Patent application title: ARTIFICIAL AND MUTATED NUCLEOTIDE SEQUENCES
Inventors:
Alexander Krichevsky (St. Louis, MO, US)
Assignees:
BIOGLOW INC.
IPC8 Class: AC12N1582FI
USPC Class:
435419
Class name: Chemistry: molecular biology and microbiology plant cell or cell line, per se (e.g., transgenic, mutant, etc.); composition thereof; process of propagating, maintaining, or preserving plant cell or cell line; process of isolating or separating a plant cell or cell line; process of regenerating plant cells into tissue, plant part, or plant, per se, where no genotypic change occurs; medium therefore plant cell or cell line, per se, contains exogenous or foreign nucleic acid
Publication date: 2014-09-18
Patent application number: 20140273224
Abstract:
The present invention relates to artificial nucleotide sequences,
including specific mutations therein.Claims:
1.-21. (canceled)
22. Any one, or more, nucleic acid(s), comprising a nucleotide sequence selected from the group consisting of SEQ ID NO:1, SEQ ID NO:2, SEQ ID NO:3, SEQ ID NO:4, SEQ ID NO:5, SEQ ID NO:6, SEQ ID NO:7, SEQ ID NO:8, SEQ ID NO:11, and SEQ ID NO:12.
23. A living cell, containing any one or more of said nucleotide sequences of claim 22.
24. (canceled)
25. (canceled)
26. (canceled)
27. (canceled)
28. The living cell of claim 23, which is selected from the group consisting of a bacterial cell and a plant cell.
29. The living cell of claim 28, which is autoluminescent.
30. A plant, a cell of which contains any one or more of said nucleotide sequences of claim 22.
31. The plant of claim 30, wherein said one or more nucleotide sequences are present in a plastid.
32. The plant of 31, wherein said plastid is a chloroplast.
33. The plant of claim 32, wherein said nucleotide sequences are expressed.
34. The plant of claim 33, which is autoluminescent.
35. Progeny of said plant of claim 34.
36. The progeny of claim 35, which are produced sexually or asexually.
37. The progeny of claim 36, which are produced asexually from cuttings.
38. A part of said plant of claim 34.
39. A part of said progeny of claim 35.
40. The part of said plant of claim 38, which is selected from the group consisting of a protoplast, a cell, a tissue, an organ, a cutting, and an explant.
41. The part of said plant of claim 40, which is selected from the group consisting of an inflorescence, a flower, a sepal, a petal, a pistil, a stigma, a style, an ovary, an ovule, an embryo, a receptacle, a seed, a fruit, a stamen, a filament, an anther, a male or female gametophyte, a pollen grain, a meristem, a terminal bud, an axillary bud, a leaf, a stem, a root, a tuberous root, a rhizome, a tuber, a stolon, a corm, a bulb, an offset, a cell of said plant in culture, a tissue of said plant in culture, an organ of said plant in culture, and a callus.
42. The part of said progeny of claim 39, which is selected from the group consisting of a protoplast, a cell, a tissue, an organ, a cutting, and an explant.
43. The part of said progeny of claim 42, which is selected from the group consisting of an inflorescence, a flower, a sepal, a petal, a pistil, a stigma, a style, an ovary, an ovule, an embryo, a receptacle, a seed, a fruit, a stamen, a filament, an anther, a male or female gametophyte, a pollen grain, a meristem, a terminal bud, an axillary bud, a leaf, a stem, a root, a tuberous root, a rhizome, a tuber, a stolon, a corm, a bulb, an offset, a cell of said plant in culture, a tissue of said plant in culture, an organ of said plant in culture, and a callus.
Description:
BACKGROUND OF THE INVENTION
[0001] 1. Field of the Invention
[0002] Artificial and synthetic DNA sequences have gained extensive use with development of the field of synthetic biology in the past decade. The present invention relates to use of artificial nucleotide sequences in the field of bioluminescence, which is emission of light by living organisms. Bioluminescence of bacterial organisms is mediated by the bacterial LUX operon. The LUX operon encodes for the bacterial luciferase, the light emitting enzyme, as well as enzymes responsible for synthesis of luciferins, substrates required for the light emission reaction. The operon contains genes named C-D-A-B-E(-G), where Lux A and Lux B code for the components of the luciferase and Lux C, D and E code for a fatty acid reductase complex producing an aldehyde necessary for the reaction. LuxG codes for an enzyme thought to participate in the turnover of the second luciferin, the flavin mononucleotide.
[0003] 2. Description of Related Art
[0004] In biotechnology, genes of the LUX operon have a wide range of applications. For instance, the LUX operon is utilized as a reporter in a variety of bacterial and plant biosensors. Bacterial cells of naturally non-glowing species such as E. coli have been engineered to contain the LUX operon inducible by pre-determined classes of chemicals. These cells start glowing in the presence of these specific compounds, reporting on the composition or toxicity of the sample. Plants engineered with a fully functional LUX operon have been contemplated for use as phytosensors, monitoring the conditions of the plant and the environment. Furthermore, ornamental plants have been engineered to contain a LUX operon to produce novel and unique types of glowing ornamental plants. The U.S. ornamentals market was sized at approximately $21 B in the early 2000's, and the entire worldwide market for ornamental plants has been estimated to be over $100 B.
[0005] The ornamental plant market is driven by innovation, where outdated varieties are inevitably replaced by new types of plants and flowers. New colors of roses and carnations, and new shapes and colors of petunias, find their way to the marketplace every year. Generation of new and esthetically pleasing varieties is known to be the key force driving the floriculture industry and stimulating its growth.
[0006] However, one of the major limitations of the applicability of LUX operon-based technologies, particularly in plants, is low levels of light emission in plants expressing LUX genes. Therefore means to engineer the LUX operon to enhance and augment plant light emission are needed.
[0007] The present invention addresses this problem, and provides several means of enhancing light emission, instrumental in producing new, exciting varieties of highly autoluminescent ornamental plants, as well as additional plant products, such as more effective autoluminescent plant phytosensors.
SUMMARY OF THE INVENTION
[0008] The present invention discloses novel artificial DNA sequences, i.e., SEQ ID NOs:1-8 and 11-12, shown in the section entitled "Nucleotide and Amino Acid Sequences of the Invention", variously encoding for LUX and other polypeptides, useful in enhancing autoluminescence.
[0009] In another aspect, the present invention discloses specific mutations in the LuxC and LuxE genes that are highly effective in enhancing light emission in an organism, such as a bacterium or plant, containing these genes in a mutated LUX operon.
[0010] More specifically, in its various aspects, the present invention provides:
[0011] [1] A nucleic acid construct, comprising the nucleotide sequences shown in SEQ ID NOs:1-5, operably linked for expression.
[0012] [2] A nucleic acid construct, comprising the nucleotide sequences shown in SEQ ID NOs:1-6, operably linked for expression.
[0013] [3] The nucleic acid construct of [1] or [2], further comprising, operably linked for expression, the nucleotide sequence shown in SEQ ID NO:7.
[0014] [4] The nucleic acid construct of any one of [1]-[3], further comprising, operably linked for expression, the nucleotide sequence shown in SEQ ID NO:8.
[0015] [5] The nucleic acid construct of any one of [1]-[4], which is an expression cassette.
[0016] [6] An expression vector, comprising the expression cassette of [5].
[0017] [7] A living cell, containing any one or more of the nucleotide sequences shown in SEQ ID NOs:1-8 or 11-12.
[0018] [8] The living cell of [7], containing the nucleotide sequences shown in SEQ ID NOs:1-5, operably linked for expression.
[0019] [9] The living cell of [7], containing the nucleotide sequences shown in SEQ ID NOs:1-6, operably linked for expression.
[0020] [10] The living cell of [8], further comprising at least one nucleotide sequence selected from the group consisting of SEQ ID NO:7 and SEQ ID NO:8, operably linked for expression.
[0021] [11] The living con of [9], further comprising at least one nucleotide sequence selected from the group consisting of SEQ ID NO:7 and SEQ 11 NO:8, operably linked for expression.
[0022] [12] The living cell of any one of [7]-[11], which is selected from the group consisting of a bacterial cell and a plant cell.
[0023] [13] The living cell of [12], which is autoluminescent.
[0024] [14] A plant, a cell of which contains said nucleic acid construct, expression cassette, expression vector, or nucleotide sequences of any one of [1]-[6].
[0025] [15] The plant of [14], wherein said nucleic acid construct, expression cassette, expression vector, or nucleotide sequences are located in a plastid.
[0026] [16] The plant of [15], wherein the plastid is a chloroplast.
[0027] [17] The plant of any one of [14]-[16], wherein said nucleotide sequences are expressed.
[0028] [18] The plant of [17], which is autoluminescent.
[0029] [19] Progeny of the plant of [18].
[0030] [20] The progeny of [19], which are produced sexually or asexually.
[0031] [21] The progeny of [20], which are produced asexually from cuttings.
[0032] [22] A part of said plant or progeny of any one of [14]-[21].
[0033] [23] The part of said plant or progeny of [22], which is selected from the group consisting of a protoplast, a cell, a tissue, an organ, a cutting, and an explant.
[0034] [24] The part of said plant or progeny of [22], which is selected from the group consisting of an inflorescence, a flower, a sepal, a petal, a pistil, a stigma, a style, an ovary, an ovule, an embryo, a receptacle, a seed, a fruit, a stamen, a filament, an anther, a male or female gametophyte, a pollen grain, a meristem, a terminal bud, an axillary bud, a leaf, a stem, a root, a tuberous root, a rhizome, a tuber, a stolon, a corm, a bulb, an offset, a cell of said plant in culture, a tissue of said plant in culture, an organ of said plant in culture, and a callus.
[0035] [25] A method of producing an autoluminescent plant, comprising asexually propagating a cutting of said plant or progeny of any one of [14]-[21].
[0036] Further scope of the applicability of the present invention will become apparent from the detailed description and drawings provided below. However, it should be understood that the detailed description and specific examples, while indicating preferred embodiments of the invention, are given by way of illustration only since various changes and modifications within the spirit and scope of the invention will become apparent to those skilled in the art from this detailed description.
BRIEF DESCRIPTION OF THE DRAWINGS
[0037] The above and other aspects, features, and advantages of the present invention will be better understood from the following detailed description taken in conjunction with the accompanying drawings, all of which are given by way of illustration only, and are not limitative of the present invention, in which:
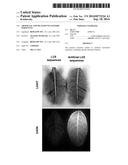
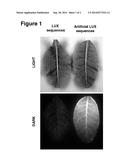
[0038] FIG. 1 shows an example of enhanced light output of transgenic autoluminescent tobacco containing a combination of artificial LUX sequences SEQ ID NOs:1, 2, 11, 4, 12, and 6 (right panel) vs. autoluminescent tobacco plants containing known wild-type LUX sequences (left panel). Light emission is detected using a ChemiDoc XRS Molecular Imager; inverse images are shown.
[0039] FIG. 2 shows the further effect of mutations in structural LUX genes in enhancing light emission of the LUX operon in E. coli. (A) Change of Ala to Gly in LuxC amino acid sequence position 389 (SEQ ID NO:3) enhances light emission; (B) Change of Gln to Glu in LuxE amino acid position 167 (SEQ ID NO:5) used in combination with SEQ ID NO:3 further enhances light emission compared to the use of SEQ ID NO:3 alone. E. coli cultures carrying the mutations imaged using ChemiDoc XRS Molecular Imager. Left panel: culture plates in light; right panel: photographic exposure of the cultures; 100 sec exposure for (A), 10 sec exposure for (B).
NUCLEOTIDE AND AMINO ACID SEQUENCES
[0040] SEQ ID NO:1: artificial Lux A nucleotide sequence;
[0041] SEQ ID NO:2: artificial Lux B nucleotide sequence;
[0042] SEQ ID NO:3: artificial Lux C nucleotide sequence, incorporating Ala Gly mutation at amino acid position 389;
[0043] SEQ ID NO:4: artificial Lux D nucleotide sequence;
[0044] SEQ ID NO:5: artificial Lux E nucleotide sequence, incorporating Gln Glu mutation at amino acid position 167;
[0045] SEQ ID NO:6: artificial Lux G nucleotide sequence;
[0046] SEQ ID NO:7: artificial E. coli Fre nucleotide sequence;
[0047] SEQ ID NO:8: artificial V. fischeri Yellow Fluorescent Protein nucleotide sequence;
[0048] SEQ ID NO:9: amino acid sequence of wild-type Photobacterium leiognathi LuxC protein;
[0049] SEQ ID NO:10: amino acid sequence of wild-type Photobacterium leiognathi LuxE protein;
[0050] SEQ ID NO:11: artificial Lux C nucleotide sequence without Ala→Gly mutation at amino acid position 389. Compare to SEQ ID NO:3;
[0051] SEQ ID NO:12: artificial Lux E nucleotide sequence without Gln→Glu mutation at amino acid position 167. Compare to SEQ ID NO:5.
[0052] Although not listed above, the present invention also encompasses the amino acid sequences of the proteins encoded by the nucleotide sequences listed. Such amino acid sequences can be deduced by, for example, conventional bioinformatics methods, including the use of publicly available and proprietary computer programs designed for this purpose.
DETAILED DESCRIPTION OF THE INVENTION
[0053] The following detailed description of the invention is provided to aid those skilled in the art in practicing the present invention. Even so, the following detailed description should not be construed to unduly limit the present invention, as modifications and variations in the embodiments herein discussed may be made by those of ordinary skill in the art without departing from the spirit or scope of the present inventive discovery.
[0054] The contents of each of the references cited herein is herein incorporated by reference in its entirety.
[0055] Methods and techniques for generating transgenic, transplastomic, and otherwise genetically modified plants are well known in the art.
[0056] The use of native LUX genes to produce autoluminescent plants has been previously described in the art. Patent applications by Krichevsky, i.e., WO 2009/017821 and WO 2011/106001, disclose the use of naturally occurring LUX genes in the form of an operon in plastids, and U.S. Pat. No. 7,663,022 by Hudkins prophetically contemplates nuclear expression of LUX genes from separate vectors.
[0057] However, none of these references either discloses or suggests the artificial LUX, E. coli Fre, or V. fischeri Yellow Fluorescent Protein (YFP) sequences SEQ ID NOs:1-8 and 11-12 of the present invention. Further, the art does not teach or suggest the mutations in LuxC and LuxE genes disclosed herein.
[0058] In one embodiment, LUX operon genes are used in variety of biotechnology applications, which can further benefit from enhancement of light output generated by the LUX operon. For example, the problem of enhancing the light output of the autoluminescent plants disclosed in the above-noted PCT applications and producing brighter glowing plants, which are appealing and attractive to the consumer, is solved by the artificial DNA sequences of the present invention. Expression of these sequences, or combinations thereof, results in autoluminescent plants that are several fold brighter than plants expressing wild-type LUX genes.
[0059] Examples of certain preferred combinations of the artificial sequences disclosed herein include, but are not limited to: SEQ ID NOs:1-5 in combination; SEQ ID NOs:1-6 in combination; or further, combination of SEQ ID NOs:1-5 in combination or SEQ ID NOs:1-6 in combination, further in combination with SEQ ID NO:7; and further, such foregoing combinations, further in combination with SEQ ID NO:8. In each of these cases, the nucleotide sequences are operably linked for expression.
[0060] One skilled in the art will recognize that the individual sequences disclosed herein can be used in combination, as indicated above, in any order, and are independent of one another.
[0061] As used herein, the phrase "operably linked for expression" and the like encompasses nucleic acid sequences linked in the 5' to 3' direction in such a way as to facilitate expression of an included nucleotide coding sequence.
[0062] The following examples are meant to be illustrative, and not limiting, of the practice or products of the present invention.
Example 1
Enhanced Light Emission By A Combination of SEQ ID NOs:1, 2, 11, 4, 12, and 6
[0063] An example of light emission enhancement in transgenic tobacco by a combination of artificial sequences of the present invention, employing SEQ ID NOs: 1, 2, 11, 4, 12, and 6 operably linked for expression, compared to that produced by a combination of wild-type LUX operon genes C-D-A-B-E(-G) operably linked for expession, is shown in FIG. 1. The methods employed to produce these plants are disclosed in the inventor's PCT International Publications WO 2009/017821 and WO 2011/106001.
[0064] Artificial sequences SEQ ID NOs:7 and 8, encoding FRE and YFP proteins, respectively, are designed to further improve light output and change the emitted light color, respectively, of the autoluminescent plants encompassed by the present invention.
Example 2
Light Emission Enhancement by LUX Structural Gene Mutants
[0065] A number of mutations in the regulatory genes governing expression of the entire LUX operon in bacteria are known in the art. However, mutations in the LUX structural genes, and particularly in LuxC and LuxE, enhancing or otherwise modulating light emission generated by the LUX operon have not been previously identified.
[0066] Disclosed herein are two novel mutations in the structural LUX genes C (encoding an Ala→Gly mutation at amino acid position 389) and E (encoding a Gln→Glu mutation at amino acid position 167), which greatly enhance light emission of the LUX operon.
[0067] The known sequence of the Photobacterium leiognathi LUX operon (GeneBank #M63594) discloses the following sequences of the wild-type LuxC and LuxE genes:
TABLE-US-00001 LuxC (Gene Bank M63594) (SEQ ID NO: 9) MIKKIPMIIGGVVQNTSGYGMRELTLNNNKVNIPIITQSDVEAIQSLNIENKLTINQIVNFLYTVGQKW KSETYSRRLTYIRDLIKFLGYSQEMAKLEANWISMILCSKSALYDIVENDLSSRHIIDEWIPQGECYVK ALPKGKSVHLLAGNVPLSGVTSILRAILTKNECIIKTSSADPFTATALVNSFIDVDAEHPITRSISVMY WSHSEDLAIPKQIMSCADVVIAWGGDDAIKWATEHAPSHADILKFGPKKSISIVDNPTDIKAAAIGVAH DICFYDQQACFSTQDIYYIGDSIDIFFDELAQQLNKYKDILPKGERNFDEKAAFSLTERECLFAKYKVQ KGESQSWLLTQSPAGSFGNQPLSRSAYIHQVNDISEVIPFVHKAVTQTVAIAPWESSFKYRDILAEHG AERIIEAGMNNIFRVGGAHDGMRPLQRLVNYISHERPSTYTTKDVSVKIEQTRYLEEDKFLVFVP LuxE (Gene Bank M63594) (SEQ ID NO: 10) MSTLLNIDATEIKVSTEIDDIIFTSSPLTLLFEDQEKIQKELILESFHYHYNHNKDYKYYCNIQGVDEN IQSIDDIPVFPTSMFKYSRLHTADESNIENWFTSSGTKGVKSHIARDRQSIERLLGSVNYGMKYLGEFH EHQLELVNMGPDRFSASNVWFKYVMSLVQLLYPTTFTVENDEIDFEQTILALKAIQRKGKGICLIGPP YFIYLLCHYMKEHNIEFNAGAHMFIITGGGWKTKQKEALNRQDFNQLLMETFSLFHESQIRDIFNQVEL NTCFFEDSLQRKHVPPWVYARALDPVTLTPVEDGQEGLMSYMDASSTSYPTFIVTDDIGIVRHLKEPDP FQGTTVEIVRRLNTREQKGCSLSMATSLK
[0068] As disclosed herein, substitution of Ala (position 389) in the amino acid sequence of LuxC (SEQ ID NO:9; emphasized in the sequence in 14 point type, bold, and underlined) with, for example Gly, strongly enhances light emission generated by the operon. Moreover, substitution of Gln (position 167) in the amino acid sequence of LuxE (SEQ ID NO:10; emphasized in the sequence in 14 point type, bold, and underlined) with, for example Glu, further increases light emission of E. coli harboring the mutated operons (FIG. 2). Methods for nucleic acid mutagenesis are known in the art (e.g., Chen et al.
[0069] (1997) "High efficiency of site-directed mutagenesis mediated by a single PCR product." Nuc. Acid Res, 25(3): 682-4). Methods for transforming E. coli are well known in the art.
[0070] In the particular example shown in FIG. 2, the mutant LuxC gene (SEQ ID NO:3) is first introduced into the operon (SEQ ID NOs: 1-4, 12, and 6) introduced into the bacteria (Panel A), followed by introduction of the mutant LuxE gene (SEQ ID NO:5) into the operon (SEQ ID NOs:1-6) introduced into the bacteria (Panel B). One skilled in the art will appreciate that these mutations can be introduced in any order, and are independent of one another. Each mutation can be used for light emission enhancement separately, in combination with one another, or as indicated above, each can be used separately, or together, in combination with any other mutations or sequences. This includes, for example, the combinations listed above.
[0071] This example demonstrates the luminescence-enhancing effect of LuxC Ala (389) substitution to Gly and LuxE Gln (167) substitution to Glu in E. coli.
[0072] Specifically, FIG. 2 shows the further effect of mutations in these structural LUX genes in enhancing light emission of the LUX operon in E. coli. Panel (A) shows that change of Ala to Gly in LuxC amino acid sequence position 389 (SEQ ID NO:3) enhances bacterial light emission; Panel (B) shows that change of Gln to Glu in LuxE amino acid position 167 (SEQ ID NO:5), used in combination with SEQ ID NO:3, further enhances light emission compared to the use of SEQ ID NO:3 alone.
[0073] In view of these results, it is fully expected that use of artificial nucleotide sequences encoding LUXC comprising the Ala→Gly mutation, e.g., SEQ ID NO:3, and LUXE, comprising the Gln→Glu mutation, e.g., SEQ ID NO:5, either alone, or together, in combination with the other artificial LUX operon sequences disclosed herein, will produce a similar light-enhancing effect in plants.
[0074] As noted above, preferred combinations of the artificial sequences disclosed herein include, but are not limited to: SEQ ID NOs:1-5 in combination; SEQ ID NOs:1-6 in combination; or further, combination of SEQ ID NOs:1-5 in combination or SEQ ID NOs:1-6 in combination, further in combination with SEQ ID NO:7; and further, such foregoing combinations, further in combination with SEQ ID NO:8. In each of these cases, the nucleotide sequences are operably linked for expression.
[0075] One skilled in the art can appreciate that substitution of LuxC with other amino acids in position (389) and LuxE in position (167) can result in different modifications of light emission, which can be further used to modulate LUX operon luminescence. Furthermore, depending on the type of bacteria from which the LUX operon is derived, positions of these critical amino acids can be shifted within close proximity to the described residues of P. leiognathi, or located in sequences with high homology to sequences surrounding position (389) in LuxC and position (167) in LuxE.
[0076] The artificial DNA sequences of the present invention incorporate the above described LuxC and LuxE mutations (SEQ ID NOs:3 and 5, respectively), designed to further enhance light output of the LUX operon. The utility and applicability of the current invention includes, for example, generating bright autoluminescent plants. Besides applications in ornamental plants, where bright plants are attractive to consumers, the present sequences have utility in producing highly effective plant biosensors emitting light in response to various types of stress or other conditions when operons containing these sequences are under the control of appropriate promoters, e.g., stress-inducible promoters, and are thus useful in agriculture for crop or environmental monitoring.
[0077] Plants encompassed by the present invention include both monocots and dicots, ornamentals as well as crop plants. Non-limiting examples include ornamental plants such as petunias, poinsettias, and roses, as well as crop plants such as corn and oil producing palms.
[0078] Also encompassed by the present invention are parts of such plants including, for example, a protoplast, a cell, a tissue, an organ, a cutting, and an explant. Such parts further include an inflorescence, a flower, a sepal, a petal, a pistil, a stigma, a style, an ovary, an ovule, an embryo, a receptacle, a seed, a fruit, a stamen, a filament, an anther, a male or female gametophyte, a pollen grain, a meristem, a terminal bud, an axillary bud, a leaf, a stem, a root, a tuberous root, a rhizome, a tuber, a stolon, a corm, a bulb, an offset, a cell of said plant in culture, a tissue of said plant in culture, an organ of said plant in culture, and a callus. The present invention also encompasses progeny, whether produced sexually or asexually, of transgsenic plants of the invention containing sequences disclosed herein.
[0079] In regard to methods of propagating autoluminescent plants encompassed by the present invention, methods of propagation and reproduction of such plants are well known in the art, and include both sexual and asexual techniques.
[0080] Asexual reproduction is the propagation of a plant to multiply the plant without the use of seeds to assure an exact genetic copy of the plant being reproduced.
[0081] Any known method of asexual reproduction which renders a true genetic copy of the plant may be employed in the present invention. Acceptable modes of asexual reproduction include, but are not limited to, rooting cuttings; grafting; explants; budding; apomictic seeds; bulbs; division; slips; layering; rhizomes; runners; corms; tissue culture; nucellar embryos; and any other conventional method of asexual propagation. The present invention encompasses all such methods of propagation and reproduction of plants encompassed by the present invention.
[0082] In additional examples, the presently disclosed DNA sequences are further useful in generating more efficient plant research systems, where their autoluminescent properties can be used as a reporter system for gene expression and other scientific assays.
[0083] The invention being thus described, it will be recognized that the same may be varied in many ways. Such variations are not to be regarded as a departure from the spirit and scope of the invention, and all such modifications as would be obvious to one skilled in the art are intended to be included within the scope of the following claims.
[0084] Nucleotide and Amino Acid Sequences of the Invention
TABLE-US-00002 SEQ ID NO: 1 ATGAAAATAAGTAATATTTGTTTCTCATATCAACCACCAGGGGAGTCCCATCAGGAGGTTATGGAAAGG TTTATACGACTAGGTGTCGCATCTGAAGAATTAAATTTTGATGGATTTTATACTTTAGAGCACCATTTT ACCGAATTTGGAATAACTGGTAATTTATATATTGCATGTGCAAACATACTAGGACGAACTAAGCGTATT CAAGTTGGCACAATGGGCATAGTTCTTCCTACAGAGCATCCGGCTCGACATGTAGAATCACTACTTGTT CTTGATCAATTGTCTAAGGGTAGATTTAATTATGGAACGGTTAGGGGTTTGTATCATAAGGATTTTCGA GTGTTTGGGACATCCCAGGAGGATTCCCGAAAAACAGCAGAAAATTTCTATTCTATGATTTTAGATGCG TCCAAGACCGGAGTGTTGCATACGGACGGGGAGGTAGTAGAATTTCCTGATGTGAATGTCTACCCAGAA GCCTATTCTAAAAAGCAGCCTACTTGTATGACTGCGGAATCTTCTGAGACTATTACTTATTTAGCGGAA AGAGGGCTACCTATGGTGTTAAGTTGGATTATCCCAGTTAGTGAAAAAGTATCTCAAATGGAGTTATAT AATGAAGTGGCCGCTGAACATGGGCATGATATAAACAATATTGAACACATTCTAACATTTATTTGCTCT GTTAATGAAGATGGGGAGAAAGCCGATAGTGTATGTAGGAATTTTTTGGAGAATTGGTATGACTCCTAC AAGAATGCCACAAACATCTTTAATGATTCCAACCAAACAAGAGGTTATGATTATTTAAAAGCTCAATGG CGAGAGTGGGTTATGAAAGGTTTAGCTGACCCACGAAGGCGTCTTGATTATTCTAATGAATTAAATCCG GTCGGTACACCTGAACGTTGTATCGAAATTATTCAAAGTAATATTGATGCAACCGGGATAAAACACATT ACCGTGGGCTTTGAAGCTAATGGTAGTGAACAGGAAATTAGAGAATCTATGGAACTTTTTATGGAAAAA GTTGCACCGCATCTTAAAGATCCCCAATAA SEQ ID NO: 2 ATGAACTTTGGATTGTTTTTCCTAAATTTCCAACCAGAAGGAATGACTTCCGAAATGGTACTAGATAAT ATGGTTGATACAGTAGCATTGGTAGACAAAGATGACTATCATTTCAAGCGTGTATTGGTGTCTGAACAT CATTTCTCCAAAAATGGCATTATAGGGGAGCCCTTAACCGCTATATCTTTCCTTTTAGGTCTAACCAAG AGAATAGAAATAGGTTCTTTGAATCAGGTTATAACGACCCACCATCCTGTAAGAATTGGCGAACAGACT GGATTATTAGATCAGATGTCTTACGGTCGTTTCGTTTTAGGTTTATCAGATTGCGTTAATGATTTCGAA ATGGATTTTTTTAAACGAAAACGTAGTTCACAACAACAACAATTCGAAGCATGTTATGAAATTTTAAAT GAAGCCTTAACTACGAATTATTGCCAAGCGGATGATGATTTTTTCAATTTTCCGAGGATCAGTGTAAAT CCCCATTGTATCTCTGAGGTTAAACAATACATTTTGGCATCTTCTATGGGTGTAGTTGAATGGGCCGCT CGAAAAGGTCTTCCTTTAACGTATAGATGGAGTGATAGTTTAGCAGAAAAAGAGAAGTATTATCAGCGT TACTTAGCGGTTGCTAAAGAGAACAATATAGATGTTTCAAATATCGATCATCAATTTCCTCTTCTTGTA AATATTAACGAAAATCGAAGAATAGCACGAGATGAAGTACGTGAGTACATTCAGAGTTATGTATCAGAA GCCTATCCCACTGACCCTAATATTGAACTTCGTGTAGAAGAATTGATCGAACAACACGCAGTCGGGAAA GTCGATGAATATTATGATTCTACGATGCACGCTGTCAAAGTTACTGGTTCTAAAAATTTATTATTATCT TTTGAATCTATGAAAAATAAAGATGACGTCACTAAACTTATCAACATGTTCAACCAAAAAATCAAGGAT AACTTAATAAAGTGA SEQ ID NO: 3 ATGATCAAAAAAATCCCTATGATAATTGGGGGAGTAGTCCAGAACACATCCGGTTATGGAATGAGAGAA TTAACATTAAACAATAATAAAGTTAACATTCCAATTATCACACAAAGTGATGTAGAAGCTATTCAATCT CTAAATATTGAGAACAAATTGACAATAAATCAGATTGTAAATTTCCTTTATACTGTAGGCCAAAAATGG AAATCTGAGACGTATAGTCGTCGATTAACTTATATCAGAGATTTAATCAAATTCTTAGGTTATAGTCAG GAAATGGCTAAATTGGAAGCTAATTGGATTAGTATGATATTATGTTCTAAAAGTGCTTTATATGACATA GTAGAAAATGATTTAAGTAGTCGTCATATCATTGATGAATGGATTCCCCAAGGTGAATGCTATGTAAAA GCATTGCCTAAGGGTAAGTCCGTACACTTGTTAGCAGGAAATGTTCCTTTATCAGGAGTAACCTCCATA CTAAGAGCAATTCTTACAAAAAATGAATGCATTATTAAAACTAGTTCAGCAGACCCATTTACTGCCACT GCACTTGTTAACTCTTTTATAGACGTTGATGCCGAACATCCTATAACACGATCCATTAGTGTAATGTAT TGGTCCCATTCTGAAGATTTAGCAATTCCCAAACAAATAATGTCTTGTGCTGACGTTGTTATAGCATGG GGAGGGGACGATGCAATAAAATGGGCAACTGAACATGCACCTTCTCACGCAGACATATTGAAATTCGGA CCGAAAAAATCCATTTCCATTGTCGATAATCCTACGGATATTAAGGCAGCTGCTATCGGAGTGGCTCAT GACATTTGTTTTTATGATCAGCAAGCATGCTTCTCAACCCAAGATATATATTATATCGGAGATTCAATT GATATTTTCTTTGATGAATTAGCTCAACAGTTAAATAAATATAAAGACATTTTACCTAAAGGGGAACGA AATTTCGATGAGAAGGCAGCTTTCTCCCTTACTGAAAGAGAGTGTCTTTTCGCAAAATATAAAGTTCAA AAAGGTGAATCCCAATCTTGGTTGCTTACCCAAAGTCCAGCGGGAAGTTTTGGAAATCAACCTTTGAGT CGTTCTGCGTATATTCATCAGGTAAATGATATAAGTGAAGTAATACCCTTCGTACATAAAGGAGTTACT CAAACTGTAGCTATCGCGCCTTGGGAATCAAGTTTTAAATACAGAGATATTTTGGCTGAGCATGGTGCT GAGCGTATCATTGAAGCAGGAATGAATAACATTTTTCGTGTAGGAGGTGCCCACGATGGGATGCGACCC TTGCAACGTTTGGTTAATTATATTTCTCATGAACGTCCTAGTACATATACAACAAAAGATGTTAGTGTA AAAATAGAACAGACAAGGTATCTTGAAGAAGATAAATTCTTAGTTTTTGTACCGTAG SEQ ID NO: 4 ATGGAAAATACACAACATAGTTTACCTATTGATCACGTAATCGACATAGGTGACAACCGTTACATCAGG GTGTGGGAAACTAAACCTAAAAACAAAGAAACTAAAAGAAATAATACCATAGTGATAGCGTCCGGTTTT GCAAGAAGAATGGATCACTTTGCTGGATTAGCTGAATATCTTGCCAACAATGGATTCCGAGTTATTAGA TACGATTCACTAAATCATGTGGGCTTGTCTAGTGGTGAAATTAAACAGTTTAGTATGTCTGTAGGTAAA CATTCTTTGCTAACGGTAATTGATTGGCTTAAAGAACGAAATATCAACAATATTGGACTAATTGCAAGT TCCTTAAGTGCCCGTATAGCCTATGAAGTAGCCGCAGAAATTGATTTATCCTTCCTTATAACAGCAGTT GGGGTTGTGAATTTACGTTCTACTCTTGAAAAAGCACTTAAATATGATTATTTGCAGATGGAAGTCAAT ACGATTCCTGAAGACTTAATATTTGAAGGGCATAATCTAGGTTCAAAAGTTTTTGTGACTGATTGTTTT GAAAACAACTGGGATTCTTTAGACTCAACTATTAATAAAATTTGTGAGCTTGATATTCCGTTCATAGCT TTCACTTCTGATGGGGATGATTGGGTTTGTCAACATGAAGTAAAACACCTAGTGTCCAATGTAAAATCT GACAAAAAAAAGATATACTCTTTAGTTGGTAGTTCCCATGATTTGGGGGAAAATTTGGTCGTTTTACGA AATTTCTATCAAAGTATGACTAAAGCTGCTGTCTCATTGGATAGGCAATTGGTTGAATTAGTTGATGAA ATCATAGAACCAAATTTTGAGGATTTAACCGTAATTACAGTCAATGAAAGAAGACTTAAAAATAAAATA GAAAATGAAATAATAAACAGACTAGCAGATCGAGTTCTTGCTTCCGTATAA SEQ ID NO: 5 ATGTCCACCTTACTAAACATCGATGCAACGGAGATTAAAGTTAGTACCGAGATAGATGATATAATCTTT ACAAGTAGTCCATTAACTTTATTATTTGAAGATCAAGAAAAAATTCAGAAAGAATTAATACTTGAAAGT TTTCATTATCATTATAACCATAATAAAGATTACAAGTATTATTGTAATATTCAGGGGGTTGATGAGAAC ATTCAATCAATTGACGACATTCCAGTATTTCCTACATCCATGTTTAAATACTCTCGTCTTCATACAGCC GATGAGAGTAATATAGAAAATTGGTTTACATCATCCGGTACTAAAGGCGTTAAGTCTCATATTGCTAGG GATAGGCAGTCAATTGAAAGATTACTAGGATCAGTTAATTATGGTATGAAATATCTTGGAGAATTTCAT GAACATCAACTTGAACTTGTAAATATGGGACCAGATCGTTTTTCCGCTTCAAACGTGTGGTTCAAATAT GTTATGAGTTTAGTAGAATTGTTATATCCTACTACTTTTACTGTGGAAAATGATGAGATAGATTTTGAA CAAACTATCTTGGCTTTAAAAGCGATACAACGAAAAGGAAAAGGAATATGTTTAATAGGACCGCCTTAT TTTATATACTTGTTATGCCATTATATGAAAGAACATAATATAGAATTTAATGCAGGGGCTCACATGTTT ATTATTACGGGAGGGGGATGGAAAACAAAACAAAAAGAGGCGTTAAATAGGCAAGATTTCAATCAACTT CTTATGGAAACATTCTCCTTATTTCATGAGTCACAAATTAGAGACATATTTAATCAAGTTGAATTGAAT ACATGTTTCTTCGAAGATTCTCTTCAACGAAAACATGTGCCACCTTGGGTATATGCTCGTGCATTAGAT CCTGTTACTTTGACTCCCGTAGAAGACGGGCAGGAAGGCTTGATGTCTTATATGGACGCCTCCAGTACA TCATATCCGACTTTCATCGTTACGGATGATATTGGCATTGTAAGGCATCTAAAAGAGCCAGATCCCTTC CAAGGTACAACCGTAGAAATTGTTAGACGTCTTAACACACGAGAGCAAAAGGGTTGTTCTTTATCTATG GCTACAAGTCTTAAATAA SEQ ID NO: 6 ATGATCTTCAACTGTAAAGTCAAAAAAGTTGAAGCATCCGATTCACATATTTATAAAGTCTTTATCAAA CCCGATAAGTGTTTCGATTTTAAAGCAGGCCAATATGTTATTGTGTACCTAAACGGGAAAAATTTACCA TTTAGTATAGCCAACTGTCCTACATGTAATGAATTATTGGAATTACATGTAGGCGGGTCTGTAAAAGAA TCTGCAATTGAAGCAATATCACACTTTATTAATGCTTTTATATATCAAAAAGAATTTACTATTGATGCT CCGCATGGAGACGCCTGGTTACGAGATGAGTCTCAATCTCCGCTTTTGTTAATAGCTGGCGGCACAGGT TTATCATATATCAATAGTATTTTAAGTTGCTGCATTTCTAAACAACTATCCCAACCGATCTATTTATAC TGGGGTGTCAACAATTGTAACCTTTTGTATGCAGATCAACAATTAAAAACTTTGGCCGCACAATATCGT AATATTAATTATATCCCTGTAGTTGAGAATCTTAATACAGATTGGCAAGGAAAAATTGGGAATGTAATA GATGCAGTAATCGAAGATTTTAGTGACCTTTCAGATTTCGACATCTATGTTTGTGGACCCTTCGGTATG TCCAGAACAGCTAAAGATATTCTAATTTCACAAAAGAAAGCAAACATAGGGAAGATGTATTCAGATGCT TTTTCTTACACGTGA SEQ ID NO: 7 ATGACTACTCTTTCTTGTAAGGTGACATCAGTGGAGGCTATAACTGACACAGTGTACAGAGTTAGAATC GTACCAGATGCAGCATTTAGTTTTAGGGCCGGTCAATATTTGATGGTTGTAATGGACGAGAGAGATAAG AGACCATTCAGCATGGCCTCTACTCCAGATGAGAAAGGGTTTATCGAACTGCACATTGGAGCATCAGAG ATCAATTTATACGCAAAAGCAGTCATGGACAGGATCTTAAAGGACCATCAGATTGTTGTTGATATTCCT CACGGCGAAGCATGGCTTAGGGATGATGAGGAAAGACCTATGATTCTCATCGCTGGCGGAACAGGGTTC TCTTACGCTAGGTCTATACTCCTCACCGCCCTAGCACGTAATCCAAATAGGGATATTACCATTTACTGG GGTGGTAGAGAAGAGCAGCACCTTTACGACCTTTGCGAATTGGAGGCCCTTAGCTTAAAGCATCCTGGT CTACAAGTTGTGCCAGTTGTCGAACAACCTGAGGCAGGATGGAGAGGGCGTACAGGAACAGTGCTAACT GCCGTTTTACAGGATCATGGCACTCTTGCTGAGCACGATATTTATATTGCCGGTAGATTCGAAATGGCT AAGATTGCACGTGACCTTTTTTGTTCTGAAAGAAATGCCAGGGAAGATAGATTGTTCGGTGATGCTTTC GCATTCATTTGA SEQ ID NO: 8 ATGTTTAAAGGAATTGTGGAAGGCATTGGAATCATTGAGAAGATAGACATATATACAGACCTTGACAAG TATGCCATCAGATTCCCTGAAAACATGTTGAACGGCATTAAAAAAGAGTCTTCCATTATGTTTAACGGC TGCTTTCTTACAGTGACCAGCGTTAATAGCAACATCGTCTGGTTTGATATTTTTGAGAAGGAAGCTAGG AAACTGGATACATTTAGAGAATATAAGGTTGGAGATAGAGTCAATTTGGGTACATTCCCAAAGTTTGGT GCTGCATCTGGAGGACATATTTTGAGTGCAAGAATATCTTGCGTTGCTAGTATTATTGAGATTATAGAG AATGAAGATTATCAACAGATGTGGATTCAGATTCCTGAGAACTTTACTGAGTTCTTAATTGACAAAGAC TATATTGCTGTCGATGGTATCTCTTTAACAATCGACACTATAAAAAACAATCAGTTTTTTATTAGTTTG CCGTTAAAAATAGCTCAAAATACCAACATGAAATGGAGGAAAAAGGGAGATAAGGTTAACGTGGAGTTG TCTAATAAGATTAACGCTAATCAGTGTTGGTGA LuxC (Gene Bank M63594) (SEQ ID NO: 9) MIKKIPMIIGGVVQNTSGYGMRELTLNNNKVNIPIITQSDVEAIQSLNIENKLTINQIVNFLYTVGQKW KSETYSRRLTYIRDLIKFLGYSQEMAKLEANWISMILCSKSALYDIVENDLSSRHIIDEWIPQGECYVK ALPKGKSVHLLAGNVPLSGVTSILRAILTKNECIIKTSSADPFTATALVNSFIDVDAEHPITRSISVMY WSHSEDLAIPKQIMSCADVVIAWGGDDAIKWATEHAPSHADILKFGPKKSISIVDNPTDIKAAAIGVAH DICFYDQQACFSTQDIYYIGDSIDIFFDELAQQLNKYKDILPKGERNFDEKAAFSLTERECLFAKYKVQ KGESQSWLLTQSPAGSFGNQPLSRSAYIHQVNDISEVIPFVHKAVTQTVAIAPWESSFKYRDILAEHG
AERIIEAGMNNIFRVGGAHDGMRPLQRLVNYISHERPSTYTTKDVSVKIEQTRYLEEDKFLVFVP LuxE (Gene Bank M63594) (SEQ ID NO: 10) MSTLLNIDATEIKVSTEIDDIIFTSSPLTLLFEDQEKIQKELILESFHYHYNHNKDYKYYCNIQGVDEN IQSIDDIFVFPTSMFKYSRLHTADESNIENWFTSSGTKGVKSHIARDRQSIERLLGSVNYGMKYLGEFH EHQLELVNMGPDRFSASNVWFKYVMSLVQLLYPTTFTVENDEIDFEQTILALKAIQRKGKGICLIGPP YFIYLLCHYMKEHNIEFNAGAHMFIITGGGWKTKQKEALNRQDFNQLLMETFSLFHESQIRDIFNQVEL NTCFFEDSLQRKHVPPWVYARALDPVTLTPVEDGQEGLMSYMDASSTSYPTFIVTDDIGIVRHLKEPDP FQGTTVEIVRRLNTREQKGCSLSMATSLK Artificial LuxC, prior to mutation: SEQ ID NO: 11 ATGATCAAAAAAATCCCTATGATAATTGGGGGAGTAGTCCAGAACACATCCGGTTATGGAATGAGAGAA TTAACATTAAACAATAATAAAGTTAACATTCCAATTATCACACAAAGTGATGTAGAAGCTATTCAATCT CTAAATATTGAGAACAAATTGACAATAAATCAGATTGTAAATTTCCTTTATACTGTAGGCCAAAAATGG AAATCTGAGACGTATAGTCGTCGATTAACTTATATCAGAGATTTAATCAAATTCTTAGGTTATAGTCAG GAAATGGCTAAATTGGAAGCTAATTGGATTAGTATGATATTATGTTCTAAAAGTGCTTTATATGACATA GTAGAAAATGATTTAAGTAGTCGTCATATCATTGATGAATGGATTCCCCAAGGTGAATGCTATGTAAAA GCATTGCCTAAGGGTAAGTCCGTACACTTGTTAGCAGGAAATGTTCCTTTATCAGGAGTAACCTCCATA CTAAGAGCAATTCTTACAAAAAATGAATGCATTATTAAAACTAGTTCAGCAGACCCATTTACTGCCACT GCACTTGTTAACTCTTTTATAGACGTTGATGCCGAACATCCTATAACACGATCCATTAGTGTAATGTAT TGGTCCCATTCTGAAGATTTAGCAATTCCCAAACAAATAATGTCTTGTGCTGACGTTGTTATAGCATGG GGAGGGGACGATGCAATAAAATGGGCAACTGAACATGCACCTTCTCACGCAGACATATTGAAATTCGGA CCGAAAAAATCCATTTCCATTGTCGATAATCCTACGGATATTAAGGCAGCTGCTATCGGAGTGGCTCAT GACATTTGTTTTTATGATCAGCAAGCATGCTTCTCAACCCAAGATATATATTATATCGGAGATTCAATT GATATTTTCTTTGATGAATTAGCTCAACAGTTAAATAAATATAAAGACATTTTACCTAAAGGGGAACGA AATTTCGATGAGAAGGCAGCTTTCTCCCTTACTGAAAGAGAGTGTCTTTTCGCAAAATATAAAGTTCAA AAAGGTGAATCCCAATCTTGGTTGCTTACCCAAAGTCCAGCGGGAAGTTTTGGAAATCAACCTTTGAGT CGTTCTGCGTATATTCATCAGGTAAATGATATAAGTGAAGTAATACCCTTCGTACATAAAGCAGTTACT CAAACTGTAGCTATCGCGCCTTGGGAATCAAGTTTTAAATACAGAGATATTTTGGCTGAGCATGGTGCT GAGCGTATCATTGAAGCAGGAATGAATAACATTTTTCGTGTAGGAGGTGCCCACGATGGGATGCGACCC TTGCAACGTTTGGTTAATTATATTTCTCATGAACGTCCTAGTACATATACAACAAAAGATGTTAGTGTA AAAATAGAACAGACAAGGTATCTTGAAGAAGATAAATTCTTAGTTTTTGTACCGTAG Artificial LuxE, prior to mutation: SEQ ID NO: 12 ATGTCCACCTTACTAAACATCGATGCAACGGAGATTAAAGTTAGTACCGAGATAGATGATATAATCTTT ACAAGTAGTCCATTAACTTTATTATTTGAAGATCAAGAAAAAATTCAGAAAGAATTAATACTTGAAAGT TTTCATTATCATTATAACCATAATAAAGATTACAAGTATTATTGTAATATTCAGGGGGTTGATGAGAAC ATTCAATCAATTGACGACATTCCAGTATTTCCTACATCCATGTTTAAATACTCTCGTCTTCATACAGCC GATGAGAGTAATATAGAAAATTGGTTTACATCATCCGGTACTAAAGGCGTTAAGTCTCATATTGCTAGG GATAGGCAGTCAATTGAAAGATTACTAGGATCAGTTAATTATGGTATGAAATATCTTGGAGAATTTCAT GAACATCAACTTGAACTTGTAAATATGGGACCAGATCGTTTTTCCGCTTCAAACGTGTGGTTCAAATAT GTTATGAGTTTAGTACAATTGTTATATCCTACTACTTTTACTGTGGAAAATGATGAGATAGATTTTGAA CAAACTATCTTGGCTTTAAAAGCGATACAACGAAAAGGAAAAGGAATATGTTTAATAGGACCGCCTTAT TTTATATACTTGTTATGCCATTATATGAAAGAACATAATATAGAATTTAATGCAGGGGCTCACATGTTT ATTATTACGGGAGGGGGATGGAAAACAAAACAAAAAGAGGCGTTAAATAGGCAAGATTTCAATCAACTT CTTATGGAAACATTCTCCTTATTTCATGAGTCACAAATTAGAGACATATTTAATCAAGTTGAATTGAAT ACATGTTTCTTCGAAGATTCTCTTCAACGAAAACATGTGCCACCTTGGGTATATGCTCGTGCATTAGAT CCTGTTACTTTGACTCCCGTAGAAGACGGGCAGGAAGGCTTGATGTCTTATATGGACGCCTCCAGTACA TCATATCCGACTTTCATCGTTACGGATGATATTGGCATTGTAAGGCATCTAAAAGAGCCAGATCCCTTC CAAGGTACAACCGTAGAAATTGTTAGACGTCTTAACACACGAGAGCAAAAGGGTTGTTCTTTATCTATG GCTACAAGTCTTAAATAA
Sequence CWU
1
1
1211065DNAArtificial SequenceSynthesized 1atgaaaataa gtaatatttg tttctcatat
caaccaccag gggagtccca tcaggaggtt 60atggaaaggt ttatacgact aggtgtcgca
tctgaagaat taaattttga tggattttat 120actttagagc accattttac cgaatttgga
ataactggta atttatatat tgcatgtgca 180aacatactag gacgaactaa gcgtattcaa
gttggcacaa tgggcatagt tcttcctaca 240gagcatccgg ctcgacatgt agaatcacta
cttgttcttg atcaattgtc taagggtaga 300tttaattatg gaacggttag gggtttgtat
cataaggatt ttcgagtgtt tgggacatcc 360caggaggatt cccgaaaaac agcagaaaat
ttctattcta tgattttaga tgcgtccaag 420accggagtgt tgcatacgga cggggaggta
gtagaatttc ctgatgtgaa tgtctaccca 480gaagcctatt ctaaaaagca gcctacttgt
atgactgcgg aatcttctga gactattact 540tatttagcgg aaagagggct acctatggtg
ttaagttgga ttatcccagt tagtgaaaaa 600gtatctcaaa tggagttata taatgaagtg
gccgctgaac atgggcatga tataaacaat 660attgaacaca ttctaacatt tatttgctct
gttaatgaag atggggagaa agccgatagt 720gtatgtagga attttttgga gaattggtat
gactcctaca agaatgccac aaacatcttt 780aatgattcca accaaacaag aggttatgat
tatttaaaag ctcaatggcg agagtgggtt 840atgaaaggtt tagctgaccc acgaaggcgt
cttgattatt ctaatgaatt aaatccggtc 900ggtacacctg aacgttgtat cgaaattatt
caaagtaata ttgatgcaac cgggataaaa 960cacattaccg tgggctttga agctaatggt
agtgaacagg aaattagaga atctatggaa 1020ctttttatgg aaaaagttgc accgcatctt
aaagatcccc aataa 10652981DNAArtificial
SequenceSynthesized 2atgaactttg gattgttttt cctaaatttc caaccagaag
gaatgacttc cgaaatggta 60ctagataata tggttgatac agtagcattg gtagacaaag
atgactatca tttcaagcgt 120gtattggtgt ctgaacatca tttctccaaa aatggcatta
taggggagcc cttaaccgct 180atatctttcc ttttaggtct aaccaagaga atagaaatag
gttctttgaa tcaggttata 240acgacccacc atcctgtaag aattggcgaa cagactggat
tattagatca gatgtcttac 300ggtcgtttcg ttttaggttt atcagattgc gttaatgatt
tcgaaatgga tttttttaaa 360cgaaaacgta gttcacaaca acaacaattc gaagcatgtt
atgaaatttt aaatgaagcc 420ttaactacga attattgcca agcggatgat gattttttca
attttccgag gatcagtgta 480aatccccatt gtatctctga ggttaaacaa tacattttgg
catcttctat gggtgtagtt 540gaatgggccg ctcgaaaagg tcttccttta acgtatagat
ggagtgatag tttagcagaa 600aaagagaagt attatcagcg ttacttagcg gttgctaaag
agaacaatat agatgtttca 660aatatcgatc atcaatttcc tcttcttgta aatattaacg
aaaatcgaag aatagcacga 720gatgaagtac gtgagtacat tcagagttat gtatcagaag
cctatcccac tgaccctaat 780attgaacttc gtgtagaaga attgatcgaa caacacgcag
tcgggaaagt cgatgaatat 840tatgattcta cgatgcacgc tgtcaaagtt actggttcta
aaaatttatt attatctttt 900gaatctatga aaaataaaga tgacgtcact aaacttatca
acatgttcaa ccaaaaaatc 960aaggataact taataaagtg a
98131437DNAArtificial SequenceSynthesized
3atgatcaaaa aaatccctat gataattggg ggagtagtcc agaacacatc cggttatgga
60atgagagaat taacattaaa caataataaa gttaacattc caattatcac acaaagtgat
120gtagaagcta ttcaatctct aaatattgag aacaaattga caataaatca gattgtaaat
180ttcctttata ctgtaggcca aaaatggaaa tctgagacgt atagtcgtcg attaacttat
240atcagagatt taatcaaatt cttaggttat agtcaggaaa tggctaaatt ggaagctaat
300tggattagta tgatattatg ttctaaaagt gctttatatg acatagtaga aaatgattta
360agtagtcgtc atatcattga tgaatggatt ccccaaggtg aatgctatgt aaaagcattg
420cctaagggta agtccgtaca cttgttagca ggaaatgttc ctttatcagg agtaacctcc
480atactaagag caattcttac aaaaaatgaa tgcattatta aaactagttc agcagaccca
540tttactgcca ctgcacttgt taactctttt atagacgttg atgccgaaca tcctataaca
600cgatccatta gtgtaatgta ttggtcccat tctgaagatt tagcaattcc caaacaaata
660atgtcttgtg ctgacgttgt tatagcatgg ggaggggacg atgcaataaa atgggcaact
720gaacatgcac cttctcacgc agacatattg aaattcggac cgaaaaaatc catttccatt
780gtcgataatc ctacggatat taaggcagct gctatcggag tggctcatga catttgtttt
840tatgatcagc aagcatgctt ctcaacccaa gatatatatt atatcggaga ttcaattgat
900attttctttg atgaattagc tcaacagtta aataaatata aagacatttt acctaaaggg
960gaacgaaatt tcgatgagaa ggcagctttc tcccttactg aaagagagtg tcttttcgca
1020aaatataaag ttcaaaaagg tgaatcccaa tcttggttgc ttacccaaag tccagcggga
1080agttttggaa atcaaccttt gagtcgttct gcgtatattc atcaggtaaa tgatataagt
1140gaagtaatac ccttcgtaca taaaggagtt actcaaactg tagctatcgc gccttgggaa
1200tcaagtttta aatacagaga tattttggct gagcatggtg ctgagcgtat cattgaagca
1260ggaatgaata acatttttcg tgtaggaggt gcccacgatg ggatgcgacc cttgcaacgt
1320ttggttaatt atatttctca tgaacgtcct agtacatata caacaaaaga tgttagtgta
1380aaaatagaac agacaaggta tcttgaagaa gataaattct tagtttttgt accgtag
14374948DNAArtificial SequenceSynthesized 4atggaaaata cacaacatag
tttacctatt gatcacgtaa tcgacatagg tgacaaccgt 60tacatcaggg tgtgggaaac
taaacctaaa aacaaagaaa ctaaaagaaa taataccata 120gtgatagcgt ccggttttgc
aagaagaatg gatcactttg ctggattagc tgaatatctt 180gccaacaatg gattccgagt
tattagatac gattcactaa atcatgtggg cttgtctagt 240ggtgaaatta aacagtttag
tatgtctgta ggtaaacatt ctttgctaac ggtaattgat 300tggcttaaag aacgaaatat
caacaatatt ggactaattg caagttcctt aagtgcccgt 360atagcctatg aagtagccgc
agaaattgat ttatccttcc ttataacagc agttggggtt 420gtgaatttac gttctactct
tgaaaaagca cttaaatatg attatttgca gatggaagtc 480aatacgattc ctgaagactt
aatatttgaa gggcataatc taggttcaaa agtttttgtg 540actgattgtt ttgaaaacaa
ctgggattct ttagactcaa ctattaataa aatttgtgag 600cttgatattc cgttcatagc
tttcacttct gatggggatg attgggtttg tcaacatgaa 660gtaaaacacc tagtgtccaa
tgtaaaatct gacaaaaaaa agatatactc tttagttggt 720agttcccatg atttggggga
aaatttggtc gttttacgaa atttctatca aagtatgact 780aaagctgctg tctcattgga
taggcaattg gttgaattag ttgatgaaat catagaacca 840aattttgagg atttaaccgt
aattacagtc aatgaaagaa gacttaaaaa taaaatagaa 900aatgaaataa taaacagact
agcagatcga gttcttgctt ccgtataa 94851122DNAArtificial
SequenceSynthesized 5atgtccacct tactaaacat cgatgcaacg gagattaaag
ttagtaccga gatagatgat 60ataatcttta caagtagtcc attaacttta ttatttgaag
atcaagaaaa aattcagaaa 120gaattaatac ttgaaagttt tcattatcat tataaccata
ataaagatta caagtattat 180tgtaatattc agggggttga tgagaacatt caatcaattg
acgacattcc agtatttcct 240acatccatgt ttaaatactc tcgtcttcat acagccgatg
agagtaatat agaaaattgg 300tttacatcat ccggtactaa aggcgttaag tctcatattg
ctagggatag gcagtcaatt 360gaaagattac taggatcagt taattatggt atgaaatatc
ttggagaatt tcatgaacat 420caacttgaac ttgtaaatat gggaccagat cgtttttccg
cttcaaacgt gtggttcaaa 480tatgttatga gtttagtaga attgttatat cctactactt
ttactgtgga aaatgatgag 540atagattttg aacaaactat cttggcttta aaagcgatac
aacgaaaagg aaaaggaata 600tgtttaatag gaccgcctta ttttatatac ttgttatgcc
attatatgaa agaacataat 660atagaattta atgcaggggc tcacatgttt attattacgg
gagggggatg gaaaacaaaa 720caaaaagagg cgttaaatag gcaagatttc aatcaacttc
ttatggaaac attctcctta 780tttcatgagt cacaaattag agacatattt aatcaagttg
aattgaatac atgtttcttc 840gaagattctc ttcaacgaaa acatgtgcca ccttgggtat
atgctcgtgc attagatcct 900gttactttga ctcccgtaga agacgggcag gaaggcttga
tgtcttatat ggacgcctcc 960agtacatcat atccgacttt catcgttacg gatgatattg
gcattgtaag gcatctaaaa 1020gagccagatc ccttccaagg tacaaccgta gaaattgtta
gacgtcttaa cacacgagag 1080caaaagggtt gttctttatc tatggctaca agtcttaaat
aa 11226705DNAArtificial SequenceSynthesized
6atgatcttca actgtaaagt caaaaaagtt gaagcatccg attcacatat ttataaagtc
60tttatcaaac ccgataagtg tttcgatttt aaagcaggcc aatatgttat tgtgtaccta
120aacgggaaaa atttaccatt tagtatagcc aactgtccta catgtaatga attattggaa
180ttacatgtag gcgggtctgt aaaagaatct gcaattgaag caatatcaca ctttattaat
240gcttttatat atcaaaaaga atttactatt gatgctccgc atggagacgc ctggttacga
300gatgagtctc aatctccgct tttgttaata gctggcggca caggtttatc atatatcaat
360agtattttaa gttgctgcat ttctaaacaa ctatcccaac cgatctattt atactggggt
420gtcaacaatt gtaacctttt gtatgcagat caacaattaa aaactttggc cgcacaatat
480cgtaatatta attatatccc tgtagttgag aatcttaata cagattggca aggaaaaatt
540gggaatgtaa tagatgcagt aatcgaagat tttagtgacc tttcagattt cgacatctat
600gtttgtggac ccttcggtat gtccagaaca gctaaagata ttctaatttc acaaaagaaa
660gcaaacatag ggaagatgta ttcagatgct ttttcttaca cgtga
7057702DNAArtificial SequenceSynthesized 7atgactactc tttcttgtaa
ggtgacatca gtggaggcta taactgacac agtgtacaga 60gttagaatcg taccagatgc
agcatttagt tttagggccg gtcaatattt gatggttgta 120atggacgaga gagataagag
accattcagc atggcctcta ctccagatga gaaagggttt 180atcgaactgc acattggagc
atcagagatc aatttatacg caaaagcagt catggacagg 240atcttaaagg accatcagat
tgttgttgat attcctcacg gcgaagcatg gcttagggat 300gatgaggaaa gacctatgat
tctcatcgct ggcggaacag ggttctctta cgctaggtct 360atactcctca ccgccctagc
acgtaatcca aatagggata ttaccattta ctggggtggt 420agagaagagc agcaccttta
cgacctttgc gaattggagg cccttagctt aaagcatcct 480ggtctacaag ttgtgccagt
tgtcgaacaa cctgaggcag gatggagagg gcgtacagga 540acagtgctaa ctgccgtttt
acaggatcat ggcactcttg ctgagcacga tatttatatt 600gccggtagat tcgaaatggc
taagattgca cgtgaccttt tttgttctga aagaaatgcc 660agggaagata gattgttcgg
tgatgctttc gcattcattt ga 7028585DNAArtificial
SequenceSynthesized 8atgtttaaag gaattgtgga aggcattgga atcattgaga
agatagacat atatacagac 60cttgacaagt atgccatcag attccctgaa aacatgttga
acggcattaa aaaagagtct 120tccattatgt ttaacggctg ctttcttaca gtgaccagcg
ttaatagcaa catcgtctgg 180tttgatattt ttgagaagga agctaggaaa ctggatacat
ttagagaata taaggttgga 240gatagagtca atttgggtac attcccaaag tttggtgctg
catctggagg acatattttg 300agtgcaagaa tatcttgcgt tgctagtatt attgagatta
tagagaatga agattatcaa 360cagatgtgga ttcagattcc tgagaacttt actgagttct
taattgacaa agactatatt 420gctgtcgatg gtatctcttt aacaatcgac actataaaaa
acaatcagtt ttttattagt 480ttgccgttaa aaatagctca aaataccaac atgaaatgga
ggaaaaaggg agataaggtt 540aacgtggagt tgtctaataa gattaacgct aatcagtgtt
ggtga 5859478PRTPhotobacterium leiognathi 9Met Ile Lys
Lys Ile Pro Met Ile Ile Gly Gly Val Val Gln Asn Thr 1 5
10 15 Ser Gly Tyr Gly Met Arg Glu Leu
Thr Leu Asn Asn Asn Lys Val Asn 20 25
30 Ile Pro Ile Ile Thr Gln Ser Asp Val Glu Ala Ile Gln
Ser Leu Asn 35 40 45
Ile Glu Asn Lys Leu Thr Ile Asn Gln Ile Val Asn Phe Leu Tyr Thr 50
55 60 Val Gly Gln Lys
Trp Lys Ser Glu Thr Tyr Ser Arg Arg Leu Thr Tyr 65 70
75 80 Ile Arg Asp Leu Ile Lys Phe Leu Gly
Tyr Ser Gln Glu Met Ala Lys 85 90
95 Leu Glu Ala Asn Trp Ile Ser Met Ile Leu Cys Ser Lys Ser
Ala Leu 100 105 110
Tyr Asp Ile Val Glu Asn Asp Leu Ser Ser Arg His Ile Ile Asp Glu
115 120 125 Trp Ile Pro Gln
Gly Glu Cys Tyr Val Lys Ala Leu Pro Lys Gly Lys 130
135 140 Ser Val His Leu Leu Ala Gly Asn
Val Pro Leu Ser Gly Val Thr Ser 145 150
155 160 Ile Leu Arg Ala Ile Leu Thr Lys Asn Glu Cys Ile
Ile Lys Thr Ser 165 170
175 Ser Ala Asp Pro Phe Thr Ala Thr Ala Leu Val Asn Ser Phe Ile Asp
180 185 190 Val Asp Ala
Glu His Pro Ile Thr Arg Ser Ile Ser Val Met Tyr Trp 195
200 205 Ser His Ser Glu Asp Leu Ala Ile
Pro Lys Gln Ile Met Ser Cys Ala 210 215
220 Asp Val Val Ile Ala Trp Gly Gly Asp Asp Ala Ile Lys
Trp Ala Thr 225 230 235
240 Glu His Ala Pro Ser His Ala Asp Ile Leu Lys Phe Gly Pro Lys Lys
245 250 255 Ser Ile Ser Ile
Val Asp Asn Pro Thr Asp Ile Lys Ala Ala Ala Ile 260
265 270 Gly Val Ala His Asp Ile Cys Phe Tyr
Asp Gln Gln Ala Cys Phe Ser 275 280
285 Thr Gln Asp Ile Tyr Tyr Ile Gly Asp Ser Ile Asp Ile Phe
Phe Asp 290 295 300
Glu Leu Ala Gln Gln Leu Asn Lys Tyr Lys Asp Ile Leu Pro Lys Gly 305
310 315 320 Glu Arg Asn Phe Asp
Glu Lys Ala Ala Phe Ser Leu Thr Glu Arg Glu 325
330 335 Cys Leu Phe Ala Lys Tyr Lys Val Gln Lys
Gly Glu Ser Gln Ser Trp 340 345
350 Leu Leu Thr Gln Ser Pro Ala Gly Ser Phe Gly Asn Gln Pro Leu
Ser 355 360 365 Arg
Ser Ala Tyr Ile His Gln Val Asn Asp Ile Ser Glu Val Ile Pro 370
375 380 Phe Val His Lys Ala Val
Thr Gln Thr Val Ala Ile Ala Pro Trp Glu 385 390
395 400 Ser Ser Phe Lys Tyr Arg Asp Ile Leu Ala Glu
His Gly Ala Glu Arg 405 410
415 Ile Ile Glu Ala Gly Met Asn Asn Ile Phe Arg Val Gly Gly Ala His
420 425 430 Asp Gly
Met Arg Pro Leu Gln Arg Leu Val Asn Tyr Ile Ser His Glu 435
440 445 Arg Pro Ser Thr Tyr Thr Thr
Lys Asp Val Ser Val Lys Ile Glu Gln 450 455
460 Thr Arg Tyr Leu Glu Glu Asp Lys Phe Leu Val Phe
Val Pro 465 470 475
10373PRTPhotobacterium leiognathi 10Met Ser Thr Leu Leu Asn Ile Asp Ala
Thr Glu Ile Lys Val Ser Thr 1 5 10
15 Glu Ile Asp Asp Ile Ile Phe Thr Ser Ser Pro Leu Thr Leu
Leu Phe 20 25 30
Glu Asp Gln Glu Lys Ile Gln Lys Glu Leu Ile Leu Glu Ser Phe His
35 40 45 Tyr His Tyr Asn
His Asn Lys Asp Tyr Lys Tyr Tyr Cys Asn Ile Gln 50
55 60 Gly Val Asp Glu Asn Ile Gln Ser
Ile Asp Asp Ile Pro Val Phe Pro 65 70
75 80 Thr Ser Met Phe Lys Tyr Ser Arg Leu His Thr Ala
Asp Glu Ser Asn 85 90
95 Ile Glu Asn Trp Phe Thr Ser Ser Gly Thr Lys Gly Val Lys Ser His
100 105 110 Ile Ala Arg
Asp Arg Gln Ser Ile Glu Arg Leu Leu Gly Ser Val Asn 115
120 125 Tyr Gly Met Lys Tyr Leu Gly Glu
Phe His Glu His Gln Leu Glu Leu 130 135
140 Val Asn Met Gly Pro Asp Arg Phe Ser Ala Ser Asn Val
Trp Phe Lys 145 150 155
160 Tyr Val Met Ser Leu Val Gln Leu Leu Tyr Pro Thr Thr Phe Thr Val
165 170 175 Glu Asn Asp Glu
Ile Asp Phe Glu Gln Thr Ile Leu Ala Leu Lys Ala 180
185 190 Ile Gln Arg Lys Gly Lys Gly Ile Cys
Leu Ile Gly Pro Pro Tyr Phe 195 200
205 Ile Tyr Leu Leu Cys His Tyr Met Lys Glu His Asn Ile Glu
Phe Asn 210 215 220
Ala Gly Ala His Met Phe Ile Ile Thr Gly Gly Gly Trp Lys Thr Lys 225
230 235 240 Gln Lys Glu Ala Leu
Asn Arg Gln Asp Phe Asn Gln Leu Leu Met Glu 245
250 255 Thr Phe Ser Leu Phe His Glu Ser Gln Ile
Arg Asp Ile Phe Asn Gln 260 265
270 Val Glu Leu Asn Thr Cys Phe Phe Glu Asp Ser Leu Gln Arg Lys
His 275 280 285 Val
Pro Pro Trp Val Tyr Ala Arg Ala Leu Asp Pro Val Thr Leu Thr 290
295 300 Pro Val Glu Asp Gly Gln
Glu Gly Leu Met Ser Tyr Met Asp Ala Ser 305 310
315 320 Ser Thr Ser Tyr Pro Thr Phe Ile Val Thr Asp
Asp Ile Gly Ile Val 325 330
335 Arg His Leu Lys Glu Pro Asp Pro Phe Gln Gly Thr Thr Val Glu Ile
340 345 350 Val Arg
Arg Leu Asn Thr Arg Glu Gln Lys Gly Cys Ser Leu Ser Met 355
360 365 Ala Thr Ser Leu Lys 370
111437DNAArtificial SequenceSynthesized 11atgatcaaaa
aaatccctat gataattggg ggagtagtcc agaacacatc cggttatgga 60atgagagaat
taacattaaa caataataaa gttaacattc caattatcac acaaagtgat 120gtagaagcta
ttcaatctct aaatattgag aacaaattga caataaatca gattgtaaat 180ttcctttata
ctgtaggcca aaaatggaaa tctgagacgt atagtcgtcg attaacttat 240atcagagatt
taatcaaatt cttaggttat agtcaggaaa tggctaaatt ggaagctaat 300tggattagta
tgatattatg ttctaaaagt gctttatatg acatagtaga aaatgattta 360agtagtcgtc
atatcattga tgaatggatt ccccaaggtg aatgctatgt aaaagcattg 420cctaagggta
agtccgtaca cttgttagca ggaaatgttc ctttatcagg agtaacctcc 480atactaagag
caattcttac aaaaaatgaa tgcattatta aaactagttc agcagaccca 540tttactgcca
ctgcacttgt taactctttt atagacgttg atgccgaaca tcctataaca 600cgatccatta
gtgtaatgta ttggtcccat tctgaagatt tagcaattcc caaacaaata 660atgtcttgtg
ctgacgttgt tatagcatgg ggaggggacg atgcaataaa atgggcaact 720gaacatgcac
cttctcacgc agacatattg aaattcggac cgaaaaaatc catttccatt 780gtcgataatc
ctacggatat taaggcagct gctatcggag tggctcatga catttgtttt 840tatgatcagc
aagcatgctt ctcaacccaa gatatatatt atatcggaga ttcaattgat 900attttctttg
atgaattagc tcaacagtta aataaatata aagacatttt acctaaaggg 960gaacgaaatt
tcgatgagaa ggcagctttc tcccttactg aaagagagtg tcttttcgca 1020aaatataaag
ttcaaaaagg tgaatcccaa tcttggttgc ttacccaaag tccagcggga 1080agttttggaa
atcaaccttt gagtcgttct gcgtatattc atcaggtaaa tgatataagt 1140gaagtaatac
ccttcgtaca taaagcagtt actcaaactg tagctatcgc gccttgggaa 1200tcaagtttta
aatacagaga tattttggct gagcatggtg ctgagcgtat cattgaagca 1260ggaatgaata
acatttttcg tgtaggaggt gcccacgatg ggatgcgacc cttgcaacgt 1320ttggttaatt
atatttctca tgaacgtcct agtacatata caacaaaaga tgttagtgta 1380aaaatagaac
agacaaggta tcttgaagaa gataaattct tagtttttgt accgtag
1437121122DNAArtificial SequenceSynthesized 12atgtccacct tactaaacat
cgatgcaacg gagattaaag ttagtaccga gatagatgat 60ataatcttta caagtagtcc
attaacttta ttatttgaag atcaagaaaa aattcagaaa 120gaattaatac ttgaaagttt
tcattatcat tataaccata ataaagatta caagtattat 180tgtaatattc agggggttga
tgagaacatt caatcaattg acgacattcc agtatttcct 240acatccatgt ttaaatactc
tcgtcttcat acagccgatg agagtaatat agaaaattgg 300tttacatcat ccggtactaa
aggcgttaag tctcatattg ctagggatag gcagtcaatt 360gaaagattac taggatcagt
taattatggt atgaaatatc ttggagaatt tcatgaacat 420caacttgaac ttgtaaatat
gggaccagat cgtttttccg cttcaaacgt gtggttcaaa 480tatgttatga gtttagtaca
attgttatat cctactactt ttactgtgga aaatgatgag 540atagattttg aacaaactat
cttggcttta aaagcgatac aacgaaaagg aaaaggaata 600tgtttaatag gaccgcctta
ttttatatac ttgttatgcc attatatgaa agaacataat 660atagaattta atgcaggggc
tcacatgttt attattacgg gagggggatg gaaaacaaaa 720caaaaagagg cgttaaatag
gcaagatttc aatcaacttc ttatggaaac attctcctta 780tttcatgagt cacaaattag
agacatattt aatcaagttg aattgaatac atgtttcttc 840gaagattctc ttcaacgaaa
acatgtgcca ccttgggtat atgctcgtgc attagatcct 900gttactttga ctcccgtaga
agacgggcag gaaggcttga tgtcttatat ggacgcctcc 960agtacatcat atccgacttt
catcgttacg gatgatattg gcattgtaag gcatctaaaa 1020gagccagatc ccttccaagg
tacaaccgta gaaattgtta gacgtcttaa cacacgagag 1080caaaagggtt gttctttatc
tatggctaca agtcttaaat aa 1122
User Contributions:
Comment about this patent or add new information about this topic: