Patent application title: TRUNCATED BARD1 PROTEIN, AND ITS DIAGNOSTIC AND THERAPEUTIC USES
Inventors:
Fabien Gautier (Avrille, FR)
Jean Harb (Nantes, FR)
Khaled Meflah (Nantes, FR)
Irmgard Irminger-Finger (Geneva, CH)
Assignees:
Ayanda Biosystems SA
IPC8 Class: AA61K3816FI
USPC Class:
514 12
Class name: Designated organic active ingredient containing (doai) peptide containing (e.g., protein, peptones, fibrinogen, etc.) doai 25 or more peptide repeating units in known peptide chain structure
Publication date: 2010-01-21
Patent application number: 20100016228
Claims:
1. A method for treating tumors which method comprises administering a
pharmaceutical composition comprising a polypeptide fragment which has a
molecular weight of approximately 67 kD, wherein the polypeptide consists
essentially of 505 to 525 contiguous amino acids from amino acid position
252 to amino acid position 777 of the C-terminal end of BARD1 of SEQ ID
NO:1 to a subject bearing a tumor.
2. An antibody specifically directed against the polypeptide which has a molecular weight of approximately 67 kD, wherein the polypeptide consists essentially of 505 to 525 contiguous amino acids from amino acid position 252 to amino acid position 777 of the C-terminal end of BARD1.
3. An in vitro method of monitoring effectiveness of an anticancer treatment in a patient treated with pro-apoptotic drugs, which method comprises detecting antibodies directed against a polypeptide fragment which has a molecular weight of approximately 67 kD, wherein the polypeptide consists essentially of 505 to 525 contiguous amino acids from amino acid position 252 to amino acid position 777 of the C-terminal end of BARD1 of SEQ ID NO:1.
4. An in vitro method of monitoring effectiveness of an anticancer treatment in a patient treated with pro-apoptotic drugs, which method comprises detecting a polypeptide fragment which has a molecular weight of approximately 67 kD, wherein the polypeptide consists essentially of 505 to 525 contiguous amino acids from amino acid position 252 to amino acid position 777 of the C-terminal end of BARD1 of SEQ ID NO:1 in a biological sample.
5. A method of in vitro detection of a polypeptide which has a molecular weight of approximately 67 kD, and the sequence of which consists of 505 to 525 amino acids from the C-terminal end of BARD1, in a biological sample, which method comprises:(i) contacting a biological sample with an antibody according to claim 2, under conditions sufficient to allow formation of immunocomplexes between the antibody and said polypeptide likely to be contained in the biological sample; and(ii) detecting the formation of the immunocomplexes, the formation of which reveals the presence of the polypeptide.
6. A method of in vitro detection of an antibody according to claim 2, in a biological sample, which method comprises:(i) contacting a biological sample with a polypeptide which has a molecular weight of approximately 67 kD, and the sequence of which consists of 505 to 525 amino acids from the C-terminal end of BARD1, under conditions sufficient to allow formation of immunocomplexes between said polypeptide and any antibody contained in the biological sample; and(ii) detecting the formation of said immunocomplexes, the formation of which reveals the presence of the antibody.
Description:
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001]This application is a continuation of U.S. application Ser. No. 10/363,285, which is a U.S. national phase application of International Patent Application No. PCT/FR01/02731 filed Sep. 3, 2001, the disclosures of which are hereby incorporated by reference in their entirety, including all figures, tables and amino acid or nucleic acid sequences.
[0002]The present invention relates to a novel polypeptide derived from cleavage of the BARD1 protein, and to its diagnostic and therapeutic uses.
[0003]The BARD1 protein is a 97 kD protein which interacts with the product of the BRCA1 tumor suppressor gene, via ring motifs present on BRCA1 and BARD1 (for "BRCA1 Associated Ring Domain") (Wu et al., 1996). The gene encoding this protein has recently been cloned (WO 98/12327).
[0004]The authors of the present invention have now identified a novel polypeptide which corresponds to a proteolytic fragment of BARD1, cleaved during the apoptotic process.
[0005]This truncated protein is very immunogenic and can be used, inter alia, in the treatment of cancers or in monitoring the effectiveness of the treatment with proapoptotic drugs of patients suffering from cancer.
[0006]A subject of the invention is therefore the isolated polypeptide which has a molecular weight of approximately 67 kD, as measured, for example, by electrophoresis under denaturing conditions (SDS-PAGE), and the sequence of which consists of the amino acid sequence of the BARD1 protein deleted of its N-terminal portion which comprises the RING domain.
[0007]More particularly, the polypeptide of the invention can be defined as consisting of approximately 505 to 525 amino acids from the C-terminal end of the human BARD1 protein (the known sequence of which is given in the annex SEQ ID No. 1). Even more particularly, the polypeptide of the invention consists of 525 to 522 C-terminal amino acids of human BARD1 (SEQ ID NO:1).
[0008]The polypeptide of the invention can, for example, be purified from the apoptotic bodies derived from colon or mammary carcinoma cell lines.
[0009]More particularly, the polypeptide of the invention can be obtained using the method consisting in: [0010]culturing cells to confluency, for example cells belonging to a cell line such as PROb, SW48 or MCF7; [0011]inducing apoptosis of these cells by treating them 15 with a culture medium containing 5 mM sodium butyrate (NaB), at 37° C. for 24 hours; [0012]adding isolated recombinant BARD1 protein; [0013]incubating for a sufficient amount of time, for example 60 minutes at 37° C., so as to observe cleavage of the BARD1 protein to a 67 kDa form.
[0014]The polypeptide of the invention is recognized by an antibody directed against a polypeptide corresponding to amino acids 255 to 265 of BARD1 (SEQ ID NO:1).
[0015]The authors of the invention have, moreover, shown that hydrolysis of BARD1 occurs at an early stage of apoptosis and in a cell cycle-dependent manner. This hydrolysis is inhibited by EGTA and the calpain inhibitor I, N-acetyl-leu-leu-norleucinal (ALLnL), but not by several caspase inhibitors, which suggests hydrolysis by calcium-dependent cysteine proteases, calpains.
[0016]Any protein consisting of a sequence homologous to said sequence of 505 to 525 amino acids of SEQ ID NO: 1 is also included in the definition of the 67 kD polypeptide. A subject of the invention is also a nucleic acid encoding this truncated protein or homologues thereof.
[0017]The expression "homologous amino acid sequence" is intended to mean a sequence at least 70%, preferably 80%, more preferably 90%, similar to said sequence of 505 to 525 amino acids of SEQ ID NO:1.
[0018]The term "similar" refers to the complete resemblance or identity between the amino acids compared, but also to the incomplete resemblance, which is referred to as similarity. This search for similarity in a polypeptide sequence takes into account the conservative substitutions, which are substitutions of amino acids of the same class, such as substitutions of amino acids with uncharged side chains (such as asparagine, glutamine, serine, threonine, or tyrosine) of amino acids with base side chains (such as lysine, arginine or histidine), of amino acids with acidic side chains (such as aspartic acid or glutamic acid), of amino acids with apolar side chains (such as glycine, alanine, valine, leucine, isoleucine, proline, phenylalanine, methionine, tryptophan or cysteine).
[0019]More generally, the expression "homologous amino acid sequence" is therefore intended to mean any amino acid sequence which differs from said sequence by substitution, deletion and/or insertion of an amino acid or of a small number of amino acids, in particular by substitution of natural amino acids with unnatural amino acids or pseudo amino acids, at positions such that these modifications do not significantly affect the biological properties mentioned above.
[0020]Homology is generally determined using sequence analysis software (for example, Sequence Analysis Software Package of the Genetics Computer Group University of Wisconsin Biotechnology Center, 1710 University Avenue, Madison, Wis. 53705). Similar amino acid sequences are aligned so as to obtain the maximum degree of homology (i.e. identity or similarity, as defined above). For this purpose, it may be necessary to artificially introduce gaps into the sequence. Once the optimal alignment has been carried out, the degree of homology is established by recording all the positions for which the amino acids of the two compared sequences are identical, with respect to the total number of positions.
[0021]The polypeptide of the present invention can be synthesized by all the methods well known to those skilled in the art. The polypeptide of the invention can, for example, be synthesized by the techniques of synthetic chemistry, such as synthesis of the Merrifield type, which is advantageous for reasons of purity, of antigenic specificity and of absence of undesired side products, and for its use of production.
[0022]The protein produced can then be recovered and purified.
[0023]The methods of purification used are known to those skilled in the art. The recombinant polypeptide obtained can be purified from cell lysates and extracts and/or from the culture medium supernatant, by methods used individually or in combination, such as fractionation, chromatography methods, immunoaffinity techniques using specific mono- or polyclonal antibodies, etc.
[0024]The polypeptide of the invention may, in particular, be purified from apoptotic bodies originating from tumor cells, by affinity chromatography on a column of antibodies specific for the C-terminal end of the BARD1 protein.
[0025]The nucleic acid sequence encoding the truncated BARD1 protein can be inserted into an expression vector, in which it is functionally linked to elements for regulating its expression, such as in particular promoters, activators and/or terminators of transcription.
[0026]A recombinant protein can then also be produced using a method in which a vector containing a nucleic acid as defined above is transferred into a host cell, which is cultured under conditions allowing the expression of the corresponding polypeptide.
[0027]The signals controlling the expression of nucleotide sequences (promoters, activators, termination sequences, etc.) are selected as a function of the cellular host used. To this effect, the nucleotide sequences according to the invention can be inserted into vectors which replicate autonomously within the selected host, or vectors which integrate in the selected host. Such vectors will be prepared according to the methods commonly used by those skilled in the art, and the clones resulting therefrom can be introduced into a suitable host by standard methods, such as, for example, electroporation or calcium phosphate precipitation.
[0028]The cloning and/or expression vectors as described above, comprising a defined nucleotide sequence according to the invention, are also part of the present invention.
[0029]The invention is also directed toward the host cells transfected, transiently, or stably with these expression vectors. These cells can be obtained by introducing, into prokaryotic or eukaryotic host cells, a nucleotide sequence inserted into a vector as defined above, and then culturing said cells under conditions allowing the replication and/or expression of the transfected nucleotide sequence.
[0030]The properties of the truncated BARD1 protein according to the invention, or of the nucleic acid encoding this protein, as a tumor repressor can be taken advantage of in the treatment of tumors. This may involve any type of tumor, but more particularly breast cancers, ovarian cancers, lung cancers or cancers of the digestive tract, such as colon carcinomas.
[0031]A subject of the invention is therefore also a pharmaceutical composition comprising a polypeptide as defined above, or a nucleic acid encoding said polypeptide, in combination with a pharmaceutically acceptable vehicle.
[0032]The methods of administration, the doses and the pharmaceutical forms of the pharmaceutical compositions according to the invention, containing at least one polypeptide, can be determined in the usual manner by those skilled in the art, in particular according to the criteria generally taken into account in establishing a therapeutic treatment suitable for a patient, such as, for example, the age or bodyweight of the patient, the seriousness of his or her general condition, the tolerance to the treatment, and the side effects noted, etc.
[0033]In general, a therapeutically or prophylactically effective amount ranging from approximately 0.1 μg to approximately 1 mg can be administered to human adults.
[0034]The subject of the invention is also a pharmaceutical composition comprising a nucleic acid as defined above, encoding the truncated BARD1 protein, and a pharmaceutically acceptable vehicle, said composition being intended to be used in gene therapy. The nucleic acid, preferably inserted into a vector, generally a viral vector (such as adenoviruses and retroviruses), can be administered in naked form, free of any vehicle promoting transfer to the target cell, such as anionic liposomes, cationic lipids, microparticles, for example gold microparticles, precipitating agents, for example calcium phosphate, or any other agent facilitating transfection. In this case, the polynucleotide can be simply diluted in a physiologically acceptable solution, such as a sterile solution or a sterile buffer solution, in the presence or absence of a vehicle.
[0035]Alternatively, a nucleic acid of the invention can be combined with agents which facilitate transfection. It may, inter alia, be (i) combined with a chemical agent which modifies cellular permeability, such as bupivacaine; (ii) encapsulated in liposomes, optionally in the presence of additional substances facilitating transfection; or (iii) combined with cationic lipids or microparticles of silica, of gold or of tungsten.
[0036]When the nucleic acid constructs of the invention coat microparticles, these microparticles can be injected intradermally or intraepidermally via the "gene gun" technique (WO 94/24263).
[0037]The amount to be used as a medicinal product depends in particular on the nucleic acid construct itself, on the individual to whom this nucleic acid is administered, on the method of administration and the type of formulation, and on the pathological condition. In general, a therapeutically or prophylactically effective amount ranging from approximately 0.1 μg to approximately 1 mg, preferably from approximately 1 μg to approximately 800 μg, and preferentially from approximately 25 μg to approximately 250 μg, can be administered to human adults.
[0038]The nucleic acid constructs of the invention can be administered via any conventional route of administration, such as in particular parenterally. The choice of the route of administration depends in particular on the formulation selected. An administration targeted to the site of the tumors targeted may be particularly advantageous.
[0039]Finally, a subject of the invention is therefore a method of therapeutic treatment, in which an effective amount of a truncated BARD1 protein as defined above, or a nucleic acid encoding this protein is administered to a patient requiring such a treatment, in the context of a gene therapy.
[0040]The intended patient is generally a human, but the application may also be extended to any mammal where appropriate.
[0041]The appearance of the truncated form of BARD1, which appears during apoptic processes, makes it possible, moreover, to follow, in vitro, the effectiveness of an anticancer treatment in a patient treated with pro-apoptopic drugs. For this, it is possible to use a method of in vitro detection of the 67 kD polypeptide of the invention in a biological sample, such as a sample of tumor tissues.
[0042]According to a variant, a method of in vitro detection of antibodies produced against the immunogenic 67 kD polypeptide, in a biological sample, such as a blood or urine sample from patients, can be used.
[0043]These methods can make use of the usual techniques of immunodetection, such as Western Blotting or immunohistochemistry for example, using an anti-BARD1 or anti-truncated BARI antibody, when it involves detecting the 67 kD polypeptide, or using said polypeptide or an epitope fragment thereof, when it involves detecting the antibodies.
[0044]More generally, the invention is also directed toward a method of in vitro detection of truncated BARD1 polypeptide or of anti-truncated BARD1 antibodies in a biological sample in which said biological sample is brought into contact with, respectively, an anti-truncated BARD1 antibody or a truncated BARD1 protein, or an epitope fragment, and the formation of immunocomplexes is observed, revealing the presence of truncated BARD1 protein or of anti-truncated BARD1 antibodies, respectively, in the biological sample.
[0045]The antibodies which are specifically directed against the truncated BARD1 protein are also part of the invention.
[0046]They may be poly- or monoclonal antibodies or fragments thereof, chimeric, in particular humanized or immunoconjugated, antibodies, or else labeled antibodies.
[0047]The polyclonal antibodies can be obtained from the serum of an animal immunized against a polypeptide according to the usual procedures.
[0048]According to one embodiment of the invention, a suitable peptide fragment of the BARD1 protein, which can be coupled via a reactor residue to a protein or another peptide, can be used as antigen. Rabbits are immunized with the equivalent of 1 mg of the peptide antigen according to the procedure described by Benoit et al. (1982). At four-week intervals, the animals are treated with injections of 200 μg of antigen, and bled 10 to 14 days later. After the third injection, the antiserum is examined in order to determine its ability to bind to the antigenic peptide, radiolabeled with iodine, prepared by the chloramine-T method, and is then purified by chromatography on a carboxymethylcellulose (CMC) ion exchange column. The antibody molecules are then collected from the mammals and isolated to the desired concentration by methods well known to those skilled in the art, for example using DEAE Sephadex to obtain the IgG fraction.
[0049]In order to improve the specificity of the polyclonal serum, the antibodies can be purified by immunoaffinity chromatography using immunizing polypeptides in solid phase. The antibody is brought into contact with the immunizing polypeptide in solid phase for a sufficient amount of time so as to immunoreact the polypeptide with the antibody molecule in order to form an immunocomplex in solid phase.
[0050]The monoclonal antibodies can be obtained according to the conventional method of hybridoma culture described by Kohler and Milstein (1975).
[0051]The following examples and figures illustrate the invention without limiting the scope thereof.
LEGENDS OF THE FIGURES
[0052]FIG. 1 shows an amino acid sequence alignment for the human (SEQ ID NO:1), rat (SEQ ID NO:6) and mouse (SEQ ID NO:5) BARD1 proteins. The sequences corresponding to the RING motif to the three ankyrin repeats and to the two BRCT domains in tandem are marked. The conserved Q564H mutation of the BARD1 human protein is also indicated. The sequences are aligned by introducing gaps so as to attain the maximum identity of the amino acid sequences. The values for the amino acids identity are then calculated by considering each space as a single dissimilarity (Table 1). The rat BARD1 cDNA sequence is available on EMBL under the accession number AF182946 (SEQ ID NO:10), that of mouse under the number AF057157 (SEQ ID NO:8) and that of humans under the Genbank number U76638 (SEQ ID NO:7).
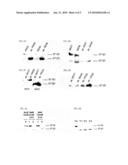
[0053]FIG. 2A represents a Western blotting analysis of proteins of rat and human carcinoma cells and of their apoptotic bodies revealed with a rat anti-BARD1 monoclonal antibody 6D10.
[0054]FIG. 2B represents the same type of analysis, with revelation using an anti-human polyclonal antibody 669D. It should be noted that a.b. signifies apoptotic body.
[0055]FIG. 2C represents a Western Blotting analysis of apoptotic body proteins, first carried out using an anti-mouse BARD1 polyclonal antibody WFS. The immunocomplexes are dissociated using the chemicon kit and the filter is further subjected to an anti-human BARD1 antibody 669D.
[0056]FIG. 2D represents an immunoprecipitation of lysates of apoptotic body derived from several carcinoma cells, with revelations in an anti-human BARD1 antibody 669D. The precipitate was subjected to electrophoresis and then immunoblotting was carried out with serum from rats immunized with PROb apoptotic bodies, diluted to 1/250.
[0057]FIG. 3A represents an analysis of the products of in vitro hydrolysis of the human BARD1 protein (SEQ ID NO:1) by SW48 cell lysates treated at confluency with 5 mM of sodium butyrate for 24 hours. The apoptotic bodies recovered from the supernatant (1) or the adherent cells (2) were solubilized in a DIV buffer and added to a human BARD1 protein labeled with 35S-methionine. After incubation for 4 hours at 37° C., the hydrolysis products were separated by SDS-PAGE and autoradiographed using phosphorimager.
[0058]FIG. 3B represents the same type of analysis, the adherent cells being solubilized in DIV buffer and incubated with a human BARD1 protein labeled with 35S-methionine for 0 to 3 hours respectively (TO to T3).
[0059]FIG. 4 is a graph representing PROb tumor growth BDIX rats after vaccination with the F1 fragment BARD 1 or a control protein (fucosyl transferase).
EXAMPLES
Example 1
Identification of a 67 kD Protein as a Product of Cleavage of the BARD1 Protein
Materials and Methods:
Cell Cultures
[0060]The REGb rat colon carcinoma cells and PROb rat colon adenocarcinoma cells used are derived from a cell line induced with dimethylhydrazine (Caignard et al., 1985). The 13762 rat breast carcinoma, the SW48 human colon carcinoma and the MCF7 breast carcinoma were obtained from the ECACC. The cells were cultured in monolayer cultures at 37° C. in RMPI 1640 medium (Gibco) with 10% of fetal calf serum and 2 mM of glutamine. The cells were subcultured with 0.025% of trypsin and 0.02% of EDTA.
Immunoscreening and Cloning of the Rat BARD1 cDNA (SEQ ID NO:10)
[0061]A cDNA library of the PROb rat carcinoma cell line was constructed in the λTriplEx expression vector (Clontech). One million plaques were screened with sera from rats vaccinated with apoptotic antibodies and IL-2 (Boisteau et al., 1997). The antibodies against E. coli were removed from the rat antisera by incubating sonicated E. coli bacteria with serum diluted to 1/10 in PBS plus 5% of dehydrated skimmed milk, for 4 hours at ambient temperature, and then centrifuged at 13,000 g for 10 minutes. The 456 base pair insert (fragment F1) was sequenced and the sequence is subjected to analysis on the NCB1 gene bank. It represented a strong homology with the human BARD1 protein. The complete rat BARD1 cDNA (SEQ ID NO:10) was cloned using the PROb cDNA library constructed with the PCR SMART kit from Clontech. The internal primers for the RACE PCR were selected based on the insert cloned according to the manufacturer's instructions.
Cloning of the Human BARD1 cDNA
[0062]Three human BARD1 cDNA fragments were amplified from total RNA extracted from SW48 human colon carcinoma cell lines. The fragments, designated A, B, and C, were obtained using the following primers: fragment A sense primer R135S/antisense primer B202N (Thai et al., 1998); fragment B, sense primer B202A (Thai et al., 1998); antisense primer: 5' CACCAATGCCTTATGCTGGAGC 3' (SEQ ID NO:2); fragment C, sense primer: 5' GAAGTAGTGACTCCTGAGAAGG 3' (SEQ ID NO:3)/antisense primer 5' TCAGCTGTCAAGAGGAAGCAACTC 3' (SEQ ID NO:4). Each fragment was cloned into a pGEM plasmid (Promega), then excised using Not1-Pst1/Pst1-Hind III/Hind III-Bst XI, respectively, purified, and then ligated into the Not1/Bst XI sites of pGEM.
Apoptotic Induction and Purification of Apoptotic Bodies
[0063]Apoptosis was induced by treatment with sodium butyrate (NaB). The cells at different stages of confluence were treated in complete medium at 37° C. and with 5 mM of NaB (Sigma) for various periods of time. The apoptotic bodies were purified as previously described (Gautier et al., 1999):
Production and Purification of F1 Fragments of Rat BARD1
[0064]The F1 fragment of rat BARD1 (SEQ ID NO:9) was excised from the plasmid λ Triplex of the cDNA library and inserted, in frame, into the Pst1 site of the plasmid pQE32 (Qiagen). The resulting fusion protein, containing a 6×His tag placed at the N-terminal end of the F1 fragment of BARD1 (SEQ ID NO:9), was expressed in E. coli, then purified by affinity chromatography on a Ni-NTA resin using the manufacturer's recommendations for the QIA a expressionist kit (Qiagen).
Immunization of the Mice and Production of Monoclonal Antibody
[0065]Balb-c mice (Iffa-credo) were given subcutaneous injections of 100 μg of the F1 fragment of rat BARD1 (SEQ ID NO:9) in 0.1 ml of incomplete Freund's adjuvant (Life Technologies) emulsified in 0.1 ml of sterile PBS buffer containing 0.5% of Triton X-100, 3 weeks apart. The splenocytes of a mouse were fused with the SP20 mouse myloma (ECACC) in the presence of polyethylene glycol 1500 (Boehringer Mannheim) The hybridomas were deposited onto 96-well plates in complete medium supplemented with 20% of fetal calf serum, hypoxanthine-aminopterin-thymidine (Sigma) and 1.5 ng/ml of recombinant IL6 (RD Systems). The hybridome supernatants were tested by ELISA using a purified BARD1 F1 fragment (SEQ ID NO:9) as antigen.
Immunoprecipitation
[0066]The apoptotic bodies were extracted on ice with 2% of Triton X-100 in PBS supplemented with a cocktail of protease inhibitors free of EDTA (Boehringer Mannheim) for 30 minutes. The extract was centrifuged for 15 minutes at 13,000 g and the supernatant was incubated with rabbit polyclonal antibodies directed against the human BARD1 protein (669D) (SEQ ID NO:1) (Wu et al., 1996 and Jin et al., 1997), diluted to 1/1000. After incubation for 4 hours, with constant stirring, the immunocomplexes were immunoprecipitated by adding 50 μl of anti-rabbit IgG agarose. The immunocomplexes bound to the agarose were washed with PBS containing 1% of Triton X-100 and protease inhibitors and were extracted from the agarose beads by heating in a reducing buffer for electrophoresis and immunoblotting as described below.
Western Blotting
[0067]The electrophoresis was carried out under denaturing conditions (SDS PAGE) (Laemmli et al., 1970). The proteins were transferred onto a 0.45 μm PVDF filter (Millipore) and brought into contact with primary antibodies. Secondary antibodies conjugated to horseradish peroxidase were used, diluted to 1/15000 (Sigma). The immunocomplexes were visualized by chemiluminescence using a Super Signal kit (Pierce).
Coupled in vitro Transcription/Translation and Determination of Cleavage of the Protein in vitro:
[0068]The human BARD1 protein (SEQ ID NO:1), labeled with 35S-methionine, was transcribed and translated in vitro using the TNT coupled reticulocytes lysate system kit (Promega). 1 μg of plasmid was used in a reaction medium for the transcription and the translation, containing 4 μl of 35S-methionine (NEN). For the in vitro cleavage, 2 μl of transcription/translation products were incubated with apoptotic or nonapoptotic cell extracts prepared in a DIV buffer (20 mM HEPES. PH 7.5, 10 mM NaCl, 1.5 mM MgCl2, 0.1% SB14, 0.5 mM PMSF) at 37° C. for the period of time indicated. The hydrolysis products were then separated by SDS-PAGE and revealed by autoradiography using the phosphorimager 445SI (Molecular Dynamics). Inhibition of the cleavage was evaluated by adding caspase inhibitors or a proteasome inhibitor (lactacystine) (Calbiochem) or the calpain inhibitor I (ALLnL) (Chemicon).
Cell Cycle Synchronization
[0069]The SW48 cells were arrested in G0 by contact inhibition in 175 cm2 flasks. After confluencing for 3 days, the cells were split 1:10 in 75 cm2 flasks at a concentration of 3×106 cells per flask. 12, 20, 28, 36 and 44 hours after seeding, the cells were treated with 5 mM of NaB for 24 hours and collected. To determine the cell cycle distribution at each time, the content of each flask was subjected to trypsinization, washed three times in 10 ml of ice-cold PBS, and fixed with 1 ml of ice-cold 70% ethanol, added dropwise, for 16 hours at -20° C. The fixed cells were pelletted, resuspended in 500 μl of PC buffer (96% 0.2M Na2HPO4, 4% 15 0.1M citric acid, pH 7.8) and left at ambient temperature for 30 minutes. They were then washed and resuspended in 500 μl of propidium iodide in a staining solution (PBS, 0.12% Triton X-100, 0.12 mM EDTA, 100 μg/ml RNase A), incubated for 30 minutes at 37° C. and analyzed on a FACScan (flow cytometer) (Beckton Dickinson). To determine the cleavage of the protein in vitro, the cells were scraped and the cell extracts were prepared as previously described.
Results
[0070]Cloning of the cDNA Encoding the 67 kD Protein
[0071]The immunoscreening of the λTriplEx cDNA library with sera from rats treated for a carcinoma with apoptotic bodies/IL-2 led to the identification of a positive insert of 456 pairs of the rat BARD1 gene (F1 fragment) (SEQ ID NO:10). This fragment extends between amino acids 460 to 611 (FIG. 1) (SEQ ID NO:9). Table 1 below gives the percentage homology between the rat (SEQ ID NO:6), human (SEQ ID NO:1) and mouse (SEQ ID NO:5) BARD1 protein.
Percentage Homology between the Rat, Human and Mouse BARD1 Protein
TABLE-US-00001 Species Total RING Ankyrin BRCT Rat/mouse 87.9 95.5 93.9 94.2 Rat/human 64.8 86.6 90.9 80.3 Mouse/human 67.5 86.6 90.9 79.7
The 67 kD Protein is a Fragment of BARD1
[0072]In order to be able to prove the identity of the 67 kD protein, as a fragment of BARD1, the authors of the invention determined its expression in the tumor cells and the apoptotic bodies. For this purpose, produce monoclonal antibodies (clone 6DI0) against F1 fragment (SEQ ID NO:9). When it was tested on PROb and REGb rat carcinoma cells, the monoclonal antibodies 6D10 recognized a 97 kD protein, but also a band of approximately 67 kD in the apoptotic bodies derived from the cells after treatment with sodium butyrate (FIG. 2A). This result was confirmed using a polyclonal antibody 669D (Wu et al., 1996 and Jin et al., 1997) against the human BARD1 protein (SEQ ID NO:1). Treatment of SV48 human colon cells or MCF7 mammary gland cells with sodium butyrate led to a similar result after immunoblotting with a polyclonal antibody 669D (FIG. 2B). Finally, immunoprecipitation of BARD1 from apoptotic bodies derived from rat human carcinomas, with the polyclonal antibody 669D, followed by immunoblotting with a treated rat serum, made it possible to detect a 67 kD protein (FIG. 2D). This set of results proves the identity of the 67 kD protein as a fragment of BARD1.
BARD1 is Cleaved During Apoptosis
[0073]When the apoptotic bodies derived either from human MCF7 carcinomas or from rat PROb carcinoma were incubated, on the same membrane, successively with the anti-human BARD1 polyclonal antibody 669D or the antibody WFS against the N-terminal portion of the mouse BARD1 protein (amino acids 101-114 of SEQ ID NO:5) (Irminger-Finger 1988), the authors of the invention observe that the antibody WFS did not recognize the 67 kD molecule, while the antibody 669D recognized the 67 kD molecule in each of the types of apoptotic body (FIG. 2C). This strongly suggests that the BARD1 cleavage site is located in the N-terminal portion, but downstream of the RING domain (amino acids 40-84 of SEQ ID NO:1, SEQ ID NO:5, and SEQ ID NO:6, FIG. 1) essential for the interaction of the BARD1 and BRCA1 proteins (Wu el al., 1996).
The Cleavage of BARD1 During Apoptosis is Cell Cycle Dependent
[0074]The authors of the invention examined the effect of the treatment with NaB for the cleavage of BARD1 in the adherent SW48 cells or in the apoptotic bodies collected from the supernatant. FIG. 3A shows that the adherent cell lysates completely cleave the radiolabeled human BARD1 protein (SEQ ID NO:1) after 4 hours of incubation, since the full length human BARD1 protein (SEQ ID NO:1) completely disappears and the 67 kD protein appears. However, incubation with lysates of apoptotic bodies has no effect on the hydrolysis of hBARD1 (SEQ ID NO:1). These results indicate that the proteolytic activity involved in the cleavage of BARD1 intervenes before the ultimate step of apoptosis which leads to the formation of apoptotic bodies and to cell detachment. Analysis of the kinetics leading to the hydrolysis of hBARD1 (SEQ ID NO:1) with lysates of the SW48 adherent cells treated for 48 hours with 5 mM of NaB showed that the cleavage is finished after one hour (FIG. 3B). Further analysis of the kinetic induction of this cleavage by NaB showed that the p67 protein appeared after 4 hours of treatment of the cells with NaB and that the cleavage was virtually complete after 12 hours. Interestingly, the hydrolysis of hBARD1 (SEQ ID NO:1) is regulated in a cell cycle-dependent manner, with a predominance during the G0/G1 phase. This was demonstrated by addition of NaB 32 hours after rupturing of the cells, a stage at which 80% of the cells were in G0/G1 phase and only 5% in G2/M phase, which led to complete conversion of hBARD1 (SEQ ID NO:1) to p67.
Example 2
Cleavage with Calpains
Materials and Methods
Determination of the Caspase Activity
[0075]For assaying the caspase activity, 10 μg of cell extracts were diluted in 5 μl of DIV buffer:
[0076]Acetyl-Asp-Glu-Val-Asp-7-amino-4-methylcoumarin (Ac-DEVD-AMC), acetyl-Val-Glu-Ile-Asp-7-amino-4-methylcoumarin (Ac-VEID-AMC) and acetyl-Ile-Glu-Ile-Asp-7-amino-4-methylcoumarin (Ac-IEID-AMC) (Bachem), substrates for caspases 3, 6 and 8, respectively, were added at a final concentration of 50 μM. The cleavage activity was controlled on Fluorolite 1000 (Dynatech laboratories).
Cellular Fraction
[0077]The cells were cultured in 75 cm2 flasks, trypsinized, and resuspended in 100 μl of CEB buffer (50 mM HEPES pH 7.4, 50 mM MgCl2, 1 mM DTT, 10 μM cytochalasin B). The resuspended cells were left on ice for 30 minutes and then homogenized with 50 strokes of a Dounce homogenizer, and cooled in ice. The nucleic fraction was prepared by centrifugation at 800×g for 10 minutes at 4° C. The pellet was resuspended in a CEB buffer and stored at -80° C. The mitochondrial and post-mitochondrial fractions were obtained after centrifugation at 13,000×g for 10 minutes at 4° C. Both the mitochondrial pellet resuspended in CEB and the post-mitochondrial fractions were divided into aliquot fractions and stored at -80° C.
Results
[0078]To define the proteolytic activity responsible for the cleavage of hBARD1 (SEQ ID NO:1), various preparations of cell extracts from SW48 cells treated with NAB were tested. It appears that both the nuclear preparations and the mitochondrial preparations were capable of cleaving hBARD1 (SEQ ID NO:1). The supernatant at 13,000×g of the organelle preparation had no effect.
[0079]The cascade of proteases which are effectors of the apoptotic process comprises cysteine proteases such as caspases or calpains. The authors of the invention then determined the caspase activities during the treatment of the SW48 cells with NaB, using specific substrates, and found that the activities of caspases 3, 6 and 8 gradually increased, caspase 3 being the most active after 6 hours and 12 hours of NaB treatment. The use of peptide inhibitors for the caspases showed that benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone (ZVAD-fmk), a caspase inhibitor, slightly inhibited the hydrolysis of hBARD1 (SEQ ID NO:1) at high concentration (100 μM). On the other hand, the inhibitors specific for caspase 3,benzyloxycarbonyl-Asp-Glu-Val-Asp-fluoromethylketone (Z-DEVD-frnk) (SEQ ID NO:11) or for caspase 6, benzyloxycarbonyl-Val-Glu-Ile-Asp-fluoromethyl ketone (Z-VEID-fmk) (SEQ ID NO:12), had no effect. This phenomenon was confirmed by the fact that purified caspase 3 had no effect on the hydrolysis of hBARD1 (SEQ ID NO:1). These results clearly show that HBARD1 (SEQ ID NO:1) is not a direct substrate for caspases. In addition, lactacystine, a proteasome inhibitor, does not block the proteolysis of hBARD1 (SEQ ID NO:1), even at concentrations of 100 μm, this excluding the possibility of the proteasome being involved in this mechanism.
[0080]A certain number of proteins which had degraded during apoptosis are targets for calpains. A possible mechanism for the activation of calpains involves the cleavage of a calpain inhibitor in vivo, calpastatin (DeMartino et al., 1984). The results of the authors of the invention show, firstly, that calpastatin was completely cleaved in the SW48 cells after 12 hours of treatment with NaB and, secondly, that the calpain inhibitor I, ALLnL, and EGTA strongly inhibited the hydrolysis of hBARD1 (SEQ ID NO:1) in a dose-dependent manner. This set of results strongly suggests that the BARD1 protein is hydrolyzed by calpains.
Example 3
Purification of the Cleaved BARD1 Protein
Production of Antibodies Specific for the Translated Protein
[0081]Poly- and monoclonal antibodies directed against peptide sequences are located at the N- and C-terminal ends of the truncated proteins are produced: [0082]against peptide 1 corresponding to amino acids 255 to 265 of the human BARD1 protein (SEQ ID NO:1); [0083]against peptide 2 corresponding to amino acids 527 to 540 of the human BARD1 protein (SEQ ID NO:1).
[0084]The peptides were coupled to KLH. Each rabbit was given three injections of 200 μg of each peptide, 15 days apart. A fourth injection was given three weeks after the last injection. The production of antibodies was tested on the serum of the rabbit by the ELISA technique using the free peptides as antigen.
[0085]The titers of the sera obtained were high (1/16000). In addition, no cross reactivity was observed between the sera of the two peptides.
Purification of the Cleaved BARD1 Protein
[0086]These antibodies allow the authors of the invention to purify the truncated protein, by affinity chromatography, either from apoptotic bodies or tumor cells or from the whole BARD1 protein produced and cleaved in vitro as described above.
[0087]The purification is carried out using standard techniques of affinity chromatography. The purified fractions are identified by western blotting with the antibodies produced.
Example 4
Vaccination of the Rats with the Truncated BARD1 Protein (SEQ ID NO:9)
Materials and Methods
[0088]Two groups of BDIX rats were given three weekly intraplantar injections of 100 μg of F1 fragment (amino acids 460-611) of BARD1 (SEQ ID NO:9) or of a controlled protein (fucosyl transferase) purified under the same conditions as the F1 fragment. The proteins were emulsified in 100 μl of complete Freund's adjuvant. Two weeks after the final immunization, the rat colon tumor cells (PROb, 50×103/rat) were injected subcutaneously and the volume of the PROb tumors was estimated.
Result
[0089]A slowing down of tumor growth is observed, thus demonstrating the protective effect of a vaccination with a BARD1 fragment derived from the 67 kD form (FIG. 4) (each point represents the mean of the tumor volumes measured on 6 rats, with the standard deviation).
BIBLIOGRAPHIC REFERENCES
[0090]Benoit et al., PNAS USA, 79, 917-921 (1982).
[0091]Boisteau et al., Apoptosis induced by sodium butyrate treatment increases immunogenicity of a rat colon tumor cell line. Apoptosis, 2:403-412, 1997.
[0092]Caignard et al., Interaction between two cellular subpopulations of a rat colonic carcinoma when inoculated to syngeneic host. Int. J. Cancer., 36: 273-279, 1985.
[0093]G. N. Demartino, and D. E. Croall Purification and characterization of a protein inhibitor of calcium-dependent protease from rat liver. Arch. Biochem. Biophys., 232; 713-720, 1984.
[0094]Gautier et al., Production and characterization of a monoclonal antibody specific for apoptotic bodies derived from several tumor cell lines. J. lmmunol. Meth., 228; 49-58, 1999.
[0095]Irminger-Finger et al., In vitro repression of Brca1-associated RING domain gene, Bard1, induces phenotypic changes in mammary epithelial cells. J. Cell Biol.: 143: 1329-1339, 1998.
[0096]Jin et al., Cell cycle-dependent colocalization of BARD1 and BRCA1 proteins in discrete nuclear domains. Proc. Natl. Acad. Sci. USA., 94: 12075-12080, 1997.
[0097]Kohler and Milstein, Nature, 256, 495-497, (1975).
[0098]Laemmli et al., Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 277:680-685, 1970.
[0099]U. K. Laemmli, Cleavage of structural proteins during the assembly of the head of bacteriophage T4, Nature, 277:680-685, 1970.
[0100]Thai, et al., Mutation in the BRCA1-associated RING domain (BARD1) gene in primary breast ovarian and uterine cancers. Human Mol. Gen., 7: 195-202, 1998.
[0101]Wu et al., Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nature Gen., 14: 430-440, 1996.
Sequence CWU
1
121777PRTHomo sapiens 1Met Pro Asp Asn Arg Gln Pro Arg Asn Arg Gln Pro Arg
Ile Arg Ser1 5 10 15Gly
Asn Glu Pro Arg Ser Ala Pro Ala Met Glu Pro Asp Gly Arg Gly 20
25 30Ala Trp Ala His Ser Arg Ala Ala
Leu Asp Arg Leu Glu Lys Leu Leu 35 40
45Arg Cys Ser Arg Cys Thr Asn Ile Leu Arg Glu Pro Val Cys Leu Gly
50 55 60Gly Cys Glu His Ile Phe Cys Ser
Asn Cys Val Ser Asp Cys Ile Gly65 70 75
80Thr Gly Cys Pro Val Cys Tyr Thr Pro Ala Trp Ile Gln
Asp Leu Lys 85 90 95Ile
Asn Arg Gln Leu Asp Ser Met Ile Gln Leu Cys Ser Lys Leu Arg
100 105 110Asn Leu Leu His Asp Asn Glu
Leu Ser Asp Leu Lys Glu Asp Lys Pro 115 120
125Arg Lys Ser Leu Phe Asn Asp Ala Gly Asn Lys Lys Asn Ser Ile
Lys 130 135 140Met Trp Phe Ser Pro Arg
Ser Lys Lys Val Arg Tyr Val Val Ser Lys145 150
155 160Ala Ser Val Gln Thr Gln Pro Ala Ile Lys Lys
Asp Ala Ser Ala Gln 165 170
175Gln Asp Ser Tyr Glu Phe Val Ser Pro Ser Pro Pro Ala Asp Val Ser
180 185 190Glu Arg Ala Lys Lys Ala
Ser Ala Arg Ser Gly Lys Lys Gln Lys Lys 195 200
205Lys Thr Leu Ala Glu Ile Asn Gln Lys Trp Asn Leu Glu Ala
Glu Lys 210 215 220Glu Asp Gly Glu Phe
Asp Ser Lys Glu Glu Ser Lys Gln Lys Leu Val225 230
235 240Ser Phe Cys Ser Gln Pro Ser Val Ile Ser
Ser Pro Gln Ile Asn Gly 245 250
255Glu Ile Asp Leu Leu Ala Ser Gly Ser Leu Thr Glu Ser Glu Cys Phe
260 265 270Gly Ser Leu Thr Glu
Val Ser Leu Pro Leu Ala Glu Gln Ile Glu Ser 275
280 285Pro Asp Thr Lys Ser Arg Asn Glu Val Val Thr Pro
Glu Lys Val Cys 290 295 300Lys Asn Tyr
Leu Thr Ser Lys Lys Ser Leu Pro Leu Glu Asn Asn Gly305
310 315 320Lys Arg Gly His His Asn Arg
Leu Ser Ser Pro Ile Ser Lys Arg Cys 325
330 335Arg Thr Ser Ile Leu Ser Thr Ser Gly Asp Phe Val
Lys Gln Thr Val 340 345 350Pro
Ser Glu Asn Ile Pro Leu Pro Glu Cys Ser Ser Pro Pro Ser Cys 355
360 365Lys Arg Lys Val Gly Gly Thr Ser Gly
Ser Lys Asn Ser Asn Met Ser 370 375
380Asp Glu Phe Ile Ser Leu Ser Pro Gly Thr Pro Pro Ser Thr Leu Ser385
390 395 400Ser Ser Ser Tyr
Arg Arg Val Met Ser Ser Pro Ser Ala Met Lys Leu 405
410 415Leu Pro Asn Met Ala Val Lys Arg Asn His
Arg Gly Glu Thr Leu Leu 420 425
430His Ile Ala Ser Ile Lys Gly Asp Ile Pro Ser Val Glu Tyr Leu Leu
435 440 445Gln Asn Gly Ser Asp Pro Asn
Val Lys Asp His Ala Gly Trp Thr Pro 450 455
460Leu His Glu Ala Cys Asn His Gly His Leu Lys Val Val Glu Leu
Leu465 470 475 480Leu Gln
His Lys Ala Leu Val Asn Thr Thr Gly Tyr Gln Asn Asp Ser
485 490 495Pro Leu His Asp Ala Ala Lys
Asn Gly His Val Asp Ile Val Lys Leu 500 505
510Leu Leu Ser Tyr Gly Ala Ser Arg Asn Ala Val Asn Ile Phe
Gly Leu 515 520 525Arg Pro Val Asp
Tyr Thr Asp Asp Glu Ser Met Lys Ser Leu Leu Leu 530
535 540Leu Pro Glu Lys Asn Glu Ser Ser Ser Ala Ser His
Cys Ser Val Met545 550 555
560Asn Thr Gly Gln Arg Arg Asp Gly Pro Leu Val Leu Ile Gly Ser Gly
565 570 575Leu Ser Ser Glu Gln
Gln Lys Met Leu Ser Glu Leu Ala Val Ile Leu 580
585 590Lys Ala Lys Lys Tyr Thr Glu Phe Asp Ser Thr Val
Thr His Val Val 595 600 605Val Pro
Gly Asp Ala Val Gln Ser Thr Leu Lys Cys Met Leu Gly Ile 610
615 620Leu Asn Gly Cys Trp Ile Leu Lys Phe Glu Trp
Val Lys Ala Cys Leu625 630 635
640Arg Arg Lys Val Cys Glu Gln Glu Glu Lys Tyr Glu Ile Pro Glu Gly
645 650 655Pro Arg Arg Ser
Arg Leu Asn Arg Glu Gln Leu Leu Pro Lys Leu Phe 660
665 670Asp Gly Cys Tyr Phe Tyr Leu Trp Gly Thr Phe
Lys His His Pro Lys 675 680 685Asp
Asn Leu Ile Lys Leu Val Thr Ala Gly Gly Gly Gln Ile Leu Ser 690
695 700Arg Lys Pro Lys Pro Asp Ser Asp Val Thr
Gln Thr Ile Asn Thr Val705 710 715
720Ala Tyr His Ala Arg Pro Asp Ser Asp Gln Arg Phe Cys Thr Gln
Tyr 725 730 735Ile Ile Tyr
Glu Asp Leu Cys Asn Tyr His Pro Glu Arg Val Arg Gln 740
745 750Gly Lys Val Trp Lys Ala Pro Ser Ser Trp
Phe Ile Asp Cys Val Met 755 760
765Ser Phe Glu Leu Leu Pro Leu Asp Ser 770
775222DNAartificialprimer 2caccaatgcc ttatgctgga gc
22322DNAartificialprimer 3gaagtagtga ctcctgagaa gg
22424DNAartificialprimer
4tcagctgtca agaggaagca actc
245765PRTMus musculus 5Met Pro Arg Arg Pro Pro Arg Val Cys Ser Gly Asn
Gln Pro Ala Pro1 5 10
15Val Pro Ala Met Glu Pro Ala Thr Asp Gly Leu Trp Ala His Ser Arg
20 25 30Ala Ala Leu Ala Arg Leu Glu
Lys Leu Leu Arg Cys Ser Arg Cys Ala 35 40
45Asn Ile Leu Lys Glu Pro Val Cys Leu Gly Gly Cys Glu His Ile
Phe 50 55 60Cys Ser Gly Cys Ile Ser
Asp Cys Val Gly Ser Gly Cys Pro Val Cys65 70
75 80Tyr Thr Pro Ala Trp Ile Leu Asp Leu Lys Ile
Asn Arg Gln Leu Asp 85 90
95Ser Met Ile Gln Leu Ser Ser Lys Leu Gln Asn Leu Leu His Asp Asn
100 105 110Lys Asp Ser Lys Asp Asn
Thr Ser Arg Ala Ser Leu Phe Gly Asp Ala 115 120
125Glu Arg Lys Lys Asn Ser Ile Lys Met Trp Phe Ser Pro Arg
Ser Lys 130 135 140Lys Val Arg Tyr Val
Val Thr Lys Val Ser Val Gln Thr Gln Pro Gln145 150
155 160Lys Ala Lys Asp Asp Lys Ala Gln Glu Ala
Ser Met Tyr Glu Phe Val 165 170
175Ser Ala Thr Pro Pro Val Ala Val Pro Lys Ser Ala Lys Thr Ala Ser
180 185 190Arg Thr Ser Ala Lys
Lys His Pro Lys Lys Ser Val Ala Lys Ile Asn 195
200 205Arg Glu Glu Asn Leu Arg Pro Glu Thr Lys Asp Ser
Arg Phe Asp Ser 210 215 220Lys Glu Glu
Leu Lys Glu Glu Lys Val Val Ser Cys Ser Gln Ile Pro225
230 235 240Val Met Glu Arg Pro Arg Val
Asn Gly Glu Ile Asp Leu Leu Ala Ser 245
250 255Gly Ser Val Val Glu Pro Glu Cys Ser Gly Ser Leu
Thr Glu Val Ser 260 265 270Leu
Pro Leu Ala Glu His Ile Val Ser Pro Asp Thr Val Ser Lys Asn 275
280 285Glu Glu Thr Pro Glu Lys Lys Val Cys
Val Lys Asp Leu Arg Ser Gly 290 295
300Gly Ser Asn Gly Asn Arg Lys Gly Cys His Arg Pro Thr Thr Ser Thr305
310 315 320Ser Asp Ser Cys
Gly Ser Asn Ile Pro Ser Thr Ser Arg Gly Ile Gly 325
330 335Glu Pro Ala Leu Leu Ala Glu Asn Val Val
Leu Val Asp Cys Ser Ser 340 345
350Leu Pro Ser Gly Gln Leu Gln Val Asp Val Thr Leu Arg Arg Lys Ser
355 360 365Asn Ala Ser Asp Asp Pro Leu
Ser Leu Ser Pro Gly Thr Pro Pro Pro 370 375
380Leu Leu Asn Asn Ser Thr His Arg Gln Met Met Ser Ser Pro Ser
Thr385 390 395 400Val Lys
Leu Ser Ser Gly Met Pro Ala Arg Lys Arg Asn His Arg Gly
405 410 415Glu Thr Leu Leu His Ile Ala
Ser Ile Lys Gly Asp Ile Pro Ser Val 420 425
430Glu Tyr Leu Leu Gln Asn Gly Asn Asp Pro Asn Val Lys Asp
His Ala 435 440 445Gly Trp Thr Pro
Leu His Glu Ala Cys Ser His Gly His Leu Lys Val 450
455 460Val Glu Leu Leu Leu Gln His Asn Ala Leu Val Asn
Thr Pro Gly Tyr465 470 475
480Gln Asn Asp Ser Pro Leu His Asp Ala Val Lys Ser Gly His Ile Asp
485 490 495Ile Val Lys Val Leu
Leu Ser His Gly Ala Ser Arg Asn Ala Val Asn 500
505 510Ile Phe Gly Val Arg Pro Val Asp Tyr Thr Asp Asn
Glu Asn Ile Arg 515 520 525Ser Leu
Leu Leu Leu Pro Glu Glu Asn Glu Ser Phe Ser Thr Ser Gln 530
535 540Cys Ser Ile Val Asn Thr Gly Gln Arg Lys Asn
Gly Pro Leu Val Phe545 550 555
560Ile Gly Ser Gly Leu Ser Ser Gln Gln Gln Lys Met Leu Ser Lys Leu
565 570 575Glu Thr Val Leu
Lys Ala Lys Lys Cys Met Glu Phe Asp Ser Thr Val 580
585 590Thr His Val Ile Val Pro Asp Glu Glu Ala Gln
Ser Thr Leu Lys Cys 595 600 605Met
Leu Gly Ile Leu Ser Gly Cys Trp Ile Leu Lys Phe Asp Trp Val 610
615 620Lys Ala Cys Leu Asp Ser Lys Val Arg Glu
Gln Glu Glu Lys Tyr Glu625 630 635
640Val Pro Gly Gly Pro Gln Arg Ser Arg Leu Asn Arg Glu Gln Leu
Leu 645 650 655Pro Lys Leu
Phe Asp Gly Cys Tyr Phe Phe Leu Gly Gly Asn Phe Lys 660
665 670His His Pro Arg Asp Asp Leu Leu Lys Leu
Ile Ala Ala Ala Gly Gly 675 680
685Lys Val Leu Ser Arg Lys Pro Lys Pro Asp Ser Asp Val Thr Gln Thr 690
695 700Ile Asn Thr Val Ala Tyr His Ala
Lys Pro Glu Ser Asp Gln Arg Phe705 710
715 720Cys Thr Gln Tyr Ile Val Tyr Glu Asp Leu Phe Asn
Cys His Pro Glu 725 730
735Arg Val Arg Gln Gly Lys Val Trp Met Ala Pro Ser Thr Trp Leu Ile
740 745 750Ser Cys Ile Met Ala Phe
Glu Leu Leu Pro Leu Asp Ser 755 760
7656768PRTRattus norvegicus 6Met Pro Arg Arg Pro Pro Arg Val Cys Ser Gly
Asn Lys Pro Pro Pro1 5 10
15Val Pro Ala Met Glu Pro Ala Thr Asp Gly Leu Trp Ala His Ser Arg
20 25 30Ala Ala Leu Ala Arg Leu Glu
Lys Leu Leu Arg Cys Ser Arg Cys Ala 35 40
45Asn Ile Leu Arg Glu Pro Val Cys Leu Gly Gly Cys Glu His Ile
Phe 50 55 60Cys Ser Gly Cys Ile Ser
Asp Cys Val Gly Ser Gly Cys Pro Val Cys65 70
75 80His Thr Pro Ala Trp Ile Leu Asp Leu Lys Ile
Asn Arg Gln Leu Asp 85 90
95Ser Met Ile Gln Leu Tyr Ser Lys Leu Gln Asn Leu Leu His Asp Asn
100 105 110Lys Gly Ser Asp Ser Lys
Asp Asp Thr Ser Arg Ala Ser Leu Phe Gly 115 120
125Asp Ala Glu Arg Lys Lys Asn Ser Val Lys Met Trp Phe Ser
Pro Arg 130 135 140Ser Lys Lys Ile Arg
Cys Val Val Asn Lys Val Ser Val Gln Thr Gln145 150
155 160Pro Gln Lys Ala Lys Asp Asp Lys Ala Gln
Glu Ala Ser Val Phe Glu 165 170
175Phe Val Ser Ala Thr Pro Pro Val Val Val Ser Thr Arg Ala Lys Thr
180 185 190Ala Ser Arg Thr Ser
Ala Lys Lys His Pro Lys Lys Ser Val Ala Lys 195
200 205Ile Asn Arg Glu Gly Asn Phe Arg Pro Glu Thr Arg
Asp Ser Arg Phe 210 215 220Asp Ser Lys
Glu Lys Leu Lys Glu Glu Lys Val Val Ser Phe Ser Gln225
230 235 240Thr Leu Val Met Glu Asn Ser
Arg Val Asn Gly Glu Ile Asp Leu Leu 245
250 255Ala Ser Gly Ser Val Val Glu Ser Val Phe Ser Gly
Ser Phe Ala Glu 260 265 270Val
Ser Leu Pro Leu Ala Glu His Ile Val Ser Pro Asp Thr Val Ser 275
280 285Lys Ser Glu Glu Ala Pro Glu Lys Lys
Val Cys Val Glu Asp Arg Cys 290 295
300Pro Val Gly Ser Asp Gly Asn Pro Lys Gly Cys His Arg Pro Pro Thr305
310 315 320Ser Thr Ser Lys
Lys Cys Gly Ser Asn Val Pro Ser Ala Ser Gly Glu 325
330 335Ile Arg Glu Pro Thr Leu Leu Ala Glu Asn
Val Val Leu Val Asp Cys 340 345
350Ser Ser Leu Pro Ser Gly Arg Leu Gln Val Asp Val Thr Leu Arg Arg
355 360 365Gln Ser Asn Ala Ser Asp Asp
Ser Leu Ser Leu Ser Pro Gly Thr Pro 370 375
380Pro Ser Leu Leu Asn Asn Ser Thr His Arg Gln Met Met Ser Lys
Pro385 390 395 400Ser Thr
Val Lys Leu Ser Ser Gly Ile Pro Ala Arg Lys Arg Asn His
405 410 415Arg Gly Glu Thr Leu Leu His
Ile Ala Ser Ile Lys Gly Asp Ile Ser 420 425
430Ser Val Glu Tyr Leu Leu Gln Asn Gly Asn Asp Pro Asn Val
Lys Asp 435 440 445His Ala Gly Trp
Thr Pro Leu His Glu Ala Cys Ser His Gly His Leu 450
455 460Lys Ile Val Glu Leu Leu Leu Gln His Asn Ala Leu
Val Asn Thr Thr465 470 475
480Gly Tyr His Asn Asp Ser Pro Leu His Asp Ala Ala Lys Asn Gly His
485 490 495Ile Asp Ile Val Lys
Val Leu Leu Ser His Gly Ala Ser Arg Asn Ala 500
505 510Val Asn Ile Phe Gly Glu Arg Pro Val Asp Tyr Thr
Asp Ala Glu Asn 515 520 525Ile Arg
Ser Leu Leu Leu Leu Pro Glu Lys Thr Asp Ser Phe Ser Thr 530
535 540Ser Gln Cys Ser Val Gln Val Asn Thr Gly Gln
Arg Lys Ser Gly Pro545 550 555
560Leu Val Leu Ile Gly Ser Gly Leu Ser Ser Gln Gln Gln Lys Leu Leu
565 570 575Ser Lys Leu Glu
Thr Val Leu Lys Ala Lys Lys Cys Ala Glu Phe Asp 580
585 590Asn Thr Val Thr His Val Ile Val Pro Asp Glu
Glu Ala Gln Ser Thr 595 600 605Leu
Lys Cys Met Leu Gly Ile Leu Asn Gly Cys Trp Val Leu Lys Phe 610
615 620Asp Trp Val Lys Ala Cys Leu Asp Ser Gln
Glu Arg Glu Gln Glu Glu625 630 635
640Lys Tyr Glu Val Pro Gly Gly Pro Gln Arg Ser Arg Leu Asn Arg
Glu 645 650 655Gln Leu Leu
Pro Lys Leu Phe Asp Gly Cys Tyr Phe Phe Leu Gly Gly 660
665 670Asn Phe Lys His His Pro Lys Glu Asp Leu
Leu Lys Leu Ile Ala Ala 675 680
685Ala Gly Gly Arg Ile Leu Ser Arg Lys Pro Lys Pro Asp Ser Asp Val 690
695 700Thr Gln Thr Ile Asn Thr Val Ala
Tyr His Ala Lys Pro Asp Ser Asp705 710
715 720Gln Arg Phe Cys Thr Gln Tyr Ile Val Tyr Glu Asp
Leu Phe Asn Cys 725 730
735His Pro Glu Arg Val Arg Gln Gly Lys Val Trp Met Ala Pro Ser Thr
740 745 750Trp Leu Ile Ser Cys Val
Met Ala Phe Glu Leu Leu Pro Leu Asp Ser 755 760
76572530DNAHomo sapiens 7cagcttccct gtggtttccc gaggcttcct
tgcttcccgc tctgcgagga gcctttcatc 60cgaaggcggg acgatgccgg ataatcggca
gccgaggaac cggcagccga ggatccgctc 120cgggaacgag cctcgttccg cgcccgccat
ggaaccggat ggtcgcggtg cctgggccca 180cagtcgcgcc gcgctcgacc gcctggagaa
gctgctgcgc tgctcgcgtt gtactaacat 240tctgagagag cctgtgtgtt taggaggatg
tgagcacatc ttctgtagta attgtgtaag 300tgactgcatt ggaactggat gtccagtgtg
ttacaccccg gcctggatac aagacttgaa 360gataaataga caactggaca gcatgattca
actttgtagt aagcttcgaa atttgctaca 420tgacaatgag ctgtcagatt tgaaagaaga
taaacctagg aaaagtttgt ttaatgatgc 480aggaaacaag aagaattcaa ttaaaatgtg
gtttagccct cgaagtaaga aagtcagata 540tgttgtgagt aaagcttcag tgcaaaccca
gcctgcaata aaaaaagatg caagtgctca 600gcaagactca tatgaatttg tttccccaag
tcctcctgca gatgtttctg agagggctaa 660aaaggcttct gcaagatctg gaaaaaagca
aaaaaagaaa actttagctg aaatcaacca 720aaaatggaat ttagaggcag aaaaagaaga
tggtgaattt gactccaaag aggaatctaa 780gcaaaagctg gtatccttct gtagccaacc
atctgttatc tccagtcctc agataaatgg 840tgaaatagac ttactagcaa gtggctcctt
gacagaatct gaatgttttg gaagtttaac 900tgaagtctct ttaccattgg ctgagcaaat
agagtctcca gacactaaga gcaggaatga 960agtagtgact cctgagaagg tctgcaaaaa
ttatcttaca tctaagaaat ctttgccatt 1020agaaaataat ggaaaacgtg gccatcacaa
tagactttcc agtcccattt ctaagagatg 1080tagaaccagc attctgagca ccagtggaga
ttttgttaag caaaccgtgc cctcagaaaa 1140tataccattg cctgaatgtt cttcaccacc
ttcatgcaaa cgtaaagttg gtggtacatc 1200agggaggaaa aacagtaaca tgtccgatga
attcattagt ctttcaccag gtacaccacc 1260ttctacatta agtagttcaa gttacaggca
agtgatgtct agtccctcag caatgaagct 1320gttgcccaat atggctgtga aaagaaatca
tagaggagag actttgctcc atattgcttc 1380tattaagggc gacatacctt ctgttgaata
ccttttacaa aatggaagtg atccaaatgt 1440taaagaccat gctggatgga caccattgca
tgaagcttgc aatcatgggc acctgaaggt 1500agtggaatta ttgctccagc ataaggcatt
ggtgaacacc accgggtatc aaaatgactc 1560accacttcac gatgcagcca agaatgggca
cgtggatata gtcaagctgt tactttccta 1620tggagcctcc agaaatgctg ttaatatatt
tggtctgcgg cctgtcgatt atacagatga 1680tgaaagtatg aaatcgctat tgctgctacc
agagaagaat gaatcatcct cagctagcca 1740ctgctcagta atgaacactg ggcagcgtag
ggatggacct cttgtactta taggcagtgg 1800gctgtcttca gaacaacaga aaatgctcag
tgagcttgca gtaattctta aggctaaaaa 1860atatactgag tttgacagta cagtaactca
tgttgttgtt cctggtgatg cagttcaaag 1920taccttgaag tgtatgcttg ggattctcaa
tggatgctgg attctaaaat ttgaatgggt 1980aaaagcatgt ctacgaagaa aagtatgtga
acaggaagaa aagtatgaaa ttcctgaagg 2040tccacgcaga agcaggctca acagagaaca
gctgttgcca aagctgtttg atggatgcta 2100cttctatttg tggggaacct tcaaacacca
tccaaaggac aaccttatta agctcgtcac 2160tgcaggtggg ggccagatcc tcagtagaaa
gcccaagcca gacagtgacg tgactcagac 2220catcaataca gtcgcatacc atgcgagacc
cgattctgat cagcgcttct gcacacagta 2280tatcatctat gaagatttgt gtaattatca
cccagagagg gttcggcagg gcaaagtctg 2340gaaggctcct tcgagctggt ttatagactg
tgtgatgtcc tttgagttgc ttcctcttga 2400cagctgaata ttataccaga tgaacatttc
aaattgaatt tgcacggttt gtgagagccc 2460agtcattgta ctgtttttaa tgttcacatt
tttacaaata ggtagagtca ttcatatttg 2520tctttgaatc
253082568DNAMus musculus 8ctccgagccg
gaggcgtccg accaattcag agactccgca cgcagcagga tccggcggtg 60accagcgggg
tgcggcgcta gagcaaacct gacctcccct cctctgtggg aatgccacgc 120cggccgccga
gggtctgctc tgggaaccag cctgctcccg tgcccgccat ggagccggct 180accgacgggc
tttgggccca cagccgcgcg gcgcttgccc gcctggagaa gctgctgcgc 240tgctcccgct
gtgctaatat tctgaaggag cccgtgtgct taggaggatg tgagcacatc 300ttctgtagtg
gttgtataag tgactgtgtt ggctcaggat gcccagtgtg ttacacccca 360gcctggatcc
tagacctcaa gataaaccga caattggaca gcatgatcca gctttctagt 420aagctccaaa
atttgctcca tgacaataaa gattcaaaag acaacacatc tagggcaagt 480ttatttggtg
atgcagaaag gaagaagaat tcaataaaaa tgtggtttag tcctcgaagt 540aagaaggtta
gatatgttgt gactaaagtg tcagtacaaa cacagcctca aaaggcaaag 600gacgacaaag
cccaggaagc ctcaatgtat gaatttgttt ccgcaactcc ccctgtagct 660gttcctaaga
gtgctaaaac agcttctaga acatctgcaa aaaagcatcc aaagaaatct 720gtagctaaga
tcaacaggga ggagaattta aggccagaaa caaaggatag tagatttgat 780tccaaagagg
agctgaagga agagaaggtt gtctcctgta gccaaatacc agttatggag 840agaccacggg
taaatggtga aatagactta ttagcaagtg gttctgttgt agaacctgaa 900tgttctggaa
gcttgactga agtctcttta ccattggctg agcatatagt gtctccagac 960actgtgagca
agaatgaaga gactccggag aagaaggtct gtgtaaaaga tcttcgttca 1020ggagggagta
atggaaatcg taaaggctgc cacaggccta ccacttctac ttctgacagt 1080tgtgggagca
acattccgag caccagcaga ggcatcggtg agccagcatt gcttgcagaa 1140aacgtagtgt
tggttgactg ctcttcactg ccttcaggcc agctacaggt tgatgtcaca 1200ctcaggagaa
agagtaacgc atcagatgac ccccttagcc tttcaccggg cacaccccca 1260cctytgctga
acaattcaac tcacagacaa atgatgtcaa gcccctccac agtgaagctg 1320tcttctggta
tgccagccag gaaaagaaat cacagaggag agaccttact tcatattgct 1380tctattaagg
gtgatatacc ttctgttgaa tacctcttgc aaaatggaaa cgacccaaat 1440gttaaagacc
atgctggatg gacaccgttg catgaagcct gcagtcatgg gcacctgaag 1500gtagtggagt
tgctgctcca gcataatgcc ctggtgaaca cccctggcta tcagaatgac 1560tcgccactcc
acgatgccgt caagagtggc cacatcgata tagtcaaggt gttactgtcc 1620cacggtgctt
ccaggaatgc tgttaacata tttggtgtgc ggcctgtgga ctatacagac 1680aatgagaata
taagatcatt attgctgctg ccagaggaga atgaatcatt ctcaactagc 1740cagtgctcta
tcgtgaacac cgggcagcga aagaatgggc cgctggtatt tataggcagt 1800gggctttctt
cacagcagca gaaaatgctc agcaaacttg agacagttct taaggctaaa 1860aagtgtatgg
aatttgacag tacagtaact catgtcattg ttcctgatga ggaagcgcag 1920agtactctga
agtgtatgct tgggattctc agtggatgct ggatcctgaa gtttgattgg 1980gtgaaagcct
gtctggacag caaagtacgt gagcaggaag aaaagtatga agttcctgga 2040ggtccacaga
ggagcaggct caacagagag cagctgttgc caaagctgtt tgatggatgc 2100tacttctttt
tggggggaaa cttcaagcat catccaaggg atgacctcct taagctcatt 2160gctgcagcag
gaggcaaagt cctgagtcgc aagcccaagc cagacagtga tgtgactcag 2220accatcaaca
cggttgcata ccatgccaag cctgagtcgg atcagcgctt ctgtacgcag 2280tacatcgtct
acgaagacct gtttaactgt cacccagaga gggttcggca gggcaaagtc 2340tggatggctc
cgtccacctg gcttatcagc tgtataatgg cctttgaatt gcttcctctt 2400gacagctgaa
tgtcatacca ggtgaacatt ttaagttgga catgcatggt ttgtaagaac 2460aacggccatt
gtgatatttc tggtcattca tgtttttatg aataggtaga accattcata 2520tttcttcccc
ctttgaattg caaaataaaa caattgatag taaaaaaa
25689152PRTRattus norvegicus 9Ser His Gly His Leu Lys Ile Val Glu Leu Leu
Leu Gln His Asn Ala1 5 10
15Leu Val Asn Thr Thr Gly Tyr His Asn Asp Ser Pro Leu His Asp Ala
20 25 30Ala Lys Asn Gly His Ile Asp
Ile Val Lys Val Leu Leu Ser His Gly 35 40
45Ala Ser Arg Asn Ala Val Asn Ile Phe Gly Glu Arg Pro Val Asp
Tyr 50 55 60Thr Asp Ala Glu Asn Ile
Arg Ser Leu Leu Leu Leu Pro Glu Lys Thr65 70
75 80Asp Ser Phe Ser Thr Ser Gln Cys Ser Val Gln
Val Asn Thr Gly Gln 85 90
95Arg Lys Ser Gly Pro Leu Val Leu Ile Gly Ser Gly Leu Ser Ser Gln
100 105 110Gln Gln Lys Leu Leu Ser
Lys Leu Glu Thr Val Leu Lys Ala Lys Lys 115 120
125Cys Ala Glu Phe Asp Asn Thr Val Thr His Val Ile Val Pro
Asp Glu 130 135 140Glu Ala Gln Ser Thr
Leu Lys Cys145 150102499DNARattus norvegicus 10gaattcacta
gtgattatgc cacgccggcc gccgagggtc tgctccggga acaagcctcc 60tcccgtgccc
gccatggaac cagctaccga cgggctttgg gcccacagcc gtgcggcgct 120tgcccgtctg
gagaagttgt tgcgctgctc ccgctgtgct aatattctga gggagcccgt 180gtgcctagga
ggatgcgagc acatcttctg tagtggttgt ataagcgact gtgttggatc 240aggatgccca
gtgtgtcata ccccagcctg gatcctagac ctcaagataa acagacagtt 300ggacagcatg
atccagcttt atagtaagct tcaaaatttg ctacatgaca ataaaggttc 360agattcaaaa
gacgacacat ctagggcaag tttatttggt gatgcagaaa ggaagaagaa 420ttcagtaaaa
atgtggttta gtcctcgaag taagaaaatt agatgtgttg tgaataaagt 480ttcagtacaa
acccagcctc aaaaggcaaa ggatgacaaa gcccaggaag cctcagtgtt 540tgaatttgtt
tccgcaactc cccctgtagt tgtttctacg agggctaaaa cagcttcaag 600aacatctgca
aaaaagcatc ccaagaaatc tgtagctaag atcaaccggg agggaaattt 660caggccagaa
acaagggata gtagatttga ttccaaagaa aagctgaagg aagagaaggt 720tgtctccttt
agccaaacac tagttatgga gaattcacgg gtaaatggcg aaatagactt 780attagcgagt
ggctctgtgg tagaatccgt cttctctggc agctttgctg aagtctcttt 840accattggct
gagcatatag tgtctccaga tactgtgagc aagagtgaag aggctcctga 900gaagaaggtc
tgtgtagaag atcgttgtcc agtagggagt gatggaaatc ccaaaggctg 960ccacaggcct
cccacttcta cttctaagaa atgcgggagc aacgttccaa gcgccagcgg 1020agaaatccgt
gagccaacat tgcttgcaga aaatgtagtg ttggttgact gttcttcact 1080gccttcaggc
cgacttcagg ttgatgtcac cctcaggaga cagagtaacg catcagatga 1140ctctcttagc
ctttcaccag gcacaccccc atctctgctg aacaattcca ctcacagaca 1200aatgatgtca
aagccctcca cagtgaagct gtcttctggt attccagcca ggaaaagaaa 1260tcacagagga
gagacgttac tgcacattgc ctctataaag ggtgatatat cttctgttga 1320atacctcttg
caaaatggaa acgacccaaa tgttaaggac catgctggat ggacaccgtt 1380gcatgaagcc
tgcagtcatg ggcacctgaa gatagtggag ctgctgctcc agcacaatgc 1440cttggtgaac
accaccggct atcacaatga ctcgccactg cacgatgccg ccaagaatgg 1500ccacatcgat
atagtcaagg tgttactgtc ccacggagct tccaggaacg ctgttaacat 1560atttggtgag
cggccagtgg attacacaga cgctgagaat ataaggtcat tattgctgct 1620gccagagaag
acggattcat tctcaactag ccagtgctct gtccaggtga acaccgggca 1680gcggaagagt
gggccgctgg tactaatagg cagtgggctt tcttcacagc agcagaaact 1740gctcagcaaa
cttgagacag tgctaaaggc taagaagtgt gctgagtttg acaacacagt 1800aactcatgtc
attgttcctg atgaggaagc tcagagtacc ttgaagtgta tgcttgggat 1860tctcaatgga
tgctgggtcc tgaagtttga ttgggtgaaa gcctgtttgg acagccaaga 1920acgtgagcag
gaagaaaagt atgaagttcc tggaggtccg cagaggagca ggctcaacag 1980agagcagctg
ctgcccaaac tgttcgatgg atgctacttc tttctggggg ggaacttcaa 2040acatcatcca
aaagaagacc tcctgaagct cattgctgca gcaggaggca gaatcctcag 2100cagaaagccc
aagccagaca gtgacgtgac tcagaccatc aacacggttg cataccatgc 2160caagcctgac
tctgatcagc gcttctgtac gcagtacatt gtctatgagg atctgtttaa 2220ctgtcaccca
gagagggttc ggcagggcaa agtctggatg gctccttcca cctggctaat 2280cagctgtgta
atggcctttg agttgcttcc tcttgacagc tgaatgtcgt accaggtgaa 2340cattttaagt
tgaaggcgcg tggtttgtga gaacacggcc attgtgatgt ttgatcactc 2400tcgcttttat
ggataggtag aaccattcat atcccccctc cctttgaatt gcaaaataaa 2460acaattgata
aaggaaaaaa aaaaaaaaaa aaaaaaaaa
2499114PRTartificial sequencesynthetic peptide 11Xaa Glu Val
Xaa1124PRTartificial sequencesynthetic peptide 12Xaa Glu Ile Xaa1
User Contributions:
Comment about this patent or add new information about this topic:
| People who visited this patent also read: | |
| Patent application number | Title |
|---|---|
| 20100304700 | SIGNAL PROCESSING DEVICE AND METHOD AND RECEPTION SYSTEM |
| 20100304699 | METHOD AND APPARATUS FOR POSITION SIGNAL ASSISTED WINDOW PLACEMENT |
| 20100304698 | RADIO WAVE RECEIVER |
| 20100304697 | APPARATUS AND METHOD FOR RECEIVING BROADCAST INFORMATION USING ZIP CODE |
| 20100304696 | TRANSMITTING AND RECEIVING SYSTEM, TRANSMITTING APPARATUS AND RECEIVING APPARATUS |