Patent application title: Traceability of Cellular Cycle Anomalies Targeting Oncology and Neurodegeneration
Inventors:
Laurence Faure (Paris, FR)
IPC8 Class: AC12Q168FI
USPC Class:
506 9
Class name: Combinatorial chemistry technology: method, library, apparatus method of screening a library by measuring the ability to specifically bind a target molecule (e.g., antibody-antigen binding, receptor-ligand binding, etc.)
Publication date: 2016-04-21
Patent application number: 20160108477
Abstract:
The present invention relates to the field of medicine and biology. It
concerns a novel test for screening and for therapeutic follow-up in
oncology. More particularly, it relates to diagnostic and/or therapeutic
tests in oncology and on neurodegenerative diseases. It is a diagnostic
test and a prognostic test for various cancers (breast cancer, bladder
cancer, ovarian cancer, lung cancer, skin cancer, prostate cancer, colon
cancer, liver cancer, glioblastoma, sarcoma, leukemia, etc.) and
therapeutics solutions for specific neurodegenerative diseases. More
particularly, the invention concerns the use of the LIV21 protein, LIV21
gene and of derivatives thereof as diagnostic and prognostic markers for
cancers. The invention therefore concerns the detection of the LIV21
protein with a kit comprising LIV21-specific antibodies.Claims:
1. A method for the detection of cancer cells in a biological sample from
a patient, comprising the detection of the product of expression of the
LIV21 gene in the nucleus and/or cytoplasm of the cells in a sample of
cells in the biological sample from said patient, localization of said
product of expression of LIV21 gene in the cytoplasm being indicative of
the presence of cancer cells and localization of said product of
expression of the LIV21 gene in the nucleus being indicative of the
presence of noncancer cells.
2. The method as claimed in claim 1, in which localization of said product of expression of the LIV21 gene in the cytoplasm is indicative of the presence of invasive and/or metastatic cancer cells.
3. The method as claimed in claim 1, also comprising the detection of the product of expression of at least three genes selected from the group consisting of the protein kinase C epsilon (PKCε) gene, the E2F1 gene, the E2F4 gene and HDAC1.
4. The method as claimed in claim 3, in which at least one of the ratios LIV21/PKCε, LIV21/E2F4 and LIV21/E2F1 is determined.
5. The method as claimed in claim 1, in which the expression product of the genes is detected at the mRNA level or a isolated polynucleotide (SEQ ID No: 119-148) related to LIV21 gene.
6. The method as claimed in claim 1, also comprising the detection of the product of expression of at least five genes or antibodies specific for a protein selected from the group consisting of RBP2, E2F4, E2F2, E2F1, SUMO1, SUMO3, HDAC1, cycE/cdk2, cdk1, CREB1, p300, Rb, PML, p107 and p130 of the pocket protein family.
7. The method as claimed in claim 1, also comprising the detection of the product of expression of at least five genes selected from NFkB, cdc2A, mdm2, p21, p53, p65, Ki67, erk, CD53 and CAF1.
8-11. (canceled)
12. The method as claimed in claim 3, in which significant increase in PKCε is indicative of the presence of cancer cells.
13. (canceled)
14. The method as claimed in claim 3, for the detection of metastasized cancer, therapeutic monitoring and/or recurrences following treatment.
15-24. (canceled)
25. The method as claimed in claim 1, in which the cancer is selected from breast cancer, bladder cancer, ovarian cancer, lung cancer, skin cancer, prostrate cancer, colon cancer, liver cancer, a sarcoma, a leukemia, and glioblastoma.
26. The use of an oligonucleotide specific probe of LIV21 mRNA or isolated polynucleotide (SEQ ID Nos. 119-198) as claimed in claim 5 or specific primers of LIV21 mRNA for the diagnosis of cancer related to the LIV21 gene.
27-33. (canceled)
Description:
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This is a Divisional Application of U.S. patent application Ser. No. 12/047,173 filed on Mar. 12, 2008 which is a continuation in part application of U.S. patent application Ser. No. 11/908,103 filed on Sep. 7, 2007 which is a National Phase Entry of PCT Application Serial No. PCT/FR2006/000510 filed on Mar. 7, 2006 which claims priority from French Patent Application Nos. 0502258 and 0502257 filed on Mar. 7, 2005.
FIELD OF THE INVENTION
[0002] The present invention relates to the field of medicine and biology. It concerns a novel test for screening and for therapeutic follow-up in oncology. More particularly, it relates to diagnostic and/or therapeutic tests in oncology and on neurodegenerative diseases.
DESCRIPTION OF THE PRIOR ART
[0003] Age-related neurodegenerative diseases and cancers both involve a modification of the physiological process of programmed cell death or apoptosis. Neuronal death is abnormally accelerated during neurodegenerative diseases such as Alzheimer's disease, Huntington's disease, Parkinson's disease, etc. On the other hand, the cancerization process corresponds to a blocking of apoptosis which results in an uncontrolled multiplication of cells. The link between these two processes has currently become a major field of investigation in research on aging.
[0004] The control of the balance between cell division (mitosis), differentation and programmed cell death (apoptosis) is fundamental during normal physiological processes, such as embryonic development, tissue regeneration and aging. An impairment of this balance can lead to major pathological situations such as the formation of tumors or certain neurodegenerative diseases.
[0005] Cancer is one of the principal causes of mortality throughout the world. Although over the course of the last generation, the percentages of deaths related to cardiac and cardiovascular diseases and a large number of other diseases has decreased, the number of deaths related to the various forms of cancer is on the increase.
[0006] Despite the rapid advance in our understanding of the various forms of cancer, the low survival rates can generally be attributed to inadequate diagnosis and inadequate treatment. Most tumors can only be detected when they reach a size of approximately 1 cm. Since there is a relatively short period of time from the continuous development of a tumor to a stage which has become incompatible with survival, this leaves little time for a therapeutic intervention. Early diagnosis therefore becomes the key to success for the treatment of cancer.
[0007] For a multitude of reasons, early diagnosis remains illusory for most forms of cancer. For certain forms of cancer, disease-specific markers are not available or are only available at an advanced stage of the disease, making diagnosis difficult. In certain other forms of cancer, the markers are available but are not always specific for the disease or they may be associated with its benign form. In yet other cases, the techniques exist but the prohibitive cost for applying them to the population in general makes them unsuitable.
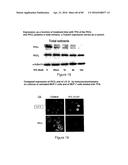
[0008] Skin cancer, for example, is the most widespread cancer in Canada. In 1992 alone, 50 300 new cases of skin cancer were reported, compared with 19 300 cases of lung cancer, 16 200 cases of colorectal cancer and 15 700 cases of breast cancer. In other words, skin cancer is as common as the three main types of cancer combined. Its incidence continues to increase, with 64 200 new cases thereof in 1997, that is an increase of 14 000 cases annually in 5 years. In particular, the incidence of malignant melanoma is increasing at a rate of 2% per year. Early diagnosis remains the key to an effective treatment. A malignant tumor is readily accessible and can be removed with minor surgery. In fact, recovery is 100% if skin cancer is detected early enough. The early diagnosis of skin cancer remains, however, difficult. The latter is not just one disease but an entire range of conditions related to one another, which appear similar in many cases upon visual inspection. A diagnosis on the basis of such an inspection is therefore subjective. In order to understand this subjectivity more fully, an abnormal skin growth should be considered. This growth may be pigmented or nonpigmented. If it is nonpigmented and malignant, it is then probably a basocellular epithelioma or a spinocellular epithelioma. However, the clinical development of these two forms of cancer is very different. A basocellular epithelioma spreads out laterally over the surface of the skin, without penetrating into the deeper skin layers. Thus, although it can be disfiguring, a basocellular epithelioma rarely develops metastases and is rarely fatal. However, a spinocellular epithelioma causes metastases and is often fatal. It therefore becomes important to be able to distinguish these two types of skin cancer. A definitive diagnosis of skin cancer requires a biopsy and histological analysis. However, the decision to send a biopsy for analysis (or even whether a patient should be referred to a dermatologist) becomes very subjective. There are several biopsies which are not taken although they should have been.
[0009] Colon cancer is the third most common cause of cancer-related mortality in men and women in North America (16 200 cases per year). Early detection, leading to an early intervention, has demonstrated that treatment success and survival rate can be improved. For example, the 5-year survival rate is 92% for a patient whose disease was detected at an early stage, whereas the rate drops to approximately 60% in patients with a localized cancer, and to approximately 6% in those with metastases. However, only a third of colon cancers are detected at an early stage. One of the reasons for this delay in diagnosis is the absence of a sensitive, relatively inexpensive, non-invasive screening test.
[0010] Breast cancer is one of the most common cancers in women, with colon cancer. The mortality rate is the highest of all the cancers affecting women.
[0011] There are very few diagnostic markers capable of detecting breast cancer and they only have a predictive value of 20%. There are no markers, either, which can detect of determine the invasiveness or the aggressiveness of metastatic cancer cells.
[0012] Over the last few years, considerable progress has been made in the understanding of the means used by oncogenes and tumor suppressor genes for regulating cell proliferation and apoptosis. One of the main targets of these regulators is the family of E2F-type transcription factors in the E2F and RB protein signaling pathway. These proteins play a central role in controlling cell division by coupling the regulation of the genes required for progression of the cell cycle with extracellular signals (mitogens, proliferation inhibitors). It behaves as an oncogene by stimulating tumor cell proliferation.
Among the expressed genes are found:
[0013] overexpression of the E2F4 transcription factor and the c-myc oncogene which induce apoptosis of post-mitotic cells by accumulation of oxygenated reactants (Tanaka, 2002);
[0014] the p53 gene, which belongs to the tumor suppressor gene family, blocks the cell cycle in the case of DNA lesion. It has now been demonstrated that this gene is also involved in the progression of apoptosis (Oren, 1994; Yonish-Rouach, 1996);
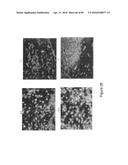
[0015] the cyclin D1, one of the proteins constituting the regulatory subunits of the kinases of the cell cycle, essential to the progression of the cell cycle. This protein is also expressed during apoptosis in various cell types (Han et al, 1996; Pardo et al, 1996).
[0016] It would be desirable to have novel diagnostic methods which would detect the presence of cancer with greater specificity and which would make it possible to distinguish between aggressive cancer cells having a tendency to metastasize and those which are more localized which have a lower probability of metastasizing. A marker which can therefore reveal cell proliferation would be of great use.
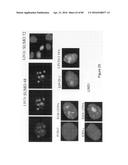
SUMMARY OF THE INVENTION
[0017] The present invention concerns a novel test for screening for reinduction of the cell cycle targeting oncology. It is a diagnostic test and a prognostic test for various cancers (breast cancer, bladder cancer, ovarian cancer, lung cancer, skin cancer, prostate cancer, colon cancer, liver cancer, glioblastoma, sarcoma, leukaemia, etc.). More particularly, the invention concerns the use of the LIV21 protein and of derivatives thereof as diagnostic and prognostic markers for cancers. The invention therefore concerns the detection of the LIV21 protein with a kit comprising LIV21-specific antibodies.
[0018] A first objective of the present invention is to demonstrate a method for the detection and prognosis of cancer and of its metastatic potential. Preferably, the cancer is selected from breast cancer, bladder cancer, ovarian cancer, lung cancer, skin cancer, prostate cancer, colon cancer, liver cancer, a sarcoma, a leukaemia and glyoblastoma, without being limited thereto.
[0019] One aspect of the present invention consists of the use of LIV21 and isoforms of LIV21 as a prognostic indicator for cancer. In fact, when LIV21 is located in the cytoplasm, the cancer cells in the tissues are aggressive. Conversely, when the LIV21 gene expression product is preferentially located in the cell nucleus, this is a prognostic indicator that the cells of the tissue are differentiated and quiescent and therefore non-invasive. The effectiveness of a cancer treatment can also be monitored by the traceability of this protein, and of its derivatives and ratios with the associated proteins.
[0020] Moreover, detection of protein kinase C epsilon (PKCε) is also advantageous since it has been determined that PKCε phosphorylates the LIV21 protein in order to maintain it in the cytoplasm. Thus, a significant increase in PKCε is indicative of the presence of cancer cells. Moreover, the LIV21/PKCε ratio increases in the cytoplasmic fraction of cancer cells.
[0021] In addition, the detection of the E2F1 and/or E2F4 proteins is advantageous. In fact, the LIV21 protein forms a complex with E2F4 which is capable of inhibiting the expression of the E2F1 gene in the nucleus, E2F1 gene expression being a sign of cell proliferation. Thus, a decrease in the association of LIV21 with the E2F4 protein is indicative of the presence of cancer cells. Similarly, the presence of the E2F1 protein in the nucleus is indicative of the presence of cancer cells.
[0022] Consequently, the present invention concerns a method for the detection (in vitro or ex vivo) of cancer cells in a biological tissue sample (for example, breast, ovary, endometrium, bladder, melanoma, prostate, glioblastoma, etc.) from patients, this method comprising the detection of the product of expression of the LIV21 gene in the nucleus and/or the cytoplasm of the cells in the biological tissue sample from said patient, localization of said product of expression of the LIV21 gene in the cytoplasm being indicative of the presence of cancer cells, and localization of said product of expression of the LIV21 gene in the nucleus being indicative of the presence of noncancer cells. Preferably, localization of said product of expression of the LIV21 gene in the cytoplasm is indicative of the presence of invasive and/or metastatic cancer cells.
[0023] Optionally, the method according to the present invention also comprises the detection of the product of expression of at least one gene selected from the group consisting of the protein kinase C epsilon (PKCε) gene, the E2F1 gene and the E2F4 gene. The method can in particular comprise the detection of the product of expression of two of these genes or of the three genes. Moreover, at least one of the ratios LIV21/PKCε, LIV21/E2F4 and LIV21/E2F1 can be determined in the present method. This ratio can be determined in the cytoplasm and/or in the nucleus. Preferably, these ratios are determined in the nucleus. Preferably, these ratios are compared with those obtained in a normal cell.
[0024] The same is true of the detection of HDAC1, which has been shown to be involved in PML/SUMO1/Rb/HDAC-1 complexes. More generally, the HDAC family plays a key role in the regulation of gene expression. When the HDACs are overexpressed, they bring about tumor suppressor gene silencing, hence the advance of using HDAC inhibitors in therapy, combined with other inhibitors which regulate the metabolic cascade involving the protein complex which contains LIV21. The level of expression of each enzyme or polypeptide of the SUMO/Rb/HDAC complex or, for certain cell types, of the PML/SUMO/Rb/HDAC complex is an additional indictor of the proliferative state of the cell. Thus, in a specific embodiment, the method according to the present invention also comprises the detection of the product of expression of at least one gene selected from the group consisting of SUMO1, Rb, HDAC, and PML.
[0025] The methods according to the present invention also consist in using the detection of the LIV21 protein in combination with all the proliferation markers and transcription factors which play a role in the cancerization and neurodegeneration process. The method therefore also comprises the detection of the product of expression of at least five genes selected from the group consisting of RBP2, E2F4, E2F1, E2F2, SUMO1, HDAC1, cycE/cdk2, cdk1, CREB1, p300, Rb, PML, p107 and p130 of the pocket protein family. Thus, the invention lies in the fabrication and the use of diagnostic antibody arrays (FIG. 2) comprising LIV21-specific antibodies and antibodies for the various proteins of the LIV21-associated complex according specific for RBP2, E2F4, E2F2, E2F1, SUMO1, SUMO3, HDAC1, cycE/cdk2, cdk1, CREB1, p300, Rb, PML, p107 and p130 of the pocket protein family (FIG. 1). In addition, the diagnostic arrays according to the present invention can comprise antibodies specific for NFkB, cdc2A, mdm2, p21, p53, p65, Ki67, erk and CAFL Ki67 and CAF1 (Amoulzy; Institut Curie) are nuclear markers which signal the proliferative state of many cancers. The protein arrays will make it possible to study protein expression, protein interactions and post-translational modifications, more particulary phosphorylations and methylations of certain proteins, which signal a characteristic state of the diseased cell. The state of expression and of silencing of certain genes is different in diseased cells and in normal cells. Moreover, the protein interactions and the metabolism of the diseased cell are different from those of the normal cell.
[0026] The other aspect of the present invention is the use of the proteins mentioned above as markers for the invasiveness and the metastatic aggressiveness of cancer cells of the prostate, colon, bladder, melanoma, ovary, endomentrium and cervix, and cancers in neurobiology, etc.
[0027] In one embodiment, the expression product of the genes is detected at the protein level. Preferably, the protein is detected using a specific antibody. For example, the protein can be detected by Western blotting analysis. In a preferred embodiment, it is detected by immunohistochemistry, immunocytochemistry or radiography, or by peroxidase labelling by microfludiic technique (sonic or light as sers or Raman effect).
[0028] In one specific embodiment of the method comprising the detection of the expression product of the PKCε gene, a significant increase in PKCε is indicative of the presence of cancer cells. Moreover, the method can also comprise the determination of the LIV21/PKCε ratio in the nucleus and/or the cytoplasm. This ratio can be compared with that observed in a normal cell. An increase in the LIV21/PKCε ratio in the cytoplasmic fraction is indicative of cancer cells.
[0029] In another specific embodiment of the method comprising the detection of the expression product of the E2F4 gene, the method comprises the detection of the association of LIV21 with the E2F4 protein, a decrease in this association in the cell nucleus being indicative of the presence of cancer cells. Moreover, the method can also comprise the determination of the LIV21/E2F4 ratio in the nucleus and/or the cytoplasm. This ratio can be compared with that observed in a normal cell. In an additional embodiment of the method comprising the detection of the expression product of the E2F1 gene, the presence of the E2F1 protein in the nucleus is indicative of the presence of cancer cells. Moreover, the method can also comprise the determination of the LIV21/E2F1 ratio in the nucleus and/or the cytoplasm. This ratio can be compared with that observed in a normal cell. The method according to the present invention allows in particular the detection of metastasized cancer, therapeutic monitoring and/or recurrences following treatment. A second aspect of the invention concerns the human LIV21 protein and also the fragments thereof. More particularly, the present invention concerns a purified or recombinant, isolated human LIV21 protein. It concerns in particular an isolated polypeptide (issue to NT032977.8.hs1 33153) having an apparent molecular weight of approximately 50-51 kD by Western blotting analysis and of approximately 60 kD when it is sumoylated and/or a polypeptide having an isoelectric point of 5.6 in its 50-51 kD form and nine polypeptides having apparent molecular weight 110 kD, 64 kD, 51 kD, 50 kD, 49 kD, 30 kD, 17 kD, 16 kD, 15 kD in hot conditions (at 100° C.) having an isoelectric point between 5.6 and 11 in its forms and a polypeptide characterized by one of the chromatograms of FIGS. 3-6 and a polypeptide comprising a peptide sequence selected from SEQ ID Nos 1-55, preferably from SEQ ID Nos 1-5, or a sequence having 70%, 80% or 90% identity to said sequences, and one of the peptide sequences obtained by MALDI (FIGS. 7 and 8) and NanoLC-ESI-MS (FIGS. 6.1, 6.2 & 9). In a preferred embodiment, the polypeptide comprises the two peptide sequences SEQ ID Nos 1 and 2. In an even more preferred embodiment, the polypeptide comprises a third peptide sequence SEQ ID No 3 and/or a fourth peptide sequence SEQ ID No 4 or a sequence having (70%), 80% or 90% identity to said sequences. Optionally, LIV21 also comprises a sequence selected from the sequences SEQ ID Nos 5-55 or a sequence having 70%, 80% or 90% identity to said sequences. Preferably, the LIV21 protein comprises a leucine zipper motif, a basic domain characteristic of DNA binding domains, a nuclearization sequence, which not only mediates DNA binding but also acts as protein interaction domain for E2F2,E2F4,E2F1 (by similarity with E2F proteins family (FIGS. 10.1 and 11.1&2), item for retinoblastoma protein, Bcl2,CD53 antigen) and comprises also a Hand Domain, Zinc finger Domain (PDZ domain) and helix loop helix domain like TCFL4. The similarity of the sequence with transferase domain showed also by similarity with TRAMIL1 (with transmembrane helix and NDST3 for the (antisens) sequences of C8T7 clones (FIG. 11.3) which matched also on chromosome 4q26 permits to understand the complex interactions of LIV21. Digestion of the LIV21 protein with trypsin gives more than 54 peptides corresponding to the monoisotopic peaks among all the peptides as specified, FIGS. 3-6; FIGS. 7 and 9; FIGS. 8.1 and 8.2 (SDS PAGE gels) and sequences (sens and antisens) of LIV21 gene are ID Nos 119 to 148.
[0030] A third aspect of the invention concerns an antibody which binds specifically to a polypeptide according to the present invention. More particulary, the antibody can bind specifically to a polypeptide comprising a peptide sequence selected from SEQ ID Nos 1-55, preferably from SEQ ID Nos 1-5, or a sequence having 70%, 80% or 90% identity to said sequences. The present invention concerns in particular an anti-LIV21 serum produced by immunizing an animal or a human with a polypeptide according to the present invention, in particular a polypeptide comprising a peptide sequence selected from SEQ ID Nos 1-55, preferably from SEQ ID Nos 1-5, or a sequence having 70%, 80% or 90% identity to said sequences.
[0031] A fourth aspect of the invention concerns a kit for the detection of cancer cells in a biological sample from a patient, this kit comprising one or more elements selected from the group consisting of an antibody which binds specifically to human LIV21 according to the present invention and an anti-LIV21 serum according to the present invention. In a specific embodiment of the invention, the kit also comprises means for detecting the product of expression of five genes selected from the group consisting of the protein kinase C epsilon (PKCε) gene, the E2F1 gene, E2F2 gene and the E2F4 gene. Preferably, the detection means is an antibody specific for the protein concerned. In another preferred embodiment, the kit also comprises a means for detecting the product of expression of a gene selected from the group consisting of RBP2, SUMO1,SUMO3, HDAC1, PML, cycE/cdk2, cdk1, CREB1, p300, Rb, p107, p130, NFkB, erk, Bcl2,CD53, cdc2A, mdm2, p21, p53, p65, Ki67 and CAFL. In a preferred embodiment, the kit comprises an antibody array comprising an LIV21-specific antibody. In a preferred embodiment, the array also comprises an antibody specific for a protein selected from PKCε, E2F1 and E2F4. In addition, it can comprise five antibodies specific for a protein selected from RBP2, SUMO, HDAC1, cycE/cdk2, cdk1, CREB1, p300, Rb, PML, p107 and p130 of the pocket protein family. The array can also comprise an antibody specific for a protein selected from NFkB, cdc2A, mdm2, p21, p53, p65, Ki67 and CAFL.
[0032] The invention concerns the use of an antibody specific for human LIV21 for the diagnosis of cancer, and/or of five or more antibodies specific for a protein complex containing LIV21, for example antibodies specific for RBP2, E2F4, E2F2, E2F1, SUMO1, SUMO3, BRCA1, HDAC1, cycE/cdk2, cdk1, CREB1, p300, Rb, PML,erk,Bcl2,CD53,p107 and p130 of the pocket protein family. Preferably, the diagnosis is performed ex vivo on samples from a patient.
DESCRIPTION OF THE FIGURES
[0033] FIG. 1: antibody array.
[0034] FIG. 2: scheme of nuclear protein interactions and consequences on the study of therapeutics.
[0035] FIG. 3: LIV21 protein profile by mass spectrometry (Maldi) M (H+) for the one-dimensional gel band corresponding to the protein doublet migrating at 50 kD. The peptides derived from the digestion are solubilized in a solvent: acetonitrile/water (1/1) containing 0.1% of TFA (trifluoroacetic acid). A saturated solution of the alpha-cyano-4-hydroxycinnamic acid matrix is prepared in the same solvent. The same volume of the two solutions is taken and mixed together, and 1 microliter is deposited onto the Maldi plate for analysis. The spectrum was determined on a Voyager with Waters software. The calibration with respect to the autolysis and trypsin digestion peaks is not excellent and needs to be looked at again.
[0036] FIG. 4 is a zoomed-in profile of the chromatogram of the 49 kD band without smoothing.
[0037] FIGS. 5 & 5A is the second chromatogram corresponding to the one-dimensional 12% acrylamide gel band migrating at 53 kD (band 2) and revealed with coomassic blue and the LIV21 antibody. The spectrum was determined on a Brucker apparatus.
[0038] FIG. 6A is the third chromatogram corresponding to the one-dimensional acrylamide gel band migrating at 49 kD (band 3) and revealed with coomassic blue and the LIV21 antibody. The spectrum was also determined on a Brucker apparatus.
[0039] FIG. 6B is an example of a chromatogram.
[0040] FIG. 6C is another example of a chromatogram.
[0041] FIG. 7A-7BB is a table of the monoisotopic peaks with a value M H+. The masses are given with three numbers after the decimal point by the proteomic platforms since they estimate that this is the acquisition precision limit of MALDI TOF machines.
[0042] FIG. 8A represents a 2D SDS PAGE gel separating the twelve polypetides bound by the LIV21 antibody and
[0043] FIG. 8B represents the sample at 100° C. few minutes before migration.
[0044] FIGS. 9A-9C describes examples of Nano LC-ESI MS characterized peptides, including the peptides of the isoforms of LIV21. The table describes a NanoLC-ESI MS experiment. NanoLC makes it possible to separate the peptides derived from the trypsin hydrolysis of the protein. The eluted peptide fragments are ionized by electrospray and the ions formed are detected by mass spectrometry (Q-TOF analyzer). Each of these ions characterizes a peptide specific for the protein. Legend illustrated by the first page (FIG. 9A) describing the ion: molecular mass 523.25 daltons. Title: elution from 29.15 to 29.23 corresponds to the peptide eluted between 29.19 and 29.23 minutes. By ionization, this molecule gives the double-charge ion (charge=2+): MH2+ of molecular mass m/2: 523.25 daltons, hence the mass of the molecule M: 1044.51. The peptide sequence of this ion is determined by MSMS. The corresponding MSMS spectrum is defined by the column of numbers between Begin ions and End ions, which corresponds to an amino acid sequence (each amino acid having a specific mass). The first column with a 4-decimal number (daughter ion: 86.1373) corresponds to the mass of the "daughter" ions. The second column (I: 45) corresponds to the intensity of these "daughter" ions.
[0045] FIGS. 9A-9C describes the MSMS analyses giving a set of polypeptides that can be assigned to the LIV21 protein and its complex or contaminants according to the various observers of the various subcontracting proteomics platforms.
[0046] Sequences common with Gallus gallus, the histatin variant HIS3-HUMAN, the HSP60 chaperonin, arginine deiminase, nucleotidyltransferase, dehydrogenase.
[0047] FIGS. 10A & 10B: Similarity between examples of peptides sequences LIV21 comparison to E2F2.
[0048] FIG. 11.1A to 11.3C: Similarity between LIV21 and other genes as E2F2 and N deacety lase/N-sulfotransferase and TRAML 1.
[0049] FIGS. 12 & 12A: cartography:localization of LIV21 gene
[0050] FIG. 13A: alignments between the histatin-3-2 variant (by Brucker Maldi Tof analysis), PATF and Q7TCL4 (turnip mosaic virus) AAN08045.2.
[0051] FIG. 13B: describes alignments between Gallus gallus (gi 50732569) and PATF and common polypeptide sequence (20 amino acids) derived from LIV21 by MALDI TOF analysis.
[0052] FIG. 14 is the morphology of MCF7 cells treated or not treated with TPA at 25 nM.
[0053] FIGS. 15A-C are an analysis by FACS; representation, for each phase of the cell cycle, of the percentage of cells as a function of treatment time: FIG. 15A. S phase, FIG. 15B. G2/M phase,
[0054] FIG. 15C. GO/G1 phase. The scale along the x-axis is not proportional to the duration of treatment.
[0055] FIGS. 16A-B are a Western blot comparing total extracts (ET) and nuclear extracts (EN) and showing the inhibition, with TP A, of the expression of LIV21 phosphorylation. FIG. 16A, as a function of the time of treatment with TPA at 25 nM; FIG. 16B, compared with LIV21 in protein extracts, at 12 h of treatment with TPA at 25 nM.
[0056] FIG. 17 is a study of the nuclear translocation of LIV21 by immunocytochemistry with an anti-LIV21 primary antibody (in green) in cultures treated or not treated with TPA at 25 nM. The nuclei are stained red with propidium iodide. The nuclei are predominantly stained yellow at 12 H until 24 H since the anti-LIV21 primary antibody (in green) is nuclear, whereas it is predominantly cytoplasmic at 72 H (red nuclei and green cytoplasms).
[0057] FIG. 18 shows the expression, as a function of time of treatment with TPA at 25 nM, of PKCε and PKCζ proteins in total extracts. α-Tubulin expression serves as a control for the amount of total proteins loaded in the wells.
[0058] FIG. 19 shows the compared expression of PKCε and of LIV21 by immunocytochemistry on MCF-7 cell cultures treated or not treated with TPA at 25 nM for 12 h, carried out with anti-LIV21 and anti-PKCε antibodies in green, and propidium iodide staining the DNA red. The LIV21 is translocated into the nucleus by specific inhibition of PKCε. The PKCε is weakly expressed at 12 h in the presence of TPA. In fact, red nuclei and little green staining in the cytoplasms are observed. On the other hand, the expression of LIV21 is strong in the nuclei, which are stained yellow (merge) both with the anti-LIV21 antibody (green) and with the nucleus-specific propidium iodide.
[0059] FIG. 20 shows the effect of the PKCε-inhibiting peptide on the LIV21 expression profile by immunocytochemistry on cultures: control or treated with 1 μM of peptide, 2 μM of peptide, or 25 nM of TPA. The treatments last 12 hours. The cells are labeled with anti-LIV21 (in green), and with propidium iodide (in red). It is observed that 2 μM of peptide (image referred to as 2 μM) have the same effectiveness as "25 nM of TPA 12 H": the nuclei (yellow) are predominantly labeled both with propidium iodide and with the anti-LIV21 antibody (in green) on these two images, whereas the control and the cells treated with only 1 μM of PKC-inhibiting peptide show red staining of the nuclei, reflecting the absence of nuclear translocation of LIV21 through its anti-LIV21 antibody (in green).
[0060] FIG. 21 shows the effect of the PKCε-inhibiting peptide on the LIV21 expression profile in cytoplasmic (C) and nuclear (N) cellular fractions, after treatment for 12 h with TPA (25 nM) or with peptide (2 μM).
[0061] FIG. 22 shows, by immunoprecipitation (IP), the coexistence between PML/SUMO and LIV21: nuclear yellow fluorescence (merge) corresponding to the colocalization of PML/SUMO and LIV21 is observed in the cell nuclei.
[0062] FIGS. 23 and 24 show, by immunocytochemistry, the coexistence between PML/SUMO and LIV21. The colabeling (by fluorescent immunolabeling) of LIV21 (green) and of SUMO-1 (red) in the nuclei of the cells treated or not treated with TPA for 24 h and 72 h show that, at 24 h, LIV21 is translocated into the nucleus where SUMO and PML coexist (merge: yellow nuclei), whereas, at 72 h, LIV21 (green) is cytoplasmic. The nuclear bodies containing the SUMO1 protein are called PML bodies.
[0063] FIG. 25 shows that, using an array of 60 biopsies, including 50 of skin cancer and 9 of normal tissues and a control T; nuclear LIV21 expression is demonstrated in the biopsies of normal tissues and cytoplasmic LIV21 expression is demonstrated in the biopsies of metastatic cancers.
[0064] Image 43: poorly differentiated skin cancer T2N0M0, image 58: normal tissue derived from the same individual suffering from a poorly differentiated skin cancer (the nuclei of the cells are stained yellow), image 40: 10 cm metastatic carcinoma, and image 17: metastatic carcinoma of 3.5 cm. The cell nuclei are stained red.
[0065] FIG. 26 is, like FIG. 25, a second example of nuclear localization of LIV21 in the control and normal tissue (No. 52) (the cell nuclei are stained yellow), of the individual No. 7 suffering from a squamous cell carcinoma of the pharynx (moderately differentiated T4N0M0). In the images No. 7 of cancerous tissue, the cell nuclei are stained red.
[0066] FIG. 27 is a sample of advanced bladder cancer on cystectomy (grade III urethral carcinoma infiltrating the chorion and the musculosa) versus normal bladder tissue from the same patient with an internal control (PI): preimmune serum PI before the rabbit has been immunized against LIV21, red labeling of the nuclei with propidium iodide.
[0067] FIG. 28 is a sample of breast cancer at confocal microscopy with alexa and propidium coloration.
[0068] FIG. 29: Describes as FIGS. 5 and 6 the MALDI TOF analyses giving a set of polypeptide (the histatin-3-2 variant) that can be assigned to LIV21 and its complex and contaminants which are sometimes different according to the observers of the various subcontracting proteomics platforms. The Mascot search parameters are: trypsin enzyme, variable modifications; carbamethylation and oxidation of methionines, without molecular mass limit, without isoelectric point restriction. Type of mass: monoisotopic. Mass error (MS); according to the observer 50 ppm or 100 ppm. Non-cleavage with trypsin: 1 The masses captured are M (H+)/real masses. For chromatogram 1, the cysteins are blocked with iodoacetamide. The possibility of digestion with Promega bovine trypsin may be incomplete with cleavage oversight.
[0069] Sequences common with Gallus gallus (gi 50732569), mouse syntaxin, the histatin-3-2 variant (PI5516-00-01-00), ZN575-Human, G6 P translocase, the HSP60 chaperonin, arginine deiminase, ferredoxin-NADP(+) reductase, pseudomonas polyribonucleotidyl transferase, dehydrolipoamide dehydrogenase.
[0070] FIG. 30: alignments between Gallus gallus (gi 50732569) and PATF and common polypeptide sequence (18 amino acids) derived from LIV21 by MALDI TOF analysis.
[0071] FIG. 31A & 31B: example of a Maldi analysis interpretation diagram for histatin 3-2, giving sequence No. 5, selecting the masses: 2383.2610 (delta at 0.005); 2539.3290 (delta: -0.02); 2511.3740 (delta at 0.02), score: 78 and expect: 0.0046.
[0072] FIG. 32: PCR with the primers showing a band of 1400 bp.
[0073] FIG. 33: gel 2 with analysis of molecular masses.
[0074] FIG. 34: gel 3 at 55° and analysis of molecular masses.
[0075] FIG. 35: gel 4 at 45° and at 55° and analysis of molecular masses.
[0076] FIG. 36: screening ligation of 400 bp band, clones B1 to B10.
[0077] FIG. 37: screening ligation of 1400 bp band, clones C1 to C10.
[0078] FIG. 38: gel 5: ligation screening on the five new clones.
[0079] FIG. 39: gel 6: screening of the S55T and S55M recombinant clones and analysis of molecular masses.
[0080] FIGS. 40A-40B: examples of nucleotide sequence comparison between the sequenced Liv21 clones.
[0081] FIGS. 41A-41B: examples of nucleotide sequence comparison between the sequenced Liv21 clones.
[0082] FIG. 41: correspondence with sequences of Liv21 peptides: transduction of clones C8 and S55M1.
DETAILED DESCRIPTION OF THE INVENTION
[0083] The invention relates to the identification of antigens in cell lysates by immunoprecipiation. The analysis of the physical interaction of various proteins associated with E2F4 and E2F1 has been studied by coimmunoprecipitation of protein complexes. This analysis has made it possible to demonstrate a novel marker which has a diagnostic and prognostic use for cancer.
[0084] A marker for PATF proliferation associated with the E2F family had been demonstrated (Crisanti) through the characterization of exons, without the gene having ever been cloned in humans nor a corresponding protein having ever been found in humans.
[0085] The discovery of this novel molecule LIV21 could have a diagnostic value for the following reasons. By carrying out a screening of the localization of LIV21 in about ten human tumors, the inventor has been able to observe that, in all proliferating tumor cells, this protein is cytoplasmic instead of nuclear. It is not therefore in the correct cellular compartment to be active on the arrest of cell multiplication.
[0086] It has thus been possible to observe the presence of LIV21 in mammals. The panel of LIV21 expression as a function of cell state (mitotic cycles, cell in the resting state, differentiation) has been studied on tissues originating from various mammals. Protein analyses on the various tissue samples have confirmed that the expression of this transcription factor appears to be associated with a progression toward a quiescent cell state (arrest of mitoses and entry into differentiation). LIV21 is present in actively proliferating tumor cell lines and its expression is essentially cytoplasmic. The same results are obtained on human mammary adenocarcinomas.
[0087] Thus, the present invention relates to a novel test for screening for anomalies of the reinduction of the cell cycle. This diagnostic test is based on the study of the mechanism of action of the novel gene, encoding a potential novel transcription factor called LIV21, which down-regulate proliferation. LIV21 is implicated in the arrest of cell proliferation. LIV21 is cytoplasmic when the cells proliferate, whereas it becomes nuclear when the cells become quiescent. The characterization of this factor suggests a new pathway for down-regulating cell proliferation, by virtue of its association with one of the members of the EF family: E2F4. The latter is known to down-regulate the cell cycle by association with the P130 protein or pocket protein of the RB family.
[0088] The localization observed for LIV21 in tumor cells (cytoplasmic localization) and in physiological cells (nuclear localization) suggests, in any event, that its function is disturbed when cell development becomes anarchical.
[0089] The characterization of this molecule and the study of the timing and the topology of its expression also indicate that the expression and the localization of this ubiquitous transcription factor are regulated as a function of cell state: greater expression and nuclear localization for cells which have exited mitotic cycles, weak expression and cytoplasmic localization for actively proliferating cells such as human tumor cells.
[0090] LIV21 appears to be a key molecule for stabilizing another transcription factor (E2F4) in the cell nucleus and thus for inducing an arrest of cell proliferation may be to inhibiting E2F2 which active entrance of viral sequences in breast cells.
[0091] Furthermore, it has been shown that the localization of LIV21 in the cytoplasmic compartment is regulated by PKCε. In fact, when LIV21 is phosphorylated by PKCε, LIV21 is located in the cytoplasmic compartment. Conversely, when the phosphorylation of LIV21 by PKCε is inhibited, LIV21 is located in the nuclear compartment.
[0092] The LIV21 gene is a human gene on chromosome Ip31 near FABP3 (MDG1),SERINC2 and TINAGL (P3ESCL) between [031,783,000-031,111, 000] with the neighbours which revealed a structure function relation (means some of genes which enter in interaction with LIV21 and the complexe LIV21 are neighbours (FIG. 12). For example, E2F2, Bcl2, CD53, HDAC1 are in the same part of chromosome 1p.
LIV21
[0093] The present invention therefore concerns the LIV21 gene (FIG. 40.1), the LIV21 protein and derivatives and fragments thereof (FIG. 40.3).
[0094] The LIV21 protein is a human protein of approximately 300 amino acids. However, depending on the alternative splicing that it undergoes, it exists as at least three forms of different sizes and nine forms in hot conditions (FIG. 8). Moreover, it can be phosphorylated or sumoylated. It has an apparent molecular weight of between 50 kD and 51 kD in Western blotting analysis. This apparent molecular weight is 60 kD when LIV21 is sumoylated and in hot temperature conditions for sample (with the most concentration of sample), the profile change and we observed apparent molecular weight for peptides (revealed helix structures) in a range of 110 kD and 15 KD (FIG. 8.2). In its 51 kD form, which may be phosphorylated or nonphosphorylated, its isoelectric point is 5.6 and its intensity is 13632 but with hot temperature and nuclear samples separated to cytoplasmic samples there are spots at basic isoelectric point (FIG. 8). This protein has been characterized by mass spectrometry (Maldi) (Example 1; FIGS. 3-13). It gives more than 54 peptides following digestion with Promega trypsin (FIG. 7). The characteristics of the LIV21 protein are also described in FIGS. 3-13. Several specific peptides of LIV21 have been characterized, and in particular the LIV21a peptide, the LIV21b peptide (SEQ ID No 2), the LIV21c peptide (SEQ ID No 3), the LIV21d peptide and the LIV21e peptide (SEQ ID No 50). The longest sequence with homology with PATF (Cortunix Japonicus Bird) is the LIV21e peptide (SEQ ID No 50 & 51) and PL MII (FIG. 30).
[0095] Other specific peptides of LIV21 (Liv21a to f) are described below in the patent in listing in ASCII form.
[0096] For the purposes of the invention, a preferred LIV21 protein comprises at least one sequence chosen from SEQ ID Nos 1-55 or a sequence having 70%, 80% or preferably 90% homology with said sequence.
[0097] The isolation of LIV21 gene by RT PCR analysis (before to choose the oligonucleotides primers for amplification) begin by using the one dimensional and two dimensional gel electrophoresis analysis (FIGS. 33 and 34), the protein samples corresponding to the putative protein and to the elements of the complex were extracted from the gels and digested with trypsin (Promega) in order to be analysed by MALDI (FIGS. 3 to 5) and ESI MS/MS mass spectrometry. The results, when put up against proteomic databanks, made it possible to reveal several peptide sequences of interest, including some given as an example (FIGS. 6 and 7), some being found in humans with very significant scores (splicing of histatin, etc., FIGS. 30 and 31), these sequences were used as primers (once reverse-transcribed to cDNA) for screening a library formed from breast cancer-specific MCF7 cells (FIGS. 32 to 39).
[0098] The cloning made it possible to bring to the fore about twenty clones out of the 150 clones obtained, of which ten clones were sequenced and characterize the new gene LIV21 (FIG. 32). Based on these sequences, siRNAs were determined in order to allow regulation of silencing type within this metabolic complex of interest so as to develop therapeutic applications (SEQ N° 122).
[0099] In post-mitotic cells, apoptosis could correspond to an aborted attempt at mitosis. It is in this context that the application of LIV21 has been developed. The inventor has identified sequences of the LIV21 gene. Using the LIV21 antibody of affinity columns it has been able to extract peptides of the LIV21 protein; it has also used a second approach by means of a coimmunoprecipitation kit (Pierce) in order to have larger amounts of proteins (Example 1). Based on peptide sequences of the LIV21 protein, obtained by mass spectrometry (Example 2), primers which make it possible to amplify a cDNA fragment were designed (Example 3). After culturing and amplification of MCF7 line cells, extraction and purification of RNAs, RT PCRs and cloning in a shuttle vector were carried out, and then screening of the resistant colonies and sequencing made it possible to reveal sequences characterizing the LIV21 gene (Examples 4 and 5). More than twenty characteristic clones out of 150 clones were studied. The cDNA of these clones was used to screen a library prepared from the total mRNA of MCF7 cells. The sequence of this new product is a new transcription factor, the nuclear translocation of which is correlated with the establishment of cell quiescence. Furthermore, it forms heterodimers with other transcription factors and it binds to DNA.
[0100] The gene of this protein is characterized by five main sequences (cf. insert of clones extracted to MCF7 cells in sequences listing) and sequences representing an alternative splicing.
[0101] Using Northern blotting, the inventor then followed the expression of this new product during development, from the embryonic stage. It was observed that the amount of the LIV21 protein increases as development progresses, i.e. as a quiescent cell state becomes established. Through the same strategy, the inventor showed that the LIV21/E2F4 complex inhibited the expression of E2F1. This complex could correspond to a new point of control in the arrest of cell proliferation.
[0102] The LIV21 protein comprises a leucine zipper motif, a basic domains characteristic of DNA binding domains, a nuclearization sequence, zinc finger domain and helix loop helix with repetitive motifs.
[0103] The present invention concerns a purified or recombinant, isolated human polypeptide having a sequence comprising the sequence SEQ ID No 1 and/or SEQ ID No 2. Preferably, the polypeptide comprises the sequences SEQ ID Nos 1 and 2. In a preferred embodiment, the polypeptide comprises (in addition) a sequence selected from SEQ ID Nos 3-55, preferably from SEQ ID Nos 3-5, or a sequence having 70%, 80% or 90% identity to said sequences. In a specific embodiment, it comprises a sequence selected from one of the peptide sequences obtained by MALDI (FIG. 7) and NanoLC-ESI-MS (FIG. 9). The invention also concerns the two peptides LIV21a (SEQ ID No 1) and LIV21b (SEQ ID No 2). The invention also concerns a peptide having a sequence selected from SEQ ID Nos 3-55, preferably from SEQ ID Nos 3-5, or a sequence having 70%, 80% or 90% identity to said sequences. It also concerns peptides comprising at least 10 consecutive amino acids of human LIV21, preferably at least 20, 30 or 50 consecutive amino acids of LIV21.
[0104] The invention also concerns LIV21 derivatives of interest which are, for example, fusion proteins in which LIV21 is fused to labeled proteins such as GFP. Moreover, the LIV21 protein can be labeled by any means known to those skilled in the art.
[0105] The present invention also concerns an antibody which binds specifically to a polypeptide according to the present invention, preferably human LIV21, or a fragment or a derivative thereof. In a specific embodiment, the antibody binds specifically to an LIV21a or LIV21b peptide. In a preferred embodiment, the antibody binds specifically to a polypeptide comprising a sequence selected from SEQ ID Nos 1-55, preferably from SEQ ID Nos 1-5, or a sequence having 70%, 80% or 90% identity to said sequences.
[0106] The antibodies may be polyclonal or monoclonal. They may be antibody fragments and derivatives having substantially the same antigenic specificity, in particular antibody fragments (for example, Fab, Fab'2, CDRs), humanized antibodies, polyfunctional antibodies, single-chain antibodies (ScFv), etc. The antibodies of the invention can be produced using conventional methods, including the immunization of an animal and the recovery of its serum (polyclonal) or of spleen cells (so as to produce hybridomas by fusion with appropriate cell lines).
[0107] Said antibodies can be obtained directly from human serum or from serum of animals immunized with the proteins or the peptides according to the present invention. Methods for producing polyclonal antibodies from varied animal species including rodents (mice, rats, etc.), primates, horses, pigs, sheep, rabbits, poultry, etc., are described, for example, in Vaitukaitis et al. Vaitukaitis, Robbins et al. 1971). The antigen is combined with an adjuvant (for example, Freund's adjuvant) and administered to an animal, typically by subcutaneous injection. Repeated injections can be carried out. Blood samples (immune serum) are collected and the immunoglobolins are separated.
[0108] The present invention concerns an anti-LIV21 serum produced by immunizing an animal with a polypeptide according to the present invention. In a specific embodiment, the animal was immunized with the LIV21a and/or LIV21b peptide. In a preferred embodiment, the animal is immunized with these two peptides. The present invention also concerns an anti-LIV21 serum produced by immunizing an animal or a human with a polypeptide according to the present invention, in particular a polypeptide comprising a peptide sequence selected from SEQ ID Nos 1-55, preferably from SEQ ID Nos 1-5, or a sequence having 70%, 80% or 90% identity to said sequences. For example, the peptides can be coupled to a carrier (protein such as hemocyanin, and then injected into an animal, for example a rabbit, for immunization. Polyclonal antibodies were obtained using these two peptides by having immunized two rabbits and having bled one rabbit so as to have a preimmune serum.
[0109] Methods for producing monoclonal antibodies from various animal species can be found, for example, in Harlow et al. (Harlow 1988) or in Kohler et al. (Kohler and Milstein 1975). These methods include the immunization of an animal with an antigen, followed by the recovery of the spleen cells, which are subsequently fused with immortalized cells, such as myeloma cells. The resulting hybridomas produce monoclonal antibodies and can be selected by limiting dilution so as to isolate the individual clones. The antibodies can also be produced by selection from combinatorial libraries of immunoglobulins, such as those disclosed, for example, in Ward et al. (Ward, Gussow et al. 1989).
[0110] The invention also includes the use of the antibodies according to the invention for the detection and/or the purification of the human LIV21 protein. In particular, the LIV21 specific antibodies can be used for the detection of these proteins in a biological sample. They thus constitute a means of immunocytochemical or immunohistochemical analysis of LIV21 expression on tissue sections. Generally for such analyses, the antibodies used are labeled in order to be detectable. As an alternative, the antibodies can be indirectly labeled.
[0111] In a preferred embodiment, the antibodies are labeled. The labels include radiolabels, enzymes, fluorescent, luminescent or chemical labels, magnetic particles, gold labeling, biotin/avidin labeling, peroxidase labeling, etc.
[0112] The invention also includes a method for detecting the LIV21 protein in a biological sample, comprising a step of suitable treatment of the cells by any appropriate means which makes it possible to render the intracellular medium accessible, a step of bringing said intracellular medium thus obtained into contact with an antibody specific for the human LIV21 protein and a step of demonstrating the LIV21-antibody complex formed, by any appropriate means. In specific embodiments, the cytoplasmic and/or nuclear extracts are prepared, and these extracts are brought into contact with the antibody specific for the human LIV21 protein.
Diagnosis
[0113] The present invention teaches the development of the diagnostic test which also makes it possible to monitor the evolution of a cell proliferation. In particular, the present invention makes it possible to monitor the evolution of a cell proliferation on fresh cells or tissues, on frozen cells or tissues and on tissues processed, inter alia, with paraffin. The applications may be the diagnosis of cancer and also the monitoring of the evolution of a cell proliferation. Preferably, the cancer is selected from breast cancer, bladder cancer, ovarian cancer, lung cancer, skin cancer, prostate cancer, colon cancer, liver cancer, a sarcoma, a leukemia and glioblastoma, without being limited thereto.
[0114] Four of these properties can be used: its passage from the cytoplasmic cellular compartment to the nuclear compartment, the property of associating with the E2F4 transcription factor in order to form a complex which inhibits the expression of the E2F1 factor, and the ability of LIV21 to translocate in the nucleus through specific inhibition of PKCε, the sumoylation of LIV21 when the latter is nuclear and integrated into PML bodies and its interaction with HDAC.
[0115] The predominantly cytoplasmic state of this protein in cases of cancer, compared with its nuclear location in normal cells, is a geographical and structural difference which makes it possible, without the need for a fluorescent label, to differentiate spectral profiles of the functional pattern of cancerous tissue versus normal tissue, and thus to make the diagnosis.
[0116] These results show that the cytoplasmic localization of LIV21 is an indicator of the aggressiveness and of the metastatic potential of the cancer. The detection of the LIV21 expression indicates the presence of cancer cells, more particularly of invasive, aggressive and/or metastatic cancer cells. These results also show that the nuclear localization of LIV21 is an indicator of normal quiescent cells or of well-differentiated tissues.
[0117] The invention concerns, moreover, methods for the diagnosis or prognosis of cancer which implement the detection of the cytoplasmic localization of a transcription factor located in the nucleus in normal cells.
[0118] The present invention concerns a method for the detection of cancer cells in a biological sample from a patient, comprising the detection of the product of expression of the LIV21 gene in the nucleus and/or the cytoplasm of the cells in the biological sample from said patient, localization of said product of expression of the LIV21 gene in the cytoplasm being indicative of the presence of cancer cells and localization of said product of expression of the LIV21 gene in the nucleus being indicative of the presence of noncancer cells. Preferably, localization of said product of expression of the LIV21 gene in the cytoplasm is indicative of the presence of invasive and/or metastatic cancer cells. The method preferably comprises a prior step of suitable treatment of the cells contained in the sample by any appropriate means which makes it possible to render the intracellular medium accessible. The method optionally comprises a step of comparison with a biological sample which does not contain cancer cells.
[0119] Optionally, the method according to the invention also comprises the detection of the product of expression of at least one gene selected from the group consisting of the protein kinase C epsilon (PKCε) gene, the E2F1 gene and the E2F4 gene. The method can in particular comprise the detection of the product of expression of two of these genes or of the three genes. Moreover, at least one of the ratios LIV21/PKCε, LIV21/E2F4 and LIV21/E2F1 can be determined in the present method. This ratio can be determined in the cytoplasm and/or in the nucleus. Preferably, these ratios are determined in the nucleus. Preferably, these ratios are compared with those obtained in a normal cell.
[0120] The method can also comprise the detection of the product of expression of at least five genes selected from the group consisting of RBP2, E2F4, E2F1, SUMO1, SUMO3, HDAC1, cycE/cdk2, cdk1, CREB1, p300, Rb, PML, p107 and p130 of the pocket protein family. It can also comprise the detection of the product of expression of at least five genes selected from the group consisting of NFkB, cdc2A, mdm2, p21, p53, p65, Ki67,erk,CF53 and CAF1. The method can comprise the detection of an interaction between some of these proteins and/or the detection of a posttranslational modification of one of these proteins.
[0121] The method may in particular comprise the detection of the product of the expression of two of these genes or of the three genes. Moreover, at least one of the ratios LIV21/PKCε, LIV21/E2F4 and LIV21/E2F1 may be determined in the present method. This ratio can be determined in the cytoplasm and/or in the nucleus. Preferably, these ratios are determined in the nucleus. Preferably, these ratios are compared with those obtained in a normal cell.
[0122] In one embodiment, the expression product of the genes is detected at the mRNA level, it being possible for the mRNA to be detected by any means known to those skilled in the art.
[0123] Thus, the method according to the present invention also relates to the detection of a polynucleotide encoding the human LIV21 protein or a fragment thereof, for example LIV21a and/or LIV21b. The polynucleotide encoding LIV21 may be an mRNA, a cDNA or a genomic DNA. The polynucleotides may be isolated from cells of the biological sample. They may also be obtained by a polymerase chain reaction (PCR) carried out on the total DNA of the cells or else by RT PCR carried out on the total RNA of the cells or polyA RNAs.
[0124] The mRNA may be detected by an RT PCR analysis. For this, the method uses a pair of primers specific for the expression product to be detected, in particular LIV21, PKCε, E2F1 or E2F4. The term "specific pair of primers" is intended to mean that at least one of the primers is specific for the expression product to be detected, i.e. that this pair of primers makes it possible to specifically amplify a fragment of the desired mRNA. Preferably, the RT PCR analysis is carried out on nuclear and/or cytoplasmic extracts of the cells contained in the sample from the patient. Optionally, the RT PCR analysis may be a quantitative analysis. A pair of primers specific for LIV21 can be prepared on the basis of the teachings of the present application. For example, the pair of primers may comprise the primers described in the sequences listing.
[0125] The pairs of primers specific for PKCε, E2F1 and E2F4 are well known by those skilled in the art (Caroll JS 2000; Mundle SD 2003; Stevaux O 2002; Cheng T 2002; Opalka B 2002).
[0126] The mRNA may also be detected by Northern blotting analysis. For this, the method uses a probe specific for the expression product to be detected, in particular LIV21, PKCε, E2F1 or E2F4. A probe specific for LIV21 can be prepared on the basis of the teachings of the present application. An example of a specific probe comprises the sequence SEQ ID No 50. Preferably, the Northern blotting analysis is carried out on nuclear and/or cytoplasmic extracts of the cells contained in the sample from the patient. The nucleic probe is labelled. The oligonucleotide labelling technique is well known to those skilled in the art. The labelling of the probes according to the invention can be carried out with radioactive elements or with non radioactive molecules. Among the radioactive isotopes used, mention may be made of 32P, 33P or 3H. The non radioactive entities are selected from ligands such as biotin, avidin, streptavidin or digoxigenin, haptens, dyes and luminescent agents such as radioluminescent, chemoluminescent, bioluminescent, fluorescent or phosphorescent agents. The probes specific for PKCε, E2F1 and E2F4 are well known to those skilled in the art.
[0127] In a preferred embodiment, the expression product of the genes is detected at the protein level. Preferably, the protein is detected using a specific antibody. Thus, the method comprises a step of bringing the cells of the biological sample into contact with an anti-human LIV21 antibody. The antibodies may be monoclonal or polyclonal. The anti-LIV21 antibody can, for example, be an anti-LIV21 serum.
[0128] When the product of expression of one of the genes PKCε, E2F1 and E2F4 must be detected, the method can use antibodies specific for the PKCε, E2F1 and E2F4 proteins, respectively. Polyclonal and monoclonal antibodies directed against PKCε, E2F1 and E2F4 are commercially available. By way of example, mention may be made of, for PKCε, a rabbit polyclonal antibody (Santa Cruz Technology, sc-214), for E2F1, a rabbit polyclonal antibody (Santa Cruz Technology, sc-860), and for E2F4, a rabbit polyclonal antibody (Santa Cruz Technology, sc-866). Preferably, the antibodies are labeled, directly or by means of a secondary antibody. The antibody labeling techniques are well known to those skilled in the art.
[0129] In a specific embodiment, the protein can be detected by Western blotting analysis. The Western blotting analysis can be carried out on nuclear and/or cytoplasmic extracts of the cells contained in the sample from the patient. Briefly, the proteins are migrated in a gel and then blotted onto a membrane. This membrane is then incubated in the presence of the antibodies and the binding of the antibodies is optionally revealed using labeled secondary antibodies.
[0130] In another embodiment, the protein is detected by immunohistochemistry, immunocytochemistry or immunoradiography. These techniques are well known to those skilled in the art. The immunocytochemical analysis can be carried out on whole cells originating from the sample or which are derived therefrom, for example by cell culture. It can also be carried out on isolated nuclei. The immunohistochemical analysis can be carried out on mammary tissue sections.
[0131] By way of illustration, an immunocytochemical analysis can include the following steps. However, it is understood that other preparatory methods can be carried out. Cells originating from the biological sample are cultured, preferably on slides (Lab Tek, Nunc, Germany), and then washed with buffer and fixed with paraformaldehyde (for example, 4%). A saturation step is preferably carried out by incubating the cells with buffer S (PBS-0.1% Triton X100-10% FCS). The cells are then incubated with a primary antibody and are then washed an incubated with a fluorescent secondary antibody, if necessary. The nuclei can be labeled with propidium iodide (Sigma). The slides are mounted in moviol for observation by fluorescence microscopy. Moreover, isolated nuclei sampled during a nuclear extraction can be fixed with paraformaldehyde (for example, 4%). The suspensions of nuclei are deposited between a slide and cover slip and the observation is carried out by fluorescence microscopy and by confocal microscopy. The primary antibodies are, for example, rabbit antibodies and the secondary antibodies are labeled antibodies directed against rabbit IgGs.
[0132] The present invention also concerns the use of a protein array for detecting the expression of one or more of these proteins, and/or an interaction between two or more of these proteins, and/or the posttranslational modification of one or more of these proteins.
[0133] In a preferred embodiment, the detection of the product of expression of one or more genes or of the interaction between several proteins is carried out by means of a protein array. Thus, a polypeptide according to the present invention, in particular LIV21 or a fragment thereof, or an antibody specific thereto or a fragment or a derivative thereof which conserves the binding specificity, can advantageously be immobilized on a support, preferably a protein array. Such a protein array is included in the invention. This array can also contain at least one polypeptide selected from the group consisting of protein kinase C epsilon (PKCε), RBP2, E2F4, E2F1, SUMO, HDAC1, cycE/cdk2, cdk1, CREB1, p300, Rb, PML, p107 and p130 of the pocket protein family or at least one antibody specific for one of these polypeptides, or a fragment or a derivative thereof which conserves the binding specificity. The array can also comprise other polypeptides well known to those skilled in the art to be advantageous for the detection and/or the prognosis of a cancer, or antibodies specific for said polypeptides. These polypeptides can, for example, be selected from the following list: NFkB, cdc2A, mdm2, p21, p53, p65, Ki67 and CAF1.
[0134] The protein arrays according to the present invention can be prepared according to the techniques well known to those skilled in the art. In practice, it is possible to synthesize the attached polypeptides directly on the protein array, or it is possible to perform an ex situ synthesis followed by a step of attachment of the synthesized polypeptide to said array. Moreover, the polypeptides or antibodies to be attached can be purified from a cell. The supports include smooth supports (for example, metal, glass, plastic, silicon, and ceramic surfaces) and also texturized and porous materials. Such supports also include, but are not limited to, gels, rubbers, polymers and other flexible materials. The supports do no need to be flat. The proteins or antibodies of the array can be attached directly to the support or can be attached by means of a spacer or a linker.
[0135] In a specific embodiment, an LIV21-specific antibody or a fragment or derivative thereof which conserves the binding specificity is immobilized on the solid support. Thus, this array provides a practical means for measuring the LIV21 expression product. Preferably, the array comprises at least one antibody specific for a polypeptide selected from the group consisting of PKCε, RBP2, E2F4, E2F1, SUMO, HDAC1, cycE/cdk2, cdk1, CREB1, p300, Rb, PML, p107 and p130 of the pocket protein family, preferably PKCε, E2F1 and E2F4. The array also comprises at least one antibody specific for a polypeptide known to those skilled in the art to be advantageous for the detection and/or the prognosis of a cancer, for example NFkB, cdc2A, mdm2, p21, p53, p65, Ki67 and CAF1. The array can comprise an antibody or a fragment or derivative thereof which has the same specificity.
[0136] The protein arrays according to the invention are also extremely useful for experiments in proteomics, which studies the interactions between the various proteins. In a simplified manner, peptides representative of the various proteins are attached to a support. Said support is then brought into contact with labeled proteins and, after an optional rinsing step, interactions between said labeled proteins and the peptides attached to the protein array are detected.
[0137] "Protein array" is intended to denote a support to which are attached polypeptides or antibodies, it being possible for each of them to be pinpointed by its geographical location. These arrays differ mainly in terms of their size, the material of the support and, optionally, the number of polypeptides which are attached thereto.
[0138] The protein arrays can also be useful for the screening of test compounds.
[0139] The present invention also relates to a method for the detection of cancer cells in a biological sample from a patient, comprising the detection of the product of expression of the LIV21 gene in the nucleus and/or the cytoplasm of the cells in a sample of cells in the biological sample from said patient, which method is characterized in that it comprises at least: (a) bringing said biological sample into contact with a protein array as defined above, and (b) revealing, by any appropriate means, antigen-antibody complexes formed in (a), for example, by EIA, ELISA or RIA or by immunofluorescence. Other detection methods are described in detail in the following document: U.S. 2004152212.
[0140] Methods applicable for the synthesis of protein arrays are described, for example, in the following patients:
[0141] WO2004/063719, WO2005/016515, U.S. 2005019828, WO03018773, U.S. 2002187464, U.S. Pat. No. 5,143,854, U.S. Pat. No. 5,242,974, U.S. Pat. No. 5,252,743, U.S. Pat. No. 5,324,633, U.S. Pat. No. 5,384,261, U.S. 2006035387, U.S. 2005100947, U.S. 2005233473, WO0198458, WO0172458, WO0004382, WO0004389, WO9015070, WO9210092, WO9310161, WO9512808 and WO9601836, the content of these patents being incorporated into the present application by way of reference. For example, these protein arrays can be fabricated according to conventional methods described (Lubman David M, QIAO TIECHENG Alex, Mathew ABY J etc.) or novel tools for the automation of hybridization and of reading, U.S. 2004152212 and Yu Xinglong U.S. 2005019828 and novel supports which attach polypeptides, Claus Peter Klages et al. (example FIG. 2).
[0142] The biological samples originate from a patient potentially suffering from cancer or for whom it has been established that said patient is suffering from cancer. "Biological sample" is intended in particular to mean a sample of the biological fluid, living tissue, tissue fragment, mucosity, organ or organ fragment type, or any culture supernatant obtained by means of taking a sample. The method according to the present invention can comprise a step of taking a biological sample from the patient. The detection step can be carried out directly on a tissue section of the sample, or on a culture of cells originating from the sample, or on total cell extracts, nuclear extracts and/or cytoplasmic extracts. The sample from the patient can come from a puncture, a biopsy, ground cellular material, a bronchial aspiration, a blood sample or a urine sample.
[0143] In a specific embodiment of the method comprising the detection of the product of expression of the PKCε gene, a significant increase in PKCε is indicative of the presence of cancer cells. More specifically, the amount of PKCε in normal cells is compared with the amount of PKCε in the cells of the sample, and the significant increase is determined by means of this comparison. The method according to the present invention can optionally comprise the measurement of the LIV21/PKCε content. This LIV21/PKCε ratio increases in the cytoplasmic fraction of cancer cells compared with normal cells.
[0144] In another specific embodiment of the method comprising the detection of the product of expression of the E2F4 gene, the method comprises the detection of the association of LIV21 with the E2F4 protein, and a decrease in this association is indicative of the presence of cancer cells. The detection of the association of LIV21 with the E2F4 protein can be carried out by concurrent detection of LIV21 and of E2F4 and/or by the concurrent measurement of HDAC1. The method according to the present invention can optionally comprise the measurement of the E2F4/LIV21 content. This E2F4/LIV21 ratio decreases in the nucleus of cancer cells compared with normal cells.
[0145] In an additional embodiment of the method comprising the detection of the product of expression of the E2F1 gene, the presence of the E2F1 protein is the nucleus is indicative of the presence of cancer cells. The method according to the present invention can optionally comprise the measurement of the E2F1/LIV21 content. This E2F1/LIV21 ratio increases in the nuclear fraction of cancer cells compared with normal cells.
[0146] The method according to the present invention allows in particular the detection of metastasized cancer, therapeutic monitoring and/or recurrences following treatment and makes it possible to determine the degree of invasiveness of a cancer. The specificity of the detection can be related to the crossing over of information obtained through the existence and the topography of LIV21 by all imaging and spectroscopy means and obtained by combination with other known cancerological indicators via protein arrays or microarrays. Thus, the detection based on LIV21 can be combined with the detection of other cancer markers, in particular breast cancer markers, known to those skilled in the art.
[0147] In fact, the present invention concerns a method for the therapeutic monitoring of an anticancer treatment in a patient suffering from cancer, comprising the administration of the anticancer treatment to said patient and the detection of cancer cells in a biological sample from the patient, according to the method of the present invention. A decrease in cancer cells will be indicative of the effectiveness of the treatment. The detection of cancer cells in a biological sample from the patient, according to the method of the present invention, can be carried out once or several times over the course of the anticancer treatment or after the anticancer treatment. Preferably, the biological sample originates from the tissue affected by the cancer treated.
[0148] Moreover, the present invention also concerns a method for the detection of recurrences subsequent to an anticancer treatment of a cancer in a patient, comprising the detection of cancer cells in a biological sample from the patient, according to the method of the present invention. The detection of cancer cells in a biological sample from the patient, according to the method of the present invention, can be carried out once or several times after the anticancer treatment. The detection of cancer cells is indicative of recurrences. Preferably, the biological sample originates from the tissue affected by the cancer treated.
[0149] The present invention also describes a kit for carrying out a method according to the invention. More particularly, the invention concerns a kit for the detection of cancer cells in a biological sample from a patient, comprising one or more elements selected from the group consisting of an antibody which binds specifically to human LIV21 according to the present invention and an anti-LIV21 serum according to the present invention, an oligonucleotide probe specific for the LIV21 mRNA and a pair of primers specific for the LIV21 mRNA. In a preferred embodiment, the kit comprises antibodies which bind specifically to human LIV21. In another preferred embodiment, the kit comprises an oligonucleotide probe specific for the LIV21 mRNA. It may also comprise a probe specific for a "housekeeping" gene. The kit according to the present invention can comprise reagents for the detection of an LIV21-antibody complex produced during an immunoreaction. Optionally, the kit according to the present invention also comprises means for detecting the product of expression of at least one gene selected from the group consisting of the protein kinase C epsilon (PKCε) gene, the E2F1 gene and the E2F4 gene. This detection means can be antibodies specific for the protein, oligonucleotide probes specific for the mRNA concerned and/or a pair of primers specific for the mRNA.
[0150] The present invention also relates to a diagnostic composition comprising one or more elements selected from the group consisting of an antibody according to the present invention and a serum according to the present invention, an oligonucleotide probe specific for the LIV21 mRNA and a pair of primers specific for the LIV21 mRNA.
[0151] The present invention also concerns a diagnostic composition comprising one or more elements selected from the group consisting of an antibody according to the present invention and a serum according to the present invention.
Anticancer Therapy
[0152] In the context of an anticancer therapy, it is possible to envision increasing the amount of LIV21 present in the nucleus. For this, the nuclear localization of LIV21 could be promoted, for example by decreasing the activity of PKCε in the cancer cells and by using HDAC inhibitors.
[0153] In another specific embodiment of anticancer therapy, it is possible to envision decreasing the activity of PKCε in the cancer cells. This decrease in activity can be produced by decreasing the activity of the PKCε protein or by decreasing its expression. A decrease in the activity of the PKCε protein can be obtained by administering PKCε-protein inhibitors to the cancer cells. The PKCε-protein inhibitors are well known to those skilled in the art. A decrease in the expression of the PKCε protein can be obtained by using antisenses or siRNA specific for the PKCε gene. Kits are commercially available. Moreover, the techniques concerning inhibition by means of antisense or siRNA are well known to those skilled in the art (Arya R 2004, Lee W 2004, Sen A 2004, Platet N 1998, Hughes 1987).
[0154] The present invention therefore concerns a pharmaceutical composition comprising a PKCε-protein inhibitor. It also concerns the use of a pharmaceutical composition comprising a PKCε-protein inhibitor as a medicament, in particular for the preparation of a medicament for use in treating cancer. Finally, it concerns a method for treating cancer in a patient, comprising the administration to the cancer cells of a PKCε-protein inhibitor, the pKCε-protein inhibitor making it possible to reduce or abolish the cancerous phenotype of the treated cells. In a first embodiment, the PKCε-protein inhibitor decreases the activity of the PKCε protein. In a second embodiment, the PKCε-protein inhibitor decreases the expression of the PKCε protein. Preferably, cancer is selected from breast cancer, bladder cancer, ovarian cancer, lung cancer, skin cancer, prostate cancer, colon cancer, liver cancer, a sarcoma, a leukaemia and glioblastoma, without being limited thereto.
[0155] In the context of a therapy for a neurodegenerative disease, it is possible to envision decreasing the amount of LIV21 present in the nucleus of the cells affected by the neurodegenerative disease. The cells affected by the neurodegenerative disease are generally neurons, motor neurons, etc. In a preferred embodiment, the neurodegenerative disease is chosen from Alzheimer's disease, Huntington's disease, Parkinson's disease and amyotrophic lateral sclerosis (ALS). For this, the nuclear localization of LIV21 could also be hindered, for example by increasing the activity of PKCε in the cells affected by the neurodegenerative disease.
[0156] The inhibition or the blocking of LIV21 expression can be carried out by any means known to those skilled in the art. In particular, by way of illustration, mention may be made of the antisense strategy, siRNA and ribozymes. Thus, an antisense oligonucleotide or an expression vector encoding this antisense oligonucleotide could be prepared and used to block the translation of the mRNA encoding LIV21 in-vivo. Moreover, a ribozyme can be prepared for cleaving and destroying, in vivo, the mRNA encoding LIV21. It is also possible to envisage a triple-helix strategy in which an oligonucleotide is designed so as to hybridize with the gene encoding LIV21 and to thus block the transcription of this gene.
[0157] Moreover, the nuclear localization of LIV21 could also be made unfavourable, for example by increasing the activity of PKCε in the cells affected by the neurodegenerative disease. We observed a strong similarity of LIV21 with ADC7 neuronal thread protein (O60448 HUMAN) which is knew to be implicated in Alzheimer disease and find in the cerebrospinal fluid. So regulate an interaction between these two proteins with therapeutic peptides or siRNA is interesting for the treatment of Alzheimer disease.
[0158] In another specific method of therapy against a neurodegenerative disease, it is possible to envisage increasing the activity of PKCε in the cells affected by the neurodegenerative disease and permits apoptosis. The cells affected by the neurodegenerative disease are generally neurons, motorneurons, etc. In a preferred embodiment, the neurodegenerative disease is chosen from Alzheimer's disease, hungtington's disease, parkinson's disease and amyotrophic lateral sclerosis (ALS). For this, the nuclear localization of Liv21 could also be hindered, for example by increasing the activity of PKCε in the cells affected by the neurodegenerative disease.
[0159] This increase in activity can be produced by increasing the activity of the PKCε protein or by increasing its expression. An increase in the activity of the PKCε protein can be obtained by administering PKCε-protein activators to the cells affected by the neurodegenerative disease. The PKCε-protein activators are well known to those skilled in the art (Toma O (2004). Activation of PKCs by DAG, AGPI: oleic acid, linoleic acid, arachidonic acid, etc. Activation and proteolysis of PKCs in gonadotropic cells; Communication 2004 by Macciano H, Junoy B, Mas JL, Drouva SV, UMR6544 Marseille). An increase in the expression of the PKCε protein can be obtained by using expression vectors encoding the PKCε protein and which make it possible to overexpress it in the cells affected by the neurodegenerative disease.
[0160] Thus, the present invention concerns a pharmaceutical composition comprising a PKCε-protein activator or an expression vector encoding the PKCε protein. It also concerns the use of a PKCε-protein activator or of an expression vector encoding the PKCε protein, for the preparation of a medicament for use in the treatment of a neurodegenerative disease.
Screening Method
[0161] The invention concerns methods for the selection, identification, characterization or optimization of active compounds which decrease cell proliferation, based on the measurement of the nuclear versus cytoplasmic localization of LIV21, or of the binding of the LIV21 protein to the E2F4 protein.
[0162] In a first embodiment, the selection, the identification, the characterization or the optimization of active compounds of therapeutic interest comprises bringing a candidate compound into contact with a cell and determining the nuclear versus cytoplasmic localization of the LIV21 expression product. An increase in the nuclear localization of LIV21 indicates that the candidate compound is active in terms of decreasing or abolishing cell proliferation. A decrease in the nuclear localization of LIV21 indicates that the candidate compound is active in terms of treating or preventing a neurodegenerative disease.
[0163] In a second embodiment, the selection, the identification, the characterization or the optimization of active compounds of therapeutic interest comprises bringing a candidate compound into contact with a cell and determining the level of expression of the gene encoding the PKCε protein. A decrease in the expression of PKCε indicates that the candidate compound is active in terms of decreasing or abolishing cell proliferation. An increase in the expression of PKCε indicates that the candidate compound is active in terms of treating or preventing a neurodegenerative disease.
[0164] In a third embodiment, the selection, the identification, the characterization or the optimization of active compounds of therapeutic interest comprises bringing a candidate compound into contact with a cell and determining the level of LIV21/E2F4 complex. An increase in the level of LIV21/E2F4 complex indicates that the candidate compound is active in terms of decreasing or abolishing cell proliferation. A decrease in the level of LIV21/E2F4 complex indicates that the candidate compound is active in terms of treating or preventing a neurodegenerative disease.
[0165] In a fourth embodiment, the selection, the identification, the characterization or the optimization of active compounds of therapeutic interest comprises bringing a candidate compound into contact with a cell and determining the level of expression of the gene encoding the E2F1 protein. A decrease in the expression of E2F1 indicates that the candidate compound is active in terms of decreasing or abolishing cell proliferation. An increase in the expression of E2F1 indicates that the candidate compound is active in terms of treating or preventing a neurodegenerative disease.
[0166] The invention also relates to a method of screening for a compound capable of interacting in vitro, directly or indirectly, with LIV21, characterized in that; in a first step, the candidate compound and LIV21 are brought into contact and, in a second step, the complex formed between said candidate compound and LIV21 is detected by any appropriate means.
[0167] The present invention also relates to a method of screening for a compound capable of modulating (activating or inhibiting) the activity of the LIV21 protein, characterized in that; in a first step, cells of a biological sample expressing the LIV21 protein are brought into contact with a candidate compound, in a second step, the effect of said candidate compound on the activity of said LIV21 protein is measured by any appropriate means, and in a third step, candidate compounds capable of modulating said activity are selected. The activity of LIV21 can, for example, be estimated by means of evaluating the ability of the cell to divide, by measuring the expression of the E2F1 gene or by the cytoplasmic and/or nuclear localization of LIV21.
[0168] The candidate compound can be a protein, a peptide, a nucleic acid (DNA or RNA), a lipid, or an organic or inorganic compound. In particular, the candidate compound could be an antibody, an antisense, a ribozyme or an siRNA.
[0169] Other advantages and characteristics of the invention will appear in the examples and the figures which follow, and which are given in a nonlimiting manner.
EXAMPLES
Example 1
[0170] The inventor performed mass spectrometry (MALDI) for the LIV21 protein based on a one-dimensional acrylamide gel. The LIV21 protein was digested with trypsin. The peptides derived from the digestion are solubilized in a solvent: acetonitrile/water (1/1) containing 0.1% of TFA (trifluoroacetic acid). A saturated solution of the alpha-cyano-4-hydroxycinnamic matrix was prepared in the same solvent. The same volume of the two solutions was taken and mixed together and 1 μl was deposited onto the MALDI plate for analysis. The mass spectrometry showed that the LIV21 protein digested with trypsin reveals 54 peptides (cf. FIGS. 3-4). The LIV21 protein was characterized by a molecular weight of 50 kD, revealed by Western blotting. Since the first MALDI results were not probative, the inventor produced a two-dimensional SDS PAGE gel (FIG. 8). More than ten proteins were revealed by silver nitrate staining, but the very small amount of material did not make it possible to test samples derived from this gel by MALDI or MSMS.
[0171] When it changes cell compartment and when it is sumoylated, the LIV21 protein has a molecular weight of approximately 60 kD. When it is phosphorylated in the cytoplasm, it exhibits two forms which differ by a few kilobases. A doublet is then observed.
[0172] The inventor performed a third MALDI analysis, which gave interesting results, especially with regard to the 49 kD gel band, on Gallus gallus and a histatin variant (FIG. 13), the inventor then examined the sequence alignments, which made it possible to confirm homologies between Gallus gallus, PATF, Q7TCL4 and the polypeptides of histatin and of Gallus gallus (FIGS. 29 to 31.2).
Only two peptides characterize in common PATF and LIV21:
[0173] The LIV21a peptide is located between a site of interaction with the Rb/p107/p130 protein (ITCCE) and a site of sumoylation by SUMO1. The sequence of this peptide is the following:
[0174] PeptideLIV21a peptide is SEQ ID No 1
[0175] The LIV21b peptide is located between a sumoylation site (PKPG) and a phospholipase C site (YVKI) followed by (KKKRK) NLS. The sequence of this peptide is SEQ ID No 2
[0176] The LIV21c peptide is SEQ ID No 3
[0177] The LIV21d peptide is SEQ ID No 4 are specific of LIV21.
[0178] Other peptides are provided in the sequences SEQ ID Nos 5-55.
Example 2: Study of the Nuclear Translocation of LIV21 in MCF-7 Cells
[0179] The study of the subcellular distribution of LIV21 in different tumor lines of various origins showed an exclusively cytoplasmic localization of this protein. The mechanism(s) by which LIV21 could be translocated into the nucleus in order to act on the cell cycle was (were) shown.
[0180] The presence of putative sites for phosphorylation by protein kinases C (PKCs), based on the possible homology that PATF is thought to have with the LIV21 sequence, directed the inventor's study toward a possible involvement of these proteins with respect to its nuclear translocation. The inventor therefore chose to study the MCF-7 line treated with TPA, which is known to modulate PKCs.
[0181] In parallel to this work, the inventor also studied the expression and the localization of cell cycle proteins implicated in the signaling pathway in which LIV21 could act.
The MCF-7 Cell Line
[0182] The MCF-7 line is a nonclonal human line of breast adenocarcinoma cells. During their differentiation induced by exogenous factors, these cells develop a hypertrophy, membrane protrusions and a tendency to dissociate from one another. They acquire a secretory phenotype which is characterized by the appearance of numerous granules and of secretory canaliculi.
[0183] In vivo, these cells are relatively nonmetastatic and this low invasiveness is thought to be due to a low constitutive activity of the protein kinases C (PKCs) and to a relatively low level of expression of protein kinase C alpha.
[0184] This line is used in many studies on proliferation, differentiation and apoptosis. These studies use appropriate drugs, such as TNF for the induction of apoptosis, or TPA (12-O-tetradecanoyl phorbol-13-SUMOate) for the induction of differentiation and therefore for the study of departure from the cell cycle.
The Effect of TPA on the MCF-7 Line
[0185] TPA is a known activator of PKCs. It activates the growth of normal breast cells, does not modify the proliferation of the cells of benign tumors from this same tissue, but drastically inhibits the proliferation of the cells of human mammary tumor lines such as the MCF-7 line. It reduces the cell growth of this line by positively controlling the c-erb-2 receptor and negatively controlling the retinoic acid receptor α, which are both expressed in particulary large amount in these cells. TPA greatly and rapidly inhibits the expression and the function of estrogen receptors (ERs) and it induces the time- and dose-dependent translocation of protein kinases C (PKCs) from the cytosol to the membranes.
[0186] Furthermore, TPA increases the migratory capacity of MCF-7 cells in vitro and a short period of treatment of these cells with TPA induces cellular expansion and microtubule organization characteristic of their differentiation.
Expression of LIV21 in MCF-7 Cells
[0187] Firstly, the inventor verified the expression of LIV21 in these cells at the protein level.
[0188] The inventor tackled the study of the expression of the LIV21 protein through the Western blotting technique, with an anti-LIV21 antibody, in MCF-7 cells compared with mammary tissues. The anti-LIV21 antibodies were obtained by the method described below. In this line, LIV21 is expressed, both in the mammary tissues and in the MCF-7 cells, in the form of a doublet which migrates at an apparent molecular weight of 50 kDa.
Production of Purified Anti-LIV21 Serum
[0189] The specific peptide sequences are the sequences No. 1 and No. 2.
[0190] These peptides were injected into rabbits (NZ W ESD 75 female, 2.3 kg at day 0), in agreement with standard immunization procedures, such as:
TABLE-US-00001 Day STEPS OF THE PROCEDURE Day 0 Collection of a control serum (20 ml) First intradermal injection (1 ml/rabbit) 1 tube for two rabbits with 1 ml of antigen + 1 ml Freund's Complete Day 14 Second intramuscular injection (1 ml per rabbit) 1 tube for two rabbits with 1 ml of antigen + 1 ml Freund's Incomplete Day 28 Third intramuscular injection (idem D14) Day 39 The test serum is collected (5 ml) and conserved at 4° C. (serum D39) Day 49 Four subcutaneous injections (1 ml) 1 tube for two rabbits with 1 ml of ag + 1 ml Freund's Incomplete Day 60 Test serum collected (25-30 ml) and storage at 4° (serum D60) Day 77 Fifth intradermal injection (1 ml/rabbit) idem D49 Day 88 Test serum collected (5 ml) and storage at 4° (serum D88) Day 99 Test serum collected (25 ml) and storage at 4° (serum D98) Day 102 Test serum collected (25 ml) and storage at 4° (serum D102) Day 104 Test serum collected (25 ml) and storage at 4° (serum D104) Day 106 Test serum collected (25 ml) and storage at 4° (serum D106) Day 109 Total collected by total collection of blood and storage at 4° C. (serum D109)
[0191] The reactivity of the serum obtained was tested with respect to binding of the peptide sequences No. 1 and 2.
[0192] At D60, the serum shows a good reactivity with each sequence. The serum produced did not bind to any member of the E2F family.
The effect of TPA in MCF-7 Cells
[0193] It has been described in the literature that TPA induces the arrest of proliferation and the differentiation of tumor cells of the MCF-7 line. In order to validate the culture conditions, the cell growth was monitored (by counting) beforehand over three days of culture in the presence or absence of TPA at a concentration of 25 nM (FIG. 14).
[0194] The cell counts demonstrate a variation in the growth kinetics between the nontreated cultures and the TPA-treated cultures. In fact, from the second day of culture onward, the number of cells is significantly different between the two treatment conditions, TPA already inducing the arrest of cell proliferation. After 3 days of treatment, the control cultures have twice as many cells as the treated cultures. TPA at the concentration of 25 nM therefore clearly inhibits proliferation under our culture conditions.
[0195] In parallel, the inventor was able to observe that the TPA-treated cells rapidly acquire characteristics of differentiated mammary gland cells (FIG. 14): hypertrophy, membrane protrusions and tendency to dissociate from one another. However, in the period of time over which the cells were studied, the secretory phenotype (appearance of granules and of secretory canaliculi) was not observed.
FACS Analysis of the Effect of TPA
[0196] These preliminary studies of the effect of TPA showed in:
[0197] S phase: The FACS study shows that the number of cells in the S phase decreases and reaches a limiting value between 12 h and 24 h of TPA treatment (FIG. 15A). However, without any further addition of TPA, the number of cells in the S phase increases again so as to return to the initial state at 72 h of treatment.
[0198] G2/M phase: The number of cells in the G2/M phase increases from the beginning of the treatment, with a maximum observed at 12 hours (FIG. 15B).
[0199] G0/G1 phase: The number of cells in the G0/G1 phase is at a minimum at 6 hours and at a maximum at 24 hours (FIG. 15C).
[0200] A maximum of cells in the S phase are therefore observed in the early periods of the kinetics (0 h to 6 h), a maximum of cells in the G2/M phase is observed at 12 h and, finally, a maximum of cells in the G0/G1 phase is observed at 24 h. Finally, without any further addition of TPA, the cells return to the S phase at 72 h.
[0201] In conclusion, these results therefore show that TPA acts rapidly on the arrest of cell proliferation and its effect on the reduction of the number of cells in the S phase is optimal at 12 h. 72 h after the single addition of TPA, the cells reinitiate the cell cycle.
The Effect of TPA on the Nuclear Localization of LIV21
[0202] The results obtained by flow cytometry led the inventor to study, in parallel, the expression of LIV21 in nuclear extracts prepared after 12 h, 24 h, 48 h and 72 h of TPA treatment (FIG. 16A). During these kinetics, maximum anti-LIV21 immunoreactivity was observed from 12 h, was maintained up to 48 h and returned to its initial intensity after 72 h of treatment. These data are to be compared with those obtained by FACS. The immunoreactivity of LIV21 significantly increases at 12 h, at which time the number of cells in the S phase is minimal. It lasts until the reinitiation of the cell cycle observed at 72 h.
[0203] Furthermore, it can be noted that this immunoreactivity is detected in the form of a single band at an apparent molecular weight of 50 kDa, whereas it was expressed predominantly in the form of a doublet in the total extracts. In order to determine the form of LIV21 to which this single band corresponds, it was compared on the same Western blot with a total extract and a nuclear extract at 12 h of treatment (FIG. 16B). The results obtained show that the nuclear form of LIV21 corresponds to the lower band of the doublet. These data suggest that the LIV21 protein might be in a different phosphorylation state according to the cell compartment.
[0204] The results of the immunocytochemical study carried out using an anti-LIV21 antibody show that the nuclear translocation of LIV21 is at a maximum at 12 h (FIG. 17) and the localization of LIV21 is predominantly cytoplasmic at 72 h, which is in agreement with the Western blotting observations. However, it is interesting to note that the expression of LIV21 already begins from 1 h of treatment in certain cells, since they are not synchronous.
[0205] All these observations show that the nuclear translocation of LIV21 is concurrent with the decreases the number of cells in the S phase.
Example 3: Study of the Influence of PKCs on the Nuclear Translocation of LIV21
[0206] Effect of TPA on PKCε expression
[0207] Western blotting study: Given that the protein sequence of LIV21 has putative PKC phosphorylation sites, including two specific for PKCε, the inventor tested the variation in the expression of this PKC as a function of the duration of TPA treatment. It was observed that TPA acts very rapidly on PKCε expression, which decreases from 30 min (FIG. 18). The expression of PKCzeta (PKCζ) is used as an internal control since it is not sensitive to TPA.
[0208] Immunocytochemistry: In parallel, immunofluorescence experiments on treated or nontreated cultures made it possible to demonstrate this decrease in PKCε concurrent with the nuclear translocation of LIV21 (FIG. 19). It can be observed that PKCε disappears from the cytoplasm when the cells are treated with TPA and that LIV21 is detected in the nucleus.
[0209] To conclude, it is interesting to note that PKCε is weakly expressed at 12 h, at which time the fewest number of cells in the S phase and the maximum nuclear translocation of LIV21 are observed.
Example 4: Study of the Specific Role of PKCε on the Nuclear Translocation of LIV21 Using a Peptide which Inhibits PKCε Function and Translocation
[0210] In order to determine the specific action of PKCε on the translocation of LIV21, the cultures were treated with a peptide which is a selective antagonist of the function and the translocation of this PKCε (EAVSLKPT (SEQ ID No 118)), and the results were compared with those obtained with TPA treatment. This peptide is recognized by the enzyme and binds as a modified substrate at the level of its catalytic site. It cannot be phosphorylated and acts as a specific inhibitor of the activity of PKCε.
[0211] The effect of the selective inhibition of the activity of PKCε on the nuclear translocation of LIV21 was studied by immunocytochemistry. These experiments were carried out on nontreated cultures or cultures treated for 12 h with TPA at 25 nM or with the peptide at two different concentrations, 1 and 2 μM (FIG. 20). The peptide used at the concentration of 2 μM has an effect identical to that of TPA on the nuclear translocation of LIV21.
[0212] These results were supported by cell fractionation experiments on cultures treated with the PKCε-inhibiting peptide at 2 μM, compared with TPA-treated cultures (FIG. 21). The same LIV21 expression profile was observed in the form of a doublet in the cytoplasm and of a single band in the nuclear fraction.
[0213] The specific inhibition of PKCε induces nuclear translocation of LIV21, thereby suggesting that LIV21 could be the target for PKCε, which would maintain it in the cytoplasm in a phosphorylated form.
Example 5: Western Blotting Analysis
[0214] This example describes the conditions used for a Western blotting analysis of cancerous breast cells.
[0215] The protein extracts are heated at 80° C. for 5 minutes in a Lacmmli buffer (pH 7.4, 0.06 M Tris, 3% SDS, 10% glycerol, 1 mM PMSF, β-mercaptoethanol). The migration is carried out by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis). 10 to 20 μg of proteins migrate in a 12% polyacrylamide gel for 1 h under denaturing conditions (migration buffer: 25 mM Tris base, 192 mM glycine, 1% SDS, pH 8.3). The proteins are then transferred onto a nitrocellulose membrane (Schleicher & Schuell) for one hour by liquid transfer, in a transfer membrane (25 mM Tris, 192 mM glycine, 20% methanol, pH 8.3). The membranes, saturated in PBS-0.1% Tween-0.1% Triton X100-5% skimmed milk for one hour, are brought into contact with the primary antibody diluted in PBS-0.1% Tween-0.1% Triton X100-1% milk at ambient temperature with gentle agitation for one hour to two hours. After washing, the peroxidase-coupled secondary antibody is incubated with the membranes for 1 h. Revelation is carried out by means of a chemiluminescence reaction using the ECL kit according to the supplier's protocol (Amersham).
The primary antibodies used are:
[0216] The anti-LIV21 serum which was produced using two synthetic peptides based on the science of LIV21: peptide LIV21a (SEQ ID No 1) and peptide LIV21b (SEQ ID No 2). The peptides were coupled to hemocyanin before being injected into rabbits for the immunization. The polyclonal antibody was obtained from these two peptides by having immunized two rabbits and having bled one rabbit so as to have a preimmune serum (in order to be sure that this antibody did not already exist in this rabbit).
[0217] The rabbit anti-CDK2 polyclonal antibody (Santa-Cruz technology sc-163) diluted to 1/200.
[0218] The mouse anti-p21 monoclonal antibody (Dako, M7202) diluted to 1/150.
[0219] The mouse anti-p27 monoclonal antibody (Santa-Cruz technology sc-1641) diluted to 1/100.
[0220] The rabbit anti-PKCε polyclonal antibody (Santa-Cruz technology sc-214) diluted to 1/200.
[0221] The rabbit anti-PKCε polyclonal antibody (Santa-Cruz technology sc-216) diluted to 1/200.
[0222] The rat anti-α tubulin polyclonal antibody (Serotec, MCAP77) diluted to 1/500.
The peroxidase-coupled secondary antibodies used (Caltag) are:
[0223] The goat anti-rabbit IgG(H+L) F(ab')2 antibody diluted to 1/2000.
[0224] The goat anti-mouse IgG(H+L) F(ab')2 antibody diluted to 1/2000.
Example 6
[0225] It was demonstrated that the LIV21 protein is associated with PML bodies and that, during sumoylation, LIV21 goes from a molecular weight of 50 kd to 60 kd. Colocalization of LIV21 with SUMO in the PML-SUMO/LIV21 complex was shown by immunoprecipitation (FIG. 22).
[0226] Antitumor role of PML bodies: At the proliferation stage, there are visualized modifications in the PML bodies since these PML bodies dissociate and degrade: (speckles), proteins then become available in the nucleus for ensuring transcription, proliferation, immune reactions and everything that is required for gene transcription. It has been shown that PML associates with SUMO and with HDAC-1 (histone deacetylase 1) and that its complex acts on the expression of E2F1 and PML thus acts on the arrest of proliferation by blocking E2F1. Thus, the PML/HDAC-1 complex down-regulates E2F1 expression. PML associated with Rb (p130) binds to the deacetylated histones and blocks E2F1 by binding to the chromatin (FIG. 2).
[0227] In acute promyelocytic leukemias, PML is truncated and becomes a fusion protein with the retinoic acid receptor. This fusion protein (PMLRARalpha) is due to a 15/17 chromosomal translocation. A new treatment for this disease by combining arsenic and retinoic acid in order to induce cancer cells into apoptosis has been reported in the literature. The PML protein is thought to regulate proliferation in cancers and lymphomas. The inventor has shown, by immunoprecipitation, the association SUMO-PML in which LIV21 is located.
[0228] In the above examples, it was shown that LIV21 is phosphorylated by PKCε and that TPA is an inhibitor of PKCs. The TPA-treated MCF-7 lines show an inhibition of cancerous proliferation and a cell differentiation, and LIV21 is translocated into the nucleus. If a PKCε-specific inhibitory peptide was used, it was the activity and not the expression of PKCε which was inhibited.
[0229] During this TPA treatment (25 nM), when E2F4, p130 and LIV21 were studied (green fluorescence) in the nuclei labeled (DNA) with propidium iodide (red fluorescence) (FIGS. 23 and 24), the following were observed:
[0230] after 12 h, intranuclear green fluorescence signals with the same pattern for E2F4, p130 and LIV21;
[0231] after 48 h, when the proliferation begins, E2F4 has a comparable localization; but at 72 h, it disappears from the nucleus (to the benefit of E2F1).
[0232] By observing, by double labeling, the colocalization of PML and of LIV21 at 24 h of TPA treatment (cf. merge: yellow fluorescence), it was observed that they are colocalized in the nuclei. At 48 h, the colocalization between LIV21 and SUMO is also observed (cf. FIG. 23). The hypothesis is that SUMO, which binds to LIV21, in fact targets LIV21 into the PML bodies and that LIV21 is involved in the PML/SUMO/Rb/HDAC-1 complexes. Two different approaches were carried out in order to demonstrate that LIV21 is physically associated with PML and SUMO in the nuclear bodies, by immunoprecipitation (FIG. 22) and by colocalization by immunocytochemistry (FIGS. 23 and 24) (Rb, p130 and p107 are pocket proteins which have the same binding site). The Rb proteins repress cell growth (Fabbro, Regazzi R, Bioch Biophys Res Comm 1986 Feb. 2; 135 (1): 65-73).
[0233] When TPA is added only once, its action is exhausted after 72 h, proliferation begins again and, at this time, the PML bodies dissociate, break up and become speckles, thus leaving the proteins to ensure transcription, proliferation, etc. LIV21 is no longer located in the nucleus since it is then rephosphorylated by PKCε and thus remains located in the cytoplasm.
Example 7: Study of the Expression of LIV21 in Breast Cancer Biopsies and Skin Cancer Biopsies
[0234] In order to determine whether the observations obtained above are applicable to human tissues, a large number of skin cancer biopsies obtained from patients were studied by immunohistochemical reaction with LIV21-specific antibodies. The immunohistochemical determination of LIV21 protein expression was carried out on 60 biopsies from patients (9 patients having a biopsy of normal tissue versus a biopsy of cancerous tissue), the other biopsies corresponding to cancers which were more or less advanced and, for some, metastatic (cf. Lame superbiochips Laboratories, Seoul, Korea). Moreover, some paraffin slides from patients suffering from bladder cancer and from breast cancer were also studied.
Immunocytochemical analysis protocol:
[0235] Deparaffinize the slides.
[0236] Rehydrate the tissues.
[0237] Saturate the nonspecific sites and permeabilize the membranes.
[0238] Add the antibody in a humid chamber.
[0239] Reveal the antibody.
[0240] Deparaffinize the slides under a hood.
[0241] Two successive baths of toluene (rectapur Prolabo) 2×30 min or 2×20 min; then dehydrate the tissues with rectapur alcohol at 100% for 15 min; then rectapur ethanol at 95% for 10 min; then rectapur 70% for a further 10 min.
[0242] Thaw the antibody at the same time.
[0243] Rehydrate the tissues gently in PBS supplemented with 10% fetal calf serum and 0.1% Triton.
[0244] Saturate the nonspecific sites (for example, with ovalbumin) and permeabilize the membranes.
[0245] Rehydrate for one hour.
[0246] Deposit one ml of this "PBS" per section in order to cover the slide without it drying out at any moment (when it is a slide with cells and not tissues, half an hour is sufficient).
[0247] Place the pane and the stainless steel cover and water below so as to create a humid chamber.
[0248] Add the antibody in the humid chamber.
[0249] Dilute the rabbit serum to 1/200 in 4 ml of PBS triton, so as to continue to permeabilize the membranes, and FCS.
[0250] Place 1 ml on each slide and keep away from the light and avoid evaporation.
[0251] Leave overnight or for a minimum of three hours.
[0252] Then rinse with 1× normal PBS pH 7, carry out two washes of 5 to 10 min so that no trace of the first antibody remains.
[0253] While preparing the Alexa 488 green probe (in the cold at 4° and in the dark) diluted to 1/250, therefore 10 μliter in 2.5 ml of PBS, still with 10% FCS and 0.1% Triton, rest the slides on the plate. Cover the section again with 2.5 ml in order to maintain a humid chamber for one hour, and then wash with 1× PBS, pH 7.
[0254] Wash with propidium iodide at 0.5 microgram per microliter to be diluted to 20 microgram per ml and then again to 1/50, but this time, diluted in 1× PBS alone (50 microliters per 2.5 ml of PBS). Drain while taking them out of the PBS and then dispense 2.5 ml of propidium iodide over the four slides for one minute, followed by two rinses with simple PBS. Mount the slides in Moviol before reading.
[0255] All the results are summarized in Table 1 below.
TABLE-US-00002 TABLE 1 Expression of the LIV2I protein determined by immunohistochemistry in 50 skin cancer biopsies and 9 normal tissue biopsies "MIXED" NUCLEAR LIV2I CYTOPLASMIC LIV2I WEAKLY LIV2I TUMOR TYPE NEGATIVE POSITIVE POSITIVE NORMAL TISSUES 6/9 3/9 0 FROM THE SAME PATIENTS WELL- 18/21 2/21 1/21 DIFFERENTIATED CARCINOMAS MODERATELY 1/7 5/7 1/7 DIFFERENTIATED CARCINOMAS POORLY 2/11 3/11 6/11 DIFFERENTIATED CARCINOMAS CARCINOMA 0 2/9 7/9 METASTASES
For example:
[0256] FIG. 25: Results: image 43: poorly differentiated cancer; image 58; normal tissue derived from the same individual suffering from a cancer; image 40: metastatic carcinoma 10 cm and image 17: metastatic carcinoma of 3.5 cm.
[0257] FIG. 26 is, like FIG. 25, a second example of nuclear localization of LIV21 in the control and normal tissue (No. 52) of individual No. 7 suffering from a squamous cell carcinoma of the pharynx (moderately differentiated T4N0M0).
[0258] FIG. 27 is a sample of advanced bladder cancer on cystectomy (grade III urothelial carcinoma infiltrating the chorion and the musculosa) versus normal bladder tissue from the same patient with an internal control (PI): preimmune serum PI before the rabbit has been immunized against LIV21, labeling of the nuclei with propidium iodide.
[0259] FIG. 28 is a sample of breast cancer which makes it possible to demonstrate the cytoplasmic labeling of the LIV21 antibody in the cancer cells.
[0260] These results show that the cytoplasmic localization of LIV21 is an indicator of the aggressiveness and of the metastatic potential of the cancer. The detection of LIV21 expression indicates the presence of invasive, aggressive and metastatic cancer cells. These results also show that the nuclear localization of LIV21 is an indicator of normal quiescent cells, that is of well-differentiated tissues.
Example 8: Physical Interaction of LIV21 with the Proteins of the E2F Family
[0261] Coimmunoprecipitation experiments carried out using anti-LIV21, anti-E2F1 and anti-E2F4 antibodies made it possible to demonstrate that LIV21 associates with E2F4.
[0262] The members of the E2F family are transcription factors whose role has been widely described in the literature as being key molecules in the positive or negative control of the cell cycle (Slansky JE and Farnham PJ 1996; Helin K 1998 and Yamasaki L, 1998), by virtue of their association with the pRb protein (Wu CL, Zukerberg LR, Lees JA 1995) or pocket proteins. E2F1 positively controls the cell cycle by transactivating the promoter of the genes responsible for cell proliferation (DNA polymerase alpha, thymidine kinase, DHFT, etc.), whereas E2F4 is described as one of the members of the EF family which negatively controls the cycle. Furthermore, a high expression of E2F1 is embryonic mammary tissues has been shown (Espanel X, Gillet G 1998), whereas it is no longer expressed in post-mitotic mammary tissues, to the benefit of a large increase in E2F4 expression (Kastner A Brun G 1998).
[0263] The identification of antigens has been carried out in cell lysates by immunoprecipitation. The analysis of the physical interaction of various proteins associated with E2F4 and E2F1 was demonstrated by coimmunoprecipitation of protein complexes. The complex was studied using μ MACS PROTEIN with MICROBEADS (MILTENYIBIOTEC). When lysates of S aureus are added, the proteins A interact with the Fc portion of the specific antibodies and the immunocomplexes become insoluble and are therefore recovered by centrifugation. After breaking of the bonds (heating) between AG/AB and protein A-rich membranes, Western blotting was carried out. These results suggest that the LIV21/E2F4 complex appears to play an important role in establishing cell quiescence.
Example 9: Functional Interaction of LIV21 with the Proteins of the E2F Family
[0264] It was demonstrated that blocking the expression of the LIV21 protein was correlated with a decrease in the expression of E2F4 and with an increase in the expression of E2F1. In parallel, the functional aspect of the increase in E2F1 was verified by studying the transcription of two of its target genes, DHFR and DNA polymerase α.
Example 10
[0265] Reverse transcriptions:
[0266] After MCF7 cells (ATCC passage 15) had been thawed and cultured up to 200 million, they were trypsinized and frozen at 80° C. The RNA was extracted from two pools of 50 million cells with the Nucleospin RNA L kit (Macherey Nagel) ref 740.962.20, resulting in a pool 1 of 318 μg and a second of 182 μg. The poly A+ RNA was extracted from 313 μg of total RNA of pool 1 using the oligo Tex mRNA Midi kit (Qiagen) ref. 70042
I Reverse transcription:
[0267] The RNA was reverse transcribed with the Fermentas Revert Aid H minus M MuLV Reverse Transctiptase, ref. EP0451 batch 1124, with 3.64 μg of total RNA and 0.45 μg of mRNA, according to the supplier's conditions, with an oligo dT primer. Reactions were carried out at 2 different temperatures at 45° C. and 55° C. so as to estimate the RNA structures that may hinder reverse transcription.
II PCR
[0268] The PCRs were carried out with the reverse transcriptions as templates, initially with the primers AI+oligo dT. Nested PCRs were subsequently carried out on these first PCRs, with the primers AI+Splicing, AI+GDBRI, or ATG+Splicing, ATG+GDBRI.
[0269] PCR amplification was then carried out with the primers specific for the genes to be detected, using the cDNAs obtained after oligo dT RT.
[0270] Enzyme: Fermentas Taq DNA polymerase. Thermocycler: Bio Rad iCycler. The quality of the cDNAs was tested by amplification of GAPDH, b actin and Histone 113.3 housekeeping genes.
TABLE-US-00003 TABLE 1 Primers Amplified fragment Reference 5'-3' sequence size Histone N 5'-GTG GTa aag cac cca gga 347 bp a-3' Histone I 5'-gct agc tgg atg tct ttt (reverse) gc-3' Hum GAPDH 5'-TGA AGG TCG GAG TCA ACG 983 bp sense G-3' Hum GAPDH 5'-CAT GTG GGC CAT GAG GTC- antisense 3' Hum 5'-GGA CTT CGA GCA AGA 234 bp b-actin GATGG-3' sense Hum 5'-AGC ACT GTG TTG GCG TAC b-actin AG-3' antisense LIV21 (A1) 5'-3' LIV21 (A2) 5'-3' odt 5'-TTTTTTTTTTTTTTTTTTT-3' ATG galgal 5'-3' Splicing 5'-3' sense Splicing 5'-3' reverse G reverse 5'-3'
TABLE-US-00004 TABLE 2 PCR mix Forward 25 mM Mg++ 10 mM primer Reverse primer 10X (1.5 mM final dNTP 10 μM 10 μM Taq (μl) cDNA H2O buffer concentration) (μl) (μl) (μl) 5 U/μl (μl) 18.3 2.5 1.5 0.5 0.5 0.5 0.2 1
PCR cycles
[0271] Denaturation: 94° C. 2 minutes
[0272] Denaturation: 94° C. 20 seconds
[0273] Annealing: 52-55° C. 1 minute 35 cycles
[0274] Elongation: 72° C. 1 minute 20
[0275] Final elongation: 72° C. 7 minutes
[0276] Conservation: 4° C.
III CONTROLS
THE PCR PRODUCTS WERE SUBSEQUENTLY CONTROLLED ON AGAROSE GELS AND ANALYSED WITH THE BIOCAPT 11.01 SOFTWARE FROM VILBER LOURMAT.
[0276]
[0277] FIG. 32: Analysis molecular masses gel 1
[0278] FIG. 33: Gel 2 with analysis of molecular masses
[0279] FIG. 34: Gel 3 at 55° and analysis of molecular masses
[0280] FIG. 35: Gel 4 at 45° and 55° and analysis of molecular masses
[0281] FIG. 36: Screening ligation of 400 bp band, clones B1 and B10
[0282] FIG. 37: PCR with the primers showing a band at 1400 bp, clones C1 to C10
[0283] FIG. 38: Gel 5: ligation, screening on the five new clones
[0284] FIG. 49: Gel 6: screening of the S55T and S55M recombinant clones and analysis of molecular masses.
Gel 2:
[0285] PCRs carried out using templates from PCRs performed with the primers AI+oligo dT on the RTs carried out at 45° C. on the total RNA and the poly A+ RNA (messenger RNA). The primers used for these PCRs are AI+G or AI+Splicing reverse.
[0286] On the RTs carried out using the total RNA, a band of 1178 1253 bp is amplified with the primers AI+G and AI+Splicing reverse. The poly A RNA was used to carry out the RT and is weakly observed at the size (FIG. 33) of 1400 bp for amplification with the primers AI+G, and of 415 bp with the primers AI+Splicing reverse. In the AI+splicing PCR product, there are other bands, of 860 and 233 bp (FIGS. 38 & 39).
Gel 3:
[0287] PCRs carried out using templates from PCRs performed with the primers AI+oligo dT on the RTs carried out at 45° C. on the total RNA and the poly A+ RNA (messenger RNA). The primers used for these PCRs are AI+GDBRI or AI+Splicing reverse (FIG. 34).
[0288] For the PCRs carried out on RTs performed at 55° C., the same overall pattern of bands as that obtained on the RTs performed at 45° C. is found.
[0289] No specific amplification is observed when the poly A RNA was used to carry out the RT. On the RTs carried out on the total RNA, a major band of 1554 1609 bp is found with the primers AI+GDBRI and AI+Splicing reverse. A band at the theoretical size of 1455 bp is expected for an amplification with the primers AI+GDBRI and a band with the theoretical size of 415 bp is expected with the primers AI+Splicing reverse. In the 2 profiles, very clear bands of 1900-2100 bp and of 1000-1300 bp are found, but with a weaker intensity than that of the band of 1500-1600 bp.
[0290] In the AI+splicing reverse PCR product, there is another major band, of 263 bp.
Gel 4:
[0291] Nested PCRs carried out using templates from PCRs performed with the primers AI+GDBRI or AI+Splicing reverse on the RTs carried out at 45° C. on the total RNA and the poly A+RNA (messenger RNA). The primers used for these PCRs are ATG+GDBRI or ATG+Splicing reverse (FIG. 35).
[0292] The nested PCRs carried out with the primers ATG+GDBRI give bands at 1213 bp (RT 45° C.) and 1559+1315 bp (RT at 55° C.); the expected theoretical size is 1455 bp. The PCRs carried with the primers ATG+Splice reverse give more varied band profiles.
[0293] These PCRs carried out on other PCRs performed with the primers AI+GDBRI or AI+Splicing reverse.
[0294] The presence of a band of 400 bp is noted in the profiles obtained from messenger RNA (the band obtained from the reverse transcription carried out at 55° C. is of greater intensity).
[0295] The profiles of the ATG+Splice reverse PCRs carried with total RNA at the start give a band of 424 437 bp of very strong intensity. Bands of 614 and 783 bp of very strong intensity are also found in the profile of the RT 45 and a greater number of bands, but of weaker intensity, is found in the profile of the RT 55, bands at 1118, 936 and 749 bp. The products of these various PCRs were cloned and sequenced.
Example 12
[0296] The PCR products of lanes 2, 4, 6, 7 and 8 were ligated with the plasmid pGEMT Easy, Promega, T7 vector and the recombinant clones were screened (FIG. 38).
[0297] Lane 2: G45T ligations
[0298] Lane 4: S45T ligations
[0299] Lane 6: G55T ligations
[0300] Lane 7: S55M ligations
[0301] Lane 8: S55T ligations
[0302] The recombinant clones obtained were screened (after extraction of the plasmid DNA) by restriction with the Eco RI enzyme, the sites of which border the site of insertion of the PCR products into the pGEMT Easy vector.
Screening of the recombinant clones:
[0303] The first experiments had been carried out using the ten clones B and the ten clones C, FIGS. 36 and 37, and the results of the sequences of clones B2 and C8 are given in the following example, and exhibit, by sequence comparison between clones, great homology with the clones of the second series of experiments.
[0304] Gel 5: screening of the S45T and G45T recombinant clones Analysis of molecular masses
[0305] The screening of the clones with Eco RI shows that, out of the 9 S45T clones, 3 have inserts of 100 bp, 216 bp and 410 bp.
[0306] On the G45T clones, out of the 6 clones tested, 3 have inserts of 57, 71 and 148 bp (FIG. 38).
[0307] FIG. 39: Screening of the S55T and S55M recombinant clones Analysis of molecular masses
[0308] The screening with Eco RI shows that, out of the 13 clones screened, 7 have inserts of sizes between 239 and 637 bp.
[0309] The clones G45T5 (148 bp), S45T9 (410 bp), S45T3 (100 bp), S55M1 (491 bp), S55T6 (251 bp) and S55T9 (637 bp) were extracted so as to be sequenced.
Primers used for determining the sequences of the various clones:
[0310] ATG galgal 5'-atgtatattatata-3' INV COMP ttagatatataata
[0311] Splicing reverse 5'-aaatat-3'
[0312] Splicing reverse 5'-t-3' inv comp
[0313] G reverse 5'-TG-3'
[0314] G reverse 5'-AT . . . -3' inv comp
Results
[0315] Clone B2, the band which is approximately 400 bp is the same fragment as S55M1, which itself is the same fragment as S45T9. These fragments are approximately between 400 and 450 bp. This is not surprising since they were obtained after a nested PCR carried out with the splicing reverse primer and the AI primer (for the B2 fragment) and the ATG galgal primer for the S45T9 and S55M1 fragments. These fragments were obtained with RTs carried out at various temperatures. On the other hand, the S55T9 fragment is nevertheless approximately 600 bp, and a part (300 bp) exhibits quite strong identity with the other fragments cloned (clone FLJ).
[0316] The inventor approached the study of the expression of the LIV21 protein by the Western blotting technique, with an anti LIV21 antibody, in MCF7 cells compared with breast tissues. The anti LIV21 antibodies were obtained by the method described. In this line, LIV21 was expressed, both in breast tissues and in the MCF 7 cells, in the form of a doublet that migrates at an apparent molecular weight of 50 kDa.
[0317] In conclusion, these results suggest that the LIV21/E2F4 complex acts as a complex which inhibits the expression of the E2F1 gene. This complex could correspond to a new point of control in the arrest of cell proliferation.
REFERENCES
[0318] Arya R, Kedar V, Hwang JR, McDonough H, Li HH, Taylor J, Patterson C. Muscle ring finger protein-1 inhibits PKC{epsilon} activation and prevents cardiomyocyte hypertrophy. J Cell Biol. 2004 Dec. 20; 167(6):1147-59. Epub 2004 Dec. 13.
[0319] Caroll JS, Prall OWJ, Musgrove EA, Sutherland RL. A pure Estrogen Antagonist Inhibits Cyclin E-Cdk2 Activity in MCF-7 Breast Cancer Cells and Induces Accumulation of p130-E2F4 Complexes Characteristic of Quiescence. (2000) J Biol. Chem, 275 (49):38221-38559.
[0320] Chan BN, Wang JY. Coordinated regulation of life and death by RB. (2003) Nat Rev Cancer, 3 (2): 130-8.
[0321] Cheng T, Scadden DT. Cell cycle entry of hematopoictic stem and progenitor cells controlled by distinct cyclin-dependent kinase inhibitors. (2002) Int J Hematol, 75 (5):460-5.
[0322] Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. (2002) Nat Rev Cancer, 2 (12): 910-7.
[0323] Coqueret O. Linking cyclins to transcriptional control. (2002) Gene. 299 (1-2):35-55.
[0324] Crisanti P, Raguenez G, Blancher C, Neron B, Mamoune A, Omri B. Cloning and characterization of a novel transcription factor involved in cellular proliferation arrest: PATF, (2001) Oncogene 20: 5475-5483.
[0325] Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB. The molecular basis of FHA domain: phosphopeptide binding specificity and implications for phospho-dependant signaling mechanisms (2000) Mol Cell, 6 (5):1169-82.
[0326] Durocher D, Jackson SP. The FHA domain. (2002) FEBS Lett, 513 (1): 58-66.
[0327] Espanel X; Le Cam L, North S, Sardet C, Bren G, Gillet G. Regulation of E2F-1 gene expression in avian cells. (1998) Oncogene, 17 (5): 585-94.
[0328] Fraering, Wenjuan Ye, Michael S Wolfe. Purification and characterization of the Human J Secretase Complex(2004) Biochemistry 43: 9774-9789.
[0329] Han EK, Begemann M, Sgambate A, Soh JW, Doki Y, Xing WQ, Liu W, Weinstein IB, Increased expression of cyclin D1 in a marine mammary epitelial cell line induces p27kip1, inhibits growth, and enhances apoptosis. Cell Growth Differ. 1996 June:7(6):699-710.
[0330] Harlow et al Antibodies: A laboratory Manual, CSH Press, 1988
[0331] He LZ, Merghoub T, Pandolfi PP. In vivo analysis of the molecular pathogenesis of acute promyclocytic leukemia in the mouse and its therapeutic implications. (1999) Oncogene, 18: 5278-5292.
[0332] Helin K. Regulation of cell proliferation by the E2F transctiption factors. (1998) Curr Opin Genel Dev, 8(1):28-35.
[0333] Horman S, Galand P, Mosselmans R, Legros N. Leclercq G, Mairesse N. Changes in the phosphorylation status of the 27 kDa heat shock protein (HSP27) associated with the modulation of growth and/or differenciation in MCF-7 cells. (1997) Cell Prolif, 30 (1):21-35.
[0334] Hughes et al. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vector. (1987) J. Virol 61: 3004-3012
[0335] Kastner A, Espanel X, Brun G. Transient accumulation of retinoblastoma/E2F-1 protein complexes correlates with the onset of neuronal differentiation in the developing quail neural retina. (1998) Cell Growth Differ, 9 (10): 857-67.
[0336] Katalin F. Medzihradszky. Characterization of Protein N-Glycosylation. (2005) Methods in Enzymology, vol 405:116-138.
[0337] Kohler G, Milstein C. Continous cultures of fused cells secreting antibody of predefined specificity. Nature, 1975 Aug. 7;256(5517):495-7.
[0338] Lee W, Bee JH, Jung MW, Park SD, Kim VH, Kim SU, Mook-Jung I. Amyloid beta peptide directly inhibits PKC activation. Mol Cell Neurosci. 2004 June;26(2) 222-31.
[0339] Mairesse N, Horman S, Musselmans R, Galand P. Antisense inhibition of the 27 kDa heat shock protein production affects growth rate and cytoskeletal organization in MCF-7 cells. (1996) Cell Biol Int, 20(3): 205-12.
[0340] Matunis MJ. On the road to repair: PCNA encounters SUMO and obiquitin modifications. (2002) Mol Cell, 10(3): 441-2.
[0341] Melchior F, Hengst L. SUMO-1 and p53. (2002) Cell Cycle, 1 (4):245-9.
[0342] Mundle SD, Saherwal G. Evolving intricacies and implications of E2F1 regulation (2003) FASEB J, 17 (6):569-74.
[0343] Opalka B, Dickopp A, Kirch HC. Apoptotic genes in cancer therapy. (2002) Cells Tissues Organs, 172 (2): 126-32.
[0344] Pardo FS, Su M, Borek C. Cyclin D1 induced apoptosis maintains the integrity of the GI/S checkpoint following ionizing radiation irradiation. Somat Cell Mol Genel 1996 March; 22(2):135-44.
[0345] Pawson T, Gish Gd, Nash P. SH2 domains, interaction modules and cellular wiring. (2001) Trends Cell Biol, 13 (12): 504-11.
[0346] Platet N. Prevostel C, Derroq D, Joebert D, Rochefort B, Garcia M. Breast cancer cell invasiveness: correlation with protein kinase C activity and differential regulation by phorbol ester in estrogen receptor-positive and -negative cells. (1998) Int J Cancer, 75(5): 750-6.
[0347] Ree AH, Bjornland K, Bruatier N, Johansen HT, Pedersen KB, Aasen AO, Podstad O. Regulation of tissue-degrading factors and in vitro invasiveness in progression of breast cancer cells. (1998) Clin Exp Metastasis, 16(3): 205-15.
[0348] Regazzi R, Fabbro D, Costa SD, Borner C, Eppenberger U. Effect of tumor promoters on growth and on cellular redistribution of phospholipid Ca2+-dependant protein kinase in human breast cancer cells. (1986) Int J Cancer, 37(5): 731-737.
[0349] Schneider SM, Offerdinger M, Huber H, Grunt TW. Involvement of nuclear steroid/thyroid/retinoid receptors and of protein kinases in the regulation of growth and of c-erbB and retinoic acid receptor expression in MCF-7 breast cancer cells. (1999) Breast Cancer Res and Treat, 58:171-181.
[0350] Senderowicz AM. Cyclin-dependent kinases as targets for cancer therapy. (2002) Cancer Chemother Biol Response Modif. 20: 169-96.
[0351] Slansky JP, and Farnham PJ. Introduction to the E2F family: protein structure and gene regulation. (1996) Curr Top Microbiol Immunol, 53:347-360.
[0352] Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, SH2 domains recognize specific phosphopeptide sequences. (1993) Cell, 72(5):767-78.
[0353] Starzec AB., Spanakis E, Nehme A, Salle V, Veber N, Mainguene C, Planchon P, Valetie A, Prevost G, Israel L. Proliferative responses of epithelial cells in 8-bromo-cyclic AMP and to a phorbol ester change during breast pathogenesis. (1994) J. Cell Physiol, 161(1):31-8.
[0354] Stevaux O, Dyson NJ. A revised picture of the E2F transcriptional network and RB function (2002) Curr Opin Cell Biol, 14(6):684-91.
[0355] Stiegler P, Giordano A. The family of retinoblastoma proteins. (2001) Crit Rev Eukaryot Gene Expr, 11 (1-3):59-76.
[0356] Toma O, Weber NC, Wolter JI, Obal D, Preckel B, Schlack W. Desflurane preconditioning induces time-dependent activation of protein kinase C epsilon and extracellular signal-regulated kinase 1 and 2 in the rat heart in vivo. Anesthesiology. 2004 December; 101(6):1372-80.
[0357] Vaitukaitis J, Robbins JB, Nieschlag E, Ross GT. A method for producing specific antisera with small doses of immunogen. J Clin Endocrinol Metab. 1971 December; 33(6):988-91.
[0358] Ward ES, Gussow D, Griffiths AD, Jones PT, Winter G. Binding activities of a repertoire of single immunoglobulin variable domains secreted from Escherichia coli. Nature. 1989 Oct. 12; 341(6242):544-6.
[0359] Wu CL, Zukerberg LR, Ngwu C, Harlow E, Lees JA. In vivo association of E2F and DP family proteins. (1995) Mol Cell Biol, 15 (5): 2536-46.
[0360] Yaffe MB. Phosphotyrosine-binding domains in signal transduction. (2002) Nat Rev Mol Cell Biol, 3(3):177-86.
[0361] Yamasaki L. Growth regulation by the E2F and DP transcription factor families. (1998) Results Probl Cell Differ, 22:199-227.
[0362] Zee-Yong Park and David H. Russell. Identification of Individual Proteins in Complex Protein Mixtures by High-Resolution. High-Mass-Accurancy MALDI TOF-Mass Spectrometry Analysis of In-Solution Thermal denaturation/Enzymatic Digestion. (2001) Anal Chem, 73(11): 2558-2564.
Sequence CWU
1
1
180113PRTHomo sapiens 1Pro Ala Ser Ala Gly Ile Thr Gly Val Ser Cys Ala Arg
1 5 10 228PRTHomo sapiens
2Gly Phe Thr Val Leu Ala Arg Met Val Ser Ile Ser Pro Arg Asp Pro 1
5 10 15 Pro Ala Ser Ala
Ser Gln Ser Val Gly Ile Ala Tyr 20 25
313PRTHomo sapiens 3Phe Tyr Cys Arg Ser Leu Gln Ser Ser Trp Asp Tyr
Arg 1 5 10 424PRTHomo
sapiens 4Ser Val Thr Gln Ala Gly Val Gln Trp Arg Asn Leu Gly Ser Arg Leu
1 5 10 15 Gln Pro
Leu Pro Pro Gly Phe Lys 20 536PRTHomo
sapiens 5Ser Val Thr Gln Ala Gly Val Gln Trp Arg Asn Leu Gly Ser Leu Gln
1 5 10 15 Pro Leu
Pro Pro Gly Phe Lys Phe Ser Cys Arg Ser Leu Gln Ser Ser 20
25 30 Trp Asp Tyr Arg 35
619PRTHomo sapiens 6Ser Ile Ser Pro His Asp Leu Pro Ala Ser Ala Ser Gln
Ser Ala Gly 1 5 10 15
Ile Thr Gly 746PRTHomo sapiensmisc_featureTranslation of clone C8T7
extracted from MCF7 cells 7Asp Phe Leu Cys Leu Thr Ile Ser Ser Phe
Ser Leu Pro Ile Ile Ile 1 5 10
15 Ile Tyr Phe Ser Asp Gly Val Ser His Cys Arg Gln Ala Gly Val
Gly 20 25 30 Gln
Trp Arg Asn Leu Gly Ser Pro Pro Pro Thr Gly Phe Lys 35
40 45 846PRTHomo
sapiensmisc_featureTranslation of clone S45T9 from MCF7 cells 8Met Glu
Ser Arg Ser Val Ala Gln Ala Gly Val Gln Trp His Asn Leu 1 5
10 15 Gly Ser Ala Leu Pro Pro Gly
Phe Met Pro Phe Ser Cys Leu Ser Leu 20 25
30 Gln Ser Ser Trp Asp Tyr Arg His Ala Pro Pro Arg
Pro Ala 35 40 45
915PRTHomo sapiens 9Pro Pro Arg Pro Ala Thr Phe Leu Tyr Phe Pro Arg Gln
Gly Phe 1 5 10 15
1015PRTHomo sapiens 10Arg Arg Phe Leu Tyr Ser Ser Asn Leu Phe Pro Ser Asn
Gly Thr 1 5 10 15
119PRTHomo sapiens 11Arg Tyr Val Pro Ser Ser Asn Leu Pro 1
5 129PRTHomo sapiens 12Arg Tyr Val Pro Ser Ser Asn Leu
Pro 1 5 139PRTHomo
sapiensmisc_featuresynthesized from de novo sequencing by ESI MS MS
13Arg Tyr Leu Val Thr Pro Val Asn Ala 1 5
148PRTHomo sapiensmisc_featurecompletely synthesized from de novo
sequencing 14Arg Tyr Val Leu Ser Pro Val Lys 1 5
159PRTHomo sapiens 15Lys Pro Ser His Pro Lys Pro Ser Lys 1
5 166PRTHomo sapiens 16Thr Phe Lys Asn Leu Cys 1
5 178PRTHomo sapiens 17Lys Ala His Asn Leu Phe Lys Thr 1
5 189PRTHomo sapiens 18Arg Tyr Leu Pro Ala Asn
Pro Arg Tyr 1 5 1913PRTHomo sapiens 19Phe
Tyr Cys Arg Ser Leu Gln Ser Ser Trp Asp Tyr Arg 1 5
10 2013PRTHomo sapiens 20Lys Thr Leu Pro Ser Ser
Ser Cys Leu Val Ala Tyr Lys 1 5 10
2114PRTHomo sapiens 21Lys Val Ile Ile Val Ala Val Asp Trp Asp Leu
Ser Lys Glu 1 5 10
2216PRTHomo sapiens 22Lys Ile Phe Ser Pro Ala Thr Val Phe Phe Thr Ser Ile
Glu Lys His 1 5 10 15
2318PRTHomo sapiens 23Lys Asn Val Trp Ile Leu Thr Gly Phe Gln Gln Gly
Gln Glu Phe Pro 1 5 10
15 Lys Phe 2412PRTHomo sapiens 24Arg Phe Asn Leu Phe Ala Gly Gly Ser
Asn Lys Ala 1 5 10 2512PRTHomo
sapiens 25Arg Ala Tyr Ser Leu Leu Gly Thr Ser Glu Arg Thr 1
5 10 2612PRTHomo sapiens 26Ala Met Ala Ala Asn
Asp Thr Gly Gly Phe Val Lys 1 5 10
2711PRTHomo sapiens 27Ala Ser Glu Glu Gly Ile Met Val Val Glu Arg 1
5 10 2818PRTHomo sapiens 28Phe Asp Val
Val Val Ile Gly Ala Gly Pro Gly Gly Tyr Val Ala Ala 1 5
10 15 Ile Lys 2919PRTHomo sapiens
29Arg Pro Val Thr Thr Asp Leu Leu Ala Ser Asp Ser Gly Val Thr Ile 1
5 10 15 Asp Glu Arg
307PRTHomo sapiens 30Phe Tyr Cys Gly Trp Asp Arg 1 5
318PRTHomo sapiens 31Asp Val Ala Gln Glu Glu Gly Lys 1 5
328PRTHomo sapiens 32Ser Gly Ile Pro Ser Glu Leu Arg 1
5 337PRTHomo sapiens 33Glu Ala His Ile Gln Met Lys
1 5 347PRTHomo sapiens 34Glu Gly Ile Trp Ile Pro
Lys 1 5 356PRTHomo sapiens 35Tyr Thr Phe Asp Ser
Arg 1 5 366PRTHomo sapiens 36Leu Thr His Glu Ile Arg
1 5 375PRTHomo sapiens 37Leu Tyr Leu Asp Lys 1
5 385PRTHomo sapiens 38Tyr Gly Leu Gln Arg 1 5
395PRTHomo sapiens 39Asp Ser Ile Ile Arg 1 5 4016PRTHomo
sapiens 40Leu Glu Ala Ile Cys Ala Ala Met Ile Glu Ser Trp Gly Tyr Asp Lys
1 5 10 15
4112PRTHomo sapiens 41Gly Asp Leu Trp Phe Met Ser His Gln Gly His Lys 1
5 10 4211PRTHomo sapiens 42Tyr Ala
Phe Asp Phe Tyr Glu Met Thr Ser Arg 1 5
10 4314PRTHomo sapiens 43Glu Val Asn Ala Gly Thr Ser Gly Thr Phe Ser
Val Pro Arg 1 5 10
449PRTHomo sapiens 44Asn Gln Asp Arg Pro Tyr Met Pro Arg 1
5 459PRTHomo sapiens 45Ile Val Ser Ile Leu Glu Trp Asp
Arg 1 5 4610PRTHomo sapiens 46Ala Pro Tyr
Ile Ala Glu Thr Ala Leu Arg 1 5 10
479PRTHomo sapiens 47Asn Met His Asn Leu Leu Gly Val Lys 1
5 489PRTHomo sapiens 48Asn Leu Thr Asp Met Ser Leu Ala
Arg 1 5 498PRTHomo sapiens 49His Thr Thr
Glu Asp Val Asn Arg 1 5 507PRTHomo sapiens
50Lys Phe Phe Val Phe Ala Leu 1 5 5115PRTHomo
sapiens 51Phe Val Phe Ala Leu Ile Leu Ala Leu Met Leu Ser Met Cys Gly 1
5 10 15 5212PRTHomo
sapiens 52Thr Leu Gln Ile Phe Asn Ile Glu Met Lys Ser Lys 1
5 10 5317PRTHomo sapiens 53Lys Asp Pro Glu Leu
Trp Ala His Val Leu Glu Glu Thr Asn Thr Ser 1 5
10 15 Arg 5412PRTHomo sapiens 54Lys Ser Trp
Glu Val Tyr Gln Gly Val Cys Gln Lys 1 5
10 5513PRTHomo sapiens 55His Thr Ser Leu Val Gly Cys Gln Val Ile
His Tyr Arg 1 5 10
5611PRTHomo sapiens 56Gln Ala Gly Val Gln Trp Arg Asn Leu Gly Ser 1
5 10 5715PRTHomo sapiens 57Ser Leu Gln Ser
Ser Trp Asp Tyr Arg Arg His Pro Pro Pro Thr 1 5
10 15 5821PRTHomo sapiens 58Phe Leu Tyr Phe Leu
Arg Trp Glu Phe Thr Gly Pro Thr Ala Ser Ala 1 5
10 15 Gly Ser Pro Pro Arg 20
5914PRTHomo sapiens 59Gly Phe Thr Met Met Ala Arg Met Val Ser Ile Ser Pro
His 1 5 10 6016PRTHomo
sapiens 60Met Glu Ser Arg Ser Val Ala Gln Ala Gly Val Gln Trp Arg Asn Leu
1 5 10 15
6111PRTHomo sapiens 61Pro Ala Ser Ala Ser Glu Ser Val Gly Ile Ala 1
5 10 6214PRTHomo sapiens 62Ile Ser Arg Ala
His Arg Thr Leu His Leu Pro Pro Cys Pro 1 5
10 6313PRTHomo sapiens 63Trp Asp Tyr Arg His Ala Leu
Pro His Pro Ala Asn Pro 1 5 10
6415PRTHomo sapiens 64Gly Phe Thr Val Leu Ala Arg Met Val Ser Ile Ser
Pro Arg Asp 1 5 10 15
6512PRTHomo sapiens 65Pro Ala Ser Ala Ser Gln Ser Ala Gly Ile Thr Gly 1
5 10 6610PRTHomo sapiens 66Arg Asp
Pro Pro Ala Ser Ala Ser Gln Ser 1 5 10
6716PRTHomo sapiens 67Met Ser His Cys Ala Arg Pro Thr Tyr Phe Lys Asn Asn
Lys Asn Ser 1 5 10 15
6821PRTHomo sapiens 68Leu Lys Cys Leu Trp Arg Ile Gln Asp Ser Ala Gly
Leu Tyr Leu Pro 1 5 10
15 Met Thr Pro Leu Ile 20 6916PRTHomo sapiens 69Gln
Phe Gln Thr Leu Leu Met Gly Leu Met Ser Phe Ile Arg Gln Cys 1
5 10 15 7014PRTHomo sapiens
70Arg Arg Phe Leu Tyr Ser Ser Asn Leu Phe Pro Phe Lys Tyr 1
5 10 7116PRTHomo sapiens 71Arg Thr Val
Arg Pro Leu Asn Ile Glu Val Gly Val Leu Pro Lys Thr 1 5
10 15 7224PRTHomo sapiens 72Lys Phe
Phe Val Phe Ala Leu Ile Leu Ala Leu Met Leu Ser Met Cys 1 5
10 15 Gly Ala Asp Ser His Ala Lys
Arg 20 7320PRTHomo sapiens 73Lys Gly Ile Thr
Glu Glu Ile Met Glu Ile Ala Leu Gly Gln Ala Leu 1 5
10 15 Glu Ala Arg Leu 20
7410PRTHomo sapiens 74Ser Gln Gln Asn Arg Trp Ala Glu Ser Lys 1
5 10 7511PRTHomo sapiens 75Met Asn Val Glu Lys Ala
Glu Phe Cys Asn Lys 1 5 10
7616PRTHomo sapiens 76Asn Ser Gln Gly Val Ala Trp Ile Thr Leu Asn Ser Ser
Ile Gln Lys 1 5 10 15
7717PRTHomo sapiens 77Val Cys Thr Lys Pro Val Glu Ser Thr Ile Glu Asp
Lys Ile Phe Gly 1 5 10
15 Lys 7813PRTHomo sapiens 78Ala Gly Leu Gln Glu Glu Ala Gln Gln Leu
Arg Asp Glu 1 5 10
7911PRTHomo sapiens 79Met Ser His Lys Gln Ile Tyr Tyr Ser Asp Lys 1
5 10 8015PRTHomo sapiens 80Tyr Asp Asp Glu
Glu Phe Glu Tyr Arg His Val Met Leu Pro Lys 1 5
10 15 8114PRTHomo sapiens 81Leu Val Pro Lys Thr
His Leu Met Ser Glu Ser Glu Trp Arg 1 5
10 8221PRTHomo sapiens 82Lys Ala Tyr Glu Cys Asp Ile Thr
Tyr Gly Thr Asn Asn Glu Phe Gly 1 5 10
15 Phe Asp Tyr Leu Arg 20 8314PRTHomo
sapiens 83Thr Ile Met Ile Thr Glu Ala Gly Ile Ser Lys Ala Glu Lys 1
5 10 8413PRTHomo sapiens 84Glu
Arg Ala Val Glu Ser Asp Cys Tyr Ala Glu Gln Val 1 5
10 8517PRTHomo sapiens 85Cys Val Ala Val Gly Gly
His Ser Gly Ser Leu Leu Ile Gln His Val 1 5
10 15 Met 8624PRTHomo sapiens 86Ala Arg Trp Thr
Phe Gly Gly Arg Asp Leu Pro Ala Glu Gln Pro Gly 1 5
10 15 Ser Phe Leu Tyr Asp Ala Arg Leu
20 8716PRTHomo sapiens 87Arg Ala Ile Cys Glu Glu
Thr Lys Ala Ser Ile Asp Ile Glu Asp Asp 1 5
10 15 8823PRTHomo sapiens 88Lys Val Thr Asp Ile
Leu Lys Glu Gly Gln Glu Val Glu Val Leu Val 1 5
10 15 Leu Asp Val Asp Asn Arg Gly
20 8915PRTHomo sapiens 89Lys Met Leu Thr Gly Val Asn Val Leu
Ala Asp Ala Val Lys Ala 1 5 10
15 9013PRTHomo sapiens 90Val Thr Asp Ile Leu Lys Glu Gly Gln Glu
Val Glu Val 1 5 10
9111PRTHomo sapiens 91Glu Thr Gln Ala Asp Pro Met Ala Phe Val Lys 1
5 10 9210PRTHomo sapiens 92Lys Thr Ala Val
Ala Pro Ile Glu Arg Val 1 5 10
9316PRTHomo sapiens 93Ser Gly Gln Val Pro Arg Tyr Thr Gly Ile Val Asn Cys
Phe Val Arg 1 5 10 15
9411PRTHomo sapiens 94Lys Asp Thr Ile Lys Gly Leu Phe Pro Lys Tyr 1
5 10 9521PRTHomo sapiens 95Gly Gly Pro
Met Ala Leu Tyr Gln Gly Phe Gly Val Ser Val Gln Gly 1 5
10 15 Ile Ile Val Tyr Arg
20 9610PRTHomo sapiens 96Lys Gly Val Leu Phe Lys Asp Glu Arg Thr 1
5 10 9712PRTHomo sapiens 97Val Ala Gln Gln
Glu Gly Met Lys Ala Phe Phe Lys 1 5 10
9812PRTHomo sapiens 98Phe Ile Asn Pro Asn Ala Val Ser Ser Ala Ser
Glu 1 5 10 9919PRTHomo sapiens
99Arg Ala Ala Val Glu Glu Gly Val Val Pro Gly Gly Gly Val Ala Leu 1
5 10 15 Ile Arg Ala
10012PRTHomo sapiens 100Phe Pro Ala Lys Leu Met Leu Gly Leu Leu Ile Phe 1
5 10 1019PRTHomo sapiens 101Phe
Leu Val Glu Thr Gly Phe His His 1 5
1027PRTHomo sapiens 102Ile Cys Asn Pro Lys Pro Gln 1 5
10312PRTHomo sapiens 103Pro Gln Ala Leu His Phe Met Leu Pro Ala Arg
Met 1 5 10 1045PRTHomo sapiens
104Pro Pro Arg Pro Pro 1 5 10513PRTHomo sapiens 105Gly
Pro Thr Ser Arg His Ala Pro Gly Phe Gly Ala Pro 1 5
10 10613PRTHomo sapiens 106Val Val Pro Ala Thr Gln
Glu Ala Glu Ala Gly Glu Trp 1 5 10
10715PRTHomo sapiens 107Pro Pro Ala Ser Arg Ala Ser Arg Gln Ser Arg
Val Glu Gly Lys 1 5 10
15 10811PRTHomo sapiens 108Ile Thr Tyr Gly Ser Arg Asn Thr Leu Val Lys 1
5 10 10912PRTHomo sapiens 109Tyr Arg
Asn Leu Leu His Val Glu Asn Thr Glu Arg 1 5
10 1106PRTHomo sapiens 110Phe Asn Phe His Glu Met 1
5 1116PRTHomo sapiens 111Pro Gly Tyr Phe Phe Tyr 1
5 1125PRTHomo sapiens 112Pro Pro Trp Arg Pro 1 5
1136PRTHomo sapiens 113Glu Trp Trp Arg Leu Asp 1 5
1145PRTHomo sapiens 114Lys Asn Ser Leu Asp 1 5
1158PRTHomo sapiens 115Met His Ile Glu Ser Leu Asp Ser 1 5
1165PRTHomo sapiens 116Leu Glu Gln Val Glu 1 5
1179PRTHomo sapiens 117Leu His Leu Glu Ser Leu Lys Asp Ser 1
5 1188PRTHomo sapiens 118Glu Ala Val Ser Leu Lys Pro
Thr 1 5 11930DNAArtificial
Sequenceoligonucleotide 119atgtatatta tatatctaaa gttataccaa
3012036DNAArtificial Sequenceoligonucleotide
120tttcctgcta agcttatgtt gggattgctt atattt
3612136DNAArtificial Sequenceoligonucleotide antisense (histatine)
121aaatataagc aatcccaaca taagcttagc aggaaa
3612219RNAArtificial Sequenceinsert of Si RNA (PKC epsilon) 122guuguagccu
ggaccuuga
19123420DNAArtificial SequenceClone S45T9T7 extracted from MCF7 cells
123taaatataag caatcccaac actttgggag gccgaggcgg gcggatcacg aggtcaggag
60atggagacca tcctggctaa cacagtgaaa ccctgtctct actgaaaata caaaaaagta
120gccgggcgtg gcggcaggcg cctgtagccc cagctactca ggaggctgag gcaggagaat
180ggcatgaacc caggaggcag agcttgcagt gagccgagat tgtgccactg cactccagcc
240tgggcaacag agcgagactc catctcaaaa aaaaaaaaaa aatcacccca aagcaataag
300gagaactaga acaggacata cactccaaca ctggtgaaac taggaaaaca tatgtaaccc
360caaaccacaa tatatacaca caaaactata cgagatgttg ggattgctta tatttaatcn
420124456DNAArtificial Sequenceantisense sequences of clone S55M1 M13
124nnnnnngntc cggccgccat ggcggccgcg ggattcgatt aaatataagc aatcccaaca
60ctttgggagg ccgaggcggg cggatcacga ggtcaggaga tggagaccat cctggctaac
120acagtgaaac cctgtctcta cggaaaatac aaaaaagtag ccgggcgtgg cggcaggcgc
180ctgtagtccc agctactcag gaggctgagg caggagaatg gcatgaaccc aggaggcaga
240gcttgcagtg agccgagatt gtgccactgc actccagcct gggcaacaga gcgagactcc
300atctcaaaaa aaaaaaaaaa tcaccccaaa gcaataagga gaactagaac aggacataca
360ctccaacact ggtgaaacta ggaaaacata tgtaacccca aaccacaata tatacacaca
420aaactatacg agatgttggg attgcttata tttaat
456125587DNAArtificial Sequenceantisense sequence of clone S55T9 M13
(MCF7 cells) 125nnnnnnnnng nnnnngnncc ntggcggccg cgggattcga ttaaatataa
gcaatcccaa 60cactttgggg gggtgaggcg gacagatcac ttgaggtcag gggtttgaga
ccagcatggc 120caacgtggtg aaaactcaac tactcaaaat agaaaaatta gctggacatg
gtggcacaca 180cctgtgaagc cagctactca ggaggctgaa gcatgagaat tgcttgaacc
ctggagatgg 240aggttacagt gagcccacgt cgcgtccctg cacgcaagcc taggcaagaa
agcaagaccc 300tgtctcaaaa aaagaaaaga gatgctgata catgctacaa catagatgaa
ccttgaggac 360attattctaa gtgaaatgag cttgtcacaa aagaacaaat attgcatgat
tccagttata 420tgaggtgccc atagttgtca aattcacaaa gacaaaaagt ggcatggtcg
ttaccaaggg 480ctgggagaaa agaggaatgg tgagttagtg tttaattggt acagagtttc
agttttgcaa 540gatgaaaaga gttctggaga tgaatgttgg gattgcttat atttaat
587126514DNAArtificial Sequenceinsert of clone extracted from
MCF7 cells culture 126nnnnnnnngn ncnnnnanng nannncnact cactnnaggg
cgaattgggc ccgacgtcgc 60atgctcccgg ccgnnannnn ggccgcggga attcgattaa
atataagcaa tcccaacact 120ttgggaggcc gaggcgggcg gatcacgagg tcaggagatg
gagaccatcc tggctaacac 180agtgaaaccc tgtctctact gaaaatacaa aaaagtagcc
gggcgtggcg gcaggcgcct 240gtagtcccag ctactcagga ggctgaggca ggagaatggc
atgaacccag gaggcagagc 300ttgcagtgag ccgagattgt gccactgcac tccagcctgg
gcaacagagc gagactccat 360ctcaaaaaaa aaaaaaaatc accccaaagc aataaggaga
actagaacag gacatacact 420ccaacactgg tgaaactagg aaaacatatg taaccccaaa
ccacaatata tacacacaaa 480actatacgag atgttgggat tgcttatatt taat
514127770DNAArtificial SequenceC8 PGEMT M13
rev1605 clone (CNCM I-3940) 127tcctatgctt gactattagc tccttttctt
tacctattat tattatttat ttttcagatg 60gagtctcaca ctgtcgccag gctggagtac
agtggcgcca tctcggctca ctgaaccctc 120cacctaccgg gttcaagtga ttctactgcc
gcagtctcca gagtagctgg gactacaggc 180atgcactacc acacccagct aattttttgt
atttttagtt gagatggggt ttcaccatga 240tggccaggat ggtctcgatc tcttgacctc
atgatctgcc tgcctcagcc tcccaaagtg 300ctgggattac gggcatgagc cactgtgcta
ggcctaccta ttttaaaaat aacaagaatt 360catccaatgg aacgctgaaa tgcctgtgga
ggatacaaga ttaatcagca ggattatatt 420tgcctatgac tccattaatt caattccaga
ccctacttat gggtctaatg tcctttatta 480ggcaatgtag gagattccta tactcctaat
aaagtaatct ttttccattt aaatatcttt 540gctttttatc aggcagtatg tatttccaca
aagttaattc aatgaaatac acctaaagct 600gagagtctta tctcgttaat gaatgaaaaa
tagaaatcaa ttattctgag atgagattct 660tcctataatt tttattagtt gttgtgaaat
ggtacatggg tttgtcttga ntatgatgat 720gtattaattc ttaatattga antaaatgnc
tctgnagtan agatgactct 77012819DNAArtificial
SequenceArtificial Sequence 128tcctatgctt gactattag
1912923DNAArtificial SequenceOligonucleotide
for the PCR 129cctgacatcc ctacatcacc gca
2313019DNAArtificial SequenceOligonucleotide for the PCR
130atgtatatta tatatctaa
1913119DNAArtificial SequenceOligonucleotide for the PCR 131ttagatatat
aatatacat
1913220DNAArtificial SequenceOligonucleotide for the PCR 132aaatataagc
aatcccaaca
2013320DNAArtificial SequenceOligonucleotide for the PCR 133tgttgggatt
gcttatattt
2013418DNAArtificial SequenceOligonucleotide for the PCR 134ctttattatt
ttgtaaat
1813518DNAArtificial SequenceOligonucleotide for the PCR 135atttacaaaa
taataaag
1813619DNAArtificial SequenceOligonucleotide for the RT-PCR 136tcctatgctt
gactattgc
1913723DNAArtificial Sequencefrom RNAm Liv21 obtained by RT-PCR
137cctgacatcc ctacatcacc cat
2313822DNAArtificial SequenceOligonucleotide for the PCR 138atgttgggat
tgcttatatt ta
2213919DNAArtificial SequenceOligonucleotide for the PCR 139tatacgagat
gttggattg
1914019DNAArtificial SequenceOligonucleotide for the PCR 140tatacgagat
gttggattg
1914119DNAArtificial SequenceOligonucleotide for the PCR 141cattttgcac
accaaggtt
19142408DNAArtificial SequenceATGS1 insert of clone extracted from MCF7
cells 142taaatataag caatcccaac actttgggag gccgangngg gcggatcacg
aggtcaggag 60atggagacca tcctggctaa cacagtgaaa ccctgtctct actgaaaata
caaaaaagta 120gccgggcgtg gcggcaggcg cctgtagccc cagctactca ggaggctgag
gcaggagaat 180ggcatgaacc caggaggcag agcttgcagt gagccgagat tgtgccactg
cactccagcc 240tgggcaacag agcgagactc catctcaaaa aaaaaaaaaa tcaccccaaa
gcaataagga 300gaactagaac aggacataca ctccaacact ggtgaaacta ggaaaacata
tgtaacccca 360aaccacaata tatacacaca aaactatacg agatgttggg attgctta
4081431005DNAArtificial SequenceT6T7 clone extracted from
MCF7 cells culture 143tccggncgcc ntggcggccg cgggantcga ttaattgntt
tttttttttt tttttttttt 60tttttttttt tttttttttt tttttnnggn nntttttttt
cctttttttt tttttttttt 120tttttttttt tttttttttt tttttttttt ttttnncaaa
aaaccaaaaa cccagnnnan 180nnnancccta ggggnttncc ggccccccgg ggggccccca
aangggaaaa ccccccancg 240ggggggngnn aaanttggga nntnnnnagg ggcccccaaa
aanntggggg aaaaccgggg 300caaaanngnt tccgggggga aanttttnnc cccccaaaat
tccccnaaaa aaaaaaaccg 360gaaanaaaaa ggggaaancc gggggggncn aaaggggggn
nnaancccnn ataantgggg 420nggnccccnn ggcccctttn cnnncgggaa anccgggngg
gcncctggnt taaagaancc 480ccccaccccg ggggnnnngn ggtttggnna ttgggggccc
ttccccttcc ccccccannn 540anccccngcc cccggnngtt cggggggggg gannggnanc
ncnnccncnn aggggggaaa 600angggtatcc ccnaaaaccg gggaaaannc annaaaaaaa
atgnggnaaa aaggnccnna 660aaagggccnn aanccnaaaa angncccggn ngggggggtt
tttccanang nccccccccc 720ccgannannn nnnnnnnnan cnncncccnn nnnnnnngng
gggnaaaccc nnnagngann 780nnnanananc nnnnnttncc ccnngnnaan nnnnnnnnnn
nnnnnnnngn nnnnnnccnn 840nnnnnnnnan annaaannnn gnnnnnnnnn nnnnnnngnn
annngggggn nntttnnnnn 900nnnnnnnnnn nnnnngnnnn gnnnnnnnnn nnnnngnnnn
nnnnntnnnn nncnnnnnnn 960nnnngnaaan nccccnnntn nnnccnnana nnnnnnnnnc
nnnnn 1005144805DNAArtificial SequenceT7 sensclone
extracted from MCF7 cells 144ctccggccgc catggcggcc gcgggaattc gatttcctat
gcttgactat tagctgagat 60gncggtttcc tctccaaaga agtaagagga cagaaaaagt
catgctggtc ttagctctac 120aagagtatga atgcaggcag gtattaccat tcaatttcca
tgagatgtat ggcctacaat 180tcttaaaagg gtatctgagg ccgggtgcag tggctgatgc
ttgtaattcc agcattttgg 240gaggccaagg caggcagatc accgatgtca ggagtttgag
actagcctgg ccaacatggt 300gaaaccccgt ctctaccaaa aataaaaaaa atagccaggc
atggtggtgc gcacttgtag 360ttccagctac tctgcttggg aggctgaggc aggagaattg
cttgaacctg ggaggcagag 420gttgcagtga gccgagatcg tgccactgca ctccagcctg
ggcgacagat tgagactctg 480tatcaaaaaa aaaaaaaaaa aaaaaaaaaa aagaaagaaa
cacaaggctg gaggttcaca 540gagtatttat attggaaaat ctcatttaag ttaatatata
acattaacat tcaaaattca 600atgcaattta atcaaacaac tgtaactcca tgacaaagag
agtcatctct acttcagagg 660catttaattc aatattaaga attaatacat catcatatct
tcaagacaaa cccatgtacc 720atttcacaac aactaataaa aattataggg aagaatctca
tctcagaata anttgatttc 780tatttttcat tcattaacga gataa
805145806DNAArtificial SequenceT7 antisense
sequence of clone extracted from MCF7 145cttatctcgt taatgaatga
aaaatagaaa tcaanttatt ctgagatgag attcttccct 60ataattttta ttagttgttg
tgaaatggta catgggtttg tcttgaagat atgatgatgt 120attaattctt aatattgaat
taaatgcctc tgaagtagag atgactctct ttgtcatgga 180gttacagttg tttgattaaa
ttgcattgaa ttttgaatgt taatgttata tattaactta 240aatgagattt tccaatataa
atactctgtg aacctccagc cttgtgtttc tttctttttt 300tttttttttt tttttttttt
ttgatacaga gtctcaatct gtcgcccagg ctggagtgca 360gtggcacgat ctcggctcac
tgcaacctct gcctcccagg ttcaagcaat tctcctgcct 420cagcctccca agcagagtag
ctggaactac aagtgcgcac caccatgcct ggctattttt 480tttatttttg gtagagacgg
ggtttcacca tgttggccag gctagtctca aactcctgac 540atcggtgatc tgcctgcctt
ggcctcccaa aatgctggaa ttacaagcat cagccactgc 600acccggcctc agataccctt
ttaagaattg taggccatac atctcatgga aattgaatgg 660taatacctgc ctgcattcat
actcttgtag agctaagacc agcatgactt tttctgtcct 720cttacttctt tggagaggaa
accgncatct cagctaatag tcaagcatag gaaatcgaat 780tcccgcggcc gccatggcgg
ccggag 806146720DNAArtificial
Sequence2006M13rev2005 clone extracted from MCF7 cells 146tcaagctatg
catccaacgc gttgggagct ctcccatatg gtcgacctgc aggcggccgc 60gaattcacta
gtgatttcct atgcttgact attagctcct tttctttacc tattattatt 120atttattttt
cagatggagt ctcacactgt cgccaggctg gagtacagtg gcgccatctc 180ggctcactga
accctccacc taccgggttc aagtgattct actgccgcag tctccagagt 240agctgggact
acaggcatgc actaccacac ccagctaatt ttttgtattt ttagttgaga 300tggggtttca
ccatgatggc caggatggtc tcgatctctt gacctcatga tctgcctgcc 360tcagcctccc
aaagtgctgg gattacgggc atgagccact gtgctaggcc tacctatttt 420aaaaataaca
agaattcatc caatggaacg ctgaaatgcc tgtggaggat acaagattaa 480tcagcaggat
tatatttgcc tatgactcca ttaattcaat tccagaccct acttatgggt 540ctaatgtcct
ttattaggca atgtaggaga ttcctatact cctantaaag taatcttttt 600ccatttaaan
atctttgctt tttatcaggc agtatgtatt ttccnnaaag ttaattcaat 660gaantacacc
ctaaagctga nagtcttatc ctcgttaatg gaatggnnaa nanaaattca
720147783DNAArtificial SequenceC8sens1605 clone extracted from MCF7 cells
culture 147catgacaaag agagtcatct ntactncaga gncatttant tcaatattaa
gaattaatac 60atcatcatan tcaagacaaa cccatgtacc atttcacaac aactaataaa
aattatagga 120agaatctcat ctcagaataa ttgatttcta tttttcattc attaacgaga
taagactctc 180agctttaggt gtatttcatt gaattaactt tgtggaaata catactgcct
gataaaaagc 240aaagatattt aaatggaaaa agattacttt attaggagta taggaatctc
ctacattgcc 300taataaagga cattagaccc ataagtaggg tctggaattg aattaatgga
gtcataggca 360aatataatcc tgctgattaa tcttgtatcc tccacaggca tttcagcgtt
ccattggatg 420aattcttgtt atttttaaaa taggtaggcc tagcacagtg gctcatgccc
gtaatcccag 480cactttggga ggctgaggca ggcagatcat gaggtcaaga gatcgagacc
atcctggcca 540tcatggtgaa accccatctc aactaaaaat acaaaaaatt agctgggtgt
ggtagtgcat 600gcctgtagtc ccagctactc tggagactgc ggcagtagaa tcacttgaac
ccggtaggtg 660gagggttcag tgagccgaga tggcgccact gtactccagc ctggcgacag
tgtgagactc 720catctgaaaa ataaataata ataataggat aagaaaagga gctaatagtc
aagcatagga 780aat
7831481200DNAArtificial Sequenceclone LIV extracted from MCF7
cells culture 148ggagaagggt ccccactgct cctgtcaagc cttgttgtgt cgggacttga
actttattct 60aagcaggtga atgcggtgca tgcaagagag acagagagaa tgtggcagga
ccaaggagga 120ggctatgcca cttatgtcac tcctggcaaa aataaggggg catggagtag
gctgtttgtg 180gtgcagatgg tgagagcagt caggtccagc acagatttta aaggttggac
ccagagaatt 240tgctgcagaa tcagatgtgg ggtgtaaggc agagaggagt caagggcaac
ttcaggattt 300ggggccggaa ctgccattag acagacaggg acactggggg agaagcaggt
taggtgggat 360taaaatcaag agttcaagtt aagtttgagc agcctgttag acctccaacg
agggccagat 420agaagaatct ggtttccagg gagaggtcag gatgagagat acacacgtgg
gaatgattgg 480cattgggcgg actttatatt ctctgggcca gtgagacagc tgggaagtga
ccacggatag 540agaagagaca aagtcacaga aaccaagaga ggtaatgttg caaggacgga
acactcaact 600ctcaaatgct gctgagacgt gggctgaggg ctgagaatgg aattgggaag
aaccgaggtc 660actggtgatc ctgagggttt cagtggcaag ggcaggtgga ctgcagtggg
gcccggtggg 720gatcggtgga gcatgggccc ctctcccgga gagttgcact gtaaacgagg
gcagacatat 780gggagtgcag ctagagggag ggaacgtagg ctcaagggag agtttattct
gaatgagaga 840gatcacagct tgtttttagg ctgacgggca tgatccatag aggggaaagt
aattaagatg 900cagaagagag gccgggggtg gtggctcacg cctgtaatct cagcactttg
ggaggctcga 960ggtgggtgga tcatttgagg acaggagttc gagaccatcc tggccagcat
ggtgaaacct 1020cgcctctact aaaaataaaa ataaaaaaaa attagctggg tgcggtgnac
gggcacctgt 1080agtaccagct acttgggagg ctgaggtaac agaatcgctt gaaccctgga
ggcaggggtt 1140gcagtgagct gagattgtgc cactgcactc tagcctgggc aacaaattga
gactccactc 120014975PRTHomo sapiens 149Met Glu Ser Arg Ser Val Ala Gln
Ala Gly Val Gln Trp Pro Asp Leu 1 5 10
15 Gly Ser Leu Gln Pro Leu Pro Pro Arg Phe Lys Arg Phe
Phe Cys Leu 20 25 30
Ser Leu Gln Ser Ser Trp Asp Tyr Arg His Ala Pro Pro Arg Pro Ala
35 40 45 Asn Phe Val Phe
Leu Val Glu Thr Gly Phe Cys His Val Ser Gln Ala 50
55 60 Gly Leu Glu Leu Leu Thr Ser Ser
Asp Pro Pro 65 70 75 15075PRTHomo
sapienstranslation clone from C18 150Met Glu Ser Arg Ser Val Ala Gln Ala
Gly Val Gln Trp His Asn Leu 1 5 10
15 Gly Ser Leu Gln Ala Leu Pro Pro Gly Phe Met Pro Phe Ser
Cys Leu 20 25 30
Ser Leu Leu Ser Ser Trp Asp Tyr Arg Arg Leu Pro Pro Arg Pro Ala
35 40 45 Thr Phe Leu Phe
Leu Pro Arg Gln Gly Phe Thr Val Leu Ser Gln Ala 50
55 60 Arg Met Val Ser Ile Ser Pro Arg
Asp Pro Pro 65 70 75 151136PRTHomo
sapienstranslation clone from C18 151Leu Asn Ile Ser Asn Pro Asn Ile Ser
Tyr Ser Phe Val Cys Ile Tyr 1 5 10
15 Cys Gly Leu Gly Leu His Met Phe Ser Phe His Gln Cys Trp
Ser Val 20 25 30
Cys Pro Val Leu Val Leu Leu Ile Ala Leu Gly Phe Phe Phe Phe Phe
35 40 45 Glu Met Glu Ser
Arg Ser Val Ala Gln Ala Gly Val Gln Trp His Asn 50
55 60 Leu Gly Ser Leu Gln Ala Leu Pro
Pro Gly Phe Met Pro Phe Ser Cys 65 70
75 80 Leu Ser Leu Leu Ser Ser Trp Asp Tyr Arg Arg Leu
Pro Pro Arg Pro 85 90
95 Ala Thr Phe Leu Tyr Phe Pro Arg Gln Gly Phe Thr Val Leu Ala Arg
100 105 110 Met Val Ser
Ile Ser Pro Arg Asp Pro Pro Ala Ser Ala Ser Gln Ser 115
120 125 Val Gly Ile Ala Tyr Ile Ser Asn
130 135 152133PRTHomo sapiens 152Leu Ser Ser Ser
Asp Pro Pro Ala Ser Ala Ser Gln Ser Val Gly Ile 1 5
10 15 Thr Gly Val Gly His His Thr Gln Leu
His Gly Tyr Phe Phe His Leu 20 25
30 Phe Tyr Phe Ile Leu Phe Tyr Tyr Leu Phe Phe Glu Thr Glu
Ser His 35 40 45
Cys Val Ala Gln Ala Gly Val Gln Trp Arg Gly Leu Gly Ser Leu Gln 50
55 60 Ala Ser Pro Pro Arg
Phe Thr Pro Phe Ser Cys Leu Ser Leu Pro Ser 65 70
75 80 Ser Trp Asp Tyr Arg Arg Leu Pro Pro Cys
Pro Ala Asn Phe Ser Tyr 85 90
95 Phe Tyr Gln Asn Gly Phe Tyr His Val Gly Gln Ala Gly Leu Glu
Phe 100 105 110 Leu
Thr Ser Gly Asp Pro Pro Ala Ser Ala Ser Gln Ser Ala Gly Ile 115
120 125 Thr Gly Met Ser His
130 15393PRTHomo sapienstranslation clone from C18 153Phe Phe
Phe Phe Phe Glu Met Glu Ser Arg Ser Val Ala Gln Ala Gly 1 5
10 15 Val Gln Trp His Asn Leu Gly
Ser Leu Gln Ala Leu Pro Pro Gly Phe 20 25
30 Met Pro Phe Ser Cys Leu Ser Leu Leu Ser Ser Trp
Asp Tyr Arg Arg 35 40 45
Leu Pro Pro Arg Pro Ala Thr Phe Leu Tyr Phe Pro Arg Gln Gly Phe
50 55 60 Thr Val Leu
Ala Arg Met Val Ser Ile Ser Pro Arg Asp Pro Pro Ala 65
70 75 80 Ser Ala Ser Gln Ser Val Gly
Ile Ala Tyr Ile Ser Asn 85 90
154100PRTHomo sapiens 154Phe Phe Phe Phe Phe Glu Thr Glu Ser Arg Ser
Val Ala Gln Ala Gly 1 5 10
15 Val Gln Trp Arg Asp Leu Gly Ser Leu Gln Ala Leu Pro Pro Gly Phe
20 25 30 Thr Pro
Phe Ser Cys Leu Ser Leu Pro Ser Ser Trp Asp Tyr Arg Arg 35
40 45 Leu Pro Pro Leu Ala Pro Ala
Asn Phe Phe Leu Ile Phe Val Phe Leu 50 55
60 Val Glu Thr Gly Phe His His Val Gly Gln Ala Gly
Leu Glu Leu Leu 65 70 75
80 Thr Ser Gly Asp Pro Pro Ala Ser Ala Ser Gln Ser Ala Gly Ile Thr
85 90 95 Gly Val Ser
His 100 155149PRTHomo sapiens 155Met Glu Ser His Ser Val Thr
Gln Ala Gly Val Gln Trp Cys Gly Leu 1 5
10 15 Gly Ser Leu Gln Pro Leu Pro Pro Gly Phe Lys
Arg Phe Phe Cys Leu 20 25
30 Ser Leu Leu Ser Ser Trp Asp Tyr Arg Arg Val Pro Pro Pro Leu
Ala 35 40 45 Asn
Phe Cys Ile Phe Phe Ser Phe Phe Phe Glu Lys Glu Ser Leu Ser 50
55 60 Val Thr Gln Ala Gly Val
Gln Trp His Asp Leu Gly Ser Leu Gln Ala 65 70
75 80 Ala Pro Pro Gly Phe Thr Pro Phe Ser Cys Leu
Ser Leu Pro Ser Ser 85 90
95 Trp Asn Tyr Arg Arg Pro Pro Pro Cys Pro Ala Asn Phe Phe Val Phe
100 105 110 Leu Val
Glu Met Gly Phe Cys His Ile Gly Gln Ala Gly Leu Glu Leu 115
120 125 Leu Thr Ser Ser Asp Pro Pro
Thr Ser Ala Ser Gln Ser Ala Gly Ile 130 135
140 Thr Gly Met Ser His 145
1561132DNAHomo sapiens 156ctagtgattt cctatgcttg actattagct ccttttcttt
acctattatt attatttatt 60tttcagatgg agtctcacac tgtcgccagg ctggagtaca
gtggcgccat ctcggctcac 120tgaaccctcc acctaccggg ttcaagtgat tctactgccg
cagtctccag agtagctggg 180actacaggca tgcactacca cacccagcta attttttgta
tttttagttg agatggggtt 240tcaccatgat ggccaggatg gtctcgatct cttgacctca
tgatctgcct gcctcagcct 300cccaaagtgc tgggattacg ggcatgagcc actgtgctag
gcctacctat tttaaaaata 360acaagaattc atccaatgga acgctgaaat gcctgtggag
gatacaagat taatcagcag 420gattatattt gcctatgact ccattaattc aattccagac
cctacttatg ggtctaatgt 480cctttattag gcaatgtagg agattcctat actcctaata
aagtaatctt tttccattta 540aatatctttg ctttttatca ggcagtatgt atttccacaa
agttaattca atgaaataca 600cctaaagctg agagtcttat ctcgttaatg aatgaaaaat
agaaatcaat tattctgaga 660aatactctgt gaacctccag ccttgtgttt ctttcttttt
tttttttttt tttttttttt 720tttgatacag agtctcaatc tgtcgcccag gctggagtgc
agtggcacga tctcggctca 780ctgcaacctc tgcctcccag gttcaagcaa ttctcctgcc
tcagcctccc aagcagagta 840gctggaacta caagtgcgca ccaccatgcc tggctatttt
ttttattttt ggtagagacg 900gggtttcacc atgttggcca ggctagtctc aaactcctga
catcggtgat ctgcctgcct 960tggcctccca aaatgctgga attacaagca tcagccactg
cacccggcct cagataccct 1020tttaagaatt ctaggccata catctcatgg aaattgaatg
gtaatacctg cctgcattca 1080tactcttgta gagctaagac cagcatgact ttttctgtcc
tcttacttct tt 11321571317DNAArtificial sequenceinsert in PGEMT
clone C18 extracted from MCF7 cells 157ctagtgattt cctatgcttg
actattagct ccttttcttt acctattatt attatttatt 60tttcagatgg agtctcacac
tgtcgccagg ctggagtaca gtggcgccat ctcggctcac 120tgaaccctcc acctaccggg
ttcaagtgat tctactgccg cagtctccag agtagctggg 180actacaggca tgcactacca
cacccagcta attttttgta tttttagttg agatggggtt 240tcaccatgat ggccaggatg
gtctcgatct cttgacctca tgatctgcct gcctcagcct 300cccaaagtgc tgggattacg
ggcatgagcc actgtgctag gcctacctat tttaaaaata 360acaagaattc atccaatgga
acgctgaaat gcctgtggag gatacaagat taatcagcag 420gattatattt gcctatgact
ccattaattc aattccagac cctacttatg ggtctaatgt 480cctttattag gcaatgtagg
agattcctat actcctaata aagtaatctt tttccattta 540aatatctttg ctttttatca
ggcagtatgt atttccacaa agttaattca atgaaataca 600cctaaagctg agagtcttat
ctcgttaatg aatgaaaaat agaaatcaat tattctgaga 660tgagattctt cctataattt
ttattagttg ttgtgaaatn nncatgggtt tgtcttgana 720natgatgatg tattaattct
naatattgna ntaaatgnct ctgnagtana gatgactctn 780ntttnntcat ggnnnnnacn
nnnnttgnnn annngcattg nnnnntgnan gnnnnngtnn 840nnnnntnnnn nnnnnnnnna
tttncaaatt cactagtgat ttcctatgct tgactattag 900ctccttttct ttacctatta
ttattattta tttttcagat ggagtctcac actgtcgcca 960ggctggagta cagtggcgcc
atctcggctc actgaaccct ccacctaccg ggttcaagtg 1020attctactgc cgcagtctcc
agagtagctg ggactacagg catgcactac cacacccagc 1080taattttttg tatttttagt
tgagatgggg tttcaccatg atggccagga tggtctcgat 1140ctcttgacct catgatctgc
ctgcctcagc ctcccaaagt gctgggatta cgggcatgag 1200ccactgtgct aggcctacct
attttaaaaa taacaagaat tcatccaatg gaacgctgaa 1260atgcctgtgg aggatacaag
attaatcagc acgattatat ttgcctatga ctccatt 131715892PRTHomo
sapienstranslation clone from C18 158Phe Phe Phe Phe Phe Glu Met Glu Ser
Arg Ser Val Ala Gln Ala Gly 1 5 10
15 Val Gln Trp His Leu Leu Gly Ser Leu Gln Ala Leu Pro Pro
Gly Phe 20 25 30
Met Pro Phe Ser Cys Leu Ser Leu Leu Ser Ser Trp Asp Tyr Arg Arg
35 40 45 Leu Pro Pro Arg
Pro Ala Thr Phe Leu Tyr Phe Pro Arg Gln Gly Phe 50
55 60 Thr Val Leu Ala Arg Met Val Ser
Ile Ser Pro Arg Asp Pro Pro Ala 65 70
75 80 Ser Ala Gln Ser Val Gly Ile Ala Tyr Ile Ser Asn
85 90 15953PRTHomo
sapienstranslation clone from C18 159Met Glu Ser Arg Ser Val Ala Gln Ala
Gly Val Gln Trp His Asn Leu 1 5 10
15 Gly Ser Leu Gln Ala Leu Pro Pro Gly Phe Met Pro Phe Ser
Cys Leu 20 25 30
Ser Leu Leu Ser Ser Trp Asp Tyr Arg Arg Leu Pro Pro Arg Pro Ala
35 40 45 Thr Phe Leu Tyr
Phe 50 160922DNAHomo sapiens 160gcttgactat tagctccttt
tctttaccta ttattattat ttattttcag atggagtcta 60cactgtcgcc aggctggagt
acagtggcgc catctcggct cactgaaccc tccacctacc 120gggttcaagt gattctactg
ccgcagtctc cagagtagtg ggactacagg catgcactac 180cacacccagc taattttttg
tatttttagt tgagatgggg tttcaccatg atggccagga 240tggtctcgat ctcttgacct
catgatctgc ctgcctcagc ctcccaaagt gctgggatta 300cgggcatgag ccactgtgct
aggcctacct attttaaaaa taacaagaat tcatccaatg 360gaacgctgaa atgcctgtgg
aggatacaag attaatcagc aggattatat ttgcctatga 420ctccattaat tcaattccag
accctactta tcacttatgg gtctaatgtc ctttattagg 480caatgtagga gattcctata
ctcctaataa agtaatcttt tccatttaaa tatctttgct 540tttatcaggc agtatgtatt
tccacaaagt taattcaatg aaatacacct aaagctgaga 600gtcttatctc gttaatgaat
gaaaaataga aatcaattat tctgagatga gattcttcct 660ataattttta ttagttgttg
tgaaatggta catgggtttg tcttgaagat atgatgatgt 720attaattctt aatattgaat
taaatgcctc tgaagtagag atgactctct ttgtcatgga 780gttacagttg tttgattaaa
ttgcattgaa ttttgaatgt taatgttata tattaactta 840aatgagattt tccaatataa
atactctgtg aacctccagc cttgtgtttc tttctttttt 900tttttttttt tttttttttt
tt 922161921DNAHomo sapiens
161gcttgactat tagctccttt tctttaccta ttattattat ttatttttca gatggagtct
60cacactgtcg ccaggctgga gtacagtggc gccatctcgg ctcactgaac cctccaccta
120ccgggttcaa gtgattctac tgccgcagtc tccagagtag ctgggactac aggcatgcac
180taccacaccc agctaatttt ttgtattttt agttgagatg gggttttcac catgatggcc
240aggatggtct cgatctcttg acctcatgat ctgcctgcct cagcctccca aagtgctggg
300attacgggca tgagccactg tgctaggcct acctatttta aaaataacaa gaattcatcc
360aatggaacgc tgaaatgcct gtggaggata caagattaat cagcaggatt atatttgcct
420atgactccat taattcaatt ccagacccta cttagggtct aatgtccttt attaggcaat
480gtaggagatt cctatacttc taataaagta atctttttcc atttaaatag tctttgcttt
540ttatcaggca gtatgtattt ccacaaagtt aattcaatga aatacaccta aagctgagag
600tcttatctcg ttaatgaatg aaaaatagaa atcaattatt ctgagatgag attcttccta
660taatttttat tagttgttgt gaaatggtac atgggtttgt cttgaagata tgatgatgta
720ttaattctta atattgaatt aaatgcctct gaagtagaga tgactctctt tgtcatggag
780ttacagttgt ttgattaaat tgcattgaat tttgaatgtt aatgttatat attaacttaa
840atgagatttt ccaatataaa tactctgtga acctccagcc ttgtgtttct ttcttttttt
900tttttttttt tttttttttt t
92116246PRTHomo sapiens 162Met Lys Phe Phe Val Phe Ala Leu Ile Leu Ala
Leu Met Leu Ser Met 1 5 10
15 Thr Gly Ala Asp Ser His Ala Lys Arg His His Gly Tyr Lys Arg Lys
20 25 30 Phe His
Glu Lys His His Ser His Gln Gly Tyr Pro Ser Asn 35
40 45 16314PRTHomo sapiens 163Met Lys Phe Phe Val
Phe Ala Leu Ile Leu Ala Leu Met Leu 1 5
10 1641199DNAHomo sapiens 164ggagaagggt ccccactgct
cctgtcaagc cttgttgtgt cgggacttga actttattct 60aagcaggtga atgcggtgca
tgcaagagag acagagagaa tgtggcagga ccaaggagga 120ggctatgcca cttatgtcac
tcctggcaaa aataaggggg catggagtag gctgtttgtg 180gtgcagatgg tgagagcagt
caggtccagc acagatttta aaggttggac ccagagaatt 240tgctgcagaa tcagatgtgg
ggtgtaaggc agagaggagt caagggcaac ttcaggattt 300ggggccggaa ctgccattag
acagacagag ggacactggg ggagaagcag gttaggtggg 360attaaaatca agagttcaag
ttaagtttga gcagcctgtt agacctccag cgagggccag 420atagaagaat ctggtttcca
ggagaggtca ggatgagaga tacacacgtg ggaatgattg 480gcattgggtg gactttatat
tctctgggtc gtgagacagc tgggaagtga ccacggatag 540agaagagaca aagtcacaga
aaccaagaga ggtaatgttg caaggacgga acactcaact 600ctcaaatgct gctgagacgt
gggctgaggg ctgagaatgg aattgggaag aaccgaggtc 660actggtgatc ctgagggttt
cagtggcaag ggcaggtgga ctgcagtggg gcccggtggg 720gatcggtgga gcatgggccc
ctctcccgga gagttgcact gtaaacgagg gcagacatat 780gggagtgcag ctagagggag
ggaacgtagg gtcaagggag agtttattct gaatgagaga 840gatcacagct tgtttttagg
ctgacgggca tgatccatag aggggaaagt aattaagatg 900cagaagagag gccgggggtg
gtggctcacg cctgtaatct cagcactttg ggaggctgag 960gtgggtggat catttgagga
caggagttcg agaccatcct ggccagcatg gtgaaaactc 1020gcctctacta aaaataaaaa
taaaaaaaaa ttagctgggt gtggtggagg gcacctgtag 1080taccagctac ttgggaggct
gaggtaacag aatcgcttga accctggagg caggggttgc 1140agtgagctga gattgtgcca
ctgcactcta gcctgggcaa caaattgaga ctccatctc 11991651199DNAHomo sapiens
165ggagaagggt ccccactgct cctgtcaagc cttgttgtgt cgggacttga actttattct
60aagcaggtga atgcggtgca tgcaagagag acagagagaa tgtggcacga ccaaggagga
120ggctatgcca cttatgtcac tcctggcaaa aataaggggg catggagtag gctgtttgtg
180gtgcagatgg tgagagcagt caggtccagc acagatttta aaggttggac ccagagaatt
240tgctgcagaa tcagatgtgg ggtgtaaggc agagaggagt caagggcaac ttcaggattt
300ggggccggaa ctgccattag acagacaggg acactggggg agaagcaggt taggtgggat
360taaaatcaag agttcaagtt aagtttgagc agcctgttag acctccaacg agggccagat
420agaagaatct ggtttccagg gagaggtcag gatgagagat acacacgtgg gaatgattgg
480cattgggcgg actttatatt ctctgggcca gtgagacagc tgggaagtga ccacggatag
540agaagagaca aagtcacaga aaccaagaga ggtaatgttg caaggacgga acactcaact
600ctcaaatgct gctgagacgt gggctgaggg ctgagaatgg aattgggaag aaccgaggtc
660actggtgatc ctgagggttt cagtggcaag ggcaggtgga ctgcagtggg gcccggtggg
720gatgggtgga gcatgggccc ctctcccgga gagttgcact gtaaacgagg gcagacatat
780gggagtgcag ctagagggag ggaacgtagg gtcaagggag agtttattct gaatgagaga
840gatcacagct tgtttttagg ctgacgggca tgatccatag aggggaaagt aattaagatg
900cagaagagag gccgggggtg gtggctcacg cctgtaatct cagcactttg ggaggctgag
960gtgggtggat catttgagga caggagttcg agaccatcct ggccagcatg gtgaaaactc
1020gcctctacta aaaataaaaa taaaaaaaaa ttagctgggt gcggtggagg gcacctgtag
1080taccagctac ttgggaggct gaggtaacag aatcgcttga accctggagg caggggttgc
1140agtgagctga gattgtgcca ctgcactcta gcctgggcaa caaattgaga ctccatctc
1199166261PRTHomo sapienstranslation clone from C18-F3 166Ser Asp Phe Leu
Cys Leu Thr Ile Ser Ser Phe Ser Leu Pro Ile Ile 1 5
10 15 Ile Ile Tyr Phe Ser Asp Gly Val Ser
His Cys Arg Gln Ala Gly Val 20 25
30 Gln Trp Arg His Leu Gly Ser Leu Asn Pro Pro Pro Thr Gly
Phe Lys 35 40 45
Phe Tyr Cys Arg Ser Leu Gln Ser Ser Trp Asp Tyr Arg His Ala Leu 50
55 60 Pro His Pro Ala Asn
Phe Leu Tyr Phe Leu Arg Trp Gly Phe Thr Met 65 70
75 80 Met Ala Arg Met Val Ser Ile Ser Pro His
Asp Leu Pro Ala Ser Ala 85 90
95 Ser Gln Ser Ala Gly Ile Thr Gly Met Ser His Cys Ala Arg Pro
Thr 100 105 110 Tyr
Phe Lys Asn Asn Lys Asn Ser Ser Asn Gly Thr Leu Lys Cys Leu 115
120 125 Trp Arg Ile Gln Asp Ser
Ala Gly Leu Tyr Leu Pro Met Thr Pro Leu 130 135
140 Ile Gln Phe Gln Thr Leu Leu Met Gly Leu Met
Ser Phe Ile Arg Gln 145 150 155
160 Cys Arg Arg Phe Leu Tyr Ser Ser Asn Leu Phe Pro Phe Lys Tyr Leu
165 170 175 Cys Phe Leu Ser
Gly Ser Met Tyr Phe His Lys Val Asn Ser Met Lys 180
185 190 Tyr Thr Ser Glu Ser Tyr Leu Val Asn Glu
Lys Ile Glu Ile Asn Tyr 195 200
205 Ser Glu Met Arg Phe Phe Leu Phe Leu Leu Val Val Val Lys
Xaa Xaa 210 215 220
Trp Val Cys Leu Xaa Xaa Cys Ile Asn Ser Xaa Tyr Xaa Xaa Lys Xaa 225
230 235 240 Leu Xaa Ser Xaa Asp
Asp Ser Xaa Xaa Xaa Met Xaa Thr Xaa His Xaa 245
250 255 Val Xaa Phe Xaa Lys 260 167144PRTHomo
sapiensmisc_featuretranslation clone from S55M-R2 167Leu Asn Ile Ser Asn
Pro Asn Ile Ser Tyr Ser Phe Val Cys Ile Tyr 1 5
10 15 Cys Gly Leu Gly Leu His Met Phe Ser Phe
His Gln Cys Trp Ser Val 20 25
30 Cys Pro Val Leu Val Leu Leu Ile Ala Leu Gly Phe Phe Phe Phe
Phe 35 40 45 Glu
Met Glu Ser Arg Ser Val Ala Gln Ala Gly Val Gln Trp His Asn 50
55 60 Leu Gly Ser Leu Gln Ala
Leu Pro Pro Gly Phe Met Pro Phe Ser Cys 65 70
75 80 Leu Ser Leu Leu Ser Ser Trp Asp Tyr Arg Arg
Leu Pro Pro Arg Pro 85 90 95
Ala Thr Phe Leu Tyr Phe Pro Arg Gln Gly Phe Thr Val Leu Ala Arg 100
105 110 Met Val Ser Ile Ser Pro
Arg Asp Pro Pro Ala Ser Ala Ser Gln Ser 115 120
125 Val Gly Ile Ala Tyr Ile Ser Asn Pro Ala Ala
Ala Met Ala Ala Gly 130 135 140
16813PRTHomo sapiens 168Ile Leu Ala Leu Met Leu Ser Met Cys Gly
Ala Asp Ser 1 5 10
169277PRTHomo sapiens 169Met Tyr Ile Ile Tyr Leu Lys Leu Tyr Gln Ala Pro
Leu Lys Lys Ser 1 5 10
15 Glu Arg Val Leu Arg Ala Val Pro Thr Ser Leu His Leu His Ile His
20 25 30 Ala Ser Leu
Tyr Pro Asp Ile Pro Thr Ser Pro His Pro Tyr Ile Thr 35
40 45 Val Ser Pro His Pro Cys Ile Leu
Ile Ser Pro His Ile Ser Leu Ser 50 55
60 Pro His Pro His Phe Pro Ala Pro Ser Ser Cys Arg Thr
Leu Leu Leu 65 70 75
80 Pro Ala Val Ser Arg Gln Gln Arg Val Pro Ser Pro Ser His Tyr Gly
85 90 95 Val His Met Asp Cys Gly
Leu Met Glu Pro Pro Ser Arg Glu Ser Ile 100 105
110 Ala Phe Pro Asp Arg Phe Pro Ala Lys Leu Met Leu Gly
Leu Leu Ile 115 120 125
Phe Thr Glu Ile Thr Cys Cys Glu Arg Arg Arg Val Tyr Gly Pro Leu 130
135 140 Thr Asn Pro Lys
Pro Gln Ser Leu Ser Ala Ile Asn Pro Lys Gly Gly 145 150
155 160 Thr Glu Asn Ser Arg Leu Phe Trp Gly
Ser Asn Cys Cys Leu Glu Val 165 170
175 Phe Leu Ser Tyr Arg Cys Val Thr Leu Ser Ala Gln Gly Arg Gly Thr
180 185 190 Pro Lys Pro Gly
Leu Ser Gly Ala Pro Cys Thr Asp Arg Arg Ala Met 195
200 205 Ser Phe Phe His Cys Tyr Arg Ser Ile
Leu His Thr Lys Val Gly Phe 210 215
220 Tyr Val Lys Ile Lys Lys Lys Lys Arg Lys Lys Lys Arg
Asn Asn Asn 225 230 235
240 Asn Asn Asn Lys Asn Asn Asn Asn Lys Tyr Ile Tyr Ile Leu Lys Arg
245 250 255 Lys Glu Asn Lys Ala Lys
Thr Cys Asn Thr Val Phe Ser Arg Arg Asn 260 265
270 Lys Ser Thr Asn Lys 275 170582PRTHomo
sapiens 170Met Thr Lys Phe Tyr Cys Gly Trp Asp Arg Leu Leu Glu Ser Leu
Pro 1 5 10 15 Asp
Gly Trp Val Tyr Cys Asp Ala Asp Gly Ser Gln Phe Asp Ser Ser
20 25 30 Leu Ser Pro Tyr Leu
Ile Asn Ala Val Leu Asn Ile Arg Leu Gly Phe 35
40 45 Met Glu Glu Trp Asp Val Gly Glu Ile
Met Leu Arg Asn Leu Tyr Thr 50 55
60 Glu Ile Val Tyr Thr Pro Ile Ser Thr Pro Asp Gly Thr
Leu Val Lys 65 70 75
80 Lys Phe Lys Gly Asn Asn Ser Gly Gln Pro Ser Thr Val Val Asp Asn
85 90 95 Thr Leu Met Val
Ile Leu Ala Val Asn Tyr Ser Leu Lys Lys Ser Gly 100
105 110 Ile Pro Ser Glu Leu Arg Asp Ser Ile
Ile Arg Phe Phe Val Asn Gly 115 120
125 Asp Asp Leu Leu Leu Ser Val His Pro Glu Tyr Glu Tyr Ile
Leu Asp 130 135 140
Thr Met Ala Asp Asn Phe Arg Glu Leu Gly Leu Lys Tyr Thr Phe Asp 145
150 155 160 Ser Arg Thr Arg Glu
Lys Gly Asp Leu Trp Phe Met Ser His Gln Gly 165
170 175 His Lys Arg Glu Gly Ile Trp Ile Pro Lys
Leu Glu Pro Glu Arg Ile 180 185
190 Val Ser Ile Leu Glu Trp Asp Arg Ser Lys Glu Pro Cys His Arg
Leu 195 200 205 Glu
Ala Ile Cys Ala Ala Met Ile Glu Ser Trp Gly Tyr Asp Lys Leu 210
215 220 Thr His Glu Ile Arg Lys
Phe Tyr Ala Trp Met Ile Glu Gln Ala Pro 225 230
235 240 Phe Ser Ser Leu Ala Gln Glu Gly Lys Ala Pro
Tyr Ile Ala Glu Thr 245 250
255 Ala Leu Arg Lys Leu Tyr Leu Asp Lys Glu Pro Ala Gln Glu Asp Leu
260 265 270 Thr His
Tyr Leu Gln Ala Ile Phe Glu Asp Tyr Glu Asp Gly Ala Glu 275
280 285 Thr Cys Val Tyr His Gln Ala
Gly Glu Thr Pro Asp Ala Gly Leu Thr 290 295
300 Asp Glu Gln Lys Gln Ala Glu Lys Glu Lys Lys Glu
Arg Glu Lys Ala 305 310 315
320 Glu Lys Glu Arg Glu Arg Gln Lys Gln Leu Ala Leu Lys Lys Gly Lys
325 330 335 Asp Val Ala
Gln Glu Glu Gly Lys Arg Asp Arg Glu Val Asn Ala Gly 340
345 350 Thr Ser Gly Thr Phe Ser Val Pro
Arg Leu Lys Ser Leu Thr Ser Lys 355 360
365 Met Arg Val Pro Arg Tyr Glu Gln Arg Val Ala Leu Asn
Leu Asp His 370 375 380
Leu Ile Leu Tyr Thr Pro Glu Gln Thr Asp Leu Ser Asn Thr Arg Ser 385
390 395 400 Thr Arg Lys Gln
Phe Asp Thr Trp Phe Glu Gly Val Met Ala Asp Tyr 405
410 415 Glu Leu Thr Glu Asp Lys Met Gln Ile
Ile Leu Asn Gly Leu Met Val 420 425
430 Trp Cys Ile Glu Asn Gly Thr Ser Pro Asn Ile Asn Gly Met
Trp Val 435 440 445
Met Met Asp Gly Asp Asp Gln Val Glu Phe Pro Ile Lys Pro Leu Ile 450
455 460 Asp His Ala Lys Pro
Thr Phe Arg Gln Ile Met Ala His Phe Ser Asp 465 470
475 480 Val Ala Glu Ala Tyr Ile Glu Lys Arg Asn
Gln Asp Arg Pro Tyr Met 485 490
495 Pro Arg Tyr Gly Leu Gln Arg Asn Leu Thr Asp Met Ser Leu Ala
Arg 500 505 510 Tyr
Ala Phe Asp Phe Tyr Glu Met Thr Ser Arg Thr Pro Ile Arg Ala 515
520 525 Arg Glu Ala His Ile Gln
Met Lys Ala Ala Ala Leu Arg Gly Ala Asn 530 535
540 Asn Asn Leu Phe Gly Leu Asp Gly Asn Val Gly
Thr Thr Val Glu Asn 545 550 555
560 Thr Glu Arg His Thr Thr Glu Asp Val Asn Arg Asn Met His Asn Leu
565 570 575 Leu Gly
Val Lys Gly Leu 580 17124PRTHomo sapiens 171Lys Phe
Phe Val Phe Ala Leu Ile Leu Ala Leu Met Leu Ser Met Cys 1 5
10 15 Gly Ala Asp Ser His Ala Lys
Arg 20 17220PRTHomo sapiens 172Lys Ser Tyr
Met Ser Met Phe Leu Leu Leu Met Ala Ile Ser Cys Val 1 5
10 15 Leu Ala Lys Asp 20
17318PRTHomo sapiens 173Pro Leu Met Ile Ile His His Leu Asp Asp Cys Pro
His Ser Gln Ala 1 5 10
15 Leu Lys 17419DNAArtificial SequenceOligonucleotide for the PCR
174gtggtaaagc acccaggaa
1917520DNAArtificial SequenceOligonucleotide for the PCR 175gctagctgga
tgtcttttgc
2017619DNAArtificial SequenceOligonucleotide for the PCR 176tgaaggtcgg
agtcaacgg
1917718DNAArtificial SequenceOligonucleotide for the PCR 177catgtgggcc
atgaggtc
1817820DNAArtificial SequenceOligonucleotide for the PCR 178ggacttcgag
caagagatgg
2017920DNAArtificial SequenceOligonucleotide for the PCR 179agcactgtgt
tggcgtacag
2018019DNAArtificial SequenceOligonucleotide for the PCR 180tttttttttt
ttttttttt 19
User Contributions:
Comment about this patent or add new information about this topic: