Patent application title: IDENTIFICATION AND CHARACTERIZATION OF RACEMASES, DEFINITION OF PROTEIN SIGNATURES, AND A TEST FOR DETECTING D-AMINO ACID AND FOR SCREENING MOLECULES CAPABLE OF INHIBITING THE ACTIVITY OF RACEMASE, ESPECIALLY PROLINE RACEMASE
Inventors:
Paola Minoprio (Villiers Sur Marne, FR)
Nathalie Chamond (Paris, FR)
Wim Degrave (Paris Cedex 15, FR)
Armand Berneman (Paris, FR)
Assignees:
INSTITUT PASTEUR
IPC8 Class: AC12Q130FI
USPC Class:
435027000
Class name:
Publication date: 2010-08-05
Patent application number: 20100196943
Claims:
1-54. (canceled)
55. A method for detecting a D-amino acid, wherein the method comprises: (A) providing a reaction medium containing the D-amino acid; (B) reacting a primary or secondary amine on the D-amino acid with a D-amino oxidase and a prosthetic group to form a reduced prosthetic group by oxidative deamination of the D-amino acid; (C) reacting the reduced prosthetic group with oxygen to form hydrogen peroxide; and (D) detecting the hydrogen peroxide thus formed.
56. The method of claim 55, wherein the prosthetic group is flavin-adenin-dinucleotide (FAD) or flavin-mononucleotide (FMN).
57. The method of claim 56, wherein the hydrogen peroxide is detected by reaction with a catalase.
58. The method of claim 55, wherein the D-amino acid is a D-Proline, D-Tyrosine, D-Valine, D-Threonine, D-Glutamic acid, D-Lysine, or D-Tryptophane.
59. The method of claim 56, wherein the hydrogen peroxide is detected by reaction with a peroxidase.
60. The method of claim 55, further comprising quantifying the D-amino acid in the reaction medium after the formation of the hydrogen peroxide.
61. The method of claim 55, wherein the reaction medium comprises a biological sample from a subject afflicted with Alzheimer's disease, Parkinson's disease, renal disease, or schizophrenia.
62. The method of claim 61, wherein the biological sample comprises a fluid or tissue sample from the subject.
63. The method of claim 61, wherein the biological sample comprises cells from the subject.
64. A method for detecting racemase activity in a reaction medium, wherein the method comprises: (A) providing a reaction medium containing a D-amino acid specific to the racemase to be detected; (B) reacting the D-amino acid with a D-amino oxidase and a prosthetic group to form a reduced prosthetic group by oxidation of the D-amino acid; (C) reacting the reduced prosthetic group with oxygen to form hydrogen peroxide; and (D) detecting the hydrogen peroxide thus formed; wherein the detection of hydrogen peroxide indicates racemase activity in the reaction medium.
65. The method of claim 64, wherein the hydrogen peroxide is detected by reaction with catalase.
66. The method of claim 64, wherein the hydrogen peroxide is detected with a chromogenic reagent.
67. The method of claim 66, wherein the chromogenic reagent is orthophenyalaninediamine (OPD), 3,3',5,5'-tetrimethylbenzadine (TMB), or 5-aminosalicyclic acid (ASA).
68. A kit for screening for inhibitors of TcPRAC, wherein the kit comprises: (A) L-proline, D-proline, and a proline-racemase; (B) a peroxidase and a substrate of a peroxidase, or a catalase and a reagent sensitive to oxygen; and (C) a D-amino acid oxidase.
69. The kit of claim 68, further comprising one or more molecules to be screened for inhibitory activity of TcPRAC.
70. A kit for detecting a D-amino acid in a sample, wherein the kit comprises: (A) a D-amino acid; (B) a peroxidase and a substrate of a peroxidase; and (C) a D-amino acid oxidase.
71. The kit according to claim 70, further comprising a L-amino acid enantiomer as control.
72. A method for detecting a D-amino acid in a sample, wherein the method comprises: (A) oxidatively deaminating a D-amino acid by reaction with a D-amino acid oxidase and a prosthetic group; and (B) detecting the hydrogen peroxide generated by the oxidative deamination; wherein the presence of hydrogen peroxide is indicative of the presence of a D-amino acid in the sample.
73. The method of claim 72, wherein the D-amino acid is D-Proline, D-Tyrosine, D-Valine, D-Threonine, D-Glutamic acid, D-Lysine, or D-Tryptophane.
74. A method for screening a molecule, which can modulate a racemase activity, wherein the method comprises: (A) modulating a racemase activity by means of a molecule being tested in the presence of an equimolar mixture of a L- and D-amino acid and of a racemase to be modulated; (B) oxidatively deaminating the D-amino acid generated in step (A) by means of a D-amino oxidase and a prosthetic group; and (C) detecting the hydrogen peroxide generated by the oxidative deamination; wherein modulation of the hydrogen peroxide is indicative of the capability of the tested molecule to modulate racemase activity.
75. The method of claim 74, wherein said molecule inhibits said racemase activity.
76. The method of claim 74, wherein said racemase is a proline racemase.
77. The method of claim 74, wherein said proline racemase is Trypanosoma cruzi proline racemase.
78. A molecule identified by the method of claim 74.
Description:
CROSS-REFERENCE TO RELATED APPLICATION
[0001] This application is based on and claims the benefit of U.S. Provisional Application Ser. No. 60/446,263, filed Feb. 11, 2003 (Attorney Docket No. 03495.6087). The entire disclosure of this Provisional application is relied upon and incorporated by reference herein
BACKGROUND OF THE INVENTION
[0002] This invention relates to the identification and characterization of racemases and definition of protein signatures of these racemases. More particularly, this invention relates to the identification of nucleic acid molecules encoding a peptide consisting of a motif characteristic of the protein signatures, and to the peptides consisting of these motifs. This invention also relates to antibodies specific for the peptides and to immune complexes of these antibodies with the peptides. Further, the invention relates to methods and kits for detecting racemases using the nucleic acid molecules of the invention, as well as the peptides consisting of the motifs and antibodies to these peptides.
[0003] D-amino acids have long been described in the cell wall of gram-positive and especially gram-negative bacteria, where they constitute essential elements of the peptidoglycan and as substitutes of cell wall techoic acids (1). Moreover, various types of D-amino acids were discovered in a number of small peptides made by a variety of microorganisms through non-ribosomal protein synthesis (2), that function mainly as antibiotic agents. However, these examples were considered exceptions to the rule of homochirality and a dogma persisted that only L-amino acid enantiomers were present in eukaryotes, apart from a very low level of D-amino acids from spontaneous racemization due to aging (3).
[0004] Recently, an increasing number of studies have reported the presence of various D-amino acids (D-aa) either as protein bound (4) or under free forms (5) in a wide variety of organisms, including mammals. The origin of free D-aa, is less clear than that of protein bound D-aa. For instance, in mammals, free D-aa may originate from exogenous sources (as described in (6), but the recent discovery of amino acid racemases in eukaryotes has also uncovered an endogenous production of D-aa, questioning their specific functions. Thus, the level of D-aspartate is developmentally regulated in rat embryos (7); the binding of D-serine to NMDA mouse brain receptors promotes neuromodulation (8), (9), and D-aspartate appears to be involved in hormonal regulation in endocrine tissues (10).
[0005] All amino acid racemases require pyridoxal phosphate as a cofactor, except proline and hydroxyproline racemases, which are cofactor-independent enzymes. For example, two reports have been published addressing the biochemical and enzymatic characteristics of the proline racemase from the gram-positive bacterium Clostridium sticklandii (11, 12). A reaction mechanism was proposed whereby the active site Cys256 forms a half-reaction site with the corresponding cysteine of the other monomer in the active, homodimeric enzyme.
[0006] Although a variety of racemases and epimerases has been demonstrated in bacteria and fungi, the first eukaryotic amino acid (proline) racemase isolated from the infective metacyclic forms of the parasitic protozoan Trypanosoma cruzi, the causative agent of Chagas' disease in humans (13), was recently described. This parasite-secreted proline racemase (TcPRAC) was shown to be a potent mitogen for host B cells and to play an important role in T. cruzi immune evasion and persistence through polyclonal lymphocyte activation (13). This protein, previously annotated as TcPA45, with monomer size of 45 kDa, is only expressed and released by infective metacyclic forms of the parasite (13).
[0007] The genomic organization and transcription of TcPRAC proline racemase gene indicated the presence of two homologous genes per haploid genome (TcPRACA and TcPRACB). Furthermore, localization studies using specific antibodies directed to 45 kDa-TcPRAC protein revealed that an intracellular and/or membrane associated isoform, with monomer size of 39 kDa is expressed in non-infective epimastigote forms of the parasite.
[0008] Computer-assisted analysis of the TcPRACA gene sequence suggested that it could give rise to both isoforms (45 kDa and 39 kDa) of parasite proline racemases through a mechanism of alternative trans-splicing, one of which would contain a signal peptide (13). In addition, preliminary analysis of putative TcPRACB gene sequences had revealed several differences that include point mutations as compared to TcPRACA, but that also suggest that TcPRACB gene could only encode an intracellular isoform of the enzyme as the gene lacks the export signal sequence. Any of these molecular mechanisms per se would ensure the differential expression of intracellular and extracellular isoforms of proline racemases produced in different T. cruzi developmental stages.
[0009] The process of production of a D-amino acid by using a L-amino acid source comprises the use of an amino acid racemase specific for the amino acid of interest, the racemase being produced from a recombinant expression system containing a vector having a polynucleotide sequence encoding the enzyme. In prokaryotic hosts, the racemases are known to be implicated in the synthesis of D-amino acids and/or in the metabolism of L-amino acids. For instance, the presence of free D-amino acids in tumors and in progressive autoimmune and degenerative diseases suggests the biological importance of eukaryotic amino acid racemases. It is well known that proteins or peptides containing D-amino acids are resistant to proteolysis by host enzymes. In addition, such proteins containing D-amino acids, at least one D-amino acid residue, can display antibiotic or immunogenic properties.
[0010] There is a growing interest in the biological role of D-amino acids, either as free molecules or within polypeptide chains in human brain, tumors, anti-microbial and neuropeptides, suggesting widespread biological implications. Research on D-amino acids in living organisms has been hampered by their difficult detection. There exists a need in the art for the identification of racemases and the identification of their enzymatic properties and their specificity for other compounds.
[0011] Although much progress has been made concerning prophylaxis of Chagas' disease, particularly vector eradication, additional cases of infection and disease development still occur every day throughout the world. Whilst infection was largely limited in the past to vector transmission in endemic areas of Latin America, its impact has increased in terms of congenital and blood transmission, transplants and recrudescence following immunosuppressive states. Prevalence of Chagas' disease in Latin America may reach 25% of the population, as is the case of Bolivia, or yet 1%, as observed in Mexico. From the 18-20 million people already infected with the parasite Trypanosoma cruzi, more than 60% live in Brazil and WHO estimates that 90 million individuals are at risk in South and Central America.
[0012] Some figures obtained from a recent census in USA, for instance, revealed that the net immigration from Mexico is about 1000 people/day, of those 5-10 individuals are infected by Chagas' disease. The disease can lie dormant for 10-30 years and as an example of many other progressive chronic pathologies it is characterized by being "asymptomatic". Although at the 1990's, blood banks increased their appeals to Hispanics (50% of Bolivian blood is contaminated), panels of Food and Drug Administration (FDA) have recommended that all donated blood be screened for Chagas. Today, FDA has not yet approved an `accurate` blood test to screen donor blood samples. This allegation seriously contrasts with the more than 30 available tests used in endemic countries. Additionally, recent reports on new insect vectors adapted to the parasite and domestic animals infected in more developed countries like USA, and the distributional predictions based on Genetic Algorithm for Rule-set Prediction models indicate a potentially broad distribution for these species and suggest additional areas of risk beyond those previously reported emphasizing the continuing worldwide public health issue.
[0013] To date, two drugs are particularly used to treat Trypanosoma cruzi infections. Nifurtimox (3-methyl-4-5'-nitrofurfurylidene-amino tetrahydro 4H-1,4-thiazine-1,1-dioxide), a nitrofurane from Bayer, known as Lampit, was the first drug to be used since 1967. After 1973, Benznidazol, a nitroimidazol derivative, known as Rochagan or Radanyl (N-benzyl-2-nitro-1-imidazol acetamide) was produced by Hoffman-La-Roche and is consensually the drug of choice. Both drugs are trypanosomicides and act against intracellular or extracellular forms of the parasite. Adverse side-effects include a localized or generalized allergic dermopathy, peripheral sensitive polyneuropathy, leucopenia, anorexia, digestive manifestations and rare cases of convulsions which are reversible by interruption of treatment. The most serious complications include agranulocytosis and trombocytopenic purpura.
[0014] Unquestionably, the treatment is efficient and should be applied in acute phases of infection, in children, and in cases where reactivation of parasitaemia is observed following therapy with immunosuppressive drugs or organ transplantation procedures. Some experts recommend that patients in indeterminate and chronic phases should also be treated. However, close to a hundred years after the discovery of the infection and its consequent disease, researchers still maintain divergent points of view concerning therapy against the chronic phases of the disease. As one of the criteria of cure is based on the absence of the parasite in the blood, it is very difficult to evaluate the efficacy of the treatment in indeterminate or chronic phases. Because the indeterminate form is asymptomatic, it is impossible to clinically evaluate the cure. Furthermore, a combination of serology and more sensitive advanced molecular techniques will be required and still may not be conclusive. The follow-up of patients for many years is then inevitable to objectively ascertain the cure.
[0015] Chagas' disease was recently considered as a neglected disease and DND-initiative (Drug for Neglected Diseases Initiative, DNDi) wishes to support drug discovery projects focused on the development of effective, safe and affordable new drugs against trypanosomiasis. Since current therapies remain a matter of debate, may be inadequate in some circumstances, are rather toxic and may be of limited effectiveness, the characterization of new formulations and the discovery of parasite molecules capable of eliciting protective immunity are absolutely required and must be considered as priorities.
SUMMARY OF THE INVENTION
[0016] This invention aids in fulfilling these needs in the art. It has been discovered that the TcPRAC genes in T. cruzi encode functional intracellular or secreted versions of the enzyme exhibiting distinct kinetic properties that may be relevant for their relative catalytic efficiency. While the KM of the enzyme isoforms were of a similar order of magnitude (29-75 mM), Vmax varied between 2×10-4 to 5.3×10-5 mol of L-proline/sec/0.125 μM of homodimeric recombinant protein. Studies with the enzyme specific inhibitor and abrogation of enzymatic activity by site-directed mutagenesis of the active site Cys330 residue, reinforced the potential of proline racemase as a critical target for drug development against Chagas' disease.
[0017] This invention provides a purified nucleic acid molecule encoding a peptide consisting of a motif selected from SEQ ID NOS: 1, 2, 3, or 4.
[0018] This invention also provides a purified nucleic acid molecule that hybridizes to either strand of a denatured, double-stranded DNA comprising this nucleic acid molecule under conditions of moderate stringency.
[0019] In addition, this invention provides a recombinant vector that directs the expression of a nucleic acid molecule selected from these purified nucleic acid molecules.
[0020] Further, this invention provides a purified polypeptide encoded by a nucleic acid molecule selected from the group consisting of a purified nucleic acid molecule coding for: [0021] (a) a purified polypeptide consisting of Motif I (SEQ ID NO:1); [0022] (b) a purified polypeptide consisting of Motif II (SEQ ID NO:2); [0023] (c) a purified polypeptide consisting of Motif III (SEQ ID NO:3); and [0024] (d) a purified polypeptide consisting of Motif III* (SEQ ID NO:4).
[0025] Purified antibodies that bind to these polypeptides are provided. The purified antibodies can be monoclonal antibodies. An immunological complex comprises a polypeptide and an antibody that specifically recognizes the polypeptide of the invention.
[0026] A host cell transfected or transduced with the recombinant vector of the invention is provided.
[0027] A method for the production of a polypeptide consisting of SEQ ID NOS: 1, 2, 3, or 4, comprises culturing a host cell of the invention under conditions promoting expression, and recovering the polypeptide from the host cell or the culture medium. The host cell can be a bacterial cell, parasite cell, or eukaryotic cell.
[0028] A method of the invention for detecting a racemase encoded by a nucleotide sequence containing a subsequence encoding a peptide selected from SEQ ID. NO: 1, 2, 3, or 4, comprises: [0029] (a) contacting the nucleotide sequence with a primer or a probe, which hybridizes with the nucleic acid molecule of the invention; [0030] (b) amplifying the nucleotide sequence using the primer or the probe; and [0031] (c) detecting a hybridized complex formed between the primer or probe and the nucleotide sequence.
[0032] This invention provides a method of detecting a racemase encoded by a nucleotide sequence containing a subsequence encoding a peptide selected from SEQ ID NO: 1, 2, 3, or 4. The method comprises: [0033] (a) contacting the racemase with antibodies of the invention; and [0034] (b) detecting the resulting immunocomplex.
[0035] A kit for detecting a racemase encoded by a nucleotide sequence containing a subsequence encoding a peptide selected from SEQ ID NO: 1, 2, 3, or 4, comprises: [0036] (a) a polynucleotide probe or primer, which hybridizes with the polynucleotide of the invention; and [0037] (b) reagents to perform a nucleic acid hybridization reaction.
[0038] This invention also provides a kit for detecting a racemase encoded by a nucleotide sequence containing a subsequence encoding a peptide selected from SEQ ID NO: 1, 2, 3, or 4. The kit comprises: [0039] (a) purified antibodies of the invention; [0040] (b) standard reagents in a purified form; and [0041] (c) detection reagents.
[0042] An in vitro method of screening for an active molecule capable of inhibiting a racemase encoded by a nucleotide sequence containing a subsequence encoding a peptide selected from SEQ ID NO: 1, 2, 3, or 4, comprises: [0043] (a) contacting the active molecule with the racemase; [0044] (b) testing the capacity of the active molecule, at various concentrations, to inhibit the activity of the racemase; and [0045] (c) choosing the active molecule that provides an inhibitory effect of at least 80% on the activity of any proline racemase.
[0046] In a preferred embodiment of the invention the racemase is a proline racemase.
[0047] An immunizing composition of the invention contains at least a purified polypeptide of the invention, capable of inducing an immune response in vivo, and a pharmaceutical carrier.
BRIEF DESCRIPTION OF THE DRAWINGS
[0048] This invention will be understood with reference to the drawings in which:
[0049] FIG. 1: Comparative analysis of sequences of T. cruzi TcPRACA and TcPRACB proline racemase isoforms. A. Alignment of TcPRACA (Tc-A) and TcPRACB (Tc-B) nucleotide sequences: non coding sequences are shown in italics; trans-splicing signals are underlined and putative spliced leader acceptor sites are double-underlined; the region encoding the computer-predicted signal peptide is indicated by double-headed arrow; initiation of translation for TcPRACA and TcPRACB are shown by single-headed arrows; nucleotides shaded in light and dark grey, represent respectively silent mutations or point mutations; box, proline racemase active site; UUA triplets are underlined in bold and precede polyadenylation sites that are double-underlined. B. Schematic representation of amino acid sequence alignments of T. cruzi TcPRACA (Tc-A), TcPRACB (Tc-B) proline racemases. The common scale is in amino acid residue positions along the linear alignment and represent the initiation codons for TcPRACA and TcPRACB proteins, respectively; ∇ represents an alternative TcPRACA putative initiation codon; Amino acid differences are indicated above and below the vertical lines and their positions in the sequence are shown in parenthesis. SP: signal peptide; the N-terminal domain of TcPRACA extends from positions 1 to 69; SPCGT: conserved active sites of TcPRACA and TcPRACB proline racemases; N-terminus and C-terminus are indicated for both proteins. C. Hydrophobicity profile of TcPRACA: dotted line depicts the cleavage site as predicted by Von Heijne's method (aa 31-32). D. Ethidium bromide-stained gel of chromosomal bands of T. cruzi CL Brener clone after separation by PFGE (lane 1) and Southern blot hybridization with TcPRAC probe (lane 2). The sizes (Mb) of chromosomal bands are indicated, as well as the region chromosome numbers in roman numerals.
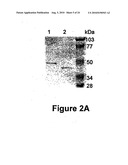
[0050] FIG. 2: Biochemical characterization of T. cruzi proline racemase isoforms and substrate specificities. A. SDS-PAGE analysis of purified rTcPRACA (lane 1) and rTcPRACB (lane 2) recombinant proteins. A 8% polyacrylamide gel was stained with Coomassie blue. Right margin, molecular weights. B. Percent of racemization of L-proline, D-proline, L-hydroxy (OH) proline and D-hydroxy (OH) proline substrates by rTcPRACB (open bar) as compared to rTcPRACA (closed bar). Racemase activity was determined with 0.25 μM of each isoform of proline racemase and 40 mM substrate in sodium acetate buffer pH 6.0. C. Percent of racemization as a function of pH: Racemase assays were performed in buffer containing 0.2 M Tris-HCl (squares), sodium acetate (triangles) and sodium phosphate (circles), 40 mM L-proline and 0.25 μM of purified rTcPRACA (closed symbols) and rTcPRACB (open symbols). After 30 min at 37° C., the reaction was stopped by heat inactivation and freezing. D. 39 kDa intracellular isoform was isolated from soluble (Ese) extracts of non-infective epimastigote forms of the parasite. Western-blots of serial dilutions of the soluble suspension was compared to known amounts of rTcPRACB protein and used for protein quantitation using Quantity One® software. Racemase assays were performed in sodium acetate buffer pH6, using 40 mM L-proline and the equivalent depicted amounts of 39 kDa (ng) contained in Ese extract.
[0051] FIG. 3: Kinetic parameters of L-proline racemization catalyzed by rTcPRACA and rTcPRACB proline racemase isoforms. The progress of racemization reaction was monitored polarimetrically, as previously described (13). A. The determination of the linear part of the curve was performed at 37° C. in medium containing 0.2 M sodium acetate, pH 6.0; 0.25 μM purified enzyme and 40 mM L-proline. rTcPRACA reactions are represented by black squares and rTcPRACB reactions by white squares. B. Initial rate of racemase activity was assayed at 37° C. in medium containing 0.2 M sodium acetate, pH 6.0, 0.125 μM of rTcPRACA (solid squares) or rTcPRACB (open squares) purified enzymes and different concentrations of L-proline. Lineweaver-Burk double reciprocal plots were used to determine values for KM and Vmax where 1N is plotted in function of 1/[S] and the slope of the curve represents KM/Vmax. Values obtained were confirmed by using the Kaleidagraph® program and Michaelis-Menten equation. The values are representative of six experiments with different enzyme preparations. C. Double reciprocal plot kinetics of 0.125 μM rTcPRACA proline racemase isoform in the presence (open) squares or absence (solid) squares of 6.7 μM PAC competitive inhibitor in function of L-proline concentration. For comparison: KM reported for the proline racemase of C. sticklandii was 2.3 mM; kinetic assays using the native protein obtained from a soluble epimastigote fraction revealed a KM of 10.7 mM and a Ki of 1.15 μM.
[0052] FIG. 4: Size exclusion chromatography of rTcPRACA protein using a Superdex 75 column. Fractions were eluted by HPLC at pH 6.0, B2 and B4 peaks correspond to rTcPRACA dimer and monomer species respectively. B1 and B5 eluted fractions were reloaded into the column (bold, see inserts) using the same conditions and compared to previous elution profile (not bold).
[0053] FIG. 5: Site-directed mutagenesis of TcPRACA proline racemase. Schematic representation of the active site mutagenesis of proline racemase of TcPRACA gene.
[0054] FIG. 6: Sequence alignments of proteins (Clustal X) obtained by screening SWISS-PROT and TrEMBL databases using motifs I, II and III. Amino acids involved in MI, MII and MIII are shaded in dark grey and light grey figures the 13-14 unspecific amino acids involved in M II. --SWISS-PROT accession numbers of the sequences are in Table IV.
[0055] FIG. 7: Cladogram of protein sequences obtained by T-coffee alignment radial tree. See Table IV for SWISS-PROT protein accession numbers.
[0056] FIG. 8 shows the percent of racemisation inhibition of different L-proline concentrations (ranging from 10-40 mM) using the D-AAO (D-MO/L-) microtest as compared to conventional detection using a polarimeter (Pol/L-).
[0057] FIG. 9 shows the comparison of D-AAO/HRP reaction using D-Proline alone or an equimolar mixture of D- and L-Proline as standard.
[0058] FIG. 10 shows optical density at 490 nm as a function of D-proline concentration under the following conditions.
[0059] FIG. 11 is a Graph obtained with the serial dilutions of D-proline, as positive reaction control Obs: OD of wells (-) average of OD obtained from blank wells.
[0060] FIG. 12 shows the loss of the enzymatic activity of proline racemase after mutagenesis of the residue Cys160 or the residue Cys330.
DETAILED DESCRIPTION OF THE INVENTION
[0061] Proline racemase catalyses the interconversion of L- and D-proline enantiomers and has to date been described in only two species. Originally found in the bacterium Clostridium sticklandii, it contains cysteine residues in the active site and does not require co-factors or other known coenzymes. The first eukaryotic amino acid (proline) racemase, after isolation and cloning of a gene from the pathogenic human parasite Trypanosoma cruzi, has been described. While this enzyme is intracellularly located in replicative non-infective forms of T. cruzi, membrane-bound and secreted forms of the enzyme are present upon differentiation of the parasite into non-dividing infective forms. The secreted isoform of proline racemase is a potent host B-cell mitogen supporting parasite evasion of specific immune responses.
[0062] Primarily it was essential to elucidate whether TcPRACB gene could encode a functional proline racemase. To answer this question, TcPRACA and TcPRACB paralogue genes were expressed in Escherichia coli and detailed studies were performed on biochemical and enzymatic characteristics of the recombinant proteins. This invention demonstrates that TcPRACB indeed encodes a functional proline racemase that exhibits slightly different kinetic parameters and biochemical characteristics when compared to TcPRACA enzyme. Enzymatic activities of the respective recombinant proteins showed that the 39 kDa intracellular isoform of proline racemase produced by TcPRACB construct is more stable and has higher rate of D/L-proline interconversion than the 45 kDa isoform produced by TcPRACA. Additionally, the dissociation constant of the enzyme-inhibitor complex (Ki) obtained with pyrrole-2-carboxylic acid, the specific inhibitor of proline racemases, is lower for the recombinant TcPRACB enzyme.
[0063] Moreover, this invention demonstrates that Cys330 and Cys160 are key amino acids of the proline racemase active site since the activity of the enzyme is totally abolished by site-direct mutagenesis of these residues. Also, multiple alignment of proline racemase amino acid sequences allowed the definition of protein signatures that can be used to identify putative proline racemases in other microorganisms. The significance of the presence of proline racemase in eukaryotes, particularly in T. cruzi, is discussed, as well as the consequences of this enzymatic activity in the biology and infectivity of the parasite.
[0064] This invention provides amino acid motifs, which are useful as signatures for proline racemaces. These amino acid motifs are as follows: TABLE-US-00001 MOTIF I [SEQ ID NO: 1] [IVL][GD]XHXXG[ENM]XX[RD]X[VI]XG MOTIF II [SEQ ID NO: 2] [NSM][VA][EP][AS][FY]X(13, 14)[GK]X[IVL]XXD[IV] [AS][YWF]GGX[FWY] MOTIF III [SEQ ID NO: 3] DRSPXGXGXXAXXA MOTIF III* [SEQ ID NO:4 ] DRSPCGXGXXAXXA
where X is an amino acid in each of these sequences.
[0065] This invention also provides polynucleotides encoding amino acid motifs, which are also referred to herein as the "polynucleotides of the invention" and the "polypeptides of the invention."
[0066] Databases were screened using these polynucleotide or polypeptide sequences of TcPRACA. Motifs I to III were searched. M I corresponds to [IVL][GD]XHXXG[ENM]XX[RD ]X[VI]XXG, M II to of [NSM][VA] [EP][AS][FY]X(13, 14)[GK]X[IVL]XXD[IV][AS][YWF] GGX[FWY] M III to DRSPXGXGXXAXXA and M III* to DRSPCGXGXX AXXA Sequences presented in the annex, where the conserved regions of 2 Cysteine residues of the active site are squared, are presented in Table V in bold with corresponding Accession numbers. The two cysteine residues are Cys330 and its homologue Cys160, where residue Cys160 mutation by a serine by site directed mutagenesis also induces a drastic loss of the enzymatic activity as for residue Cys330.
[0067] Proline racemase, an enzyme previously only described in protobacterium Clostridium sticklandii (11), was shown to be encoded also by the eukaryote Trypanosoma cruzi, a highly pathogenic protozoan parasite (13). The Trypanosoma cruzi proline racemase (TcPRAC), formerly called TcPA45, is an efficient mitogen for host B cells and is secreted by the metacyclic forms of the parasite upon infection, contributing to its immune-evasion and persistence through non-specific polyclonal lymphocyte activation (13). Previous results suggested that TcPRAC is encoded by two paralogous genes per haploid genome. Protein localization studies have also indicated that T. cruzi can differentially express intracellular and secreted versions of TcPRAC during cell cycle and differentiation, as the protein is found in the cytoplasm of non-infective replicative (epimastigote) forms of the parasite, and bound to the membrane or secreted in the infective, non-replicative (metacyclic trypomastigote) parasites (13).
[0068] This invention characterizes the two TcPRAC paralogues and demonstrates that both TcPRACA and TcPRACB give rise to functional isoforms of co-factor independent proline racemases, which display different biochemical properties that may well have important implications in the efficiency of the respective enzymatic activities. As suggested before by biochemical and theoretical studies for the bacterial proline racemase (11, 17, 18), TcPRAC activities rely on two monomeric enzyme subunits that perform interconversion of L- and/or D-proline enantiomers by a two base mechanism reaction in which the enzyme removes an α-hydrogen from the substrate and donates a proton to the opposite side of the α-carbon. It has been predicted that each subunit of the homodimer furnishes one of the sulphydryl groups (18).
[0069] The present invention demonstrates that TcPRAC enzymatic activities are bona fide dependent on the Cys330 residue of the active site, as site-specific 330Cys>Ser mutation totally abrogates L- and D-proline racemization, in agreement with a previous demonstration that TcPRAC enzymatic activity is abolished through alkylation with iodoacetate or iodoacetamide (13), similarly to the Clostridium proline racemase, where carboxymethylation was shown to occur specifically with the two cysteines of the reactive site leading to enzyme inactivation (12). The present invention demonstrates also that the residue Cys160 is also a critical residue of the active site and that TcPRAC possesses two active sites in its homodimer. These observations make it possible to search for inhibitors by means of assays based on the native and mutated sequences.
[0070] While gene sequence analysis predicted that, by a mechanism of alternative splicing, TcPRACA could generate both intracellular and secreted versions of parasite proline racemase, the present invention demonstrates that TcPRACB gene sequence per se codes for a protein lacking the amino acids involved in peptide signal formation and an extra N-terminal domain present in TcPRACA protein, resembling more closely the CsPR. Thus, TcPRACB can only generate an intracellular version of TcPRAC proline racemase. This discovery makes it possible to carry out a search of one putative inhibitor of an intracellular enzyme should penetrate the cell.
[0071] Interestingly, the presence of two homologous copies of TcPRAC genes in the T. cruzi genome, coding for two similar polypeptides but with distinct specific biochemical properties, could reflect an evolutionary mechanism of gene duplication and a parasite strategy to ensure a better environmental flexibility. This assumption is comforted by the potential of TcPRACA gene to generate two related protein isoforms by alternative splicing, a mechanism that is particularly adept for cells that must respond rapidly to environmental stimuli. Primarily, trans-splicing appears indeed to be an ancient process that may constitute a selective advantage for split genes in higher organisms (19) and alternative trans-splicing was only recently proven to occur in T. cruzi (20). As an alternative for promoter selection, the regulated production of intracellular and/or secreted isoforms of proline racemase in T. cruzi by alternative trans-splicing of TcPRACA gene would allow the stringent conservation of a constant protein domain and/or the possibility of acquisition of an additional secretory region domain. As a matter of fact, recent investigations using RT-PCR based strategy and a common 3' probe to TcPRACA and TcPRACB sequences combined to a 5' spliced leader oligonucleotide followed by cloning and sequencing of the resulting fragments have indeed proved that an intracellular version of TcPRAC may also originate from the TcPRACA gene, corroborating this hypothesis.
[0072] Gene duplication is a relatively common event in T. cruzi that adds complexity to parasite genomic studies. Moreover, TcPRAC chromosomal mapping revealed two chromosomal bands that possess more than 3 chromosomes each and that may indicate that proline racemase genes are mapped in size-polymorphic homologous chromosomes, an important finding for proline racemase gene family characterization. Preliminary results have, for instance, revealed that T. cruzi DM28c type I strain maps proline racemase genes to the same chromoblot regions identified with T. cruzi CL type II strain used in the present invention.
[0073] It is well known that proline constitutes an important source of energy for several organisms, such as several hemoflagellates (21), (22), (23), and for flight muscles in insects (24). Furthermore, a proline oxidase system was suggested in trypanosomes (25) and the studies reporting the abundance of proline in triatominae guts (26) have implicated proline in metabolic pathways of Trypanosoma cruzi parasites as well as in its differentiation in the digestive tract of the insect vector (27). Thus, it is well accepted that T. cruzi can use L-proline as a principal source of carbon (25).
[0074] Moreover, preliminary results using parasites cultured in defined media indicate that both epimastigotes, found in the vector, and infective metacyclic trypomastigote forms can efficiently metabolize L- or D-proline as the sole source of carbon. While certain reports indicate that biosynthesis of proline occurs in trypanosomes, i.e. via reduction of glutamate carbon chains or transamination reactions, an additional and direct physiological regulation of proline might exist in the parasite to control amino acid oxidation and its subsequent degradation or yet to allow proline utilization. In fact, a recent report showed two active proline transporter systems in T. cruzi (28). T. cruzi proline racemase may possibly play a consequential role in the regulation of intracellular proline metabolic pathways, or else, it could participate in mechanisms of post-translational addition of D-amino acid to polypeptide chains.
[0075] On one hand, these hypotheses would allow for an energy gain and, on the other hand, would permit the parasite to evade host responses. In this respect, it was reported that a single D-amino acid addition in the N-terminus of a protein is sufficient to confer general resistance to lytic reactions involving host proteolytic enzymes (29). The expression of proteins containing D-amino acids in the parasite membrane would benefit the parasite inside host cell lysosomes, in addition to the contribution to the initiation of polyclonal activation, as already described for polymers composed of D-enantiomers (30), (31). Although D-amino acid inclusion in T. cruzi proteins would benefit the parasite, this hypothesis remains to be proven and direct evidence is technically difficult to obtain.
[0076] It is worth noting that metacyclogenesis of epimastigotes into infective metacyclic forms involves parasite morphologic changes that include the migration of the kinetoplast, a structure that is physically linked to the parasite flagellum, and many other significant metabolic alterations that combine to confer infectivity/virulence to the parasite (13, 32). Proline racemase was shown to be preferentially localized in the flagellar pocket of infective parasite forms after metacyclogenesis (13), as are many other known proteins secreted and involved in early infection (33).
[0077] It is also conceivable that parasite proline racemase may function as an early mediator for T. cruzi differentiation through intracellular modification of internalized environmental free proline, as suggested above and already observed in some bacterial systems. As an illustration, exogenous alanine has been described as playing an important role in bacterial transcriptional regulation by controlling an operon formed by genes coding for alanine racemase and a smaller subunit of bacterial dehydrogenase (34).
[0078] In bacteria, membrane alanine receptors are responsible for alanine and proline entry into the bacterial cell (35). It can then be hypothesized that the availability of proline in the insect gut milieu associated to a mechanism of environmental sensing by specific receptors in the parasite membrane would stand for parasite proline uptake and its further intracellular racemization. Proline racemase would then play a fundamental role in the regulation of parasite growth and differentiation by its participation in both metabolic energetic pathways and the expression of proteins containing D-proline, as described above, consequently conferring parasite infectivity and its ability to escape host specific responses.
[0079] Thus far, and contrasting to the intracellular isoform of TcPRAC found in epimastigote forms of T. cruzi, the ability of metacyclic and bloodstream forms of the parasite to express and secrete proline racemase may have further implications in host/parasite interaction. In fact, the parasite-secreted isoform of proline racemase participates actively in the induction of non-specific polyclonal B-cell responses upon host infection (13) and favors parasite evasion, thus ensuring its persistence in the host.
[0080] As described for other mitogens and parasite antigens (36), (37), (38), and in addition to its mitogenic property, TcPRAC could also be involved in modifications of host cell targets enabling better parasite attachment to host cell membranes in turn assuring improved infectivity. Since several reports associate accumulation of L-proline with muscular dysfunction (39) and inhibition of muscle contraction (40), the release of proline racemase by intracellular parasites could alternatively contribute to the maintenance of infection through regulation of L-proline concentration inside host cells, as proline was described as essential for the integrity of muscular cell targets. Therefore, it has recently been demonstrated that transgenic parasites hyperexpressing TcPRACA or TcPRACB genes, but not functional knock outs, are 5-10 times more infective to host target cells pointing to a critical role of proline racemases in the ongoing of the infectious process. Likewise, previous reports demonstrated that genetic inactivation of Lysteria monocytogenes alanine racemase and D-amino acid oxidase genes abolishes bacterial pathogenicity, since the presence of D-alanine is required for the synthesis of the mucopeptide component of the cell wall that protects virtually all bacteria from the external milieu (41).
[0081] Present analysis using identified critical conserved residues in TcPRAC and C. sticklandii proline racemase genes and the screening of SWISS-PROT and TrEMBL databases led to the discovery of a minimal signature for proline racemases, DRSPXGX[GA]XXAXXA, and to confirm the presence of putative proteins in at least 10 distinct organisms. Screening of unfinished genome sequences showed highly homologous proline racemase candidate genes in an additional 8 organisms, amongst which are the fungus Aspergillus fumigatus and the bacteria Bacillus anthracis and Clostridium botulinum. This is of particular interest, since racemases, but not proline racemases, are widespread in bacteria and only recently described in more complex organisms such as T. cruzi, 42,43). These findings may possibly reflect cell adaptative responses to extracellular stimuli and uncover more general mechanisms for the regulation of gene expression by D-amino acids in eukaryotes. The finding of similar genes in human and mouse genome databases using less stringent signatures for proline racemase is striking. However, the absence of the crucial amino acid cysteine in the putative active site of those predicted proteins suggests a different functionality than that of a proline racemase.
[0082] This invention shows that TcPRAC isoforms are highly stable and have the capacity to perform their activities across a large spectrum of pH. In addition, the affinity of pyrrol-carboxylic acid, a specific inhibitor of proline racemase, is higher for TcPRAC enzymes than for CsPR.
[0083] The invention also provides amino acid or nucleic acid sequences substantially similar to specific sequences disclosed herein.
[0084] The term "substantially similar" when used to define either amino acid or nucleic acid sequences means that a particular subject sequence, for example, a mutant sequence, varies from a reference sequence by one or more substitutions, deletions, or additions, the net effect of which is to retain activity. Alternatively, nucleic acid subunits and analogs are "substantially similar" to the specific DNA sequences disclosed herein if: (a) the DNA sequence is derived from a region of the invention; (b) the DNA sequence is capable of hybridization to DNA sequences of (a) and/or which encodes active molecules; or DNA sequences that are degenerate as a result of the genetic code to the DNA sequences defined in (a) or (b) and/or which encode active molecules.
[0085] In order to preserve the activity, deletions and substitutions will preferably result in homologously or conservatively substituted sequences, meaning that a given residue is replaced by a biologically similar residue. Examples of conservative substitutions include substitution of one aliphatic residue for another, such as IIe, Val, Leu, or Ala for one another, or substitution of one polar residue for another, such as between Lys and Arg; Glu and Asp; or Gln and Asn. Other such conservative substitutions, for example, substitutions of entire regions having similar hydrophobicity characteristics, are well known. When said activity is proline racemase activity, Cys330 and Cys160 must be present.
[0086] The polynucleotides of the invention can be used as probes or to select nucleotide primers notably for an amplification reaction. PCR is described in the U.S. Pat. No. 4,683,202 granted to Cetus Corp. The amplified fragments can be identified by agarose or polyacrylamide gel electrophoresis, or by a capillary electrophoresis, or alternatively by a chromatography technique (gel filtration, hydrophobic chromatography, or ion exchange chromatography). The specificity of the amplification can be ensured by a molecular hybridization using as nucleic acid probes the polynucleotides of the invention, oligonucleotides that are complementary to these polynucleotides, or their amplification products themselves.
[0087] Amplified nucleotide fragments are useful as probes in hybridization reactions in order to detect the presence of one polynucleotide according to the present invention or in order to detect the presence of a gene encoding racemase activity, such as in a biological sample. This invention also provides the amplified nucleic acid fragments ("amplicons") defined herein above. These probes and amplicons can be radioactively or non-radioactively labeled using, for example, enzymes or fluorescent compounds.
[0088] Other techniques related to nucleic acid amplification can also be used alternatively to the PCR technique. The Strand Displacement Amplification (SDA) technique (Walker et al., 1992) is an isothermal amplification technique based on the ability of a restriction enzyme to cleave one of the strands at a recognition site (which is under a hemiphosphorothioate form), and on the property of a DNA polymerase to initiate the synthesis of a new strand from the 3' OH end generated by the restriction enzyme, and on the property of this DNA polymerase to displace the previously synthesized strand being localized downstream.
[0089] The SDA amplification technique is more easily performed than PCR (a single thermostated water bath device is necessary), and is faster than the other amplification methods. Thus, the present invention also comprises using the nucleic acid fragments according to the invention (primers) in a method of DNA or RNA amplification, such as the SDA technique.
[0090] The polynucleotides of the invention, especially the primers according to the invention, are useful as technical means for performing different target nucleic acid amplification methods, such as:
[0091] TAS (Transcription-based Amplification System), described by Kwoh et al. in 1989;
[0092] SR (Self-Sustained Sequence Replication), described by Guatelli et al. in 1990;
[0093] NASBA (Nucleic acid Sequence Based Amplification), described by Kievitis et al. in 1991; and
[0094] TMA (Transcription Mediated Amplification).
[0095] The polynucleotides of the invention, especially the primers according to the invention, are also useful as technical means for performing methods for amplification or modification of a nucleic acid used as a probe, such as:
[0096] LCR (Ligase Chain Reaction), described by Landegren et al. in 1988 and improved by Barany et al. in 1991, who employ a thermostable ligase;
[0097] RCR (Repair Chain Reaction), described by Segev et al. in 1992;
[0098] CPR (Cycling Probe Reaction), described by Duck et al. in 1990; and
[0099] Q-beta replicase reaction, described by Miele et al. in 1983 and improved by Chu et al. in 1986, Lizardi et al. in 1988, and by Burg et al. and Stone et al. in 1996.
[0100] When the target polynucleotide to be detected is RNA, for example mRNA, a reverse transcriptase enzyme can be used before the amplification reaction in order to obtain a cDNA from the RNA contained in the biological sample. The generated cDNA can be subsequently used as the nucleic acid target for the primers or the probes used in an amplification process or a detection process according to the present invention.
[0101] The oligonucleotide probes according to the present invention hybridize specifically with a DNA or RNA molecule comprising all or part of the polynucleotide of the invention under stringent conditions. As an illustrative embodiment, the stringent hybridization conditions used in order to specifically detect a polynucleotide according to the present invention are advantageously the following:
[0102] Prehybridization and hybridization are performed as follows in order to increase the probability for heterologous hybridization: [0103] The prehybridization and hybridization are done at 50° C. in a solution containing 5×SSC and 1×Denhardt's solution.
[0104] The washings are performed as follows: [0105] 2×SSC at 60° C. 3 times during 20 minutes each.
[0106] The non-labeled polynucleotides of the invention can be directly used as probes. Nevertheless, the polynucleotides can generally be labeled with a radioactive element (32P, 35S, 3H, 125I) or by a non-isotopic molecule (for example, biotin, acetylaminofluorene, digoxigenin, 5-bromodesoxyuridin, fluorescein) in order to generate probes that are useful for numerous applications. Examples of non-radioactive labeling of nucleic acid fragments are described in the French Patent No. FR 78 10975 or by Urdea et al. or Sanchez-Pescador et al. 1988.
[0107] Other labeling techniques can also be used, such as those described in the French patents 2 422 956 and 2 518 755. The hybridization step can be performed in different ways. A general method comprises immobilizing the nucleic acid that has been extracted from the biological sample on a substrate (nitrocellulose, nylon, polystyrene) and then incubating, in defined conditions, the target nucleic acid with the probe. Subsequent to the hybridization step, the excess amount of the specific probe is discarded, and the hybrid molecules formed are detected by an appropriate method (radioactivity, fluorescence, or enzyme activity measurement).
[0108] Advantageously, the probes according to the present invention can have structural characteristics such that they allow signal amplification, such structural characteristics being, for example, branched DNA probes as those described by Urdea et al. in 1991 or in the European Patent No. 0 225 807 (Chiron).
[0109] In another advantageous embodiment of the present invention, the probes described herein can be used as "capture probes", and are for this purpose immobilized on a substrate in order to capture the target nucleic acid contained in a biological sample. The captured target nucleic acid is subsequently detected with a second probe, which recognizes a sequence of the target nucleic acid that is different from the sequence recognized by the capture probe.
[0110] The oligonucleotide probes according to the present invention can also be used in a detection device comprising a matrix library of probes immobilized on a substrate, the sequence of each probe of a given length being localized in a shift of one or several bases, one from the other, each probe of the matrix library thus being complementary to a distinct sequence of the target nucleic acid. Optionally, the substrate of the matrix can be a material able to act as an electron donor, the detection of the matrix positions in which hybridization has occurred being subsequently determined by an electronic device. Such matrix libraries of probes and methods of specific detection of a target nucleic acid are described in European patent application No. 0 713 016, or PCT Application No. WO 95 33846, or also PCT Application No. WO 95 11995 (Affymax Technologies), PCT Application No. WO 97 02357 (Affymetrix Inc.), and also in U.S. Pat. No. 5,202,231 (Drmanac), said patents and patent applications being herein incorporated by reference.
[0111] The present invention also pertains to recombinant plasmids containing at least a nucleic acid according to the invention. A suitable vector for the expression in bacteria, and in particular in E. coli, is pET-28 (Novagen), which allows the production of a recombinant protein containing a 6×His affinity tag. The 6×His tag is placed at the C-terminus or N-terminus of the recombinant polypeptide.
[0112] The polypeptides according to the invention can also be prepared by conventional methods of chemical synthesis, either in a homogenous solution or in solid phase. As an illustrative embodiment of such chemical polypeptide synthesis techniques, the homogenous solution technique described by Houbenweyl in 1974 may be cited.
[0113] The polypeptides of the invention are useful for the preparation of polyclonal or monoclonal antibodies that recognize the polypeptides (SEQ ID NOS: 1, 2, 3, and 4) or fragments thereof. The monoclonal antibodies can be prepared from hybridomas according to the technique described by Kohler and Milstein in 1975. The polyclonal antibodies can be prepared by immunization of a mammal, especially a mouse or a rabbit, with a polypeptide according to the invention, which is combined with an adjuvant, and then by purifying specific antibodies contained in the serum of the immunized animal on a affinity chromatography column on which has previously been immobilized the polypeptide that has been used as the antigen.
[0114] A method of detecting a racemase encoded by a nucleotide sequence containing a subsequence encoding a peptide selected from SEQ ID NOS: 1, 2, 3, or 4.
[0115] Consequently, the invention is also directed to a method for detecting specifically the presence of a polypeptide according to the invention in a biological sample. The method comprises: [0116] a) bringing into contact the biological sample with an antibody according to the invention; and [0117] b) detecting antigen-antibody complex formed.
[0118] Also part of the invention is a diagnostic kit for in vitro detecting the presence of a polypeptide according to the present invention in a biological sample. The kit comprises: [0119] a polyclonal or monoclonal antibody as described above, optionally labeled; and [0120] a reagent allowing the detection of the antigen-antibody complexes formed, wherein the reagent carries optionally a label, or being able to be recognized itself by a labeled reagent, more particularly in the case when the above-mentioned monoclonal or polyclonal antibody is not labeled by itself.
[0121] The present invention is also directed to bioinformatic searches in data banks using the whole sequences of the polypeptides using the whole sequences of the polypeptides (SEQ ID NOS: 1, 2, 3, or 4). In this case the method detects the presence of at least a subsequence encoding a peptide selected from SEQ ID NOS: 1, 2, 3, or 4 wherein the said at least subsequence is indicative of a racemase.
[0122] The invention also pertains to:
[0123] A purified polypeptide or a peptide fragment having at least 10 amino acids, which is recognized by antibodies directed against a polynucleotide or peptide sequence according to the invention.
[0124] A monoclonal or polyclonal antibody directed against a polypeptide or a peptide fragment encoded by the polynucleotide sequences according to the invention.
[0125] A method of detecting a racemase in a biological sample comprising: [0126] a) contacting DNA or RNA of the biological sample with a primer or a probe from a polynucleotide according to the invention, which hybridizes with a nucleotide sequence; [0127] b) amplifying the nucleotide sequence using the primer or said probe; and [0128] c) detecting the hybridized complex formed between the primer or probe with the DNA or RNA.
[0129] A kit for detecting the presence of a racemase in a biological sample, comprises: [0130] a) a polynucleotide primer or probe according to the invention; and [0131] b) reagents necessary to perform a nucleic acid hybridization reaction.
[0132] An in vitro method of screening for an active molecule capable of inhibiting a racemase encoded by a nucleic acid containing a polynucleotide according to the invention, wherein the inhibiting activity of the molecule is tested on at least said racemase, comprises: [0133] a) providing racemase containing a polypeptide according to the invention; [0134] b) contacting the active molecule with said racemase; [0135] c) testing the capacity of the active molecule, at various concentrations, to inhibit the activity of the racemase; and [0136] d) choosing the active molecule that provides an inhibitory effect of at least 80% on the activity of the racemase.
[0137] The term "recombinant" as used herein means that a protein or polypeptide employed in the invention is derived from recombinant (e.g., microbial or mammalian) expression systems. "Microbial" refers to recombinant proteins or polypeptides made in bacterial or fungal (e.g., yeast) expression systems. As a product, "recombinant microbial" defines a protein or polypeptide produced in a microbial expression system, which is essentially free of native endogenous substances. Proteins or polypeptides expressed in most bacterial cultures, e.g. E. coli, will be free of glycan. Proteins or polypeptides expressed in yeast may have a glycosylation pattern different from that expressed in mammalian cells.
[0138] The polypeptide or polynucleotide of this invention can be in isolated or purified form. The terms "isolated" or "purified", as used in the context of this specification to define the purity of protein or polypeptide compositions, means that the protein or polypeptide composition is substantially free of other proteins of natural or endogenous origin and contains less than about 1% by mass of protein contaminants residual of production processes. Such compositions, however, can contain other proteins added as stabilizers, excipients, or co-therapeutics. These properties similarly apply to polynucleotides of the invention.
[0139] The platform of the invention relates to reagents, systems and devices for performing the process of screening of D-amino acid tests.
[0140] Appropriate carriers, diluents, and adjuvants can be combined with the polypeptides and polynucleotides described herein in order to prepare the compositions of the invention. The compositions of this invention contain the polypeptides or polynucleotides together with a solid or liquid acceptable nontoxic carrier. Such carriers can be sterile liquids, such as water an oils, including those of petroleum, animal, vegetable, or synthetic origin. Examples of suitable liquids are peanut oil, soybean oil, mineral oil, sesame oil and the like. Water is a preferred carrier. Physiological solutions can also be employed as liquid carriers.
[0141] This invention will now be described with reference to the following Examples.
Example 1
Cloning and Automated Sequencing
[0142] Lambda phage and plasmid. DNA were prepared using standard techniques and direct sequencing was accomplished with the Big dye Terminator Kit (Perkin Elmer, Montigny-le Bretonneux, France) according to the manufacturer's instructions. Extension products were run for 7 h in an ABI 377 automated sequencer. Briefly, to obtain the full length of the TcPRAC gene, 32P-labeled 239 bp PCR product was used as a probe to screen a T. cruzi clone CL-Brener lamba Fix II genomic library (see details in (13)). There were isolated 4 independent positive phages. Restriction analysis and Southern blot hybridization showed two types of genomic fragments, each represented by 2 phages. Complete sequence and flanking regions of representative phages for each pattern was done. Complete characterization of TcPRACA gene, representing the first phage type, was previously described in (13). Full sequence of the putative TcPRACB gene, representing the second phage type was then performed and primers internal to the sequence were used for sequencing, as described before (13).
Example 2
Chromoblots
[0143] Epimastigote forms T. cruzi (clone CL Brener) are maintained by weekly passage in LIT medium. Agarose (0.7%) blocks containing 1×107 cultured parasites were lysed with 0.5 M EDTA/10 mM Tris/1% sarcosyl pH 8.0, digested by proteinase K and washed in 10 mM Tris/1 mM EDTA, pH 8.0. Pulsed field gel electrophoresis (PFGE) was carried out at 18° C. using the Gene Navigator apparatus (Pharmacia, Upsala, Sweden) in 0.5×TBE. Electrophoresis were performed, as described in (14). Gels were then stained with ethidium bromide, photographed, exposed to UV light (265 nm) for 5 min and further blotted under alkaline conditions to a nylon filter (HybondN+, Amersham Life Science Inc., Cleveland, USA). DNA probe, obtained by PCR amplification of TcPRACA gene with Hi-45 (5' CTC TCC CAT GGG GCA GGA AAA GCT TCT G 3') [SEQ ID NO:5] and Bg-45 (5'CTG AGC TCG ACC AGA T(CA)T ACT GC 3') [SEQ ID NO:6] oligonucleotides (as described in (13)) was labelled with αdATP32 using Megaprime DNA labelling system (Amersham). The chromoblot was hybridized overnight in 2×Denhart's/5×SSPE/1.5% SDS at 55° C. and washed in 2×SSPE/0.1% SDS followed by 1×SSPE at 60° C. Autoradiography was obtained by overnight exposure of the chromoblot using a Phosphorimager cassette (Molecular Dynamics, UK).
Example 3
Plasmid Construction and Protein Purification
[0144] TcPRACA gene fragment starting at codon 30 was obtained by PCR, using Hi- and Bg-45 primers, and cloned in frame with a C-terminal six-histidine tag into the pET28b(+) expression vector (Novagen-Tebu, Le Perray en Yvelines, France). The fragment encoding for the TcPRACB consisted of a HindIII digestion of TcPRACB gene fragment obtained by similar PCR and cloned in frame with a C-terminal six-histidine tag into the pET28b(+) expression vector. Respective recombinant proteins TcPRACA and TcPRACB were produced in E. coli BL21 (DE3) (Invitrogen, Cergy Pontoise, France) and purified using Immobilized Metal Affinity Chromatography on nickel columns (Novagen-Tebu, Le Parrayen Yvelines, France) following the manufacturer's instructions.
Example 4
Size Exclusion Chromatography
[0145] rTcPRACA and rTcPRACB proteins were purified as described here above and dialysed against PBS pH 7.4 or 0.2 M NaOAc pH 6.0 elution buffers in dialysis cassettes (Slide-A-lyzer 7K Pierce), overnight at 4° C. The final protein concentration was adjusted to 2 mg/ml and 0.5 ml of the solution were loaded onto Pharmacia Superdex 75 column (HR10×30), previously calibrated with a medium range protein calibration kit (Pharmacia). Size exclusion chromatography (SEC) was carried out using an FPLC system (AKTA Purifier, Pharmacia). Elution was performed at a constant flow rate of 0.5 ml/min, protein fractions of 0.5 ml were collected and the absorbance was monitored at 280 nm. Each fraction was assayed in racemization assays as described here below. Fractions B1 and B5, were reloaded in the Superdex 75 column and submitted to a further SEC to verify the purity of the fractions.
Example 5
Racemization Assays
[0146] The percent of racemization with different concentrations of L-proline, D-proline, L-hydroxy (OH)-proline, D-hydroxy (OH)-proline was calculated, as described in (13), by incubating a 500 μl mixture of 0.25 μM of dimeric protein and 40 mM substrate in 0.2 M sodium acetate pH 6.0 for 30 min or 1 h at 37° C. The reaction was stopped by incubating for 10 min at 80° C. and freezing. Water (1 ml) was then added, and the optical rotation was measured in a polarimeter 241 MC (Perkin Elmer, Montigny le Bretonneux, France) at a wavelength of 365 nm, in a cell with a path length of 10 cm, at a precision of 0.001 degree. The percent of racemization of 40 mM L-proline as a function of pH was determined using 0.2 M sodium acetate, potassium phosphate and Tris-HCl buffers; reactions were incubated 30 min at 37° C., as described above. All reagents were purchased from Sigma.
Example 6
Kinetic Assays
[0147] Concentrations of L- and D-proline were determined polarimetrically from the optical rotation of the solution at 365 nm in a cell of 10 cm path length, thermostated at 37° C. Preliminary assays were done with 40 mM of L-proline in 0.2 M sodium acetate pH 6 in a final volume of 1.5 ml. Optical rotation was measured every 5 sec during 10 min and every 5 min to 1 hour. After determination of the linear part of the curve, velocity in 5-160 mM substrate was measured every 30 sec during 10 min to determine KM and Vmax. Calculations were done using the Kaleidagraph program. Inhibition assays were done by incubating 0.125 μM dimeric protein, 6, 7 μM-6 mM pyrrole-2-carboxylic acid (PAC), 20 to 160 mM L-proline, as described above. Graphic representation and linear curve regression allowed the determination of Ki as [PAC]/[(slope with PAC/slope without PAC)-1]. All reagents were purchased from Sigma.
Example 7
Site-Directed Mutagenesis of .sup.C330STcPRACA
[0148] Site-directed mutagenesis was performed by PCR, adapting the method of Higuchi et al. (15). Briefly, mutation of Cys330 of the proline racemase active site was produced by two successive polymerase chain reactions based on site-directed mutagenesis using two overlapping mutagenic primers: (act-1) 5' GCG GAT CGC TCT CCA AGC GGG ACA GGC ACC 3' [SEQ ID NO:7] and (act-2) 5' GGT GCC TGT CCC GCT TGG AGA GCG ATC CGC 3', [SEQ ID NO:8] designed to introduce a single codon mutation in the active site by replacement of the cysteine (TGT) at the position 330 by a serin (AGC). A first step standard PCR amplification was performed using the TcPRACA DNA as template and a mixture of act-1 primer and the reverse C-terminus primer (Bg-45) 5' CTG AGC TCG ACC AGA T(CA)T ACT GC 3' (codon 423), or a mixture of act-2 primer and the forward N-terminus primer (Hi-45) 5' CTC TCC CAT GGG GCA GGA AAA GCT TCT G 3' (codon-53) (see FIG. 5). Resulting amplified fragments of, respectively, 316 bp and 918 bp were purified by Qiagen PCR extraction kit (Qiagen, Courtaboeuf, France), as prescribed, and further ligated by T4 ligase to generate a template consisting of the full length of a potentially mutated TcPRACA* coding sequence used for the second step PCR. Amplification of this template was performed using forward Hi-45 and reverse Bg-45 primers and the resulting TcPRACA* fragment encoding for the mature proline racemase was purified and cloned in pCR®2.1-TOPO® vector (Invitrogen). TOP10 competent E. coli were transformed with the pCR®2.1-TOPO®-TcPRACA* construct and plasmid DNA isolated from individual clones prepared for DNA sequencing. Positive mutants were then sub-cloned in frame with a C-terminal six-histidine tag into the Nco I/Sac I sites of the pET 28b(+) expression vector (Novagen-Tebu, Le Parrayen Yvelines, France). Sub-clones of pET28b(+)-TcPRACA* produced in E. coli (DH5α) were sequenced again to confirm the presence of the mutation. Soluble recombinant .sup.C330STcPRACA protein was produced in E. coli BL21(DE3) (Invitrogen) and purified using a nickel column (Novagen-Tebu), according the manufacturer's instructions.
Example 8
Mutagenesis
[0149] To verify the implication of the residue Cys160 in the reaction mechanism of the proline racemase, a site specific mutagenesis was performed to replace the residue Cys160 by a Serine, similarly to mutation described for Cys330 residue (see Example 7). Briefly, the site specific mutagenesis was performed by PCR using the following primers: TABLE-US-00002 Ser160-Forward: 5'GGCTATTTAAATATGTCTGGACATAACTCAATTGCAGCG3' Ser160-Reverse: 5'CGCTGCAATTGAGTTATGTCCAGACATATTTAAATAGC3'
[0150] The presence of the mutation Cystein-Serine was verified by sequencing of the respective plasmids containing the PCR products, as shown here below. The plasmid pET-C160S was used to transform E. coli BL21(DE3) and to produce the corresponding recombinant mutated protein. TABLE-US-00003 139 M D T C G Y L N M C G H N G I A A 145 pET-TcPRAC 499 ATCGATACCGCTGGCTATTTAAATATGTGTGGACATAACTCAATTGCAGCG 550 Ser16O-F/R GGCTATTTAAATATGTCTGGACATAACTCAATTGCAGCG 550 pET-C160S 499 ATGGATACCGGTGGCTATTTAAATATGTCTGGACATAACTCAATTGCAGCG 550 pET-C330S 499 ATGGATACCGGTGGCTATTTAAATATGTGTGGACATAACTCAATTGCAGCG 550 139 M D T C G Y L N M S G H N G I A A 145 318 V I F G N R Q A D R S P C G T C T 334 pET-TcPRAC 999 GTGATATTTGGCAATCGCCAGGCGGATCGCTCTCCATGTGGGACAGGCACC 1050 Ser330-F/R GCGGATCGCTCTCCAAGCGGGACAGGCACC 1050 pET-C160S 999 GTGATATTTGGCAATCGCCAGGCGGATCGCTCTCCATGTGGGACAGGCACC 1050 PET-C330S 999 GTGATATTTGGCAATCGCCAGGCGGATCGCTCTCCAAGCGGGACAGGCACC 1050 318 V I F G N R Q A D R S P S G T C T 334
[0151] Underlined are the primer sequences used for the site specific mutageneses. The mutations Cys→Ser are represented in bold and underlined for both Cys160 and Cys330 residues.
Example 9
Expression of a Functional Intracellular Isoform of Proline Racemase
[0152] Previously characterized was a TcPRAC gene from T. cruzi, and it was demonstrated in vivo and in vitro that it encodes a proline racemase enzyme (13). Analysis of the genomic organization and transcription of the TcPRAC gene indicated the presence of two paralogue gene copies per haploid genome, named TcPRACA 1 and TcPRACB 2. It was shown that TcPRACA encodes a functional co-factor independent proline racemase, closely resembling the C. sticklandii proline racemase (CsPR) (11). Now sequenced was the full length of TcPRACB and, as can be observed in FIG. 1A, TcPRACA and TcPRACB genes both possess the characteristic trypanosome polypyrimidine-rich motifs in the intergenic region that are crucial trans-splicing signals when located upstream of an (AG)-dinucleotide used as acceptor site. As in other T. cruzi genes, UUA triplets are found at the end of the 3' untranslated region preceding the polyadenylation site. Comparison between the two sequences revealed 14 point mutations (resulting in 96% identity) giving rise to 7 amino acid differences. When expressed, the TcPRACB is predicted to produce a shorter protein (39 kDa) whose translation would start at the ATG codon at position 274 located downstream of the (AG)-spliced leader acceptor site (at position 175). In comparison, TcPRACA has an open reading frame that encodes a peptide with an apparent molecular mass of 45 kDa. The schematic protein sequence alignment of the two proteins TcPRACA and TcPRACB depicted in FIG. 1B reveals that TcPRACB proline racemase lacks the amino acid sequence corresponding to the signal peptide observed in the TcPRACA protein (hatched box in the figure; see predicted cleavage site in FIG. 1c). Therefore the TcPRACB would produce a 39 kDa, intracellular and non-secreted isoform of the protein. As with CsPR (11) and TcPRACA (13 and FIG. 1B), the active site of proline racemase is conserved in TcPRACB sequence. Furthermore, while differing by only 7 amino acids, both the TcPRACA and TcPRACB sequences display around 50% homology to the CsPR (13). In accordance with other protein-coding genes in T. cruzi, TcPRAC genes are located on two different chromosomal bands of which one contains three or more chromosomes of similar size, see FIG. 1D. Thus, hybridization of blots containing T. cruzi CL Brener chromosomal bands separated by pulsed field gel electrophoresis revealed that sequences recognized by an homologous probe to both TcPRACA and TcPRACB are mapped in neighboring migrating bands of approximately 0.9 Mb and 0.8 Mb, corresponding respectively to regions VII and V, according to Cano et al. numbering system (14).
[0153] In order to verify if the TcPRACB gene could encode a functional proline racemase, both T. cruzi paralogues were expressed in E. coli to produce C-terminal His6-tagged recombinant proteins. After purification by affinity chromatography on nickel-nitrilotriacetic acid agarose column, recombinant proteins were separated by SDS gel electrophoresis revealing single bands with the expected sizes of 45.8 and 40.1 kDa, respectively, for the rTcPRACA and rTcPRACB proteins (FIG. 2A). To determine whether rTcPRACB displays proline racemase enzymatic activity, biochemical assays were employed to measure the shift in optical rotation of L- and D-proline substrates, as described (13). As can be seen in FIG. 2B, rTcPRACB racemizes both L- and D-proline but not L-hydroxy-proline, like rTcPRACA. In a similar manner, rTcPRACB is a co-factor independent proline racemase as described for CsPR (11) and rTcPRACA (13) proline racemases. The rate of conversion of L- into D-proline was measured at various pH values using both recombinant enzymes. As illustrated in FIG. 2c, rTcPRACA activity clearly shows a pH dependency with an optimal activity from pH 5.5 to 7.0. In contrast, the optimum activity of rTcPRACB can be observed in a large pH spectrum varying from pH 4.5 to 8.5. These results revealed that translation of the open reading frame of both TcPRAC genes copies result in functional proline racemase isoforms. As previously described, Western blot analysis of non-infective epimastigote parasite extracts using antibodies raised against the 45 kDa secreted proline racemase had previously revealed a 39 kDa protein mostly in the soluble cellular fraction, only weakly in the cellular insoluble fraction and absent from culture medium (13). To demonstrate that the intracellular 39 kDa isoform of the protein was equally functional in vivo, soluble cellular extracts were obtained from 5×108 epimastigotes, non-infective parasites and the levels of 39 kDa soluble protein quantified by Western blot comparatively to known amounts of rTcPRACB enzyme. As can be observed in FIG. 2D, the intracellular isoform of the protein is indeed functional in vivo, since proline racemase enzymatic activity was displayed and levels of racemization were dependent on protein concentration. This discovery is useful for specific inhibitors reaching the intracellular compartment.
Example 10
Functional Analysis and Kinetic Properties of Recombinant T. cruzi Proline Racemases
[0154] Since the TcPRAC gene copies encode for secreted and non-secreted isoforms of proline racemase with distinct pH requirements for activity, our investigation was made to determine whether other biochemical properties differ between rTcPRACA and rTcPRACB proteins. Such differences might reflect the cellular localization of the protein during parasite differentiation and survival in the host. Both rTcPRACA and rTcPRACB enzyme activities are maximal at 37° C. and can be abolished by heating for 5 min at 80° C. However, the stability of the two recombinant enzymes differs considerably, when analyzed under different storage conditions. Thus, as shown in Table 1, purified rTcPRACB is highly stable, since its activity is maintained for at least 10 days at room temperature in 0.5 M imidazol buffer pH 8.0, as compared to rTcPRACA that loses 84% of its activity under such conditions. In contrast, most of the enzymatic activity of rTcPRACA is maintained at 4° C. (65%), compared to that of rTcPRACB (34%). Both enzymes can be preserved in 50% glycerol at -20° C., or diluted in sodium acetate buffer at pH 6.0, but under these storage conditions rTcPRACA activity is impaired. However, best preservation of both recombinant proline racemases was undoubtedly obtained when proteins were kept at -20° C. as ammonium sulfate precipitates. Preservation is important for a kit. TABLE-US-00004 TABLE I Stability of recombinant TcPRACA and TcPRACB proline racemases under different storage conditions % of preservation of proline racemase activity NaOAc Column pH 6 (NH4)2SO4 Protein CTRL RT +4° C. Gly/-20° C. 4° C. 4° C. -20° C. rTcPRACA 100.0 16.0 66.5 62.9 31.0 53.9 100.0 rTcPRACB 100.0 100.0 34.0 93.6 77.6 98.4 100.0
[0155] After purification on nickel-nitrilotriacteic acid agarose column, recombinant proteins were kept for 10 days in nickel column buffer (20 mM Tris/500 mM NaCl/500 mM imidazol, pH 8.0) at room temperature (RT) or at +4° C., or either diluted in 50% glycerol and maintained at -20° C. (Gly/-20° C.) or in optimum pH buffer (NaOAc, pH 6.0) at 4° C. Recombinant enzymes were precipitated in (NH4)2SO4 and kept in solution at 4° C. or pellet dried at -20° C. Racemase assays were performed for 30 min at 37° C. Percent of preservation was determined polarimetrically using 0.25 μM of either purified rTcPRACA or rTcPRACB enzymes and 40 mM of L-proline, as compared to results obtained with freshly purified proteins (CTRL). These results are representative of at least two independent experiments.
[0156] Both recombinant enzymes exhibited Michaelis-Menten kinetics (FIG. 3A) and rTcPRACB had a higher activity than rTcPRACA. Indeed, as can be observed in FIG. 3B, analysis of L>D conversion of serial dilutions of L-proline catalyzed by a constant amount of each enzyme showed that rTcPRACB enzyme (KM of 75 mM and Vmax of 2×10-4 mol.sec-1) has a higher velocity as compared to rTcPRACA (KM of 29 mM and Vmax of 5.3×10-5 mol.sec-1). In order to determine the Ki values for pyrrole-2-carboxylic acid (PAC), the specific and competitive inhibitor of CsPR (16), assays were performed with both recombinant proteins. These assays revealed that PAC is comparably effective as inhibitor of rTcPRACA (FIG. 3C) and rTcPRACB, and Ki values obtained were, respectively, 5.7 μM and 3.6 μM. The difference in Ki values reflects almost perfectly the difference in KM values reported for both enzymes, which are similar to that of the native protein. These Ki values indicate that the affinity of PAC inhibitor is higher for rTcPRACA and rTcPRACB than for CsPR (Ki of 18 μM). The Km and Ki values are important for an inhibitor.
Example 11
Requirement of a Dimeric Structure for Proline Racemase Activity
[0157] When rTcPRACA was submitted to size exclusion chromatography on a Superdex 75 column at pH 6.0, two peaks of protein were eluted, respectively, around 80 kDa (B2 fraction) and 43 kDa (B4 fraction), presumably corresponding to dimeric and monomeric forms of the enzyme (FIG. 4). Western blot analysis of whole T. cruzi epimastigote extracts using non-denaturing PAGE had previously indicated a molecular mass of 80 kDa for the native protein while a 45 kDa band was obtained by SDS-PAGE (13). In order to eliminate cross-contamination, B1 and B5 fractions, eluted, respectively, at the start and at the end of the predicted dimer (B2) or monomer (B4) peaks, were reloaded on the column and the profiles obtained (see FIG. 4 inserts) confirmed the purity of the fractions. Enzyme activity resides in the 80 kDa peak, but not in the 43 kDa peak (Table II). These results corroborated that two subunits of the protein are necessary for racemase activity. At neutral pH (7.4 or above), the rTcPRACA gives rise to high molecular weight aggregates which are not observed with rTcPRACB, consistently with its broader optimal pH spectrum. The enzyme should be in optimal pH conditions for a kit buffer, for example. TABLE-US-00005 TABLE II Racemase activity of recombinant TcPRACA fractions after size exclusion chromatography Fractions A15 B1 B2 B3 B4 B5 B6 B7 % racemization 1.3 35.5 62.9 42.8 0.7 0 0 0
[0158] After elution from Superdex 75 column, 20 μl of each peak (A15 to B7, see FIG. 4) corresponding to 1 μg of protein were incubated 1 h at 37° C. with 40 mM of L-proline in 0.2 M NaOAc, pH 6.0. Optical rotation was measured and % of racemization was determined as described in Example 5.
Example 11
Abrogation of Proline Racemase Activity by Mutation of Cys330 and Alternately Cys160 of the Catalytic Site
[0159] C. sticklandii proline racemase is described as a homodimeric enzyme with subunits of 38 kDa and a single proline binding site for every two subunits, where two cysteines at position 256 might play a crucial role in catalysis by the transfer of protons from and to the bound substrate (12). It has previously been shown that mitogenic properties of the T. cruzi proline racemase are dependent on the integrity of the enzyme active site, as inhibition of B-cell proliferation is obtained by substrate competition and specific use of analogues (PAC) resembling the structure assumed by the substrate proline in its transition state (16). To verify the potential role of the cysteine residues at the active site of the T. cruzi proline racemase, Cys330 and alternately Cys160 were replaced by a serine residue through site specific mutation of TcPRACA. The choice of serine as the substituting amino acid was made to avoid further major disturbances on three dimensional structure of the protein (see strategy in FIG. 5 above). After confirmation of the single codon mutation through sequencing of the construct, the C330S or C160S rTcPRACA mutant proline racemase was expressed in E. coli and purified in the same manner as wild type rTcPRACA. Then used were C330S or C160S rTcPRACA in racemization assays to verify the effects of the mutation on the enzymatic activity of the protein. As can be observed in Table III (and in FIG. 12) a total loss of proline racemase activity is observed as compared to the wild type enzyme, establishing that proton transfer during proline racemization is specifically dependent on the presence of the cysteine residue in the active site. TABLE-US-00006 TABLE III Loss of racemase enzymatic activity on the site direct .sup.C330S rTcPRACA Data set rTcPRACA .sup.C330S rTcPRACA Time (min) 0 10 30 60 0 10 30 60 Optical rotation -0.385 -0.300 -0.162 -0.088 -0.385 -0.382 -0.391 -0.387 % racemization 0 22 58 77 0 0 0 0
[0160] After purification, 5 μg of rTcPRACA or .sup.C330SrTcPRACA were incubated at 37° C. with 40 mM of L-proline in NaOAc buffer, pH 6.0. Optical rotation was measured at different times and % of racemization was determined as described in Example 5.
Example 12
Proline Racemase Protein Signatures and Putative Proline Racemases in Sequence Databases
[0161] The conservation of critical residues between parasite and bacterial proline racemases prompted a search for similarities between TcPRAC and other protein sequences in SWISS-PROT and TrEMBL databases. Twenty one protein sequences yielded significant homologies, from 11 organisms, such as several proteobacteria of the alpha subdivision (Agrobacterium, Brucella, Rhizobium) and gamma subdivision (Xanthomonas and Pseudomonas), as well as of the fermicutes (Streptomyces and Clostridium). Within the eukaryota, besides in T. cruzi, homologous genes were detected in the human and mouse genomes, where predicted proteins show overall similarities with proline racemase. Except for Clostridium sticklandii and Xantomonas campestri, each other organism encodes 2 paralogues, and Agrobacterium tumefaciens contains 3 genes. The multiple alignment also allowed for the definition of three signatures of proline racemase, which are described here in PROSITE format. As can be seen in Table IV, when using a minimal motif of proline racemase protein (M I), [IVL][GD]XHXXG[ENM]XX[RD]X[VI]XXG, located immediately after the start codon at position 79, the inventors obtained 9 hits. A second motif (M II), consisting of [NSM][VA][EP][AS][FY]X(13, 14)[GK]X[IVL]XXD[IV][AS][YWF]GGX[FWY], starting at position 218, gave 14 hits; however, the first or the second half of this motif is not sufficiently stringent to be restrictive for putative proline racemases, but gives hits for different protein families. A third motif (M III), from positions 326 to 339, namely DRSPXGX[GA]XXAXXA, was considered as a minimal pattern. Note that in position 330, the cysteine of the active site was replaced by an X. As shown in Table IV, this minimal pattern yields all 21 hits. Curiously, both genes in human as well as in mouse encode threonine instead of cysteine at the X position in motif III, while in Brucella, Rhizobium and Agrobacterium species each encode one protein with C and one with T in this position. One cannot hypothesize the implications of this substitution for the functionality of these putative proteins. If the residue at position 330 is maintained as a cysteine in motif III, a reduced number of 12 hits from 9 organisms is thus obtained, which can probably be considered as true proline racemases. The alignment of the 21 protein sequences and derived cladogram are shown in FIG. 6 and FIG. 7, respectively, the three boxes depicted correspond to motifs I, II and III described here above. This invention thus shows that DRSPCGXGXXAXXA is the minimal signature for proline racemases. Blast searches against unfinished genomes yielded, at present, an additional 13 predicted protein sequences from 8 organisms, with high similarity to proline racemases, all containing motif Ill. Organisms are Clostridium difficile, C. botulinum, Bacillus anthracis, Brucella suis, Pseudomonas putida, Rhodobacter sphaeroides, Burkholderia pseudomallei, B. mallei, and the fungus Aspergillus fumigatus. These results indicate that proline racemases might be quite widespread. TABLE-US-00007 TABLE IV SWISS-PROT and TrEMBL databases screening using PROSITE motifs Motif Organism Seq Access. nb M I M II M III M III* Agrobacterium tumefaciens 1 Q8UIA0 + + + + Agrobacterium tumefaciens 2 Q8U6X2 - - + - Agrobacterium tumefaciens 3 Q8U8Y5 - - + - Brucella melitensis 1 Q8YJ29 - + + + Brucella melitensis 2 Q8YFD6 + - + - Clostridium stickilandii Q9L4Q3 - + + + Homo sapiens 1 Q96EM0 + + + - Homo sapiens 2 Q96LJ5 + + + - Mus musculus 1 Q9CXA2 + + + - Mus musculus 2 Q99KB5 + + + - Pseudomonas aeruginosa 1 Q9I476 - + + + Pseudomonas aeruginosa 2 Q9I489 - - + + Rhizobium loti 1 Q98F20 - + + + Rhizobium loti 2 Q988B5 + + + - Rhizobium meliloti 1 Q92WR9 - - + - Rhizobium meliloti 2 Q92WS1 - + + + Streptomyces coelicolor Q93RX9 + - + + Trypanosoma cruzi 1 Q9NCP4 + + + + Trypanosoma cruzi 2 + + + + Xanthomonas axonopodis 1 Q8PJI1 - + + + Xanthomonas axonopodis 2 Q8PKE4 - - + + Xanthomonas campestris Q8P833 - + + + Bacillus anthracis (Ames) 1 Q81UH1 + - + + Bacillus anthracis (Ames) 2 Q81PH1 - - + + Bacillus cereus 1 Q81HB1 + - + + Bacillus cereus 2 Q81CD7 - - + + Brucella suis 1 Q8FYSO + + + + Brucella suis 2 Q8G213 + - + - Chromobacterium violaceum Q7NU77 + + + + Photorhabdus luminescens Q7N4S6 + + + + Pseudomonas putida Q88NF3 + + + + Rhodopirella baltica Q7UWF3 - - + + Streptomyces avermitilis Q82MDO + - + + Vibrio parahaemolyticus Q87Q20 + + + + SWISS-PROT and TrEMBL databases were screened using motifs I to III (M I, M II and M III). M I corresponds to [VL][GD]XHXXG[ENM]XX[RD]X[VI]XXG, M II to of [NSM][VA][EP][AS]FY]X(13, 14)[GK]X[IVL]XXD[IV][AS][YWF]GGX[FWY] M III to DRSPXGXGXXAXXA and M III* to DRSPCGXGXXAXXA. Access. nb, SWISS-PROT accession number of the sequence: seq, sequence number according to FIG. 6; + and -, presence or absence respectively of hit using # the corresponding motif.
[0162] Finally, Table V summarizes the genes in which the proline racemase signature has been identified and the sequences including both crucial residues Cys330 and Cys160 of the catalytic site are present. TABLE-US-00008 TABLE V Results of screening using nucleotide or peptide sequence of TcPRACA common Motifs sequence M III* MCGH Organism Accession number Database M I M II M III Cys130 Cys160 EPRGH Aspergillus fumigatus Af0787f05.p1c TIGR + - + - + + Aspergillus fumigatus TIGR 5085 TIGR + + + + ? + Bacillus anthracis str. Ames AE017027 EMBL + + + + + + Bacillus anthracis str. Ames AE017033 EMBL + + + + + + (minus strand) Bacillus anthracis TIGR 1392 TIGR + + + + + + Bacillus cereus ATCC14579 AE017007 EMBL + + + + + + minus strand Brucella suis 1330 AE014469 EMBL + + + + + + (minus strand) Brucella suis TIGR 29461 TIGR + + + + + + Burkholderia mallei contig:33162:b_mallei TIGR + + + + + EPRGSD Burkholderia mallei TIGR 13373 TIGR + ? + + + EPRGSD Burkholderia pseudomallei SANGER 28450 Sanger + ? + + + EPRGSD Clostridium botulinum Cbot12g05.q1c Sanger ? + + + + + Clostridium botulinum SANGER 36826 Sanger + + + + + + Clostridium difficile Clostridium difficile Sanger ? + + + + + 630 Clostridium difficile SANGER 1496 Sanger + + + + + + Clostridium sticklandii CST130879 EMBL + + + + + + Leishmania major LM16BINcontig2054 Sanger ? + + + + EPRGND Leishmania major LM16W5b02.q1c Sanger ? + ? ? + EPRGND Pseudomonas putida KT2440 AE016778 EMBL + + + + + EPRGND Pseudomonas putida KT2440 TIGRpputida 13538 TIGR + ? + + + EPRGND Rhodobacter sphaeroides UTHSC 1063 UTHSC + ? - - + + Trypanosoma brucei TbKIX28b06.qlc Sanger ? + ? ? + + Trypanosoma brucei TbKIX28b06.plc Sanger ? + ? ? + + Trypanosoma vivax Tviv655d02 Sanger ? + + + ? ? Trypanosoma vivax Tviv380d6 Sanger + ? ? ? + + Trypanosoma congolense congo208e06 Sanger ? + + + ? ? Vibrio parahaemolyticus AP005077 EMBL + + + + + + Databases were screened using nucleotide or peptide sequences of TcPRACA. Motifs I to III (M I, M II and M III) were searched. M I corresponds to [IVL][GD]XHXXG[ENM]XX[RD]X[VI]XXG, M II to of [NSM][VA][EP][AS][FY]X(13, 14)[GK]X[IVL]XXD[IV][AS][YWF]GGX[FWY] M III to DRSPXGXGXXAXXA and M III* to DRSPCGXGXXAXXA. Access. nbs, TIGR, EMBL or SANGER accession numbers of the sequence; + and -, presence or absence respectively # of the corresponding motif. Others, extremely conserved regions outside the motifs, including NMCGH which contains one of the active site cysteine. Sequences presented in annexe pages where the conserved regions of 2 Cysteine residues of the active site are squared, are presented in the table in bold with corresponding Accession numbers.
[0163] A variety of free D-amino acids can be found in different mammalian tissues in naturally occurring conditions. Some examples include the presence of D-serine in mammalian brain, peripheral and physiological fluids, or else D-asp that can be also detected in endocrine glands, testis, adrenals and pituitary gland. D-pro and D-leu levels are also very high in some brain regions, pineal and pituitary glands. Some reports attribute to D-amino acids a crucial role as neuromodulators (receptor-mediated neurotransmission), as is the case of D-ser, or as regulators of hormonal secretion, oncogeny and differentiation (i.e. D-asp). It is believed that the most probable origin of naturally occurring D-amino acids in mammalian tissues and fluids is the synthesis by direct racemization of free L-enantiomers present in situ. However, a part from the cloning of serine racemase genes from rat brain and human no other amino acid racemases were identified until now in man. Some others report that D-amino acids present in mammalian tissues are derived from nutrition and bacteria.
[0164] The increasing number of reports associating the presence of D-amino acids and pathological processes indicate that the alteration of their level in biological samples would be of some diagnostic value as, for instance, the identification of changes in free levels of D-asp and D-Ala in brain regions of individuals presenting Alzheimer. The amounts of D-asp seems to decrease in brain regions bearing neuropathological changes and is paralleled by an increase of D-ala. Overall, total amounts of D-amino acids increase in the brain of individuals presenting memory deficits in Alzheimer, as compared to normal brains, offering new insights towards the development of new simple methods of D-amino acid detection. In the same line, D-ser concentrations in the brain are altered in Parkinson disease and schizophrenia but other findings clearly associate significant higher concentrations of D-amino acids in plasma of patients with renal diseases or else in plasma of elderly people.
[0165] Previous results determined that the polyclonal B cell activation by parasite mitogens contributes to the mechanisms leading to parasite evasion and persistence in the mammalian host. It has also been demonstrated that TcPRAC is a potent B cell mitogen released by the infective forms of the parasite. The TcPRAC inhibition by pyrrole carboxylic acid induces a total loss of TcPRAC B cell mitogenic ability.
[0166] It has also been shown that the overexpression of TcPRACA and TcPRACB genes by mutant parasites are able to confer to these mutants a better invasion ability of host cells in vitro. This contrasts to the inability of parasites to survive if these TcPRAC genes are inactivated by genetic manipulation. In addition, the immunization of mice with sub-mitogenic doses of TcPRAC, or with appropriate TcPRAC-DNA vector vaccine preparations, was shown to trigger high levels of specific antibody responses directed to TcPRAC and high levels of immunoprotection against an infectious challenge with live Trypanosoma cruzi.
[0167] Altogether, these data suggest that TcPRAC enzyme isoforms are essential elements for parasite survival and fate and also support that parasite proline racemase is a good target for both vaccination and chemotherapy. In fact, the addition of pyrrole carboxylic acid at TcPRAC neutralizing doses to non-infected monkey cell cultures do not interfere with cellular growth. Besides, the utilization of a proline racemase inhibitor in humans would be a priori possible since the absence of the two critical active site cysteine residues (Cys 330 and Cys 160) for the PRAC enzyme activity has been observed in the single sequence that displays some peptide homologies with TcPRAC that was identified by blasting the Human Genome available data with the TcPRAC gene sequence.
[0168] As observed by data mining using TcPRAC gene sequences, it has been possible to identify putative proline racemases in other microrganisms of medical and agricultural interest. As can be seen in FIG. 8, the presence of MI, MII and most particularly MIII stringent motif (the signature for proline racemases) indicates the potentiality of those proteins to be functional proline racemases. On the one hand, it can be observed that critical residues necessary for the enzyme activity are displayed in those sequences and, on the other hand, that the open reading frames (ORF) are highly homologous to the ORF of the parasite PRAC.
[0169] In order to search for putative molecules that could be used as inhibitors of TcPRAC, or other proline racemases, it would be necessary to develop a microtest able to specifically reveal the inhibition of proline racemization performed by TcPRAC and consequently the blockage of a given proline stereoisomer generation. For instance, this could be done by analysing the ability of any potential inhibitory molecule to hinder the generation of D-proline in a reaction where L-proline is submitted to TcPRAC enzymatic activity.
[0170] At present, the available analyses to detect D- (or L-) amino acids are very challenging and methods to differentiate L-stereoisomers from D-stereoisomers are time-consuming, i.e. gas chromatography, thin layer chromatography using chiral plates, high-performance capillary electrophoretic methods, HPLC, and some enzymatic methods. Some of those techniques also require the use of columns and/or heavy equipment, such as polarimeters or fluorescence detectors.
[0171] With the aim of developing a simple test that is useful to rapidly screen putative inhibitors of TcPRAC, TcPRAC constructs allowing for the production of high amounts of the recombinant active enzyme were used together with the knowledge of a specific inhibitor of proline racemases (pyrrole carboxylic acid, PAC) to develop a medium/high throughput microplate test that can be used to easily screen a high number of inhibitor candidates (i.e. 100-1000). Such a test is based on colorimetric reactions that are certainly a simpler alternative to polarimetry and other time-consuming tests. Thus, the evaluation of light deviation of L- or D-proline enantiomers by a polarimeter to quantify the inhibition of proline racemization to test such an elevated number of molecules is impracticable, offers a low sensibility, and would require greater amounts of reagents as compared to a microplate test that would additionally be of an affordable price.
[0172] Accordingly, this invention is based on the detection of D-proline originated through racemization of L-proline by TcPRAC, in the presence or in the absence of known concentrations of PAC inhibitor as positive and negative controls of racemization, respectively. For that purpose, this invention utilizes another enzyme, D-amino acid oxidase (D-AAO), that has the ability to specifically oxidize D-amino acids in the presence of a donor/acceptor of electrons and yield hydrogen peroxide. The advantage of this strategy is that hydrogen peroxide can be classically quantified by peroxidase in a very sensitive reaction involving ortho-phenylenediamine, for example, ultimately offering a chromogenic reaction that is visualized by colorimetry at 490 nm.
[0173] Since D-amino acid oxidase reacts indiscriminately with any "D-amino acid", and not with their L-stereoisomers, such a test is not only helpful to identify proline racemase inhibitors, but also applicable, if slightly modified, to detect any alterations in levels of free D-aa in various fluids to make a diagnosis of some pathogenic processes.
I-Basics for a D-Amino-Acid Quantitative Test
[0174] The following method of the invention allows detection and quantitation of D-Amino acids. A first reaction involves a D-amino-oxidase. This enzyme specifically catalyses an oxidative deamination of D-amino-acids, together with a prosthetic group, either Flavin-Adenin-Dinucleotide (FAD) or Flavin-Mononucleotide (FMN), according to the origin of the Enzyme. (Obs. FAD if the enzyme comes from porcine kidney).
[0175] The general reaction is as follows: ##STR1## Either a catalase or a peroxidase can decompose hydrogen peroxide. A catalase activity is written as: 2H2O2→2H2O+O2(O═O) [0176] Oxygen whereas a peroxidase activity is H2O2+HO--R'--OH→2H2O+O═R'═O [0177] wherein R' is any carbon chain
[0178] Thus, detection of hydrogen peroxide can be done with the use of catalase and a reagent sensitive to oxygen such as by destaining reduced methylene blue for instance with oxygen or with the use of peroxidase with a change in color of the reagent indicated by: HO--R'--OH→O═R'═O II-Application of such a test for evaluating the T. cruzi racemase activity and the inhibition of this racemase.
II-1--Test for Racemase Activity
[0179] The T. cruzi racemase activity converts reversibly L-Pro into D-Pro. Since these two forms can induce polarized light deviation, this conversion can be measured by optical polarized light deviation. But the presence of the D-form allows also the use of D-amino-acid oxidase in order to assess the amount of D-Proline in racemase kinetics. In this test the following reactions are involved:
1) Proline-Racemase Activity. L-Proline⇄D-Proline 2) D-Amino-Acid Oxidase D-Proline+FAD⇄1-Pyrroline 2-carboxylic acid+FADH2 (1) (Obs: There is no ammonia formed in the case of Proline, because the nitrogen of Proline is involved in a secondary amine.) FADH2+O2⇄FAD+H2O2 (2) 3) Detection of Hydrogen Peroxide with Peroxidase ##STR2##
[0180] The chromogenic reagent can be, for example, orthophenylenediamine (OPD), or 3,3',5,5' tetramethyl benzidine (TMB), or 5-aminosalicylic acid (ASA).
[0181] These reactions can be carried out using the following exemplary, but preferred, materials and methods.
[0182] II-1-1--Materials TABLE-US-00009 Materials Comments Proline-racemase (TcPRAC) (1 mg/ml Stock) L-Proline, Sigma, Ref. P-0380 (1 M An equimolar D- and L-Proline Stock) D-Proline, Aldrich, ref. 85 is made by mixing equal volumes 891-9 (1 M Stock) of 2 M D-Proline with 2 M L- Proline Orhtophenylenediamine (OPD) 10 mg tablets. Extemporaneously Sigma ref P-8287 lot 119H8200 used as a 20 mg/ml stock solution in water. D-AAO from swine kidney (Sigma) Powder dissolved into 1 ml ref. A-5222 lot 102K1287 Buffer* + 1 ml 100% glycerol. The resulting activity is 50 U/ml. Stored at -20° C. Horse radish peroxidase (HRP) Powder dissolved into 2.5 ml Sigma ref P8375 lot 69F95002 Buffer* + 2.5 ml 100% glycerol. The resulting activity is 5042 U/ml. Stored at -20° C. Sodium acetate 0.2 M Ph 6.0 Flavine-adenine-dinucleotide (FAD) Stock solution of 10-1 M in water. (Sigma) ref. F-6625 Stored at -20° C. Used as a 10-3 M sub-stock solution. Sodium pyrophosphate (Pop) Not soluble at a higher 0.235 M concentration. Must be stored at 4° C. and gently heated before use in order to solubilize crystals which may occur. Buffer* = 10 ml of 0.2 M sodium The final pH us 8.3. acetate buffer pH 6.0 +680 μl 0.235 M Pop Microplates (96 wells) With adhesive coverlid ELISA reader for microplates With a wavelength filter at 490 nm for OPD substrate.
II-1-2--Methods II-1-2.1--Racemisation in Microplates:
[0183] (1) The volumes are indicated for a single well, but duplicates are mandatory. Leave enough raws of the microplate empty for standard and controls to be used in further steps. Distribute the following volumes per well reactions:
[0184] a) without inhibitor (Vol=QS 81 μl) TABLE-US-00010 TcPRAC 1 mg/ml 2 μl 2 μl 2 μl 2 μl L-Proline 0.1 M 32 μl 16 μl 8 μl 4 μl Proline Final (40 mM) (20 mM) (10 mM) (5 mM) concentration Sodium acetate 47 μl 63 μl 71 μl 75 μl buffer 0.2 M pH 6
b) with inhibitor (Vol=QS 81 μl)
[0185] A range of concentrations between 5 mM and 1 mM can be planned for the inhibitor. It should be diluted in sodium acetate buffer 0.2 M pH 6.0. Hence, the volume of inhibitor is substracted from the volume of buffer added in order to reach a final volume of 81 μl. For instance, 50% inhibition of racemisation of 10 mM L-proline is obtained with 45 μM Pyrrole carboxylic acid (PAC, specific inhibitor of proline racemase), when 36.5 μl PAC+44.5 μl buffer are used (see results in FIG. 8).
[0186] Table VI is provided for 10 mM L-Proline as a substrate. TABLE-US-00011 TABLE VI TcPrac 1 mg/ml 2 μl 2 μl 2 μl 2 μl 2 μl 2 μl 2 μl 2 μl 2 μl 2 μl L-Proline 0.1 M 8 μl 8 μl 8 μl 8 μl 8 μl 8 μl 8 μl 8 μl 8 μl 8 μl PAC 0 μl 5.4 μl 11 μl 22 μl 43 μl 9 μl** 17 μl** 35 μl** 69 μl** 14 μl*** 0.1 mM/1 mM**/ 10 mM*** Final 0 6.7 13.5 27 54 107 214 429 858 1715 concentration (μM) Sodium acetate buffer 0.2 M 71 μl 65.6 μl 60 μl 49 μl 28 μl 62 μl 54 μl 36 μl 2 μl 57 μl pH 6 QS 81 μl
[0187] (2) Cover the microplate with an adhesive coverlid and leave for 30 mn at 37° C.
[0188] (3) At the end of racemisation, 5.5 μl of 0.235M Pop are added in each reaction well of the microplate in order to shift pH from pH6.0 to pH 8.3.
II-1-2.1-2--Quantitation of Formed D-Proline: Standards and Controls.
(1) Prepare Standard and Controls:
[0189] Standard: An equimolar mixture of L- and D-Proline is used as a standard in a range from 0.05 mM to 50 mM (final concentration in the assay). It is used for assessing the amount of D-Proline formed after racemization. The standard range is made in microtubes, as follows:
[0190] In tube 1, mix Proline and buffer according to the described proportions.
[0191] Then, add 500 μl of the obtained mixture to 500 μl of buffer in next tube, and so on. TABLE-US-00012 ##STR3##
[0192] Negative control: is prepared in an other microtube, as follows: TABLE-US-00013 L-Proline (1 M) 200 μl Buffer* 800 μl Final concentration 40 ml Blank = Buffer*.
[0193] (2) Dispense in the Empty Wells of the Microplate (See Step II-1-2.1): TABLE-US-00014 Buffer* 67 μl Standard dilutions 20 μl or negative control Obs: For the blank dispense 87 μl of Buffer* only
(3) Prepare a Mixture Containing the Enzymes (D-AAO/HRP Mix), as Follows:
[0194] The amounts are given for one well, provided that the final volume will be 100 μl with the racemase products or the substrate: TABLE-US-00015 For 13 μl: Buffer* 6.5 μl D-AAO 50 U/ml 1.7 μl OPD (20 mg/ml) 2.5 μl HRP 5000 U/ml 0.75 μl FAD 10-3 M (4.5 μl 10-1 M +446 μl buffer) 1.5 μl
This mixture is kept in the ice until use. (4) The quantitation reaction starts when 13 μl of D-AAO/HRP mix is added to the reaction well. (5) The microplate is covered with an adhesive coverlid and it is left in the dark at 37° C. between 30 nm and 2 hours. The reaction can be monitored by eye whenever a color gradient matches the D-amino acid concentration of the standard dilutions. (6) The microplate is read with a microplate spectrophotometer using a filter of at 490 nm.
Example 13
D-AOO Microplate Test is More Sensitive than D-Amino Acid Detection by Detection in Polarimeter
[0195] In order to compare the D-Proline quantitation by polarimeter and by D-amino-oxidase/HRP a comparison was performed between the two tests using different concentrations of L-proline and different concentrations of PAC, the specific inhibitor of proline racemases. FIG. 8 shows the percent of racemisation inhibition of different L-proline concentrations (ranging from 10-40 mM) using the D-MO (D-AA0/L-) microtest as compared to conventional detection using a polarimeter (Pol/L-).
[0196] With the polarimeter, there seems to be no difference of PAC inhibition of TcPRAC with the three concentrations of L-Proline. Therefore, 50% inhibition is obtained with 1 mM PAC, whether 10 mM or 40 mM L-Proline is used. In contrast, when using D-AAO/HRP test, it can be seen that inhibition by PAC is somewhat higher with a low concentration of L-Proline (10 mM for example) than with an increased one (20 mM or 40 mM). Therefore, 50% inhibition is obtained:
[0197] with 50 μM PAC when 10 mM L-Proline is used,
[0198] with 170 μM PAC when 20 mM L-Proline is used and
[0199] with 220 μM PAC when 40 mM L-Proline is used.
[0200] In conclusion, D-AAO/HRP evaluation is more sensitive since it can discriminate PAC inhibition at a lower concentration than evaluation with the polarimeter. Furthermore, inhibition is logically conversely proportional to L-Proline concentration, which can be assessed with the D-MO/HRP method, but not with the polarimeter measurement. Such a test is useful for the screening of new inhibitors of TcPRAC in a medium/high throughput test.
[0201] A preferred technological platform to perform the above test and to select appropriate inhibitors contains at least the following products:
[0202] L-Proline, D-Proline, a proline-racemase
[0203] A peroxidase, a substrate of a peroxidase
[0204] A D-amino-acid oxidase
[0205] And optionally a battery of potential inhibitory molecules.
Example 14
L-Proline Inhibits D-Amino-Oxidase Activity
[0206] FIG. 9 shows the comparison of D-MO/HRP reaction using D-Proline alone or an equimolar mixture of D- and L-Proline as standard. It can be seen that the amount of D-Proline required to obtain a given optical density is higher when a mixture of L- and D-Proline are used as compared to a standard using D-proline alone. Since Proline-racemase activity ends when both L- and D-Proline are in equal amounts, it was also adequate to use an equimolar mixture of both enantiomers of Proline as standard for D-Proline determination.
Example 15
PAC does not Interfere with DAAO/HRPactivity
[0207] FIG. 10 shows optical density at 490 nm as a function of D-proline concentration under the following conditions.
[0208] Conditions in μl wells, TABLE-US-00016 [D-Proline] range between 0.1 mM and 40 mM [D-AAO] 0.89 U/ml [HRP} 37.5 U/ml [OPD] 0.5 U/ml [FAD] 1.5 × 10-5 M Buffer*
[0209] The presence of PAC does not influence DAAO/HRP reaction.
Example 16
A Medium/High Throughput Test Using the D-AAO Microplate Test
[0210] Table VII is an Example of a medium/high throughput test using the D-AAO microplate test.
Blue: D-proline-standard (column 1)
Green: Positive control of racemization using avec 10 mM substrate (column 2, line A and B)
Orange: control for inhibition of racemization reaction by PAC using 10 mM substrate (column 2, line C and D)
Blank 1: mix with racemase (column 2, line E)
Blank 2: mix without racemase (column 2, line F)
Yellow: Negative control for specificity of (without racemase+40 mM L-proline) (column 2, line G and H)
[0211] Other wells: with Inhibitors (T1, T2, T3, . . . T40): in duplicates TABLE-US-00017 TABLE VII 1 D-Pro (mM) 2 3 4 5 6 7 8 9 10 11 12 A 10 L-Pro T1 T2 T3 T4 T5 T6 T7 T8 T9 T10 L-Pro '' '' '' '' '' '' '' '' T11 T12 T13 T14 T15 T16 T17 T18 T19 T20 '' '' '' '' '' '' '' '' '' '' Blanc 1 T21 T22 T23 T24 T25 T26 T27 T28 T29 T30 Blanc 2 '' '' '' '' '' '' '' '' '' L-Pro T31 T32 T33 T34 T35 T36 T37 T38 T39 T40 H 0.07 L-Pro '' '' '' '' '' '' '' ''
Example 17
Application of Such a Test for General Detection of D-Amino Acids in Samples
[0212] The use of a microplate test based on D-amino-acid oxidase together with a peroxidase, such as horseradish peroxidase, can be used to detect and quantitate any D-amino acid in any biological or chemical sample. For example, since D-amino acids are described to be involved in several pathological processes or neurological diseases, such as Alzheimer disease, Parkinson, or renal diseases, their detection can be an important marker or parameter for the diagnosis and the follow-up of these pathologies. This technology can be also extended to the detection and quantification of D-amino acids in eukaryotic organisms, such as plants or fungi, and in bacteria.
[0213] The D-AAO/HRP test described here above can also be used for this purpose with slight modifications. For that purpose, the racemase reaction step should be skipped and the microplate test should start straightforward at the II-1-2.1-2 step described above with the following remarks:
[0214] 1) Standard: It should not be an equimolar mixture of D- and L-amino acid, but rather a serial dilution of D-Amino acids. The choice of amino acid is made according to the interest of the D-amino acid under investigation. The final volume in wells should be of 87 μl.
[0215] 2) Negative control: It is made with the L-enantiomer of the D-amino acid under investigation. The final volume should be 87 μl.
[0216] 3) Blank: It is made with 87 μl buffer*. (See paragraph II.1.1 Materials.)
[0217] 4) Samples: The samples to be tested should be adjusted to pH 8, 3 with buffer* and their final volumes should be of 87 μl per well.
[0218] Obs: Standards, negative controls, samples to test and blanks should be made in duplicates. They are dispensed into the wells of the microplate.
[0219] 5) Then, the procedure follows steps 3) to 6), as above.
[0220] Several D-amino acids and their L-counterparts have been tested using the microplate test described above. Tables VIII and IX show that D-forms of Tyrosine, Valine, Threonine, Glutamic acid, Lysine and Tryptophane are indeed substrates for the D-AA0/HRP and are detected by the test, as described for D-Proline. The results also show that no L-amino acid is detected by such a methodology. TABLE-US-00018 TABLE VIII A Blank 49.5 24.75 12.37 6.19 3.09 1.55 0.77 0.39 0.19 0.09 0.05 D-pro B Blank 49.5 24.75 12.37 6.19 3.09 1.55 0.77 0.39 0.19 0.09 0.05 mM C Blank L-Tyr L-Val L-Thr L-Glu L-Lys L-Try D Blank 12.5 12.5 12.5 12.5 12.5 12.5 mm E Blank D-Tyr D-Val D-Thr D-Glu D-Lys D-Try F Blank 6.25 6.25 6.25 6.25 6.25 6.25 mm
[0221] Optical Densities at 490 nm Obtained after D-AAO Reaction. (Raw OD Data). TABLE-US-00019 TABLE IX A 0.105 1.961 1.757 1.814 1.983 1.716 1.234 0.809 0.496 0.308 0.213 0.173 D-pro B 0.118 2.004 1.885 1.976 1.949 1.879 1.221 0.824 0.504 0.32 0.215 0.159 mM C 0.123 0.193 0.135 0.124 0.131 0.125 0.131 L- D 0.125 0.141 0.129 0.128 0.141 0.131 0.138 L- E 0.120 1.317 1.683 0.215 0.147 0.243 0.615 D- F 0.105 0.991 1.612 0.157 0.116 0.157 0.662 D-
[0222] Template of microplate, where, a serial dilution of D-Proline (mM) was made as positive control of the D-AAO reaction. Blank wells containing buffer* are shown. Different L- and D-amino acids were tested, namely Tyrosine (Tyr), Valine (Val), Threonine (Thr), Glutamic acid (Glu), Lysine (Lys) and Tryptophan (Try). To highlight the sensitivity of the D-AAO microtest, higher concentrations of L-enantiomers (12.5 mM) were used in the reactions as compared to the concentrations used for D-enantiomers (6.25 mM):
[0223] FIG. 11 is a Graph obtained with the serial dilutions of D-proline, as positive reaction control Obs: OD of wells (-) average of OD obtained from blank wells.
[0224] A preferred platform to search and quantitate the presence of a D-Amino acid in samples contains at least the following products:
[0225] A D-amino acid,
[0226] A peroxidase and a substrate of a peroxidase
[0227] A D-amino-acid oxidase
[0228] And optionally, a L-amino acid enantiomer, as control.
[0229] Finally, this invention relates to a method for screening a molecule, which can modulate a racemase activity, wherein the method comprises: [0230] (A) modulating a racemase activity by means of a molecule being tested in the presence of an equimolar mixture of a L- and D-amino acid and of a racemase to be modulated; [0231] (B) oxidatively deaminating the D-amino acid generated in step (A) by means of a D-amino oxidase in a prosthetic group; and [0232] (C) detecting the hydrogen peroxide generated by the oxidative deamination; wherein modulation of the hydrogen peroxide is indicative of the capability of the tested molecule to modulate racemase activity. Preferably the molecule inhibits racemase activity, and more preferably the racemase is a proline racemase, for example, Tripanosoma curzi proline racemase. A molecule identified by a method is also part of this invention.
[0233] Further, this invention relates to technological platform and all reagents and devices necessary to perform the methods of the invention. The technological platform comprises: [0234] a) L-amino acid, D-amino acid, and a racemase; [0235] b) a peroxydase and a substrate of a peroxydase, or a catalase and a reagent sensitive to oxygen; [0236] c) a D-amino acid oxidase; and [0237] d) optionally, one or more molecules to be screened for inhibitory activity of said racemase.
[0238] Preferably, the racemase is a proline racemase and the L-amino acid and D-amino acid are L-proline and D-proline, respectively.
[0239] A molecule inhibits a proline racemase containing a subsequence selected from the SEQ ID NO: 1, 2, 3 or 4.
Example 18
Signature of Proline Racemases
[0240] TABLE-US-00020 DRSPGXGXXAXXA
[0241] The signature of proline racemases DRSPGXGXXAXXA defined here as Motif III* contains the residue Cy330 that is also observed in the sequences here above. Fragments of the different sequences and contigs contain also the NMGH motif, corresponding to the sequence around residue Cys160 of TcPRAC, shown to be important for the enzymatic activity. Some examples are depicted here below. The sequences related to the crucial Cys residues for proline racemase activity are squared. TABLE-US-00021 Squared: NMCGH (Cys160) residues and DRSPCGTGTSAKMA (Motif III, signature containing Cys330) residues 1-Bacillus anthracis >gnl|TIGR 1392|banth_4742 Bacillus anthracis unfinished fragment of complete genome Length = 11981 Score = 141 bits (302), Expect(3) = 4e-69 Identities = 60/146 (41%), Positives = 91/146 (62%) Frame = +1 / -3 Query: 763 GEVRVDIAFGGNFFAIVPAEQLGIDISVQNLSRLQEAGELLRTEINRSVKVQHPQLPHIN 942 G V DIA+GGNF+AI+ A+ +G+++ ++ S + + +R IN ++ HP+ I Sbjct: 8379 GTVEADIAYGGNFYAIIDAKSVGLELVPEHASTIIDKAIHIRNIINERFEIIHPEYSFIR 8200 ##STR4## Query: 1123 SILGSLFQGRVLGEERIPGVKVPVTK 1200 SI+GSLF+G V+ + ++ VTK Sbjct: 8019 SIVGSLFKGCVINTTNVANMEAVVTK 7942 Score = 137 bits (294), Expect(3) = 4e-69 Identities = 54/117 (46%), Positives = 79/117 (67%) Frame = +1 / -3 Query: 262 MRFKKSFTCIDMHTEGEAARIVTSGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEPRGHD 441 MR +K FT ID HT G R + SGLP + G MAEK ++++ D++R+ +M EPRGHD Sbjct: 8859 MRTQKVFTTIDTHTGGNPTRTLISGLPKLLGETMAEKMLHMKKEYDWIRKLLMNEPRGHD 8680 ##STR5## >gnl|TIGR_1392|banth_4799 Bacillus anthracis unfinished fragment of complete genome Length = 22506 Score = 125 bits (267), Expect(4) = 4e-68 Identities = 56/145 (38%), Positives = 86/145 (59%) Frame = +1 / -3 Query: 766 EVRVDIAFGGNFFAIVPAEQLGIDISVQNLSRLQEAGELLRTEINRSVKVQHPQLPHINT 945 E +VDIAFGG F+A+V +++ G+ + ++LS +Q+ G ++ I ++V+HP + Sbjct: 5188 EFQVDIAFGGAFYAVVDSKEFGLKVDFKDLSAIQQWGGKIKHYIESKMEVKHPLEEGLKG 5009 ##STR6## Query: 1126 ILGSLFQGRVLGEERIPGVKVPVTK 1200 I F+G VL + + V K Sbjct: 4828 ITDGEFEGEVLSVTAVHTYEAVVPK 4754 Score = 124 bits (266), Expect(4) = 4e-68 Identities = 48/113 (42%), Positives = 65/113 (57%) Frame = +1 / -3 Query: 262 MRFKKSFTCIDMHTEGEAARIVTSGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEPRGHD 441 M+ K +T ID H GE RI+T G+P I G E++ Y E++DYLR +M EPRGH Sbjct: 5662 MKVSKVYTTIDAHVAGEPLRIITGGVPEIKGETQLERRWYCMEHLDYLREVLMYEPRGHH 5483 ##STR7## 2-Clostridium botulinum >gnl|SANGER_36826|sbotul_Contig173 Clostridium botulinum A unfinished fragment of complete genome Length = 97750 Score = 178 bits (383), Expect(4) = 3e-98 Identities = 70/138 (50%), Positives = 102/138 (73%) Frame = +1 / -2 Query: 760 YGEVRVDIAFGGNFFAIVPAEQLGIDISVQNLSRLQEAGELLRTEINRSVKVQHPQLPHI 939 YG++ +DI+FGG+FFA+V AE++GIDIS N +L + G + +N V+++HP L HI Sbjct: 70443 YGKLTLDISFGGSFFAMVDAEKVGIDISPANSQKLNDLGMKIVHAVNEQVEIKHPVLEHI 70264 ##STR8## Query: 1120 ESILGSLFQGRVLGEERI 1173 ESI+ + F+G++L E ++ Sbjct: 70083 ESIICTKFKGKILEETKV 70030 Score = 166 bits (357), Expect(4) = 3e-98 Identities = 70/118 (59%), Positives = 81/118 (68%) Frame = +1 / -2 Query: 259 IMRFKKSFTCIDMHTEGEAARIVTSGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEPRGH 438 IMR K+ I+ HT GE RIV GLP +PG MAEK YL+EN D LR +M EPRGH Sbjct: 70926 IMRAIKTIQTIESHTMGEPTRIVIGGLPKVPGKTMAEKMEYLEENNDSLRTMLMSEPRGH 70747 ##STR9## > SANGER Cbot12g05.q1c Score = 584 (210.6 bits), Expect = 7.7e-57, P = 7.7e-57 Identities = 115/224 (51%), Positives = 156/224 (69%), Frame = -2 ##STR10## Query: 135 QSGTESEVSNASIINVPSFLYQQDVVIVLPKPYGEVRVDIAFGGNFFAIVPAEHLGIDIS 194 + G E S I+NVP+FLY++DV I +P YG++ +DI+FGG+FFA+V AE +GIDIS Sbjct: 480 EDGKAKETS---IVNVPAFLYKKDVEIDVPD-YGKLTLDISFGGSFFAMVDAEKVGIDIS 313 Query: 195 VQNLSRLQEAGELLRTEINRSVKVQHPQLPHINTVDCVEIYGNATNPEAKYKNVVIFGNR 254 N +L + G + +N V+++HP L HI TVD E YG A + +A +NVV+FG Sbjct: 312 PANSQKLNDLGMKIVHAVNEQVEIKHPVLEHIKTVDLCEFYGPAKSEDADVQNVVVFGQG 133 ##STR11## 3-Aspergillus fumigatus ×gnl|TIGR_5085|afumi_1044 Aspergillus fumigatus unfinished fragment of complete genome Length = 7621 Score = 46.0 bits (94), Expect(4) = 3e-16 Identities = 21/72 (29%), Positives = 34/72 (47%) Frame = +1 / +2 ##STR12## Query: 1153 VLGEERIPGVKV 1188 E + V + Sbjct: 6407 AFSAEIVEEVTI 6442 Score = 40.9 bits (83), Expect(4) = 3e-16 Identities = 13/34 (38%), Positives = 26/34 (76%) Frame = +1 / +2 Query: 361 MAEKKAYLQENMDYLRRGIMLEPRGHDDMFGAFL 462 + E++ +++ D++R+ +MLEPRGH+ M+GA + Sbjct: 5513 LLEQRDQAKQHHDHIRKCLMLEPRGHNGMYGAII 5614 Score =40.0 bits (81), Expect(4) = 3e-16 Identities = 14/29 (48%), Positives = 20/29 (68%) Frame = +1 / +2 Query: 286 CIDMHTEGEAARIVTSGLPHIPGSNMAEK 372 CIDMHT GE RI+ SG P + G+ + ++ Sbjct: 5441 CIDMHTTGEPTRIIYSGFPPLSGTLLEQR 5527 Score = 32.2 bits (64), Expect(4) = 3e-16 Identities = 12/27 (44%), Positives = 20/27 (74%) Frame = +1 / +2 Query: 775 VDIAFGGNFFAIVPAEQLGIDISVQNL 855 +DI++GG F+AIV A +LG +++L Sbjct: 5996 LDISYGGAFYAIVQASELGFSGGLRDL 6076 Score = 25.8 bits (50), Expect(4) = 5e-04 Identities = 12/21 (57%), Positives = 13/21 (61%) Frame = -2 / -2 Query: 479 SSIGSNKKAPNISS*PRGSSI 417 SS+ AP I *PRGSSI Sbjct: 5631 SSVSGRMMAPYIPL*PRGSSI 5569 4-Clostridium difficile >gnl|Sanger_1496|cdifficile_1080 Clostridium difficile unfinished fragment of complete genome Length = 204145 Score = 209 bits (451), Expect(4) = e-109 Identities = 86/146 (58%), Positives = 107/146 (73%) Frame = +1 / -2 Query: 763 GEVRVDIAFGGNFFAIVPAEQLGIDISVQNLSRLQEAGELLRTEINRSVKVQHPQLPHIN 942 G V+ DI+FGG+FFAI+ A QLG+ I QN +L E LR IN +++QHP L HI Sbjct: 88224 GTVKFDISFGGSFFAIIHASQLGLKIEPQNAGKLTELAMKLRDIINEKIEIQHPTLAHIK 88045 ##STR13## Query: 1123 SILGSLFQGRVLGEERIPGVKVPVTK 1200 SILG+LF+G ++ E ++ V K Sbjct: 87864 SILGTLFKGEIVEETKVADFNAVVPK 87787 Score = 173 bits (373), Expect(4) = e-109 Identities = 68/117 (58%), Positives = 86/117 (73) Frame = +1 / -2 Query: 262 MRFKKSFTCIDMHTEGEAARIVTSGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEPRGHD 441 M+F +S ID HT GEA RIV G+P+I G++M EKK YL+EN+DYLR IMLEPRGH+ Sbjct: 88707 MKFSRSIQAIDSHTAGEATRIVVGGIPNIKGNSMPEKKEYLEENLDYLRTAIMLEPRGHN 88528 ##STR14## 5-Brucella suis >gnl|TIGR_29461|bsuis_1327 Brucella suis unfinished fragment of complete genome Length = 69104 Score = 150 bits (323), Expect(5) = 3e-73 Identities = 62/139 (44%), Positives = 92/139 (66%) Frame = +1 / -2 Query: 763 GEVRVDIAFGGNFFAIVPAEQLGIDISVQNLSRLQEAGELLRTEINRSVKVQHPQLPHIN 942 G ++VD+A+GGNF+AIV ++ D+ + +L +LR +N K QHP+LP IN Sbjct: 24931 GPIKVDVAYGGNFYAIVEPQENYTDMDDYSALQLIAWSPVLRQRLNEKYKFQHPELPDIN 24752 ##STR15## Query: 1123 SILGSLFQGRVLGEERIPG 1179 SI+GSLF GRV + G Sbjct: 24571 SIIGSLFHGRVERAAEVAG 24515 Score = 122 bits (262), Expect(5) = 3e-73 Identities = 47/106 (44%), Positives = 68/106 (64%) Frame = +1 / -2 Query: 271 KKSFTCIDMHTEGEAARIVTSGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEPRGHDDMF 450 + SF C+D HT G R+V G P++ GS M EK+A+ D++R G+M EPRGHD M Sbjct: 25402 RHSFFCVDGHTCGNPVRLVAGGGPNLNGSTMMEKRAHFLAEYDWIRTGLMFEPRGHDMMS 25223 ##STR16## 6-Rhodobacter sphaeroides >gnl|UTHSC_1063|rsphaer_x8758Contig3 Length = 2326 Score = 124 bits (265), Expect(5) = 8e-41 Identities = 50/109 (45%), Positives = 70/109 (64%) Frame = +1 / +2 Query: 262 MRFKKSFTCIDMHTEGEAARIVTSGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEPRGHD 441 MR + + I HTEGE I+ SG+P+ GS + EK+A+L+EN D+LR+ +M EPRGH Sbjct: 1448 MRVQDVYNVIYTHTEGEPLCIIYSGVPYPAGSTILEKRAFLEENYDWLRKALMREPRGHA 1627 ##STR17## Score = 65.2 bits (136), Expect = 4e-09 Identities = 18/47 (38%), Positives = 51/95 (53%) Frame = -2 / -2 Score = 34.1 bits (68), Expect(5) = 8e-41 Identities = 18/47 (38%), Positives = 23/47 (48%) Frame = +1 / +2 ##STR18## 7-Burkholderia pseudomallei >gnl|Sanger_28450|bpsmalle_Contig394 Burkholderia pseudomallei unfinished fragment of complete genome Length = 3107 Score = 105 bits (224), Expect(3) = 1e-33 Identities = 47/118 (39%), Positives = 59/118 (50%) Frame = +1 / +1 Query: 265 RFKKSFTCIDMHTEGEAARIVTSGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEPRGHDD 444 R K ID HT GE R+V SG P + G MAE+ A L D R +LEPRG D Sbjct: 1033 RDMKHIHIIDSHTGGEPTRVVVSGFPALGGGTMAERLAVLAREHDRYRAACILEPRGSDV
1212 ##STR19## Score = 61.5 bits (128), Expect(3) = 1e-33 Identities = 27/63 (42%), Positives = 38/63 (60%) Frame = +1 / +1 ##STR20## Query: 1159 GEE 1167 E Sbjct: 1861 AAE 1869 8-Burkholderia mallei >gnl|TIGR_13373|bmallei_191 Burkholderia mallei unfinished fragment of complete genome Length = 4017 Score = 105 bits (224), Expect(3) = 4e-33 Identities = 47/118 (39%), Positives 59/118 (50%) Frame = +1 / -1 Query: 265 RFKKSFTCIDMHTEGEAARIVTSGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEPRGHDD 444 R K ID HT GE R+V SG P + G MAE+ A L D R +LEPRG D Sbjct: 2601 RDMKHIHIIDSHTGGEPTRVVVSGFPALGGGTMAERLAVLAREHDRYRAACILEPRGSDV 2422 ##STR21## Score = 60.6 bits (126), Expect(3) = 4e-33 Identities = 27/63 (42%), Positives = 38/63 (60%) Frame = +1 / -1 ##STR22## Query: 1159 GEE 1167 E Sbjct: 1773 AAE 1765 9-Pseudomonas putida >gnl|TIGR|pputida_13538 Pseudomonas putida KT2440 unfinished fragment of complete genome Length = 6184039 Score = 108 bits (230), Expect(2) = 1e-32 Identities = 45/115 (39%), Positives = 64/115 (55%) Frame = +1 / -2 Query: 274 KSFTCIDMHTEGEAARIVTSGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEPRGHDDMFG 453 K ID HT GE R+V G P + G +MAE++ L+E D RR +LEPRG+D + G Sbjct: 909066 KQIHVIDSHTGGEPTRLVMKGFPQLRGRSMAEQRDELRELHDRWRRACLLEPRGNDVLVG 908887 ##STR23## Score = 71.2 bits (149), Expect(2) = 1e-32 Identities = 31/58 (53%), Positives = 40/58 (68%) Frame = +1 / -2 ##STR24## 10-Leishmania major SANGER LM16 BIN Contig2054 L. major Friedlin contig not yet a . . . 2.5e-31 3 LM16W5b02.q1c 2.3e-19 1 LM16B3d03.p1c 3.1e-05 3 >LM16_BIN_Contig2054 L. major Friedlin contig not yet assigned to chromosome from LM16 bin, unfinished whole chromosome shotgun data sequenced by the Wellcome Trust Sanger Institute, Contig number Contig2054, length 873 bp Length = 873 Score = 242 (90.2 bits), Expect = 2.5e-31, Sum p(3) = 2.5e-31 Identities = 61/180 (33%), Positives = 91/180 (50%), Frame = +2 [HSP Sequence] Query: 93 SGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEPRGHDDMFGAFLFDPIEEGADLGIVFMD 152 +G P + G +A+K L+ D RR +LEPRG+D + GA P+ A G++F + Sbjct: 2 TGFPELAGETIADKLDNLRTQHDQWRRACLLEPRGNDVLVGALYCAPCSADATCGVIFFN 181 ##STR25## Query: 213 NASIINVPSFLYQQDVVVVLPKPYGEVRVDIAFGGNFFAIVPAEQLGIDISVQNLSRLQE 272 ++ NVP++ Y+Q V V +P +G V DIA+GGN+F +V G + + N+ L + Sbjct: 329 --TLRNVPAYRYRQQVPVDVPG-HGRVYGDIAWGGNWFFLVSDH--GQALQMDNVEALTD 493 Score = 91 (37.1 bits), Expect = 2.5e-31, Sum P(3) = 2.5e-31 Identities = 24/69 (34%), Positives = 34/69 (49%), Frame = +3 [HSP Sequence] ##STR26## Query: 367 VLGE-ERIP 374 E +R+P Sbjct: 759 YQWECKRVP 785 Score = 48 (22.0 bits), Expect = 2.5e-31, Sum P(3) = 2.5e-31 Identities = 11/28 (39%), Positives = 16/28 (57%), Frame = +3 [HSP Sequence] Query: 391 VTAEITGKAFIMGFNTMLFDPTDPFKNG 418 V IT +A++ +T+L D DPF G Sbjct: 780 VPPSITRRAYMTADSTLLID*QDPFAWG 863 >LM16W5b02.q1c Score = 245 (91.3 bits), Expect = 2.3e-19, P = 2.3e-19 Identities = 62/182 (34%), Positives = 92/182 (50%), Frame = +1 [HSP Sequence] Query: 91 VTSGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEPRGHDDMFGAFLFDPIEEGADLGIVF 150 V +G P + G +A+K L+ D RR +LEPRG+D + GA P+ A G++F Sbjct: 1 VMTGFPELAGETIADKLDNLRTQHDQWRRACLLEPRGNDVLVGALYCAPVSADATCGVIF 180 ##STR27## Query: 211 VSNASIINVPSFLYQQDVVVVLPKPYGEVRVDIAFGGNFFAIVPAEQLGIDISVQNLSRL 270 ++ NVP++ Y+Q V V +P +G V DIA+GGN+F +V G + + N+ L Sbjct: 334 ----TLRNVPAYRYRQQVPVDVPG-HGRVYGDIAWGGNWFFLVSDH--GQALQMDNVEAL 492 Query: 271 QE 272 + Sbjct: 493 TD 498 11. Trypanosoma brucei SANGER >tryp_IXb-28b06.q1c Score = 305 (112.4 bits), Expect = 5.4e-27, P = 5.4e-27 Identities = 61/142 (42%), Positives = 84/142 (59%), Frame = -2 [HSP Sequence] Query: 20 RIVTSGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEPRGHDDMFGAFLFDPIEEGADLGI 79 RI+T G+P I G E++AY E++DYLR +M EPRGH M+G + P AD G+ Sbjct: 421 RIITGGVPEIKGETQLERRAYCMEHLDYLREILMYEPRGHHGMYGCIITPPASAHADFGV 242 ##STR28## Query: 140 SEVSNASIINVPSFLYQQDVVI 161 SEV + S NVPSF+Y++DV I Sbjct: 76 SEVESVSFENVPSFVYKKDVPI 11 >tryp_IXb-28b06.p1c [Full Sequence] Score = 296 (109.3 bits), Expect = 4.8e-26, P = 4.8e-26 Identities = 59/140 (42%), Positives = 82/140 (58%), Frame = +1 [HSP Sequence] Query: 22 VTSGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEPRGHDDMFGAFLFDPIEEGADLGIVF 81 +T G+P I G E++AY E++DYLR +M EPRGH M+G + P AD G++F Sbjct: 10 ITGGVPEIKGETQLERRAYCMEHLDYLREILMYEPRGHHGMYGCIITPPASAHADFGVLF 189 ##STR29## Query: 142 VSNASIINVPSFLYQQDVVI 161 V + S NVPSF+Y++DV I Sbjct: 355 VESVSFENVPSFVYKKDVPI 414 12. Trypansoma congolense SANGER>congo208e06.p1kw [Full Sequence] Length = 478 Plus Strand HSPs: Score = 104 (41.7 bits), Expect = 0.00070, P = 0.00070 Identities = 31/103 (30%), Positives = 56/103 (54%), Frame = +3 [HSP Sequence] Query: 187 FGGNFFAIVPAEQLGIDISVQNLSRLQEAGELLRTEINRSVKVQHPQLPHINTVDCVEIY 246 +GGN+F +V G ++ + N+ L + + +N +++ Q + +D +E++ Sbjct: 54 WGGNWFFLVSDH--GHELQMDNVEALTDYTWAM---LN-ALEAQGIRGADGALIDHIELF 215 ##STR30## SANGER>congo208e06.p1k [Full Sequence] Length = 164 Plus Strand HSPs: Score = 78 (32.5 bits), Expect = 0.085, P = 0.082 Identities = 19/55 (34%), Positives = 34/55 (61%), Frame = +3 [HSP Sequence] Query: 160 NVPSFLYQQDVVVVLPKPYGEVRVDIAFGGNFFAIVPAEQLGIDISVQNLSRLQE 214 +VP++ Y++ V V +P +G V DIA+GGN+F +V G ++ + N+ L + Sbjct: 3 HVPAYRYRKQVPVEVPG-HGVVLGDIAWGGNWFFLVSDH--GHELQMDNVEALTD 158 13. Trypanosoma vivax SANGER>Tviv655d02.p1k 4405 bp, 11 reads, 51.90 AT [Full Sequence] Length = 4405 Plus Strand HSPs: Score = 403 (146.9 bits), Expect = 4.3e-37, P = 4.3e-37 Identities = 77/117 (65%), Positives = 91/117 (77%), Frame = +1 [HSP Sequence] Query: 174 LPKPYGEVRVDIAFGGNFFAIVPAEQLGIDISVQNLSRLQEAGELLRTEINRSVKVQHPQ 233 LP PYG+ V I+FGG+FFA++ A QL + + +LS LQ G LLR +NR V VQHPQ Sbjct: 34 LPHPYGKYAV-ISFGGSFFALIDAAQLQLTVDKGHLSTLQHVGGLLRDTLNRNVSVQHPQ 210 ##STR31## SANGER>Tviv380d6.p1k Score = 156 bits (395), Expect = 1e-36 Identities = 70/106 (66%), Positives = 88/106 (82%) Query: 67 REIMRFKKSFTCIDMHTEGEAARIVTSGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEPR 126 R +M+F + TCIDMHT GE ARIVTSG P+IPG+++ EK+ +LQ +MD++RR +MLEPR Sbjct: 41 RVVMQFTGTMTCIDMHTAGEPARIVTSGFPNIPGASLVEKRDHLQRHMDHIRRRVMLEPR 100 ##STR32## 14. Vibrio parahaemolyticus >EM_PRO:AP005077 AP005077.1 Vibrio parahaemolyticus DNA, chromosome 1, complete sequence, 5/11. Length = 299, 130 Minus Strand HSPs: Score = 616 (221.9 bits), Expect = 6.2e-57, P = 6.2e-57 Identities = 134/357 (37%), Positives = 207/357 (57%), Frame = -2 I Query: 66 KREIMRFKKSFTCIDMHTEGEAARIVTSGLPHIPGSNMAEKKAYLQENMDYLRRGIMLEP 125 K MR + +F CID HT G R+V G+P + G+ M+EK+ Y E+ D++R+ +M EP Sbjct: 210923 KERKMR-QGTFFCIDAHTCGNPVRLVAGGVPPLEGNTMSEKRQYFLEHYDWIRQALMFEP 210747 ##STR33## Query: 186 VPVVLDTPAGLVRGTAHLQSGTESEVSNASIINVPSFLYQQDVVVVLPKPYGEVRVDIAF 245 + +LD PAG + H Q+ + +V++ I NVP++L QDV V + + GE+ VD+A+ Sbjct: 210569 L--ILDVPAGQIE--VHYQTKGD-KVTSVKIFNVPAYLAHQDVTVEI-EGLGEITVDVAY 210408 Query: 246 GGNFFAIVPAEQLGIDISVQNLSRLQEAGELLRTEINRSVKVQHPQLPHINTVDCVEIYG 305 GGN++ IV ++ + + + +RT ++++V+ HP P + V V G Sbjct: 210407 GGNYYVIVDPQENTAGLEHYSPDEILMLSPKVRTAVSKAVECIHPNDPTVCGVSHVLWTG 210228 ##STR34## Query: 366 RVLGEERIPGVKVPVTKDAEEGMLVVTAEITGKAFIMGFNTMLFDPTDPFKNGFTLK 422 R+ G +T+ G + I G A + G NT+ D DP+ GF +K Sbjct: 210047 RIEG----------ITE--VNGQTAILPSIEGWAQVYGHNTIWVDDEDPYAYGFEVK 209913
REFERENCES
[0242] The following references are incorporated by reference, in their entirety, herein. [0243] 1. Lamzin, V. S., Dauter, Z., and Wilson, K. S. (1995) Curr Opin Struct Biol 5, 830-836. [0244] 2. Kleinkauf, H., and von Dohren, H. (1987) Annu. Rev. Microbiol. 41, 259-289 [0245] 3. Fisher, G. H. (1998) Exs 85, 109-118 [0246] 4. Nagata, Y., Fujiwara, T., Kawaguchi-Nagata, K., Fukumori, Y., and Yamanaka, T. (1998) Biochim Biophys Acta 1379, 76-82. [0247] 5. Nagata, Y., Tanaka, K., Iida, T., Kera, Y., Yamada, R., Nakajima, Y., Fujiwara, T., Fukumori, Y., Yamanaka, T., Koga, Y., Tsuji, S., and Kawaguchi-Nagata, K. (1999) Biochim Biophys Acta 1435, 160-166. [0248] 6. Oguri, S., Kumazaki, M., Kitou, R., Nonoyama, H., and Tooda, N. (1999) Biochim Biophys Acta 1472, 107-114. [0249] 7. Neidle, A., and Dunlop, D. S. (1990) Life sci. 46, 1512-1522 [0250] 8. Schell, M. J., Molliver, M. E., and Snyder, S. H. (1995) Proc. Nat. Acad. Sci. 92, 3948-3952 [0251] 9. Wolosker, H., Blackshaw, S., and Snyder, S. H. (1999) Proc Natl Acad Sci USA 96, 13409-13414. [0252] 10. Nagata, Y., Homma, H., Matsumoto, M., and Imai, K. (1999) FEBS 454, 317-320 [0253] 11. Cardinale, G. J., and Abeles, R. H. (1968) Biochemistry 7, 3970-3978 [0254] 12. Rudnick, G., and Abeles, R. H. (1975) Biochemistry 14, 4515-4522 [0255] 13. Reina-San-Martin, B., Degrave, W., Rougeot, C., Cosson, A., Chamond, N., Cordeiro-da-Silva, A., Arala-Chaves, M., and Minoprio, P. (2000) Nature Medicine 6, 890-897 [0256] 14. Cano, M. I., Gruber, A., Vazquez, M., Cortes, A., Levin, M. J., Gonzalez, A., Degrave, W., Rondinelli, E., Zingales, B., Ramirez, J. L., Alonso, C., Requena, J. M., and Silveira, J. F. d. (1995) Mol. Bio. Par. 71, 273-278 [0257] 15. Higuchi, R., Krummel, B., and Saiki, K. K. (1988) Nuc. Ac. Res. 16, 7351-7367 [0258] 16. Keenan, M. V., and Alworth, W. L. (1974) Biochem Biophys Res Commun 57, 500-504. [0259] 17. Fisher, L. M., Albery, W. J., and Knowles, J. R. (1986) Biochemistry 25, 2529-2537. [0260] 18. Albery, W. J., and Knowled, J. R. (1986) Biochemistry 25, 2572-2577. [0261] 19. Breitbart, R. E., Andreadis, A., and Nadal-Ginard, B. (1987) Annu. Rev. Biochem. 56, 467-495 [0262] 20. Manning-Cela, R., Gonzalez, A., and Swindle, J. (2002) Infect Immun. 70, 4726-4728 [0263] 21. Krassner, S. M., and Flory, B. (1972) J. Protozool. 19, 917-920 [0264] 22. Bowman, I. B. R., Srivastava, H. K., and Flynn, I. W. (1972) Adaptation in oxidative metabolism during the transformation of Trypanosoma rhodesiense from bloodstream into culture forms, Van den Bossche H. Ed. Comparative Biochemistry of parasites, Academic Press, New York [0265] 23. Evans, D. A., and Brown, R. C. (1972) J. Protozool. 19, 686-690 [0266] 24. Auerswald, L., Schneider, P., and Gade, G. (1998) J. Exp. Biol. 201, 2333-2342 [0267] 25. Sylvester, D., and Krassner, S. M. (1976) Comp. Biochem. Physiol. 55B, 443-447 [0268] 26. de Isola, E. L., Lammel, E. M., Katzin, V. J., and Gonzalez Cappa, S. M. (1981) J. Parasitol. 67, 53-58 [0269] 27. Contreras, V. T., Salles, J. M., Thomas, N., Morel, C. M., and Goldenberg, S. (1985) Mol. Biochem. Parasitol. 16, 315-327 [0270] 28. Silber, A. M., Tonelli, R. R., Martinelli, M., Colli, W., and Alves, M. J. (2002) J. Eukaryot. Microbiol. 49, 441-446 [0271] 29. Janeway, C. A., and Humphrey, J. H. (1970) Folia Biol. 16, 156-172 [0272] 30. Mozes, E., Kohn, L. D., Hakim, F., and Singer, D. S. (1993) Science 261, 91-92 [0273] 31. Sela, M., and Zisman, E. (1997) FASEB J. 11, 449-456 [0274] 32. Contreras, V. T., Morel, C. M., and Goldenberg, S. (1985) Mol Biochem Parasitol 14, 83-96 [0275] 33. Souto-Padron, T., Reyes, M. B., Leguizamon, S., Campetella, O. E., Frash, A. C., and de Souza, W. (1989) Eur. J. Cell Biol. 50, 272-278 [0276] 34. Janes, B. K., and Bender, R. A. (1998) J Bacteriol 180, 563-570. [0277] 35. de Jong, M. H., van der Drift, C., and Vogels, G. D. (1975) J. Bacteriol. 123, 824-827 [0278] 36. Shakibaei, M., and Frevert, U. (1996) J. Exp. Med. 184, 1699-1711 [0279] 37. Burleigh, B. A., and Andrews, N. W. (1.998) Cur. Op. Microbiol. 1, 461-465 [0280] 38. Gao, W., Wortis, H. H., and Pereira, M. A. (2002) Internat. Immunol. 14, 299-308 [0281] 39. Martin, D., Ault, B., and Nadler, J. V. (1992) Eur. J. Pharmacol. 219, 59-66 [0282] 40. Van Harreveld, A. (1980) J. Neurobiol. 11, 519-529 [0283] 41. Thompson, R. J., Bouwer, H. G., Portnoy, D. A., and Frankel, F. R. (1998) Infect Immun 66, 3552-3561. [0284] 42. Wolosker, H., Sheth, K. N., Takahashi, M., Mothet, J. P., Brady, R. O., Jr., Ferris, C. D., and Snyder, S. H. (1999) Proc Natl Acad Sci USA 96, 721-725. [0285] 43. Watanabe, T., Shibata, K., Kera, Y., and Yamada, R. (1998) Amino Acids 14, 353-360 [0286] 1 GenBank accession number AF1 95522 [0287] 2 GenBank accession number AY140947 [0288] 3 EMBL accession number E10199. [0289] 4 The proline racemase/B-cell mitogen of Trypanosoma cruzi is a virulence factor whose mRNA is differentially regulated through development by alternative splicing. N. Chamond, N. Coatnoan, J. C. Barale, A. Cosson, A. Berneman, W. Degrave and P. Minoprio. Manuscript in preparation. Bibliography Related to the Microtest Using D-Amino Acid Oxidase According to the Invention: [0290] 1. Reina-San-Martin B., Degrave W., Rougeot C., Cosson A., Chamond N., Cordeiro-da-Silva A., Arala-Chaves M. & Minoprio P. (2000) A B-cell mitogen from a pathogenic trypanosome is a eukaryotic proline racemase. Nature Medicine, 6, 890. [0291] 2. Chamond N., Gregoire C., Coatnoan N., Rougeot C., Freitas-Junior L. H., da Silveira J. F., Degrave W. M. & Minoprio P. (2003) Biochemical characterization of proline racemases from the human protozoan parasite Trypanosoma cruzi and definition of putative protein signatures. J Biol Chem, 278, 15484. [0292] 3. Chamond N., Coatnoan N. & Minoprio P. (2002) Immunotherapy of Trypanosoma cruzi infections. Current Drug Targets, 2, 247. [0293] 4. Rassi A. & Luquetti A. O. (1992) Therapy of Chagas Disease. In: Chagas Disease (American Trypanosomiasis): its impact on transfusion and clinical medicine (ed. S. Wendel, Z. Brener, M. E. Camargo & A. Rassi), p. 237. ISBT Brazil'92-SBHH, Sao Paulo. [0294] 5. Cancado J. R. (1999) Criteria of Chagas disease cure. Mem Inst Oswaldo Cruz, 94 Suppl 1, 331. [0295] 6. Urbina J. A. (2001) Specific treatment of Chagas disease: current status and new developments. Curr Opin Infect Dis, 14, 733. [0296] 7. Urbina J. A. (2002) Chemotherapy of Chagas disease. Curr Pharm Des, 8, 287. [0297] 8. Donald, G. & McNeil Jr. (2003) Rare Infection Threatens to Spread in Blood Supply, in New York Times, Nov. 18, 2003 [0298] 9. Beard, C., Pye, G. Steurer, F. J., Rodriguez, R., Campman, R., Townsend Peterson, A., Ramsey, J., Wirtz, R. A. & Robinson, L. E. Chagas Disease in a Domestic Transmission Cycle, Southern Texas, USA. (2003) Emerging Infectious Diseases, 9, 103. [0299] 10. Hamase K., Morikawa A. & Zaitsu K. (2002) D-amino acids in mammals and their diagnostic value. J Chromatogr B Analyt Technol Biomed Life Sci, 781, 73. [0300] 11. D'Aniello A., Lee J. M., Petrucelli L. & Di Fiore M. M. (1998) Regional decreases of free D-aspartate levels in Alzheimer's disease. Neurosci Lett, 250, 131. [0301] 12. D'Aniello A., Di Fiore M. M., Fisher G. H., Milone A., Seleni A., D'Aniello S., Perna A. F. & Ingrosso D. (2000) Occurence of D-aspartic acid and N-methyl-D-aspartic acid in rat neuroendocrine tissues and their role in the modulation of luteinizing hormone and growth hormone release. FASEB J, 14, 699. [0302] 13. Fisher G. H., D'Aniello A., Vetere A., Padula L., Cusano G. P. & Man E. H. (1991) Free D-aspartate and D-alanine in normal and Alzheimer brain. Brain Res Bull, 26, 983. [0303] 14. Fisher G.H., Torres D., Bruna J., Cerwinski S., Martin T., Bergljung C., Gruneiro A., Chou S. J., Man E. H. & Pappatheodorou S. (1995) Presence of D-aspartate and D-glutamate in tumor proteins. Cancer Biochem Biophys, 15, 79. [0304] 15. Fisher G., Lorenzo N., Abe H., Fujita E., Frey W. H., Emory C., Di Fiore M. M. & A D. A. (1998) Free D- and L-amino acids in ventricular cerebrospinal fluid from Alzheimer and normal subjects. Amino Acids, 15, 263. [0305] 16. Fisher G.H. (1998) Appearance of D-amino acids during aging: D-amino acids in tumor proteins. Exs, 85, 109. [0306] 17. Nagata Y., Akino T., Ohno K., Kataoka Y., Ueda T., Sakurai T., Shiroshita K. & Yasuda T. (1987) Free D-amino acids in human plasma in relation to senescence and renal diseases. Clin Sci (Colch), 73, 105. [0307] 18. Nagata Y., Masui R. & Akino T. (1992) The presence of free D-serine, D-alanine and D-proline in human plasma. Experientia, 48, 986. [0308] 19. Chouinard M. L., Gaitan D. & Wood P.L. (1993) Presence of the N-methyl-D-aspartate-associated glycine receptor agonist, D-serine, in human temporal cortex: comparison of normal, Parkinson, and Alzheimer tissues. J Neurochem, 61, 1561. [0309] 20. Kumashiro S., Hashimoto A. & Nishikawa T. (1995) Free D-serine in post-mortem brains and spinal cords of individuals with and without neuropsychiatric diseases. Brain Res, 681, 117. [0310] 21. Wellner D. and L. A. Lichtenberg, (1968); Assay of Amino acid oxidase, Methods in Enzymology XVII, "metabolism of Amino acids", 593 [0311] 22. Scannone H., D. Wellner and A. Novogrodsky, (1964), A study of amino acid oxidase specificity, using a new sensitive assay, Biochemistry, 3, 1742. [0312] 23. Kishimoto M. and Takahashi T., (2001), A spectrophotometric microplate Assay for L-amino acid oxidase, Analytical Biochemistry, 298, 136. [0313] 24. Wolosker H., Sheth K. N., Takahashi M., Mothet J.-P., Brady R. O. [0314] Jr, Ferris, C.D. and Snyder S. H., (1999), Purification of serine racemase: Biosynthesis of the neuromodulator D-Serine, Proc. Nat. Acad. Sci. USA, 96, 721
Sequence CWU
1
129 1 15 PRT Artificial Sequence Description of Artificial Sequence
Synthetic peptide motif MOD_RES (1) Ile, Val, or Leu MOD_RES (2)
Gly or Asp MOD_RES (3) Variable amino acid MOD_RES (5)..(6) Variable
amino acid MOD_RES (8) Glu, Asn, or Met MOD_RES (9)..(10) Variable amino
acid MOD_RES (11) Arg or Asp MOD_RES (12) Variable amino acid MOD_RES
(13) Val or Ile MOD_RES (14) Variable amino acid 1 Xaa Xaa Xaa His Xaa
Xaa Gly Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly 1 5
10 15 2 32 PRT Artificial Sequence Description of
Artificial Sequence Synthetic peptide motif MOD_RES (1) Asn, Ser,
or Met MOD_RES (2) Val or Ala MOD_RES (3) Glu or Pro MOD_RES (4) Ala or
Ser MOD_RES (5) Phe or Tyr MOD_RES (6)..(19) This region may encompass 13
to 14 variable amino acids MOD_RES (20) Gly or Lys MOD_RES (21)
Variable amino acid MOD_RES (22) Ile, Val, or Leu MOD_RES (23)..(24)
Variable amino acid MOD_RES (26) Ile or Val MOD_RES (27) Ala or Ser
MOD_RES (28) Tyr, Trp, or Phe MOD_RES (31) Variable amino acid MOD_RES
(32) Phe, Trp, or Tyr 2 Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 1 5 10
15 Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asp Xaa Xaa Xaa Gly Gly Xaa Xaa
20 25 30 3 14 PRT Artificial
Sequence Description of Artificial Sequence Synthetic peptide motif
MOD_RES (5) Variable amino acid MOD_RES (7) Variable amino acid MOD_RES
(9)..(10) Variable amino acid MOD_RES (12)..(13) Variable amino acid 3
Asp Arg Ser Pro Xaa Gly Xaa Gly Xaa Xaa Ala Xaa Xaa Ala 1
5 10 4 14 PRT Artificial Sequence Description of
Artificial Sequence Synthetic peptide motif MOD_RES (7) Variable
amino acid MOD_RES (9)..(10) Variable amino acid MOD_RES (12)..(13)
Variable amino acid 4 Asp Arg Ser Pro Cys Gly Xaa Gly Xaa Xaa Ala Xaa
Xaa Ala 1 5 10 5 28 DNA Artificial
Sequence Description of Artificial Sequence Synthetic
oligonucleotide 5 ctctcccatg gggcaggaaa agcttctg
28 6 23 DNA Artificial Sequence Description of Artificial
Sequence Synthetic oligonucleotide 6 ctgagctcga ccagatmtac tgc
23 7 30 DNA Artificial Sequence
Description of Artificial Sequence Synthetic oligonucleotide 7
gcggatcgct ctccaagcgg gacaggcacc 30
8 30 DNA Artificial Sequence Description of Artificial Sequence Synthetic
oligonucleotide 8 ggtgcctgtc ccgcttggag agcgatccgc
30 9 39 DNA Artificial Sequence Description of
Artificial Sequence Synthetic oligonucleotide 9 ggctatttaa
atatgtctgg acataactca attgcagcg 39 10 38 DNA
Artificial Sequence Description of Artificial Sequence Synthetic
oligonucleotide 10 cgctgcaatt gagttatgtc cagacatatt taaatagc
38 11 51 DNA Artificial Sequence Description of
Artificial Sequence Synthetic oligonucleotide CDS (1)..(51) 11 atg
gat acc ggt ggc tat tta aat atg tgt gga cat aac tca att gca 48 Met
Asp Thr Gly Gly Tyr Leu Asn Met Cys Gly His Asn Ser Ile Ala 1
5 10 15 gcg
51 Ala 12 17 PRT Artificial
Sequence Description of Artificial Sequence Synthetic peptide 12
Met Asp Thr Gly Gly Tyr Leu Asn Met Cys Gly His Asn Ser Ile Ala 1
5 10 15 Ala 13 39 DNA
Artificial Sequence Description of Artificial Sequence Synthetic
oligonucleotide 13 ggctatttaa atatgtctgg acataactca attgcagcg
39 14 51 DNA Artificial Sequence Description of
Artificial Sequence Synthetic oligonucleotide CDS (1)..(51) 14 atg
gat acc ggt ggc tat tta aat atg tct gga cat aac tca att gca 48 Met
Asp Thr Gly Gly Tyr Leu Asn Met Ser Gly His Asn Ser Ile Ala 1
5 10 15 gcg
51 Ala 15 51 DNA Artificial
Sequence Description of Artificial Sequence Synthetic
oligonucleotide 15 atggataccg gtggctattt aaatatgtgt ggacataact
caattgcagc g 51 16 17 PRT Artificial Sequence Description of
Artificial Sequence Synthetic peptide 16 Met Asp Thr Gly Gly Tyr
Leu Asn Met Ser Gly His Asn Ser Ile Ala 1 5
10 15 Ala 17 51 DNA Artificial Sequence Description
of Artificial Sequence Synthetic oligonucleotide CDS (1)..(51) 17
gtg ata ttt ggc aat cgc cag gcg gat cgc tct cca tgt ggg aca ggc 48
Val Ile Phe Gly Asn Arg Gln Ala Asp Arg Ser Pro Cys Gly Thr Gly 1
5 10 15 acc
51 Thr 18 17 PRT Artificial
Sequence Description of Artificial Sequence Synthetic peptide 18
Val Ile Phe Gly Asn Arg Gln Ala Asp Arg Ser Pro Cys Gly Thr Gly 1
5 10 15 Thr 19 30 DNA
Artificial Sequence Description of Artificial Sequence Synthetic
oligonucleotide 19 gcggatcgct ctccaagcgg gacaggcacc
30 20 51 DNA Artificial Sequence Description of
Artificial Sequence Synthetic oligonucleotide 20 gtgatatttg
gcaatcgcca ggcggatcgc tctccatgtg ggacaggcac c 51 21 51 DNA
Artificial Sequence Description of Artificial Sequence Synthetic
oligonucleotide CDS (1)..(51) 21 gtg ata ttt ggc aat cgc cag gcg gat cgc
tct cca agc ggg aca ggc 48 Val Ile Phe Gly Asn Arg Gln Ala Asp Arg
Ser Pro Ser Gly Thr Gly 1 5 10
15 acc
51 Thr 22 17 PRT Artificial Sequence Description of Artificial
Sequence Synthetic peptide 22 Val Ile Phe Gly Asn Arg Gln Ala Asp
Arg Ser Pro Ser Gly Thr Gly 1 5 10
15 Thr 23 16 PRT Artificial Sequence Description of Artificial
Sequence Synthetic peptide motif MOD_RES (1) Ile, Val, or Leu
MOD_RES (2) Gly or Asp MOD_RES (3) Variable amino acid MOD_RES (5)..(6)
Variable amino acid MOD_RES (8) Glu, Asn, or Met MOD_RES (9)..(10)
Variable amino acid MOD_RES (11) Arg or Asp MOD_RES (12) Variable amino
acid MOD_RES (13) Val or Ile MOD_RES (14)..(15) Variable amino acid 23
Xaa Xaa Xaa His Xaa Xaa Gly Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly 1
5 10 15 24 14 PRT Artificial
Sequence Description of Artificial Sequence Synthetic peptide motif
MOD_RES (5) Variable amino acid MOD_RES (7) Variable amino acid MOD_RES
(8) Gly or Ala MOD_RES (9)..(10) Variable amino acid MOD_RES (12)..(13)
Variable amino acid 24 Asp Arg Ser Pro Xaa Gly Xaa Xaa Xaa Xaa Ala Xaa
Xaa Ala 1 5 10 25 5 PRT Artificial
Sequence Description of Artificial Sequence Synthetic peptide 25
Glu Pro Arg Gly His 1 5 26 6 PRT Artificial Sequence
Description of Artificial Sequence Synthetic peptide 26 Glu Pro
Arg Gly Ser Asp 1 5 27 6 PRT Artificial Sequence
Description of Artificial Sequence Synthetic peptide 27 Glu Pro
Arg Gly Asn Asp 1 5 28 1598 DNA Trypanosoma cruzi 28
cctttttctt tttaaaaaca aaaaaaattc cggggggaat atggaacagg gtatatgcgt 60
aaaagtgtct gtcccaaaca aaaatttttt ttttccgcct tcccattttt tttttttttt 120
tgtgtgtttc ccttgatctc tcgaacaggg caggaaaagc ttctgtttga ccaaaaatat 180
aaaattatta agggcgagaa aaaagaaaag aaaaaaaatc aacgagcaaa caggagagaa 240
caccaacaaa aaagggaaat tatgcgattt aagaaatcat tcacatgcat cgacatgcat 300
acggaaggtg aagcagcacg gattgtgacg agtggtttgc cacacattcc aggttcgaat 360
atggcggaga agaaagcata cctgcaggaa aacatggatt atttgaggcg tggcataatg 420
ctggaaccac gtggtcatga tgatatgttt ggagcctttt tatttgaccc tattgaagaa 480
ggcgctgact tgggcatggt attcatggat accggtggct atttaaatat gtgtggacat 540
aactcaattg cagcggttac ggcggcagtt gaaacgggaa ttgtgagcgt gccggcgaag 600
gcaacaaatg ttccggttgt cctggacaca cctgcggggt tggtgcgcgg tacggcacac 660
cttcagagtg gtactgagag tgaggtgtca aatgcgagta ttatcaatgt accctcattt 720
ttgtatcagc aggatgtggt ggttgtgttg ccaaagccct atggtgaagt acgggttgat 780
attgcatttg gaggcaattt tttcgccatt gttcccgcgg agcagttggg aattgatatc 840
tccgttcaaa acctctccag gctgcaggag gcaggagaac ttctgcgtac tgaaatcaat 900
cgcagtgtga aggttcagca ccctcagctg ccccatatta acactgtgga ctgtgttgag 960
atatacggtc cgccaacgaa cccggaggca aactacaaga acgttgtgat atttggcaat 1020
cgccaggcgg atcgctctcc atgtgggaca ggcaccagcg ccaagatggc aacactttat 1080
gccaaaggcc agcttcgcat cggagagact tttgtgtacg agagcatact cggctcactc 1140
ttccagggca gggtacttgg ggaggagcga ataccggggg tgaaggtgcc ggtgaccaaa 1200
gatgccgagg aagggatgct cgttgtaacg gcagaaatta ctggaaaggc ttttatcatg 1260
ggtttcaaca ccatgctgtt tgacccaacg gatccgttta agaacggatt cacattaaag 1320
cagtagatct ggtagagcac agaaactatt ggggaacacg tgcgaacagg tgctgctacg 1380
tgaagggtat tgaatgaatc gttttttttt atttttattt tttattttta ttagtgcatt 1440
attattaaat tttttttttg ttttggggtt tcaacggtac cgcgttggga gcagggaagc 1500
gatagcggcc ggacaatttt ttgcttttat tttcattttc atcttcctac ccaaccccct 1560
tggttccacc ggtcgcggcg gggtcttgtg ggtggagg 1598
29 1582 DNA Trypanosoma cruzi 29 gtgtgttcaa cagttttgtt tccttttttc
tctttttctc tttccatcat acatacatac 60 atacatacat atatatatct gcgtagatat
gcacatgcgt atatgcgtga agagtgtctg 120 tcccaacatt tttttttttt ttttgtgtgt
tttcccttga ttcccgaacg ggcaggaaaa 180 gcttctgttt gaccaaaaat ataaaattat
taagggcgag aaaagaaaaa aaaaatcaac 240 cgaggagaca acaccaacaa aaaagggaaa
ttatgcgatt taagaaatca ttgacatgca 300 tcgacatgca tacggaaggt gaagcagcac
ggattgtgac gagtggtttg ccacacattc 360 caggttcgaa tatggcggag aagaaagcat
acctgcagga aaacatggat tatttgaggc 420 gtggcataat gctggagcca cgtggtcatg
atgatatgtt tggagccttt ttatttgacc 480 ctattgaaga aggcgctgac ttgggcatcg
tattcatgga taccggtggc tatttaaata 540 tgtgtggaca taactcaatt gcagcggtta
cggcggcagt ggaaacggga attttgagcg 600 tgccggcgaa ggcaacaaat gttccggttg
tcctggacac acctgcgggg ttggtgcgcg 660 gtacggcaca ccttcagagt ggtactgaga
gtgaggtgtc aaatgcgagt attatcaatg 720 tgccctcatt tttgtatcag caggatgtgg
tgattgtttt gccaaagccc tatggtgagg 780 tacgggttga tattgcattt ggaggcaatt
ttttcgccat tgttcccgcg gagcacttgg 840 gaattgatat ctccgttcaa aacctctcca
ggctgcagga ggcaggagaa cttctgcgta 900 ctgaaatcaa tcgcagtgtg aaggttcagc
accctcagct gccccatatt aacactgtgg 960 actgtgttga gatatacggt ccgccaacga
acccggaggc aaaatacaag aacgttgtga 1020 tatttggcaa tcgccaggcg gatcgctctc
catgtgggac aggcaccagc gccaagatgg 1080 caacacttta tgccaaaggc cagcttcgca
tcggagagac ttttgtgtac gagagcatac 1140 tcggctcact cttccagggc agggtacttg
gggaggagcg aataccgggg gtgaaggtgc 1200 cggtgaccaa agatgccgag gaagggatgc
tcgttgtaac gacagaaatt actggaaagg 1260 cttttatcat gggtttcaac accatgctgt
ttgacccaac ggatccgttc ttaaacggat 1320 tcacactaaa gcggtagatc tggtagagca
cagaaactat tggggaacac gtgcgaacag 1380 gtgctgctac gtaaagggta ttgaatgaat
cgtttttttt tttttttttt tattagtgca 1440 ttattttttt ttttttttgt tttggggttt
caacggtacc acgttgggag cagggaaacg 1500 atagcggccg gacaattttt tacttttatt
ttcattttca ccttcctacc caaccccctt 1560 ggttccaccg gtcgcggcgg gg
1582 30 423 PRT Trypanosoma cruzi 30
Met Arg Lys Ser Val Cys Pro Lys Gln Lys Phe Phe Phe Ser Ala Phe 1
5 10 15 Pro Phe Phe Phe Phe Phe
Cys Val Phe Pro Leu Ile Ser Arg Thr Gly 20
25 30 Gln Glu Lys Leu Leu Phe Asp Gln Lys Tyr Lys Ile
Ile Lys Gly Glu 35 40 45 Lys
Lys Glu Lys Lys Lys Asn Gln Arg Ala Asn Arg Arg Glu His Gln 50
55 60 Gln Lys Arg Glu Ile Met Arg Phe Lys Lys
Ser Phe Thr Cys Ile Asp 65 70 75
80 Met His Thr Glu Gly Glu Ala Ala Arg Ile Val Thr Ser Gly Leu
Pro 85 90 95 His Ile
Pro Gly Ser Asn Met Ala Glu Lys Lys Ala Tyr Leu Gln Glu 100
105 110 Asn Met Asp Tyr Leu Arg Arg Gly Ile
Met Leu Glu Pro Arg Gly His 115 120
125 Asp Asp Met Phe Gly Ala Phe Leu Phe Asp Pro Ile Glu Glu Gly Ala
130 135 140 Asp Leu Gly Met Val Phe Met
Asp Thr Gly Gly Tyr Leu Asn Met Cys 145 150
155 160 Gly His Asn Ser Ile Ala Ala Val Thr Ala Ala Val
Glu Thr Gly Ile 165 170
175 Val Ser Val Pro Ala Lys Ala Thr Asn Val Pro Val Val Leu Asp Thr
180 185 190 Pro Ala Gly Leu Val Arg
Gly Thr Ala His Leu Gln Ser Gly Thr Glu 195 200
205 Ser Glu Val Ser Asn Ala Ser Ile Ile Asn Val Pro Ser Phe
Leu Tyr 210 215 220 Gln Gln Asp Val
Val Val Val Leu Pro Lys Pro Tyr Gly Glu Val Arg 225 230
235 240 Val Asp Ile Ala Phe Gly Gly Asn Phe
Phe Ala Ile Val Pro Ala Glu 245 250
255 Gln Leu Gly Ile Asp Ile Ser Val Gln Asn Leu Ser Arg Leu Gln
Glu 260 265 270 Ala Gly Glu
Leu Leu Arg Thr Glu Ile Asn Arg Ser Val Lys Val Gln 275
280 285 His Pro Gln Leu Pro His Ile Asn Thr Val Asp
Cys Val Glu Ile Tyr 290 295 300 Gly
Pro Pro Thr Asn Pro Glu Ala Asn Tyr Lys Asn Val Val Ile Phe 305
310 315 320 Gly Asn Arg Gln Ala Asp
Arg Ser Pro Cys Gly Thr Gly Thr Ser Ala 325
330 335 Lys Met Ala Thr Leu Tyr Ala Lys Gly Gln Leu Arg
Ile Gly Glu Thr 340 345 350
Phe Val Tyr Glu Ser Ile Leu Gly Ser Leu Phe Gln Gly Arg Val Leu
355 360 365 Gly Glu Glu Arg Ile Pro Gly
Val Lys Val Pro Val Thr Lys Asp Ala 370 375
380 Glu Glu Gly Met Leu Val Val Thr Ala Glu Ile Thr Gly Lys Ala Phe
385 390 395 400 Ile Met
Gly Phe Asn Thr Met Leu Phe Asp Pro Thr Asp Pro Phe Lys
405 410 415 Asn Gly Phe Thr Leu Lys Gln
420 31 354 PRT Trypanosoma cruzi 31 Met Arg Phe Lys Lys Ser
Leu Thr Cys Ile Asp Met His Thr Glu Gly 1 5
10 15 Glu Ala Ala Arg Ile Val Thr Ser Gly Leu Pro His
Ile Pro Gly Ser 20 25 30
Asn Met Ala Glu Lys Lys Ala Tyr Leu Gln Glu Asn Met Asp Tyr Leu
35 40 45 Arg Arg Gly Ile Met Leu Glu
Pro Arg Gly His Asp Asp Met Phe Gly 50 55
60 Ala Phe Leu Phe Asp Pro Ile Glu Glu Gly Ala Asp Leu Gly Ile Val
65 70 75 80 Phe Met
Asp Thr Gly Gly Tyr Leu Asn Met Cys Gly His Asn Ser Ile
85 90 95 Ala Ala Val Thr Ala Ala Val
Glu Thr Gly Ile Leu Ser Val Pro Ala 100 105
110 Lys Ala Thr Asn Val Pro Val Val Leu Asp Thr Pro Ala Gly
Leu Val 115 120 125 Arg Gly Thr
Ala His Leu Gln Ser Gly Thr Glu Ser Glu Val Ser Asn 130
135 140 Ala Ser Ile Ile Asn Val Pro Ser Phe Leu Tyr Gln
Gln Asp Val Val 145 150 155
160 Ile Val Leu Pro Lys Pro Tyr Gly Glu Val Arg Val Asp Ile Ala Phe
165 170 175 Gly Gly Asn Phe
Phe Ala Ile Val Pro Ala Glu His Leu Gly Ile Asp 180
185 190 Ile Ser Val Gln Asn Leu Ser Arg Leu Gln Glu
Ala Gly Glu Leu Leu 195 200 205
Arg Thr Glu Ile Asn Arg Ser Val Lys Val Gln His Pro Gln Leu Pro 210
215 220 His Ile Asn Thr Val Asp Cys Val Glu
Ile Tyr Gly Asn Ala Thr Asn 225 230 235
240 Pro Glu Ala Lys Tyr Lys Asn Val Val Ile Phe Gly Asn Arg
Gln Ala 245 250 255 Asp
Arg Ser Pro Cys Gly Thr Gly Thr Ser Ala Lys Met Ala Thr Leu
260 265 270 Tyr Ala Lys Gly Gln Leu Arg
Ile Gly Glu Thr Phe Val Tyr Glu Ser 275 280
285 Ile Leu Gly Ser Leu Phe Gln Gly Arg Val Leu Gly Glu Glu Arg
Ile 290 295 300 Pro Gly Val Lys Val
Pro Val Thr Lys Asp Ala Glu Glu Gly Met Leu 305 310
315 320 Val Val Thr Thr Glu Ile Thr Gly Lys Ala
Phe Ile Met Gly Phe Asn 325 330
335 Thr Met Leu Phe Asp Pro Thr Asp Pro Phe Leu Asn Gly Phe Thr Leu
340 345 350 Lys Arg 32 335
PRT Clostridium sticklandii 32 Met Lys Phe Ser Lys Gly Ile His Ala Ile
Asp Ser His Thr Met Gly 1 5 10
15 Glu Pro Thr Arg Ile Val Val Gly Gly Ile Pro Gln Ile Asn Gly Glu
20 25 30 Thr Met Ala Asp
Lys Lys Lys Tyr Leu Glu Asp Asn Leu Asp Tyr Val 35
40 45 Arg Thr Ala Leu Met His Glu Pro Arg Gly His Asn
Asp Met Phe Gly 50 55 60 Ser Ile
Ile Thr Ser Ser Asn Asn Lys Glu Ala Asp Phe Gly Ile Ile 65
70 75 80 Phe Met Asp Gly Gly Gly Tyr
Leu Asn Met Cys Gly His Gly Ser Ile 85
90 95 Gly Ala Ala Thr Val Ala Val Glu Thr Gly Met Val
Glu Met Val Glu 100 105 110
Pro Val Thr Asn Ile Asn Met Glu Ala Pro Ala Gly Leu Ile Lys Ala
115 120 125 Lys Val Met Val Glu Asn Glu
Lys Val Lys Glu Val Ser Ile Thr Asn 130 135
140 Val Pro Ser Phe Leu Tyr Met Glu Asp Ala Lys Leu Glu Val Pro Ser
145 150 155 160 Leu Asn
Lys Thr Ile Thr Phe Asp Ile Ser Phe Gly Gly Ser Phe Phe
165 170 175 Ala Ile Ile His Ala Lys Glu
Leu Gly Val Lys Val Glu Thr Ser Gln 180 185
190 Val Asp Val Leu Lys Lys Leu Gly Ile Glu Ile Arg Asp Leu
Ile Asn 195 200 205 Glu Lys Ile
Lys Val Gln His Pro Glu Leu Glu His Ile Lys Thr Val 210
215 220 Asp Leu Val Glu Ile Tyr Asp Glu Pro Ser Asn Pro
Glu Ala Thr Tyr 225 230 235
240 Lys Asn Val Val Ile Phe Gly Gln Gly Gln Val Asp Arg Ser Pro Cys
245 250 255 Gly Thr Gly Thr
Ser Ala Lys Leu Ala Thr Leu Tyr Lys Lys Gly His 260
265 270 Leu Lys Ile Asp Glu Lys Phe Val Tyr Glu Ser
Ile Thr Gly Thr Met 275 280 285
Phe Lys Gly Arg Val Leu Glu Glu Thr Lys Val Gly Glu Phe Asp Ala 290
295 300 Ile Ile Pro Glu Ile Thr Gly Gly Ala
Tyr Ile Thr Gly Phe Asn His 305 310 315
320 Phe Val Ile Asp Pro Glu Asp Pro Leu Lys Tyr Gly Phe Thr
Val 325 330 335 33 354
PRT Homo sapiens 33 Met Glu Ser Ala Leu Ala Val Pro Trp Leu Pro Pro His
Asp Pro Gly 1 5 10 15
Thr Pro Val Leu Ser Val Val Asp Met His Thr Gly Gly Glu Pro Leu
20 25 30 Arg Ile Val Leu Ala Gly Cys
Pro Glu Val Ser Gly Pro Thr Leu Leu 35 40
45 Ala Lys Arg Arg Tyr Met Arg Gln His Leu Asp His Val Arg Arg
Arg 50 55 60 Leu Met Phe Glu Pro
Arg Gly His Arg Asp Met Tyr Gly Ala Val Leu 65 70
75 80 Val Pro Ser Glu Leu Pro Asp Ala His Leu
Gly Val Leu Phe Leu His 85 90
95 Asn Glu Gly Tyr Ser Ser Met Cys Gly His Ala Val Leu Ala Leu Gly
100 105 110 Arg Phe Ala Leu
Asp Phe Gly Leu Val Pro Ala Pro Pro Ala Gly Thr 115
120 125 Arg Glu Ala Arg Val Asn Ile His Cys Pro Cys Gly
Leu Val Thr Ala 130 135 140 Phe Val
Ala Cys Glu Asp Gly Arg Ser His Gly Pro Val Arg Phe His 145
150 155 160 Ser Val Pro Ala Phe Val Leu
Ala Thr Asp Leu Met Val Asp Val Pro 165
170 175 Gly His Gly Lys Val Met Val Asp Ile Ala Tyr Gly
Gly Ala Phe Tyr 180 185 190
Ala Phe Val Thr Ala Glu Lys Leu Gly Leu Asp Ile Cys Ser Ala Lys
195 200 205 Thr Arg Asp Leu Val Asp Ala
Ala Ser Ala Val Thr Glu Ala Val Lys 210 215
220 Ala Gln Phe Lys Ile Asn His Pro Asp Ser Glu Asp Leu Ala Phe Leu
225 230 235 240 Tyr Gly
Thr Ile Leu Thr Asp Gly Lys Asp Ala Tyr Thr Lys Glu Pro
245 250 255 Thr Thr Asn Ile Cys Val Phe
Ala Asp Glu Gln Val Asp Arg Ser Pro 260 265
270 Thr Gly Ser Gly Val Thr Ala Arg Ile Ala Leu Gln Tyr His
Lys Gly 275 280 285 Leu Leu Glu
Leu Asn Gln Met Arg Ala Phe Lys Ser Ser Ala Thr Gly 290
295 300 Ser Val Phe Thr Gly Lys Ala Val Arg Glu Ala Lys
Cys Gly Asp Phe 305 310 315
320 Lys Ala Val Ile Val Glu Val Ser Gly Gln Ala His Tyr Thr Gly Thr
325 330 335 Ala Ser Phe Ile
Ile Glu Asp Asp Asp Pro Leu Arg Asp Gly Phe Leu 340
345 350 Leu Lys 34 354 PRT Homo sapiens 34 Met Glu
Ser Ala Leu Ala Val Pro Arg Leu Pro Pro His Asp Pro Gly 1
5 10 15 Thr Pro Val Leu Ser Val Val Asp
Met His Thr Gly Gly Glu Pro Leu 20 25
30 Arg Ile Val Leu Ala Gly Cys Pro Glu Val Ser Gly Pro Thr Leu
Leu 35 40 45 Ala Lys Arg Arg
Tyr Met Arg Gln His Leu Asp His Val Arg Arg Arg 50
55 60 Leu Met Phe Glu Pro Arg Gly His Arg Asp Met Tyr
Gly Ala Val Leu 65 70 75
80 Val Pro Ser Glu Leu Pro Asp Ala His Leu Gly Val Leu Phe Leu His
85 90 95 Asn Glu Gly Tyr
Ser Ser Met Cys Gly His Ala Val Leu Ala Leu Gly 100
105 110 Arg Phe Ala Leu Asp Phe Gly Leu Val Pro Ala
Pro Pro Ala Gly Thr 115 120 125
Arg Glu Ala Arg Val Asn Ile His Cys Pro Cys Gly Leu Val Thr Ala 130
135 140 Phe Val Ala Cys Glu Asp Gly Arg Ser
His Gly Pro Val Arg Phe His 145 150 155
160 Ser Val Pro Ala Phe Val Leu Ala Thr Asp Leu Met Val Asp
Val Pro 165 170 175 Gly
His Gly Lys Val Met Val Asp Ile Ala Tyr Gly Gly Ala Phe Tyr
180 185 190 Ala Phe Val Thr Ala Glu Lys
Leu Gly Leu Asp Ile Cys Ser Ala Lys 195 200
205 Thr Arg Asp Leu Val Asp Ala Ala Ser Ala Val Thr Glu Ala Val
Lys 210 215 220 Ala Gln Phe Lys Ile
Asn His Pro Asp Ser Glu Asp Leu Ala Phe Leu 225 230
235 240 Tyr Gly Thr Ile Leu Thr Asp Gly Lys Asp
Ala Tyr Thr Lys Glu Pro 245 250
255 Thr Thr Asn Ile Cys Val Phe Ala Asp Glu Gln Val Asp Arg Ser Pro
260 265 270 Thr Gly Ser Gly
Val Thr Ala Arg Ile Ala Leu Gln Tyr His Lys Gly 275
280 285 Leu Leu Glu Leu Asn Gln Met Arg Ala Phe Lys Ser
Ser Ala Thr Gly 290 295 300 Ser Val
Phe Thr Gly Lys Ala Val Arg Glu Ala Lys Cys Gly Asp Phe 305
310 315 320 Lys Ala Val Ile Val Glu Val
Ser Gly Gln Ala His Tyr Thr Gly Thr 325
330 335 Ala Ser Phe Ile Ile Glu Asp Asp Asp Pro Leu Arg
Asp Gly Phe Leu 340 345 350
Leu Lys 35 354 PRT Mus musculus 35 Met Glu Ala Ala Leu Ala Val Thr Arg
Leu Pro Pro Asn Asp Pro Arg 1 5 10
15 Thr Pro Ala Leu Ser Val Val Asp Met His Thr Gly Gly Glu Pro
Leu 20 25 30 Arg Ile Val
His Ala Gly Cys Pro Glu Val Ala Gly Pro Thr Leu Leu 35
40 45 Ala Lys Arg Arg Tyr Met Arg Gln His Leu Asp
Tyr Ile Arg Arg Arg 50 55 60 Leu
Val Phe Glu Pro Arg Gly His Arg Asp Met Tyr Gly Ala Ile Leu 65
70 75 80 Val Pro Ser Glu Leu Pro
Asp Ala His Leu Gly Val Leu Phe Leu His 85
90 95 Asn Glu Gly Tyr Ser Ser Met Cys Gly His Ala Val
Leu Ala Leu Gly 100 105 110
Arg Phe Ala Leu Asp Phe Gly Leu Val Pro Ala Pro Pro Lys Gly Ala
115 120 125 Arg Glu Ala Gln Val Asn Ile
His Cys Pro Cys Gly Leu Val Thr Ala 130 135
140 Phe Val Glu Cys Glu Gly Gly Arg Ser Cys Gly Pro Val Arg Phe His
145 150 155 160 Ser Val
Pro Ala Phe Val Leu Ala Ser Asp Leu Thr Val Asp Val Pro
165 170 175 Gly His Gly Lys Val Leu Val
Asp Ile Ala Tyr Gly Gly Ala Phe Tyr 180 185
190 Ala Phe Val Ser Ala Glu Lys Leu Gly Leu Asp Val Cys Ser
Ala Lys 195 200 205 Thr Arg Asp
Leu Val Asp Ala Ala Ser Ala Leu Thr Gly Ala Val Lys 210
215 220 Ala Gln Phe Lys Ile Asn His Pro Glu Ser Glu Asp
Leu Gly Phe Leu 225 230 235
240 Tyr Gly Ser Ile Leu Thr Asp Gly Lys Asp Ala Tyr Ser Glu Glu Ala
245 250 255 Thr Thr Asn Ile
Cys Val Phe Ala Asp Glu Gln Val Asp Arg Ser Pro 260
265 270 Thr Gly Ser Gly Val Thr Ala Arg Ile Ala Leu
Gln Tyr His Lys Gly 275 280 285
Leu Leu Gln Leu Asn Gln Thr Arg Ala Phe Lys Ser Ser Ala Thr Gly 290
295 300 Ser Val Phe Thr Gly Cys Ala Val Arg
Glu Ala Lys Cys Gly Asp Phe 305 310 315
320 Lys Ala Val Ile Val Glu Val Ala Gly Gln Ala His Tyr Thr
Gly Thr 325 330 335 Ala
Asn Leu Thr Val Glu Asp Gly Asp Pro Leu Arg Asp Gly Phe Leu
340 345 350 Leu Lys 36 354 PRT Mus
musculus 36 Met Glu Ala Ala Leu Ala Val Thr Arg Leu Pro Pro His Asp Ser
Arg 1 5 10 15 Thr Pro
Ala Leu Ser Val Val Asp Met His Thr Gly Gly Glu Pro Leu 20
25 30 Arg Ile Val His Ala Gly Cys Pro Glu
Val Ala Gly Pro Thr Leu Leu 35 40
45 Ala Lys Arg Arg Tyr Met Arg Gln His Leu Asp Tyr Ile Arg Arg Arg
50 55 60 Leu Val Phe Glu Pro Arg Gly
His Arg Asp Met Tyr Gly Ala Ile Leu 65 70
75 80 Met Pro Ser Glu Leu Pro Asp Ala His Leu Gly Val
Leu Phe Leu His 85 90
95 Asn Glu Gly Tyr Ser Ser Met Cys Gly His Ala Val Leu Ala Leu Gly
100 105 110 Arg Phe Ala Leu Asp Phe
Gly Leu Val Pro Ala Pro Pro Glu Gly Ala 115 120
125 Arg Glu Ala Gln Val Asn Ile His Cys Pro Cys Gly Leu Val
Thr Ala 130 135 140 Phe Val Glu Cys
Glu Gly Gly Arg Ser Cys Gly Pro Val Arg Phe His 145 150
155 160 Ser Val Pro Ala Phe Val Leu Ala Ser
Asp Leu Thr Val Asp Val Pro 165 170
175 Gly His Gly Lys Val Leu Val Asp Ile Ala Tyr Gly Gly Ala Phe
Tyr 180 185 190 Ala Phe Val
Ser Ala Glu Lys Leu Gly Leu Asp Val Cys Ser Ala Lys 195
200 205 Thr Arg Asp Leu Val Asp Ala Ala Ser Ala Leu
Thr Gly Ala Val Lys 210 215 220 Ala
Gln Phe Lys Ile Asn His Pro Glu Ser Glu Asp Leu Gly Phe Leu 225
230 235 240 Tyr Gly Ser Ile Leu Thr
Asp Gly Lys Asp Ala Tyr Ser Glu Glu Ala 245
250 255 Thr Thr Asn Ile Cys Val Phe Ala Asp Glu Gln Val
Asp Arg Ser Pro 260 265 270
Thr Gly Ser Gly Val Thr Ala Arg Ile Ala Leu Gln Tyr His Lys Gly
275 280 285 Leu Leu Gln Leu Asn Gln Thr
Arg Ala Phe Lys Ser Ser Ala Thr Gly 290 295
300 Ser Val Phe Thr Gly Cys Ala Val Arg Glu Ala Lys Cys Gly Asp Phe
305 310 315 320 Lys Ala
Val Ile Val Glu Val Ala Gly Gln Ala His Tyr Thr Gly Thr
325 330 335 Ala Asn Leu Thr Val Glu Asp
Gly Asp Pro Leu Arg Asp Gly Phe Leu 340 345
350 Leu Lys 37 333 PRT Rhizobium loti 37 Met Ala Lys Lys
Ser Phe Phe Cys Ile Asp Gly His Thr Cys Gly Asn 1 5
10 15 Pro Val Arg Leu Val Ala Gly Gly Gly Pro
Leu Leu Glu Gly Ser Thr 20 25
30 Met Met Glu Arg Arg Ala His Phe Leu Ala Glu Tyr Asp Trp Ile Arg
35 40 45 Thr Gly Leu Met Phe Glu
Pro Arg Gly His Asp Val Met Ser Gly Ser 50 55
60 Ile Leu Tyr Pro Pro Thr Arg Glu Asp Cys Asp Ile Ala Ile Leu
Phe 65 70 75 80 Ile
Glu Thr Ser Gly Cys Leu Pro Met Cys Gly His Gly Thr Ile Gly
85 90 95 Thr Val Thr Met Ala Ile Glu
His Gly Leu Ile Lys Pro Lys Thr Pro 100 105
110 Gly Val Leu Arg Leu Asp Thr Pro Ala Gly Leu Val Ile Ala
Glu Tyr 115 120 125 Lys Gln Val
Gly Glu Tyr Val Glu Glu Val Arg Ile Thr Asn Val Pro 130
135 140 Ser Phe Leu His Ala Glu Gly Leu Thr Val Glu Cys
Pro Gly Leu Gly 145 150 155
160 Glu Ile Thr Val Asp Val Ala Tyr Gly Gly Asn Phe Tyr Ala Ile Val
165 170 175 Glu Pro Gln Glu
Asn Tyr Arg Asp Met Ala Asp His Ser Ala Gly Asp 180
185 190 Leu Ile Ala Trp Ser Pro Val Val Arg Gln Arg
Leu Asn Glu Lys Tyr 195 200 205
Ser Phe Val His Pro Glu Asn Pro Gly Ile Asn Arg Leu Ser His Met 210
215 220 Leu Trp Thr Gly Lys Pro Thr Val Glu
Gly Ala Asp Ala Arg Asn Ala 225 230 235
240 Val Phe Tyr Gly Asp Lys Ala Ile Asp Arg Ser Pro Cys Gly
Thr Gly 245 250 255 Thr
Ser Ala Arg Met Ala Gln Leu His Ala Lys Gly Lys Leu Lys Ala
260 265 270 Gly Asp Ala Phe Val His Glu
Ser Ile Ile Gly Ser Leu Phe Lys Gly 275 280
285 Lys Val Glu Lys Glu Val Thr Val Ala Gly Lys Pro Ala Ile Ile
Pro 290 295 300 Ser Ile Gly Gly Trp
Ala Arg Leu Thr Gly Leu Asn Thr Ile Phe Ile 305 310
315 320 Asp Asp Arg Asp Pro Phe Ala His Gly Phe
Val Val Thr 325 330 38 333 PRT Brucella
melitensis 38 Met Ala Arg His Ser Phe Phe Cys Val Asp Gly His Thr Cys
Gly Asn 1 5 10 15 Pro
Val Arg Leu Val Ala Gly Gly Gly Pro Asn Leu Asn Gly Ser Thr
20 25 30 Met Met Glu Lys Cys Ala His
Phe Leu Ala Glu Tyr Asp Trp Ile Arg 35 40
45 Thr Gly Leu Met Phe Glu Pro Arg Gly His Asp Met Met Ser Gly
Ser 50 55 60 Ile Leu Tyr Pro Pro
Thr Arg Pro Asp Cys Asp Val Ala Val Leu Phe 65 70
75 80 Ile Glu Thr Ser Gly Cys Leu Pro Met Cys
Gly His Gly Thr Ile Gly 85 90
95 Thr Val Thr Met Ala Ile Glu Gln Gly Leu Val Thr Pro Lys Thr Pro
100 105 110 Gly Lys Leu Asn
Leu Asp Thr Pro Ala Gly Leu Val Ala Ile Glu Tyr 115
120 125 Glu Gln Asp Gly Gln Tyr Val Glu Arg Val Arg Leu
Thr Asn Val Pro 130 135 140 Ala Phe
Leu Tyr Ala Glu Gly Leu Glu Val Glu Cys Pro Asp Leu Gly 145
150 155 160 Pro Ile Lys Val Asp Val Ala
Tyr Gly Gly Asn Phe Tyr Ala Ile Val 165
170 175 Glu Pro Gln Glu Asn Tyr Thr Asp Met Asp Asp Tyr
Ser Ala Leu Gln 180 185 190
Leu Ile Ala Trp Ser Pro Val Leu Arg Gln Arg Leu Asn Glu Lys Tyr
195 200 205 Lys Phe Gln His Pro Glu Leu
Pro Asp Ile Asn Arg Leu Ser His Ile 210 215
220 Leu Trp Thr Gly Lys Pro Lys His Pro Gln Ala His Ala Arg Asn Ala
225 230 235 240 Val Phe
Tyr Gly Asp Lys Ala Ile Asp Arg Ser Pro Cys Gly Thr Gly
245 250 255 Thr Ser Ala Arg Met Ala Gln
Leu Ala Ala Lys Gly Lys Leu Lys Pro 260 265
270 Gly Asp Glu Phe Ile His Glu Ser Ile Ile Gly Ser Leu Phe
His Gly 275 280 285 Arg Val Glu
Arg Ala Ala Glu Val Ala Gly Arg Pro Ala Ile Val Pro 290
295 300 Ser Ile Ala Gly Trp Ala Arg Met Thr Gly Tyr Asn
Thr Ile Phe Ile 305 310 315
320 Asp Asp Arg Asp Pro Phe Ala His Gly Phe Ser Ala Ala
325 330 39 333 PRT Rhizobium meliloti 39 Met Ala Thr His
Thr Phe Ser Cys Ile Asp Gly His Thr Cys Gly Asn 1 5
10 15 Pro Val Arg Leu Val Ser Gly Gly Gly Pro
Arg Leu Glu Gly Ala Asn 20 25
30 Met Leu Glu Lys Arg Ala His Phe Leu Lys Glu Phe Asp Trp Ile Arg
35 40 45 Thr Gly Leu Met Phe Glu
Pro Arg Gly His Asp Met Met Ser Gly Ser 50 55
60 Ile Leu Tyr Pro Pro Thr Arg Pro Asp Cys Asp Val Ala Val Leu
Phe 65 70 75 80 Ile
Glu Thr Ser Gly Cys Leu Pro Met Cys Gly His Gly Thr Ile Gly
85 90 95 Thr Ile Thr Met Gly Ile Glu
Asn Gly Leu Ile Thr Pro Arg Glu Pro 100 105
110 Gly Lys Leu Ser Ile Asp Ala Pro Ala Gly Lys Val Asp Ile
Thr Tyr 115 120 125 Arg Gln Glu
Gly Arg Phe Val Glu Glu Val Arg Leu Thr Asn Val Pro 130
135 140 Ser Phe Leu Tyr Ala Glu Gly Leu Ala Ala Glu Val
Glu Gly Leu Gly 145 150 155
160 Glu Ile Val Val Asp Val Ala Tyr Gly Gly Asn Phe Tyr Ala Ile Val
165 170 175 Glu Pro Gln Lys
Asn Phe Arg Asp Met Ala Asp His Thr Ala Gly Glu 180
185 190 Leu Val Gly Trp Ser Pro Lys Leu Arg Ala Ala
Leu Asn Ala Lys Tyr 195 200 205
Glu Phe Val His Pro Glu His Pro Glu Ile Arg Gly Leu Ser His Ile 210
215 220 Gln Trp Thr Gly Lys Pro Thr Gln Pro
Glu Ala His Ala Arg Asn Ala 225 230 235
240 Val Phe Tyr Gly Glu Lys Ala Ile Asp Arg Ser Pro Cys Gly
Thr Gly 245 250 255 Thr
Ser Ala Arg Ile Ala Gln Leu Ala Ala Lys Gly Lys Leu Lys Val
260 265 270 Gly Asp Glu Phe Val His Glu
Ser Ile Ile Gly Ser Leu Phe Lys Gly 275 280
285 Arg Val Glu Ala Ala Ala Lys Val Ala Asp Arg Asp Ala Ile Ile
Pro 290 295 300 Ser Ile Ala Gly Trp
Ala Arg Met Thr Gly Ile Asn Thr Ile Phe Ile 305 310
315 320 Asp Asp Arg Asp Pro Phe Ala His Gly Phe
Val Val Arg 325 330 40 332 PRT
Agrobacterium tumefaciens 40 Met Arg His Ser Phe Phe Cys Ile Asp Ser His
Thr Cys Gly Asn Pro 1 5 10
15 Val Arg Leu Val Ala Gly Gly Gly Pro Leu Leu Pro His Leu Pro Ile
20 25 30 Ser Glu Arg Arg Asp
Leu Phe Val Arg Asn His Asp Trp Val Arg Gln 35
40 45 Ala Leu Met Phe Glu Pro Arg Gly His Asp Ile Met
Ser Gly Ala Val 50 55 60 Ile Tyr
Pro Ala Tyr Arg Asp Asp Cys Asp Phe Ala Val Ile Phe Ile 65
70 75 80 Glu Val Ser Gly Cys Leu Pro
Met Cys Gly Ala Gly Thr Ile Gly Leu 85
90 95 Val Thr Ala Ala Ile Glu Glu Gly Leu Val Thr Pro
Arg Ile Pro Gly 100 105 110
Arg Leu Ser Ile Glu Thr Pro Ala Gly Lys Val Asp Ile Gln Tyr Asp
115 120 125 Lys Pro Gly Glu Phe Val Glu
Ser Val Arg Ile Phe Asn Val Ala Ser 130 135
140 Tyr Leu His Ala Ala Asp Val Glu Val Asn Val Pro Gly Leu Gly Lys
145 150 155 160 Leu Val
Val Asp Ile Ala Tyr Gly Gly Asn Tyr Tyr Ala Val Ile Glu
165 170 175 Pro Gln Val Gly Trp Pro Gly
Leu Asp Gly Met Thr Ala Gly Asp Val 180 185
190 Val Asp Leu Ser Gln Lys Leu Arg Asp Ala Leu Gly Thr Ile
Cys Asp 195 200 205 Pro Val His
Pro Asp Asp Glu Arg Ile Arg Gly Val His His Ala Ile 210
215 220 Trp Cys Asp Arg Pro Val Ser Ala Glu Ala Asp Gly
Arg Gly Ala Val 225 230 235
240 Phe Tyr Gly Asp Lys Ala Ile Asp Arg Ser Pro Gly Gly Thr Gly Thr
245 250 255 Ser Ala Arg Met
Ala Gln Leu His Gly Lys Gly Arg Leu Lys Ala Gly 260
265 270 Glu Thr Phe Arg Gln Glu Ser Leu Ile Gly Thr
Ile Phe Glu Gly Lys 275 280 285
Val Glu Glu Glu Thr Thr Val Gly Ser Phe Ser Gly Ile Arg Pro Ser 290
295 300 Ile Gly Gly Trp Ala Arg Ile Ile Gly
His Asn Thr Ile Phe Val Asp 305 310 315
320 Asp Arg Asp Pro Leu Ala His Gly Phe Gln Val Arg
325 330 41 312 PRT Xanthomonas campestris 41 Met
His Thr Ile Asp Val Ile Asp Ser His Thr Ala Gly Glu Pro Thr 1
5 10 15 Arg Val Val Leu Ala Gly Phe
Pro Asp Leu Gly Asp Gly Asp Leu Ala 20 25
30 Gln Cys Arg Glu Arg Phe Arg Ser Asp Phe Asp His Trp Arg
Ser Ala 35 40 45 Ile Ala Cys
Glu Pro Arg Gly Ser Asp Thr Met Val Gly Ala Leu Leu 50
55 60 Leu Pro Pro Arg Asp Pro Ser Ala Cys Thr Gly Val
Ile Phe Phe Asn 65 70 75
80 Asn Val Gly Tyr Leu Gly Met Cys Gly His Gly Thr Ile Gly Val Val
85 90 95 Arg Thr Leu Ala
Glu Leu Gly Arg Ile Ala Pro Gly Gln His Arg Ile 100
105 110 Glu Thr Pro Val Gly Thr Val Gly Val Ala Leu
Ala Asp Asp Gly Thr 115 120 125
Val Ser Ile Asp Asn Val Glu Ser Tyr Arg His Ala Ala Gly Val Glu 130
135 140 Val Asp Val Pro Gly His Gly Arg Val
Arg Gly Asp Val Ala Trp Gly 145 150 155
160 Gly Asn Trp Phe Phe Ile Thr Glu Gln Ala Pro Cys Ala Leu
Gly Leu 165 170 175 Ala
Gln Gln Arg Glu Leu Thr Ala Tyr Thr Glu Ala Ile Arg Leu Ala
180 185 190 Leu Glu Ala Ala Gly Ile Thr
Gly Glu Ala Gly Gly Glu Ile Asp His 195 200
205 Ile Glu Ile Ser Gly Val Ala Pro Asp Gly Ser Gly Ala Ala Arg
Asn 210 215 220 Phe Val Leu Cys Pro
Gly Leu Ala Tyr Asp Arg Ser Pro Cys Gly Thr 225 230
235 240 Gly Thr Ser Ala Lys Leu Ala Cys Leu Ala
Ala Asp Gly Lys Leu Ala 245 250
255 Glu Gly Glu Arg Trp Leu Gln Gln Gly Ile Leu Gly Ser Ala Phe Glu
260 265 270 Gly Ser Tyr Arg
His Ser Gly Arg Gly Ile Ala Pro Arg Ile Ser Gly 275
280 285 His Ala Phe Ile Thr Ala Arg Ser Gln Leu Leu Ile
Asp Pro Ala Asp 290 295 300 Pro Phe
Ala Trp Gly Ile Val Ala 305 310 42 312 PRT Xanthomonas
axonopodis 42 Met His Thr Ile Asp Val Ile Asp Ser His Thr Ala Gly Glu
Pro Thr 1 5 10 15 Arg
Val Val Leu Ser Gly Phe Pro Asp Leu Gly Asp Gly Asp Leu Ala
20 25 30 Gln Cys Arg Glu Arg Phe Arg
Ser Glu Phe Asp His Trp Arg Ser Ala 35 40
45 Ile Ala Cys Glu Pro Arg Gly Ser Asp Thr Met Val Gly Ala Leu
Leu 50 55 60 Leu Pro Pro Arg Asp
Pro Ser Ala Cys Thr Gly Val Ile Phe Phe Asn 65 70
75 80 Asn Val Gly Tyr Leu Gly Met Cys Gly His
Gly Thr Ile Gly Val Val 85 90
95 Arg Thr Leu Ala Glu Leu Gly Arg Ile Ala Pro Gly Gln His Arg Ile
100 105 110 Glu Thr Pro Val
Gly Thr Val Gly Val Glu Leu Ala Asp Asp Gly Thr 115
120 125 Val Ser Val Asp Asn Val Glu Ser Tyr Arg Phe Ala
Ser Gly Val Glu 130 135 140 Val Glu
Val Pro Gly His Gly Arg Val Cys Gly Asp Val Ala Trp Gly 145
150 155 160 Gly Asn Trp Phe Phe Ile Thr
Glu His Ala Pro Cys Ala Leu Asp Leu 165
170 175 Ala His Gln Arg Glu Leu Thr Ala Tyr Thr Glu Ala
Ile Arg Leu Ala 180 185 190
Leu Glu Ala Ala Gly Ile Thr Gly Glu Ala Gly Gly Glu Ile Asp His
195 200 205 Ile Glu Val Asn Gly Ala Ala
Pro Asp Gly Ser Gly Val Ala Arg Asn 210 215
220 Phe Val Leu Cys Pro Gly Leu Ala Tyr Asp Arg Ser Pro Cys Gly Thr
225 230 235 240 Gly Thr
Ser Ala Lys Leu Ala Cys Leu Ala Ala Asp Gly Lys Leu Ala
245 250 255 Glu Gly Glu Arg Trp Val Gln
Gln Gly Ile Leu Gly Ser Ala Phe Glu 260 265
270 Gly Asn Tyr Arg Leu Ser Gly Arg Gly Ile Ala Pro Arg Ile
Ser Gly 275 280 285 Arg Ala Tyr
Ile Thr Ala Arg Ala Gln Leu Val Ile Asp Pro Ala Asp 290
295 300 Pro Phe Ala Trp Gly Ile Val Ala 305
310 43 314 PRT Pseudomonas aeruginosa 43 Met Gln Arg Ile Arg Ile Ile
Asp Ser His Thr Gly Gly Glu Pro Thr 1 5
10 15 Arg Leu Val Ile Gly Gly Phe Pro Asp Leu Gly Gln
Gly Asp Met Ala 20 25 30
Glu Arg Arg Arg Leu Leu Gly Glu Arg His Asp Ala Trp Arg Ala Ala
35 40 45 Cys Ile Leu Glu Pro Arg Gly
Ser Asp Val Leu Val Gly Ala Leu Leu 50 55
60 Cys Ala Pro Val Asp Pro Glu Ala Cys Ala Gly Val Ile Phe Phe Asn
65 70 75 80 Asn Ser
Gly Tyr Leu Gly Met Cys Gly His Gly Thr Ile Gly Leu Val
85 90 95 Ala Ser Leu Ala His Leu Gly
Arg Ile Gly Pro Gly Val His Arg Ile 100 105
110 Glu Thr Pro Val Gly Glu Val Glu Ala Thr Leu His Glu Asp
Gly Ser 115 120 125 Val Ser Val
Arg Asn Val Pro Ala Tyr Arg Tyr Arg Arg Gln Val Ser 130
135 140 Val Glu Val Pro Gly Ile Gly Arg Val Ser Gly Asp
Ile Ala Trp Gly 145 150 155
160 Gly Asn Trp Phe Phe Leu Val Ala Gly His Gly Gln Arg Leu Ala Gly
165 170 175 Asp Asn Leu Asp
Ala Leu Thr Ala Tyr Thr Val Ala Val Gln Gln Ala 180
185 190 Leu Asp Asp Gln Asp Ile Arg Gly Glu Asp Gly
Gly Ala Ile Asp His 195 200 205
Ile Glu Leu Phe Ala Asp Asp Pro His Ala Asp Ser Arg Asn Phe Val 210
215 220 Leu Cys Pro Gly Lys Ala Tyr Asp Arg
Ser Pro Cys Gly Thr Gly Thr 225 230 235
240 Ser Ala Lys Leu Ala Cys Leu Ala Ala Asp Gly Lys Leu Leu
Pro Gly 245 250 255 Gln
Pro Trp Arg Gln Ala Ser Val Ile Gly Ser Gln Phe Glu Gly Arg
260 265 270 Tyr Glu Trp Leu Asp Gly Gln
Pro Gly Gly Pro Ile Val Pro Thr Ile 275 280
285 Arg Gly Arg Ala His Val Ser Ala Glu Ala Thr Leu Leu Leu Ala
Asp 290 295 300 Asp Asp Pro Phe Ala
Trp Gly Ile Arg Arg 305 310 44 344 PRT Pseudomonas
aeruginosa 44 Met Arg Ser Gln Arg Ile Val His Ile Val Ser Cys His Ala
Glu Gly 1 5 10 15 Glu
Val Gly Asp Val Ile Val Gly Gly Val Ala Ala Pro Pro Gly Ala
20 25 30 Thr Leu Trp Glu Gln Ser Arg
Trp Ile Ala Arg Asp Gln Asp Leu Arg 35 40
45 Asn Phe Val Leu Asn Glu Pro Arg Gly Gly Val Phe Arg His Ala
Asn 50 55 60 Leu Leu Val Pro Ala
Lys Asp Pro Arg Ala Gln Met Gly Trp Ile Ile 65 70
75 80 Met Glu Pro Ala Asp Thr Pro Pro Met Ser
Gly Ser Asn Ser Leu Cys 85 90
95 Val Ala Thr Val Leu Leu Asp Ser Gly Ile Leu Pro Met Arg Glu Pro
100 105 110 Leu Thr Arg Leu
Leu Leu Glu Ala Pro Gly Gly Leu Ile Glu Ala Arg 115
120 125 Ala Glu Cys Arg Asp Gly Lys Ala Glu Arg Val Glu
Ile Arg Asn Val 130 135 140 Pro Ser
Phe Ala Asp Arg Leu Asp Ala Trp Ile Glu Val Glu Gly Leu 145
150 155 160 Gly Ser Leu Gln Val Asp Thr
Ala Tyr Gly Gly Asp Ser Phe Val Ile 165
170 175 Ala Asp Ala Arg Arg Leu Gly Phe Ala Leu Arg Ala
Asp Glu Ala Ala 180 185 190
Glu Leu Val Ala Thr Gly Leu Lys Ile Thr His Ala Ala Asn Glu Gln
195 200 205 Leu Gly Phe Arg His Pro Thr
Asn Pro Asp Trp Asp His Leu Ser Phe 210 215
220 Cys Gln Leu Ala Ala Pro Pro Glu Arg Arg Asp Gly Val Leu Gly Ala
225 230 235 240 Asn Asn
Ala Val Val Ile Arg Pro Gly Lys Ile Asp Arg Ser Pro Cys
245 250 255 Gly Thr Gly Cys Ser Ala Arg
Met Ala Val Leu Gln Ala Lys Gly Gln 260 265
270 Leu Arg Val Gly Glu Arg Phe Val Gly Arg Ser Ile Ile Gly
Ser Glu 275 280 285 Phe His Cys
His Ile Glu Ser Leu Thr Glu Leu Gly Gly Arg Pro Ala 290
295 300 Ile Leu Pro Cys Leu Ser Gly Arg Ala Trp Ile Thr
Gly Ile His Gln 305 310 315
320 Tyr Leu Leu Asp Pro Asp Asp Pro Trp Pro Gln Gly Tyr Arg Leu Ser
325 330 335 Asp Thr Trp Pro
Gly Gly His Cys 340 45 342 PRT Brucella melitensis 45 Met
Arg Ser Thr Lys Val Ile His Ile Val Gly Cys His Ala Glu Gly 1
5 10 15 Glu Val Gly Asp Val Ile Val
Gly Gly Val Ala Pro Pro Pro Gly Glu 20 25
30 Thr Val Trp Glu Gln Ser Arg Phe Ile Ala Asn Asp Glu Thr
Leu Arg 35 40 45 Asn Phe Val
Leu Asn Lys Pro Arg Gly Gly Val Phe Arg His Val Asn 50
55 60 Leu Leu Val Pro Pro Lys Asp Pro Arg Ala Gln Met
Gly Phe Ile Ile 65 70 75
80 Met Glu Pro Ala Asp Thr Pro Pro Met Ser Gly Ser Asn Ser Ile Cys
85 90 95 Val Ser Thr Val
Leu Leu Asp Ser Gly Ile Ile Ala Met Gln Glu Pro 100
105 110 Val Thr His Met Val Leu Glu Ala Pro Gly Gly
Ile Ile Glu Val Glu 115 120 125
Ala Glu Cys Arg Asn Gly Lys Ala Glu Arg Ile Ser Val Arg Asn Val 130
135 140 Pro Ser Phe Ala Asp Arg Leu Asp Ala
Pro Leu Asp Val Thr Gly Leu 145 150 155
160 Gly Thr Ile Met Val Asp Thr Ala Tyr Gly Gly Asp Ser Phe
Val Ile 165 170 175 Val
Asp Ala Ala Gln Ile Gly Met Lys Ile Glu Pro Gly Gln Ala Arg
180 185 190 Glu Leu Ala Glu Ile Gly Val
Lys Ile Thr Lys Ala Ala Asn Glu Gln 195 200
205 Leu Gly Phe Arg His Pro Glu Arg Asp Trp Arg His Ile Ser Phe
Cys 210 215 220 Gln Ile Thr Glu Pro
Val Thr Arg Glu Gly Asp Val Leu Thr Gly Val 225 230
235 240 Asn Thr Val Ala Ile Arg Pro Ala Lys Phe
Asp Arg Ser Pro Thr Gly 245 250
255 Thr Gly Cys Ser Ala Arg Met Ala Val Leu His Ala Lys Gly Gln Met
260 265 270 Lys Ala Gly Glu
Arg Phe Ile Gly Lys Ser Val Leu Gly Thr Glu Phe 275
280 285 His Cys Arg Leu Asp Lys Val Leu Glu Leu Gly Gly
Lys Pro Ala Ile 290 295 300 Ser Pro
Ile Ile Ser Gly Arg Ala Trp Val Thr Gly Thr Ser Gln Leu 305
310 315 320 Met Leu Asp Pro Ser Asp Pro
Phe Pro His Gly Tyr Arg Leu Ser Asp 325
330 335 Thr Trp Pro Arg Asp Glu 340 46 342
PRT Agrobacterium tumefaciens 46 Met Arg Ser Ile Lys Thr Val His Val Ile
Ser Ala His Ala Glu Gly 1 5 10
15 Glu Val Gly Asp Val Ile Val Gly Gly Val Lys Pro Pro Pro Gly Glu
20 25 30 Thr Ile Trp Glu
Gln Ser Arg Phe Ile Ala Arg Asp Glu Thr Leu Arg 35
40 45 Asn Phe Val Leu Asn Glu Pro Arg Gly Gly Val Phe
Arg His Val Asn 50 55 60 Leu Leu
Val Pro Pro Lys His Pro Asp Ala Asp Ala Ala Phe Ile Ile 65
70 75 80 Met Glu Pro Glu Asp Thr Pro
Pro Met Ser Gly Ser Asn Ser Ile Cys 85
90 95 Val Ser Thr Val Leu Leu Asp Gly Gly Ile Val Pro
Met Gln Glu Pro 100 105 110
Glu Thr His Met Leu Leu Glu Ala Pro Gly Gly Leu Val Lys Val Arg
115 120 125 Ala Glu Cys Arg Asn Gly Lys
Ala Glu Arg Ile Phe Val Gln Asn Leu 130 135
140 Pro Ser Phe Ala Ala Lys Leu Asp Ala Glu Leu Glu Val Glu Gly Leu
145 150 155 160 Gly Lys
Leu Lys Val Asp Thr Ala Tyr Gly Gly Asp Ser Phe Val Ile
165 170 175 Val Asp Ala Glu Ala Met Gly
Phe Ser Leu Lys Pro Glu Glu Ala His 180 185
190 Glu Ile Ala Arg Leu Gly Val Arg Ile Thr Asn Ala Ala Asn
Lys Ala 195 200 205 Leu Gly Phe
Asp His Pro Glu Asn Pro Asp Trp Arg His Phe Ser Phe 210
215 220 Cys Leu Phe Ala Gly Lys Val Glu Arg Thr Ala Glu
Gly Leu Arg Ala 225 230 235
240 Gly Ala Ala Val Ala Ile Gln Pro Gly Lys Val Asp Arg Ser Pro Thr
245 250 255 Gly Thr Ala Leu
Ser Ala Arg Met Ala Val Leu His Ala Arg Gly Glu 260
265 270 Met Lys Glu Gly Glu Thr Leu Thr Ala Val Ser
Leu Ile Gly Ser Thr 275 280 285
Phe Thr Gly Arg Ile Leu Gly Thr Thr Thr Val Gly Asp Arg Pro Ala 290
295 300 Ile Leu Pro Glu Ile Ser Gly Arg Gly
Trp Ile Thr Gly Ile His Gln 305 310 315
320 His Met Leu Asp Pro Ser Asp Pro Trp Pro Glu Gly Tyr Arg
Leu Thr 325 330 335 Asp
Thr Trp Gly Ala Arg 340 47 342 PRT Rhizobium meliloti 47 Met
Arg Ser Thr Lys Thr Ile His Val Ile Ser Ala His Ala Glu Gly 1
5 10 15 Glu Val Gly Asp Val Ile Val
Gly Gly Val Ala Pro Pro Pro Gly Asp 20 25
30 Thr Ile Trp Glu Gln Ser Arg Trp Ile Ala Arg Glu Gln Thr
Leu Arg 35 40 45 Asn Phe Val
Leu Asn Glu Pro Arg Gly Gly Val Phe Arg His Val Asn 50
55 60 Leu Leu Val Pro Pro Lys His Pro Asp Ala Asp Ala
Ala Phe Ile Ile 65 70 75
80 Met Glu Pro Glu Asp Thr Pro Pro Met Ser Gly Ser Asn Ser Ile Cys
85 90 95 Val Ser Thr Val
Leu Leu Asp Ser Gly Ile Leu Pro Met Lys Glu Pro 100
105 110 Val Thr Glu Ile Thr Leu Glu Ala Pro Gly Gly
Leu Val Arg Val Arg 115 120 125
Ala Glu Cys Arg Asp Gly Lys Ala Glu Arg Ile Phe Val Glu Asn Leu 130
135 140 Pro Ser Phe Ala Glu Arg Leu Asp Ala
Lys Leu Glu Val Glu Gly Leu 145 150 155
160 Gly Thr Leu Thr Val Asp Thr Ala Tyr Gly Gly Asp Ser Phe
Val Ile 165 170 175 Val
Asp Ala Ala Ala Met Gly Phe Ala Leu Lys Pro Asp Glu Ala His
180 185 190 Asp Ile Ala Arg Leu Gly Val
Arg Ile Thr Asn Ala Ala Asn Ala Lys 195 200
205 Leu Gly Phe His His Pro Glu Asn Pro Asp Trp Arg His Phe Ser
Phe 210 215 220 Cys Leu Phe Ala Gly
Pro Val Glu Arg Thr Ala Glu Gly Leu Arg Ala 225 230
235 240 Gly Ala Ala Val Ala Ile Gln Pro Gly Lys
Val Asp Arg Ser Pro Thr 245 250
255 Gly Thr Ala Leu Ser Ala Arg Met Ala Val Leu His Ala Arg Gly Gln
260 265 270 Met Gly Leu Ser
Asp Arg Leu Thr Ala Val Ser Leu Ile Gly Ser Thr 275
280 285 Phe Ser Gly Arg Ile Leu Gly Thr Thr Glu Val Gly
Gly Arg Pro Ala 290 295 300 Val Leu
Pro Glu Ile Ser Gly Arg Ala Trp Ile Thr Gly Thr His Gln 305
310 315 320 His Met Leu Asp Pro Ser Asp
Pro Trp Pro Glu Gly Tyr Arg Leu Thr 325
330 335 Asp Thr Trp Gly Ala Arg 340 48 346
PRT Rhizobium loti 48 Met Arg Ser Lys Thr Ser Ile Arg Val Val Gly Cys
His Ala Glu Gly 1 5 10
15 Glu Val Gly Asp Val Ile Ile Gly Gly Val Leu Pro Pro Ala Gly Arg
20 25 30 Thr Met Met Asp Lys Met
Ile Thr Met Glu Arg Asp His Asp His Ile 35 40
45 Arg Arg Met Leu Ile Cys Glu Pro Arg Gly Ser Val Ala Arg
His Val 50 55 60 Asn Leu Leu Val
Pro Ser Thr Arg Glu Asp Cys Ala Ala Gly Ala Ile 65 70
75 80 Ile Met Glu Pro Thr Glu Tyr Pro Pro
Met Ser Gly Ser Asn Thr Ile 85 90
95 Cys Val Ala Thr Val Leu Leu Glu Thr Gly Met Val Pro Met Gln
Glu 100 105 110 Pro Glu Thr
Arg Phe Lys Leu Asp Met Pro Gly Gly Val Ile Glu Val 115
120 125 Arg Ala Gln Cys Arg Asp Gly Lys Cys Val Ser
Ile Thr Leu Arg Asn 130 135 140 Ala
Pro Ala Phe Val Asp Arg Leu Asp Ala Ser Ile Glu Val Glu Gly 145
150 155 160 Leu Gly Thr Leu Thr Val
Asp Ile Ala Tyr Gly Gly Met Phe Tyr Ala 165
170 175 Ile Val Asp Ala Lys Ala Leu Gly Phe Ser Ile Ala
Pro Asp Glu Ala 180 185 190
Arg Glu Leu Ala Val Ala Gly Glu Lys Ile Arg Arg Ala Ala Arg Glu
195 200 205 Gln Leu Asp Val Val His Pro
Gln Phe Asp His Val Arg Gly Val Ser 210 215
220 Ile Val Gln Phe Ala Met Pro Phe Gln Gly Pro Gly Asn Val Thr Arg
225 230 235 240 Asn Thr
Cys Ile Val Ser Pro Gly Arg Ser Asp Arg Ser Pro Thr Gly
245 250 255 Thr Gly Thr Ser Ala Arg Met
Ala Val Leu Gln Ala Arg Gly Leu Met 260 265
270 Gly Val Gly Asp Val Leu Ile His Glu Ser Ile Ile Gly Ser
Arg Phe 275 280 285 Thr Gly Arg
Ile Val Glu Leu Ala Glu Ile Ala Gly Arg Lys Ala Ile 290
295 300 Val Pro Glu Ile Thr Gly Arg Ala Trp Ile Thr Gly
Glu His Ser Tyr 305 310 315
320 Tyr Leu Asp Pro Thr Asp Pro Tyr Pro Gln Gly Tyr Val Leu Ser Asp
325 330 335 Thr Trp Gly Thr
Ser Thr Ser Val Lys Gln 340 345 49 339 PRT
Xanthomonas axonopodis 49 Met Arg Trp Ser Lys Gln Phe Ser Val Val Asp
Cys His Ala Glu Gly 1 5 10
15 Glu Val Gly Lys Val Ile Val Gly Gly Val Gly Asn Ile Pro Gly Thr
20 25 30 Thr Met Phe Glu Lys
Lys Leu Tyr Leu Glu Gln His Arg Asp Asp Ile 35
40 45 Arg Lys Leu Val Leu Gln Glu Pro Arg Gly Ala Thr
Trp His Asn Ala 50 55 60 Asn Ile
Ile Leu Pro Pro Ser His Pro Glu Ala Ser Met Gly Phe Val 65
70 75 80 Ile Leu Glu Ala Thr Glu Tyr
Pro Ala Met Ser Gly Ser Asn Thr Ile 85
90 95 Cys Val Ala Thr Val Leu Leu Glu Thr Gly Ile Leu
Pro Met Gln Glu 100 105 110
Pro Ile Thr Asp Leu Val Leu Glu Ala Pro Ala Gly Leu Ile Arg Val
115 120 125 Arg Cys Asp Cys Lys Asp Gly
Lys Val Thr Arg Val Lys Leu Val Asn 130 135
140 Gln Pro Ala Phe Val Tyr His Leu Asp Ala Lys Val Glu Val Ala Gly
145 150 155 160 Ile Gly
Thr Val Ser Ala Asp Ile Ala Phe Gly Gly Met Thr Phe Ala
165 170 175 Leu Val Asp Ala Ser Ser Leu
Gly Phe Glu Ile Val Pro Ala Glu Ala 180 185
190 Arg Glu Leu Cys Glu Tyr Gly Gln Lys Ile Lys Ala Ala Ala
Ala Glu 195 200 205 Gln Leu Asp
Val Ala Phe Pro Gly Asn Pro Asp Met Pro Gly Ile Thr 210
215 220 Met Thr Gln Phe Thr Gly Pro Leu Ser Arg Ala Asp
Gly Lys Phe Phe 225 230 235
240 Ser Arg Asn Thr Thr Ile Val Ser Pro Gly Arg Cys Asp Arg Ser Pro
245 250 255 Cys Gly Ala Gly
Ser Ser Ala Arg Leu Ala Ala Leu His Ala Lys Gly 260
265 270 Val Leu Ala Lys Gly Asp Thr Leu Val His Glu
Ser Ile Ile Gly Ser 275 280 285
Arg Phe Glu Cys Gly Ile Glu Asp Met Ser Asn Val Gly Asp Tyr Pro 290
295 300 Ala Val Val Pro Ser Ile Ala Gly Gln
Ala Trp Ile Ser Gly Leu Ser 305 310 315
320 Gln Leu Gly Leu Asp Pro Ser Asp Pro Tyr Ala Glu Gly Phe
Thr Leu 325 330 335 Ala
Asp Arg 50 333 PRT Streptomyces coelicolor 50 Met Arg Ser Thr Val Cys
Tyr His Ala Val Asp Ser His Thr Glu Gly 1 5
10 15 Met Pro Thr Arg Val Ile Thr Gly Gly Val Gly Val
Leu Pro Gly Ala 20 25 30
Thr Met Phe Glu Arg Arg Gln Arg Phe Val Ala Glu Arg Asp His Leu
35 40 45 Arg Thr Leu Leu Met Cys Glu
Pro Arg Gly His Ala Ser Met Ser Gly 50 55
60 Ala Ile Leu Gln Pro Pro Thr Arg Pro Asp Ala Asp Tyr Gly Val Leu
65 70 75 80 Phe Ile
Glu Val Ser Gly Cys Leu Pro Met Cys Gly His Gly Thr Ile
85 90 95 Gly Val Ala Thr Val Leu Val
Glu Thr Gly Met Val Glu Val Thr Glu 100 105
110 Pro Glu Thr Thr Val Arg Leu Asp Thr Pro Ala Gly Leu Val
Thr Ala 115 120 125 Arg Val Arg
Val Arg Asp Gly His Ala Glu Ser Val Thr Leu Glu Asn 130
135 140 Val Ala Ser Tyr Ser His Ala Leu Asp Gln Val Val
Asp Val Pro Gly 145 150 155
160 His Gly Glu Val Arg Tyr Asp Ile Ala Tyr Gly Gly Asn Phe Tyr Ala
165 170 175 Phe Val Arg Thr
Asp Asp Leu Gly Ile Pro Phe Glu Arg Ala His Lys 180
185 190 Gln Pro Leu Leu Asp Ala Gly Leu Ala Val Met
Asp Ala Ile Asn Lys 195 200 205
Gln Asn Pro Val Ser His Pro Glu Asn Pro Asp Ile Asp Val Cys His 210
215 220 His Val Tyr Leu Glu Ala Pro Gly Ser
Thr Ala Glu His Ser Arg His 225 230 235
240 Ala Met Ala Ile His Pro Gly Trp Phe Asp Arg Ser Pro Cys
Gly Thr 245 250 255 Gly
Thr Ser Ala Arg Met Ala Gln Leu His Ala Arg Gly Leu Leu Pro
260 265 270 Ala Gly Arg Asp Phe Val Asn
Glu Ser Phe Ile Gly Ser Arg Phe Val 275 280
285 Gly Arg Val Leu Gly Glu Thr Thr Val Gly Gly Arg Pro Ala Val
Leu 290 295 300 Pro Ser Val Thr Gly
Arg Ala Trp Ile Thr Gly Thr Ala Gln Tyr Leu 305 310
315 320 Leu Asp Pro Ser Asp Pro Tyr Pro Ala Gly
Phe Thr Leu 325 330 51 345 PRT
Agrobacterium tumefaciens 51 Met Arg Trp Lys Arg Thr Leu Gln Leu Leu Asp
Val His Cys Glu Gly 1 5 10
15 Glu Ile Gly Arg Val Val Thr Gly Gly Ala Pro Lys Ile Pro Gly Asn
20 25 30 Thr Val Ala Glu Gln
Leu His Trp Met Asn Thr Asp Pro Gln Gly Glu 35
40 45 Ala Leu Arg Arg Phe Leu Thr Leu Glu Pro Arg Gly
Thr Pro Met Gly 50 55 60 Ser Val
Asp Leu Leu Leu Pro Pro Lys His Pro Asp Ala His Ala Ala 65
70 75 80 Phe Val Ile Leu Gln Pro Asp
Gln Ala His Ala Ser Ser Gly Ser Asn 85
90 95 Ser Ile Cys Ala Thr Thr Ala Leu Leu Glu Ser Gly
Met Val Glu Met 100 105 110
Gln Glu Pro Glu Thr Val Ile Ile Leu Glu Thr Ala Ala Gly Leu Val
115 120 125 Lys Ala Thr Ala Thr Cys Arg
Asp Gly Arg Cys Glu Lys Val Lys Leu 130 135
140 Thr Met Val Pro Ser Phe Val His Glu Leu Asp Val Ser Ile Asp Thr
145 150 155 160 Pro Glu
Trp Gly Arg Val Thr Met Asp Ile Ser Tyr Gly Gly Ile Phe
165 170 175 Tyr Ala Leu Val Asp Val Arg
Gln Ile Gly Leu Thr Ile Glu Lys Ala 180 185
190 Asn Ala Ala Lys Leu Val Ala Ala Gly Met Thr Leu Lys Asp
Leu Val 195 200 205 Asn Arg Glu
Met Thr Val Val His Pro Glu Ile Pro Ala Ile Ser Gly 210
215 220 Val Ala Tyr Val Met Phe Arg Asp Val Asp Ala Asp
Gly Ser Ile Arg 225 230 235
240 Thr Cys Thr Thr Met Trp Pro Gly Arg Ala Asp Arg Ser Pro Cys Gly
245 250 255 Thr Gly Asn Ser
Ala Asn Leu Ala Thr Leu Tyr Ala Arg Gly Lys Val 260
265 270 Lys Val Gly Asp Glu Tyr Lys Ser Arg Ser Ile
Ile Gly Ser Glu Phe 275 280 285
Asp Val Gly Leu Ser Ala Val Thr Glu Val Ala Gly Arg Pro Ala Val 290
295 300 Ile Pro Thr Ile Ala Gly Arg Gly Phe
Thr Phe Gly Leu His Gln Val 305 310 315
320 Gly Leu Asp Pro Phe Asp Pro Leu Gly Asp Gly Phe Ala Met
Thr Asp 325 330 335 Val
Trp Gly Pro Glu Ala Gly Asn Ile 340 345 52
354 PRT Trypanosoma cruzi 52 Met Arg Phe Lys Lys Ser Leu Thr Cys Ile Asp
Met His Thr Glu Gly 1 5 10
15 Glu Ala Ala Arg Ile Val Thr Ser Gly Leu Pro His Ile Pro Gly Ser
20 25 30 Asn Met Ala Glu Lys
Lys Ala Tyr Leu Gln Glu Asn Met Asp Tyr Leu 35
40 45 Arg Arg Gly Ile Met Leu Glu Pro Arg Gly His Asp
Asp Met Phe Gly 50 55 60 Ala Phe
Leu Phe Asp Pro Ile Glu Glu Gly Ala Asp Leu Gly Ile Val 65
70 75 80 Phe Met Asp Thr Gly Gly Tyr
Leu Asn Met Cys Gly His Asn Ser Ile 85
90 95 Ala Ala Val Thr Ala Ala Val Glu Thr Gly Ile Leu
Ser Val Pro Ala 100 105 110
Lys Ala Thr Asn Val Pro Val Val Leu Asp Thr Pro Ala Gly Leu Val
115 120 125 Arg Gly Thr Ala His Leu Gln
Ser Gly Thr Glu Ser Glu Val Ser Asn 130 135
140 Ala Ser Ile Ile Asn Val Pro Ser Phe Leu Tyr Gln Gln Asp Val Val
145 150 155 160 Ile Val
Leu Pro Lys Pro Tyr Gly Glu Val Arg Val Asp Ile Ala Phe
165 170 175 Gly Gly Asn Phe Phe Ala Ile
Val Pro Ala Glu His Leu Gly Ile Asp 180 185
190 Ile Ser Val Gln Asn Leu Ser Arg Leu Gln Glu Ala Gly Glu
Leu Leu 195 200 205 Arg Thr Glu
Ile Asn Arg Ser Val Lys Val Gln His Pro Gln Leu Pro 210
215 220 His Ile Asn Thr Val Asp Cys Val Glu Ile Tyr Gly
Pro Pro Thr Asn 225 230 235
240 Pro Glu Ala Lys Tyr Lys Asn Val Val Ile Phe Gly Asn Arg Gln Ala
245 250 255 Asp Arg Ser Pro
Cys Gly Thr Gly Thr Ser Ala Lys Met Ala Thr Leu 260
265 270 Tyr Ala Lys Gly Gln Leu Arg Ile Gly Glu Thr
Phe Val Tyr Glu Ser 275 280 285
Ile Leu Gly Ser Leu Phe Gln Gly Arg Val Leu Gly Glu Glu Arg Ile 290
295 300 Pro Gly Val Lys Val Pro Val Thr Lys
Asp Ala Glu Glu Gly Met Leu 305 310 315
320 Val Val Thr Thr Glu Ile Thr Gly Lys Ala Phe Ile Met Gly
Phe Asn 325 330 335 Thr
Met Leu Phe Asp Pro Thr Asp Pro Phe Leu Asn Gly Phe Thr Leu
340 345 350 Lys Arg 53 15 DNA Artificial
Sequence Description of Artificial Sequence Synthetic
oligonucleotide CDS (1)..(15) 53 tct cca tgt ggg aca
15 Ser Pro Cys Gly Thr 1 5
54 5 PRT Artificial Sequence Description of Artificial Sequence Synthetic
peptide 54 Ser Pro Cys Gly Thr 1 5 55 15 DNA
Artificial Sequence Description of Artificial Sequence Synthetic
oligonucleotide CDS (1)..(15) 55 tct cca agc ggg aca
15 Ser Pro Ser Gly Thr 1 5
56 5 PRT Artificial Sequence Description of Artificial Sequence Synthetic
peptide 56 Ser Pro Ser Gly Thr 1 5 57 146 PRT
Trypanosoma cruzi 57 Gly Glu Val Arg Val Asp Ile Ala Phe Gly Gly Asn Phe
Phe Ala Ile 1 5 10 15
Val Pro Ala Glu Gln Leu Gly Ile Asp Ile Ser Val Gln Asn Leu Ser
20 25 30 Arg Leu Gln Glu Ala Gly Glu
Leu Leu Arg Thr Glu Ile Asn Arg Ser 35 40
45 Val Lys Val Gln His Pro Gln Leu Pro His Ile Asn Thr Val Asp
Cys 50 55 60 Val Glu Ile Tyr Gly
Pro Pro Thr Asn Pro Glu Ala Asn Tyr Lys Asn 65 70
75 80 Val Val Ile Phe Gly Asn Arg Gln Ala Asp
Arg Ser Pro Cys Gly Thr 85 90
95 Gly Thr Ser Ala Lys Met Ala Thr Leu Tyr Ala Lys Gly Gln Leu Arg
100 105 110 Ile Gly Glu Thr
Phe Val Tyr Glu Ser Ile Leu Gly Ser Leu Phe Gln 115
120 125 Gly Arg Val Leu Gly Glu Glu Arg Ile Pro Gly Val
Lys Val Pro Val 130 135 140 Thr Lys
145 58 146 PRT Bacillus anthracis 58 Gly Thr Val Glu Ala Asp Ile Ala Tyr
Gly Gly Asn Phe Tyr Ala Ile 1 5 10
15 Ile Asp Ala Lys Ser Val Gly Leu Glu Leu Val Pro Glu His Ala
Ser 20 25 30 Thr Ile Ile
Asp Lys Ala Ile His Ile Arg Asn Ile Ile Asn Glu Arg 35
40 45 Phe Glu Ile Ile His Pro Glu Tyr Ser Phe Ile
Arg Gly Leu Thr His 50 55 60 Val
Glu Phe Tyr Thr Asp Pro Thr His Glu Ser Ala His Val Lys Asn 65
70 75 80 Thr Val Val Val Pro Pro
Gly Gly Ile Asp Arg Ser Pro Cys Gly Thr 85
90 95 Gly Thr Ser Ala Lys Leu Ala Val Leu Tyr Ala Asn
Gln Lys Ile Glu 100 105 110
Met Asn Glu Glu Phe Val His Glu Ser Ile Val Gly Ser Leu Phe Lys
115 120 125 Gly Cys Val Ile Asn Thr Thr
Asn Val Ala Asn Met Glu Ala Val Val 130 135
140 Thr Lys 145 59 117 PRT Trypanosoma cruzi 59 Met Arg Phe Lys
Lys Ser Phe Thr Cys Ile Asp Met His Thr Glu Gly 1 5
10 15 Glu Ala Ala Arg Ile Val Thr Ser Gly Leu
Pro His Ile Pro Gly Ser 20 25
30 Asn Met Ala Glu Lys Lys Ala Tyr Leu Gln Glu Asn Met Asp Tyr Leu
35 40 45 Arg Arg Gly Ile Met Leu
Glu Pro Arg Gly His Asp Asp Met Phe Gly 50 55
60 Ala Phe Leu Phe Asp Pro Ile Glu Glu Gly Ala Asp Leu Gly Met
Val 65 70 75 80 Phe
Met Asp Thr Gly Gly Tyr Leu Asn Met Cys Gly His Asn Ser Ile
85 90 95 Ala Ala Val Thr Ala Ala Val
Glu Thr Gly Ile Val Ser Val Pro Ala 100 105
110 Lys Ala Thr Asn Val 115 60 117 PRT Bacillus
anthracis 60 Met Arg Thr Gln Lys Val Phe Thr Thr Ile Asp Thr His Thr Gly
Gly 1 5 10 15 Asn Pro
Thr Arg Thr Leu Ile Ser Gly Leu Pro Lys Leu Leu Gly Glu 20
25 30 Thr Met Ala Glu Lys Met Leu His Met
Lys Lys Glu Tyr Asp Trp Ile 35 40
45 Arg Lys Leu Leu Met Asn Glu Pro Arg Gly His Asp Val Met Ser Gly
50 55 60 Ala Leu Leu Thr Asp Pro Cys
His Pro Asp Ala Asp Ile Gly Val Ile 65 70
75 80 Tyr Ile Glu Thr Gly Gly Tyr Leu Pro Met Cys Gly
His Asp Thr Ile 85 90
95 Gly Val Cys Thr Ala Leu Ile Glu Ser Gly Leu Ile Pro Val Val Glu
100 105 110 Pro Ile Thr Ser Leu
115 61 145 PRT Trypanosoma cruzi 61 Glu Val Arg Val Asp Ile Ala Phe
Gly Gly Asn Phe Phe Ala Ile Val 1 5 10
15 Pro Ala Glu Gln Leu Gly Ile Asp Ile Ser Val Gln Asn Leu
Ser Arg 20 25 30 Leu Gln
Glu Ala Gly Glu Leu Leu Arg Thr Glu Ile Asn Arg Ser Val 35
40 45 Lys Val Gln His Pro Gln Leu Pro His Ile
Asn Thr Val Asp Cys Val 50 55 60
Glu Ile Tyr Gly Pro Pro Thr Asn Pro Glu Ala Asn Tyr Lys Asn Val 65
70 75 80 Val Ile Phe Gly Asn
Arg Gln Ala Asp Arg Ser Pro Cys Gly Thr Gly 85
90 95 Thr Ser Ala Lys Met Ala Thr Leu Tyr Ala Lys
Gly Gln Leu Arg Ile 100 105
110 Gly Glu Thr Phe Val Tyr Glu Ser Ile Leu Gly Ser Leu Phe Gln Gly
115 120 125 Arg Val Leu Gly Glu Glu Arg
Ile Pro Gly Val Lys Val Pro Val Thr 130 135
140 Lys 145 62 145 PRT Bacillus anthracis 62 Glu Phe Gln Val Asp
Ile Ala Phe Gly Gly Ala Phe Tyr Ala Val Val 1 5
10 15 Asp Ser Lys Glu Phe Gly Leu Lys Val Asp Phe
Lys Asp Leu Ser Ala 20 25
30 Ile Gln Gln Trp Gly Gly Lys Ile Lys His Tyr Ile Glu Ser Lys Met
35 40 45 Glu Val Lys His Pro Leu Glu
Glu Gly Leu Lys Gly Ile Tyr Gly Val 50 55
60 Ile Phe Ser Asp Asp Pro Lys Gly Glu Gly Ala Thr Leu Arg Asn Val
65 70 75 80 Thr Ile
Phe Ala Asp Gly Gln Val Asp Arg Ser Pro Cys Gly Thr Gly
85 90 95 Thr Ser Ala Arg Ile Ala Thr
Leu Phe Glu Lys Gly Ile Leu Gln Lys 100 105
110 Gly Glu Ile Phe Ile His Glu Cys Ile Thr Asp Gly Glu Phe
Glu Gly 115 120 125 Glu Val Leu
Ser Val Thr Ala Val His Thr Tyr Glu Ala Val Val Pro 130
135 140 Lys 145 63 113 PRT Trypanosoma cruzi 63 Met
Arg Phe Lys Lys Ser Phe Thr Cys Ile Asp Met His Thr Glu Gly 1
5 10 15 Glu Ala Ala Arg Ile Val Thr
Ser Gly Leu Pro His Ile Pro Gly Ser 20 25
30 Asn Met Ala Glu Lys Lys Ala Tyr Leu Gln Glu Asn Met Asp
Tyr Leu 35 40 45 Arg Arg Gly
Ile Met Leu Glu Pro Arg Gly His Asp Asp Met Phe Gly 50
55 60 Ala Phe Leu Phe Asp Pro Ile Glu Glu Gly Ala Asp
Leu Gly Met Val 65 70 75
80 Phe Met Asp Thr Gly Gly Tyr Leu Asn Met Cys Gly His Asn Ser Ile
85 90 95 Ala Ala Val Thr
Ala Ala Val Glu Thr Gly Ile Val Ser Val Pro Ala 100
105 110 Lys 64 113 PRT Bacillus anthracis 64 Met
Lys Val Ser Lys Val Tyr Thr Thr Ile Asp Ala His Val Ala Gly 1
5 10 15 Glu Pro Leu Arg Ile Ile Thr
Gly Gly Val Pro Glu Ile Lys Gly Glu 20 25
30 Thr Gln Leu Glu Arg Arg Trp Tyr Cys Met Glu His Leu Asp
Tyr Leu 35 40 45 Arg Glu Val
Leu Met Tyr Glu Pro Arg Gly His His Gly Met Tyr Gly 50
55 60 Cys Ile Ile Thr Pro Pro Ala Ser Ala His Ala Asp
Phe Gly Val Leu 65 70 75
80 Phe Met His Asn Glu Gly Trp Ser Thr Met Cys Gly His Gly Ile Ile
85 90 95 Ala Val Ile Thr
Val Gly Ile Glu Thr Gly Met Phe Glu Thr Lys Gln 100
105 110 Lys 65 138 PRT Trypanosoma cruzi 65 Tyr
Gly Glu Val Arg Val Asp Ile Ala Phe Gly Gly Asn Phe Phe Ala 1
5 10 15 Ile Val Pro Ala Glu Gln Leu
Gly Ile Asp Ile Ser Val Gln Asn Leu 20 25
30 Ser Arg Leu Gln Glu Ala Gly Glu Leu Leu Arg Thr Glu Ile
Asn Arg 35 40 45 Ser Val Lys
Val Gln His Pro Gln Leu Pro His Ile Asn Thr Val Asp 50
55 60 Cys Val Glu Ile Tyr Gly Pro Pro Thr Asn Pro Glu
Ala Asn Tyr Lys 65 70 75
80 Asn Val Val Ile Phe Gly Asn Arg Gln Ala Asp Arg Ser Pro Cys Gly
85 90 95 Thr Gly Thr Ser
Ala Lys Met Ala Thr Leu Tyr Ala Lys Gly Gln Leu 100
105 110 Arg Ile Gly Glu Thr Phe Val Tyr Glu Ser Ile
Leu Gly Ser Leu Phe 115 120 125
Gln Gly Arg Val Leu Gly Glu Glu Arg Ile 130 135 66
138 PRT Clostridium botulinum 66 Tyr Gly Lys Leu Thr Leu Asp Ile Ser Phe
Gly Gly Ser Phe Phe Ala 1 5 10
15 Met Val Asp Ala Glu Lys Val Gly Ile Asp Ile Ser Pro Ala Asn Ser
20 25 30 Gln Lys Leu Asn
Asp Leu Gly Met Lys Ile Val His Ala Val Asn Glu 35
40 45 Gln Val Glu Ile Lys His Pro Val Leu Glu His Ile
Lys Thr Val Asp 50 55 60 Leu Cys
Glu Phe Tyr Gly Pro Ala Lys Ser Glu Asp Ala Asp Val Gln 65
70 75 80 Asn Val Val Val Phe Gly Gln
Gly Gln Val Asp Arg Ser Pro Cys Gly 85
90 95 Thr Gly Thr Ser Ala Lys Met Ala Leu Leu Tyr Ala
Gln Gly Lys Met 100 105 110
Lys Val Gly Glu Glu Ile Val Asn Glu Ser Ile Ile Cys Thr Lys Phe
115 120 125 Lys Gly Lys Ile Leu Glu Glu
Thr Lys Val 130 135 67 118 PRT Trypanosoma cruzi 67
Ile Met Arg Phe Lys Lys Ser Phe Thr Cys Ile Asp Met His Thr Glu 1
5 10 15 Gly Glu Ala Ala Arg Ile
Val Thr Ser Gly Leu Pro His Ile Pro Gly 20
25 30 Ser Asn Met Ala Glu Lys Lys Ala Tyr Leu Gln Glu
Asn Met Asp Tyr 35 40 45 Leu
Arg Arg Gly Ile Met Leu Glu Pro Arg Gly His Asp Asp Met Phe 50
55 60 Gly Ala Phe Leu Phe Asp Pro Ile Glu Glu
Gly Ala Asp Leu Gly Met 65 70 75
80 Val Phe Met Asp Thr Gly Gly Tyr Leu Asn Met Cys Gly His Asn
Ser 85 90 95 Ile Ala
Ala Val Thr Ala Ala Val Glu Thr Gly Ile Val Ser Val Pro 100
105 110 Ala Lys Ala Thr Asn Val 115
68 118 PRT Clostridium botulinum 68 Ile Met Arg Ala Ile Lys Thr Ile Gln
Thr Ile Glu Ser His Thr Met 1 5 10
15 Gly Glu Pro Thr Arg Ile Val Ile Gly Gly Leu Pro Lys Val Pro
Gly 20 25 30 Lys Thr Met
Ala Glu Lys Met Glu Tyr Leu Glu Glu Asn Asn Asp Ser 35
40 45 Leu Arg Thr Met Leu Met Ser Glu Pro Arg Gly
His Asn Asp Met Phe 50 55 60 Gly
Ala Ile Tyr Thr Glu Pro Ala Asp Glu Thr Ala Asp Leu Gly Ile 65
70 75 80 Ile Phe Met Asp Gly Gly
Gly Tyr Leu Asn Met Cys Gly His Gly Ser 85
90 95 Ile Gly Ala Ala Thr Cys Ala Val Glu Met Gly Ile
Val Lys Val Glu 100 105 110
Glu Pro Tyr Thr Asn Ile 115 69 224 PRT Trypanosoma cruzi 69 Ala
Asp Leu Gly Ile Val Phe Met Asp Thr Gly Gly Tyr Leu Asn Met 1
5 10 15 Cys Gly His Asn Ser Ile Ala
Ala Val Thr Ala Ala Val Glu Thr Gly 20 25
30 Ile Leu Ser Val Pro Ala Lys Ala Thr Asn Val Pro Val Val
Leu Asp 35 40 45 Thr Pro Ala
Gly Leu Val Arg Gly Thr Ala His Leu Gln Ser Gly Thr 50
55 60 Glu Ser Glu Val Ser Asn Ala Ser Ile Ile Asn Val
Pro Ser Phe Leu 65 70 75
80 Tyr Gln Gln Asp Val Val Ile Val Leu Pro Lys Pro Tyr Gly Glu Val
85 90 95 Arg Val Asp Ile
Ala Phe Gly Gly Asn Phe Phe Ala Ile Val Pro Ala 100
105 110 Glu His Leu Gly Ile Asp Ile Ser Val Gln Asn
Leu Ser Arg Leu Gln 115 120 125
Glu Ala Gly Glu Leu Leu Arg Thr Glu Ile Asn Arg Ser Val Lys Val 130
135 140 Gln His Pro Gln Leu Pro His Ile Asn
Thr Val Asp Cys Val Glu Ile 145 150 155
160 Tyr Gly Asn Ala Thr Asn Pro Glu Ala Lys Tyr Lys Asn Val
Val Ile 165 170 175 Phe
Gly Asn Arg Gln Ala Asp Arg Ser Pro Cys Gly Thr Gly Thr Ser
180 185 190 Ala Lys Met Ala Thr Leu Tyr
Ala Lys Gly Gln Leu Arg Ile Gly Glu 195 200
205 Thr Phe Val Tyr Glu Ser Ile Leu Gly Ser Leu Phe Gln Gly Arg
Val 210 215 220 70 218 PRT
Clostridium botulinum 70 Ala Asp Leu Gly Ile Ile Phe Met Asp Gly Gly Gly
Tyr Leu Asn Met 1 5 10
15 Cys Gly His Gly Ser Ile Gly Ala Ala Thr Cys Ala Val Glu Met Gly
20 25 30 Ile Val Lys Val Glu Glu
Pro Tyr Thr Asn Ile Lys Leu Glu Ala Pro 35 40
45 Ala Gly Met Ile Asn Ala Arg Val Lys Val Glu Asp Gly Lys
Ala Lys 50 55 60 Glu Thr Ser Ile
Val Asn Val Pro Ala Phe Leu Tyr Lys Lys Asp Val 65 70
75 80 Glu Ile Asp Val Pro Asp Tyr Gly Lys
Leu Thr Leu Asp Ile Ser Phe 85 90
95 Gly Gly Ser Phe Phe Ala Met Val Asp Ala Glu Lys Val Gly Ile
Asp 100 105 110 Ile Ser Pro
Ala Asn Ser Gln Lys Leu Asn Asp Leu Gly Met Lys Ile 115
120 125 Val His Ala Val Asn Glu Gln Val Glu Ile Lys
His Pro Val Leu Glu 130 135 140 His
Ile Lys Thr Val Asp Leu Cys Glu Phe Tyr Gly Pro Ala Lys Ser 145
150 155 160 Glu Asp Ala Asp Val Gln
Asn Val Val Val Phe Gly Gln Gly Gln Val 165
170 175 Asp Arg Ser Pro Cys Gly Thr Gly Thr Ser Ala Lys
Met Ala Leu Leu 180 185 190
Tyr Ala Gln Gly Lys Met Lys Val Gly Glu Glu Ile Val Asn Glu Ser
195 200 205 Ile Ile Cys Thr Lys Phe Lys
Gly Lys Ile 210 215 71 72 PRT Trypanosoma cruzi 71
Pro Thr Asn Pro Glu Ala Asn Tyr Lys Asn Val Val Ile Phe Gly Asn 1
5 10 15 Arg Gln Ala Asp Arg Ser
Pro Cys Gly Thr Gly Thr Ser Ala Lys Met 20
25 30 Ala Thr Leu Tyr Ala Lys Gly Gln Leu Arg Ile Gly
Glu Thr Phe Val 35 40 45 Tyr
Glu Ser Ile Leu Gly Ser Leu Phe Gln Gly Arg Val Leu Gly Glu 50
55 60 Glu Arg Ile Pro Gly Val Lys Val 65
70 72 72 PRT Aspergillus fumigatus 72 Pro Asp Asp Val Gln
Gly Ala Glu Thr Gly Leu Cys Tyr Phe Ala Glu 1 5
10 15 Asn Gln Ile Asp Arg Ser Pro Thr Gly Ser Cys
Val Ile Ala Arg Met 20 25
30 Ala Leu Ala Tyr Ala Lys Gly Leu Arg Ser Leu Gly Gln Arg Trp Ala
35 40 45 Tyr Asn Ser Leu Val Ser Asn
Arg Phe Gly Thr Gly Ala Phe Ser Ala 50 55
60 Glu Ile Val Glu Glu Val Thr Ile 65 70 73 34
PRT Trypanosoma cruzi 73 Met Ala Glu Lys Lys Ala Tyr Leu Gln Glu Asn Met
Asp Tyr Leu Arg 1 5 10
15 Arg Gly Ile Met Leu Glu Pro Arg Gly His Asp Asp Met Phe Gly Ala
20 25 30 Phe Leu 74 34 PRT
Aspergillus fumigatus 74 Leu Leu Glu Gln Arg Asp Gln Ala Lys Gln His His
Asp His Ile Arg 1 5 10
15 Lys Cys Leu Met Leu Glu Pro Arg Gly His Asn Gly Met Tyr Gly Ala
20 25 30 Ile Ile 75 29 PRT
Trypanosoma cruzi 75 Cys Ile Asp Met His Thr Glu Gly Glu Ala Ala Arg Ile
Val Thr Ser 1 5 10 15
Gly Leu Pro His Ile Pro Gly Ser Asn Met Ala Glu Lys 20
25 76 29 PRT Aspergillus fumigatus 76 Cys Ile Asp Met His Thr
Thr Gly Glu Pro Thr Arg Ile Ile Tyr Ser 1 5
10 15 Gly Phe Pro Pro Leu Ser Gly Thr Leu Leu Glu Gln
Arg 20 25 77 27 PRT Trypanosoma cruzi 77
Val Asp Ile Ala Phe Gly Gly Asn Phe Phe Ala Ile Val Pro Ala Glu 1
5 10 15 Gln Leu Gly Ile Asp Ile
Ser Val Gln Asn Leu 20 25 78 27 PRT
Aspergillus fumigatus 78 Leu Asp Ile Ser Tyr Gly Gly Ala Phe Tyr Ala Ile
Val Gln Ala Ser 1 5 10
15 Glu Leu Gly Phe Ser Gly Gly Leu Arg Asp Leu 20
25 79 20 PRT Trypanosoma cruzi 79 Ser Ser Ile Gly Ser Asn Lys Lys
Ala Pro Asn Ile Ser Ser Pro Arg 1 5 10
15 Gly Ser Ser Ile 20 80 20 PRT Aspergillus
fumigatus 80 Ser Ser Val Ser Gly Arg Met Met Ala Pro Tyr Ile Pro Leu Pro
Arg 1 5 10 15 Gly Ser
Ser Ile 20 81 146 PRT Trypanosoma cruzi 81 Gly Glu Val Arg
Val Asp Ile Ala Phe Gly Gly Asn Phe Phe Ala Ile 1 5
10 15 Val Pro Ala Glu Gln Leu Gly Ile Asp Ile
Ser Val Gln Asn Leu Ser 20 25
30 Arg Leu Gln Glu Ala Gly Glu Leu Leu Arg Thr Glu Ile Asn Arg Ser
35 40 45 Val Lys Val Gln His Pro
Gln Leu Pro His Ile Asn Thr Val Asp Cys 50 55
60 Val Glu Ile Tyr Gly Pro Pro Thr Asn Pro Glu Ala Asn Tyr Lys
Asn 65 70 75 80 Val
Val Ile Phe Gly Asn Arg Gln Ala Asp Arg Ser Pro Cys Gly Thr
85 90 95 Gly Thr Ser Ala Lys Met Ala
Thr Leu Tyr Ala Lys Gly Gln Leu Arg 100 105
110 Ile Gly Glu Thr Phe Val Tyr Glu Ser Ile Leu Gly Ser Leu
Phe Gln 115 120 125 Gly Arg Val
Leu Gly Glu Glu Arg Ile Pro Gly Val Lys Val Pro Val 130
135 140 Thr Lys 145 82 146 PRT Clostridium difficile
82 Gly Thr Val Lys Phe Asp Ile Ser Phe Gly Gly Ser Phe Phe Ala Ile 1
5 10 15 Ile His Ala Ser Gln
Leu Gly Leu Lys Ile Glu Pro Gln Asn Ala Gly 20
25 30 Lys Leu Thr Glu Leu Ala Met Lys Leu Arg Asp Ile
Ile Asn Glu Lys 35 40 45 Ile
Glu Ile Gln His Pro Thr Leu Ala His Ile Lys Thr Val Asp Leu 50
55 60 Val Glu Ile Tyr Asp Glu Pro Thr His Pro
Glu Ala Thr Tyr Lys Asn 65 70 75
80 Val Val Ile Phe Gly Gln Gly Gln Val Asp Arg Ser Pro Cys Gly
Thr 85 90 95 Gly Thr
Ser Ala Lys Leu Ala Thr Leu His Ala Lys Gly Glu Leu Lys 100
105 110 Val Gly Glu Lys Phe Val Tyr Glu Ser
Ile Leu Gly Thr Leu Phe Lys 115 120
125 Gly Glu Ile Val Glu Glu Thr Lys Val Ala Asp Phe Asn Ala Val Val
130 135 140 Pro Lys 145 83 117 PRT
Trypanosoma cruzi 83 Met Arg Phe Lys Lys Ser Phe Thr Cys Ile Asp Met His
Thr Glu Gly 1 5 10 15
Glu Ala Ala Arg Ile Val Thr Ser Gly Leu Pro His Ile Pro Gly Ser
20 25 30 Asn Met Ala Glu Lys Lys Ala
Tyr Leu Gln Glu Asn Met Asp Tyr Leu 35 40
45 Arg Arg Gly Ile Met Leu Glu Pro Arg Gly His Asp Asp Met Phe
Gly 50 55 60 Ala Phe Leu Phe Asp
Pro Ile Glu Glu Gly Ala Asp Leu Gly Met Val 65 70
75 80 Phe Met Asp Thr Gly Gly Tyr Leu Asn Met
Cys Gly His Asn Ser Ile 85 90
95 Ala Ala Val Thr Ala Ala Val Glu Thr Gly Ile Val Ser Val Pro Ala
100 105 110 Lys Ala Thr Asn
Val 115 84 117 PRT Clostridium difficile 84 Met Lys Phe Ser Arg
Ser Ile Gln Ala Ile Asp Ser His Thr Ala Gly 1 5
10 15 Glu Ala Thr Arg Ile Val Val Gly Gly Ile Pro
Asn Ile Lys Gly Asn 20 25
30 Ser Met Pro Glu Lys Lys Glu Tyr Leu Glu Glu Asn Leu Asp Tyr Leu
35 40 45 Arg Thr Ala Ile Met Leu Glu
Pro Arg Gly His Asn Asp Met Phe Gly 50 55
60 Ser Val Met Thr Gln Pro Cys Cys Pro Asp Ala Asp Phe Gly Ile Ile
65 70 75 80 Phe Met
Asp Gly Gly Gly Tyr Leu Asn Met Cys Gly His Gly Thr Ile
85 90 95 Gly Ala Met Thr Ala Ala Ile
Glu Thr Gly Val Val Pro Ala Val Glu 100 105
110 Pro Val Thr His Val 115 85 139 PRT Trypanosoma
cruzi 85 Gly Glu Val Arg Val Asp Ile Ala Phe Gly Gly Asn Phe Phe Ala Ile
1 5 10 15 Val Pro Ala
Glu Gln Leu Gly Ile Asp Ile Ser Val Gln Asn Leu Ser 20
25 30 Arg Leu Gln Glu Ala Gly Glu Leu Leu Arg
Thr Glu Ile Asn Arg Ser 35 40
45 Val Lys Val Gln His Pro Gln Leu Pro His Ile Asn Thr Val Asp Cys
50 55 60 Val Glu Ile Tyr Gly Pro Pro
Thr Asn Pro Glu Ala Asn Tyr Lys Asn 65 70
75 80 Val Val Ile Phe Gly Asn Arg Gln Ala Asp Arg Ser
Pro Cys Gly Thr 85 90
95 Gly Thr Ser Ala Lys Met Ala Thr Leu Tyr Ala Lys Gly Gln Leu Arg
100 105 110 Ile Gly Glu Thr Phe Val
Tyr Glu Ser Ile Leu Gly Ser Leu Phe Gln 115 120
125 Gly Arg Val Leu Gly Glu Glu Arg Ile Pro Gly 130
135 86 139 PRT Brucella suis 86 Gly Pro Ile Lys Val Asp Val
Ala Tyr Gly Gly Asn Phe Tyr Ala Ile 1 5
10 15 Val Glu Pro Gln Glu Asn Tyr Thr Asp Met Asp Asp
Tyr Ser Ala Leu 20 25 30
Gln Leu Ile Ala Trp Ser Pro Val Leu Arg Gln Arg Leu Asn Glu Lys
35 40 45 Tyr Lys Phe Gln His Pro Glu
Leu Pro Asp Ile Asn Arg Leu Ser His 50 55
60 Ile Leu Trp Thr Gly Lys Pro Lys His Pro Gln Ala His Ala Arg Asn
65 70 75 80 Ala Val
Phe Tyr Gly Asp Lys Ala Ile Asp Arg Ser Pro Cys Gly Thr
85 90 95 Gly Thr Ser Ala Arg Met Ala
Gln Leu Ala Ala Lys Gly Lys Leu Lys 100 105
110 Pro Gly Asp Glu Phe Ile His Glu Ser Ile Ile Gly Ser Leu
Phe His 115 120 125 Gly Arg Val
Glu Arg Ala Ala Glu Val Ala Gly 130 135 87 106 PRT
Trypanosoma cruzi 87 Lys Lys Ser Phe Thr Cys Ile Asp Met His Thr Glu Gly
Glu Ala Ala 1 5 10 15
Arg Ile Val Thr Ser Gly Leu Pro His Ile Pro Gly Ser Asn Met Ala
20 25 30 Glu Lys Lys Ala Tyr Leu Gln
Glu Asn Met Asp Tyr Leu Arg Arg Gly 35 40
45 Ile Met Leu Glu Pro Arg Gly His Asp Asp Met Phe Gly Ala Phe
Leu 50 55 60 Phe Asp Pro Ile Glu
Glu Gly Ala Asp Leu Gly Met Val Phe Met Asp 65 70
75 80 Thr Gly Gly Tyr Leu Asn Met Cys Gly His
Asn Ser Ile Ala Ala Val 85 90
95 Thr Ala Ala Val Glu Thr Gly Ile Val Ser 100
105 88 106 PRT Brucella suis 88 Arg His Ser Phe Phe Cys Val Asp
Gly His Thr Cys Gly Asn Pro Val 1 5 10
15 Arg Leu Val Ala Gly Gly Gly Pro Asn Leu Asn Gly Ser Thr
Met Met 20 25 30 Glu Lys
Arg Ala His Phe Leu Ala Glu Tyr Asp Trp Ile Arg Thr Gly 35
40 45 Leu Met Phe Glu Pro Arg Gly His Asp Met
Met Ser Gly Ser Ile Leu 50 55 60
Tyr Pro Pro Thr Arg Pro Asp Cys Asp Val Ala Val Leu Phe Ile Glu 65
70 75 80 Thr Ser Gly Cys Leu
Pro Met Cys Gly His Gly Thr Ile Gly Thr Val 85
90 95 Thr Met Ala Ile Glu Gln Gly Leu Val Thr
100 105 89 109 PRT Trypanosoma cruzi 89 Met Arg
Phe Lys Lys Ser Phe Thr Cys Ile Asp Met His Thr Glu Gly 1
5 10 15 Glu Ala Ala Arg Ile Val Thr Ser
Gly Leu Pro His Ile Pro Gly Ser 20 25
30 Asn Met Ala Glu Lys Lys Ala Tyr Leu Gln Glu Asn Met Asp Tyr
Leu 35 40 45 Arg Arg Gly Ile
Met Leu Glu Pro Arg Gly His Asp Asp Met Phe Gly 50
55 60 Ala Phe Leu Phe Asp Pro Ile Glu Glu Gly Ala Asp
Leu Gly Met Val 65 70 75
80 Phe Met Asp Thr Gly Gly Tyr Leu Asn Met Cys Gly His Asn Ser Ile
85 90 95 Ala Ala Val Thr
Ala Ala Val Glu Thr Gly Ile Val Ser 100 105
90 109 PRT Rhodobacter sphaeroides 90 Met Arg Val Gln Asp Val Tyr Asn
Val Ile Tyr Thr His Thr Glu Gly 1 5 10
15 Glu Pro Leu Cys Ile Ile Tyr Ser Gly Val Pro Tyr Pro Ala
Gly Ser 20 25 30 Thr Ile
Leu Glu Lys Arg Ala Phe Leu Glu Glu Asn Tyr Asp Trp Leu 35
40 45 Arg Lys Ala Leu Met Arg Glu Pro Arg Gly
His Ala Asp Met Phe Gly 50 55 60
Val Phe Leu Thr Pro Pro Ser Ser Arg Asp Tyr Asp Ala Gly Leu Ile 65
70 75 80 Tyr Ile Asp Gly Lys
Glu Tyr Ser His Met Cys Gly His Gly Thr Ile 85
90 95 Ala Val Ala Met Ala Met Val Ala Asn Gly Leu
Val Ala 100 105 91 47 PRT Trypanosoma cruzi
91 Lys Val Gln His Pro Gln Leu Pro His Ile Asn Thr Val Asp Cys Val 1
5 10 15 Glu Ile Tyr Gly Pro
Pro Thr Asn Pro Glu Ala Asn Tyr Lys Asn Val 20
25 30 Val Ile Phe Gly Asn Arg Gln Ala Asp Arg Ser Pro
Cys Gly Thr 35 40 45 92 47 PRT
Rhodobacter sphaeroides 92 Lys Ser Ser Thr Pro Thr Glu Ala His Ile Asn
Asn Leu Asn Phe Val 1 5 10
15 Thr Leu Trp His Lys Pro Pro Ser Arg Gly Trp Leu Tyr Lys Asn Val
20 25 30 His Cys Phe Leu Glu
Gly Gln Leu Asp Arg Leu Pro Gly Gly Thr 35 40
45 93 118 PRT Trypanosoma cruzi 93 Arg Phe Lys Lys Ser
Phe Thr Cys Ile Asp Met His Thr Glu Gly Glu 1 5
10 15 Ala Ala Arg Ile Val Thr Ser Gly Leu Pro His
Ile Pro Gly Ser Asn 20 25
30 Met Ala Glu Lys Lys Ala Tyr Leu Gln Glu Asn Met Asp Tyr Leu Arg
35 40 45 Arg Gly Ile Met Leu Glu Pro
Arg Gly His Asp Asp Met Phe Gly Ala 50 55
60 Phe Leu Phe Asp Pro Ile Glu Glu Gly Ala Asp Leu Gly Met Val Phe
65 70 75 80 Met Asp
Thr Gly Gly Tyr Leu Asn Met Cys Gly His Asn Ser Ile Ala
85 90 95 Ala Val Thr Ala Ala Val Glu
Thr Gly Ile Val Ser Val Pro Ala Lys 100 105
110 Ala Thr Asn Val Pro Val 115 94 118 PRT
Burkholderia pseudomallei 94 Arg Asp Met Lys His Ile His Ile Ile Asp Ser
His Thr Gly Gly Glu 1 5 10
15 Pro Thr Arg Val Val Val Ser Gly Phe Pro Ala Leu Gly Gly Gly Thr
20 25 30 Met Ala Glu Arg Leu
Ala Val Leu Ala Arg Glu His Asp Arg Tyr Arg 35
40 45 Ala Ala Cys Ile Leu Glu Pro Arg Gly Ser Asp Val
Leu Val Gly Ala 50 55 60 Leu Leu
Cys Glu Pro Val Ser Ala Gly Ala Ala Ala Gly Val Ile Phe 65
70 75 80 Phe Asn Asn Ala Gly Tyr Leu
Gly Met Cys Gly His Gly Thr Ile Gly 85
90 95 Leu Val Arg Thr Leu His His Met Gly Arg Ile Gly
Pro Gly Val His 100 105 110
Arg Ile Glu Thr Pro Val 115 95 63 PRT Trypanosoma cruzi 95 Asn
Pro Glu Ala Asn Tyr Lys Asn Val Val Ile Phe Gly Asn Arg Gln 1
5 10 15 Ala Asp Arg Ser Pro Cys Gly
Thr Gly Thr Ser Ala Lys Met Ala Thr 20 25
30 Leu Tyr Ala Lys Gly Gln Leu Arg Ile Gly Glu Thr Phe Val
Tyr Glu 35 40 45 Ser Ile Leu
Gly Ser Leu Phe Gln Gly Arg Val Leu Gly Glu Glu 50
55 60 96 63 PRT Burkholderia pseudomallei 96 Asp Pro
Glu Tyr Asp Ser Arg Ser Phe Val Leu Cys Pro Gly His Ala 1
5 10 15 Tyr Asp Arg Ser Pro Cys Gly Thr
Gly Thr Ser Ala Lys Leu Ala Cys 20 25
30 Leu Ala Ala Asp Gly Lys Leu Ala Ala Gly Val Thr Trp Arg Gln
Ala 35 40 45 Ser Val Ile Gly
Ser Val Phe Ser Ala Ser Tyr Ala Ala Ala Glu 50 55
60 97 118 PRT Trypanosoma cruzi 97 Arg Phe Lys Lys Ser
Phe Thr Cys Ile Asp Met His Thr Glu Gly Glu 1 5
10 15 Ala Ala Arg Ile Val Thr Ser Gly Leu Pro His
Ile Pro Gly Ser Asn 20 25
30 Met Ala Glu Lys Lys Ala Tyr Leu Gln Glu Asn Met Asp Tyr Leu Arg
35 40 45 Arg Gly Ile Met Leu Glu Pro
Arg Gly His Asp Asp Met Phe Gly Ala 50 55
60 Phe Leu Phe Asp Pro Ile Glu Glu Gly Ala Asp Leu Gly Met Val Phe
65 70 75 80 Met Asp
Thr Gly Gly Tyr Leu Asn Met Cys Gly His Asn Ser Ile Ala
85 90 95 Ala Val Thr Ala Ala Val Glu
Thr Gly Ile Val Ser Val Pro Ala Lys 100 105
110 Ala Thr Asn Val Pro Val 115 98 118 PRT
Burkholderia mallei 98 Arg Asp Met Lys His Ile His Ile Ile Asp Ser His
Thr Gly Gly Glu 1 5 10
15 Pro Thr Arg Val Val Val Ser Gly Phe Pro Ala Leu Gly Gly Gly Thr
20 25 30 Met Ala Glu Arg Leu Ala
Val Leu Ala Arg Glu His Asp Arg Tyr Arg 35 40
45 Ala Ala Cys Ile Leu Glu Pro Arg Gly Ser Asp Val Leu Val
Gly Ala 50 55 60 Leu Leu Cys Glu
Pro Val Ser Ala Gly Ala Ala Ala Gly Val Ile Phe 65 70
75 80 Phe Asn Asn Ala Gly Tyr Leu Gly Met
Cys Gly His Gly Thr Ile Gly 85 90
95 Leu Val Arg Thr Leu His His Met Gly Arg Ile Gly Pro Gly Val
His 100 105 110 Arg Ile Glu
Thr Pro Val 115 99 63 PRT Trypanosoma cruzi 99 Asn Pro Glu Ala
Asn Tyr Lys Asn Val Val Ile Phe Gly Asn Arg Gln 1 5
10 15 Ala Asp Arg Ser Pro Cys Gly Thr Gly Thr
Ser Ala Lys Met Ala Thr 20 25
30 Leu Tyr Ala Lys Gly Gln Leu Arg Ile Gly Glu Thr Phe Val Tyr Glu
35 40 45 Ser Ile Leu Gly Ser Leu
Phe Gln Gly Arg Val Leu Gly Glu Glu 50 55
60 100 63 PRT Burkholderia mallei 100 Asp Pro Glu Tyr Asp Ser Arg
Ser Phe Val Leu Cys Pro Gly His Ala 1 5
10 15 Tyr Asp Arg Ser Pro Cys Gly Thr Gly Thr Ser Ala
Lys Leu Ala Cys 20 25 30
Leu Ala Ala Asp Gly Lys Leu Val Ala Gly Val Thr Trp Arg Gln Ala
35 40 45 Ser Val Ile Gly Ser Val Phe
Ser Ala Ser Tyr Ala Ala Ala Glu 50 55
60 101 115 PRT Trypanosoma cruzi 101 Lys Ser Phe Thr Cys Ile Asp Met
His Thr Glu Gly Glu Ala Ala Arg 1 5 10
15 Ile Val Thr Ser Gly Leu Pro His Ile Pro Gly Ser Asn Met
Ala Glu 20 25 30 Lys Lys
Ala Tyr Leu Gln Glu Asn Met Asp Tyr Leu Arg Arg Gly Ile 35
40 45 Met Leu Glu Pro Arg Gly His Asp Asp Met
Phe Gly Ala Phe Leu Phe 50 55 60
Asp Pro Ile Glu Glu Gly Ala Asp Leu Gly Met Val Phe Met Asp Thr 65
70 75 80 Gly Gly Tyr Leu Asn
Met Cys Gly His Asn Ser Ile Ala Ala Val Thr 85
90 95 Ala Ala Val Glu Thr Gly Ile Val Ser Val Pro
Ala Lys Ala Thr Asn 100 105
110 Val Pro Val 115 102 115 PRT Pseudomonas putida 102 Lys Gln
Ile His Val Ile Asp Ser His Thr Gly Gly Glu Pro Thr Arg 1
5 10 15 Leu Val Met Lys Gly Phe Pro Gln
Leu Arg Gly Arg Ser Met Ala Glu 20 25
30 Gln Arg Asp Glu Leu Arg Glu Leu His Asp Arg Trp Arg Arg Ala
Cys 35 40 45 Leu Leu Glu Pro
Arg Gly Asn Asp Val Leu Val Gly Ala Leu Tyr Cys 50
55 60 Pro Pro Val Ser Ala Asp Ala Thr Cys Gly Val Ile
Phe Phe Asn Asn 65 70 75
80 Ala Gly Tyr Leu Asn Met Cys Gly His Gly Thr Ile Gly Leu Val Ala
85 90 95 Ser Leu Gln His
Met Gly Leu Ile Thr Pro Gly Val His Lys Ile Asp 100
105 110 Thr Pro Val 115 103 58 PRT
Trypanosoma cruzi 103 Asn Pro Glu Ala Asn Tyr Lys Asn Val Val Ile Phe
Gly Asn Arg Gln 1 5 10
15 Ala Asp Arg Ser Pro Cys Gly Thr Gly Thr Ser Ala Lys Met Ala Thr
20 25 30 Leu Tyr Ala Lys Gly Gln
Leu Arg Ile Gly Glu Thr Phe Val Tyr Glu 35 40
45 Ser Ile Leu Gly Ser Leu Phe Gln Gly Arg 50
55 104 58 PRT Pseudomonas putida 104 Asp Pro Asn Ala Asp Ser
Arg Asn Phe Val Met Cys Pro Gly Lys Ala 1 5
10 15 Tyr Asp Arg Ser Pro Cys Gly Thr Gly Thr Ser Ala
Lys Leu Ala Cys 20 25 30
Leu Ala Ala Asp Gly Lys Leu Ala Glu Gly Gln Thr Trp Val Gln Ala
35 40 45 Ser Ile Thr Gly Ser Gln Phe
His Gly Arg 50 55 105 180 PRT Trypanosoma cruzi
105 Ser Gly Leu Pro His Ile Pro Gly Ser Asn Met Ala Glu Lys Lys Ala 1
5 10 15 Tyr Leu Gln Glu Asn
Met Asp Tyr Leu Arg Arg Gly Ile Met Leu Glu 20
25 30 Pro Arg Gly His Asp Asp Met Phe Gly Ala Phe Leu
Phe Asp Pro Ile 35 40 45 Glu
Glu Gly Ala Asp Leu Gly Ile Val Phe Met Asp Thr Gly Gly Tyr 50
55 60 Leu Asn Met Cys Gly His Asn Ser Ile Ala
Ala Val Thr Ala Ala Val 65 70 75
80 Glu Thr Gly Ile Val Ser Val Pro Ala Lys Ala Thr Asn Val Pro
Val 85 90 95 Val Leu
Asp Thr Pro Ala Gly Leu Val Arg Gly Thr Ala His Leu Gln 100
105 110 Ser Gly Thr Glu Ser Glu Val Ser Asn
Ala Ser Ile Ile Asn Val Pro 115 120
125 Ser Phe Leu Tyr Gln Gln Asp Val Val Val Val Leu Pro Lys Pro Tyr
130 135 140 Gly Glu Val Arg Val Asp Ile
Ala Phe Gly Gly Asn Phe Phe Ala Ile 145 150
155 160 Val Pro Ala Glu Gln Leu Gly Ile Asp Ile Ser Val
Gln Asn Leu Ser 165 170
175 Arg Leu Gln Glu 180 106 164 PRT Leishmania major 106 Thr
Gly Phe Pro Glu Leu Ala Gly Glu Thr Ile Ala Asp Lys Leu Asp 1
5 10 15 Asn Leu Arg Thr Gln His Asp
Gln Trp Arg Arg Ala Cys Leu Leu Glu 20 25
30 Pro Arg Gly Asn Asp Val Leu Val Gly Ala Leu Tyr Cys Ala
Pro Val 35 40 45 Ser Ala Asp
Ala Thr Cys Gly Val Ile Phe Phe Asn Asn Ala Gly Tyr 50
55 60 Leu Gly Met Cys Gly His Gly Thr Ile Gly Leu Val
Ala Ser Leu His 65 70 75
80 His Leu Gly Arg Ile Ala Pro Gly Val His Lys Ile Asp Thr Pro Val
85 90 95 Gly Pro Val Ser
Ala Thr Leu His Ala Asp Gly Ala Val Thr Leu Arg 100
105 110 Asn Val Pro Ala Tyr Arg Tyr Arg Gln Gln Val
Pro Val Asp Val Pro 115 120 125
Gly His Gly Arg Val Tyr Gly Asp Ile Ala Trp Gly Gly Asn Trp Phe 130
135 140 Phe Leu Val Ser Asp His Gly Gln Ala
Leu Gln Met Asp Asn Val Glu 145 150 155
160 Ala Leu Thr Asp 107 68 PRT Trypanosoma cruzi 107 Pro
Thr Asn Pro Glu Ala Asn Tyr Lys Asn Val Val Ile Phe Gly Asn 1
5 10 15 Arg Gln Ala Asp Arg Ser Pro
Cys Gly Thr Gly Thr Ser Ala Lys Met 20 25
30 Ala Thr Leu Tyr Ala Lys Gly Gln Leu Arg Ile Gly Glu Thr
Phe Val 35 40 45 Tyr Glu Ser
Ile Leu Gly Ser Leu Phe Gln Gly Arg Val Leu Gly Glu 50
55 60 Glu Arg Ile Pro 65 108 68 PRT Leishmania major
108 Pro Thr Thr Pro Thr Pro Thr Ala Thr Ser Ser Cys Ala Gln Gly Lys 1
5 10 15 Ala Tyr Asp Arg
Ser Pro Cys Gly Thr Gly Thr Asn Ala Lys Leu Ala 20
25 30 Cys Leu Ala Gly Asp Ser Lys Leu Ala Ala Gly
Glu Pro Trp Leu Gln 35 40 45
Val Thr Ile Thr Cys Arg Gln Phe Lys Arg Ser Tyr Gln Trp Glu Cys 50
55 60 Lys Arg Val Pro 65 109 28 PRT
Trypanosoma cruzi 109 Val Thr Ala Glu Ile Thr Gly Lys Ala Phe Ile Met
Gly Phe Asn Thr 1 5 10
15 Met Leu Phe Asp Pro Thr Asp Pro Phe Lys Asn Gly 20
25 110 27 PRT Leishmania major 110 Val Pro Pro Ser Ile Thr Arg
Arg Ala Tyr Met Thr Ala Asp Ser Thr 1 5
10 15 Leu Leu Ile Asp Gln Asp Pro Phe Ala Trp Gly
20 25 111 182 PRT Trypanosoma cruzi 111 Val Thr
Ser Gly Leu Pro His Ile Pro Gly Ser Asn Met Ala Glu Lys 1
5 10 15 Lys Ala Tyr Leu Gln Glu Asn Met
Asp Tyr Leu Arg Arg Gly Ile Met 20 25
30 Leu Glu Pro Arg Gly His Asp Asp Met Phe Gly Ala Phe Leu Phe
Asp 35 40 45 Pro Ile Glu Glu
Gly Ala Asp Leu Gly Ile Val Phe Met Asp Thr Gly 50
55 60 Gly Tyr Leu Asn Met Cys Gly His Asn Ser Ile Ala
Ala Val Thr Ala 65 70 75
80 Ala Val Glu Thr Gly Ile Val Ser Val Pro Ala Lys Ala Thr Asn Val
85 90 95 Pro Val Val Leu
Asp Thr Pro Ala Gly Leu Val Arg Gly Thr Ala His 100
105 110 Leu Gln Ser Gly Thr Glu Ser Glu Val Ser Asn
Ala Ser Ile Ile Asn 115 120 125
Val Pro Ser Phe Leu Tyr Gln Gln Asp Val Val Val Val Leu Pro Lys 130
135 140 Pro Tyr Gly Glu Val Arg Val Asp Ile
Ala Phe Gly Gly Asn Phe Phe 145 150 155
160 Ala Ile Val Pro Ala Glu Gln Leu Gly Ile Asp Ile Ser Val
Gln Asn 165 170 175 Leu
Ser Arg Leu Gln Glu 180 112 166 PRT Leishmania major 112 Val
Met Thr Gly Phe Pro Glu Leu Ala Gly Glu Thr Ile Ala Asp Lys 1
5 10 15 Leu Asp Asn Leu Arg Thr Gln
His Asp Gln Trp Arg Arg Ala Cys Leu 20 25
30 Leu Glu Pro Arg Gly Asn Asp Val Leu Val Gly Ala Leu Tyr
Cys Ala 35 40 45 Pro Val Ser
Ala Asp Ala Thr Cys Gly Val Ile Phe Phe Asn Asn Ala 50
55 60 Gly Tyr Leu Gly Met Cys Gly His Gly Thr Ile Gly
Leu Val Ala Ser 65 70 75
80 Leu His His Leu Gly Arg Ile Ala Pro Gly Val His Lys Ile Asp Thr
85 90 95 Pro Val Gly Pro
Val Ser Ala Thr Leu His Ala Asp Gly Ala Val Thr 100
105 110 Leu Arg Asn Val Pro Ala Tyr Arg Tyr Arg Gln
Gln Val Pro Val Asp 115 120 125
Val Pro Gly His Gly Arg Val Tyr Gly Asp Ile Ala Trp Gly Gly Asn 130
135 140 Trp Phe Phe Leu Val Ser Asp His Gly
Gln Ala Leu Gln Met Asp Asn 145 150 155
160 Val Glu Ala Leu Thr Asp 165 113 142 PRT
Trypanosoma cruzi 113 Arg Ile Val Thr Ser Gly Leu Pro His Ile Pro Gly
Ser Asn Met Ala 1 5 10
15 Glu Lys Lys Ala Tyr Leu Gln Glu Asn Met Asp Tyr Leu Arg Arg Gly
20 25 30 Ile Met Leu Glu Pro Arg
Gly His Asp Asp Met Phe Gly Ala Phe Leu 35 40
45 Phe Asp Pro Ile Glu Glu Gly Ala Asp Leu Gly Ile Val Phe
Met Asp 50 55 60 Thr Gly Gly Tyr
Leu Asn Met Cys Gly His Asn Ser Ile Ala Ala Val 65 70
75 80 Thr Ala Ala Val Glu Thr Gly Ile Leu
Ser Val Pro Ala Lys Ala Thr 85 90
95 Asn Val Pro Val Val Leu Asp Thr Pro Ala Gly Leu Val Arg Gly
Thr 100 105 110 Ala His Leu
Gln Ser Gly Thr Glu Ser Glu Val Ser Asn Ala Ser Ile 115
120 125 Ile Asn Val Pro Ser Phe Leu Tyr Gln Gln Asp
Val Val Ile 130 135 140 114 137 PRT
Trypanosoma brucei 114 Arg Ile Ile Thr Gly Gly Val Pro Glu Ile Lys Gly
Glu Thr Gln Leu 1 5 10
15 Glu Arg Arg Ala Tyr Cys Met Glu His Leu Asp Tyr Leu Arg Glu Ile
20 25 30 Leu Met Tyr Glu Pro Arg
Gly His His Gly Met Tyr Gly Cys Ile Ile 35 40
45 Thr Pro Pro Ala Ser Ala His Ala Asp Phe Gly Val Leu Phe
Met His 50 55 60 Asn Glu Gly Trp
Ser Thr Met Cys Gly His Gly Ile Ile Ala Val Ile 65 70
75 80 Thr Val Gly Ile Glu Thr Gly Met Phe
Glu Val Lys Gly Glu Lys Gln 85 90
95 Asn Phe Ile Ile Asp Ser Pro Ala Gly Glu Val Ile Ala Tyr Ala
Lys 100 105 110 Tyr Asn Gly
Ser Glu Val Glu Ser Val Ser Phe Glu Asn Val Pro Ser 115
120 125 Phe Val Tyr Lys Lys Asp Val Pro Ile 130
135 115 140 PRT Trypanosoma cruzi 115 Val Thr Ser Gly
Leu Pro His Ile Pro Gly Ser Asn Met Ala Glu Lys 1 5
10 15 Lys Ala Tyr Leu Gln Glu Asn Met Asp Tyr
Leu Arg Arg Gly Ile Met 20 25
30 Leu Glu Pro Arg Gly His Asp Asp Met Phe Gly Ala Phe Leu Phe Asp
35 40 45 Pro Ile Glu Glu Gly Ala
Asp Leu Gly Ile Val Phe Met Asp Thr Gly 50 55
60 Gly Tyr Leu Asn Met Cys Gly His Asn Ser Ile Ala Ala Val Thr
Ala 65 70 75 80 Ala
Val Glu Thr Gly Ile Leu Ser Val Pro Ala Lys Ala Thr Asn Val
85 90 95 Pro Val Val Leu Asp Thr Pro
Ala Gly Leu Val Arg Gly Thr Ala His 100 105
110 Leu Gln Ser Gly Thr Glu Ser Glu Val Ser Asn Ala Ser Ile
Ile Asn 115 120 125 Val Pro Ser
Phe Leu Tyr Gln Gln Asp Val Val Ile 130 135
140 116 135 PRT Trypanosoma brucei 116 Ile Thr Gly Gly Val Pro Glu
Ile Lys Gly Glu Thr Gln Leu Glu Arg 1 5
10 15 Arg Ala Tyr Cys Met Glu His Leu Asp Tyr Leu Arg
Glu Ile Leu Met 20 25 30
Tyr Glu Pro Arg Gly His His Gly Met Tyr Gly Cys Ile Ile Thr Pro
35 40 45 Pro Ala Ser Ala His Ala Asp
Phe Gly Val Leu Phe Met His Asn Glu 50 55
60 Gly Trp Ser Thr Met Cys Gly His Gly Ile Ile Ala Val Ile Thr Val
65 70 75 80 Gly Ile
Glu Thr Gly Met Phe Glu Val Lys Gly Glu Lys Gln Asn Phe
85 90 95 Ile Ile Asp Ser Pro Ala Gly
Glu Val Ile Ala Tyr Ala Lys Tyr Asn 100 105
110 Gly Ser Glu Val Glu Ser Val Ser Phe Glu Asn Val Pro Ser
Phe Val 115 120 125 Tyr Lys Lys
Asp Val Pro Ile 130 135 117 103 PRT Trypanosoma cruzi
117 Phe Gly Gly Asn Phe Phe Ala Ile Val Pro Ala Glu Gln Leu Gly Ile 1
5 10 15 Asp Ile Ser Val
Gln Asn Leu Ser Arg Leu Gln Glu Ala Gly Glu Leu 20
25 30 Leu Arg Thr Glu Ile Asn Arg Ser Val Lys Val
Gln His Pro Gln Leu 35 40 45
Pro His Ile Asn Thr Val Asp Cys Val Glu Ile Tyr Gly Pro Pro Thr 50
55 60 Asn Pro Glu Ala Asn Tyr Lys Asn Val
Val Ile Phe Gly Asn Arg Gln 65 70 75
80 Ala Asp Arg Ser Pro Cys Gly Thr Gly Thr Ser Ala Lys Met
Ala Thr 85 90 95 Leu
Tyr Ala Lys Gly Gln Leu 100 118 95 PRT Trypanosoma congolense
118 Trp Gly Gly Asn Trp Phe Phe Leu Val Ser Asp His Gly His Glu Leu 1
5 10 15 Gln Met Asp Asn
Val Glu Ala Leu Thr Asp Tyr Thr Trp Ala Met Leu 20
25 30 Asn Ala Leu Glu Ala Gln Gly Ile Arg Gly Ala
Asp Gly Ala Leu Ile 35 40 45
Asp His Ile Glu Leu Phe Ala Asp Asp Ala His Ala Asp Ser Arg Asn 50
55 60 Phe Val Met Cys Pro Gly Lys Ala Tyr
Asp Arg Ser Pro Cys Gly Thr 65 70 75
80 Gly Thr Ser Ala Lys Leu Ala Cys Leu Ala Ala Asp Ala Lys
Leu 85 90 95 119 55
PRT Trypanosoma cruzi 119 Asn Val Pro Ser Phe Leu Tyr Gln Gln Asp Val
Val Val Val Leu Pro 1 5 10
15 Lys Pro Tyr Gly Glu Val Arg Val Asp Ile Ala Phe Gly Gly Asn Phe
20 25 30 Phe Ala Ile Val Pro
Ala Glu Gln Leu Gly Ile Asp Ile Ser Val Gln 35
40 45 Asn Leu Ser Arg Leu Gln Glu 50
55 120 52 PRT Trypanosoma congolense 120 His Val Pro Ala Tyr Arg Tyr
Arg Lys Gln Val Pro Val Glu Val Pro 1 5
10 15 Gly His Gly Val Val Leu Gly Asp Ile Ala Trp Gly
Gly Asn Trp Phe 20 25 30
Phe Leu Val Ser Asp His Gly His Glu Leu Gln Met Asp Asn Val Glu
35 40 45 Ala Leu Thr Asp 50 121
117 PRT Trypanosoma cruzi 121 Leu Pro Lys Pro Tyr Gly Glu Val Arg Val
Asp Ile Ala Phe Gly Gly 1 5 10
15 Asn Phe Phe Ala Ile Val Pro Ala Glu Gln Leu Gly Ile Asp Ile Ser
20 25 30 Val Gln Asn Leu
Ser Arg Leu Gln Glu Ala Gly Glu Leu Leu Arg Thr 35
40 45 Glu Ile Asn Arg Ser Val Lys Val Gln His Pro Gln
Leu Pro His Ile 50 55 60 Asn Thr
Val Asp Cys Val Glu Ile Tyr Gly Pro Pro Thr Asn Pro Glu 65
70 75 80 Ala Asn Tyr Lys Asn Val Val
Ile Phe Gly Asn Arg Gln Ala Asp Arg 85
90 95 Ser Pro Cys Gly Thr Gly Thr Ser Ala Lys Met Ala
Thr Leu Tyr Ala 100 105 110
Lys Gly Gln Leu Arg 115 122 116 PRT Trypanosoma vivax 122 Leu
Pro His Pro Tyr Gly Lys Tyr Ala Val Ile Ser Phe Gly Gly Ser 1
5 10 15 Phe Phe Ala Leu Ile Asp Ala
Ala Gln Leu Gln Leu Thr Val Asp Lys 20 25
30 Gly His Leu Ser Thr Leu Gln His Val Gly Gly Leu Leu Arg
Asp Thr 35 40 45 Leu Asn Arg
Asn Val Ser Val Gln His Pro Gln Leu Pro His Ile Asn 50
55 60 Arg Ile Asp Cys Val Glu Ile Tyr Asp Pro Pro Thr
Asn Pro Ala Ala 65 70 75
80 Ser Cys Lys Asn Val Val Ile Phe Gly Asn Ser Gln Val Asp Arg Ser
85 90 95 Pro Cys Gly Thr
Gly Thr Cys Ala Lys Met Ala Leu Leu Tyr Ala Lys 100
105 110 Gly Lys Leu Lys 115 123 106 PRT
Trypanosoma cruzi 123 Arg Glu Ile Met Arg Phe Lys Lys Ser Phe Thr Cys
Ile Asp Met His 1 5 10
15 Thr Glu Gly Glu Ala Ala Arg Ile Val Thr Ser Gly Leu Pro His Ile
20 25 30 Pro Gly Ser Asn Met Ala
Glu Lys Lys Ala Tyr Leu Gln Glu Asn Met 35 40
45 Asp Tyr Leu Arg Arg Gly Ile Met Leu Glu Pro Arg Gly His
Asp Asp 50 55 60 Met Phe Gly Ala
Phe Leu Phe Asp Pro Ile Glu Glu Gly Ala Asp Leu 65 70
75 80 Gly Met Val Phe Met Asp Thr Gly Gly
Tyr Leu Asn Met Cys Gly His 85 90
95 Asn Ser Ile Ala Ala Val Thr Ala Ala Val 100
105 124 106 PRT Trypanosoma vivax 124 Arg Val Val Met Gln
Phe Thr Gly Thr Met Thr Cys Ile Asp Met His 1 5
10 15 Thr Ala Gly Glu Pro Ala Arg Ile Val Thr Ser
Gly Phe Pro Asn Ile 20 25
30 Pro Gly Ala Ser Leu Val Glu Lys Arg Asp His Leu Gln Arg His Met
35 40 45 Asp His Ile Arg Arg Arg Val
Met Leu Glu Pro Arg Gly His Asp Asn 50 55
60 Met Phe Gly Ala Phe Leu Phe Tyr Pro Leu Thr Asp Gly Ala Asp Phe
65 70 75 80 Ser Val
Ile Phe Met Asp Ala Gly Gly Tyr Leu Asn Met Cys Gly His
85 90 95 Asn Ser Ile Ala Ile Ala Thr
Ala Ala Val 100 105 125 357 PRT Trypanosoma
cruzi 125 Lys Arg Glu Ile Met Arg Phe Lys Lys Ser Phe Thr Cys Ile Asp
Met 1 5 10 15 His Thr
Glu Gly Glu Ala Ala Arg Ile Val Thr Ser Gly Leu Pro His 20
25 30 Ile Pro Gly Ser Asn Met Ala Glu Lys
Lys Ala Tyr Leu Gln Glu Asn 35 40
45 Met Asp Tyr Leu Arg Arg Gly Ile Met Leu Glu Pro Arg Gly His Asp
50 55 60 Asp Met Phe Gly Ala Phe Leu
Phe Asp Pro Ile Glu Glu Gly Ala Asp 65 70
75 80 Leu Gly Met Val Phe Met Asp Thr Gly Gly Tyr Leu
Asn Met Cys Gly 85 90
95 His Asn Ser Ile Ala Ala Val Thr Ala Ala Val Glu Thr Gly Ile Val
100 105 110 Ser Val Pro Ala Lys Ala
Thr Asn Val Pro Val Val Leu Asp Thr Pro 115 120
125 Ala Gly Leu Val Arg Gly Thr Ala His Leu Gln Ser Gly Thr
Glu Ser 130 135 140 Glu Val Ser Asn
Ala Ser Ile Ile Asn Val Pro Ser Phe Leu Tyr Gln 145 150
155 160 Gln Asp Val Val Val Val Leu Pro Lys
Pro Tyr Gly Glu Val Arg Val 165 170
175 Asp Ile Ala Phe Gly Gly Asn Phe Phe Ala Ile Val Pro Ala Glu
Gln 180 185 190 Leu Gly Ile
Asp Ile Ser Val Gln Asn Leu Ser Arg Leu Gln Glu Ala 195
200 205 Gly Glu Leu Leu Arg Thr Glu Ile Asn Arg Ser
Val Lys Val Gln His 210 215 220 Pro
Gln Leu Pro His Ile Asn Thr Val Asp Cys Val Glu Ile Tyr Gly 225
230 235 240 Pro Pro Thr Asn Pro Glu
Ala Asn Tyr Lys Asn Val Val Ile Phe Gly 245
250 255 Asn Arg Gln Ala Asp Arg Ser Pro Cys Gly Thr Gly
Thr Ser Ala Lys 260 265 270
Met Ala Thr Leu Tyr Ala Lys Gly Gln Leu Arg Ile Gly Glu Thr Phe
275 280 285 Val Tyr Glu Ser Ile Leu Gly
Ser Leu Phe Gln Gly Arg Val Leu Gly 290 295
300 Glu Glu Arg Ile Pro Gly Val Lys Val Pro Val Thr Lys Asp Ala Glu
305 310 315 320 Glu Gly
Met Leu Val Val Thr Ala Glu Ile Thr Gly Lys Ala Phe Ile
325 330 335 Met Gly Phe Asn Thr Met Leu
Phe Asp Pro Thr Asp Pro Phe Lys Asn 340 345
350 Gly Phe Thr Leu Lys 355 126 337 PRT Vibrio
parahaemolyticus 126 Lys Glu Arg Lys Met Arg Gln Gly Thr Phe Phe Cys Ile
Asp Ala His 1 5 10 15
Thr Cys Gly Asn Pro Val Arg Leu Val Ala Gly Gly Val Pro Pro Leu
20 25 30 Glu Gly Asn Thr Met Ser Glu
Lys Arg Gln Tyr Phe Leu Glu His Tyr 35 40
45 Asp Trp Ile Arg Gln Ala Leu Met Phe Glu Pro Arg Gly His Ser
Met 50 55 60 Met Ser Gly Ser Val
Val Leu Pro Pro Cys Ser Asp Asn Ala Asp Ala 65 70
75 80 Ser Ile Leu Phe Ile Glu Thr Ser Gly Cys
Leu Pro Met Cys Gly His 85 90
95 Gly Thr Ile Gly Thr Val Thr Thr Ala Ile Glu Asn Arg Leu Ile Thr
100 105 110 Pro Lys Glu Glu
Gly Arg Leu Ile Leu Asp Val Pro Ala Gly Gln Ile 115
120 125 Glu Val His Tyr Gln Thr Lys Gly Asp Lys Val Thr
Ser Val Lys Ile 130 135 140 Phe Asn
Val Pro Ala Tyr Leu Ala His Gln Asp Val Thr Val Glu Ile 145
150 155 160 Glu Gly Leu Gly Glu Ile Thr
Val Asp Val Ala Tyr Gly Gly Asn Tyr 165
170 175 Tyr Val Ile Val Asp Pro Gln Glu Asn Tyr Ala Gly
Leu Glu His Tyr 180 185 190
Ser Pro Asp Glu Ile Leu Met Leu Ser Pro Lys Val Arg Thr Ala Val
195 200 205 Ser Lys Ala Val Glu Cys Ile
His Pro Asn Asp Pro Thr Val Cys Gly 210 215
220 Val Ser His Val Leu Trp Thr Gly Lys Pro Thr Gln Glu Gly Ala Thr
225 230 235 240 Ala Arg
Asn Ala Val Phe Tyr Gly Asp Lys Ala Leu Asp Arg Ser Pro
245 250 255 Cys Gly Thr Gly Thr Ser Ala
Arg Met Ala Gln Trp His Ala Lys Gly 260 265
270 Lys Leu Lys Ser Gly Glu Asp Phe Val His Glu Ser Ile Ile
Gly Ser 275 280 285 Leu Phe Asn
Gly Arg Ile Glu Gly Ile Thr Glu Val Asn Gly Gln Thr 290
295 300 Ala Ile Leu Pro Ser Ile Glu Gly Trp Ala Gln Val
Tyr Gly His Asn 305 310 315
320 Thr Ile Trp Val Asp Asp Glu Asp Pro Tyr Ala Tyr Gly Phe Glu Val
325 330 335 Lys 127 14 PRT
Artificial Sequence Description of Artificial Sequence Synthetic
peptide motif 127 Asp Arg Ser Pro Cys Gly Thr Gly Thr Ser Ala Lys Met
Ala 1 5 10 128 5 PRT Artificial Sequence
Description of Artificial Sequence Synthetic peptide motif 128 Asn
Met Cys Gly His 1 5 129 6 PRT Artificial Sequence
Description of Artificial Sequence Synthetic 6xHis tag 129 His His
His His His His 1 5
User Contributions:
Comment about this patent or add new information about this topic:
| People who visited this patent also read: | |
| Patent application number | Title |
|---|---|
| 20140200417 | MENTAL STATE ANALYSIS USING BLINK RATE |
| 20140200416 | MENTAL STATE ANALYSIS USING HEART RATE COLLECTION BASED ON VIDEO IMAGERY |
| 20140200415 | SYSTEM FOR CALIBRATING A PTT-BASED BLOOD PRESSURE MEASUREMENT USING ARM HEIGHT |
| 20140200414 | SYSTEMS APPROACH TO COMORBIDITY ASSESSMENT |
| 20140200413 | BIOLOGICAL INFORMATION MANAGEMENT MODULE, SLEEP MONITOR, AND CONTROL APPARATUS |