Patent application title: Methods for Identifying Compounds That Modulate Ion Channel Activity of a Kir Channel
Inventors:
Roderick Mackinnon (New York, NY, US)
IPC8 Class: AG01N3368FI
USPC Class:
435 61
Class name: Chemistry: molecular biology and microbiology measuring or testing process involving enzymes or micro-organisms; composition or test strip therefore; processes of forming such composition or test strip involving nucleic acid
Publication date: 2015-11-12
Patent application number: 20150323549
Abstract:
Methods for identifying compounds that modulate the ion channel activity
of a Kir channel are provided. Methods for identifying compounds that
selectively modulate the ion channel activity of specific types of Kir
channels based on the turret region of a Kir channel are also provided.
Methods for identifying compounds to treat conditions associated with
abnormal ion channel activity are also provided. Compounds including
purified antibodies and methods of making antibodies which bind to the
turret region of a Kir channel are provided. Purified polypeptides
including at least a portion of the turret region of a Kir channel and
nucleic acid sequences encoding these polypeptides are also provided.Claims:
1. A method for identifying a compound that modulates ion channel
activity of a Kir channel comprising: identifying a compound which binds
the turret region of a Kir channel; and determining if the compound
modulates ion channel activity of the Kir channel.
2. The method of claim 1, wherein the compound is an antibody.
3. The method of claim 2, wherein the antibody is human, chimeric, or humanized.
4. The method of claim 2, wherein the antibody is selected from the group consisting of polyclonal antibodies, monoclonal antibodies, an intact immunoglobulin molecule, an antibody fragment, a scFv, a Fab, a F(ab)2, a Fv, and a disulfide linked Fv.
5. The method of claim 1, wherein the compound is a nucleic acid.
6. The method of claim 5, wherein the nucleic acid is selected from the group consisting of DNA and RNA.
7. The method of claim 5, wherein the identifying includes in vitro selection of the nucleic acid.
8. The method of claim 1, wherein the compound is a protein/peptide.
9. The method of claim 8, wherein the protein/peptide is attached to a protein scaffold.
10. The method of claim 8, wherein the protein/peptide is displayed on the surface of a phage.
11. The method of claim 1, wherein the compound is a small molecule.
12. The method of claim 1, wherein the Kir channel is human.
13. The method of claim 1, wherein the Kir channel is a chicken/human hybrid.
14. The method of claim 13, wherein the chicken/human hybrid Kir channel comprises a human Kir channel turret region.
15. The method of claim 1, wherein the identifying step comprises an ELISA and a Western blot to determine if the compound binds to a properly folded Kir channel but not to a denatured Kir channel.
16. The method of claim 1, wherein the identifying step comprises determining if the compound binds to a Kir channel with a normal turret region but not a mutated turret region.
17. The method of claim 1 wherein the determining step comprises an electrophysiological assay.
18. The method of claim 17, wherein the electrophysiological assay is selected from the group consisting of two-electrode voltage clamp, patch clamp, and planar lipid bilayer assays.
19. The method of claim 1, wherein the determining step comprises a fluorescent assay.
20. The method of claim 19, wherein the fluorescent assay utilizes a thallium specific fluorescent dye.
21. A method for identifying a compound that selectively modulates ion channel activity of a specific type of Kir channel comprising: identifying a compound which binds the turret region of a specific type of Kir channel but does not bind to other types of Kir channels; and determining if the compound modulates the activity of the Kir channel.
22. The method of claim 21, wherein the identifying comprises deter pining if the compound binds the turret region of the specific type of Kir channel but does not bind the turret region of other type of Kir channels.
23. A method of identifying a compound to treat a condition associated with abnormal ion channel activity by a Kir channel comprising: identifying a compound which binds the turret region of a Kir channel; determining if said compound modulates ion channel activity of the Kir channel; and administering said compound which modulates ion channel activity to a subject to determine if the compound is able to treat said condition.
24. The method of claim 23, wherein the condition is selected from the group consisting of diabetes mellitus, hypertension, cardiac arrhythmia, and epilepsy.
25.-43. (canceled)
Description:
FIELD OF INVENTION
[0001] The present invention relates to Kir channel proteins and methods for identifying compounds that modulate ion channel activity by Kir channels. In particular, the present invention relates to identifying compounds which are useful for treating diseases related to the function of Kir channel proteins.
BACKGROUND OF THE INVENTION
[0002] Inward rectifier K+ charnels (Kir channel proteins) are involved in the control of many physiological processes that are important to human health. Kir channel proteins normally function as K+ (potassium) selective pores that span cell membranes. The Kir channels are referred to as inward rectifier K+ (Kir) channels based on a fundamental ion conduction property of these channels: given an equal but opposite electrochemical driving force K+ conductance into the cell far exceeds conductance out of the cell.
[0003] Among their many functions Kir channel proteins control the pace of the heart, regulate secretion of hormones into the blood stream, generate electrical impulses underlying information transfer in the nervous system and control airway and vascular smooth muscle tone. It is believed that various disease states are directly related to the function of Kir channel proteins. Members of this channel family include Kir1-Kir7, (Kubo et al., Pharmacological Rev., 57:509-526, 2005) Hypertension, atrial fibrillation, and type 2 diabetes are related to Kir channel protein function and are serious conditions for which new therapies are needed. Specific links between Kir channel proteins and disease have been found. Kir1.1 channels are present in the kidney and regulate salt secretion into the urine. Heritable mutations involving Kir1.1 cause Barter's syndrome and hypotension. Compounds which selectively inhibit Kir1.1 have the potential to serve as a new form of anti-hypertensive agent in which hypokalemia, a major side-effect of currently used diuretics, should in principle not be a problem. Thus, hypertensive individuals could benefit from Kir1.1 inhibitor-based therapies. Kir3.1 and Kir3.4 channels, which assemble to form a heteromultimer, are called G-protein-gated K+ channels (GIRK). These channels control heart rate through stimulation by the parasympathetic nervous system. GIRK channel knock-out mice do not develop atrial fibrillation under any of the usual stimuli that induce this arrhythmia in mice. (Claphan et al., JACC 37, 2136-2143 (Jun. 15, 2001)) Accordingly, inhibition of GIRK channels in humans might provide effective treatment for atrial fibrillation. Kir6 channels are expressed in beta cells of the pancreas and control insulin secretion. With the identification of compounds that selectively inhibit the Kir6 channel new therapies could be realized for the treatment of type 2 diabetes. Accordingly, Kir channel proteins are good targets for the treatment of various diseases.
[0004] The Kir channel family of proteins are very similar to each other in both sequence and, by inference, structure; thus, it has been very difficult to identify compounds that can specifically modulate one kind of Kir channel protein without cross-reacting with other types of Kir channel proteins.
[0005] For the first time the structure of a eukaryotic Kir channel has been determined, and a structural feature "the turret region" has been identified that is highly ordered in structure and, based on the amino acid sequences will differ among Kir channels. Prior to this structure, only the structure of a prokaryotic Kir channel had been determined. (Nishida et al., EMBO, vol. 26, pp. 4005-4015 (2007)) The turret is an important functional region of the protein and faces the outside of the cell making this region an attractive target for identifying potential therapeutic compounds. Given the identification of the turret region in the various Kir channel proteins and the structure in a prototype, the present invention provides a variety of methods by which the turret region may be used to identify compounds having therapeutic utility for treating the various diseases related to the function of Kir channels.
[0006] The present invention provides for the first time the expression and purification of a eukaryotic Kir channel as explained in detail below. Study of the structure of this eukaryotic Kir channel resulted in a realization of the importance of the turret region and the invention of methods which allow identification of therapeutic compounds that can selectively bind to different members of the Kir channel family of proteins.
SUMMARY OF THE INVENTION
[0007] The present invention relates to methods for identifying a compound that modulates ion channel activity of a Kir channel including identifying a compound which binds the turret region of a Kir channel; and determining if the compound modulates ion channel activity of the Kir channel.
[0008] In particular embodiments, the method may be used to identify an antibody that binds the turret region of a Kir channel and modulates the Kir channel's activity. The antibody may be human, chimeric or humanized. The antibody may also be a polyclonal antibody, monoclonal antibody, an intact immunoglobulin molecule, an antibody fragment, a scFv, a Fab, a F(ab)2, a Fv, or a disulfide linked Fv.
[0009] In another embodiment, the method may be used to identify suitable nucleic acid molecules that can modulate a Kir channel's activity by binding to its turret region. In such an embodiment, the nucleic acid may be a DNA or RNA molecule. In certain embodiments the nucleic acid is an aptamer. The method may also include identifying a suitable nucleic acid by using in vitro selection techniques.
[0010] In another embodiment, the method is used to identify a protein or peptide that can bind a turret region of a Kir channel and modulate the Kir channel. In this embodiment, the protein/peptide may be attached to a protein scaffold or displayed on the surface of a phage.
[0011] In another embodiment, the method discussed above is used to screen for small molecules that can modulate Kir channel activity by binding to the Kir channel's turret region.
[0012] In any of the methods discussed above, the Kir channel may be a human Kir channel or a chicken/human hybrid Kir channel. Typically, the chicken/human hybrid Kir channel will comprise a human Kir channel turret region.
[0013] Various standard biochemical assays may be used to identify whether a compound binds to the turret region of a Kir channel in the method of the present invention. For example, the identifying step may comprise an ELISA and a Western blot to determine if the compound binds to a properly folded Kir channel but not to a denatured Kir channel. Moreover, the identifying step may comprise determining if the compound binds to a Kir channel with a normal turret region but not a mutated turret region.
[0014] Regarding the determining whether a compound modulates the activity of a Kir channel, various electrophysiological assays may be used such as two-electrode voltage clamp, patch clamp, and planar lipid bilayer assays. Alternatively or additionally, the determining step may include a fluorescent assay such as one utilizing a thallium specific fluorescent dye.
[0015] In another aspect, the present invention relates to a method for identifying a compound that selectively modulates ion channel activity of a specific type of Kir channel including identifying a compound which binds the turret region of a specific type of Kir channel but does not bind to other types of Kir channels; and determining if the compound modulates the activity of the Kir channel.
[0016] In another embodiment, the present invention relates to a method of identifying a compound to treat a condition associated with abnormal ion channel activity by a Kir channel including identifying a compound which binds the turret region of a Kir channel; determining if the compound modulates ion channel activity of the Kir channel; and administering the compound which modulates ion channel activity to a subject to determine if the compound is able to treat the condition. In such a method, the condition may be diabetes mellitus, hypertension, cardiac arrhythmia, or epilepsy.
[0017] In another aspect, the present invention relates to a purified antibody that specifically binds to an epitope in the turret region of a Kir channel. In this embodiment, the purified antibody may be a polyclonal antibody, a monoclonal antibody, an intact immunoglobulin molecule, an antibody fragment, a scFv, a Fab, a F(ab)2, a Fv, or a disulfide linked Fv. The antibody may specifically bind to a human Kir channel such as a Kir1, Kir2, Kir3, Kir4, Kir5, Kir6, or Kir7 channel. The antibody preferably binds to an epitope within the turret region of a human Kir channel such as Kir1.1, Kir1.2, Kir2.1, Kir2.2, Kir2.3, Kir2.4, Kir3.1, Kir3.4, Kir4.1, Kir4.2, Kir5.1, Kir6.1, or Kir6.2 channel. Even more preferably, the antibody binds to the variable portion of the turret region of a human Kir channel.
[0018] In another embodiment, the present invention relates to a method of making an antibody that specifically binds to an epitope in the turret region of a human Kir channel, including providing a chicken/human hybrid Kir channel, wherein the chicken/human hybrid comprises a human Kir channel turret region; immunizing a non-human animal with the chicken/human hybrid Kir channel; and determining whether the antibody is binding to the human Kir channel turret region. In this embodiment, the chicken portion of the chicken/human hybrid Kir channel may be derived from a chicken Kir2.2 channel. Moreover, in this embodiment, the human Kir channel turret region may be derived from Kir1, Kir2, Kir3, Kir4, Kir5, Kir6, or Kir7. Preferably, the human Kir channel turret region is derived from a human Kir1.1, Kir1.2, Kir2.1, Kir2.2, Kir2.3, Kir2.4, Kir3.1, Kir3.4, Kir4.1, Kir4.2, Kir5.1, Kir6.1, or Kir6.2 channel.
[0019] In another embodiment, the present invention relates to a method of making an antibody that specifically binds to an epitope in the turret region of a human Kir channel, including providing a human Kir channel; immunizing a non-human animal with the Kir channel; and determining whether the antibody is binding to the human Kir channel turret region.,
[0020] In another embodiment, the present invention relates to a purified polypeptide that consists of the turret region of human Kir channels such as Kir1.1, Kir1.2, Kir2.1, Kir2.2, Kir2.3, Kir2.4, Kir3.1, Kir3.4, Kir4.1, Kir4.2, Kir5.1, Kir6.1, or Kir6.2. In another aspect, the present invention relates to an isolated nucleic acid comprising a nucleotide sequence that encodes a polypeptide that consists of the turret region of human Kir channels such as Kir1.1, Kir1.2, Kir2.1, Kir2.2, Kir2.3, Kir2.4, Kir3.1, Kir3.4, Kir4.1, Kir4.2, Kir5.1, Kir6.1, or Kir6.2.
BRIEF DESCRIPTION OF THE DRAWINGS
[0021] FIG. 1A-1C shows key residues in eukaryotic Kir channels. A sequence alignment of chicken Kir2.2 (GI: 118097849), human Kir2.2 (GI:23110982), human Kir2.1 (GI:8132301), human Kir1.1 (GI:1352479), human Kir3.1 (GI:1352482), human Kir3.4 (GI:1352484), human Kir6.1 (GI:2493600), human Kir7.1 (GI:3150184), KirBac1.1 (GI:33357898), KcsA (GI:39654804), and rat Kv1.2 (G1:73536156) is shown. For all the Kir sequences only the core region corresponding to the expressed protein and atomic structure of Kir2.2 is included in the alignment. For Kv1.2 only the transmembrane pore region is shown. Secondary structure elements are indicated above the sequences and the turret is shown in small dotted lines above the sequence. Residues discussed in the text are boxed in a series of alternating dashes and dots (acidic residues), a series of large dots (two disulfide-bonded cysteines), alternating dashes and pairs of dots (the inner helix bundle activation gate), series of small dashes (conserved residues among the turrets of eukaryotic Kir channels), a series of small dots (the selectivity filter and E139), and a series of large dashes (critical residues for channel-PIP2 interactions).
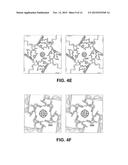
[0022] FIG. 2A-2E illustrates a structure of Kir2.2. (FIG. 2A) Stereoview of a ribbon representation of the Kir2.2 tetramer from the side with the extracellular solution above. Four subunits of the channel are shown. Approximate boundaries of the lipid bilayer are shown as bars. (FIG. 2B) A close-up view of the pore-region of a single subunit (in ribbon representation) with the turret, pore helix and selectivity filter labeled. Side chains of residues E139, R149 and a pair of disulfide-bonded cysteines (C123 and C155) are shown as sticks. Ionized hydrogen-bonds are indicated by dashed black lines. The region flanked by the two disulfide-bonded cysteines is stippled. (FIG. 2C) Electron density (wire mesh, 2Fo-Fc, calculated from 50-3.1 Å using phases from the final model and contoured at 1.0σ) is shown for the side chains of E139 and R149 forming a salt-bridge. (FIG. 2D) (FIG. 2E) K+ selectivity filter of the Kir2.2 channel (FIG. 2 D) compared with that of the Kv1.2-Kv2.1 paddle chimera channel (FIG. 2E, PDB ID 2R9R). For clarity, only two of the four subunits are shown. K+(cross hatched circles), water molecules (solid spheres), and hydrogen bonds between R149 and E139 (Kir, dashed black lines), or between D379, M380 and waters (Kv, dashed black lines) are shown,
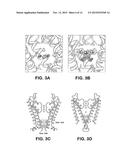
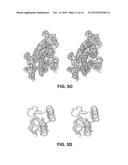
[0023] FIG. 3A-3G illustrates the cavity and gates region of a Kir channel. (FIG. 3A) (FIG. 3B) Electron density in the cavity of the Kir2.2 channel (A, Fo-Fc omit map, calculated from 50-3.1 Å using phases from the final model and contoured at 2.0σ) and of the KcsA channel (FIG. 3B, PDB ID 1K4C, Fo-Fc omit map, calculated from 50-3.1 Å using phases from the final model and contoured at 2.8σ). The channels are shown as ribbon representations with the subunit closest to the viewer removed. Only the side chains facing the cavity are shown (sticks). (FIG. 3C) (FIG. 3D) Comparison of the transmembrane inner helix bundle activation gate of Kir2.2 (FIG. 3C) with the KcsA structure (FIG. 3D, PDB ID 1K4C). For clarity, only two of the four subunits (ribbon) are shown. Side chains of the residues in the bundle crossing are shown as sticks and van der Waals surfaces. K+ ions are shown as cross hatched spheres. Inner and Outer helices are indicated. (FIG. 3E) Superposition of the chicken Kir2.2 cytoplasmic domain (α-carbon trace) and the mouse Kir2.1 cytoplasmic domain (α-carbon trace, PDB ID 1U4F) in stereo viewed from the extracellular side. (FIG. 3F) (FIG. 30) Comparison of the apex (G-loop) of the cytoplasmic pores of Kir2.2 (FIG. 3F) and mouse Kir2.1 (FIG. 3G), with the same view as FIG. 3E. The cytoplasmic domains are shown as α-carbon traces, with residues 303-309 (Kir2.2) and 302-308 (Kir2.1) shown as CPK models.
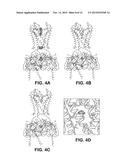
[0024] FIG. 4A-4F illustration of ion binding sites. (FIG. 4A) (FIG. 4B) (FIG. 4C) Electron density (wire mesh) of Rb+ (FIG. 4A, Fo-Fc map calculated to 4.0 Å, contoured at 3.5σ for density in the filter and 2.0σ for density elsewhere), Sr2+ (FIG. 4B, 10 mM, Fo-Fc map calculated to 3.3 Å, contoured at 1.5σ for density in the cavity and 3.0σ for density elsewhere) and Eu3+ (FIG. 4C, 10 mM, anomalous difference map calculated to 6.0 Å, contoured at 2.8σ) inside the Kir2.2 channel ion conduction pathway. Kir2.2 is represented as a α-carbon trace with the transmembrane domain and cytoplasmic domain closest to viewer removed for clarity. The ions are shown as spheres. (FIG. 4D) Electron density (200 mM Sr2+, Fo-Fc map calculated from 50-3.8 Å, contoured at 2.5σ, wire mesh) of Sr2+ (spheres) in the cavity of Kir2.2. The channel is shown as a ribbon with the subunit closest to the viewer removed. Only the side chains facing the cavity are shown (sticks). (FIG. 4E) Stereoview of the ion binding site near the upper ring of charges in the cytoplasmic domain of Kir2.2, viewed from the extracellular side. Residues E225, H227, E300, and Q311 are shown as sticks, and hydrogen bonds between them are indicated as dashed black lines. Electron density (200 mM Sr2+, Fo-Fc map calculated from 50-3.8 Å, contoured at 4.5σ) of Sr2+ (spheres) is shown as wire mesh. (FIG. 4F) Stereoview of the ion binding site at the lower ring of charges in the cytoplasmic domain of Kir2.2, viewed from the intracellular side. Residues F255, D256, and K257 are shown as sticks, and hydrogen bonds between D256 from different subunits are indicated as dashed black lines. Electron density (200 mM Sr2+, Fo-Fc map calculated from 50-3.8 Å, contoured at 4.5 a) of Sr2+ (spheres) is shown as wire mesh.
[0025] FIG. 5A-5D illustrates the unique structure of the extracellular entryway. (FIG. 5A) (FIG. 5B) Surface representation of chicken Kir2.2 (FIG. 5A) and Kv1.2-Kv2.1 paddle chimera (FIG. 5B, PDB ID 2R9R) in stereo, viewed from the extracellular side. The four protrusions formed by the top of the turrets are highlighted with a black perimeter and F148 in Kir2.2 is labeled. (FIG. 5C) Stereo representation of electron density (wire mesh) for the turret region (2Fo-Fc, calculated from 50-3.1 Å using phases from the final model and contoured at 1.0σ). The turret is shown as sticks (colored according to atom types), and residues corresponding to the highlighted protrusions in panel A are labeled. (FIG. 5D) A close-up view of the turret region in a single subunit in stereo. Side chains of those conserved residues among the turrets of eukaryotic Kir channels, as well as C155 are shown as sticks. Hydrogen bonds between H108, D110 and C123 are indicated as dashed black lines.
[0026] FIG. 6A-6D results showing the chicken Kir2.2 channel is a strong inward rectifier. (FIG. 6A) (FIG. 6B) Macroscopic currents are shown from an uninjected oocyte (FIG. 6A) and a chicken Kir2.2 channel injected oocyte (FIG. 6B) without subtracting leak and capacitive currents. The currents were recorded from oocytes using two-electrode voltage-clamp. Voltage pulses: holding potential (h.p.) 0 mV, depolarizing steps: -80 mV to +80 mV, ΔV=10 mV, stepping back to 0 mV. (FIG. 6C) Macroscopic currents recorded from oocytes using patch-clamp. The three current traces show a current trace recorded on-cell (labeled B), a trace recorded immediately after excision of the inside-out patch (labeled C), and a trace recorded approximately 10 minutes after the excision (labeled A) Voltage pulses: ramp from -80 mV to =80 mV over 10 seconds duration, (FIG. 6D) I-V curve from a patch containing only a few channels. The single channel current is graphed as a function of voltage (inset).
[0027] FIG. 7 provides a surface representation of Kir2.2, viewed from the side with the extracellular side above. The surface is shaded for qualitative assessment of the negative and positive electrostatic potential at the surface.
[0028] FIG. 8 illustrates ion binding sites of Kir2.2 in the selectivity filter, central cavity, upper and lower rings of charges are shown as sticks (oxygens and stippled). The channel is represented as a a-carbon trace with the transmembrane domain and cytoplasmic domain closest to viewer removed for clarity.
DETAILED DESCRIPTION OF THE INVENTION
[0029] The present invention is based in part on the discovery of an important structural feature present in Kir channel proteins. in particular, the present invention relates to the discovery of a "turret region" which is highly ordered in structure and which differs in sequence among different Kir channel proteins. In addition, this turret region faces the outside of the cell making the protein accessible to compounds that bind or otherwise interact with this turret region thereby affecting the ability of the Kir channel to function. The discovery of the fact that this turret region, which differs in sequence among members of the Kir channel family, is structured provides a basis to identify compounds which can treat disease states related to Kir channel functions.
[0030] Example 1 provided below presents a determination of the crystal structure of a eukaryotic Kir channel protein. In particular, the crystal structure of the chicken Kir channel protein, Kir2.2 is presented. Excluding unstructured amino and carboxy termini, the chicken Kir2.2 protein is 90% identical to human Kir2.2. More importantly, for the purposes of the present invention, these structural studies demonstrate that Kir channels have a large structured turret region which provide the basis for the development of compounds that may be used to bind and interact with these turrets and treat disease states related to the functioning of Kir channels. In particular, these turret regions suggest approaches to the development of inhibitory compounds which will bind to a specific member of the Kir channel family of proteins and inhibit Kir channel function.
[0031] The turret region of a variety of Kir channel proteins are identified in FIGS. 1A-1C and in the sequence listings of the present application. In particular, FIGS. 1A-1C illustrates that a sequence alignment of various human Kir channels indicates that the turret region begins with a consensus sequence HGDL (or minor sequence variations thereof) and extends six amino acid residues after a highly conserved cysteine residue labeled as C123 in FIGS. 1A-1C. This turret region is highly conserved and most of the variation that occurs in the sequence is located in a variable portion located after the sequence HGDL up to the cysteine labeled as C123. This variable portion within the turret region constitutes a basis for differentiating Kir channels from one another and provides a target for mutagenesis assays to identify compounds capable of turret specific binding.
[0032] Given the identification of the structured turret region in the crystal structure, the turret region of other Kir channels may be identified using sequence alignment programs and the teachings of the present invention relating to the consensus sequence and structural features of the Kir channels.
[0033] Based on this structural information, methods are presented below in which the identification of the turret region and knowledge of the amino acid sequence of the turret may be used to develop assays to identify therapeutic compounds which include, but are not limited to, antibodies, nucleic acids, peptides and small molecules that are capable of selective binding to Kir channel proteins.
[0034] In general the methods of the present invention for identifying a compound that modulates ion channel activity of a Kir channel comprises a two step process: a first step of identifying a compound which binds the turret region of a Kir channel; and a second step of determining if the compound modulates the ion channel activity of the Kir channel.
[0035] Production of Antibodies
[0036] In a first method for identifying compounds that modulate the ion channel activity of a Kir Channel, antibodies are prepared against a Kir channel. A variety of Kir channels are known and the methods described below may be used to prepare antibodies against any Kir channel.
[0037] Given the present discovery of the importance of the turret region in distinguishing one Kir channel from another, it is particularly useful to obtain antibodies which bind the turret region,
[0038] The Antigens and Assay Reagents
[0039] Two types of Kir channel proteins may be of particular utility in preparing antibodies. The first type are human Kir channel proteins. The second type are chimeric constructs which utilize a non-human sequence, preferably a eukaryotic sequence, such as a chicken sequence, in particular a chicken Kir 2.2 sequence into which a human Kir channel turret region has been inserted, thereby replacing the native turret region. As an example, chimeric proteins which utilize a chicken Kir2.2 "scaffold" into which the turret region from a given human Kir channel is inserted may be used to prepare antibodies which are specific for different human Kir channel proteins. Both human and chimeric Kir channels may be full length proteins or may contain deletions at the amino and/or carboxy termini of the protein if desired.
[0040] Conventional molecular biology, microbiology, and recombinant DNA techniques within the skill of the art may be used in order to prepare human and chimeric Kir channel proteins. Such techniques are explained fully in the literature. See, e.g., Sambrook, Fritsch & Maniatis, Molecular Cloning: A Laboratory Manual, Second Edition (1989) Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. (herein "Sambrook et al., 1989"); DNA Cloning: A Practical Approach, Volumes I and II (D. N. Glover ed. 1985); Oligonucleotide Synthesis (M. J. Gait ed. 1984); Nucleic Acid Hybridization [B. D. Hames & S. J. Higgins eds. (1985)]; Transcription And Translation [B. D. Hames & S. J. Higgins, eds. (1984)]; Animal Cell Culture [R. I. Freshney, ed. (1986)]; Immobilized Cells And Enzymes [IRL Press, (1986)]; B. Perbal, A Practical Guide To Molecular Cloning (1984); F. M. Ausubel et al, (eds.), Current Protocols in Molecular Biology, John Wiley & Sons, Inc. (1994); Molecular Cloning: A Laboratory Manual Third Edition [Joseph Sambrook and David W, Russell Cold Spring Harbor Laboratory Press (2001)]; The Condensed Protocols from Molecular Cloning: A Laboratory Manual [Joseph Sambrook and David W. Russell, Cold Spring Harbor Laboratory Press (2006)]; Gene Cloning and Manipulation Second Edition [Christopher Howe, Cambridge University Press (2007)].
[0041] The cDNA sequences for exemplary human Kir channels are presented in SEQ ID NOS 30-43. The cDNA sequence of the chicken Kir2.2 channel is presented in SEQ ID NO: 45. DNA and cDNA sequences for other types of Kir channels are available in public databases, The turret regions of exemplary human Kir proteins are identified in SEQ ID NOs: 46-56.
[0042] Expression of Chicken Kir 2.2
[0043] As an example of expression and purification of a eukaryotic Kir channel a protocol for preparing a chicken Kir 2.2 channel protein is provided below. Using standard techniques this procedure may be modified to prepare any of the human Kir channel proteins or a desired chimeric Kir channel protein.
[0044] To prepare the chicken Kir 2.2 channel, a synthetic gene fragment (Bio Basic, Inc.) encoding residues 38 to 369 of chicken Kir2.2 channel (GI:118097849) was ligated into the XhoI/EcoRI cloning sites of a modified pPICZ-B vector (Invitrogen). The resulting protein has green fluorescent protein (GFP) and a 1D4 antibody recognition sequence (TETSQVAPA) on the C terminus (I), separated by a PreScission protease cleavage site (SNSLEVLFQ/GP).
[0045] The construct was linearized using PmeI and transformed into a HIS+ strain of SMD1163 of Pichia pastoris (Invitrogen) by electroporation (BioRad Micropulser). Transformants were selected on YPDS plates containing 400-1200 μ/ml Zeocin (Invitrogen). Resistant colonies were tested for expression by anti-1D4 tag Western Blot. For large-scale expression, small cultures grown from the best expressing colony were diluted into BMGY media (Invitrogen) and inoculated at 29° C. overnight, until OD600 reached between 20-30. Cells were then pelleted, resuspended in BMM media (Invitrogen) and expressed overnight at 24° C. Cells were harvested, flash-frozen in liquid N2, and stored at -80° C. until needed.
[0046] Cells were lysed in a Retsch, Inc. Model MM301 mixer mill (5×3.0 minutes at 25 cps). The lysis buffer contained 150 mM KCl, 50 mM TRIS-HCl pH 8.0, 0.1 mg/ml deoxyribonuclease I, 0.1 μg/ml pepstatin, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 0.1 mg/ml soy trypsin inhibitor, 1 mM benzamidine, 0.1 mg/ml AEBSF, with 1 mM phenylmethysulfonyl fluoride added just before lysis (3.0 ml lysis buffer/g cells). pH of the lysate was adjusted to 8.0 with KOH. The lysate was extracted with 100 mM DM (n2 decyl-β-D-maltopyranoside, Anatrace, solgrade) at room temperature for 1 hour with stirring, and then centrifuged for 40 minutes at 30,000 g, 10° C. Supernatant was added to 1D4-affinity resin pre-equilibrated with 150 mM KCl, 50 mM TRIS-HCl pH 8.0, and 4 mM DM. Suspension was layered with Argon and mixed by inversion for 2 hours at room temperature. Beads were collected on a column by gravity, washed with 2 column volumes of buffer (150 mM KCl, 50 mM TRIS-HCl pH 8.0, 1 mM EDTA pH 8.0, and 4 mM DM), and eluted with buffer plus 1 mg/ml 1D4 peptide (AnaSpec, Inc.) over 1 hour at room temperature. 20 mM DTT (Dithiothreitol) and 3 mM TECP were added to eluted protein. The protein was then digested with PreScission protease (20:1 w/w ratio) overnight at 4° C. Concentrated protein was further purified on a Superdex-200 gel filtration column in 150 mM KCl, 20 mM TRIS-HCl pH 8.0, 4 mM DM (anagrade), 3 mM TCEP, 20 mM DTT and 1 mM EDTA at 4° C. In a preferred embodiment, the protein extract is maintained in a mild detergent, such as DM, which will maintain the three-dimensional structure of the Kir channel.
[0047] Preparation of Human/Chicken Hybrid Kir Channels
[0048] Using standard techniques in molecular biology, chimeric Kir channel protein may be prepared by inserting the turret region of a human Kir channel protein into the Kir2.2 chicken sequence described above. The location of the turret regions are identified in FIGS. 1A-1C.
[0049] By way of example, site-directed mutagenesis procedures may be used to insert the coding sequence for a human turret region into a eukaryotic "scaffold" Kir channel coding region. In a preferred embodiment, Strategene's QuickChange® is used. QuickChange® utilizes a supercoiled double-stranded DNA (dsDNA) vector with an insert of interest and two synthetic oligonucleotide primers containing the desired mutation. The oligonucleotide primers, each complementary to opposite strands of the vector, are extended during temperature cycling by PfuTurbo DNA polymerase. The desired mutation (in this case--the insertion of the human turret region) should be in the middle of the primer with about 10-15 bases of correct sequence on both sides. Incorporation of the oligonucleotide primers generates a mutated plasmid containing staggered nicks. Following temperature cycling, the product is treated with Dpn I. The Dpn I endonuclease (target sequence: 5'-Gm6ATC-3') is specific for methylated and hemimethylated DNA and is used to digest the parental DNA template and to select for mutation-containing synthesized DNA. DNA isolated from almost all E. coli strains is Dam methylated and therefore susceptible to Dpn I digestion. The nicked vector DNA containing the desired mutations is then transformed into XL1-Blue supercompetent cells. See, e.g. U.S. Pat. Nos. 5,789,166, 5,932,419, and 6,391,548.
[0050] As an example, the chicken Kir2.2 protein may be used as a scaffold and the human Kir2.2 channel turret region synthesized for insertion. This methodology can be repeated with any combination of scaffold protein and human turret region.
[0051] Preparation of Mutated Turret Regions
[0052] Given the identification of the turret regions in the human Kir channels site directed mutagenesis or other techniques known in the art may be used to prepare proteins having mutations in the DNA sequence of the turret. In a preferred embodiment, Strategene's QuickChange® is used. Such mutations should be non-silent mutations--that is the mutations should result in amino acid changes at positions within the turret region.
[0053] Generation of Antibodies
[0054] A human Kir protein or a chimeric Kir channel protein is prepared using standard techniques such as those outlined herein, and used in standard techniques to obtain antibodies.
[0055] A variety of antibodies may be used in the present invention and such antibodies include but are not limited to polyclonal, monoclonal, human, humanized chimeric, an intact immunoglobulin molecule, an antibody fragment, single chain, ScFv, Fab fragments, F(ab)2 Fab, Fv and a disulfide linked Fv.
[0056] Various procedures known in the art may be used for the production of antibodies. Host animals can be immunized by injection with a human Kir channel protein, or chimeric Kir protein or fragments of these proteins. Animals which may be used to generate antibodies include, but are not limited to, rabbits, mice, rats, sheep, goats, and others known in the art. The human and chimeric proteins of the present invention may also be conjugated to an immunogenic carrier, e.g., bovine serum albumin (BSA) or keyhole limpet hemocyanin (KLH). Various adjuvants may be used to increase the immunological response, depending on the host species, including but not limited to Freund's (complete and incomplete), mineral gels such as aluminum hydroxide, surface active substances such as lysolecithin, pluronic polyols, polyanions, peptides, oil emulsions, keyhole limpet hernocyanins, dinitrophenol, and potentially useful human adjuvants such as BCG (Bacille Calmette-Guerin) and Corynebacterium parvum.
[0057] For preparation of monoclonal antibodies directed toward a Kir channel protein of the present invention, any technique that provides for the production of antibody molecules by continuous cell lines in culture may be used, These include but are not limited to the hybridoma technique originally developed by Kohler and Milstein [Nature 256:495-497 (1975)], as well as the trioma technique, the human B-cell hybridoma technique [Kozbor et al., Immunology Today 4:72 1983); Cote et al., Proc. Nail. Acad. Sci. U.S.A. 80:2026-2030 (1983)], and the EBV-hybridoma technique to produce human monoclonal antibodies [Cole et al., in Monoclonal Antibodies and Cancer Therapy, Alan R. Liss, Inc., pp. 77-96 (1985)]. In addition, techniques developed for the production of "chimeric antibodies" [Morrison et al., J. Bacterial. 159:870 (1984); Neuberger et al., Nature 312:604-608 (1984); Takeda et al., Nature 314:452-454 (1985)] by splicing the genes from a mouse antibody molecule specific for an isolated Kir channel protein of the present invention, or conserved variants thereof, together with a fragment of a human antibody molecule of appropriate biological activity can be used.
[0058] Human antibodies can be prepared using any technique. Examples of techniques for human monoclonal antibody production include those described by Cole et al, (Monoclonal Antibodies and Cancer Therapy, Alan R. Liss, p. 77, 1985) and by Boemer et al, (J. Immunol., 147(1):86-95, 1991). Human antibodies (and fragments thereof) can also be produced using phage display libraries (Hoogenboom et al., J. Mol. Biol., 227:331, 1991; Marks et al., J. Mol, Biol., 222:581, 1991), Information on monoclonal and other types of therapeutic antibodies can also be found in Cellular and Molecular Immunology, 6th Edition, [A. K. Abbas, A. H. Lichtman, S. Pillai (Saunders Elsevier Press, 2007)], and U.S. Pat. Nos. 7,390,887 and 7,629,171. For a discussion of various types of therapeutic antibodies, see Strategies and Challenges for the Next Generation of Therapeutic Antibodies, A. Beck, T. Wurch, C. Bailly and N. Corvaia, Nature Rev. Immuno. 10, 345-352 (2010).
[0059] Human antibodies can also be obtained from transgenic animals. For example, transgenic, mutant mice that are capable of producing a full repertoire of human antibodies, in response to immunization, have been described (see, e.g., Jakobovits et al., Proc. Natl. Acad. Sci. USA, 90:2551-255 (1993); Jakobovits et al., Nature, 362:255-258 (1993); Bruggennann et al., Year in Immunol., 7:33 (1993)).
[0060] Humanized antibodies may also be used in the present invention. Antibody humanization techniques generally involve the use of recombinant DNA technology to manipulate the DNA sequence encoding one or more polypeptide chains of an antibody molecule. Accordingly, a humanized form of a non-human antibody (or a fragment thereof) is a chimeric antibody or antibody chain (or a fragment thereof, such as an Fv, Fab, Fab', or other antigen-binding portion of an antibody) which contains a portion of an antigen binding site from a non-human (donor) antibody integrated into the framework of a human (recipient) antibody. Methods for humanizing non-human antibodies are well known in the art. For example, humanized antibodies can be generated according to the methods of Winter and co-workers (Jones et al., Nature, 321:522-525 (1986), Riechmann et al., Nature, 332:323-327 (1988), Verhoeyen et al., Science, 239:1534-1536 (1988)), by substituting rodent CDRs or CDR sequences for the corresponding sequences of a human antibody. Methods that can be used to produce humanized antibodies are also described in U.S. Pat. No. 4,816,567 (Cabilly et al.), U.S. Pat. No. 5,565,332 (1-Hoogenboom et al.), U.S. Pat. No. 5,721,367 (Kay et al.), U.S. Pat. No. 5,837,243 (Deo et al.), U.S. Pat. No. 5,939,598 (Kucherlapati et al.), U.S. Pat. No. 6,130,364 (Jakobovits et al.), and U.S. Pat. No. 6,180,377 (Morgan et al.).
[0061] Techniques described for the production of single chain antibodies [U.S. Pat. Nos. 5,476,786 and 5,132,405 to Huston; U.S. Pat. No. 4,946,778] can be adapted to produce single chain antibodies specific for a Kir channel protein. An additional embodiment of the invention utilizes the techniques described for the construction of Fab expression libraries [Huse et al., Science 246:1275-1281(1989)] to allow rapid and easy identification of monoclonal Fab fragments with the desired specificity for the Kir channel proteins.
[0062] Antibody fragments which contain the idiotype of the antibody molecule can be generated by known techniques. For example, such fragments include but are not limited to: the F(ab')2 fragment which can be produced by pepsin digestion of the antibody molecule; the Fab' fragments which can be generated by reducing the disulfide bridges of the F(ab')2 fragment, and the Fab fragments which can be generated by treating the antibody molecule with papain and a reducing agent.
[0063] Antibodies or fragments thereof, whether attached to other sequences or not, can also include insertions, deletions, substitutions, or other selected modifications of particular regions or specific amino acids residues, provided the activity of the antibody or antibody fragment is not significantly altered or impaired compared to the non-modified antibody or antibody fragment. Such methods are readily apparent to a skilled practitioner in the art and can include site-specific mutagenesis of the nucleic acid encoding the antibody or antibody fragment.
[0064] Nucleic Acids
[0065] The compounds of the present invention include nucleic acids. In particular, nucleic acid sequences capable of binding to a Kir channel may be used in the practice of the present invention. These nucleic acids may be identified using in vitro selection of sequences which bind Kir channel proteins, in particular the turret region of the Kir channel proteins. One type of nucleic acid that is of particular interest is an aptamer. Typically aptamers are small nucleic acid sequences ranging from 15-50 bases in length that fold into defined secondary and tertiary structures that bind to another molecule. This binding is not the typical nucleic acid to nucleic acid hydrogen bond formation but the binding of aptamers can include all other types of covalent and noncovalent binding. In a preferred embodiment, the nucleic acid is DNA, however, other nucleic acids such as RNA may be used. The nucleic acids may be modified or prepared using techniques known in the art to increase the stability of nucleic acids. Representative examples of how to make and use aptamers to bind a variety of different target molecules can be found in the following U.S. Pat. Nos. 5,582,981; 5,595,877; 5,637,459; 6,020,130; 6,028,186; 6,030,776; and 6,051,698. See also Published U.S. patent application Ser. No. 11/917,884 (publication No. US2009/0155779A1); Bock L C, Griffin L C, Latham J A, Vermaas E H, Took J J (February 1992). "Selection of single-stranded DNA molecules that bind and inhibit human thrombin" Nature 355(6360): 564-6; Bunka D H, Stockley P G (August 2006) "Aptamers come of age--at last" Nat Rev Microbio. 4(8): 588-96.
[0066] Small Protein/Peptide Compounds
[0067] Small Protein/Peptide Compounds may also be used in the practice of the present invention. In particular, small proteins may be prepared and screened for the ability to bind to a Kir channel protein based on binding assays disclosed herein and known in the art. Small molecules such as toxins may also be used in the practice of the invention. In particular, small proteins/peptides modeled on toxins which bind to Kir channel proteins may be prepared and tested for the ability to bind and modulate the activity of Kir channel proteins. (Ramu, et al. (2008) Engineered specific and high affinity inhibition for a subtype of inward rectifier Kir channels Proc. Nat'l Acid Sci USA 105:10774-10778)
[0068] A variety of toxins may provide information useful in designing a compound useful in the practice of the present invention. In particular, scorpion toxins (Lu and Mackinnon, 1997 Biochemistry, vol. 36, no, 23, pp 6936 to 6940) snake toxins (for example, the 57 amino acid δ-dendrotoxin from the green mamba snake which inhibits Kir 1.1 channels, (J. P. Imredy, C. Chen, R. Mackinnon, BioChemistry 37, 14867 (Oct. 20, 1998)) and bee venom toxins (Ramu, et al. (2008) Engineered specific and high affinity inhibition for a subtype of inward rectifier Kir channels Proc. Nat'l Acid Sci USA 105:10774-10778) may be helpful in synthesizing libraries of protein/peptide compounds that can bind and effect a Kir channel. Known toxins are often small proteins typically between 20 and 50 amino acids in size containing disulfide bridges. For some of these toxins, the surface important for binding to a Kir channel protein is known and stretches of amino acids of less than 10 amino acids are believed to be important for binding specificity.
[0069] A library of these toxin-based compounds may be prepared while maintaining the key amino acids such as cysteine residues that are important for the folding and structure of the proteins. The amino acid residues important for binding to a Kir channel may be randomized to generate proteins/peptides with enhanced binding strength and turret based specificity for the different members of the Kir channel family of proteins.
[0070] Phage Display
[0071] One method known in the art to rapidly screen a large variety of potential binding proteins/peptides is a phage display assay.
[0072] Phage display libraries may be prepared using known protocols. "Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface". Science 288 (4705): 1315-1317. Smith G P, Petrenko V A (1997). "Phage display". Chem. Rev. 97 (2): 391-410. Kehoe J W, Kay B K (2005), "Filamentous phage display in the new millennium". Chem. Rev. 105 (11): 4056-4072. Hufton S E, Moerkerk P T, Meulemans E V, de Bruine A, Arends J W, Hoogenboom H R (1999). "Phage display of cDNA repertoires: the pVI display system and its applications for the selection of immunogenic ligands." J. Immunol. Methods 231 (1-2): 39-51, Lunder M, Bratkovic T, Doljak B, Kreft S, Urleb U, Strukelj B, Plazar N. (2005). "Comparison of bacterial and phage display peptide libraries in search of target-binding motif". Appl. Biochem. Biotechnol. 127 (2): 125-31. Lunder M, Bratkovic T, Kreft S, Strukelj B (2005). "Peptide inhibitor of pancreatic lipase selected by phage display using different elution strategies". J. Lipid Res. 2005 46 (7): 1512-6.)
[0073] Using phage display the protein/peptide constructs may be expressed on the outer coat of the phage. To identify useful sequences a Kir channel protein may be immobilized on the surface of a well of a standard assay plate, and a phage that displays a protein that binds to Kir channels will bind the target Kir channel protein and remain bound while non-binding phage are removed by washing the plates. The bound phage can be eluted and used to produce more phage for further binding assays. These binding assays may be performed with Kir channels having mutated turrets and wild type turrets to select for proteins/peptides that bind in the turret region. Repeated cycles of these binding assays (`panning`) results in the identification of phage containing potentially strong binding sequences.
[0074] Phage that contain these strong binding sequences can be used to infect a suitable bacterial host, and phagemids are collected and the DNA sequence of interest encoding the binding region excised and sequenced to identify the protein/peptide compound which be further tested using the assays described below.
[0075] Small Molecules
[0076] Small molecules may also be used in the practice of the present invention. In particular, small molecules may be prepared and screened for the ability to bind to a Kir channel protein based on binding assays disclosed herein and known in the art. See for example U.S. Pat. No. 6,641,997. Additionally, small molecule libraries may also be screened.
[0077] Immunoassays
[0078] Once an antibody has been generated by the methods described above, a variety of different immunoassays may be performed to identify antibodies that bind property folded Kir channels, are specific for the turret region of the Kir channel and can differentiate between different members of the Kir family based on the turret region.
[0079] It is believed that ELISA and Western blot assays are straightforward and efficient assays to identify such antibodies before performing functional assays such as electrophysiological assays.
[0080] In general, immunoassays involve contacting a Kir channel protein with an anti-Kir channel antibody under conditions effective, and for a period of time sufficient, to allow the formation of immune complexes (primary immune complexes). Forming such complexes is generally a matter of simply bringing into contact the antibody and the Kir channel protein sample and incubating the mixture for a period of time long enough for the antibodies to form immune complexes with, i.e., to bind to, any molecule (e.g., antigens) present to which the antibodies can bind.
[0081] In many forms of immunoassay, the sample-antibody composition, such as an ELISA plate or Western blot, can then be washed to remove any non-specifically bound antibody species, allowing only those antibodies specifically bound within the primary immune complexes to be detected. Immunoassays can include methods for detecting or quantifying the amount of a molecule of interest (such as the disclosed biomarkers or their antibodies) in a sample. In general, the detection of an immunocomplex formation is well known in the art and can be achieved by numerous approaches. These methods are generally based upon the detection of a label or marker, such as any radioactive, fluorescent, biological or enzymatic tags or any other known label. Such assays include but are not limited to ELISA, western blots, radioimmunoassay, (enzyme-linked immunosorbant assay), "sandwich" immunoassays, immunoradiometric assays, gel diffusion precipitin reactions, immunodiffusion assays, in situ immunoassays (using colloidal gold, enzyme or radioisotope labels, for example), precipitation reactions, agglutination assays (e.g., gel agglutination assays, hemagglutination assays), complement fixation assays, immunofluorescence assays, protein A assays, and immunoelectrophoresis assays, and other assays known in the art.
[0082] Antibody binding can be detected by detecting a label on the primary antibody or the primary antibody is detected by detecting binding of a secondary antibody or reagent to the primary antibody. For some assays, the secondary antibody is labeled. Many means are known in the art for detecting binding in an immunoassay and are within the scope of the present invention.
[0083] The use of immunoassays to detect a specific protein can involve the separation of the proteins by electrophoresis. Electrophoresis is the migration of charged molecules in solution in response to an electric field. Their rate of migration depends on the strength of the field; on the net charge, size and shape of the molecules and also on the ionic strength, viscosity and temperature of the medium in which the molecules are moving. As an analytical tool, electrophoresis is simple, rapid and highly sensitive. It is used analytically to study the properties of a single charged species, and as a separation technique. Electrophoresis is used in the Western blots described below.
[0084] ELISA
[0085] Enzyme-Linked Immunosorbent Assay (ELISA), or more generically termed EIA (Enzyme ImmunoAssay), is an immunoassay that can detect an antibody specific for a protein. In such an assay, a detectable label bound to either an antibody-binding or antigen-binding reagent is an enzyme. When exposed to its substrate, this enzyme reacts in such a manner as to produce a chemical moiety which can be detected, for example, by spectrophotometric, fluorometric or visual means. Enzymes which can be used to detectably label reagents useful for detection include, but are not limited to, horseradish peroxidase, alkaline phosphatase, glucose oxidase, galactosidase, ribonuclease, urease, catalase, malate dehydrogenase, staphylococcal nuclease, asparaginase, yeast alcohol dehydrogenase, alpha.-glycerophosphate dehydrogenase, triose phosphate isomerase, glucose-6-phosphate dehydrogenase, glucoamylase and acetylcholinesterase. For descriptions of ELISA procedures, see Voller, A. et al., J. Clin. Pathol. 31:507-520 (1978); Butler, J. E., Meth. Enzymol. 73:482-523 (1981); Maggio, E. (ed.), Enzyme Immunoassay, CRC Press, Boca Raton, 1980; Butler, J. E., In: Structure of Antigens, Vol. 1 (Van Regenmortel, M., CRC Press, Boca Raton, 1992, pp. 209-259; Butler, J. E., In: van Oss, C. J. et al., (eds), Immunochemistry, Marcel Dekker, Inc., New York, 1994, pp. 759-803; Butler, J. E. (ed.), Immunochemistry of Solid-Phase Immunoassay, CRC Press, Boca Raton, 1991); Crowther, "ELISA: Theory and Practice," In: Methods in Molecule Biology, Vol. 42, Humana Press; New Jersey, 1995; U.S. Pat. No. 4,376,110, each of which is incorporated herein by reference in its entirety and specifically for teachings regarding ELISA methods.
[0086] In preferred embodiments of the present invention, ELISA assays are used to identify antibodies that bind to the Kir channel proteins and are specific to the turret region.
[0087] As illustrated in FLOWCHART I, a first ELISA assay is performed to identify antibodies that bind to the Kir channel protein. As an example, if a human Kir 2.2 channel protein is used as an antigen, an ELISA Assay is performed to identify antibodies that bind to human Kir 2.2 channel protein.
[0088] By way of an example ELISA assay, solutions are prepared as follows:
[0089] Buffer A: Protein buffer containing detergent slightly above CMC
[0090] Coating Solution--Buffer A+20 ug/ml Protein (50 ul/well, 5.0 ml./plate)
[0091] Wash Solution--Buffer A (200 ul/well×14 washes; 2.8 ml/well, 280 ml total/plate)
[0092] Blocking Solution--Buffer A+5% BSA (0.45u filtered) (400 u/well, 40 ml./plate)
[0093] Primary Ab solution--Cell culture supernatant (diluted 1:1 with 2×Buffer A/2% BSA) or control sera (1:100 in Buffer A+2% BSA)
[0094] Secondary Ab Solution--1:10,000 goat α mouse-Horseradish peroxidase conjugate in Buffer A+2% BSA (100ul/well, 10 ml/plate)
[0095] Substrate Solution--1:1 TMB:H2O2 (100 ul/well, 10 ml/plate)
[0096] Stop Solution--2M H2SO4 (100 ul/well, 10 ml./plate) The following steps are then performed:
[0097] a) Add 50 ul of coating solution to each well. Prepare coated plates the day before the assay and store at 4° C. overnight, or prepare on day of assay and allow to shake for 1 hour at room temperature. Add solution directly to the bottom of the well, avoiding the sides as much as possible. Coat at least 2 more wells than you have samples for (+) and (-) controls, Leave at least 2 wells uncoated (just wash solution) as negative controls.
[0098] b) Remove coating solution by pouring out and smacking plate face down on a paper towel. Wash wells 3× by adding 200 ul of wash solution to each well, shaking for 1 minute, pouring out wash solution and smacking plates face down on paper towels.
[0099] c) Add 300 ul of blocking solution to each well and let plates sit at room temperature for 2 hour.
[0100] d) Remove blocking solution and wash wells 3× with 200 ul of washing solution.
[0101] e) Add 50 ul 2×Buffer to each well (except controls)
[0102] f) Add 50 ul primary antibody solution to the 50 ul 2×buffer in each well and mix. Add diluted (+) control serum to a coated and uncoated well and diluted (-) control serum to coated and uncoated well. Let plates sit at room temperature for 1 hour on orbital shaker.
[0103] g) Remove Primary Ab solution and wash wells 3× with 200 ul washing solution.
[0104] h) Add 100 ul secondary Ab solution to each well and let plates sit at room temperature for 1 hour.
[0105] The plates are then examined to determine if antibodies for a Kir channel protein are present using standard techniques as described above.
Western Blot
[0106] Western blotting or immunoblotting allows the determination of the molecular mass of a protein and the measurement of relative amounts of the protein present in different samples. Detection methods include chemiluminescence and chromagenic detection. Standard methods for Western blot analysis can be found in, for example, D. M. Bollag et al., Protein Methods (2d edition 1996) and E. Harlow & D. Lane, Antibodies, a Laboratory Manual (1988), U.S. Pat. No. 4,452,901, each of which is herein incorporated by reference in their entirety for teachings regarding Western blot methods. Generally, proteins are separated by gel electrophoresis, usually SDS-PAGE. For the assays used in the present invention for initial antibody screening, it is preferred to use an SDS-PAGE so as to denature the Kir channel proteins used in the blot. The proteins are transferred to a sheet of special blotting paper, e.g., nitrocellulose, though other types of paper, or membranes, can be used. The proteins retain the same pattern of separation they had on the gel. The blot is incubated with a generic protein (such as milk proteins) to bind to any remaining sticky places on the nitrocellulose. An antibody is then added to the solution which is able to bind to its specific protein.
[0107] The attachment of specific antibodies to specific immobilized antigens can be readily visualized by indirect enzyme immunoassay techniques, usually using a chromogenic substrate (e.g. alkaline phosphatase or horseradish peroxidase) or chemiluminescent substrates. Other possibilities for probing include the use of fluorescent or radioisotope labels (e.g., fluorescein, 125I). Probes for the detection of antibody binding can be conjugated anti-immunoglobulins, conjugated staphylococcal Protein A (binds IgG), or probes to biotinylated primary antibodies (e.g., conjugated avidin/streptavidin).
[0108] As illustrated in FLOWCHART 1, a western blot is performed to determine if an antibody binds to the denatured form of a Kir channel protein, The combination of the ELISA and the Western blot Kir channel assays as illustrated in FLOWCHART 1 facilitates the identification of antibodies that recognize the properly folded native Kir channel (ELISA Positive) but not the denatured (Western Negative) form of the protein.
[0109] In particular, if a given antibody binds to a Kir channel protein in an ELISA assay ("ELISA Positive"), but fails to bind to the same Kir channel protein in a Western blot (Western Negative), then the antibody is binding to the native conformation of the protein but not the denatured form.
[0110] Identification of Antibodies with Turret Specificity
[0111] Given the discovery in the present invention of the importance of the turret regions and the identification of the region of the Kir proteins which constitute the turret region it is possible to prepare Kir channel proteins which contain mutations located in the turret region. This in turn provides the basis for identification of antibodies that are specific for the turret region of the Kir channel proteins. In particular, the determination of the atomic structure of what constitutes the turret region of the Kir channels identifies where to introduce such mutations so as to selectively identify anti-Kir Channel antibodies that are directed against the turret. Such mutations may be made in a variety of places, such as following the L residue in the sequence HGDL (or slight variations of that sequence) and up to but not including the conserved cysteine labeled C123 in the structure of the proteins (see FIGS. 1A-1C). Examples of these variable portions of certain turret regions is provided in SEQ ID NOs 1-13. Kir channel proteins which contain mutations in the turret region, or preferably in the variable portion, may be used in assays described below.
[0112] To identify antibodies that are specific for the turret region of the Kir channel further ELISA assays may be performed as illustrated in FLOWCHART 1. These ELISA assays utilize Kir channels with mutated turret regions. Antibodies that bind a normal Kir channel in an ELISA assay but do not bind a channel with a mutated turret will be isolated since these antibodies may be considered turret specific--that is the epitope for the antibody is located in the turret region of the protein. The source of these antibodies will be used to prepare monoclonal antibodies using standard techniques as described above. An additional assay described below and presented in FLOWCHART 2 identifies antibodies or other compounds with the ability to bind the turret region using a fluorescent assay.
[0113] Assays for Kir Channel Activity
[0114] Even if an antibody or other type of compound binds the turret region its utility as a therapeutic compound is based on its functional effect on a Kir channel, Accordingly, the next step is to determine if an antibody which binds the turret region of a Kir channel is capable of modulating electrolyte processing. Monoclonal antibodies prepared from turret specific antibodies identified above can be used in electrophysiological assays as can other compounds found to have binding specificity for the turret region of Kir channels.
[0115] There are a variety of electrophysiological assays known to those with skill in the art which may be used to determine whether the compounds of the present invention have an effect on the electrophysiological state of a Kir Channel.
[0116] Useful electrophysiological assays include a variety of in vitro and in vivo assays, e.g., measuring voltage, current, measuring membrane potential, measuring ion flux, e.g., potassium or rubidium, measuring potassium concentration, measuring second messengers and transcription levels, and using e.g., voltage-sensitive dyes, radioactive tracers, electrode voltage clamps and patch-clamp electrophysiology. Such assays can be used to test for both inhibitors and activators of Kir channels.
[0117] Modulators of the Kir channels may be tested using biologically active, functional Kir channels, either recombinant or naturally occurring. In recombinantly based assays, the subunits are typically expressed and modulation is tested using one of the in vitro or in vivo assays described herein.
[0118] In brief, samples or assays that are treated with a potential Kir channel turret binding compounds inhibitors or activators are compared to control samples without the test compound, to examine the extent of modulation. Control samples e.g. those untreated with the compounds are assigned a relative Kir channel activity value of 100. Inhibition is present when Kir channel activity value relative to the control is about 90%, preferably 50%, more preferably 25%.
[0119] It should be noted that the compounds may also result in activation of Kir channels. Activation of channels is achieved when the select Kir channel activity value relative to the control is 110%, more preferably 150%, more preferable 200% higher. It is possible that for treating some diseases states such activating compounds may be useful alone or in combination with inhibitors.
[0120] Changes in ion flux may be assessed by determining changes in polarization (i.e., electrical potential) of the cell or membrane expressing the Kir channels of this invention. A preferred means to determine changes in cellular polarization is by measuring changes in current (thereby measuring changes in polarization) with voltage-clamp and patch-clamp techniques, e.g., the "outside-out" mode, and the "whole cell" mode (see, e.g., Ackerman et al., New Engl. J. Med. 336:1575-1595 (1997) and Single Channel Recording, Plenum Press, B. Sakmann and E. Neher eds). Whole cell currents are conveniently determined using the standard methodology (see, e.g., Hamil et al., P Fingers. Archly. 391:85 (1981). Other known assays include: radiolabeled rubidium flux assays and fluorescence assays using voltage-sensitive dyes (see, e.g., Vestergarrd-Bogind et al., J. Membrane Biol. 88:67-75 (1988); Daniel et al., J. Pharmacol. Meth. 25:185-193 (1991); Holevinsky et al., J. Membrane Biology 137:59-70 (1994)). Assays for compounds capable of inhibiting or increasing potassium flux through the channel proteins can be performed by application of the compounds to a bath solution in contact with and comprising cells having an channel of the present invention (see e.g., Blatz et al., Nature 323:718-720 (1986); Park, J. Physiol. 481:555-570 (1994)). Generally, the compounds to be tested are present in the range from 1 pM to 100 μM.
[0121] The effects of the test compounds upon the function of the Kir channels can be measured by changes in the electrical currents or ionic flux or by the consequences of changes in currents and flux. Changes in electrical current or ionic flux are measured by either increases or decreases in flux of cations such as potassium or rubidium ions. The cations can be measured in a variety of standard ways. They can be measured directly by concentration changes of the ions or indirectly by membrane potential or by radiolabeling of the ions. Consequences of the test compound on ion flux can be quite varied. Accordingly, any suitable physiological change can be used to assess the influence of a test compound on the Kir channels of this invention. The effects of a test compound can be measured by a toxin binding assay. When the functional consequences are determined using intact cells or animals, one can also measure a variety of effects such as transmitter release (e.g., dopamine), hormone release (e.g., insulin), transcriptional changes to both known and uncharacterized genetic markers (e.g., northern blots), cell volume changes (e.g., in red blood cells), immunoresponses (e.g., T cell activation), changes in cell metabolism such as cell growth or pH changes, and changes in intracellular second messengers such as [Ca2+].
[0122] Two Electrode Voltage Clamp Assay
[0123] One assay that may be of particular use in the present invention is a two electrode voltage clamp assay. This assay may be performed using any of these Kir channel proteins and compounds of the present invention using modifications readily known to those in the art. In the example below, this assay was conducted using the chicken Kir 2.2 channel. To perform this assay, Xenopus oocytes will be harvested from mature female Xenopus laevis and defolliculated by collagenase treatment for 1-2 hours. Oocytes will then rinsed thoroughly and stored in ND96 solution (96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, 5 mM HEPES, 50 μg/ml gentamycin, pH 7.6 with NaOH). Defolliculated oocytes will be selected 2-4 hours after collagenase treatment and injected with cRNA the next day. The injected oocytes will be incubated in ND96 solution for 1-5 days before recording. All oocytes will be stored in an incubator at 18° C.
[0124] The desired human or chimeric Kir channel protein DNA will be sub-cloned into the pGEM vector (Promega). cRNA will be prepared using T7 RNA polymerase (Promega) from NdeI-linearized plasmid DNA.
[0125] All recordings will be performed at room temperature. For two-electrode voltage-clamp experiments, oocytes will be held at 0 mV and pulsed from -80 mV to +80 mV with 10 mV increment steps. Recording solution will contain 98 mM KCl, 0.3 mM CaCl2, 1 mM MgCl2, and 5 mM HEPES pH 7.6. The ionic currents will be recorded with an oocyte clamp amplifier (OC-725C, Warner Instrument Corp.). The recorded signal will be filtered at 1 kHz and sampled at 10 kHz using an analogue-to-digital converter (Digidata 1440A, Axon Instruments, Inc) interfaced with a computer. pClamp10.1 software (Axon Instruments, Inc) will be used for controlling the amplifier and data acquisition.
[0126] Patch Clamp Assays
[0127] Patch clamp assays use a micropipette attached to a cell membrane to allow recording from a single ion channel in the cell membrane.
[0128] To perform this type of assay a micropipette which serves as a microelectrode is positioned next to a cell, and a piece of the cell membrane (the `patch`) is drawn into the microelectrode tip; the glass tip of the micropipette forms a high resistance `seal` with the cell membrane, then whole cell mode is entered by applying suction. Next, the pipette is moved away from the cell to form an outside-out patch. Examples of useful protocols may be found in Single Channel Recording, Plenum Press, B. Sakmann and E. Neher eds. This configuration can be used to study Kir channels present in the isolated patch of membrane. Variations of this technique include the "perforated patch" technique, or the patch of membrane can be pulled away from the rest of the cell.
[0129] As an example, for patch-clamp experiments in the outside-out mode, each oocyte will be incubated in a hypertonic solution containing 200 mM NaCl, 130 mM KCl, 5 mM K2EDTA, 5 mM K2HPO4, 5 mM KH2PO4 pH 7.2 for 5-10 minutes and the vitelline membrane will be removed before seal formation. Currents will be recorded in either cell-attached or outside-out configuration with an Axopatch 200B amplifier, Digidata 1440A analogue-to-digital converter and pClamp10.1 software to control membrane voltage and record. During the current recordings, the membrane will be first held at 0 mV followed by a 10-second voltage ramp from +80 mV to -80 mV. The pipette solution will contain 140 mM KCl, 5 mM K2HPO4, 5 mM KH2PO4, 0.3 mM CaCl2, 1 mM MgCl2, pH 7.2 with KOH. The bath solution will contain 130 mM KCl, 5 mM K2EDTA, 5 mM K2HPO4, 5 mM KH2PO4, pH 7.2 with KOH.
[0130] To measure a compound for activity, first a control current is measured while perfusing with the recording solution without the compound present. Then, a second current is recorded while perfusing with the solution and the compound of interest. Any difference in current levels indicates that the compound acts to modulate the activity of the Kir channel. An example of this type of assay may be found in Lu and MacKinnon 1997 Biochemistry, vol. 36, no. 23, pp. 6936-6940 or Namba et al., 1996 FEBS Letters vol. 386, pp. 211-214.
[0131] Planar Lipid Assay
[0132] Another electrophysiological assay which may be used in the present invention is a planar lipid bilayer assay. In this type of assay a lipid bilayer is created and the Kir channel protein is introduced into the lipid bilayer. A hydrophobic material such as Teflon is used to prepare the lipid bilayer by making a small hole (an aperture) in a sheet of Teflon. A syringe containing a solution of lipids dissolved in an organic solvent is introduced to the hole and a bilayer is formed in the center of the aperture, with solvent forming the perimeter of the newly formed bilayer.
[0133] The Teflon sheet provides a partition between two chambers allowing the placement of electrodes on both sides of the sheet. Preferably, the purified Kir channel is reconstituted into lipid vesicles and then fused with the bilayer after it is formed. The detergent coating facilitates insertion into the bilayer. (See U.S. Pat. No. 6,191,254 and Guillermo et al J. Membrane Biol (2008) 223: 13-26).
[0134] The amount of lipid desired (preferably PE:PG 3:1) is pipetted into a glass vial (about 5-10 mg). The lipid is dried under argon and then further under a room temperature vacuum for about 3 hours.
[0135] The lipid is rehydrated with hydration buffer (10 mM HEPES 7.4 (KOH), 450 mM KCl, 4 mM N-methylglucamine, 2 mM DM to a final lipid concentration of 10 mg/ml and vortexed briefly. The glass vial is flushed with argon and the lipid mixture is sonicated mildly, i.e., with short pulses of no longer than 30 seconds each. In between sonication pulses, the lipid mixture is cooled in a room temperature water bath to ensure that the lipid mixture does not get too hot. This procedure is repeated until the lipid mixture becomes translucent with a distinct pink shade.
[0136] A solution containing 50 mM DM in the hydration buffer is prepared. The DM solution is added to the lipid mixture to give a final concentration of 10 mM of DM and rotated at room temperature for 2 hours. To the detergent/lipid mixture, the Kir channel is added to the desired ratios (e.g., about 0.05-0.1). The concentration of DM is then raised to 17.5 mM and the mixture is rotated at room temperature for 1 hour. The detergent/lipid mixture is then put into dialysis tubing and dialysed against the hydration buffer.
[0137] Fluorescent Dye Assay
[0138] Another assay which may be used to identify compounds which specifically bind the Kir channel turret region is an assay utilizing a fluorescent dye. An example of a suitable dye is FIuxOR® available from Invitrogen (catalog nos, F10016, E10017), An example of this method is shown in FLOWCHART 2.
[0139] The FluxOR® reagent is a fluorogenic indicator dye, which is loaded into cells as a mernbrance-permeable Acetoxymethanol (AM) ester. According to the protocol, the FluxOR® reagent is dissolved in DMSO and further diluted with the FluxOR® assay buffer, a physiological Hank's balanced salt solution, for loading into cells. Pluronic® surfactants, which disperse and stabilize the dye are used to facilitate loading in aqueous solution.
[0140] Mammalian cells such as HEK, COS or CHO cells are grown in culture and incubated with the dye. Inside the cell, the non-fluorescent AM ester form of the FluxOR® dye is cleaved by endogenous esterases into a flourogenic thallium-sensitive indicator. The thallium-sensitive form is retained in the cytosol and its extrusion is inhibited by water-soluble Probenecid, which blocks organic anion pumps. The dye-loading buffer is replaced with fresh, dye-free assay buffer, composed of physiological HBSS containing Probenecid, before the assay. During the assay, a small amount of thallium is added to the cells with a stimulus solution that opens potassium-permeant ion channels with a mild depolarization or agonist addition. Thallium then passes into cells through open potassium channels according to a strong inward driving force. Upon binding cytosolic thallium, the de-esterified FluxOR® dye exhibits a strong increase in fluorescence intensity at its peak emission of 525 mm Baseline and stimulated fluorescence is monitored in real time to give a dynamic, functional readout of thallium redistribution across the membrance with no interference from quencher dyes.
[0141] inhibitors such as, for example, the compounds of the present invention may slow the rate of entry of thallium and thus reduce the onset of a fluorescent signal. This assay may be used for the selection of compounds that specifically bind to the turret regions of the Kir proteins. To identify such compounds a first group of cells would be transfected with wild type Kir channels and a second group of cells transfected with Kir channel having mutated turrets as illustrated in FLOWCHART 2. Test compounds such as the antibodies identified in the assays above would be added to the cells to screen for those compounds which inhibit or reduce the onset of fluorescence upon addition of the thallium dye due to inhibition of the channel. Compounds which reduced the rate of thallium intake in cells with normal turrets but had no effect on cells with mutant turrets would be classified as turret specific inhibitor compounds.
[0142] This assay may also be used to determine the specificity of the compounds for given turrets, in other words, the compounds may be introduced into cells which have been transfected with different versions of the Kir Channel to determine if the compound is specific for a given type of Kir Channel protein.
[0143] Assay for Selective Binding to Specific Types of Kir Channels
[0144] In order to determine whether a given antibody is specific for a given type of Kir Channel, assays such as an ELISA assay may be performed in which an antibody is tested against a variety of different Kir Channels to determine if the antibody is specific for a single type of Kir Channel. Ideally, antibodies that would be used as therapeutic compounds will bind to only one type of Kir channel in the turret region.
[0145] Methods to Identify Compounds to Treat Conditions
[0146] Compounds that bind to the turret region of a Kir channel and which modulate the ion channel activity of a Kir channel may be administered to a subject to determine if such compounds are able to treat a given condition. As an example, a compound may be administered to a subject such as a mammal with a given disease state using known methods of administration and the subject is then monitored clinically and tested using biochemical assays to determine if the compound is able to treat the condition using known assays for the disease state. It is believed that a variety of conditions may be treated with the compounds of the present invention, including, but not limited to, diabetes mellitus, hypertension, cardiac arrhythmia and epilepsy.
[0147] The present invention may be better understood by reference to the following non-limiting example, which is provided as exemplary of the invention. This example should in no way be construed, however, as limiting the broad scope of the invention. Example 1, which follows below, provides the first determination of the crystal structure of a euraryotic Kir channel protein and identification of the structured turret region present in Kir channel proteins. (See Crystal Structure of the Eukaryotic Strong Inward-Rectifier K+ Channel Kir2.2 at 3.1 Å Resolution, X. Tao, J. L. Avalos, J. Chen and R. MacKinnon, Science 2009 December 18; 326 (5960); 1668.)
Example 1
Crystal Structure of the Eukaryotic Strong Inward-Rectifier K+ Channel Kir2.2 at 3.1 Å Resolution
[0148] Inward-rectifier K+ channels conduct K+ ions most efficiently in one direction, into the cell. Kir2 channels control the resting membrane voltage in many electrically excitable cells and heritable mutations cause periodic paralysis and cardiac arrhythmia. We present the crystal structure of Kir2.2 from chicken, which, excluding the unstructured N- and C-termini, is 90% identical to human Kir2.2, Crystals containing Rb+, Sr2+ and Eu3+ reveal binding sites along the ion conduction pathway that are both conductive and inhibitory. The sites correlate with extensive electrophysiological data and provide a structural basis for understanding rectification. The channel's extracellular surface, with large structured turrets and an unusual selectivity filter entryway, might explain the relative insensitivity of eukaryotic inward rectifiers to toxins. These same surface features also suggest a possible approach to the development of inhibitory agents specific to each member of the inward-rectifier K+ channel family. A crystal structure reveals the structural basis of diode-like conduction properties and relative toxin insensitivity in inward rectifier K+ channels.
Introduction
[0149] In 1949 Bernard Katz introduced the term `anomalous rectification` to distinguish the K+ currents he observed in frog skeletal muscle from the `delayed rectification` K+ currents of the squid axon action potential (1, 2). Today we know that `delayed rectifiers` are a subset of the large family of voltage-dependent K+ (Kv) channels, while `anomalous rectifiers` are members of a different family of channels more commonly known as inward rectifier K+ (Kir) channels (3). The name inward rectifier refers to a fundamental ion conduction property exhibited to a greater or lesser degree by all members of the family: given an equal but opposite electrochemical driving force, K+ conductance into the cell far exceeds conductance out of the cell. Thus, Kir channels are analogous to one-way conductors, or diodes, in solid-state electronic devices.
[0150] Electrophysiological experiments have shown that inward rectification is a consequence of voltage-dependent pore blockage by intracellular multivalent cations, especially Mg2+ and polyamines (4-8). At internal negative (hyperpolarizing) membrane voltages the blocking ions are cleared from the pore so that K+conducts, whereas at internal positive (depolarizing) membrane voltages the blocking ions are driven into the pore from the cytoplasm so that K+ conduction is blocked. As a result, Kir channels are conductive when an excitable cell is at rest and non-conductive during excitation. This property is thought to foster energy efficiency because it enables Kir channels to regulate the resting membrane potential, but not dissipate the K+ gradient during an action potential (3).
[0151] A central mechanistic question is why are Kir channels blocked by intracellular multivalent cations? Mutational studies have identified several amino acids that confer sensitivity to blocking ions (9-19), but a structural description of these sites has remained elusive. Structures of prokaryotic Kir channels, due to their low sequence similarity to eukaryotic Kir channels, do not contain the specific amino acids that are known to underlie blockage and rectification (20, 21).
[0152] Another longstanding puzzle in eukaryotic Kir channel studies is their relative insensitivity to natural toxins that typically inhibit other K+ channels (22-24). Snake, spider and scorpion venoms, for example, contain numerous toxins against various Kv channels and Ca2+-activated K+ channels (25-27). By contrast, Kir channel toxins are rare, and no specific toxins against Kir2 channels have been discovered.
Results and Discussion
Eukaryotic Kir Channels as a Molecular Family
[0153] The eukaryotic Kir channels contain several amino acid sequence motifs and conserved amino acids that are essential to their functional properties (FIGS. 1A-1C). For example, in most other K+ channels the selectivity filter comprises the `canonical` filter sequence TXGYGDX, where X represents an aliphatic amino acid (FIGS. 1A-1C). The corresponding sequence in eukaryotic Kir channels is TXGYGFR, with F sometimes replaced by another amino acid. In light of the structural importance of DX in the canonical sequence, the amino acids FR signify a marked variation on the filter sequence. Eukaryotic Kir channels also contain an absolutely conserved pair of cysteine residues flanking the pore-region, which is the re-entrant peptide segment that forms the pore-helix and selectivity filter of K+ channels. Between the outer helix (the first transmembrane segment) and pore-region the `turret`, though varied amongst inward rectifiers, contains the sequence HGDL that could be considered a signature of eukaryotic Kir channels, Finally, through extensive studies combining electrophysiology and mutagenesis several acidic amino acids (D and E) are known to be critical to inward rectification (9-19), and motifs containing basic amino acids (e.g. PKKR) are critical to PIP2 activation of Kir channels (28-35). These positions are enclosed in boxes on the sequences in FIGS. 1A-1C.
[0154] The Kir2.2 channel from chicken is 90% identical to the human ortholog (excluding the N- and C-termini) and contains all of the sequence characteristics of a strong inward rectifier (36). FIGS. 6A-6D shows that the chicken Kir2.2 channel expressed in Xenopus oocytes indeed functions as a strong rectifier. In oocyte two-electrode voltage clamp recordings with 98 mM KCl in the bath solution inward currents are much larger than outward currents (FIG. 6B). In on-cell and excised gigaseal patch recordings channel activity is observed at hyperpolarizing (negative internal) membrane voltages but not at depolarizing (positive internal) voltages (FIG. 6C). The single channel conductance measured near -80 mV is approximately 40 pS, which is very similar to the values reported for the guinea pig and mouse Kir2.2 channels (37, 38) (FIG. 6D, inset). The sharp transition between channel conductance and non-conductance as a function of membrane voltage is characteristic of a strong rectifier (36). Note that upon patch excision from the oocyte surface some outward current is observed at voltages slightly positive to the reversal potential because the concentration of intracellular blockers is decreased (FIG. 6C, trace labeled (C)). However, the current still decreases with further depolarization (negative conductance) as channels become blocked in a voltage-dependent manner: this behavior reflects the inherent difficulty in washing away trace yet still active concentrations of polyamine molecules due to their very high affinity for the pore in strong rectifiers (39, 40). Several minutes following patch excision the currents decrease (FIG. 6C, trace labeled (A)). This `run-down` reflects altered channel regulation mediated by kinases, phosphatases and lipid signaling (34, 36, 41, 42).
[0155] In order to obtain diffracting crystals the intrinsically disordered N- and C-terminal regions were removed, The electrophysiological recordings shown in FIGS. 6A-6D were made using a similar construct with N- and C-terminal truncations, confirming that the crystal structure corresponds to a functional channel unit with strong rectifying properties. The Kir2.2 model, consisting of the cytoplasmic domain and transmembrane channel, was refined at 3.1 Å to a free R-factor of 0.27. A ribbon diagram in stereo shows the transmembrane pore (above) and the cytoplasmic pore (below) (FIG. 2A). Lateral openings between the transmembrane and cytoplasmic pores, at the level of the lipid membrane headgroup layer, contain many arginine and lysine residues. The high density of positive charges makes it unlikely that K+ ions would pass through these openings (FIG. 7). In FIG. 7 the shading at the top of the Figure illustrates a negative electrostatic potential at the surface and the darker shaded region in the center of the Figure illustrates regions of positive electrostatic potential. The structure is therefore consistent with mutagenesis studies, which support the conclusion that the ion pathway extends across the full length of the transmembrane and cytoplasmic pores (9-19). The overall architecture is similar to prokaryotic Kir channels but with a notable difference: the Kir2.2 channel contains prominent, highly structured turrets on the extracellular face of the channel. These surround as if to protect the pore entryway.
The Selectivity Filter
[0156] At a detailed structural level Kir2.2 is quite different from prokaryotic Kir channels owing to minimal (<20%) sequence conservation. The cysteine pair that is absolutely conserved among eukaryotic Kir channels creates a circularized pore region through covalent linkage of the segment preceding the pore helix (C123) to the segment following the selectivity filter (C155) (FIG. 2B). The existence of a disulfide bond was correctly predicted on the basis of mutagenesis studies: mutation of the corresponding cysteines in Kir2.1 led to the absence of currents even though expressed protein was detectable by Western Blot analysis (43, 44). Application of 10 mM DTT or reduced glutathione to the outside of cells expressing the wild-type channels did not affect currents. From these two observations it was concluded that a disulfide bridge must be essential for proper folding, but apparently not for function (43, 44), The structure provides an alternative interpretation. The disulfide bridge is buried beneath the protein surface at the level of the membrane interface. Furthermore, the Kir2.2 channel was purified and crystallized in the presence of 20 mM DTT and 3 mM TCEP, and yet the disulfide bridge remained intact. It is therefore possible that the disulfide bridge remains intact upon exposure to moderate concentrations of DTT, and that the bridge may be important for channel function.
[0157] The pore region is further stapled together by an ionized hydrogen bond between R149 in the filter sequence TXGYGFR and E139 (FIGS. 2B and 2C). The Glu O-ε to Arg distance is 2.4 Å, compatible with an energetically strong interaction. Mutations altering this interaction are known to alter channel function (45, 46). On the basis of studies with concatenated subunits the salt bridge was thought to be inter-subunit, but the crystal structure shows that this interaction ties together two segments of the pore-region within a single subunit (46).
[0158] Despite the presence of substantially different protein contacts surrounding the selectivity filter, the main-chain structure of the filter in Kir2.2 is the same as in other K+ channels (47). For example, the main chain RMSD between Kv1.2 and Kir2.2 is 0.4 Å, which is within the margin of certainty to discriminate atomic positions with 3.1 Å diffraction data (FIGS. 2D and 2E) (20, 48, 49), One structural difference near the filter could possibly account for important pharmacological differences between Kir and other K+ channels. In the canonical filter sequence the Asp (D) residue in the filter sequence is buried, creating a flat surface surrounding the filter opening. By contrast, in Kir channels the Phe (F) residue at the corresponding position projects directly into aqueous solution, creating four protrusions on the perimeter where the filter opens to the extracellular solution.
The Cavity and Gates
[0159] The pore lining on the intracellular side of the selectivity filter is mainly hydrophobic in nearly all K+ channels. Eukaryotic Kir channels are an exception in which the central region of the pore--known as the central cavity--contains four polar amino acids (one from each subunit) projecting toward the ion pathway (FIG. 3A). In Kir2.2 and other strong rectifiers these polar amino acids are Asp (D173), whereas in weak rectifiers such as Kir1.1 and Kir6.1 they are Asn (FIG. 1A-1C). On the basis of electrophysiological studies, Asp residues in the central cavity of strong rectifiers are hypothesized to influence the affinity of Mg2+ and polyamines by an electrostatic mechanism (12, 18).
[0160] Beneath the central cavity, residues I177 and M181 on the inner helices form two hydrophobic seals that close off the pore leading to the cytoplasm (FIG. 3C). Kir2.2 is therefore physically shut at the `activation gate` (50). Amino acids corresponding to positions 177 and 181 are also large and hydrophobic in most other eukaryotic Kir channels, but not in many other K+ channels (FIGS. 1A-1C). For example, in KcsA, Kv channels and prokaryotic Kir channels, the position corresponding to 177 usually contains a small and sometimes polar amino acid, typically Ala or Thr. In KcsA both seal positions contain small amino acids (FIG. 3D). Because of the large hydrophobic residues at positions 177 and 181, the inner helices of Kir2.2 do not come as close together in the closed conformation as in KcsA (FIGS. 3C and 3D).
[0161] FIG. 3E shows the cytoplasmic domain tetramer from the Kir2.2 channel superimposed onto the domain from Kir2.1, which was solved by crystallography in the absence of a transmembrane channel (11). Over most of the domain these structures are nearly identical. This observation supports the expectation (based on 80% sequence identity) that Kir2.2 should represent an excellent model for the complete Kir2.1 channel. In addition to the activation gate formed by the transmembrane inner helices, Kir channels have been proposed to have a second gate (G-loop) at the apex of the cytoplasmic domain tetramer (11, 51). The G-loop is physically open in Kir2.2 and closed in the Kir2.1 domain (FIGS. 3F and 3G). The differences in conformation are due to local movements of the G-loop rather than rigid body motions of the cytoplasmic domains. Local G-loop movements contrast observations on the cytoplasmic domain of Kir3.1, in which G-loop opening appears associated with rigid body movements of domains in the tetramer (20).
Ion Binding Sites for Conduction and Inward Rectification
[0162] FIG. 4A-F shows the locations of ions in difference Fourier maps from crystals containing Rb+, Sr2+, and Eu3+. Rb+ is a K+ analog that conducts. Density for this ion is observed at multiple sites in the selectivity filter and at three positions within the pore on the intracellular side of the selectivity filter, but is absent in the central cavity (FIG. 4A). The three occupied intracellular positions are: immediately internal to the activation gate in the transmembrane pore, in the cytoplasmic pore internal to the G-loop, and at the entryway to the cytoplasmic pore. We refer to the two sites in the cytoplasmic pore as the upper and lower rings of charges, respectively (FIG. 8). The presence of multiple sites along the pore occupied by conducting ions area prerequisite for strong voltage-dependent block by intracellular cations that cannot pass through the selectivity filter (12, 52-57).
[0163] Crystals of Kir2.2 were grown in the presence of 650 mM Rb+ and yet electron density for Rh+ is not observed in the cavity (FIG. 4A). This finding is noteworthy because under similar conditions a strong monovalent cation peak is observed in the cavity of KcsA (47, 58). Native crystals of Kir2.2, grown in the presence of 150 mM K+ and 500 mM Na+, show a weak electron density peak at the cavity center with additional peaks on the perimeter, apparently bridging toward the D173 side-chain (FIG. 3A). We can not discern whether these peaks represent a disordered ion, multiple ions, or a low occupancy K+ (or Na+) in the center, perhaps surrounded by water molecules hydrogen bonded to the Asp carboxylate. We can conclude, however, that the central cavity in Kir2.2, at least in the closed conformation, has cation attractive properties that are different from KcsA.
[0164] The divalent cation Sr2+ should behave as an electron dense mimic of Mg2+, a biologically important metal ion inhibitor of eukaryotic Kir channels (7, 8). In Fo-Fc Fourier maps from crystals with 10 mM Si2+, 500 mM Na+ and 150 mM K+, density peaks due to Sr2+ are observed at three sites inside the pore intracellular to the selectivity filter: in the cavity, at the upper ring and at the lower ring of charges (FIGS. 8 and 4B). The magnitude of the Sr2+ peak is small in the cavity (3.4σ) compared to the peaks at the upper (9.6σ) and lower (7.2σ) rings of charges. Separate experiments with crystals containing 200 mM Sr2+ support that the weak cavity peak is indeed due to Sr2+, which is present apparently at relatively low occupancy. Detailed views of these sites are shown (FIGS. 4D-F). They each consist of planar rings of acidic amino acids arranged on the pore's perimeter. All three sites exhibit a preference for Sr2+:10 mM Sr2+ out competes 150 mM K+. This selectivity is likely to be electrostatic in origin. The sites are too wide (10.5 Å, 8.9 Å and 9.3 Å diameter for the cavity, upper and lower ring of charges) to mediate direct coordination of an ion at the center. Presumably ions at the center of these sites interact through bridging water molecules. Since each site has the potential to contain multiple negatively charged carboxyl groups, the resulting strong electric field is expected to create a good match for a multivalent cation. Crystals containing the lanthanide Eu3+, which we assume to be trivalent (59), provide support for this hypothesis. An anomalous difference Fourier map shows that Eu3+ binds at only one site, the upper ring of charges. This site appears to be more electronegative than the others because it contains two concentric rings of acidic amino acids, E225 and E300.
[0165] Mutagenesis studies have identified several amino acids that, when mutated, affect the affinity of Mg2+ and polyamines in strong rectifiers. D173 in the cavity, E225 and E300 forming the upper ring of charges, and D256 forming the lower ring of charges are among those known to be important (9-19). The weak Sr2+ peak in the cavity might seem incompatible with the large influence that mutations of the cavity Asp (D173) have on Mg2+ affinity. However, the channel in the crystal is not in an applied electric field: in an electric field imposed by a depolarized (positive inside) membrane we expect that the distribution of blocker occupancies among the multiple sites will change. Specifically, we expect the blocking cations to be driven deeper into the pore toward the cavity. In correlating the crystallographic with electrophysiological data, it is most significant that the amino acids forming the Sr2+ sites in the crystal are the same amino acids that are known to affect blockage and rectification in electrophysiology experiments (36). Beyond providing a structural basis with which to explain past electrophysiological studies, the Kir2.2 structure also suggests many new experiments. For example, most studies on the mechanism of rectification have focused on electrostatic interactions between the positively charged blocker and negatively charged groups on the protein. But hydrophobic interactions between methylene groups of polyamine molecules and hydrophobic residues in the channel may be important. In particular, we might anticipate that when the pore opens polyamines could interact strongly with the large hydrophobic amino acids at positions 177 and 181 when the leading amino group of the polyamine reaches into the central cavity (FIG. 3C) (54).
[0166] Since the earliest investigations of strong inward rectifiers two important properties have been noted: a sharp transition from a conductive state to a non-conductive (blocked) state over a very narrow voltage range, and a dependence of the transition on the extracellular K+ concentration (60-63). Specifically, the voltage at which the transition occurs shifts to more depolarizing values as extracellular K4 concentration is increased. Both properties, the sharp transition (i.e. strong voltage dependence) and its dependence on extracellular K+, have been attributed to the simple notion that conducting ions and blocking ions compete for sites in the pore (12, 52-57, 64-66). The crystallographic data presented here support this conclusion. We observe in the crystal Rb+binding at the same sites that can bind multivalent blocking ions. Therefore a high extracellular K+ (or Rb+) concentration should favor occupation of the sites by conducting ions, and a more depolarizing voltage should be required to drive blocking ions into the pore from the cytoplasm to replace the conducting ions. Moreover, as blocking ions enter the pore from the intracellular side, the displaced conducting ions must move through the selectivity filter to the extracellular side. This is to say that movements of blocking and conducting ions must be coupled. Such coupling would have energetic consequences because movement of an ion across the membrane voltage difference constitutes work. In other words a blocking ion entering the pore will exhibit a voltage dependence that results from a combination of its own charge and the charge of the displaced ions. This can be the origin of strong voltage dependent block, which can be the origin of a biologically important property of strong rectifiers--their diode property of a sharp transition from a conductive to a non-conductive state as a function of membrane voltage (12, 52-55, 64).
The Extracellular Pore Entryway and Pharmacology of Kir Channels
[0167] Two aspects of the structure may account for the fact that eukaryotic Kir channels, especially members of the Kir2 subfamily, are relatively insensitive to K+ channel toxins (22-24). The turrets in Kir2.2 are larger and come closer together, constricting the pore entryway compared to Kv1.2; and F148 in the sequence TXGYGFR creates four protrusions on the surface at the pore opening (FIGS. 5A and 5B). Thus, in Kv channels the entryway is wider and the pore opens onto intersecting grooves with a flat base, which form the docking surface for pore-blocking scorpion toxins (FIG. 5B). In Kir2.2 the entryway is constricted and the grooves are absent (FIG. 5A).
[0168] Though the shape of the eukaryotic Kir channel pore entryway might offer fewer opportunities for inhibitory protein-protein interactions, inhibition might occur by a somewhat different strategy. Inhibitors of Kir1.1 and Kir3.4 channels have been identified. A bee venom toxin, tertiapin, inhibits both of these channels (22). At 21 amino acids tertiapin is smaller than most other venom toxins so it might fit between the turrets more effectively, Alternatively, the turrets themselves might form the binding site for tertiapin (67-69). At 57 amino acids δ-dendrotoxin from the green mamba snake is rather large and yet it inhibits Kir1.1 channels (23). Compared to tertiapin less is known about the binding site on the channel for δ-dendrotoxin, but one aspect of its inhibition is intriguing: the blocked state reduces single channel conductance to about 10% rather than inhibiting all the way. δ-dendrotoxin most likely binds to the turrets but is too large to fit tightly over the pore, which would imply that binding to the turret may be sufficient to alter the channel's function.
[0169] The idea that binding to the turrets could alter function is not surprising when one considers that the turret in Kir2.2 is not a loop, but forms a highly ordered structure (FIG. 5C). The base of the turret is formed and pinned together by the HGDL sequence, which with only minor variation is found in all eukaryotic Kir channels (FIGS. 1A-1C and FIG. 5D.) H108 stabilizes D110 through a hydrogen bond. The Asp (D) itself is hydrogen bonded to the amide nitrogen of C123, which effectively holds the two ends of the turret together. L111 projects from the surface of a short 310 helix into the protein interior to make stabilizing hydrophobic interactions. Thus, the turrets are structurally important elements of the channel. Between the sequence HGDL and the first Cys of the disulfide bridge the turret sequence is highly variable among Kir channel subtypes. The Kir2.1 channel becomes sensitive to tertiapin if the variable sequence is mutated to be Kir3.4-like (68). Therefore, the turrets appear to be structures through which specific inhibition of Kir channel subtypes might be achievable through directed evolution of specific protein binding partners.
SUMMARY
[0170] This presents the atomic structure of a eukaryotic Kir channel, Kir2.2, a strong inward rectifier. The sequence TXGYGFR gives rise to a K+ selectivity filter stabilized by disulfide bridges and salt bridges that distinguish eukaryotic Kir channels. Multiple ion binding sites on the intracellular side of the selectivity filter can be occupied by conducting ions but exhibit higher affinity for multivalent blocking ions. Thus, blocking ions entering from the cytoplasm must displace conducting ions through the pore. This situation is expected to give rise to strong voltage-dependent block and diode-like conduction properties. Structural features of the extracellular pore entryway offer an explanation for the relative insensitivity of Kir channels to venomous toxins and a possible approach to the development of selective Kir channel inhibitors,
REFERENCES AND NOTES
[0171] 1. H. A, Hodgkin A L, Katz B, Archives des Sciences Physiologiques 3, 129 (1949).
[0172] 2. B. Katz, Arch. Sci. Physiol. (Paris) 3, 285 (1949).
[0173] 3. B. Hille, Ion Channels of Excitable Membranes. (Sinauer Associates, Inc., Sunderland, Mass., 2001).
[0174] 4. B. Fakler et al., Cell 80, 149 (Jan. 13, 1995).
[0175] 5. A. N, Lopatin, E. N. Makhina, C. G. Nichols, Nature 372, 366 (Nov. 24, 1994).
[0176] 6. M. Haile, H. Irisawa, A. Noma, J Physiol 387, 251 (June 1987).
[0177] 7. H. Matsuda, A. Saigusa, H. Irisawa, Nature 325, 156 (Jan. 8-14, 1987).
[0178] 8. C. A. Vandenberg, Proc Natl Acad Sci USA 84, 2560 (April 1987).
[0179] 9. H. T. Kuraia, W. W. Cheng, C. Ambit, P. A. Slesinger, C. G. Nichols, J Gen Physiol 130, 145 (August 2007).
[0180] 10. Y. Fujiwara, Y. Kubo, J Gen Physiol 127, 401 (April 2006).
[0181] 11. S. Pegan et al., Nat Neurosci 8, 279 (March 2005).
[0182] 12. D. Guo, Z. Lu, J Gen Physiol 122, 485 (November 2003).
[0183] 13. Y, Kubo, Y. Murata, J Physiol 531, 645 (Mar. 15, 2001),
[0184] 14. J. Yang, Y. N. Jan, L. Y. Jan, Neuron 14, 1047 (May, 1995).
[0185] 15. M. Taglialatela, E. Ficker, B. A. Wible, A. M. Brown, EMBO J 14, 5532 (Nov. 15, 1995).
[0186] 16. B. A. Wible, M. Taglialatela, E. Ficker, A. M. Brown, Nature 371, 246 (Sep. 15, 1994).
[0187] 17. P. R. Stanfield et al., J Physiol 478 (Pt 1), 1 (Jul. 1, 1994).
[0188] 18. Z. Lu, R. MacKinnon, Nature 371, 243 (Sep. 15, 1994).
[0189] 19. B. Fakler et al., FEBS Lett 356, 199 (Dec. 19, 1994),
[0190] 20. M. Nishida, M. Cadene, B. T. Chait, R. MacKinnon, EMBO J 26, 4005 (Sep. 5, 2007).
[0191] 21. A. Kuo et al., Science 300, 1922 (Jun. 20, 2003),
[0192] 22. W. Jin, Z. Lu, Biochemistry 37, 13291 (Sep. 22, 1998).
[0193] 23. J. P. Imredy, C. Chen, R. MacKinnon, Biochemistry 37, 14867 (Oct. 20, 1998).
[0194] 24. Z. Lu, R. MacKinnon, Biochemistry 36, 6936 (Jun. 10, 1997).
[0195] 25. A. L. Harvey, B. Robertson, Curr Med Chem 11, 3065 (December 2004).
[0196] 26. K. J. Swartz, R. MacKinnon, Neuron 15, 941 (October 1995).
[0197] 27. M. L. Garcia, M. Hanner, H. G. Knaus, R. Slaughter, G. J. Kaczorowski, Methods Enzymol 294, 624 (1999).
[0198] 28. D. Schulze, T. Krauter, H. Fritzenschaft, M. Soom, T. Baukrowitz, J Biol Chem 278, 10500 (Mar. 21, 2003).
[0199] 29. W. Z. Zeng, H. H. Lieu, U, M. Krishna, J. R. Faick, C. L. Huang, Am J Physiol Renal Physiol 282, F826 (May, 2002).
[0200] 30. C. M. Lopes et al., Neuron 34, 933 (Jun. 13, 2002).
[0201] 31. M. Scorn et at, FEBS Lett 490, 49 (Feb. 9, 2001).
[0202] 32. S. L. Shyng, C. A. Cukras, J. Harwood, C. G. Nichols, J Gen Physiol 116, 599 (November 2000).
[0203] 33. H. Zhang, C. He, X. Yan, T. Mirshahi, D. E. Logothetis, Nat Cell Biol 1, 183 (July 1999).
[0204] 34. C. L. Huang, S. Feng, D. W. Hilgemann, Nature 391, 803 (Feb. 19, 1998).
[0205] 35, Z. Fan, J. C. Makielski, J Biol Chem 272, 5388 (Feb 28, 1997).
[0206] 36. P. R. Stanfield, S. Nakajima, Y. Nakajima, Rev. Physiol Biochem. Pharmacol. 145, 47 (2002).
[0207] 37. N, Takahashi et al., J Biol Chem 269, 23274 (Sep. 16, 1994).
[0208] 38. G. X. Liu et al., J Physiol 532, 115 (Apr. 1, 2001).
[0209] 39. Z. Lu, Annu Rev Physiol 66, 103 (2004).
[0210] 40. C. G. Nichols, A. N. Lopatin, Annu Rev Physiol 59, 171 (1997).
[0211] 41. P. Henry, W. L. Pearson, C. G. Nichols, J Physiol 495 (Pt 3), 681 (Sep. 15, 1996),
[0212] 42. B. Fakler, U. Brandle, E. Glowatzki, H. P. Zenner, J. P. Ruppersberg, Neuron 13, 1413 (December 1994).
[0213] 43. H, C, Cho, R, G. Tsushima, T. T, Nguyen, H. R. Guy, P. H. Backx, Biochemistry 39, 4649 (Apr. 25, 2000).
[0214] 44. M. L. Leyland, C. Dart, P. J. Spencer, M. J. Sutcliffe, P. R. Stanfield, Pflugers Arch 438, 778 (November 1999).
[0215] 45. K. M. Dibb et al., J Biol Chem 278, 49537 (Dec. 5, 2003).
[0216] 46. J. Yang, M. Yu, Y. N. Jan, L. Y. Jan, Proc Natl Acad Sci USA 94, 1568 (Feb. 18, 1997).
[0217] 47. Y. Thou, J. H. Morais-Cabral, A. Kaufman, R. MacKinnon, Nature 414, 43 (Nov. 1, 2001).
[0218] 48. S. B. Long, X. Tao, E. B. Campbell, R. MacKinnon, Nature 450, 376 (Nov. 15, 2007).
[0219] 49. S. B. Long, E, B. Campbell, R. Mackinnon, Science 309, 897 (Aug. 5, 2005).
[0220] 50. Y. Jiang et al., Nature 417, 523 (May 30, 2002).
[0221] 51. S. Pegan, C. Arrabit, P. A, Slesinger, S. Choe, Biochemistry 45, 8599 (Jul. 18, 2006).
[0222] 52. H. G. Shin, Z. Lu, J Gen Physiol 125, 413 (April 2005).
[0223] 53. H. G. Shin, Y. Xu, Z. Lu, J Gen Physiol 126, 123 (August 2005).
[0224] 54. D. Guo, Y. Rarnu, A. M. Klein, Z. Lu, J Gen Physiol 121, 261 (April 2003).
[0225] 55. D. Guo, Z. Lu, J Gen Physiol 115, 799 (June 2000).
[0226] 56. M. Spassova, Z. Lu, J Gen Physiol 112, 211 (August 1998).
[0227] 57. W. L. Pearson, C. G. Nichols, J Gen Physiol 112, 351 (September 1998).
[0228] 58. Y. Thou, R. MacKinnon, Biochemistry 43, 4978 (May 4, 2004).
[0229] 59. A. Cotton, G. Wilkinson, Wiley interscience.
[0230] 60. D. Noble, R. W. Tsien, J Physiol 195, 185 (March 1968).
[0231] 61. C. A. Leech, P. R. Stanfield, J Physiol 319, 295 (1981).
[0232] 62. S. Hagiwara, M. Yoshii, J Physiol 292, 251 (July 1979).
[0233] 63. A. L, Hodgkin, P. Horowicz, J Physiol 148, 127 (October 1959).
[0234] 64. D. Guo, Z. Lu, J Gen Physiol 117, 395 (May, 2001).
[0235] 65. D. Oliver, H. Hahn, C. Antz, J. P. Ruppersberg, B. Faller, Biophys J 74, 2318 (May, 1998).
[0236] 66. M. Spassova, Z. Lu, J Gen Physiol 114, 415 (September 1999).
[0237] 67. J. P. Felix et al., Biochemistry 45, 10129 (Aug. 22, 2006),
[0238] 68. Y. Ramu, A. M. Klem, Z. Lu, Biochemistry 43, 10701 (Aug. 24, 2004).
[0239] 69. W. Jin, A. M. Klem, J. H. Lewis, Z. Lu, Biochemistry 38, 14294 (Oct. 26, 1999).
[0240] 70. The X-ray crystallographic coordinates and structure factor files have been deposited in the Protein Data Bank with accession ID 3JYC.
Materials and Methods
Cloning, Expression and Purification
[0241] A synthetic gene fragment (Bio Basic, Inc) encoding residues 38 to 369 of chicken Kir2.2 channel (GI:118097849) was ligated into the XhoI/EcoRI cloning sites of a modified pPICZ-B vector (Invitrogen). The resulting protein has green fluorescent protein (GFP) and a 1D4 antibody recognition sequence (TETSQVAPA) on the C-terminus (1), separated by a PreScission protease cleavage site (SNSLEVLPQ/GP).
[0242] The construct was linearized using PmeI and transformed into a HIS+ strain of SMD1163 of Pichia pastoris (Invitrogen) by electroporation (BioRad Micropulser). Transformants were selected on YPDS plates containing 400-1200 μg/ml Zeocin (Invitrogen). Resistant colonies were tested for expression by anti-1D4 tag Western Blot. For large-scale expression, small cultures grown from the best expressing colony were diluted into BMGY media (Invitrogen) and inoculated at 29° C. overnight, until OD600 reached between 20-30. Cells were then pelleted, resuspended in BMM media (Invitrogen) and expressed overnight at 24° C. Cells were harvested, flash-frozen in liquid N2, and stored at -80° C. until needed.
[0243] Cells were lysed in a Retsch, Inc. Model MM301 mixer mill (5×3.0 minutes at 25 cps). The lysis buffer contained 150 mM KCl, 50 mM TRIS-HCl pH 8.0, 0.1 mg/ml deoxyribonuclease I, 0.1 μg/ml pepstatin, 1 μl/ml leupeptin, 1 μg/ml aprotinin, 0.1 mg/ml soy trypsin inhibitor, 1 mM benzamidine, 0.1 mg/ml AEBSF, with 1 mM phenylmethysulfonyl fluoride added just before lysis (3.0 ml lysis buffer/g cells). pH of the lysate was adjusted to 8.0 with KOH. The lysate was extracted with 100 mM DM (n-decyl-β-D-maltopyranoside, Anatrace, solgrade) at room temperature for 1 hour with stirring, and then centrifuged for 40 minutes at 30,000 g, 10° C. Supernatant was added to 1D4-affinity resin pre-equilibrated with 150 mM KCl, 50 mM TRIS-HCl pH 8.0, and 4 mM DM. Suspension was layered with Argon and mixed by inversion for 2 hours at room temperature. Beads were collected on a column by gravity, washed with 2 column volumes of buffer (150 mM KCl, 50 mM TRIS-HCl pH 8.0, 1 mM EDTA pH 8.0, and 4 mM DM), and eluted with buffer plus 1 mg/ml 1D4 peptide (AnaSpec, Inc.) over 1 hour at room temperature. 20 mM DTT (Dithiothreitol) and 3 mM TECP were added to eluted protein. The protein was then digested with PreScission protease (20:1 w/w ratio) overnight at 4° C. Concentrated protein was further purified on a Superdex-200 gel filtration column in 150 mM KCl, 20 mM TRIS-HCl pH 8.0, 4 mM DM (anagrade), 3 mM TCEP, 20 mM DTT and 1 mM EDTA at 4° C.
[0244] The fraction corresponding to the tetramer peak was concentrated to about 8 mg/ml, mixed 1:1 with crystallization solution and set up as hanging drops over reservoirs containing 0.1 ml crystallization solution. Crystals appeared in 7-20% PEG400 or 2-10% PEG4000, with 500 mM KCl or NaCl, and 50 mM buffer pH 6.0-9.5 at 4° C. overnight and grew to full size within 2-3 days.
[0245] For studies with RbCl, the protein was purified in a similar fashion except that KCl was replaced with RbCl in all buffer solutions and crystals were grown in 10-20% PEG400, 500 mM RbCl, and 50 mM MES pH 6.5. For studies with 10 mM EuCl3, crystals were grown in 7-20% PEG400, 1 M ammonium formate, 50 mM TRIS-HCl pH 8.5, and 10 mM EuCl3. For studies with 10 mM SrCl2, crystals were grown in 10-20% PEG400, 500 mM NaCl, 50 mM HEPES pH 7.5, and 10 mM SrCl2. For studies with 200 mM SrCl2, crystals were grown in 3-7% PEG4000, 200 mM SrCl2, and 50 mM Na Citrate pH 5.6.
Structure Determination
[0246] Crystals were cryo-protected in reservoir plus 25% glycerol (v/v), 4 mM DM, 20 mM DTT, 3 mM TCEP, and 1 mM EDTA in a step-wise manner (5% glycerol increase each step) and flash-frozen in liquid nitrogen. Diffraction data from native crystals were collected to 3.1 Å at beamline 24ID-C (APS) and for crystals in various metal ions (Rb+, Sr+, and Eu3+) at beamline X29 (Brookhaven NSLS). Images were processed with DENZO and intensities merged with SCALEPACK (2). Data were further processed using the CCP4 suite (3). The crystals belong to the 14 space group. The structure was solved by molecular replacement using the program MOLREP (4), with the 2.4 Å resolution structure of the cytoplasmic domain of mouse Kir2.1 (PDB 1U4F) as a search model. There is one copy of the subunit in the asymmetric unit. The model was built using O (5) and refined with CNS (50-3.1 Å) to Rfree=27.2% (6). The final model contains residues 43-60 and 70-369 (residues 70 to 78 are modeled as alanines) of chicken Kir2.2, three additional residues SNS on the C-terminus corresponding to the PreScission cleavage site, and five K+ ions. During the final minimization refinement step in CNS, occupancies of the K+ ions were set to 0.5 (which gave rise to a lower R free compared to occupancy of 1.0) and B-factors of the K+ ions were set to 85 (roughly the average B-factor of surrounding protein atoms). Crystallographic data and refinement statistics are shown in Table S1. Figures were made using PYMOL (www.pymol.org) (7).
[0247] Ion binding was assessed by calculating anomalous difference Fourier maps for data with Eu3+ and Fo-Fc maps for data with Rb+ and Sr2+ using fft in the CCP4 suite (3), Sr2+ was analyzed at two different concentrations to discern whether the weak cavity peak was due to Sr2+. This peak became stronger when Sr2+ was increased from 10 mM to 200 mM while the monovalent cation concentration was decreased, consistent with Sr2+ being present in the cavity but probably at low occupancy. Phases used to calculate Fo-Fc omit maps were derived from a channel model devoid of ions in the cavity or cytoplasmic domain throughout refinement,
Electrophysiology
[0248] Xenopus oocytes were harvested from mature female Xenopus laevis and defolliculated by collagenase treatment for 1-2 hours. Oocytes were then rinsed thoroughly and stored in ND96 solution (96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, 5 mM HEPES, 50 μg/ml gentamycin, pH 7.6 with NaOH). Defolliculated oocytes were selected 2-4 hours after collagenase treatment and injected with cRNA the next day. The injected oocytes were incubated in ND96 solution for 1-5 days before recording. All oocytes were stored in an incubator at 18° C.
[0249] The chicken Kir2.2 (residues 38 to 369) gene was sub-cloned into the pGEM vector (Promega). cRNA was prepared using T7 RNA polymerase (Promega) from NdeI-linearized plasmid DNA.
[0250] All recordings were performed at room temperature. For two-electrode voltage-clamp experiments, oocytes were held at 0 mV and pulsed from -80 mV to +80 mV with 10 mV increment steps. Recording solution contained 98 mM KCl, 0.3 mM CaCl2, 1 mM MgCl2, and 5 mM HEPES pH 7.6. The ionic currents were recorded with an oocyte clamp amplifier (OC-725C, Warner Instrument Corp.). The recorded signal was filtered at 1 kHz and sampled at 10 kHz using an analogue-to-digital converter (Digidata 1440A, Axon Instruments, Inc) interfaced with a computer. pClamp10.1 software (Axon Instruments, Inc) was used for controlling the amplifier and data acquisition. For patch-clamp experiments, each oocyte was incubated in a hypertonic solution containing 200 mM NaCl, 130 mM KCl, 5 mM K2EDTA, 5 mM K2HPO4, 5 mM KH2PO4 pH 7.2 for 5-10 minutes and the vitelline membrane was removed before seal formation. Currents were recorded in either cell-attached or inside-out configuration with an Axopatch 200B amplifier, Digidata 1440A analogue-to-digital converter and pClamp10.1 software to control membrane voltage and record. During the current recordings, the membrane was first held at 0 mV followed by a 10-second voltage ramp from +80 mV to -80 mV .The pipette solution contained 140 mM KCl, 5 mM K2HPO4, 5 mM KH2PO4, 0.3 mM CaCl2, 1 mM MgCl2, pH 7.2 with KOH. The bath solution contained 130 mM KCl, 5 mM K2EDTA, 5 mM K2HPO4, 5 mM KH2PO4, pH 7.2 with KOH.
[0251] For the single channel I-V curve shown in Figure S1D inset, each data point represents the current difference at a given voltage associated with the opening of a single channel.
REFERENCES
[0252] 1. J. P. Wong, E. Reboul, R. S. Molday, J. Kast, J Proteome Res 8, 2388 (May, 2009).
[0253] 2. Z. Otwinowski, W. Minor, Methods Enzymol. 276, 307 (1997).
[0254] 3. N. Collaborative Computational Project, Acta Cryst. D50, 60 (1994).
[0255] 4. A. Vagin, A. Teplyakov, Acta Crystallogr. D. Biol. Crystallogr. 56 Pt 12:1622-4., 1622 (2000).
[0256] 5. T. A. Jones, J. Y. Zola, S. W. Cowan, M. Kjeldgaard, Acta Cryst. A47, 110 (1991).
[0257] 6. A. T. Brunger et al., Acta Cryst. D54, 905 (1998).
[0258] 7. W. L. DeLano, DeLano Scientific, Palo Alto, Calif., USA. http://www.pymol.org, (2002).
[0259] The present invention is not to be limited in scope by the specific embodiments described herein. Indeed, various modifications of the invention in addition to those described herein will become apparent to those skilled in the art from the foregoing description. Such modifications are intended to fall within the scope of the appended claims.
[0260] It is further to be understood that all values are approximate, and are provided for description.
[0261] All patents, applications, publications, test methods, literature, and other materials cited herein are hereby incorporated by reference.
TABLE-US-00001 TABLE S1 Crystallographic data and refinement statistics Data Collection Data set Native Rb+ (650 mM) Sr2+ (10 mM) Sr2+ (200 mM) Eu3+ (10 mM) Space group I4 I4 I4 I4 I4 Lattice constants (Å) a = b = 84.018, a = b = 82.714, a = b = 82.806, a = b = 83.268, a = b = 84.221, c = 196.121 c =196.142 c = 195.605 c = 197.143 c = 195.915 α = β = γ = 90° α = β = γ = 90° α = β = γ = 90° α = β = γ = 90° α = β = γ = 90° Source APS 24ID-C BNL X29 BNL X29 BNL X29 BNL X29 Wavelength (Å) 0.97949 1.0809 1.0809 1.0809 1.5222 Resolution (Å) 50-3.1 50-4.0 50-3.3 50-3.8 50-6.0 Total/unique observations 65,452/12,153 25,333/5,231 60,844/9,910 31,462/6,571 5,649/1,517 I/sigma (I) a 33.1 (23) 20.0 (1.8) 24.3 (2.4) 20.8 (2.7) 24.7 (1.6) Redundancy a 5.4 (4.6) 4.8 (4.3) 6.1 (5.9) 4.8 (4.7) 3.7 (1.4) Completeness (%) a 99.7 (99.9) 93.3 (83.8) 99.8 (100.0) 99.7 (100.0) 87.9 (49.4) Rsym (%) a,b 6.8 (73.6) 10.1 (78.3) 11.5 (89.4) 9.6 (69.3) 7.0 (35.7) Model refinement Resolution (Å) 50-3.1 50-4.0 50-3.3 50-3.8 50-6.0 Number of reflections 12,122 (609) 5,221 (272) 9,895 (504) 6,569 (346) 1,517 (76) Rwork/Rfree 24.4/27.2 29.6/35.0 24.9/28.6 25.8/30.4 36.5/39.9 R.m.s. deviation of bond length (Å) 0.010 R.m.s. deviation of bond angles (°) 1.59 Protein atoms/K+ ions 2.530/5 Mean B value 112.1 Ramachandran plot c 76.0/21.9/2.1 R.m.s., root mean-squared. a Number in the parentheses represents statistics for data in the highest resolution shell. b Rsym = Σ|Ii - <Ii>|/ΣIi, where <Ii> is the average intensity of symmetry equivalent reflections. c The three numbers represent the percentage of residues in most favored/additionally allowed/generously allowed regions.
TABLE-US-00002 SEQUENCES: 1. human Kir 1.1- variable portion of turret region PEFHPSANHTP 2. human Kir 1.2- variable portion of turret region LELDPPANHTP 3. human Kir 2.1- variable portion of turret region DASKEGKA 4. human Kir 2.2- variable portion of turret region EPAEGRGRTP 5. human Kir 2.3- variable portion of turret region EASPGVPAAGGPAAGGGGAAPVAPKP 6. human Kir 2.4- variable portion of turret region AAPPPPAP 7. human Kir 3.1- variable portion of turret region NKAHVGNYTP 8. human Kir 3.4- variable portion of turret region DHVGDQEWIP 9. human Kir 6.1- variable portion of turret region YAYMEKSGMEKSGLESTV 10. human Kir 6.2- variable portion of turret region APSEGTAEP 11. human Kir 7.1- variable portion of turret region ELDHDAPPENHTI 12. chicken Kir 2.1- variable portion of turret region ENQENNKP 13. chicken Kir 2.2- variable portion of turret region ENPGGDDTFKP 14. human Kir 1.1- amino acid sequence 1 MNASSRNVFD TLIRVLTESM FKHLRKWVVT REFGHSRQRA RLVSKDGRCN IEFGNVEAQS 61 RFIFFVDIWT TVLDLKWRYK MTIFITAFLG SWEFFGLLWY AVAYIHKDLP EFHPSANHTP 121 CVENINGLTS AFLFSLETQV TIGYGFRCVT EQCATAIFLL IFQSILGVII NSFMCGAILA 181 KISRPKKRAK TITFSKNAVI SKRGGKLCLL IRVANLRKSL LIGSHIYGKL LKTTVTPEGE 241 TIILDQININ FVVDAGNENL FFISPLTIYH VIDHNSPFFH MAAETLLQQD FELVVFLDGT 301 VESTSATCQV RTSYVPEEVL WGYRFAPLVS KTKEGKYRVD FHNFSKTVEV ETPHCAMCLY 361 NEKDVRARMK RGYDNPNFIL SEVNETDDTK M 15. human Kir 1.2- amino acid sequence 1 MTSVAKVYYS QTTQTESRPL MGPGIRRRRV LTKDGRSNVR MEHIADKRFL YLKDLWTTFI 61 DMQWRYKLLL FSATFAGTWF LFGVVWYLVA VAHGDLLELD PPANHTPCVV QVHTLTGAFL 121 FSLESQTTIG YGFRYISEEC PLAIVLLIAQ LVLTTILEIF ITGTFLAKIA RPKKRAETIR 181 FSQHAVVASH NGKFCLMIRV ANMRKSLLIG CQVTGKLLQT HQTKEGENIR LNQVNVTFQV 241 DTASDSPFLI LPLTFYHVVD ETSPLKDLPL RSGEGDFELV LILSGTVEST SATCQVRTSY 301 LPEEILWGYE FTPAISLSAS GKYIADFSLF DQVVKVASPS GLRDSTVRYG DPEKLKLEES 361 LREQAEKEGS ALSVRISNV 16. human Kir 2.1- amino acid sequence 1 MGSVRTNRYS IVSSEEDGMK LATMAVANGF GNGKSKVHTR QQCRSRFVKK DGHCNVQFIN 61 VGEKGQRYLA DTFTTCVDIR WRWMLVIFCL AFVLSWLFFG CVFWLIALLH GDLDASKFGK 121 ACVSEVNSFT AAFLFSIETQ TTIGYGFRCV TDECPIAVFM VVFQSIVGCI IDAFIIGAVM 181 AKMAKPKKRN ETLVFSHNAV IAMRDGKLCL MWRVGNLRKS HLVEAHVRAQ LLKSRITSEG 241 EYIPLDQIDI NVGFDSGIDR IFLVSPITIV HEIDED5PLY DLSKQDIDNA DFEIVVILEG 301 MVEATAMITQ CRSSYLANEI LVGHRYEPVL FEEKHYYKVD YSRFHKTYEV PNTPLCSARD 361 LAEKKYILSN ANSFCYENEV ALTSKEEDDS ENGVPESTST DTPPDIDLHN QASVPLEPRP 421 LRRESEI 17. human Kir 2.2- amino acid sequence 1 MTAASRANPY SIVSSEEDGL HLVTMSGANG FGNGKVHTRR RCRNRFVKKN GQCNIEFANM 61 DEKSQRYLAD MFTTCVDIRW RYMLLIFSLA FLASWLLEGI IFWVIAVAHG DLEPAEGRGR 121 TPCVMQVHGE MAAFLPSIET QTTIGYGLRC VTEECPVAVF MVVAQSIVGC IIDSFMIGAI 181 MAKMARPKKR AQTLLFSHNA VVALRDGKLC LMWRVGNLRK SHIVEAHVRA QLIKPRVTEE 241 GEYIPLDQID IDVGFDKGLD RIFLVSPITI LHEIDEASPL FGISRQDLET DDFEIVVILE 301 GMVEATAMTT QARSSYLANE ILWGHRFEPV LFEEKNQYKI DYSHFHKTYE VPSTPRCSAX 361 DLVENKFLLP SANSFCYENE LAFLSRDEED EADGDQDGRS RDGLSPQARH DFDRLQAGGG 421 VLEQRPYRRE SEI 18. human Kir 2.3- amino acid sequence 1 MHGHSRNGQA HVPRRKRRNR FVKKNGQCNV YFANLSNKSQ RYMADIFTTC VDTRWRYMLM 61 IFSAAFLVSW LFFGLLFWCI AFFHGDLEAS PGVPAAGGPA AGGGGAAPVA PKPCIMHVNG 121 FLCAFLFSVE TQTTIGYGFR CVTEECPLAV IAVVVQSIVG CVIDSFMIGT IMAKMARPKK 181 RAQTLLFSHH AVISVRDGKL CLMWRVGNLR KSHIVEAHVR AQLIKPYMTQ EGEYLPLDQR 241 DLNVGYDIGL DRIFLVSPII IVHEIDEDSP LYGMGKEELE SEDFEIVVIL EGMVEATAMT 301 TQARSSYLAS EILWGHRFEP VVFEEKSHYK VDYSRFHKTY EVAGTPCCSA RELQESKITV 361 LPAPPPPPSA FCYENELALM SQEEEEMEEE AAAAAAVAAG LGLEAGSKEE AGIIRMLEFG 421 SHLDLERMQA SLPLDNISYR RESAI 19. human Kir 2.4- sequence 1 MGLARALRRL SGALDSGDSR AGDEEEAGPG LCRNGWAPAP VQSPVGRRRG RFVKKDGHCN 61 VREVNLGGQG ARYLSDLFTT CVDVRWRWMC LLFSCSFLAS WLLFGLAFWL IASLHGDLAA 121 PPPPAPCFSH VASFLAAFLF ALETQTSIGY GVRSVTEECP AAVAAVVLQC IAGCVLDAFV 181 VGAVMAXMAK PKKRNEILVF SENAVVALRD HRLCLMWRVG NLRRSHLVEA HVRAQLLQPR 241 VTPEGEYIPL DHQDVDVGFD GGTDRIFLVS PITIVHEIDS ASPLYELGRA ELARADFELV 301 VILEGMVEAT AMTTQCRSSY LPGELLWGHR FEPVLFQRGS QYEVDYRHFH RTYEVPGTPV 361 CSAKELDERA EQASHSLKSS FPGSLTAFCY ENELALSCCQ EEDEDDETEE GNGVETEDGA 421 ASPRVLTPTL ALTLPP 20. human Kir 3.1- amino acid sequence 1 MSALRRKFGD DYQVVTTSSS GSGLQPQGPG QDPQQQLVPK KKRQRFVDKN GRCNVQHGNL 61 GSETSRYLSD LFTTLVDLKW RWNLFIFILT YTVAWLFMAS MNWVIAYTRG DLNKAHVGNY 121 TPCVANVYNF PSAFLEFIET EATIGYGYRY ITDKCPEGII LFLFQSILGS IVDAFLIGCM 181 FIKMSQPKKR AETLMFSEHA VISMRDGKLT LMERVGNERN SHMVSAQIRC KLLKSRQTPE 241 GEFLPLDQLE LDVGFSTGAD QLFLVSPLTI CHVIDAKSPF YDLSQRSMQT EQFEIVVILE 301 GIVETTGMTC QARTSYTEDE VLWGHRFFPV ISLEEGFFKV DYSQFHATFE VPTPPYSVKE 361 QEEMLLMSSP LIAPAITNSK ERHNSVECLD GLDDITITLP SKLQKITGRE DFPKKLLRMS 421 STTSEKAYSL GDLPMKLQRI SSVPGNSEEK LVSKTTKMLS DPMSQSVADL PPKLQKMAGG 481 AARMEGNLPA KLRKMNSDRF T 21. human Kir 3.4- amino acid sequence 1 MAGDSRNAMN QDKEIGVTPW DPKKIPKQAR DYVPIATDRT RLLAEGKKPR QRYNEKSGKC 61 NVHHGNVQET YRYLSDLFTT LVDLKWRFNL LVFTMVYTVT WLFEGFIWWL IAYIRGDLDH 121 VGDQEWIPCV ENLSGFVSAF LFSIETETTI GYGFRVITEK CPEGIILLLV QAILGSIVNA 181 FMVGCMFVKI SQPKKRAETL MESNNAVISM RDEKLCLMFR VGDLRNSHIV EASIRAKLIK 241 SRQTKEGEFI PLNQTDINVG FDTGDDRLFL VSPLIISHEI NQKSPFWEMS QAQLHQEEFE 301 VVVILEGMVE ATGMTCQARS SYMDTEVLWG HRFTPVLTLE KGFYEVDYNT FHDTYETNTP 361 SCCAKELAEM KREGRLLQYL PSPPLLGGCA EAGLDAEAEQ NEEDEPKGLG GSREARGSV 22. human Kir 4.1- amino acid sequence 1 MTSVARVYYS QTTQTESRPL MGPGIRRRRV LTKDGRSNVR MEHIADKRFL YLKDLWTTFI 61 DMQWRYKLLL FSATFAGTWF LFGVVWYLVA VAHGDLLELD PPANHTPCVV QVHTLTGAFL 121 FSLESQTTIG YGFRYISEEC PLAIVLLIAQ LVLTTILEIF ITGTFLAKIA RPKKRAETIR 181 FSQHAVVASH NGKPCLMIRV ANMRKSLLIG CQVTGKLLQT HQTKEGENIR LNQVNVTFQV 241 DTASDSPFLI LPLTFYHVVD ETSPLKDLPL RSGEGDFELV LILSGTVEST SATCQVRTSY 301 LPEEILWGYE FTPAISLSAS GKYIADFSLF DQVVKVASPS GLRDSTVRYG DPEKLKLEES 361 LREQAEKEGS ALSVRISNV 23. human Kir 4.2- amino acid sequence 1 MDAIHIGMSS TPLVKHTAGA GLKANRPRVM SKSGHSNVRI DKVDGIYLLY LQDLWTTVID 61 NKWRYKLTLF AATFVMTWFL FGVIYYAIAF IHGDLEPGEP ISNHTPCINE VDSLTGAFLF 121 SLESQTTIGY GVRSITEECP HAIFILVAQL VITTLIEIFI TGTFLAKIAR PKKRAETIKF 181 SHCAVITXQN GKLCLVIQVA NMRKSLLIQC QLSGKLLQTH VTKEGERILL NQATVKFHVD 241 SSSESPFLIL PMTFYHVLDE TSPLRDLTPQ NLKEKEFELV VLLNATVEST SAVCQSRTSY 301 IPEEIYWGFE FVPVVSLSKN GKYVADFSQF EQIRKSPDCT FYCADSEKQQ LEEKYRQEDQ 361 RERELRTLLL QQSNV 24. human Kir 5.1- amino acid sequence 1 MSYYGSSYHI INADAKYPGY PPEHIIAEKR RARRRLLHKD GSCNVYFKHI FGEWGSYVVD 61 IFTTLVDTKW RHMEVIFSLS YILSWLIFGS VFWLIAFHHG OLLNDPDITP CVDNVHSFTG 121 AFLFSLETQT TIGYGYRCVT EECSVAVLMV ILQSILSCII NTFIIGAALA KMATARKRAQ 181 TIRESYFALI GMRDGKLCLM WRIGDFRPNH VVEGTVRAQL LRYTEDSEGR MTMAFKDLKL 241 VNDQIILVTP VTIVHEIDHE SPLYALDRKA VAKDNFEILV TFIYTGDSTG TSHQSRSSYV 301 PREILWOHRF NOVLEVKRKY YKVNCLQFEG SVEVYAPFCS AKQLDWKDQQ LHIEKAPPVR 361 ESCTSDTKAR RRSFSAVAIV SSCENPEETT TSATHEYRET PYQKALLTLN RISVESQM 25. human Kir 6.1- amino acid sequence 1 MLARKSIIPE EYVLARIAAE NLRKPRIRDR LPKARFIAKS GACNLAHKNI REQGRFLQDI 61 FTTLVDLKWR HTLVIFTMSF LCSWLLFAIM WWLVAFAHGD IYAYMEKSGM EKSGLFSTVC 121 VTNVRSFTSA FLFSIEVQVT IGFGGRMMTE ECPLAITVLI LQNIVGLIIN AVMLGCIFMK 181 TAQAHRRAET LIFSRHAVIA VRNGKLCFMF RVGDLRKSMI ISASVRIQVV KKTTTPEGEV 241 VPIHQLDIPV DNPIESNNIF LVAPLIICHV IDKRSPLYDI SATDLANQDL EVIVILEGVV 301 ETTGITTQAR TSYIAEEIQW GHRFVSIVTE EEGVYSVDYS KFGNTVKVAA PRCSARELDE 361 KPSILIQTLQ KSELSHQNSL RKRNSMRRNN SMRRNNSIRR NNSSLMVPKV QFMTPEGNQN 421 TSES 26. human Kir 6.2- amino acid sequence 1 MLSRKGIIPE EYVLTRLAED PAKPRYRARQ RRARFVSKKG NCNVAHKNIR EQGRFLQDVF 61 TTLVDLKWPH TLLIFTMSFL CSWLLFAMAW WLEAFAHGDL APSEGTAEPC VTSIHSFSSA 121 FLFSIEVQVT IGFGGRMVTE ECPLAILILI VQNIVGLMIN AIMLGCIFMK TAQAHRRAET 181 LIFSKHAVIA LRHGRLCFML RVGDLRKSMI ISATIHMQVV RKTTSPEGEV VPLHQVDIPM 241 ENGVGGNSIF LVAPLIIYHV IDANSPLYDL APSDLHHHQD LEIIVILEGV VETTGITTQA 301 RTSYLADEIL WGQRFVPIVA EEDGRYSVDY SKFGNTVKVP TPLCTARQLD EDHSLLEALT
361 LASARGPLRK RSVPMAKAKP KFSISPDSLS 27. human Kir 7.1- amino acid sequence 1 mdssnckvia pllsgryrrm vtkdghstlq mdgagrglay lrdawgilmd mrwrwmmlvf 61 sasfvvhwlv favlwyvlae mngdleldhd appenhticv kyitsftaaf sfsletqlti 121 gygtmfpsgd cpsaiallai gmllglmlea fitgafvaki arpknrafsi rftdtavvah 181 mdgkpnlifq vantrpsplt svrvsavlyq erengklygt svdfhldgis sdecpffifp 241 ltyyhsitps sp1atllghe npshfelvvf lsamgegtge icgrrtsylp seimlhhcfa 301 slltrgskge ygikmenfdk typefptply skspnrtdld ihinggsidn fqisetglte 28. chicken Kir 2.1- amino acid sequence 1 MGSVRTNRYS IVSSEEDGMK LATMAVANGF GNGKSKVHTR QQCRSRFVXK DGHCNVQFIN 61 VGEKGQRYLA DIFTTCVDIR WRWMLVIFCL TEILSWLFFG CVFWLIALLH GDLENQENNK 121 PCVSQVSSFT AAFLFSIETQ TTIGYGFRCV TDECPIAVFM VVFQSIVGCI IDAFIIGAVM 181 AKMAKPKKRN ETLVFSHNAV VAMRDGKLCL MWRVGNLRKS HLVEAHVRAQ LLKSRITSEG 241 EYIPLDEIDI NVGFDSGIDR IFLVSPITIV HEIDEDSPLY DLSKQDMDNA DFEIVVILEG 301 MVEATAMTTQ CRSSYLANEI LWGHRYEPVL FEEKNYYKVD YSRFHKTYEV PNTPICSARD 361 LAEKKYILSN ANSFCYENEV ALTSKEEDEI DTGVPESTST DTHPDMDHHN QAGVPLEPRP 421 LRRESEI 29. chicken Kir 2.2- amino acid sequence 1 mtagrvnpys ivsseedglr lttmpgingf gngkihtrrk crnrfvkkng qcnveftnmd 61 dkpqryiadm fttcvdirwr ymlllfslaf lvswllfgli fwlialihgd lenpggddtf 121 kpcvlqvngf vaallfsiet qttigygfrc vteecplavf mvvvqsivgc iidsfmigai 181 makmarpkkr aqtllfshna vvamrdgklc lmwrvgnlrk shiveahvra qlikpritee 241 geyipldgid idvgfdkgld riflvspiti lheinedspl fgisrqdlet ddfeivvile 301 gmveatamtt qarssylase ilwghrfepv lfeeknqykv dyshfhktye vpstprcsak 361 dlvenkfllp stnsfcyene lafmsrdede edddsrgldd lspdnrhefd rlqatialdq 421 rsyrresei 30. human Kir 1.1- cDNA 1 atgaatgctt ccagtcggaa tgtgtttgac acgttgatca gggtgttgac agaaagtatg 61 ttcaaacatc ttcggaaatg ggtcgtcact cgcttttttg ggcattctcg gcaaagagca 121 aggctagtct ccaaagatgg aaggtgcaac atagaatttg gcaatgtgga ggcacagtca 181 aggtttatat tctttgtgga catctggaca acggtacttg acctcaagtg gagatacaaa 241 atgaccattt tcatcacagc cttcttgggg agttggtttt tctttggtct cctgtggtat 301 gcagtagcgt acattcacaa agacctcccg gaattccatc cttctgccaa tcacactccc 361 tgtgtggaga atattaatgg cttgacctca gcttttctgt tttctctgga gactcaagtg 421 accattggat atggattcag gtgtgtgaca gaacagtgtg ccactgccat ttttctgctt 481 atctttcagt ctatacttgg agttataatc aattctttca tgtgtggggc catcttagcc 541 aagatctcca ggcccaaaaa acgtgccaag accattacgt tcagcaagaa cgcagtgatc 601 agcaaacggg gagggaagct ttgcctccta atccgagcgg ctaatctcag gaagagcctt 661 cttattggca gtcacattta tggaaagctt ctgaagacca cagtcactcc tgaaggagag 721 accattattt tggaccagat caatatcaac tttgtagttg acgctgggaa tgaaaattta 781 ttcttcatct ccccattgac aatttaccat gtcattgatc acaacagccc tttcttccac 841 atggcagcgg agacccttct ccagcaggac tttgaattag tggtgttttt agatggcaca 901 gtggagtcca ccagtgctac ctgccaagtc cggacatcct atgtcccaga ggaggtgctt 961 tggggctacc gttttgctcc catagtatcc aagacaaagg aagggaaata ccgagtggat 1021 ttccataact ttagcaagac agtggaagtg gagacccctc actgtgccat gtgcctttat 1081 aatgagaaag atgttagagc caggatgaag agaggctatg acaaccccaa cttcatcttg 1141 tcagaagtca atgaaacaga tgacaccaaa atgtaa 31. human Kir 1.2- cDNA 1 cttttctgat cccagctccg ggtttaagag tcctggcacg gcccgtcgca cagctctgct 61 cctaactcct gcccgccccg tccgtccatc tgtcccgctg ccccgcggcc catccaaggg 121 gccactccac ctcggaccca agatgacgtc agttgccaag gtgtattaca gtcagaccac 181 tcagacagaa agccggcccc taatgggccc agggatacga cggcggagag tcctgacaaa 241 agatggtcgc agcaacgtga gaatggagca cattgccgac aagcgcttcc tctacctcaa 301 ggacctgtgg acaaccttca ttgacatgca gtggcgctac aagcttctgc tcttctctgc 361 gacctttgca ggcacatggt tcctctttgt cgtggtgtgg tatctggtag ctgtggcaca 421 tggggacctg ctggagctgg accccccggc caaccacacc ccctgtgtgg tacaggtgca 481 cacactcact ggagccttcc tcttctccct tgaatcccaa accaccattg gctatggctt 541 ccgctacatc agtgaggaat gtccactagc cattgtgctt cttattgccc agctggtgct 601 caccaccatc ctggaaatct tcatcacagg taccttcctg gcgaagattg cccggcccaa 661 gaagcgggct gagaccattc gtttcagcca gcatgcagtt gtggcctccc acaatggcaa 721 gccctgcctc atgatccgag ttgccaatat gcgcaaaagc ctcctcattg gctgccaggt 781 gacaggaaaa ctgcttcaga cccaccaaac caaggaaggg gagaacaccc ggctcaacca 841 ggtcaatgtg actttccaag tagacacagc ccctgacagc cccttcctta ttctacccct 901 taccttctat catgtggtag atgagaccag tcccttgaaa gatctccctc ttcgcagcgg 961 tgagggtgac tttgagctgg tgctgatcct aagtgggaca gtggagtcca ccagtgccac 1021 ctgtcaggtg cgcacttcct acctgccaga ggagatcctt tggggctacg agttcacacc 1081 tgccatctca ctgtcagcca gtggtaaata catagctgac tttagccttt ttgaccaagt 1141 tgtgaaagtg gcctctccta gtggcctccg tgacagcact gtacgctacg gagaccctga 1201 aaagctcaag ttggaggagt cattaaggga gcaagctgag aaggagggca gtgcccttag 1261 tgtgcgcatc agcaatgtct ga 32. human Kir 2.1- cDNA 1 atgggcagtg tgcgaaccaa ccgctacagc atcgtctctt cagaagaaga cggtatgaag 61 ttggccacca tggcagttgc aaatggcttt gggaacggga agagtaaagt ccacacccga 121 caacagtgca ggagccgctt tgtgaagaaa gatggccact gtaatgttca gttcatcaat 181 gtgggtgaga aggggcaacg gtacctcgca gacatcttca ccacgtgtgt ggacattcgc 241 tggcggtgga tgctggttat cttctgcctg gctttcgtcc tgtcatggct gttttttggc 301 Lgtgtgcttt ggttgatagc tctgctccat ggggacctgg atgcatccaa agagggcaaa 361 gcttgtgtgt ccgaggtcaa cagcttcacg gctgccttcc tcttctccat tgagacccag 421 acaaccatag gctatggttt cagatgtgtc acggatgaat gcccaattgc tgttttcatg 481 gtggtgttcc agtcaatcgt gggctgcatc atcgatgctt tcatcattgg cgcagtcatg 541 gccaagatgg caaagccaaa gaagagaaac gagactcttg tcttcagtca caatgccgtg 601 attgccatga gagacggcaa gctgtgtttg atgtggcgag tgggcaatct tcggaaaagc 661 cacttggtgg aagctcatgt tcgagcacag ctcctcaaat ccagaattac ttctgaaggg 721 gagtatatcc ctctggatca aatagacatc aatgttgggt ttgagagcgg aatcgatcgt 781 atatttctgg tgtccccaat cactatagtc catgaaatag atgaagacag tcctttatat 841 gatttgagta aacaggacat tgacaacgca gactttgaaa tcgtggtcat actggaaggc 901 atggtggaag ccactgccat gacgacacag tgccgtagct cttatctagc aaatgaaatc 961 ctgtggggcc accgctatga gcctgtgctc tttgaagaga agcactacta caaagtggac 1021 tattccaggt tccacaaaac ttacgaagtc cccaacactc ccctttgtag tgccagagac 1081 ttagcagaaa agaaatatat cctctcaaat gcaaattcat tttgctatga aaatgaagtt 1141 gccctcacaa gcaaagagga agacgacagt gaaaatggag ttccagaaag cactagtacg 1201 gacacgcccc ctgacataga ccttcacaac caggcaagtg tacctctaga gcccaggccc 1261 ttacggcgag agtcggagat atga 33. human Kir 2.2- cDNA 1 atgaccgcgg ccagccgggc caacccctac agcatcgtgt catcggagga ggacgggctg 61 cacctggtca ccatgtcggg cgccaacggc ttcggcaacg gcaaggtgca cacgcgccgc 121 aggtgccgca accgcttcgt caagaagaat ggccagtgca acattgagtt cgccaacatg 181 gacgagaagt cacagcgcta cctggctgac atgtccacca cctgtgcgga catccgctgg 241 cggtacatgc tgctcatctt ctcgctggcc ttccttgcct cctggctgct gttcggcatc 301 atcttctggg tcatcgcggt ggcacacggt gacctggagc cggctgaggg ccggggccgc 361 acaccctgtg tgatgcaggt gcacggcttc atggcggcct tcctcttctc catcgagacg 421 cagaccacca tcggctacgg gctgcgctgt gtgacggagg agtgcccggt ggccgtcttc 481 atggtggtgg cccagtccat cgtgggctgc atcatcgact cctccatgat tggtgccatc 541 atggccaaga tggcaaggcc caagaagcgg gcacagacgc tgctgttgag ccacaacgcc 601 gtggtggccc tgcgtgacgg caagctctgc ctcatgtggc gtgtgggtaa cctgcgcaag 661 agccacattg tggaggccca tgtgcgcgcg cagctcatca agccgcgggt caccgaggag 721 ggcgagtaca tcccgctgga ccagatcgac atcgatgtgg gcttcgacaa gggcctggac 781 cgcatctttc tggtgtcgcc catcaccatc ttgcatgaga ttgacgaggc caggccgctc 841 ttcggcatca gccggcagga cctggagacg gacgactttg agatcgtggt catcctggaa 901 ggcatggtgg aggccacagc catgaccacc caggcccgca gctcctacct ggccaatgag 961 atcttctggg gtcaccgctt tgagcccgtg ctcttcgagg agaagaacca gtacaagatt 1021 gactactcgc acttccacaa gacctatgag gtgccctcta cgccccgctg cagtgcgaag 1081 gatctggtag agaacaagtt cctgctgccc agcgccaact ccttctgcta cgagaacgag 1141 ctggccttcc tgagccgtga cgaggaggat gaggcggacg gagaccagga cggccgaagc 1201 cgggacggcc tcagccccca ggccaggcat gactttgaca gactccaggc tggcggcggg 1261 gacctggagc agcggcccta cagacgggag tcagagatct ga 34. human Kir 2.3- cDNA 1 atgcacggac acagccgcaa cggccaggcc cacgtgcccc ggcggaagcg ccgcaaccgc 61 ttcgtcaaga agaacggcca atgcaacgtg tacttcgcca acctgagcaa caagtcgcag 121 cgctacatgg cggacatctt caccacctgc gtggacacgc gctggcgcta catgctcatg 181 atcttctccg cggccttcct tgtctcctgg ctctttttcg gcctcctctt ctggtgtatc 241 gccttcttcc acggtgacct ggaggccagc ccaggggtgc ctgcggcggg gggcccggcg 301 gcgggtggtg gcggaggagc cccggtggcc cccaagccct gcatcatgca cgtgaacggc 361 ttcctgggtg ccttcctgtt ctcggtggag acgcagacga ccatcggcta tgggttccgg 421 tgcgtgacag aggagtgccc gctggcagtc atcgctgtgg tggtccagtc catcgtgggc 481 tgcgtcatcg actccttcat gattggcacc atcatggcca agatggcgcg gcccaagaag 541 cgggcgcaga cgttgctgtt cagccaccac gcggtcattt cggtgcgcga cggcaagctc 601 tgcctcatgt ggcgcgtggg caacctgcgc aagagccaca ttgtggaggc ccacgtgcgg 661 gcccagctca tcaagccata catgacccag gagggcgagt acctgcccct ggaccagcgg 721 gacctcaacg tgggctatga catcggcctg gaccgcatct tcctggtgtc gcccatcatc
781 attgtccacg agatcgacga ggacagcccg ctttatggca tgggcaagga ggagctggag 841 tcggaggact ttgagatcgt ggtcatcctg gagggcatgg tggaggccac ggccatgacc 901 acccaggccc gcagctccta cctggccagc gagatcctgt ggggccaccg ctttgagcct 961 gtggtcttcg aggagaagag ccactacaag gtggactact cacgttttca caagacctac 1021 gaggtggccg gcacgccctg ctgctcggcc cgggagctgc aggagagtaa gatcaccgtg 1081 ctgcccgccc caccgccccc tcccagtgcc ttctgctacg agaacgagct ggcccttatg 1141 agccaggagg aagaggagat ggaggaggag gcagctgcgg cggccgcggt ggccgcaggc 1201 ctgggcctgg aggcgggttc caaggaggag gcgggcatca tccggatgct ggagttcggc 1261 agccacctgg acctggagcg catgcaggct tccctcccgc tggacaacat ctcctaccgc 1321 agggagtctg ccatctga 35. human Kir 2.4- cDNA 1 atgggcctgg ccagggccct acgccgcctc agcggcgccc tggattcggg agacagccgg 61 gcgggcgatg aagaggaggc cgggcccggg ttgtgccgca acgggtgggc gccggcaccg 121 gtgcagtcac ccgtgggccg gcgccgcggt cgcttcgtca agaaagacgg gcactgcaac 181 gtgcgtttcg taaacctggg tggccagggc gcgcgctacc tgagcgacct gttcaccaca 241 tgcgtggacg tgcgctggcg ctggatgtgc ctgctcttct cctgctcctt cctcgcctcc 301 tggctgctct tcggcctggc cttctggctc attgcctcgc tgcacggcga cctggccgcc 361 ccgccaccgc ccgcgccctg cttctcacac gtggccagct tcctggccgc cttcctcttc 421 gcgctggaga cgcagacgtc catcggctac ggcgtgcgca gcgtcaccga ggagtgcccg 481 gccgctgtgg ccgccgtggt gctgcagtgc attgccggct gcgtgctcga cgccttcgtc 541 gtgggtgctg tcatggccaa gatggccaaa cccaagaagc gcaacgagac gctggtcttc 601 agcgagaacg ccgtcgtggc gctgcgcgac caccgcctct gcctcatgtg gcgcgtcggc 661 aacctgcgcc gcagccacct ggtcgagacc cacgtgcgtg cccagctgct gcagccccgt 721 gtgaccccag agggtgagta catcccgctg gaccaccagg atgtggatgt gggctttgat 781 ggaggcaccg atcgtatctt cctcgtgtcc cccatcacca tcgtccatga gatcgactct 841 gccagtcctc tgtatgagct aggacgtgcc gagctggcca gggctgactt tgagctggtg 901 gtcattctcg aggggatggt tgaggccaca gccatgacca cacagtgtcg ctcgtcctac 961 ctccctggtg aactgctctg gggccatcgt tttgagccag ttctcttcca gcgtggctcc 1021 cagtatgagg tcgactatcg ccacttccat cgcacttatg aggtcccagg gacaccggtc 1081 tgcagtgcta aggagctgga tgaacgggca gagcaggctt cccacagcct caagtctagt 1141 ttccccggct ctctgactgc attttgttat gagaatgaac ttgctctgag ctgctgccag 1201 gaggaagatg aggacgatga gactgaggaa gggaatgggg tggaaacaga agatggggct 1261 gctagccccc gagttctcac accaaccctg gcgctgaccc tgcctccatg a 36. human Kir 3.1- cDNA 1 atgtctgcac tccgaaggaa atttggggac gattatcagg tagtgaccac atcgtccagc 61 ggctcgggct tgcagcccca ggggccaggc caggaccctc aggaggagct tgtgcccaag 121 aagaagcggc agcggttcgt ggacaagaac ggccggtgca atgtacagca cggcaacctg 181 ggcagcgaga caagccgcta cctctcggac ctcttcacca cgctggtgga cctcaagtgg 241 cgctggaacc tcttcatctt cattctcacc tacaccgtgg cctggctttt catggcgtcc 301 atgtggtggg tgatcgccta cactcggggc gacctgaaca aagcccacgt cggtaactac 361 acgccttgcg tggccaatgt ctataacttc ccttctgcct tcctcttctt catcgagacg 421 gaggccacca tcggctatgg ctaccgatac atcacagaca agtgccccga gggcatcatc 481 ctcttcctct tccagtccat cctgggctcc atcgtggacg ccttcctcat cggctgcatg 541 ttcatcaaga tgtcccagcc caagaagcgc gccgagaccc tcatgttcag cgagcacgcg 601 gtgatctcca tgagggacgg aaaactcacg cttatgttcc gggtgggcaa cctgcgcaac 661 agccacatgg tctccgcgca gattcgctgc aagctgctca aatctcggca gacacctgag 721 ggtgagttcc ttccccttga ccaacttgaa ctggatgtag gttttagtac aggggcagat 781 caactttttc ttgtgtcccc cctcacaatt tgccacgtga tcgatgccaa aagccccttt 841 tatgacctat cccagcgaag catgcaaact gaacagttcg agattgtcgt catcctagaa 901 ggcattgtgg aaacaactgg gatgacttgt caagctcgaa catcatatac tgaagatgaa 961 gttctttggg gtcatcgttt ttttcctgta atttccttag aagagggatt ctttaaagtt 1021 gattactccc agttccatgc aacatttgaa gtccccaccc caccttacag tgtgaaagag 1081 caggaggaaa tgcttctcat gtcgtcccct ttaatagcac cagccataac taacagcaaa 1141 gaaagacata attctgtgga atgcttagat ggactagatg atattactac aaaactacca 1201 tctaagctgc agaaaattac tggaagagaa gactttccca aaaaactctt gaggatgagt 1261 tctacaactt cagaaaaagc ctacagcttg ggagacttgc ccatgaaact tcaacgaata 1321 agttcagttc cgggcaactc agaagaaaaa ctggtatcta aaaccaccaa gatgttatct 1381 gatcccatga gccagtctgt ggctgatttg ccaccaaagc ttcaaaagat ggctggagga 1441 gcagctagga tggaagggaa ccttccagcc aaattaagaa aaatgaactc tgatcgcttc 1501 acataa 37. human Kir 3.4- cDNA 1 atggctggcg attctaggaa tgccatgaac caggacatgg agattggagt cactccctgg 61 gaccccaaga agattccaaa acaggcccgc gattatgtcc ccattgccac agaccgtacg 121 cgcctgctgg ccgagggcaa gaagccacgc cagcgctaca tggagaagag tggcaagtgc 181 aacgtgcacc acggcaacgt ccaggagacc taccggtacc tgagtgacct cttcaccacc 241 ctggtggacc tcaagtggcg cttcaacttg ctcgtcttca ccatggttta cactgtcacc 301 tggctgttct tcggcttcat ttggtggctc attgcttata tccggggtga cctggaccat 361 gttggcgacc aagagtggat tccttgtgtt gaaaacctca gtggcttcgt gtccgctttc 421 ctgttctcca ttgagaccga aacaaccatt gggtatggct tccgagtcat cacagagaag 481 tgtccagagg ggattatact cctcttggtc caggccaLcc tgggctccat cgtcaatgcc 541 ttcatggtgg ggtgcatgtt tgtcaagatc agccagccca agaagagagc ggagaccctc 601 atgttttcca acaacgcagt catctccatg cgggacgaga agctgtgcct catgttccgg 661 gttggcgacc tccgcaactC CCacatcgtg gaggcctcca tccgggccaa gctcatcaag 721 tcccggcaga ccaaagaggg ggagttcatc cccctgaacc agacagacat caacgtgggc 781 tttgacacgg gcgacgaccg cctcttcctt gtgtctcctc tgatcatctc ccatgagatc 841 aaccagaaga gccctttctg ggagatgtct caggctcagc tgcatcagga agagtttgaa 901 gttgtggtca ttctagaagg gatggtggaa gccacaggca tgacctgcca agcccggagc 961 tcctacatgg atacagaggt gctctggggc caccgattca ccaccatcct caccttggaa 1021 aagggcttct atgaggtgga ctacaacacc ttccatgata cctatgagac caacacaccc 1081 agctgctgtg ccaaggagct ggcagaaatg aagagggaag gccggctcct ccagtacctc 1141 cccagccccc cactgctggg gggctgtgct gaggcagggc tggatgcaga ggctgagcag 1201 aatgaagaag atgagcccaa ggggctgggt gggtccaggg aggccagggg ctcggtgtga 38. human Kir 4.1- cDNA 1 atgacgtcag ttgccaaggt gtattacagt cagaccactc agacagaaag ccggccccta 61 atgggcccag ggatacgacg gcggagagtc ctgacaaaag atggtcgcag caacgtgaga 121 atggagcaca ttgccgacaa gcgcttcctc tacctcaagg acctgtggac aaccttcatt 181 gacatgcagt ggcgctacaa gcttctgctc ttctctgcga cctttgcagg cacatggttc 241 ctctttggcg tggtgtggta tctggtagct gtggcacatg gggacctgct ggagctggac 301 cccccggcca accacacccc ctgtgtggta caggtgcaca cactcactgg agccttcctc 361 ttctcccttg aatcccaaac caccattggc tatggcttcc gctacatcag tgaggaatgt 421 ccactggcca ttgtgcttct tattgcccag ctggtgctca ccaccatcct ggaaatcttc 481 atcacaggta ccttcctggc gaagattgcc cggcccaaga agcgggctga gaccattcgt 541 ttcagccagc atgcagttgt ggcctcccac aatggcaagc cctgcctcat gatccgagtt 601 gccaatatgc gcaaaagcct cctcattggc tgccaggtga caggaaaact gcttcagacc 661 caccaaacca aggaagggga gaacatccgg ctcaaccagg tcaatgtgac tttccaagta 721 gacacagcct ctgacagccc cttccttatt ctacccctta ccttctatca tgtggtagat 781 gagaccagtc ccttgaaaga tctccctctt cgcagtggtg agggtgactt tgagctggtg 841 ctgatcctaa gtgggacagt ggagtccacc agtgccacct gtcaggtgcg cacttcctac 901 ctgccagagg agatcctttg gggctacgag ttcacacctg ccatctcact gtcagccagt 961 ggtaaataca tagctgactt tagccttttt gaccaagttg tgaaagtggc ctctcctagt 1021 ggcctccgtg acagcactgt acgctacgga gaccctgaaa agctcaagtt ggaggagtca 1081 ttaagggagc aagctgagaa ggagggcagt gcccttagtg tgcgcatcag caatgtctga 39. human Kir 4.2- cDNA 1 atggatgcca ttcacatcgg catgtccagc acccccctgg tgaagcacac tgctggggct 61 gggctcaagg ccaacagacc ccgcgtcatg tccaagagtg ggcacagcaa cgtgagaatt 121 gacaaagtgg atggcatata cctactctac ctgcaagacc tgtggaccac agttatcgac 181 atgaagtgga gatacaaact caccctgttc gotgccactt ttgtgatgac ctggttcctt 241 tttggagtca tctactatgc catcgcgttt attcatgggg acttagaacc cggtgagccc 301 atttcaaatc ataccccctg catcatgaaa gtggactctc tcactggggc gtttctcttt 361 tccctggaat cccagacaac cattggctat ggagtccgtt ccatcacaga ggaatgtcct 421 catgccatct tcctgttggt tgctcagttg gtcatcacga ccttgattga gatcttcatc 481 accggaacct tcctggccaa aatcgccaga cccaaaaagc gggctgagac catcaagttc 541 agccactgtg cagtcatcac caagcagaat gggaagctgt gcttggtgat tcaggtagcc 601 aatatgagga agagcctctt gattcagtgc cagctctctg gcaagctcct gcagacccac 661 gtcaccaagg agggggagcg gattctcctc aaccaagcca ctgtcaaatt ccacgtggac 721 tcctcctctg agagcccctt cctcattctg cccatgacat tctaccatgt gctggatgag 781 acgagccccc tgagagacct cacaccccaa aacctaaagg agaaggagtt tgagcttgtg 841 gtcctcctca atgccactgt ggaatccacc agcgctgtct gccagagccg aacatcttat 901 atcccagagg aaatctactg gggttttgag tttgtgcctg tggtatctct ctccaaaaat 961 ggaaaatatg tggctgattt cagtcagttt gaacagattc ggaaaagccc agattgcaca 1021 ttttactgtg cagattctga gaaacagcaa ctcgaggaga agtacaggca ggaggatcag 1081 agggaaagag aactgaggac acttttatta caacagagca atgtctga 40. human Kir 5.1- cDNA 1 atgagctatt acggcagcag ctatcatatt atcaatgcgg acgcaaaata cccaggctac 61 ccgccagagc acattatagc tgagaagaga agagcaagaa gacgattact tcacaaagat 121 ggcagctgta atgtctactt caagcacatt tttggagaat ggggaagcta tgtggttgac 181 atcttcacca ctcttgtgga caccaagtgg cgccatatgt ttgtgatatt ttctttatct 241 tatattctct cgtggttgat atttggctct gtcttttggc tcatagcctt tcatcatggc
301 gatctattaa atgatccaga catcacacct tgtgttgaca acgtccattc tttcacaggg 361 gcctttttgt tctccctaga gacccaaacc accataggat atggttatcg ctgtgttact 421 gaagaatgtt ctgtggccgt gctcatggtg atcctccagt ccatcttaag ttgcatcata 481 aataccttta tcattggagc tgccttggcc aaaatggcaa ctgctcgaaa gagagcccaa 541 accattcgtt tcagctactt tgcacttata ggtatgagag atgggaagct ttgcctcatg 601 tggcgcattg gtgattttcg gccaaaccac gtggtagaag gaacagttag agcccaactt 661 ctccgctata cagaagacag tgaagggagg atgacgatgg catttaaaga cctcaaatta 721 gtcaacgacc aaatcatcct ggtcaccccg gtaactattg tccatgaaat tgaccatgag 781 agccctctgt atgcccttga ccgCaaagca gtagccaaag ataactttga gattttggtg 841 acatttatct atactggtga ttccactgga acatctcacc aatctagaag ctcctatgtt 901 ccccgagaaa ttctctgggg ccataggttt aatgatgtct tggaagttaa gaggaagtat 961 tacaaagtga actgcttaca gtttgaagga agtgtggaag tatatgcccc cttttgcagt 1021 gccaagcaat tggactggaa agaccagcag ctccacatag aaaaagcacc accagttcga 1081 gaatcctgca cgtcggacac caaggcgaga cgaaggtcat ttagtgcagt tgccattgtc 1141 agcagctgtg aaaaccctga ggagaccacc acttccgcca cacatgaata tagggaaaca 1201 ccttatcaga aagctctcct gactttaaac agaatctctg tagaatccca aatgtag 41. human Kir 6.1- cDNA 1 atgttggcca gaaagagtat catcccggag gagaatgtgc tggcgcgcat cgccgcagag 61 aacctgcgca agccgcgcat ccgagaccgc ctccccaaag cccgcttcat cgccaagagc 121 ggggcctgca acctggcgca taagaacatc cgtgagcaag gacgctttct acaggacatc 181 ttcaccacct tggtggacct gaaatggcgc cacacgctgg tcatctttac catgtccttc 241 ctctgcagct ggctgctctt cgctatcatg tggtggctgg tggcctttgc ccatggggac 301 atctatgctt acatggagaa aagtggaatg gagaaaagtg gtttggagtc cactgtgtgt 361 gtgactaatg tcaggtcttt cacttctgct tttctcttct ccattgaagt tcaagttacc 421 attgggtttg gagggaggat gatgacagag gaatgccctt tggccatcac ggttttgatt 481 ctccagaata ttgtgggttt gatcatcaat gcagtcatgt taggctgcat tttcatgaaa 541 acagctcagg ctcacagaag ggcagaaact ttgattttca gccgccatgc tgtgattgcc 601 gtccgaaatg gcaagctgtg cttcatgttc cgagtgggtg acctgaggaa aagcatgatc 661 attagtgcct ctgtgcgcat ccaggtggtc aagaaaacaa ctacacctga aggggaggtg 721 gttcctattc accaactgga cattcctgtt gataacccaa tcgagagcaa taacattttt 781 ctggtggccc ctttgatcat ctgccacgtg attgacaagc gcagtcccct gtatgacatc 841 tcagcaactg acctggccaa ccaagacttg gaggtcatag ttattctgga aggagtggtt 901 gaaactactg gcatcaccac acaagcacga acctcctaca ttgctgagga gatccaatgg 961 ggccaccgct ttgtgtccat tgtgactgag gaagaaggag tgtattctgt ggattactcc 1021 aaatttggca acactgttaa agtagctgct ccacggtgca gtgcccgaga gctggatgag 1081 aaaccttcca tccttattca gaccctccaa aagagtgaac tgtctcatca aaattctctg 1141 aggaagcgca actccatgag aagaaacaat tccatgagga ggaacaattc tatccgaagg 1201 aacaattctt ccctcatggt accaaaggtg caatttatga ctccagaagg aaatcaaaac 1261 acatcggaat catga 42. human Kir 6.2- cDNA 1 atgctgtccc gcaagggcat catccccgag gaatacgtgc tgacacgcct ggcagaggac 61 cctgccaagc ccaggtaccg tgcccgccag cggagggccc gctttgcgtc caagaaaggc 121 aactgcaacg tggcccacaa gaacatccgg gagcagggcc gcttcctgca ggacgtgttc 181 accacgctgg tggacctcaa gtggccacac acattgctca tcctCaccat gtccttcctg 241 tgcagctggc tgctcttcgc catggcctgg tggctcatcg ccttcgccca cggtgacctg 301 gcccccagcg agggcactgc tgagccctgt gtcaccagca tccactcctt ctcgtctgcc 361 ttccttttct ccattgaggt ccaagtgact attggctttg gggggcgcat ggtgactgag 421 gagtgcccac tggccatcct gatcctcatc gtgcagaaca tcgtggggct catgatcaac 481 gccatcatgc ttggctgcat cttcatgaag actgcccaag cccaccgcag ggctgagacc 541 ctcatcttca gcaagcatgc ggtgatcgcc ctgcgccacg gccgcctctg catcatgcta 601 cgtgtgggtg acctccgcaa gagcatgatc atcagcgcca ccatccacat gcaggtggta 661 cgcaagacca ccagccccga gggcgaggtg gtgcccctcc accaggtgga catccccatg 721 gagaacggcg tgggtggcaa cagcatcttc ctggtggccc cgctgatcat ctaccatgtc 781 attgatgcca acagcccact ctacgacctg gcacccagcg acctgcacca ccaccaggac 841 ctcgagatca tcgtcatcct ggaaggcgtg gtggaaacca cgggcatcac cacccaggcc 901 cgcacctcct acctggccga tgagatcctg tggggccagc gctttgtgcc cattgtagct 961 gaggaggacg gacgttactc tgtggactac tccaagtttg gcaacaccgt caaagtgccc 1021 acaccactct gcacggcccg ccagcttgat gaggaccaca gcctactgga agctctgacc 1081 ctcgcctcag cccgcgggcc cctgcgcaag cgcagcgtgc ccatggccaa ggccaagccc 1141 aagttcagca tctctccaga ttccctgtcc tga 43. human Kir 7.1- cDNA 1 atggacagca gtaattgcaa agttattgct cctctcctaa gtcaaagata ccggaggatg 61 gtcaccaagg atggccacag cacacttcaa atggatggcg ctcaaagagg tcttgcatat 121 cttcgagatg cttggggaat cctaatggac atgcgctggc gttggatgat gttggtcttt 181 tctgcttctt ttgttgtcca ctggcttgtc tttgcagtgc tctggtatgt tctggctgag 241 atgaatggtg atctggaact agatcatgat gccccacctg aaaaccacac tatctgtgtc 301 aagtatatca ccagtttcac agctgcattc tccttctccc tggagacaca actcacaatt 361 ggttatggta ccatgttccc cagcggcgac tgtccaagtg caatcgcctt acttgccata 421 caaatgctcc taggcctcat gctagaggct tttatcacag gtgcttttgt ggcgaagatt 481 gcccggccaa aaaatcgagc tttttcaatt cgctttactg acacagcagt agtagctcac 541 atggatggca aacctaatct tatcttccaa gtggccaaca cccgacctag ccctctaacc 601 agtgtccggg tctcagctgt actctatcag gaaagagaaa atggcaaact ctaccagacc 661 agtgtggatt tccaccttga tggcatcagt tctgacgaat gtccattctt catctttcca 721 ctaacgtact atcactccat tacaccatca agtcctctgg ctactctgct ccagcatgaa 781 aatccttctc actttgaatt agtagtattc ctttcagcaa tgcaggaggg cactggagaa 841 atatgccaaa ggaggacatc ctacctaccg tctgaaatca tgttacatca ctgttttgca 901 tctctgttga cccgaggttc caaaggtgaa tatcaaatca agatggagaa ttttgacaag 961 actgtccctg aatttccaac tcctctggtt Cctaaaagcc caaacaggac tgacctggat 1021 atccacatca atggacaaag cattgacaat tttcagatct ctgaaacagg actgacagaa 1081 taa 44. chicken Kir 2.1- cDNA 1 atgggcagcg tgcgaaccaa ccgctacagc atcgtgtctt cggaagagga cggcatgaag 61 ctggcaacca tggccgttgc caatggcttt gggaatggaa aaagtaaggt acacaccagg 121 cagcagtgca ggagccgctt tgtcaaaaaa gatggccact gcaacgtcca gtttattaat 181 gtgggtgaga agggacagcg atacctcgca gacatcttca ccacttgcgt ggacatccgc 241 tggaggtgga tgctggttat cttctgcctg acattcatcc tctcctggct tttctttggc 301 tgtgtgtttt ggttgattgc gctgttgcac ggggatctgg agaaccaaga aaataacaaa 361 ccgtgtgtct cgcaagtgag cagcttcact gcagcctttc tgttctccat tgagacccag 421 accacgatcg gctatggctt caggtgcgtc acagacgagt gccccattgc tgttttcatg 481 gtggttttcc agtctatagt aggctgcatc attgacgcct tcatcattgg tgccgtcatg 541 gcaaagatgg ctaagccaaa aaagagaaac gaaactcttg tcttcagcca caatgccgtg 601 gtggccatga gagatgggaa actgtgcctg atgtggcgtg tcggaaacct gaggaaaagc 661 cacttggttg aggcacacgt gcgagcacag ctcctcaagt ccaggatcac gtcagaaggg 721 gagtacatcc ctttggatga aatagacatc aatgtagggt ttgacagcgg gatagaccgc 781 atattcctgg tctccccaat tacaatagta cacgaaatag atgaagatag tcctttgtat 841 gacttgagca aacaagacat ggacaatgct gactttgaaa ttgtagtcat tttagagggc 901 atggtggaag ccactgccat gactacccag tgccgcagct catatctggc aaatgaaatc 961 ctctggggcc accgctatga gcccgtactc tttgaagaaa aaaactacta caaagtggac 1021 tattcaaggt tccacaaaac atacgaagtg cccaacacac ccatctgcag tgccagagac 1081 ttagcagaaa agaaatacat tctctcgaac gcaaactcct tttgctacga gaacgaagtg 1141 gccctcacca gcaaggagga ggacgagatc gacacggggg tgcccgagag cacaagcaca 1201 gacacccacc ccgacatgga ccaccacaac caggcaggag tgcccctaga gccacggccg 1261 ctgcggcgtg agtcggaaat atga 45. chicken Kir 2.2- cDNA 1 atgactgcag gcagggtcaa cccttacagc atagtgtcct ccgaggaaga cggactgagg 61 ttgaccacca tgccagggat taacggcttt ggcaatggga aaatccacac caggaggaaa 121 tgcaggaaca ggtttgtaaa gaagaacggt cagtgcaatg tggagttcac caacatggat 181 gacaagccac agaggtacat tgcagacatg ttcaccacgt gcgttgacat ccgttggagg 241 tatatgctct cgctcttctc cctggcattt ctggtatcct ggttattgtt tgggctgatt 301 ttctggctaa ttgcactcat tcatggagat ctagaaaacc caggtggaga tgataccttc 361 aagccttgcg ttctgcagga caatggcttt gtggctgctt ttctgttctc catcgagacc 421 caaacgacta ttggttatgg cttccgctgt gtgacagagg agtgcccgct cgcagtcttc 481 atggtggtgg ttcagtccat cgtggggtgt ataatcgact ctttcatgat tggtgcaata 541 atggcaaaga tggccaggcc caaaaaaagg gcccagacat tgcttttcag ccataatgca 601 gtagtggcaa tgagagatgg aaaactctgc ctgatgtgga gagttgggaa tctccggaaa 661 agccacatag tagaagccca cgtacgagct caattaatta agcccagaat cacagaagaa 721 ggggagtaca tcccactcga ccaaatagac atcgacgtgg ggtttgacaa aggcttggac 781 cgaatcttct tggtgtcccc cattaccatt ctccatgaga tcaacgaaga cagcccgctg 841 ttcgggatca gccgccagga cttggagacg gatgactttg agattgtggt catcctcgaa 901 ggcatggtag aagccaccgc gatgacgaca caagctcgga gctcctacct ggccagcgag 961 atcctgtggg gccaccgctt cgagcccgtc ttgttcgagg agaaaaacca gtacaaagta 1021 gactattccc acttccacaa aacatacgag gtcccgtcca caccccgctg cagcgccaag 1081 gacttggtgg agaacaaatt cctgctgccc agcaccaact ccttctgcta cgagaatgag 1141 ctggccttca tgagccgcga tgaggatgag gaggatgatg acagcagggg tttggacgac 1201 ctgagcccag acaacaggca cgagttcgac cggcttcagg caacgatagc gttggatcag 1261 aggtcatacc ggagggagtc agaaatatga 46. human Kir 1.1- turret region HKDLPEFHPSANHTPCVENING 47. human Kir 1.2- turret region
HGDLLELDPPANHTPCVVQVHT 48. human Kir 2.1- turret region HGDLDASKEGKACVSEVNS 49. human Kir 2.2- turret region HGDLEPAEGRGRTPCVMQVHG 50. human Kir 2.3- turret region HGDLEASPGVPAAGGPAAGGGGAAPVAPKPCIMHVNG 51. human Kir 2.4- turret region HGDLAAPPPPAPCFSHVAS 52. human Kir 3.1- turret region RGDLNKAHVGNYTPCVANVYN 53. human Kir 3.4- turret region RGDLDHVGDQEWIPCVENLSG 54. human Kir 6.1- turret region HGDIYAYMEKSGMEKSGLESTVCVTNVRS 55. human Kir 6.2- turret region HGDLAPSEGTAEPCVTSIHS 56. human Kir 7.1- turret region NGDLELDHDAPPENHTICVKYITS 57. chicken Kir 2.1- turret region HGDLENQENNKPCVSQVSS 58. chicken Kir 2.2- turret region HGDLENPGGDDTFKPCVLQVNG 59. Kir Bac 1.1- turret region SPARKPPRGGRRIWSGTREVIAYGMPASVWRDLYYWALKVSWPVFFASLAALF VVNNTLFALLYQLGDAPIANQSPPGFVGAFFFSVETLATVGYGDMHPQTVYAH AIATLEIFVGMSGIALSTGLVFARFARPRAKIMFARHAIVRPFNGRMTLMVRA ANARQNVIAEARAKMRLMRREHSSEGYSLMKIHDLKLVRNEHPIFLLGWNMMH VIDESSPLFGETPESLAEGRAMLLVMIEGSDETTAQVMQARHAWEHDDIRWHH RYVDLMSDVDGMTHIDYTRFNDTEPVEPPGAAPDAQAFAAKPGE 60. KcsA- turret region MAPMLSGLLARLVKLLLGRHGSALHWRAAGAATVLLVIVLLAGSYLAVLAERG APGAQLITYPRALWWSVETATTVGYGDLYPVTLWGRCVAVVVMVAGITSFGLV TAALATWFVGREQERRGH 61. rKv1.2- turret region MRELGLLIFFLFIGVILFSSAVYFAEADERDSQFPSIPDAFWWAVVSMTTVGY GDMVPTTIGGKIVGSLCAIAGVLTIALPVPVIVSNFNYFYHRETEGE
Sequence CWU
1
1
61111PRTHomo sapiens 1Pro Glu Phe His Pro Ser Ala Asn His Thr Pro 1
5 10 211PRTHomo sapiens 2Leu Glu Leu Asp
Pro Pro Ala Asn His Thr Pro 1 5 10
38PRTHomo sapiens 3Asp Ala Ser Lys Glu Gly Lys Ala 1 5
410PRTHomo sapiens 4Glu Pro Ala Glu Gly Arg Gly Arg Thr Pro 1
5 10 526PRTHomo sapiens 5Glu Ala Ser Pro
Gly Val Pro Ala Ala Gly Gly Pro Ala Ala Gly Gly 1 5
10 15 Gly Gly Ala Ala Pro Val Ala Pro Lys
Pro 20 25 68PRTHomo sapiens 6Ala Ala
Pro Pro Pro Pro Ala Pro 1 5 710PRTHomo
sapiens 7Asn Lys Ala His Val Gly Asn Tyr Thr Pro 1 5
10 810PRTHomo sapiens 8Asp His Val Gly Asp Gln Glu Trp Ile
Pro 1 5 10 918PRTHomo sapiens 9Tyr Ala
Tyr Met Glu Lys Ser Gly Met Glu Lys Ser Gly Leu Glu Ser 1 5
10 15 Thr Val 109PRTHomo sapiens
10Ala Pro Ser Glu Gly Thr Ala Glu Pro 1 5
1113PRTHomo sapiens 11Glu Leu Asp His Asp Ala Pro Pro Glu Asn His Thr Ile
1 5 10 128PRTGallus gallus
12Glu Asn Gln Glu Asn Asn Lys Pro 1 5
1311PRTGallus gallus 13Glu Asn Pro Gly Gly Asp Asp Thr Phe Lys Pro 1
5 10 14391PRTHomo sapiens 14Met Asn Ala
Ser Ser Arg Asn Val Phe Asp Thr Leu Ile Arg Val Leu 1 5
10 15 Thr Glu Ser Met Phe Lys His Leu
Arg Lys Trp Val Val Thr Arg Phe 20 25
30 Phe Gly His Ser Arg Gln Arg Ala Arg Leu Val Ser Lys
Asp Gly Arg 35 40 45
Cys Asn Ile Glu Phe Gly Asn Val Glu Ala Gln Ser Arg Phe Ile Phe 50
55 60 Phe Val Asp Ile
Trp Thr Thr Val Leu Asp Leu Lys Trp Arg Tyr Lys 65 70
75 80 Met Thr Ile Phe Ile Thr Ala Phe Leu
Gly Ser Trp Phe Phe Phe Gly 85 90
95 Leu Leu Trp Tyr Ala Val Ala Tyr Ile His Lys Asp Leu Pro
Glu Phe 100 105 110
His Pro Ser Ala Asn His Thr Pro Cys Val Glu Asn Ile Asn Gly Leu
115 120 125 Thr Ser Ala Phe
Leu Phe Ser Leu Glu Thr Gln Val Thr Ile Gly Tyr 130
135 140 Gly Phe Arg Cys Val Thr Glu Gln
Cys Ala Thr Ala Ile Phe Leu Leu 145 150
155 160 Ile Phe Gln Ser Ile Leu Gly Val Ile Ile Asn Ser
Phe Met Cys Gly 165 170
175 Ala Ile Leu Ala Lys Ile Ser Arg Pro Lys Lys Arg Ala Lys Thr Ile
180 185 190 Thr Phe Ser
Lys Asn Ala Val Ile Ser Lys Arg Gly Gly Lys Leu Cys 195
200 205 Leu Leu Ile Arg Val Ala Asn Leu
Arg Lys Ser Leu Leu Ile Gly Ser 210 215
220 His Ile Tyr Gly Lys Leu Leu Lys Thr Thr Val Thr Pro
Glu Gly Glu 225 230 235
240 Thr Ile Ile Leu Asp Gln Ile Asn Ile Asn Phe Val Val Asp Ala Gly
245 250 255 Asn Glu Asn Leu
Phe Phe Ile Ser Pro Leu Thr Ile Tyr His Val Ile 260
265 270 Asp His Asn Ser Pro Phe Phe His Met
Ala Ala Glu Thr Leu Leu Gln 275 280
285 Gln Asp Phe Glu Leu Val Val Phe Leu Asp Gly Thr Val Glu
Ser Thr 290 295 300
Ser Ala Thr Cys Gln Val Arg Thr Ser Tyr Val Pro Glu Glu Val Leu 305
310 315 320 Trp Gly Tyr Arg Phe
Ala Pro Ile Val Ser Lys Thr Lys Glu Gly Lys 325
330 335 Tyr Arg Val Asp Phe His Asn Phe Ser Lys
Thr Val Glu Val Glu Thr 340 345
350 Pro His Cys Ala Met Cys Leu Tyr Asn Glu Lys Asp Val Arg Ala
Arg 355 360 365 Met
Lys Arg Gly Tyr Asp Asn Pro Asn Phe Ile Leu Ser Glu Val Asn 370
375 380 Glu Thr Asp Asp Thr Lys
Met 385 390 15379PRTHomo sapiens 15Met Thr Ser Val
Ala Lys Val Tyr Tyr Ser Gln Thr Thr Gln Thr Glu 1 5
10 15 Ser Arg Pro Leu Met Gly Pro Gly Ile
Arg Arg Arg Arg Val Leu Thr 20 25
30 Lys Asp Gly Arg Ser Asn Val Arg Met Glu His Ile Ala Asp
Lys Arg 35 40 45
Phe Leu Tyr Leu Lys Asp Leu Trp Thr Thr Phe Ile Asp Met Gln Trp 50
55 60 Arg Tyr Lys Leu Leu
Leu Phe Ser Ala Thr Phe Ala Gly Thr Trp Phe 65 70
75 80 Leu Phe Gly Val Val Trp Tyr Leu Val Ala
Val Ala His Gly Asp Leu 85 90
95 Leu Glu Leu Asp Pro Pro Ala Asn His Thr Pro Cys Val Val Gln
Val 100 105 110 His
Thr Leu Thr Gly Ala Phe Leu Phe Ser Leu Glu Ser Gln Thr Thr 115
120 125 Ile Gly Tyr Gly Phe Arg
Tyr Ile Ser Glu Glu Cys Pro Leu Ala Ile 130 135
140 Val Leu Leu Ile Ala Gln Leu Val Leu Thr Thr
Ile Leu Glu Ile Phe 145 150 155
160 Ile Thr Gly Thr Phe Leu Ala Lys Ile Ala Arg Pro Lys Lys Arg Ala
165 170 175 Glu Thr
Ile Arg Phe Ser Gln His Ala Val Val Ala Ser His Asn Gly 180
185 190 Lys Pro Cys Leu Met Ile Arg
Val Ala Asn Met Arg Lys Ser Leu Leu 195 200
205 Ile Gly Cys Gln Val Thr Gly Lys Leu Leu Gln Thr
His Gln Thr Lys 210 215 220
Glu Gly Glu Asn Ile Arg Leu Asn Gln Val Asn Val Thr Phe Gln Val 225
230 235 240 Asp Thr Ala
Ser Asp Ser Pro Phe Leu Ile Leu Pro Leu Thr Phe Tyr 245
250 255 His Val Val Asp Glu Thr Ser Pro
Leu Lys Asp Leu Pro Leu Arg Ser 260 265
270 Gly Glu Gly Asp Phe Glu Leu Val Leu Ile Leu Ser Gly
Thr Val Glu 275 280 285
Ser Thr Ser Ala Thr Cys Gln Val Arg Thr Ser Tyr Leu Pro Glu Glu 290
295 300 Ile Leu Trp Gly
Tyr Glu Phe Thr Pro Ala Ile Ser Leu Ser Ala Ser 305 310
315 320 Gly Lys Tyr Ile Ala Asp Phe Ser Leu
Phe Asp Gln Val Val Lys Val 325 330
335 Ala Ser Pro Ser Gly Leu Arg Asp Ser Thr Val Arg Tyr Gly
Asp Pro 340 345 350
Glu Lys Leu Lys Leu Glu Glu Ser Leu Arg Glu Gln Ala Glu Lys Glu
355 360 365 Gly Ser Ala Leu
Ser Val Arg Ile Ser Asn Val 370 375
16427PRTHomo sapiens 16Met Gly Ser Val Arg Thr Asn Arg Tyr Ser Ile Val
Ser Ser Glu Glu 1 5 10
15 Asp Gly Met Lys Leu Ala Thr Met Ala Val Ala Asn Gly Phe Gly Asn
20 25 30 Gly Lys Ser
Lys Val His Thr Arg Gln Gln Cys Arg Ser Arg Phe Val 35
40 45 Lys Lys Asp Gly His Cys Asn Val
Gln Phe Ile Asn Val Gly Glu Lys 50 55
60 Gly Gln Arg Tyr Leu Ala Asp Ile Phe Thr Thr Cys Val
Asp Ile Arg 65 70 75
80 Trp Arg Trp Met Leu Val Ile Phe Cys Leu Ala Phe Val Leu Ser Trp
85 90 95 Leu Phe Phe Gly
Cys Val Phe Trp Leu Ile Ala Leu Leu His Gly Asp 100
105 110 Leu Asp Ala Ser Lys Glu Gly Lys Ala
Cys Val Ser Glu Val Asn Ser 115 120
125 Phe Thr Ala Ala Phe Leu Phe Ser Ile Glu Thr Gln Thr Thr
Ile Gly 130 135 140
Tyr Gly Phe Arg Cys Val Thr Asp Glu Cys Pro Ile Ala Val Phe Met 145
150 155 160 Val Val Phe Gln Ser
Ile Val Gly Cys Ile Ile Asp Ala Phe Ile Ile 165
170 175 Gly Ala Val Met Ala Lys Met Ala Lys Pro
Lys Lys Arg Asn Glu Thr 180 185
190 Leu Val Phe Ser His Asn Ala Val Ile Ala Met Arg Asp Gly Lys
Leu 195 200 205 Cys
Leu Met Trp Arg Val Gly Asn Leu Arg Lys Ser His Leu Val Glu 210
215 220 Ala His Val Arg Ala Gln
Leu Leu Lys Ser Arg Ile Thr Ser Glu Gly 225 230
235 240 Glu Tyr Ile Pro Leu Asp Gln Ile Asp Ile Asn
Val Gly Phe Asp Ser 245 250
255 Gly Ile Asp Arg Ile Phe Leu Val Ser Pro Ile Thr Ile Val His Glu
260 265 270 Ile Asp
Glu Asp Ser Pro Leu Tyr Asp Leu Ser Lys Gln Asp Ile Asp 275
280 285 Asn Ala Asp Phe Glu Ile Val
Val Ile Leu Glu Gly Met Val Glu Ala 290 295
300 Thr Ala Met Thr Thr Gln Cys Arg Ser Ser Tyr Leu
Ala Asn Glu Ile 305 310 315
320 Leu Trp Gly His Arg Tyr Glu Pro Val Leu Phe Glu Glu Lys His Tyr
325 330 335 Tyr Lys Val
Asp Tyr Ser Arg Phe His Lys Thr Tyr Glu Val Pro Asn 340
345 350 Thr Pro Leu Cys Ser Ala Arg Asp
Leu Ala Glu Lys Lys Tyr Ile Leu 355 360
365 Ser Asn Ala Asn Ser Phe Cys Tyr Glu Asn Glu Val Ala
Leu Thr Ser 370 375 380
Lys Glu Glu Asp Asp Ser Glu Asn Gly Val Pro Glu Ser Thr Ser Thr 385
390 395 400 Asp Thr Pro Pro
Asp Ile Asp Leu His Asn Gln Ala Ser Val Pro Leu 405
410 415 Glu Pro Arg Pro Leu Arg Arg Glu Ser
Glu Ile 420 425 17433PRTHomo sapiens
17Met Thr Ala Ala Ser Arg Ala Asn Pro Tyr Ser Ile Val Ser Ser Glu 1
5 10 15 Glu Asp Gly Leu
His Leu Val Thr Met Ser Gly Ala Asn Gly Phe Gly 20
25 30 Asn Gly Lys Val His Thr Arg Arg Arg
Cys Arg Asn Arg Phe Val Lys 35 40
45 Lys Asn Gly Gln Cys Asn Ile Glu Phe Ala Asn Met Asp Glu
Lys Ser 50 55 60
Gln Arg Tyr Leu Ala Asp Met Phe Thr Thr Cys Val Asp Ile Arg Trp 65
70 75 80 Arg Tyr Met Leu Leu
Ile Phe Ser Leu Ala Phe Leu Ala Ser Trp Leu 85
90 95 Leu Phe Gly Ile Ile Phe Trp Val Ile Ala
Val Ala His Gly Asp Leu 100 105
110 Glu Pro Ala Glu Gly Arg Gly Arg Thr Pro Cys Val Met Gln Val
His 115 120 125 Gly
Phe Met Ala Ala Phe Leu Phe Ser Ile Glu Thr Gln Thr Thr Ile 130
135 140 Gly Tyr Gly Leu Arg Cys
Val Thr Glu Glu Cys Pro Val Ala Val Phe 145 150
155 160 Met Val Val Ala Gln Ser Ile Val Gly Cys Ile
Ile Asp Ser Phe Met 165 170
175 Ile Gly Ala Ile Met Ala Lys Met Ala Arg Pro Lys Lys Arg Ala Gln
180 185 190 Thr Leu
Leu Phe Ser His Asn Ala Val Val Ala Leu Arg Asp Gly Lys 195
200 205 Leu Cys Leu Met Trp Arg Val
Gly Asn Leu Arg Lys Ser His Ile Val 210 215
220 Glu Ala His Val Arg Ala Gln Leu Ile Lys Pro Arg
Val Thr Glu Glu 225 230 235
240 Gly Glu Tyr Ile Pro Leu Asp Gln Ile Asp Ile Asp Val Gly Phe Asp
245 250 255 Lys Gly Leu
Asp Arg Ile Phe Leu Val Ser Pro Ile Thr Ile Leu His 260
265 270 Glu Ile Asp Glu Ala Ser Pro Leu
Phe Gly Ile Ser Arg Gln Asp Leu 275 280
285 Glu Thr Asp Asp Phe Glu Ile Val Val Ile Leu Glu Gly
Met Val Glu 290 295 300
Ala Thr Ala Met Thr Thr Gln Ala Arg Ser Ser Tyr Leu Ala Asn Glu 305
310 315 320 Ile Leu Trp Gly
His Arg Phe Glu Pro Val Leu Phe Glu Glu Lys Asn 325
330 335 Gln Tyr Lys Ile Asp Tyr Ser His Phe
His Lys Thr Tyr Glu Val Pro 340 345
350 Ser Thr Pro Arg Cys Ser Ala Lys Asp Leu Val Glu Asn Lys
Phe Leu 355 360 365
Leu Pro Ser Ala Asn Ser Phe Cys Tyr Glu Asn Glu Leu Ala Phe Leu 370
375 380 Ser Arg Asp Glu Glu
Asp Glu Ala Asp Gly Asp Gln Asp Gly Arg Ser 385 390
395 400 Arg Asp Gly Leu Ser Pro Gln Ala Arg His
Asp Phe Asp Arg Leu Gln 405 410
415 Ala Gly Gly Gly Val Leu Glu Gln Arg Pro Tyr Arg Arg Glu Ser
Glu 420 425 430 Ile
18445PRTHomo sapiens 18Met His Gly His Ser Arg Asn Gly Gln Ala His Val
Pro Arg Arg Lys 1 5 10
15 Arg Arg Asn Arg Phe Val Lys Lys Asn Gly Gln Cys Asn Val Tyr Phe
20 25 30 Ala Asn Leu
Ser Asn Lys Ser Gln Arg Tyr Met Ala Asp Ile Phe Thr 35
40 45 Thr Cys Val Asp Thr Arg Trp Arg
Tyr Met Leu Met Ile Phe Ser Ala 50 55
60 Ala Phe Leu Val Ser Trp Leu Phe Phe Gly Leu Leu Phe
Trp Cys Ile 65 70 75
80 Ala Phe Phe His Gly Asp Leu Glu Ala Ser Pro Gly Val Pro Ala Ala
85 90 95 Gly Gly Pro Ala
Ala Gly Gly Gly Gly Ala Ala Pro Val Ala Pro Lys 100
105 110 Pro Cys Ile Met His Val Asn Gly Phe
Leu Gly Ala Phe Leu Phe Ser 115 120
125 Val Glu Thr Gln Thr Thr Ile Gly Tyr Gly Phe Arg Cys Val
Thr Glu 130 135 140
Glu Cys Pro Leu Ala Val Ile Ala Val Val Val Gln Ser Ile Val Gly 145
150 155 160 Cys Val Ile Asp Ser
Phe Met Ile Gly Thr Ile Met Ala Lys Met Ala 165
170 175 Arg Pro Lys Lys Arg Ala Gln Thr Leu Leu
Phe Ser His His Ala Val 180 185
190 Ile Ser Val Arg Asp Gly Lys Leu Cys Leu Met Trp Arg Val Gly
Asn 195 200 205 Leu
Arg Lys Ser His Ile Val Glu Ala His Val Arg Ala Gln Leu Ile 210
215 220 Lys Pro Tyr Met Thr Gln
Glu Gly Glu Tyr Leu Pro Leu Asp Gln Arg 225 230
235 240 Asp Leu Asn Val Gly Tyr Asp Ile Gly Leu Asp
Arg Ile Phe Leu Val 245 250
255 Ser Pro Ile Ile Ile Val His Glu Ile Asp Glu Asp Ser Pro Leu Tyr
260 265 270 Gly Met
Gly Lys Glu Glu Leu Glu Ser Glu Asp Phe Glu Ile Val Val 275
280 285 Ile Leu Glu Gly Met Val Glu
Ala Thr Ala Met Thr Thr Gln Ala Arg 290 295
300 Ser Ser Tyr Leu Ala Ser Glu Ile Leu Trp Gly His
Arg Phe Glu Pro 305 310 315
320 Val Val Phe Glu Glu Lys Ser His Tyr Lys Val Asp Tyr Ser Arg Phe
325 330 335 His Lys Thr
Tyr Glu Val Ala Gly Thr Pro Cys Cys Ser Ala Arg Glu 340
345 350 Leu Gln Glu Ser Lys Ile Thr Val
Leu Pro Ala Pro Pro Pro Pro Pro 355 360
365 Ser Ala Phe Cys Tyr Glu Asn Glu Leu Ala Leu Met Ser
Gln Glu Glu 370 375 380
Glu Glu Met Glu Glu Glu Ala Ala Ala Ala Ala Ala Val Ala Ala Gly 385
390 395 400 Leu Gly Leu Glu
Ala Gly Ser Lys Glu Glu Ala Gly Ile Ile Arg Met 405
410 415 Leu Glu Phe Gly Ser His Leu Asp Leu
Glu Arg Met Gln Ala Ser Leu 420 425
430 Pro Leu Asp Asn Ile Ser Tyr Arg Arg Glu Ser Ala Ile
435 440 445 19436PRTHomo sapiens
19Met Gly Leu Ala Arg Ala Leu Arg Arg Leu Ser Gly Ala Leu Asp Ser 1
5 10 15 Gly Asp Ser Arg
Ala Gly Asp Glu Glu Glu Ala Gly Pro Gly Leu Cys 20
25 30 Arg Asn Gly Trp Ala Pro Ala Pro Val
Gln Ser Pro Val Gly Arg Arg 35 40
45 Arg Gly Arg Phe Val Lys Lys Asp Gly His Cys Asn Val Arg
Phe Val 50 55 60
Asn Leu Gly Gly Gln Gly Ala Arg Tyr Leu Ser Asp Leu Phe Thr Thr 65
70 75 80 Cys Val Asp Val Arg
Trp Arg Trp Met Cys Leu Leu Phe Ser Cys Ser 85
90 95 Phe Leu Ala Ser Trp Leu Leu Phe Gly Leu
Ala Phe Trp Leu Ile Ala 100 105
110 Ser Leu His Gly Asp Leu Ala Ala Pro Pro Pro Pro Ala Pro Cys
Phe 115 120 125 Ser
His Val Ala Ser Phe Leu Ala Ala Phe Leu Phe Ala Leu Glu Thr 130
135 140 Gln Thr Ser Ile Gly Tyr
Gly Val Arg Ser Val Thr Glu Glu Cys Pro 145 150
155 160 Ala Ala Val Ala Ala Val Val Leu Gln Cys Ile
Ala Gly Cys Val Leu 165 170
175 Asp Ala Phe Val Val Gly Ala Val Met Ala Lys Met Ala Lys Pro Lys
180 185 190 Lys Arg
Asn Glu Thr Leu Val Phe Ser Glu Asn Ala Val Val Ala Leu 195
200 205 Arg Asp His Arg Leu Cys Leu
Met Trp Arg Val Gly Asn Leu Arg Arg 210 215
220 Ser His Leu Val Glu Ala His Val Arg Ala Gln Leu
Leu Gln Pro Arg 225 230 235
240 Val Thr Pro Glu Gly Glu Tyr Ile Pro Leu Asp His Gln Asp Val Asp
245 250 255 Val Gly Phe
Asp Gly Gly Thr Asp Arg Ile Phe Leu Val Ser Pro Ile 260
265 270 Thr Ile Val His Glu Ile Asp Ser
Ala Ser Pro Leu Tyr Glu Leu Gly 275 280
285 Arg Ala Glu Leu Ala Arg Ala Asp Phe Glu Leu Val Val
Ile Leu Glu 290 295 300
Gly Met Val Glu Ala Thr Ala Met Thr Thr Gln Cys Arg Ser Ser Tyr 305
310 315 320 Leu Pro Gly Glu
Leu Leu Trp Gly His Arg Phe Glu Pro Val Leu Phe 325
330 335 Gln Arg Gly Ser Gln Tyr Glu Val Asp
Tyr Arg His Phe His Arg Thr 340 345
350 Tyr Glu Val Pro Gly Thr Pro Val Cys Ser Ala Lys Glu Leu
Asp Glu 355 360 365
Arg Ala Glu Gln Ala Ser His Ser Leu Lys Ser Ser Phe Pro Gly Ser 370
375 380 Leu Thr Ala Phe Cys
Tyr Glu Asn Glu Leu Ala Leu Ser Cys Cys Gln 385 390
395 400 Glu Glu Asp Glu Asp Asp Glu Thr Glu Glu
Gly Asn Gly Val Glu Thr 405 410
415 Glu Asp Gly Ala Ala Ser Pro Arg Val Leu Thr Pro Thr Leu Ala
Leu 420 425 430 Thr
Leu Pro Pro 435 20501PRTHomo sapiens 20Met Ser Ala Leu Arg
Arg Lys Phe Gly Asp Asp Tyr Gln Val Val Thr 1 5
10 15 Thr Ser Ser Ser Gly Ser Gly Leu Gln Pro
Gln Gly Pro Gly Gln Asp 20 25
30 Pro Gln Gln Gln Leu Val Pro Lys Lys Lys Arg Gln Arg Phe Val
Asp 35 40 45 Lys
Asn Gly Arg Cys Asn Val Gln His Gly Asn Leu Gly Ser Glu Thr 50
55 60 Ser Arg Tyr Leu Ser Asp
Leu Phe Thr Thr Leu Val Asp Leu Lys Trp 65 70
75 80 Arg Trp Asn Leu Phe Ile Phe Ile Leu Thr Tyr
Thr Val Ala Trp Leu 85 90
95 Phe Met Ala Ser Met Trp Trp Val Ile Ala Tyr Thr Arg Gly Asp Leu
100 105 110 Asn Lys
Ala His Val Gly Asn Tyr Thr Pro Cys Val Ala Asn Val Tyr 115
120 125 Asn Phe Pro Ser Ala Phe Leu
Phe Phe Ile Glu Thr Glu Ala Thr Ile 130 135
140 Gly Tyr Gly Tyr Arg Tyr Ile Thr Asp Lys Cys Pro
Glu Gly Ile Ile 145 150 155
160 Leu Phe Leu Phe Gln Ser Ile Leu Gly Ser Ile Val Asp Ala Phe Leu
165 170 175 Ile Gly Cys
Met Phe Ile Lys Met Ser Gln Pro Lys Lys Arg Ala Glu 180
185 190 Thr Leu Met Phe Ser Glu His Ala
Val Ile Ser Met Arg Asp Gly Lys 195 200
205 Leu Thr Leu Met Phe Arg Val Gly Asn Leu Arg Asn Ser
His Met Val 210 215 220
Ser Ala Gln Ile Arg Cys Lys Leu Leu Lys Ser Arg Gln Thr Pro Glu 225
230 235 240 Gly Glu Phe Leu
Pro Leu Asp Gln Leu Glu Leu Asp Val Gly Phe Ser 245
250 255 Thr Gly Ala Asp Gln Leu Phe Leu Val
Ser Pro Leu Thr Ile Cys His 260 265
270 Val Ile Asp Ala Lys Ser Pro Phe Tyr Asp Leu Ser Gln Arg
Ser Met 275 280 285
Gln Thr Glu Gln Phe Glu Ile Val Val Ile Leu Glu Gly Ile Val Glu 290
295 300 Thr Thr Gly Met Thr
Cys Gln Ala Arg Thr Ser Tyr Thr Glu Asp Glu 305 310
315 320 Val Leu Trp Gly His Arg Phe Phe Pro Val
Ile Ser Leu Glu Glu Gly 325 330
335 Phe Phe Lys Val Asp Tyr Ser Gln Phe His Ala Thr Phe Glu Val
Pro 340 345 350 Thr
Pro Pro Tyr Ser Val Lys Glu Gln Glu Glu Met Leu Leu Met Ser 355
360 365 Ser Pro Leu Ile Ala Pro
Ala Ile Thr Asn Ser Lys Glu Arg His Asn 370 375
380 Ser Val Glu Cys Leu Asp Gly Leu Asp Asp Ile
Thr Thr Lys Leu Pro 385 390 395
400 Ser Lys Leu Gln Lys Ile Thr Gly Arg Glu Asp Phe Pro Lys Lys Leu
405 410 415 Leu Arg
Met Ser Ser Thr Thr Ser Glu Lys Ala Tyr Ser Leu Gly Asp 420
425 430 Leu Pro Met Lys Leu Gln Arg
Ile Ser Ser Val Pro Gly Asn Ser Glu 435 440
445 Glu Lys Leu Val Ser Lys Thr Thr Lys Met Leu Ser
Asp Pro Met Ser 450 455 460
Gln Ser Val Ala Asp Leu Pro Pro Lys Leu Gln Lys Met Ala Gly Gly 465
470 475 480 Ala Ala Arg
Met Glu Gly Asn Leu Pro Ala Lys Leu Arg Lys Met Asn 485
490 495 Ser Asp Arg Phe Thr
500 21419PRTHomo sapiens 21Met Ala Gly Asp Ser Arg Asn Ala Met Asn
Gln Asp Met Glu Ile Gly 1 5 10
15 Val Thr Pro Trp Asp Pro Lys Lys Ile Pro Lys Gln Ala Arg Asp
Tyr 20 25 30 Val
Pro Ile Ala Thr Asp Arg Thr Arg Leu Leu Ala Glu Gly Lys Lys 35
40 45 Pro Arg Gln Arg Tyr Met
Glu Lys Ser Gly Lys Cys Asn Val His His 50 55
60 Gly Asn Val Gln Glu Thr Tyr Arg Tyr Leu Ser
Asp Leu Phe Thr Thr 65 70 75
80 Leu Val Asp Leu Lys Trp Arg Phe Asn Leu Leu Val Phe Thr Met Val
85 90 95 Tyr Thr
Val Thr Trp Leu Phe Phe Gly Phe Ile Trp Trp Leu Ile Ala 100
105 110 Tyr Ile Arg Gly Asp Leu Asp
His Val Gly Asp Gln Glu Trp Ile Pro 115 120
125 Cys Val Glu Asn Leu Ser Gly Phe Val Ser Ala Phe
Leu Phe Ser Ile 130 135 140
Glu Thr Glu Thr Thr Ile Gly Tyr Gly Phe Arg Val Ile Thr Glu Lys 145
150 155 160 Cys Pro Glu
Gly Ile Ile Leu Leu Leu Val Gln Ala Ile Leu Gly Ser 165
170 175 Ile Val Asn Ala Phe Met Val Gly
Cys Met Phe Val Lys Ile Ser Gln 180 185
190 Pro Lys Lys Arg Ala Glu Thr Leu Met Phe Ser Asn Asn
Ala Val Ile 195 200 205
Ser Met Arg Asp Glu Lys Leu Cys Leu Met Phe Arg Val Gly Asp Leu 210
215 220 Arg Asn Ser His
Ile Val Glu Ala Ser Ile Arg Ala Lys Leu Ile Lys 225 230
235 240 Ser Arg Gln Thr Lys Glu Gly Glu Phe
Ile Pro Leu Asn Gln Thr Asp 245 250
255 Ile Asn Val Gly Phe Asp Thr Gly Asp Asp Arg Leu Phe Leu
Val Ser 260 265 270
Pro Leu Ile Ile Ser His Glu Ile Asn Gln Lys Ser Pro Phe Trp Glu
275 280 285 Met Ser Gln Ala
Gln Leu His Gln Glu Glu Phe Glu Val Val Val Ile 290
295 300 Leu Glu Gly Met Val Glu Ala Thr
Gly Met Thr Cys Gln Ala Arg Ser 305 310
315 320 Ser Tyr Met Asp Thr Glu Val Leu Trp Gly His Arg
Phe Thr Pro Val 325 330
335 Leu Thr Leu Glu Lys Gly Phe Tyr Glu Val Asp Tyr Asn Thr Phe His
340 345 350 Asp Thr Tyr
Glu Thr Asn Thr Pro Ser Cys Cys Ala Lys Glu Leu Ala 355
360 365 Glu Met Lys Arg Glu Gly Arg Leu
Leu Gln Tyr Leu Pro Ser Pro Pro 370 375
380 Leu Leu Gly Gly Cys Ala Glu Ala Gly Leu Asp Ala Glu
Ala Glu Gln 385 390 395
400 Asn Glu Glu Asp Glu Pro Lys Gly Leu Gly Gly Ser Arg Glu Ala Arg
405 410 415 Gly Ser Val
22379PRTHomo sapiens 22Met Thr Ser Val Ala Lys Val Tyr Tyr Ser Gln Thr
Thr Gln Thr Glu 1 5 10
15 Ser Arg Pro Leu Met Gly Pro Gly Ile Arg Arg Arg Arg Val Leu Thr
20 25 30 Lys Asp Gly
Arg Ser Asn Val Arg Met Glu His Ile Ala Asp Lys Arg 35
40 45 Phe Leu Tyr Leu Lys Asp Leu Trp
Thr Thr Phe Ile Asp Met Gln Trp 50 55
60 Arg Tyr Lys Leu Leu Leu Phe Ser Ala Thr Phe Ala Gly
Thr Trp Phe 65 70 75
80 Leu Phe Gly Val Val Trp Tyr Leu Val Ala Val Ala His Gly Asp Leu
85 90 95 Leu Glu Leu Asp
Pro Pro Ala Asn His Thr Pro Cys Val Val Gln Val 100
105 110 His Thr Leu Thr Gly Ala Phe Leu Phe
Ser Leu Glu Ser Gln Thr Thr 115 120
125 Ile Gly Tyr Gly Phe Arg Tyr Ile Ser Glu Glu Cys Pro Leu
Ala Ile 130 135 140
Val Leu Leu Ile Ala Gln Leu Val Leu Thr Thr Ile Leu Glu Ile Phe 145
150 155 160 Ile Thr Gly Thr Phe
Leu Ala Lys Ile Ala Arg Pro Lys Lys Arg Ala 165
170 175 Glu Thr Ile Arg Phe Ser Gln His Ala Val
Val Ala Ser His Asn Gly 180 185
190 Lys Pro Cys Leu Met Ile Arg Val Ala Asn Met Arg Lys Ser Leu
Leu 195 200 205 Ile
Gly Cys Gln Val Thr Gly Lys Leu Leu Gln Thr His Gln Thr Lys 210
215 220 Glu Gly Glu Asn Ile Arg
Leu Asn Gln Val Asn Val Thr Phe Gln Val 225 230
235 240 Asp Thr Ala Ser Asp Ser Pro Phe Leu Ile Leu
Pro Leu Thr Phe Tyr 245 250
255 His Val Val Asp Glu Thr Ser Pro Leu Lys Asp Leu Pro Leu Arg Ser
260 265 270 Gly Glu
Gly Asp Phe Glu Leu Val Leu Ile Leu Ser Gly Thr Val Glu 275
280 285 Ser Thr Ser Ala Thr Cys Gln
Val Arg Thr Ser Tyr Leu Pro Glu Glu 290 295
300 Ile Leu Trp Gly Tyr Glu Phe Thr Pro Ala Ile Ser
Leu Ser Ala Ser 305 310 315
320 Gly Lys Tyr Ile Ala Asp Phe Ser Leu Phe Asp Gln Val Val Lys Val
325 330 335 Ala Ser Pro
Ser Gly Leu Arg Asp Ser Thr Val Arg Tyr Gly Asp Pro 340
345 350 Glu Lys Leu Lys Leu Glu Glu Ser
Leu Arg Glu Gln Ala Glu Lys Glu 355 360
365 Gly Ser Ala Leu Ser Val Arg Ile Ser Asn Val 370
375 23375PRTHomo sapiens 23Met Asp Ala
Ile His Ile Gly Met Ser Ser Thr Pro Leu Val Lys His 1 5
10 15 Thr Ala Gly Ala Gly Leu Lys Ala
Asn Arg Pro Arg Val Met Ser Lys 20 25
30 Ser Gly His Ser Asn Val Arg Ile Asp Lys Val Asp Gly
Ile Tyr Leu 35 40 45
Leu Tyr Leu Gln Asp Leu Trp Thr Thr Val Ile Asp Met Lys Trp Arg 50
55 60 Tyr Lys Leu Thr
Leu Phe Ala Ala Thr Phe Val Met Thr Trp Phe Leu 65 70
75 80 Phe Gly Val Ile Tyr Tyr Ala Ile Ala
Phe Ile His Gly Asp Leu Glu 85 90
95 Pro Gly Glu Pro Ile Ser Asn His Thr Pro Cys Ile Met Lys
Val Asp 100 105 110
Ser Leu Thr Gly Ala Phe Leu Phe Ser Leu Glu Ser Gln Thr Thr Ile
115 120 125 Gly Tyr Gly Val
Arg Ser Ile Thr Glu Glu Cys Pro His Ala Ile Phe 130
135 140 Leu Leu Val Ala Gln Leu Val Ile
Thr Thr Leu Ile Glu Ile Phe Ile 145 150
155 160 Thr Gly Thr Phe Leu Ala Lys Ile Ala Arg Pro Lys
Lys Arg Ala Glu 165 170
175 Thr Ile Lys Phe Ser His Cys Ala Val Ile Thr Lys Gln Asn Gly Lys
180 185 190 Leu Cys Leu
Val Ile Gln Val Ala Asn Met Arg Lys Ser Leu Leu Ile 195
200 205 Gln Cys Gln Leu Ser Gly Lys Leu
Leu Gln Thr His Val Thr Lys Glu 210 215
220 Gly Glu Arg Ile Leu Leu Asn Gln Ala Thr Val Lys Phe
His Val Asp 225 230 235
240 Ser Ser Ser Glu Ser Pro Phe Leu Ile Leu Pro Met Thr Phe Tyr His
245 250 255 Val Leu Asp Glu
Thr Ser Pro Leu Arg Asp Leu Thr Pro Gln Asn Leu 260
265 270 Lys Glu Lys Glu Phe Glu Leu Val Val
Leu Leu Asn Ala Thr Val Glu 275 280
285 Ser Thr Ser Ala Val Cys Gln Ser Arg Thr Ser Tyr Ile Pro
Glu Glu 290 295 300
Ile Tyr Trp Gly Phe Glu Phe Val Pro Val Val Ser Leu Ser Lys Asn 305
310 315 320 Gly Lys Tyr Val Ala
Asp Phe Ser Gln Phe Glu Gln Ile Arg Lys Ser 325
330 335 Pro Asp Cys Thr Phe Tyr Cys Ala Asp Ser
Glu Lys Gln Gln Leu Glu 340 345
350 Glu Lys Tyr Arg Gln Glu Asp Gln Arg Glu Arg Glu Leu Arg Thr
Leu 355 360 365 Leu
Leu Gln Gln Ser Asn Val 370 375 24418PRTHomo sapiens
24Met Ser Tyr Tyr Gly Ser Ser Tyr His Ile Ile Asn Ala Asp Ala Lys 1
5 10 15 Tyr Pro Gly Tyr
Pro Pro Glu His Ile Ile Ala Glu Lys Arg Arg Ala 20
25 30 Arg Arg Arg Leu Leu His Lys Asp Gly
Ser Cys Asn Val Tyr Phe Lys 35 40
45 His Ile Phe Gly Glu Trp Gly Ser Tyr Val Val Asp Ile Phe
Thr Thr 50 55 60
Leu Val Asp Thr Lys Trp Arg His Met Phe Val Ile Phe Ser Leu Ser 65
70 75 80 Tyr Ile Leu Ser Trp
Leu Ile Phe Gly Ser Val Phe Trp Leu Ile Ala 85
90 95 Phe His His Gly Asp Leu Leu Asn Asp Pro
Asp Ile Thr Pro Cys Val 100 105
110 Asp Asn Val His Ser Phe Thr Gly Ala Phe Leu Phe Ser Leu Glu
Thr 115 120 125 Gln
Thr Thr Ile Gly Tyr Gly Tyr Arg Cys Val Thr Glu Glu Cys Ser 130
135 140 Val Ala Val Leu Met Val
Ile Leu Gln Ser Ile Leu Ser Cys Ile Ile 145 150
155 160 Asn Thr Phe Ile Ile Gly Ala Ala Leu Ala Lys
Met Ala Thr Ala Arg 165 170
175 Lys Arg Ala Gln Thr Ile Arg Phe Ser Tyr Phe Ala Leu Ile Gly Met
180 185 190 Arg Asp
Gly Lys Leu Cys Leu Met Trp Arg Ile Gly Asp Phe Arg Pro 195
200 205 Asn His Val Val Glu Gly Thr
Val Arg Ala Gln Leu Leu Arg Tyr Thr 210 215
220 Glu Asp Ser Glu Gly Arg Met Thr Met Ala Phe Lys
Asp Leu Lys Leu 225 230 235
240 Val Asn Asp Gln Ile Ile Leu Val Thr Pro Val Thr Ile Val His Glu
245 250 255 Ile Asp His
Glu Ser Pro Leu Tyr Ala Leu Asp Arg Lys Ala Val Ala 260
265 270 Lys Asp Asn Phe Glu Ile Leu Val
Thr Phe Ile Tyr Thr Gly Asp Ser 275 280
285 Thr Gly Thr Ser His Gln Ser Arg Ser Ser Tyr Val Pro
Arg Glu Ile 290 295 300
Leu Trp Gly His Arg Phe Asn Asp Val Leu Glu Val Lys Arg Lys Tyr 305
310 315 320 Tyr Lys Val Asn
Cys Leu Gln Phe Glu Gly Ser Val Glu Val Tyr Ala 325
330 335 Pro Phe Cys Ser Ala Lys Gln Leu Asp
Trp Lys Asp Gln Gln Leu His 340 345
350 Ile Glu Lys Ala Pro Pro Val Arg Glu Ser Cys Thr Ser Asp
Thr Lys 355 360 365
Ala Arg Arg Arg Ser Phe Ser Ala Val Ala Ile Val Ser Ser Cys Glu 370
375 380 Asn Pro Glu Glu Thr
Thr Thr Ser Ala Thr His Glu Tyr Arg Glu Thr 385 390
395 400 Pro Tyr Gln Lys Ala Leu Leu Thr Leu Asn
Arg Ile Ser Val Glu Ser 405 410
415 Gln Met 25424PRTHomo sapiens 25Met Leu Ala Arg Lys Ser Ile
Ile Pro Glu Glu Tyr Val Leu Ala Arg 1 5
10 15 Ile Ala Ala Glu Asn Leu Arg Lys Pro Arg Ile
Arg Asp Arg Leu Pro 20 25
30 Lys Ala Arg Phe Ile Ala Lys Ser Gly Ala Cys Asn Leu Ala His
Lys 35 40 45 Asn
Ile Arg Glu Gln Gly Arg Phe Leu Gln Asp Ile Phe Thr Thr Leu 50
55 60 Val Asp Leu Lys Trp Arg
His Thr Leu Val Ile Phe Thr Met Ser Phe 65 70
75 80 Leu Cys Ser Trp Leu Leu Phe Ala Ile Met Trp
Trp Leu Val Ala Phe 85 90
95 Ala His Gly Asp Ile Tyr Ala Tyr Met Glu Lys Ser Gly Met Glu Lys
100 105 110 Ser Gly
Leu Glu Ser Thr Val Cys Val Thr Asn Val Arg Ser Phe Thr 115
120 125 Ser Ala Phe Leu Phe Ser Ile
Glu Val Gln Val Thr Ile Gly Phe Gly 130 135
140 Gly Arg Met Met Thr Glu Glu Cys Pro Leu Ala Ile
Thr Val Leu Ile 145 150 155
160 Leu Gln Asn Ile Val Gly Leu Ile Ile Asn Ala Val Met Leu Gly Cys
165 170 175 Ile Phe Met
Lys Thr Ala Gln Ala His Arg Arg Ala Glu Thr Leu Ile 180
185 190 Phe Ser Arg His Ala Val Ile Ala
Val Arg Asn Gly Lys Leu Cys Phe 195 200
205 Met Phe Arg Val Gly Asp Leu Arg Lys Ser Met Ile Ile
Ser Ala Ser 210 215 220
Val Arg Ile Gln Val Val Lys Lys Thr Thr Thr Pro Glu Gly Glu Val 225
230 235 240 Val Pro Ile His
Gln Leu Asp Ile Pro Val Asp Asn Pro Ile Glu Ser 245
250 255 Asn Asn Ile Phe Leu Val Ala Pro Leu
Ile Ile Cys His Val Ile Asp 260 265
270 Lys Arg Ser Pro Leu Tyr Asp Ile Ser Ala Thr Asp Leu Ala
Asn Gln 275 280 285
Asp Leu Glu Val Ile Val Ile Leu Glu Gly Val Val Glu Thr Thr Gly 290
295 300 Ile Thr Thr Gln Ala
Arg Thr Ser Tyr Ile Ala Glu Glu Ile Gln Trp 305 310
315 320 Gly His Arg Phe Val Ser Ile Val Thr Glu
Glu Glu Gly Val Tyr Ser 325 330
335 Val Asp Tyr Ser Lys Phe Gly Asn Thr Val Lys Val Ala Ala Pro
Arg 340 345 350 Cys
Ser Ala Arg Glu Leu Asp Glu Lys Pro Ser Ile Leu Ile Gln Thr 355
360 365 Leu Gln Lys Ser Glu Leu
Ser His Gln Asn Ser Leu Arg Lys Arg Asn 370 375
380 Ser Met Arg Arg Asn Asn Ser Met Arg Arg Asn
Asn Ser Ile Arg Arg 385 390 395
400 Asn Asn Ser Ser Leu Met Val Pro Lys Val Gln Phe Met Thr Pro Glu
405 410 415 Gly Asn
Gln Asn Thr Ser Glu Ser 420 26390PRTHomo
sapiens 26Met Leu Ser Arg Lys Gly Ile Ile Pro Glu Glu Tyr Val Leu Thr Arg
1 5 10 15 Leu Ala
Glu Asp Pro Ala Lys Pro Arg Tyr Arg Ala Arg Gln Arg Arg 20
25 30 Ala Arg Phe Val Ser Lys Lys
Gly Asn Cys Asn Val Ala His Lys Asn 35 40
45 Ile Arg Glu Gln Gly Arg Phe Leu Gln Asp Val Phe
Thr Thr Leu Val 50 55 60
Asp Leu Lys Trp Pro His Thr Leu Leu Ile Phe Thr Met Ser Phe Leu 65
70 75 80 Cys Ser Trp
Leu Leu Phe Ala Met Ala Trp Trp Leu Ile Ala Phe Ala 85
90 95 His Gly Asp Leu Ala Pro Ser Glu
Gly Thr Ala Glu Pro Cys Val Thr 100 105
110 Ser Ile His Ser Phe Ser Ser Ala Phe Leu Phe Ser Ile
Glu Val Gln 115 120 125
Val Thr Ile Gly Phe Gly Gly Arg Met Val Thr Glu Glu Cys Pro Leu 130
135 140 Ala Ile Leu Ile
Leu Ile Val Gln Asn Ile Val Gly Leu Met Ile Asn 145 150
155 160 Ala Ile Met Leu Gly Cys Ile Phe Met
Lys Thr Ala Gln Ala His Arg 165 170
175 Arg Ala Glu Thr Leu Ile Phe Ser Lys His Ala Val Ile Ala
Leu Arg 180 185 190
His Gly Arg Leu Cys Phe Met Leu Arg Val Gly Asp Leu Arg Lys Ser
195 200 205 Met Ile Ile Ser
Ala Thr Ile His Met Gln Val Val Arg Lys Thr Thr 210
215 220 Ser Pro Glu Gly Glu Val Val Pro
Leu His Gln Val Asp Ile Pro Met 225 230
235 240 Glu Asn Gly Val Gly Gly Asn Ser Ile Phe Leu Val
Ala Pro Leu Ile 245 250
255 Ile Tyr His Val Ile Asp Ala Asn Ser Pro Leu Tyr Asp Leu Ala Pro
260 265 270 Ser Asp Leu
His His His Gln Asp Leu Glu Ile Ile Val Ile Leu Glu 275
280 285 Gly Val Val Glu Thr Thr Gly Ile
Thr Thr Gln Ala Arg Thr Ser Tyr 290 295
300 Leu Ala Asp Glu Ile Leu Trp Gly Gln Arg Phe Val Pro
Ile Val Ala 305 310 315
320 Glu Glu Asp Gly Arg Tyr Ser Val Asp Tyr Ser Lys Phe Gly Asn Thr
325 330 335 Val Lys Val Pro
Thr Pro Leu Cys Thr Ala Arg Gln Leu Asp Glu Asp 340
345 350 His Ser Leu Leu Glu Ala Leu Thr Leu
Ala Ser Ala Arg Gly Pro Leu 355 360
365 Arg Lys Arg Ser Val Pro Met Ala Lys Ala Lys Pro Lys Phe
Ser Ile 370 375 380
Ser Pro Asp Ser Leu Ser 385 390 27360PRTHomo sapiens
27Met Asp Ser Ser Asn Cys Lys Val Ile Ala Pro Leu Leu Ser Gln Arg 1
5 10 15 Tyr Arg Arg Met
Val Thr Lys Asp Gly His Ser Thr Leu Gln Met Asp 20
25 30 Gly Ala Gln Arg Gly Leu Ala Tyr Leu
Arg Asp Ala Trp Gly Ile Leu 35 40
45 Met Asp Met Arg Trp Arg Trp Met Met Leu Val Phe Ser Ala
Ser Phe 50 55 60
Val Val His Trp Leu Val Phe Ala Val Leu Trp Tyr Val Leu Ala Glu 65
70 75 80 Met Asn Gly Asp Leu
Glu Leu Asp His Asp Ala Pro Pro Glu Asn His 85
90 95 Thr Ile Cys Val Lys Tyr Ile Thr Ser Phe
Thr Ala Ala Phe Ser Phe 100 105
110 Ser Leu Glu Thr Gln Leu Thr Ile Gly Tyr Gly Thr Met Phe Pro
Ser 115 120 125 Gly
Asp Cys Pro Ser Ala Ile Ala Leu Leu Ala Ile Gln Met Leu Leu 130
135 140 Gly Leu Met Leu Glu Ala
Phe Ile Thr Gly Ala Phe Val Ala Lys Ile 145 150
155 160 Ala Arg Pro Lys Asn Arg Ala Phe Ser Ile Arg
Phe Thr Asp Thr Ala 165 170
175 Val Val Ala His Met Asp Gly Lys Pro Asn Leu Ile Phe Gln Val Ala
180 185 190 Asn Thr
Arg Pro Ser Pro Leu Thr Ser Val Arg Val Ser Ala Val Leu 195
200 205 Tyr Gln Glu Arg Glu Asn Gly
Lys Leu Tyr Gln Thr Ser Val Asp Phe 210 215
220 His Leu Asp Gly Ile Ser Ser Asp Glu Cys Pro Phe
Phe Ile Phe Pro 225 230 235
240 Leu Thr Tyr Tyr His Ser Ile Thr Pro Ser Ser Pro Leu Ala Thr Leu
245 250 255 Leu Gln His
Glu Asn Pro Ser His Phe Glu Leu Val Val Phe Leu Ser 260
265 270 Ala Met Gln Glu Gly Thr Gly Glu
Ile Cys Gln Arg Arg Thr Ser Tyr 275 280
285 Leu Pro Ser Glu Ile Met Leu His His Cys Phe Ala Ser
Leu Leu Thr 290 295 300
Arg Gly Ser Lys Gly Glu Tyr Gln Ile Lys Met Glu Asn Phe Asp Lys 305
310 315 320 Thr Val Pro Glu
Phe Pro Thr Pro Leu Val Ser Lys Ser Pro Asn Arg 325
330 335 Thr Asp Leu Asp Ile His Ile Asn Gly
Gln Ser Ile Asp Asn Phe Gln 340 345
350 Ile Ser Glu Thr Gly Leu Thr Glu 355
360 28427PRTGallus gallus 28Met Gly Ser Val Arg Thr Asn Arg Tyr Ser
Ile Val Ser Ser Glu Glu 1 5 10
15 Asp Gly Met Lys Leu Ala Thr Met Ala Val Ala Asn Gly Phe Gly
Asn 20 25 30 Gly
Lys Ser Lys Val His Thr Arg Gln Gln Cys Arg Ser Arg Phe Val 35
40 45 Lys Lys Asp Gly His Cys
Asn Val Gln Phe Ile Asn Val Gly Glu Lys 50 55
60 Gly Gln Arg Tyr Leu Ala Asp Ile Phe Thr Thr
Cys Val Asp Ile Arg 65 70 75
80 Trp Arg Trp Met Leu Val Ile Phe Cys Leu Thr Phe Ile Leu Ser Trp
85 90 95 Leu Phe
Phe Gly Cys Val Phe Trp Leu Ile Ala Leu Leu His Gly Asp 100
105 110 Leu Glu Asn Gln Glu Asn Asn
Lys Pro Cys Val Ser Gln Val Ser Ser 115 120
125 Phe Thr Ala Ala Phe Leu Phe Ser Ile Glu Thr Gln
Thr Thr Ile Gly 130 135 140
Tyr Gly Phe Arg Cys Val Thr Asp Glu Cys Pro Ile Ala Val Phe Met 145
150 155 160 Val Val Phe
Gln Ser Ile Val Gly Cys Ile Ile Asp Ala Phe Ile Ile 165
170 175 Gly Ala Val Met Ala Lys Met Ala
Lys Pro Lys Lys Arg Asn Glu Thr 180 185
190 Leu Val Phe Ser His Asn Ala Val Val Ala Met Arg Asp
Gly Lys Leu 195 200 205
Cys Leu Met Trp Arg Val Gly Asn Leu Arg Lys Ser His Leu Val Glu 210
215 220 Ala His Val Arg
Ala Gln Leu Leu Lys Ser Arg Ile Thr Ser Glu Gly 225 230
235 240 Glu Tyr Ile Pro Leu Asp Glu Ile Asp
Ile Asn Val Gly Phe Asp Ser 245 250
255 Gly Ile Asp Arg Ile Phe Leu Val Ser Pro Ile Thr Ile Val
His Glu 260 265 270
Ile Asp Glu Asp Ser Pro Leu Tyr Asp Leu Ser Lys Gln Asp Met Asp
275 280 285 Asn Ala Asp Phe
Glu Ile Val Val Ile Leu Glu Gly Met Val Glu Ala 290
295 300 Thr Ala Met Thr Thr Gln Cys Arg
Ser Ser Tyr Leu Ala Asn Glu Ile 305 310
315 320 Leu Trp Gly His Arg Tyr Glu Pro Val Leu Phe Glu
Glu Lys Asn Tyr 325 330
335 Tyr Lys Val Asp Tyr Ser Arg Phe His Lys Thr Tyr Glu Val Pro Asn
340 345 350 Thr Pro Ile
Cys Ser Ala Arg Asp Leu Ala Glu Lys Lys Tyr Ile Leu 355
360 365 Ser Asn Ala Asn Ser Phe Cys Tyr
Glu Asn Glu Val Ala Leu Thr Ser 370 375
380 Lys Glu Glu Asp Glu Ile Asp Thr Gly Val Pro Glu Ser
Thr Ser Thr 385 390 395
400 Asp Thr His Pro Asp Met Asp His His Asn Gln Ala Gly Val Pro Leu
405 410 415 Glu Pro Arg Pro
Leu Arg Arg Glu Ser Glu Ile 420 425
29429PRTGallus gallus 29Met Thr Ala Gly Arg Val Asn Pro Tyr Ser Ile Val
Ser Ser Glu Glu 1 5 10
15 Asp Gly Leu Arg Leu Thr Thr Met Pro Gly Ile Asn Gly Phe Gly Asn
20 25 30 Gly Lys Ile
His Thr Arg Arg Lys Cys Arg Asn Arg Phe Val Lys Lys 35
40 45 Asn Gly Gln Cys Asn Val Glu Phe
Thr Asn Met Asp Asp Lys Pro Gln 50 55
60 Arg Tyr Ile Ala Asp Met Phe Thr Thr Cys Val Asp Ile
Arg Trp Arg 65 70 75
80 Tyr Met Leu Leu Leu Phe Ser Leu Ala Phe Leu Val Ser Trp Leu Leu
85 90 95 Phe Gly Leu Ile
Phe Trp Leu Ile Ala Leu Ile His Gly Asp Leu Glu 100
105 110 Asn Pro Gly Gly Asp Asp Thr Phe Lys
Pro Cys Val Leu Gln Val Asn 115 120
125 Gly Phe Val Ala Ala Phe Leu Phe Ser Ile Glu Thr Gln Thr
Thr Ile 130 135 140
Gly Tyr Gly Phe Arg Cys Val Thr Glu Glu Cys Pro Leu Ala Val Phe 145
150 155 160 Met Val Val Val Gln
Ser Ile Val Gly Cys Ile Ile Asp Ser Phe Met 165
170 175 Ile Gly Ala Ile Met Ala Lys Met Ala Arg
Pro Lys Lys Arg Ala Gln 180 185
190 Thr Leu Leu Phe Ser His Asn Ala Val Val Ala Met Arg Asp Gly
Lys 195 200 205 Leu
Cys Leu Met Trp Arg Val Gly Asn Leu Arg Lys Ser His Ile Val 210
215 220 Glu Ala His Val Arg Ala
Gln Leu Ile Lys Pro Arg Ile Thr Glu Glu 225 230
235 240 Gly Glu Tyr Ile Pro Leu Asp Gln Ile Asp Ile
Asp Val Gly Phe Asp 245 250
255 Lys Gly Leu Asp Arg Ile Phe Leu Val Ser Pro Ile Thr Ile Leu His
260 265 270 Glu Ile
Asn Glu Asp Ser Pro Leu Phe Gly Ile Ser Arg Gln Asp Leu 275
280 285 Glu Thr Asp Asp Phe Glu Ile
Val Val Ile Leu Glu Gly Met Val Glu 290 295
300 Ala Thr Ala Met Thr Thr Gln Ala Arg Ser Ser Tyr
Leu Ala Ser Glu 305 310 315
320 Ile Leu Trp Gly His Arg Phe Glu Pro Val Leu Phe Glu Glu Lys Asn
325 330 335 Gln Tyr Lys
Val Asp Tyr Ser His Phe His Lys Thr Tyr Glu Val Pro 340
345 350 Ser Thr Pro Arg Cys Ser Ala Lys
Asp Leu Val Glu Asn Lys Phe Leu 355 360
365 Leu Pro Ser Thr Asn Ser Phe Cys Tyr Glu Asn Glu Leu
Ala Phe Met 370 375 380
Ser Arg Asp Glu Asp Glu Glu Asp Asp Asp Ser Arg Gly Leu Asp Asp 385
390 395 400 Leu Ser Pro Asp
Asn Arg His Glu Phe Asp Arg Leu Gln Ala Thr Ile 405
410 415 Ala Leu Asp Gln Arg Ser Tyr Arg Arg
Glu Ser Glu Ile 420 425
301176DNAHomo sapiens 30atgaatgctt ccagtcggaa tgtgtttgac acgttgatca
gggtgttgac agaaagtatg 60ttcaaacatc ttcggaaatg ggtcgtcact cgcttttttg
ggcattctcg gcaaagagca 120aggctagtct ccaaagatgg aaggtgcaac atagaatttg
gcaatgtgga ggcacagtca 180aggtttatat tctttgtgga catctggaca acggtacttg
acctcaagtg gagatacaaa 240atgaccattt tcatcacagc cttcttgggg agttggtttt
tctttggtct cctgtggtat 300gcagtagcgt acattcacaa agacctcccg gaattccatc
cttctgccaa tcacactccc 360tgtgtggaga atattaatgg cttgacctca gcttttctgt
tttctctgga gactcaagtg 420accattggat atggattcag gtgtgtgaca gaacagtgtg
ccactgccat ttttctgctt 480atctttcagt ctatacttgg agttataatc aattctttca
tgtgtggggc catcttagcc 540aagatctcca ggcccaaaaa acgtgccaag accattacgt
tcagcaagaa cgcagtgatc 600agcaaacggg gagggaagct ttgcctccta atccgagtgg
ctaatctcag gaagagcctt 660cttattggca gtcacattta tggaaagctt ctgaagacca
cagtcactcc tgaaggagag 720accattattt tggaccagat caatatcaac tttgtagttg
acgctgggaa tgaaaattta 780ttcttcatct ccccattgac aatttaccat gtcattgatc
acaacagccc tttcttccac 840atggcagcgg agacccttct ccagcaggac tttgaattag
tggtgttttt agatggcaca 900gtggagtcca ccagtgctac ctgccaagtc cggacatcct
atgtcccaga ggaggtgctt 960tggggctacc gttttgctcc catagtatcc aagacaaagg
aagggaaata ccgagtggat 1020ttccataact ttagcaagac agtggaagtg gagacccctc
actgtgccat gtgcctttat 1080aatgagaaag atgttagagc caggatgaag agaggctatg
acaaccccaa cttcatcttg 1140tcagaagtca atgaaacaga tgacaccaaa atgtaa
1176311282DNAHomo sapiens 31cttttctgat cccagctccg
ggtttaagag tcctggcacg gcccgtcgca cagctctgct 60cctaactcct gcccgccccg
tccgtccatc tgtcccgctg ccccgcggcc catccaaggg 120gccactccac ctcggaccca
agatgacgtc agttgccaag gtgtattaca gtcagaccac 180tcagacagaa agccggcccc
taatgggccc agggatacga cggcggagag tcctgacaaa 240agatggtcgc agcaacgtga
gaatggagca cattgccgac aagcgcttcc tctacctcaa 300ggacctgtgg acaaccttca
ttgacatgca gtggcgctac aagcttctgc tcttctctgc 360gacctttgca ggcacatggt
tcctctttgt cgtggtgtgg tatctggtag ctgtggcaca 420tggggacctg ctggagctgg
accccccggc caaccacacc ccctgtgtgg tacaggtgca 480cacactcact ggagccttcc
tcttctccct tgaatcccaa accaccattg gctatggctt 540ccgctacatc agtgaggaat
gtccactagc cattgtgctt cttattgccc agctggtgct 600caccaccatc ctggaaatct
tcatcacagg taccttcctg gcgaagattg cccggcccaa 660gaagcgggct gagaccattc
gtttcagcca gcatgcagtt gtggcctccc acaatggcaa 720gccctgcctc atgatccgag
ttgccaatat gcgcaaaagc ctcctcattg gctgccaggt 780gacaggaaaa ctgcttcaga
cccaccaaac caaggaaggg gagaacatcc ggctcaacca 840ggtcaatgtg actttccaag
tagacacagc ctctgacagc cccttcctta ttctacccct 900taccttctat catgtggtag
atgagaccag tcccttgaaa gatctccctc ttcgcagtgg 960tgagggtgac tttgagctgg
tgctgatcct aagtgggaca gtggagtcca ccagtgccac 1020ctgtcaggtg cgcacttcct
acctgccaga ggagatcctt tggggctacg agttcacacc 1080tgccatctca ctgtcagcca
gtggtaaata catagctgac tttagccttt ttgaccaagt 1140tgtgaaagtg gcctctccta
gtggcctccg tgacagcact gtacgctacg gagaccctga 1200aaagctcaag ttggaggagt
cattaaggga gcaagctgag aaggagggca gtgcccttag 1260tgtgcgcatc agcaatgtct
ga 1282321326DNAHomo sapiens
32atgggcagtg tgcgaaccaa ccgctacagc atcgtctctt cagaagaaga cggtatgaag
60ttggccacca tggcagttgc aaatggcttt gggaacggga agagtaaagt ccacacccga
120caacagtgca ggagccgctt tgtgaagaaa gatggccact gtaatgttca gttcatcaat
180gtgggtgaga aggggcaacg gtacctcgca gacatcttca ccacgtgtgt ggacattcgc
240tggcggtgga tgctggttat cttctgcctg gctttcgtcc tgtcatggct gttttttggc
300tgtgtgtttt ggttgatagc tctgctccat ggggacctgg atgcatccaa agagggcaaa
360gcttgtgtgt ccgaggtcaa cagcttcacg gctgccttcc tcttctccat tgagacccag
420acaaccatag gctatggttt cagatgtgtc acggatgaat gcccaattgc tgttttcatg
480gtggtgttcc agtcaatcgt gggctgcatc atcgatgctt tcatcattgg cgcagtcatg
540gccaagatgg caaagccaaa gaagagaaac gagactcttg tcttcagtca caatgccgtg
600attgccatga gagacggcaa gctgtgtttg atgtggcgag tgggcaatct tcggaaaagc
660cacttggtgg aagctcatgt tcgagcacag ctcctcaaat ccagaattac ttctgaaggg
720gagtatatcc ctctggatca aatagacatc aatgttgggt ttgacagtgg aatcgatcgt
780atatttctgg tgtccccaat cactatagtc catgaaatag atgaagacag tcctttatat
840gatttgagta aacaggacat tgacaacgca gactttgaaa tcgtggtcat actggaaggc
900atggtggaag ccactgccat gacgacacag tgccgtagct cttatctagc aaatgaaatc
960ctgtggggcc accgctatga gcctgtgctc tttgaagaga agcactacta caaagtggac
1020tattccaggt tccacaaaac ttacgaagtc cccaacactc ccctttgtag tgccagagac
1080ttagcagaaa agaaatatat cctctcaaat gcaaattcat tttgctatga aaatgaagtt
1140gccctcacaa gcaaagagga agacgacagt gaaaatggag ttccagaaag cactagtacg
1200gacacgcccc ctgacataga ccttcacaac caggcaagtg tacctctaga gcccaggccc
1260ttacggcgag agtcggagat atgagtcctg gagcagcggc cctacagacg ggagtcagag
1320atctga
1326331302DNAHomo sapiens 33atgaccgcgg ccagccgggc caacccctac agcatcgtgt
catcggagga ggacgggctg 60cacctggtca ccatgtcggg cgccaacggc ttcggcaacg
gcaaggtgca cacgcggcgc 120aggtgccgca accgcttcgt caagaagaat ggccagtgca
acattgagtt cgccaacatg 180gacgagaagt cacagcgcta cctggctgac atgttcacca
cctgtgtgga catccgctgg 240cggtacatgc tgctcatctt ctcgctggcc ttccttgcct
cctggctgct gttcggcatc 300atcttctggg tcatcgcggt ggcacacggt gacctggagc
cggctgaggg ccggggccgc 360acaccctgtg tgatgcaggt gcacggcttc atggcggcct
tcctcttctc catcgagacg 420cagaccacca tcggctacgg gctgcgctgt gtgacggagg
agtgcccggt ggccgtcttc 480atggtggtgg cccagtccat cgtgggctgc atcatcgact
ccttcatgat tggtgccatc 540atggccaaga tggcaaggcc caagaagcgg gcacagacgc
tgctgttcag ccacaacgcc 600gtggtggccc tgcgtgacgg caagctctgc ctcatgtggc
gtgtgggtaa cctgcgcaag 660agccacattg tggaggccca tgtgcgcgcg cagctcatca
agccgcgggt caccgaggag 720ggcgagtaca tcccgctgga ccagatcgac atcgatgtgg
gcttcgacaa gggcctggac 780cgcatctttc tggtgtcgcc catcaccatc ttgcatgaga
ttgacgaggc cagcccgctc 840ttcggcatca gccggcagga cctggagacg gacgactttg
agatcgtggt catcctggaa 900ggcatggtgg aggccacagc catgaccacc caggcccgca
gctcctacct ggccaatgag 960atcctgtggg gtcaccgctt tgagcccgtg ctcttcgagg
agaagaacca gtacaagatt 1020gactactcgc acttccacaa gacctatgag gtgccctcta
cgccccgctg cagtgcgaag 1080gatctggtag agaacaagtt cctgctgccc agcgccaact
ccttctgcta cgagaacgag 1140ctggccttcc tgagccgtga cgaggaggat gaggcggacg
gagaccagga cggccgaagc 1200cgggacggcc tcagccccca ggccaggcat gactttgaca
gactccaggc tggcggcggg 1260gtcctggagc agcggcccta cagacgggag tcagagatct
ga 1302341338DNAHomo sapiens 34atgcacggac acagccgcaa
cggccaggcc cacgtgcccc ggcggaagcg ccgcaaccgc 60ttcgtcaaga agaacggcca
atgcaacgtg tacttcgcca acctgagcaa caagtcgcag 120cgctacatgg cggacatctt
caccacctgc gtggacacgc gctggcgcta catgctcatg 180atcttctccg cggccttcct
tgtctcctgg ctctttttcg gcctcctctt ctggtgtatc 240gccttcttcc acggtgacct
ggaggccagc ccaggggtgc ctgcggcggg gggcccggcg 300gcgggtggtg gcggagcagc
cccggtggcc cccaagccct gcatcatgca cgtgaacggc 360ttcctgggtg ccttcctgtt
ctcggtggag acgcagacga ccatcggcta tgggttccgg 420tgcgtgacag aggagtgccc
gctggcagtc atcgctgtgg tggtccagtc catcgtgggc 480tgcgtcatcg actccttcat
gattggcacc atcatggcca agatggcgcg gcccaagaag 540cgggcgcaga cgttgctgtt
cagccaccac gcggtcattt cggtgcgcga cggcaagctc 600tgcctcatgt ggcgcgtggg
caacctgcgc aagagccaca ttgtggaggc ccacgtgcgg 660gcccagctca tcaagcccta
catgacccag gagggcgagt acctgcccct ggaccagcgg 720gacctcaacg tgggctatga
catcggcctg gaccgcatct tcctggtgtc gcccatcatc 780attgtccacg agatcgacga
ggacagcccg ctttatggca tgggcaagga ggagctggag 840tcggaggact ttgagatcgt
ggtcatcctg gagggcatgg tggaggccac ggccatgacc 900acccaggccc gcagctccta
cctggccagc gagatcctgt ggggccaccg ctttgagcct 960gtggtcttcg aggagaagag
ccactacaag gtggactact cacgttttca caagacctac 1020gaggtggccg gcacgccctg
ctgctcggcc cgggagctgc aggagagtaa gatcaccgtg 1080ctgcccgccc caccgccccc
tcccagtgcc ttctgctacg agaacgagct ggcccttatg 1140agccaggagg aagaggagat
ggaggaggag gcagctgcgg cggccgcggt ggccgcaggc 1200ctgggcctgg aggcgggttc
caaggaggag gcgggcatca tccggatgct ggagttcggc 1260agccacctgg acctggagcg
catgcaggct tccctcccgc tggacaacat ctcctaccgc 1320agggagtctg ccatctga
1338351311DNAHomo sapiens
35atgggcctgg ccagggccct acgccgcctc agcggcgccc tggattcggg agacagccgg
60gcgggcgatg aagaggaggc cgggcccggg ttgtgccgca acgggtgggc gccggcaccg
120gtgcagtcac ccgtgggccg gcgccgcggt cgcttcgtca agaaagacgg gcactgcaac
180gtgcgtttcg taaacctggg tggccagggc gcgcgctacc tgagcgacct gttcaccaca
240tgcgtggacg tgcgctggcg ctggatgtgc ctgctcttct cctgctcctt cctcgcctcc
300tggctgctct tcggcctggc cttctggctc attgcctcgc tgcacggcga cctggccgcc
360ccgccaccgc ccgcgccctg cttctcacac gtggccagct tcctggccgc cttcctcttc
420gcgctggaga cgcagacgtc catcggctac ggcgtgcgca gcgtcaccga ggagtgcccg
480gccgctgtgg ccgccgtggt gctgcagtgc attgccggct gcgtgctcga cgccttcgtc
540gtgggtgctg tcatggccaa gatggccaaa cccaagaagc gcaacgagac gctggtcttc
600agcgagaacg ccgtcgtggc gctgcgcgac caccgcctct gcctcatgtg gcgcgtcggc
660aacctgcgcc gcagccacct ggtcgaggcc cacgtgcgtg cccagctgct gcagccccgt
720gtgaccccag agggtgagta catcccgctg gaccaccagg atgtggatgt gggctttgat
780ggaggcaccg atcgtatctt cctcgtgtcc cccatcacca tcgtccatga gatcgactct
840gccagtcctc tgtatgagct aggacgtgcc gagctggcca gggctgactt tgagctggtg
900gtcattctcg aggggatggt tgaggccaca gccatgacca cacagtgtcg ctcgtcctac
960ctccctggtg aactgctctg gggccatcgt tttgagccag ttctcttcca gcgtggctcc
1020cagtatgagg tcgactatcg ccacttccat cgcacttatg aggtcccagg gacaccggtc
1080tgcagtgcta aggagctgga tgaacgggca gagcaggctt cccacagcct caagtctagt
1140ttccccggct ctctgactgc attttgttat gagaatgaac ttgctctgag ctgctgccag
1200gaggaagatg aggacgatga gactgaggaa gggaatgggg tggaaacaga agatggggct
1260gctagccccc gagttctcac accaaccctg gcgctgaccc tgcctccatg a
1311361506DNAHomo sapiens 36atgtctgcac tccgaaggaa atttggggac gattatcagg
tagtgaccac atcgtccagc 60ggctcgggct tgcagcccca ggggccaggc caggaccctc
agcagcagct tgtgcccaag 120aagaagcggc agcggttcgt ggacaagaac ggccggtgca
atgtacagca cggcaacctg 180ggcagcgaga caagccgcta cctctcggac ctcttcacca
cgctggtgga cctcaagtgg 240cgctggaacc tcttcatctt cattctcacc tacaccgtgg
cctggctttt catggcgtcc 300atgtggtggg tgatcgccta cactcggggc gacctgaaca
aagcccacgt cggtaactac 360acgccttgcg tggccaatgt ctataacttc ccttctgcct
tcctcttctt catcgagacg 420gaggccacca tcggctatgg ctaccgatac atcacagaca
agtgccccga gggcatcatc 480ctcttcctct tccagtccat cctgggctcc atcgtggacg
ccttcctcat cggctgcatg 540ttcatcaaga tgtcccagcc caagaagcgc gccgagaccc
tcatgttcag cgagcacgcg 600gtgatctcca tgagggacgg aaaactcacg cttatgttcc
gggtgggcaa cctgcgcaac 660agccacatgg tctccgcgca gattcgctgc aagctgctca
aatctcggca gacacctgag 720ggtgagttcc ttccccttga ccaacttgaa ctggatgtag
gttttagtac aggggcagat 780caactttttc ttgtgtcccc cctcacaatt tgccacgtga
tcgatgccaa aagccccttt 840tatgacctat cccagcgaag catgcaaact gaacagttcg
agattgtcgt catcctagaa 900ggcattgtgg aaacaactgg gatgacttgt caagctcgaa
catcatatac tgaagatgaa 960gttctttggg gtcatcgttt ttttcctgta atttccttag
aagagggatt ctttaaagtt 1020gattactccc agttccatgc aacatttgaa gtccccaccc
caccttacag tgtgaaagag 1080caggaggaaa tgcttctcat gtcgtcccct ttaatagcac
cagccataac taacagcaaa 1140gaaagacata attctgtgga atgcttagat ggactagatg
atattactac aaaactacca 1200tctaagctgc agaaaattac tggaagagaa gactttccca
aaaaactctt gaggatgagt 1260tctacaactt cagaaaaagc ctacagcttg ggagacttgc
ccatgaaact tcaacgaata 1320agttcagttc cgggcaactc agaagaaaaa ctggtatcta
aaaccaccaa gatgttatct 1380gatcccatga gccagtctgt ggctgatttg ccaccaaagc
ttcaaaagat ggctggagga 1440gcagctagga tggaagggaa ccttccagcc aaattaagaa
aaatgaactc tgatcgcttc 1500acataa
1506371260DNAHomo sapiens 37atggctggcg attctaggaa
tgccatgaac caggacatgg agattggagt cactccctgg 60gaccccaaga agattccaaa
acaggcccgc gattatgtcc ccattgccac agaccgtacg 120cgcctgctgg ccgagggcaa
gaagccacgc cagcgctaca tggagaagag tggcaagtgc 180aacgtgcacc acggcaacgt
ccaggagacc taccggtacc tgagtgacct cttcaccacc 240ctggtggacc tcaagtggcg
cttcaacttg ctcgtcttca ccatggttta cactgtcacc 300tggctgttct tcggcttcat
ttggtggctc attgcttata tccggggtga cctggaccat 360gttggcgacc aagagtggat
tccttgtgtt gaaaacctca gtggcttcgt gtccgctttc 420ctgttctcca ttgagaccga
aacaaccatt gggtatggct tccgagtcat cacagagaag 480tgtccagagg ggattatact
cctcttggtc caggccatcc tgggctccat cgtcaatgcc 540ttcatggtgg ggtgcatgtt
tgtcaagatc agccagccca agaagagagc ggagaccctc 600atgttttcca acaacgcagt
catctccatg cgggacgaga agctgtgcct catgttccgg 660gtgggcgacc tccgcaactc
ccacatcgtg gaggcctcca tccgggccaa gctcatcaag 720tcccggcaga ccaaagaggg
ggagttcatc cccctgaacc agacagacat caacgtgggc 780tttgacacgg gcgacgaccg
cctcttcctt gtgtctcctc tgatcatctc ccatgagatc 840aaccagaaga gccctttctg
ggagatgtct caggctcagc tgcatcagga agagtttgaa 900gttgtggtca ttctagaagg
gatggtggaa gccacaggca tgacctgcca agcccggagc 960tcctacatgg atacagaggt
gctctggggc caccgattca caccagtcct caccttggaa 1020aagggcttct atgaggtgga
ctacaacacc ttccatgata cctatgagac caacacaccc 1080agctgctgtg ccaaggagct
ggcagaaatg aagagggaag gccggctcct ccagtacctc 1140cccagccccc cactgctggg
gggctgtgct gaggcagggc tggatgcaga ggctgagcag 1200aatgaagaag atgagcccaa
ggggctgggt gggtccaggg aggccagggg ctcggtgtga 1260381140DNAHomo sapiens
38atgacgtcag ttgccaaggt gtattacagt cagaccactc agacagaaag ccggccccta
60atgggcccag ggatacgacg gcggagagtc ctgacaaaag atggtcgcag caacgtgaga
120atggagcaca ttgccgacaa gcgcttcctc tacctcaagg acctgtggac aaccttcatt
180gacatgcagt ggcgctacaa gcttctgctc ttctctgcga cctttgcagg cacatggttc
240ctctttggcg tggtgtggta tctggtagct gtggcacatg gggacctgct ggagctggac
300cccccggcca accacacccc ctgtgtggta caggtgcaca cactcactgg agccttcctc
360ttctcccttg aatcccaaac caccattggc tatggcttcc gctacatcag tgaggaatgt
420ccactggcca ttgtgcttct tattgcccag ctggtgctca ccaccatcct ggaaatcttc
480atcacaggta ccttcctggc gaagattgcc cggcccaaga agcgggctga gaccattcgt
540ttcagccagc atgcagttgt ggcctcccac aatggcaagc cctgcctcat gatccgagtt
600gccaatatgc gcaaaagcct cctcattggc tgccaggtga caggaaaact gcttcagacc
660caccaaacca aggaagggga gaacatccgg ctcaaccagg tcaatgtgac tttccaagta
720gacacagcct ctgacagccc cttccttatt ctacccctta ccttctatca tgtggtagat
780gagaccagtc ccttgaaaga tctccctctt cgcagtggtg agggtgactt tgagctggtg
840ctgatcctaa gtgggacagt ggagtccacc agtgccacct gtcaggtgcg cacttcctac
900ctgccagagg agatcctttg gggctacgag ttcacacctg ccatctcact gtcagccagt
960ggtaaataca tagctgactt tagccttttt gaccaagttg tgaaagtggc ctctcctagt
1020ggcctccgtg acagcactgt acgctacgga gaccctgaaa agctcaagtt ggaggagtca
1080ttaagggagc aagctgagaa ggagggcagt gcccttagtg tgcgcatcag caatgtctga
1140391128DNAHomo sapiens 39atggatgcca ttcacatcgg catgtccagc acccccctgg
tgaagcacac tgctggggct 60gggctcaagg ccaacagacc ccgcgtcatg tccaagagtg
ggcacagcaa cgtgagaatt 120gacaaagtgg atggcatata cctactctac ctgcaagacc
tgtggaccac agttatcgac 180atgaagtgga gatacaaact caccctgttc gctgccactt
ttgtgatgac ctggttcctt 240tttggagtca tctactatgc catcgcgttt attcatgggg
acttagaacc cggtgagccc 300atttcaaatc ataccccctg catcatgaaa gtggactctc
tcactggggc gtttctcttt 360tccctggaat cccagacaac cattggctat ggagtccgtt
ccatcacaga ggaatgtcct 420catgccatct tcctgttggt tgctcagttg gtcatcacga
ccttgattga gatcttcatc 480accggaacct tcctggccaa aatcgccaga cccaaaaagc
gggctgagac catcaagttc 540agccactgtg cagtcatcac caagcagaat gggaagctgt
gcttggtgat tcaggtagcc 600aatatgagga agagcctctt gattcagtgc cagctctctg
gcaagctcct gcagacccac 660gtcaccaagg agggggagcg gattctcctc aaccaagcca
ctgtcaaatt ccacgtggac 720tcctcctctg agagcccctt cctcattctg cccatgacat
tctaccatgt gctggatgag 780acgagccccc tgagagacct cacaccccaa aacctaaagg
agaaggagtt tgagcttgtg 840gtcctcctca atgccactgt ggaatccacc agcgctgtct
gccagagccg aacatcttat 900atcccagagg aaatctactg gggttttgag tttgtgcctg
tggtatctct ctccaaaaat 960ggaaaatatg tggctgattt cagtcagttt gaacagattc
ggaaaagccc agattgcaca 1020ttttactgtg cagattctga gaaacagcaa ctcgaggaga
agtacaggca ggaggatcag 1080agggaaagag aactgaggac acttttatta caacagagca
atgtctga 1128401257DNAHomo sapiens 40atgagctatt acggcagcag
ctatcatatt atcaatgcgg acgcaaaata cccaggctac 60ccgccagagc acattatagc
tgagaagaga agagcaagaa gacgattact tcacaaagat 120ggcagctgta atgtctactt
caagcacatt tttggagaat ggggaagcta tgtggttgac 180atcttcacca ctcttgtgga
caccaagtgg cgccatatgt ttgtgatatt ttctttatct 240tatattctct cgtggttgat
atttggctct gtcttttggc tcatagcctt tcatcatggc 300gatctattaa atgatccaga
catcacacct tgtgttgaca acgtccattc tttcacaggg 360gcctttttgt tctccctaga
gacccaaacc accataggat atggttatcg ctgtgttact 420gaagaatgtt ctgtggccgt
gctcatggtg atcctccagt ccatcttaag ttgcatcata 480aataccttta tcattggagc
tgccttggcc aaaatggcaa ctgctcgaaa gagagcccaa 540accattcgtt tcagctactt
tgcacttata ggtatgagag atgggaagct ttgcctcatg 600tggcgcattg gtgattttcg
gccaaaccac gtggtagaag gaacagttag agcccaactt 660ctccgctata cagaagacag
tgaagggagg atgacgatgg catttaaaga cctcaaatta 720gtcaacgacc aaatcatcct
ggtcaccccg gtaactattg tccatgaaat tgaccatgag 780agccctctgt atgcccttga
ccgcaaagca gtagccaaag ataactttga gattttggtg 840acatttatct atactggtga
ttccactgga acatctcacc aatctagaag ctcctatgtt 900ccccgagaaa ttctctgggg
ccataggttt aatgatgtct tggaagttaa gaggaagtat 960tacaaagtga actgcttaca
gtttgaagga agtgtggaag tatatgcccc cttttgcagt 1020gccaagcaat tggactggaa
agaccagcag ctccacatag aaaaagcacc accagttcga 1080gaatcctgca cgtcggacac
caaggcgaga cgaaggtcat ttagtgcagt tgccattgtc 1140agcagctgtg aaaaccctga
ggagaccacc acttccgcca cacatgaata tagggaaaca 1200ccttatcaga aagctctcct
gactttaaac agaatctctg tagaatccca aatgtag 1257411275DNAHomo sapiens
41atgttggcca gaaagagtat catcccggag gagtatgtgc tggcgcgcat cgccgcagag
60aacctgcgca agccgcgcat ccgagaccgc ctccccaaag cccgcttcat cgccaagagc
120ggggcctgca acctggcgca taagaacatc cgtgagcaag gacgctttct acaggacatc
180ttcaccacct tggtggacct gaaatggcgc cacacgctgg tcatctttac catgtccttc
240ctctgcagct ggctgctctt cgctatcatg tggtggctgg tggcctttgc ccatggggac
300atctatgctt acatggagaa aagtggaatg gagaaaagtg gtttggagtc cactgtgtgt
360gtgactaatg tcaggtcttt cacttctgct tttctcttct ccattgaagt tcaagttacc
420attgggtttg gagggaggat gatgacagag gaatgccctt tggccatcac ggttttgatt
480ctccagaata ttgtgggttt gatcatcaat gcagtcatgt taggctgcat tttcatgaaa
540acagctcagg ctcacagaag ggcagaaact ttgattttca gccgccatgc tgtgattgcc
600gtccgaaatg gcaagctgtg cttcatgttc cgagtgggtg acctgaggaa aagcatgatc
660attagtgcct ctgtgcgcat ccaggtggtc aagaaaacaa ctacacctga aggggaggtg
720gttcctattc accaactgga cattcctgtt gataacccaa tcgagagcaa taacattttt
780ctggtggccc ctttgatcat ctgccacgtg attgacaagc gcagtcccct gtatgacatc
840tcagcaactg acctggccaa ccaagacttg gaggtcatag ttattctgga aggagtggtt
900gaaactactg gcatcaccac acaagcacga acctcctaca ttgctgagga gatccaatgg
960ggccaccgct ttgtgtccat tgtgactgag gaagaaggag tgtattctgt ggattactcc
1020aaatttggca acactgttaa agtagctgct ccacggtgca gtgcccgaga gctggatgag
1080aaaccttcca tccttattca gaccctccaa aagagtgaac tgtctcatca aaattctctg
1140aggaagcgca actccatgag aagaaacaat tccatgagga ggaacaattc tatccgaagg
1200aacaattctt ccctcatggt accaaaggtg caatttatga ctccagaagg aaatcaaaac
1260acatcggaat catga
1275421173DNAHomo sapiens 42atgctgtccc gcaagggcat catccccgag gaatacgtgc
tgacacgcct ggcagaggac 60cctgccaagc ccaggtaccg tgcccgccag cggagggccc
gctttgtgtc caagaaaggc 120aactgcaacg tggcccacaa gaacatccgg gagcagggcc
gcttcctgca ggacgtgttc 180accacgctgg tggacctcaa gtggccacac acattgctca
tcttcaccat gtccttcctg 240tgcagctggc tgctcttcgc catggcctgg tggctcatcg
ccttcgccca cggtgacctg 300gcccccagcg agggcactgc tgagccctgt gtcaccagca
tccactcctt ctcgtctgcc 360ttccttttct ccattgaggt ccaagtgact attggctttg
gggggcgcat ggtgactgag 420gagtgcccac tggccatcct gatcctcatc gtgcagaaca
tcgtggggct catgatcaac 480gccatcatgc ttggctgcat cttcatgaag actgcccaag
cccaccgcag ggctgagacc 540ctcatcttca gcaagcatgc ggtgatcgcc ctgcgccacg
gccgcctctg cttcatgcta 600cgtgtgggtg acctccgcaa gagcatgatc atcagcgcca
ccatccacat gcaggtggta 660cgcaagacca ccagccccga gggcgaggtg gtgcccctcc
accaggtgga catccccatg 720gagaacggcg tgggtggcaa cagcatcttc ctggtggccc
cgctgatcat ctaccatgtc 780attgatgcca acagcccact ctacgacctg gcacccagcg
acctgcacca ccaccaggac 840ctcgagatca tcgtcatcct ggaaggcgtg gtggaaacca
cgggcatcac cacccaggcc 900cgcacctcct acctggccga tgagatcctg tggggccagc
gctttgtgcc cattgtagct 960gaggaggacg gacgttactc tgtggactac tccaagtttg
gcaacaccgt caaagtgccc 1020acaccactct gcacggcccg ccagcttgat gaggaccaca
gcctactgga agctctgacc 1080ctcgcctcag cccgcgggcc cctgcgcaag cgcagcgtgc
ccatggccaa ggccaagccc 1140aagttcagca tctctccaga ttccctgtcc tga
1173431083DNAHomo sapiens 43atggacagca gtaattgcaa
agttattgct cctctcctaa gtcaaagata ccggaggatg 60gtcaccaagg atggccacag
cacacttcaa atggatggcg ctcaaagagg tcttgcatat 120cttcgagatg cttggggaat
cctaatggac atgcgctggc gttggatgat gttggtcttt 180tctgcttctt ttgttgtcca
ctggcttgtc tttgcagtgc tctggtatgt tctggctgag 240atgaatggtg atctggaact
agatcatgat gccccacctg aaaaccacac tatctgtgtc 300aagtatatca ccagtttcac
agctgcattc tccttctccc tggagacaca actcacaatt 360ggttatggta ccatgttccc
cagtggtgac tgtccaagtg caatcgcctt acttgccata 420caaatgctcc taggcctcat
gctagaggct tttatcacag gtgcttttgt ggcgaagatt 480gcccggccaa aaaatcgagc
tttttcaatt cgctttactg acacagcagt agtagctcac 540atggatggca aacctaatct
tatcttccaa gtggccaaca cccgacctag ccctctaacc 600agtgtccggg tctcagctgt
actctatcag gaaagagaaa atggcaaact ctaccagacc 660agtgtggatt tccaccttga
tggcatcagt tctgacgaat gtccattctt catctttcca 720ctaacgtact atcactccat
tacaccatca agtcctctgg ctactctgct ccagcatgaa 780aatccttctc actttgaatt
agttgtattc ctttcagcaa tgcaggaggg cactggagaa 840atatgccaaa ggaggacatc
ctacctaccg tctgaaatca tgttacatca ctgttttgca 900tctctgttga cccgaggttc
caaaggtgaa tatcaaatca agatggagaa ttttgacaag 960actgtccctg aatttccaac
tcctctggtt tctaaaagcc caaacaggac tgacctggat 1020atccacatca atggacaaag
cattgacaat tttcagatct ctgaaacagg actgacagaa 1080taa
1083441284DNAGallus gallus
44atgggcagcg tgcgaaccaa ccgctacagc atcgtgtctt cggaagagga cggcatgaag
60ctggcaacca tggccgttgc caatggcttt gggaatggaa aaagtaaggt acacaccagg
120cagcagtgca ggagccgctt tgtcaaaaaa gatggccact gcaacgtcca gtttattaat
180gtgggtgaga agggacagcg atacctcgca gacatcttca ccacttgcgt ggacatccgc
240tggaggtgga tgctggttat cttctgcctg acattcatcc tctcctggct tttctttggc
300tgtgtgtttt ggttgattgc gctgttgcac ggggatctgg agaaccaaga aaataacaaa
360ccgtgtgtct cgcaagtgag cagcttcact gcagcctttc tgttctccat tgagacccag
420accacgatcg gctatggctt caggtgcgtc acagacgagt gccccattgc tgttttcatg
480gtggttttcc agtctatagt aggctgcatc attgacgcct tcatcattgg tgccgtcatg
540gcaaagatgg ctaagccaaa aaagagaaac gaaactcttg tcttcagcca caatgccgtg
600gtggccatga gagatgggaa actgtgcctg atgtggcgtg tcggaaacct gaggaaaagc
660cacttggttg aggcacacgt gcgagcacag ctcctcaagt ccaggatcac gtcagaaggg
720gagtacatcc ctttggatga aatagacatc aatgtagggt ttgacagcgg gatagaccgc
780atattcctgg tctccccaat tacaatagta cacgaaatag atgaagatag tcctttgtat
840gacttgagca aacaagacat ggacaatgct gactttgaaa ttgtagtcat tttagagggc
900atggtggaag ccactgccat gactacccag tgccgcagct catatctggc aaatgaaatc
960ctctggggcc accgctatga gcccgtactc tttgaagaaa aaaactacta caaagtggac
1020tattcaaggt tccacaaaac atacgaagtg cccaacacac ccatctgcag tgccagagac
1080ttagcagaaa agaaatacat tctctcgaac gcaaactcct tttgctacga gaacgaagtg
1140gccctcacca gcaaggagga ggacgagatc gacacggggg tgcccgagag cacaagcaca
1200gacacccacc ccgacatgga ccaccacaac caggcaggag tgcccctaga gccacggccg
1260ctgcggcgtg agtcggaaat atga
1284451290DNAGallus gallus 45atgactgcag gcagggtcaa cccttacagc atagtgtcct
ccgaggaaga cggactgagg 60ttgaccacca tgccagggat taacggcttt ggcaatggga
aaatccacac caggaggaaa 120tgcaggaaca ggtttgtaaa gaagaacggt cagtgcaatg
tggagttcac caacatggat 180gacaagccac agaggtacat tgcagacatg ttcaccacgt
gcgttgacat ccgttggagg 240tatatgctct tgctcttctc cctggcattt ctggtatcct
ggttattgtt tgggctgatt 300ttctggctaa ttgcactcat tcatggagat ctagaaaacc
caggtggaga tgataccttc 360aagccttgcg ttctgcaggt caatggcttt gtggctgctt
ttctgttctc catcgagacc 420caaacgacta ttggttatgg cttccgctgt gtgacagagg
agtgcccgct cgcagtcttc 480atggtggtgg ttcagtccat cgtggggtgt ataatcgact
ctttcatgat tggtgcaata 540atggcaaaga tggccaggcc caaaaaaagg gcccagacat
tgcttttcag ccataatgca 600gtagtggcaa tgagagatgg aaaactctgc ctgatgtgga
gagttgggaa tctccggaaa 660agccacatag tagaagccca cgtacgagct caattaatta
agcccagaat cacagaagaa 720ggggagtaca tcccactcga ccaaatagac atcgacgtgg
ggtttgacaa aggcttggac 780cgaatcttct tggtgtcccc cattaccatt ctccatgaga
tcaacgaaga cagcccgctg 840ttcgggatca gccgccagga cttggagacg gatgactttg
agattgtggt catcctcgaa 900ggcatggtag aagccaccgc gatgacgaca caagctcgga
gctcctacct ggccagcgag 960atcctgtggg gccaccgctt cgagcccgtc ttgttcgagg
agaaaaacca gtacaaagta 1020gactattccc acttccacaa aacatacgag gtcccgtcca
caccccgctg cagcgccaag 1080gacttggtgg agaacaaatt cctgctgccc agcaccaact
ccttctgcta cgagaatgag 1140ctggccttca tgagccgcga tgaggatgag gaggatgatg
acagcagggg tttggacgac 1200ctgagcccag acaacaggca cgagttcgac cggcttcagg
caacgatagc gttggatcag 1260aggtcatacc ggagggagtc agaaatatga
12904622PRTHomo sapiens 46His Lys Asp Leu Pro Glu
Phe His Pro Ser Ala Asn His Thr Pro Cys 1 5
10 15 Val Glu Asn Ile Asn Gly 20
4722PRTHomo sapiens 47His Gly Asp Leu Leu Glu Leu Asp Pro Pro Ala Asn
His Thr Pro Cys 1 5 10
15 Val Val Gln Val His Thr 20 4819PRTHomo
sapiens 48His Gly Asp Leu Asp Ala Ser Lys Glu Gly Lys Ala Cys Val Ser Glu
1 5 10 15 Val Asn
Ser 4921PRTHomo sapiens 49His Gly Asp Leu Glu Pro Ala Glu Gly Arg Gly Arg
Thr Pro Cys Val 1 5 10
15 Met Gln Val His Gly 20 5037PRTHomo sapiens 50His
Gly Asp Leu Glu Ala Ser Pro Gly Val Pro Ala Ala Gly Gly Pro 1
5 10 15 Ala Ala Gly Gly Gly Gly
Ala Ala Pro Val Ala Pro Lys Pro Cys Ile 20
25 30 Met His Val Asn Gly 35
5119PRTHomo sapiens 51His Gly Asp Leu Ala Ala Pro Pro Pro Pro Ala Pro Cys
Phe Ser His 1 5 10 15
Val Ala Ser 5221PRTHomo sapiens 52Arg Gly Asp Leu Asn Lys Ala His Val
Gly Asn Tyr Thr Pro Cys Val 1 5 10
15 Ala Asn Val Tyr Asn 20 5321PRTHomo
sapiens 53Arg Gly Asp Leu Asp His Val Gly Asp Gln Glu Trp Ile Pro Cys Val
1 5 10 15 Glu Asn
Leu Ser Gly 20 5429PRTHomo sapiens 54His Gly Asp Ile Tyr
Ala Tyr Met Glu Lys Ser Gly Met Glu Lys Ser 1 5
10 15 Gly Leu Glu Ser Thr Val Cys Val Thr Asn
Val Arg Ser 20 25
5520PRTHomo sapiens 55His Gly Asp Leu Ala Pro Ser Glu Gly Thr Ala Glu Pro
Cys Val Thr 1 5 10 15
Ser Ile His Ser 20 5624PRTHomo sapiens 56Asn Gly Asp Leu
Glu Leu Asp His Asp Ala Pro Pro Glu Asn His Thr 1 5
10 15 Ile Cys Val Lys Tyr Ile Thr Ser
20 5719PRTGallus gallus 57His Gly Asp Leu Glu
Asn Gln Glu Asn Asn Lys Pro Cys Val Ser Gln 1 5
10 15 Val Ser Ser 5822PRTGallus gallus 58His
Gly Asp Leu Glu Asn Pro Gly Gly Asp Asp Thr Phe Lys Pro Cys 1
5 10 15 Val Leu Gln Val Asn Gly
20 59309PRTUnknownDescription of Artificial Sequence
Note = Synthetic Construct 59Ser Pro Ala Arg Lys Pro Pro Arg Gly Gly
Arg Arg Ile Trp Ser Gly 1 5 10
15 Thr Arg Glu Val Ile Ala Tyr Gly Met Pro Ala Ser Val Trp Arg
Asp 20 25 30 Leu
Tyr Tyr Trp Ala Leu Lys Val Ser Trp Pro Val Phe Phe Ala Ser 35
40 45 Leu Ala Ala Leu Phe Val
Val Asn Asn Thr Leu Phe Ala Leu Leu Tyr 50 55
60 Gln Leu Gly Asp Ala Pro Ile Ala Asn Gln Ser
Pro Pro Gly Phe Val 65 70 75
80 Gly Ala Phe Phe Phe Ser Val Glu Thr Leu Ala Thr Val Gly Tyr Gly
85 90 95 Asp Met
His Pro Gln Thr Val Tyr Ala His Ala Ile Ala Thr Leu Glu 100
105 110 Ile Phe Val Gly Met Ser Gly
Ile Ala Leu Ser Thr Gly Leu Val Phe 115 120
125 Ala Arg Phe Ala Arg Pro Arg Ala Lys Ile Met Phe
Ala Arg His Ala 130 135 140
Ile Val Arg Pro Phe Asn Gly Arg Met Thr Leu Met Val Arg Ala Ala 145
150 155 160 Asn Ala Arg
Gln Asn Val Ile Ala Glu Ala Arg Ala Lys Met Arg Leu 165
170 175 Met Arg Arg Glu His Ser Ser Glu
Gly Tyr Ser Leu Met Lys Ile His 180 185
190 Asp Leu Lys Leu Val Arg Asn Glu His Pro Ile Phe Leu
Leu Gly Trp 195 200 205
Asn Met Met His Val Ile Asp Glu Ser Ser Pro Leu Phe Gly Glu Thr 210
215 220 Pro Glu Ser Leu
Ala Glu Gly Arg Ala Met Leu Leu Val Met Ile Glu 225 230
235 240 Gly Ser Asp Glu Thr Thr Ala Gln Val
Met Gln Ala Arg His Ala Trp 245 250
255 Glu His Asp Asp Ile Arg Trp His His Arg Tyr Val Asp Leu
Met Ser 260 265 270
Asp Val Asp Gly Met Thr His Ile Asp Tyr Thr Arg Phe Asn Asp Thr
275 280 285 Glu Pro Val Glu
Pro Pro Gly Ala Ala Pro Asp Ala Gln Ala Phe Ala 290
295 300 Ala Lys Pro Gly Glu 305
60124PRTUnknownDescription of Artificial Sequence Note =
Synthetic Construct 60Met Ala Pro Met Leu Ser Gly Leu Leu Ala Arg Leu Val
Lys Leu Leu 1 5 10 15
Leu Gly Arg His Gly Ser Ala Leu His Trp Arg Ala Ala Gly Ala Ala
20 25 30 Thr Val Leu Leu
Val Ile Val Leu Leu Ala Gly Ser Tyr Leu Ala Val 35
40 45 Leu Ala Glu Arg Gly Ala Pro Gly Ala
Gln Leu Ile Thr Tyr Pro Arg 50 55
60 Ala Leu Trp Trp Ser Val Glu Thr Ala Thr Thr Val Gly
Tyr Gly Asp 65 70 75
80 Leu Tyr Pro Val Thr Leu Trp Gly Arg Cys Val Ala Val Val Val Met
85 90 95 Val Ala Gly Ile
Thr Ser Phe Gly Leu Val Thr Ala Ala Leu Ala Thr 100
105 110 Trp Phe Val Gly Arg Glu Gln Glu Arg
Arg Gly His 115 120
61100PRTUnknownDescription of Artificial Sequence Note = Synthetic
Contruct 61Met Arg Glu Leu Gly Leu Leu Ile Phe Phe Leu Phe Ile Gly Val
Ile 1 5 10 15 Leu
Phe Ser Ser Ala Val Tyr Phe Ala Glu Ala Asp Glu Arg Asp Ser
20 25 30 Gln Phe Pro Ser Ile
Pro Asp Ala Phe Trp Trp Ala Val Val Ser Met 35
40 45 Thr Thr Val Gly Tyr Gly Asp Met Val
Pro Thr Thr Ile Gly Gly Lys 50 55
60 Ile Val Gly Ser Leu Cys Ala Ile Ala Gly Val Leu Thr
Ile Ala Leu 65 70 75
80 Pro Val Pro Val Ile Val Ser Asn Phe Asn Tyr Phe Tyr His Arg Glu
85 90 95 Thr Glu Gly Glu
100
User Contributions:
Comment about this patent or add new information about this topic:
| People who visited this patent also read: | |
| Patent application number | Title |
|---|---|
| 20180244678 | SMALL MOLECULE INHIBITORS OF KU70/80 AND USES THEREOF |
| 20180244677 | IRAK4 INHIBITOR AND USE THEREOF |
| 20180244676 | LRRK2 INHIBITORS AND METHODS OF MAKING AND USING THE SAME |
| 20180244675 | PURINONE DERIVATIVE HYDROCHLORIDE |
| 20180244674 | SUBSTITUTED XANTHINES AND METHODS OF USE THEREOF |