disclosures of which are herein incorporated by reference in their entireties. In a preferred embodiment, metabolic pathway comprises a KARI and/or an ADH enzyme of the present invention.
[0387]The metabolite 2-methyl-1-butanol can be produced by a recombinant microorganism engineered to express or over-express a metabolic pathway that converts pyruvate to 2-methyl-1-butanol. An exemplary metabolic pathway that converts pyruvate to 2-methyl-1-butanol has been described in WO/2008/098227 and by Atsumi et al. (Atsumi et al., 2008, Nature 451(7174): 86-9), the disclosures of which are herein incorporated by reference in their entireties. In a preferred embodiment, metabolic pathway comprises a KARI and/or an ADH enzyme of the present invention.
[0388]The disclosure identifies specific genes useful in the methods, compositions and organisms of the disclosure; however it will be recognized that absolute identity to such genes is not necessary. For example, changes in a particular gene or polynucleotide comprising a sequence encoding a polypeptide or enzyme can be performed and screened for activity. Typically such changes comprise conservative mutation and silent mutations. Such modified or mutated polynucleotides and polypeptides can be screened for expression of a functional enzyme using methods known in the art. In addition, homologs of enzymes useful for generating metabolites are encompassed by the microorganisms and methods provided herein.
Method of Using Microorganism for Anaerobic Isobutanol Fermentation
[0389]In a method to produce a target compound from a carbon source at high yield a modified microorganism subject to this invention is cultured in an appropriate culture medium containing a carbon source.
[0390]An exemplary embodiment provide a method for producing isobutanol comprising a modified microorganism of the invention in a suitable culture medium containing a carbon source that can be converted to isobutanol by the microorganism of the invention.
[0391]In certain embodiments, the method further includes isolating said target compound from the culture medium. For example, isobutanol may be isolated from the culture medium by any method, in particular a method known to those skilled in the art, such as distillation, pervaporation, or liquid-liquid extraction.
[0392]This invention is further illustrated by the following examples that should not be construed as limiting. The contents of all references, patents, and published patent applications cited throughout this application, as well as the Figures and the Sequence Listing, are incorporated herein by reference for all purposes.
EXAMPLES
[0393]The following provides examples that demonstrate that microorganisms modified to resolve a cofactor imbalance produce a target compound at higher yield under conditions that include anaerobic conditions. One compound to be produced by the recombinant microorganism according to the present invention is isobutanol. The present invention is not limited to isobutanol. The invention may be applicable to any metabolic pathway that is imbalanced with respect to cofactor usage. One skilled in the art is able identify pathways that are imbalanced with respect to cofactor usage and apply this invention to provide recombinant microorganisms in which the same pathway is balanced with respect to cofactor usage.
Sample Preparation
[0394]Generally, samples (2 mL) from fermentation experiments performed in shake flasks were stored at 4° C. for later substrate and product analysis. Prior to analysis, samples were centrifuged at 14,000×g for 10 min. The supernatant was filtered through a 0.2 μm filter. Analysis of substrates and products was performed using authentic standards (>99%, obtained from Sigma-Aldrich), and a 5-point calibration curve (with 1-pentanol as an internal standard for analysis by gas chromatography).
Determination of Optical Density
[0395]The optical density of the yeast cultures was determined at 600 nm using a DU 800 spectrophotometer (Beckman-Coulter, Fullerton, Calif., USA). Samples were diluted as necessary to yield an optical density of between 0.1 and 0.8.
Gas Chromatography
[0396]Analysis of volatile organic compounds, including ethanol and isobutanol was performed on a HP 5890 gas chromatograph fitted with an HP 7673 Autosampler, a DB-FFAP column (J&W; 30 m length, 0.32 mm ID, 0.25 μM film thickness) or equivalent connected to a flame ionization detector (FID). The temperature program was as follows: 200° C. for the injector, 300° C. for the detector, 100° C. oven for 1 minute, 70° C./minute gradient to 235° C., and then hold for 2.5 min.
[0397]Analysis was performed using authentic standards (>99%, obtained from Sigma-Aldrich), and a 5-point calibration curve with 1-pentanol as the internal standard.
High Performance Liquid Chromatography
[0398]Analysis of glucose and organic acids was performed on a HP-1100 High Performance Liquid Chromatography system equipped with an Aminex HPX-87H Ion Exclusion column (Bio-Rad, 300×7.8 mm) or equivalent and an H.sup.+ cation guard column (Bio-Rad) or equivalent. Organic acids were detected using an HP-1100 UV detector (210 nm, 8 nm 360 nm reference) while glucose was detected using an HP-1100 refractive index detector. The column temperature was 60° C. This method was Isocratic with 0.008N sulfuric acid in water as mobile phase. Flow was set at 0.6 mL/min. Injection size was 20 μL and the run time was 30 minutes.
Molecular Biology and Bacterial Cell Culture
[0399]Standard molecular biology methods for cloning and plasmid construction were generally used, unless otherwise noted (Sambrook, J., Russel, D. W. Molecular Cloning, A Laboratory Manual. 3 ed. 2001, Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press).
[0400]Standard recombinant DNA and molecular biology techniques used in the Examples are well known in the art and are described by Sambrook, J., Russel, D. W. Molecular Cloning, A Laboratory Manual. 3 ed. 2001, Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; and by T. J. Silhavy, M. L. Bennan, and L. W. Enquist, Experiments with Gene Fusions, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. (1984) and by Ausubel, F. M. et al., Current Protocols in Molecular Biology, pub. by Greene Publishing Assoc. and Wiley-Interscience (1987).
[0401]General materials and methods suitable for the routine maintenance and growth of bacterial cultures are well known in the art. Techniques suitable for use in the following examples may be found as set out in Manual of Methods for General Bacteriology (Phillipp Gerhardt, R. G. E. Murray, Ralph N. Costilow, Eugene W. Nester, Willis A. Wood, Noel R. Krieg and G. Briggs Phillips, eds.), American Society for Microbiology, Washington, D.C. (1994)) or by Thomas D. Brock in Biotechnology: A Textbook of Industrial Microbiology, Second Edition, Sinauer Associates, Inc., Sunderland, Mass. (1989).
Preparation of Electrocompetent E. coli Cells and Transformation
[0402]The acceptor strain culture was grown in SOB-medium (Sambrook, J., Russel, D. W. Molecular Cloning, A Laboratory Manual. 3 ed. 2001, Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press) to an OD600 of about 0.6 to 0.8. The culture was concentrated 100-fold, washed once with ice cold water and 3 times with ice cold 10% glycerol. The cells were then resuspended in 150 μL of ice-cold 10% glycerol and aliquoted into 50 μL portions. These aliquots were used immediately for standard transformation or stored at -80° C. These cells were transformed with the desired plasmid(s) via electroporation. After electroporation, SOC medium (Sambrook, J., Russel, D. W. Molecular Cloning, A Laboratory Manual. 3 ed. 2001, Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press) was immediately added to the cells. After incubation for an hour at 37° C. the cells were plated onto LB-plates containing the appropriate antibiotics and incubated overnight at 37° C.
Transformation of S. cerevisiae Cells
[0403]S. cerevisiae strains were transformed by the Lithium Acetate method (Gietz et al., Nucleic Acids Res. 27:69-74 (1992): Cells from 50 mL YPD cultures (YPGaI for valine auxotrophs) were collected by centrifugation (2700 rcf, 2 minutes, 25° C.) once the cultures reached an OD600 of 1.0. The cells were washed cells with 50 mL sterile water and collected by centrifugation at 2700 rcf for 2 minutes at 25° C. The cells were washed again with 25 mL sterile water and collected cells by centrifugation at 2700 rcf for 2 minutes at 25° C. The cells were resuspended in 1 mL of 100 mM lithium acetate and transferred to a 1.5 mL eppendorf tube. The cells were collected cells by centrifugation for 20 sec at 18,000 rcf, 25° C. The cells were resuspended cells in a volume of 100 mM lithium acetate that was approximately 4× the volume of the cell pellet. A mixture of DNA (final volume of 15 μl with sterile water), 72 μl 50% PEG, 10 μl 1 M lithium acetate, and 3 μl denatured salmon sperm DNA was prepared for each transformation. In a 1.5 mL tube, 15 μl of the cell suspension was added to the DNA mixture (85 μl), and the transformation suspension was vortexed with 5 short pulses. The transformation was incubated at 30 minutes at 30° C., followed by incubation for 22 minutes at 42° C. The cells were collected by centrifugation for 20 sec at 18,000 rcf, 25° C. The cells were resuspended in 100 μl SOS (1 M sorbitol, 34% (v/v) YP (1% yeast extract, 2% peptone), 6.5 mM CaCl2) or 100 μl YP (1% yeast extract, 2% peptone) and spread over an appropriate selective plate.
Sporulation of Diploid S. cerevisiae and Germination to Obtain Haploids
[0404]Random spore analysis was used to identify desired haploid segregants of relevant diploid strains. Diploid strains were sporulated by pre-culturing in YPD for 24 hrs and then transferring the cells into 5 mL of sporulation medium (1% wt/vol potassium acetate). After 4-5 days, the culture was examined microscopically for the presence of visible spore-containing asci. To the 5 mL sporulation culture, 0.5 mL of 1 mg/mL Zymolyase-T (Seikagaku Biobusiness, Tokyo, Japan) and 10 μL of β-mercaptoethanol were added, and the cells were incubated overnight at 30° C. while shaking slowly (60 rpm). The next day, 5 mL of 1.5% IGEPAL-CA-630 [reference] were added and the mixture incubated on ice for 15 minutes. The cell suspension was then sonicated (3 rounds, 30 seconds per round, 50% power) with 2 minutes on ice between sonications. The suspension was centrifuged (1200×g, 10 min), the liquid poured off, 5 mL of 1.5% IGEPAL-CA-630 (Sigma-Aldrich Co., St. Louis, Mo.) were added, and the centrifugation and resuspension step repeated once more. The cell suspension was again sonicated as described above, after which it was centrifuged and washed as described above except that instead of IGEPAL, sterile water was used to resuspend the cells. The cells were finally resuspended in 1 mL of sterile water, and 0.1 mL of a 1:10, 1:100, 1:100, and 1:10,000 dilution of the initial 1 mL cell suspension were plated onto SCE-Trp, Leu, Ura (for full-pathway integrants strains) or SCD-Trp, Ura (for partial-pathway integrant strains) media and the plates incubated at 30° C. until colonies appeared (typically, 4-5 days).
Yeast Colony PCR
[0405]Colony PCR was carried out using the FailSafe mix (Epicentre Biotechnologies, Madison, Wis.). Specifically, 15 L of FailSafe Mix "E" were combined with 13 μL sterile water, 0.35 μL of each primer (from a 100 μM solution), and 0.6 μL FailSafe polymerase. For template, a small dab of yeast cells sufficient to just turn the solution turbid was swirled into each individual reaction mixture. The PCR reactions were incubated as follows: 1 cycle of 94° C.×2 min; 40 cycles of 94° C.×15 sec, 53° C.×15 sec, 72° C.×1 min; 1 cycle of 72° C.×8 min.
qRT-PCR
[0406]Performed by isolating RNA, synthesizing cDNA by reverse transcription and performing qPCR using protocols described below.
RNA Isolation for Reverse Transcription (RT)
[0407]3 ml YPD cell cultures were incubated at 30° C., 250 RPM until they reached OD600's of 0.7 to 1.5. 2 OD600's (e.g. 1 mL of a culture at 2 OD600) of cells were then harvested from each culture in 1.5 ml tubes by centrifugation at full speed in a microfuge for 2 minutes. The cell pellet was stored overnight at -20° C. RNA was isolated using the YeaStar RNAKit® (Zymo Research Corp. Orange, Calif. 92867 USA). Following the protocol provided with the kit, cells were resuspended in 80 μl of YR Digestion Buffer and 5 μl of Zymolyase®. The pellet was completely resuspended by repeated pipetting. The suspension was incubated at 37° C. for 60 minutes. 160 μl of YR Lysis Buffer was added to the suspension, which was then mixed thoroughly by vortexing. The mixture was centrifuged at >4,000×g for 2 minutes in the microfuge, and the supernatant was transferred to a Zymo-Spin Column in a Collection Tube. The column was centrifuged at >10,000×g for 1 minute in the microfuge. To the column, 200 μl RNA Wash Buffer was added, and the column was centrifuged for 1 minute at 14,000 RPM in the microfuge. The flow-through was discarded and 200 μl RNA Wash Buffer was added to the column. The column was centrifuged for 1 minute at >10,000×g. The Zymo-Spin Column was transferred to a new RNase-free 1.5 ml centrifuge tube, and 60 μl of DNase/RNase-Free Water was added directly to the column membrane. The RNA was eluted by centrifugation for 30 seconds at >10,000×g in the microfuge.
cDNA Synthesis (Reverse Transcription) for qPCR
[0408]Using the qScript® cDNA SuperMix kit provided by Quanta Biosciences® (Gaithersburg, Md.), cDNA was prepared following the protocol provided with the kit. First, the concentration of RNA was measured for the preparations from each transformant candidate and control strain. A final solution of 300 ng of RNA in sterile water was prepared in a volume of 16 μl in 0.2 ml PCR tube (RNase-free). To each sample, 4 μl of qScript cDNA Supermix was added. The reactions were incubated on a thermocycler for 5 minutes at 25° C., 30 minutes at 42° C., and 5 minutes at 85° C.
qPCR:
[0409]Each reaction contained: 10 μL of PerfeCTa® SYBR® Green SuperMix kit (Quanta Biosciences® Gaithersburg, Md.), 1 μl of cDNA, 1 μl of a 5 μM (each) mix of forward and reverse primers and 8 μl of sterile water. Each reaction was assembled in a well of a 0.2 ml 96-well plate, and a clear plastic sheet was carefully (to avoid the introduction of warped surface or fingerprints or smudges) and firmly placed over the 96-well plate. The reactions were incubated in an Eppendorf Mastercycler ep thermocycler (Eppendorf, Hamburg, Germany) using the following conditions: 95° C. for 2 minutes, 40 cycles of 95° C. for 15 seconds and 60° C. for 45 seconds, 95° C. for 15 seconds, 60° C. for 15 seconds, and a 20 minute slow ramping up of the temperature until it reaches 95° C. Finally, it was incubated at 95° for 15 seconds. The fluorescence emitted by the SYBR dye was measured at the 60° C. incubation step during each of the 40 cycles, as well as during the ramping up to 95° C. for melting curve analysis of the primer sets.
Construction of E. coli Strains
[0410]GEVO1385 was constructed by integrating the Z1 module into the chromosome of JCL260 by P1 transduction from the strain E. coli W3110,Z1 (Lutz, R, Bujard, H Nucleic Acids Research (1997) 25, 1203-1210).
[0411]GEVO1399: The gene zwf was deleted according to the standard protocol for gene deletion using the Wanner method (Datsenko, K. and Wanner, B. One-step Inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. PNAS 2000). Primers 73 and 74 were used to amplify the Kan resistance cassette from pKD13. The linear PCR product was transformed into E. coli W3110 pKD46 electro competent cells and the knock-out of zwf was verified by PCR. Lysate of the new strain (E. coli W3110, Δzwf::FRT::Kan::FRT) was prepared and the knock-out was transferred into JCL260 by P1 transduction. Removal of the Kan resistance cassette from this strain using transient expression of FLP recombinase yielded GEVO1399.
[0412]GEVO1608: The gene Ec_yqhD (SEQ ID NO: 68) was deleted according to the standard protocol for gene deletion using the Wanner method (Datsenko, K and Wanner, B, "One-step Inactivation of chromosomal genes in Escherichia coli K-12 using PCR products," PNAS 2000, 97:6640-6645). Primers 1155 and 1156 were used to amplify the Kan resistance cassette from pKD13. The linear PCR product was transformed into E. coli W3110 pKD46 electro competent cells and the knock-out of Ec_yqhD was verified by PCR. A lysate of the new strain (E. coli W3110, ΔyqhD::FRT::Kan::FRT) was prepared and the knock-out was transferred into JCL260 by P1 transduction yielding GEVO1608.
[0413]GEVO1745: Removal of the Kan resistance cassette from GEVO1608 using transient expression of FLP recombinase yielded GEVO1745.
[0414]GEVO1748 and GEVO1749 are derivatives of JCL260. For the construction of GEVO1748, PLlacO1:Ll_kiv1:Ec_ilvD_coEc was integrated into the ilvC locus on the E. coli chromosome. In particular primers 869 and 1030 were used to amplify the kanamycin resistance cassette (Kan) from pKD13, and primers 1031 and 1032 were used to amplify PLlacO1::Ll_kivdt:Ec_ilvD_coEc from pGV1655 (SEQ ID NO: 109). For the construction of GEVO1749 PLlacO1:Ll_kivd1::Ec_ilvD_coEc was integrated into the adhE locus on the E. coli chromosome. In particular primers 50 and 1030 were used to amplify the kanamycin resistance cassette from pKD13, and primers 1031 and 1205 were used to amplify PLlacO1:Ll_kivd1::Ec_ilvD_coEc from pGV1655 (SEQ ID NO: 109). Afterwards, SOE (splicing by overlap extension) (Horton, R M, Cai, Z L, Ho, S N, et al. Biotechniques Vol. 8 (1990) pp 528) reactions were done to connect the gene expression cassettes to the resistance cassette using primers 1032 and 869 for the ilvC locus and primers 1205 and 50 for the adhE locus. The linear PCR products were transformed into W3110 pKD46 electro competent cells and the knock ins of PLlacO1:Ll_kivd1::Ec_ilvD_coEc::FRT::Kan::FRT were verified by PCR. The knock ins were further verified by sequencing. Lysates of the new strains E. coli W3110, ΔilvC::PLlacO1::Ll_kivd1::Ec_ilvD_coEc::FRT::Kan::FRT) and E. coli W3110, ΔadhE::PLlacO1:Ll_kivd1:: Ec_ilvD_coEc::FRT::Kan::FRT) were prepared and the knock ins were transferred to JCL260 by P1 transduction. Removal of the Kan resistance cassette from this strain using expression of FLP recombinase yielded GEVO1748 and GEVO1749.
[0415]GEVO1725, GEVO1750, GEVO1751: The gene maeA was deleted according to the standard protocol for gene deletion using the Wanner method (Datsenko, K. and Wanner, B. One-step Inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. PNAS 2000). Primers 116 and 117 were used to amplify the Kan resistance cassette from pKD13. The linear PCR product was transformed into E. coli W3110 pKD46 electro competent cells and the knock-out of maeA was verified by PCR. Lysate of the new strain (E. coli W3110, ΔmaeA::FRT::Kan::FRT) was prepared and the knock-out was transferred into JCL260 by P1 transduction. The Kan resistance cassette was removed from this strain using transient expression of FLP recombinase. The resulting strain was transduced with the Z1 cassette yielding GEVO1750, and the same strain was transduced with a lysate conferring a pykA deletion. The pykA deletion lysate was prepared from W3110, L\pykA::FRT::Kan::FRT, which was created using homologous recombination according to the Wanner method using primers 1187 and 1188 for the amplification of the Kan cassette from pKD13. The Kan resistance cassette was removed from this strain using transient expression of FLP recombinase. The resulting strain was transduced with a lysate conferring a pykF deletion. The pykF deletion lysate was prepared from W3110, ΔpykF::FRT::Kan::FRT, which was created using homologous recombination according to the Wanner method using primers 1191 and 1192 for the amplification of the Kan cassette from pKD13. Removal of the Kan resistance cassette from this strain using transient expression of FLP recombinase yielded GEVO1725. For the construction of GEVO1751 strain GEVO1725 was transduced with a lysate of W3110Z1. The resulting strain was GEVO1751.
[0416]For the construction of GEVO1777 ilvC was deleted according to the standard protocol for gene deletion using the Wanner method. Primers 868 and 869 were used to amplify the Kan resistance cassette from pKD13. The linear PCR product was transformed into E. coli W3110 pKD46 electro competent cells and the knock-out of ilvC was verified by PCR. The Kan resistance cassette was removed from this strain using transient expression of FLP recombinase. The resulting strain was transduced with the Z1 cassette yielding GEVO1777.
[0417]GEVO1780 was constructed by transforming JCL260 with plasmids pGV1655 (SEQ ID NO: 109) and pGV1698 (SEQ ID NO: 112).
[0418]GEVO1844: An E. coli sthA deletion strain was obtained from the Keio collection and the deletion of sthA was verified. The sthA deletion was transferred to GEVO1748 by P1 phage transduction and after removal of the Kan resistance cassette by transient expression of FLP recombinase the resulting strain GEVO1844 was verified for the sthA deletion.
[0419]GEVO1846 was constructed by transforming strain GEVO1748 with plasmids pGV1745 (SEQ ID NO: 117) and pGV1698 (SEQ ID NO: 112).
[0420]GEVO1859 was constructed according to the standard protocol for gene integration using the Wanner method (Datsenko, K. and Wanner, B. One-step Inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. PNAS 2000). Primers 1219 and 1485 were used to amplify PLlacO1::Bs_alsS1::Ec_ilvC_coEc from pGV1698 (SEQ ID NO: 112). Primers 1218 and 1486 were used to amplify the Kan resistance cassette from pKD13. SOE (splicing by overlap extension) was used to combine the two pieces to one integration cassette. The linear PCR product was transformed into E. coli W3110 pKD46 electro competent cells and the knock-in of PLlacO1::Bs_alsS1::Ec_ilvC_coEc::FRT::Kan::FRT into the pflB locus was verified by PCR. The knock-in was further verified by sequencing. Lysate of the new strain (E. coli W3110, ΔpflB:: PLlacO1::Bs_alsS1::Ec_ilvC_coEc::FRT::Kan::FRT) was prepared and the knock-in was transferred into GEVO1749 by P1 transduction. Removal of the Kan resistance cassette from this strain using transient expression of FLP recombinase yielded GEVO1859.
[0421]GEVO1886 was constructed according to the standard protocol for gene integration using the Wanner method (Datsenko, K. and Wanner, B. One-step Inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. PNAS 2000). Primers 1562 and 1539 were used to amplify PLlacO1::pntAB from pGV1745 (SEQ ID NO: 117). Primers 1479 and 1561 were used to amplify the Kan resistance cassette from pKD13. SOE was used to combine the two pieces to one integration cassette. The linear PCR product was transformed into E. coli W3110 pKD46 electro competent cells and the knock-in of PLlacO1::pntAB::FRT::Kan::FRT into the sthA locus was verified by PCR. The knock-in was further verified by sequencing. Lysate of the new strain (E. coli W3110, ΔsthA:: PLlacO1::pntAB::FRT::Kan::FRT) was prepared and the knock-in was transferred into GEVO1859 by P1 transduction. Removal of the Kan resistance cassette from this strain using transient expression of FLP recombinase yielded GEVO1886.
[0422]GEVO1993 is a derivative of GEVO1748. For the construction of GEVO1993, PLlacO1::Bs_alsS1 was integrated into the pta locus on the E. coli chromosome. In particular primers 1526 and 474 were used to amplify the kanamycin resistance cassette (Kan) from pKD13, and primers 1563 and 1527 were used to amplify PLlacO1:: Bs_alsS1 from pGV1698. Afterwards, SOE (splicing by overlap extension) reactions were done to connect the gene expression cassette to the resistance cassette using primers 1563 and 474. The linear PCR products were transformed into E. coli W3110 pKD46 electro competent cells and the knock-ins of PLlacO1::Bsa/sS1::FRT::Kan::FRT were verified by PCR. The knock-ins were further verified by sequencing. Lysate of the new strain E. coli W3110, Δpta::PLlacO1::Bs_alsS1::FRT::Kan::FRT was prepared and the knock-in was transferred to GEVO1748 by P1 transduction yielding GEVO1993. The integration into the pta locus in GEVO1993 was verified by PCR.
Construction of Saccharomyces cerevisiae Strains
[0423]A PDC deletion variant S. cerevisiae, GEVO2302, was evolved so that it does not have the requirement for a two-carbon molecule and has a growth rate similar to the parental strain on glucose.
[0424]GEVO1186 is S. cerevisiae CEN.PK2
[0425]GEVO1803 was made by transforming GEVO1186 with the 6.7 kb pGV1730 (SEQ ID NO: 116) (contains S. cerevisiae TRP1 marker and the CUP1 promoter-driven Bs_alsS2) that had been linearized by digestion with NruI. Completion of the digest was confirmed by running a small sample on a gel. The digested DNA was then purified using Zymo Research DNA Clean and Concentrator and used in the transformation. Trp+clones were confirmed for the correct integration into the PDC1 locus by colony PCR using primer pairs 1440+1441 and 1442+1443 for the 5' and 3' junctions, respectively. Expression of Bs_alsS2 was confirmed by qRT-PCR using primer pairs 1323+1324.
[0426]GEVO2107 was made by transforming GEVO1803 with linearized, Hpal-digested pGV1914 (SEQ ID NO: 119). Correct integration of pGV1914 at the PDC6 locus was confirmed by analyzing candidate Ura+colonies by colony PCR using primers 1440 plus 1441, or 1443 plus 1633, to detect the 5' and 3' junctions of the integrated construct, respectively. Expression of all transgenes were confirmed by qRT-PCR using primer pairs 1321 plus 1322, 1587 plus 1588, and 1633 plus 1634 to examine Bs_alsS2, Ll_kivd2 coEc, and Dm_ADH transcript levels, respectively.
[0427]GEVO2158 was made by transforming GEVO2107 with NruI-digested pGV1936 (SEQ ID NO: 120). Correct integration of pGV1936 at the PDC5 locus was confirmed by analyzing candidate Ura+, Leu+colonies by colony PCR using primers primers 1436 plus 1437, or 1595 plus 1439, to detect the 5' and 3' junctions of the integrated construct, respectively. Expression of all transgenes were confirmed by qRT-PCR using primer pairs 1321 plus 1322, 1597 plus 1598, 1566 plus 1567, 1587 plus 1588, 1633 plus 1634, and 1341 plus 1342 to examine levels of Bs_alsS2, Ec_ilvC_coSc.sup.Q110V, Sc_ilv3ΔN, Ll_kivd2_coEc, Dm_ADH, and ACT1, respectively.
[0428]GEVO2302 was constructed by sporulating GEVO2158. Haploid spores were prepared for random spores analysis (as described above), and the spores were plated onto SCE-Trp,Leu,Ura medium (14 g/L Sigma® Synthetic Dropout Media supplement (includes amino acids and nutrients excluding histidine, tryptophan, uracil, and leucine), 6.7 g/L Difco® Yeast Nitrogen Base without amino acids. 0.076 g/L histidine and 25 mL/L 100% ethanol). Candidate colonies were patched onto SCE-Trp, Leu, Ura plates (Plate version of the above medium was prepared using 20 g/L agar) and then replica plated onto YPD (10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose) and YPE (10 g/L yeast extract, 20 g/L peptone, 25 mL/L 100% ethanol) plates. Patches that grew on YPE but failed to grow on YPD were further analyzed by colony PCR to confirm mating type (and, hence, their status as haploid). Several verified haploid candidates were further analyzed for transgene expression by qRT-PCR. GEVO2302 contains the full isobutanol pathway with ALS, KARI, DHAD, KIVD, and ADH being encoded by Bs_alsS2, Ec_ilvC_coSc.sup.Q110V, Sc_ilv3ΔN, Ll_kivd2_coEc, Dm_ADH, respectively.
[0429]GEVO2710, GEVO2711, GEVO2712 and GEVO2799 are C2-independent, glucose de-repressed derivatives of GEVO2302, which were constructed via chemostat evolution: A DasGip fermentor vessel was sterilized and filled with 1 L of YNB+histidine medium (Yeast Nitrogen Base+histidine, containing per liter of distilled water: 6.7 g YNB without amino acids from Difco and 0.076 g histidine; the medium was adjusted to pH 5 by adding a few drops of HCL or KOH) and contained 2% w/v ethanol. The vessel was installed and all probes were calibrated according to DasGip instructions. The vessel was also attached to an off-gas analyzer of the DasGip system, as well as to a mass spectrometer. Online measurements of oxygen, carbon dioxide, isobutanol, and ethanol were taken throughout the experiment. The two probes that were inside the vessel measured pH and dissolved oxygen levels at all times. A medium inlet and an outlet were also set up on the vessel. The outlet tube was placed at a height just above the 1 L level, and the pump rate was set to maximum. This arrangement helped maintain the volume in the vessel at 1 L. Air was sparged into the fermentor at 12 standard liters per hour (slph) at all times. The temperature of the vessel was held constant at 30.0° C. and the agitation rate was set at a minimum of 500 rpm, with a cascade control to adjust the agitation to maintain 50% dissolved oxygen in the culture. The off-gas was analyzed for CO2, O2, ethanol and isobutanol concentrations. The amount of carbon dioxide (XcO2) and oxygen (XO2) levels in the off-gas were used to assess the metabolic state of the cells. An increase in XCO2 levels and decrease in XO2 levels indicated an increase in growth rate and glucose consumption rate. The ethanol levels were monitored to ensure that there was no contamination, either from other yeast cells or from potential revertants of the mutant strain because the S. cerevisiae PDC triple-mutant (GEVO2302) does not produce ethanol. The minimum pH in the vessel was set to 5, and a base control was set up to pump in potassium hydroxide into the vessel when the pH dropped below 5.
[0430]GEVO2302 was inoculated into 10 ml of YNB+histidine medium with 2% w/v ethanol as the carbon source. The culture was incubated at 30° C. overnight with shaking. The overnight culture was used to inoculate the DasGip vessel. Initially, the vessel was run in batch mode, to build up a high cell density. When about a cell biomass of OD600=8 was reached, the vessel was switched to chemostat mode and the dilution of the culture began. The medium pumped into the vessel was YNB+histidine with 6.357 g/L glucose and 0.364 g/L of acetate (5% carbon equivalent). The initial dilution rate was set to 0.06 h-1 to avoid washout.
[0431]After the culture in the chemostat was stabilized at the 0.06 h-1 dilution rate, the concentration of acetate was slowly decreased. This was achieved by using a two pump system, effectively producing a gradient pumping scheme. Initially pump A was pumping YNB+histidine medium with 10 g/L glucose at a rate of 35.5 mL/h and pump B was pumping YNB+histidine medium with only 1 g/L acetate at a rate of 20.3 mL/h. The total acetate going into the vessel was 0.364 g/L. Then, over a period of 5 days, the rate of pump B was slowly decreased and the rate of pump A was increased so that the combined rate of feeding increased from 56 mL/h to 74 ml/h. Over this period, the rate of pump B was finally reduced to 0, resulting in no (0 g/L) acetate addition to the chemostat. The glucose feed to the chemostat over this period was increased from 6.4 g/L to 10 g/L and the evolved strain was able to grow on glucose only.
[0432]Evolution of the strain for growth on increased glucose concentration was performed by slowly increasing the concentration of glucose in the chemostat with the evolved strain that no longer required a 2-carbon supplement. The concentration of glucose in the feed medium was increased from 10 g/L to 38 g/L over a period of 31 days. This was achieved by using a two pump system, effectively producing a gradient pumping scheme. Initially pump A was pumping YNB+histidine medium with 10 g/L glucose at a rate of 35.2 mL/h and pump B was pumping YNB+histidine medium with 15 g/L glucose at a rate of 32.9 mL/h. The total glucose going into the vessel was 12.4 g/L. Then, over a period of 18 days, the medium reservoirs for pump A and pump B were replaced with reservoirs containing increased concentrations of glucose until the reservoir for pump A contained 80 g/L glucose and the reservoir for pump B contained 100 g/L glucose. During this period, the combined rate of feeding maintained a dilution rate of 0.04 h-1. At the end of this period, the rate of pump A was finally reduced to 0, resulting in a feed of 100 g/L glucose. This dilution rate resulted in a biomass of OD600=4.8 at this glucose concentration and increasing the dilution rate to 0.09 h-1 over a period of 4 days lowered the biomass to an OD600=2.6. The dilution rate was lowered to 0.03 h-1 and gradually raised to 0.04 h-1 at 100 g/L glucose feed to raise the biomass to an OD600=4.4 over a period of 5 days. The glucose feed was then lowered by replacing the medium reservoir for pump A with a reservoir containing 0 g/L glucose, pumping initially at a rate of 33.4 ml/h, and pumping the 100 g/L glucose feed from pump B at 2.4 ml/h. This resulted in a dilution rate of 0.04 h-1, a glucose feed of 6.7 g/L and a biomass of OD600=6.0. Over a period of 4 days, the glucose concentration in the feed was gradually increased to 37.8 g/L by increasing the rate of pump B and decreasing the rate of pump A while maintaining a dilution rate of 0.04 h-1 and resulting in a biomass under these conditions of an OD600=6.6 and a glucose level in the chemostat of 18.8 g/L.
[0433]Evolution of the strain for increased growth rate was performed by slowly increasing the dilution rate in the chemostat with the evolved strain that no longer required a 2-carbon supplement and could grow with a feed of 37.8 g/L glucose with a glucose level in the chemostat of 18.8 g/L. Over a period of 13 days, the dilution rate was gradually increased from 0.04 h-1 to 0.14 h-1 by alternately increasing the rates of pump A and pump B to maintain a glucose feed concentration of 21-24 g/L glucose. A biomass of OD500=1.6 to an OD600=2.0 was maintained at dilution rates of 0.13 h-1 to 0.14 h-1.
[0434]Over the period of evolution, a sample was occasionally removed from the vessel directly. Samples were analyzed for glucose, acetate, and pyruvate using HPLC. Samples were plated onto YNB+histidine medium with 2% w/v ethanol as carbon source, YNB+histidine medium with different glucose concentrations (5 g/L, 10 g/L, 15 g/L, 20 g/L, 25 g/L and 50 g/L glucose), and YPD medium (containing 10 g/L yeast extract, 20 g/L peptone and 20 g/L dextrose) agar plates (plates contain the indicated medium+20 g/L agar). OD600 measurements were taken regularly to make sure the chemostat did not wash out. Freezer stocks of samples of the culture were made regularly for future characterization of the strains.
[0435]The chemostat with the evolved strain that no longer required a 2-carbon supplement and could grow with a feed of 37.8 g/L glucose with a glucose level in the chemostat of 18.8 g/L and could grow at a dilution rate >0:13 h-1 was maintained for another 23 days with varying dilution rates from 0.07 h-1 to 0.11 h-1 to allow further evolution for improved growth rate. At the end of this period, a sample from the chemostat was plated onto YNB+histidine medium with 50 g/L glucose agar plates and allowed to form colonies at 30° C. Ten colonies were picked for further characterization and re-streaked onto YNB+histidine medium with 50 g/L glucose agar plates for purification. None of these 10 evolved strains isolated from the chemostat sample grew when streaked onto SC-histidine medium (Synthetic complete medium lacking histidine, containing per liter of distilled water: 6.7 g YNB without amino acids from Difco, 100 ml of a solution of 14 g Yeast Synthetic Drop-out Medium Supplements without histidine, leucine, tryptophan and uracil from Sigma dissolved in 1 L water, 20 ml of a solution of 3.8 g/L tryptophan, 20 ml of a solution of 19 g/L leucine and 40 ml of a solution of 1.9 g/L uracil) containing 20 g/L glucose plates but did grow on SC-leucine medium (Synthetic complete medium lacking leucine, containing per liter of distilled water: 6.7 g YNB without amino acids from Difco, 100 ml of a solution of 14 g Yeast Synthetic Drop-out Medium Supplements without histidine, leucine, tryptophan and uracil from Sigma dissolved in 1 L water, 20 ml of a solution of 3.8 g/L tryptophan, 20 ml of a solution of 3.8 g/L histidine and 40 ml of a solution of 1.9 g/L uracil) containing 20 g/L glucose plates, indicating that they were still auxotrophic for histidine.
[0436]To characterize growth of the evolved strains, single colonies from each of the 10 evolved isolates purified on YNB+histidine medium with 50 g/L glucose agar plates were inoculated into 3 ml of YNB+histidine medium with 50 g/L glucose and YPD medium in 14 ml round-bottom snap-cap tubes and incubated overnight at 30° C. as a pre-culture. The next day the pre-cultures were used to inoculate 5 ml of the same medium as the pre-cultures in 50 ml conical plastic screw-cap centrifuge tubes to an OD600 of 0.01. The cultures were incubated shaking upright at 250 rpm at 30° C. and sampled periodically for OD600 measurement. Growth rates were calculated from plots of the OD600 measurements vs. time of incubation. Evolved isolates GEVO2710, GEVO2711, GEVO2712 and GEVO2799 were selected because of high growth rates in both YNB+histidine medium with 50 g/L glucose and YPD medium.
[0437]GEVO2792 is a C2-independent, PDC-minus S. cerevisiae strain carrying a control plasmid encoding no genes for an isobutanol metabolic pathway. To generate this strain, GEVO2710 was transformed with plasmid pGV2020 (SEQ ID NO: 121).
[0438]GEVO2844 is a C2-independent, PDC-minus S. cerevisiae strain carrying a control plasmid encoding no genes for an isobutanol metabolic pathway. To generate this strain, GEVO2799 was transformed with plasmid pGV2020 (SEQ ID NO: 121).
[0439]GEVO2847 is a C2-independent, PDC-minus S. cerevisiae strain carrying a partially NADH-utilizing isobutanol metabolic pathway. To generate this strain, GEVO2799 was transformed with plasmid pGV2082 (SEQ ID NO: 122), carrying the genes encoding NADPH-dependent KARI and the NADH-dependent ADH, Ec_ilvC_coSc.sup.Q110V (SEQ ID NO: 24), and Dm_ADH (SEQ ID NO: 60), respectively.
[0440]GEVO2848 is a O2-independent, PDC-minus S. cerevisiae strain carrying a partially NADH-utilizing isobutanol metabolic pathway. To generate this strain, GEVO2799 was transformed with plasmid pGV2227 (SEQ ID NO: 123), carrying the genes encoding NADPH-dependent KARI and the NADH-dependent ADH, Ec_ilvC_coSc.sup.Q110V(SEQ ID NO: 24), and Ll_adhA (SEQ ID NO: 66), respectively.
[0441]GEVO2849 is a C2-independent, PDC-minus S. cerevisiae strain carrying an NADH-utilizing isobutanol metabolic pathway. To generate this strain, GEVO2799 was transformed with plasmid pGV2242 (SEQ ID NO: 125), carrying the genes encoding NADH-dependent KARI and ADH, Ec_ilvC_coSc.sup.P2D1 (SEQ ID NO: 39) and Ll_adhA (SEQ ID NO: 66), respectively.
[0442]GEVO2851 is a C2-independent, PDC-minus S. cerevisiae strain carrying a partially NADH-utilizing isobutanol metabolic pathway. To generate this strain, GEVO2711 was transformed with plasmid pGV2227 (SEQ ID NO: 123), carrying the genes encoding NADPH-dependent KARI and the NADH-dependent ADH, Ec_ilvC_coSc.sup.Q110V (SEQ ID NO: 24), and Ll_adhA (SEQ ID NO: 66), respectively.
[0443]GEVO2852 is a C2-independent, PDC-minus S. cerevisiae strain carrying an NADH-utilizing isobutanol metabolic pathway. To generate this strain, GEVO2711 was transformed with plasmid pGV2242 (SEQ ID NO: 125), carrying the genes encoding NADH-dependent KARI and ADH, Ec_ilvC_coSc.sup.P2D1 (SEQ ID NO: 39) and Ll_adhA (SEQ ID NO: 66), respectively.
[0444]GEVO2854 is a C2-independent, PDC-minus S. cerevisiae strain carrying a partially NADH-utilizing isobutanol metabolic pathway. To generate this strain, GEVO2710 was transformed with plasmid pGV2082 (SEQ ID NO: 122), carrying the genes encoding NADPH-dependent KARI and the NADH-dependent ADH, Ec_ilvC_coSc.sup.Q110V, and Dm_ADH (SEQ ID NO: 60), respectively.
[0445]GEVO2855 is a C2-independent, PDC-minus S. cerevisiae strain carrying a partially NADH-utilizing isobutanol metabolic pathway. To generate this strain, GEVO2710 was transformed with plasmid pGV2227 (SEQ ID NO: 123), carrying the genes encoding NADPH-dependent KARI and the NADH-dependent ADH Ec_ilvC_coSc.sup.Q110V, and Ll_adhA (SEQ ID NO: 66), respectively.
[0446]GEVO2856 is a C2-independent, PDC-minus S. cerevisiae strain carrying an NADH-utilizing isobutanol metabolic pathway. To generate this strain, GEVO2710 was transformed with plasmid pGV2242 (SEQ ID NO: 125), carrying the genes encoding NADH-dependent KARI and ADH, Ec_ilvC_coSc.sup.P2D1 (SEQ ID NO: 39) and Ll_adhA (SEQ ID NO: 66), respectively.
Construction of E. coli Expression Plasmids
[0447]pGV1631: The adh2 gene was cut out of plasmid pSA55 using appropriate restriction enzymes. Re-ligation yielded plasmid pGV1631 featuring only Ll_kivd1 (SEQ ID NO: 45) under the control of the PLlacO1 promoter. The plasmid was verified by sequencing prior to its use.
[0448]pGV1705A: The Ec_yqhD gene (SEQ ID NO: 68) contained on plasmid pGV1705 was cloned into plasmid pGV1711 (SEQ ID NO: 113) using the primers XX3 and XX4. These primers added additional sequences surrounding the ADH coding sequence. Specifically, the 5'-end of the PCR product contains an EcoRI site, a BamHI site, a RBS (aggaga), a 7 nucleotide space sequence, and the initiating ATG codon. The 3' end of the product, following the stop codon, contains a NotI site followed by an AvrlI site. The amplified product was digested with EcoRI and NotI and ligated into pGV1711 (SEQ ID NO: 113) which had been cut with both EcoRI and AvrlI and gel purified to generate plasmid pGV1705-A,
[0449]ADH genes, whether PCR amplified or ordered as synthetic DNA sequences were cloned into plasmid pGV1716 (SEQ ID NO: 114), a derivative of plasmid pGV1698 carrying an in vitro-synthesized gene for S. cerevisiae ADH2, codon-optimized for expression in E. coli (="ADH2co"). ADH2co gene was amplified from plasmid pGV1527 in a PCR reaction using KOD polymerase (Novagen, Gibbstown, N.J.) and primers 1296 and 1297. These primers add additional sequences surrounding the ADH2co coding sequence. Specifically, the 5'-end of the PCR product contains a SalI site, a BamHI site, an RBS (aggaga), a 7 nucleotide space sequence, and the initiating ATG codon. The 3' end of the product, following the stop codon, contains a NotI site followed by a SalI site. The amplified product was digested SalI and was ligated into pGV1698 (SEQ ID NO: 112) which had been cut with SalI and gel purified. DNA constructs were analyzed by multiple restriction digests, and also by DNA sequencing to confirm integrity and to correct construction. Oligonucleotides 1220 and 1365 were used as primers in standard DNA sequencing reactions to sequence all of the aforementioned clones.
[0450]Plasmid pGV1748, which contains the ORF for Ec_fucO (SEQ ID NO: 64) expressed under the control of the IPTG-inducible promoter PLlacO1, was generated by amplifying the Ec_fucO gene in a PCR reaction, using primers 1470 and 1471 and E. coli genomic DNA as a template. The ˜1.2 kb PCR product so generated was digested with BamHI plus NotI, purified using a Zymo Research DNA Gel Extraction kit (Zymo Research, Orange, Calif.) according to manufacturer's protocol, and ligated into the vector pGV1716 (SEQ ID NO: 114) which had been digested with BamHI plus NotI and purified using a Zymo Research DNA Gel Extraction kit (Zymo Research, Orange, Calif.).
[0451]Plasmid pGV1748-A: The Ec_fucO gene contained on plasmid pGV1748 was cloned into plasmid pGV1711 (SEQ ID NO: 113) using the primers XX1 and XX2. These primers add additional sequences into the vector backbone upstream of the AvrlI restriction site and downstream of the EcoRI restriction site. Specifically, the 5'-end of the PCR product contains a NotI site followed by an AvrlI site and the 3' end of the product, contains an AgeI site followed by an EcoRI site. The amplified product was digested with AgeI and Non and ligated with the similarly digested pGV1711 to generate plasmid 1748-A.
[0452]Plasmid pGV1749, which contains the ORF for Dm_ADH (SEQ ID NO: 60) expressed under the control of the IPTG-inducible promoter PLlacO1, was generated by amplifying the Dm_ADH gene in a PCR reaction, using primers 1469 and 1364 and the clone RH54514 (Drosophila Genome Resource Center) as a template. The ˜0.8 kb PCR product was digested with BgIII plus NotI, was purified using a Zymo Research DNA Gel Extraction kit according to manufacturer's protocol, and was ligated into the vector pGV1716 (SEQ ID NO: 114) which had been digested with BamHI plus NotI and purified using a Zymo Research DNA Gel Extraction kit.
[0453]Plasmid pGV1749-A: The Dm_ADH gene (SEQ ID NO: 60) contained on plasmid pGV1749 was cloned into plasmid pGV1711 (SEQ ID NO: 113) using the primers XX1 and XX2. These primers add additional sequences into the vector backbone 5' of the AvrlI restriction site and 3' of the EcoRI restriction site. Specifically, the 5'-end of the PCR product contains a NotI site followed by an AvrlI site and the 3' end of the product, contains an AgeI site followed by an EcorI site. The amplified product was digested with AgeI and NotI and ligated with the product of the ADH gene similarly digested with AgeI and NotI to generate plasmid pGV1749-A.
[0454]Plasmid pGV1778, which contains the ORF for Kp_dhaT (SEQ ID NO: 62) expressed under the control of the IPTG-inducible promoter PLlacO1, was generated by excising the Kp_dhaT gene from an in vitro synthesized plasmid (generated by DNA2.0, Menlo Park, Calif.) by digestion with BamHI plus NotI. The released 1.16 kb fragment was purified using a Zymo Research DNA Gel Extraction kit according to manufacturer's protocol, and was ligated into the vector pGV1716 (SEQ ID NO: 114) which had been digested with BamHI plus NotI and purified using a Zymo Research DNA Gel Extraction kit.
[0455]Plasmid pGV1778-A: The Kp_dhaT gene (SEQ ID NO: 62) contained on plasmid pGV1778 was cloned into plasmid pGV1711 (SEQ ID NO: 113) using the primers XX1 and XX2. These primers add additional sequences into the vector backbone 5' of the AvrlI restriction site and 3' of the EcoRI restriction site. Specifically, the 5'-end of the PCR product contains a NotI site followed by an AvrlI site and the 3' end of the product, contains an AgeI site followed by an EcoRI site. The amplified product was digested with AgeI and NotI and ligated with the product of the ADH gene similarly digested with AgeI and NotI to generate plasmid pGV1778-A.
[0456]Plasmids pGV1655 (SEQ ID NO: 109) and pGV1711 (SEQ ID NO: 113) have been described previously. Briefly, pGV1655 is a low-copy, KanR-selected plasmid that expresses E. coli Ec_ilvD_coEc (SEQ ID NO: 51) and Ll_kivd1 (SEQ ID NO: 41) under the control of the PLlac promoter.
[0457]Plasmid pGV1938 was constructed by inserting the gene coding for Ec_llvC_coEc.sup.S78D into pGV1711 (SEQ ID NO: 113). The KARI variant gene was amplified with primers Not_in_for and AvrlI_in_rev introducing the 5' NotI and the 3' AvrlI restriction sites, DpnI digested for 1 h at 37° C., and then cleaned up using the Zymo PCR clean up kit. The fragment and the vector pGV1711 were restriction digested with NotI and AvrlI and run out on a 1% agarose gel. After cutting out the fragments, they were cleaned up using the Freeze'n'Squeeze and pellet paint procedure. Ligation was performed with the rapid ligation kit from Roche according to the manufacturer's instructions.
[0458]Plasmid pGV1939 was generated using primers XX3 and XX4 to amplify the Ec_fucO gene from plasmid pGV1748-A. The forward primer adds a new RBS (aggaga), a 7 nucleotide space sequence, and the initiating ATG codon. The amplified product was digested with EcoRI and NotI and ligated with the similarly digested pGV1711 (SEQ. ID NO: 113) to generate plasmid pGV1939 containing the modified RBS.
[0459]The genes coding for KARI variants Ec_ilvC_coEc.sup.his6 (SEQ ID NO: 14), Ec_ilvC_coEc.sup.S78D-his6 (SEQ ID NO: 16), Ec_ilvC_coEc6E6-his6 (SEQ ID NO: 32) and Ec_ilvC_coEc2H10-his6 (SEQ ID NO: 30) were cloned into pGV1939 generating plasmids pGV1925, pGV1927, pGV1975 and pGV1976, respectively using primers NotI_in_for and AvrlI_in_rev. The PCR products were DpnI digested for 1 h and cleaned over a 1% agarose gel. After a sequential restriction digestion of vector and insert with NotI for 1 h followed by 1 h with AvrlI, ligation was performed using rapid ligase (Roche). Ligation mixture was desalted using the Zymo PCR clean up kit and used to transform E. coli DH5α. DNA constructs were analyzed by restriction digests, and also by DNA sequencing to confirm integrity and correct construction. Primers pETup and KARIpETrev were used as primers in standard DNA sequencing reactions to sequence pET22b(+) derivatives, primer seq_ilvc_pGV was used to sequence pGV1925, pGV1927, pGV1975 and pGV1976.
Construction of Saccharomyces cerevisiae Expression Plasmids
[0460]pGV1824: The gene coding for Ec_llvC (SEQ ID NO: 13) was codon optimized for S. cerevisiae and synthesized (DNA2.0, Menlo Park, Calif.), resulting in Ec_ilvC_coSc (SEQ ID NO: 12). To generate pGV1824, the Ec_ilvC_coSc gene was excised from plasmid pGV1774 using BglII and XhoI. Plasmid pGV1662 was digested with SalI and BamHI. The pGV1662 vector backbone and Ec_ilvC_coSc insert were ligated using standard methods resulting in plasmid pGV1824 containing the gene Ec_ilvC_coSc.
[0461]pGV1914 (SEQ ID NO: 119) is a yeast integrating vector (YIp) that utilizes the S. cerevisiae URA3 gene as a selection marker and contains homologous sequence for targeting the HpaI-digested, linearized plasmid for integration at the PDC6 locus of S. cerevisiae. This plasmid does not carry a yeast replication origin, thus is unable to replicate episomally. This plasmid carries the Dm_ADH (SEQ ID NO: 60) and Ll_kivd2_coEc (SEQ ID NO: 48) genes, expressed under the control of the S. cerevisiae TDH3 and TEF1 promoters, respectively. pGV1914 was generated in two steps. First, the Dm_ADH-containing E. coli expression plasmid pGV1749 was digested with SalI plus NotI, and the 0.78 kb fragment containing the Dm_ADH ORF released by digestion was gel purified and ligated into pGV1635, which had been digested with XhoI plus NotI and gel purified. Plasmid pGV1635 is a yeast expression plasmid which has as its salient feature a TDH3 promoter followed by several restriction enzyme recognition sites, into which the Dm_ADH sequence was cloned as described above. A correct recombinant plasmid was named pGV1913. In the second step of pGV1914 construction, pGV1913 was digested with BamHI plus NotI and the 1.45 kb fragment, containing the TDH3 promoter-Dm_ADH ORF sequence was gel purified and ligated into pGV1733, which had been digested with BamHI plus NotI and similarly gel purified, yielding pGV1914. Thus, the ScADH7 ORF in pGV1733 is replaced by the Dm_ADH ORF in the pGV1914, both under the control of the TDH3 promoter; both plasmids also contain the PTEF1-Ll_kivd2_coEc cassette as well as the URA3 selection marker and ScPDC6 5' and 3' regions suitable for homologous recombination targeting following linearization of the plasmid with HpaI.
[0462]pGV1936 (SEQ ID NO: 120) is a yeast integrating vector (YIp) that utilizes the S. cerevisiae LEU2 gene as a selection marker and contains homologous sequence for targeting the linearized (by HpaI digestion) plasmid for integration at the PDC5 locus of S. cerevisiae. This plasmid does not carry a yeast replication origin, thus is unable to replicate episomally. This plasmid carries the Ec_ilvC_coSc.sup.Q110V (SEQ ID NO: 24) mutant (i.e. codon optimized for expression in S. cerevisiae) and S. cerevisiae ILV3ΔN genes, expressed under the control of the S. cerevisiae TDH3 and TEF1 promoters, respectively. pGV1936 was constructed using SOE PCR method that amplified the Ec_ilvC_coSc gene while simultaneously introducing the nucleotide changes coding for a Q110V mutation. Specifically, primers 1624 and 1814 were used to amplify a portion of plasmid pGV1774 containing the Ec_ilvC_coSc gene; primers 1813 and 1798 were used to amplify a portion of plasmid pGV1824 that also contained the Ec_ilvC coSc gene. The two separate PCR products were gel purified, eluted in 15 μL, and 3 μL of each were used as a template along with primers 1624 and 1798. The resulting PCR product was digested with XhoI plus NotI and ligated into pGV1765 that had been digested with XhoI plus NotI, yielding pGV1936. Candidate clones of pGV1936 were confirmed by sequencing, using primers 350, 1595, and 1597.
[0463]pGV1994: Mutations found in variant Ec_llvC6E6-his6 were introduced into pGV1824 by SOE PCR. The 5' PCR used primers 1898 and 2037 and the 3' PCR used primers 1893 and 2036. Each of these primer pairs were used with pGV1894 as the template in two separate PCR reactions. The product was used in a second PCR with the end primers 1898 and 1893 to yield a final PCR product. This final PCR product has a 5' SalI restriction site and 3' BglII followed by NotI restriction sites. These were cloned into pGV1662 using the SalI and NotI site and yielding plasmid pGV1994 which carries Ec_ilvC_coSc6E6 (SEQ ID NO: 35).
[0464]pGV2020 (SEQ ID NO: 121) is an empty G418 resistant 2-micron yeast vector that was generated by removing the Ll_kivd2_coEc sequence from pGV2017. This was carried out by amplifying the TDH3 promoter from pGV2017 using primers 1926 and 1927, digesting with SalI and NotI and cloning into the same sites of pGV2017.
[0465]pGV2082 (SEQ ID NO: 122) is a G418 resistant yeast 2-micron plasmid for the expressions of Ec_ilvC_coSc.sup.Q110V (SEQ ID NO: 24), Ll_ilvD_coSc (SEQ ID NO: 54), Ll_kivd2_coEc (SEQ ID NO: 48), and Dm_ADH (SEQ ID NO: 60). A fragment carrying the PGK1 promoter, Ll_kivd2_coEc and a short region of the PDC1 terminator sequence was obtained by cutting pGV2047 with AvrlI and NcoI. This fragment was treated with Klenow to generate blunt ends then cloned into pGV2044 that had been digested with EcoRI and SbfI and the overhangs filled in with Klenow. This construction replaced the CUP1 promoter and the Bs_alsS1_coSc (SEQ ID NO: 6) in pGV2044 with the PGK1 promoter and Ll_kivd2 coEc.
[0466]pGV2193: The Ec_llvC variant encoded by Ec_ilvC_coSc6E6-his6 (SEQ ID NO: 33) encoded on pGV2241 (SEQ ID NO: 124) served as template for error-prone PCR using primers pGV1994ep_for and pGV1994ep_rev yielding variant Ec_llvC.sup.P2D1-his6 (SEQ ID NO: 38) which is encoded by Ec_ilvC_coSc.sup.P2D1-his6(SEQ ID NO: 37) on construct pGV2193.
[0467]pGV2227 (SEQ ID NO: 123) is a G418 resistant yeast 2-micron plasmid for the expressions of Ec_ilvC_coSc.sup.Q110V (SEQ ID NO: 24), Ll_ilvD_coSc (SEQ ID NO: 54), Ll_kivd2 coEc (SEQ ID NO: 48), and Ll_adhA (SEQ ID NO: 66). pGV2227 is a derivative of pGV2201 where the BamHI and XhoI sites at the 3' end of the Ll_adhA were removed and replaced with an AvrlI site. This construction was carried out by cloning into the NheI-MluI sites of pGV2202 a fragment carrying the 3' end of the Ll_adhA sequence, an AvrlI site, and the 5' part of the CYC1 terminator. This fragment was generated by SOE PCR combining a PCR product using primers 2091 and 2352 with pGV2201 as template and a PCR product using primers 2353 and 772 with pGV2201 as template. The sequences of primers 2352 and 2353 overlap and introduce an AvrlI site. This SOE PCR product was digested with NheI and MluI for cloning into pGV2201.
[0468]pGV2238: The Ec_llvC variant encoded by Ec_ilvC_coSc.sup.P201-his6 (SEQ ID NO: 37) encoded on pGV2193 served as parent for an additional error-prone PCR round using the same primers as described before on template DNA pGV2193 yielding an improved KARI variant named Ec_llvC.sup.P2D1-A1-his6 (SEQ ID NO: 42) which is encoded by the gene Ec_ilvC_coSc2D1-A1-his6 (SEQ ID NO: 41) on plasmid pGV2238.
[0469]pGV2241 (SEQ ID NO: 124): The gene Ec_ilvC_coSc6E6 (SEQ ID NO: 35) was his-tagged using primers pGV1994_ep_for and 1994hisrev, cleaned with the Zymo PCR clean up kit (Zymo Research), NotI and SalI digested, and ligated into similarly digested pGV1994, resulting in construct pGV2241 coding for Ec_ilvC_coSc6E6-his6 (SEQ ID NO: 33).
[0470]pGV2242 (SEQ ID NO: 125) is a G418 resistant yeast 2-micron plasmid for the expressions of Ec_ilvC_coSc.sup.P2D1 (SEQ ID NO: 39), Ll_ilvD_coSc (SEQ ID NO: 54), Ll_kivd2_coEc (SEQ ID NO: 48), and Ll_adhA (SEQ ID NO: 66). This plasmid was generated by cloning the SalI-BspEI fragment of pGV2193 carrying the region encoding for Ec_llvC with the relevant mutations for the Ec_ilvC_coSc.sup.P2D1 allele into the XhoI-BspEI sites of pGV2227 (SEQ ID NO: 123).
TABLE-US-00003 TABLE 3 Strains disclosed herein Strain No. Description GEVO1186 S. cerevisiae CEN.PK2 (MATa/α ura3/ura3 leu2/leu2 his3/his3 trp1/trp1 PDC1/PDC1 PDC5/PDC5 PDC6/PDC6) GEVO1385 E. coli BW25113, ΔldhA-fnr::FRT, ΔadhE::FRT, Δfrd::FRT, Δpta::FRT, ΔpflB::FRT, F' (laclq+), attB::(Sp.sup.+ laclq.sup.+ tetR.sup.+) GEVO1399 E. coli BW25113, ΔldhA-fnr::FRT, ΔadhE::FRT, Δfrd::FRT, Δpta::FRT, pflB::FRT, Δzwf::FRT F' (laclq+) GEVO1608 E. coli BW25113, ΔldhA-fnr::FRT, ΔadhE::FRT, Δfrd::FRT, ΔpflB::FRT, Δpta::FRT, ΔyqhD::FRT-Kan-FRT, F' (laclq+) GEVO1725 E. coli BW25113, ΔldhA-fnr::FRT, ΔadhE::FRT, Δfrd::FRT, Δpta::FRT, ΔpflB::FRT, ΔmaeA::FRT, ΔpykA::FRT, ΔpykF::FRT, F' (laclq+) GEVO1745 E. coli BW25113, ΔldhA-fnr::FRT, ΔadhE::FRT, Δfrd::FRT, ΔpflB::FRT, Δpta::FRT, ΔyqhD::FRT GEVO1748 E. coli BW25113, ΔldhA-fnr::FRT, ΔadhE::FRT, Δfrd::FRT, Δpta::FRT, pflB::FRT, F' (laclq+), ΔilvC::PLlacO1::Ll_kivd1::Ec_ilvD_coEc::FRT GEVO1749 E. coli BW25113, ΔldhA-fnr::FRT, Δfrd::FRT, Δpta::FRT, pflB::FRT, F' (laclq+), ΔadhE::[PLlacO1::Ll_kivd1::Ec_ilvD_coEc::FRT] GEVO1750 E. coli BW25113, ΔldhA-fnr::FRT, ΔadhE::FRT, Δfrd::FRT, Δpta::FRT, ΔpflB::FRT, ΔmaeA::FRT, F' (laclq+), attB::(Sp+ laclq+ tetR+) GEVO1751 E. coli BW25113, ΔldhA-fnr::FRT, ΔadhE::FRT, Δfrd::FRT, Δpta::FRT, ΔpflB::FRT, ΔmaeA::FRT, ΔpykA::FRT, ΔpykF::FRT, F' (laclq+), attB::(Sp+ laclq+ tetR+) GEVO1777 E. coli W3110, ΔilvC::FRT, attB::(Sp+ laclq+ tetR+) GEVO1780 JCL260 transformed with pGV1655 and pGV1698 GEVO1803 S. cerevisiae CEN.PK2, MATa/alpha ura3/ura3 leu2/leu2 his3/his3 trp1/trp1 pdc1::Bs_alsS2, TRP1/PDC1 GEVO1844 E. coli BW25113, Δ(ldhA-fnr::FRT) ΔadhE::FRT Δfrd::FRT Δpta::FRT ΔpflB::FRT ΔilvC::P.sub.LlacO1::Ll_kivd1::Ec_ilvD_coEc::FRT ΔsthA::FRT GEVO1846 E. coli BW25113, ΔldhA-fnr::FRT, ΔadhE::FRT, Δfrd::FRT, Δpta::FRT, pflB::FRT, F' (laclq+), ΔilvC::PLlacO1::Ll_kivd1::Ec_ilvD_coEc::FRT, pGV1745, pGV1698 GEVO1859 E. coli BW25113, ΔldhA-fnr::FRT, Δfrd::FRT, Δpta::FRT, F' (laclq+), ΔadhE::[pLlacO1::Ll_kivd1::Ec_ilvD_coEc::FRT], pflB::[pLlacO1::Bs_alsS1::Ec_ilvC_coEc::FRT] GEVO1886 E. coli BW25113, ΔldhA-fnr::FRT, Δfrd::FRT, Δpta::FRT, F' (laclq+), ΔadhE::[pLlacO1::Ll_kivd1::Ec_ilvD_coEc::FRT], ΔpflB::[pLlacO1::Bs_alsS1:: Ec_ilvC_coEc::FRT] ΔsthA::[pLlacO1::pntA::pntB::FRT] GEVO1993 E. coli BW25113, ΔldhA-fnr::FRT, ΔadhE::FRT, Δfrd::FRT, DpflB::FRT, F' (laclq+), ΔilvC::PLlacO1::Ll_kivd1::Ec_ilvD_coEc:.FRT, Δpta::PLlacO1::Bs_alsS1, FRT::KAN::FRT GEVO2107 S. cerevisiae CEN.PK2, MATa/alpha ura3/ura3 leu2/leu2 his3/his3 trp1/trp1 pdc1::Bs_alsS2, TRP1/PDC1 pdc6::{ScTEF1p-Ll_kivd2_coEc ScTDH3p-Dm_ADH URA3}/PDC6 GEVO2158 S. cerevisiae CEN. PK2; MATa/α ura3/ura3 leu2/leu2 his3/his3 trp1/trp1 pdc1::Bs_alsS2, TRP1/PDC1 pdc5:{ScTEF1prom- Sc_ILV3ΔN ScTDH3prom-Ec_ilvC_coSc.sup.Q110V LEU2}/PDC5 pdc6::{ScTEF1p-Ll_kivd2_coEc ScTDH3p-Dm_ADH URA3}/PDC6 GEVO2302 S. cerevisiae CEN.PK2; MATa ura3 leu2 his3 trp1 pdc1::Bs_alsS2, TRP1 pdc5::{PTEF1:Sc_ILV3ΔN PTDH3:Ec_ilvC_coSc.sup.Q110V LEU2} pdc6::{PTEF1: Ll_kivd2_coEc PTDH3:Dm_ADH URA3} GEVO2710 S. cerevisiae CEN.PK2; MATa ura3 leu2 his3 trp1 pdc1::{PCUP1- Bs_alsS2, TRP1} pdc5::{PTEF1:Sc_ILV3ΔN PTDH3:Ec_ilvC_coSc.sup.Q110V, LEU2} pdc6::{PTEF1: Ll_kivd2_coEc PTDH3:Dm_ADH, URA3}, evolved for C2 supplement-independence, glucose tolerance and faster growth GEVO2711 S. cerevisiae CEN.PK2; MATa ura3 leu2 his3 trp1 pdc1::{PCUP1- Bs_alsS2, TRP1} pdc5::{PTEF1:Sc_ILV3ΔN PTDH3:Ec_ilvC_coSc.sup.Q110V, LEU2} pdc6::{PTEF1: Ll_kivd2_coEc PTDH3:Dm_ADH, URA3}, evolved for C2 supplement-independence, glucose tolerance and faster growth GEVO2712 S. cerevisiae CEN.PK2; MATa ura3 leu2 his3 trp1 pdc1::{PCUP1- Bs_alsS2, TRP1} pdc5::{PTEF1:Sc_ILV3ΔN PTDH3:Ec_ilvC_coSc.sup.Q110V, LEU2} pdc6::{PTEF1: Ll_kivd2_coEc PTDH3:Dm_ADH, URA3}, evolved for C2 supplement-independence, glucose tolerance and faster growth GEVO2799 S. cerevisiae CEN.PK2; MATa ura3 leu2 his3 trp1 pdc1::{PCUP1- Bs_alsS2, TRP1} pdc5::{PTEF1:Sc_ILV3ΔN PTDH3:Ec_ilvC_coSc.sup.Q110V, LEU2} pdc6::{PTEF1: Ll_kivd2_coEc PTDH3:Dm_ADH, URA3}, evolved for C2 supplement-independence, glucose tolerance and faster growth GEVO2792 GEVO2710 transformed with pGV2020 GEVO2844 GEVO2799 transformed with pGV2020 GEVO2847 GEVO2799 transformed with pGV2082 GEVO2848 GEVO2799 transformed with pGV2227 GEVO2849 GEVO2799 transformed with pGV2242 GEVO2851 GEVO2711 transformed with pGV2227 GEVO2052 GEVO2711 transformed with pGV2242 GEVO2854 GEVO2710 transformed with pGV2082 GEVO2855 GEVO2710 transformed with pGV2227 GEVO2856 GEVO2710 transformed with pGV2242 GEVO5001 S. cerevisiae CEN.PK2, Δpdc1 Δpdc5 Δpdc6 expressing an isobutanol pathway (ALS, KARI, DHAD, KIVD, ADH) GEVO5002 GEVO5001 PTEF1:NADH kinase PTDH3:NADP.sup.+ phosphatase HPH GEVO5003 GEVO5001, PTDH3:Kl_GDP1 HPH GEVO5004 GEVO5001 PTEF1:ess:pntA PTDH3:ess:pntB HPH GEVO5005 GEVO5001 PTEF1:mts:pntA PTDH3:mts:pntB HPH GEVO5006 GEVO5001 PADH1:PYC1 PTEF1:MDH2 PTDH3:maeB HPH E. coli BL21 Lucigen Corporation (Middleton, WI) (DE3) E. coli Lutz, R. and Bujard, H, Nucleic Acids Research (1997) 25 1203-1210 DH5αZ1 JCL260* E. coli BW25113, ΔldhA-fnr::FRT, ΔadhE::FRT, Δfrd::FRT, ΔpflB::FRT, Δpta::FRT, F' (lacIq+) *These strains are described in PCT/US2008/053514
TABLE-US-00004 TABLE 4 Plasmids disclosed herein SEQ ID GEVO No. FIG. NO Genotype or Reference pKD13 n/a Datsenko, K and Wanner, B. PNAS 2000, 97: 6640-5 pKD46 n/a Datsenko, K and Wanner, B. PNAS 2000, 97: 6640-5 pSA55* n/a pLlacO1::Ll_kivd1::ADH2, ColE1, Amp pSA69* n/a pLlacO1::Bs_alsS1::Ec_ilvC::Ec_ilvD, p15A, Kan pET22b(+) n/a Novagen, Gibbstown, NJ pET22b[ilvCco] n/a Novagen, Gibbstown, NJ pGV1102 101 PTEF1-HA-tag-MCS-TCYC1, URA3,2-micron, bla, pUC-ori pGV1323 102 pGV1485 103 PLlacO1::Ll_kivd1::ADH2, pSC101, Km pGV1490 104 pLtetO1:: p15A, Cm pGV1527 PLtetO1::Ll_kivd1_coEc::S. cerevisiae ADH2 ColE1, bla pGV1572 105 PLlacO1::empty, p15A, CmR pGV1573 106 PLlacO1::GDP1, p15A, CmR pGV1575 107 PLlacO1::gapC, p15A, CmR pGV1609 108 PLlacO1::Bs_alsS1::ilvC::Ec_ilvD, p15A, Cm pGV1631 PLlacO1::Ll_kivd1, ColE1, Amp pGV1655 109 PLlacO1::Ll_kivd1::Ec_ilvD_coEc,, pSC101, Km pGV1661 110 pLtetO1::maeB::ppc::mdh, p15A, Cm pGV1662 pGV1685 111 PLtetO1::pntAB, p15A, Cm pGV1698 112 PLlacO1::Bs_alsS1::ilvC, bla, ColE1 ORI pGV1705-A PLlacO1::Ec_yqhD bla, ColE1 ORI pGV1711 113 PLlacO1::(no ORF) bla, ColE1 ORI pGV1716 114 PLlacO1::Bs_alsS1::Saccharomyces cerevisiae ADH2::ilvC bla, ColE1 ORI pGV1720 115 pLlacO1::empty, pSC101, Km pGV1730 116 PCUP1-Bs_alsS2-PDC1 3' region-PDC1 5' region, TRP1, bla, pUC ori pGV1745 117 pLlacO1::pntAB, pSC101, Km pGV1748 PLlacO1::Bs_alsS1::Ec_fucO::Ec_ilvC_coEc bla, ColE1 ORI pGV1748-A PLlacO1::Ec_fucO:: bla, ColE1 ORI pGV1749 PLlacO1:: Bs_alsS1::Dm_ADH: Ec_ilvC_coEc bla, ColE1 ORI pGV1749-A PLlacO1::Dm_ADH:: bla, ColE1 ORI pGV1772 pLtetO1::maeB::pck::mdh, p15A, Cm pGV1777 118 PLlacO1::Ec_ilvC_coEc, bla, ColE1 ORI pGV1778 PLlacO1:: Bs_alsS1::Kp_dhaT::Ec_ilvC_coEc bla, ColE1 ORI pGV1778-A PLlacO1::Kp_dhaT::bla, ColE1 ORI pGV1824 PTEF1::Ec_ilvC_coSc:TCYC1, pUC ORI, URA3, 2μ ORI, bla pGV1914 119 PTEF1:Ll_kivd2: PTDH3:Dm_ADH PDC6 5',3' targeting homology URA3 pUC ori bla(ampR) pGV1925 pLlacO1::Ec_fucO ::Ec_ilvC_coEc::bla, ColE1 ORI pGV1927 pLlacO1::Ec_fucO::Ec_ilvC_coEc.sup.S78D bla, ColE1 ORI pGV1936 120 PTEF1:Sc_ILV3ΔN PTDH3:Ec_ilvC_coSc.sup.Q110V PDC5 5',3' targeting homology LEU2 pGV1938 pLlac01::ilvC_coS78D bla, ColE1 ORI pGV1939 pLlacO1::E. coli fucO bla, ColE1 ORI pGV1975 pLlacO1::Ec_fucO::Ec_ilvC_coEc6E6 bla, ColE1 ORI pGV1976 pLlacO1::Ec_fucO::Ec_ilvC_coEC2H10 bla, ColE1 ORI pGV1994 PTEF1::Ec_ilvC_coSc6E6:TCYC1, bla, pUC ORI, URA3, 2μ ORI pGV2020 121 PSc--TEF1, PSc--TPI1, PSc--TPI1G418R, APr, 2μ --Vector Control pGV2082 122 PTEF1-Ll_ilvD_coSc-PTDH3-Ec_ilvC_coSc.sup.Q110V-PTPI1- G418R-P.sub.PGK1-Ll_kivd2_coEc-PDC1-3'region-PENO2- Dm_ADH 2μ bla, pUC-ori pGV2193 PTEF1::Ec_ilvC_coSc.sup.P2D1-his6:TCYC1, bla, pUC ORI, URA3, 2μ ORI pGV2227 123 PTEF1-Ll_ilvD_coSc-PTDH3-Ec_ilvC_coSc.sup.Q110V-PTPI1- G418R-P.sub.PGK1-Ll_kivd2_coEc-PDC1-3'region-PENO2- Ll_adhA 2μ bla, pUC-ori pGV2238 PTEF1::Ec_ilvC_coSc.sup.P2D1-A1-his6:TCYC1, bla, pUC ORI, URA3, 2μ ORI. pGV2241 124 PTEF1::Ec_ilvC_coSc6E6-his6:TCYC1, bla, pUC ORI, URA3, 2μ ORI. pGV2242 125 PTEF1-Ll_ilvD_coSc-PTDH3-Ec_ilvC_coSc.sup.P2D1-PTPI1- G418R-P.sub.PGK1-Ll_kivd2_coEc-PDC1-3'region-PENO2- Ll_adhA 2μ bla, pUC-ori pGV6000 PTEF1:NADH kinase PTDH3:NADP.sup.+ phosphatase HPH pGV6001 PTDH3:Kl_GDP1 HPH pGV6002 PTEF1:ess:pntA PTDH3:ess:pntB HPH pGV6003 PTEF1:mts:pntA PTDH3:mts:pntB HPH pGV6004 PADH1:PYC1 PTEF1:MDH2 PTDH3:maeB HPH *These plasmids are described in PCT/US2008/053514
TABLE-US-00005 TABLE 5 Amino acid and nucleotide sequences of enzymes and genes disclosed herein Enz. Source Gene (SEQ ID NO) Corresponding Protein (SEQ ID NO) pntA E. coli E. coli pntA (SEQ ID NO: 1) E. coli PntA (SEQ ID NO: 2) pntB E. coli E. coli pntB (SEQ ID NO: 3) E. coli PntB (SEQ ID NO: 4) ALS B. subtilis Bs_alsS1 (SEQ ID NO: 5) Bs_AlsS1 (SEQ ID NO: 7) Bs_alsS1_coSc (SEQ ID NO: 6) Bs_alsS2 (SEQ ID NO: 8) Bs_AlsS2 (SEQ ID NO: 9) KARI E. coli Ec_ilvC (SEQ ID NO: 10) Ec_llvC (SEQ ID NO: 13) Ec_ilvC-coEc (SEQ ID NO: 11) Ec_ilvC_coSc (SEQ ID NO: 12) Ec_ilvC_coEc.sup.his6 (SEQ ID NO: 14) Ec_llvC.sup.his6 (SEQ ID NO: 15) Ec_ilvC_coEc.sup.S78D-his6 (SEQ ID NO: 16) Ec_llvC.sup.S78D-his6 (SEQ ID NO: 17) Ec_ilvC_coEc.sup.S78D (SEQ ID NO: 18) Ec_llvC.sup.S78D (SEQ ID NO: 19) Ec_ilvC-coEc.sup.Q110A-his6 (SEQ ID NO: 20) Ec_llvC.sup.Q110A-his6 (SEQ ID NO: 21) Ec_ilvC-coEc.sup.Q110V-his6 (SEQ ID NO: 22) Ec_llvC.sup.Q110V-his6 (SEQ ID NO: 23) Ec_ilvC-coSc.sup.Q110V (SEQ ID NO: 24) Ec_llvC.sup.Q110V (SEQ ID NO: 25) Ec_ilvC_coEc.sup.B8-his6 (SEQ ID NO: 26) Ec_llvC.sup.B8-his6 (SEQ ID NO: 27) Ec_ilvC-coEc.sup.B8A71S-his6 (SEQ ID NO: 28) Ec_llvC.sup.B8A71S-his6 (SEQ ID NO: 29) Ec_ilvC_coEc2H10-his6 (SEQ ID NO: 30) Ec-llvC2H10-his6 (SEQ ID NO: 31) Ec_ilvC_coEc6E6-his6 (SEQ ID NO: 32) Ec_llvC6E6-his6 (SEQ ID NO: 34) Ec_ilvC-coSc6e8-his6 (SEQ ID NO: 33) Ec_ilvC-coSc6E6 (SEQ ID NO: 35) Ec_llvC6E6 (SEQ ID NO: 36) Ec_ilvC_coSc.sup.P2D1-his6 (SEQ ID NO: 37) Ec_llvC.sup.P2D1-his6 (SEQ ID NO: 38) Ec_ilvC_coSc.sup.P2D1 (SEQ ID NO: 39) Ec_llvC.sup.P2D1 (SEQ ID NO: 40) Ec_ilvC_coSc.sup.P2D1-A1-his6 (SEQ ID NO: 41) Ec_llvC.sup.P2D1-A1-his6 (SEQ ID NO: 42) Ec_ilvC_coSc.sup.P2D1-A1 (SEQ ID NO: 43) Ec_llvC.sup.P2D1-A1 (SEQ ID NO: 44) KIVD L. lactis Ll_kivd1 (SEQ ID NO: 45) Ll_Kivd1 (SEQ ID NO: 47) Ll_kivd1_coEc (SEQ ID NO: 46) Ll_kivd2_coEc (SEQ ID NO: 48) Ll_Kivd2 (SEQ ID NO: 49) DHAD E. coli Ec_ilvD (SEQ ID NO: 50) Ec_llvD (SEQ ID NO: 52) Ec_ilvD_coEc (SEQ ID NO: 51) L. lactis Ll_ilvD_coSc (SEQ ID NO: 54) Ll_llvD (SEQ ID NO: 55) S. cerevisiae Sc_lLV3 (SEQ ID NO: 56) Sc_llv3 (SEQ ID NO: 57) Sc_lLV3ΔN (SEQ ID NO: 58) Sc_llv3ΔN (SEQ ID NO: 59) ADH D. melanogaster Dm_ADH (SEQ ID NO: 60) Dm_Adh (SEQ ID NO: 61) K. pneumoniae Kp_dhaT (SEQ ID NO: 62) Kp_DhaT (SEQ ID NO: 63) E. coli Ec_fucO (SEQ ID NO: 64) Ec_FucO (SEQ ID NO: 65) L. lactis Ll_adhA (SEQ ID NO: 66) Ll_AdhA (SEQ ID NO: 67) E. coli Ec_yqhD (SEQ ID NO: 68) Ec_YqhD (SEQ ID NO: 69)
TABLE-US-00006 TABLE 6 Primers sequences disclosed herein No. (SEQ ID NO) Sequence (listed as 5' to 3') XX1 CGCACCGGTTTTCTCCTCTTTAATGAATTCGGTC (SEQ ID NO: 201) AGTGCGTCCTGC XX2 GCGGCCGCCCTAGGGCGTTCGGCTGCGGCGAGCG (SEQ ID NO: 202) GT XX3 CGCGAATTCGGATCCGAGGAGAAAATAGTTATGA (SEQ ID NO: 203) ACAACTTTAATCTGCACACCCC XX4 GCGCCTAGGGCGGCCGCTTAGCGGGCGGCTTCGT (SEQ ID NO: 204) ATATACGG 50 GCAGTTTCACCTTCTACATAATCACGACCGTAGT (SEQ ID NO: 205) AGGTATCATTCCGGGGATCCGTCGACC 73 CTGGCTTAAGTACCGGGTTAGTTAACTTAAGGAG (SEQ ID NO: 206) AATGACGTGTAGGCTGGAGCTGCTTC 74 CTCAAACTCATTCCAGGAACGACCATCACGGGTA (SEQ ID NO: 207) ATCATCATTCCGGGGATCCGTCGACC 116 CAGCGTTCGCTTTATATCCCTTACGCTGGCCCTG (SEQ ID NO: 208) TACTGCTGGAAGTGTAGGCTGGAGCTGCTTC 117 TTCGGCTTGCCAGAAATTATCGTCAATGGCCTGT (SEQ ID NO: 209) TGCAGGGCTTCATTCCGGGGATCCGTCGACC 350 CTTAAATTCTACTTTTATAGTTAGTC (SEQ ID NO: 210) 474 CAAAGCTGCGGATGATGACGAGATTACTGCTGCT (SEQ ID NO: 211) GTGCAGACTGAATTCCGGGGATCCGTCGACC 772 AGGAAGGAGCACAGACTTAG (SEQ ID NO: 212) 868 CACAACATCACGAGGAATCACCATGGCTAACTAC (SEQ ID NO: 213) TTCAATACACGTGTAGGCTGGAGCTGCTTC 869 CTTAACCCGCAACAGCAATACGTTTCATATCTGT (SEQ ID NO: 214) CATATAGCCGCATTCCGGGGATCCGTCGACC 1030 GTCGGTGAACGCTCTCCTGAGTAGGGTGTAGGCT (SEQ ID NO: 215) GGAGCTGCTTC 1031 GAAGCAGCTCCAGCCTACACCCTACTCAGGAGAG (SEQ ID NO: 216) CGTTCACCGAC 1032 CACAACATCACGAGGAATCACCATGGCTAACTAC (SEQ ID NO: 217) TTCAATACACCACGAGGCCCTTTCGTCTTCACCT C 1155 CCCAACCCGCATTCTGTTTGGTAAAGGCGCAATC (SEQ ID NO: 218) GCTGGTTTACGGTGTAGGCTGGAGCTGCTTC 1156 CAATCGCGGCGTCAATACGCTCATCATCGGAACC (SEQ ID NO: 219) TTCAGTGATGTATTCCGGGGATCCGTCGACC 1187 CGGATAAAGTTCGTGAGATTGCCGCAAAACTGGG (SEQ ID NO: 220) GCGTCATGTGGGTGTAGGCTGGAGCTGCTTC 1188 CAGACATCAAGTAACCTTTATCGCGCAGCAGATT (SEQ ID NO: 221) AACCGCTTCGCATTCCGGGGATCCGTCGACC 1191 GGCACTCACGTTGGGCTGAGACACAAGCACACAT (SEQ ID NO: 222) TCCTCTGCACGGTGTAGGCTGGAGCTGCTTC 1192 GCACCAGAAACCATAACTACAACGTCACCTTTGT (SEQ ID NO: 223) GTGCCAGACCGATTCCGGGGATCCGTCGACC 1205 GTTATCTAGTTGTGCAAAACATGCTAATGTAGCC (SEQ ID NO: 224) ACCAAATCCACGAGGCCCTTTCGTCTTCACCTC 1218 GCTCACTCAAAGGCGGTAATACGTGTAGGCTGGA (SEQ ID NO: 225) GCTGCTTC 1219 GAAGCAGCTCCAGCCTACACGTATTACCGCCTTT (SEQ ID NO: 226) GAGTGAGC 1220 CGTAGAATCACCAGACCAGC (SEQ ID NO: 227) 1296 TTTTGTCGACGGATCCAGGAGACAACATTATGTC (SEQ ID NO: 228) TATTCCAGAAACTCAAAAAGCG 1297 TTTTGTCGACGCGGCCGCTTATTTAGAGGTGTCC (SEQ ID NO: 229) ACCACGTAACGG 1321 AATCATATCGAACACGATGC (SEQ ID NO: 230) 1322 TCAGAAAGGATCTTCTGCTC (SEQ ID NO: 231) 1323 ATCGATATCGTGAAATACGC (SEQ ID NO: 232) 1324 AGCTGGTCTGGTGATTCTAC (SEQ ID NO: 233) 1341 TGCTGAAAGAGAAATTGTCC (SEQ ID NO: 234) 1342 TTTCTTGTTCGAAGTCCAAG (SEQ ID NO: 235) 1364 TTTTGCGGCCGCTTAGATGCCGGAGTCCCAGTGC (SEQ ID NO: 236) TTG 1365 AGTTGTTGACGCAGGTTCAGAG (SEQ ID NO: 237) 1436 AAATGACGACGAGCCTGAAG (SEQ ID NO: 238) 1437 GACCTGACCATTTGATGGAG (SEQ ID NO: 239) 1439 CAATTGGCGAAGCAGAACAAG (SEQ ID NO: 240) 1469 TTTTAGATCTAGGAGATACCGGTATGTCGTTTAC (SEQ ID NO: 241) TTTGACCAACAAG 1440 ATCGTACATCTTCCAAGCATC (SEQ ID NO: 242) 1441 AATCGGAACCCTAAAGGGAG (SEQ ID NO: 243) 1442 AATGGGCAAGCTGTTTGCTG (SEQ ID NO: 244) 1443 TGCAGATGCAGATGTGAGAC (SEQ ID NO: 245) 1470 TTTTGGATCCAGGAAATAGATCTATGATGGCTAA (SEQ ID NO: 246) CAGAATGATTCTGAACG 1471 TTTTGCGGCCGCTTACCAGGCGGTATGGTAAAGC (SEQ ID NO: 247) TC 1479 CCGATAGGCTTCCGCCATCGTCGGGTAGTTAAAG (SEQ ID NO: 248) GTGGTGTTGAGTGTAGGCTGGAGCTGCTTC 1485 GCCTTTATTGTACGCTTTTTACTGTACGATTTCA (SEQ ID NO: 249) GTCAAATCTAACACGAGGCCCTTTCGTCTTCACC TC 1486 AAGTACGCAGTAAATAAAAAATCCACTTAAGAAG (SEQ ID NO: 250) GTAGGTGTTACATTCCGGGGATCCGTCGACC 1526 TCGACGAGGAGACAACATTGTGTAGGCTGGAGCT (SEQ ID NO: 251) GCTTC 1527 GAAGCAGCTCCAGCCTACACAATGTTGTCTCCTC (SEQ ID NO: 252) GTCGA 1539 CCATTCTGTTGCTTTTATGTATAAGAACAGGTAA (SEQ ID NO: 253) GCCCTACCATGGAGAATTGTGAGCGGATAAC 1561 GCAATCCTGAAAGCTCTGTAACATTCCGGGGATC (SEQ ID NO: 254) CGTCGACC 1562 GGTCGACGGATCCCCGGAATGTTACAGAGCTTTC (SEQ ID NO: 255) AGGATTGC 1563 CAAATCGGCGGTAACGAAAGAGGATAAACCGTGT (SEQ ID NO: 256) CCCGTATTATTCACGAGGCCCTTTCGTCTTCACC TC 1566 TCCCACCCAATCAAGGCCAACG (SEQ ID NO: 257) 1567 TCCACCTGGTGCCAATGAACCG (SEQ ID NO: 258) 1587 CGGCTGCCAGAACTCTACTAACTG (SEQ ID NO: 259) 1588 GCGACGTCTACTGGCAGGTTAAT (SEQ ID NO: 260) 1595 CAACCTGGTGATTTGGGGAAG (SEQ ID NO: 261) 1597 GAATGATGGCAGATTGGGCA (SEQ ID NO: 262) 1598 TATTGTGGGGCTGTCTCGAATG (SEQ ID NO: 263) 1624 CCCTCATGTTGTCTAACGG (SEQ ID NO: 264) 1633 TCCGTCACTGGATTCAATGCCATC (SEQ ID NO: 265) 1634 TTCGCCAGGGAGCTGGTGAA (SEQ ID NO: 266) 1798 GCAAATTAAAGCCTTCGAGCG (SEQ ID NO: 267) 1926 TTTTTGTCGACGGATCCAGTTTATCATTATCAAT (SEQ ID NO: 268) ACTCG 1927 TTTTGCGGCCGCAGATCTCTCGAGTCGAAACTAA (SEQ ID NO: 269) GTTCTGGTGTT 2091 CTTTTCTTCCCTTGTCTCAATC (SEQ ID NO: 270) 2352 GACTCGACCTAGGTTATTTAGTAAAATCAATGAC (SEQ ID NO: 271) CATTC 2353 CTAAATAACCTAGGTCGAGTCATGTAATTAGTTA (SEQ ID NO: 272) TGTC KARIpETfor ATTCATATGGCGAATTATTTCAACACTCTG (SEQ ID NO: 273) KARIpETrev TAATCTCGAGGCCAGCCACCGCGATGCG (SEQ ID NO: 274) pETup ATGCGTCCGGCGTAGA (SEQ ID NO: 275) seq_ilvC_pGV GCGGCCGCGTCGACGAGGAGACAACATTATGGCG (SEQ ID NO: 276) A pGV1994ep_for CGGTCTTCAATTTCTCAAGTTTCAGTTTCATTTT (SEQ ID NO: 277) TCTTGTTCTATTACAAC pGV1994ep_rev CTAACTCCTTCCTTTTCGGTTAGAGCGGATGTGG (SEQ ID NO: 278) G Not_in_for CCTCTAGAAATAATTTGCGGCCGCGTTAAGAAGG (SEQ ID NO: 279) AGATATACATATG AvrII_in_rev CCGAACGCCCTAGGTCAGTGGTGGTGGTGGTGGT (SEQ ID NO: 280) GCTCGAG R68DK69Lfor TAGCTATGCGCTGGACCTGGAGGCTATC (SEQ ID NO: 281)
R68DK69Lrev GATAGCCTCCAGGTCCAGCGCATAGCTA (SEQ ID NO: 282) K75VR76Dfor AGGCTATCGCGGAAGTTGACGCTAGCTG (SEQ ID NO: 283) K75VR76Drev CAGCTAGCGTCAACTTCCGCGATAGCCT (SEQ ID NO: 284) R69NNKfor TAGCTATGCGCTGCGCNNKGAGGCTATC (SEQ ID NO: 285) R69NNKrev GATAGCCTCMNNGCGCAGCGCATAGCTA (SEQ ID NO: 286) K75NNKfor AGGCTATCGCGGAANNKCGTGCTAGCTG (SEQ ID NO: 287) K75NNKrev CAGCTAGCACGMNNTTCCGCGATAGCCT (SEQ ID NO: 288) R76NNKfor AGGCTATCGCGGAAAAANNKGCTAGCTGGC (SEQ ID NO: 289) R76NNKrev GCCAGCTAGCMNNTTTTTCCGCGATAGCCT (SEQ ID NO: 290) R68NNK_for TAGCTATGCGCTGNNKAAGGAGGCTATC (SEQ ID NO: 291) R68NNK_rev GATAGCCTCCTTMNNCAGCGCATAGCTA (SEQ ID NO: 292) S78NNK_for GCGGAAAAACGTGCTNNKTGGCGCAAGGCTACT (SEQ ID NO: 293) S78NNK_rev AGTAGCCTTGCGCCAMNNAGCACGTTTTTCCGC (SEQ ID NO: 294) A71NNK_for GCGCTGCGCAAGGAGNNKATCGCGGAAAAAC (SEQ ID NO: 295) A71NNK_rev GTTTTTCCGCGATMNNCTCCTTGCGCAGCGC (SEQ ID NO: 296) Gln110NNK_for CTGACCCCAGATAAANNKCATAGCGACGTTG (SEQ ID NO: 297) Gln110NNK_rev CAACGTCGCTATGMNNTTTATCTGGGGTCAG (SEQ ID NO: 298) seq_ilvC_pGV GCGGCCGCGTCGACGAGGAGACAACATTATGGC (SEQ ID NO: 299) GA Q110Qfor GACCCCAGATAAACAACATAGCGACGTTGTT (SEQ ID NO: 300) Q110Qrev AACAACGTCGCTATGTTGTTTATCTGGGGTC (SEQ ID NO: 301) Q110Afor GACCCCAGATAAAGCACATAGCGACGTTGTT (SEQ ID NO: 302) Q110Arev AACAACGTCGCTATGTGCTTTATCTGGGGTC (SEQ ID NO: 303) Q110Vfor GACCCCAGATAAAGTACATAGCGACGTTGTT (SEQ ID NO: 304) Q110Vrev AACAACGTCGCTATGTACTTTATCTGGGGTC (SEQ ID NO: 305) R68A71recombfor GCTATGCGCTGCKAAAGGAGDCAATCGCGG (SEQ ID NO: 306) R68A71recombrev CGGCGATTGHCTCCTTTMGCAGCGCATAGC (SEQ ID NO: 307) R76S78recombfor GAAAAACGTGCTAGCTGGCGCAAGGCTACT (SEQ ID NO: 308) R76S78recombrev AGTAGCCTTGCGCCAGCTAGCACGTTTTTC (SEQ ID NO: 309) G76S78recombfor GAAAAAGGTGCTAGCTGGCGCAAGGCTACT (SEQ ID NO: 310) G76S78recombrev AGTAGCCTTGCGCCAGCTAGCACCTTTTTC (SEQ ID NO: 311) S76S78recombfor GAAAAAAGTGCTAGCTGGCGCAAGGCTACT (SEQ ID NO: 312) S76S78recombrev AGTAGCCTTGCGCCAGCTAGCACTTTTTTC (SEQ ID NO: 313) T76S78recombfor GAAAAAACTGCTAGCTGGCGCAAGGCTACT (SEQ ID NO: 314) T76S78recombrev AGTAGCCTTGCGCCAGCTAGCAGTTTTTTC (SEQ ID NO: 315) D76S78recombfor GAAAAAGATGCTAGCTGGCGCAAGGCTACT (SEQ ID NO: 316) D76S78recombrev AGTAGCCTTGCGCCAGCTAGCATCTTTTTC (SEQ ID NO: 317) R76D78recombfor GAAAAACGTGCTGACTGGCGCAAGGCTACT (SEQ ID NO: 318) R76D78recombrev AGTAGCCTTGCGCCAGTCAGCACGTTTTTC (SEQ ID NO: 319) G76D78recombfor GAAAAAGGTGCTGACTGGCGCAAGGCTACT (SEQ ID NO: 320) G76D78recombrev AGTAGCCTTGCGCCAGTCAGCACCTTTTTC (SEQ ID NO: 321) S76D78recombfor GAAAAAAGTGCTGACTGGCGCAAGGCTACT (SEQ ID NO: 322) S76D78recombrev AGTAGCCTTGCGCCAGTCAGCACTTTTTTC (SEQ ID NO: 323) T76D78recombfor GAAAAAACTGCTGACTGGCGCAAGGCTACT (SEQ ID NO: 324) T76D78recombrev AGTAGCCTTGCGCCAGTCAGCAGTTTTTTC (SEQ ID NO: 325) D76D78recombfor GAAAAAGATGCTGACTGGCGCAAGGCTACT (SEQ ID NO: 326) D76D78recombrev AGTAGCCTTGCGCCAGTCAGCATCTTTTTC (SEQ ID NO: 327) 1994hisrev TGACTCGAGCGGCCGCGGATCCTTAGTGGTGGTG (SEQ ID NO: 328) GTGGTGGTGTCCTGCCACTGCA pGV1994ep_for CGGTCTTCAATTTCTCAAGTTTCAGTTTCATTTT (SEQ ID NO: 329) TCTTGTTCTATTACAAC pGV1994ep_rev CTAACTCCTTCCTTTTCGGTTAGAGCGGATGTGG (SEQ ID NO: 330) G
EXAMPLE 1
Low-Level Anaerobic Production of Isobutanol
[0471]This example illustrates that a modified microorganism which is engineered to overexpress an isobutanol producing pathway produces a low amount of isobutanol under anaerobic conditions.
[0472]Overnight cultures of GEVO1859 were started from glycerol stocks stored at -80° C. of previously transformed strains. These cultures were started in 3 mL M9 minimal medium (Miller, J. H. A Short Course in Bacterial Genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. 1992. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.), supplemented with 10 g/L yeast extract, 10 μM ferric citrate and trace metals, containing 8.5% glucose and the appropriate antibiotics in snap cap tubes about 14 h prior to the start of the fermentation. Isobutanol fermentations were then carried out in screw cap flasks containing 20 mL of the same medium that was inoculated with 0.2 mL of the overnight culture. The cells were incubated at 37° C./250 rpm until the strains had grown to an OD600 of 0.6-0.8 and were then induced with Isopropyl 13-D-1-thiogalactopyranoside at 1 mM final concentration.
[0473]Three hours after induction the cultures were either kept under the current conditions (micro-aerobic conditions) or shifted to anaerobic conditions by loosening the cap of the flasks and placing the flasks into to a Coy Laboratory Products Type B Vinyl anaerobic chamber (Coy Laboratory Products, Grass Lakes, Mich.) through an airlock in which the flasks were cycled three times with nitrogen and vacuum, and then filled with the a hydrogen gas mix (95% Nitrogen, 5% Hydrogen).
[0474]Once the flasks were inside the anaerobic chamber, the flasks were closed again and incubated without shaking at 30° C. The flasks in the anaerobic chamber were swirled twice a day. Samples (2 mL) were taken at the time of the shift and at 24 h and 48 h after inoculation, spun down at 22,000 g for 1 min to separate the cell pellet from the supernatant and stored frozen at -20° C. until analysis. The samples were analyzed using High performance liquid chromatography (HPLC) and gas chromatography (GC).
[0475]GEVO1859 was run in triplicate. Stable OD values can be observed for all strains under anaerobic shift conditions over the course of the fermentation (FIG. 8). The complete pathway integrant strain showed low-level anaerobic isobutanol production over the course of the fermentation (FIG. 9, Table 7).
TABLE-US-00007 TABLE 7 Volumetric productivity, specific productivity titer and yield reached in an anaerobic fermentation for the tested strains and plasmid systems Volumetric Specific Productivity Productivity [g/ [g/L/ Titer Yield Samples L/h] ± h/OD] ± [g/L] ± [g/g] ± GEVO1859 0.088 0.028 0.019 0.005 4.22 1.35 0.140 0.029
[0476]In the period from 6 h to 48 h, i.e. under anaerobic conditions GEVO1859 demonstrated limited production of isobutanol (Table 8).
TABLE-US-00008 TABLE 8 Volumetric productivity, specific productivity titer and yield reached in the period from 6 to 48 h for the tested strain Volumetric Specific Productivity Productivity Titer Yield Samples Condition [g/L/h] ± [g/L/h/OD] ± [g/L] ± [g/g] ± GEVO1859 Micro- 0.266 0.010 0.040 0.004 11.2 0.4 0.33 0.016 aerobic GEVO1859 Anaerobic 0.086 0.026 0.019 0.005 3.60 1.1 0.14 0.032
EXAMPLE 2
Determination of Transhydrogenase Activity
[0477]This example illustrates that an isobutanol producing microorganism which carries a plasmid for the expression of the E. coli PntAB transhydrogenase (SEQ ID NO: 2 and SEQ ID NO: 4) contains increased transhydrogenase activity.
[0478]A fermentation was performed with a strain expressing the tet repressor (GEVO1385) and carrying the plasmids pGV1655 (SEQ ID NO: 109) and pGV1698 (SEQ ID NO: 112) for expression of the isobutanol pathway. The E. coli transhydrogenase PntAB was expressed from a third plasmid pGV1685 (SEQ ID NO: 111), which contained the E. coli pntAB genes under control of the PLtet promoter. The appropriate empty vector control carries the plasmid pGV1490 (SEQ ID NO: 104).
[0479]GEVO1385 was transformed with pGV1698, pGV1655, and either pGV1685 or pGV1490. Transformed cells were plated on LB-plates containing the appropriate antibiotics and the plates were incubated overnight at 37° C. Overnight cultures were started in 3 mL EZ-Rich Defined Medium (Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974, Culture medium for enterobacteria, J Bacteriol. 119:736-47) containing 5% glucose and the appropriate antibiotics in snap cap tubes about 14 h prior to the start of the fermentation. Isobutanol fermentations were then carried out in EZ-Rich containing 5% glucose and the appropriate antibiotics. 250 mL screw cap flasks with 20 mL EZ-Rich containing 5% glucose and the appropriate antibiotics were inoculated with 1% of the grown overnight culture. The cells were incubated at 37° C./250 rpm until the strains were grown to an OD600 of 0.6-0.8 and these strains were then induced with Isopropyl β-D-1-thiogalactopyranoside (IPTG (Gold BioTechnology, Inc, 12481C100) 1 mM) and anhydrotetracycline (aTc (Sigma, 37919-100 mg) 100 ng/mL). Samples were taken of the medium 48 h after inoculation. 15 mL of cell culture from each flask were centrifuged at 5,000×g for 5 min to separate the cell pellet from the supernatant. The cell pellets were stored frozen at -80° C. until analysis. The cultures grew to a comparable OD in this experiment.
[0480]To confirm that the transhydrogenase was actually expressed from the plasmids and to assess their enzymatic activity levels, enzyme assays were done with lysates prepared from the fermentation cultures. Frozen cell pellets were thawed on ice. The pellets were resuspended in 1.2 mL lysis buffer (50 mM potassium phosphate buffer at pH 7.5, MgCl2 2 mM). The suspensions were sonicated on ice for twice 2 min. The transhydrogenase enzyme assay was done in potassium phosphate buffer (50 mM pH 7.5, MgCl2 2 mM, 1 mM acetylpyridine-AD, 0.5 mM NADPH). The assay was run at 25° C. in a 96 well plate. Absorbance at 375 nm was followed in a kinetic assay format. To measure PntAB activity lysates were not cleared by centrifugation. The activity obtained for the samples featuring over-expressed E. coli pntAB show at least a 10 fold increase in transhydrogenase activity (Table 9).
TABLE-US-00009 TABLE 9 Shown are the enzymatic activities of the independent E. coli pntAB overexpressing strains and the amount of isobutanol production that would be supported by that activity calculated from Vmax values obtained from the enzyme assay specific activity protein [u/mg average stdev. conc. units in (total cell Samples Vmax Vmax [mg/mL] reaction protein)] pntAB-1 33.81 3.87 1.17 0.0010 0.1646 pntAB-2 45.06 1.51 1.89 0.0013 0.1355 empty vector-1 2.24 0.21 0.89 0.0001 0.0142 empty vector-2 -0.01 2.00 0.71 0.0000 -0.0001
EXAMPLE 3
Overexpression of pntAB Improves Isobutanol Fermentation Performance
[0481]This example illustrates that overexpression of a transhydrogenase, exemplified by the E. coli pntAB operon (SEQ ID NO: 1 and SEQ ID NO: 3) on a low copy plasmid improves isobutanol production under micro-aerobic conditions.
[0482]GEVO1748 was transformed with plasmids pGV1698 (SEQ ID NO: 112) and one of either pGV1720 (SEQ ID NO: 115) (control) or pGV1745 (SEQ ID NO: 117) (E. coli pntAB).
[0483]The aforementioned strains were plated on LB-plates containing the appropriate antibiotics and incubated overnight at 37° C. Overnight cultures were started in 3 ml. EZ-Rich medium (Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J Bacteriol. 119:736-47) containing 5% glucose and the appropriate antibiotics in snap cap tubes about 14 h prior to the start of the fermentation. Isobutanol fermentations were then carried out in EZ-Rich Medium containing 5% glucose and the appropriate antibiotics. 250 mL screw cap flasks with 20 mL EZ-Rich medium containing 5% glucose and the appropriate antibiotics were inoculated with 1% of the grown overnight culture. The cells were incubated at 37° C./250 rpm until they reached an OD600 of 0.6-0.8 followed by induction with Isopropyl β-D-1-thiogalactopyranoside (IPTG, 1 mM) and anhydrotetracycline (aTc, 100 ng/mL). Samples (2 mL) were taken 24 h and 48 h post inoculation, centrifuged at 22,000×g for 1 min and stored frozen at -20° C. until via Gas Chromatography (GC) and High Performance Liquid Chromatography (HPLC). Fermentations were run with two biological replicates.
[0484]All cultures grew to an OD of 5.5 to 6.5. Volumetric productivity and titer were improved by 45%, specific productivity even by 51%. Yield was improved by 8% (Table 10).
TABLE-US-00010 TABLE 10 Overexpression of E. coli pntAB improves isobutanol fermentation performance Volumetric Specific Productivity Productivity Titer Yield Strain [g/L/h] ± [g/L/h/OD] ± [g/L] ± [g/g] ± GEVO1748 + 0.205 0.001 0.035 0.001 9.86 0.04 0.311 0.001 pGV1698 + pGV1720 GEVO1748 + 0.298 0.006 0.053 0.003 14.29 0.28 0.337 0.001 pGV1698 + pGV1745
EXAMPLE 4
Overexpression of pnfAB Enables Anaerobic Isobutanol Production
[0485]This example illustrates that overexpression of a transhydrogenase, exemplified by the E. coli pntAB operon product (SEQ ID NO: 2 and SEQ ID NO: 4), improves anaerobic isobutanol production. This is surprising because it was previously not known that isobutanol could be produced anaerobically. In addition, this result was achieved without modifying the isobutanol biosynthetic pathway itself.
[0486]GEVO1748 was transformed with plasmids pGV1698 (SEQ ID NO: 112) and pGV1720 (SEQ ID NO: 115) (control) or pGV1745 (SEQ ID NO: 117) (E. coli pntAB).
[0487]Overnight cultures of the aforementioned strains were started from glycerol stocks stored at -80° C. of previously transformed strains. These cultures were started in 3 mL M9 minimal medium (Miller, J. H. A Short Course in Bacterial Genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. 1992. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.), supplemented with 10 g/L yeast extract, 10 μM ferric citrate and trace metals, containing 8.5% glucose and the appropriate antibiotics in snap cap tubes about 14 h prior to the start of the fermentation. Isobutanol fermentations were then carried out in 250 mL screw cap flasks containing 20 mL of the same medium that was inoculated with 0.2 mL of the overnight culture. The cells were incubated at 37° C./250 rpm until the strains had grown to an OD600 of 0.6-0.8 and were then induced with Isopropyl β-D-1-thiogalactopyranoside at 1 mM final concentration.
[0488]Three hours after induction the cultures were shifted to anaerobic fermentation conditions by loosening the cap of the flasks and placing the flasks into to a Coy Laboratory Products Type B Vinyl anaerobic chamber (Coy Laboratory Products, Grass Lakes, Mich.) through an airlock in which the flasks were cycled three times with nitrogen and vacuum, and then filled with the a hydrogen gas mix (95% Nitrogen, 5% Hydrogen). Once the flasks were inside the anaerobic chamber, the flasks were closed again and incubated without shaking at 30° C. Inside the chamber, an anaerobic atmosphere (<5 ppm oxygen) was maintained through the hydrogen gas mix (95% Nitrogen, 5% Hydrogen) reacting with a palladium catalyst to remove oxygen. The flasks in the anaerobic chamber were swirled twice a day. Samples (2 mL) were taken at the time of the shift and at 24 h and 48 h after inoculation, spun down at 22,000×g for 1 min to separate the cell pellet from the supernatant and stored frozen at -20° C. until analysis. The samples were analyzed using High performance liquid chromatography (HPLC) and gas chromatography (GC). All experiments for the E. coli pntAB-expressing strain were performed in duplicate while the control strain was only run in a single experiment.
[0489]At the time of shifting the cultures to anaerobic conditions all samples had an OD600 ranging between 2.3 and 3.3. All samples featuring an overexpressed E. coli pntAB operon (pGV1745) increased in OD600 from 6 h to 24 h by 0.2-1.1, all samples lacking pntAB (pGV1720) decreased in OD600 by 0.5-1.2 (FIG. 10), indicating that overexpression of E. coli pntAB is beneficial under anaerobic conditions.
[0490]Furthermore, pntAB over-expression is beneficial for anaerobic isobutanol production. All samples featuring E. coli PntAB continued isobutanol production under anaerobic conditions until the fermentation was stopped at 48 hours whereas the samples lacking E. coli PntAB did not produce isobutanol between 24 and 48 hours (FIG. 11)
[0491]In the strain overexpressing E. coli pntAB, volumetric productivity and titer are increased 2.4-fold, specific productivity by 85% and yield by 9% (Table 11).
TABLE-US-00011 TABLE 11 Shown are the results for volumetric productivity, specific productivity titer and yield reached in an anaerobic fermentation for the tested strains and plasmid systems after 48 h Volumetric Specific Productivity Productivity Titer Yield Samples [g/L/h] ± [g/L/h/OD] ± [g/L] ± [g/g] ± GEVO1748 + 0.047 0.022 2.24 0.279 pGV1720 + pGV1698 GEVO1748 + 0.111 0.002 0.041 0.012 5.32 0.10 0.304 0.004 pGV1745 + pGV1698
[0492]In the period from 6 h to 48 h, (i.e. under anaerobic conditions), GEVO1748 transformed with plasmids pGV1698 and pGV1745 (carrying E. coli pntAB) demonstrated significantly higher productivity, titer, and yield of isobutanol compared to the control strain carrying pGV1720 (without E. coli pntAB) (Table 12).
TABLE-US-00012 TABLE 12 Shown are the results for volumetric productivity, specific productivity titer and yield reached in the period from 6 to 48 h for the tested strains and plasmid systems Volumetric Specific Productivity Productivity Titer Yield sample [g/L/h] ± [g/L/h/OD] ± [g/L] ± [g/g] ± GEVO1748 + 0.029 0.014 1.21 0.171 pGV1720 + pGV1698 GEVO1748 + 0.096 0.003 0.035 0.015 4.01 0.15 0.246 0.002 pGV1745 + pGV1698
EXAMPLE 5
Chromosomal Integration of pntAB Improves Anaerobic Isobutanol Production
[0493]This example illustrates that overexpression of a transhydrogenase, exemplified by the E. coli pntAB operon product (SEQ ID NO: 2 and SEQ ID NO: 4), from the chromosome improves isobutanol production under anaerobic conditions compared to the case in which E. coli pntAB is expressed from a low copy plasmid. This strain reaches the same titer aerobically as anaerobically.
[0494]Overnight cultures of GEVO1846, GEVO1859, GEVO1886 were started from glycerol stocks stored at -80° C. of previously transformed strains. These cultures were started in 3 mL M9 minimal medium (Miller, J. H. A Short Course in Bacterial Genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. 1992. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.), supplemented with 10 g/L yeast extract, 10 μM ferric citrate and trace metals, containing 8.5% glucose and the appropriate antibiotics in snap cap tubes about 14 h prior to the start of the fermentation. Isobutanol fermentations were then carried out in screw cap flasks containing 20 mL of the same medium that was inoculated with 0.2 mL of the overnight culture. The cells were incubated at 37° C./250 rpm until the strains had grown to an OD600 of 0.6-0.8 and were then induced with Isopropyl β-D-1-thiogalactopyranoside at 1 mM final concentration.
[0495]Three hours after induction the cultures were either kept under the current conditions (micro-aerobic conditions) or shifted to anaerobic conditions by loosening the cap of the flasks and placing the flasks into to a Coy Laboratory Products Type B Vinyl anaerobic chamber (Coy Laboratory Products, Grass Lakes, Mich.) through an airlock in which the flasks were cycled three times with nitrogen and vacuum, and then filled with the a hydrogen gas mix (95% Nitrogen, 5% Hydrogen). Once the flasks were inside the anaerobic chamber, the flasks were closed again and incubated without shaking at 30° C. The flasks in the anaerobic chamber were swirled twice a day. Samples (2 mL) were taken at the time of the shift and at 24 h and 48 h after inoculation, spun down at 22,000×g for 1 min to separate the cell pellet from the supernatant and stored frozen at -20° C. until analysis. The samples were analyzed using High performance liquid chromatography (HPLC) and gas chromatography (GC). All experiments were performed in duplicate.
[0496]GEVO1886, GEVO1859 and GEVO1846 were run in parallel. Each strain was run in triplicate. Stable OD values can be observed for all strains under anaerobic shift conditions over the course of the fermentation (FIG. 12). The over-expression of E. coli pntAB in the complete pathway integrant strain again showed improvement for isobutanol production over the course of the fermentation (FIG. 13).
[0497]Compared to the complete pathway integrant strain without E. coli pntAB knock-in (GEVO1859), volumetric productivity and titer are increased 3.8-fold, specific productivity is increased 2.8-fold and the yield is 2.2-fold higher in GEVO1886. In addition, GEVO1886 shows superior performance compared to the plasmid system strain (GEVO1846) under anaerobic conditions. Volumetric productivity and titer are increased by 48%, specific productivity is increased by 18% and yield is 12% higher (Table 13).
TABLE-US-00013 TABLE 13 Shown are the results for volumetric productivity, specific productivity titer and yield reached in an anaerobic fermentation for the tested strains and plasmid systems Volumetric Specific Productivity Productivity [g/ [g/L/ Titer Yield Samples L/h] ± h/OD] ± [g/L] ± [g/g] ± GEVO1886 0.335 0.002 0.053 0.001 16.08 0.08 0.307 0.004 GEVO1859 0.088 0.028 0.019 0.005 4.22 1.35 0.140 0.029 GEVO1846 0.227 0.021 0.045 0.005 10.88 1.01 0.274 0.003
[0498]The performance numbers in the period from 6 to 48 demonstrate that most of isobutanol production occurred under anaerobic conditions. Highest values for yield and specific productivity were reached by the strain featuring the complete pathway integration and the E. coli pntAB knock-in (GEVO1886) under anaerobic conditions. In addition this strain reached the highest values for volumetric productivity and titer under both conditions anaerobic and micro-aerobic (Table 14).
TABLE-US-00014 TABLE 14 Shown are the results for volumetric productivity, specific productivity titer and yield reached in the period from 6 to 48 h for the tested strains and plasmid systems Volumetric Specific Productivity Productivity Titer Yield Samples Condition [g/L/h] ± [g/L/h/OD] ± [g/L] ± [g/g] ± GEVO1886 Micro- 0.355 0.004 0.042 0.001 149 0.2 0.33 0.012 aerobic GEVO1859 Micro- 0.266 0.010 0.040 0.004 11.2 0.4 0.33 0.016 aerobic GEVO1846 Micro- 0.344 0.007 0.051 0.004 14.4 0.3 0.33 0.005 aerobic GEVO1886 Anaerobic 0.355 0.008 0.056 0.001 14.9 0.1 0.35 0.004 GEVO1859 Anaerobic 0.086 0.026 0.019 0.005 3.60 1.1 0.14 0.032 GEVO1846 Anaerobic 0.209 0.019 0.041 0.004 8.79 0.8 0.27 0.006
[0499]The performance numbers in the period from 6 to 48 demonstrate that most of isobutanol production occurred under anaerobic conditions. Highest values for yield and specific productivity were reached by the strain featuring the complete pathway integration and the E. coli pntAB knock-in (GEVO1886) under anaerobic conditions.
EXAMPLE 6
Anaerobic Batch Fermentation of GEVO1886 and GEVO1859
[0500]This example illustrates that an engineered microorganism which overexpresses a transhydrogenase, exemplified by the E. coli pntAB gene product (SEQ ID NO: 2 and SEQ ID NO: 4), from the chromosome produces isobutanol at a higher rate, titer and productivity compared to the a strain that does not overexpress a transhydrogenase. This is surprising because the increase in rate, titer, and productivity was achieved without modifying the isobutanol biosynthetic pathway itself.
[0501]Overnight cultures were started in 250 mL Erlenmeyer flasks with strain GEVO1886 and strain GEVO1859 cells from fresh streak plates with a 40 mL volume of M9 medium (Miller, J. H. A Short Course in Bacterial Genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. 1992. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.) containing 85 g/L glucose, 20 g/L yeast extract, 20 μM ferric citrate, trace metals, an additional 1 g/L NH4Cl, an additional 1 mM MgSO4 and an additional 1 mM CaCl2 and at a culture OD600 of 0.02 to 0.05. The overnight cultures were grown for approximately 14 hours at 30° C. at 250 rpm.
[0502]Some of the overnight cultures were then transferred to 400 mL DasGip fermenter vessels containing about 200 mL of M9 medium (Miller, J. H. A Short Course in Bacterial Genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. 1992. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.) containing 85 g/L glucose, 20 g/L yeast extract, 20 μM ferric citrate, trace metals, an additional 1 g/L NH4Cl, an additional 1 mM MgSO4 and an additional 1 mM CaCl2 to achieve a starting cell concentration by optical density at 600 nm of 0.1. The vessels were attached to a computer control system to monitor and control pH at 6.5 through addition of base, temperature at 30° C., dissolved oxygen, and agitation. The vessels were agitated, with a minimum agitation of 200 rpm and agitation was varied to maintain a dissolved oxygen content of about 50% using a 12 sL/h air sparge until the OD600 was about 1.0. The vessels were then induced with 1 mM IPTG.
[0503]After continuing growth for 3 hrs, the dissolved oxygen content was decreased to 0% with 200 rpm agitation and 2.5 sL/h sparge with nitrogen (N2) gas. Measurement of the fermenter vessel off-gas for isobutanol and ethanol was performed throughout the experiment by passage of the off-gas stream through a mass spectrometer. Continuous measurement of off-gas concentrations of carbon dioxide and oxygen were also measured by a DasGip off-gas analyzer throughout the experiment. Samples were aseptically removed from the fermenter vessel throughout the experiment and used to measure OD600, glucose concentration by HPLC, and isobutanol concentration in the broth by GC. Each strain was run in three independent fermentations.
[0504]Strain GEVO1886 reached an average isobutanol total titer of 21.6 g/L. The average yield of the fermentation, calculated when the titer of isobutanol was between 1 g/L and 15 g/L, was 88% of theoretical. The average productivity of the fermentation was 0.4 g/L/h. As described in Example 5, GEVO1886 performs at least equally well in terms of isobutanol productivity, titer, yield under anaerobic and aerobic conditions.
[0505]By comparison, strain GEVO1859 reached an average isobutanol total titer of 1.8 g/L. The average yield of the fermentation was 56% of theoretical, and the average productivity of the fermentation was 0.02 g/l/h.
EXAMPLE 7
PntAB Overexpression Rescues a zwf-deletion Phenotype
[0506]This example illustrates that a strain that has a growth defect and does not produce isobutanol because of the deletion in a native pathway that reduces the strains ability to produce the redox cofactor NADPH can surprisingly be rescued by overexpression of E. coli pntAB.
[0507]Overnight cultures of GEVO1399 transformed with plasmids pSA55, pGV1609 (SEQ ID NO: 108), and pGV1745 (SEQ ID NO: 117) and GEVO1399 transformed with plasmids pSA55, pGV1609, and pGV1720 (SEQ ID NO: 115) were started from glycerol stock cultures stored at -80° C. in 3 mL fermentation medium (M9 minimal medium according to Miller (Miller, J. H. A Short Course in Bacterial Genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. 1992. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.), supplemented with 10 g/L yeast extract, 10 μM ferric citrate and trace metals) containing 8.5% glucose and the appropriate antibiotics in snap cap tubes about 14 h prior to the start of the fermentation.
[0508]Isobutanol fermentations were then carried out in fermentation medium containing 8.5% glucose and the appropriate antibiotics. Two 250 mL screw cap flasks with 20 mL fermentation medium containing 8.5% glucose and the appropriate antibiotics were inoculated with 1% of each grown overnight culture. The cells were incubated at 37° C./250 rpm until the strains were grown to an OD600 of 0.6-0.8 and were then induced with Isopropyl β-D-1-thiogalactopyranoside at 1 mM final concentration. Three hours after induction one flask per overnight culture was shifted to anaerobic fermentation conditions. This was done by loosening the cap of the flasks and introducing the flasks into the anaerobic chamber. Once the flasks were flushed with oxygen free atmosphere (while going through the airlock), the flasks were closed again and incubated without shaking at 30° C. in the anaerobic chamber. The flasks in the anaerobic chamber were swirled twice a day. Samples were taken from the medium at the time of the shift and at 24 h and 48 h after inoculation, spun down at 22,000×g for 1 min to separate the cell pellet from the supernatant and stored frozen at -20° C. until analysis. The samples were analyzed using High performance liquid chromatography (HPLC) and gas chromatography (GC).
[0509]The strain lacking zwf without E. coli pntAB grew to an OD of about 3, whereas the samples featuring E. coli pntAB reached OD values of about 5-6. This OD was not significantly different from normal growth and thus the over-expression of E. coli pntAB rescues the zwf growth phenotype (FIG. 14).
[0510]Isobutanol production was rescued under micro-aerobic conditions by the overexpression of E. coli pntAB. Volumetric productivity and titer are improved 7.4 fold, specific productivity was improved 3.3 fold and yield 2.5 fold (Table 15).
TABLE-US-00015 TABLE 15 Volumetric productivity, specific productivity titer and yield in a micro- aerobic fermentation for the tested strains and plasmid systems Volumetric Specific Productivity Productivity Titer Yield Samples [g/L/h] ± [g/L/h/OD] ± [g/L] ± [g/g] ± GEVO1399 + 0.170 0.001 0.030 0.003 8.18 0.02 0.248 0.012 pGV1745 + pSA55 + pGV1609 GEVO1399 + 0.023 0.004 0.009 0.002 1.10 0.18 0.100 0.013 pGV1720 + pSA55 + pGV1609
[0511]For the anaerobic shift experiment the same trend was observed as under micro-aerobic conditions. Isobutanol production was rescued by the over-expression of E. coli pntAB. Volumetric productivity and titer are improved 3.4 fold, specific productivity was improved 2.1 fold and yield by 43% (Table 16).
TABLE-US-00016 TABLE 16 Volumetric productivity, specific productivity titer and yield in an anaerobic fermentation for the tested strains and plasmid systems Volumetric Specific Productivity Productivity Titer Yield Samples [g/L/h] ± [g/L/h/OD] ± [g/L] ± [g/g] ± GEVO1399 + 0.125 0.038 0.035 0.003 6.00 1.84 0.297 0.008 pGV1745 + pSA55 + pGV1609 GEVO1399 + 0.037 0.001 0.017 0.001 1.78 0.04 0.207 0.005 pGV1720 + pSA55 + pGV1609
EXAMPLE 8
sthA Does Not Contribute to Improvement in Anaerobic Isobutanol Production
[0512]This example illustrates that an isobutanol production strain with a deletion of the soluble transhydrogenase sthA produces low amounts of isobutanol anaerobically. This shows that the introduction of the sthA deletion does not provide cofactor balance to the isobutanol production strain and does not enable anaerobic isobutanol production above the levels seen for strains without redox engineering. The deletion of sthA has no significant effect on anaerobic performance of a production strain that overexpresses E. coli pntAB.
[0513]GEVO1748 and GEVO1844 were transformed with plasmids pGV1698 (SEQ ID NO: 112) and one of either pGV1720 (SEQ ID NO: 115) (control) or pGV1745 (SEQ ID NO: 117) (E. coli pntAB).
[0514]Overnight cultures of the strains to be tested were started either using fresh transformants (for all combinations featuring strain GEVO1844) or using frozen stocks (all other samples). The cultures were started in 3 mL fermentation medium (M9 minimal medium according to Miller (Miller, J. H. A Short Course in Bacterial Genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. 1992. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.), supplemented with 10 g/L yeast extract, 10 μM ferric citrate and trace metals) containing 8.5% glucose and the appropriate antibiotics in snap cap tubes about 14 h prior to the start of the fermentation.
[0515]Isobutanol fermentations were then carried out in fermentation medium containing 8.5% glucose and the appropriate antibiotics. Two 250 mL screw cap flasks with 20 mL fermentation medium containing 8.5% glucose and the appropriate antibiotics were inoculated with 1% of each grown overnight culture. The cells were incubated at 37° C./250 rpm until the strains were grown to an OD600 of 0.6-0.8 and were then induced with Isopropyl β-D-1-thiogalactopyranoside at 1 mM final concentration. Three hours after induction the flasks were shifted to anaerobic fermentation conditions. This was done by loosening the cap of the flasks and introducing the flasks into the anaerobic chamber. Once the flasks were flushed with oxygen free atmosphere (while going through the airlock), the flasks were closed again and incubated without shaking at 30° C. in the anaerobic chamber. The flasks in the anaerobic chamber were swirled twice a day. Samples were taken of the medium at the time of the shift and at 24 h and 48 h after inoculation, spun down at 22,000×g for 1 min to separate the cell pellet from the supernatant and stored frozen at -20° C. until analysis. The samples were analyzed using High performance liquid chromatography (HPLC) and gas chromatography (GC).
[0516]Strain GEVO1844 showed similar isobutanol production compared to non redox cofactor engineered strain GEVO1748 (Table 17).
TABLE-US-00017 TABLE 17 Shown are the results for volumetric productivity, specific productivity titer and yield reached in an anaerobic fermentation for the tested strains and plasmid systems Volumetric Specific Productivity Productivity Titer Yield Samples [g/L/h] ± [g/L/h/OD] ± [g/L] ± [g/g] ± GEVO1844 + 0.039 0.004 0.036 0.006 1.89 0.20 0.236 0.025 pGV1720 + pGV1698 (i.e. ΔsthA without PntAB) GEVO1748 + 0.047 0.022 2.24 0.279 pGV1720 + pGV1698 (i.e. Control without PntAB) GEVO1844 + 0.127 0.004 0.033 0.002 6.11 0.19 0.310 0.007 pGV1745 + pGV1698 (i.e. ΔsthA with PntAB) GEVO1748 + 0.111 0.002 0.041 0.012 5.32 0.10 0.304 0.004 pGV1745 + pGV1698 (i.e. control with PntAB)
[0517]The strains with the sthA deletion exhibited similar isobutanol production compared to the strains without the sthA deletion. This was independent on the presence or absence of overexpression of E. coli pntAB. It can thus be concluded that the sthA deletion has no significant effect on isobutanol production.
EXAMPLE 9
pntAB in Yeast
[0518]This example illustrates an isobutanol producing yeast which is engineered to express a transhydrogenase.
[0519]Yeast strain, GEVO5001, which is deficient in pyruvate decarboxylase activity and expresses the isobutanol biosynthetic pathway is further engineered to express a transhydrogenase. The E. coli pntA (SEQ ID NO: 1) and pntB (SEQ ID NO: 3) genes are expressed in yeast with the modifications of (1) N-terminal addition of amino acids to target the proteins to the plasma membrane (export signal sequence (ess)) and (2) N-terminal modifications to target the proteins to the mitochondrial outer membrane (mitochondrial targeting sequence (mts)). pGV6002 is a yeast integration plasmid that carries versions of pntA and pntB with modifications to target them to the plasma membrane. pGV6003 is a yeast integration plasmid that carries versions of pntA and pntB with modifications to target them to the mitochondrial outer membrane. In both cases, the pntA and pntB genes are under the control of the strong constitutive promoters from TEF1 and TDH3, respectively. pGV6002 and pGV6003 are linearized and transformed into GEVO5001 to generate GEVO5004 and GEVO5005, respectively. Expression of pntA and pntB is confirmed by qRT-PCR and once confirmed; GEVO5004 and GEVO5005 are used in fermentations for the production of isobutanol.
EXAMPLE 10
Native E. coli Alcohol Dehydrogenase Activity Converts Isobutyraldehyde to Isobutanol
[0520]This example illustrates that native E. coli alcohol dehydrogenase activity converts isobutyraldehyde to isobutanol.
[0521]Strain JCL260 transformed with pGV1631 and pSA69 (strain without S. cerevisiae ADH2) and JCL260 transformed with pSA55 and pSA69 (strain with S. cerevisiae ADH2) were plated onto LB-plates containing the appropriate antibiotics and incubated overnight at 37° C. Plates were taken out of the incubator and kept at room temperature until further use. Overnight cultures were started in 3 mL EZ-Rich medium containing 7.2% glucose and the appropriate antibiotics in snap cap tubes about 14 hours prior to the start of the fermentation. Isobutanol fermentations were then carried out in EZ-Rich defined medium containing 7.2% glucose and the appropriate antibiotics. Screw cap flasks with 20 mL EZ-Rich medium containing 7.2% glucose and the appropriate antibiotics were inoculated with 1% of the grown overnight culture. The cells were incubated at 37° C./250 rpm until they were grown to an OD600 of 0.6-0.8 and induced with Isopropyl β-D-1-thiogalactopyranoside (IPTG, 1 mM).
[0522]After induction the cells were incubated at 30° C./250 rpm. Samples were taken from the medium before induction, and 24 and 48 hours after inoculation, spun down at 22,000×g for 1 min to separate the cell pellet from the supernatant and stored frozen at -20° C. until analysis.
[0523]The ADH2 gene product is expected to be functionally expressed from pSA55 and required for isobutanol production. Thus, no isobutanol should be produced with the plasmid combination lacking ADH2 as adhE is deleted in JCL260. However, isobutanol production for the system lacking ADH2 was higher than for the system with ADH2 expression. Table 18 shows the results for the isobutanol fermentation comparing the pathway including Adh2 expression with the exact same system excluding Adh2 expression. Both systems feature Bs_AlsS1, Ec_llvC and Ec_ilvD expressed from the same medium copy plasmid and Ll_Kivd1 expressed from a high copy plasmid. Volumetric productivity and titer showed 42% increase, specific productivity 18% and yield 12% increase. This suggests strongly that a native E. coli dehydrogenase is responsible for the conversion of isobutyraldehyde to isobutanol, and that Adh2 is not expressed and not necessary for isobutanol production in E. coli.
TABLE-US-00018 TABLE 18 Isobutanol fermentation with and without Adh2 expression Volumetric Productivity Specific [g/ Productivity Titer Yield samples L/h] ± [g/L/h/OD] ± [g/L] ± [g/g] ± without 0.175 0.006 0.039 0.003 8.40 0.26 0.207 0.009 Adh2 with 0.123 0.004 0.033 0.001 5.88 0.17 0.185 0.004 Adh2
EXAMPLE 11
Identification of Native ADH
[0524]This example illustrates that the native E. coli alcohol dehydrogenase is encoded by the Ec_yqhD gene (SEQ ID NO: 68).
[0525]Several E. coli genes predicted or known to code for alcohol dehydrogenases were knocked out of strain JCL260 to determine whether any of them are involved in isobutyraldehyde reduction. Fermentations were carried out with GEVO1608 and with JCL260, each transformed with plasmids pGV1609 (SEQ ID NO: 108) and pGV1631 by electroporation. Single colonies were grown and two colonies from each strain were started in a 3 mL overnight culture, with appropriate antibiotics. Each 250 mL fermentation flask was filled with 20 mL of EZ-Rich medium (Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J Bacteriol. 119:736-47) supplemented with 5% glucose, Ampicillin (100 mg/mL), and Chloramphenical (100 mg/mL).
[0526]The cell densities of the overnight cultures were normalized and 2% inoculum was added to each fermentation flask and incubated at 270 rpm/37° C. The cultures were induced with 20 μL 0.1 M IPTG after they reached an OD600 of 0.6-0.8 at which time the temperature was lowered to 30° C. Samples were taken from the medium before induction, and 24 hours after inoculation, spun down at 22,000×g for 1 min to separate the cell pellet from the supernatant and stored frozen at -20° C. until analysis. A second fermentation was performed in the same way with the best candidate, GEVO1608 containing the yqhD deletion, and samples were taken at 24 and 48 hours.
[0527]While both GEVO1608 and JCL260 grew to similar cell densities, GEVO1608 produced ˜80% less isobutanol than the control strain (Table 19), indicating that the Ec_yqhD gene product is primarily responsible for isobutyraldehyde reduction.
TABLE-US-00019 TABLE 19 Specific Productivity and Titer of Fermentation Strain Plasmids Time Titer (g/L) GEVO1608 pGV1609, pGV1631 24 h 0.33 JCL260 pGV1609, pGV1631 24 h 2.45 GEVO1608 pGV1609, pGV1631 48 h 0.83 JCL260 pGV1609, pGV1631 48 h 4.00
EXAMPLE 12
Overexpression of NADH-Dependent Alcohol Dehydrogenase and Propanediol Dehydrogenases
[0528]This example demonstrates that overexpression of an NADH-dependent alcohol dehydrogenase or propanediol dehydrogenases increases isobutanol production.
[0529]Relevant E. coli strains were transformed with the appropriate plasmids (Table 20).
TABLE-US-00020 TABLE 20 Plasmid and strain combinations used in isobutanol fermentations # Plasmid 1 Plasmid 2 Strain Comments 1 pGV1655 pGV1698 GEVO1745 No ADH on plasmid 2 pGV1655 pGV1698 JCL260 GEVO1780 3 pGV1655 pGV1748 GEVO1745 Ec_fucO 4 pGV1655 pGV1749 GEVO1745 Dm_ADH 5 pGV1655 pGV1778 GEVO1745 Kp_dhaT
[0530]Following transformation, the strains were plated on LB-plates containing the appropriate antibiotics and incubated overnight at 37° C. Overnight cultures Were started in 3 mL EZ-Rich medium (Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for Enterobacteria. J Bacteriol. 119:736-47) containing 8% glucose and the appropriate antibiotics in snap cap tubes about 14 h prior to the start of the fermentation. Isobutanol fermentations were then carried out in EZ-Rich Medium containing 8% glucose and the appropriate antibiotics. Screw cap flasks with 25 mL EZ-Rich medium containing 8% glucose and the appropriate antibiotics were inoculated with a sufficient volume of the grown overnight culture to obtain a starting OD600 of 0.1. The cells were incubated at 37° C./250 rpm until they reached an OD600 of 0.6-0.8 followed by induction with Isopropyl β-D-1-thiogalactopyranoside (IPTG, 1 mM). After induction, cultures were capped, sealed and placed in 30° C. shaker, 225 rpm to start fermentation. Samples (2 mL) were taken 24 h and 48 h post induction, centrifuged at 22,000×g for 1 min and the supernatant stored at 4° C. until analyzed. Prior to analysis, the supernatants were filtered and then analyzed via Gas Chromatography and High Performance Liquid Chromatography. All experiments were carried out in triplicate.
[0531]Results are presented in Table 21, below. Expression of either 1,2-propanediol dehydrogenase Ec_fucO or 1,3-propanediol dehydrogenase Kp_dhaT significantly and reproducibly increases titer in the ΔyqhD background of strain GEVO1745. Expression of Dm_ADH enhances titer and yield of the fermentations in the ΔyqhD background of strain GEVO1745.
TABLE-US-00021 TABLE 21 Summary of isobutanol titer, and yield data from fermentations after 48 hours # Comments titer [g/L] ± Yield [% theor.] ± 1 no ADH 1.91 0.50 38.5 10.30 2 GEVO1780 3.39 0.15 65.0 2.83 3 Ec_FucO 6.30 0.10 79.9 1.79 4 Dm_Adh 4.86 0.29 67.0 4.54 5 Kp_DhaT 6.22 0.16 75.3 2.04
EXAMPLE 13
Characterization of Alcohol Dehydrogenases
[0532]This example demonstrates that the alcohol dehydrogenases Ec_FucO (SEQ ID NO: 65), Kp_DhaT (SEQ ID NO: 63), and Dm_Adh (SEQ ID NO: 61) catalyze the NADH-dependent reduction of isobutyraldehyde.
[0533]E. coli strain GEVO1745 was transformed by electroporation with one of plasmids pGV1705-A, pGV1748-A, pGV1749-A, or pGV1778-A. 50 mL of TB medium (23.1 g/L KH2PO4, 125.4 g/L K2HPO4, 12 g/L Bacto-tryptone, 24 g/L yeast extract, 4 ml/L glycerol) were inoculated to an initial OD600 of 0.2 using a 3 mL overnight LB culture of a single colony. The 50 mL culture was allowed to grow for 3-4 hrs at 250 rpm and 37° C. Protein expression was induced at an OD600 of 2-2.5 by the addition of IPTG to a final concentration of 1 mM. After the addition of IPTG, protein expression was allowed to continue for 20-24 hours at 225 rpm and 25° C.
[0534]Alcohol dehydrogenase (ADH) activity was assayed kinetically by monitoring the decrease in NAD(P)H concentration by measuring the absorbance at 340 nm. A reaction buffer was prepared containing 0.1 M potassium phosphate, 0.4 mM NAD(P)H, 10 mM isobutyraldehyde, 1 mM DTT, and 1 mM PMSF. Cell pellets were resuspended in 0.1 M potassium phosphate buffer containing 1 mM DTT and 1 mM PMSF at one fifth of the culture volume, i.e. 10 mL resuspension buffer for cell pellet from a 50 mL culture. The resuspended cells were lysed by sonication for 1 min with a 50% duty cycle. The reaction was initiated by the addition of 0.5 mL of the reaction buffer to 0.5 mL of clarified lysate in a cuvette. Dilution of the clarified lysate was necessary for ADHs that were highly active. A substrate free control was conducted using reaction buffer without the addition of aldehyde.
[0535]Kinetic parameters were determined for Ec_YghD, Ec_FucO, Dm_Adh, and Kp_DhaT (Table 22).
TABLE-US-00022 TABLE 22 Kinetic parameters for the conversion of isobutyraldehyde to isobutanol by Ec_YqhD, Ec_FucO, Dm_Adh, and Kp_DhaT NADH NADPH Activity Activity KM (U/min-1 mg-1 KM (U/min-1 mg-1 Plasmid ADH (mM) crude lysate) (mM) crude lysate) pGV1705-A Ec_YqhD n.d. n.d. 0.25 0.09 pGV1748-A Ec_FucO 0.8 0.23 0.2 0.04 pGV1749-A Dm_Adh 0.9 6.60 2.7 1.70 pGV1778-A Kp_DhaT 1.3 0.56 0.6 0.08
The kinetic properties of the Ll_AdhA enzyme were described by Atsumi et al. (Atsumi et al., Appl. Microbiol. Biotechnol., 2009, DOI 10.1007/s00253-009-2085-6), and are shown in Table 23.
TABLE-US-00023 TABLE 23 Kinetic parameters for Ll_AdhA (Atsumi et al., Appl. Microbiol. Biotechnol., 2009, DOI 10.1007/s00253-009-2085-6) NADH NADPH KM kcat Kcat/ KM kcat Kcat/ ADH Substrate (mM) (s-1) KM (mM) (s-1) KM Ll_AdhA Acetaldehyde 0.5 10 20.9 n.d.a Ll_AdhA isobutyraldehyde 9.1 6.6 0.8 adid not show any detectably activity when tested with NADPH as a cofactor
EXAMPLE 14
KARI Engineering by Saturation Mutagenesis
[0536]Construction of KARI-containing plasmids: Standard molecular biology procedures (Sambrook and Russell, Molecular Cloning, A Laboratory Manual, 3rd Edition, Vol. 3, 2001) were utilized to make plasmid pGV1711 (SEQ ID NO: 113) (pLlacO1::(no ORF) bla, ColE1 OR1). Plasmid pGV1711 is a high-copy, AmpR vector that serves as an "empty vector" control, i.e. it contains no open reading frames under the control of the PLlac promoter. The E. coli KARI gene Ec_ilvC (SEQ ID NO: 10) was codon optimized for E. coli resulting in gene Ec_ilvC coEc (SEQ ID NO: 11)
[0537]The codon optimized gene Ec_ilvC_coEc was cloned into pET22b(+) using primers KARIpETfor and KARIpETrev introducing a 5' NdeI and a 3' XhoI restriction site and a C-terminal his6-tag, resulting in plasmid pET22b[ilvCco] carrying Ec_ilvC_coEc.sup.his6(SEQ ID NO: 14).
[0538]DNA constructs were analyzed by restriction digests, and also by DNA sequencing to confirm integrity and correct construction. Primers pETup and KARIpETrev were used as primers in standard DNA sequencing reactions to sequence pET22b(+) derivatives.
[0539]Construction of NNK libraries: NNK libraries were constructed using site directed mutagenesis overlap extension (SOE) PCR. First, the fragments containing the mutations were created allowing for at least 15 by of overlap using KARIpET_for and KARIpET_rev and the respective NNK primers listed in Table 6 (SEQ ID NO 285 through SEQ ID NO 298). After digesting traces of template DNA with DpnI, the fragments were separated on a 1% TAE agarose gel, extracted, and the PCR products were precipitated using pellet paint (Novagen). The clean products were used as templates in a subsequent assembly PCR. The PCR product was cleaned up (Zymo Research, Orange, Calif.), restriction digested with NdeI and XhoI for 1.5 h at 37° C., cleaned on a 1% agarose gel, and ligated into pET22b(+).
[0540]Site directed mutagenesis mutants were generated as described above. The successful mutagenesis was confirmed by DNA sequencing.
[0541]Cell growth and protein expression in shake flasks: Flasks containing 25 mL of Luria-Bertani. (LB) medium (10 g tryptone, 10 g NaCl, 5 g yeast extract) with ampicillin (final concentration 0.1 mg/mL) were inoculated to an initial OD600 of 0.1 using 0.25 mL overnight LB culture of a single colony. The 25 mL LB expression culture was allowed to grow for 3-4 h at 250 rpm and 37° C. Protein expression was induced at OD600 of 1 by the addition of IPTG to a final concentration of 0.5 mM. Protein expression was allowed to continue for 20-24 h at 225 rpm and 25° C. Cells were harvested at 5300×g and 4° C. for 10 min and the cell pellets were frozen at -20° C. until further use.
[0542]Cell growth and protein expression in microplates: In order to grow and express KARI variants in deep well plates, sterile toothpicks were used to pick single colonies into shallow 96 well plates filled with 300 μl LBamp. 75 μl of these overnight cultures were used to inoculate deep well plates filled with 600 μl of LBamp per well. The plates were grown at 37° C. and 210 rpm for 4 h. One hour before induction with IPTG (final concentration 0.5 mM), the temperature of the incubator was reduced to 25° C. After induction, growth and expression continued for 20 h at 25° C. and 210 rpm. Cells were harvested at 5300×g and 4° C. and stored at -20° C.
[0543]KARI cuvette assay: KARI activity was assayed kinetically by monitoring the decrease in NAD(P)H concentration by measuring the absorbance at 340 nm. A reaction buffer was prepared containing 250 mM potassium phosphate pH 7, 1 mM DTT and 10 mM MgCl2. Cell pellets were resuspended (0.25 g wet weight/mL buffer) in 250 mM potassium phosphate (KPi) buffer containing 1 mM DTT and 10 mM MgCl2. The resuspended cells were lysed by sonication for 1 min with a 50% duty cycle and pelleted at 11000×g and 4° C. for 15 min. A reaction mixture consisting of 910 μl reaction buffer, 50 μl acetolactate, and 20 μl lysate was prepared in a cuvette. The reaction was initiated by addition of 20 μL of 10 mM NAD(P)H. A substrate free control was conducted using reaction buffer without the addition of acetolactate.
[0544]KARI high-throughput assay: Frozen cell pellets were thawed at room temperature for 20 min and then 100 μL of lysis buffer (250 mM Kpi, 750 mg/L lysozyme, 10 mg/L DNasel, pH 7) were added. Plates were vortexed to resuspend the cell pellets. After a 30 min incubation at 37° C., plates were centrifuged at 5300×g and 4° C. for 10 min. 20 μL of the resulting crude extract were transferred into assay plates (flat bottom, Rainin) using a liquid handling robot. 10 mL assay buffer per plate were prepared (250 mM Kpi, pH 7, 500 μL acetolactate, 1 mM DTT, 10 mM NAD(P)H, and 10 mM MgCl2) and 90 μL thereof were added to each well to start the reaction. The depletion of NAD(P)H was monitored at 340 nm in a plate reader (TECAN) over 1.5 min.
[0545]Purification of KARI: Cell pellets used for purification were resuspended in purification buffer A (20 mM Tris, ±20 mM imidazol, 100 mM NaCl, 10 mM MgCl2, pH 7.4). KARI was purified by IMAC (Immobilized metal affinity chromatography) over a 1 ml Histrap High Performance (histrap HP) column pre-charged with Nickel (GE Healthcare) using an Akta FPLC system (GE Healthcare). The column was equilibrated with four column volumes (cv) of buffer A. After injecting the crude extract, the column was washed with buffer A for 2 cv, followed by a wash step with a mixture of 10% elution buffer B (20 mM Tris, 300 mM imidazol, 100 mM NaCl, 10 mM MgCl2, pH 7.4) for 5 cv. KARI variants were eluted at 40% buffer B and stored at 4° C.
[0546]Homology modeling was performed with pymol and x-ray structures of E. coli KARI (PDB ID: 1YRL) and spinach KARI (PDB ID: 1YVE), the latter containing NADPH co-crystallized.
[0547]A KARI expression construct (pGV1777 (SEQ ID NO: 118)) (pLlacO1::Ec_ilvC_coEc::bla, ColE1 ORI) was tested in E. coli strain GEVO1777 and yielded KARI activity in lysates. On this plasmid, the ilvC gene was not his-tagged and therefore no purification was attempted. In order to obtain higher expression levels for a high-throughput screen (HTS) in 96-well plate format, ilvC_co was sub-cloned into pET22b(+). This plasmid also ads a his-tag to the C-terminus of the protein to facilitate purification. E. coli BL21 (DE3) (Lucigen, Middleton, Wis.) cells were transformed with pET22[ilvCco] and protein expression was performed in LB medium with ampicillin at 25° C. SDS PAGE analysis (FIG. 15) shows a comparison of crude extracts of BL21 (DE3) and GEVO1777 expressing KAR1.
[0548]Table 24 shows the specific activities in U/mg of KARI in lysates of GEVO1777 and BL21(DE3) being 15-fold higher in BL21 crude extract, mirroring the results shown in the SDS PAGE.
TABLE-US-00024 TABLE 24 Specific Activities of KARI in U/mg Expressed in GEVO1777 and BL21 (DE) measured with NADPH Strain/Construct U/mg Crude Extract pGV1777 in GEVO1777 0.03 pET22b[ilvCco] in BL21 (DE3) 0.45
[0549]Purification of his-tagged KARI expressed from pET22[ilvCco] in BL21(DE3) cells was first performed over a linear gradient to determine the proper amount of imidazol to elute KARI. Then, a step gradient was implemented and the protein was eluted at 40% elution buffer B (140 mM imidazol). A SDS PAGE documented the purity of the enriched protein (FIG. 16).
[0550]A quadruplet E. coli llvC mutant (R68D:K69L:K75V:R76D), which was described previously by Rane and coworkers (Rane et al., 1997, Arch Biochem Biophys 338: 83-89) was constructed using the respective primers listed in Table 6 (SEQ ID NO: 281 through SEQ ID NO 284) and cloned into pET22b(+) as described, but did not yield the cofactor switch that was described in the paper, although the ratio NADH/NADPH was 2.5 (wild-type 0.08). In fact, the specific activity of the quadruplet mutant on NADH was even worse than wild-type (Table 25), suggesting this mutant enzyme is not suited for the aforementioned aims.
TABLE-US-00025 TABLE 25 Comparison of specific activities from purified Ec_IlvC.sup.his6 and purified IlvC.sup.quadruplet-his6 quadruplet in U/mg measured on NAD(P)H U/mg with Variant U/mg with NADH NADPH NADH/NADPH Ec_IlvC.sup.his6 0.03 1 0.08 IlvC.sup.quadruplet-his6 0.45 0.02 2.5
[0551]Since the quadruplet KARI mutant did not yield the promised activity, the Ec_ilvC_coEc.sup.his6 gene (SEQ ID NO: 14) was used as starting point for engineering a cofactor switch. A structure alignment of E. coli KARI with spinach KARI was generated (FIG. 17) because spinach KARI was co-crystallized with NADPH. The position of the cofactor in the spinach KARI structure was in good agreement with the NADPH phosphate group in the E. coli KARI structure. Based on this, amino acid residues R68, A71, R76, S78, and Q110 seemed likely to be interacting with NADPH and therefore were chosen as targets in a site saturation mutagenesis experiment. Only residues R68 and R76 were found in the aforementioned quadruplet mutant. Residues K69 and K75 seemed less likely to be involved in cofactor binding.
[0552]Five individual site saturation libraries were generated and electro-competent E. coli BL21(DE3) cells were transformed with the desalted ligation mixtures. 88 clones of each library were screened for NAD(P)H depletion at 340 nm in microplates. Clones with an improved NADH/NADPH consumption ratio while maintaining or increasing their NADH activity were chosen for a rescreen. Variants that passed the rescreen were sequenced, expressed in shake flasks, purified, and characterized.
[0553]The first screening round resulted in several improved variants in terms of their specific activity on NADH (and NADPH for most of them) (Table 26). The first variant to favor NADH over NADPH was Ec_llvC.sup.S78D-his6 which showed a specific activity for NADH that equals the specific activity of Ec_llvC.sup.his6 for NAPDH (1 U/mg). Table 26 shows the variants resulting from the first round of site saturation mutagenesis compared to the parent Ec_llvC.sup.his6. All proteins were purified over a histrap column.
TABLE-US-00026 TABLE 26 Specific Activities for NADH and NADPH in U/mg Variant U/mg NADH U/mg NADPH NADH/NADPH No mutation 0.08 1 0.08 (Ec_IlvC.sup.his6) Ec_IlvCR68L-his6 0.27 1.15 0.23 Ec_IlvC.sup.A71T-his6 0.48 1.81 0.27 Ec_IlvC.sup.A71S-his6 0.57 2.65 0.22 Ec_IlvCR76G-his6 0.64 2.73 0.23 Ec_IlvCR76S-his6 0.59 1.51 0.39 Ec_IlvCR76T-his6 0.25 1 0.25 Ec_IlvCR76D-his6 0.26 0.69 0.38 Ec_IlvC.sup.S78D-his6 1 0.61 1.64 Ec_IlvC.sup.Q110A-his6 0.85 2 0.43 Ec_IlvC.sup.Q110V-his6 0.93 2 0.47
[0554]The three best variants Ec_llvC.sup.S78D-his6, Ec_llvC.sup.Q110A-his6, and Ec_llvC.sup.Q110V-his6 were characterized according to their specific activities [U/mg], kcal values [s-1], catalytic efficiencies [M-1*s-1] (FIG. 18), and KM values (Table 27).
TABLE-US-00027 TABLE 27 KM values of Ec_IlvC.sup.his6 compared to three variants resulting from the site saturation library KM [mM] KM [mM] Variant NADPH NADH Ec_IlvC.sup.his6 41 1075 Ec_IlvC.sup.S78D-his6 658 130 Ec_IlvC.sup.Q110V-his6 13 135 Ec_IlvC.sup.Q110A-his6 24 277
[0555]All three variants were improved compared to the parent Ec_llvC.sup.his6. Ec_llvC.sup.S78D-his6 was the first variant to show an actual preference of NADH over NADPH, while variants Ec_llvC.sup.Q110A-his6 and Ec_llvC.sup.Q110V-his6 showed drastic improvements in their overall catalytic efficiencies (FIG. 18). Table 28 contains a comparison of the KM values of Ec_llvC.sup.his6 with the three best variants resulting from the site saturation mutagenesis library on both cofactors. All variants showed improved KM values on NADH. While Ec_llvC.sup.Q110V-his6 and Ec_llvC.sup.Q110A-his6 had improved KM values on NADPH compared to wild-type, the KM value of variant Ec_llvC.sup.S78D-his6 on NADPH was decreased 16-fold from 1075 μM to 130 μM. The catalytic efficiencies on NADH were greatly improved as well. Ec_llvC.sup.his6 showed 1,000 M-1*s-1, while Ec_llvC.sup.S78D-his6 yielded 27,600 M-1*s-1.
TABLE-US-00028 TABLE 28 Catalytic efficiencies [M-1*s-1] for Ec_IlvC.sup.his6 and variants Ec_IlvC.sup.Q110V-his6, Ec_IlvC.sup.Q110A-his6, and Ec_IlvC.sup.S78D-his6 on NADPH (kcat/KM with NADH)/(kcat/KM of kcat/KM kcat/KM Ec_IlvC.sup.his6 with with NADH with NADH NADPH) Variant [M-1*s-1] [M-1*s-1] [%] Ec_IlvC.sup.his6 1000 87300 1% Ec_IlvC.sup.Q110V-his6 24800 569000 28% Ec_IlvC.sup.Q110A-his6 11063 301800 13% Ec_IlvC.sup.S78D-his6 27600 3770 32%
[0556]As a next step, the gene encoding variant Ec_llvC.sup.Q110V-his6 (SEQ ID NO: 23) was used as template to generate individual combinations of the mutation Q110V with other mutations: R68L, A71T, A71S, R76G, R76S, R76T, S78D, and R76D. After screening the variants as described above, the most promising ones were expressed, purified, and characterized. Table 29 lists the KM values in μM on NADPH and NADH for Ec_llvC.sup.his6, Ec_llvC.sup.Q110V-his6, and variants of Ec_llvC.sup.Q110V-his6, Variant Ec_llvC88-his6 containing amino acid mutations Q110V and S78D, showed the same KM value for NADH and for NADPH with 65 μM. The A71S mutation was introduced into Ec_llvC.sup.B8-his6 resulting in a variant Ec_llvC.sup.B8A71S-his6, which yielded 44% catalytic efficiency on NADH compared to the catalytic efficiency of wild-type KARI on NADPH (FIG. 19 and Table 30).
TABLE-US-00029 TABLE 29 KM values for Ec_IlvC.sup.his6, Ec_IlvC.sup.Q110V-his6, and variants of Ec_IlvC.sup.Q110V-his6 on NADPH and on NADH Variant KM for NADPH [mM] KM for NADH [mM] Ec_IlvC.sup.his6 41 1075 Ec_IlvC.sup.Q110V-his6 13 135 Ec_IlvC.sup.Q110VA71T-his6 37 80 Ec_IlvC.sup.Q110VA71S-his6 30 70 Ec_IlvC.sup.Q110VR76G-his6 47 87 Ec_IlvC.sup.Q110VR76S-his6 n.d. 223 Ec_IlvC.sup.B8-his6 65 65
TABLE-US-00030 TABLE 30 Catalytic efficiencies [M-1*s-1] for wild-type Ec_IlvC.sup.his6 and variants Ec_IlvC.sup.Q110V-his6, Ec_IlvC.sup.Q110A-his6, and Ec_IlvC.sup.S78D-his6 on NAD(P)H compared to Ec_IlvC.sup.B8-his6 and Ec_IlvC.sup.B8A71S-his6 kcat/KM kcat/KM (kcat/KM with NADH)/ with with (kcat/KM of NADH NADH Ec_IlvC.sup.his6 with NADPH) Variant [M-1*s-1] [M-1*s-1] [%] Ec_IlvC.sup.his6 1000 87300 1% Ec_IlvC.sup.Q110V-his6 24800 569000 28% Ec_IlvC.sup.Q110A-his6 11063 301800 13% Ec_IlvC.sup.S78D-his6 27600 3770 32% Ec_IlvC.sup.B8-his6 31775 34188 36% Ec_IlvC.sup.B8A71S-his6 38330 37459 44%
EXAMPLE 15
KARI Engineering by Recombination
[0557]The codon optimized gene Ec_ilvC_coEc.sup.his6 (SEQ ID NO: 14) and libraries thereof were cloned into pET22b(+) using primers KARIpETfor and KARIpETrev (Table 6). DNA constructs were analyzed by restriction digests, and also by DNA sequencing to confirm integrity and correct construction. Primers pETup and KARIpETrev (Table 6) were used as primers in standard DNA sequencing reactions to sequence pET22b(+) derivatives.
[0558]The recombination library was constructed using SOE PCR introducing mutations found at the five targeted sites while allowing for wild-type sequence as well. The first fragments were generated using degenerate primers R68A71recombfor and R68A71recombrev which covered the gene sequence coding for the region at amino acid positions 68/71 (Table 6). After assembling the long and the short fragment, the assembly product was DpnI digested for 1 h, separated on an agarose gel, freeze'n' squeeze (BioRad, Hercules, Calif.) treated, and finally pellet painted (Novagen, Gibbstown, N.J.). The clean assembly product served as template for the second round of SOE PCR introducing mutations at amino acid positions 76/78 using the following primers: R68A71recombfor, R68A71recombrev, R76S78recombfor, R76S78recombrev, G76S78recombfor, G76S78recombrev, S76S78recombfor, S76S78recombrev, T76S78recombfor, T76S78recombrev, D76S78recombfor, D76S78recombrev, R76D78recombfor, R76D78recombrev, G76D78recombfor, G76D78recombrev, S76D78recombfor, S76D78recombrev, T76D78recombfor, T76D78recombrev, D76D78recombfor, D76D78recombrev (Table 6). The mixture of primers was used, since degenerate codons would have expanded the library size immensely. Again, the assembly product served as template to complete the recombination library with amino acid position 110. The same procedure was applied as described for the first two rounds of SOE PCR. Primers used were again a mixture prepared out of equimolar concentrations of Q110Qfor, Q110Qrev, Q110Afor, Q110Arev, Q110Vfor, and Q110Vrev. After all sites were recombined, the insert was restriction digested with NdeI and XhoI, ligated into pET22b(+), and electro-competent BL21(D3) (Lucigen, Middleton, Wis.) were transformed. In order to oversample the library by approximately five-fold, one thousand clones were picked and cultured as described below. In order to check for possible biases (i.e. certain mutations occurring more frequently than others), 20 clones were randomly chosen for DNA sequence analysis.
[0559]As described in Example 14, the first screening round identified several individual point mutations within the KARI cofactor binding region that either improved NADH-dependent activity or were at least neutral (i.e. had neither a beneficial nor deleterious effect). These mutations, along with the wild-type amino acid residue are listed in Table 31.
TABLE-US-00031 TABLE 31 Amino Acid Mutations Included in the Recombinatorial Library Amino Acid Neutral or beneficial Total # (including Position Wild-type mutations identified wild-type) 68 R L 2 71 A T, S 3 76 R G, S, T, D 5 78 S D 2 110 Q A, V 3
[0560]A complete recombination library was constructed allowing for all beneficial and some neutral mutations (and including the wild-type residues) at each of the five sites. The total number of unique combinations was 180.
[0561]Generating all mutations using a single primer would result in a large library of ˜4,000. Thus, the present inventors built the library stepwise in three SOE reactions using primers mixed in equimolar amounts for each of three SOE reactions:
TABLE-US-00032 SOE 1: R68/A71, R68/T71, R68/S71, L68/A71, L68/ T71, L68/S71 SOE 2: A76/S78, G76/S78, S76/S78, T76/S78, D76/ S78, A76/D78, G76/D78, S76/D78, T76/D78, D76/D78, SOE3: Q11O, A11O, V11O
[0562]First, mutations at amino acid sites 68 and 71 were introduced into the Ec_ilvC_coEc.sup.his6 gene, followed by mutations at site 76 and finally, by mutations at site 110. After the library had been generated, it was ligated into pET22b(+). The resulting plasmid library was used to transform E. coli BL21(DE3) electro-competent cells. Cells were grown in 96-well plates according to the protocol for cell growth and protein expression in microplates as described in Example 14. The KARI enzyme activity of each of 1,000 individual transformants was determined using the high-throughput assay as described in Example 14.
[0563]Only 20% of the enzymes of the recombination library were active on NADH. After screening 1,000 clones using the NADH depletion assay at 340 nm, 26 KARI variants were selected for a rescreen by the high-throughput assay described in Example 14 and eight thereof were expressed in 25 ml LBamp medium in shake flasks according to the protocol for cell growth and protein expression in shake flasks as described in Example 14, purified according to the protocol for purification of KARI enzymes as described in Example 14, and NAD(P)H depletion at 340 nm was measured again. Two candidates Ec_llvC2H10-his6 (containing the amino acid substitutions A71S, R76D, S78D, and Q110A) and Ec_llvC6E6-his6 (containing the amino acid substitutions A71S, R76D, S78D, and Q110V) showed good specific activity on NADH and were only marginally active on NADPH. The other six variants showed lower specific activities on NADH (ranging from 0.44-0.55 U/mg) compared to the two favored variants Ec_llvC2H10-his6 and Ec_llvC6E6-his6 and higher specific activities on NADPH (0.72-2.62 U/mg). The KM values of variants Ec_llvC2H10-his6 and Ec_llvC6E6-his6 were measured and the catalytic efficiencies were calculated.
[0564]The kinetic parameters of the recombination variants and previously described KARI mutants are shown in Table 32. Both variants found in the recombination library showed an almost complete switch in cofactor preference from NADPH to NADH. The KM values of the mutants on NADH rival the KM value of KARI Ec_llvC.sup.his6 on NADPH (44.2 and 31.6 μM on NADH vs. 41 μM for Ec_llvC.sup.his6 on NADPH). The catalytic efficiencies of Ec_llvC2H10-his6 and Ec_llvC6E6-his6 on NADH (60322 and 74045 M-1*s-1, respectively) came very close to the catalytic efficiency of Ec_llvC.sup.his6 on NADPH (87300 M-1*s-1). The mutants described herein exhibit a complete reversal in cofactor specificity and the NADH-dependent activity approaches the NADPH-dependent activity of the wild-type enzyme. The best variant exhibited 85% activity (in terms of kcat/KM) on NADH compared to wild-type activity on NADPH.
TABLE-US-00033 TABLE 32 Kinetic parameters of Ec_IlvC.sup.his6, two of the enzymes described previously (Ec_IlvC.sup.B8-his6 and Ec_IlvC.sup.B8A71S-his6), as well as the two mutants Ec_IlvC2H10-his6 and Ec_IlvC6E6-his6 U/mg KM [μM] kcat [s-1] kcat/KM [M-1 * s-1] Variant NADH NADPH NADH NADPH NADH NADPH NADH NADPH Ec_IlvC.sup.his6 0.08 1.00 1,075 41 1.0 3.6 1,000 87,300 Ec_IlvC.sup.B8-his6 0.57 0.62 65 65 2.0 2.2 31,775 34,188 Ec_IlvC.sup.B8A71S-his6 0.57 0.66 53.5 63.4 2.0 2.4 38,330 37,459 Ec_IlvC2H10-his6 0.74 0.17 44.2 568 2.6 0.61 60,322 1,078 Ec_IlvC6E6-his6 0.65 0.07 31.6 653 2.3 0.2 74,045 386
[0565]The above data demonstrates the effects brought on by the beneficial mutations at positions 71 and 110. Moreover, aspartic acids at positions 76 and 78 electrostatically repel the phosphate of NADPH. It is noted that the electrostatic attraction of arginine to the NADPH phosphate is lost when R76 is mutated to an aspartic acid residue.
EXAMPLE 16
KARI Engineering by Random Mutagenesis in Yeast
[0566]The following example demonstrates increases in specific, NADH-dependent KARI activity.
[0567]Methods: Plasmid pGV2241 (SEQ ID NO: 124) carrying the Ec_ilvC_coSc6E6-his6 gene (SEQ ID NO: 33) served as template for generating the first error-prone FOR library using forward primer pGV1994ep_for and reverse primer pGV1994_rev. These primers are specific to the backbone pGV1102 (SEQ ID NO: 101) and bind 50 by upstream and downstream of the KARI insert to create an overlap for homologous recombination in yeast. Generally, three different MnCl2 concentrations were tested (100, 200, and 300 μM MnCl2) and the PCR compositions are summarized in Table 33.
TABLE-US-00034 TABLE 33 PCR set up for different concentrations of MnCl2 that were tested. The final volumes were 100 μL and amounts of ingredients are in μL final MnCl2 concentration [μM] 100 150 200 250 300 Template 1 1 1 1 1 primer forward 2 2 2 2 2 primer reverse 2 2 2 2 2 dNTP's 4 4 4 4 4 Taq buffer 10 10 10 10 10 MgCl2 28 28 28 28 28 Taq polymerase 1.6 1.6 1.6 1.6 1.6 MnCl2 (1 mM stock) 10 15 20 25 30 PCR grade water 41.4 36.4 31.4 26.4 21.4
[0568]The temperature profile was the following: 95° C. 3 min initial denaturation, 95° C. 30 s denaturation, 55° C. 30 s annealing, 72° C. 2 min elongation, 25 cycles, 5 min final elongation at 72° C.
[0569]The PCR products were checked on a 1% analytical TAE agarose gel, DpnI digested for 1 h at 37° C. to remove traces of template DNA, and then cleaned up using a 1% preparative TAE agarose gel. The agarose pieces containing the PCR products were put into Freeze'n' Squeeze tubes (BIORAD, catalog #732-6166) and frozen for 10 min at -20° C. Then, they were spun down at room temperature and 10,000 rpm to "squeeze" the buffer with the soluble DNA out of the agarose mesh. The volume of the eluted DNA/buffer mixture was estimated and then subjected to the pellet paint procedure (Novagen, catalog #69049-3), which was performed according to the manufacturer's manual. The dried pink DNA pellets were resuspended in 50 μL PCR grade water. In the meantime, the restriction digest of the backbone pGV1102 (SEQ ID NO: 101) was performed as follows: 10 μL of DNA, 32 μL PCR grade water, 5 μL NEB buffer 3 (10×), 2 μL NotI, and 1 μL SalI. After an incubation time of 3 h at 37° C., the digest was run out on an agarose gel and then pellet painted as described above. After determining the DNA concentration of cut vector and insert, 500 ng of each were mixed together, precipitated with pellet paint, and resuspended in 6 μL of PCR grade water. This mixture can be prepared a day before the transformation.
[0570]In the evening before the planned transformation, YPD medium (10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose) was inoculated with a single colony of GEVO1186 and incubated at 30° C. and 250 rpm over night. The next morning, a 20 mL YPD culture was started in a 250 ml Erlenmeyer flask without baffles with the overnight culture at an OD600 of 0.1. This culture was incubated at 30° C. and 250 rpm until it reached an OD600 of 1.3-1.5. When the culture had reached the desired OD600, 200 μL of freshly prepared sterile-filtered Tris-DTT (0.39 g 1,4-dithiothreitol per 1 mL of 1 M Tris, pH 8.0) were added and the culture was allowed to incubate at 30° C. and 250 rpm for another 15 min. The cells were then pelleted at 4° C. and 2,500×g for 3 min. After removing the supernatant, the pellet was resuspended in 10 mL of ice-cold buffer E and spun down again as described above. Then, the cell pellet was resuspended in 1 mL of sterile-filtered ice-cold buffer E (1.2 g Tris base, 92.4 g glucose, and 0.2 g MgCl2 per 1 L deionized water, adjusted to pH 7.5) and spun down one more time as before. After removal of the supernatant with a pipette, 200 μL of ice-cold buffer E (1.2 g/L Tris, 92.4 g/L glucose, and 0.2 g/L MgCl2, pH 7.5) were added and the pellet was gently resuspended. The 6 μL of insert/backbone mixture were split in half and added to 50 μL of electrocompetent GEVO1186 cells. The DNA/cell mixtures were transferred into 0.2 cm electroporation cuvettes (BioRad) and electroporated without a pulse controller at 0.54 kV and 25 μF. 1 mL of pre-warmed YPD medium was added immediately and the transformed cells were allowed to regenerate at 30° C. and 250 rpm in 15 mL round bottom culture tubes (Falcon). After 1 hour, the cells were spun down at 4° C. and 2,500×g for 3 min, and the pellets were resuspended in 1 mL pre-warmed SD-URA medium (1.7 g/L yeast nitrogen base, 5 g/L ammonium sulfate, 20 g/L glucose, with casamino acids but without uracil (CSM-URA). Different amounts of transformed cells were plated on SD-URA agar plats plates and incubated at 30° C. for 1.5 days or until the colonies were large enough to be picked with sterile toothpicks.
[0571]Single yeast colonies were picked with sterile toothpicks into shallow 96-well plates containing 300 μL of SC-URA medium (6.7 g/L Difco® Yeast Nitrogen Base, 14 g/L Sigma® Synthetic Dropout Media supplement (includes amino acids and nutrients excluding histidine, tryptophan, uracil, and leucine), 10 g/L casamino acids, 20 g/L glucose, 0.018 g/L adenine hemisulfate, and 0.076 g/L tryptophan) per well. Each plate encompassed 88 wells with variants, four wells with parent, three wells with GEVO1886 carrying pGV1102 as background control, and one well with medium only, which served as a sterility control. The plates were incubated at 250 rpm and 30° C. in a humidified plate shaker (Kuhner) over night. On the next morning, 50 μL of the overnight culture were transferred into 600 μL SC-URA medium in 96 well deep well plates (2 mL capacity per well). The cultures were allowed to grow for another 8 h at the same conditions, before they were spun down at 4° C. and 5000 rpm for 5 min. The supernatants were removed and the pellets were frozen at -20° C. until they were screened for activity as described in Example 14 above.
[0572]Improved variants were expressed and purified from GEVO1186. 20 mL SC-URA medium overnight cultures were grown at 30° C. and 250 rpm in 250 mL flasks and were then used to inoculate 96 well deep well plates on the next morning. 50 μL of the overnight cultures were transferred into 600 μL SC-URA medium per well. The plates were then grown at 30° C. and 250 rpm in a humidified plate shaker for 8 h. In order to the harvest, the cultures were transferred into 50 mL Falcon tubes and then spun down at 4° C. and 5,000 rpm for 10 min. The pellets were frozen until they were processed and purified as described in Example 14 above.
[0573]Results: Two rounds of error-prone PCR and screening were carried out. The libraries (˜2400 clones per library) were screened using the KARI high-throughput assay. KARI variants that exhibited an improved activity compared to their parent (total of 88 variants) were picked and rescreened in triplicate and five clones were selected for sequencing and purification. In the first round variant Ec_llvC.sup.P2D1-his6 (SEQ ID NO: 38), encoded by Ec_ilvC_coSc.sup.P201-his6 (SEQ ID NO: 37) was identified carrying the following mutations: D146G and G185R. This variant served as parent for the second round of error-prone PCR and screening which yielded variant Ec_llvC.sup.P201-A1-his6 (SEQ ID NO: 42), encoded by Ec_ilvC_coSc.sup.P2D1-A1-his6 (SEQ ID NO: 41) with one additional mutation (K433E). The biochemical properties were determined and are summarized in Table 34. A two-fold improvement of the specific activity in lysate and in the purified enzyme was observed after two rounds of error-prone PCR.
TABLE-US-00035 TABLE 34 Comparison of the biochemical properties of the parent Ec_IlvC6E6-his-6 with the variants found in round 1 (Ec_IlvC.sup.P2D1-his6) and 2 (Ec_IlvC.sup.P2D1-A1-his6). The variants were purified before characterization U/mg KM [μM] kcat [s-1] kcat/KM [M-1 * s-1] Variant NADH NADPH NADH NADPH NADH NADPH NADH NADPH Ec_IlvC6E6-his6 0.69 39 2.4 63,000 Ec_IlvC.sup.P2D1-his6 0.92 0.15 40 1432 3.3 0.54 82,650 377 Ec_IlvC.sup.P2D1-A1-his6 1.2 0.15 26 >1432 4.3 0.54 167,687 <377
EXAMPLE 17
NADH-Dependent Anaerobic Isobutanol Production
[0574]This example illustrates that an isobutanol producing microorganism which is engineered to carry NADH-dependent KARI and ADH enzymes produces isobutanol at higher yield compared to strains engineered to carry NADPH-dependent KARI and ADH enzymes. These strains also acquire the ability to produce isobutanol anaerobically.
[0575]A first set of anaerobic fermentations with isobutanol producing strains according to Table 35 were performed. Strain GEVO1993 is an E. coli strain in which the native ilvC gene was deleted and the other three steps of the isobutanol pathway (Bs_alsS1, Ec_ilvD_coEc and Ll_kivd1) were integrated into the chromosome.
TABLE-US-00036 TABLE 35 Strain/Plasmid combinations described herein. Cofactor usage of the isobutanol Plasmid Strain KARI gene ADH gene pathway pGV1777 GEVO1993 Ec_ilvC_coEc Ec_yqhD NADPH/ (native) NADPH pGV1925 GEVO1993 Ec_ilvC_coEc Ec_fucO NADPH/ NADH pGV1938 GEVO1993 Ec_ilvC_coEc.sup.S78D Ec_yqhD NADH/ (native) NADPH pGV1927 GEVO1993 Ec_ilvC_coEc.sup.S78D Ec_fucO NADH/ NADH
[0576]Overnight cultures of the GEVO1993 transformed with pGV1777 (SEQ ID NO: 118), pGV1925, pGV1938, or pGV1927 were started from individual colonies of previously transformed strains. These cultures were started in 3 mL M9 minimal medium (Miller, J. H. A Short Course in Bacterial Genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. 1992. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.), supplemented with 10 g/L yeast extract, 10 μM ferric citrate and trace metals, containing 8.5% glucose and the appropriate antibiotics in snap cap tubes about 14 h prior to the start of the fermentation. Isobutanol fermentations were then carried out in screw cap flasks containing 20 mL of the same medium that was inoculated with 0.2 mL of the overnight culture. The cells were incubated at 37° C./250 rpm until the strains had grown to an OD500 of 0.6-0.8 and were then induced with Isopropyl (β-D-1-thiogalactopyranoside at 1 mM final concentration.
[0577]Three hours after induction the cultures were shifted to anaerobic fermentation conditions by loosening the cap of the flasks and placing the flasks into to a Coy Laboratory Products Type B Vinyl anaerobic chamber (Coy Laboratory Products, Grass Lakes, Mich.) through an airlock in which the flasks were cycled three times with nitrogen and vacuum, and then filled with the a hydrogen gas mix (95% Nitrogen, 5% Hydrogen). Once the flasks were inside the anaerobic chamber, the flasks were closed again and incubated without shaking at 30° C. Inside the chamber, an anaerobic atmosphere (<5 ppm oxygen) was maintained through the hydrogen gas mix (95% Nitrogen, 5% Hydrogen) reacting with a palladium catalyst to remove oxygen. The flasks in the anaerobic chamber were swirled twice a day. Samples (2 mL) were taken at the time of the shift and at 21 h and 45 h after shifting to anaerobic conditions, spun down at 22,000 g for 1 min to separate the cell pellet from the supernatant and stored frozen at -20° C. until analysis. The samples were analyzed using High performance liquid chromatography (HPLC) and gas chromatography GC. All experiments were performed in triplicate.
[0578]The OD600 values of the cultures were similar amongst the three replicates. Notably, after 45 h, GEVO1993+pGV1927 (i.e. expressing NADH-dependent KARI and ADH) produced isobutanol at approximately twice the volumetric productivity, specific productivity, and titer. Surprisingly the theoretical yield increased from about 70% of theoretical to 96% of theoretical. Expressing only one NADH-dependent enzyme with the other enzyme being NADPH-dependent did not have an effect (Table 36).
TABLE-US-00037 TABLE 36 45 h performance parameters Vol. Spec. Anaerobic Productivity Productivity Yielda Titer Sample KARI/ADH [g/L/h] ± [g/L/h/OD] ± % theor. ± [g/L] ± GEVO1993 + Ec_IlvC/ 0.044 0.019 0.018 0.003 72 3 2.4 1.0 pGV1777 Ec_YqhD GEVO1993 + Ec_IlvC/ 0.031 0.002 0.017 0.003 55 4 1.9 0.1 pGV1925 Ec_FucO GEVO1993 + Ec_IlvC.sup.S78D/ 0.040 0.015 0.021 0.002 78 10 2.1 0.9 pGV1938 Ec_YqhD GEVO1993 + Ec_IlvC.sup.S78D/ 0.078 0.006 0.030 0.003 96 5 3.8 0.2 pGV1927 Ec_FucO a The anaerobic yield is calculated by dividing the isobutanol produced from time of anaerobic shift until 45 hours after the shift by the amount of glucose consumed during this time period
[0579]A second set of anaerobic fermentations with isobutanol producing strains according to Table 37 were performed to demonstrate that the of improved KARI variants correlates with an improvement of isobutanol production under anaerobic conditions.
TABLE-US-00038 TABLE 37 Strain/Plasmid combinations used for the second set of anaerobic fermentations. KARI ADH KARI (kcat/KM,NADH)/ # Plasmid Strain KARI gene gene kcat/KM,NADH (kcat/KM,NADPH) 1 pGV1927 GEVO1993 Ec_ilvC_coEc.sup.S78D Ec_fucO 27,600 7 2 pGV1976 GEVO1993 Ec_ilvC_coEc2H10 Ec_fucO 60,300 56 3 pGV1975 GEVO1993 Ec_ilvC_coEc6E6 Ec_fucO 74,000 192
[0580]The experiment was carried out as described above except that the cell cultures were induced at an OD600 of 0.8-1.0 instead of 0.6-0.8 and shifted to anaerobic conditions at and OD OD600 of 4.0-6.0 instead of 3 hours after induction. In addition, samples were taken at the time of the anaerobic shift and 24 h and 48 h after induction (i.e. 20 h and 44 h after the anaerobic shift, respectively).
[0581]44 hours after shift to anaerobic fermentation conditions, the trend for volumetric and specific productivity is the same as observed 20 hours after shift to anaerobic conditions: strains carrying improved KARI variants Ec_llvC2H10 and Ec_llvC6E6 produced isobutanol at higher volumetric and specific productivity as well as yield compared to strains carrying KARI variant Ec_llvC.sup.S78D (Table 38).
TABLE-US-00039 TABLE 38 44 h performance parameters Vol. Spec. anaerobic Productivity Productivity Yielda Titer Sample KARI/ADH [g/L/h] ± [g/L/h/OD] ± % theor. ± [g/L] ± GEVO1993 + Ec_IlvC.sup.S78D/ 0.215 0.005 0.037 0.002 79 12 10.9 0.3 pGV1927 Ec_FucO GEVO1993 + Ec_IlvC2H10/ 0.274 0.008 0.047 0.002 107 15 13.0 0.6 pGV1976 Ec_FucO GEVO1993 + Ec_IlvC6E6/ 0.270 0.032 0.047 0.005 97 2 12.5 1.5 pGV1975 Ec_FucO aThe anaerobic yield is calculated by dividing the isobutanol produced from time of anaerobic shift until 44 hours after the shift by the amount of glucose consumed during this time period
EXAMPLE 18
NADH-Dependent Anaerobic Isobutanol Production in Yeast
[0582]This example illustrates that isobutanol producing yeast microorganisms engineered to carry NADH-dependent KARI and ADH enzymes produce isobutanol at higher yields compared to isobutanol producing yeast microorganisms engineered to carry NADPH-dependent KARI and/or ADH enzymes. These strains also produce isobutanol anaerobically.
[0583]Cultures of GEVO2710, GEVO2711 and GEVO2799 transformed with pGV2227 (SEQ ID NO: 123) or pGV2242 (SEQ ID NO: 125) and cultures of GEVO2710, and GEVO2799 transformed with pGV2020 (SEQ ID NO: 121) or pGV2082 (SEQ ID NO: 122) were started from individual colonies of previously transformed and purified strains. These cultures were started in 14 ml round-bottom snap-cap test tubes containing 3 ml of YPD medium supplemented with 0.2 g/L G418 antibiotic, and 1% (v/v) of a stock solution containing 3 g/L ergosterol and 66 g/L Tween 80 dissolved in ethanol. The snap-cap test tubes were not closed completely so that air would vent in/out of the tubes. After growth for about 10 hours at 30° C. shaking at 250 rpm, these cultures were added to 47 ml of the same medium in 250 ml non-baffled flasks with sleeve closures and incubated for about 14 hours at 30° C. shaking at 250 rpm. Isobutanol fermentations were then carried out after harvesting the cells from the 50 ml cultures by centrifugation, and resuspending the cell pellets in f 50 ml of the same medium in 250 ml non-baffled flasks to an initial optical density (OD600) of 3-6.
[0584]Anaerobic fermentations were carried out by inoculating flasks with screw-cap closures as above and placing the flasks with loose caps into to a Coy Laboratory Products Type B Vinyl anaerobic chamber (Coy Laboratory Products, Grass Lakes, Mich.) through an airlock in which the flasks were cycled three times with nitrogen and vacuum, and then filled with a hydrogen gas mix (95% Nitrogen, 5% Hydrogen). The flasks were moved inside the anaerobic chamber from the airlock and the screw-caps on the flasks were closed inside the anaerobic chamber. Inside the chamber, an anaerobic atmosphere (<5 ppm oxygen) was maintained through the hydrogen gas mix (95% Nitrogen, 5% Hydrogen) reacting with a palladium catalyst to remove oxygen. The flasks were then removed from the anaerobic chamber and incubated outside the anaerobic chamber at 30° C. shaking at 75 rpm. Samples (2 ml) were taken at the beginning of the incubation of the anaerobic fermentations and after 24 hours, 48 hours and 72 hours of incubation. The samples taken at the beginning of the incubation were taken before moving the flasks into the anaerobic chamber. The 24 hour and 48 hour samples were taken by moving the flasks into the anaerobic chamber through the airlock as above, opening the flasks in the anaerobic chamber to remove the samples, re-closing the flasks in the anaerobic chamber and removing the flasks from the anaerobic chamber for continued incubation. The 72 hour samples were taken outside of the anaerobic chamber because these were the final samples from the flasks.
[0585]Samples from fermentations were centrifuged for 10 minutes at 18,000 g to separate the cells from the supernatant. The supernatant was removed and stored under refrigeration until analyzed by gas chromatography and high performance liquid chromatography as described above. All experiments were performed in triplicate.
[0586]In the anaerobic fermentations the OD600 values of the cultures were similar amongst the three replicates. Notably, after 72 hours in anaerobic fermentations, GEVO2710+pGV2242, GEVO2711+pGV2242 and GEVO2799+pGV2242 (i.e. strains expressing an NADH-dependent KARI) produced isobutanol at an approximately 1.25- to 2-fold higher volumetric productivity, specific productivity, and titer than the same strains containing pGV2227 (i.e. strains expressing an NADPH-dependent KARI). The anaerobic yield increased from about 16-25% of theoretical to 22-35% of theoretical (Table 39).
TABLE-US-00040 TABLE 39 72 hour performance parameters from anaerobic fermentations KARI/ADH Vol. Spec. Specific overexpressed Productivity Productivity Yield Titer Sample from plasmid [g/L/h] ± [g/L/h/OD] ± % theor. ± [g/L/OD] ± GEVO2710 + None/ 0.000 0.000 0.0001 0.0000 1 0 0.01 0.00 pGV2020 None GEVO2710 + Ec_IlvC.sup.Q110V/ 0.006 0.001 0.0014 0.0001 21 2 0.10 0.01 pGV2082 Dm_Adh GEVO2710 + Ec_IlvC.sup.Q110V/ 0.006 0.001 0.0017 0.0003 17 9 0.12 0.02 pGV2227 Ll_AdhA GEVO2710 + Ec_IlvC.sup.P2D1/ 0.011 0.001 0.0029 0.0003 22 2 0.21 0.02 pGV2242 Ll_AdhA GEVO2799 + None/ 0.001 0.000 0.0002 0.0000 6 1 0.01 0.00 pGV2020 None GEVO2799 + Ec_IlvC.sup.Q110V/ 0.010 0.000 0.0019 0.0003 38 2 0.14 0.02 pGV2082 Dm_Adh GEVO2799 + Ec_IlvC.sup.Q110V/ 0.009 0.001 0.0014 0.0002 20 2 0.10 0.01 pGV2227 Ll_AdhA GEVO2799 + Ec_IlvC.sup.P2D1/ 0.014 0.003 0.0026 0.0003 33 10 0.19 0.03 pGV2242 Ll_AdhA GEVO2711 + Ec_IlvC.sup.Q110V/ 0.008 0.000 0.0020 0.0000 24 2 0.14 0.00 pGV2227 Ll_AdhA GEVO2711 + Ec_IlvC.sup.P2D1/ 0.014 0.004 0.0025 0.0008 37 8 0.18 0.06 pGV2242 Ll_AdhA
EXAMPLE 19
Overexpression of an NADPH-Dependent GAPDH, GDP1
[0587]The purpose of this example is to describe how overexpression of an NADPH-dependent GAPDH can improve isobutanol production under anaerobic conditions.
[0588]GDP1 is expressed from plasmid pGV1573 (SEQ ID NO: 106) together with an isobutanol biosynthetic pathway expressed from pGV1485 (SEQ ID NO: 103) and pSA69. As a control the plasmid pGV1573 is replaced by the empty version of this plasmid pGV1572 (SEQ ID NO: 105). These plasmids are transformed into GEVO1859ΔgapA. Overnight cultures of Strain 1: GEVO1859 ΔgapA, pGV1573, pGV1485, pSA69 and Strain 2: GEVO1859ΔgapA, pGV1572, pGV1485, pSA69 are started from individual colonies of previously transformed strains. These cultures are started in 3 mL M9 minimal medium (Miller, J. H. A Short Course in Bacterial Genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. 1992. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.), supplemented with 10 g/L yeast extract, 10 μM ferric citrate and trace metals, containing 8.5% glucose and the appropriate antibiotics in snap cap tubes about 14 h prior to the start of the fermentation. Isobutanol fermentations are then carried out in screw cap flasks containing 20 mL of the same medium that was inoculated with 0.2 mL of the overnight culture. The cells are incubated at 37° C./250 rpm until the strains had grown to an OD600 of 0.6-0.8 and are then induced with Isopropyl β-D-1-thiogalactopyranoside at 1 mM final concentration.
[0589]Three hours after induction the cultures are shifted to anaerobic fermentation conditions by loosening the cap of the flasks and placing the flasks into to a Coy Laboratory Products Type B Vinyl anaerobic chamber (Coy Laboratory Products, Grass Lakes, Mich.) through an airlock in which the flasks are cycled three times with nitrogen and vacuum, and then filled with the a hydrogen gas mix (95% Nitrogen, 5% Hydrogen). Once the flasks are inside the anaerobic chamber, the flasks are closed again and incubated without shaking at 30° C. Inside the chamber, an anaerobic atmosphere (<5 ppm oxygen) was maintained through the hydrogen gas mix (95% Nitrogen, 5% Hydrogen) reacting with a palladium catalyst to remove oxygen. The flasks in the anaerobic chamber are swirled twice a day. Samples (2 mL) are taken at the time of the shift and at 24 h and 48 h after inoculation, spun down at 22,000 g for 1 min to separate the cell pellet from the supernatant and stored frozen at -20° C. until analysis. The samples are analyzed using High performance liquid chromatography (HPLC) and gas chromatography GC. All experiments are performed in duplicate.
EXAMPLE 20
Overexpression of NADPH-Dependent GADPHs GDP1 and gapC
[0590]pGV1572 (SEQ ID NO: 105) (PLlacO, p15A, CmR) was constructed as an empty vector compatible with the plasmids pGV1698 (SEQ ID NO: 112) and pGV1655 (SEQ ID NO: 109) for the expression of the isobutanol pathway. The GAPDHs from Kluyveromyces lactis, and Clostridium acetobutylicum were cloned into pGV1572 to make pGV1573 (SEQ ID NO: 106) (PLlacO1::GDP1, p15A, CmR), and pGV1573 (SEQ ID NO: 107) (PLlacO1::GapC, p15A, CmR) respectively. K. lactis GAPDH was subcloned from pGV1323 (SEQ ID NO: 102), which contains the GDP1 gene cloned from genomic DNA of K. lactis. GapC (C. acetobutylicum) was cloned from genomic DNA using primers 1049 and 1050.
[0591]E. coli DH5αZ1 (Lutz, R. and Bujard, H, Nucleic Acids Research (1997) 25 1203-1210) was chosen as the host strain. This strain contains the Z1 integration which provides overexpression of lacl from a lacIq expression cassette. DH5aZ1 was transformed with pGV1572, pGV1573, and pGV1575. Transformants were used to inoculate 5 mL cultures, which were incubated at 37° C., 250 rpm overnight. 50 mL cultures were inoculated with 1 mL overnight culture and incubated at 37° C., 250 rpm. The cultures were induced with IPTG when OD500 was approximately 0.6 and incubated at 30° C., 250 rpm for 2 hours. The cultures were centrifuged at 2700×g at 4° C. for 10 min and the pellets were frozen at -80° C.
[0592]Pellets were resuspended with lysis buffer to 40% (w/v). (lysis buffer was the same as the reaction buffer but without substrate and cofactors). Cells were lysed in a bead mill using 3 times 1 min intervals, placing them on ice for 2 min in between each run. The lysate was centrifuged at 25000×g at 4° C. for 10 min, the supernatant was kept on ice and it was used as whole cell lysate for the enzyme assays.
[0593]The total reaction volume was 100 μL consisting of 90 μL of Reaction Buffer: 50 mM glycine buffer pH 9.5, 5 mM EDTA, 40 mM triethanolamine, 3 mM beta-mercaptoethanol, 6 mM NAD+ or NADP+, and 10 μL lysate. 10 μL of lysate were pipette into a UV permeable 96 well plate. 90 μL of reaction buffer was added to the lysate and mixed well by pipetting up and down. The plate was read for 5 min at 340 nm. Results are shown in Table 40.
TABLE-US-00041 TABLE 40 Volumetric and specific activity of various GAPDH with NADP.sup.+ NADP.sup.+ Sp. Activity Volumetric (nmol/min/ Activity μg total Lysate Name (mU/ml) cell protein) pGV# organism gapC 10.022 0.010 1575 C. acetobutylicum GDP1 26.849 0.031 1573 K. lactis Control 3.819 0.005 1572 (DH5az1)
[0594]DH5aZ1 was the host strain for all the plasmids and has its own indigenous GAPDH. The results show that the GAPDH enzymes are expressed and active in E. coli. The strain expressing GDP1 had more than 6 times higher in vitro GAPDH specific activity with the cofactor NADPH than the control strain not overexpressing GAPDH. The strain overexpressing gapC had twice the in vitro GAPDH specific activity with the cofactor NADPH than the control strain not overexpressing GAPDH.
EXAMPLE 21
NADPH-Dependent GAPDH in Yeast
[0595]The purpose of this example is to describe how an isobutanol producing yeast which is engineered to express NADPH-dependent GAPDH and produce isobutanol anaerobically.
[0596]A yeast strain, GEVO5001, which expresses the isobutanol biosynthetic pathway and is deficient in pyruvate decarboxylase activity, is engineered to overproduce the K. lactis Gdp1. pGV6001 is a yeast integration plasmid carrying a hygromycin resistance marker and the GDP1 gene under the strong constitutive promoter from TDH3. This plasmid is linearized and transformed into GEVO5001 to generate GEVO5003. Expression of GDP1 is confirmed by qRT-PCR. Once confirmed, GEVO5003 and the parent strain GEVO5001 are used in fermentations for the production of isobutanol, Two fermentations are performed with the two strains. Fermentation 1 is an aerobic fermentation and Fermentation 2 is an anaerobic fermentation.
EXAMPLE 22
pyk Bypass 1
[0597]This example illustrates that an isobutanol producing microorganism which is engineered to bypass the pyruvate kinase reaction shows increased productivity, titer and yield of isobutanol compared to the control strain without said engineering.
[0598]For the pyk bypass experiment, GEVO1385, GEVO1725 (triple deletion strain-tet repressor), and GEVO1751 were transformed with pGV1655 (SEQ ID NO: 109), pGV1698 (SEQ ID NO: 112), and pGV1490 (SEQ ID NO: 104) or pGV1661 (SEQ ID NO: 110). Strains GEVO1725 and GEVO1751 contain the deletions of pyruvate kinase and of the NADH dependent malic enzyme which are part of the pyruvate bypass engineering. All of these transformants were tested in isobutanol fermentations.
[0599]The aforementioned strains were grown overnight in two biological replicates for each strain in M9+A5 salts+FeCl3+10 g/L YE media and the appropriate antibiotics in 14 ml snap cap tubes and incubated at 37° C., 250 rpm. Screw cap flasks with 20 ml M9+A5 salts+FeCl3+10 g/L YE media and the appropriate antibiotics were inoculated with overnight culture to an OD600 of 0.1. The cells were incubated at 37° C., 250 rpm until they were grown to an OD600 of 0.6-0.8 and induced with IPTG [1 mM] and aTc [100 ng/ml]. Afterwards the cultures were incubated at 30° C., 250 rpm. Samples were taken of the medium, at 24 h and 48 h after inoculation. Samples were centrifuged at 15000 g for 1 min to separate the cell pellet from the supernatant and stored in -20° C. until sample submission. The samples were analyzed using High performance liquid chromatography (HPLC) and gas chromatography (GC).
[0600]The triple deletion strains GEVO1725 and GEVO1751 have a severe growth defect which is partially rescued by introduction of pGV1661.
[0601]The analysis of the fermentation data shows that the partial deletion strain, GEVO1750, with pGV1661 only has negative effects on isobutanol production (Tables 41, 42). However, at the 24 h time point the triple deletion strain with and without the tet repressor (GEVO1725 and GEVO1751 respectively) shows increased yield (Table 41). GEVO1725 shows a 20% increase in yield, with specific productivity similar to the control strain. GEVO1751 shows a 13% increase in yield and specific productivity.
TABLE-US-00042 TABLE 41 Analysis of the second pyk bypass fermentation from the 24 hour time point Volumetric Specific Productivity Productivity Titer Yield Samples 24 h [g/L/h] ± [g/L/h/OD] ± [g/L] ± [g/g] ± GEVO1385 + pGV1655, 0.205 0.008 0.031 0.001 4.93 0.18 0.277 0.002 pGV1698, pGV1490 (control) GEVO1385 + pGV1655, 0.197 0.003 0.028 0.002 4.65 0.01 0.285 0.035 pGV1698, pGV1661 (control) GEVO1725 + pGV1655, 0.125 0.009 0.034 0.005 2.83 0.19 0.331 0.029 pGV1698, pGV1490 GEVO1725 + pGV1655, 0.184 0.002 0.031 0.001 4.16 0.04 0.333 0.004 pGV1698, pGV1661 GEVO1750 + pGV1655, 0.144 0.004 0.022 0.001 3.30 0.14 0.267 0.001 pGV1698, pGV1490 GEVO1750 + pGV1655, 0.080 0.005 0.013 0.001 1.84 0.09 0.305 pGV1698, pGV1661 GEVO1751 + pGV1655, 0.138 0.006 0.031 0.001 3.09 0.13 0.303 0.008 pGV1698, pGV1490 GEVO1751 + pGV1655, 0.204 0.004 0.035 0.001 4.55 0.08 0.318 0.006 pGV1698, pGV1661
TABLE-US-00043 TABLE 42 Analysis of the second pyk bypass fermentation from the 48 hour time point Volumetric Specific Productivity Productivity Titer Yield samples 48 h [g/L/h] ± [g/L/h/OD] ± [g/L] ± [g/g] ± GEVO1385 + pGV1655, 0.128 0.011 0.023 0.002 6.14 0.53 0.271 0.004 pGV1698, pGV1490 (control) GEVO1385 + pGV1655, 0.141 0.029 0.023 0.005 6.75 1.41 0.263 0.002 pGV1698, pGV1661 (control) GEVO1725 + pGV1655, 0.070 0.002 0.024 0.002 3.25 0.10 0.299 0.009 pGV1698, pGV1490 GEVO1725 + pGV1655, 0.101 0.006 0.024 0.002 4.72 0.28 0.309 0.005 pGV1698, pGV1661 GEVO1750 + pGV1655, 0.102 0.013 0.018 0.002 4.77 0.54 0.277 0.013 pGV1698, pGV1490 GEVO1750 + pGV1655, 0.085 0.003 0.015 0.001 4.02 0.13 0.261 0.018 pGV1698, pGV1661 GEVO1751 + pGV1655, 0.093 0.004 0.029 0.001 4.29 0.16 0.267 0.006 pGV1698, pGV1490 GEVO1751 + pGV1655, 0.123 0.002 0.041 0.001 5.68 0.06 0.302 0.009 pGV1698, pGV1661
[0602]To verify that maeB, ppc, and mdh were expressed, cell lysates were made from GEVO1780 transformed with the above plasmids and run on a protein gel (FIG. 20).
[0603]The gel shows that all pathway enzymes are expressed in GEVO1780 with pGV1490 (Ec_llvD=65.5 kD, Ll_Kivd1/Bs_AlsS1=60.9 kD, Ec_llvC=54.1 kD). The gel also shows that all pathway enzymes and Ppc (99 kD), MaeB (82 kD), and Mdh (32 kD) are expressed in GEVO1780 with pGV1661.
EXAMPLE 23
pyk Bypass 2
[0604]This example illustrates that an isobutanol producing microorganism which is engineered to bypass the pyruvate kinase reaction shows increased productivity, titer and yield of isobutanol compared to the control strain without overexpression of ppc or pck.
[0605]Both plasmid constructs (pGV1661 (SEQ ID NO: 110) and pGV1772) were sequence verified. GEVO1725, and GEVO1751 were transformed with isobutanol pathway plasmids pGV1655 (SEQ ID NO: 109) and pGV1698 (SEQ ID NO: 112), and pyk bypass plasmids pGV1661 (ppc) or pGV1772 (pck). The controls were the same strains and pathway plasmids, but with the empty vector, pGV1490 (SEQ ID NO: 104), in place of pGV1661 or pGV1772. Strains GEVO1725 and GEVO1751 have deletions of pyruvate kinase (pykAF) and of the NADH dependent malic enzyme, maeA, which are part of the pyruvate kinase bypass engineering. The difference between GEVO1725 and GEVO1751 is that GEVO1725 does not have the tet repressor, and therefore, pGV1490, pGV1661, and pGV1772 are constitutively expressed in this strain.
[0606]All of these transformants were tested in isobutanol fermentations.
[0607]Overnight cultures were started in duplicate for each transformation in 3 mL M9+A5 salts+FeCl3+10 g/L YE media and the appropriate antibiotics in 14 mL snap cap tubes and incubated at 37° C., 250 rpm. Screw cap flasks with 20 mL M9+A5 salts+FeCl3+10 g/L YE media and the appropriate antibiotics were inoculated to a starting OD600 of 0.1 with overnight culture. The cells were incubated at 37° C., 250 rpm until they reached an OD600 of 0.6-0.8 and were then induced with IPTG [1 mM] and aTc [1 ng/mL]. After induction, the cultures were switched to incubation at 30° C., 250 rpm. Samples were taken of the cultures at 24 and 48 hours after inoculation and OD600 and pH were measured. Samples were centrifuged at 22,000×g for 5 min and the supernatant was collected and stored at -20° C. until sample submission. After 48 hour samples were taken, the remainder of the culture was transferred to a 50 ml tube, centrifuged at 4000×g, for 10 min at 4° C. The supernatant was removed, and the cell pellet was stored at -80° C. The samples were analyzed using High performance liquid chromatography (HPLC) and gas chromatography (GC).
[0608]The deletion strains with pck (pGV1772) had greater specific productivities than the strains with ppc (pGV1661). When ppc is used in the pyk bypass system in GEVO1725 and GEVO1751, the specific productivity of these strains increased by 3% in GEVO1751 and by 13% in GEVO1725 compared to GEVO1385 with the empty vector. When pck is used instead of ppc, the specific productivity increased by 43% in GEVO1725 and by 50% in GEVO1751. Both of the deletion strains show improved volumetric and specific productivity, titer, and yield when pGV1661 and pGV1772 are expressed compared to the empty vector (Table 43).
TABLE-US-00044 TABLE 43 Isobutanol production at 24 hours for pyk bypass system with ppc or pck Volumetric Specific Productivity Productivity Titer Yield samples 24 h [g/L/h] ± [g/L/h/OD] ± [g/L] ± [g/g] ± GEVO1725 empty 0.126 0.001 0.033 0.001 3.03 0.03 0.224 0.005 vector GEVO1725 pGV1661 0.266 0.003 0.045 0.001 6.38 0.07 0.304 0.022 GEVO1725 pGV1772 0.311 0.021 0.057 0.003 7.46 0.49 0.306 0.006 GEVO1751 empty 0.159 0.005 0.033 0.001 3.83 0.1 0.218 0.002 vector GEVO1751 pGV1661 0.262 0.054 0.041 0.005 6.29 1.29 0.236 0.035 GEVO1751 pGV1772 0.309 0.049 0.06 0.002 7.41 1.18 0.292 0.005
EXAMPLE 24
NADH Kinase and NADP+ Phosphatase in Yeast
[0609]The purpose of this example is to describe how an isobutanol producing yeast which is engineered to express NADPH biosynthesis enzymes to convert NADH into NADPH can produce isobutanol under anaerobic conditions.
[0610]A yeast strain GEVO5001 which expresses the isobutanol biosynthetic pathway and is deficient in pyruvate decarboxylase activity is engineered to express NADH kinase and NADP+ phosphatase. pGV6000, which is a yeast integration plasmid carrying an hygromycin resistance marker, NADH kinase and NADP+ phosphatase, is linearized by restriction digestion and transformed into GEVO5001. NADH kinase and NADP+ phosphatase are expressed using the strong constitutive promoters from TEF1 and TDH3, respectively. Clones in which the NADH kinase and NADP+ phosphatase are first identified by resistance to hygromycin. The clones are confirmed to be expressing NADH kinase and NADP+ phosphatase by qRT-PCR. The resulting strain, GEVO5002, along with the parent strain, GEVO5001, is used in fermentations for production of isobutanol.
EXAMPLE 25
Metabolic Transhydrogenation in Yeast
[0611]This example describes an isobutanol producing yeast which is engineered to convert NADH into NADPH through the combination of two redox enzymes that are catalyzing a conversion that is part of the same pathway wherein one redox enzyme oxidizes NADH and the other redox enzyme reduces NADP+.
[0612]The yeast strain, GEVO5001, is a yeast strain that has been engineered to be deficient in pyruvate decarboxylase activity and also to express the isobutanol pathway. A pyruvate bypass is generated by overexpressing in this yeast the genes for (a) pyruvate carboxylase (PYC1 or PYC2), (b) malate dehydrogenase, MDH2, and (c) malic enzyme (maeB). These genes are cloned to generate the yeast integration plasmid, pGV6004. This plasmid carries the hygromycin resistance marker and expresses PYC1, MDH2 and maeB under the strong promoters from ADH1, TEF1 and TDH3, respectively. pGV6004 is linearized and transformed into GEVO5001 to generate GEVO5006. Over-expressions of PYC1, MDH2 and maeB are confirmed by qRT-PCR.
[0613]The foregoing detailed description has been given for clearness of understanding only and no unnecessary limitations should be understood there from as modifications will be obvious to those skilled in the art.
[0614]While the invention has been described in connection with specific embodiments thereof, it will be understood that it is capable of further modifications and this application is intended to cover any variations, uses, or adaptations of the invention following, in general, the principles of the invention and including such departures from the present disclosure as come within known or customary practice within the art to which the invention pertains and as may be applied to the essential features hereinbefore set forth and as follows in the scope of the appended claims.
[0615]The disclosures, including the claims, figures and/or drawings, of each and every patent, patent application, and publication cited herein are hereby incorporated herein by reference in their entireties.
Sequence CWU
1
SEQUENCE LISTING
<160> NUMBER OF SEQ ID NOS: 330
<210> SEQ ID NO 1
<211> LENGTH: 1533
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 1
atgcgaattg gcataccaag agaacggtta accaatgaaa cccgtgttgc agcaacgcca 60
aaaacagtgg aacagctgct gaaactgggt tttaccgtcg cggtagagag cggcgcgggt 120
caactggcaa gttttgacga taaagcgttt gtgcaagcgg gcgctgaaat tgtagaaggg 180
aatagcgtct ggcagtcaga gatcattctg aaggtcaatg cgccgttaga tgatgaaatt 240
gcgttactga atcctgggac aacgctggtg agttttatct ggcctgcgca gaatccggaa 300
ttaatgcaaa aacttgcgga acgtaacgtg accgtgatgg cgatggactc tgtgccgcgt 360
atctcacgcg cacaatcgct ggacgcacta agctcgatgg cgaacatcgc cggttatcgc 420
gccattgttg aagcggcaca tgaatttggg cgcttcttta ccgggcaaat tactgcggcc 480
gggaaagtgc caccggcaaa agtgatggtg attggtgcgg gtgttgcagg tctggccgcc 540
attggcgcag caaacagtct cggcgcgatt gtgcgtgcat tcgacacccg cccggaagtg 600
aaagaacaag ttcaaagtat gggcgcggaa ttcctcgagc tggattttaa agaggaagct 660
ggcagcggcg atggctatgc caaagtgatg tcggacgcgt tcatcaaagc ggaaatggaa 720
ctctttgccg cccaggcaaa agaggtcgat atcattgtca ccaccgcgct tattccaggc 780
aaaccagcgc cgaagctaat tacccgtgaa atggttgact ccatgaaggc gggcagtgtg 840
attgtcgacc tggcagccca aaacggcggc aactgtgaat acaccgtgcc gggtgaaatc 900
ttcactacgg aaaatggtgt caaagtgatt ggttataccg atcttccggg ccgtctgccg 960
acgcaatcct cacagcttta cggcacaaac ctcgttaatc tgctgaaact gttgtgcaaa 1020
gagaaagacg gcaatatcac tgttgatttt gatgatgtgg tgattcgcgg cgtgaccgtg 1080
atccgtgcgg gcgaaattac ctggccggca ccgccgattc aggtatcagc tcagccgcag 1140
gcggcacaaa aagcggcacc ggaagtgaaa actgaggaaa aatgtacctg ctcaccgtgg 1200
cgtaaatacg cgttgatggc gctggcaatc attctttttg gctggatggc aagcgttgcg 1260
ccgaaagaat tccttgggca cttcaccgtt ttcgcgctgg cctgcgttgt cggttattac 1320
gtggtgtgga atgtatcgca cgcgctgcat acaccgttga tgtcggtcac caacgcgatt 1380
tcagggatta ttgttgtcgg agcactgttg cagattggcc agggcggctg ggttagcttc 1440
cttagtttta tcgcggtgct tatagccagc attaatattt tcggtggctt caccgtgact 1500
cagcgcatgc tgaaaatgtt ccgcaaaaat taa 1533
<210> SEQ ID NO 2
<211> LENGTH: 510
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 2
Met Arg Ile Gly Ile Pro Arg Glu Arg Leu Thr Asn Glu Thr Arg Val
1 5 10 15
Ala Ala Thr Pro Lys Thr Val Glu Gln Leu Leu Lys Leu Gly Phe Thr
20 25 30
Val Ala Val Glu Ser Gly Ala Gly Gln Leu Ala Ser Phe Asp Asp Lys
35 40 45
Ala Phe Val Gln Ala Gly Ala Glu Ile Val Glu Gly Asn Ser Val Trp
50 55 60
Gln Ser Glu Ile Ile Leu Lys Val Asn Ala Pro Leu Asp Asp Glu Ile
65 70 75 80
Ala Leu Leu Asn Pro Gly Thr Thr Leu Val Ser Phe Ile Trp Pro Ala
85 90 95
Gln Asn Pro Glu Leu Met Gln Lys Leu Ala Glu Arg Asn Val Thr Val
100 105 110
Met Ala Met Asp Ser Val Pro Arg Ile Ser Arg Ala Gln Ser Leu Asp
115 120 125
Ala Leu Ser Ser Met Ala Asn Ile Ala Gly Tyr Arg Ala Ile Val Glu
130 135 140
Ala Ala His Glu Phe Gly Arg Phe Phe Thr Gly Gln Ile Thr Ala Ala
145 150 155 160
Gly Lys Val Pro Pro Ala Lys Val Met Val Ile Gly Ala Gly Val Ala
165 170 175
Gly Leu Ala Ala Ile Gly Ala Ala Asn Ser Leu Gly Ala Ile Val Arg
180 185 190
Ala Phe Asp Thr Arg Pro Glu Val Lys Glu Gln Val Gln Ser Met Gly
195 200 205
Ala Glu Phe Leu Glu Leu Asp Phe Lys Glu Glu Ala Gly Ser Gly Asp
210 215 220
Gly Tyr Ala Lys Val Met Ser Asp Ala Phe Ile Lys Ala Glu Met Glu
225 230 235 240
Leu Phe Ala Ala Gln Ala Lys Glu Val Asp Ile Ile Val Thr Thr Ala
245 250 255
Leu Ile Pro Gly Lys Pro Ala Pro Lys Leu Ile Thr Arg Glu Met Val
260 265 270
Asp Ser Met Lys Ala Gly Ser Val Ile Val Asp Leu Ala Ala Gln Asn
275 280 285
Gly Gly Asn Cys Glu Tyr Thr Val Pro Gly Glu Ile Phe Thr Thr Glu
290 295 300
Asn Gly Val Lys Val Ile Gly Tyr Thr Asp Leu Pro Gly Arg Leu Pro
305 310 315 320
Thr Gln Ser Ser Gln Leu Tyr Gly Thr Asn Leu Val Asn Leu Leu Lys
325 330 335
Leu Leu Cys Lys Glu Lys Asp Gly Asn Ile Thr Val Asp Phe Asp Asp
340 345 350
Val Val Ile Arg Gly Val Thr Val Ile Arg Ala Gly Glu Ile Thr Trp
355 360 365
Pro Ala Pro Pro Ile Gln Val Ser Ala Gln Pro Gln Ala Ala Gln Lys
370 375 380
Ala Ala Pro Glu Val Lys Thr Glu Glu Lys Cys Thr Cys Ser Pro Trp
385 390 395 400
Arg Lys Tyr Ala Leu Met Ala Leu Ala Ile Ile Leu Phe Gly Trp Met
405 410 415
Ala Ser Val Ala Pro Lys Glu Phe Leu Gly His Phe Thr Val Phe Ala
420 425 430
Leu Ala Cys Val Val Gly Tyr Tyr Val Val Trp Asn Val Ser His Ala
435 440 445
Leu His Thr Pro Leu Met Ser Val Thr Asn Ala Ile Ser Gly Ile Ile
450 455 460
Val Val Gly Ala Leu Leu Gln Ile Gly Gln Gly Gly Trp Val Ser Phe
465 470 475 480
Leu Ser Phe Ile Ala Val Leu Ile Ala Ser Ile Asn Ile Phe Gly Gly
485 490 495
Phe Thr Val Thr Gln Arg Met Leu Lys Met Phe Arg Lys Asn
500 505 510
<210> SEQ ID NO 3
<211> LENGTH: 1389
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 3
atgtctggag gattagttac agctgcatac attgttgccg cgatcctgtt tatcttcagt 60
ctggccggtc tttcgaaaca tgaaacgtct cgccagggta acaacttcgg tatcgccggg 120
atggcgattg cgttaatcgc aaccattttt ggaccggata cgggtaatgt tggctggatc 180
ttgctggcga tggtcattgg tggggcaatt ggtatccgtc tggcgaagaa agttgaaatg 240
accgaaatgc cagaactggt ggcgatcctg catagcttcg tgggtctggc ggcagtgctg 300
gttggcttta acagctatct gcatcatgac gcgggaatgg caccgattct ggtcaatatt 360
cacctgacgg aagtgttcct cggtatcttc atcggggcgg taacgttcac gggttcggtg 420
gtggcgttcg gcaaactgtg tggcaagatt tcgtctaaac cattgatgct gccaaaccgt 480
cacaaaatga acctggcggc tctggtcgtt tccttcctgc tgctgattgt atttgttcgc 540
acggacagcg tcggcctgca agtgctggca ttgctgataa tgaccgcaat tgcgctggta 600
ttcggctggc atttagtcgc ctccatcggt ggtgcagata tgccagtggt ggtgtcgatg 660
ctgaactcgt actccggctg ggcggctgcg gctgcgggct ttatgctcag caacgacctg 720
ctgattgtga ccggtgcgct ggtcggttct tcgggggcta tcctttctta cattatgtgt 780
aaggcgatga accgttcctt tatcagcgtt attgcgggtg gtttcggcac cgacggctct 840
tctactggcg atgatcagga agtgggtgag caccgcgaaa tcaccgcaga agagacagcg 900
gaactgctga aaaactccca ttcagtgatc attactccgg ggtacggcat ggcagtcgcg 960
caggcgcaat atcctgtcgc tgaaattact gagaaattgc gcgctcgtgg tattaatgtg 1020
cgtttcggta tccacccggt cgcggggcgt ttgcctggac atatgaacgt attgctggct 1080
gaagcaaaag taccgtatga catcgtgctg gaaatggacg agatcaatga tgactttgct 1140
gataccgata ccgtactggt gattggtgct aacgatacgg ttaacccggc ggcgcaggat 1200
gatccgaaga gtccgattgc tggtatgcct gtgctggaag tgtggaaagc gcagaacgtg 1260
attgtcttta aacgttcgat gaacactggc tatgctggtg tgcaaaaccc gctgttcttc 1320
aaggaaaaca cccacatgct gtttggtgac gccaaagcca gcgtggatgc aatcctgaaa 1380
gctctgtaa 1389
<210> SEQ ID NO 4
<211> LENGTH: 462
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 4
Met Ser Gly Gly Leu Val Thr Ala Ala Tyr Ile Val Ala Ala Ile Leu
1 5 10 15
Phe Ile Phe Ser Leu Ala Gly Leu Ser Lys His Glu Thr Ser Arg Gln
20 25 30
Gly Asn Asn Phe Gly Ile Ala Gly Met Ala Ile Ala Leu Ile Ala Thr
35 40 45
Ile Phe Gly Pro Asp Thr Gly Asn Val Gly Trp Ile Leu Leu Ala Met
50 55 60
Val Ile Gly Gly Ala Ile Gly Ile Arg Leu Ala Lys Lys Val Glu Met
65 70 75 80
Thr Glu Met Pro Glu Leu Val Ala Ile Leu His Ser Phe Val Gly Leu
85 90 95
Ala Ala Val Leu Val Gly Phe Asn Ser Tyr Leu His His Asp Ala Gly
100 105 110
Met Ala Pro Ile Leu Val Asn Ile His Leu Thr Glu Val Phe Leu Gly
115 120 125
Ile Phe Ile Gly Ala Val Thr Phe Thr Gly Ser Val Val Ala Phe Gly
130 135 140
Lys Leu Cys Gly Lys Ile Ser Ser Lys Pro Leu Met Leu Pro Asn Arg
145 150 155 160
His Lys Met Asn Leu Ala Ala Leu Val Val Ser Phe Leu Leu Leu Ile
165 170 175
Val Phe Val Arg Thr Asp Ser Val Gly Leu Gln Val Leu Ala Leu Leu
180 185 190
Ile Met Thr Ala Ile Ala Leu Val Phe Gly Trp His Leu Val Ala Ser
195 200 205
Ile Gly Gly Ala Asp Met Pro Val Val Val Ser Met Leu Asn Ser Tyr
210 215 220
Ser Gly Trp Ala Ala Ala Ala Ala Gly Phe Met Leu Ser Asn Asp Leu
225 230 235 240
Leu Ile Val Thr Gly Ala Leu Val Gly Ser Ser Gly Ala Ile Leu Ser
245 250 255
Tyr Ile Met Cys Lys Ala Met Asn Arg Ser Phe Ile Ser Val Ile Ala
260 265 270
Gly Gly Phe Gly Thr Asp Gly Ser Ser Thr Gly Asp Asp Gln Glu Val
275 280 285
Gly Glu His Arg Glu Ile Thr Ala Glu Glu Thr Ala Glu Leu Leu Lys
290 295 300
Asn Ser His Ser Val Ile Ile Thr Pro Gly Tyr Gly Met Ala Val Ala
305 310 315 320
Gln Ala Gln Tyr Pro Val Ala Glu Ile Thr Glu Lys Leu Arg Ala Arg
325 330 335
Gly Ile Asn Val Arg Phe Gly Ile His Pro Val Ala Gly Arg Leu Pro
340 345 350
Gly His Met Asn Val Leu Leu Ala Glu Ala Lys Val Pro Tyr Asp Ile
355 360 365
Val Leu Glu Met Asp Glu Ile Asn Asp Asp Phe Ala Asp Thr Asp Thr
370 375 380
Val Leu Val Ile Gly Ala Asn Asp Thr Val Asn Pro Ala Ala Gln Asp
385 390 395 400
Asp Pro Lys Ser Pro Ile Ala Gly Met Pro Val Leu Glu Val Trp Lys
405 410 415
Ala Gln Asn Val Ile Val Phe Lys Arg Ser Met Asn Thr Gly Tyr Ala
420 425 430
Gly Val Gln Asn Pro Leu Phe Phe Lys Glu Asn Thr His Met Leu Phe
435 440 445
Gly Asp Ala Lys Ala Ser Val Asp Ala Ile Leu Lys Ala Leu
450 455 460
<210> SEQ ID NO 5
<211> LENGTH: 1716
<212> TYPE: DNA
<213> ORGANISM: Bacillus subtilis
<400> SEQUENCE: 5
atgttgacaa aagcaacaaa agaacaaaaa tcccttgtga aaaacagagg ggcggagctt 60
gttgttgatt gcttagtgga gcaaggtgtc acacatgtat ttggcattcc aggtgcaaaa 120
attgatgcgg tatttgacgc tttacaagat aaaggacctg aaattatcgt tgcccggcac 180
gaacaaaacg cagcattcat ggcccaagca gtcggccgtt taactggaaa accgggagtc 240
gtgttagtca catcaggacc gggtgcctct aacttggcaa caggcctgct gacagcgaac 300
actgaaggag accctgtcgt tgcgcttgct ggaaacgtga tccgtgcaga tcgtttaaaa 360
cggacacatc aatctttgga taatgcggcg ctattccagc cgattacaaa atacagtgta 420
gaagttcaag atgtaaaaaa tataccggaa gctgttacaa atgcatttag gatagcgtca 480
gcagggcagg ctggggccgc ttttgtgagc tttccgcaag atgttgtgaa tgaagtcaca 540
aatacgaaaa acgtgcgtgc tgttgcagcg ccaaaactcg gtcctgcagc agatgatgca 600
atcagtgcgg ccatagcaaa aatccaaaca gcaaaacttc ctgtcgtttt ggtcggcatg 660
aaaggcggaa gaccggaagc aattaaagcg gttcgcaagc ttttgaaaaa ggttcagctt 720
ccatttgttg aaacatatca agctgccggt accctttcta gagatttaga ggatcaatat 780
tttggccgta tcggtttgtt ccgcaaccag cctggcgatt tactgctaga gcaggcagat 840
gttgttctga cgatcggcta tgacccgatt gaatatgatc cgaaattctg gaatatcaat 900
ggagaccgga caattatcca tttagacgag attatcgctg acattgatca tgcttaccag 960
cctgatcttg aattgatcgg tgacattccg tccacgatca atcatatcga acacgatgct 1020
gtgaaagtgg aatttgcaga gcgtgagcag aaaatccttt ctgatttaaa acaatatatg 1080
catgaaggtg agcaggtgcc tgcagattgg aaatcagaca gagcgcaccc tcttgaaatc 1140
gttaaagagt tgcgtaatgc agtcgatgat catgttacag taacttgcga tatcggttcg 1200
cacgccattt ggatgtcacg ttatttccgc agctacgagc cgttaacatt aatgatcagt 1260
aacggtatgc aaacactcgg cgttgcgctt ccttgggcaa tcggcgcttc attggtgaaa 1320
ccgggagaaa aagtggtttc tgtctctggt gacggcggtt tcttattctc agcaatggaa 1380
ttagagacag cagttcgact aaaagcacca attgtacaca ttgtatggaa cgacagcaca 1440
tatgacatgg ttgcattcca gcaattgaaa aaatataacc gtacatctgc ggtcgatttc 1500
ggaaatatcg atatcgtgaa atatgcggaa agcttcggag caactggctt gcgcgtagaa 1560
tcaccagacc agctggcaga tgttctgcgt caaggcatga acgctgaagg tcctgtcatc 1620
atcgatgtcc cggttgacta cagtgataac attaatttag caagtgacaa gcttccgaaa 1680
gaattcgggg aactcatgaa aacgaaagct ctctag 1716
<210> SEQ ID NO 6
<211> LENGTH: 1716
<212> TYPE: DNA
<213> ORGANISM: Bacillus subtilis
<400> SEQUENCE: 6
atgttgacta aagctacaaa agagcagaaa tcattggtga aaaatagggg tgcagaactt 60
gttgtggact gtttggtaga acagggcgta acacatgttt ttggtatccc aggtgcaaaa 120
atcgacgccg tgtttgatgc attacaagac aagggtccag aaattattgt tgctagacat 180
gagcaaaatg ccgcatttat ggcgcaagct gtaggtaggc ttacaggtaa acctggtgtt 240
gtcctagtta cgtctggccc aggagcctcc aatttagcaa ctggtctatt gacagctaat 300
actgagggag atcctgtagt tgcgttagcc ggtaatgtaa ttagagctga taggcttaag 360
agaactcacc agtctctaga caacgctgct ttattccaac cgatcaccaa gtactcagta 420
gaggtacaag acgtaaagaa tatacctgaa gctgtgacaa acgcatttcg tatagcttct 480
gctggtcagg ctggtgccgc gtttgtttct tttcctcaag acgttgtcaa tgaagtgacc 540
aatactaaaa acgttagagc ggttgcagcc cctaaactag gtccagccgc agacgacgca 600
attagcgctg caattgctaa aattcagacg gcgaaactac cagtagtcct tgtcggtatg 660
aagggcggaa gaccagaagc aataaaagct gttcgtaagt tattgaagaa agtccaatta 720
cctttcgttg agacttacca agcagcaggt actttatcta gagatttaga ggatcagtat 780
tttggaagga taggtctatt tagaaaccaa ccaggagatt tactattaga acaagctgat 840
gttgtactta ctatcggtta tgatcctata gagtatgacc caaagttttg gaacataaat 900
ggggatagaa caattataca tctagacgag ataatcgccg acatcgatca cgcttatcaa 960
ccagatttag aactaatcgg agatatcccg tcaacaatca atcatattga acatgatgct 1020
gtaaaggttg agttcgctga acgtgagcag aaaatcttat ctgatctaaa gcaatatatg 1080
catgagggtg aacaagttcc agcagactgg aaatctgacc gtgcacatcc tttggaaatc 1140
gttaaggaac taagaaatgc ggtcgatgat catgtgactg ttacatgtga tatcggttca 1200
catgcaattt ggatgtcacg ttattttagg agctacgaac cattaacttt aatgatatct 1260
aacgggatgc aaactctggg ggttgcactt ccttgggcta ttggcgctag tttagttaag 1320
cccggtgaga aggtggtatc ggtatcaggt gatggtggct ttctgttttc ggctatggaa 1380
ttagaaactg cagtccgttt aaaagctccc attgtgcata ttgtctggaa tgattctact 1440
tacgacatgg ttgcttttca acagttgaag aaatacaata gaacttcggc tgtagacttt 1500
ggtaacatcg atattgtgaa atatgctgag tcttttggcg caacaggcct gagggtggaa 1560
agtccagatc agttagctga tgtgttgaga caagggatga atgccgaggg accggtaatc 1620
atagatgtgc cagttgacta ctcagacaat attaatttgg cttctgataa acttcctaaa 1680
gagtttggcg agctaatgaa gaccaaagcc ttataa 1716
<210> SEQ ID NO 7
<211> LENGTH: 571
<212> TYPE: PRT
<213> ORGANISM: Bacillus subtilis
<400> SEQUENCE: 7
Met Leu Thr Lys Ala Thr Lys Glu Gln Lys Ser Leu Val Lys Asn Arg
1 5 10 15
Gly Ala Glu Leu Val Val Asp Cys Leu Val Glu Gln Gly Val Thr His
20 25 30
Val Phe Gly Ile Pro Gly Ala Lys Ile Asp Ala Val Phe Asp Ala Leu
35 40 45
Gln Asp Lys Gly Pro Glu Ile Ile Val Ala Arg His Glu Gln Asn Ala
50 55 60
Ala Phe Met Ala Gln Ala Val Gly Arg Leu Thr Gly Lys Pro Gly Val
65 70 75 80
Val Leu Val Thr Ser Gly Pro Gly Ala Ser Asn Leu Ala Thr Gly Leu
85 90 95
Leu Thr Ala Asn Thr Glu Gly Asp Pro Val Val Ala Leu Ala Gly Asn
100 105 110
Val Ile Arg Ala Asp Arg Leu Lys Arg Thr His Gln Ser Leu Asp Asn
115 120 125
Ala Ala Leu Phe Gln Pro Ile Thr Lys Tyr Ser Val Glu Val Gln Asp
130 135 140
Val Lys Asn Ile Pro Glu Ala Val Thr Asn Ala Phe Arg Ile Ala Ser
145 150 155 160
Ala Gly Gln Ala Gly Ala Ala Phe Val Ser Phe Pro Gln Asp Val Val
165 170 175
Asn Glu Val Thr Asn Thr Lys Asn Val Arg Ala Val Ala Ala Pro Lys
180 185 190
Leu Gly Pro Ala Ala Asp Asp Ala Ile Ser Ala Ala Ile Ala Lys Ile
195 200 205
Gln Thr Ala Lys Leu Pro Val Val Leu Val Gly Met Lys Gly Gly Arg
210 215 220
Pro Glu Ala Ile Lys Ala Val Arg Lys Leu Leu Lys Lys Val Gln Leu
225 230 235 240
Pro Phe Val Glu Thr Tyr Gln Ala Ala Gly Thr Leu Ser Arg Asp Leu
245 250 255
Glu Asp Gln Tyr Phe Gly Arg Ile Gly Leu Phe Arg Asn Gln Pro Gly
260 265 270
Asp Leu Leu Leu Glu Gln Ala Asp Val Val Leu Thr Ile Gly Tyr Asp
275 280 285
Pro Ile Glu Tyr Asp Pro Lys Phe Trp Asn Ile Asn Gly Asp Arg Thr
290 295 300
Ile Ile His Leu Asp Glu Ile Ile Ala Asp Ile Asp His Ala Tyr Gln
305 310 315 320
Pro Asp Leu Glu Leu Ile Gly Asp Ile Pro Ser Thr Ile Asn His Ile
325 330 335
Glu His Asp Ala Val Lys Val Glu Phe Ala Glu Arg Glu Gln Lys Ile
340 345 350
Leu Ser Asp Leu Lys Gln Tyr Met His Glu Gly Glu Gln Val Pro Ala
355 360 365
Asp Trp Lys Ser Asp Arg Ala His Pro Leu Glu Ile Val Lys Glu Leu
370 375 380
Arg Asn Ala Val Asp Asp His Val Thr Val Thr Cys Asp Ile Gly Ser
385 390 395 400
His Ala Ile Trp Met Ser Arg Tyr Phe Arg Ser Tyr Glu Pro Leu Thr
405 410 415
Leu Met Ile Ser Asn Gly Met Gln Thr Leu Gly Val Ala Leu Pro Trp
420 425 430
Ala Ile Gly Ala Ser Leu Val Lys Pro Gly Glu Lys Val Val Ser Val
435 440 445
Ser Gly Asp Gly Gly Phe Leu Phe Ser Ala Met Glu Leu Glu Thr Ala
450 455 460
Val Arg Leu Lys Ala Pro Ile Val His Ile Val Trp Asn Asp Ser Thr
465 470 475 480
Tyr Asp Met Val Ala Phe Gln Gln Leu Lys Lys Tyr Asn Arg Thr Ser
485 490 495
Ala Val Asp Phe Gly Asn Ile Asp Ile Val Lys Tyr Ala Glu Ser Phe
500 505 510
Gly Ala Thr Gly Leu Arg Val Glu Ser Pro Asp Gln Leu Ala Asp Val
515 520 525
Leu Arg Gln Gly Met Asn Ala Glu Gly Pro Val Ile Ile Asp Val Pro
530 535 540
Val Asp Tyr Ser Asp Asn Ile Asn Leu Ala Ser Asp Lys Leu Pro Lys
545 550 555 560
Glu Phe Gly Glu Leu Met Lys Thr Lys Ala Leu
565 570
<210> SEQ ID NO 8
<211> LENGTH: 1716
<212> TYPE: DNA
<213> ORGANISM: Bacillus subtilis
<400> SEQUENCE: 8
atgttgacaa aagcaacaaa agaacaaaaa tcccttgtga aaagcagagg ggcggagctt 60
gttgttgatt gcttagcgga gcaaggtgtc acacatgtat ttggcattcc aggtgcaaaa 120
attgatgcgg tatttgacgc tttacaagat aaagggcctg aaattatcgt tgcccggcat 180
gaacaaaatg cagcatttat ggcgcaagca gtcggccgtt taactggaaa accgggagtc 240
gtgttagtca catcaggacc aggtgcttcg aacttggcaa caggactgct gacagcaaac 300
actgaaggtg accctgtcgt tgcgcttgct gggaacgtga tccgtgcaga tcgtttaaaa 360
cggacacatc aatctttgga taatgcggcg ctattccagc cgattacaaa atacagtgta 420
gaagttcaag atgtaaaaaa tataccggaa gctgttacaa atgcgtttag gatagcgtca 480
gcagggcagg ctggggccgc ttttgtgagt tttccgcaag atgttgtgaa tgaagtcaca 540
aatacaaaaa acgtacgtgc tgtcgcagcg ccaaaacttg gtcccgcagc agatgacgca 600
atcagtatgg ccattgcaaa aattcaaaca gcaaaacttc ctgtcgtttt agtcggcatg 660
aagggcggaa gaccggaagc gattaaagcg gttcgcaagc tattgaaaaa agtgcagctt 720
ccattcgttg aaacatatca agctgccggt actcttacga gagatttaga ggatcagtat 780
tttggccgga tcggtttatt ccgcaaccag cctggcgatc tgctgcttga gcaggctgat 840
gttgttctga caatcggcta tgacccaatt gaatatgatc cgaaattctg gaatgtcaat 900
ggagaccgga cgatcatcca tttagacgag attctggctg acattgatca tgcttaccag 960
ccggatcttg aactgatcgg tgatattcca tctacgatca atcatatcga acacgatgct 1020
gtgaaagtag actttgcgga acgtgagcag aagatccttt ctgatttaaa acaatatatg 1080
catgagggtg agcaggtgcc tgcagattgg aaatcagaca gagtgcatcc tcttgaaatc 1140
gttaaagaat tgcgaaacgc agtcgatgat catgttacag tgacttgcga tatcggttca 1200
cacgcgattt ggatgtcacg ttatttccgc agctacgagc cgttaacatt aatgattagt 1260
aacggtatgc aaacactcgg cgttgcgctt ccttgggcaa tcggcgcttc attggtgaaa 1320
ccgggagaaa aagtagtatc agtctccggt gatggcggtt tcttattctc agctatggaa 1380
ttagagacag cagttcgttt aaaagcacca attgtacaca ttgtatggaa cgacagcaca 1440
tatgacatgg ttgcattcca gcaattgaaa aaatataatc gtacatctgc ggtcgatttc 1500
ggaaatatcg atatcgtgaa atacgcggaa agcttcggag caactggctt acgcgtagaa 1560
tcaccagacc agctggcaga tgttctgcgt caaggcatga acgctgaggg gcctgtcatc 1620
attgatgtcc cggttgacta cagtgataac gttaatttag caagtgacaa gcttccgaaa 1680
gaattcgggg aactcatgaa aacgaaagct ctctag 1716
<210> SEQ ID NO 9
<211> LENGTH: 571
<212> TYPE: PRT
<213> ORGANISM: Bacillus subtilis
<400> SEQUENCE: 9
Met Leu Thr Lys Ala Thr Lys Glu Gln Lys Ser Leu Val Lys Ser Arg
1 5 10 15
Gly Ala Glu Leu Val Val Asp Cys Leu Ala Glu Gln Gly Val Thr His
20 25 30
Val Phe Gly Ile Pro Gly Ala Lys Ile Asp Ala Val Phe Asp Ala Leu
35 40 45
Gln Asp Lys Gly Pro Glu Ile Ile Val Ala Arg His Glu Gln Asn Ala
50 55 60
Ala Phe Met Ala Gln Ala Val Gly Arg Leu Thr Gly Lys Pro Gly Val
65 70 75 80
Val Leu Val Thr Ser Gly Pro Gly Ala Ser Asn Leu Ala Thr Gly Leu
85 90 95
Leu Thr Ala Asn Thr Glu Gly Asp Pro Val Val Ala Leu Ala Gly Asn
100 105 110
Val Ile Arg Ala Asp Arg Leu Lys Arg Thr His Gln Ser Leu Asp Asn
115 120 125
Ala Ala Leu Phe Gln Pro Ile Thr Lys Tyr Ser Val Glu Val Gln Asp
130 135 140
Val Lys Asn Ile Pro Glu Ala Val Thr Asn Ala Phe Arg Ile Ala Ser
145 150 155 160
Ala Gly Gln Ala Gly Ala Ala Phe Val Ser Phe Pro Gln Asp Val Val
165 170 175
Asn Glu Val Thr Asn Thr Lys Asn Val Arg Ala Val Ala Ala Pro Lys
180 185 190
Leu Gly Pro Ala Ala Asp Asp Ala Ile Ser Met Ala Ile Ala Lys Ile
195 200 205
Gln Thr Ala Lys Leu Pro Val Val Leu Val Gly Met Lys Gly Gly Arg
210 215 220
Pro Glu Ala Ile Lys Ala Val Arg Lys Leu Leu Lys Lys Val Gln Leu
225 230 235 240
Pro Phe Val Glu Thr Tyr Gln Ala Ala Gly Thr Leu Thr Arg Asp Leu
245 250 255
Glu Asp Gln Tyr Phe Gly Arg Ile Gly Leu Phe Arg Asn Gln Pro Gly
260 265 270
Asp Leu Leu Leu Glu Gln Ala Asp Val Val Leu Thr Ile Gly Tyr Asp
275 280 285
Pro Ile Glu Tyr Asp Pro Lys Phe Trp Asn Val Asn Gly Asp Arg Thr
290 295 300
Ile Ile His Leu Asp Glu Ile Leu Ala Asp Ile Asp His Ala Tyr Gln
305 310 315 320
Pro Asp Leu Glu Leu Ile Gly Asp Ile Pro Ser Thr Ile Asn His Ile
325 330 335
Glu His Asp Ala Val Lys Val Asp Phe Ala Glu Arg Glu Gln Lys Ile
340 345 350
Leu Ser Asp Leu Lys Gln Tyr Met His Glu Gly Glu Gln Val Pro Ala
355 360 365
Asp Trp Lys Ser Asp Arg Val His Pro Leu Glu Ile Val Lys Glu Leu
370 375 380
Arg Asn Ala Val Asp Asp His Val Thr Val Thr Cys Asp Ile Gly Ser
385 390 395 400
His Ala Ile Trp Met Ser Arg Tyr Phe Arg Ser Tyr Glu Pro Leu Thr
405 410 415
Leu Met Ile Ser Asn Gly Met Gln Thr Leu Gly Val Ala Leu Pro Trp
420 425 430
Ala Ile Gly Ala Ser Leu Val Lys Pro Gly Glu Lys Val Val Ser Val
435 440 445
Ser Gly Asp Gly Gly Phe Leu Phe Ser Ala Met Glu Leu Glu Thr Ala
450 455 460
Val Arg Leu Lys Ala Pro Ile Val His Ile Val Trp Asn Asp Ser Thr
465 470 475 480
Tyr Asp Met Val Ala Phe Gln Gln Leu Lys Lys Tyr Asn Arg Thr Ser
485 490 495
Ala Val Asp Phe Gly Asn Ile Asp Ile Val Lys Tyr Ala Glu Ser Phe
500 505 510
Gly Ala Thr Gly Leu Arg Val Glu Ser Pro Asp Gln Leu Ala Asp Val
515 520 525
Leu Arg Gln Gly Met Asn Ala Glu Gly Pro Val Ile Ile Asp Val Pro
530 535 540
Val Asp Tyr Ser Asp Asn Val Asn Leu Ala Ser Asp Lys Leu Pro Lys
545 550 555 560
Glu Phe Gly Glu Leu Met Lys Thr Lys Ala Leu
565 570
<210> SEQ ID NO 10
<211> LENGTH: 1476
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 10
atggctaact acttcaatac actgaatctg cgccagcagc tggcacagct gggcaaatgt 60
cgctttatgg gccgcgatga attcgccgat ggcgcgagct accttcaggg taaaaaagta 120
gtcatcgtcg gctgtggcgc acagggtctg aaccagggcc tgaacatgcg tgattctggt 180
ctcgatatct cctacgctct gcgtaaagaa gcgattgccg agaagcgcgc gtcctggcgt 240
aaagcgaccg aaaatggttt taaagtgggt acttacgaag aactgatccc acaggcggat 300
ctggtgatta acctgacgcc ggacaagcag cactctgatg tagtgcgcac cgtacagcca 360
ctgatgaaag acggcgcggc gctgggctac tcgcacggtt tcaacatcgt cgaagtgggc 420
gagcagatcc gtaaagatat caccgtagtg atggttgcgc cgaaatgccc aggcaccgaa 480
gtgcgtgaag agtacaaacg tgggttcggc gtaccgacgc tgattgccgt tcacccggaa 540
aacgatccga aaggcgaagg catggcgatt gccaaagcct gggcggctgc aaccggtggt 600
caccgtgcgg gtgtgctgga atcgtccttc gttgcggaag tgaaatctga cctgatgggc 660
gagcaaacca tcctgtgcgg tatgttgcag gctggctctc tgctgtgctt cgacaagctg 720
gtggaagaag gtaccgatcc agcatacgca gaaaaactga ttcagttcgg ttgggaaacc 780
atcaccgaag cactgaaaca gggcggcatc accctgatga tggaccgtct ctctaacccg 840
gcgaaactgc gtgcttatgc gctttctgaa cagctgaaag agatcatggc acccctgttc 900
cagaaacata tggacgacat catctccggc gaattctctt ccggtatgat ggcggactgg 960
gccaacgatg ataagaaact gctgacctgg cgtgaagaga ccggcaaaac cgcgtttgaa 1020
accgcgccgc agtatgaagg caaaatcggc gagcaggagt acttcgataa aggcgtactg 1080
atgattgcga tggtgaaagc gggcgttgaa ctggcgttcg aaaccatggt cgattccggc 1140
atcattgaag agtctgcata ttatgaatca ctgcacgagc tgccgctgat tgccaacacc 1200
atcgcccgta agcgtctgta cgaaatgaac gtggttatct ctgataccgc tgagtacggt 1260
aactatctgt tctcttacgc ttgtgtgccg ttgctgaaac cgtttatggc agagctgcaa 1320
ccgggcgacc tgggtaaagc tattccggaa ggcgcggtag ataacgggca actgcgtgat 1380
gtgaacgaag cgattcgcag ccatgcgatt gagcaggtag gtaagaaact gcgcggctat 1440
atgacagata tgaaacgtat tgctgttgcg ggttaa 1476
<210> SEQ ID NO 11
<211> LENGTH: 1476
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 11
atggcgaatt atttcaacac tctgaacctg cgtcaacaac tggcgcaact gggtaagtgc 60
cgtttcatgg gtcgtgacga gtttgcggac ggtgcttctt atctgcaagg caagaaggtt 120
gttattgttg gttgcggtgc gcaaggcctg aatcaaggtc tgaatatgcg cgacagcggc 180
ctggacatta gctatgcgct gcgcaaggag gctatcgcgg aaaaacgtgc tagctggcgc 240
aaggctactg agaacggctt caaggttggc acctatgagg agctgattcc gcaagctgac 300
ctggttatca atctgacccc agataaacaa catagcgacg ttgttcgtac tgttcaaccg 360
ctgatgaagg atggtgctgc tctgggttat agccacggct ttaacattgt tgaggtaggt 420
gaacaaattc gcaaggacat tactgttgtt atggtggctc caaagtgtcc gggtactgag 480
gttcgcgagg aatataagcg cggttttggt gttccaaccc tgatcgcggt gcatccagag 540
aatgacccaa agggtgaggg tatggctatc gcgaaggcgt gggctgcggc gactggcggc 600
catcgcgctg gcgttctgga gagcagcttt gtggctgagg ttaagagcga tctgatgggt 660
gaacagacta ttctgtgtgg tatgctgcaa gcgggtagcc tgctgtgttt tgataaactg 720
gttgaggagg gcactgaccc ggcgtatgcg gagaagctga tccaatttgg ctgggagact 780
attactgagg cgctgaagca aggtggtatt actctgatga tggatcgcct gagcaatcca 840
gctaagctgc gcgcgtacgc tctgagcgag caactgaagg aaattatggc accgctgttt 900
caaaagcaca tggatgatat cattagcggt gagtttagca gcggcatgat ggctgattgg 960
gcgaatgacg acaaaaagct gctgacttgg cgcgaggaaa ctggtaagac tgctttcgag 1020
actgctccac aatacgaggg taagattggt gaacaagaat attttgacaa gggtgttctg 1080
atgatcgcta tggttaaggc tggtgtggag ctggcttttg agactatggt tgacagcggt 1140
attatcgagg aaagcgcgta ctacgagagc ctgcatgaac tgccactgat cgcgaatact 1200
attgcgcgca aacgcctgta tgagatgaat gttgtgatta gcgacactgc ggaatatggc 1260
aattacctgt ttagctatgc gtgcgttcca ctgctgaagc cattcatggc ggaactgcag 1320
ccaggtgatc tgggcaaggc gatcccagag ggtgctgttg acaatggtca gctgcgcgac 1380
gttaatgagg ctatccgttc tcacgctatc gaacaagttg gcaaaaagct gcgtggttac 1440
atgaccgaca tgaagcgcat cgcggtggct ggctaa 1476
<210> SEQ ID NO 12
<211> LENGTH: 1476
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 12
atggccaact attttaacac attaaatttg agacaacaat tggctcaact gggtaagtgc 60
agatttatgg gaagggacga gtttgctgat ggtgcttctt atctgcaagg aaagaaagta 120
gtaattgttg gctgcggtgc tcagggtcta aaccaaggtt taaacatgag agattcaggt 180
ctggatattt cgtatgcatt gaggaaagag gcaattgcag aaaagagggc ctcctggcgt 240
aaagcgacgg aaaatgggtt caaagttggt acttacgaag aactgatccc tcaggcagat 300
ttagtgatta acctaacacc agataagcaa cactcagacg tagtaagaac agttcaaccg 360
ctgatgaagg atggggcagc tttaggttac tctcatggct ttaatatcgt tgaagtgggc 420
gagcagatca gaaaagatat aacagtcgta atggttgcac caaagtgccc aggtacggaa 480
gtcagagagg agtacaagag gggttttggt gtacctacat tgatcgccgt acatcctgaa 540
aatgacccca aaggtgaagg tatggcaatt gcgaaggcat gggcagccgc aaccggaggt 600
catagagcgg gtgtgttaga gagttctttc gtagctgagg tcaagagtga cttaatgggt 660
gaacaaacca ttctgtgcgg aatgttgcag gcagggtctt tactatgctt tgataaattg 720
gtcgaagagg gtacagatcc tgcctatgct gaaaagttga tacaatttgg ttgggagaca 780
atcaccgagg cacttaaaca aggtggcata acattgatga tggatagact ttcaaatccg 840
gccaagctaa gagcctacgc cttatctgag caactaaaag agatcatggc accattattc 900
caaaagcaca tggacgatat tatctccggt gagttttcct caggaatgat ggcagattgg 960
gcaaacgatg ataaaaagtt attgacgtgg agagaagaaa ccggcaagac ggcattcgag 1020
acagccccac aatacgaagg taaaattggt gaacaagaat actttgataa gggagtattg 1080
atgatagcta tggtgaaggc aggggtagaa cttgcattcg aaactatggt tgactccggt 1140
atcattgaag aatctgcata ctatgagtct ttgcatgaat tgcctttgat agcaaatact 1200
attgcaagaa aaagacttta cgagatgaat gttgtcatat cagacactgc agaatatggt 1260
aattacttat ttagctacgc atgtgtcccg ttgttaaagc ccttcatggc cgagttacaa 1320
cctggtgatt tggggaaggc tattccggaa ggagcggttg acaatggcca actgagagac 1380
gtaaatgaag ctattcgttc acatgctata gaacaggtgg gtaaaaagct gagaggatat 1440
atgaccgata tgaaaagaat tgcagtggca ggatga 1476
<210> SEQ ID NO 13
<211> LENGTH: 491
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 13
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ala Ile Ala Glu Lys Arg Ala Ser Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Gln His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Asp Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Gly Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Lys Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly
485 490
<210> SEQ ID NO 14
<211> LENGTH: 1500
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 14
atggcgaatt atttcaacac tctgaacctg cgtcaacaac tggcgcaact gggtaagtgc 60
cgtttcatgg gtcgtgacga gtttgcggac ggtgcttctt atctgcaagg caagaaggtt 120
gttattgttg gttgcggtgc gcaaggcctg aatcaaggtc tgaatatgcg cgacagcggc 180
ctggacatta gctatgcgct gcgcaaggag gctatcgcgg aaaaacgtgc tagctggcgc 240
aaggctactg agaacggctt caaggttggc acctatgagg agctgattcc gcaagctgac 300
ctggttatca atctgacccc agataaacaa catagcgacg ttgttcgtac tgttcaaccg 360
ctgatgaagg atggtgctgc tctgggttat agccacggct ttaacattgt tgaggtaggt 420
gaacaaattc gcaaggacat tactgttgtt atggtggctc caaagtgtcc gggtactgag 480
gttcgcgagg aatataagcg cggttttggt gttccaaccc tgatcgcggt gcatccagag 540
aatgacccaa agggtgaggg tatggctatc gcgaaggcgt gggctgcggc gactggcggc 600
catcgcgctg gcgttctgga gagcagcttt gtggctgagg ttaagagcga tctgatgggt 660
gaacagacta ttctgtgtgg tatgctgcaa gcgggtagcc tgctgtgttt tgataaactg 720
gttgaggagg gcactgaccc ggcgtatgcg gagaagctga tccaatttgg ctgggagact 780
attactgagg cgctgaagca aggtggtatt actctgatga tggatcgcct gagcaatcca 840
gctaagctgc gcgcgtacgc tctgagcgag caactgaagg aaattatggc accgctgttt 900
caaaagcaca tggatgatat cattagcggt gagtttagca gcggcatgat ggctgattgg 960
gcgaatgacg acaaaaagct gctgacttgg cgcgaggaaa ctggtaagac tgctttcgag 1020
actgctccac aatacgaggg taagattggt gaacaagaat attttgacaa gggtgttctg 1080
atgatcgcta tggttaaggc tggtgtggag ctggcttttg agactatggt tgacagcggt 1140
attatcgagg aaagcgcgta ctacgagagc ctgcatgaac tgccactgat cgcgaatact 1200
attgcgcgca aacgcctgta tgagatgaat gttgtgatta gcgacactgc ggaatatggc 1260
aattacctgt ttagctatgc gtgcgttcca ctgctgaagc cattcatggc ggaactgcag 1320
ccaggtgatc tgggcaaggc gatcccagag ggtgctgttg acaatggtca gctgcgcgac 1380
gttaatgagg ctatccgttc tcacgctatc gaacaagttg gcaaaaagct gcgtggttac 1440
atgaccgaca tgaagcgcat cgcggtggct ggcctcgagc accaccacca ccaccactga 1500
<210> SEQ ID NO 15
<211> LENGTH: 499
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 15
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ala Ile Ala Glu Lys Arg Ala Ser Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Gln His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Asp Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Gly Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Lys Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly Leu Glu His His His
485 490 495
His His His
<210> SEQ ID NO 16
<211> LENGTH: 1494
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 16
atggcgaatt atttcaacac tctgaacctg cgtcaacaac tggcgcaact gggtaagtgc 60
cgtttcatgg gtcgtgacga gtttgcggac ggtgcttctt atctgcaagg caagaaggtt 120
gttattgttg gttgcggtgc gcaaggcctg aatcaaggtc tgaatatgcg cgacagcggc 180
ctggacatta gctatgcgct gcgcaaggag gctatcgcgg aaaaacgtgc tgactggcgc 240
aaggctactg agaacggctt caaggttggc acctatgagg agctgattcc gcaagctgac 300
ctggttatca atctgacccc agataaacaa catagcgacg ttgttcgtac tgttcaaccg 360
ctgatgaagg atggtgctgc tctgggttat agccacggct ttaacattgt tgaggtaggt 420
gaacaaattc gcaaggacat tactgttgtt atggtggctc caaagtgtcc gggtactgag 480
gttcgcgagg aatataagcg cggttttggt gttccaaccc tgatcgcggt gcatccagag 540
aatgacccaa agggtgaggg tatggctatc gcgaaggcgt gggctgcggc gactggcggc 600
catcgcgctg gcgttctgga gagcagcttt gtggctgagg ttaagagcga tctgatgggt 660
gaacagacta ttctgtgtgg tatgctgcaa gcgggtagcc tgctgtgttt tgataaactg 720
gttgaggagg gcactgaccc ggcgtatgcg gagaagctga tccaatttgg ctgggagact 780
attactgagg cgctgaagca aggtggtatt actctgatga tggatcgcct gagcaatcca 840
gctaagctgc gcgcgtacgc tctgagcgag caactgaagg aaattatggc accgctgttt 900
caaaagcaca tggatgatat cattagcggt gagtttagca gcggcatgat ggctgattgg 960
gcgaatgacg acaaaaagct gctgacttgg cgcgaggaaa ctggtaagac tgctttcgag 1020
actgctccac aatacgaggg taagattggt gaacaagaat attttgacaa gggtgttctg 1080
atgatcgcta tggttaaggc tggtgtggag ctggcttttg agactatggt tgacagcggt 1140
attatcgagg aaagcgcgta ctacgagagc ctgcatgaac tgccactgat cgcgaatact 1200
attgcgcgca aacgcctgta tgagatgaat gttgtgatta gcgacactgc ggaatatggc 1260
aattacctgt ttagctatgc gtgcgttcca ctgctgaagc cattcatggc ggaactgcag 1320
ccaggtgatc tgggcaaggc gatcccagag ggtgctgttg acaatggtca gctgcgcgac 1380
gttaatgagg ctatccgttc tcacgctatc gaacaagttg gcaaaaagct gcgtggttac 1440
atgaccgaca tgaagcgcat cgcggtggct ggccaccacc accaccacca ctaa 1494
<210> SEQ ID NO 17
<211> LENGTH: 499
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 17
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ala Ile Ala Glu Lys Arg Ala Asp Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Gln His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Asp Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Gly Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Lys Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly Leu Glu His His His
485 490 495
His His His
<210> SEQ ID NO 18
<211> LENGTH: 1476
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 18
atggcgaatt atttcaacac tctgaacctg cgtcaacaac tggcgcaact gggtaagtgc 60
cgtttcatgg gtcgtgacga gtttgcggac ggtgcttctt atctgcaagg caagaaggtt 120
gttattgttg gttgcggtgc gcaaggcctg aatcaaggtc tgaatatgcg cgacagcggc 180
ctggacatta gctatgcgct gcgcaaggag gctatcgcgg aaaaacgtgc tgactggcgc 240
aaggctactg agaacggctt caaggttggc acctatgagg agctgattcc gcaagctgac 300
ctggttatca atctgacccc agataaacaa catagcgacg ttgttcgtac tgttcaaccg 360
ctgatgaagg atggtgctgc tctgggttat agccacggct ttaacattgt tgaggtaggt 420
gaacaaattc gcaaggacat tactgttgtt atggtggctc caaagtgtcc gggtactgag 480
gttcgcgagg aatataagcg cggttttggt gttccaaccc tgatcgcggt gcatccagag 540
aatgacccaa agggtgaggg tatggctatc gcgaaggcgt gggctgcggc gactggcggc 600
catcgcgctg gcgttctgga gagcagcttt gtggctgagg ttaagagcga tctgatgggt 660
gaacagacta ttctgtgtgg tatgctgcaa gcgggtagcc tgctgtgttt tgataaactg 720
gttgaggagg gcactgaccc ggcgtatgcg gagaagctga tccaatttgg ctgggagact 780
attactgagg cgctgaagca aggtggtatt actctgatga tggatcgcct gagcaatcca 840
gctaagctgc gcgcgtacgc tctgagcgag caactgaagg aaattatggc accgctgttt 900
caaaagcaca tggatgatat cattagcggt gagtttagca gcggcatgat ggctgattgg 960
gcgaatgacg acaaaaagct gctgacttgg cgcgaggaaa ctggtaagac tgctttcgag 1020
actgctccac aatacgaggg taagattggt gaacaagaat attttgacaa gggtgttctg 1080
atgatcgcta tggttaaggc tggtgtggag ctggcttttg agactatggt tgacagcggt 1140
attatcgagg aaagcgcgta ctacgagagc ctgcatgaac tgccactgat cgcgaatact 1200
attgcgcgca aacgcctgta tgagatgaat gttgtgatta gcgacactgc ggaatatggc 1260
aattacctgt ttagctatgc gtgcgttcca ctgctgaagc cattcatggc ggaactgcag 1320
ccaggtgatc tgggcaaggc gatcccagag ggtgctgttg acaatggtca gctgcgcgac 1380
gttaatgagg ctatccgttc tcacgctatc gaacaagttg gcaaaaagct gcgtggttac 1440
atgaccgaca tgaagcgcat cgcggtggct ggctaa 1476
<210> SEQ ID NO 19
<211> LENGTH: 493
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 19
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ala Ile Ala Glu Lys Arg Ala Asp Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Gln His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Asp Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Gly Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Lys Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly Leu Glu
485 490
<210> SEQ ID NO 20
<211> LENGTH: 1476
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 20
atggcgaatt atttcaacac tctgaacctg cgtcaacaac tggcgcaact gggtaagtgc 60
cgtttcatgg gtcgtgacga gtttgcggac ggtgcttctt atctgcaagg caagaaggtt 120
gttattgttg gttgcggtgc gcaaggcctg aatcaaggtc tgaatatgcg cgacagcggc 180
ctggacatta gctatgcgct gcgcaaggag gctatcgcgg aaaaacgtgc tagctggcgc 240
aaggctactg agaacggctt caaggttggc acctatgagg agctgattcc gcaagctgac 300
ctggttatca atctgacccc agataaagca catagcgacg ttgttcgtac tgttcaaccg 360
ctgatgaagg atggtgctgc tctgggttat agccacggct ttaacattgt tgaggtaggt 420
gaacaaattc gcaaggacat tactgttgtt atggtggctc caaagtgtcc gggtactgag 480
gttcgcgagg aatataagcg cggttttggt gttccaaccc tgatcgcggt gcatccagag 540
aatgacccaa agggtgaggg tatggctatc gcgaaggcgt gggctgcggc gactggcggc 600
catcgcgctg gcgttctgga gagcagcttt gtggctgagg ttaagagcga tctgatgggt 660
gaacagacta ttctgtgtgg tatgctgcaa gcgggtagcc tgctgtgttt tgataaactg 720
gttgaggagg gcactgaccc ggcgtatgcg gagaagctga tccaatttgg ctgggagact 780
attactgagg cgctgaagca aggtggtatt actctgatga tggatcgcct gagcaatcca 840
gctaagctgc gcgcgtacgc tctgagcgag caactgaagg aaattatggc accgctgttt 900
caaaagcaca tggatgatat cattagcggt gagtttagca gcggcatgat ggctgattgg 960
gcgaatgacg acaaaaagct gctgacttgg cgcgaggaaa ctggtaagac tgctttcgag 1020
actgctccac aatacgaggg taagattggt gaacaagaat attttgacaa gggtgttctg 1080
atgatcgcta tggttaaggc tggtgtggag ctggcttttg agactatggt tgacagcggt 1140
attatcgagg aaagcgcgta ctacgagagc ctgcatgaac tgccactgat cgcgaatact 1200
attgcgcgca aacgcctgta tgagatgaat gttgtgatta gcgacactgc ggaatatggc 1260
aattacctgt ttagctatgc gtgcgttcca ctgctgaagc cattcatggc ggaactgcag 1320
ccaggtgatc tgggcaaggc gatcccagag ggtgctgttg acaatggtca gctgcgcgac 1380
gttaatgagg ctatccgttc tcacgctatc gaacaagttg gcaaaaagct gcgtggttac 1440
atgaccgaca tgaagcgcat cgcggtggct ggctaa 1476
<210> SEQ ID NO 21
<211> LENGTH: 498
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 21
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ala Ile Ala Glu Lys Arg Ala Ser Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Ala His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Asp Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Gly Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Lys Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly Leu Glu His His His
485 490 495
His His
<210> SEQ ID NO 22
<211> LENGTH: 1500
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 22
atggcgaatt atttcaacac tctgaacctg cgtcaacaac tggcgcaact gggtaagtgc 60
cgtttcatgg gtcgtgacga gtttgcggac ggtgcttctt atctgcaagg caagaaggtt 120
gttattgttg gttgcggtgc gcaaggcctg aatcaaggtc tgaatatgcg cgacagcggc 180
ctggacatta gctatgcgct gcgcaaggag gctatcgcgg aaaaacgtgc tagctggcgc 240
aaggctactg agaacggctt caaggttggc acctatgagg agctgattcc gcaagctgac 300
ctggttatca atctgacccc agataaagtg catagcgacg ttgttcgtac tgttcaaccg 360
ctgatgaagg atggtgctgc tctgggttat agccacggct ttaacattgt tgaggtaggt 420
gaacaaattc gcaaggacat tactgttgtt atggtggctc caaagtgtcc gggtactgag 480
gttcgcgagg aatataagcg cggttttggt gttccaaccc tgatcgcggt gcatccagag 540
aatgacccaa agggtgaggg tatggctatc gcgaaggcgt gggctgcggc gactggcggc 600
catcgcgctg gcgttctgga gagcagcttt gtggctgagg ttaagagcga tctgatgggt 660
gaacagacta ttctgtgtgg tatgctgcaa gcgggtagcc tgctgtgttt tgataaactg 720
gttgaggagg gcactgaccc ggcgtatgcg gagaagctga tccaatttgg ctgggagact 780
attactgagg cgctgaagca aggtggtatt actctgatga tggatcgcct gagcaatcca 840
gctaagctgc gcgcgtacgc tctgagcgag caactgaagg aaattatggc accgctgttt 900
caaaagcaca tggatgatat cattagcggt gagtttagca gcggcatgat ggctgattgg 960
gcgaatgacg acaaaaagct gctgacttgg cgcgaggaaa ctggtaagac tgctttcgag 1020
actgctccac aatacgaggg taagattggt gaacaagaat attttgacaa gggtgttctg 1080
atgatcgcta tggttaaggc tggtgtggag ctggcttttg agactatggt tgacagcggt 1140
attatcgagg aaagcgcgta ctacgagagc ctgcatgaac tgccactgat cgcgaatact 1200
attgcgcgca aacgcctgta tgagatgaat gttgtgatta gcgacactgc ggaatatggc 1260
aattacctgt ttagctatgc gtgcgttcca ctgctgaagc cattcatggc ggaactgcag 1320
ccaggtgatc tgggcaaggc gatcccagag ggtgctgttg acaatggtca gctgcgcgac 1380
gttaatgagg ctatccgttc tcacgctatc gaacaagttg gcaaaaagct gcgtggttac 1440
atgaccgaca tgaagcgcat cgcggtggct ggcctcgagc accaccacca ccaccactaa 1500
<210> SEQ ID NO 23
<211> LENGTH: 499
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 23
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ala Ile Ala Glu Lys Arg Ala Ser Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Val His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Asp Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Gly Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Lys Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly Leu Glu His His His
485 490 495
His His His
<210> SEQ ID NO 24
<211> LENGTH: 1476
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 24
atggcgaatt atttcaacac tctgaacctg cgtcaacaac tggcgcaact gggtaagtgc 60
cgtttcatgg gtcgtgacga gtttgcggac ggtgcttctt atctgcaagg caagaaggtt 120
gttattgttg gttgcggtgc gcaaggcctg aatcaaggtc tgaatatgcg cgacagcggc 180
ctggacatta gctatgcgct gcgcaaggag gctatcgcgg aaaaacgtgc tagctggcgc 240
aaggctactg agaacggctt caaggttggc acctatgagg agctgattcc gcaagctgac 300
ctggttatca atctgacccc agataaagtg catagcgacg ttgttcgtac tgttcaaccg 360
ctgatgaagg atggtgctgc tctgggttat agccacggct ttaacattgt tgaggtaggt 420
gaacaaattc gcaaggacat tactgttgtt atggtggctc caaagtgtcc gggtactgag 480
gttcgcgagg aatataagcg cggttttggt gttccaaccc tgatcgcggt gcatccagag 540
aatgacccaa agggtgaggg tatggctatc gcgaaggcgt gggctgcggc gactggcggc 600
catcgcgctg gcgttctgga gagcagcttt gtggctgagg ttaagagcga tctgatgggt 660
gaacagacta ttctgtgtgg tatgctgcaa gcgggtagcc tgctgtgttt tgataaactg 720
gttgaggagg gcactgaccc ggcgtatgcg gagaagctga tccaatttgg ctgggagact 780
attactgagg cgctgaagca aggtggtatt actctgatga tggatcgcct gagcaatcca 840
gctaagctgc gcgcgtacgc tctgagcgag caactgaagg aaattatggc accgctgttt 900
caaaagcaca tggatgatat cattagcggt gagtttagca gcggcatgat ggctgattgg 960
gcgaatgacg acaaaaagct gctgacttgg cgcgaggaaa ctggtaagac tgctttcgag 1020
actgctccac aatacgaggg taagattggt gaacaagaat attttgacaa gggtgttctg 1080
atgatcgcta tggttaaggc tggtgtggag ctggcttttg agactatggt tgacagcggt 1140
attatcgagg aaagcgcgta ctacgagagc ctgcatgaac tgccactgat cgcgaatact 1200
attgcgcgca aacgcctgta tgagatgaat gttgtgatta gcgacactgc ggaatatggc 1260
aattacctgt ttagctatgc gtgcgttcca ctgctgaagc cattcatggc ggaactgcag 1320
ccaggtgatc tgggcaaggc gatcccagag ggtgctgttg acaatggtca gctgcgcgac 1380
gttaatgagg ctatccgttc tcacgctatc gaacaagttg gcaaaaagct gcgtggttac 1440
atgaccgaca tgaagcgcat cgcggtggct ggctaa 1476
<210> SEQ ID NO 25
<211> LENGTH: 493
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 25
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ala Ile Ala Glu Lys Arg Ala Ser Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Val His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Asp Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Gly Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Lys Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly Leu Glu
485 490
<210> SEQ ID NO 26
<211> LENGTH: 1494
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 26
atggcgaatt atttcaacac tctgaacctg cgtcaacaac tggcgcaact gggtaagtgc 60
cgtttcatgg gtcgtgacga gtttgcggac ggtgcttctt atctgcaagg caagaaggtt 120
gttattgttg gttgcggtgc gcaaggcctg aatcaaggtc tgaatatgcg cgacagcggc 180
ctggacatta gctatgcgct gcgcaaggag gctatcgcgg aaaaacgtgc tgactggcgc 240
aaggctactg agaacggctt caaggttggc acctatgagg agctgattcc gcaagctgac 300
ctggttatca atctgacccc agataaagtg catagcgacg ttgttcgtac tgttcaaccg 360
ctgatgaagg atggtgctgc tctgggttat agccacggct ttaacattgt tgaggtaggt 420
gaacaaattc gcaaggacat tactgttgtt atggtggctc caaagtgtcc gggtactgag 480
gttcgcgagg aatataagcg cggttttggt gttccaaccc tgatcgcggt gcatccagag 540
aatgacccaa agggtgaggg tatggctatc gcgaaggcgt gggctgcggc gactggcggc 600
catcgcgctg gcgttctgga gagcagcttt gtggctgagg ttaagagcga tctgatgggt 660
gaacagacta ttctgtgtgg tatgctgcaa gcgggtagcc tgctgtgttt tgataaactg 720
gttgaggagg gcactgaccc ggcgtatgcg gagaagctga tccaatttgg ctgggagact 780
attactgagg cgctgaagca aggtggtatt actctgatga tggatcgcct gagcaatcca 840
gctaagctgc gcgcgtacgc tctgagcgag caactgaagg aaattatggc accgctgttt 900
caaaagcaca tggatgatat cattagcggt gagtttagca gcggcatgat ggctgattgg 960
gcgaatgacg acaaaaagct gctgacttgg cgcgaggaaa ctggtaagac tgctttcgag 1020
actgctccac aatacgaggg taagattggt gaacaagaat attttgacaa gggtgttctg 1080
atgatcgcta tggttaaggc tggtgtggag ctggcttttg agactatggt tgacagcggt 1140
attatcgagg aaagcgcgta ctacgagagc ctgcatgaac tgccactgat cgcgaatact 1200
attgcgcgca aacgcctgta tgagatgaat gttgtgatta gcgacactgc ggaatatggc 1260
aattacctgt ttagctatgc gtgcgttcca ctgctgaagc cattcatggc ggaactgcag 1320
ccaggtgatc tgggcaaggc gatcccagag ggtgctgttg acaatggtca gctgcgcgac 1380
gttaatgagg ctatccgttc tcacgctatc gaacaagttg gcaaaaagct gcgtggttac 1440
atgaccgaca tgaagcgcat cgcggtggct ggccaccacc accaccacca ctaa 1494
<210> SEQ ID NO 27
<211> LENGTH: 499
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 27
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ala Ile Ala Glu Lys Arg Ala Asp Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Val His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Asp Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Gly Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Lys Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly Leu Glu His His His
485 490 495
His His His
<210> SEQ ID NO 28
<211> LENGTH: 1494
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 28
atggcgaatt atttcaacac tctgaacctg cgtcaacaac tggcgcaact gggtaagtgc 60
cgtttcatgg gtcgtgacga gtttgcggac ggtgcttctt atctgcaagg caagaaggtt 120
gttattgttg gttgcggtgc gcaaggcctg aatcaaggtc tgaatatgcg cgacagcggc 180
ctggacatta gctatgcgct gcgcaaggag tctatcgcgg aaaaacgtgc tgactggcgc 240
aaggctactg agaacggctt caaggttggc acctatgagg agctgattcc gcaagctgac 300
ctggttatca atctgacccc agataaagtg catagcgacg ttgttcgtac tgttcaaccg 360
ctgatgaagg atggtgctgc tctgggttat agccacggct ttaacattgt tgaggtaggt 420
gaacaaattc gcaaggacat tactgttgtt atggtggctc caaagtgtcc gggtactgag 480
gttcgcgagg aatataagcg cggttttggt gttccaaccc tgatcgcggt gcatccagag 540
aatgacccaa agggtgaggg tatggctatc gcgaaggcgt gggctgcggc gactggcggc 600
catcgcgctg gcgttctgga gagcagcttt gtggctgagg ttaagagcga tctgatgggt 660
gaacagacta ttctgtgtgg tatgctgcaa gcgggtagcc tgctgtgttt tgataaactg 720
gttgaggagg gcactgaccc ggcgtatgcg gagaagctga tccaatttgg ctgggagact 780
attactgagg cgctgaagca aggtggtatt actctgatga tggatcgcct gagcaatcca 840
gctaagctgc gcgcgtacgc tctgagcgag caactgaagg aaattatggc accgctgttt 900
caaaagcaca tggatgatat cattagcggt gagtttagca gcggcatgat ggctgattgg 960
gcgaatgacg acaaaaagct gctgacttgg cgcgaggaaa ctggtaagac tgctttcgag 1020
actgctccac aatacgaggg taagattggt gaacaagaat attttgacaa gggtgttctg 1080
atgatcgcta tggttaaggc tggtgtggag ctggcttttg agactatggt tgacagcggt 1140
attatcgagg aaagcgcgta ctacgagagc ctgcatgaac tgccactgat cgcgaatact 1200
attgcgcgca aacgcctgta tgagatgaat gttgtgatta gcgacactgc ggaatatggc 1260
aattacctgt ttagctatgc gtgcgttcca ctgctgaagc cattcatggc ggaactgcag 1320
ccaggtgatc tgggcaaggc gatcccagag ggtgctgttg acaatggtca gctgcgcgac 1380
gttaatgagg ctatccgttc tcacgctatc gaacaagttg gcaaaaagct gcgtggttac 1440
atgaccgaca tgaagcgcat cgcggtggct ggccaccacc accaccacca ctaa 1494
<210> SEQ ID NO 29
<211> LENGTH: 499
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 29
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ser Ile Ala Glu Lys Arg Ala Asp Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Val His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Asp Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Gly Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Lys Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly Leu Glu His His His
485 490 495
His His His
<210> SEQ ID NO 30
<211> LENGTH: 1494
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 30
atggcgaatt atttcaacac tctgaacctg cgtcaacaac tggcgcaact gggtaagtgc 60
cgtttcatgg gtcgtgacga gtttgcggac ggtgcttctt atctgcaagg caagaaggtt 120
gttattgttg gttgcggtgc gcaaggcctg aatcaaggtc tgaatatgcg cgacagcggc 180
ctggacatta gctatgcgct gcgcaaggag tctatcgcgg aaaaagatgc tgattggcgc 240
aaggctactg agaacggctt caaggttggc acctatgagg agctgattcc gcaagctgac 300
ctggttatca atctgacccc agataaagca catagcgacg ttgttcgtac tgttcaaccg 360
ctgatgaagg atggtgctgc tctgggttat agccacggct ttaacattgt tgaggtaggt 420
gaacaaattc gcaaggacat tactgttgtt atggtggctc caaagtgtcc gggtactgag 480
gttcgcgagg aatataagcg cggttttggt gttccaaccc tgatcgcggt gcatccagag 540
aatgacccaa agggtgaggg tatggctatc gcgaaggcgt gggctgcggc gactggcggc 600
catcgcgctg gcgttctgga gagcagcttt gtggctgagg ttaagagcga tctgatgggt 660
gaacagacta ttctgtgtgg tatgctgcaa gcgggtagcc tgctgtgttt tgataaactg 720
gttgaggagg gcactgaccc ggcgtatgcg gagaagctga tccaatttgg ctgggagact 780
attactgagg cgctgaagca aggtggtatt actctgatga tggatcgcct gagcaatcca 840
gctaagctgc gcgcgtacgc tctgagcgag caactgaagg aaattatggc accgctgttt 900
caaaagcaca tggatgatat cattagcggt gagtttagca gcggcatgat ggctgattgg 960
gcgaatgacg acaaaaagct gctgacttgg cgcgaggaaa ctggtaagac tgctttcgag 1020
actgctccac aatacgaggg taagattggt gaacaagaat attttgacaa gggtgttctg 1080
atgatcgcta tggttaaggc tggtgtggag ctggcttttg agactatggt tgacagcggt 1140
attatcgagg aaagcgcgta ctacgagagc ctgcatgaac tgccactgat cgcgaatact 1200
attgcgcgca aacgcctgta tgagatgaat gttgtgatta gcgacactgc ggaatatggc 1260
aattacctgt ttagctatgc gtgcgttcca ctgctgaagc cattcatggc ggaactgcag 1320
ccaggtgatc tgggcaaggc gatcccagag ggtgctgttg acaatggtca gctgcgcgac 1380
gttaatgagg ctatccgttc tcacgctatc gaacaagttg gcaaaaagct gcgtggttac 1440
atgaccgaca tgaagcgcat cgcggtggct ggccaccacc accaccacca ctaa 1494
<210> SEQ ID NO 31
<211> LENGTH: 499
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 31
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ser Ile Ala Glu Lys Asp Ala Asp Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Ala His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Asp Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Gly Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Lys Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly Leu Glu His His His
485 490 495
His His His
<210> SEQ ID NO 32
<211> LENGTH: 1494
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 32
atggcgaatt atttcaacac tctgaacctg cgtcaacaac tggcgcaact gggtaagtgc 60
cgtttcatgg gtcgtgacga gtttgcggac ggtgcttctt atctgcaagg caagaaggtt 120
gttattgttg gttgcggtgc gcaaggcctg aatcaaggtc tgaatatgcg cgacagcggc 180
ctggacatta gctatgcgct gcgcaaggag tctatcgcgg aaaaagatgc tgattggcgc 240
aaggctactg agaacggctt caaggttggc acctatgagg agctgattcc gcaagctgac 300
ctggttatca atctgacccc agataaagta catagcgacg ttgttcgtac tgttcaaccg 360
ctgatgaagg atggtgctgc tctgggttat agccacggct ttaacattgt tgaggtaggt 420
gaacaaattc gcaaggacat tactgttgtt atggtggctc caaagtgtcc gggtactgag 480
gttcgcgagg aatataagcg cggttttggt gttccaaccc tgatcgcggt gcatccagag 540
aatgacccaa agggtgaggg tatggctatc gcgaaggcgt gggctgcggc gactggcggc 600
catcgcgctg gcgttctgga gagcagcttt gtggctgagg ttaagagcga tctgatgggt 660
gaacagacta ttctgtgtgg tatgctgcaa gcgggtagcc tgctgtgttt tgataaactg 720
gttgaggagg gcactgaccc ggcgtatgcg gagaagctga tccaatttgg ctgggagact 780
attactgagg cgctgaagca aggtggtatt actctgatga tggatcgcct gagcaatcca 840
gctaagctgc gcgcgtacgc tctgagcgag caactgaagg aaattatggc accgctgttt 900
caaaagcaca tggatgatat cattagcggt gagtttagca gcggcatgat ggctgattgg 960
gcgaatgacg acaaaaagct gctgacttgg cgcgaggaaa ctggtaagac tgctttcgag 1020
actgctccac aatacgaggg taagattggt gaacaagaat attttgacaa gggtgttctg 1080
atgatcgcta tggttaaggc tggtgtggag ctggcttttg agactatggt tgacagcggt 1140
attatcgagg aaagcgcgta ctacgagagc ctgcatgaac tgccactgat cgcgaatact 1200
attgcgcgca aacgcctgta tgagatgaat gttgtgatta gcgacactgc ggaatatggc 1260
aattacctgt ttagctatgc gtgcgttcca ctgctgaagc cattcatggc ggaactgcag 1320
ccaggtgatc tgggcaaggc gatcccagag ggtgctgttg acaatggtca gctgcgcgac 1380
gttaatgagg ctatccgttc tcacgctatc gaacaagttg gcaaaaagct gcgtggttac 1440
atgaccgaca tgaagcgcat cgcggtggct ggccaccacc accaccacca ctaa 1494
<210> SEQ ID NO 33
<211> LENGTH: 1494
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 33
atggccaact attttaacac attaaatttg agacaacaat tggctcaact gggtaagtgc 60
agatttatgg gaagggacga gtttgctgat ggtgcttctt atctgcaagg aaagaaagta 120
gtaattgttg gctgcggtgc tcagggtcta aaccaaggtt taaacatgag agattcaggt 180
ctggatattt cgtatgcatt gaggaaagag tctattgcag aaaaggatgc cgattggcgt 240
aaagcgacgg aaaatgggtt caaagttggt acttacgaag aactgatccc tcaggcagat 300
ttagtgatta acctaacacc agataaggtt cactcagacg tagtaagaac agttcaaccg 360
ctgatgaagg atggggcagc tttaggttac tctcatggct ttaatatcgt tgaagtgggc 420
gagcagatca gaaaagatat aacagtcgta atggttgcac caaagtgccc aggtacggaa 480
gtcagagagg agtacaagag gggttttggt gtacctacat tgatcgccgt acatcctgaa 540
aatgacccca aaggtgaagg tatggcaatt gcgaaggcat gggcagccgc aaccggaggt 600
catagagcgg gtgtgttaga gagttctttc gtagctgagg tcaagagtga cttaatgggt 660
gaacaaacca ttctgtgcgg aatgttgcag gcagggtctt tactatgctt tgataaattg 720
gtcgaagagg gtacagatcc tgcctatgct gaaaagttga tacaatttgg ttgggagaca 780
atcaccgagg cacttaaaca aggtggcata acattgatga tggatagact ttcaaatccg 840
gccaagctaa gagcctacgc cttatctgag caactaaaag agatcatggc accattattc 900
caaaagcaca tggacgatat tatctccggt gagttttcct caggaatgat ggcagattgg 960
gcaaacgatg ataaaaagtt attgacgtgg agagaagaaa ccggcaagac ggcattcgag 1020
acagccccac aatacgaagg taaaattggt gaacaagaat actttgataa gggagtattg 1080
atgatagcta tggtgaaggc aggggtagaa cttgcattcg aaactatggt tgactccggt 1140
atcattgaag aatctgcata ctatgagtct ttgcatgaat tgcctttgat agcaaatact 1200
attgcaagaa aaagacttta cgagatgaat gttgtcatat cagacactgc agaatatggt 1260
aattacttat ttagctacgc atgtgtcccg ttgttaaagc ccttcatggc cgagttacaa 1320
cctggtgatt tggggaaggc tattccggaa ggagcggttg acaatggcca actgagagac 1380
gtaaatgaag ctattcgttc acatgctata gaacaggtgg gtaaaaagct gagaggatat 1440
atgaccgata tgaaaagaat tgcagtggca ggacaccacc accaccacca ctga 1494
<210> SEQ ID NO 34
<211> LENGTH: 499
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 34
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ser Ile Ala Glu Lys Asp Ala Asp Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Val His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Asp Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Gly Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Lys Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly Leu Glu His His His
485 490 495
His His His
<210> SEQ ID NO 35
<211> LENGTH: 1476
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 35
atggccaact attttaacac attaaatttg agacaacaat tggctcaact gggtaagtgc 60
agatttatgg gaagggacga gtttgctgat ggtgcttctt atctgcaagg aaagaaagta 120
gtaattgttg gctgcggtgc tcagggtcta aaccaaggtt taaacatgag agattcaggt 180
ctggatattt cgtatgcatt gaggaaagag tctattgcag aaaaggatgc cgattggcgt 240
aaagcgacgg aaaatgggtt caaagttggt acttacgaag aactgatccc tcaggcagat 300
ttagtgatta acctaacacc agataaggtt cactcagacg tagtaagaac agttcaaccg 360
ctgatgaagg atggggcagc tttaggttac tctcatggct ttaatatcgt tgaagtgggc 420
gagcagatca gaaaagatat aacagtcgta atggttgcac caaagtgccc aggtacggaa 480
gtcagagagg agtacaagag gggttttggt gtacctacat tgatcgccgt acatcctgaa 540
aatgacccca aaggtgaagg tatggcaatt gcgaaggcat gggcagccgc aaccggaggt 600
catagagcgg gtgtgttaga gagttctttc gtagctgagg tcaagagtga cttaatgggt 660
gaacaaacca ttctgtgcgg aatgttgcag gcagggtctt tactatgctt tgataaattg 720
gtcgaagagg gtacagatcc tgcctatgct gaaaagttga tacaatttgg ttgggagaca 780
atcaccgagg cacttaaaca aggtggcata acattgatga tggatagact ttcaaatccg 840
gccaagctaa gagcctacgc cttatctgag caactaaaag agatcatggc accattattc 900
caaaagcaca tggacgatat tatctccggt gagttttcct caggaatgat ggcagattgg 960
gcaaacgatg ataaaaagtt attgacgtgg agagaagaaa ccggcaagac ggcattcgag 1020
acagccccac aatacgaagg taaaattggt gaacaagaat actttgataa gggagtattg 1080
atgatagcta tggtgaaggc aggggtagaa cttgcattcg aaactatggt tgactccggt 1140
atcattgaag aatctgcata ctatgagtct ttgcatgaat tgcctttgat agcaaatact 1200
attgcaagaa aaagacttta cgagatgaat gttgtcatat cagacactgc agaatatggt 1260
aattacttat ttagctacgc atgtgtcccg ttgttaaagc ccttcatggc cgagttacaa 1320
cctggtgatt tggggaaggc tattccggaa ggagcggttg acaatggcca actgagagac 1380
gtaaatgaag ctattcgttc acatgctata gaacaggtgg gtaaaaagct gagaggatat 1440
atgaccgata tgaaaagaat tgcagtggca ggatga 1476
<210> SEQ ID NO 36
<211> LENGTH: 491
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 36
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ser Ile Ala Glu Lys Asp Ala Asp Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Val His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Asp Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Gly Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Lys Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly
485 490
<210> SEQ ID NO 37
<211> LENGTH: 1494
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 37
atggccaact attttaacac attaaatttg agacaacaat tggctcaact gggtaagtgc 60
agatttatgg gaagggacga gtttgctgat ggtgcttctt atctgcaagg aaagaaagta 120
gtaattgttg gctgcggtgc tcagggtcta aaccaaggtt taaacatgag agattcaggt 180
ctggatattt cgtatgcatt gaggaaagag tctattgcag aaaaggatgc cgattggcgt 240
aaagcgacgg aaaatgggtt caaagttggt acttacgaag aactgatccc tcaggcagat 300
ttagtgatta acctaacacc agataaggtt cactcagacg tagtaagaac agttcaaccg 360
ctgatgaagg atggggcagc tttaggttac tctcatggct ttaatatcgt tgaagtgggc 420
gagcagatca gaaaaggtat aacagtcgta atggttgcgc caaagtgccc aggtacggaa 480
gtcagagagg agtacaagag gggttttggt gtacctacat tgatcgccgt acatcctgaa 540
aatgacccca aacgtgaagg tatggcaatt gcgaaggcat gggcagccgc aaccggaggt 600
catagagcgg gtgtgttaga gagttctttc gtagctgagg tcaagagtga cttaatgggt 660
gaacaaacca ttctgtgcgg aatgttgcag gcagggtctt tactatgctt tgataaattg 720
gtcgaagagg gtacagatcc tgcctatgct gaaaagttga tacaatttgg ttgggagaca 780
atcaccgagg cacttaaaca aggtggcata acattgatga tggatagact ttcaaatccg 840
gccaagctaa gagcctacgc cttatctgag caactaaaag agatcatggc accattattc 900
caaaagcaca tggacgatat tatctccggt gagttttcct caggaatgat ggcagattgg 960
gcaaacgatg ataaaaagtt attgacgtgg agagaagaaa ccggcaagac ggcattcgag 1020
acagccccac aatacgaagg taaaattggt gaacaagaat actttgataa gggagtattg 1080
atgatagcta tggtgaaggc aggggtagaa cttgcattcg aaactatggt tgactccggt 1140
atcattgaag aatctgcata ctatgagtct ttgcatgaat tgcctttgat agcaaatact 1200
attgcaagaa aaagacttta cgagatgaat gttgtcatat cagacactgc agaatatggt 1260
aattacttat ttagctacgc atgtgtcccg ttgttaaagc ccttcatggc cgagttacaa 1320
cctggtgatt tggggaaggc tattccggaa ggagcggttg acaatggcca actgagagac 1380
gtaaatgaag ctattcgttc acatgctata gaacaggtgg gtaaaaagct gagaggatat 1440
atgaccgata tgaaaagaat tgcagtggca ggacaccacc accaccacca ctga 1494
<210> SEQ ID NO 38
<211> LENGTH: 497
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 38
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ser Ile Ala Glu Lys Asp Ala Asp Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Val His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Gly Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Arg Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Lys Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly His His His His His
485 490 495
His
<210> SEQ ID NO 39
<211> LENGTH: 1476
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 39
atggccaact attttaacac attaaatttg agacaacaat tggctcaact gggtaagtgc 60
agatttatgg gaagggacga gtttgctgat ggtgcttctt atctgcaagg aaagaaagta 120
gtaattgttg gctgcggtgc tcagggtcta aaccaaggtt taaacatgag agattcaggt 180
ctggatattt cgtatgcatt gaggaaagag tctattgcag aaaaggatgc cgattggcgt 240
aaagcgacgg aaaatgggtt caaagttggt acttacgaag aactgatccc tcaggcagat 300
ttagtgatta acctaacacc agataaggtt cactcagacg tagtaagaac agttcaaccg 360
ctgatgaagg atggggcagc tttaggttac tctcatggct ttaatatcgt tgaagtgggc 420
gagcagatca gaaaaggtat aacagtcgta atggttgcgc caaagtgccc aggtacggaa 480
gtcagagagg agtacaagag gggttttggt gtacctacat tgatcgccgt acatcctgaa 540
aatgacccca aacgtgaagg tatggcaatt gcgaaggcat gggcagccgc aaccggaggt 600
catagagcgg gtgtgttaga gagttctttc gtagctgagg tcaagagtga cttaatgggt 660
gaacaaacca ttctgtgcgg aatgttgcag gcagggtctt tactatgctt tgataaattg 720
gtcgaagagg gtacagatcc tgcctatgct gaaaagttga tacaatttgg ttgggagaca 780
atcaccgagg cacttaaaca aggtggcata acattgatga tggatagact ttcaaatccg 840
gccaagctaa gagcctacgc cttatctgag caactaaaag agatcatggc accattattc 900
caaaagcaca tggacgatat tatctccggt gagttttcct caggaatgat ggcagattgg 960
gcaaacgatg ataaaaagtt attgacgtgg agagaagaaa ccggcaagac ggcattcgag 1020
acagccccac aatacgaagg taaaattggt gaacaagaat actttgataa gggagtattg 1080
atgatagcta tggtgaaggc aggggtagaa cttgcattcg aaactatggt tgactccggt 1140
atcattgaag aatctgcata ctatgagtct ttgcatgaat tgcctttgat agcaaatact 1200
attgcaagaa aaagacttta cgagatgaat gttgtcatat cagacactgc agaatatggt 1260
aattacttat ttagctacgc atgtgtcccg ttgttaaagc ccttcatggc cgagttacaa 1320
cctggtgatt tggggaaggc tattccggaa ggagcggttg acaatggcca actgagagac 1380
gtaaatgaag ctattcgttc acatgctata gaacaggtgg gtaaaaagct gagaggatat 1440
atgaccgata tgaaaagaat tgcagtggca ggatga 1476
<210> SEQ ID NO 40
<211> LENGTH: 491
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 40
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ser Ile Ala Glu Lys Asp Ala Asp Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Val His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Gly Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Arg Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Lys Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly
485 490
<210> SEQ ID NO 41
<211> LENGTH: 1494
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 41
atggccaact attttaacac attaaatttg agacaacaat tggctcaact gggtaagtgc 60
agatttatgg gaagggacga gtttgctgat ggtgcttctt atctgcaagg aaagaaagta 120
gtaattgttg gctgcggtgc tcagggtcta aaccaaggtt taaacatgag agattcaggt 180
ctggatattt cgtatgcatt gaggaaagag tctattgcag aaaaggatgc cgattggcgt 240
aaagcgacgg aaaatgggtt caaagttggt acttacgaag aactgatccc tcaggcagat 300
ttagtgatta acctaacacc agataaggtt cactcagacg tagtaagaac agttcaaccg 360
ctgatgaagg atggggcagc tttaggttac tctcatggct ttaatatcgt tgaagtgggc 420
gagcagatca gaaaaggtat aacagtcgta atggttgcgc caaagtgccc aggtacggaa 480
gtcagagagg agtacaagag gggttttggt gtacctacat tgatcgccgt acatcctgaa 540
aatgacccca aacgtgaagg tatggcaata gcgaaggcat gggcagccgc aaccggaggt 600
catagagcgg gtgtgttaga gagttctttc gtagctgagg tcaagagtga cttaatgggt 660
gaacaaacca ttctgtgcgg aatgttgcag gcagggtctt tactatgctt tgataaattg 720
gtcgaagagg gtacagatcc tgcctatgct gaaaagttga tacaatttgg ttgggagaca 780
atcaccgagg cacttaaaca aggtggcata acattgatga tggatagact ttcaaatccg 840
gccaagctaa gagcctacgc cttatctgag caactaaaag agatcatggc accattattc 900
caaaagcaca tggacgatat tatctccggt gagttttcct caggaatgat ggcagattgg 960
gcaaacgatg ataaaaagtt attgacgtgg agagaagaaa ccggcaagac ggcattcgag 1020
acagccccac aatacgaagg taaaattggt gaacaagaat actttgataa gggagtattg 1080
atgatagcta tggtgaaggc aggggtagaa cttgcattcg aaactatggt tgactccggt 1140
atcattgaag aatctgcata ctatgagtct ttgcatgaat tgcctttgat agcaaatact 1200
attgcaagaa aaagacttta cgagatgaat gttgtcatat cagacactgc agaatatggt 1260
aattacttat ttagctacgc gtgtgtcccg ttgttagagc ccttcatggc cgagttacaa 1320
cctggtgatt tggggaaggc tattccggaa ggagcggttg acaatggcca actgagagac 1380
gtaaatgaag ctattcgttc gcatgctata gaacaggtgg gtaaaaagct gagaggatat 1440
atgaccgata tgaaaagaat tgcagtggca ggacaccacc accaccacca ctga 1494
<210> SEQ ID NO 42
<211> LENGTH: 497
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 42
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ser Ile Ala Glu Lys Asp Ala Asp Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Val His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Gly Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Arg Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Glu Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly His His His His His
485 490 495
His
<210> SEQ ID NO 43
<211> LENGTH: 1476
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 43
atggccaact attttaacac attaaatttg agacaacaat tggctcaact gggtaagtgc 60
agatttatgg gaagggacga gtttgctgat ggtgcttctt atctgcaagg aaagaaagta 120
gtaattgttg gctgcggtgc tcagggtcta aaccaaggtt taaacatgag agattcaggt 180
ctggatattt cgtatgcatt gaggaaagag tctattgcag aaaaggatgc cgattggcgt 240
aaagcgacgg aaaatgggtt caaagttggt acttacgaag aactgatccc tcaggcagat 300
ttagtgatta acctaacacc agataaggtt cactcagacg tagtaagaac agttcaaccg 360
ctgatgaagg atggggcagc tttaggttac tctcatggct ttaatatcgt tgaagtgggc 420
gagcagatca gaaaaggtat aacagtcgta atggttgcgc caaagtgccc aggtacggaa 480
gtcagagagg agtacaagag gggttttggt gtacctacat tgatcgccgt acatcctgaa 540
aatgacccca aacgtgaagg tatggcaata gcgaaggcat gggcagccgc aaccggaggt 600
catagagcgg gtgtgttaga gagttctttc gtagctgagg tcaagagtga cttaatgggt 660
gaacaaacca ttctgtgcgg aatgttgcag gcagggtctt tactatgctt tgataaattg 720
gtcgaagagg gtacagatcc tgcctatgct gaaaagttga tacaatttgg ttgggagaca 780
atcaccgagg cacttaaaca aggtggcata acattgatga tggatagact ttcaaatccg 840
gccaagctaa gagcctacgc cttatctgag caactaaaag agatcatggc accattattc 900
caaaagcaca tggacgatat tatctccggt gagttttcct caggaatgat ggcagattgg 960
gcaaacgatg ataaaaagtt attgacgtgg agagaagaaa ccggcaagac ggcattcgag 1020
acagccccac aatacgaagg taaaattggt gaacaagaat actttgataa gggagtattg 1080
atgatagcta tggtgaaggc aggggtagaa cttgcattcg aaactatggt tgactccggt 1140
atcattgaag aatctgcata ctatgagtct ttgcatgaat tgcctttgat agcaaatact 1200
attgcaagaa aaagacttta cgagatgaat gttgtcatat cagacactgc agaatatggt 1260
aattacttat ttagctacgc gtgtgtcccg ttgttagagc ccttcatggc cgagttacaa 1320
cctggtgatt tggggaaggc tattccggaa ggagcggttg acaatggcca actgagagac 1380
gtaaatgaag ctattcgttc gcatgctata gaacaggtgg gtaaaaagct gagaggatat 1440
atgaccgata tgaaaagaat tgcagtggca ggatga 1476
<210> SEQ ID NO 44
<211> LENGTH: 491
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 44
Met Ala Asn Tyr Phe Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln
1 5 10 15
Leu Gly Lys Cys Arg Phe Met Gly Arg Asp Glu Phe Ala Asp Gly Ala
20 25 30
Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Lys Glu Ser Ile Ala Glu Lys Asp Ala Asp Trp Arg
65 70 75 80
Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Val His Ser
100 105 110
Asp Val Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Ile Val Glu Val Gly Glu Gln Ile Arg
130 135 140
Lys Gly Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Arg Glu Gly Met Ala Ile Ala Lys
180 185 190
Ala Trp Ala Ala Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu
225 230 235 240
Val Glu Glu Gly Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Gln Gly Gly Ile Thr Leu
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr Ala Leu
275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp Arg Glu Glu Thr Gly Lys
325 330 335
Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Asp Ser Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu
420 425 430
Glu Pro Phe Met Ala Glu Leu Gln Pro Gly Asp Leu Gly Lys Ala Ile
435 440 445
Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp Val Asn Glu Ala
450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr
465 470 475 480
Met Thr Asp Met Lys Arg Ile Ala Val Ala Gly
485 490
<210> SEQ ID NO 45
<211> LENGTH: 1647
<212> TYPE: DNA
<213> ORGANISM: Lactococcus lactis
<400> SEQUENCE: 45
atgtatacag taggagatta cctattagac cgattacacg agttaggaat tgaagaaatt 60
tttggagtcc ctggagacta taacttacaa tttttagatc aaattatttc ccgcaaggat 120
atgaaatggg tcggaaatgc taatgaatta aatgcttcat atatggctga tggctatgct 180
cgtactaaaa aagctgccgc atttcttaca acctttggag taggtgaatt gagtgcagtt 240
aatggattag caggaagtta cgccgaaaat ttaccagtag tagaaatagt gggatcacct 300
acatcaaaag ttcaaaatga aggaaaattt gttcatcata cgctggctga cggtgatttt 360
aaacacttta tgaaaatgca cgaacctgtt acagcagctc gaactttact gacagcagaa 420
aatgcaaccg ttgaaattga ccgagtactt tctgcactat taaaagaaag aaaacctgtc 480
tatatcaact taccagttga tgttgctgct gcaaaagcag agaaaccctc actccctttg 540
aaaaaagaaa actcaacttc aaatacaagt gaccaagaga tcttgaacaa aattcaagaa 600
agcttgaaaa atgccaaaaa accaatcgtg attacaggac atgaaataat tagttttggc 660
ttagaaaaaa cagtctctca atttatttca aagacaaaac tacctattac gacattaaac 720
tttggaaaaa gttcagttga tgaagctctc ccttcatttt taggaatcta taatggtaaa 780
ctctcagagc ctaatcttaa agaattcgtg gaatcagccg acttcatcct gatgcttgga 840
gttaaactca cagactcttc aacaggagcc ttcactcatc atttaaatga aaataaaatg 900
atttcactga atatagatga aggaaaaata tttaacgaaa gcatccaaaa ttttgatttt 960
gaatccctca tctcctctct cttagaccta agcgaaatag aatacaaagg aaaatatatc 1020
gataaaaagc aagaagactt tgttccatca aatgcgcttt tatcacaaga ccgcctatgg 1080
caagcagttg aaaacctaac tcaaagcaat gaaacaatcg ttgctgaaca agggacatca 1140
ttctttggcg cttcatcaat tttcttaaaa ccaaagagtc attttattgg tcaaccctta 1200
tggggatcaa ttggatatac attcccagca gcattaggaa gccaaattgc agataaagaa 1260
agcagacacc ttttatttat tggtgatggt tcacttcaac ttacggtgca agaattagga 1320
ttagcaatca gagaaaaaat taatccaatt tgctttatta tcaataatga tggttataca 1380
gtcgaaagag aaattcatgg accaaatcaa agctacaatg atattccaat gtggaattac 1440
tcaaaattac cagaatcatt tggagcaaca gaagaacgag tagtctcgaa aatcgttaga 1500
actgaaaatg aatttgtgtc tgtcatgaaa gaagctcaag cagatccaaa tagaatgtac 1560
tggattgagt taattttggc aaaagaagat gcaccaaaag tactgaaaaa aatgggcaaa 1620
ctatttgctg aacaaaataa atcataa 1647
<210> SEQ ID NO 46
<211> LENGTH: 1647
<212> TYPE: DNA
<213> ORGANISM: Lactococcus lactis
<400> SEQUENCE: 46
atgtatactg ttggtgatta tctgctggat cgtctgcatg aactgggtat tgaggagatc 60
tttggtgttc cgggcgacta caacctgcag ttcctggatc agatcatttc ccgtaaggat 120
atgaaatggg ttggcaacgc caacgagctg aatgctagct atatggctga tggttatgcg 180
cgtaccaaaa aggcggctgc cttcctgacc acgttcggtg ttggcgaact gtctgccgtc 240
aacggcctgg ctggtagcta tgctgagaac ctgccagtgg ttgaaattgt tggttctcct 300
acctctaaag ttcagaacga aggtaaattc gtgcatcaca ctctggctga cggtgatttc 360
aaacacttca tgaaaatgca cgagccggtg accgctgccc gtactctgct gacggctgag 420
aacgcgactg tggagatcga ccgtgtgctg tctgcactgc tgaaagagcg taaaccggtg 480
tacattaacc tgccggtgga tgtcgccgca gctaaagcag agaaaccgtc tctgccgctg 540
aaaaaggaga acagcacgtc taacacgtcc gatcaggaga tcctgaacaa aatccaggag 600
tccctgaaaa acgcgaagaa accgatcgta atcactggtc atgaaattat cagctttggc 660
ctggaaaaga ctgtaagcca gtttatctct aaaaccaaac tgccgatcac cactctgaat 720
ttcggcaaaa gcagcgttga tgaggcactg ccttccttcc tgggcattta taacggtaaa 780
ctgtccgagc cgaacctgaa agagttcgtt gagtccgccg atttcattct gatgctgggc 840
gtcaaactga ctgactcttc tactggtgcc ttcacccacc acctgaacga aaacaaaatg 900
atttccctga acattgatga gggtaaaatc ttcaacgaaa gcatccagaa cttcgacttc 960
gaatctctga tctcctctct gctggatctg agcgagatcg aatacaaggg caaatacatt 1020
gataagaaac aggaggactt cgttccgtct aacgctctgc tgagccagga ccgtctgtgg 1080
caggcagtcg aaaacctgac ccagtccaac gaaaccatcg ttgcagagca gggtacttcc 1140
ttcttcggtg cctcttctat cttcctgaaa ccgaagtccc acttcattgg ccagccgctg 1200
tggggtagca tcggctatac cttccctgca gctctgggtt ctcagattgc ggataaagaa 1260
tctcgccatc tgctgttcat cggcgacggc agcctgcagc tgaccgttca ggaactgggc 1320
ctggctatcc gtgaaaagat caacccaatt tgcttcatca tcaataacga cggttacact 1380
gtggaacgcg agatccacgg tccgaaccag tcttacaacg atatcccgat gtggaactac 1440
tccaagctgc cagagagctt cggtgctact gaggaacgtg tcgttagcaa gatcgtacgc 1500
accgaaaatg agttcgtaag cgttatgaaa gaagctcaag ctgatccgaa ccgcatgtat 1560
tggatcgagc tgatcctggc aaaagaggat gccccaaaag ttctgaagaa aatgggcaaa 1620
ctgttcgccg agcaaaacaa atcataa 1647
<210> SEQ ID NO 47
<211> LENGTH: 548
<212> TYPE: PRT
<213> ORGANISM: Lactococcus lactis
<400> SEQUENCE: 47
Met Tyr Thr Val Gly Asp Tyr Leu Leu Asp Arg Leu His Glu Leu Gly
1 5 10 15
Ile Glu Glu Ile Phe Gly Val Pro Gly Asp Tyr Asn Leu Gln Phe Leu
20 25 30
Asp Gln Ile Ile Ser Arg Lys Asp Met Lys Trp Val Gly Asn Ala Asn
35 40 45
Glu Leu Asn Ala Ser Tyr Met Ala Asp Gly Tyr Ala Arg Thr Lys Lys
50 55 60
Ala Ala Ala Phe Leu Thr Thr Phe Gly Val Gly Glu Leu Ser Ala Val
65 70 75 80
Asn Gly Leu Ala Gly Ser Tyr Ala Glu Asn Leu Pro Val Val Glu Ile
85 90 95
Val Gly Ser Pro Thr Ser Lys Val Gln Asn Glu Gly Lys Phe Val His
100 105 110
His Thr Leu Ala Asp Gly Asp Phe Lys His Phe Met Lys Met His Glu
115 120 125
Pro Val Thr Ala Ala Arg Thr Leu Leu Thr Ala Glu Asn Ala Thr Val
130 135 140
Glu Ile Asp Arg Val Leu Ser Ala Leu Leu Lys Glu Arg Lys Pro Val
145 150 155 160
Tyr Ile Asn Leu Pro Val Asp Val Ala Ala Ala Lys Ala Glu Lys Pro
165 170 175
Ser Leu Pro Leu Lys Lys Glu Asn Ser Thr Ser Asn Thr Ser Asp Gln
180 185 190
Glu Ile Leu Asn Lys Ile Gln Glu Ser Leu Lys Asn Ala Lys Lys Pro
195 200 205
Ile Val Ile Thr Gly His Glu Ile Ile Ser Phe Gly Leu Glu Lys Thr
210 215 220
Val Ser Gln Phe Ile Ser Lys Thr Lys Leu Pro Ile Thr Thr Leu Asn
225 230 235 240
Phe Gly Lys Ser Ser Val Asp Glu Ala Leu Pro Ser Phe Leu Gly Ile
245 250 255
Tyr Asn Gly Lys Leu Ser Glu Pro Asn Leu Lys Glu Phe Val Glu Ser
260 265 270
Ala Asp Phe Ile Leu Met Leu Gly Val Lys Leu Thr Asp Ser Ser Thr
275 280 285
Gly Ala Phe Thr His His Leu Asn Glu Asn Lys Met Ile Ser Leu Asn
290 295 300
Ile Asp Glu Gly Lys Ile Phe Asn Glu Ser Ile Gln Asn Phe Asp Phe
305 310 315 320
Glu Ser Leu Ile Ser Ser Leu Leu Asp Leu Ser Glu Ile Glu Tyr Lys
325 330 335
Gly Lys Tyr Ile Asp Lys Lys Gln Glu Asp Phe Val Pro Ser Asn Ala
340 345 350
Leu Leu Ser Gln Asp Arg Leu Trp Gln Ala Val Glu Asn Leu Thr Gln
355 360 365
Ser Asn Glu Thr Ile Val Ala Glu Gln Gly Thr Ser Phe Phe Gly Ala
370 375 380
Ser Ser Ile Phe Leu Lys Pro Lys Ser His Phe Ile Gly Gln Pro Leu
385 390 395 400
Trp Gly Ser Ile Gly Tyr Thr Phe Pro Ala Ala Leu Gly Ser Gln Ile
405 410 415
Ala Asp Lys Glu Ser Arg His Leu Leu Phe Ile Gly Asp Gly Ser Leu
420 425 430
Gln Leu Thr Val Gln Glu Leu Gly Leu Ala Ile Arg Glu Lys Ile Asn
435 440 445
Pro Ile Cys Phe Ile Ile Asn Asn Asp Gly Tyr Thr Val Glu Arg Glu
450 455 460
Ile His Gly Pro Asn Gln Ser Tyr Asn Asp Ile Pro Met Trp Asn Tyr
465 470 475 480
Ser Lys Leu Pro Glu Ser Phe Gly Ala Thr Glu Glu Arg Val Val Ser
485 490 495
Lys Ile Val Arg Thr Glu Asn Glu Phe Val Ser Val Met Lys Glu Ala
500 505 510
Gln Ala Asp Pro Asn Arg Met Tyr Trp Ile Glu Leu Ile Leu Ala Lys
515 520 525
Glu Asp Ala Pro Lys Val Leu Lys Lys Met Gly Lys Leu Phe Ala Glu
530 535 540
Gln Asn Lys Ser
545
<210> SEQ ID NO 48
<211> LENGTH: 1647
<212> TYPE: DNA
<213> ORGANISM: Lactococcus lactis
<400> SEQUENCE: 48
atgtatactg ttggtgatta tctgctggac cgtctgcatg aactgggtat cgaagaaatc 60
ttcggcgttc cgggtgatta caatctgcag ttcctggatc agatcatctc tcataaagac 120
atgaaatggg tgggtaacgc taacgaactg aacgcaagct acatggcaga tggttatgca 180
cgtaccaaga aagccgcggc atttctgacc actttcggtg ttggcgaact gagcgccgtc 240
aacggtctgg cgggctccta cgccgaaaac ctgccggtgg tggagatcgt aggcagccca 300
acgagcaaag ttcagaacga aggtaaattc gtccaccaca ctctggctga cggcgatttc 360
aaacacttca tgaaaatgca tgaacctgtg actgcggcac gtacgctgct gactgcagag 420
aacgctactg tggaaatcga ccgcgttctg tctgcgctgc tgaaagaacg caaaccagtt 480
tacatcaacc tgcctgtgga tgttgcggca gctaaagcgg aaaaaccgag cctgccgctg 540
aagaaagaaa actccacttc taacactagc gaccaggaaa tcctgaacaa aatccaggag 600
tctctgaaaa acgcaaagaa accaatcgtg atcaccggcc acgaaatcat ttcttttggt 660
ctggagaaga ccgtgaccca attcatcagc aaaaccaaac tgccgattac caccctgaac 720
ttcggcaagt cctctgttga cgaggctctg ccgtctttcc tgggcatcta caacggtact 780
ctgagcgaac cgaacctgaa agaatttgtt gaatctgcgg acttcatcct gatgctgggc 840
gttaaactga ccgactcttc taccggtgca ttcactcacc atctgaacga aaacaaaatg 900
attagcctga acatcgacga gggtaaaatc ttcaacgagc gtatccagaa cttcgacttc 960
gaaagcctga tcagctctct gctggacctg tccgaaatcg agtataaagg caaatacatt 1020
gacaaaaagc aagaagattt cgtaccatct aacgcactgc tgtcccagga tcgcctgtgg 1080
caggccgtgg agaacctgac ccagagcaat gaaaccatcg tggcggaaca aggtacgagc 1140
tttttcggcg cgtcttctat ctttctgaaa tccaaaagcc attttatcgg tcagccgctg 1200
tggggtagca ttggctatac tttcccggca gcgctgggct ctcagatcgc tgataaagaa 1260
tctcgtcatc tgctgttcat cggtgacggt tccctgcagc tgaccgtaca ggaactgggt 1320
ctggcaattc gtgaaaagat caacccgatt tgcttcatta ttaacaatga cggctacacc 1380
gttgagcgtg agatccacgg tccgaaccag tcttacaacg atatccctat gtggaactac 1440
tctaaactgc cggagtcctt cggcgcaact gaggaccgtg ttgtgtctaa aattgtgcgt 1500
accgaaaacg aatttgtgag cgtgatgaaa gaggcccagg ccgatccgaa ccgtatgtac 1560
tggatcgaac tgatcctggc gaaagaaggc gcaccgaagg tactgaagaa aatgggcaag 1620
ctgtttgctg aacagaataa atcctaa 1647
<210> SEQ ID NO 49
<211> LENGTH: 548
<212> TYPE: PRT
<213> ORGANISM: Lactococcus lactis
<400> SEQUENCE: 49
Met Tyr Thr Val Gly Asp Tyr Leu Leu Asp Arg Leu His Glu Leu Gly
1 5 10 15
Ile Glu Glu Ile Phe Gly Val Pro Gly Asp Tyr Asn Leu Gln Phe Leu
20 25 30
Asp Gln Ile Ile Ser His Lys Asp Met Lys Trp Val Gly Asn Ala Asn
35 40 45
Glu Leu Asn Ala Ser Tyr Met Ala Asp Gly Tyr Ala Arg Thr Lys Lys
50 55 60
Ala Ala Ala Phe Leu Thr Thr Phe Gly Val Gly Glu Leu Ser Ala Val
65 70 75 80
Asn Gly Leu Ala Gly Ser Tyr Ala Glu Asn Leu Pro Val Val Glu Ile
85 90 95
Val Gly Ser Pro Thr Ser Lys Val Gln Asn Glu Gly Lys Phe Val His
100 105 110
His Thr Leu Ala Asp Gly Asp Phe Lys His Phe Met Lys Met His Glu
115 120 125
Pro Val Thr Ala Ala Arg Thr Leu Leu Thr Ala Glu Asn Ala Thr Val
130 135 140
Glu Ile Asp Arg Val Leu Ser Ala Leu Leu Lys Glu Arg Lys Pro Val
145 150 155 160
Tyr Ile Asn Leu Pro Val Asp Val Ala Ala Ala Lys Ala Glu Lys Pro
165 170 175
Ser Leu Pro Leu Lys Lys Glu Asn Ser Thr Ser Asn Thr Ser Asp Gln
180 185 190
Glu Ile Leu Asn Lys Ile Gln Glu Ser Leu Lys Asn Ala Lys Lys Pro
195 200 205
Ile Val Ile Thr Gly His Glu Ile Ile Ser Phe Gly Leu Glu Lys Thr
210 215 220
Val Thr Gln Phe Ile Ser Lys Thr Lys Leu Pro Ile Thr Thr Leu Asn
225 230 235 240
Phe Gly Lys Ser Ser Val Asp Glu Ala Leu Pro Ser Phe Leu Gly Ile
245 250 255
Tyr Asn Gly Thr Leu Ser Glu Pro Asn Leu Lys Glu Phe Val Glu Ser
260 265 270
Ala Asp Phe Ile Leu Met Leu Gly Val Lys Leu Thr Asp Ser Ser Thr
275 280 285
Gly Ala Phe Thr His His Leu Asn Glu Asn Lys Met Ile Ser Leu Asn
290 295 300
Ile Asp Glu Gly Lys Ile Phe Asn Glu Arg Ile Gln Asn Phe Asp Phe
305 310 315 320
Glu Ser Leu Ile Ser Ser Leu Leu Asp Leu Ser Glu Ile Glu Tyr Lys
325 330 335
Gly Lys Tyr Ile Asp Lys Lys Gln Glu Asp Phe Val Pro Ser Asn Ala
340 345 350
Leu Leu Ser Gln Asp Arg Leu Trp Gln Ala Val Glu Asn Leu Thr Gln
355 360 365
Ser Asn Glu Thr Ile Val Ala Glu Gln Gly Thr Ser Phe Phe Gly Ala
370 375 380
Ser Ser Ile Phe Leu Lys Ser Lys Ser His Phe Ile Gly Gln Pro Leu
385 390 395 400
Trp Gly Ser Ile Gly Tyr Thr Phe Pro Ala Ala Leu Gly Ser Gln Ile
405 410 415
Ala Asp Lys Glu Ser Arg His Leu Leu Phe Ile Gly Asp Gly Ser Leu
420 425 430
Gln Leu Thr Val Gln Glu Leu Gly Leu Ala Ile Arg Glu Lys Ile Asn
435 440 445
Pro Ile Cys Phe Ile Ile Asn Asn Asp Gly Tyr Thr Val Glu Arg Glu
450 455 460
Ile His Gly Pro Asn Gln Ser Tyr Asn Asp Ile Pro Met Trp Asn Tyr
465 470 475 480
Ser Lys Leu Pro Glu Ser Phe Gly Ala Thr Glu Asp Arg Val Val Ser
485 490 495
Lys Ile Val Arg Thr Glu Asn Glu Phe Val Ser Val Met Lys Glu Ala
500 505 510
Gln Ala Asp Pro Asn Arg Met Tyr Trp Ile Glu Leu Ile Leu Ala Lys
515 520 525
Glu Gly Ala Pro Lys Val Leu Lys Lys Met Gly Lys Leu Phe Ala Glu
530 535 540
Gln Asn Lys Ser
545
<210> SEQ ID NO 50
<211> LENGTH: 1851
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 50
atgcctaagt accgttccgc caccaccact catggtcgta atatggcggg tgctcgtgcg 60
ctgtggcgcg ccaccggaat gaccgacgcc gatttcggta agccgattat cgcggttgtg 120
aactcgttca cccaatttgt accgggtcac gtccatctgc gcgatctcgg taaactggtc 180
gccgaacaaa ttgaagcggc tggcggcgtt gccaaagagt tcaacaccat tgcggtggat 240
gatgggattg ccatgggcca cggggggatg ctttattcac tgccatctcg cgaactgatc 300
gctgattccg ttgagtatat ggtcaacgcc cactgcgccg acgccatggt ctgcatctct 360
aactgcgaca aaatcacccc ggggatgctg atggcttccc tgcgcctgaa tattccggtg 420
atctttgttt ccggcggccc gatggaggcc gggaaaacca aactttccga tcagatcatc 480
aagctcgatc tggttgatgc gatgatccag ggcgcagacc cgaaagtatc tgactcccag 540
agcgatcagg ttgaacgttc cgcgtgtccg acctgcggtt cctgctccgg gatgtttacc 600
gctaactcaa tgaactgcct gaccgaagcg ctgggcctgt cgcagccggg caacggctcg 660
ctgctggcaa cccacgccga ccgtaagcag ctgttcctta atgctggtaa acgcattgtt 720
gaattgacca aacgttatta cgagcaaaac gacgaaagtg cactgccgcg taatatcgcc 780
agtaaggcgg cgtttgaaaa cgccatgacg ctggatatcg cgatgggtgg atcgactaac 840
accgtacttc acctgctggc ggcggcgcag gaagcggaaa tcgacttcac catgagtgat 900
atcgataagc tttcccgcaa ggttccacag ctgtgtaaag ttgcgccgag cacccagaaa 960
taccatatgg aagatgttca ccgtgctggt ggtgttatcg gtattctcgg cgaactggat 1020
cgcgcggggt tactgaaccg tgatgtgaaa aacgtacttg gcctgacgtt gccgcaaacg 1080
ctggaacaat acgacgttat gctgacccag gatgacgcgg taaaaaatat gttccgcgca 1140
ggtcctgcag gcattcgtac cacacaggca ttctcgcaag attgccgttg ggatacgctg 1200
gacgacgatc gcgccaatgg ctgtatccgc tcgctggaac acgcctacag caaagacggc 1260
ggcctggcgg tgctctacgg taactttgcg gaaaacggct gcatcgtgaa aacggcaggc 1320
gtcgatgaca gcatcctcaa attcaccggc ccggcgaaag tgtacgaaag ccaggacgat 1380
gcggtagaag cgattctcgg cggtaaagtt gtcgccggag atgtggtagt aattcgctat 1440
gaaggcccga aaggcggtcc ggggatgcag gaaatgctct acccaaccag cttcctgaaa 1500
tcaatgggtc tcggcaaagc ctgtgcgctg atcaccgacg gtcgtttctc tggtggcacc 1560
tctggtcttt ccatcggcca cgtctcaccg gaagcggcaa gcggcggcag cattggcctg 1620
attgaagatg gtgacctgat cgctatcgac atcccgaacc gtggcattca gttacaggta 1680
agcgatgccg aactggcggc gcgtcgtgaa gcgcaggacg ctcgaggtga caaagcctgg 1740
acgccgaaaa atcgtgaacg tcaggtctcc tttgccctgc gtgcttatgc cagcctggca 1800
accagcgccg acaaaggcgc ggtgcgcgat aaatcgaaac tggggggtta a 1851
<210> SEQ ID NO 51
<211> LENGTH: 1851
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 51
atgcctaaat atcgcagcgc aactactacc cacggccgca acatggcagg cgcgcgtgct 60
ctgtggcgtg cgactggtat gactgatgcg gactttggca aaccaatcat tgctgtggtt 120
aatagcttta ctcagttcgt tccaggccat gttcacctgc gtgacctggg caagctggtt 180
gcggagcaga tcgaggctgc gggtggtgtg gcgaaggaat ttaacaccat cgctgttgac 240
gacggtatcg cgatgggtca tggtggtatg ctgtacagcc tgccgagccg tgagctgatt 300
gcggacagcg tggaatacat ggttaatgcg cattgtgcgg atgcgatggt ttgtattagc 360
aactgtgata agattactcc aggtatgctg atggcgagcc tgcgtctgaa catcccagtt 420
attttcgtga gcggtggtcc aatggaagcg ggtaagacta agctgagcga ccagattatc 480
aaactggacc tggtggacgc tatgattcaa ggtgctgatc caaaggttag cgatagccaa 540
tctgaccaag tggagcgcag cgcttgccca acttgtggca gctgtagcgg tatgttcact 600
gcgaatagca tgaattgtct gactgaggct ctgggtctga gccaaccagg taatggtagc 660
ctgctggcga ctcatgcgga tcgcaaacaa ctgtttctga acgcgggcaa gcgtatcgtg 720
gagctgacta agcgctacta tgaacagaat gatgagtccg cgctgccacg caacattgcg 780
tccaaagctg ctttcgagaa tgcgatgacc ctggacattg ctatgggcgg tagcaccaat 840
actgttctgc atctgctggc tgctgctcaa gaggctgaga ttgattttac tatgtccgac 900
attgacaaac tgagccgtaa agtgccgcaa ctgtgcaagg tggctccatc tactcaaaag 960
tatcacatgg aggacgtgca tcgcgcgggt ggcgtgattg gcatcctggg tgagctggac 1020
cgtgctggtc tgctgaatcg cgacgttaag aatgttctgg gtctgaccct gccacagacc 1080
ctggagcagt atgatgtgat gctgactcaa gacgatgctg ttaagaacat gtttcgtgct 1140
ggtccggcgg gtatccgcac tacccaagcg tttagccagg actgtcgctg ggacaccctg 1200
gatgatgacc gtgcgaacgg ttgcattcgt agcctggaac atgcgtattc taaggatggt 1260
ggtctggctg ttctgtatgg caatttcgct gagaatggtt gtattgttaa gaccgcgggt 1320
gttgacgatt ctattctgaa gtttactggt ccagctaagg tttatgagtc tcaagatgac 1380
gctgttgagg ctatcctggg tggcaaggtg gttgcgggtg acgttgttgt tatccgttac 1440
gagggtccaa agggtggccc aggtatgcaa gagatgctgt atccgacttc ttttctgaag 1500
agcatgggcc tgggtaaggc gtgcgctctg attactgatg gccgctttag cggcggtact 1560
agcggcctga gcattggtca tgttagccca gaggctgcgt ctggtggttc tatcggtctg 1620
atcgaggacg gcgatctgat tgcgattgat attccaaatc gcggtatcca actgcaagtt 1680
tctgacgcgg agctggctgc tcgccgcgag gctcaagatg cgcgtggcga taaggcgtgg 1740
accccaaaga accgcgagcg ccaagttagc ttcgcgctgc gcgcgtacgc ctctctggcg 1800
acttctgcgg ataagggtgc tgttcgtgac aagagcaagc tgggtggcta a 1851
<210> SEQ ID NO 52
<211> LENGTH: 616
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 52
Met Pro Lys Tyr Arg Ser Ala Thr Thr Thr His Gly Arg Asn Met Ala
1 5 10 15
Gly Ala Arg Ala Leu Trp Arg Ala Thr Gly Met Thr Asp Ala Asp Phe
20 25 30
Gly Lys Pro Ile Ile Ala Val Val Asn Ser Phe Thr Gln Phe Val Pro
35 40 45
Gly His Val His Leu Arg Asp Leu Gly Lys Leu Val Ala Glu Gln Ile
50 55 60
Glu Ala Ala Gly Gly Val Ala Lys Glu Phe Asn Thr Ile Ala Val Asp
65 70 75 80
Asp Gly Ile Ala Met Gly His Gly Gly Met Leu Tyr Ser Leu Pro Ser
85 90 95
Arg Glu Leu Ile Ala Asp Ser Val Glu Tyr Met Val Asn Ala His Cys
100 105 110
Ala Asp Ala Met Val Cys Ile Ser Asn Cys Asp Lys Ile Thr Pro Gly
115 120 125
Met Leu Met Ala Ser Leu Arg Leu Asn Ile Pro Val Ile Phe Val Ser
130 135 140
Gly Gly Pro Met Glu Ala Gly Lys Thr Lys Leu Ser Asp Gln Ile Ile
145 150 155 160
Lys Leu Asp Leu Val Asp Ala Met Ile Gln Gly Ala Asp Pro Lys Val
165 170 175
Ser Asp Ser Gln Ser Asp Gln Val Glu Arg Ser Ala Cys Pro Thr Cys
180 185 190
Gly Ser Cys Ser Gly Met Phe Thr Ala Asn Ser Met Asn Cys Leu Thr
195 200 205
Glu Ala Leu Gly Leu Ser Gln Pro Gly Asn Gly Ser Leu Leu Ala Thr
210 215 220
His Ala Asp Arg Lys Gln Leu Phe Leu Asn Ala Gly Lys Arg Ile Val
225 230 235 240
Glu Leu Thr Lys Arg Tyr Tyr Glu Gln Asn Asp Glu Ser Ala Leu Pro
245 250 255
Arg Asn Ile Ala Ser Lys Ala Ala Phe Glu Asn Ala Met Thr Leu Asp
260 265 270
Ile Ala Met Gly Gly Ser Thr Asn Thr Val Leu His Leu Leu Ala Ala
275 280 285
Ala Gln Glu Ala Glu Ile Asp Phe Thr Met Ser Asp Ile Asp Lys Leu
290 295 300
Ser Arg Lys Val Pro Gln Leu Cys Lys Val Ala Pro Ser Thr Gln Lys
305 310 315 320
Tyr His Met Glu Asp Val His Arg Ala Gly Gly Val Ile Gly Ile Leu
325 330 335
Gly Glu Leu Asp Arg Ala Gly Leu Leu Asn Arg Asp Val Lys Asn Val
340 345 350
Leu Gly Leu Thr Leu Pro Gln Thr Leu Glu Gln Tyr Asp Val Met Leu
355 360 365
Thr Gln Asp Asp Ala Val Lys Asn Met Phe Arg Ala Gly Pro Ala Gly
370 375 380
Ile Arg Thr Thr Gln Ala Phe Ser Gln Asp Cys Arg Trp Asp Thr Leu
385 390 395 400
Asp Asp Asp Arg Ala Asn Gly Cys Ile Arg Ser Leu Glu His Ala Tyr
405 410 415
Ser Lys Asp Gly Gly Leu Ala Val Leu Tyr Gly Asn Phe Ala Glu Asn
420 425 430
Gly Cys Ile Val Lys Thr Ala Gly Val Asp Asp Ser Ile Leu Lys Phe
435 440 445
Thr Gly Pro Ala Lys Val Tyr Glu Ser Gln Asp Asp Ala Val Glu Ala
450 455 460
Ile Leu Gly Gly Lys Val Val Ala Gly Asp Val Val Val Ile Arg Tyr
465 470 475 480
Glu Gly Pro Lys Gly Gly Pro Gly Met Gln Glu Met Leu Tyr Pro Thr
485 490 495
Ser Phe Leu Lys Ser Met Gly Leu Gly Lys Ala Cys Ala Leu Ile Thr
500 505 510
Asp Gly Arg Phe Ser Gly Gly Thr Ser Gly Leu Ser Ile Gly His Val
515 520 525
Ser Pro Glu Ala Ala Ser Gly Gly Ser Ile Gly Leu Ile Glu Asp Gly
530 535 540
Asp Leu Ile Ala Ile Asp Ile Pro Asn Arg Gly Ile Gln Leu Gln Val
545 550 555 560
Ser Asp Ala Glu Leu Ala Ala Arg Arg Glu Ala Gln Asp Ala Arg Gly
565 570 575
Asp Lys Ala Trp Thr Pro Lys Asn Arg Glu Arg Gln Val Ser Phe Ala
580 585 590
Leu Arg Ala Tyr Ala Ser Leu Ala Thr Ser Ala Asp Lys Gly Ala Val
595 600 605
Arg Asp Lys Ser Lys Leu Gly Gly
610 615
<210> SEQ ID NO 53
<400> SEQUENCE: 53
000
<210> SEQ ID NO 54
<211> LENGTH: 1713
<212> TYPE: DNA
<213> ORGANISM: Lactococcus lactis
<400> SEQUENCE: 54
atggagttta agtataacgg caaagttgaa tctgttgaac tgaataagta cagcaaaacg 60
ttgacacaag atcccacaca acccgccaca caggcaatgt attacggcat cgggtttaaa 120
gacgaagatt tcaagaaagc tcaagtgggt atagtgtcga tggactggga tggaaatcca 180
tgcaacatgc atttaggaac ccttggatca aagattaaaa gctcagtaaa tcagacagat 240
ggtctgatcg gcttacaatt tcatacgata ggagtttctg atgggatagc aaatggaaag 300
ttgggaatga gatactccct tgtttccaga gaagttatag ctgactctat tgaaaccaac 360
gctggcgctg aatactatga tgcaattgta gccatcccag gttgtgacaa aaatatgcca 420
ggttctatta ttggtatggc aagacttaat aggccaagca ttatggtgta tggaggaaca 480
atagaacacg gtgaatataa aggtgagaaa ttgaacatcg tatcggcttt tgaatctcta 540
ggccagaaaa ttaccggcaa tatctctgat gaagattatc acggtgttat ttgtaatgct 600
attcctggtc aaggggcatg tggggggatg tacacagcta ataccttagc tgccgctatc 660
gaaacactag gtatgtcatt gccgtattct tcttcgaacc ctgcagtatc tcaagaaaaa 720
caagaagaat gtgatgagat tggattagcc attaagaatc ttttggaaaa agacatcaag 780
cctagtgata taatgactaa ggaggcgttc gagaacgcta ttaccattgt gatggtcttg 840
gggggtagta ctaatgctgt cttgcatatt attgcaatgg ctaacgcgat aggtgtcgaa 900
ataactcagg atgacttcca aagaattagt gacattactc cagtactagg tgattttaaa 960
ccttcaggta aatatatgat ggaagatttg cataaaattg gaggcttgcc agcagtgctt 1020
aagtaccttc taaaggaagg aaaattgcat ggtgactgcc ttactgtgac gggtaaaaca 1080
ttagccgaga atgtcgagac tgccctagac ttggatttcg actcacaaga tatcatgagg 1140
ccactaaaga atcctatcaa ggccaccggc cacttgcaga ttctgtacgg taatttagct 1200
caagggggtt ccgtagcaaa aattagcggt aaagaaggag agttcttcaa aggcactgcc 1260
agagtctttg atggtgaaca acattttatc gacggcatag aatctggtcg tttgcatgct 1320
ggagatgtag cggtaattag gaatataggt cccgtcggcg gacctggtat gcccgaaatg 1380
ctgaagccta catcagcatt aattggtgcg ggtttaggga aaagttgcgc gttaattacg 1440
gatggtagat tctccggtgg cactcacggt tttgttgtcg gccatattgt gcctgaagcc 1500
gttgagggtg gactaatcgg cttagttgaa gatgacgata taatagagat agatgcagtc 1560
aacaactcta tatccctgaa agtttccgat gaagaaatcg caaagagaag agctaattat 1620
cagaagccaa ctccgaaagc caccagggga gttttggcaa aattcgctaa attaacccgt 1680
cctgcatcgg aagggtgtgt tactgatctg taa 1713
<210> SEQ ID NO 55
<211> LENGTH: 570
<212> TYPE: PRT
<213> ORGANISM: Lactococcus lactis
<400> SEQUENCE: 55
Met Glu Phe Lys Tyr Asn Gly Lys Val Glu Ser Val Glu Leu Asn Lys
1 5 10 15
Tyr Ser Lys Thr Leu Thr Gln Asp Pro Thr Gln Pro Ala Thr Gln Ala
20 25 30
Met Tyr Tyr Gly Ile Gly Phe Lys Asp Glu Asp Phe Lys Lys Ala Gln
35 40 45
Val Gly Ile Val Ser Met Asp Trp Asp Gly Asn Pro Cys Asn Met His
50 55 60
Leu Gly Thr Leu Gly Ser Lys Ile Lys Ser Ser Val Asn Gln Thr Asp
65 70 75 80
Gly Leu Ile Gly Leu Gln Phe His Thr Ile Gly Val Ser Asp Gly Ile
85 90 95
Ala Asn Gly Lys Leu Gly Met Arg Tyr Ser Leu Val Ser Arg Glu Val
100 105 110
Ile Ala Asp Ser Ile Glu Thr Asn Ala Gly Ala Glu Tyr Tyr Asp Ala
115 120 125
Ile Val Ala Ile Pro Gly Cys Asp Lys Asn Met Pro Gly Ser Ile Ile
130 135 140
Gly Met Ala Arg Leu Asn Arg Pro Ser Ile Met Val Tyr Gly Gly Thr
145 150 155 160
Ile Glu His Gly Glu Tyr Lys Gly Glu Lys Leu Asn Ile Val Ser Ala
165 170 175
Phe Glu Ser Leu Gly Gln Lys Ile Thr Gly Asn Ile Ser Asp Glu Asp
180 185 190
Tyr His Gly Val Ile Cys Asn Ala Ile Pro Gly Gln Gly Ala Cys Gly
195 200 205
Gly Met Tyr Thr Ala Asn Thr Leu Ala Ala Ala Ile Glu Thr Leu Gly
210 215 220
Met Ser Leu Pro Tyr Ser Ser Ser Asn Pro Ala Val Ser Gln Glu Lys
225 230 235 240
Gln Glu Glu Cys Asp Glu Ile Gly Leu Ala Ile Lys Asn Leu Leu Glu
245 250 255
Lys Asp Ile Lys Pro Ser Asp Ile Met Thr Lys Glu Ala Phe Glu Asn
260 265 270
Ala Ile Thr Ile Val Met Val Leu Gly Gly Ser Thr Asn Ala Val Leu
275 280 285
His Ile Ile Ala Met Ala Asn Ala Ile Gly Val Glu Ile Thr Gln Asp
290 295 300
Asp Phe Gln Arg Ile Ser Asp Ile Thr Pro Val Leu Gly Asp Phe Lys
305 310 315 320
Pro Ser Gly Lys Tyr Met Met Glu Asp Leu His Lys Ile Gly Gly Leu
325 330 335
Pro Ala Val Leu Lys Tyr Leu Leu Lys Glu Gly Lys Leu His Gly Asp
340 345 350
Cys Leu Thr Val Thr Gly Lys Thr Leu Ala Glu Asn Val Glu Thr Ala
355 360 365
Leu Asp Leu Asp Phe Asp Ser Gln Asp Ile Met Arg Pro Leu Lys Asn
370 375 380
Pro Ile Lys Ala Thr Gly His Leu Gln Ile Leu Tyr Gly Asn Leu Ala
385 390 395 400
Gln Gly Gly Ser Val Ala Lys Ile Ser Gly Lys Glu Gly Glu Phe Phe
405 410 415
Lys Gly Thr Ala Arg Val Phe Asp Gly Glu Gln His Phe Ile Asp Gly
420 425 430
Ile Glu Ser Gly Arg Leu His Ala Gly Asp Val Ala Val Ile Arg Asn
435 440 445
Ile Gly Pro Val Gly Gly Pro Gly Met Pro Glu Met Leu Lys Pro Thr
450 455 460
Ser Ala Leu Ile Gly Ala Gly Leu Gly Lys Ser Cys Ala Leu Ile Thr
465 470 475 480
Asp Gly Arg Phe Ser Gly Gly Thr His Gly Phe Val Val Gly His Ile
485 490 495
Val Pro Glu Ala Val Glu Gly Gly Leu Ile Gly Leu Val Glu Asp Asp
500 505 510
Asp Ile Ile Glu Ile Asp Ala Val Asn Asn Ser Ile Ser Leu Lys Val
515 520 525
Ser Asp Glu Glu Ile Ala Lys Arg Arg Ala Asn Tyr Gln Lys Pro Thr
530 535 540
Pro Lys Ala Thr Arg Gly Val Leu Ala Lys Phe Ala Lys Leu Thr Arg
545 550 555 560
Pro Ala Ser Glu Gly Cys Val Thr Asp Leu
565 570
<210> SEQ ID NO 56
<211> LENGTH: 1758
<212> TYPE: DNA
<213> ORGANISM: Saccharomyces cerevisiae
<400> SEQUENCE: 56
atgggcttgt taacgaaagt tgctacatct agacaattct ctacaacgag atgcgttgca 60
aagaagctca acaagtactc gtatatcatc actgaaccta agggccaagg tgcgtcccag 120
gccatgcttt atgccaccgg tttcaagaag gaagatttca agaagcctca agtcggggtt 180
ggttcctgtt ggtggtccgg taacccatgt aacatgcatc tattggactt gaataacaga 240
tgttctcaat ccattgaaaa agcgggtttg aaagctatgc agttcaacac catcggtgtt 300
tcagacggta tctctatggg tactaaaggt atgagatact cgttacaaag tagagaaatc 360
attgcagact cctttgaaac catcatgatg gcacaacact acgatgctaa catcgccatc 420
ccatcatgtg acaaaaacat gcccggtgtc atgatggcca tgggtagaca taacagacct 480
tccatcatgg tatatggtgg tactatcttg cccggtcatc caacatgtgg ttcttcgaag 540
atctctaaaa acatcgatat cgtctctgcg ttccaatcct acggtgaata tatttccaag 600
caattcactg aagaagaaag agaagatgtt gtggaacatg catgcccagg tcctggttct 660
tgtggtggta tgtatactgc caacacaatg gcttctgccg ctgaagtgct aggtttgacc 720
attccaaact cctcttcctt cccagccgtt tccaaggaga agttagctga gtgtgacaac 780
attggtgaat acatcaagaa gacaatggaa ttgggtattt tacctcgtga tatcctcaca 840
aaagaggctt ttgaaaacgc cattacttat gtcgttgcaa ccggtgggtc cactaatgct 900
gttttgcatt tggtggctgt tgctcactct gcgggtgtca agttgtcacc agatgatttc 960
caaagaatca gtgatactac accattgatc ggtgacttca aaccttctgg taaatacgtc 1020
atggccgatt tgattaacgt tggtggtacc caatctgtga ttaagtatct atatgaaaac 1080
aacatgttgc acggtaacac aatgactgtt accggtgaca ctttggcaga acgtgcaaag 1140
aaagcaccaa gcctacctga aggacaagag attattaagc cactctccca cccaatcaag 1200
gccaacggtc acttgcaaat tctgtacggt tcattggcac caggtggagc tgtgggtaaa 1260
attaccggta aggaaggtac ttacttcaag ggtagagcac gtgtgttcga agaggaaggt 1320
gcctttattg aagccttgga aagaggtgaa atcaagaagg gtgaaaaaac cgttgttgtt 1380
atcagatatg aaggtccaag aggtgcacca ggtatgcctg aaatgctaaa gccttcctct 1440
gctctgatgg gttacggttt gggtaaagat gttgcattgt tgactgatgg tagattctct 1500
ggtggttctc acgggttctt aatcggccac attgttcccg aagccgctga aggtggtcct 1560
atcgggttgg tcagagacgg cgatgagatt atcattgatg ctgataataa caagattgac 1620
ctattagtct ctgataagga aatggctcaa cgtaaacaaa gttgggttgc acctccacct 1680
cgttacacaa gaggtactct atccaagtat gctaagttgg tttccaacgc ttccaacggt 1740
tgtgttttag atgcttga 1758
<210> SEQ ID NO 57
<211> LENGTH: 585
<212> TYPE: PRT
<213> ORGANISM: Saccharomyces cerevisiae
<400> SEQUENCE: 57
Met Gly Leu Leu Thr Lys Val Ala Thr Ser Arg Gln Phe Ser Thr Thr
1 5 10 15
Arg Cys Val Ala Lys Lys Leu Asn Lys Tyr Ser Tyr Ile Ile Thr Glu
20 25 30
Pro Lys Gly Gln Gly Ala Ser Gln Ala Met Leu Tyr Ala Thr Gly Phe
35 40 45
Lys Lys Glu Asp Phe Lys Lys Pro Gln Val Gly Val Gly Ser Cys Trp
50 55 60
Trp Ser Gly Asn Pro Cys Asn Met His Leu Leu Asp Leu Asn Asn Arg
65 70 75 80
Cys Ser Gln Ser Ile Glu Lys Ala Gly Leu Lys Ala Met Gln Phe Asn
85 90 95
Thr Ile Gly Val Ser Asp Gly Ile Ser Met Gly Thr Lys Gly Met Arg
100 105 110
Tyr Ser Leu Gln Ser Arg Glu Ile Ile Ala Asp Ser Phe Glu Thr Ile
115 120 125
Met Met Ala Gln His Tyr Asp Ala Asn Ile Ala Ile Pro Ser Cys Asp
130 135 140
Lys Asn Met Pro Gly Val Met Met Ala Met Gly Arg His Asn Arg Pro
145 150 155 160
Ser Ile Met Val Tyr Gly Gly Thr Ile Leu Pro Gly His Pro Thr Cys
165 170 175
Gly Ser Ser Lys Ile Ser Lys Asn Ile Asp Ile Val Ser Ala Phe Gln
180 185 190
Ser Tyr Gly Glu Tyr Ile Ser Lys Gln Phe Thr Glu Glu Glu Arg Glu
195 200 205
Asp Val Val Glu His Ala Cys Pro Gly Pro Gly Ser Cys Gly Gly Met
210 215 220
Tyr Thr Ala Asn Thr Met Ala Ser Ala Ala Glu Val Leu Gly Leu Thr
225 230 235 240
Ile Pro Asn Ser Ser Ser Phe Pro Ala Val Ser Lys Glu Lys Leu Ala
245 250 255
Glu Cys Asp Asn Ile Gly Glu Tyr Ile Lys Lys Thr Met Glu Leu Gly
260 265 270
Ile Leu Pro Arg Asp Ile Leu Thr Lys Glu Ala Phe Glu Asn Ala Ile
275 280 285
Thr Tyr Val Val Ala Thr Gly Gly Ser Thr Asn Ala Val Leu His Leu
290 295 300
Val Ala Val Ala His Ser Ala Gly Val Lys Leu Ser Pro Asp Asp Phe
305 310 315 320
Gln Arg Ile Ser Asp Thr Thr Pro Leu Ile Gly Asp Phe Lys Pro Ser
325 330 335
Gly Lys Tyr Val Met Ala Asp Leu Ile Asn Val Gly Gly Thr Gln Ser
340 345 350
Val Ile Lys Tyr Leu Tyr Glu Asn Asn Met Leu His Gly Asn Thr Met
355 360 365
Thr Val Thr Gly Asp Thr Leu Ala Glu Arg Ala Lys Lys Ala Pro Ser
370 375 380
Leu Pro Glu Gly Gln Glu Ile Ile Lys Pro Leu Ser His Pro Ile Lys
385 390 395 400
Ala Asn Gly His Leu Gln Ile Leu Tyr Gly Ser Leu Ala Pro Gly Gly
405 410 415
Ala Val Gly Lys Ile Thr Gly Lys Glu Gly Thr Tyr Phe Lys Gly Arg
420 425 430
Ala Arg Val Phe Glu Glu Glu Gly Ala Phe Ile Glu Ala Leu Glu Arg
435 440 445
Gly Glu Ile Lys Lys Gly Glu Lys Thr Val Val Val Ile Arg Tyr Glu
450 455 460
Gly Pro Arg Gly Ala Pro Gly Met Pro Glu Met Leu Lys Pro Ser Ser
465 470 475 480
Ala Leu Met Gly Tyr Gly Leu Gly Lys Asp Val Ala Leu Leu Thr Asp
485 490 495
Gly Arg Phe Ser Gly Gly Ser His Gly Phe Leu Ile Gly His Ile Val
500 505 510
Pro Glu Ala Ala Glu Gly Gly Pro Ile Gly Leu Val Arg Asp Gly Asp
515 520 525
Glu Ile Ile Ile Asp Ala Asp Asn Asn Lys Ile Asp Leu Leu Val Ser
530 535 540
Asp Lys Glu Met Ala Gln Arg Lys Gln Ser Trp Val Ala Pro Pro Pro
545 550 555 560
Arg Tyr Thr Arg Gly Thr Leu Ser Lys Tyr Ala Lys Leu Val Ser Asn
565 570 575
Ala Ser Asn Gly Cys Val Leu Asp Ala
580 585
<210> SEQ ID NO 58
<211> LENGTH: 1701
<212> TYPE: DNA
<213> ORGANISM: Saccharomyces cerevisiae
<400> SEQUENCE: 58
atgaagaagc tcaacaagta ctcgtatatc atcactgaac ctaagggcca aggtgcgtcc 60
caggccatgc tttatgccac cggtttcaag aaggaagatt tcaagaagcc tcaagtcggg 120
gttggttcct gttggtggtc cggtaaccca tgtaacatgc atctattgga cttgaataac 180
agatgttctc aatccattga aaaagcgggt ttgaaagcta tgcagttcaa caccatcggt 240
gtttcagacg gtatctctat gggtactaaa ggtatgagat actcgttaca aagtagagaa 300
atcattgcag actcctttga aaccatcatg atggcacaac actacgatgc taacatcgcc 360
atcccatcat gtgacaaaaa catgcccggt gtcatgatgg ccatgggtag acataacaga 420
ccttccatca tggtatatgg tggtactatc ttgcccggtc atccaacatg tggttcttcg 480
aagatctcta aaaacatcga tatcgtctct gcgttccaat cctacggtga atatatttcc 540
aagcaattca ctgaagaaga aagagaagat gttgtggaac atgcatgccc aggtcctggt 600
tcttgtggtg gtatgtatac tgccaacaca atggcttctg ccgctgaagt gctaggtttg 660
accattccaa actcctcttc cttcccagcc gtttccaagg agaagttagc tgagtgtgac 720
aacattggtg aatacatcaa gaagacaatg gaattgggta ttttacctcg tgatatcctc 780
acaaaagagg cttttgaaaa cgccattact tatgtcgttg caaccggtgg gtccactaat 840
gctgttttgc atttggtggc tgttgctcac tctgcgggtg tcaagttgtc accagatgat 900
ttccaaagaa tcagtgatac tacaccattg atcggtgact tcaaaccttc tggtaaatac 960
gtcatggccg atttgattaa cgttggtggt acccaatctg tgattaagta tctatatgaa 1020
aacaacatgt tgcacggtaa cacaatgact gttaccggtg acactttggc agaacgtgca 1080
aagaaagcac caagcctacc tgaaggacaa gagattatta agccactctc ccacccaatc 1140
aaggccaacg gtcacttgca aattctgtac ggttcattgg caccaggtgg agctgtgggt 1200
aaaattaccg gtaaggaagg tacttacttc aagggtagag cacgtgtgtt cgaagaggaa 1260
ggtgccttta ttgaagcctt ggaaagaggt gaaatcaaga agggtgaaaa aaccgttgtt 1320
gttatcagat atgaaggtcc aagaggtgca ccaggtatgc ctgaaatgct aaagccttcc 1380
tctgctctga tgggttacgg tttgggtaaa gatgttgcat tgttgactga tggtagattc 1440
tctggtggtt ctcacgggtt cttaatcggc cacattgttc ccgaagccgc tgaaggtggt 1500
cctatcgggt tggtcagaga cggcgatgag attatcattg atgctgataa taacaagatt 1560
gacctattag tctctgataa ggaaatggct caacgtaaac aaagttgggt tgcacctcca 1620
cctcgttaca caagaggtac tctatccaag tatgctaagt tggtttccaa cgcttccaac 1680
ggttgtgttt tagatgcttg a 1701
<210> SEQ ID NO 59
<211> LENGTH: 566
<212> TYPE: PRT
<213> ORGANISM: Saccharomyces cerevisiae
<400> SEQUENCE: 59
Met Lys Lys Leu Asn Lys Tyr Ser Tyr Ile Ile Thr Glu Pro Lys Gly
1 5 10 15
Gln Gly Ala Ser Gln Ala Met Leu Tyr Ala Thr Gly Phe Lys Lys Glu
20 25 30
Asp Phe Lys Lys Pro Gln Val Gly Val Gly Ser Cys Trp Trp Ser Gly
35 40 45
Asn Pro Cys Asn Met His Leu Leu Asp Leu Asn Asn Arg Cys Ser Gln
50 55 60
Ser Ile Glu Lys Ala Gly Leu Lys Ala Met Gln Phe Asn Thr Ile Gly
65 70 75 80
Val Ser Asp Gly Ile Ser Met Gly Thr Lys Gly Met Arg Tyr Ser Leu
85 90 95
Gln Ser Arg Glu Ile Ile Ala Asp Ser Phe Glu Thr Ile Met Met Ala
100 105 110
Gln His Tyr Asp Ala Asn Ile Ala Ile Pro Ser Cys Asp Lys Asn Met
115 120 125
Pro Gly Val Met Met Ala Met Gly Arg His Asn Arg Pro Ser Ile Met
130 135 140
Val Tyr Gly Gly Thr Ile Leu Pro Gly His Pro Thr Cys Gly Ser Ser
145 150 155 160
Lys Ile Ser Lys Asn Ile Asp Ile Val Ser Ala Phe Gln Ser Tyr Gly
165 170 175
Glu Tyr Ile Ser Lys Gln Phe Thr Glu Glu Glu Arg Glu Asp Val Val
180 185 190
Glu His Ala Cys Pro Gly Pro Gly Ser Cys Gly Gly Met Tyr Thr Ala
195 200 205
Asn Thr Met Ala Ser Ala Ala Glu Val Leu Gly Leu Thr Ile Pro Asn
210 215 220
Ser Ser Ser Phe Pro Ala Val Ser Lys Glu Lys Leu Ala Glu Cys Asp
225 230 235 240
Asn Ile Gly Glu Tyr Ile Lys Lys Thr Met Glu Leu Gly Ile Leu Pro
245 250 255
Arg Asp Ile Leu Thr Lys Glu Ala Phe Glu Asn Ala Ile Thr Tyr Val
260 265 270
Val Ala Thr Gly Gly Ser Thr Asn Ala Val Leu His Leu Val Ala Val
275 280 285
Ala His Ser Ala Gly Val Lys Leu Ser Pro Asp Asp Phe Gln Arg Ile
290 295 300
Ser Asp Thr Thr Pro Leu Ile Gly Asp Phe Lys Pro Ser Gly Lys Tyr
305 310 315 320
Val Met Ala Asp Leu Ile Asn Val Gly Gly Thr Gln Ser Val Ile Lys
325 330 335
Tyr Leu Tyr Glu Asn Asn Met Leu His Gly Asn Thr Met Thr Val Thr
340 345 350
Gly Asp Thr Leu Ala Glu Arg Ala Lys Lys Ala Pro Ser Leu Pro Glu
355 360 365
Gly Gln Glu Ile Ile Lys Pro Leu Ser His Pro Ile Lys Ala Asn Gly
370 375 380
His Leu Gln Ile Leu Tyr Gly Ser Leu Ala Pro Gly Gly Ala Val Gly
385 390 395 400
Lys Ile Thr Gly Lys Glu Gly Thr Tyr Phe Lys Gly Arg Ala Arg Val
405 410 415
Phe Glu Glu Glu Gly Ala Phe Ile Glu Ala Leu Glu Arg Gly Glu Ile
420 425 430
Lys Lys Gly Glu Lys Thr Val Val Val Ile Arg Tyr Glu Gly Pro Arg
435 440 445
Gly Ala Pro Gly Met Pro Glu Met Leu Lys Pro Ser Ser Ala Leu Met
450 455 460
Gly Tyr Gly Leu Gly Lys Asp Val Ala Leu Leu Thr Asp Gly Arg Phe
465 470 475 480
Ser Gly Gly Ser His Gly Phe Leu Ile Gly His Ile Val Pro Glu Ala
485 490 495
Ala Glu Gly Gly Pro Ile Gly Leu Val Arg Asp Gly Asp Glu Ile Ile
500 505 510
Ile Asp Ala Asp Asn Asn Lys Ile Asp Leu Leu Val Ser Asp Lys Glu
515 520 525
Met Ala Gln Arg Lys Gln Ser Trp Val Ala Pro Pro Pro Arg Tyr Thr
530 535 540
Arg Gly Thr Leu Ser Lys Tyr Ala Lys Leu Val Ser Asn Ala Ser Asn
545 550 555 560
Gly Cys Val Leu Asp Ala
565
<210> SEQ ID NO 60
<211> LENGTH: 771
<212> TYPE: DNA
<213> ORGANISM: Drosophila melanogaster
<400> SEQUENCE: 60
atgtcgttta ctttgaccaa caagaacgtg attttcgttg ccggtctggg aggcattggt 60
ctggacacca gcaaggagct gctcaagcgc gatctgaaga acctggtgat cctcgaccgc 120
attgagaacc cggctgccat tgccgagctg aaggcaatca atccaaaggt gaccgtcacc 180
ttctacccct atgatgtgac cgtgcccatt gccgagacca ccaagctgct gaagaccatc 240
ttcgcccagc tgaagaccgt cgatgtcctg atcaacggag ctggtatcct ggacgatcac 300
cagatcgagc gcaccattgc cgtcaactac actggcctgg tcaacaccac gacggccatt 360
ctggacttct gggacaagcg caagggcggt cccggtggta tcatctgcaa cattggatcc 420
gtcactggat tcaatgccat ctaccaggtg cccgtctact ccggcaccaa ggccgccgtg 480
gtcaacttca ccagctccct ggcgaaactg gcccccatta ccggcgtgac ggcttacact 540
gtgaaccccg gcatcacccg caccaccctg gtgcacacgt tcaactcctg gttggatgtt 600
gagcctcagg ttgccgagaa gctcctggct catcccaccc agccctcgtt ggcctgcgcc 660
gagaacttcg tcaaggctat cgagctgaac cagaacggag ccatctggaa actggacttg 720
ggcaccctgg aggccatcca gtggaccaag cactgggact ccggcatcta a 771
<210> SEQ ID NO 61
<211> LENGTH: 256
<212> TYPE: PRT
<213> ORGANISM: Drosophila melanogaster
<400> SEQUENCE: 61
Met Ser Phe Thr Leu Thr Asn Lys Asn Val Ile Phe Val Ala Gly Leu
1 5 10 15
Gly Gly Ile Gly Leu Asp Thr Ser Lys Glu Leu Leu Lys Arg Asp Leu
20 25 30
Lys Asn Leu Val Ile Leu Asp Arg Ile Glu Asn Pro Ala Ala Ile Ala
35 40 45
Glu Leu Lys Ala Ile Asn Pro Lys Val Thr Val Thr Phe Tyr Pro Tyr
50 55 60
Asp Val Thr Val Pro Ile Ala Glu Thr Thr Lys Leu Leu Lys Thr Ile
65 70 75 80
Phe Ala Gln Leu Lys Thr Val Asp Val Leu Ile Asn Gly Ala Gly Ile
85 90 95
Leu Asp Asp His Gln Ile Glu Arg Thr Ile Ala Val Asn Tyr Thr Gly
100 105 110
Leu Val Asn Thr Thr Thr Ala Ile Leu Asp Phe Trp Asp Lys Arg Lys
115 120 125
Gly Gly Pro Gly Gly Ile Ile Cys Asn Ile Gly Ser Val Thr Gly Phe
130 135 140
Asn Ala Ile Tyr Gln Val Pro Val Tyr Ser Gly Thr Lys Ala Ala Val
145 150 155 160
Val Asn Phe Thr Ser Ser Leu Ala Lys Leu Ala Pro Ile Thr Gly Val
165 170 175
Thr Ala Tyr Thr Val Asn Pro Gly Ile Thr Arg Thr Thr Leu Val His
180 185 190
Thr Phe Asn Ser Trp Leu Asp Val Glu Pro Gln Val Ala Glu Lys Leu
195 200 205
Leu Ala His Pro Thr Gln Pro Ser Leu Ala Cys Ala Glu Asn Phe Val
210 215 220
Lys Ala Ile Glu Leu Asn Gln Asn Gly Ala Ile Trp Lys Leu Asp Leu
225 230 235 240
Gly Thr Leu Glu Ala Ile Gln Trp Thr Lys His Trp Asp Ser Gly Ile
245 250 255
<210> SEQ ID NO 62
<211> LENGTH: 1164
<212> TYPE: DNA
<213> ORGANISM: Klebsiella pneumoniae
<400> SEQUENCE: 62
atgagctacc gtatgtttga ctatctggtc cctaacgtga acttcttcgg cccgaatgca 60
atctctgtgg ttggcgaacg ttgccaactg ctgggtggta aaaaggcgct gctggtgacg 120
gataaaggtc tgcgtgcaat taaagacggt gccgttgata aaaccctgca ctatctgcgt 180
gaggccggca ttgaggttgc catcttcgat ggtgtagaac cgaacccgaa agatacgaac 240
gtgcgcgacg gtctggctgt tttccgtcgt gaacaatgtg acattatcgt taccgtgggt 300
ggtggctctc cgcatgattg cggtaaaggc atcggtatcg cggctaccca cgaaggtgat 360
ctgtaccagt atgcgggcat cgagactctg accaacccgc tgccgccgat cgttgctgta 420
aacaccacgg ccggcaccgc ctccgaagtt acccgtcatt gtgtgctgac taacaccgag 480
acgaaagtga aattcgttat tgtgtcctgg cgcaatctgc ctagcgtaag cattaacgat 540
ccgctgctga tgatcggcaa accagcggca ctgaccgctg caactggtat ggacgccctg 600
actcacgcag tcgaagcata tatctccaaa gatgctaacc cggtaaccga cgcggcagct 660
atgcaggcga ttcgtctgat tgcccgtaac ctgcgtcagg cagtggctct gggcagcaac 720
ctgcaggctc gtgagaacat ggcctacgcg agcctgctgg ccggcatggc attcaacaac 780
gctaacctgg gttacgttca tgcgatggct catcagctgg gcggcctgta cgacatgccg 840
cacggtgtag ctaacgcagt tctgctgcca catgttgctc gttataacct gatcgctaat 900
ccggaaaaat tcgcagacat cgcagaactg atgggcgaga acatcacggg tctgagcact 960
ctggatgccg cggaaaaagc gatcgcagcg attacgcgtc tgtctatgga cattggtatt 1020
ccgcaacacc tgcgtgacct gggtgtaaaa gaagctgatt tcccttacat ggcggaaatg 1080
gcactgaaag atggtaatgc gttttccaac ccacgtaaag gtaacgaaca ggagattgcg 1140
gctattttcc gtcaagcatt ctga 1164
<210> SEQ ID NO 63
<211> LENGTH: 387
<212> TYPE: PRT
<213> ORGANISM: Klebsiella pneumoniae
<400> SEQUENCE: 63
Met Ser Tyr Arg Met Phe Asp Tyr Leu Val Pro Asn Val Asn Phe Phe
1 5 10 15
Gly Pro Asn Ala Ile Ser Val Val Gly Glu Arg Cys Gln Leu Leu Gly
20 25 30
Gly Lys Lys Ala Leu Leu Val Thr Asp Lys Gly Leu Arg Ala Ile Lys
35 40 45
Asp Gly Ala Val Asp Lys Thr Leu His Tyr Leu Arg Glu Ala Gly Ile
50 55 60
Glu Val Ala Ile Phe Asp Gly Val Glu Pro Asn Pro Lys Asp Thr Asn
65 70 75 80
Val Arg Asp Gly Leu Ala Val Phe Arg Arg Glu Gln Cys Asp Ile Ile
85 90 95
Val Thr Val Gly Gly Gly Ser Pro His Asp Cys Gly Lys Gly Ile Gly
100 105 110
Ile Ala Ala Thr His Glu Gly Asp Leu Tyr Gln Tyr Ala Gly Ile Glu
115 120 125
Thr Leu Thr Asn Pro Leu Pro Pro Ile Val Ala Val Asn Thr Thr Ala
130 135 140
Gly Thr Ala Ser Glu Val Thr Arg His Cys Val Leu Thr Asn Thr Glu
145 150 155 160
Thr Lys Val Lys Phe Val Ile Val Ser Trp Arg Asn Leu Pro Ser Val
165 170 175
Ser Ile Asn Asp Pro Leu Leu Met Ile Gly Lys Pro Ala Ala Leu Thr
180 185 190
Ala Ala Thr Gly Met Asp Ala Leu Thr His Ala Val Glu Ala Tyr Ile
195 200 205
Ser Lys Asp Ala Asn Pro Val Thr Asp Ala Ala Ala Met Gln Ala Ile
210 215 220
Arg Leu Ile Ala Arg Asn Leu Arg Gln Ala Val Ala Leu Gly Ser Asn
225 230 235 240
Leu Gln Ala Arg Glu Asn Met Ala Tyr Ala Ser Leu Leu Ala Gly Met
245 250 255
Ala Phe Asn Asn Ala Asn Leu Gly Tyr Val His Ala Met Ala His Gln
260 265 270
Leu Gly Gly Leu Tyr Asp Met Pro His Gly Val Ala Asn Ala Val Leu
275 280 285
Leu Pro His Val Ala Arg Tyr Asn Leu Ile Ala Asn Pro Glu Lys Phe
290 295 300
Ala Asp Ile Ala Glu Leu Met Gly Glu Asn Ile Thr Gly Leu Ser Thr
305 310 315 320
Leu Asp Ala Ala Glu Lys Ala Ile Ala Ala Ile Thr Arg Leu Ser Met
325 330 335
Asp Ile Gly Ile Pro Gln His Leu Arg Asp Leu Gly Val Lys Glu Ala
340 345 350
Asp Phe Pro Tyr Met Ala Glu Met Ala Leu Lys Asp Gly Asn Ala Phe
355 360 365
Ser Asn Pro Arg Lys Gly Asn Glu Gln Glu Ile Ala Ala Ile Phe Arg
370 375 380
Gln Ala Phe
385
<210> SEQ ID NO 64
<211> LENGTH: 1152
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 64
atgatggcta acagaatgat tctgaacgaa acggcatggt ttggtcgggg tgctgttggg 60
gctttaaccg atgaggtgaa acgccgtggt tatcagaagg cgctgatcgt caccgataaa 120
acgctggtgc aatgcggcgt ggtggcgaaa gtgaccgata agatggatgc tgcagggctg 180
gcatgggcga tttacgacgg cgtagtgccc aacccaacaa ttactgtcgt caaagaaggg 240
ctcggtgtat tccagaatag cggcgcggat tacctgatcg ctattggtgg tggttctcca 300
caggatactt gtaaagcgat tggcattatc agcaacaacc cggagtttgc cgatgtgcgt 360
agcctggaag ggctttcccc gaccaataaa cccagtgtac cgattctggc aattcctacc 420
acagcaggta ctgcggcaga agtgaccatt aactacgtga tcactgacga agagaaacgg 480
cgcaagtttg tttgcgttga tccgcatgat atcccgcagg tggcgtttat tgacgctgac 540
atgatggatg gtatgcctcc agcgctgaaa gctgcgacgg gtgtcgatgc gctcactcat 600
gctattgagg ggtatattac ccgtggcgcg tgggcgctaa ccgatgcact gcacattaaa 660
gcgattgaaa tcattgctgg ggcgctgcga ggatcggttg ctggtgataa ggatgccgga 720
gaagaaatgg cgctcgggca gtatgttgcg ggtatgggct tctcgaatgt tgggttaggg 780
ttggtgcatg gtatggcgca tccactgggc gcgttttata acactccaca cggtgttgcg 840
aacgccatcc tgttaccgca tgtcatgcgt tataacgctg actttaccgg tgagaagtac 900
cgcgatatcg cgcgcgttat gggcgtgaaa gtggaaggta tgagcctgga agaggcgcgt 960
aatgccgctg ttgaagcggt gtttgctctc aaccgtgatg tcggtattcc gccacatttg 1020
cgtgatgttg gtgtacgcaa ggaagacatt ccggcactgg cgcaggcggc actggatgat 1080
gtttgtaccg gtggcaaccc gcgtgaagca acgcttgagg atattgtaga gctttaccat 1140
accgcctggt aa 1152
<210> SEQ ID NO 65
<211> LENGTH: 383
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 65
Met Met Ala Asn Arg Met Ile Leu Asn Glu Thr Ala Trp Phe Gly Arg
1 5 10 15
Gly Ala Val Gly Ala Leu Thr Asp Glu Val Lys Arg Arg Gly Tyr Gln
20 25 30
Lys Ala Leu Ile Val Thr Asp Lys Thr Leu Val Gln Cys Gly Val Val
35 40 45
Ala Lys Val Thr Asp Lys Met Asp Ala Ala Gly Leu Ala Trp Ala Ile
50 55 60
Tyr Asp Gly Val Val Pro Asn Pro Thr Ile Thr Val Val Lys Glu Gly
65 70 75 80
Leu Gly Val Phe Gln Asn Ser Gly Ala Asp Tyr Leu Ile Ala Ile Gly
85 90 95
Gly Gly Ser Pro Gln Asp Thr Cys Lys Ala Ile Gly Ile Ile Ser Asn
100 105 110
Asn Pro Glu Phe Ala Asp Val Arg Ser Leu Glu Gly Leu Ser Pro Thr
115 120 125
Asn Lys Pro Ser Val Pro Ile Leu Ala Ile Pro Thr Thr Ala Gly Thr
130 135 140
Ala Ala Glu Val Thr Ile Asn Tyr Val Ile Thr Asp Glu Glu Lys Arg
145 150 155 160
Arg Lys Phe Val Cys Val Asp Pro His Asp Ile Pro Gln Val Ala Phe
165 170 175
Ile Asp Ala Asp Met Met Asp Gly Met Pro Pro Ala Leu Lys Ala Ala
180 185 190
Thr Gly Val Asp Ala Leu Thr His Ala Ile Glu Gly Tyr Ile Thr Arg
195 200 205
Gly Ala Trp Ala Leu Thr Asp Ala Leu His Ile Lys Ala Ile Glu Ile
210 215 220
Ile Ala Gly Ala Leu Arg Gly Ser Val Ala Gly Asp Lys Asp Ala Gly
225 230 235 240
Glu Glu Met Ala Leu Gly Gln Tyr Val Ala Gly Met Gly Phe Ser Asn
245 250 255
Val Gly Leu Gly Leu Val His Gly Met Ala His Pro Leu Gly Ala Phe
260 265 270
Tyr Asn Thr Pro His Gly Val Ala Asn Ala Ile Leu Leu Pro His Val
275 280 285
Met Arg Tyr Asn Ala Asp Phe Thr Gly Glu Lys Tyr Arg Asp Ile Ala
290 295 300
Arg Val Met Gly Val Lys Val Glu Gly Met Ser Leu Glu Glu Ala Arg
305 310 315 320
Asn Ala Ala Val Glu Ala Val Phe Ala Leu Asn Arg Asp Val Gly Ile
325 330 335
Pro Pro His Leu Arg Asp Val Gly Val Arg Lys Glu Asp Ile Pro Ala
340 345 350
Leu Ala Gln Ala Ala Leu Asp Asp Val Cys Thr Gly Gly Asn Pro Arg
355 360 365
Glu Ala Thr Leu Glu Asp Ile Val Glu Leu Tyr His Thr Ala Trp
370 375 380
<210> SEQ ID NO 66
<211> LENGTH: 1023
<212> TYPE: DNA
<213> ORGANISM: Lactococcus lactis
<400> SEQUENCE: 66
atgaaagcag cagtagtaag acacaatcca gatggttatg cggaccttgt tgaaaaggaa 60
cttcgagcaa tcaaacctaa tgaagctttg cttgacatgg agtattgtgg agtctgtcat 120
accgatttgc acgttgcagc aggtgattat ggcaacaaag cagggactgt tcttggtcat 180
gaaggaattg gaattgtcaa agaaattgga gctgatgtaa gctcgcttca agttggtgat 240
cgggtttcag tggcttggtt ctttgaagga tgtggtcact gtgaatactg tgtatctggt 300
aatgaaactt tttgtcgaga agttaaaaat gcaggatatt cagttgatgg cggaatggct 360
gaagaagcaa ttgttgttgc cgattatgct gtcaaagttc ctgacggact tgacccaatt 420
gaagctagct caattacttg tgctggagta acaacttaca aagcaatcaa agtatcagga 480
gtaaaacctg gtgattggca agtaattttt ggtgctggag gacttggaaa tttagcaatt 540
caatatgcta aaaatgtttt tggagcaaaa gtaattgctg ttgatattaa tcaagataaa 600
ttaaatttag ctaaaaaaat tggagctgat gtgattatca attctggtga tgtaaatcca 660
gttgatgaaa ttaaaaaaat aactggcggc ttaggggtgc aaagtgcaat agtttgtgct 720
gttgcaagga ttgcttttga acaagcggtt gcttctttga aacctatggg caaaatggtt 780
gctgtggcac ttcccaatac tgagatgact ttatcagttc caacagttgt ttttgacgga 840
gtggaggttg caggttcact tgtcggaaca agacttgact tggcagaagc ttttcaattt 900
ggagcagaag gtaaggtaaa accaattgtt gcgacacgca aactggaaga aatcaatgat 960
attattgatg aaatgaaggc aggaaaaatt gaaggccgaa tggtcattga ttttactaaa 1020
taa 1023
<210> SEQ ID NO 67
<211> LENGTH: 340
<212> TYPE: PRT
<213> ORGANISM: Lactococcus lactis
<400> SEQUENCE: 67
Met Lys Ala Ala Val Val Arg His Asn Pro Asp Gly Tyr Ala Asp Leu
1 5 10 15
Val Glu Lys Glu Leu Arg Ala Ile Lys Pro Asn Glu Ala Leu Leu Asp
20 25 30
Met Glu Tyr Cys Gly Val Cys His Thr Asp Leu His Val Ala Ala Gly
35 40 45
Asp Tyr Gly Asn Lys Ala Gly Thr Val Leu Gly His Glu Gly Ile Gly
50 55 60
Ile Val Lys Glu Ile Gly Ala Asp Val Ser Ser Leu Gln Val Gly Asp
65 70 75 80
Arg Val Ser Val Ala Trp Phe Phe Glu Gly Cys Gly His Cys Glu Tyr
85 90 95
Cys Val Ser Gly Asn Glu Thr Phe Cys Arg Glu Val Lys Asn Ala Gly
100 105 110
Tyr Ser Val Asp Gly Gly Met Ala Glu Glu Ala Ile Val Val Ala Asp
115 120 125
Tyr Ala Val Lys Val Pro Asp Gly Leu Asp Pro Ile Glu Ala Ser Ser
130 135 140
Ile Thr Cys Ala Gly Val Thr Thr Tyr Lys Ala Ile Lys Val Ser Gly
145 150 155 160
Val Lys Pro Gly Asp Trp Gln Val Ile Phe Gly Ala Gly Gly Leu Gly
165 170 175
Asn Leu Ala Ile Gln Tyr Ala Lys Asn Val Phe Gly Ala Lys Val Ile
180 185 190
Ala Val Asp Ile Asn Gln Asp Lys Leu Asn Leu Ala Lys Lys Ile Gly
195 200 205
Ala Asp Val Ile Ile Asn Ser Gly Asp Val Asn Pro Val Asp Glu Ile
210 215 220
Lys Lys Ile Thr Gly Gly Leu Gly Val Gln Ser Ala Ile Val Cys Ala
225 230 235 240
Val Ala Arg Ile Ala Phe Glu Gln Ala Val Ala Ser Leu Lys Pro Met
245 250 255
Gly Lys Met Val Ala Val Ala Leu Pro Asn Thr Glu Met Thr Leu Ser
260 265 270
Val Pro Thr Val Val Phe Asp Gly Val Glu Val Ala Gly Ser Leu Val
275 280 285
Gly Thr Arg Leu Asp Leu Ala Glu Ala Phe Gln Phe Gly Ala Glu Gly
290 295 300
Lys Val Lys Pro Ile Val Ala Thr Arg Lys Leu Glu Glu Ile Asn Asp
305 310 315 320
Ile Ile Asp Glu Met Lys Ala Gly Lys Ile Glu Gly Arg Met Val Ile
325 330 335
Asp Phe Thr Lys
340
<210> SEQ ID NO 68
<211> LENGTH: 1164
<212> TYPE: DNA
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 68
atgaacaact ttaatctgca caccccaacc cgcattctgt ttggtaaagg cgcaatcgct 60
ggtttacgcg aacaaattcc tcacgatgct cgcgtattga ttacctacgg cggcggcagc 120
gtgaaaaaaa ccggcgttct cgatcaagtt ctggatgccc tgaaaggcat ggacgtgctg 180
gaatttggcg gtattgagcc aaacccggct tatgaaacgc tgatgaacgc cgtgaaactg 240
gttcgcgaac agaaagtgac tttcctgctg gcggttggcg gcggttctgt actggacggc 300
accaaattta tcgccgcagc ggctaactat ccggaaaata tcgatccgtg gcacattctg 360
caaacgggcg gtaaagagat taaaagcgcc atcccgatgg gctgtgtgct gacgctgcca 420
gcaaccggtt cagaatccaa cgcaggcgcg gtgatctccc gtaaaaccac aggcgacaag 480
caggcgttcc attctgccca tgttcagccg gtatttgccg tgctcgatcc ggtttatacc 540
tacaccctgc cgccgcgtca ggtggctaac ggcgtagtgg acgcctttgt acacaccgtg 600
gaacagtatg ttaccaaacc ggttgatgcc aaaattcagg accgtttcgc agaaggcatt 660
ttgctgacgc taatcgaaga tggtccgaaa gccctgaaag agccagaaaa ctacgatgtg 720
cgcgccaacg tcatgtgggc ggcgactcag gcgctgaacg gtttgattgg cgctggcgta 780
ccgcaggact gggcaacgca tatgctgggc cacgaactga ctgcgatgca cggtctggat 840
cacgcgcaaa cactggctat cgtcctgcct gcactgtgga atgaaaaacg cgataccaag 900
cgcgctaagc tgctgcaata tgctgaacgc gtctggaaca tcactgaagg ttccgatgat 960
gagcgtattg acgccgcgat tgccgcaacc cgcaatttct ttgagcaatt aggcgtgccg 1020
acccacctct ccgactacgg tctggacggc agctccatcc cggctttgct gaaaaaactg 1080
gaagagcacg gcatgaccca actgggcgaa aatcatgaca ttacgttgga tgtcagccgc 1140
cgtatatacg aagccgcccg ctaa 1164
<210> SEQ ID NO 69
<211> LENGTH: 387
<212> TYPE: PRT
<213> ORGANISM: Escherichia coli
<400> SEQUENCE: 69
Met Asn Asn Phe Asn Leu His Thr Pro Thr Arg Ile Leu Phe Gly Lys
1 5 10 15
Gly Ala Ile Ala Gly Leu Arg Glu Gln Ile Pro His Asp Ala Arg Val
20 25 30
Leu Ile Thr Tyr Gly Gly Gly Ser Val Lys Lys Thr Gly Val Leu Asp
35 40 45
Gln Val Leu Asp Ala Leu Lys Gly Met Asp Val Leu Glu Phe Gly Gly
50 55 60
Ile Glu Pro Asn Pro Ala Tyr Glu Thr Leu Met Asn Ala Val Lys Leu
65 70 75 80
Val Arg Glu Gln Lys Val Thr Phe Leu Leu Ala Val Gly Gly Gly Ser
85 90 95
Val Leu Asp Gly Thr Lys Phe Ile Ala Ala Ala Ala Asn Tyr Pro Glu
100 105 110
Asn Ile Asp Pro Trp His Ile Leu Gln Thr Gly Gly Lys Glu Ile Lys
115 120 125
Ser Ala Ile Pro Met Gly Cys Val Leu Thr Leu Pro Ala Thr Gly Ser
130 135 140
Glu Ser Asn Ala Gly Ala Val Ile Ser Arg Lys Thr Thr Gly Asp Lys
145 150 155 160
Gln Ala Phe His Ser Ala His Val Gln Pro Val Phe Ala Val Leu Asp
165 170 175
Pro Val Tyr Thr Tyr Thr Leu Pro Pro Arg Gln Val Ala Asn Gly Val
180 185 190
Val Asp Ala Phe Val His Thr Val Glu Gln Tyr Val Thr Lys Pro Val
195 200 205
Asp Ala Lys Ile Gln Asp Arg Phe Ala Glu Gly Ile Leu Leu Thr Leu
210 215 220
Ile Glu Asp Gly Pro Lys Ala Leu Lys Glu Pro Glu Asn Tyr Asp Val
225 230 235 240
Arg Ala Asn Val Met Trp Ala Ala Thr Gln Ala Leu Asn Gly Leu Ile
245 250 255
Gly Ala Gly Val Pro Gln Asp Trp Ala Thr His Met Leu Gly His Glu
260 265 270
Leu Thr Ala Met His Gly Leu Asp His Ala Gln Thr Leu Ala Ile Val
275 280 285
Leu Pro Ala Leu Trp Asn Glu Lys Arg Asp Thr Lys Arg Ala Lys Leu
290 295 300
Leu Gln Tyr Ala Glu Arg Val Trp Asn Ile Thr Glu Gly Ser Asp Asp
305 310 315 320
Glu Arg Ile Asp Ala Ala Ile Ala Ala Thr Arg Asn Phe Phe Glu Gln
325 330 335
Leu Gly Val Pro Thr His Leu Ser Asp Tyr Gly Leu Asp Gly Ser Ser
340 345 350
Ile Pro Ala Leu Leu Lys Lys Leu Glu Glu His Gly Met Thr Gln Leu
355 360 365
Gly Glu Asn His Asp Ile Thr Leu Asp Val Ser Arg Arg Ile Tyr Glu
370 375 380
Ala Ala Arg
385
<210> SEQ ID NO 70
<211> LENGTH: 395
<212> TYPE: PRT
<213> ORGANISM: Saccharomyces cerevisiae
<400> SEQUENCE: 70
Met Leu Arg Thr Gln Ala Ala Arg Leu Ile Cys Asn Ser Arg Val Ile
1 5 10 15
Thr Ala Lys Arg Thr Phe Ala Leu Ala Thr Arg Ala Ala Ala Tyr Ser
20 25 30
Arg Pro Ala Ala Arg Phe Val Lys Pro Met Ile Thr Thr Arg Gly Leu
35 40 45
Lys Gln Ile Asn Phe Gly Gly Thr Val Glu Thr Val Tyr Glu Arg Ala
50 55 60
Asp Trp Pro Arg Glu Lys Leu Leu Asp Tyr Phe Lys Asn Asp Thr Phe
65 70 75 80
Ala Leu Ile Gly Tyr Gly Ser Gln Gly Tyr Gly Gln Gly Leu Asn Leu
85 90 95
Arg Asp Asn Gly Leu Asn Val Ile Ile Gly Val Arg Lys Asp Gly Ala
100 105 110
Ser Trp Lys Ala Ala Ile Glu Asp Gly Trp Val Pro Gly Lys Asn Leu
115 120 125
Phe Thr Val Glu Asp Ala Ile Lys Arg Gly Ser Tyr Val Met Asn Leu
130 135 140
Leu Ser Asp Ala Ala Gln Ser Glu Thr Trp Pro Ala Ile Lys Pro Leu
145 150 155 160
Leu Thr Lys Gly Lys Thr Leu Tyr Phe Ser His Gly Phe Ser Pro Val
165 170 175
Phe Lys Asp Leu Thr His Val Glu Pro Pro Lys Asp Leu Asp Val Ile
180 185 190
Leu Val Ala Pro Lys Gly Ser Gly Arg Thr Val Arg Ser Leu Phe Lys
195 200 205
Glu Gly Arg Gly Ile Asn Ser Ser Tyr Ala Val Trp Asn Asp Val Thr
210 215 220
Gly Lys Ala His Glu Lys Ala Gln Ala Leu Ala Val Ala Ile Gly Ser
225 230 235 240
Gly Tyr Val Tyr Gln Thr Thr Phe Glu Arg Glu Val Asn Ser Asp Leu
245 250 255
Tyr Gly Glu Arg Gly Cys Leu Met Gly Gly Ile His Gly Met Phe Leu
260 265 270
Ala Gln Tyr Asp Val Leu Arg Glu Asn Gly His Ser Pro Ser Glu Ala
275 280 285
Phe Asn Glu Thr Val Glu Glu Ala Thr Gln Ser Leu Tyr Pro Leu Ile
290 295 300
Gly Lys Tyr Gly Met Asp Tyr Met Tyr Asp Ala Cys Ser Thr Thr Ala
305 310 315 320
Arg Arg Gly Ala Leu Asp Trp Tyr Pro Ile Phe Lys Asn Ala Leu Lys
325 330 335
Pro Val Phe Gln Asp Leu Tyr Glu Ser Thr Lys Asn Gly Thr Glu Thr
340 345 350
Lys Arg Ser Leu Glu Phe Asn Ser Gln Pro Asp Tyr Arg Glu Lys Leu
355 360 365
Glu Lys Glu Leu Asp Thr Ile Arg Asn Met Glu Ile Trp Lys Val Gly
370 375 380
Lys Glu Val Arg Lys Leu Arg Pro Glu Asn Gln
385 390 395
<210> SEQ ID NO 71
<211> LENGTH: 330
<212> TYPE: PRT
<213> ORGANISM: Methanococcus maripaludis
<400> SEQUENCE: 71
Met Lys Val Phe Tyr Asp Ser Asp Phe Lys Leu Asp Ala Leu Lys Glu
1 5 10 15
Lys Thr Ile Ala Val Ile Gly Tyr Gly Ser Gln Gly Arg Ala Gln Ser
20 25 30
Leu Asn Met Lys Asp Ser Gly Leu Asn Val Val Val Gly Leu Arg Lys
35 40 45
Asn Gly Ala Ser Trp Glu Asn Ala Lys Ala Asp Gly His Asn Val Met
50 55 60
Thr Ile Glu Glu Ala Ala Glu Lys Ala Asp Ile Ile His Ile Leu Ile
65 70 75 80
Pro Asp Glu Leu Gln Ala Glu Val Tyr Glu Ser Gln Ile Lys Pro Tyr
85 90 95
Leu Lys Glu Gly Lys Thr Leu Ser Phe Ser His Gly Phe Asn Ile His
100 105 110
Tyr Gly Phe Ile Val Pro Pro Lys Gly Val Asn Val Val Leu Val Ala
115 120 125
Pro Lys Ser Pro Gly Lys Met Val Arg Arg Thr Tyr Glu Glu Gly Phe
130 135 140
Gly Val Pro Gly Leu Ile Cys Ile Glu Ile Asp Ala Thr Asn Asn Ala
145 150 155 160
Phe Asp Ile Val Ser Ala Met Ala Lys Gly Ile Gly Leu Ser Arg Ala
165 170 175
Gly Val Ile Gln Thr Thr Phe Lys Glu Glu Thr Glu Thr Asp Leu Phe
180 185 190
Gly Glu Gln Ala Val Leu Cys Gly Gly Val Thr Glu Leu Ile Lys Ala
195 200 205
Gly Phe Glu Thr Leu Val Glu Ala Gly Tyr Ala Pro Glu Met Ala Tyr
210 215 220
Phe Glu Thr Cys His Glu Leu Lys Leu Ile Val Asp Leu Ile Tyr Gln
225 230 235 240
Lys Gly Phe Lys Asn Met Trp Asn Asp Val Ser Asn Thr Ala Glu Tyr
245 250 255
Gly Gly Leu Thr Arg Arg Ser Arg Ile Val Thr Ala Asp Ser Lys Ala
260 265 270
Ala Met Lys Glu Ile Leu Lys Glu Ile Gln Asp Gly Arg Phe Thr Lys
275 280 285
Glu Phe Val Leu Glu Lys Gln Val Asn His Ala His Leu Lys Ala Met
290 295 300
Arg Arg Ile Glu Gly Asp Leu Gln Ile Glu Glu Val Gly Ala Lys Leu
305 310 315 320
Arg Lys Met Cys Gly Leu Glu Lys Glu Glu
325 330
<210> SEQ ID NO 72
<211> LENGTH: 342
<212> TYPE: PRT
<213> ORGANISM: Bacillus subtilis
<400> SEQUENCE: 72
Met Val Lys Val Tyr Tyr Asn Gly Asp Ile Lys Glu Asn Val Leu Ala
1 5 10 15
Gly Lys Thr Val Ala Val Ile Gly Tyr Gly Ser Gln Gly His Ala His
20 25 30
Ala Leu Asn Leu Lys Glu Ser Gly Val Asp Val Ile Val Gly Val Arg
35 40 45
Gln Gly Lys Ser Phe Thr Gln Ala Gln Glu Asp Gly His Lys Val Phe
50 55 60
Ser Val Lys Glu Ala Ala Ala Gln Ala Glu Ile Ile Met Val Leu Leu
65 70 75 80
Pro Asp Glu Gln Gln Gln Lys Val Tyr Glu Ala Glu Ile Lys Asp Glu
85 90 95
Leu Thr Ala Gly Lys Ser Leu Val Phe Ala His Gly Phe Asn Val His
100 105 110
Phe His Gln Ile Val Pro Pro Ala Asp Val Asp Val Phe Leu Val Ala
115 120 125
Pro Lys Gly Pro Gly His Leu Val Arg Arg Thr Tyr Glu Gln Gly Ala
130 135 140
Gly Val Pro Ala Leu Phe Ala Ile Tyr Gln Asp Val Thr Gly Glu Ala
145 150 155 160
Arg Asp Lys Ala Leu Ala Tyr Ala Lys Gly Ile Gly Gly Ala Arg Ala
165 170 175
Gly Val Leu Glu Thr Thr Phe Lys Glu Glu Thr Glu Thr Asp Leu Phe
180 185 190
Gly Glu Gln Ala Val Leu Cys Gly Gly Leu Ser Ala Leu Val Lys Ala
195 200 205
Gly Phe Glu Thr Leu Thr Glu Ala Gly Tyr Gln Pro Glu Leu Ala Tyr
210 215 220
Phe Glu Cys Leu His Glu Leu Lys Leu Ile Val Asp Leu Met Tyr Glu
225 230 235 240
Glu Gly Leu Ala Gly Met Arg Tyr Ser Ile Ser Asp Thr Ala Gln Trp
245 250 255
Gly Asp Phe Val Ser Gly Pro Arg Val Val Asp Ala Lys Val Lys Glu
260 265 270
Ser Met Lys Glu Val Leu Lys Asp Ile Gln Asn Gly Thr Phe Ala Lys
275 280 285
Glu Trp Ile Val Glu Asn Gln Val Asn Arg Pro Arg Phe Asn Ala Ile
290 295 300
Asn Ala Ser Glu Asn Glu His Gln Ile Glu Val Val Gly Arg Lys Leu
305 310 315 320
Arg Glu Met Met Pro Phe Val Lys Gln Gly Lys Lys Lys Glu Ala Val
325 330 335
Val Ser Val Ala Gln Asn
340
<210> SEQ ID NO 73
<211> LENGTH: 352
<212> TYPE: PRT
<213> ORGANISM: Piromyces sp.
<400> SEQUENCE: 73
Met Val Lys Val Ile Asn Phe Gly Gly Val Asp Glu Thr Val Tyr Glu
1 5 10 15
Arg Ala Asp Phe Pro Gln Glu Lys Leu Asn Glu Ile Phe Lys Asp Asp
20 25 30
Val Phe Val Val Ile Gly Tyr Gly Thr Gln Gly Arg Asn Gln Ser Arg
35 40 45
Asn Leu Arg Asp Lys Gly Phe Lys Val Ile Val Gly Leu Arg Lys Gly
50 55 60
Pro Ser Trp Asp Leu Ala Lys Glu Asp Gly Trp Val Glu Ser Glu Ser
65 70 75 80
Leu Phe Glu Ile Thr Glu Ala Cys Gln Lys Gly Thr Ile Ile Met Tyr
85 90 95
Leu Leu Ser Asp Ala Gly Gln Lys Ala Cys Trp Asn Thr Ile Lys Glu
100 105 110
Leu Val His Gly Lys Thr Leu Tyr Phe Ser His Gly Phe Ser Ile Val
115 120 125
Phe Lys Glu Lys Thr Gly Val Val Pro Pro Glu Asp Cys Asp Val Ile
130 135 140
Met Val Ala Pro Lys Gly Ser Gly Thr Thr Val Arg Thr Leu Phe Leu
145 150 155 160
Glu Gly Arg Gly Ile Asn Ser Ser Val Ala Val Phe Gln Asn Trp Ser
165 170 175
Gly Lys Ala Glu Glu Arg Ala Tyr Ala Ala Gly Ile Ala Ile Gly Ser
180 185 190
Gly Tyr Leu Tyr Pro Thr Thr Phe Glu Arg Glu Thr Tyr Ser Asp Leu
195 200 205
Thr Gly Glu Arg Gly Thr Leu Met Gly Cys Ile Gln Gly Cys Phe Lys
210 215 220
Ala Gln Phe Glu Val Leu Ile Ala Asn Gly His Thr Pro Ser Glu Ala
225 230 235 240
Phe Ser Glu Thr Val Glu Glu Ala Thr Gln Ser Leu Tyr Pro Leu Ile
245 250 255
Gly Lys Asp Gly Met Asp Trp Met Tyr Asp Asn Cys Ser Thr Thr Ala
260 265 270
Arg Arg Gly Ala Leu Asp Trp Met Asp Lys Phe Tyr Ala Ala Thr Lys
275 280 285
Pro Val Phe Glu Glu Leu Tyr Glu Ser Val Arg Asn Gly Thr Glu Ala
290 295 300
Glu Asn Thr Leu Val Ala Asn Ser Lys Pro Asp Tyr Arg Glu Asn Leu
305 310 315 320
Ala Lys Glu Leu Lys Glu Leu Arg Glu Ser Gln Met Trp Gln Thr Ala
325 330 335
Val Thr Val Arg Ser Leu Arg Pro Glu Asn Gln Lys Val Glu Lys Asn
340 345 350
<210> SEQ ID NO 74
<211> LENGTH: 490
<212> TYPE: PRT
<213> ORGANISM: Buchnera aphidicola
<400> SEQUENCE: 74
Met Lys Asn Tyr Phe Asn Ser Leu Asn Phe Arg Gln Lys Leu Ile Asn
1 5 10 15
Leu Gln Lys Cys Lys Leu Ile Asp Asn Gln Phe Leu Ser Glu Lys Asn
20 25 30
Asn Val Leu Lys Gly Lys Asn Ile Val Ile Val Gly Cys Gly Ser Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asn Ile Ser
50 55 60
Tyr Ala Leu Arg Asp Asp Ser Ile Phe Asn Lys Asn Gln Ser Trp Ile
65 70 75 80
Asn Ala Thr Ser Asn Gly Phe Phe Val Gly Thr Tyr Glu Asn Ile Ile
85 90 95
Pro Thr Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Gln His Glu
100 105 110
Gln Val Val Asn Val Leu Gln Lys Phe Met Lys Pro Asn Ser Val Leu
115 120 125
Gly Phe Ser His Gly Phe Asn Ile Val Glu Val Gly Gln Leu Ile Arg
130 135 140
Asn Asp Ile Thr Val Ile Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Ala Leu Ile Ala
165 170 175
Val His Ser Glu Asn Asp Pro His Asp Ile Gly Phe Glu Ile Ala Lys
180 185 190
Ser Trp Ala Ile Ser Ile Gly Ser His His Ala Gly Ile Leu His Ser
195 200 205
Ser Phe Ile Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Ser Ser Leu Val Cys Tyr Asn Gln Leu
225 230 235 240
Ile Phe Gln Gly Val Asn Pro Ser Tyr Ala Gly Lys Leu Ile Gln Thr
245 250 255
Gly Trp Glu Val Ile Thr Glu Ser Val Lys His Gly Gly Ile Thr Leu
260 265 270
Met Leu Asp Arg Leu Ser Asn Thr Ala Lys Ile Arg Ala Tyr Phe Leu
275 280 285
Ser Lys Lys Leu Lys Lys Ile Phe Phe Pro Leu Phe Arg Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Lys Asn Met Met Phe Asp Trp
305 310 315 320
Lys Asn Asn Asp Gln Gln Leu Lys Glu Trp Arg Thr Glu Ile Gln Asn
325 330 335
Thr Asp Phe Glu Lys Cys Asn Ile Tyr Tyr Lys Gln Ile Pro Glu Gln
340 345 350
Glu Tyr Phe Asp Asn Gly Leu Leu Met Val Ala Ile Leu Lys Ala Gly
355 360 365
Ile Glu Leu Ser Phe Glu Ile Met Ile Glu Thr Gly Ile Lys Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Leu Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Ser Tyr Leu Phe Ser His Ala Ala Ile Pro Leu Leu
420 425 430
Lys Lys Phe Met Asn Glu Leu Gln Pro Gly Asp Leu Gly Asn Lys Ile
435 440 445
Ser Thr Ser Glu Leu Asp Asn Ile Thr Leu Tyr Lys Val Asn Ala Lys
450 455 460
Ile Glu Ser His Pro Ile Glu Ile Ile Gly Lys Lys Leu Arg Leu Tyr
465 470 475 480
Met Thr Ser Met Val Pro Ile Lys Thr Lys
485 490
<210> SEQ ID NO 75
<211> LENGTH: 595
<212> TYPE: PRT
<213> ORGANISM: Spinacia oleracea
<400> SEQUENCE: 75
Met Ala Ala Thr Ala Ala Thr Thr Phe Ser Leu Ser Ser Ser Ser Ser
1 5 10 15
Thr Ser Ala Ala Ala Ser Lys Ala Leu Lys Gln Ser Pro Lys Pro Ser
20 25 30
Ala Leu Asn Leu Gly Phe Leu Gly Ser Ser Ser Thr Ile Lys Ala Cys
35 40 45
Arg Ser Leu Lys Ala Ala Arg Val Leu Pro Ser Gly Ala Asn Gly Gly
50 55 60
Gly Ser Ala Leu Ser Ala Gln Met Val Ser Ala Pro Ser Ile Asn Thr
65 70 75 80
Pro Ser Ala Thr Thr Phe Asp Phe Asp Ser Ser Val Phe Lys Lys Glu
85 90 95
Lys Val Thr Leu Ser Gly His Asp Glu Tyr Ile Val Arg Gly Gly Arg
100 105 110
Asn Leu Phe Pro Leu Leu Pro Asp Ala Phe Lys Gly Ile Lys Gln Ile
115 120 125
Gly Val Ile Gly Trp Gly Ser Gln Ala Pro Ala Gln Ala Gln Asn Leu
130 135 140
Lys Asp Ser Leu Thr Glu Ala Lys Ser Asp Val Val Val Lys Ile Gly
145 150 155 160
Leu Arg Lys Gly Ser Asn Ser Phe Ala Glu Ala Arg Ala Ala Gly Phe
165 170 175
Ser Glu Glu Asn Gly Thr Leu Gly Asp Met Trp Glu Thr Ile Ser Gly
180 185 190
Ser Asp Leu Val Leu Leu Leu Ile Ser Asp Ser Ala Gln Ala Asp Asn
195 200 205
Tyr Glu Lys Val Phe Ser His Met Lys Pro Asn Ser Ile Leu Gly Leu
210 215 220
Ser His Gly Phe Leu Leu Gly His Leu Gln Ser Leu Gly Gln Asp Phe
225 230 235 240
Pro Lys Asn Ile Ser Val Ile Ala Val Cys Pro Lys Gly Met Gly Pro
245 250 255
Ser Val Arg Arg Leu Tyr Val Gln Gly Lys Glu Val Asn Gly Ala Gly
260 265 270
Ile Asn Ser Ser Phe Ala Val His Gln Asp Val Asp Gly Arg Ala Thr
275 280 285
Asp Val Ala Leu Gly Trp Ser Ile Ala Leu Gly Ser Pro Phe Thr Phe
290 295 300
Ala Thr Thr Leu Glu Gln Glu Tyr Lys Ser Asp Ile Phe Gly Glu Arg
305 310 315 320
Gly Ile Leu Leu Gly Ala Val His Gly Ile Val Glu Cys Leu Phe Arg
325 330 335
Arg Tyr Thr Glu Ser Gly Met Ser Glu Asp Leu Ala Tyr Lys Asn Thr
340 345 350
Val Glu Cys Ile Thr Gly Val Ile Ser Lys Thr Ile Ser Thr Lys Gly
355 360 365
Met Leu Ala Leu Tyr Asn Ser Leu Ser Glu Glu Gly Lys Lys Asp Phe
370 375 380
Gln Ala Ala Tyr Ser Ala Ser Tyr Tyr Pro Ser Met Asp Ile Leu Tyr
385 390 395 400
Glu Cys Tyr Glu Asp Val Ala Ser Gly Ser Glu Ile Arg Ser Val Val
405 410 415
Leu Ala Gly Arg Arg Phe Tyr Glu Lys Glu Gly Leu Pro Ala Phe Pro
420 425 430
Met Gly Lys Ile Asp Gln Thr Arg Met Trp Lys Val Gly Glu Lys Val
435 440 445
Arg Ser Val Arg Pro Ala Gly Asp Leu Gly Pro Leu Tyr Pro Phe Thr
450 455 460
Ala Gly Val Tyr Val Ala Leu Met Met Ala Gln Ile Glu Ile Leu Arg
465 470 475 480
Lys Lys Gly His Ser Tyr Ser Glu Ile Ile Asn Glu Ser Val Ile Glu
485 490 495
Ala Val Asp Ser Leu Asn Pro Phe Met His Ala Arg Gly Val Ser Phe
500 505 510
Met Val Asp Asn Cys Ser Thr Thr Ala Arg Leu Gly Ser Arg Lys Trp
515 520 525
Ala Pro Arg Phe Asp Tyr Ile Leu Ser Gln Gln Ala Leu Val Ala Val
530 535 540
Asp Asn Gly Ala Pro Ile Asn Gln Asp Leu Ile Ser Asn Phe Leu Ser
545 550 555 560
Asp Pro Val His Glu Ala Ile Gly Val Cys Ala Gln Leu Arg Pro Ser
565 570 575
Val Asp Ile Ser Val Thr Ala Asp Ala Asp Phe Val Arg Pro Glu Leu
580 585 590
Arg Gln Ala
595
<210> SEQ ID NO 76
<211> LENGTH: 578
<212> TYPE: PRT
<213> ORGANISM: Oryza sativa
<400> SEQUENCE: 76
Met Ala Ala Ser Thr Thr Leu Ala Leu Ser His Pro Lys Thr Leu Ala
1 5 10 15
Ala Ala Ala Ala Ala Ala Pro Lys Ala Pro Thr Ala Pro Ala Ala Val
20 25 30
Ser Phe Pro Val Ser His Ala Ala Cys Ala Pro Leu Ala Ala Arg Arg
35 40 45
Arg Ala Val Thr Ala Met Val Ala Ala Pro Pro Ala Val Gly Ala Ala
50 55 60
Met Pro Ser Leu Asp Phe Asp Thr Ser Val Phe Asn Lys Glu Lys Val
65 70 75 80
Ser Leu Ala Gly His Glu Glu Tyr Ile Val Arg Gly Gly Arg Asn Leu
85 90 95
Phe Pro Leu Leu Pro Glu Ala Phe Lys Gly Ile Lys Gln Ile Gly Val
100 105 110
Ile Gly Trp Gly Ser Gln Gly Pro Ala Gln Ala Gln Asn Leu Arg Asp
115 120 125
Ser Leu Ala Glu Ala Lys Ser Asp Ile Val Val Lys Ile Gly Leu Arg
130 135 140
Lys Gly Ser Lys Ser Phe Asp Glu Ala Arg Ala Ala Gly Phe Thr Glu
145 150 155 160
Glu Ser Gly Thr Leu Gly Asp Ile Trp Glu Thr Val Ser Gly Ser Asp
165 170 175
Leu Val Leu Leu Leu Ile Ser Asp Ala Ala Gln Ala Asp Asn Tyr Glu
180 185 190
Lys Ile Phe Ser His Met Lys Pro Asn Ser Ile Leu Gly Leu Ser His
195 200 205
Gly Phe Leu Leu Gly His Leu Gln Ser Ala Gly Leu Asp Phe Pro Lys
210 215 220
Asn Ile Ser Val Ile Ala Val Cys Pro Lys Gly Met Gly Pro Ser Val
225 230 235 240
Arg Arg Leu Tyr Val Gln Gly Lys Glu Ile Asn Gly Ala Gly Ile Asn
245 250 255
Ser Ser Phe Ala Val His Gln Asp Val Asp Gly Arg Ala Thr Asp Val
260 265 270
Ala Leu Gly Trp Ser Val Ala Leu Gly Ser Pro Phe Thr Phe Ala Thr
275 280 285
Thr Leu Glu Gln Glu Tyr Lys Ser Asp Ile Phe Gly Glu Arg Gly Ile
290 295 300
Leu Leu Gly Ala Val His Gly Ile Val Glu Ala Leu Phe Arg Arg Tyr
305 310 315 320
Thr Glu Gln Gly Met Asp Glu Glu Met Ala Tyr Lys Asn Thr Val Glu
325 330 335
Gly Ile Thr Gly Ile Ile Ser Lys Thr Ile Ser Lys Lys Gly Met Leu
340 345 350
Glu Val Tyr Asn Ser Leu Thr Glu Glu Gly Lys Lys Glu Phe Asn Lys
355 360 365
Ala Tyr Ser Ala Ser Phe Tyr Pro Cys Met Asp Ile Leu Tyr Glu Cys
370 375 380
Tyr Glu Asp Val Ala Ser Gly Ser Glu Ile Arg Ser Val Val Leu Ala
385 390 395 400
Gly Arg Arg Phe Tyr Glu Lys Glu Gly Leu Pro Ala Phe Pro Met Gly
405 410 415
Asn Ile Asp Gln Thr Arg Met Trp Lys Val Gly Glu Lys Val Arg Ser
420 425 430
Thr Arg Pro Glu Asn Asp Leu Gly Pro Leu His Pro Phe Thr Ala Gly
435 440 445
Val Tyr Val Ala Leu Met Met Ala Gln Ile Glu Val Leu Arg Lys Lys
450 455 460
Gly His Ser Tyr Ser Glu Ile Ile Asn Glu Ser Val Ile Glu Ser Val
465 470 475 480
Asp Ser Leu Asn Pro Phe Met His Ala Arg Gly Val Ala Phe Met Val
485 490 495
Asp Asn Cys Ser Thr Thr Ala Arg Leu Gly Ser Arg Lys Trp Ala Pro
500 505 510
Arg Phe Asp Tyr Ile Leu Thr Gln Gln Ala Phe Val Thr Val Asp Lys
515 520 525
Asp Ala Pro Ile Asn Gln Asp Leu Ile Ser Asn Phe Met Ser Asp Pro
530 535 540
Val His Gly Ala Ile Glu Val Cys Ala Glu Leu Arg Pro Thr Val Asp
545 550 555 560
Ile Ser Val Pro Ala Asn Ala Asp Phe Val Arg Pro Glu Leu Arg Gln
565 570 575
Ser Ser
<210> SEQ ID NO 77
<211> LENGTH: 555
<212> TYPE: PRT
<213> ORGANISM: Chlamydomonas reinhardtii
<400> SEQUENCE: 77
Met Gln Leu Leu Asn Ser Lys Ser Arg Val Leu Ser Gly Ser Arg Gln
1 5 10 15
Gln Ala Ala Ala Lys Ala Val Arg Val Ala Pro Ser Gly Arg Arg Ser
20 25 30
Ala Val Arg Val Ser Ala Ala Val His Leu Asp Phe Asn Thr Lys Val
35 40 45
Phe Gln Lys Glu His Ala Lys Phe Gly Pro Thr Glu Glu Tyr Ile Val
50 55 60
Arg Gly Gly Arg Asp Lys Tyr Pro Leu Leu Lys Glu Ala Phe Lys Gly
65 70 75 80
Ile Lys Lys Val Ser Val Ile Gly Trp Gly Ser Gln Ala Pro Ala Gln
85 90 95
Ala Gln Asn Leu Arg Asp Ser Ile Ala Glu Ala Gly Met Asp Ile Lys
100 105 110
Val Ala Ile Gly Leu Arg Pro Asp Ser Pro Ser Trp Ala Glu Ala Glu
115 120 125
Ala Cys Gly Phe Ser Lys Thr Asp Gly Thr Leu Gly Glu Val Phe Glu
130 135 140
Gln Ile Ser Ser Ser Asp Phe Val Ile Leu Leu Ile Ser Asp Ala Ala
145 150 155 160
Gln Ala Lys Leu Tyr Pro Arg Ile Leu Ala Ala Met Lys Pro Gly Ala
165 170 175
Thr Leu Gly Leu Ser His Gly Phe Leu Leu Gly Val Met Arg Asn Asp
180 185 190
Gly Val Asp Phe Arg Lys Asp Ile Asn Val Val Leu Val Ala Pro Lys
195 200 205
Gly Met Gly Pro Ser Val Arg Arg Leu Tyr Glu Gln Gly Lys Ser Val
210 215 220
Asn Gly Ala Gly Ile Asn Cys Ser Phe Ala Ile Gln Gln Asp Ala Thr
225 230 235 240
Gly Gln Ala Ala Asp Ile Ala Ile Gly Trp Ala Ile Gly Val Gly Ala
245 250 255
Pro Phe Ala Phe Pro Thr Thr Leu Glu Ser Glu Tyr Lys Ser Asp Ile
260 265 270
Tyr Gly Glu Arg Cys Val Leu Leu Gly Ala Val His Gly Ile Val Glu
275 280 285
Ala Leu Phe Arg Arg Tyr Thr Arg Gln Gly Met Ser Asp Glu Glu Ala
290 295 300
Phe Lys Gln Ser Val Glu Ser Ile Thr Gly Pro Ile Ser Arg Thr Ile
305 310 315 320
Ser Thr Lys Gly Met Leu Ser Val Tyr Asn Ser Phe Asn Glu Ala Asp
325 330 335
Lys Lys Ile Phe Glu Gln Ala Tyr Ser Ala Ser Tyr Lys Pro Ala Leu
340 345 350
Asp Ile Cys Phe Glu Ile Tyr Glu Asp Val Ala Ser Gly Asn Glu Ile
355 360 365
Lys Ser Val Val Gln Ala Val Gln Arg Phe Asp Arg Phe Pro Met Gly
370 375 380
Lys Ile Asp Gln Thr Tyr Met Trp Lys Val Gly Gln Lys Val Arg Ala
385 390 395 400
Glu Arg Asp Glu Ser Lys Ile Pro Val Asn Pro Phe Thr Ala Gly Val
405 410 415
Tyr Val Ala Val Met Met Ala Thr Val Glu Val Leu Arg Glu Lys Gly
420 425 430
His Pro Phe Ser Glu Ile Cys Asn Glu Ser Ile Ile Glu Ala Val Asp
435 440 445
Ser Leu Asn Pro Tyr Met His Ala Arg Gly Val Ala Phe Met Val Asp
450 455 460
Asn Cys Ser Tyr Thr Ala Arg Leu Gly Ser Arg Lys Trp Ala Pro Arg
465 470 475 480
Phe Asp Tyr Ile Ile Glu Gln Gln Ala Phe Val Asp Ile Asp Ser Gly
485 490 495
Lys Ala Ala Asp Lys Glu Val Met Ala Glu Phe Leu Ala His Pro Val
500 505 510
His Ser Ala Leu Ala Thr Cys Ser Ser Met Arg Pro Ser Val Asp Ile
515 520 525
Ser Val Gly Gly Glu Asn Ser Ser Val Gly Val Gly Ala Gly Ala Ala
530 535 540
Arg Thr Glu Phe Arg Ser Thr Ala Ala Lys Val
545 550 555
<210> SEQ ID NO 78
<211> LENGTH: 402
<212> TYPE: PRT
<213> ORGANISM: Neurospora crassa
<400> SEQUENCE: 78
Met Ala Ala Arg Asn Cys Thr Lys Ala Leu Arg Pro Leu Ala Arg Gln
1 5 10 15
Leu Ala Thr Pro Ala Val Gln Arg Arg Thr Phe Val Ala Ala Ala Ser
20 25 30
Ala Val Arg Ala Ser Val Ala Val Lys Ala Val Ala Ala Pro Ala Arg
35 40 45
Gln Gln Val Arg Gly Val Lys Thr Met Asp Phe Ala Gly His Lys Glu
50 55 60
Glu Val His Glu Arg Ala Asp Trp Pro Ala Glu Lys Leu Leu Asp Tyr
65 70 75 80
Phe Lys Asn Asp Thr Leu Ala Leu Ile Gly Tyr Gly Ser Gln Gly His
85 90 95
Gly Gln Gly Leu Asn Leu Arg Asp Asn Gly Leu Asn Val Ile Val Gly
100 105 110
Val Arg Lys Asn Gly Lys Ser Trp Glu Asp Ala Ile Gln Asp Gly Trp
115 120 125
Val Pro Gly Lys Asn Leu Phe Asp Val Asp Glu Ala Ile Ser Arg Gly
130 135 140
Thr Ile Val Met Asn Leu Leu Ser Asp Ala Ala Gln Ser Glu Thr Trp
145 150 155 160
Pro His Ile Lys Pro Gln Ile Thr Lys Gly Lys Thr Leu Tyr Phe Ser
165 170 175
His Gly Phe Ser Pro Val Phe Lys Asp Leu Thr Lys Val Glu Val Pro
180 185 190
Thr Asp Val Asp Val Ile Leu Val Ala Pro Lys Gly Ser Gly Arg Thr
195 200 205
Val Arg Ser Leu Phe Arg Glu Gly Arg Gly Ile Asn Ser Ser Phe Ala
210 215 220
Val Tyr Gln Asp Val Thr Gly Lys Ala Lys Glu Lys Ala Val Ala Leu
225 230 235 240
Gly Val Ala Val Gly Ser Gly Tyr Leu Tyr Glu Thr Thr Phe Glu Lys
245 250 255
Glu Val Tyr Ser Asp Leu Tyr Gly Glu Arg Gly Cys Leu Met Gly Gly
260 265 270
Ile His Gly Met Phe Leu Ala Gln Tyr Glu Val Leu Arg Glu Arg Gly
275 280 285
His Ser Pro Ser Glu Ala Phe Asn Glu Thr Val Glu Glu Ala Thr Gln
290 295 300
Ser Leu Tyr Pro Leu Ile Gly Ala His Gly Met Asp Trp Met Phe Asp
305 310 315 320
Ala Cys Ser Thr Thr Ala Arg Arg Gly Ala Ile Asp Trp Thr Pro Lys
325 330 335
Phe Lys Asp Ala Leu Lys Pro Val Phe Asn Asn Leu Tyr Asp Ser Val
340 345 350
Lys Asn Gly Asp Glu Thr Lys Arg Ser Leu Glu Tyr Asn Ser Gln Pro
355 360 365
Asp Tyr Arg Glu Arg Tyr Glu Ala Glu Leu Asp Glu Ile Arg Asn Leu
370 375 380
Glu Ile Trp Arg Ala Gly Lys Ala Val Arg Ser Leu Arg Pro Glu Asn
385 390 395 400
Gln Lys
<210> SEQ ID NO 79
<211> LENGTH: 404
<212> TYPE: PRT
<213> ORGANISM: Schizosaccharomyces pombe
<400> SEQUENCE: 79
Met Ser Phe Arg Asn Ser Ser Arg Met Ala Met Lys Ala Leu Arg Thr
1 5 10 15
Met Gly Ser Arg Arg Leu Ala Thr Arg Ser Met Ser Val Met Ala Arg
20 25 30
Thr Ile Ala Ala Pro Ser Met Arg Phe Ala Pro Arg Met Thr Ala Pro
35 40 45
Leu Met Gln Thr Arg Gly Met Arg Val Met Asp Phe Ala Gly Thr Lys
50 55 60
Glu Asn Val Trp Glu Arg Ser Asp Trp Pro Arg Glu Lys Leu Val Asp
65 70 75 80
Tyr Phe Lys Asn Asp Thr Leu Ala Ile Ile Gly Tyr Gly Ser Gln Gly
85 90 95
His Gly Gln Gly Leu Asn Ala Arg Asp Gln Gly Leu Asn Val Ile Val
100 105 110
Gly Val Arg Lys Asp Gly Ala Ser Trp Lys Gln Ala Ile Glu Asp Gly
115 120 125
Trp Val Pro Gly Lys Thr Leu Phe Pro Val Glu Glu Ala Ile Lys Lys
130 135 140
Gly Ser Ile Ile Met Asn Leu Leu Ser Asp Ala Ala Gln Thr Glu Thr
145 150 155 160
Trp Pro Lys Ile Ala Pro Leu Ile Thr Lys Gly Lys Thr Leu Tyr Phe
165 170 175
Ser His Gly Phe Ser Val Ile Phe Lys Asp Gln Thr Lys Ile His Pro
180 185 190
Pro Lys Asp Val Asp Val Ile Leu Val Ala Pro Lys Gly Ser Gly Arg
195 200 205
Thr Val Arg Thr Leu Phe Lys Glu Gly Arg Gly Ile Asn Ser Ser Phe
210 215 220
Ala Val Tyr Gln Asp Val Thr Gly Lys Ala Gln Glu Lys Ala Ile Gly
225 230 235 240
Leu Ala Val Ala Val Gly Ser Gly Phe Ile Tyr Gln Thr Thr Phe Lys
245 250 255
Lys Glu Val Ile Ser Asp Leu Val Gly Glu Arg Gly Cys Leu Met Gly
260 265 270
Gly Ile Asn Gly Leu Phe Leu Ala Gln Tyr Gln Val Leu Arg Glu Arg
275 280 285
Gly His Ser Pro Ala Glu Ala Phe Asn Glu Thr Val Glu Glu Ala Thr
290 295 300
Gln Ser Leu Tyr Pro Leu Ile Gly Lys Tyr Gly Leu Asp Tyr Met Phe
305 310 315 320
Ala Ala Cys Ser Thr Thr Ala Arg Arg Gly Ala Ile Asp Trp Thr Pro
325 330 335
Arg Phe Leu Glu Ala Asn Lys Lys Val Leu Asn Glu Leu Tyr Asp Asn
340 345 350
Val Glu Asn Gly Asn Glu Ala Lys Arg Ser Leu Glu Tyr Asn Ser Ala
355 360 365
Pro Asn Tyr Arg Glu Leu Tyr Asp Lys Glu Leu Glu Glu Ile Arg Asn
370 375 380
Leu Glu Ile Trp Lys Ala Gly Glu Val Val Arg Ser Leu Arg Pro Glu
385 390 395 400
His Asn Lys His
<210> SEQ ID NO 80
<211> LENGTH: 415
<212> TYPE: PRT
<213> ORGANISM: Laccaria bicolor
<400> SEQUENCE: 80
Met Ala Ser Leu Ala Arg Ser Ala Ser Gln Ser Leu Arg Ala Ser Ala
1 5 10 15
Arg Arg Ala Pro Arg Ser Leu Ala Lys Ser Ala Val Arg Pro Thr Gln
20 25 30
Ala Ala Ser Tyr Ser Leu Phe Ala Arg Ala Ala Ala Ala Lys Val Ala
35 40 45
Gln Thr Ser Thr Ala Lys Gly Val Arg Gly Val Lys Thr Leu Asp Phe
50 55 60
Ala Gly Thr Lys Glu Val Val Tyr Glu Arg Ser Asp Trp Pro Leu Ala
65 70 75 80
Lys Leu Gln Asp Tyr Phe Lys Asn Asp Thr Leu Ala Leu Ile Gly Tyr
85 90 95
Gly Ser Gln Gly His Gly Gln Gly Leu Asn Ala Arg Asp Asn Gly Leu
100 105 110
Asn Val Ile Val Gly Val Arg Lys Asp Gly Glu Ser Trp Arg Gln Ala
115 120 125
Leu Glu Asp Gly Trp Glu Ser Phe Ser Pro Val Pro Gly Glu Thr Leu
130 135 140
Phe Pro Ile Glu Glu Ala Ile Asn Lys Gly Thr Ile Ile Met Asn Leu
145 150 155 160
Leu Ser Asp Ala Ala Gln Ser Gln Thr Trp Pro Gln Leu Ala Pro Leu
165 170 175
Ile Thr Lys Gly Lys Thr Leu Tyr Phe Ser His Gly Phe Ser Val Val
180 185 190
Tyr Lys Asp Asp Thr His Val Ile Pro Pro Lys Asp Val Asp Val Ile
195 200 205
Leu Val Ala Pro Lys Gly Ser Gly Arg Thr Val Arg Thr Leu Phe Lys
210 215 220
Glu Gly Arg Gly Ile Asn Ser Ser Ile Ala Val Trp Gln Asp Val Thr
225 230 235 240
Gly Lys Ala Lys Glu Lys Ala Ile Ala Leu Gly Val Gly Ile Gly Ser
245 250 255
Gly Tyr Met Tyr Glu Thr Thr Phe Glu Lys Glu Val Tyr Ser Asp Leu
260 265 270
Tyr Gly Glu Arg Gly Val Leu Met Gly Gly Ile Gln Gly Leu Phe Leu
275 280 285
Ala Gln Tyr Gln Val Leu Arg Lys Asn Gly His Ser Pro Ser Glu Ala
290 295 300
Phe Asn Glu Thr Val Glu Glu Ala Thr Gln Ser Leu Tyr Pro Leu Ile
305 310 315 320
Gly Gln Lys Gly Met Asp Tyr Met Tyr Asn Ala Cys Ser Thr Thr Ala
325 330 335
Arg Arg Gly Ala Leu Asp Trp Ala Pro Ile Phe Glu Lys Ala Asn Val
340 345 350
Pro Val Phe Glu Ala Leu Tyr Glu Ser Val Arg Asn Gly Thr Glu Thr
355 360 365
Arg Lys Ser Leu Glu Phe Asn Gly Arg Ala Thr Tyr Arg Glu Asp Leu
370 375 380
Ala Lys Glu Leu Ala Val Ile Asp Asn Gln Glu Ile Trp Arg Ala Gly
385 390 395 400
Lys Thr Val Arg Ser Leu Arg Pro Asp Tyr Lys Pro Glu Ser Glu
405 410 415
<210> SEQ ID NO 81
<211> LENGTH: 343
<212> TYPE: PRT
<213> ORGANISM: Ignicoccus hospitalis
<400> SEQUENCE: 81
Met Gly Leu Asn Ala Gly Ala Leu Arg Arg Val Gly Val Thr Val Ala
1 5 10 15
Gln Ile Trp Lys Asp Ser Asp Val Ser Leu Glu Pro Leu Lys Gly Arg
20 25 30
Lys Val Ala Ile Ile Gly Tyr Gly Ser Gln Gly Arg Ala Trp Ala Leu
35 40 45
Asn Ile Arg Asp Ser Gly Val Asp Val Val Val Gly Leu Arg Pro Gly
50 55 60
Gly Lys Ser Trp Glu Leu Ala Thr Lys Asp Gly Phe Glu Pro Lys Pro
65 70 75 80
Ile Pro Glu Ala Ala Lys Glu Gly Asp Val Ile Ala Met Leu Ile Pro
85 90 95
Asp Met Ala Gln Pro Glu Ile Tyr Glu Lys Tyr Val Glu Pro Asn Leu
100 105 110
His Glu Gly Asn Ala Leu Val Phe Ala His Gly Phe Asn Ile His Tyr
115 120 125
Gly Leu Ile Lys Pro Pro Lys Asn Val Asp Val Ile Met Val Ala Pro
130 135 140
Lys Ser Pro Gly Pro Lys Val Arg Glu Ala Phe Leu Ser Gly Arg Gly
145 150 155 160
Val Pro Ala Leu Val Ala Val His Gln Asp Tyr Thr Gly Lys Ala Trp
165 170 175
Asp Leu Val Leu Ala Leu Ala Lys Ala Leu Gly Cys Thr Arg Ala Gly
180 185 190
Val Ile Lys Thr Thr Phe Lys Glu Glu Thr Glu Ser Asp Leu Ile Gly
195 200 205
Glu Gln Thr Val Leu Val Gly Gly Leu Met Glu Leu Leu Lys Lys Gly
210 215 220
Phe Glu Asn Leu Val Glu Leu Gly Tyr Gln Pro Glu Val Ala Tyr Phe
225 230 235 240
Glu Ala Ile Asn Glu Ala Lys Leu Ile Met Asp Leu Ile Trp Gln Tyr
245 250 255
Gly Phe Tyr Gly Met Leu Leu Arg Val Ser Asp Thr Ala Lys Tyr Gly
260 265 270
Gly Leu Thr Val Gly Pro Lys Val Ile Asp Glu His Val Lys Glu Asn
275 280 285
Met Lys Lys Ala Ser Glu Arg Val Ile Ser Gly Glu Phe Ala Lys Glu
290 295 300
Trp Val Glu Glu Tyr Lys Lys Gly Met Pro Thr Leu Lys Glu Leu Met
305 310 315 320
Glu Lys Val Lys Glu His Gln Ala Glu Lys Val Gly Lys Glu Leu Arg
325 330 335
Lys Leu Met Gly Leu Glu Glu
340
<210> SEQ ID NO 82
<211> LENGTH: 329
<212> TYPE: PRT
<213> ORGANISM: Picrophilus torridus
<400> SEQUENCE: 82
Met Glu Lys Val Tyr Thr Glu Asn Asp Leu Lys Glu Asn Leu Met Arg
1 5 10 15
Asn Lys Lys Ile Ala Val Leu Gly Tyr Gly Ser Gln Gly Arg Ala Trp
20 25 30
Ala Leu Asn Met Arg Asp Ser Gly Leu Asn Val Thr Val Gly Leu Glu
35 40 45
Arg Gln Gly Lys Ser Trp Glu Lys Ala Val Ala Asp Gly Phe Lys Pro
50 55 60
Leu Lys Ser Arg Asp Ala Val Arg Asp Ala Asp Ala Val Ile Phe Leu
65 70 75 80
Val Pro Asp Met Ala Gln Arg Glu Leu Tyr Lys Asn Ile Met Asn Asp
85 90 95
Ile Lys Asp Asp Ala Asp Ile Val Phe Ala His Gly Phe Asn Val His
100 105 110
Tyr Gly Leu Ile Asn Pro Lys Asn His Asp Val Tyr Met Val Ala Pro
115 120 125
Lys Ala Pro Gly Pro Ser Val Arg Glu Phe Tyr Glu Arg Gly Gly Gly
130 135 140
Val Pro Val Leu Ile Ala Val Ala Asn Asp Val Ser Gly Arg Ser Lys
145 150 155 160
Glu Lys Ala Leu Ser Ile Ala Tyr Ser Leu Gly Ala Leu Arg Ala Gly
165 170 175
Ala Ile Glu Thr Thr Phe Lys Glu Glu Thr Glu Thr Asp Leu Ile Gly
180 185 190
Glu Gln Leu Asp Leu Val Gly Gly Ile Thr Glu Leu Leu Arg Ser Thr
195 200 205
Phe Asn Ile Met Val Glu Met Gly Tyr Lys Pro Glu Met Ala Tyr Phe
210 215 220
Glu Ala Ile Asn Glu Met Lys Leu Ile Val Asp Gln Val Phe Glu Lys
225 230 235 240
Gly Ile Ser Gly Met Leu Arg Ala Val Ser Asp Thr Ala Lys Tyr Gly
245 250 255
Gly Leu Thr Thr Gly Lys Tyr Ile Ile Asn Asp Asp Val Arg Lys Arg
260 265 270
Met Arg Glu Arg Ala Glu Tyr Ile Val Ser Gly Lys Phe Ala Glu Glu
275 280 285
Trp Ile Glu Glu Tyr Gly Glu Gly Ser Lys Asn Leu Glu Ser Met Met
290 295 300
Leu Asp Ile Asp Asn Ser Leu Glu Glu Gln Val Gly Lys Gln Leu Arg
305 310 315 320
Glu Ile Val Leu Arg Gly Arg Pro Lys
325
<210> SEQ ID NO 83
<211> LENGTH: 339
<212> TYPE: PRT
<213> ORGANISM: Acidiphilium cryptum
<400> SEQUENCE: 83
Met Arg Val Tyr Tyr Asp Ser Asp Ala Asp Val Asn Leu Ile Lys Ala
1 5 10 15
Lys Lys Val Ala Val Val Gly Tyr Gly Ser Gln Gly His Ala His Ala
20 25 30
Leu Asn Leu Lys Glu Ser Gly Val Lys Glu Leu Val Val Ala Leu Arg
35 40 45
Lys Gly Ser Ala Ala Val Ala Lys Ala Glu Ala Ala Gly Leu Arg Val
50 55 60
Met Thr Pro Glu Glu Ala Ala Ala Trp Ala Asp Val Val Met Ile Leu
65 70 75 80
Thr Pro Asp Glu Gly Gln Gly Asp Leu Tyr Arg Asp Ser Leu Ala Ala
85 90 95
Asn Leu Lys Pro Gly Ala Ala Ile Ala Phe Ala His Gly Leu Asn Ile
100 105 110
His Phe Asn Leu Ile Glu Pro Arg Ala Asp Ile Asp Val Phe Met Ile
115 120 125
Ala Pro Lys Gly Pro Gly His Thr Val Arg Ser Glu Tyr Gln Arg Gly
130 135 140
Gly Gly Val Pro Cys Leu Val Ala Val Ala Gln Asn Pro Ser Gly Asn
145 150 155 160
Ala Leu Asp Ile Ala Leu Ser Tyr Ala Ser Ala Ile Gly Gly Gly Arg
165 170 175
Ala Gly Ile Ile Glu Thr Thr Phe Lys Glu Glu Cys Glu Thr Asp Leu
180 185 190
Phe Gly Glu Gln Thr Val Leu Cys Gly Gly Leu Val Glu Leu Ile Lys
195 200 205
Ala Gly Phe Glu Thr Leu Val Glu Ala Gly Tyr Ala Pro Glu Met Ala
210 215 220
Tyr Phe Glu Cys Leu His Glu Val Lys Leu Ile Val Asp Leu Ile Tyr
225 230 235 240
Glu Gly Gly Ile Ala Asn Met Asn Tyr Ser Ile Ser Asn Thr Ala Glu
245 250 255
Tyr Gly Glu Tyr Val Thr Gly Pro Arg Met Ile Thr Pro Glu Thr Lys
260 265 270
Ala Glu Met Lys Arg Val Leu Asp Asp Ile Gln Lys Gly Arg Phe Thr
275 280 285
Arg Asp Trp Met Leu Glu Asn Lys Val Asn Gln Thr Asn Phe Lys Ala
290 295 300
Met Arg Arg Ala Asn Ala Ala His Pro Ile Glu Glu Val Gly Glu Lys
305 310 315 320
Leu Arg Ala Met Met Pro Trp Ile Lys Lys Gly Ala Leu Val Asp Lys
325 330 335
Thr Arg Asn
<210> SEQ ID NO 84
<211> LENGTH: 330
<212> TYPE: PRT
<213> ORGANISM: Cyanobacteria/Synechococcus sp.
<400> SEQUENCE: 84
Met Ala Arg Leu Tyr Tyr Asp Thr Asp Ala Asn Leu Asp Leu Leu Asp
1 5 10 15
Gly Lys Thr Val Ala Ile Ile Gly Tyr Gly Ser Gln Gly His Ala His
20 25 30
Ala Leu Asn Leu Arg Asp Ser Gly Val Asn Val Leu Val Gly Leu Tyr
35 40 45
Pro Gly Ser Pro Ser Trp Pro Lys Ala Glu Arg Asp Gly Leu Thr Val
50 55 60
Lys Thr Val Ala Asp Ala Ala Ala Ala Ala Asp Trp Val Met Ile Leu
65 70 75 80
Leu Pro Asp Glu Val Gln Lys Thr Val Phe Gln Ser Glu Ile Arg Pro
85 90 95
His Leu Lys Pro Gly Lys Val Leu Leu Phe Ala His Gly Phe Asn Ile
100 105 110
His Phe Gly Gln Ile Gln Pro Pro Pro Asp Ile Asp Val Ile Met Val
115 120 125
Ala Pro Lys Gly Pro Gly His Leu Val Arg Arg Thr Tyr Leu Glu Gly
130 135 140
Gln Gly Val Pro Cys Leu Phe Ala Val Tyr Gln Asp Ala Ser Gly Met
145 150 155 160
Ala Arg Glu Arg Ala Met Ala Tyr Ala Lys Ala Ile Gly Gly Thr Arg
165 170 175
Ala Gly Ile Leu Glu Thr Ser Phe Arg Glu Glu Thr Glu Thr Asp Leu
180 185 190
Phe Gly Glu Gln Val Val Leu Cys Gly Gly Leu Thr Ala Leu Ile Lys
195 200 205
Ala Gly Phe Glu Thr Leu Val Glu Ala Gly Tyr Gln Pro Glu Leu Ala
210 215 220
Tyr Phe Glu Cys Leu His Glu Val Lys Leu Ile Val Asp Leu Ile Val
225 230 235 240
Glu Gly Gly Leu Glu Lys Met Arg His Ser Ile Ser Asn Thr Ala Glu
245 250 255
Tyr Gly Asp Tyr Thr Arg Gly Pro Arg Ile Ile Thr Glu Gln Thr Arg
260 265 270
Ala Glu Met Lys Arg Ile Leu Ser Glu Ile Gln Ser Gly Gln Phe Ala
275 280 285
Arg Glu Phe Val Leu Glu Asn Gln Ala Gly Lys Pro Val Leu Thr Ala
290 295 300
Met Arg Arg Arg Glu Ala Glu His Pro Ile Glu Lys Val Gly Lys Glu
305 310 315 320
Leu Arg Ala Met Phe Ser Trp Leu Lys Lys
325 330
<210> SEQ ID NO 85
<211> LENGTH: 339
<212> TYPE: PRT
<213> ORGANISM: Zymomonas mobilis
<400> SEQUENCE: 85
Met Lys Val Tyr Tyr Asp Ser Asp Ala Asp Leu Gly Leu Ile Lys Ser
1 5 10 15
Lys Lys Ile Ala Ile Leu Gly Tyr Gly Ser Gln Gly His Ala His Ala
20 25 30
Gln Asn Leu Arg Asp Ser Gly Val Ala Glu Val Ala Ile Ala Leu Arg
35 40 45
Pro Asp Ser Ala Ser Val Lys Lys Ala Gln Asp Ala Gly Phe Lys Val
50 55 60
Leu Thr Asn Ala Glu Ala Ala Lys Trp Ala Asp Ile Leu Met Ile Leu
65 70 75 80
Ala Pro Asp Glu His Gln Ala Ala Ile Tyr Ala Glu Asp Leu Lys Asp
85 90 95
Asn Leu Arg Pro Gly Ser Ala Ile Ala Phe Ala His Gly Leu Asn Ile
100 105 110
His Phe Gly Leu Ile Glu Pro Arg Lys Asp Ile Asp Val Phe Met Ile
115 120 125
Ala Pro Lys Gly Pro Gly His Thr Val Arg Ser Glu Tyr Val Arg Gly
130 135 140
Gly Gly Val Pro Cys Leu Val Ala Val Asp Gln Asp Ala Ser Gly Asn
145 150 155 160
Ala His Asp Ile Ala Leu Ala Tyr Ala Ser Gly Ile Gly Gly Gly Arg
165 170 175
Ser Gly Val Ile Glu Thr Thr Phe Arg Glu Glu Val Glu Thr Asp Leu
180 185 190
Phe Gly Glu Gln Ala Val Leu Cys Gly Gly Leu Thr Ala Leu Ile Thr
195 200 205
Ala Gly Phe Glu Thr Leu Thr Glu Ala Gly Tyr Ala Pro Glu Met Ala
210 215 220
Phe Phe Glu Cys Met His Glu Met Lys Leu Ile Val Asp Leu Ile Tyr
225 230 235 240
Glu Ala Gly Ile Ala Asn Met Arg Tyr Ser Ile Ser Asn Thr Ala Glu
245 250 255
Tyr Gly Asp Ile Val Ser Gly Pro Arg Val Ile Asn Glu Glu Ser Lys
260 265 270
Lys Ala Met Lys Ala Ile Leu Asp Asp Ile Gln Ser Gly Arg Phe Val
275 280 285
Ser Lys Phe Val Leu Asp Asn Arg Ala Gly Gln Pro Glu Leu Lys Ala
290 295 300
Ala Arg Lys Arg Met Ala Ala His Pro Ile Glu Gln Val Gly Ala Arg
305 310 315 320
Leu Arg Lys Met Met Pro Trp Ile Ala Ser Asn Lys Leu Val Asp Lys
325 330 335
Ala Arg Asn
<210> SEQ ID NO 86
<211> LENGTH: 359
<212> TYPE: PRT
<213> ORGANISM: Bacteroides thetaiotaomicron
<400> SEQUENCE: 86
Met Ala Gln Val Ile Lys Thr Lys Lys Gln Lys Lys Met Ala Gln Leu
1 5 10 15
Asn Phe Gly Gly Thr Val Glu Asn Val Val Ile Arg Asp Glu Phe Pro
20 25 30
Leu Glu Lys Ala Arg Glu Val Leu Lys Asn Glu Thr Ile Ala Val Ile
35 40 45
Gly Tyr Gly Val Gln Gly Pro Gly Gln Ala Leu Asn Leu Arg Asp Asn
50 55 60
Gly Phe Asn Val Ile Val Gly Gln Arg Gln Gly Lys Thr Tyr Asp Lys
65 70 75 80
Ala Val Ala Asp Gly Trp Val Pro Gly Glu Thr Leu Phe Gly Ile Glu
85 90 95
Glu Ala Cys Glu Lys Gly Thr Ile Ile Met Cys Leu Leu Ser Asp Ala
100 105 110
Ala Val Met Ser Val Trp Pro Thr Ile Lys Pro Tyr Leu Thr Ala Gly
115 120 125
Lys Ala Leu Tyr Phe Ser His Gly Phe Ala Ile Thr Trp Ser Asp Arg
130 135 140
Thr Gly Val Val Pro Pro Ala Asp Ile Asp Val Ile Met Val Ala Pro
145 150 155 160
Lys Gly Ser Gly Thr Ser Leu Arg Thr Met Phe Leu Glu Gly Arg Gly
165 170 175
Leu Asn Ser Ser Tyr Ala Ile Tyr Gln Asp Ala Thr Gly Asn Ala Met
180 185 190
Asp Arg Thr Ile Ala Leu Gly Ile Gly Ile Gly Ser Gly Tyr Leu Phe
195 200 205
Glu Thr Thr Phe Ile Arg Glu Ala Thr Ser Asp Leu Thr Gly Glu Arg
210 215 220
Gly Ser Leu Met Gly Ala Ile Gln Gly Leu Leu Leu Ala Gln Tyr Glu
225 230 235 240
Val Leu Arg Glu Asn Gly His Thr Pro Ser Glu Ala Phe Asn Glu Thr
245 250 255
Val Glu Glu Leu Thr Gln Ser Leu Met Pro Leu Phe Ala Lys Asn Gly
260 265 270
Met Asp Trp Met Tyr Ala Asn Cys Ser Thr Thr Ala Gln Arg Gly Ala
275 280 285
Leu Asp Trp Met Gly Pro Phe His Asp Ala Ile Lys Pro Val Val Glu
290 295 300
Lys Leu Tyr His Ser Val Lys Thr Gly Asn Glu Ala Gln Ile Ser Ile
305 310 315 320
Asp Ser Asn Ser Lys Pro Asp Tyr Arg Glu Lys Leu Glu Glu Glu Leu
325 330 335
Lys Ala Leu Arg Glu Ser Glu Met Trp Gln Thr Ala Val Thr Val Arg
340 345 350
Lys Leu Arg Pro Glu Asn Asn
355
<210> SEQ ID NO 87
<211> LENGTH: 494
<212> TYPE: PRT
<213> ORGANISM: Vibrio fischeri
<400> SEQUENCE: 87
Met Ser Asn Tyr Phe Asn Thr Leu Asn Leu Arg Glu Gln Leu Asp Gln
1 5 10 15
Leu Gly Arg Cys Arg Phe Met Asp Arg Glu Glu Phe Ala Thr Glu Ala
20 25 30
Asp Tyr Leu Lys Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Val Ala
50 55 60
Tyr Ala Leu Arg Gln Ala Ala Ile Asp Glu Gln Arg Gln Ser Tyr Lys
65 70 75 80
Asn Ala Lys Glu Asn Gly Phe Glu Val Ala Ser Tyr Glu Thr Leu Ile
85 90 95
Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Gln His Thr
100 105 110
Asn Val Val Glu Thr Val Met Pro Leu Met Lys Glu Gly Ala Ala Leu
115 120 125
Gly Tyr Ser His Gly Phe Asn Val Val Glu Glu Gly Met Gln Ile Arg
130 135 140
Lys Asp Leu Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Lys Gly Glu Gly Trp Asp Ile Ala Lys
180 185 190
Ala Trp Ala Ala Gly Thr Gly Gly His Arg Ala Gly Cys Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Ile Val Ser Tyr Glu Lys Met
225 230 235 240
Ile Ala Asp Gly Ile Glu Pro Gly Tyr Ala Gly Lys Leu Leu Gln Tyr
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys Phe Gly Gly Val Thr His
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Val Lys Ala Phe Glu Leu
275 280 285
Ser Glu Glu Leu Lys Glu Leu Met Arg Pro Leu Tyr Asn Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Arg Thr Met Met Ala Asp Trp
305 310 315 320
Ala Asn Asp Asp Val Asn Leu Phe Gly Trp Arg Glu Glu Thr Gly Gln
325 330 335
Thr Ala Phe Glu Asn Tyr Pro Glu Ser Asp Val Glu Ile Ser Glu Gln
340 345 350
Glu Tyr Phe Asp Asn Gly Ile Leu Leu Val Ala Met Val Arg Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Ala Met Thr Ala Ser Gly Ile Ile Asp Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro Leu Ile Ala Asn Thr
385 390 395 400
Val Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Asn Tyr Leu Phe Ala Asn Val Ala Thr Pro Leu Leu
420 425 430
Arg Glu Lys Phe Met Pro Ser Val Glu Thr Asp Val Ile Gly Arg Gly
435 440 445
Leu Gly Glu Ala Ser Asn Gln Val Asp Asn Ala Thr Leu Ile Ala Val
450 455 460
Asn Asp Ala Ile Arg Asn His Pro Val Glu Tyr Ile Gly Glu Glu Leu
465 470 475 480
Arg Ser Tyr Met Ser Asp Met Lys Arg Ile Ala Val Gly Gly
485 490
<210> SEQ ID NO 88
<211> LENGTH: 492
<212> TYPE: PRT
<213> ORGANISM: Shewanella sp.
<400> SEQUENCE: 88
Met Ala Asn Tyr Phe Asn Ser Leu Asn Leu Arg Gln Gln Leu Glu Gln
1 5 10 15
Leu Gly Gln Cys Arg Phe Met Asp Arg Ser Glu Phe Ser Asp Gly Cys
20 25 30
Asn Tyr Ile Lys Asp Trp Asn Ile Val Ile Leu Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asn Ile Ala
50 55 60
Tyr Ala Leu Arg Pro Glu Ala Ile Ala Gln Lys Arg Ala Ser Trp Gln
65 70 75 80
Lys Ala Thr Asp Asn Gly Phe Lys Val Gly Thr Phe Glu Glu Leu Ile
85 90 95
Pro Thr Ala Asp Leu Val Leu Asn Leu Thr Pro Asp Lys Gln His Ser
100 105 110
Asn Val Val Ser Ala Val Met Pro Leu Met Lys Gln Gly Ala Thr Leu
115 120 125
Ser Tyr Ser His Gly Phe Asn Ile Val Glu Glu Gly Met Gln Ile Arg
130 135 140
Pro Asp Ile Thr Val Val Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Asn Gly Asp Gly Leu Glu Ile Ala Lys
180 185 190
Ala Tyr Ala Ser Ala Thr Gly Gly Asp Arg Ala Gly Val Leu Gln Ser
195 200 205
Ser Phe Ile Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Thr Gly Ala Ile Leu Gly Tyr Asp Lys Met
225 230 235 240
Val Ala Asp Gly Val Glu Pro Gly Tyr Ala Ala Lys Leu Ile Gln Gln
245 250 255
Gly Trp Glu Thr Val Thr Glu Ala Leu Lys His Gly Gly Ile Thr Asn
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Ile Lys Ala Phe Glu Ile
275 280 285
Ala Glu Asp Leu Lys Glu Ile Leu Gln Pro Leu Phe Glu Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Arg Thr Met Met Gln Asp Trp
305 310 315 320
Ala Asn Asp Asp Ala Asn Leu Leu Arg Trp Arg Ala Glu Thr Ala Glu
325 330 335
Thr Gly Phe Glu Asn Ala Pro Val Ser Ser Glu His Ile Asp Glu Gln
340 345 350
Thr Tyr Phe Asp Lys Gly Ile Phe Leu Val Ala Met Ile Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Asp Thr Met Val Ser Ala Gly Ile Val Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Thr Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Cys Tyr Leu Phe Asn His Ala Ala Val Pro Met Leu
420 425 430
Arg Asp Tyr Val Asn Ala Met Ser Pro Glu Tyr Leu Gly Ala Gly Leu
435 440 445
Lys Asp Ser Ser Asn Asn Val Asp Asn Leu Gln Leu Ile Ala Ile Asn
450 455 460
Asp Ala Ile Arg His Thr Ser Val Glu Tyr Ile Gly Ala Glu Leu Arg
465 470 475 480
Gly Tyr Met Thr Asp Met Lys Ser Ile Val Gly Ala
485 490
<210> SEQ ID NO 89
<211> LENGTH: 491
<212> TYPE: PRT
<213> ORGANISM: Gramella forsetti
<400> SEQUENCE: 89
Met Thr Asn Tyr Phe Asn Ser Leu Ser Leu Arg Asp Gln Leu Ala Gln
1 5 10 15
Leu Gly Thr Cys Arg Phe Met Glu Leu Asp Glu Phe Ser Asn Glu Val
20 25 30
Ala Val Leu Lys Asp Lys Lys Ile Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser
50 55 60
Tyr Ala Leu Arg Glu Gly Ala Ile Lys Glu Lys Arg Gln Ser Trp Lys
65 70 75 80
Asn Ala Thr Glu Asn Asn Phe Asn Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Lys Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Gln His Thr
100 105 110
Ser Val Ile Lys Ala Ile Gln Pro His Ile Lys Lys Asp Ala Val Leu
115 120 125
Ser Tyr Ser His Gly Phe Asn Ile Val Glu Glu Gly Thr Lys Ile Arg
130 135 140
Glu Asp Ile Thr Val Ile Met Val Ala Pro Lys Cys Pro Gly Thr Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro His Gly Ile Gly Leu Asp Trp Ala Lys
180 185 190
Ala Tyr Ala Tyr Ala Thr Gly Gly His Arg Ala Gly Val Leu Glu Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Met
210 215 220
Leu Cys Gly Val Leu Gln Thr Gly Ser Ile Leu Thr Phe Asp Lys Met
225 230 235 240
Val Ala Asp Gly Val Glu Pro Asn Tyr Ala Ala Lys Leu Ile Gln Tyr
245 250 255
Gly Trp Glu Thr Ile Thr Glu Ala Leu Lys His Gly Gly Ile Thr Asn
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Asn Glu Ile
275 280 285
Ala Glu Glu Leu Lys Glu Lys Met Arg Pro Leu Phe Gln Lys His Met
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Ser Arg Met Met Arg Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Glu Leu Leu Thr Trp Arg Ala Glu Thr Glu Asn
325 330 335
Thr Ala Phe Glu Lys Thr Glu Ala Thr Ser Glu Glu Ile Lys Glu Gln
340 345 350
Glu Tyr Phe Asp Lys Gly Val Leu Met Val Ala Phe Val Arg Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Val Glu Ala Gly Ile Ile Glu Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Thr Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Lys Leu Tyr Glu Met Asn Arg Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Cys Tyr Leu Phe Asp His Ala Ala Lys Pro Leu Val
420 425 430
Lys Asp Tyr Val Asn Ser Leu Glu Pro Glu Val Ala Gly Lys Lys Phe
435 440 445
Gly Thr Asp Cys Asn Gly Val Asp Asn Gln Lys Leu Ile His Val Asn
450 455 460
Asp Asp Leu Arg Ser His Pro Val Glu Lys Val Gly Ala Arg Leu Arg
465 470 475 480
Thr Ala Met Thr Ala Met Lys Lys Ile Tyr Ala
485 490
<210> SEQ ID NO 90
<211> LENGTH: 493
<212> TYPE: PRT
<213> ORGANISM: Psychromonas ingrhamaii
<400> SEQUENCE: 90
Met Ala Asn Tyr Phe Asn Thr Leu Ser Leu Arg Glu Lys Leu Asn Gln
1 5 10 15
Leu Gly Gln Cys Arg Phe Met Asp Arg Ser Glu Phe Thr Asp Gly Cys
20 25 30
Asp Ala Leu Lys Gly Lys Lys Val Val Ile Ile Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Val Ser
50 55 60
Tyr Thr Leu Arg Ala Gln Ala Ile Ala Glu Lys Arg Gln Ser Trp Lys
65 70 75 80
Asn Ala Thr Glu Asn Gly Phe Val Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Glu Ala Asp Leu Leu Cys Asn Leu Thr Pro Asp Lys Gln His Thr
100 105 110
Ala Val Val Gly Ala Val Met Pro Leu Met Lys Glu Gly Ala Thr Leu
115 120 125
Ser Tyr Ser His Gly Phe Asn Ile Val Glu Glu Gly Met Gln Val Arg
130 135 140
Glu Asp Leu Thr Val Ile Met Cys Ala Pro Lys Cys Pro Gly Ser Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Ala Asn Asp Pro Gln Gly Gln Gly Leu Val Trp Ala Lys
180 185 190
Ala Tyr Ala Ser Ala Thr Gly Gly Asp Arg Ala Gly Val Leu Met Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Met Leu Gln Thr Gly Ala Ile Ile Gly Tyr Glu Lys Met
225 230 235 240
Val Ala Asp Gly Ile Glu Pro Gly Tyr Ala Ser Lys Leu Ile Gln Tyr
245 250 255
Gly Trp Glu Thr Val Thr Glu Gly Met Lys Tyr Gly Gly Ile Thr Asn
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Ile Lys Ala Phe Asp Met
275 280 285
Ser Leu Glu Leu Lys Glu Ile Leu Arg Pro Leu Phe Asn Lys His Met
290 295 300
Asp Asp Ile Ile Glu Gly Glu Phe Ser Arg Thr Met Met Glu Asp Trp
305 310 315 320
Ala Asn Asp Asp Lys Asn Leu Leu Gln Trp Arg Ala Glu Thr Ala Glu
325 330 335
Thr Gly Phe Glu Lys Gln Pro Ala Gly Asp Met Lys Ile Asp Glu Gln
340 345 350
Glu Phe Tyr Asp Asn Gly Ile Phe Leu Ile Ala Met Ile Lys Ala Gly
355 360 365
Val Glu Leu Ala Phe Asp Ala Met Thr Ala Ser Gly Ile Ile Ala Asp
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Thr Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Lys Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Cys Tyr Leu Phe Asp His Ala Ala Lys Pro Leu Leu
420 425 430
Ala Asp Phe Val Lys Ala Leu Asp Pro Glu Met Leu Gly Lys Pro Leu
435 440 445
Thr Val Lys Asn Asn Ala Val Asp Asn Ala Arg Leu Ile Glu Val Asn
450 455 460
Glu Ala Ile Arg Ser His Pro Val Glu Ile Val Gly Lys Lys Leu Arg
465 470 475 480
Gly Tyr Met Thr Glu Met Lys Thr Ile Ile Thr Ala Ser
485 490
<210> SEQ ID NO 91
<211> LENGTH: 492
<212> TYPE: PRT
<213> ORGANISM: Cytophaga hutchinsonii
<400> SEQUENCE: 91
Met Ala Asn Tyr Phe Asn Thr Leu Ser Leu Arg Glu Lys Leu Asp Gln
1 5 10 15
Leu Gly Val Cys Glu Phe Met Asp Arg Ser Glu Phe Ser Asp Gly Val
20 25 30
Ala Ala Leu Lys Gly Lys Lys Ile Val Ile Val Gly Cys Gly Ala Gln
35 40 45
Gly Leu Asn Gln Gly Leu Asn Leu Arg Asp Ser Gly Leu Asp Val Ser
50 55 60
Tyr Thr Leu Arg Lys Glu Ala Ile Asp Ser Lys Arg Gln Ser Phe Leu
65 70 75 80
Asn Ala Ser Glu Asn Gly Phe Lys Val Gly Thr Tyr Glu Glu Leu Ile
85 90 95
Pro Thr Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Gln His Thr
100 105 110
Ala Val Val Ser Ala Val Met Pro Leu Met Lys Lys Gly Ser Thr Leu
115 120 125
Ser Tyr Ser His Gly Phe Asn Ile Val Glu Glu Gly Met Gln Ile Arg
130 135 140
Lys Asp Ile Thr Val Ile Met Val Ala Pro Lys Ser Pro Gly Ser Glu
145 150 155 160
Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175
Val His Pro Glu Asn Asp Pro Glu Gly Lys Gly Trp Asp Tyr Ala Lys
180 185 190
Ala Tyr Cys Val Gly Thr Gly Gly Asp Arg Ala Gly Val Leu Lys Ser
195 200 205
Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu Gln Thr Ile
210 215 220
Leu Cys Gly Leu Leu Gln Thr Gly Ser Ile Leu Cys Phe Asp Lys Met
225 230 235 240
Val Glu Lys Gly Ile Asp Lys Gly Tyr Ala Ser Lys Leu Ile Gln Tyr
245 250 255
Gly Trp Glu Val Ile Thr Glu Ser Leu Lys His Gly Gly Ile Ser Gly
260 265 270
Met Met Asp Arg Leu Ser Asn Pro Ala Lys Ile Lys Ala Phe Gln Val
275 280 285
Ser Glu Glu Leu Lys Asp Ile Met Arg Pro Leu Phe Arg Lys His Gln
290 295 300
Asp Asp Ile Ile Ser Gly Glu Phe Ser Arg Ile Met Met Glu Asp Trp
305 310 315 320
Ala Asn Gly Asp Lys Asn Leu Leu Thr Trp Arg Ala Ala Thr Gly Glu
325 330 335
Thr Ala Phe Glu Lys Thr Pro Ala Gly Asp Val Lys Ile Ala Glu Gln
340 345 350
Glu Tyr Tyr Asp Asn Gly Leu Leu Met Val Ala Met Val Arg Ala Gly
355 360 365
Val Glu Leu Ala Phe Glu Thr Met Thr Glu Ser Gly Ile Ile Asp Glu
370 375 380
Ser Ala Tyr Tyr Glu Ser Leu His Glu Thr Pro Leu Ile Ala Asn Thr
385 390 395 400
Ile Ala Arg Lys Lys Leu Phe Glu Met Asn Arg Val Ile Ser Asp Thr
405 410 415
Ala Glu Tyr Gly Cys Tyr Leu Phe Asp His Ala Cys Lys Pro Leu Leu
420 425 430
Ala Asn Phe Met Lys Thr Val Asp Thr Asp Ile Ile Gly Lys Asn Phe
435 440 445
Asn Ala Gly Lys Asp Asn Gly Val Asp Asn Gln Met Leu Ile Ala Val
450 455 460
Asn Glu Val Leu Arg Ser His Pro Ile Glu Ile Val Gly Ala Glu Leu
465 470 475 480
Arg Glu Ala Met Thr Glu Met Lys Ala Ile Val Ser
485 490
<210> SEQ ID NO 92
<400> SEQUENCE: 92
000
<210> SEQ ID NO 93
<400> SEQUENCE: 93
000
<210> SEQ ID NO 94
<400> SEQUENCE: 94
000
<210> SEQ ID NO 95
<400> SEQUENCE: 95
000
<210> SEQ ID NO 96
<400> SEQUENCE: 96
000
<210> SEQ ID NO 97
<400> SEQUENCE: 97
000
<210> SEQ ID NO 98
<400> SEQUENCE: 98
000
<210> SEQ ID NO 99
<400> SEQUENCE: 99
000
<210> SEQ ID NO 100
<400> SEQUENCE: 100
000
<210> SEQ ID NO 101
<211> LENGTH: 6362
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1102
<400> SEQUENCE: 101
tcgcgcgttt cggtgatgac ggtgaaaacc tctgacacat gcagctcccg gagacggtca 60
cagcttgtct gtaagcggat gccgggagca gacaagcccg tcagggcgcg tcagcgggtg 120
ttggcgggtg tcggggctgg cttaactatg cggcatcaga gcagattgta ctgagagtgc 180
accataccac agcttttcaa ttcaattcat catttttttt ttattctttt ttttgatttc 240
ggtttctttg aaattttttt gattcggtaa tctccgaaca gaaggaagaa cgaaggaagg 300
agcacagact tagattggta tatatacgca tatgtagtgt tgaagaaaca tgaaattgcc 360
cagtattctt aacccaactg cacagaacaa aaacctgcag gaaacgaaga taaatcatgt 420
cgaaagctac atataaggaa cgtgctgcta ctcatcctag tcctgttgct gccaagctat 480
ttaatatcat gcacgaaaag caaacaaact tgtgtgcttc attggatgtt cgtaccacca 540
aggaattact ggagttagtt gaagcattag gtcccaaaat ttgtttacta aaaacacatg 600
tggatatctt gactgatttt tccatggagg gcacagttaa gccgctaaag gcattatccg 660
ccaagtacaa ttttttactc ttcgaagaca gaaaatttgc tgacattggt aatacagtca 720
aattgcagta ctctgcgggt gtatacagaa tagcagaatg ggcagacatt acgaatgcac 780
acggtgtggt gggcccaggt attgttagcg gtttgaagca ggcggcagaa gaagtaacaa 840
aggaacctag aggccttttg atgttagcag aattgtcatg caagggctcc ctatctactg 900
gagaatatac taagggtact gttgacattg cgaagagcga caaagatttt gttatcggct 960
ttattgctca aagagacatg ggtggaagag atgaaggtta cgattggttg attatgacac 1020
ccggtgtggg tttagatgac aagggagacg cattgggtca acagtataga accgtggatg 1080
atgtggtctc tacaggatct gacattatta ttgttggaag aggactattt gcaaagggaa 1140
gggatgctaa ggtagagggt gaacgttaca gaaaagcagg ctgggaagca tatttgagaa 1200
gatgcggcca gcaaaactaa aaaactgtat tataagtaaa tgcatgtata ctaaactcac 1260
aaattagagc ttcaatttaa ttatatcagt tattacccta tgcggtgtga aataccgcac 1320
agatgcgtaa ggagaaaata ccgcatcagg aaattgtaaa cgttaatatt ttgttaaaat 1380
tcgcgttaaa tttttgttaa atcagctcat tttttaacca ataggccgaa atcggcaaaa 1440
tcccttataa atcaaaagaa tagaccgaga tagggttgag tgttgttcca gtttggaaca 1500
agagtccact attaaagaac gtggactcca acgtcaaagg gcgaaaaacc gtctatcagg 1560
gcgatggccc actacgtgaa ccatcaccct aatcaagttt tttggggtcg aggtgccgta 1620
aagcactaaa tcggaaccct aaagggagcc cccgatttag agcttgacgg ggaaagccgg 1680
cgaacgtggc gagaaaggaa gggaagaaag cgaaaggagc gggcgctagg gcgctggcaa 1740
gtgtagcggt cacgctgcgc gtaaccacca cacccgccgc gcttaatgcg ccgctacagg 1800
gcgcgtcgcg ccattcgcca ttcaggctgc gcaactgttg ggaagggcga tcggtgcggg 1860
cctcttcgct attacgccag ctggcgaaag ggggatgtgc tgcaaggcga ttaagttggg 1920
taacgccagg gttttcccag tcacgacgtt gtaaaacgac ggccagtgag cgcgcgtaat 1980
acgactcact atagggcgaa ttgggtaccg gccgcaaatt aaagccttcg agcgtcccaa 2040
aaccttctca agcaaggttt tcagtataat gttacatgcg tacacgcgtc tgtacagaaa 2100
aaaaagaaaa atttgaaata taaataacgt tcttaatact aacataacta taaaaaaata 2160
aatagggacc tagacttcag gttgtctaac tccttccttt tcggttagag cggatgtggg 2220
gggagggcgt gaatgtaagc gtgacataac taattacatg actcgagcgg ccgcggatcc 2280
cgggaattcg tcgacacccg catagtcagg aacatcgtat gggtacatgc tagttctaga 2340
aaacttagat tagattgcta tgctttcttt ctaatgagca agaagtaaaa aaagttgtaa 2400
tagaacaaga aaaatgaaac tgaaacttga gaaattgaag accgtttatt aacttaaata 2460
tcaatgggag gtcatcgaaa gagaaaaaaa tcaaaaaaaa aattttcaag aaaaagaaac 2520
gtgataaaaa tttttattgc ctttttcgac gaagaaaaag aaacgaggcg gtctcttttt 2580
tcttttccaa acctttagta cgggtaatta acgacaccct agaggaagaa agaggggaaa 2640
tttagtatgc tgtgcttggg tgttttgaag tggtacggcg atgcgcggag tccgagaaaa 2700
tctggaagag taaaaaagga gtagaaacat tttgaagcta tgagctccag cttttgttcc 2760
ctttagtgag ggttaattgc gcgcttggcg taatcatggt catagctgtt tcctgtgtga 2820
aattgttatc cgctcacaat tccacacaac ataggagccg gaagcataaa gtgtaaagcc 2880
tggggtgcct aatgagtgag gtaactcaca ttaattgcgt tgcgctcact gcccgctttc 2940
cagtcgggaa acctgtcgtg ccagctgcat taatgaatcg gccaacgcgc ggggagaggc 3000
ggtttgcgta ttgggcgctc ttccgcttcc tcgctcactg actcgctgcg ctcggtcgtt 3060
cggctgcggc gagcggtatc agctcactca aaggcggtaa tacggttatc cacagaatca 3120
ggggataacg caggaaagaa catgtgagca aaaggccagc aaaaggccag gaaccgtaaa 3180
aaggccgcgt tgctggcgtt tttccatagg ctccgccccc ctgacgagca tcacaaaaat 3240
cgacgctcaa gtcagaggtg gcgaaacccg acaggactat aaagatacca ggcgtttccc 3300
cctggaagct ccctcgtgcg ctctcctgtt ccgaccctgc cgcttaccgg atacctgtcc 3360
gcctttctcc cttcgggaag cgtggcgctt tctcatagct cacgctgtag gtatctcagt 3420
tcggtgtagg tcgttcgctc caagctgggc tgtgtgcacg aaccccccgt tcagcccgac 3480
cgctgcgcct tatccggtaa ctatcgtctt gagtccaacc cggtaagaca cgacttatcg 3540
ccactggcag cagccactgg taacaggatt agcagagcga ggtatgtagg cggtgctaca 3600
gagttcttga agtggtggcc taactacggc tacactagaa ggacagtatt tggtatctgc 3660
gctctgctga agccagttac cttcggaaaa agagttggta gctcttgatc cggcaaacaa 3720
accaccgctg gtagcggtgg tttttttgtt tgcaagcagc agattacgcg cagaaaaaaa 3780
ggatctcaag aagatccttt gatcttttct acggggtctg acgctcagtg gaacgaaaac 3840
tcacgttaag ggattttggt catgagatta tcaaaaagga tcttcaccta gatcctttta 3900
aattaaaaat gaagttttaa atcaatctaa agtatatatg agtaaacttg gtctgacagt 3960
taccaatgct taatcagtga ggcacctatc tcagcgatct gtctatttcg ttcatccata 4020
gttgcctgac tccccgtcgt gtagataact acgatacggg agggcttacc atctggcccc 4080
agtgctgcaa tgataccgcg agacccacgc tcaccggctc cagatttatc agcaataaac 4140
cagccagccg gaagggccga gcgcagaagt ggtcctgcaa ctttatccgc ctccatccag 4200
tctattaatt gttgccggga agctagagta agtagttcgc cagttaatag tttgcgcaac 4260
gttgttgcca ttgctacagg catcgtggtg tcacgctcgt cgtttggtat ggcttcattc 4320
agctccggtt cccaacgatc aaggcgagtt acatgatccc ccatgttgtg caaaaaagcg 4380
gttagctcct tcggtcctcc gatcgttgtc agaagtaagt tggccgcagt gttatcactc 4440
atggttatgg cagcactgca taattctctt actgtcatgc catccgtaag atgcttttct 4500
gtgactggtg agtactcaac caagtcattc tgagaatagt gtatgcggcg accgagttgc 4560
tcttgcccgg cgtcaatacg ggataatacc gcgccacata gcagaacttt aaaagtgctc 4620
atcattggaa aacgttcttc ggggcgaaaa ctctcaagga tcttaccgct gttgagatcc 4680
agttcgatgt aacccactcg tgcacccaac tgatcttcag catcttttac tttcaccagc 4740
gtttctgggt gagcaaaaac aggaaggcaa aatgccgcaa aaaagggaat aagggcgaca 4800
cggaaatgtt gaatactcat actcttcctt tttcaatatt attgaagcat ttatcagggt 4860
tattgtctca tgagcggata catatttgaa tgtatttaga aaaataaaca aataggggtt 4920
ccgcgcacat ttccccgaaa agtgccacct gaacgaagca tctgtgcttc attttgtaga 4980
acaaaaatgc aacgcgagag cgctaatttt tcaaacaaag aatctgagct gcatttttac 5040
agaacagaaa tgcaacgcga aagcgctatt ttaccaacga agaatctgtg cttcattttt 5100
gtaaaacaaa aatgcaacgc gagagcgcta atttttcaaa caaagaatct gagctgcatt 5160
tttacagaac agaaatgcaa cgcgagagcg ctattttacc aacaaagaat ctatacttct 5220
tttttgttct acaaaaatgc atcccgagag cgctattttt ctaacaaagc atcttagatt 5280
actttttttc tcctttgtgc gctctataat gcagtctctt gataactttt tgcactgtag 5340
gtccgttaag gttagaagaa ggctactttg gtgtctattt tctcttccat aaaaaaagcc 5400
tgactccact tcccgcgttt actgattact agcgaagctg cgggtgcatt ttttcaagat 5460
aaaggcatcc ccgattatat tctataccga tgtggattgc gcatactttg tgaacagaaa 5520
gtgatagcgt tgatgattct tcattggtca gaaaattatg aacggtttct tctattttgt 5580
ctctatatac tacgtatagg aaatgtttac attttcgtat tgttttcgat tcactctatg 5640
aatagttctt actacaattt ttttgtctaa agagtaatac tagagataaa cataaaaaat 5700
gtagaggtcg agtttagatg caagttcaag gagcgaaagg tggatgggta ggttatatag 5760
ggatatagca cagagatata tagcaaagag atacttttga gcaatgtttg tggaagcggt 5820
attcgcaata ttttagtagc tcgttacagt ccggtgcgtt tttggttttt tgaaagtgcg 5880
tcttcagagc gcttttggtt ttcaaaagcg ctctgaagtt cctatacttt ctagagaata 5940
ggaacttcgg aataggaact tcaaagcgtt tccgaaaacg agcgcttccg aaaatgcaac 6000
gcgagctgcg cacatacagc tcactgttca cgtcgcacct atatctgcgt gttgcctgta 6060
tatatatata catgagaaga acggcatagt gcgtgtttat gcttaaatgc gtacttatat 6120
gcgtctattt atgtaggatg aaaggtagtc tagtacctcc tgtgatatta tcccattcca 6180
tgcggggtat cgtatgcttc cttcagcact accctttagc tgttctatat gctgccactc 6240
ctcaattgga ttagtctcat ccttcaatgc tatcatttcc tttgatattg gatcatacta 6300
agaaaccatt attatcatga cattaaccta taaaaatagg cgtatcacga ggccctttcg 6360
tc 6362
<210> SEQ ID NO 102
<211> LENGTH: 7314
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1323
<400> SEQUENCE: 102
tcgcgcgttt cggtgatgac ggtgaaaacc tctgacacat gcagctcccg gagacggtca 60
cagcttgtct gtaagcggat gccgggagca gacaagcccg tcagggcgcg tcagcgggtg 120
ttggcgggtg tcggggctgg cttaactatg cggcatcaga gcagattgta ctgagagtgc 180
accataaacg acattactat atatataata taggaagcat ttaatagaca gcatcgtaat 240
atatgtgtac tttgcagtta tgacgccaga tggcagtagt ggaagatatt ctttattgaa 300
aaatagcttg tcaccttacg tacaatcttg atccggagct tttctttttt tgccgattaa 360
gaattaattc ggtcgaaaaa agaaaaggag agggccaaga gggagggcat tggtgactat 420
tgagcacgtg agtatacgtg attaagcaca caaaggcagc ttggagtatg tctgttatta 480
atttcacagg tagttctggt ccattggtga aagtttgcgg cttgcagagc acagaggccg 540
cagaatgtgc tctagattcc gatgctgact tgctgggtat tatatgtgtg cccaatagaa 600
agagaacaat tgacccggtt attgcaagga aaatttcaag tcttgtaaaa gcatataaaa 660
atagttcagg cactccgaaa tacttggttg gcgtgtttcg taatcaacct aaggaggatg 720
ttttggctct ggtcaatgat tacggcattg atatcgtcca actgcatgga gatgagtcgt 780
ggcaagaata ccaagagttc ctcggtttgc cagttattaa aagactcgta tttccaaaag 840
actgcaacat actactcagt gcagcttcac agaaacctca ttcgtttatt cccttgtttg 900
attcagaagc aggtgggaca ggtgaacttt tggattggaa ctcgatttct gactgggttg 960
gaaggcaaga gagccccgaa agcttacatt ttatgttagc tggtggactg acgccagaaa 1020
atgttggtga tgcgcttaga ttaaatggcg ttattggtgt tgatgtaagc ggaggtgtgg 1080
agacaaatgg tgtaaaagac tctaacaaaa tagcaaattt cgtcaaaaat gctaagaaat 1140
aggttattac tgagtagtat ttatttaagt attgtttgtg cacttgccta tgcggtgtga 1200
aataccgcac agatgcgtaa ggagaaaata ccgcatcagg aaattgtaaa cgttaatatt 1260
ttgttaaaat tcgcgttaaa tttttgttaa atcagctcat tttttaacca ataggccgaa 1320
atcggcaaaa tcccttataa atcaaaagaa tagaccgaga tagggttgag tgttgttcca 1380
gtttggaaca agagtccact attaaagaac gtggactcca acgtcaaagg gcgaaaaacc 1440
gtctatcagg gcgatggccc actacgtgaa ccatcaccct aatcaagttt tttggggtcg 1500
aggtgccgta aagcactaaa tcggaaccct aaagggagcc cccgatttag agcttgacgg 1560
ggaaagccgg cgaacgtggc gagaaaggaa gggaagaaag cgaaaggagc gggcgctagg 1620
gcgctggcaa gtgtagcggt cacgctgcgc gtaaccacca cacccgccgc gcttaatgcg 1680
ccgctacagg gcgcgtcgcg ccattcgcca ttcaggctgc gcaactgttg ggaagggcga 1740
tcggtgcggg cctcttcgct attacgccag ctggcgaaag ggggatgtgc tgcaaggcga 1800
ttaagttggg taacgccagg gttttcccag tcacgacgtt gtaaaacgac ggccagtgag 1860
cgcgcgtaat acgactcact atagggcgaa ttgggtaccg gccgcaaatt aaagccttcg 1920
agcgtcccaa aaccttctca agcaaggttt tcagtataat gttacatgcg tacacgcgtc 1980
tgtacagaaa aaaaagaaaa atttgaaata taaataacgt tcttaatact aacataacta 2040
taaaaaaata aatagggacc tagacttcag gttgtctaac tccttccttt tcggttagag 2100
cggatgtggg gggagggcgt gaatgtaagc gtgacataac taattacatg actcgagcgg 2160
ccgcggatcc ttaaacacca gcttcgaagt ccttttgagc catgaaaatg gataaatcaa 2220
ccactcttga agagtaacca tattcattat cataccaaga aaggaccttg aaaaaatggt 2280
cgttcaattc aataccggcc ttggcatcaa caatagatga acgtgaatcg gatgtgaagt 2340
cagaggacac aacggcgtct ttggtaacac ccaaaacacc cttcatatcg ctgcgagatc 2400
tttgttctag ggccttcata atgtcatcgt aagaagtttt ctttgctgta cggaatgtca 2460
agtcaaccag ggaaatatta attgttggga ctcttataga cataccggtg atcttaccat 2520
taagttcagg caagattttc cctacagcct tagctgcacc agtagatgaa ggaatgatat 2580
ttccctggca agatctaccg cctctccagt ccttaccacc agaactggta ccatcgacag 2640
tcttttgaga agcagtagtt gcatgaatag ttgtcatcaa ggcttcttcg ataccgaact 2700
catcgtccaa agccttaacc aacggagcca aacagttggt agtacaggag gcattagaga 2760
ccacgtgatc cgtcaatggg ttgtatttaa cgtggttaac accatagacg tacattggcg 2820
cggtctttga tggagcagta atgataactt ttttgacacc tttatgtcta gaggctgtat 2880
cgacttcctt gaagacaccg gttgagtcaa ttacataatc gacgttgtag gaagcccatg 2940
ggatacgctc tggttcccta aaatgagata gagggatatg agccgaaaca tggtcatttt 3000
gaatgatgat acgttcatcg tcgaattcaa cttcaccacg atacttgccg tgagtagaat 3060
cgtatttgaa caaataagca gcgtattctg gtgttgtgga tggattattg attaatctga 3120
ccttaacttc tgggtgcgtc aaagcagcac gtagaaccaa tctaccgatt ctaccaaaac 3180
cattgatacc aatgttaatt tgagctggct tagaagaaga ttcgtttgtc atatcgggca 3240
tgtcgacacc cgcatagtca ggaacatcgt atgggtacat gctagttcta gaaaacttag 3300
attagattgc tatgctttct ttctaatgag caagaagtaa aaaaagttgt aatagaacaa 3360
gaaaaatgaa actgaaactt gagaaattga agaccgttta ttaacttaaa tatcaatggg 3420
aggtcatcga aagagaaaaa aatcaaaaaa aaaattttca agaaaaagaa acgtgataaa 3480
aatttttatt gcctttttcg acgaagaaaa agaaacgagg cggtctcttt tttcttttcc 3540
aaacctttag tacgggtaat taacgacacc ctagaggaag aaagagggga aatttagtat 3600
gctgtgcttg ggtgttttga agtggtacgg cgatgcgcgg agtccgagaa aatctggaag 3660
agtaaaaaag gagtagaaac attttgaagc tatgagctcc agcttttgtt ccctttagtg 3720
agggttaatt gcgcgcttgg cgtaatcatg gtcatagctg tttcctgtgt gaaattgtta 3780
tccgctcaca attccacaca acataggagc cggaagcata aagtgtaaag cctggggtgc 3840
ctaatgagtg aggtaactca cattaattgc gttgcgctca ctgcccgctt tccagtcggg 3900
aaacctgtcg tgccagctgc attaatgaat cggccaacgc gcggggagag gcggtttgcg 3960
tattgggcgc tcttccgctt cctcgctcac tgactcgctg cgctcggtcg ttcggctgcg 4020
gcgagcggta tcagctcact caaaggcggt aatacggtta tccacagaat caggggataa 4080
cgcaggaaag aacatgtgag caaaaggcca gcaaaaggcc aggaaccgta aaaaggccgc 4140
gttgctggcg tttttccata ggctccgccc ccctgacgag catcacaaaa atcgacgctc 4200
aagtcagagg tggcgaaacc cgacaggact ataaagatac caggcgtttc cccctggaag 4260
ctccctcgtg cgctctcctg ttccgaccct gccgcttacc ggatacctgt ccgcctttct 4320
cccttcggga agcgtggcgc tttctcatag ctcacgctgt aggtatctca gttcggtgta 4380
ggtcgttcgc tccaagctgg gctgtgtgca cgaacccccc gttcagcccg accgctgcgc 4440
cttatccggt aactatcgtc ttgagtccaa cccggtaaga cacgacttat cgccactggc 4500
agcagccact ggtaacagga ttagcagagc gaggtatgta ggcggtgcta cagagttctt 4560
gaagtggtgg cctaactacg gctacactag aaggacagta tttggtatct gcgctctgct 4620
gaagccagtt accttcggaa aaagagttgg tagctcttga tccggcaaac aaaccaccgc 4680
tggtagcggt ggtttttttg tttgcaagca gcagattacg cgcagaaaaa aaggatctca 4740
agaagatcct ttgatctttt ctacggggtc tgacgctcag tggaacgaaa actcacgtta 4800
agggattttg gtcatgagat tatcaaaaag gatcttcacc tagatccttt taaattaaaa 4860
atgaagtttt aaatcaatct aaagtatata tgagtaaact tggtctgaca gttaccaatg 4920
cttaatcagt gaggcaccta tctcagcgat ctgtctattt cgttcatcca tagttgcctg 4980
actccccgtc gtgtagataa ctacgatacg ggagggctta ccatctggcc ccagtgctgc 5040
aatgataccg cgagacccac gctcaccggc tccagattta tcagcaataa accagccagc 5100
cggaagggcc gagcgcagaa gtggtcctgc aactttatcc gcctccatcc agtctattaa 5160
ttgttgccgg gaagctagag taagtagttc gccagttaat agtttgcgca acgttgttgc 5220
cattgctaca ggcatcgtgg tgtcacgctc gtcgtttggt atggcttcat tcagctccgg 5280
ttcccaacga tcaaggcgag ttacatgatc ccccatgttg tgcaaaaaag cggttagctc 5340
cttcggtcct ccgatcgttg tcagaagtaa gttggccgca gtgttatcac tcatggttat 5400
ggcagcactg cataattctc ttactgtcat gccatccgta agatgctttt ctgtgactgg 5460
tgagtactca accaagtcat tctgagaata gtgtatgcgg cgaccgagtt gctcttgccc 5520
ggcgtcaata cgggataata ccgcgccaca tagcagaact ttaaaagtgc tcatcattgg 5580
aaaacgttct tcggggcgaa aactctcaag gatcttaccg ctgttgagat ccagttcgat 5640
gtaacccact cgtgcaccca actgatcttc agcatctttt actttcacca gcgtttctgg 5700
gtgagcaaaa acaggaaggc aaaatgccgc aaaaaaggga ataagggcga cacggaaatg 5760
ttgaatactc atactcttcc tttttcaata ttattgaagc atttatcagg gttattgtct 5820
catgagcgga tacatatttg aatgtattta gaaaaataaa caaatagggg ttccgcgcac 5880
atttccccga aaagtgccac ctgaacgaag catctgtgct tcattttgta gaacaaaaat 5940
gcaacgcgag agcgctaatt tttcaaacaa agaatctgag ctgcattttt acagaacaga 6000
aatgcaacgc gaaagcgcta ttttaccaac gaagaatctg tgcttcattt ttgtaaaaca 6060
aaaatgcaac gcgagagcgc taatttttca aacaaagaat ctgagctgca tttttacaga 6120
acagaaatgc aacgcgagag cgctatttta ccaacaaaga atctatactt cttttttgtt 6180
ctacaaaaat gcatcccgag agcgctattt ttctaacaaa gcatcttaga ttactttttt 6240
tctcctttgt gcgctctata atgcagtctc ttgataactt tttgcactgt aggtccgtta 6300
aggttagaag aaggctactt tggtgtctat tttctcttcc ataaaaaaag cctgactcca 6360
cttcccgcgt ttactgatta ctagcgaagc tgcgggtgca ttttttcaag ataaaggcat 6420
ccccgattat attctatacc gatgtggatt gcgcatactt tgtgaacaga aagtgatagc 6480
gttgatgatt cttcattggt cagaaaatta tgaacggttt cttctatttt gtctctatat 6540
actacgtata ggaaatgttt acattttcgt attgttttcg attcactcta tgaatagttc 6600
ttactacaat ttttttgtct aaagagtaat actagagata aacataaaaa atgtagaggt 6660
cgagtttaga tgcaagttca aggagcgaaa ggtggatggg taggttatat agggatatag 6720
cacagagata tatagcaaag agatactttt gagcaatgtt tgtggaagcg gtattcgcaa 6780
tattttagta gctcgttaca gtccggtgcg tttttggttt tttgaaagtg cgtcttcaga 6840
gcgcttttgg ttttcaaaag cgctctgaag ttcctatact ttctagagaa taggaacttc 6900
ggaataggaa cttcaaagcg tttccgaaaa cgagcgcttc cgaaaatgca acgcgagctg 6960
cgcacataca gctcactgtt cacgtcgcac ctatatctgc gtgttgcctg tatatatata 7020
tacatgagaa gaacggcata gtgcgtgttt atgcttaaat gcgtacttat atgcgtctat 7080
ttatgtagga tgaaaggtag tctagtacct cctgtgatat tatcccattc catgcggggt 7140
atcgtatgct tccttcagca ctacccttta gctgttctat atgctgccac tcctcaattg 7200
gattagtctc atccttcaat gctatcattt cctttgatat tggatcatat taagaaacca 7260
ttattatcat gacattaacc tataaaaata ggcgtatcac gaggcccttt cgtc 7314
<210> SEQ ID NO 103
<211> LENGTH: 6294
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1485
<400> SEQUENCE: 103
taagaaacca ttattatcat gacattaacc tataaaaata ggcgtatcac gaggcccttt 60
cgtcttcacc tcgagaattg tgagcggata acaattgaca ttgtgagcgg ataacaagat 120
actgagcaca tcagcaggac gcactgaccg aattcattaa agaggagaaa ggtaccatgt 180
atacagtagg agattaccta ttagaccgat tacacgagtt aggaattgaa gaaatttttg 240
gagtccctgg agactataac ttacaatttt tagatcaaat tatttcccgc aaggatatga 300
aatgggtcgg aaatgctaat gaattaaatg cttcatatat ggctgatggc tatgctcgta 360
ctaaaaaagc tgccgcattt cttacaacct ttggagtagg tgaattgagt gcagttaatg 420
gattagcagg aagttacgcc gaaaatttac cagtagtaga aatagtggga tcacctacat 480
caaaagttca aaatgaagga aaatttgttc atcatacgct ggctgacggt gattttaaac 540
actttatgaa aatgcacgaa cctgttacag cagctcgaac tttactgaca gcagaaaatg 600
caaccgttga aattgaccga gtactttctg cactattaaa agaaagaaaa cctgtctata 660
tcaacttacc agttgatgtt gctgctgcaa aagcagagaa accctcactc cctttgaaaa 720
aagaaaactc aacttcaaat acaagtgacc aagagatctt gaacaaaatt caagaaagct 780
tgaaaaatgc caaaaaacca atcgtgatta caggacatga aataattagt tttggcttag 840
aaaaaacagt ctctcaattt atttcaaaga caaaactacc tattacgaca ttaaactttg 900
gaaaaagttc agttgatgaa gctctccctt catttttagg aatctataat ggtaaactct 960
cagagcctaa tcttaaagaa ttcgtggaat cagccgactt catcctgatg cttggagtta 1020
aactcacaga ctcttcaaca ggagccttca ctcatcattt aaatgaaaat aaaatgattt 1080
cactgaatat agatgaagga aaaatattta acgaaagcat ccaaaatttt gattttgaat 1140
ccctcatctc ctctctctta gacctaagcg aaatagaata caaaggaaaa tatatcgata 1200
aaaagcaaga agactttgtt ccatcaaatg cgcttttatc acaagaccgc ctatggcaag 1260
cagttgaaaa cctaactcaa agcaatgaaa caatcgttgc tgaacaaggg acatcattct 1320
ttggcgcttc atcaattttc ttaaaaccaa agagtcattt tattggtcaa cccttatggg 1380
gatcaattgg atatacattc ccagcagcat taggaagcca aattgcagat aaagaaagca 1440
gacacctttt atttattggt gatggttcac ttcaacttac ggtgcaagaa ttaggattag 1500
caatcagaga aaaaattaat ccaatttgct ttattatcaa taatgatggt tatacagtcg 1560
aaagagaaat tcatggacca aatcaaagct acaatgatat tccaatgtgg aattactcaa 1620
aattaccaga atcatttgga gcaacagaag aacgagtagt ctcgaaaatc gttagaactg 1680
aaaatgaatt tgtgtctgtc atgaaagaag ctcaagcaga tccaaataga atgtactgga 1740
ttgagttaat tttggcaaaa gaagatgcac caaaagtact gaaaaaaatg ggcaaactat 1800
ttgctgaaca aaataaatca taagcatgca ggagatatac catgtctatt ccagaaactc 1860
aaaaagccat tatcttctac gaatccaacg gcaagttgga gcataaggat atcccagttc 1920
caaagccaaa gcccaacgaa ttgttaatca acgtcaagta ctctggtgtc tgccacaccg 1980
atttgcacgc ttggcatggt gactggccat tgccaactaa gttaccatta gttggtggtc 2040
acgaaggtgc cggtgtcgtt gtcggcatgg gtgaaaacgt taagggctgg aagatcggtg 2100
actacgccgg tatcaaatgg ttgaacggtt cttgtatggc ctgtgaatac tgtgaattgg 2160
gtaacgaatc caactgtcct cacgctgact tgtctggtta cacccacgac ggttctttcc 2220
aagaatacgc taccgctgac gctgttcaag ccgctcacat tcctcaaggt actgacttgg 2280
ctgaagtcgc gccaatcttg tgtgctggta tcaccgtata caaggctttg aagtctgcca 2340
acttgagagc aggccactgg gcggccattt ctggtgctgc tggtggtcta ggttctttgg 2400
ctgttcaata tgctaaggcg atgggttaca gagtcttagg tattgatggt ggtccaggaa 2460
aggaagaatt gtttacctcg ctcggtggtg aagtattcat cgacttcacc aaagagaagg 2520
acattgttag cgcagtcgtt aaggctacca acggcggtgc ccacggtatc atcaatgttt 2580
ccgtttccga agccgctatc gaagcttcta ccagatactg tagggcgaac ggtactgttg 2640
tcttggttgg tttgccagcc ggtgcaaagt gctcctctga tgtcttcaac cacgttgtca 2700
agtctatctc cattgtcggc tcttacgtgg ggaacagagc tgataccaga gaagccttag 2760
atttctttgc cagaggtcta gtcaagtctc caataaaggt agttggctta tccagtttac 2820
cagaaattta cgaaaagatg gagaagggcc aaattgctgg tagatacgtt gttgacactt 2880
ctaaataatc tagaggcatc aaataaaacg aaaggctcag tcgaaagact gggcctttcg 2940
ttttatctgt tgtttgtcgg tgaacgctct cctgagtagg acaaatccgc cgccctagac 3000
ctagggtacg ggttttgctg cccgcaaacg ggctgttctg gtgttgctag tttgttatca 3060
gaatcgcaga tccggcttca ggtttgccgg ctgaaagcgc tatttcttcc agaattgcca 3120
tgattttttc cccacgggag gcgtcactgg ctcccgtgtt gtcggcagct ttgattcgat 3180
aagcagcatc gcctgtttca ggctgtctat gtgtgactgt tgagctgtaa caagttgtct 3240
caggtgttca atttcatgtt ctagttgctt tgttttactg gtttcacctg ttctattagg 3300
tgttacatgc tgttcatctg ttacattgtc gatctgttca tggtgaacag ctttaaatgc 3360
accaaaaact cgtaaaagct ctgatgtatc tatctttttt acaccgtttt catctgtgca 3420
tatggacagt tttccctttg atatctaacg gtgaacagtt gttctacttt tgtttgttag 3480
tcttgatgct tcactgatag atacaagagc cataagaacc tcagatcctt ccgtatttag 3540
ccagtatgtt ctctagtgtg gttcgttgtt tttgcgtgag ccatgagaac gaaccattga 3600
gatcatgctt actttgcatg tcactcaaaa attttgcctc aaaactggtg agctgaattt 3660
ttgcagttaa agcatcgtgt agtgtttttc ttagtccgtt acgtaggtag gaatctgatg 3720
taatggttgt tggtattttg tcaccattca tttttatctg gttgttctca agttcggtta 3780
cgagatccat ttgtctatct agttcaactt ggaaaatcaa cgtatcagtc gggcggcctc 3840
gcttatcaac caccaatttc atattgctgt aagtgtttaa atctttactt attggtttca 3900
aaacccattg gttaagcctt ttaaactcat ggtagttatt ttcaagcatt aacatgaact 3960
taaattcatc aaggctaatc tctatatttg ccttgtgagt tttcttttgt gttagttctt 4020
ttaataacca ctcataaatc ctcatagagt atttgttttc aaaagactta acatgttcca 4080
gattatattt tatgaatttt tttaactgga aaagataagg caatatctct tcactaaaaa 4140
ctaattctaa tttttcgctt gagaacttgg catagtttgt ccactggaaa atctcaaagc 4200
ctttaaccaa aggattcctg atttccacag ttctcgtcat cagctctctg gttgctttag 4260
ctaatacacc ataagcattt tccctactga tgttcatcat ctgagcgtat tggttataag 4320
tgaacgatac cgtccgttct ttccttgtag ggttttcaat cgtggggttg agtagtgcca 4380
cacagcataa aattagcttg gtttcatgct ccgttaagtc atagcgacta atcgctagtt 4440
catttgcttt gaaaacaact aattcagaca tacatctcaa ttggtctagg tgattttaat 4500
cactatacca attgagatgg gctagtcaat gataattact agtccttttc ccgggagatc 4560
tgggtatctg taaattctgc tagacctttg ctggaaaact tgtaaattct gctagaccct 4620
ctgtaaattc cgctagacct ttgtgtgttt tttttgttta tattcaagtg gttataattt 4680
atagaataaa gaaagaataa aaaaagataa aaagaataga tcccagccct gtgtataact 4740
cactacttta gtcagttccg cagtattaca aaaggatgtc gcaaacgctg tttgctcctc 4800
tacaaaacag accttaaaac cctaaaggct taagtagcac cctcgcaagc tcgggcaaat 4860
cgctgaatat tccttttgtc tccgaccatc aggcacctga gtcgctgtct ttttcgtgac 4920
attcagttcg ctgcgctcac ggctctggca gtgaatgggg gtaaatggca ctacaggcgc 4980
cttttatgga ttcatgcaag gaaactaccc ataatacaag aaaagcccgt cacgggcttc 5040
tcagggcgtt ttatggcggg tctgctatgt ggtgctatct gactttttgc tgttcagcag 5100
ttcctgccct ctgattttcc agtctgacca cttcggatta tcccgtgaca ggtcattcag 5160
actggctaat gcacccagta aggcagcggt atcatcaaca ggcttacccg tcttactgtc 5220
cctagtgctt ggattctcac caataaaaaa cgcccggcgg caaccgagcg ttctgaacaa 5280
atccagatgg agttctgagg tcattactgg atctatcaac aggagtccaa gcgagctctc 5340
gaaccccaga gtcccgctca gaagaactcg tcaagaaggc gatagaaggc gatgcgctgc 5400
gaatcgggag cggcgatacc gtaaagcacg aggaagcggt cagcccattc gccgccaagc 5460
tcttcagcaa tatcacgggt agccaacgct atgtcctgat agcggtccgc cacacccagc 5520
cggccacagt cgatgaatcc agaaaagcgg ccattttcca ccatgatatt cggcaagcag 5580
gcatcgccat gggtcacgac gagatcctcg ccgtcgggca tgcgcgcctt gagcctggcg 5640
aacagttcgg ctggcgcgag cccctgatgc tcttcgtcca gatcatcctg atcgacaaga 5700
ccggcttcca tccgagtacg tgctcgctcg atgcgatgtt tcgcttggtg gtcgaatggg 5760
caggtagccg gatcaagcgt atgcagccgc cgcattgcat cagccatgat ggatactttc 5820
tcggcaggag caaggtgaga tgacaggaga tcctgccccg gcacttcgcc caatagcagc 5880
cagtcccttc ccgcttcagt gacaacgtcg agcacagctg cgcaaggaac gcccgtcgtg 5940
gccagccacg atagccgcgc tgcctcgtcc tgcagttcat tcagggcacc ggacaggtcg 6000
gtcttgacaa aaagaaccgg gcgcccctgc gctgacagcc ggaacacggc ggcatcagag 6060
cagccgattg tctgttgtgc ccagtcatag ccgaatagcc tctccaccca agcggccgga 6120
gaacctgcgt gcaatccatc ttgttcaatc atgcgaaacg atcctcatcc tgtctcttga 6180
tcagatcttg atcccctgcg ccatcagatc cttggcggca agaaagccat ccagtttact 6240
ttgcagggct tcccaacctt accagagggc gccccagctg gcaattccga cgtc 6294
<210> SEQ ID NO 104
<211> LENGTH: 1980
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1490
<400> SEQUENCE: 104
ctcgagtccc tatcagtgat agagattgac atccctatca gtgatagaga tactgagcac 60
atcagcagga cgcactgacc gaattcatta aagaggagaa aggtacctgc acgtcgactc 120
cgtcctaggg gatatattcc gcttcctcgc tcactgactc gctacgctcg gtcgttcgac 180
tgcggcgagc ggaaatggct tacgaacggg gcggagattt cctggaagat gccaggaaga 240
tacttaacag ggaagtgaga gggccgcggc aaagccgttt ttccataggc tccgcccccc 300
tgacaagcat cacgaaatct gacgctcaaa tcagtggtgg cgaaacccga caggactata 360
aagataccag gcgtttcccc ctggcggctc cctcgtgcgc tctcctgttc ctgcctttcg 420
gtttaccggt gtcattccgc tgttatggcc gcgtttgtct cattccacgc ctgacactca 480
gttccgggta ggcagttcgc tccaagctgg actgtatgca cgaacccccc gttcagtccg 540
accgctgcgc cttatccggt aactatcgtc ttgagtccaa cccggaaaga catgcaaaag 600
caccactggc agcagccact ggtaattgat ttagaggagt tagtcttgaa gtcatgcgcc 660
ggttaaggct aaactgaaag gacaagtttt ggtgactgcg ctcctccaag ccagttacct 720
cggttcaaag agttggtagc tcagagaacc ttcgaaaaac cgccctgcaa ggcggttttt 780
tcgttttcag agcaagagat tacgcgcaga ccaaaacgat ctcaagaaga tcatcttatt 840
aatcagataa aatatttcta gatttcagtg caatttatct cttcaaatgt agcacctgaa 900
gtcagcccca tacgatataa gttgttacta gtgcttggat tctcaccaat aaaaaacgcc 960
cggcggcaac cgagcgttct gaacaaatcc agatggagtt ctgaggtcat tactggatct 1020
atcaacagga gtccaagcga gctcgatatc aaattacgcc ccgccctgcc actcatcgca 1080
gtactgttgt aattcattaa gcattctgcc gacatggaag ccatcacaga cggcatgatg 1140
aacctgaatc gccagcggca tcagcacctt gtcgccttgc gtataatatt tgcccatggt 1200
gaaaacgggg gcgaagaagt tgtccatatt ggccacgttt aaatcaaaac tggtgaaact 1260
cacccaggga ttggctgaga cgaaaaacat attctcaata aaccctttag ggaaataggc 1320
caggttttca ccgtaacacg ccacatcttg cgaatatatg tgtagaaact gccggaaatc 1380
gtcgtggtat tcactccaga gcgatgaaaa cgtttcagtt tgctcatgga aaacggtgta 1440
acaagggtga acactatccc atatcaccag ctcaccgtct ttcattgcca tacgaaactc 1500
cggatgagca ttcatcaggc gggcaagaat gtgaataaag gccggataaa acttgtgctt 1560
atttttcttt acggtcttta aaaaggccgt aatatccagc tgaacggtct ggttataggt 1620
acattgagca actgactgaa atgcctcaaa atgttcttta cgatgccatt gggatatatc 1680
aacggtggta tatccagtga tttttttctc cattttagct tccttagctc ctgaaaatct 1740
cgataactca aaaaatacgc ccggtagtga tcttatttca ttatggtgaa agttggaacc 1800
tcttacgtgc cgatcaacgt ctcattttcg ccagatatcg acgtctaaga aaccattatt 1860
atcatgacat taacctataa aaataggcgt atcacgaggc cctttcgtct tcacgaaacc 1920
attattatca tgacattaac ctataaaaat aggcgtatca cgaggccctt tcgtcttcac 1980
<210> SEQ ID NO 105
<211> LENGTH: 2077
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1572
<400> SEQUENCE: 105
ctagtgcttg gattctcacc aataaaaaac gcccggcggc aaccgagcgt tctgaacaaa 60
tccagatgga gttctgaggt cattactgga tctatcaaca ggagtccaag cgagctcgat 120
atcaaattac gccccgccct gccactcatc gcagtactgt tgtaattcat taagcattct 180
gccgacatgg aagccatcac agacggcatg atgaacctga atcgccagcg gcatcagcac 240
cttgtcgcct tgcgtataat atttgcccat ggtgaaaacg ggggcgaaga agttgtccat 300
attggccacg tttaaatcaa aactggtgaa actcacccag ggattggctg agacgaaaaa 360
catattctca ataaaccctt tagggaaata ggccaggttt tcaccgtaac acgccacatc 420
ttgcgaatat atgtgtagaa actgccggaa atcgtcgtgg tattcactcc agagcgatga 480
aaacgtttca gtttgctcat ggaaaacggt gtaacaaggg tgaacactat cccatatcac 540
cagctcaccg tctttcattg ccatacgaaa ctccggatga gcattcatca ggcgggcaag 600
aatgtgaata aaggccggat aaaacttgtg cttatttttc tttacggtct ttaaaaaggc 660
cgtaatatcc agctgaacgg tctggttata ggtacattga gcaactgact gaaatgcctc 720
aaaatgttct ttacgatgcc attgggatat atcaacggtg gtatatccag tgattttttt 780
ctccatttta gcttccttag ctcctgaaaa tctcgataac tcaaaaaata cgcccggtag 840
tgatcttatt tcattatggt gaaagttgga acctcttacg tgccgatcaa cgtctcattt 900
tcgccagata tcgacgtcta agaaaccatt attatcatga cattaaccta taaaaatagg 960
cgtatcacga ggccctttcg tcttcacctc gagaaatgtg agcggataac aattgacatt 1020
gtgagcggat aacaagatac tgagcacatc agcaggacgc actgaccggg aattcattaa 1080
agaggagaaa gtcgacatta tgcggccgcg gatccataag gaggattaat taagacttcc 1140
cgggtgatcc catggtacgc gtgctagagg catcaaataa aacgaaaggc tcagtcgaaa 1200
gactgggcct ttcgttttat ctgttgtttg tcggtgaacg ctctcctgag taggacaaat 1260
ccgccgccct agacctaggg gatatattcc gcttcctcgc tcactgactc gctacgctcg 1320
gtcgttcgac tgcggcgagc ggaaatggct tacgaacggg gcggagattt cctggaagat 1380
gccaggaaga tacttaacag ggaagtgaga gggccgcggc aaagccgttt ttccataggc 1440
tccgcccccc tgacaagcat cacgaaatct gacgctcaaa tcagtggtgg cgaaacccga 1500
caggactata aagataccag gcgtttcccc ctggcggctc cctcgtgcgc tctcctgttc 1560
ctgcctttcg gtttaccggt gtcattccgc tgttatggcc gcgtttgtct cattccacgc 1620
ctgacactca gttccgggta ggcagttcgc tccaagctgg actgtatgca cgaacccccc 1680
gttcagtccg accgctgcgc cttatccggt aactatcgtc ttgagtccaa cccggaaaga 1740
catgcaaaag caccactggc agcagccact ggtaattgat ttagaggagt tagtcttgaa 1800
gtcatgcgcc ggttaaggct aaactgaaag gacaagtttt ggtgactgcg ctcctccaag 1860
ccagttacct cggttcaaag agttggtagc tcagagaacc ttcgaaaaac cgccctgcaa 1920
ggcggttttt tcgttttcag agcaagagat tacgcgcaga ccaaaacgat ctcaagaaga 1980
tcatcttatt aatcagataa aatatttcta gatttcagtg caatttatct cttcaaatgt 2040
agcacctgaa gtcagcccca tacgatataa gttgtta 2077
<210> SEQ ID NO 106
<211> LENGTH: 3135
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1573
<400> SEQUENCE: 106
ctagtgcttg gattctcacc aataaaaaac gcccggcggc aaccgagcgt tctgaacaaa 60
tccagatgga gttctgaggt cattactgga tctatcaaca ggagtccaag cgagctcgat 120
atcaaattac gccccgccct gccactcatc gcagtactgt tgtaattcat taagcattct 180
gccgacatgg aagccatcac agacggcatg atgaacctga atcgccagcg gcatcagcac 240
cttgtcgcct tgcgtataat atttgcccat ggtgaaaacg ggggcgaaga agttgtccat 300
attggccacg tttaaatcaa aactggtgaa actcacccag ggattggctg agacgaaaaa 360
catattctca ataaaccctt tagggaaata ggccaggttt tcaccgtaac acgccacatc 420
ttgcgaatat atgtgtagaa actgccggaa atcgtcgtgg tattcactcc agagcgatga 480
aaacgtttca gtttgctcat ggaaaacggt gtaacaaggg tgaacactat cccatatcac 540
cagctcaccg tctttcattg ccatacgaaa ctccggatga gcattcatca ggcgggcaag 600
aatgtgaata aaggccggat aaaacttgtg cttatttttc tttacggtct ttaaaaaggc 660
cgtaatatcc agctgaacgg tctggttata ggtacattga gcaactgact gaaatgcctc 720
aaaatgttct ttacgatgcc attgggatat atcaacggtg gtatatccag tgattttttt 780
ctccatttta gcttccttag ctcctgaaaa tctcgataac tcaaaaaata cgcccggtag 840
tgatcttatt tcattatggt gaaagttgga acctcttacg tgccgatcaa cgtctcattt 900
tcgccagata tcgacgtcta agaaaccatt attatcatga cattaaccta taaaaatagg 960
cgtatcacga ggccctttcg tcttcacctc gagaaatgtg agcggataac aattgacatt 1020
gtgagcggat aacaagatac tgagcacatc agcaggacgc actgaccggg aattcattaa 1080
agaggagaaa gtcgacatgc ccgatatgac aaacgaatct tcttctaagc cagctcaaat 1140
taacattggt atcaatggtt ttggtagaat cggtagattg gttctacgtg ctgctttgac 1200
gcacccagaa gttaaggtca gattaatcaa taatccatcc acaacaccag aatacgctgc 1260
ttatttgttc aaatacgatt ctactcacgg caagtatcgt ggtgaagttg aattcgacga 1320
tgaacgtatc atcattcaaa atgaccatgt ttcggctcat atccctctat ctcattttag 1380
ggaaccagag cgtatcccat gggcttccta caacgtcgat tatgtaattg actcaaccgg 1440
tgtcttcaag gaagtcgata cagcctctag acataaaggt gtcaaaaaag ttatcattac 1500
tgctccatca aagaccgcgc caatgtacgt ctatggtgtt aaccacgtta aatacaaccc 1560
attgacggat cacgtggtct ctaatgcctc ctgtactacc aactgtttgg ctccgttggt 1620
taaggctttg gacgatgagt tcggtatcga agaagccttg atgacaacta ttcatgcaac 1680
tactgcttct caaaagactg tcgatggtac cagttctggt ggtaaggact ggagaggcgg 1740
tagatcttgc cagggaaata tcattccttc atctactggt gcagctaagg ctgtagggaa 1800
aatcttgcct gaacttaatg gtaagatcac cggtatgtct ataagagtcc caacaattaa 1860
tatttccctg gttgacttga cattccgtac agcaaagaaa acttcttacg atgacattat 1920
gaaggcccta gaacaaagat ctcgcagcga tatgaagggt gttttgggtg ttaccaaaga 1980
cgccgttgtg tcctctgact tcacatccga ttcacgttca tctattgttg atgccaaggc 2040
cggtattgaa ttgaacgacc attttttcaa ggtcctttct tggtatgata atgaatatgg 2100
ttactcttca agagtggttg atttatccat tttcatggct caaaaggact tcgaagctgg 2160
tgtttaagga tccataagga ggattaatta agacttcccg ggtgatccca tggtacgcgt 2220
gctagaggca tcaaataaaa cgaaaggctc agtcgaaaga ctgggccttt cgttttatct 2280
gttgtttgtc ggtgaacgct ctcctgagta ggacaaatcc gccgccctag acctagggga 2340
tatattccgc ttcctcgctc actgactcgc tacgctcggt cgttcgactg cggcgagcgg 2400
aaatggctta cgaacggggc ggagatttcc tggaagatgc caggaagata cttaacaggg 2460
aagtgagagg gccgcggcaa agccgttttt ccataggctc cgcccccctg acaagcatca 2520
cgaaatctga cgctcaaatc agtggtggcg aaacccgaca ggactataaa gataccaggc 2580
gtttccccct ggcggctccc tcgtgcgctc tcctgttcct gcctttcggt ttaccggtgt 2640
cattccgctg ttatggccgc gtttgtctca ttccacgcct gacactcagt tccgggtagg 2700
cagttcgctc caagctggac tgtatgcacg aaccccccgt tcagtccgac cgctgcgcct 2760
tatccggtaa ctatcgtctt gagtccaacc cggaaagaca tgcaaaagca ccactggcag 2820
cagccactgg taattgattt agaggagtta gtcttgaagt catgcgccgg ttaaggctaa 2880
actgaaagga caagttttgg tgactgcgct cctccaagcc agttacctcg gttcaaagag 2940
ttggtagctc agagaacctt cgaaaaaccg ccctgcaagg cggttttttc gttttcagag 3000
caagagatta cgcgcagacc aaaacgatct caagaagatc atcttattaa tcagataaaa 3060
tatttctaga tttcagtgca atttatctct tcaaatgtag cacctgaagt cagccccata 3120
cgatataagt tgtta 3135
<210> SEQ ID NO 107
<211> LENGTH: 3069
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1575
<400> SEQUENCE: 107
ctagtgcttg gattctcacc aataaaaaac gcccggcggc aaccgagcgt tctgaacaaa 60
tccagatgga gttctgaggt cattactgga tctatcaaca ggagtccaag cgagctcgat 120
atcaaattac gccccgccct gccactcatc gcagtactgt tgtaattcat taagcattct 180
gccgacatgg aagccatcac agacggcatg atgaacctga atcgccagcg gcatcagcac 240
cttgtcgcct tgcgtataat atttgcccat ggtgaaaacg ggggcgaaga agttgtccat 300
attggccacg tttaaatcaa aactggtgaa actcacccag ggattggctg agacgaaaaa 360
catattctca ataaaccctt tagggaaata ggccaggttt tcaccgtaac acgccacatc 420
ttgcgaatat atgtgtagaa actgccggaa atcgtcgtgg tattcactcc agagcgatga 480
aaacgtttca gtttgctcat ggaaaacggt gtaacaaggg tgaacactat cccatatcac 540
cagctcaccg tctttcattg ccatacgaaa ctccggatga gcattcatca ggcgggcaag 600
aatgtgaata aaggccggat aaaacttgtg cttatttttc tttacggtct ttaaaaaggc 660
cgtaatatcc agctgaacgg tctggttata ggtacattga gcaactgact gaaatgcctc 720
aaaatgttct ttacgatgcc attgggatat atcaacggtg gtatatccag tgattttttt 780
ctccatttta gcttccttag ctcctgaaaa tctcgataac tcaaaaaata cgcccggtag 840
tgatcttatt tcattatggt gaaagttgga acctcttacg tgccgatcaa cgtctcattt 900
tcgccagata tcgacgtcta agaaaccatt attatcatga cattaaccta taaaaatagg 960
cgtatcacga ggccctttcg tcttcacctc gagaaatgtg agcggataac aattgacatt 1020
gtgagcggat aacaagatac tgagcacatc agcaggacgc actgaccggg aattcattaa 1080
agaggagaaa gtcgacatgg caaagatagc tattaatggt tttggaagaa taggaagatt 1140
agctttaaga agaattcttg aagtacctgg attggaagtt gttgcaataa acgacttaac 1200
tgatgcaaaa atgttagcac acttatttaa atatgattca tcacaaggaa gattcaatgg 1260
agaaattgaa gttaaagaag gagctttcgt agtaaacgga aaagaagtta aagttttcgc 1320
tgaagcagat cctgaaaaat taccttgggg agatcttgga atagacgttg ttcttgagtg 1380
cacaggtttc ttcacaaaga aagaaaaagc agaagctcac gtaagagcag gcgctaaaaa 1440
agttgttata tcagctccag ctggaaacga cttaaagaca atagttttca acgttaataa 1500
tgaagatctt gatggaacag aaacagttat atcaggtgca tcatgcacaa ctaactgctt 1560
agctccaatg gctaaagtat taaatgataa atttggaata gaaaaaggat tcatgactac 1620
aattcatgcg ttcactaatg accaaaacac attagatggt ccacacagaa aaggagattt 1680
aagaagagct agagctgctg ctgtaagtat catccctaac tcaactggtg ctgctaaagc 1740
tataagccaa gttattcctg acttagctgg aaaattagac ggaaacgctc aaagagttcc 1800
agttccaact ggttcaataa ctgaattagt ttcagttctt aagaaaaaag ttacagttga 1860
agaaatcaac gctgctatga aagaagctgc tgatgaatca tttggataca ctgaagatcc 1920
aatcgtttca gctgacgtag taggaatcaa ctacggatca ttatttgatg caactttaac 1980
taaaattgtt gatgttaacg gatcacaatt agttaaaaca gctgcttggt atgataatga 2040
aatgtcatac acttcacaat tagttagaac tttagcttac tttgcaaaaa tagcaaaata 2100
gggatccata aggaggatta attaagactt cccgggtgat cccatggtac gcgtgctaga 2160
ggcatcaaat aaaacgaaag gctcagtcga aagactgggc ctttcgtttt atctgttgtt 2220
tgtcggtgaa cgctctcctg agtaggacaa atccgccgcc ctagacctag gggatatatt 2280
ccgcttcctc gctcactgac tcgctacgct cggtcgttcg actgcggcga gcggaaatgg 2340
cttacgaacg gggcggagat ttcctggaag atgccaggaa gatacttaac agggaagtga 2400
gagggccgcg gcaaagccgt ttttccatag gctccgcccc cctgacaagc atcacgaaat 2460
ctgacgctca aatcagtggt ggcgaaaccc gacaggacta taaagatacc aggcgtttcc 2520
ccctggcggc tccctcgtgc gctctcctgt tcctgccttt cggtttaccg gtgtcattcc 2580
gctgttatgg ccgcgtttgt ctcattccac gcctgacact cagttccggg taggcagttc 2640
gctccaagct ggactgtatg cacgaacccc ccgttcagtc cgaccgctgc gccttatccg 2700
gtaactatcg tcttgagtcc aacccggaaa gacatgcaaa agcaccactg gcagcagcca 2760
ctggtaattg atttagagga gttagtcttg aagtcatgcg ccggttaagg ctaaactgaa 2820
aggacaagtt ttggtgactg cgctcctcca agccagttac ctcggttcaa agagttggta 2880
gctcagagaa ccttcgaaaa accgccctgc aaggcggttt tttcgttttc agagcaagag 2940
attacgcgca gaccaaaacg atctcaagaa gatcatctta ttaatcagat aaaatatttc 3000
tagatttcag tgcaatttat ctcttcaaat gtagcacctg aagtcagccc catacgatat 3060
aagttgtta 3069
<210> SEQ ID NO 108
<211> LENGTH: 7093
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1609
<400> SEQUENCE: 108
cgatatcaaa ttacgccccg ccctgccact catcgcagta ctgttgtaat tcattaagca 60
ttctgccgac atggaagcca tcacagacgg catgatgaac ctgaatcgcc agcggcatca 120
gcaccttgtc gccttgcgta taatatttgc ccatggtgaa aacgggggcg aagaagttgt 180
ccatattggc cacgtttaaa tcaaaactgg tgaaactcac ccagggattg gctgagacga 240
aaaacatatt ctcaataaac cctttaggga aataggccag gttttcaccg taacacgcca 300
catcttgcga atatatgtgt agaaactgcc ggaaatcgtc gtggtattca ctccagagcg 360
atgaaaacgt ttcagtttgc tcatggaaaa cggtgtaaca agggtgaaca ctatcccata 420
tcaccagctc accgtctttc attgccatac gaaactccgg atgagcattc atcaggcggg 480
caagaatgtg aataaaggcc ggataaaact tgtgcttatt tttctttacg gtctttaaaa 540
aggccgtaat atccagctga acggtctggt tataggtaca ttgagcaact gactgaaatg 600
cctcaaaatg ttctttacga tgccattggg atatatcaac ggtggtatat ccagtgattt 660
ttttctccat tttagcttcc ttagctcctg aaaatctcga taactcaaaa aatacgcccg 720
gtagtgatct tatttcatta tggtgaaagt tggaacctct tacgtgccga tcaacgtctc 780
attttcgcca gatatcgacg tctaagaaac cattattatc atgacattaa cctataaaaa 840
taggcgtatc acgaggccct ttcgtcttca cctcgagaat tgtgagcgga taacaattga 900
cattgtgagc ggataacaag atactgagca catcagcagg acgcactgac cgaattcatt 960
aaagaggaga aaggtacaat gttgacaaaa gcaacaaaag aacaaaaatc ccttgtgaaa 1020
aacagagggg cggagcttgt tgttgattgc ttagtggagc aaggtgtcac acatgtattt 1080
ggcattccag gtgcaaaaat tgatgcggta tttgacgctt tacaagataa aggacctgaa 1140
attatcgttg cccggcacga acaaaacgca gcattcatgg cccaagcagt cggccgttta 1200
actggaaaac cgggagtcgt gttagtcaca tcaggaccgg gtgcctctaa cttggcaaca 1260
ggcctgctga cagcgaacac tgaaggagac cctgtcgttg cgcttgctgg aaacgtgatc 1320
cgtgcagatc gtttaaaacg gacacatcaa tctttggata atgcggcgct attccagccg 1380
attacaaaat acagtgtaga agttcaagat gtaaaaaata taccggaagc tgttacaaat 1440
gcatttagga tagcgtcagc agggcaggct ggggccgctt ttgtgagctt tccgcaagat 1500
gttgtgaatg aagtcacaaa tacgaaaaac gtgcgtgctg ttgcagcgcc aaaactcggt 1560
cctgcagcag atgatgcaat cagtgcggcc atagcaaaaa tccaaacagc aaaacttcct 1620
gtcgttttgg tcggcatgaa aggcggaaga ccggaagcaa ttaaagcggt tcgcaagctt 1680
ttgaaaaagg ttcagcttcc atttgttgaa acatatcaag ctgccggtac cctttctaga 1740
gatttagagg atcaatattt tggccgtatc ggtttgttcc gcaaccagcc tggcgattta 1800
ctgctagagc aggcagatgt tgttctgacg atcggctatg acccgattga atatgatccg 1860
aaattctgga atatcaatgg agaccggaca attatccatt tagacgagat tatcgctgac 1920
attgatcatg cttaccagcc tgatcttgaa ttgatcggtg acattccgtc cacgatcaat 1980
catatcgaac acgatgctgt gaaagtggaa tttgcagagc gtgagcagaa aatcctttct 2040
gatttaaaac aatatatgca tgaaggtgag caggtgcctg cagattggaa atcagacaga 2100
gcgcaccctc ttgaaatcgt taaagagttg cgtaatgcag tcgatgatca tgttacagta 2160
acttgcgata tcggttcgca cgccatttgg atgtcacgtt atttccgcag ctacgagccg 2220
ttaacattaa tgatcagtaa cggtatgcaa acactcggcg ttgcgcttcc ttgggcaatc 2280
ggcgcttcat tggtgaaacc gggagaaaaa gtggtttctg tctctggtga cggcggtttc 2340
ttattctcag caatggaatt agagacagca gttcgactaa aagcaccaat tgtacacatt 2400
gtatggaacg acagcacata tgacatggtt gcattccagc aattgaaaaa atataaccgt 2460
acatctgcgg tcgatttcgg aaatatcgat atcgtgaaat atgcggaaag cttcggagca 2520
actggcttgc gcgtagaatc accagaccag ctggcagatg ttctgcgtca aggcatgaac 2580
gctgaaggtc ctgtcatcat cgatgtcccg gttgactaca gtgataacat taatttagca 2640
agtgacaagc ttccgaaaga attcggggaa ctcatgaaaa cgaaagctct ctaggtcgac 2700
gaggaatcac catggctaac tacttcaata cactgaatct gcgccagcag ctggcacagc 2760
tgggcaaatg tcgctttatg ggccgcgatg aattcgccga tggcgcgagc taccttcagg 2820
gtaaaaaagt agtcatcgtc ggctgtggcg cacagggtct gaaccagggc ctgaacatgc 2880
gtgattctgg tctcgatatc tcctacgctc tgcgtaaaga agcgattgcc gagaagcgcg 2940
cgtcctggcg taaagcgacc gaaaatggtt ttaaagtggg tacttacgaa gaactgatcc 3000
cacaggcgga tctggtgatt aacctgacgc cggacaagca gcactctgat gtagtgcgca 3060
ccgtacagcc actgatgaaa gacggcgcgg cgctgggcta ctcgcacggt ttcaacatcg 3120
tcgaagtggg cgagcagatc cgtaaagata tcaccgtagt gatggttgcg ccgaaatgcc 3180
caggcaccga agtgcgtgaa gagtacaaac gtgggttcgg cgtaccgacg ctgattgccg 3240
ttcacccgga aaacgatccg aaaggcgaag gcatggcgat tgccaaagcc tgggcggctg 3300
caaccggtgg tcaccgtgcg ggtgtgctgg aatcgtcctt cgttgcggaa gtgaaatctg 3360
acctgatggg cgagcaaacc atcctgtgcg gtatgttgca ggctggctct ctgctgtgct 3420
tcgacaagct ggtggaagaa ggtaccgatc cagcatacgc agaaaaactg attcagttcg 3480
gttgggaaac catcaccgaa gcactgaaac agggcggcat caccctgatg atggaccgtc 3540
tctctaaccc ggcgaaactg cgtgcttatg cgctttctga acagctgaaa gagatcatgg 3600
cacccctgtt ccagaaacat atggacgaca tcatctccgg cgaattctct tccggtatga 3660
tggcggactg ggccaacgat gataagaaac tgctgacctg gcgtgaagag accggcaaaa 3720
ccgcgtttga aaccgcgccg cagtatgaag gcaaaatcgg cgagcaggag tacttcgata 3780
aaggcgtact gatgattgcg atggtgaaag cgggcgttga actggcgttc gaaaccatgg 3840
tcgattccgg catcattgaa gagtctgcat attatgaatc actgcacgag ctgccgctga 3900
ttgccaacac catcgcccgt aagcgtctgt acgaaatgaa cgtggttatc tctgataccg 3960
ctgagtacgg taactatctg ttctcttacg cttgtgtgcc gttgctgaaa ccgtttatgg 4020
cagagctgca accgggcgac ctgggtaaag ctattccgga aggcgcggta gataacgggc 4080
aactgcgtga tgtgaacgaa gcgattcgca gccatgcgat tgagcaggta ggtaagaaac 4140
tgcgcggcta tatgacagat atgaaacgta ttgctgttgc gggttaaccc ggaaggagat 4200
ataccatgcc taagtaccgt tccgccacca ccactcatgg tcgtaatatg gcgggtgctc 4260
gtgcgctgtg gcgcgccacc ggaatgaccg acgccgattt cggtaagccg attatcgcgg 4320
ttgtgaactc gttcacccaa tttgtaccgg gtcacgtcca tctgcgcgat ctcggtaaac 4380
tggtcgccga acaaattgaa gcggctggcg gcgttgccaa agagttcaac accattgcgg 4440
tggatgatgg gattgccatg ggccacgggg ggatgcttta ttcactgcca tctcgcgaac 4500
tgatcgctga ttccgttgag tatatggtca acgcccactg cgccgacgcc atggtctgca 4560
tctctaactg cgacaaaatc accccgggga tgctgatggc ttccctgcgc ctgaatattc 4620
cggtgatctt tgtttccggc ggcccgatgg aggccgggaa aaccaaactt tccgatcaga 4680
tcatcaagct cgatctggtt gatgcgatga tccagggcgc agacccgaaa gtatctgact 4740
cccagagcga tcaggttgaa cgttccgcgt gtccgacctg cggttcctgc tccgggatgt 4800
ttaccgctaa ctcaatgaac tgcctgaccg aagcgctggg cctgtcgcag ccgggcaacg 4860
gctcgctgct ggcaacccac gccgaccgta agcagctgtt ccttaatgct ggtaaacgca 4920
ttgttgaatt gaccaaacgt tattacgagc aaaacgacga aagtgcactg ccgcgtaata 4980
tcgccagtaa ggcggcgttt gaaaacgcca tgacgctgga tatcgcgatg ggtggatcga 5040
ctaacaccgt acttcacctg ctggcggcgg cgcaggaagc ggaaatcgac ttcaccatga 5100
gtgatatcga taagctttcc cgcaaggttc cacagctgtg taaagttgcg ccgagcaccc 5160
agaaatacca tatggaagat gttcaccgtg ctggtggtgt tatcggtatt ctcggcgaac 5220
tggatcgcgc ggggttactg aaccgtgatg tgaaaaacgt acttggcctg acgttgccgc 5280
aaacgctgga acaatacgac gttatgctga cccaggatga cgcggtaaaa aatatgttcc 5340
gcgcaggtcc tgcaggcatt cgtaccacac aggcattctc gcaagattgc cgttgggata 5400
cgctggacga cgatcgcgcc aatggctgta tccgctcgct ggaacacgcc tacagcaaag 5460
acggcggcct ggcggtgctc tacggtaact ttgcggaaaa cggctgcatc gtgaaaacgg 5520
caggcgtcga tgacagcatc ctcaaattca ccggcccggc gaaagtgtac gaaagccagg 5580
acgatgcggt agaagcgatt ctcggcggta aagttgtcgc cggagatgtg gtagtaattc 5640
gctatgaagg cccgaaaggc ggtccgggga tgcaggaaat gctctaccca accagcttcc 5700
tgaaatcaat gggtctcggc aaagcctgtg cgctgatcac cgacggtcgt ttctctggtg 5760
gcacctctgg tctttccatc ggccacgtct caccggaagc ggcaagcggc ggcagcattg 5820
gcctgattga agatggtgac ctgatcgcta tcgacatccc gaaccgtggc attcagttac 5880
aggtaagcga tgccgaactg gcggcgcgtc gtgaagcgca ggacgctcga ggtgacaaag 5940
cctggacgcc gaaaaatcgt gaacgtcagg tctcctttgc cctgcgtgct tatgccagcc 6000
tggcaaccag cgccgacaaa ggcgcggtgc gcgataaatc gaaactgggg ggttaaacgc 6060
gtgctagagg catcaaataa aacgaaaggc tcagtcgaaa gactgggcct ttcgttttat 6120
ctgttgtttg tcggtgaacg ctctcctgag taggacaaat ccgccgccct agacctaggg 6180
gatatattcc gcttcctcgc tcactgactc gctacgctcg gtcgttcgac tgcggcgagc 6240
ggaaatggct tacgaacggg gcggagattt cctggaagat gccaggaaga tacttaacag 6300
ggaagtgaga gggccgcggc aaagccgttt ttccataggc tccgcccccc tgacaagcat 6360
cacgaaatct gacgctcaaa tcagtggtgg cgaaacccga caggactata aagataccag 6420
gcgtttcccc ctggcggctc cctcgtgcgc tctcctgttc ctgcctttcg gtttaccggt 6480
gtcattccgc tgttatggcc gcgtttgtct cattccacgc ctgacactca gttccgggta 6540
ggcagttcgc tccaagctgg actgtatgca cgaacccccc gttcagtccg accgctgcgc 6600
cttatccggt aactatcgtc ttgagtccaa cccggaaaga catgcaaaag caccactggc 6660
agcagccact ggtaattgat ttagaggagt tagtcttgaa gtcatgcgcc ggttaaggct 6720
aaactgaaag gacaagtttt ggtgactgcg ctcctccaag ccagttacct cggttcaaag 6780
agttggtagc tcagagaacc ttcgaaaaac cgccctgcaa ggcggttttt tcgttttcag 6840
agcaagagat tacgcgcaga ccaaaacgat ctcaagaaga tcatcttatt aatcagataa 6900
aatatttcta gatttcagtg caatttatct cttcaaatgt agcacctgaa gtcagcccca 6960
tacgatataa gttgttacta gtgcttggat tctcaccaat aaaaaacgcc cggcggcaac 7020
cgagcgttct gaacaaatcc agatggagtt ctgaggtcat tactggatct atcaacagga 7080
gtccaagcga gct 7093
<210> SEQ ID NO 109
<211> LENGTH: 7112
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1655
<400> SEQUENCE: 109
taagaaacca ttattatcat gacattaacc tataaaaata ggcgtatcac gaggcccttt 60
cgtcttcacc tcgagaattg tgagcggata acaattgaca ttgtgagcgg ataacaagat 120
actgagcaca tcagcaggac gcactgaccg aattcattaa agaggagaaa ggtaccatgt 180
atacagtagg agattaccta ttagaccgat tacacgagtt aggaattgaa gaaatttttg 240
gagtccctgg agactataac ttacaatttt tagatcaaat tatttcccgc aaggatatga 300
aatgggtcgg aaatgctaat gaattaaatg cttcatatat ggctgatggc tatgctcgta 360
ctaaaaaagc tgccgcattt cttacaacct ttggagtagg tgaattgagt gcagttaatg 420
gattagcagg aagttacgcc gaaaatttac cagtagtaga aatagtggga tcacctacat 480
caaaagttca aaatgaagga aaatttgttc atcatacgct ggctgacggt gattttaaac 540
actttatgaa aatgcacgaa cctgttacag cagctcgaac tttactgaca gcagaaaatg 600
caaccgttga aattgaccga gtactttctg cactattaaa agaaagaaaa cctgtctata 660
tcaacttacc agttgatgtt gctgctgcaa aagcagagaa accctcactc cctttgaaaa 720
aagaaaactc aacttcaaat acaagtgacc aagagatctt gaacaaaatt caagaaagct 780
tgaaaaatgc caaaaaacca atcgtgatta caggacatga aataattagt tttggcttag 840
aaaaaacagt ctctcaattt atttcaaaga caaaactacc tattacgaca ttaaactttg 900
gaaaaagttc agttgatgaa gctctccctt catttttagg aatctataat ggtaaactct 960
cagagcctaa tcttaaagaa ttcgtggaat cagccgactt catcctgatg cttggagtta 1020
aactcacaga ctcttcaaca ggagccttca ctcatcattt aaatgaaaat aaaatgattt 1080
cactgaatat agatgaagga aaaatattta acgaaagcat ccaaaatttt gattttgaat 1140
ccctcatctc ctctctctta gacctaagcg aaatagaata caaaggaaaa tatatcgata 1200
aaaagcaaga agactttgtt ccatcaaatg cgcttttatc acaagaccgc ctatggcaag 1260
cagttgaaaa cctaactcaa agcaatgaaa caatcgttgc tgaacaaggg acatcattct 1320
ttggcgcttc atcaattttc ttaaaaccaa agagtcattt tattggtcaa cccttatggg 1380
gatcaattgg atatacattc ccagcagcat taggaagcca aattgcagat aaagaaagca 1440
gacacctttt atttattggt gatggttcac ttcaacttac ggtgcaagaa ttaggattag 1500
caatcagaga aaaaattaat ccaatttgct ttattatcaa taatgatggt tatacagtcg 1560
aaagagaaat tcatggacca aatcaaagct acaatgatat tccaatgtgg aattactcaa 1620
aattaccaga atcatttgga gcaacagaag aacgagtagt ctcgaaaatc gttagaactg 1680
aaaatgaatt tgtgtctgtc atgaaagaag ctcaagcaga tccaaataga atgtactgga 1740
ttgagttaat tttggcaaaa gaagatgcac caaaagtact gaaaaaaatg ggcaaactat 1800
ttgctgaaca aaataaatca taaggtcgac aggagatata ctatgcctaa atatcgcagc 1860
gcaactacta cccacggccg caacatggca ggcgcgcgtg ctctgtggcg tgcgactggt 1920
atgactgatg cggactttgg caaaccaatc attgctgtgg ttaatagctt tactcagttc 1980
gttccaggcc atgttcacct gcgtgacctg ggcaagctgg ttgcggagca gatcgaggct 2040
gcgggtggtg tggcgaagga atttaacacc atcgctgttg acgacggtat cgcgatgggt 2100
catggtggta tgctgtacag cctgccgagc cgtgagctga ttgcggacag cgtggaatac 2160
atggttaatg cgcattgtgc ggatgcgatg gtttgtatta gcaactgtga taagattact 2220
ccaggtatgc tgatggcgag cctgcgtctg aacatcccag ttattttcgt gagcggtggt 2280
ccaatggaag cgggtaagac taagctgagc gaccagatta tcaaactgga cctggtggac 2340
gctatgattc aaggtgctga tccaaaggtt agcgatagcc aatctgacca agtggagcgc 2400
agcgcttgcc caacttgtgg cagctgtagc ggtatgttca ctgcgaatag catgaattgt 2460
ctgactgagg ctctgggtct gagccaacca ggtaatggta gcctgctggc gactcatgcg 2520
gatcgcaaac aactgtttct gaacgcgggc aagcgtatcg tggagctgac taagcgctac 2580
tatgaacaga atgatgagtc cgcgctgcca cgcaacattg cgtccaaagc tgctttcgag 2640
aatgcgatga ccctggacat tgctatgggc ggtagcacca atactgttct gcatctgctg 2700
gctgctgctc aagaggctga gattgatttt actatgtccg acattgacaa actgagccgt 2760
aaagtgccgc aactgtgcaa ggtggctcca tctactcaaa agtatcacat ggaggacgtg 2820
catcgcgcgg gtggcgtgat tggcatcctg ggtgagctgg accgtgctgg tctgctgaat 2880
cgcgacgtta agaatgttct gggtctgacc ctgccacaga ccctggagca gtatgatgtg 2940
atgctgactc aagacgatgc tgttaagaac atgtttcgtg ctggtccggc gggtatccgc 3000
actacccaag cgtttagcca ggactgtcgc tgggacaccc tggatgatga ccgtgcgaac 3060
ggttgcattc gtagcctgga acatgcgtat tctaaggatg gtggtctggc tgttctgtat 3120
ggcaatttcg ctgagaatgg ttgtattgtt aagaccgcgg gtgttgacga ttctattctg 3180
aagtttactg gtccagctaa ggtttatgag tctcaagatg acgctgttga ggctatcctg 3240
ggtggcaagg tggttgcggg tgacgttgtt gttatccgtt acgagggtcc aaagggtggc 3300
ccaggtatgc aagagatgct gtatccgact tcttttctga agagcatggg cctgggtaag 3360
gcgtgcgctc tgattactga tggccgcttt agcggcggta ctagcggcct gagcattggt 3420
catgttagcc cagaggctgc gtctggtggt tctatcggtc tgatcgagga cggcgatctg 3480
attgcgattg atattccaaa tcgcggtatc caactgcaag tttctgacgc ggagctggct 3540
gctcgccgcg aggctcaaga tgcgcgtggc gataaggcgt ggaccccaaa gaaccgcgag 3600
cgccaagtta gcttcgcgct gcgcgcgtac gcctctctgg cgacttctgc ggataagggt 3660
gctgttcgtg acaagagcaa gctgggtggc taaacgcgtg ctagaggcat caaataaaac 3720
gaaaggctca gtcgaaagac tgggcctttc gttttatctg ttgtttgtcg gtgaacgctc 3780
tcctgagtag gacaaatccg ccgccctaga cctagctagg gtacgggttt tgctgcccgc 3840
aaacgggctg ttctggtgtt gctagtttgt tatcagaatc gcagatccgg cttcagccgg 3900
tttgccggct gaaagcgcta tttcttccag aattgccatg attttttccc cacgggaggc 3960
gtcactggct cccgtgttgt cggcagcttt gattcgataa gcagcatcgc ctgtttcagg 4020
ctgtctatgt gtgactgttg agctgtaaca agttgtctca ggtgttcaat ttcatgttct 4080
agttgctttg ttttactggt ttcacctgtt ctattaggtg ttacatgctg ttcatctgtt 4140
acattgtcga tctgttcatg gtgaacagct ttaaatgcac caaaaactcg taaaagctct 4200
gatgtatcta tcttttttac accgttttca tctgtgcata tggacagttt tccctttgat 4260
atctaacggt gaacagttgt tctacttttg tttgttagtc ttgatgcttc actgatagat 4320
acaagagcca taagaacctc agatccttcc gtatttagcc agtatgttct ctagtgtggt 4380
tcgttgtttt tgcgtgagcc atgagaacga accattgaga tcatgcttac tttgcatgtc 4440
actcaaaaat tttgcctcaa aactggtgag ctgaattttt gcagttaaag catcgtgtag 4500
tgtttttctt agtccgttac gtaggtagga atctgatgta atggttgttg gtattttgtc 4560
accattcatt tttatctggt tgttctcaag ttcggttacg agatccattt gtctatctag 4620
ttcaacttgg aaaatcaacg tatcagtcgg gcggcctcgc ttatcaacca ccaatttcat 4680
attgctgtaa gtgtttaaat ctttacttat tggtttcaaa acccattggt taagcctttt 4740
aaactcatgg tagttatttt caagcattaa catgaactta aattcatcaa ggctaatctc 4800
tatatttgcc ttgtgagttt tcttttgtgt tagttctttt aataaccact cataaatcct 4860
catagagtat ttgttttcaa aagacttaac atgttccaga ttatatttta tgaatttttt 4920
taactggaaa agataaggca atatctcttc actaaaaact aattctaatt tttcgcttga 4980
gaacttggca tagtttgtcc actggaaaat ctcaaagcct ttaaccaaag gattcctgat 5040
ttccacagtt ctcgtcatca gctctctggt tgctttagct aatacaccat aagcattttc 5100
cctactgatg ttcatcatct gagcgtattg gttataagtg aacgataccg tccgttcttt 5160
ccttgtaggg ttttcaatcg tggggttgag tagtgccaca cagcataaaa ttagcttggt 5220
ttcatgctcc gttaagtcat agcgactaat cgctagttca tttgctttga aaacaactaa 5280
ttcagacata catctcaatt ggtctaggtg attttaatca ctataccaat tgagatgggc 5340
tagtcaatga taattactag tccttttccc gggagatctg ggtatctgta aattctgcta 5400
gacctttgct ggaaaacttg taaattctgc tagaccctct gtaaattccg ctagaccttt 5460
gtgtgttttt tttgtttata ttcaagtggt tataatttat agaataaaga aagaataaaa 5520
aaagataaaa agaatagatc ccagccctgt gtataactca ctactttagt cagttccgca 5580
gtattacaaa aggatgtcgc aaacgctgtt tgctcctcta caaaacagac cttaaaaccc 5640
taaaggctta agtagcaccc tcgcaagctc gggcaaatcg ctgaatattc cttttgtctc 5700
cgaccatcag gcacctgagt cgctgtcttt ttcgtgacat tcagttcgct gcgctcacgg 5760
ctctggcagt gaatgggggt aaatggcact acaggcgcct tttatggatt catgcaagga 5820
aactacccat aatacaagaa aagcccgtca cgggcttctc agggcgtttt atggcgggtc 5880
tgctatgtgg tgctatctga ctttttgctg ttcagcagtt cctgccctct gattttccag 5940
tctgaccact tcggattatc ccgtgacagg tcattcagac tggctaatgc acccagtaag 6000
gcagcggtat catcaacagg cttacccgtc ttactgtccc tagtgcttgg attctcacca 6060
ataaaaaacg cccggcggca accgagcgtt ctgaacaaat ccagatggag ttctgaggtc 6120
attactggat ctatcaacag gagtccaagc gagctctcga accccagagt cccgctcaga 6180
agaactcgtc aagaaggcga tagaaggcga tgcgctgcga atcgggagcg gcgataccgt 6240
aaagcacgag gaagcggtca gcccattcgc cgccaagctc ttcagcaata tcacgggtag 6300
ccaacgctat gtcctgatag cggtccgcca cacccagccg gccacagtcg atgaatccag 6360
aaaagcggcc attttccacc atgatattcg gcaagcaggc atcgccatgg gtcacgacga 6420
gatcctcgcc gtcgggcatg cgcgccttga gcctggcgaa cagttcggct ggcgcgagcc 6480
cctgatgctc ttcgtccaga tcatcctgat cgacaagacc ggcttccatc cgagtacgtg 6540
ctcgctcgat gcgatgtttc gcttggtggt cgaatgggca ggtagccgga tcaagcgtat 6600
gcagccgccg cattgcatca gccatgatgg atactttctc ggcaggagca aggtgagatg 6660
acaggagatc ctgccccggc acttcgccca atagcagcca gtcccttccc gcttcagtga 6720
caacgtcgag cacagctgcg caaggaacgc ccgtcgtggc cagccacgat agccgcgctg 6780
cctcgtcctg cagttcattc agggcaccgg acaggtcggt cttgacaaaa agaaccgggc 6840
gcccctgcgc tgacagccgg aacacggcgg catcagagca gccgattgtc tgttgtgccc 6900
agtcatagcc gaatagcctc tccacccaag cggccggaga acctgcgtgc aatccatctt 6960
gttcaatcat gcgaaacgat cctcatcctg tctcttgatc agatcttgat cccctgcgcc 7020
atcagatcct tggcggcaag aaagccatcc agtttacttt gcagggcttc ccaaccttac 7080
cagagggcgc cccagctggc aattccgacg tc 7112
<210> SEQ ID NO 110
<211> LENGTH: 7884
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1661
<400> SEQUENCE: 110
ctcgagtccc tatcagtgat agagattgac atccctatca gtgatagaga tactgagcac 60
atcagcagga cgcactgacc gaattcatta aagaggaaca accaaatgga tgaccagtta 120
aaacaaagtg cacttgattt ccatgaattt ccagttccag ggaaaatcca ggtttctcca 180
accaagcctc tggcaacaca gcgcgatctg gcgctggcct actcaccagg cgttgccgca 240
ccttgtcttg aaatcgaaaa agacccgtta aaagcctaca aatataccgc ccgaggtaac 300
ctggtggcgg tgatctctaa cggtacggcg gtgctggggt taggcaacat tggcgcgctg 360
gcaggcaaac cggtgatgga aggcaagggc gttctgttta agaaattcgc cgggattgat 420
gtatttgaca ttgaagttga cgaactcgac ccggacaaat ttattgaagt tgtcgccgcg 480
ctcgaaccaa ccttcggcgg catcaacctc gaagacatta aagcgccaga atgtttctat 540
attgaacaga aactgcgcga gcggatgaat attccggtat tccacgacga tcagcacggc 600
acggcaatta tcagcactgc cgccatcctc aacggcttgc gcgtggtgga gaaaaacatc 660
tccgacgtgc ggatggtggt ttccggcgcg ggtgccgcag caatcgcctg tatgaacctg 720
ctggtagcgc tgggtctgca aaaacataac atcgtggttt gcgattcaaa aggcgttatc 780
tatcagggcc gtgagccaaa catggcggaa accaaagccg catatgcggt ggtggatgac 840
ggcaaacgta ccctcgatga tgtgattgaa ggcgcggata ttttcctggg ctgttccggc 900
ccgaaagtgc tgacccagga aatggtgaag aaaatggctc gtgcgccaat gatcctggcg 960
ctggcgaacc cggaaccgga aattctgccg ccgctggcga aagaagtgcg tccggatgcc 1020
atcatttgca ccggtcgttc tgactatccg aaccaggtga acaacgtcct gtgcttcccg 1080
ttcatcttcc gtggcgcgct ggacgttggc gcaaccgcca tcaacgaaga gatgaaactg 1140
gcggcggtac gtgcgattgc agaactcgcc catgcggaac agagcgaagt ggtggcttca 1200
gcgtatggcg atcaggatct gagctttggt ccggaataca tcattccaaa accgtttgat 1260
ccgcgcttga tcgttaagat cgctcctgcg gtcgctaaag ccgcgatgga gtcgggcgtg 1320
gcgactcgtc cgattgctga tttcgacgtc tacatcgaca agctgactga gttcgtttac 1380
aaaaccaacc tgtttatgaa gccgattttc tcccaggctc gcaaagcgcc gaagcgcgtt 1440
gttctgccgg aaggggaaga ggcgcgcgtt ctgcatgcca ctcaggaact ggtaacgctg 1500
ggactggcga aaccgatcct tatcggtcgt ccgaacgtga tcgaaatgcg cattcagaaa 1560
ctgggcttgc agatcaaagc gggcgttgat tttgagatcg tcaataacga atccgatccg 1620
cgctttaaag agtactggac cgaatacttc cagatcatga agcgtcgcgg cgtcactcag 1680
gaacaggcgc agcgggcgct gatcagtaac ccgacagtga tcggcgcgat catggttcag 1740
cgtggggaag ccgatgcaat gatttgcggt acggtgggtg attatcatga acattttagc 1800
gtggtgaaaa atgtctttgg ttatcgcgat ggcgttcaca ccgcaggtgc catgaacgcg 1860
ctgctgctgc cgagtggtaa cacctttatt gccgatacat atgttaatga tgaaccggat 1920
gcagaagagc tggcggagat caccttgatg gcggcagaaa ctgtccgtcg ttttggtatt 1980
gagccgcgcg ttgctttgtt gtcgcactcc aactttggtt cttctgactg cccgtcgtcg 2040
agcaaaatgc gtcaggcgct ggaactggtc agggaacgtg caccagaact gatgattgat 2100
ggtgaaatgc acggcgatgc agcgctggtg gaagcgattc gcaacgaccg tatgccggac 2160
agctctttga aaggttccgc caatattctg gtgatgccga acatggaagc tgcccgcatt 2220
agttacaact tactgcgtgt ttccagctcg gaaggtgtga ctgtcggccc ggtgctgatg 2280
ggtgtggcga aaccggttca cgtgttaacg ccgatcgcat cggtgcgtcg tatcgtcaac 2340
atggtggcgc tggccgtggt agaagcgcaa acccaaccgc tgtaaggtac cattaaagag 2400
gagaaacgta gcatgaacga acaatattcc gcattgcgta gtaatgtcag tatgctcggc 2460
aaagtgctgg gagaaaccat caaggatgcg ttgggagaac acattcttga acgcgtagaa 2520
actatccgta agttgtcgaa atcttcacgc gctggcaatg atgctaaccg ccaggagttg 2580
ctcaccacct tacaaaattt gtcgaacgac gagctgctgc ccgttgcgcg tgcgtttagt 2640
cagttcctga acctggccaa caccgccgag caataccaca gcatttcgcc gaaaggcgaa 2700
gctgccagca acccggaagt gatcgcccgc accctgcgta aactgaaaaa ccagccggaa 2760
ctgagcgaag acaccatcaa aaaagcagtg gaatcgctgt cgctggaact ggtcctcacg 2820
gctcacccaa ccgaaattac ccgtcgtaca ctgatccaca aaatggtgga agtgaacgcc 2880
tgtttaaaac agctcgataa caaagatatc gctgactacg aacacaacca gctgatgcgt 2940
cgcctgcgcc agttgatcgc ccagtcatgg cataccgatg aaatccgtaa gctgcgtcca 3000
agcccggtag atgaagccaa atggggcttt gccgtagtgg aaaacagcct gtggcaaggc 3060
gtaccaaatt acctgcgcga actgaacgaa caactggaag agaacctcgg ctacaaactg 3120
cccgtcgaat ttgttccggt ccgttttact tcgtggatgg gcggcgaccg cgacggcaac 3180
ccgaacgtca ctgccgatat cacccgccac gtcctgctac tcagccgctg gaaagccacc 3240
gatttgttcc tgaaagatat tcaggtgctg gtttctgaac tgtcgatggt tgaagcgacc 3300
cctgaactgc tggcgctggt tggcgaagaa ggtgccgcag aaccgtatcg ctatctgatg 3360
aaaaacctgc gttctcgcct gatggcgaca caggcatggc tggaagcgcg cctgaaaggc 3420
gaagaactgc caaaaccaga aggcctgctg acacaaaacg aagaactgtg ggaaccgctc 3480
tacgcttgct accagtcact tcaggcgtgt ggcatgggta ttatcgccaa cggcgatctg 3540
ctcgacaccc tgcgccgcgt gaaatgtttc ggcgtaccgc tggtccgtat tgatatccgt 3600
caggagagca cgcgtcatac cgaagcgctg ggcgagctga cccgctacct cggtatcggc 3660
gactacgaaa gctggtcaga ggccgacaaa caggcgttcc tgatccgcga actgaactcc 3720
aaacgtccgc ttctgccgcg caactggcaa ccaagcgccg aaacgcgcga agtgctcgat 3780
acctgccagg tgattgccga agcaccgcaa ggctccattg ccgcctacgt gatctcgatg 3840
gcgaaaacgc cgtccgacgt actggctgtc cacctgctgc tgaaagaagc gggtatcggg 3900
tttgcgatgc cggttgctcc gctgtttgaa accctcgatg atctgaacaa cgccaacgat 3960
gtcatgaccc agctgctcaa tattgactgg tatcgtggcc tgattcaggg caaacagatg 4020
gtgatgattg gctattccga ctcagcaaaa gatgcgggag tgatggcagc ttcctgggcg 4080
caatatcagg cacaggatgc attaatcaaa acctgcgaaa aagcgggtat tgagctgacg 4140
ttgttccacg gtcgcggcgg ttccattggt cgcggcggcg cacctgctca tgcggcgctg 4200
ctgtcacaac cgccaggaag cctgaaaggc ggcctgcgcg taaccgaaca gggcgagatg 4260
atccgcttta aatatggtct gccagaaatc accgtcagca gcctgtcgct ttataccggg 4320
gcgattctgg aagccaacct gctgccaccg ccggagccga aagagagctg gcgtcgcatt 4380
atggatgaac tgtcagtcat ctcctgcgat gtctaccgcg gctacgtacg tgaaaacaaa 4440
gattttgtgc cttacttccg ctccgctacg ccggaacaag aactgggcaa actgccgttg 4500
ggttcacgtc cggcgaaacg tcgcccaacc ggcggcgtcg agtcactacg cgccattccg 4560
tggatcttcg cctggacgca aaaccgtctg atgctccccg cctggctggg tgcaggtacg 4620
gcgctgcaaa aagtggtcga agacggcaaa cagagcgagc tggaggctat gtgccgcgat 4680
tggccattct tctcgacgcg tctcggcatg ctggagatgg tcttcgccaa agcagacctg 4740
tggctggcgg aatactatga ccaacgcctg gtagacaaag cactgtggcc gttaggtaaa 4800
gagttacgca acctgcaaga agaagacatc aaagtggtgc tggcgattgc caacgattcc 4860
catctgatgg ccgatctgcc gtggattgca gagtctattc agctacggaa tatttacacc 4920
gacccgctga acgtattgca ggccgagttg ctgcaccgct cccgccaggc agaaaaagaa 4980
ggccaggaac cggatcctcg cgtcgaacaa gcgttaatgg tcactattgc cgggattgcg 5040
gcaggtatgc gtaataccgg ctaagtcgac attaaagagg agattactta tgaaagttgc 5100
tgttctgggt gctgcaggtg gtattggtca ggcactggcc ctgctgctga aaactcagct 5160
gccgagcggt tctgaactgt ccctgtacga tattgcgcct gttactccgg gtgtcgctgt 5220
agacctgtct catatcccta cggcagtaaa aatcaaaggc tttagcggtg aagatgcaac 5280
tccggcgctg gaaggtgccg acgttgtact gatctctgcg ggcgtggctc gtaaaccggg 5340
catggaccgt tctgatctgt tcaacgtgaa cgctggcatt gttaaaaatc tggtgcagca 5400
ggttgcaaaa acctgtccga aagcgtgcat tggcatcatc actaacccag ttaacaccac 5460
cgtcgcgatc gcggcagaag tcctgaagaa agcaggcgtg tacgataaaa acaaactgtt 5520
cggtgttact accctggaca tcatccgttc taatactttc gtagctgagc tgaaaggcaa 5580
acagccgggt gaagttgaag ttccggttat cggtggccac agcggtgtta ccatcctgcc 5640
tctgctgagc caggttccgg gtgtgtcttt caccgaacaa gaagtagcgg acctgaccaa 5700
acgtatccaa aacgctggca ccgaagttgt tgaagccaaa gcaggtggtg gctctgctac 5760
cctgtctatg ggtcaagcgg cagcacgctt tggcctgtct ctggttcgcg ctctgcaggg 5820
tgaacaaggt gtggtagaat gtgcttacgt tgaaggcgat ggccagtatg cacgcttctt 5880
ctcccaacct ctgctgctgg gcaaaaacgg tgttgaggaa cgtaaatcta tcggcactct 5940
gtccgcgttc gaacaaaacg cgctggaagg catgctggat actctgaaga aagatatcgc 6000
tctgggtgag gaatttgtta acaaatgacc tagggatata ttccgcttcc tcgctcactg 6060
actcgctacg ctcggtcgtt cgactgcggc gagcggaaat ggcttacgaa cggggcggag 6120
atttcctgga agatgccagg aagatactta acagggaagt gagagggccg cggcaaagcc 6180
gtttttccat aggctccgcc cccctgacaa gcatcacgaa atctgacgct caaatcagtg 6240
gtggcgaaac ccgacaggac tataaagata ccaggcgttt ccccctggcg gctccctcgt 6300
gcgctctcct gttcctgcct ttcggtttac cggtgtcatt ccgctgttat ggccgcgttt 6360
gtctcattcc acgcctgaca ctcagttccg ggtaggcagt tcgctccaag ctggactgta 6420
tgcacgaacc ccccgttcag tccgaccgct gcgccttatc cggtaactat cgtcttgagt 6480
ccaacccgga aagacatgca aaagcaccac tggcagcagc cactggtaat tgatttagag 6540
gagttagtct tgaagtcatg cgccggttaa ggctaaactg aaaggacaag ttttggtgac 6600
tgcgctcctc caagccagtt acctcggttc aaagagttgg tagctcagag aaccttcgaa 6660
aaaccgccct gcaaggcggt tttttcgttt tcagagcaag agattacgcg cagaccaaaa 6720
cgatctcaag aagatcatct tattaatcag ataaaatatt tctagatttc agtgcaattt 6780
atctcttcaa atgtagcacc tgaagtcagc cccatacgat ataagttgtt actagtgctt 6840
ggattctcac caataaaaaa cgcccggcgg caaccgagcg ttctgaacaa atccagatgg 6900
agttctgagg tcattactgg atctatcaac aggagtccaa gcgagctcga tatcaaatta 6960
cgccccgccc tgccactcat cgcagtactg ttgtaattca ttaagcattc tgccgacatg 7020
gaagccatca cagacggcat gatgaacctg aatcgccagc ggcatcagca ccttgtcgcc 7080
ttgcgtataa tatttgccca tggtgaaaac gggggcgaag aagttgtcca tattggccac 7140
gtttaaatca aaactggtga aactcaccca gggattggct gagacgaaaa acatattctc 7200
aataaaccct ttagggaaat aggccaggtt ttcaccgtaa cacgccacat cttgcgaata 7260
tatgtgtaga aactgccgga aatcgtcgtg gtattcactc cagagcgatg aaaacgtttc 7320
agtttgctca tggaaaacgg tgtaacaagg gtgaacacta tcccatatca ccagctcacc 7380
gtctttcatt gccatacgaa actccggatg agcattcatc aggcgggcaa gaatgtgaat 7440
aaaggccgga taaaacttgt gcttattttt ctttacggtc tttaaaaagg ccgtaatatc 7500
cagctgaacg gtctggttat aggtacattg agcaactgac tgaaatgcct caaaatgttc 7560
tttacgatgc cattgggata tatcaacggt ggtatatcca gtgatttttt tctccatttt 7620
agcttcctta gctcctgaaa atctcgataa ctcaaaaaat acgcccggta gtgatcttat 7680
ttcattatgg tgaaagttgg aacctcttac gtgccgatca acgtctcatt ttcgccagat 7740
atcgacgtct aagaaaccat tattatcatg acattaacct ataaaaatag gcgtatcacg 7800
aggccctttc gtcttcacga aaccattatt atcatgacat taacctataa aaataggcgt 7860
atcacgaggc cctttcgtct tcac 7884
<210> SEQ ID NO 111
<211> LENGTH: 4895
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1685
<400> SEQUENCE: 111
ctcgagtccc tatcagtgat agagattgac atccctatca gtgatagaga tactgagcac 60
atcagcagga cgcactgacc gaattcatta aagaggagaa aggtaccatg cgaattggca 120
taccaagaga acggttaacc aatgaaaccc gtgttgcagc aacgccaaaa acagtggaac 180
agctgctgaa actgggtttt accgtcgcgg tagagagcgg cgcgggtcaa ctggcaagtt 240
ttgacgataa agcgtttgtg caagcgggcg ctgaaattgt agaagggaat agcgtctggc 300
agtcagagat cattctgaag gtcaatgcgc cgttagatga tgaaattgcg ttactgaatc 360
ctgggacaac gctggtgagt tttatctggc ctgcgcagaa tccggaatta atgcaaaaac 420
ttgcggaacg taacgtgacc gtgatggcga tggactctgt gccgcgtatc tcacgcgcac 480
aatcgctgga cgcactaagc tcgatggcga acatcgccgg ttatcgcgcc attgttgaag 540
cggcacatga atttgggcgc ttctttaccg ggcaaattac tgcggccggg aaagtgccac 600
cggcaaaagt gatggtgatt ggtgcgggtg ttgcaggtct ggccgccatt ggcgcagcaa 660
acagtctcgg cgcgattgtg cgtgcattcg acacccgccc ggaagtgaaa gaacaagttc 720
aaagtatggg cgcggaattc ctcgagctgg attttaaaga ggaagctggc agcggcgatg 780
gctatgccaa agtgatgtcg gacgcgttca tcaaagcgga aatggaactc tttgccgccc 840
aggcaaaaga ggtcgatatc attgtcacca ccgcgcttat tccaggcaaa ccagcgccga 900
agctaattac ccgtgaaatg gttgactcca tgaaggcggg cagtgtgatt gtcgacctgg 960
cagcccaaaa cggcggcaac tgtgaataca ccgtgccggg tgaaatcttc actacggaaa 1020
atggtgtcaa agtgattggt tataccgatc ttccgggccg tctgccgacg caatcctcac 1080
agctttacgg cacaaacctc gttaatctgc tgaaactgtt gtgcaaagag aaagacggca 1140
atatcactgt tgattttgat gatgtggtga ttcgcggcgt gaccgtgatc cgtgcgggcg 1200
aaattacctg gccggcaccg ccgattcagg tatcagctca gccgcaggcg gcacaaaaag 1260
cggcaccgga agtgaaaact gaggaaaaat gtacctgctc accgtggcgt aaatacgcgt 1320
tgatggcgct ggcaatcatt ctttttggct ggatggcaag cgttgcgccg aaagaattcc 1380
ttgggcactt caccgttttc gcgctggcct gcgttgtcgg ttattacgtg gtgtggaatg 1440
tatcgcacgc gctgcataca ccgttgatgt cggtcaccaa cgcgatttca gggattattg 1500
ttgtcggagc actgttgcag attggccagg gcggctgggt tagcttcctt agttttatcg 1560
cggtgcttat agccagcatt aatattttcg gtggcttcac cgtgactcag cgcatgctga 1620
aaatgttccg caaaaattaa ggggtaacat atgtctggag gattagttac agctgcatac 1680
attgttgccg cgatcctgtt tatcttcagt ctggccggtc tttcgaaaca tgaaacgtct 1740
cgccagggta acaacttcgg tatcgccggg atggcgattg cgttaatcgc aaccattttt 1800
ggaccggata cgggtaatgt tggctggatc ttgctggcga tggtcattgg tggggcaatt 1860
ggtatccgtc tggcgaagaa agttgaaatg accgaaatgc cagaactggt ggcgatcctg 1920
catagcttcg tgggtctggc ggcagtgctg gttggcttta acagctatct gcatcatgac 1980
gcgggaatgg caccgattct ggtcaatatt cacctgacgg aagtgttcct cggtatcttc 2040
atcggggcgg taacgttcac gggttcggtg gtggcgttcg gcaaactgtg tggcaagatt 2100
tcgtctaaac cattgatgct gccaaaccgt cacaaaatga acctggcggc tctggtcgtt 2160
tccttcctgc tgctgattgt atttgttcgc acggacagcg tcggcctgca agtgctggca 2220
ttgctgataa tgaccgcaat tgcgctggta ttcggctggc atttagtcgc ctccatcggt 2280
ggtgcagata tgccagtggt ggtgtcgatg ctgaactcgt actccggctg ggcggctgcg 2340
gctgcgggct ttatgctcag caacgacctg ctgattgtga ccggtgcgct ggtcggttct 2400
tcgggggcta tcctttctta cattatgtgt aaggcgatga accgttcctt tatcagcgtt 2460
attgcgggtg gtttcggcac cgacggctct tctactggcg atgatcagga agtgggtgag 2520
caccgcgaaa tcaccgcaga agagacagcg gaactgctga aaaactccca ttcagtgatc 2580
attactccgg ggtacggcat ggcagtcgcg caggcgcaat atcctgtcgc tgaaattact 2640
gagaaattgc gcgctcgtgg tattaatgtg cgtttcggta tccacccggt cgcggggcgt 2700
ttgcctggac atatgaacgt attgctggct gaagcaaaag taccgtatga catcgtgctg 2760
gaaatggacg agatcaatga tgactttgct gataccgata ccgtactggt gattggtgct 2820
aacgatacgg ttaacccggc ggcgcaggat gatccgaaga gtccgattgc tggtatgcct 2880
gtgctggaag tgtggaaagc gcagaacgtg attgtcttta aacgttcgat gaacactggc 2940
tatgctggtg tgcaaaaccc gctgttcttc aaggaaaaca cccacatgct gtttggtgac 3000
gccaaagcca gcgtggatgc aatcctgaaa gctctgtaac ctagggatat attccgcttc 3060
ctcgctcact gactcgctac gctcggtcgt tcgactgcgg cgagcggaaa tggcttacga 3120
acggggcgga gatttcctgg aagatgccag gaagatactt aacagggaag tgagagggcc 3180
gcggcaaagc cgtttttcca taggctccgc ccccctgaca agcatcacga aatctgacgc 3240
tcaaatcagt ggtggcgaaa cccgacagga ctataaagat accaggcgtt tccccctggc 3300
ggctccctcg tgcgctctcc tgttcctgcc tttcggttta ccggtgtcat tccgctgtta 3360
tggccgcgtt tgtctcattc cacgcctgac actcagttcc gggtaggcag ttcgctccaa 3420
gctggactgt atgcacgaac cccccgttca gtccgaccgc tgcgccttat ccggtaacta 3480
tcgtcttgag tccaacccgg aaagacatgc aaaagcacca ctggcagcag ccactggtaa 3540
ttgatttaga ggagttagtc ttgaagtcat gcgccggtta aggctaaact gaaaggacaa 3600
gttttggtga ctgcgctcct ccaagccagt tacctcggtt caaagagttg gtagctcaga 3660
gaaccttcga aaaaccgccc tgcaaggcgg ttttttcgtt ttcagagcaa gagattacgc 3720
gcagaccaaa acgatctcaa gaagatcatc ttattaatca gataaaatat ttctagattt 3780
cagtgcaatt tatctcttca aatgtagcac ctgaagtcag ccccatacga tataagttgt 3840
tactagtgct tggattctca ccaataaaaa acgcccggcg gcaaccgagc gttctgaaca 3900
aatccagatg gagttctgag gtcattactg gatctatcaa caggagtcca agcgagctcg 3960
atatcaaatt acgccccgcc ctgccactca tcgcagtact gttgtaattc attaagcatt 4020
ctgccgacat ggaagccatc acagacggca tgatgaacct gaatcgccag cggcatcagc 4080
accttgtcgc cttgcgtata atatttgccc atggtgaaaa cgggggcgaa gaagttgtcc 4140
atattggcca cgtttaaatc aaaactggtg aaactcaccc agggattggc tgagacgaaa 4200
aacatattct caataaaccc tttagggaaa taggccaggt tttcaccgta acacgccaca 4260
tcttgcgaat atatgtgtag aaactgccgg aaatcgtcgt ggtattcact ccagagcgat 4320
gaaaacgttt cagtttgctc atggaaaacg gtgtaacaag ggtgaacact atcccatatc 4380
accagctcac cgtctttcat tgccatacga aactccggat gagcattcat caggcgggca 4440
agaatgtgaa taaaggccgg ataaaacttg tgcttatttt tctttacggt ctttaaaaag 4500
gccgtaatat ccagctgaac ggtctggtta taggtacatt gagcaactga ctgaaatgcc 4560
tcaaaatgtt ctttacgatg ccattgggat atatcaacgg tggtatatcc agtgattttt 4620
ttctccattt tagcttcctt agctcctgaa aatctcgata actcaaaaaa tacgcccggt 4680
agtgatctta tttcattatg gtgaaagttg gaacctctta cgtgccgatc aacgtctcat 4740
tttcgccaga tatcgacgtc taagaaacca ttattatcat gacattaacc tataaaaata 4800
ggcgtatcac gaggcccttt cgtcttcacg aaaccattat tatcatgaca ttaacctata 4860
aaaataggcg tatcacgagg ccctttcgtc ttcac 4895
<210> SEQ ID NO 112
<211> LENGTH: 5336
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1698
<400> SEQUENCE: 112
taagaaacca ttattatcat gacattaacc tataaaaata ggcgtatcac gaggcccttt 60
cgtcttcacc tcgagaattg tgagcggata acaattgaca ttgtgagcgg ataacaagat 120
actgagcaca tcagcaggac gcactgaccg aattcattaa agaggagaaa ggtacaatgt 180
tgacaaaagc aacaaaagaa caaaaatccc ttgtgaaaaa cagaggggcg gagcttgttg 240
ttgattgctt agtggagcaa ggtgtcacac atgtatttgg cattccaggt gcaaaaattg 300
atgcggtatt tgacgcttta caagataaag gacctgaaat tatcgttgcc cggcacgaac 360
aaaacgcagc attcatggcc caagcagtcg gccgtttaac tggaaaaccg ggagtcgtgt 420
tagtcacatc aggaccgggt gcctctaact tggcaacagg cctgctgaca gcgaacactg 480
aaggagaccc tgtcgttgcg cttgctggaa acgtgatccg tgcagatcgt ttaaaacgga 540
cacatcaatc tttggataat gcggcgctat tccagccgat tacaaaatac agtgtagaag 600
ttcaagatgt aaaaaatata ccggaagctg ttacaaatgc atttaggata gcgtcagcag 660
ggcaggctgg ggccgctttt gtgagctttc cgcaagatgt tgtgaatgaa gtcacaaata 720
cgaaaaacgt gcgtgctgtt gcagcgccaa aactcggtcc tgcagcagat gatgcaatca 780
gtgcggccat agcaaaaatc caaacagcaa aacttcctgt cgttttggtc ggcatgaaag 840
gcggaagacc ggaagcaatt aaagcggttc gcaagctttt gaaaaaggtt cagcttccat 900
ttgttgaaac atatcaagct gccggtaccc tttctagaga tttagaggat caatattttg 960
gccgtatcgg tttgttccgc aaccagcctg gcgatttact gctagagcag gcagatgttg 1020
ttctgacgat cggctatgac ccgattgaat atgatccgaa attctggaat atcaatggag 1080
accggacaat tatccattta gacgagatta tcgctgacat tgatcatgct taccagcctg 1140
atcttgaatt gatcggtgac attccgtcca cgatcaatca tatcgaacac gatgctgtga 1200
aagtggaatt tgcagagcgt gagcagaaaa tcctttctga tttaaaacaa tatatgcatg 1260
aaggtgagca ggtgcctgca gattggaaat cagacagagc gcaccctctt gaaatcgtta 1320
aagagttgcg taatgcagtc gatgatcatg ttacagtaac ttgcgatatc ggttcgcacg 1380
ccatttggat gtcacgttat ttccgcagct acgagccgtt aacattaatg atcagtaacg 1440
gtatgcaaac actcggcgtt gcgcttcctt gggcaatcgg cgcttcattg gtgaaaccgg 1500
gagaaaaagt ggtttctgtc tctggtgacg gcggtttctt attctcagca atggaattag 1560
agacagcagt tcgactaaaa gcaccaattg tacacattgt atggaacgac agcacatatg 1620
acatggttgc attccagcaa ttgaaaaaat ataaccgtac atctgcggtc gatttcggaa 1680
atatcgatat cgtgaaatat gcggaaagct tcggagcaac tggcttgcgc gtagaatcac 1740
cagaccagct ggcagatgtt ctgcgtcaag gcatgaacgc tgaaggtcct gtcatcatcg 1800
atgtcccggt tgactacagt gataacatta atttagcaag tgacaagctt ccgaaagaat 1860
tcggggaact catgaaaacg aaagctctct aggtcgacga ggagacaaca ttatggcgaa 1920
ttatttcaac actctgaacc tgcgtcaaca actggcgcaa ctgggtaagt gccgtttcat 1980
gggtcgtgac gagtttgcgg acggtgcttc ttatctgcaa ggcaagaagg ttgttattgt 2040
tggttgcggt gcgcaaggcc tgaatcaagg tctgaatatg cgcgacagcg gcctggacat 2100
tagctatgcg ctgcgcaagg aggctatcgc ggaaaaacgt gctagctggc gcaaggctac 2160
tgagaacggc ttcaaggttg gcacctatga ggagctgatt ccgcaagctg acctggttat 2220
caatctgacc ccagataaac aacatagcga cgttgttcgt actgttcaac cgctgatgaa 2280
ggatggtgct gctctgggtt atagccacgg ctttaacatt gttgaggtag gtgaacaaat 2340
tcgcaaggac attactgttg ttatggtggc tccaaagtgt ccgggtactg aggttcgcga 2400
ggaatataag cgcggttttg gtgttccaac cctgatcgcg gtgcatccag agaatgaccc 2460
aaagggtgag ggtatggcta tcgcgaaggc gtgggctgcg gcgactggcg gccatcgcgc 2520
tggcgttctg gagagcagct ttgtggctga ggttaagagc gatctgatgg gtgaacagac 2580
tattctgtgt ggtatgctgc aagcgggtag cctgctgtgt tttgataaac tggttgagga 2640
gggcactgac ccggcgtatg cggagaagct gatccaattt ggctgggaga ctattactga 2700
ggcgctgaag caaggtggta ttactctgat gatggatcgc ctgagcaatc cagctaagct 2760
gcgcgcgtac gctctgagcg agcaactgaa ggaaattatg gcaccgctgt ttcaaaagca 2820
catggatgat atcattagcg gtgagtttag cagcggcatg atggctgatt gggcgaatga 2880
cgacaaaaag ctgctgactt ggcgcgagga aactggtaag actgctttcg agactgctcc 2940
acaatacgag ggtaagattg gtgaacaaga atattttgac aagggtgttc tgatgatcgc 3000
tatggttaag gctggtgtgg agctggcttt tgagactatg gttgacagcg gtattatcga 3060
ggaaagcgcg tactacgaga gcctgcatga actgccactg atcgcgaata ctattgcgcg 3120
caaacgcctg tatgagatga atgttgtgat tagcgacact gcggaatatg gcaattacct 3180
gtttagctat gcgtgcgttc cactgctgaa gccattcatg gcggaactgc agccaggtga 3240
tctgggcaag gcgatcccag agggtgctgt tgacaatggt cagctgcgcg acgttaatga 3300
ggctatccgt tctcacgcta tcgaacaagt tggcaaaaag ctgcgtggtt acatgaccga 3360
catgaagcgc atcgcggtgg ctggctaacc tagggcgttc ggctgcggcg agcggtatca 3420
gctcactcaa aggcggtaat acggttatcc acagaatcag gggataacgc aggaaagaac 3480
atgtgagcaa aaggccagca aaaggccagg aaccgtaaaa aggccgcgtt gctggcgttt 3540
ttccataggc tccgcccccc tgacgagcat cacaaaaatc gacgctcaag tcagaggtgg 3600
cgaaacccga caggactata aagataccag gcgtttcccc ctggaagctc cctcgtgcgc 3660
tctcctgttc cgaccctgcc gcttaccgga tacctgtccg cctttctccc ttcgggaagc 3720
gtggcgcttt ctcatagctc acgctgtagg tatctcagtt cggtgtaggt cgttcgctcc 3780
aagctgggct gtgtgcacga accccccgtt cagcccgacc gctgcgcctt atccggtaac 3840
tatcgtcttg agtccaaccc ggtaagacac gacttatcgc cactggcagc agccactggt 3900
aacaggatta gcagagcgag gtatgtaggc ggtgctacag agttcttgaa gtggtggcct 3960
aactacggct acactagaag gacagtattt ggtatctgcg ctctgctgaa gccagttacc 4020
ttcggaaaaa gagttggtag ctcttgatcc ggcaaacaaa ccaccgctgg tagcggtggt 4080
ttttttgttt gcaagcagca gattacgcgc agaaaaaaag gatctcaaga agatcctttg 4140
atcttttcta cggggtctga cgctcagtgg aacgaaaact cacgttaagg gattttggtc 4200
atgactagtg cttggattct caccaataaa aaacgcccgg cggcaaccga gcgttctgaa 4260
caaatccaga tggagttctg aggtcattac tggatctatc aacaggagtc caagcgagct 4320
cgtaaacttg gtctgacagt taccaatgct taatcagtga ggcacctatc tcagcgatct 4380
gtctatttcg ttcatccata gttgcctgac tccccgtcgt gtagataact acgatacggg 4440
agggcttacc atctggcccc agtgctgcaa tgataccgcg agacccacgc tcaccggctc 4500
cagatttatc agcaataaac cagccagccg gaagggccga gcgcagaagt ggtcctgcaa 4560
ctttatccgc ctccatccag tctattaatt gttgccggga agctagagta agtagttcgc 4620
cagttaatag tttgcgcaac gttgttgcca ttgctacagg catcgtggtg tcacgctcgt 4680
cgtttggtat ggcttcattc agctccggtt cccaacgatc aaggcgagtt acatgatccc 4740
ccatgttgtg caaaaaagcg gttagctcct tcggtcctcc gatcgttgtc agaagtaagt 4800
tggccgcagt gttatcactc atggttatgg cagcactgca taattctctt actgtcatgc 4860
catccgtaag atgcttttct gtgactggtg agtactcaac caagtcattc tgagaatagt 4920
gtatgcggcg accgagttgc tcttgcccgg cgtcaatacg ggataatacc gcgccacata 4980
gcagaacttt aaaagtgctc atcattggaa aacgttcttc ggggcgaaaa ctctcaagga 5040
tcttaccgct gttgagatcc agttcgatgt aacccactcg tgcacccaac tgatcttcag 5100
catcttttac tttcaccagc gtttctgggt gagcaaaaac aggaaggcaa aatgccgcaa 5160
aaaagggaat aagggcgaca cggaaatgtt gaatactcat actcttcctt tttcaatatt 5220
attgaagcat ttatcagggt tattgtctca tgagcggata catatttgaa tgtatttaga 5280
aaaataaaca aataggggtt ccgcgcacat ttccccgaaa agtgccacct gacgtc 5336
<210> SEQ ID NO 113
<211> LENGTH: 2289
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1711
<400> SEQUENCE: 113
ctagtgcttg gattctcacc aataaaaaac gcccggcggc aaccgagcgt tctgaacaaa 60
tccagatgga gttctgaggt cattactgga tctatcaaca ggagtccaag cgagctcgta 120
aacttggtct gacagttacc aatgcttaat cagtgaggca cctatctcag cgatctgtct 180
atttcgttca tccatagttg cctgactccc cgtcgtgtag ataactacga tacgggaggg 240
cttaccatct ggccccagtg ctgcaatgat accgcgagac ccacgctcac cggctccaga 300
tttatcagca ataaaccagc cagccggaag ggccgagcgc agaagtggtc ctgcaacttt 360
atccgcctcc atccagtcta ttaattgttg ccgggaagct agagtaagta gttcgccagt 420
taatagtttg cgcaacgttg ttgccattgc tacaggcatc gtggtgtcac gctcgtcgtt 480
tggtatggct tcattcagct ccggttccca acgatcaagg cgagttacat gatcccccat 540
gttgtgcaaa aaagcggtta gctccttcgg tcctccgatc gttgtcagaa gtaagttggc 600
cgcagtgtta tcactcatgg ttatggcagc actgcataat tctcttactg tcatgccatc 660
cgtaagatgc ttttctgtga ctggtgagta ctcaaccaag tcattctgag aatagtgtat 720
gcggcgaccg agttgctctt gcccggcgtc aatacgggat aataccgcgc cacatagcag 780
aactttaaaa gtgctcatca ttggaaaacg ttcttcgggg cgaaaactct caaggatctt 840
accgctgttg agatccagtt cgatgtaacc cactcgtgca cccaactgat cttcagcatc 900
ttttactttc accagcgttt ctgggtgagc aaaaacagga aggcaaaatg ccgcaaaaaa 960
gggaataagg gcgacacgga aatgttgaat actcatactc ttcctttttc aatattattg 1020
aagcatttat cagggttatt gtctcatgag cggatacata tttgaatgta tttagaaaaa 1080
taaacaaata ggggttccgc gcacatttcc ccgaaaagtg ccacctgacg tctaagaaac 1140
cattattatc atgacattaa cctataaaaa taggcgtatc acgaggccct ttcgtcttca 1200
cctcgagaat tgtgagcgga taacaattga cattgtgagc ggataacaag atactgagca 1260
catcagcagg acgcactgac cgaattcatt agtcgacatt atgcggccgc ggatccataa 1320
ggaggattaa ttaagacttc ccgggtgatc ccatggtacg cgtgctagag gcatcaaata 1380
aaacgaaagg ctcagtcgaa agactgggcc tttcgtttta tctgttgttt gtcggtgaac 1440
gctctcctga gtaggacaaa tccgccgccc tagacctagg cgttcggctg cggcgagcgg 1500
tatcagctca ctcaaaggcg gtaatacggt tatccacaga atcaggggat aacgcaggaa 1560
agaacatgtg agcaaaaggc cagcaaaagg ccaggaaccg taaaaaggcc gcgttgctgg 1620
cgtttttcca taggctccgc ccccctgacg agcatcacaa aaatcgacgc tcaagtcaga 1680
ggtggcgaaa cccgacagga ctataaagat accaggcgtt tccccctgga agctccctcg 1740
tgcgctctcc tgttccgacc ctgccgctta ccggatacct gtccgccttt ctcccttcgg 1800
gaagcgtggc gctttctcaa tgctcacgct gtaggtatct cagttcggtg taggtcgttc 1860
gctccaagct gggctgtgtg cacgaacccc ccgttcagcc cgaccgctgc gccttatccg 1920
gtaactatcg tcttgagtcc aacccggtaa gacacgactt atcgccactg gcagcagcca 1980
ctggtaacag gattagcaga gcgaggtatg taggcggtgc tacagagttc ttgaagtggt 2040
ggcctaacta cggctacact agaaggacag tatttggtat ctgcgctctg ctgaagccag 2100
ttaccttcgg aaaaagagtt ggtagctctt gatccggcaa acaaaccacc gctggtagcg 2160
gtggtttttt tgtttgcaag cagcagatta cgcgcagaaa aaaaggatct caagaagatc 2220
ctttgatctt ttctacgggg tctgacgctc agtggaacga aaactcacgt taagggattt 2280
tggtcatga 2289
<210> SEQ ID NO 114
<211> LENGTH: 6416
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1716
<400> SEQUENCE: 114
taagaaacca ttattatcat gacattaacc tataaaaata ggcgtatcac gaggcccttt 60
cgtcttcacc tcgagaattg tgagcggata acaattgaca ttgtgagcgg ataacaagat 120
actgagcaca tcagcaggac gcactgaccg aattcattaa agaggagaaa ggtacaatgt 180
tgacaaaagc aacaaaagaa caaaaatccc ttgtgaaaaa cagaggggcg gagcttgttg 240
ttgattgctt agtggagcaa ggtgtcacac atgtatttgg cattccaggt gcaaaaattg 300
atgcggtatt tgacgcttta caagataaag gacctgaaat tatcgttgcc cggcacgaac 360
aaaacgcagc attcatggcc caagcagtcg gccgtttaac tggaaaaccg ggagtcgtgt 420
tagtcacatc aggaccgggt gcctctaact tggcaacagg cctgctgaca gcgaacactg 480
aaggagaccc tgtcgttgcg cttgctggaa acgtgatccg tgcagatcgt ttaaaacgga 540
cacatcaatc tttggataat gcggcgctat tccagccgat tacaaaatac agtgtagaag 600
ttcaagatgt aaaaaatata ccggaagctg ttacaaatgc atttaggata gcgtcagcag 660
ggcaggctgg ggccgctttt gtgagctttc cgcaagatgt tgtgaatgaa gtcacaaata 720
cgaaaaacgt gcgtgctgtt gcagcgccaa aactcggtcc tgcagcagat gatgcaatca 780
gtgcggccat agcaaaaatc caaacagcaa aacttcctgt cgttttggtc ggcatgaaag 840
gcggaagacc ggaagcaatt aaagcggttc gcaagctttt gaaaaaggtt cagcttccat 900
ttgttgaaac atatcaagct gccggtaccc tttctagaga tttagaggat caatattttg 960
gccgtatcgg tttgttccgc aaccagcctg gcgatttact gctagagcag gcagatgttg 1020
ttctgacgat cggctatgac ccgattgaat atgatccgaa attctggaat atcaatggag 1080
accggacaat tatccattta gacgagatta tcgctgacat tgatcatgct taccagcctg 1140
atcttgaatt gatcggtgac attccgtcca cgatcaatca tatcgaacac gatgctgtga 1200
aagtggaatt tgcagagcgt gagcagaaaa tcctttctga tttaaaacaa tatatgcatg 1260
aaggtgagca ggtgcctgca gattggaaat cagacagagc gcaccctctt gaaatcgtta 1320
aagagttgcg taatgcagtc gatgatcatg ttacagtaac ttgcgatatc ggttcgcacg 1380
ccatttggat gtcacgttat ttccgcagct acgagccgtt aacattaatg atcagtaacg 1440
gtatgcaaac actcggcgtt gcgcttcctt gggcaatcgg cgcttcattg gtgaaaccgg 1500
gagaaaaagt ggtttctgtc tctggtgacg gcggtttctt attctcagca atggaattag 1560
agacagcagt tcgactaaaa gcaccaattg tacacattgt atggaacgac agcacatatg 1620
acatggttgc attccagcaa ttgaaaaaat ataaccgtac atctgcggtc gatttcggaa 1680
atatcgatat cgtgaaatat gcggaaagct tcggagcaac tggcttgcgc gtagaatcac 1740
cagaccagct ggcagatgtt ctgcgtcaag gcatgaacgc tgaaggtcct gtcatcatcg 1800
atgtcccggt tgactacagt gataacatta atttagcaag tgacaagctt ccgaaagaat 1860
tcggggaact catgaaaacg aaagctctct aggtcgacgg atccaggaga caacattatg 1920
tctattccag aaactcaaaa agcgattatt ttctacgagt ccaacggcaa actggaacac 1980
aaagatatcc cggtgccgaa accgaagccg aacgagctgc tgattaacgt aaaatactct 2040
ggtgtgtgcc acactgatct gcacgcttgg cacggtgatt ggcctctgcc gaccaaactg 2100
ccgctggttg gtggtcatga gggtgcgggc gttgtagtag gcatgggtga aaacgtgaag 2160
ggctggaaaa tcggtgacta cgcaggtatc aagtggctga acggttcttg catggcctgc 2220
gaatactgcg agctgggtaa cgaatctaac tgcccgcacg cagacctgtc tggctatacc 2280
catgatggtt cctttcagga atacgctact gcagacgcag tgcaggctgc acatattcca 2340
cagggcaccg atctggcgga ggtagctcct attctgtgcg ctggtattac ggtttacaag 2400
gcgctgaaaa gcgccaacct gcgtgccggc cactgggcag cgatctctgg tgcggcaggc 2460
ggtctgggtt ctctggcagt ccaatatgca aaagcgatgg gttaccgcgt tctgggcatc 2520
gacggtggtc cgggtaagga ggaactgttc acttctctgg gcggcgaggt gtttatcgac 2580
ttcactaagg agaaagatat cgtttccgcg gttgttaaag cgaccaacgg tggcgcgcac 2640
ggcattatca acgtatctgt gtccgaggct gcaatcgagg cgtctactcg ttactgccgt 2700
gctaacggca ctgtggtcct ggtaggtctg ccggctggtg ctaaatgttc tagcgatgtt 2760
ttcaaccacg tagtaaaaag catcagcatc gtgggttcct acgttggcaa ccgtgcagac 2820
actcgtgagg ctctggactt cttcgcacgc ggcctggtga aatctccgat taaggttgtt 2880
ggtctgtcta gcctgccgga aatctatgag aaaatggaaa aaggtcagat tgcgggccgt 2940
tacgtggtgg acacctctaa ataagcggcc gcgtcgacga ggagacaaca ttatggcgaa 3000
ttatttcaac actctgaacc tgcgtcaaca actggcgcaa ctgggtaagt gccgtttcat 3060
gggtcgtgac gagtttgcgg acggtgcttc ttatctgcaa ggcaagaagg ttgttattgt 3120
tggttgcggt gcgcaaggcc tgaatcaagg tctgaatatg cgcgacagcg gcctggacat 3180
tagctatgcg ctgcgcaagg aggctatcgc ggaaaaacgt gctagctggc gcaaggctac 3240
tgagaacggc ttcaaggttg gcacctatga ggagctgatt ccgcaagctg acctggttat 3300
caatctgacc ccagataaac aacatagcga cgttgttcgt actgttcaac cgctgatgaa 3360
ggatggtgct gctctgggtt atagccacgg ctttaacatt gttgaggtag gtgaacaaat 3420
tcgcaaggac attactgttg ttatggtggc tccaaagtgt ccgggtactg aggttcgcga 3480
ggaatataag cgcggttttg gtgttccaac cctgatcgcg gtgcatccag agaatgaccc 3540
aaagggtgag ggtatggcta tcgcgaaggc gtgggctgcg gcgactggcg gccatcgcgc 3600
tggcgttctg gagagcagct ttgtggctga ggttaagagc gatctgatgg gtgaacagac 3660
tattctgtgt ggtatgctgc aagcgggtag cctgctgtgt tttgataaac tggttgagga 3720
gggcactgac ccggcgtatg cggagaagct gatccaattt ggctgggaga ctattactga 3780
ggcgctgaag caaggtggta ttactctgat gatggatcgc ctgagcaatc cagctaagct 3840
gcgcgcgtac gctctgagcg agcaactgaa ggaaattatg gcaccgctgt ttcaaaagca 3900
catggatgat atcattagcg gtgagtttag cagcggcatg atggctgatt gggcgaatga 3960
cgacaaaaag ctgctgactt ggcgcgagga aactggtaag actgctttcg agactgctcc 4020
acaatacgag ggtaagattg gtgaacaaga atattttgac aagggtgttc tgatgatcgc 4080
tatggttaag gctggtgtgg agctggcttt tgagactatg gttgacagcg gtattatcga 4140
ggaaagcgcg tactacgaga gcctgcatga actgccactg atcgcgaata ctattgcgcg 4200
caaacgcctg tatgagatga atgttgtgat tagcgacact gcggaatatg gcaattacct 4260
gtttagctat gcgtgcgttc cactgctgaa gccattcatg gcggaactgc agccaggtga 4320
tctgggcaag gcgatcccag agggtgctgt tgacaatggt cagctgcgcg acgttaatga 4380
ggctatccgt tctcacgcta tcgaacaagt tggcaaaaag ctgcgtggtt acatgaccga 4440
catgaagcgc atcgcggtgg ctggctaacc tagggcgttc ggctgcggcg agcggtatca 4500
gctcactcaa aggcggtaat acggttatcc acagaatcag gggataacgc aggaaagaac 4560
atgtgagcaa aaggccagca aaaggccagg aaccgtaaaa aggccgcgtt gctggcgttt 4620
ttccataggc tccgcccccc tgacgagcat cacaaaaatc gacgctcaag tcagaggtgg 4680
cgaaacccga caggactata aagataccag gcgtttcccc ctggaagctc cctcgtgcgc 4740
tctcctgttc cgaccctgcc gcttaccgga tacctgtccg cctttctccc ttcgggaagc 4800
gtggcgcttt ctcatagctc acgctgtagg tatctcagtt cggtgtaggt cgttcgctcc 4860
aagctgggct gtgtgcacga accccccgtt cagcccgacc gctgcgcctt atccggtaac 4920
tatcgtcttg agtccaaccc ggtaagacac gacttatcgc cactggcagc agccactggt 4980
aacaggatta gcagagcgag gtatgtaggc ggtgctacag agttcttgaa gtggtggcct 5040
aactacggct acactagaag gacagtattt ggtatctgcg ctctgctgaa gccagttacc 5100
ttcggaaaaa gagttggtag ctcttgatcc ggcaaacaaa ccaccgctgg tagcggtggt 5160
ttttttgttt gcaagcagca gattacgcgc agaaaaaaag gatctcaaga agatcctttg 5220
atcttttcta cggggtctga cgctcagtgg aacgaaaact cacgttaagg gattttggtc 5280
atgactagtg cttggattct caccaataaa aaacgcccgg cggcaaccga gcgttctgaa 5340
caaatccaga tggagttctg aggtcattac tggatctatc aacaggagtc caagcgagct 5400
cgtaaacttg gtctgacagt taccaatgct taatcagtga ggcacctatc tcagcgatct 5460
gtctatttcg ttcatccata gttgcctgac tccccgtcgt gtagataact acgatacggg 5520
agggcttacc atctggcccc agtgctgcaa tgataccgcg agacccacgc tcaccggctc 5580
cagatttatc agcaataaac cagccagccg gaagggccga gcgcagaagt ggtcctgcaa 5640
ctttatccgc ctccatccag tctattaatt gttgccggga agctagagta agtagttcgc 5700
cagttaatag tttgcgcaac gttgttgcca ttgctacagg catcgtggtg tcacgctcgt 5760
cgtttggtat ggcttcattc agctccggtt cccaacgatc aaggcgagtt acatgatccc 5820
ccatgttgtg caaaaaagcg gttagctcct tcggtcctcc gatcgttgtc agaagtaagt 5880
tggccgcagt gttatcactc atggttatgg cagcactgca taattctctt actgtcatgc 5940
catccgtaag atgcttttct gtgactggtg agtactcaac caagtcattc tgagaatagt 6000
gtatgcggcg accgagttgc tcttgcccgg cgtcaatacg ggataatacc gcgccacata 6060
gcagaacttt aaaagtgctc atcattggaa aacgttcttc ggggcgaaaa ctctcaagga 6120
tcttaccgct gttgagatcc agttcgatgt aacccactcg tgcacccaac tgatcttcag 6180
catcttttac tttcaccagc gtttctgggt gagcaaaaac aggaaggcaa aatgccgcaa 6240
aaaagggaat aagggcgaca cggaaatgtt gaatactcat actcttcctt tttcaatatt 6300
attgaagcat ttatcagggt tattgtctca tgagcggata catatttgaa tgtatttaga 6360
aaaataaaca aataggggtt ccgcgcacat ttccccgaaa agtgccacct gacgtc 6416
<210> SEQ ID NO 115
<211> LENGTH: 3644
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1720
<400> SEQUENCE: 115
taagaaacca ttattatcat gacattaacc tataaaaata ggcgtatcac gaggcccttt 60
cgtcttcacc tcgagaattg tgagcggata acaattgaca ttgtgagcgg ataacaagat 120
actgagcaca tcagcaggac gcactgaccg aattcattag tcgacattat gcggccgcgg 180
atccataagg aggattaatt aagacttccc gggtgatccc atggtacgcg tgctagaggc 240
atcaaataaa acgaaaggct cagtcgaaag actgggcctt tcgttttatc tgttgtttgt 300
cggtgaacgc tctcctgagt aggacaaatc cgccgcccta gacctagcta gggtacgggt 360
tttgctgccc gcaaacgggc tgttctggtg ttgctagttt gttatcagaa tcgcagatcc 420
ggcttcagcc ggtttgccgg ctgaaagcgc tatttcttcc agaattgcca tgattttttc 480
cccacgggag gcgtcactgg ctcccgtgtt gtcggcagct ttgattcgat aagcagcatc 540
gcctgtttca ggctgtctat gtgtgactgt tgagctgtaa caagttgtct caggtgttca 600
atttcatgtt ctagttgctt tgttttactg gtttcacctg ttctattagg tgttacatgc 660
tgttcatctg ttacattgtc gatctgttca tggtgaacag ctttaaatgc accaaaaact 720
cgtaaaagct ctgatgtatc tatctttttt acaccgtttt catctgtgca tatggacagt 780
tttccctttg atatctaacg gtgaacagtt gttctacttt tgtttgttag tcttgatgct 840
tcactgatag atacaagagc cataagaacc tcagatcctt ccgtatttag ccagtatgtt 900
ctctagtgtg gttcgttgtt tttgcgtgag ccatgagaac gaaccattga gatcatgctt 960
actttgcatg tcactcaaaa attttgcctc aaaactggtg agctgaattt ttgcagttaa 1020
agcatcgtgt agtgtttttc ttagtccgtt acgtaggtag gaatctgatg taatggttgt 1080
tggtattttg tcaccattca tttttatctg gttgttctca agttcggtta cgagatccat 1140
ttgtctatct agttcaactt ggaaaatcaa cgtatcagtc gggcggcctc gcttatcaac 1200
caccaatttc atattgctgt aagtgtttaa atctttactt attggtttca aaacccattg 1260
gttaagcctt ttaaactcat ggtagttatt ttcaagcatt aacatgaact taaattcatc 1320
aaggctaatc tctatatttg ccttgtgagt tttcttttgt gttagttctt ttaataacca 1380
ctcataaatc ctcatagagt atttgttttc aaaagactta acatgttcca gattatattt 1440
tatgaatttt tttaactgga aaagataagg caatatctct tcactaaaaa ctaattctaa 1500
tttttcgctt gagaacttgg catagtttgt ccactggaaa atctcaaagc ctttaaccaa 1560
aggattcctg atttccacag ttctcgtcat cagctctctg gttgctttag ctaatacacc 1620
ataagcattt tccctactga tgttcatcat ctgagcgtat tggttataag tgaacgatac 1680
cgtccgttct ttccttgtag ggttttcaat cgtggggttg agtagtgcca cacagcataa 1740
aattagcttg gtttcatgct ccgttaagtc atagcgacta atcgctagtt catttgcttt 1800
gaaaacaact aattcagaca tacatctcaa ttggtctagg tgattttaat cactatacca 1860
attgagatgg gctagtcaat gataattact agtccttttc ccgggagatc tgggtatctg 1920
taaattctgc tagacctttg ctggaaaact tgtaaattct gctagaccct ctgtaaattc 1980
cgctagacct ttgtgtgttt tttttgttta tattcaagtg gttataattt atagaataaa 2040
gaaagaataa aaaaagataa aaagaataga tcccagccct gtgtataact cactacttta 2100
gtcagttccg cagtattaca aaaggatgtc gcaaacgctg tttgctcctc tacaaaacag 2160
accttaaaac cctaaaggct taagtagcac cctcgcaagc tcgggcaaat cgctgaatat 2220
tccttttgtc tccgaccatc aggcacctga gtcgctgtct ttttcgtgac attcagttcg 2280
ctgcgctcac ggctctggca gtgaatgggg gtaaatggca ctacaggcgc cttttatgga 2340
ttcatgcaag gaaactaccc ataatacaag aaaagcccgt cacgggcttc tcagggcgtt 2400
ttatggcggg tctgctatgt ggtgctatct gactttttgc tgttcagcag ttcctgccct 2460
ctgattttcc agtctgacca cttcggatta tcccgtgaca ggtcattcag actggctaat 2520
gcacccagta aggcagcggt atcatcaaca ggcttacccg tcttactgtc cctagtgctt 2580
ggattctcac caataaaaaa cgcccggcgg caaccgagcg ttctgaacaa atccagatgg 2640
agttctgagg tcattactgg atctatcaac aggagtccaa gcgagctctc gaaccccaga 2700
gtcccgctca gaagaactcg tcaagaaggc gatagaaggc gatgcgctgc gaatcgggag 2760
cggcgatacc gtaaagcacg aggaagcggt cagcccattc gccgccaagc tcttcagcaa 2820
tatcacgggt agccaacgct atgtcctgat agcggtccgc cacacccagc cggccacagt 2880
cgatgaatcc agaaaagcgg ccattttcca ccatgatatt cggcaagcag gcatcgccat 2940
gggtcacgac gagatcctcg ccgtcgggca tgcgcgcctt gagcctggcg aacagttcgg 3000
ctggcgcgag cccctgatgc tcttcgtcca gatcatcctg atcgacaaga ccggcttcca 3060
tccgagtacg tgctcgctcg atgcgatgtt tcgcttggtg gtcgaatggg caggtagccg 3120
gatcaagcgt atgcagccgc cgcattgcat cagccatgat ggatactttc tcggcaggag 3180
caaggtgaga tgacaggaga tcctgccccg gcacttcgcc caatagcagc cagtcccttc 3240
ccgcttcagt gacaacgtcg agcacagctg cgcaaggaac gcccgtcgtg gccagccacg 3300
atagccgcgc tgcctcgtcc tgcagttcat tcagggcacc ggacaggtcg gtcttgacaa 3360
aaagaaccgg gcgcccctgc gctgacagcc ggaacacggc ggcatcagag cagccgattg 3420
tctgttgtgc ccagtcatag ccgaatagcc tctccaccca agcggccgga gaacctgcgt 3480
gcaatccatc ttgttcaatc atgcgaaacg atcctcatcc tgtctcttga tcagatcttg 3540
atcccctgcg ccatcagatc cttggcggca agaaagccat ccagtttact ttgcagggct 3600
tcccaacctt accagagggc gccccagctg gcaattccga cgtc 3644
<210> SEQ ID NO 116
<211> LENGTH: 6654
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1730
<400> SEQUENCE: 116
caggcaagtg cacaaacaat acttaaataa atactactca gtaataacct atttcttagc 60
atttttgacg aaatttgcta ttttgttaga gtcttttaca ccatttgtct ccacacctcc 120
gcttacatca acaccaataa cgccatttaa tctaagcgca tcaccaacat tttctggcgt 180
cagtccacca gctaacataa aatgtaagct ttcggggctc tcttgccttc caacccagtc 240
agaaatcgag ttccaatcca aaagttcacc tgtcccacct gcttctgaat caaacaaggg 300
aataaacgaa tgaggtttct gtgaagctgc actgagtagt atgttgcagt cttttggaaa 360
tacgagtctt ttaataactg gcaaaccgag gaactcttgg tattcttgcc acgactcatc 420
tccatgcagt tggacgatat caatgccgta atcattgacc agagccaaaa catcctcctt 480
aggttgatta cgaaacacgc caaccaagta tttcggagtg cctgaactat ttttatatgc 540
ttttacaaga cttgaaattt tccttgcaat aaccgggtca attgttctct ttctattggg 600
cacacatata atacccagca agtcagcatc ggaatctaga gcacattctg cggcctctgt 660
gctctgcaag ccgcaaactt tcaccaatgg accagaacta cctgtgaaat taataacaga 720
catactccaa gctgcctttg tgtgcttaat cacgtatact cacgtgctca atagtcacca 780
atgccctccc tcttggccct ctccttttct tttttcgacc gaattaattc ttaatcggca 840
aaaaaagaaa agctccggat caagattgta cgtaaggtga caagctattt ttcaataaag 900
aatatcttcc actactgcca tctggcgtca taactgcaaa gtacacatat attacgatgc 960
tgtctattaa atgcttccta tattatatat atagtaatgt cgttgacgtc gccggcagga 1020
gagtgaaaga gccttgttta tatatttttt tttcctatgt tcaacgagga cagctaggtt 1080
tatgcaaaaa tgtgccatca ccataagctg attcaaatga gctaaaaaaa aaatagttag 1140
aaaataaggt ggtgttgaac gatagcaagt agatcaagac accgtctaac agaaaaaggg 1200
gcagcggaca atattatgca attatgaaga aaagtactca aagggtcgga aaaatattca 1260
aacgatattt gcattaaatc ctcaattgat tgattattcc atagtaaaat accgtaacaa 1320
cacaaaattg ttctcaaatt cataaattat tcattttttc cacgagcctc atcacacgaa 1380
aagtcagaag agcatacata atcttttaaa tgcataggtt atgcattttg caaatgccac 1440
caggcaacaa aaatatgcgt ttagcgggcg gaatcgggaa ggaagccgga accaccaaaa 1500
actggaagct acgtttttaa ggaaggtatg ggtgcagtgt gcttatctca agaaatatta 1560
gttatgatat aaggtgttga agtttagaga taggtaaata aacgcggggt gtgtttatta 1620
catgaagaag aagttagttt ctgccttgct tgtttatctt gcacatcaca tcagcggaac 1680
atatgctcac ccagtcgcga catccaattt atagaaatca gcttgtgggt attgttcaga 1740
gaatttttca atcattggag caatcatttt acatggaccg caccaagtgg cgtagaaatc 1800
tacgacaact agcttgtctt gagcaattgc agagtcgaat tcgctggcag ttttgaattg 1860
agtaaccatt atttgtatcg aggtgtctag tcttctatta cactaatgca gtttcagggt 1920
tttggaaacc acactgttta aacagtgttc cttaatcaag gatacctctt tttttttcct 1980
tggttccact aattcatcgg tttttttttt ggaagacatc ttttccaacg aaaagaatat 2040
acatatcgtt taagagaaat tctccaaatt tgtaaagaag cggacccaga cttaagccta 2100
accaggccaa ttcaacagac tgtcggcaac ttcttgtctg gtctttccat ggtaagtgac 2160
agtgcagtaa taatatgaac caatttattt ttcgttacat aaaaatgctt ataaaacttt 2220
aactaataat tagagattaa atcgcggccg cggatcccta gagagctttc gttttcatga 2280
gttccccgaa ttctttcgga agcttgtcac ttgctaaatt aacgttatca ctgtagtcaa 2340
ccgggacatc aatgatgaca ggcccctcag cgttcatgcc ttgacgcaga acatctgcca 2400
gctggtctgg tgattctacg cgtaagccag ttgctccgaa gctttccgcg tatttcacga 2460
tatcgatatt tccgaaatcg accgcagatg tacgattata ttttttcaat tgctggaatg 2520
caaccatgtc atatgtgctg tcgttccata caatgtgtac aattggtgct tttaaacgaa 2580
ctgctgtctc taattccata gctgagaata agaaaccgcc atcaccggag actgatacta 2640
ctttttctcc cggtttcacc aatgaagcgc cgattgccca aggaagcgca acgccgagtg 2700
tttgcatacc gttactaatc attaatgtta acggctcgta gctgcggaaa taacgtgaca 2760
tccaaatcgc gtgtgaaccg atatcgcaag tcactgtaac atgatcatcg actgcgtttc 2820
gcaattcttt aacgatttca agaggatgca ctctgtctga tttccaatct gcaggcacct 2880
gctcaccctc atgcatatat tgttttaaat cagaaaggat cttctgctca cgttccgcaa 2940
agtctacttt cacagcatcg tgttcgatat gattgatcgt agatggaata tcaccgatca 3000
gttcaagatc cggctggtaa gcatgatcaa tgtcagccag aatctcgtct aaatggatga 3060
tcgtccggtc tccattgaca ttccagaatt tcggatcata ttcaattggg tcatagccga 3120
ttgtcagaac aacatcagcc tgctcaagca gcagatcgcc aggctggttg cggaataaac 3180
cgatccggcc aaaatactga tcctctaaat ctctcgtaag agtaccggca gcttgatatg 3240
tttcaacgaa tggaagctgc acttttttca atagcttgcg aaccgcttta atcgcttccg 3300
gtcttccgcc cttcatgccg actaaaacga caggaagttt tgctgtttga atttttgcaa 3360
tggccatact gattgcgtca tctgctgcgg gaccaagttt tggcgctgcg acagcacgta 3420
cgttttttgt atttgtgact tcattcacaa catcttgcgg aaaactcaca aaagcggccc 3480
cagcctgccc tgctgacgct atcctaaacg catttgtaac agcttccggt atatttttta 3540
catcttgaac ttctacactg tattttgtaa tcggctggaa tagcgccgca ttatccaaag 3600
attgatgtgt ccgttttaaa cgatctgcac ggatcacgtt cccagcaagc gcaacgacag 3660
ggtcaccttc agtgtttgct gtcagcagtc ctgttgccaa gttcgaagca cctggtcctg 3720
atgtgactaa cacgactccc ggttttccag ttaaacggcc gactgcttgc gccataaatg 3780
ctgcattttg ttcatgccgg gcaacgataa tttcaggccc tttatcttgt aaagcgtcaa 3840
ataccgcatc aatttttgca cctggaatgc caaatacatg tgtgacacct tgctccgcta 3900
agcaatcaac aacaagctcc gcccctctgc ttttcacaag ggatttttgt tcttttgttg 3960
cttttgtcaa catgtcgact ttatgtgatg attgattgat tgattgtaca gtttgttttt 4020
cttaatatct atttcgatga cttctatatg atattgcact aacaagaaga tattataatg 4080
caattgatac aagacaagga gttatttgct tctcttttat atgattctga caatccatat 4140
tgcgttggta gtcttttttg ctggaacggt tcagcggaaa agacgcatcg ctctttttgc 4200
ttctagaaga aatgccagca aaagaatctc ttgacagtga ctgacagcaa aaatgtcttt 4260
ttctaactag taacaaggct aagatatcag cctgaaataa agggtggtga agtaataatt 4320
aaatcatccg tataaaccta tacacatata tgaggaaaaa taatacaaaa gtgttttaaa 4380
tacagataca tacatgaaca tatgcacgta tagcgcccaa atgtcggtaa tgggatcggc 4440
gagctccagc ttttgttccc tttagtgagg gttaattgcg cgcttggcgt aatcatggtc 4500
atagctgttt cctgtgtgaa attgttatcc gctcacaatt ccacacaaca taggagccgg 4560
aagcataaag tgtaaagcct ggggtgccta atgagtgagg taactcacat taattgcgtt 4620
gcgctcactg cccgctttcc agtcgggaaa cctgtcgtgc cagctgcatt aatgaatcgg 4680
ccaacgcgcg gggagaggcg gtttgcgtat tgggcgctct tccgcttcct cgctcactga 4740
ctcgctgcgc tcggtcgttc ggctgcggcg agcggtatca gctcactcaa aggcggtaat 4800
acggttatcc acagaatcag gggataacgc aggaaagaac atgtgagcaa aaggccagca 4860
aaaggccagg aaccgtaaaa aggccgcgtt gctggcgttt ttccataggc tccgcccccc 4920
tgacgagcat cacaaaaatc gacgctcaag tcagaggtgg cgaaacccga caggactata 4980
aagataccag gcgtttcccc ctggaagctc cctcgtgcgc tctcctgttc cgaccctgcc 5040
gcttaccgga tacctgtccg cctttctccc ttcgggaagc gtggcgcttt ctcatagctc 5100
acgctgtagg tatctcagtt cggtgtaggt cgttcgctcc aagctgggct gtgtgcacga 5160
accccccgtt cagcccgacc gctgcgcctt atccggtaac tatcgtcttg agtccaaccc 5220
ggtaagacac gacttatcgc cactggcagc agccactggt aacaggatta gcagagcgag 5280
gtatgtaggc ggtgctacag agttcttgaa gtggtggcct aactacggct acactagaag 5340
gacagtattt ggtatctgcg ctctgctgaa gccagttacc ttcggaaaaa gagttggtag 5400
ctcttgatcc ggcaaacaaa ccaccgctgg tagcggtggt ttttttgttt gcaagcagca 5460
gattacgcgc agaaaaaaag gatctcaaga agatcctttg atcttttcta cggggtctga 5520
cgctcagtgg aacgaaaact cacgttaagg gattttggtc atgagattat caaaaaggat 5580
cttcacctag atccttttaa attaaaaatg aagttttaaa tcaatctaaa gtatatatga 5640
gtaaacttgg tctgacagtt accaatgctt aatcagtgag gcacctatct cagcgatctg 5700
tctatttcgt tcatccatag ttgcctgact ccccgtcgtg tagataacta cgatacggga 5760
gggcttacca tctggcccca gtgctgcaat gataccgcga gacccacgct caccggctcc 5820
agatttatca gcaataaacc agccagccgg aagggccgag cgcagaagtg gtcctgcaac 5880
tttatccgcc tccatccagt ctattaattg ttgccgggaa gctagagtaa gtagttcgcc 5940
agttaatagt ttgcgcaacg ttgttgccat tgctacaggc atcgtggtgt cacgctcgtc 6000
gtttggtatg gcttcattca gctccggttc ccaacgatca aggcgagtta catgatcccc 6060
catgttgtgc aaaaaagcgg ttagctcctt cggtcctccg atcgttgtca gaagtaagtt 6120
ggccgcagtg ttatcactca tggttatggc agcactgcat aattctctta ctgtcatgcc 6180
atccgtaaga tgcttttctg tgactggtga gtactcaacc aagtcattct gagaatagtg 6240
tatgcggcga ccgagttgct cttgcccggc gtcaatacgg gataataccg cgccacatag 6300
cagaacttta aaagtgctca tcattggaaa acgttcttcg gggcgaaaac tctcaaggat 6360
cttaccgctg ttgagatcca gttcgatgta acccactcgt gcacccaact gatcttcagc 6420
atcttttact ttcaccagcg tttctgggtg agcaaaaaca ggaaggcaaa atgccgcaaa 6480
aaagggaata agggcgacac ggaaatgttg aatactcata ctcttccttt ttcaatatta 6540
ttgaagcatt tatcagggtt attgtctcat gagcggatac atatttgaat gtatttagaa 6600
aaataaacaa ataggggttc cgcgcacatt tccccgaaaa gtgccacctg acgt 6654
<210> SEQ ID NO 117
<211> LENGTH: 6597
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1745
<400> SEQUENCE: 117
taagaaacca ttattatcat gacattaacc tataaaaata ggcgtatcac gaggcccttt 60
cgtcttcacc tcgagaattg tgagcggata acaattgaca ttgtgagcgg ataacaagat 120
actgagcaca tcagcaggac gcactgaccg aattcattag tcgacaggag aaaggtacta 180
tgcgaattgg cataccaaga gaacggttaa ccaatgaaac ccgtgttgca gcaacgccaa 240
aaacagtgga acagctgctg aaactgggtt ttaccgtcgc ggtagagagc ggcgcgggtc 300
aactggcaag ttttgacgat aaagcgtttg tgcaagcggg cgctgaaatt gtagaaggga 360
atagcgtctg gcagtcagag atcattctga aggtcaatgc gccgttagat gatgaaattg 420
cgttactgaa tcctgggaca acgctggtga gttttatctg gcctgcgcag aatccggaat 480
taatgcaaaa acttgcggaa cgtaacgtga ccgtgatggc gatggactct gtgccgcgta 540
tctcacgcgc acaatcgctg gacgcactaa gctcgatggc gaacatcgcc ggttatcgcg 600
ccattgttga agcggcacat gaatttgggc gcttctttac cgggcaaatt actgcggccg 660
ggaaagtgcc accggcaaaa gtgatggtga ttggtgcggg tgttgcaggt ctggccgcca 720
ttggcgcagc aaacagtctc ggcgcgattg tgcgtgcatt cgacacccgc ccggaagtga 780
aagaacaagt tcaaagtatg ggcgcggaat tcctcgagct ggattttaaa gaggaagctg 840
gcagcggcga tggctatgcc aaagtgatgt cggacgcgtt catcaaagcg gaaatggaac 900
tctttgccgc ccaggcaaaa gaggtcgata tcattgtcac caccgcgctt attccaggca 960
aaccagcgcc gaagctaatt acccgtgaaa tggttgactc catgaaggcg ggcagtgtga 1020
ttgttgacct ggcagcccaa aacggcggca actgtgaata caccgtgccg ggtgaaatct 1080
tcactacgga aaatggtgtc aaagtgattg gttataccga tcttccgggc cgtctgccga 1140
cgcaatcctc acagctttac ggcacaaacc tcgttaatct gctgaaactg ttgtgcaaag 1200
agaaagacgg caatatcact gttgattttg atgatgtggt gattcgcggc gtgaccgtga 1260
tccgtgcggg cgaaattacc tggccggcac cgccgattca ggtatcagct cagccgcagg 1320
cggcacaaaa agcggcaccg gaagtgaaaa ctgaggaaaa atgtacctgc tcaccgtggc 1380
gtaaatacgc gttgatggcg ctggcaatca ttctttttgg ctggatggca agcgttgcgc 1440
cgaaagaatt ccttgggcac ttcaccgttt tcgcgctggc ctgcgttgtc ggttattacg 1500
tggtgtggaa tgtatcgcac gcgctgcata caccgttgat gtcggtcacc aacgcgattt 1560
cagggattat tgttgtcgga gcactgttgc agattggcca gggcggctgg gttagcttcc 1620
ttagttttat cgcggtgctt atagccagca ttaatatttt cggtggcttc accgtgactc 1680
agcgcatgct gaaaatgttc cgcaaaaatt aaggggtaac atatgtctgg aggattagtt 1740
acagctgcat acattgttgc cgcgatcctg tttatcttca gtctggccgg tctttcgaaa 1800
catgaaacgt ctcgccaggg taacaacttc ggtatcgccg ggatggcgat tgcgttaatc 1860
gcaaccattt ttggaccgga tacgggtaat gttggctgga tcttgctggc gatggtcatt 1920
ggtggggcaa ttggtatccg tctggcgaag aaagttgaaa tgaccgaaat gccagaactg 1980
gtggcgatcc tgcatagctt cgtgggtctg gcggcagtgc tggttggctt taacagctat 2040
ctgcatcatg acgcgggaat ggcaccgatt ctggtcaata ttcacctgac ggaagtgttc 2100
ctcggtatct tcatcggggc ggtaacgttc acgggttcgg tggtggcgtt cggcaaactg 2160
tgtggcaaga tttcgtctaa accattgatg ctgccaaacc gtcacaaaat gaacctggcg 2220
gctctggtcg tttccttcct gctgctgatt gtatttgttc gcacggacag cgtcggcctg 2280
caagtgctgg cattgctgat aatgaccgca attgcgctgg tattcggctg gcatttagtc 2340
gcctccatcg gtggtgcaga tatgccagtg gtggtgtcga tgctgaactc gtactccggc 2400
tgggcggctg cggctgcggg ctttatgctc agcaacgacc tgctgattgt gaccggtgcg 2460
ctggtcggtt cttcgggggc tatcctttct tacattatgt gtaaggcgat gaaccgttcc 2520
tttatcagcg ttattgcggg tggtttcggc accgacggct cttctactgg cgatgatcag 2580
gaagtgggtg agcaccgcga aatcaccgca gaagagacag cggaactgct gaaaaactcc 2640
cattcagtga tcattactcc ggggtacggc atggcagtcg cgcaggcgca atatcctgtc 2700
gctgaaatta ctgagaaatt gcgcgctcgt ggtattaatg tgcgtttcgg tatccacccg 2760
gtcgcggggc gtttgcctgg acatatgaac gtattgctgg ctgaagcaaa agtaccgtat 2820
gacatcgtgc tggaaatgga cgagatcaat gatgactttg ctgataccga taccgtactg 2880
gtgattggtg ctaacgatac ggttaacccg gcggcgcagg atgatccgaa gagtccgatt 2940
gctggtatgc ctgtgctgga agtgtggaaa gcgcagaacg tgattgtctt taaacgttcg 3000
atgaacactg gctatgctgg tgtgcaaaac ccgctgttct tcaaggaaaa cacccacatg 3060
ctgtttggtg acgccaaagc cagcgtggat gcaatcctga aagctctgta acgtcgacat 3120
tatgcggccg cggatccata aggaggatta attaagactt cccgggtgat cccatggtac 3180
gcgtgctaga ggcatcaaat aaaacgaaag gctcagtcga aagactgggc ctttcgtttt 3240
atctgttgtt tgtcggtgaa cgctctcctg agtaggacaa atccgccgcc ctagacctag 3300
ctagggtacg ggttttgctg cccgcaaacg ggctgttctg gtgttgctag tttgttatca 3360
gaatcgcaga tccggcttca gccggtttgc cggctgaaag cgctatttct tccagaattg 3420
ccatgatttt ttccccacgg gaggcgtcac tggctcccgt gttgtcggca gctttgattc 3480
gataagcagc atcgcctgtt tcaggctgtc tatgtgtgac tgttgagctg taacaagttg 3540
tctcaggtgt tcaatttcat gttctagttg ctttgtttta ctggtttcac ctgttctatt 3600
aggtgttaca tgctgttcat ctgttacatt gtcgatctgt tcatggtgaa cagctttaaa 3660
tgcaccaaaa actcgtaaaa gctctgatgt atctatcttt tttacaccgt tttcatctgt 3720
gcatatggac agttttccct ttgatatcta acggtgaaca gttgttctac ttttgtttgt 3780
tagtcttgat gcttcactga tagatacaag agccataaga acctcagatc cttccgtatt 3840
tagccagtat gttctctagt gtggttcgtt gtttttgcgt gagccatgag aacgaaccat 3900
tgagatcatg cttactttgc atgtcactca aaaattttgc ctcaaaactg gtgagctgaa 3960
tttttgcagt taaagcatcg tgtagtgttt ttcttagtcc gttacgtagg taggaatctg 4020
atgtaatggt tgttggtatt ttgtcaccat tcatttttat ctggttgttc tcaagttcgg 4080
ttacgagatc catttgtcta tctagttcaa cttggaaaat caacgtatca gtcgggcggc 4140
ctcgcttatc aaccaccaat ttcatattgc tgtaagtgtt taaatcttta cttattggtt 4200
tcaaaaccca ttggttaagc cttttaaact catggtagtt attttcaagc attaacatga 4260
acttaaattc atcaaggcta atctctatat ttgccttgtg agttttcttt tgtgttagtt 4320
cttttaataa ccactcataa atcctcatag agtatttgtt ttcaaaagac ttaacatgtt 4380
ccagattata ttttatgaat ttttttaact ggaaaagata aggcaatatc tcttcactaa 4440
aaactaattc taatttttcg cttgagaact tggcatagtt tgtccactgg aaaatctcaa 4500
agcctttaac caaaggattc ctgatttcca cagttctcgt catcagctct ctggttgctt 4560
tagctaatac accataagca ttttccctac tgatgttcat catctgagcg tattggttat 4620
aagtgaacga taccgtccgt tctttccttg tagggttttc aatcgtgggg ttgagtagtg 4680
ccacacagca taaaattagc ttggtttcat gctccgttaa gtcatagcga ctaatcgcta 4740
gttcatttgc tttgaaaaca actaattcag acatacatct caattggtct aggtgatttt 4800
aatcactata ccaattgaga tgggctagtc aatgataatt actagtcctt ttcccgggag 4860
atctgggtat ctgtaaattc tgctagacct ttgctggaaa acttgtaaat tctgctagac 4920
cctctgtaaa ttccgctaga cctttgtgtg ttttttttgt ttatattcaa gtggttataa 4980
tttatagaat aaagaaagaa taaaaaaaga taaaaagaat agatcccagc cctgtgtata 5040
actcactact ttagtcagtt ccgcagtatt acaaaaggat gtcgcaaacg ctgtttgctc 5100
ctctacaaaa cagaccttaa aaccctaaag gcttaagtag caccctcgca agctcgggca 5160
aatcgctgaa tattcctttt gtctccgacc atcaggcacc tgagtcgctg tctttttcgt 5220
gacattcagt tcgctgcgct cacggctctg gcagtgaatg ggggtaaatg gcactacagg 5280
cgccttttat ggattcatgc aaggaaacta cccataatac aagaaaagcc cgtcacgggc 5340
ttctcagggc gttttatggc gggtctgcta tgtggtgcta tctgactttt tgctgttcag 5400
cagttcctgc cctctgattt tccagtctga ccacttcgga ttatcccgtg acaggtcatt 5460
cagactggct aatgcaccca gtaaggcagc ggtatcatca acaggcttac ccgtcttact 5520
gtccctagtg cttggattct caccaataaa aaacgcccgg cggcaaccga gcgttctgaa 5580
caaatccaga tggagttctg aggtcattac tggatctatc aacaggagtc caagcgagct 5640
ctcgaacccc agagtcccgc tcagaagaac tcgtcaagaa ggcgatagaa ggcgatgcgc 5700
tgcgaatcgg gagcggcgat accgtaaagc acgaggaagc ggtcagccca ttcgccgcca 5760
agctcttcag caatatcacg ggtagccaac gctatgtcct gatagcggtc cgccacaccc 5820
agccggccac agtcgatgaa tccagaaaag cggccatttt ccaccatgat attcggcaag 5880
caggcatcgc catgggtcac gacgagatcc tcgccgtcgg gcatgcgcgc cttgagcctg 5940
gcgaacagtt cggctggcgc gagcccctga tgctcttcgt ccagatcatc ctgatcgaca 6000
agaccggctt ccatccgagt acgtgctcgc tcgatgcgat gtttcgcttg gtggtcgaat 6060
gggcaggtag ccggatcaag cgtatgcagc cgccgcattg catcagccat gatggatact 6120
ttctcggcag gagcaaggtg agatgacagg agatcctgcc ccggcacttc gcccaatagc 6180
agccagtccc ttcccgcttc agtgacaacg tcgagcacag ctgcgcaagg aacgcccgtc 6240
gtggccagcc acgatagccg cgctgcctcg tcctgcagtt cattcagggc accggacagg 6300
tcggtcttga caaaaagaac cgggcgcccc tgcgctgaca gccggaacac ggcggcatca 6360
gagcagccga ttgtctgttg tgcccagtca tagccgaata gcctctccac ccaagcggcc 6420
ggagaacctg cgtgcaatcc atcttgttca atcatgcgaa acgatcctca tcctgtctct 6480
tgatcagatc ttgatcccct gcgccatcag atccttggcg gcaagaaagc catccagttt 6540
actttgcagg gcttcccaac cttaccagag ggcgccccag ctggcaattc cgacgtc 6597
<210> SEQ ID NO 118
<211> LENGTH: 3625
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1777
<400> SEQUENCE: 118
taagaaacca ttattatcat gacattaacc tataaaaata ggcgtatcac gaggcccttt 60
cgtcttcacc tcgagaattg tgagcggata acaattgaca ttgtgagcgg ataacaagat 120
actgagcaca tcagcaggac gcactgaccg aattcattaa agaggagaaa ggtacctgca 180
cgtcgacgag gagacaacat tatggcgaat tatttcaaca ctctgaacct gcgtcaacaa 240
ctggcgcaac tgggtaagtg ccgtttcatg ggtcgtgacg agtttgcgga cggtgcttct 300
tatctgcaag gcaagaaggt tgttattgtt ggttgcggtg cgcaaggcct gaatcaaggt 360
ctgaatatgc gcgacagcgg cctggacatt agctatgcgc tgcgcaagga ggctatcgcg 420
gaaaaacgtg ctagctggcg caaggctact gagaacggct tcaaggttgg cacctatgag 480
gagctgattc cgcaagctga cctggttatc aatctgaccc cagataaaca acatagcgac 540
gttgttcgta ctgttcaacc gctgatgaag gatggtgctg ctctgggtta tagccacggc 600
tttaacattg ttgaggtagg tgaacaaatt cgcaaggaca ttactgttgt tatggtggct 660
ccaaagtgtc cgggtactga ggttcgcgag gaatataagc gcggttttgg tgttccaacc 720
ctgatcgcgg tgcatccaga gaatgaccca aagggtgagg gtatggctat cgcgaaggcg 780
tgggctgcgg cgactggcgg ccatcgcgct ggcgttctgg agagcagctt tgtggctgag 840
gttaagagcg atctgatggg tgaacagact attctgtgtg gtatgctgca agcgggtagc 900
ctgctgtgtt ttgataaact ggttgaggag ggcactgacc cggcgtatgc ggagaagctg 960
atccaatttg gctgggagac tattactgag gcgctgaagc aaggtggtat tactctgatg 1020
atggatcgcc tgagcaatcc agctaagctg cgcgcgtacg ctctgagcga gcaactgaag 1080
gaaattatgg caccgctgtt tcaaaagcac atggatgata tcattagcgg tgagtttagc 1140
agcggcatga tggctgattg ggcgaatgac gacaaaaagc tgctgacttg gcgcgaggaa 1200
actggtaaga ctgctttcga gactgctcca caatacgagg gtaagattgg tgaacaagaa 1260
tattttgaca agggtgttct gatgatcgct atggttaagg ctggtgtgga gctggctttt 1320
gagactatgg ttgacagcgg tattatcgag gaaagcgcgt actacgagag cctgcatgaa 1380
ctgccactga tcgcgaatac tattgcgcgc aaacgcctgt atgagatgaa tgttgtgatt 1440
agcgacactg cggaatatgg caattacctg tttagctatg cgtgcgttcc actgctgaag 1500
ccattcatgg cggaactgca gccaggtgat ctgggcaagg cgatcccaga gggtgctgtt 1560
gacaatggtc agctgcgcga cgttaatgag gctatccgtt ctcacgctat cgaacaagtt 1620
ggcaaaaagc tgcgtggtta catgaccgac atgaagcgca tcgcggtggc tggctaacct 1680
agggcgttcg gctgcggcga gcggtatcag ctcactcaaa ggcggtaata cggttatcca 1740
cagaatcagg ggataacgca ggaaagaaca tgtgagcaaa aggccagcaa aaggccagga 1800
accgtaaaaa ggccgcgttg ctggcgtttt tccataggct ccgcccccct gacgagcatc 1860
acaaaaatcg acgctcaagt cagaggtggc gaaacccgac aggactataa agataccagg 1920
cgtttccccc tggaagctcc ctcgtgcgct ctcctgttcc gaccctgccg cttaccggat 1980
acctgtccgc ctttctccct tcgggaagcg tggcgctttc tcatagctca cgctgtaggt 2040
atctcagttc ggtgtaggtc gttcgctcca agctgggctg tgtgcacgaa ccccccgttc 2100
agcccgaccg ctgcgcctta tccggtaact atcgtcttga gtccaacccg gtaagacacg 2160
acttatcgcc actggcagca gccactggta acaggattag cagagcgagg tatgtaggcg 2220
gtgctacaga gttcttgaag tggtggccta actacggcta cactagaagg acagtatttg 2280
gtatctgcgc tctgctgaag ccagttacct tcggaaaaag agttggtagc tcttgatccg 2340
gcaaacaaac caccgctggt agcggtggtt tttttgtttg caagcagcag attacgcgca 2400
gaaaaaaagg atctcaagaa gatcctttga tcttttctac ggggtctgac gctcagtgga 2460
acgaaaactc acgttaaggg attttggtca tgactagtgc ttggattctc accaataaaa 2520
aacgcccggc ggcaaccgag cgttctgaac aaatccagat ggagttctga ggtcattact 2580
ggatctatca acaggagtcc aagcgagctc gtaaacttgg tctgacagtt accaatgctt 2640
aatcagtgag gcacctatct cagcgatctg tctatttcgt tcatccatag ttgcctgact 2700
ccccgtcgtg tagataacta cgatacggga gggcttacca tctggcccca gtgctgcaat 2760
gataccgcga gacccacgct caccggctcc agatttatca gcaataaacc agccagccgg 2820
aagggccgag cgcagaagtg gtcctgcaac tttatccgcc tccatccagt ctattaattg 2880
ttgccgggaa gctagagtaa gtagttcgcc agttaatagt ttgcgcaacg ttgttgccat 2940
tgctacaggc atcgtggtgt cacgctcgtc gtttggtatg gcttcattca gctccggttc 3000
ccaacgatca aggcgagtta catgatcccc catgttgtgc aaaaaagcgg ttagctcctt 3060
cggtcctccg atcgttgtca gaagtaagtt ggccgcagtg ttatcactca tggttatggc 3120
agcactgcat aattctctta ctgtcatgcc atccgtaaga tgcttttctg tgactggtga 3180
gtactcaacc aagtcattct gagaatagtg tatgcggcga ccgagttgct cttgcccggc 3240
gtcaatacgg gataataccg cgccacatag cagaacttta aaagtgctca tcattggaaa 3300
acgttcttcg gggcgaaaac tctcaaggat cttaccgctg ttgagatcca gttcgatgta 3360
acccactcgt gcacccaact gatcttcagc atcttttact ttcaccagcg tttctgggtg 3420
agcaaaaaca ggaaggcaaa atgccgcaaa aaagggaata agggcgacac ggaaatgttg 3480
aatactcata ctcttccttt ttcaatatta ttgaagcatt tatcagggtt attgtctcat 3540
gagcggatac atatttgaat gtatttagaa aaataaacaa ataggggttc cgcgcacatt 3600
tccccgaaaa gtgccacctg acgtc 3625
<210> SEQ ID NO 119
<211> LENGTH: 8870
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1914
<400> SEQUENCE: 119
tcgcgcgttt cggtgatgac ggtgaaaacc tctgacacat gcagctcccg gagacggtca 60
cagcttgtct gtaagcggat gccgggagca gacaagcccg tcagggcgcg tcagcgggtg 120
ttggcgggtg tcggggctgg cttaactatg cggcatcaga gcagattgta ctgagagtgc 180
accatatgcg gtgtgaaata ccgcacagat gcgtaaggag aaaataccgc atcaggcgcc 240
attcgccatt caggctgcgc aactgttggg aagggcgatc ggtgcgggcc tcttcgctat 300
tacgccagct ggcgaaaggg ggatgtgctg caaggcgatt aagttgggta acgccagggt 360
tttcccagtc acgacgttgt aaaacgacgg ccagtgaatt cataccacag cttttcaatt 420
caattcatca tttttttttt attctttttt ttgatttcgg tttccttgaa atttttttga 480
ttcggtaatc tccgaacaga aggaagaacg aaggaaggag cacagactta gattggtata 540
tatacgcata tgtagtgttg aagaaacatg aaattgccca gtattcttaa cccaactgca 600
cagaacaaaa acctgcagga aacgaagata aatcatgtcg aaagctacat ataaggaacg 660
tgctgctact catcctagtc ctgttgctgc caagctattt aatatcatgc acgaaaagca 720
aacaaacttg tgtgcttcat tggatgttcg taccaccaag gaattactgg agttagttga 780
agcattaggt cccaaaattt gtttactaaa aacacatgtg gatatcttga ctgatttttc 840
catggagggc acagttaagc cgctaaaggc attatccgcc aagtacaatt ttttactctt 900
cgaagacaga aaatttgctg acattggtaa tacagtcaaa ttgcagtact ctgcgggtgt 960
atacagaata gcagaatggg cagacattac gaatgcacac ggtgtggtgg gcccaggtat 1020
tgttagcggt ttgaagcagg cggcagaaga agtaacaaag gaacctagag gccttttgat 1080
gttagcagaa ttgtcatgca agggctccct atctactgga gaatatacta agggtactgt 1140
tgacattgcg aagagcgaca aagattttgt tatcggcttt attgctcaaa gagacatggg 1200
tggaagagat gaaggttacg attggttgat tatgacaccc ggtgtgggtt tagatgacaa 1260
gggagacgca ttgggtcaac agtatagaac cgtggatgat gtggtctcta caggatctga 1320
cattattatt gttggaagag gactatttgc aaagggaagg gatgctaagg tagagggtga 1380
acgttacaga aaagcaggct gggaagcata tttgagaaga tgcggccagc aaaactaaaa 1440
aactgtatta taagtaaatg catgtatact aaactcacaa attagagctt caatttaatt 1500
atatcagtta ttaccctatg cggtgtgaaa taccgcacag atgcgtaagg agaaaatacc 1560
gcatcaggaa attgtaaacg ttaatatttt gttaaaattc gcgttaaatt tttgttaaat 1620
cagctcattt tttaaccaat aggccgaaat cggcaaaatc ccttataaat caaaagaata 1680
gaccgagata gggttgagtg ttgttccagt ttggaacaag agtccactat taaagaacgt 1740
ggactccaac gtcaaagggc gaaaaaccgt ctatcagggc gatggcccac tacgtgaacc 1800
atcaccctaa tcaagttttt tggggtcgag gtgccgtaaa gcactaaatc ggaaccctaa 1860
agggagcccc cgatttagag cttgacgggg aaagccggcg aggactgcaa tagcacaaga 1920
ttaagataga atggcttcaa acagccgcct tttatacata ttggtaaaag ctcgcgaatc 1980
gcaccatatc ccttatcctg taatcaaatc gatctaggtg cagatacaga tcaattcata 2040
aaaagaaatt gaagcaccag tttatcacta ctacactatc tttttctttt tttttttttt 2100
ttgcgcagtt tcgccctttg ttcaatatca cttgataagt tgtgggcttt ttctgtcact 2160
cattcggctt aaaaagtatt cgttcttttg tgttttatga aaagggaacg tgatataaaa 2220
aaacatcctt tggtgtggga catgggcttt tgtttagaga atggttatca ctaccgcccc 2280
cacccttgaa agccacagaa aatgaaaaag tatgtgaata aggtgtgaac tctataacat 2340
tttggccaaa tgccacagcc gatctgcata ttccaatgga catgatgcaa caacaattga 2400
tgtcacattc tcttacacac ttcgattggt ccgtacgtag tactttttac ataactgact 2460
caggcgtttc cttcattgaa atgctcatct attgccaagt acatagaatc cacagtgcat 2520
aggttaacgc attgtaccca aacgacggga aacaaggaag gatgcagaat gagcacttgt 2580
tatttataaa aagacacggg agggggaatc ccgtctttcg tccgtcggag ccaaagagat 2640
gagccaaagc agaaaaacag gggacgccgc ccttcttccg tcccgtgcgt gaggggggcg 2700
cggccattcg gtttttgcaa tatgacctgt gggccaaaaa tcgaaaaaaa aaaaaaaaat 2760
aagaggcggc tgcggaattt tataagacaa gcgcagggcc aaagaaaaaa taataattga 2820
cgtggctgaa caacagtctc tccccacccc tttccaaaaa ggggaatgaa atacgagttc 2880
tttttcccaa ttggtagata ttcaacaaga gacgcgcagt acgtaacatg cgaattgcgt 2940
aattcacggc gataacgtag tatttagatt tagtataatt tgaaccgatg tatttatttg 3000
tctgattgat ttatgtattc aaactgtgta agtttattta tttgcaacaa taattcgttt 3060
gagtacacta ctaatggcgg ccgcttagat gccggagtcc cagtgcttgg tccactggat 3120
ggcctccagg gtgcccaagt ccagtttcca gatggctccg ttctggttca gctcgatagc 3180
cttgacgaag ttctcggcgc aggccaacga gggctgggtg ggatgagcca ggagcttctc 3240
ggcaacctga ggctcaacat ccaaccagga gttgaacgtg tgcaccaggg tggtgcgggt 3300
gatgccgggg ttcacagtgt aagccgtcac gccggtaatg ggggccagtt tcgccaggga 3360
gctggtgaag ttgaccacgg cggccttggt gccggagtag acgggcacct ggtagatggc 3420
attgaatcca gtgacggatc caatgttgca gatgatacca ccgggaccgc ccttgcgctt 3480
gtcccagaag tccagaatgg ccgtcgtggt gttgaccagg ccagtgtagt tgacggcaat 3540
ggtgcgctcg atctggtgat cgtccaggat accagctccg ttgatcagga catcgacggt 3600
cttcagctgg gcgaagatgg tcttcagcag cttggtggtc tcggcaatgg gcacggtcac 3660
atcatagggg tagaaggtga cggtcacctt tggattgatt gccttcagct cggcaatggc 3720
agccgggttc tcaatgcggt cgaggatcac caggttcttc agatcgcgct tgagcagctc 3780
cttgctggtg tccagaccaa tgcctcccag accggcaacg aaaatcacgt tcttgttggt 3840
caaagtaaac gacataccgg tatctcctag atccgtcgaa gtcgaaacta agttctggtg 3900
ttttaaaact aaaaaaaaga ctaactataa aagtagaatt taagaagttt aagaaataga 3960
tttacagaat tacaatcaat acctaccgtc tttatatact tattagtcaa gtaggggaat 4020
aatttcaggg aactggtttc aacctttttt ttcagctttt tccaaatcag agagagcaga 4080
aggtaataga aggtgtaaga aaatgagata gatacatgcg tgggtcaatt gccttgtgtc 4140
atcatttact ccaggcaggt tgcatcactc cattgaggtt gtgcccgttt tttgcctgtt 4200
tgtgcccctg ttctctgtag ttgcgctaag agaatggacc tatgaactga tggttggtga 4260
agaaaacaat attttggtgc tgggattctt tttttttctg gatgccagct taaaaagcgg 4320
gctccattat atttagtgga tgccaggaat aaactgttca cccagacacc tacgatgtta 4380
tatattctgt gtaacccgcc ccctattttg ggcatgtacg ggttacagca gaattaaaag 4440
gctaattttt tgactaaata aagttaggaa aatcactact attaattatt tacgtattct 4500
ttgaaatggc gagtattgat aatgataaac tggatcctta ggatttattc tgttcagcaa 4560
acagcttgcc cattttcttc agtaccttcg gtgcgccttc tttcgccagg atcagttcga 4620
tccagtacat acggttcgga tcggcctggg cctctttcat cacgctcaca aattcgtttt 4680
cggtacgcac aattttagac acaacacggt cctcagttgc gccgaaggac tccggcagtt 4740
tagagtagtt ccacataggg atatcgttgt aagactggtt cggaccgtgg atctcacgct 4800
caacggtgta gccgtcattg ttaataatga agcaaatcgg gttgatcttt tcacgaattg 4860
ccagacccag ttcctgtacg gtcagctgca gggaaccgtc accgatgaac agcagatgac 4920
gagattcttt atcagcgatc tgagagccca gcgctgccgg gaaagtatag ccaatgctac 4980
cccacagcgg ctgaccgata aaatggcttt tggatttcag aaagatagaa gacgcgccga 5040
aaaagctcgt accttgttcc gccacgatgg tttcattgct ctgggtcagg ttctccacgg 5100
cctgccacag gcgatcctgg gacagcagtg cgttagatgg tacgaaatct tcttgctttt 5160
tgtcaatgta tttgccttta tactcgattt cggacaggtc cagcagagag ctgatcaggc 5220
tttcgaagtc gaagttctgg atacgctcgt tgaagatttt accctcgtcg atgttcaggc 5280
taatcatttt gttttcgttc agatggtgag tgaatgcacc ggtagaagag tcggtcagtt 5340
taacgcccag catcaggatg aagtccgcag attcaacaaa ttctttcagg ttcggttcgc 5400
tcagagtacc gttgtagatg cccaggaaag acggcagagc ctcgtcaaca gaggacttgc 5460
cgaagttcag ggtggtaatc ggcagtttgg ttttgctgat gaattgggtc acggtcttct 5520
ccagaccaaa agaaatgatt tcgtggccgg tgatcacgat tggtttcttt gcgtttttca 5580
gagactcctg gattttgttc aggatttcct ggtcgctagt gttagaagtg gagttttctt 5640
tcttcagcgg caggctcggt ttttccgctt tagctgccgc aacatccaca ggcaggttga 5700
tgtaaactgg tttgcgttct ttcagcagcg cagacagaac gcggtcgatt tccacagtag 5760
cgttctctgc agtcagcagc gtacgtgccg cagtcacagg ttcatgcatt ttcatgaagt 5820
gtttgaaatc gccgtcagcc agagtgtggt ggacgaattt accttcgttc tgaactttgc 5880
tcgttgggct gcctacgatc tccaccaccg gcaggttttc ggcgtaggag cccgccagac 5940
cgttgacggc gctcagttcg ccaacaccga aagtggtcag aaatgccgcg gctttcttgg 6000
tacgtgcata accatctgcc atgtagcttg cgttcagttc gttagcgtta cccacccatt 6060
tcatgtcttt atgagagatg atctgatcca ggaactgcag attgtaatca cccggaacgc 6120
cgaagatttc ttcgataccc agttcatgca gacggtccag cagataatca ccaacagtat 6180
acatgtcgac aaacttagat tagattgcta tgctttcttt ctaatgagca agaagtaaaa 6240
aaagttgtaa tagaacaaga aaaatgaaac tgaaacttga gaaattgaag accgtttatt 6300
aacttaaata tcaatgggag gtcatcgaaa gagaaaaaaa tcaaaaaaaa aattttcaag 6360
aaaaagaaac gtgataaaaa tttttattgc ctttttcgac gaagaaaaag aaacgaggcg 6420
gtctcttttt tcttttccaa acctttagta cgggtaatta acgacaccct agaggaagaa 6480
agaggggaaa tttagtatgc tgtgcttggg tgttttgaag tggtacggcg atgcgcggag 6540
tccgagaaaa tctggaagag taaaaaagga gtagaaacat tttgaagcta tgagctccag 6600
cttttgttcc ctttagtgag ggttaattgc gcgcttggcg taatcatggt catagctgtt 6660
tcctgtgtga aattgttatc cgctcacaat tccacacaac ataggagccg gaagcataaa 6720
gtgtaaagcc tggggtgcct aatgagtgag gtaactcaca ttaattgcgt tgcgctcact 6780
gcccgctttc cagtcgggaa acctgtcgtg ccagctgcat taatgaatcg gccaacgcgc 6840
ggggagaggc ggtttgcgta ttgggcgctc ttccgcttcc tcgctcactg actcgctgcg 6900
ctcggtcgtt cggctgcggc gagcggtatc agctcactca aaggcggtaa tacggttatc 6960
cacagaatca ggggataacg caggaaagaa catgtgagca aaaggccagc aaaaggccag 7020
gaaccgtaaa aaggccgcgt tgctggcgtt tttccatagg ctccgccccc ctgacgagca 7080
tcacaaaaat cgacgctcaa gtcagaggtg gcgaaacccg acaggactat aaagatacca 7140
ggcgtttccc cctggaagct ccctcgtgcg ctctcctgtt ccgaccctgc cgcttaccgg 7200
atacctgtcc gcctttctcc cttcgggaag cgtggcgctt tctcatagct cacgctgtag 7260
gtatctcagt tcggtgtagg tcgttcgctc caagctgggc tgtgtgcacg aaccccccgt 7320
tcagcccgac cgctgcgcct tatccggtaa ctatcgtctt gagtccaacc cggtaagaca 7380
cgacttatcg ccactggcag cagccactgg taacaggatt agcagagcga ggtatgtagg 7440
cggtgctaca gagttcttga agtggtggcc taactacggc tacactagaa ggacagtatt 7500
tggtatctgc gctctgctga agccagttac cttcggaaaa agagttggta gctcttgatc 7560
cggcaaacaa accaccgctg gtagcggtgg tttttttgtt tgcaagcagc agattacgcg 7620
cagaaaaaaa ggatctcaag aagatccttt gatcttttct acggggtctg acgctcagtg 7680
gaacgaaaac tcacgttaag ggattttggt catgagatta tcaaaaagga tcttcaccta 7740
gatcctttta aattaaaaat gaagttttaa atcaatctaa agtatatatg agtaaacttg 7800
gtctgacagt taccaatgct taatcagtga ggcacctatc tcagcgatct gtctatttcg 7860
ttcatccata gttgcctgac tccccgtcgt gtagataact acgatacggg agggcttacc 7920
atctggcccc agtgctgcaa tgataccgcg agacccacgc tcaccggctc cagatttatc 7980
agcaataaac cagccagccg gaagggccga gcgcagaagt ggtcctgcaa ctttatccgc 8040
ctccatccag tctattaatt gttgccggga agctagagta agtagttcgc cagttaatag 8100
tttgcgcaac gttgttgcca ttgctacagg catcgtggtg tcacgctcgt cgtttggtat 8160
ggcttcattc agctccggtt cccaacgatc aaggcgagtt acatgatccc ccatgttgtg 8220
caaaaaagcg gttagctcct tcggtcctcc gatcgttgtc agaagtaagt tggccgcagt 8280
gttatcactc atggttatgg cagcactgca taattctctt actgtcatgc catccgtaag 8340
atgcttttct gtgactggtg agtactcaac caagtcattc tgagaatagt gtatgcggcg 8400
accgagttgc tcttgcccgg cgtcaatacg ggataatacc gcgccacata gcagaacttt 8460
aaaagtgctc atcattggaa aacgttcttc ggggcgaaaa ctctcaagga tcttaccgct 8520
gttgagatcc agttcgatgt aacccactcg tgcacccaac tgatcttcag catcttttac 8580
tttcaccagc gtttctgggt gagcaaaaac aggaaggcaa aatgccgcaa aaaagggaat 8640
aagggcgaca cggaaatgtt gaatactcat actcttcctt tttcaatatt attgaagcat 8700
ttatcagggt tattgtctca tgagcggata catatttgaa tgtatttaga aaaataaaca 8760
aataggggtt ccgcgcacat ttccccgaaa agtgccacct gacgtctaag aaaccattat 8820
tatcatgaca ttaacctata aaaataggcg tatcacgagg ccctttcgtc 8870
<210> SEQ ID NO 120
<211> LENGTH: 9516
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV1936
<400> SEQUENCE: 120
ccagttaact gtgggaatac tcaggtatcg taagatgcaa gagttcgaat ctcttagcaa 60
ccattatttt tttcctcaac ataacgagaa cacacagggg cgctatcgca cagaatcaaa 120
ttcgatgact ggaaattttt tgttaatttc agaggtcgcc tgacgcatat acctttttca 180
actgaaaaat tgggagaaaa aggaaaggtg agagcgccgg aaccggcttt tcatatagaa 240
tagagaagcg ttcatgacta aatgcttgca tcacaatact tgaagttgac aatattattt 300
aaggacctat tgttttttcc aataggtggt tagcaatcgt cttactttct aacttttctt 360
accttttaca tttcagcaat atatatatat atatttcaag gatataccat tctaatgtct 420
gcccctaaga agatcgtcgt tttgccaggt gaccacgttg gtcaagaaat cacagccgaa 480
gccattaagg ttcttaaagc tatttctgat gttcgttcca atgtcaagtt cgatttcgaa 540
aatcatttaa ttggtggtgc tgctatcgat gctacaggtg ttccacttcc agatgaggcg 600
ctggaagcct ccaagaaggc tgatgccgtt ttgttaggtg ctgtgggtgg tcctaaatgg 660
ggtaccggta gtgttagacc tgaacaaggt ttactaaaaa tccgtaaaga acttcaattg 720
tacgccaact taagaccatg taactttgca tccgactctc ttttagactt atctccaatc 780
aagccacaat ttgctaaagg tactgacttc gttgttgtca gagaattagt gggaggtatt 840
tactttggta agagaaagga agacgatggt gatggtgtcg cttgggatag tgaacaatac 900
accgttccag aagtgcaaag aatcacaaga atggccgctt tcatggccct acaacatgag 960
ccaccattgc ctatttggtc cttggataaa gctaatgttt tggcctcttc aagattatgg 1020
agaaaaactg tggaggaaac catcaagaac gaattcccta cattgaaggt tcaacatcaa 1080
ttgattgatt ctgccgccat gatcctagtt aagaacccaa cccacctaaa tggtattata 1140
atcaccagca acatgtttgg tgatatcatc tccgatgaag cctccgttat cccaggttcc 1200
ttgggtttgt tgccatctgc gtccttggcc tctttgccag acaagaacac cgcatttggt 1260
ttgtacgaac catgccacgg ttctgctcca gatttgccaa agaataaggt caaccctatc 1320
gccactatct tgtctgctgc aatgatgttg aaattgtcat tgaacttgcc tgaagaaggt 1380
aaggccattg aagatgcagt taaaaaggtt ttggatgcag gtatcagaac tggtgattta 1440
ggtggttcca acagtaccac cgaagtcggt gatgctgtcg ccgaagaagt taagaaaatc 1500
cttgcttaaa aagattctct ttttttatga tatttgtaca taaactttat aaatgaaatt 1560
cataatagaa acgacacgaa attacaaaat ggaatatgtt catagggtag acgaaactat 1620
atacgcaatc tacatacatt tatcaagaag gagaaaaagg aggatgtaaa ggaatacagg 1680
taagcaaatt gatactaatg gctcaacgtg ataaggaaaa agaattgcac tttaacatta 1740
atattgacaa ggaggagggc accacacaaa aagttaggtg taacagaaaa tcatgaaact 1800
atgattccta atttatatat tggaggattt tctctaaaaa aaaaaaaata caacaaataa 1860
aaaacactca atgacctgac catttgatgg agttgccggc ttgatcgaga atggcagctc 1920
ttatatacaa gttcttttag caagcgccgc tgcattattc aagtctcatc atatgaaatt 1980
tctttcgaga gattgtcata atcaaaaaat tgcataatgc atttcttgca acacattttc 2040
tgatataatc ttaccttaat gcaggtttac gtattagttt ttctaaaaga aacgcgacct 2100
ttggatatgg aggcttttcc cataaacgca tgtagtatgc atttacgatg agaatcaatt 2160
tttttccaag gggcgcaaaa cgcataaacg cataaagtat gcatcagaag gattctcacc 2220
tggttgcaac catacaggtg ttagcgacag taatagaaaa aaaattaaaa taatggtgtt 2280
attgttattt gctttatttc cttggccttt gttgaaggaa ttcgtatacg tattacaaat 2340
aactagtatc gaggaacttg aaagagctga aatttttgca ttcttcttcg gtgattatgc 2400
ctaagccaat gaggtcgccc caaaagaccg caatcttgtc acgaccataa gccatataat 2460
cgcgaacaaa aacccgtttt taggaaggac agaggtccat atcaatataa ttaagaaggc 2520
atgttggcct ctgtttctta atatattcta aataagatgt aaggccttgt aattcagttt 2580
gttcacaaaa ttaaaaactg tttaatgttt tttgttttgt tgtagtattc gagcattaag 2640
gataaaaaaa gcttgtgaat aaaaatcttt cgctaaaaat caatataaga aaatggtaag 2700
cagctgaaag ataataaggt atggttaaag atcacaccac cctcttcaat tagctaagat 2760
catagctaaa ggtacaaaac cgaatacgaa agtaaataaa ttaatcagca taaaattaaa 2820
taataaacca cctaaaatat tagaagctaa tctttaacct ggaagacagg acagaaaagt 2880
aattacaaga acatatgtga aaaaaaatag ttgatatttt aaaccaaatc agaaatttat 2940
tatactaaaa ctatatctat gccaattatt tacctaaaca tctataacct tcaaaagtaa 3000
aaaaatacac aaacgttgaa tcatgagttt tatgttaatt aggcggccgc ggatcttcat 3060
cctgccactg caattctttt catatcggtc atatatcctc tcagcttttt acccacctgt 3120
tctatagcat gtgaacgaat agcttcattt acgtctctca gttggccatt gtcaaccgct 3180
ccttccggaa tagccttccc caaatcacca ggttgtaact cggccatgaa gggctttaac 3240
aacgggacac atgcgtagct aaataagtaa ttaccatatt ctgcagtgtc tgatatgaca 3300
acattcatct cgtaaagtct ttttcttgca atagtatttg ctatcaaagg caattcatgc 3360
aaagactcat agtatgcaga ttcttcaatg ataccggagt caaccatagt ttcgaatgca 3420
agttctaccc ctgccttcac catagctatc atcaatactc ccttatcaaa gtattcttgt 3480
tcaccaattt taccttcgta ttgtggggct gtctcgaatg ccgtcttgcc ggtttcttct 3540
ctccacgtca ataacttttt atcatcgttt gcccaatctg ccatcattcc tgaggaaaac 3600
tcaccggaga taatatcgtc catgtgcttt tggaataatg gtgccatgat ctcttttagt 3660
tgctcagata aggcgtaggc tcttagcttg gccggatttg aaagtctatc catcatcaat 3720
gttatgccac cttgtttaag tgcctcggtg attgtctccc aaccaaattg tatcaacttt 3780
tcagcatagg caggatctgt accctcttcg accaatttat caaagcatag taaagaccct 3840
gcctgcaaca ttccgcacag aatggtttgt tcacccatta agtcactctt gacctcagct 3900
acgaaagaac tctctaacac acccgctcta tgacctccgg ttgcggctgc ccatgccttc 3960
gcaattgcca taccttcacc tttggggtca ttttcaggat gtacggcgat caatgtaggt 4020
acaccaaaac ccctcttgta ctcctctctg acttccgtac ctgggcactt tggtgcaacc 4080
attacgactg ttatatcttt tctgatctgc tcgcccactt caacgatatt aaagccatga 4140
gagtaaccta aagctgcccc atccttcatc agcggttgaa ctgttcttac tacgtctgag 4200
tgaaccttat ctggtgttag gttaatcact aaatctgcct gagggatcag ttcttcgtaa 4260
gtaccaactt tgaacccatt ttccgtcgct ttacgccagg aggccctctt ttctgcaatt 4320
gcctctttcc tcaatgcata cgaaatatcc agacctgaat ctctcatgtt taaaccttgg 4380
tttagaccct gagcaccgca gccaacaatt actactttct ttccttgcag ataagaagca 4440
ccatcagcaa actcgtccct tcccataaat ctgcacttac ccagttgagc caattgttgt 4500
ctcaaattta atgtgttaaa atagttggcc atctcgagtc gaaactaagt tctggtgttt 4560
taaaactaaa aaaaagacta actataaaag tagaatttaa gaagtttaag aaatagattt 4620
acagaattac aatcaatacc taccgtcttt atatacttat tagtcaagta ggggaataat 4680
ttcagggaac tggtttcaac cttttttttc agctttttcc aaatcagaga gagcagaagg 4740
taatagaagg tgtaagaaaa tgagatagat acatgcgtgg gtcaattgcc ttgtgtcatc 4800
atttactcca ggcaggttgc atcactccat tgaggttgtg cccgtttttt gcctgtttgt 4860
gcccctgttc tctgtagttg cgctaagaga atggacctat gaactgatgg ttggtgaaga 4920
aaacaatatt ttggtgctgg gattcttttt ttttctggat gccagcttaa aaagcgggct 4980
ccattatatt tagtggatgc caggaataaa ctgttcaccc agacacctac gatgttatat 5040
attctgtgta acccgccccc tattttgggc atgtacgggt tacagcagaa ttaaaaggct 5100
aattttttga ctaaataaag ttaggaaaat cactactatt aattatttac gtattctttg 5160
aaatggcgag tattgataat gataaactgg atcctcaagc atctaaaaca caaccgttgg 5220
aagcgttgga aaccaactta gcatacttgg atagagtacc tcttgtgtaa cgaggtggag 5280
gtgcaaccca actttgttta cgttgagcca tttccttatc agagactaat aggtcaatct 5340
tgttattatc agcatcaatg ataatctcat cgccgtctct gaccaacccg ataggaccac 5400
cttcagcggc ttcgggaaca atgtggccga ttaagaaccc gtgagaacca ccagagaatc 5460
taccatcagt caacaatgca acatctttac ccaaaccgta acccatcaga gcagaggaag 5520
gctttagcat ttcaggcata cctggtgcac ctcttggacc ttcatatctg ataacaacaa 5580
cggttttttc acccttcttg atttcacctc tttccaaggc ttcaataaag gcaccttcct 5640
cttcgaacac acgtgctcta cccttgaagt aagtaccttc cttaccggta attttaccca 5700
cagctccacc tggtgccaat gaaccgtaca gaatttgcaa gtgaccgttg gccttgattg 5760
ggtgggagag tggcttaata atctcttgtc cttcaggtag gcttggtgct ttctttgcac 5820
gttctgccaa agtgtcaccg gtaacagtca ttgtgttacc gtgcaacatg ttgttttcat 5880
atagatactt aatcacagat tgggtaccac caacgttaat caaatcggcc atgacgtatt 5940
taccagaagg tttgaagtca ccgatcaatg gtgtagtatc actgattctt tggaaatcat 6000
ctggtgacaa cttgacaccc gcagagtgag caacagccac caaatgcaaa acagcattag 6060
tggacccacc ggttgcaacg acataagtaa tggcgttttc aaaagcctct tttgtgagga 6120
tatcacgagg taaaataccc aattccattg tcttcttgat gtattcacca atgttgtcac 6180
actcagctaa cttctccttg gaaacggctg ggaaggaaga ggagtttgga atggtcaaac 6240
ctagcacttc agcggcagaa gccattgtgt tggcagtata cataccacca caagaaccag 6300
gacctgggca tgcatgttcc acaacatctt ctctttcttc ttcagtgaat tgcttggaaa 6360
tatattcacc gtaggattgg aacgcagaga cgatatcgat gtttttagag atcttcgaag 6420
aaccacatgt tggatgaccg ggcaagatag taccaccata taccatgatg gaaggtctgt 6480
tatgtctacc catggccatc atgacaccgg gcatgttttt gtcacatgat gggatggcga 6540
tgttagcatc gtagtgttgt gccatcatga tggtttcaaa ggagtctgca atgatttctc 6600
tactttgtaa cgagtatctc atacctttag tacccataga gataccgtct gaaacaccga 6660
tggtgttgaa ctgcatagct ttcaaacccg ctttttcaat ggattgagaa catctgttat 6720
tcaagtccaa tagatgcatg ttacatgggt taccggacca ccaacaggaa ccaaccccga 6780
cttgaggctt cttgaaatct tccttcttga aaccggtggc ataaagcatg gcctgggacg 6840
caccttggcc cttaggttca gtgatgatat acgagtactt gttgagcttc ttcatgtcga 6900
caaacttaga ttagattgct atgctttctt tctaatgagc aagaagtaaa aaaagttgta 6960
atagaacaag aaaaatgaaa ctgaaacttg agaaattgaa gaccgtttat taacttaaat 7020
atcaatggga ggtcatcgaa agagaaaaaa atcaaaaaaa aaattttcaa gaaaaagaaa 7080
cgtgataaaa atttttattg cctttttcga cgaagaaaaa gaaacgaggc ggtctctttt 7140
ttcttttcca aacctttagt acgggtaatt aacgacaccc tagaggaaga aagaggggaa 7200
atttagtatg ctgtgcttgg gtgttttgaa gtggtacggc gatgcgcgga gtccgagaaa 7260
atctggaaga gtaaaaaagg agtagaaaca ttttgaagct atgagctcca gcttttgttc 7320
cctttagtga gggttaattg cgcgcttggc gtaatcatgg tcatagctgt ttcctgtgtg 7380
aaattgttat ccgctcacaa ttccacacaa cataggagcc ggaagcataa agtgtaaagc 7440
ctggggtgcc taatgagtga ggtaactcac attaattgcg ttgcgctcac tgcccgcttt 7500
ccagtcggga aacctgtcgt gccagctgca ttaatgaatc ggccaacgcg cggggagagg 7560
cggtttgcgt attgggcgct cttccgcttc ctcgctcact gactcgctgc gctcggtcgt 7620
tcggctgcgg cgagcggtat cagctcactc aaaggcggta atacggttat ccacagaatc 7680
aggggataac gcaggaaaga acatgtgagc aaaaggccag caaaaggcca ggaaccgtaa 7740
aaaggccgcg ttgctggcgt ttttccatag gctccgcccc cctgacgagc atcacaaaaa 7800
tcgacgctca agtcagaggt ggcgaaaccc gacaggacta taaagatacc aggcgtttcc 7860
ccctggaagc tccctcgtgc gctctcctgt tccgaccctg ccgcttaccg gatacctgtc 7920
cgcctttctc ccttcgggaa gcgtggcgct ttctcatagc tcacgctgta ggtatctcag 7980
ttcggtgtag gtcgttcgct ccaagctggg ctgtgtgcac gaaccccccg ttcagcccga 8040
ccgctgcgcc ttatccggta actatcgtct tgagtccaac ccggtaagac acgacttatc 8100
gccactggca gcagccactg gtaacaggat tagcagagcg aggtatgtag gcggtgctac 8160
agagttcttg aagtggtggc ctaactacgg ctacactaga aggacagtat ttggtatctg 8220
cgctctgctg aagccagtta ccttcggaaa aagagttggt agctcttgat ccggcaaaca 8280
aaccaccgct ggtagcggtg gtttttttgt ttgcaagcag cagattacgc gcagaaaaaa 8340
aggatctcaa gaagatcctt tgatcttttc tacggggtct gacgctcagt ggaacgaaaa 8400
ctcacgttaa gggattttgg tcatgagatt atcaaaaagg atcttcacct agatcctttt 8460
aaattaaaaa tgaagtttta aatcaatcta aagtatatat gagtaaactt ggtctgacag 8520
ttaccaatgc ttaatcagtg aggcacctat ctcagcgatc tgtctatttc gttcatccat 8580
agttgcctga ctccccgtcg tgtagataac tacgatacgg gagggcttac catctggccc 8640
cagtgctgca atgataccgc gagacccacg ctcaccggct ccagatttat cagcaataaa 8700
ccagccagcc ggaagggccg agcgcagaag tggtcctgca actttatccg cctccatcca 8760
gtctattaat tgttgccggg aagctagagt aagtagttcg ccagttaata gtttgcgcaa 8820
cgttgttgcc attgctacag gcatcgtggt gtcacgctcg tcgtttggta tggcttcatt 8880
cagctccggt tcccaacgat caaggcgagt tacatgatcc cccatgttgt gcaaaaaagc 8940
ggttagctcc ttcggtcctc cgatcgttgt cagaagtaag ttggccgcag tgttatcact 9000
catggttatg gcagcactgc ataattctct tactgtcatg ccatccgtaa gatgcttttc 9060
tgtgactggt gagtactcaa ccaagtcatt ctgagaatag tgtatgcggc gaccgagttg 9120
ctcttgcccg gcgtcaatac gggataatac cgcgccacat agcagaactt taaaagtgct 9180
catcattgga aaacgttctt cggggcgaaa actctcaagg atcttaccgc tgttgagatc 9240
cagttcgatg taacccactc gtgcacccaa ctgatcttca gcatctttta ctttcaccag 9300
cgtttctggg tgagcaaaaa caggaaggca aaatgccgca aaaaagggaa taagggcgac 9360
acggaaatgt tgaatactca tactcttcct ttttcaatat tattgaagca tttatcaggg 9420
ttattgtctc atgagcggat acatatttga atgtatttag aaaaataaac aaataggggt 9480
tccgcgcaca tttccccgaa aagtgccacc tgacgt 9516
<210> SEQ ID NO 121
<211> LENGTH: 6679
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV2020
<400> SEQUENCE: 121
ttggatcata ctaagaaacc attattatca tgacattaac ctataaaaat aggcgtatca 60
cgaggccctt tcgtctcgcg cgtttcggtg atgacggtga aaacctctga cacatgcagc 120
tcccggagac ggtcacagct tgtctgtaag cggatgccgg gagcagacaa gcccgtcagg 180
gcgcgtcagc gggtgttggc gggtgtcggg gctggcttaa ctatgcggca tcagagcaga 240
ttgtactgag agtgcaccat accacagctt ttcaattcaa ttcatcattt tttttttatt 300
cttttttttg atttcggttt ctttgaaatt tttttgattc ggtaatctcc gaacagaagg 360
aagaacgaag gaaggagcac agacttagat tggtatatat acgcatatgg caaattaaag 420
ccttcgagcg tcccaaaacc ttctcaagca aggttttcag tataatgtta catgcgtaca 480
cgcgtctgta cagaaaaaaa agaaaaattt gaaatataaa taacgttctt aatactaaca 540
taactataaa aaaataaata gggacctaga cttcaggttg tctaactcct tccttttcgg 600
ttagagcgga tgtgggggga gggcgtgaat gtaagcgtga cataagaatt cttagaaaaa 660
ctcatcgagc atcaaatgaa actgcaattt attcatatca ggattatcaa taccatattt 720
ttgaaaaagc cgtttctgta atgaaggaga aaactcaccg aggcagttcc ataggatggc 780
aagatcctgg tatcggtctg cgatcccgac tcgtccaaca tcaatacaac ctattaattt 840
cccctcgtca aaaataaggt tatcaagtga gaaatcacca tgagtgacga ctgaatccgg 900
tgagaatggc aaaagcttat gcatttcttt ccagacttgt tcaacaggcc agccattacg 960
ctcgtcatca aaatcactcg cgtcaaccaa accgttattc attcgtgatt gcgcctgagc 1020
gaggcgaaat acgcgatcgc tgttaaaagg acaattacaa acaggaatcg aatgcaaccg 1080
gcgcaggaac actgccagcg catcaacaat attttcacct gaatcaggat attcttctaa 1140
tacctggaat gctgttttgc cggggatcgc agtggtgagt aaccatgcat catcaggagt 1200
acggacaaaa tgcttgatgg tcggaagagg cataaattcc gtcagccagt ttagtctgac 1260
catctcatct gcaacatcat tggcaacgct acctttgcca tgtttcagaa acaactctgg 1320
cgcatcgggc ttcccataca atcgatagat tgtcgcacct gattgcccga cattatcgcg 1380
agcccattta tacccatata aatcagcatc catgttggaa tttaatcgcg gcctcgaaac 1440
gtgagtcttt tccttaccca tactagtttt tagtttatgt atgtgttttt tgtagttata 1500
gatttaagca agaaaagaat acaaacaaaa aattgaaaaa gattgattta gaattaaaaa 1560
gaaaaatatt tacgtaagaa gggaaaatag taaatgttgc aagttcacta aactcctaaa 1620
ttatgctgcc ctttatattc cctgttacag cagccgagcc aaaggtatat aggctccttt 1680
gcattagcat gcgtaacaaa ccacctgtca gtttcaaccg aggtggtatc cgagagaatt 1740
gtgtgattgc tttaattaat ttcggagaat ctcacatgcc actgaagatt aaaaactgga 1800
tgccagaaaa ggggtgtcca ggtgtaacat caatagagga agctgaaaag tcttagaacg 1860
ggtaatcttc caccaacctg atgggttcct agatataatc tcgaagggaa taagtagggt 1920
gataccgcag aagtgtctga atgtattaag gtcctcacag tttaaatccc gctcacacta 1980
acgtaggatt attataactc aaaaaaatgg cattattcta agtaagttaa atatccgtaa 2040
tctttaaaca gcggccgcag atctctcgag tcgaaactaa gttctggtgt tttaaaacta 2100
aaaaaaagac taactataaa agtagaattt aagaagttta agaaatagat ttacagaatt 2160
acaatcaata cctaccgtct ttatatactt attagtcaag taggggaata atttcaggga 2220
actggtttca accttttttt tcagcttttt ccaaatcaga gagagcagaa ggtaatagaa 2280
ggtgtaagaa aatgagatag atacatgcgt gggtcaattg ccttgtgtca tcatttactc 2340
caggcaggtt gcatcactcc attgaggttg tgcccgtttt ttgcctgttt gtgcccctgt 2400
tctctgtagt tgcgctaaga gaatggacct atgaactgat ggttggtgaa gaaaacaata 2460
ttttggtgct gggattcttt ttttttctgg atgccagctt aaaaagcggg ctccattata 2520
tttagtggat gccaggaata aactgttcac ccagacacct acgatgttat atattctgtg 2580
taacccgccc cctattttgg gcatgtacgg gttacagcag aattaaaagg ctaatttttt 2640
gactaaataa agttaggaaa atcactacta ttaattattt acgtattctt tgaaatggcg 2700
agtattgata atgataaact ggatccgtcg acaaacttag attagattgc tatgctttct 2760
ttctaatgag caagaagtaa aaaaagttgt aatagaacaa gaaaaatgaa actgaaactt 2820
gagaaattga agaccgttta ttaacttaaa tatcaatggg aggtcatcga aagagaaaaa 2880
aatcaaaaaa aaaattttca agaaaaagaa acgtgataaa aatttttatt gcctttttcg 2940
acgaagaaaa agaaacgagg cggtctcttt tttcttttcc aaacctttag tacgggtaat 3000
taacgacacc ctagaggaag aaagagggga aatttagtat gctgtgcttg ggtgttttga 3060
agtggtacgg cgatgcgcgg agtccgagaa aatctggaag agtaaaaaag gagtagaaac 3120
attttgaagc tatgagctcc agcttttgtt ccctttagtg agggttaatt gcgcgcttgg 3180
cgtaatcatg gtcatagctg tttcctgtgt gaaattgtta tccgctcaca attccacaca 3240
acataggagc cggaagcata aagtgtaaag cctggggtgc ctaatgagtg aggtaactca 3300
cattaattgc gttgcgctca ctgcccgctt tccagtcggg aaacctgtcg tgccagctgc 3360
attaatgaat cggccaacgc gcggggagag gcggtttgcg tattgggcgc tcttccgctt 3420
cctcgctcac tgactcgctg cgctcggtcg ttcggctgcg gcgagcggta tcagctcact 3480
caaaggcggt aatacggtta tccacagaat caggggataa cgcaggaaag aacatgtgag 3540
caaaaggcca gcaaaaggcc aggaaccgta aaaaggccgc gttgctggcg tttttccata 3600
ggctccgccc ccctgacgag catcacaaaa atcgacgctc aagtcagagg tggcgaaacc 3660
cgacaggact ataaagatac caggcgtttc cccctggaag ctccctcgtg cgctctcctg 3720
ttccgaccct gccgcttacc ggatacctgt ccgcctttct cccttcggga agcgtggcgc 3780
tttctcatag ctcacgctgt aggtatctca gttcggtgta ggtcgttcgc tccaagctgg 3840
gctgtgtgca cgaacccccc gttcagcccg accgctgcgc cttatccggt aactatcgtc 3900
ttgagtccaa cccggtaaga cacgacttat cgccactggc agcagccact ggtaacagga 3960
ttagcagagc gaggtatgta ggcggtgcta cagagttctt gaagtggtgg cctaactacg 4020
gctacactag aaggacagta tttggtatct gcgctctgct gaagccagtt accttcggaa 4080
aaagagttgg tagctcttga tccggcaaac aaaccaccgc tggtagcggt ggtttttttg 4140
tttgcaagca gcagattacg cgcagaaaaa aaggatctca agaagatcct ttgatctttt 4200
ctacggggtc tgacgctcag tggaacgaaa actcacgtta agggattttg gtcatgagat 4260
tatcaaaaag gatcttcacc tagatccttt taaattaaaa atgaagtttt aaatcaatct 4320
aaagtatata tgagtaaact tggtctgaca gttaccaatg cttaatcagt gaggcaccta 4380
tctcagcgat ctgtctattt cgttcatcca tagttgcctg actccccgtc gtgtagataa 4440
ctacgatacg ggagggctta ccatctggcc ccagtgctgc aatgataccg cgagacccac 4500
gctcaccggc tccagattta tcagcaataa accagccagc cggaagggcc gagcgcagaa 4560
gtggtcctgc aactttatcc gcctccatcc agtctattaa ttgttgccgg gaagctagag 4620
taagtagttc gccagttaat agtttgcgca acgttgttgc cattgctaca ggcatcgtgg 4680
tgtcacgctc gtcgtttggt atggcttcat tcagctccgg ttcccaacga tcaaggcgag 4740
ttacatgatc ccccatgttg tgcaaaaaag cggttagctc cttcggtcct ccgatcgttg 4800
tcagaagtaa gttggccgca gtgttatcac tcatggttat ggcagcactg cataattctc 4860
ttactgtcat gccatccgta agatgctttt ctgtgactgg tgagtactca accaagtcat 4920
tctgagaata gtgtatgcgg cgaccgagtt gctcttgccc ggcgtcaata cgggataata 4980
ccgcgccaca tagcagaact ttaaaagtgc tcatcattgg aaaacgttct tcggggcgaa 5040
aactctcaag gatcttaccg ctgttgagat ccagttcgat gtaacccact cgtgcaccca 5100
actgatcttc agcatctttt actttcacca gcgtttctgg gtgagcaaaa acaggaaggc 5160
aaaatgccgc aaaaaaggga ataagggcga cacggaaatg ttgaatactc atactcttcc 5220
tttttcaata ttattgaagc atttatcagg gttattgtct catgagcgga tacatatttg 5280
aatgtattta gaaaaataaa caaatagggg ttccgcgcac atttccccga aaagtgccac 5340
ctgaacgaag catctgtgct tcattttgta gaacaaaaat gcaacgcgag agcgctaatt 5400
tttcaaacaa agaatctgag ctgcattttt acagaacaga aatgcaacgc gaaagcgcta 5460
ttttaccaac gaagaatctg tgcttcattt ttgtaaaaca aaaatgcaac gcgagagcgc 5520
taatttttca aacaaagaat ctgagctgca tttttacaga acagaaatgc aacgcgagag 5580
cgctatttta ccaacaaaga atctatactt cttttttgtt ctacaaaaat gcatcccgag 5640
agcgctattt ttctaacaaa gcatcttaga ttactttttt tctcctttgt gcgctctata 5700
atgcagtctc ttgataactt tttgcactgt aggtccgtta aggttagaag aaggctactt 5760
tggtgtctat tttctcttcc ataaaaaaag cctgactcca cttcccgcgt ttactgatta 5820
ctagcgaagc tgcgggtgca ttttttcaag ataaaggcat ccccgattat attctatacc 5880
gatgtggatt gcgcatactt tgtgaacaga aagtgatagc gttgatgatt cttcattggt 5940
cagaaaatta tgaacggttt cttctatttt gtctctatat actacgtata ggaaatgttt 6000
acattttcgt attgttttcg attcactcta tgaatagttc ttactacaat ttttttgtct 6060
aaagagtaat actagagata aacataaaaa atgtagaggt cgagtttaga tgcaagttca 6120
aggagcgaaa ggtggatggg taggttatat agggatatag cacagagata tatagcaaag 6180
agatactttt gagcaatgtt tgtggaagcg gtattcgcaa tattttagta gctcgttaca 6240
gtccggtgcg tttttggttt tttgaaagtg cgtcttcaga gcgcttttgg ttttcaaaag 6300
cgctctgaag ttcctatact ttctagagaa taggaacttc ggaataggaa cttcaaagcg 6360
tttccgaaaa cgagcgcttc cgaaaatgca acgcgagctg cgcacataca gctcactgtt 6420
cacgtcgcac ctatatctgc gtgttgcctg tatatatata tacatgagaa gaacggcata 6480
gtgcgtgttt atgcttaaat gcgtacttat atgcgtctat ttatgtagga tgaaaggtag 6540
tctagtacct cctgtgatat tatcccattc catgcggggt atcgtatgct tccttcagca 6600
ctacccttta gctgttctat atgctgccac tcctcaattg gattagtctc atccttcaat 6660
gctatcattt cctttgata 6679
<210> SEQ ID NO 122
<211> LENGTH: 13805
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV2082
<400> SEQUENCE: 122
ttggatcata ctaagaaacc attattatca tgacattaac ctataaaaat aggcgtatca 60
cgaggccctt tcgtctcgcg cgtttcggtg atgacggtga aaacctctga cacatgcagc 120
tcccggagac ggtcacagct tgtctgtaag cggatgccgg gagcagacaa gcccgtcagg 180
gcgcgtcagc gggtgttggc gggtgtcggg gctggcttaa ctatgcggca tcagagcaga 240
ttgtactgag agtgcaccat accacagctt ttcaattcaa ttcatcattt tttttttatt 300
cttttttttg atttcggttt ctttgaaatt tttttgattc ggtaatctcc gaacagaagg 360
aagaacgaag gaaggagcac agacttagat tggtatatat acgcatatgg caaattaaag 420
ccttcgagcg tcccaaaacc ttctcaagca aggttttcag tataatgtta catgcgtaca 480
cgcgtctgta cagaaaaaaa agaaaaattt gaaatataaa taacgttctt aatactaaca 540
taactataaa aaaataaata gggacctaga cttcaggttg tctaactcct tccttttcgg 600
ttagagcgga tgtgggggga gggcgtgaat gtaagcgtga cataactaat tacatgactc 660
gagcggccgc ttagatgccg gagtcccagt gcttggtcca ctggatggcc tccagggtgc 720
ccaagtccag tttccagatg gctccgttct ggttcagctc gatagccttg acgaagttct 780
cggcgcaggc caacgagggc tgggtgggat gagccaggag cttctcggca acctgaggct 840
caacatccaa ccaggagttg aacgtgtgca ccagggtggt gcgggtgatg ccggggttca 900
cagtgtaagc cgtcacgccg gtaatggggg ccagtttcgc cagggagctg gtgaagttga 960
ccacggcggc cttggtgccg gagtagacgg gcacctggta gatggcattg aatccagtga 1020
cggatccaat gttgcagatg ataccaccgg gaccgccctt gcgcttgtcc cagaagtcca 1080
gaatggccgt cgtggtgttg accaggccag tgtagttgac ggcaatggtg cgctcgatct 1140
ggtgatcgtc caggatacca gctccgttga tcaggacatc gacggtcttc agctgggcga 1200
agatggtctt cagcagcttg gtggtctcgg caatgggcac ggtcacatca taggggtaga 1260
aggtgacggt cacctttgga ttgattgcct tcagctcggc aatggcagcc gggttctcaa 1320
tgcggtcgag gatcaccagg ttcttcagat cgcgcttgag cagctccttg ctggtgtcca 1380
gaccaatgcc tcccagaccg gcaacgaaaa tcacgttctt gttggtcaaa gtaaacgaca 1440
tggtacctat tattgtatgt tatagtatta gttgcttggt gttatgaaag aaactaagaa 1500
aagaaaaata aaataaaaat aaaagattga gacaagggaa gaaaagatac aaaataagaa 1560
ttaattacaa ttgcgtttgc tataaatacg tttttaacaa tcaactctgg taggaagata 1620
atgctttttt tttttatata tgcttggtgc cacttgtcac atacaattct acaaccttcg 1680
acaaaaatcc aaatgatagt aagatcaaag ccagaaagca atggagaaaa aaaattaatg 1740
aaccacgatg aaccaaatga tcaatacaac caaagaaact accctagtga ggtgtatgct 1800
gacttggtat cacacttcat gaattttgca tatggcaaag tccacgaaag tgggcttcag 1860
aaaaaaggcg tgcggtgtgt agatgtatca attagtggat gccagttttg gaacgggatt 1920
ccactttccg caagttggtg cacgtcgtta gtgacataac gccgcgttca tctttgggaa 1980
gaagcagatg ctgagcgagg aggtactata gagtaaagaa ccctttctat acccgcagcc 2040
ccatggtaag tgacagtgca gtaataatat gaaccaattt atttttcgtt acataaaaat 2100
gcttataaaa ctttaactaa taattagaga ttaaatcgcg gccgcaaaag atccttagga 2160
tttattctgt tcagcaaaca gcttgcccat tttcttcagt accttcggtg cgccttcttt 2220
cgccaggatc agttcgatcc agtacatacg gttcggatcg gcctgggcct ctttcatcac 2280
gctcacaaat tcgttttcgg tacgcacaat tttagacaca acacggtcct cagttgcgcc 2340
gaaggactcc ggcagtttag agtagttcca catagggata tcgttgtaag actggttcgg 2400
accgtggatc tcacgctcaa cggtgtagcc gtcattgtta ataatgaagc aaatcgggtt 2460
gatcttttca cgaattgcca gacccagttc ctgtacggtc agctgcaggg aaccgtcacc 2520
gatgaacagc agatgacgag attctttatc agcgatctga gagcccagcg ctgccgggaa 2580
agtatagcca atgctacccc acagcggctg accgataaaa tggcttttgg atttcagaaa 2640
gatagaagac gcgccgaaaa agctcgtacc ttgttccgcc acgatggttt cattgctctg 2700
ggtcaggttc tccacggcct gccacaggcg atcctgggac agcagtgcgt tagatggtac 2760
gaaatcttct tgctttttgt caatgtattt gcctttatac tcgatttcgg acaggtccag 2820
cagagagctg atcaggcttt cgaagtcgaa gttctggata cgctcgttga agattttacc 2880
ctcgtcgatg ttcaggctaa tcattttgtt ttcgttcaga tggtgagtga atgcaccggt 2940
agaagagtcg gtcagtttaa cgcccagcat caggatgaag tccgcagatt caacaaattc 3000
tttcaggttc ggttcgctca gagtaccgtt gtagatgccc aggaaagacg gcagagcctc 3060
gtcaacagag gacttgccga agttcagggt ggtaatcggc agtttggttt tgctgatgaa 3120
ttgggtcacg gtcttctcca gaccaaaaga aatgatttcg tggccggtga tcacgattgg 3180
tttctttgcg tttttcagag actcctggat tttgttcagg atttcctggt cgctagtgtt 3240
agaagtggag ttttctttct tcagcggcag gctcggtttt tccgctttag ctgccgcaac 3300
atccacaggc aggttgatgt aaactggttt gcgttctttc agcagcgcag acagaacgcg 3360
gtcgatttcc acagtagcgt tctctgcagt cagcagcgta cgtgccgcag tcacaggttc 3420
atgcattttc atgaagtgtt tgaaatcgcc gtcagccaga gtgtggtgga cgaatttacc 3480
ttcgttctga actttgctcg ttgggctgcc tacgatctcc accaccggca ggttttcggc 3540
gtaggagccc gccagaccgt tgacggcgct cagttcgcca acaccgaaag tggtcagaaa 3600
tgccgcggct ttcttggtac gtgcataacc atctgccatg tagcttgcgt tcagttcgtt 3660
agcgttaccc acccatttca tgtctttatg agagatgatc tgatccagga actgcagatt 3720
gtaatcaccc ggaacgccga agatttcttc gatacccagt tcatgcagac ggtccagcag 3780
ataatcacca acagtataca tgtcgagctt gttttatatt tgttgtaaaa agtagataat 3840
tacttccttg atgatctgta aaaaagagaa aaagaaagca tctaagaact tgaaaaacta 3900
cgaattagaa aagaccaaat atgtatttct tgcattgacc aatttatgca agtttatata 3960
tatgtaaatg taagtttcac gaggttctac taaactaaac cacccccttg gttagaagaa 4020
aagagtgtgt gagaacaggc tgttgttgtc acacgattcg gacaattctg tttgaaagag 4080
agagagtaac agtacgatcg aacgaacttt gctctggaga tcacagtggg catcatagca 4140
tgtggtacta aaccctttcc cgccattcca gaaccttcga ttgcttgtta caaaacctgt 4200
gagccgtcgc taggaccttg ttgtgtgacg aaattggaag ctgcaatcaa taggaagaca 4260
ggaagtcgag cgtgtctggg ttttttcagt tttgttcttt ttgcaaacaa atcacgagcg 4320
acggtaattt ctttctcgat aagaggccac gtgctttatg agggtaacat caattcaaga 4380
aggagggaaa cacttccttt ttctggccct gataatagta tgagggtgaa gccaaaataa 4440
aggattcgcg cccaaatcgg catctttaaa tgcaggtatg cgatagttcc tcactctttc 4500
cttactcacg agtaattctt gcaaatgcct attatgcaga tgttataata tctgtgcgtc 4560
ttgagttgag cctagaattc ttagaaaaac tcatcgagca tcaaatgaaa ctgcaattta 4620
ttcatatcag gattatcaat accatatttt tgaaaaagcc gtttctgtaa tgaaggagaa 4680
aactcaccga ggcagttcca taggatggca agatcctggt atcggtctgc gatcccgact 4740
cgtccaacat caatacaacc tattaatttc ccctcgtcaa aaataaggtt atcaagtgag 4800
aaatcaccat gagtgacgac tgaatccggt gagaatggca aaagcttatg catttctttc 4860
cagacttgtt caacaggcca gccattacgc tcgtcatcaa aatcactcgc gtcaaccaaa 4920
ccgttattca ttcgtgattg cgcctgagcg aggcgaaata cgcgatcgct gttaaaagga 4980
caattacaaa caggaatcga atgcaaccgg cgcaggaaca ctgccagcgc atcaacaata 5040
ttttcacctg aatcaggata ttcttctaat acctggaatg ctgttttgcc ggggatcgca 5100
gtggtgagta accatgcatc atcaggagta cggacaaaat gcttgatggt cggaagaggc 5160
ataaattccg tcagccagtt tagtctgacc atctcatctg caacatcatt ggcaacgcta 5220
cctttgccat gtttcagaaa caactctggc gcatcgggct tcccatacaa tcgatagatt 5280
gtcgcacctg attgcccgac attatcgcga gcccatttat acccatataa atcagcatcc 5340
atgttggaat ttaatcgcgg cctcgaaacg tgagtctttt ccttacccat actagttttt 5400
agtttatgta tgtgtttttt gtagttatag atttaagcaa gaaaagaata caaacaaaaa 5460
attgaaaaag attgatttag aattaaaaag aaaaatattt acgtaagaag ggaaaatagt 5520
aaatgttgca agttcactaa actcctaaat tatgctgccc tttatattcc ctgttacagc 5580
agccgagcca aaggtatata ggctcctttg cattagcatg cgtaacaaac cacctgtcag 5640
tttcaaccga ggtggtatcc gagagaattg tgtgattgct ttaattaatt tcggagaatc 5700
tcacatgcca ctgaagatta aaaactggat gccagaaaag gggtgtccag gtgtaacatc 5760
aatagaggaa gctgaaaagt cttagaacgg gtaatcttcc accaacctga tgggttccta 5820
gatataatct cgaagggaat aagtagggtg ataccgcaga agtgtctgaa tgtattaagg 5880
tcctcacagt ttaaatcccg ctcacactaa cgtaggatta ttataactca aaaaaatggc 5940
attattctaa gtaagttaaa tatccgtaat ctttaaacag cggccgcgga tcttcatcct 6000
gccactgcaa ttcttttcat atcggtcata tatcctctca gctttttacc cacctgttct 6060
atagcatgtg aacgaatagc ttcatttacg tctctcagtt ggccattgtc aaccgctcct 6120
tccggaatag ccttccccaa atcaccaggt tgtaactcgg ccatgaaggg ctttaacaac 6180
gggacacatg cgtagctaaa taagtaatta ccatattctg cagtgtctga tatgacaaca 6240
ttcatctcgt aaagtctttt tcttgcaata gtatttgcta tcaaaggcaa ttcatgcaaa 6300
gactcatagt atgcagattc ttcaatgata ccggagtcaa ccatagtttc gaatgcaagt 6360
tctacccctg ccttcaccat agctatcatc aatactccct tatcaaagta ttcttgttca 6420
ccaattttac cttcgtattg tggggctgtc tcgaatgccg tcttgccggt ttcttctctc 6480
cacgtcaata actttttatc atcgtttgcc caatctgcca tcattcctga ggaaaactca 6540
ccggagataa tatcgtccat gtgcttttgg aataatggtg ccatgatctc ttttagttgc 6600
tcagataagg cgtaggctct tagcttggcc ggatttgaaa gtctatccat catcaatgtt 6660
atgccacctt gtttaagtgc ctcggtgatt gtctcccaac caaattgtat caacttttca 6720
gcataggcag gatctgtacc ctcttcgacc aatttatcaa agcatagtaa agaccctgcc 6780
tgcaacattc cgcacagaat ggtttgttca cccattaagt cactcttgac ctcagctacg 6840
aaagaactct ctaacacacc cgctctatga cctccggttg cggctgccca tgccttcgca 6900
attgccatac cttcaccttt ggggtcattt tcaggatgta cggcgatcaa tgtaggtaca 6960
ccaaaacccc tcttgtactc ctctctgact tccgtacctg ggcactttgg tgcaaccatt 7020
acgactgtta tatcttttct gatctgctcg cccacttcaa cgatattaaa gccatgagag 7080
taacctaaag ctgccccatc cttcatcagc ggttgaactg ttcttactac gtctgagtga 7140
accttatctg gtgttaggtt aatcactaaa tctgcctgag ggatcagttc ttcgtaagta 7200
ccaactttga acccattttc cgtcgcttta cgccaggagg ccctcttttc tgcaattgcc 7260
tctttcctca atgcatacga aatatccaga cctgaatctc tcatgtttaa accttggttt 7320
agaccctgag caccgcagcc aacaattact actttctttc cttgcagata agaagcacca 7380
tcagcaaact cgtcccttcc cataaatctg cacttaccca gttgagccaa ttgttgtctc 7440
aaatttaatg tgttaaaata gttggccatc tcgagtcgaa actaagttct ggtgttttaa 7500
aactaaaaaa aagactaact ataaaagtag aatttaagaa gtttaagaaa tagatttaca 7560
gaattacaat caatacctac cgtctttata tacttattag tcaagtaggg gaataatttc 7620
agggaactgg tttcaacctt ttttttcagc tttttccaaa tcagagagag cagaaggtaa 7680
tagaaggtgt aagaaaatga gatagataca tgcgtgggtc aattgccttg tgtcatcatt 7740
tactccaggc aggttgcatc actccattga ggttgtgccc gttttttgcc tgtttgtgcc 7800
cctgttctct gtagttgcgc taagagaatg gacctatgaa ctgatggttg gtgaagaaaa 7860
caatattttg gtgctgggat tctttttttt tctggatgcc agcttaaaaa gcgggctcca 7920
ttatatttag tggatgccag gaataaactg ttcacccaga cacctacgat gttatatatt 7980
ctgtgtaacc cgccccctat tttgggcatg tacgggttac agcagaatta aaaggctaat 8040
tttttgacta aataaagtta ggaaaatcac tactattaat tatttacgta ttctttgaaa 8100
tggcgagtat tgataatgat aaactggatc cgcggccgct tacagatcag taacacaccc 8160
ttccgatgca ggacgggtta atttagcgaa ttttgccaaa actcccctgg tggctttcgg 8220
agttggcttc tgataattag ctcttctctt tgcgatttct tcatcggaaa ctttcaggga 8280
tatagagttg ttgactgcat ctatctctat tatatcgtca tcttcaacta agccgattag 8340
tccaccctca acggcttcag gcacaatatg gccgacaaca aaaccgtgag tgccaccgga 8400
gaatctacca tccgtaatta acgcgcaact tttccctaaa cccgcaccaa ttaatgctga 8460
tgtaggcttc agcatttcgg gcataccagg tccgccgacg ggacctatat tcctaattac 8520
cgctacatct ccagcatgca aacgaccaga ttctatgccg tcgataaaat gttgttcacc 8580
atcaaagact ctggcagtgc ctttgaagaa ctctccttct ttaccgctaa tttttgctac 8640
ggaaccccct tgagctaaat taccgtacag aatctgcaag tggccggtgg ccttgatagg 8700
attctttagt ggcctcatga tatcttgtga gtcgaaatcc aagtctaggg cagtctcgac 8760
attctcggct aatgttttac ccgtcacagt aaggcagtca ccatgcaatt ttccttcctt 8820
tagaaggtac ttaagcactg ctggcaagcc tccaatttta tgcaaatctt ccatcatata 8880
tttacctgaa ggtttaaaat cacctagtac tggagtaatg tcactaattc tttggaagtc 8940
atcctgagtt atttcgacac ctatcgcgtt agccattgca ataatatgca agacagcatt 9000
agtactaccc cccaagacca tcacaatggt aatagcgttc tcgaacgcct ccttagtcat 9060
tatatcacta ggcttgatgt ctttttccaa aagattctta atggctaatc caatctcatc 9120
acattcttct tgtttttctt gagatactgc agggttcgaa gaagaatacg gcaatgacat 9180
acctagtgtt tcgatagcgg cagctaaggt attagctgtg tacatccccc cacatgcccc 9240
ttgaccagga atagcattac aaataacacc gtgataatct tcatcagaga tattgccggt 9300
aattttctgg cctagagatt caaaagccga tacgatgttc aatttctcac ctttatattc 9360
accgtgttct attgttcctc catacaccat aatgcttggc ctattaagtc ttgccatacc 9420
aataatagaa cctggcatat ttttgtcaca acctgggatg gctacaattg catcatagta 9480
ttcagcgcca gcgttggttt caatagagtc agctataact tctctggaaa caagggagta 9540
tctcattccc aactttccat ttgctatccc atcagaaact cctatcgtat gaaattgtaa 9600
gccgatcaga ccatctgtct gatttactga gcttttaatc tttgatccaa gggttcctaa 9660
atgcatgttg catggatttc catcccagtc catcgacact atacccactt gagctttctt 9720
gaaatcttcg tctttaaacc cgatgccgta atacattgcc tgtgtggcgg gttgtgtggg 9780
atcttgtgtc aacgttttgc tgtacttatt cagttcaaca gattcaactt tgccgttata 9840
cttaaactcc atgtcgacaa acttagatta gattgctatg ctttctttct aatgagcaag 9900
aagtaaaaaa agttgtaata gaacaagaaa aatgaaactg aaacttgaga aattgaagac 9960
cgtttattaa cttaaatatc aatgggaggt catcgaaaga gaaaaaaatc aaaaaaaaaa 10020
ttttcaagaa aaagaaacgt gataaaaatt tttattgcct ttttcgacga agaaaaagaa 10080
acgaggcggt ctcttttttc ttttccaaac ctttagtacg ggtaattaac gacaccctag 10140
aggaagaaag aggggaaatt tagtatgctg tgcttgggtg ttttgaagtg gtacggcgat 10200
gcgcggagtc cgagaaaatc tggaagagta aaaaaggagt agaaacattt tgaagctatg 10260
agctccagct tttgttccct ttagtgaggg ttaattgcgc gcttggcgta atcatggtca 10320
tagctgtttc ctgtgtgaaa ttgttatccg ctcacaattc cacacaacat aggagccgga 10380
agcataaagt gtaaagcctg gggtgcctaa tgagtgaggt aactcacatt aattgcgttg 10440
cgctcactgc ccgctttcca gtcgggaaac ctgtcgtgcc agctgcatta atgaatcggc 10500
caacgcgcgg ggagaggcgg tttgcgtatt gggcgctctt ccgcttcctc gctcactgac 10560
tcgctgcgct cggtcgttcg gctgcggcga gcggtatcag ctcactcaaa ggcggtaata 10620
cggttatcca cagaatcagg ggataacgca ggaaagaaca tgtgagcaaa aggccagcaa 10680
aaggccagga accgtaaaaa ggccgcgttg ctggcgtttt tccataggct ccgcccccct 10740
gacgagcatc acaaaaatcg acgctcaagt cagaggtggc gaaacccgac aggactataa 10800
agataccagg cgtttccccc tggaagctcc ctcgtgcgct ctcctgttcc gaccctgccg 10860
cttaccggat acctgtccgc ctttctccct tcgggaagcg tggcgctttc tcatagctca 10920
cgctgtaggt atctcagttc ggtgtaggtc gttcgctcca agctgggctg tgtgcacgaa 10980
ccccccgttc agcccgaccg ctgcgcctta tccggtaact atcgtcttga gtccaacccg 11040
gtaagacacg acttatcgcc actggcagca gccactggta acaggattag cagagcgagg 11100
tatgtaggcg gtgctacaga gttcttgaag tggtggccta actacggcta cactagaagg 11160
acagtatttg gtatctgcgc tctgctgaag ccagttacct tcggaaaaag agttggtagc 11220
tcttgatccg gcaaacaaac caccgctggt agcggtggtt tttttgtttg caagcagcag 11280
attacgcgca gaaaaaaagg atctcaagaa gatcctttga tcttttctac ggggtctgac 11340
gctcagtgga acgaaaactc acgttaaggg attttggtca tgagattatc aaaaaggatc 11400
ttcacctaga tccttttaaa ttaaaaatga agttttaaat caatctaaag tatatatgag 11460
taaacttggt ctgacagtta ccaatgctta atcagtgagg cacctatctc agcgatctgt 11520
ctatttcgtt catccatagt tgcctgactc cccgtcgtgt agataactac gatacgggag 11580
ggcttaccat ctggccccag tgctgcaatg ataccgcgag acccacgctc accggctcca 11640
gatttatcag caataaacca gccagccgga agggccgagc gcagaagtgg tcctgcaact 11700
ttatccgcct ccatccagtc tattaattgt tgccgggaag ctagagtaag tagttcgcca 11760
gttaatagtt tgcgcaacgt tgttgccatt gctacaggca tcgtggtgtc acgctcgtcg 11820
tttggtatgg cttcattcag ctccggttcc caacgatcaa ggcgagttac atgatccccc 11880
atgttgtgca aaaaagcggt tagctccttc ggtcctccga tcgttgtcag aagtaagttg 11940
gccgcagtgt tatcactcat ggttatggca gcactgcata attctcttac tgtcatgcca 12000
tccgtaagat gcttttctgt gactggtgag tactcaacca agtcattctg agaatagtgt 12060
atgcggcgac cgagttgctc ttgcccggcg tcaatacggg ataataccgc gccacatagc 12120
agaactttaa aagtgctcat cattggaaaa cgttcttcgg ggcgaaaact ctcaaggatc 12180
ttaccgctgt tgagatccag ttcgatgtaa cccactcgtg cacccaactg atcttcagca 12240
tcttttactt tcaccagcgt ttctgggtga gcaaaaacag gaaggcaaaa tgccgcaaaa 12300
aagggaataa gggcgacacg gaaatgttga atactcatac tcttcctttt tcaatattat 12360
tgaagcattt atcagggtta ttgtctcatg agcggataca tatttgaatg tatttagaaa 12420
aataaacaaa taggggttcc gcgcacattt ccccgaaaag tgccacctga acgaagcatc 12480
tgtgcttcat tttgtagaac aaaaatgcaa cgcgagagcg ctaatttttc aaacaaagaa 12540
tctgagctgc atttttacag aacagaaatg caacgcgaaa gcgctatttt accaacgaag 12600
aatctgtgct tcatttttgt aaaacaaaaa tgcaacgcga gagcgctaat ttttcaaaca 12660
aagaatctga gctgcatttt tacagaacag aaatgcaacg cgagagcgct attttaccaa 12720
caaagaatct atacttcttt tttgttctac aaaaatgcat cccgagagcg ctatttttct 12780
aacaaagcat cttagattac tttttttctc ctttgtgcgc tctataatgc agtctcttga 12840
taactttttg cactgtaggt ccgttaaggt tagaagaagg ctactttggt gtctattttc 12900
tcttccataa aaaaagcctg actccacttc ccgcgtttac tgattactag cgaagctgcg 12960
ggtgcatttt ttcaagataa aggcatcccc gattatattc tataccgatg tggattgcgc 13020
atactttgtg aacagaaagt gatagcgttg atgattcttc attggtcaga aaattatgaa 13080
cggtttcttc tattttgtct ctatatacta cgtataggaa atgtttacat tttcgtattg 13140
ttttcgattc actctatgaa tagttcttac tacaattttt ttgtctaaag agtaatacta 13200
gagataaaca taaaaaatgt agaggtcgag tttagatgca agttcaagga gcgaaaggtg 13260
gatgggtagg ttatataggg atatagcaca gagatatata gcaaagagat acttttgagc 13320
aatgtttgtg gaagcggtat tcgcaatatt ttagtagctc gttacagtcc ggtgcgtttt 13380
tggttttttg aaagtgcgtc ttcagagcgc ttttggtttt caaaagcgct ctgaagttcc 13440
tatactttct agagaatagg aacttcggaa taggaacttc aaagcgtttc cgaaaacgag 13500
cgcttccgaa aatgcaacgc gagctgcgca catacagctc actgttcacg tcgcacctat 13560
atctgcgtgt tgcctgtata tatatataca tgagaagaac ggcatagtgc gtgtttatgc 13620
ttaaatgcgt acttatatgc gtctatttat gtaggatgaa aggtagtcta gtacctcctg 13680
tgatattatc ccattccatg cggggtatcg tatgcttcct tcagcactac cctttagctg 13740
ttctatatgc tgccactcct caattggatt agtctcatcc ttcaatgcta tcatttcctt 13800
tgata 13805
<210> SEQ ID NO 123
<211> LENGTH: 14056
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV2227
<400> SEQUENCE: 123
ttggatcata ctaagaaacc attattatca tgacattaac ctataaaaat aggcgtatca 60
cgaggccctt tcgtctcgcg cgtttcggtg atgacggtga aaacctctga cacatgcagc 120
tcccggagac ggtcacagct tgtctgtaag cggatgccgg gagcagacaa gcccgtcagg 180
gcgcgtcagc gggtgttggc gggtgtcggg gctggcttaa ctatgcggca tcagagcaga 240
ttgtactgag agtgcaccat accacagctt ttcaattcaa ttcatcattt tttttttatt 300
cttttttttg atttcggttt ctttgaaatt tttttgattc ggtaatctcc gaacagaagg 360
aagaacgaag gaaggagcac agacttagat tggtatatat acgcatatgg caaattaaag 420
ccttcgagcg tcccaaaacc ttctcaagca aggttttcag tataatgtta catgcgtaca 480
cgcgtctgta cagaaaaaaa agaaaaattt gaaatataaa taacgttctt aatactaaca 540
taactataaa aaaataaata gggacctaga cttcaggttg tctaactcct tccttttcgg 600
ttagagcgga tgtgggggga gggcgtgaat gtaagcgtga cataactaat tacatgactc 660
gacctaggtt atttagtaaa atcaatgacc attcggcctt caatttttcc tgccttcatt 720
tcatcaataa tatcattgat ttcttccagt ttgcgtgtcg caacaattgg ttttacctta 780
ccttctgctc caaattgaaa agcttctgcc aagtcaagtc ttgttccgac aagtgaacct 840
gcaacctcca ctccgtcaaa aacaactgtt ggaactgata aagtcatctc agtattggga 900
agtgccacag caaccatttt gcccataggt ttcaaagaag caaccgcttg ttcaaaagca 960
atccttgcaa cagcacaaac tattgcactt tgcaccccta agccgccagt tattttttta 1020
atttcatcaa ctggatttac atcaccagaa ttgataatca catcagctcc aattttttta 1080
gctaaattta atttatcttg attaatatca acagcaatta cttttgctcc aaaaacattt 1140
ttagcatatt gaattgctaa atttccaagt cctccagcac caaaaattac ttgccaatca 1200
ccaggtttta ctcctgatac tttgattgct ttgtaagttg ttactccagc acaagtaatt 1260
gagctagctt caattgggtc aagtccgtca ggaactttga cagcataatc ggcaacaaca 1320
attgcttctt cagccattcc gccatcaact gaatatcctg catttttaac ttctcgacaa 1380
aaagtttcat taccagatac acagtattca cagtgaccac atccttcaaa gaaccaagcc 1440
actgaaaccc gatcaccaac ttgaagcgag cttacatcag ctccaatttc tttgacaatt 1500
ccaattcctt catgaccaag aacagtccct gctttgttgc cataatcacc tgctgcaacg 1560
tgcaaatcgg tatgacagac tccacaatac tccatgtcaa gcaaagcttc attaggtttg 1620
attgctcgaa gttccttttc aacaaggtcc gcataaccat ctggattgtg tcttactact 1680
gctgctttca ttggtaccta ttattgtatg ttatagtatt agttgcttgg tgttatgaaa 1740
gaaactaaga aaagaaaaat aaaataaaaa taaaagattg agacaaggga agaaaagata 1800
caaaataaga attaattaca attgcgtttg ctataaatac gtttttaaca atcaactctg 1860
gtaggaagat aatgcttttt ttttttatat atgcttggtg ccacttgtca catacaattc 1920
tacaaccttc gacaaaaatc caaatgatag taagatcaaa gccagaaagc aatggagaaa 1980
aaaaattaat gaaccacgat gaaccaaatg atcaatacaa ccaaagaaac taccctagtg 2040
aggtgtatgc tgacttggta tcacacttca tgaattttgc atatggcaaa gtccacgaaa 2100
gtgggcttca gaaaaaaggc gtgcggtgtg tagatgtatc aattagtgga tgccagtttt 2160
ggaacgggat tccactttcc gcaagttggt gcacgtcgtt agtgacataa cgccgcgttc 2220
atctttggga agaagcagat gctgagcgag gaggtactat agagtaaaga accctttcta 2280
tacccgcagc cccatggtaa gtgacagtgc agtaataata tgaaccaatt tatttttcgt 2340
tacataaaaa tgcttataaa actttaacta ataattagag attaaatcgc ggccgcaaaa 2400
gatccttagg atttattctg ttcagcaaac agcttgccca ttttcttcag taccttcggt 2460
gcgccttctt tcgccaggat cagttcgatc cagtacatac ggttcggatc ggcctgggcc 2520
tctttcatca cgctcacaaa ttcgttttcg gtacgcacaa ttttagacac aacacggtcc 2580
tcagttgcgc cgaaggactc cggcagttta gagtagttcc acatagggat atcgttgtaa 2640
gactggttcg gaccgtggat ctcacgctca acggtgtagc cgtcattgtt aataatgaag 2700
caaatcgggt tgatcttttc acgaattgcc agacccagtt cctgtacggt cagctgcagg 2760
gaaccgtcac cgatgaacag cagatgacga gattctttat cagcgatctg agagcccagc 2820
gctgccggga aagtatagcc aatgctaccc cacagcggct gaccgataaa atggcttttg 2880
gatttcagaa agatagaaga cgcgccgaaa aagctcgtac cttgttccgc cacgatggtt 2940
tcattgctct gggtcaggtt ctccacggcc tgccacaggc gatcctggga cagcagtgcg 3000
ttagatggta cgaaatcttc ttgctttttg tcaatgtatt tgcctttata ctcgatttcg 3060
gacaggtcca gcagagagct gatcaggctt tcgaagtcga agttctggat acgctcgttg 3120
aagattttac cctcgtcgat gttcaggcta atcattttgt tttcgttcag atggtgagtg 3180
aatgcaccgg tagaagagtc ggtcagttta acgcccagca tcaggatgaa gtccgcagat 3240
tcaacaaatt ctttcaggtt cggttcgctc agagtaccgt tgtagatgcc caggaaagac 3300
ggcagagcct cgtcaacaga ggacttgccg aagttcaggg tggtaatcgg cagtttggtt 3360
ttgctgatga attgggtcac ggtcttctcc agaccaaaag aaatgatttc gtggccggtg 3420
atcacgattg gtttctttgc gtttttcaga gactcctgga ttttgttcag gatttcctgg 3480
tcgctagtgt tagaagtgga gttttctttc ttcagcggca ggctcggttt ttccgcttta 3540
gctgccgcaa catccacagg caggttgatg taaactggtt tgcgttcttt cagcagcgca 3600
gacagaacgc ggtcgatttc cacagtagcg ttctctgcag tcagcagcgt acgtgccgca 3660
gtcacaggtt catgcatttt catgaagtgt ttgaaatcgc cgtcagccag agtgtggtgg 3720
acgaatttac cttcgttctg aactttgctc gttgggctgc ctacgatctc caccaccggc 3780
aggttttcgg cgtaggagcc cgccagaccg ttgacggcgc tcagttcgcc aacaccgaaa 3840
gtggtcagaa atgccgcggc tttcttggta cgtgcataac catctgccat gtagcttgcg 3900
ttcagttcgt tagcgttacc cacccatttc atgtctttat gagagatgat ctgatccagg 3960
aactgcagat tgtaatcacc cggaacgccg aagatttctt cgatacccag ttcatgcaga 4020
cggtccagca gataatcacc aacagtatac atgtcgagct tgttttatat ttgttgtaaa 4080
aagtagataa ttacttcctt gatgatctgt aaaaaagaga aaaagaaagc atctaagaac 4140
ttgaaaaact acgaattaga aaagaccaaa tatgtatttc ttgcattgac caatttatgc 4200
aagtttatat atatgtaaat gtaagtttca cgaggttcta ctaaactaaa ccaccccctt 4260
ggttagaaga aaagagtgtg tgagaacagg ctgttgttgt cacacgattc ggacaattct 4320
gtttgaaaga gagagagtaa cagtacgatc gaacgaactt tgctctggag atcacagtgg 4380
gcatcatagc atgtggtact aaaccctttc ccgccattcc agaaccttcg attgcttgtt 4440
acaaaacctg tgagccgtcg ctaggacctt gttgtgtgac gaaattggaa gctgcaatca 4500
ataggaagac aggaagtcga gcgtgtctgg gttttttcag ttttgttctt tttgcaaaca 4560
aatcacgagc gacggtaatt tctttctcga taagaggcca cgtgctttat gagggtaaca 4620
tcaattcaag aaggagggaa acacttcctt tttctggccc tgataatagt atgagggtga 4680
agccaaaata aaggattcgc gcccaaatcg gcatctttaa atgcaggtat gcgatagttc 4740
ctcactcttt ccttactcac gagtaattct tgcaaatgcc tattatgcag atgttataat 4800
atctgtgcgt cttgagttga gcctagaatt cttagaaaaa ctcatcgagc atcaaatgaa 4860
actgcaattt attcatatca ggattatcaa taccatattt ttgaaaaagc cgtttctgta 4920
atgaaggaga aaactcaccg aggcagttcc ataggatggc aagatcctgg tatcggtctg 4980
cgatcccgac tcgtccaaca tcaatacaac ctattaattt cccctcgtca aaaataaggt 5040
tatcaagtga gaaatcacca tgagtgacga ctgaatccgg tgagaatggc aaaagcttat 5100
gcatttcttt ccagacttgt tcaacaggcc agccattacg ctcgtcatca aaatcactcg 5160
cgtcaaccaa accgttattc attcgtgatt gcgcctgagc gaggcgaaat acgcgatcgc 5220
tgttaaaagg acaattacaa acaggaatcg aatgcaaccg gcgcaggaac actgccagcg 5280
catcaacaat attttcacct gaatcaggat attcttctaa tacctggaat gctgttttgc 5340
cggggatcgc agtggtgagt aaccatgcat catcaggagt acggacaaaa tgcttgatgg 5400
tcggaagagg cataaattcc gtcagccagt ttagtctgac catctcatct gcaacatcat 5460
tggcaacgct acctttgcca tgtttcagaa acaactctgg cgcatcgggc ttcccataca 5520
atcgatagat tgtcgcacct gattgcccga cattatcgcg agcccattta tacccatata 5580
aatcagcatc catgttggaa tttaatcgcg gcctcgaaac gtgagtcttt tccttaccca 5640
tactagtttt tagtttatgt atgtgttttt tgtagttata gatttaagca agaaaagaat 5700
acaaacaaaa aattgaaaaa gattgattta gaattaaaaa gaaaaatatt tacgtaagaa 5760
gggaaaatag taaatgttgc aagttcacta aactcctaaa ttatgctgcc ctttatattc 5820
cctgttacag cagccgagcc aaaggtatat aggctccttt gcattagcat gcgtaacaaa 5880
ccacctgtca gtttcaaccg aggtggtatc cgagagaatt gtgtgattgc tttaattaat 5940
ttcggagaat ctcacatgcc actgaagatt aaaaactgga tgccagaaaa ggggtgtcca 6000
ggtgtaacat caatagagga agctgaaaag tcttagaacg ggtaatcttc caccaacctg 6060
atgggttcct agatataatc tcgaagggaa taagtagggt gataccgcag aagtgtctga 6120
atgtattaag gtcctcacag tttaaatccc gctcacacta acgtaggatt attataactc 6180
aaaaaaatgg cattattcta agtaagttaa atatccgtaa tctttaaaca gcggccgcgg 6240
atcttcatcc tgccactgca attcttttca tatcggtcat atatcctctc agctttttac 6300
ccacctgttc tatagcatgt gaacgaatag cttcatttac gtctctcagt tggccattgt 6360
caaccgctcc ttccggaata gccttcccca aatcaccagg ttgtaactcg gccatgaagg 6420
gctttaacaa cgggacacat gcgtagctaa ataagtaatt accatattct gcagtgtctg 6480
atatgacaac attcatctcg taaagtcttt ttcttgcaat agtatttgct atcaaaggca 6540
attcatgcaa agactcatag tatgcagatt cttcaatgat accggagtca accatagttt 6600
cgaatgcaag ttctacccct gccttcacca tagctatcat caatactccc ttatcaaagt 6660
attcttgttc accaatttta ccttcgtatt gtggggctgt ctcgaatgcc gtcttgccgg 6720
tttcttctct ccacgtcaat aactttttat catcgtttgc ccaatctgcc atcattcctg 6780
aggaaaactc accggagata atatcgtcca tgtgcttttg gaataatggt gccatgatct 6840
cttttagttg ctcagataag gcgtaggctc ttagcttggc cggatttgaa agtctatcca 6900
tcatcaatgt tatgccacct tgtttaagtg cctcggtgat tgtctcccaa ccaaattgta 6960
tcaacttttc agcataggca ggatctgtac cctcttcgac caatttatca aagcatagta 7020
aagaccctgc ctgcaacatt ccgcacagaa tggtttgttc acccattaag tcactcttga 7080
cctcagctac gaaagaactc tctaacacac ccgctctatg acctccggtt gcggctgccc 7140
atgccttcgc aattgccata ccttcacctt tggggtcatt ttcaggatgt acggcgatca 7200
atgtaggtac accaaaaccc ctcttgtact cctctctgac ttccgtacct gggcactttg 7260
gtgcaaccat tacgactgtt atatcttttc tgatctgctc gcccacttca acgatattaa 7320
agccatgaga gtaacctaaa gctgccccat ccttcatcag cggttgaact gttcttacta 7380
cgtctgagtg aaccttatct ggtgttaggt taatcactaa atctgcctga gggatcagtt 7440
cttcgtaagt accaactttg aacccatttt ccgtcgcttt acgccaggag gccctctttt 7500
ctgcaattgc ctctttcctc aatgcatacg aaatatccag acctgaatct ctcatgttta 7560
aaccttggtt tagaccctga gcaccgcagc caacaattac tactttcttt ccttgcagat 7620
aagaagcacc atcagcaaac tcgtcccttc ccataaatct gcacttaccc agttgagcca 7680
attgttgtct caaatttaat gtgttaaaat agttggccat ctcgagtcga aactaagttc 7740
tggtgtttta aaactaaaaa aaagactaac tataaaagta gaatttaaga agtttaagaa 7800
atagatttac agaattacaa tcaataccta ccgtctttat atacttatta gtcaagtagg 7860
ggaataattt cagggaactg gtttcaacct tttttttcag ctttttccaa atcagagaga 7920
gcagaaggta atagaaggtg taagaaaatg agatagatac atgcgtgggt caattgcctt 7980
gtgtcatcat ttactccagg caggttgcat cactccattg aggttgtgcc cgttttttgc 8040
ctgtttgtgc ccctgttctc tgtagttgcg ctaagagaat ggacctatga actgatggtt 8100
ggtgaagaaa acaatatttt ggtgctggga ttcttttttt ttctggatgc cagcttaaaa 8160
agcgggctcc attatattta gtggatgcca ggaataaact gttcacccag acacctacga 8220
tgttatatat tctgtgtaac ccgcccccta ttttgggcat gtacgggtta cagcagaatt 8280
aaaaggctaa ttttttgact aaataaagtt aggaaaatca ctactattaa ttatttacgt 8340
attctttgaa atggcgagta ttgataatga taaactggat ccgcggccgc ttacagatca 8400
gtaacacacc cttccgatgc aggacgggtt aatttagcga attttgccaa aactcccctg 8460
gtggctttcg gagttggctt ctgataatta gctcttctct ttgcgatttc ttcatcggaa 8520
actttcaggg atatagagtt gttgactgca tctatctcta ttatatcgtc atcttcaact 8580
aagccgatta gtccaccctc aacggcttca ggcacaatat ggccgacaac aaaaccgtga 8640
gtgccaccgg agaatctacc atccgtaatt aacgcgcaac ttttccctaa acccgcacca 8700
attaatgctg atgtaggctt cagcatttcg ggcataccag gtccgccgac gggacctata 8760
ttcctaatta ccgctacatc tccagcatgc aaacgaccag attctatgcc gtcgataaaa 8820
tgttgttcac catcaaagac tctggcagtg cctttgaaga actctccttc tttaccgcta 8880
atttttgcta cggaaccccc ttgagctaaa ttaccgtaca gaatctgcaa gtggccggtg 8940
gccttgatag gattctttag tggcctcatg atatcttgtg agtcgaaatc caagtctagg 9000
gcagtctcga cattctcggc taatgtttta cccgtcacag taaggcagtc accatgcaat 9060
tttccttcct ttagaaggta cttaagcact gctggcaagc ctccaatttt atgcaaatct 9120
tccatcatat atttacctga aggtttaaaa tcacctagta ctggagtaat gtcactaatt 9180
ctttggaagt catcctgagt tatttcgaca cctatcgcgt tagccattgc aataatatgc 9240
aagacagcat tagtactacc ccccaagacc atcacaatgg taatagcgtt ctcgaacgcc 9300
tccttagtca ttatatcact aggcttgatg tctttttcca aaagattctt aatggctaat 9360
ccaatctcat cacattcttc ttgtttttct tgagatactg cagggttcga agaagaatac 9420
ggcaatgaca tacctagtgt ttcgatagcg gcagctaagg tattagctgt gtacatcccc 9480
ccacatgccc cttgaccagg aatagcatta caaataacac cgtgataatc ttcatcagag 9540
atattgccgg taattttctg gcctagagat tcaaaagccg atacgatgtt caatttctca 9600
cctttatatt caccgtgttc tattgttcct ccatacacca taatgcttgg cctattaagt 9660
cttgccatac caataataga acctggcata tttttgtcac aacctgggat ggctacaatt 9720
gcatcatagt attcagcgcc agcgttggtt tcaatagagt cagctataac ttctctggaa 9780
acaagggagt atctcattcc caactttcca tttgctatcc catcagaaac tcctatcgta 9840
tgaaattgta agccgatcag accatctgtc tgatttactg agcttttaat ctttgatcca 9900
agggttccta aatgcatgtt gcatggattt ccatcccagt ccatcgacac tatacccact 9960
tgagctttct tgaaatcttc gtctttaaac ccgatgccgt aatacattgc ctgtgtggcg 10020
ggttgtgtgg gatcttgtgt caacgttttg ctgtacttat tcagttcaac agattcaact 10080
ttgccgttat acttaaactc catgtcgaca aacttagatt agattgctat gctttctttc 10140
taatgagcaa gaagtaaaaa aagttgtaat agaacaagaa aaatgaaact gaaacttgag 10200
aaattgaaga ccgtttatta acttaaatat caatgggagg tcatcgaaag agaaaaaaat 10260
caaaaaaaaa attttcaaga aaaagaaacg tgataaaaat ttttattgcc tttttcgacg 10320
aagaaaaaga aacgaggcgg tctctttttt cttttccaaa cctttagtac gggtaattaa 10380
cgacacccta gaggaagaaa gaggggaaat ttagtatgct gtgcttgggt gttttgaagt 10440
ggtacggcga tgcgcggagt ccgagaaaat ctggaagagt aaaaaaggag tagaaacatt 10500
ttgaagctat gagctccagc ttttgttccc tttagtgagg gttaattgcg cgcttggcgt 10560
aatcatggtc atagctgttt cctgtgtgaa attgttatcc gctcacaatt ccacacaaca 10620
taggagccgg aagcataaag tgtaaagcct ggggtgccta atgagtgagg taactcacat 10680
taattgcgtt gcgctcactg cccgctttcc agtcgggaaa cctgtcgtgc cagctgcatt 10740
aatgaatcgg ccaacgcgcg gggagaggcg gtttgcgtat tgggcgctct tccgcttcct 10800
cgctcactga ctcgctgcgc tcggtcgttc ggctgcggcg agcggtatca gctcactcaa 10860
aggcggtaat acggttatcc acagaatcag gggataacgc aggaaagaac atgtgagcaa 10920
aaggccagca aaaggccagg aaccgtaaaa aggccgcgtt gctggcgttt ttccataggc 10980
tccgcccccc tgacgagcat cacaaaaatc gacgctcaag tcagaggtgg cgaaacccga 11040
caggactata aagataccag gcgtttcccc ctggaagctc cctcgtgcgc tctcctgttc 11100
cgaccctgcc gcttaccgga tacctgtccg cctttctccc ttcgggaagc gtggcgcttt 11160
ctcatagctc acgctgtagg tatctcagtt cggtgtaggt cgttcgctcc aagctgggct 11220
gtgtgcacga accccccgtt cagcccgacc gctgcgcctt atccggtaac tatcgtcttg 11280
agtccaaccc ggtaagacac gacttatcgc cactggcagc agccactggt aacaggatta 11340
gcagagcgag gtatgtaggc ggtgctacag agttcttgaa gtggtggcct aactacggct 11400
acactagaag gacagtattt ggtatctgcg ctctgctgaa gccagttacc ttcggaaaaa 11460
gagttggtag ctcttgatcc ggcaaacaaa ccaccgctgg tagcggtggt ttttttgttt 11520
gcaagcagca gattacgcgc agaaaaaaag gatctcaaga agatcctttg atcttttcta 11580
cggggtctga cgctcagtgg aacgaaaact cacgttaagg gattttggtc atgagattat 11640
caaaaaggat cttcacctag atccttttaa attaaaaatg aagttttaaa tcaatctaaa 11700
gtatatatga gtaaacttgg tctgacagtt accaatgctt aatcagtgag gcacctatct 11760
cagcgatctg tctatttcgt tcatccatag ttgcctgact ccccgtcgtg tagataacta 11820
cgatacggga gggcttacca tctggcccca gtgctgcaat gataccgcga gacccacgct 11880
caccggctcc agatttatca gcaataaacc agccagccgg aagggccgag cgcagaagtg 11940
gtcctgcaac tttatccgcc tccatccagt ctattaattg ttgccgggaa gctagagtaa 12000
gtagttcgcc agttaatagt ttgcgcaacg ttgttgccat tgctacaggc atcgtggtgt 12060
cacgctcgtc gtttggtatg gcttcattca gctccggttc ccaacgatca aggcgagtta 12120
catgatcccc catgttgtgc aaaaaagcgg ttagctcctt cggtcctccg atcgttgtca 12180
gaagtaagtt ggccgcagtg ttatcactca tggttatggc agcactgcat aattctctta 12240
ctgtcatgcc atccgtaaga tgcttttctg tgactggtga gtactcaacc aagtcattct 12300
gagaatagtg tatgcggcga ccgagttgct cttgcccggc gtcaatacgg gataataccg 12360
cgccacatag cagaacttta aaagtgctca tcattggaaa acgttcttcg gggcgaaaac 12420
tctcaaggat cttaccgctg ttgagatcca gttcgatgta acccactcgt gcacccaact 12480
gatcttcagc atcttttact ttcaccagcg tttctgggtg agcaaaaaca ggaaggcaaa 12540
atgccgcaaa aaagggaata agggcgacac ggaaatgttg aatactcata ctcttccttt 12600
ttcaatatta ttgaagcatt tatcagggtt attgtctcat gagcggatac atatttgaat 12660
gtatttagaa aaataaacaa ataggggttc cgcgcacatt tccccgaaaa gtgccacctg 12720
aacgaagcat ctgtgcttca ttttgtagaa caaaaatgca acgcgagagc gctaattttt 12780
caaacaaaga atctgagctg catttttaca gaacagaaat gcaacgcgaa agcgctattt 12840
taccaacgaa gaatctgtgc ttcatttttg taaaacaaaa atgcaacgcg agagcgctaa 12900
tttttcaaac aaagaatctg agctgcattt ttacagaaca gaaatgcaac gcgagagcgc 12960
tattttacca acaaagaatc tatacttctt ttttgttcta caaaaatgca tcccgagagc 13020
gctatttttc taacaaagca tcttagatta ctttttttct cctttgtgcg ctctataatg 13080
cagtctcttg ataacttttt gcactgtagg tccgttaagg ttagaagaag gctactttgg 13140
tgtctatttt ctcttccata aaaaaagcct gactccactt cccgcgttta ctgattacta 13200
gcgaagctgc gggtgcattt tttcaagata aaggcatccc cgattatatt ctataccgat 13260
gtggattgcg catactttgt gaacagaaag tgatagcgtt gatgattctt cattggtcag 13320
aaaattatga acggtttctt ctattttgtc tctatatact acgtatagga aatgtttaca 13380
ttttcgtatt gttttcgatt cactctatga atagttctta ctacaatttt tttgtctaaa 13440
gagtaatact agagataaac ataaaaaatg tagaggtcga gtttagatgc aagttcaagg 13500
agcgaaaggt ggatgggtag gttatatagg gatatagcac agagatatat agcaaagaga 13560
tacttttgag caatgtttgt ggaagcggta ttcgcaatat tttagtagct cgttacagtc 13620
cggtgcgttt ttggtttttt gaaagtgcgt cttcagagcg cttttggttt tcaaaagcgc 13680
tctgaagttc ctatactttc tagagaatag gaacttcgga ataggaactt caaagcgttt 13740
ccgaaaacga gcgcttccga aaatgcaacg cgagctgcgc acatacagct cactgttcac 13800
gtcgcaccta tatctgcgtg ttgcctgtat atatatatac atgagaagaa cggcatagtg 13860
cgtgtttatg cttaaatgcg tacttatatg cgtctattta tgtaggatga aaggtagtct 13920
agtacctcct gtgatattat cccattccat gcggggtatc gtatgcttcc ttcagcacta 13980
ccctttagct gttctatatg ctgccactcc tcaattggat tagtctcatc cttcaatgct 14040
atcatttcct ttgata 14056
<210> SEQ ID NO 124
<211> LENGTH: 7795
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV2241
<400> SEQUENCE: 124
ttggatcata ctaagaaacc attattatca tgacattaac ctataaaaat aggcgtatca 60
cgaggccctt tcgtctcgcg cgtttcggtg atgacggtga aaacctctga cacatgcagc 120
tcccggagac ggtcacagct tgtctgtaag cggatgccgg gagcagacaa gcccgtcagg 180
gcgcgtcagc gggtgttggc gggtgtcggg gctggcttaa ctatgcggca tcagagcaga 240
ttgtactgag agtgcaccat accacagctt ttcaattcaa ttcatcattt tttttttatt 300
cttttttttg atttcggttt ctttgaaatt tttttgattc ggtaatctcc gaacagaagg 360
aagaacgaag gaaggagcac agacttagat tggtatatat acgcatatgt agtgttgaag 420
aaacatgaaa ttgcccagta ttcttaaccc aactgcacag aacaaaaacc tgcaggaaac 480
gaagataaat catgtcgaaa gctacatata aggaacgtgc tgctactcat cctagtcctg 540
ttgctgccaa gctatttaat atcatgcacg aaaagcaaac aaacttgtgt gcttcattgg 600
atgttcgtac caccaaggaa ttactggagt tagttgaagc attaggtccc aaaatttgtt 660
tactaaaaac acatgtggat atcttgactg atttttccat ggagggcaca gttaagccgc 720
taaaggcatt atccgccaag tacaattttt tactcttcga agacagaaaa tttgctgaca 780
ttggtaatac agtcaaattg cagtactctg cgggtgtata cagaatagca gaatgggcag 840
acattacgaa tgcacacggt gtggtgggcc caggtattgt tagcggtttg aagcaggcgg 900
cagaagaagt aacaaaggaa cctagaggcc ttttgatgtt agcagaattg tcatgcaagg 960
gctccctatc tactggagaa tatactaagg gtactgttga cattgcgaag agcgacaaag 1020
attttgttat cggctttatt gctcaaagag acatgggtgg aagagatgaa ggttacgatt 1080
ggttgattat gacacccggt gtgggtttag atgacaaggg agacgcattg ggtcaacagt 1140
atagaaccgt ggatgatgtg gtctctacag gatctgacat tattattgtt ggaagaggac 1200
tatttgcaaa gggaagggat gctaaggtag agggtgaacg ttacagaaaa gcaggctggg 1260
aagcatattt gagaagatgc ggccagcaaa actaaaaaac tgtattataa gtaaatgcat 1320
gtatactaaa ctcacaaatt agagcttcaa tttaattata tcagttatta ccctatgcgg 1380
tgtgaaatac cgcacagatg cgtaaggaga aaataccgca tcaggaaatt gtaaacgtta 1440
atattttgtt aaaattcgcg ttaaattttt gttaaatcag ctcatttttt aaccaatagg 1500
ccgaaatcgg caaaatccct tataaatcaa aagaatagac cgagataggg ttgagtgttg 1560
ttccagtttg gaacaagagt ccactattaa agaacgtgga ctccaacgtc aaagggcgaa 1620
aaaccgtcta tcagggcgat ggcccactac gtgaaccatc accctaatca agttttttgg 1680
ggtcgaggtg ccgtaaagca ctaaatcgga accctaaagg gagcccccga tttagagctt 1740
gacggggaaa gccggcgaac gtggcgagaa aggaagggaa gaaagcgaaa ggagcgggcg 1800
ctagggcgct ggcaagtgta gcggtcacgc tgcgcgtaac caccacaccc gccgcgctta 1860
atgcgccgct acagggcgcg tcgcgccatt cgccattcag gctgcgcaac tgttgggaag 1920
ggcgatcggt gcgggcctct tcgctattac gccagctggc gaaaggggga tgtgctgcaa 1980
ggcgattaag ttgggtaacg ccagggtttt cccagtcacg acgttgtaaa acgacggcca 2040
gtgagcgcgc gtaatacgac tcactatagg gcgaattggg taccggccgc aaattaaagc 2100
cttcgagcgt cccaaaacct tctcaagcaa ggttttcagt ataatgttac atgcgtacac 2160
gcgtctgtac agaaaaaaaa gaaaaatttg aaatataaat aacgttctta atactaacat 2220
aactataaaa aaataaatag ggacctagac ttcaggttgt ctaactcctt ccttttcggt 2280
tagagcggat gtggggggag ggcgtgaatg taagcgtgac ataactaatt acatgacgcc 2340
gcggatcctt agtggtggtg gtggtggtgt cctgccactg caattctttt catatcggtc 2400
atatatcctc tcagcttttt acccacctgt tctatagcat gtgaacgaat agcttcattt 2460
acgtctctca gttggccatt gtcaaccgct ccttccggaa tagccttccc caaatcacca 2520
ggttgtaact cggccatgaa gggctttaac aacgggacac atgcgtagct aaataagtaa 2580
ttaccatatt ctgcagtgtc tgatatgaca acattcatct cgtaaagtct ttttcttgca 2640
atagtatttg ctatcaaagg caattcatgc aaagactcat agtatgcaga ttcttcaatg 2700
ataccggagt caaccatagt ttcgaatgca agttctaccc ctgccttcac catagctatc 2760
atcaatactc ccttatcaaa gtattcttgt tcaccaattt taccttcgta ttgtggggct 2820
gtctcgaatg ccgtcttgcc ggtttcttct ctccacgtca ataacttttt atcatcgttt 2880
gcccaatctg ccatcattcc tgaggaaaac tcaccggaga taatatcgtc catgtgcttt 2940
tggaataatg gtgccatgat ctcttttagt tgctcagata aggcgtaggc tcttagcttg 3000
gccggatttg aaagtctatc catcatcaat gttatgccac cttgtttaag tgcctcggtg 3060
attgtctccc aaccaaattg tatcaacttt tcagcatagg caggatctgt accctcttcg 3120
accaatttat caaagcatag taaagaccct gcctgcaaca ttccgcacag aatggtttgt 3180
tcacccatta agtcactctt gacctcagct acgaaagaac tctctaacac acccgctcta 3240
tgacctccgg ttgcggctgc ccatgccttc gcaattgcca taccttcacc tttggggtca 3300
ttttcaggat gtacggcgat caatgtaggt acaccaaaac ccctcttgta ctcctctctg 3360
acttccgtac ctgggcactt tggtgcaacc attacgactg ttatatcttt tctgatctgc 3420
tcgcccactt caacgatatt aaagccatga gagtaaccta aagctgcccc atccttcatc 3480
agcggttgaa ctgttcttac tacgtctgag tgaaccttat ctggtgttag gttaatcact 3540
aaatctgcct gagggatcag ttcttcgtaa gtaccaactt tgaacccatt ttccgtcgct 3600
ttacgccaat cggcatcctt ttctgcaata gactctttcc tcaatgcata cgaaatatcc 3660
agacctgaat ctctcatgtt taaaccttgg tttagaccct gagcaccgca gccaacaatt 3720
actactttct ttccttgcag ataagaagca ccatcagcaa actcgtccct tcccataaat 3780
ctgcacttac ccagttgagc caattgttgt ctcaaattta atgtgttaaa atagttggcc 3840
atgtcgacaa acttagatta gattgctatg ctttctttct aatgagcaag aagtaaaaaa 3900
agttgtaata gaacaagaaa aatgaaactg aaacttgaga aattgaagac cgtttattaa 3960
cttaaatatc aatgggaggt catcgaaaga gaaaaaaatc aaaaaaaaaa ttttcaagaa 4020
aaagaaacgt gataaaaatt tttattgcct ttttcgacga agaaaaagaa acgaggcggt 4080
ctcttttttc ttttccaaac ctttagtacg ggtaattaac gacaccctag aggaagaaag 4140
aggggaaatt tagtatgctg tgcttgggtg ttttgaagtg gtacggcgat gcgcggagtc 4200
cgagaaaatc tggaagagta aaaaaggagt agaaacattt tgaagctatg agctccagct 4260
tttgttccct ttagtgaggg ttaattgcgc gcttggcgta atcatggtca tagctgtttc 4320
ctgtgtgaaa ttgttatccg ctcacaattc cacacaacat aggagccgga agcataaagt 4380
gtaaagcctg gggtgcctaa tgagtgaggt aactcacatt aattgcgttg cgctcactgc 4440
ccgctttcca gtcgggaaac ctgtcgtgcc agctgcatta atgaatcggc caacgcgcgg 4500
ggagaggcgg tttgcgtatt gggcgctctt ccgcttcctc gctcactgac tcgctgcgct 4560
cggtcgttcg gctgcggcga gcggtatcag ctcactcaaa ggcggtaata cggttatcca 4620
cagaatcagg ggataacgca ggaaagaaca tgtgagcaaa aggccagcaa aaggccagga 4680
accgtaaaaa ggccgcgttg ctggcgtttt tccataggct ccgcccccct gacgagcatc 4740
acaaaaatcg acgctcaagt cagaggtggc gaaacccgac aggactataa agataccagg 4800
cgtttccccc tggaagctcc ctcgtgcgct ctcctgttcc gaccctgccg cttaccggat 4860
acctgtccgc ctttctccct tcgggaagcg tggcgctttc tcatagctca cgctgtaggt 4920
atctcagttc ggtgtaggtc gttcgctcca agctgggctg tgtgcacgaa ccccccgttc 4980
agcccgaccg ctgcgcctta tccggtaact atcgtcttga gtccaacccg gtaagacacg 5040
acttatcgcc actggcagca gccactggta acaggattag cagagcgagg tatgtaggcg 5100
gtgctacaga gttcttgaag tggtggccta actacggcta cactagaagg acagtatttg 5160
gtatctgcgc tctgctgaag ccagttacct tcggaaaaag agttggtagc tcttgatccg 5220
gcaaacaaac caccgctggt agcggtggtt tttttgtttg caagcagcag attacgcgca 5280
gaaaaaaagg atctcaagaa gatcctttga tcttttctac ggggtctgac gctcagtgga 5340
acgaaaactc acgttaaggg attttggtca tgagattatc aaaaaggatc ttcacctaga 5400
tccttttaaa ttaaaaatga agttttaaat caatctaaag tatatatgag taaacttggt 5460
ctgacagtta ccaatgctta atcagtgagg cacctatctc agcgatctgt ctatttcgtt 5520
catccatagt tgcctgactc cccgtcgtgt agataactac gatacgggag ggcttaccat 5580
ctggccccag tgctgcaatg ataccgcgag acccacgctc accggctcca gatttatcag 5640
caataaacca gccagccgga agggccgagc gcagaagtgg tcctgcaact ttatccgcct 5700
ccatccagtc tattaattgt tgccgggaag ctagagtaag tagttcgcca gttaatagtt 5760
tgcgcaacgt tgttgccatt gctacaggca tcgtggtgtc acgctcgtcg tttggtatgg 5820
cttcattcag ctccggttcc caacgatcaa ggcgagttac atgatccccc atgttgtgca 5880
aaaaagcggt tagctccttc ggtcctccga tcgttgtcag aagtaagttg gccgcagtgt 5940
tatcactcat ggttatggca gcactgcata attctcttac tgtcatgcca tccgtaagat 6000
gcttttctgt gactggtgag tactcaacca agtcattctg agaatagtgt atgcggcgac 6060
cgagttgctc ttgcccggcg tcaatacggg ataataccgc gccacatagc agaactttaa 6120
aagtgctcat cattggaaaa cgttcttcgg ggcgaaaact ctcaaggatc ttaccgctgt 6180
tgagatccag ttcgatgtaa cccactcgtg cacccaactg atcttcagca tcttttactt 6240
tcaccagcgt ttctgggtga gcaaaaacag gaaggcaaaa tgccgcaaaa aagggaataa 6300
gggcgacacg gaaatgttga atactcatac tcttcctttt tcaatattat tgaagcattt 6360
atcagggtta ttgtctcatg agcggataca tatttgaatg tatttagaaa aataaacaaa 6420
taggggttcc gcgcacattt ccccgaaaag tgccacctga acgaagcatc tgtgcttcat 6480
tttgtagaac aaaaatgcaa cgcgagagcg ctaatttttc aaacaaagaa tctgagctgc 6540
atttttacag aacagaaatg caacgcgaaa gcgctatttt accaacgaag aatctgtgct 6600
tcatttttgt aaaacaaaaa tgcaacgcga gagcgctaat ttttcaaaca aagaatctga 6660
gctgcatttt tacagaacag aaatgcaacg cgagagcgct attttaccaa caaagaatct 6720
atacttcttt tttgttctac aaaaatgcat cccgagagcg ctatttttct aacaaagcat 6780
cttagattac tttttttctc ctttgtgcgc tctataatgc agtctcttga taactttttg 6840
cactgtaggt ccgttaaggt tagaagaagg ctactttggt gtctattttc tcttccataa 6900
aaaaagcctg actccacttc ccgcgtttac tgattactag cgaagctgcg ggtgcatttt 6960
ttcaagataa aggcatcccc gattatattc tataccgatg tggattgcgc atactttgtg 7020
aacagaaagt gatagcgttg atgattcttc attggtcaga aaattatgaa cggtttcttc 7080
tattttgtct ctatatacta cgtataggaa atgtttacat tttcgtattg ttttcgattc 7140
actctatgaa tagttcttac tacaattttt ttgtctaaag agtaatacta gagataaaca 7200
taaaaaatgt agaggtcgag tttagatgca agttcaagga gcgaaaggtg gatgggtagg 7260
ttatataggg atatagcaca gagatatata gcaaagagat acttttgagc aatgtttgtg 7320
gaagcggtat tcgcaatatt ttagtagctc gttacagtcc ggtgcgtttt tggttttttg 7380
aaagtgcgtc ttcagagcgc ttttggtttt caaaagcgct ctgaagttcc tatactttct 7440
agagaatagg aacttcggaa taggaacttc aaagcgtttc cgaaaacgag cgcttccgaa 7500
aatgcaacgc gagctgcgca catacagctc actgttcacg tcgcacctat atctgcgtgt 7560
tgcctgtata tatatataca tgagaagaac ggcatagtgc gtgtttatgc ttaaatgcgt 7620
acttatatgc gtctatttat gtaggatgaa aggtagtcta gtacctcctg tgatattatc 7680
ccattccatg cggggtatcg tatgcttcct tcagcactac cctttagctg ttctatatgc 7740
tgccactcct caattggatt agtctcatcc ttcaatgcta tcatttcctt tgata 7795
<210> SEQ ID NO 125
<211> LENGTH: 14056
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Plasmid pGV2242
<400> SEQUENCE: 125
ttggatcata ctaagaaacc attattatca tgacattaac ctataaaaat aggcgtatca 60
cgaggccctt tcgtctcgcg cgtttcggtg atgacggtga aaacctctga cacatgcagc 120
tcccggagac ggtcacagct tgtctgtaag cggatgccgg gagcagacaa gcccgtcagg 180
gcgcgtcagc gggtgttggc gggtgtcggg gctggcttaa ctatgcggca tcagagcaga 240
ttgtactgag agtgcaccat accacagctt ttcaattcaa ttcatcattt tttttttatt 300
cttttttttg atttcggttt ctttgaaatt tttttgattc ggtaatctcc gaacagaagg 360
aagaacgaag gaaggagcac agacttagat tggtatatat acgcatatgg caaattaaag 420
ccttcgagcg tcccaaaacc ttctcaagca aggttttcag tataatgtta catgcgtaca 480
cgcgtctgta cagaaaaaaa agaaaaattt gaaatataaa taacgttctt aatactaaca 540
taactataaa aaaataaata gggacctaga cttcaggttg tctaactcct tccttttcgg 600
ttagagcgga tgtgggggga gggcgtgaat gtaagcgtga cataactaat tacatgactc 660
gacctaggtt atttagtaaa atcaatgacc attcggcctt caatttttcc tgccttcatt 720
tcatcaataa tatcattgat ttcttccagt ttgcgtgtcg caacaattgg ttttacctta 780
ccttctgctc caaattgaaa agcttctgcc aagtcaagtc ttgttccgac aagtgaacct 840
gcaacctcca ctccgtcaaa aacaactgtt ggaactgata aagtcatctc agtattggga 900
agtgccacag caaccatttt gcccataggt ttcaaagaag caaccgcttg ttcaaaagca 960
atccttgcaa cagcacaaac tattgcactt tgcaccccta agccgccagt tattttttta 1020
atttcatcaa ctggatttac atcaccagaa ttgataatca catcagctcc aattttttta 1080
gctaaattta atttatcttg attaatatca acagcaatta cttttgctcc aaaaacattt 1140
ttagcatatt gaattgctaa atttccaagt cctccagcac caaaaattac ttgccaatca 1200
ccaggtttta ctcctgatac tttgattgct ttgtaagttg ttactccagc acaagtaatt 1260
gagctagctt caattgggtc aagtccgtca ggaactttga cagcataatc ggcaacaaca 1320
attgcttctt cagccattcc gccatcaact gaatatcctg catttttaac ttctcgacaa 1380
aaagtttcat taccagatac acagtattca cagtgaccac atccttcaaa gaaccaagcc 1440
actgaaaccc gatcaccaac ttgaagcgag cttacatcag ctccaatttc tttgacaatt 1500
ccaattcctt catgaccaag aacagtccct gctttgttgc cataatcacc tgctgcaacg 1560
tgcaaatcgg tatgacagac tccacaatac tccatgtcaa gcaaagcttc attaggtttg 1620
attgctcgaa gttccttttc aacaaggtcc gcataaccat ctggattgtg tcttactact 1680
gctgctttca ttggtaccta ttattgtatg ttatagtatt agttgcttgg tgttatgaaa 1740
gaaactaaga aaagaaaaat aaaataaaaa taaaagattg agacaaggga agaaaagata 1800
caaaataaga attaattaca attgcgtttg ctataaatac gtttttaaca atcaactctg 1860
gtaggaagat aatgcttttt ttttttatat atgcttggtg ccacttgtca catacaattc 1920
tacaaccttc gacaaaaatc caaatgatag taagatcaaa gccagaaagc aatggagaaa 1980
aaaaattaat gaaccacgat gaaccaaatg atcaatacaa ccaaagaaac taccctagtg 2040
aggtgtatgc tgacttggta tcacacttca tgaattttgc atatggcaaa gtccacgaaa 2100
gtgggcttca gaaaaaaggc gtgcggtgtg tagatgtatc aattagtgga tgccagtttt 2160
ggaacgggat tccactttcc gcaagttggt gcacgtcgtt agtgacataa cgccgcgttc 2220
atctttggga agaagcagat gctgagcgag gaggtactat agagtaaaga accctttcta 2280
tacccgcagc cccatggtaa gtgacagtgc agtaataata tgaaccaatt tatttttcgt 2340
tacataaaaa tgcttataaa actttaacta ataattagag attaaatcgc ggccgcaaaa 2400
gatccttagg atttattctg ttcagcaaac agcttgccca ttttcttcag taccttcggt 2460
gcgccttctt tcgccaggat cagttcgatc cagtacatac ggttcggatc ggcctgggcc 2520
tctttcatca cgctcacaaa ttcgttttcg gtacgcacaa ttttagacac aacacggtcc 2580
tcagttgcgc cgaaggactc cggcagttta gagtagttcc acatagggat atcgttgtaa 2640
gactggttcg gaccgtggat ctcacgctca acggtgtagc cgtcattgtt aataatgaag 2700
caaatcgggt tgatcttttc acgaattgcc agacccagtt cctgtacggt cagctgcagg 2760
gaaccgtcac cgatgaacag cagatgacga gattctttat cagcgatctg agagcccagc 2820
gctgccggga aagtatagcc aatgctaccc cacagcggct gaccgataaa atggcttttg 2880
gatttcagaa agatagaaga cgcgccgaaa aagctcgtac cttgttccgc cacgatggtt 2940
tcattgctct gggtcaggtt ctccacggcc tgccacaggc gatcctggga cagcagtgcg 3000
ttagatggta cgaaatcttc ttgctttttg tcaatgtatt tgcctttata ctcgatttcg 3060
gacaggtcca gcagagagct gatcaggctt tcgaagtcga agttctggat acgctcgttg 3120
aagattttac cctcgtcgat gttcaggcta atcattttgt tttcgttcag atggtgagtg 3180
aatgcaccgg tagaagagtc ggtcagttta acgcccagca tcaggatgaa gtccgcagat 3240
tcaacaaatt ctttcaggtt cggttcgctc agagtaccgt tgtagatgcc caggaaagac 3300
ggcagagcct cgtcaacaga ggacttgccg aagttcaggg tggtaatcgg cagtttggtt 3360
ttgctgatga attgggtcac ggtcttctcc agaccaaaag aaatgatttc gtggccggtg 3420
atcacgattg gtttctttgc gtttttcaga gactcctgga ttttgttcag gatttcctgg 3480
tcgctagtgt tagaagtgga gttttctttc ttcagcggca ggctcggttt ttccgcttta 3540
gctgccgcaa catccacagg caggttgatg taaactggtt tgcgttcttt cagcagcgca 3600
gacagaacgc ggtcgatttc cacagtagcg ttctctgcag tcagcagcgt acgtgccgca 3660
gtcacaggtt catgcatttt catgaagtgt ttgaaatcgc cgtcagccag agtgtggtgg 3720
acgaatttac cttcgttctg aactttgctc gttgggctgc ctacgatctc caccaccggc 3780
aggttttcgg cgtaggagcc cgccagaccg ttgacggcgc tcagttcgcc aacaccgaaa 3840
gtggtcagaa atgccgcggc tttcttggta cgtgcataac catctgccat gtagcttgcg 3900
ttcagttcgt tagcgttacc cacccatttc atgtctttat gagagatgat ctgatccagg 3960
aactgcagat tgtaatcacc cggaacgccg aagatttctt cgatacccag ttcatgcaga 4020
cggtccagca gataatcacc aacagtatac atgtcgagct tgttttatat ttgttgtaaa 4080
aagtagataa ttacttcctt gatgatctgt aaaaaagaga aaaagaaagc atctaagaac 4140
ttgaaaaact acgaattaga aaagaccaaa tatgtatttc ttgcattgac caatttatgc 4200
aagtttatat atatgtaaat gtaagtttca cgaggttcta ctaaactaaa ccaccccctt 4260
ggttagaaga aaagagtgtg tgagaacagg ctgttgttgt cacacgattc ggacaattct 4320
gtttgaaaga gagagagtaa cagtacgatc gaacgaactt tgctctggag atcacagtgg 4380
gcatcatagc atgtggtact aaaccctttc ccgccattcc agaaccttcg attgcttgtt 4440
acaaaacctg tgagccgtcg ctaggacctt gttgtgtgac gaaattggaa gctgcaatca 4500
ataggaagac aggaagtcga gcgtgtctgg gttttttcag ttttgttctt tttgcaaaca 4560
aatcacgagc gacggtaatt tctttctcga taagaggcca cgtgctttat gagggtaaca 4620
tcaattcaag aaggagggaa acacttcctt tttctggccc tgataatagt atgagggtga 4680
agccaaaata aaggattcgc gcccaaatcg gcatctttaa atgcaggtat gcgatagttc 4740
ctcactcttt ccttactcac gagtaattct tgcaaatgcc tattatgcag atgttataat 4800
atctgtgcgt cttgagttga gcctagaatt cttagaaaaa ctcatcgagc atcaaatgaa 4860
actgcaattt attcatatca ggattatcaa taccatattt ttgaaaaagc cgtttctgta 4920
atgaaggaga aaactcaccg aggcagttcc ataggatggc aagatcctgg tatcggtctg 4980
cgatcccgac tcgtccaaca tcaatacaac ctattaattt cccctcgtca aaaataaggt 5040
tatcaagtga gaaatcacca tgagtgacga ctgaatccgg tgagaatggc aaaagcttat 5100
gcatttcttt ccagacttgt tcaacaggcc agccattacg ctcgtcatca aaatcactcg 5160
cgtcaaccaa accgttattc attcgtgatt gcgcctgagc gaggcgaaat acgcgatcgc 5220
tgttaaaagg acaattacaa acaggaatcg aatgcaaccg gcgcaggaac actgccagcg 5280
catcaacaat attttcacct gaatcaggat attcttctaa tacctggaat gctgttttgc 5340
cggggatcgc agtggtgagt aaccatgcat catcaggagt acggacaaaa tgcttgatgg 5400
tcggaagagg cataaattcc gtcagccagt ttagtctgac catctcatct gcaacatcat 5460
tggcaacgct acctttgcca tgtttcagaa acaactctgg cgcatcgggc ttcccataca 5520
atcgatagat tgtcgcacct gattgcccga cattatcgcg agcccattta tacccatata 5580
aatcagcatc catgttggaa tttaatcgcg gcctcgaaac gtgagtcttt tccttaccca 5640
tactagtttt tagtttatgt atgtgttttt tgtagttata gatttaagca agaaaagaat 5700
acaaacaaaa aattgaaaaa gattgattta gaattaaaaa gaaaaatatt tacgtaagaa 5760
gggaaaatag taaatgttgc aagttcacta aactcctaaa ttatgctgcc ctttatattc 5820
cctgttacag cagccgagcc aaaggtatat aggctccttt gcattagcat gcgtaacaaa 5880
ccacctgtca gtttcaaccg aggtggtatc cgagagaatt gtgtgattgc tttaattaat 5940
ttcggagaat ctcacatgcc actgaagatt aaaaactgga tgccagaaaa ggggtgtcca 6000
ggtgtaacat caatagagga agctgaaaag tcttagaacg ggtaatcttc caccaacctg 6060
atgggttcct agatataatc tcgaagggaa taagtagggt gataccgcag aagtgtctga 6120
atgtattaag gtcctcacag tttaaatccc gctcacacta acgtaggatt attataactc 6180
aaaaaaatgg cattattcta agtaagttaa atatccgtaa tctttaaaca gcggccgcgg 6240
atcttcatcc tgccactgca attcttttca tatcggtcat atatcctctc agctttttac 6300
ccacctgttc tatagcatgt gaacgaatag cttcatttac gtctctcagt tggccattgt 6360
caaccgctcc ttccggaata gccttcccca aatcaccagg ttgtaactcg gccatgaagg 6420
gctttaacaa cgggacacat gcgtagctaa ataagtaatt accatattct gcagtgtctg 6480
atatgacaac attcatctcg taaagtcttt ttcttgcaat agtatttgct atcaaaggca 6540
attcatgcaa agactcatag tatgcagatt cttcaatgat accggagtca accatagttt 6600
cgaatgcaag ttctacccct gccttcacca tagctatcat caatactccc ttatcaaagt 6660
attcttgttc accaatttta ccttcgtatt gtggggctgt ctcgaatgcc gtcttgccgg 6720
tttcttctct ccacgtcaat aactttttat catcgtttgc ccaatctgcc atcattcctg 6780
aggaaaactc accggagata atatcgtcca tgtgcttttg gaataatggt gccatgatct 6840
cttttagttg ctcagataag gcgtaggctc ttagcttggc cggatttgaa agtctatcca 6900
tcatcaatgt tatgccacct tgtttaagtg cctcggtgat tgtctcccaa ccaaattgta 6960
tcaacttttc agcataggca ggatctgtac cctcttcgac caatttatca aagcatagta 7020
aagaccctgc ctgcaacatt ccgcacagaa tggtttgttc acccattaag tcactcttga 7080
cctcagctac gaaagaactc tctaacacac ccgctctatg acctccggtt gcggctgccc 7140
atgccttcgc aattgccata ccttcacgtt tggggtcatt ttcaggatgt acggcgatca 7200
atgtaggtac accaaaaccc ctcttgtact cctctctgac ttccgtacct gggcactttg 7260
gcgcaaccat tacgactgtt ataccttttc tgatctgctc gcccacttca acgatattaa 7320
agccatgaga gtaacctaaa gctgccccat ccttcatcag cggttgaact gttcttacta 7380
cgtctgagtg aaccttatct ggtgttaggt taatcactaa atctgcctga gggatcagtt 7440
cttcgtaagt accaactttg aacccatttt ccgtcgcttt acgccaatcg gcatcctttt 7500
ctgcaataga ctctttcctc aatgcatacg aaatatccag acctgaatct ctcatgttta 7560
aaccttggtt tagaccctga gcaccgcagc caacaattac tactttcttt ccttgcagat 7620
aagaagcacc atcagcaaac tcgtcccttc ccataaatct gcacttaccc agttgagcca 7680
attgttgtct caaatttaat gtgttaaaat agttggccat gtcgagtcga aactaagttc 7740
tggtgtttta aaactaaaaa aaagactaac tataaaagta gaatttaaga agtttaagaa 7800
atagatttac agaattacaa tcaataccta ccgtctttat atacttatta gtcaagtagg 7860
ggaataattt cagggaactg gtttcaacct tttttttcag ctttttccaa atcagagaga 7920
gcagaaggta atagaaggtg taagaaaatg agatagatac atgcgtgggt caattgcctt 7980
gtgtcatcat ttactccagg caggttgcat cactccattg aggttgtgcc cgttttttgc 8040
ctgtttgtgc ccctgttctc tgtagttgcg ctaagagaat ggacctatga actgatggtt 8100
ggtgaagaaa acaatatttt ggtgctggga ttcttttttt ttctggatgc cagcttaaaa 8160
agcgggctcc attatattta gtggatgcca ggaataaact gttcacccag acacctacga 8220
tgttatatat tctgtgtaac ccgcccccta ttttgggcat gtacgggtta cagcagaatt 8280
aaaaggctaa ttttttgact aaataaagtt aggaaaatca ctactattaa ttatttacgt 8340
attctttgaa atggcgagta ttgataatga taaactggat ccgcggccgc ttacagatca 8400
gtaacacacc cttccgatgc aggacgggtt aatttagcga attttgccaa aactcccctg 8460
gtggctttcg gagttggctt ctgataatta gctcttctct ttgcgatttc ttcatcggaa 8520
actttcaggg atatagagtt gttgactgca tctatctcta ttatatcgtc atcttcaact 8580
aagccgatta gtccaccctc aacggcttca ggcacaatat ggccgacaac aaaaccgtga 8640
gtgccaccgg agaatctacc atccgtaatt aacgcgcaac ttttccctaa acccgcacca 8700
attaatgctg atgtaggctt cagcatttcg ggcataccag gtccgccgac gggacctata 8760
ttcctaatta ccgctacatc tccagcatgc aaacgaccag attctatgcc gtcgataaaa 8820
tgttgttcac catcaaagac tctggcagtg cctttgaaga actctccttc tttaccgcta 8880
atttttgcta cggaaccccc ttgagctaaa ttaccgtaca gaatctgcaa gtggccggtg 8940
gccttgatag gattctttag tggcctcatg atatcttgtg agtcgaaatc caagtctagg 9000
gcagtctcga cattctcggc taatgtttta cccgtcacag taaggcagtc accatgcaat 9060
tttccttcct ttagaaggta cttaagcact gctggcaagc ctccaatttt atgcaaatct 9120
tccatcatat atttacctga aggtttaaaa tcacctagta ctggagtaat gtcactaatt 9180
ctttggaagt catcctgagt tatttcgaca cctatcgcgt tagccattgc aataatatgc 9240
aagacagcat tagtactacc ccccaagacc atcacaatgg taatagcgtt ctcgaacgcc 9300
tccttagtca ttatatcact aggcttgatg tctttttcca aaagattctt aatggctaat 9360
ccaatctcat cacattcttc ttgtttttct tgagatactg cagggttcga agaagaatac 9420
ggcaatgaca tacctagtgt ttcgatagcg gcagctaagg tattagctgt gtacatcccc 9480
ccacatgccc cttgaccagg aatagcatta caaataacac cgtgataatc ttcatcagag 9540
atattgccgg taattttctg gcctagagat tcaaaagccg atacgatgtt caatttctca 9600
cctttatatt caccgtgttc tattgttcct ccatacacca taatgcttgg cctattaagt 9660
cttgccatac caataataga acctggcata tttttgtcac aacctgggat ggctacaatt 9720
gcatcatagt attcagcgcc agcgttggtt tcaatagagt cagctataac ttctctggaa 9780
acaagggagt atctcattcc caactttcca tttgctatcc catcagaaac tcctatcgta 9840
tgaaattgta agccgatcag accatctgtc tgatttactg agcttttaat ctttgatcca 9900
agggttccta aatgcatgtt gcatggattt ccatcccagt ccatcgacac tatacccact 9960
tgagctttct tgaaatcttc gtctttaaac ccgatgccgt aatacattgc ctgtgtggcg 10020
ggttgtgtgg gatcttgtgt caacgttttg ctgtacttat tcagttcaac agattcaact 10080
ttgccgttat acttaaactc catgtcgaca aacttagatt agattgctat gctttctttc 10140
taatgagcaa gaagtaaaaa aagttgtaat agaacaagaa aaatgaaact gaaacttgag 10200
aaattgaaga ccgtttatta acttaaatat caatgggagg tcatcgaaag agaaaaaaat 10260
caaaaaaaaa attttcaaga aaaagaaacg tgataaaaat ttttattgcc tttttcgacg 10320
aagaaaaaga aacgaggcgg tctctttttt cttttccaaa cctttagtac gggtaattaa 10380
cgacacccta gaggaagaaa gaggggaaat ttagtatgct gtgcttgggt gttttgaagt 10440
ggtacggcga tgcgcggagt ccgagaaaat ctggaagagt aaaaaaggag tagaaacatt 10500
ttgaagctat gagctccagc ttttgttccc tttagtgagg gttaattgcg cgcttggcgt 10560
aatcatggtc atagctgttt cctgtgtgaa attgttatcc gctcacaatt ccacacaaca 10620
taggagccgg aagcataaag tgtaaagcct ggggtgccta atgagtgagg taactcacat 10680
taattgcgtt gcgctcactg cccgctttcc agtcgggaaa cctgtcgtgc cagctgcatt 10740
aatgaatcgg ccaacgcgcg gggagaggcg gtttgcgtat tgggcgctct tccgcttcct 10800
cgctcactga ctcgctgcgc tcggtcgttc ggctgcggcg agcggtatca gctcactcaa 10860
aggcggtaat acggttatcc acagaatcag gggataacgc aggaaagaac atgtgagcaa 10920
aaggccagca aaaggccagg aaccgtaaaa aggccgcgtt gctggcgttt ttccataggc 10980
tccgcccccc tgacgagcat cacaaaaatc gacgctcaag tcagaggtgg cgaaacccga 11040
caggactata aagataccag gcgtttcccc ctggaagctc cctcgtgcgc tctcctgttc 11100
cgaccctgcc gcttaccgga tacctgtccg cctttctccc ttcgggaagc gtggcgcttt 11160
ctcatagctc acgctgtagg tatctcagtt cggtgtaggt cgttcgctcc aagctgggct 11220
gtgtgcacga accccccgtt cagcccgacc gctgcgcctt atccggtaac tatcgtcttg 11280
agtccaaccc ggtaagacac gacttatcgc cactggcagc agccactggt aacaggatta 11340
gcagagcgag gtatgtaggc ggtgctacag agttcttgaa gtggtggcct aactacggct 11400
acactagaag gacagtattt ggtatctgcg ctctgctgaa gccagttacc ttcggaaaaa 11460
gagttggtag ctcttgatcc ggcaaacaaa ccaccgctgg tagcggtggt ttttttgttt 11520
gcaagcagca gattacgcgc agaaaaaaag gatctcaaga agatcctttg atcttttcta 11580
cggggtctga cgctcagtgg aacgaaaact cacgttaagg gattttggtc atgagattat 11640
caaaaaggat cttcacctag atccttttaa attaaaaatg aagttttaaa tcaatctaaa 11700
gtatatatga gtaaacttgg tctgacagtt accaatgctt aatcagtgag gcacctatct 11760
cagcgatctg tctatttcgt tcatccatag ttgcctgact ccccgtcgtg tagataacta 11820
cgatacggga gggcttacca tctggcccca gtgctgcaat gataccgcga gacccacgct 11880
caccggctcc agatttatca gcaataaacc agccagccgg aagggccgag cgcagaagtg 11940
gtcctgcaac tttatccgcc tccatccagt ctattaattg ttgccgggaa gctagagtaa 12000
gtagttcgcc agttaatagt ttgcgcaacg ttgttgccat tgctacaggc atcgtggtgt 12060
cacgctcgtc gtttggtatg gcttcattca gctccggttc ccaacgatca aggcgagtta 12120
catgatcccc catgttgtgc aaaaaagcgg ttagctcctt cggtcctccg atcgttgtca 12180
gaagtaagtt ggccgcagtg ttatcactca tggttatggc agcactgcat aattctctta 12240
ctgtcatgcc atccgtaaga tgcttttctg tgactggtga gtactcaacc aagtcattct 12300
gagaatagtg tatgcggcga ccgagttgct cttgcccggc gtcaatacgg gataataccg 12360
cgccacatag cagaacttta aaagtgctca tcattggaaa acgttcttcg gggcgaaaac 12420
tctcaaggat cttaccgctg ttgagatcca gttcgatgta acccactcgt gcacccaact 12480
gatcttcagc atcttttact ttcaccagcg tttctgggtg agcaaaaaca ggaaggcaaa 12540
atgccgcaaa aaagggaata agggcgacac ggaaatgttg aatactcata ctcttccttt 12600
ttcaatatta ttgaagcatt tatcagggtt attgtctcat gagcggatac atatttgaat 12660
gtatttagaa aaataaacaa ataggggttc cgcgcacatt tccccgaaaa gtgccacctg 12720
aacgaagcat ctgtgcttca ttttgtagaa caaaaatgca acgcgagagc gctaattttt 12780
caaacaaaga atctgagctg catttttaca gaacagaaat gcaacgcgaa agcgctattt 12840
taccaacgaa gaatctgtgc ttcatttttg taaaacaaaa atgcaacgcg agagcgctaa 12900
tttttcaaac aaagaatctg agctgcattt ttacagaaca gaaatgcaac gcgagagcgc 12960
tattttacca acaaagaatc tatacttctt ttttgttcta caaaaatgca tcccgagagc 13020
gctatttttc taacaaagca tcttagatta ctttttttct cctttgtgcg ctctataatg 13080
cagtctcttg ataacttttt gcactgtagg tccgttaagg ttagaagaag gctactttgg 13140
tgtctatttt ctcttccata aaaaaagcct gactccactt cccgcgttta ctgattacta 13200
gcgaagctgc gggtgcattt tttcaagata aaggcatccc cgattatatt ctataccgat 13260
gtggattgcg catactttgt gaacagaaag tgatagcgtt gatgattctt cattggtcag 13320
aaaattatga acggtttctt ctattttgtc tctatatact acgtatagga aatgtttaca 13380
ttttcgtatt gttttcgatt cactctatga atagttctta ctacaatttt tttgtctaaa 13440
gagtaatact agagataaac ataaaaaatg tagaggtcga gtttagatgc aagttcaagg 13500
agcgaaaggt ggatgggtag gttatatagg gatatagcac agagatatat agcaaagaga 13560
tacttttgag caatgtttgt ggaagcggta ttcgcaatat tttagtagct cgttacagtc 13620
cggtgcgttt ttggtttttt gaaagtgcgt cttcagagcg cttttggttt tcaaaagcgc 13680
tctgaagttc ctatactttc tagagaatag gaacttcgga ataggaactt caaagcgttt 13740
ccgaaaacga gcgcttccga aaatgcaacg cgagctgcgc acatacagct cactgttcac 13800
gtcgcaccta tatctgcgtg ttgcctgtat atatatatac atgagaagaa cggcatagtg 13860
cgtgtttatg cttaaatgcg tacttatatg cgtctattta tgtaggatga aaggtagtct 13920
agtacctcct gtgatattat cccattccat gcggggtatc gtatgcttcc ttcagcacta 13980
ccctttagct gttctatatg ctgccactcc tcaattggat tagtctcatc cttcaatgct 14040
atcatttcct ttgata 14056
<210> SEQ ID NO 126
<400> SEQUENCE: 126
000
<210> SEQ ID NO 127
<400> SEQUENCE: 127
000
<210> SEQ ID NO 128
<400> SEQUENCE: 128
000
<210> SEQ ID NO 129
<400> SEQUENCE: 129
000
<210> SEQ ID NO 130
<400> SEQUENCE: 130
000
<210> SEQ ID NO 131
<400> SEQUENCE: 131
000
<210> SEQ ID NO 132
<400> SEQUENCE: 132
000
<210> SEQ ID NO 133
<400> SEQUENCE: 133
000
<210> SEQ ID NO 134
<400> SEQUENCE: 134
000
<210> SEQ ID NO 135
<400> SEQUENCE: 135
000
<210> SEQ ID NO 136
<400> SEQUENCE: 136
000
<210> SEQ ID NO 137
<400> SEQUENCE: 137
000
<210> SEQ ID NO 138
<400> SEQUENCE: 138
000
<210> SEQ ID NO 139
<400> SEQUENCE: 139
000
<210> SEQ ID NO 140
<400> SEQUENCE: 140
000
<210> SEQ ID NO 141
<400> SEQUENCE: 141
000
<210> SEQ ID NO 142
<400> SEQUENCE: 142
000
<210> SEQ ID NO 143
<400> SEQUENCE: 143
000
<210> SEQ ID NO 144
<400> SEQUENCE: 144
000
<210> SEQ ID NO 145
<400> SEQUENCE: 145
000
<210> SEQ ID NO 146
<400> SEQUENCE: 146
000
<210> SEQ ID NO 147
<400> SEQUENCE: 147
000
<210> SEQ ID NO 148
<400> SEQUENCE: 148
000
<210> SEQ ID NO 149
<400> SEQUENCE: 149
000
<210> SEQ ID NO 150
<400> SEQUENCE: 150
000
<210> SEQ ID NO 151
<400> SEQUENCE: 151
000
<210> SEQ ID NO 152
<400> SEQUENCE: 152
000
<210> SEQ ID NO 153
<400> SEQUENCE: 153
000
<210> SEQ ID NO 154
<400> SEQUENCE: 154
000
<210> SEQ ID NO 155
<400> SEQUENCE: 155
000
<210> SEQ ID NO 156
<400> SEQUENCE: 156
000
<210> SEQ ID NO 157
<400> SEQUENCE: 157
000
<210> SEQ ID NO 158
<400> SEQUENCE: 158
000
<210> SEQ ID NO 159
<400> SEQUENCE: 159
000
<210> SEQ ID NO 160
<400> SEQUENCE: 160
000
<210> SEQ ID NO 161
<400> SEQUENCE: 161
000
<210> SEQ ID NO 162
<400> SEQUENCE: 162
000
<210> SEQ ID NO 163
<400> SEQUENCE: 163
000
<210> SEQ ID NO 164
<400> SEQUENCE: 164
000
<210> SEQ ID NO 165
<400> SEQUENCE: 165
000
<210> SEQ ID NO 166
<400> SEQUENCE: 166
000
<210> SEQ ID NO 167
<400> SEQUENCE: 167
000
<210> SEQ ID NO 168
<400> SEQUENCE: 168
000
<210> SEQ ID NO 169
<400> SEQUENCE: 169
000
<210> SEQ ID NO 170
<400> SEQUENCE: 170
000
<210> SEQ ID NO 171
<400> SEQUENCE: 171
000
<210> SEQ ID NO 172
<400> SEQUENCE: 172
000
<210> SEQ ID NO 173
<400> SEQUENCE: 173
000
<210> SEQ ID NO 174
<400> SEQUENCE: 174
000
<210> SEQ ID NO 175
<400> SEQUENCE: 175
000
<210> SEQ ID NO 176
<400> SEQUENCE: 176
000
<210> SEQ ID NO 177
<400> SEQUENCE: 177
000
<210> SEQ ID NO 178
<400> SEQUENCE: 178
000
<210> SEQ ID NO 179
<400> SEQUENCE: 179
000
<210> SEQ ID NO 180
<400> SEQUENCE: 180
000
<210> SEQ ID NO 181
<400> SEQUENCE: 181
000
<210> SEQ ID NO 182
<400> SEQUENCE: 182
000
<210> SEQ ID NO 183
<400> SEQUENCE: 183
000
<210> SEQ ID NO 184
<400> SEQUENCE: 184
000
<210> SEQ ID NO 185
<400> SEQUENCE: 185
000
<210> SEQ ID NO 186
<400> SEQUENCE: 186
000
<210> SEQ ID NO 187
<400> SEQUENCE: 187
000
<210> SEQ ID NO 188
<400> SEQUENCE: 188
000
<210> SEQ ID NO 189
<400> SEQUENCE: 189
000
<210> SEQ ID NO 190
<400> SEQUENCE: 190
000
<210> SEQ ID NO 191
<400> SEQUENCE: 191
000
<210> SEQ ID NO 192
<400> SEQUENCE: 192
000
<210> SEQ ID NO 193
<400> SEQUENCE: 193
000
<210> SEQ ID NO 194
<400> SEQUENCE: 194
000
<210> SEQ ID NO 195
<400> SEQUENCE: 195
000
<210> SEQ ID NO 196
<400> SEQUENCE: 196
000
<210> SEQ ID NO 197
<400> SEQUENCE: 197
000
<210> SEQ ID NO 198
<400> SEQUENCE: 198
000
<210> SEQ ID NO 199
<400> SEQUENCE: 199
000
<210> SEQ ID NO 200
<400> SEQUENCE: 200
000
<210> SEQ ID NO 201
<211> LENGTH: 46
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer XX1
<400> SEQUENCE: 201
cgcaccggtt ttctcctctt taatgaattc ggtcagtgcg tcctgc 46
<210> SEQ ID NO 202
<211> LENGTH: 36
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer XX2
<400> SEQUENCE: 202
gcggccgccc tagggcgttc ggctgcggcg agcggt 36
<210> SEQ ID NO 203
<211> LENGTH: 56
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer XX3
<400> SEQUENCE: 203
cgcgaattcg gatccgagga gaaaatagtt atgaacaact ttaatctgca cacccc 56
<210> SEQ ID NO 204
<211> LENGTH: 42
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer XX4
<400> SEQUENCE: 204
gcgcctaggg cggccgctta gcgggcggct tcgtatatac gg 42
<210> SEQ ID NO 205
<211> LENGTH: 61
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 50
<400> SEQUENCE: 205
gcagtttcac cttctacata atcacgaccg tagtaggtat cattccgggg atccgtcgac 60
c 61
<210> SEQ ID NO 206
<211> LENGTH: 60
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 73
<400> SEQUENCE: 206
ctggcttaag taccgggtta gttaacttaa ggagaatgac gtgtaggctg gagctgcttc 60
<210> SEQ ID NO 207
<211> LENGTH: 60
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 74
<400> SEQUENCE: 207
ctcaaactca ttccaggaac gaccatcacg ggtaatcatc attccgggga tccgtcgacc 60
<210> SEQ ID NO 208
<211> LENGTH: 65
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 116
<400> SEQUENCE: 208
cagcgttcgc tttatatccc ttacgctggc cctgtactgc tggaagtgta ggctggagct 60
gcttc 65
<210> SEQ ID NO 209
<211> LENGTH: 65
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 117
<400> SEQUENCE: 209
ttcggcttgc cagaaattat cgtcaatggc ctgttgcagg gcttcattcc ggggatccgt 60
cgacc 65
<210> SEQ ID NO 210
<211> LENGTH: 26
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 350
<400> SEQUENCE: 210
cttaaattct acttttatag ttagtc 26
<210> SEQ ID NO 211
<211> LENGTH: 65
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 474
<400> SEQUENCE: 211
caaagctgcg gatgatgacg agattactgc tgctgtgcag actgaattcc ggggatccgt 60
cgacc 65
<210> SEQ ID NO 212
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 772
<400> SEQUENCE: 212
aggaaggagc acagacttag 20
<210> SEQ ID NO 213
<211> LENGTH: 64
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 868
<400> SEQUENCE: 213
cacaacatca cgaggaatca ccatggctaa ctacttcaat acacgtgtag gctggagctg 60
cttc 64
<210> SEQ ID NO 214
<211> LENGTH: 65
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 869
<400> SEQUENCE: 214
cttaacccgc aacagcaata cgtttcatat ctgtcatata gccgcattcc ggggatccgt 60
cgacc 65
<210> SEQ ID NO 215
<211> LENGTH: 45
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1030
<400> SEQUENCE: 215
gtcggtgaac gctctcctga gtagggtgta ggctggagct gcttc 45
<210> SEQ ID NO 216
<211> LENGTH: 45
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1031
<400> SEQUENCE: 216
gaagcagctc cagcctacac cctactcagg agagcgttca ccgac 45
<210> SEQ ID NO 217
<211> LENGTH: 69
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1032
<400> SEQUENCE: 217
cacaacatca cgaggaatca ccatggctaa ctacttcaat acaccacgag gccctttcgt 60
cttcacctc 69
<210> SEQ ID NO 218
<211> LENGTH: 65
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1155
<400> SEQUENCE: 218
cccaacccgc attctgtttg gtaaaggcgc aatcgctggt ttacggtgta ggctggagct 60
gcttc 65
<210> SEQ ID NO 219
<211> LENGTH: 65
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1156
<400> SEQUENCE: 219
caatcgcggc gtcaatacgc tcatcatcgg aaccttcagt gatgtattcc ggggatccgt 60
cgacc 65
<210> SEQ ID NO 220
<211> LENGTH: 65
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1187
<400> SEQUENCE: 220
cggataaagt tcgtgagatt gccgcaaaac tggggcgtca tgtgggtgta ggctggagct 60
gcttc 65
<210> SEQ ID NO 221
<211> LENGTH: 65
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1188
<400> SEQUENCE: 221
cagacatcaa gtaaccttta tcgcgcagca gattaaccgc ttcgcattcc ggggatccgt 60
cgacc 65
<210> SEQ ID NO 222
<211> LENGTH: 65
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1191
<400> SEQUENCE: 222
ggcactcacg ttgggctgag acacaagcac acattcctct gcacggtgta ggctggagct 60
gcttc 65
<210> SEQ ID NO 223
<211> LENGTH: 65
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1192
<400> SEQUENCE: 223
gcaccagaaa ccataactac aacgtcacct ttgtgtgcca gaccgattcc ggggatccgt 60
cgacc 65
<210> SEQ ID NO 224
<211> LENGTH: 67
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1205
<400> SEQUENCE: 224
gttatctagt tgtgcaaaac atgctaatgt agccaccaaa tccacgaggc cctttcgtct 60
tcacctc 67
<210> SEQ ID NO 225
<211> LENGTH: 42
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1218
<400> SEQUENCE: 225
gctcactcaa aggcggtaat acgtgtaggc tggagctgct tc 42
<210> SEQ ID NO 226
<211> LENGTH: 42
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1219
<400> SEQUENCE: 226
gaagcagctc cagcctacac gtattaccgc ctttgagtga gc 42
<210> SEQ ID NO 227
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1220
<400> SEQUENCE: 227
cgtagaatca ccagaccagc 20
<210> SEQ ID NO 228
<211> LENGTH: 56
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1296
<400> SEQUENCE: 228
ttttgtcgac ggatccagga gacaacatta tgtctattcc agaaactcaa aaagcg 56
<210> SEQ ID NO 229
<211> LENGTH: 46
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1297
<400> SEQUENCE: 229
ttttgtcgac gcggccgctt atttagaggt gtccaccacg taacgg 46
<210> SEQ ID NO 230
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1321
<400> SEQUENCE: 230
aatcatatcg aacacgatgc 20
<210> SEQ ID NO 231
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1322
<400> SEQUENCE: 231
tcagaaagga tcttctgctc 20
<210> SEQ ID NO 232
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1323
<400> SEQUENCE: 232
atcgatatcg tgaaatacgc 20
<210> SEQ ID NO 233
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1324
<400> SEQUENCE: 233
agctggtctg gtgattctac 20
<210> SEQ ID NO 234
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1341
<400> SEQUENCE: 234
tgctgaaaga gaaattgtcc 20
<210> SEQ ID NO 235
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1342
<400> SEQUENCE: 235
tttcttgttc gaagtccaag 20
<210> SEQ ID NO 236
<211> LENGTH: 37
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1364
<400> SEQUENCE: 236
ttttgcggcc gcttagatgc cggagtccca gtgcttg 37
<210> SEQ ID NO 237
<211> LENGTH: 22
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1365
<400> SEQUENCE: 237
agttgttgac gcaggttcag ag 22
<210> SEQ ID NO 238
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 238
<400> SEQUENCE: 238
aaatgacgac gagcctgaag 20
<210> SEQ ID NO 239
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1437
<400> SEQUENCE: 239
gacctgacca tttgatggag 20
<210> SEQ ID NO 240
<211> LENGTH: 21
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1439
<400> SEQUENCE: 240
caattggcga agcagaacaa g 21
<210> SEQ ID NO 241
<211> LENGTH: 47
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1469
<400> SEQUENCE: 241
ttttagatct aggagatacc ggtatgtcgt ttactttgac caacaag 47
<210> SEQ ID NO 242
<211> LENGTH: 21
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1440
<400> SEQUENCE: 242
atcgtacatc ttccaagcat c 21
<210> SEQ ID NO 243
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1441
<400> SEQUENCE: 243
aatcggaacc ctaaagggag 20
<210> SEQ ID NO 244
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1442
<400> SEQUENCE: 244
aatgggcaag ctgtttgctg 20
<210> SEQ ID NO 245
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1443
<400> SEQUENCE: 245
tgcagatgca gatgtgagac 20
<210> SEQ ID NO 246
<211> LENGTH: 51
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1470
<400> SEQUENCE: 246
ttttggatcc aggaaataga tctatgatgg ctaacagaat gattctgaac g 51
<210> SEQ ID NO 247
<211> LENGTH: 36
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1471
<400> SEQUENCE: 247
ttttgcggcc gcttaccagg cggtatggta aagctc 36
<210> SEQ ID NO 248
<211> LENGTH: 64
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1479
<400> SEQUENCE: 248
ccgataggct tccgccatcg tcgggtagtt aaaggtggtg ttgagtgtag gctggagctg 60
cttc 64
<210> SEQ ID NO 249
<211> LENGTH: 70
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1485
<400> SEQUENCE: 249
gcctttattg tacgcttttt actgtacgat ttcagtcaaa tctaacacga ggccctttcg 60
tcttcacctc 70
<210> SEQ ID NO 250
<211> LENGTH: 65
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1486
<400> SEQUENCE: 250
aagtacgcag taaataaaaa atccacttaa gaaggtaggt gttacattcc ggggatccgt 60
cgacc 65
<210> SEQ ID NO 251
<211> LENGTH: 39
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1526
<400> SEQUENCE: 251
tcgacgagga gacaacattg tgtaggctgg agctgcttc 39
<210> SEQ ID NO 252
<211> LENGTH: 39
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1527
<400> SEQUENCE: 252
gaagcagctc cagcctacac aatgttgtct cctcgtcga 39
<210> SEQ ID NO 253
<211> LENGTH: 65
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1539
<400> SEQUENCE: 253
ccattctgtt gcttttatgt ataagaacag gtaagcccta ccatggagaa ttgtgagcgg 60
ataac 65
<210> SEQ ID NO 254
<211> LENGTH: 42
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1561
<400> SEQUENCE: 254
gcaatcctga aagctctgta acattccggg gatccgtcga cc 42
<210> SEQ ID NO 255
<211> LENGTH: 42
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1562
<400> SEQUENCE: 255
ggtcgacgga tccccggaat gttacagagc tttcaggatt gc 42
<210> SEQ ID NO 256
<211> LENGTH: 70
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1563
<400> SEQUENCE: 256
caaatcggcg gtaacgaaag aggataaacc gtgtcccgta ttattcacga ggccctttcg 60
tcttcacctc 70
<210> SEQ ID NO 257
<211> LENGTH: 22
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1566
<400> SEQUENCE: 257
tcccacccaa tcaaggccaa cg 22
<210> SEQ ID NO 258
<211> LENGTH: 22
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1567
<400> SEQUENCE: 258
tccacctggt gccaatgaac cg 22
<210> SEQ ID NO 259
<211> LENGTH: 24
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1587
<400> SEQUENCE: 259
cggctgccag aactctacta actg 24
<210> SEQ ID NO 260
<211> LENGTH: 23
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1588
<400> SEQUENCE: 260
gcgacgtcta ctggcaggtt aat 23
<210> SEQ ID NO 261
<211> LENGTH: 21
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1595
<400> SEQUENCE: 261
caacctggtg atttggggaa g 21
<210> SEQ ID NO 262
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1597
<400> SEQUENCE: 262
gaatgatggc agattgggca 20
<210> SEQ ID NO 263
<211> LENGTH: 22
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1598
<400> SEQUENCE: 263
tattgtgggg ctgtctcgaa tg 22
<210> SEQ ID NO 264
<211> LENGTH: 19
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1624
<400> SEQUENCE: 264
ccctcatgtt gtctaacgg 19
<210> SEQ ID NO 265
<211> LENGTH: 24
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1633
<400> SEQUENCE: 265
tccgtcactg gattcaatgc catc 24
<210> SEQ ID NO 266
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1634
<400> SEQUENCE: 266
ttcgccaggg agctggtgaa 20
<210> SEQ ID NO 267
<211> LENGTH: 21
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1798
<400> SEQUENCE: 267
gcaaattaaa gccttcgagc g 21
<210> SEQ ID NO 268
<211> LENGTH: 39
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1926
<400> SEQUENCE: 268
tttttgtcga cggatccagt ttatcattat caatactcg 39
<210> SEQ ID NO 269
<211> LENGTH: 45
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1927
<400> SEQUENCE: 269
ttttgcggcc gcagatctct cgagtcgaaa ctaagttctg gtgtt 45
<210> SEQ ID NO 270
<211> LENGTH: 22
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 2091
<400> SEQUENCE: 270
cttttcttcc cttgtctcaa tc 22
<210> SEQ ID NO 271
<211> LENGTH: 39
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 2352
<400> SEQUENCE: 271
gactcgacct aggttattta gtaaaatcaa tgaccattc 39
<210> SEQ ID NO 272
<211> LENGTH: 38
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 2353
<400> SEQUENCE: 272
ctaaataacc taggtcgagt catgtaatta gttatgtc 38
<210> SEQ ID NO 273
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer KARIpETfor
<400> SEQUENCE: 273
attcatatgg cgaattattt caacactctg 30
<210> SEQ ID NO 274
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer KARIpETrev
<400> SEQUENCE: 274
taatctcgag gccagccacc gcgatgcg 28
<210> SEQ ID NO 275
<211> LENGTH: 16
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer pETup
<400> SEQUENCE: 275
atgcgtccgg cgtaga 16
<210> SEQ ID NO 276
<211> LENGTH: 35
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer seq_ilvC_pGV
<400> SEQUENCE: 276
gcggccgcgt cgacgaggag acaacattat ggcga 35
<210> SEQ ID NO 277
<211> LENGTH: 51
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer pGV1994ep_for
<400> SEQUENCE: 277
cggtcttcaa tttctcaagt ttcagtttca tttttcttgt tctattacaa c 51
<210> SEQ ID NO 278
<211> LENGTH: 35
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer pGV1994ep_rev
<400> SEQUENCE: 278
ctaactcctt ccttttcggt tagagcggat gtggg 35
<210> SEQ ID NO 279
<211> LENGTH: 47
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer Not_in_for
<400> SEQUENCE: 279
cctctagaaa taatttgcgg ccgcgttaag aaggagatat acatatg 47
<210> SEQ ID NO 280
<211> LENGTH: 41
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer AvrII_in_rev
<400> SEQUENCE: 280
ccgaacgccc taggtcagtg gtggtggtgg tggtgctcga g 41
<210> SEQ ID NO 281
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer R68DK69Lfor
<400> SEQUENCE: 281
tagctatgcg ctggacctgg aggctatc 28
<210> SEQ ID NO 282
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer R68DK69Lrev
<400> SEQUENCE: 282
gatagcctcc aggtccagcg catagcta 28
<210> SEQ ID NO 283
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer K75VR76Dfor
<400> SEQUENCE: 283
aggctatcgc ggaagttgac gctagctg 28
<210> SEQ ID NO 284
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer K75VR76Drev
<400> SEQUENCE: 284
cagctagcgt caacttccgc gatagcct 28
<210> SEQ ID NO 285
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer R69NNKfor
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (17)..(18)
<223> OTHER INFORMATION: n is a, c, g, or t
<400> SEQUENCE: 285
tagctatgcg ctgcgcnnkg aggctatc 28
<210> SEQ ID NO 286
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer R69NNKrev
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (11)..(12)
<223> OTHER INFORMATION: n is a, c, g, or t
<400> SEQUENCE: 286
gatagcctcm nngcgcagcg catagcta 28
<210> SEQ ID NO 287
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer K75NNKfor
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (15)..(16)
<223> OTHER INFORMATION: n is a, c, g, or t
<400> SEQUENCE: 287
aggctatcgc ggaannkcgt gctagctg 28
<210> SEQ ID NO 288
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer K75NNKrev
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (13)..(14)
<223> OTHER INFORMATION: n is a, c, g, or t
<400> SEQUENCE: 288
cagctagcac gmnnttccgc gatagcct 28
<210> SEQ ID NO 289
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer R76NNKfor
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (18)..(19)
<223> OTHER INFORMATION: n is a, c, g, or t
<400> SEQUENCE: 289
aggctatcgc ggaaaaannk gctagctggc 30
<210> SEQ ID NO 290
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer R76NNKrev
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (12)..(13)
<223> OTHER INFORMATION: n is a, c, g, or t
<400> SEQUENCE: 290
gccagctagc mnntttttcc gcgatagcct 30
<210> SEQ ID NO 291
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer R68NNK_for
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (14)..(15)
<223> OTHER INFORMATION: n is a, c, g, or t
<400> SEQUENCE: 291
tagctatgcg ctgnnkaagg aggctatc 28
<210> SEQ ID NO 292
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer R68NNK_rev
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (14)..(15)
<223> OTHER INFORMATION: n is a, c, g, or t
<400> SEQUENCE: 292
gatagcctcc ttmnncagcg catagcta 28
<210> SEQ ID NO 293
<211> LENGTH: 33
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer S78NNK_for
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (16)..(17)
<223> OTHER INFORMATION: n is a, c, g, or t
<400> SEQUENCE: 293
gcggaaaaac gtgctnnktg gcgcaaggct act 33
<210> SEQ ID NO 294
<211> LENGTH: 33
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer S78NNK_rev
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (17)..(18)
<223> OTHER INFORMATION: n is a, c, g, or t
<400> SEQUENCE: 294
agtagccttg cgccamnnag cacgtttttc cgc 33
<210> SEQ ID NO 295
<211> LENGTH: 31
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer A71NNK_for
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (16)..(17)
<223> OTHER INFORMATION: n is a, c, g, or t
<400> SEQUENCE: 295
gcgctgcgca aggagnnkat cgcggaaaaa c 31
<210> SEQ ID NO 296
<211> LENGTH: 31
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer A71NNK_rev
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (15)..(16)
<223> OTHER INFORMATION: n is a, c, g, or t
<400> SEQUENCE: 296
gtttttccgc gatmnnctcc ttgcgcagcg c 31
<210> SEQ ID NO 297
<211> LENGTH: 31
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer Gln110NNK_for
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (16)..(17)
<223> OTHER INFORMATION: n is a, c, g, or t
<400> SEQUENCE: 297
ctgaccccag ataaannkca tagcgacgtt g 31
<210> SEQ ID NO 298
<211> LENGTH: 31
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Gln110NNK_rev
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (15)..(16)
<223> OTHER INFORMATION: n is a, c, g, or t
<400> SEQUENCE: 298
caacgtcgct atgmnnttta tctggggtca g 31
<210> SEQ ID NO 299
<211> LENGTH: 35
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer seq_ilvC_pGV
<400> SEQUENCE: 299
gcggccgcgt cgacgaggag acaacattat ggcga 35
<210> SEQ ID NO 300
<211> LENGTH: 31
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer Q110Qfor
<400> SEQUENCE: 300
gaccccagat aaacaacata gcgacgttgt t 31
<210> SEQ ID NO 301
<211> LENGTH: 31
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer Q110Qrev
<400> SEQUENCE: 301
aacaacgtcg ctatgttgtt tatctggggt c 31
<210> SEQ ID NO 302
<211> LENGTH: 31
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer Q110Afor
<400> SEQUENCE: 302
gaccccagat aaagcacata gcgacgttgt t 31
<210> SEQ ID NO 303
<211> LENGTH: 31
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer Q110Arev
<400> SEQUENCE: 303
aacaacgtcg ctatgtgctt tatctggggt c 31
<210> SEQ ID NO 304
<211> LENGTH: 31
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer Q110Vfor
<400> SEQUENCE: 304
gaccccagat aaagtacata gcgacgttgt t 31
<210> SEQ ID NO 305
<211> LENGTH: 31
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer Q110Vrev
<400> SEQUENCE: 305
aacaacgtcg ctatgtactt tatctggggt c 31
<210> SEQ ID NO 306
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer R68A71recombfor
<400> SEQUENCE: 306
gctatgcgct gckaaaggag dcaatcgcgg 30
<210> SEQ ID NO 307
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer R68A71recombrev
<400> SEQUENCE: 307
ccgcgattgh ctcctttmgc agcgcatagc 30
<210> SEQ ID NO 308
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer R76S78recombfor
<400> SEQUENCE: 308
gaaaaacgtg ctagctggcg caaggctact 30
<210> SEQ ID NO 309
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer R76S78recombrev
<400> SEQUENCE: 309
agtagccttg cgccagctag cacgtttttc 30
<210> SEQ ID NO 310
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer G76S78recombfor
<400> SEQUENCE: 310
gaaaaaggtg ctagctggcg caaggctact 30
<210> SEQ ID NO 311
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer G76S78recombrev
<400> SEQUENCE: 311
agtagccttg cgccagctag cacctttttc 30
<210> SEQ ID NO 312
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer S76S78recombfor
<400> SEQUENCE: 312
gaaaaaagtg ctagctggcg caaggctact 30
<210> SEQ ID NO 313
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer S76S78recombrev
<400> SEQUENCE: 313
agtagccttg cgccagctag cacttttttc 30
<210> SEQ ID NO 314
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer T76S78recombfor
<400> SEQUENCE: 314
gaaaaaactg ctagctggcg caaggctact 30
<210> SEQ ID NO 315
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer T76S78recombrev
<400> SEQUENCE: 315
agtagccttg cgccagctag cagttttttc 30
<210> SEQ ID NO 316
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer D76S78recombfor
<400> SEQUENCE: 316
gaaaaagatg ctagctggcg caaggctact 30
<210> SEQ ID NO 317
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer D76S78recombrev
<400> SEQUENCE: 317
agtagccttg cgccagctag catctttttc 30
<210> SEQ ID NO 318
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Unknown R76D78recombfor
<400> SEQUENCE: 318
gaaaaacgtg ctgactggcg caaggctact 30
<210> SEQ ID NO 319
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer R76D78recombrev
<400> SEQUENCE: 319
agtagccttg cgccagtcag cacgtttttc 30
<210> SEQ ID NO 320
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer G76D78recombfor
<400> SEQUENCE: 320
gaaaaaggtg ctgactggcg caaggctact 30
<210> SEQ ID NO 321
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer G76D78recombrev
<400> SEQUENCE: 321
agtagccttg cgccagtcag cacctttttc 30
<210> SEQ ID NO 322
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer S76D78recombfor
<400> SEQUENCE: 322
gaaaaaagtg ctgactggcg caaggctact 30
<210> SEQ ID NO 323
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer S76D78recombrev
<400> SEQUENCE: 323
agtagccttg cgccagtcag cacttttttc 30
<210> SEQ ID NO 324
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer T76D78recombfor
<400> SEQUENCE: 324
gaaaaaactg ctgactggcg caaggctact 30
<210> SEQ ID NO 325
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer T76D78recombrev
<400> SEQUENCE: 325
agtagccttg cgccagtcag cagttttttc 30
<210> SEQ ID NO 326
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer D76D78recombfor
<400> SEQUENCE: 326
gaaaaagatg ctgactggcg caaggctact 30
<210> SEQ ID NO 327
<211> LENGTH: 30
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer D76D78recombrev
<400> SEQUENCE: 327
agtagccttg cgccagtcag catctttttc 30
<210> SEQ ID NO 328
<211> LENGTH: 56
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer 1994hisrev
<400> SEQUENCE: 328
tgactcgagc ggccgcggat ccttagtggt ggtggtggtg gtgtcctgcc actgca 56
<210> SEQ ID NO 329
<211> LENGTH: 51
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer pGV1994ep_for
<400> SEQUENCE: 329
cggtcttcaa tttctcaagt ttcagtttca tttttcttgt tctattacaa c 51
<210> SEQ ID NO 330
<211> LENGTH: 35
<212> TYPE: DNA
<213> ORGANISM: UNKNOWN
<220> FEATURE:
<223> OTHER INFORMATION: Primer pGV1994ep_rev
<400> SEQUENCE: 330
ctaactcctt ccttttcggt tagagcggat gtggg 35
User Contributions:
Comment about this patent or add new information about this topic:
| People who visited this patent also read: | |
| Patent application number | Title |
|---|---|
| 20150127320 | METHOD AND APPARATUS FOR TRANSLATION |
| 20150127319 | Filled Translation for Bootstrapping Language Understanding of Low-Resourced Languages |
| 20150127318 | APPARATUS AND METHOD FOR SIMULATING AN OPERATION OF AN OUT-OF-ORDER PROCESSOR |
| 20150127317 | Method for in silico Modeling of Gene Product Expression and Metabolism |
| 20150127316 | METHOD AND SYSTEM FOR SIMULATING SURGICAL PROCEDURES |