Patent application title: Process and Systems for the Fermentative Production of Alcohols
Inventors:
IPC8 Class: AC12P716FI
USPC Class:
1 1
Class name:
Publication date: 2017-05-25
Patent application number: 20170145445
Abstract:
The present invention relates to the fermentative production of alcohols
including ethanol and butanol, and processes for improving alcohol
fermentation employing in situ product removal methods.Claims:
1-21. (canceled)
22. A method for recovering a product alcohol from a fermentation broth comprising providing a fermentation broth comprising a microorganism, wherein the microorganism produces product alcohol in a fermentor; providing a feedstock slurry comprising fermentable carbon source, undissolved solids, oil, and water; separating the feedstock slurry whereby (i) an aqueous solution comprising fermentable carbon source, (ii) a wet cake comprising solids, and (iii) an oil are formed; adding the aqueous solution to the fermentation broth; contacting the fermentation broth with at least one extractant; and recovering the product alcohol.
23. The method of claim 22, wherein the oil is hydrolyzed to form fatty acids.
24. The method of claim 23, wherein the fermentation broth is contacted with the fatty acids.
25. The method of claim 23, wherein the oil is hydrolyzed by an enzyme.
26. The method of claim 25, wherein the enzyme is one or more lipases or phospholipases.
27. The method of claim 22, wherein the feedstock slurry is generated by hydrolysis of feedstock.
28. The method of claim 27, wherein feedstock is selected from rye, wheat, corn, cane, barley, cellulosic or lignocellulosic material, or combinations thereof.
29. The method of claim 22, wherein the feedstock slurry is separated by decanter bowl centrifugation, three-phase centrifugation, disk stack centrifugation, filtering centrifugation, decanter centrifugation, filtration, vacuum filtration, beltfilter, pressure filtration, filtration using a screen, screen separation, grating, porous grating, flotation, hydroclone, filter press, screwpress, gravity settler, vortex separator, or combination thereof.
30. The method of claim 22, wherein separating the feedstock is a single step process.
31. The method of claim 22, wherein the wet cake is combined with the aqueous solution.
32. The method of claim 22, further comprising contacting the aqueous solution with a catalyst converting oil in the aqueous solution to fatty acids.
33. The method of claim 32, wherein the aqueous solution and fatty acids are added to the fermentation broth.
34. The method of claim 32, wherein the catalyst is deactivated.
35. A system comprising one or more fermentors comprising: an inlet for receiving feedstock slurry; and an outlet for discharging fermentation broth comprising product alcohol; and one or more extractors comprising: a first inlet for receiving the fermentation broth; a second inlet for receiving extractant; a first outlet for discharging a lean fermentation broth; and a second outlet for discharging a rich extractant.
36. The system of claim 35, further comprising one or more liquefaction units; one or more separation means; and optionally one or more wash systems.
37. The system of claim 36, wherein the separation means is selected from decanter bowl centrifugation, three-phase centrifugation, disk stack centrifugation, filtering centrifugation, decanter centrifugation, filtration, vacuum filtration, belt filter, pressure filtration, membrane filtration, filtration using a screen, screen separation, grating, porous grating, flotation, hydroclone, filter press, screwpress, gravity settler, vortex separator, and combinations thereof.
38. The system of claim 35, wherein the system further comprises on-line measurement devices.
39. The system of claim 38, wherein the on-line measurement devices are selected from particle size analyzers, Fourier transform infrared spectroscopes, near-infrared spectroscopes, Raman spectroscopes, high pressure liquid chromatography, viscometers, densitometers, tensiometers, droplet size analyzers, pH meters, dissolved oxygen probes, or combinations thereof.
Description:
[0001] This application is a continuation of U.S. patent application Ser.
No. 13/828,353, filed on Mar. 14, 2013 which claims the benefit of U.S.
Provisional Application No. 61/699,976, filed on Sep. 12, 2012; the
entire contents of each are herein incorporated by reference.
[0002] The Sequence Listing associated with this application is filed in electronic form via EFS-Web and hereby incorporated by reference into the specification in its entirety.
FIELD OF THE INVENTION
[0003] The present invention relates to the fermentative production of alcohols including ethanol and butanol, and processes for improving alcohol fermentation employing in situ product removal methods.
BACKGROUND OF THE INVENTION
[0004] Alcohols have a variety of industrial and scientific applications such as a beverage (i.e., ethanol), fuel, reagents, solvents, and antiseptics. For example, butanol is an important industrial chemical and drop-in fuel component with a variety of applications including use as a renewable fuel additive, a feedstock chemical in the plastics industry, and a food-grade extractant in the food and flavor industry. Accordingly, there is a high demand for alcohols such as butanol, as well as for efficient and environmentally-friendly production methods.
[0005] Production of alcohol utilizing fermentation by microorganisms is one such environmentally-friendly production method. However, in the production of butanol, for example, some microorganisms that produce butanol in high yields also have low butanol toxicity thresholds. Removal of butanol from the fermentation as it is being produced is a means to manage these low butanol toxicity thresholds. Thus, there is a continuing need to develop efficient methods and systems for producing butanol in high yields despite the low butanol toxicity thresholds of the butanol-producing microorganisms.
[0006] In situ product removal (ISPR) (also referred to as extractive fermentation) can be used to remove butanol or other fermentative alcohols from the fermentation as it is produced, thereby allowing the microorganism to produce butanol at high yields. One ISPR method for removing fermentative alcohol that has been described in the art is liquid-liquid extraction (see, e.g., U.S. Patent Application Publication No. 2009/0305370). In general, with regard to butanol fermentation, the fermentation broth which includes the microorganism is contacted with an extractant at a time before the butanol concentration reaches, for example, a toxic level. The butanol partitions into the extractant decreasing the concentration of butanol in the fermentation broth containing the microorganism, thereby limiting the exposure of the microorganism to the inhibitory butanol.
[0007] In order to be technically and economically viable, liquid-liquid extraction requires contact between the extractant and the fermentation broth for efficient mass transfer of the alcohol into the extractant; phase separation of the extractant from the fermentation broth (during and/or after fermentation); efficient recovery and recycle of the extractant; and minimal decrease of the partition coefficient of the extractant over long-term operation. Extractant can become contaminated over time with each recycle, for example, by the build-up of lipids present in the biomass used as feedstock for fermentation, and this contamination can lead to a concomitant reduction in the partition coefficient of the extractant.
[0008] In addition, the presence of undissolved solids during extractive fermentation can negatively affect the efficiency of alcohol production. For example, the presence of the undissolved solids may lower the mass transfer coefficient, impede phase separation, result in the accumulation of oil from the undissolved solids in the extractant leading to reduced extraction efficiency over time, slow the disengagement of extractant drops from the fermentation broth, result in a lower fermentation vessel volume efficiency, and increase the loss of extractant because it becomes trapped in the solids and ultimately removed as Dried Distillers' Grains with Solubles (DDGS).
[0009] Thus, there is a continuing need for alternative extractive fermentation processes that reduce the toxic effect of the fermentative alcohol such as butanol on the microorganism, and which can also reduce the degradation of the partition coefficient of an extractant. The present invention satisfies the above needs and provides processes and systems for the fermentative production of alcohols such as ethanol and butanol.
SUMMARY OF THE INVENTION
[0010] The present invention is directed to a method method for recovering a product alcohol from a fermentation broth comprising providing a fermentation broth comprising a microorganism, wherein the microorganism produces product alcohol in a fermentor; contacting the fermentation broth with at least one extractant; and recovering the product alcohol. In some embodiments, the contacting of the fermentation broth with at least one extractant occurs in the fermentor, an external unit, or both. In some embodiments, the external unit is an extractor. In some embodiments, the extractor is selected from siphon, decanter, centrifuge, gravity settler, phase splitter, mixer-settler, column extractor, centrifugal extractor, agitated extractor, hydrocyclone, spray tower, or combinations thereof. In some embodiments, the extractant is selected from C.sub.7 to C.sub.22 fatty alcohols, C.sub.7 to C.sub.22 fatty acids, esters of C.sub.7 to C.sub.22 fatty acids, C.sub.7 to C.sub.22 fatty aldehydes, C.sub.7 to C.sub.22 fatty amides, and mixtures thereof. In some embodiments, the extractant is selected from oleyl alcohol, behenyl alcohol, cetyl alcohol, lauryl alcohol, myristyl alcohol, stearyl alcohol, oleic acid, lauric acid, linoleic acid, linolenic acid, myristic acid, stearic acid, octanoic acid, decanoic acid, undecanoic acid, methyl myristate, methyl oleate, 1-nonanol, 1-decanol, 2-undecanol, 1-nonanal, 1-undecanol, undecanal, lauric aldehyde, 2-methylundecanal, oleamide, linoleamide, palmitamide, stearylamide, 2-ethyl-1-hexanol, 2-hexyl-1-decanol, 2-octyl-1-dodecanol, and mixtures thereof. In some embodiments, a hydrophilic solute is added to the fermentation broth. In some embodiments, the hydrophilic solute is selected from the group consisting of polyhydroxlated compounds, polycarboxylic acids, polyol compounds, ionic salts, or mixtures thereof. In some embodiments, the contacting of the fermentation broth with at least one extractant occurs in two or more external units. In some embodiments, the contacting of the fermentation broth with at least one extractant occurs in two or more fermentors. In some embodiments, the fermentors comprise internals or devices to improve phase separation. In some embodiments, the internals or devices are selected from the group consisting of coalescers, baffles, perforated plates, wells, lamella separators, cones, or combinations thereof. In some embodiments, real-time measurements are used to monitor extraction of product alcohol. In some embodiments, extraction of product alcohol is monitored by real-time measurements of phase separation. In some embodiments, phase separation is monitored by measuring rate of phase separation, extractant droplet size, and/or composition of fermentation broth. In some embodiments, phase separation is monitored by conductivity measurements, dielectric measurements, viscoelastic measurements, or ultrasonic measurements. In some embodiments, providing a fermentation broth comprising a microorganism occurs in two or more fermentors. In some embodiments, the product alcohol is selected from ethanol, propanol, butanol, pentanol, hexanol, and fusel alcohols. In some embodiments, the microorganism comprises a butanol biosynthetic pathway. In some embodiments, the butanol biosynthetic pathway is a 1-butanol biosynthetic pathway, a 2-butanol biosynthetic pathway, or an isobutanol biosynthetic pathway. In some embodiments, the microorganism is a recombinant microorganism. In some embodiments, the method further comprises the steps of providing a feedstock slurry comprising fermentable carbon source, undissolved solids, oil, and water; separating the feedstock slurry whereby (i) an aqueous solution comprising fermentable carbon source, (ii) a wet cake comprising solids, and (iii) an oil are formed; and adding the aqueous solution to the fermentation broth. In some embodiments, the oil is hydrolyzed to form fatty acids. In some embodiments, the fermentation broth is contacted with the fatty acids. In some embodiments, the oil is hydrolyzed by an enzyme. In some embodiments, the enzyme is one or more lipases or phospholipases. In some embodiments, the feedstock slurry is generated by hydrolysis of feedstock. In some embodiments, feedstock is selected from rye, wheat, corn, cane, barley, cellulosic or lignocellulosic material, or combinations thereof. In some embodiments, the feedstock slurry is separated by decanter bowl centrifugation, three-phase centrifugation, disk stack centrifugation, filtering centrifugation, decanter centrifugation, filtration, vacuum filtration, beltfilter, pressure filtration, filtration using a screen, screen separation, grating, porous grating, flotation, hydroclone, filter press, screwpress, gravity settler, vortex separator, or combination thereof. In some embodiments, separating the feedstock is a single step process. In some embodiments, the wet cake is combined with the aqueous solution. In some embodiments, the method further comprises contacting the aqueous solution with a catalyst converting oil in the aqueous solution to fatty acids. In some embodiments, the aqueous solution and fatty acids are added to the fermentation broth. In some embodiments, the catalyst is deactivated.
[0011] The present invention is also directed to a system comprising one or more fermentors comprising: an inlet for receiving feedstock slurry; and an outlet for discharging fermentation broth comprising product alcohol; and one or more extractors comprising: a first inlet for receiving the fermentation broth; a second inlet for receiving extractant; a first outlet for discharging a lean fermentation broth; and a second outlet for discharging a rich extractant. In some embodiments, the system further comprises one or more liquefaction units; one or more separation means; and optionally one or more wash systems. In some embodiments, the separation means is selected from decanter bowl centrifugation, three-phase centrifugation, disk stack centrifugation, filtering centrifugation, decanter centrifugation, filtration, vacuum filtration, belt filter, pressure filtration, membrane filtration, filtration using a screen, screen separation, grating, porous grating, flotation, hydroclone, filter press, screwpress, gravity settler, vortex separator, and combinations thereof. In some embodiments, the system also comprises on-line measurement devices. In some embodiments, the on-line measurement devices are selected from particle size analyzers, Fourier transform infrared spectroscopes, near-infrared spectroscopes, Raman spectroscopes, high pressure liquid chromatography, viscometers, densitometers, tensiometers, droplet size analyzers, pH meters, dissolved oxygen probes, or combinations thereof.
DESCRIPTION OF THE DRAWINGS
[0012] The accompanying drawings, which are incorporated herein and form a part of the specification, illustrate the present invention and, together with the description, further serve to explain the principles of the invention and to enable a person skilled in the pertinent art to make and use the invention.
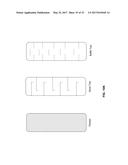
[0013] FIG. 1 schematically illustrates an exemplary process and system of the present invention, in which undissolved solids are removed via separation after liquefaction and before fermentation.
[0014] FIG. 2 schematically illustrates an exemplary process and system of the present invention, in which ISPR is conducted downstream of fermentation.
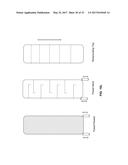
[0015] FIG. 3 schematically illustrates another exemplary alternative process and system of the present invention, in which an oil stream is discharged.
[0016] FIG. 4 schematically illustrates another exemplary alternative process and system of the present invention, in which the wet cake is subjected to wash cycles.
[0017] FIG. 5 schematically illustrates another exemplary alternative process and system of the present invention, in which an oil stream is discharged and wet cake is subjected to wash cycles.
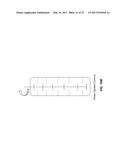
[0018] FIGS. 6A and 6B schematically illustrates another exemplary alternative process and system of the present invention, in which the aqueous solution and wet cake are combined and conducted to fermentation.
[0019] FIGS. 7A-7C schematically illustrates exemplary alternative processes and systems of the present invention, in which the aqueous solution is subjected to hydrolysis and/or deactivation.
[0020] FIG. 8 schematically illustrates an exemplary fermentation process of the present invention including downstream processing.
[0021] FIG. 9 schematically illustrates an exemplary fermentation process of the present invention including downstream processing.
[0022] FIGS. 10A-10M illustrated various systems that may be used in the processes described herein. FIG. 10A illustrates a mixer-settler using a pump as the source of mixing. FIG. 10B illustrates a mixer-settler using a mixer as the source of mixing. FIG. 10C illustrates a mixer-settler using a static mixer as the source of mixing. FIG. 10D illustrates a mixer-settler using a mixing tee as the source of mixing. FIG. 10E illustrates a mixer-settler using an impingement mixer as the source of mixing. FIG. 10F illustrates a mixer-settler using a raining bucket or meshed screen as the source of mixing. FIG. 10G illustrates a mixer-settler using a centrifuge as a settler. FIG. 10H illustrates a mixer-settler using a hydrocyclone or vortex separator as a settler. FIGS. 10I and 10J illustrate one or more mixer-settlers arranged in series or in countercurrent mode. FIG. 10K illustrates examples of non-mechanical extractors such as packed towers, sieve trays, and baffle trays. FIG. 10L illustrates pulsed-agitated extractors and FIG. 10M illustrates rotary-agitated or rotating disc contactors.
[0023] FIGS. 11A and 11B schematically illustrate multiple pass extractant flow systems.
[0024] FIG. 12 schematically illustrates an exemplary fermentation process of the present invention utilizing on-line, in-line, at-line, and/or real-time measurements for monitoring fermentation processes.
[0025] FIGS. 13A and 13B schematically illustrates exemplary processes of the present invention for mitigating formation of a rag layer. FIG. 13A exemplifies the use of a static mixer in combination with an agitation unit downstream of the settler or decanter for the treatment of a rag layer, and FIG. 13B exemplifies the use of a static mixer in combination with an agitation unit upstream of the settler or decanter for the treatment of a rag layer.
[0026] FIG. 14 schematically illustrates an exemplary process of the present invention including fermentation, extraction, and distillation processes.
[0027] FIG. 15 shows the effects of the fermentation broth to extractant ratios (aq/org) on extraction column efficiency.
[0028] FIGS. 16A and 16B show the effects of ISPR using an external extraction column on isobutanol concentrations and glucose profiles. FIG. 16A shows the amount of isobutanol produced in the fermentation and FIG. 16B shows the glucose profile of the fermentation.
[0029] FIG. 17 show the effects of ISPR using a mixer-settler on isobutanol removal rates.
[0030] FIG. 18 shows an FTIR spectra of the range of starch concentrations using in-line measurements.
[0031] FIG. 19 shows an FTIR spectra of the starch concentration of wet cake during processing of corn mash.
[0032] FIG. 20 shows an FTIR spectra of corn oil during processing of corn mash.
[0033] FIG. 21 demonstrates a real-time measurement of isobutanol in COFA.
DETAILED DESCRIPTION OF THE INVENTION
[0034] Unless defined otherwise, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this invention belongs. In case of conflict, the present application including the definitions will control. Also, unless otherwise required by context, singular terms shall include pluralities and plural terms shall include the singular. All publications, patents, and other references mentioned herein are incorporated by reference in their entireties for all purposes.
[0035] In order to further define this invention, the following terms and definitions are herein provided.
[0036] As used herein, the terms "comprises," "comprising," "includes," "including," "has," "having," "contains," or "containing," or any other variation thereof, will be understood to imply the inclusion of a stated integer or group of integers but not the exclusion of any other integer or group of integers. For example, a composition, a mixture, a process, a method, an article, or an apparatus that comprises a list of elements is not necessarily limited to only those elements but can include other elements not expressly listed or inherent to such composition, mixture, process, method, article, or apparatus. Further, unless expressly stated to the contrary, "or" refers to an inclusive or and not to an exclusive or. For example, a condition A or B is satisfied by any one of the following: A is true (or present) and B is false (or not present), A is false (or not present) and B is true (or present), and both A and B are true (or present).
[0037] Also, the indefinite articles "a" and "an" preceding an element or component of the invention are intended to be nonrestrictive regarding the number of instances, i.e., occurrences of the element or component. Therefore, "a" or "an" should be read to include one or at least one, and the singular word form of the element or component also includes the plural unless the number is obviously meant to be singular.
[0038] The term "invention" or "present invention" as used herein is a non-limiting term and is not intended to refer to any single embodiment of the invention but encompasses all possible embodiments as described in the application.
[0039] As used herein, the term "about" modifying the quantity of an ingredient or reactant of the invention employed refers to variation in the numerical quantity that can occur, for example, through typical measuring and liquid handling procedures used for making concentrates or solutions in the real world; through inadvertent error in these procedures; through differences in the manufacture, source, or purity of the ingredients employed to make the compositions or to carry out the methods; and the like. The term "about" also encompasses amounts that differ due to different equilibrium conditions for a composition resulting from a particular initial mixture. Whether or not modified by the term "about," the claims include equivalents to the quantities. In one embodiment, the term "about" means within 10% of the reported numerical value, alternatively within 5% of the reported numerical value.
[0040] "Biomass" as used herein refers to a natural product containing hydrolyzable polysaccharides that provide fermentable sugars including any sugar and starch derived from natural resources such as corn, sugar cane, wheat, cellulosic or lignocellulosic material, and materials comprising cellulose, hemicellulose, lignin, starch, oligosaccharides, disaccharides, and/or monosaccharides, and mixtures thereof. Biomass may also comprise additional components such as protein and/or lipids. Biomass may be derived from a single source or biomass may comprise a mixture derived from more than one source. For example, biomass may comprise a mixture of corn cobs and corn stover, or a mixture of grass and leaves. Biomass includes, but is not limited to, bioenergy crops, agricultural residues, municipal solid waste, industrial solid waste, sludge from paper manufacture, yard waste, wood and forestry waste (e.g., forest thinnings). Examples of biomass include, but are not limited to, corn, corn cobs, crop residues such as corn husks, corn stover, grasses, wheat, rye, wheat straw, spelt, triticale, barley, barley straw, oats, hay, rice, rice straw, switchgrass, potato, sweet potato, cassava, Jerusalem artichoke, sugar cane bagasse, sorghum, sugar cane, sugar beet, fodder beet, soy, palm, coconut, rapeseed, safflower, sunflower, millet, eucalyptus, miscanthus, components obtained from milling of grains, trees (e.g., branches, roots, leaves), wood chips, sawdust, shrubs and bushes, vegetables, fruits, flowers, manure, and mixtures thereof. For example, mash, juice, molasses, or hydrolysate may be formed from biomass by any processing known in the art for processing biomass for purposes of fermentation such as milling and liquefaction. For example, cellulosic and/or lignocellulosic biomass may be processed to obtain a hydrolysate containing fermentable sugars by any method known to one skilled in the art, such as low ammonia pretreatment disclosed in U.S. Patent Application Publication No. 2007/0031918, which is herein incorporated by reference. Enzymatic saccharification of cellulosic and/or lignocellulosic biomass typically makes use of an enzyme consortium (e.g., cellulases, xylanases, glucosidases, glucanases, lyases) for breaking down cellulose and hemicellulose to produce a hydrolysate containing sugars including glucose, xylose, and arabinose. Saccharification enzymes suitable for cellulosic and/or lignocellulosic biomass are reviewed in Lynd, et al. (Microbiol. Mol. Biol. Rev. 66:506-577, 2002).
[0041] "Fermentable carbon source" or "fermentable carbon substrate" as used herein refers to a carbon source capable of being metabolized by microorganisms. Suitable fermentable carbon sources include, but are not limited to, monosaccharides such as glucose or fructose; disaccharides such as lactose or sucrose; oligosaccharides; polysaccharides such as starch or cellulose; one carbon substrates; and mixtures thereof.
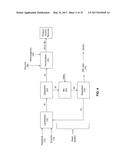
[0042] "Fermentable sugar" as used herein refers to one or more sugars capable of being metabolized by the microorganisms disclosed herein for the production of fermentative alcohol.
[0043] "Feedstock" as used herein refers to a feed in a fermentation process, the feed containing a fermentable carbon source with or without undissolved solids and oil, and where applicable, the feed containing the fermentable carbon source before or after the fermentable carbon source has been removed from starch or obtained from the breakdown of complex sugars by further processing such as by liquefaction, saccharification, or other process. Feedstock includes or may be derived from biomass. Suitable feedstocks include, but are not limited to, rye, wheat, corn, corn mash, cane, cane mash, barley, cellulosic material, lignocellulosic material, or mixtures thereof. Where reference is made to "feedstock oil," it will be appreciated that the term encompasses the oil produced from a given feedstock.
[0044] "Fermentation broth" as used herein refers to a mixture of water, fermentable carbon sources (e.g., sugars), dissolved solids, optionally microorganisms producing alcohol, optionally product alcohol, and all other constituents of the material held in the fermentor in which product alcohol is being made by the metabolism of fermentable carbon sources by the microorganisms. From time to time as used herein, the term "fermentation broth" may be used synonymously with "fermentation medium" or "fermented mixture." In some embodiments, fermentation broth comprising product alcohol may be referred to as fermentation beer or beer.
[0045] "Fermentor" or "fermentation vessel" as used herein refers to the unit in which the fermentation reaction is carried out whereby product alcohol such as ethanol or butanol is produced from fermentable carbon sources. The term "fermentor" may be used synonymously herein with "fermentation vessel."
[0046] "Liquefaction unit" as used herein refers to the unit in which liquefaction is carried out. Liquefaction is the process in which oligosaccharides are released from feedstock. In some embodiments where the feedstock is corn, oligosaccharides are released from the corn starch content during liquefaction.
[0047] "Saccharification unit" as used herein refers to the unit in which saccharification (i.e., the breakdown of oligosaccharides into monosaccharides) is carried out. Where fermentation and saccharification occur simultaneously, the saccharification unit and the fermentor may be the same unit.
[0048] "Sugar" as used herein refers to oligosaccharides, disaccharides, monosaccharides, and/or mixtures thereof. The term "saccharide" also includes carbohydrates including starches, dextrans, glycogens, cellulose, pentosans, as well as sugars.
[0049] As used herein, "saccharification enzyme" refers to one or more enzymes that are capable of hydrolyzing polysaccharides and/or oligosaccharides, for example, alpha-1,4-glucosidic bonds of glycogen or starch. Saccharification enzymes may include enzymes capable of hydrolyzing cellulosic or lignocellulosic materials as well.
[0050] "Undissolved solids" as used herein refers to non-fermentable portions of feedstock, for example, germ, fiber, gluten, and any additional components that do not dissolve in aqueous media. For example, the non-fermentable portions of feedstock include the portion of feedstock that remains as solids and can absorb liquid from the fermentation broth.
[0051] "Oil" as used herein refers to lipids obtained from plants (e.g., biomass) or animals. Examples of oils include, but are not limited to, tallow, corn, canola, capric/caprylic triglycerides, castor, coconut, cottonseed, fish, jojoba, lard, linseed, neetsfoot, oiticica, palm, peanut, rapeseed, rice, safflower, soya, sunflower, tung, jatropha, and vegetable oil blends.
[0052] "Product alcohol" as used herein refers to any alcohol that can be produced by a microorganism in a fermentation process that utilizes biomass as a source of fermentable carbon substrate. Product alcohols include, but are not limited to, C.sub.1 to C.sub.8 alkyl alcohols. In some embodiments, the product alcohols are C.sub.2 to C.sub.8 alkyl alcohols. In other embodiments, the product alcohols are C.sub.2 to C.sub.5 alkyl alcohols. It will be appreciated that C.sub.1 to C.sub.8 alkyl alcohols include, but are not limited to, methanol, ethanol, propanol, butanol, and pentanol. Likewise C.sub.2 to C.sub.8 alkyl alcohols include, but are not limited to, ethanol, propanol, butanol, and pentanol. In some embodiments, product alcohol may also include fusel alcohols (or fusel oils). "Alcohol" is also used herein with reference to a product alcohol.
[0053] "Butanol" as used herein refers to the butanol isomers 1-butanol (1-BuOH), 2-butanol (2-BuOH), tert-butanol (t-BuOH), and/or isobutanol (iBuOH, i-BuOH, I-BUOH, iB also known as 2-methyl-1-propanol), either individually or as mixtures thereof. From time to time, when referring to esters of butanol, the terms "butyl esters" and "butanol esters" may be used interchangeably.
[0054] "Propanol" as used herein refers to the propanol isomers isopropanol or 1-propanol.
[0055] "Pentanol" as used herein refers to the pentanol isomers 1-pentanol, 3-methyl-1-butanol, 2-methyl-1-butanol, 2,2-dimethyl-1-propanol, 3-pentanol, 2-pentanol, 3-methyl-2-butanol, or 2-methyl-2-butanol.
[0056] "In Situ Product Removal (ISPR)" as used herein refers to the selective removal of a specific fermentation product from a biological process such as fermentation to control the product concentration in the biological process as the product is produced.
[0057] "Extractant" as used herein refers to a solvent used to extract a product alcohol. From time to time as used herein, the term "extractant" may be used synonymously with "solvent."
[0058] "Water-immiscible" as used herein refers to a chemical component such as an extractant or solvent, which is incapable of mixing with an aqueous solution such as fermentation broth, in such a manner as to form one liquid phase.
[0059] "Carboxylic acid" as used herein refers to any organic compound with the general chemical formula --COOH in which a carbon atom is bonded to an oxygen atom by a double bond to make a carbonyl group (--C.dbd.O) and to a hydroxyl group (--OH) by a single bond. A carboxylic acid may be in the form of the protonated carboxylic acid, in the form of a salt of a carboxylic acid (e.g., an ammonium, sodium, or potassium salt), or as a mixture of protonated carboxylic acid and salt of a carboxylic acid. The term carboxylic acid may describe a single chemical species (e.g., oleic acid) or a mixture of carboxylic acids as can be produced, for example, by the hydrolysis of biomass-derived fatty acid esters or triglycerides, diglycerides, monoglycerides, and phospholipids.
[0060] "Fatty acid" as used herein refers to a carboxylic acid (e.g., aliphatic monocarboxylic acid) having C.sub.4 to C.sub.28 carbon atoms (most commonly C.sub.12 to C.sub.24 carbon atoms), which is either saturated or unsaturated. Fatty acids may also be branched or unbranched. Fatty acids may be derived from, or contained in esterified form, an animal or vegetable fat, oil, or wax. Fatty acids may occur naturally in the form of glycerides in fats and fatty oils or may be obtained by hydrolysis of fats or by synthesis. The term fatty acid may describe a single chemical species or a mixture of fatty acids. In addition, the term fatty acid also encompasses free fatty acids.
[0061] "Fatty alcohol" as used herein refers to an alcohol having an aliphatic chain of C.sub.4 to C.sub.22 carbon atoms, which is either saturated or unsaturated.
[0062] "Fatty aldehyde" as used herein refers to an aldehyde having an aliphatic chain of C.sub.4 to C.sub.22 carbon atoms, which is either saturated or unsaturated.
[0063] "Fatty amide" as used herein refers to an amide having an aliphatic chain of C.sub.4 to C.sub.22 carbon atoms, which is either saturated or unsaturated.
[0064] "Fatty ester" as used herein refers to an ester having an aliphatic chain of C.sub.4 to C.sub.22 carbon atoms, which is either saturated or unsaturated.
[0065] "Aqueous phase" as used herein refers to the aqueous phase of, for example, a biphasic mixture containing, for example, a liquid phase and a vapor phase, to the aqueous phase of a triphasic mixture containing two liquid phases (e.g., an organic phase and an aqueous phase) and a vapor phase, to the aqueous phase of either a biphasic or triphasic mixture where the aqueous phase contains some amount of suspended solids, or to a quartphasic mixture comprising a vapor phase, an organic phase, an aqueous phase and a solid phase. In some embodiments, a triphasic mixture may comprise a vapor phase, a liquid phase, and a solid phase. In some embodiments, an aqueous phase may be obtained by contacting a fermentation broth with a water-immiscible organic extractant. In an embodiment of a process described herein that includes fermentative extraction, the term "fermentation broth" then may refer to the aqueous phase in biphasic fermentative extraction.
[0066] "Organic phase" as used herein refers to the non-aqueous phase of a mixture (e.g., biphasic mixture, triphasic mixture, quartphasic mixture) obtained by contacting a fermentation broth with a water-immiscible organic extractant. From time to time as used herein, the terms "organic phase" may be used synonymously with "extractant phase."
[0067] "Effective titer" as used herein refers to the total amount of a particular product alcohol produced by fermentation per liter of fermentation broth.
[0068] "Portion" as used herein with reference to a process stream refers to any fractional part of the stream which retains the composition of the stream, including the entire stream, as well as any component or components of the stream, including all components of the stream.
[0069] The present invention provides processes and systems for producing a product alcohol by fermentative processes and recovering a product alcohol produced by a fermentative process. As an example of an embodiment of the processes described herein, fermentation may be initiated by introducing feedstock directly into a fermentor. In some embodiments, one or more fermentors may be used in the processes described herein. Suitable feedstocks include, but are not limited to, rye, wheat, corn, corn mash, cane, cane mash, barley, cellulosic material, lignocellulosic material, or mixtures thereof. These feedstocks may be processed using methods such as dry milling or wet milling. In some embodiments, prior to the introduction to the fermentor, the feedstock may be liquefied to create feedstock slurry which may comprise undissolved solids, a fermentable carbon source (e.g., sugar), and oil. Liquefaction of the feedstock may be accomplished by any known liquefying processes including, but not limited to, acid process, enzyme process (e.g., alpha-amylase), acid-enzyme process, or combinations thereof. In some embodiments, liquefaction may take place in a liquefaction unit.
[0070] If the feedstock slurry is fed directly to the fermentor, the undissolved solids and/or oil may interfere with efficient removal and recovery of a product alcohol. In particular, when liquid-liquid extraction is utilized to extract a product alcohol from the fermentation broth, the presence of the undissolved solids (e.g., particulates) may cause system inefficiencies including, but not limited to, decreasing the mass transfer rate of the product alcohol to the extractant by interfering with the contact between the extractant and the fermentation broth; creating or promoting an emulsion in the fermentor and thereby interfering with phase separation of the extractant and the fermentation broth; reducing the efficiency of recovering and recycling the extractant because at least a portion of the extractant and product alcohol becomes "trapped" in the solids which may be removed as Distillers' Dried Grains with Solubles (DDGS); lowering fermentor volume efficiency because there are solids taking up volume in the fermentor and because there is a slower disengagement of the extractant from the fermentation broth; and shortening the life cycle of the extractant by contamination with oil. These effects may result in higher capital and operating costs. In addition, extractant "trapped" in the DDGS may detract from the DDGS value and qualification for sale as animal feed. Thus, in order to avoid and/or minimize these problems, at least a portion of the undissolved solids may be removed from the feedstock slurry prior to the addition of the feedstock slurry to the fermentor. Extraction activity and efficiency of product alcohol production may be increased when extraction is performed on a fermentation broth where the undissolved solids have been removed.
[0071] Processes and systems to process feedstock generating a feedstock slurry and to separate feedstock slurry generating an aqueous phase comprising fermentable carbon source and a solid phase (e.g., wet cake) are described herein with reference to the Figures. As shown in FIG. 1, in some embodiments, the system includes liquefaction 10 configured to liquefy feedstock to create a feedstock slurry. For example, feedstock 12 may be introduced to liquefaction 10 (e.g., via an inlet in the liquefaction unit). Feedstock 12 can be any suitable biomass material known in the industry including, but not limited to, barley, oat, rye, sorghum, wheat, triticale, spelt, millet, cane, corn, or combinations thereof that contains a fermentable carbon source such as sugar and/or starch. Water may also be introduced to liquefaction 10.
[0072] The process of liquefying feedstock 12 involves hydrolysis of starch in feedstock 12 to water-soluble sugars. Any known liquefying processes, as well as liquefaction unit, utilized by the industry can be used including, but not limited to, an acid process, an enzyme process, or an acid-enzyme process. Such processes can be used alone or in combination. In some embodiments, the enzyme process may be utilized and an appropriate enzyme 14, for example, alpha-amylase, is introduced to liquefaction 10. Examples of alpha-amylases that may be used in the systems and processes of the present invention are described in U.S. Pat. No. 7,541,026; U.S. Patent Application Publication No. 2009/0209026; U.S. Patent Application Publication No. 2009/0238923; U.S. Patent Application Publication No. 2009/0252828; U.S. Patent Application Publication No. 2009/0314286; U.S. Patent Application Publication No. 2010/02278970; U.S. Patent Application Publication No. 2010/0048446; U.S. Patent Application Publication No. 2010/0021587, the entire contents of each are herein incorporated by reference.
[0073] In some embodiments, the enzymes for liquefaction and/or saccharification may be produced by the microorganism. Examples of microorganisms producing such enzymes are described in U.S. Pat. No. 7,498,159; U.S. Patent Application Publication No. 2012/0003701; U.S. Patent Application Publication No. 2012/0129229; PCT International Publication No. WO 2010/096562; and PCT International Publication No. WO 2011/153516, the entire contents of each are herein incorporated by reference. In some embodiments, enzymes for liquefaction and/or saccharification may be expressed by a microorganism that also produces a product alcohol. In some embodiments, enzymes for liquefaction and/or saccharification may be expressed by a microorganism that also expresses a butanol biosynthetic pathway.
[0074] The process of liquefying feedstock 12 creates feedstock slurry 16 (also referred to as mash or thick mash) that includes fermentable carbon source (e.g., sugar) and undissolved solids. In some embodiments, feedstock slurry 16 may include fermentable carbon source (e.g., sugar), oil, and undissolved solids. The undissolved solids may be non-fermentable portions of feedstock 12. In some embodiments, feedstock 12 may be corn, such as dry milled, unfractionated corn kernels, and feedstock slurry 16 is corn mash slurry. Feedstock slurry 16 may be discharged from an outlet of liquefaction 10, and may be conducted to separation 20.
[0075] Separation 20 has an inlet for receiving feedstock slurry 16, and may be configured to remove undissolved solids from feedstock slurry 16. Separation 20 may also be configured to remove oil, and/or oil and undissolved solids. Separation 20 may agitate or spin feedstock slurry 16 to create a liquid phase or aqueous solution 22 and a solid phase or wet cake 24.
[0076] Aqueous solution 22 may include sugar, for example, in the form of oligosaccharides, and water. Aqueous solution 22 may comprise at least about 10% by weight oligosaccharides, at least about 20% by weight of oligosaccharides, or at least about 30% by weight of oligosaccharides. Aqueous solution 22 may be discharged from separation 20 via an outlet. In some embodiments, the outlet may be located near the top of separation 20.
[0077] Wet cake 24 may include undissolved solids. Wet cake 24 may be discharged from separation 20 via an outlet. In some embodiments, the outlet may be located near the bottom of separation 20. Wet cake 24 may also include a portion of sugar and water. Wet cake 24 may be washed with additional water in separation 20 after aqueous solution 22 has been discharged from separation 20. Alternatively, wet cake 24 may be washed with additional water by additional separation devices. Washing wet cake 24 will recover the sugar (e.g., oligosaccharides) present in the wet cake, and the recovered sugar and water may be recycled to liquefaction 10. After washing, wet cake 24 may be further processed to form Dried Distillers' Grains with Solubles (DDGS) through any suitable known process. The formation of the DDGS from wet cake 24 formed in separation 20 has several benefits. Since the undissolved solids do not go to the fermentor, DDGS is not subjected to the conditions of the fermentor. For example, DDGS does not contact the microorganisms present in the fermentor or any other substances that may be present in the fermentor (e.g., extractant and/or product alcohol) and therefore, the microorganism and/or other substances are not trapped in the DDGS. These effects provide benefits to subsequent processing and selling of DDGS, for example, as animal feed.
[0078] Separation 20 may be any conventional separation device utilized in the industry, including, for example, centrifuges such as a decanter bowl centrifuge, three-phase centrifugation, disk stack centrifuge, filtering centrifuge, or decanter centrifuge. In some embodiments, removal of the undissolved solids from feedstock slurry 16 may be accomplished by filtration, vacuum filtration, belt filter, pressure filtration, membrane filtration, filtration using a screen, screen separation, grates or grating, porous grating, flotation, hydrocyclone, filter press, screwpress, gravity settler, vortex separator, or any method or device that may be used to separate solids from liquids. In some embodiments, separation 20 is a single step process. In one embodiment, undissolved solids may be removed from feedstock slurry 16 to form two product streams, for example, an aqueous solution of oligosaccharides which contains a lower concentration of solids as compared to feedstock slurry 16 and a wet cake which contains a higher concentration of solids as compared to feedstock slurry 16. In addition, a third stream containing oil may be generated, for example, if three-phase centrifugation is utilized for solids removal from feedstock slurry 16. As such, a number of product streams may be generated by using different separation techniques or combinations thereof.
[0079] As mentioned, a three-phase centrifuge may be used for three-phase separation of feedstock slurry such as separation of the feedstock slurry to generate two liquid phases (e.g., aqueous stream and oil stream) and a solid phase (e.g., solids or wet cake) (see, e.g., Flottweg Tricanter.RTM., Flottweg AG, Vilsibiburg, Germany). The two liquid phases may be separated and decanted, for example, from the bowl of the centrifuge via two discharge systems to prevent cross contamination and the solids phase may be removed via a separate discharge system.
[0080] In some embodiments using corn as feedstock, a three-phase centrifuge may be used to remove solids and corn oil simultaneously from liquefied corn mash. The solids may be the undissolved solids remaining after starch is hydrolyzed to soluble oligosaccharides during liquefaction. The corn oil may be released from the germ of the corn kernel during grinding and/or liquefaction. In some embodiments, the three-phase centrifuge may have one feed stream and three outlet streams. The feed stream may consist of liquefied corn mash produced during liquefaction. The mash may consist of an aqueous solution of oligosaccharides (e.g., liquefied starch); undissolved solids which consist of insoluble, non-starch components from the corn; and corn oil which consists of glycerides and free fatty acids. The three outlet streams from the three-phase centrifuge may be a wet cake which contains most of the undissolved solids from the mash; a heavy centrate stream which contains most of the liquefied starch from the mash; and a light centrate stream which contains most of the corn oil from the mash. The heavy centrate stream may be fed to fermentation. The wet cake may be washed with process recycle water, such as evaporator condensate and/or backset as described herein, to recover soluble starch from the wet cake. The light centrate stream may be sold as a co-product, converted to another co-product, or used in processing such as the case in converting the corn oil to corn oil fatty acids (COFA). In some embodiments, COFA may be used as an extractant.
[0081] Referring to FIG. 1, fermentation 30 (or fermentor 30), configured to ferment aqueous solution 22 to produce a product alcohol, has an inlet for receiving aqueous solution 22. Fermentation 30 may be any suitable fermentor known in the art. Fermentation 30 may include fermentation broth. In some embodiments, simultaneous saccharification and fermentation (SSF) may occur inside fermentation 30. Any known saccharification process utilized by the industry may be used including, but not limited to, an acid process, an enzyme process, or an acid-enzyme process. In some embodiments, enzyme 38 (e.g., such as glucoamylase) may be introduced to an inlet in fermentation 30 in order to hydrolyze oligosaccharides in aqueous solution 22 forming monosaccharides. Examples of glucoamylases that may be used in the systems and processes of the present invention are described in U.S. Pat. No. 7,413,887; U.S. Pat. No. 7,723,079; U.S. Patent Application Publication No. 2009/0275080; U.S. Patent Application Publication No. 2010/0267114; U.S. Patent Application Publication No. 2011/0014681; and U.S. Patent Application Publication No. 2011/0020899, the entire contents of each are herein incorporated by reference. In some embodiments, glucoamylase may be expressed by the microorganism. In some embodiments, glucoamylase may be expressed by a microorganism that also produces a product alcohol. In some embodiments, glucoamylase may be expressed by a microorganism that also expresses a butanol biosynthetic pathway.
[0082] In some embodiments, enzymes such as glucoamylases may be added to liquefaction. The addition of enzymes such as glucoamylases to liquefaction may reduce the viscosity of the feedstock slurry or liquefied mash and may improve separation efficiency. In some embodiments, any enzyme capable of reducing the viscosity of the feedstock slurry may be used (e.g., Viscozyme.RTM., Sigma-Aldrich, St. Louis, Mo.). Viscosity of the feedstock may be measured by any method known in the art (e.g., viscometers, rheometers).
[0083] Microorganism 32 may be introduced to fermentation 30. In some embodiments, microorganism 32 may be included in the fermentation broth. In some embodiments, microorganism 32 may be propagated in a separate vessel or tank (e.g., propagation tank). In some embodiments, microorganisms from the propagation tank may be used to inoculate one or more fermentors. In some embodiments, one or more propagation tanks may be used in the processes and systems described herein. In some embodiments, the propagation tank may be about 2% to about 5% the size of the fermentor. In some embodiments, the propagation tank may comprise one or more of the following mash, water, enzymes, nutrients, and microorganisms. In some embodiments, product alcohol may be produced in the propagation tank.
[0084] In some embodiments, microorganism 32 may be bacteria, cyanobacteria, filamentous fungi, or yeast. In some embodiments, microorganism 32 metabolizes the sugar in aqueous solution 22 and produces product alcohol. In some embodiments, microorganism 32 may be a recombinant microorganism. In some embodiments, microorganism 32 may be immobilized, such as by adsorption, covalent bonding, crosslinking, entrapment, and encapsulation. Methods for encapsulating cells are known in the art, for example, as described in U.S. Patent Application Publication No. 2011/0306116, which is incorporated herein by reference.
[0085] In some embodiments, in situ product removal (ISPR) may be utilized to remove product alcohol from fermentation 30 as the product alcohol is produced by microorganism 32. In some embodiments, liquid-liquid extraction may be utilized for ISPR. In some embodiments, fermentation 30 may have an inlet for receiving extractant 34. In some embodiments, extractant 34 may be added to the fermentation broth downstream of fermentation 30. Alternative means of additions of extractant 34 to fermentation 30 or downstream of fermentation 30 are represented by the dotted lines. In some embodiments, ISPR may be conducted in a propagation tank. In some embodiments, ISPR may be conducted in the fermentor and the propagation tank. In some embodiments, ISPR may be performed at the initiation (e.g., time 0) of fermentation and/or propagation. By initiating ISPR at the beginning of fermentation and/or propagation, the concentration of product alcohol in the fermentor and propagation tank may be maintained at low levels, and thereby minimize the effects of product alcohol on the microorganism and allowing the microorganism to achieve increased cell mass. Examples of liquid-liquid extraction are described herein. Processes for producing and recovering alcohols from fermentation broth using extractive fermentation are described in U.S. Patent Application Publication No. 2009/0305370; U.S. Patent Application Publication No. 2010/0221802; U.S. Patent Application Publication No. 2011/0097773; U.S. Patent Application Publication No. 2011/0312044; U.S. Patent Application Publication No. 2011/0312043; and PCT International Publication No. WO 2011/159998; the entire contents of each are herein incorporated by reference.
[0086] Extractant 34 contacts the fermentation broth forming stream 36 comprising, for example, a biphasic mixture (e.g., extractant-rich phase with product alcohol and aqueous phase depleted of product alcohol). In some embodiments, stream 36 may be a quartphasic mixture comprising, for example, a vapor phase, an organic phase, an aqueous phase, and a solid phase. Product alcohol in the fermentation broth is transferred to extractant 34. In some embodiments, stream 36 may be discharged through an outlet in fermentation 30. Product alcohol may be separated from the extractant in stream 36 using conventional techniques.
[0087] In some embodiments, fermentor internals or devices may be used to improve phase separation between fermentation broth and extractant. For example, the internal or device may serve as a coalescer to promote phase separation between fermentation broth and extractant and/or act as a physical barrier to improve phase separation. These fermentor internals or devices may also prevent solids from settling in the extractant phase (or layer), promote coalescensce of aqueous droplets that may be entrained in the extractant layer, and promote removal of off-gases (e.g., CO.sub.2, air), and thereby minimize disturbance of the extractant phase and/or liquid-liquid interface. Examples of internals or devices that may be used in the processes and systems described herein include, but are not limited to, baffles, perforated plates, deep wells, lamella separators, cones, and the like. In some embodiments, the perforated plate may be a flat horizontal perforated plate. In some embodiments, the cone may be an inverted cone or concentric cone(s). In some embodiments, the internals may be rotating. In some embodiments, the internals or devices may be located at or about the level of the liquid-liquid interface of fermentation broth and extractant.
[0088] In some embodiments prior to ISPR and/or completion of fermentation, stream 35 may be discharged from an outlet in fermentation 30. Discharged stream 35 may include microorganism 32. Microorganism 32 may be separated from stream 35, for example, by centrifugation or membrane filtration. In some embodiments, by removing the microorganism prior to addition of extractant to the fermentation broth, the microorganism is not exposed to the extractant and therefore, not exposed to any negative impact that the extractant may have on the microorganism. In addition, by removing the microorganism upstream of the extraction process, a more aggressive extraction process (e.g., heating or cooling the mixture to enhance separation, using a higher K.sub.D and/or higher selectivity extractant, or an extractant with improved properties but lower biocompatibility) may be employed to recover the product alcohol. In some embodiments, microorganism 32 may be recycled to fermentation 30 which can increase the production rate of product alcohol, thereby resulting in an increase in the efficiency of product alcohol production.
[0089] Referring to FIG. 2, in some embodiments, ISPR may be conducted downstream of fermentation 30. In some embodiments, stream 33 including product alcohol and microorganism 32 may be discharged from an outlet in fermentation 30 and conducted downstream, for example, to an extraction column for recovery of product alcohol. In some embodiments, stream 33 may be processed by separating microorganism 32 prior to ISPR. For example, removal of microorganism 32 from stream 33 may be accomplished by centrifugation, filtration, vacuum filtration, belt filter, pressure filtration, membrane filtration, filtration using a screen, screen separation, grates or grating, porous grating, flotation, hydrocyclone, filter press, screwpress, gravity settler, vortex separator, or any method or separation device that may be used to separate solids (e.g., microorganisms) from liquids. Following removal of microorganism 32, stream 33 may be conducted to an extraction column for recovery of product alcohol.
[0090] Additional embodiments of the processes and systems described herein are illustrated in FIGS. 3 to 6. FIGS. 3 to 6, including the options for the addition of extractant to the fermentor (e.g., generating stream 36) or extraction conducted downstream of the fermentor (e.g., generating stream 33), are similar to FIGS. 1 and 2, respectively, and therefore will not be described in detail again.
[0091] Referring to FIG. 3, the systems and processes of the present invention may include discharging oil 26 from an outlet of separation 20. Feedstock slurry 16 may be separated into a first liquid phase or aqueous solution 22 comprising a fermentable sugar, a solid phase or wet cake 24 comprising undissolved solids, and a second liquid phase comprising oil 26 which may exit separation 20. In some embodiments, feedstock 12 is corn and oil 26 is corn oil. In some embodiments, oil 26 may be conducted to a storage tank or any unit that is suitable for oil storage. Any suitable separation device may be used to discharge aqueous solution 22, wet cake 24, and oil 26, for example, a three-phase centrifuge. In some embodiments, a portion of the oil from feedstock 12 such as corn oil when the feedstock is corn, remains in wet cake 24. In some embodiments, when oil 26 is removed via separation 20 from feedstock 12 (e.g., corn), the fermentation broth in fermentation 30 includes a reduced amount of corn oil.
[0092] As described herein, in some embodiments, oil may be separated from the feedstock or feedstock slurry and may be stored in an oil storage unit. For example, oil may be separated from the feedstock or feedstock slurry using any suitable means for separation including a three-phase centrifuge or mechanical extraction. To improve the removal of oil from the feedstock or feedstock slurry, oil extraction aids such surfactants, anti-emulsifiers, or flocculents as well as enzymes may be utilized. Examples of oil extraction aids include, but are not limited to, non-polymeric, liquid surfactants; talcum powder; microtalcum powder; salts (NaOH); calcium carbonate; and enzymes such as Pectinex.RTM. Ultra SP-L, Celluclast.RTM., and Viscozyme.RTM. L (Sigma-Aldrich, St. Louis, Mo.), and NZ 33095 (Novozymes, Franklinton, N.C.).
[0093] As illustrated in FIG. 4, if oil is not discharged separately it may be removed with wet cake 24. When wet cake 24 is removed via separation 20, in some embodiments, a portion of the oil from feedstock 12, such as corn oil when the feedstock is corn, remains in wet cake 24. Wet cake 24 may be conducted to mix 60 and combined with water or other solvents forming wet cake mixture 65. In some embodiments, water may be fresh water, backset, cook water, process water, lutter water, evaporation water, or any water source available in the fermentation processing facility, or any combination thereof. Wet cake mixture 65 may be conducted to separation 70 producing wash centrate 75 comprising fermentable sugars recovered from wet cake 24, and wet cake 74. Wash centrate 75 may be recycled to liquefaction 10.
[0094] In some embodiments, separation 70 may be any separation device capable of separating solids and liquids including, for example, decanter bowl centrifugation, three-phase centrifugation, disk stack centrifugation, filtering centrifugation, decanter centrifugation, filtration, vacuum filtration, belt filter, pressure filtration, membrane filtration, filtration using a screen, screen separation, grating, porous grating, flotation, hydrocyclone, filter press, screwpress, gravity settler, vortex separator, or combinations thereof.
[0095] In some embodiments, wet cake may be subjected to one or more wash cycles or wash systems. For example, wet cake 74 may be further processed by conducting wet cake 74 to a second wash system. In some embodiments, wet cake 74 may be conducted to a second mix 60' forming wet cake mixture 65'. Wet cake mixture 65' may be conducted to a second separation 70' producing wash centrate 75' and wet cake 74'. Wash centrate 75' may be recycled to liquefaction 10, and wet cake 74' may be combined with wet cake 74 for further processing as described herein. In some embodiments, separation 70' may be any separation device capable of separating solids and liquids including, for example, decanter bowl centrifugation, three-phase centrifugation, disk stack centrifugation, filtering centrifugation, decanter centrifugation, filtration, vacuum filtration, belt filter, pressure filtration, membrane filtration, filtration using a screen, screen separation, grating, porous grating, flotation, hydrocyclone, filter press, screwpress, gravity settler, vortex separator, or combination thereof. In some embodiments, the wet cake may be subjected to one, two, three, four, five or more wash cycles or wash systems.
[0096] Wet cake 74 may be combined with syrup and then dried to form DDGS through any suitable known process. The formation of the DDGS from wet cake 74 has several benefits. Since the undissolved solids do not go to the fermentor, the DDGS does not have trapped extractant and/or product alcohol, it is not subjected to the conditions of the fermentor, and it does not contact the microorganisms present in the fermentor. These benefits make it easier to process DDGS, for example, as animal feed.
[0097] In some embodiments, a portion of undissolved solids may be conducted to fermentation 30. In some embodiments, this portion of undissolved solids may have smaller particle sizes (e.g., fines). In some embodiments, this portion of undissolved solids may form whole stillage. In some embodiments, this whole stillage may be processed to form thin stillage and a wet cake. In some embodiments, the wet cake formed from whole stillage and wet cake 74 and/or 74' may be combined and further processed to produce DDGS.
[0098] As shown in FIG. 4, oil is not discharged separately from the wet cake, but rather oil is included as part of the wet cake and is ultimately present in the DDGS. If corn is utilized as feedstock, corn oil contains triglycerides, diglycerides, monoglycerides, fatty acids, and phospholipids, which provide a source of metabolizable energy for animals. The presence of oil (e.g., corn oil) in the wet cake and ultimately DDGS may provide a desirable animal feed, for example, a high fat content animal feed.
[0099] In some embodiments, oil may be separated from wet cake and DDGS and converted to an ISPR extractant for subsequent use in the same or different alcohol fermentation processes. Methods for deriving extractants from biomass are described in U.S. Patent Application Publication No. 2011/0312044 and PCT International Publication No. WO 2011/159998; the entire contents of each are herein incorporated by reference. Oil may be separated from wet cake and DDGS using any suitable process including, for example, a solvent extraction process. In one embodiment of the invention, wet cake or DDGS may be added to an extraction unit and washed with a solvent such as hexane to remove oil. Other solvents that may be utilized include, for example, butanol, isohexane, ethanol, petroleum distillates such as petroleum ether, or mixtures thereof. Following oil extraction, wet cake or DDGS may be treated to remove any residual solvent. For example, wet cake or DDGS may be heated to vaporize any residual solvent using any method known in the art. Following solvent removal, wet cake or DDGS may be subjected to a drying process to remove any residual water. The processed wet cake may be used to generate DDGS. The processed DDGS may be used as a feed supplement for animals such as dairy and beef cattle, poultry, swine, livestock, equine, aquaculture, and domestic pets.
[0100] In some embodiments, extractant may be used as a means to modify the color of the wet cake. For example, feedstocks such as corn contain pigments (e.g., xanthophylls) which may be used as a coloring agent in food products including animal feeds (e.g., poultry feed). Exposure to extractants can modify these pigments resulting in a wet cake that is, for example, lighter in color. A lighter color wet cake may produce DDGS with a lighter color, which may be a desirable quality for certain animal feeds.
[0101] In some embodiments, where corn is used as the feedstock, xanthophylls may be isolated from corn and/or undissolved solids and used as a pigment ingredient in DDGS or animal feed, or as a supplement for pharmaceutical and nutraceutical applications. Methods for isolating xanthophylls include, but are not limited to, chromatography such as size exclusion chromatography, solvent extraction such as ethanol extraction, and enzyme treatment such as alcalase hydrolysis (see, e.g., Tsui, et al., J. Food Eng. 83:590-595, 2007; Li, et al., Food Science 31: 72-77, 2010: U.S. Pat. No. 5,648,564; U.S. Pat. No. 6,169,217; U.S. Pat. No. 6,329,557; U.S. Pat. No. 8,236,929; the entire contents of each are herein incorporated by reference). In some embodiments, xanthophylls may be isolated from corn and/or undissolved solids and added to COFA. In some embodiments, COFA and/or xanthophylls may be used for food, pharmaceutical, and nutraceutical applications.
[0102] After extraction from wet cake or DDGS, the resulting oil and solvent mixture may be collected for separation of oil and solvent. In one embodiment, the oil/solvent mixture may be processed by evaporation whereby the solvent is evaporated and may be collected and recycled. The recovered oil may be converted to an ISPR extractant for subsequent use in the same or different alcohol fermentation processes.
[0103] Removal of the oil component of the feedstock is advantageous to product alcohol production because oil present in the fermentor can break down into fatty acids and glycerin. Glycerin can accumulate in water and reduce the amount of water that is available for recycling throughout the system. Thus, removal of the oil component of feedstock can increase the efficiency of product alcohol production by increasing the amount of water that can be recycled through the system.
[0104] Referring to FIG. 5, oil may be removed at various points during the processes described herein. Feedstock slurry 16 may be separated, for example, using a three-phase centrifuge, into a first liquid phase or aqueous solution 22, a second liquid phase comprising oil 26, and a solid phase or wet cake 24. Wet cake 24 may be further processed to recover fermentable sugars and oil. Wet cake 24 may be conducted to mix 60 and combined with water or other solvents forming wet cake mixture 65. In some embodiments, water may be backset, cook water, process water, lutter water, water collected from evaporation, or any water source available in the fermentation processing facility, or any combination thereof. Wet cake mixture 65 may be conducted to separation 70 (e.g., three-phase centrifuge) producing wash centrate 75 comprising fermentable sugars, oil stream 76, and wet cake 74. Wash centrate 75 may be recycled to liquefaction 10.
[0105] As described herein, wet cake may be subjected to one or more wash cycles or wash systems. In some embodiments, wet cake 74 may be conducted to a second mix 60' forming wet cake mixture 65'. Wet cake mixture 65' may be conducted to a second separation 70' producing wash centrate 75', oil stream 76' and wet cake 74'. Wash centrate 75' may be recycled to liquefaction 10, and wet cake 74' may be combined with wet cake 74 for further processing as described below. Oil stream 76' and oil 26 may be combined and further processed for the generation of extractant that may be used in the fermentation process or oil stream 76' and oil 26 may be combined and further processed for the manufacture of consumer products.
[0106] Wet cake 74 may be combined with syrup and then dried to form Dried Distillers' Grains with Solubles (DDGS) utilizing any suitable process. The formation of DDGS from wet cake 74 has several benefits. Since the undissolved solids do not go to the fermentor, the DDGS does not contain extractant and/or product alcohol, it is not subjected to the conditions of the fermentor, and it does not contact the microorganisms present in the fermentor. These benefits make it easier to process DDGS, for example, as animal feed. As described above, in some embodiments, wet cake 74, 74', and wet cake formed from whole stillage may be combined and further processed to produce DDGS.
[0107] As illustrated in FIG. 6A, aqueous solution 22 and wet cake 24 may be combined, cooled, and conducted to fermentation 30. Feedstock slurry 16 may be separated, for example, using a three-phase centrifuge, into a first liquid phase or aqueous solution 22, a second liquid phase comprising oil 26, and a solid phase or wet cake 24. In some embodiments, oil 26 may be conducted to a storage tank or any unit that is suitable for oil storage. Aqueous solution 22 and wet cake 24 may be conducted to mix 80 and re-slurried forming aqueous solution/wet cake mixture 82. Mixture 82 may be conducted to cooler 90 producing cooled mixture 92 which may be conducted to fermentation 30. In some embodiments, when oil 26 is removed via separation 20 from feedstock 12, mixtures 82 and 92 include a reduced amount of oil.
[0108] In another embodiment, as illustrated in FIG. 6B, feedstock slurry 16 may be separated using a three-phase centrifuge to generate a first liquid phase or aqueous solution 22, a second liquid phase comprising oil 26, and a solid phase or wet cake 24. Aqueous solution 22, wet cake 24, and oil 26, or portions thereof, may be conducted to fermentation 30. In some embodiments, aqueous solution 22, wet cake 24, and oil 26, or portions thereof, may be combined, for example, by mixing, forming an aqueous solution, wet cake, and oil mixture, and the mixture may be conducted to fermentation 30. In some embodiments, aqueous solution 22 and wet cake 24 may be combined forming an aqueous solution and wet cake mixture, then oil 26 may be added to the mixture forming an aqueous solution, wet cake, and oil mixture and this mixture may be conducted to fermentation 30. In some embodiments, aqueous solution 22 and wet cake 24 may be combined forming an aqueous solution and wet cake mixture, and this mixture and oil 26, or a portion thereof, may be conducted to fermentation 30 as separate streams.
[0109] In additional embodiments of the processes and systems described herein, saccharification may occur in a separate saccharification system. In some embodiments, a saccharification system may be located between liquefaction 10 and separation 20 or between separation 20 and fermentation 30. In some embodiments, liquefaction and/or saccharification may be conducted utilizing raw starch enzymes or low temperature hydrolysis enzymes such as Stargen.TM. (Genencor International, Palo Alto, Calif.) and BPX.TM. (Novozymes, Franklinton, N.C.). In some embodiments, feedstock slurry may be subjected to raw starch hydrolysis (also known as cold cooking or cold hydrolysis).
[0110] In some embodiments, the systems and processes of the present invention may include a series of two or more separation devices (e.g., centrifuges) for the removal of undissolved solids and/or oil. For example, aqueous solution discharged from a first separation unit may be conducted to an inlet of a second separation unit. The first separation unit and second separation unit may be identical (e.g., two three-phase centrifuges) or may be different (e.g., a three-phase centrifuge and a decanter centrifuge). Separation may be accomplished by a number of means including, but not limited to, decanter bowl centrifugation, three-phase centrifugation, disk stack centrifugation, filtering centrifugation, decanter centrifugation, filtration, vacuum filtration, belt filter, pressure filtration, membrane filtration, filtration using a screen, screen separation, grating, porous grating, flotation, hydrocyclone, filter press, screwpress, gravity settler, vortex separator, or combinations thereof.
[0111] The absence or minimization of undissolved solids in the fermentation broth has several benefits. For example, the need for units of operation in the downstream processing may be eliminated, thereby resulting in an increased efficiency for product alcohol production. Also, some or all of the centrifuges used to process whole stillage may be eliminated as a result of less undissolved solids in the fermentation broth exiting the fermentor. Removal of undissolved solids from feedstock slurry may improve the processing productivity of biomass and cost effectiveness. Improved productivity may include increased efficiency of product alcohol production and/or increased extraction activity relative to processes and systems that do not remove undissolved solids prior to fermentation. For additional description of processes and systems for separating undissolved solids from feedstock slurry see, for example, U.S. Patent Application Publication No. 2012/0164302 and U.S. Provisional Patent Application No. 61/674,607, the entire contents of each are herein incorporated by reference.
[0112] As described herein, product alcohol may be recovered from fermentation broth using a number of methods including liquid-liquid extraction. In some embodiments of the processes and systems described herein, an extractant may be used to recover product alcohol from fermentation broth. Extractants used herein may have, for example, one or more of the following properties and/or characteristics: (i) biocompatible with the microorganisms, (ii) immiscible with the fermentation broth, (iii) a high partition coefficient (Kp) for the extraction of product alcohol, (iv) a low partition coefficient for the extraction of nutrients, (v) low viscosity (.mu.), (vi) high selectivity for product alcohol as compared to, for example, water, (vii) low density (.rho.) relative to the fermentation broth or a density that is different as compared to the density of the fermentation broth, (viii) a boiling point suitable for downstream processing of the extractant and product alcohol, (ix) a melting point lower than ambient temperature, (x) minimal solubility in solids, (xi) a low tendency to form emulsions with the fermentation broth, (xii) stability throughout the fermentation process, (xiii) low cost, and (xiv) nonhazardous.
[0113] In some embodiments, the extractant may be selected based upon certain properties and/or characteristics as described herein. For example, viscosity of the extractant can influence the mass transfer properties of the system, that is, the efficiency with which the product alcohol may be extracted from the aqueous phase to the extractant phase (i.e., organic phase). The density of the extractant can affect phase separation. In some embodiments, selectivity refers to the relative amounts of product alcohol to water taken up by the extractant. The boiling point can affect the cost and method of product alcohol recovery. For example, in the case where butanol is recovered from the extractant phase by distillation, the boiling point of the extractant should be sufficiently low as to enable separation of butanol while minimizing any thermal degradation or side reactions of the extractant, or the need for vacuum in the distillation process.
[0114] The extractant may be biocompatible with the microorganism, that is, nontoxic to the microorganism or toxic only to such an extent that the microorganism is impaired to an acceptable level. In some embodiments, biocompatible refers to the measure of the ability of a microorganism to utilize fermentable carbon sources in the presence of an extractant. The extent of biocompatibility of an extractant may be determined, for example, by the glucose utilization rate of the microorganism in the presence of the extractant and product alcohol. In some embodiments, a non-biocompatible extractant refers to an extractant that interferes with the ability of a microorganism to utilize fermentable carbon sources. For example, a non-biocompatible extractant does not permit the microorganism to utilize glucose at a rate greater than about 25%, greater than about 30%, greater than about 35%, greater than about 40%, greater than about 45%, or greater than about 50% of the rate when the extractant is not present.
[0115] One skilled in the art may select an extractant to maximize the desired properties and/or characteristics as described herein and to optimize recovery of a product alcohol. One of skill in the art can also appreciate that it may be advantageous to use a mixture of extractants. For example, extractant mixtures may be used to increase the partition coefficient for the product alcohol. Additionally, extractant mixtures may be used to adjust and optimize physical characteristics of the extractant, such as the density, boiling point, and viscosity. For example, the appropriate combination may provide an extractant which has a sufficient partition coefficient for the product alcohol and sufficient biocompatibility to enable its economical use for removing product alcohol from fermentative broth.
[0116] In some embodiments, extractants useful in the processes and systems described herein may be organic solvents. In some embodiments, extractants useful in the processes and systems described herein may be water-immiscible organic solvents. The extractant may be an organic extractant selected from the group consisting of saturated, mono-unsaturated, poly-unsaturated C.sub.12 to C.sub.22 fatty alcohols, C.sub.12 to C.sub.22 fatty acids, esters of C.sub.12 to C.sub.22 fatty acids, C.sub.12 to C.sub.22 fatty aldehydes, C.sub.12 to C.sub.22 fatty amides, and mixtures thereof. The extractant may also be an organic extractant selected from the group consisting of saturated, mono-unsaturated, poly-unsaturated C.sub.4 to C.sub.22 fatty alcohols, C.sub.4 to C.sub.28 fatty acids, esters of C.sub.4 to C.sub.28 fatty acids, C.sub.4 to C.sub.22 fatty aldehydes, and mixtures thereof. In some embodiments, the extractant may include a first extractant selected from C.sub.12 to C.sub.22 fatty alcohols, C.sub.12 to C.sub.22 fatty acids, esters of C.sub.12 to C.sub.22 fatty acids, C.sub.12 to C.sub.22 fatty aldehydes, C.sub.12 to C.sub.22 fatty amides, and mixtures thereof; and a second extractant selected from C.sub.7 to C.sub.11 fatty alcohols, C.sub.7 to C.sub.11 fatty acids, esters of C.sub.7 to C.sub.11 fatty acids, C.sub.7 to C.sub.11 fatty aldehydes, and mixtures thereof. In some embodiments, the extractant may comprise carboxylic acids. In some embodiments, the extractant may be an organic extractant such as oleyl alcohol, behenyl alcohol, cetyl alcohol, lauryl alcohol (also referred to as 1-dodecanol), myristyl alcohol, stearyl alcohol, oleic acid, lauric acid, linoleic acid, linolenic acid, myristic acid, stearic acid, octanoic acid, decanoic acid, undecanoic acid, methyl myristate, methyl oleate, 1-nonanol, 1-decanol, 2-undecanol, 1-nonanal, 1-undecanol, undecanal, lauric aldehyde, 2-methylundecanal, oleamide, linoleamide, palmitamide, stearylamide, 2-ethyl-1-hexanol, 2-hexyl-1-decanol, 2-octyl-1-dodecanol, and mixtures thereof.
[0117] In some embodiments, the extractant may be a mixture of biocompatible and non-biocompatible extractants. Examples of mixtures of biocompatible and non-biocompatible extractants include, but are not limited to, oleyl alcohol and nonanol, oleyl alcohol and 1-undecanol, oleyl alcohol and 2-undecanol, oleyl alcohol and 1-nonanal, oleyl alcohol and decanol, and oleyl alcohol and dodecanol. Additional examples of biocompatible and non-biocompatible extractants are described in U.S. Patent Application Publication No. 2009/0305370 and U.S. Patent Application Publication No. 2011/0097773; the entire contents of each herein incorporated by reference. In some embodiments, biocompatible extractants may have high atmospheric boiling points. For example, biocompatible extractants may have atmospheric boiling points greater than the atmospheric boiling point of water.
[0118] In some embodiments, a hydrophilic solute may be added to fermentation broth that is contacted with an extractant. The presence of a hydrophilic solute in the aqueous phase may improve phase separation and may increase the fraction of product alcohol that partitions into the organic phase. Examples of a hydrophilic solute may include, but are not limited to, polyhydroxylated compounds, polycarboxylic compounds, polyol compounds, and dissociating ionic salts. Sugars such as glucose, fructose, sucrose, maltose, and oligosaccharides may serve as a hydrophilic solute. Other polyhydroxylated compounds may include glycerol, ethylene glycol, propanediol, polyglycerol, and hydroxylated fullerene. Polycarboxylic compounds may include citric acid, tartaric acid, maleic acid, succinic acid, polyacrylic acid, and sodium, potassium, or ammonium salts thereof. Ionic salts that may be used as a hydrophilic solute in fermentation broth comprise cations that include sodium, potassium, ammonium, magnesium, calcium, and zinc; and anions that include sulfate, phosphate, chloride, and nitrate. The amount of hydrophilic solute in the fermentation broth may be selected by one skilled in the art to maximize the transfer of product alcohol from the aqueous phase (e.g., fermentation broth) to the organic phase (e.g., extractant) while not having a negative impact on the growth and/or productivity of the product alcohol-producing microorganism. High levels of hydrophilic solute may impose osmotic stress and/or toxicity on the microorganism. One skilled in the art may use any number of known methods to determine an optimal amount of hydrophilic solute to minimize the effects of osmotic stress and/or toxicity on the microorganism.
[0119] In some embodiments where the product alcohol is butanol, the extractant may be selected for attracting the alkyl portion of butanol and for providing little or no affinity to water. An extractant that offers no hydrogen bonding, for example, to water will absorb the alcohol selectively. In some embodiments, the extractant may comprise an aromatic compound. In some embodiments, the extractant may comprise alkyl substituted benzenes including, but not limited to, cumene, para-cymene (also known as 1-methyl-4-(1-methylethyl)benzene), meta-cymene (also known as 1-methyl-3-(1-methylethyl)benzene), meta-diisopropylbenzene, para-diisopropylbenzene, triethylbenzene, ethyl butyl benzene, and tert-butylstyrene. An advantage of using an alkyl-substituted benzene is the comparatively higher butanol affinity relative to other hydrocarbons. In addition, isopropyl-substituted or isobutyl-substituted benzenes may offer a particular advantage in butanol affinity over other substituted benzenes. Another advantage is the lower viscosity, lower surface tension, lower density, higher thermal stability, and higher chemical stability that aids in phase separability and long-term reuse. In some embodiments, an extractant that attracts the alkyl portion of butanol may be combined with another extractant that offers affinity in the form of hydrogen bonding, for example, to the hydroxyl portion of butanol such that the mixture provides an optimal balance between selectivity and partitioning over water. In some embodiments, an extractant containing butanol may be phase separated from fermentation broth and distilled in a column operating under vacuum. This distillation may operate with reflux in order to maintain a distillate of high purity butanol that contains very little extractant. The bottoms may comprise a portion of the butanol contained in the distillation feed such that the reboiling temperature under vacuum is suitable for delivering heat indirectly from available steam. Distillation may be carried out with a partial condenser where only reflux liquid is condensed, and a vapor distillate of substantially butanol composition may be directed into the bottom of a rectification column that is simultaneously fed a butanol stream decanted from condensed beer column overhead vapor. An advantage of this type of distillation is that the need for a reboiler to purify the decanted butanol stream is eliminated by heat integrating the vapor generated from stripping butanol out of the extractant.
[0120] In some embodiments, extractant may be generated from feedstock. For example, oils such as corn oil present in feedstock may be used for the generation of extractant for extractive fermentation. The glycerides in oil may be chemically or enzymatically converted into a reaction product, such as fatty acids which may be used an extractant for the recovery of the product alcohol. Using corn oil as an example, corn oil triglycerides may be reacted with a base such as ammonia hydroxide or sodium hydroxide to obtain fatty amides, fatty acids, and glycerol. In some embodiments, oil in the feedstock may be hydrolyzed by a catalyst to generate fatty acids. In some embodiments, at least a portion of the acyl glycerides in oil may be hydrolyzed to carboxylic acid by contacting the oil with catalyst. In some embodiments, the resulting acid/oil composition includes monoglycerides and/or diglycerides from the partial hydrolysis of the acyl glycerides in the oil. In some embodiments, the resulting acid/oil composition includes glycerol, a by-product of acyl glyceride hydrolysis. In some embodiments, the resulting acid/oil composition includes lysophospholipids from the partial hydrolysis of phospholipids in the oil. Methods for deriving extractants from biomass are described in U.S. Patent Application Publication No. 2011/0312044 and PCT International Publication No. WO 2011/159998, the entire contents of which are all herein incorporated by reference.
[0121] In some embodiments, the hydrolysis of oil in the feedstock or feedstock slurry may occur in the fermentor by the addition of a catalyst to the fermentor. In some embodiments, the hydrolysis of oil in the feedstock or feedstock slurry may occur in a separate unit. For example, the feedstock or feedstock slurry may be conducted to a unit, and a catalyst such as lipase may be added to the unit, converting the oil present in the feedstock or feedstock slurry to fatty acids. In some embodiments, the feedstock or feedstock slurry comprising the fatty acids generated by hydrolysis may be added to the fermentor. In some embodiments, the fatty acids generated by hydrolysis of feedstock or feedstock slurry may be added to an external extractor or extractant column.
[0122] In some embodiments, oil may be separated from feedstock slurry and the oil may be conducted to a unit, and a catalyst such as lipase may be added to the unit, generating a fatty acid stream. The fatty acid stream may be heated to deactivate the lipase and then the fatty acid stream may conducted to an external extractor or a storage tank. Fatty acids from the storage tank may be conducted to an external extractor for extraction of product alcohol from fermentation broth. In some embodiments, oil separated from feedstock slurry may be stored in a storage tank. A catalyst such as lipase may be added to the storage tank, generating a fatty acid stream. The fatty acid stream may be heated to deactivate the lipase, cooled, and then conducted to an external extractor for extraction of product alcohol from fermentation broth. In some embodiments, oil separated from feedstock slurry may be conducted to a unit, and a catalyst such as lipase may be added to the unit, generating a fatty acid stream. The fatty acid stream may be heated to deactivate the lipase, cooled, and then the fatty acid stream may conducted to a fermentor.
[0123] In some embodiments, the one or more catalysts may be one or more enzymes, for example, hydrolase enzymes. In some embodiments, the one or more catalysts may be one or more enzymes, for example, lipase enzymes. Lipase enzymes may be derived from any source including, for example, Absidia, Achromobacter, Aeromonas, Alcaligenes, Alternaria, Aspergillus, Achromobacter, Aureobasidium, Bacillus, Beauveria, Brochothrix, Candida, Chromobacter, Coprinus, Fusarium, Geotricum, Hansenula, Humicola, Hyphozyma, Lactobacillus, Metarhizium, Mucor, Nectria, Neurospora, Paecilomyces, Penicillium, Pseudomonas, Rhizoctonia, Rhizomucor, Rhizopus, Rhodosporidium, Rhodotorula, Saccharomyces, Sus, Sporobolomyces, Thermomyces, Thiarosporella, Trichoderma, Verticillium, and/or Yarrowia. In some embodiments, the source of the lipase may be selected from the group consisting of Absidia blakesleena, Absidia corymbifera, Achromobacter iophagus, Alcaligenes sp., Alternaria brassiciola, Aspergillus flavus, Aspergillus niger, Aspergillus tubingensis, Aureobasidium pullulans, Bacillus coagulans, Bacillus pumilus, Bacillus strearothermophilus, Bacillus subtilis, Brochothrix thermosohata, Burkholderia cepacia, Candida cylindracea (Candida rugosa), Candida paralipolytica, Candida antarctica lipase A, Candida antarctica lipase B, Candida ernobii, Candida deformans, Candida rugosa, Candida parapsilosis, Chromobacter viscosum, Coprinus cinerius, Fusarium heterosporum, Fusarium oxysporum, Fusarium solani, Fusarium solani pisi, Fusarium roseum culmorum, Geotrichum candidum, Geotricum penicillatum, Hansenula anomala, Humicola brevispora, Humicola brevis var. thermoidea, Humicola insolens, Lactobacillus curvatus, Rhizopus oryzae, Mucor javanicus, Neurospora crassa, Nectria haematococca, Penicillium cyclopium, Penicillium crustosum, Penicillium expansum, Penicillium roqueforti, Penicillium camembertii, Penicillium sp. I, Penicillium sp. II, Pseudomonas aeruginosa, Pseudomonas alcaligenes, Pseudomonas cepacia (syn. Burkholderia cepacia), Pseudomonas fluorescens, Pseudomonas fragi, Pseudomonas maltophilia, Pseudomonas mendocina, Pseudomonas mephitica lipolytica, Pseudomonas alcaligenes, Pseudomonas plantari, Pseudomonas pseudoalcaligenes, Pseudomonas putida, Pseudomonas stutzeri, and Pseudomonas wisconsinensis, Rhizoctonia solani, Rhizomucor miehei, Rhizopus arrhizus, Rhizopus delemar, Rhizopus japonicus, Rhizopus microsporus, Rhizopus nodosus, Rhizopus oryzae, Rhodosporidium toruloides, Rhodotorula glutinis, Saccharomyces cerevisiae, Sporobolomyces shibatanus, Sus scrofa, Thermomyces lanuginosus (formerly Humicola lanuginose), Thiarosporella phaseolina, Trichoderma harzianum, Trichoderma reesei, and Yarrowia lipolytica.
[0124] In some embodiments, hydrolase and/or lipase may be expressed by the microorganism. In some embodiments, the microorganism may be engineered to express homologous or heterologous hydrolase and/or lipase. In some embodiments, hydrolase and/or lipase may be expressed by a microorganism that also produces a product alcohol. In some embodiments, hydrolase and/or lipase may be expressed by a microorganism that also expresses a butanol biosynthetic pathway.
[0125] Commercial lipase preparations suitable as a catalyst include, but are not limited to, Lipolase.RTM. 100 L, Lipex.RTM. 100L, Lipoclean.RTM. 2000T, Lipozyme.RTM. CALB L, Novozyme.RTM. CALA L, and Palatase 20000L, available from Novozymes (Franklinton, N.C.), or lipases from Pseudomonas fluorescens, Pseudomonas cepacia, Mucor miehei, hog pancreas, Candida cylindracea, Candida rugosa, Rhizopus niveus, Candida antarctica, Rhizopus arrhizus or Aspergillus available from Sigma Aldrich. In some embodiments, the lipase may be thermostable and/or thermotolerant, and/or solvent tolerant.
[0126] In some embodiments, the one or more catalysts may be phospholipases. A phospholipase useful in the present invention may be obtained from a variety of biological sources, for example, but not limited to, filamentous fungal species within the genus Fusarium, such as a strain of Fusarium culmorum, Fusarium heterosporum, Fusarium solani, or Fusarium oxysporum; or a filamentous fungal species within the genus Aspergillus, such as a strain of Aspergillus awamori, Aspergillus foetidus, Aspergillus japonicus, Aspergillus niger or Aspergillus oryzae. Also useful in the present invention are Thermomyces lanuginosus phospholipase variants such as the commercial product Lecitase.RTM. Ultra (Novozymes A'S, Denmark). One or more phospholipases may be applied as lyophilized powder, immobilized, or in aqueous solution.
[0127] In some embodiments, phospholipase may be expressed by the microorganism. In some embodiments, the microorganism may be engineered to express homologous or heterologous phospholipases. In some embodiments, phospholipase may be expressed by a microorganism that also produces a product alcohol. In some embodiments, phospholipase may be expressed by a microorganism that also expresses a butanol biosynthetic pathway.
[0128] By-products of fermentation such as isobutyric acid, phenylethanol, 3-methyl-1-butanol, 2-methyl-1-butanol, isobutyraldehyde, acetic acid, ketoisovaleric acid, pyruvic acid, and dihydroxyisovaleric acid may have an inhibitory effect on the microorganism. In some embodiments, these by-products may be modified by esterification. For example, the by-products may be esterified with carboxylic acids, alcohols, fatty acids, or other by-products. In some embodiments, these esterification reactions may be catalyzed by lipases or phospholipases. As an example, lipase present in the fermentation broth may catalyze the esterification of by-products generated during fermentation. Esterification of these by-products may minimize their inhibitory effects on the microorganism.
[0129] Referring to FIG. 7a, feedstock 12 may be processed as described in, for example, FIGS. 1 to 6, and therefore will not be described in detail. Aqueous solution 22 may then be further treated to remove any residual oil. In some embodiments, aqueous solution 22 may be subjected to centrifugation, decantation, or any other method that may be used for oil removal. In some embodiments, aqueous solution 22 may be conducted to unit 25 and catalyst 23 (e.g., lipase) may be added to the unit 25, converting the oil present in aqueous solution 22 to fatty acids, generating stream 27. Stream 27 may then be conducted to fermentation 30 and microorganism 32 may also be added to fermentation 30 for the production of product alcohol. Following fermentation 30, stream 31 comprising product alcohol and fatty acids may be conducted to an external unit, for example, an external extractor or external extraction loop for the recovery of product alcohol.
[0130] Referring to FIG. 7b, in some embodiments, catalyst 23 may be deactivated, for example, by heating. In some embodiments, stream 27 comprising catalyst 23 may be heated (q) to deactivate catalyst 23 prior to addition to fermentation 30. Referring to FIG. 7c, in some embodiments, deactivation may be conducted in a separate unit, for example, a deactivation unit. In some embodiments, stream 27 may be conducted to deactivation 28. Following deactivation, stream 27' may be conducted to fermentation 30 and microorganism 32 may also be added to fermentation 30 for production of product alcohol.
[0131] Removing oil from aqueous solution 22 by converting the oil to fatty acids can result in energy savings for the production plant due to more efficient fermentation, less fouling of the equipment due to the removal of the oil, decreased energy requirements, for example, the energy needed to dry distillers grains, and improved operation of evaporators or evaporation train. In addition, removal of the oil component of the feedstock is advantageous to product alcohol production because oil present in the fermentor can break down into fatty acids and glycerin. The glycerin can accumulate in the water and reduce the amount of water that is available for recycling throughout the system. Thus, removal of the oil component of the feedstock increases the efficiency of the product alcohol production by increasing the amount of water that can be recycled through the system. Also, stable emulsions are less likely to occur by removal of oil. In some embodiments of the present invention, in the event that an emulsion forms, emulsions may be readily broken by mechanical processing, addition of protic solvents, or by other conventional means.
[0132] The present invention also provides processes and systems for recovering a product alcohol produced by a fermentative process. One such process for product alcohol recovery is liquid-liquid extraction. Using liquid-liquid extraction as an ISPR technique is best served by a liquid-liquid extraction process that maximizes the net present value of the capital investment required to practice the technology. An aspect of maximizing the net present value of a liquid-liquid extraction process is to avoid large capital and operating cost expenditures associated with separating extractant from fermentation broth.
[0133] In one embodiment of a liquid-liquid extraction process, extractant may be added directly to the fermentor, and fermentation broth and extractant may be mixed together in a way that effects mass transfer (e.g., transfer of product alcohol from fermentation broth to extractant) and allows the fermentation to proceed to high effective product alcohol titer. In such a process, if mixing is too intense or vigorous, the fermentation broth and extractant may have to be separated using a separation device such as a centrifuge. If mixing is not too intense, phase separation may be achieved through gravity settling brought on by the density difference between the extractant and the fermentation broth. In either case, additional fermentors may be required to overcome the loss of fermentor volume taken up by extractant added to the fermentor. Adding extractant directly to the fermentor may be carried out in batch, semi-batch, or continuous modes irrespective of phase separation within the fermentor. If continuous mode is employed and gravity separation of fermentation broth and extractant is not possible, then a separation device such as a centrifuge may be required for the separation of product alcohol from extractant. If the separation process employed to remove product alcohol from extractant is such that the microorganism present in the fermentation broth survives the separation process, then separation of fermentation broth from product alcohol/extractant may not be required.
[0134] Another embodiment of a liquid-liquid extraction process may include an external extractor or extraction column. For example, fermentation broth from the fermentor may be conducted to an external extractor where the fermentation broth is mixed with extractant. The mixture of fermentation broth and extractant may then be separated, generating a fermentation broth stream leaner in product alcohol and an extractant stream richer in product alcohol. The leaner fermentation broth stream may be returned to the fermentor. The richer extractant stream may be processed further to separate at least a portion of product alcohol from the extractant for product alcohol recovery. In some embodiments, the rate of product alcohol recovery from the extractant stream may be set at a rate to maintain plant production. In some embodiments, the liquid-liquid extraction process may comprise one or more external liquid-liquid extractors.
[0135] In some embodiments, fermentation may occur in the fermentor and the external extractor. The additional volume of fermentation broth present in the external extractor may serve to increase the overall fermentor volume and therefore, may increase the overall production of product alcohol.
[0136] The performance of the external extractor with regard to removing product alcohol may depend on the surface area available for interfacial contact, the physical nature of the fermentation broth and extractant, the relative amounts of the two phases (e.g., fermentation broth phase and extractant phase) present in the external extractor, and the concentration driving force difference between the fermentation broth and extractant phases. Maximizing the efficiency of the external extractor for a given product alcohol concentration driving force may be accomplished by reducing the droplet size of the dispersed phase in the external extractor, for example, via nozzle design, internals design, and/or agitation. In some embodiments, the design and operation of the external extractor may provide enough mixing to effect adequate product alcohol transfer between the fermentation broth and extractant phases to maintain product alcohol productivity requirements.
[0137] Conditions to separate product alcohol from fermentation broth may be deleterious to the microorganism present in the fermentation broth. In some embodiments, the microorganism may be separated from fermentation broth prior to contacting the fermentation broth with the extractant. In some embodiments, the microorganism may be separated from a mixture of fermentation broth and extractant prior to the separation (or processing) of this mixture. Any separation method capable of separating the microorganism from fermentation broth or mixture of fermentation broth and extractant may be used including, for example, centrifugation. By separating the microorganism prior to contacting the fermentation broth with extractant, it may be possible to use more rigorous extraction conditions such as higher temperatures and/or non-biocompatible extractants. If a separation method was used that was not deleterious to the microorganism, then separating the fermentation broth and extractant prior to product alcohol removal would not be required.
[0138] If extractant and fermentation broth are not separated, then the extractant may be included in the evaporator train feed and therefore, become a component of the syrup formed during evaporation, and possibly incorporated in animal feed. In some embodiments, extractant may be separated from the syrup using any separation means including, for example, centrifugation. A low boiling point (e.g., comparable to water) biocompatible extractant may not require such separation because the extractant and water may be recycled for use in the production process.
[0139] In a typical corn-to-product alcohol production plant, the water balance of the overall production process may be maintained by recycling water of the production plant with recycled water distilled in an evaporator train to remove salts and other dissolved solids of the beer. The resulting syrup from the evaporator train may be mixed with undissolved solids, and the mixture may be dried and sold as animal feed. Processes and systems for processing undissolved solids for animal feed are described, for example, in U.S. Patent Application Publication No. 2012/0164302; U.S. Patent Application Publication No. 2011/0315541; U.S. patent application Ser. No. 13/524,990; and U.S. Provisional Patent Application No. 61/674,607, the entire contents of each are herein incorporated by reference.
[0140] As described herein, undissolved solids may be removed from feedstock (or feedstock slurry) prior to the addition of the feedstock to fermentation. If undissolved solids are not removed upstream of fermentation, then centrifugation of the beer to remove undissolved solids may be necessary to avoid fouling of the evaporators. For example, in a commercial corn-to-product alcohol dry-grind production plant, undissolved solids content in an evaporator train feed may operate at about 3% total suspended solids, and may be as high as 3.5-4% total suspended solids. An upstream process that removes enough solids to maintain the percentage of total suspended solids at or below these percentage values may eliminate the need for centrifugation, for example, prior to conducting the beer to the evaporators (or evaporation train). The elimination of this centrifugation would result in a savings on the capital required to retrofit a dry-grind corn-to-product alcohol production plant.
[0141] By removing at least a portion of the undissolved solids present in the feedstock slurry prior to fermentation, the interfacial surface area between the fermentation broth and extractant phases in an external extractor may be increased by reducing the amount of undissolved solids at the interface, enhancing product alcohol transfer between the fermentation broth and the extractant and providing for a clean phase separation between the fermentation broth and extractant. A clean phase separation may also eliminate the need for additional separation steps (e.g., centrifugation) and therefore, a savings on capital expenses.
[0142] The separation of fermentation broth and extractant leaving the external extractor may be influenced by the solids content and particle size distribution of the solids content in the fermentation broth, the gas content and gas bubble size distribution in the fermentation broth, the physical properties of the fermentation broth and extractant including, but not limited to, viscosity, density, and surface tension as well as the design and operation of the external extractor and the design and operation of the fermentor. These properties may determine the need for separation devices (e.g., centrifuges) to separate the fermentation broth and extractant leaving the external extractor or the fermentor. Operating under conditions that eliminate the need for separation devices may minimize the capital expenditure to practice liquid-liquid extraction ISPR. In addition, minimizing the size of the extractors by maximizing the interfacial area between fermentation broth and extractant phases for a given set of fermentation broth and extractant physical properties can maintain the ability to inexpensively phase separate fermentation broth and extractant. By eliminating the capital and operating cost of separation devices such as centrifuges, the net present value of a dry grind corn-to-product alcohol production plant employing a liquid-liquid extraction ISPR process may be improved.
[0143] In another embodiment of the processes and systems described herein, the extractor design including phase separation capacity may be tailored to accommodate the physical properties of the fermentation broth and extractant. If undissolved solids are not removed from feedstock slurry or if the concentration of product alcohol in the fermentation broth is too low, it may not be possible to remove enough product alcohol to maintain plant productivity employing an extractor that does not include phase separation equipment. Therefore, the present invention provides for processes and systems that include solids removal as well as recovery of product alcohol utilizing an external extractor wherein the extractor has been designed to improve phase separation capacity for maximum product alcohol recovery.
[0144] An exemplary process of the present invention is described in FIG. 8. Some processes and streams in FIG. 8 have been identified using the same name and numbering as used in FIGS. 1-7 and represent the same or similar processes and streams as described in FIGS. 1-7.
[0145] Feedstock 12 may be processed and solids separated (100) as described herein with reference to FIGS. 1-7. Briefly, feedstock 12 may be liquefied to generate feedstock slurry comprising undissolved solids, fermentable sugars (or fermentable carbon source), and depending on the feedstock, oil. The feedstock slurry may then be subjected to separation methods to remove suspended solids, generating a wet cake, an aqueous solution 22 (or centrate) comprising dissolved fermentable sugars, and optionally an oil stream. Solids separation may be accomplished by a number of means including, but not limited to, decanter bowl centrifugation, three-phase centrifugation, disk stack centrifugation, filtering centrifugation, decanter centrifugation, filtration, vacuum filtration, belt filter, pressure filtration, membrane filtration, filtration using a screen, screen separation, grating, porous grating, flotation, hydrocyclone, filter press, screwpress, gravity settler, vortex separator, or combination thereof.
[0146] Aqueous solution 22 and microorganism 32 may be added to fermentation 30 where the fermentable sugars are fermented by microorganism 32 to produce stream 105 comprising product alcohol. In some embodiments, during fermentation, a portion of stream 105 may be transferred to extractor 120 (or extraction 120) where stream 105 is contacted with extractant 124. In some embodiments, stream 105 may be removed from fermentation 30 when the concentration of product alcohol and/or other metabolic products reach a predetermined concentration. In some embodiments, the predetermined concentration may be a concentration of product alcohol and/or other metabolic products which negatively impact the metabolism of the microorganism. In some embodiments, stream 105 may be removed from fermentation 30 when fermentation is initiated. In some embodiments, stream 105 may be removed from fermentation 30 to minimize the effects of product alcohol on microorganism 32. In some embodiments, fermentation 30 may comprise one, two, three, four, five, six, seven, eight, or more fermentors.
[0147] In some embodiments, extractant may be added to fermentation 30. In some embodiments, a portion of fermentation broth comprising extractant may be transferred to extractor 120, and in some embodiments, extractant may be recovered from the fermentation broth comprising extractant. By adding extractant to fermentation 30, ISPR may be initiated in fermentation 30.
[0148] Product alcohol transfers from stream 105 to extractant 124, and stream 122 comprising extractant richer in product alcohol may be conducted to separation 130. Stream 127 comprising fermentation broth leaner in product alcohol may be returned to fermentation 30. Separation 130 removes a portion of product alcohol from extractant 124 and stream 125 comprising leaner extractant may be returned to extractor 120. In some embodiments, extractor 120 may be external to fermentation 30. In some embodiments, fermentation 30 may comprise an extractor. In some embodiments, extractant, fermentation broth, or both may be at least partially immiscible. Stream 135 may be conducted downstream for further processing including recovery of product alcohol.
[0149] In some embodiments, phase separation of fermentation broth and extractant after passing through an extractor may be insufficient such that an unacceptable level of dispersed extractant remains in the fermentation broth returning to the fermentor and/or an unacceptable level of fermentation broth droplets remain in the extractant advancing to distillation. In some embodiments, the phase separation of fermentation broth and extractant may be enhanced by processing a heterogeneous mixture exiting the top or bottom of an extractor through one or more hydrocyclones or similar vortex device. In some embodiments, a static mixer may be used in place of an extractor to bring fermentation broth and extractant into contact with each other and the heterogeneous mixture that is formed may be pumped through one or more hydrocyclones or similar vortex device to effect a separation of the aqueous (e.g., fermentation broth) and organic (e.g., extractant) phases. In some embodiments, one or more hydrocyclones or similar vortex device may be used to remove liquid or liquid droplets from a gas stream. In some embodiments, the gas stream may be from the fermentor. In some embodiments, the gas stream may be from a degassing device.
[0150] In a batch or semi-batch fermentation process, when a portion of the fermentable sugars has been metabolized by microorganism 32, stream 103 comprising beer may be conducted downstream to separation 140 to separate product alcohol from the beer. Stream 145 comprising product alcohol may be conducted downstream for further processing including recovery of product alcohol (e.g., distillation). In a continuous fermentation process, stream 103 comprising beer may be conducted downstream to separation 140 to separate product alcohol from the beer. Stream 142 comprising whole stillage may be conducted downstream for further processing including solids removal and generation of thin stillage.
[0151] In some embodiments, fermentation 30 may comprise two or more fermentors, and stream 105 may comprise combined multiple streams from the two or more fermentors. In some embodiments, the combined multiple streams may be conducted to extractor 120. In some embodiments, stream 127 may be split and portions of stream 127 may be returned to the multiple fermentors. In some embodiments, extractor 120 may be a series of units connected together in parallel or in series.
[0152] In some embodiments, extraction may be conducted for a certain period of time. Extraction may be conducted, for example, until the concentration of product alcohol in fermentation 30 is low enough that separation 140 is not required. In some embodiments, extraction may be conducted for an extended period of time.
[0153] In some embodiments of the processes and systems described herein, a decanter may be used for phase separation. In some embodiments, a decanter may be used in combination with an extractor. In some embodiments, the surfaces of the decanter may be modified to improve phase separation. For example, surfaces of the decanter may be modified by the addition of hydrophilic and/or hydrophobic surfaces.
[0154] In some embodiments, oxygen, air, and/or nutrients may be added to stream 125 and/or stream 127. In some embodiments, the nutrients may be soluble in extractant. In some embodiments, the concentration of oxygen may be measured in the various streams, and may be used as part of a control loop to vary the flow of oxygen into the process. In some embodiments, mash may be added to extractor 120 to allow for higher effective titers. In some embodiments, separation 130 and 140 may be extractors. In some embodiments, these extractors may use water to extract product alcohol from extractant, and product alcohol may be subsequently separated from an aqueous phase. In some embodiments, extractant may be infused with solutes that enhance its capacity to extract product alcohol from fermentation broth. In some embodiments, a surge tank may be located between extractor 120 and separation 130 as a means to equilibrate the concentration of product alcohol in the extractant prior to separation (e.g., distillation).
[0155] In some embodiments, extractor 120 may be designed to utilize CO.sub.2 generated during fermentation for the purpose of mixing fermentation broth and extractant. In some embodiments, extractor 120 may be designed to allow for ready disengagement of CO.sub.2 in the fermentation broth. This design would facilitate the control of the level of mixing by CO.sub.2 bubbles rising through extractor 120. In some embodiments, fermentation broth may be removed from fermentation 30 to minimize the concentration of CO.sub.2 in stream 105. In some embodiments, the design of extractor disengagement zones may include surfaces to promote phase separation between fermentation broth and extractant. In some embodiments, hydrophilic and/or hydrophobic surfaces may be installed in the disengagement zones to improve phase separation.
[0156] In some embodiments to minimize CO.sub.2 mixing, the extractor may be designed with a small diameter at the bottom of the extractor, graduating to a large diameter at the top of the extractor (e.g., conical shape). In some embodiments, the extractor may be designed with a stepwise increase in diameter. For example, the extractor may comprise a first region of constant diameter flowed by a stepwise increase of diameter to a second region of constant diameter. In some embodiments, the extractor may further comprise a second stepwise increase of diameter to a third region of constant diameter. In some embodiments, the extractor may comprise one or more stepwise increases of diameter. In some embodiments, the extractor may comprise one or more regions of constant diameter.
[0157] Over the course of fermentation, the gas content (e.g., CO.sub.2) of the fermentation broth changes, and these gases may be removed from the fermentation broth by utilizing a gas stripper. The amount of gas stripped from the fermentation broth may be adjusted by varying the flow through the gas stripper and/or the pressure of the gas stripper. In some embodiments, the amount of CO.sub.2 in the fermentation broth may be reduced prior to transferring the fermentation broth to an extractor. For example, CO.sub.2 may be stripped from the fermentation broth using a gas stripper or any means known to those skilled in the art. In some embodiments, removal of CO.sub.2 may be performed at or below ambient pressure. In some embodiments, fermentation may continue in the extractor, and CO.sub.2 may be produced by the microorganism. In some embodiments to minimize CO.sub.2 mixing in the extractor, the residence time of the fermentation broth in the extractor may be reduced. In some embodiments, residence time may be reduced by modifying the height of the extractor. In some embodiments, the height of the extractor may be reduced. Reducing the height of the extractor may reduce the number of theoretical extraction stages. In some embodiments, to maintain the number of theoretical extraction stages, the extractor may be replaced with two or more extractors of reduced height. In some embodiments, the two or more extractors may be in series. In some embodiments, the two or more extractors may be connected. In some embodiments, the two or more extractors may be connected in such a way to maintain countercurrent flow. In some embodiments, a degassing stage may be added to one or more extraction stages.
[0158] Referring to FIG. 8, in some embodiments, the size of dispersed phase droplets in extractor 120 may be measured and adjusted through various means to enhance the rate of mass transfer. For example, droplet size may be measured using particle size analysis such as focused beam reflectance measurement (FBRM.RTM.) or particle vision and measurement (PVM.RTM.) technologies (Mettler-Toledo, LLC, Columbus Ohio). In some embodiments, the fermentation broth may be the dispersed phase and extractant may be the continuous phase, and under these conditions solids present in the fermentation broth may interact to a lesser degree with the extractant. In some embodiments, conditions of separation 130 may be controlled to minimize oxidative and thermal instabilities effects on the extractant.
[0159] In some embodiments, the quality of the extractant may be monitored and extractant replenished at a frequency necessary for successful production of product alcohol. In some embodiments, extractant may be taken up by whole stillage solids. The whole stillage may be separated into liquid (e.g., thin stillage) and solid streams, and the solids may be washed to recover the extractant. In some embodiments, the temperature of extractor 120 may be adjusted to improve the efficiency of the overall process. In some embodiments, the flows of fermentation broth and extractant to extractor 120 may be co-current or countercurrent. In some embodiments, membranes may be used to minimize the mixing of fermentation broth and extractant. In some embodiments, extractant may be polymer beads or inorganic beads that absorb product alcohol. In some embodiments, the polymer beads or inorganic beads may be preferentially absorb product alcohol.
[0160] In some embodiments, measurements such as in-line, on-line, at-line, or real-time measurements may be used to measure the concentration of product alcohol and/or metabolic by-products in the various streams. These measurements may be used as part of a control loop to vary the flow between the various units or vessels (e.g., fermentation 30, extractor 120, separations 130 and 140, etc.) and to improve the overall process.
[0161] Another exemplary process of the present invention is described in FIG. 9. Some processes and streams in FIG. 9 have been identified using the same name and numbering as used in FIGS. 1-8 and represent the same or similar processes and streams as described in FIGS. 1-8.
[0162] Feedstock 12 may be processed and solids separated (100) as described above with reference to FIGS. 1-7. In some embodiments, feedstock 12 may be mixed with recycled water (e.g., stream 162) generated by evaporation 160. As described herein, feedstock slurry may be subjected to separation methods to remove suspended solids, generating a wet cake 24, an aqueous solution 22 (or centrate) comprising dissolved fermentable sugars, and depending on the feedstock, oil. Wet cake 24 may be dried in dryer 170 and used to produce DDGS. In some embodiments, wet cake 24 may be re-slurried with water (e.g., recycled water/stream 162) and subjected to separation to remove additional fermentable sugars, generating washed wet cake (e.g., 74, 74' as described in FIGS. 4 and 5). In some embodiments, wet cake streams 24, 74, and 74' may be combined and the combined wet cake streams may be dried in a dryer 170 and used to produce DDGS.
[0163] Aqueous solution 22 and microorganism 32 may be added to fermentation 30 where the fermentable sugars are metabolized by microorganism 32 to produce stream 105 comprising product alcohol. In some embodiments, enzyme may be added to fermentation 30. Stream 105 may be conducted to extractor 120, and may be contacted with extractant 124. Stream 127 comprising fermentation broth leaner in product alcohol may be returned to the fermentation 30 and stream 122 comprising extractant richer in product alcohol may be conducted to separation 130. In some embodiments, extractor 120 may be operated in such a way that stream 122 contains minimal cell mass and minimal substrate. Separation 130 may damage microorganism 32 or substrate resulting in a decrease in the fermentation rate. Operating extractor 120 with minimal cell mass and substrate may minimize any potential damage by separation 130. Stream 125 comprising leaner extractant may be returned to extractor 120. Stream 135 from separation 130 may be conducted to purification 150 for further processing including recovery of product alcohol. In some embodiments, extractant may be added to fermentation 30. In some embodiments, a portion of fermentation broth comprising extractant may be transferred to extractor 120, and in some embodiments, extractant may be recovered from the fermentation broth comprising extractant. In some embodiments, the flow rates of fermentation broth and extractant to extractor may be modified to improve phase separation. For example, lower overall flow rates entering the extractor in the early or later stages of fermentation can improve the phase separation of fermentation broth and extractant.
[0164] As described herein, after a batch fermentation process or as a steady effluent stream in a continuous fermentation process, stream 103 comprising beer may be conducted downstream to separation 140 to separate product alcohol from the whole stillage 142. Utilizing an upstream solids removal process may lower the undissolved solids content in the thin mash and therefore, it may not be necessary to centrifuge whole stillage 142 to remove solids. Thus, whole stillage 142 may be conducted directly to evaporation 160. Syrup 165 generated by evaporation 160 may be mixed with wet cake 24, 74, 74' in dryer 170 to form DDGS.
[0165] In some embodiments, backset comprising total suspended solids from whole stillage may be used (or recycled) for feedstock slurry preparation. In some embodiments, whole stillage or a portion of whole stillage may be processed through a solids separation system including, but not limited to, turbo filtration or ultracentrifugation prior to evaporation, or whole stillage or a portion of whole stillage may be processed for self-cleaning water purification.
[0166] In some embodiments where coarse grain solids are removed from liquefied mash, the whole stillage that is produced may contain fine solids and insoluble microorganism fragments, and these dispersed solids may be removed using turbo filtration. Turbo filtration may include subjecting a feed suspension to centrifugal motion through a strainer that can retain fine solids. These fine solids when formed into a wet cake may contain some extractant that is absorbed both on the surface of and inside the pores of fine grain particles. In some instances, washing the wet cake with water is insufficient for recovering extractant from the wet cake. In some embodiments, a concentrated product alcohol stream such as the organic phase may be used to recover extractant from whole stillage wet cake. In some embodiments, this organic phase may be formed in an aqueous product alcohol decanter. In some embodiments, the wet cake that has been washed with product alcohol may be subsequently washed with water to recover the product alcohol from the wet cake.
[0167] In some embodiments, the processes and systems described herein may include an extractant reservoir (or tank or vessel). Extractant may be added to the extractant reservoir and this extractant may be circulated to an extractor. In some embodiments, extractant may be conducted to an extractor and a stream from the extractor may be returned to the extractant reservoir. In some embodiments, extractant from an extractant reservoir may be circulated to an extractor and/or fermentor. In some embodiments, an extractant stream may be circulated between an extractant reservoir, an extractor, and a fermentor. In some embodiments, at the completion of fermentation, the contents of the extractant reservoir and the fermentor may be further processed to recover product alcohol.
[0168] Separation or extraction of product alcohol from extractant may be accomplished using methods known in the art, including but not limited to, siphoning, decantation, centrifugation, gravity settler, membrane-assisted phase splitting, and the like. In some embodiments, extraction may be performed using, for example, mixer-settlers. Mixer-settlers are stage-wise extractors and are available with various elements for mixing such as, pumps, agitators, static mixers, mixing tees, impingement devices, circulating screens, or raining buckets. Examples of mixer-settlers are shown in FIGS. 10A-10H. For example, FIG. 10A illustrates a mixer-settler using a pump as the source of mixing. FIG. 10B illustrates a mixer-settler using a mixer as the source of mixing. FIG. 10C illustrates a mixer-settler using a static mixer as the source of mixing. FIG. 10D illustrates a mixer-settler using a mixing tee as the source of mixing. FIG. 10E illustrates a mixer-settler using an impingement mixer as the source of mixing. FIG. 10F illustrates a mixer-settler using a raining bucket or meshed screen as the source of mixing. FIG. 10G illustrates a mixer-settler using a centrifuge as a settler. FIG. 10H illustrates a mixer-settler using a hydrocyclone or vortex separator as a settler. In some embodiments, one or more mixing devices may be used in the processes and systems as described herein.
[0169] In some embodiments, mixers may comprise agitators such as, for example, flat blades, pitched blade turbines, or curved propellers. Droplet size produced by agitated mixers may be controlled by agitator design, tank design, agitator speed, and mode of operation. For static mixers, droplet size may be controlled by the diameter of the mixer and flow rate. For example, droplet size may be controlled by varying the flow through the mixer over the course of the fermentation. In some embodiments, gases and mixers may be used for mixing purposes.
[0170] In some embodiments, one or more mixer-settlers may be used in the processes and systems as described herein. In some embodiments, the one or more mixer-settlers may be arranged in series or in countercurrent mode as illustrated in FIGS. 10I and 10J. In some embodiments, mixer-settlers may be stacked in a column arrangement, providing multiple mixing and settling zones. In some embodiments, the settler may comprise hydrophilic or hydrophobic surfaces to promote phase separation.
[0171] In another embodiment, column extractors or centrifugal extractors may be used in the processes and systems as described herein. Column extractors are differential extractors providing conditions for mass transfer over their length with a steadily changing concentration profile. The different types of differential extractors may be divided into non-mechanical, pulse-agitated, and rotary-agitated. Centrifugal extractors are a separate class of differential extractors with the Podbielniak.RTM. centrifugal contactor being one such type.
[0172] In some embodiments, non-mechanical spray towers may be used in the processes and systems as described herein. One example of a non-mechanical spray tower includes a non-mechanical spray tower without column internals. The number of nozzles and nozzle diameter may be used to determine droplet size. In some embodiments, the spray tower may have internals. In some embodiments, a spray tower may comprise helical piping. Helical piping may allow for droplet rise and additional mixing of fermentation broth and extractant. In some embodiments, non-mechanical extractors such as packed towers, sieve trays, and baffle trays may be used in the processes and systems as described herein. Examples of these extractors are shown in FIG. 10K. In some embodiments, the packing of such extractors may be random or structured.
[0173] In some embodiments, pulsed-agitated extractors may be used in the processes and systems as described herein. Pulsed-agitated extractors have different designs as well including reciprocating trays or vibrating plates where the trays move in vertical fashion. The entire packed and/or sieve tray column can also vibrate in a vertical fashion to promote smaller dispersed phase droplets and more mass transfer. Examples of these extractors are shown in FIG. 10L. In some embodiments, rotary-agitated or rotating disc contactors may be used in the processes and systems as described herein. Examples of these extractors are shown in FIG. 10M.
[0174] In some embodiments, agitated extractors may be used in the processes and systems as described herein. For example, agitated extractors with centrifuges may provide high mass transfer rates and clean phase separation. In some embodiments, agitated columns may be used in the processes and systems as described herein. For example, agitated columns with internals may provide high mass transfer rates.
[0175] One aspect of a liquid-liquid extraction process is determining successful operating conditions for the extractor over the course of the constantly changing fermentation. For example, a typical corn-to-product alcohol batch fermentation employs an initial inoculum of microorganism (or cell mass) added to a certain volume of fermentation broth in the fermentor, followed by further filling of the fermentor to a specified volume. The fermentation is permitted to proceed until a pre-determined amount of the fermentable carbon source (e.g., sugar) is consumed. Over the course of batch fermentation, the concentrations of cell mass, reaction intermediates, reaction by-products, and substrate components change with time as do the physical properties of the fermentation broth including viscosity, density, and surface tension. To improve performance parameters of the fermentation, for example, rate, titer, and yield parameters of production and plant economics such as sales volume, return on investment, and profit, the extractor may be operated in a variable way to compensate for the changing fermentation broth. In addition, properties of a dynamic fermentation may impact the size limits of the extractor. Proper integration of the operation of the extractor and the fermentor may be benefit by use of mathematical models of the process (see, e.g., Daugulis and Kollerup, Biotechnology and Bioengineering 27:1345-1356, 1985). Augmenting the mathematical model, for example, setting the key model parameters with experimental data is also valuable. Design parameters for differential extractors to consider for improved rate, titer, and yield of the fermentation process include the maximum total flow to the extractor per cross-sectional area of the extractor column as well as the height of the extractor required to remove enough product alcohol at a given fermentation broth to extractant ratio. It may be necessary to change the maximum flow per unit area and extractor height during a batch fermentation. Another consideration for differential extractors is droplet size of the dispersed phase. Appropriate droplet size may be a balance between small enough to provide adequate mass transfer but large enough to allow for clean phase separation exiting the extractor. In stage-wise extractors, the mixing intensity required for efficient mass transfer, the corresponding time needed to settle, and/or energy needed to separate the phases are additional elements to consider. In either type of extractor, stage-wise or differential, the ratio of fermentation broth to extractant fed to the extractor plays a role in determining the size of the extractor.
[0176] In some embodiments, if an extractor of a fixed size were utilized and the maximum allowable flow that avoids flooding to the extractor varied from a low value to a high value (e.g., from 1/3 to 2/3 the maximum for a given extractor design) over the course of the fermentation owing to changes in the physical properties and concentrations of the fermentation broth, then the flows to the extractor may be varied, not exceeding the maximum flow, while still completing the fermentation in a reasonable time. In some embodiments, if an extractor is agitated, the speed of the agitation may be varied over the course of the fermentation to offset changes in the fermentation broth. Droplet size may be measured within the extractor, and the speed to maintain a fixed droplet size may be controlled throughout the fermentation to offset changes in the fermentation broth. The amount of mass transfer occurring at any time point may be assessed by measuring the concentrations of product alcohol in the inlet and outlet streams and adjusting conditions (e.g., flow, flow ratio, agitation) to control the mass transfer over the course of the fermentation.
[0177] In some embodiments, multiple extractors of different sizes may be utilized and conditions (e.g., flow, flow ratio, agitation) in each extractor may be adjusted to provide improved control of the fermentation process. In some embodiments, the ratio of fermentation broth to extractant may be modified to improve extraction efficiency, increase the concentration of product alcohol in the extractant (equivalent to increased efficiency), and reduce the required flows through the extractor.
[0178] In additional embodiments of the processes and systems described herein, there may be two or more fermentation broth or aqueous streams. An extractant phase that has absorbed product alcohol from a first aqueous stream may be brought into contact with a second aqueous stream that contains less product alcohol than the first aqueous stream or fermentation broth, enabling the transfer of product alcohol from the rich extractant phase to the second aqueous phase. In some embodiments, contacting the rich extractant with a dilute aqueous stream may take place in a multi-stage contacting device or in a static mixer followed by a settler. In some embodiments, contacting the rich extractant with a dilute aqueous stream may take place in the same device where lean extractant is contacted with fermentation broth. An extractor with perforated baffles would allow downflow of both fermentation broth and a dilute aqueous stream in separate compartments while an extractant that is lean in product alcohol may form a continuous phase throughout all compartments. An advantage of this configuration is a reduced amount of extractant would be needed in the production plant if the extractant remains confined to the closed volume of an extractor. Another advantage of this configuration is that the extractant is not subjected to potential degradation during distillation and therefore, may exhibit a longer service life. By transferring product alcohol to a homogeneous aqueous stream, the product alcohol may be conducted to more than one stripping column via partitioning of the dilute aqueous stream, taking into consideration column capacities and heat integration. The need to clean equipment that is exposed to an extractant may be reduced when product alcohol is extracted into an aqueous medium during or immediately after the product alcohol is extracted from fermentation broth.
[0179] In some embodiments, product alcohol may be transferred from fermentation broth to a second aqueous stream or an extractant across a barrier that is selective for product alcohol transport. In some embodiments, this barrier may be provided by a membrane material. The membrane material may be either organic or inorganic. Examples of membrane material include polymers and ceramics. In some embodiments, product alcohol may be separated from fermentation broth utilizing a hydrogel. In some embodiments, the hydrogel may comprise functional elements that promote interaction with a product alcohol such as, but not limited to, hydroxyl functionality, hydrocarbon character, network size, and the like. In some embodiments, a hydrogel may comprise a polymeric network structure or polymer formulations. Examples of polymer formulations include, but are not limited to, one or more of the following: acrylic acid, sodium acrylate, hydroxyethyl acrylate, methacrylate, hydroxybutyl acrylate, butylacrylate, vinylated polyethylene oxide, vinylated polypropylene oxide, vinylated polytetratmethylene oxide, acrylates and diacrylates of polyglycols, polyvinyl alcohol and hydrocarbon derivatized polyvinyl alcohol, and styrene and styrene derivatives. In some embodiments, the hydrogel may comprise hydroxyethyl acrylate and methacrylate, hydroxybutyl acrylate and methacrylate, or butylacrylate and methacrylate.
[0180] In other embodiments of the processes and systems described herein, fermentation broth may be removed from the bottom of the fermentor at above atmospheric pressure and passed through a first flash tank operating at atmospheric pressure to release dissolved gases such as CO.sub.2. This first flash tank may be a degassing cyclone and the vapors from this first flash tank may be combined with vapors from the fermentor and directed to a scrubber. In some embodiments, the fermentation broth from the first flash tank may be passed through a second flash tank operating below atmospheric pressure to release more dissolved gases such as CO.sub.2. This second flash tank may be a degassing cyclone and the vapors from this second flash tank may be re-compressed to atmospheric pressure, cooled, and partially condensed prior to being combined with vapors from the fermentor and being directed to a scrubber. The fermentation broth exiting this second flash tank may be pumped to an extraction column operating at above atmospheric pressure so that any remaining or newly formed dissolved gases will not lead to formation of a vapor phase in the extraction column.
[0181] In another embodiment of the processes and systems described herein, fermentation broth may be conducted to an extractor and contacted with extractant generating an aqueous stream and organic stream comprising extractant and product alcohol. This organic stream may be conducted to a flash tank (e.g., vacuum flash) for separation of product alcohol from extractant. In some embodiments, the extractant stream from the flash tank may be recycled to the extractor and/or the fermentor. In some embodiments, the organic stream may be conducted to a second extractor prior to the flash tank. This second extractor may be used to remove, for example, any residual water in the organic stream. The extractors may be siphons, decanters, centrifuges, gravity settlers, mixer-settlers, or combinations thereof. In some embodiments, the extractant may be an oil such as, but are not limited to, tallow, corn, canola, capric/caprylic triglycerides, castor, coconut, cottonseed, fish, jojoba, lard, linseed, neetsfoot, oiticica, palm, peanut, rapeseed, rice, safflower, soya, sunflower, tung, jatropha, and vegetable oil blends.
[0182] In some embodiments of the processes and systems described herein, automatic self-cleaning filtration may be used in these processes and systems. Fermentation broth may be removed from a fermentor and may be cooled using a cooler (e.g., an existing cooler in a fermentation production facility) before entering an automatic self-cleaning filter. Some particulates may be retained on the screen medium of the filter as clarified mash passes through the filter. Additional filters may be simultaneously undergoing backflush where a portion of the clarified mash flows back through the screen carrying the particulates with it, discharging a concentrated solids stream. In some embodiments, a portion of the clarified mash may enter the top of an extractor while an extractant is fed in the bottom of the extractor. The clarified mash and extractant may be brought into contact either passively by density differences or with the aid of mechanical motion (e.g., a Karr.RTM. column) by means commonly used in the art. In some embodiments, an organic liquid stream of extractant containing product alcohol emerges from the top of the extractor and an aqueous liquid stream of fermentation broth that has been at least partially depleted of product alcohol relative to clarified mash emerges from the bottom of the extractor. The aqueous liquid stream and concentrated solids stream may be combined and returned to the fermentor. The extractant stream rich in product alcohol may be heated in a heat exchanger that transfers heat from an extractant stream that is lean in product alcohol and that originates from the bottom of the extractor. After releasing some heat, the lean extractant may be further cooled with water in a heat exchanger to reach a temperature that is suitable for fermentation. Circulation of fermentation broth may include a pathway through a heat transfer device and mass transfer device enabling the removal of heat and product alcohol per pass through an external cooling loop. Moreover, in some embodiments, the rate of heat and product alcohol removal may be balanced with the rate of heat and product alcohol production during fermentation by adjusting the circulation flow through the external cooling loop, adjusting the flow of cooling fluid in a heat exchanger, and/or adjusting the flow of extractant.
[0183] In some embodiments of the processes and systems described herein, phase separation of extractant from fermentation broth may be enhanced by modifying the temperature and/or pH of the process. For example, the process may be operated at temperatures and/or pH that are different than the temperature and/or pH of the fermentor. In some embodiments, the process may be operated at a reduced pH as compared to the fermentor. In some embodiments, the process may be operated at a higher temperature as compared to the fermentor. In some embodiments, the process may be operated at a reduced pH and a higher temperature as compared to the fermentor. A higher temperature can increase the kinetics of mass transfer of product alcohol between the aqueous and organic phases and may increase the kinetics of coalescence for extractant droplets dispersed in the aqueous phase and for aqueous droplets dispersed in the organic phase. In some embodiments, the temperature inside an extractor containing fermentation broth and extractant may be increased by heating the fermentation broth and/or extractant entering the extractor. The fermentation broth may be heated either directly with injection of water vapor or steam or indirectly via a heat exchanger. In some embodiments, the extractant feeding the extractor may originate from distillation where its temperature may already be elevated. In some embodiments, the extractant may be cooled to a temperature higher than the fermentation temperature.
[0184] In some embodiments, a reduced pH can minimize the solubility and dispersibility of extractant in the aqueous broth phase. In some embodiments, the extractant may be a fatty acid with a known associated pKa value. In some embodiments, the pH of the fermentation broth may be reduced to below the pKa of the extractant such that the carboxylic acid groups of the fatty acid are substantially protonated. In some embodiments, the pH may be reduced by introducing CO.sub.2 gas into the fermentation broth or by injecting a small amount of liquid acids such as sulfuric acid or any other organic or inorganic acid into the fermentation broth. In some embodiments, the pH of the fermentation broth after separating from the extractant may be adjusted to the pH of fermentation.
[0185] In some embodiments where the extractant phase is the continuous phase, the aqueous phase may be distributed or dispersed in the extractant phase. For example, fermentation broth comprising product alcohol may be conducted to an extractor (e.g., external extractor) via a distributor or dispersal device. In some embodiments, the distributor or dispersal device may be a nozzle such as a spray nozzle. In some embodiments, the distributor or dispersal device may be a spray tower. As an example, droplets of fermentation broth may be passed through extractant, and product alcohol is transferred to the extractant. Droplets of fermentation broth coalesce at the bottom of the extractor and may be returned to the fermentor. Extractant comprising product alcohol may be further processed for recovery of product alcohol as described herein. In addition, at the completion of fermentation, residual product alcohol in the fermentor may also be further processed for recovery of product alcohol. In some embodiments, the extractant phase may be countercurrent.
[0186] In some embodiments where the extractant phase is the continuous phase and the aqueous phase is the dispersed phase, mass transfer rates may be improved by using electrostatic spraying to disperse the aqueous phase in the extractant phase. In some embodiments, one or more spray nozzles may be utilized for electrostatic spraying. In some embodiments, the one or more spray nozzles may be an anode. In some embodiments, the one or more spray nozzles may be a cathode.
[0187] In some embodiments, extractor effluent may be used to enhance phase separation. For example, a portion of rich extractant (i.e., extractant rich in product alcohol) from the top of the extractor may be returned to the top of the extractor as reflux, and the remaining rich extractant may be further processed for recovery of product alcohol. Also, a portion of lean fermentation broth from the bottom of the extractor may be returned to the bottom of the extractor as reflux and the remaining lean fermentation broth may be returned to the fermentor. In another embodiment, rich extractant may exit the top of the extractor into a decanter and separated into a heavy phase and light phase. The heavy phase from the decanter may be conducted to the top of the extractor to enhance phase separation. The light phase from the decanter may be may be further processed for recovery of product alcohol.
[0188] In some embodiments of the processes and systems described herein, multiple pass extractant flow may be utilized for product alcohol recovery. For example, multiple fermentors and extractors may be used, where the fermentation cycle of each fermentor is at a different timepoint. Referring to FIG. 11A as an example, fermentor 300 is at an earlier timepoint as compared to fermentor 400 which is at an earlier timepoint as compared to fermentor 500. Fermentation broth comprising product alcohol 302 from fermentor 300 may be contacted with extractant 307 in extractor 305, and product alcohol may be transferred to extractant generating product alcohol-rich extractant 309. Product alcohol-rich extractant 309 from extractor 305 may be conducted to extractor 405. Fermentation broth comprising product alcohol 402 from fermentor 400 may be conducted to extractor 405, producing product alcohol-rich extractant 409. Product alcohol-rich extractant 409 may be conducted to extractor 505. Fermentation broth comprising product alcohol 502 from fermentor 500 may be conducted to extractor 505. Product alcohol-rich extractant 509 from extractor 505 may be processed for recovery of product alcohol. Product alcohol-lean fermentation broth (304, 404, 504) may be returned to fermentors 300, 400, and 500, respectively. The number of fermentors and extractors may vary depending on the operational facility. A benefit of this process is, for example, the reduction in total extractant processing and the size of the extractor.
[0189] In another embodiment of this example, there may be an additional fermentor F' and an additional extractor E' (FIG. 11B). In this embodiment, when fermentor 500 (which is at a later timepoint compared to fermentors 300 and 400) has completed fermentation, fermentor 500 may be taken off-line, and in some embodiments, fermentor 500 may undergo sanitation and/or sterilization procedures such as clean-in-place (CIP) and sterilization-in-place (SIP) procedures. When fermentor 500 is taken off-line, fermentor F' may be brought on-line. In this embodiment, fermentor F' is at an earlier timepoint as compared to fermentor 300 which is at an earlier timepoint as compared to fermentor 400. Similar to the description for FIG. 11A, fermentation broth comprising product alcohol F'-02 from fermentor F' may be contacted with extractant in extractor E', and product alcohol may be transferred to extractant generating product alcohol-rich extractant E'-09. Product alcohol-rich extractant E'-09 from extractor E' may be conducted to extractor 305. Fermentation broth comprising product alcohol 302 from fermentor 300 may be conducted to extractor 305, producing product alcohol-rich extractant 309. Product alcohol-rich extractant 309 may be conducted to extractor 405. Fermentation broth comprising product alcohol 402 from fermentor 400 may be conducted to extractor 405. Product alcohol-rich extractant 409 from extractor 405 may be processed for recovery of product alcohol. Product alcohol-lean fermentation broth (F'-04, 304, 404) may be returned to fermentors F', 300, and 400, respectively. In some embodiments, this process may be repeated for multiple cycles, for example, at least one, at least two, at least three, at least four, at least five, at least ten, at least fifteen, at least twenty, or more cycles. In some embodiments, the process of taking fermentors off-line and putting additional fermentors on-line may be manual or automated. A benefit of this process is reduced extractor flow to product recovery (e.g., distillation).
[0190] In some embodiments, an extractant may reduce the flashpoint (i.e., flammability) of the product alcohol. Flashpoint refers to the lowest temperature at which flame propagation occurs across the surface of a liquid. Flashpoint may be measured, for example, using the ASTM D93-02 method ("Standard Test Methods for Flash Point by Pensky-Martens Closed Tester"). Reduction of the flashpoint of the product alcohol can improve the safety conditions of an alcohol production plant, for example, by minimizing the fire hazard of the potentially flammable product alcohol. By improving safety conditions, the risk of injury is minimized as well as the risk of property damage and revenue loss. In some embodiments where inactivation of the microorganism is required, an extractant may improve the inactivation of the microorganism.
[0191] In some embodiments, the processes described herein may be integrated extraction fermentation processes using on-line, in-line, at-line, and/or real-time measurements, for example, of concentrations and other physical properties of the fermentation broth and extractant. These measurements may be used, for example, in feed-back loops to adjust and control the conditions of the fermentation and/or the conditions of the extractor. In some embodiments, the concentration of product alcohol and/or other metabolites and substrates in the fermentation broth may be measured using any suitable measurement device for on-line, in-line, at-line, and/or real-time measurements. In some embodiments, the measurement device may be one or more of the following: Fourier transform infrared spectroscope (FTIR), near-infrared spectroscope (NIR), Raman spectroscope, high pressure liquid chromatography (HPLC), viscometer, densitometer, tensiometer, droplet size analyzer, pH meter, dissolved oxygen (DO) probe, and the like. In some embodiments, off-gas venting from the fermentor may be analyzed, for example, by an in-line mass spectrometer. Measuring off-gas venting from the fermentor may be used as a means to identify species present in the fermentation reaction. The concentration of product alcohol and other metabolites and substrates dissolved in the extractant may also be measured using the techniques and devices described herein.
[0192] In some embodiments, measured inputs may be sent to a controller and/or control system, and conditions within the fermentor (temperature, pH, nutrients, enzyme and/or substrate concentration) may be varied to maintain a concentration, concentration profile, and/or conditions within the extractor (fermentation broth flow, fermentation broth to extractant flow, agitation rate, droplet size, temperature, pH, DO content). Similarly, conditions within the extractor may be varied to maintain a concentration and/or concentration profile within the fermentor. By utilizing such a control system, process parameters may be maintained in such a way to improve overall plant productivity and economic goals. In some embodiments, real-time control of fermentation may be achieved by variation of concentrations of components (e.g., sugars, enzymes, nutrients, and the like) in the fermentor, variation of conditions within the extractor, or both.
[0193] As an example of an isobutanol fermentation process, the efficiency of isobutanol extraction in a Karr.RTM. column is continuously changing as the concentrations of starch, sugars and isobutanol change in the fermentation broth. In order to maximize the efficiency of the extractor, it may be advantageous to alter the rate at which isobutanol is removed from the fermentation broth to match the production profile of the isobutanol fermentation. Isobutanol concentrations in the extractant may be maximized resulting in more energy efficient distillation operations.
[0194] As part of a process control strategy, real-time measurements of isobutanol in the fermentation broth (e.g., column feed) may be coupled with real-time measurements of isobutanol in the extractant and in the lean fermentation broth. These measurements may be used to adjust the fermentation broth to extractant ratio (flows) to the extractor. The flexibility to match the rate of isobutanol extraction with the rate of isobutanol generation may allow the extractor to be operated efficiently throughout the extraction. In addition, by maintaining a high concentration of isobutanol in the extractant, the volumetric flow rate to the distillation columns can be minimized, resulting in an energy savings for distillation operations. Phase separation may also be monitored using real-time measurements, for example, by monitoring the rate of phase separation, extractant droplet size, and/or composition of fermentation broth. In some embodiments, phase separation may be monitored by conductivity measurements, dielectric measurements, viscoelastic measurements, or ultrasonic measurements. In some embodiments, an automated phase separation detection system may be used to monitor phase separation. This automated system may be used to adjust the flow rates of fermentation broth and extractant to and from the extractor and/or adjust the droplet size of extractant, for example, after mixing of fermentation broth and extractant. By using these real-time monitoring systems, clean phase separation of aqueous and organic phases may be accomplished.
[0195] As another example of process control strategy, droplet size may be measured using particle size analysis such as a process particle analyzer (JM Canty, Inc., Buffalo, N.Y.), focused beam reflectance measurement (FBRM.RTM.), or particle vision and measurement (PVM.RTM.) technologies (Mettler-Toledo, LLC, Columbus Ohio). In some embodiments, these measurements may be real-time in situ particle system characterizations. By monitoring droplet size in real time, changes in droplet shape and dimensions may be detected and process steps may be adjusted to modify droplet size and enhance the rate of mass transfer. For example, droplet size may be used to monitor the amount of extractant in fermentation broth. Following phase separation, some extractant may be present in the fermentation broth, and in some embodiments where the fermentation broth is recycled to the fermentor, monitoring droplet size would provide a means to minimize the amount of extractant in the fermentation broth returning to the fermentor. If the amount of extractant in the fermentation broth is too high, then phase separation may be improved, for example, by adjusting the droplet size of extractant in the extractor and/or adjusting the flow rates of fermentation broth and extractant to the extractor. These adjustment in the process steps can minimize the amount of extractant in the fermentation broth, as well as minimize the amount of extractant in thin stillage and DDGS.
[0196] In one embodiment of this control strategy, isobutanol in the fermentation broth would not exceed a concentration or setpoint at which the concentration of isobutanol becomes deleterious to the microorganism. The isobutanol fermentation broth setpoint may be adjusted higher or lower as the fermentation progresses based upon the trajectory of the fermentation. For example, continuous comparison of the concentration of isobutanol in the fermentation broth to a setpoint concentration of isobutanol can be utilized to modify fermentation broth to extractant ratios or flow rates of fermentation broth and extractant to an extractor. To monitor isobutanol concentrations in the fermentation broth, in situ measurements of the fermentation broth may be performed using Fourier transform infrared spectroscopy (FTIR), near infrared spectroscopy (NIR), and/or Raman spectroscopy. In addition, measurements of the fermentor headspace may be performed using FTIR, Raman spectroscopy, and/or mass spectrometry.
[0197] In some embodiments, efficient extractor operation may occur close to the point of extractor flooding. The use of real-time process control that utilizes concentration data from inlet and outlet streams may allow the extractor to be operated reliably near the point of flooding. In some embodiments, real-time extractant monitoring may be used to detect the partitioning of by-products from the fermentation broth or contaminants into the extractant. By-products such as alcohols, lipids, oils, and other fermentation components may reduce the extraction efficiency of the extractant. Numerous process monitoring techniques may be applied to this measurement including, but are not limited to, Fourier transform infrared spectroscopy (FTIR), near infrared spectroscopy (NIR), high performance liquid chromatography (HPLC), and nuclear magnetic resonance (NMR). The analytical technique selected to monitor the extractant for the presence of by-products or contamination may be a different technique than employed for real-time alcohol determination. Real-time data may be used to trigger the remediation of contaminated extractant or the purge of contaminated extractant from the process. These techniques as well as gas chromatography (GC) and supercritical fluid chromatography (SFC) may also be utilized to monitor thermal breakdown of extractant.
[0198] Referring to FIG. 12, the systems and processes of the present invention may include means for on-line, in-line, at-line, and/or real-time measurements (circles represent measurement devices and dotted lines represent feedback loops). FIG. 12 is similar to FIG. 9, except for the addition of measurement devices for on-line, in-line, at-line, and/or real-time measurements, and therefore will not be described in detail again.
[0199] As an example, on-line measurements of aqueous stream 22 may be utilized to monitor the concentration of fermentable carbon sources (e.g., polysaccharides), oil, and/or dissolved oxygen. For example, FTIR may be used to monitor the dispersion of oil in aqueous stream 22, and process imaging may be used to monitor the concentration and size of oil droplets in the aqueous stream 22. In some embodiments, on-line measurements of fermentation 30 may be utilized to monitor removal rates of product alcohol. Measurements of fermentable carbon sources, dissolved oxygen, product alcohol, and by-products may be used to adjust the removal rate of product alcohol in order to maintain a concentration of product alcohol in fermentation 30 that is tolerable to microorganisms. By maintaining a setpoint product alcohol concentration, product inhibition and toxicity may be minimized.
[0200] On-line measurements of stream 105 and stream 122 may be used to operate process control feedback loops. For example, the concentration of product alcohol in stream 105 may be used to control the flow rate of this stream to extractor 120; and the concentration of product alcohol in stream 122 may be used to control the flow rate of this stream to separation 130 and to set the ratio of fermentation broth to extractant. In addition, on-line measurements of stream 105 and stream 122 may also be utilized to establish real-time product alcohol mass balance. Process control feedback loops for extractor 120 and separation 130 may be used to monitor the quality of phase separation of extractant and fermentation broth. For example, on-line measurement devices may be used to detect the balance of the separation of extractant and fermentation broth, and feed rates of extractant and fermentation broth may be adjusted accordingly to improve phase separation. On-line devices such as optical devices may be used to detect the presence of a rag layer (e.g., mixture of oil, aqueous solution, and solids) in, for example, extractor 120, and the ratio of fermentation broth to extractant may be adjusted to minimize the formation of a rag layer. On-line measurements of stream 135 from separation 130 may be used to monitor the presence of fermentation broth in this stream, and the presence of fermentation broth in stream 135 may indicate poor phase separation. If the concentration of fermentation broth in stream 135 exceeds a certain setpoint, process changes such as flow rate adjustments or adjustments to the ratio of fermentation broth to extractant may be implemented to improve phase separation. In addition, the concentration of product alcohol in stream 135 may be used as a process control feedback loop to ensure efficient operation of separation 130.
[0201] As another example, on-line measurements of the concentration of product alcohol in stream 127 may be used to monitor extraction efficiency and to maintain a concentration of product alcohol in fermentation 30 that is tolerable to microorganisms. In addition, stream 127 may be monitored for the presence of extractant as a means to minimize the amount of extractant returning to fermentation 30. For example, spectroscopic and process imaging techniques may be used to monitor the presence of extractant in stream 127. Furthermore, a certain concentration of extractant in stream 127 may be maintained to improve extraction efficiency and phase separation.
[0202] In another embodiment, stream 135 from separation 130 may be conducted to purification 150 for further processing including recovery of product alcohol and extractant 152. Extractant 152 may be conducted to extractor 120. On-line measurements may be used to monitor stream 152 for contaminants and degradation products. By monitoring stream 152, the potential for contamination of extractor 120 and fermentation 30 is minimized. If there is an increase in contaminants in stream 152, this stream may be further processed to remove these contaminants, for example, by absorption or chemical reaction.
[0203] During the extraction process, a rag layer may form at the interface of the aqueous and organic phases, and the rag layer, composed of solids and extractant (e.g., droplets of extractant), can accumulate and possibly interfere with phase separation. To mitigate the formation of rag layer, agitation of the aqueous and organic phases may be employed. For example, an impeller may be used to disperse the rag layer at the aqueous-organic interface. Also, fluid flow such as a recirculating loop or vibrations/oscillations may be used to disrupt rag formation. FIGS. 13A and 13B illustrate exemplary processes for mitigating formation of a rag layer. FIG. 13A exemplifies the use of a static mixer in combination with an agitation unit downstream of the settler or decanter for the treatment of a rag layer, and FIG. 13B exemplifies the use of a static mixer in combination with an agitation unit upstream of the settler or decanter for the treatment of a rag layer. In some embodiments, other devices such as coalescers or sonic agitation may be used to disperse the rag layer. In some embodiments, these devices may be integrated into the settler or decanter.
[0204] The processes and systems described herein may be conducted using batch, fed-batch, or continuous fermentation. Batch fermentation is a closed system in which the composition of the fermentation broth is established at the beginning of the fermentation and is not subjected to artificial alterations during the fermentation process. In some embodiments of batch fermentation, extractant may be added to the fermentor. In some embodiments, the volume of extractant may be about 20% to about 60% of the fermentor working volume.
[0205] Fed-batch fermentation is a variation of batch fermentation, in which substrates (e.g., fermentable sugars) are added in increments during the fermentation process. Fed-batch systems are useful when catabolite repression may inhibit the metabolism of the microorganism and where it is desirable to have limited amounts of substrate in the media. In some embodiments, concentrations of substrate and/or nutrients may be monitored during fermentation. In some embodiments, parameters such as pH, dissolved oxygen, and gases (e.g., CO.sub.2) may be monitored during fermentation. From these measurements, the rate or amount of substrate and/or nutrients addition may be determined. In some embodiments, as the level or amount of fermentation broth decreases during fermentation, additional mash may be added to the fermentor to maintain the level or amount of fermentation broth, for example, maintain the level or amount of fermentation broth at the initiation of the fermentation process. In some embodiments of fed-batch fermentation, extractant may be added to the fermentor.
[0206] Continuous fermentation is an open system where fermentation broth is added continuously to a fermentor and an amount of fermentation broth is removed for further processing (e.g., recovery of product alcohol). In some embodiments, addition and removal of fermentation broth may be simultaneous. In some embodiments, equal amounts of fermentation broth may be added and removed from the fermentor. In some embodiments of continuous fermentation, extractant may be added to the fermentor. In some embodiments, the volume of extractant may be about 3% to about 50% of the fermentor working volume. In some embodiments, the volume of extractant may be about 3% to about 20% of the fermentor working volume. In some embodiments, the volume of extractant may be about 3% to about 10% of the fermentor working volume.
[0207] In some embodiments of the processes and systems described herein, gas stripping may be used to remove product alcohol from the fermentation broth. Gas stripping may be performed by providing one or more gases such as air, nitrogen, or carbon dioxide to the fermentation broth, thereby forming a product alcohol-containing gas phase. For example, gas stripping may be performed by sparging one or more gases through the fermentation broth. In some embodiments, the gas may be provided by the fermentation reaction. As an example, carbon dioxide may be provided as a by-product of the metabolism of a fermentable carbon source by the microorganism. In some embodiments, gas stripping may be used concurrently with extractant to remove product alcohol from the fermentation broth. Product alcohol may be recovered from the product alcohol-containing gas phase using methods known in the art, such as using a chilled water trap to condense the product alcohol, or scrubbing the gas phase with a solvent.
Recombinant Microorganisms and Biosynthetic Pathways
[0208] While not wishing to be bound by theory, it is believed that the processes described herein are useful in conjunction with any alcohol-producing microorganism, particularly recombinant microorganisms which produce alcohol at titers above their tolerance levels.
[0209] Alcohol-producing microorganisms are known in the art. For example, fermentative oxidation of methane by methanotrophic bacteria (e.g., Methylosinus trichosporium) produces methanol, and the yeast strain CEN.PK113-7D (CBS 8340, the Centraal Buro voor Schimmelculture; van Dijken, et al., Enzyme Microb. Techno. 26:706-714, 2000) produces ethanol. Recombinant microorganisms which produce alcohol are also known in the art (e.g., Ohta, et al., Appl. Environ. Microbiol. 57:893-900, 1991; Underwood, et al., Appl. Environ. Microbiol. 68:1071-1081, 2002; Shen and Liao, Metab. Eng. 10:312-320, 2008; Hahnai, et al., Appl. Environ. Microbiol. 73:7814-7818, 2007; U.S. Pat. No. 5,514,583; U.S. Pat. No. 5,712,133; PCT Application Publication No. WO 1995/028476; Feldmann, et al., Appl. Microbiol. Biotechnol. 38: 354-361, 1992; Zhang, et al., Science 267:240-243, 1995; U.S. Patent Application Publication No. 2007/0031918 A1; U.S. Pat. No. 7,223,575; U.S. Pat. No. 7,741,119; U.S. Pat. No. 7,851,188; U.S. Patent Application Publication No. 2009/0203099 A1; U.S. Patent Application Publication No. 2009/0246846 A1; and PCT Application Publication No. WO 2010/075241, which are all herein incorporated by reference).
[0210] In addition, microorganisms may be modified using recombinant technologies to generate recombinant microorganisms capable of producing product alcohols such as ethanol and butanol. Microorganisms that may be recombinantly modified to produce a product alcohol via a biosynthetic pathway include members of the genera Clostridium, Zymomonas, Escherichia, Salmonella, Serratia, Erwinia, Klebsiella, Shigella, Rhodococcus, Pseudomonas, Bacillus, Lactobacillus, Enterococcus, Alcaligenes, Klebsiella, Paenibacillus, Arthrobacter, Corynebacterium, Brevibacterium, Schizosaccharomyces, Kluyveromyces, Yarrowia, Pichia, Candida, Hansenula, Issatchenkia, or Saccharomyces. In some embodiments, recombinant microorganisms may be selected from the group consisting of Escherichia coli, Lactobacillus plantarum, Kluyveromyces lactis, Kluyveromyces marxianus and Saccharomyces cerevisiae. In some embodiments, the recombinant microorganism is yeast. In some embodiments, the recombinant microorganism is crabtree-positive yeast selected from Saccharomyces, Zygosaccharomyces, Schizosaccharomyces, Dekkera, Torulopsis, Brettanomyces, and some species of Candida. Species of crabtree-positive yeast include, but are not limited to, Saccharomyces cerevisiae, Saccharomyces kluyveri, Schizosaccharomyces pombe, Saccharomyces bayanus, Saccharomyces mikitae, Saccharomyces paradoxus, Zygosaccharomyces rouxii, and Candida glabrata.
[0211] Saccharomyces cerevisiae are known in the art and are available from a variety of sources including, but not limited to, American Type Culture Collection (Rockville, Md.), Centraalbureau voor Schimmelcultures (CBS) Fungal Biodiversity Centre, LeSaffre, Gert Strand AB, Ferm Solutions, North American Bioproducts, Martrex, and Lallemand. Saccharomyces cerevisiae include, but are not limited to, BY4741, CEN.PK 113-7D, Ethanol Red.RTM. yeast, Ferm Pro.TM. yeast, Bio-Ferm.RTM. XR yeast, Gert Strand Prestige Batch Turbo alcohol yeast, Gert Strand Pot Distillers yeast, Gert Strand Distillers Turbo yeast, FerMax.TM. Green yeast, FerMax.TM. Gold yeast, Thermosacc.RTM. yeast, BG-1, PE-2, CAT-1, CBS7959, CBS7960, and CBS7961.
[0212] In some embodiments, the microorganism may be immobilized or encapsulated. For example, the microorganism may be immobilized or encapsulated using alginate, calcium alginate, or polyacrylamide gels, or through the induction of biofilm formation onto a variety of high surface area support matrices such as diatomite, celite, diatomaceous earth, silica gels, plastics, or resins. In some embodiments, ISPR may be used in combination with immobilized or encapsulated microorganisms. This combination may improve productivity such as specific volumetric productivity, metabolic rate, product alcohol yields, tolerance to product alcohol. In addition, immobilization and encapsulation may minimize the effects of the process conditions such as shearing on the microorganisms.
[0213] The production of butanol utilizing fermentation, as well as microorganisms which produce butanol, is disclosed, for example, in U.S. Pat. No. 7,851,188, and U.S. Patent Application Publication Nos. 2007/0092957; 2007/0259410; 2007/0292927; 2008/0182308; 2008/0274525; 2009/0155870; 2009/0305363; and 2009/0305370, the entire contents of each are herein incorporated by reference. In some embodiments, the microorganism is engineered to contain a biosynthetic pathway. In some embodiments, the biosynthetic pathway is an engineered butanol biosynthetic pathway. In some embodiments, the biosynthetic pathway converts pyruvate to a fermentative product. In some embodiments, the biosynthetic pathway converts pyruvate as well as amino acids to a fermentative product. In some embodiments, at least one, at least two, at least three, or at least four polypeptides catalyzing substrate to product conversions of a pathway are encoded by heterologous polynucleotides in the microorganism. In some embodiments, all polypeptides catalyzing substrate to product conversions of a pathway are encoded by heterologous polynucleotides in the microorganism. In some embodiments, the polypeptide catalyzing the substrate to product conversions of acetolactate to 2,3-dihydroxyisovalerate and/or the polypeptide catalyzing the substrate to product conversion of isobutyraldehyde to isobutanol are capable of utilizing reduced nicotinamide adenine dinucleotide (NADH) as a cofactor.
Biosynthetic Pathways
[0214] Biosynthetic pathways for the production of isobutanol that may be used include those described in U.S. Pat. No. 7,851,188, which is incorporated herein by reference. In one embodiment, the isobutanol biosynthetic pathway comprises the following substrate to product conversions:
[0215] a) pyruvate to acetolactate, which may be catalyzed, for example, by acetolactate synthase;
[0216] b) acetolactate to 2,3-dihydroxyisovalerate, which may be catalyzed, for example, by acetohydroxy acid reductoisomerase;
[0217] c) 2,3-dihydroxyisovalerate to .alpha.-ketoisovalerate, which may be catalyzed, for example, by acetohydroxy acid dehydratase;
[0218] d) .alpha.-ketoisovalerate to isobutyraldehyde, which may be catalyzed, for example, by a branched-chain .alpha.-keto acid decarboxylase; and
[0219] e) isobutyraldehyde to isobutanol, which may be catalyzed, for example, by a branched-chain alcohol dehydrogenase.
[0220] In another embodiment, the isobutanol biosynthetic pathway comprises the following substrate to product conversions:
[0221] a) pyruvate to acetolactate, which may be catalyzed, for example, by acetolactate synthase;
[0222] b) acetolactate to 2,3-dihydroxyisovalerate, which may be catalyzed, for example, by ketol-acid reductoisomerase;
[0223] c) 2,3-dihydroxyisovalerate to .alpha.-ketoisovalerate, which may be catalyzed, for example, by dihydroxyacid dehydratase;
[0224] d) .alpha.-ketoisovalerate to valine, which may be catalyzed, for example, by transaminase or valine dehydrogenase;
[0225] e) valine to isobutylamine, which may be catalyzed, for example, by valine decarboxylase;
[0226] f) isobutylamine to isobutyraldehyde, which may be catalyzed by, for example, omega transaminase; and
[0227] g) isobutyraldehyde to isobutanol, which may be catalyzed, for example, by a branched-chain alcohol dehydrogenase.
[0228] In another embodiment, the isobutanol biosynthetic pathway comprises the following substrate to product conversions:
[0229] a) pyruvate to acetolactate, which may be catalyzed, for example, by acetolactate synthase;
[0230] b) acetolactate to 2,3-dihydroxyisovalerate, which may be catalyzed, for example, by acetohydroxy acid reductoisomerase;
[0231] c) 2,3-dihydroxyisovalerate to .alpha.-ketoisovalerate, which may be catalyzed, for example, by acetohydroxy acid dehydratase;
[0232] d) .alpha.-ketoisovalerate to isobutyryl-CoA, which may be catalyzed, for example, by branched-chain keto acid dehydrogenase;
[0233] e) isobutyryl-CoA to isobutyraldehyde, which may be catalyzed, for example, by acylating aldehyde dehydrogenase; and
[0234] f) isobutyraldehyde to isobutanol, which may be catalyzed, for example, by a branched-chain alcohol dehydrogenase.
[0235] Biosynthetic pathways for the production of 1-butanol that may be used include those described in U.S. Patent Application Publication No. 2008/0182308, which is incorporated herein by reference. In one embodiment, the 1-butanol biosynthetic pathway comprises the following substrate to product conversions:
[0236] a) acetyl-CoA to acetoacetyl-CoA, which may be catalyzed, for example, by acetyl-CoA acetyltransferase;
[0237] b) acetoacetyl-CoA to 3-hydroxybutyryl-CoA, which may be catalyzed, for example, by 3-hydroxybutyryl-CoA dehydrogenase;
[0238] c) 3-hydroxybutyryl-CoA to crotonyl-CoA, which may be catalyzed, for example, by crotonase;
[0239] d) crotonyl-CoA to butyryl-CoA, which may be catalyzed, for example, by butyryl-CoA dehydrogenase;
[0240] e) butyryl-CoA to butyraldehyde, which may be catalyzed, for example, by butyraldehyde dehydrogenase; and
[0241] f) butyraldehyde to 1-butanol, which may be catalyzed, for example, by butanol dehydrogenase.
[0242] Biosynthetic pathways for the production of 2-butanol that may be used include those described in U.S. Patent Application Publication No. 2007/0259410 and U.S. Patent Application Publication No. 2009/0155870, which are incorporated herein by reference. In one embodiment, the 2-butanol biosynthetic pathway comprises the following substrate to product conversions:
[0243] a) pyruvate to alpha-acetolactate, which may be catalyzed, for example, by acetolactate synthase;
[0244] b) alpha-acetolactate to acetoin, which may be catalyzed, for example, by acetolactate decarboxylase;
[0245] c) acetoin to 3-amino-2-butanol, which may be catalyzed, for example, acetonin aminase;
[0246] d) 3-amino-2-butanol to 3-amino-2-butanol phosphate, which may be catalyzed, for example, by aminobutanol kinase;
[0247] e) 3-amino-2-butanol phosphate to 2-butanone, which may be catalyzed, for example, by aminobutanol phosphate phosphorylase; and
[0248] f) 2-butanone to 2-butanol, which may be catalyzed, for example, by butanol dehydrogenase.
[0249] In another embodiment, the 2-butanol biosynthetic pathway comprises the following substrate to product conversions:
[0250] a) pyruvate to alpha-acetolactate, which may be catalyzed, for example, by acetolactate synthase;
[0251] b) alpha-acetolactate to acetoin, which may be catalyzed, for example, by acetolactate decarboxylase;
[0252] c) acetoin to 2,3-butanediol, which may be catalyzed, for example, by butanediol dehydrogenase;
[0253] d) 2,3-butanediol to 2-butanone, which may be catalyzed, for example, by dial dehydratase; and
[0254] e) 2-butanone to 2-butanol, which may be catalyzed, for example, by butanol dehydrogenase.
[0255] Biosynthetic pathways for the production of 2-butanone that may be used include those described in U.S. Patent Application Publication No. 2007/0259410 and U.S. Patent Application Publication No. 2009/0155870, which are incorporated herein by reference. In one embodiment, the 2-butanone biosynthetic pathway comprises the following substrate to product conversions:
[0256] a) pyruvate to alpha-acetolactate, which may be catalyzed, for example, by acetolactate synthase;
[0257] b) alpha-acetolactate to acetoin, which may be catalyzed, for example, by acetolactate decarboxylase;
[0258] c) acetoin to 3-amino-2-butanol, which may be catalyzed, for example, acetonin aminase;
[0259] d) 3-amino-2-butanol to 3-amino-2-butanol phosphate, which may be catalyzed, for example, by aminobutanol kinase; and
[0260] e) 3-amino-2-butanol phosphate to 2-butanone, which may be catalyzed, for example, by aminobutanol phosphate phosphorylase.
[0261] In another embodiment, the 2-butanone biosynthetic pathway comprises the following substrate to product conversions:
[0262] a) pyruvate to alpha-acetolactate, which may be catalyzed, for example, by acetolactate synthase;
[0263] b) alpha-acetolactate to acetoin which may be catalyzed, for example, by acetolactate decarboxylase;
[0264] c) acetoin to 2,3-butanediol, which may be catalyzed, for example, by butanediol dehydrogenase; and
[0265] d) 2,3-butanediol to 2-butanone, which may be catalyzed, for example, by diol dehydratase.
[0266] The terms "acetohydroxyacid synthase," "acetolactate synthase," and "acetolactate synthetase" (abbreviated "ALS") are used interchangeably herein to refer to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of pyruvate to acetolactate and CO.sub.2. Example acetolactate synthases are known by the EC number 2.2.1.6 (Enzyme Nomenclature 1992, Academic Press, San Diego). These unmodified enzymes are available from a number of sources, including, but not limited to, Bacillus subtilis (GenBank Nos: CAB15618 (SEQ ID NO: 1), Z99122 (SEQ ID NO: 2), NCBI (National Center for Biotechnology Information) amino acid sequence, NCBI nucleotide sequence, respectively), Klebsiella pneumoniae (GenBank Nos: AAA25079 (SEQ ID NO: 3), M73842 (SEQ ID NO: 4)), and Lactococcus lactis (GenBank Nos: AAA25161 (SEQ ID NO: 5), L16975 (SEQ ID NO: 6)).
[0267] The term "ketol-acid reductoisomerase" ("KARI"), "acetohydroxy acid isomeroreductase," and "acetohydroxy acid reductoisomerase" will be used interchangeably and refer to a polypeptide (or polypeptides) having enzyme activity that catalyzes the reaction of (S)-acetolactate to 2,3-dihydroxyisovalerate. Example KARI enzymes may be classified as EC number EC 1.1.1.86 (Enzyme Nomenclature 1992, Academic Press, San Diego), and are available from a vast array of microorganisms, including, but not limited to, Escherichia coli (GenBank Nos: NP_418222 (SEQ ID NO: 7), NC_000913 (SEQ ID NO: 8)), Saccharomyces cerevisiae (GenBank Nos: NP_013459 (SEQ ID NO: 9), NC_001144 (SEQ ID NO: 10)), Methanococcus maripaludis (GenBank Nos: CAF30210 (SEQ ID NO: 11), BX957220 (SEQ ID NO: 12)), and Bacillus subtilis (GenBank Nos: CAB14789 (SEQ ID NO: 13), Z99118 (SEQ ID NO: 14)). KARIs include Anaerostipes caccae KARI variants "K9G9" and "K9D3" (SEQ ID NOs: 15 and 16, respectively). Ketol-acid reductoisomerase (KARI) enzymes are described in U.S. Patent Application Publication Nos. 2008/0261230, 2009/0163376, and 2010/0197519, and PCT Application Publication No. WO/2011/041415, which are incorporated herein by reference. Examples of KARIs disclosed therein are those from Lactococcus lactis, Vibrio cholera, Pseudomonas aeruginosa PAO1, and Pseudomonas fluorescens PF5 mutants In some embodiments, the KARI utilizes NADH. In some embodiments, the KARI utilizes reduced nicotinamide adenine dinucleotide phosphate (NADPH).
[0268] The term "acetohydroxy acid dehydratase" and "dihydroxyacid dehydratase" ("DHAD") refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion the conversion of 2,3-dihydroxyisovalerate to .alpha.-ketoisovalerate. Example acetohydroxy acid dehydratases are known by the EC number 4.2.1.9. Such enzymes are available from a vast array of microorganisms, including, but not limited to, Escherichia coli (GenBank Nos: YP_026248 (SEQ ID NO: 17), NC000913 (SEQ ID NO: 18)), Saccharomyces cerevisiae (GenBank Nos: NP_012550 (SEQ ID NO: 19), NC 001142 (SEQ ID NO: 20), M. maripaludis (GenBank Nos: CAF29874 (SEQ ID NO: 21), BX957219 (SEQ ID NO: 22)), B. subtilis (GenBank Nos: CAB14105 (SEQ ID NO: 23), Z99115 (SEQ ID NO: 24)), L. lactis, and N. crassa. U.S. Patent Application Publication No. 2010/0081154, and U.S. Pat. No. 7,851,188, which are incorporated herein by reference, describe dihydroxyacid dehydratases (DHADs), including a DHAD from Streptococcus mutans.
[0269] The term "branched-chain .alpha.-keto acid decarboxylase," ".alpha.-ketoacid decarboxylase," ".alpha.-ketoisovalerate decarboxylase," or "2-ketoisovalerate decarboxylase" ("KIVD") refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of .alpha.-ketoisovalerate to isobutyraldehyde and CO.sub.2. Example branched-chain .alpha.-keto acid decarboxylases are known by the EC number 4.1.1.72 and are available from a number of sources, including, but not limited to, Lactococcus lactis (GenBank Nos: AAS49166 (SEQ ID NO: 25), AY548760 (SEQ ID NO: 26); CAG34226 (SEQ ID NO: 27), AJ746364 (SEQ ID NO: 28), Salmonella typhimurium (GenBank Nos: NP_461346 (SEQ ID NO: 29), NC_003197 (SEQ ID NO: 30)), Clostridium acetobutylicum (GenBank Nos: NP_149189 (SEQ ID NO: 31), NC_001988 (SEQ ID NO: 32)), M. caseolyticus (SEQ ID NO: 33), and L. grayi (SEQ ID NO: 34).
[0270] The term "branched-chain alcohol dehydrogenase" ("ADH") refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of isobutyraldehyde to isobutanol. Example branched-chain alcohol dehydrogenases are known by the EC number 1.1.1.265, but may also be classified under other alcohol dehydrogenases (specifically, EC 1.1.1.1 or 1.1.1.2). Alcohol dehydrogenases may be NADPH-dependent or NADH-dependent. Such enzymes are available from a number of sources, including, but not limited to, Saccharomyces cerevisiae (GenBank Nos: NP_010656 (SEQ ID NO: 35), NC_001136 (SEQ ID NO: 36), NP_014051 (SEQ ID NO: 37), NC_001145 (SEQ ID NO: 38)), Escherichia coli (GenBank Nos: NP_417484 (SEQ ID NO: 39), NC_000913 (SEQ ID NO: 40)), C. acetobutylicum (GenBank Nos: NP_349892 (SEQ ID NO: 41), NC_003030 (SEQ ID NO: 42); NP_349891 (SEQ ID NO: 43), NC_003030 (SEQ ID NO: 44)). U.S. Patent Application Publication No. 2009/0269823 describes SadB, an alcohol dehydrogenase (ADH) from Achromobacter xylosoxidans. Alcohol dehydrogenases also include horse liver ADH and Beijerinkia indica ADH (as described by U.S. Patent Application Publication No. 2011/0269199, which is incorporated herein by reference).
[0271] The term "butanol dehydrogenase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of isobutyraldehyde to isobutanol or the conversion of 2-butanone and 2-butanol. Butanol dehydrogenases are a subset of a broad family of alcohol dehydrogenases. Butanol dehydrogenase may be NAD- or NADP-dependent. The NAD-dependent enzymes are known as EC 1.1.1.1 and are available, for example, from Rhodococcus ruber (GenBank Nos: CAD36475, AJ491307). The NADP dependent enzymes are known as EC 1.1.1.2 and are available, for example, from Pyrococcus furiosus (GenBank Nos: AAC25556, AF013169). Additionally, a butanol dehydrogenase is available from Escherichia coli (GenBank Nos: NP 417484, NC_000913) and a cyclohexanol dehydrogenase is available from Acinetobacter sp. (GenBank Nos: AAG10026, AF282240). The term "butanol dehydrogenase" also refers to an enzyme that catalyzes the conversion of butyraldehyde to 1-butanol, using either NADH or NADPH as cofactor. Butanol dehydrogenases are available from, for example, C. acetobutylicum (GenBank Nos: NP_149325, NC_001988; note: this enzyme possesses both aldehyde and alcohol dehydrogenase activity); NP_349891, NC_003030; and NP_349892, NC_003030) and Escherichia coli (GenBank Nos: NP_417-484, NC_000913).
[0272] The term "branched-chain keto acid dehydrogenase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of .alpha.-ketoisovalerate to isobutyryl-CoA (isobutyryl-coenzyme A), typically using NAD.sup.+ (nicotinamide adenine dinucleotide) as an electron acceptor. Example branched-chain keto acid dehydrogenases are known by the EC number 1.2.4.4. Such branched-chain keto acid dehydrogenases are comprised of four subunits and sequences from all subunits are available from a vast array of microorganisms, including, but not limited to, Bacillus subtilis (GenBank Nos: CAB14336 (SEQ ID NO: 45), Z99116 (SEQ ID NO: 46); CAB14335 (SEQ ID NO: 47), Z99116 (SEQ ID NO: 48); CAB14334 (SEQ ID NO: 49), Z99116 (SEQ ID NO: 50); and CAB14337 (SEQ ID NO: 51), Z99116 (SEQ ID NO: 52)) and Pseudomonas putida (GenBank Nos: AAA65614 (SEQ ID NO: 53), M57613 (SEQ ID NO: 54); AAA65615 (SEQ ID NO: 55), M57613 (SEQ ID NO: 56); AAA65617 (SEQ ID NO: 57), M57613 (SEQ ID NO: 58); and AAA65618 (SEQ ID NO: 59), M57613 (SEQ ID NO: 60)).
[0273] The term "acylating aldehyde dehydrogenase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of isobutyryl-CoA to isobutyraldehyde, typically using either NADH or NADPH as an electron donor. Example acylating aldehyde dehydrogenases are known by the EC numbers 1.2.1.10 and 1.2.1.57. Such enzymes are available from multiple sources, including, but not limited to, Clostridium beijerinckii (GenBank Nos: AAD31841 (SEQ ID NO: 61), AF157306 (SEQ ID NO: 62)), Clostridium acetobutylicum (GenBank Nos: NP_149325 (SEQ ID NO: 63), NC_001988 (SEQ ID NO: 64); NP_149199 (SEQ ID NO: 65), NC_001988 (SEQ ID NO: 66)), Pseudomonas putida (GenBank Nos: AAA89106 (SEQ ID NO: 67), U13232 (SEQ ID NO: 68)), and Thermus thermophilus (GenBank Nos: YP_145486 (SEQ ID NO: 69), NC_006461 (SEQ ID NO: 70)).
[0274] The term "transaminase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of .alpha.-ketoisovalerate to L-valine, using either alanine or glutamate as an amine donor. Example transaminases are known by the EC numbers 2.6.1.42 and 2.6.1.66. Such enzymes are available from a number of sources. Examples of sources for alanine-dependent enzymes include, but are not limited to, Escherichia coli (GenBank Nos: YP_026231 (SEQ ID NO: 71), NC_000913 (SEQ ID NO: 72)) and Bacillus licheniformis (GenBank Nos: YP_093743 (SEQ ID NO: 73), NC_006322 (SEQ ID NO: 74)). Examples of sources for glutamate-dependent enzymes include, but are not limited to, Escherichia coli (GenBank Nos: YP_026247 (SEQ ID NO: 75), NC_000913 (SEQ ID NO: 76)), Saccharomyces cerevisiae (GenBank Nos: NP_012682 (SEQ ID NO: 77), NC_001142 (SEQ ID NO: 78)) and Methanobacterium thermoautotrophicum (GenBank Nos: NP_276546 (SEQ ID NO: 79), NC_000916 (SEQ ID NO: 80)).
[0275] The term "valine dehydrogenase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of .alpha.-ketoisovalerate to L-valine, typically using NAD(P)H as an electron donor and ammonia as an amine donor. Example valine dehydrogenases are known by the EC numbers 1.4.1.8 and 1.4.1.9 and such enzymes are available from a number of sources, including, but not limited to, Streptomyces coelicolor (GenBank Nos: NP_628270 (SEQ ID NO: 81), NC_003888 (SEQ ID NO: 82)) and Bacillus subtilis (GenBank Nos: CAB14339 (SEQ ID NO: 83), Z99116 (SEQ ID NO: 84)).
[0276] The term "valine decarboxylase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of L-valine to isobutylamine and CO.sub.2. Example valine decarboxylases are known by the EC number 4.1.1.14. Such enzymes are found in Streptomyces, such as for example, Streptomyces viridifaciens (GenBank Nos: AAN10242 (SEQ ID NO: 85), AY116644 (SEQ ID NO: 86)).
[0277] The term "omega transaminase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of isobutylamine to isobutyraldehyde using a suitable amino acid as an amine donor. Example omega transaminases are known by the EC number 2.6.1.18 and are available from a number of sources, including, but not limited to, Alcaligenes denitrificans (AAP92672 (SEQ ID NO: 87), AY330220 (SEQ ID NO: 88)), Ralstonia eutropha (GenBank Nos: YP_294474 (SEQ ID NO: 89), NC_007347 (SEQ ID NO: 90)), Shewanella oneidensis (GenBank Nos: NP_719046 (SEQ ID NO: 91), NC_004347 (SEQ ID NO: 92)), and Pseudomonas putida (GenBank Nos: AAN66223 (SEQ ID NO: 93), AE016776 (SEQ ID NO: 94)).
[0278] The term "acetyl-CoA acetyltransferase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of two molecules of acetyl-CoA to acetoacetyl-CoA and coenzyme A (CoA). Example acetyl-CoA acetyltransferases are acetyl-CoA acetyltransferases with substrate preferences (reaction in the forward direction) for a short chain acyl-CoA and acetyl-CoA and are classified as E.C. 2.3.1.9 [Enzyme Nomenclature 1992, Academic Press, San Diego]; although, enzymes with a broader substrate range (E.C. 2.3.1.16) will be functional as well. Acetyl-CoA acetyltransferases are available from a number of sources, for example, Escherichia coli (GenBank Nos: NP_416728, NC_000913; NCBI (National Center for Biotechnology Information) amino acid sequence, NCBI nucleotide sequence), Clostridium acetobutylicum (GenBank Nos: NP_349476.1, NC_003030; NP_149242, NC_001988, Bacillus subtilis (GenBank Nos: NP_390297, NC_000964), and Saccharomyces cerevisiae (GenBank Nos: NP_015297, NC_001148).
[0279] The term "3-hydroxybutyryl-CoA dehydrogenase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of acetoacetyl-CoA to 3-hydroxybutyryl-CoA. Example 3-hydroxybutyryl-CoA dehydrogenases may be NADH-dependent, with a substrate preference for (S)-3-hydroxybutyryl-CoA or (R)-3-hydroxybutyryl-CoA. Examples may be classified as E.C. 1.1.1.35 and E.C. 1.1.1.30, respectively. Additionally, 3-hydroxybutyryl-CoA dehydrogenases may be NADPH-dependent, with a substrate preference for (S)-3-hydroxybutyryl-CoA or (R)-3-hydroxybutyryl-CoA and are classified as E.C. 1.1.1.157 and E.C. 1.1.1.36, respectively. 3-Hydroxybutyryl-CoA dehydrogenases are available from a number of sources, for example, Clostridium acetobutylicum (GenBank Nos: NP_349314, NC_003030), Bacillus subtilis (GenBank Nos: AAB09614, U29084), Ralstonia eutropha (GenBank Nos: YP_294481, NC_007347), and Alcaligenes eutrophus (GenBank Nos: AAA21973, J04987).
[0280] The term "crotonase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of 3-hydroxybutyryl-CoA to crotonyl-CoA and H.sub.2O. Example crotonases may have a substrate preference for (S)-3-hydroxybutyryl-CoA or (R)-3-hydroxybutyryl-CoA and may be classified as E.C. 4.2.1.17 and E.C. 4.2.1.55, respectively. Crotonases are available from a number of sources, for example, Escherichia coli (GenBank Nos: NP_415911, NC_000913), Clostridium acetobutylicum (GenBank Nos: NP_349318, NC_003030), Bacillus subtilis (GenBank Nos: CAB13705, Z99113), and Aeromonas caviae (GenBank Nos: BAA21816, D88825).
[0281] The term "butyryl-CoA dehydrogenase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of crotonyl-CoA to butyryl-CoA. Example butyryl-CoA dehydrogenases may be NADH-dependent, NADPH-dependent, or flavin-dependent and may be classified as E.C. 1.3.1.44, E.C. 1.3.1.38, and E.C. 1.3.99.2, respectively. Butyryl-CoA dehydrogenases are available from a number of sources, for example, Clostridium acetobutylicum (GenBank Nos: NP_347102, NC.sub.-- 003030), Euglena gracilis (GenBank Nos: Q5EU90, AY741582), Streptomyces collinus (GenBank Nos: AAA92890, U37135), and Streptomyces coelicolor (GenBank Nos: CAA22721, AL939127).
[0282] The term "butyraldehyde dehydrogenase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of butyryl-CoA to butyraldehyde, using NADH or NADPH as cofactor. Butyraldehyde dehydrogenases with a preference for NADH are known as E.C. 1.2.1.57 and are available from, for example, Clostridium beijerinckii (GenBank Nos: AAD31841, AF157306) and Clostridium acetobutylicum (GenBank Nos: NP.sub.-149325, NC.sub.-001988).
[0283] The term "isobutyryl-CoA mutase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of butyryl-CoA to isobutyryl-CoA. This enzyme uses coenzyme B.sub.12 as cofactor. Example isobutyryl-CoA mutases are known by the EC number 5.4.99.13. These enzymes are found in a number of Streptomyces, including, but not limited to, Streptomyces cinnamonensis (GenBank Nos: AAC08713 (SEQ ID NO: 95), U67612 (SEQ ID NO: 96); CAB59633 (SEQ ID NO: 97), AJ246005 (SEQ ID NO: 98)), Streptomyces coelicolor (GenBank Nos: CAB70645 (SEQ ID NO: 99), AL939123 (SEQ ID NO: 100); CAB92663 (SEQ ID NO: 101), AL939121 (SEQ ID NO: 102)), and Streptomyces avermitilis (GenBank Nos: NP_824008 (SEQ ID NO: 103), NC_003155 (SEQ ID NO: 104); NP_824637 (SEQ ID NO: 105), NC_003155 (SEQ ID NO: 106)).
[0284] The term "acetolactate decarboxylase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of alpha-acetolactate to acetoin. Example acetolactate decarboxylases are known as EC 4.1.1.5 and are available, for example, from Bacillus subtilis (GenBank Nos: AAA22223, L04470), Klebsiella terrigena (GenBank Nos: AAA25054, L04507) and Klebsiella pneumoniae (GenBank Nos: AAU43774, AY722056).
[0285] The term "acetoin aminase" or "acetoin transaminase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of acetoin to 3-amino-2-butanol. Acetoin aminase may utilize the cofactor pyridoxal 5'-phosphate or NADH or NADPH. The resulting product may have (R) or (S) stereochemistry at the 3-position. The pyridoxal phosphate-dependent enzyme may use an amino acid such as alanine or glutamate as the amino donor. The NADH- and NADPH-dependent enzymes may use ammonia as a second substrate. A suitable example of an NADH-dependent acetoin aminase, also known as amino alcohol dehydrogenase, is described by Ito, et al. (U.S. Pat. No. 6,432,688). An example of a pyridoxal-dependent acetoin aminase is the amine:pyruvate aminotransferase (also called amine:pyruvate transaminase) described by Shin and Kim (J. Org. Chem. 67:2848-2853, 2002).
[0286] The term "acetoin kinase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of acetoin to phosphoacetoin. Acetoin kinase may utilize ATP (adenosine triphosphate) or phosphoenolpyruvate as the phosphate donor in the reaction. Enzymes that catalyze the analogous reaction on the similar substrate dihydroxyacetone, for example, include enzymes known as EC 2.7.1.29 (Garcia-Alles, et al., Biochemistry 43:13037-13046, 2004).
[0287] The term "acetoin phosphate aminase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of phosphoacetoin to 3-amino-2-butanol O-phosphate. Acetoin phosphate aminase may use the cofactor pyridoxal 5'-phosphate, NADH, or NADPH. The resulting product may have (R) or (S) stereochemistry at the 3-position. The pyridoxal phosphate-dependent enzyme may use an amino acid such as alanine or glutamate. The NADH-dependent and NADPH-dependent enzymes may use ammonia as a second substrate. Although there are no reports of enzymes catalyzing this reaction on phosphoacetoin, there is a pyridoxal phosphate-dependent enzyme that is proposed to carry out the analogous reaction on the similar substrate serinol phosphate (Yasuta, et al., Appl. Environ. Microbial. 67:4999-5009, 2001).
[0288] The term "aminobutanol phosphate phospholyase," also called "amino alcohol O-phosphate lyase," refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of 3-amino-2-butanol O-phosphate to 2-butanone. Amino butanol phosphate phospho-lyase may utilize the cofactor pyridoxal 5'-phosphate. There are reports of enzymes that catalyze the analogous reaction on the similar substrate 1-amino-2-propanol phosphate (Jones, et al., Biochem J. 134:167-182, 1973). U.S. Patent Application Publication No. 2007/0259410 describes an aminobutanol phosphate phospho-lyase from the organism Erwinia carotovora.
[0289] The term "aminobutanol kinase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of 3-amino-2-butanol to 3-amino-2-butanol O-phosphate. Amino butanol kinase may utilize ATP as the phosphate donor. Although there are no reports of enzymes catalyzing this reaction on 3-amino-2-butanol, there are reports of enzymes that catalyze the analogous reaction on the similar substrates ethanolamine and 1-amino-2-propanol (Jones, et al., supra). U.S. Patent Application Publication No. 2009/0155870 describes, in Example 14, an amino alcohol kinase of Erwinia carotovora subsp. Atroseptica.
[0290] The term "butanediol dehydrogenase" also known as "acetoin reductase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of acetoin to 2,3-butanediol. Butanedial dehydrogenases are a subset of the broad family of alcohol dehydrogenases. Butanediol dehydrogenase enzymes may have specificity for production of (R)- or (S)-stereochemistry in the alcohol product. (S)-specific butanediol dehydrogenases are known as EC 1.1.1.76 and are available, for example, from Klebsiella pneumoniae (GenBank Nos: BBA13085, D86412). (R)-specific butanediol dehydrogenases are known as EC 1.1.1.4 and are available, for example, from Bacillus cereus (GenBank Nos. NP 830481, NC_004722; AAP07682, AE017000), and Lactococcus lactis (GenBank Nos. AAK04995, AE006323).
[0291] The term "butanediol dehydratase," also known as "dial dehydratase" or "propanediol dehydratase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the conversion of 2,3-butanediol to 2-butanone. Butanediol dehydratase may utilize the cofactor adenosyl cobalamin (also known as coenzyme Bw or vitamin B12; although vitamin B12 may refer also to other forms of cobalamin that are not coenzyme B12). Adenosyl cobalamin-dependent enzymes are known as EC 4.2.1.28 and are available, for example, from Klebsiella oxytoca (GenBank Nos: AA08099 (alpha subunit), D45071; BAA08100 (beta subunit), D45071; and BBA08101 (gamma subunit), D45071 (Note all three subunits are required for activity), and Klebsiella pneumonia (GenBank Nos: AAC98384 (alpha subunit), AF102064; GenBank Nos: AAC98385 (beta subunit), AF102064, GenBank Nos: AAC98386 (gamma subunit), AF102064). Other suitable dial dehydratases include, but are not limited to, B12-dependent dial dehydratases available from Salmonella typhimurium (GenBank Nos: AAB84102 (large subunit), AF026270; GenBank Nos: AAB84103 (medium subunit), AF026270; GenBank Nos: AAB84104 (small subunit), AF026270); and Lactobacillus collinoides (GenBank Nos: CAC82541 (large subunit), AJ297723; GenBank Nos: CAC82542 (medium subunit); AJ297723; GenBank Nos: CAD01091 (small subunit), AJ297723); and enzymes from Lactobacillus brevis (particularly strains CNRZ 734 and CNRZ 735, Speranza, et al., J. Agric. Food Chem. 45:3476-3480, 1997), and nucleotide sequences that encode the corresponding enzymes. Methods of dial dehydratase gene isolation are well known in the art (e.g., U.S. Pat. No. 5,686,276).
[0292] The term "pyruvate decarboxylase" refers to a polypeptide (or polypeptides) having enzyme activity that catalyzes the decarboxylation of pyruvic acid to acetaldehyde and carbon dioxide. Pyruvate dehydrogenases are known by the EC number 4.1.1.1. These enzymes are found in a number of yeast, including Saccharomyces cerevisiae (GenBank Nos: CAA97575 (SEQ ID NO: 107), CAA97705 (SEQ ID NO: 109), CAA97091 (SEQ ID NO: 111)).
[0293] It will be appreciated that microorganisms comprising an isobutanol biosynthetic pathway as provided herein may further comprise one or more additional modifications. U.S. Patent Application Publication No. 2009/0305363 (incorporated by reference) discloses increased conversion of pyruvate to acetolactate by engineering yeast for expression of a cytosol-localized acetolactate synthase and substantial elimination of pyruvate decarboxylase activity. In some embodiments, the microorganisms may comprise modifications to reduce glycerol-3-phosphate dehydrogenase activity and/or disruption in at least one gene encoding a polypeptide having pyruvate decarboxylase activity or a disruption in at least one gene encoding a regulatory element controlling pyruvate decarboxylase gene expression as described in U.S. Patent Application Publication No. 2009/0305363 (incorporated herein by reference), and/or modifications that provide for increased carbon flux through an Entner-Doudoroff Pathway or reducing equivalents balance as described in U.S. Patent Application Publication No. 2010/0120105 (incorporated herein by reference). Other modifications include integration of at least one polynucleotide encoding a polypeptide that catalyzes a step in a pyruvate-utilizing biosynthetic pathway. Other modifications include at least one deletion, mutation, and/or substitution in an endogenous polynucleotide encoding a polypeptide having acetolactate reductase activity. In some embodiments, the polypeptide having acetolactate reductase activity is YMR226C (SEQ ID NOs: 127, 128) of Saccharomyces cerevisiae or a homolog thereof. Additional modifications include a deletion, mutation, and/or substitution in an endogenous polynucleotide encoding a polypeptide having aldehyde dehydrogenase and/or aldehyde oxidase activity. In some embodiments, the polypeptide having aldehyde dehydrogenase activity is ALD6 from Saccharomyces cerevisiae or a homolog thereof. A genetic modification which has the effect of reducing glucose repression wherein the yeast production host cell is pdc- is described in U.S. Patent Application Publication No. 2011/0124060, incorporated herein by reference. In some embodiments, the pyruvate decarboxylase that is deleted or down-regulated is selected from the group consisting of: PDC1, PDC5, PDC6, and combinations thereof. In some embodiments, the pyruvate decarboxylase is selected from those enzymes in Table 1. In some embodiments, microorganisms may contain a deletion or down-regulation of a polynucleotide encoding a polypeptide that catalyzes the conversion of glyceraldehyde-3-phosphate to glycerate 1,3, bisphosphate. In some embodiments, the enzyme that catalyzes this reaction is glyceraldehyde-3-phosphate dehydrogenase.
TABLE-US-00001 TABLE 1 SEQ ID Numbers of PDC Target Gene coding regions and Proteins SEQ ID NO: SEQ ID NO: Description (Amino Acid) (Nucleic Acid) PDC1 pyruvate 107 108 decarboxylase from Saccharomyces cerevisiae PDC5 pyruvate 109 110 decarboxylase from Saccharomyces cerevisiae PDC6 pyruvate 111 112 decarboxylase Saccharomyces cerevisiae pyruvate decarboxylase 113 114 from Candida glabrata PDC1 pyruvate 115 116 decarboxylase from Pichia stipitis PDC2 pyruvate 117 118 decarboxylase from Pichia stipitis pyruvate decarboxylase 119 120 from Kluyveromyces lactis pyruvate decarboxylase 121 122 from Yarrowia lipolytica pyruvate decarboxylase 123 124 from Schizosaccharomyces pombe pyruvate decarboxylase 125 126 from Zygosaccharomyces rouxii
[0294] In some embodiments, any particular nucleic acid molecule or polypeptide may be at least 80%, 85%, 90%, 95%, 96%, 97%, 98%, or 99% identical to a nucleotide sequence or polypeptide sequence described herein. The term "percent identity" as known in the art, is a relationship between two or more polypeptide sequences or two or more polynucleotide sequences, as determined by comparing the sequences. In the art, "identity" also means the degree of sequence relatedness between polypeptide or polynucleotide sequences, as the case may be, as determined by the match between strings of such sequences. Identity and similarity can be readily calculated by known methods, including but not limited to those disclosed in: Computational Molecular Biology (Lesk, A. M., Ed.) Oxford University: NY (1988); Biocomputing: Informatics and Genome Projects (Smith, D. W., Ed.) Academic: NY (1993); Computer Analysis of Sequence Data, Part I (Griffin, A. M., and Griffin, H. G., Eds.) Humania: NJ (1994); Sequence Analysis in Molecular Biology (von Heinje, G., Ed.) Academic (1987); and Sequence Analysis Primer (Gribskov, M. and Devereux, J., Eds.) Stockton: NY (1991).
[0295] Methods to determine identity are designed to give the best match between the sequences tested. Methods to determine identity and similarity are codified in publicly available computer programs. Sequence alignments and percent identity calculations may be performed using the MegAlign.TM. program of the LASERGENE bioinformatics computing suite (DNASTAR Inc., Madison, Wis.). Multiple alignment of the sequences may be performed using the Clustal method of alignment which encompasses several varieties of the algorithm including the Clustal V method of alignment corresponding to the alignment method labeled Clustal V (disclosed by Higgins and Sharp, CABIOS. 5:151-153, 1989; Higgins, et al., Comput. Appl. Biosci. 8:189-191, 1992) and found in the MegAlign.TM. program of the LASERGENE bioinformatics computing suite (DNASTAR Inc.). For multiple alignments, the default values correspond to GAP PENALTY=10 and GAP LENGTH PENALTY=10. Default parameters for pairwise alignments and calculation of percent identity of protein sequences using the Clustal method are KTUPLE=1, GAP PENALTY=3, WINDOW=5 and DIAGONALS SAVED=5. For nucleic acids these parameters are KTUPLE=2, GAP PENALTY=5, WINDOW=4 and DIAGONALS SAVED=4. After alignment of the sequences using the Clustal V program, it is possible to obtain a percent identity by viewing the sequence distances table in the same program. Additionally the Clustal W method of alignment is available and corresponds to the alignment method labeled Clustal W (described by Higgins and Sharp, CABIOS. 5:151-153, 1989; Higgins, et al., Comput. Appl. Biosci. 8:189-191, 1992) and found in the MegAlign.TM. v6.1 program of the LASERGENE bioinformatics computing suite (DNASTAR Inc.). Default parameters for multiple alignment (GAP PENALTY=10, GAP LENGTH PENALTY=0.2, Delay Divergen Seqs(%)=30, DNA Transition Weight=0.5, Protein Weight Matrix=Gonnet Series, DNA Weight Matrix=IUB). After alignment of the sequences using the Clustal W program, it is possible to obtain a percent identity by viewing the sequence distances table in the same program.
[0296] Standard recombinant DNA and molecular cloning techniques are well known in the art and are described by Sambrook, et al. (Sambrook, J., Fritsch, E. F. and Maniatis, T. (Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1989, here in referred to as Maniatis) and by Ausubel, et al. (Ausubel, et al., Current Protocols in Molecular Biology, pub. by Greene Publishing Assoc. and Wiley-Interscience, 1987). Examples of methods to construct microorganisms that comprise a butanol biosynthetic pathway are disclosed, for example, in U.S. Pat. No. 7,851,188, and U.S. Patent Application Publication Nos. 2007/0092957; 2007/0259410; 2007/0292927; 2008/0182308; 2008/0274525; 2009/0155870; 2009/0305363; and 2009/0305370, the entire contents of each are herein incorporated by reference.
[0297] Further, while various embodiments of the present invention have been described above, it should be understood that they have been presented by way of example only, and not limitation. It will be apparent to persons skilled in the relevant art that various changes in form and detail can be made therein without departing from the spirit and scope of the invention. Thus, the breadth and scope of the present invention should not be limited by any of the above-described exemplary embodiments, but should be defined only in accordance with the claims and their equivalents.
[0298] All publications, patents, and patent applications mentioned in this specification are indicative of the level of skill of those skilled in the art to which this invention pertains, and are herein incorporated by reference to the same extent as if each individual publication, patent, or patent application was specifically and individually indicated to be incorporated by reference.
EXAMPLES
[0299] The following nonlimiting examples will further illustrate the invention. It should be understood that, while the following examples involve corn as feedstock, other biomass sources such as cane may be used for feedstock without departing from the present invention. Moreover, while the following examples involve ethanol and butanol, other alcohols may be produced without departing from the present invention.
[0300] The meaning of abbreviations is as follows: "atm" means atmosphere, "ccm" means cubic centimeter(s) per minute, "g/L" means gram(s) per liter, "g" means gram(s), "gpl" means gram(s) per liter, "gpm" means gallon(s) per minute, "h" means hour(s), "HPLC" means high performance liquid chromatography, "kg" means kilogram(s), "L" means liter(s), "min" means minute(s), "mL" means milliliter(s), "ppm" means parts per million, "psig" means pound(s) per square inch, gauge, and "wt %" means weight percent.
Example 1
Process for Production and Recovery of Butanol Produced by Fermentation
[0301] The processes described herein may be demonstrated using computational modeling such as Aspen modeling (see, e.g., U.S. Pat. No. 7,666,282). For example, the commercial modeling software Aspen Plus.RTM. (Aspen Technology, Inc., Burlington, Mass.) may be used in conjunction with physical property databases such as DIPPR, available from American Institute of Chemical Engineers, Inc. (New York, N.Y.) to develop an Aspen model for an integrated butanol fermentation, purification, and water management process. This process modeling can perform many fundamental engineering calculations, for example, mass and energy balances, vapor/liquid equilibrium, and reaction rate computations. In order to generate an Aspen model, information input may include, for example, experimental data, water content and composition of feedstock, temperature for mash cooking and flashing, saccharification conditions (e.g., enzyme feed, starch conversion, temperature, pressure), fermentation conditions (e.g., microorganism feed, glucose conversion, temperature, pressure), degassing conditions, solvent columns, pre-flash columns, condensers, evaporators, centrifuges, etc.
[0302] An Aspen model was developed with rigorous material and energy balance in which 53400 kg/h of corn was mashed and fermented to produce isobutanol and in which most of the isobutanol was extracted during fermentation and distilled. This model included an approximation of sequenced batch fermentations as continuous processes. An example of this fermentation, extraction, and distillation process is illustrated in FIG. 14.
[0303] Liquefied corn mash 601 that was clarified to comprise 1.5 wt % suspended solids was pumped at 170.7 tonnes/h and 85.degree. C. through a heat exchanger and a water cooler and fed to fermentation 600 at 32.degree. C. Vapor stream 602 was vented at 17.2 tonnes/h at atmospheric pressure from fermentation 600 to a scrubber with an average continuous molar composition of 95.8% carbon dioxide, 3.4% water, and 0.8% isobutanol. An average beer stream 603 comprising 12.6 gpl isobutanol was discharged continuously from fermentation 600 and pre-heated through a heat exchanger by the mash 601 prior to being distilled for isobutanol recovery.
[0304] Stream 604 with 3875 tonnes/h combined average flow is removed from fermentation 600 at an average isobutanol concentration of 11.1 gpl and an average temperature of 32.degree. C. and circulated through an extractor 610 for partial removal of isobutanol. The exiting aqueous broth 605 containing 7.9 gpl isobutanol is cooled by heat exchange with cooler tower water (CTW) to 30.degree. C. prior to re-entering fermentation 600. A solvent comprising diisopropylbenzene enters the extractor 610 and exits as stream 606 comprising 30.1 gpl isobutanol. The extractor 610 provides effectively five theoretical liquid-liquid equilibrium stages for contacting fermentation broth with solvent. Stream 606 passes at 340 tonnes/h through a heat exchanger and enters the middle of twelve theoretical stages of distillation column 620. A reboiler is operating at 0.6 atm and 183.degree. C. using 150 psig steam to produce solvent stream 607 comprising diisopropylbenzene and essentially no isobutanol that exchanges heat with solvent stream 606 through a heat exchanger and is further cooled by cooling water CTW prior to re-entering extractor 610. The overhead vapor of distillation column 620 is cooled CTW and condensed 630 to form 23.1 tonnes/h of reflux, 0.2 tonnes/h of a residual vapor off-gas 608, and 13.2 tonnes/h of product distillate 609 that comprises 99.2% isobutanol, 0.6% water, and 0.2% diisopropylbenzene.
Example 2
Process for Recovery of Ethanol Using an Extractant Column
[0305] A 1'' diameter Karr.RTM. extraction column (Koch Modular Process Systems, Paramus, N.J.) was used to process fermentation broth that was produced during ethanol fermentation. The column contains a series of plates that run down the length of the column and which are attached to a central shaft. The shaft is attached to a drive which can move the perforated plates (1/4'' diameter perforations) up and down in a reciprocating motion. The frequency of the motion was a variable during testing, but both the stroke length of the oscillation (0.75'') and the spacing of the trays (2'') were fixed. The column used had a plate stack height of 3000 mm.
[0306] The top of the column was provided with an aqueous feed consisting of fermentation broth, while the bottom of the column was provided with a feed of corn oil fatty acids (COFA) as the extractant. The two feeds flowed countercurrent to one another through the column, and were collected as product at opposite ends of the column.
[0307] The fermentation broth was obtained using a fermentation protocol for production of ethanol from liquefied and saccharified corn mash from which, in some cases, some of the solids had been removed via centrifugation. In some cases, the extraction testing was done over the course of several days, such that a portion of the testing was done while CO.sub.2 off-gassing was at or near its maximum, while another portion was done when off-gassing had effectively stopped. The COFA used in this work was distilled grade from Emery Oleochemicals (Cincinnati, Ohio).
[0308] Some experiments were run with COFA as the continuous phase in the column, while others were run with continuous aqueous phase. Experiments were also conducted with or without internals in place. Two types of internals were tested: stainless steel and polytetrafluoroethylene (PTFE). A range of flow rates were examined, in order to determine the flow regimes under which the column could be operated without flooding.
Impact of Dynamic Feed from Fermentation
[0309] During the course of the testing, it was determined that in some cases the column performance varied as the fermentation progressed. Early in the fermentation, the fermentation broth comprising the feed is high in sugar, at intermediate times a considerable amount of CO.sub.2 (which can impact fluid flows) evolves from the fermentation broth, while at later times the concentration of ethanol in the fermentation broth is high. This temporal variation in the feed was reflected in variations in the capacity of the extraction column.
[0310] With conditions using PTFE plates and continuous COFA phase (no agitation), a difference was noted in performance when using fermentation broth collected when the fermentation was near the period of peak gas evolution ("intermediate broth") and toward the end of fermentation ("end broth"). Using end broth, a liquid throughput rate of 14 gpm/ft.sup.2 (Case 3E) was achieved without flooding the column. The maximum throughput for intermediate broth that could be achieved prior to flooding was less than to 9 gpm/ft.sup.2 (case 4D), with noticeable differences in the size and appearance of the aqueous droplets. The droplet size of the aqueous phase was larger (with the formation of globules) in end broth as compared to intermediate broth.
Continuous Phase
[0311] The maximum column throughput was also impacted by the nature of the continuous phase. For end of fermentation conditions, running with continuous aqueous phase and stainless steel (S. Steel) internals, a total liquid capacity of almost 14 gpm/ft.sup.2 was achieved (Case 2B). For continuous organic phase and PTFE internals, the rate was less than 9 gpm/ft2 (Case 4D). Results are shown in Table 2. The abbreviation AQ refers to the aqueous phase and the abbreviation ORG refers to the organic phase. Referring to Table 2, the Phase was continuous, Sample refers to run conditions, Internals refer to the material of the internals, Nom. AQ refers to the nominal aqueous flow rate, Nom. ORG refers to nominal organic flow rate, Total Flow (ccm) refers to the total flow of the aqueous and organic feeds, and Total Flow (gpm/ft.sup.2) refers to total flow per unit cross-sectional area.
TABLE-US-00002 TABLE 2 Nom. Nom. AQ ORG Total Flow Total Flow Phase Sample Internals (ccm) (ccm) (ccm) (gpm/ft.sup.2) AQ 1A S. Steel 160 60 220 10.7 AQ 1B S. Steel 100 30 130 6.3 AQ 1C S. Steel 89 20 109 5.3 AQ 1D S. Steel 120 65-101 -- -- AQ 1E S. Steel 135 60 195 9.4 AQ 1F S. Steel 132 50 182 8.8 AQ 1G S. Steel 150 50 200 9.7 AQ 1G' S. Steel 120 50 170 8.2 AQ 1H S. Steel 210 75 285 13.8 AQ 1I S. Steel 210 75 285 13.8 AQ 2A S. Steel 210 75 285 13.8 AQ 2B S. Steel 210 75 285 13.8 AQ 2C S. Steel 87 85 172 4.1 ORG 3A PTFE 100 50 150 7.3 ORG 3B PTFE 100 100 200 9.7 ORG 3C PTFE 200 100 300 14.5 ORG 3D PTFE 180 75 255 12.4 ORG 3E PTFE 180 120 300 14.5 ORG 3F PTFE 180 170 350 17.0 ORG 3G PTFE 180 60 240 11.6 ORG 3G PTFE 180 60 240 11.6 ORG 4A PTFE 110 70 180 8.7 ORG 4B PTFE 100 30 130 6.3 ORG 4C PTFE 100 60 160 7.7 ORG 4D PTFE 100 80 180 8.7 ORG 4E PTFE 85 60 145 7.0 ORG 4F PTFE 90 40 130 6.3 ORG 4G PTFE 70 40 110 5.3 ORG 4M PTFE 60 60 120 5.8 ORG 4N PTFE 80 80 160 7.7
[0312] When the column was operated without internals using feed comprised of fermentation broth near the end of fermentation, the choice of the continuous phase affected the column capacity. For continuous aqueous phase, it was possible to operate at approximately 25 gpm/ft.sup.2 (cases 2G and 2H). With continuous COFA phase, however, problems with flooding occurred at 18 gpm/ft.sup.2 (Case 2I). Results are shown in Table 3.
TABLE-US-00003 TABLE 3 Nom. Aq Nom. Org. Total Flow Total Flow Phase (ccm) (ccm) (ccm) (gpm/ft.sup.2) AQ 200 75 275 13.3 AQ 200 75 275 13.3 AQ 200 75 275 13.3 AQ 200 120 320 15.5 AQ 240 160 400 19.4 AQ 240 160 400 19.4 AQ 320 170 490 23.7 AQ 390 170 560 27.1 ORG 210 170 380 18.4 ORG 210 170 380 18.4 ORG 60 60 120 5.8 ORG 80 60 140 6.8 ORG 80 80 160 7.7 ORG 90 90 180 8.7 ORG 100 100 200 9.7 ORG 150 50 200 9.7 ORG 170 60 230 11.1
Example 3
Effect of Fermentation Conditions on Extraction Column Capacity
[0313] The nature of fermentation broth is not static, but changes as the fermentation process progresses. In fermentation, the concentration of carbohydrates decreases as the carbohydrates are metabolized by microorganisms. This compositional change in the fermentation broth will alter physical parameters such as viscosity and surface tension of the fermentation broth, which have an effect on the extraction process. In addition to the changes in concentration, at intermediate times a considerable amount of CO.sub.2 is evolved; and this CO.sub.2 will impact the flow of the aqueous and organic liquids through the column.
[0314] A 1'' diameter glass Karr.RTM. extraction column (Koch Modular Process Systems, Paramus, N.J.), outfitted with PTFE internals, was used to process fermentation broth from an ethanol fermentation. The processing was done at several discrete times during the course of the fermentation. Organic extractant (COFA) was the continuous phase in the column, with the fermentation broth passing through the column as droplets. Prior to the introduction of the fermentation broth to the column, the fermentation broth was passed through a tee in the line where CO.sub.2 bubbles present in the feed were removed through a vent.
[0315] With static internals (no agitation), a difference was noted in performance when using fermentation broth taken during the period of peak gas evolution ("intermediate broth") compared to broth taken toward the end of fermentation ("end broth"). Using intermediate broth, a liquid throughput rate of 14 gpm/ft.sup.2 was achieved. The maximum throughput (before column flooding) for the end broth was less than to 9 gpm/ft.sup.2. There were noticeable differences in the size and appearance of the aqueous droplets. The droplet size of the aqueous phase was visibly larger for end broth as compared to intermediate broth.
Example 4
Effect of Isobutanol Concentration on Extraction Column Efficiency
[0316] During a typical fermentation process, the levels of product change with time. This dynamic concentration change can affect the mass transfer in an extraction process.
[0317] To demonstrate the effect of isobutanol concentration, a 1'' diameter glass Karr.RTM. extraction column (Koch Modular Process Systems, Paramus, N.J.), outfitted with stainless steel internals, was used to process fermentation broth from a fermentation that contained approximately 3 g/L of isobutanol. The fermentation broth formed the continuous phase in the extractor, while the organic extractant (COFA) passed through the column as droplets. Although CO.sub.2 production had essentially ceased, the fermentation broth was passed through a tee in the line where any CO.sub.2 bubbles present in the feed were removed prior to the feed entering the extraction column.
[0318] Samples of the feed and exiting streams were analyzed for isobutanol by liquid chromatography (LC) or gas chromatography (GC). Results are shown in (see Table 4). Mass balances were done, and the height of an equilibrium transfer stage (HETS) calculated using Kremser equations. For the two data points on as-is fermentation broth, the HETS values were 10 and 13 feet.
[0319] Isobutanol was then added to the fermentation broth to bring the concentration to 20 g/L. An extraction test was conducted and from the data, the HETS was found to be 18 feet. This value was some 50% higher than the values obtained on plain broth, and is in line with data obtained using thin stillage spiked with approximately 20 g/L isobutanol (see FIG. 15).
TABLE-US-00004 TABLE 4 Isobutanol Isobutanol Isobutanol in the rich in the lean in the rich Isobutanol Flow of Flow of aqueous aqueous organic in the lean aqueous organic phase phase phase organic phase phase (LC) (LC) (GC) phase HETS Aqueous phase mL/min mL/min g/L g/L g/L g/L ft Broth 192.5 85 2.80 1.21 3.3 0 10 Broth 247.5 105 3.14 1.65 3.6 0 13 Broth with added 187.5 89.7 20.6 11.5 20.6 0 18 iBuOH
Example 5
ISPR Using an External Extraction Column
[0320] Fermentation broth from an isobutanol fermentation (10-liter scale) was circulated to a 5/8'' diameter bench top Karr.RTM. column. The extraction solvent (COFA) was recycled from an extractant reservoir to the Karr.RTM. column. A control fermentation was run in which a volume of COFA was added to the fermentor to continuously extract isobutanol from the fermentation broth.
[0321] The Karr.RTM. column was run twice during the fermentation. The first run was at timepoint 4 to 7 hours of the fermentation and the second run was at timepoint 22 to 33 hours of the fermentation. Parameters such as pO2 and pH were monitored for both fermentations. The measured pO2 was lower for the run in which the Karr.RTM. column was used, as compared to the control run that did not use the Karr.RTM. column. Absolute pH values were similar for the Karr.RTM. column and the control, but the pH profiles were different for the two runs. The pH in the Karr.RTM. column run peaked early, flattened, then peaked again, versus a single gradual peak for the control.
[0322] Two aliquots of extraction solvent (1.8 liters each) were analyzed from the Karr.RTM. column. Samples were taken from each aliquot and analyzed for isobutanol content. The amount of isobutanol produced in the fermentation with the Karr.RTM. column was comparable to that produced in the control fermentation. The fermentation using the Karr.RTM. column produced a total of 82.4 grams isobutanol: approximately 34 grams were in 3.6 liters of organic phase and 48 grams in the aqueous phase. The control (30% by volume organic phase added to the fermentor) produced 90 g/L, 60 grams in 3 liters of organic phase and 30 grams in the aqueous phase. Isobutanol concentration in the aqueous phase was lower in the control due to the presence of COFA in the control fermentor from time zero, versus a non-zero start of extraction in the Karr.RTM. column run. For the Karr.RTM. column at 22 hours, isobutanol was extracted from the fermentor more quickly than it was being produced. Glucose profiles were generated for the control and Karr.RTM. column. The profiles were similar, indicating cell growth and metabolism were comparable. Results are shown in FIGS. 16A and 16B. Brackets indicate the timepoints (4 to 7 hours and 22 to 33 hours) when the Karr.RTM. column was in operation.
Example 6
ISPR Using Mixer-Settler
[0323] An external mixer settler system was used to continuously remove isobutanol from an active fermentation broth containing a microorganism that produced isobutanol (i.e., isobutanologen). The study used approximately 100 liters of fermentation broth inoculated with an isobutanologen. The contents of the fermentor were re-circulated from the fermentor through the mixer-settler extraction system. The extractant, comprising distilled COFA which contained no isobutanol, was used on a once-through basis.
[0324] Two static mixers were tested. The majority of the test used a Kenics.RTM. stainless steel static mixer (1/2'' in diameter with 36 mixing elements). Between hours 12 and 24 of the run, a plastic mixer was used (StaMixCo HT-11-12.6-24, StaMixCo LLC, Brooklyn, N.Y.). Fermentation broth and COFA were fed to opposite sides of a tee, from which the mixture flowed through the static mixer. The material exiting the static mixer was fed to the settler. The settler was made from a five-liter glass tank. A dip tube passed through the top of the settler, near the perimeter, and extended approximately halfway down the settler. The organic phase was withdrawn through a port at the top of the settler, while fermentation broth was removed from the bottom of the settler. The settler was fitted with an agitator that provided gentle mixing to the aqueous-organic interface in order to aid disengagement of the two liquid phases and thereby minimize accumulation of solids at the interface. Data collected during the run is presented in Table 5, and FIG. 17 shows the isobutanol removal rates that were achieved during the course of the fermentation. As can be seen from the data, isobutanol levels in the aqueous broth remained relatively constant, indicating that isobutanol was removed from the fermentation broth at about the same rate as it is being produced. Referring to Table 5, Elapsed Time is time from start of fermentation, AQ Flow is aqueous feed flow, ORG flow is organic feed flow, iB in AQ feed is isobutanol in the aqueous feed, and iB in ORG product is isobutanol in the rich organic product.
TABLE-US-00005 TABLE 5 Elapsed iB in AQ iB in ORG Time Type of AQ Flow ORG flow feed product (hr) Mixer* (ccm) (ccm) (g/L) (g/L) 0.0 A 648 100 5.97 10.14 4.0 A 648 100 5.30 9.22 8.0 A 648 100 4.56 8.20 12.0 A 648 100 4.03 7.63 16.0 B 648 100 4.23 5.36 20.0 B 842.2 130 4.00 7.15 24.0 B 572.4 170 4.53 6.15 28.0 A 648 100 4.65 9.82 32.0 A 648 100 4.92 9.00 36.0 A 648 100 5.27 11.45 41.0 A 648 100 5.65 10.16 45.0 A 648 100 5.40 6.92 *A: 1/2'' stainless Kenics .RTM. mixer, 32 elements B: StaMixCo HT-11-12.6-24, plastic mixer
Example 7
On-Line, at-Line, and Real-Time Measurements
[0325] A mash stream prepared from corn feedstock was conducted to a three-phase centrifuge generating three streams: thin mash, corn oil, and wet cake. On-line or at-line process measurements are employed, for example, to improve the recovery of starch/sugars and the quality of corn oil, and to maximize the amount of starch/sugars extracted from wet cake. Real-time measurements are used, for example, to control the addition of backset, cookwater, or water to slurry tanks to maintain a set starch/sugar concentration set-point. The amount of starch/sugar extracted from the wet cake is maximized using the minimum amount of added water, reducing the hydraulic load on the three-phase centrifuge.
[0326] Corn mash samples were analyzed using Fourier transform infrared spectroscopy (FTIR) with a diamond attenuated total reflectance (ATR) probe that allows for measurements in the presence of solids. The FTIR was calibrated by collecting spectra of standard samples in which total starch/sugar determinations using HPLC had been completed. The HPLC data was used to create a multivariate partial least squares (PLS) model for the FTIR. FTIR spectra were collected and a total starch concentration generated. FIG. 18 shows the range of starch concentrations used to calibrate the FTIR.
[0327] Corn mash with an average starting concentration of 250 g/L was fed to a three-phase centrifuge. The subsequent wet cake was re-slurried and the concentration of starch was measured on two samples: 80 g/L and 70 g/L. This slurry was then separated using a three-phase centrifuge and the wet cake re-slurried. The starch concentration of this slurry was determined to be 28.9 g/L. Results are shown in FIG. 19. These measurements were used to determine the correct amount of water to re-slurry the wet cake at each stage. Optimizing the water addition maximized the starch concentration and minimized the hydraulic load on the separation step. Moisture content of the wet cake was measured using near-infrared spectroscopy (NIR).
[0328] Corn oil quality is monitored in real-time and the data is used to control the three-phase centrifuge variables (e.g., feed rate, g forces, inlet flow rate, scroll speed). The quality of corn oil generated by the three-phase centrifuge was measured by monitoring the concentration of water carried into the corn oil during the separation. FTIR with a diamond ATR probe was used to collect corn oil spectra as it exited the three-phase centrifuge. The detection limit for water using the diamond ATR probe approach was approximately 500 ppm. Lower detection limits are achieved with the use of a flow cell with a longer effective path length.
[0329] FIG. 20 contains a series of infrared spectra of corn oils that contain a range of water concentrations in excess of percent level concentrations down to 100's of ppm. Water concentration was determined using the --OH stretching region between 3700 cm-1 and 3050 cm-1. The data indicated that a process FTIR may be used to generate real-time water concentration in oil data. Real-time water concentration data may be used to control the process variables of the three-phase centrifuge (e.g., feed rate, g forces, inlet flow rate, scroll speed). The operation of the three-phase centrifuge may be controlled to yield the highest quality corn oil or to maximize throughput while not exceeding a water set point.
[0330] Real-time extractant monitoring was used to detect and monitor thermal breakdown of the extractant. Detection of these thermal breakdown products in real-time is used to trigger remediation of the extractant or purging of the contaminated extractant from the process.
[0331] FIG. 21 is an example of the real-time measurement of isobutanol-rich COFA. The data was collected using a Metter-Toledo ReactIR.TM. 247 using a diamond ATR sampling probe in a flow cell. The COFA stream was collected from the outlet of a 1-inch diameter Karr.RTM. column and pumped to the FTIR using a peristaltic pump. The FTIR was calibrated by creating COFA standards spiked with isobutanol and generating a multivariate PLS model.
Example 8
Droplet Size Analysis
[0332] This example describes the analysis of liquid extractant droplets after conducting a process stream containing fermentation broth and extractant (COFA) to a static mixer. A PVM.RTM. probe (Mettler-Toledo, LLC, Columbus Ohio) was inserted into the process stream approximately 24 hr after the process stream exited the static mixer. The PVM.RTM. probe was used to collect images every two minutes during a fermentation run. The images showed the presence of both COFA droplets ranging in size from 50 to 80 .mu.m in diameter and CO.sub.2 bubbles ranging in size from 200 and 400 .mu.m in diameter. Monitoring droplet size in the process stream containing fermentation broth and COFA after the static mixer is used to ensure that the droplets remain below a particular average diameter to ensure good mass transfer of isobutanol into the COFA droplets
[0333] The PVM.RTM. probe was also used to image the COFA droplets in the lean broth stream prior to return of the stream to the fermentor. The detection of COFA droplets in this stream is an indication of the amount of COFA returning to the fermentor. The PVM.RTM. probe was used to collect an image of the stream every two minutes during a fermentation. Unlike the stream exiting the static mixer, the lean broth stream had fewer and smaller droplets (10-40 .mu.m). These measurements demonstrate the feasibility of using process imaging to monitor the amount of COFA returning to the fermentor.
[0334] Real-time average droplet size data from both sample points are used to monitor the phase separation of fermentation broth and COFA. An increase in the concentration or number of small COFA droplets detected in the lean fermentation broth recycle stream (after isobutanol extraction) can be an indicator that the phase separation of fermentation broth and COFA has degraded and too much COFA is exiting the extractor. To improve the quality of the phase separation and reduce the number or concentration of the COFA droplets returning to the fermentor in the lean broth stream, the average COFA droplet size is increased post static mixer.
[0335] Additional process variables that can impact average COFA droplet size include the concentration of polysaccharides in the fermentation broth, the ratio of fermentation broth to COFA, and total flow rate through the static mixer. As the fermentation progresses, flow rate and/or fermentation broth to COFA ratios may be changed to maintain a constant average COFA droplet size.
Example 9
Extractor Design
[0336] This example describes a method to design a large-scale extractor unit. Data from a pilot-scale extraction is used to estimate the size of the large-scale extractor unit. The effects of flow rate, agitation rate, and the presence or absence of internals on phase separation of the streams of the extractor unit from a pilot-scale extraction are determined. The total flow and ratio of fermentation broth flow to extractant flow is varied at fixed temperature over the course of the fermentation, and the conditions under which phase separation discontinues are observed. The maximum achievable flow to the extractor unit per square foot of extractor unit flow surface area is recorded. The following equation is used to determine flow per unit area:
U = F A ( Equation 1 ) ##EQU00001##
[0337] U=flow per unit area (gallons/minute/square foot)
[0338] F=total flow of fermentation broth and extractant to the extractor unit (gallons/minute)
[0339] A=cross-sectional area in direction of flow (square feet)
[0340] for an extraction column this is given by
[0340] .pi. D 2 4 ##EQU00002##
[0341] D=column diameter (feet).
[0342] The diameter of a large-scale extractor unit is estimated by the expected flow of fermentation broth and extractant to the extractor unit using the following equation:
D = 4 F large - scale .pi. U ( Equation 2 ) ##EQU00003##
[0343] F.sub.large-scale=Total flow of fermentation broth and extractant to the large-scale extractor (gallons/minute).
[0344] The height of the pilot-scale extractor unit is measured under different flow regimes including different flow rates, with and without internals present, different agitation rates, and at different concentrations of the product alcohol. Using this data, the number of theoretical stages achieved by the height of the extractor unit is estimated using the Kremser Equation (Seader and Henley, Separation Process Principles, 2.sup.nd edition, John Wiley & Sons, 2006, pp. 358-359):
n = ln [ ( x f - y s m x n - y s m ) ( 1 - 1 E ) + 1 E ] ln ( E ) ( Equation 3 ) ##EQU00004##
E = extraction factor = m F broth F extractant ##EQU00005##
[0345] F.sub.broth=flow of broth to the extractor unit (gallons/minute)
[0346] F.sub.extractant=flow of extractant to the extractor unit (gallons/minute)
[0347] m=partition coefficient for product alcohol in fermentation broth and extractant phases (g/L per g/L)
[0348] Xf=concentration of product alcohol in fermentation broth feed (g/L)
[0349] Xn=concentration of product alcohol in fermentation broth leaving the extractor unit (g/L)
[0350] Ys=concentration of product alcohol in extractant entering the extractor unit (g/L)
[0351] n=number of theoretical stages achieved by the height of the extractor unit Equation 3 is only valid when E.noteq.1.
[0352] The height of a theoretical stage for the extractor unit is given by the height of the extraction column used in the pilot-scale extraction divided by the number of theoretical stages realized in a given experiment. The number of theoretical stages required to achieve the separation at large-scale is estimated using the operating conditions expected at large-scale in Equation 4:
n = ln [ ( x f ' - y s ' m x n ' - y s ' m ) ( 1 - 1 E ' ) + 1 E ' ] ln ( E ' ) ( Equation 4 ) ##EQU00006##
[0353] where ' indicates the condition of the large-scale extractor unit.
[0354] The product of the number of theoretical stages and height of a theoretical stage measured for similar flow conditions provides an estimate of the total height of the large-scale extractor unit. The flows and concentrations expected at a large-scale extractor unit are estimated using a dynamic fermentation model (e.g., Daugulis, et al., Biotech. Bioeng. 27:1345-1356, 1985).
Sequence CWU
1
1
1281570PRTBacillus subtilis 1Met Thr Lys Ala Thr Lys Glu Gln Lys Ser Leu
Val Lys Asn Arg Gly 1 5 10
15 Ala Glu Leu Val Val Asp Cys Leu Val Glu Gln Gly Val Thr His Val
20 25 30 Phe Gly
Ile Pro Gly Ala Lys Ile Asp Ala Val Phe Asp Ala Leu Gln 35
40 45 Asp Lys Gly Pro Glu Ile Ile
Val Ala Arg His Glu Gln Asn Ala Ala 50 55
60 Phe Met Ala Gln Ala Val Gly Arg Leu Thr Gly Lys
Pro Gly Val Val 65 70 75
80 Leu Val Thr Ser Gly Pro Gly Ala Ser Asn Leu Ala Thr Gly Leu Leu
85 90 95 Thr Ala Asn
Thr Glu Gly Asp Pro Val Val Ala Leu Ala Gly Asn Val 100
105 110 Ile Arg Ala Asp Arg Leu Lys Arg
Thr His Gln Ser Leu Asp Asn Ala 115 120
125 Ala Leu Phe Gln Pro Ile Thr Lys Tyr Ser Val Glu Val
Gln Asp Val 130 135 140
Lys Asn Ile Pro Glu Ala Val Thr Asn Ala Phe Arg Ile Ala Ser Ala 145
150 155 160 Gly Gln Ala Gly
Ala Ala Phe Val Ser Phe Pro Gln Asp Val Val Asn 165
170 175 Glu Val Thr Asn Thr Lys Asn Val Arg
Ala Val Ala Ala Pro Lys Leu 180 185
190 Gly Pro Ala Ala Asp Asp Ala Ile Ser Ala Ala Ile Ala Lys
Ile Gln 195 200 205
Thr Ala Lys Leu Pro Val Val Leu Val Gly Met Lys Gly Gly Arg Pro 210
215 220 Glu Ala Ile Lys Ala
Val Arg Lys Leu Leu Lys Lys Val Gln Leu Pro 225 230
235 240 Phe Val Glu Thr Tyr Gln Ala Ala Gly Thr
Leu Ser Arg Asp Leu Glu 245 250
255 Asp Gln Tyr Phe Gly Arg Ile Gly Leu Phe Arg Asn Gln Pro Gly
Asp 260 265 270 Leu
Leu Leu Glu Gln Ala Asp Val Val Leu Thr Ile Gly Tyr Asp Pro 275
280 285 Ile Glu Tyr Asp Pro Lys
Phe Trp Asn Ile Asn Gly Asp Arg Thr Ile 290 295
300 Ile His Leu Asp Glu Ile Ile Ala Asp Ile Asp
His Ala Tyr Gln Pro 305 310 315
320 Asp Leu Glu Leu Ile Gly Asp Ile Pro Ser Thr Ile Asn His Ile Glu
325 330 335 His Asp
Ala Val Lys Val Glu Phe Ala Glu Arg Glu Gln Lys Ile Leu 340
345 350 Ser Asp Leu Lys Gln Tyr Met
His Glu Gly Glu Gln Val Pro Ala Asp 355 360
365 Trp Lys Ser Asp Arg Ala His Pro Leu Glu Ile Val
Lys Glu Leu Arg 370 375 380
Asn Ala Val Asp Asp His Val Thr Val Thr Cys Asp Ile Gly Ser His 385
390 395 400 Ala Ile Trp
Met Ser Arg Tyr Phe Arg Ser Tyr Glu Pro Leu Thr Leu 405
410 415 Met Ile Ser Asn Gly Met Gln Thr
Leu Gly Val Ala Leu Pro Trp Ala 420 425
430 Ile Gly Ala Ser Leu Val Lys Pro Gly Glu Lys Val Val
Ser Val Ser 435 440 445
Gly Asp Gly Gly Phe Leu Phe Ser Ala Met Glu Leu Glu Thr Ala Val 450
455 460 Arg Leu Lys Ala
Pro Ile Val His Ile Val Trp Asn Asp Ser Thr Tyr 465 470
475 480 Asp Met Val Ala Phe Gln Gln Leu Lys
Lys Tyr Asn Arg Thr Ser Ala 485 490
495 Val Asp Phe Gly Asn Ile Asp Ile Val Lys Tyr Ala Glu Ser
Phe Gly 500 505 510
Ala Thr Gly Leu Arg Val Glu Ser Pro Asp Gln Leu Ala Asp Val Leu
515 520 525 Arg Gln Gly Met
Asn Ala Glu Gly Pro Val Ile Ile Asp Val Pro Val 530
535 540 Asp Tyr Ser Asp Asn Ile Asn Leu
Ala Ser Asp Lys Leu Pro Lys Glu 545 550
555 560 Phe Gly Glu Leu Met Lys Thr Lys Ala Leu
565 570 21716DNABacillus subtilis 2atgttgacaa
aagcaacaaa agaacaaaaa tcccttgtga aaaacagagg ggcggagctt 60gttgttgatt
gcttagtgga gcaaggtgtc acacatgtat ttggcattcc aggtgcaaaa 120attgatgcgg
tatttgacgc tttacaagat aaaggacctg aaattatcgt tgcccggcac 180gaacaaaacg
cagcattcat ggcccaagca gtcggccgtt taactggaaa accgggagtc 240gtgttagtca
catcaggacc gggtgcctct aacttggcaa caggcctgct gacagcgaac 300actgaaggag
accctgtcgt tgcgcttgct ggaaacgtga tccgtgcaga tcgtttaaaa 360cggacacatc
aatctttgga taatgcggcg ctattccagc cgattacaaa atacagtgta 420gaagttcaag
atgtaaaaaa tataccggaa gctgttacaa atgcatttag gatagcgtca 480gcagggcagg
ctggggccgc ttttgtgagc tttccgcaag atgttgtgaa tgaagtcaca 540aatacgaaaa
acgtgcgtgc tgttgcagcg ccaaaactcg gtcctgcagc agatgatgca 600atcagtgcgg
ccatagcaaa aatccaaaca gcaaaacttc ctgtcgtttt ggtcggcatg 660aaaggcggaa
gaccggaagc aattaaagcg gttcgcaagc ttttgaaaaa ggttcagctt 720ccatttgttg
aaacatatca agctgccggt accctttcta gagatttaga ggatcaatat 780tttggccgta
tcggtttgtt ccgcaaccag cctggcgatt tactgctaga gcaggcagat 840gttgttctga
cgatcggcta tgacccgatt gaatatgatc cgaaattctg gaatatcaat 900ggagaccgga
caattatcca tttagacgag attatcgctg acattgatca tgcttaccag 960cctgatcttg
aattgatcgg tgacattccg tccacgatca atcatatcga acacgatgct 1020gtgaaagtgg
aatttgcaga gcgtgagcag aaaatccttt ctgatttaaa acaatatatg 1080catgaaggtg
agcaggtgcc tgcagattgg aaatcagaca gagcgcaccc tcttgaaatc 1140gttaaagagt
tgcgtaatgc agtcgatgat catgttacag taacttgcga tatcggttcg 1200cacgccattt
ggatgtcacg ttatttccgc agctacgagc cgttaacatt aatgatcagt 1260aacggtatgc
aaacactcgg cgttgcgctt ccttgggcaa tcggcgcttc attggtgaaa 1320ccgggagaaa
aagtggtttc tgtctctggt gacggcggtt tcttattctc agcaatggaa 1380ttagagacag
cagttcgact aaaagcacca attgtacaca ttgtatggaa cgacagcaca 1440tatgacatgg
ttgcattcca gcaattgaaa aaatataacc gtacatctgc ggtcgatttc 1500ggaaatatcg
atatcgtgaa atatgcggaa agcttcggag caactggctt gcgcgtagaa 1560tcaccagacc
agctggcaga tgttctgcgt caaggcatga acgctgaagg tcctgtcatc 1620atcgatgtcc
cggttgacta cagtgataac attaatttag caagtgacaa gcttccgaaa 1680gaattcgggg
aactcatgaa aacgaaagct ctctag
17163559PRTKlebsiella pneumoniae 3Met Asp Lys Gln Tyr Pro Val Arg Gln Trp
Ala His Gly Ala Asp Leu 1 5 10
15 Val Val Ser Gln Leu Glu Ala Gln Gly Val Arg Gln Val Phe Gly
Ile 20 25 30 Pro
Gly Ala Lys Ile Asp Lys Val Phe Asp Ser Leu Leu Asp Ser Ser 35
40 45 Ile Arg Ile Ile Pro Val
Arg His Glu Ala Asn Ala Ala Phe Met Ala 50 55
60 Ala Ala Val Gly Arg Ile Thr Gly Lys Ala Gly
Val Ala Leu Val Thr 65 70 75
80 Ser Gly Pro Gly Cys Ser Asn Leu Ile Thr Gly Met Ala Thr Ala Asn
85 90 95 Ser Glu
Gly Asp Pro Val Val Ala Leu Gly Gly Ala Val Lys Arg Ala 100
105 110 Asp Lys Ala Lys Gln Val His
Gln Ser Met Asp Thr Val Ala Met Phe 115 120
125 Ser Pro Val Thr Lys Tyr Ala Ile Glu Val Thr Ala
Pro Asp Ala Leu 130 135 140
Ala Glu Val Val Ser Asn Ala Phe Arg Ala Ala Glu Gln Gly Arg Pro 145
150 155 160 Gly Ser Ala
Phe Val Ser Leu Pro Gln Asp Val Val Asp Gly Pro Val 165
170 175 Ser Gly Lys Val Leu Pro Ala Ser
Gly Ala Pro Gln Met Gly Ala Ala 180 185
190 Pro Asp Asp Ala Ile Asp Gln Val Ala Lys Leu Ile Ala
Gln Ala Lys 195 200 205
Asn Pro Ile Phe Leu Leu Gly Leu Met Ala Ser Gln Pro Glu Asn Ser 210
215 220 Lys Ala Leu Arg
Arg Leu Leu Glu Thr Ser His Ile Pro Val Thr Ser 225 230
235 240 Thr Tyr Gln Ala Ala Gly Ala Val Asn
Gln Asp Asn Phe Ser Arg Phe 245 250
255 Ala Gly Arg Val Gly Leu Phe Asn Asn Gln Ala Gly Asp Arg
Leu Leu 260 265 270
Gln Leu Ala Asp Leu Val Ile Cys Ile Gly Tyr Ser Pro Val Glu Tyr
275 280 285 Glu Pro Ala Met
Trp Asn Ser Gly Asn Ala Thr Leu Val His Ile Asp 290
295 300 Val Leu Pro Ala Tyr Glu Glu Arg
Asn Tyr Thr Pro Asp Val Glu Leu 305 310
315 320 Val Gly Asp Ile Ala Gly Thr Leu Asn Lys Leu Ala
Gln Asn Ile Asp 325 330
335 His Arg Leu Val Leu Ser Pro Gln Ala Ala Glu Ile Leu Arg Asp Arg
340 345 350 Gln His Gln
Arg Glu Leu Leu Asp Arg Arg Gly Ala Gln Leu Asn Gln 355
360 365 Phe Ala Leu His Pro Leu Arg Ile
Val Arg Ala Met Gln Asp Ile Val 370 375
380 Asn Ser Asp Val Thr Leu Thr Val Asp Met Gly Ser Phe
His Ile Trp 385 390 395
400 Ile Ala Arg Tyr Leu Tyr Thr Phe Arg Ala Arg Gln Val Met Ile Ser
405 410 415 Asn Gly Gln Gln
Thr Met Gly Val Ala Leu Pro Trp Ala Ile Gly Ala 420
425 430 Trp Leu Val Asn Pro Glu Arg Lys Val
Val Ser Val Ser Gly Asp Gly 435 440
445 Gly Phe Leu Gln Ser Ser Met Glu Leu Glu Thr Ala Val Arg
Leu Lys 450 455 460
Ala Asn Val Leu His Leu Ile Trp Val Asp Asn Gly Tyr Asn Met Val 465
470 475 480 Ala Ile Gln Glu Glu
Lys Lys Tyr Gln Arg Leu Ser Gly Val Glu Phe 485
490 495 Gly Pro Met Asp Phe Lys Ala Tyr Ala Glu
Ser Phe Gly Ala Lys Gly 500 505
510 Phe Ala Val Glu Ser Ala Glu Ala Leu Glu Pro Thr Leu Arg Ala
Ala 515 520 525 Met
Asp Val Asp Gly Pro Ala Val Val Ala Ile Pro Val Asp Tyr Arg 530
535 540 Asp Asn Pro Leu Leu Met
Gly Gln Leu His Leu Ser Gln Ile Leu 545 550
555 42055DNAKlebsiella pneumoniae 4tcgaccacgg
ggtgctgacc ttcggcgaaa ttcacaagct gatgatcgac ctgcccgccg 60acagcgcgtt
cctgcaggct aatctgcatc ccgataatct cgatgccgcc atccgttccg 120tagaaagtta
agggggtcac atggacaaac agtatccggt acgccagtgg gcgcacggcg 180ccgatctcgt
cgtcagtcag ctggaagctc agggagtacg ccaggtgttc ggcatccccg 240gcgccaaaat
cgacaaggtc tttgattcac tgctggattc ctccattcgc attattccgg 300tacgccacga
agccaacgcc gcatttatgg ccgccgccgt cggacgcatt accggcaaag 360cgggcgtggc
gctggtcacc tccggtccgg gctgttccaa cctgatcacc ggcatggcca 420ccgcgaacag
cgaaggcgac ccggtggtgg ccctgggcgg cgcggtaaaa cgcgccgata 480aagcgaagca
ggtccaccag agtatggata cggtggcgat gttcagcccg gtcaccaaat 540acgccatcga
ggtgacggcg ccggatgcgc tggcggaagt ggtctccaac gccttccgcg 600ccgccgagca
gggccggccg ggcagcgcgt tcgttagcct gccgcaggat gtggtcgatg 660gcccggtcag
cggcaaagtg ctgccggcca gcggggcccc gcagatgggc gccgcgccgg 720atgatgccat
cgaccaggtg gcgaagctta tcgcccaggc gaagaacccg atcttcctgc 780tcggcctgat
ggccagccag ccggaaaaca gcaaggcgct gcgccgtttg ctggagacca 840gccatattcc
agtcaccagc acctatcagg ccgccggagc ggtgaatcag gataacttct 900ctcgcttcgc
cggccgggtt gggctgttta acaaccaggc cggggaccgt ctgctgcagc 960tcgccgacct
ggtgatctgc atcggctaca gcccggtgga atacgaaccg gcgatgtgga 1020acagcggcaa
cgcgacgctg gtgcacatcg acgtgctgcc cgcctatgaa gagcgcaact 1080acaccccgga
tgtcgagctg gtgggcgata tcgccggcac tctcaacaag ctggcgcaaa 1140atatcgatca
tcggctggtg ctctccccgc aggcggcgga gatcctccgc gaccgccagc 1200accagcgcga
gctgctggac cgccgcggcg cgcagctcaa ccagtttgcc ctgcatcccc 1260tgcgcatcgt
tcgcgccatg caggatatcg tcaacagcga cgtcacgttg accgtggaca 1320tgggcagctt
ccatatctgg attgcccgct acctgtacac gttccgcgcc cgtcaggtga 1380tgatctccaa
cggccagcag accatgggcg tcgccctgcc ctgggctatc ggcgcctggc 1440tggtcaatcc
tgagcgcaaa gtggtctccg tctccggcga cggcggcttc ctgcagtcga 1500gcatggagct
ggagaccgcc gtccgcctga aagccaacgt gctgcatctt atctgggtcg 1560ataacggcta
caacatggtc gctatccagg aagagaaaaa atatcagcgc ctgtccggcg 1620tcgagtttgg
gccgatggat tttaaagcct atgccgaatc cttcggcgcg aaagggtttg 1680ccgtggaaag
cgccgaggcg ctggagccga ccctgcgcgc ggcgatggac gtcgacggcc 1740cggcggtagt
ggccatcccg gtggattatc gcgataaccc gctgctgatg ggccagctgc 1800atctgagtca
gattctgtaa gtcatcacaa taaggaaaga aaaatgaaaa aagtcgcact 1860tgttaccggc
gccggccagg ggattggtaa agctatcgcc cttcgtctgg tgaaggatgg 1920atttgccgtg
gccattgccg attataacga cgccaccgcc aaagcggtcg cctccgaaat 1980caaccaggcc
ggcggccgcg ccatggcggt gaaagtggat gtttctgacc gcgaccaggt 2040atttgccgcc
gtcga
20555554PRTLactococcus lactis 5Met Ser Glu Lys Gln Phe Gly Ala Asn Leu
Val Val Asp Ser Leu Ile 1 5 10
15 Asn His Lys Val Lys Tyr Val Phe Gly Ile Pro Gly Ala Lys Ile
Asp 20 25 30 Arg
Val Phe Asp Leu Leu Glu Asn Glu Glu Gly Pro Gln Met Val Val 35
40 45 Thr Arg His Glu Gln Gly
Ala Ala Phe Met Ala Gln Ala Val Gly Arg 50 55
60 Leu Thr Gly Glu Pro Gly Val Val Val Val Thr
Ser Gly Pro Gly Val 65 70 75
80 Ser Asn Leu Ala Thr Pro Leu Leu Thr Ala Thr Ser Glu Gly Asp Ala
85 90 95 Ile Leu
Ala Ile Gly Gly Gln Val Lys Arg Ser Asp Arg Leu Lys Arg 100
105 110 Ala His Gln Ser Met Asp Asn
Ala Gly Met Met Gln Ser Ala Thr Lys 115 120
125 Tyr Ser Ala Glu Val Leu Asp Pro Asn Thr Leu Ser
Glu Ser Ile Ala 130 135 140
Asn Ala Tyr Arg Ile Ala Lys Ser Gly His Pro Gly Ala Thr Phe Leu 145
150 155 160 Ser Ile Pro
Gln Asp Val Thr Asp Ala Glu Val Ser Ile Lys Ala Ile 165
170 175 Gln Pro Leu Ser Asp Pro Lys Met
Gly Asn Ala Ser Ile Asp Asp Ile 180 185
190 Asn Tyr Leu Ala Gln Ala Ile Lys Asn Ala Val Leu Pro
Val Ile Leu 195 200 205
Val Gly Ala Gly Ala Ser Asp Ala Lys Val Ala Ser Ser Leu Arg Asn 210
215 220 Leu Leu Thr His
Val Asn Ile Pro Val Val Glu Thr Phe Gln Gly Ala 225 230
235 240 Gly Val Ile Ser His Asp Leu Glu His
Thr Phe Tyr Gly Arg Ile Gly 245 250
255 Leu Phe Arg Asn Gln Pro Gly Asp Met Leu Leu Lys Arg Ser
Asp Leu 260 265 270
Val Ile Ala Val Gly Tyr Asp Pro Ile Glu Tyr Glu Ala Arg Asn Trp
275 280 285 Asn Ala Glu Ile
Asp Ser Arg Ile Ile Val Ile Asp Asn Ala Ile Ala 290
295 300 Glu Ile Asp Thr Tyr Tyr Gln Pro
Glu Arg Glu Leu Ile Gly Asp Ile 305 310
315 320 Ala Ala Thr Leu Asp Asn Leu Leu Pro Ala Val Arg
Gly Tyr Lys Ile 325 330
335 Pro Lys Gly Thr Lys Asp Tyr Leu Asp Gly Leu His Glu Val Ala Glu
340 345 350 Gln His Glu
Phe Asp Thr Glu Asn Thr Glu Glu Gly Arg Met His Pro 355
360 365 Leu Asp Leu Val Ser Thr Phe Gln
Glu Ile Val Lys Asp Asp Glu Thr 370 375
380 Val Thr Val Asp Val Gly Ser Leu Tyr Ile Trp Met Ala
Arg His Phe 385 390 395
400 Lys Ser Tyr Glu Pro Arg His Leu Leu Phe Ser Asn Gly Met Gln Thr
405 410 415 Leu Gly Val Ala
Leu Pro Trp Ala Ile Thr Ala Ala Leu Leu Arg Pro 420
425 430 Gly Lys Lys Val Tyr Ser His Ser Gly
Asp Gly Gly Phe Leu Phe Thr 435 440
445 Gly Gln Glu Leu Glu Thr Ala Val Arg Leu Asn Leu Pro Ile
Val Gln 450 455 460
Ile Ile Trp Asn Asp Gly His Tyr Asp Met Val Lys Phe Gln Glu Glu 465
470 475 480 Met Lys Tyr Gly Arg
Ser Ala Ala Val Asp Phe Gly Tyr Val Asp Tyr 485
490 495 Val Lys Tyr Ala Glu Ala Met Arg Ala Lys
Gly Tyr Arg Ala His Ser 500 505
510 Lys Glu Glu Leu Ala Glu Ile Leu Lys Ser Ile Pro Asp Thr Thr
Gly 515 520 525 Pro
Val Val Ile Asp Val Pro Leu Asp Tyr Ser Asp Asn Ile Lys Leu 530
535 540 Ala Glu Lys Leu Leu Pro
Glu Glu Phe Tyr 545 550
63220DNALactococcus lactis 6tagatccgga aacaactgat tacctgagtt aacttagcag
aaattgcaga agataacggt 60aatttggatg aagcattaaa ttacctttat caaattccgg
tgaatgatga aaattatatt 120gctgctttaa tcaaaattgc tgacttatat caatttgaag
ttgattttga aacagcaatt 180tctaagttag aagaagcaag agaattatcg gattctcctc
tgattacttt tgctttggct 240gagtcctact ttgaacaagg tgattattca gctgccatta
ccgaatatgc aaaactttca 300gaacgaaaaa ttttacatga aacaaaaatt tctatttatc
aaagaattgg tgactcttat 360gcccaattag gtaattttga gaatgccata tcatttcttg
aaaaatcact tgaatttgat 420gaaaaaccgg aaaccttgta taaaattgct cttctttatg
gagaaactca taatgaaaca 480agagccattg ctaatttcaa acggttagaa aaaatggatg
ttgaattttt gaactatgaa 540ttagcctatg cccaaaccct agaagctaat caagaattta
aagctgcact agaaatggca 600aagaaaggga tgaaaaaaaa tcctaatgcc gttcctctct
tacacttcgc ttcaaaaatt 660tgtttcaaac ttaaggacaa agctgcagca gaacgttatc
tcgtggatgc tttaaattta 720ccagaattac atgacgaaac agtctttttg cttgctaatt
tatacttcaa cgaagaagat 780tttgaagctg tcattaatct tgaagagctt ttagaagatg
aacatttatt agctaaatgg 840ctttttgcag gagcacataa agctttggaa aatgattctg
aagcggctgc tttgtatgaa 900gaactcattc aaaccaatct gtcagagaat ccagagtttt
tagaagacta tattgatttt 960cttaaagaaa ttggtcaaat ttctaaaaca gaaccaatta
ttgaacaata tttggaactt 1020gttccagatg atgaaaatat gagaaattta ctgacagact
taaaaaataa ttactgacaa 1080agctgtcagt aattattttt attgtaagct agaaaattca
aaaacttgcg tcaaaataat 1140tgtaaaaggt tctattatct gataaaatga ttgtgaagta
atccaagaga ttatgaaata 1200tgaattagaa caaatagagg taaaataaaa aatgtctgag
aaacaatttg gggcgaactt 1260ggttgtcgat agtttgatta accataaagt gaagtatgta
tttgggattc caggagcaaa 1320aattgaccgg gtttttgatt tattagaaaa tgaagaaggc
cctcaaatgg tcgtgactcg 1380tcatgagcaa ggagctgctt tcatggctca agctgtcggt
cgtttaactg gcgaacctgg 1440tgtagtagtt gttacgagtg ggcctggtgt atcaaacctt
gcgactccgc ttttgaccgc 1500gacatcagaa ggtgatgcta ttttggctat cggtggacaa
gttaaacgaa gtgaccgtct 1560taaacgtgcg caccaatcaa tggataatgc tggaatgatg
caatcagcaa caaaatattc 1620agcagaagtt cttgacccta atacactttc tgaatcaatt
gccaacgctt atcgtattgc 1680aaaatcagga catccaggtg caactttctt atcaatcccc
caagatgtaa cggatgccga 1740agtatcaatc aaagccattc aaccactttc agaccctaaa
atggggaatg cctctattga 1800tgacattaat tatttagcac aagcaattaa aaatgctgta
ttgccagtaa ttttggttgg 1860agctggtgct tcagatgcta aagtcgcttc atccttgcgt
aatctattga ctcatgttaa 1920tattcctgtc gttgaaacat tccaaggtgc aggggttatt
tcacatgatt tagaacatac 1980tttttatgga cgtatcggtc ttttccgcaa tcaaccaggc
gatatgcttc tgaaacgttc 2040tgaccttgtt attgctgttg gttatgaccc aattgaatat
gaagctcgta actggaatgc 2100agaaattgat agtcgaatta tcgttattga taatgccatt
gctgaaattg atacttacta 2160ccaaccagag cgtgaattaa ttggtgatat cgcagcaaca
ttggataatc ttttaccagc 2220tgttcgtggc tacaaaattc caaaaggaac aaaagattat
ctcgatggcc ttcatgaagt 2280tgctgagcaa cacgaatttg atactgaaaa tactgaagaa
ggtagaatgc accctcttga 2340tttggtcagc actttccaag aaatcgtcaa ggatgatgaa
acagtaaccg ttgacgtagg 2400ttcactctac atttggatgg cacgtcattt caaatcatac
gaaccacgtc atctcctctt 2460ctcaaacgga atgcaaacac tcggagttgc acttccttgg
gcaattacag ccgcattgtt 2520gcgcccaggt aaaaaagttt attcacactc tggtgatgga
ggcttccttt tcacagggca 2580agaattggaa acagctgtac gtttgaatct tccaatcgtt
caaattatct ggaatgacgg 2640ccattatgat atggttaaat tccaagaaga aatgaaatat
ggtcgttcag cagccgttga 2700ttttggctat gttgattacg taaaatatgc tgaagcaatg
agagcaaaag gttaccgtgc 2760acacagcaaa gaagaacttg ctgaaattct caaatcaatc
ccagatacta ctggaccggt 2820ggtaattgac gttcctttgg actattctga taacattaaa
ttagcagaaa aattattgcc 2880tgaagagttt tattgattac aatcaagcaa tttgtggcat
aacaaaataa aagaagaagg 2940ccttgaacac ctaagcgttc agggcctttt tttgtgaaat
aaattagatg aaatttacaa 3000tgagttttgt gaaactagct tctagtttgt gaaaaattgc
ctataattgc cgaataaaaa 3060tacccattta ccactccaag aggatgcttc aaattagcta
aatacccgtt ttagaggatg 3120cgtaaaaaca acaaaagagg atgagtatag aacgataaaa
cttttttatg ataggttgag 3180agaattgaat ataaaatata ataagtagaa ggcagcaatt
32207491PRTEscherichia coli 7Met Ala Asn Tyr Phe
Asn Thr Leu Asn Leu Arg Gln Gln Leu Ala Gln 1 5
10 15 Leu Gly Lys Cys Arg Phe Met Gly Arg Asp
Glu Phe Ala Asp Gly Ala 20 25
30 Ser Tyr Leu Gln Gly Lys Lys Val Val Ile Val Gly Cys Gly Ala
Gln 35 40 45 Gly
Leu Asn Gln Gly Leu Asn Met Arg Asp Ser Gly Leu Asp Ile Ser 50
55 60 Tyr Ala Leu Arg Lys Glu
Ala Ile Ala Glu Lys Arg Ala Ser Trp Arg 65 70
75 80 Lys Ala Thr Glu Asn Gly Phe Lys Val Gly Thr
Tyr Glu Glu Leu Ile 85 90
95 Pro Gln Ala Asp Leu Val Ile Asn Leu Thr Pro Asp Lys Gln His Ser
100 105 110 Asp Val
Val Arg Thr Val Gln Pro Leu Met Lys Asp Gly Ala Ala Leu 115
120 125 Gly Tyr Ser His Gly Phe Asn
Ile Val Glu Val Gly Glu Gln Ile Arg 130 135
140 Lys Asp Ile Thr Val Val Met Val Ala Pro Lys Cys
Pro Gly Thr Glu 145 150 155
160 Val Arg Glu Glu Tyr Lys Arg Gly Phe Gly Val Pro Thr Leu Ile Ala
165 170 175 Val His Pro
Glu Asn Asp Pro Lys Gly Glu Gly Met Ala Ile Ala Lys 180
185 190 Ala Trp Ala Ala Ala Thr Gly Gly
His Arg Ala Gly Val Leu Glu Ser 195 200
205 Ser Phe Val Ala Glu Val Lys Ser Asp Leu Met Gly Glu
Gln Thr Ile 210 215 220
Leu Cys Gly Met Leu Gln Ala Gly Ser Leu Leu Cys Phe Asp Lys Leu 225
230 235 240 Val Glu Glu Gly
Thr Asp Pro Ala Tyr Ala Glu Lys Leu Ile Gln Phe 245
250 255 Gly Trp Glu Thr Ile Thr Glu Ala Leu
Lys Gln Gly Gly Ile Thr Leu 260 265
270 Met Met Asp Arg Leu Ser Asn Pro Ala Lys Leu Arg Ala Tyr
Ala Leu 275 280 285
Ser Glu Gln Leu Lys Glu Ile Met Ala Pro Leu Phe Gln Lys His Met 290
295 300 Asp Asp Ile Ile Ser
Gly Glu Phe Ser Ser Gly Met Met Ala Asp Trp 305 310
315 320 Ala Asn Asp Asp Lys Lys Leu Leu Thr Trp
Arg Glu Glu Thr Gly Lys 325 330
335 Thr Ala Phe Glu Thr Ala Pro Gln Tyr Glu Gly Lys Ile Gly Glu
Gln 340 345 350 Glu
Tyr Phe Asp Lys Gly Val Leu Met Ile Ala Met Val Lys Ala Gly 355
360 365 Val Glu Leu Ala Phe Glu
Thr Met Val Asp Ser Gly Ile Ile Glu Glu 370 375
380 Ser Ala Tyr Tyr Glu Ser Leu His Glu Leu Pro
Leu Ile Ala Asn Thr 385 390 395
400 Ile Ala Arg Lys Arg Leu Tyr Glu Met Asn Val Val Ile Ser Asp Thr
405 410 415 Ala Glu
Tyr Gly Asn Tyr Leu Phe Ser Tyr Ala Cys Val Pro Leu Leu 420
425 430 Lys Pro Phe Met Ala Glu Leu
Gln Pro Gly Asp Leu Gly Lys Ala Ile 435 440
445 Pro Glu Gly Ala Val Asp Asn Gly Gln Leu Arg Asp
Val Asn Glu Ala 450 455 460
Ile Arg Ser His Ala Ile Glu Gln Val Gly Lys Lys Leu Arg Gly Tyr 465
470 475 480 Met Thr Asp
Met Lys Arg Ile Ala Val Ala Gly 485 490
81476DNAEscherichia coli 8atggctaact acttcaatac actgaatctg cgccagcagc
tggcacagct gggcaaatgt 60cgctttatgg gccgcgatga attcgccgat ggcgcgagct
accttcaggg taaaaaagta 120gtcatcgtcg gctgtggcgc acagggtctg aaccagggcc
tgaacatgcg tgattctggt 180ctcgatatct cctacgctct gcgtaaagaa gcgattgccg
agaagcgcgc gtcctggcgt 240aaagcgaccg aaaatggttt taaagtgggt acttacgaag
aactgatccc acaggcggat 300ctggtgatta acctgacgcc ggacaagcag cactctgatg
tagtgcgcac cgtacagcca 360ctgatgaaag acggcgcggc gctgggctac tcgcacggtt
tcaacatcgt cgaagtgggc 420gagcagatcc gtaaagatat caccgtagtg atggttgcgc
cgaaatgccc aggcaccgaa 480gtgcgtgaag agtacaaacg tgggttcggc gtaccgacgc
tgattgccgt tcacccggaa 540aacgatccga aaggcgaagg catggcgatt gccaaagcct
gggcggctgc aaccggtggt 600caccgtgcgg gtgtgctgga atcgtccttc gttgcggaag
tgaaatctga cctgatgggc 660gagcaaacca tcctgtgcgg tatgttgcag gctggctctc
tgctgtgctt cgacaagctg 720gtggaagaag gtaccgatcc agcatacgca gaaaaactga
ttcagttcgg ttgggaaacc 780atcaccgaag cactgaaaca gggcggcatc accctgatga
tggaccgtct ctctaacccg 840gcgaaactgc gtgcttatgc gctttctgaa cagctgaaag
agatcatggc acccctgttc 900cagaaacata tggacgacat catctccggc gaattctctt
ccggtatgat ggcggactgg 960gccaacgatg ataagaaact gctgacctgg cgtgaagaga
ccggcaaaac cgcgtttgaa 1020accgcgccgc agtatgaagg caaaatcggc gagcaggagt
acttcgataa aggcgtactg 1080atgattgcga tggtgaaagc gggcgttgaa ctggcgttcg
aaaccatggt cgattccggc 1140atcattgaag agtctgcata ttatgaatca ctgcacgagc
tgccgctgat tgccaacacc 1200atcgcccgta agcgtctgta cgaaatgaac gtggttatct
ctgataccgc tgagtacggt 1260aactatctgt tctcttacgc ttgtgtgccg ttgctgaaac
cgtttatggc agagctgcaa 1320ccgggcgacc tgggtaaagc tattccggaa ggcgcggtag
ataacgggca actgcgtgat 1380gtgaacgaag cgattcgcag ccatgcgatt gagcaggtag
gtaagaaact gcgcggctat 1440atgacagata tgaaacgtat tgctgttgcg ggttaa
14769395PRTSaccharomyces cerevisiae 9Met Leu Arg
Thr Gln Ala Ala Arg Leu Ile Cys Asn Ser Arg Val Ile 1 5
10 15 Thr Ala Lys Arg Thr Phe Ala Leu
Ala Thr Arg Ala Ala Ala Tyr Ser 20 25
30 Arg Pro Ala Ala Arg Phe Val Lys Pro Met Ile Thr Thr
Arg Gly Leu 35 40 45
Lys Gln Ile Asn Phe Gly Gly Thr Val Glu Thr Val Tyr Glu Arg Ala 50
55 60 Asp Trp Pro Arg
Glu Lys Leu Leu Asp Tyr Phe Lys Asn Asp Thr Phe 65 70
75 80 Ala Leu Ile Gly Tyr Gly Ser Gln Gly
Tyr Gly Gln Gly Leu Asn Leu 85 90
95 Arg Asp Asn Gly Leu Asn Val Ile Ile Gly Val Arg Lys Asp
Gly Ala 100 105 110
Ser Trp Lys Ala Ala Ile Glu Asp Gly Trp Val Pro Gly Lys Asn Leu
115 120 125 Phe Thr Val Glu
Asp Ala Ile Lys Arg Gly Ser Tyr Val Met Asn Leu 130
135 140 Leu Ser Asp Ala Ala Gln Ser Glu
Thr Trp Pro Ala Ile Lys Pro Leu 145 150
155 160 Leu Thr Lys Gly Lys Thr Leu Tyr Phe Ser His Gly
Phe Ser Pro Val 165 170
175 Phe Lys Asp Leu Thr His Val Glu Pro Pro Lys Asp Leu Asp Val Ile
180 185 190 Leu Val Ala
Pro Lys Gly Ser Gly Arg Thr Val Arg Ser Leu Phe Lys 195
200 205 Glu Gly Arg Gly Ile Asn Ser Ser
Tyr Ala Val Trp Asn Asp Val Thr 210 215
220 Gly Lys Ala His Glu Lys Ala Gln Ala Leu Ala Val Ala
Ile Gly Ser 225 230 235
240 Gly Tyr Val Tyr Gln Thr Thr Phe Glu Arg Glu Val Asn Ser Asp Leu
245 250 255 Tyr Gly Glu Arg
Gly Cys Leu Met Gly Gly Ile His Gly Met Phe Leu 260
265 270 Ala Gln Tyr Asp Val Leu Arg Glu Asn
Gly His Ser Pro Ser Glu Ala 275 280
285 Phe Asn Glu Thr Val Glu Glu Ala Thr Gln Ser Leu Tyr Pro
Leu Ile 290 295 300
Gly Lys Tyr Gly Met Asp Tyr Met Tyr Asp Ala Cys Ser Thr Thr Ala 305
310 315 320 Arg Arg Gly Ala Leu
Asp Trp Tyr Pro Ile Phe Lys Asn Ala Leu Lys 325
330 335 Pro Val Phe Gln Asp Leu Tyr Glu Ser Thr
Lys Asn Gly Thr Glu Thr 340 345
350 Lys Arg Ser Leu Glu Phe Asn Ser Gln Pro Asp Tyr Arg Glu Lys
Leu 355 360 365 Glu
Lys Glu Leu Asp Thr Ile Arg Asn Met Glu Ile Trp Lys Val Gly 370
375 380 Lys Glu Val Arg Lys Leu
Arg Pro Glu Asn Gln 385 390 395
101188DNASaccharomyces cerevisiae 10atgttgagaa ctcaagccgc cagattgatc
tgcaactccc gtgtcatcac tgctaagaga 60acctttgctt tggccacccg tgctgctgct
tacagcagac cagctgcccg tttcgttaag 120ccaatgatca ctacccgtgg tttgaagcaa
atcaacttcg gtggtactgt tgaaaccgtc 180tacgaaagag ctgactggcc aagagaaaag
ttgttggact acttcaagaa cgacactttt 240gctttgatcg gttacggttc ccaaggttac
ggtcaaggtt tgaacttgag agacaacggt 300ttgaacgtta tcattggtgt ccgtaaagat
ggtgcttctt ggaaggctgc catcgaagac 360ggttgggttc caggcaagaa cttgttcact
gttgaagatg ctatcaagag aggtagttac 420gttatgaact tgttgtccga tgccgctcaa
tcagaaacct ggcctgctat caagccattg 480ttgaccaagg gtaagacttt gtacttctcc
cacggtttct ccccagtctt caaggacttg 540actcacgttg aaccaccaaa ggacttagat
gttatcttgg ttgctccaaa gggttccggt 600agaactgtca gatctttgtt caaggaaggt
cgtggtatta actcttctta cgccgtctgg 660aacgatgtca ccggtaaggc tcacgaaaag
gcccaagctt tggccgttgc cattggttcc 720ggttacgttt accaaaccac tttcgaaaga
gaagtcaact ctgacttgta cggtgaaaga 780ggttgtttaa tgggtggtat ccacggtatg
ttcttggctc aatacgacgt cttgagagaa 840aacggtcact ccccatctga agctttcaac
gaaaccgtcg aagaagctac ccaatctcta 900tacccattga tcggtaagta cggtatggat
tacatgtacg atgcttgttc caccaccgcc 960agaagaggtg ctttggactg gtacccaatc
ttcaagaatg ctttgaagcc tgttttccaa 1020gacttgtacg aatctaccaa gaacggtacc
gaaaccaaga gatctttgga attcaactct 1080caacctgact acagagaaaa gctagaaaag
gaattagaca ccatcagaaa catggaaatc 1140tggaaggttg gtaaggaagt cagaaagttg
agaccagaaa accaataa 118811330PRTMethanococcus maripaludis
11Met Lys Val Phe Tyr Asp Ser Asp Phe Lys Leu Asp Ala Leu Lys Glu 1
5 10 15 Lys Thr Ile Ala
Val Ile Gly Tyr Gly Ser Gln Gly Arg Ala Gln Ser 20
25 30 Leu Asn Met Lys Asp Ser Gly Leu Asn
Val Val Val Gly Leu Arg Lys 35 40
45 Asn Gly Ala Ser Trp Asn Asn Ala Lys Ala Asp Gly His Asn
Val Met 50 55 60
Thr Ile Glu Glu Ala Ala Glu Lys Ala Asp Ile Ile His Ile Leu Ile 65
70 75 80 Pro Asp Glu Leu Gln
Ala Glu Val Tyr Glu Ser Gln Ile Lys Pro Tyr 85
90 95 Leu Lys Glu Gly Lys Thr Leu Ser Phe Ser
His Gly Phe Asn Ile His 100 105
110 Tyr Gly Phe Ile Val Pro Pro Lys Gly Val Asn Val Val Leu Val
Ala 115 120 125 Pro
Lys Ser Pro Gly Lys Met Val Arg Arg Thr Tyr Glu Glu Gly Phe 130
135 140 Gly Val Pro Gly Leu Ile
Cys Ile Glu Ile Asp Ala Thr Asn Asn Ala 145 150
155 160 Phe Asp Ile Val Ser Ala Met Ala Lys Gly Ile
Gly Leu Ser Arg Ala 165 170
175 Gly Val Ile Gln Thr Thr Phe Lys Glu Glu Thr Glu Thr Asp Leu Phe
180 185 190 Gly Glu
Gln Ala Val Leu Cys Gly Gly Val Thr Glu Leu Ile Lys Ala 195
200 205 Gly Phe Glu Thr Leu Val Glu
Ala Gly Tyr Ala Pro Glu Met Ala Tyr 210 215
220 Phe Glu Thr Cys His Glu Leu Lys Leu Ile Val Asp
Leu Ile Tyr Gln 225 230 235
240 Lys Gly Phe Lys Asn Met Trp Asn Asp Val Ser Asn Thr Ala Glu Tyr
245 250 255 Gly Gly Leu
Thr Arg Arg Ser Arg Ile Val Thr Ala Asp Ser Lys Ala 260
265 270 Ala Met Lys Glu Ile Leu Arg Glu
Ile Gln Asp Gly Arg Phe Thr Lys 275 280
285 Glu Phe Leu Leu Glu Lys Gln Val Ser Tyr Ala His Leu
Lys Ser Met 290 295 300
Arg Arg Leu Glu Gly Asp Leu Gln Ile Glu Glu Val Gly Ala Lys Leu 305
310 315 320 Arg Lys Met Cys
Gly Leu Glu Lys Glu Glu 325 330
12993DNAMethanococcus maripaludis 12atgaaggtat tctatgactc agattttaaa
ttagatgctt taaaagaaaa aacaattgca 60gtaatcggtt atggaagtca aggtagggca
cagtccttaa acatgaaaga cagcggatta 120aacgttgttg ttggtttaag aaaaaacggt
gcttcatgga acaacgctaa agcagacggt 180cacaatgtaa tgaccattga agaagctgct
gaaaaagcgg acatcatcca catcttaata 240cctgatgaat tacaggcaga agtttatgaa
agccagataa aaccatacct aaaagaagga 300aaaacactaa gcttttcaca tggttttaac
atccactatg gattcattgt tccaccaaaa 360ggagttaacg tggttttagt tgctccaaaa
tcacctggaa aaatggttag aagaacatac 420gaagaaggtt tcggtgttcc aggtttaatc
tgtattgaaa ttgatgcaac aaacaacgca 480tttgatattg tttcagcaat ggcaaaagga
atcggtttat caagagctgg agttatccag 540acaactttca aagaagaaac agaaactgac
cttttcggtg aacaagctgt tttatgcggt 600ggagttaccg aattaatcaa ggcaggattt
gaaacactcg ttgaagcagg atacgcacca 660gaaatggcat actttgaaac ctgccacgaa
ttgaaattaa tcgttgactt aatctaccaa 720aaaggattca aaaacatgtg gaacgatgta
agtaacactg cagaatacgg cggacttaca 780agaagaagca gaatcgttac agctgattca
aaagctgcaa tgaaagaaat cttaagagaa 840atccaagatg gaagattcac aaaagaattc
cttctcgaaa aacaggtaag ctatgctcat 900ttaaaatcaa tgagaagact cgaaggagac
ttacaaatcg aagaagtcgg cgcaaaatta 960agaaaaatgt gcggtcttga aaaagaagaa
taa 99313342PRTBacillus subtilis 13Met
Val Lys Val Tyr Tyr Asn Gly Asp Ile Lys Glu Asn Val Leu Ala 1
5 10 15 Gly Lys Thr Val Ala Val
Ile Gly Tyr Gly Ser Gln Gly His Ala His 20
25 30 Ala Leu Asn Leu Lys Glu Ser Gly Val Asp
Val Ile Val Gly Val Arg 35 40
45 Gln Gly Lys Ser Phe Thr Gln Ala Gln Glu Asp Gly His Lys
Val Phe 50 55 60
Ser Val Lys Glu Ala Ala Ala Gln Ala Glu Ile Ile Met Val Leu Leu 65
70 75 80 Pro Asp Glu Gln Gln
Gln Lys Val Tyr Glu Ala Glu Ile Lys Asp Glu 85
90 95 Leu Thr Ala Gly Lys Ser Leu Val Phe Ala
His Gly Phe Asn Val His 100 105
110 Phe His Gln Ile Val Pro Pro Ala Asp Val Asp Val Phe Leu Val
Ala 115 120 125 Pro
Lys Gly Pro Gly His Leu Val Arg Arg Thr Tyr Glu Gln Gly Ala 130
135 140 Gly Val Pro Ala Leu Phe
Ala Ile Tyr Gln Asp Val Thr Gly Glu Ala 145 150
155 160 Arg Asp Lys Ala Leu Ala Tyr Ala Lys Gly Ile
Gly Gly Ala Arg Ala 165 170
175 Gly Val Leu Glu Thr Thr Phe Lys Glu Glu Thr Glu Thr Asp Leu Phe
180 185 190 Gly Glu
Gln Ala Val Leu Cys Gly Gly Leu Ser Ala Leu Val Lys Ala 195
200 205 Gly Phe Glu Thr Leu Thr Glu
Ala Gly Tyr Gln Pro Glu Leu Ala Tyr 210 215
220 Phe Glu Cys Leu His Glu Leu Lys Leu Ile Val Asp
Leu Met Tyr Glu 225 230 235
240 Glu Gly Leu Ala Gly Met Arg Tyr Ser Ile Ser Asp Thr Ala Gln Trp
245 250 255 Gly Asp Phe
Val Ser Gly Pro Arg Val Val Asp Ala Lys Val Lys Glu 260
265 270 Ser Met Lys Glu Val Leu Lys Asp
Ile Gln Asn Gly Thr Phe Ala Lys 275 280
285 Glu Trp Ile Val Glu Asn Gln Val Asn Arg Pro Arg Phe
Asn Ala Ile 290 295 300
Asn Ala Ser Glu Asn Glu His Gln Ile Glu Val Val Gly Arg Lys Leu 305
310 315 320 Arg Glu Met Met
Pro Phe Val Lys Gln Gly Lys Lys Lys Glu Ala Val 325
330 335 Val Ser Val Ala Gln Asn
340 14 1476DNABacillus subtilis 14atggctaact acttcaatac
actgaatctg cgccagcagc tggcacagct gggcaaatgt 60cgctttatgg gccgcgatga
attcgccgat ggcgcgagct accttcaggg taaaaaagta 120gtcatcgtcg gctgtggcgc
acagggtctg aaccagggcc tgaacatgcg tgattctggt 180ctcgatatct cctacgctct
gcgtaaagaa gcgattgccg agaagcgcgc gtcctggcgt 240aaagcgaccg aaaatggttt
taaagtgggt acttacgaag aactgatccc acaggcggat 300ctggtgatta acctgacgcc
ggacaagcag cactctgatg tagtgcgcac cgtacagcca 360ctgatgaaag acggcgcggc
gctgggctac tcgcacggtt tcaacatcgt cgaagtgggc 420gagcagatcc gtaaagatat
caccgtagtg atggttgcgc cgaaatgccc aggcaccgaa 480gtgcgtgaag agtacaaacg
tgggttcggc gtaccgacgc tgattgccgt tcacccggaa 540aacgatccga aaggcgaagg
catggcgatt gccaaagcct gggcggctgc aaccggtggt 600caccgtgcgg gtgtgctgga
atcgtccttc gttgcggaag tgaaatctga cctgatgggc 660gagcaaacca tcctgtgcgg
tatgttgcag gctggctctc tgctgtgctt cgacaagctg 720gtggaagaag gtaccgatcc
agcatacgca gaaaaactga ttcagttcgg ttgggaaacc 780atcaccgaag cactgaaaca
gggcggcatc accctgatga tggaccgtct ctctaacccg 840gcgaaactgc gtgcttatgc
gctttctgaa cagctgaaag agatcatggc acccctgttc 900cagaaacata tggacgacat
catctccggc gaattctctt ccggtatgat ggcggactgg 960gccaacgatg ataagaaact
gctgacctgg cgtgaagaga ccggcaaaac cgcgtttgaa 1020accgcgccgc agtatgaagg
caaaatcggc gagcaggagt acttcgataa aggcgtactg 1080atgattgcga tggtgaaagc
gggcgttgaa ctggcgttcg aaaccatggt cgattccggc 1140atcattgaag agtctgcata
ttatgaatca ctgcacgagc tgccgctgat tgccaacacc 1200atcgcccgta agcgtctgta
cgaaatgaac gtggttatct ctgataccgc tgagtacggt 1260aactatctgt tctcttacgc
ttgtgtgccg ttgctgaaac cgtttatggc agagctgcaa 1320ccgggcgacc tgggtaaagc
tattccggaa ggcgcggtag ataacgggca actgcgtgat 1380gtgaacgaag cgattcgcag
ccatgcgatt gagcaggtag gtaagaaact gcgcggctat 1440atgacagata tgaaacgtat
tgctgttgcg ggttaa 147615343PRTAnaerostipes
caccae 15Met Glu Glu Cys Lys Met Ala Lys Ile Tyr Tyr Gln Glu Asp Cys Asn
1 5 10 15 Leu Ser
Leu Leu Asp Gly Lys Thr Ile Ala Val Ile Gly Tyr Gly Ser 20
25 30 Gln Gly His Ala His Ala Leu
Asn Ala Lys Glu Ser Gly Cys Asn Val 35 40
45 Ile Ile Gly Leu Tyr Glu Gly Ala Lys Glu Trp Lys
Arg Ala Glu Glu 50 55 60
Gln Gly Phe Glu Val Tyr Thr Ala Ala Glu Ala Ala Lys Lys Ala Asp 65
70 75 80 Ile Ile Met
Ile Leu Ile Asn Asp Glu Lys Gln Ala Thr Met Tyr Lys 85
90 95 Asn Asp Ile Glu Pro Asn Leu Glu
Ala Gly Asn Met Leu Met Phe Ala 100 105
110 His Gly Phe Asn Ile His Phe Gly Cys Ile Val Pro Pro
Lys Asp Val 115 120 125
Asp Val Thr Met Ile Ala Pro Lys Gly Pro Gly His Thr Val Arg Ser 130
135 140 Glu Tyr Glu Glu
Gly Lys Gly Val Pro Cys Leu Val Ala Val Glu Gln 145 150
155 160 Asp Ala Thr Gly Lys Ala Leu Asp Met
Ala Leu Ala Tyr Ala Leu Ala 165 170
175 Ile Gly Gly Ala Arg Ala Gly Val Leu Glu Thr Thr Phe Arg
Thr Glu 180 185 190
Thr Glu Thr Asp Leu Phe Gly Glu Gln Ala Val Leu Cys Gly Gly Val
195 200 205 Cys Ala Leu Met
Gln Ala Gly Phe Glu Thr Leu Val Glu Ala Gly Tyr 210
215 220 Asp Pro Arg Asn Ala Tyr Phe Glu
Cys Ile His Glu Met Lys Leu Ile 225 230
235 240 Val Asp Leu Ile Tyr Gln Ser Gly Phe Ser Gly Met
Arg Tyr Ser Ile 245 250
255 Ser Asn Thr Ala Glu Tyr Gly Asp Tyr Ile Thr Gly Pro Lys Ile Ile
260 265 270 Thr Glu Asp
Thr Lys Lys Ala Met Lys Lys Ile Leu Ser Asp Ile Gln 275
280 285 Asp Gly Thr Phe Ala Lys Asp Phe
Leu Val Asp Met Ser Asp Ala Gly 290 295
300 Ser Gln Val His Phe Lys Ala Met Arg Lys Leu Ala Ser
Glu His Pro 305 310 315
320 Ala Glu Val Val Gly Glu Glu Ile Arg Ser Leu Tyr Ser Trp Ser Asp
325 330 335 Glu Asp Lys Leu
Ile Asn Asn 340 16343PRTAnaerostipes caccae 16Met
Glu Glu Cys Lys Met Ala Lys Ile Tyr Tyr Gln Glu Asp Cys Asn 1
5 10 15 Leu Ser Leu Leu Asp Gly
Lys Thr Ile Ala Val Ile Gly Tyr Gly Ser 20
25 30 Gln Gly His Ala His Ala Leu Asn Ala Lys
Glu Ser Gly Cys Asn Val 35 40
45 Ile Ile Gly Leu Tyr Glu Gly Ala Lys Asp Trp Lys Arg Ala
Glu Glu 50 55 60
Gln Gly Phe Glu Val Tyr Thr Ala Ala Glu Ala Ala Lys Lys Ala Asp 65
70 75 80 Ile Ile Met Ile Leu
Ile Asn Asp Glu Lys Gln Ala Thr Met Tyr Lys 85
90 95 Asn Asp Ile Glu Pro Asn Leu Glu Ala Gly
Asn Met Leu Met Phe Ala 100 105
110 His Gly Phe Asn Ile His Phe Gly Cys Ile Val Pro Pro Lys Asp
Val 115 120 125 Asp
Val Thr Met Ile Ala Pro Lys Gly Pro Gly His Thr Val Arg Ser 130
135 140 Glu Tyr Glu Glu Gly Lys
Gly Val Pro Cys Leu Val Ala Val Glu Gln 145 150
155 160 Asp Ala Thr Gly Lys Ala Leu Asp Met Ala Leu
Ala Tyr Ala Leu Ala 165 170
175 Ile Gly Gly Ala Arg Ala Gly Val Leu Glu Thr Thr Phe Arg Thr Glu
180 185 190 Thr Glu
Thr Asp Leu Phe Gly Glu Gln Ala Val Leu Cys Gly Gly Val 195
200 205 Cys Ala Leu Met Gln Ala Gly
Phe Glu Thr Leu Val Glu Ala Gly Tyr 210 215
220 Asp Pro Arg Asn Ala Tyr Phe Glu Cys Ile His Glu
Met Lys Leu Ile 225 230 235
240 Val Asp Leu Ile Tyr Gln Ser Gly Phe Ser Gly Met Arg Tyr Ser Ile
245 250 255 Ser Asn Thr
Ala Glu Tyr Gly Asp Tyr Ile Thr Gly Pro Lys Ile Ile 260
265 270 Thr Glu Asp Thr Lys Lys Ala Met
Lys Lys Ile Leu Ser Asp Ile Gln 275 280
285 Asp Gly Thr Phe Ala Lys Asp Phe Leu Val Asp Met Ser
Asp Ala Gly 290 295 300
Ser Gln Val His Phe Lys Ala Met Arg Lys Leu Ala Ser Glu His Pro 305
310 315 320 Ala Glu Val Val
Gly Glu Glu Ile Arg Ser Leu Tyr Ser Trp Ser Asp 325
330 335 Glu Asp Lys Leu Ile Asn Asn
340 17616PRTEscherichia coli 17Met Pro Lys Tyr Arg Ser
Ala Thr Thr Thr His Gly Arg Asn Met Ala 1 5
10 15 Gly Ala Arg Ala Leu Trp Arg Ala Thr Gly Met
Thr Asp Ala Asp Phe 20 25
30 Gly Lys Pro Ile Ile Ala Val Val Asn Ser Phe Thr Gln Phe Val
Pro 35 40 45 Gly
His Val His Leu Arg Asp Leu Gly Lys Leu Val Ala Glu Gln Ile 50
55 60 Glu Ala Ala Gly Gly Val
Ala Lys Glu Phe Asn Thr Ile Ala Val Asp 65 70
75 80 Asp Gly Ile Ala Met Gly His Gly Gly Met Leu
Tyr Ser Leu Pro Ser 85 90
95 Arg Glu Leu Ile Ala Asp Ser Val Glu Tyr Met Val Asn Ala His Cys
100 105 110 Ala Asp
Ala Met Val Cys Ile Ser Asn Cys Asp Lys Ile Thr Pro Gly 115
120 125 Met Leu Met Ala Ser Leu Arg
Leu Asn Ile Pro Val Ile Phe Val Ser 130 135
140 Gly Gly Pro Met Glu Ala Gly Lys Thr Lys Leu Ser
Asp Gln Ile Ile 145 150 155
160 Lys Leu Asp Leu Val Asp Ala Met Ile Gln Gly Ala Asp Pro Lys Val
165 170 175 Ser Asp Ser
Gln Ser Asp Gln Val Glu Arg Ser Ala Cys Pro Thr Cys 180
185 190 Gly Ser Cys Ser Gly Met Phe Thr
Ala Asn Ser Met Asn Cys Leu Thr 195 200
205 Glu Ala Leu Gly Leu Ser Gln Pro Gly Asn Gly Ser Leu
Leu Ala Thr 210 215 220
His Ala Asp Arg Lys Gln Leu Phe Leu Asn Ala Gly Lys Arg Ile Val 225
230 235 240 Glu Leu Thr Lys
Arg Tyr Tyr Glu Gln Asn Asp Glu Ser Ala Leu Pro 245
250 255 Arg Asn Ile Ala Ser Lys Ala Ala Phe
Glu Asn Ala Met Thr Leu Asp 260 265
270 Ile Ala Met Gly Gly Ser Thr Asn Thr Val Leu His Leu Leu
Ala Ala 275 280 285
Ala Gln Glu Ala Glu Ile Asp Phe Thr Met Ser Asp Ile Asp Lys Leu 290
295 300 Ser Arg Lys Val Pro
Gln Leu Cys Lys Val Ala Pro Ser Thr Gln Lys 305 310
315 320 Tyr His Met Glu Asp Val His Arg Ala Gly
Gly Val Ile Gly Ile Leu 325 330
335 Gly Glu Leu Asp Arg Ala Gly Leu Leu Asn Arg Asp Val Lys Asn
Val 340 345 350 Leu
Gly Leu Thr Leu Pro Gln Thr Leu Glu Gln Tyr Asp Val Met Leu 355
360 365 Thr Gln Asp Asp Ala Val
Lys Asn Met Phe Arg Ala Gly Pro Ala Gly 370 375
380 Ile Arg Thr Thr Gln Ala Phe Ser Gln Asp Cys
Arg Trp Asp Thr Leu 385 390 395
400 Asp Asp Asp Arg Ala Asn Gly Cys Ile Arg Ser Leu Glu His Ala Tyr
405 410 415 Ser Lys
Asp Gly Gly Leu Ala Val Leu Tyr Gly Asn Phe Ala Glu Asn 420
425 430 Gly Cys Ile Val Lys Thr Ala
Gly Val Asp Asp Ser Ile Leu Lys Phe 435 440
445 Thr Gly Pro Ala Lys Val Tyr Glu Ser Gln Asp Asp
Ala Val Glu Ala 450 455 460
Ile Leu Gly Gly Lys Val Val Ala Gly Asp Val Val Val Ile Arg Tyr 465
470 475 480 Glu Gly Pro
Lys Gly Gly Pro Gly Met Gln Glu Met Leu Tyr Pro Thr 485
490 495 Ser Phe Leu Lys Ser Met Gly Leu
Gly Lys Ala Cys Ala Leu Ile Thr 500 505
510 Asp Gly Arg Phe Ser Gly Gly Thr Ser Gly Leu Ser Ile
Gly His Val 515 520 525
Ser Pro Glu Ala Ala Ser Gly Gly Ser Ile Gly Leu Ile Glu Asp Gly 530
535 540 Asp Leu Ile Ala
Ile Asp Ile Pro Asn Arg Gly Ile Gln Leu Gln Val 545 550
555 560 Ser Asp Ala Glu Leu Ala Ala Arg Arg
Glu Ala Gln Asp Ala Arg Gly 565 570
575 Asp Lys Ala Trp Thr Pro Lys Asn Arg Glu Arg Gln Val Ser
Phe Ala 580 585 590
Leu Arg Ala Tyr Ala Ser Leu Ala Thr Ser Ala Asp Lys Gly Ala Val
595 600 605 Arg Asp Lys Ser
Lys Leu Gly Gly 610 615 181851DNAEscherichia coli
18atgcctaagt accgttccgc caccaccact catggtcgta atatggcggg tgctcgtgcg
60ctgtggcgcg ccaccggaat gaccgacgcc gatttcggta agccgattat cgcggttgtg
120aactcgttca cccaatttgt accgggtcac gtccatctgc gcgatctcgg taaactggtc
180gccgaacaaa ttgaagcggc tggcggcgtt gccaaagagt tcaacaccat tgcggtggat
240gatgggattg ccatgggcca cggggggatg ctttattcac tgccatctcg cgaactgatc
300gctgattccg ttgagtatat ggtcaacgcc cactgcgccg acgccatggt ctgcatctct
360aactgcgaca aaatcacccc ggggatgctg atggcttccc tgcgcctgaa tattccggtg
420atctttgttt ccggcggccc gatggaggcc gggaaaacca aactttccga tcagatcatc
480aagctcgatc tggttgatgc gatgatccag ggcgcagacc cgaaagtatc tgactcccag
540agcgatcagg ttgaacgttc cgcgtgtccg acctgcggtt cctgctccgg gatgtttacc
600gctaactcaa tgaactgcct gaccgaagcg ctgggcctgt cgcagccggg caacggctcg
660ctgctggcaa cccacgccga ccgtaagcag ctgttcctta atgctggtaa acgcattgtt
720gaattgacca aacgttatta cgagcaaaac gacgaaagtg cactgccgcg taatatcgcc
780agtaaggcgg cgtttgaaaa cgccatgacg ctggatatcg cgatgggtgg atcgactaac
840accgtacttc acctgctggc ggcggcgcag gaagcggaaa tcgacttcac catgagtgat
900atcgataagc tttcccgcaa ggttccacag ctgtgtaaag ttgcgccgag cacccagaaa
960taccatatgg aagatgttca ccgtgctggt ggtgttatcg gtattctcgg cgaactggat
1020cgcgcggggt tactgaaccg tgatgtgaaa aacgtacttg gcctgacgtt gccgcaaacg
1080ctggaacaat acgacgttat gctgacccag gatgacgcgg taaaaaatat gttccgcgca
1140ggtcctgcag gcattcgtac cacacaggca ttctcgcaag attgccgttg ggatacgctg
1200gacgacgatc gcgccaatgg ctgtatccgc tcgctggaac acgcctacag caaagacggc
1260ggcctggcgg tgctctacgg taactttgcg gaaaacggct gcatcgtgaa aacggcaggc
1320gtcgatgaca gcatcctcaa attcaccggc ccggcgaaag tgtacgaaag ccaggacgat
1380gcggtagaag cgattctcgg cggtaaagtt gtcgccggag atgtggtagt aattcgctat
1440gaaggcccga aaggcggtcc ggggatgcag gaaatgctct acccaaccag cttcctgaaa
1500tcaatgggtc tcggcaaagc ctgtgcgctg atcaccgacg gtcgtttctc tggtggcacc
1560tctggtcttt ccatcggcca cgtctcaccg gaagcggcaa gcggcggcag cattggcctg
1620attgaagatg gtgacctgat cgctatcgac atcccgaacc gtggcattca gttacaggta
1680agcgatgccg aactggcggc gcgtcgtgaa gcgcaggacg ctcgaggtga caaagcctgg
1740acgccgaaaa atcgtgaacg tcaggtctcc tttgccctgc gtgcttatgc cagcctggca
1800accagcgccg acaaaggcgc ggtgcgcgat aaatcgaaac tggggggtta a
185119585PRTSaccharomyces cerevisiae 19Met Gly Leu Leu Thr Lys Val Ala
Thr Ser Arg Gln Phe Ser Thr Thr 1 5 10
15 Arg Cys Val Ala Lys Lys Leu Asn Lys Tyr Ser Tyr Ile
Ile Thr Glu 20 25 30
Pro Lys Gly Gln Gly Ala Ser Gln Ala Met Leu Tyr Ala Thr Gly Phe
35 40 45 Lys Lys Glu Asp
Phe Lys Lys Pro Gln Val Gly Val Gly Ser Cys Trp 50
55 60 Trp Ser Gly Asn Pro Cys Asn Met
His Leu Leu Asp Leu Asn Asn Arg 65 70
75 80 Cys Ser Gln Ser Ile Glu Lys Ala Gly Leu Lys Ala
Met Gln Phe Asn 85 90
95 Thr Ile Gly Val Ser Asp Gly Ile Ser Met Gly Thr Lys Gly Met Arg
100 105 110 Tyr Ser Leu
Gln Ser Arg Glu Ile Ile Ala Asp Ser Phe Glu Thr Ile 115
120 125 Met Met Ala Gln His Tyr Asp Ala
Asn Ile Ala Ile Pro Ser Cys Asp 130 135
140 Lys Asn Met Pro Gly Val Met Met Ala Met Gly Arg His
Asn Arg Pro 145 150 155
160 Ser Ile Met Val Tyr Gly Gly Thr Ile Leu Pro Gly His Pro Thr Cys
165 170 175 Gly Ser Ser Lys
Ile Ser Lys Asn Ile Asp Ile Val Ser Ala Phe Gln 180
185 190 Ser Tyr Gly Glu Tyr Ile Ser Lys Gln
Phe Thr Glu Glu Glu Arg Glu 195 200
205 Asp Val Val Glu His Ala Cys Pro Gly Pro Gly Ser Cys Gly
Gly Met 210 215 220
Tyr Thr Ala Asn Thr Met Ala Ser Ala Ala Glu Val Leu Gly Leu Thr 225
230 235 240 Ile Pro Asn Ser Ser
Ser Phe Pro Ala Val Ser Lys Glu Lys Leu Ala 245
250 255 Glu Cys Asp Asn Ile Gly Glu Tyr Ile Lys
Lys Thr Met Glu Leu Gly 260 265
270 Ile Leu Pro Arg Asp Ile Leu Thr Lys Glu Ala Phe Glu Asn Ala
Ile 275 280 285 Thr
Tyr Val Val Ala Thr Gly Gly Ser Thr Asn Ala Val Leu His Leu 290
295 300 Val Ala Val Ala His Ser
Ala Gly Val Lys Leu Ser Pro Asp Asp Phe 305 310
315 320 Gln Arg Ile Ser Asp Thr Thr Pro Leu Ile Gly
Asp Phe Lys Pro Ser 325 330
335 Gly Lys Tyr Val Met Ala Asp Leu Ile Asn Val Gly Gly Thr Gln Ser
340 345 350 Val Ile
Lys Tyr Leu Tyr Glu Asn Asn Met Leu His Gly Asn Thr Met 355
360 365 Thr Val Thr Gly Asp Thr Leu
Ala Glu Arg Ala Lys Lys Ala Pro Ser 370 375
380 Leu Pro Glu Gly Gln Glu Ile Ile Lys Pro Leu Ser
His Pro Ile Lys 385 390 395
400 Ala Asn Gly His Leu Gln Ile Leu Tyr Gly Ser Leu Ala Pro Gly Gly
405 410 415 Ala Val Gly
Lys Ile Thr Gly Lys Glu Gly Thr Tyr Phe Lys Gly Arg 420
425 430 Ala Arg Val Phe Glu Glu Glu Gly
Ala Phe Ile Glu Ala Leu Glu Arg 435 440
445 Gly Glu Ile Lys Lys Gly Glu Lys Thr Val Val Val Ile
Arg Tyr Glu 450 455 460
Gly Pro Arg Gly Ala Pro Gly Met Pro Glu Met Leu Lys Pro Ser Ser 465
470 475 480 Ala Leu Met Gly
Tyr Gly Leu Gly Lys Asp Val Ala Leu Leu Thr Asp 485
490 495 Gly Arg Phe Ser Gly Gly Ser His Gly
Phe Leu Ile Gly His Ile Val 500 505
510 Pro Glu Ala Ala Glu Gly Gly Pro Ile Gly Leu Val Arg Asp
Gly Asp 515 520 525
Glu Ile Ile Ile Asp Ala Asp Asn Asn Lys Ile Asp Leu Leu Val Ser 530
535 540 Asp Lys Glu Met Ala
Gln Arg Lys Gln Ser Trp Val Ala Pro Pro Pro 545 550
555 560 Arg Tyr Thr Arg Gly Thr Leu Ser Lys Tyr
Ala Lys Leu Val Ser Asn 565 570
575 Ala Ser Asn Gly Cys Val Leu Asp Ala 580
585 20 1131DNASaccharomyces cerevisiae 20atgaccttgg
cacccctaga cgcctccaaa gttaagataa ctaccacaca acatgcatct 60aagccaaaac
cgaacagtga gttagtgttt ggcaagagct tcacggacca catgttaact 120gcggaatgga
cagctgaaaa agggtggggt accccagaga ttaaacctta tcaaaatctg 180tctttagacc
cttccgcggt ggttttccat tatgcttttg agctattcga agggatgaag 240gcttacagaa
cggtggacaa caaaattaca atgtttcgtc cagatatgaa tatgaagcgc 300atgaataagt
ctgctcagag aatctgtttg ccaacgttcg acccagaaga gttgattacc 360ctaattggga
aactgatcca gcaagataag tgcttagttc ctgaaggaaa aggttactct 420ttatatatca
ggcctacatt aatcggcact acggccggtt taggggtttc cacgcctgat 480agagccttgc
tatatgtcat ttgctgccct gtgggtcctt attacaaaac tggatttaag 540gcggtcagac
tggaagccac tgattatgcc acaagagctt ggccaggagg ctgtggtgac 600aagaaactag
gtgcaaacta cgccccctgc gtcctgccac aattgcaagc tgcttcaagg 660ggttaccaac
aaaatttatg gctatttggt ccaaataaca acattactga agtcggcacc 720atgaatgctt
ttttcgtgtt taaagatagt aaaacgggca agaaggaact agttactgct 780ccactagacg
gtaccatttt ggaaggtgtt actagggatt ccattttaaa tcttgctaaa 840gaaagactcg
aaccaagtga atggaccatt agtgaacgct acttcactat aggcgaagtt 900actgagagat
ccaagaacgg tgaactactt gaagcctttg gttctggtac tgctgcgatt 960gtttctccca
ttaaggaaat cggctggaaa ggcgaacaaa ttaatattcc gttgttgccc 1020ggcgaacaaa
ccggtccatt ggccaaagaa gttgcacaat ggattaatgg aatccaatat 1080ggcgagactg
agcatggcaa ttggtcaagg gttgttactg atttgaactg a
113121550PRTMethanococcus maripaludis 21Met Ile Ser Asp Asn Val Lys Lys
Gly Val Ile Arg Thr Pro Asn Arg 1 5 10
15 Ala Leu Leu Lys Ala Cys Gly Tyr Thr Asp Glu Asp Met
Glu Lys Pro 20 25 30
Phe Ile Gly Ile Val Asn Ser Phe Thr Glu Val Val Pro Gly His Ile
35 40 45 His Leu Arg Thr
Leu Ser Glu Ala Ala Lys His Gly Val Tyr Ala Asn 50
55 60 Gly Gly Thr Pro Phe Glu Phe Asn
Thr Ile Gly Ile Cys Asp Gly Ile 65 70
75 80 Ala Met Gly His Glu Gly Met Lys Tyr Ser Leu Pro
Ser Arg Glu Ile 85 90
95 Ile Ala Asp Ala Val Glu Ser Met Ala Arg Ala His Gly Phe Asp Gly
100 105 110 Leu Val Leu
Ile Pro Thr Cys Asp Lys Ile Val Pro Gly Met Ile Met 115
120 125 Gly Ala Leu Arg Leu Asn Ile Pro
Phe Ile Val Val Thr Gly Gly Pro 130 135
140 Met Leu Pro Gly Glu Phe Gln Gly Lys Lys Tyr Glu Leu
Ile Ser Leu 145 150 155
160 Phe Glu Gly Val Gly Glu Tyr Gln Val Gly Lys Ile Thr Glu Glu Glu
165 170 175 Leu Lys Cys Ile
Glu Asp Cys Ala Cys Ser Gly Ala Gly Ser Cys Ala 180
185 190 Gly Leu Tyr Thr Ala Asn Ser Met Ala
Cys Leu Thr Glu Ala Leu Gly 195 200
205 Leu Ser Leu Pro Met Cys Ala Thr Thr His Ala Val Asp Ala
Gln Lys 210 215 220
Val Arg Leu Ala Lys Lys Ser Gly Ser Lys Ile Val Asp Met Val Lys 225
230 235 240 Glu Asp Leu Lys Pro
Thr Asp Ile Leu Thr Lys Glu Ala Phe Glu Asn 245
250 255 Ala Ile Leu Val Asp Leu Ala Leu Gly Gly
Ser Thr Asn Thr Thr Leu 260 265
270 His Ile Pro Ala Ile Ala Asn Glu Ile Glu Asn Lys Phe Ile Thr
Leu 275 280 285 Asp
Asp Phe Asp Arg Leu Ser Asp Glu Val Pro His Ile Ala Ser Ile 290
295 300 Lys Pro Gly Gly Glu His
Tyr Met Ile Asp Leu His Asn Ala Gly Gly 305 310
315 320 Ile Pro Ala Val Leu Asn Val Leu Lys Glu Lys
Ile Arg Asp Thr Lys 325 330
335 Thr Val Asp Gly Arg Ser Ile Leu Glu Ile Ala Glu Ser Val Lys Tyr
340 345 350 Ile Asn
Tyr Asp Val Ile Arg Lys Val Glu Ala Pro Val His Glu Thr 355
360 365 Ala Gly Leu Arg Val Leu Lys
Gly Asn Leu Ala Pro Asn Gly Cys Val 370 375
380 Val Lys Ile Gly Ala Val His Pro Lys Met Tyr Lys
His Asp Gly Pro 385 390 395
400 Ala Lys Val Tyr Asn Ser Glu Asp Glu Ala Ile Ser Ala Ile Leu Gly
405 410 415 Gly Lys Ile
Val Glu Gly Asp Val Ile Val Ile Arg Tyr Glu Gly Pro 420
425 430 Ser Gly Gly Pro Gly Met Arg Glu
Met Leu Ser Pro Thr Ser Ala Ile 435 440
445 Cys Gly Met Gly Leu Asp Asp Ser Val Ala Leu Ile Thr
Asp Gly Arg 450 455 460
Phe Ser Gly Gly Ser Arg Gly Pro Cys Ile Gly His Val Ser Pro Glu 465
470 475 480 Ala Ala Ala Gly
Gly Val Ile Ala Ala Ile Glu Asn Gly Asp Ile Ile 485
490 495 Lys Ile Asp Met Ile Glu Lys Glu Ile
Asn Val Asp Leu Asp Glu Ser 500 505
510 Val Ile Lys Glu Arg Leu Ser Lys Leu Gly Glu Phe Glu Pro
Lys Ile 515 520 525
Lys Lys Gly Tyr Leu Ser Arg Tyr Ser Lys Leu Val Ser Ser Ala Asp 530
535 540 Glu Gly Ala Val Leu
Lys 545 550 221653DNAMethanococcus maripaludis
22atgataagtg ataacgtcaa aaagggagtt ataagaactc caaaccgagc tcttttaaag
60gcttgcggat atacagacga agacatggaa aaaccattta ttggaattgt aaacagcttt
120acagaagttg ttcccggcca cattcactta agaacattat cagaagcggc taaacatggt
180gtttatgcaa acggtggaac accatttgaa tttaatacca ttggaatttg cgacggtatt
240gcaatgggcc acgaaggtat gaaatactct ttaccttcaa gagaaattat tgcagacgct
300gttgaatcaa tggcaagagc acatggattt gatggtcttg ttttaattcc tacgtgtgat
360aaaatcgttc ctggaatgat aatgggtgct ttaagactaa acattccatt tattgtagtt
420actggaggac caatgcttcc cggagaattc caaggtaaaa aatacgaact tatcagcctt
480tttgaaggtg tcggagaata ccaagttgga aaaattactg aagaagagtt aaagtgcatt
540gaagactgtg catgttcagg tgctggaagt tgtgcagggc tttacactgc aaacagtatg
600gcctgcctta cagaagcttt gggactctct cttccaatgt gtgcaacaac gcatgcagtt
660gatgcccaaa aagttaggct tgctaaaaaa agtggctcaa aaattgttga tatggtaaaa
720gaagacctaa aaccaacaga catattaaca aaagaagctt ttgaaaatgc tattttagtt
780gaccttgcac ttggtggatc aacaaacaca acattacaca ttcctgcaat tgcaaatgaa
840attgaaaata aattcataac tctcgatgac tttgacaggt taagcgatga agttccacac
900attgcatcaa tcaaaccagg tggagaacac tacatgattg atttacacaa tgctggaggt
960attcctgcgg tattgaacgt tttaaaagaa aaaattagag atacaaaaac agttgatgga
1020agaagcattt tggaaatcgc agaatctgtt aaatacataa attacgacgt tataagaaaa
1080gtggaagctc cggttcacga aactgctggt ttaagggttt taaagggaaa tcttgctcca
1140aacggttgcg ttgtaaaaat cggtgcagta catccgaaaa tgtacaaaca cgatggacct
1200gcaaaagttt acaattccga agatgaagca atttctgcga tacttggcgg aaaaattgta
1260gaaggggacg ttatagtaat cagatacgaa ggaccatcag gaggccctgg aatgagagaa
1320atgctctccc caacttcagc aatctgtgga atgggtcttg atgacagcgt tgcattgatt
1380actgatggaa gattcagtgg tggaagtagg ggcccatgta tcggacacgt ttctccagaa
1440gctgcagctg gcggagtaat tgctgcaatt gaaaacgggg atatcatcaa aatcgacatg
1500attgaaaaag aaataaatgt tgatttagat gaatcagtca ttaaagaaag actctcaaaa
1560ctgggagaat ttgagcctaa aatcaaaaaa ggctatttat caagatactc aaaacttgtc
1620tcatctgctg acgaaggggc agttttaaaa taa
165323558PRTBacillus subtilis 23Met Ala Glu Leu Arg Ser Asn Met Ile Thr
Gln Gly Ile Asp Arg Ala 1 5 10
15 Pro His Arg Ser Leu Leu Arg Ala Ala Gly Val Lys Glu Glu Asp
Phe 20 25 30 Gly
Lys Pro Phe Ile Ala Val Cys Asn Ser Tyr Ile Asp Ile Val Pro 35
40 45 Gly His Val His Leu Gln
Glu Phe Gly Lys Ile Val Lys Glu Ala Ile 50 55
60 Arg Glu Ala Gly Gly Val Pro Phe Glu Phe Asn
Thr Ile Gly Val Asp 65 70 75
80 Asp Gly Ile Ala Met Gly His Ile Gly Met Arg Tyr Ser Leu Pro Ser
85 90 95 Arg Glu
Ile Ile Ala Asp Ser Val Glu Thr Val Val Ser Ala His Trp 100
105 110 Phe Asp Gly Met Val Cys Ile
Pro Asn Cys Asp Lys Ile Thr Pro Gly 115 120
125 Met Leu Met Ala Ala Met Arg Ile Asn Ile Pro Thr
Ile Phe Val Ser 130 135 140
Gly Gly Pro Met Ala Ala Gly Arg Thr Ser Asp Gly Arg Lys Ile Ser 145
150 155 160 Leu Ser Ser
Val Phe Glu Gly Val Gly Ala Tyr Gln Ala Gly Lys Ile 165
170 175 Asn Glu Asn Glu Leu Gln Glu Leu
Glu Gln Phe Gly Cys Pro Thr Cys 180 185
190 Gly Ser Cys Ser Gly Met Phe Thr Ala Asn Ser Met Asn
Cys Leu Ser 195 200 205
Glu Ala Leu Gly Leu Ala Leu Pro Gly Asn Gly Thr Ile Leu Ala Thr 210
215 220 Ser Pro Glu Arg
Lys Glu Phe Val Arg Lys Ser Ala Ala Gln Leu Met 225 230
235 240 Glu Thr Ile Arg Lys Asp Ile Lys Pro
Arg Asp Ile Val Thr Val Lys 245 250
255 Ala Ile Asp Asn Ala Phe Ala Leu Asp Met Ala Leu Gly Gly
Ser Thr 260 265 270
Asn Thr Val Leu His Thr Leu Ala Leu Ala Asn Glu Ala Gly Val Glu
275 280 285 Tyr Ser Leu Glu
Arg Ile Asn Glu Val Ala Glu Arg Val Pro His Leu 290
295 300 Ala Lys Leu Ala Pro Ala Ser Asp
Val Phe Ile Glu Asp Leu His Glu 305 310
315 320 Ala Gly Gly Val Ser Ala Ala Leu Asn Glu Leu Ser
Lys Lys Glu Gly 325 330
335 Ala Leu His Leu Asp Ala Leu Thr Val Thr Gly Lys Thr Leu Gly Glu
340 345 350 Thr Ile Ala
Gly His Glu Val Lys Asp Tyr Asp Val Ile His Pro Leu 355
360 365 Asp Gln Pro Phe Thr Glu Lys Gly
Gly Leu Ala Val Leu Phe Gly Asn 370 375
380 Leu Ala Pro Asp Gly Ala Ile Ile Lys Thr Gly Gly Val
Gln Asn Gly 385 390 395
400 Ile Thr Arg His Glu Gly Pro Ala Val Val Phe Asp Ser Gln Asp Glu
405 410 415 Ala Leu Asp Gly
Ile Ile Asn Arg Lys Val Lys Glu Gly Asp Val Val 420
425 430 Ile Ile Arg Tyr Glu Gly Pro Lys Gly
Gly Pro Gly Met Pro Glu Met 435 440
445 Leu Ala Pro Thr Ser Gln Ile Val Gly Met Gly Leu Gly Pro
Lys Val 450 455 460
Ala Leu Ile Thr Asp Gly Arg Phe Ser Gly Ala Ser Arg Gly Leu Ser 465
470 475 480 Ile Gly His Val Ser
Pro Glu Ala Ala Glu Gly Gly Pro Leu Ala Phe 485
490 495 Val Glu Asn Gly Asp His Ile Ile Val Asp
Ile Glu Lys Arg Ile Leu 500 505
510 Asp Val Gln Val Pro Glu Glu Glu Trp Glu Lys Arg Lys Ala Asn
Trp 515 520 525 Lys
Gly Phe Glu Pro Lys Val Lys Thr Gly Tyr Leu Ala Arg Tyr Ser 530
535 540 Lys Leu Val Thr Ser Ala
Asn Thr Gly Gly Ile Met Lys Ile 545 550
555 241677DNABacillus subtilis 24atggcagaat tacgcagtaa
tatgatcaca caaggaatcg atagagctcc gcaccgcagt 60ttgcttcgtg cagcaggggt
aaaagaagag gatttcggca agccgtttat tgcggtgtgt 120aattcataca ttgatatcgt
tcccggtcat gttcacttgc aggagtttgg gaaaatcgta 180aaagaagcaa tcagagaagc
agggggcgtt ccgtttgaat ttaataccat tggggtagat 240gatggcatcg caatggggca
tatcggtatg agatattcgc tgccaagccg tgaaattatc 300gcagactctg tggaaacggt
tgtatccgca cactggtttg acggaatggt ctgtattccg 360aactgcgaca aaatcacacc
gggaatgctt atggcggcaa tgcgcatcaa cattccgacg 420atttttgtca gcggcggacc
gatggcggca ggaagaacaa gttacgggcg aaaaatctcc 480ctttcctcag tattcgaagg
ggtaggcgcc taccaagcag ggaaaatcaa cgaaaacgag 540cttcaagaac tagagcagtt
cggatgccca acgtgcgggt cttgctcagg catgtttacg 600gcgaactcaa tgaactgtct
gtcagaagca cttggtcttg ctttgccggg taatggaacc 660attctggcaa catctccgga
acgcaaagag tttgtgagaa aatcggctgc gcaattaatg 720gaaacgattc gcaaagatat
caaaccgcgt gatattgtta cagtaaaagc gattgataac 780gcgtttgcac tcgatatggc
gctcggaggt tctacaaata ccgttcttca tacccttgcc 840cttgcaaacg aagccggcgt
tgaatactct ttagaacgca ttaacgaagt cgctgagcgc 900gtgccgcact tggctaagct
ggcgcctgca tcggatgtgt ttattgaaga tcttcacgaa 960gcgggcggcg tttcagcggc
tctgaatgag ctttcgaaga aagaaggagc gcttcattta 1020gatgcgctga ctgttacagg
aaaaactctt ggagaaacca ttgccggaca tgaagtaaag 1080gattatgacg tcattcaccc
gctggatcaa ccattcactg aaaagggagg ccttgctgtt 1140ttattcggta atctagctcc
ggacggcgct atcattaaaa caggcggcgt acagaatggg 1200attacaagac acgaagggcc
ggctgtcgta ttcgattctc aggacgaggc gcttgacggc 1260attatcaacc gaaaagtaaa
agaaggcgac gttgtcatca tcagatacga agggccaaaa 1320ggcggacctg gcatgccgga
aatgctggcg ccaacatccc aaatcgttgg aatgggactc 1380gggccaaaag tggcattgat
tacggacgga cgtttttccg gagcctcccg tggcctctca 1440atcggccacg tatcacctga
ggccgctgag ggcgggccgc ttgcctttgt tgaaaacgga 1500gaccatatta tcgttgatat
tgaaaaacgc atcttggatg tacaagtgcc agaagaagag 1560tgggaaaaac gaaaagcgaa
ctggaaaggt tttgaaccga aagtgaaaac cggctacctg 1620gcacgttatt ctaaacttgt
gacaagtgcc aacaccggcg gtattatgaa aatctag 167725547PRTLactococcus
lactis 25Met Tyr Thr Val Gly Asp Tyr Leu Leu Asp Arg Leu His Glu Leu Gly
1 5 10 15 Ile Glu
Glu Ile Phe Gly Val Pro Gly Asp Tyr Asn Leu Gln Phe Leu 20
25 30 Asp Gln Ile Ile Ser Arg Glu
Asp Met Lys Trp Ile Gly Asn Ala Asn 35 40
45 Glu Leu Asn Ala Ser Tyr Met Ala Asp Gly Tyr Ala
Arg Thr Lys Lys 50 55 60
Ala Ala Ala Phe Leu Thr Thr Phe Gly Val Gly Glu Leu Ser Ala Ile 65
70 75 80 Asn Gly Leu
Ala Gly Ser Tyr Ala Glu Asn Leu Pro Val Val Glu Ile 85
90 95 Val Gly Ser Pro Thr Ser Lys Val
Gln Asn Asp Gly Lys Phe Val His 100 105
110 His Thr Leu Ala Asp Gly Asp Phe Lys His Phe Met Lys
Met His Glu 115 120 125
Pro Val Thr Ala Ala Arg Thr Leu Leu Thr Ala Glu Asn Ala Thr Tyr 130
135 140 Glu Ile Asp Arg
Val Leu Ser Gln Leu Leu Lys Glu Arg Lys Pro Val 145 150
155 160 Tyr Ile Asn Leu Pro Val Asp Val Ala
Ala Ala Lys Ala Glu Lys Pro 165 170
175 Ala Leu Ser Leu Glu Lys Glu Ser Ser Thr Thr Asn Thr Thr
Glu Gln 180 185 190
Val Ile Leu Ser Lys Ile Glu Glu Ser Leu Lys Asn Ala Gln Lys Pro
195 200 205 Val Val Ile Ala
Gly His Glu Val Ile Ser Phe Gly Leu Glu Lys Thr 210
215 220 Val Thr Gln Phe Val Ser Glu Thr
Lys Leu Pro Ile Thr Thr Leu Asn 225 230
235 240 Phe Gly Lys Ser Ala Val Asp Glu Ser Leu Pro Ser
Phe Leu Gly Ile 245 250
255 Tyr Asn Gly Lys Leu Ser Glu Ile Ser Leu Lys Asn Phe Val Glu Ser
260 265 270 Ala Asp Phe
Ile Leu Met Leu Gly Val Lys Leu Thr Asp Ser Ser Thr 275
280 285 Gly Ala Phe Thr His His Leu Asp
Glu Asn Lys Met Ile Ser Leu Asn 290 295
300 Ile Asp Glu Gly Ile Ile Phe Asn Lys Val Val Glu Asp
Phe Asp Phe 305 310 315
320 Arg Ala Val Val Ser Ser Leu Ser Glu Leu Lys Gly Ile Glu Tyr Glu
325 330 335 Gly Gln Tyr Ile
Asp Lys Gln Tyr Glu Glu Phe Ile Pro Ser Ser Ala 340
345 350 Pro Leu Ser Gln Asp Arg Leu Trp Gln
Ala Val Glu Ser Leu Thr Gln 355 360
365 Ser Asn Glu Thr Ile Val Ala Glu Gln Gly Thr Ser Phe Phe
Gly Ala 370 375 380
Ser Thr Ile Phe Leu Lys Ser Asn Ser Arg Phe Ile Gly Gln Pro Leu 385
390 395 400 Trp Gly Ser Ile Gly
Tyr Thr Phe Pro Ala Ala Leu Gly Ser Gln Ile 405
410 415 Ala Asp Lys Glu Ser Arg His Leu Leu Phe
Ile Gly Asp Gly Ser Leu 420 425
430 Gln Leu Thr Val Gln Glu Leu Gly Leu Ser Ile Arg Glu Lys Leu
Asn 435 440 445 Pro
Ile Cys Phe Ile Ile Asn Asn Asp Gly Tyr Thr Val Glu Arg Glu 450
455 460 Ile His Gly Pro Thr Gln
Ser Tyr Asn Asp Ile Pro Met Trp Asn Tyr 465 470
475 480 Ser Lys Leu Pro Glu Thr Phe Gly Ala Thr Glu
Asp Arg Val Val Ser 485 490
495 Lys Ile Val Arg Thr Glu Asn Glu Phe Val Ser Val Met Lys Glu Ala
500 505 510 Gln Ala
Asp Val Asn Arg Met Tyr Trp Ile Glu Leu Val Leu Glu Lys 515
520 525 Glu Asp Ala Pro Lys Leu Leu
Lys Lys Met Gly Lys Leu Phe Ala Glu 530 535
540 Gln Asn Lys 545 261828DNALactococcus
lactis 26tttaaataag tcaatatcgt tgacttattt agaagaaaga gttattcttt
aaatgtcaag 60ttagttgact aaattaaata taaaatatgg aggaatgtga tgtatacagt
aggagattac 120ctgttagacc gattacacga gttgggaatt gaagaaattt ttggagttcc
tggtgactat 180aacttacaat ttttagatca aattatttca cgcgaagata tgaaatggat
tggaaatgct 240aatgaattaa atgcttctta tatggctgat ggttatgctc gtactaaaaa
agctgccgca 300tttctcacca catttggagt cggcgaattg agtgcgatca atggactggc
aggaagttat 360gccgaaaatt taccagtagt agaaattgtt ggttcaccaa cttcaaaagt
acaaaatgac 420ggaaaatttg tccatcatac actagcagat ggtgatttta aacactttat
gaagatgcat 480gaacctgtta cagcagcgcg gactttactg acagcagaaa atgccacata
tgaaattgac 540cgagtacttt ctcaattact aaaagaaaga aaaccagtct atattaactt
accagtcgat 600gttgctgcag caaaagcaga gaagcctgca ttatctttag aaaaagaaag
ctctacaaca 660aatacaactg aacaagtgat tttgagtaag attgaagaaa gtttgaaaaa
tgcccaaaaa 720ccagtagtga ttgcaggaca cgaagtaatt agttttggtt tagaaaaaac
ggtaactcag 780tttgtttcag aaacaaaact accgattacg acactaaatt ttggtaaaag
tgctgttgat 840gaatctttgc cctcattttt aggaatatat aacgggaaac tttcagaaat
cagtcttaaa 900aattttgtgg agtccgcaga ctttatccta atgcttggag tgaagcttac
ggactcctca 960acaggtgcat tcacacatca tttagatgaa aataaaatga tttcactaaa
catagatgaa 1020ggaataattt tcaataaagt ggtagaagat tttgatttta gagcagtggt
ttcttcttta 1080tcagaattaa aaggaataga atatgaagga caatatattg ataagcaata
tgaagaattt 1140attccatcaa gtgctccctt atcacaagac cgtctatggc aggcagttga
aagtttgact 1200caaagcaatg aaacaatcgt tgctgaacaa ggaacctcat tttttggagc
ttcaacaatt 1260ttcttaaaat caaatagtcg ttttattgga caacctttat ggggttctat
tggatatact 1320tttccagcgg ctttaggaag ccaaattgcg gataaagaga gcagacacct
tttatttatt 1380ggtgatggtt cacttcaact taccgtacaa gaattaggac tatcaatcag
agaaaaactc 1440aatccaattt gttttatcat aaataatgat ggttatacag ttgaaagaga
aatccacgga 1500cctactcaaa gttataacga cattccaatg tggaattact cgaaattacc
agaaacattt 1560ggagcaacag aagatcgtgt agtatcaaaa attgttagaa cagagaatga
atttgtgtct 1620gtcatgaaag aagcccaagc agatgtcaat agaatgtatt ggatagaact
agttttggaa 1680aaagaagatg cgccaaaatt actgaaaaaa atgggtaaat tatttgctga
gcaaaataaa 1740tagatatcaa cggatgatga aaagtaaaat agacaaagtc caataatttt
ataaaaagta 1800aaaacattag gattttccta atgttttt
182827548PRTLactococcus lactis 27Met Tyr Thr Val Gly Asp Tyr
Leu Leu Asp Arg Leu His Glu Leu Gly 1 5
10 15 Ile Glu Glu Ile Phe Gly Val Pro Gly Asp Tyr
Asn Leu Gln Phe Leu 20 25
30 Asp Gln Ile Ile Ser His Lys Asp Met Lys Trp Val Gly Asn Ala
Asn 35 40 45 Glu
Leu Asn Ala Ser Tyr Met Ala Asp Gly Tyr Ala Arg Thr Lys Lys 50
55 60 Ala Ala Ala Phe Leu Thr
Thr Phe Gly Val Gly Glu Leu Ser Ala Val 65 70
75 80 Asn Gly Leu Ala Gly Ser Tyr Ala Glu Asn Leu
Pro Val Val Glu Ile 85 90
95 Val Gly Ser Pro Thr Ser Lys Val Gln Asn Glu Gly Lys Phe Val His
100 105 110 His Thr
Leu Ala Asp Gly Asp Phe Lys His Phe Met Lys Met His Glu 115
120 125 Pro Val Thr Ala Ala Arg Thr
Leu Leu Thr Ala Glu Asn Ala Thr Val 130 135
140 Glu Ile Asp Arg Val Leu Ser Ala Leu Leu Lys Glu
Arg Lys Pro Val 145 150 155
160 Tyr Ile Asn Leu Pro Val Asp Val Ala Ala Ala Lys Ala Glu Lys Pro
165 170 175 Ser Leu Pro
Leu Lys Lys Glu Asn Ser Thr Ser Asn Thr Ser Asp Gln 180
185 190 Glu Ile Leu Asn Lys Ile Gln Glu
Ser Leu Lys Asn Ala Lys Lys Pro 195 200
205 Ile Val Ile Thr Gly His Glu Ile Ile Ser Phe Gly Leu
Glu Lys Thr 210 215 220
Val Thr Gln Phe Ile Ser Lys Thr Lys Leu Pro Ile Thr Thr Leu Asn 225
230 235 240 Phe Gly Lys Ser
Ser Val Asp Glu Ala Leu Pro Ser Phe Leu Gly Ile 245
250 255 Tyr Asn Gly Thr Leu Ser Glu Pro Asn
Leu Lys Glu Phe Val Glu Ser 260 265
270 Ala Asp Phe Ile Leu Met Leu Gly Val Lys Leu Thr Asp Ser
Ser Thr 275 280 285
Gly Ala Phe Thr His His Leu Asn Glu Asn Lys Met Ile Ser Leu Asn 290
295 300 Ile Asp Glu Gly Lys
Ile Phe Asn Glu Arg Ile Gln Asn Phe Asp Phe 305 310
315 320 Glu Ser Leu Ile Ser Ser Leu Leu Asp Leu
Ser Glu Ile Glu Tyr Lys 325 330
335 Gly Lys Tyr Ile Asp Lys Lys Gln Glu Asp Phe Val Pro Ser Asn
Ala 340 345 350 Leu
Leu Ser Gln Asp Arg Leu Trp Gln Ala Val Glu Asn Leu Thr Gln 355
360 365 Ser Asn Glu Thr Ile Val
Ala Glu Gln Gly Thr Ser Phe Phe Gly Ala 370 375
380 Ser Ser Ile Phe Leu Lys Ser Lys Ser His Phe
Ile Gly Gln Pro Leu 385 390 395
400 Trp Gly Ser Ile Gly Tyr Thr Phe Pro Ala Ala Leu Gly Ser Gln Ile
405 410 415 Ala Asp
Lys Glu Ser Arg His Leu Leu Phe Ile Gly Asp Gly Ser Leu 420
425 430 Gln Leu Thr Val Gln Glu Leu
Gly Leu Ala Ile Arg Glu Lys Ile Asn 435 440
445 Pro Ile Cys Phe Ile Ile Asn Asn Asp Gly Tyr Thr
Val Glu Arg Glu 450 455 460
Ile His Gly Pro Asn Gln Ser Tyr Asn Asp Ile Pro Met Trp Asn Tyr 465
470 475 480 Ser Lys Leu
Pro Glu Ser Phe Gly Ala Thr Glu Asp Arg Val Val Ser 485
490 495 Lys Ile Val Arg Thr Glu Asn Glu
Phe Val Ser Val Met Lys Glu Ala 500 505
510 Gln Ala Asp Pro Asn Arg Met Tyr Trp Ile Glu Leu Ile
Leu Ala Lys 515 520 525
Glu Gly Ala Pro Lys Val Leu Lys Lys Met Gly Lys Leu Phe Ala Glu 530
535 540 Gln Asn Lys Ser
545 281954DNALactococcus lactis 28ctagagtttt ctttagtcat
aattcactcc ttttattagt ctattatact tgataattca 60aataagtcaa tatcgttgac
ttatttaaag aaaagcgtta ttctataaat gtcaagttga 120ttgaccaata tataataaaa
tatggaggaa tgcgatgtat acagtaggag attacctatt 180agaccgatta cacgagttag
gaattgaaga aatttttgga gtccctggag actataactt 240acaattttta gatcaaatta
tttcccacaa ggatatgaaa tgggtcggaa atgctaatga 300attaaatgct tcatatatgg
ctgatggcta tgctcgtact aaaaaagctg ccgcatttct 360tacaaccttt ggagtaggtg
aattgagtgc agttaatgga ttagcaggaa gttacgccga 420aaatttacca gtagtagaaa
tagtgggatc acctacatca aaagttcaaa atgaaggaaa 480atttgttcat catacgctgg
ctgacggtga ttttaaacac tttatgaaaa tgcacgaacc 540tgttacagca gctcgaactt
tactgacagc agaaaatgca accgttgaaa ttgaccgagt 600actttctgca ctattaaaag
aaagaaaacc tgtctatatc aacttaccag ttgatgttgc 660tgctgcaaaa gcagagaaac
cctcactccc tttgaaaaag gaaaactcaa cttcaaatac 720aagtgaccaa gaaattttga
acaaaattca agaaagcttg aaaaatgcca aaaaaccaat 780cgtgattaca ggacatgaaa
taattagttt tggcttagaa aaaacagtca ctcaatttat 840ttcaaagaca aaactaccta
ttacgacatt aaactttggt aaaagttcag ttgatgaagc 900cctcccttca tttttaggaa
tctataatgg tacactctca gagcctaatc ttaaagaatt 960cgtggaatca gccgacttca
tcttgatgct tggagttaaa ctcacagact cttcaacagg 1020agccttcact catcatttaa
atgaaaataa aatgatttca ctgaatatag atgaaggaaa 1080aatatttaac gaaagaatcc
aaaattttga ttttgaatcc ctcatctcct ctctcttaga 1140cctaagcgaa atagaataca
aaggaaaata tatcgataaa aagcaagaag actttgttcc 1200atcaaatgcg cttttatcac
aagaccgcct atggcaagca gttgaaaacc taactcaaag 1260caatgaaaca atcgttgctg
aacaagggac atcattcttt ggcgcttcat caattttctt 1320aaaatcaaag agtcatttta
ttggtcaacc cttatgggga tcaattggat atacattccc 1380agcagcatta ggaagccaaa
ttgcagataa agaaagcaga caccttttat ttattggtga 1440tggttcactt caacttacag
tgcaagaatt aggattagca atcagagaaa aaattaatcc 1500aatttgcttt attatcaata
atgatggtta tacagtcgaa agagaaattc atggaccaaa 1560tcaaagctac aatgatattc
caatgtggaa ttactcaaaa ttaccagaat cgtttggagc 1620aacagaagat cgagtagtct
caaaaatcgt tagaactgaa aatgaatttg tgtctgtcat 1680gaaagaagct caagcagatc
caaatagaat gtactggatt gagttaattt tggcaaaaga 1740aggtgcacca aaagtactga
aaaaaatggg caaactattt gctgaacaaa ataaatcata 1800atttataaat agtaaaaaac
attaggaaat acctaatgtt tttttgttga ctaaatcaat 1860ccctctttat atagaaaacc
ttagtttctc aaagacaact taattaagcc tgccaaattg 1920gaactcgcaa aatgtaatct
atcctctgct ccta 195429550PRTSalmonella
typhimurium 29Met Gln Asn Pro Tyr Thr Val Ala Asp Tyr Leu Leu Asp Arg Leu
Ala 1 5 10 15 Gly
Cys Gly Ile Gly His Leu Phe Gly Val Pro Gly Asp Tyr Asn Leu
20 25 30 Gln Phe Leu Asp His
Val Ile Asp His Pro Thr Leu Arg Trp Val Gly 35
40 45 Cys Ala Asn Glu Leu Asn Ala Ala Tyr
Ala Ala Asp Gly Tyr Ala Arg 50 55
60 Met Ser Gly Ala Gly Ala Leu Leu Thr Thr Phe Gly Val
Gly Glu Leu 65 70 75
80 Ser Ala Ile Asn Gly Ile Ala Gly Ser Tyr Ala Glu Tyr Val Pro Val
85 90 95 Leu His Ile Val
Gly Ala Pro Cys Ser Ala Ala Gln Gln Arg Gly Glu 100
105 110 Leu Met His His Thr Leu Gly Asp Gly
Asp Phe Arg His Phe Tyr Arg 115 120
125 Met Ser Gln Ala Ile Ser Ala Ala Ser Ala Ile Leu Asp Glu
Gln Asn 130 135 140
Ala Cys Phe Glu Ile Asp Arg Val Leu Gly Glu Met Leu Ala Ala Arg 145
150 155 160 Arg Pro Gly Tyr Ile
Met Leu Pro Ala Asp Val Ala Lys Lys Thr Ala 165
170 175 Ile Pro Pro Thr Gln Ala Leu Ala Leu Pro
Val His Glu Ala Gln Ser 180 185
190 Gly Val Glu Thr Ala Phe Arg Tyr His Ala Arg Gln Cys Leu Met
Asn 195 200 205 Ser
Arg Arg Ile Ala Leu Leu Ala Asp Phe Leu Ala Gly Arg Phe Gly 210
215 220 Leu Arg Pro Leu Leu Gln
Arg Trp Met Ala Glu Thr Pro Ile Ala His 225 230
235 240 Ala Thr Leu Leu Met Gly Lys Gly Leu Phe Asp
Glu Gln His Pro Asn 245 250
255 Phe Val Gly Thr Tyr Ser Ala Gly Ala Ser Ser Lys Glu Val Arg Gln
260 265 270 Ala Ile
Glu Asp Ala Asp Arg Val Ile Cys Val Gly Thr Arg Phe Val 275
280 285 Asp Thr Leu Thr Ala Gly Phe
Thr Gln Gln Leu Pro Ala Glu Arg Thr 290 295
300 Leu Glu Ile Gln Pro Tyr Ala Ser Arg Ile Gly Glu
Thr Trp Phe Asn 305 310 315
320 Leu Pro Met Ala Gln Ala Val Ser Thr Leu Arg Glu Leu Cys Leu Glu
325 330 335 Cys Ala Phe
Ala Pro Pro Pro Thr Arg Ser Ala Gly Gln Pro Val Arg 340
345 350 Ile Asp Lys Gly Glu Leu Thr Gln
Glu Ser Phe Trp Gln Thr Leu Gln 355 360
365 Gln Tyr Leu Lys Pro Gly Asp Ile Ile Leu Val Asp Gln
Gly Thr Ala 370 375 380
Ala Phe Gly Ala Ala Ala Leu Ser Leu Pro Asp Gly Ala Glu Val Val 385
390 395 400 Leu Gln Pro Leu
Trp Gly Ser Ile Gly Tyr Ser Leu Pro Ala Ala Phe 405
410 415 Gly Ala Gln Thr Ala Cys Pro Asp Arg
Arg Val Ile Leu Ile Ile Gly 420 425
430 Asp Gly Ala Ala Gln Leu Thr Ile Gln Glu Met Gly Ser Met
Leu Arg 435 440 445
Asp Gly Gln Ala Pro Val Ile Leu Leu Leu Asn Asn Asp Gly Tyr Thr 450
455 460 Val Glu Arg Ala Ile
His Gly Ala Ala Gln Arg Tyr Asn Asp Ile Ala 465 470
475 480 Ser Trp Asn Trp Thr Gln Ile Pro Pro Ala
Leu Asn Ala Ala Gln Gln 485 490
495 Ala Glu Cys Trp Arg Val Thr Gln Ala Ile Gln Leu Ala Glu Val
Leu 500 505 510 Glu
Arg Leu Ala Arg Pro Gln Arg Leu Ser Phe Ile Glu Val Met Leu 515
520 525 Pro Lys Ala Asp Leu Pro
Glu Leu Leu Arg Thr Val Thr Arg Ala Leu 530 535
540 Glu Ala Arg Asn Gly Gly 545
550 301653DNASalmonella typhimurium 30ttatcccccg ttgcgggctt ccagcgcccg
ggtcacggta cgcagtaatt ccggcagatc 60ggcttttggc aacatcactt caataaatga
cagacgttgt gggcgcgcca accgttcgag 120gacctctgcc agttggatag cctgcgtcac
ccgccagcac tccgcctgtt gcgccgcgtt 180tagcgccggt ggtatctgcg tccagttcca
gctcgcgatg tcgttatacc gctgggccgc 240gccgtgaatg gcgcgctcta cggtatagcc
gtcattgttg agcagcagga tgaccggcgc 300ctgcccgtcg cgtaacatcg agcccatctc
ctgaatcgtg agctgcgccg cgccatcgcc 360gataatcaga atcacccgcc gatcgggaca
ggcggtttgc gcgccaaacg cggcgggcaa 420ggaatagccg atagaccccc acagcggctg
taacacaact tccgcgccgt caggaagcga 480cagcgcggca gcgccaaaag ctgctgtccc
ctggtcgaca aggataatat ctccgggttt 540gagatactgc tgtaaggttt gccagaagct
ttcctgggtc agttctcctt tatcaatccg 600cactggctgt ccggcggaac gcgtcggcgg
cggcgcaaaa gcgcattcca ggcacagttc 660gcgcagcgta gacaccgcct gcgccatcgg
gaggttgaac caggtttcgc cgatgcgcga 720cgcgtaaggc tgaatctcca gcgtgcgttc
cgccggtaat tgttgggtaa atccggccgt 780aagggtatcg acaaaacggg tgccgacgca
gataacccta tcggcgtcct ctatggcctg 840acgcacttct ttgctgctgg cgccagcgct
ataggtgcca acgaagttcg ggtgctgttc 900atcaaaaagc cccttcccca tcagtagtgt
cgcatgagcg atgggcgttt ccgccatcca 960gcgctgcaac agtggtcgta aaccaaaacg
cccggcaaga aagtcggcca atagcgcaat 1020gcgccgactg ttcatcaggc actgacgggc
gtgataacga aaggccgtct ccacgccgct 1080ttgcgcttca tgcacgggca acgccagcgc
ctgcgtaggt gggatggccg tttttttcgc 1140cacatcggcg ggcaacatga tgtatcctgg
cctgcgtgcg gcaagcattt cacccaacac 1200gcggtcaatc tcgaaacagg cgttctgttc
atctaatatt gcgctggcag cggatatcgc 1260ctgactcatg cgataaaaat gacgaaaatc
gccgtcaccg agggtatggt gcatcaattc 1320gccacgctgc tgcgcagcgc tacagggcgc
gccgacgata tgcaagaccg ggacatattc 1380cgcgtaactg cccgcgatac cgttaatagc
gctaagttct cccacgccaa aggtggtgag 1440tagcgctcca gcgcccgaca tgcgcgcata
gccgtccgcg gcataagcgg cgttcagctc 1500attggcgcat cccacccaac gcagggtcgg
gtggtcaatc acatggtcaa gaaactgcaa 1560gttataatcg cccggtacgc caaaaagatg
gccaatgccg catcctgcca gtctgtccag 1620caaatagtcg gccacggtat aggggttttg
cat 165331554PRTClostridium acetobutylicum
31Met Lys Ser Glu Tyr Thr Ile Gly Arg Tyr Leu Leu Asp Arg Leu Ser 1
5 10 15 Glu Leu Gly Ile
Arg His Ile Phe Gly Val Pro Gly Asp Tyr Asn Leu 20
25 30 Ser Phe Leu Asp Tyr Ile Met Glu Tyr
Lys Gly Ile Asp Trp Val Gly 35 40
45 Asn Cys Asn Glu Leu Asn Ala Gly Tyr Ala Ala Asp Gly Tyr
Ala Arg 50 55 60
Ile Asn Gly Ile Gly Ala Ile Leu Thr Thr Phe Gly Val Gly Glu Leu 65
70 75 80 Ser Ala Ile Asn Ala
Ile Ala Gly Ala Tyr Ala Glu Gln Val Pro Val 85
90 95 Val Lys Ile Thr Gly Ile Pro Thr Ala Lys
Val Arg Asp Asn Gly Leu 100 105
110 Tyr Val His His Thr Leu Gly Asp Gly Arg Phe Asp His Phe Phe
Glu 115 120 125 Met
Phe Arg Glu Val Thr Val Ala Glu Ala Leu Leu Ser Glu Glu Asn 130
135 140 Ala Ala Gln Glu Ile Asp
Arg Val Leu Ile Ser Cys Trp Arg Gln Lys 145 150
155 160 Arg Pro Val Leu Ile Asn Leu Pro Ile Asp Val
Tyr Asp Lys Pro Ile 165 170
175 Asn Lys Pro Leu Lys Pro Leu Leu Asp Tyr Thr Ile Ser Ser Asn Lys
180 185 190 Glu Ala
Ala Cys Glu Phe Val Thr Glu Ile Val Pro Ile Ile Asn Arg 195
200 205 Ala Lys Lys Pro Val Ile Leu
Ala Asp Tyr Gly Val Tyr Arg Tyr Gln 210 215
220 Val Gln His Val Leu Lys Asn Leu Ala Glu Lys Thr
Gly Phe Pro Val 225 230 235
240 Ala Thr Leu Ser Met Gly Lys Gly Val Phe Asn Glu Ala His Pro Gln
245 250 255 Phe Ile Gly
Val Tyr Asn Gly Asp Val Ser Ser Pro Tyr Leu Arg Gln 260
265 270 Arg Val Asp Glu Ala Asp Cys Ile
Ile Ser Val Gly Val Lys Leu Thr 275 280
285 Asp Ser Thr Thr Gly Gly Phe Ser His Gly Phe Ser Lys
Arg Asn Val 290 295 300
Ile His Ile Asp Pro Phe Ser Ile Lys Ala Lys Gly Lys Lys Tyr Ala 305
310 315 320 Pro Ile Thr Met
Lys Asp Ala Leu Thr Glu Leu Thr Ser Lys Ile Glu 325
330 335 His Arg Asn Phe Glu Asp Leu Asp Ile
Lys Pro Tyr Lys Ser Asp Asn 340 345
350 Gln Lys Tyr Phe Ala Lys Glu Lys Pro Ile Thr Gln Lys Arg
Phe Phe 355 360 365
Glu Arg Ile Ala His Phe Ile Lys Glu Lys Asp Val Leu Leu Ala Glu 370
375 380 Gln Gly Thr Cys Phe
Phe Gly Ala Ser Thr Ile Gln Leu Pro Lys Asp 385 390
395 400 Ala Thr Phe Ile Gly Gln Pro Leu Trp Gly
Ser Ile Gly Tyr Thr Leu 405 410
415 Pro Ala Leu Leu Gly Ser Gln Leu Ala Asp Gln Lys Arg Arg Asn
Ile 420 425 430 Leu
Leu Ile Gly Asp Gly Ala Phe Gln Met Thr Ala Gln Glu Ile Ser 435
440 445 Thr Met Leu Arg Leu Gln
Ile Lys Pro Ile Ile Phe Leu Ile Asn Asn 450 455
460 Asp Gly Tyr Thr Ile Glu Arg Ala Ile His Gly
Arg Glu Gln Val Tyr 465 470 475
480 Asn Asn Ile Gln Met Trp Arg Tyr His Asn Val Pro Lys Val Leu Gly
485 490 495 Pro Lys
Glu Cys Ser Leu Thr Phe Lys Val Gln Ser Glu Thr Glu Leu 500
505 510 Glu Lys Ala Leu Leu Val Ala
Asp Lys Asp Cys Glu His Leu Ile Phe 515 520
525 Ile Glu Val Val Met Asp Arg Tyr Asp Lys Pro Glu
Pro Leu Glu Arg 530 535 540
Leu Ser Lys Arg Phe Ala Asn Gln Asn Asn 545 550
321665DNAClostridium acetobutylicum 32ttgaagagtg aatacacaat
tggaagatat ttgttagacc gtttatcaga gttgggtatt 60cggcatatct ttggtgtacc
tggagattac aatctatcct ttttagacta tataatggag 120tacaaaggga tagattgggt
tggaaattgc aatgaattga atgctgggta tgctgctgat 180ggatatgcaa gaataaatgg
aattggagcc atacttacaa catttggtgt tggagaatta 240agtgccatta acgcaattgc
tggggcatac gctgagcaag ttccagttgt taaaattaca 300ggtatcccca cagcaaaagt
tagggacaat ggattatatg tacaccacac attaggtgac 360ggaaggtttg atcacttttt
tgaaatgttt agagaagtaa cagttgctga ggcattacta 420agcgaagaaa atgcagcaca
agaaattgat cgtgttctta tttcatgctg gagacaaaaa 480cgtcctgttc ttataaattt
accgattgat gtatatgata aaccaattaa caaaccatta 540aagccattac tcgattatac
tatttcaagt aacaaagagg ctgcatgtga atttgttaca 600gaaatagtac ctataataaa
tagggcaaaa aagcctgtta ttcttgcaga ttatggagta 660tatcgttacc aagttcaaca
tgtgcttaaa aacttggccg aaaaaaccgg atttcctgtg 720gctacactaa gtatgggaaa
aggtgttttc aatgaagcac accctcaatt tattggtgtt 780tataatggtg atgtaagttc
tccttattta aggcagcgag ttgatgaagc agactgcatt 840attagcgttg gtgtaaaatt
gacggattca accacagggg gattttctca tggattttct 900aaaaggaatg taattcacat
tgatcctttt tcaataaagg caaaaggtaa aaaatatgca 960cctattacga tgaaagatgc
tttaacagaa ttaacaagta aaattgagca tagaaacttt 1020gaggatttag atataaagcc
ttacaaatca gataatcaaa agtattttgc aaaagagaag 1080ccaattacac aaaaacgttt
ttttgagcgt attgctcact ttataaaaga aaaagatgta 1140ttattagcag aacagggtac
atgctttttt ggtgcgtcaa ccatacaact acccaaagat 1200gcaactttta ttggtcaacc
tttatgggga tctattggat acacacttcc tgctttatta 1260ggttcacaat tagctgatca
aaaaaggcgt aatattcttt taattgggga tggtgcattt 1320caaatgacag cacaagaaat
ttcaacaatg cttcgtttac aaatcaaacc tattattttt 1380ttaattaata acgatggtta
tacaattgaa cgtgctattc atggtagaga acaagtatat 1440aacaatattc aaatgtggcg
atatcataat gttccaaagg ttttaggtcc taaagaatgc 1500agcttaacct ttaaagtaca
aagtgaaact gaacttgaaa aggctctttt agtggcagat 1560aaggattgtg aacatttgat
ttttatagaa gttgttatgg atcgttatga taaacccgag 1620cctttagaac gtctttcgaa
acgttttgca aatcaaaata attag 1665331641DNAClostridium
acetobutylicum 33atgaaacaac gtatcgggca atacttgatc gatgccctac acgttaatgg
tgtcgataag 60atctttggag tcccaggtga tttcacttta gcctttttgg acgatatcat
aagacatgac 120aacgtggaat gggtgggaaa tactaatgag ttgaacgccg cttacgccgc
tgatggttac 180gctagagtta atggattagc cgctgtatct accacttttg gggttggcga
gttatctgct 240gtgaatggta ttgctggaag ttacgcagag cgtgttcctg taatcaaaat
ctcaggcggt 300ccttcatcag ttgctcaaca agagggtaga tatgtccacc attcattggg
tgaaggaatc 360tttgattcat attcaaagat gtacgctcac ataaccgcaa caactacaat
cttatccgtt 420gacaacgcag tcgacgaaat tgatagagtt attcattgtg ctttgaagga
aaagaggcca 480gtgcatattc atttgcctat tgacgtagcc ttaactgaga ttgaaatccc
tcatgcacca 540aaagtttaca cacacgaatc ccagaacgtc gatgcttaca ttcaagctgt
tgagaaaaag 600ttaatgtctg caaaacaacc agtaatcata gcaggtcatg aaatcaattc
attcaagttg 660cacgaacaac tggaacagtt tgtcaatcag acaaacatcc ctgttgcaca
actttccttg 720ggtaagtctg ctttcaatga agagaatgaa cattaccttg gtatctacga
tggcaaaatc 780gcaaaggaaa atgtgagaga gtacgtcgac aatgctgatg tcatattgaa
cataggtgcc 840aaactgactg attctgctac agctggattt tcctacaagt tcgatacaaa
caacataatc 900tacattaacc ataatgactt caaagctgaa gatgtgattt ctgataatgt
ttcactgatt 960gatcttgtga atggcctgaa ttctattgac tatagaaatg aaacacacta
cccatcttat 1020caaagatctg atatgaaata cgaattgaat gacgcaccac ttacacaatc
taactatttc 1080aaaatgatga acgcttttct agaaaaagat gacatcctac tagctgaaca
aggtacatcc 1140tttttcggcg catatgactt atccctatac aagggaaatc agtttatcgg
tcagccttta 1200tgggggtcaa tagggtatac ttttccatct ttactaggaa gtcaactagc
agacatgcat 1260aggagaaaca ttttgcttat aggcgatggt agtttacaac ttactgttca
agccctaagt 1320acaatgatta gaaaggatat caaaccaatc attttcgtta tcaataacga
cggttacacc 1380gtcgaaagac ttatccacgg catggaagag ccatacaatg atatccaaat
gtggaactac 1440aagcaattgc cagaagtatt tggtggaaaa gatactgtaa aagttcatga
tgctaaaacc 1500tccaacgaac tgaaaactgt aatggattct gttaaagcag acaaagatca
catgcatttc 1560attgaagtgc atatggcagt agaggacgcc ccaaagaagt tgattgatat
agctaaagcc 1620tttagtgatg ctaacaagta a
1641341647DNAListeria grayi 34atgtacaccg tcggccaata cttagtagac
cgcttagaag agatcggcat cgataaggtt 60tttggtgtcc cgggtgacta caacctgacc
tttttggact acatccagaa ccacgaaggt 120ctgagctggc aaggtaatac gaatgaactg
aatgccgcgt acgcagctga tggctatgct 180cgtgaacgcg gtgttagcgc tttggtcacg
accttcggcg ttggtgagct gtccgcaatc 240aatggcaccg caggtagctt cgcggagcaa
gttccggtga ttcatatcgt gggcagcccg 300accatgaatg ttcagagcaa caagaaactg
gttcatcaca gcctgggtat gggcaacttt 360cacaacttca gcgagatggc gaaagaagtc
accgccgcaa ccacgatgct gacggaagag 420aatgcggcgt cggagattga tcgtgttctg
gaaaccgccc tgctggagaa acgcccagtg 480tacatcaatc tgccgatcga cattgctcac
aaggcgatcg tcaagccggc gaaagccctg 540caaaccgaga agagctctgg cgagcgtgag
gcacaactgg cggagatcat tctgagccat 600ctggagaagg ctgcacagcc gattgtgatt
gcgggtcacg agatcgcgcg cttccagatc 660cgtgagcgtt tcgagaattg gattaatcaa
acgaaactgc cggtgaccaa tctggcctac 720ggcaagggta gcttcaacga agaaaacgag
catttcattg gtacctatta tcctgcattt 780agcgataaga acgtgctgga ctacgtggat
aactccgact ttgtcctgca ctttggtggt 840aaaatcattg ataacagcac ctccagcttc
tcccaaggct tcaaaaccga gaacaccctg 900actgcggcga acgatatcat tatgctgccg
gacggtagca cgtattctgg tattagcctg 960aatggcctgc tggccgagct ggaaaaactg
aatttcacgt ttgccgacac cgcagcaaag 1020caggcggagt tggcggtgtt tgagccgcag
gctgaaaccc cgttgaaaca ggaccgtttt 1080caccaggcgg tgatgaattt tctgcaagct
gacgatgtcc tggttacgga acagggcacc 1140tcttcttttg gcttgatgct ggcgcctctg
aaaaagggta tgaacttgat ctcgcaaacg 1200ctgtggggta gcattggtta cacgttgccg
gcgatgattg gtagccaaat tgcggcaccg 1260gagcgtcgtc atatcctgag cattggtgat
ggtagctttc agctgactgc gcaggaaatg 1320agcaccattt tccgtgagaa actgacccca
gtcatcttca tcattaacaa tgatggctat 1380accgttgagc gtgcgatcca tggcgaagat
gaaagctata acgacattcc gacgtggaac 1440ttgcaactgg tggcggaaac cttcggtggt
gacgccgaaa ccgtcgacac tcacaatgtg 1500ttcacggaga ctgatttcgc caacaccctg
gcggcaattg acgcgacgcc gcagaaagca 1560cacgttgtgg aagttcacat ggaacaaatg
gatatgccgg agagcctgcg ccagatcggt 1620ctggcactgt ccaagcagaa tagctaa
164735312PRTSaccharomyces cerevisiae
35Met Pro Ala Thr Leu Lys Asn Ser Ser Ala Thr Leu Lys Leu Asn Thr 1
5 10 15 Gly Ala Ser Ile
Pro Val Leu Gly Phe Gly Thr Trp Arg Ser Val Asp 20
25 30 Asn Asn Gly Tyr His Ser Val Ile Ala
Ala Leu Lys Ala Gly Tyr Arg 35 40
45 His Ile Asp Ala Ala Ala Ile Tyr Leu Asn Glu Glu Glu Val
Gly Arg 50 55 60
Ala Ile Lys Asp Ser Gly Val Pro Arg Glu Glu Ile Phe Ile Thr Thr 65
70 75 80 Lys Leu Trp Gly Thr
Glu Gln Arg Asp Pro Glu Ala Ala Leu Asn Lys 85
90 95 Ser Leu Lys Arg Leu Gly Leu Asp Tyr Val
Asp Leu Tyr Leu Met His 100 105
110 Trp Pro Val Pro Leu Lys Thr Asp Arg Val Thr Asp Gly Asn Val
Leu 115 120 125 Cys
Ile Pro Thr Leu Glu Asp Gly Thr Val Asp Ile Asp Thr Lys Glu 130
135 140 Trp Asn Phe Ile Lys Thr
Trp Glu Leu Met Gln Glu Leu Pro Lys Thr 145 150
155 160 Gly Lys Thr Lys Ala Val Gly Val Ser Asn Phe
Ser Ile Asn Asn Ile 165 170
175 Lys Glu Leu Leu Glu Ser Pro Asn Asn Lys Val Val Pro Ala Thr Asn
180 185 190 Gln Ile
Glu Ile His Pro Leu Leu Pro Gln Asp Glu Leu Ile Ala Phe 195
200 205 Cys Lys Glu Lys Gly Ile Val
Val Glu Ala Tyr Ser Pro Phe Gly Ser 210 215
220 Ala Asn Ala Pro Leu Leu Lys Glu Gln Ala Ile Ile
Asp Met Ala Lys 225 230 235
240 Lys His Gly Val Glu Pro Ala Gln Leu Ile Ile Ser Trp Ser Ile Gln
245 250 255 Arg Gly Tyr
Val Val Leu Ala Lys Ser Val Asn Pro Glu Arg Ile Val 260
265 270 Ser Asn Phe Lys Ile Phe Thr Leu
Pro Glu Asp Asp Phe Lys Thr Ile 275 280
285 Ser Asn Leu Ser Lys Val His Gly Thr Lys Arg Val Val
Asp Met Lys 290 295 300
Trp Gly Ser Phe Pro Ile Phe Gln 305 310
36939DNASaccharomyces cerevisiae 36atgcctgcta cgttaaagaa ttcttctgct
acattaaaac taaatactgg tgcctccatt 60ccagtgttgg gtttcggcac ttggcgttcc
gttgacaata acggttacca ttctgtaatt 120gcagctttga aagctggata cagacacatt
gatgctgcgg ctatctattt gaatgaagaa 180gaagttggca gggctattaa agattccgga
gtccctcgtg aggaaatttt tattactact 240aagctttggg gtacggaaca acgtgatccg
gaagctgctc taaacaagtc tttgaaaaga 300ctaggcttgg attatgttga cctatatctg
atgcattggc cagtgccttt gaaaaccgac 360agagttactg atggtaacgt tctgtgcatt
ccaacattag aagatggcac tgttgacatc 420gatactaagg aatggaattt tatcaagacg
tgggagttga tgcaagagtt gccaaagacg 480ggcaaaacta aagccgttgg tgtctctaat
ttttctatta acaacattaa agaattatta 540gaatctccaa ataacaaggt ggtaccagct
actaatcaaa ttgaaattca tccattgcta 600ccacaagacg aattgattgc cttttgtaag
gaaaagggta ttgttgttga agcctactca 660ccatttggga gtgctaatgc tcctttacta
aaagagcaag caattattga tatggctaaa 720aagcacggcg ttgagccagc acagcttatt
atcagttgga gtattcaaag aggctacgtt 780gttctggcca aatcggttaa tcctgaaaga
attgtatcca attttaagat tttcactctg 840cctgaggatg atttcaagac tattagtaac
ctatccaaag tgcatggtac aaagagagtc 900gttgatatga agtggggatc cttcccaatt
ttccaatga 93937360PRTSaccharomyces cerevisiae
37Met Ser Tyr Pro Glu Lys Phe Glu Gly Ile Ala Ile Gln Ser His Glu 1
5 10 15 Asp Trp Lys Asn
Pro Lys Lys Thr Lys Tyr Asp Pro Lys Pro Phe Tyr 20
25 30 Asp His Asp Ile Asp Ile Lys Ile Glu
Ala Cys Gly Val Cys Gly Ser 35 40
45 Asp Ile His Cys Ala Ala Gly His Trp Gly Asn Met Lys Met
Pro Leu 50 55 60
Val Val Gly His Glu Ile Val Gly Lys Val Val Lys Leu Gly Pro Lys 65
70 75 80 Ser Asn Ser Gly Leu
Lys Val Gly Gln Arg Val Gly Val Gly Ala Gln 85
90 95 Val Phe Ser Cys Leu Glu Cys Asp Arg Cys
Lys Asn Asp Asn Glu Pro 100 105
110 Tyr Cys Thr Lys Phe Val Thr Thr Tyr Ser Gln Pro Tyr Glu Asp
Gly 115 120 125 Tyr
Val Ser Gln Gly Gly Tyr Ala Asn Tyr Val Arg Val His Glu His 130
135 140 Phe Val Val Pro Ile Pro
Glu Asn Ile Pro Ser His Leu Ala Ala Pro 145 150
155 160 Leu Leu Cys Gly Gly Leu Thr Val Tyr Ser Pro
Leu Val Arg Asn Gly 165 170
175 Cys Gly Pro Gly Lys Lys Val Gly Ile Val Gly Leu Gly Gly Ile Gly
180 185 190 Ser Met
Gly Thr Leu Ile Ser Lys Ala Met Gly Ala Glu Thr Tyr Val 195
200 205 Ile Ser Arg Ser Ser Arg Lys
Arg Glu Asp Ala Met Lys Met Gly Ala 210 215
220 Asp His Tyr Ile Ala Thr Leu Glu Glu Gly Asp Trp
Gly Glu Lys Tyr 225 230 235
240 Phe Asp Thr Phe Asp Leu Ile Val Val Cys Ala Ser Ser Leu Thr Asp
245 250 255 Ile Asp Phe
Asn Ile Met Pro Lys Ala Met Lys Val Gly Gly Arg Ile 260
265 270 Val Ser Ile Ser Ile Pro Glu Gln
His Glu Met Leu Ser Leu Lys Pro 275 280
285 Tyr Gly Leu Lys Ala Val Ser Ile Ser Tyr Ser Ala Leu
Gly Ser Ile 290 295 300
Lys Glu Leu Asn Gln Leu Leu Lys Leu Val Ser Glu Lys Asp Ile Lys 305
310 315 320 Ile Trp Val Glu
Thr Leu Pro Val Gly Glu Ala Gly Val His Glu Ala 325
330 335 Phe Glu Arg Met Glu Lys Gly Asp Val
Arg Tyr Arg Phe Thr Leu Val 340 345
350 Gly Tyr Asp Lys Glu Phe Ser Asp 355
360 38 1083DNASaccharomyces cerevisiae 38ctagtctgaa aattctttgt
cgtagccgac taaggtaaat ctatatctaa cgtcaccctt 60ttccatcctt tcgaaggctt
catggacgcc ggcttcacca acaggtaatg tttccaccca 120aattttgata tctttttcag
agactaattt caagagttgg ttcaattctt tgatggaacc 180taaagcactg taagaaatgg
agacagcctt taagccatat ggctttagcg ataacatttc 240gtgttgttct ggtatagaga
ttgagacaat tctaccacca accttcatag cctttggcat 300aatgttgaag tcaatgtcgg
taagggagga agcacagact acaatcaggt cgaaggtgtc 360aaagtacttt tcaccccaat
caccttcttc taatgtagca atgtagtgat cggcgcccat 420cttcattgca tcttctcttt
ttctcgaaga acgagaaata acatacgtct ctgcccccat 480ggctttggaa atcaatgtac
ccatactgcc gataccacca agaccaacta taccaacttt 540tttacctgga ccgcaaccgt
tacgaaccaa tggagagtac acagtcaaac caccacataa 600tagtggagca gccaaatgtg
atggaatatt ctctgggata ggcaccacaa aatgttcatg 660aactctgacg tagtttgcat
agccaccctg cgacacatag ccgtcttcat aaggctgact 720gtatgtggta acaaacttgg
tgcagtatgg ttcattatca ttcttacaac ggtcacattc 780caagcatgaa aagacttgag
cacctacacc aacacgttga ccgactttca acccactgtt 840tgacttgggc cctagcttga
caactttacc aacgatttca tgaccaacga ctagcggcat 900cttcatattg ccccaatgac
cagctgcaca atgaatatca ctaccgcaga caccacatgc 960ttcgatctta atgtcaatgt
catgatcgta aaatggtttt gggtcatact ttgtcttctt 1020tgggtttttc caatcttcgt
gtgattgaat agcgatacct tcaaatttct caggataaga 1080cat
108339387PRTEscherichia coli
39Met Asn Asn Phe Asn Leu His Thr Pro Thr Arg Ile Leu Phe Gly Lys 1
5 10 15 Gly Ala Ile Ala
Gly Leu Arg Glu Gln Ile Pro His Asp Ala Arg Val 20
25 30 Leu Ile Thr Tyr Gly Gly Gly Ser Val
Lys Lys Thr Gly Val Leu Asp 35 40
45 Gln Val Leu Asp Ala Leu Lys Gly Met Asp Val Leu Glu Phe
Gly Gly 50 55 60
Ile Glu Pro Asn Pro Ala Tyr Glu Thr Leu Met Asn Ala Val Lys Leu 65
70 75 80 Val Arg Glu Gln Lys
Val Thr Phe Leu Leu Ala Val Gly Gly Gly Ser 85
90 95 Val Leu Asp Gly Thr Lys Phe Ile Ala Ala
Ala Ala Asn Tyr Pro Glu 100 105
110 Asn Ile Asp Pro Trp His Ile Leu Gln Thr Gly Gly Lys Glu Ile
Lys 115 120 125 Ser
Ala Ile Pro Met Gly Cys Val Leu Thr Leu Pro Ala Thr Gly Ser 130
135 140 Glu Ser Asn Ala Gly Ala
Val Ile Ser Arg Lys Thr Thr Gly Asp Lys 145 150
155 160 Gln Ala Phe His Ser Ala His Val Gln Pro Val
Phe Ala Val Leu Asp 165 170
175 Pro Val Tyr Thr Tyr Thr Leu Pro Pro Arg Gln Val Ala Asn Gly Val
180 185 190 Val Asp
Ala Phe Val His Thr Val Glu Gln Tyr Val Thr Lys Pro Val 195
200 205 Asp Ala Lys Ile Gln Asp Arg
Phe Ala Glu Gly Ile Leu Leu Thr Leu 210 215
220 Ile Glu Asp Gly Pro Lys Ala Leu Lys Glu Pro Glu
Asn Tyr Asp Val 225 230 235
240 Arg Ala Asn Val Met Trp Ala Ala Thr Gln Ala Leu Asn Gly Leu Ile
245 250 255 Gly Ala Gly
Val Pro Gln Asp Trp Ala Thr His Met Leu Gly His Glu 260
265 270 Leu Thr Ala Met His Gly Leu Asp
His Ala Gln Thr Leu Ala Ile Val 275 280
285 Leu Pro Ala Leu Trp Asn Glu Lys Arg Asp Thr Lys Arg
Ala Lys Leu 290 295 300
Leu Gln Tyr Ala Glu Arg Val Trp Asn Ile Thr Glu Gly Ser Asp Asp 305
310 315 320 Glu Arg Ile Asp
Ala Ala Ile Ala Ala Thr Arg Asn Phe Phe Glu Gln 325
330 335 Leu Gly Val Pro Thr His Leu Ser Asp
Tyr Gly Leu Asp Gly Ser Ser 340 345
350 Ile Pro Ala Leu Leu Lys Lys Leu Glu Glu His Gly Met Thr
Gln Leu 355 360 365
Gly Glu Asn His Asp Ile Thr Leu Asp Val Ser Arg Arg Ile Tyr Glu 370
375 380 Ala Ala Arg 385
40387PRTEscherichia coli 40Met Asn Asn Phe Asn Leu His Thr Pro Thr
Arg Ile Leu Phe Gly Lys 1 5 10
15 Gly Ala Ile Ala Gly Leu Arg Glu Gln Ile Pro His Asp Ala Arg
Val 20 25 30 Leu
Ile Thr Tyr Gly Gly Gly Ser Val Lys Lys Thr Gly Val Leu Asp 35
40 45 Gln Val Leu Asp Ala Leu
Lys Gly Met Asp Val Leu Glu Phe Gly Gly 50 55
60 Ile Glu Pro Asn Pro Ala Tyr Glu Thr Leu Met
Asn Ala Val Lys Leu 65 70 75
80 Val Arg Glu Gln Lys Val Thr Phe Leu Leu Ala Val Gly Gly Gly Ser
85 90 95 Val Leu
Asp Gly Thr Lys Phe Ile Ala Ala Ala Ala Asn Tyr Pro Glu 100
105 110 Asn Ile Asp Pro Trp His Ile
Leu Gln Thr Gly Gly Lys Glu Ile Lys 115 120
125 Ser Ala Ile Pro Met Gly Cys Val Leu Thr Leu Pro
Ala Thr Gly Ser 130 135 140
Glu Ser Asn Ala Gly Ala Val Ile Ser Arg Lys Thr Thr Gly Asp Lys 145
150 155 160 Gln Ala Phe
His Ser Ala His Val Gln Pro Val Phe Ala Val Leu Asp 165
170 175 Pro Val Tyr Thr Tyr Thr Leu Pro
Pro Arg Gln Val Ala Asn Gly Val 180 185
190 Val Asp Ala Phe Val His Thr Val Glu Gln Tyr Val Thr
Lys Pro Val 195 200 205
Asp Ala Lys Ile Gln Asp Arg Phe Ala Glu Gly Ile Leu Leu Thr Leu 210
215 220 Ile Glu Asp Gly
Pro Lys Ala Leu Lys Glu Pro Glu Asn Tyr Asp Val 225 230
235 240 Arg Ala Asn Val Met Trp Ala Ala Thr
Gln Ala Leu Asn Gly Leu Ile 245 250
255 Gly Ala Gly Val Pro Gln Asp Trp Ala Thr His Met Leu Gly
His Glu 260 265 270
Leu Thr Ala Met His Gly Leu Asp His Ala Gln Thr Leu Ala Ile Val
275 280 285 Leu Pro Ala Leu
Trp Asn Glu Lys Arg Asp Thr Lys Arg Ala Lys Leu 290
295 300 Leu Gln Tyr Ala Glu Arg Val Trp
Asn Ile Thr Glu Gly Ser Asp Asp 305 310
315 320 Glu Arg Ile Asp Ala Ala Ile Ala Ala Thr Arg Asn
Phe Phe Glu Gln 325 330
335 Leu Gly Val Pro Thr His Leu Ser Asp Tyr Gly Leu Asp Gly Ser Ser
340 345 350 Ile Pro Ala
Leu Leu Lys Lys Leu Glu Glu His Gly Met Thr Gln Leu 355
360 365 Gly Glu Asn His Asp Ile Thr Leu
Asp Val Ser Arg Arg Ile Tyr Glu 370 375
380 Ala Ala Arg 385 41389PRTClostridium
acetobutylicum 41Met Leu Ser Phe Asp Tyr Ser Ile Pro Thr Lys Val Phe Phe
Gly Lys 1 5 10 15
Gly Lys Ile Asp Val Ile Gly Glu Glu Ile Lys Lys Tyr Gly Ser Arg
20 25 30 Val Leu Ile Val Tyr
Gly Gly Gly Ser Ile Lys Arg Asn Gly Ile Tyr 35
40 45 Asp Arg Ala Thr Ala Ile Leu Lys Glu
Asn Asn Ile Ala Phe Tyr Glu 50 55
60 Leu Ser Gly Val Glu Pro Asn Pro Arg Ile Thr Thr Val
Lys Lys Gly 65 70 75
80 Ile Glu Ile Cys Arg Glu Asn Asn Val Asp Leu Val Leu Ala Ile Gly
85 90 95 Gly Gly Ser Ala
Ile Asp Cys Ser Lys Val Ile Ala Ala Gly Val Tyr 100
105 110 Tyr Asp Gly Asp Thr Trp Asp Met Val
Lys Asp Pro Ser Lys Ile Thr 115 120
125 Lys Val Leu Pro Ile Ala Ser Ile Leu Thr Leu Ser Ala Thr
Gly Ser 130 135 140
Glu Met Asp Gln Ile Ala Val Ile Ser Asn Met Glu Thr Asn Glu Lys 145
150 155 160 Leu Gly Val Gly His
Asp Asp Met Arg Pro Lys Phe Ser Val Leu Asp 165
170 175 Pro Thr Tyr Thr Phe Thr Val Pro Lys Asn
Gln Thr Ala Ala Gly Thr 180 185
190 Ala Asp Ile Met Ser His Thr Phe Glu Ser Tyr Phe Ser Gly Val
Glu 195 200 205 Gly
Ala Tyr Val Gln Asp Gly Ile Ala Glu Ala Ile Leu Arg Thr Cys 210
215 220 Ile Lys Tyr Gly Lys Ile
Ala Met Glu Lys Thr Asp Asp Tyr Glu Ala 225 230
235 240 Arg Ala Asn Leu Met Trp Ala Ser Ser Leu Ala
Ile Asn Gly Leu Leu 245 250
255 Ser Leu Gly Lys Asp Arg Lys Trp Ser Cys His Pro Met Glu His Glu
260 265 270 Leu Ser
Ala Tyr Tyr Asp Ile Thr His Gly Val Gly Leu Ala Ile Leu 275
280 285 Thr Pro Asn Trp Met Glu Tyr
Ile Leu Asn Asp Asp Thr Leu His Lys 290 295
300 Phe Val Ser Tyr Gly Ile Asn Val Trp Gly Ile Asp
Lys Asn Lys Asp 305 310 315
320 Asn Tyr Glu Ile Ala Arg Glu Ala Ile Lys Asn Thr Arg Glu Tyr Phe
325 330 335 Asn Ser Leu
Gly Ile Pro Ser Lys Leu Arg Glu Val Gly Ile Gly Lys 340
345 350 Asp Lys Leu Glu Leu Met Ala Lys
Gln Ala Val Arg Asn Ser Gly Gly 355 360
365 Thr Ile Gly Ser Leu Arg Pro Ile Asn Ala Glu Asp Val
Leu Glu Ile 370 375 380
Phe Lys Lys Ser Tyr 385 421170DNAClostridium
acetobutylicum 42ttaataagat tttttaaata tctcaagaac atcctctgca tttattggtc
ttaaacttcc 60tattgttcct ccagaatttc taacagcttg ctttgccatt agttctagtt
tatcttttcc 120tattccaact tctctaagct ttgaaggaat acccaatgaa ttaaagtatt
ctctcgtatt 180tttaatagcc tctcgtgcta tttcatagtt atctttgttc ttgtctattc
cccaaacatt 240tattccataa gaaacaaatt tatgaagtgt atcgtcattt agaatatatt
ccatccaatt 300aggtgttaaa attgcaagtc ctacaccatg tgttatatca taatatgcac
ttaactcgtg 360ttccatagga tgacaactcc attttctatc cttaccaagt gataatagac
catttatagc 420taaacttgaa gcccacatca aattagctct agcctcgtaa tcatcagtct
tctccattgc 480tatttttcca tactttatac atgttcttaa gattgcttct gctataccgt
cctgcacata 540agcaccttca acaccactaa agtaagattc aaaggtgtga ctcataatgt
cagctgttcc 600cgctgctgtt tgatttttag gtactgtaaa agtatatgta ggatctaaca
ctgaaaattt 660aggtctcata tcatcatgtc ctactccaag cttttcatta gtctccatat
ttgaaattac 720tgcaatttga tccatttcag accctgttgc tgaaagagta agtatacttg
caattggaag 780aactttagtt attttagatg gatctttaac catgtcccat gtatcgccat
cataataaac 840tccagctgca attaccttag aacagtctat tgcacttcct ccccctattg
ctaatactaa 900atccacatta ttttctctac atatttctat gccttttttt actgttgtta
tcctaggatt 960tggctctact cctgaaagtt catagaaagc tatattgttt tcttttaata
tagctgttgc 1020tctatcatat ataccgttcc tttttatact tcctccgcca taaactataa
gcactcttga 1080gccatatttc ttaatttctt ctccaattac gtctattttt ccttttccaa
aaaaaacttt 1140agttggtatt gaataatcaa aacttagcat
117043390PRTClostridium acetobutylicum 43Met Val Asp Phe Glu
Tyr Ser Ile Pro Thr Arg Ile Phe Phe Gly Lys 1 5
10 15 Asp Lys Ile Asn Val Leu Gly Arg Glu Leu
Lys Lys Tyr Gly Ser Lys 20 25
30 Val Leu Ile Val Tyr Gly Gly Gly Ser Ile Lys Arg Asn Gly Ile
Tyr 35 40 45 Asp
Lys Ala Val Ser Ile Leu Glu Lys Asn Ser Ile Lys Phe Tyr Glu 50
55 60 Leu Ala Gly Val Glu Pro
Asn Pro Arg Val Thr Thr Val Glu Lys Gly 65 70
75 80 Val Lys Ile Cys Arg Glu Asn Gly Val Glu Val
Val Leu Ala Ile Gly 85 90
95 Gly Gly Ser Ala Ile Asp Cys Ala Lys Val Ile Ala Ala Ala Cys Glu
100 105 110 Tyr Asp
Gly Asn Pro Trp Asp Ile Val Leu Asp Gly Ser Lys Ile Lys 115
120 125 Arg Val Leu Pro Ile Ala Ser
Ile Leu Thr Ile Ala Ala Thr Gly Ser 130 135
140 Glu Met Asp Thr Trp Ala Val Ile Asn Asn Met Asp
Thr Asn Glu Lys 145 150 155
160 Leu Ile Ala Ala His Pro Asp Met Ala Pro Lys Phe Ser Ile Leu Asp
165 170 175 Pro Thr Tyr
Thr Tyr Thr Val Pro Thr Asn Gln Thr Ala Ala Gly Thr 180
185 190 Ala Asp Ile Met Ser His Ile Phe
Glu Val Tyr Phe Ser Asn Thr Lys 195 200
205 Thr Ala Tyr Leu Gln Asp Arg Met Ala Glu Ala Leu Leu
Arg Thr Cys 210 215 220
Ile Lys Tyr Gly Gly Ile Ala Leu Glu Lys Pro Asp Asp Tyr Glu Ala 225
230 235 240 Arg Ala Asn Leu
Met Trp Ala Ser Ser Leu Ala Ile Asn Gly Leu Leu 245
250 255 Thr Tyr Gly Lys Asp Thr Asn Trp Ser
Val His Leu Met Glu His Glu 260 265
270 Leu Ser Ala Tyr Tyr Asp Ile Thr His Gly Val Gly Leu Ala
Ile Leu 275 280 285
Thr Pro Asn Trp Met Glu Tyr Ile Leu Asn Asn Asp Thr Val Tyr Lys 290
295 300 Phe Val Glu Tyr Gly
Val Asn Val Trp Gly Ile Asp Lys Glu Lys Asn 305 310
315 320 His Tyr Asp Ile Ala His Gln Ala Ile Gln
Lys Thr Arg Asp Tyr Phe 325 330
335 Val Asn Val Leu Gly Leu Pro Ser Arg Leu Arg Asp Val Gly Ile
Glu 340 345 350 Glu
Glu Lys Leu Asp Ile Met Ala Lys Glu Ser Val Lys Leu Thr Gly 355
360 365 Gly Thr Ile Gly Asn Leu
Arg Pro Val Asn Ala Ser Glu Val Leu Gln 370 375
380 Ile Phe Lys Lys Ser Val 385
390 441173DNAClostridium acetobutylicum 44gtggttgatt tcgaatattc
aataccaact agaatttttt tcggtaaaga taagataaat 60gtacttggaa gagagcttaa
aaaatatggt tctaaagtgc ttatagttta tggtggagga 120agtataaaga gaaatggaat
atatgataaa gctgtaagta tacttgaaaa aaacagtatt 180aaattttatg aacttgcagg
agtagagcca aatccaagag taactacagt tgaaaaagga 240gttaaaatat gtagagaaaa
tggagttgaa gtagtactag ctataggtgg aggaagtgca 300atagattgcg caaaggttat
agcagcagca tgtgaatatg atggaaatcc atgggatatt 360gtgttagatg gctcaaaaat
aaaaagggtg cttcctatag ctagtatatt aaccattgct 420gcaacaggat cagaaatgga
tacgtgggca gtaataaata atatggatac aaacgaaaaa 480ctaattgcgg cacatccaga
tatggctcct aagttttcta tattagatcc aacgtatacg 540tataccgtac ctaccaatca
aacagcagca ggaacagctg atattatgag tcatatattt 600gaggtgtatt ttagtaatac
aaaaacagca tatttgcagg atagaatggc agaagcgtta 660ttaagaactt gtattaaata
tggaggaata gctcttgaga agccggatga ttatgaggca 720agagccaatc taatgtgggc
ttcaagtctt gcgataaatg gacttttaac atatggtaaa 780gacactaatt ggagtgtaca
cttaatggaa catgaattaa gtgcttatta cgacataaca 840cacggcgtag ggcttgcaat
tttaacacct aattggatgg agtatatttt aaataatgat 900acagtgtaca agtttgttga
atatggtgta aatgtttggg gaatagacaa agaaaaaaat 960cactatgaca tagcacatca
agcaatacaa aaaacaagag attactttgt aaatgtacta 1020ggtttaccat ctagactgag
agatgttgga attgaagaag aaaaattgga cataatggca 1080aaggaatcag taaagcttac
aggaggaacc ataggaaacc taagaccagt aaacgcctcc 1140gaagtcctac aaatattcaa
aaaatctgtg taa 117345330PRTBacillus
subtilis 45Met Ser Thr Asn Arg His Gln Ala Leu Gly Leu Thr Asp Gln Glu
Ala 1 5 10 15 Val
Asp Met Tyr Arg Thr Met Leu Leu Ala Arg Lys Ile Asp Glu Arg
20 25 30 Met Trp Leu Leu Asn
Arg Ser Gly Lys Ile Pro Phe Val Ile Ser Cys 35
40 45 Gln Gly Gln Glu Ala Ala Gln Val Gly
Ala Ala Phe Ala Leu Asp Arg 50 55
60 Glu Met Asp Tyr Val Leu Pro Tyr Tyr Arg Asp Met Gly
Val Val Leu 65 70 75
80 Ala Phe Gly Met Thr Ala Lys Asp Leu Met Met Ser Gly Phe Ala Lys
85 90 95 Ala Ala Asp Pro
Asn Ser Gly Gly Arg Gln Met Pro Gly His Phe Gly 100
105 110 Gln Lys Lys Asn Arg Ile Val Thr Gly
Ser Ser Pro Val Thr Thr Gln 115 120
125 Val Pro His Ala Val Gly Ile Ala Leu Ala Gly Arg Met Glu
Lys Lys 130 135 140
Asp Ile Ala Ala Phe Val Thr Phe Gly Glu Gly Ser Ser Asn Gln Gly 145
150 155 160 Asp Phe His Glu Gly
Ala Asn Phe Ala Ala Val His Lys Leu Pro Val 165
170 175 Ile Phe Met Cys Glu Asn Asn Lys Tyr Ala
Ile Ser Val Pro Tyr Asp 180 185
190 Lys Gln Val Ala Cys Glu Asn Ile Ser Asp Arg Ala Ile Gly Tyr
Gly 195 200 205 Met
Pro Gly Val Thr Val Asn Gly Asn Asp Pro Leu Glu Val Tyr Gln 210
215 220 Ala Val Lys Glu Ala Arg
Glu Arg Ala Arg Arg Gly Glu Gly Pro Thr 225 230
235 240 Leu Ile Glu Thr Ile Ser Tyr Arg Leu Thr Pro
His Ser Ser Asp Asp 245 250
255 Asp Asp Ser Ser Tyr Arg Gly Arg Glu Glu Val Glu Glu Ala Lys Lys
260 265 270 Ser Asp
Pro Leu Leu Thr Tyr Gln Ala Tyr Leu Lys Glu Thr Gly Leu 275
280 285 Leu Ser Asp Glu Ile Glu Gln
Thr Met Leu Asp Glu Ile Met Ala Ile 290 295
300 Val Asn Glu Ala Thr Asp Glu Ala Glu Asn Ala Pro
Tyr Ala Ala Pro 305 310 315
320 Glu Ser Ala Leu Asp Tyr Val Tyr Ala Lys 325
330 46993DNABacillus subtilis 46atgagtacaa accgacatca agcactaggg
ctgactgatc aggaagccgt tgatatgtat 60agaaccatgc tgttagcaag aaaaatcgat
gaaagaatgt ggctgttaaa ccgttctggc 120aaaattccat ttgtaatctc ttgtcaagga
caggaagcag cacaggtagg agcggctttc 180gcacttgacc gtgaaatgga ttatgtattg
ccgtactaca gagacatggg tgtcgtgctc 240gcgtttggca tgacagcaaa ggacttaatg
atgtccgggt ttgcaaaagc agcagatccg 300aactcaggag gccgccagat gccgggacat
ttcggacaaa agaaaaaccg cattgtgacg 360ggatcatctc cggttacaac gcaagtgccg
cacgcagtcg gtattgcgct tgcgggacgt 420atggagaaaa aggatatcgc agcctttgtt
acattcgggg aagggtcttc aaaccaaggc 480gatttccatg aaggggcaaa ctttgccgct
gtccataagc tgccggttat tttcatgtgt 540gaaaacaaca aatacgcaat ctcagtgcct
tacgataagc aagtcgcatg tgagaacatt 600tccgaccgtg ccataggcta tgggatgcct
ggcgtaactg tgaatggaaa tgatccgctg 660gaagtttatc aagcggttaa agaagcacgc
gaaagggcac gcagaggaga aggcccgaca 720ttaattgaaa cgatttctta ccgccttaca
ccacattcca gtgatgacga tgacagcagc 780tacagaggcc gtgaagaagt agaggaagcg
aaaaaaagtg atcccctgct tacttatcaa 840gcttacttaa aggaaacagg cctgctgtcc
gatgagatag aacaaaccat gctggatgaa 900attatggcaa tcgtaaatga agcgacggat
gaagcggaga acgccccata tgcagctcct 960gagtcagcgc ttgattatgt ttatgcgaag
tag 99347327PRTBacillus subtilis 47Met
Ser Val Met Ser Tyr Ile Asp Ala Ile Asn Leu Ala Met Lys Glu 1
5 10 15 Glu Met Glu Arg Asp Ser
Arg Val Phe Val Leu Gly Glu Asp Val Gly 20
25 30 Arg Lys Gly Gly Val Phe Lys Ala Thr Ala
Gly Leu Tyr Glu Gln Phe 35 40
45 Gly Glu Glu Arg Val Met Asp Thr Pro Leu Ala Glu Ser Ala
Ile Ala 50 55 60
Gly Val Gly Ile Gly Ala Ala Met Tyr Gly Met Arg Pro Ile Ala Glu 65
70 75 80 Met Gln Phe Ala Asp
Phe Ile Met Pro Ala Val Asn Gln Ile Ile Ser 85
90 95 Glu Ala Ala Lys Ile Arg Tyr Arg Ser Asn
Asn Asp Trp Ser Cys Pro 100 105
110 Ile Val Val Arg Ala Pro Tyr Gly Gly Gly Val His Gly Ala Leu
Tyr 115 120 125 His
Ser Gln Ser Val Glu Ala Ile Phe Ala Asn Gln Pro Gly Leu Lys 130
135 140 Ile Val Met Pro Ser Thr
Pro Tyr Asp Ala Lys Gly Leu Leu Lys Ala 145 150
155 160 Ala Val Arg Asp Glu Asp Pro Val Leu Phe Phe
Glu His Lys Arg Ala 165 170
175 Tyr Arg Leu Ile Lys Gly Glu Val Pro Ala Asp Asp Tyr Val Leu Pro
180 185 190 Ile Gly
Lys Ala Asp Val Lys Arg Glu Gly Asp Asp Ile Thr Val Ile 195
200 205 Thr Tyr Gly Leu Cys Val His
Phe Ala Leu Gln Ala Ala Glu Arg Leu 210 215
220 Glu Lys Asp Gly Ile Ser Ala His Val Val Asp Leu
Arg Thr Val Tyr 225 230 235
240 Pro Leu Asp Lys Glu Ala Ile Ile Glu Ala Ala Ser Lys Thr Gly Lys
245 250 255 Val Leu Leu
Val Thr Glu Asp Thr Lys Glu Gly Ser Ile Met Ser Glu 260
265 270 Val Ala Ala Ile Ile Ser Glu His
Cys Leu Phe Asp Leu Asp Ala Pro 275 280
285 Ile Lys Arg Leu Ala Gly Pro Asp Ile Pro Ala Met Pro
Tyr Ala Pro 290 295 300
Thr Met Glu Lys Tyr Phe Met Val Asn Pro Asp Lys Val Glu Ala Ala 305
310 315 320 Met Arg Glu Leu
Ala Glu Phe 325 48984DNABacillus subtilis
48atgtcagtaa tgtcatatat tgatgcaatc aatttggcga tgaaagaaga aatggaacga
60gattctcgcg ttttcgtcct tggggaagat gtaggaagaa aaggcggtgt gtttaaagcg
120acagcgggac tctatgaaca atttggggaa gagcgcgtta tggatacgcc gcttgctgaa
180tctgcaatcg caggagtcgg tatcggagcg gcaatgtacg gaatgagacc gattgctgaa
240atgcagtttg ctgatttcat tatgccggca gtcaaccaaa ttatttctga agcggctaaa
300atccgctacc gcagcaacaa tgactggagc tgtccgattg tcgtcagagc gccatacggc
360ggaggcgtgc acggagccct gtatcattct caatcagtcg aagcaatttt cgccaaccag
420cccggactga aaattgtcat gccatcaaca ccatatgacg cgaaagggct cttaaaagcc
480gcagttcgtg acgaagaccc cgtgctgttt tttgagcaca agcgggcata ccgtctgata
540aagggcgagg ttccggctga tgattatgtc ctgccaatcg gcaaggcgga cgtaaaaagg
600gaaggcgacg acatcacagt gatcacatac ggcctgtgtg tccacttcgc cttacaagct
660gcagaacgtc tcgaaaaaga tggcatttca gcgcatgtgg tggatttaag aacagtttac
720ccgcttgata aagaagccat catcgaagct gcgtccaaaa ctggaaaggt tcttttggtc
780acagaagata caaaagaagg cagcatcatg agcgaagtag ccgcaattat atccgagcat
840tgtctgttcg acttagacgc gccgatcaaa cggcttgcag gtcctgatat tccggctatg
900ccttatgcgc cgacaatgga aaaatacttt atggtcaacc ctgataaagt ggaagcggcg
960atgagagaat tagcggagtt ttaa
98449424PRTBacillus subtilis 49Met Ala Ile Glu Gln Met Thr Met Pro Gln
Leu Gly Glu Ser Val Thr 1 5 10
15 Glu Gly Thr Ile Ser Lys Trp Leu Val Ala Pro Gly Asp Lys Val
Asn 20 25 30 Lys
Tyr Asp Pro Ile Ala Glu Val Met Thr Asp Lys Val Asn Ala Glu 35
40 45 Val Pro Ser Ser Phe Thr
Gly Thr Ile Thr Glu Leu Val Gly Glu Glu 50 55
60 Gly Gln Thr Leu Gln Val Gly Glu Met Ile Cys
Lys Ile Glu Thr Glu 65 70 75
80 Gly Ala Asn Pro Ala Glu Gln Lys Gln Glu Gln Pro Ala Ala Ser Glu
85 90 95 Ala Ala
Glu Asn Pro Val Ala Lys Ser Ala Gly Ala Ala Asp Gln Pro 100
105 110 Asn Lys Lys Arg Tyr Ser Pro
Ala Val Leu Arg Leu Ala Gly Glu His 115 120
125 Gly Ile Asp Leu Asp Gln Val Thr Gly Thr Gly Ala
Gly Gly Arg Ile 130 135 140
Thr Arg Lys Asp Ile Gln Arg Leu Ile Glu Thr Gly Gly Val Gln Glu 145
150 155 160 Gln Asn Pro
Glu Glu Leu Lys Thr Ala Ala Pro Ala Pro Lys Ser Ala 165
170 175 Ser Lys Pro Glu Pro Lys Glu Glu
Thr Ser Tyr Pro Ala Ser Ala Ala 180 185
190 Gly Asp Lys Glu Ile Pro Val Thr Gly Val Arg Lys Ala
Ile Ala Ser 195 200 205
Asn Met Lys Arg Ser Lys Thr Glu Ile Pro His Ala Trp Thr Met Met 210
215 220 Glu Val Asp Val
Thr Asn Met Val Ala Tyr Arg Asn Ser Ile Lys Asp 225 230
235 240 Ser Phe Lys Lys Thr Glu Gly Phe Asn
Leu Thr Phe Phe Ala Phe Phe 245 250
255 Val Lys Ala Val Ala Gln Ala Leu Lys Glu Phe Pro Gln Met
Asn Ser 260 265 270
Met Trp Ala Gly Asp Lys Ile Ile Gln Lys Lys Asp Ile Asn Ile Ser
275 280 285 Ile Ala Val Ala
Thr Glu Asp Ser Leu Phe Val Pro Val Ile Lys Asn 290
295 300 Ala Asp Glu Lys Thr Ile Lys Gly
Ile Ala Lys Asp Ile Thr Gly Leu 305 310
315 320 Ala Lys Lys Val Arg Asp Gly Lys Leu Thr Ala Asp
Asp Met Gln Gly 325 330
335 Gly Thr Phe Thr Val Asn Asn Thr Gly Ser Phe Gly Ser Val Gln Ser
340 345 350 Met Gly Ile
Ile Asn Tyr Pro Gln Ala Ala Ile Leu Gln Val Glu Ser 355
360 365 Ile Val Lys Arg Pro Val Val Met
Asp Asn Gly Met Ile Ala Val Arg 370 375
380 Asp Met Val Asn Leu Cys Leu Ser Leu Asp His Arg Val
Leu Asp Gly 385 390 395
400 Leu Val Cys Gly Arg Phe Leu Gly Arg Val Lys Gln Ile Leu Glu Ser
405 410 415 Ile Asp Glu Lys
Thr Ser Val Tyr 420 501275DNABacillus
subtilis 50atggcaattg aacaaatgac gatgccgcag cttggagaaa gcgtaacaga
ggggacgatc 60agcaaatggc ttgtcgcccc cggtgataaa gtgaacaaat acgatccgat
cgcggaagtc 120atgacagata aggtaaatgc agaggttccg tcttctttta ctggtacgat
aacagagctt 180gtgggagaag aaggccaaac cctgcaagtc ggagaaatga tttgcaaaat
tgaaacagaa 240ggcgcgaatc cggctgaaca aaaacaagaa cagccagcag catcagaagc
cgctgagaac 300cctgttgcaa aaagtgctgg agcagccgat cagcccaata aaaagcgcta
ctcgccagct 360gttctccgtt tggccggaga gcacggcatt gacctcgatc aagtgacagg
aactggtgcc 420ggcgggcgca tcacacgaaa agatattcag cgcttaattg aaacaggcgg
cgtgcaagaa 480cagaatcctg aggagctgaa aacagcagct cctgcaccga agtctgcatc
aaaacctgag 540ccaaaagaag agacgtcata tcctgcgtct gcagccggtg ataaagaaat
ccctgtcaca 600ggtgtaagaa aagcaattgc ttccaatatg aagcgaagca aaacagaaat
tccgcatgct 660tggacgatga tggaagtcga cgtcacaaat atggttgcat atcgcaacag
tataaaagat 720tcttttaaga agacagaagg ctttaattta acgttcttcg ccttttttgt
aaaagcggtc 780gctcaggcgt taaaagaatt cccgcaaatg aatagcatgt gggcggggga
caaaattatt 840cagaaaaagg atatcaatat ttcaattgca gttgccacag aggattcttt
atttgttccg 900gtgattaaaa acgctgatga aaaaacaatt aaaggcattg cgaaagacat
taccggccta 960gctaaaaaag taagagacgg aaaactcact gcagatgaca tgcagggagg
cacgtttacc 1020gtcaacaaca caggttcgtt cgggtctgtt cagtcgatgg gcattatcaa
ctaccctcag 1080gctgcgattc ttcaagtaga atccatcgtc aaacgcccgg ttgtcatgga
caatggcatg 1140attgctgtca gagacatggt taatctgtgc ctgtcattag atcacagagt
gcttgacggt 1200ctcgtgtgcg gacgattcct cggacgagtg aaacaaattt tagaatcgat
tgacgagaag 1260acatctgttt actaa
127551474PRTBacillus subtilis 51Met Ala Thr Glu Tyr Asp Val
Val Ile Leu Gly Gly Gly Thr Gly Gly 1 5
10 15 Tyr Val Ala Ala Ile Arg Ala Ala Gln Leu Gly
Leu Lys Thr Ala Val 20 25
30 Val Glu Lys Glu Lys Leu Gly Gly Thr Cys Leu His Lys Gly Cys
Ile 35 40 45 Pro
Ser Lys Ala Leu Leu Arg Ser Ala Glu Val Tyr Arg Thr Ala Arg 50
55 60 Glu Ala Asp Gln Phe Gly
Val Glu Thr Ala Gly Val Ser Leu Asn Phe 65 70
75 80 Glu Lys Val Gln Gln Arg Lys Gln Ala Val Val
Asp Lys Leu Ala Ala 85 90
95 Gly Val Asn His Leu Met Lys Lys Gly Lys Ile Asp Val Tyr Thr Gly
100 105 110 Tyr Gly
Arg Ile Leu Gly Pro Ser Ile Phe Ser Pro Leu Pro Gly Thr 115
120 125 Ile Ser Val Glu Arg Gly Asn
Gly Glu Glu Asn Asp Met Leu Ile Pro 130 135
140 Lys Gln Val Ile Ile Ala Thr Gly Ser Arg Pro Arg
Met Leu Pro Gly 145 150 155
160 Leu Glu Val Asp Gly Lys Ser Val Leu Thr Ser Asp Glu Ala Leu Gln
165 170 175 Met Glu Glu
Leu Pro Gln Ser Ile Ile Ile Val Gly Gly Gly Val Ile 180
185 190 Gly Ile Glu Trp Ala Ser Met Leu
His Asp Phe Gly Val Lys Val Thr 195 200
205 Val Ile Glu Tyr Ala Asp Arg Ile Leu Pro Thr Glu Asp
Leu Glu Ile 210 215 220
Ser Lys Glu Met Glu Ser Leu Leu Lys Lys Lys Gly Ile Gln Phe Ile 225
230 235 240 Thr Gly Ala Lys
Val Leu Pro Asp Thr Met Thr Lys Thr Ser Asp Asp 245
250 255 Ile Ser Ile Gln Ala Glu Lys Asp Gly
Glu Thr Val Thr Tyr Ser Ala 260 265
270 Glu Lys Met Leu Val Ser Ile Gly Arg Gln Ala Asn Ile Glu
Gly Ile 275 280 285
Gly Leu Glu Asn Thr Asp Ile Val Thr Glu Asn Gly Met Ile Ser Val 290
295 300 Asn Glu Ser Cys Gln
Thr Lys Glu Ser His Ile Tyr Ala Ile Gly Asp 305 310
315 320 Val Ile Gly Gly Leu Gln Leu Ala His Val
Ala Ser His Glu Gly Ile 325 330
335 Ile Ala Val Glu His Phe Ala Gly Leu Asn Pro His Pro Leu Asp
Pro 340 345 350 Thr
Leu Val Pro Lys Cys Ile Tyr Ser Ser Pro Glu Ala Ala Ser Val 355
360 365 Gly Leu Thr Glu Asp Glu
Ala Lys Ala Asn Gly His Asn Val Lys Ile 370 375
380 Gly Lys Phe Pro Phe Met Ala Ile Gly Lys Ala
Leu Val Tyr Gly Glu 385 390 395
400 Ser Asp Gly Phe Val Lys Ile Val Ala Asp Arg Asp Thr Asp Asp Ile
405 410 415 Leu Gly
Val His Met Ile Gly Pro His Val Thr Asp Met Ile Ser Glu 420
425 430 Ala Gly Leu Ala Lys Val Leu
Asp Ala Thr Pro Trp Glu Val Gly Gln 435 440
445 Thr Ile His Pro His Pro Thr Leu Ser Glu Ala Ile
Gly Glu Ala Ala 450 455 460
Leu Ala Ala Asp Gly Lys Ala Ile His Phe 465 470
521425DNABacillus subtilis 52atggcaactg agtatgacgt agtcattctg
ggcggcggta ccggcggtta tgttgcggcc 60atcagagccg ctcagctcgg cttaaaaaca
gccgttgtgg aaaaggaaaa actcggggga 120acatgtctgc ataaaggctg tatcccgagt
aaagcgctgc ttagaagcgc agaggtatac 180cggacagctc gtgaagccga tcaattcgga
gtggaaacgg ctggcgtgtc cctcaacttt 240gaaaaagtgc agcagcgtaa gcaagccgtt
gttgataagc ttgcagcggg tgtaaatcat 300ttaatgaaaa aaggaaaaat tgacgtgtac
accggatatg gacgtatcct tggaccgtca 360atcttctctc cgctgccggg aacaatttct
gttgagcggg gaaatggcga agaaaatgac 420atgctgatcc cgaaacaagt gatcattgca
acaggatcaa gaccgagaat gcttccgggt 480cttgaagtgg acggtaagtc tgtactgact
tcagatgagg cgctccaaat ggaggagctg 540ccacagtcaa tcatcattgt cggcggaggg
gttatcggta tcgaatgggc gtctatgctt 600catgattttg gcgttaaggt aacggttatt
gaatacgcgg atcgcatatt gccgactgaa 660gatctagaga tttcaaaaga aatggaaagt
cttcttaaga aaaaaggcat ccagttcata 720acaggggcaa aagtgctgcc tgacacaatg
acaaaaacat cagacgatat cagcatacaa 780gcggaaaaag acggagaaac cgttacctat
tctgctgaga aaatgcttgt ttccatcggc 840agacaggcaa atatcgaagg catcggccta
gagaacaccg atattgttac tgaaaatggc 900atgatttcag tcaatgaaag ctgccaaacg
aaggaatctc atatttatgc aatcggagac 960gtaatcggtg gcctgcagtt agctcacgtt
gcttcacatg agggaattat tgctgttgag 1020cattttgcag gtctcaatcc gcatccgctt
gatccgacgc ttgtgccgaa gtgcatttac 1080tcaagccctg aagctgccag tgtcggctta
accgaagacg aagcaaaggc gaacgggcat 1140aatgtcaaaa tcggcaagtt cccatttatg
gcgattggaa aagcgcttgt atacggtgaa 1200agcgacggtt ttgtcaaaat cgtggctgac
cgagatacag atgatattct cggcgttcat 1260atgattggcc cgcatgtcac cgacatgatt
tctgaagcgg gtcttgccaa agtgctggac 1320gcaacaccgt gggaggtcgg gcaaacgatt
cacccgcatc caacgctttc tgaagcaatt 1380ggagaagctg cgcttgccgc agatggcaaa
gccattcatt tttaa 142553410PRTPseudomonas putida 53Met
Asn Glu Tyr Ala Pro Leu Arg Leu His Val Pro Glu Pro Thr Gly 1
5 10 15 Arg Pro Gly Cys Gln Thr
Asp Phe Ser Tyr Leu Arg Leu Asn Asp Ala 20
25 30 Gly Gln Ala Arg Lys Pro Pro Val Asp Val
Asp Ala Ala Asp Thr Ala 35 40
45 Asp Leu Ser Tyr Ser Leu Val Arg Val Leu Asp Glu Gln Gly
Asp Ala 50 55 60
Gln Gly Pro Trp Ala Glu Asp Ile Asp Pro Gln Ile Leu Arg Gln Gly 65
70 75 80 Met Arg Ala Met Leu
Lys Thr Arg Ile Phe Asp Ser Arg Met Val Val 85
90 95 Ala Gln Arg Gln Lys Lys Met Ser Phe Tyr
Met Gln Ser Leu Gly Glu 100 105
110 Glu Ala Ile Gly Ser Gly Gln Ala Leu Ala Leu Asn Arg Thr Asp
Met 115 120 125 Cys
Phe Pro Thr Tyr Arg Gln Gln Ser Ile Leu Met Ala Arg Asp Val 130
135 140 Ser Leu Val Glu Met Ile
Cys Gln Leu Leu Ser Asn Glu Arg Asp Pro 145 150
155 160 Leu Lys Gly Arg Gln Leu Pro Ile Met Tyr Ser
Val Arg Glu Ala Gly 165 170
175 Phe Phe Thr Ile Ser Gly Asn Leu Ala Thr Gln Phe Val Gln Ala Val
180 185 190 Gly Trp
Ala Met Ala Ser Ala Ile Lys Gly Asp Thr Lys Ile Ala Ser 195
200 205 Ala Trp Ile Gly Asp Gly Ala
Thr Ala Glu Ser Asp Phe His Thr Ala 210 215
220 Leu Thr Phe Ala His Val Tyr Arg Ala Pro Val Ile
Leu Asn Val Val 225 230 235
240 Asn Asn Gln Trp Ala Ile Ser Thr Phe Gln Ala Ile Ala Gly Gly Glu
245 250 255 Ser Thr Thr
Phe Ala Gly Arg Gly Val Gly Cys Gly Ile Ala Ser Leu 260
265 270 Arg Val Asp Gly Asn Asp Phe Val
Ala Val Tyr Ala Ala Ser Arg Trp 275 280
285 Ala Ala Glu Arg Ala Arg Arg Gly Leu Gly Pro Ser Leu
Ile Glu Trp 290 295 300
Val Thr Tyr Arg Ala Gly Pro His Ser Thr Ser Asp Asp Pro Ser Lys 305
310 315 320 Tyr Arg Pro Ala
Asp Asp Trp Ser His Phe Pro Leu Gly Asp Pro Ile 325
330 335 Ala Arg Leu Lys Gln His Leu Ile Lys
Ile Gly His Trp Ser Glu Glu 340 345
350 Glu His Gln Ala Thr Thr Ala Glu Phe Glu Ala Ala Val Ile
Ala Ala 355 360 365
Gln Lys Glu Ala Glu Gln Tyr Gly Thr Leu Ala Asn Gly His Ile Pro 370
375 380 Ser Ala Ala Ser Met
Phe Glu Asp Val Tyr Lys Glu Met Pro Asp His 385 390
395 400 Leu Arg Arg Gln Arg Gln Glu Leu Gly Val
405 410 546643DNAPseudomonas putida
54gcatgcctgc aggccgccga tgaaatggtg gaaggtatcg gtaggctggc cctgctcatc
60gctgaacacg ttacgcccgc tgccggtatc gaccaggctc tggtgaatat gcatggaact
120gccaggcgtg cgcgccagcg gtttggccat gcacaccacg gtcagcccgt gcttgagtgc
180cacttccttg agcaggtgtt tgaacaggaa ggtctggtcg gccagcagca gcgggtcgcc
240atgtagcaag ttgatctcga actggctgac gcccatttcg tgcatgaagg tgtcgcgcgg
300caggccgagc gcggccatgc actggtacac ctcattgaag aacgggcgca ggccgttgtt
360ggaactgaca ctgaacgccg aatggcccag ctcgcggcgg ccgtcggtgc ccagcggtgg
420ctggaacggc tgctgcgggt cactgttggg ggcaaacacg aagaactcaa gctcggtcgc
480cactaccggt gccagaccca acgctgcgta gcgggcgatc acggccttca gctggccccg
540ggtggacagt gccgagggcc ggccatccag ttcattggca tcgcagatgg ccagggcgcg
600accgtcatcg ctccagggca agcgatgaac ctggctgggt tccgctacca acgccaggtc
660gccgtcgtcg cagccgtaga atttcgccgg cgggtagccg cccatgatgc attgcagcag
720caccccacgg gccatctgca ggcggcggcc ttcgagaaag ccttcggcgg tcatcacctt
780gccgcgtggg acgccgttga ggtcgggggt gacgcattcg atttcatcga tgccctggag
840ctgagcgatg ctcatgacgc ttgtccttgt tgttgtaggc tgacaacaac ataggctggg
900ggtgtttaaa atatcaagca gcctctcgaa cgcctggggc ctcttctatt cgcgcaaggt
960catgccattg gccggcaacg gcaaggctgt cttgtagcgc acctgtttca aggcaaaact
1020cgagcggata ttcgccacac ccggcaaccg ggtcaggtaa tcgagaaacc gctccagcgc
1080ctggatactc ggcagcagta cccgcaacag gtagtccggg tcgcccgtca tcaggtagca
1140ctccatcacc tcgggccgtt cggcaatttc ttcctcgaag cggtgcagcg actgctctac
1200ctgtttttcc aggctgacat ggatgaacac attcacatcc agccccaacg cctcgggcga
1260caacaaggtc acctgctggc ggatcacccc cagttcttcc atggcccgca cccggttgaa
1320acagggcgtg ggcgacaggt tgaccgagcg tgccagctcg gcgttggtga tgcgggcgtt
1380ttcctgcagg ctgttgagaa tgccgatatc ggtacgatcg agtttgcgca tgagacaaaa
1440tcaccggttt tttgtgttta tgcggaatgt ttatctgccc cgctcggcaa aggcaatcaa
1500cttgagagaa aaattctcct gccggaccac taagatgtag gggacgctga cttaccagtc
1560acaagccggt actcagcggc ggccgcttca gagctcacaa aaacaaatac ccgagcgagc
1620gtaaaaagca tgaacgagta cgcccccctg cgtttgcatg tgcccgagcc caccggccgg
1680ccaggctgcc agaccgattt ttcctacctg cgcctgaacg atgcaggtca agcccgtaaa
1740ccccctgtcg atgtcgacgc tgccgacacc gccgacctgt cctacagcct ggtccgcgtg
1800ctcgacgagc aaggcgacgc ccaaggcccg tgggctgaag acatcgaccc gcagatcctg
1860cgccaaggca tgcgcgccat gctcaagacg cggatcttcg acagccgcat ggtggttgcc
1920cagcgccaga agaagatgtc cttctacatg cagagcctgg gcgaagaagc catcggcagc
1980ggccaggcgc tggcgcttaa ccgcaccgac atgtgcttcc ccacctaccg tcagcaaagc
2040atcctgatgg cccgcgacgt gtcgctggtg gagatgatct gccagttgct gtccaacgaa
2100cgcgaccccc tcaagggccg ccagctgccg atcatgtact cggtacgcga ggccggcttc
2160ttcaccatca gcggcaacct ggcgacccag ttcgtgcagg cggtcggctg ggccatggcc
2220tcggcgatca agggcgatac caagattgcc tcggcctgga tcggcgacgg cgccactgcc
2280gaatcggact tccacaccgc cctcaccttt gcccacgttt accgcgcccc ggtgatcctc
2340aacgtggtca acaaccagtg ggccatctca accttccagg ccatcgccgg tggcgagtcg
2400accaccttcg ccggccgtgg cgtgggctgc ggcatcgctt cgctgcgggt ggacggcaac
2460gacttcgtcg ccgtttacgc cgcttcgcgc tgggctgccg aacgtgcccg ccgtggtttg
2520ggcccgagcc tgatcgagtg ggtcacctac cgtgccggcc cgcactcgac ctcggacgac
2580ccgtccaagt accgccctgc cgatgactgg agccacttcc cgctgggtga cccgatcgcc
2640cgcctgaagc agcacctgat caagatcggc cactggtccg aagaagaaca ccaggccacc
2700acggccgagt tcgaagcggc cgtgattgct gcgcaaaaag aagccgagca gtacggcacc
2760ctggccaacg gtcacatccc gagcgccgcc tcgatgttcg aggacgtgta caaggagatg
2820cccgaccacc tgcgccgcca acgccaggaa ctgggggttt gagatgaacg accacaacaa
2880cagcatcaac ccggaaaccg ccatggccac cactaccatg accatgatcc aggccctgcg
2940ctcggccatg gatgtcatgc ttgagcgcga cgacaatgtg gtggtgtacg gccaggacgt
3000cggctacttc ggcggcgtgt tccgctgcac cgaaggcctg cagaccaagt acggcaagtc
3060ccgcgtgttc gacgcgccca tctctgaaag cggcatcgtc ggcaccgccg tgggcatggg
3120tgcctacggc ctgcgcccgg tggtggaaat ccagttcgct gactacttct acccggcctc
3180cgaccagatc gtttctgaaa tggcccgcct gcgctaccgt tcggccggcg agttcatcgc
3240cccgctgacc ctgcgtatgc cctgcggtgg cggtatctat ggcggccaga cacacagcca
3300gagcccggaa gcgatgttca ctcaggtgtg cggcctgcgc accgtaatgc catccaaccc
3360gtacgacgcc aaaggcctgc tgattgcctc gatcgaatgc gacgacccgg tgatcttcct
3420ggagcccaag cgcctgtaca acggcccgtt cgacggccac catgaccgcc cggttacgcc
3480gtggtcgaaa cacccgcaca gcgccgtgcc cgatggctac tacaccgtgc cactggacaa
3540ggccgccatc acccgccccg gcaatgacgt gagcgtgctc acctatggca ccaccgtgta
3600cgtggcccag gtggccgccg aagaaagtgg cgtggatgcc gaagtgatcg acctgcgcag
3660cctgtggccg ctagacctgg acaccatcgt cgagtcggtg aaaaagaccg gccgttgcgt
3720ggtagtacac gaggccaccc gtacttgtgg ctttggcgca gaactggtgt cgctggtgca
3780ggagcactgc ttccaccacc tggaggcgcc gatcgagcgc gtcaccggtt gggacacccc
3840ctaccctcac gcgcaggaat gggcttactt cccagggcct tcgcgggtag gtgcggcatt
3900gaaaaaggtc atggaggtct gaatgggcac gcacgtcatc aagatgccgg acattggcga
3960aggcatcgcg caggtcgaat tggtggaatg gttcgtcaag gtgggcgaca tcatcgccga
4020ggaccaagtg gtagccgacg tcatgaccga caaggccacc gtggaaatcc cgtcgccggt
4080cagcggcaag gtgctggccc tgggtggcca gccaggtgaa gtgatggcgg tcggcagtga
4140gctgatccgc atcgaagtgg aaggcagcgg caaccatgtg gatgtgccgc aagccaagcc
4200ggccgaagtg cctgcggcac cggtagccgc taaacctgaa ccacagaaag acgttaaacc
4260ggcggcgtac caggcgtcag ccagccacga ggcagcgccc atcgtgccgc gccagccggg
4320cgacaagccg ctggcctcgc cggcggtgcg caaacgcgcc ctcgatgccg gcatcgaatt
4380gcgttatgtg cacggcagcg gcccggccgg gcgcatcctg cacgaagacc tcgacgcgtt
4440catgagcaaa ccgcaaagcg ctgccgggca aacccccaat ggctatgcca ggcgcaccga
4500cagcgagcag gtgccggtga tcggcctgcg ccgcaagatc gcccagcgca tgcaggacgc
4560caagcgccgg gtcgcgcact tcagctatgt ggaagaaatc gacgtcaccg ccctggaagc
4620cctgcgccag cagctcaaca gcaagcacgg cgacagccgc ggcaagctga cactgctgcc
4680gttcctggtg cgcgccctgg tcgtggcact gcgtgacttc ccgcagataa acgccaccta
4740cgatgacgaa gcgcagatca tcacccgcca tggcgcggtg catgtgggca tcgccaccca
4800aggtgacaac ggcctgatgg tacccgtgct gcgccacgcc gaagcgggca gcctgtgggc
4860caatgccggt gagatttcac gcctggccaa cgctgcgcgc aacaacaagg ccagccgcga
4920agagctgtcc ggttcgacca ttaccctgac cagcctcggc gccctgggcg gcatcgtcag
4980cacgccggtg gtcaacaccc cggaagtggc gatcgtcggt gtcaaccgca tggttgagcg
5040gcccgtggtg atcgacggcc agatcgtcgt gcgcaagatg atgaacctgt ccagctcgtt
5100cgaccaccgc gtggtcgatg gcatggacgc cgccctgttc atccaggccg tgcgtggcct
5160gctcgaacaa cccgcctgcc tgttcgtgga gtgagcatgc aacagactat ccagacaacc
5220ctgttgatca tcggcggcgg ccctggcggc tatgtggcgg ccatccgcgc cgggcaactg
5280ggcatcccta ccgtgctggt ggaaggccag gcgctgggcg gtacctgcct gaacatcggc
5340tgcattccgt ccaaggcgct gatccatgtg gccgagcagt tccaccaggc ctcgcgcttt
5400accgaaccct cgccgctggg catcagcgtg gcttcgccac gcctggacat cggccagagc
5460gtggcctgga aagacggcat cgtcgatcgc ctgaccactg gtgtcgccgc cctgctgaaa
5520aagcacgggg tgaaggtggt gcacggctgg gccaaggtgc ttgatggcaa gcaggtcgag
5580gtggatggcc agcgcatcca gtgcgagcac ctgttgctgg ccacgggctc cagcagtgtc
5640gaactgccga tgctgccgtt gggtgggccg gtgatttcct cgaccgaggc cctggcaccg
5700aaagccctgc cgcaacacct ggtggtggtg ggcggtggct acatcggcct ggagctgggt
5760atcgcctacc gcaagctcgg cgcgcaggtc agcgtggtgg aagcgcgcga gcgcatcctg
5820ccgacttacg acagcgaact gaccgccccg gtggccgagt cgctgaaaaa gctgggtatc
5880gccctgcacc ttggccacag cgtcgaaggt tacgaaaatg gctgcctgct ggccaacgat
5940ggcaagggcg gacaactgcg cctggaagcc gaccgggtgc tggtggccgt gggccgccgc
6000ccacgcacca agggcttcaa cctggaatgc ctggacctga agatgaatgg tgccgcgatt
6060gccatcgacg agcgctgcca gaccagcatg cacaacgtct gggccatcgg cgacgtggcc
6120ggcgaaccga tgctggcgca ccgggccatg gcccagggcg agatggtggc cgagatcatc
6180gccggcaagg cacgccgctt cgaacccgct gcgatagccg ccgtgtgctt caccgacccg
6240gaagtggtcg tggtcggcaa gacgccggaa caggccagtc agcaaggcct ggactgcatc
6300gtcgcgcagt tcccgttcgc cgccaacggc cgggccatga gcctggagtc gaaaagcggt
6360ttcgtgcgcg tggtcgcgcg gcgtgacaac cacctgatcc tgggctggca agcggttggc
6420gtggcggttt ccgagctgtc cacggcgttt gcccagtcgc tggagatggg tgcctgcctg
6480gaggatgtgg ccggtaccat ccatgcccac ccgaccctgg gtgaagcggt acaggaagcg
6540gcactgcgtg ccctgggcca cgccctgcat atctgacact gaagcggccg aggccgattt
6600ggcccgccgc gccgagaggc gctgcgggtc ttttttatac ctg
664355352PRTPseudomonas putida 55Met Asn Asp His Asn Asn Ser Ile Asn Pro
Glu Thr Ala Met Ala Thr 1 5 10
15 Thr Thr Met Thr Met Ile Gln Ala Leu Arg Ser Ala Met Asp Val
Met 20 25 30 Leu
Glu Arg Asp Asp Asn Val Val Val Tyr Gly Gln Asp Val Gly Tyr 35
40 45 Phe Gly Gly Val Phe Arg
Cys Thr Glu Gly Leu Gln Thr Lys Tyr Gly 50 55
60 Lys Ser Arg Val Phe Asp Ala Pro Ile Ser Glu
Ser Gly Ile Val Gly 65 70 75
80 Thr Ala Val Gly Met Gly Ala Tyr Gly Leu Arg Pro Val Val Glu Ile
85 90 95 Gln Phe
Ala Asp Tyr Phe Tyr Pro Ala Ser Asp Gln Ile Val Ser Glu 100
105 110 Met Ala Arg Leu Arg Tyr Arg
Ser Ala Gly Glu Phe Ile Ala Pro Leu 115 120
125 Thr Leu Arg Met Pro Cys Gly Gly Gly Ile Tyr Gly
Gly Gln Thr His 130 135 140
Ser Gln Ser Pro Glu Ala Met Phe Thr Gln Val Cys Gly Leu Arg Thr 145
150 155 160 Val Met Pro
Ser Asn Pro Tyr Asp Ala Lys Gly Leu Leu Ile Ala Ser 165
170 175 Ile Glu Cys Asp Asp Pro Val Ile
Phe Leu Glu Pro Lys Arg Leu Tyr 180 185
190 Asn Gly Pro Phe Asp Gly His His Asp Arg Pro Val Thr
Pro Trp Ser 195 200 205
Lys His Pro His Ser Ala Val Pro Asp Gly Tyr Tyr Thr Val Pro Leu 210
215 220 Asp Lys Ala Ala
Ile Thr Arg Pro Gly Asn Asp Val Ser Val Leu Thr 225 230
235 240 Tyr Gly Thr Thr Val Tyr Val Ala Gln
Val Ala Ala Glu Glu Ser Gly 245 250
255 Val Asp Ala Glu Val Ile Asp Leu Arg Ser Leu Trp Pro Leu
Asp Leu 260 265 270
Asp Thr Ile Val Glu Ser Val Lys Lys Thr Gly Arg Cys Val Val Val
275 280 285 His Glu Ala Thr
Arg Thr Cys Gly Phe Gly Ala Glu Leu Val Ser Leu 290
295 300 Val Gln Glu His Cys Phe His His
Leu Glu Ala Pro Ile Glu Arg Val 305 310
315 320 Thr Gly Trp Asp Thr Pro Tyr Pro His Ala Gln Glu
Trp Ala Tyr Phe 325 330
335 Pro Gly Pro Ser Arg Val Gly Ala Ala Leu Lys Lys Val Met Glu Val
340 345 350 56
6643DNAPseudomonas putida 56gcatgcctgc aggccgccga tgaaatggtg gaaggtatcg
gtaggctggc cctgctcatc 60gctgaacacg ttacgcccgc tgccggtatc gaccaggctc
tggtgaatat gcatggaact 120gccaggcgtg cgcgccagcg gtttggccat gcacaccacg
gtcagcccgt gcttgagtgc 180cacttccttg agcaggtgtt tgaacaggaa ggtctggtcg
gccagcagca gcgggtcgcc 240atgtagcaag ttgatctcga actggctgac gcccatttcg
tgcatgaagg tgtcgcgcgg 300caggccgagc gcggccatgc actggtacac ctcattgaag
aacgggcgca ggccgttgtt 360ggaactgaca ctgaacgccg aatggcccag ctcgcggcgg
ccgtcggtgc ccagcggtgg 420ctggaacggc tgctgcgggt cactgttggg ggcaaacacg
aagaactcaa gctcggtcgc 480cactaccggt gccagaccca acgctgcgta gcgggcgatc
acggccttca gctggccccg 540ggtggacagt gccgagggcc ggccatccag ttcattggca
tcgcagatgg ccagggcgcg 600accgtcatcg ctccagggca agcgatgaac ctggctgggt
tccgctacca acgccaggtc 660gccgtcgtcg cagccgtaga atttcgccgg cgggtagccg
cccatgatgc attgcagcag 720caccccacgg gccatctgca ggcggcggcc ttcgagaaag
ccttcggcgg tcatcacctt 780gccgcgtggg acgccgttga ggtcgggggt gacgcattcg
atttcatcga tgccctggag 840ctgagcgatg ctcatgacgc ttgtccttgt tgttgtaggc
tgacaacaac ataggctggg 900ggtgtttaaa atatcaagca gcctctcgaa cgcctggggc
ctcttctatt cgcgcaaggt 960catgccattg gccggcaacg gcaaggctgt cttgtagcgc
acctgtttca aggcaaaact 1020cgagcggata ttcgccacac ccggcaaccg ggtcaggtaa
tcgagaaacc gctccagcgc 1080ctggatactc ggcagcagta cccgcaacag gtagtccggg
tcgcccgtca tcaggtagca 1140ctccatcacc tcgggccgtt cggcaatttc ttcctcgaag
cggtgcagcg actgctctac 1200ctgtttttcc aggctgacat ggatgaacac attcacatcc
agccccaacg cctcgggcga 1260caacaaggtc acctgctggc ggatcacccc cagttcttcc
atggcccgca cccggttgaa 1320acagggcgtg ggcgacaggt tgaccgagcg tgccagctcg
gcgttggtga tgcgggcgtt 1380ttcctgcagg ctgttgagaa tgccgatatc ggtacgatcg
agtttgcgca tgagacaaaa 1440tcaccggttt tttgtgttta tgcggaatgt ttatctgccc
cgctcggcaa aggcaatcaa 1500cttgagagaa aaattctcct gccggaccac taagatgtag
gggacgctga cttaccagtc 1560acaagccggt actcagcggc ggccgcttca gagctcacaa
aaacaaatac ccgagcgagc 1620gtaaaaagca tgaacgagta cgcccccctg cgtttgcatg
tgcccgagcc caccggccgg 1680ccaggctgcc agaccgattt ttcctacctg cgcctgaacg
atgcaggtca agcccgtaaa 1740ccccctgtcg atgtcgacgc tgccgacacc gccgacctgt
cctacagcct ggtccgcgtg 1800ctcgacgagc aaggcgacgc ccaaggcccg tgggctgaag
acatcgaccc gcagatcctg 1860cgccaaggca tgcgcgccat gctcaagacg cggatcttcg
acagccgcat ggtggttgcc 1920cagcgccaga agaagatgtc cttctacatg cagagcctgg
gcgaagaagc catcggcagc 1980ggccaggcgc tggcgcttaa ccgcaccgac atgtgcttcc
ccacctaccg tcagcaaagc 2040atcctgatgg cccgcgacgt gtcgctggtg gagatgatct
gccagttgct gtccaacgaa 2100cgcgaccccc tcaagggccg ccagctgccg atcatgtact
cggtacgcga ggccggcttc 2160ttcaccatca gcggcaacct ggcgacccag ttcgtgcagg
cggtcggctg ggccatggcc 2220tcggcgatca agggcgatac caagattgcc tcggcctgga
tcggcgacgg cgccactgcc 2280gaatcggact tccacaccgc cctcaccttt gcccacgttt
accgcgcccc ggtgatcctc 2340aacgtggtca acaaccagtg ggccatctca accttccagg
ccatcgccgg tggcgagtcg 2400accaccttcg ccggccgtgg cgtgggctgc ggcatcgctt
cgctgcgggt ggacggcaac 2460gacttcgtcg ccgtttacgc cgcttcgcgc tgggctgccg
aacgtgcccg ccgtggtttg 2520ggcccgagcc tgatcgagtg ggtcacctac cgtgccggcc
cgcactcgac ctcggacgac 2580ccgtccaagt accgccctgc cgatgactgg agccacttcc
cgctgggtga cccgatcgcc 2640cgcctgaagc agcacctgat caagatcggc cactggtccg
aagaagaaca ccaggccacc 2700acggccgagt tcgaagcggc cgtgattgct gcgcaaaaag
aagccgagca gtacggcacc 2760ctggccaacg gtcacatccc gagcgccgcc tcgatgttcg
aggacgtgta caaggagatg 2820cccgaccacc tgcgccgcca acgccaggaa ctgggggttt
gagatgaacg accacaacaa 2880cagcatcaac ccggaaaccg ccatggccac cactaccatg
accatgatcc aggccctgcg 2940ctcggccatg gatgtcatgc ttgagcgcga cgacaatgtg
gtggtgtacg gccaggacgt 3000cggctacttc ggcggcgtgt tccgctgcac cgaaggcctg
cagaccaagt acggcaagtc 3060ccgcgtgttc gacgcgccca tctctgaaag cggcatcgtc
ggcaccgccg tgggcatggg 3120tgcctacggc ctgcgcccgg tggtggaaat ccagttcgct
gactacttct acccggcctc 3180cgaccagatc gtttctgaaa tggcccgcct gcgctaccgt
tcggccggcg agttcatcgc 3240cccgctgacc ctgcgtatgc cctgcggtgg cggtatctat
ggcggccaga cacacagcca 3300gagcccggaa gcgatgttca ctcaggtgtg cggcctgcgc
accgtaatgc catccaaccc 3360gtacgacgcc aaaggcctgc tgattgcctc gatcgaatgc
gacgacccgg tgatcttcct 3420ggagcccaag cgcctgtaca acggcccgtt cgacggccac
catgaccgcc cggttacgcc 3480gtggtcgaaa cacccgcaca gcgccgtgcc cgatggctac
tacaccgtgc cactggacaa 3540ggccgccatc acccgccccg gcaatgacgt gagcgtgctc
acctatggca ccaccgtgta 3600cgtggcccag gtggccgccg aagaaagtgg cgtggatgcc
gaagtgatcg acctgcgcag 3660cctgtggccg ctagacctgg acaccatcgt cgagtcggtg
aaaaagaccg gccgttgcgt 3720ggtagtacac gaggccaccc gtacttgtgg ctttggcgca
gaactggtgt cgctggtgca 3780ggagcactgc ttccaccacc tggaggcgcc gatcgagcgc
gtcaccggtt gggacacccc 3840ctaccctcac gcgcaggaat gggcttactt cccagggcct
tcgcgggtag gtgcggcatt 3900gaaaaaggtc atggaggtct gaatgggcac gcacgtcatc
aagatgccgg acattggcga 3960aggcatcgcg caggtcgaat tggtggaatg gttcgtcaag
gtgggcgaca tcatcgccga 4020ggaccaagtg gtagccgacg tcatgaccga caaggccacc
gtggaaatcc cgtcgccggt 4080cagcggcaag gtgctggccc tgggtggcca gccaggtgaa
gtgatggcgg tcggcagtga 4140gctgatccgc atcgaagtgg aaggcagcgg caaccatgtg
gatgtgccgc aagccaagcc 4200ggccgaagtg cctgcggcac cggtagccgc taaacctgaa
ccacagaaag acgttaaacc 4260ggcggcgtac caggcgtcag ccagccacga ggcagcgccc
atcgtgccgc gccagccggg 4320cgacaagccg ctggcctcgc cggcggtgcg caaacgcgcc
ctcgatgccg gcatcgaatt 4380gcgttatgtg cacggcagcg gcccggccgg gcgcatcctg
cacgaagacc tcgacgcgtt 4440catgagcaaa ccgcaaagcg ctgccgggca aacccccaat
ggctatgcca ggcgcaccga 4500cagcgagcag gtgccggtga tcggcctgcg ccgcaagatc
gcccagcgca tgcaggacgc 4560caagcgccgg gtcgcgcact tcagctatgt ggaagaaatc
gacgtcaccg ccctggaagc 4620cctgcgccag cagctcaaca gcaagcacgg cgacagccgc
ggcaagctga cactgctgcc 4680gttcctggtg cgcgccctgg tcgtggcact gcgtgacttc
ccgcagataa acgccaccta 4740cgatgacgaa gcgcagatca tcacccgcca tggcgcggtg
catgtgggca tcgccaccca 4800aggtgacaac ggcctgatgg tacccgtgct gcgccacgcc
gaagcgggca gcctgtgggc 4860caatgccggt gagatttcac gcctggccaa cgctgcgcgc
aacaacaagg ccagccgcga 4920agagctgtcc ggttcgacca ttaccctgac cagcctcggc
gccctgggcg gcatcgtcag 4980cacgccggtg gtcaacaccc cggaagtggc gatcgtcggt
gtcaaccgca tggttgagcg 5040gcccgtggtg atcgacggcc agatcgtcgt gcgcaagatg
atgaacctgt ccagctcgtt 5100cgaccaccgc gtggtcgatg gcatggacgc cgccctgttc
atccaggccg tgcgtggcct 5160gctcgaacaa cccgcctgcc tgttcgtgga gtgagcatgc
aacagactat ccagacaacc 5220ctgttgatca tcggcggcgg ccctggcggc tatgtggcgg
ccatccgcgc cgggcaactg 5280ggcatcccta ccgtgctggt ggaaggccag gcgctgggcg
gtacctgcct gaacatcggc 5340tgcattccgt ccaaggcgct gatccatgtg gccgagcagt
tccaccaggc ctcgcgcttt 5400accgaaccct cgccgctggg catcagcgtg gcttcgccac
gcctggacat cggccagagc 5460gtggcctgga aagacggcat cgtcgatcgc ctgaccactg
gtgtcgccgc cctgctgaaa 5520aagcacgggg tgaaggtggt gcacggctgg gccaaggtgc
ttgatggcaa gcaggtcgag 5580gtggatggcc agcgcatcca gtgcgagcac ctgttgctgg
ccacgggctc cagcagtgtc 5640gaactgccga tgctgccgtt gggtgggccg gtgatttcct
cgaccgaggc cctggcaccg 5700aaagccctgc cgcaacacct ggtggtggtg ggcggtggct
acatcggcct ggagctgggt 5760atcgcctacc gcaagctcgg cgcgcaggtc agcgtggtgg
aagcgcgcga gcgcatcctg 5820ccgacttacg acagcgaact gaccgccccg gtggccgagt
cgctgaaaaa gctgggtatc 5880gccctgcacc ttggccacag cgtcgaaggt tacgaaaatg
gctgcctgct ggccaacgat 5940ggcaagggcg gacaactgcg cctggaagcc gaccgggtgc
tggtggccgt gggccgccgc 6000ccacgcacca agggcttcaa cctggaatgc ctggacctga
agatgaatgg tgccgcgatt 6060gccatcgacg agcgctgcca gaccagcatg cacaacgtct
gggccatcgg cgacgtggcc 6120ggcgaaccga tgctggcgca ccgggccatg gcccagggcg
agatggtggc cgagatcatc 6180gccggcaagg cacgccgctt cgaacccgct gcgatagccg
ccgtgtgctt caccgacccg 6240gaagtggtcg tggtcggcaa gacgccggaa caggccagtc
agcaaggcct ggactgcatc 6300gtcgcgcagt tcccgttcgc cgccaacggc cgggccatga
gcctggagtc gaaaagcggt 6360ttcgtgcgcg tggtcgcgcg gcgtgacaac cacctgatcc
tgggctggca agcggttggc 6420gtggcggttt ccgagctgtc cacggcgttt gcccagtcgc
tggagatggg tgcctgcctg 6480gaggatgtgg ccggtaccat ccatgcccac ccgaccctgg
gtgaagcggt acaggaagcg 6540gcactgcgtg ccctgggcca cgccctgcat atctgacact
gaagcggccg aggccgattt 6600ggcccgccgc gccgagaggc gctgcgggtc ttttttatac
ctg 664357423PRTPseudomonas putida 57Met Gly Thr His
Val Ile Lys Met Pro Asp Ile Gly Glu Gly Ile Ala 1 5
10 15 Gln Val Glu Leu Val Glu Trp Phe Val
Lys Val Gly Asp Ile Ile Ala 20 25
30 Glu Asp Gln Val Val Ala Asp Val Met Thr Asp Lys Ala Thr
Val Glu 35 40 45
Ile Pro Ser Pro Val Ser Gly Lys Val Leu Ala Leu Gly Gly Gln Pro 50
55 60 Gly Glu Val Met Ala
Val Gly Ser Glu Leu Ile Arg Ile Glu Val Glu 65 70
75 80 Gly Ser Gly Asn His Val Asp Val Pro Gln
Ala Lys Pro Ala Glu Val 85 90
95 Pro Ala Ala Pro Val Ala Ala Lys Pro Glu Pro Gln Lys Asp Val
Lys 100 105 110 Pro
Ala Ala Tyr Gln Ala Ser Ala Ser His Glu Ala Ala Pro Ile Val 115
120 125 Pro Arg Gln Pro Gly Asp
Lys Pro Leu Ala Ser Pro Ala Val Arg Lys 130 135
140 Arg Ala Leu Asp Ala Gly Ile Glu Leu Arg Tyr
Val His Gly Ser Gly 145 150 155
160 Pro Ala Gly Arg Ile Leu His Glu Asp Leu Asp Ala Phe Met Ser Lys
165 170 175 Pro Gln
Ser Ala Ala Gly Gln Thr Pro Asn Gly Tyr Ala Arg Arg Thr 180
185 190 Asp Ser Glu Gln Val Pro Val
Ile Gly Leu Arg Arg Lys Ile Ala Gln 195 200
205 Arg Met Gln Asp Ala Lys Arg Arg Val Ala His Phe
Ser Tyr Val Glu 210 215 220
Glu Ile Asp Val Thr Ala Leu Glu Ala Leu Arg Gln Gln Leu Asn Ser 225
230 235 240 Lys His Gly
Asp Ser Arg Gly Lys Leu Thr Leu Leu Pro Phe Leu Val 245
250 255 Arg Ala Leu Val Val Ala Leu Arg
Asp Phe Pro Gln Ile Asn Ala Thr 260 265
270 Tyr Asp Asp Glu Ala Gln Ile Ile Thr Arg His Gly Ala
Val His Val 275 280 285
Gly Ile Ala Thr Gln Gly Asp Asn Gly Leu Met Val Pro Val Leu Arg 290
295 300 His Ala Glu Ala
Gly Ser Leu Trp Ala Asn Ala Gly Glu Ile Ser Arg 305 310
315 320 Leu Ala Asn Ala Ala Arg Asn Asn Lys
Ala Ser Arg Glu Glu Leu Ser 325 330
335 Gly Ser Thr Ile Thr Leu Thr Ser Leu Gly Ala Leu Gly Gly
Ile Val 340 345 350
Ser Thr Pro Val Val Asn Thr Pro Glu Val Ala Ile Val Gly Val Asn
355 360 365 Arg Met Val Glu
Arg Pro Val Val Ile Asp Gly Gln Ile Val Val Arg 370
375 380 Lys Met Met Asn Leu Ser Ser Ser
Phe Asp His Arg Val Val Asp Gly 385 390
395 400 Met Asp Ala Ala Leu Phe Ile Gln Ala Val Arg Gly
Leu Leu Glu Gln 405 410
415 Pro Ala Cys Leu Phe Val Glu 420
586643DNAPseudomonas putida 58gcatgcctgc aggccgccga tgaaatggtg gaaggtatcg
gtaggctggc cctgctcatc 60gctgaacacg ttacgcccgc tgccggtatc gaccaggctc
tggtgaatat gcatggaact 120gccaggcgtg cgcgccagcg gtttggccat gcacaccacg
gtcagcccgt gcttgagtgc 180cacttccttg agcaggtgtt tgaacaggaa ggtctggtcg
gccagcagca gcgggtcgcc 240atgtagcaag ttgatctcga actggctgac gcccatttcg
tgcatgaagg tgtcgcgcgg 300caggccgagc gcggccatgc actggtacac ctcattgaag
aacgggcgca ggccgttgtt 360ggaactgaca ctgaacgccg aatggcccag ctcgcggcgg
ccgtcggtgc ccagcggtgg 420ctggaacggc tgctgcgggt cactgttggg ggcaaacacg
aagaactcaa gctcggtcgc 480cactaccggt gccagaccca acgctgcgta gcgggcgatc
acggccttca gctggccccg 540ggtggacagt gccgagggcc ggccatccag ttcattggca
tcgcagatgg ccagggcgcg 600accgtcatcg ctccagggca agcgatgaac ctggctgggt
tccgctacca acgccaggtc 660gccgtcgtcg cagccgtaga atttcgccgg cgggtagccg
cccatgatgc attgcagcag 720caccccacgg gccatctgca ggcggcggcc ttcgagaaag
ccttcggcgg tcatcacctt 780gccgcgtggg acgccgttga ggtcgggggt gacgcattcg
atttcatcga tgccctggag 840ctgagcgatg ctcatgacgc ttgtccttgt tgttgtaggc
tgacaacaac ataggctggg 900ggtgtttaaa atatcaagca gcctctcgaa cgcctggggc
ctcttctatt cgcgcaaggt 960catgccattg gccggcaacg gcaaggctgt cttgtagcgc
acctgtttca aggcaaaact 1020cgagcggata ttcgccacac ccggcaaccg ggtcaggtaa
tcgagaaacc gctccagcgc 1080ctggatactc ggcagcagta cccgcaacag gtagtccggg
tcgcccgtca tcaggtagca 1140ctccatcacc tcgggccgtt cggcaatttc ttcctcgaag
cggtgcagcg actgctctac 1200ctgtttttcc aggctgacat ggatgaacac attcacatcc
agccccaacg cctcgggcga 1260caacaaggtc acctgctggc ggatcacccc cagttcttcc
atggcccgca cccggttgaa 1320acagggcgtg ggcgacaggt tgaccgagcg tgccagctcg
gcgttggtga tgcgggcgtt 1380ttcctgcagg ctgttgagaa tgccgatatc ggtacgatcg
agtttgcgca tgagacaaaa 1440tcaccggttt tttgtgttta tgcggaatgt ttatctgccc
cgctcggcaa aggcaatcaa 1500cttgagagaa aaattctcct gccggaccac taagatgtag
gggacgctga cttaccagtc 1560acaagccggt actcagcggc ggccgcttca gagctcacaa
aaacaaatac ccgagcgagc 1620gtaaaaagca tgaacgagta cgcccccctg cgtttgcatg
tgcccgagcc caccggccgg 1680ccaggctgcc agaccgattt ttcctacctg cgcctgaacg
atgcaggtca agcccgtaaa 1740ccccctgtcg atgtcgacgc tgccgacacc gccgacctgt
cctacagcct ggtccgcgtg 1800ctcgacgagc aaggcgacgc ccaaggcccg tgggctgaag
acatcgaccc gcagatcctg 1860cgccaaggca tgcgcgccat gctcaagacg cggatcttcg
acagccgcat ggtggttgcc 1920cagcgccaga agaagatgtc cttctacatg cagagcctgg
gcgaagaagc catcggcagc 1980ggccaggcgc tggcgcttaa ccgcaccgac atgtgcttcc
ccacctaccg tcagcaaagc 2040atcctgatgg cccgcgacgt gtcgctggtg gagatgatct
gccagttgct gtccaacgaa 2100cgcgaccccc tcaagggccg ccagctgccg atcatgtact
cggtacgcga ggccggcttc 2160ttcaccatca gcggcaacct ggcgacccag ttcgtgcagg
cggtcggctg ggccatggcc 2220tcggcgatca agggcgatac caagattgcc tcggcctgga
tcggcgacgg cgccactgcc 2280gaatcggact tccacaccgc cctcaccttt gcccacgttt
accgcgcccc ggtgatcctc 2340aacgtggtca acaaccagtg ggccatctca accttccagg
ccatcgccgg tggcgagtcg 2400accaccttcg ccggccgtgg cgtgggctgc ggcatcgctt
cgctgcgggt ggacggcaac 2460gacttcgtcg ccgtttacgc cgcttcgcgc tgggctgccg
aacgtgcccg ccgtggtttg 2520ggcccgagcc tgatcgagtg ggtcacctac cgtgccggcc
cgcactcgac ctcggacgac 2580ccgtccaagt accgccctgc cgatgactgg agccacttcc
cgctgggtga cccgatcgcc 2640cgcctgaagc agcacctgat caagatcggc cactggtccg
aagaagaaca ccaggccacc 2700acggccgagt tcgaagcggc cgtgattgct gcgcaaaaag
aagccgagca gtacggcacc 2760ctggccaacg gtcacatccc gagcgccgcc tcgatgttcg
aggacgtgta caaggagatg 2820cccgaccacc tgcgccgcca acgccaggaa ctgggggttt
gagatgaacg accacaacaa 2880cagcatcaac ccggaaaccg ccatggccac cactaccatg
accatgatcc aggccctgcg 2940ctcggccatg gatgtcatgc ttgagcgcga cgacaatgtg
gtggtgtacg gccaggacgt 3000cggctacttc ggcggcgtgt tccgctgcac cgaaggcctg
cagaccaagt acggcaagtc 3060ccgcgtgttc gacgcgccca tctctgaaag cggcatcgtc
ggcaccgccg tgggcatggg 3120tgcctacggc ctgcgcccgg tggtggaaat ccagttcgct
gactacttct acccggcctc 3180cgaccagatc gtttctgaaa tggcccgcct gcgctaccgt
tcggccggcg agttcatcgc 3240cccgctgacc ctgcgtatgc cctgcggtgg cggtatctat
ggcggccaga cacacagcca 3300gagcccggaa gcgatgttca ctcaggtgtg cggcctgcgc
accgtaatgc catccaaccc 3360gtacgacgcc aaaggcctgc tgattgcctc gatcgaatgc
gacgacccgg tgatcttcct 3420ggagcccaag cgcctgtaca acggcccgtt cgacggccac
catgaccgcc cggttacgcc 3480gtggtcgaaa cacccgcaca gcgccgtgcc cgatggctac
tacaccgtgc cactggacaa 3540ggccgccatc acccgccccg gcaatgacgt gagcgtgctc
acctatggca ccaccgtgta 3600cgtggcccag gtggccgccg aagaaagtgg cgtggatgcc
gaagtgatcg acctgcgcag 3660cctgtggccg ctagacctgg acaccatcgt cgagtcggtg
aaaaagaccg gccgttgcgt 3720ggtagtacac gaggccaccc gtacttgtgg ctttggcgca
gaactggtgt cgctggtgca 3780ggagcactgc ttccaccacc tggaggcgcc gatcgagcgc
gtcaccggtt gggacacccc 3840ctaccctcac gcgcaggaat gggcttactt cccagggcct
tcgcgggtag gtgcggcatt 3900gaaaaaggtc atggaggtct gaatgggcac gcacgtcatc
aagatgccgg acattggcga 3960aggcatcgcg caggtcgaat tggtggaatg gttcgtcaag
gtgggcgaca tcatcgccga 4020ggaccaagtg gtagccgacg tcatgaccga caaggccacc
gtggaaatcc cgtcgccggt 4080cagcggcaag gtgctggccc tgggtggcca gccaggtgaa
gtgatggcgg tcggcagtga 4140gctgatccgc atcgaagtgg aaggcagcgg caaccatgtg
gatgtgccgc aagccaagcc 4200ggccgaagtg cctgcggcac cggtagccgc taaacctgaa
ccacagaaag acgttaaacc 4260ggcggcgtac caggcgtcag ccagccacga ggcagcgccc
atcgtgccgc gccagccggg 4320cgacaagccg ctggcctcgc cggcggtgcg caaacgcgcc
ctcgatgccg gcatcgaatt 4380gcgttatgtg cacggcagcg gcccggccgg gcgcatcctg
cacgaagacc tcgacgcgtt 4440catgagcaaa ccgcaaagcg ctgccgggca aacccccaat
ggctatgcca ggcgcaccga 4500cagcgagcag gtgccggtga tcggcctgcg ccgcaagatc
gcccagcgca tgcaggacgc 4560caagcgccgg gtcgcgcact tcagctatgt ggaagaaatc
gacgtcaccg ccctggaagc 4620cctgcgccag cagctcaaca gcaagcacgg cgacagccgc
ggcaagctga cactgctgcc 4680gttcctggtg cgcgccctgg tcgtggcact gcgtgacttc
ccgcagataa acgccaccta 4740cgatgacgaa gcgcagatca tcacccgcca tggcgcggtg
catgtgggca tcgccaccca 4800aggtgacaac ggcctgatgg tacccgtgct gcgccacgcc
gaagcgggca gcctgtgggc 4860caatgccggt gagatttcac gcctggccaa cgctgcgcgc
aacaacaagg ccagccgcga 4920agagctgtcc ggttcgacca ttaccctgac cagcctcggc
gccctgggcg gcatcgtcag 4980cacgccggtg gtcaacaccc cggaagtggc gatcgtcggt
gtcaaccgca tggttgagcg 5040gcccgtggtg atcgacggcc agatcgtcgt gcgcaagatg
atgaacctgt ccagctcgtt 5100cgaccaccgc gtggtcgatg gcatggacgc cgccctgttc
atccaggccg tgcgtggcct 5160gctcgaacaa cccgcctgcc tgttcgtgga gtgagcatgc
aacagactat ccagacaacc 5220ctgttgatca tcggcggcgg ccctggcggc tatgtggcgg
ccatccgcgc cgggcaactg 5280ggcatcccta ccgtgctggt ggaaggccag gcgctgggcg
gtacctgcct gaacatcggc 5340tgcattccgt ccaaggcgct gatccatgtg gccgagcagt
tccaccaggc ctcgcgcttt 5400accgaaccct cgccgctggg catcagcgtg gcttcgccac
gcctggacat cggccagagc 5460gtggcctgga aagacggcat cgtcgatcgc ctgaccactg
gtgtcgccgc cctgctgaaa 5520aagcacgggg tgaaggtggt gcacggctgg gccaaggtgc
ttgatggcaa gcaggtcgag 5580gtggatggcc agcgcatcca gtgcgagcac ctgttgctgg
ccacgggctc cagcagtgtc 5640gaactgccga tgctgccgtt gggtgggccg gtgatttcct
cgaccgaggc cctggcaccg 5700aaagccctgc cgcaacacct ggtggtggtg ggcggtggct
acatcggcct ggagctgggt 5760atcgcctacc gcaagctcgg cgcgcaggtc agcgtggtgg
aagcgcgcga gcgcatcctg 5820ccgacttacg acagcgaact gaccgccccg gtggccgagt
cgctgaaaaa gctgggtatc 5880gccctgcacc ttggccacag cgtcgaaggt tacgaaaatg
gctgcctgct ggccaacgat 5940ggcaagggcg gacaactgcg cctggaagcc gaccgggtgc
tggtggccgt gggccgccgc 6000ccacgcacca agggcttcaa cctggaatgc ctggacctga
agatgaatgg tgccgcgatt 6060gccatcgacg agcgctgcca gaccagcatg cacaacgtct
gggccatcgg cgacgtggcc 6120ggcgaaccga tgctggcgca ccgggccatg gcccagggcg
agatggtggc cgagatcatc 6180gccggcaagg cacgccgctt cgaacccgct gcgatagccg
ccgtgtgctt caccgacccg 6240gaagtggtcg tggtcggcaa gacgccggaa caggccagtc
agcaaggcct ggactgcatc 6300gtcgcgcagt tcccgttcgc cgccaacggc cgggccatga
gcctggagtc gaaaagcggt 6360ttcgtgcgcg tggtcgcgcg gcgtgacaac cacctgatcc
tgggctggca agcggttggc 6420gtggcggttt ccgagctgtc cacggcgttt gcccagtcgc
tggagatggg tgcctgcctg 6480gaggatgtgg ccggtaccat ccatgcccac ccgaccctgg
gtgaagcggt acaggaagcg 6540gcactgcgtg ccctgggcca cgccctgcat atctgacact
gaagcggccg aggccgattt 6600ggcccgccgc gccgagaggc gctgcgggtc ttttttatac
ctg 664359459PRTPseudomonas putida 59Met Gln Gln Thr
Ile Gln Thr Thr Leu Leu Ile Ile Gly Gly Gly Pro 1 5
10 15 Gly Gly Tyr Val Ala Ala Ile Arg Ala
Gly Gln Leu Gly Ile Pro Thr 20 25
30 Val Leu Val Glu Gly Gln Ala Leu Gly Gly Thr Cys Leu Asn
Ile Gly 35 40 45
Cys Ile Pro Ser Lys Ala Leu Ile His Val Ala Glu Gln Phe His Gln 50
55 60 Ala Ser Arg Phe Thr
Glu Pro Ser Pro Leu Gly Ile Ser Val Ala Ser 65 70
75 80 Pro Arg Leu Asp Ile Gly Gln Ser Val Ala
Trp Lys Asp Gly Ile Val 85 90
95 Asp Arg Leu Thr Thr Gly Val Ala Ala Leu Leu Lys Lys His Gly
Val 100 105 110 Lys
Val Val His Gly Trp Ala Lys Val Leu Asp Gly Lys Gln Val Glu 115
120 125 Val Asp Gly Gln Arg Ile
Gln Cys Glu His Leu Leu Leu Ala Thr Gly 130 135
140 Ser Ser Ser Val Glu Leu Pro Met Leu Pro Leu
Gly Gly Pro Val Ile 145 150 155
160 Ser Ser Thr Glu Ala Leu Ala Pro Lys Ala Leu Pro Gln His Leu Val
165 170 175 Val Val
Gly Gly Gly Tyr Ile Gly Leu Glu Leu Gly Ile Ala Tyr Arg 180
185 190 Lys Leu Gly Ala Gln Val Ser
Val Val Glu Ala Arg Glu Arg Ile Leu 195 200
205 Pro Thr Tyr Asp Ser Glu Leu Thr Ala Pro Val Ala
Glu Ser Leu Lys 210 215 220
Lys Leu Gly Ile Ala Leu His Leu Gly His Ser Val Glu Gly Tyr Glu 225
230 235 240 Asn Gly Cys
Leu Leu Ala Asn Asp Gly Lys Gly Gly Gln Leu Arg Leu 245
250 255 Glu Ala Asp Arg Val Leu Val Ala
Val Gly Arg Arg Pro Arg Thr Lys 260 265
270 Gly Phe Asn Leu Glu Cys Leu Asp Leu Lys Met Asn Gly
Ala Ala Ile 275 280 285
Ala Ile Asp Glu Arg Cys Gln Thr Ser Met His Asn Val Trp Ala Ile 290
295 300 Gly Asp Val Ala
Gly Glu Pro Met Leu Ala His Arg Ala Met Ala Gln 305 310
315 320 Gly Glu Met Val Ala Glu Ile Ile Ala
Gly Lys Ala Arg Arg Phe Glu 325 330
335 Pro Ala Ala Ile Ala Ala Val Cys Phe Thr Asp Pro Glu Val
Val Val 340 345 350
Val Gly Lys Thr Pro Glu Gln Ala Ser Gln Gln Gly Leu Asp Cys Ile
355 360 365 Val Ala Gln Phe
Pro Phe Ala Ala Asn Gly Arg Ala Met Ser Leu Glu 370
375 380 Ser Lys Ser Gly Phe Val Arg Val
Val Ala Arg Arg Asp Asn His Leu 385 390
395 400 Ile Leu Gly Trp Gln Ala Val Gly Val Ala Val Ser
Glu Leu Ser Thr 405 410
415 Ala Phe Ala Gln Ser Leu Glu Met Gly Ala Cys Leu Glu Asp Val Ala
420 425 430 Gly Thr Ile
His Ala His Pro Thr Leu Gly Glu Ala Val Gln Glu Ala 435
440 445 Ala Leu Arg Ala Leu Gly His Ala
Leu His Ile 450 455
606643DNAPseudomonas putida 60gcatgcctgc aggccgccga tgaaatggtg gaaggtatcg
gtaggctggc cctgctcatc 60gctgaacacg ttacgcccgc tgccggtatc gaccaggctc
tggtgaatat gcatggaact 120gccaggcgtg cgcgccagcg gtttggccat gcacaccacg
gtcagcccgt gcttgagtgc 180cacttccttg agcaggtgtt tgaacaggaa ggtctggtcg
gccagcagca gcgggtcgcc 240atgtagcaag ttgatctcga actggctgac gcccatttcg
tgcatgaagg tgtcgcgcgg 300caggccgagc gcggccatgc actggtacac ctcattgaag
aacgggcgca ggccgttgtt 360ggaactgaca ctgaacgccg aatggcccag ctcgcggcgg
ccgtcggtgc ccagcggtgg 420ctggaacggc tgctgcgggt cactgttggg ggcaaacacg
aagaactcaa gctcggtcgc 480cactaccggt gccagaccca acgctgcgta gcgggcgatc
acggccttca gctggccccg 540ggtggacagt gccgagggcc ggccatccag ttcattggca
tcgcagatgg ccagggcgcg 600accgtcatcg ctccagggca agcgatgaac ctggctgggt
tccgctacca acgccaggtc 660gccgtcgtcg cagccgtaga atttcgccgg cgggtagccg
cccatgatgc attgcagcag 720caccccacgg gccatctgca ggcggcggcc ttcgagaaag
ccttcggcgg tcatcacctt 780gccgcgtggg acgccgttga ggtcgggggt gacgcattcg
atttcatcga tgccctggag 840ctgagcgatg ctcatgacgc ttgtccttgt tgttgtaggc
tgacaacaac ataggctggg 900ggtgtttaaa atatcaagca gcctctcgaa cgcctggggc
ctcttctatt cgcgcaaggt 960catgccattg gccggcaacg gcaaggctgt cttgtagcgc
acctgtttca aggcaaaact 1020cgagcggata ttcgccacac ccggcaaccg ggtcaggtaa
tcgagaaacc gctccagcgc 1080ctggatactc ggcagcagta cccgcaacag gtagtccggg
tcgcccgtca tcaggtagca 1140ctccatcacc tcgggccgtt cggcaatttc ttcctcgaag
cggtgcagcg actgctctac 1200ctgtttttcc aggctgacat ggatgaacac attcacatcc
agccccaacg cctcgggcga 1260caacaaggtc acctgctggc ggatcacccc cagttcttcc
atggcccgca cccggttgaa 1320acagggcgtg ggcgacaggt tgaccgagcg tgccagctcg
gcgttggtga tgcgggcgtt 1380ttcctgcagg ctgttgagaa tgccgatatc ggtacgatcg
agtttgcgca tgagacaaaa 1440tcaccggttt tttgtgttta tgcggaatgt ttatctgccc
cgctcggcaa aggcaatcaa 1500cttgagagaa aaattctcct gccggaccac taagatgtag
gggacgctga cttaccagtc 1560acaagccggt actcagcggc ggccgcttca gagctcacaa
aaacaaatac ccgagcgagc 1620gtaaaaagca tgaacgagta cgcccccctg cgtttgcatg
tgcccgagcc caccggccgg 1680ccaggctgcc agaccgattt ttcctacctg cgcctgaacg
atgcaggtca agcccgtaaa 1740ccccctgtcg atgtcgacgc tgccgacacc gccgacctgt
cctacagcct ggtccgcgtg 1800ctcgacgagc aaggcgacgc ccaaggcccg tgggctgaag
acatcgaccc gcagatcctg 1860cgccaaggca tgcgcgccat gctcaagacg cggatcttcg
acagccgcat ggtggttgcc 1920cagcgccaga agaagatgtc cttctacatg cagagcctgg
gcgaagaagc catcggcagc 1980ggccaggcgc tggcgcttaa ccgcaccgac atgtgcttcc
ccacctaccg tcagcaaagc 2040atcctgatgg cccgcgacgt gtcgctggtg gagatgatct
gccagttgct gtccaacgaa 2100cgcgaccccc tcaagggccg ccagctgccg atcatgtact
cggtacgcga ggccggcttc 2160ttcaccatca gcggcaacct ggcgacccag ttcgtgcagg
cggtcggctg ggccatggcc 2220tcggcgatca agggcgatac caagattgcc tcggcctgga
tcggcgacgg cgccactgcc 2280gaatcggact tccacaccgc cctcaccttt gcccacgttt
accgcgcccc ggtgatcctc 2340aacgtggtca acaaccagtg ggccatctca accttccagg
ccatcgccgg tggcgagtcg 2400accaccttcg ccggccgtgg cgtgggctgc ggcatcgctt
cgctgcgggt ggacggcaac 2460gacttcgtcg ccgtttacgc cgcttcgcgc tgggctgccg
aacgtgcccg ccgtggtttg 2520ggcccgagcc tgatcgagtg ggtcacctac cgtgccggcc
cgcactcgac ctcggacgac 2580ccgtccaagt accgccctgc cgatgactgg agccacttcc
cgctgggtga cccgatcgcc 2640cgcctgaagc agcacctgat caagatcggc cactggtccg
aagaagaaca ccaggccacc 2700acggccgagt tcgaagcggc cgtgattgct gcgcaaaaag
aagccgagca gtacggcacc 2760ctggccaacg gtcacatccc gagcgccgcc tcgatgttcg
aggacgtgta caaggagatg 2820cccgaccacc tgcgccgcca acgccaggaa ctgggggttt
gagatgaacg accacaacaa 2880cagcatcaac ccggaaaccg ccatggccac cactaccatg
accatgatcc aggccctgcg 2940ctcggccatg gatgtcatgc ttgagcgcga cgacaatgtg
gtggtgtacg gccaggacgt 3000cggctacttc ggcggcgtgt tccgctgcac cgaaggcctg
cagaccaagt acggcaagtc 3060ccgcgtgttc gacgcgccca tctctgaaag cggcatcgtc
ggcaccgccg tgggcatggg 3120tgcctacggc ctgcgcccgg tggtggaaat ccagttcgct
gactacttct acccggcctc 3180cgaccagatc gtttctgaaa tggcccgcct gcgctaccgt
tcggccggcg agttcatcgc 3240cccgctgacc ctgcgtatgc cctgcggtgg cggtatctat
ggcggccaga cacacagcca 3300gagcccggaa gcgatgttca ctcaggtgtg cggcctgcgc
accgtaatgc catccaaccc 3360gtacgacgcc aaaggcctgc tgattgcctc gatcgaatgc
gacgacccgg tgatcttcct 3420ggagcccaag cgcctgtaca acggcccgtt cgacggccac
catgaccgcc cggttacgcc 3480gtggtcgaaa cacccgcaca gcgccgtgcc cgatggctac
tacaccgtgc cactggacaa 3540ggccgccatc acccgccccg gcaatgacgt gagcgtgctc
acctatggca ccaccgtgta 3600cgtggcccag gtggccgccg aagaaagtgg cgtggatgcc
gaagtgatcg acctgcgcag 3660cctgtggccg ctagacctgg acaccatcgt cgagtcggtg
aaaaagaccg gccgttgcgt 3720ggtagtacac gaggccaccc gtacttgtgg ctttggcgca
gaactggtgt cgctggtgca 3780ggagcactgc ttccaccacc tggaggcgcc gatcgagcgc
gtcaccggtt gggacacccc 3840ctaccctcac gcgcaggaat gggcttactt cccagggcct
tcgcgggtag gtgcggcatt 3900gaaaaaggtc atggaggtct gaatgggcac gcacgtcatc
aagatgccgg acattggcga 3960aggcatcgcg caggtcgaat tggtggaatg gttcgtcaag
gtgggcgaca tcatcgccga 4020ggaccaagtg gtagccgacg tcatgaccga caaggccacc
gtggaaatcc cgtcgccggt 4080cagcggcaag gtgctggccc tgggtggcca gccaggtgaa
gtgatggcgg tcggcagtga 4140gctgatccgc atcgaagtgg aaggcagcgg caaccatgtg
gatgtgccgc aagccaagcc 4200ggccgaagtg cctgcggcac cggtagccgc taaacctgaa
ccacagaaag acgttaaacc 4260ggcggcgtac caggcgtcag ccagccacga ggcagcgccc
atcgtgccgc gccagccggg 4320cgacaagccg ctggcctcgc cggcggtgcg caaacgcgcc
ctcgatgccg gcatcgaatt 4380gcgttatgtg cacggcagcg gcccggccgg gcgcatcctg
cacgaagacc tcgacgcgtt 4440catgagcaaa ccgcaaagcg ctgccgggca aacccccaat
ggctatgcca ggcgcaccga 4500cagcgagcag gtgccggtga tcggcctgcg ccgcaagatc
gcccagcgca tgcaggacgc 4560caagcgccgg gtcgcgcact tcagctatgt ggaagaaatc
gacgtcaccg ccctggaagc 4620cctgcgccag cagctcaaca gcaagcacgg cgacagccgc
ggcaagctga cactgctgcc 4680gttcctggtg cgcgccctgg tcgtggcact gcgtgacttc
ccgcagataa acgccaccta 4740cgatgacgaa gcgcagatca tcacccgcca tggcgcggtg
catgtgggca tcgccaccca 4800aggtgacaac ggcctgatgg tacccgtgct gcgccacgcc
gaagcgggca gcctgtgggc 4860caatgccggt gagatttcac gcctggccaa cgctgcgcgc
aacaacaagg ccagccgcga 4920agagctgtcc ggttcgacca ttaccctgac cagcctcggc
gccctgggcg gcatcgtcag 4980cacgccggtg gtcaacaccc cggaagtggc gatcgtcggt
gtcaaccgca tggttgagcg 5040gcccgtggtg atcgacggcc agatcgtcgt gcgcaagatg
atgaacctgt ccagctcgtt 5100cgaccaccgc gtggtcgatg gcatggacgc cgccctgttc
atccaggccg tgcgtggcct 5160gctcgaacaa cccgcctgcc tgttcgtgga gtgagcatgc
aacagactat ccagacaacc 5220ctgttgatca tcggcggcgg ccctggcggc tatgtggcgg
ccatccgcgc cgggcaactg 5280ggcatcccta ccgtgctggt ggaaggccag gcgctgggcg
gtacctgcct gaacatcggc 5340tgcattccgt ccaaggcgct gatccatgtg gccgagcagt
tccaccaggc ctcgcgcttt 5400accgaaccct cgccgctggg catcagcgtg gcttcgccac
gcctggacat cggccagagc 5460gtggcctgga aagacggcat cgtcgatcgc ctgaccactg
gtgtcgccgc cctgctgaaa 5520aagcacgggg tgaaggtggt gcacggctgg gccaaggtgc
ttgatggcaa gcaggtcgag 5580gtggatggcc agcgcatcca gtgcgagcac ctgttgctgg
ccacgggctc cagcagtgtc 5640gaactgccga tgctgccgtt gggtgggccg gtgatttcct
cgaccgaggc cctggcaccg 5700aaagccctgc cgcaacacct ggtggtggtg ggcggtggct
acatcggcct ggagctgggt 5760atcgcctacc gcaagctcgg cgcgcaggtc agcgtggtgg
aagcgcgcga gcgcatcctg 5820ccgacttacg acagcgaact gaccgccccg gtggccgagt
cgctgaaaaa gctgggtatc 5880gccctgcacc ttggccacag cgtcgaaggt tacgaaaatg
gctgcctgct ggccaacgat 5940ggcaagggcg gacaactgcg cctggaagcc gaccgggtgc
tggtggccgt gggccgccgc 6000ccacgcacca agggcttcaa cctggaatgc ctggacctga
agatgaatgg tgccgcgatt 6060gccatcgacg agcgctgcca gaccagcatg cacaacgtct
gggccatcgg cgacgtggcc 6120ggcgaaccga tgctggcgca ccgggccatg gcccagggcg
agatggtggc cgagatcatc 6180gccggcaagg cacgccgctt cgaacccgct gcgatagccg
ccgtgtgctt caccgacccg 6240gaagtggtcg tggtcggcaa gacgccggaa caggccagtc
agcaaggcct ggactgcatc 6300gtcgcgcagt tcccgttcgc cgccaacggc cgggccatga
gcctggagtc gaaaagcggt 6360ttcgtgcgcg tggtcgcgcg gcgtgacaac cacctgatcc
tgggctggca agcggttggc 6420gtggcggttt ccgagctgtc cacggcgttt gcccagtcgc
tggagatggg tgcctgcctg 6480gaggatgtgg ccggtaccat ccatgcccac ccgaccctgg
gtgaagcggt acaggaagcg 6540gcactgcgtg ccctgggcca cgccctgcat atctgacact
gaagcggccg aggccgattt 6600ggcccgccgc gccgagaggc gctgcgggtc ttttttatac
ctg 664361468PRTClostridium beijerinckii 61Met Asn
Lys Asp Thr Leu Ile Pro Thr Thr Lys Asp Leu Lys Leu Lys 1 5
10 15 Thr Asn Val Glu Asn Ile Asn
Leu Lys Asn Tyr Lys Asp Asn Ser Ser 20 25
30 Cys Phe Gly Val Phe Glu Asn Val Glu Asn Ala Ile
Asn Ser Ala Val 35 40 45
His Ala Gln Lys Ile Leu Ser Leu His Tyr Thr Lys Glu Gln Arg Glu
50 55 60 Lys Ile Ile
Thr Glu Ile Arg Lys Ala Ala Leu Glu Asn Lys Glu Val 65
70 75 80 Leu Ala Thr Met Ile Leu Glu
Glu Thr His Met Gly Arg Tyr Glu Asp 85
90 95 Lys Ile Leu Lys His Glu Leu Val Ala Lys Tyr
Thr Pro Gly Thr Glu 100 105
110 Asp Leu Thr Thr Thr Ala Trp Ser Gly Asp Asn Gly Leu Thr Val
Val 115 120 125 Glu
Met Ser Pro Tyr Gly Val Ile Gly Ala Ile Thr Pro Ser Thr Asn 130
135 140 Pro Thr Glu Thr Val Ile
Cys Asn Ser Ile Gly Met Ile Ala Ala Gly 145 150
155 160 Asn Ala Val Val Phe Asn Gly His Pro Gly Ala
Lys Lys Cys Val Ala 165 170
175 Phe Ala Ile Glu Met Ile Asn Lys Ala Ile Ile Ser Cys Gly Gly Pro
180 185 190 Glu Asn
Leu Val Thr Thr Ile Lys Asn Pro Thr Met Glu Ser Leu Asp 195
200 205 Ala Ile Ile Lys His Pro Leu
Ile Lys Leu Leu Cys Gly Thr Gly Gly 210 215
220 Pro Gly Met Val Lys Thr Leu Leu Asn Ser Gly Lys
Lys Ala Ile Gly 225 230 235
240 Ala Gly Ala Gly Asn Pro Pro Val Ile Val Asp Asp Thr Ala Asp Ile
245 250 255 Glu Lys Ala
Gly Lys Ser Ile Ile Glu Gly Cys Ser Phe Asp Asn Asn 260
265 270 Leu Pro Cys Ile Ala Glu Lys Glu
Val Phe Val Phe Glu Asn Val Ala 275 280
285 Asp Asp Leu Ile Ser Asn Met Leu Lys Asn Asn Ala Val
Ile Ile Asn 290 295 300
Glu Asp Gln Val Ser Lys Leu Ile Asp Leu Val Leu Gln Lys Asn Asn 305
310 315 320 Glu Thr Gln Glu
Tyr Phe Ile Asn Lys Lys Trp Val Gly Lys Asp Ala 325
330 335 Lys Leu Phe Ser Asp Glu Ile Asp Val
Glu Ser Pro Ser Asn Ile Lys 340 345
350 Cys Ile Val Cys Glu Val Asn Ala Asn His Pro Phe Val Met
Thr Glu 355 360 365
Leu Met Met Pro Ile Leu Pro Ile Val Arg Val Lys Asp Ile Asp Glu 370
375 380 Ala Val Lys Tyr Thr
Lys Ile Ala Glu Gln Asn Arg Lys His Ser Ala 385 390
395 400 Tyr Ile Tyr Ser Lys Asn Ile Asp Asn Leu
Asn Arg Phe Glu Arg Glu 405 410
415 Ile Asp Thr Thr Ile Phe Val Lys Asn Ala Lys Ser Phe Ala Gly
Val 420 425 430 Gly
Tyr Glu Ala Glu Gly Phe Thr Thr Phe Thr Ile Ala Gly Ser Thr 435
440 445 Gly Glu Gly Ile Thr Ser
Ala Arg Asn Phe Thr Arg Gln Arg Arg Cys 450 455
460 Val Leu Ala Gly 465
626558DNAClostridium beijerinckii 62aagcttaaaa tatccatagg ctattgttaa
taagactata gcgcttaata ctctaagcgc 60accatctaaa aaattataca taggaattgg
ataaactcca atcttctcca taatactttt 120cataggtgaa attgatatta taattaaggc
tgcataaagc aaactcatat ctccaataaa 180tgtcactatc ataggtaatt tccttttcat
gttcacatac tcccccgtat tctataataa 240tttacaataa acttccatca caaataaatt
ataacatata ttgtaaatat agttttatta 300ttcgcatatt tatagataaa caatatataa
cttagactta tgcaataacc taccgaaagt 360aaaaacattg ttatttcaca ggactatgaa
aatttcgctg aaagtactaa attgcaggtt 420gcaccactaa tgcttgctcc caatttcatt
gtgacaagca ttagttgaac aacctacaat 480taagagcctt taacagctca tttccaatgc
ctgctacata aaaatgtttc tactttctaa 540tatggtattt acttcaaagt gtaaatcaaa
ttcttaagtt gtttatctat atagggttcc 600atattgataa caatacatat ctactgtttc
aatttcatga ataccagcga tattgaaatt 660ttgtaagttt gaatatcatt gtgaaaccct
atatatcaaa tacaatctca aaattataca 720aaaagatccc aacttcacaa tatgaatttg
agatcttcta tcttaacttc ttcaatattt 780ttaagattat ttatactgcg tgcaaatctt
ctataagtat caattatcag ctatgtacat 840tgatagtgga cttgtaatct gaacagctta
tatatttaca cttttaagtt ctcttccact 900aacccctgct ttgaaacttc tcataaacaa
atcgaaagta cctttaatta gctgtgcatt 960ctcaaactca gaatttaact taagtacttc
aattgccgct tcgatagtat agagctctcc 1020ttctgaagca ccttttctta aagtatattc
tgatttatta attggattta atgaaattct 1080tggaagcttc tttaagtagt cgctctttct
tagtatcttc tctgcttctt tccatgtgcc 1140atctaagata ataaatgctg gaattttttc
tgaatcttta catttagact ttctttctag 1200aggttcatca tcatccatag gaaataatac
acgtatttca taatcatcgc tattaatata 1260ttcaattaat ttttcaggag tctttactct
ctcccaaaga attaactcag ttgattctgg 1320attcaccaat ttcaataatc tagcggtatt
tgaaggccta ctaaattctc tttctgttga 1380taatatcaat atctttgctt ttgtctctat
tttaggcaca atatcgcaga tacaatttat 1440tattggcaac ccacatttat tgcagctctc
atataactta gtaatttgct taactttaaa 1500ttcagactcc attttacctc cattattagt
tggttagtgt gtcatatctt cttgctatta 1560ctaactgatt ataacatatg tattcaatat
atcactccta gttttcaaag cactggcaat 1620acgaattaca aattaatttc tggatttatg
tcagtatttc attaataaaa ggtcggactt 1680ttaagatact tgttttagct attgatcata
tttattaaag actatgcatt taatgtataa 1740ttataatgaa tattatcaat aatatttatt
ttatattaca atcttacagt ctttattcta 1800aatttcactc aaataccaaa cgagctttat
tcataaacaa tatataacaa taattccaaa 1860ataatacgat attttatctg taacagccat
ataaaaaaaa tatcatatag tcttgtcatt 1920tgataacgtt ttgtcttcct tatatttact
ttttcggttt aataggttga ttctgtaaat 1980tttagtgata acatatattt gatgacatta
aaaatttaat atttcatata aatttttaat 2040gtctattaat ttttaaatca caaggaggaa
tagttcatga ataaagacac actaatacct 2100acaactaaag atttaaaatt aaaaacaaat
gttgaaaaca ttaatttaaa gaactacaag 2160gataattctt catgtttcgg agtattcgaa
aatgttgaaa atgctataaa cagcgctgta 2220cacgcgcaaa agatattatc ccttcattat
acaaaagaac aaagagaaaa aatcataact 2280gagataagaa aggccgcatt agaaaataaa
gaggttttag ctaccatgat tctggaagaa 2340acacatatgg gaaggtatga agataaaata
ttaaagcatg aattagtagc taaatatact 2400cctggtacag aagatttaac tactactgct
tggtcaggtg ataatggtct tacagttgta 2460gaaatgtctc catatggcgt tataggtgca
ataactcctt ctacgaatcc aactgaaact 2520gtaatatgta atagcatcgg catgatagct
gctggaaatg ctgtagtatt taacggacac 2580ccaggcgcta aaaaatgtgt tgcttttgct
attgaaatga taaataaagc aattatttca 2640tgtggcggtc ctgagaattt agtaacaact
ataaaaaatc caactatgga atccctagat 2700gcaattatta agcatccttt aataaaactt
ctttgcggaa ctggaggtcc aggaatggta 2760aaaaccctct taaattctgg caagaaagct
ataggtgctg gtgctggaaa tccaccagtt 2820attgtagatg ataccgctga tatagaaaag
gctggtaaga gtatcattga aggctgttct 2880tttgataata atttaccttg tattgcagaa
aaagaagtat ttgtttttga gaatgttgca 2940gatgatttaa tatctaacat gctaaaaaat
aatgctgtaa ttataaatga agatcaagta 3000tcaaaattaa tagatttagt attacaaaaa
aataatgaaa ctcaagaata ctttataaac 3060aaaaaatggg taggaaaaga tgcaaaatta
ttctcagatg aaatagatgt tgagtctcct 3120tcaaatatta aatgcatagt ctgcgaagta
aatgcaaatc atccatttgt catgacagaa 3180ctcatgatgc caatattacc aattgtaaga
gttaaagata tagatgaagc tgttaaatat 3240acaaagatag cagaacaaaa tagaaaacat
agtgcctata tttattctaa aaatatagac 3300aacctaaata gatttgaaag agaaattgat
actactattt ttgtaaagaa tgctaaatct 3360tttgctggtg ttggttatga agctgaagga
tttacaactt tcactattgc tggatctact 3420ggtgaaggca taacctctgc aagaaatttt
acaagacaaa gaagatgtgt acttgccggc 3480taacttcttg ctaaatttat acatttattc
acataacttt aatatgcaat gttcccacaa 3540aatattaaaa actatttaga agggagatat
taaatgaata aattagtaaa attaacagat 3600ttaaagcgca ttttcaaaga tggtatgaca
attatggttg ggggtttttt agattgtgga 3660actcctgaaa atattataga tatgctagtt
gatttaaata taaaaaatct gactattata 3720agcaatgata cagcttttcc taataaagga
ataggaaaac ttattgtaaa tggtcaagtt 3780tctaaagtaa ttgcttcaca tattggaact
aatcctgaaa ctgggaaaaa aatgagctct 3840ggtgaactta aagttgagct ttctccacaa
ggaacactga tcgaaagaat tcgtgcagct 3900ggatctggac tcggaggtgt attaactcca
accggacttg ggactatcgt tgaagaaggt 3960aagaaaaaag ttactatcgg tggcaaagaa
tatctattag aacttccttt atccgctgat 4020gtttcattaa taaaaggtag cattgtagat
gaatttggaa ataccttcta tagagctgct 4080actaaaaatt tcaatccata tatggcaatg
gctgcaaaaa cagttatagt tgaagcagaa 4140aatttagtta aatgtgaaga tttaaaaaga
gatgccataa tgactcctgg cgtattagta 4200gattatatcg ttaaggaggc ggcttaattg
attgtagata aagttttagc aaaagagata 4260attgccaaaa gagttgcaaa agaactaaaa
aaaggccaac tcgtaaacct tggaatagga 4320cttccaactt tagtagctaa ttatgtgcca
aaagaaatga acattacttt cgaatcagaa 4380aatggcatgg ttggcatggc acaaatggcc
tcatcaggtg aaaatgaccc agatataata 4440aatgctggtg gggaatatgt aacattatta
cctcaaggtg cattttttga tagttcaacg 4500tcttttgcac taataagagg aggacatgtt
gatgttgctg ttcttggtgc tctagaagtt 4560gatgaagaag gtaatttagc taactggatt
gttccaaata aaattgtccc aggtatggga 4620ggcgccatgg atttggcaat aggcgcaaaa
aaaataatag tggcaatgca acatacagga 4680aaaggtaaac ctaaaatcgt aaaaaaatgt
actctcccac ttactgctaa ggctcaggta 4740gatttaattg ttacagaact ttgtgtaatt
gatgtaacaa atgatggttt acttttcaga 4800gaaattcata aagatacaac tattgatgaa
ataaaatttt taacagatgc agatttaatt 4860attcccgaca acttaaaaat tatggatatc
taaatcattc tattttaaat atataacttt 4920aaaaatctta tgtattaaaa actaagaaaa
gaggttgatt attttatgtt agaaagtgaa 4980gtatctaaac aaattacaac tccacttgct
gctccagcgt ttcctagagg accatataga 5040tttcacaata gagaatatct aaacattatt
tatcgaactg atttagatgc tcttcgaaaa 5100atagtaccag agccacttga attagatgga
gcatatgtta ggtttgagat gatggctatg 5160cctgatacaa ccggactagg ctcatatact
gagtgtggtc aagccattcc agtaaaatat 5220aatgaggtta aaggtgacta cttgcatatg
atgtacctag ataatgaacc tgctattgct 5280gttggaagag aaagcagtgc ttatcccaaa
aagttcggct atccaaagct atttgttgat 5340tcagacgccc tagttggcgc ccttaagtat
ggtgcattac cggtagttac tgcgacgatg 5400ggatataagc atgagcccct agatcttaaa
gaagcctata ctcaaattgc aagacccaat 5460ttcatgctaa aaatcattca aggttatgat
ggtaagccaa gaatttgtga actcatctgt 5520gcagaaaata ctgatataac tatccacggt
gcttggactg gaagtgcacg cctacaatta 5580tttagccatg cactagctcc tcttgctgat
ttacctgtat tagagatcgt atcagcatct 5640catatcctaa cagatttaac tcttggaaca
cctaaggttg tacatgatta tctttcagta 5700aaataaaagc aatatagaat aaccactaca
aaagtagtgg ttattctata ttttaaatca 5760aactgtaaaa cttaagtttt atagtaccta
ataatatttt actaccagca ttagattagt 5820taaaatacaa agtttgtggt aaaagtattt
tagattgcat aatagccttc tatactttta 5880acaatataac caattgctca ccatctgctt
agaatatgct tctttaagct ctaaaataca 5940tataaaaaag taggaatttc ttattaaaat
tcctacttat attatatata aatttaatcg 6000ttaggtttta ttcgcattgt tcctctttaa
tttatctctt ataacatttt attataattg 6060ttcatataat taattcaata tactattata
tattttcaag cattaataat tattcagcat 6120ctgtcattac atatgcttcc atactttgac
ttcttattaa atcatagcta atccatccat 6180agccattgat tccccagtct ttaccccatg
aatttattat ttttacagct tttttactat 6240catcataacc aactacgcaa actgcatgac
cacctctatt ttctccatca atctggtcat 6300aaattggatt atcagaattt aaattatcaa
aatctggata tactgatatt ccaataacta 6360ctggatttcc agctgctatt tgtgccttta
ttgcattata gtcaccatct ggaagttgac 6420tccaactttt tgctttatat ttggctgcat
tagccttttg ttcatctgta ggtgtaacct 6480cccaactata ttcactacca tcataaggca
tatcagataa tgtagtacaa ccttgttctt 6540ctaataattt aaatgcat
655863862PRTClostridium acetobutylicum
63Met Lys Val Thr Thr Val Lys Glu Leu Asp Glu Lys Leu Lys Val Ile 1
5 10 15 Lys Glu Ala Gln
Lys Lys Phe Ser Cys Tyr Ser Gln Glu Met Val Asp 20
25 30 Glu Ile Phe Arg Asn Ala Ala Met Ala
Ala Ile Asp Ala Arg Ile Glu 35 40
45 Leu Ala Lys Ala Ala Val Leu Glu Thr Gly Met Gly Leu Val
Glu Asp 50 55 60
Lys Val Ile Lys Asn His Phe Ala Gly Glu Tyr Ile Tyr Asn Lys Tyr 65
70 75 80 Lys Asp Glu Lys Thr
Cys Gly Ile Ile Glu Arg Asn Glu Pro Tyr Gly 85
90 95 Ile Thr Lys Ile Ala Glu Pro Ile Gly Val
Val Ala Ala Ile Ile Pro 100 105
110 Val Thr Asn Pro Thr Ser Thr Thr Ile Phe Lys Ser Leu Ile Ser
Leu 115 120 125 Lys
Thr Arg Asn Gly Ile Phe Phe Ser Pro His Pro Arg Ala Lys Lys 130
135 140 Ser Thr Ile Leu Ala Ala
Lys Thr Ile Leu Asp Ala Ala Val Lys Ser 145 150
155 160 Gly Ala Pro Glu Asn Ile Ile Gly Trp Ile Asp
Glu Pro Ser Ile Glu 165 170
175 Leu Thr Gln Tyr Leu Met Gln Lys Ala Asp Ile Thr Leu Ala Thr Gly
180 185 190 Gly Pro
Ser Leu Val Lys Ser Ala Tyr Ser Ser Gly Lys Pro Ala Ile 195
200 205 Gly Val Gly Pro Gly Asn Thr
Pro Val Ile Ile Asp Glu Ser Ala His 210 215
220 Ile Lys Met Ala Val Ser Ser Ile Ile Leu Ser Lys
Thr Tyr Asp Asn 225 230 235
240 Gly Val Ile Cys Ala Ser Glu Gln Ser Val Ile Val Leu Lys Ser Ile
245 250 255 Tyr Asn Lys
Val Lys Asp Glu Phe Gln Glu Arg Gly Ala Tyr Ile Ile 260
265 270 Lys Lys Asn Glu Leu Asp Lys Val
Arg Glu Val Ile Phe Lys Asp Gly 275 280
285 Ser Val Asn Pro Lys Ile Val Gly Gln Ser Ala Tyr Thr
Ile Ala Ala 290 295 300
Met Ala Gly Ile Lys Val Pro Lys Thr Thr Arg Ile Leu Ile Gly Glu 305
310 315 320 Val Thr Ser Leu
Gly Glu Glu Glu Pro Phe Ala His Glu Lys Leu Ser 325
330 335 Pro Val Leu Ala Met Tyr Glu Ala Asp
Asn Phe Asp Asp Ala Leu Lys 340 345
350 Lys Ala Val Thr Leu Ile Asn Leu Gly Gly Leu Gly His Thr
Ser Gly 355 360 365
Ile Tyr Ala Asp Glu Ile Lys Ala Arg Asp Lys Ile Asp Arg Phe Ser 370
375 380 Ser Ala Met Lys Thr
Val Arg Thr Phe Val Asn Ile Pro Thr Ser Gln 385 390
395 400 Gly Ala Ser Gly Asp Leu Tyr Asn Phe Arg
Ile Pro Pro Ser Phe Thr 405 410
415 Leu Gly Cys Gly Phe Trp Gly Gly Asn Ser Val Ser Glu Asn Val
Gly 420 425 430 Pro
Lys His Leu Leu Asn Ile Lys Thr Val Ala Glu Arg Arg Glu Asn 435
440 445 Met Leu Trp Phe Arg Val
Pro His Lys Val Tyr Phe Lys Phe Gly Cys 450 455
460 Leu Gln Phe Ala Leu Lys Asp Leu Lys Asp Leu
Lys Lys Lys Arg Ala 465 470 475
480 Phe Ile Val Thr Asp Ser Asp Pro Tyr Asn Leu Asn Tyr Val Asp Ser
485 490 495 Ile Ile
Lys Ile Leu Glu His Leu Asp Ile Asp Phe Lys Val Phe Asn 500
505 510 Lys Val Gly Arg Glu Ala Asp
Leu Lys Thr Ile Lys Lys Ala Thr Glu 515 520
525 Glu Met Ser Ser Phe Met Pro Asp Thr Ile Ile Ala
Leu Gly Gly Thr 530 535 540
Pro Glu Met Ser Ser Ala Lys Leu Met Trp Val Leu Tyr Glu His Pro 545
550 555 560 Glu Val Lys
Phe Glu Asp Leu Ala Ile Lys Phe Met Asp Ile Arg Lys 565
570 575 Arg Ile Tyr Thr Phe Pro Lys Leu
Gly Lys Lys Ala Met Leu Val Ala 580 585
590 Ile Thr Thr Ser Ala Gly Ser Gly Ser Glu Val Thr Pro
Phe Ala Leu 595 600 605
Val Thr Asp Asn Asn Thr Gly Asn Lys Tyr Met Leu Ala Asp Tyr Glu 610
615 620 Met Thr Pro Asn
Met Ala Ile Val Asp Ala Glu Leu Met Met Lys Met 625 630
635 640 Pro Lys Gly Leu Thr Ala Tyr Ser Gly
Ile Asp Ala Leu Val Asn Ser 645 650
655 Ile Glu Ala Tyr Thr Ser Val Tyr Ala Ser Glu Tyr Thr Asn
Gly Leu 660 665 670
Ala Leu Glu Ala Ile Arg Leu Ile Phe Lys Tyr Leu Pro Glu Ala Tyr
675 680 685 Lys Asn Gly Arg
Thr Asn Glu Lys Ala Arg Glu Lys Met Ala His Ala 690
695 700 Ser Thr Met Ala Gly Met Ala Ser
Ala Asn Ala Phe Leu Gly Leu Cys 705 710
715 720 His Ser Met Ala Ile Lys Leu Ser Ser Glu His Asn
Ile Pro Ser Gly 725 730
735 Ile Ala Asn Ala Leu Leu Ile Glu Glu Val Ile Lys Phe Asn Ala Val
740 745 750 Asp Asn Pro
Val Lys Gln Ala Pro Cys Pro Gln Tyr Lys Tyr Pro Asn 755
760 765 Thr Ile Phe Arg Tyr Ala Arg Ile
Ala Asp Tyr Ile Lys Leu Gly Gly 770 775
780 Asn Thr Asp Glu Glu Lys Val Asp Leu Leu Ile Asn Lys
Ile His Glu 785 790 795
800 Leu Lys Lys Ala Leu Asn Ile Pro Thr Ser Ile Lys Asp Ala Gly Val
805 810 815 Leu Glu Glu Asn
Phe Tyr Ser Ser Leu Asp Arg Ile Ser Glu Leu Ala 820
825 830 Leu Asp Asp Gln Cys Thr Gly Ala Asn
Pro Arg Phe Pro Leu Thr Ser 835 840
845 Glu Ile Lys Glu Met Tyr Ile Asn Cys Phe Lys Lys Gln Pro
850 855 860
641665DNAClostridium acetobutylicum 64ttgaagagtg aatacacaat tggaagatat
ttgttagacc gtttatcaga gttgggtatt 60cggcatatct ttggtgtacc tggagattac
aatctatcct ttttagacta tataatggag 120tacaaaggga tagattgggt tggaaattgc
aatgaattga atgctgggta tgctgctgat 180ggatatgcaa gaataaatgg aattggagcc
atacttacaa catttggtgt tggagaatta 240agtgccatta acgcaattgc tggggcatac
gctgagcaag ttccagttgt taaaattaca 300ggtatcccca cagcaaaagt tagggacaat
ggattatatg tacaccacac attaggtgac 360ggaaggtttg atcacttttt tgaaatgttt
agagaagtaa cagttgctga ggcattacta 420agcgaagaaa atgcagcaca agaaattgat
cgtgttctta tttcatgctg gagacaaaaa 480cgtcctgttc ttataaattt accgattgat
gtatatgata aaccaattaa caaaccatta 540aagccattac tcgattatac tatttcaagt
aacaaagagg ctgcatgtga atttgttaca 600gaaatagtac ctataataaa tagggcaaaa
aagcctgtta ttcttgcaga ttatggagta 660tatcgttacc aagttcaaca tgtgcttaaa
aacttggccg aaaaaaccgg atttcctgtg 720gctacactaa gtatgggaaa aggtgttttc
aatgaagcac accctcaatt tattggtgtt 780tataatggtg atgtaagttc tccttattta
aggcagcgag ttgatgaagc agactgcatt 840attagcgttg gtgtaaaatt gacggattca
accacagggg gattttctca tggattttct 900aaaaggaatg taattcacat tgatcctttt
tcaataaagg caaaaggtaa aaaatatgca 960cctattacga tgaaagatgc tttaacagaa
ttaacaagta aaattgagca tagaaacttt 1020gaggatttag atataaagcc ttacaaatca
gataatcaaa agtattttgc aaaagagaag 1080ccaattacac aaaaacgttt ttttgagcgt
attgctcact ttataaaaga aaaagatgta 1140ttattagcag aacagggtac atgctttttt
ggtgcgtcaa ccatacaact acccaaagat 1200gcaactttta ttggtcaacc tttatgggga
tctattggat acacacttcc tgctttatta 1260ggttcacaat tagctgatca aaaaaggcgt
aatattcttt taattgggga tggtgcattt 1320caaatgacag cacaagaaat ttcaacaatg
cttcgtttac aaatcaaacc tattattttt 1380ttaattaata acgatggtta tacaattgaa
cgtgctattc atggtagaga acaagtatat 1440aacaatattc aaatgtggcg atatcataat
gttccaaagg ttttaggtcc taaagaatgc 1500agcttaacct ttaaagtaca aagtgaaact
gaacttgaaa aggctctttt agtggcagat 1560aaggattgtg aacatttgat ttttatagaa
gttgttatgg atcgttatga taaacccgag 1620cctttagaac gtctttcgaa acgttttgca
aatcaaaata attag 166565858PRTClostridium acetobutylicum
65Met Lys Val Thr Asn Gln Lys Glu Leu Lys Gln Lys Leu Asn Glu Leu 1
5 10 15 Arg Glu Ala Gln
Lys Lys Phe Ala Thr Tyr Thr Gln Glu Gln Val Asp 20
25 30 Lys Ile Phe Lys Gln Cys Ala Ile Ala
Ala Ala Lys Glu Arg Ile Asn 35 40
45 Leu Ala Lys Leu Ala Val Glu Glu Thr Gly Ile Gly Leu Val
Glu Asp 50 55 60
Lys Ile Ile Lys Asn His Phe Ala Ala Glu Tyr Ile Tyr Asn Lys Tyr 65
70 75 80 Lys Asn Glu Lys Thr
Cys Gly Ile Ile Asp His Asp Asp Ser Leu Gly 85
90 95 Ile Thr Lys Val Ala Glu Pro Ile Gly Ile
Val Ala Ala Ile Val Pro 100 105
110 Thr Thr Asn Pro Thr Ser Thr Ala Ile Phe Lys Ser Leu Ile Ser
Leu 115 120 125 Lys
Thr Arg Asn Ala Ile Phe Phe Ser Pro His Pro Arg Ala Lys Lys 130
135 140 Ser Thr Ile Ala Ala Ala
Lys Leu Ile Leu Asp Ala Ala Val Lys Ala 145 150
155 160 Gly Ala Pro Lys Asn Ile Ile Gly Trp Ile Asp
Glu Pro Ser Ile Glu 165 170
175 Leu Ser Gln Asp Leu Met Ser Glu Ala Asp Ile Ile Leu Ala Thr Gly
180 185 190 Gly Pro
Ser Met Val Lys Ala Ala Tyr Ser Ser Gly Lys Pro Ala Ile 195
200 205 Gly Val Gly Ala Gly Asn Thr
Pro Ala Ile Ile Asp Glu Ser Ala Asp 210 215
220 Ile Asp Met Ala Val Ser Ser Ile Ile Leu Ser Lys
Thr Tyr Asp Asn 225 230 235
240 Gly Val Ile Cys Ala Ser Glu Gln Ser Ile Leu Val Met Asn Ser Ile
245 250 255 Tyr Glu Lys
Val Lys Glu Glu Phe Val Lys Arg Gly Ser Tyr Ile Leu 260
265 270 Asn Gln Asn Glu Ile Ala Lys Ile
Lys Glu Thr Met Phe Lys Asn Gly 275 280
285 Ala Ile Asn Ala Asp Ile Val Gly Lys Ser Ala Tyr Ile
Ile Ala Lys 290 295 300
Met Ala Gly Ile Glu Val Pro Gln Thr Thr Lys Ile Leu Ile Gly Glu 305
310 315 320 Val Gln Ser Val
Glu Lys Ser Glu Leu Phe Ser His Glu Lys Leu Ser 325
330 335 Pro Val Leu Ala Met Tyr Lys Val Lys
Asp Phe Asp Glu Ala Leu Lys 340 345
350 Lys Ala Gln Arg Leu Ile Glu Leu Gly Gly Ser Gly His Thr
Ser Ser 355 360 365
Leu Tyr Ile Asp Ser Gln Asn Asn Lys Asp Lys Val Lys Glu Phe Gly 370
375 380 Leu Ala Met Lys Thr
Ser Arg Thr Phe Ile Asn Met Pro Ser Ser Gln 385 390
395 400 Gly Ala Ser Gly Asp Leu Tyr Asn Phe Ala
Ile Ala Pro Ser Phe Thr 405 410
415 Leu Gly Cys Gly Thr Trp Gly Gly Asn Ser Val Ser Gln Asn Val
Glu 420 425 430 Pro
Lys His Leu Leu Asn Ile Lys Ser Val Ala Glu Arg Arg Glu Asn 435
440 445 Met Leu Trp Phe Lys Val
Pro Gln Lys Ile Tyr Phe Lys Tyr Gly Cys 450 455
460 Leu Arg Phe Ala Leu Lys Glu Leu Lys Asp Met
Asn Lys Lys Arg Ala 465 470 475
480 Phe Ile Val Thr Asp Lys Asp Leu Phe Lys Leu Gly Tyr Val Asn Lys
485 490 495 Ile Thr
Lys Val Leu Asp Glu Ile Asp Ile Lys Tyr Ser Ile Phe Thr 500
505 510 Asp Ile Lys Ser Asp Pro Thr
Ile Asp Ser Val Lys Lys Gly Ala Lys 515 520
525 Glu Met Leu Asn Phe Glu Pro Asp Thr Ile Ile Ser
Ile Gly Gly Gly 530 535 540
Ser Pro Met Asp Ala Ala Lys Val Met His Leu Leu Tyr Glu Tyr Pro 545
550 555 560 Glu Ala Glu
Ile Glu Asn Leu Ala Ile Asn Phe Met Asp Ile Arg Lys 565
570 575 Arg Ile Cys Asn Phe Pro Lys Leu
Gly Thr Lys Ala Ile Ser Val Ala 580 585
590 Ile Pro Thr Thr Ala Gly Thr Gly Ser Glu Ala Thr Pro
Phe Ala Val 595 600 605
Ile Thr Asn Asp Glu Thr Gly Met Lys Tyr Pro Leu Thr Ser Tyr Glu 610
615 620 Leu Thr Pro Asn
Met Ala Ile Ile Asp Thr Glu Leu Met Leu Asn Met 625 630
635 640 Pro Arg Lys Leu Thr Ala Ala Thr Gly
Ile Asp Ala Leu Val His Ala 645 650
655 Ile Glu Ala Tyr Val Ser Val Met Ala Thr Asp Tyr Thr Asp
Glu Leu 660 665 670
Ala Leu Arg Ala Ile Lys Met Ile Phe Lys Tyr Leu Pro Arg Ala Tyr
675 680 685 Lys Asn Gly Thr
Asn Asp Ile Glu Ala Arg Glu Lys Met Ala His Ala 690
695 700 Ser Asn Ile Ala Gly Met Ala Phe
Ala Asn Ala Phe Leu Gly Val Cys 705 710
715 720 His Ser Met Ala His Lys Leu Gly Ala Met His His
Val Pro His Gly 725 730
735 Ile Ala Cys Ala Val Leu Ile Glu Glu Val Ile Lys Tyr Asn Ala Thr
740 745 750 Asp Cys Pro
Thr Lys Gln Thr Ala Phe Pro Gln Tyr Lys Ser Pro Asn 755
760 765 Ala Lys Arg Lys Tyr Ala Glu Ile
Ala Glu Tyr Leu Asn Leu Lys Gly 770 775
780 Thr Ser Asp Thr Glu Lys Val Thr Ala Leu Ile Glu Ala
Ile Ser Lys 785 790 795
800 Leu Lys Ile Asp Leu Ser Ile Pro Gln Asn Ile Ser Ala Ala Gly Ile
805 810 815 Asn Lys Lys Asp
Phe Tyr Asn Thr Leu Asp Lys Met Ser Glu Leu Ala 820
825 830 Phe Asp Asp Gln Cys Thr Thr Ala Asn
Pro Arg Tyr Pro Leu Ile Ser 835 840
845 Glu Leu Lys Asp Ile Tyr Ile Lys Ser Phe 850
855 662589DNAClostridium acetobutylicum 66atgaaagtca
caacagtaaa ggaattagat gaaaaactca aggtaattaa agaagctcaa 60aaaaaattct
cttgttactc gcaagaaatg gttgatgaaa tctttagaaa tgcagcaatg 120gcagcaatcg
acgcaaggat agagctagca aaagcagctg ttttggaaac cggtatgggc 180ttagttgaag
acaaggttat aaaaaatcat tttgcaggcg aatacatcta taacaaatat 240aaggatgaaa
aaacctgcgg tataattgaa cgaaatgaac cctacggaat tacaaaaata 300gcagaaccta
taggagttgt agctgctata atccctgtaa caaaccccac atcaacaaca 360atatttaaat
ccttaatatc ccttaaaact agaaatggaa ttttcttttc gcctcaccca 420agggcaaaaa
aatccacaat actagcagct aaaacaatac ttgatgcagc cgttaagagt 480ggtgccccgg
aaaatataat aggttggata gatgaacctt caattgaact aactcaatat 540ttaatgcaaa
aagcagatat aacccttgca actggtggtc cctcactagt taaatctgct 600tattcttccg
gaaaaccagc aataggtgtt ggtccgggta acaccccagt aataattgat 660gaatctgctc
atataaaaat ggcagtaagt tcaattatat tatccaaaac ctatgataat 720ggtgttatat
gtgcttctga acaatctgta atagtcttaa aatccatata taacaaggta 780aaagatgagt
tccaagaaag aggagcttat ataataaaga aaaacgaatt ggataaagtc 840cgtgaagtga
tttttaaaga tggatccgta aaccctaaaa tagtcggaca gtcagcttat 900actatagcag
ctatggctgg cataaaagta cctaaaacca caagaatatt aataggagaa 960gttacctcct
taggtgaaga agaacctttt gcccacgaaa aactatctcc tgttttggct 1020atgtatgagg
ctgacaattt tgatgatgct ttaaaaaaag cagtaactct aataaactta 1080ggaggcctcg
gccatacctc aggaatatat gcagatgaaa taaaagcacg agataaaata 1140gatagattta
gtagtgccat gaaaaccgta agaacctttg taaatatccc aacctcacaa 1200ggtgcaagtg
gagatctata taattttaga ataccacctt ctttcacgct tggctgcgga 1260ttttggggag
gaaattctgt ttccgagaat gttggtccaa aacatctttt gaatattaaa 1320accgtagctg
aaaggagaga aaacatgctt tggtttagag ttccacataa agtatatttt 1380aagttcggtt
gtcttcaatt tgctttaaaa gatttaaaag atctaaagaa aaaaagagcc 1440tttatagtta
ctgatagtga cccctataat ttaaactatg ttgattcaat aataaaaata 1500cttgagcacc
tagatattga ttttaaagta tttaataagg ttggaagaga agctgatctt 1560aaaaccataa
aaaaagcaac tgaagaaatg tcctccttta tgccagacac tataatagct 1620ttaggtggta
cccctgaaat gagctctgca aagctaatgt gggtactata tgaacatcca 1680gaagtaaaat
ttgaagatct tgcaataaaa tttatggaca taagaaagag aatatatact 1740ttcccaaaac
tcggtaaaaa ggctatgtta gttgcaatta caacttctgc tggttccggt 1800tctgaggtta
ctccttttgc tttagtaact gacaataaca ctggaaataa gtacatgtta 1860gcagattatg
aaatgacacc aaatatggca attgtagatg cagaacttat gatgaaaatg 1920ccaaagggat
taaccgctta ttcaggtata gatgcactag taaatagtat agaagcatac 1980acatccgtat
atgcttcaga atacacaaac ggactagcac tagaggcaat acgattaata 2040tttaaatatt
tgcctgaggc ttacaaaaac ggaagaacca atgaaaaagc aagagagaaa 2100atggctcacg
cttcaactat ggcaggtatg gcatccgcta atgcatttct aggtctatgt 2160cattccatgg
caataaaatt aagttcagaa cacaatattc ctagtggcat tgccaatgca 2220ttactaatag
aagaagtaat aaaatttaac gcagttgata atcctgtaaa acaagcccct 2280tgcccacaat
ataagtatcc aaacaccata tttagatatg ctcgaattgc agattatata 2340aagcttggag
gaaatactga tgaggaaaag gtagatctct taattaacaa aatacatgaa 2400ctaaaaaaag
ctttaaatat accaacttca ataaaggatg caggtgtttt ggaggaaaac 2460ttctattcct
cccttgatag aatatctgaa cttgcactag atgatcaatg cacaggcgct 2520aatcctagat
ttcctcttac aagtgagata aaagaaatgt atataaattg ttttaaaaaa 2580caaccttaa
258967307PRTPseudomonas putida 67Met Ser Lys Lys Leu Lys Ala Ala Ile Ile
Gly Pro Gly Asn Ile Gly 1 5 10
15 Thr Asp Leu Val Met Lys Met Leu Arg Ser Glu Trp Ile Glu Pro
Val 20 25 30 Trp
Met Val Gly Ile Asp Pro Asn Ser Asp Gly Leu Lys Arg Ala Arg 35
40 45 Asp Phe Gly Met Lys Thr
Thr Ala Glu Gly Val Asp Gly Leu Leu Pro 50 55
60 His Val Leu Asp Asp Asp Ile Arg Ile Ala Phe
Asp Ala Thr Ser Ala 65 70 75
80 Tyr Val His Ala Glu Asn Ser Arg Lys Leu Asn Ala Leu Gly Val Leu
85 90 95 Met Val
Asp Leu Thr Pro Ala Ala Ile Gly Pro Tyr Cys Val Pro Pro 100
105 110 Val Asn Leu Lys Gln His Val
Gly Arg Leu Glu Met Asn Val Asn Met 115 120
125 Val Thr Cys Gly Gly Gln Ala Thr Ile Pro Met Val
Ala Ala Val Ser 130 135 140
Arg Val Gln Pro Val Ala Tyr Ala Glu Ile Val Ala Thr Val Ser Ser 145
150 155 160 Arg Ser Val
Gly Pro Gly Thr Arg Lys Asn Ile Asp Glu Phe Thr Arg 165
170 175 Thr Thr Ala Gly Ala Ile Glu Gln
Val Gly Gly Ala Arg Glu Gly Lys 180 185
190 Ala Ile Ile Val Ile Asn Pro Ala Glu Pro Pro Leu Met
Met Arg Asp 195 200 205
Thr Ile His Cys Leu Thr Asp Ser Glu Pro Asp Gln Ala Ala Ile Thr 210
215 220 Ala Ser Val His
Ala Met Ile Ala Glu Val Gln Lys Tyr Val Pro Gly 225 230
235 240 Tyr Arg Leu Lys Asn Gly Pro Val Phe
Asp Gly Asn Arg Val Ser Ile 245 250
255 Phe Met Glu Val Glu Gly Leu Gly Asp Tyr Leu Pro Lys Tyr
Ala Gly 260 265 270
Asn Leu Asp Ile Met Thr Ala Ala Ala Leu Arg Thr Gly Glu Met Phe
275 280 285 Ala Glu Glu Ile
Ala Ala Gly Thr Ile Gln Leu Pro Arg Arg Asp Ile 290
295 300 Ala Leu Ala 305
682180DNAPseudomonas putida 68ggtacccctg gagccggtca aggccggcga cttcatgcgc
gtcgagatcg gcggcatcgg 60cagcgcctcc gtgcgcttca cctgatcgaa cagaggacaa
acccatgagc aagaaactca 120aggcggccat cataggcccc ggcaatatcg gtaccgatct
ggtgatgaag atgctccgtt 180ccgagtggat tgagccggtg tggatggtcg gcatcgaccc
caactccgac ggcctcaaac 240gcgcccgcga tttcggcatg aagaccacag ccgaaggcgt
cgacggcctg ctcccgcacg 300tgctggacga cgacatccgc atcgccttcg acgccacctc
ggcctatgtg catgccgaga 360atagccgcaa gctcaacgcg cttggcgtgc tgatggtcga
cctgaccccg gcggccatcg 420gcccctactg cgtgccgccg gtcaacctca agcagcatgt
cggccgcctg gaaatgaacg 480tcaacatggt cacctgcggc ggccaggcca ccatccccat
ggtcgccgcg gtgtcccgcg 540tgcagccggt ggcctacgcc gagatcgtcg ccaccgtctc
ctcgcgctcg gtcggcccgg 600gcacgcgcaa gaacatcgac gagttcaccc gcaccaccgc
cggcgccatc gagcaggtcg 660gcggcgccag ggaaggcaag gcgatcatcg tcatcaaccc
ggccgagccg ccgctgatga 720tgcgcgacac catccactgc ctgaccgaca gcgagccgga
ccaggctgcg atcaccgctt 780cggttcacgc gatgatcgcc gaggtgcaga aatacgtgcc
cggctaccgc ctgaagaacg 840gcccggtgtt cgacggcaac cgcgtgtcga tcttcatgga
agtcgaaggc ctgggcgact 900acctgcccaa gtacgccggc aacctcgaca tcatgaccgc
cgccgcgctg cgtaccggcg 960agatgttcgc cgaggaaatc gccgccggca ccattcaact
gccgcgtcgc gacatcgcgc 1020tggcttgagg agtagcacca tgaatttgca cggcaagagc
gtcatcctgc acgacatgag 1080cctgcgcgac ggcatgcacg ccaagcgcca ccagatcagc
ctggagcaga tggtcgcggt 1140cgccaccggc ctcgatcaag ccggtatgcc gctgatcgag
atcacccacg gcgacggcct 1200cggcggtcgt tcgatcaact acggcttccc ggcccacagt
gacgaggagt acctgcgcgc 1260ggtgatcccg cagctcaagc aggccaaagt ctcggcgctg
ctgctgcccg gcatcggcac 1320cgtcgaccac ctgaagatgg ccctggactg cggcgtctcg
actattcgcg tggccaccca 1380ctgtaccgag gcggatgtct ccgagcagca catcggcatg
gcgcgcaagc tgggggtcga 1440caccgtcggc ttcctgatga tggcgcacat gatcagcgcc
gagaaagtcc tggagcaggc 1500caagctgatg gaaagctatg gtgccaactg catctactgc
accgactcgg ccggctacat 1560gctgcctgat gaagtcagcg agaaaatcgg cctcctgcgc
gccgagctga acccggccac 1620cgaagtcggc ttccacggcc accacaacat gggcatggct
atcgccaact cgctggccgc 1680catcgaagcc ggtgccgcgc gcatcgacgg ctcggtcgcc
ggcctcggcg ccggtgccgg 1740caacaccccg ctggaagtgt tcgtcgcagt gtgcaaacgc
atgggcgtgg agaccggcat 1800cgacctgtac aagatcatgg acgtggccga ggacctggtg
gtgccgatga tggatcagcc 1860gatccgcgtc gaccgcgacg ccctgaccct gggctacgcc
ggggtgtaca gctcgttcct 1920gctgttcgcc cagcgcgccg agaagaaata tggcgtgtcg
gcccgcgaca tcctggtcga 1980actgggccgg cgcggcaccg tcggtggcca ggaagacatg
atcgaagacc tcgccctgga 2040catggcccgg gcccgtcagc agcagaaggt gagcgcatga
accgtaccct gacccgcgaa 2100caggtgctgg ccctggccga gcacatcgaa aacgccgagc
tgaatgtcca cgacatcggc 2160aaggtgacca acgattttcc
218069307PRTThermus thermophilus 69Met Ser Glu Arg
Val Lys Val Ala Ile Leu Gly Ser Gly Asn Ile Gly 1 5
10 15 Thr Asp Leu Met Tyr Lys Leu Leu Lys
Asn Pro Gly His Met Glu Leu 20 25
30 Val Ala Val Val Gly Ile Asp Pro Lys Ser Glu Gly Leu Ala
Arg Ala 35 40 45
Arg Ala Leu Gly Leu Glu Ala Ser His Glu Gly Ile Ala Tyr Ile Leu 50
55 60 Glu Arg Pro Glu Ile
Lys Ile Val Phe Asp Ala Thr Ser Ala Lys Ala 65 70
75 80 His Val Arg His Ala Lys Leu Leu Arg Glu
Ala Gly Lys Ile Ala Ile 85 90
95 Asp Leu Thr Pro Ala Ala Arg Gly Pro Tyr Val Val Pro Pro Val
Asn 100 105 110 Leu
Lys Glu His Leu Asp Lys Asp Asn Val Asn Leu Ile Thr Cys Gly 115
120 125 Gly Gln Ala Thr Ile Pro
Leu Val Tyr Ala Val His Arg Val Ala Pro 130 135
140 Val Leu Tyr Ala Glu Met Val Ser Thr Val Ala
Ser Arg Ser Ala Gly 145 150 155
160 Pro Gly Thr Arg Gln Asn Ile Asp Glu Phe Thr Phe Thr Thr Ala Arg
165 170 175 Gly Leu
Glu Ala Ile Gly Gly Ala Lys Lys Gly Lys Ala Ile Ile Ile 180
185 190 Leu Asn Pro Ala Glu Pro Pro
Ile Leu Met Thr Asn Thr Val Arg Cys 195 200
205 Ile Pro Glu Asp Glu Gly Phe Asp Arg Glu Ala Val
Val Ala Ser Val 210 215 220
Arg Ala Met Glu Arg Glu Val Gln Ala Tyr Val Pro Gly Tyr Arg Leu 225
230 235 240 Lys Ala Asp
Pro Val Phe Glu Arg Leu Pro Thr Pro Trp Gly Glu Arg 245
250 255 Thr Val Val Ser Met Leu Leu Glu
Val Glu Gly Ala Gly Asp Tyr Leu 260 265
270 Pro Lys Tyr Ala Gly Asn Leu Asp Ile Met Thr Ala Ser
Ala Arg Arg 275 280 285
Val Gly Glu Val Phe Ala Gln His Leu Leu Gly Lys Pro Val Glu Glu 290
295 300 Val Val Ala 305
70924DNAThermus thermophilus 70atgtccgaaa gggttaaggt agccatcctg
ggctccggca acatcgggac ggacctgatg 60tacaagctcc tgaagaaccc gggccacatg
gagcttgtgg cggtggtggg gatagacccc 120aagtccgagg gcctggcccg ggcgcgggcc
ttagggttag aggcgagcca cgaagggatc 180gcctacatcc tggagaggcc ggagatcaag
atcgtctttg acgccaccag cgccaaggcc 240cacgtgcgcc acgccaagct cctgagggag
gcggggaaga tcgccataga cctcacgccg 300gcggcccggg gcccttacgt ggtgcccccg
gtgaacctga aggaacacct ggacaaggac 360aacgtgaacc tcatcacctg cggggggcag
gccaccatcc ccctggtcta cgcggtgcac 420cgggtggccc ccgtgctcta cgcggagatg
gtctccacgg tggcctcccg ctccgcgggc 480cccggcaccc ggcagaacat cgacgagttc
accttcacca ccgcccgggg cctggaggcc 540atcggggggg ccaagaaggg gaaggccatc
atcatcctga acccggcgga accccccatc 600ctcatgacca acaccgtgcg ctgcatcccc
gaggacgagg gctttgaccg ggaggccgtg 660gtggcgagcg tccgggccat ggagcgggag
gtccaggcct acgtgcccgg ctaccgcctg 720aaggcggacc cggtgtttga gaggcttccc
accccctggg gggagcgcac cgtggtctcc 780atgctcctgg aggtggaggg ggcgggggac
tatttgccca aatacgccgg caacctggac 840atcatgacgg cttctgcccg gagggtgggg
gaggtcttcg cccagcacct cctggggaag 900cccgtggagg aggtggtggc gtga
92471417PRTEscherichia coli 71Met Thr
Phe Ser Leu Phe Gly Asp Lys Phe Thr Arg His Ser Gly Ile 1 5
10 15 Thr Leu Leu Met Glu Asp Leu
Asn Asp Gly Leu Arg Thr Pro Gly Ala 20 25
30 Ile Met Leu Gly Gly Gly Asn Pro Ala Gln Ile Pro
Glu Met Gln Asp 35 40 45
Tyr Phe Gln Thr Leu Leu Thr Asp Met Leu Glu Ser Gly Lys Ala Thr
50 55 60 Asp Ala Leu
Cys Asn Tyr Asp Gly Pro Gln Gly Lys Thr Glu Leu Leu 65
70 75 80 Thr Leu Leu Ala Gly Met Leu
Arg Glu Lys Leu Gly Trp Asp Ile Glu 85
90 95 Pro Gln Asn Ile Ala Leu Thr Asn Gly Ser Gln
Ser Ala Phe Phe Tyr 100 105
110 Leu Phe Asn Leu Phe Ala Gly Arg Arg Ala Asp Gly Arg Val Lys
Lys 115 120 125 Val
Leu Phe Pro Leu Ala Pro Glu Tyr Ile Gly Tyr Ala Asp Ala Gly 130
135 140 Leu Glu Glu Asp Leu Phe
Val Ser Ala Arg Pro Asn Ile Glu Leu Leu 145 150
155 160 Pro Glu Gly Gln Phe Lys Tyr His Val Asp Phe
Glu His Leu His Ile 165 170
175 Gly Glu Glu Thr Gly Met Ile Cys Val Ser Arg Pro Thr Asn Pro Thr
180 185 190 Gly Asn
Val Ile Thr Asp Glu Glu Leu Leu Lys Leu Asp Ala Leu Ala 195
200 205 Asn Gln His Gly Ile Pro Leu
Val Ile Asp Asn Ala Tyr Gly Val Pro 210 215
220 Phe Pro Gly Ile Ile Phe Ser Glu Ala Arg Pro Leu
Trp Asn Pro Asn 225 230 235
240 Ile Val Leu Cys Met Ser Leu Ser Lys Leu Gly Leu Pro Gly Ser Arg
245 250 255 Cys Gly Ile
Ile Ile Ala Asn Glu Lys Ile Ile Thr Ala Ile Thr Asn 260
265 270 Met Asn Gly Ile Ile Ser Leu Ala
Pro Gly Gly Ile Gly Pro Ala Met 275 280
285 Met Cys Glu Met Ile Lys Arg Asn Asp Leu Leu Arg Leu
Ser Glu Thr 290 295 300
Val Ile Lys Pro Phe Tyr Tyr Gln Arg Val Gln Glu Thr Ile Ala Ile 305
310 315 320 Ile Arg Arg Tyr
Leu Pro Glu Asn Arg Cys Leu Ile His Lys Pro Glu 325
330 335 Gly Ala Ile Phe Leu Trp Leu Trp Phe
Lys Asp Leu Pro Ile Thr Thr 340 345
350 Lys Gln Leu Tyr Gln Arg Leu Lys Ala Arg Gly Val Leu Met
Val Pro 355 360 365
Gly His Asn Phe Phe Pro Gly Leu Asp Lys Pro Trp Pro His Thr His 370
375 380 Gln Cys Met Arg Met
Asn Tyr Val Pro Glu Pro Glu Lys Ile Glu Ala 385 390
395 400 Gly Val Lys Ile Leu Ala Glu Glu Ile Glu
Arg Ala Trp Ala Glu Ser 405 410
415 His 72417PRTEscherichia coli 72Met Thr Phe Ser Leu Phe Gly
Asp Lys Phe Thr Arg His Ser Gly Ile 1 5
10 15 Thr Leu Leu Met Glu Asp Leu Asn Asp Gly Leu
Arg Thr Pro Gly Ala 20 25
30 Ile Met Leu Gly Gly Gly Asn Pro Ala Gln Ile Pro Glu Met Gln
Asp 35 40 45 Tyr
Phe Gln Thr Leu Leu Thr Asp Met Leu Glu Ser Gly Lys Ala Thr 50
55 60 Asp Ala Leu Cys Asn Tyr
Asp Gly Pro Gln Gly Lys Thr Glu Leu Leu 65 70
75 80 Thr Leu Leu Ala Gly Met Leu Arg Glu Lys Leu
Gly Trp Asp Ile Glu 85 90
95 Pro Gln Asn Ile Ala Leu Thr Asn Gly Ser Gln Ser Ala Phe Phe Tyr
100 105 110 Leu Phe
Asn Leu Phe Ala Gly Arg Arg Ala Asp Gly Arg Val Lys Lys 115
120 125 Val Leu Phe Pro Leu Ala Pro
Glu Tyr Ile Gly Tyr Ala Asp Ala Gly 130 135
140 Leu Glu Glu Asp Leu Phe Val Ser Ala Arg Pro Asn
Ile Glu Leu Leu 145 150 155
160 Pro Glu Gly Gln Phe Lys Tyr His Val Asp Phe Glu His Leu His Ile
165 170 175 Gly Glu Glu
Thr Gly Met Ile Cys Val Ser Arg Pro Thr Asn Pro Thr 180
185 190 Gly Asn Val Ile Thr Asp Glu Glu
Leu Leu Lys Leu Asp Ala Leu Ala 195 200
205 Asn Gln His Gly Ile Pro Leu Val Ile Asp Asn Ala Tyr
Gly Val Pro 210 215 220
Phe Pro Gly Ile Ile Phe Ser Glu Ala Arg Pro Leu Trp Asn Pro Asn 225
230 235 240 Ile Val Leu Cys
Met Ser Leu Ser Lys Leu Gly Leu Pro Gly Ser Arg 245
250 255 Cys Gly Ile Ile Ile Ala Asn Glu Lys
Ile Ile Thr Ala Ile Thr Asn 260 265
270 Met Asn Gly Ile Ile Ser Leu Ala Pro Gly Gly Ile Gly Pro
Ala Met 275 280 285
Met Cys Glu Met Ile Lys Arg Asn Asp Leu Leu Arg Leu Ser Glu Thr 290
295 300 Val Ile Lys Pro Phe
Tyr Tyr Gln Arg Val Gln Glu Thr Ile Ala Ile 305 310
315 320 Ile Arg Arg Tyr Leu Pro Glu Asn Arg Cys
Leu Ile His Lys Pro Glu 325 330
335 Gly Ala Ile Phe Leu Trp Leu Trp Phe Lys Asp Leu Pro Ile Thr
Thr 340 345 350 Lys
Gln Leu Tyr Gln Arg Leu Lys Ala Arg Gly Val Leu Met Val Pro 355
360 365 Gly His Asn Phe Phe Pro
Gly Leu Asp Lys Pro Trp Pro His Thr His 370 375
380 Gln Cys Met Arg Met Asn Tyr Val Pro Glu Pro
Glu Lys Ile Glu Ala 385 390 395
400 Gly Val Lys Ile Leu Ala Glu Glu Ile Glu Arg Ala Trp Ala Glu Ser
405 410 415 His
73425PRTBacillus licheniformis 73Met Lys Pro Pro Leu Ser Lys Ile Gly Glu
Lys Met Ile Glu Lys Thr 1 5 10
15 Gly Val Arg Ala Val Met Ser Asp Ile Gln Glu Val Leu Ala Gly
Gly 20 25 30 Glu
Arg Ser Tyr Ile Asn Leu Ser Ala Gly Asn Pro Met Ile Leu Pro 35
40 45 Gly Val Ser Ala Met Trp
Lys Ser Ala Leu Ala Asp Leu Leu Asp Asp 50 55
60 Asp Arg Phe Ser Ser Val Ile Gly Gln Tyr Gly
Ser Ser Tyr Gly Thr 65 70 75
80 Asp Glu Leu Ile Ala Ser Val Val Arg Phe Phe Ser Glu Arg Tyr Ser
85 90 95 Ala Gly
Ile Arg Lys Glu Asn Val Leu Ile Thr Ala Gly Ser Gln Gln 100
105 110 Leu Phe Phe Leu Ala Ile Asn
Ser Phe Cys Gly Met Gly Ser Gly Ser 115 120
125 Val Met Lys Lys Ala Leu Ile Pro Met Leu Pro Asp
Tyr Ser Gly Tyr 130 135 140
Ser Gly Ala Ala Leu Glu Arg Glu Met Ile Glu Gly Ile Pro Pro Leu 145
150 155 160 Ile Ser Lys
Leu Asp Asp His Thr Phe Arg Tyr Glu Leu Asp Arg Lys 165
170 175 Gly Phe Leu Glu Arg Met Arg Ile
Gly Ala Val Leu Leu Ser Arg Pro 180 185
190 Asn Asn Pro Cys Gly Asn Ile Leu Pro Lys Glu Asp Val
Ala Phe Ile 195 200 205
Ser Asp Ala Cys Arg Glu Ala Asn Val Pro Leu Phe Ile Asp Ser Ala 210
215 220 Tyr Ala Pro Pro
Phe Pro Ala Ile His Phe Ile Asp Met Glu Pro Ile 225 230
235 240 Phe Asn Glu Gln Ile Ile His Cys Met
Ser Leu Ser Lys Ala Gly Leu 245 250
255 Pro Gly Glu Arg Ile Gly Ile Ala Ile Gly Pro Ser Arg Tyr
Ile Gln 260 265 270
Ala Met Glu Ala Phe Gln Ser Asn Ala Ala Ile His Ser Ser Arg Leu
275 280 285 Gly Gln Tyr Met
Ala Ala Ser Val Leu Asn Asp Gly Arg Leu Ala Asp 290
295 300 Val Ser Leu Asn Glu Val Arg Pro
Tyr Tyr Arg Asn Lys Phe Met Leu 305 310
315 320 Leu Lys Glu Thr Leu Leu Cys Lys Met Pro Glu Asp
Ile Lys Trp Tyr 325 330
335 Leu His Gln Gly Glu Gly Ser Leu Phe Gly Trp Leu Trp Phe Glu Asp
340 345 350 Leu Pro Val
Thr Asp Ala Ala Leu Tyr Glu Tyr Met Lys Ala Asp Gly 355
360 365 Val Ile Ile Val Pro Gly Ser Ser
Phe Phe His Arg Gln Ser Arg Arg 370 375
380 Leu Ala His Ser His Gln Cys Ile Arg Ile Ser Leu Thr
Ala Ala Asp 385 390 395
400 Glu Asp Ile Ile Arg Gly Ile Asp Val Leu Ala Lys Ile Ala Lys Gly
405 410 415 Val Tyr Glu Lys
Gln Val Glu Tyr Leu 420 425 741278DNABacillus
licheniformis 74ttataagtat tcaacctgtt tctcatatac acccttcgca attttagcta
aaacatcgat 60tccccttata atatcttcat ccgccgcggt taggctgatt cgtatacact
ggtgtgaatg 120cgccaggcgc cgggattgac ggtgaaagaa agatgatccg ggaacgataa
tgactccatc 180cgctttcata tactcataca gcgctgcatc ggtcaccggc aggtcttcaa
accacagcca 240tccgaaaagc gatccttccc cttgatgcag ataccatttg atgtcttcag
gcatcttgca 300taaaagcgtt tccttgagca gcatgaattt attgcggtaa tatggcctga
cttcattcag 360cgacacgtcg gcgaggcgcc cgtcattcaa tactgatgca gccatatact
gccccagcct 420tgaagaatgg atcgccgcat tcgactgaaa agcttccatt gcctgaatat
accgggacgg 480cccgatggcg attccgatcc tttcgccagg caggccggct tttgaaaggc
tcatacagtg 540aatgatctgc tcgttgaaaa tcggttccat gtcgataaag tgaatcgccg
gaaaaggcgg 600agcatatgcg gaatcaatga acagcggaac attcgcttct cggcatgcgt
ctgaaatgaa 660tgctacatct tctttaggca agatgtttcc gcaaggattg ttcgggcgcg
atagcaagac 720agcaccgatg cgcatcctct ctaaaaaccc cttacggtcg agctcatatc
gaaacgtatg 780atcatccaat ttcgatatga gcggagggat cccctcaatc atctcccgct
ccagtgccgc 840cccgctgtat cccgaatagt caggcagcat cgggatcaag gcttttttca
tcacagatcc 900gcttcccatt ccgcaaaacg aattgatcgc cagaaaaaac agctgctggc
ttccggctgt 960aatcaacacg ttctcttttc gaatgccggc gctataccgc tctgaaaaga
agcggacaac 1020acttgcaatc agttcatcgg ttccatagct cgatccgtat tggccgatca
ccgaagaaaa 1080cctgtcatcg tcaaggagat cggcaagagc cgacttccac atggctgaca
cgccgggcaa 1140aatcatcgga ttgcccgcac ttaaattaat gtatgaccgt tcaccgccgg
ccaggacttc 1200ctgaatatcg ctcatcacag ccctgacccc tgttttctca atcattttct
ctccgatttt 1260gcttaatggc ggcttcac
127875309PRTEscherichia coli 75Met Thr Thr Lys Lys Ala Asp Tyr
Ile Trp Phe Asn Gly Glu Met Val 1 5 10
15 Arg Trp Glu Asp Ala Lys Val His Val Met Ser His Ala
Leu His Tyr 20 25 30
Gly Thr Ser Val Phe Glu Gly Ile Arg Cys Tyr Asp Ser His Lys Gly
35 40 45 Pro Val Val Phe
Arg His Arg Glu His Met Gln Arg Leu His Asp Ser 50
55 60 Ala Lys Ile Tyr Arg Phe Pro Val
Ser Gln Ser Ile Asp Glu Leu Met 65 70
75 80 Glu Ala Cys Arg Asp Val Ile Arg Lys Asn Asn Leu
Thr Ser Ala Tyr 85 90
95 Ile Arg Pro Leu Ile Phe Val Gly Asp Val Gly Met Gly Val Asn Pro
100 105 110 Pro Ala Gly
Tyr Ser Thr Asp Val Ile Ile Ala Ala Phe Pro Trp Gly 115
120 125 Ala Tyr Leu Gly Ala Glu Ala Leu
Glu Gln Gly Ile Asp Ala Met Val 130 135
140 Ser Ser Trp Asn Arg Ala Ala Pro Asn Thr Ile Pro Thr
Ala Ala Lys 145 150 155
160 Ala Gly Gly Asn Tyr Leu Ser Ser Leu Leu Val Gly Ser Glu Ala Arg
165 170 175 Arg His Gly Tyr
Gln Glu Gly Ile Ala Leu Asp Val Asn Gly Tyr Ile 180
185 190 Ser Glu Gly Ala Gly Glu Asn Leu Phe
Glu Val Lys Asp Gly Val Leu 195 200
205 Phe Thr Pro Pro Phe Thr Ser Ser Ala Leu Pro Gly Ile Thr
Arg Asp 210 215 220
Ala Ile Ile Lys Leu Ala Lys Glu Leu Gly Ile Glu Val Arg Glu Gln 225
230 235 240 Val Leu Ser Arg Glu
Ser Leu Tyr Leu Ala Asp Glu Val Phe Met Ser 245
250 255 Gly Thr Ala Ala Glu Ile Thr Pro Val Arg
Ser Val Asp Gly Ile Gln 260 265
270 Val Gly Glu Gly Arg Cys Gly Pro Val Thr Lys Arg Ile Gln Gln
Ala 275 280 285 Phe
Phe Gly Leu Phe Thr Gly Glu Thr Glu Asp Lys Trp Gly Trp Leu 290
295 300 Asp Gln Val Asn Gln 305
761476DNAEscherichia coli 76atggctaact acttcaatac
actgaatctg cgccagcagc tggcacagct gggcaaatgt 60cgctttatgg gccgcgatga
attcgccgat ggcgcgagct accttcaggg taaaaaagta 120gtcatcgtcg gctgtggcgc
acagggtctg aaccagggcc tgaacatgcg tgattctggt 180ctcgatatct cctacgctct
gcgtaaagaa gcgattgccg agaagcgcgc gtcctggcgt 240aaagcgaccg aaaatggttt
taaagtgggt acttacgaag aactgatccc acaggcggat 300ctggtgatta acctgacgcc
ggacaagcag cactctgatg tagtgcgcac cgtacagcca 360ctgatgaaag acggcgcggc
gctgggctac tcgcacggtt tcaacatcgt cgaagtgggc 420gagcagatcc gtaaagatat
caccgtagtg atggttgcgc cgaaatgccc aggcaccgaa 480gtgcgtgaag agtacaaacg
tgggttcggc gtaccgacgc tgattgccgt tcacccggaa 540aacgatccga aaggcgaagg
catggcgatt gccaaagcct gggcggctgc aaccggtggt 600caccgtgcgg gtgtgctgga
atcgtccttc gttgcggaag tgaaatctga cctgatgggc 660gagcaaacca tcctgtgcgg
tatgttgcag gctggctctc tgctgtgctt cgacaagctg 720gtggaagaag gtaccgatcc
agcatacgca gaaaaactga ttcagttcgg ttgggaaacc 780atcaccgaag cactgaaaca
gggcggcatc accctgatga tggaccgtct ctctaacccg 840gcgaaactgc gtgcttatgc
gctttctgaa cagctgaaag agatcatggc acccctgttc 900cagaaacata tggacgacat
catctccggc gaattctctt ccggtatgat ggcggactgg 960gccaacgatg ataagaaact
gctgacctgg cgtgaagaga ccggcaaaac cgcgtttgaa 1020accgcgccgc agtatgaagg
caaaatcggc gagcaggagt acttcgataa aggcgtactg 1080atgattgcga tggtgaaagc
gggcgttgaa ctggcgttcg aaaccatggt cgattccggc 1140atcattgaag agtctgcata
ttatgaatca ctgcacgagc tgccgctgat tgccaacacc 1200atcgcccgta agcgtctgta
cgaaatgaac gtggttatct ctgataccgc tgagtacggt 1260aactatctgt tctcttacgc
ttgtgtgccg ttgctgaaac cgtttatggc agagctgcaa 1320ccgggcgacc tgggtaaagc
tattccggaa ggcgcggtag ataacgggca actgcgtgat 1380gtgaacgaag cgattcgcag
ccatgcgatt gagcaggtag gtaagaaact gcgcggctat 1440atgacagata tgaaacgtat
tgctgttgcg ggttaa 147677376PRTSaccharomyces
cerevisiae 77Met Thr Leu Ala Pro Leu Asp Ala Ser Lys Val Lys Ile Thr Thr
Thr 1 5 10 15 Gln
His Ala Ser Lys Pro Lys Pro Asn Ser Glu Leu Val Phe Gly Lys
20 25 30 Ser Phe Thr Asp His
Met Leu Thr Ala Glu Trp Thr Ala Glu Lys Gly 35
40 45 Trp Gly Thr Pro Glu Ile Lys Pro Tyr
Gln Asn Leu Ser Leu Asp Pro 50 55
60 Ser Ala Val Val Phe His Tyr Ala Phe Glu Leu Phe Glu
Gly Met Lys 65 70 75
80 Ala Tyr Arg Thr Val Asp Asn Lys Ile Thr Met Phe Arg Pro Asp Met
85 90 95 Asn Met Lys Arg
Met Asn Lys Ser Ala Gln Arg Ile Cys Leu Pro Thr 100
105 110 Phe Asp Pro Glu Glu Leu Ile Thr Leu
Ile Gly Lys Leu Ile Gln Gln 115 120
125 Asp Lys Cys Leu Val Pro Glu Gly Lys Gly Tyr Ser Leu Tyr
Ile Arg 130 135 140
Pro Thr Leu Ile Gly Thr Thr Ala Gly Leu Gly Val Ser Thr Pro Asp 145
150 155 160 Arg Ala Leu Leu Tyr
Val Ile Cys Cys Pro Val Gly Pro Tyr Tyr Lys 165
170 175 Thr Gly Phe Lys Ala Val Arg Leu Glu Ala
Thr Asp Tyr Ala Thr Arg 180 185
190 Ala Trp Pro Gly Gly Cys Gly Asp Lys Lys Leu Gly Ala Asn Tyr
Ala 195 200 205 Pro
Cys Val Leu Pro Gln Leu Gln Ala Ala Ser Arg Gly Tyr Gln Gln 210
215 220 Asn Leu Trp Leu Phe Gly
Pro Asn Asn Asn Ile Thr Glu Val Gly Thr 225 230
235 240 Met Asn Ala Phe Phe Val Phe Lys Asp Ser Lys
Thr Gly Lys Lys Glu 245 250
255 Leu Val Thr Ala Pro Leu Asp Gly Thr Ile Leu Glu Gly Val Thr Arg
260 265 270 Asp Ser
Ile Leu Asn Leu Ala Lys Glu Arg Leu Glu Pro Ser Glu Trp 275
280 285 Thr Ile Ser Glu Arg Tyr Phe
Thr Ile Gly Glu Val Thr Glu Arg Ser 290 295
300 Lys Asn Gly Glu Leu Leu Glu Ala Phe Gly Ser Gly
Thr Ala Ala Ile 305 310 315
320 Val Ser Pro Ile Lys Glu Ile Gly Trp Lys Gly Glu Gln Ile Asn Ile
325 330 335 Pro Leu Leu
Pro Gly Glu Gln Thr Gly Pro Leu Ala Lys Glu Val Ala 340
345 350 Gln Trp Ile Asn Gly Ile Gln Tyr
Gly Glu Thr Glu His Gly Asn Trp 355 360
365 Ser Arg Val Val Thr Asp Leu Asn 370
375 78376PRTSaccharomyces cerevisiae 78Met Thr Leu Ala Pro Leu
Asp Ala Ser Lys Val Lys Ile Thr Thr Thr 1 5
10 15 Gln His Ala Ser Lys Pro Lys Pro Asn Ser Glu
Leu Val Phe Gly Lys 20 25
30 Ser Phe Thr Asp His Met Leu Thr Ala Glu Trp Thr Ala Glu Lys
Gly 35 40 45 Trp
Gly Thr Pro Glu Ile Lys Pro Tyr Gln Asn Leu Ser Leu Asp Pro 50
55 60 Ser Ala Val Val Phe His
Tyr Ala Phe Glu Leu Phe Glu Gly Met Lys 65 70
75 80 Ala Tyr Arg Thr Val Asp Asn Lys Ile Thr Met
Phe Arg Pro Asp Met 85 90
95 Asn Met Lys Arg Met Asn Lys Ser Ala Gln Arg Ile Cys Leu Pro Thr
100 105 110 Phe Asp
Pro Glu Glu Leu Ile Thr Leu Ile Gly Lys Leu Ile Gln Gln 115
120 125 Asp Lys Cys Leu Val Pro Glu
Gly Lys Gly Tyr Ser Leu Tyr Ile Arg 130 135
140 Pro Thr Leu Ile Gly Thr Thr Ala Gly Leu Gly Val
Ser Thr Pro Asp 145 150 155
160 Arg Ala Leu Leu Tyr Val Ile Cys Cys Pro Val Gly Pro Tyr Tyr Lys
165 170 175 Thr Gly Phe
Lys Ala Val Arg Leu Glu Ala Thr Asp Tyr Ala Thr Arg 180
185 190 Ala Trp Pro Gly Gly Cys Gly Asp
Lys Lys Leu Gly Ala Asn Tyr Ala 195 200
205 Pro Cys Val Leu Pro Gln Leu Gln Ala Ala Ser Arg Gly
Tyr Gln Gln 210 215 220
Asn Leu Trp Leu Phe Gly Pro Asn Asn Asn Ile Thr Glu Val Gly Thr 225
230 235 240 Met Asn Ala Phe
Phe Val Phe Lys Asp Ser Lys Thr Gly Lys Lys Glu 245
250 255 Leu Val Thr Ala Pro Leu Asp Gly Thr
Ile Leu Glu Gly Val Thr Arg 260 265
270 Asp Ser Ile Leu Asn Leu Ala Lys Glu Arg Leu Glu Pro Ser
Glu Trp 275 280 285
Thr Ile Ser Glu Arg Tyr Phe Thr Ile Gly Glu Val Thr Glu Arg Ser 290
295 300 Lys Asn Gly Glu Leu
Leu Glu Ala Phe Gly Ser Gly Thr Ala Ala Ile 305 310
315 320 Val Ser Pro Ile Lys Glu Ile Gly Trp Lys
Gly Glu Gln Ile Asn Ile 325 330
335 Pro Leu Leu Pro Gly Glu Gln Thr Gly Pro Leu Ala Lys Glu Val
Ala 340 345 350 Gln
Trp Ile Asn Gly Ile Gln Tyr Gly Glu Thr Glu His Gly Asn Trp 355
360 365 Ser Arg Val Val Thr Asp
Leu Asn 370 375 79330PRTMethanobacterium
thermoautotrophicum 79Met Arg Leu Trp Arg Ala Leu Tyr Arg Pro Pro Thr Ile
Thr Tyr Pro 1 5 10 15
Ser Lys Ser Pro Glu Val Ile Ile Met Ser Cys Glu Ala Ser Gly Lys
20 25 30 Ile Trp Leu Asn
Gly Glu Met Val Glu Trp Glu Glu Ala Thr Val His 35
40 45 Val Leu Ser His Val Val His Tyr Gly
Ser Ser Val Phe Glu Gly Ile 50 55
60 Arg Cys Tyr Arg Asn Ser Lys Gly Ser Ala Ile Phe Arg
Leu Arg Glu 65 70 75
80 His Val Lys Arg Leu Phe Asp Ser Ala Lys Ile Tyr Arg Met Asp Ile
85 90 95 Pro Tyr Thr Gln
Glu Gln Ile Cys Asp Ala Ile Val Glu Thr Val Arg 100
105 110 Glu Asn Gly Leu Glu Glu Cys Tyr Ile
Arg Pro Val Val Phe Arg Gly 115 120
125 Tyr Gly Glu Met Gly Val His Pro Val Asn Cys Pro Val Asp
Val Ala 130 135 140
Val Ala Ala Trp Glu Trp Gly Ala Tyr Leu Gly Ala Glu Ala Leu Glu 145
150 155 160 Val Gly Val Asp Ala
Gly Val Ser Thr Trp Arg Arg Met Ala Pro Asn 165
170 175 Thr Met Pro Asn Met Ala Lys Ala Gly Gly
Asn Tyr Leu Asn Ser Gln 180 185
190 Leu Ala Lys Met Glu Ala Val Arg His Gly Tyr Asp Glu Ala Ile
Met 195 200 205 Leu
Asp Tyr His Gly Tyr Ile Ser Glu Gly Ser Gly Glu Asn Ile Phe 210
215 220 Leu Val Ser Glu Gly Glu
Ile Tyr Thr Pro Pro Val Ser Ser Ser Leu 225 230
235 240 Leu Arg Gly Ile Thr Arg Asp Ser Val Ile Lys
Ile Ala Arg Thr Glu 245 250
255 Gly Val Thr Val His Glu Glu Pro Ile Thr Arg Glu Met Leu Tyr Ile
260 265 270 Ala Asp
Glu Ala Phe Phe Thr Gly Thr Ala Ala Glu Ile Thr Pro Ile 275
280 285 Arg Ser Val Asp Gly Ile Glu
Ile Gly Ala Gly Arg Arg Gly Pro Val 290 295
300 Thr Lys Leu Leu Gln Asp Glu Phe Phe Arg Ile Ile
Arg Ala Glu Thr 305 310 315
320 Glu Asp Ser Phe Gly Trp Leu Thr Tyr Ile 325
330 80993DNAMethanobacterium thermoautotrophicum 80tcagatgtag
gtgagccatc cgaagctgtc ctctgtctct gccctgatta tcctgaagaa 60ctcatcctgc
agcagctttg taacgggacc ccttcgcccg gcacctatct ctataccatc 120aactgatctg
atgggtgtta tctctgcggc tgtacctgtg aagaaggcct catctgcgat 180gtagagcatc
tccctggtta tgggttcctc atgcacggta acaccctcgg tcctggctat 240ctttattacg
gagtcccttg ttatccccct cagaagggat gatgaaacag ggggggtgta 300aatttcaccc
tcactgacga ggaatatgtt ctccccgcta ccctcactta tgtagccatg 360gtagtccagc
attatggcct catcatagcc gtgtctcaca gcctccatct tggcaagctg 420tgagttgagg
tagttaccgc cggcctttgc catgttgggc attgtgtttg gtgccatcct 480ccgccaggtt
gaaacaccag catcgacacc aacctcaagg gcctctgcac ccagataggc 540cccccattcc
caggcagcca cagcgacgtc cactgggcag ttcaccgggt gaacacccat 600ctcaccgtat
cccctgaata ccacgggtct tatatagcac tcctcaagtc cgttctccct 660gacggtctca
actatggcat cacatatctg ctcctgggtg tagggtatgt ccatccggta 720tatctttgca
gaatcaaaaa ggcgtttaac atgctcccgc aaacggaaga tggctgaccc 780cttactgttc
ctgtagcacc ttattccctc aaagacagat gatccataat gcacaacatg 840tgagagtacg
tggacggtgg cttcttccca ttcaaccatt tcaccgttta accatatctt 900tccactggct
tcgcatgaca tgataataac ctcaggtgat ttactaggat aggttatggt 960tggaggccta
tataatgctc tccataaccg caa
99381364PRTStreptomyces coelicolor 81Met Thr Asp Val Asn Gly Ala Pro Ala
Asp Val Leu His Thr Leu Phe 1 5 10
15 His Ser Asp Gln Gly Gly His Glu Gln Val Val Leu Cys Gln
Asp Arg 20 25 30
Ala Ser Gly Leu Lys Ala Val Ile Ala Leu His Ser Thr Ala Leu Gly
35 40 45 Pro Ala Leu Gly
Gly Thr Arg Phe Tyr Pro Tyr Ala Ser Glu Ala Glu 50
55 60 Ala Val Ala Asp Ala Leu Asn Leu
Ala Arg Gly Met Ser Tyr Lys Asn 65 70
75 80 Ala Met Ala Gly Leu Asp His Gly Gly Gly Lys Ala
Val Ile Ile Gly 85 90
95 Asp Pro Glu Gln Ile Lys Ser Glu Glu Leu Leu Leu Ala Tyr Gly Arg
100 105 110 Phe Val Ala
Ser Leu Gly Gly Arg Tyr Val Thr Ala Cys Asp Val Gly 115
120 125 Thr Tyr Val Ala Asp Met Asp Val
Val Ala Arg Glu Cys Arg Trp Thr 130 135
140 Thr Gly Arg Ser Pro Glu Asn Gly Gly Ala Gly Asp Ser
Ser Val Leu 145 150 155
160 Thr Ser Phe Gly Val Tyr Gln Gly Met Arg Ala Ala Ala Gln His Leu
165 170 175 Trp Gly Asp Pro
Thr Leu Arg Asp Arg Thr Val Gly Ile Ala Gly Val 180
185 190 Gly Lys Val Gly His His Leu Val Glu
His Leu Leu Ala Glu Gly Ala 195 200
205 His Val Val Val Thr Asp Val Arg Lys Asp Val Val Arg Gly
Ile Thr 210 215 220
Glu Arg His Pro Ser Val Val Ala Val Ala Asp Thr Asp Ala Leu Ile 225
230 235 240 Arg Val Glu Asn Leu
Asp Ile Tyr Ala Pro Cys Ala Leu Gly Gly Ala 245
250 255 Leu Asn Asp Asp Thr Val Pro Val Leu Thr
Ala Lys Val Val Cys Gly 260 265
270 Ala Ala Asn Asn Gln Leu Ala His Pro Gly Val Glu Lys Asp Leu
Ala 275 280 285 Asp
Arg Gly Ile Leu Tyr Ala Pro Asp Tyr Val Val Asn Ala Gly Gly 290
295 300 Val Ile Gln Val Ala Asp
Glu Leu His Gly Phe Asp Phe Asp Arg Cys 305 310
315 320 Lys Ala Lys Ala Ser Lys Ile Tyr Asp Thr Thr
Leu Ala Ile Phe Ala 325 330
335 Arg Ala Lys Glu Asp Gly Ile Pro Pro Ala Ala Ala Ala Asp Arg Ile
340 345 350 Ala Glu
Gln Arg Met Ala Glu Ala Arg Pro Arg Pro 355 360
82 1095DNAStreptomyces coelicolor 82tcacggccgg
ggacgggcct ccgccatccg ctgctcggcg atccggtcgg ccgccgcggc 60cggcggaata
ccgtcctcct tcgcacgtgc gaatatggcc agcgtggtgt cgtagatctt 120cgaggccttc
gccttgcacc ggtcgaagtc gaacccgtgc agctcgtcgg cgacctggat 180gacaccgccg
gcgttcacca catagtccgg cgcgtagagg atcccgcggt cggcgaggtc 240cttctcgacg
cccgggtggg cgagctggtt gttggccgcg ccgcacacca ccttggcggt 300cagcaccggc
acggtgtcgt cgttcagcgc gccgccgagc gcgcagggcg cgtagatgtc 360caggttctcc
acccggatca gcgcgtcggt gtcggcgacg gcgaccaccg acgggtgccg 420ctccgtgatc
ccgcgcacca cgtccttgcg cacgtccgtg acgacgacgt gggcgccctc 480ggcgagcagg
tgctcgacca ggtggtggcc gaccttgccg acgcccgcga tgccgacggt 540gcggtcgcgc
agcgtcgggt cgccccacag gtgctgggcg gcggcccgca tgccctggta 600gacgccgaag
gaggtgagca cggaggagtc gcccgcgccg ccgttctccg gggaacgccc 660ggtcgtccag
cggcactcgc gggccacgac gtccatgtcg gcgacgtagg tgccgacgtc 720gcacgcggtg
acgtagcggc cgcccagcga ggcgacgaac cggccgtagg cgaggagcag 780ctcctcgctc
ttgatctgct ccggatcgcc gatgatcacg gccttgccgc caccgtggtc 840cagaccggcc
atggcgttct tgtacgacat cccgcgggcg aggttcagcg cgtcggcgac 900ggcctccgcc
tcgctcgcgt acgggtagaa gcgggtaccg ccgagcgccg ggcccagggc 960ggtggagtgg
agggcgatca cggccttgag gccgctggca cggtcctggc agagcacgac 1020ttgctcatgt
cccccctgat ccgagtggaa cagggtgtgc agtacatcag caggtgcgcc 1080gtttacgtcg
gtcac
109583364PRTBacillus subtilis 83Met Glu Leu Phe Lys Tyr Met Glu Lys Tyr
Asp Tyr Glu Gln Leu Val 1 5 10
15 Phe Cys Gln Asp Glu Gln Ser Gly Leu Lys Ala Ile Ile Ala Ile
His 20 25 30 Asp
Thr Thr Leu Gly Pro Ala Leu Gly Gly Thr Arg Met Trp Thr Tyr 35
40 45 Glu Asn Glu Glu Ala Ala
Ile Glu Asp Ala Leu Arg Leu Ala Arg Gly 50 55
60 Met Thr Tyr Lys Asn Ala Ala Ala Gly Leu Asn
Leu Gly Gly Gly Lys 65 70 75
80 Thr Val Ile Ile Gly Asp Pro Arg Lys Asp Lys Asn Glu Glu Met Phe
85 90 95 Arg Ala
Phe Gly Arg Tyr Ile Gln Gly Leu Asn Gly Arg Tyr Ile Thr 100
105 110 Ala Glu Asp Val Gly Thr Thr
Val Glu Asp Met Asp Ile Ile His Asp 115 120
125 Glu Thr Asp Tyr Val Thr Gly Ile Ser Pro Ala Phe
Gly Ser Ser Gly 130 135 140
Asn Pro Ser Pro Val Thr Ala Tyr Gly Val Tyr Arg Gly Met Lys Ala 145
150 155 160 Ala Ala Lys
Ala Ala Phe Gly Thr Asp Ser Leu Glu Gly Lys Thr Ile 165
170 175 Ala Val Gln Gly Val Gly Asn Val
Ala Tyr Asn Leu Cys Arg His Leu 180 185
190 His Glu Glu Gly Ala Asn Leu Ile Val Thr Asp Ile Asn
Lys Gln Ser 195 200 205
Val Gln Arg Ala Val Glu Asp Phe Gly Ala Arg Ala Val Asp Pro Asp 210
215 220 Asp Ile Tyr Ser
Gln Asp Cys Asp Ile Tyr Ala Pro Cys Ala Leu Gly 225 230
235 240 Ala Thr Ile Asn Asp Asp Thr Ile Lys
Gln Leu Lys Ala Lys Val Ile 245 250
255 Ala Gly Ala Ala Asn Asn Gln Leu Lys Glu Thr Arg His Gly
Asp Gln 260 265 270
Ile His Glu Met Gly Ile Val Tyr Ala Pro Asp Tyr Val Ile Asn Ala
275 280 285 Gly Gly Val Ile
Asn Val Ala Asp Glu Leu Tyr Gly Tyr Asn Ala Glu 290
295 300 Arg Ala Leu Lys Lys Val Glu Gly
Ile Tyr Gly Asn Ile Glu Arg Val 305 310
315 320 Leu Glu Ile Ser Gln Arg Asp Gly Ile Pro Ala Tyr
Leu Ala Ala Asp 325 330
335 Arg Leu Ala Glu Glu Arg Ile Glu Arg Met Arg Arg Ser Arg Ser Gln
340 345 350 Phe Leu Gln
Asn Gly His Ser Val Leu Ser Arg Arg 355 360
84364PRTBacillus subtilis 84Met Glu Leu Phe Lys Tyr Met Glu Lys
Tyr Asp Tyr Glu Gln Leu Val 1 5 10
15 Phe Cys Gln Asp Glu Gln Ser Gly Leu Lys Ala Ile Ile Ala
Ile His 20 25 30
Asp Thr Thr Leu Gly Pro Ala Leu Gly Gly Thr Arg Met Trp Thr Tyr
35 40 45 Glu Asn Glu Glu
Ala Ala Ile Glu Asp Ala Leu Arg Leu Ala Arg Gly 50
55 60 Met Thr Tyr Lys Asn Ala Ala Ala
Gly Leu Asn Leu Gly Gly Gly Lys 65 70
75 80 Thr Val Ile Ile Gly Asp Pro Arg Lys Asp Lys Asn
Glu Glu Met Phe 85 90
95 Arg Ala Phe Gly Arg Tyr Ile Gln Gly Leu Asn Gly Arg Tyr Ile Thr
100 105 110 Ala Glu Asp
Val Gly Thr Thr Val Glu Asp Met Asp Ile Ile His Asp 115
120 125 Glu Thr Asp Tyr Val Thr Gly Ile
Ser Pro Ala Phe Gly Ser Ser Gly 130 135
140 Asn Pro Ser Pro Val Thr Ala Tyr Gly Val Tyr Arg Gly
Met Lys Ala 145 150 155
160 Ala Ala Lys Ala Ala Phe Gly Thr Asp Ser Leu Glu Gly Lys Thr Ile
165 170 175 Ala Val Gln Gly
Val Gly Asn Val Ala Tyr Asn Leu Cys Arg His Leu 180
185 190 His Glu Glu Gly Ala Asn Leu Ile Val
Thr Asp Ile Asn Lys Gln Ser 195 200
205 Val Gln Arg Ala Val Glu Asp Phe Gly Ala Arg Ala Val Asp
Pro Asp 210 215 220
Asp Ile Tyr Ser Gln Asp Cys Asp Ile Tyr Ala Pro Cys Ala Leu Gly 225
230 235 240 Ala Thr Ile Asn Asp
Asp Thr Ile Lys Gln Leu Lys Ala Lys Val Ile 245
250 255 Ala Gly Ala Ala Asn Asn Gln Leu Lys Glu
Thr Arg His Gly Asp Gln 260 265
270 Ile His Glu Met Gly Ile Val Tyr Ala Pro Asp Tyr Val Ile Asn
Ala 275 280 285 Gly
Gly Val Ile Asn Val Ala Asp Glu Leu Tyr Gly Tyr Asn Ala Glu 290
295 300 Arg Ala Leu Lys Lys Val
Glu Gly Ile Tyr Gly Asn Ile Glu Arg Val 305 310
315 320 Leu Glu Ile Ser Gln Arg Asp Gly Ile Pro Ala
Tyr Leu Ala Ala Asp 325 330
335 Arg Leu Ala Glu Glu Arg Ile Glu Arg Met Arg Arg Ser Arg Ser Gln
340 345 350 Phe Leu
Gln Asn Gly His Ser Val Leu Ser Arg Arg 355 360
85594PRTStreptomyces viridifaciens 85Met Ser Thr Ser Ser
Ala Ser Ser Gly Pro Asp Leu Pro Phe Gly Pro 1 5
10 15 Glu Asp Thr Pro Trp Gln Lys Ala Phe Ser
Arg Leu Arg Ala Val Asp 20 25
30 Gly Val Pro Arg Val Thr Ala Pro Ser Ser Asp Pro Arg Glu Val
Tyr 35 40 45 Met
Asp Ile Pro Glu Ile Pro Phe Ser Lys Val Gln Ile Pro Pro Asp 50
55 60 Gly Met Asp Glu Gln Gln
Tyr Ala Glu Ala Glu Ser Leu Phe Arg Arg 65 70
75 80 Tyr Val Asp Ala Gln Thr Arg Asn Phe Ala Gly
Tyr Gln Val Thr Ser 85 90
95 Asp Leu Asp Tyr Gln His Leu Ser His Tyr Leu Asn Arg His Leu Asn
100 105 110 Asn Val
Gly Asp Pro Tyr Glu Ser Ser Ser Tyr Thr Leu Asn Ser Lys 115
120 125 Val Leu Glu Arg Ala Val Leu
Asp Tyr Phe Ala Ser Leu Trp Asn Ala 130 135
140 Lys Trp Pro His Asp Ala Ser Asp Pro Glu Thr Tyr
Trp Gly Tyr Val 145 150 155
160 Leu Thr Met Gly Ser Ser Glu Gly Asn Leu Tyr Gly Leu Trp Asn Ala
165 170 175 Arg Asp Tyr
Leu Ser Gly Lys Leu Leu Arg Arg Gln His Arg Glu Ala 180
185 190 Gly Gly Asp Lys Ala Ser Val Val
Tyr Thr Gln Ala Leu Arg His Glu 195 200
205 Gly Gln Ser Pro His Ala Tyr Glu Pro Val Ala Phe Phe
Ser Gln Asp 210 215 220
Thr His Tyr Ser Leu Thr Lys Ala Val Arg Val Leu Gly Ile Asp Thr 225
230 235 240 Phe His Ser Ile
Gly Ser Ser Arg Tyr Pro Asp Glu Asn Pro Leu Gly 245
250 255 Pro Gly Thr Pro Trp Pro Thr Glu Val
Pro Ser Val Asp Gly Ala Ile 260 265
270 Asp Val Asp Lys Leu Ala Ser Leu Val Arg Phe Phe Ala Ser
Lys Gly 275 280 285
Tyr Pro Ile Leu Val Ser Leu Asn Tyr Gly Ser Thr Phe Lys Gly Ala 290
295 300 Tyr Asp Asp Val Pro
Ala Val Ala Gln Ala Val Arg Asp Ile Cys Thr 305 310
315 320 Glu Tyr Gly Leu Asp Arg Arg Arg Val Tyr
His Asp Arg Ser Lys Asp 325 330
335 Ser Asp Phe Asp Glu Arg Ser Gly Phe Trp Ile His Ile Asp Ala
Ala 340 345 350 Leu
Gly Ala Gly Tyr Ala Pro Tyr Leu Gln Met Ala Arg Asp Ala Gly 355
360 365 Met Val Glu Glu Ala Pro
Pro Val Phe Asp Phe Arg Leu Pro Glu Val 370 375
380 His Ser Leu Thr Met Ser Gly His Lys Trp Met
Gly Thr Pro Trp Ala 385 390 395
400 Cys Gly Val Tyr Met Thr Arg Thr Gly Leu Gln Met Thr Pro Pro Lys
405 410 415 Ser Ser
Glu Tyr Ile Gly Ala Ala Asp Thr Thr Phe Ala Gly Ser Arg 420
425 430 Asn Gly Phe Ser Ser Leu Leu
Leu Trp Asp Tyr Leu Ser Arg His Ser 435 440
445 Tyr Asp Asp Leu Val Arg Leu Ala Ala Asp Cys Asp
Arg Leu Ala Gly 450 455 460
Tyr Ala His Asp Arg Leu Leu Thr Leu Gln Asp Lys Leu Gly Met Asp 465
470 475 480 Leu Trp Val
Ala Arg Ser Pro Gln Ser Leu Thr Val Arg Phe Arg Gln 485
490 495 Pro Cys Ala Asp Ile Val Arg Lys
Tyr Ser Leu Ser Cys Glu Thr Val 500 505
510 Tyr Glu Asp Asn Glu Gln Arg Thr Tyr Val His Leu Tyr
Ala Val Pro 515 520 525
His Leu Thr Arg Glu Leu Val Asp Glu Leu Val Arg Asp Leu Arg Gln 530
535 540 Pro Gly Ala Phe
Thr Asn Ala Gly Ala Leu Glu Gly Glu Ala Trp Ala 545 550
555 560 Gly Val Ile Asp Ala Leu Gly Arg Pro
Asp Pro Asp Gly Thr Tyr Ala 565 570
575 Gly Ala Leu Ser Ala Pro Ala Ser Gly Pro Arg Ser Glu Asp
Gly Gly 580 585 590
Gly Ser 861785DNAStreptomyces viridifaciens 86gtgtcaactt cctccgcttc
ttccgggccg gacctcccct tcgggcccga ggacacgcca 60tggcagaagg ccttcagcag
gctgcgggcg gtggatggcg tgccgcgcgt caccgcgccg 120tccagtgatc cgcgtgaggt
ctacatggac atcccggaga tccccttctc caaggtccag 180atccccccgg acggaatgga
cgagcagcag tacgcagagg ccgagagcct cttccgccgc 240tacgtagacg cccagacccg
caacttcgcg ggataccagg tcaccagcga cctcgactac 300cagcacctca gtcactatct
caaccggcat ctgaacaacg tcggcgatcc ctatgagtcc 360agctcctaca cgctgaactc
caaggtcctt gagcgagccg ttctcgacta cttcgcctcc 420ctgtggaacg ccaagtggcc
ccatgacgca agcgatccgg aaacgtactg gggttacgtg 480ctgaccatgg gctccagcga
aggcaacctg tacgggttgt ggaacgcacg ggactatctg 540tcgggcaagc tgctgcggcg
ccagcaccgg gaggccggcg gcgacaaggc ctcggtcgtc 600tacacgcaag cgctgcgaca
cgaagggcag agtccgcatg cctacgagcc ggtggcgttc 660ttctcgcagg acacgcacta
ctcgctcacg aaggccgtgc gggttctggg catcgacacc 720ttccacagca tcggcagcag
tcggtatccg gacgagaacc cgctgggccc cggcactccg 780tggccgaccg aagtgccctc
ggttgacggt gccatcgatg tcgacaaact cgcctcgttg 840gtccgcttct tcgccagcaa
gggctacccg atactggtca gcctcaacta cgggtcaacg 900ttcaagggcg cctacgacga
cgtcccggcc gtggcacagg ccgtgcggga catctgcacg 960gaatacggtc tggatcggcg
gcgggtatac cacgaccgca gtaaggacag tgacttcgac 1020gagcgcagcg gcttctggat
ccacatcgat gccgccctgg gggcgggcta cgctccctac 1080ctgcagatgg cccgggatgc
cggcatggtc gaggaggcgc cgcccgtttt cgacttccgg 1140ctcccggagg tgcactcgct
gaccatgagc ggccacaagt ggatgggaac accgtgggca 1200tgcggtgtct acatgacacg
gaccgggctg cagatgaccc cgccgaagtc gtccgagtac 1260atcggggcgg ccgacaccac
cttcgcgggc tcccgcaacg gcttctcgtc actgctgctg 1320tgggactacc tgtcccggca
ttcgtatgac gatctggtgc gcctggccgc cgactgcgac 1380cggctggccg gctacgccca
cgaccggttg ctgaccttgc aggacaaact cggcatggat 1440ctgtgggtcg cccgcagccc
gcagtccctc acggtgcgct tccgtcagcc atgtgcagac 1500atcgtccgca agtactcgct
gtcgtgtgag acggtctacg aagacaacga gcaacggacc 1560tacgtacatc tctacgccgt
tccccacctc actcgggaac tcgtggatga gctcgtgcgc 1620gatctgcgcc agcccggagc
cttcaccaac gctggtgcac tggaggggga ggcctgggcc 1680ggggtgatcg atgccctcgg
ccgcccggac cccgacggaa cctatgccgg cgccttgagc 1740gctccggctt ccggcccccg
ctccgaggac ggcggcggga gctga 178587440PRTAlcaligenes
denitrificans 87Met Ser Ala Ala Lys Leu Pro Asp Leu Ser His Leu Trp Met
Pro Phe 1 5 10 15
Thr Ala Asn Arg Gln Phe Lys Ala Asn Pro Arg Leu Leu Ala Ser Ala
20 25 30 Lys Gly Met Tyr Tyr
Thr Ser Phe Asp Gly Arg Gln Ile Leu Asp Gly 35
40 45 Thr Ala Gly Leu Trp Cys Val Asn Ala
Gly His Cys Arg Glu Glu Ile 50 55
60 Val Ser Ala Ile Ala Ser Gln Ala Gly Val Met Asp Tyr
Ala Pro Gly 65 70 75
80 Phe Gln Leu Gly His Pro Leu Ala Phe Glu Ala Ala Thr Ala Val Ala
85 90 95 Gly Leu Met Pro
Gln Gly Leu Asp Arg Val Phe Phe Thr Asn Ser Gly 100
105 110 Ser Glu Ser Val Asp Thr Ala Leu Lys
Ile Ala Leu Ala Tyr His Arg 115 120
125 Ala Arg Gly Glu Ala Gln Arg Thr Arg Leu Ile Gly Arg Glu
Arg Gly 130 135 140
Tyr His Gly Val Gly Phe Gly Gly Ile Ser Val Gly Gly Ile Ser Pro 145
150 155 160 Asn Arg Lys Thr Phe
Ser Gly Ala Leu Leu Pro Ala Val Asp His Leu 165
170 175 Pro His Thr His Ser Leu Glu His Asn Ala
Phe Thr Arg Gly Gln Pro 180 185
190 Glu Trp Gly Ala His Leu Ala Asp Glu Leu Glu Arg Ile Ile Ala
Leu 195 200 205 His
Asp Ala Ser Thr Ile Ala Ala Val Ile Val Glu Pro Met Ala Gly 210
215 220 Ser Thr Gly Val Leu Val
Pro Pro Lys Gly Tyr Leu Glu Lys Leu Arg 225 230
235 240 Glu Ile Thr Ala Arg His Gly Ile Leu Leu Ile
Phe Asp Glu Val Ile 245 250
255 Thr Ala Tyr Gly Arg Leu Gly Glu Ala Thr Ala Ala Ala Tyr Phe Gly
260 265 270 Val Thr
Pro Asp Leu Ile Thr Met Ala Lys Gly Val Ser Asn Ala Ala 275
280 285 Val Pro Ala Gly Ala Val Ala
Val Arg Arg Glu Val His Asp Ala Ile 290 295
300 Val Asn Gly Pro Gln Gly Gly Ile Glu Phe Phe His
Gly Tyr Thr Tyr 305 310 315
320 Ser Ala His Pro Leu Ala Ala Ala Ala Val Leu Ala Thr Leu Asp Ile
325 330 335 Tyr Arg Arg
Glu Asp Leu Phe Ala Arg Ala Arg Lys Leu Ser Ala Ala 340
345 350 Phe Glu Glu Ala Ala His Ser Leu
Lys Gly Ala Pro His Val Ile Asp 355 360
365 Val Arg Asn Ile Gly Leu Val Ala Gly Ile Glu Leu Ser
Pro Arg Glu 370 375 380
Gly Ala Pro Gly Ala Arg Ala Ala Glu Ala Phe Gln Lys Cys Phe Asp 385
390 395 400 Thr Gly Leu Met
Val Arg Tyr Thr Gly Asp Ile Leu Ala Val Ser Pro 405
410 415 Pro Leu Ile Val Asp Glu Asn Gln Ile
Gly Gln Ile Phe Glu Gly Ile 420 425
430 Gly Lys Val Leu Lys Glu Val Ala 435
440 88 1947DNAAlcaligenes denitrificans 88ttcgatggcg cgctgcacgg
cggccaccag ctgctccacc aggggtgggc gcctgcccgc 60gcgcgcggtc gggctggaaa
tcgatcatgg atgaatctat acagttgtca tgattgcaac 120tatacagtta gcccgttttg
cggcaattgt atattttcat tcgctcgtgg acgtccgaga 180atcggtttga tcgcgccgcc
cgcccctttc cgcgcagcgg cgtttctttt cctccggagt 240ctccccatga gcgctgccaa
actgcccgac ctgtcccacc tctggatgcc ctttaccgcc 300aaccggcagt tcaaggcgaa
cccccgcctg ctggcctcgg ccaagggcat gtactacacg 360tctttcgacg gccgccagat
cctggacggc acggccggcc tgtggtgcgt gaacgccggc 420cactgccgcg aagaaatcgt
ctccgccatc gccagccagg ccggcgtcat ggactacgcg 480ccggggttcc agctcggcca
cccgctggcc ttcgaggccg ccaccgccgt ggccggcctg 540atgccgcagg gcctggaccg
cgtgttcttc accaattcgg gctccgaatc ggtggacacc 600gcgctgaaga tcgccctggc
ctaccaccgc gcgcgcggcg aggcgcagcg cacccgcctc 660atcgggcgcg agcgcggcta
ccacggcgtg ggcttcggcg gcatttccgt gggcggcatc 720tcgcccaacc gcaagacctt
ctccggcgcg ctgctgccgg ccgtggacca cctgccgcac 780acccacagcc tggaacacaa
cgccttcacg cgcggccagc ccgagtgggg cgcgcacctg 840gccgacgagt tggaacgcat
catcgccctg cacgacgcct ccaccatcgc ggccgtgatc 900gtcgagccca tggccggctc
caccggcgtg ctcgtcccgc ccaagggcta tctcgaaaaa 960ctgcgcgaaa tcaccgcccg
ccacggcatt ctgctgatct tcgacgaagt catcaccgcg 1020tacggccgcc tgggcgaggc
caccgccgcg gcctatttcg gcgtaacgcc cgacctcatc 1080accatggcca agggcgtgag
caacgccgcc gttccggccg gcgccgtcgc ggtgcgccgc 1140gaagtgcatg acgccatcgt
caacggaccg caaggcggca tcgagttctt ccacggctac 1200acctactcgg cccacccgct
ggccgccgcc gccgtgctcg ccacgctgga catctaccgc 1260cgcgaagacc tgttcgcccg
cgcccgcaag ctgtcggccg cgttcgagga agccgcccac 1320agcctcaagg gcgcgccgca
cgtcatcgac gtgcgcaaca tcggcctggt ggccggcatc 1380gagctgtcgc cgcgcgaagg
cgccccgggc gcgcgcgccg ccgaagcctt ccagaaatgc 1440ttcgacaccg gcctcatggt
gcgctacacg ggcgacatcc tcgcggtgtc gcctccgctc 1500atcgtcgacg aaaaccagat
cggccagatc ttcgagggca tcggcaaggt gctcaaggaa 1560gtggcttagg gtgaacacgc
cctgagccgg ccccggcagg aaacgcgccg ccgcgcggcg 1620gcgcgtccat cgaactcccg
catcgagctt ttgcattcat gaagaaaatc acgcatttca 1680tcaacggcca gccccacgaa
ggccgcagca accgctacac cgagggcttc aacccggcca 1740cgggcgagtc gtctcctcga
tctgcctggg cggggccgaa gaagtggacc tggccgtggc 1800ggccgcccgc gcggcctttc
ccgcctggtc cgaaacgccg gcgctcaagc gcgcgcgcgt 1860gctgttcaac ttcaaggcgc
tgctggacaa gcaccaggac gagctggccg cgctcatcac 1920gcgcgagcac ggcaaggtgt
tttccga 194789443PRTRalstonia
eutropha 89Met Asp Ala Ala Lys Thr Val Ile Pro Asp Leu Asp Ala Leu Trp
Met 1 5 10 15 Pro
Phe Thr Ala Asn Arg Gln Tyr Lys Ala Ala Pro Arg Leu Leu Ala
20 25 30 Ser Ala Ser Gly Met
Tyr Tyr Thr Thr His Asp Gly Arg Gln Ile Leu 35
40 45 Asp Gly Cys Ala Gly Leu Trp Cys Val
Ala Ala Gly His Cys Arg Lys 50 55
60 Glu Ile Ala Glu Ala Val Ala Arg Gln Ala Ala Thr Leu
Asp Tyr Ala 65 70 75
80 Pro Pro Phe Gln Met Gly His Pro Leu Ser Phe Glu Ala Ala Thr Lys
85 90 95 Val Ala Ala Ile
Met Pro Gln Gly Leu Asp Arg Ile Phe Phe Thr Asn 100
105 110 Ser Gly Ser Glu Ser Val Asp Thr Ala
Leu Lys Ile Ala Leu Ala Tyr 115 120
125 His Arg Ala Arg Gly Glu Gly Gln Arg Thr Arg Phe Ile Gly
Arg Glu 130 135 140
Arg Gly Tyr His Gly Val Gly Phe Gly Gly Met Ala Val Gly Gly Ile 145
150 155 160 Gly Pro Asn Arg Lys
Ala Phe Ser Ala Asn Leu Met Pro Gly Thr Asp 165
170 175 His Leu Pro Ala Thr Leu Asn Ile Ala Glu
Ala Ala Phe Ser Lys Gly 180 185
190 Gln Pro Thr Trp Gly Ala His Leu Ala Asp Glu Leu Glu Arg Ile
Val 195 200 205 Ala
Leu His Asp Pro Ser Thr Ile Ala Ala Val Ile Val Glu Pro Leu 210
215 220 Ala Gly Ser Ala Gly Val
Leu Val Pro Pro Val Gly Tyr Leu Asp Lys 225 230
235 240 Leu Arg Glu Ile Thr Thr Lys His Gly Ile Leu
Leu Ile Phe Asp Glu 245 250
255 Val Ile Thr Ala Phe Gly Arg Leu Gly Thr Ala Thr Ala Ala Glu Arg
260 265 270 Phe Lys
Val Thr Pro Asp Leu Ile Thr Met Ala Lys Ala Ile Asn Asn 275
280 285 Ala Ala Val Pro Met Gly Ala
Val Ala Val Arg Arg Glu Val His Asp 290 295
300 Thr Val Val Asn Ser Ala Ala Pro Gly Ala Ile Glu
Leu Ala His Gly 305 310 315
320 Tyr Thr Tyr Ser Gly His Pro Leu Ala Ala Ala Ala Ala Ile Ala Thr
325 330 335 Leu Asp Leu
Tyr Gln Arg Glu Asn Leu Phe Gly Arg Ala Ala Glu Leu 340
345 350 Ser Pro Val Phe Glu Ala Ala Val
His Ser Val Arg Ser Ala Pro His 355 360
365 Val Lys Asp Ile Arg Asn Leu Gly Met Val Ala Gly Ile
Glu Leu Glu 370 375 380
Pro Arg Pro Gly Gln Pro Gly Ala Arg Ala Tyr Glu Ala Phe Leu Lys 385
390 395 400 Cys Leu Glu Arg
Gly Val Leu Val Arg Tyr Thr Gly Asp Ile Leu Ala 405
410 415 Phe Ser Pro Pro Leu Ile Ile Ser Glu
Ala Gln Ile Ala Glu Leu Phe 420 425
430 Asp Thr Val Lys Gln Ala Leu Gln Glu Val Gln 435
440 90 1341DNARalstonia eutropha
90atggccgact cacccaacaa cctcgctcac gaacatcctt cacttgaaca ctattggatg
60ccttttaccg ccaatcgcca attcaaagcg agccctcgtt tactcgccca agctgaaggt
120atgtattaca cagatatcaa tggcaacaag gtattagact ctacagcggg cttatggtgt
180tgtaatgctg gccatggtcg ccgtgagatc agtgaagccg tcagcaaaca aattcggcag
240atggattacg ctccctcctt ccaaatgggc catcccatcg cttttgaact ggccgaacgt
300ttaaccgaac tcagcccaga aggactcaac aaagtattct ttaccaactc aggctctgag
360tcggttgata ccgcgctaaa aatggctctt tgctaccata gagccaatgg ccaagcgtca
420cgcacccgct ttattggccg tgaaatgggt taccatggcg taggatttgg tgggatctcg
480gtgggtggtt taagcaataa ccgtaaagcc ttcagcggcc agctattgca aggcgtggat
540cacctgcccc acaccttaga cattcaacat gccgccttta gtcgtggctt accgagcctc
600ggtgctgaaa aagctgaggt attagaacaa ttagtcacac tccatggcgc cgaaaatatt
660gccgccgtta ttgttgaacc catgtcaggt tctgcagggg taattttacc acctcaaggc
720tacttaaaac gcttacgtga aatcactaaa aaacacggca tcttattgat tttcgatgaa
780gtcattaccg catttggccg tgtaggtgca gcattcgcca gccaacgttg gggcgttatt
840ccagacataa tcaccacggc taaagccatt aataatggcg ccatccccat gggcgcagtg
900tttgtacagg attatatcca cgatacttgc atgcaagggc caaccgaact gattgaattt
960ttccacggtt atacctattc gggccaccca gtcgccgcag cagcagcact cgccacgctc
1020tccatctacc aaaacgagca actgtttgag cgcagttttg agcttgagcg gtatttcgaa
1080gaagccgttc atagcctcaa agggttaccg aatgtgattg atattcgcaa caccggatta
1140gtcgcgggtt tccagctagc accgaatagc caaggtgttg gtaaacgcgg atacagcgtg
1200ttcgagcatt gtttccatca aggcacactc gtgcgggcaa cgggcgatat tatcgccatg
1260tccccaccac tcattgttga gaaacatcag attgaccaaa tggtaaatag ccttagcgat
1320gcaattcacg ccgttggatg a
134191446PRTShewanella oneidensis 91Met Ala Asp Ser Pro Asn Asn Leu Ala
His Glu His Pro Ser Leu Glu 1 5 10
15 His Tyr Trp Met Pro Phe Thr Ala Asn Arg Gln Phe Lys Ala
Ser Pro 20 25 30
Arg Leu Leu Ala Gln Ala Glu Gly Met Tyr Tyr Thr Asp Ile Asn Gly
35 40 45 Asn Lys Val Leu
Asp Ser Thr Ala Gly Leu Trp Cys Cys Asn Ala Gly 50
55 60 His Gly Arg Arg Glu Ile Ser Glu
Ala Val Ser Lys Gln Ile Arg Gln 65 70
75 80 Met Asp Tyr Ala Pro Ser Phe Gln Met Gly His Pro
Ile Ala Phe Glu 85 90
95 Leu Ala Glu Arg Leu Thr Glu Leu Ser Pro Glu Gly Leu Asn Lys Val
100 105 110 Phe Phe Thr
Asn Ser Gly Ser Glu Ser Val Asp Thr Ala Leu Lys Met 115
120 125 Ala Leu Cys Tyr His Arg Ala Asn
Gly Gln Ala Ser Arg Thr Arg Phe 130 135
140 Ile Gly Arg Glu Met Gly Tyr His Gly Val Gly Phe Gly
Gly Ile Ser 145 150 155
160 Val Gly Gly Leu Ser Asn Asn Arg Lys Ala Phe Ser Gly Gln Leu Leu
165 170 175 Gln Gly Val Asp
His Leu Pro His Thr Leu Asp Ile Gln His Ala Ala 180
185 190 Phe Ser Arg Gly Leu Pro Ser Leu Gly
Ala Glu Lys Ala Glu Val Leu 195 200
205 Glu Gln Leu Val Thr Leu His Gly Ala Glu Asn Ile Ala Ala
Val Ile 210 215 220
Val Glu Pro Met Ser Gly Ser Ala Gly Val Ile Leu Pro Pro Gln Gly 225
230 235 240 Tyr Leu Lys Arg Leu
Arg Glu Ile Thr Lys Lys His Gly Ile Leu Leu 245
250 255 Ile Phe Asp Glu Val Ile Thr Ala Phe Gly
Arg Val Gly Ala Ala Phe 260 265
270 Ala Ser Gln Arg Trp Gly Val Ile Pro Asp Ile Ile Thr Thr Ala
Lys 275 280 285 Ala
Ile Asn Asn Gly Ala Ile Pro Met Gly Ala Val Phe Val Gln Asp 290
295 300 Tyr Ile His Asp Thr Cys
Met Gln Gly Pro Thr Glu Leu Ile Glu Phe 305 310
315 320 Phe His Gly Tyr Thr Tyr Ser Gly His Pro Val
Ala Ala Ala Ala Ala 325 330
335 Leu Ala Thr Leu Ser Ile Tyr Gln Asn Glu Gln Leu Phe Glu Arg Ser
340 345 350 Phe Glu
Leu Glu Arg Tyr Phe Glu Glu Ala Val His Ser Leu Lys Gly 355
360 365 Leu Pro Asn Val Ile Asp Ile
Arg Asn Thr Gly Leu Val Ala Gly Phe 370 375
380 Gln Leu Ala Pro Asn Ser Gln Gly Val Gly Lys Arg
Gly Tyr Ser Val 385 390 395
400 Phe Glu His Cys Phe His Gln Gly Thr Leu Val Arg Ala Thr Gly Asp
405 410 415 Ile Ile Ala
Met Ser Pro Pro Leu Ile Val Glu Lys His Gln Ile Asp 420
425 430 Gln Met Val Asn Ser Leu Ser Asp
Ala Ile His Ala Val Gly 435 440
445 92 1341DNAShewanella oneidensis 92atggccgact cacccaacaa
cctcgctcac gaacatcctt cacttgaaca ctattggatg 60ccttttaccg ccaatcgcca
attcaaagcg agccctcgtt tactcgccca agctgaaggt 120atgtattaca cagatatcaa
tggcaacaag gtattagact ctacagcggg cttatggtgt 180tgtaatgctg gccatggtcg
ccgtgagatc agtgaagccg tcagcaaaca aattcggcag 240atggattacg ctccctcctt
ccaaatgggc catcccatcg cttttgaact ggccgaacgt 300ttaaccgaac tcagcccaga
aggactcaac aaagtattct ttaccaactc aggctctgag 360tcggttgata ccgcgctaaa
aatggctctt tgctaccata gagccaatgg ccaagcgtca 420cgcacccgct ttattggccg
tgaaatgggt taccatggcg taggatttgg tgggatctcg 480gtgggtggtt taagcaataa
ccgtaaagcc ttcagcggcc agctattgca aggcgtggat 540cacctgcccc acaccttaga
cattcaacat gccgccttta gtcgtggctt accgagcctc 600ggtgctgaaa aagctgaggt
attagaacaa ttagtcacac tccatggcgc cgaaaatatt 660gccgccgtta ttgttgaacc
catgtcaggt tctgcagggg taattttacc acctcaaggc 720tacttaaaac gcttacgtga
aatcactaaa aaacacggca tcttattgat tttcgatgaa 780gtcattaccg catttggccg
tgtaggtgca gcattcgcca gccaacgttg gggcgttatt 840ccagacataa tcaccacggc
taaagccatt aataatggcg ccatccccat gggcgcagtg 900tttgtacagg attatatcca
cgatacttgc atgcaagggc caaccgaact gattgaattt 960ttccacggtt atacctattc
gggccaccca gtcgccgcag cagcagcact cgccacgctc 1020tccatctacc aaaacgagca
actgtttgag cgcagttttg agcttgagcg gtatttcgaa 1080gaagccgttc atagcctcaa
agggttaccg aatgtgattg atattcgcaa caccggatta 1140gtcgcgggtt tccagctagc
accgaatagc caaggtgttg gtaaacgcgg atacagcgtg 1200ttcgagcatt gtttccatca
aggcacactc gtgcgggcaa cgggcgatat tatcgccatg 1260tccccaccac tcattgttga
gaaacatcag attgaccaaa tggtaaatag ccttagcgat 1320gcaattcacg ccgttggatg a
134193448PRTPseudomonas
putida 93Met Asn Met Pro Glu Thr Gly Pro Ala Gly Ile Ala Ser Gln Leu Lys
1 5 10 15 Leu Asp
Ala His Trp Met Pro Tyr Thr Ala Asn Arg Asn Phe Gln Arg 20
25 30 Asp Pro Arg Leu Ile Val Ala
Ala Glu Gly Asn Tyr Leu Val Asp Asp 35 40
45 His Gly Arg Lys Ile Phe Asp Ala Leu Ser Gly Leu
Trp Thr Cys Gly 50 55 60
Ala Gly His Thr Arg Lys Glu Ile Ala Asp Ala Val Thr Arg Gln Leu 65
70 75 80 Ser Thr Leu
Asp Tyr Ser Pro Ala Phe Gln Phe Gly His Pro Leu Ser 85
90 95 Phe Gln Leu Ala Glu Lys Ile Ala
Glu Leu Val Pro Gly Asn Leu Asn 100 105
110 His Val Phe Tyr Thr Asn Ser Gly Ser Glu Cys Ala Asp
Thr Ala Leu 115 120 125
Lys Met Val Arg Ala Tyr Trp Arg Leu Lys Gly Gln Ala Thr Lys Thr 130
135 140 Lys Ile Ile Gly
Arg Ala Arg Gly Tyr His Gly Val Asn Ile Ala Gly 145 150
155 160 Thr Ser Leu Gly Gly Val Asn Gly Asn
Arg Lys Met Phe Gly Gln Leu 165 170
175 Leu Asp Val Asp His Leu Pro His Thr Val Leu Pro Val Asn
Ala Phe 180 185 190
Ser Lys Gly Leu Pro Glu Glu Gly Gly Ile Ala Leu Ala Asp Glu Met
195 200 205 Leu Lys Leu Ile
Glu Leu His Asp Ala Ser Asn Ile Ala Ala Val Ile 210
215 220 Val Glu Pro Leu Ala Gly Ser Ala
Gly Val Leu Pro Pro Pro Lys Gly 225 230
235 240 Tyr Leu Lys Arg Leu Arg Glu Ile Cys Thr Gln His
Asn Ile Leu Leu 245 250
255 Ile Phe Asp Glu Val Ile Thr Gly Phe Gly Arg Met Gly Ala Met Thr
260 265 270 Gly Ser Glu
Ala Phe Gly Val Thr Pro Asp Leu Met Cys Ile Ala Lys 275
280 285 Gln Val Thr Asn Gly Ala Ile Pro
Met Gly Ala Val Ile Ala Ser Ser 290 295
300 Glu Ile Tyr Gln Thr Phe Met Asn Gln Pro Thr Pro Glu
Tyr Ala Val 305 310 315
320 Glu Phe Pro His Gly Tyr Thr Tyr Ser Ala His Pro Val Ala Cys Ala
325 330 335 Ala Gly Leu Ala
Ala Leu Asp Leu Leu Gln Lys Glu Asn Leu Val Gln 340
345 350 Ser Ala Ala Glu Leu Ala Pro His Phe
Glu Lys Leu Leu His Gly Val 355 360
365 Lys Gly Thr Lys Asn Ile Val Asp Ile Arg Asn Tyr Gly Leu
Ala Gly 370 375 380
Ala Ile Gln Ile Ala Ala Arg Asp Gly Asp Ala Ile Val Arg Pro Tyr 385
390 395 400 Glu Ala Ala Met Lys
Leu Trp Lys Ala Gly Phe Tyr Val Arg Phe Gly 405
410 415 Gly Asp Thr Leu Gln Phe Gly Pro Thr Phe
Asn Thr Lys Pro Gln Glu 420 425
430 Leu Asp Arg Leu Phe Asp Ala Val Gly Glu Thr Leu Asn Leu Ile
Asp 435 440 445
94930DNAPseudomonas putida 94atgaccacga agaaagctga ttacatttgg ttcaatgggg
agatggttcg ctgggaagac 60gcgaaggtgc atgtgatgtc gcacgcgctg cactatggca
cttcggtttt tgaaggcatc 120cgttgctacg actcgcacaa aggaccggtt gtattccgcc
atcgtgagca tatgcagcgt 180ctgcatgact ccgccaaaat ctatcgcttc ccggtttcgc
agagcattga tgagctgatg 240gaagcttgtc gtgacgtgat ccgcaaaaac aatctcacca
gcgcctatat ccgtccgctg 300atcttcgtcg gtgatgttgg catgggagta aacccgccag
cgggatactc aaccgacgtg 360attatcgctg ctttcccgtg gggagcgtat ctgggcgcag
aagcgctgga gcaggggatc 420gatgcgatgg tttcctcctg gaaccgcgca gcaccaaaca
ccatcccgac ggcggcaaaa 480gccggtggta actacctctc ttccctgctg gtgggtagcg
aagcgcgccg ccacggttat 540caggaaggta tcgcgctgga tgtgaacggt tatatctctg
aaggcgcagg cgaaaacctg 600tttgaagtga aagatggtgt gctgttcacc ccaccgttca
cctcctccgc gctgccgggt 660attacccgtg atgccatcat caaactggcg aaagagctgg
gaattgaagt acgtgagcag 720gtgctgtcgc gcgaatccct gtacctggcg gatgaagtgt
ttatgtccgg tacggcggca 780gaaatcacgc cagtgcgcag cgtagacggt attcaggttg
gcgaaggccg ttgtggcccg 840gttaccaaac gcattcagca agccttcttc ggcctcttca
ctggcgaaac cgaagataaa 900tggggctggt tagatcaagt taatcaataa
93095566PRTStreptomyces cinnamonensis 95Met Asp
Ala Asp Ala Ile Glu Glu Gly Arg Arg Arg Trp Gln Ala Arg 1 5
10 15 Tyr Asp Lys Ala Arg Lys Arg
Asp Ala Asp Phe Thr Thr Leu Ser Gly 20 25
30 Asp Pro Val Asp Pro Val Tyr Gly Pro Arg Pro Gly
Asp Thr Tyr Asp 35 40 45
Gly Phe Glu Arg Ile Gly Trp Pro Gly Glu Tyr Pro Phe Thr Arg Gly
50 55 60 Leu Tyr Ala
Thr Gly Tyr Arg Gly Arg Thr Trp Thr Ile Arg Gln Phe 65
70 75 80 Ala Gly Phe Gly Asn Ala Glu
Gln Thr Asn Glu Arg Tyr Lys Met Ile 85
90 95 Leu Ala Asn Gly Gly Gly Gly Leu Ser Val Ala
Phe Asp Met Pro Thr 100 105
110 Leu Met Gly Arg Asp Ser Asp Asp Pro Arg Ser Leu Gly Glu Val
Gly 115 120 125 His
Cys Gly Val Ala Ile Asp Ser Ala Ala Asp Met Glu Val Leu Phe 130
135 140 Lys Asp Ile Pro Leu Gly
Asp Val Thr Thr Ser Met Thr Ile Ser Gly 145 150
155 160 Pro Ala Val Pro Val Phe Cys Met Tyr Leu Val
Ala Ala Glu Arg Gln 165 170
175 Gly Val Asp Pro Ala Val Leu Asn Gly Thr Leu Gln Thr Asp Ile Phe
180 185 190 Lys Glu
Tyr Ile Ala Gln Lys Glu Trp Leu Phe Gln Pro Glu Pro His 195
200 205 Leu Arg Leu Ile Gly Asp Leu
Met Glu His Cys Ala Arg Asp Ile Pro 210 215
220 Ala Tyr Lys Pro Leu Ser Val Ser Gly Tyr His Ile
Arg Glu Ala Gly 225 230 235
240 Ala Thr Ala Ala Gln Glu Leu Ala Tyr Thr Leu Ala Asp Gly Phe Gly
245 250 255 Tyr Val Glu
Leu Gly Leu Ser Arg Gly Leu Asp Val Asp Val Phe Ala 260
265 270 Pro Gly Leu Ser Phe Phe Phe Asp
Ala His Val Asp Phe Phe Glu Glu 275 280
285 Ile Ala Lys Phe Arg Ala Ala Arg Arg Ile Trp Ala Arg
Trp Leu Arg 290 295 300
Asp Glu Tyr Gly Ala Lys Thr Glu Lys Ala Gln Trp Leu Arg Phe His 305
310 315 320 Thr Gln Thr Ala
Gly Val Ser Leu Thr Ala Gln Gln Pro Tyr Asn Asn 325
330 335 Val Val Arg Thr Ala Val Glu Ala Leu
Ala Ala Val Leu Gly Gly Thr 340 345
350 Asn Ser Leu His Thr Asn Ala Leu Asp Glu Thr Leu Ala Leu
Pro Ser 355 360 365
Glu Gln Ala Ala Glu Ile Ala Leu Arg Thr Gln Gln Val Leu Met Glu 370
375 380 Glu Thr Gly Val Ala
Asn Val Ala Asp Pro Leu Gly Gly Ser Trp Tyr 385 390
395 400 Ile Glu Gln Leu Thr Asp Arg Ile Glu Ala
Asp Ala Glu Lys Ile Phe 405 410
415 Glu Gln Ile Arg Glu Arg Gly Arg Arg Ala Cys Pro Asp Gly Gln
His 420 425 430 Pro
Ile Gly Pro Ile Thr Ser Gly Ile Leu Arg Gly Ile Glu Asp Gly 435
440 445 Trp Phe Thr Gly Glu Ile
Ala Glu Ser Ala Phe Gln Tyr Gln Arg Ser 450 455
460 Leu Glu Lys Gly Asp Lys Arg Val Val Gly Val
Asn Cys Leu Glu Gly 465 470 475
480 Ser Val Thr Gly Asp Leu Glu Ile Leu Arg Val Ser His Glu Val Glu
485 490 495 Arg Glu
Gln Val Arg Glu Leu Ala Gly Arg Lys Gly Arg Arg Asp Asp 500
505 510 Ala Arg Val Arg Ala Ser Leu
Asp Ala Met Leu Ala Ala Ala Arg Asp 515 520
525 Gly Ser Asn Met Ile Ala Pro Met Leu Glu Ala Val
Arg Ala Glu Ala 530 535 540
Thr Leu Gly Glu Ile Cys Gly Val Leu Arg Asp Glu Trp Gly Val Tyr 545
550 555 560 Val Glu Pro
Pro Gly Phe 565 964362DNAStreptomyces cinnamonensis
96tgaggcgctg gatcgcctcg gagagcagct ggtaacggtc cgcgtggtac tcggccgggg
60tgcagccgtc cacgatgtgc gggatcgcgt cgggctcgag gatcaccagg gcgggggcgt
120cgccgatcgc gtcggcgaac gtgtccaccc agctccggta ggcctccgca ctggccgcgc
180cgcccgcgga gtgctgaccg cagtcgcggt gcgggatgtt gtacgcgacg agtacggcgg
240tgcggtcctc cttgaccgcg ccccgcgtcg ccttcgcgac gtcgggcgcc ggatcgtccc
300cggccggcca cacggccatg gcccgttcgg agatgcgcct gagcgtctcg gcgtcctcgg
360cgcggccctg ttcctcccac tgcctgacct ggcgcgcggc ggggctgtcg gggtcgaccc
420agaaggtgcc ggcggggggc ccggcgctcg cggtggcggg cttgcgcacg gccgcctcct
480ccttcgtgcc gtcggacccc gggtctgagg aggagcagcc tgccgggagc ccgagggcgg
540cgagggccgc gagtgccgtg aacgtgcgga gcagccggtg catccagccc ccttgggcga
600tggtgacagt gacggtcagt cagcccggca atcgttacat aaaggactat tcaagctctt
660gtgccacacc gcctccggtg ccgagcgcga acccggcggg caccagagcc ccgccgcggc
720cgcggagccg tacgtacgac cgaattgcga gacggggctg accaccatat gaccggcggg
780taaggtcgat gccgtgccga agccgctcag cctccccttc gatcccatcg cccgcgccga
840cgagctctgg aagcagcgct ggggatcggt cccggccatg ggcgcgatca cctcgatcat
900gcgggcgcac cagatcctgc tcgccgaggt cgacgcggtc gtcaagccgt acggactgac
960cttcgcgcgc tacgaggcgc tggtgctcct caccttctcg caggccggcg agttgccgat
1020gtcgaagatc ggcgagcggc tcatggtgca cccgacctcg gtcacgaaca ccgtggaccg
1080cctggtgaag tccggcctgg tcgacaagcg cccgaacccc aacgacggcc gcggcacgct
1140cgcctccatc acggagaagg gccgcgaggt cgtcgaggcg gccacccgcg agctgatggc
1200gatggacttc gggctcgggg tgtacgacgc ggaggagtgc ggggagatct tcgcgatgct
1260gcggcccctg cgggtggcgg cgcgcgattt cgaggagcag tagggcccgc ccggtgagaa
1320gtgggatcgg gtcgtcccgg tacgggcggg ggcggcgaag atcgcgtgaa aagggcggtt
1380acgctcgtag ccatgaaacg cagcgtgctg acccgctacc gggtgatggc ctacgtcacc
1440gccgtcatgc tcctcatcct gtgcgcctgc atggtggcca agtacggctt cgacaagggc
1500gagggtctga ccctcgtcgt gtcgcaggtg cacggcgtgc tctacatcat ctacctgatc
1560ttcgccttcg acctgggctc caaggcgaag tggccgttcg gcaagctgct ctgggtgctg
1620gtctcgggca cgatcccgac cgccgccttc ttcgtcgagc gcaaggtcgc ccgtgacgtc
1680gagccgctga tcgccgacgg ctccccggtc accgcgaagg cgtaacccgc accgccacgg
1740acaggtccgt ggcggttggc catcgacttt tactaggacg tcctagtaaa ttcgatggta
1800tggacgctga cgcgatcgag gaaggccgcc gacgctggca ggcccgttac gacaaggccc
1860gcaagcgcga cgcggacttc accacgctct ccggggaccc cgtcgacccc gtctacggcc
1920cccggcccgg ggacacgtac gacgggttcg agcggatcgg ctggccgggg gagtacccct
1980tcacccgcgg gctctacgcc accgggtacc gcggccgcac ctggaccatc cgccagttcg
2040ccggcttcgg caacgccgag cagacgaacg agcgctacaa gatgatcctg gccaacggcg
2100gcggcggcct ctccgtcgcc ttcgacatgc cgaccctcat gggccgcgac tccgacgacc
2160cgcgctcgct cggcgaggtc ggccactgcg gtgtcgccat cgactccgcc gccgacatgg
2220aggtcctctt caaggacatc ccgctcggcg acgtcacgac gtccatgacc atcagcgggc
2280ccgccgtgcc cgtcttctgc atgtacctcg tcgcggccga gcgccagggc gtcgacccgg
2340ccgtcctcaa cggcacgctg cagaccgaca tcttcaagga gtacatcgcc cagaaggagt
2400ggctcttcca gcccgagccg cacctgcgcc tcatcggcga cctgatggag cactgcgcgc
2460gcgacatccc cgcgtacaag ccgctctcgg tctccggcta ccacatccgc gaggccgggg
2520cgacggccgc gcaggagctc gcgtacaccc tcgcggacgg cttcgggtac gtggaactgg
2580gcctctcgcg cggcctggac gtggacgtct tcgcgcccgg cctctccttc ttcttcgacg
2640cgcacgtcga cttcttcgag gagatcgcga agttccgcgc cgcacgccgc atctgggcgc
2700gctggctccg ggacgagtac ggagcgaaga ccgagaaggc acagtggctg cgcttccaca
2760cgcagaccgc gggggtctcg ctcacggccc agcagccgta caacaacgtg gtgcggacgg
2820cggtggaggc cctcgccgcg gtgctcggcg gcacgaactc cctgcacacc aacgctctcg
2880acgagaccct tgccctcccc agcgagcagg ccgcggagat cgcgctgcgc acccagcagg
2940tgctgatgga ggagaccggc gtcgccaacg tcgcggaccc gctgggcggc tcctggtaca
3000tcgagcagct caccgaccgc atcgaggccg acgccgagaa gatcttcgag cagatcaggg
3060agcgggggcg gcgggcctgc cccgacgggc agcacccgat cgggccgatc acctccggca
3120tcctgcgcgg catcgaggac ggctggttca ccggcgagat cgccgagtcc gccttccagt
3180accagcggtc cctggagaag ggcgacaagc gggtcgtcgg cgtcaactgc ctcgaaggct
3240ccgtcaccgg cgacctggag atcctgcgcg tcagccacga ggtcgagcgc gagcaggtgc
3300gggagcttgc ggggcgcaag gggcggcgtg acgatgcgcg ggtgcgggcc tcgctcgacg
3360cgatgctcgc cgctgcgcgg gacgggtcga acatgattgc ccccatgctg gaggcggtgc
3420gggccgaggc gaccctcggg gagatctgcg gggtgcttcg cgatgagtgg ggggtctacg
3480tggagccgcc cgggttctga gggcgcgctc cctttgcctg cgggtctgct gtggctggtc
3540gcgcagttcc ccgcacccct gaaagacccc ggcgctttcc cttcctggct cgcctcgtcg
3600ctgtctgcgg ggccgtgggg gctggtcgcg cagttccccg cgcccctgcc cgcacctgcg
3660ccccgccgcc tgcatgccgc ccccaccctg acgggggcgt tcggggccca ccctgacggg
3720tgcggtcggg gcgtgccggg gtcttttagg ggcgcgggga actgcgcgag caacccccac
3780ccacccgcag gtgcacgcgg agcggcggac gccccgcaga cgggggcaaa acgggcggag
3840tgcccccgcc cgccgggcgg cgcgaattcg taggtttaag gggcaggggt cagggcaggc
3900gccgagccgg tcaaccgccc ccgtcccagg agaccccgtg acctcgaccg gccacgcccg
3960caccgccgcc atcgccatcg gagccgccac cgccaccgtc ctcggcgcgc tgctggtcgg
4020cggctccggc gaggtgagtg cgagcccgcc gcccgagccc aaggtccagg acgacttcga
4080ctccctcggc cccgaggtgc gcgccgcgaa gctctccgac gggcggacgg cccactactc
4140ggacacgggc gacaaggacg gcaagccggc cctgttcatc ggcggcaccg gcacgagcgc
4200ccgcgcctcc cacatgaccg acttcttccg ctcgacgcgc gaggacctgg gcctgcgcct
4260catctccgtg gagcgcaacg gcttcggcga caccgcgttc gacgagaagc tgggcaccgc
4320cgacttcgcg aaggacgccc tcgaagtcct cgaccggctc gg
436297136PRTStreptomyces cinnamonensis 97Met Gly Val Ala Ala Gly Pro Ile
Arg Val Val Val Ala Lys Pro Gly 1 5 10
15 Leu Asp Gly His Asp Arg Gly Ala Lys Val Ile Ala Arg
Ala Leu Arg 20 25 30
Asp Ala Gly Met Glu Val Ile Tyr Thr Gly Leu His Gln Thr Pro Glu
35 40 45 Gln Val Val Asp
Thr Ala Ile Gln Glu Asp Ala Asp Ala Ile Gly Leu 50
55 60 Ser Ile Leu Ser Gly Ala His Asn
Thr Leu Phe Ala Arg Val Leu Glu 65 70
75 80 Leu Leu Lys Glu Arg Asp Ala Glu Asp Ile Lys Val
Phe Gly Gly Gly 85 90
95 Ile Ile Pro Glu Ala Asp Ile Ala Pro Leu Lys Glu Lys Gly Val Ala
100 105 110 Glu Ile Phe
Thr Pro Gly Ala Thr Thr Thr Ser Ile Val Glu Trp Val 115
120 125 Arg Gly Asn Val Arg Gln Ala Val
130 135 981643DNAStreptomyces cinnamonensis
98gtcgacctcc cgtttggcgc acggaaggga ggctctgtcc cccgtgtgcc ctagggggag
60tcgtggtcga ggagtcggct gtgcgatggc gatcccggcc accgccctgc ggtgactccg
120tgcccccgtt gcatcgccga tgcgcggtgt caccacgccg tgcggctgcc ggcgcggtgg
180cccggcgtct cgttgcggct cccctcgcgc ctggtccgga tgcggagcgt gaacccctgg
240gttacggacg ggcgcgcagc gaacgtgtcc cacgtgtgat ttccccctcg ctctccaccg
300cgaaactgcc gcttgcgcga tgctggggat aacgttcgtt cacttccccg gccggtgcgg
360tgcggggtat ctgtgccggg acagactttg tcggtacgga tatcggtaca tggaggcagt
420gatgggtgtg gcagccgggc cgatccgcgt ggtggtcgcc aagccggggc tcgacgggca
480cgatcgcggg gccaaggtga tcgcgcgggc gttgcgtgac gcgggtatgg aggtcatcta
540caccgggctg caccagacgc ccgagcaggt ggtggacacc gcgatccagg aggacgccga
600cgcgatcggc ctctccatcc tctccggagc gcacaacacg ctgttcgcgc gcgtgttgga
660gctcttgaag gagcgggacg cggaggacat caaggtgttt ggtggcggca tcatcccgga
720ggcggacatc gcgccgctga aggagaaggg cgtcgcggag atcttcacgc ccggggccac
780caccacgtcg atcgtggagt gggttcgggg gaacgtgcga caggccgtct gaggcattcc
840ccgtcgcccg tctgccgtgg tcggcgtcat atcggcggac atcgtctcgg tggacgtcat
900ggcggcgggg ggagttcgtc gcgtatcgcc gcgcggaggc gcagggtggt gaccaggcgc
960tggaacgctt ccgaccagta gctgcccgcg ccgggtgacg cgtcctccgc ttcgtcgggg
1020accgcggtga gcgcttccag gcggaccgcc tcggccgggt ccagacagcg ttccgccagg
1080cccatcactc cgctgaagct ccatgggtaa ctgcccgcgt cgcgcgcgat gttcagggcg
1140tccaccacgg cccggccgag agggccggcc cagggcaccg cgcagacgcc gagcagttgg
1200aacgcctccg acaggccgtg tgccgctatg aaccccgcca cccagtccgc gcgctcggcg
1260gcaggcatgg aggcgagcag tttggcccgc tcggcgaggg acacggcgcc aggccccgcc
1320gcgtcgggtg aggcgggggc gccgagcagc gctctggacc aggcgacgtc acgctggcgt
1380acggccgcgc ggcaccatgc ggcgtgcagt tcgccccgcc agtcgtcggc caccgggagc
1440gccacgatct ccgccggggt gcggttgccg agccggggcg gccaggtggc gagcggggcc
1500gattccacga gctggccgag ccaccaggag cgctcgcccc ggccggtggg gggcttcggg
1560acgacgccgt cccgctccat gcccgcgtcg cactcgtgcg gcgcctcgac ggtgagggtc
1620ggcgtgctcg atgtgtggtc gac
164399566PRTStreptomyces coelicolor 99Met Asp Ala His Ala Ile Glu Glu Gly
Arg Leu Arg Trp Gln Ala Arg 1 5 10
15 Tyr Asp Ala Ala Arg Lys Arg Asp Ala Asp Phe Thr Thr Leu
Ser Gly 20 25 30
Asp Pro Val Glu Pro Val Tyr Gly Pro Arg Pro Gly Asp Glu Tyr Glu
35 40 45 Gly Phe Glu Arg
Ile Gly Trp Pro Gly Glu Tyr Pro Phe Thr Arg Gly 50
55 60 Leu Tyr Pro Thr Gly Tyr Arg Gly
Arg Thr Trp Thr Ile Arg Gln Phe 65 70
75 80 Ala Gly Phe Gly Asn Ala Glu Gln Thr Asn Glu Arg
Tyr Lys Met Ile 85 90
95 Leu Arg Asn Gly Gly Gly Gly Leu Ser Val Ala Phe Asp Met Pro Thr
100 105 110 Leu Met Gly
Arg Asp Ser Asp Asp Pro Arg Ser Leu Gly Glu Val Gly 115
120 125 His Cys Gly Val Ala Ile Asp Ser
Ala Ala Asp Met Glu Val Leu Phe 130 135
140 Lys Asp Ile Pro Leu Gly Asp Val Thr Thr Ser Met Thr
Ile Ser Gly 145 150 155
160 Pro Ala Val Pro Val Phe Cys Met Tyr Leu Val Ala Ala Glu Arg Gln
165 170 175 Gly Val Asp Ala
Ser Val Leu Asn Gly Thr Leu Gln Thr Asp Ile Phe 180
185 190 Lys Glu Tyr Ile Ala Gln Lys Glu Trp
Leu Phe Gln Pro Glu Pro His 195 200
205 Leu Arg Leu Ile Gly Asp Leu Met Glu Tyr Cys Ala Ala Gly
Ile Pro 210 215 220
Ala Tyr Lys Pro Leu Ser Val Ser Gly Tyr His Ile Arg Glu Ala Gly 225
230 235 240 Ala Thr Ala Ala Gln
Glu Leu Ala Tyr Thr Leu Ala Asp Gly Phe Gly 245
250 255 Tyr Val Glu Leu Gly Leu Ser Arg Gly Leu
Asp Val Asp Val Phe Ala 260 265
270 Pro Gly Leu Ser Phe Phe Phe Asp Ala His Leu Asp Phe Phe Glu
Glu 275 280 285 Ile
Ala Lys Phe Arg Ala Ala Arg Arg Ile Trp Ala Arg Trp Met Arg 290
295 300 Asp Val Tyr Gly Ala Arg
Thr Asp Lys Ala Gln Trp Leu Arg Phe His 305 310
315 320 Thr Gln Thr Ala Gly Val Ser Leu Thr Ala Gln
Gln Pro Tyr Asn Asn 325 330
335 Val Val Arg Thr Ala Val Glu Ala Leu Ala Ala Val Leu Gly Gly Thr
340 345 350 Asn Ser
Leu His Thr Asn Ala Leu Asp Glu Thr Leu Ala Leu Pro Ser 355
360 365 Glu Gln Ala Ala Glu Ile Ala
Leu Arg Thr Gln Gln Val Leu Met Glu 370 375
380 Glu Thr Gly Val Ala Asn Val Ala Asp Pro Leu Gly
Gly Ser Trp Phe 385 390 395
400 Ile Glu Gln Leu Thr Asp Arg Ile Glu Ala Asp Ala Glu Lys Ile Phe
405 410 415 Glu Gln Ile
Lys Glu Arg Gly Leu Arg Ala His Pro Asp Gly Gln His 420
425 430 Pro Val Gly Pro Ile Thr Ser Gly
Leu Leu Arg Gly Ile Glu Asp Gly 435 440
445 Trp Phe Thr Gly Glu Ile Ala Glu Ser Ala Phe Arg Tyr
Gln Gln Ser 450 455 460
Leu Glu Lys Asp Asp Lys Lys Val Val Gly Val Asn Val His Thr Gly 465
470 475 480 Ser Val Thr Gly
Asp Leu Glu Ile Leu Arg Val Ser His Glu Val Glu 485
490 495 Arg Glu Gln Val Arg Val Leu Gly Glu
Arg Lys Asp Ala Arg Asp Asp 500 505
510 Ala Ala Val Arg Gly Ala Leu Asp Ala Met Leu Ala Ala Ala
Arg Ser 515 520 525
Gly Gly Asn Met Ile Gly Pro Met Leu Asp Ala Val Arg Ala Glu Ala 530
535 540 Thr Leu Gly Glu Ile
Cys Gly Val Leu Arg Asp Glu Trp Gly Val Tyr 545 550
555 560 Thr Glu Pro Ala Gly Phe
565 1001701DNAStreptomyces coelicolor 100atggacgctc atgccataga
ggagggccgc cttcgctggc aggcccggta cgacgcggcg 60cgcaagcgcg acgcggactt
caccacgctc tccggagacc ccgtggagcc ggtgtacggg 120ccccgccccg gggacgagta
cgagggcttc gagcggatcg gctggccggg cgagtacccc 180ttcacccgcg gcctgtatcc
gaccgggtac cgggggcgta cgtggaccat ccggcagttc 240gccgggttcg gcaacgccga
gcagaccaac gagcgctaca agatgatcct ccgcaacggc 300ggcggcgggc tctcggtcgc
cttcgacatg ccgaccctga tgggccgcga ctccgacgac 360ccgcgctcgc tgggcgaggt
cgggcactgc ggggtggcca tcgactcggc cgccgacatg 420gaagtgctgt tcaaggacat
cccgctcggg gacgtgacga cctccatgac gatcagcggg 480cccgccgtgc ccgtgttctg
catgtacctc gtcgccgccg agcgccaggg cgtcgacgca 540tccgtgctca acggcacgct
gcagaccgac atcttcaagg agtacatcgc ccagaaggag 600tggctcttcc agcccgagcc
ccacctccgg ctcatcggcg acctcatgga gtactgcgcg 660gccggcatcc ccgcctacaa
gccgctctcc gtctccggct accacatccg cgaggcgggc 720gcgacggccg cgcaggagct
ggcgtacacg ctcgccgacg gcttcggata cgtggagctg 780ggcctcagcc gcgggctcga
cgtggacgtc ttcgcgcccg gcctctcctt cttcttcgac 840gcgcacctcg acttcttcga
ggagatcgcc aagttccgcg cggcccgcag gatctgggcc 900cgctggatgc gcgacgtgta
cggcgcgcgg accgacaagg cccagtggct gcggttccac 960acccagaccg ccggagtctc
gctcaccgcg cagcagccgt acaacaacgt cgtacgcacc 1020gcggtggagg cgctggcggc
cgtgctcggc ggcaccaact ccctgcacac caacgcgctc 1080gacgagaccc tcgccctgcc
cagcgagcag gccgccgaga tcgccctgcg cacccagcag 1140gtgctgatgg aggagaccgg
cgtcgccaac gtcgccgacc cgctgggcgg ttcctggttc 1200atcgagcagc tgaccgaccg
catcgaggcc gacgccgaga agatcttcga gcagatcaag 1260gagcgggggc tgcgcgccca
ccccgacggg cagcaccccg tcggaccgat cacctccggc 1320ctgctgcgcg gcatcgagga
cggctggttc accggcgaga tcgccgagtc cgccttccgc 1380taccagcagt ccttggagaa
ggacgacaag aaggtggtcg gcgtcaacgt ccacaccggc 1440tccgtcaccg gcgacctgga
gatcctgcgg gtcagccacg aggtcgagcg cgagcaggtg 1500cgggtcctgg gcgagcgcaa
ggacgcccgg gacgacgccg ccgtgcgcgg cgccctggac 1560gccatgctgg ccgcggcccg
ctccggcggc aacatgatcg ggccgatgct ggacgcggtg 1620cgcgcggagg cgacgctggg
cgagatctgc ggtgtgctgc gcgacgagtg gggggtgtac 1680acggaaccgg cggggttctg a
1701101138PRTStreptomyces
coelicolor 101Met Gly Val Ala Ala Gly Pro Ile Arg Val Val Val Ala Lys Pro
Gly 1 5 10 15 Leu
Asp Gly His Asp Arg Gly Ala Lys Val Ile Ala Arg Ala Leu Arg
20 25 30 Asp Ala Gly Met Glu
Val Ile Tyr Thr Gly Leu His Gln Thr Pro Glu 35
40 45 Gln Ile Val Asp Thr Ala Ile Gln Glu
Asp Ala Asp Ala Ile Gly Leu 50 55
60 Ser Ile Leu Ser Gly Ala His Asn Thr Leu Phe Ala Ala
Val Ile Glu 65 70 75
80 Leu Leu Arg Glu Arg Asp Ala Ala Asp Ile Leu Val Phe Gly Gly Gly
85 90 95 Ile Ile Pro Glu
Ala Asp Ile Ala Pro Leu Lys Glu Lys Gly Val Ala 100
105 110 Glu Ile Phe Thr Pro Gly Ala Thr Thr
Ala Ser Ile Val Asp Trp Val 115 120
125 Arg Ala Asn Val Arg Glu Pro Ala Gly Ala 130
135 102417DNAStreptomyces coelicolor 102atgggtgtgg
cagccggtcc gatccgcgtg gtggtggcca agccggggct cgacggccac 60gatcgcgggg
ccaaggtgat cgcgagggcc ctgcgtgacg ccggtatgga ggtgatctac 120accgggctcc
accagacgcc cgagcagatc gtcgacaccg cgatccagga ggacgccgac 180gcgatcgggc
tgtccatcct ctccggtgcg cacaacacgc tcttcgccgc cgtgatcgag 240ctgctccggg
agcgggacgc cgcggacatc ctggtcttcg gcggcgggat catccccgag 300gcggacatcg
ccccgctgaa ggagaagggc gtcgcggaga tcttcacgcc cggcgccacc 360acggcgtcca
tcgtggactg ggtccgggcg aacgtgcggg agcccgcggg agcatag
417103566PRTStreptomyces avermitilis 103Met Asp Ala Asp Ala Ile Glu Glu
Gly Arg Arg Arg Trp Gln Ala Arg 1 5 10
15 Tyr Asp Ala Ser Arg Lys Arg Glu Ala Asp Phe Thr Thr
Leu Ser Gly 20 25 30
Asp Pro Val Glu Pro Ala Tyr Gly Pro Arg Pro Gly Asp Ala Tyr Glu
35 40 45 Gly Phe Glu Arg
Ile Gly Trp Pro Gly Glu Tyr Pro Phe Thr Arg Gly 50
55 60 Leu Tyr Pro Thr Gly Tyr Arg Gly
Arg Thr Trp Thr Ile Arg Gln Phe 65 70
75 80 Ala Gly Phe Gly Asn Ala Glu Gln Thr Asn Glu Arg
Tyr Lys Lys Ile 85 90
95 Leu Ala Asn Gly Gly Gly Gly Leu Ser Val Ala Phe Asp Met Pro Thr
100 105 110 Leu Met Gly
Arg Asp Ser Asp Asp Arg Arg Ala Leu Gly Glu Val Gly 115
120 125 His Cys Gly Val Ala Ile Asp Ser
Ala Ala Asp Met Glu Val Leu Phe 130 135
140 Lys Asp Ile Pro Leu Gly Asp Val Thr Thr Ser Met Thr
Ile Ser Gly 145 150 155
160 Pro Ala Val Pro Val Phe Cys Met Tyr Leu Val Ala Ala Glu Arg Gln
165 170 175 Gly Val Asp Pro
Ser Val Leu Asn Gly Thr Leu Gln Thr Asp Ile Phe 180
185 190 Lys Glu Tyr Ile Ala Gln Lys Glu Trp
Leu Phe Gln Pro Glu Pro His 195 200
205 Leu Arg Leu Ile Gly Asp Leu Met Glu His Cys Ala Ser Lys
Ile Pro 210 215 220
Ala Tyr Lys Pro Leu Ser Val Ser Gly Tyr His Ile Arg Glu Ala Gly 225
230 235 240 Ala Thr Ala Ala Gln
Glu Leu Ala Tyr Thr Leu Ala Asp Gly Phe Gly 245
250 255 Tyr Val Glu Leu Gly Leu Ser Arg Gly Leu
Asp Val Asp Val Phe Ala 260 265
270 Pro Gly Leu Ser Phe Phe Phe Asp Ala His Val Asp Phe Phe Glu
Glu 275 280 285 Ile
Ala Lys Phe Arg Ala Ala Arg Arg Ile Trp Ala Arg Trp Leu Arg 290
295 300 Asp Val Tyr Gly Ala Lys
Ser Glu Lys Ala Gln Trp Leu Arg Phe His 305 310
315 320 Thr Gln Thr Ala Gly Val Ser Leu Thr Ala Gln
Gln Pro Tyr Asn Asn 325 330
335 Val Val Arg Thr Ala Val Glu Ala Leu Ala Ala Val Leu Gly Gly Thr
340 345 350 Asn Ser
Leu His Thr Asn Ala Leu Asp Glu Thr Leu Ala Leu Pro Ser 355
360 365 Glu Gln Ala Ala Glu Ile Ala
Leu Arg Thr Gln Gln Val Leu Met Glu 370 375
380 Glu Thr Gly Val Ala Asn Val Ala Asp Pro Leu Gly
Gly Ser Trp Tyr 385 390 395
400 Val Glu Gln Leu Thr Asp Arg Ile Glu Ala Asp Ala Glu Lys Ile Phe
405 410 415 Glu Gln Ile
Arg Glu Arg Gly Leu Arg Ala His Pro Asp Gly Arg His 420
425 430 Pro Ile Gly Pro Ile Thr Ser Gly
Ile Leu Arg Gly Ile Glu Asp Gly 435 440
445 Trp Phe Thr Gly Glu Ile Ala Glu Ser Ala Phe Gln Tyr
Gln Gln Ala 450 455 460
Leu Glu Lys Gly Asp Lys Arg Val Val Gly Val Asn Val His His Gly 465
470 475 480 Ser Val Thr Gly
Asp Leu Glu Ile Leu Arg Val Ser His Glu Val Glu 485
490 495 Arg Glu Gln Val Arg Val Leu Gly Glu
Arg Lys Ser Gly Arg Asp Asp 500 505
510 Thr Ala Val Thr Ala Ala Leu Asp Ala Met Leu Ala Ala Ala
Arg Asp 515 520 525
Gly Ser Asn Met Ile Ala Pro Met Leu Asp Ala Val Arg Ala Glu Ala 530
535 540 Thr Leu Gly Glu Ile
Cys Asp Val Leu Arg Glu Glu Trp Gly Val Tyr 545 550
555 560 Thr Glu Pro Ala Gly Phe
565 1041701DNAStreptomyces avermitilis 104tcagaaaccg gcgggctccg
tgtagacccc ccactcctcc cggaggacat cgcagatctc 60gcccagcgtg gcctccgcgc
ggaccgcgtc cagcatcggg gcgatcatgt tcgacccgtc 120gcgcgcggcg gcgagcatcg
cgtccagggc cgcggttacg gccgtgtcgt cgcgccccga 180cttccgctcg cccagcaccc
gcacctgctc gcgctccacc tcgtggctga cgcgcaggat 240ctccaggtcg cccgtcacgg
acccgtggtg gacgttgacg ccgacgaccc gcttgtcgcc 300cttctccagc gcctgctggt
actggaaggc cgactcggcg atctccccgg tgaaccagcc 360gtcctcgatg ccgcgcagga
tgccggaggt gatgggcccg atcgggtgcc gcccgtccgg 420gtgggcccgc agcccgcgct
ccctgatctg ttcgaagatc ttctcggcgt cggcctcgat 480ccggtcggtc agctgctcca
cgtaccagga accgcccagc ggatcggcca cgttggcgac 540gcccgtctcc tccatcagca
cctgctgggt gcgcagggcg atctcggccg cctgctcgga 600cggcagggcg agggtctcgt
cgagggcgtt ggtgtgcagc gagttcgtcc cgccgagcac 660cgcggcgagg gcctccacgg
ccgtccgtac gacgttgttg tacggctgct gcgcggtgag 720cgagacgccc gcggtctggg
tgtggaagcg cagccactgc gccttctccg acttcgcccc 780gtacacgtcc cgcagccagc
gcgcccagat gcgccgcgcc gcacggaact tggcgatctc 840ctcgaagaag tcgacgtgcg
cgtcgaagaa gaaggagagc ccgggcgcga acacgtccac 900gtccaggccg cggctcagcc
ccagctccac gtatccgaaa ccgtcggcga gggtgtacgc 960cagctcctgg gcggccgtgg
caccggcctc ccggatgtgg tacccggaga cggacagcgg 1020cttgtacgcg gggatcttcg
aggcgcagtg ctccatcagg tcgccgatga gccgcagatg 1080gggctcgggc tggaagagcc
actccttctg cgcgatgtac tccttgaaga tgtcggtctg 1140gagggtgccg ttgaggacgg
aggggtcgac gccctgccgc tcggccgcga ccaggtacat 1200gcagaagacg ggcacggcgg
gcccgctgat cgtcatcgac gtcgtcacgt cacccagcgg 1260gatgtccttg aacaggacct
ccatgtcggc cgccgagtcg atcgcgaccc cgcagtgccc 1320gacctcgccg agcgcgcggc
ggtcgtcgga gtcgcgcccc atgagcgtcg gcatgtcgaa 1380ggccacggac agcccaccgc
cgccgttggc gaggatcttc ttgtagcgct cgttggtctg 1440ctcggcgttg ccgaacccgg
cgaactgccg gatggtccag gtccggcccc ggtagccggt 1500cggatacaga ccgcgcgtga
aggggtactc acccggccag ccgatccgct cgaaaccctc 1560gtacgcgtcc ccgggccggg
gcccgtacgc cggctccacg ggatcgccgg agagcgtggt 1620gaaatcggcc tcgcgcttgc
gtgaggcgtc gtagcgggcc tgccagcgtc ggcggccttc 1680ctcgatggcg tcagcgtcca t
1701105138PRTStreptomyces
avermitilis 105Met Gly Val Ala Ala Gly Pro Ile Arg Val Val Val Ala Lys
Pro Gly 1 5 10 15
Leu Asp Gly His Asp Arg Gly Ala Lys Val Ile Ala Arg Ala Leu Arg
20 25 30 Asp Ala Gly Met Glu
Val Ile Tyr Thr Gly Leu His Gln Thr Pro Glu 35
40 45 Gln Ile Val Gly Thr Ala Ile Gln Glu
Asp Ala Asp Ala Ile Gly Leu 50 55
60 Ser Ile Leu Ser Gly Ala His Asn Thr Leu Phe Ala Ala
Val Ile Asp 65 70 75
80 Leu Leu Lys Glu Arg Asp Ala Glu Asp Ile Lys Val Phe Gly Gly Gly
85 90 95 Ile Ile Pro Glu
Ala Asp Ile Ala Pro Leu Lys Glu Lys Gly Val Ala 100
105 110 Glu Ile Phe Thr Pro Gly Ala Thr Thr
Ala Ser Ile Val Glu Trp Val 115 120
125 Arg Ala Asn Val Arg Gln Pro Ala Gly Ala 130
135 1061701DNAStreptomyces avermitilis 106tcagaaaccg
gcgggctccg tgtagacccc ccactcctcc cggaggacat cgcagatctc 60gcccagcgtg
gcctccgcgc ggaccgcgtc cagcatcggg gcgatcatgt tcgacccgtc 120gcgcgcggcg
gcgagcatcg cgtccagggc cgcggttacg gccgtgtcgt cgcgccccga 180cttccgctcg
cccagcaccc gcacctgctc gcgctccacc tcgtggctga cgcgcaggat 240ctccaggtcg
cccgtcacgg acccgtggtg gacgttgacg ccgacgaccc gcttgtcgcc 300cttctccagc
gcctgctggt actggaaggc cgactcggcg atctccccgg tgaaccagcc 360gtcctcgatg
ccgcgcagga tgccggaggt gatgggcccg atcgggtgcc gcccgtccgg 420gtgggcccgc
agcccgcgct ccctgatctg ttcgaagatc ttctcggcgt cggcctcgat 480ccggtcggtc
agctgctcca cgtaccagga accgcccagc ggatcggcca cgttggcgac 540gcccgtctcc
tccatcagca cctgctgggt gcgcagggcg atctcggccg cctgctcgga 600cggcagggcg
agggtctcgt cgagggcgtt ggtgtgcagc gagttcgtcc cgccgagcac 660cgcggcgagg
gcctccacgg ccgtccgtac gacgttgttg tacggctgct gcgcggtgag 720cgagacgccc
gcggtctggg tgtggaagcg cagccactgc gccttctccg acttcgcccc 780gtacacgtcc
cgcagccagc gcgcccagat gcgccgcgcc gcacggaact tggcgatctc 840ctcgaagaag
tcgacgtgcg cgtcgaagaa gaaggagagc ccgggcgcga acacgtccac 900gtccaggccg
cggctcagcc ccagctccac gtatccgaaa ccgtcggcga gggtgtacgc 960cagctcctgg
gcggccgtgg caccggcctc ccggatgtgg tacccggaga cggacagcgg 1020cttgtacgcg
gggatcttcg aggcgcagtg ctccatcagg tcgccgatga gccgcagatg 1080gggctcgggc
tggaagagcc actccttctg cgcgatgtac tccttgaaga tgtcggtctg 1140gagggtgccg
ttgaggacgg aggggtcgac gccctgccgc tcggccgcga ccaggtacat 1200gcagaagacg
ggcacggcgg gcccgctgat cgtcatcgac gtcgtcacgt cacccagcgg 1260gatgtccttg
aacaggacct ccatgtcggc cgccgagtcg atcgcgaccc cgcagtgccc 1320gacctcgccg
agcgcgcggc ggtcgtcgga gtcgcgcccc atgagcgtcg gcatgtcgaa 1380ggccacggac
agcccaccgc cgccgttggc gaggatcttc ttgtagcgct cgttggtctg 1440ctcggcgttg
ccgaacccgg cgaactgccg gatggtccag gtccggcccc ggtagccggt 1500cggatacaga
ccgcgcgtga aggggtactc acccggccag ccgatccgct cgaaaccctc 1560gtacgcgtcc
ccgggccggg gcccgtacgc cggctccacg ggatcgccgg agagcgtggt 1620gaaatcggcc
tcgcgcttgc gtgaggcgtc gtagcgggcc tgccagcgtc ggcggccttc 1680ctcgatggcg
tcagcgtcca t
1701107139PRTSaccharomyces cerevisiae 107Met Ser Glu Ile Thr Leu Gly Lys
Tyr Leu Phe Glu Arg Leu Lys Gln 1 5 10
15 Val Asn Val Asn Thr Val Phe Gly Leu Pro Gly Asp Phe
Asn Leu Ser 20 25 30
Leu Leu Asp Lys Ile Tyr Glu Val Glu Gly Met Arg Trp Ala Gly Asn
35 40 45 Ala Asn Glu Leu
Asn Ala Ala Tyr Ala Ala Asp Gly Tyr Ala Arg Ile 50
55 60 Lys Gly Met Ser Cys Ile Ile Thr
Thr Phe Gly Val Gly Glu Leu Ser 65 70
75 80 Ala Leu Asn Gly Ile Ala Gly Ser Tyr Ala Glu His
Val Gly Val Leu 85 90
95 His Val Val Gly Val Pro Ser Ile Ser Ala Gln Ala Lys Gln Leu Leu
100 105 110 Leu His His
Thr Leu Gly Asn Gly Asp Phe Thr Val Phe His Arg Met 115
120 125 Ser Ala Asn Ile Ser Glu Thr Thr
Ala Met Ile 130 135
1081689DNASaccharomyces cerevisiae 108atgtctgaaa ttactttggg taaatatttg
ttcgaaagat taaagcaagt caacgttaac 60accgttttcg gtttgccagg tgacttcaac
ttgtccttgt tggacaagat ctacgaagtt 120gaaggtatga gatgggctgg taacgccaac
gaattgaacg ctgcttacgc cgctgatggt 180tacgctcgta tcaagggtat gtcttgtatc
atcaccacct tcggtgtcgg tgaattgtct 240gctttgaacg gtattgccgg ttcttacgct
gaacacgtcg gtgttttgca cgttgttggt 300gtcccatcca tctctgctca agctaagcaa
ttgttgttgc accacacctt gggtaacggt 360gacttcactg ttttccacag aatgtctgcc
aacatttctg aaaccactgc tatgatcact 420gacattgcta ccgccccagc tgaaattgac
agatgtatca gaaccactta cgtcacccaa 480agaccagtct acttaggttt gccagctaac
ttggtcgact tgaacgtccc agctaagttg 540ttgcaaactc caattgacat gtctttgaag
ccaaacgatg ctgaatccga aaaggaagtc 600attgacacca tcttggcttt ggtcaaggat
gctaagaacc cagttatctt ggctgatgct 660tgttgttcca gacacgacgt caaggctgaa
actaagaagt tgattgactt gactcaattc 720ccagctttcg tcaccccaat gggtaagggt
tccattgacg aacaacaccc aagatacggt 780ggtgtttacg tcggtacctt gtccaagcca
gaagttaagg aagccgttga atctgctgac 840ttgattttgt ctgtcggtgc tttgttgtct
gatttcaaca ccggttcttt ctcttactct 900tacaagacca agaacattgt cgaattccac
tccgaccaca tgaagatcag aaacgccact 960ttcccaggtg tccaaatgaa attcgttttg
caaaagttgt tgaccactat tgctgacgcc 1020gctaagggtt acaagccagt tgctgtccca
gctagaactc cagctaacgc tgctgtccca 1080gcttctaccc cattgaagca agaatggatg
tggaaccaat tgggtaactt cttgcaagaa 1140ggtgatgttg tcattgctga aaccggtacc
tccgctttcg gtatcaacca aaccactttc 1200ccaaacaaca cctacggtat ctctcaagtc
ttatggggtt ccattggttt caccactggt 1260gctaccttgg gtgctgcttt cgctgctgaa
gaaattgatc caaagaagag agttatctta 1320ttcattggtg acggttcttt gcaattgact
gttcaagaaa tctccaccat gatcagatgg 1380ggcttgaagc catacttgtt cgtcttgaac
aacgatggtt acaccattga aaagttgatt 1440cacggtccaa aggctcaata caacgaaatt
caaggttggg accacctatc cttgttgcca 1500actttcggtg ctaaggacta tgaaacccac
agagtcgcta ccaccggtga atgggacaag 1560ttgacccaag acaagtcttt caacgacaac
tctaagatca gaatgattga aatcatgttg 1620ccagtcttcg atgctccaca aaacttggtt
gaacaagcta agttgactgc tgctaccaac 1680gctaagcaa
1689109563PRTSaccharomyces cerevisiae
109Met Ser Glu Ile Thr Leu Gly Lys Tyr Leu Phe Glu Arg Leu Ser Gln 1
5 10 15 Val Asn Cys Asn
Thr Val Phe Gly Leu Pro Gly Asp Phe Asn Leu Ser 20
25 30 Leu Leu Asp Lys Leu Tyr Glu Val Lys
Gly Met Arg Trp Ala Gly Asn 35 40
45 Ala Asn Glu Leu Asn Ala Ala Tyr Ala Ala Asp Gly Tyr Ala
Arg Ile 50 55 60
Lys Gly Met Ser Cys Ile Ile Thr Thr Phe Gly Val Gly Glu Leu Ser 65
70 75 80 Ala Leu Asn Gly Ile
Ala Gly Ser Tyr Ala Glu His Val Gly Val Leu 85
90 95 His Val Val Gly Val Pro Ser Ile Ser Ser
Gln Ala Lys Gln Leu Leu 100 105
110 Leu His His Thr Leu Gly Asn Gly Asp Phe Thr Val Phe His Arg
Met 115 120 125 Ser
Ala Asn Ile Ser Glu Thr Thr Ala Met Ile Thr Asp Ile Ala Asn 130
135 140 Ala Pro Ala Glu Ile Asp
Arg Cys Ile Arg Thr Thr Tyr Thr Thr Gln 145 150
155 160 Arg Pro Val Tyr Leu Gly Leu Pro Ala Asn Leu
Val Asp Leu Asn Val 165 170
175 Pro Ala Lys Leu Leu Glu Thr Pro Ile Asp Leu Ser Leu Lys Pro Asn
180 185 190 Asp Ala
Glu Ala Glu Ala Glu Val Val Arg Thr Val Val Glu Leu Ile 195
200 205 Lys Asp Ala Lys Asn Pro Val
Ile Leu Ala Asp Ala Cys Ala Ser Arg 210 215
220 His Asp Val Lys Ala Glu Thr Lys Lys Leu Met Asp
Leu Thr Gln Phe 225 230 235
240 Pro Val Tyr Val Thr Pro Met Gly Lys Gly Ala Ile Asp Glu Gln His
245 250 255 Pro Arg Tyr
Gly Gly Val Tyr Val Gly Thr Leu Ser Arg Pro Glu Val 260
265 270 Lys Lys Ala Val Glu Ser Ala Asp
Leu Ile Leu Ser Ile Gly Ala Leu 275 280
285 Leu Ser Asp Phe Asn Thr Gly Ser Phe Ser Tyr Ser Tyr
Lys Thr Lys 290 295 300
Asn Ile Val Glu Phe His Ser Asp His Ile Lys Ile Arg Asn Ala Thr 305
310 315 320 Phe Pro Gly Val
Gln Met Lys Phe Ala Leu Gln Lys Leu Leu Asp Ala 325
330 335 Ile Pro Glu Val Val Lys Asp Tyr Lys
Pro Val Ala Val Pro Ala Arg 340 345
350 Val Pro Ile Thr Lys Ser Thr Pro Ala Asn Thr Pro Met Lys
Gln Glu 355 360 365
Trp Met Trp Asn His Leu Gly Asn Phe Leu Arg Glu Gly Asp Ile Val 370
375 380 Ile Ala Glu Thr Gly
Thr Ser Ala Phe Gly Ile Asn Gln Thr Thr Phe 385 390
395 400 Pro Thr Asp Val Tyr Ala Ile Val Gln Val
Leu Trp Gly Ser Ile Gly 405 410
415 Phe Thr Val Gly Ala Leu Leu Gly Ala Thr Met Ala Ala Glu Glu
Leu 420 425 430 Asp
Pro Lys Lys Arg Val Ile Leu Phe Ile Gly Asp Gly Ser Leu Gln 435
440 445 Leu Thr Val Gln Glu Ile
Ser Thr Met Ile Arg Trp Gly Leu Lys Pro 450 455
460 Tyr Ile Phe Val Leu Asn Asn Asn Gly Tyr Thr
Ile Glu Lys Leu Ile 465 470 475
480 His Gly Pro His Ala Glu Tyr Asn Glu Ile Gln Gly Trp Asp His Leu
485 490 495 Ala Leu
Leu Pro Thr Phe Gly Ala Arg Asn Tyr Glu Thr His Arg Val 500
505 510 Ala Thr Thr Gly Glu Trp Glu
Lys Leu Thr Gln Asp Lys Asp Phe Gln 515 520
525 Asp Asn Ser Lys Ile Arg Met Ile Glu Val Met Leu
Pro Val Phe Asp 530 535 540
Ala Pro Gln Asn Leu Val Lys Gln Ala Gln Leu Thr Ala Ala Thr Asn 545
550 555 560 Ala Lys Gln
1101689DNASaccharomyces cerevisiae 110atgtctgaaa taaccttagg taaatattta
tttgaaagat tgagccaagt caactgtaac 60accgtcttcg gtttgccagg tgactttaac
ttgtctcttt tggataagct ttatgaagtc 120aaaggtatga gatgggctgg taacgctaac
gaattgaacg ctgcctatgc tgctgatggt 180tacgctcgta tcaagggtat gtcctgtatt
attaccacct tcggtgttgg tgaattgtct 240gctttgaatg gtattgccgg ttcttacgct
gaacatgtcg gtgttttgca cgttgttggt 300gttccatcca tctcttctca agctaagcaa
ttgttgttgc atcatacctt gggtaacggt 360gacttcactg ttttccacag aatgtctgcc
aacatttctg aaaccactgc catgatcact 420gatattgcta acgctccagc tgaaattgac
agatgtatca gaaccaccta cactacccaa 480agaccagtct acttgggttt gccagctaac
ttggttgact tgaacgtccc agccaagtta 540ttggaaactc caattgactt gtctttgaag
ccaaacgacg ctgaagctga agctgaagtt 600gttagaactg ttgttgaatt gatcaaggat
gctaagaacc cagttatctt ggctgatgct 660tgtgcttcta gacatgatgt caaggctgaa
actaagaagt tgatggactt gactcaattc 720ccagtttacg tcaccccaat gggtaagggt
gctattgacg aacaacaccc aagatacggt 780ggtgtttacg ttggtacctt gtctagacca
gaagttaaga aggctgtaga atctgctgat 840ttgatattgt ctatcggtgc tttgttgtct
gatttcaata ccggttcttt ctcttactcc 900tacaagacca aaaatatcgt tgaattccac
tctgaccaca tcaagatcag aaacgccacc 960ttcccaggtg ttcaaatgaa atttgccttg
caaaaattgt tggatgctat tccagaagtc 1020gtcaaggact acaaacctgt tgctgtccca
gctagagttc caattaccaa gtctactcca 1080gctaacactc caatgaagca agaatggatg
tggaaccatt tgggtaactt cttgagagaa 1140ggtgatattg ttattgctga aaccggtact
tccgccttcg gtattaacca aactactttc 1200ccaacagatg tatacgctat cgtccaagtc
ttgtggggtt ccattggttt cacagtcggc 1260gctctattgg gtgctactat ggccgctgaa
gaacttgatc caaagaagag agttatttta 1320ttcattggtg acggttctct acaattgact
gttcaagaaa tctctaccat gattagatgg 1380ggtttgaagc catacatttt tgtcttgaat
aacaacggtt acaccattga aaaattgatt 1440cacggtcctc atgccgaata taatgaaatt
caaggttggg accacttggc cttattgcca 1500acttttggtg ctagaaacta cgaaacccac
agagttgcta ccactggtga atgggaaaag 1560ttgactcaag acaaggactt ccaagacaac
tctaagatta gaatgattga agttatgttg 1620ccagtctttg atgctccaca aaacttggtt
aaacaagctc aattgactgc cgctactaac 1680gctaaacaa
1689111533PRTSaccharomyces cerevisiae
111Met Ser Glu Ile Thr Leu Gly Lys Tyr Leu Phe Glu Arg Leu Lys Gln 1
5 10 15 Val Asn Val Asn
Thr Ile Phe Gly Leu Pro Gly Asp Phe Asn Leu Ser 20
25 30 Leu Leu Asp Lys Ile Tyr Glu Val Asp
Gly Leu Arg Trp Ala Gly Asn 35 40
45 Ala Asn Glu Leu Asn Ala Ala Tyr Ala Ala Asp Gly Tyr Ala
Arg Ile 50 55 60
Lys Gly Leu Ser Val Leu Val Thr Thr Phe Gly Val Gly Glu Leu Ser 65
70 75 80 Ala Leu Asn Gly Ile
Ala Gly Ser Tyr Ala Glu His Val Gly Val Leu 85
90 95 His Val Val Gly Val Pro Ser Ile Ser Ala
Gln Ala Lys Gln Leu Leu 100 105
110 Leu His His Thr Leu Gly Asn Gly Asp Phe Thr Val Phe His Arg
Met 115 120 125 Ser
Ala Asn Ile Ser Glu Thr Thr Ser Met Ile Thr Asp Ile Ala Thr 130
135 140 Ala Pro Ser Glu Ile Asp
Arg Leu Ile Arg Thr Thr Phe Ile Thr Gln 145 150
155 160 Arg Pro Ser Tyr Leu Gly Leu Pro Ala Asn Leu
Val Asp Leu Lys Val 165 170
175 Pro Gly Ser Leu Leu Glu Lys Pro Ile Asp Leu Ser Leu Lys Pro Asn
180 185 190 Asp Pro
Glu Ala Glu Lys Glu Val Ile Asp Thr Val Leu Glu Leu Ile 195
200 205 Gln Asn Ser Lys Asn Pro Val
Ile Leu Ser Asp Ala Cys Ala Ser Arg 210 215
220 His Asn Val Lys Lys Glu Thr Gln Lys Leu Ile Asp
Leu Thr Gln Phe 225 230 235
240 Pro Ala Phe Val Thr Pro Leu Gly Lys Gly Ser Ile Asp Glu Gln His
245 250 255 Pro Arg Tyr
Gly Gly Val Tyr Val Gly Thr Leu Ser Lys Gln Asp Val 260
265 270 Lys Gln Ala Val Glu Ser Ala Asp
Leu Ile Leu Ser Val Gly Ala Leu 275 280
285 Leu Ser Asp Phe Asn Thr Gly Ser Phe Ser Tyr Ser Tyr
Lys Thr Lys 290 295 300
Asn Val Val Glu Phe His Ser Asp Tyr Val Lys Val Lys Asn Ala Thr 305
310 315 320 Phe Leu Gly Val
Gln Met Lys Phe Ala Leu Gln Asn Leu Leu Lys Val 325
330 335 Ile Pro Asp Val Val Lys Gly Tyr Lys
Ser Val Pro Val Pro Thr Lys 340 345
350 Thr Pro Ala Asn Lys Gly Val Pro Ala Ser Thr Pro Leu Lys
Gln Glu 355 360 365
Trp Leu Trp Asn Glu Leu Ser Lys Phe Leu Gln Glu Gly Asp Val Ile 370
375 380 Ile Ser Glu Thr Gly
Thr Ser Ala Phe Gly Ile Asn Gln Thr Ile Phe 385 390
395 400 Pro Lys Asp Ala Tyr Gly Ile Ser Gln Val
Leu Trp Gly Ser Ile Gly 405 410
415 Phe Thr Thr Gly Ala Thr Leu Gly Ala Ala Phe Ala Ala Glu Glu
Ile 420 425 430 Asp
Pro Asn Lys Arg Val Ile Leu Phe Ile Gly Asp Gly Ser Leu Gln 435
440 445 Leu Thr Val Gln Glu Ile
Ser Thr Met Ile Arg Trp Gly Leu Lys Pro 450 455
460 Tyr Leu Phe Val Leu Asn Asn Asp Gly Tyr Thr
Ile Glu Lys Leu Ile 465 470 475
480 His Gly Pro His Ala Glu Tyr Asn Glu Ile Gln Thr Trp Asp His Leu
485 490 495 Ala Leu
Leu Pro Ala Phe Gly Ala Lys Lys Tyr Glu Asn His Lys Ile 500
505 510 Ala Thr Thr Gly Glu Trp Asp
Ala Leu Thr Thr Asp Ser Glu Phe Gln 515 520
525 Lys Asn Ser Val Ile 530
1121599DNASaccharomyces cerevisiae 112atgtctgaaa ttactcttgg aaaatactta
tttgaaagat tgaagcaagt taatgttaac 60accatttttg ggctaccagg cgacttcaac
ttgtccctat tggacaagat ttacgaggta 120gatggattga gatgggctgg taatgcaaat
gagctgaacg ccgcctatgc cgccgatggt 180tacgcacgca tcaagggttt atctgtgctg
gtaactactt ttggcgtagg tgaattatcc 240gccttgaatg gtattgcagg atcgtatgca
gaacacgtcg gtgtactgca tgttgttggt 300gtcccctcta tctccgctca ggctaagcaa
ttgttgttgc atcatacctt gggtaacggt 360gattttaccg tttttcacag aatgtccgcc
aatatctcag aaactacatc aatgattaca 420gacattgcta cagccccttc agaaatcgat
aggttgatca ggacaacatt tataacacaa 480aggcctagct acttggggtt gccagcgaat
ttggtagatc taaaggttcc tggttctctt 540ttggaaaaac cgattgatct atcattaaaa
cctaacgatc ccgaagctga aaaggaagtt 600attgataccg tactagaatt gatccagaat
tcgaaaaacc ctgttatact atcggatgcc 660tgtgcttcta ggcacaacgt taaaaaagaa
acccagaagt taattgattt gacgcaattc 720ccagcttttg tgacacctct aggtaaaggg
tcaatagatg aacagcatcc cagatatggc 780ggtgtttatg tgggaacgct gtccaaacaa
gacgtgaaac aggccgttga gtcggctgat 840ttgatccttt cggtcggtgc tttgctctct
gattttaaca caggttcgtt ttcctactcc 900tacaagacta aaaatgtagt ggagtttcat
tccgattacg taaaggtgaa gaacgctacg 960ttcctcggtg tacaaatgaa atttgcacta
caaaacttac tgaaggttat tcccgatgtt 1020gttaagggct acaagagcgt tcccgtacca
accaaaactc ccgcaaacaa aggtgtacct 1080gctagcacgc ccttgaaaca agagtggttg
tggaacgaat tgtccaaatt cttgcaagaa 1140ggtgatgtta tcatttccga gaccggcacg
tctgccttcg gtatcaatca aactatcttt 1200cctaaggacg cctacggtat ctcgcaggtg
ttgtgggggt ccatcggttt tacaacagga 1260gcaactttag gtgctgcctt tgccgctgag
gagattgacc ccaacaagag agtcatctta 1320ttcataggtg acgggtcttt gcagttaacc
gtccaagaaa tctccaccat gatcagatgg 1380gggttaaagc cgtatctttt tgtccttaac
aacgacggct acactatcga aaagctgatt 1440catgggcctc acgcagagta caacgaaatc
cagacctggg atcacctcgc cctgttgccc 1500gcatttggtg cgaaaaagta cgaaaatcac
aagatcgcca ctacgggtga gtgggatgcc 1560ttaaccactg attcagagtt ccagaaaaac
tcggtgatc 1599113564PRTCandida glabrata 113Met
Ser Glu Ile Thr Leu Gly Arg Tyr Leu Phe Glu Arg Leu Asn Gln 1
5 10 15 Val Asp Val Lys Thr Ile
Phe Gly Leu Pro Gly Asp Phe Asn Leu Ser 20
25 30 Leu Leu Asp Lys Ile Tyr Glu Val Glu Gly
Met Arg Trp Ala Gly Asn 35 40
45 Ala Asn Glu Leu Asn Ala Ala Tyr Ala Ala Asp Gly Tyr Ala
Arg Ile 50 55 60
Lys Gly Met Ser Cys Ile Ile Thr Thr Phe Gly Val Gly Glu Leu Ser 65
70 75 80 Ala Leu Asn Gly Ile
Ala Gly Ser Tyr Ala Glu His Val Gly Val Leu 85
90 95 His Val Val Gly Val Pro Ser Ile Ser Ser
Gln Ala Lys Gln Leu Leu 100 105
110 Leu His His Thr Leu Gly Asn Gly Asp Phe Thr Val Phe His Arg
Met 115 120 125 Ser
Ala Asn Ile Ser Glu Thr Thr Ala Met Val Thr Asp Ile Ala Thr 130
135 140 Ala Pro Ala Glu Ile Asp
Arg Cys Ile Arg Thr Thr Tyr Ile Thr Gln 145 150
155 160 Arg Pro Val Tyr Leu Gly Leu Pro Ala Asn Leu
Val Asp Leu Lys Val 165 170
175 Pro Ala Lys Leu Leu Glu Thr Pro Ile Asp Leu Ser Leu Lys Pro Asn
180 185 190 Asp Pro
Glu Ala Glu Thr Glu Val Val Asp Thr Val Leu Glu Leu Ile 195
200 205 Lys Ala Ala Lys Asn Pro Val
Ile Leu Ala Asp Ala Cys Ala Ser Arg 210 215
220 His Asp Val Lys Ala Glu Thr Lys Lys Leu Ile Asp
Ala Thr Gln Phe 225 230 235
240 Pro Ser Phe Val Thr Pro Met Gly Lys Gly Ser Ile Asp Glu Gln His
245 250 255 Pro Arg Phe
Gly Gly Val Tyr Val Gly Thr Leu Ser Arg Pro Glu Val 260
265 270 Lys Glu Ala Val Glu Ser Ala Asp
Leu Ile Leu Ser Val Gly Ala Leu 275 280
285 Leu Ser Asp Phe Asn Thr Gly Ser Phe Ser Tyr Ser Tyr
Lys Thr Lys 290 295 300
Asn Ile Val Glu Phe His Ser Asp Tyr Ile Lys Ile Arg Asn Ala Thr 305
310 315 320 Phe Pro Gly Val
Gln Met Lys Phe Ala Leu Gln Lys Leu Leu Asn Ala 325
330 335 Val Pro Glu Ala Ile Lys Gly Tyr Lys
Pro Val Pro Val Pro Ala Arg 340 345
350 Val Pro Glu Asn Lys Ser Cys Asp Pro Ala Thr Pro Leu Lys
Gln Glu 355 360 365
Trp Met Trp Asn Gln Val Ser Lys Phe Leu Gln Glu Gly Asp Val Val 370
375 380 Ile Thr Glu Thr Gly
Thr Ser Ala Phe Gly Ile Asn Gln Thr Pro Phe 385 390
395 400 Pro Asn Asn Ala Tyr Gly Ile Ser Gln Val
Leu Trp Gly Ser Ile Gly 405 410
415 Phe Thr Thr Gly Ala Cys Leu Gly Ala Ala Phe Ala Ala Glu Glu
Ile 420 425 430 Asp
Pro Lys Lys Arg Val Ile Leu Phe Ile Gly Asp Gly Ser Leu Gln 435
440 445 Leu Thr Val Gln Glu Ile
Ser Thr Met Ile Arg Trp Gly Leu Lys Pro 450 455
460 Tyr Leu Phe Val Leu Asn Asn Asp Gly Tyr Thr
Ile Glu Arg Leu Ile 465 470 475
480 His Gly Glu Lys Ala Gly Tyr Asn Asp Ile Gln Asn Trp Asp His Leu
485 490 495 Ala Leu
Leu Pro Thr Phe Gly Ala Lys Asp Tyr Glu Asn His Arg Val 500
505 510 Ala Thr Thr Gly Glu Trp Asp
Lys Leu Thr Gln Asp Lys Glu Phe Asn 515 520
525 Lys Asn Ser Lys Ile Arg Met Ile Glu Val Met Leu
Pro Val Met Asp 530 535 540
Ala Pro Thr Ser Leu Ile Glu Gln Ala Lys Leu Thr Ala Ser Ile Asn 545
550 555 560 Ala Lys Gln
Glu 1141692DNACandida glabrata 114atgtctgaga ttactttggg tagatacttg
ttcgagagat tgaaccaagt cgacgttaag 60accatcttcg gtttgccagg tgacttcaac
ttgtccctat tggacaagat ctacgaagtt 120gaaggtatga gatgggctgg taacgctaac
gaattgaacg ctgcttacgc tgctgacggt 180tacgctagaa tcaagggtat gtcctgtatc
atcaccacct tcggtgtcgg tgaattgtct 240gccttgaacg gtattgccgg ttcttacgct
gaacacgtcg gtgtcttgca cgtcgtcggt 300gtcccatcca tctcctctca agctaagcaa
ttgttgttgc accacacctt gggtaacggt 360gacttcactg tcttccacag aatgtccgct
aacatctctg agaccaccgc tatggtcact 420gacatcgcta ccgctccagc tgagatcgac
agatgtatca gaaccaccta catcacccaa 480agaccagtct acttgggtct accagctaac
ttggtcgacc taaaggtccc agccaagctt 540ttggaaaccc caattgactt gtccttgaag
ccaaacgacc cagaagccga aactgaagtc 600gttgacaccg tcttggaatt gatcaaggct
gctaagaacc cagttatctt ggctgatgct 660tgtgcttcca gacacgacgt caaggctgaa
accaagaagt tgattgacgc cactcaattc 720ccatccttcg ttaccccaat gggtaagggt
tccatcgacg aacaacaccc aagattcggt 780ggtgtctacg tcggtacctt gtccagacca
gaagttaagg aagctgttga atccgctgac 840ttgatcttgt ctgtcggtgc tttgttgtcc
gatttcaaca ctggttcttt ctcttactct 900tacaagacca agaacatcgt cgaattccac
tctgactaca tcaagatcag aaacgctacc 960ttcccaggtg tccaaatgaa gttcgctttg
caaaagttgt tgaacgccgt cccagaagct 1020atcaagggtt acaagccagt ccctgtccca
gctagagtcc cagaaaacaa gtcctgtgac 1080ccagctaccc cattgaagca agaatggatg
tggaaccaag tttccaagtt cttgcaagaa 1140ggtgatgttg ttatcactga aaccggtacc
tccgcttttg gtatcaacca aaccccattc 1200ccaaacaacg cttacggtat ctcccaagtt
ctatggggtt ccatcggttt caccaccggt 1260gcttgtttgg gtgccgcttt cgctgctgaa
gaaatcgacc caaagaagag agttatcttg 1320ttcattggtg acggttcttt gcaattgact
gtccaagaaa tctccaccat gatcagatgg 1380ggcttgaagc catacttgtt cgtcttgaac
aacgacggtt acaccatcga aagattgatt 1440cacggtgaaa aggctggtta caacgacatc
caaaactggg accacttggc tctattgcca 1500accttcggtg ctaaggacta cgaaaaccac
agagtcgcca ccaccggtga atgggacaag 1560ttgacccaag acaaggaatt caacaagaac
tccaagatca gaatgatcga agttatgttg 1620ccagttatgg acgctccaac ttccttgatt
gaacaagcta agttgaccgc ttccatcaac 1680gctaagcaag aa
1692115596PRTPichia stipitis 115Met Ala
Glu Val Ser Leu Gly Arg Tyr Leu Phe Glu Arg Leu Tyr Gln 1 5
10 15 Leu Gln Val Gln Thr Ile Phe
Gly Val Pro Gly Asp Phe Asn Leu Ser 20 25
30 Leu Leu Asp Lys Ile Tyr Glu Val Glu Asp Ala His
Gly Lys Asn Ser 35 40 45
Phe Arg Trp Ala Gly Asn Ala Asn Glu Leu Asn Ala Ser Tyr Ala Ala
50 55 60 Asp Gly Tyr
Ser Arg Val Lys Arg Leu Gly Cys Leu Val Thr Thr Phe 65
70 75 80 Gly Val Gly Glu Leu Ser Ala
Leu Asn Gly Ile Ala Gly Ser Tyr Ala 85
90 95 Glu His Val Gly Leu Leu His Val Val Gly Val
Pro Ser Ile Ser Ser 100 105
110 Gln Ala Lys Gln Leu Leu Leu His His Thr Leu Gly Asn Gly Asp
Phe 115 120 125 Thr
Val Phe His Arg Met Ser Asn Asn Ile Ser Gln Thr Thr Ala Phe 130
135 140 Ile Ser Asp Ile Asn Ser
Ala Pro Ala Glu Ile Asp Arg Cys Ile Arg 145 150
155 160 Glu Ala Tyr Val Lys Gln Arg Pro Val Tyr Ile
Gly Leu Pro Ala Asn 165 170
175 Leu Val Asp Leu Asn Val Pro Ala Ser Leu Leu Glu Ser Pro Ile Asn
180 185 190 Leu Ser
Leu Glu Lys Asn Asp Pro Glu Ala Gln Asp Glu Val Ile Asp 195
200 205 Ser Val Leu Asp Leu Ile Lys
Lys Ser Ser Asn Pro Ile Ile Leu Val 210 215
220 Asp Ala Cys Ala Ser Arg His Asp Cys Lys Ala Glu
Val Thr Gln Leu 225 230 235
240 Ile Glu Gln Thr Gln Phe Pro Val Phe Val Thr Pro Met Gly Lys Gly
245 250 255 Thr Val Asp
Glu Gly Gly Val Asp Gly Glu Leu Leu Glu Asp Asp Pro 260
265 270 His Leu Ile Ala Lys Val Ala Ala
Arg Leu Ser Ala Gly Lys Asn Ala 275 280
285 Ala Ser Arg Phe Gly Gly Val Tyr Val Gly Thr Leu Ser
Lys Pro Glu 290 295 300
Val Lys Asp Ala Val Glu Ser Ala Asp Leu Ile Leu Ser Val Gly Ala 305
310 315 320 Leu Leu Ser Asp
Phe Asn Thr Gly Ser Phe Ser Tyr Ser Tyr Arg Thr 325
330 335 Lys Asn Ile Val Glu Phe His Ser Asp
Tyr Thr Lys Ile Arg Gln Ala 340 345
350 Thr Phe Pro Gly Val Gln Met Lys Glu Ala Leu Gln Glu Leu
Asn Lys 355 360 365
Lys Val Ser Ser Ala Ala Ser His Tyr Glu Val Lys Pro Val Pro Lys 370
375 380 Ile Lys Leu Ala Asn
Thr Pro Ala Thr Arg Glu Val Lys Leu Thr Gln 385 390
395 400 Glu Trp Leu Trp Thr Arg Val Ser Ser Trp
Phe Arg Glu Gly Asp Ile 405 410
415 Ile Ile Thr Glu Thr Gly Thr Ser Ser Phe Gly Ile Val Gln Ser
Arg 420 425 430 Phe
Pro Asn Asn Thr Ile Gly Ile Ser Gln Val Leu Trp Gly Ser Ile 435
440 445 Gly Phe Ser Val Gly Ala
Thr Leu Gly Ala Ala Met Ala Ala Gln Glu 450 455
460 Leu Asp Pro Asn Lys Arg Thr Ile Leu Phe Val
Gly Asp Gly Ser Leu 465 470 475
480 Gln Leu Thr Val Gln Glu Ile Ser Thr Ile Ile Arg Trp Gly Thr Thr
485 490 495 Pro Tyr
Leu Phe Val Leu Asn Asn Asp Gly Tyr Thr Ile Glu Arg Leu 500
505 510 Ile His Gly Val Asn Ala Ser
Tyr Asn Asp Ile Gln Pro Trp Gln Asn 515 520
525 Leu Glu Ile Leu Pro Thr Phe Ser Ala Lys Asn Tyr
Asp Ala Val Arg 530 535 540
Ile Ser Asn Ile Gly Glu Ala Glu Asp Ile Leu Lys Asp Lys Glu Phe 545
550 555 560 Gly Lys Asn
Ser Lys Ile Arg Leu Ile Glu Val Met Leu Pro Arg Leu 565
570 575 Asp Ala Pro Ser Asn Leu Ala Lys
Gln Ala Ala Ile Thr Ala Ala Thr 580 585
590 Asn Ala Glu Ala 595 1161788DNAPichia
stipitis 116atggctgaag tctcattagg aagatatctc ttcgagagat tgtaccaatt
gcaagtgcag 60accatcttcg gtgtccctgg tgatttcaac ttgtcgcttt tggacaagat
ctacgaagtg 120gaagatgccc atggcaagaa ttcgtttaga tgggctggta atgccaacga
attgaatgca 180tcgtacgctg ctgacggtta ctcgagagtc aagcgtttag ggtgtttggt
cactaccttt 240ggtgtcggtg aattgtctgc tttgaatggt attgccggtt cttatgccga
acatgttggt 300ttgcttcatg tcgtaggtgt tccatcgatt tcctcgcaag ctaagcaatt
gttacttcac 360cacactttgg gtaatggtga tttcactgtt ttccatagaa tgtccaacaa
catttctcag 420accacagcct ttatctccga tatcaactcg gctccagctg aaattgatag
atgtatcaga 480gaggcctacg tcaaacaaag accagtttat atcgggttac cagctaactt
agttgatttg 540aatgttccgg cctctttgct tgagtctcca atcaacttgt cgttggaaaa
gaacgaccca 600gaggctcaag atgaagtcat tgactctgtc ttagacttga tcaaaaagtc
gctgaaccca 660atcatcttgg tcgatgcctg tgcctcgaga catgactgta aggctgaagt
tactcagttg 720attgaacaaa cccaattccc agtatttgtc actccaatgg gtaaaggtac
cgttgatgag 780ggtggtgtag acggagaatt gttagaagat gatcctcatt tgattgccaa
ggtcgctgct 840aggttgtctg ctggcaagaa cgctgcctct agattcggag gtgtttatgt
cggaaccttg 900tcgaagcccg aagtcaagga cgctgtagag agtgcagatt tgattttgtc
tgtcggtgcc 960cttttgtctg atttcaacac tggttcattt tcctactcct acagaaccaa
gaacatcgtc 1020gaattccatt ctgattacac taagattaga caagccactt tcccaggtgt
gcagatgaag 1080gaagccttgc aagaattgaa caagaaagtt tcatctgctg ctagtcacta
tgaagtcaag 1140cctgtgccca agatcaagtt ggccaataca ccagccacca gagaagtcaa
gttaactcag 1200gaatggttgt ggaccagagt gtcttcgtgg ttcagagaag gtgatattat
tatcaccgaa 1260accggtacat cctccttcgg tatagttcaa tccagattcc caaacaacac
catcggtatc 1320tcccaagtat tgtggggttc tattggtttc tctgttggtg ccactttggg
tgctgccatg 1380gctgcccaag aactcgaccc taacaagaga accatcttgt ttgttggaga
tggttctttg 1440caattgaccg ttcaggaaat ctccaccata atcagatggg gtaccacacc
ttaccttttc 1500gtgttgaaca atgacggtta caccatcgag cgtttgatcc acggtgtaaa
tgcctcatat 1560aatgacatcc aaccatggca aaacttggaa atcttgccta ctttctcggc
caagaactac 1620gacgctgtga gaatctccaa catcggagaa gcagaagata tcttgaaaga
caaggaattc 1680ggaaagaact ccaagattag attgatagaa gtcatgttac caagattgga
tgcaccatct 1740aaccttgcca aacaagctgc cattacagct gccaccaacg ccgaagct
1788117569PRTPichia stipitis 117Met Val Ser Thr Tyr Pro Glu
Ser Glu Val Thr Leu Gly Arg Tyr Leu 1 5
10 15 Phe Glu Arg Leu His Gln Leu Lys Val Asp Thr
Ile Phe Gly Leu Pro 20 25
30 Gly Asp Phe Asn Leu Ser Leu Leu Asp Lys Val Tyr Glu Val Pro
Asp 35 40 45 Met
Arg Trp Ala Gly Asn Ala Asn Glu Leu Asn Ala Ala Tyr Ala Ala 50
55 60 Asp Gly Tyr Ser Arg Ile
Lys Gly Leu Ser Cys Leu Val Thr Thr Phe 65 70
75 80 Gly Val Gly Glu Leu Ser Ala Leu Asn Gly Val
Gly Gly Ala Tyr Ala 85 90
95 Glu His Val Gly Leu Leu His Val Val Gly Val Pro Ser Ile Ser Ser
100 105 110 Gln Ala
Lys Gln Leu Leu Leu His His Thr Leu Gly Asn Gly Asp Phe 115
120 125 Thr Val Phe His Arg Met Ser
Asn Ser Ile Ser Gln Thr Thr Ala Phe 130 135
140 Leu Ser Asp Ile Ser Ile Ala Pro Gly Gln Ile Asp
Arg Cys Ile Arg 145 150 155
160 Glu Ala Tyr Val His Gln Arg Pro Val Tyr Val Gly Leu Pro Ala Asn
165 170 175 Met Val Asp
Leu Lys Val Pro Ser Ser Leu Leu Glu Thr Pro Ile Asp 180
185 190 Leu Lys Leu Lys Gln Asn Asp Pro
Glu Ala Gln Glu Val Val Glu Thr 195 200
205 Val Leu Lys Leu Val Ser Gln Ala Thr Asn Pro Ile Ile
Leu Val Asp 210 215 220
Ala Cys Ala Leu Arg His Asn Cys Lys Glu Glu Val Lys Gln Leu Val 225
230 235 240 Asp Ala Thr Asn
Phe Gln Val Phe Thr Thr Pro Met Gly Lys Ser Gly 245
250 255 Ile Ser Glu Ser His Pro Arg Leu Gly
Gly Val Tyr Val Gly Thr Met 260 265
270 Ser Ser Pro Gln Val Lys Lys Ala Val Glu Asn Ala Asp Leu
Ile Leu 275 280 285
Ser Val Gly Ser Leu Leu Ser Asp Phe Asn Thr Gly Ser Phe Ser Tyr 290
295 300 Ser Tyr Lys Thr Lys
Asn Val Val Glu Phe His Ser Asp Tyr Met Lys 305 310
315 320 Ile Arg Gln Ala Thr Phe Pro Gly Val Gln
Met Lys Glu Ala Leu Gln 325 330
335 Gln Leu Ile Lys Arg Val Ser Ser Tyr Ile Asn Pro Ser Tyr Ile
Pro 340 345 350 Thr
Arg Val Pro Lys Arg Lys Gln Pro Leu Lys Ala Pro Ser Glu Ala 355
360 365 Pro Leu Thr Gln Glu Tyr
Leu Trp Ser Lys Val Ser Gly Trp Phe Arg 370 375
380 Glu Gly Asp Ile Ile Val Thr Glu Thr Gly Thr
Ser Ala Phe Gly Ile 385 390 395
400 Ile Gln Ser His Phe Pro Ser Asn Thr Ile Gly Ile Ser Gln Val Leu
405 410 415 Trp Gly
Ser Ile Gly Phe Thr Val Gly Ala Thr Val Gly Ala Ala Met 420
425 430 Ala Ala Gln Glu Ile Asp Pro
Ser Arg Arg Val Ile Leu Phe Val Gly 435 440
445 Asp Gly Ser Leu Gln Leu Thr Val Gln Glu Ile Ser
Thr Leu Cys Lys 450 455 460
Trp Asp Cys Asn Asn Thr Tyr Leu Tyr Val Leu Asn Asn Asp Gly Tyr 465
470 475 480 Thr Ile Glu
Arg Leu Ile His Gly Lys Ser Ala Ser Tyr Asn Asp Ile 485
490 495 Gln Pro Trp Asn His Leu Ser Leu
Leu Arg Leu Phe Asn Ala Lys Lys 500 505
510 Tyr Gln Asn Val Arg Val Ser Thr Ala Gly Glu Leu Asp
Ser Leu Phe 515 520 525
Ser Asp Lys Lys Phe Ala Ser Pro Asp Arg Ile Arg Met Ile Glu Val 530
535 540 Met Leu Ser Arg
Leu Asp Ala Pro Ala Asn Leu Val Ala Gln Ala Lys 545 550
555 560 Leu Ser Glu Arg Val Asn Leu Glu Asn
565 1181707DNAPichia stipitis
118atggtatcaa cctacccaga atcagaggtt actctaggaa ggtacctctt tgagcgactc
60caccaattga aagtggacac cattttcggc ttgccgggtg acttcaacct ttccttattg
120gacaaagtgt atgaagttcc ggatatgagg tgggctggaa atgccaacga attgaatgct
180gcctatgctg ccgatggtta ctccagaata aagggattgt cttgcttggt cacaactttt
240ggtgttggtg aattgtctgc tttaaacgga gttggtggtg cctatgctga acacgtagga
300cttctacatg tcgttggagt tccatccata tcgtcacagg ctaaacagtt gttgctccac
360cataccttgg gtaatggtga cttcactgtt tttcacagaa tgtccaatag catttctcaa
420actacagcat ttctctcaga tatctctatt gcaccaggtc aaatagatag atgcatcaga
480gaagcatatg ttcatcagag accagtttat gttggtttac cggcaaatat ggttgatctc
540aaggttcctt ctagtctctt agaaactcca attgatttga aattgaaaca aaatgatcct
600gaagctcaag aagttgttga aacagtcctg aagttggtgt cccaagctac aaaccccatt
660atcttggtag acgcttgtgc cctcagacac aattgcaaag aggaagtcaa acaattggtt
720gatgccacta attttcaagt ctttacaact ccaatgggta aatctggtat ctccgaatct
780catccaagat tgggcggtgt ctatgtcggg acaatgtcga gtcctcaagt caaaaaagcc
840gttgaaaatg ccgatcttat actatctgtt ggttcgttgt tatcggactt caatacaggt
900tcattttcat actcctacaa gacgaagaat gttgttgaat tccactctga ctatatgaaa
960atcagacagg ccaccttccc aggagttcaa atgaaagaag ccttgcaaca gttgataaaa
1020agggtctctt cttacatcaa tccaagctac attcctactc gagttcctaa aaggaaacag
1080ccattgaaag ctccatcaga agctcctttg acccaagaat atttgtggtc taaagtatcc
1140ggctggttta gagagggtga tattatcgta accgaaactg gtacatctgc tttcggaatt
1200attcaatccc attttcccag caacactatc ggtatatccc aagtcttgtg gggctcaatt
1260ggtttcacag taggtgcaac agttggtgct gccatggcag cccaggaaat cgaccctagc
1320aggagagtaa ttttgttcgt cggtgatggt tcattgcagt tgacggttca ggaaatctct
1380acgttgtgta aatgggattg taacaatact tatctttacg tgttgaacaa tgatggttac
1440actatagaaa ggttgatcca cggcaaaagt gccagctaca acgatataca gccttggaac
1500catttatcct tgcttcgctt attcaatgct aagaaatacc aaaatgtcag agtatcgact
1560gctggagaat tggactcttt gttctctgat aagaaatttg cttctccaga taggataaga
1620atgattgagg tgatgttatc gagattggat gcaccagcaa atcttgttgc tcaagcaaag
1680ttgtctgaac gggtaaacct tgaaaat
1707119563PRTKluyveromyces lactis 119Met Ser Glu Ile Thr Leu Gly Arg Tyr
Leu Phe Glu Arg Leu Lys Gln 1 5 10
15 Val Glu Val Gln Thr Ile Phe Gly Leu Pro Gly Asp Phe Asn
Leu Ser 20 25 30
Leu Leu Asp Asn Ile Tyr Glu Val Pro Gly Met Arg Trp Ala Gly Asn
35 40 45 Ala Asn Glu Leu
Asn Ala Ala Tyr Ala Ala Asp Gly Tyr Ala Arg Leu 50
55 60 Lys Gly Met Ser Cys Ile Ile Thr
Thr Phe Gly Val Gly Glu Leu Ser 65 70
75 80 Ala Leu Asn Gly Ile Ala Gly Ser Tyr Ala Glu His
Val Gly Val Leu 85 90
95 His Val Val Gly Val Pro Ser Val Ser Ser Gln Ala Lys Gln Leu Leu
100 105 110 Leu His His
Thr Leu Gly Asn Gly Asp Phe Thr Val Phe His Arg Met 115
120 125 Ser Ser Asn Ile Ser Glu Thr Thr
Ala Met Ile Thr Asp Ile Asn Thr 130 135
140 Ala Pro Ala Glu Ile Asp Arg Cys Ile Arg Thr Thr Tyr
Val Ser Gln 145 150 155
160 Arg Pro Val Tyr Leu Gly Leu Pro Ala Asn Leu Val Asp Leu Thr Val
165 170 175 Pro Ala Ser Leu
Leu Asp Thr Pro Ile Asp Leu Ser Leu Lys Pro Asn 180
185 190 Asp Pro Glu Ala Glu Glu Glu Val Ile
Glu Asn Val Leu Gln Leu Ile 195 200
205 Lys Glu Ala Lys Asn Pro Val Ile Leu Ala Asp Ala Cys Cys
Ser Arg 210 215 220
His Asp Ala Lys Ala Glu Thr Lys Lys Leu Ile Asp Leu Thr Gln Phe 225
230 235 240 Pro Ala Phe Val Thr
Pro Met Gly Lys Gly Ser Ile Asp Glu Lys His 245
250 255 Pro Arg Phe Gly Gly Val Tyr Val Gly Thr
Leu Ser Ser Pro Ala Val 260 265
270 Lys Glu Ala Val Glu Ser Ala Asp Leu Val Leu Ser Val Gly Ala
Leu 275 280 285 Leu
Ser Asp Phe Asn Thr Gly Ser Phe Ser Tyr Ser Tyr Lys Thr Lys 290
295 300 Asn Ile Val Glu Phe His
Ser Asp Tyr Thr Lys Ile Arg Ser Ala Thr 305 310
315 320 Phe Pro Gly Val Gln Met Lys Phe Ala Leu Gln
Lys Leu Leu Thr Lys 325 330
335 Val Ala Asp Ala Ala Lys Gly Tyr Lys Pro Val Pro Val Pro Ser Glu
340 345 350 Pro Glu
His Asn Glu Ala Val Ala Asp Ser Thr Pro Leu Lys Gln Glu 355
360 365 Trp Val Trp Thr Gln Val Gly
Glu Phe Leu Arg Glu Gly Asp Val Val 370 375
380 Ile Thr Glu Thr Gly Thr Ser Ala Phe Gly Ile Asn
Gln Thr His Phe 385 390 395
400 Pro Asn Asn Thr Tyr Gly Ile Ser Gln Val Leu Trp Gly Ser Ile Gly
405 410 415 Phe Thr Thr
Gly Ala Thr Leu Gly Ala Ala Phe Ala Ala Glu Glu Ile 420
425 430 Asp Pro Lys Lys Arg Val Ile Leu
Phe Ile Gly Asp Gly Ser Leu Gln 435 440
445 Leu Thr Val Gln Glu Ile Ser Thr Met Ile Arg Trp Gly
Leu Lys Pro 450 455 460
Tyr Leu Phe Val Leu Asn Asn Asp Gly Tyr Thr Ile Glu Arg Leu Ile 465
470 475 480 His Gly Glu Thr
Ala Gln Tyr Asn Cys Ile Gln Asn Trp Gln His Leu 485
490 495 Glu Leu Leu Pro Thr Phe Gly Ala Lys
Asp Tyr Glu Ala Val Arg Val 500 505
510 Ser Thr Thr Gly Glu Trp Asn Lys Leu Thr Thr Asp Glu Lys
Phe Gln 515 520 525
Asp Asn Thr Arg Ile Arg Leu Ile Glu Val Met Leu Pro Thr Met Asp 530
535 540 Ala Pro Ser Asn Leu
Val Lys Gln Ala Gln Leu Thr Ala Ala Thr Asn 545 550
555 560 Ala Lys Asn 1201689DNAKluyveromyces
lactis 120atgtctgaaa ttacattagg tcgttacttg ttcgaaagat taaagcaagt
cgaagttcaa 60accatctttg gtctaccagg tgatttcaac ttgtccctat tggacaatat
ctacgaagtc 120ccaggtatga gatgggctgg taatgccaac gaattgaacg ctgcttacgc
tgctgatggt 180tacgccagat taaagggtat gtcctgtatc atcaccacct tcggtgtcgg
tgaattgtct 240gctttgaacg gtattgccgg ttcttacgct gaacacgttg gtgtcttgca
cgttgtcggt 300gttccatccg tctcttctca agctaagcaa ttgttgttgc accacacctt
gggtaacggt 360gacttcactg ttttccacag aatgtcctcc aacatttctg aaaccactgc
tatgatcacc 420gatatcaaca ctgccccagc tgaaatcgac agatgtatca gaaccactta
cgtttcccaa 480agaccagtct acttgggttt gccagctaac ttggtcgact tgactgtccc
agcttctttg 540ttggacactc caattgattt gagcttgaag ccaaatgacc cagaagccga
agaagaagtc 600atcgaaaacg tcttgcaact gatcaaggaa gctaagaacc cagttatctt
ggctgatgct 660tgttgttcca gacacgatgc caaggctgag accaagaagt tgatcgactt
gactcaattc 720ccagccttcg ttaccccaat gggtaagggt tccattgacg aaaagcaccc
aagattcggt 780ggtgtctacg tcggtaccct atcttctcca gctgtcaagg aagccgttga
atctgctgac 840ttggttctat cggtcggtgc tctattgtcc gatttcaaca ctggttcttt
ctcttactct 900tacaagacca agaacattgt cgaattccac tctgactaca ccaagatcag
aagcgctacc 960ttcccaggtg tccaaatgaa gttcgcttta caaaaattgt tgactaaggt
tgccgatgct 1020gctaagggtt acaagccagt tccagttcca tctgaaccag aacacaacga
agctgtcgct 1080gactccactc cattgaagca agaatgggtc tggactcaag tcggtgaatt
cttgagagaa 1140ggtgatgttg ttatcactga aaccggtacc tctgccttcg gtatcaacca
aactcatttc 1200ccaaacaaca catacggtat ctctcaagtt ttatggggtt ccattggttt
caccactggt 1260gctaccttgg gtgctgcctt cgctgccgaa gaaattgatc caaagaagag
agttatctta 1320ttcattggtg acggttcttt gcaattgact gttcaagaaa tctccaccat
gatcagatgg 1380ggcttgaagc catacttgtt cgtattgaac aacgacggtt acaccattga
aagattgatt 1440cacggtgaaa ccgctcaata caactgtatc caaaactggc aacacttgga
attattgcca 1500actttcggtg ccaaggacta cgaagctgtc agagtttcca ccactggtga
atggaacaag 1560ttgaccactg acgaaaagtt ccaagacaac accagaatca gattgatcga
agttatgttg 1620ccaactatgg atgctccatc taacttggtt aagcaagctc aattgactgc
tgctaccaac 1680gctaagaac
1689121571PRTYarrowia lipolytica 121Met Ser Asp Ser Glu Pro
Gln Met Val Asp Leu Gly Asp Tyr Leu Phe 1 5
10 15 Ala Arg Phe Lys Gln Leu Gly Val Asp Ser Val
Phe Gly Val Pro Gly 20 25
30 Asp Phe Asn Leu Thr Leu Leu Asp His Val Tyr Asn Val Asp Met
Arg 35 40 45 Trp
Val Gly Asn Thr Asn Glu Leu Asn Ala Gly Tyr Ser Ala Asp Gly 50
55 60 Tyr Ser Arg Val Lys Arg
Leu Ala Cys Leu Val Thr Thr Phe Gly Val 65 70
75 80 Gly Glu Leu Ser Ala Val Ala Ala Val Ala Gly
Ser Tyr Ala Glu His 85 90
95 Val Gly Val Val His Val Val Gly Val Pro Ser Thr Ser Ala Glu Asn
100 105 110 Lys His
Leu Leu Leu His His Thr Leu Gly Asn Gly Asp Phe Arg Val 115
120 125 Phe Ala Gln Met Ser Lys Leu
Ile Ser Glu Tyr Thr His His Ile Glu 130 135
140 Asp Pro Ser Glu Ala Ala Asp Val Ile Asp Thr Ala
Ile Arg Ile Ala 145 150 155
160 Tyr Thr His Gln Arg Pro Val Tyr Ile Ala Val Pro Ser Asn Phe Ser
165 170 175 Glu Val Asp
Ile Ala Asp Gln Ala Arg Leu Asp Thr Pro Leu Asp Leu 180
185 190 Ser Leu Gln Pro Asn Asp Pro Glu
Ser Gln Tyr Glu Val Ile Glu Glu 195 200
205 Ile Cys Ser Arg Ile Lys Ala Ala Lys Lys Pro Val Ile
Leu Val Asp 210 215 220
Ala Cys Ala Ser Arg Tyr Arg Cys Val Asp Glu Thr Lys Glu Leu Ala 225
230 235 240 Lys Ile Thr Asn
Phe Ala Tyr Phe Val Thr Pro Met Gly Lys Gly Ser 245
250 255 Val Asp Glu Asp Thr Asp Arg Tyr Gly
Gly Thr Tyr Val Gly Ser Leu 260 265
270 Thr Ala Pro Ala Thr Ala Glu Val Val Glu Thr Ala Asp Leu
Ile Ile 275 280 285
Ser Val Gly Ala Leu Leu Ser Asp Phe Asn Thr Gly Ser Phe Ser Tyr 290
295 300 Ser Tyr Ser Thr Lys
Asn Val Val Glu Leu His Ser Asp His Val Lys 305 310
315 320 Ile Lys Ser Ala Thr Tyr Asn Asn Val Gly
Met Lys Met Leu Phe Pro 325 330
335 Pro Leu Leu Glu Ala Val Lys Lys Leu Val Ala Glu Thr Pro Asp
Phe 340 345 350 Ala
Ser Lys Ala Leu Ala Val Pro Asp Thr Thr Pro Lys Ile Pro Glu 355
360 365 Val Pro Asp Asp His Ile
Thr Thr Gln Ala Trp Leu Trp Gln Arg Leu 370 375
380 Ser Tyr Phe Leu Arg Pro Thr Asp Ile Val Val
Thr Glu Thr Gly Thr 385 390 395
400 Ser Ser Phe Gly Ile Ile Gln Thr Lys Phe Pro His Asn Val Arg Gly
405 410 415 Ile Ser
Gln Val Leu Trp Gly Ser Ile Gly Tyr Ser Val Gly Ala Ala 420
425 430 Cys Gly Ala Ser Ile Ala Ala
Gln Glu Ile Asp Pro Gln Gln Arg Val 435 440
445 Ile Leu Phe Val Gly Asp Gly Ser Leu Gln Leu Thr
Val Thr Glu Ile 450 455 460
Ser Cys Met Ile Arg Asn Asn Val Lys Pro Tyr Ile Phe Val Leu Asn 465
470 475 480 Asn Asp Gly
Tyr Thr Ile Glu Arg Leu Ile His Gly Glu Asn Ala Ser 485
490 495 Tyr Asn Asp Val His Met Trp Lys
Tyr Ser Lys Ile Leu Asp Thr Phe 500 505
510 Asn Ala Lys Ala His Glu Ser Ile Val Val Asn Thr Lys
Gly Glu Met 515 520 525
Asp Ala Leu Phe Asp Asn Glu Glu Phe Ala Lys Pro Asp Lys Ile Arg 530
535 540 Leu Ile Glu Val
Met Cys Asp Lys Met Asp Ala Pro Ala Ser Leu Ile 545 550
555 560 Lys Gln Ala Glu Leu Ser Ala Lys Thr
Asn Val 565 570 1221713DNAYarrowia
lipolytica 122atgagcgact ccgaacccca aatggtcgac ctgggcgact atctctttgc
ccgattcaag 60cagctaggcg tggactccgt ctttggagtg cccggcgact tcaacctcac
cctgttggac 120cacgtgtaca atgtcgacat gcggtgggtt gggaacacaa acgagctgaa
tgccggctac 180tcggccgacg gctactcccg ggtcaagcgg ctggcatgtc ttgtcaccac
ctttggcgtg 240ggagagctgt ctgccgtggc tgctgtggca ggctcgtacg ccgagcatgt
gggcgtggtg 300catgttgtgg gcgttcccag cacctctgct gagaacaagc atctgctgct
gcaccacaca 360ctcggtaacg gcgacttccg ggtctttgcc cagatgtcca aactcatctc
cgagtacacc 420caccatattg aggaccccag cgaggctgcc gacgtaatcg acaccgccat
ccgaatcgcc 480tacacccacc agcggcccgt ttacattgct gtgccctcca acttctccga
ggtcgatatt 540gccgaccagg ctagactgga tacccccctg gacctttcgc tgcagcccaa
cgaccccgag 600agccagtacg aggtgattga ggagatttgc tcgcgtatca aggccgccaa
gaagcccgtg 660attctcgtcg acgcctgcgc ttcgcgatac agatgtgtgg acgagaccaa
ggagctggcc 720aagatcacca actttgccta ctttgtcact cccatgggta agggttctgt
ggacgaggat 780actgaccggt acggaggaac atacgtcgga tcgctgactg ctcctgctac
tgccgaggtg 840gttgagacag ctgatctcat catctccgta ggagctcttc tgtcggactt
caacaccggt 900tccttctcgt actcctactc caccaaaaac gtggtggaat tgcattcgga
ccacgtcaaa 960atcaagtccg ccacctacaa caacgtcggc atgaaaatgc tgttcccgcc
cctgctcgaa 1020gccgtcaaga aactggttgc cgagacccct gactttgcat ccaaggctct
ggctgttccc 1080gacaccactc ccaagatccc cgaggtaccc gatgatcaca ttacgaccca
ggcatggctg 1140tggcagcgtc tcagttactt tctgaggccc accgacatcg tggtcaccga
gaccggaacc 1200tcgtcctttg gaatcatcca gaccaagttc ccccacaacg tccgaggtat
ctcgcaggtg 1260ctgtggggct ctattggata ctcggtggga gcagcctgtg gagcctccat
tgctgcacag 1320gagattgacc cccagcagcg agtgattctg tttgtgggcg acggctctct
tcagctgacg 1380gtgaccgaga tctcgtgcat gatccgcaac aacgtcaagc cgtacatttt
tgtgctcaac 1440aacgacggct acaccatcga gaggctcatt cacggcgaaa acgcctcgta
caacgatgtg 1500cacatgtgga agtactccaa gattctcgac acgttcaacg ccaaggccca
cgagtcgatt 1560gtggtcaaca ccaagggcga gatggacgct ctgttcgaca acgaagagtt
tgccaagccc 1620gacaagatcc ggctcattga ggtcatgtgc gacaagatgg acgcgcctgc
ctcgttgatc 1680aagcaggctg agctctctgc caagaccaac gtt
1713123571PRTSchizosaccharomyces pombe 123Met Ser Gly Asp Ile
Leu Val Gly Glu Tyr Leu Phe Lys Arg Leu Glu 1 5
10 15 Gln Leu Gly Val Lys Ser Ile Leu Gly Val
Pro Gly Asp Phe Asn Leu 20 25
30 Ala Leu Leu Asp Leu Ile Glu Lys Val Gly Asp Glu Lys Phe Arg
Trp 35 40 45 Val
Gly Asn Thr Asn Glu Leu Asn Gly Ala Tyr Ala Ala Asp Gly Tyr 50
55 60 Ala Arg Val Asn Gly Leu
Ser Ala Ile Val Thr Thr Phe Gly Val Gly 65 70
75 80 Glu Leu Ser Ala Ile Asn Gly Val Ala Gly Ser
Tyr Ala Glu His Val 85 90
95 Pro Val Val His Ile Val Gly Met Pro Ser Thr Lys Val Gln Asp Thr
100 105 110 Gly Ala
Leu Leu His His Thr Leu Gly Asp Gly Asp Phe Arg Thr Phe 115
120 125 Met Asp Met Phe Lys Lys Val
Ser Ala Tyr Ser Ile Met Ile Asp Asn 130 135
140 Gly Asn Asp Ala Ala Glu Lys Ile Asp Glu Ala Leu
Ser Ile Cys Tyr 145 150 155
160 Lys Lys Ala Arg Pro Val Tyr Ile Gly Ile Pro Ser Asp Ala Gly Tyr
165 170 175 Phe Lys Ala
Ser Ser Ser Asn Leu Gly Lys Arg Leu Lys Leu Glu Glu 180
185 190 Asp Thr Asn Asp Pro Ala Val Glu
Gln Glu Val Ile Asn His Ile Ser 195 200
205 Glu Met Val Val Asn Ala Lys Lys Pro Val Ile Leu Ile
Asp Ala Cys 210 215 220
Ala Val Arg His Arg Val Val Pro Glu Val His Glu Leu Ile Lys Leu 225
230 235 240 Thr His Phe Pro
Thr Tyr Val Thr Pro Met Gly Lys Ser Ala Ile Asp 245
250 255 Glu Thr Ser Gln Phe Phe Asp Gly Val
Tyr Val Gly Ser Ile Ser Asp 260 265
270 Pro Glu Val Lys Asp Arg Ile Glu Ser Thr Asp Leu Leu Leu
Ser Ile 275 280 285
Gly Ala Leu Lys Ser Asp Phe Asn Thr Gly Ser Phe Ser Tyr His Leu 290
295 300 Ser Gln Lys Asn Ala
Val Glu Phe His Ser Asp His Met Arg Ile Arg 305 310
315 320 Tyr Ala Leu Tyr Pro Asn Val Ala Met Lys
Tyr Ile Leu Arg Lys Leu 325 330
335 Leu Lys Val Leu Asp Ala Ser Met Cys His Ser Lys Ala Ala Pro
Thr 340 345 350 Ile
Gly Tyr Asn Ile Lys Pro Lys His Ala Glu Gly Tyr Ser Ser Asn 355
360 365 Glu Ile Thr His Cys Trp
Phe Trp Pro Lys Phe Ser Glu Phe Leu Lys 370 375
380 Pro Arg Asp Val Leu Ile Thr Glu Thr Gly Thr
Ala Asn Phe Gly Val 385 390 395
400 Leu Asp Cys Arg Phe Pro Lys Asp Val Thr Ala Ile Ser Gln Val Leu
405 410 415 Trp Gly
Ser Ile Gly Tyr Ser Val Gly Ala Met Phe Gly Ala Val Leu 420
425 430 Ala Val His Asp Ser Lys Glu
Pro Asp Arg Arg Thr Ile Leu Val Val 435 440
445 Gly Asp Gly Ser Leu Gln Leu Thr Ile Thr Glu Ile
Ser Thr Cys Ile 450 455 460
Arg His Asn Leu Lys Pro Ile Ile Phe Ile Ile Asn Asn Asp Gly Tyr 465
470 475 480 Thr Ile Glu
Arg Leu Ile His Gly Leu His Ala Ser Tyr Asn Glu Ile 485
490 495 Asn Thr Lys Trp Gly Tyr Gln Gln
Ile Pro Lys Phe Phe Gly Ala Ala 500 505
510 Glu Asn His Phe Arg Thr Tyr Cys Val Lys Thr Pro Thr
Asp Val Glu 515 520 525
Lys Leu Phe Ser Asp Lys Glu Phe Ala Asn Ala Asp Val Ile Gln Val 530
535 540 Val Glu Leu Val
Met Pro Met Leu Asp Ala Pro Arg Val Leu Val Glu 545 550
555 560 Gln Ala Lys Leu Thr Ser Lys Ile Asn
Lys Gln 565 570
1241713DNASchizosaccharomyces pombe 124atgagtgggg atattttagt cggtgaatat
ctattcaaaa ggcttgaaca attaggggtc 60aagtccattc ttggtgttcc aggagatttc
aatttagctc tacttgactt aattgagaaa 120gttggagatg agaaatttcg ttgggttggc
aataccaatg agttgaatgg tgcttatgcc 180gctgatggtt atgctcgtgt taatggtctt
tcagccattg ttacaacgtt cggcgtggga 240gagctttccg ctattaatgg agtggcaggt
tcttatgcgg agcatgtccc agtagttcat 300attgttggaa tgccttccac aaaggtgcaa
gatactggag ctttgcttca tcatacttta 360ggagatggag actttcgcac tttcatggat
atgtttaaga aagtttctgc ctacagtata 420atgatcgata acggaaacga tgcagctgaa
aagatcgatg aagccttgtc gatttgttat 480aaaaaggcta ggcctgttta cattggtatt
ccttctgatg ctggctactt caaagcatct 540tcatcaaatc ttgggaaaag actaaagctc
gaggaggata ctaacgatcc agcagttgag 600caagaagtca tcaatcatat ctcggaaatg
gttgtcaatg caaagaaacc agtgatttta 660attgacgctt gtgctgtaag acatcgtgtc
gttccagaag tacatgagct gattaaattg 720acccatttcc ctacatatgt aactcccatg
ggtaaatctg caattgacga aacttcgcaa 780ttttttgacg gcgtttatgt tggttcaatt
tcagatcctg aagttaaaga cagaattgaa 840tccactgatc tgttgctatc catcggtgct
ctcaaatcag actttaacac gggttccttc 900tcttaccacc tcagccaaaa gaatgccgtt
gagtttcatt cagaccacat gcgcattcga 960tatgctcttt atccaaatgt agccatgaag
tatattcttc gcaaactgtt gaaagtactt 1020gatgcttcta tgtgtcattc caaggctgct
cctaccattg gctacaacat caagcctaag 1080catgcggaag gatattcttc caacgagatt
actcattgct ggttttggcc taaatttagt 1140gaatttttga agccccgaga tgttttgatc
accgagactg gaactgcaaa ctttggtgtc 1200cttgattgca ggtttccaaa ggatgtaaca
gccatttccc aggtattatg gggatctatt 1260ggatactccg ttggtgcaat gtttggtgct
gttttggccg tccacgattc taaagagccc 1320gatcgtcgta ccattcttgt agtaggtgat
ggatccttac aactgacgat tacagagatt 1380tcaacctgca ttcgccataa cctcaaacca
attattttca taattaacaa cgacggttac 1440accattgagc gtttaattca tggtttgcat
gctagctata acgaaattaa cactaaatgg 1500ggctaccaac agattcccaa gtttttcgga
gctgctgaaa accacttccg cacttactgt 1560gttaaaactc ctactgacgt tgaaaagttg
tttagcgaca aggagtttgc aaatgcagat 1620gtcattcaag tagttgagct tgtaatgcct
atgttggatg cacctcgtgt cctagttgag 1680caagccaagt tgacgtctaa gatcaataag
caa 1713125563PRTZygosaccharomyces rouxii
125Met Ser Glu Ile Thr Leu Gly Arg Tyr Leu Phe Glu Arg Leu Lys Gln 1
5 10 15 Val Asp Thr Asn
Thr Ile Phe Gly Val Pro Gly Asp Phe Asn Leu Ser 20
25 30 Leu Leu Asp Lys Val Tyr Glu Val Gln
Gly Leu Arg Trp Ala Gly Asn 35 40
45 Ala Asn Glu Leu Asn Ala Ala Tyr Ala Ala Asp Gly Tyr Ala
Arg Val 50 55 60
Lys Gly Leu Ala Ala Leu Ile Thr Thr Phe Gly Val Gly Glu Leu Ser 65
70 75 80 Ala Leu Asn Gly Ile
Ala Gly Ser Tyr Ala Glu His Val Gly Val Leu 85
90 95 His Ile Val Gly Val Pro Ser Val Ser Ser
Gln Ala Lys Gln Leu Leu 100 105
110 Leu His His Thr Leu Gly Asn Gly Asp Phe Thr Val Phe His Arg
Met 115 120 125 Ser
Ala Asn Ile Ser Glu Thr Thr Ala Met Leu Thr Asp Ile Thr Ala 130
135 140 Ala Pro Ala Glu Ile Asp
Arg Cys Ile Arg Val Ala Tyr Val Asn Gln 145 150
155 160 Arg Pro Val Tyr Leu Gly Leu Pro Ala Asn Leu
Val Asp Gln Lys Val 165 170
175 Pro Ala Ser Leu Leu Asn Thr Pro Ile Asp Leu Ser Leu Lys Glu Asn
180 185 190 Asp Pro
Glu Ala Glu Thr Glu Val Val Asp Thr Val Leu Glu Leu Ile 195
200 205 Lys Glu Ala Lys Asn Pro Val
Ile Leu Ala Asp Ala Cys Cys Ser Arg 210 215
220 His Asp Val Lys Ala Glu Thr Lys Lys Leu Ile Asp
Leu Thr Gln Phe 225 230 235
240 Pro Ser Phe Val Thr Pro Met Gly Lys Gly Ser Ile Asp Glu Gln Asn
245 250 255 Pro Arg Phe
Gly Gly Val Tyr Val Gly Thr Leu Ser Ser Pro Glu Val 260
265 270 Lys Glu Ala Val Glu Ser Ala Asp
Leu Val Leu Ser Val Gly Ala Leu 275 280
285 Leu Ser Asp Phe Asn Thr Gly Ser Phe Ser Tyr Ser Tyr
Lys Thr Lys 290 295 300
Asn Val Val Glu Phe His Ser Asp His Ile Lys Ile Arg Asn Ala Thr 305
310 315 320 Phe Pro Gly Val
Gln Met Lys Phe Val Leu Lys Lys Leu Leu Gln Ala 325
330 335 Val Pro Glu Ala Val Lys Asn Tyr Lys
Pro Gly Pro Val Pro Ala Pro 340 345
350 Pro Ser Pro Asn Ala Glu Val Ala Asp Ser Thr Thr Leu Lys
Gln Glu 355 360 365
Trp Leu Trp Arg Gln Val Gly Ser Phe Leu Arg Glu Gly Asp Val Val 370
375 380 Ile Thr Glu Thr Gly
Thr Ser Ala Phe Gly Ile Asn Gln Thr His Phe 385 390
395 400 Pro Asn Gln Thr Tyr Gly Ile Ser Gln Val
Leu Trp Gly Ser Ile Gly 405 410
415 Tyr Thr Thr Gly Ser Thr Leu Gly Ala Ala Phe Ala Ala Glu Glu
Ile 420 425 430 Asp
Pro Lys Lys Arg Val Ile Leu Phe Ile Gly Asp Gly Ser Leu Gln 435
440 445 Leu Thr Val Gln Glu Ile
Ser Thr Met Ile Arg Trp Gly Leu Lys Pro 450 455
460 Tyr Leu Phe Val Leu Asn Asn Asp Gly Tyr Thr
Ile Glu Arg Leu Ile 465 470 475
480 His Gly Glu Thr Ala Glu Tyr Asn Cys Ile Gln Pro Trp Lys His Leu
485 490 495 Glu Leu
Leu Asn Thr Phe Gly Ala Lys Asp Tyr Glu Asn His Arg Val 500
505 510 Ser Thr Val Gly Glu Trp Asn
Lys Leu Thr Gln Asp Pro Lys Phe Asn 515 520
525 Glu Asn Ser Arg Ile Arg Met Ile Glu Val Met Leu
Glu Val Met Asp 530 535 540
Ala Pro Ser Ser Leu Val Ala Gln Ala Gln Leu Thr Ala Ala Thr Asn 545
550 555 560 Ala Lys Gln
1261689DNAZygosaccharomyces rouxii 126atgtctgaaa ttactctagg tcgttacttg
ttcgaaagat taaagcaagt tgacactaac 60accatcttcg gtgttccagg tgacttcaac
ttgtccttgt tggacaaggt ctacgaagtg 120caaggtctaa gatgggctgg taacgctaac
gaattgaacg ctgcctacgc tgctgacggt 180tacgccagag ttaagggttt ggctgctttg
atcaccacct tcggtgtcgg tgaattgtct 240gctttgaacg gtattgcagg ttcttacgct
gaacacgttg gtgttttgca cattgttggt 300gttccatctg tctcttctca agctaagcaa
ttgttgttgc accacacctt gggtaacggt 360gacttcactg ttttccacag aatgtccgcc
aacatctctg aaaccaccgc tatgttgacc 420gacatcactg ctgctccagc tgaaattgac
cgttgcatca gagttgctta cgtcaaccaa 480agaccagtct acttgggtct accagctaac
ttggttgacc aaaaggtccc agcttctttg 540ttgaacactc caattgatct atctctaaag
gagaacgacc cagaagctga aaccgaagtt 600gttgacaccg ttttggaatt gatcaaggaa
gctaagaacc cagttatctt ggctgatgct 660tgctgctcca gacacgacgt caaggctgaa
accaagaagt tgatcgactt gactcaattc 720ccatctttcg ttactcctat gggtaagggt
tccatcgacg aacaaaaccc aagattcggt 780ggtgtctacg tcggtactct atccagccca
gaagttaagg aagctgttga atctgctgac 840ttggttctat ctgtcggtgc tctattgtcc
gatttcaaca ctggttcttt ctcttactct 900tacaagacca agaacgttgt tgaattccac
tctgaccaca tcaagatcag aaacgctacc 960ttcccaggtg ttcaaatgaa attcgttttg
aagaaactat tgcaagctgt cccagaagct 1020gtcaagaact acaagccagg tccagtccca
gctccgccat ctccaaacgc tgaagttgct 1080gactctacca ccttgaagca agaatggtta
tggagacaag tcggtagctt cttgagagaa 1140ggtgatgttg ttattaccga aactggtacc
tctgctttcg gtatcaacca aactcacttc 1200cctaaccaaa cttacggtat ctctcaagtc
ttgtggggtt ctattggtta caccactggt 1260tccactttgg gtgctgcctt cgctgctgaa
gaaattgacc ctaagaagag agttatcttg 1320ttcattggtg acggttctct acaattgacc
gttcaagaaa tctccaccat gatcagatgg 1380ggtctaaagc catacttgtt cgttttgaac
aacgatggtt acaccattga aagattgatt 1440cacggtgaaa ccgctgaata caactgtatc
caaccatgga agcacttgga attgttgaac 1500accttcggtg ccaaggacta cgaaaaccac
agagtctcca ctgtcggtga atggaacaag 1560ttgactcaag atccaaaatt caacgaaaac
tctagaatta gaatgatcga agttatgctt 1620gaagtcatgg acgctccatc ttctttggtc
gctcaagctc aattgaccgc tgctactaac 1680gctaagcaa
1689127267PRTSaccharomyces cerevisiae
127Met Ser Gln Gly Arg Lys Ala Ala Glu Arg Leu Ala Lys Lys Thr Val 1
5 10 15 Leu Ile Thr Gly
Ala Ser Ala Gly Ile Gly Lys Ala Thr Ala Leu Glu 20
25 30 Tyr Leu Glu Ala Ser Asn Gly Asp Met
Lys Leu Ile Leu Ala Ala Arg 35 40
45 Arg Leu Glu Lys Leu Glu Glu Leu Lys Lys Thr Ile Asp Gln
Glu Phe 50 55 60
Pro Asn Ala Lys Val His Val Ala Gln Leu Asp Ile Thr Gln Ala Glu 65
70 75 80 Lys Ile Lys Pro Phe
Ile Glu Asn Leu Pro Gln Glu Phe Lys Asp Ile 85
90 95 Asp Ile Leu Val Asn Asn Ala Gly Lys Ala
Leu Gly Ser Asp Arg Val 100 105
110 Gly Gln Ile Ala Thr Glu Asp Ile Gln Asp Val Phe Asp Thr Asn
Val 115 120 125 Thr
Ala Leu Ile Asn Ile Thr Gln Ala Val Leu Pro Ile Phe Gln Ala 130
135 140 Lys Asn Ser Gly Asp Ile
Val Asn Leu Gly Ser Ile Ala Gly Arg Asp 145 150
155 160 Ala Tyr Pro Thr Gly Ser Ile Tyr Cys Ala Ser
Lys Phe Ala Val Gly 165 170
175 Ala Phe Thr Asp Ser Leu Arg Lys Glu Leu Ile Asn Thr Lys Ile Arg
180 185 190 Val Ile
Leu Ile Ala Pro Gly Leu Val Glu Thr Glu Phe Ser Leu Val 195
200 205 Arg Tyr Arg Gly Asn Glu Glu
Gln Ala Lys Asn Val Tyr Lys Asp Thr 210 215
220 Thr Pro Leu Met Ala Asp Asp Val Ala Asp Leu Ile
Val Tyr Ala Thr 225 230 235
240 Ser Arg Lys Gln Asn Thr Val Ile Ala Asp Thr Leu Ile Phe Pro Thr
245 250 255 Asn Gln Ala
Ser Pro His His Ile Phe Arg Gly 260 265
128804DNASaccharomyces cerevisiae 128atgtcccaag gtagaaaagc tgcagaaaga
ttggctaaga agactgtcct cattacaggt 60gcatctgctg gtattggtaa ggcgaccgca
ttagagtact tggaggcatc caatggtgat 120atgaaactga tcttggctgc tagaagatta
gaaaagctcg aggaattgaa gaagaccatt 180gatcaagagt ttccaaacgc aaaagttcat
gtggcccagc tggatatcac tcaagcagaa 240aaaatcaagc ccttcattga aaacttgcca
caagagttca aggatattga cattctggtg 300aacaatgccg gaaaggctct tggcagtgac
cgtgtgggcc agatcgcaac ggaggatatc 360caggacgtgt ttgacaccaa cgtcacggct
ttaatcaata tcacacaagc tgtactgccc 420atattccaag ccaagaattc aggagatatt
gtaaatttgg gttcaatcgc tggcagagac 480gcatacccaa caggttctat ctattgtgcc
tctaagtttg ccgtgggggc gttcactgat 540agtttgagaa aggagctcat caacactaaa
attagagtca ttctaattgc accagggcta 600gtcgagactg aattttcact agttagatac
agaggtaacg aggaacaagc caagaatgtt 660tacaaggata ctaccccatt gatggctgat
gacgtggctg atctgatcgt ctatgcaact 720tccagaaaac aaaatactgt aattgcagac
actttaatct ttccaacaaa ccaagcgtca 780cctcatcata tcttccgtgg ataa
804
User Contributions:
Comment about this patent or add new information about this topic: