Patent application title: CHIMERIC AUTOPROCESSING POLYPEPTIDES AND USES THEREOF
Inventors:
Shmuel Pietrokovski (Rehovot, IL)
Gil Amitai (Rehovot, IL)
Assignees:
YEDA RESEARCH AND DEVELOPMENT CO. LTD.
IPC8 Class: AC12N1563FI
USPC Class:
4353201
Class name: Chemistry: molecular biology and microbiology vector, per se (e.g., plasmid, hybrid plasmid, cosmid, viral vector, bacteriophage vector, etc.) bacteriophage vector, etc.)
Publication date: 2011-01-06
Patent application number: 20110003379
Claims:
1. A chimeric polypeptide comprising an autoproces sing segment having an
amino acid sequence set forth by SEQ ID NO: 37.
2. The chimeric polypeptide of claim 1, wherein said autoprocessing segment is derived from a protein of an organism belonging to a genus selected from the group consisting of Leptospira interrogans.
3. The chimeric polypeptide of claim 1, wherein said auto-cleavage results in removal of a segment of the chimeric polypeptide adjacent to an amino terminal end or a carboxy terminal end of said autoprocessing segment.
4. The chimeric polypeptide of claim 1, wherein said auto-cleavage results in auto-splicing.
5. The chimeric polypeptide of claim 4, wherein said auto-splicing is auto-splicing of segments of the chimeric polypeptide flanking said autoprocessing segment.
6. The chimeric polypeptide of claim 1, further comprising an affinity tag capable of specifically binding a substrate.
7. The chimeric polypeptide of claim 6, wherein said affinity tag is a maltose-binding domain or a chitin-binding domain.
8. The chimeric polypeptide of claim 6, wherein said substrate is selected from the group consisting of a molecule, a compound, a virus, and a cell.
9. A polynucleotide encoding a chimeric polypeptide comprising an autoprocessing segment having an amino acid sequence set forth by SEQ ID NO: 37, the polypeptide being capable of auto-cleavage.
10. The polynucleotide of claim 9, further comprising a promoter sequence being for directing expression of the chimeric polypeptide in an expression system.
11. The polynucleotide of claim 10, wherein said promoter sequence is inducible by isopropyl beta-D-thiogalactoside.
12. A nucleic acid construct comprising the polynucleotide of claim 10.
13. A method of purifying a protein, the method comprising:(a) generating a chimeric polypeptide including an autoproces sing segment having an amino acid sequence set forth by SEQ ID NO: 37, said autoprocessing segment being terminally attached to, or flanked by, an amino acid sequence of the protein, said chimeric polypeptide being capable of auto-cleavage when subjected to suitable conditions to thereby remove said amino acid sequence of the protein from said chimeric polypeptide thereby generating the protein;(b) immobilizing said chimeric polypeptide to a support; and(c) subjecting said chimeric polypeptide to said suitable conditions, thereby purifying the protein.
14. The method of claim 13, further comprising the step of separating the protein from said autoprocessing segment following step (c).
15. The method of claim 13, wherein said support includes an antibody or antibody fragment capable of specifically binding said autoprocessing segment, and whereas said immobilizing is via said autoprocessing segment.
16. The method of claim 13, wherein said chimeric polypeptide further includes an affinity tag sequence, and whereas said immobilizing is via said affinity tag sequence.
17. The method of claim 16, wherein said support includes a specific ligand of said affinity tag sequence, and whereas said immobilizing is via said specific ligand of said affinity tag sequence.
Description:
RELATED APPLICATIONS
[0001]This application is a divisional of U.S. patent application Ser. No.:10/534,544, filed on May 10, 2005 which is a National Phase of PCT Patent Application No. PCT/IL03/00956, having International Filing Date of Nov. 12, 2003, which claims the benefit of U.S. Provisional Patent Application No. 60/425,295 filed on Nov. 12, 2002.
FIELD AND BACKGROUND OF THE INVENTION
[0002]The present invention relates to polypeptides having the capacity to display auto-cleavage, polynucleotides encoding such polypeptides, and uses of such polypeptides and polynucleotides for reversibly binding proteins to specific substrates, reversibly binding specific substrates to each other, and for splicing amino acid sequences. More particularly, the present invention relates to chimeric polypeptides capable of auto-cleaving at defined locations, including auto-cleaving resulting in defined auto-splicing, to polynucleotides suitable for expressing such polypeptides, and to methods of using such polypeptides and polynucleotides for protein purification, affinity selection of display phages, and post-translational ligation of proteins.
[0003]Autoprocessing protein domains, such as inteins and Hogs, have the capacity to post-translationally auto-cleave or auto-splice flanking polypeptide sequences and thereby serve as unique and potent protein engineering tools useful in various applications, including protein purification, affinity selection of display phages, generation of cytotoxic proteins, segmental modification or labeling of proteins, protein or peptide cyclization, and generation of reactive polypeptide termini in expressed proteins for various biochemical reactions, including protein ligation (Perler and Adam, 2000. Curr Opin Biotechnol. 11, 377-83). However, the usefulness of the presently available repertoire of autoprocessing polypeptides is hampered by various limitations, as described in further detail hereinbelow. Inteins are internal protein domains naturally occurring in a variety of host proteins (Hirata et al., 1990. J. Biol. Chem. 265, 6726-6733; Kane et al., 1990. Science 250, 651-657; Perler et al., 1994. Nucl. Acids Res. 22, 1125-1127; Noren et al., 2000. Angew. Chem. Int. Ed. 39, 450). Inteins have been found in organisms from all three domains of life, including in yeast and algal chloroplasts (eukaryotes), mycobacteria and cyanobacteria (bacteria), and thermophilic archaea (archaea). So far, no essential biological role has been shown for inteins, and all of their identified functions involve their own preservation and maintenance, with no apparent benefit to the host protein and organism (reviewed in Pietrokovski, 2000. Trends in Genetics 17, 465-472). At least some inteins are multifunctional, being able to both catalyze their own protein splicing and to home a copy of their gene into intein-less alleles (Gimble and Thorner, 1992. Nature 357, 301; Chong et al., 1996. J Biol Chem. 271, 22159). Hogs are protein domains found in Hedgehogs which are proteins composed of an amino terminal Hedge protein domain and a carboxy terminal Hog protein region (Aspock G., 1999. Genome Res. 9, 909; Hammerschmidt et al., 1997. Trends Genet. 13, 14). Other protein domains, such as various Caenorhabditis elegans carboxy terminal domains, are believed to autocatalytically cleave themselves from host proteins, thereby modulating the activity of the amino terminal parts (Burglin, 1996. Curr Biol. 6, 1047; Porter et al., 1996. Cell 86, 21), similarly to Hogs.
[0004]Members of the intein and Hog protein domain families share the capacity to autocatalytically cleave the peptide bond joining them to polypeptides flanking their amino terminal ends ("amino terminal cleavage"). Inteins have the further capacity to cleave the peptide bond joining them to polypeptides flanking their carboxy terminal ends ("carboxy terminal cleavage") while splicing polypeptides flanking their amino and carboxy terminal ends (termed "exteins"), resulting in self-excision of the intein from the host protein, and concomitant ligation of the flanking extein domains with a peptide bond. Thus, intein-containing host proteins undergo a switch from an intein-containing state to an intein-less state via such a process. Most reported inteins furthermore also contain an endonuclease domain whose function is to mediate the copying of the intein gene into specific unoccupied genomic insertion points, thereby enabling intein propagation.
[0005]Both inteins and Hogs share a similar structure fold and contain characteristic "Hint" consensus motifs which mediate the biochemical reactions involved in the autocatalytic activities of these protein domains (Hall T M., 1997. Cell 91, 85; Pietrokovski S., 1994. Protein Sci. 3, 2340; Pietrokovski S., 1998. Protein Sci. 7, 64; Paulus, 2000. Annu. Rev. Biochem. 69, 447). These Hint motif-mediated biochemical reactions are similar in both inteins and Hogs, but are involved in different biological processes (Dalgaard et al., 1997. J Comput Biol. 4, 193; Hall et al., 1997. Cell 91, 85; Pietrokovski S., 1998. Protein Sci. 7, 64; Xu and Perler, 1996. EMBO J. 15, 5146). The initial biochemical reactions of intein and Hog amino terminal cleavage are identical; the peptide bond attaching the amino terminal end of the Hint domain to an amino terminal-flanking sequence is converted into a thioester (or ester) bond, a transesterification reaction then covalently attaches the sequence flanking the carboxy terminal end of the intein, or a cholesterol molecule in the case of Hog proteins, to the amino terminal flanking sequence, thereby cleaving the bond attaching the amino terminal sequence to the Hint domain. In a process essential for organismal development, Hint-mediated autocatalytic excision of the carboxy terminal Hog protein domain from the amino terminal Hedge protein domain in Hedgehogs leads to covalent attachment of a cholesterol molecule to the carboxy end of the Hedge domain, leading to its activation and secretion from the cell (Porter J A. et al., 1996. Cell 86, 21; Porter, J A. et al., 1996. Science 274, 255). In the case of inteins, protein splicing is effected sequentially by cleavage of the bond attaching the intein amino terminal end to the carboxy terminal extein, ligation of the amino and carboxy terminal exteins, and cleavage of the bond attaching the intein carboxy terminal end to the carboxy terminal extein.
[0006]Mechanistic studies have determined the roles of highly conserved residues positioned near the intein/extein junctions in the splicing reaction (Chong et al., 1996. J. Biol. Chem. 271, 22159-22168; Xu et al., 1996. EMBO J. 15, 5146-5153; Stoddard et al., 1998. Nat. Struct. Biol. 5, 3). These residues include: the Cys, Ser or Thr residue forming the amino terminal end of the intein, which initiates splicing with an acyl shift; the conserved Cys, Ser or Thr residue flanking the carboxy terminal end of the intein, which ligates the exteins through nucleophilic attack; and the conserved Asn forming the carboxy terminal end of the intein, which releases the intein from the ligated exteins via succinimide formation. The amino terminal acyl shift and the carboxy terminal succinimide formation cleavage activities of the intein are separable. The amino terminal cleavage takes place in two separate steps. In the first step, as described above, the peptide bond between the intein and the amino terminal extein is converted to a thioester (or ester in some cases). In the second step, the thioester bond is cleaved by a nucleophilic attack from the side-chain of the residue flanking the carboxy terminal end of the intein, causing a transesterification reaction.
[0007]Because the structural information required for splicing exists entirely within inteins, and since the process of splicing has no energy requirements (for example hydrolysis of ATP), such protein domains can be used in a variety of applications involving intein insertion into foreign contexts. Various methods have been used in attempts to control and alter intein-mediated functions. Since endonuclease activity is not required for protein splicing, mini-inteins with accurate splicing activity have been generated by deletion of this central domain (Derbyshire et al., 1997. Proc. Natl. Acad. Sci. USA. 94, 11466; Chong et al., 1997. J. Biol. Chem. 272, 15587; and Shingledecker et al., 1998. Gene 207, 187). Also, mutation of residues near the intein/extein junctions has been used to alter intein activity, for example, to yield isolated cleavage at one or both of the intein-extein junctions (Chong et al., 1998. J. Biol. Chem. 273, 10567).
[0008]Thus, the ability to modulate the function of autoprocessing polypeptides such as inteins has broad potential application, as described above. In the case of protein purification where an autoproces sing polypeptide is used in conjunction with an affinity group to purify a desired target protein (Chong et al., 1997. Gene 192, 271-281; Chong et al., 1998. Nucl. Acids Res. 26, 5109), purification of a target protein is effected by co-expressing the target protein as a fusion protein containing a purification tag in one terminal segment, an internal autoprocessing polypeptide, and a target protein forming the other terminal segment. Such fusion proteins are exposed to affinity purification matrices designed to capture the tagged molecule. The target protein is then selectively released from the purification matrix by inducing autoprocessing polypeptide-mediated auto-cleavage of the peptide bond attaching the target protein to the autoprocessing polypeptide. Such a procedure is advantageous since autoprocessing polypeptide cleavage affects the fusion protein only, and thus non-specifically bound contaminant proteins are not released into the product stream. Furthermore, such a method does not employ contaminating and expensive proteases, such as those used in technologies employing protease-mediated cleavage of purification-tagged target proteins. The aforementioned strategy forms the basis of the protein purification systems such as the commercially available IMPACT-CN system (New England Biolabs, Beverly, Mass.).
[0009]However, prior art methods of using such autoprocessing polypeptides for applied uses have numerous drawbacks. In applied systems such as IMPACT-CN, the accessory molecule involved in cleavage of the thioester bond between the intein and the extein following amino terminal cleavage must be effected with a strong thiol-containing nucleophile such as 2-mercaptoethanol or dithiothreitol (DTT), both of which are strong reducing agents which modify the carboxy terminal end of the extein. In such systems, although initial thioester formation is mediated by the intein, the actual cleavage of the extein is effected via non-enzymatic chemical cleavage of a thioester bond by a small nucleophilic molecule, thereby severely limiting the maximal reaction rates achievable. While such systems allow carboxy terminal cleavage, such cleavage has the drawback of resulting in undesirable amino terminal cleavage, thereby requiring the amino terminal fragment to be removed in an additional purification step. Furthermore, despite insights into intein structure and function, modifications often result in unacceptably low activity, poor precursor stability, or insolubility (Derbyshire et al., 1997. Proc. Natl. Acad. Sci. USA. 94, 11466; Chong et al., 1997. Gene 192, 271-281; Shingledecker et al., 1998. Gene 207, 187; Chong et al., 1998. Nucl. Acids Res. 26, 5109).
[0010]Thus, all prior art approaches have failed to provide an adequate solution for providing autoprocessing polypeptides optimal for protein engineering applications.
[0011]There is thus a widely recognized need for, and it would be highly advantageous to have, autoproces sing polypeptides devoid of the above limitation.
SUMMARY OF THE INVENTION
[0012]According to one aspect of the present invention there is provided an chimeric polypeptide comprising an autoprocessing segment having an amino acid sequence set forth by SEQ ID NO: 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105 or 106, the polypeptide being capable of auto-cleavage.
[0013]According to another aspect of the present invention there is provided a polynucleotide encoding a chimeric polypeptide comprising an autoprocessing segment having an amino acid sequence set forth by SEQ ID NO: 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105 or 106, the polypeptide being capable of auto-cleavage.
[0014]According to further features in preferred embodiments of the invention described below, the chimeric polypeptide further comprises an affinity tag capable of specifically binding a substrate.
[0015]According to still further features in preferred embodiments, the substrate is selected from the group consisting of a molecule, a compound, a virus, and a cell.
[0016]According to yet another aspect of the present invention there is provided a nucleic acid construct comprising a nucleic acid sequence encoding a chimeric polypeptide comprising an autoprocessing segment having an amino acid sequence set forth by SEQ ID NO: 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105 or 106, the chimeric polypeptide being capable of auto-cleavage.
[0017]According to further features in preferred embodiments of the invention described below, the nucleic acid construct further comprises a promoter sequence being for directing expression of the chimeric polypeptide in an expression system.
[0018]According to still further features in preferred embodiments, the chimeric polypeptide further comprises an affinity tag capable of specifically binding a specific substrate.
[0019]According to still another aspect of the present invention there is provided a method of generating a chimeric polypeptide capable of displaying auto-cleavage, the method comprising generating a chimeric amino acid sequence including an autoprocessing segment, the autoprocessing segment having an amino acid sequence set forth by SEQ ID NO: 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105 or 106, thereby producing the chimeric polypeptide capable of displaying auto-cleavage.
[0020]According to further features in preferred embodiments of the invention described below, the chimeric polypeptide includes an affinity tag.
[0021]According to a further aspect of the present invention there is provided a method of purifying a protein, the method comprising: (a) generating a chimeric polypeptide including an autoprocessing segment having an amino acid sequence set forth by SEQ ID NO: 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105 or 106, the autoprocessing segment being terminally attached to, or flanked by, an amino acid sequence of the protein, the chimeric polypeptide being capable of auto-cleavage when subjected to suitable conditions to thereby remove the amino acid sequence of the protein from the chimeric polypeptide thereby generating the protein; (b) immobilizing the chimeric polypeptide to a support; and (c) subjecting the chimeric polypeptide to the suitable conditions, thereby purifying the protein.
[0022]According to further features in preferred embodiments of the invention described below, the method of purifying a protein further comprises the step of separating the protein from the autoprocessing segment following step (c).
[0023]According to still further features in preferred embodiments, the support includes an antibody or antibody fragment capable of specifically binding the autoprocessing segment, and the immobilizing is via the autoprocessing segment.
[0024]According to still further features in preferred embodiments, the chimeric polypeptide further includes an affinity tag sequence, and the immobilizing is via the affinity tag sequence.
[0025]According to still further features in preferred embodiments, the support includes a specific ligand of the affinity tag sequence, and the immobilizing is via the specific ligand of the affinity tag sequence.
[0026]According to still further features in preferred embodiments, the generating the chimeric polypeptide is effected by synthesizing a polynucleotide encoding the chimeric polypeptide and expressing the polynucleotide in an expression system. According to still further features in preferred embodiments, the expression system is a cellular expression system or a cell-free expression system.
[0027]According to still further features in preferred embodiments, the cellular expression system is an E. coli cellular expression system.
[0028]According to still further features in preferred embodiments, the cell-free expression system is an E. coli S30 extract expression system.
[0029]According to still further features in preferred embodiments, the polynucleotide comprises a promoter sequence being for directing the expression of the chimeric polypeptide.
[0030]According to still further features in preferred embodiments, the promoter sequence is inducible by isopropyl beta-D-thiogalactoside.
[0031]According to still further features in preferred embodiments, the auto-cleavage results in auto-splicing.
[0032]According to still further features in preferred embodiments, the auto-splicing is auto-splicing of segments of the chimeric polypeptide flanking the autoprocessing segment.
[0033]According to a further aspect of the present invention there is provided a method of reversibly attaching a first substrate to a second substrate, the method comprising: (a) providing a chimeric polypeptide including an autoproces sing segment having an amino acid sequence set forth by SEQ ID NO: 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105 or 106 flanked by a first amino acid sequence capable of binding the first substrate and a second amino acid sequence capable of binding the second substrate, the chimeric polypeptide being capable of auto-cleavage when subjected to suitable conditions, to thereby release the first amino acid sequence from the second amino acid sequence; (b) exposing the first substrate and the second substrate to the chimeric polypeptide, thereby generating a complex including the first substrate attached via the chimeric polypeptide to the second substrate; and (c) subjecting the complex to the suitable conditions, thereby detaching the first substrate from the second substrate.
[0034]According to further features in preferred embodiments of the invention described below, each of the first and second substrates is independently selected from the group consisting of a molecule, a compound, a virus, and a cell.
[0035]According to still further features in preferred embodiments, the molecule is amylose or chitin.
[0036]According to still further features in preferred embodiments, the virus is a bacteriophage.
[0037]According to still further features in preferred embodiments, the chimeric polypeptide includes an affinity tag sequence, and the binding the first substrate or the binding the second substrate is via the affinity tag sequence.
[0038]According to still further features in preferred embodiments, the affinity tag sequence is a maltose-binding domain or a chitin-binding domain.
[0039]According to still further features in preferred embodiments, the autoprocessing segment is selected from the group consisting of BIL1_cloth, BIL2_cloth, BIL3_cloth, BIL4_cloth, BIL5_cloth, BIL6_cloth, BIL7_cloth, BIL9_cloth, BIL10_cloth, BIL11_cloth, 3875--87_magma, FhaB_manha, BIL2_neigo, BIL3_neigo, BIL5_neigo, BIL6_neigo, MafB1_neigo, MafB2_neigo, B0369+_neimeB, B0372+_neimeB, B0655+_neimeB, A2115_neime, BIL2_neimeC, BIL3_neimeC, BIL4_neimeC, BIL5_neimeC, BIL6_neimeC, MafB 1_neimeC, FhaB1_psefl-PfO-1, FhaB1_psefl-SBW25, FhaB_psesy, SCP1.201_strco, 39--9_thefus, BIL1_gemob, BIL2_gemob, 0709_lepin, 3725_lepin, 3719_lepin, o665_myxxa, o1078_myxxa, o1070_myxxa, BIL1_strav, BIL2_strav, BIL3_strav, BIL1_pirsp, BIL1_chrvi, BIL1_glovi, BIL2_glovi, BIL3_glovi, BIL4_glovi, BIL5_glovi, BIL6_glovi, BIL7_glovi, o649_versp, o5687_versp, o3395_versp, II0519_brume, BIL2_magma, BIL3_magma, BIL4_magma, BIL5_magma, BIL6_magma, 06786_metex, 00126_rhoca, 00199_rhoca, 00459_rhoca, 00460_rhoca, 00746_rhoca, 00949_rhoca, 01216_rhoca, 01374_rhoca, 01523_rhoca, 01524_rhoca, 02710_rhoca, 03530_rhoca, 4825_rhosp, BIL2_rhosp, BIL1_silpo, BIL2_silpo, BIL3_silpo, BIL4_silpo, BIL5_silpo, BIL6_silpo, BIL7_silpo, BIL8_silpo, BIL9_silpo, BIL10_silpo, BIL11_silpo, BIL12_silpo, BIL13_silpo, BIL14_silpo, BIL15_silpo, BIL16_silpo, Bil1_rhile, BIL1_unknwn, and BIL2_unknwn.
[0040]According to still further features in preferred embodiments, the autoprocessing segment is derived from a protein of an organism belonging to a genus selected from the group consisting of Brucella, Clostridium, Magnetospirillum, Mannheimia, Methylobacterium, Neisseria, Pseudomonas, Rhodobacter, Silicibacter, Streptomyces, Thermobifida, Rhizobium, Chromobacterium, Myxococcus, Leptospira, Pirellula, Gemmata, Gloeobacter and Verrucomicrobium.
[0041]According to still further features in preferred embodiments, the organism is selected from the group consisting of Rhodobacter capsulatus, Rhodobacter sphaeroides, Silicibacter pomeroyi, Brucella melitensis, Brucella suis, Magnetospirillum magnetotacticum, Methylobacterium extorquens, Rhizobium leguminosarum, Neisseria meningitidis, Neisseria meningitidis, Neisseria meningitidis, Neisseria gonorrhoeae, Chromobacterium violaceum, Pseudomonas syringae, Pseudomonas fluorescens, Pseudomonas fluorescens, Mannheimia haemolytica, Myxococcus xanthus, Leptospira interrogans, Streptomyces coelicolor, Streptomyces avermitilis, Thermobifida fusca, Clostridium thermocellum, Pirellula species 1, Gemmata obscuriglobus, Gloeobacter violaceus, and Verrucomicrobium spinosum.
[0042]According to still further features in preferred embodiments, the auto-cleavage results in removal of a segment of the chimeric polypeptide adjacent to an amino terminal end or a carboxy terminal end of the autoproces sing segment.
[0043]According to still further features in preferred embodiments, the segment of the chimeric polypeptide adjacent to the autoproces sing segment is an amino terminal segment or a carboxy terminal segment of the chimeric polypeptide.
[0044]According to still further features in preferred embodiments, the segment of the chimeric polypeptide adjacent to the carboxy terminal end of the autoprocessing segment includes an amino acid residue comprising a nucleophilic group at an amino terminal end thereof.
[0045]According to still further features in preferred embodiments, the nucleophilic group is a hydroxyl group.
[0046]According to still further features in preferred embodiments, the amino acid residue is a threonine residue.
[0047]According to still further features in preferred embodiments, the segment of the chimeric polypeptide adjacent to the amino terminal end of the autoprocessing segment includes a serine amino acid residue at a carboxy terminal end thereof.
[0048]According to still further features in preferred embodiments, the chimeric polypeptide is capable of the auto-cleavage under a condition selected from the group consisting of a temperature selected from a range of 33° C. to 41° C., a pH selected from a range of pH 7.8 to pH 8.2, and a concentration of dithiothreitol selected from a range of 0.1 mM to 20 mM.
[0049]The present invention successfully addresses the shortcomings of the presently known configurations by providing novel chimeric polypeptides capable of defined auto-cleaving, including auto-cleaving resulting in defined auto-splicing, polynucleotides suitable for expressing such polypeptides, and methods of using such polypeptides and polynucleotides to purify proteins, affinity-select display phages, and post-translationally ligate proteins together.
[0050]Unless otherwise defined, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this invention belongs. Although methods and materials similar or equivalent to those described herein can be used in the practice or testing of the present invention, suitable methods and materials are described below. All publications, patent applications, patents, and other references mentioned herein are incorporated by reference in their entirety. In case of conflict, the patent specification, including definitions, will control. In addition, the materials, methods, and examples are illustrative only and not intended to be limiting.
BRIEF DESCRIPTION OF THE DRAWINGS
[0051]The invention is herein described, by way of example only, with reference to the accompanying drawings. With specific reference now to the drawings in detail, it is stressed that the particulars shown are by way of example and for purposes of illustrative discussion of the preferred embodiments of the present invention only, and are presented in the cause of providing what is believed to be the most useful and readily understood description of the principles and conceptual aspects of the invention. In this regard, no attempt is made to show structural details of the invention in more detail than is necessary for a fundamental understanding of the invention, the description taken with the drawings making apparent to those skilled in the art how the several forms of the invention may be embodied in practice.
[0052]In the drawings:
[0053]FIG. 1a is a sequence alignment diagram of the amino acid sequences of various Type A BIL domains (SEQ ID NOs: 8-62). The names of the domains and the amino acid sequence coordinates of their amino terminal residues in their host proteins are indicated to the left of the sequence. Numbers indicated in parentheses indicate the amino acid residue length of an intervening amino acid sequence which may have any amino acid sequence. Dashed lines have been inserted to assist in visualizing the alignments.
[0054]FIG. 1b is a sequence alignment diagram of the amino acid sequences of various Type B BIL domains (SEQ ID NOs: 63-104). The names of the domains and the amino acid sequence coordinates of their amino terminal residues in their host proteins are indicated to the left of the sequence. Numbers indicated in parentheses indicate the amino acid residue length of an intervening amino acid sequence which may have any amino acid sequence. Dashed lines have been inserted to assist in visualizing the alignments.
[0055]FIG. 2a is a sequence diagram depicting an amino acid sequence motif (SEQ ID NO: 105) exclusively defining amino acid sequences of a subset of Type A BIL domains, including: 39--9_thefus, SCP1.201_strco, 3875--87_magma, B0372+_neimeB, B0655+_neimeB, A2115_neime, MafB1_neimeC, BIL2_neimeC, BIL4_neimeC, BIL5_neimeC, BIL6_neimeC, MafB 1_neigo, BIL2_neigo, BIL3_neigo, MafB2_neigo, BIL5_neigo, BIL6_neigo, FhaB_psesy, FhaB_manha, FhaB1_psefl-PfO-1, FhaB1_psefl-SBW25, BIL6_cloth, BIL5_cloth, BIL2_cloth, BIL4_cloth, BIL1_cloth, BIL8_cloth, BIL9_cloth, BIL1_gemob, BIL2_gemob, 0709_lepin, 3725_lepin, 01078_myxxa, o1070_myxxa, BIL1_strav, BIL2_strav, BIL3_strav, BIL1_pirsp, BIL1_chrvi, o3395_versp, o5687_versp, o649_versp, BIL1_glovi, BIL2_glovi, BIL3_glovi, and BIL4_glovi. Amino acid residues are indicated in standard single-letter code. X - any amino acid; X(1-100)--any amino acid sequence composed of 1 to 100 amino acid residues.
[0056]FIG. 2b is a sequence diagram depicting an amino acid sequence motif (SEQ ID NO: 106) exclusively defining amino acid sequences of a subset of Type B BIL domains, including: 4825_rhosp, BIL2_rhosp, 00588_rhoca, 02710_rhoca, 01524_rhoca, 01523_rhoca, 00126_rhoca, 01216_rhoca, 00949_rhoca, 01374_rhoca, 00459_rhoca, 00460_rhoca, 00746_rhoca, 03530_rhoca, 00199_rhoca, BIL3_magma, BIL4_magma, BIL1_brusu, BIL1_unknwn, BIL2_unknwn, 06786_metex, BIL1_silpo, BIL2_silpo, BIL3_silpo, BIL4_silpo, BIL5_silpo, BIL6_silpo, BIL7_silpo, BIL8_silpo, BIL9_silpo, BIL10_silpo, BIL11_silpo, BIL12_silpo, BIL13_silpo, BIL14_silpo, BIL15_silpo, BIL16_silpo, BIL1_rhile, and II0519_brume. Amino acid residues are indicated in standard single-letter code. X--any amino acid; X(1-100)--any amino acid sequence composed of 1 to 100 amino acid residues.
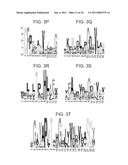
[0057]FIGS. 3a-z are block-logo diagrams depicting conserved amino acids of Hint-like motifs of Type A BIL domains (FIGS. 3a-g) and Type B BIL domains (FIGS. 3h-o) and of homologous Hint motifs of Hog proteins (FIGS. 3p-t) and inteins (FIGS. 3u-z). Unique BIL domain motifs are underlined with hatched lines. Motifs are ordered left to right in the amino to carboxy terminal positions along the protein sequences. Similar motifs are vertically aligned. The motifs are shown as sequence logos in which the heights of amino acid letter designations are proportional to their degree of conservation in each position. Protein splicing active site residues of intein Hint domains are indicated by asterisks. Motifs were identified and are displayed as previously described (Pietrokovski S., 1998. Protein Sci. 7, 64). The BIL domain motifs shown include sequences described in FIGS. 1a-b and Tables 1-2. The intein and hedgehog Hint domain sequences were obtained from previously published sources (Aspock G., 1999. Genome Res. 9, 909; Pietrokovski S., 2001. Trends Genet. 17, 465). The amino acid residue position shown forming the carboxy terminal end of the motif depicted in FIG. 3z actually represents the amino acid residue forming the amino terminal end of the carboxy terminal extein. Only intein and hedgehog motifs common to Hint domains are shown.
[0058]FIGS. 4a-b are dendrograms depicting phylogenetic relationships of Type A and Type B BIL domains, respectively.
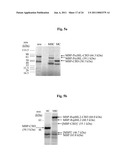
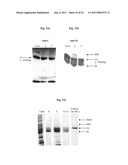
[0059]FIGS. 5a-b are autoradiographs depicting SDS-PAGE separation of in-vitro translated, [35S]-methionine-labeled, protein products of the MBP-BIL-CBD expression constructs pC2C-PsyBIL, and pC2C-RspBIL2 (FIGS. 5a and 5b, respectively, "MBC" lane). Translation of control constructs pC2C ("MC" lane) was used as a positive control and no DNA template ("-" lane) were used as a negative control. Molecular weights were estimated using translation products of pBESTluc as reference standards. In FIG. 5a, molecular weights ("mw" lane) were further estimated using unlabeled protein markers (dotted lines). In FIG. 5b [MBP-CBD]' and [MBP]' indicate splicing and amino terminal cleavage products derived from MBP-RspBIL2-CBD. These fragments migrated at molecular weights greater than those the corresponding control fragments due to their containing residual BIL2_rhosp-flanking amino acid residues as a result of the pC2C-RspBIL2 cloning scheme described under Materials and Methods. Expected molecular weights of identified fragments are indicated in parentheses. The expected molecular weights of MBP-PsyBIL-CBD and of its carboxy terminal cleavage product MBP-PsyBIL, and its MBP-CBD splicing product were 66.3 and 59.1, and 50.6 kDa, respectively.
[0060]FIG. 5c is a diagram of the amino acid sequence of FhaB_psesy (SEQ ID NO: 107) depicting the putative protein splicing motifs (underlined) and catalytic residues (double-underlined) responsible for auto-cleavage-splicing activity by this Type A BIL domain.
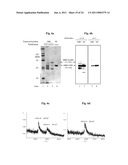
[0061]FIGS. 6a-b are photographs depicting SDS-PAGE analysis of autocatalytically processed protein products overexpressed in-vivo in E. coli transformed with the MBP-PsyBIL-CBD ("MBC" lane) expression construct pC2C-PsyBIL. FIG. 6a depicts Coomassie Blue staining of chitin (lane 2) or amylose (lane 3) affinity column separated protein. Lane 4 is a control showing protein from E. coli transformed with pC2C to overexpress MBP-CBD ("MC" lane) chimeric protein. Fragments corresponding to the MBP-PsyBIL carboxy terminal auto-cleavage product and the MBP-CBD auto-splicing product of the chimera are indicated. FIG. 6b is an autoradiograph depicting Western immunoblotting analysis of separated protein following purification on chitin beads. Both chitin purified samples from E. coli transformed with pC2C-PsyBIL or control plasmid pC2C to overexpress MBP-PsyBIL-CBD or MBP-CBD chimeric proteins, respectively, were separated in duplicate lanes and blotted onto a single nitrocellulose membrane. The membrane was cut in half and each sample was reacted in duplicate with either anti-MBP (anti-M) or anti-CBD (anti-C) antibodies. Both anti-M and anti-C antibodies reacted with the protein band corresponding to the mass of the MBP-CBD product. Protein bands corresponding to MBP-PsyBIL and MBP products, that appear following purification on chitin beads result from non specific binding by excess amounts of overexpressed protein. Expected molecular weights of identified fragments are indicated in parentheses. The expected molecular weights of MBP-PsyBIL-CBD and of its carboxy terminal cleavage product MBP-PsyBIL, and its MBP-CBD splicing product were 66.3 and 59.1, and 50.6 kDa, respectively.
[0062]FIGS. 6c-d are data plots depicting MALDI mass spectra of MBP-CBD ligation product (FIG. 6c) and MBP-PsyBIL carboxy terminal cleavage product (FIG. 6d) electroeluted from SDS-PAGE gels. The expected molecular weights of MBP-PsyBIL-CBD and of its carboxy terminal cleavage product MBP-PsyBIL, and its MBP-CBD splicing product were 66.3 and 59.1, and 50.6 kDa, respectively.
[0063]FIG. 7 is an amino acid sequence diagram depicting positioning of peptide sequences identified by MALDI peptide mass mapping analysis within the amino acid sequence of the MBP-CBD splicing product (SEQ ID NO: 108). Twenty-five tryptic peptide masses (underlined) were assigned to the amino acid sequence of the MBP-CBD protein, corresponding to 49% coverage of the MBP-CBD sequence. Lettering in non-bold/italic font indicates the amino acid sequence of the MBP tag, and the amino acid sequence of the CBD tag (amino acids 394-461) is indicated in bold+italic font. The peptide corresponding to amino acids 388-396 contains the BIL domain splice site between amino acids Ser393 and Thr394.
[0064]FIG. 8 is an amino acid sequence diagram depicting MALDI peptide mapping of the 59.3 kDa MBP-PsyBIL carboxy terminal cleavage product (SEQ ID NO: 109). Underlined sequences correspond to peptides detected by MALDI. Lettering in non-bold/italic font indicates the amino acid sequence of the MBP tag and that in bold/italic font indicates the amino acid sequence of the PsyBIL domain. The carboxy terminal end of the protein, asparagine N541 represents the carboxy terminal of the PsyBIL domain. The expected molecular weights of MBP-PsyBIL-CBD and of its carboxy terminal cleavage product MBP-PsyBIL, and its MBP-CBD splicing product were 66.3 and 59.1, and 50.6 kDa, respectively.
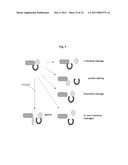
[0065]FIG. 9 is a schematic diagram depicting functions of BIL domains. Hint domains are shown as dark gray horseshoes with their flanks as ovals. Proteins are depicted with amino termini positioned on the left.
[0066]FIGS. 10a-c are electrophoretic analyses depicting C-terminal auto-cleavage by MBP-RspBIL2a-CBD chimera. FIG. 10a is a photograph of a Coomassie blue stained electrophoretic separation of in-vivo expressed MBP-RspBIL2a-CBD chimera affinity purified on amylose depicting C-terminal cleavage activity ("MB" product). FIG. 10b is a photograph of a Western immunoblotting analysis depicting C-terminal cleavage activity ("MB" product) using anti-MBP antibodies (anti-M).
[0067]FIG. 10c is an autoradiograph of an SDS-PAGE separation of the in-vitro translated, [35S]-methionine-labeled chimera depicting C-terminal cleavage activity ("MB" product). M--MBP specific fragment, MB--MBP-BIL specific fragment, MBC--intact chimera.
[0068]FIGS. 11a-b are electrophoretic analyses depicting N-terminal auto-cleavage by in-vivo expressed MBP-4825rhosp-CBD chimera. FIG. 11a is a photograph of a Coomassie blue stained electrophoretic separation of chimera protein products depicting N-terminal cleavage activity of protein products affinity purified on chitin ("BC" product) and affinity purified on amylose ("M" product). FIG. 11b is a photograph of a Western immunoblotting analysis depicting N-terminal cleavage activity using anti-CBD antibody as a probe ("BC" product) and anti-MBP antibody as a probe ("M" product). BC--BIL-CBD specific fragment, M--MBP specific fragment, MB--MBP-BIL specific fragment, MBC--intact chimera.
[0069]FIGS. 12a-c are electrophoretic analyses depicting auto-processing and by in-vivo expressed MBP-BIL4_cloth-CBD chimera. FIG. 12a is a photograph of a Western immunoblotting analysis of amylose (lane "A") or chitin (lane "C") purified protein products depicting auto-splicing activity using anti-CBD antibody as a probe ("MC" product). FIG. 12b is a photograph of a Western immunoblotting analysis of amylose (lane "A") or chitin (lane "C") purified protein products depicting auto-splicing activity ("MC" product). FIG. 12b also shows carboxy terminal auto-cleavage of protein products affinity purified via amylose based affinity chromatography (lane "A", "MB" product). FIG. 12c is a photograph of a Coomassie blue stained electrophoretic separation of protein products isolated via amylose based (lane "A") or chitin based (lane "C") affinity chromatography depicting auto-splicing activity ("MC" species). FIG. 12c also shows carboxy terminal auto-cleavage of protein products affinity purified via amylose based affinity chromatography (lane "A", "MB" species). Note the very small amounts of the uncleaved precursor (lane "C", "MBC" species) suggesting very efficient autoprocessing activity by this chimera. M--MBP specific fragment, MB--MBP-BIL specific fragment, MBC--intact chimera.
DESCRIPTION OF THE PREFERRED EMBODIMENTS
[0070]The present invention is of chimeric autoproces sing polypeptides, polynucleotides encoding such polypeptides, and uses of such polypeptides and polynucleotides for reversibly binding proteins to specific substrates, reversibly binding specific substrates to each other, and auto-splicing amino acid sequences. Specifically, the present invention can be used to purify proteins, to affinity-select display phages, and to post-translationally ligate proteins together.
[0071]Before explaining at least one embodiment of the invention in detail, it is to be understood that the invention is not limited in its application to the details set forth in the following description or exemplified by the Examples. The invention is capable of other embodiments or of being practiced or carried out in various ways. Also, it is to be understood that the phraseology and terminology employed herein is for the purpose of description and should not be regarded as limiting.
[0072]Autoprocessing polypeptides, polypeptides having the capacity to post-translationally auto-cleave and/or auto-splice, can be used to greatly facilitate various industrially and scientifically important biochemical procedures, as described above. For example, such autoproces sing polypeptides can be used in applications which involve reversible binding of proteins to specific substrates, such as protein purification, and reversible binding of specific substrates to each other, such as affinity-selection of display phages.
[0073]Various chimeric autoproces sing polypeptides and methods of using such have been described in the prior art (reviewed in: Perler and Adam, 2000. Curr Opin Biotechnol. 11, 377-83; Paulus H., 2000. Annu Rev Biochem. 69, 447).
[0074]However, all such prior art chimeric autoproces sing polypeptides suffer from various drawbacks. As described above, these drawbacks include suboptimal activity, poor stability, insolubility, requirement for strong auxiliary nucleophiles causing undesirable modifications at carboxy termini of cleaved amino terminal fragments, and undesirable amino terminal cleavages.
[0075]Thus, all prior art approaches have failed to provide optimal chimeric autoprocessing polypeptides for use in protein engineering.
[0076]While reducing the present invention to practice it was uncovered that the chimeric polypeptides of the present invention display efficient auto-cleavage, including auto-cleavage resulting in auto-splicing.
[0077]The chimeric polypeptides of the present invention comprise novel autoprocessing domains characterized by unique amino acid sequences, unique host protein/organism type origins, and unique natural biological capacities. Thus, the chimeric polypeptides of the present invention are highly novel and significantly enlarge and enhance the prior art spectrum of available types of chimeric autoprocessing polypeptides and their possible applications.
[0078]Thus, according to one aspect of the present invention, there are provided chimeric polypeptides having efficient auto-cleavage activity and comprising an autoprocessing segment having an amino acid sequence set forth by SEQ ID NO: 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105 or 106.
[0079]Preferably, the amino acid sequence of the autoprocessing segment is set forth by SEQ ID NO: 12, 31, 76, or 77.
[0080]As used herein, the phrase "auto-cleavage activity" refers to cleavage of a polypeptide of the present invention in a region adjacent to the autoproces sing segment. Auto-cleavage occurs following exposure of the polypeptide of the present invention to suitable conditions in the absence of any other protein. Suitable auto-cleavage conditions are described hereinbelow.
[0081]Depending on the purpose, application and configuration, the polypeptides of the present invention may display different types of auto-cleavage activity.
[0082]Preferably, the auto-cleavage activity of the polypeptides of the present invention results in removal of a segment of the polypeptide adjacent to the amino terminal end or the carboxy terminal end of the autoprocessing segment. Preferably, the segment of the polypeptide adjacent to the autoprocessing segment is an amino terminal segment or a carboxy terminal segment of the polypeptide.
[0083]According to one embodiment, the polypeptides of the present invention may comprise the autoproces sing domain as an amino terminal segment thereof. In this configuration, the polypeptides of the present invention may display removal of the segment thereof adjacent to the carboxy terminal end of the autoproces sing segment, i.e., the segment removed is the carboxy terminal segment of the polypeptide.
[0084]According to further embodiments, the polypeptides of the present invention may comprise the autoprocessing domain as a carboxy terminal segment thereof. In this configuration, the polypeptides of the present invention may display removal of the segment thereof adjacent to the amino terminal end of the autoproces sing segment, i.e., the segment removed is the amino terminal segment of the polypeptide.
[0085]According to yet further embodiments, the polypeptides of the present invention may comprise the autoprocessing domain as an internal segment of the polypeptide. In this configuration, the polypeptides of the present invention may display one or more of the following: removal of a segment adjacent to the amino terminal end of the autoprocessing segment, (i.e., the amino terminal segment of the polypeptide); removal of a segment adjacent to the carboxy terminal end thereof (i.e., the carboxy terminal segment of the polypeptide); removal of both the carboxy and amino terminal segments.
[0086]In some cases, removal of both segments may result in subsequent covalent fusion between the removed segments and, as such, auto-splicing of the polypeptide.
[0087]As used herein, the term "auto-splicing" refers to covalent bond formation between the amino acid residue forming the carboxy terminal end of a segment of the polypeptide adjacent to the amino terminal end of the autoprocessing domain and the amino acid residue forming the amino terminal end of a segment of the polypeptide adjacent to the carboxy terminal end of the autoproces sing domain.
[0088]As is illustrated in the Examples section which follows, the polypeptides of the present invention demonstrate such auto-splicing activity.
[0089]The polypeptides of the present invention may be advantageously used to post-translationally ligate essentially any protein to essentially any other protein via formation of a covalent bond between amino acid residues forming complementary terminal ends thereof. This may be effected by using a polypeptide of the present invention comprising the proteins to be ligated in the configuration described hereinabove enabling auto-splicing to yield the desired ligation product.
[0090]Preferably, the covalent bond is a peptide bond. Alternately, as described above, the covalent bond may be an ester bond, such as the ester bond formed during auto-cleavage of prior art autoprocessing polypeptides.
[0091]Depending on the application and purpose, auto-cleavage of the polypeptide may be specifically induced under suitable conditions, preferably a specific temperature, pH, or concentration of dithiothreitol (DTT).
[0092]Preferably, the temperature is in the range of 33 to 41° C., more preferably the temperature is in the range of 34 to 40° C., more preferably the temperature is in the range of 35 to 39° C., more preferably the temperature is in the range of 36 to 38° C., more preferably the temperature is in the range of 36.5 to 37.5° C., and most preferably the temperature is 37.0° C.
[0093]Preferably, the pH is in the range of pH 7.8 to 8.2, more preferably the pH is in the range of pH 7.9 to 8.1, and most preferably the pH is 8.0.
[0094]Preferably, the concentration of dithiothreitol is in the range of 0.1-20 millimolar, more preferably the concentration of dithiothreitol is in the range of 0.2-10 millimolar, more preferably the concentration of dithiothreitol is in the range of 0.5-5 millimolar, and most preferably the concentration of dithiothreitol is in the range of 1.0-2.0 millimolar.
[0095]Without being bound to a paradigm, the present inventors are of the opinion that auto-cleavage activity, in addition to being governed by the amino acid sequence of the autoprocessing segment, is also influenced by the amino acid sequence of the segments adjacent to the autoproces sing segment.
[0096]For example, the present inventors are of the opinion that in configurations in which the polypeptide comprises a carboxy terminal segment adjacent to the carboxy terminal end of the autoproces sing segment, the efficiency of cleavage may be enhanced if such a carboxy terminal segment includes at its amino terminal end, an amino acid residue comprising a nucleophilic group such as a sulfhydryl group, or more preferably a hydroxyl group.
[0097]As such, in cases wherein the polypeptide of the present invention includes a carboxy terminal segment adjacent to the autoprocessing segment, the amino acid residue forming the amino terminal end of such a carboxy terminal segment is preferably cysteine, serine, or more preferably threonine. As is illustrated in the Examples section below, polypeptides of the present invention having such a carboxy terminal segment display auto-cleavage of the carboxy terminal segment, including auto-cleavage resulting in auto-splicing.
[0098]In configurations wherein the polypeptide of the present invention includes an amino terminal segment adjacent to the autoproces sing segment, the amino acid residue forming the carboxy terminal end of such an amino terminal segment is preferably serine. As is illustrated in the Examples section below, polypeptides of the present invention having such an amino terminal segment display auto-cleavage of the amino terminal segment, including auto-cleavage resulting in auto-splicing.
[0099]It is recognized in the art that certain prior art autoproces sing segments lack the capacity to auto-cleave the bond attaching a given terminal end of the autoprocessing segment to particular flanking amino acid residues (Perler and Adam, 2000. Curr Opin Biotechnol. 11, 377-83).
[0100]In particular, certain prior art autoprocessing amino acid sequences lack the capacity to auto-cleave the bond attaching their amino terminal end to a flanking serine residue. As is illustrated in the Examples section which follows, and in sharp contrast to polypeptides containing such prior art autoprocessing amino acid sequences, the polypeptides of the present invention possess the capacity to auto-cleave such a bond.
[0101]Specific amino acid sequences of autoproces sing domains comprised in the polypeptides of the present invention may be obtained by referring to Table 1, FIGS. 1a-b, and FIGS. 2a-b of the Examples section below. Table 1 provides database coordinates which can be used to retrieve the nucleic acid sequences encoding such amino acid sequences. Such amino acid sequences may easily be determined from such nucleic acid sequences by the ordinarily skilled artisan using a suitable nucleic acid-to-amino acid sequence translation software, such as, for example translation software made publicly available by the National Center for Biotechnology Information (NCBI) or the European Molecular Biology Laboratory (EMBL) on the World Wide Web (WWW). FIGS. 1a-b provide specific amino acid sequences of various autoproces sing domains comprised in the polypeptides of the present invention. FIGS. 2a-b provide amino acid sequence motifs which define autoprocessing domains comprised in the polypeptides of the present invention.
[0102]For example, as is shown in the Examples section which follows, polypeptides of the present invention comprising autoprocessing segments FhaB_psesy (SEQ ID NO: 12), BIL4_cloth (SEQ ID NO: 31), 4825_rhosp (SEQ ID NO: 76) or BIL2_rhosp (SEQ ID NO: 77) display auto-cleavage activity, including auto-cleavage activity resulting in auto-splicing.
[0103]The polypeptides of the present invention may further comprise at least one affinity tag capable of specifically binding a substrate. As is further described hereinbelow, the affinity tag type depends on the intended use of the polypeptide.
[0104]As used herein, the phrase "affinity tag" refers to any moiety (preferably a peptide or polypeptide moiety) which is capable of specifically binding a substrate.
[0105]While the substrate can be essentially any substance or particle which can be specifically bound by the affinity tag, the substrate is preferably a molecule, a compound, a virus, or a cell.
[0106]The polypeptides of the present invention may comprise essentially any affinity tag.
[0107]Examples of peptide/polypeptide affinity tags include streptavidin, His-tags, strep-tags, epitope tags, maltose-binding proteins, and chitin-binding domains.
[0108]His-tags (histidine tags) consist of a chain of 2 to 10, most preferably 6, contiguous histidine amino acid residues. His-tags have the capacity to specifically bind substrates including nickel. Ample guidance regarding tagging polypeptides with His-tags is available in the literature of the art (for example, refer to: Sheibani N. 1999. Prep Biochem Biotechnol. 29:77). Purification of molecules comprising histidine tags is routinely effected using nickel-based automatic affinity column purification techniques. A suitable capture ligand for histidine-tagged molecules is the anti histidine tag single chain antibody 3D5 (Kaufmann, M. et al., 2002. J Mol Biol. 318. 135-47).
[0109]Examples of epitope tags include an 11-mer Herpes simplex virus glycoprotein D peptide, and an 11-mer N-terminal bacteriophage t7 peptide, being commercially known as HSVTag and t7Tag, respectively (Novagen, Madison, Wis., USA), and 10- or 9-amino acid c-myc or Hemophilus influenza hemagglutinin (HA) peptides, which are recognized by the variable regions of monoclonal antibodies 9E10 and 12Ca5, respectively.
[0110]Strep-tags are peptides having the capacity to specifically bind streptavidin. Ample guidance regarding the use of strep-tags is provided in the literature of the art (see, for example: Schmidt, TGM. and Skerra, A. 1993. Protein Eng. 6, 109; Schmidt TGM. et al., 1996. Journal of Molecular Biology 255, 753-766; Skerra A. and Schmidt T G M., 1999. Biomolecular Engineering 16, 79-86; Sano T. and Cantor C R. 2000. Methods Enzymol. 326, 305-11; Sano T. et al., 1998. Journal of Chromatography B 715, 85-91).
[0111]Preferably, the affinity tag is a maltose-binding domain or a chitin-binding domain.
[0112]Preferably, the maltose-binding domain is malE-encoded maltose-binding protein (MBP). Ample guidance regarding the use of maltose-binding protein as an affinity tag is provided in the Examples section which follows and in the literature of the art (see, for example: Guan M. et al., 2002. Protein Expr Purif. 26, 229-34; Cattoli F. and Sarti G C., 2002. Biotechnol Prog. 18, 94-100).
[0113]In cases where the affinity tag is a maltose-binding protein, the substrate is preferably amylose, a specific ligand of such an affinity tag. Alternately, the substrate may be maltose, also a specific ligand of such an affinity tag.
[0114]As is shown in the Examples section below, polypeptides of the present invention comprising maltose-binding protein (MBP) can specifically bind a support including amylose.
[0115]Preferably, the chitin-binding domain is B. circulans cbd-encoded chitin binding domain (CBD). Ample guidance regarding the use of chitin-binding domain as an affinity tag is provided in the Examples section which follows and in the literature of the art (see, for example: Humphries H E. et al., 2002. Protein Expr Purif. 26, 243-8; Chong S. et al., 1997. Gene 192, 271-81).
[0116]In cases where the affinity tag is cbd-encoded chitin binding domain (CBD), the substrate is preferably chitin, a specific ligand of such an affinity tag.
[0117]As is illustrated in the Examples section which follows, polypeptides of the present invention comprising cbd-encoded chitin-binding domain can specifically bind a support including chitin.
[0118]The polypeptides of the present invention can be generated using chemical synthesis approaches or preferably recombinant techniques.
[0119]While reducing the present invention to practice, nucleic acid sequences encoding polypeptides having putative autoproces sing segments were identified in nucleic acid sequence databases (see Examples section below for further detail). The sequences were analyzed and the autoprocessing segment encoding regions were identified and used to generate polynucleotides encoding the polypeptides of the present invention.
[0120]Thus, according to another aspect of the present invention there is provided a polynucleotide sequence which encodes the auto-cleavable polypeptide of the present invention.
[0121]The polynucleotides of the present invention can be assembled from genomic, and/or complementary sequences.
[0122]As used herein, the phrase "complementary sequence" refers to a polynucleotide having a nucleic acid sequence resulting from reverse transcription of messenger RNA using a reverse transcriptase or any other RNA dependent DNA polymerase. Such sequences can be subsequently amplified in-vivo or in-vitro using a DNA dependent DNA polymerase.
[0123]As used herein, the phrase "genomic sequence" refers to a polynucleotide derived from a chromosome which thus reflects a contiguous portion of a chromosome.
[0124]In the case of a polynucleotide of the present invention encoding an autoprocessing domain expressed in a prokaryotic organism, the nucleic acid sequence encoding the autoproces sing domain may be conveniently generated via polymerase chain reaction (PCR) amplification using genomic DNA of the prokaryotic organism as a template.
[0125]Alternately, in the case of a polynucleotide of the present invention encoding an autoproces sing domain expressed in a eukaryotic organism, the nucleic acid sequence encoding the autoproces sing domain may be conveniently generated via PCR amplification using a cDNA library derived from the organism as a template.
[0126]Suitable oligonucleotide primers for PCR amplifying nucleic acid sequences encoding specific autoprocessing domains comprised in polypeptides of the present invention can be designed using the nucleic acid sequences identified by the database coordinates provided in Table 1 of the Examples section which follows.
[0127]For example, as is illustrated in the Examples section below, oligonucleotide primers suitable for PCR amplifying nucleic acid sequences encoding the autoprocessing domains FhaB_psesy, or BIL2_rhosp were derived from nucleic acid sequences retrieved using the relevant database coordinates provided in Table 1 of the Examples section below. The nucleic acid sequences of such primers are set forth by SEQ ID NOs: 4-5, or 6-7, respectively. Ample guidance for determining suitable reaction conditions for amplifying nucleic acid sequences encoding the aforementioned autoprocessing domains is provided in the Examples section which follows. PCR amplification of nucleic acid sequences is a commonly performed procedure and suitable primers and reaction conditions for a broad range of such procedures can generally be routinely determined by one of ordinary skill in the art, for example via suitable software, such as, for example, OLIGO 4.0 (National Biosciences, Plymouth, Minn.).
[0128]It will be appreciated that since autoprocessing activity is a characteristic of the amino acid sequence, one of ordinary skill in the art may alternatively use the amino acid sequences provided herein as a template for designing nucleic acid sequences which encode such amino acid sequences, and which take into consideration parameters such as codon usage which may increase the efficiency of expression of such sequences in specific organisms.
[0129]As described above, the polypeptides of the present invention may further comprise at least one affinity tag. In order to produce polypeptides of the present invention comprising affinity tags, coding nucleotides are formed comprising nucleic acid sequences encoding such affinity tags.
[0130]As described hereinabove, methods of generating nucleic acid sequences encoding affinity tags are well known to one of ordinary skill in the art. For example, nucleic acid sequences encoding affinity tags can be advantageously generated by PCR amplification of nucleic acid sequences encoding such affinity tags.
[0131]For example, as described and demonstrated in the Examples section which follows, suitable oligonucleotide primers for amplification of nucleic acid sequences encoding B. circulans cbd-encoded chitin-binding domain are set forth in SEQ ID NOs: 1-2.
[0132]Alternately, autoproces sing domains may be conveniently cloned into nucleic acid constructs configured for expressing fusion proteins comprising an affinity tag fused to a polypeptide encoded by a nucleic acid insert cloned into such a construct.
[0133]As is shown in the Examples section below, polynucleotides of the present invention comprising nucleic acid sequences encoding the affinity tag maltose-binding protein were assembled by cloning a nucleic acid sequence encoding an autoprocessing domain of the polypeptides of the present invention into the expression construct pMALC2 (New England Biolabs) which is designed to express fusion proteins comprising maltose-binding protein fused to a polypeptide encoded by a cloned insert.
[0134]Such nucleic acid constructs can be advantageously used to insert and/or express a chimeric polynucleotide within a host cell.
[0135]Thus, according to yet another aspect of the present invention there is provided a nucleic acid construct comprising a polynucleotide of the present invention.
[0136]The nucleic acid constructs of the present invention preferably comprise suitable promoter sequences so as to enable efficient expression of the polynucleotide of the present invention in an expression system.
[0137]Expression of the polynucleotides of the present invention may be conveniently controlled using an inducible promoter. A suitable inducible promoter is an isopropyl beta-D-thiogalactoside (IPTG)-inducible promoter, such as a T7 promoter. As described in the Examples section which follows, a T7 promoter can be used to drive IPTG-inducible expression of a polynucleotide of the present invention in a suitable cell-free expression system or in a cellular expression system.
[0138]IPTG-induced expression of polynucleotides under the regulatory control of T7 promoters is widely practiced in the art by the ordinarily skilled practitioner and ample guidance regarding the use of such promoters is available in the literature of the art (see, for example, Sambrook et al., infra).
[0139]The nucleic acid constructs of the present invention can be used to produce the polypeptides of the present invention in a suitable expression system.
[0140]Thus, according to still another aspect of the present invention there is provided a method of generating a polypeptide of the present invention.
[0141]The method is effected by generating a chimeric amino acid sequence including an autoprocessing segment of the present invention. Preferably the chimeric amino acid sequence is generated by expressing a polynucleotide of the present invention in an expression system suitable for generating the chimeric amino acid sequence from the chimeric polynucleotide.
[0142]The nucleic acid constructs of the present invention can be used to express the polypeptides of the present invention in various expression systems, including any cellular or cell-free expression systems suitable for expressing recombinant proteins such as the polypeptides of the present invention.
[0143]When used to express the polypeptides of the present invention in a cell-free expression system, the constructs of the present invention may be advantageously expressed in any suitable in-vitro transcription/translation system.
[0144]Numerous in-vitro transcription/translation systems are commercially available for expressing recombinant proteins such as the polypeptides of the present invention.
[0145]For example, a suitable cell-free expression system for expressing nucleic acid constructs of the present invention is an E. coli S30 extract expression system, as described and as demonstrated in the Examples section below.
[0146]Numerous cellular expression systems, including yeast, bacterial, insect, and mammalian cells can be employed to express the nucleic acid constructs of the present invention.
[0147]As described and illustrated in the Examples section which follows, the polypeptides of the present invention may be advantageously expressed in E. coli by transforming E. coli with the nucleic acid constructs.
[0148]Transformation of E. coli with nucleic acid constructs is a routine procedure widely practiced in the art (see, for example, Sambrook et al., infra).
[0149]For example, for expression of the polypeptides of the present invention using the nucleic acid constructs of the present invention in E. coli,competent cells capable of DNA uptake may be prepared from cells harvested in exponential growth phase and rendered competent via the widely practiced CaCl2 method. Addition of MgCl2 or RbCl to the transformation reaction medium may be employed to increase transformation efficiency. Alternative transformation methods include methods such as electroporation or host cell protoplast transformation.
[0150]As described hereinabove, the capacities of the polypeptides of the present invention to specifically bind substrates and to auto-cleave can be advantageously used in various practical applications involving reversible binding of substrates, such as protein purification.
[0151]Thus, according to a further aspect of the present invention there is provided a method of purifying a protein.
[0152]The method is effected by generating a polypeptide of the present invention comprising an autoprocessing segment being terminally attached to, or flanked by, an amino acid sequence of the protein, immobilizing the polypeptide to a support, and subjecting the immobilized polypeptide to suitable conditions for enabling auto-cleavage resulting in removal of the protein from the polypeptide.
[0153]According to a preferred embodiment, the polypeptide of the present invention is configured such that one terminal end of the autoprocessing segment is adjacent to a terminal segment of the polypeptide being the protein to be purified and the other terminal end of the autoprocessing segment is adjacent to a terminal segment of the polypeptide comprising an affinity tag.
[0154]According to this embodiment, the polypeptide of the present invention is preferably immobilized via a specific binding of the affinity tag to a specific ligand thereof included in the support. Optionally, in cases where auto-cleavage further results in detachment of the autoproces sing segment from the terminal segment of the polypeptide comprising the affinity tag, the method may advantageously further comprise the step of separating the protein from the autoprocessing segment so as to further facilitate purification of the protein. Such separation may be effected as described further hereinbelow.
[0155]According to another embodiment, the polypeptide of the present invention consists of a chimera comprising the autoprocessing segment fused to the protein to be purified.
[0156]According to this embodiment, the polypeptide of the present invention is preferably immobilized via a specific binding of the autoproces sing segment to a specific ligand of the autoproces sing segment included in the support.
[0157]According to yet another embodiment, the polypeptide of the present invention consists of an autoproces sing segment being flanked at its amino terminal end with an amino terminal segment of the amino acid sequence of the protein to be purified, and being flanked at its carboxy terminal end with the carboxy terminal segment of the amino acid sequence of the protein to be purified complementing the amino terminal segment of the amino acid sequence of the protein to be purified.
[0158]According to this embodiment, the polypeptide of the present invention is preferably immobilized via a specific binding of the autoproces sing segment to a specific ligand of the autoprocessing segment included in the support, and auto-cleavage results in auto-splicing of the complementary amino and carboxy terminal segments of the protein to be purified to thereby release the protein.
[0159]In embodiments in which the polypeptide of the present invention is immobilized via the autoprocessing segment, the specific ligand included in the support is preferably an antibody or antibody fragment capable of specifically binding the autoproces sing segment.
[0160]Purification of a protein according to the method of the present invention may be advantageously effected via standard affinity chromatography techniques. For example, a suitable support for immobilizing a polypeptide of the present invention may be an affinity resin coupled to a specific ligand of the polypeptide of the present invention packed in a standard affinity purification column. Following subjecting of the support-bound polypeptide of the present invention to conditions suitable for auto-cleavage thereof, the highly purified protein released from the support-bound autoprocessing segment may be conveniently recovered as a flow-through fraction eluted from the column.
[0161]In the case described hereinabove, wherein auto-cleavage further results in detachment of the autoproces sing segment from the support-bound segment of the polypeptide of the present invention, separating the protein from the autoproces sing segment may be effected analogously to the method described hereinabove by using such standard affinity chromatography techniques wherein the affinity resin includes a specific ligand of the autoproces sing segment.
[0162]Alternately, various methods suitable for separating such mixtures of polypeptides may be practiced by the ordinarily skilled artisan. Such techniques include, for example high-performance liquid chromatography (HPLC), size-exclusion chromatography, and similar methodologies.
[0163]Ample guidance regarding chromatographic isolation of proteins is widely available in the literature of the art (see, for example: Wilchek M. and Chaiken I., 2000. Methods Mol Biol 147, 1-6; Jack G W., 1994. Mol Biotechnol 1, 59-86; Narayanan S R., 1994. Journal of Chromatography A 658, 237-258; Nisnevitch M. and Firer M A., 2001. J Biochem Biophys Methods 49, 467-80; Janson J C. and Kristiansen T. in Packings and Stationary Phases in Chromatography Techniques. Unger K K. (ed.), Marcel Dekker, New York, pp. 747 (1990); Clonis YD: HPLC of Macromolecules: A Practical Approach, IRL Press, Oxford, pp. 157 (1989); Nilsson J. et al., 1997. Protein Expr Purif. 11, 1-16).
[0164]As described in the Examples section which follows, amylose affinity ligand based column chromatography of a polypeptide of the present invention comprising the autoprocessing domain BIL2_rhosp flanked at its amino terminal end with an amino terminal segment including the affinity tag MBP, and flanked at its carboxy terminal end with the amino acid sequence of a protein to be purified resulted in column-retention of a segment of the polypeptide of the present invention lacking the amino acid sequence of the protein to be purified (FIG. 10a), thereby demonstrating the utility of the method of the present invention for purifying proteins.
[0165]As described in the Examples section which follows, chitin affinity ligand based column chromatography of a polypeptide of the present invention comprising the autoprocessing domain 4825_rhosp flanked at its carboxy terminal end with an carboxy terminal segment including the affinity tag CBD, and flanked at its amino terminal end with the amino acid sequence of a protein to be purified resulted in column-retention of a segment of the polypeptide of the present invention lacking the amino acid sequence of the protein to be purified (FIG. 11b), thereby demonstrating the utility of the method of the present invention for purifying proteins.
[0166]According to the teachings of the present invention, the polypeptides of the present invention may include affinity tags flanking the autoprocessing segment.
[0167]Thus, according to yet a further aspect of the present invention there is provided a method of reversibly attaching a first substrate to a second substrate.
[0168]The first and second substrates may be the same or may be different.
[0169]The method is effected using a polypeptide of the present invention in which the autoprocessing segment is flanked by a first amino acid sequence capable of binding the first substrate and a second amino acid sequence capable of binding the second substrate, such that auto-cleavage releases the first amino acid sequence from the second amino acid sequence. Exposing the first substrate and the second substrate to the polypeptide of the present invention generates a complex including the first substrate attached via the polypeptide to the second substrate. Following generation thereof, the complex is subjected to suitable conditions for auto-cleavage, thereby detaching the first substrate from the second substrate.
[0170]Complex generation may be effected in various ways, depending on the application and purpose. For example, complex generation may be effected wherein neither, one or both substrates is included in, or consists of, a support specifically binding an affinity tag comprised in the polypeptide of the present invention.
[0171]According to a preferred embodiment, the method is used for reversibly attaching a first substrate being a protein displayed by a bacteriophage to a second substrate specifically binding the phage-displayed protein, which substrate being included in a support.
[0172]According to this embodiment, the method is effected using a polypeptide of the present invention comprising a first amino acid sequence having the capacity to specifically bind the substrate and a second amino acid sequences having the capacity to bind the phage-displayed protein.
[0173]The method according to this aspect of the present invention may be advantageously employed with phage-display libraries for selecting bacteriophages displaying a protein having high affinity to a specific substrate. This may be effected by exposing a phage display library to a support including a substrate being a target molecule to which a high affinity ligand is desired. Elements of the phage display library not being bound with high affinity to the support may be washed and recovery of phages specifically binding the substrate with high affinity via a displayed protein capable of specifically binding the target molecule with high affinity may be conveniently recovered by subjecting the support-bound phages to conditions suitable for auto-cleavage of the polypeptide of the present invention so as to effect the detachment thereof from the support.
[0174]Additional objects, advantages, and novel features of the present invention will become apparent to one ordinarily skilled in the art upon examination of the following examples, which are not intended to be limiting. Additionally, each of the various embodiments and aspects of the present invention as delineated hereinabove and as claimed in the claims section below finds experimental support in the following examples.
EXAMPLES
[0175]Reference is now made to the following examples, which together with the above descriptions, illustrate the invention in a non limiting fashion.
[0176]Generally, the nomenclature used herein and the laboratory procedures utilized in the present invention include molecular, biochemical, microbiological and recombinant DNA techniques. Such techniques are thoroughly explained in the literature. See, for example, "Molecular Cloning: A laboratory Manual" Sambrook et al., (1989); "Current Protocols in Molecular Biology" Volumes I-III Ausubel, R. M., ed. (1994); Ausubel et al., "Current Protocols in Molecular Biology", John Wiley and Sons, Baltimore, Md. (1989); Perbal, "A Practical Guide to Molecular Cloning", John Wiley & Sons, New York (1988); Watson et al., "Recombinant DNA", Scientific American Books, New York; Birren et al. (eds) "Genome Analysis: A Laboratory Manual Series", Vols. 1-4, Cold Spring Harbor Laboratory Press, New York (1998); methodologies as set forth in U.S. Pat. Nos. 4,666,828; 4,683,202; 4,801,531; 5,192,659 and 5,272,057; "Cell Biology: A Laboratory Handbook", Volumes I-III Cellis, J. E., ed. (1994); "Current Protocols in Immunology" Volumes I-III Coligan J. E., ed. (1994); Stites et al. (eds), "Basic and Clinical Immunology" (8th Edition), Appleton & Lange, Norwalk, Conn. (1994); Mishell and Shiigi (eds), "Selected Methods in Cellular Immunology", W. H. Freeman and Co., New York (1980); available immunoassays are extensively described in the patent and scientific literature, see, for example, U.S. Pat. Nos. 3,791,932; 3,839,153; 3,850,752; 3,850,578; 3,853,987; 3,867,517; 3,879,262; 3,901,654; 3,935,074; 3,984,533; 3,996,345; 4,034,074; 4,098,876; 4,879,219; 5,011,771 and 5,281,521; "Oligonucleotide Synthesis" Gait, M. J., ed. (1984); "Nucleic Acid Hybridization" Hames, B. D., and Higgins S. J., eds. (1985); "Transcription and Translation" Hames, B. D., and Higgins S. J., eds. (1984); "Animal Cell Culture" Freshney, R. I., ed. (1986); "Immobilized Cells and Enzymes" IRL Press, (1986); "A Practical Guide to Molecular Cloning" Perbal, B., (1984) and "Methods in Enzymology" Vol. 1-317, Academic Press; "PCR Protocols: A Guide To Methods And Applications", Academic Press, San Diego, Calif. (1990); Marshak et al., "Strategies for Protein Purification and Characterization--A Laboratory Course Manual" CSHL Press (1996); all of which are incorporated by reference as if fully set forth herein. Other general references are provided throughout this document. The procedures therein are believed to be well known in the art and are provided for the convenience of the reader.
[0177]Unless otherwise defined, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this invention belongs. Although methods and materials similar or equivalent to those described herein can be used in the practice or testing of the present invention, suitable methods and materials are described below.
Example I
Bacterial-Intein Like (BIL) Domains: Novel Auto-Cleaving/Auto-Splicing Protein Domains
[0178]Autoprocessing polypeptides capable of auto-cleavage have been shown to be uniquely useful in a wide range of protein engineering applications, for example for protein purification without the requirement for proteases. However, all prior art autoprocessing polypeptides suffer from various drawbacks, including suboptimal activity, stability, solubility, and requirement for auxiliary molecules causing undesirable protein modifications. In order to enlarge and enhance the current repertoire of autoprocessing polypeptides, the present inventors have identified, generated and demonstrated the functionality of novel autoprocessing polypeptides, as follows.
[0179]Materials and Methods:
[0180]In order to identify novel auto-cleaving/-splicing proteins, databases storing genomic sequences of various organisms, including bacterial pathogens were searched for open reading frames (ORFs) coding for protein sequences containing Hint domains. Following identification of protein sequences containing Hint domains, such protein sequences were cloned and tested for auto-cleaving/-splicing activity, as described below.
[0181]Data sources: BILs were identified in bacterial genomes by searching the following databases: National Center for Biotechnology Information (NCBI) sequence databases for Brucella melitensis (B. melitensis) 16M, Streptomyces coelicolor (S. coelicolor) A3(2), Neisseria meningitidis (N. meningitidis) MC58, N. meningitidis Z2491, Pseudomonas fluorescens (P. fluorescens) PfO-1, Leptospira interrogans (L. interrogans) 56601, Streptomyces avermitilis (S. avermitilis) MA-468, Pirellula species 1, and Chromobacterium violaceum (C. violaceum) ATCC 12472 sequences; Integrated Genomics (http://www.integratedgenomics.com) sequence databases for Rhodobacter capsulatus (R. capsulatus) SB1003 genomic sequences (Haselkorn et al., 2001. Photosynthesis Research 70, 43-52) and Methylobacterium extorquens (M. extorquens) AM1; Joint Genome Institute (http://www.jgi.doe.gov) sequence databases for Rhodobacter sphaeroides (R. sphaeroides) 2.4.1 (Mackenzie et al., 2001. Proc Natl Acad Sci U S A. 99, 2275-2280), Magnetospirillum magnetotacticum (M. magnetotacticum) MS-1, Clostridium thermocellum (C. thermocellum) ATCC 27405, and Thermobifida fusca (T. fusca) YX genomic sequences; The Institute for Genomic Research (http://www.tigr.org) sequence databases for Pseudomonas syringae (P. syringae) DC3000 (Fouts et al., 2002. Proc Natl Acad Sci U S A 99, 2275-2280), Silicibacter pomeroyi (S. pomeroyi) DSS-3, Gemmata obscuriglobus (G. obscuriglobus) UQM 2246, Myxococcus xanthus (M. xanthus) DK1622, and Verrucomicrobium spinosum (V. spinosum) DSM 4136 sequences; The Sanger Institute (http://www.sanger.ac.uk) sequence databases for Neisseria meningitidis (N. meningitidis) FAM18, P. fluorescens SBW25, and Rhizobium leguminosarum (R. leguminosarum) bv. viciae 3841 sequences; University of Oklahoma, Advanced Center for Genome Technology (http://www.genome.ou.edu) sequence databases for Neisseria gonorrhoeae (N. gonorrhoeae) FA1090 genomic sequences; Baylor College of Medicine Human Genome Sequencing Center (http://www.hgsc.bcm.tmc.edu) sequence databases for Mannheimia haemolytica (M. haemolytica) PHL213 genomic sequences; and Kazusa DNA Research Institute database (Japan; http://www.kazusa.or.jp) for Gloeobacter violaceus (G. violaceus) PCC 7421 sequences.
[0182]BIL domain nomenclature: BIL domains were named using the format "a_b" where "b" is an abbreviation of the bacterial species of origin and where "a" is either host protein name (e.g., "FhaB" or "MafB"); an arbitrary BIL# designation; an Integratedgenomics Database (http://ergo.integratedgenomics.com/R_capsulatus.html) number for R. capsulatus; a Computational Biology Program at ORNL (http://genome.ornl.gov/microbial/rsph) analysis code for R. sphaeroides; or a gene number for B.melitensis strain 16M and N.meningitidis strains MC58 and Z2491. Available gene identifier accession numbers and further information relevant to identified BIL domains are provided in Table 1, below.
[0183]Computational sequence analysis: The BLAST software package of the NCBI was used for sequence-to-sequence searches (Altschul, SF. et al., 1997. Nucleic Acids Res. 25, 3389) of BIL domains with BIL domains and with intein sequences, and the BLIMPS software was employed for block-to-sequence searches (Henikoff S. et al., 1995. Gene 163, GC17). Multiple block sequence alignments were constructed using BLOCKMAKER (Henikoff S. et al., 1995. Gene 163, GC17) and MACAW (Schuler GD. et al., 1991. Structure, Function and Genetics 9, 180) software, as previously described (Pietrokovski S., 1998. Protein Science 7, 64). BIL domains were aligned with other BIL domains having higher scores than with intein sequences and alignments of BIL domains with each other was across their whole, or almost whole, lengths (results not shown). This is also a practical way to classify BIL domains as such. Phylogenetic analysis was performed using the PHYLIP software package (Felsenstein J., 1989. Cladistics 5, 164) version 3.55.
[0184]Generation of BIL domain phylogeny dendrograms: BIL domain phylogeny dendrograms were computed from DNA multiple sequence alignment of 49 mostly complete BIL domains aligned across 201 positions, coding for 67 amino acids which could be confidently aligned across BIL domains. Nodes with bootstrap values below 440/1000 were collapsed, and bootstrap values above 800/1000 are shown. Bootstrap values of the nodes grouping all A-type and B-type BILs are 441 and 519, respectively. The D. melanogaster Hedgehog Hint domain (Porter J A., et al., 1996. Cell 86, 21) was used as an outgroup to root the tree. The dendrogram was calculated using the DNADIST program (version 3.5) of the PHYLIP software package (Felsenstein J., 1989. Cladistics 5, 164). Results were verified against those obtained using CLUSTALW software (Thompson J D. et al., 1994. Nucl Acid Res. 22, 4673) and from protein multiple sequence alignments obtained using PHYLIP, PROTDIST, and CLUSTALW software.
[0185]BIL functional activity assays:
[0186]In order to analyze the capacity of BIL domains to auto-cleave/auto-splice flanking sequences, genetic sequences encoding BIL domains and portions of flanking sequences were cloned for expression as chimeric proteins tagged at their amino terminal ends with the malE gene-encoded maltose-binding protein (MBP) affinity tag, and at the carboxy terminal end with the B. circulans cbd gene-encoded chitin-binding domain (CBD) affinity tag. These chimeras were expressed in an in-vitro transcription/translation system, or overexpressed in-vivo in E. coli,and resulting protein products were analyzed for evidence of BIL domain-mediated autoproces sing activity.
[0187]Such chimeras were cloned using BIL domain genetic sequences encoding:
[0188](i) the Type A BIL domain FhaB_psesy (Table 1, FIG. 1a) and its downstream-flanking threonine residue to generate the chimera "MBP-PsyBIL-CBD"; and
[0189](ii) the Type B BIL domain BIL2_rhosp (Table 1, FIG. 1b) including 32 amino terminal-flanking and 11 carboxy terminal-flanking amino acids to generate the chimera "MBP-RspBIL2-CBD".
[0190]Constructs: B. circulans cbd gene sequences encoding CBD were cloned by PCR from expression vector pTYB2 (New England Biolabs, Beverly, Mass.) using the primers 5'-AAATGTCGACTGCGGTGGCCTGACC-3' (SEQ ID NO: 1) and 5'-TGTCGTATTGCTTCCTTTCGGGCTT-3' (SEQ ID NO: 2), and inserted, including the upstream linker 5'-TGCGGTGGCCTGACCGGTCTGAACTCAGGCCTC-3' (SEQ ID NO: 3), into the SalI/PstI linearized, isopropyl beta-D-thiogalactoside (IPTG)-inducible MBP-fusion protein expression construct pMALC2 to generate the MBP-CBD fusion protein expression construct pC2C. Construct pC2C was used in functional assays as a positive control for expression of MBP-CBD.
[0191]For MBP-PsyBIL-CBD expression, genetic sequences encoding PsyBIL and flanking sequences were PCR amplified from P. syringae DC3000 strain genomic DNA (kindly provided by Dr. G. Sessa, Tel-Aviv University) using the primers 5'-AAAAGGATCCTGCTTTGCGGCCGGAACGA-3' (SEQ ID NO: 4) and 5'-AAAATCTAGAGGTATTATGCACCCATGTCTTG-3' (SEQ ID NO: 5), and cloned in BamHI/XbaI linearized pC2C, between the malE MBP-encoding sequences and the CBD expressing cbd sequences to generate the MBP-PsyBIL-CBD expression construct pC2C-PsyBIL.
[0192]For MBP-RspBIL2-CBD expression, genetic sequences encoding RspBIL2 and flanking sequences were PCR-amplified from R. sphaeroides 2.4.1 strain genomic DNA (supplied by Dr. Steven L. Porter, Department of Biochemistry, University of Oxford) using the primers 5'-GAATTCGGTGATTCATCCTTGGGGCGA-3' (SEQ ID NO: 6) and 5'-TCTAGAAAAACACGGCAAGGGCGAGCGG-3' (SEQ ID NO: 7), and cloned in EcoRI/XbaI linearized pC2C, between the MBP-encoding malE sequences and the CBD-encoding cbd sequences to generate the MBP-RspBIL2-CBD expression construct pC2C-RspBIL2.
the MBP-BIL4cloth-CBD expression construct pC2C-BIL4cloth.
[0193]Polymerase chain reactions were performed using a Biometra thermal cycler in a 50 μl reaction mixture containing Taq polymerase buffer (Sigma, St. Louis, Mich.), 1 μl Taq DNA polymerase, 200 mM dNTP, 10 mM of each primer and 100 ng genomic DNA
[0194]The chimeras were constructed such that carboxy terminal or amino terminal cleavage thereof was expected to generate [MBP-BIL+CBD] or [MBP+BIL-CBD] specific protein products, respectively. The MBP- or CBD-containing protein products were expected to vary in size according to the size of the BIL domain flanking sequences included in the BIL sequences cloned in the chimeric proteins. Auto-splicing by the chimeras was expected to generate MBP-CBD protein products having a molecular weight varying according to the size of the BIL domain flanking sequences included in the cloned BIL segment.
[0195]Chimeras were expressed and analyzed for evidence of BIL processing activity, as described below.
[0196]In-vitro BIL protein expression and activity assays: In-vitro transcription-translation of MBP-BIL-CBD and MBP-CBD chimeric proteins was achieved using chimera expression constructs pC2C-RspBIL2 and pC2C-RspBIL2a, and expression construct pC2C as DNA templates, respectively, using E. coli S30 extract for circular DNA system (Promega Kit #L1030, Promega, Madison WI). Reactions were carried out according to the manufacturer's instructions, using a reaction containing 0.25 mM [35S]-methionine, 220 nmol of expression construct DNA as template, 1-2 mM dithiothreitol, and having a pH of about 8.0. Reactions were incubated at 37° C. for 90-120 minutes. Prior to electrophoresis of expressed protein, 5 μl or 10 μl aliquots of reaction mixtures were mixed with four volumes of acetone in order to remove polyethylene glycol. Acetone precipitation was followed by centrifugation at 12,000×g for 5 minutes. The supernatant was discarded and the protein-containing pellet was mixed with gel loading buffer to give a final concentration of 0.06 M Tris-Cl, 2% SDS, 10% (v/v) glycerol, and 0.01% bromophenol blue. Proteins were separated via 7.5% or 10% SDS-PAGE, and the separated proteins were visualized using a phosphor-imaging screen. Phosphor-imaging signals were quantified using NIH IMAGE 1.62 software. Product quantities were derived from values of three independent experiments averaged for each sample together with their standard deviation of the means. The molar percentage of each product was calculated.
[0197]The expected molecular weights of MBP-RspBIL2-CBD, and of its splicing product MBP-CBD, its carboxy terminal cleavage product MBP-RspBIL2, and its MBP-containing amino terminal cleavage product are 68.0, 55.1, 60.5 and 46.7 kDa, respectively.
[0198]The expected molecular weights of control vector pC2C-expressed MBP-CBD and of its MBP portion were 50.3 and 43.0 kDa, respectively.
[0199]In-vivo Type A BIL domain protein expression, purification and activity assay: Competent TB1 E. coli cells (NEB, Beverly, Mass.) were transformed with constructs for expression of MBP-BIL-CBD chimeras. Transformants were plated on LB agar supplemented with ampicillin (100 μg/ml). Single colonies were used to inoculate 3 ml aliquots of LB medium supplemented with ampicillin (100 μg/ml). Following incubation at 37° C. for 16 hours with shaking, 1 ml of culture was used to inoculate a 2 liter flask containing 500 ml of LB supplemented with ampicillin (100 μg/ml). Incubation was continued at 37° C. with shaking until the optical density at 600 nm was 0.6, at which point IPTG was added to a final concentration of 0.3 mM. After further incubation for 3 hours, cells were harvested by centrifugation at 5,000×g for 20 minutes, re-suspended in solution containing 20 mM Tris (pH 7.4), 200 mM NaCl, and protease inhibitor cocktail (Sigma, St. Louis, Mich.), and lysed by sonication. Lysates were centrifuged at 17,000×g for 20 minutes to remove cell debris, and supernatants were harvested for subsequent analyses. Proteins were then affinity purified with either chitin (NEB, Beverly, Mass.) or amylose beads (NEB, Beverly, Mass.) which bind to the CBD or MBP affinity tags included in the chimeric protein. Elution of protein from beads prior to electrophoresis was performed by mixing the protein-bound beads with SDS-PAGE sample loading buffer.
[0200]Western immunoblotting assays and protein staining: Products generated by expression of MBP-BIL-CBD chimeras were separated by SDS-PAGE. Briefly, protein samples were mixed with protein loading buffer to give a final concentration of 0.06 M Tris-C1, 2% SDS, 10% (v/v) glycerol, 0.1 M dithiothreitol, and 0.01% bromophenol blue. All samples were boiled for 3 minutes prior to electrophoresis. Separated proteins were analyzed by Western immunoblotting using either monoclonal mouse anti-MBP (Novus Biologicals, Inc. Littleton, Colo.) antibody for identification of the MBP tag, or polyclonal rabbit anti-CBD (NEB, Beverly, Mass.) for identification of the CBD tag. Secondary antibodies used were HRP conjugated goat anti-mouse IgG or goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories Inc., West Grove, Pa.). Relative apparent molecular weights were calculated using TriChromoRanger (Pierce, Rockford Ill.) prestained markers.
[0201]Electrophoretic gels containing separated proteins were fixed in 40% methanol/7% acetic acid and stained with PhastGel Blue R stain (Pharmacia Biotech AB, Sweden). Gels were destained in 40% methanol/7% acetic acid and then in deionized water, and visualized protein bands were excised and electroeluted for MALDI mass spectroscopy (MS) analysis.
[0202]Electroelution of protein: Protein was electroeluted from gels at 150 volts for 2 hours in GeBAflex tubes (Gene Bio Application Ltd., Israel) using elution buffer containing 0.025% SDS, Tris and Tricine (pH 8.5). Following electroelution SDS was removed from electroeluted protein using cold TCA:acetone precipitation in the presence of 0.5% sodium deoxycholate (NaDOC; T. Mehlman and A. Shainskaya, unpublished).
[0203]In-gel proteolysis: Protein bands from PhastGel Blue R stained gels were destained using multiple washes in 50% acetonitrile in 50 mM ammonium bicarbonate. Destained protein bands were subsequently reduced, alkylated and in-gel proteolysed using either bovine trypsin (sequencing grade, Roche Diagnostics, Germany) or chymotrypsin, (Boehringer Mannheim, Germany) by incubation with 12.5 ng/μl protease in 50 mM ammonium bicarbonate at 37° C., as previously described (Shevchenko et al., 1996. Analytical Chemistry 68, 850). Extracted peptide solutions were dried for subsequent MALDI-MS analysis.
[0204]Mass Spectrometry: Intact molecular mass measurement and peptide mass mapping were performed using a Bruker Reflex III MALDI time-of-flight (TOF) mass spectrometer (Bruker, Bremen, Germany) equipped with SCOUT source, delayed ion extraction, reflector and a 337 nm nitrogen laser. Each mass spectrum was generated using data accumulated from 200 laser shots. Both external and nearby calibrations for proteins were performed using BSA and myoglobin (Sigma). For peptide mapping, internal calibration with molecular ions of regularly occurring matrix ions and peptides derived from trypsin was additionally performed to consolidate further peptide assignment.
[0205]Intact molecular weight measurements by MALDI MS: Gel electroeluted proteins were further purified by cold acetone precipitation. The dried extract from one lane of the gel was re-dissolved in 0.5 ml of 80% formic acid and immediately diluted with water to yield a solution containing 20% formic acid, and 50% of this solution was applied to a target plate.
[0206]Peptide mass mapping by MALDI mass spectrometry: Aliquots of one tenth of the extracted peptide mixture volume, dissolved in 0.1% TFA or formic acid/isopropanol/water (1:3:2), were used for MALDI-MS using the fast evaporation or dry droplet method. Matrix surfaces of a-cyano-4-hydroxycinnamic acid (4-HCCA) or 2,5-dihydroxybenzoic acid (DHB) were utilized for the fast evaporation (Jensen O N. et al., 1996. Rapid Commun in Mass Spectrom. 10, 1371; Vorm 0. et al., 1994. Analyt Chem. 66, 3281) or dry droplet method (Kussmann K. et al., 1997. J Mass Spectrom. 32, 593), respectively.
[0207]Experimental Results:
[0208]Identification of two novel bacterial intein-like domains containing Hint-like motifs: Searches of sequence databases of diverse bacterial species for Hint-like motif-containing putative ORFs identified open reading frames coding for proteins comprising two related types of novel intein-like protein domains termed by the present inventors Type A and Type B bacterial intein-like (BIL) domains. Novel type A and B BIL domain sequences identified are shown aligned in FIGS. 1a (SEQ ID NOs: 8-62) and 1b (SEQ ID NOs: 63-104), respectively. The bioinformatic sources used to identify these BIL domains are shown in Table 1.
[0209]An amino acid sequence motif (SEQ ID NO: 105) was identified (FIG. 2a) which exclusively defines a subset of Type A BIL domains, including domains: 39--9_thefus, SCP1.201_strco, 3875--87_magma, B0372+_neimeB, B0655+_neimeB, A2115_neime, MafB1_neimeC, BIL2_neimeC, BIL4_neimeC, BIL5_neimeC, BIL6_neimeC, MafB1_neigo, BIL2_neigo, BIL3_neigo, MafB2_neigo, BIL5_neigo, BIL6_neigo, FhaB_psesy, FhaB_manha, FhaB1_psefl-PfO-1, FhaB1_psefl-SBW25, BIL6_cloth, BIL5_cloth, BIL2_cloth, BIL4_cloth, BILl_cloth, BIL8_cloth, BIL9_cloth, BIL1_gemob, BIL2_gemob, 0709_lepin, 3725_lepin, 01078_myxxa, o1070_myxxa, BIL1_strav, BIL2_strav, BIL3_strav, BIL1_pirsp, BIL1_chrvi, o3395_versp, o5687_versp, o649_versp, BIL1_glovi, BIL2_glovi, BIL3_glovi, and BIL4_glovi.
[0210]An amino acid sequence motif (SEQ ID NO: 106) was identified (FIG. 2b) which exclusively defines a subset of Type B BIL domains, including: 4825_rhosp, BIL2_rhosp, 00588_rhoca, 02710_rhoca, 01524_rhoca, 01523_rhoca, 00126_rhoca, 01216_rhoca, 00949_rhoca, 01374_rhoca, 00459_rhoca, 00460_rhoca, 00746_rhoca, 03530_rhoca, 00199_rhoca, BIL3_magma, BIL4_magma, BIL1_brusu, BIL1_unknwn, BIL2_unknwn, 06786_metex, BIL1_silpo, BIL2_silpo, BIL3_silpo, BIL4_silpo, BIL5_silpo, BIL6_silpo, BIL7_silpo, BIL8_silpo, BIL9_silpo, BIL10_silpo, BIL11_silpo, BIL12_silpo, BIL13_silpo, BIL14_silpo, BIL15_silpo, BIL16_silpo, BIL1_rhile, and II0519_brume.
[0211]This new type of domain appears in non-conserved regions of hyper-variable proteins. Thus, these domains are distinct from the Hint domains of inteins and Hog-proteins by the species and proteins in which they appear. An analysis of amino acid residue conservation within Hint-like motifs of BIL domains and within homologous Hint domain motifs of Hog proteins and inteins is shown in (FIGS. 3a-z). Examination of BLAST sequence alignments (Altschul, S F. et al., 1997. Nucleic Acids Result. 25, 3389) of BIL domains with BIL domains and with intein sequences showed that BIL domains aligned with each other with higher scores than with intein sequences across their whole, or almost whole, lengths (results not shown). Therefore, BIL domains were found to be distinct from inteins by their global sequence features.
TABLE-US-00001 TABLE 1 Databases used to identify BIL domains. BIL BIL domain Type name* Source** Date*** Contig/Entry Coordinates A BIL1_cloth NCBI 23022619 -- A BIL2_cloth NCBI 28N'aa+3023020813+ -- 59aa+23020812 A BIL3_cloth NCBI 23022239+14N'aa -- A BIL4_cloth NCBI 23020817+5N'amin 311-445 o acid/gil23020817 A BIL5_cloth NCBI 23020815+13N'aa -- A BIL6_cloth NCBI 23022237 -- A BIL7_cloth NCBI 23022587+7N'aa -- A BIL9_cloth NCBI 23022893+59aa+ -- 23022892 A BIL10_cloth NCBI 22262017 34594-34986 A BIL11_cloth NCBI 22262176 416-728 A 3875_87_magma NCBI 21614488 76532-75165 A FhaB_manha BCM 4 Oct. 2001 C78-C85 11046-20977 A BIL2_neigo OU- 26 Sep. 2000 AE004969 1563413-1564129 ACGT A BIL3_neigo OU- 26 Sep. 2000 AE004969 1565033-1565809 ACGT A BIL5_neigo OU- 26 Sep. 2000 AE004969 1351509-1350766 ACGT A BIL6_neigo OU- 26 Sep. 2000 AE004969 1349978-1349310 ACGT A MafB1_neigo OU- 26 Sep. 2000 AE004969 1560214-1561941 ACGT A MafB2_neigo OU- 26 Sep. 2000 AE004969 1355876-1354062 ACGT A B0369+_neimeB NCBI 7225591 +34 N' aa -- A B0372+_neimeB NCBI 7225594 +11 N' aa -- A B0655+_neimeB NCBI 7225882 +14 N' aa -- A BIL2_neimeC Sanger 15 May 2002 NmC 1836857-1837573 A BIL3_neimeC Sanger 15 May 2002 NmC 1838418-1838981 A BIL4_neimeC Sanger 15 May2002 NmC 1839771-1840439 A BIL5_neimeC Sanger 15 May 2002 NmC 627204-627920 A BIL6_neimeC Sanger 15 May 2002 NmC 628395-629102 A MafB1_neimeC Sanger 15 May 2002 NmC 1833717-1835480 A FhaB1_psefl- NCBI- 205922-575 3-6313 PfO-1 TIGR A FhaB1_psefl- Sanger 2 Sep. 2002 Pflu552a01 41728-29387 SBW25 A FhaB_psesy TIGR 30 Aug. 2002 5668 5148986-5149429 A SCP1.201_strco NCBI 13620683 +32 N' aa A 39_9_thefus JGI 1 Nov. 2000 39 13655-15508 A BIL1_gemob TIGR 23 Sep. 2002 14 32-20392 A BIL2_gemob TIGR 14 Feb. 2003 354 16692-11734 A 0709_lepin NCBI 10 Nov. 2002 24213409 -- A 3725_lepin NCBI 10 Nov. 2002 24216424 -- A 3719_lepin NCBI 10 Nov. 2002 24197710 -- A o665_myxxa TIGR 23 Apr. 2003 168 495-3704 A o1078_myxxa TIGR 23 Apr. 2003 157 103477-106710 A o1070_myxxa TIGR 23 Apr. 2003 168 495-3704 A BIL1_strav NCBI 20 Jul. 2003 29826740 -- A BIL2_strav NCBI 20 Jul. 2003 29826826 -- A BIL3_strav NCBI 20 Jul. 2003 29831835 -- A BIL1_pirsp NCBI 20 Jul. 2003 32470666 -- A BIL1_chrvi NCBI 07 Sep. 2003 34104178 -- A BIL1_glovi Kazusa 04 Sep. 2003 gll0211 -- A BIL2_glovi Kazusa 04 Sep. 2003 gll0207 -- A BIL3_glovi Kazusa 04 Sep. 2003 gll0213 -- A BIL4_glovi Kazusa 04 Sep. 2003 gll0212 -- A BIL5_glovi Kazusa 04 Sep. 2003 gll0205 -- A BIL6_glovi Kazusa 04 Sep. 2003 gll0208 -- A BIL7_glovi Kazusa 04 Sep. 2003 gsl3615 -- A o649_versp TIGR 02 Sep. 2003 65738 3-1949 A o5687_versp TIGR 02 Sep. 2003 65921 18348-1288 A o3395_versp TIGR 02 Sep. 2003 65925 56853-46669 B II0519_brume.sup. NCBI 17988864 B BIL2_magma NCBI 21613062 922-1590 B BIL3_magma NCBI 21614112 2449-1475 B BIL4_magma NCBI 21614173 2216-3187 B BIL5_magma NCBI 21612572 3-338 B BIL6_magma NCBI 21613847 2033-1774 B 06786_metex IG June 2002 1507 6076-7113 B 00126_rhoca IG December 2001 2G06-2D11 114767-113670 B 00199_rhoca IG December 2001 2G06-2D11 178648-177806 B 00459_rhoca IG December 2001 2G06-2D11 434469-435083 B 00460_rhoca IG December 2001 2G06-2D11 435094-436191 B 00746_rhoca IG December 2001 2D10-2D06 2243-3079 B 00949_rhoca IG December 2001 2A12-2D05 325707-326651 B 01216_rhoca IG December 2001 2A12-2D05 222590-223555 B 01374_rhoca IG December 2001 2A12-2D05 148470-149462 B 01523_rhoca IG December 2001 1A01-1C09 279638-280444 B 01524_rhoca IG December 2001 1A01-1C09 280569-281423 B 02710_rhoca IG December 2001 1D09-1F02 197288-199588 B 03530_rhoca IG December 2001 1A01-1C09 521700-521173 B 4825_rhosp JGI 26 Mar. 2001 184 67785-67165 B BIL2_rhosp JGI 26 Mar. 2001 177 9673-10194 B BIL1_silpo TIGR 18 Jun. 2002 50 10679-14440 B BIL2_silpo TIGR 18 Jun. 2002 50 1-2688 B BIL3_silpo TIGR 18 Jun. 2002 4 10260-12356 B BIL4_silpo TIGR 18 Jun. 2002 290 18579-19640 B BIL5_silpo TIGR 18 Jun. 2002 11 3224-2217 B BIL6_silpo TIGR 18 Jun. 2002 199 13687-15303 B BIL7_silpo TIGR 18 Jun. 2002 199 15104-16399 B BIL8_silpo TIGR 18 Jun. 2002 32 1329-2600 B BIL9_silpo TIGR 18 Jun. 2002 32 28672-26588 B BIL10_silpo TIGR 18 Jun. 2002 60 19338-18265 B BIL11_silpo TIGR 18 Jun. 2002 89 10027-11181 B BIL12_silpo TIGR 18 Jun. 2002 126 9020-10066 B BIL13_silpo TIGR 18 Jun. 2002 110 13640-14440 B BIL14_silpo TIGR 18 Jun. 2002 125 9769-10353 B BIL15_silpo TIGR 18 Jun. 2002 129 7727-8257 B BIL16_silpo TIGR 18 Jun. 2002 195 7271-8050 B Bil1_rhile Sanger 14 Jul. 2003 RHIZ10E3Cb12.s1 -- k B BIL1_unknwn TIGR 13 Mar. 2001 14712 1124-51 B BIL2_unknwn TIGR 13 Mar. 2001 12703 2-1369 *the bacterial species origin of the BIL domains is identified by the bacterial species code following the underscore in the BIL name (refer to Table 2 below for the code-species correspondance). **The sources are named as follows JGI--Joint Genome Institute (http://www.jgi.doe.gov), NCBI--(http://www.ncbi.nlm.nih.gov), Sanger--The Sanger Institute (http://www.sanger.ac.uk), OU-ACGT--University of Oklahoma, Advanced Center for Genome Technology (http://www.genome.ou.edu), TIGR--The Institute for Genomic Research (http://www.tigr.org), BCM--Baylor College of Medicine Human Genome Sequencing Center (http://www.hgsc.bcm.tmc.edu), IG--IntegratedGenomics (http://www.integratedgenomics.com), and Kazusa--Kazusa DNA Research Institute database (Japan; http://www.kazusa.or.jp). ***Dates refer to the data release dates used. The NCBI entries were extended as noted. Coordinates of the BIL host protein ORF are given for nucleotide contigs/entries. The positions of BILs within these ORFs are provided in FIGS. 1a-b. .sup. An identical sequence was identified in Brucella suis
[0212]Conserved motifs in BIL domains, Hog proteins and inteins: Alignments of conserved motifs in Type A BIL domains, Type B BIL domains, Hog proteins, and inteins are shown in FIGS. 3a-g, 3h-o, 3p-t, and 3u-z, respectively. Type A BIL domains were found to share 7 Hint-like consensus sequence motifs (FIGS. 3a-d and 3f-g) and one novel motif (FIG. 3e) having no known Hog or intein counterpart.
[0213]Almost all Type A BIL domains were found to comprise apparent functional protein splicing active sites corresponding to those present in inteins (marked by asterisks in FIGS. 3u, 3w and 3z), and several are also flanked at their carboxy terminal ends with serine or threonine amino acid residues, similarly to the carboxy terminal ends of inteins.
[0214]Type A BIL domains were found to contain an invariant His-Asn amino acid residue pair adjacent to the carboxy terminal end thereof (FIG. 3g), similarly to the His-Asn amino acid residue pair typically forming the carboxy terminal ends of inteins (FIG. 3z, positions 7-8). Conservation of the Type A BIL domain carboxy terminal end with that of inteins suggests that, similarly to inteins, Type A BILs undergo cyclization of the Asn residue forming the carboxy terminal end thereof. However, the residue at the carboxy terminal end of Type A BIL domains (FIG. 3g, position 8), which corresponds to the residue forming the amino terminal end of polypeptide segments flanking the carboxy terminal ends of inteins which is always Cys, Ser or Thr (FIG. 3z, position 9), is not conserved, since only a few Type A BIL domains have a serine or threonine residue in that position. Other Type A BIL domains have aspartate, glutamate, asparagine, tyrosine or alanine residues in that position, which are not found in any intein.
[0215]Type B BIL domains were also found to share 6 Hint-like consensus sequence motifs (FIGS. 3h-i and 31-o) and two novel motifs (FIGS. 3j-k) having no known Hog protein or intein Hint domain counterpart. Type B BIL domain carboxy terminal ends were found to have a conserved position comprising Cys, Ser or Thr residues (FIG. 3o, position 6), potentially corresponding to the carboxy terminal flanking position of inteins (FIG. 3z, position 9), however this carboxy terminal residue of
[0216]Type B BIL domains (FIG. 3o, position 7) is not preceded by the His-Asn motif typically found in inteins (FIG. 3z, positions 7-8) and in Type A BIL domains (FIG. 3g, positions 6-7). The --SH/--OH groups on the side chains of the aforementioned Cys, Ser or Thr residues in intein host proteins have been found to be essential for ligation of the intein carboxy and amino flanks in the protein splicing reaction (Xu M Q. and Perler F B., 1996. EMBO J. 15, 5146).
[0217]In Type A BIL domains whose carboxy terminal ends are not flanked by Thr or Ser residues, Asn cyclization may nevertheless occur without trans-esterification by the flanking residue. Alternately, trans-esterification may occur by the mildly nucleophilic residues found in this position. In the first case the BIL domain would be cleaved from a segment flanking its carboxy terminal end, and in the second case protein splicing would occur. Since Type B BIL domains do not have any conserved Asn or Gln residue at their carboxy terminal end, cleavage of this end could then proceed by a mechanism different from the Asn and Gln cyclizations of inteins (Paulus H., 2000. Annu Rev Biochem. 69, 447).
[0218]Key amino acid residues corresponding to protein splicing active sites (marked by asterisks in FIGS. 3u, 3w, and 3z-position 9) were found to be conserved in Type B BIL domains.
[0219]Both Type A and Type B BIL domains were found to be distinct from inteins in having additional unique sequence motifs, in not being integrated in highly conserved sites of essential proteins, and in not comprising endonuclease domains.
[0220]Phylogenetic distribution of BIL domains: The phylogenetic distribution of the BIL domains identified is shown in Table 2. BIL domains were identified in 3 evolutionarily distant bacterial types--alpha, beta and gamma proteobacteria (gram-negative bacteria), actinobacteria (high GC gram-positive bacteria), and Bacillus/Clostridium group bacteria (low GC gram-positive bacteria).
[0221]Both presence and genomic distribution were found to be variable, even in closely related species and strains. For example, 1, 3 and 6 ORFs encoding BIL domains were identified, respectively, in N. meningitidis strains whose genomes have been completely or almost completely sequenced; 2 and 14 ORFs encoding BIL domains were identified in 2 different Rhodobacter species; and while one such ORF was identified in P. syringae, none were found in P. aeruginosa and P. putida.
[0222]BIL domains and inteins were found to coexist in certain species. For example, the genome of M. magnetotacticum was found to comprise ORFs encoding both Type A and Type B BIL domains, and that of T. fusca was found to comprise ORFs encoding both BIL domains and inteins.
[0223]The variability observed in the number of BIL domain ORFs in different species is probably due to gene duplications. As shown in a dendrogram demonstrating phylogenetic relationships of Type A BIL domains (FIGS. 4a), all BIL domains derived from Neisseria species cluster together, and BIL domains from different species sub-cluster as well, implying that all Neisseria BIL domains arose by duplication from a single ancestor and that some are paralogs within different species. The latter is corroborated by the apparent duplication of some gene loci containing BIL domains in these species (not shown). Clustering of BIL domains from the same species was also observed in C. thermocellum (FIG. 4a) and M. Magnetotacticum (FIG. 4b).
[0224]BIL domain host proteins: BIL domains were identified in putative ORFs coding for a few hundred to a few thousand amino acids. Several BIL domains were found to be flanked by domains present in secreted bacterial proteins. In P. syringae and M. haemolytica, BIL domains were identified near the carboxy terminal end of FhaB-like ORFs. FhaB is a very large Bordetella gene coding for a secreted filamentous hemagglutinin protein, which functions as an adhesin important for B. pertussis virulence (Smith A M. et al., 2001. FEMS Microbiol Rev. 25, 309). Three of the R. capsulatus BIL domain-containing ORFs include RTX repeats--calcium binding repeats found in various secreted bacterial proteins, including many toxins (Coote J G., 1992. FEMS Microbiol Rev. 8, 137). In N. meningitidis and N. gonorrhoeae, BIL domains were identified in MafB proteins. These are part of multiple adhesin family possibly involved in glycolipid adhesion to cells (Naumann et al., 1999. Curr Opin Microbiol 2, 62-70; Paruchuri et al., 1990. Proc Natl Acad Sci U S A. 87, 333-7). Three other Neisseria BIL domains were found to have an HNH nuclease domain in amino acid sequences flanking their carboxy terminal ends. HNH domains are found in various DNase and endonuclease proteins including secreted toxins (Belfort M. and Roberts R J., 1997. Nucleic Acids Res. 25, 3379; James R. et al., 1996. Microbiology 142, 1569). A domain present in the amino acid sequence flanking the carboxy terminal end of a BIL domain in the gram-positive bacterium T. fusca is also found in a short, conserved Salmonella ORF (GenBank accession NP--454902) and in an amino acid sequence flanking the carboxy terminal end of a N. meningitidis FhaB/hemolysin protein (gene NMA0688). Both of these proteins are from gram-negative bacteria and are likely to be secreted.
TABLE-US-00002 TABLE 2 Phylogenetic distribution of BIL domains identified. No. of BIL domains identified BIL in Taxonomic group Species Species code type organism alpha proteobacteria Rhodobacter capsulatus SB1003 rhoca B 14.sup.* alpha proteobacteria Rhodobacter sphaeroides 2.4.1 rhosp B 2.sup.* alpha proteobacteria Silicibacter pomeroyi DSS-3 silpo B 16.sup.* alpha proteobacteria Brucella melitensis 16M/Brucella suis brume B 1 alpha proteobacteria Magnetospirillum magnetotacticum MS-1 magma A 1 alpha proteobacteria B 5.sup.* alpha proteobacteria Methylobacterium extorquens AM1 metex B 1.sup.* alpha proteobacteria Rhizobium leguminosarum by. viciae 3841 rhile B 1 beta proteobacteria Neisseria meningitidis Z2491 neimeA A 1 beta proteobacteria Neisseria meningitidis MC58 neimeB A 3 beta proteobacteria Neisseria meningitidis FAM18 neimeC A 6.sup.* beta proteobacteria Neisseria gonorrhoeae FA1090 neigo A 6 beta proteobacteria Chromobacterium violaceum ATCC 12472 chrvi A 1 gamma Pseudomonas syringae DC3000 psesy A 1.sup.* proteobacteria gamma Pseudomonas fluorescens PfO-1 psefl-PfO-1 A 1.sup.* proteobacteria gamma Pseudomonas fluorescens PfSBW25 psefl-SBW25 A 1.sup.* proteobacteria gamma Mannheimia haemolytica PHL213 manha A 1.sup.* proteobacteria delta proteobacteria Myxococcus xanthus DK1622 myxxa A 3.sup.* spirochaetes Leptospira interrogans 56601 lepin A 3 actinobacteria Streptomyces coelicolor A3(2) strco A 1 actinobacteria Streptomyces avermitilis MA-468 stray A 3 actinobacteria Thermobifida fitsca YX thefu A 1.sup.* Bacillus/Clostridium Clostridium thermocellum ATCC 27405 cloth A 10.sup.* group planctomycetes Pirellula species 1 pirsp A 1 planctomycetes Gemmata obscuriglobus UQM 2246 gemob A 2.sup.* cyanobacteria Gloeobacter violaceus PCC 7421 glovi A 7 verrucomicrobium Verrucomicrobium spinosum DSM 4136 versp A 3.sup.* unknown Unknown B 2.sup.* .sup.*Genome not fully sequenced--total number of BILs may be greater.
[0225]BIL domain-mediated auto-cleavage/auto-splicing activity:
[0226]Type A BIL domain-mediated auto-cleavage/auto-splicing activity in an in-vitro transcription/translation system: Electrophoretic analysis (FIG. 5a) showed that protein products generated following in-vitro transcription/translation of the MBP-PsyBIL-CBD expression construct pC2C-PsyBIL displayed molecular weights corresponding to the uncleaved precursor MBP-PsyBIL-CBD, the splicing product MBP-CBD, and the carboxy terminal cleavage product MBP-PsyBIL. Two additional protein products displayed molecular weights of 43 and 45 kDa. Control reactions using the MBP-CBD expression construct pC2C as a transcription template produced two protein products, one corresponding in weight to MBP-CBD and the other to the MBP portion thereof. The latter may be observed in chimeric proteins having MBP as an amino terminal tag (refer to: NEB instruction manual "pMAL protein fusion and purification system", Catalog #E8000S). The 43 kDa protein product may represent a premature transcription or translation stop side product unrelated to BIL domain-mediated activity. Appearance of the 45 kDa band, not seen in the control reaction and slightly larger than the expected weight of MBP, may be due to an additional termination point introduced in the BIL domain. As radioactive methionine was used to label the reaction products, and as, unlike the MBP and BIL domains, the CBD domain lacks a methionine residue, its isolated product cannot be visualized according to the protocol employed.
[0227]The relative amounts of the MBP-PsyBIL-CBD, MBP-PsyBIL and MBP-CBD protein products were found to be 15%, 57% and 28%, respectively.
[0228]These results therefore demonstrated the capacity of Type A BIL domains to auto-cleave and auto-splice flanking sequences.
[0229]Type B BIL domain-mediated auto-cleavage and auto-splicing activity in an in-vitro transcription/translation system: Electrophoretic analysis (FIG. 5b) showed that protein products generated following in-vitro translation of the MBP-RspBIL2-CBD expression construct pC2C-RspBIL2 included proteins with sizes corresponding to the unprocessed MBP-RspBIL2-CBD precursor, the carboxy terminal cleavage product MBP-RspBIL2, the MBP-containing amino terminal cleavage product, and the splicing product MBP-CBD. Also apparent was a 43 kDa MBP-containing fragment which also appeared in the control reaction, as described above.
[0230]These results therefore demonstrated the capacity of Type B BIL domains to auto-cleave and auto-splice flanking sequences. The putative protein splicing motifs and catalytic residues of the FhaB_psesy amino acid sequence responsible for the observed autoprocessing activity are shown in FIG. 5c (SEQ ID NO: 107).
[0231]Recombinant BIL domains expressed in-vivo in E. coli display auto-cleavage and auto-splicing activity--mass spectrometric confirmation of BIL-mediated autoprocessing activity: In order to examine the possibility of industrially producing functional recombinant Type A BIL domain, pC2C-PsyBIL was overexpressed in-vivo in E. coli,and the recombinant protein products were analyzed for BIL domain-mediated autoprocessing activity.
[0232]Similarly to the in-vitro results described above, purified protein from E. coli transformed with pC2C-PsyBIL was found to include a protein product having a molecular weight corresponding to the MBP-PsyBIL carboxy terminal cleavage product, as well as a protein product having a molecular weight corresponding to the MBP-CBD auto-splicing product, as determined by both Coomassie Blue staining and Western immunoblotting analysis of SDS-PAGE separated proteins (FIGS. 6a and 6b, respectively). The main product was again observed to be MBP-PsyBIL protein, as displayed by comparing the quantity thereof produced to that of the MBP-CBD protein product when both were purified using amylose beads (FIG. 6a, lane 3).
[0233]The identities of the PsyBIL reaction products, the MBP-CBD and MBP-BIL protein products were also confirmed by mass spectrometry analysis (FIGS. 6c and 6d, respectively). The measured mass of the MBP-CBD protein (50,602.07 Da) was found to be in close agreement to the expected mass of the unmodified protein (50,266.39 Da). The measured and expected masses of MBP-BIL protein were also found to be in close agreement: 59,332.79 and 59,070.11 Da, respectively. A prominent peak with a mass of 43,303 Da, was also observed in MALDI spectra of electroeluted 50 kDa MBP-CBD protein. Such cross-contamination can be observed in gel purified protein bands (A. Shainskaya, unpublished). Reactivity of the 43 kDa band with anti MBP tag antibody (FIG. 6b) indicates that this band is a truncated product.
[0234]Peptide mass mapping of the 50.1 and 59.3 kDa protein products proteins by MALDI analysis (Tables 3 and 4, respectively) confirmed their assigned identity as MBP-CBD (FIG. 7; SEQ ID NO: 108) and MBP-BIL (FIG. 8; SEQ ID NO: 109), respectively, in particular by identifying the splice junction of the MBP-CBD protein (peptide position 388-396; FIG. 7; Table 3) and the carboxy terminal end of the MBP-BIL protein (peptide position 535-541; FIG. 8; Table 4) with accuracies of 27 and 100 ppm, respectively.
[0235]Thus, following extensive experimentation, the splicing junction and cleavage points were found to precisely correspond to those predicted from the sequence similarity of the BIL and intein domains, thereby unambiguously demonstrating BIL domain autoproces sing capacity.
TABLE-US-00003 TABLE 3 MALDI identification of peptides of the 50.1 kDa MBP-CBD splicing product of MBP-PsyBIL-CBD. [M + H].sup.+ [M + H].sup.+ Mass Peptide calculated measured accuracy position.sup.* mass (Da) mass (Da) (ppm) 1-2 278.1538 278.1460 28 3-7 563.2677 563.2599 13 8-16 1057.604 1057.596 7 8-26 2047.363 2047.35 6 27-35 1064.532 1064.586 50 28-35 936.442 936.491 52 28-30 423.2244 423.215 22 90-99 1267.647 1267.6 37 129-138 1201.522 1201.602 66 129-141 1571.732 1571.823 57 191-201 1188.642 1188.711 58 172-180 1129.55 1129.57 17 253-274 2137.972 2138.147 81 279-296 2095.812 2096.030 104 297-306 1010.472 1010.612 138 328-345 2109.02 2109.017 1 356-387 3461.516 3461.33 53 364-387 2576.53 2576.41 46 388-396 983.48 983.55 71 397-435 3986.34.sup.* 3986.13 52 397-438 4380.80.sup.* 4379.90 205 439-461 2634.93.sup.* 2634.72 79 .sup.*mass corresponds to peptide with an alkylated cysteine residue
[0236]These results therefore fully and clearly demonstrate the capacity of BIL domains to auto-cleave and auto-splice flanking sequences, similarly to inteins. The presently described BIL domains therefore represent a novel and highly useful class of autoprocessing proteins which can be harnessed for manipulating and modifying proteins, for example as depicted in FIG. 9. These results furthermore demonstrate the suitability of utilizing genetically transformed host cells, such as E. coli,to industrially express chimeric proteins which comprise functional BIL domains.
TABLE-US-00004 TABLE 4 MALDI-identification of carboxy terminal peptides of the 59.3 kDa MBP-PsyBIL carboxy terminal cleavage product of MBP-PsyBIL-CBD. Peptide [M + H].sup.+ calculated [M + H].sup.+ measured Mass accuracy position mass (Da) mass (Da) (ppm) 535-541 897.4947 897.47 27 537-541 656.3156 656.25 100
[0237]Discussion: BIL domains are present in several hyper-variable bacterial proteins, such as FhaB adhesins and MafB proteins of Neisseria strains. Their immediate flanks are the most variable portions of the proteins and they themselves are not always present in these proteins, even in closely related strains of the same species. Some, and perhaps all, proteins with BIL domains seem to be secreted proteins. BIL domains might enhance the variability of secreted proteins by their protein splicing and cleavage activity as detailed below.
[0238]As described above, the amino terminal ends of BIL domains, and of Hint domains of inteins and Hog proteins are very similar (FIGS. 3a-z). Thus all these domains probably form labile ester bonds on their amino terminal ends. In proteins with BIL domains these ester bonds could be attacked by various nucleophilic molecules, including peptides, proteins and small reactive compounds, such as glutathione or cysteine. Such reactions would ligate the attacking nucleophiles to a carboxy terminal position of the host protein and release the BIL domain and the host protein region downstream to it. This is analogous to Hedgehog protein maturation where the Hint domain mediates the attachment of a cholesterol molecule to the cleaved Hedge domain. In adhesins with BIL domains this putative ligation might serve to covalently attach the bacteria to its adhesion target. Additionally, released BIL and carboxy terminal domains could have a function of their own. For example, in pathogenic bacteria that have such proteins, the released domains could serve as decoys to the immune system.
[0239]In Neisseria strains, sequences encoding BIL domains appear as either as short open reading frames downstream of MafB genes and in the carboxy terminal ends of these proteins upstream of a variable domain. This suggests that, at least in such Neisseria strains, BIL domains function as cassettes which can be fused to genes by genetic rearrangement to promote the variability of the encoded proteins. Other microevolutionary processes in Neisseria and Ralstonia solanacearum, a plant pathogen bacterium with a wide host range, are known to generate different carboxy terminal ends for surface-exposed and virulence proteins (Parkhill et al., 2000. Nature, 404, 502-506; Salanoubat et al., 2002. Nature 415, 497-502).
[0240]Not all species with BIL domains are pathogens and many pathogenic bacteria with fully sequenced genomes do not have BIL domains. BIL domains might be used in different processes not connected with pathogenicity. For example, BIL domain activity might be one way for bacteria to attach to diverse surfaces.
[0241]In summary, two novel types of Hint domain-containing proteins, BIL Types A and B, were identified. Both types have the active site sequence features of the Hint domains but also possess sequence features that distinguish them from the known Hint domains and from each other. BIL domains appear in different proteins from diverse bacteria, including pathogenic species of humans and plants, such as Neisseria meningitidis and P. syringae. These domains are present in variable protein regions and are typically flanked by domains that also appear in secreted proteins such as filamentous hemagglutinin and calcium binding RTX repeats. Phylogenetic and genomic analysis of BIL domain sequences suggests that they were positively selected for in different lineages. Type A and Type B BIL domains were cloned and shown to display auto-cleavage and auto-splicing of flanking polypeptide sequences in an in-vitro transcription/translation system, as well as when overexpressed in E. coli, thereby indicating the capacity of BIL domains to autocatalyze post-translational modifications of host proteins.
[0242]Conclusion: The above-described experimental results demonstrate the capacity of the autoprocessing polypeptides of the present invention to efficiently auto-cleave and auto-splice flanking sequences. The presently described experimental results furthermore demonstrate the feasibility of utilizing genetically transformed host cells, such as E. coli,for efficient industrial production of such autoprocessing polypeptides. Thus, the autoprocessing polypeptides and the polynucleotides encoding such polypeptides of the present invention significantly expand and enhance the available repertoire of available autoproces sing polypeptides having utility in numerous commercially important protein engineering applications, such as protein purification, affinity selection of display phages and post-translational protein ligation.
Example 2
Supplementary Evidence Demonstrating C-Terminal In-Vitro and In-Vivo Autocleavage by Chimeric Protein Including the Type B BIL Domain BIL2_rhosp
[0243]Materials and Methods:
[0244]In order to analyze the capacity of Type B BIL domain BIL2_rhosp (Table 1, FIG. 1b) to display autoprocessing activity, genetic sequences encoding this BIL domain including one flanking amino acid residue at each terminus were cloned for expression as a chimeric protein tagged at its amino terminal end with the malE gene-encoded maltose-binding protein (MBP) affinity tag, and at the carboxy terminal end with the B. circulans cbd gene-encoded chitin-binding domain (CBD) affinity tag. The resultant "MBP-RspBIL2a-CBD" chimera was expressed in-vivo and in-vitro and resulting protein products were analyzed for evidence of BIL domain-mediated autoprocessing activity according to methods described in Example 1 above.
[0245]Experimental Results:
[0246]The in-vivo expressed MBP-RspBIL2a-CBD chimera was shown to display C-terminal auto-cleavage activity via amylose-based affinity purification, electrophoretic separation, and Coomassie blue staining of the electrophoretically separated proteins (FIG. 10a). The in-vivo expressed MBP-RspBIL2a-CBD chimera was also shown to display C-terminal cleavage via Western immunoblotting analysis of SDS-PAGE separated proteins (FIG. 10b). The in-vitro expressed MBP-RspBIL2a-CBD chimera was shown to display C-terminal cleavage following [35S]-methionine-labeling and autoradiography of electrophoretically separated protein (FIG. 10c). Evidence from intact mass mass-spectrometry of the MBP-BIL specific carboxy terminal cleavage product indicates that the C-terminal end of this product is located 6-11 amino acid residues from the predicted carboxy terminal end of the BIL domain towards the N-terminus. Evidence for the MBP-RspBIL2a identity of the cleavage product was obtained from Western Blot, and affinity column analysis. The presence of the protein chaperone DnaK was detected during affinity purification of the BIL products. This chaperone may bind the BIL domain, and may also be involved in its activity.
[0247]Conclusion: There is ample evidence that a chimeric protein which comprises the type B BIL domain BIL2_rhosp has the capacity to efficiently display carboxy terminal auto-cleavage activity. Hence, such a BIL domain can therefore be advantageously exploited in applications benefiting from an auto-cleaving chimeric protein.
Example 3
N-Terminal Autocleavage of In-Vivo Expressed Chimeric Polypeptide Comprising the Type B BIL Domain 4825_rhosp
[0248]Materials and Methods:
[0249]In order to analyze the capacity of Type B BIL domain 4825_rhosp (Table 1, FIG. 1b) to display autoprocessing activity, genetic sequences encoding this BIL domain with 14 amino terminal-flanking and 51 carboxy terminal-flanking amino acids were cloned for expression as a chimeric protein tagged at its amino terminal end with the malE gene-encoded maltose-binding protein (MBP) affinity tag, and at the carboxy terminal end with the B. circulans cbd gene-encoded chitin-binding domain (CBD) affinity tag. The resultant "MBP-4825rhosp-CBD" chimera was expressed in-vivo and resulting protein products were analyzed for evidence of BIL domain-mediated autoprocessing activity according to methods described in Example 1 above.
[0250]Experimental Results:
[0251]The in-vivo expressed MBP-4825rhosp-CBD chimera was shown to display N-terminal auto-cleavage activity via chitin or amylose based affinity purification of expressed protein and electrophoretic separation, and Coomassie blue staining of electrophoretically separated protein (FIG. 11a). The chimera was also shown to display N-terminal cleavage via Western immunoblotting analysis of SDS-PAGE separated proteins using anti-CBD and anti-MBP antibodies (FIG. 11b). The cleavage site was found to be located exactly between the predicted N-terminal residue of the BIL domain and the amino terminal flanking sequence as demonstrated by amino terminal end sequencing. Large amounts of the chaperone GroEL were detected during the purification of the protein products when overexpres sing the MBP-4825rhosp-CBD chimera the whereas no GroEL was detected when overexpressing control chimera lacking the BIL domain.
[0252]Conclusion: A chimeric protein which comprises the type B BIL domain 4825_rhosp has the capacity to efficiently display N-terminal auto-cleavage activity. Hence, such a BIL domain can therefore be advantageously exploited in applications benefiting from an auto-cleaving chimeric protein.
Example 4
Auto-Splicing and C-Terminal Auto-Cleavage of In-Vivo Expressed Chimeric Protein Including the Type A BIL Domain BIL4_cloth
[0253]Materials and Methods:
[0254]In order to analyze the capacity of Type A BIL domain BIL4_cloth (Table 1, FIG. 1a) to display autoprocessing activity, genetic sequences encoding this BIL domain were cloned, including one flanking amino acid residue of the host protein at the C-terminal end, for expression as a chimeric protein tagged at its amino terminal end with the malE gene-encoded maltose-binding protein (MBP) affinity tag, and at the carboxy terminal end with the B. circulans cbd gene-encoded chitin-binding domain (CBD) affinity tag. The resultant "MBP-BIL4cloth-CBD" chimera was expressed in-vivo and resulting protein products were analyzed for evidence of BIL domain-mediated autoprocessing activity according to methods described in Example 1 above.
[0255]Experimental Results:
[0256]The in-vivo expressed MBP-BIL4cloth-CBD chimera was shown to display auto-splicing activity (MBP-CBD specific fragment) as shown by Western immunoblotting assay using anti CBD and anti MBP antibody probes (FIGS. 12a and 12b, respectively), and as shown by Coomassie blue staining of electrophoretically separated protein products isolated via amylose based or chitin based affinity chromatography depicting auto-splicing activity. The identity of the auto-splicing product was verified via mass-spectrometry peptide mapping (data not shown). Carboxy terminal auto-cleavage activity (MBP-BIL domain specific fragment) was also detected by Western immunoblotting assay using an anti MBP antibody probe (FIG. 12b) and by Coomassie blue staining of electrophoretically separated protein products isolated via amylose based affinity chromatography (FIG. 12c).
[0257]Conclusion: A chimeric protein which comprises A type a BIL domain, such as the type A BIL domain BIL4_cloth, has the capacity to efficiently display auto-splicing and carboxy terminal auto-cleaving activity. Hence, such a BIL domain can therefore be advantageously exploited in applications benefiting from an auto-splicing and/or carboxy terminal auto-cleaving chimeric protein.
[0258]It is appreciated that certain features of the invention, which are, for clarity, described in the context of separate embodiments, may also be provided in combination in a single embodiment. Conversely, various features of the invention, which are, for brevity, described in the context of a single embodiment, may also be provided separately or in any suitable subcombination.
[0259]Although the invention has been described in conjunction with specific embodiments thereof, it is evident that many alternatives, modifications and variations will be apparent to those skilled in the art. Accordingly, it is intended to embrace all such alternatives, modifications and variations that fall within the spirit and broad scope of the appended claims. All publications, patents, patent applications and sequences identified by their accession numbers mentioned in this specification are herein incorporated in their entirety by reference into the specification, to the same extent as if each individual publication, patent, patent application or sequence identified by their accession number was specifically and individually indicated to be incorporated herein by reference. In addition, citation or identification of any reference in this application shall not be construed as an admission that such reference is available as prior art to the present invention.
Sequence CWU
1
109125DNAartificial sequenceSingle strand DNA oligonucleotide 1aaatgtcgac
tgcggtggcc tgacc
25225DNAartificial sequenceSingle strand DNA oligonucleotide 2tgtcgtattg
cttcctttcg ggctt
25333DNAartificial sequenceSingle strand DNA oligonucleotide 3tgcggtggcc
tgaccggtct gaactcaggc ctc
33429DNAartificial sequenceSingle strand DNA oligonucleotide 4aaaaggatcc
tgctttgcgg ccggaacga
29532DNAartificial sequenceSingle strand DNA oligonucleotide 5aaaatctaga
ggtattatgc acccatgtct tg
32627DNAartificial sequenceSingle strand DNA oligonucleotide 6gaattcggtg
attcatcctt ggggcga
27728DNAartificial sequenceSingle strand DNA oligonucleotide 7tctagaaaaa
cacggcaagg gcgagcgg
288143PRTNeisseria meningitidismisc_feature(36)..(62)Xaa can be any
naturally occurring amino acid 8Ser Phe His Gly Ser Thr Leu Val Lys Thr
Ala Asp Gly Tyr Lys Ala1 5 10
15Ile Ala His Ile Gln Ala Gly Asp Arg Val Phe Ala Lys Asp Glu Thr
20 25 30Ser Gly Lys Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Asn Asn 50 55 60Gln Thr Leu Ile Ser
Asn Lys Ile His Pro Phe Tyr Ser Xaa Xaa Xaa65 70
75 80Trp Ile Gln Ala Gly Arg Leu Lys Lys Gly
Asp Thr Leu Leu Ser Glu 85 90
95Ser Gly Ala Lys Gln Thr Val Gln Asn Ile Thr Leu Lys Xaa Xaa Xaa
100 105 110Xaa Lys Ala Tyr Asn
Leu Thr Val Ala Asp Trp His Thr Tyr Phe Val 115
120 125Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Gly Val Trp Val
His Asn Glu 130 135
1409143PRTNeisseria meningitidis (group B)misc_feature(36)..(62)Xaa can
be any naturally occurring amino acid 9Ser Phe His Gly Ser Thr Leu Val
Lys Thr Ala Asp Gly Tyr Lys Ala1 5 10
15Ile Ala Arg Ile Arg Thr Gly Asp Arg Val Phe Ala Lys Asp
Glu Ala 20 25 30Ser Gly Lys
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Asn Asn 50 55 60Gln Thr
Leu Ile Ser Asn Lys Ile His Pro Phe Tyr Ser Xaa Xaa Xaa65
70 75 80Trp Ile Gln Ala Gly Arg Leu
Lys Lys Gly Asp Thr Leu Leu Ser Glu 85 90
95Ser Gly Ala Lys Gln Thr Val Gln Asn Ile Thr Leu Lys
Xaa Xaa Xaa 100 105 110Xaa Lys
Ala Tyr Asn Leu Thr Val Ala Asp Trp His Thr Tyr Phe Val 115
120 125Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Gly Val
Trp Val His Asn Asp 130 135
14010143PRTNeisseria meningitidis (group B)misc_feature(36)..(62)Xaa can
be any naturally occurring amino acid 10Ser Phe His Gly Ser Thr Leu Val
Lys Thr Ala Asp Gly Tyr Lys Ala1 5 10
15Ile Ala Arg Ile Arg Thr Gly Asp Arg Val Phe Ala Lys Asp
Glu Ala 20 25 30Ser Gly Lys
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Asn Asn 50 55 60Gln Thr
Leu Ile Ser Asn Lys Ile His Pro Phe Tyr Ser Xaa Xaa Xaa65
70 75 80Trp Ile Gln Ala Gly Arg Leu
Lys Lys Gly Asp Thr Leu Leu Ser Glu 85 90
95Ser Gly Ala Lys Gln Thr Val Gln Asn Ile Thr Phe Lys
Xaa Xaa Xaa 100 105 110Xaa Lys
Ala Tyr Asn Leu Thr Val Ala Asp Trp His Thr Tyr Phe Val 115
120 125Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Gly Val
Trp Val His Asn Asp 130 135
14011142PRTMagnetospirillum magnetotacticummisc_feature(14)..(14)Xaa can
be any naturally occurring amino acid 11Cys Phe Val Ala Gly Thr Pro Val
Arg Met Ala Asp Gly Xaa Glu Lys1 5 10
15Ala Ile Glu Thr Val Glu Ile Gly Glu Gln Val Gln Gly Thr
Asp Gly 20 25 30Thr Ile Asn
Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Asn Ser Leu Asp Phe Phe
Val Thr Ala Asp His 50 55 60Pro Phe
Leu Thr Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa65
70 75 80Xaa Xaa Xaa Xaa Xaa Ala Leu
Asn Val Thr Gln Leu Val Ile Gly Asp 85 90
95Thr Leu Ile Thr Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa 100 105 110Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Val Val Tyr Asn Leu His Leu Ile Gly 115
120 125Asn Asn Thr Tyr Val Ala Ser Gly Tyr Tyr
Val His Asn Tyr 130 135
14012149PRTPseudomonas syringaemisc_feature(36)..(66)Xaa can be any
naturally occurring amino acid 12Cys Phe Ala Ala Gly Thr Met Val Ser Thr
Pro Asp Gly Glu Arg Ala1 5 10
15Ile Asp Thr Leu Lys Val Gly Asp Ile Val Trp Ser Lys Pro Glu Gly
20 25 30Gly Gly Lys Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa 50 55 60Xaa Xaa Glu Asp Glu
Ser Leu Leu Val Thr Pro Gly His Pro Phe Tyr65 70
75 80Val Xaa Xaa Xaa Xaa Xaa Phe Val Pro Val
Ile Asp Leu Lys Pro Gly 85 90
95Asp Arg Leu Gln Ser Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
100 105 110Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Lys Thr Tyr Asn 115
120 125Leu Thr Val Asp Val Gly His Thr Phe Tyr Val Xaa
Xaa Leu Lys Thr 130 135 140Trp Val His
Asn Thr14513149PRTPseudomonas fluorescensmisc_feature(36)..(66)Xaa can be
any naturally occurring amino acid 13Cys Phe Ala Ala Gly Thr Met Val Ala
Thr Pro Lys Gly Glu Arg Ala1 5 10
15Ile Glu Thr Leu Lys Ile Gly Asp Val Val Trp Ser Lys Pro Glu
Gln 20 25 30Gly Gly Glu Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 50 55 60Xaa Xaa Ser
Ser Glu Thr Leu Glu Val Thr Pro Gly His Pro Phe Tyr65 70
75 80Val Xaa Xaa Xaa Xaa Xaa Phe Val
Pro Leu Ile Glu Leu Gln Pro Gly 85 90
95Asp Arg Leu Gln Ser Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa 100 105 110Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Arg Thr Tyr Asn 115
120 125Leu Thr Val Asp Ile Gly His Thr Phe Tyr Val
Xaa Xaa Leu Gly Thr 130 135 140Trp Val
His Asn Val14514149PRTPseudomonas fluorescensmisc_feature(36)..(66)Xaa
can be any naturally occurring amino acid 14Cys Phe Ala Ala Gly Thr Met
Val Ala Thr Pro Ser Gly Asp Arg Ala1 5 10
15Ile Asp Thr Leu Lys Val Gly Glu Ile Val Trp Ser Lys
Pro Glu His 20 25 30Gly Gly
Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa 50 55 60Xaa
Xaa Glu Gly Glu Thr Leu Leu Val Thr Pro Ser His Pro Phe Tyr65
70 75 80Val Xaa Xaa Xaa Xaa Xaa
Phe Val Pro Ala Ile Asn Leu Lys Pro Gly 85
90 95Asp Leu Leu Gln Ser Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 100 105 110Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Lys Thr Phe Asn 115
120 125Leu Thr Val Asp Ile Gly His Thr Phe
Tyr Val Xaa Xaa Leu Lys Thr 130 135
140Trp Val His Asn Thr14515140PRTStreptomyces
coelicolormisc_feature(14)..(14)Xaa can be any naturally occurring amino
acid 15Ser Phe Pro Ala Gly Thr Arg Val Leu Met Ala Asp Gly Xaa Arg Arg1
5 10 15Ser Ile Glu Gln Ile
Glu Ala Gly Asp Leu Val Thr Ala Thr Asp Pro 20
25 30Thr Thr Gly Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asp Gly Ser Thr 50
55 60Leu Thr Ser Thr Thr His His Pro Tyr Trp Ser
Xaa Xaa Xaa Xaa Xaa65 70 75
80Trp Lys Asn Ala Gly Asp Leu Glu Ala Gly Asp Thr Leu Arg Thr Pro
85 90 95Gln Asn Thr Ala Val
Val Ile Ala Ala Thr His Asp Trp Xaa Xaa Xaa 100
105 110Xaa Asp Ala Tyr Asp Leu Thr Val Asp Gly Phe His
Ser Tyr Tyr Val 115 120 125Xaa Xaa
Xaa Xaa Thr Asp Val Leu Val His Asn Asn 130 135
14016145PRTThermobifida fuscamisc_feature(14)..(14)Xaa can be
any naturally occurring amino acid 16Ser Phe Val Pro Gly Thr Leu Val Leu
Leu Ala Asp Gly Xaa Tyr Ala1 5 10
15Pro Ile Glu Thr Ile Thr Val Gly Asp Asp Val Trp Ala Phe Asp
Pro 20 25 30Arg Thr Gly Thr
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 50 55 60His Gly Gly
Val Val Val Ala Thr Asp Ala His Pro Phe Trp Val Xaa65 70
75 80Xaa Xaa Xaa Xaa Trp Val Ala Ala
Ile Asp Leu Glu Pro Gly Thr Trp 85 90
95Leu Arg Thr Ser Ala Gly Thr Trp Val Gln Val Arg Ala Val
Ala Val 100 105 110Arg Xaa Xaa
Xaa Xaa Xaa Arg Val His Asn Leu Thr Val Ala Asp Leu 115
120 125His Thr Tyr Tyr Val Xaa Xaa Xaa Xaa Ala Asp
Ala Leu Val His Asn 130 135
140Glu14517130PRTNeisseria gonorrhoeaemisc_feature(23)..(49)Xaa can be
any naturally occurring amino acid 17Tyr Lys Ala Ile Ala His Ile Gln Ala
Gly Asp Arg Val Leu Ser Lys1 5 10
15Asp Glu Ala Ser Gly Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa 20 25 30Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35
40 45Xaa Asn Ser Gln Thr Leu Ile Ser Asn Arg Ile His
Pro Phe Tyr Ser 50 55 60Xaa Xaa Xaa
Trp Ile Lys Ala Glu Asp Leu Lys Ala Gly Ser Arg Leu65 70
75 80Leu Ser Glu Ser Gly Lys Thr Gln
Thr Val Arg Asn Ile Val Val Lys 85 90
95Xaa Xaa Xaa Xaa Lys Ala Tyr Asn Leu Thr Val Ala Asp Trp
His Thr 100 105 110Tyr Phe Val
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Gly Val Trp Val His 115
120 125Asn Asp 13018143PRTNeisseria
gonorrhoeaemisc_feature(36)..(62)Xaa can be any naturally occurring amino
acid 18Pro Phe His Gly Ser Thr Leu Val Lys Thr Ala Asp Gly Tyr Lys Ala1
5 10 15Ile Ala Arg Ile Arg
Val Gly Asp His Val Phe Ala Lys Asp Glu Ala 20
25 30Ser Gly Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asn Asn 50
55 60Gln Thr Leu Ile Ser Asn Arg Ile His Pro Phe
Tyr Ser Xaa Xaa Xaa65 70 75
80Trp Ile Lys Ala Glu Asp Leu Lys Ala Gly Ser Arg Leu Leu Ser Glu
85 90 95Ser Gly Arg Thr Gln
Thr Val Arg Asn Ile Ile Val Lys Xaa Xaa Xaa 100
105 110Xaa Lys Ala Tyr Asn Leu Thr Val Ala Asp Trp His
Thr Tyr Phe Val 115 120 125Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Glu Gly Val Trp Val His Asn Ala 130
135 14019143PRTNeisseria
gonorrhoeaemisc_feature(36)..(62)Xaa can be any naturally occurring amino
acid 19Ser Phe His Gly Ser Thr Leu Val Arg Thr Ala Asp Gly Tyr Lys Ala1
5 10 15Ile Ala His Ile Gln
Ala Gly Asp Arg Val Leu Ser Lys Asp Glu Ala 20
25 30Ser Gly Lys Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asn Ser 50
55 60Gln Thr Leu Ile Ser Asn Arg Ile His Pro Phe
Tyr Ser Xaa Xaa Xaa65 70 75
80Trp Ile Lys Ala Glu Asp Leu Lys Ala Gly Asn Arg Leu Phe Ala Glu
85 90 95Ser Gly Lys Thr Gln
Thr Val Arg Asn Ile Val Val Lys Xaa Xaa Xaa 100
105 110Xaa Lys Ala Tyr Asn Leu Thr Val Ala Asp Trp His
Thr Tyr Phe Val 115 120 125Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Glu Gly Val Trp Val His Asn Ser 130
135 14020143PRTNeisseria
gonorrhoeaemisc_feature(36)..(62)Xaa can be any naturally occurring amino
acid 20Ser Phe His Gly Ser Thr Leu Val Lys Thr Ala Asp Gly Tyr Lys Ala1
5 10 15Ile Ala His Ile Gln
Ala Gly Asp Arg Val Leu Ser Lys Asp Glu Ala 20
25 30Ser Gly Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asn Ser 50
55 60Gln Thr Leu Ile Ser Asn Arg Ile His Pro Phe
Tyr Ser Xaa Xaa Xaa65 70 75
80Trp Ile Lys Ala Glu Asp Leu Lys Ala Gly Ser Arg Leu Leu Ser Glu
85 90 95Ser Gly Lys Thr Gln
Thr Val Arg Asn Ile Val Val Lys Xaa Xaa Xaa 100
105 110Xaa Lys Ala Tyr Asn Leu Thr Val Ala Asp Trp His
Thr Tyr Phe Val 115 120 125Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Glu Gly Val Trp Val His Asn Asp 130
135 14021143PRTNeisseria
gonorrhoeaemisc_feature(36)..(62)Xaa can be any naturally occurring amino
acid 21Ser Phe His Gly Ser Thr Leu Val Lys Thr Ala Asp Gly Tyr Lys Ala1
5 10 15Ile Ala His Ile Gln
Ala Gly Asp Arg Val Leu Ser Lys Asp Glu Ala 20
25 30Ser Gly Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asn Ser 50
55 60Gln Thr Leu Ile Ser Asn Arg Ile His Pro Phe
Tyr Ser Xaa Xaa Xaa65 70 75
80Trp Ile Lys Ala Glu Asp Leu Lys Ala Gly Ser Arg Leu Phe Ala Glu
85 90 95Ser Gly Lys Thr Gln
Thr Val Arg Asn Ile Ile Val Lys Xaa Xaa Xaa 100
105 110Xaa Lys Ala Tyr Asn Leu Thr Val Ala Asp Trp His
Thr Tyr Phe Val 115 120 125Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Glu Gly Val Trp Val His Asn Asp 130
135 14022143PRTNeisseria
gonorrhoeaemisc_feature(36)..(62)Xaa can be any naturally occurring amino
acid 22Pro Phe His Gly Ser Thr Leu Val Lys Thr Ala Asp Gly Tyr Lys Ala1
5 10 15Ile Ala His Ile Gln
Thr Gly Glu His Val Phe Ala Lys Asp Glu Thr 20
25 30Ser Gly Lys Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asn Ser 50
55 60Gln Thr Leu Ile Ser Asn Arg Ile His Pro Phe
Tyr Ser Xaa Xaa Xaa65 70 75
80Trp Ile Lys Ala Glu Asp Leu Lys Ala Gly Ser Arg Leu Leu Ser Glu
85 90 95Ser Gly Arg Thr Gln
Thr Val Arg Asn Thr Val Val Lys Xaa Xaa Xaa 100
105 110Xaa Lys Ala Tyr Asn Leu Thr Val Ala Asp Trp His
Thr Tyr Phe Val 115 120 125Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Glu Gly Val Trp Val His Asn Ser 130
135 14023143PRTNeisseria meningitidis (group
C)misc_feature(36)..(62)Xaa can be any naturally occurring amino acid
23Ser Phe His Gly Ser Thr Leu Val Lys Thr Ala Asp Gly Tyr Lys Ala1
5 10 15Ile Ala His Ile Arg Val
Gly Glu Ser Val Phe Ala Lys Asp Glu Thr 20 25
30Ser Gly Lys Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa 35 40 45Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asn Ser 50
55 60Gln Thr Leu Ile Ser Asn Arg Ile His Pro Phe Tyr
Ser Xaa Xaa Xaa65 70 75
80Trp Ile Gln Ala Gly Arg Leu Lys Lys Gly Asp Thr Leu Leu Ser Glu
85 90 95Ser Gly Ala Lys Gln Thr
Val Gln Asn Ile Thr Leu Lys Xaa Xaa Xaa 100
105 110Xaa Lys Ala Tyr Asn Leu Thr Val Ala Asp Trp His
Thr Tyr Phe Val 115 120 125Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Glu Gly Val Trp Val His Asn Asp 130
135 14024143PRTNeisseria meningitidis (group
C)misc_feature(36)..(62)Xaa can be any naturally occurring amino acid
24Pro Leu Tyr Val Gly Ala Leu Val Lys Thr Ala Asp Gly Tyr Lys Ala1
5 10 15Ile Ala His Ile Arg Val
Gly Glu Ser Val Leu Ser Lys Asp Glu Ala 20 25
30Ser Gly Lys Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa 35 40 45Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asn Ser 50
55 60Gln Thr Leu Ile Ser Asn Arg Ile His Pro Phe Tyr
Ser Xaa Xaa Xaa65 70 75
80Trp Ile Gln Ala Gly Arg Leu Lys Lys Gly Asp Thr Leu Leu Ser Glu
85 90 95Ser Gly Ala Lys Gln Thr
Val Gln Asn Ile Thr Phe Lys Xaa Xaa Xaa 100
105 110Xaa Lys Ala Tyr Asn Leu Thr Val Ala Asp Trp His
Thr Tyr Phe Val 115 120 125Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Glu Gly Val Trp Val His Asn Ala 130
135 14025143PRTNeisseria meningitidis (group
C)misc_feature(36)..(62)Xaa can be any naturally occurring amino acid
25Ser Phe His Gly Ser Thr Leu Val Lys Thr Ala Asp Gly Tyr Lys Ala1
5 10 15Ile Ala His Ile Arg Val
Gly Glu Ser Val Leu Ser Lys Asp Glu Ala 20 25
30Ser Gly Lys Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa 35 40 45Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asn Ser 50
55 60Gln Thr Leu Val Ser Asn Lys Ile His Pro Phe Tyr
Ser Xaa Xaa Xaa65 70 75
80Trp Ile Lys Ala Glu Asp Leu Lys Ala Gly Ser Arg Leu Leu Ser Glu
85 90 95Ser Gly Lys Thr Gln Thr
Val Arg Asn Ile Val Val Lys Xaa Xaa Xaa 100
105 110Xaa Lys Ala Tyr Asn Leu Thr Val Ala Asp Trp His
Thr Tyr Phe Val 115 120 125Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Glu Gly Val Trp Val His Asn Ala 130
135 14026143PRTNeisseria meningitidis (group
C)misc_feature(36)..(62)Xaa can be any naturally occurring amino acid
26Ser Phe His Gly Ser Thr Leu Val Lys Thr Ala Asp Gly Tyr Lys Ala1
5 10 15Ile Ala Arg Ile Arg Thr
Gly Asp Arg Val Phe Ala Lys Asp Glu Ala 20 25
30Ser Gly Lys Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa 35 40 45Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asn Asn 50
55 60Gln Thr Leu Ile Ser Asn Lys Ile His Pro Phe Tyr
Ser Xaa Xaa Xaa65 70 75
80Trp Ile Gln Ala Gly Arg Leu Lys Lys Gly Asp Thr Leu Leu Ser Glu
85 90 95Ser Gly Ala Lys Gln Thr
Val Gln Asn Ile Thr Phe Lys Xaa Xaa Xaa 100
105 110Xaa Lys Ala Tyr Asn Leu Thr Val Ala Asp Trp His
Thr Tyr Phe Val 115 120 125Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Glu Gly Val Trp Val His Asn Ala 130
135 14027143PRTNeisseria meningitidis (group
C)misc_feature(36)..(62)Xaa can be any naturally occurring amino acid
27Ser Phe His Gly Ser Thr Leu Val Lys Thr Ala Asp Gly Tyr Lys Ala1
5 10 15Ile Ala His Ile Gln Ala
Gly Asp Arg Val Leu Ser Lys Asp Glu Ala 20 25
30Ser Gly Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa 35 40 45Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asn Ser 50
55 60Gln Thr Leu Val Ser Asn Lys Ile His Pro Phe Tyr
Ser Xaa Xaa Xaa65 70 75
80Trp Ile Gln Ala Gly Arg Leu Lys Lys Gly Asp Thr Leu Leu Ser Glu
85 90 95Ser Gly Ala Lys Gln Thr
Val Gln Asn Ile Thr Leu Lys Xaa Xaa Xaa 100
105 110Xaa Lys Ala Tyr Asn Leu Thr Val Ala Asp Trp His
Thr Tyr Phe Val 115 120 125Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Glu Gly Val Trp Val His Asn Ser 130
135 14028164PRTMannheimia
haemolyticamisc_feature(36)..(62)Xaa can be any naturally occurring amino
acid 28Ser Phe His Gly Asp Met Glu Val Lys Thr Asp Lys Gly Tyr Arg Gln1
5 10 15Ile Ser Ser Ile Lys
Val Gly Asp Lys Val Leu Ala Lys Asn Glu Arg 20
25 30Thr Gly Ile Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Lys Tyr 50
55 60His Thr Ile Val Ser Asn Lys Ile His Pro Phe
Phe Thr Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
85 90 95Xaa Xaa Xaa Xaa Xaa
Trp Val Asp Ala Gln His Leu Gln Lys Gly Tyr 100
105 110Arg Leu Leu Ala Glu Ser Gly Glu Trp Gln Thr Val
Thr Lys Val Lys 115 120 125Ile Lys
Xaa Xaa Xaa Xaa Lys Ala Tyr Asn Met Thr Val Glu Lys Asp 130
135 140His Thr Tyr Phe Ile Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Glu Gly Val Trp145 150 155
160Val His Asn Asp29136PRTClostridium
thermocellummisc_feature(36)..(57)Xaa can be any naturally occurring
amino acid 29Cys Phe Val Ala Gly Thr Leu Ile Leu Thr Val Ala Gly Leu Val
Ala1 5 10 15Ile Glu Asn
Ile Lys Ala Gly Asp Lys Val Ile Ala Thr Asn Leu Glu 20
25 30Thr Phe Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asn Gly Glu Val Ile Lys Thr 50
55 60Thr Phe Glu His Pro Phe Tyr Val Xaa
Xaa Xaa Xaa Phe Val Glu Ala65 70 75
80Lys Glu Leu Gln Val Gly Asp Lys Leu Leu Asp Ser Lys Gly
Asn Val 85 90 95Leu Val
Val Glu Glu Lys Lys Leu Glu Xaa Xaa Xaa Xaa Xaa Xaa Lys 100
105 110Val Tyr Asn Phe His Val Asp Asp Phe
Tyr Thr Tyr His Val Xaa Xaa 115 120
125Asn Gly Ile Leu Val His Asn Ala 130
13530136PRTClostridium thermocellummisc_feature(36)..(57)Xaa can be any
naturally occurring amino acid 30Cys Phe Val Ala Gly Thr Met Val Leu Thr
Ala Ala Gly Leu Val Ala1 5 10
15Ile Glu Asn Ile Lys Val Gly Asp Lys Val Ile Ala Ala Asn Pro Glu
20 25 30Thr Phe Glu Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Gly Glu Val Ile
Lys Thr 50 55 60Thr Val Asp His Pro
Phe Tyr Val Xaa Xaa Xaa Xaa Phe Val Glu Ala65 70
75 80Val Asn Leu Gln Val Gly Asp Lys Leu Val
Asp Ser Lys Gly Asn Val 85 90
95Leu Val Val Glu Glu Lys Lys Leu Lys Xaa Xaa Xaa Xaa Xaa Xaa Lys
100 105 110Val Tyr Asn Phe Lys
Val Asp Asp Phe His Thr Tyr His Val Xaa Xaa 115
120 125Lys Gly Ile Leu Val His Asn Ala 130
13531136PRTClostridium thermocellummisc_feature(36)..(57)Xaa can be
any naturally occurring amino acid 31Cys Phe Val Ala Gly Thr Met Ile Leu
Thr Ala Thr Gly Leu Val Ala1 5 10
15Ile Glu Asn Ile Lys Ala Gly Asp Lys Val Ile Ala Thr Asn Pro
Glu 20 25 30Thr Phe Glu Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Gly Glu
Val Ile Lys Thr 50 55 60Thr Phe Asp
His Pro Phe Tyr Val Xaa Xaa Xaa Xaa Phe Val Glu Ala65 70
75 80Gly Lys Leu Gln Val Gly Asp Lys
Leu Leu Asp Ser Arg Gly Asn Val 85 90
95Leu Val Val Glu Glu Lys Lys Leu Glu Xaa Xaa Xaa Xaa Xaa
Xaa Lys 100 105 110Val Tyr Asn
Phe Lys Val Asp Asp Phe His Thr Tyr His Val Xaa Xaa 115
120 125Asn Glu Val Leu Val His Asn Ala 130
13532136PRTClostridium thermocellummisc_feature(36)..(57)Xaa can
be any naturally occurring amino acid 32Cys Phe Val Ala Gly Thr Met Ile
Leu Thr Thr Thr Gly Leu Val Ala1 5 10
15Ile Glu Asn Ile Lys Ala Gly Asp Lys Val Ile Ala Thr Asn
Pro Glu 20 25 30Thr Phe Glu
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Gly
Glu Val Ile Lys Thr 50 55 60Thr Phe
Asp His Pro Phe Tyr Val Xaa Xaa Xaa Xaa Phe Val Glu Ala65
70 75 80Lys Gln Leu His Val Gly Asp
Lys Leu Leu Asp Ser Lys Gly Asn Val 85 90
95Leu Val Val Glu Asp Lys Lys Ile Lys Xaa Xaa Xaa Xaa
Xaa Xaa Lys 100 105 110Val Tyr
Asn Phe Gln Val Ala Asp Phe His Thr Tyr His Val Xaa Xaa 115
120 125Asn Gly Val Leu Val His Asn Val 130
13533136PRTClostridium thermocellummisc_feature(36)..(57)Xaa
can be any naturally occurring amino acid 33Cys Phe Val Ala Gly Thr Met
Ile Leu Thr Val Ala Gly Leu Val Ala1 5 10
15Ile Glu Asn Ile Lys Ala Gly Asp Lys Val Ile Ala Thr
Asn Pro Glu 20 25 30Thr Phe
Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asn
Gly Asp Val Ile Lys Thr 50 55 60Thr
Phe Glu His Leu Phe Tyr Ala Xaa Xaa Xaa Xaa Phe Val Glu Ala65
70 75 80Lys Glu Leu Gln Val Gly
Asp Lys Leu Leu Asp Ser Lys Gly Asn Val 85
90 95Leu Val Val Glu Asp Lys Lys Ile Lys Xaa Xaa Xaa
Xaa Xaa Xaa Lys 100 105 110Val
Tyr Asn Phe Gln Val Asp Asp Phe His Thr Tyr His Val Xaa Xaa 115
120 125Asn Gly Val Leu Val His Asn Val
130 13534136PRTClostridium
thermocellummisc_feature(36)..(57)Xaa can be any naturally occurring
amino acid 34Cys Phe Val Ala Gly Thr Met Ile Leu Thr Ala Thr Gly Leu Val
Ala1 5 10 15Ile Glu Asn
Ile Lys Ala Gly Asp Lys Val Ile Ala Thr Asn Pro Glu 20
25 30Thr Phe Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Xaa Glu Ile Ile Lys Thr 50
55 60Thr Leu Gly His Leu Phe Tyr Val Xaa
Xaa Xaa Xaa Phe Val Glu Ala65 70 75
80Val Lys Leu Gln Pro Thr Asp Lys Leu Val Asp Ser Gly Gly
Asn Val 85 90 95Leu Val
Val Glu Xaa Lys Lys Phe Glu Xaa Xaa Xaa Xaa Xaa Xaa Lys 100
105 110Val Tyr Asn Phe Lys Val Asn Asp Phe
Tyr Thr Tyr His Val Xaa Xaa 115 120
125Asn Gly Ile Leu Val His Asn Val 130
13535136PRTClostridium thermocellummisc_feature(36)..(57)Xaa can be any
naturally occurring amino acid 35Cys Phe Val Ala Gly Thr Met Ile Leu Thr
Ala Thr Gly Leu Val Ala1 5 10
15Ile Glu Asn Ile Lys Ala Gly Asp Lys Val Ile Ala Thr Asn Pro Glu
20 25 30Thr Phe Glu Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Xaa Glu Ile Ile
Lys Thr 50 55 60Thr Leu Gly His Leu
Phe Tyr Val Xaa Xaa Xaa Xaa Phe Val Glu Ala65 70
75 80Val Lys Leu Gln Pro Thr Asp Lys Leu Val
Asp Ser Gly Gly Asn Val 85 90
95Leu Val Val Glu Xaa Lys Lys Phe Glu Xaa Xaa Xaa Xaa Xaa Xaa Lys
100 105 110Val Tyr Asn Phe Lys
Val Asn Asp Phe Tyr Thr Tyr His Val Xaa Xaa 115
120 125Asn Gly Ile Leu Val His Asn Val 130
13536214PRTLeptospira interrogansmisc_feature(36)..(58)Xaa can be any
naturally occurring amino acid 36Cys Phe Thr Ala Gly Ser Lys Val Thr Lys
Leu Lys Asn Phe Ala Asn1 5 10
15Ile Glu Glu Ile Lys Ile Gly Asp Ile Val Arg Ser Trp Asn Glu Asn
20 25 30Thr Asn Thr Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Glu Glu Glu
Ile His 50 55 60Thr Thr Trp Asn His
Pro Phe Arg Arg Xaa Xaa Xaa Xaa Xaa Xaa Xaa65 70
75 80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa 85 90
95Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
100 105 110Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 115
120 125Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 130 135 140Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Trp Val Lys Val Glu Asp Leu Arg Leu145
150 155 160Lys Asp Gln Val Leu Arg Ser
Asp Gly Ser Trp Gly Thr Val Thr Gly 165
170 175Ile Tyr Tyr Tyr Xaa Xaa Xaa Xaa Xaa Lys Val Tyr
Asn Leu Glu Val 180 185 190Glu
Asp Asn His Thr Tyr Val Val Xaa Xaa Xaa Xaa Xaa Xaa Ile Gly 195
200 205Tyr Val Val His Asn Tyr
21037239PRTLeptospira interrogansmisc_feature(14)..(42)Xaa can be any
naturally occurring amino acid 37Cys Phe Val Ala Gly Ser Lys Val Thr Lys
Leu Lys Asn Xaa Xaa Xaa1 5 10
15Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
20 25 30Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Phe Ala Asn Ile Glu Glu 35 40
45Ile Arg Ile Gly Asp Val Val Arg Ser Trp Asn Glu Asn Thr
Asn Thr 50 55 60Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa65 70
75 80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Glu Glu
Glu Ile His Thr Thr Trp 85 90
95Asn His Pro Phe Arg Arg Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
100 105 110Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 115
120 125Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 130 135 140Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa145
150 155 160Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 165
170 175Trp Val Lys Val Glu Asp Leu Arg Leu Arg Asp Gln
Val Leu Arg Ser 180 185 190Asp
Gly Ser Trp Gly Thr Val Thr Gly Ile Tyr Tyr Tyr Xaa Xaa Xaa 195
200 205Xaa Xaa Lys Val Tyr Asn Leu Glu Val
Glu Asp Asn His Thr Tyr Val 210 215
220Val Xaa Xaa Xaa Xaa Xaa Xaa Lys Gly Tyr Val Val His Asn Tyr225
230 23538144PRTGemmata
obscuriglobusmisc_feature(36)..(59)Xaa can be any naturally occurring
amino acid 38Cys Phe Ala Ala Gly Thr Lys Leu Leu Thr Arg Arg Gly Trp Val
Ala1 5 10 15Val Glu Leu
Leu Gly Ile Gly Asp Glu Val Ala Ser Arg Thr Glu His 20
25 30Asp Leu Thr Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Gly Glu Leu Ile 50
55 60Arg Thr Thr Pro Glu His Pro Phe Trp
Val Xaa Xaa Xaa Xaa Trp Thr65 70 75
80Ala Ala Gly Ser Leu Ala Ala Gly Asp Arg Ile Ala Thr Xaa
Xaa Xaa 85 90 95Xaa Leu
Ser Gly Glu Trp Val Pro Ile Ala Glu Val Phe Asp Thr Xaa 100
105 110Xaa Xaa Xaa Pro Val Tyr Asn Leu Arg
Val Ala Asp His His Thr Tyr 115 120
125Phe Val Xaa Xaa Xaa Xaa Xaa Xaa Phe Ala Ala Trp Ala His Asn Ala
130 135 14039139PRTGemmata
obscuriglobusmisc_feature(36)..(58)Xaa can be any naturally occurring
amino acid 39Cys Phe Ala Ser Gly Thr Pro Met Arg Thr Pro Gly Gly Trp Cys
Asn1 5 10 15Ile Glu Asn
Leu Arg Val Gly Asp Phe Val Leu Ser Arg Asp Glu Phe 20
25 30Ser Pro Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Leu Gly Gln Thr Ile Arg 50
55 60Ser Thr Asp Glu His Pro Phe Phe Val
Xaa Xaa Xaa Xaa Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa 85 90 95Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Thr Val 100
105 110Tyr Asn Phe Arg Val Ala Asp His His
Thr Tyr Phe Val Xaa Xaa Xaa 115 120
125Xaa Xaa Xaa Phe Ser Val Trp Ala His Asn Ile 130
13540132PRTPirellula sp.misc_feature(36)..(57)Xaa can be any naturally
occurring amino acid 40Cys Leu Val Ala Gly Thr Leu Val Trp Thr Asp Arg
Gly Met Arg Pro1 5 10
15Val Glu Ser Leu Arg Leu Gly Asp Gln Val Leu Ser Cys Asp Val Gln
20 25 30Thr Gly Ser Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Ser Asp Glu Ile Val
Ala 50 55 60Ser Lys Gly His Pro Phe
Trp Val Xaa Xaa Xaa Xaa Trp Thr Thr Thr65 70
75 80Glu Gln Leu Val Pro Gly Asp Ala Leu His Gly
Xaa Xaa Xaa Xaa Xaa 85 90
95Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Lys Thr Tyr Asn Leu
100 105 110Val Val Glu Gln Thr His
Ser Tyr Phe Val Xaa Xaa Ser Arg Ile Leu 115 120
125Ser His Asp Ala 13041144PRTStreptomyces
avermitilismisc_feature(14)..(14)Xaa can be any naturally occurring amino
acid 41Ser Phe Lys Pro Thr Thr Arg Val Leu Met Lys Asp Gly Xaa Thr Lys1
5 10 15Pro Leu Gly Lys Ile
Lys Pro Gly Asp Leu Val Glu Ala Ala Asp Pro 20
25 30Thr Ser Gly His Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 50
55 60Arg Ile Gln Thr Leu His Thr Thr Ala Arg His
Arg Ile Trp Asp Xaa65 70 75
80Xaa Xaa Xaa Xaa Trp Glu Gln Ala Gly Arg Leu Ile Thr Gly His Lys
85 90 95Val Asn Thr Ser Gly
Asn Gln His Ala Thr Ile Thr Ser Val Leu Ala 100
105 110Gln Xaa Xaa Xaa Xaa Asp Met Tyr Asp Leu Thr Val
Glu Gly Leu His 115 120 125Thr Tyr
Tyr Val Xaa Xaa Xaa Xaa Thr Pro Val Leu Val His Asn Gly 130
135 14042143PRTStreptomyces
avermitilismisc_feature(14)..(14)Xaa can be any naturally occurring amino
acid 42Cys Phe Leu Ala Gly Thr Asp Ile Leu Met Ala Asp Gly Xaa Thr Lys1
5 10 15Asp Ile Glu Glu Val
Glu Leu Gly Asp Lys Val Gln Ala Thr Asp Pro 20
25 30Lys Thr Gly Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly 50
55 60Ala Glu Glu Leu Thr Ala Thr His Glu His Pro
Phe Trp Ser Xaa Xaa65 70 75
80Xaa Xaa Xaa Trp Ile Thr Ala Gly Ser Leu Glu Pro Gly Met Thr Leu
85 90 95Leu Thr Asp Asp Gly
Asp Thr Val Ile Val Thr Gly Asn Arg Ala Phe 100
105 110Xaa Xaa Xaa Xaa Thr Thr Tyr Asn Leu Thr Val Asn
Asp Leu His Thr 115 120 125Tyr Tyr
Ala Xaa Xaa Xaa Xaa Thr Pro Val Leu Val His Asn Ser 130
135 14043146PRTStreptomyces
avermitilismisc_feature(14)..(15)Xaa can be any naturally occurring amino
acid 43Ser Phe Pro Ala Gly Thr Arg Val Leu Met Gly Asp Gly Xaa Xaa Thr1
5 10 15Leu Pro Ile Glu Gln
Ile Thr Val Gly Asp Ser Val Leu Ala Thr Asp 20
25 30Pro Glu Ala Gly Thr Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 50
55 60Xaa Xaa Gly Pro Pro Ala Leu Thr Ala Thr Asp
Arg His Pro Phe Trp65 70 75
80Val Xaa Xaa Xaa Xaa Xaa Trp Ala Asp Ala Arg Asp Leu Asn Ser Gly
85 90 95Asp Thr Leu Arg Thr
Pro Asp Gly Thr Gly Val Arg Ile Asp Lys Val 100
105 110Thr His Trp Xaa Xaa Xaa Xaa Gly Ala Tyr Asn Leu
Thr Val Asn Asp 115 120 125Leu His
Thr Tyr Tyr Val Xaa Xaa Xaa Xaa Val Pro Val Leu Val His 130
135 140Asn Ala14544129PRTMyxococcus
xanthusmisc_feature(14)..(14)Xaa can be any naturally occurring amino
acid 44Cys Val Ala Pro Trp Glu Leu Val Leu Leu Gly Asp Gly Xaa Glu Val1
5 10 15Pro Ala Glu Met Leu
Arg Pro Gly Met Arg Val Leu Thr Met His Glu 20
25 30His Glu Arg Asp Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asp Gly Arg Val Leu Val 50
55 60Val Thr Pro Asp His Arg Trp Arg Thr Xaa Xaa
Xaa Xaa Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
85 90 95Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Asp Val Met Arg Ile Thr 100
105 110Val Arg Phe Ala Met Thr Tyr Ile Val Gln Gly Leu
Leu Ala His Asn 115 120 125Leu
45131PRTMyxococcus xanthusmisc_feature(14)..(14)Xaa can be any naturally
occurring amino acid 45Cys Val Ala Pro Trp Glu Pro Val Leu Leu Ser Asp
Gly Xaa Glu Val1 5 10
15Pro Ala Glu Met Leu Arg Pro Gly Met Lys Val Leu Thr Met His Glu
20 25 30His Glu Arg Asp Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asp Gly Arg Ala Val
Val 50 55 60Val Thr Pro Asp His Arg
Trp Arg Thr Xaa Xaa Xaa Xaa Xaa Xaa Xaa65 70
75 80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa 85 90
95Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asp Val Met Lys
100 105 110Ile Ser Val Arg Phe Ala
Lys Thr Tyr Val Val Gln Gly Leu Leu Ala 115 120
125His Asn Leu 13046137PRTVerrucomicrobium
spinosummisc_feature(36)..(57)Xaa can be any naturally occurring amino
acid 46Cys Phe Pro Ser Gly Thr Met Val Gln Thr Ala Arg Gly Lys Val Ala1
5 10 15Ile Glu Thr Leu Lys
Glu Gly Asp Val Val Leu Ala Tyr Asp Phe Leu 20
25 30Ser Glu Cys Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Asp Ser Lys Ile Ser Ala 50
55 60Thr Arg Phe His Leu Phe Trp Val Xaa Xaa Xaa
Xaa Xaa Trp Val Pro65 70 75
80Ala Val Asp Leu Gln Pro Gly Met Val Leu Arg Leu Glu Ser Gly Ala
85 90 95Leu Thr Val Val Thr
Leu Ala Lys Leu Arg Xaa Xaa Xaa Xaa Xaa Xaa 100
105 110Ala Thr His Asn Phe Glu Val Ala Asp Leu His Asn
Tyr Phe Val Xaa 115 120 125Xaa Gln
Gly Phe Leu Val His Asn Gly 130
13547140PRTVerrucomicrobium spinosummisc_feature(14)..(14)Xaa can be any
naturally occurring amino acid 47Cys Phe Pro Ala Gly Thr Met Val Leu Met
Ala Asp Gly Xaa Ser Val1 5 10
15Pro Ile Glu Gln Val Val Glu Gly Asp Ile Val Leu Ala Ala Glu Pro
20 25 30Glu Thr Glu Ser Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Thr Gly
Ser Val 50 55 60Leu Lys Val Thr Gly
Glu His Pro Ile Trp Thr Xaa Xaa Xaa Xaa Trp65 70
75 80Gln His Ala Asp Asp Leu Val Glu Gly Asp
Leu Leu Leu Lys Xaa Xaa 85 90
95Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
100 105 110Xaa Asp Thr Phe Asn
Leu Cys Val Glu Gly Val His Thr Phe Tyr Val 115
120 125Xaa Xaa Xaa Xaa Asp Ala Val Leu Val His Asn Thr
130 135 14048145PRTVerrucomicrobium
spinosummisc_feature(14)..(14)Xaa can be any naturally occurring amino
acid 48Cys Phe Ala Pro Gly Thr Pro Val Leu Met Gly Asp Gly Xaa Thr Arg1
5 10 15Pro Val Glu Thr Ile
Arg Glu Gly Asp Trp Ile Met Ala Asp Asp Pro 20
25 30Glu Asp Glu Arg Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 50
55 60Xaa Xaa Pro Asp Gly Ala Leu Lys Ala Thr Gly
Gly His Pro Phe Trp65 70 75
80Thr Xaa Xaa Xaa Xaa Trp Ile Lys Val Cys Asn Leu Gln Pro Asn Asp
85 90 95Ile Leu Ala Asp Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 100
105 110Xaa Xaa Xaa Xaa Xaa Xaa Ala Thr Tyr Asn Leu Ser
Val Ala Asn Ile 115 120 125His Thr
Phe Phe Val Xaa Xaa Xaa Xaa Val Pro Val Leu Val His Asn 130
135 140Thr14549141PRTGloeobacter
violaceusmisc_feature(36)..(64)Xaa can be any naturally occurring amino
acid 49Cys Phe Ala Glu Gly Thr Glu Val Gln Thr Glu Thr Gly Thr Lys Ala1
5 10 15Ile Glu Lys Val Glu
Pro Gly Glu Lys Val Leu Ala Arg Asn Glu Lys 20
25 30Thr Gly Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 50
55 60Glu Arg Asp Thr Leu Thr Val Thr Gly Glu His
Pro Phe Phe Leu Xaa65 70 75
80Xaa Xaa Xaa Trp Thr Ala Ala Glu Arg Leu Arg Ser Gly Glu Arg Val
85 90 95Gln Ala Val Asp Gly
Lys Trp Leu Arg Val Val Gly Leu Gln Pro Gln 100
105 110Xaa Xaa Xaa Xaa Arg Thr Tyr Asn Leu Glu Val Glu
Gly Glu His Thr 115 120 125Phe Phe
Val Xaa Xaa Thr Arg Ala Trp Val His Asn Glu 130 135
14050141PRTGloeobacter violaceusmisc_feature(36)..(64)Xaa
can be any naturally occurring amino acid 50Cys Phe Ala Glu Gly Thr Glu
Val Gln Thr Glu Thr Gly Ala Lys Pro1 5 10
15Ile Glu Leu Val Ala Pro Gly Glu Lys Val Leu Ala Arg
Asn Glu Gln 20 25 30Thr Gly
Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa 50 55 60Asp
Arg Asp Val Leu Thr Val Thr Gly Glu His Pro Phe Phe Leu Xaa65
70 75 80Xaa Xaa Xaa Trp Thr Ala
Ala Asp Lys Leu Gln Val Gly Glu Arg Val 85
90 95Gln Thr Val Asp Gly Gln Trp Leu Arg Val Ala Gly
Leu Gln Ala Gln 100 105 110Xaa
Xaa Xaa Xaa Arg Thr Tyr Asn Leu Glu Val Glu Arg Asp His Thr 115
120 125Phe Phe Val Xaa Xaa Ser Lys Ala Trp
Val His Asn Glu 130 135
14051141PRTGloeobacter violaceusmisc_feature(36)..(64)Xaa can be any
naturally occurring amino acid 51Cys Phe Ser Glu Gly Thr Glu Val Gln Thr
Glu Ala Gly Ala Lys Pro1 5 10
15Ile Glu Leu Val Glu Pro Gly Glu Lys Val Leu Ala Arg Asn Glu Gln
20 25 30Thr Gly Glu Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa 50 55 60Glu Arg Asp Thr Leu
Thr Val Thr Gly Glu His Pro Phe Phe Leu Xaa65 70
75 80Xaa Xaa Xaa Trp Thr Ala Ala Glu Arg Leu
Lys Ser Gly Glu Arg Val 85 90
95Gln Ala Ala Asp Gly Lys Trp Leu Arg Val Ala Gly Leu Glu Ala Gln
100 105 110Xaa Xaa Xaa Xaa Arg
Thr Tyr Asn Leu Glu Val Glu Gly Asp His Thr 115
120 125Phe Phe Val Xaa Xaa Asn Gln Ala Trp Val His Asn
Glu 130 135 14052139PRTChromobacterium
violaceummisc_feature(36)..(62)Xaa can be any naturally occurring amino
acid 52Cys Phe Val Ala Gly Thr Gln Val Leu Thr Asp Lys Gly Leu Lys Ala1
5 10 15Ile Glu Thr Phe Val
Gly Gly Glu Trp Val Trp Ser Arg Ser Asp Gln 20
25 30Thr Gly Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Arg Gln 50
55 60Glu Thr Phe Arg Thr Thr Ala Glu His Pro Phe
Trp Val Xaa Xaa Xaa65 70 75
80Xaa Trp Leu Lys Ala Ser Leu Leu Gln Ala Gly Val Ile Leu Val Asp
85 90 95Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 100
105 110Xaa Xaa Thr Val Phe Asn Ile Gln Val Ala Glu Phe
Gln Thr Tyr His 115 120 125Val Xaa
Xaa Leu Gly Val Trp Val His Asn Ala 130
13553141PRTGloeobacter violaceusmisc_feature(36)..(64)Xaa can be any
naturally occurring amino acid 53Cys Phe Ala Glu Gly Thr Glu Val Gln Thr
Glu Thr Gly Thr Lys Ala1 5 10
15Ile Glu Lys Val Glu Pro Gly Glu Lys Val Leu Ala Arg Asn Glu Lys
20 25 30Thr Gly Glu Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa 50 55 60Glu Arg Asp Thr Leu
Thr Val Thr Gly Glu His Pro Phe Phe Leu Xaa65 70
75 80Xaa Xaa Xaa Trp Thr Ala Ala Asp Lys Leu
Gln Ala Gly Asp Arg Val 85 90
95Gln Ala Val Asp Gly Arg Trp Leu Arg Val Val Gly Leu Ala Ala Gln
100 105 110Xaa Xaa Xaa Xaa Arg
Thr Tyr Asn Leu Glu Ile Glu Gly Glu His Thr 115
120 125Phe Phe Val Xaa Xaa Asn Gln Ala Trp Val His Asn
Glu 130 135 1405493PRTGloeobacter
violaceusmisc_feature(36)..(78)Xaa can be any naturally occurring amino
acid 54Cys Phe Gly Glu Gly Thr Ala Val Gln Thr Glu Thr Arg Ala Lys Pro1
5 10 15Ile Glu Gln Ile Glu
Pro Gly Glu Lys Val Leu Ala Arg Ser Glu Arg 20
25 30Thr Gly Gln Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 50
55 60Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Glu Arg65 70 75
80Asp Thr Ser Thr Val Thr Gly Glu His Pro Phe Tyr Leu
85 905581PRTNeisseria meningitidis (group
C)misc_feature(16)..(18)Xaa can be any naturally occurring amino acid
55Asn Ser Gln Ile Leu Ile Ser Asn Arg Ile His Pro Phe Tyr Ser Xaa1
5 10 15Xaa Xaa Trp Ile Lys Ala
Glu Asp Leu Lys Ala Gly Ser Arg Leu Leu 20 25
30Ser Glu Ser Gly Lys Thr Gln Thr Val Arg Asn Ile Val
Val Lys Xaa 35 40 45Xaa Xaa Xaa
Lys Ala Tyr Asn Leu Thr Val Ala Asp Trp His Thr Tyr 50
55 60Phe Val Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Gly Val
Trp Val His Asn65 70 75
80Asp5691PRTClostridium thermocellummisc_feature(36)..(57)Xaa can be any
naturally occurring amino acid 56Cys Phe Val Ala Gly Thr Met Ile Leu Thr
Ala Thr Gly Leu Val Ala1 5 10
15Ile Glu Asn Ile Lys Ala Gly Asp Lys Val Ile Ala Thr Asn Pro Glu
20 25 30Thr Phe Glu Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asn Gly Glu Val Ile
Lys Thr 50 55 60Thr Phe Glu His Pro
Phe Tyr Val Xaa Xaa Xaa Xaa Phe Val Glu Ala65 70
75 80Gly Lys Leu Gln Ile Gly Asp Arg Leu Val
Asp 85 905745PRTClostridium
thermocellummisc_feature(15)..(20)Xaa can be any naturally occurring
amino acid 57Ser Lys Gly Asn Val Leu Val Val Glu Glu Lys Lys Leu Glu Xaa
Xaa1 5 10 15Xaa Xaa Xaa
Xaa Lys Val Tyr Asn Phe Lys Val Asn Asp Phe His Thr 20
25 30Tyr His Val Xaa Xaa Asp Gly Ile Leu Val
His Asn Ala 35 40
455824PRTClostridium thermocellummisc_feature(15)..(16)Xaa can be any
naturally occurring amino acid 58Val Tyr Asn Phe Lys Val Asp Asn Phe His
Thr Tyr His Val Xaa Xaa1 5 10
15Asn Arg Val Leu Val His Asn Ala 205939PRTClostridium
thermocellummisc_feature(16)..(34)Xaa can be any naturally occurring
amino acid 59Phe Val Lys Glu Met Lys Leu Gln Pro Gly Asn Arg Leu Val Asp
Xaa1 5 10 15Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 20
25 30Xaa Xaa Lys Val Tyr Asn Phe
356063PRTLeptospira interrogansmisc_feature(30)..(34)Xaa can be any
naturally occurring amino acid 60Trp Val Lys Val Glu Asp Leu Arg Leu Arg
Asp Gln Val Leu Arg Ser1 5 10
15Asp Gly Ser Trp Gly Thr Val Thr Gly Ile Tyr Tyr Tyr Xaa Xaa Xaa
20 25 30Xaa Xaa Lys Val Tyr Asn
Leu Glu Val Glu Asp Asn His Thr Tyr Ile 35 40
45Val Xaa Xaa Xaa Xaa Xaa Xaa Ile Gly Tyr Val Val His Asn
Tyr 50 55 606110PRTGloeobacter
violaceus 61Cys Phe Ala Glu Gly Thr Glu Val Gln Thr1 5
106215PRTGloeobacter violaceus 62Trp Thr Ala Ala Glu Arg Leu
Glu Pro Gly Asp Arg Val Gln Ala1 5 10
1563153PRTRhodobacter capsulatusmisc_feature(31)..(33)Xaa
can be any naturally occurring amino acid 63Cys Phe Thr Pro Gly Thr Leu
Ile Asp Thr Pro Ala Gly Pro Arg Pro1 5 10
15Val Glu Ala Leu Arg Pro Gly Asp Arg Val Ser Thr Arg
Asp Xaa Xaa 20 25 30Xaa Gln
Glu Ile Leu Trp Ile Gly Ser Arg Arg Met Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Pro Val
Arg Leu Gly Ala Val Arg 50 55 60Leu
Gly Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa65
70 75 80Ala Ala Asp Leu Leu Val
Ser Pro Gln His Arg Val Leu Val Xaa Xaa 85
90 95Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Val
Leu Val Gln Ala 100 105 110Cys
Asp Leu Val Asp Asp Ala Ala Val Xaa Xaa Xaa Xaa Xaa Xaa Xaa 115
120 125Xaa Val Thr Tyr Leu His Leu Leu Phe
Ala Arg His Gln Val Ile Arg 130 135
140Ala Asn Gly Val Glu Thr Glu Ser Phe145
15064141PRTRhodobacter capsulatusmisc_feature(31)..(33)Xaa can be any
naturally occurring amino acid 64Gly Phe Tyr Gly Glu Thr Val Leu Gln Thr
Ala Arg Gly Leu Arg Arg1 5 10
15Val Ser Ser Ile Leu Glu Gly Glu Lys Met Arg Thr Phe Thr Xaa Xaa
20 25 30Xaa Ala Pro Val Leu Ser
Ile Glu Arg Phe Ala Leu Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Pro Leu Ser Leu Pro Ala
Gly Leu 50 55 60Phe Gly Xaa Thr Arg
Asn Arg Phe Val Ala Pro Glu Gln Cys Leu Leu65 70
75 80Leu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Leu Leu Leu 85 90
95Val Pro Ala Lys Val Leu Gly Leu Leu Pro Gln Val Xaa Xaa Xaa Xaa
100 105 110Xaa Xaa Xaa Xaa Ala
Val Leu Tyr Arg Leu Leu Phe Glu Arg Pro Glu 115
120 125Leu Val Val Thr Asp Xaa Gly Ala Val Met Leu Cys
Asp 130 135 14065134PRTRhodobacter
capsulatusmisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 65Gly Phe Ala Ala Gly Thr Arg Val Arg Thr Pro Ala Gly Leu Arg Arg1
5 10 15Ile Glu Thr Leu Lys
Pro Gly Asp Leu Val Glu Thr Gln Glu Xaa Xaa 20
25 30Xaa Gln Pro Val Val Ala Val Glu Arg Thr Arg Leu
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Pro Ile Arg Phe Ala Ala Gly Ala His Gly Xaa Glu Arg 50
55 60Pro Val Leu Val Ala Pro Gln Gln Arg Val Leu
Val Xaa Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Val Leu Val Ala Ala Arg Thr
85 90 95Leu Val Asp Gly Glu
Met Val Xaa Xaa Xaa Xaa Xaa Xaa Xaa Val Asp 100
105 110Tyr Val Arg Leu Val Phe Asp Cys Ala His Met Val
Phe Ala Glu Gly 115 120 125Leu Ala
Val Glu Cys Phe 13066135PRTRhodobacter
capsulatusmisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 66Cys Phe Ala Pro Ser Thr Pro Ile Ala Thr Pro Gly Gly Asp Cys Pro1
5 10 15Ala Ala Ser Leu Lys
Ala Gly Asp Leu Val Leu Thr Ala Asp Xaa Xaa 20
25 30Xaa Gln Pro Ile Leu Trp Ser Gly Arg Ile Ala Leu
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Pro Val Arg Leu Cys Ala Pro Ala Phe Gly Xaa Thr Arg 50
55 60Asp Leu Trp Val Leu Pro Gln His Arg Val Ala
Leu Xaa Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Val Leu Val Pro Ala His His
85 90 95Leu Val Asp Gly Ile
Ser Ala Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Leu 100
105 110Ser Trp His Gly Leu Leu Leu Gln Gly His His Leu
Leu Ile Ala Asp 115 120 125Gly Cys
Arg Val Glu Ser Leu 130 13567135PRTRhodobacter
capsulatusmisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 67Cys Phe Thr Ala Gly Thr Leu Ile Glu Thr Pro Arg Gly Pro Val Pro1
5 10 15Val Glu Ser Leu Arg
Ala Gly Asp Leu Val Val Thr Arg Asp Xaa Xaa 20
25 30Xaa Val Pro Val Leu Trp Ser Gly Gly Arg Ser Leu
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Pro Val Ala Ile Arg Glu Asn Ala 50
55 60Leu Gly Xaa His Gly Ala Leu Leu Leu Ser Pro
Gln His Ala Val Leu65 70 75
80Ala Xaa Xaa Xaa Xaa Xaa Glu Arg Leu Val Arg Ala Arg His Leu Ala
85 90 95Gly Leu Asn Asp Pro
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Val 100
105 110Ser Tyr His His Ile Leu Leu Glu Arg His Gly Ile
Val Thr Ala Asn 115 120 125Gly Leu
Ala Cys Glu Ser Leu 130 13568146PRTRhodobacter
capsulatusmisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 68Ala Leu Ala Arg Gly Ser Val Leu Met Thr Glu Asp Gly Pro Val Ala1
5 10 15Ile Glu Asp Leu Gln
Pro Gly Gln Gly Val Leu Thr Ala Glu Xaa Xaa 20
25 30Xaa Glu Arg Val Cys Trp Ile Gly Ser Met Val Ile
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Leu Thr Arg Ile Thr 50
55 60Ala Glu Ala Phe Gly Xaa Xaa Xaa Xaa Ala Leu
Asp Leu Val Leu Gly65 70 75
80Pro Arg Ala Arg Leu Cys Leu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
85 90 95Xaa Xaa Xaa Ala Ala
Asp Val Pro Ala Arg Ala Phe Leu Asp Gly Ile 100
105 110Ser Val Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Val Thr
Val Tyr His Val 115 120 125Val Leu
Glu Gln His Gly Ser Leu Arg Val Ala Gly Leu Glu Val Glu 130
135 140Ala Phe14569136PRTRhodobacter
capsulatusmisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 69Cys Leu Gly Thr Gly Thr Met Ile Ala Thr Ala Glu Gly Pro Ala Pro1
5 10 15Ile Asp Trp Leu Arg
Pro Gly Asp Arg Val Leu Thr Arg Asp Xaa Xaa 20
25 30Xaa Gln Pro Leu Leu Trp Val Gly Gln His Thr Met
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Pro Leu Leu Leu Ser Ala Ala Cys Phe Gly Xaa 50
55 60Xaa Xaa Xaa Glu Arg Asp Val Leu Leu Ser Pro
Gly Thr Gly Val Leu65 70 75
80Leu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Met Phe
85 90 95Ala Lys Ala Arg His
Ala Leu Pro Lys Ala Glu Ala Xaa Xaa Xaa Xaa 100
105 110Gln Lys Leu Tyr Ser Met Leu Leu Ala Thr Pro Glu
Val Val Leu Ala 115 120 125Glu Gly
Met Trp Val Gly Ser Val 130 13570132PRTRhodobacter
capsulatusmisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 70Cys Phe Ala Ala Gly Thr Leu Ile Ala Thr Arg Arg Gly Pro Lys Pro1
5 10 15Val Glu Asp Leu Gly
Pro Glu Asp Arg Leu Gln Thr Ser Asp Xaa Xaa 20
25 30Xaa Arg Pro Val Gln Trp Val Gly Arg Trp Arg Val
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Pro Val Arg Phe Ala Pro Gly Val Leu Gly Xaa Asp Arg 50
55 60Ala Leu Phe Leu Ser Gly Gln His Arg Val Leu
Ile Xaa Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Glu Val Leu Val Ala Ala Lys Ala Leu Val
85 90 95Gly Leu Pro Gly Ile
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Val Asp Trp Val 100
105 110His Val Met Met Pro Thr His Glu Val Ile Phe Ala
Glu Asn Ala Arg 115 120 125Ala Glu
Thr Met 13071135PRTRhodobacter capsulatusmisc_feature(14)..(14)Xaa can
be any naturally occurring amino acid 71Ala Phe Thr Thr Gly Thr Leu Ile
Thr Met Ala Gly Gly Xaa Gln Arg1 5 10
15Pro Ile Glu Thr Leu Ala Pro Gly Asp Arg Val Leu Thr Arg
Asp Xaa 20 25 30Xaa Xaa Gln
Pro Val Arg Leu Val Ala Arg Ala Thr Leu Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Pro Val Val Ile Ser Ala Gly
Thr Leu Gly Xaa Glu 50 55 60Ser Asp
Leu Val Val Ala Pro His His Arg Val Phe Leu Xaa Xaa Xaa65
70 75 80Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Glu Ile Leu Val Gln Ala Lys 85 90
95His Leu Val Asp Gly Glu His Val Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Val 100 105 110Asp Tyr
Phe Ala Leu Val Phe Asp Arg His Glu Ile Val Tyr Ala Glu 115
120 125Gly Val Pro Val Glu Ser Leu 130
13572139PRTRhodobacter capsulatusmisc_feature(31)..(33)Xaa can
be any naturally occurring amino acid 72Cys Phe Thr Ala Thr Ser Leu Ile
Ala Thr Gly Gln Gly Gly Val Pro1 5 10
15Val Ser Glu Leu Val Pro Gly Ala Arg Val Ile Thr Arg Asp
Xaa Xaa 20 25 30Xaa Gln Glu
Leu Leu Trp Val Gly Arg Arg Arg Phe Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Pro Val Arg
Ile Ala Ala Gly Ala 50 55 60Leu Gly
Xaa Xaa Xaa Xaa Glu Arg Asp Met Leu Val Ser Pro Asn His65
70 75 80Arg Phe Leu Thr Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Glu Arg Leu 85 90
95Thr Met Ala Arg Asp Leu Val Gly Leu Asp Gly Ile Xaa
Xaa Xaa Xaa 100 105 110Xaa Xaa
Xaa Val Asp Tyr Trp Gln Leu Leu Phe Ala His His Glu Leu 115
120 125Val Leu Ala Asp Gly Ala Trp Ser Glu Ser
Phe 130 13573142PRTRhodobacter
capsulatusmisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 73Cys Leu Thr Pro Gly Thr Leu Ile Glu Thr Lys Arg Gly Gln Val Pro1
5 10 15Val Glu Lys Leu Arg
Pro Gly Asp Arg Val Leu Thr Arg Asp Xaa Xaa 20
25 30Xaa Gln Pro Ile Arg Trp Ile Gly Arg Arg Arg Leu
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Pro Val Arg Ile Ala Ala Gly Ala 50
55 60Leu Gly Xaa Xaa Xaa Xaa Glu Thr Asp Met Leu
Val Ser Pro Gln His65 70 75
80Arg Met Leu Ile Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
85 90 95Glu Val Leu Ala Ala
Ala Leu His Met Leu Gly Gln Pro Gly Ile Xaa 100
105 110Xaa Xaa Xaa Xaa Xaa Xaa Val Thr Tyr Leu His Leu
Met Leu Asp Ala 115 120 125His Glu
Ile Ile Arg Ala Asn Gly Ala Trp Thr Glu Ser Phe 130
135 14074136PRTRhodobacter
capsulatusmisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 74Cys Leu Val Ala Gly Ser Arg Val Ser Thr Pro Arg Gly Pro Val Pro1
5 10 15Val Glu Asp Leu Arg
Pro Glu Asp Leu Val Thr Val Arg Asp Xaa Xaa 20
25 30Xaa Leu Pro Val Leu Trp Ile Gly Arg Arg Arg Val
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Pro Val Glu Ile Gly Ala Gly Arg 50
55 60Leu Gly Xaa Ala Ala Pro Val Arg Leu Ser Ala
Leu His Gly Ile Ala65 70 75
80Val Xaa Xaa Gly Phe Leu Ala Arg Ala Gly His Leu Ala Ala Thr Gly
85 90 95Trp Gly Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 100
105 110Val Leu Tyr Leu His Leu Leu Leu Pro Arg His Ala
Leu Leu Ser Val 115 120 125Glu Gly
Leu Trp Val Glu Ser Phe 130 13575132PRTRhodobacter
capsulatusmisc_feature(31)..(46)Xaa can be any naturally occurring amino
acid 75Gly Phe Ala Met Gly Ser Arg Val Ala Thr Met Asp Gly Leu Leu Pro1
5 10 15Val Glu Phe Leu Asn
Leu Gly Asp Arg Ile Val Thr Arg Ser Xaa Xaa 20
25 30Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Leu Val 35 40 45Gly Ile
Ala Pro Gly Ala Leu Gly Xaa Xaa Xaa Xaa Gly Gln Ala Met 50
55 60Val Leu Gly Ser Gly Thr Gln Val Leu Leu Xaa
Xaa Xaa Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Gln Ala Leu Val Ala Val Glu Arg Leu Ile
85 90 95Asp Gly Gln Phe Ile
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Ile Arg Ile Phe 100
105 110Ala Leu His Phe Glu Ala Pro Glu Val Ile Tyr Ala
Asp Gly Val Glu 115 120 125Ile Gly
Cys Lys 13076142PRTRhodobacter sphaeroidesmisc_feature(31)..(33)Xaa
can be any naturally occurring amino acid 76Cys Phe Thr Pro Gly Thr Leu
Ile Ala Thr Val Arg Gly Glu Val Ala1 5 10
15Val Glu Ala Leu Ala Ala Gly Asp Arg Ile Val Thr Arg
Asp Xaa Xaa 20 25 30Xaa Gln
Pro Leu Arg Trp Ile Ser Arg Arg Arg Leu Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Pro Val
Leu Ile Glu Lys Gly Ser 50 55 60Leu
Gly Xaa Xaa Xaa Xaa Asp Arg Asp Met Met Val Ser Pro Asn His65
70 75 80Arg Ile Leu Val Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 85
90 95Glu Val Leu Val Ala Ala Lys His Leu Val Gly Pro
Arg Gly Ile Xaa 100 105 110Xaa
Xaa Xaa Xaa Xaa Xaa Thr Thr Tyr Leu His Leu Met Phe Asp Arg 115
120 125His Glu Val Val Leu Ala Asn Gly Ala
Trp Thr Glu Ser Phe 130 135
14077133PRTRhodobacter sphaeroidesmisc_feature(31)..(47)Xaa can be any
naturally occurring amino acid 77Ser Leu Thr Ala Gly Thr Pro Val Leu Thr
Leu Ala Gly Ile Arg Pro1 5 10
15Ala Glu Gly Ile Arg Pro Gly Asp Arg Leu Val Ala Arg Ser Xaa Xaa
20 25 30Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Met 35 40
45Val Ala Ile Gly Ala Ser Thr Leu Ala Xaa Xaa Xaa Xaa Asp
Glu Thr 50 55 60Leu Leu Val Pro Ala
Asp Gln Pro Leu Leu Leu Xaa Xaa Xaa Xaa Xaa65 70
75 80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Pro Val Val
Leu Pro Ala Arg Arg Leu 85 90
95Val Asp Gly Gln Leu Thr Xaa Xaa Xaa Xaa Xaa Xaa Xaa Val Asp Leu
100 105 110Val Thr Leu Thr Phe
Ala Ala Pro Ala Ala Ile Tyr Ala Ser Glu Leu 115
120 125His Pro Val Thr Arg 13078132PRTBrucella
suismisc_feature(31)..(33)Xaa can be any naturally occurring amino acid
78Cys Leu Leu Lys Gly Thr Leu Val Thr Thr Pro Asn Gly Pro Val Ala1
5 10 15Val Glu Lys Leu Cys Val
Gly Asp Leu Val Thr Thr Val Ser Xaa Xaa 20 25
30Xaa Leu Pro Ile Lys Trp Ile Gly Trp Gln Asn Tyr Xaa
Xaa Xaa Xaa 35 40 45Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Pro Ile Arg Val Arg Arg His Ala 50
55 60Leu Asp Xaa Xaa Xaa Xaa His Arg Asp Leu Tyr Leu
Ser Pro Asn His65 70 75
80Ala Leu Phe Ile Xaa Gly Val Leu Ile Arg Val Lys Asp Leu Val Asn
85 90 95Gly Arg Ser Ile Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Leu Asp Tyr Tyr 100
105 110Asn Ile Val Leu Asp Arg His Ala Val Val Leu Ala
Glu Gly Ala Ala 115 120 125Val Glu
Thr Phe 13079132PRTBrucella melitensismisc_feature(31)..(33)Xaa can be
any naturally occurring amino acid 79Cys Leu Leu Lys Gly Thr Leu Val Thr
Thr Pro Asn Gly Pro Val Ala1 5 10
15Val Glu Lys Leu Cys Val Gly Asp Leu Val Thr Thr Val Ser Xaa
Xaa 20 25 30Xaa Leu Pro Ile
Lys Trp Ile Gly Trp Gln Asn Tyr Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Pro Ile Arg Val
Arg Arg His Ala 50 55 60Leu Asp Xaa
Xaa Xaa Xaa His Arg Asp Leu Tyr Leu Ser Pro Asn His65 70
75 80Ala Leu Phe Ile Xaa Gly Val Leu
Ile Arg Val Lys Asp Leu Val Asn 85 90
95Gly Arg Ser Ile Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Leu Asp
Tyr Tyr 100 105 110Asn Ile Val
Leu Asp Arg His Ala Val Val Leu Ala Glu Gly Ala Ala 115
120 125Val Glu Thr Phe 13080138PRTUnknownBIL1
domain sequence from an unknown origin 80Cys Phe Leu Pro Gly Thr Met Ile
Lys Thr Pro Ser Gly Glu Arg Pro1 5 10
15Val Glu Asp Ile Gln Ile Asn Asp Glu Val Ile Thr Phe Asp
Xaa Xaa 20 25 30Xaa Xaa Xaa
Xaa Xaa Xaa Ser Lys Ile Lys Trp Val Gly Ser Lys Thr 35
40 45Ile Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Pro Val 50 55 60Arg Ile
Leu Lys Asn Ala Ile Ser Xaa Xaa Xaa Xaa His Lys Asp Leu65
70 75 80Leu Val Thr Pro Glu His Cys
Leu Phe Phe Xaa Gly Lys Phe Ile Pro 85 90
95Val Arg Met Leu Val Asn His Gln Thr Ile Xaa Xaa Xaa
Xaa Xaa Xaa 100 105 110Xaa Xaa
Tyr Thr Tyr Tyr His Ile Glu Thr Glu Asn His Ser Val Ile 115
120 125Tyr Ser Asp Gly Met Leu Thr Glu Ser Tyr
130 13581138PRTUnknownBIL2 domain sequence from an
unknown origin 81Cys Phe Leu Ser Gly Thr Gln Ile Lys Thr Lys Leu Gly Val
Lys Asn1 5 10 15Ile Glu
Ala Leu Gln Val Gly Asp Phe Val Thr Thr Tyr Asp Xaa Xaa 20
25 30Xaa Xaa Xaa Xaa Xaa Xaa Arg Glu Val
Thr Trp Val Gly Xaa Lys Tyr 35 40
45Cys Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Pro Val 50
55 60Arg Ile Val Lys Asp Ala Ile Ala Xaa
Xaa Xaa Xaa Tyr Lys Asp Leu65 70 75
80Leu Val Thr Ala Glu His Cys Leu Phe Phe Xaa Asp Lys Phe
Ile Pro 85 90 95Ala Arg
Met Leu Val Asn Gly Ser Thr Ile Xaa Xaa Xaa Xaa Xaa Xaa 100
105 110Xaa Xaa Tyr Glu Tyr Tyr His Leu Glu
Thr Gln Asp His Ala Val Ile 115 120
125Ile Ala Asp Gly Val Arg Thr Glu Ser Tyr 130
13582138PRTMethylobacterium extorquensmisc_feature(31)..(33)Xaa can be
any naturally occurring amino acid 82Cys Phe Thr Thr Gly Thr Leu Ile Arg
Thr Ala Arg Gly Ser Val Ala1 5 10
15Val Glu Asp Leu Ile Val Gly Asp Leu Ala Val Thr Ala Ser Xaa
Xaa 20 25 30Xaa Arg Pro Ile
Thr Trp Ile Gly Asn Arg Ala Leu Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Pro Ile Arg Ile
Arg Ala Gly Ala 50 55 60Phe Gly Xaa
Xaa Xaa Xaa Ala Arg Asp Leu Arg Leu Ser His Gly His65 70
75 80Pro Val Leu Val Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Gly Val Leu Val 85 90
95Pro Val Met Cys Leu Ile Asn Gly Thr Ser Val Xaa Xaa Xaa
Xaa Xaa 100 105 110Xaa Xaa Val
Thr Tyr Trp His Ile Glu Leu Asp Ala His Asp Ile Leu 115
120 125Leu Ala Glu Gly Leu Ala Ala Glu Ser Tyr
130 13583142PRTSilicibacter
pomeroyimisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 83Cys Phe Thr Pro Gly Thr Lys Ile Ala Thr Pro Lys Gly Glu Arg Leu1
5 10 15Val Glu Asp Leu Glu
Val Gly Asp Arg Val Ile Thr Arg Asp Xaa Xaa 20
25 30Xaa Gln Glu Ile Arg Trp Val Gly Ser Arg Thr Leu
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Pro Val Leu Ile Arg Gln Gly Ala 50
55 60Leu Gly Xaa Xaa Xaa Xaa Glu Arg Asp Met Ile
Val Ser Pro Asn His65 70 75
80Arg Ile Leu Val Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
85 90 95Glu Val Leu Val Ala
Ala Lys His Leu Ile Gly Leu Glu Gly Val Xaa 100
105 110Xaa Xaa Xaa Xaa Xaa Xaa Val Thr Tyr Ile His Phe
Met Phe Asp Gln 115 120 125His Glu
Val Val Leu Ser Asp Gly Ala Trp Thr Glu Ser Phe 130
135 14084134PRTSilicibacter
pomeroyimisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 84Cys Phe Cys Arg Gly Thr Leu Ile Ala Thr Ala Gly Gly Glu Ile Pro1
5 10 15Val Glu Lys Leu Arg
Pro Gly Asp Arg Val Ile Thr Arg Asp Xaa Xaa 20
25 30Xaa Gln Arg Ile Arg Trp Ile Gly Gly Thr Ser Arg
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Pro Ile Arg Ile Arg Thr Gly Val Leu Lys Xaa Thr Arg 50
55 60Asp Leu Leu Val Ser Pro Asn His Arg Ile Leu
Met Xaa Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Val Leu Val Ala Ala Lys Phe
85 90 95Leu Val Asp Gly Arg
Ala Ile Xaa Xaa Xaa Xaa Xaa Xaa Xaa Val Asp 100
105 110Tyr Tyr His Met Leu Phe Asp Gln His Glu Leu Val
Leu Ser Glu Gln 115 120 125Ala Trp
Ser Glu Ser Phe 13085134PRTSilicibacter
pomeroyimisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 85Cys Phe Ala Ala Gly Thr Arg Ile Glu Thr Asp Arg Gly Gly Arg Ala1
5 10 15Ile Glu Asp Ile Ala
Val Gly Asp Leu Val Leu Thr Arg Asp Xaa Xaa 20
25 30Xaa Gln Pro Val Arg Trp Thr Gly Arg Arg Ser Val
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Pro Ile Arg Ile Ala Ser Gly Lys Leu Gly Xaa Leu Arg 50
55 60Asp Leu Leu Val Ser Pro Gln His Arg Leu Leu
Leu Xaa Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Val Leu Ala Ala Ala Val His
85 90 95Leu Arg Asp Asp Arg
His Ile Xaa Xaa Xaa Xaa Xaa Xaa Xaa Val Thr 100
105 110Tyr Val His Leu Met Phe Asp Arg His Glu Ile Ile
Tyr Ala Glu Gly 115 120 125Val Ala
Ser Glu Ser Phe 13086143PRTSilicibacter
pomeroyimisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 86Cys Phe Thr Pro Gly Thr Arg Ile Ala Thr Pro Thr Gly Pro Arg Leu1
5 10 15Ile Glu Glu Leu Arg
Glu Gly Asp Lys Val Gln Thr Arg Asp Xaa Xaa 20
25 30Xaa Gln Glu Ile Gln Trp Ile Gly Gln Arg Arg Met
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Pro Ile Arg Met Arg Val Gly Ala 50
55 60Leu Gly Xaa Xaa Xaa Xaa Asp Ala Glu Leu Leu
Val Ser Pro Glu His65 70 75
80Arg Met Leu Leu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
85 90 95Glu Val Leu Val Pro
Ala Arg Asp Leu Val Asn Asp Ser Thr Ile Xaa 100
105 110Xaa Xaa Xaa Xaa Xaa Xaa Xaa Val Thr Tyr Val His
Leu Leu Leu Pro 115 120 125Ser His
Gln Ile Leu Trp Ala Asn Gly Ile Glu Thr Glu Ser Phe 130
135 14087134PRTSilicibacter
pomeroyimisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 87Cys Phe Ala Ala Gly Thr Phe Ile Glu Ile Glu Ser Gly Pro Ile Pro1
5 10 15Val Glu Thr Leu Arg
Pro Gly Asp Leu Val Gln Thr Leu Asp Xaa Xaa 20
25 30Xaa Gln Pro Leu Leu Gln Leu Ala Lys Thr Thr Val
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Pro Val Leu Phe Arg Ala Gly Val Leu Gly Xaa Phe Arg 50
55 60Asp Leu Tyr Val Ser Gln Gln His Arg Met Leu
Ile Xaa Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Val Phe Val Pro Ala Arg Met
85 90 95Leu Val Asn Gly Ser
Thr Ile Xaa Xaa Xaa Xaa Xaa Xaa Xaa Leu Thr 100
105 110Tyr Tyr His Leu Leu Phe Ala Arg His Glu Ile Val
Phe Ser Glu Gly 115 120 125Ile Pro
Thr Glu Ser Tyr 13088134PRTSilicibacter
pomeroyimisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 88Cys Phe Val Ala Gly Thr Leu Ile Asp Thr Pro Tyr Gly Glu Arg Gln1
5 10 15Val Glu Arg Leu Thr
Pro Gly Asp Gln Val Phe Thr Arg Asp Xaa Xaa 20
25 30Xaa Gln Glu Val Arg Trp Val Gly Glu Arg Thr Val
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Pro Ile Leu Ile Arg Ala Gly Thr Tyr Gly Xaa Gln Arg 50
55 60Asp Leu Met Val Ser Pro Gln His Arg Ile Leu
Ile Xaa Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Val Leu Val Ala Ala Lys Asp
85 90 95Leu Val Asp Gly Arg
Arg Val Xaa Xaa Xaa Xaa Xaa Xaa Xaa Ile Thr 100
105 110Tyr Val His Val Met Phe Asp Ser His Gln Val Ile
Tyr Ser Glu Gly 115 120 125Leu Ala
Ser Glu Ser Phe 13089135PRTSilicibacter
pomeroyimisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 89Ser Leu His Pro Glu Thr Pro Ile Ala Thr Pro Asp Gly Tyr Arg Pro1
5 10 15Leu Ser Lys Ile Arg
Arg Gly Asp Thr Val Ile Val Ala Ser Xaa Xaa 20
25 30Xaa Val Pro Val Leu His Arg Val Ser Arg Thr Met
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Pro Leu Thr Ile Arg Arg Pro Tyr Phe Gly Xaa Arg Gln 50
55 60Asp Ile Gln Ala Ala Pro Ser Gln Arg Leu Leu
Leu Xaa Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Ser Val Leu Val Pro Ala Arg His
85 90 95Leu Thr Gly Gly His
Ser Val Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Ala 100
105 110Thr Tyr Ala Gln Leu Leu Leu Pro Thr Asn Glu Ala
Met Ile Thr Ala 115 120 125Gly Ala
Leu Ala Glu Ser Leu 130 13590143PRTSilicibacter
pomeroyimisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 90Cys Phe Val Ala Gly Ser Leu Ile Asp Thr Val Glu Gly Pro Arg Pro1
5 10 15Val Glu Thr Leu Ala
Val Gly Asp Leu Val Pro Val Glu Asp Xaa Xaa 20
25 30Xaa Gln Pro Ile Leu Trp Ile Gly Lys Arg Thr Leu
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Pro Val Arg Ile Arg Arg Asp Ala 50
55 60Leu Gly Xaa Xaa Xaa Xaa His Arg Thr Leu Trp
Val Ser Pro Gln His65 70 75
80Arg Ile Val Leu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
85 90 95Gln Val Phe Ala Ala
Ala Ile His Leu Thr Asn Asp Asp Thr Ile Xaa 100
105 110Xaa Xaa Xaa Xaa Xaa Xaa Xaa Val Thr Tyr Tyr His
Leu Ala Phe Glu 115 120 125Arg His
Leu Leu Leu Arg Ala His Gly Leu Leu Ser Glu Ser Ile 130
135 14091135PRTSilicibacter
pomeroyimisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 91Cys Phe Thr Pro Gly Thr Leu Ile Ala Thr Ala His Gly Pro Arg Ala1
5 10 15Ile Glu Thr Leu Arg
Pro Gly Asp Leu Ile Val Thr Arg Asp Xaa Xaa 20
25 30Xaa Gln Pro Leu Arg Trp Val Gly Ser Arg Thr Val
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Pro Ile Arg Leu Asp Pro Thr Leu Leu Gln Xaa Xaa Ser 50
55 60Ala Pro Leu Leu Val Ser Pro Gln His Arg Met
Leu Trp Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Val Leu Val Ala Ala Thr
85 90 95His Leu Leu Gly Ser
Pro Ala Ala Xaa Xaa Xaa Xaa Xaa Xaa Xaa Val 100
105 110Thr Tyr Met His Leu Met Leu Asp Arg His Glu Val
Ile Tyr Ala Asn 115 120 125Asp Ala
Ala Thr Glu Ser Phe 130 13592134PRTSilicibacter
pomeroyimisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 92Cys Phe Thr Pro Gly Thr Ile Ile Asp Thr Glu Asp Gly Pro Arg Leu1
5 10 15Ile Glu Glu Leu Gln
Pro Gly Asp Leu Ile Arg Thr Leu Asp Xaa Xaa 20
25 30Xaa Gln Pro Leu Arg Trp Ile Gly Arg Thr Thr Val
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Pro Val Leu Ile Arg Ala Gly Ala Leu Asp Xaa Arg Arg 50
55 60Asp Leu Ile Val Ser Pro Gln His Arg Met Leu
Ile Xaa Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gln Ala Leu Val Ala Ala Lys His
85 90 95Leu Val Asn Ala Arg
Asp Ile Xaa Xaa Xaa Xaa Xaa Xaa Xaa Val Thr 100
105 110Tyr Ile His Leu Leu Phe Asp Arg His Glu Ile Ile
Trp Ala Glu Gly 115 120 125Cys Pro
Thr Glu Ser Phe 13093143PRTSilicibacter
pomeroyimisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 93Cys Phe Ala Ala Gly Thr Arg Ile Ala Thr Pro Lys Gly Ala Arg Pro1
5 10 15Val Glu Thr Leu Ala
Val Gly Asp Leu Val Gln Thr Leu Asp Xaa Xaa 20
25 30Xaa Gln Pro Ile Arg Trp Ile Gly Thr Arg Arg Val
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Pro Val Val Ile Pro Ala His Ser 50
55 60Phe Ala Xaa Xaa Xaa Xaa Thr His Pro Leu Leu
Leu Ser Gln Gln His65 70 75
80Arg Val Leu Leu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
85 90 95Glu Ile Leu Ile Ala
Ala Arg Arg Leu Thr Gly Leu His Gly Ile Xaa 100
105 110Xaa Xaa Xaa Xaa Xaa Xaa Xaa Val Arg Tyr Ile His
Phe Ala Leu Asp 115 120 125Arg His
Glu Ile Val Phe Ala Asn Gly Leu Pro Ala Glu Thr Leu 130
135 14094135PRTSilicibacter
pomeroyimisc_feature(14)..(14)Xaa can be any naturally occurring amino
acid 94Ser Phe Thr Arg Gly Thr His Ile Thr Leu Gly Ser Gly Xaa Gln Val1
5 10 15Arg Ile Glu Asp Leu
Lys Val Gly Asp Arg Val Leu Thr Arg Asp Xaa 20
25 30Xaa Xaa Arg Glu Val Arg Trp Ile Gly Gln Thr Thr
Val Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Pro Ile Val Ile Arg Ala Gly Thr Leu Asn Xaa Glu 50
55 60Asn Asp Leu Val Val Ser Pro Asp His Arg Leu
Phe Val Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Glu Leu Leu Leu Lys Ala Arg
85 90 95His Leu Val Asn Gly
Asp Thr Val Xaa Xaa Xaa Xaa Xaa Xaa Xaa Val 100
105 110Asp Tyr Phe Gln Leu Leu Phe Asp Arg His His Ile
Ile Tyr Ala Glu 115 120 125Gly Ile
Ala Ala Glu Thr Met 130 13595144PRTSilicibacter
pomeroyimisc_feature(31)..(33)Xaa can be any naturally occurring amino
acid 95Ala Phe Ser Arg Gly Ser Leu Ile Asp Thr Asp Cys Gly Pro Met Ala1
5 10 15Ile Glu Asp Leu Leu
Pro Gly Asp Arg Val Ile Thr Gln Asp Xaa Xaa 20
25 30Xaa Gln Glu Val Val Trp Lys Gly Ser Thr Val Ile
Xaa Xaa Xaa Xaa 35 40 45Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Leu Thr Arg Ile Met Ala Asp 50
55 60Ala Phe Gly Xaa Xaa Xaa Xaa Met Ser Gly Val
Ile Ala Gly Pro Ser65 70 75
80Ala Arg Leu Leu Ala Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
85 90 95Xaa Pro Met Leu Thr
Pro Val Gln His Phe Val Asp Gly Met Gly Ile 100
105 110Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Ile Glu Val Phe
His Ile Cys Leu 115 120 125Arg Arg
His Ala Val Ile Asn Val Asp Gly Leu Gln Phe Glu Thr Tyr 130
135 14096125PRTSilicibacter
pomeroyimisc_feature(31)..(49)Xaa can be any naturally occurring amino
acid 96Gly Leu Pro Ala Gly Thr Met Leu Glu Thr Glu Ala Gly Trp Ser Pro1
5 10 15Val Glu Glu Ile Arg
Pro Gly Thr Arg Val Ala Thr Ile Asp Xaa Xaa 20
25 30Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 35 40 45Xaa Leu
Trp Arg Ile Pro Gly Gly Thr Leu Gly Xaa Cys Ser Asp Leu 50
55 60Leu Leu Pro Glu Gly His Phe Leu Ala Leu Xaa
Xaa Xaa Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Thr Val Leu Ala Pro Val Ala Ala Leu Ala
85 90 95Gly Phe Glu Gly Ile
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Leu Pro Ala His 100
105 110Ser Leu Arg Phe Ala Glu Glu Glu Val Val Trp Ala
Gln 115 120
12597126PRTSilicibacter pomeroyimisc_feature(31)..(33)Xaa can be any
naturally occurring amino acid 97Gly Phe Leu Ala Gly Thr Ile Leu Leu Thr
Gln Asp Gly Glu Met Pro1 5 10
15Val Glu Phe Leu Ser Pro Gly Asp Arg Ile Ile Thr Arg Asp Xaa Xaa
20 25 30Xaa Val Pro Leu His His
Ile Thr Arg Ala Pro Gln Xaa Xaa Xaa Ala 35 40
45Ile Arg Ile Ala Ala Gly Ser Leu Gly Xaa Xaa Xaa Xaa Asp
Cys Asp 50 55 60Leu Ile Leu Pro Ala
Gly Gln Pro Val Leu Ile Xaa Xaa Xaa Xaa Xaa65 70
75 80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gln Ala Met
Val Arg Ala Asp Ala Leu 85 90
95Val Asp Gly Glu Phe Ile Xaa Xaa Xaa Xaa Xaa Xaa Xaa Met Gln Leu
100 105 110Phe Gln Leu His Phe
Asp Ser Ala His Val Leu Tyr Ala Gly 115 120
12598132PRTSilicibacter pomeroyimisc_feature(31)..(33)Xaa can be
any naturally occurring amino acid 98Gly Leu Leu Ala Gly Thr Ser Val Ala
Ser Asn Phe Gly Trp Gln Pro1 5 10
15Val Glu Ala Leu Lys Val Gly Asp Lys Val Leu Thr Phe Asp Xaa
Xaa 20 25 30Xaa Gln Thr Val
Ala Asp Ile Gln Arg Glu Thr Val Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Pro Val Arg Leu Pro
Glu Gly Val Cys 50 55 60His Xaa Arg
Arg Asp Leu Trp Met Met Pro Asp Gln Gly Leu Leu Val65 70
75 80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Tyr Ala Val Val 85 90
95Pro Ala Arg Met Leu Arg Gly Tyr Arg Gly Ile Xaa Xaa Xaa
Xaa Xaa 100 105 110Xaa Xaa Xaa
Val Glu Val Thr Thr Leu Ala Phe His Gln Asp Glu Val 115
120 125Ile Tyr Val Glu 13099134PRTRhizobium
leguminosarummisc_feature(31)..(33)Xaa can be any naturally occurring
amino acid 99Cys Phe Leu Arg Gly Thr Ala Ile Leu Thr Asp Cys Gly Glu Lys
Pro1 5 10 15Val Glu Asn
Leu Ser Ile Gly Asp Arg Val Ala Leu Pro Asp Xaa Xaa 20
25 30Xaa Arg Pro Ile Lys Trp Val Gly Arg Gln
Ser Phe Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Pro Ile Arg Val Ser Arg His Ala 50
55 60Leu Asp Xaa Xaa Xaa Xaa His Ser Asp
Leu Tyr Leu Ser Pro Gly His65 70 75
80Ala Leu Tyr Leu Xaa Gly Ile Leu Ile Gln Val Lys Asp Leu
Val Asn 85 90 95Gly Lys
Thr Ile Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Ile Glu 100
105 110Tyr Tyr Ala Val Met Leu Asp Thr His
Glu Val Ile Leu Ala Gly Gly 115 120
125Ala Glu Thr Glu Ser Phe 130100138PRTMagnetospirillum
magnetotacticummisc_feature(31)..(33)Xaa can be any naturally occurring
amino acid 100Cys Tyr Val Thr Gly Thr Arg Ile Arg Thr Glu Arg Gly Glu Ile
Ala1 5 10 15Val Glu Asp
Leu Gln Val Gly Asp Phe Ala Val Thr Ala Ser Xaa Xaa 20
25 30Xaa Arg Pro Ile Thr Trp Ile Gly His Arg
Glu Ile Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Pro Val Arg Val Arg Ala Gly Ala 50
55 60Phe Gly Xaa Xaa Xaa Xaa Val Asn Asp
Leu Phe Leu Ser Pro Gly His65 70 75
80Pro Val Leu Val Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Val
Leu Val 85 90 95Pro Val
Met Cys Leu Ile Asn Gly Thr Thr Ile Xaa Xaa Xaa Xaa Xaa 100
105 110Xaa Xaa Val Thr Tyr Trp His Val Glu
Leu Asp Ala His Asp Ile Leu 115 120
125Leu Ala Glu Gly Leu Pro Ala Glu Ser Tyr 130
135101138PRTMagnetospirillum magnetotacticummisc_feature(31)..(33)Xaa can
be any naturally occurring amino acid 101Cys Phe Val Ser Gly Thr Arg Ile
Ser Val Glu Arg Gly Ser Ile Pro1 5 10
15Val Glu Leu Leu Arg Ile Gly Glu Lys Ala Arg Leu Ala Ser
Xaa Xaa 20 25 30Xaa Arg Thr
Ile Thr Trp Ile Gly His Arg Glu Ile Xaa Xaa Xaa Xaa 35
40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Pro Val Arg
Val Arg Ala Gly Ala 50 55 60Phe Gly
Xaa Xaa Xaa Xaa Ala Arg Asp Leu Phe Leu Ser Pro Gly His65
70 75 80Pro Val Leu Ile Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Gly Val Leu Val 85 90
95Pro Val Met Cys Leu Ile Asn Gly Thr Ser Ile Xaa Xaa
Xaa Xaa Xaa 100 105 110Xaa Xaa
Val Thr Tyr Trp His Val Glu Leu Asp Arg His Asp Ile Leu 115
120 125Leu Ala Glu Gly Leu Pro Ala Glu Ser Tyr
130 13510298PRTMagnetospirillum
magnetotacticummisc_feature(31)..(33)Xaa can be any naturally occurring
amino acid 102Cys Phe Val Thr Gly Thr Met Ile Ala Thr Ala Arg Gly Glu Val
Ala1 5 10 15Val Glu Asp
Leu Arg Ala Gly Asp Phe Ala Arg Thr Ala Glu Xaa Xaa 20
25 30Xaa Arg Pro Ile Val Trp Ile Gly His Arg
Glu Ile Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Pro Val Arg Val Arg Thr Gly Ala 50
55 60Phe Gly Xaa Xaa Xaa Xaa Ala Arg Asp
Leu Tyr Leu Ser Pro Gly His65 70 75
80Pro Val Leu Val Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Thr
Leu Val 85 90 95Pro
Ile10324PRTMagnetospirillum magnetotacticum 103Val Thr Tyr Trp His Val
Glu Leu Asp Ala His Asp Ile Leu Leu Ala1 5
10 15Glu Gly Leu Pro Ala Glu Ser Tyr
201047PRTMagnetospirillum magnetotacticum 104Arg Leu Pro Ala Glu Ser Tyr1
510557PRTartificial sequenceType B BIL domain consensus
sequence 105Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa1 5 10 15Xaa Xaa Xaa
Xaa Gly Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 20
25 30Xaa His Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa 35 40
45Xaa Xaa Xaa Xaa Xaa Xaa His Xaa Xaa 50
5510688PRTartificial sequenceType B BIL domain consensus sequence 106Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Xaa Xaa Xaa1
5 10 15Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 20 25
30Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa 35 40 45Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 50 55
60Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa65 70 75
80Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 85107149PRTPseudomonas
syringae 107Cys Phe Ala Ala Gly Thr Met Val Ser Thr Pro Asp Gly Glu Arg
Ala1 5 10 15Ile Asp Thr
Leu Lys Val Gly Asp Ile Val Trp Ser Lys Pro Glu Gly 20
25 30Gly Gly Lys Pro Phe Ala Ala Ala Ile Leu
Ala Thr His Ile Arg Thr 35 40
45Asp Gln Pro Ile Tyr Arg Leu Lys Leu Lys Gly Lys Gln Glu Asn Gly 50
55 60Gln Ala Glu Asp Glu Ser Leu Leu Val
Thr Pro Gly His Pro Phe Tyr65 70 75
80Val Pro Ala Gln His Gly Phe Val Pro Val Ile Asp Leu Lys
Pro Gly 85 90 95Asp Arg
Leu Gln Ser Leu Ala Asp Gly Ala Ser Glu Asn Thr Ser Ser 100
105 110Glu Val Glu Ser Leu Glu Leu Tyr Leu
Pro Val Gly Lys Thr Tyr Asn 115 120
125Leu Thr Val Asp Val Gly His Thr Phe Tyr Val Gly Lys Leu Lys Thr
130 135 140Trp Val His Asn
Thr145108461PRTartificial sequenceMBP-CBD splicing product 108Met Lys Thr
Glu Glu Gly Lys Leu Val Ile Trp Ile Asn Gly Asp Lys1 5
10 15Gly Tyr Asn Gly Leu Ala Glu Val Gly
Lys Lys Phe Glu Lys Asp Thr 20 25
30Gly Ile Lys Val Thr Val Glu His Pro Asp Lys Leu Glu Glu Lys Phe
35 40 45Pro Gln Val Ala Ala Thr Gly
Asp Gly Pro Asp Ile Ile Phe Trp Ala 50 55
60His Asp Arg Phe Gly Gly Tyr Ala Gln Ser Gly Leu Leu Ala Glu Ile65
70 75 80Thr Pro Asp Lys
Ala Phe Gln Asp Lys Leu Tyr Pro Phe Thr Trp Asp 85
90 95Ala Val Arg Tyr Asn Gly Lys Leu Ile Ala
Tyr Pro Ile Ala Val Glu 100 105
110Ala Leu Ser Leu Ile Tyr Asn Lys Asp Leu Leu Pro Asn Pro Pro Lys
115 120 125Thr Trp Glu Glu Ile Pro Ala
Leu Asp Lys Glu Leu Lys Ala Lys Gly 130 135
140Lys Ser Ala Leu Met Phe Asn Leu Gln Glu Pro Tyr Phe Thr Trp
Pro145 150 155 160Leu Ile
Ala Ala Asp Gly Gly Tyr Ala Phe Lys Tyr Glu Asn Gly Lys
165 170 175Tyr Asp Ile Lys Asp Val Gly
Val Asp Asn Ala Gly Ala Lys Phe Leu 180 185
190Val Asp Leu Ile Ala Gly Leu Thr Lys Asn Lys His Met Asn
Ala Asp 195 200 205Thr Asp Tyr Ser
Ile Ala Glu Ala Ala Phe Asn Lys Gly Glu Thr Ala 210
215 220Met Thr Ile Asn Gly Pro Trp Ala Trp Ser Asn Ile
Asp Thr Ser Lys225 230 235
240Val Asn Tyr Gly Val Thr Val Leu Pro Thr Phe Lys Gly Gln Pro Ser
245 250 255Lys Pro Phe Val Gly
Val Leu Ser Ala Gly Ile Asn Ala Ala Ser Pro 260
265 270Asn Lys Glu Leu Ala Lys Glu Phe Leu Glu Asn Tyr
Leu Leu Thr Asp 275 280 285Glu Gly
Leu Glu Ala Val Asn Lys Asp Lys Pro Leu Gly Ala Val Ala 290
295 300Leu Lys Ser Tyr Glu Glu Glu Leu Ala Lys Asp
Pro Arg Ile Ala Ala305 310 315
320Thr Met Glu Asn Ala Gln Lys Gly Glu Ile Met Pro Asn Ile Pro Gln
325 330 335Met Ser Ala Phe
Trp Tyr Ala Val Arg Thr Ala Val Ile Asn Ala Ala 340
345 350Ser Gly Arg Gln Thr Val Asp Glu Ala Leu Lys
Asp Ala Gln Thr Asn 355 360 365Ser
Ser Ser Asn Asn Asn Asn Asn Asn Asn Asn Asn Asn Leu Gly Ile 370
375 380Glu Gly Arg Ile Ser Glu Phe Gly Ser Thr
Ser Arg Val Asp Cys Gly385 390 395
400Gly Leu Thr Gly Leu Asn Ser Gly Leu Thr Thr Asn Pro Gly Val
Ser 405 410 415Ala Trp Gln
Val Asn Thr Ala Tyr Thr Ala Gly Gln Leu Val Thr Tyr 420
425 430Asn Gly Lys Thr Tyr Lys Cys Leu Gln Pro
His Thr Ser Leu Ala Gly 435 440
445Trp Glu Pro Ser Asn Val Pro Ala Leu Trp Gln Leu Gln 450
455 460109541PRTartificial sequenceMBP-PsyBIL carboxy
terminal cleavage product 109Met Lys Thr Glu Glu Gly Lys Leu Val Ile Trp
Ile Asn Gly Asp Lys1 5 10
15Gly Tyr Asn Gly Leu Ala Glu Val Gly Lys Lys Phe Glu Lys Asp Thr
20 25 30Gly Ile Lys Val Thr Val Glu
His Pro Asp Lys Leu Glu Glu Lys Phe 35 40
45Pro Gln Val Ala Ala Thr Gly Asp Gly Pro Asp Ile Ile Phe Trp
Ala 50 55 60His Asp Arg Phe Gly Gly
Tyr Ala Gln Ser Gly Leu Leu Ala Glu Ile65 70
75 80Thr Pro Asp Lys Ala Phe Gln Asp Lys Leu Tyr
Pro Phe Thr Trp Asp 85 90
95Ala Val Arg Tyr Asn Gly Lys Leu Ile Ala Tyr Pro Ile Ala Val Glu
100 105 110Ala Leu Ser Leu Ile Tyr
Asn Lys Asp Leu Leu Pro Asn Pro Pro Lys 115 120
125Thr Trp Glu Glu Ile Pro Ala Leu Asp Lys Glu Leu Lys Ala
Lys Gly 130 135 140Lys Ser Ala Leu Met
Phe Asn Leu Gln Glu Pro Tyr Phe Thr Trp Pro145 150
155 160Leu Ile Ala Ala Asp Gly Gly Tyr Ala Phe
Lys Tyr Glu Asn Gly Lys 165 170
175Tyr Asp Ile Lys Asp Val Gly Val Asp Asn Ala Gly Ala Lys Ala Gly
180 185 190Leu Thr Phe Leu Val
Asp Leu Ile Lys Asn Lys His Met Asn Ala Asp 195
200 205Thr Asp Tyr Ser Ile Ala Glu Ala Ala Phe Asn Lys
Gly Glu Thr Ala 210 215 220Met Thr Ile
Asn Gly Pro Trp Ala Trp Ser Asn Ile Asp Thr Ser Lys225
230 235 240Val Asn Tyr Gly Val Thr Val
Leu Pro Thr Phe Lys Gly Gln Pro Ser 245
250 255Lys Pro Phe Val Gly Val Leu Ser Ala Gly Ile Asn
Ala Ala Ser Pro 260 265 270Asn
Lys Glu Leu Ala Lys Glu Phe Leu Glu Asn Tyr Leu Leu Thr Asp 275
280 285Glu Gly Leu Glu Ala Val Asn Lys Asp
Lys Pro Leu Gly Ala Val Ala 290 295
300Leu Lys Ser Tyr Glu Glu Glu Leu Ala Lys Asp Pro Arg Ile Ala Ala305
310 315 320Thr Met Glu Asn
Ala Gln Lys Gly Glu Ile Met Pro Asn Ile Pro Gln 325
330 335Met Ser Ala Phe Trp Tyr Ala Val Arg Thr
Ala Val Ile Asn Ala Ala 340 345
350Ser Gly Arg Gln Thr Val Asp Glu Ala Leu Lys Asp Ala Gln Thr Asn
355 360 365Ser Ser Ser Asn Asn Asn Asn
Asn Asn Asn Asn Asn Asn Leu Gly Ile 370 375
380Glu Gly Arg Ile Ser Glu Phe Gly Ser Cys Phe Ala Ala Gly Thr
Met385 390 395 400Val Ser
Thr Pro Asp Gly Glu Arg Ala Ile Asp Thr Leu Lys Val Gly
405 410 415Asp Ile Val Trp Ser Lys Pro
Glu Gly Gly Gly Lys Pro Phe Ala Ala 420 425
430Ala Ile Leu Ala Thr His Ile Arg Thr Asp Gln Pro Ile Tyr
Arg Leu 435 440 445Lys Leu Lys Gly
Lys Gln Glu Asn Gly Gln Ala Glu Asp Glu Ser Leu 450
455 460Leu Val Thr Pro Gly His Pro Phe Tyr Val Pro Ala
Gln His Gly Phe465 470 475
480Val Pro Val Ile Asp Leu Lys Pro Gly Asp Arg Leu Gln Ser Leu Ala
485 490 495Asp Gly Ala Ser Glu
Asn Thr Ser Ser Glu Val Glu Ser Leu Glu Leu 500
505 510Tyr Leu Pro Val Gly Lys Thr Tyr Asn Leu Thr Val
Asp Val Gly His 515 520 525Thr Phe
Tyr Val Gly Lys Leu Lys Thr Trp Val His Asn 530 535
540
User Contributions:
Comment about this patent or add new information about this topic:
| People who visited this patent also read: | |
| Patent application number | Title |
|---|---|
| 20110171452 | PROCEDURE FOR MAKING PRE-IMPREGNATED REINFORCED COMPOSITE, AS WELL AS FIBER REINFORCED COMPOSITE, AND THEIR APPLICATION |
| 20110171451 | DARK TYPE FLUORORESIN FILM AND BACK SHEET FOR SOLAR CELL MODULE |
| 20110171450 | Process for preparing a blown film from a polyethylene molding composition |
| 20110171449 | PAPER-LIKE FILM AND PROCESS FOR MAKING IT |
| 20110171448 | PREPARATION OF HYPERBRANCHED POLY(TRIAZOLE)S BY IN SITU CLICK POLYMERIZATION AND ADHESIVE CONTAINING THE SAME |