Patent application title: METHODS AND COMPOSITIONS TO GENERATE AND CONTROL THE EFFECTOR PROFILE OF T CELLS BY SIMULTANEOUS LOADING AND ACTIVATION OF SELECTED SUBSETS OF ANTIGEN PRESENTING CELLS
Inventors:
Adrian Bot (Valencia, CA, US)
Lilin Wang (San Diego, CA, US)
Lilin Wang (San Diego, CA, US)
Dan Smith (San Diego, CA, US)
Bill Phillips (San Diego, CA, US)
IPC8 Class: AA61K3817FI
USPC Class:
4241791
Class name: Drug, bio-affecting and body treating compositions conjugate or complex of monoclonal or polyclonal antibody, immunoglobulin, or fragment thereof with nonimmunoglobulin material conjugated via claimed linking group, bond, chelating agent, or coupling agent (e.g., conjugated to proteinaceous toxin via claimed linking group, bond, coupling agent, etc.)
Publication date: 2011-11-10
Patent application number: 20110274705
Abstract:
The present invention is directed to novel compositions that cause
effective redirection of class I-immunity to Tc1 effectors, that take
advantage of the unexpected loading of MHC I by peptide within IgG
backbone combined with appropriate instruction of antigen presenting
cells. Such compositions are able to transform a seemingly ineffective
therapeutics into a highly effective one, associated with generation of
class I-restricted cytolytic cells and IFN-γ, IL-2 producing T
cells, further associated with protection against a highly virulent
microbe or recovery from malignant tumoral process.Claims:
1) A method of generating an enhanced T cell response to an antigen in a
patient, the method comprising, administering to the patient: a) a
polypeptide comprising at least one MHC-class I restricted T cell
epitope, and; b) a double stranded RNA; wherein the double-stranded RNA
is pA:pU, and wherein said polypeptide and said double-stranded RNA are
administered in an amount sufficient to generate a Tc1 response in the
patient to the antigen.
2) The method of claim 1, wherein the polypeptide comprises at least one MHC-class I restricted T cell epitope covalently attached to an IgG backbone without modification of the Fc portion.
3) The method of claim 2, wherein the MHC-class I restricted T cell epitope of the antigen is covalently attached within the Complementarity Determining Region (CDR) of the IgG.
4) The method of claim 1, wherein the pA:pU is provided in an amount sufficient to induce MHC class I-restricted Tc1 cells thereby producing IFN-.gamma..
5) The method of claim 1, wherein the double-stranded RNA has a molecular weight from 10 to 50 Kd.
6) The method of claim 1, wherein the double-stranded RNA are from 100 to 2000 base pairs in length.
7) The method of claim 2, wherein the immunoglobulin backbone of the IgG is derived from human IgG, or is a humanized IgG.
8) The method of claim 1, wherein the patient is human.
9) The method of claim 1, wherein the antigen is a virus.
10) The method of claim 9, wherein the virus is influenza virus.
11) The method of claim 1, wherein the T cell epitope is selected from: influenza virus MI or M2; hepatitis C virus NS3; hepatitis B virus core antigen; human papilloma virus HPV 18-E7, HPV 16-E7, HPV 18-E6, HPV 16-E6; HIV-I: reverse transcriptase; HIV-I: gag; herpes simplex antigens; and respiratory syncytial virus antigens.
12) The method of claim 1, wherein the T-cell epitope is a tumor associated T cell epitope.
13) The method of claim 1, wherein the T cell epitope is selected from: melanoma-gp100; MART-1; TRP-2; carcinoembryonic antigen precursor; Her-2; prostate tumor antigens; carcinoembryonic antigen precursor XP064845/NCB1; MUC 1; and mucin 1.
14) The method of claim 1, wherein the polypeptide and double-stranded RNA are admixed together.
15) The method of claim 1, wherein the polypeptide and double-stranded RNA are administered separately.
Description:
RELATED CASES
[0001] The present application claims priority to U.S. patent application Ser. No. 60/412,219 filed Sep. 20, 2002 and international application number PCT/US 03/07995 filed on Mar. 14, 2003, both of which are hereby incorporated by reference in their entireties.
FIELD OF THE INVENTION
[0002] The present invention is generally directed to methods and compositions to generate an immune response. More specifically, the present invention is directed to methods and compositions of loading an antigen presenting cell to display a delivered epitope on a MHC class I molecule in a context appropriate for the generation of desired T cell responses.
BACKGROUND OF THE INVENTION
[0003] No direct evidence has been shown that delivery of antigen via Fc gamma receptors ("FcγR") triggers an effective antitumoral or antiinfectious response. For example, it was previously shown that delivery of a viral NP (nucleoprotein) derived epitope within an immunoglobulin or IgG backbone did not result in detectable induction of cytotoxic immunity (Zaghouani et al., Eur J Immunol. 1993 November; 23(11):2746-50). In contrast, delivery of the same epitope in context of NP expressing cells (transfectomas) resulted in significant cytolytic activity. It was therefore concluded at that time that "APC (antigen presenting cells) are unable to present an influenza nucleoprotein [NP] peptide from the same context (1 microM Ig-NP) to an MHC class I-restricted T cell" and thus, "the endocytic compartment, when offered MHC class I- and II-restricted peptides within the same carrier protein context, favors presentation by class II by at least 1000-fold".
[0004] Access of the NP epitope to MHC class I presentation pathway is dependent on delivery strategy and was thus believed to be severely limited subsequent to FcγR internalization. More recently, it has been proposed that cross-linking or simultaneous engagement of FcγR on antigen presenting cells ("APC") may greatly optimize signal transduction and result in stimulation of cross-priming and APC stimulation, resulting in effective loading of MHC class I molecules (Regnault et al., J Exp Med. 1999, Jan. 18; 189(2):371-80). This could be achieved using immune complexes (multivalent antigen-antibody non-covalent complexes); however, due to the potential of C ("complement") mediated disease, the complexes could only be administered to the APC ex vivo (Naama et al., J Clin Lab Immunol. 1985 June; 17(2):59-67; Rafiq et al., J Clin Invest. 2002 July; 110(1):71-9). Alternatively, (Fab)2-antigen recombinant fusion constructs directed to receptors onto APC, can result in receptor cross-linking internalization, and presentation in context of MHC class II molecules (Lunde et al., Biochem Soc Trans 2002; 30(4):500-6). The insertion of antigen, however, modifies the Fc portion of the constant domains (CH2 and CH3) of the immunoglobulin ("Ig") that can result in serious and unpredictable effects on the half life and pharmacokinetics, two parameters that are tightly associated with the integrity of this segment (Spiegelberg H L, J Clin Invest 1975 September; 56(3):588-94). Finally, there is no conclusive evidence to date that either one of the strategies described above, when applied in vivo, induce protective or therapeutic anti-tumoral or anti-microbial immunity that would be associated with the generation of optimal MHC class I and II-restricted T cells that produce specific cytokines such as IFN-γ. Even when applied ex vivo, the immune complex strategy has displayed limited efficacy due to the balance in the activity of ITAM+ and ITIM+ FcγR (Kalergis and Ravetch, J Exp Med 2002 Jun. 17; 195(12):1653-9). Thus, it has yet to be determined whether in vivo delivery of antigen to APC via the monovalent ligation of Fcγ receptors can be used to induce effective anti-tumoral or antiviral immunity.
[0005] PCT Application Serial Number PCT/US03/07995 filed Mar. 14, 2002 and U.S. patent application Ser. No. 60/364,490 filed Apr. 30, 2002 are hereby incorporated by reference. Swiss-Protein/Trembl Protein Knowledgebase® on CD-ROM, available from Geneva BioInformatics, is hereby incorporated by reference in its entirety.
SUMMARY OF THE INVENTION
[0006] The present invention demonstrates, contrary to expectations, that in vivo and ex vivo loading of APC via monovalent engagement of FcγR, using peptide epitopes covalently attached to the IgG backbone without modification of the Fc portion, results in access of the epitope to the MHC I processing and presentation pathway, with effective loading of MHC class I molecules. Unexpectedly, this results in generation of robust Tc2 responses characterized by IL-4, but not IL-2 or IFN-γ-producing, MHC class I restricted T cells that recognize the epitope within IgG backbone.
[0007] In addition, the generation of this "deviated" response was not effective in controlling a pathologic process associated with tumor growth, nor was it associated with significant priming of cytolytic T cells. This explains largely the previous failure to detect induction of immunity in similar context previously and demonstrates, unexpectedly, that cross-linking or multivalent engagement of FcγR on APC (such as in context of immune complexes or Fab2-antigen compounds) is not a prerequisite for effective loading of the peptide onto MHC class I molecules. This is important since the concept could be applied in vivo (in contrast to immune complexes) and the integrity of Fc portion and thus PK profile could be retained (in contrast to Fab2-antigen recombinant molecules). Despite effective loading of MHC class I molecules, the APC were not able to trigger protective anti-tumoral and anti-microbial immunity when loaded in vivo by peptide epitope within IgG backbone.
[0008] Further, the present application discloses novel compositions that result in effective redirection of class I-immunity to Tc1 effectors that take advantage of the unexpected loading of MHC I by peptide within IgG backbone. Such compositions are able to transform seemingly ineffective MHC class II and class I-restricted peptides into highly effective ones. FcγR-mediated loading of APC associated with stimulation of APC by novel synthetic polynucleotides, result in generation of class I-restricted cytolytic cells and IFN-γ, IL-2 producing T cells, further associated with protection against a highly virulent microbe or recovery from malignant tumoral process. It is also shown that variants of the technology, applied incorrectly or as previously proposed, are not optimal in generation of immunity protective against viruses or tumors, in particular of MHC class I-restricted nature. The present application demonstrates the reason for past failures and teaches how to obtain and apply the different components of the technology in order to obtain optimal effect.
[0009] Various embodiments of the invention include: [0010] 1. A method of loading an antigen presenting cell and generating a T cell response against an antigen or peptide epitope by use of at least one peptide epitope attached to an Ig, Ig backbone backbone or portion thereof thereby forming an Ig-peptide molecule/complex or portion thereof wherein when administered to a patient in vivo or ex vivo, the epitope is effectively processed and presented by the MHC I pathway of the antigen presenting cell resulting in effective loading of MHC class, I molecules on the antigen presenting cell thereby resulting in an MHC class I-peptide complex. [0011] 2. The method of paragraph 1 wherein the Ig-peptide molecule/complex or portion thereof is administered with RNA strands. [0012] 3. The method of paragraph 2 wherein the RNA is dsRNA strand and is pA:pU. [0013] 4. The method of paragraph 3 wherein the dsRNA is pA:pU and the dsRNA is between approximately 20-100 base pairs in size. [0014] 5.The method of paragraphs 1, 2, 3 or 4 wherein the Ig backbone is derived from human Ig. [0015] 6. The method of paragraphs 1, 2, 3 or 4 wherein the Ig backbone is derived from human IgG. [0016] 7. The method of paragraph 1, 2, 3, or 4 wherein the Ig backbone is humanized Ig. [0017] 8. The method of paragraph 1 wherein the antigen presenting cell is loaded via monovalent engagement of FcγR. [0018] 9. The method of paragraph 1 wherein the antigen presenting cell may be loaded in vivo or ex vivo. [0019] 10. The method of paragraph 1 wherein the peptide epitopes are covalently attached to the Ig backbone. [0020] 11. The method of paragraph 1 wherein the peptide epitope is attached to the Ig backbone without modification of the Fc portion of the Ig. [0021] 12. The method of paragraph 1 wherein the peptide epitope is inserted within a CDR region of the immunoglobulin molecule. [0022] 13. The method of paragraphs 1, 2, 3 or 4 wherein the peptide epitope is inserted within a CDR region of the immunoglobulin molecule by insertion or deletion. [0023] 14. The method of paragraphs 1, 2, 3 or 4 wherein the MHC class I-peptide complex results in generation of robust Tc2 responses characterized by IL-4 but not IL-2 or IFN-γ-production. [0024] 15. The method of paragraph 1 wherein the peptide epitope is selected from the group consisting of: influenza virus M1 or M2; hepatitis C virus NS3; hepatitis B virus core antigen; human papilloma virus HPV 18-E7, HPV 16-E7, HPV 18 E6, HPV 16 E6; melanoma-gp100; MART-1; TRP-2; carcinoembryonic antigen precursor; Her-2; tetanus toxin universal T helper epitope; HIV-1: reverse transcriptase; HIV1: gag; insulin precursor-human; human Gad 65; prostate tumor antigens; mucin 1; herpes simplex antigens; and, respiratory syncytial virus antigens. [0025] 16. The method of paragraph 1 wherein the negative effects of sera are avoided. [0026] 17. The method of paragraphs 1, 2, 3 or 4 wherein the Ig peptide molecule and dsRNA are administered by subcutaneous or intraperitoneal injection. [0027] 18. The method of paragraph 1 wherein the antigen presenting cell is selected from the group consisting of dendritic cells, monocytes, macrophages and B cells. [0028] 19. The method of paragraph 1 wherein the antigen presenting cell is selected from the group consisting of CD11c+ and CD11b+ APC. [0029] 20. The method of paragraph 1 wherein the resulting MHC-peptide complexes formed by in vivo delivery are expressed for up to 1 to 2 weeks. [0030] 21. The method of paragraphs 1, 2, 3 or 4 wherein the MHC-peptide complex results in activation of T cells. [0031] 22. The method of paragraph 21 wherein the T cell response is determined by ITAM+ and ITIM+ Fcgamma receptors on APC. [0032] 23. The method of paragraph 21 wherein expression of the gamma chain of ITAM+ FcγR isoforms induces the T cell response wherein ITIM+ FcγRII limits the T cell response. [0033] 24. The method of paragraphs 18 or 19 wherein monocytes induce Th2 and Tr1 cells, both dendritic cells and monocytes induce Th3 cells, and wherein CD11b+ monocytes are more potent than dendritic cells in triggering a regulatory response following IgG-mediated delivery of T cell epitope. [0034] 25. The method of paragraphs 1, 2, 3 or 4 wherein the loading of APC with a peptide delivered within an Ig backbone in vivo results in induction of Th2 immunity. [0035] 26. The method of paragraphs 1, 2, 3 or 4 wherein the loading of APC with a peptide delivered within an Ig backbone in vivo results in induction of Th3 and Tr1 immunity. [0036] 27. The method of paragraph 1 wherein the T cell response is enhanced by co-stimulation with one of the following selected from the group consisting of anti-CD40mAb, recombinant IL-12 or synthetic dsRNA. [0037] 28. The method of paragraphs 1, 2, 3 or 4 wherein IL-2, IFN-γ and IL-4 are down-regulated in a dose dependent manner and IL-10 and TGF-beta are upregulated in a dose-dependent manner. [0038] 29. The method of paragraphs 1, 2, 3, or 4 wherein the peptide epitope is recNP and induces NP-specific MHC class I-restricted T cell immunity consisting of IL-4 producing Tc2 cells. [0039] 30. The method of paragraph 1 further comprising the use of RNA motifs thereby resulting in a modified immune response. [0040] 31. The method of paragraph 30 wherein the RNA motifs are dsRNA. [0041] 32. The method of paragraph 27 wherein the IgG1 and IgG2a antibody responses were increased and associated with an enhanced Th1 and Th2 response. [0042] 33. The method of paragraph 2, 27 or 30 wherein the dsRNA was selected from the group consisting of pA:pU, pI:pC and pC:pG. [0043] 34. The method of paragraphs 27 or 30 wherein the dsRNA is pA:pU and induced MHC class I-restricted Tc1 cells thereby producing IFN-γ. [0044] 35. The method of paragraphs 33 or 34 wherein the dsRNA are from 10-50 Kd. [0045] 36. The method of paragraphs 2 or 30 wherein the RNA motifs are ssRNA selected from the group consisting of p(A), p(C), p(G), p(I) and p(U). [0046] 37. The method of paragraph 1 wherein the peptide-epitope is NP and further comprising the coadministration of dsRNA motifs thereby resulting in effective induction of IL-2 and IFN-gamma. [0047] 38. The method of paragraph 1 wherein the APC are loaded ex vivo resulting in the formation of MHC class I-peptide complexes and generation of a Tc response. [0048] 39. The method of paragraph 38 wherein the APC are administered to the patient by adoptive transfer. [0049] 40. The method of paragraph 38 wherein the formation of MHC class I-peptide complexes results in differentiation of Tc2 cells producing IL-4 but not IFN-gamma. [0050] 41. The method of paragraph 38 wherein further comprising the step of administering RNA motifs thereby resulting in a broadening of the T cell profile to include IFN-gamma producing Tc1 cells. [0051] 42. A method of immunization of a patient comprised of loading an antigen presenting cell by use of at least one peptide epitope of an antigen attached to an Ig backbone or portion thereof thereby forming an Ig-peptide molecule and administering to the patient in vivo the Ig-peptide molecule in conjunction with a dsRNA motif wherein the epitope is effectively processed and presented by the MHC I pathway resulting in effective loading of MHC class I molecules and thereby resulting in an effective secondary expansion of MHC class I-restricted T cells subsequent to in vivo exposure to the antigen. [0052] 43. The method of paragraph 42 wherein the antigen is a virus. [0053] 44. The method of paragraph 43 wherein the virus is the influenza virus. [0054] 45. The method of paragraph 42 wherein the peptide-epitope is recIgG-NP(Kd). [0055] 46. The method of paragraph 42 wherein the dsRNA is pA:pU. [0056] 47. The method of paragraph 42 wherein the T cells are cytotoxic T lymphocytes. [0057] 48. The method of paragraph 42 wherein the secondary expansion of MHC class I-restricted T cells subsequent to in vivo exposure to the antigen is greater than administration of the recombinant antigen in sterile saline only. [0058] 49. A method of controlling and treatment of a tumor after clinical diagnosis, by loading an antigen presenting cell by use of at least one tumor associated T cell epitope attached to an IgG backbone or portion thereof thereby forming an IgG-peptide molecule and administering the Ig-peptide molecule in vivo in conjunction with dsRNA. [0059] 50. The method of paragraph 49 wherein the tumor associated T cell epitope is effectively processed and presented by the MHC I pathway resulting in effective loading of MHC class I molecules on the antigen presenting cell thereby resulting in an MHC class I-peptide complex. [0060] 51. The method of paragraph 49 wherein the method results in an immune response to the tumor associated T cell epitope and tumor rejection. [0061] 52. The method of paragraphs 49, 50 or 51 wherein the dsRNA is pA:pU. [0062] 53. The method of paragraph 49 wherein the Ig-G peptide complex and dsRNA are administered repeatedly as an anti-tumor therapy. [0063] 54. The method of paragraph 49 wherein upon tumor rejection, Tc1 immunity is developed against the tumor associated epitope. [0064] 55. The method of paragraph 49 where upon administration of IgG-peptide and dsRNA, Tc2 immunity is developed against the tumor associated epitope. [0065] 56. The method of paragraph 49 wherein the method further induces an effective memory response to the same tumor associated epitope. [0066] 57. The method of paragraph 49 wherein the method results in continued immunity to tumor cell variants. [0067] 58. The method of paragraphs 49, 50, 51, 52, 53, 54, 55, 56, or 57 wherein the tumor associated T cell epitope is selected from the group consisting of melanoma-gp100, MART-1, TRP-2, carcinoembryonic antigen precursor XP 064845/NCB1, Her-2, prostate tumor antigens, and MUC 1. [0068] 59. A recombinant human Ig molecule or portion thereof capable of binding to an FcγR of an APC, comprising of a CH3 region adjacent to a CH2 region whereby a hinge region attaches an antigen to the CH2 region wherein the antigen has an oligo-glycine linker attached to the hinge region. [0069] 60. The recombinant human Ig molecule of paragraph 59 whereby the antigen has a flanking sequence extending therefrom followed by a leader. [0070] 61. The recombinant human Ig molecule of paragraph 59 wherein the human Ig molecule is an IgG molecule. [0071] 62. The recombinant human Ig molecule of paragraph 59 wherein the antigen is a viral or tumor antigen. [0072] 63.The recombinant human Ig molecule of paragraph 59 wherein the amino acid sequence of the CH3 region is: GQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNY KTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSP GK and conservatively modified variants thereof. [Seq. I.D. No. 1]. [0073] 64. The recombinant human Ig molecule of paragraph 59 wherein the amino acid sequence of the CH2 region is: APELLGGPSVFLFPPKPKDTLMISRTPEVTCV VVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWL NGKEYKCKVFNKALPAPIEKTISKAK and conservatively modified variants thereof. [Seq. I.D. No. 2]. [0074] 65. The recombinant human Ig molecule of paragraph 59 wherein the amino acid sequence of the hinge region is: EPKSCDKTHTCPPCP and conservatively modified variants thereof. [Seq. I.D. No. 3]. [0075] 66. The recombinant human Ig molecule of paragraph 53 wherein the amino acid sequence of the flanking sequence is: QVQLQ and conservatively modified variants thereof. [Seq. I.D. No. 4]. [0076] 67. A composition for enhancing an immune response to an antigen wherein the composition is a polynucleotide wherein the polynucleotide is made up of compounds selected from the group consisting of adenine, uracil, guanine, cytosine and inosine. [0077] 68. The composition of paragraph 67 wherein the polynucleotide is dsRNA. [0078] 69. The composition of paragraph 68 wherein the dsRNA is selected from the group consisting of pA:pU and pI:pC. [0079] 70. The composition of paragraph 69 wherein the dsRNA is pA:pU and wherein some of the adenine and uracil is occasionally replaced by guanine, cytosine or inosine along the polynucleotide chain. [0080] 71. The composition of paragraph 69 wherein the antigen is a virus. [0081] 72. The composition of paragraph 69 wherein the antigen is attached to an inununoglobulin or portion thereof and administered in vivo. [0082] 73. The composition of paragraph 72 wherein the antigen is protein or a peptide. [0083] 74. The composition of paragraphs 67, 68, 69 or 70 wherein the antigen is a tumor associated epitope. [0084] 75. The composition of paragraph 74 wherein the antigen is a T cell epitope. [0085] 76. The composition of paragraphs 67, 68, 69 or 70 wherein the dsRNA is administered together with said antigen. [0086] 77. The composition of paragraph 67 wherein the polynucleotide is dsRNA and is coadministererd with the antigen. [0087] 78. The composition of paragraph 67 wherein the antigen is already present in the body. [0088] 79. The composition of paragraph 67 wherein the antigen is administered in a pharmaceutically acceptable carrier. [0089] 80. Use of dsRNA in the manufacture of a medicament for enhancing an immune response to an antigen in a patient, comprising administering said dsRNA to a patient in conjunction with said antigen. [0090] 81. The use of paragraph 80 wherein an epitope of said antigen is delivered to the patient in an immunoglobulin or portion thereof. [0091] 82. The use of paragraphs 80 or 81 wherein the dsRNA is comprised of pA:pU. [0092] 83. The use of paragraphs 80 or 81 wherein the dsRNA is comprised of pI:pC. [0093] 84. The use of paragraph 81 wherein the dsRNA consists of bases selected from the group consisting of adenine, cytosine, uracil, guanine and inosine. [0094] 85. The use of paragraphs 81, 82 or 83 wherein the use enhances the Th1 and/or Tc1 response to the antigen. [0095] 86. The use of paragraphs 81, 82 or 83 wherein the use induces a Tc1 cell response to the antigen. [0096] 87. The use of paragraphs 81, 82 or 83 wherein the immune response includes an enhanced B cell response. [0097] 88. The use of paragraphs 81, 82 or 83 wherein the antigen is administered with additional antigen. [0098] 89. The use of paragraphs 81, 82 or 83 wherein the use induces expression of CXC and CC chemokines. [0099] 90. The use of paragraphs 81, 82 or 83 wherein the administering of dsRNA enhances T or B cell responses or both T and B cell responses by recruitment and activation of CD11b+ monocytes. [0100] 91. The use of paragraphs 81, 82 or 83 wherein the administering of dsRNA enhances T or B cell responses or both T and B cell responses by recruitment and activation of dendritic cells. [0101] 92. The use of paragraphs 81, 82 or 83 wherein the dsRNA compositions enhance an immune response by recruiting antigen presenting cells. [0102] 93. The use of paragraph 92 wherein the antigen presenting cell is a professional antigen presenting cell.
[0103] 94. The use of paragraph 92 wherein the antigen presenting cell is a naive antigen presenting cell. [0104] 95. The use of paragraphs 81, 82 or 83 wherein the antigen is a non-infectious antigen and wherein the MHC Class 1 restricted T cells are cross-primed by the dsRNA. [0105] 96. The use of paragraphs 81, 82 or 83 wherein the composition and antigens are administered by one of the following selected from the group consisting of mucosal administration, respiratory administration, intravenous administration, subcutaneous administration, and intramuscular administration. [0106] 97. The use of paragraph 81 wherein the antigen is administered in an immunoglobulin or portion thereof or in an immunoglobulin backbone. [0107] 98. The use of paragraph 97 wherein the wherein the antigen is a peptide epitope. [0108] 99. A method of preventing high zone tolerance in a patient to an antigen comprising administering said antigen together with a dsRNA composition wherein the dsRNA composition comprises at least one compound selected from the group consisting of poly-adenine, poly-uracil, poly-guanine, poly-cytosine, poly-inosine. [0109] 100. The method of paragraph 99 wherein the antigen is non-infectious. [0110] 101. The method of paragraph 99 wherein the antigen is administered in high doses or already present in the body. [0111] 102. The method of paragraphs 99, 100 or 101 wherein the dsRNA is selected from the group consisting of pA:pU and pI:pC. [0112] 103. The method of paragraphs 99, 100, 101 or 102 wherein the method prevents B cell unresponsiveness. [0113] 104. A method of enhancing the immune system in a patient exposed to a pathogen comprising the administration of dsRNA to the patient. [0114] 105. The method of paragraph 104 wherein the dsRNA is selected from the group consisting of pA:pU and pI:pC. [0115] 106. The method of paragraphs 104 or 105 wherein the dsRNA is administered to a patient in concentrations ranging from 100 ug/ml to 1 mg/ml. [0116] 107. The method of paragraphs 104, 105 or 106 wherein the pathogen is unknown. [0117] 108. The method of paragraphs 104, 105, 106 or 107 wherein the dsRNA is administered in a pharmaceutically acceptable carrier. [0118] 109. The method of paragraph 104 wherein a T cell response to the pathogen is enhanced. [0119] 110. A method of enhancing an immune response in a patient in need thereof comprising loading an antigen presenting cell by use of at least one peptide epitope of an antigen attached to an Ig backbone thereby forming an Ig-peptide complex or molecule and administering the Ig-peptide complex or molecule in vivo in conjunction with a dsRNA motif wherein the epitope is effectively processed and presented by the MHC pathway of the antigen presenting cell resulting in effective loading of MHC molecules and thereby resulting in an effective secondary expansion of MHC molecules subsequent to in vivo exposure to the antigen. [0120] 111. The method of paragraph 110 wherein the MHC pathway is the MHC I pathway. [0121] 112. The method of paragraph 110 wherein the MHC pathway is the MHC II pathway. [0122] 113. The method of paragraph 111 wherein the method results in effective loading of MHC Class I molecules on the antigen presenting cell. [0123] 114. The method of paragraph 112 wherein the method results in effective loading of MHC Class II molecules on the antigen presenting cell. [0124] 115. The method paragraphs 110, 111 or 112 wherein the dsRNA is pA:pU. [0125] 116. The method of paragraphs 110, 111 or 113 wherein the method results in secondary expansion of MHC Class I restricted T cells. [0126] 117. The method of paragraph 115 wherein the antigen is a virus. [0127] 118. The method of paragraph 117 wherein the virus is an influenza virus. [0128] 119. The method of paragraph 115 wherein the antigen is a tumor associated epitope. [0129] 120. The method of paragraph 115 wherein the T cell is a cytotoxic T lymphocyte. [0130] 121. A method of generating an immune response to an antigen in a patient comprising: [0131] administering to the patient an immunoglobulin or portion thereof wherein said immunoglobulin has at least one peptide epitope of said antigen attached to said immunoglobulin or portion thereof and administering said immunoglobulin or portion thereof in conjunction with a dsRNA segment. [0132] 122. The method of paragraph 121 wherein the immunoglobulin or portion thereof and said dsRNA segment are administered together. [0133] 123. The method of paragraph 121 wherein the immunoglobulin or portion thereof and said dsRNA segment are administered separately. [0134] 124. The method of paragraph 121 wherein said patient is human. [0135] 125. The method of paragraph 121 wherein upon administration of said immunoglobulin or portion thereof to said patient the immunoglobulin or portion thereof loads the antigen presenting cell by engagement with the antigen presenting cell's FcγR said peptide epitope is effectively processed and presented by the MHC I pathway of the antigen presenting cell resulting in effective loading of the MHC class I molecules. [0136] 126. The method of paragraph 121 wherein the peptide epitope is attached within the CDR region of the immunoglobulin or portion thereof. [0137] 127. The method of paragraph 121 wherein the immune response generates an effective T cell response to the antigen. [0138] 128. The method of paragraph 121 wherein the T cells are cytotoxic T lymphocytes. [0139] 129. The method of paragraph 121 wherein the dsRNA segment is selected from the group consisting of pA:pU and pI:pC. [0140] 130. The method of paragraph 121 wherein the peptide epitope is a T cell epitope. [0141] 131. The method of paragraph 121 wherein the peptide epitope is selected from the group consisting of influenza virus M1 or M2; hepatitis C virus NS3; hepatitis B virus core antigen; human papilloma virus HPV 18-E7, HPV 16-E7, HPV 18 E6, HPV 16 E6; melanoma-gp100; MART-1; TRP-2; carcinoembryonic antigen precursor; Her-2; tetanus toxin universal T helper epitope; HIV-1: reverse transcriptase; HIV1: gag; insulin precursor-human; human Gad 65; prostate tumor antigens; mucin 1; herpes simplex antigens; and, respiratory syncytial virus antigens. [0142] 132. The method of paragraph 121 wherein the immunoglobulin or portion and dsRNA segment thereof is administered by one of the methods selected from the group consisting of intravenous administration and bolus injection. [0143] 133. The method of paragraph 121 wherein the immunoglobulin or portion thereof and the dsRNA are administered in a pharmaceutically acceptable carrier. [0144] 134. The method of paragraph 121 wherein the method induces an effective memory response to the peptide epitope.
BRIEF DESCRIPTION OF THE DRAWINGS
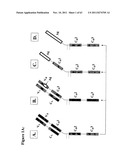
[0145] FIG. 1A shows (a) representation of natural IgG (light chain-heavy chain heterodimer); (B) antigen (Ag) derived peptide inserted within CDR (complementarity determining region) 3, 2, 1 or framework region; (C) VH (heavy chain, variable region) segment replaced with an antigen or fragment; (D) VH and CH1 segments replaced with antigen or antigen fragment;
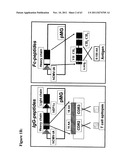
[0146] FIG. 1B illustrates diagramatically the IgG-peptide and Fc peptide;
[0147] FIG. 1C shows properties of selected human IgG backbone;
[0148] FIG. 1D shows the sequence of the constant region of the heavy chain as well as schematic depiction of a prospective construct;
[0149] FIGS. 1E-1M show the sequences of various antigens and epitopes discussed in the present application and which can be inserted into an immunoglobulin [sequences can be accessed on the internet at ncbi.nlm.nih.gov (add the proper address prefix: http://www.) by searching the "proteins" section by use of the provided accession number. The content of this database is hereby incorporated by reference in its entirety.];
[0150] FIGS. 2A-2B show that while the injection of the peptide epitope in saline was not immunogenic, a similar dose of peptide used for ex vivo loading of APC effectively triggered a substantial immune response upon adoptive transfer;
[0151] FIG. 3 shows that delivery of epitope within Ig backbone considerably favored its stability in the systemic circulation;
[0152] FIGS. 4A-4B show that pre-incubation of peptide with serum resulted in decreased TcH activation;
[0153] FIGS. 5A-5B show that the relative efficiency of MHC-peptide complex formation greatly varied depending on the nature of antigen and APC;
[0154] FIGS. 6A-6B show that the peptide epitope within IgG backbone was more effective on a molar basis (1 order of magnitude) than the peptide alone in inducing TcH activation when handled by blood-derived APC;
[0155] FIGS. 7A-7B show that the use of oil-in-water adjuvant (incomplete Freund's adjuvant, IFA) only modestly enhanced the in vivo formation of MHC-peptide complexes on APC of lymph nodes but not the spleen or thymus;
[0156] FIGS. 8A-8D show that use of FcγR mediated delivery of peptides results in preferential formation of immunogenic MHC II-peptide complexes on CD11c+ and CD11b+ APC;
[0157] FIGS. 9A-9C show long lasting expression of peptide onto endogenous MHC II, on both DC (dendritic cells) and monocytes;
[0158] FIG. 10 shows that formation of MHC II-peptide complexes on dendritic cells and monocytes, subsequent to IgG mediated delivery of peptide epitope, is critically dependent on ITAM+ FcγR that encompass the gamma chain;
[0159] FIG. 11 shows that results show that the expression of the gamma chain of ITAM+ FcγR isoforms is necessary for the induction of T cell response to APC loaded with peptide within the IgG backbone;
[0160] FIGS. 12A-12D show that unexpectedly and in contrast with the potency/cell basis (Example 8), at the organism level, the CD11b.sup.+ monocytes have the highest impact on the immune response to a peptide epitope delivered within the IgG backbone;
[0161] FIGS. 13A-13B shows that FcγR-mediated delivery of a T cell epitope within the recombinant Ig backbone results in Th2 rather than Th1 response;
[0162] FIG. 14 shows that FcγR-mediated delivery of T cell epitope within recombinant Ig backbone results in Th2 rather than Th1 response;
[0163] FIG. 15 shows that a peptide epitope within the IgG backbone triggers a cellular response of Th2 profile that is enhanced but not switched by a conventional adjuvant (CFA);
[0164] FIG. 16 shows that peptide presentation by APC, subsequent to loading with antigen by using recombinant IgG as delivery platform, occurs in context of limited co-stimulation;
[0165] FIGS. 17A-17B show that the activity of HA (110-120 hemagglutinin peptide) specific IL-4 producing T cells triggered by administration of recHA(I-Ed)-IgG is dependent on CD4 rather than CD8;
[0166] FIG. 18 shows that the IgG mediated delivery of T cell epitope has a profound and differential effect on the expansion and cytokine production by activated T cells: IL-2, IFN-γ and surprisingly IL-4, were down-regulated in a dose-related manner;
[0167] FIGS. 19A-19B show that in contrast to viral immunization with an influenza virus strain bearing the cognate peptide, Ig-mediated peptide delivery was ineffective in triggering cytotoxic response;
[0168] FIGS. 20A-20D show that co-administration of MBP and PLP epitopes by using recombinant IgG curbed the chronic progression of disease;
[0169] FIG. 21 summarizes the impact of IgG/FcγR-mediated delivery of epitopes on the T cell response, based on data provided in Examples 2-20;
[0170] FIG. 22 shows that shows that natural, non-infectious double stranded RNA produced during infection with influenza virus, has substantial effects on the specific immune response to a protein antigen;
[0171] FIG. 23A shows an extensive library of synthetic RNA motifs;
[0172] FIGS. 23B-23D show that different synthetic RNAs have an enhancing effect on the B and T cell response to a prototype protein antigen;
[0173] FIGS. 24A-24B show effects of selected RNA motifs on the innate immune response;
[0174] FIG. 25 shows that distinct RNA motifs bind to different receptors on antigen presenting cells;
[0175] FIG. 26 shows that distinct RNA motifs induce differential upregulation of chemokines;
[0176] FIG. 27 shows that the control of replication of influenza virus can be achieved by using selected synthetic RNA motifs;
[0177] FIG. 28 shows that selected synthetic RNA motifs pI:pC and pA:pU largely prevent high zone tolerance that is usually associated with administration of large amounts of purified protein;
[0178] FIG. 29 shows that selected synthetic RNA motifs effect on human monocytic cells;
[0179] FIGS. 30A-30B show that non-tagged pA:pU, but not non-tagged pI:pC, was able to compete out the binding of tagged pA:pU to human THP-1 monocytic cells;
[0180] FIG. 31 shows the purification and fractionation steps of dsRNA;
[0181] FIG. 32 shows that lower molecular weight fractions of a selected synthetic RNA compounds are endowed with different biological activity;
[0182] FIG. 33 shows that pI:pC but not pA:pU induced antibody response against itself, with a cross-reactive component against another RNA motif;
[0183] FIGS. 34A-34B show that co-use of selected synthetic RNAs promote effective induction of IL-2 and IFN-gamma subsequent to IgG mediated delivery of an MHC class I-restricted epitope;
[0184] FIG. 35 shows that ex vivo APC loading by recombinant IgG is more effective in formation of MHC class I-peptide complexes and generation of Tc response, compared to use of free peptide itself;
[0185] FIG. 36 show that IgG mediated delivery of a class I restricted epitope is most effective in priming class I restricted Tc1 responses when co-administration of selected synthetic RNA was carried out;
[0186] FIG. 37 shows that effective priming of anti-viral cytotoxic T cells requires both effective in vivo loading of APC with class I restricted epitope delivered via IgG, together with appropriate instruction by selected synthetic RNA motif;
[0187] FIG. 38 shows that immunization with a recombinant IgG bearing a viral class I restricted epitope together with selected synthetic dsRNA, resulted in priming of an immune response capable of limiting the replication of a virus subsequent to infectious challenge;
[0188] FIG. 39 describes the tumor models used for testing the efficiency of Ig-peptide-based molecules;
[0189] FIG. 40 shows that both effective in vivo loading of APC with tumor associated antigen, together with simultaneous activation by selected synthetic RNA motifs, are necessary and sufficient for effective control of tumor growth and induction of tumor rejection;
[0190] FIG. 41 shows that both effective in vivo loading of APC with tumor associated antigen, together with simultaneous activation by selected synthetic RNA, can trigger an effective immune response to tumor-associated antigens;
[0191] FIG. 42 shows that tumor infiltrating lymphocytes displaying the T cell receptor marker TCRβ acquired expression of the activation marker CD25 upon treatment with recombinant immunoglobulin bearing tumor associated epitope, together with selected synthetic dsRNA motif;
[0192] FIG. 43 shows that the treated mice that successfully rejected the tumor developed Tc1 responses against the tumor-associated epitopeon the therapeutic Ig, along with Tc2 immunity;
[0193] FIG. 44 shows that successful rejection of tumor induced by indicated treatment is followed by effective protection against subsequent challenge with the same tumor, indicating development of effective immune memory; and,
[0194] FIGS. 45A-45B show that the emerging immunity, subsequent to the indicated treatment that results in tumor rejection, protects against challenge with loss of antigen variants and is associated with overall expansion of cytokine producing cells.
DETAILED DESCRIPTION OF THE INVENTION
[0195] Definitions:
[0196] The following definitions are intended to act as a guide and are not to be considered limiting of terms found throughout the specification: [0197] adjuvant--a substance that enhances the adaptive arm of the immune response to an antigen; [0198] adoptive transfer--transfer of a cell population from one animal to another of the same haplotype; [0199] antigen--a molecule that can be specifically recognized by the adaptive elements of the immune system (B cells, T cells or both); [0200] antigen presenting cell--heterogeneous population of leukocytes with very efficient immunostimulatory capacity; [0201] BALB/C mouse--Widely distributed and among the most widely used inbred mouse strains; [0202] B cell--a type of lymphocyte developed in the bone marrow. Each B cell encodes a surface receptor specific for a particular antigen. Upon recognition of a specific antigen, B cells multiply and produce large amounts of antibodies which in turn bind to the antigen which activated the B cell; [0203] B cell unresponsiveness--antigen-specific lack of response by B cell; [0204] CDR--Complementarity Determining Region; hypervariable regions in an immunoglobulin which create the antigen binding site. There are three CDR regions: CDR1, CDR2 and CDR3; [0205] chemokines--a group of at least 25 small cytokines, all of which bind to heparin; [0206] complete Freund's adjuvant--an oil-in-water emulsion containing mycobacterial cell wall components; [0207] cross primed--antigen presenting cells that have acquired antigens from infected tissues and then present them to cognate T cells; [0208] Dendritic Cells--A subtype of antigen presenting cells (i.e. CD11c+); [0209] downregulation--decreasing the expression or activity of a particular compound or effect; [0210] epitope--parts of an antigen which contact the antigen binding site of the antibody or T cell receptor; [0211] FcγR--Ig receptors on cell surfaces of which there are three recognized groups: FcγRI (CD64), FcγRII (CD32) and FcγRIII (CD16); [0212] heterodimer--dimeric protein consisting of 2 different protein sequences; [0213] high zone tolerance--a state of unresponsiveness specific to a particular antigen that is induced upon challenge with a high concentration of said antigen; [0214] IL-2--refers to interleukin-2; [0215] IL-4--refers to interleukin-4; [0216] Immunoglobulin--a group of glycoproteins present in the serum and tissue fluids of all mammals and are located on the surface of B cells and serve as antibodies free in the blood or lymph. There are five classes of immunoglobulins: IgG (70-75%), IgM (10%), IgA (15-20%), IgD (>1%) and IgE (found on basophils and mast cells in all individuals). IgG has four human subclasses (IgG1, IgG2, IgG3 and IgG4); [0217] Immunoglobulin backbone--refers to an immunoglobulin molecule or portion thereof wherein at least one CDR region is able to receive an inserted peptide epitope; [0218] immunoglobulin isotype switching--stimulation of B cells to switch production from one immunoglobulin isotype to another; [0219] incomplete Freund's adjuvant--an oil-in-water emulsion not containing mycobacterial cell wall components; [0220] innate immunity--The innate immune system provides broad relatively nonspecific host defenses that lack antigenic specificity but have the ability to guide acquired immunity. Among the cells types involved are dendritic cells and macrophages; [0221] intraperitoneally--within peritoneal cavity; [0222] intravenously--within vasculature; [0223] isoforms--different glycosylation, phosphorylation, deamidation and other posttranslational modifications of proteins; [0224] ITAM--immunoreceptor tyrosine-based activation motifs; [0225] ITIM--immunoreceptor tyrosine-based inhibitory motifs; [0226] macrophages--Any mononuclear, actively phagocytic cell arising from monocytic stem cells in the bone marrow; [0227] MHC--refers to the Major Histocompatibility Complex; [0228] modified immune response--enhanced or diminished immune response; [0229] monocytes--Mononuclear leukocytes found in lymph nodes, spleen, bone marrow and loose connective tissue; [0230] naive--non-differentiated, non-activated cell; [0231] peptide--a compound consisting of two or more amino acids joined together by a peptide bond; [0232] polynucleotide--a polymer of nucleotides; [0233] professional antigen presenting cell--mature, able to present antigenic epitope; [0234] recruitment--attraction of a cell population to inflammatory site; [0235] secondary expansion--immune response which follows a second or subsequent encounter with a particular antigen; [0236] self-antigens--antigens that are derived from the host; [0237] subcutaneously--beneath the skin; [0238] Tc1 immunity--Cytotoxic T cell type 1, CD 8+; [0239] Th1 cells--T helper 1 cells which are involved in cell mediated inflammatory reactions, identified by production of IFNγ, TNFβ and IL-2; [0240] Th2 cells--T helper 2 cells which encourage production of antibodies and are identified by production of IL-4 and IL-5; [0241] Th3 cells--T helper regulatory cell, known to produce transforming growth factor (TGF)-beta; [0242] TR1 cells--T regulatory, cell, known to produce interleukin `10; and, [0243] upregulation--enhancement of expression or activity of a particular compound or effect;
Materials and Methods
[0244] For selective in vivo loading of antigen presenting cell subsets, the use of compounds described schematically in the FIG. 1A are used: (A) representation of natural IgG (light chain--heavy chain heterodimer); (B) antigen (Ag) derived peptide inserted within CDR 3, 2, 1 or framework region; (C) VII segment replaced with an antigen or fragment; and, (D) VII and CH1 segments replaced with antigen or antigen fragment. This type of molecules are engineered using methods known in the art and as stated as follows:
Construction of Model Recombinant IgG.
[0245] Polymerase chain reaction (PCR) mutagenesis was used to replace the CDR3 region of VII chain with the stated epitopes. Briefly, a pUC19 plasmid harboring the 5.5-kb EcoRI fragment carrying the VH gene of the murine anti-arsonate antibody, 91A3, was used as template DNA in two PCRs to delete the diversity segment (D) of the complementarity-determining region 3' (CDR3) loop and inserted DNA fragments encoding various antigen epitopes. These chimeric VII and as well as wild type VH genes were then ligated with Ig gamma 1 heavy chain constant region within the plasmid pSV2ΔHgptDNSVH-hCgamma1 from which the EcoRI dansyl (dns)-conjugated VH gene was cut out. The sequences of VH and inserted epitopes were confirmed by. DNA sequencing. To express these chimeric IgGs with murine 91A3 VH-human C gamma1 heavy chain genes and a mouse-human chimeric k light chain gene, an 8-kb BamHI fragment encoding the entire murine 91A3 kappa light chain gene was subcloned into the BamHI site of pUC19 plasmid. Subsequently, a HindIII fragment with the kappa light chain promoter and the V kappa region coding sequences was cut out from this plasmid and subcloned into the HindIII site of pSV184ΔHneoDNSVk-hCk upstream of the gene encoding a human k light chain C region (Ck) from which the dns-conjugated Vk (dnsVk) had been excised. This plasmid, which will encode a murine 91A3 Vk-human Ck light chain, is designated pSV184Δhneo91A3'Vk-hCk.
Construction of Human Recombinant IgG.
[0246] The human IgG backbone was obtained from IgGA1 myeloma cell line by RT-PCR. The recombinant human IgG was cloned by inserting the stated epitopes to replace the CDR2 or CDR3 regions of the human IgG1 backbone. Briefly, T cell epitopes were created by PCR mutagenesis and subcloned into the CDR2/CDR3 region. The recombinant heavy chains were then subcloned into pMG vector (Invivogen, San Diego, Calif.) by BamHI and XbaI sites. The heavy chain expression was controlled by the hCMV promoter. In parallel, the human kappa light chain was subcloned into the pMG vector by StuI and NheI sites. The expression of the light chain was controlled by an EF-1 alpha and HTLV-1 LTR hybrid promoter. The double expression vector carrying both the recombinant heavy chain and light chain were then transfected into expression cell lines.
[0247] The Fc-peptides were constructed by cutting off the VH and CH1 fragment and replacing it with stated viral or tumor antigens (8-150 Aas). Briefly, the human IgG1 heavy chain was subcloned into pCDNA3 vector by EcoRI and XhoI sites. Then the stated antigens are inserted between the leader sequence and hinge region of IgG1 by PCR mutagenesis. To increase the flexibility of the fused antigens, an oligo-glycine linker (5 glycines) was added after the antigen. The expression of human IgG recombinant molecules can be performed by using either one of the strategies displayed in FIG. 1B.
[0248] The human IgG backbone has been selected rationally, based on the ability to bind to FcγR, complement and cytokine activation in various states. Properties of selected human IgG backbone are shown in the FIG. 1C and the sequence of the constant region of the heavy chain as well as the schematic depiction of a prospective construct, is shown in FIG. 1D.
[0249] Epitopes used for model recombinant IgG are shown in FIG. 1E (mouse MHC class II-restricted HA epitope and mouse MHC class I restricted NP epitope). The nomenclature of recombinant constructs is recIgG-epitope (HA or NP)-restriction element (I-Ed or Kd, respectively). In short, they may be referred to as IgHA or IgNP. Model molecules comprising defined mouse self epitopes (MBP or PLP derived) were similarly constructed. The sequence of the variable region of the heavy chain of anti-arsonate antibody used as the backbone has been depicted in FIG. 1E and the technology is well known in the art (Zaghouani et al., Science 1993 Jan. 8; 259(5092):224-7) the contents of which is hereby incorporated by reference.
[0250] In FIGS. 1E-1M, examples of antigens and epitopes (in bold) are provided that could be inserted (larger parts up to 150 AA spanning one or multiple epitopes) or attached to the backbone. Such constructs comprising the shown antigens/epitopes may be used as drugs against infectious or tumoral diseases. In FIG. 1I there is the HLA-A2 anchor motif displayed, that allows the prediction of location of potentially therapeutic cytotoxic epitopes in any protein, facilitating the selection of the antigen fragment to be used in the recombinant immunoglobulin.
[0251] In FIG. 1J, examples of "universal" T helper epitopes (Kumar et al. J Immunol 1992 Mar. 1; 148(5):1499-505) are provided, both dominant and promiscuous from the point of view of MHC restriction, that could be used for construction of composite molecules for the purpose of inducing or enhancing immunity to MHC class I-restricted epitopes, using compounds such as:
[0252] [antigen fragment]-[universal Th epitope]-Fc(IgG).
[0253] Examples of such constructs are schematically represented in FIG. 1K (bottom).
[0254] In FIG. 1K top, examples of human self antigens with epitopes bolded are shown, that could be used to generate recombinant IgG molecules against autoimmune/inflammatory disorders.
[0255] In FIG. 1L and 1M other antigen sequences that could be used for the construction of above mentioned immunoglobulin constructs are shown. The antigen fragments of interest could be defined by using methods to predict MHC class I epitopes (Lim et al., Mol Immunol. 1996 February; 33[2]:221-30).
Production of Recombinant IgG
[0256] The SP2/0 cell line (American Type Culture Collection) is used for the production of all the recombinant IgGs (rIgG) discussed in this patent application. Stable expressing cell lines (i.e. transfectomas) were produced using a double transfection protocol with plasmids encoding the heavy and light chains of an anti-arsenate mouse IgG. Each transfectoma differs only in the sequence of the CDR3 region of the heavy chain. Methods for growing the cell lines as well as producing the different purified rIgG used in the experiments reported in this application are identical in all cases.
[0257] The SP2/0 transfectomas were initially grown in Quantum Yield media (BD Biosciences) supplemented with 5% (v/v) heat-inactivated fetal bovine serum, 0.5 mg/mM gentamicin and 2.5 μg/mL Fugizone. Cultures were maintained at 37° C. in a humidified CO2 incubator. Efforts were made to adapt each of the cell lines to growth in different commercially available serum-free medias (Lymphocyte Growth Media 2, Clonetics; Cell MAb Growth Media Serum Free, BD Biosciences; Animal Component Free Cell Media, BD Biosciences). Each of the serum-free medias was supplemented with antibiotics as above. Culture media containing secreted IgG was produced from each media noted above. No difference in the IgGs produced in the different medias was observed over the course of this work (molecular weight analysis by SDS PAGE [see below], ELISPOT assays, and immune responses in mice).
[0258] The amount of secreted rIgG was quantitated using an ELISA: capture antibody was a goat anti-mouse IgG (Sigma) and secondary antibody was an anti-mouse IgG HRP conjugate (Sigma). Purified mouse IgG (Sigma) was used as a standard.
[0259] Four different methods have been used to produce media containing the different rIgGs (i.e. conditioned media, "CM"): flasks, stirred vessels, packed bed bioreactors (New Brunswick Cellagen), CELLine flasks (BD Biosciences). In the case of CM produced in flasks, the cells were fed and/or harvested twice a week and maintained at least 50% viability, but viability was generally greater than 70%. Collected media was filtered and held at 4 C. Stirred vessels (1 L) were seeded at 106 cells per mL in 200 mL starting volume. Media was added weekly to keep the cell number between 107 and 106 per mL until 800 mL of total volume was reached. At this point cell viability was determined (typically greater than 80%), and the run was continued until such time that the viability fell below 50%. Media was then collected and sterile filtered to remove cells and held at 4° C. For the packed bed bioreactors: each unit was seeded with approximately 108 cells in 400 mL of media; maintained in a CO2 incubator at 37° C. with constant stirring; media was changed every 3-4 days and CM was filtered as above; production of rIgGs in the CM was monitored with ELISA. Bioreactor runs were continued until production of rIgGs began to decline or the vessel became contaminated. The 1 L CELLine flasks were used according to manufacturer's instructions.: each flask was seeded with 107 to 108 cells in a total volume 40 mL in the cell compartment; 1 L of media was added to the feed compartment; CM was harvested from the cell chamber after 2 to 3 weeks, or when viability of the cells fell below 20%.
Purification of rIgG
[0260] The rIgGs produced by the above methods were purified by one of two methods. For CM that contained FBS, an anti-mouse IgG immunoaffinity resin was used. The immunoaffinity resin was synthesized using the following protocol: 10 mL of cyanogen bromide-activated Sepharose 4B (Sigma) was washed with 1 mM HCl as per manufacturer's instructions; 10-20 mg of goat anti-mouse IgG (Sigma) was dissolved in coupling buffer (0.1 M sodium carbonate [pH 8.4]/0.5 M NaCl) at a concentration of 2 mg/mL; the IgG solution was added to the washed resin, and the slurry was mixed end-over-end at room temperature; the extent of coupling was monitored using the Bradford assay to determine the amount of remaining soluble IgG; the coupling was quenched by addition of ethanolamine to a final concentration of 10 mM when the amount of soluble IgG was less than 10% of the starting concentration (approximately 45 minutes). The immunoaffinity resin was then washed with the following buffers: PBS, 10 mM glycine (pH 2.4), 20 mM Tris/1 M NaCl (pH 8.0), PBS. The resin was stored at 4° C. in PBS. The protocol for purifying rIgG with this resin was initiated by passing CM through the column at 1 to 2 mL/min. The resin was then washed free of nonbound protein using the following protocol: 100 mL PBS/0.5M NaCl followed by 50 mL 1 mM Tris (pH 8). Fractions were monitored for protein using the Bradford assay. Specifically bound rIgG was eluted with a low pH buffer (5 mM glycine (pH 2.4)/0.5 M NaCl). The eluted protein was collected and held at 4° C. for further processing (see below).
[0261] The rIgG produced in serum-free culture media was purified using Protein A affinity chromatography. Typically, a 5 mL rProtein A column (HiTrap rProtein A FF from Amersham Pharmacia Biotech) was equilibrated with PBS and the sample was run through the column at 2 mL/min using a FPLC unit (Pharmacia). The resin was washed free of nonspecifically bound protein with PBS, followed by 20 mM Tris (pH 8.0)/1 M NaCl, then water. The specifically bound rIgG was eluted with 1 mM glycine (pH 2.4). The eluted peak was collected and held at 4 C for further processing.
[0262] Generally, the rIgG fractions were pooled and concentrated using Centricon . ultrafiltration units (Amicon) to a final concentration of 1 to 4 mg/mL (Bradford assay with IgG as standard). The concentrated fraction was then dialyzed into 1 mM glycine (pH 2.4), the final concentration determined by A280 using an extinction coefficient of 1.4 for a 1 mg/mL IgG solution, and aliquoted into 100 μl fractions that were stored in the -80° C. freezer. The purified rIgGs were analyzed for structural integrity and purity by SDS gel electrophoresis. The gels were stained with Coomassie blue (Pierce Chemical). In all cases the rIgGs used in the reported experiments displayed their expected molecular weight (reduced and nonreduced) as compared to protein standards and control IgG. Generally, the purified rIgG was greater than 95% pure as determined by visual inspection of the stained bands relative to the bands of known amounts of control IgG run on the same gel.
RNA Segments
[0263] The double stranded RNA (dsRNA) or single stranded RNA (ssRNA) segments of the present invention can be made according to the following method (and are available commercially): 1) ssRNA: The polynucleotides (polyA, polyU) are enzymatically prepared, using nucleotides and polynucleotide-phosphorylase, with no animal-sourced material entering into its preparation process. 2) dsRNA: Annealing of polyadenylic acid (polyA or pA) with polyuridylic acid (polyU or pU).
[0264] In general, the dsRNA and ssRNA of the present invention are homopolymers with, in the case of dsRNA, a single base or nucleotide (e.g., adenine) consistently forming one strand with its complement consistently forming the other strand. In the case of ssRNA, the single strand is consistently made of the same nucleotide. However, it is within the scope of the invention to use dsRNA or ssRNA compositions that are made up of mixed nucleotides (and without or without their complements in the case of dsRNA). For example, a polyA:polyU dsRNA segment with occasional substitution by an a non-complementary nucleotide (e.g., guanine, cytosine or inosine). The dsRNA and ssRNA compositions of the present invention are comprised of the bases/nucleotides adenine (A), guanine (G), cytosine (C), uracil (U) and inosine (I) and could also be comprised of a small percentage of the DNA base thymine (T). The RNA compositions in Table I and FIG. 8A is descriptive of various RNA compositions used in the Examples. The RNA compositions of the present invention were prepared and purified according to Example 30.
[0265] The various RNA strands used in the present invention are generally between 100-2000 base pairs in length but may be between 1-20, 20-40, 40-60, 60-80, 80-100, 1-100, 100-200, 200-300, 300-400, 400-500, 500-600, 600-700, 800-900, 1000-1100, 1100-1200, 1200-1300, 1300-1400, 1400-1500, 1500-1600, 1600-1700, 1700-1800, 1800-1900, 1900-2000, 2000-2100, 2100-2200, 2300-2400, 2400-2500, 2500-3000, 3000-4000, 4000-5000, 5000-10,000 base pairs and greater than 10,000 base pairs in length and/or mixtures thereof.
EXAMPLE 1
[0266] Shows that a significant factor limiting the activity, of peptides that encompass T cell epitopes is the poor pharmacokinetics resulting in reduced in vivo loading of APC.
[0267] Antigen presenting cells ("APCs") from 1 naive BALB/c mouse were obtained from splenic tissue. Following washing, three million APC were incubated with 13.5 nM HA 110-120 peptide for 3 hours at 37° C., in 1 ml of HL-1 medium. The cells were washed, divided into three equal inoculi and injected (1/2 subcutaneously+1/2 intraperitoneally) into 3 naive BALB/c mice. The mice were sacrificed 2 weeks later and the immune response measured against HA 110-120 peptide, by. ELISPOT analysis as follows: the ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/ml for anti-IL2 and anti-IL4, and 8 μg/mg for anti-IFN gamma, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media and blocked with 200μl/well of DMEM complete containing FBS, for an hour at 37° C. Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with 20 μg/ml HA 110-120 peptide or just with media, to assess the background.
[0268] Plates were incubated 72 hours at 37° C., 5% CO2. After 3 days, plates were washed 5 times with PBS--tween 20 0.05% (washing buffer), and incubated with 100 μl/well of biotinylated anti-cytokine Abs, 2 μg/ml in PBS--tween 20 0.05% --FBS 0.1% (ELISPOT buffer) overnight at 4° C. The next day, the plates were washed five times with washing buffer, and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. Plates were then allowed to dry at room temperature for 24 hours. The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.). In parallel, 3 naive BALB/c mice were each injected with 4.5 nM of HA peptide in sterile PBS, half of it administered subcutaneously and half of it intraperitoneally. The mice were sacrificed 2 weeks later and the T cell response characterized as above, by ELISPOT analysis.
[0269] In FIG. 2(A), the experimental protocol is described. In FIG. 2(B), the results of the experiment are shown: they were expressed as number of IFN-γ, IL-2 and IL-4 spot forming colonies/spleen, after the subtraction of the background (mean±SEM). "HA-APC" corresponds to antigen presenting cells (dendritic cells) loaded ex vivo prior to adoptive transfer. "HA" corresponds to peptide directly injected into animals.
[0270] The results described in the FIGS. 2A -2B show that while the injection of the peptide epitope in saline was not immunogenic, a similar dose of peptide used for ex vivo loading of APC effectively triggered a substantial immune response upon adoptive transfer. This shows that if directly injected, the peptide does not effectively reach APC, a prerequisite for effective induction of an immune response.
EXAMPLE 2
[0271] Demonstrates that incorporation of a peptide epitope within the IgG ameliorated its pharmacokinetics profile.
[0272] BALB/c Scid mice (3/group) were injected intravenously with 60 nM of SFERFEIFPKE ("HA") [Seq. I.D. No. 5] peptide or 2.4 nM of recHA (I-Ed)-IgG ("Ig-HA") and blood was harvested at various intervals. Serum was immediately separated and promptly frozen at -70° C. Later, the serum samples were incubated with 2×104 cells/well/50 μl HA-specific T cell hybridoma (TcH) and 1×104 cells/well/50 μl M12 B cell lymphoma APC, in serum free HL-1 medium at 37° C. and 5% CO2 for 24 hours. The next day the plate was centrifuged for 15 min/4° C./1500 RPM, then the supernatant was flicked, the cells were fixed with cold freshly made fixing solution (2% Formaldehyde, 0.2% Glutaraldehyde in 1× PBS) and the plate was again centrifuged for 3 min/4° C./1500 RPM. Fixing solution was flicked off the plate, cells washed once with PBS 200 ul/well, centrifuging the plate for 3min/4° C./1500 RPM. PBS was flicked off the plate and cells were incubated overnight at 37° C. with 200 μl/well of the X-gal substrate freshly prepared as follows: 200 μl of the X-gal stock solution, (40 mg/ml in DMSO) in 10 ml of substrate buffer (5 mM Potassium Ferrocyanide, 5 mM Potassium Ferricyanide, 2 mM MgCl 2 in 1× PBS). The blue activated TcH were scored visually using the microscope.
[0273] The activation of TcH was represented as function of time post-injection. The epitope could be detected in the blood only in the case of mice injected with recHA(I-Ed)-IgG, for an interval of about one day. In contrast, the HA peptide injected as is, was not detected in the periphery despite being used in large molar excess (25 fold).
[0274] Thus, the results described in the FIG. 3 show that delivery of epitope within Ig backbone considerably favored its stability in the systemic circulation.
EXAMPLE 3
[0275] Shows that a peptide encompassing a T cell epitope is ineffectively presented by APC to specific T cells in the presence of serum and this is corrected by incorporation of the peptide epitope within the IgG backbone
[0276] FIG. 4(A) shows the detrimental effect of serum on the presentation of a T cell epitope peptide: M12 B cell lymphoma APC were incubated with TcH in the presence of various amounts of SFERFEIFPKE (HA) peptide in serum-free HL-1 medium ("HA+HL-1") or HL-1 medium supplemented with 20% mouse serum from BALB/c scid mice ("HA+serum"). The number of cells incubated was 2×104 M12 and 1×104 TcH/100 μl of HL-1 medium supplemented or not with serum. The next day the plate was centrifuged for 15 min/4° C./1500 RPM, then the supernatant was flicked, the cells were fixed with cold freshly made fixing solution (2% Formaldehyde, 0.2% Glutaraldehyde in 1× PBS) and the plate was again centrifuged for 3 min/4° C./1500 RPM. Fixing solution was flicked off the plate, cells washed once with PBS 200 μl/well, centrifuging the plate for 3 min/4° C./1500 RPM. PBS was flicked off the plate and cells were incubated overnight at 37° C. with 200 μl/well of the X-gal substrate freshly prepared as follows: 200 ul of the X-gal stock solution, (40 mg/ml in DMSO) in 10 ml of substrate buffer (5 mM Potassium Ferrocyanide, 5 mM Potassium Ferricyanide, 2 mM MgCl 2 in 1× PBS). The blue activated TcH were scored visually using the microscope.
[0277] The serum negatively interfered with the formation and/or presentation of immunogenic MHC-peptide complexes.
[0278] FIG. 4B: the serum negatively interfered with the formation and/or presentation of immunogenic MHC-peptide complexes.
[0279] This phenomenon was further studied by sequential incubation of peptide ("HA peptide") or recHA (I-Ed)-IgG ("IgHA") first with APC or serum, followed by addition after 1 hour of TcH and serum, or APC and TcH, respectively. Control corresponds to cells incubated with antigens in the absence of added serum ("Ctrl"). The number of cells incubated was 2×104 M12 and 1×104 TcH/100 μl of HL-1 medium supplemented or not with serum. The next day the plate was centrifuged for 15 min/4° C./1500 RPM, then the supernatant was flicked, the cells were fixed with cold freshly made fixing solution (2% Formaldehyde, 0.2% Glutaraldehyde in 1× PBS) and the plate was again centrifuged for 3 min/4° C./1500 RPM. Fixing solution was flicked off the plate, cells washed once with PBS 200 μl/well, centrifuging the plate for 3 min/4° C./1500 RPM. PBS was flicked off the plate and cells were incubated overnight at 37° C. with 200 μl/well of the X-gal substrate freshly prepared as follows: 200 μl of the X-gal stock solution, (40 mg/ml in DMSO) in 10 ml substrate buffer (5 mM Potassium Ferrocyanide, 5 mM Potassium Ferricyanide, 2 mM MgCl 2 in 1× PBS). The blue activated TcH were scored visually using the microscope.
[0280] The results were represented as percentage of activated T cells (beta-gal.sup.+ TcH)/well at concentrations of 2 μg/ml of recHA (I-Ed)-IgG ("IgHA") or 40 μg/ml of HA peptide (1,000 molar excess relative to the recombinant Ig).
[0281] The results described in the FIG. 4 show that pre-incubation of peptide with serum resulted in decreased TcH activation. Addition of serum after APC pulsing did not have an effect on TcH activation. In contrast, the formation of MHC-peptide complexes was not impaired by serum when the recombinant immunoglobulin carrying the peptide was used instead of the peptide alone.
EXAMPLE 4
[0282] Shows that incorporation of a T cell peptide epitope within an IgG backbone improves its presentation to specific T cells by APC, with a rate depending on the nature of APC.
[0283] As shown in FIG. 5A, ex vivo formation of MHC-peptide complexes on antigen presenting cells (APCs) from spleen was measured as follows: splenic APC were isolated by magnetic sorting using anti-MHC II antibodies. Separation by using magnetic beads coupled with anti-MHC II was carried out using magnetic cell separators and reagents from Miltenyi Biotec, Germany as follows: spleens were processed to single cell suspension, red blood cells lysed, then cells washed, counted and resuspended in MACS buffer (PBS supplemented with 2 mM EDTA and 0.5% BSA). Magnetically labeled cells were passed through a separation column which is placed in the magnetic field of a MACS separator. The magnetically labeled positive fraction is retained in the column while the negative fraction runs through. After removal of the column from the magnetic field, the magnetically retained positive cells are eluted from the column, cells are washed, counted, resuspended in HL1 complete media and they were incubated with specific T cell hybridoma recognizing I-Ed+SFERFEIFPKE overnight, in the presence of various amounts of SFERFEIFPKE ("HA") peptide or recHA(I-Ed)-IgG ("IgHA"). Per well, 2×104 APC were incubated with 1×104TcH. Next day the plate was centrifuged for 15 min/4° C./1500 RPM, then the supernatant was flicked, the cells were fixed with cold freshly made fixing solution (2% Formaldehyde, 0.2% Glutaraldehyde in 1× PBS) and the plate was again centrifuged for 3 min/4° C./1500 RPM. Fixing solution was flicked off the plate, cells washed once with PBS 200 μl /well, centrifuging the plate for 3 min/4° C./1500 RPM. PBS was flicked off the plate and cells were incubated overnight at 37° C. with 200 μI/well of the X-gal substrate freshly prepared as follows: 200 μl of the X-gal stock solution, (40 mg/ml in DMSO) in 10 ml of substrate buffer (5 mM Potassium Ferrocyanide, 5 mM Potassium Ferricyanide, 2 mM MgCl 2 in 1× PBS). The blue activated TcH were scored visually using the microscope. The number of activated TcH was quantified and the results expressed as activation versus molar amount of epitope. [0284] (B) A protocol similar to that described above has been applied to M12 B cell lymphoma APC.
[0285] Thus, the results described in the FIG. 5B show that the relative efficiency of MHC-peptide complex formation greatly varied depending on the nature of antigen and APC. On a molar basis, the peptide epitope within the IgG backbone was 10 times more effectively handled by MHC II+APC from lymphoid organs and 1000 times more effectively handled by transformed B cell lymphoma cells, as compared to the free peptide itself. Thus, the cellular handling of the epitope and formation of MHC-peptide complexes subsequent to delivery within IgG, greatly varies with the nature of APC.
EXAMPLE 5
[0286] Shows that FcγR-mediated delivery of a peptide encompassing a T cell epitope results in more effective cellular handling and presentation by cell populations (peripheral blood white Cell) containing reduced numbers of professional APC. [0287] (A) To quantify the APC, peripheral blood mononuclear cells (PBMC) were separated by Ficoll gradient centrifugation from BALB/c mice and FACS analysis for expression of CD11 c, CD11b and B220 was carried out. The results are represented in FIG. 6A as percentage of APC and T cells in blood versus a prototype secondary lymphoid organ (spleen). The number of professional APC such as CD11c+ cells is tremendously (2 logs) decreased in blood as compared to spleen. B220+ and CD11b+ cells were decreased as well (1 order of magnitude). The following materials and methods were used.
Materials:
[0287] [0288] Ficoll: Ficoll-hypaque (1.077, Amersham, cat #17-1440-02) [0289] Antibodies: CD11b cat #01715A, CD11c cat #557401, 13220 cat #01125A, all PE conjugated (BD PharMingen) [0290] Flow Cytometer: FACSCalibur, Becton Dickinson [0291] FACS Buffer: PBS, 1% FCS, 0.1% sodium azide.
Methods:
[0291] [0292] 1. Animal blood was harvested and mononuclear cells were separated by Ficoll gradient separation. [0293] 2. Cells were suspended and labeled with fluorescently-tagged anti-mouse CD-11c, CD11b or B220 at 2 ug/ml for 20 minutes on ice [0294] 3. Cells were washed once and resuspended in 300 ul of FACS buffer [0295] 4. Flow cytometric analysis was carried out to determine fractions of total cell population which labeled with each specific antibody [0296] (B) PBMC were used as APC with SFERFEIFPKE (HA)-specific TcH, in the presence of cognate peptide or recHA (I-Ed)-IgG. The cells were co-incubated for 24 hours (2×104 APC+1×104 TcH). The next day the plate was centrifuged for 15 min/4 C/1500 RPM, then the supernatant was flicked, the cells were fixed with cold freshly made fixing solution (2% Formaldehyde, 0.2% Glutaraldehyde in 1× PBS) and the plate was again centrifuged for 3 min/4° C./1500 RPM. Fixing solution was flicked off the plate, cells washed once with PBS 200 μl/well, centrifuging the plate for 3 min/4° C./1500 RPM. PBS was flicked off the plate and cells were incubated overnight at 37° C. with 200 μl/well of the X-gal substrate freshly prepared as follows: 200 μl of the X-gal stock solution, (40 mg/ml in DMSO) in 10 ml of substrate buffer (5 mM Potassium Ferrocyanide, 5 mM Potassium Ferricyanide, 2 mM MgCl 2 in 1× PBS). The blue activated TcH were scored visually using the microscope. The results are expressed as number of activated TcH/well, at different molar concentrations of epitope.
[0297] The results described in the FIGS. 6A-6B show that the peptide epitope within IgG backbone was more effective on a molar basis (1 order of magnitude) than the peptide alone in inducing TcH activation when handled by blood-derived APC, suggesting that in suboptimal conditions associated with limiting numbers of professional APC, the Ig backbone greatly facilitates the creation of MHC-peptide complexes.
EXAMPLE 6
[0298] Shows that delivery of a T cell epitope within IgG backbone dramatically improves the loading and presentation of epitope by APC in the secondary (draining lymph nodes+spleen) but not central lymphoid organs. The emulsification of the peptide epitope in IFA or increase of dose 100 fold could not reproduce the same degree of loading. Thus, epitope insertion within the IgG backbone removes limiting factors associated with peptide-based strategy, that cannot be otherwise compensated by dose escalation or depot effect.
[0299] Assessment of in vivo formation of MHC-peptide complexes and a comparison with peptide in saline or standard oil-in-water emulsion were carried out in I-Ed.sup.+ BALB/c mice. BALB/c mice were treated with recHA (I-Ed)-IgG, peptide in saline or peptide emulsified in incomplete Freund's adjuvant (WA), by subcutaneous and intraperitoneal injection (doses depicted in FIG. 7B). At 24 hours, the local (mesenteric) lymphoid nodes (LN), spleen and thymus were harvested, single cell suspensions were made, red blood cells lysed from the spleens, LN and thymus were collagenase digested. All cells were washed, counted and incubated with TcH recognizing I-Ed+SFERFEIFPKE (MHC class II-HA) complexes. The number of TcH was 1×104/well. The formation of such MHC--peptide complexes was evaluated by titrating the number of APC with constant number of Tell and measuring TcH activation after overnight incubation. The next day the plate was centrifuged for 15 min/4° C./1500 RPM, then the supernatant was flicked, the cells were fixed with cold freshly made fixing solution (2% Formaldehyde, 0.2% Glutaraldehyde in 1× PBS) and the plate was again centrifuged for 3 min/4° C./1500 RPM. Fixing solution was flicked off the plate, cells washed once with PBS 200 μl/well, centrifuging the plate for 3 min/4° C./1500 RPM. PBS was flicked off the plate and cells were incubated overnight at 37° C. with 200 μl/well of the X-gal substrate freshly prepared as follows: 200 μl of the X-gal stock solution, (40 mg/mg in DMSO) in 10 ml of substrate buffer (5 mM Potassium Ferrocyanide, 5 mM Potassium Ferricyanide, 2 mM MgCl 2 in 1× PBS). The blue activated TcH were scored visually using the microscope.
[0300] The data are expressed as TcH activation versus APC number (FIG. 7A) and as estimated percentage of APC expressing MHC-peptide complexes (FIG. 7B), based on in vitro standard curve obtained as depicted in the previous Examples, 5 and 6.
[0301] The data presented in the FIGS. 7A -7B show that the use of oil-in-water adjuvant (IFA) modestly enhanced the in vivo formation of MHC-peptide complexes on APC of lymph nodes but not spleen or thymus. Substantial dose escalation of peptide in saline or in emulsion is not paralleled by proportional enhancement in the generation of loaded APC and/or MHC--peptide complexes on APC in vivo. In contrast, use of peptide within Ig backbone enhances the formation of MHC peptide complexes considerably, on APC from secondary lymphoid organs such as lymph nodes and spleen. The formation of MHC II-peptide complexes on APC from thymus remained limited, similar to that conferred by peptide alone. The enhancement factor conferred by incorporation of peptide within the IgG was unexpectedly high (approximately 2-3 orders of magnitude), indicating that other factors, in addition to cellular handling (e.g. the above described pharmacokinetics and protective effects), were involved. Even 100 fold dose escalation of peptide alone, in saline or IFA, could not restore the in vivo loading of APC noted with peptide within IgG backbone.
EXAMPLE 7
[0302] Shows that among the three major APC subsets (DC, monocytes/macrophages and B cells) that express FcγR, the CD11c+ (DC) and CD11b+ (mostly monocytes) rather than B cells are the most potent on a per cell basis in presenting the peptide epitope subsequent to in vivo delivery via IgG backbone. The efficiency of APC loading and resulting presentation is substantially higher than that resulting from delivery of free peptide.
[0303] In vivo formation of MHC--peptide complexes on APC has been assessed subsequent to the administration of peptide epitope within IgG backbone followed by separation of various subsets of APC. [0304] (A) Separation by using magnetic beads coupled with anti-MHC II or anti-CD11c mAb is carried out using magnetic cell separators and reagents from Miltenyi Biotec, Germany as follows: spleens were processed to single cell suspension, red blood cells lysed, then cells washed, counted and resuspended in MACS buffer (PBS supplemented with 2 mM EDTA and 0.5% BSA). Magnetically labeled cells were passed through a separation column which is placed in the magnetic field of a MACS separator. The magnetically labeled positive fraction is retained in the column while the negative fraction runs through. After removal of the column from the magnetic field, the magnetically retained positive cells are eluted from the column, cells are washed, counted, resuspended in HL1 complete media and incubated in ELISPOT plates. Usually, from the total number of approximately 90 million splenocytes separated/1 BALB/c mouse approximately 20 millions bind to magnetic beads coupled to anti-MHC II antibody and 3 millions interact with anti-CD11c mAb. Thus, less than 20 percent of splenocytes are able to present MHC class II restricted epitopes and approximately 2-3 percent are dendritic cells (see FIG. 8A). These figures were confirmed by FACS analysis using specific antibodies. [0305] (B) The in vivo loading of APC and formation of MHC II- peptide complexes on MHC II+ splenocytes has been assessed comparatively in Balb/c mice injected intravenously with 0.72 uM of recHA (I-Ed)-IgG ("IgHA") or 18 uM of HA peptide. At 24 hours, MHC class II+ APC were isolated from spleen by MACS as above, and incubated with peptide specific TcH (1×104/well), in dose response manner. The next day the plate was centrifuged for 15 min/4° C./1500 RPM, then the supernatant was flicked, the cells were fixed with cold freshly made fixing solution (2% Formaldehyde, 0.2% Glutaraldehyde in 1× PBS) and the plate was again centrifuged for 3 min/4° C./1500 RPM. Fixing solution was flicked off the plate, cells washed once with PBS 200 μl/well, centrifuging the plate for 3 min/4° C./1500 RPM. PBS was flicked off the plate and cells were incubated overnight at 37° C. with 200 μl of the X-gal substrate freshly prepared as follows: 200 μl of the X-gal stock solution, (40 mg/ml in DMSO) in 10 ml of substrate buffer (5 mM Potassium Ferrocyanide, 5 mM Potassium Ferricyanide, 2 mM MgCl 2 in 1× PBS). The blue activated TcH were scored visually using the microscope.
[0306] The results are expressed in FIG. 8B as number of activated TcH/well. As a control, MHC II+ APC from naive BALB/c mice were incubated in vitro, overnight, with an optimal concentration of HA peptide (50 ug/ml), extensively washed and incubated in different numbers with TcH as above. The results show that the formation of MHC II- peptide complexes on splenic APC is at least 2 orders of magnitude more effective when the epitope is delivered within IgG backbone. [0307] (C) A comparative assessment of the in vivo loading of various APC subsets after administration of recHA (I-Ed)-IgG has been carried out by magnetic separation of CD11c+, CD11b+ and CD19+ APC using the same protocol as above, using CD11c, CD11b and CD19 microbeads from Miltenyi Biotec. At 24 hours after intravenous injection with 0.72 uM of recombinant immunoglobulin, the APC were isolated and incubated in a dose effect manner with a constant number of peptide specific Tell After additional 24 hours, the assay was developed as above and results expressed as number of activated TcH/well. The results in FIG. 8C show that on a per cell basis, use of peptide within IgG backbone led to predominant formation of immunogenic MHC II- peptide complexes on CD11c+ APC (dendritic cells), followed by CD11b+ monocytes and very ineffectively on CD 19+ B cells. [0308] (D) A comparison between the efficiency of in vivo formation of MHC II- peptide complexes on CD11c+ APC subsequent to peptide versus recombinant Ig delivery has been carried out following treatment of mice as described in the section B above. The CD11c+ splenic DC were isolated by MACS using CD 11c microbeads and incubated in different numbers with 1×104TcH/well. Activated TcH were quantified as above and the results expressed as number of X-gal+T cells/well. As a control, CD11c+ APC from naive mice loaded ex vivo with peptide were used as described in section B. The results in FIG. 8D show that formation of MHC II peptide complexes was at least three orders of magnitude more effective when the peptide epitope was delivered within IgG backbone.
[0309] In conclusion, delivery of a peptide epitope within an IgG backbone resulted in more effective formation of MHC II- peptide complexes on CD11c+ DC. In addition, the efficiency of APC loading and formation of MHC II- peptide complexes was substantially higher when the peptide was delivered within IgG backbone. The results in FIGS. 8A-8D show that use of FcgR mediated delivery of peptides results in preferential formation of immunogenic MHC II- peptide complexes on CD11c+ and CD11b+ APC.
EXAMPLE 8
[0310] Shows a prolonged persistence in vivo of MHC-peptide complexes on APC (DC and monocytes) following administration via an IgG backbone.
[0311] The persistence of MHC II- peptide complexes on specific APC subsets was measured by magnetic separation of CD11c+ DC and CD11b+ monocytes at various intervals subsequent to intravenous injection of 2 uM of recHA (I-Ed)-IgG. In brief, magnetic separation was carried out using magnetic cell separators and reagents from Miltenyi Biotec, Germany as follows: spleens were processed to single cell suspension, red blood cells lysed, then cells washed, counted and resuspended in MACS buffer (PBS supplemented with 2 mM EDTA and 0.5% BSA). Magnetically labeled cells were passed through a separation column which is placed in the magnetic field of a MACS separator. The magnetically labeled positive fraction is retained in the column while the negative fraction runs through. After removal of the column from the magnetic field, the magnetically retained positive cells are eluted from the column, cells are washed, counted, resuspended in HL1 complete media and incubated. Different numbers of separated APC (A--CD11b+ monocytes, B--CD11c+ dendritic cells, C--whole splenocyte population) were incubated overnight with 1×104 TcH specific for the HA peptide.
[0312] As a control, APC from naive mice were used that were in vitro loaded with optimal amounts of HA peptide (50 μg/ml), overnight and washed prior to incubation ("ctrl"). The next day the plate was centrifuged for 15 min/4° C./1500 RPM, then the supernatant was flicked, the cells were fixed with cold freshly made fixing solution (2% Formaldehyde, 0.2% Glutaraldehyde in 1× PBS) and the plate was again centrifuged for 3 min/4° C./1500 RPM. Fixing solution was flicked off the plate, cells washed once with PBS 200 μl/well, centrifuging the plate for 3 min/4° C./1500 RPM. PBS was flicked off the plate and cells were incubated overnight at 37° C. with 200 μl/well of the X-gal substrate freshly prepared as follows: 200 μl of the X-gal stock solution, (40 mg/ml in DMSO) in 10 ml of substrate buffer (5 mM Potassium Ferrocyanide, 5 mM Potassium Ferricyanide, 2 mM MgCl 2 in 1× PBS). The blue activated TcH were scored visually using the microscope and the number of activated TcH/well was plotted against the number of APC harvested at various intervals after treatment.
[0313] The results show long lasting expression of peptide onto endogenous MHC II, on both DC and monocytes. The complexes persisted between 1 and 2 weeks on these two APC subsets, in the conditions employed in this assay (strategy of APC separation and detection of MHC II- peptides).
[0314] Thus, the results in FIGS. 9A-9C show that the MHC-peptide complexes on selected APC formed subsequent to in vivo delivery of epitope via Ig are long-lived.
EXAMPLE 9
[0315] Shows that the γ chain of the Fc receptors (I and III) is essential for effective in vivo loading and presentation of a T cell epitope delivered within IgG backbone, by DC and monocytes.
[0316] The dependency of APC loading on the interaction with FcγR was studied by administration of 2 uM of recHA(I-Ed)-IgG to BALB/c, mice that lack a functional FcR gamma gene. One day after intravenous treatment, the CD 11 c+ and CD 11b+ APC from spleen were separated by MACS. Separation by using magnetic beads coupled with anti-CD11c and anti-CD11b antibodies was carried out using magnetic cell separators and reagents from Miltenyi Biotec, Germany, as follows: spleens were processed to single cell suspension, red blood cells lysed, then cells washed, counted and resuspended in MACS buffer (PBS supplemented with 2 mM EDTA and 0.5% BSA). Magnetically labeled cells were passed through a separation column which is placed in the magnetic field of a MACS separator. The magnetically labeled positive fraction is retained in the column while the negative fraction runs through. After removal of the column from the magnetic field, the magnetically retained positive cells are eluted from the column, cells are washed, counted, resuspended in HL1 complete media and they were incubated in different numbers with 1×104TcH specific for the HA peptide, overnight. As a control, APC from FcR gamma competent BALB/c mice were used. The next day the plate was centrifuged for 15 min/4° C./1500 RPM, then the supernatant was flicked, the cells were fixed with cold freshly made fixing solution (2% Formaldehyde, 0.2% Glutaraldehyde in 1× PBS) and the plate was again centrifuged for 3 min/4° C./1500 RPM. Fixing solution was flicked off the plate, cells washed once with PBS 200 μl/well, centrifuging the plate for 3 min/4° C./1500 RPM. PBS was flicked off the plate and cells Were incubated overnight at 37° C. with 200 μl/well of the X-gal substrate freshly prepared as follows: 200 μl of the X-gal stock solution, (40 mg/mg in DMSO) in 10 ml of substrate buffer (5 mM Potassium Ferrocyanide, 5 mM Potassium Ferricyanide, 2 mM MgCl 2 in 1× PBS). The blue activated TcH were scored visually using the microscope. The results are expressed as number of activated TcH/well for different APC subsets: CD11c+ DC (A) and CD11b+ monocytes (B), or as control, whole splenic population (C).
[0317] The results (FIG. 10) clearly show that the formation of MHC II- peptide complexes on DC and monocytes, subsequent to IgG mediated delivery of peptide epitope, is critically dependent on ITAM+ FcgR that encompass the gamma chain. In addition, gamma chain negative FcR isoforms cannot compensate for the absence of gamma chain+ FcR isoforms, in that regard.
EXAMPLE 10
[0318] Shows that the efficiency of T cell activation by a peptide delivered within the IgG backbone is dependent on the expression of γ chain+ FcγR (that promote activity) and FcγRIIB (that limit the activity) on APC. In addition, this experiment shows that ITIM-bearing FcγRIIB keeps in check the immune response to a peptide delivered within IgG backbone.
[0319] The differential role of FcR gamma+ versus gamma- isoforms to the immune response triggered by peptide epitope within IgG backbone, was studied by ex vivo loading of APC followed by adoptive transfer. Splenocytes from wild type, FcR gamma- or FcRIIB- BALB/c mice were incubated for 3 hours at 370° C. as follows: 10 million cells/1 ml of serum free HL-1 medium were admixed with 50 ug/ml of HA 110-120 peptide or 10 ug/ml of recHA(I-Ed)-IgG. Subsequently, the cells were washed and adoptively transferred into naive BALB/c mice (1 million cells suspended in 200u1 serum free HL-1 and divided into 2 equal inoculi administered subcutaneously and intraperitoneally). After 2 weeks, the recipient mice were sacrificed, spleens harvested and the T cell response to the HA 110-120 peptide measured by ELISPOT analysis as follows: the ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/mg for anti-IL2 and anti-IL4, and 8 μg/mg for anti-IFN gamma, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media and blocked with 200 μl/well of DMEM complete containing FBS, for an hour at 37° C. Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with 50 μg/ml HA 110-120 peptide or just with media, to assess the background.
[0320] Plates were incubated 72 hours at 3-7° C., 5% CO2. After 3 days, the plates were washed 5 times with PBS--tween20 0.05% (washing buffer), and incubated with 100 μl/well of biotinylated anti-cytokine Abs, 2 μg/ml in PBS--tween20 0.05%--FBS 0.1% (ELISPOT buffer) overnight at 4° C.
[0321] The next day plates were washed five times with washing buffer, and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. Plates were then allowed to dry at room temperature for 24 hours. The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.). The results are expressed in FIG. 11 as frequency of cytokine producing (A: IL-2, B: IL-4, and C: IFN-gamma) spot forming colonies obtained by incubation with medium only, or medium supplemented with HA 110-120 peptide (10 ug/ml) (mean+SEM of triplicates, corresponding to 3 mice/group).
[0322] The results (FIG. 11) show that the expression of the gamma chain of ITAM+ FcgR isoforms is necessary for the induction of T cell response to APC loaded with peptide within IgG backbone. This was not necessary for the immunogenic effect of APC pulsed with peptide. Conversely, absence of ITIM+ FcgRII results in profound increase of the T cell response to APC pulsed with recombinant IgG but not HA peptide. Together, these data show that the T cell response to recombinant IgG bearing a peptide epitope is determined by a complex interplay between ITAM+ and ITIM+ Fcgamma receptors on APC.
EXAMPLE 11
[0323] Shows that unexpectedly, various subsets of APC in vivo loaded with epitope inserted within IgG backbone, differentially induce distinct regulatory subsets: while monocytes induce Th2 and Tr1 cells more effectively, both dendritic cells and monocytes induce Th3 cells. In addition, on a cell population level, the CD11b+ monocytes are more potent than the dendritic cells in triggering a regulatory response following IgG-mediated delivery of T cell epitope.
[0324] Four BALB/c mice were injected intravenously with 2 μM of recHA (I-Ed)-IgG. One day later, the spleens were harvested and APC were isolated by MACS using anti-CD11 c, anti-CD11b or anti-CD19 monoclonal antibodies coupled with magnetic beads. Separation by using magnetic beads coupled with anti-CD11b, anti-CD11c and anti-CD19 mAb is carried out using magnetic cell separators and reagents from Miltenyi Biotec, Germany as follows: spleens were processed to single cell suspension, red blood cells lysed, then cells washed, counted and resuspended in MACS buffer (PBS supplemented with 2 mM EDTA and 0.5% BSA). Magnetically labeled cells were passed through a separation column which is placed in the magnetic field of a MACS separator. The magnetically labeled positive fraction is retained in the column while the negative fraction runs through. After removal of the column from the magnetic field, the magnetically retained positive cells are eluted from the column, cells are washed, counted, resuspended in serum free HL-1 medium as follows: 3×106/mg CD11c.sup.+ DC, 28×106/ml CD11b.sup.+ or 84×106/ml of CD19.sup.+ B cells. This numerical distribution respects the proportion of the APC subsets isolated from the splenic tissue. Cells were transferred into naive BALB/c mice by subcutaneous and intraperitoneal injection (100+100 μl/mouse, n=2 mice/group). At 2 weeks after the adoptive transfer, mice were sacrificed and T cell response measured by ELISPOT (IL-4 and IFN-γ) or measurement of cytokine production in cell culture supernatants, by ELISA TGF-β1 kit (R&D Systems, cat #DY240) and IL-10 kit (Biosource international, cat #KMC0104).
[0325] The results are expressed in FIG. 12 as number of spot forming colonies/spleen (average of duplicates; panels A, B) or amount of cytokine measured in supernatants (pg/ml, average of duplicates; panels C, D) at various concentrations of HA peptide used for restimulation.
[0326] The results (FIG. 12, panels A-D) clearly show that unexpectedly, and in contrast with the potency/cell basis (Example 8), at the organism level, the CD 11b.sup.+ monocytes `have the highest impact on the immune response to a peptide epitope delivered within the IgG backbone. Thus, the CD11b.sup.+ APC subset induced both Th2, Tr1 and Th3 cells. In contrast, the CD11c.sup.+ DC induced Th3 cells and more reduced Th2 response. Finally, despite their substantial number, the CD19.sup.+ B cells were poor inducers of T cell immunity to the peptide epitope within the IgG backbone. No significant Th1 responses were induced by either of the APC subsets tested.
EXAMPLE 12
[0327] Shows that the loading of APC in vivo with a peptide delivered within IgG backbone results in induction of Th2 but not Th1 immunity.
[0328] BALB/c mice were immunized with 100 μg of recHA. (I-Ed)-IgG ("IgHA"), or a, molar equivalent amount of HA peptide epitope (2 μg), by subcutaneous injection and sacrificed 2 weeks later. The immune response was measured by ELISPOT analysis using splenocytes from treated mice as responders, and mitomycin-treated splenocytes from naive mice as stimulators, as follows: the ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/ml for anti-IL2 and anti-IL4, and 8 μg/ml for anti-IFN gamma, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media and blocked with 200 μl/well of DMEM complete containing FBS, for an hour at 37° C. Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with 20 μg/ml HA 110-120 peptide or just with media, to assess the background.
[0329] Stimulator cells were prepared from naive mice as follows: single cell suspension was prepared from spleens, red blood cells were lysed, cells were washed, resuspended in HL1 complete and mitomycin treated for 30 minutes. Afterwards, cells were washed 3 times, counted and resuspended in serum free HL1 media. The plates were incubated 72 hours at 37° C., 5% CO2. After 3 days, the plates were washed 5 times with PBS--tween20 0.05% (washing buffer), and incubated with 100 μl/well of biotinylated. anti-cytokine Abs, 2 μg/ml in PBS--tween20 0.05% --FBS 0.1%(ELISPOT buffer) overnight at 4° C.
[0330] The next day, the plates were washed five times with washing buffer and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. The plates were then allowed to dry at room temperature for 24 hours. The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.).
[0331] The results are expressed in FIG. 13 as number of IL-4-producing (A) or IFN-γ producing (B) T cell colonies/spleen (mean±SEM of triplicates) when splenocytes were restimulated with 10 μg/ml of HA peptide or cell culture medium alone. Thus, this Example shows that FcgR-mediated delivery of T cell epitope within recombinant Ig backbone results in Th2 rather than Th1 response.
EXAMPLE 13
[0332] Shows that the repeated loading of APC in vivo with a peptide delivered within IgG backbone results in induction of Th3 and Tr1 immunity.
[0333] BALB/c mice were immunized with 40 ug of heat aggregated (15 ruins at 63° C.) of recHA (I-Ed)-IgG ("IgHA") administered by intranasal instillation boosted 2 weeks later by subcutaneous injection with 100 ug of recombinant immunoglobulin in saline. As controls, mice primed with heat aggregated IgG2b isotype control were used. After an additional 2 weeks, the mice were sacrificed and T cell response assessed by in vitro restimulation of splenocytes with HA peptide by ELISPOT analysis as follows: the ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/ml for anti-IL2 and anti-IL4, and 8 μg/ml for anti-IFN gamma, from ED Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media and blocked with 200 μl/well of DMEM complete containing FBS, for an hour at 37° C.
[0334] Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with 20 μg/ml HA 110-120 peptide or just with media, to assess the background. Plates were incubated 72 hours at 37° C., 5% CO2. After 3 days, plates were washed 5 times with PBS--tween20 0.05% (washing buffer), and incubated with 100 μl/well of biotinylated anti-cytokine Abs, 2 μg/ml in PBS--tween20 0.05%--FBS 0.1%(ELISPOT buffer) overnight at 4° C.
[0335] The next day, plates were washed five times with washing buffer and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. Plates were then allowed to dry at room temperature for 24 hours.
[0336] The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.). The TGF-beta and IL-10 production were measured by ELISA TGF-β1 kit (R&D Systems, cat #DY240) and IL-10 kit (Biosource international, cat #KMC0104). The results are expressed as cytokine concentration (average of triplicates) after subtraction of background.
[0337] The data, as shown in FIG. 14, show that mucosal priming with epitope bearing recombinant immunoglobulin resulted in differentiation of Th3 and Tr1 cells that were expanded subsequently by systemic boosting.
EXAMPLE 14
[0338] Shows that only a virus, but not the conventional adjuvant CFA, was able to trigger significant Th1 response to a peptide epitope inserted within the IgG backbone.
[0339] BALB/c mice were immunized intraperitoneally with 100 ug of recHA (I-Ed)-IgG in saline, emulsified in Complete Freund's Adjuvant ("CFA") or with 105 TCID50 of influenza virus strain WSN, that bears the HA epitope. At 2 weeks after immunization, the mice (n=3/group) were sacrificed and the T cell response to HA peptide measured by ELISPOT analysis as follows: the ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/ml for anti-IL2 and anti-IL4, and 8 μg/ml for anti-IFN gamma, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media,and blocked with 200 μl/well of DMEM complete containing FBS, for an hour at 37° C.
[0340] Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with 20 μg/ml HA 110-120 peptide or just with media, to assess the background.
[0341] Plates were incubated 72 hours at 37° C., 5% CO2. After 3 days, the plates were washed 5 times with PBS--tween20 0.05% (washing buffer), and incubated with 100 μl/well of biotinylated anti-cytokine Abs, 2 μg/ml in PBS--tween20 0.05%--FBS 0.1% (ELISPOT buffer) overnight at 4° C. The next day; plates were washed five times with washing buffer, and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. The plates were then allowed to dry at room temperature for 24 hours.
[0342] The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.). The results are represented as mean±SEM of frequency of cytokine producing colonies in the spleen.
[0343] The results in FIG. 15 show that a peptide epitope within the IgG backbone triggers a cellular response of Th2 profile that is enhanced but not switched by a conventional adjuvant (CFA). In contrast, the profile afforded by live virus immunization was Th1 biased.
EXAMPLE 15
[0344] Shows that the presentation of peptide epitope subsequent to IgG mediated delivery results in a T cell response that could be further manipulated by increasing co-stimulation with anti-CD40mAb, recombinant IL-12 or synthetic dsRNA.
[0345] Dendritic cells from naive BALB/c mice were harvested by MACS from splenic cell suspensions as follows: Separation by using magnetic beads coupled with anti-CD11c was carried out using magnetic cell separators and reagents from Miltenyi Biotec, Germany as follows: spleens were processed to single cell suspension, red blood cells lysed, the cells washed, counted and resuspended in MACS buffer (PBS supplemented with 2 mM EDTA and 0.5% BSA). Magnetically labeled cells were passed through a separation column which is placed in the magnetic field of a MACS separator. The magnetically labeled positive fraction is retained in the column while the negative fraction runs through. After removal of the column from the magnetic field, the magnetically retained positive cells are eluted from the column, cells are washed, counted, resuspended in HL1 complete media and were pulsed ex vivo in serum free HL-1 medium for 2 hours, at a concentration of 3 million/ml, with 50 ug/ml of recHA(I-Ed)-IgG alone or supplemented with 5 ng/ml of recIL-12, 50 ug/ml of double stranded RNAs (pA:pU or pI:pC). Alternatively, the cells were incubated with recombinant Ig and wells precoated with 10 ug/ml of anti-CD40 mAb. The cells were harvested, washed and adoptively transferred to naive BALB/c mice (300,000 delivered half subcutaneously and half intraperitoneally) in serum free HL-1 medium.
[0346] At 2 weeks, the mice were sacrificed and T cell responses measured against HA by ELISPOT analysis as follows: the ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/mg for anti-IL2 and anti-IL4, and 8 μg/ml for anti-IFN gamma, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media and blocked with 200 μl/well of DMEM complete containing FBS, for an hour at 37° C.
[0347] Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with 50 μg/ml HA 110-120 peptide or just with media, to assess the background. Plates were incubated 72 hours at 37° C., 5% CO2. After 3 days, plates were washed 5 times with PBS--tween20 0.05% (washing buffer) and incubated with 100 μl/well of biotinylated anti-cytokine Abs, 2 μg/ml in PBS--tween20 0.05%--FBS 0.1%(ELISPOT buffer) overnight at 4° C. The next day plates were washed five times with washing buffer and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. The plates were then allowed to dry at room temperature for 24 hours.
[0348] The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.). The results are shown as mean+SEM (n=3) of the frequency of spot forming colonies associated with IL-2 or IL-4 production, after subtraction of the background, for each ex vivo stimulatory combination.
[0349] The results in FIG. 16 show that peptide presentation by APC, subsequent to loading with antigen by using recombinant IgG as delivery platform, occurs in context of limited co-stimulation. IL-12, anti-CD40 or synthetic dsRNA can all enable APC loaded with antigen via FcgR, to prime IL-2 and enhanced IL-4 producing T cell immunity against the cognate (HA) peptide.
EXAMPLE 16
[0350] The activity of the long-lived IL-4 producing Th2 cells triggered by in vivo loading of APC with IgG-peptide is dependent on the continuous interaction with endogenous APC and requires competent CD4.
[0351] BALB/c mice were immunized with 100 ug of recHA (I-Ed)-IgG or HA peptide subcutaneously, sacrificed at 2 weeks and the T cell response measured by ELISPOT analysis as follows: the ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/ml anti-IL4, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plate was washed 2 times with DMEM media and blocked with 200 μl/well of DMEM complete containing FBS, for an hour at 37° C.
[0352] Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with 20 μg/ml HA 110-120 peptide or just with media, to assess the background. The plate was incubated 72 hours at 37° C., 5% CO2. After 3 days, the plate was washed 5 times with PBS--tween20 0.05% (washing buffer) and incubated with 100 μl/well of biotinylated anti-cytokine Abs, 2 μg/mg in PBS--tween20 0.05%--FBS 0.1%(ELISPOT buffer) overnight at 4° C.
[0353] The next day, the plate was washed five times with washing buffer and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. The plate was then allowed to dry at room temperature for 24 hours. The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.). [0354] (A) During the HA stimulation phase, blocking anti-CD4 or anti-CD8 mAb was added at 10 ug/mg in selected wells. The results are expressed in FIG. 17A as mean+SEM of number of HA-stimulated IL-4 producing colonies per spleen, after subtraction of background (n=3 mice/group). [0355] (B) Splenocytes from mice immunized with recombinant Ig as above, were incubated in elispot plate as is or after magnetic depletion of endogenous MHC II+ APC with MHC II+ from naive BALB/c mice, with medium alone or in the presence of 10 ug/ml of HA peptide. Separation by using magnetic beads coupled with anti-MHC II was carried out using magnetic cell separators and reagents from Miltenyi Biotec, Germany as follows: spleens were processed to single cell suspension, red blood cells lysed, then cells washed, counted and resuspended in MACS buffer (PBS supplemented with 2 mM EDTA and 0.5% BSA). Magnetically labeled cells were passed through a separation column which is placed in the magnetic field of a MACS separator. The magnetically labeled positive fraction is retained in the column while the negative fraction runs through. After removal of the column from the magnetic field, the magnetically retained positive cells are eluted from the column, cells are washed, counted, resuspended in HL I complete media and were incubated in the ELISPOT assay, protocol to follow. The ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/ml for anti-IL2 and anti-IL4, and 8 μg/ml for anti-IFN gamma, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media and blocked with 200 μl/well of DMEM complete containing FBS, for an hour at 37° C. Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with 50 μg/ml HA 110-120 peptide or just with media, to assess the background.
[0356] Plates were incubated 72 hours at 37° C., 5% CO2. After 3 days, the plates were washed 5 times with PBS--tween 20 0.05% (washing buffer) and incubated with 100 μl/well well of biotinylated anti-cytokine Abs, 2 μg/ml in PBS--tween 20 0.05%--FBS 0.1% (ELISPOT buffer) overnight at 4° C.
[0357] The next day, plates were washed five times with washing buffer, and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. Plates were then allowed to dry at room temperature for 24 hours.
[0358] The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.) and the results expressed as mean±SEM of the frequency of IL-4 producing T cells. The results in FIGS. 17A-17B show that the activity of HA specific IL-4 producing T cells triggered by administration of recHA(I-Ed)-IgG is dependent on CD4 rather CD8. In addition, the long lived IL-4 production by primed T cells depends on stable interaction with endogenous APC.
EXAMPLE 17
[0359] Shows that FcγR-mediated delivery of a T cell epitope is more effective than the peptide in differentially affecting the phenotype of activated, specific T cells: dose-dependent down regulation of IL-2, IFN-γ, and IL-4, with up-regulation of IL-10 and TGF-β.
[0360] Activated SFERFEIFPKE-specific T cells were separated from BALB/c mice immunized 2 weeks previously with 100 μg peptide in CFA. They were incubated with mitomycin treated splenocytes in the presence of various amounts of recHA(I-Ed)-IgG or corresponding peptide. The expansion and cytokine production (IFN-γ, IL-4, IL-2) was estimated by ELISPOT analysis as follows: the ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/ml for anti-IL2 and anti-IL4, and 8 μg/ml for anti-IFN gamma, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media and blocked with 200 μl/well of DMEM complete containing FBS, for an hour, at 37° C. Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with 20 μs/ml HA 110-120 peptide or just with media, to assess the background.
[0361] The plates were incubated 72 hours at 37° C., 5% CO2. After 3 days, the plates were washed 5 times with PBS--tween 20 0.05% (washing buffer) and incubated with 100 μl/well of biotinylated anti-cytokine Abs, 2 μg/ml in PBS--tween 20 0.05%--FBS 0.1% (ELISPOT buffer) overnight at 4° C. The next day, the plates were washed five times with washing buffer, and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. The plates were then allowed to dry at room temperature for 24 hours.
[0362] The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.). In addition, TGF-β and IL-10 production were measured by ELISA at 48 hours after incubation using TGF-β1 kit (R&D Systems, cat #DY240) and IL-10 kit (Biosource international, cat #KMC0104).The results are expressed as frequency of spot forming cells (SFC) or concentration of cytokine versus amount of antigen added in vitro.
[0363] The results in FIG. 18 show that the IgG mediated delivery of a T cell epitope has a profound and differential effect on the expansion and cytokine production by activated T cells: IL-2, IFN-γ and surprisingly EL-4, were down-regulated in a dose-related manner. The Ig-peptide was substantially more effective in modulating the cytokine production, as compared to the peptide itself. In contrast, only the Ig-peptide turned on effectively the production of IL-10 and TGF-beta in a dose-dependent manner. Thus, the T cell epitope in context of Ig backbone, but not separately, differentially modulated the function of activated cells.
EXAMPLE 18
[0364] Shows that surprisingly, a peptide delivered within the IgG backbone, that is not an immune complex nor is a receptor cross-linking antibody, results in induction of a class I restricted immune response. This response had a different profile from that triggered by live virus (Tc2 type consisting in IL-4 but not IFN-γ production).
[0365] BALB/c mice were injected with 50 μg of recNP(Kd)-IgG encompassing the MHC class I-restricted peptide TYTQTRALV (Seq. I.D. No. 6) by subcutaneous injection. The mice were sacrificed 2 weeks later and peptide-specific cytokine production was measured by ELISPOT analysis as follows: the ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/ml for anti-IL2 and anti-IL4, and 8 μg/ml for anti-IFN gamma, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media and blocked with 200 μl/well of DMEM complete containing PBS, for an hour at 37° C.
[0366] Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with various concentrations of NP peptide. Plates were incubated 72 hours at 37° C., 5% CO2. After 3 days, the plates were washed 5 times with PBS--tween 20 0.05% (washing buffer) and incubated with 100 μl/well of biotinylated anti-cytokine Abs, 2 μg/ml in PBS--tween 20 0.05%--FBS 0.1% (ELISPOT buffer) overnight at 4° C. The next day the plates were washed five times with washing buffer and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. The plates were then allowed to dry at room temperature for 24 hours.
[0367] The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.). The results are expressed in FIG. 19A as total number of spot forming colonies (SFC)/spleen (mean of n=3). As controls, naive mice or mice injected intraperitoneally with 105 TCID50 of live WSN influenza virus were used.
[0368] The results in FIG. 19A-19B show that in contrast to viral immunization with an influenza virus strain bearing the cognate peptide, Ig-mediated peptide delivery was ineffective in triggering IFN-γ producing Tc1 cells. However, Ig-peptide administration still resulted in formation of MHC class I-peptide complexes and induced significant NP-specific MHC class I-restricted T cell immunity consisting in IL-4 producing Tc2 cells.
EXAMPLE 19
[0369] Shows that in vivo loading of selected APC with disease associated epitopes suppressed an aggravated form of autoimmunity by expanding rather than ablating, epitope-specific autoreactive T.
[0370] SJL mice were injected subcutaneously with 200 μl of rat brain homogenate emulsified in Complete Freund's Adjuvant and boosted with 50 ng of pertussis toxin at 6 hours and 2 days. The mice developed an aggravated, progressive form of paralytic disease. Half of the mice received via subcutaneous injection a combination of recombinant immunoglobulins bearing the MBP and the PLP epitopes (recMBP(I-As)-IgG; recPLP(I-As)-IgG), respectively (150 μg/molecule, on day 8, 12, 18 after induction of disease). In panel A, the mean clinical score for treated and non-treated mice is represented, respectively (n=8).
[0371] After a period of observation of 70 days, the mice were sacrificed, spleens harvested and elispot analysis carried out as follows: the ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/ml for and anti-IL4, and 8 μg/mg for anti-IFN gamma, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media and blocked with 200 μl/well of DMEM complete containing FBS, for an hour at 37° C. Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 1×106/well together with 20 μg/ml of peptides (PLP or MBP) or just with media, to assess the background.
[0372] Plates were incubated 72 hours at 37° C., 5% CO2. After 3 days, the plates were Washed 5 times with PBS--tween 20 0.05% (washing buffer) and incubated with 100 μl/well of biotinylated anti-cytokine Abs, 2 μg/ml in PBS--tween 20 0.05% FBS 0.1% (ELISPOT buffer) overnight at 4° C. The next day, the plates were washed five times with washing buffer, and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. Plates were then allowed to dry at room temperature for 24 hours.
[0373] The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.). The results (FIG. 20B) were expressed as frequency of IL-4 producing T cell colonies in the absence of added PLP peptide plotted against the frequency of IFN-γ-producing T cells in condition of peptide stimulation. Mice progressing to full-blown limb paralysis (score equal to or higher than 1.5) were represented with closed symbols. Mice that did not progress to limb paralysis were represented with open symbols. In FIG. 20C, the total number of IL-4 spot forming colonies/spleen (mean±SEM) in condition of in vitro stimulation was represented with nil, MBP or PLP peptide. An additional control; consisting of splenocytes from mice treated with IgG2b isotype control, has been included. In parallel, in vitro culture was carried out in the presence of neutralizing anti-IL-4 mAb (40 μg/ml) and the number of IFN-γ-producing T cells was represented in the panel D.
[0374] The results in FIGS. 20A-D show that co-administration of MBP and PLP epitopes by using recombinant IgG significantly curbed the chronic progression of disease. The mice protected from paralysis developed unexpectedly, an enhanced reactivity to self-epitopes MBP and PLP, manifested by increased basal and peptide-stimulated IL-4 or IFN-γ production, respectively. Finally, the reactivity of IFN-γ-producing T cells is kept in check by IL-4 suggesting a complex immunomodulatory mechanism triggered by IgG-mediated delivery of epitopes.
EXAMPLE 20
[0375] Summarizes the impact of IgG/FcγR-mediated delivery of epitopes on the T cell response, based on data provided in the Examples 1-19.
[0376] First, the loading of APC T cell response to IgG-mediated delivery of T cell epitopes is controlled by two functionally opposing receptors: ITIM and ITAM Fc (gamma.sup.+)-bearing receptors on APC. ITIM.sup.+ FcγRIIB limits the degree of activation of T cells and gamma.sup.+ FcRs are required for effective formation of MHC-peptide complexes when epitopes are delivered via the IgG backbone. Such in vivo delivery of epitope results in effective formation of MHC--peptide complexes on peripheral CD11c.sup.+ and CD11b.sup.+ APC, but not thymic APC. However, the interplay between ITIM.sup.+ and ITAM.sup.+ FcγRs makes the nature and magnitude of resulting T cell response difficult to predict without experimentation.
[0377] The data in FIG. 21 show that IgG-delivery of peptide epitope results in exposure of T cells to peptide-loaded APC in context of limited co-stimulation, having a differential effect on naive versus activated T cells: 1) de novo induction of Th2, Tc2, Th3, Tr1 cells and, 2) downregulation of activated Th1; Th2 cells with stimulation of activated Tr1 and Th3 cells. The overall effect is immunomodulatory, rather than pro-inflammatory (associated with Th1 and Tc1 immunity).
EXAMPLE 21
[0378] Naturally occurring dsRNA bridges the innate with adaptive immune response. Example 21 shows that natural, non-infectious double stranded RNA produced during infection with influenza virus, has substantial effects on the specific immune response to a protein antigen.
[0379] Permissive MDCK cells were infected with WSN influenza virus (108' TCID50/1×109 cells) and after 24 hours, the cells were harvested, washed and the total RNA extracted using an RNA separation kit (Qiagen, Valencia, Calif.). The RNA was further purified by treatment with RNAse-free DNAseI (Stratagene, San Diego, Calif.). The single stranded RNA in the samples was then removed by 30 minutes incubation at 37° C. with 5U of S1 nuclease (Ambion, Inc., Austin-Tex.)/μg of RNA. The RNA was analyzed prior to and subsequent to the digestion by gel electrophoresis. The absence of infectious properties of the purified dsRNA was confirmed by standard influenza virus titration. As a control, material purified and treated similarly, from 109 non-infected MDCK cells was used. The concentration of nucleic acid was measured by spectrophotometry (A260 nm) and the absence of endotoxin confirmed by Limulus assay. The purified dsRNA and control RNA were used individually, or as a mixture with gp140 recombinant antigen (25 μg of RNA and 2 μg of antigen in 25 ml of sterile PBS).
[0380] After demonstrating lack of infectivity, 40 μg of dsRNA or control RNA were admixed with 40 μg of recombinant truncated antigen (gp140 of HIV envelope) and were administered to BALB/c mice by intranasal instillation (n=3/group). Additional controls were animals immunized with 40 μg of gp140 protein in saline (n=3/group). The mice were boosted once, at 2 weeks after priming. Blood was harvested 2 weeks after the boost, sera prepared and the antibody response against gp140 measured by ELISA. In brief, wells,were coated with antigen (2 μg/ml of gp140) and blocked with SeaBlock (Pierce, Rockford-Ill., catalog #37527). Serial dilutions of serum and bronchoalveolar lavage fluid were incubated for at least 2 hours at room temperature. After washing, the assay was developed with anti-mouse IgG antibody coupled with alkaline phosphatase (Sigma, cat #A7434) followed by addition of substrate (pNPP, Sigma, cat #N2765) and measurement by using an automatic microtiter plate reader (Molecular Devices, ThermoMax) equipped with SoftMax software.
[0381] In FIG. 22A, the general principle of the experiment is illustrated. In FIG. 22B, the absorption after assay development is represented, corresponding to various serum dilutions, in case of whole IgG. In FIG. 22B, the absorption at 1/50 serum dilution, in case of IgG2a and IgG1 antibody isotypes, is represented.
[0382] Overall, the data in FIGS. 22A-B show that natural, non-infectious dsRNA from influenza virus-infected MDCK cells, has an unexpected enhancing effect on the adaptive response to a prototype antigen. Both IgG1 and IgG2a antibody responses were increased showing that a strong T helper1 and T helper 2 response was induced.
EXAMPLE 22
[0383] Effects of selected RNA motifs on the innate immune response: heterogeneous motifs. This Example shows, unexpectedly, that different synthetic RNA motifs have a distinct effect on the adaptive specific immune response to a protein antigen.
[0384] FIG. 23A shows an extensive library of synthetic RNA motifs, that was grouped in pools and used for a two-tier screening process as follows: [0385] (A) The mice were immunized intratracheally with RNA pools, followed by 2 boosts two weeks apart, carried out by intranasal instillation. The antibody response measured (FIG. 23 B) by ELISA was expressed as mean±SEM of IgG endpoint titers (n=4/group). As controls, dose-matched OVA in sterile PBS was used, OVA with cholera toxin subunit B (CTB) and PBS alone, respectively. In brief, wells were coated with antigen (10 μg/mg of OVA) and blocked with SeaBlock (Pierce, Rockford-Ill., catalog #37527). Serial dilutions of serum and bronchoalveolar lavage fluid were incubated for at least 2 hours at room temperature. After washing, the assay was developed with anti-mouse IgG antibody coupled with alkaline phosphatase (Sigma, cat #A7434) followed by addition of substrate (pNPP, Sigma, cat #N2765) and measurement by using an automatic microtiter plate reader (Molecular Devices, ThermoMax) equipped with SoftMax software. [0386] (B) The effect of various dsRNA motifs on the induction of antibody response to OVA: the results are expressed as in FIG. 23 C. The data are representative for two independent experiments. INSET: the ratio between mean IgG2a and IgG1 titers to OVA. For this purpose, biotin-conjugated anti-mouse IgG1 and IgG2a antibodies were used followed by incubation with streptavidin-AKP conjugate. The order from left to right is similar as in the main panel in FIG. 23C: PBS OVA, CTB OVA, pC:pG OVA, pI:pC OVA and pA:pU OVA. [0387] (C) The magnitude and profile of T cell response induced by OVA together with various dsRNA motifs, in female C57BL/6 mice. For the measurement of cellular response, splenic cell suspensions were obtained by passing the organ through 70 micron nylon Falcon strainers (Becton Dickinson, cat #352350) followed by lysis of red blood cells with red blood cell lysis buffer (Sigma, cat #R7757). The lymphocytes from the pulmonary associated lymphoid tissue were isolated by collagenase (Sigma, cat #C9891) digestion of lung tissue followed by Ficoll-Paque (Amersham Pharmacia, cat #17-1440-02) gradient centrifugation. The T cell response was measured by ELISPOT analysis as follows: 96-well 45 micron mixed cellulose ester plates (Millipore, cat #MAHA S4510) were coated with 4 μg/ml of rat anti-mouse anti-IFNγ, IL-2 or IL-4 monoclonal antibodies (BD-PharMingen, cat #554430, cat #18161D, cat #554387 respectively). After blocking with 10% FCS in sterile saline for 1 hour at 37° C., spleen cell suspensions were added at 5×105 cells/well, with or without antigens/peptides. For stimulation, graded amounts of antigen (OVA) were used. At 72 hours after stimulation, the assay was developed with biotinylated rat anti-mouse cytokine antibodies (BD-PharMingen) followed by streptavidin-HRP (BioSource Int., Camarillo, Calif.) and insoluble AEC substrate. The results were measured using an automatic imaging system (Navitar/Micromate) equipped with multiparametric-analysis software (Image Pro, Media Cybernetics). The results are expressed in FIG. 23 D as mean±SEM of the number of IFN-γ and IL-4 spot-forming-colonies (SFC) per spleen (n=4/group). The results are representative for two independent experiments.
[0388] The results in FIGS. 23B-D show that different synthetic RNAs have an enhancing effect on the B and T cell response to a prototype protein antigen. In addition, different motifs, comprising specific nucleotide combinations, have specific effects in terms of T1 versus T2 induction and subsequently, immunoglobulin isotype switching.
EXAMPLE 23
[0389] Use of selected synthetic RNA motifs facilitates the induction of MHC class I-restricted Tc1 cells, producing IFN-γ. [0390] (A) Cross-priming stimulated by dsRNA motifs was studied in BALB/c mice treated (priming plus 2 boosts) with 10 μg of recombinant-engineered HIV gp140 antigen together with pA:pU. The response was measured by ELISPOT analysis as described in Example 22, using in vitro stimulation with the MHC class I-restricted cognate peptide R10K derived from the V3 domain. As a control, dose-matched gp140 antigen was used. The results are expressed in FIG. 24A as mean±SEM of the number of IFN-γ and IL-4 SFC/spleen (n=4/group). [0391] (B) Cross-priming stimulated by dsRNA motifs was studied in C57BL/6 mice treated with 100 μg of whole OVA together with pA:pU by ELISPOT analysis as described in Example 22, using in vitro stimulation with the MHC class I-restricted peptide SIINFEKL (Seq. 1D No.). As a control, dose-matched OVA antigen in saline or sterile PBS was used. The results are expressed in FIG. 24B as mean±SEM of the number of IFN-γ and IL-4 SFC/spleen (n=4/group).
[0392] The results in FIGS. 24A-B show that a selected synthetic RNA motif was able to promote increased T cell immunity to different MHC class I-restricted peptides encompassed within larger antigens (polypeptides). This immune response comprised a Tc1 component, consisting in IFN-γ-producing MHC class I-restricted T cells.
EXAMPLE 24
[0393] Shows that unexpectedly, different synthetic RNA motifs bind to different receptors; in other words, there are multiple receptors that discriminate among RNA motifs.
[0394] In vitro binding of CD 11b.sup.+ APC by fluorescently-tagged pA:pU was measured by FACS analysis. The MACS-separated APC were incubated at 4° C. for 30 minutes with 10 μg/mg of tagged pA:pU ([pA:pU]-F), washed and analyzed. Alternatively, APC were preincubated for 10 minutes with 20 or 100 μg/mg of non-tagged pA:pU, pA or pI:pC respectively, before staining with tagged pA:pU and FACS analysis. The profiles of stained (open area), non-stained (filled area) cells and the percentage of highly stained APC were represented in each panel, with logarithmic x axis. The data are representative of two independent measurements with 10,000 events acquired for each sample.
Materials:
[0395] 1. Mouse CD11b, CD11c Magnetic Separation Beads: Miltenyi Biotec, cat #130-049-601, cat #130-052-001 respectively; [0396] 2. ULYSIS Nucleic Acid Labeling Kit: Alexa 488, Molecular Probes cat #U21650; [0397] 3. RNA Motifs: [0398] pA:pU, (Sigma, Lot #22K4068); [0399] pI:pC, (Sigma, Lot#52K4047); [0400] pA, (Sigma, Lot#22K4022); [0401] 4. FACS Buffer: PBS, 1% FCS, 0.1% sodium azide; [0402] 5. MACs buffer: PBS, 2 mM EDTA, 0.5% BSA; [0403] 6. Collagenase Buffer: 0.225 mg BSA, 0.0062 mg collagenase in 50 ml RPMI; and, [0404] 7. 70 um cell strainer: (Falcon/Becton Dickinson, cat #352350.
Methods:
I. Labeling of RNA Motifs:
[0404] [0405] 1. In the following protocol, each RNA motif was tagged with the ULYSIS Alexa 488 label.
II. Splenocyte Preparation:
[0405] [0406] 1. Isolate splenocytes and lung cells from 4 female C57 BL/6 mice; [0407] Lung cells, in contrast to splenocytes, must be minced and incubated in collagenase buffer for 30 minutes at 37° C. prior to the following step; [0408] Pass through 70 um falcon cell strainer; [0409] Wash and resuspend in MACS buffer: [0410] 2. Label with either CD11b or CD 11c specific MACS beads following suggested protocol; [0411] 3. Cells were then treated with: [0412] Non-tagged pA, pA:pU, or pI:pC (20 or 100 ug/ml) for 10 minutes at room temperature; [0413] ULYSIS tagged pA or pA:pU was added at 1.5 ug/tube and 10 ug/tube, respectively, to match dye:dsRNA ratio of each motif. [0414] 4. Mix and incubate 30 minutes on ice. [0415] 5. Wash once and resuspend in FACS buffer
III. Flow Cytometry:
[0415] [0416] Run flow cytometric analysis to determine/compare competitive inhibition of tagged versus non-tagged RNA motifs and cell receptor binding.
[0417] The results in FIG. 25 show that pA:pU and pI:pC bind to different cellular receptors. Since pI:pC binds to TLR3, it results that additional receptors distinct from TLR3 are involved in RNA recognition immune function.
EXAMPLE 25
[0418] Shows that selected synthetic RNA motifs trigger in vivo expression of chemokine genes, of importance for immunological activity.
[0419] Local up-regulation of chemokine gene-expression by dsRNA motifs was measured by DNA array technique using RNA from the pulmonary tissue, extracted one day after the administration via the respiratory tract. Total RNA was isolated from lungs using an RNeasy kit (Qiagen, Valencia, Calif.). The RNAs were further purified by treatment with RNase-free DNase I (Stratagene, San Diego, Calif.). DNA array was performed by using the Nonrad-GEArray kit from SuperArray Inc. (Bethesda, Md.). Briefly, cDNA probes were synthesized using MMLV reverse transcriptase with dNTP mix containing biotin-16-dUTP. The GEArray membranes were prehybridized at 68° C. for 1-2 hours. The hybridization was carried out by incubation of the membranes with biotin-labeled cDNA. The hybridized membranes were washed in 2×SSC-1% SDS twice and 0.1×SSC-0.5% SDS twice. The membranes were further incubated with alkaline phosphatase-conjugated streptavidin (BioSource Int., Camarillo, Calif.) and finally developed with CDP-Star chemiluminescent substrate. The intensity of signal was measured with Image-Pro analysis system equipped with Gel-Pro software (Media Cybernetics, Silver Springs, Md.).
[0420] The results are expressed as fold-increase of gene expression, over expression levels measured in the pulmonary tissue of non-treated mice. The pattern of chemokine expression triggered by dsRNAs (50 μg of pA:pU and pI:pC, respectively) was compared to that induced by 1 μg of LPS. The chemokines that selectively bind to receptors on Th1 and Th2 cells were indicated with continuous and interrupted contours, respectively.
[0421] The results in FIG. 26 show that pA:pU and pI:pC trigger expression of a wide range of chemokines and that the expression pattern is motif-dependent and different from that elicited by LPS (endotoxin).
EXAMPLE 26
[0422] Shows that selected synthetic RNA motifs mobilize an immune defense that is capable to control infection with a pulmonary virus.
[0423] dsRNA motifs display differential ability to mobilize immune defense against influenza virus infection. C3H/HeJ mice were treated via the respiratory route with 50 μg of pI:pC, pA:pU or 50 μl of saline one day before and after pulmonary infection with a sublethal dose of influenza virus. For virus challenge, C57BL/6 and TLR4-/- C3H/HeJ mice under Metofane anesthesia were infected with sublethal doses (104 tissue culture infective doses 50%--TCID50) of live WSN virus, via the nasal route. On day 5 after infection, the mice were sacrificed, lungs retrieved, homogenized and stored at -70° C. The virus titers were measured by 48-hour incubation of serial dilutions of samples with permissive MDCK cells, followed by standard hemagglutination with chicken red blood cells (From Animal Technologies). The endpoint titers were estimated in triplicate measurements by interpolation and expressed as TCID50/organ (means±SEM; n=6/group; results are representative of two independent studies in C3H/HeJ TLR-4-/- and competent mice). Similar results were obtained in TLR4 competent, C57BL/6 mice.
[0424] Thus, the results depicted in FIG. 27 show that the control of replication of influenza virus can be achieved by using selected synthetic RNA motifs (dsRNA1 is pA:pU and dsRNA2 is pI:pC).
EXAMPLE 27
[0425] Shows that co-administration of selected synthetic RNA motifs breaks tolerance to high dose standard antigen.
[0426] dsRNA motifs prevent high-zone tolerance in mice injected with human IgG. The mice (C57BL/6) were initially injected intravenously with a toleragenic dose of 200 μg of hIgG alone (closed symbols) or together with 100 μg of pI:pC or pA:pU (open symbols) and subsequently boosted subcutaneously with an immunogenic dose of 100 μg of hIgG emulsified in CFA. The titer of antibodies against hIgG was measured by ELISA (as detailed in the Example 23, with the difference consisting in use of 10 μg/ml of hIgG for coating) at various intervals after the first injection. As a control, mice immunized with 100 μg of hIgG emulsified in CFA were included and represented the maximal titer on the graph (interrupted line).
[0427] The results are represented in FIG. 28 as means±SEM of endpoint titers (n=5/group). Similar results were obtained in TLR4 deficient (C3H/HeJ) and LPS-responsive C3H/SnJ mice. Thus, the results in FIG. 28 show that selected synthetic RNA motifs pI:pC and pA:pU largely prevent high zone tolerance that is usually associated with administration of large amounts of purified protein.
EXAMPLE 28
[0428] Shows that selected RNA motifs induce differential cytokine production by human APC.
[0429] Human THP-1 monocytic cells, following differentiation,.Were incubated with different concentrations of synthetic RNA (pA:pU, pI:pC or pA) for 24 hours, and the cell supernatants collected. The concentration of IL-12 and TNF-α were measured by ELISA. The results are expressed in FIG. 29 as pg/ml (concentration) for each cytokine and culture condition.
Materials:
[0430] 1. THP-1 Human monocytic cell line: ATCC, cat #TIB-202; [0431] 2. IL-12 Cytokine: Human ELISA, IL-12 ultra sensitive (US) cat #KHC0123; [0432] 3. TNF alpha Cytokine: Human ELISA, TNF alpha cat #KHC3012; [0433] 4. RNA Motifs: [0434] pA:pU, (Sigma, Lot #22K4068); [0435] pI:pC, (Sigma, Lot #52K4047); and, [0436] pA, (Sigma, Lot #22K4022).
Method:
[0436] [0437] 1. The THP-1 cells were allowed to differentiate following addition of 10 ng/ml PMA in media containing 10% FCS. [0438] 2. After gently washing cells and adding non-FCS containing Media (HL-1), treatments (RNA motifs and controls) were added at concentrations of from 3 to 100 μg/mg on top of adherent THP-1 cells. [0439] 3. After 24 hours incubation, cell supernatants were harvested and IL-12 and TNF alpha concentrations were measured by ELISA.
[0440] The results in FIG. 29 show selected synthetic RNA motifs effect on human monocytic cells; in addition, this effect is heterogeneous, depending on the chemical structure of the motifs (nucleotide composition). Selected but not all synthetic RNA motifs are able to trigger IL-12 production, an important T1 regulatory cytokine, by human monocytic cells.
EXAMPLE 29
[0441] Shows that two distinct synthetic RNA motifs bind to human THP-1 monocytic cells in a manner demonstrating interaction with different receptors.
[0442] THP-1 cells were incubated at for 15 minutes at room temperature with different amounts of non-labeled synthetic RNA. Subsequently, tagged pA:pU was added for 30 minutes at 4° C., cells washed and the fluorescence quantified by FACS analysis. The results are expressed in FIGS. 30A-30B as histograms corresponding to the large cell subset (A) and total cell population (B). Percentages of stained cells were represented on each Figure.
Materials:
[0443] 1. ULYSIS: Nucleic acid fluorescent label (Molecular Probes, cat #U-21650). [0444] 2. RNA Motifs: [0445] pA:pU, (Sigma, Lot #22K4068); [0446] pI:pC, (Sigma, Lot # 52K4047); [0447] 3. Detoxi-Gel column: (Pierce, cat #20344).
Method:
Labeling of Polyadenylic-Polyuridylic Acid (pA:pU):
[0447] [0448] 1. Following removal of endotoxin using a Detoxi-Gel column, pA:pU was labeled with the Alexa Fluor 488 fluorescent dye using the ULYSIS nucleic acid labeling system. [0449] 2. Briefly: [0450] The pA:pU was precipitated using sodium acetate and ethanol at "-70° C.; [0451] The pA:pU was heat denatured and labeled with the Alexa Fluor 488 reagent at 90° C.; and, [0452] The reaction was stopped and the labeled pA:pU was ethanol precipitated.
Cell Treatment:
[0452] [0453] 1. THP-1 cells were suspended at 2×106 cells/ml; [0454] 2. 50 μl of above suspension (5×104 cells) were placed in 12×75 mm tubes; [0455] 3. Non-tagged pA:pU or pI:pC were added to the THP-1 cells at a concentration of either 20 or 100 μg/mg and incubated 15 minutes; ULYSIS labeled pA:pU was added at a concentration of 100 ug/ml for 30 minutes on ice. [0456] 4. The THP-1 cells were washed once and suspended in FACS buffer followed by flowcytometric analysis to determine relative fluorescent differences between different treatment populations.
[0457] The results in FIGS. 30A-30B show that non-tagged pA:pU but not non-tagged pI:pC was able to compete out the binding of tagged pA:pU to human THP-1 monocytic cells, both at the level of large cell subset and whole population.
EXAMPLE 30
[0458] Shows how the adjuvant synthetic RNA should be prepared and purified prior to use in its most effective format.
[0459] The bulk synthetic RNA material is obtained by standard methods of organic synthesis. Afterwards, the material is dissolved in sterile endotoxin-free saline, passed through endotoxin removal columns until the concentration of LPS is below 0.005EU/μg. The measurement of LPS is carried out by standard Limulus assay. Subsequently, the material is fractionated by a series of centrifugation steps through filters of defined porosity (see FIG. 31).
[0460] A useful fraction comprises synthetic RNA of less than 20 to maximum 100 bp size, however, larger RNA fragments may be used. After purification, the material is measured and Validated on standard assays: spectrophotometry (OD260 nm); gel electrophoresis; endotoxin quantitation by Limulus assay; bioactivity on human THP-1 cells (as in Example 28).
EXAMPLE 31
[0461] Shows that unexpectedly, different fractions of a selected synthetic RNA compound are endowed with different biological activity, based on size.
[0462] Differentiated human THP-1 monocytic cells were incubated with different concentrations of synthetic RNA (pA:pU, fractionated as described in the Example 30) for 24 hours, and the supernatants collected. The concentration of TNF-α was measured by ELISA using BioSource International kits (Camarillo, Calif.). The results are expressed in FIG. 32 as pg/ml (concentration) for each culture condition.
[0463] The results depicted in FIG. 32 show that lower molecular weight fractions of a selected synthetic RNA compound are endowed with higher biological activity, in terms of cytokine production, by human monocytic THP-1 cells.
EXAMPLE 32
[0464] Selected synthetic RNA motifs have, unexpectedly, a different immune profile in regard to generation of anti-RNA antibodies.
[0465] BALB/c mice were immunized intraperitoneally and subcutaneously with 50 μg+50 μg of hIgG and synthetic RNA (pI:pC or pA:pU) and serum samples were prepared 1 week later. As a control, mice injected with hIgG in saline were used. The anti-hIgG, and dsRNA IgG antibody titers against pA:pU, pI:pC, pA and hIgG were measured by ELISA. In brief, wells were coated with antigen (10 μg/mg of hIgG or synthetic RNAs) and blocked with SeaBlock (Pierce, Rockford, Ill., catalog #37527). Serial dilutions of serum and bronchoalveolar lavage fluid were incubated for at least 2 hours at room temperature. After washing, the assay was developed with anti-mouse IgG antibody coupled with alkaline phosphatase (Sigma, cat #A7434) followed by addition of substrate (pNPP, Sigma, cat #N2765) and measurement by using an automatic microtiter plate reader (Molecular Devices, ThermoMax) equipped with SoftMax software.
[0466] The results are expressed in FIG. 33 as mean±SEM of endpoint titers (n=3/group). The results in FIG. 33 show that pI:pC but not pA:pU induced antibody response against itself, with a cross-reactive component against another RNA motif.
EXAMPLE 33
[0467] In vivo loading of APC by recombinant IgG results in generation of Tc1 type of MHC class I responses only when additional conditions are satisfied.
[0468] BALB/c mice were immunized with 50 ug of recIgG-NP(Kd) subcutaneously, admixed with 50 ug of selected synthetic RNA (pA:pU or pI:pC). As a control, naive mice or mice immunized with recombinant IgG only were used. At 3 weeks after immunization, the T cell response was measured by ELISPOT analysis as follows: the ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/ml for anti-IL4, and 8 μg/mg for anti-IFN gamma, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media and blocked with 200 μl/well of DMEM complete containing FBS, for an hour at 37° C. Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with NP 147-155 peptide or just with media, to assess the background. Plates were incubated 72 hours at 37° C., 5% CO2. After 3 days, the plates were washed 5 times with PBS--tween 20 0.05% (washing buffer) and incubated with 100 μl/well of biotinylated anti-cytokine Abs, 2 μg/ml in PBS--tween 20 0.05%--FBS 0.1%(ELISPOT buffer) overnight at 4° C.
[0469] The next day, the plates were washed five times with washing buffer and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. The plates were then allowed to dry at room temperature for 24 hours.
[0470] The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.). The frequency of cytokine producing T cells reacting to NP peptide was measured and expressed against the amount of peptide used for stimulation. The results are expressed as means±SEM of triplicates (n=3 mice/group).
[0471] As shown previously in FIG. 19, the administration of recombinant IgG bearing the NP MHC class I-restricted epitope resulted in generation of Tc2 immunity but not Tc1 response, implying in vivo formation of class I-peptide complexes with a specific co-stimulation profile. The results in FIGS. 34A and 34B show that co-use of selected synthetic RNAs promoted effective induction of IL-2 and IFN-gamma subsequent to IgG mediated delivery of an MHC class I-restricted epitope (dsRNA1 is pA:pU and dsRNA2 is pI:pC).
EXAMPLE 34
[0472] Effective formation of MHC class I-peptides and instruction of the resulting T cell response by simultaneous manipulation of APC loading via Fcgamma R and activation via RNA receptors.
[0473] Splenic APC were isolated from naive BALBc mice and pulsed ex vivo overnight with 1 ug NP peptide, or 50 μg recIgG-NP (Kd) with or without 50 μg/mg selected synthetic dsRNA (pA: pU). The cells were washed and 5×106 cells were administered by s.c. and i.p. injection equal amount, to naive BALB/c mice. The response was measured 3 weeks later by ELISPOT analysis as follows: the ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 μg/mg for anti-IL4, and 8 μg/mg for anti-IFN gamma, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media and blocked with 200 μl/well of DMEM complete containing FBS, for an hour at 37° C. Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with 30 μg/ml, 10 μg/ml, or 3 μg/ml NP peptide. or just with media, to assess the background. Plates were incubated 72 hours at 37° C., 5% CO2. After 3 days the plates were washed 5 times with PBS--tween 20 0.05% (washing buffer) and incubated with 100 μl/well of biotinylated anti-cytokine Abs, 2 μg/ml in PBS--tween 20 0.05%--FBS 0.1%(ELISPOT buffer) overnight at 4° C. The next day the plates were washed five times with washing buffer and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. The plates were then allowed to dry at room temperature for 24 hours.
[0474] The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.). The results are expressed in FIG. 35 as frequency of cytokine producing spot forming colonies against the concentration of peptide used for ex vivo stimulation (mean±SEM, n=3 mice/group). In addition, the mean area/colony versus the concentration of peptide used for stimulation is plotted, for both IFN-gamma and IL-4 (arbitrary units).
[0475] The results in FIG. 35 show that ex vivo APC loading by recombinant IgG is significantly more effective in formation of MHC class I-peptide complexes and generation of Tc response, compared to use of peptide itself. In addition, the mere formation of MHC class I-peptide complexes subsequent to epitope delivery via IgG/FcgammaR results in differentiation of Tc2 cells producing IL-4 but not MN-gamma. Simultaneous treatment of APC with selected synthetic RNA results in broadening of the T cell profile, to IFN-gamma producing Tc1 cells.
EXAMPLE 35
[0476] Shows that co-priming with IgG-peptide together with a selected co-stimulatory motif resulted in more effective secondary expansion of MHC class I-restricted T cells subsequent of virus infection.
[0477] BALB/c mice were injected with recIgG-NP(Kd), pA:pU separately, or in combination (50 ug/injection). As a control, naive mice were used; Three weeks after treatment, the mice were infected with 104 TCID50 of A/WSN/32 H1N1 influenza virus, via the respiratory tract. Four days after infection, the T cell profile in the spleen was measured by ELISPOT analysis subsequent to ex vivo stimulation with NP peptide as follows: the ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/ml for anti-IL2 and anti-IL4, and 8 μg/mg for anti-IFN gamma, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media and blocked with 200 μl/well of DMEM complete containing FBS, for an hour at 37° C. Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with 20 μg/ml NP peptide or just with media, to assess the background. Plates were incubated 72 hours at 37° C., 5% CO2. After 3 days, the plates were washed 5 times with PBS--tween 20 0.05% (washing buffer) and incubated with 100 μl/well of biotinylated anti-cytokine Abs, 2 μg/ml in PBS--tween 20 0.05%--FBS 0.1% (ELISPOT buffer) overnight at 4° C. The next day the plates were, washed five times with washing buffer and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. The plates were then allowed to dry at room temperature for 24 hours.
[0478] The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.). The results are expressed in FIG. 36 as frequency of NP-specific MHC class I-restricted T cells forming cytokine producing colonies (means±SEM, n=4 mice/group).
[0479] The results in FIG. 36 show that IgG mediated delivery of a class I restricted epitope is most effective in priming class I restricted Tc1 responses when co-administration of selected synthetic RNA was carried out. Such primed precursors were rapidly expanded subsequent to infection with influenza virus.
EXAMPLE 36
[0480] Shows that the most effective priming of cytotoxic lymphocytes recognizing an MHC class I-restricted epitope occurs by co-administration of selected RNA motif together with peptide epitope inserted within the IgG backbone.
[0481] BALBc mice were immunized and challenged with recIgG-NP (Kd) as in the previous Example and sacrificed 4 days after influenza virus infection. The splenocytes were prepared, suspended in HL-1 medium at 5 million/ml and co-incubated for 5 days with 10 μg/mg of NP 147-155 peptide and in presence of 5U/mg of recombinant IL-2. Splenocytes from 4 mice/group were pooled and incubated in flasks.
[0482] After expansion, viable cells were recovered by Ficoll gradient centrifugation, washed and incubated for 5 hours in V-bottom plates, in various numbers, with a fixed number of sp20 target cells with or without NP peptide (20 μg/ml). The supernatants were harvested after plate centrifugation, and the concentration of LDH measured by using a Promega kit (cat #G1780). The results are expressed as percent specific lysis at different E: T ratios (Effector to Target ratio).
[0483] The results in FIG. 37 show that effective priming of anti-viral cytotoxic T cells requires both effective in vivo loading of APC with class. I restricted epitope delivered via IgG, together with appropriate instruction by selected synthetic RNA motif, namely pA:pU.
EXAMPLE 37
[0484] Shows that vaccination with an IgG bearing a viral MHC class I-restricted epitope, together with selected synthetic RNA motif, provided protection against infectious challenge with a prototype virus.
[0485] BALB/c mice were immunized with 50 ug of recIgG-NP (Kd) together with 50 ug of selected synthetic RNA (pA: pU), by subcutaneous injection. Three weeks after immunization, the mice were challenged with 104 TCID 50 of infectious WSN influenza virus and sacrificed 5 days later. The pulmonary virus was titrated in lung homogenates by standard MDCK hemagglutination assay as follows: on day one MDCK cells were plated in 96 well plates at 2×104/well/200 ul and incubated for 24 hours at 37° C., 5CO2. The next day, 25 μl of the 10 fold dilutions in DMEM media of the lung homogenates were incubates in briefly tripsinized MDCK plates (1 minute) in triplicates and incubated at 37° C. After one hour, 175 ul of the DMEM complete media was added and plates were incubated for 48 hours at 37° C., 5% CO2. After two days, the hemagglutination-inhibition was done with chicken red blood cells incubated with the cell culture supernatants from the MDCK plate for 30 minutes at room temperature and the results were expressed as means±SEM'of total pulmonary virus (n=4 mice/group). As a control, non-immunized mice were used.
[0486] The results in FIG. 38 show that immunization with a recombinant IgG bearing a viral class I restricted epitope together with selected synthetic dsRNA (pA:pU) resulted in priming of an immune response capable to limit the replication of a virus subsequent to infectious challenge.
EXAMPLE 38
[0487] FIG. 39 describes the tumor models used for testing the efficiency of a Ig-peptide-based molecules.
[0488] Balb-c mice (Kd restricted) have been used to establish a tumor model. Tumor cells (1 to 15 million in 100 μL) were typically injected in the flank to the mouse (see arrow in upper photo in FIG. 39). Primary tumors (i.e. those at the sight of injection) were first detected by palpating the area and then quantitated by measuring the tumor size with a caliper (see FIG. 39). In one series of experiments, the mouse myeloma cell line (SP2/0), either untransfected cells or cells stable transfected expressing heterologous protein (recombinant IgG expressing different epitope peptides in the CDR3 region of the heavy chain or the complete NP protein), was used to induce tumors in the mice. Expression of heterologous proteins in the SP2/0 cells provided specific tumor associated antigens (TAA) for testing various anti-tumor strategies in the immunocompetent mice. Typically, untreated mice developed palpable solid primary tumors 1 week post injection that led to morbidity and death over the next 4 weeks. Postmortem examination of the injected mice revealed metastatic lesions (see FIG. 39). Sp2/0 cells were cultured from primary tumor tissue as well as spleen taken from tumor-bearing mice (data not shown). SP2/0 cells were stably transfected with a recombinant IgG-expressing plasmids that were all identical except for the specific epitope sequence introduced into the CDR3 region of the heavy chain, for example, the MHC I restricted NP epitope (amino acids 147-155, see FIG. 39). SP2/0 cells were also stably transfected with a plasmid containing the coding sequence for the entire NP protein of WSN virus under control of the CMV promoter. All transfected cell lines produced primary tumors over the same frame as wild type SP2/0 cells.
[0489] This tumor model was extended to include an adenocarcinoma cell line (4T1, ATCC CRL-2539, Kd restricted), previously shown to induce metastatic tumors in Balb-c mice. The 4T-1 cell line was similar to that described above for the SP/0 line. Injection of 1 to 15 million 4T-1 cells into the flank of Balb-c mice produced a palpable primary tumor over a time frame similar to injections of SP2/0 cells eventually leading to death. Postmortem collection of tissue from various organs showed that 4T-1 could be recovered from spleen, lungs as well as the primary tumor (not shown). 4T-1 cells were stably transfected with a NP-expressing plasmid described above. As with SP2/0 cells, transfection of the 4T-1 cell did not affect the course of tumor growth and lethality of disease.
EXAMPLE 39
[0490] Demonstrates successful control and treatment of a tumor after clinical diagnosis, by using a tumor associate T cell epitope within a recombinant IgG together with a selected co-stimulatory RNA motif.
[0491] Balb/c mice were injected with SP2/0 cells (15 million in 100 μL) stably expressing recombinant IgG carrying the MHC I (Kd) NP epitope peptide in the CDR3 region of the heavy chain (IgNP). At day 7 post injection all mice had palpable tumors and the mice were randomized into 3 groups: co-stimulatory motif (i.e. dsRNA comprised of polymeric pApU) alone; purified IgTAA protein (IgNP); and both dsRNA pA:pU and purified IgTAA protein. The time of treatment is indicated by the arrows in FIG. 40, and each injection contained 50 μg of the indicated compound. The mice that developed metastatic disease and died are represented with a "D" in the figure.
[0492] The data show that the combination of dsRNA (co-stimulatory motif) and IgTAA (IgNP) produced a dramatic protective response in mice that all had primary tumors at the start of therapy. While all mice treated with either the dsRNA or IgTAA compound alone succumbed to disease, 100% of the mice treated with both were still alive 3 weeks after initiation of treatment and were in good clinical condition at the time of sacrifice for measurement of T cell response. These data show that in vivo loading of APC with TAA (accomplished by uptake of IgNP via the Fc receptor of APC) is not sufficient for a potent anti-tumor response. The tumor rejection and survival displayed by, mice treated with IgNP in combination with pApU dsRNA highlights the important role co-stimulation plays in treatment of tumors with tumor-associated antigens.
[0493] In conclusion, the results in FIG. 40 show that both effective in vivo loading of APC with tumor associated antigen, together with simultaneous activation by selected synthetic RNA motifs, are necessary and sufficient for effective control of tumor growth and induction of tumor rejection.
EXAMPLE 40
[0494] This Example, in context of sublethal inoculation of tumor cells, shows that the suboptimal response to tumor antigens could be corrected by therapy with peptide epitope within an IgG backbone, together with co-stimulatory motif.
[0495] Balb/c mice were injected with SP2/0 cells stably expressing recombinant IgG (IgNP) that contains the MHC I (Kd)epitope (amino acids 147-155) of WSN virus nucleoprotein in the CDR3 of the heavy chain. The cell inoculum was 1 million cells (in 100 μL) per mouse. The mice were observed until such time as palpable tumors were detected at the site of injection. At this point the tumors were measured and 8 mice were left untreated (control) while 6 were injected intratumorally with purified IgTAA (i.e. purified IgNP, 2 mg/kg) and dsRNA (pApU, 4 mg/kg) weekly. Weekly measurements of the tumors were taken.
[0496] Panel A of FIG. 41 shows that in 6 of 8 of the control mice the induced tumor was progressive and ultimately lethal whereas 2 of the mice completely rejected the tumor spontaneously. Panel B of FIG. 41 shows that the 3 weekly treatments with IgNP/dsRNA (indicated by the arrows) stimulated complete tumor rejection in 4 of the 6 mice and significant remission in another.
[0497] The results in FIG. 41 shows that both effective in vivo loading of APC with tumor associated antigen, together with simultaneous activation by selected synthetic RNA, can trigger an effective immune response to tumor-associated antigens.
EXAMPLE 41
[0498] Shows that therapy of tumor-bearing mice with a tumor epitope within an IgG backbone together with co-stimulatory synthetic dsRNA results in the restoration of the activatory status of tumor infiltrating lymphocytes.
[0499] Two BALB/c mice were injected with 10 million sp20 transfectoma expressing the NP-Kd epitope. After tumors developed, one mouse was injected intratumorally with 50 μg of selected dsRNA motif (pApU) plus 50 μg of "IgNP"--recIgG-NP(10 in saline. The mice were sacrificed 24 hours later, tumors excised, digested with collagenase, filtered through 70 um filter and viable cells isolated on Ficoll gradient. Cells were stained with mAbs against TCRβ, CD25 or isotype control and assessed by FACS analysis. The results were expressed as histograms, with percentage stained cells indicated.
Materials:
[0500] 1. SP20 cell line (ATCC); [0501] BALB/c mice (Harland Sprague Dawley); [0502] 2. Falcon 70 micron filter(Becton Dickinson, cat #352350); [0503] 3. Collagenase (Sigma, cat #C-9891); [0504] 4. BSA, fraction V (Sigma, cat #A-4503); [0505] 5. Collagenase buffer: 0.225 gm BSA +0.00625 gm in 50 ml RPMI; [0506] 6. Ficoll-hypaque (1.077, Amersham, cat #17-1440-02); [0507] 7. FACS Buffer:1% fetal calf serum+0.1% azide in PBS; [0508] 8. Antibodies: All from BD Pharmingen; and, [0509] 9. Flow Cytometer: FACSCalibur (Becton Dickinson).
Method: Tumor Cell Isolation and FACS Analysis:
[0509] [0510] 1. Tumor was induced as stated above 6 weeks prior; [0511] 2. Tumor was isolated from BALB/c mouse; [0512] 3. Tumor was minced with sterile scissors and 10 ml of collagenase buffer added; [0513] 4. Incubate 40 minutes, 37° C.; [0514] 5. Force tumor through a 70 μm Falcon filler with a 3 ml syringe plunger into a 50 ml tube while washing with RPMI; [0515] 6. Wash 1× and resuspend in 4 mls warm RPMI buffer; [0516] 7. With equal volume of cell suspension layered over Ficoll, centrifuge at RT, 2000 RPM, for 15 minutes; [0517] 8. Isolate layer and wash once in HL-1 buffer and resuspend in FACS buffer to 2×106/ml and run flow cytometry analysis; [0518] 9. `Remaining cells were used for ELISPOT analysis; [0519] 10. Cells were placed in 12×75mm tubes, 50 μl/tube and stained with FITC labeled anti-mouse antibody, 2 μg/tube plus 1 μl/tube mouse serum: [0520] Isotypic Control; [0521] Anti -CD40; [0522] Anti -CD8; [0523] Anti -CD4; [0524] Anti -CD25; [0525] Anti -TCR gamma delta; [0526] Anti -TCR Beta; [0527] 11. Incubate 30 minutes on ice; and, [0528] 12. Wash once with FACS buffer and resuspend in 300 μl FACS buffer.
[0529] The results in FIG. 42 show that tumor infiltrating lymphocytes displaying the T cell receptor marker TCRβ acquired expression of the activation marker CD25 upon treatment with recombinant immunoglobulin bearing tumor associated epitope, together with selected synthetic dsRNA motif.
EXAMPLE 42
[0530] Shows that successful therapy of tumor bearing mice with a peptide epitope within the IgG backbone together with a selected co-stimulatory molecule is associated with a specific differentiation pattern of Tc, comprising Tc1 in addition to Tc2.
[0531] Mice that successfully rejected the tumor following treatment with recombinant Ig carrying a tumor associated epitope together with selected synthetic dsRNA motif as explained in Example 40, were sacrificed and the T cell response against tumor associated epitope measured by ELISPOT analysis. The ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/mg for anti-IL2 and anti-IL4, and 8 μg/ml for anti-IFN gamma, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media and blocked with 200 μl/well of DMEM complete containing FBS, for an hour at 37° C.
[0532] Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with various concentrations of NP peptide. Plates were incubated 72 hours at 37° C., 5% CO2. After 3 days, plates were washed 5 times with PBS--tween 20 0.05% (washing buffer), and incubated with 100 μl/well of biotinylated anti-cytokine Abs, 2 μg/ml in PBS--tween 20 0.05%--FBS 0.1% (ELISPOT buffer) overnight at 4° C. The next day the plates were washed five times with washing buffer, and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. Plates were then allowed to dry at room temperature for 24 hours.
[0533] The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.). The results were expressed as number (mean±SEM) of spot forming colonies corresponding to IL-4, IL-2 and IFN-γ. As a control, non-treated mice were used, which failed to reject tumor (n=4/group).
[0534] The results in FIG. 43 show that the treated mice that successfully rejected the tumor, developed Tc1 responses against the tumor associated epitope on the therapeutic Ig, along with Tc2 immunity. In contrast, the mice that failed to reject the tumor developed only Tc2 immunity.
EXAMPLE 43
[0535] Shows induction of effective memory response subsequent to specific treatment of tumor bearing mice with a T cell epitope within the IgG backbone, together with a selected co-stimulatory motif.
[0536] Mice bearing sp2/0 tumors expressing the NP-Kd TAA were treated as described in the Example 40, by injection with recombinant Ig bearing TAA together with selected synthetic RNA motifs. After tumor rejection, the mice were challenged by subcutaneous injection administered contralaterally, with 15 million SP2/0 cells expressing NP-Kd epitope. In parallel, 4 control naive mice were similarly injected with a tumorigenic/lethal dose of same type of cells. The development and size of the tumors was monitored and represented as diameter (mm) versus time since challenge.
[0537] The results in FIG. 44 show that successful rejection of the tumor induced by indicated treatment is followed by effective protection against subsequent challenge with the same tumor, indicating development of effective immune memory.
EXAMPLE 44
[0538] Shows that surprisingly, the induction of tumor rejection by an IgG bearing a TAA together with a costimulator dsRNA motif, results in cross-protection against a range of tumor cell variants lacking the TAA or displaying variants of TAA.
[0539] The mice protected against homologous challenge as described in Example 43, were subjected to sequential challenge with 15 million tumor cells representing the same tumor cells devoid of TAA (loss of antigen mutants) or bearing variants of TAA lacking the NP-Kd epitope. In addition, mice were challenged with a different type of tumor cell line (4T-1 adenocarcinoma) as a control, displayed in the table attached to FIG. 45A. In every case, naive controls were included.
[0540] The status of T cell immunity of mice protected against multiple challenges with tumor variants, has been assessed by ELISPOT `analysis using splenic cell suspensions stimulated with TAA (NP-Kd peptide), HA (MHC class II-restricted peptide), or protein extracts from cell lysates. The ELISPOT plates (Millipore, Molsheim, France) were incubated with purified anti-cytokine Abs (4 ug/ml for anti-IL2 and anti-IL4, and 8 μg/mg for anti-IFN gamma, from BD Pharmingen) in sterile PBS (50 μl/well) at 4° C. overnight. The next day, the plates were washed 2 times with DMEM media and blocked with 200 μl/well of DMEM complete containing FBS, for an hour at 37° C.
[0541] Single cell suspension was made from the spleens, red blood cells were lysed, cells washed, counted and incubated at 5×105/well together with various concentrations of antigen. Plates were incubated 72 hours at 37° C., 5% CO2. After 3 days, the plates were washed 5 times with PBS--tween 20 0.05% (washing buffer) and incubated with 100 μl/well of biotinylated anti-cytokine Abs, 2 μg/ml in PBS--tween 20 0.05%--FBS 0.1% (ELISPOT buffer) overnight at 4° C. The next day the plates were washed five ` times with washing buffer, and incubated for an hour with 1:1000 Streptavidin-HRP diluted in ELISPOT buffer. The reaction was developed with 3-amino-9-ethylcarbazole substrate (Sigma, St. Luis, Mo.) and stopped by washing the plate twice with tap water. The plates were then allowed to dry at room temperature for 24 hours.
[0542] The data were acquired using an automated system (Navitar, Rochester, N.Y.) with ImagePro-Plus) software (Media Cybernetics, Silver Spring, Md.). The results were expressed as number (mean±SEM) of spot forming colonies corresponding to IL-4, IL-2 and IFN-γ. As a control, non-treated mice that failed to reject tumor (n=4/group) were used. As a control, naive mice were included. The data are expressed as number (mean±SEM) of cytokine producing cells/organ (n=3/group).
[0543] The results in FIG. 45A-45B (including the table in FIG. 45 A) show that the emerging immunity, subsequent to the indicated treatment that results in tumor rejection, protects against challenge with loss of antigen variants and is associated with overall expansion of cytokine producing cells. This indicates a broadening of the repertoire of anti-tumor lymphocytes, promoted by the proposed regimen, to tumor associated antigens that are not borne by the immunotherapeutic molecule.
Sequence CWU
1
531107PRTHomo sapiens 1Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro
Ser Arg Glu1 5 10 15Glu
Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe 20
25 30Tyr Pro Ser Asp Ile Ala Val Glu
Trp Glu Ser Asn Gly Gln Pro Glu 35 40
45Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe
50 55 60Phe Leu Tyr Ser Lys Leu Thr Val
Asp Lys Ser Arg Trp Gln Gln Gly65 70 75
80Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu His
Asn His Tyr 85 90 95Thr
Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 100
1052110PRTHomo sapiens 2Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu
Phe Pro Pro Lys1 5 10
15Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val
20 25 30Val Val Asp Val Ser His Glu
Asp Pro Glu Val Lys Phe Asn Trp Tyr 35 40
45Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu
Glu 50 55 60Gln Tyr Asn Ser Thr Tyr
Arg Val Val Ser Val Leu Thr Val Leu His65 70
75 80Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys
Lys Val Phe Asn Lys 85 90
95Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys 100
105 110315PRTHomo sapiens 3Glu Pro Lys
Ser Cys Asp Lys Thr His Thr Cys Pro Pro Cys Pro1 5
10 1545PRTHomo sapiens 4Gln Val Gln Leu Gln1
5511PRTInfluenza A virus 5Ser Phe Glu Arg Phe Glu Ile Phe Pro
Lys Glu1 5 1069PRTUnknownDescription of
Unknown MHC class I-restricted peptide 6Thr Tyr Thr Gln Thr Arg Ala
Leu Val1 57237PRTArtificial SequenceDescription of
Artificial Sequence Synthetic polypeptide 7Gly Gly Gly Gly Gly Glu
Pro Lys Ser Cys Asp Lys Thr His Thr Cys1 5
10 15Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro
Ser Val Phe Leu 20 25 30Phe
Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu 35
40 45Val Thr Cys Val Val Val Asp Val Ser
His Glu Asp Pro Glu Val Lys 50 55
60Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys65
70 75 80Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu 85
90 95Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys
Glu Tyr Lys Cys Lys 100 105
110Val Phe Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys
115 120 125Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val Tyr Thr Leu Pro Pro Ser 130 135
140Arg Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val
Lys145 150 155 160Gly Phe
Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln
165 170 175Pro Glu Asn Asn Tyr Lys Thr
Thr Pro Pro Val Leu Asp Ser Asp Gly 180 185
190Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg
Trp Gln 195 200 205Gln Gly Asn Val
Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn 210
215 220His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly
Lys225 230 2358566PRTInfluenza A virus
8Met Lys Ala Asn Leu Leu Val Leu Leu Cys Ala Leu Ala Ala Ala Asp1
5 10 15Ala Asp Thr Ile Cys Ile
Gly Tyr His Ala Asn Asn Ser Thr Asp Thr 20 25
30Val Asp Thr Val Leu Glu Lys Asn Val Thr Val Thr His
Ser Val Asn 35 40 45Leu Leu Glu
Asp Ser His Asn Gly Lys Leu Cys Arg Leu Lys Gly Ile 50
55 60Ala Pro Leu Gln Leu Gly Lys Cys Asn Ile Ala Gly
Trp Leu Leu Gly65 70 75
80Asn Pro Glu Cys Asp Pro Leu Leu Pro Val Arg Ser Trp Ser Tyr Ile
85 90 95Val Glu Thr Pro Asn Ser
Glu Asn Gly Ile Cys Tyr Pro Gly Asp Phe 100
105 110Ile Asp Tyr Glu Glu Leu Arg Glu Gln Leu Ser Ser
Val Ser Ser Phe 115 120 125Glu Arg
Phe Glu Ile Phe Pro Lys Glu Ser Ser Trp Pro Asn His Asn 130
135 140Thr Thr Lys Gly Val Thr Ala Ala Cys Ser His
Ala Gly Lys Ser Ser145 150 155
160Phe Tyr Arg Asn Leu Leu Trp Leu Thr Glu Lys Glu Gly Ser Tyr Pro
165 170 175Lys Leu Lys Asn
Ser Tyr Val Asn Lys Lys Gly Lys Glu Val Leu Val 180
185 190Leu Trp Gly Ile His His Pro Ser Asn Ser Lys
Asp Gln Gln Asn Ile 195 200 205Tyr
Gln Asn Glu Asn Ala Tyr Val Ser Val Val Thr Ser Asn Tyr Asn 210
215 220Arg Arg Phe Thr Pro Glu Ile Ala Glu Arg
Pro Lys Val Arg Asp Gln225 230 235
240Ala Gly Arg Met Asn Tyr Tyr Trp Thr Leu Leu Lys Pro Gly Asp
Thr 245 250 255Ile Ile Phe
Glu Ala Asn Gly Asn Leu Ile Ala Pro Arg Tyr Ala Phe 260
265 270Ala Leu Ser Arg Gly Phe Gly Ser Gly Ile
Ile Thr Ser Asn Ala Ser 275 280
285Met His Glu Cys Asn Thr Lys Cys Gln Thr Pro Leu Gly Ala Ile Asn 290
295 300Ser Ser Leu Pro Phe Gln Asn Ile
His Pro Val Thr Ile Gly Glu Cys305 310
315 320Pro Lys Tyr Val Arg Ser Ala Lys Leu Arg Met Val
Thr Gly Leu Arg 325 330
335Asn Ile Pro Ser Ile Gln Ser Arg Gly Leu Phe Gly Ala Ile Ala Gly
340 345 350Phe Ile Glu Gly Gly Trp
Thr Gly Met Ile Asp Gly Trp Tyr Gly Tyr 355 360
365His His Gln Asn Glu Gln Gly Ser Gly Tyr Ala Ala Asp Gln
Lys Ser 370 375 380Thr Gln Asn Ala Ile
Asn Gly Ile Thr Asn Lys Val Asn Ser Val Ile385 390
395 400Glu Lys Met Asn Ile Gln Phe Thr Ala Val
Gly Lys Glu Phe Asn Lys 405 410
415Leu Glu Lys Arg Met Glu Asn Leu Asn Lys Lys Val Asp Asp Gly Phe
420 425 430Leu Asp Ile Trp Thr
Tyr Asn Ala Glu Leu Leu Val Leu Leu Glu Asn 435
440 445Glu Arg Thr Leu Asp Phe His Asp Ser Asn Val Lys
Asn Leu Tyr Glu 450 455 460Lys Val Lys
Ser Gln Leu Lys Asn Asn Ala Lys Glu Ile Gly Asn Gly465
470 475 480Cys Phe Glu Phe Tyr His Lys
Cys Asp Asn Glu Cys Met Glu Ser Val 485
490 495Arg Asn Gly Thr Tyr Asp Tyr Pro Lys Tyr Ser Glu
Glu Ser Lys Leu 500 505 510Asn
Arg Glu Lys Val Asp Gly Val Lys Leu Glu Ser Met Gly Ile Tyr 515
520 525Gln Ile Leu Ala Ile Tyr Ser Thr Val
Ala Ser Ser Leu Val Leu Leu 530 535
540Val Ser Leu Gly Ala Ile Ser Phe Trp Met Cys Ser Asn Gly Ser Leu545
550 555 560Gln Cys Arg Ile
Cys Ile 5659498PRTInfluenza A virus 9Met Ala Ser Gln Gly
Thr Lys Arg Ser Tyr Glu Gln Met Glu Thr Gly1 5
10 15Gly Glu Arg Gln Asn Ala Thr Glu Ile Arg Ala
Ser Val Gly Arg Met 20 25
30Val Gly Gly Ile Gly Arg Phe Tyr Ile Gln Met Cys Thr Glu Leu Gln
35 40 45Leu Ser Asp Tyr Glu Gly Arg Leu
Ile Gln Asn Ser Ile Thr Ile Glu 50 55
60Arg Met Val Leu Ser Ala Phe Asp Glu Arg Arg Asn Lys Tyr Leu Glu65
70 75 80Glu His Pro Ser Ala
Gly Lys Asp Pro Lys Lys Thr Gly Gly Pro Ile 85
90 95Tyr Lys Lys Arg Asp Gly Lys Trp Met Arg Glu
Leu Ile Leu Tyr Asp 100 105
110Lys Asp Glu Ile Arg Arg Ile Trp Arg Gln Ala Asn Asn Gly Glu Asp
115 120 125Ala Thr Ala Gly Leu Thr His
Leu Met Ile Trp His Ser Asn Leu Asn 130 135
140Asp Ala Thr Tyr Gln Arg Thr Arg Ala Leu Val Arg Thr Gly Met
Asp145 150 155 160Pro Arg
Met Cys Ser Leu Met Gln Gly Ser Thr Leu Pro Arg Arg Ser
165 170 175Gly Ala Ala Gly Ala Ala Val
Lys Gly Ile Gly Thr Met Val Met Glu 180 185
190Leu Ile Arg Met Ile Lys Arg Gly Ile Asn Asp Arg Asn Phe
Trp Arg 195 200 205Gly Glu Asn Gly
Arg Arg Thr Arg Ile Ala Tyr Glu Arg Met Cys Asn 210
215 220Ile Leu Lys Gly Lys Phe Gln Thr Ala Ala Gln Arg
Ala Met Met Asp225 230 235
240Gln Val Arg Glu Ser Arg Asn Pro Gly Asn Ala Glu Ile Glu Asp Leu
245 250 255Ile Phe Leu Ala Arg
Ser Ala Leu Ile Leu Arg Gly Ser Val Ala His 260
265 270Lys Ser Cys Leu Pro Ala Cys Ile Tyr Gly Leu Val
Val Ala Ser Gly 275 280 285Tyr Asp
Phe Glu Arg Glu Gly Tyr Ser Leu Val Gly Ile Asp Pro Phe 290
295 300Arg Leu Leu Gln Asn Ser Gln Val Phe Ser Leu
Ile Arg Pro Asn Glu305 310 315
320Asn Pro Val His Lys Ser Gln Leu Ile Trp Met Ala Cys His Ser Ala
325 330 335Ala Phe Glu Asp
Leu Arg Val Ser Ser Phe Ile Arg Gly Thr Lys Val 340
345 350Val Pro Arg Gly Gln Leu Thr Thr Arg Gly Val
Gln Ile Ala Ser Asn 355 360 365Glu
Asn Met Glu Thr Met Asp Ser Ile Thr Leu Glu Leu Arg Ser Lys 370
375 380Tyr Trp Ala Ile Arg Thr Arg Ser Gly Gly
Asn Thr Asn Gln Gln Arg385 390 395
400Ala Ser Ala Gly Gln Ile Ser Val Gln Pro Thr Phe Ser Val Gln
Arg 405 410 415Asn Leu Pro
Phe Glu Arg Ala Thr Ile Met Ala Ala Phe Thr Gly Asn 420
425 430Asn Glu Gly Arg Thr Ser Asp Met Arg Thr
Glu Ile Ile Arg Met Met 435 440
445Glu Ser Ala Arg Pro Asp Asp Val Ser Phe Gln Gly Arg Gly Val Phe 450
455 460Glu Leu Ser Asp Glu Lys Ala Thr
Asn Pro Ile Val Pro Ser Phe Asp465 470
475 480Met Ser Asn Glu Gly Ser Tyr Phe Phe Gly Asp Asn
Ala Glu Glu Tyr 485 490
495Asp Asn10114PRTMus sp. 10Glu Val Gln Leu Gln Gln Ser Gly Ala Glu Leu
Val Lys Ala Gly Ser1 5 10
15Ser Val Lys Met Ser Cys Lys Ala Thr Gly Tyr Thr Phe Ser Ser Tyr
20 25 30Glu Leu Tyr Trp Val Arg Gln
Ala Pro Gly Gln Gly Leu Glu Asp Leu 35 40
45Gly Tyr Ile Ser Ser Ser Ser Ala Tyr Pro Asn Tyr Ala Gln Lys
Phe 50 55 60Gln Gly Arg Val Thr Ile
Thr Ala Asp Glu Ser Thr Asn Thr Ala Tyr65 70
75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr
Ala Val Tyr Phe Cys 85 90
95Ala Val Arg Val Ile Ser Arg Tyr Phe Asp Gly Trp Gly Gln Gly Thr
100 105 110Leu Val11252PRTInfluenza A
virus 11Met Ser Leu Leu Thr Glu Val Glu Thr Tyr Val Leu Ser Ile Ile Pro1
5 10 15Ser Gly Pro Leu
Lys Ala Glu Ile Ala Gln Arg Leu Glu Asp Val Phe 20
25 30Ala Gly Lys Asn Thr Asp Leu Glu Val Leu Met
Glu Trp Leu Lys Thr 35 40 45Arg
Pro Ile Leu Ser Pro Leu Thr Lys Gly Val Leu Gly Phe Val Phe 50
55 60Thr Leu Thr Val Pro Ser Glu Arg Gly Leu
Gln Arg Arg Arg Phe Val65 70 75
80Gln Asn Ala Leu Asn Gly Asn Gly Asp Pro Asn Asn Met Asp Lys
Ala 85 90 95Val Lys Leu
Tyr Arg Lys Leu Lys Arg Glu Ile Thr Phe Tyr Gly Ala 100
105 110Lys Glu Val Ala Leu Ser Tyr Ser Thr Gly
Ala Leu Ala Ser Cys Met 115 120
125Gly Leu Ile Tyr Asn Arg Met Gly Thr Val Thr Thr Glu Val Ala Phe 130
135 140Gly Leu Val Cys Ala Thr Cys Glu
Gln Ile Ala Asp Ser Gln His Arg145 150
155 160Ser His Arg Gln Met Val Thr Thr Thr Asn Pro Leu
Ile Arg His Glu 165 170
175Asn Arg Met Val Leu Ala Ser Thr Thr Ala Lys Ala Met Glu Gln Met
180 185 190Ala Gly Ser Ser Glu Gln
Ala Ala Glu Ala Met Glu Val Ala Ser Gln 195 200
205Ala Arg Gln Met Val Gln Ala Met Arg Thr Val Gly Thr His
Pro Ser 210 215 220Ser Ser Ala Gly Leu
Lys Asp Asp Leu Leu Glu Asn Leu Gln Ala Tyr225 230
235 240Gln Lys Arg Met Gly Val Gln Leu Gln Arg
Phe Lys 245 2501297PRTInfluenza A virus
12Met Ser Leu Leu Thr Glu Val Glu Thr Pro Thr Arg Asn Gly Trp Glu1
5 10 15Cys Ser Cys Ser Asp Ser
Ser Asp Pro Leu Val Ile Ala Ala Ser Ile 20 25
30Ile Gly Ile Leu His Phe Ile Leu Trp Ile Leu Asp Arg
Leu Phe Phe 35 40 45Lys Cys Ile
Tyr Arg Arg Leu Lys Tyr Gly Leu Lys Arg Gly Pro Ser 50
55 60Thr Glu Gly Val Pro Lys Ser Met Arg Glu Glu Tyr
Arg Gln Glu Gln65 70 75
80Gln Asn Ala Val Asp Val Asp Asp Gly His Phe Val Asn Ile Glu Leu
85 90 95Glu13181PRTHepatitis C
virus 13Ala Pro Ile Thr Ala Tyr Ala Gln Gln Thr Arg Gly Leu Leu Gly Cys1
5 10 15Ile Ile Thr Ser
Leu Thr Gly Arg Asp Arg Asn Gln Val Glu Gly Glu 20
25 30Val Gln Val Val Ser Thr Ala Thr Gln Ser Phe
Leu Ala Thr Cys Ile 35 40 45Asn
Gly Val Cys Trp Thr Val Phe His Gly Ala Gly Ser Lys Thr Leu 50
55 60Ala Gly Pro Lys Gly Pro Ile Thr Gln Met
Tyr Thr Asn Val Asp Gln65 70 75
80Asp Leu Val Gly Trp Pro Ala Pro Pro Gly Ala Arg Ser Leu Thr
Pro 85 90 95Cys Thr Cys
Gly Ser Ser Asp Leu Tyr Leu Val Thr Arg His Ala Asp 100
105 110Val Val Pro Val Arg Arg Arg Ser Asp Ser
Arg Gly Ser Leu Leu Ser 115 120
125Pro Arg Pro Ile Ser Tyr Leu Lys Gly Ser Ser Gly Gly Pro Leu Leu 130
135 140Cys Pro Ser Gly His Ala Val Gly
Ile Phe Arg Ala Ala Val Cys Thr145 150
155 160Arg Gly Val Ala Lys Ala Val Asp Phe Val Pro Val
Glu Ser Met Glu 165 170
175Thr Thr Met Arg Ser 18014139PRTHepatitis B virus 14Met Asp
Ile Asp Pro Tyr Lys Glu Phe Gly Ala Ser Val Glu Leu Leu1 5
10 15Ser Phe Leu Pro Ser Asp Phe Phe
Pro Ser Ile Arg Asp Leu Leu Asp 20 25
30Thr Ala Ser Ala Leu Tyr Arg Glu Ala Leu Glu Ser Pro Glu His
Cys 35 40 45Ser Pro His His Thr
Ala Leu Arg Gln Ala Ile Leu Cys Trp Gly Glu 50 55
60Leu Met Asn Leu Ala Thr Trp Val Gly Ser Asn Leu Glu Asp
Pro Ala65 70 75 80Ser
Arg Glu Leu Val Val Ser Tyr Val Asn Val Asn Met Gly Leu Lys
85 90 95Ile Arg Gln Leu Leu Arg Phe
His Ile Ser Cys Leu Thr Phe Gly Arg 100 105
110Glu Thr Val Leu Glu Tyr Leu Val Ser Phe Gly Val Trp Ile
Arg Thr 115 120 125Pro Pro Ala Tyr
Arg Pro Pro Asn Ala Pro Ile 130 13515105PRTHuman
papillomavirus 15Met His Gly Pro Lys Ala Thr Leu Gln Asp Ile Val Leu His
Leu Glu1 5 10 15Pro Gln
Asn Glu Ile Pro Val Asp Leu Leu Cys His Glu Gln Leu Ser 20
25 30Asp Ser Glu Glu Glu Asn Asp Glu Ile
Asp Gly Val Asn His Gln His 35 40
45Leu Pro Ala Arg Arg Ala Glu Pro Gln Arg His Thr Met Leu Cys Met 50
55 60Cys Cys Lys Cys Glu Ala Arg Ile Lys
Leu Val Val Glu Ser Ser Ala65 70 75
80Asp Asp Leu Arg Ala Phe Gln Gln Leu Phe Leu Asn Thr Leu
Ser Phe 85 90 95Val Cys
Pro Trp Cys Ala Ser Gln Gln 100
1051698PRTHuman papillomavirus 16Met His Gly Asp Thr Pro Thr Leu His Glu
Tyr Met Leu Asp Leu Gln1 5 10
15Pro Glu Thr Thr Asp Leu Tyr Cys Tyr Glu Gln Leu Asn Asp Ser Ser
20 25 30Glu Glu Glu Asp Glu Ile
Asp Gly Pro Ala Gly Gln Ala Glu Pro Asp 35 40
45Arg Ala His Tyr Asn Ile Val Thr Phe Cys Cys Lys Cys Asp
Ser Thr 50 55 60Leu Arg Leu Cys Val
Gln Ser Thr His Val Asp Ile Arg Thr Leu Glu65 70
75 80Asp Leu Leu Met Gly Thr Leu Gly Ile Val
Cys Pro Ile Cys Ser Gln 85 90
95Lys Pro17158PRTHuman papillomavirus 17Met Ala Arg Phe Glu Asp Pro
Thr Arg Arg Pro Tyr Lys Leu Pro Asp1 5 10
15Leu Cys Thr Glu Leu Asn Thr Ser Leu Gln Asp Ile Glu
Ile Thr Cys 20 25 30Val Tyr
Cys Lys Thr Val Leu Glu Leu Thr Glu Val Phe Glu Phe Ala 35
40 45Phe Lys Asp Leu Phe Val Val Tyr Arg Asp
Ser Ile Pro His Ala Ala 50 55 60Cys
His Lys Cys Ile Asp Phe Tyr Ser Arg Ile Arg Glu Leu Arg His65
70 75 80Tyr Ser Asp Ser Val Tyr
Gly Asp Thr Leu Glu Lys Leu Thr Asn Thr 85
90 95Gly Leu Tyr Asn Leu Leu Ile Arg Cys Leu Arg Cys
Gln Lys Pro Leu 100 105 110Asn
Pro Ala Glu Lys Leu Arg His Leu Asn Glu Lys Arg Arg Phe His 115
120 125Lys Ile Ala Gly His Tyr Arg Gly Gln
Cys His Ser Cys Cys Asn Arg 130 135
140Ala Arg Gln Glu Arg Leu Gln Arg Arg Arg Glu Thr Gln Val145
150 15518158PRTHuman papillomavirus 18Met His Gln Lys
Arg Thr Ala Met Phe Gln Asp Pro Gln Glu Arg Pro1 5
10 15Arg Lys Leu Pro His Leu Cys Thr Glu Leu
Gln Thr Thr Ile His Asp 20 25
30Ile Ile Leu Glu Cys Val Tyr Cys Lys Gln Gln Leu Leu Arg Arg Glu
35 40 45Val Tyr Asp Phe Ala Phe Arg Asp
Leu Cys Ile Val Tyr Arg Asp Gly 50 55
60Asn Pro Tyr Ala Val Cys Asp Lys Cys Leu Lys Phe Tyr Ser Lys Ile65
70 75 80Ser Glu Tyr Arg Tyr
Tyr Cys Tyr Ser Val Tyr Gly Thr Thr Leu Glu 85
90 95Gln Gln Tyr Asn Lys Pro Leu Cys Asp Leu Leu
Ile Arg Cys Ile Asn 100 105
110Cys Gln Lys Pro Leu Cys Pro Glu Glu Lys Gln Arg His Leu Asp Lys
115 120 125Lys Gln Arg Phe His Asn Ile
Arg Gly Arg Trp Thr Gly Arg Cys Met 130 135
140Ser Cys Cys Arg Ser Ser Arg Thr Arg Arg Glu Thr Gln Leu145
150 15519661PRTHomo sapiens 19Met Asp Leu Val
Leu Lys Arg Cys Leu Leu His Leu Ala Val Ile Gly1 5
10 15Ala Leu Leu Ala Val Gly Ala Thr Lys Val
Pro Arg Asn Gln Asp Trp 20 25
30Leu Gly Val Ser Arg Gln Leu Arg Thr Lys Ala Trp Asn Arg Gln Leu
35 40 45Tyr Pro Glu Trp Thr Glu Ala Gln
Arg Leu Asp Cys Trp Arg Gly Gly 50 55
60Gln Val Ser Leu Lys Val Ser Asn Asp Gly Pro Thr Leu Ile Gly Ala65
70 75 80Asn Ala Ser Phe Ser
Ile Ala Leu Asn Phe Pro Gly Ser Gln Lys Val 85
90 95Leu Pro Asp Gly Gln Val Ile Trp Val Asn Asn
Thr Ile Ile Asn Gly 100 105
110Ser Gln Val Trp Gly Gly Gln Pro Val Tyr Pro Gln Glu Thr Asp Asp
115 120 125Ala Cys Ile Phe Pro Asp Gly
Gly Pro Cys Pro Ser Gly Ser Trp Ser 130 135
140Gln Lys Arg Ser Phe Val Tyr Val Trp Lys Thr Trp Gly Gln Tyr
Trp145 150 155 160Gln Val
Leu Gly Gly Pro Val Ser Gly Leu Ser Ile Gly Thr Gly Arg
165 170 175Ala Met Leu Gly Thr His Thr
Met Glu Val Thr Val Tyr His Arg Arg 180 185
190Gly Ser Arg Ser Tyr Val Pro Leu Ala His Ser Ser Ser Ala
Phe Thr 195 200 205Ile Thr Asp Gln
Val Pro Phe Ser Val Ser Val Ser Gln Leu Arg Ala 210
215 220Leu Asp Gly Gly Asn Lys His Phe Leu Arg Asn Gln
Pro Leu Thr Phe225 230 235
240Ala Leu Gln Leu His Asp Pro Ser Gly Tyr Leu Ala Glu Ala Asp Leu
245 250 255Ser Tyr Thr Trp Asp
Phe Gly Asp Ser Ser Gly Thr Leu Ile Ser Arg 260
265 270Ala Leu Val Val Thr His Thr Tyr Leu Glu Pro Gly
Pro Val Thr Ala 275 280 285Gln Val
Val Leu Gln Ala Ala Ile Pro Leu Thr Ser Cys Gly Ser Ser 290
295 300Pro Val Pro Gly Thr Thr Asp Gly His Arg Pro
Thr Ala Glu Ala Pro305 310 315
320Asn Thr Thr Ala Gly Gln Val Pro Thr Thr Glu Val Val Gly Thr Thr
325 330 335Pro Gly Gln Ala
Pro Thr Ala Glu Pro Ser Gly Thr Thr Ser Val Gln 340
345 350Val Pro Thr Thr Glu Val Ile Ser Thr Ala Pro
Val Gln Met Pro Thr 355 360 365Ala
Glu Ser Thr Gly Met Thr Pro Glu Lys Val Pro Val Ser Glu Val 370
375 380Met Gly Thr Thr Leu Ala Glu Met Ser Thr
Pro Glu Ala Thr Gly Met385 390 395
400Thr Pro Ala Glu Val Ser Ile Val Val Leu Ser Gly Thr Thr Ala
Ala 405 410 415Gln Val Thr
Thr Thr Glu Trp Val Glu Thr Thr Ala Arg Glu Leu Pro 420
425 430Ile Pro Glu Pro Glu Gly Pro Asp Ala Ser
Ser Ile Met Ser Thr Glu 435 440
445Ser Ile Thr Gly Ser Leu Gly Pro Leu Leu Asp Gly Thr Ala Thr Leu 450
455 460Arg Leu Val Lys Arg Gln Val Pro
Leu Asp Cys Val Leu Tyr Arg Tyr465 470
475 480Gly Ser Phe Ser Val Thr Leu Asp Ile Val Gln Gly
Ile Glu Ser Ala 485 490
495Glu Ile Leu Gln Ala Val Pro Ser Gly Glu Gly Asp Ala Phe Glu Leu
500 505 510Thr Val Ser Cys Gln Gly
Gly Leu Pro Lys Glu Ala Cys Met Glu Ile 515 520
525Ser Ser Pro Gly Cys Gln Pro Pro Ala Gln Arg Leu Cys Gln
Pro Val 530 535 540Leu Pro Ser Pro Ala
Cys Gln Leu Val Leu His Gln Ile Leu Lys Gly545 550
555 560Gly Ser Gly Thr Tyr Cys Leu Asn Val Ser
Leu Ala Asp Thr Asn Ser 565 570
575Leu Ala Val Val Ser Thr Gln Leu Ile Met Pro Gly Gln Glu Ala Gly
580 585 590Leu Gly Gln Val Pro
Leu Ile Val Gly Ile Leu Leu Val Leu Met Ala 595
600 605Val Val Leu Ala Ser Leu Ile Tyr Arg Arg Arg Leu
Met Lys Gln Asp 610 615 620Phe Ser Val
Pro Gln Leu Pro His Ser Ser Ser His Trp Leu Arg Leu625
630 635 640Pro Arg Ile Phe Cys Ser Cys
Pro Ile Gly Glu Asn Ser Pro Leu Leu 645
650 655Ser Gly Gln Gln Val 66020118PRTHomo
sapiens 20Met Pro Arg Glu Asp Ala His Phe Ile Tyr Gly Tyr Pro Lys Lys
Gly1 5 10 15His Gly His
Ser Tyr Thr Thr Ala Glu Glu Ala Ala Gly Ile Gly Ile 20
25 30Leu Thr Val Ile Leu Gly Val Leu Leu Leu
Ile Gly Cys Trp Tyr Cys 35 40
45Arg Arg Arg Asn Gly Tyr Arg Ala Leu Met Asp Lys Ser Leu His Val 50
55 60Gly Thr Gln Cys Ala Leu Thr Arg Arg
Cys Pro Gln Glu Gly Phe Asp65 70 75
80His Arg Asp Ser Lys Val Ser Leu Gln Glu Lys Asn Cys Glu
Pro Val 85 90 95Val Pro
Asn Ala Pro Pro Ala Tyr Glu Lys Leu Ser Ala Glu Gln Ser 100
105 110Pro Pro Pro Tyr Ser Pro
11521237PRTHomo sapiens 21Met Ser Pro Leu Trp Trp Gly Phe Leu Leu Ser Cys
Leu Gly Cys Lys1 5 10
15Ile Leu Pro Gly Ala Gln Gly Gln Phe Pro Arg Val Cys Met Thr Val
20 25 30Asp Ser Leu Val Asn Lys Glu
Cys Cys Pro Arg Leu Gly Ala Glu Ser 35 40
45Ala Asn Val Cys Gly Ser Gln Gln Gly Arg Gly Gln Cys Thr Glu
Val 50 55 60Arg Ala Asp Thr Arg Pro
Trp Ser Gly Pro Tyr Ile Leu Arg Asn Gln65 70
75 80Asp Asp Arg Glu Leu Trp Pro Arg Lys Phe Phe
His Arg Thr Cys Lys 85 90
95Cys Thr Gly Asn Phe Ala Gly Tyr Asn Cys Gly Asp Cys Lys Phe Gly
100 105 110Trp Thr Gly Pro Asn Cys
Glu Arg Lys Lys Pro Pro Val Ile Arg Gln 115 120
125Asn Ile His Ser Leu Ser Pro Gln Glu Arg Glu Gln Phe Leu
Gly Ala 130 135 140Leu Asp Leu Ala Lys
Lys Arg Val His Pro Asp Tyr Val Ile Thr Thr145 150
155 160Gln His Trp Val Gly Leu Leu Gly Pro Asn
Gly Thr Gln Pro Gln Phe 165 170
175Ala Asn Cys Ser Val Tyr Asp Phe Phe Val Trp Leu His Tyr Tyr Ser
180 185 190Val Arg Asp Thr Leu
Leu Gly Gly Phe Phe Pro Trp Leu Lys Val Tyr 195
200 205Tyr Tyr Arg Phe Val Ile Gly Leu Arg Val Trp Gln
Trp Glu Val Ile 210 215 220Ser Cys Lys
Leu Ile Lys Arg Ala Thr Thr Arg Gln Pro225 230
23522337PRTHomo sapiens 22Met Asp Leu Ser Arg Pro Arg Trp Ser Leu
Trp Arg Arg Val Phe Leu1 5 10
15Met Ala Ser Leu Leu Ala Cys Gly Ile Cys Gln Ala Ser Gly Gln Ile
20 25 30Phe Ile Thr Gln Thr Leu
Gly Ile Lys Gly Tyr Arg Thr Val Val Ala 35 40
45Leu Asp Lys Val Pro Glu Asp Val Gln Glu Tyr Ser Trp Tyr
Trp Gly 50 55 60Ala Asn Asp Ser Ala
Gly Asn Met Ile Ile Ser His Lys Pro Pro Ser65 70
75 80Ala Gln Gln Pro Gly Pro Met Tyr Thr Gly
Arg Glu Arg Val Asn Arg 85 90
95Glu Gly Ser Leu Leu Ile Arg Pro Thr Ala Leu Asn Asp Thr Gly Asn
100 105 110Tyr Thr Val Arg Val
Val Ala Gly Asn Glu Thr Gln Arg Ala Thr Gly 115
120 125Trp Leu Glu Val Leu Glu Leu Gly Ser Asn Leu Gly
Ile Ser Val Asn 130 135 140Ala Ser Ser
Leu Val Glu Asn Met Asp Ser Val Ala Ala Asp Cys Leu145
150 155 160Thr Asn Val Thr Asn Ile Thr
Trp Tyr Val Asn Asp Val Pro Thr Ser 165
170 175Ser Ser Asp Arg Met Thr Ile Ser Pro Asp Gly Lys
Thr Leu Val Ile 180 185 190Leu
Arg Val Ser Arg Tyr Asp Arg Thr Ile Gln Cys Met Ile Glu Ser 195
200 205Phe Pro Glu Ile Phe Gln Arg Ser Glu
Arg Ile Ser Leu Thr Val Ala 210 215
220Tyr Gly Pro Asp Tyr Val Leu Leu Arg Ser Asn Pro Asp Asp Phe Asn225
230 235 240Gly Ile Val Thr
Ala Glu Ile Gly Ser Gln Val Glu Met Glu Cys Ile 245
250 255Cys Tyr Ser Phe Leu Asp Leu Lys Tyr His
Trp Ile His Asn Gly Ser 260 265
270Leu Leu Asn Phe Ser Asp Ala Lys Met Asn Leu Ser Ser Leu Ala Trp
275 280 285Glu Gln Met Gly Arg Tyr Arg
Cys Thr Val Glu Asn Pro Val Thr Gln 290 295
300Leu Ile Met Tyr Met Asp Val Arg Ile Gln Ala Pro His Glu Cys
Ser305 310 315 320Ser Ser
Pro Pro Gly Ser Cys Phe Ala His Leu Pro Ala Ser Met Pro
325 330 335Cys231255PRTHomo sapiens 23Met
Glu Leu Ala Ala Leu Cys Arg Trp Gly Leu Leu Leu Ala Leu Leu1
5 10 15Pro Pro Gly Ala Ala Ser Thr
Gln Val Cys Thr Gly Thr Asp Met Lys 20 25
30Leu Arg Leu Pro Ala Ser Pro Glu Thr His Leu Asp Met Leu
Arg His 35 40 45Leu Tyr Gln Gly
Cys Gln Val Val Gln Gly Asn Leu Glu Leu Thr Tyr 50 55
60Leu Pro Thr Asn Ala Ser Leu Ser Phe Leu Gln Asp Ile
Gln Glu Val65 70 75
80Gln Gly Tyr Val Leu Ile Ala His Asn Gln Val Arg Gln Val Pro Leu
85 90 95Gln Arg Leu Arg Ile Val
Arg Gly Thr Gln Leu Phe Glu Asp Asn Tyr 100
105 110Ala Leu Ala Val Leu Asp Asn Gly Asp Pro Leu Asn
Asn Thr Thr Pro 115 120 125Val Thr
Gly Ala Ser Pro Gly Gly Leu Arg Glu Leu Gln Leu Arg Ser 130
135 140Leu Thr Glu Ile Leu Lys Gly Gly Val Leu Ile
Gln Arg Asn Pro Gln145 150 155
160Leu Cys Tyr Gln Asp Thr Ile Leu Trp Lys Asp Ile Phe His Lys Asn
165 170 175Asn Gln Leu Ala
Leu Thr Leu Ile Asp Thr Asn Arg Ser Arg Ala Cys 180
185 190His Pro Cys Ser Pro Met Cys Lys Gly Ser Arg
Cys Trp Gly Glu Ser 195 200 205Ser
Glu Asp Cys Gln Ser Leu Thr Arg Thr Val Cys Ala Gly Gly Cys 210
215 220Ala Arg Cys Lys Gly Pro Leu Pro Thr Asp
Cys Cys His Glu Gln Cys225 230 235
240Ala Ala Gly Cys Thr Gly Pro Lys His Ser Asp Cys Leu Ala Cys
Leu 245 250 255His Phe Asn
His Ser Gly Ile Cys Glu Leu His Cys Pro Ala Leu Val 260
265 270Thr Tyr Asn Thr Asp Thr Phe Glu Ser Met
Pro Asn Pro Glu Gly Arg 275 280
285Tyr Thr Phe Gly Ala Ser Cys Val Thr Ala Cys Pro Tyr Asn Tyr Leu 290
295 300Ser Thr Asp Val Gly Ser Cys Thr
Leu Val Cys Pro Leu His Asn Gln305 310
315 320Glu Val Thr Ala Glu Asp Gly Thr Gln Arg Cys Glu
Lys Cys Ser Lys 325 330
335Pro Cys Ala Arg Val Cys Tyr Gly Leu Gly Met Glu His Leu Arg Glu
340 345 350Val Arg Ala Val Thr Ser
Ala Asn Ile Gln Glu Phe Ala Gly Cys Lys 355 360
365Lys Ile Phe Gly Ser Leu Ala Phe Leu Pro Glu Ser Phe Asp
Gly Asp 370 375 380Pro Ala Ser Asn Thr
Ala Pro Leu Gln Pro Glu Gln Leu Gln Val Phe385 390
395 400Glu Thr Leu Glu Glu Ile Thr Gly Tyr Leu
Tyr Ile Ser Ala Trp Pro 405 410
415Asp Ser Leu Pro Asp Leu Ser Val Phe Gln Asn Leu Gln Val Ile Arg
420 425 430Gly Arg Ile Leu His
Asn Gly Ala Tyr Ser Leu Thr Leu Gln Gly Leu 435
440 445Gly Ile Ser Trp Leu Gly Leu Arg Ser Leu Arg Glu
Leu Gly Ser Gly 450 455 460Leu Ala Leu
Ile His His Asn Thr His Leu Cys Phe Val His Thr Val465
470 475 480Pro Trp Asp Gln Leu Phe Arg
Asn Pro His Gln Ala Leu Leu His Thr 485
490 495Ala Asn Arg Pro Glu Asp Glu Cys Val Gly Glu Gly
Leu Ala Cys His 500 505 510Gln
Leu Cys Ala Arg Gly His Cys Trp Gly Pro Gly Pro Thr Gln Cys 515
520 525Val Asn Cys Ser Gln Phe Leu Arg Gly
Gln Glu Cys Val Glu Glu Cys 530 535
540Arg Val Leu Gln Gly Leu Pro Arg Glu Tyr Val Asn Ala Arg His Cys545
550 555 560Leu Pro Cys His
Pro Glu Cys Gln Pro Gln Asn Gly Ser Val Thr Cys 565
570 575Phe Gly Pro Glu Ala Asp Gln Cys Val Ala
Cys Ala His Tyr Lys Asp 580 585
590Pro Pro Phe Cys Val Ala Arg Cys Pro Ser Gly Val Lys Pro Asp Leu
595 600 605Ser Tyr Met Pro Ile Trp Lys
Phe Pro Asp Glu Glu Gly Ala Cys Gln 610 615
620Pro Cys Pro Ile Asn Cys Thr His Ser Cys Val Asp Leu Asp Asp
Lys625 630 635 640Gly Cys
Pro Ala Glu Gln Arg Ala Ser Pro Leu Thr Ser Ile Ile Ser
645 650 655Ala Val Val Gly Ile Leu Leu
Val Val Val Leu Gly Val Val Phe Gly 660 665
670Ile Leu Ile Lys Arg Arg Gln Gln Lys Ile Arg Lys Tyr Thr
Met Arg 675 680 685Arg Leu Leu Gln
Glu Thr Glu Leu Val Glu Pro Leu Thr Pro Ser Gly 690
695 700Ala Met Pro Asn Gln Ala Gln Met Arg Ile Leu Lys
Glu Thr Glu Leu705 710 715
720Arg Lys Val Lys Val Leu Gly Ser Gly Ala Phe Gly Thr Val Tyr Lys
725 730 735Gly Ile Trp Ile Pro
Asp Gly Glu Asn Val Lys Ile Pro Val Ala Ile 740
745 750Lys Val Leu Arg Glu Asn Thr Ser Pro Lys Ala Asn
Lys Glu Ile Leu 755 760 765Asp Glu
Ala Tyr Val Met Ala Gly Val Gly Ser Pro Tyr Val Ser Arg 770
775 780Leu Leu Gly Ile Cys Leu Thr Ser Thr Val Gln
Leu Val Thr Gln Leu785 790 795
800Met Pro Tyr Gly Cys Leu Leu Asp His Val Arg Glu Asn Arg Gly Arg
805 810 815Leu Gly Ser Gln
Asp Leu Leu Asn Trp Cys Met Gln Ile Ala Lys Gly 820
825 830Met Ser Tyr Leu Glu Asp Val Arg Leu Val His
Arg Asp Leu Ala Ala 835 840 845Arg
Asn Val Leu Val Lys Ser Pro Asn His Val Lys Ile Thr Asp Phe 850
855 860Gly Leu Ala Arg Leu Leu Asp Ile Asp Glu
Thr Glu Tyr His Ala Asp865 870 875
880Gly Gly Lys Val Pro Ile Lys Trp Met Ala Leu Glu Ser Ile Leu
Arg 885 890 895Arg Arg Phe
Thr His Gln Ser Asp Val Trp Ser Tyr Gly Val Thr Val 900
905 910Trp Glu Leu Met Thr Phe Gly Ala Lys Pro
Tyr Asp Gly Ile Pro Ala 915 920
925Arg Glu Ile Pro Asp Leu Leu Glu Lys Gly Glu Arg Leu Pro Gln Pro 930
935 940Pro Ile Cys Thr Ile Asp Val Tyr
Met Ile Met Val Lys Cys Trp Met945 950
955 960Ile Asp Ser Glu Cys Arg Pro Arg Phe Arg Glu Leu
Val Ser Glu Phe 965 970
975Ser Arg Met Ala Arg Asp Pro Gln Arg Phe Val Val Ile Gln Asn Glu
980 985 990Asp Leu Gly Pro Ala Ser
Pro Leu Asp Ser Thr Phe Tyr Arg Ser Leu 995 1000
1005Leu Glu Asp Asp Asp Met Gly Asp Leu Val Asp Ala
Glu Glu Tyr 1010 1015 1020Leu Val Pro
Gln Gln Gly Phe Phe Cys Pro Asp Pro Ala Pro Gly 1025
1030 1035Ala Gly Gly Met Val His His Arg His Arg Ser
Ser Ser Thr Arg 1040 1045 1050Ser Gly
Gly Gly Asp Leu Thr Leu Gly Leu Glu Pro Ser Glu Glu 1055
1060 1065Glu Ala Pro Arg Ser Pro Leu Ala Pro Ser
Glu Gly Ala Gly Ser 1070 1075 1080Asp
Val Phe Asp Gly Asp Leu Gly Met Gly Ala Ala Lys Gly Leu 1085
1090 1095Gln Ser Leu Pro Thr His Asp Pro Ser
Pro Leu Gln Arg Tyr Ser 1100 1105
1110Glu Asp Pro Thr Val Pro Leu Pro Ser Glu Thr Asp Gly Tyr Val
1115 1120 1125Ala Pro Leu Thr Cys Ser
Pro Gln Pro Glu Tyr Val Asn Gln Pro 1130 1135
1140Asp Val Arg Pro Gln Pro Pro Ser Pro Arg Glu Gly Pro Leu
Pro 1145 1150 1155Ala Ala Arg Pro Ala
Gly Ala Thr Leu Glu Arg Pro Lys Thr Leu 1160 1165
1170Ser Pro Gly Lys Asn Gly Val Val Lys Asp Val Phe Ala
Phe Gly 1175 1180 1185Gly Ala Val Glu
Asn Pro Glu Tyr Leu Thr Pro Gln Gly Gly Ala 1190
1195 1200Ala Pro Gln Pro His Pro Pro Pro Ala Phe Ser
Pro Ala Phe Asp 1205 1210 1215Asn Leu
Tyr Tyr Trp Asp Gln Asp Pro Pro Glu Arg Gly Ala Pro 1220
1225 1230Pro Ser Thr Phe Lys Gly Thr Pro Thr Ala
Glu Asn Pro Glu Tyr 1235 1240 1245Leu
Gly Leu Asp Val Pro Val 1250
1255249PRTUnknownDescription of Unknown HLA-A2 anchor motif peptide
24Phe Leu Xaa Xaa Xaa Xaa Xaa Xaa Val1 525875PRTClostridium
tetani 25Lys Ile Ile Pro Pro Thr Asn Ile Arg Glu Asn Leu Tyr Asn Arg Thr1
5 10 15Ala Ser Leu Thr
Asp Leu Gly Gly Glu Leu Cys Ile Lys Ile Lys Asn 20
25 30Glu Asp Leu Thr Phe Ile Ala Glu Lys Asn Ser
Phe Ser Glu Glu Pro 35 40 45Phe
Gln Asp Glu Ile Val Ser Tyr Asn Thr Lys Asn Lys Pro Leu Asn 50
55 60Phe Asn Tyr Ser Leu Asp Lys Ile Ile Val
Asp Tyr Asn Leu Gln Ser65 70 75
80Lys Ile Thr Leu Pro Asn Asp Arg Thr Thr Pro Val Thr Lys Gly
Ile 85 90 95Pro Tyr Ala
Pro Glu Tyr Lys Ser Asn Ala Ala Ser Thr Ile Glu Ile 100
105 110His Asn Ile Asp Asp Asn Thr Ile Tyr Gln
Tyr Leu Tyr Ala Gln Lys 115 120
125Ser Pro Thr Thr Leu Gln Arg Ile Thr Met Thr Asn Ser Val Asp Asp 130
135 140Ala Leu Ile Asn Ser Thr Lys Ile
Tyr Ser Tyr Phe Pro Ser Val Ile145 150
155 160Ser Lys Val Asn Gln Gly Ala Gln Gly Ile Leu Phe
Leu Gln Trp Val 165 170
175Arg Asp Ile Ile Asp Asp Phe Thr Asn Glu Ser Ser Gln Lys Thr Thr
180 185 190Ile Asp Lys Ile Ser Asp
Val Ser Thr Ile Val Pro Tyr Ile Gly Pro 195 200
205Ala Leu Asn Ile Val Lys Gln Gly Tyr Glu Gly Asn Phe Ile
Gly Ala 210 215 220Leu Glu Thr Thr Gly
Val Val Leu Leu Leu Glu Tyr Ile Pro Glu Ile225 230
235 240Thr Leu Pro Val Ile Ala Ala Leu Ser Ile
Ala Glu Ser Ser Thr Gln 245 250
255Lys Glu Lys Ile Ile Lys Thr Ile Asp Asn Phe Leu Glu Lys Arg Tyr
260 265 270Glu Lys Trp Ile Glu
Val Tyr Lys Leu Val Lys Ala Lys Trp Leu Gly 275
280 285Thr Val Asn Thr Gln Phe Gln Lys Arg Ser Tyr Gln
Met Tyr Arg Ser 290 295 300Leu Glu Tyr
Gln Val Asp Ala Ile Lys Lys Ile Ile Asp Tyr Glu Tyr305
310 315 320Lys Ile Tyr Ser Gly Pro Asp
Lys Glu Gln Ile Ala Asp Glu Ile Asn 325
330 335Asn Leu Lys Asn Lys Leu Glu Glu Lys Ala Asn Lys
Ala Met Ile Asn 340 345 350Ile
Asn Ile Phe Met Arg Glu Ser Ser Arg Ser Phe Leu Val Asn Gln 355
360 365Met Ile Asn Glu Ala Lys Lys Gln Leu
Leu Glu Phe Asp Thr Gln Ser 370 375
380Lys Asn Ile Leu Met Gln Tyr Ile Lys Ala Asn Ser Lys Phe Ile Gly385
390 395 400Ile Thr Glu Leu
Lys Lys Leu Glu Ser Lys Ile Asn Lys Val Phe Ser 405
410 415Thr Pro Ile Pro Phe Ser Tyr Ser Lys Asn
Leu Asp Cys Trp Val Asp 420 425
430Asn Glu Glu Asp Ile Asp Val Ile Leu Lys Lys Ser Thr Ile Leu Asn
435 440 445Leu Asp Ile Asn Asn Asp Ile
Ile Ser Asp Ile Ser Gly Phe Asn Ser 450 455
460Ser Val Ile Thr Tyr Pro Asp Ala Gln Leu Val Pro Gly Ile Asn
Gly465 470 475 480Lys Ala
Ile His Leu Val Asn Asn Glu Ser Ser Glu Val Ile Val His
485 490 495Lys Ala Met Asp Ile Glu Tyr
Asn Asp Met Phe Asn Asn Phe Thr Val 500 505
510Ser Phe Trp Leu Arg Val Pro Lys Val Ser Ala Ser His Leu
Glu Gln 515 520 525Tyr Gly Thr Asn
Glu Tyr Ser Ile Ile Ser Ser Met Lys Lys His Ser 530
535 540Leu Ser Ile Gly Ser Gly Trp Ser Val Ser Leu Lys
Gly Asn Asn Leu545 550 555
560Ile Trp Thr Leu Lys Asp Ser Ala Gly Glu Val Arg Gln Ile Thr Phe
565 570 575Arg Asp Leu Pro Asp
Lys Phe Asn Ala Tyr Leu Ala Asn Lys Trp Val 580
585 590Phe Ile Thr Ile Thr Asn Asp Arg Leu Ser Ser Ala
Asn Leu Tyr Ile 595 600 605Asn Gly
Val Leu Met Gly Ser Ala Glu Ile Thr Gly Leu Gly Ala Ile 610
615 620Arg Glu Asp Asn Asn Ile Thr Leu Lys Leu Asp
Arg Cys Asn Asn Asn625 630 635
640Asn Gln Tyr Val Ser Ile Asp Lys Phe Arg Ile Phe Cys Lys Ala Leu
645 650 655Asn Pro Lys Glu
Ile Glu Lys Leu Tyr Thr Ser Tyr Leu Ser Ile Thr 660
665 670Phe Leu Arg Asp Phe Trp Gly Asn Pro Leu Arg
Tyr Asp Thr Glu Tyr 675 680 685Tyr
Leu Ile Pro Val Ala Ser Ser Ser Lys Asp Val Gln Leu Lys Asn 690
695 700Ile Thr Asp Tyr Met Tyr Leu Thr Asn Ala
Pro Ser Tyr Thr Asn Gly705 710 715
720Lys Leu Asn Ile Tyr Tyr Arg Arg Leu Tyr Asn Gly Leu Lys Phe
Ile 725 730 735Ile Lys Arg
Tyr Thr Pro Asn Asn Glu Ile Asp Ser Phe Val Lys Ser 740
745 750Gly Asp Phe Ile Lys Leu Tyr Val Ser Tyr
Asn Asn Asn Glu His Ile 755 760
765Val Gly Tyr Pro Lys Asp Gly Asn Ala Phe Asn Asn Leu Asp Arg Ile 770
775 780Leu Arg Val Gly Tyr Asn Ala Pro
Gly Ile Pro Leu Tyr Lys Lys Met785 790
795 800Glu Ala Val Lys Leu Arg Asp Leu Lys Thr Tyr Ser
Val Gln Leu Lys 805 810
815Leu Tyr Asp Asp Lys Asn Ala Ser Leu Gly Leu Val Gly Thr His Asn
820 825 830Gly Gln Ile Gly Asn Asp
Pro Asn Arg Asp Ile Leu Ile Ala Ser Asn 835 840
845Trp Tyr Phe Asn His Leu Lys Asp Lys Ile Leu Gly Cys Asp
Trp Tyr 850 855 860Phe Val Pro Thr Asp
Glu Gly Trp Thr Asn Asp865 870
8752615PRTUnknownDescription of Unknown Universal T helper epitope
peptide 26Gln Tyr Ile Lys Ala Asn Ser Lys Phe Ile Gly Ile Thr Glu Leu1
5 10 1527190PRTHuman
immunodeficiency virus 1 27Glu Ile Cys Thr Glu Met Glu Lys Glu Gly Lys
Ile Ser Lys Ile Gly1 5 10
15Pro Glu Asn Pro Tyr Asn Thr Pro Val Phe Ala Ile Lys Lys Lys Asp
20 25 30Ser Thr Lys Trp Arg Lys Leu
Val Asp Phe Arg Glu Leu Asn Lys Arg 35 40
45Thr Gln Asp Phe Trp Glu Val Gln Leu Gly Ile Pro His Pro Ala
Gly 50 55 60Leu Lys Lys Lys Lys Ser
Val Thr Val Leu Asp Val Gly Asp Ala Tyr65 70
75 80Phe Ser Val Pro Leu Asp Lys Asp Phe Arg Lys
Tyr Thr Ala Phe Thr 85 90
95Ile Pro Ser Thr Asn Asn Glu Thr Pro Gly Ile Arg Tyr Gln Tyr Asn
100 105 110Val Leu Pro Gln Gly Trp
Lys Gly Ser Pro Ala Ile Phe Gln Ser Ser 115 120
125Met Thr Lys Ile Leu Glu Pro Phe Arg Lys Gln Asn Pro Glu
Ile Val 130 135 140Ile Tyr Gln Tyr Met
Asp Asp Leu Tyr Ile Gly Ser Asp Leu Glu Ile145 150
155 160Gly Gln His Arg Thr Lys Ile Glu Glu Leu
Arg Gln His Leu Leu Lys 165 170
175Trp Gly Leu Thr Thr Pro Asp Lys Lys His Gln Lys Glu Pro
180 185 1902820PRTUnknownDescription of
Unknown Universal T helper epitope peptide 28Phe Arg Lys Gln Asn Pro
Asp Ile Val Ile Tyr Gln Tyr Met Asp Asp1 5
10 15Leu Tyr Val Gly 2029236PRTHuman
immunodeficiency virus 1 29Trp Asp Arg Leu His Pro Ala Gln Ala Gly Pro
Ile Ala Pro Gly Gln1 5 10
15Ile Arg Glu Pro Arg Gly Ser Asp Ile Ala Gly Thr Thr Ser Thr Leu
20 25 30Gln Glu Gln Ile Thr Trp Met
Thr Asn Asn Pro Pro Ile Pro Val Gly 35 40
45Glu Ile Tyr Lys Arg Trp Ile Ile Leu Gly Leu Asn Lys Ile Val
Arg 50 55 60Met Tyr Ser Pro Val Ser
Ile Leu Asp Ile Lys Gln Gly Pro Lys Glu65 70
75 80Pro Phe Arg Asp Tyr Val Asp Arg Phe Phe Lys
Ala Leu Arg Ala Glu 85 90
95Gln Ala Thr Gln Asp Val Lys Asn Trp Met Thr Asp Thr Leu Leu Val
100 105 110Gln Asn Ala Asn Pro Asp
Cys Lys Ser Ile Leu Arg Gly Leu Gly Pro 115 120
125Gly Ala Ser Leu Glu Glu Met Met Thr Ala Cys Gln Gly Val
Gly Gly 130 135 140Pro Ser His Lys Ala
Arg Val Leu Ala Glu Ala Met Ser Gln Ala Asn145 150
155 160Ser Val Asn Met Met Gln Arg Ser Asn Phe
Lys Gly Pro Lys Arg Thr 165 170
175Val Lys Cys Phe Asn Cys Gly Lys Glu Gly His Ile Ala Arg Asn Cys
180 185 190Arg Ala Pro Arg Lys
Lys Gly Cys Trp Lys Cys Gly Gln Glu Gly His 195
200 205Gln Met Lys Asp Cys Thr Glu Arg Gln Ala Asn Phe
Leu Gly Lys Ile 210 215 220Trp Pro Ser
His Lys Gly Arg Pro Gly Asn Phe Leu225 230
23530110PRTHomo sapiens 30Met Ala Leu Trp Met Arg Leu Leu Pro Leu Leu
Ala Leu Leu Ala Leu1 5 10
15Trp Gly Pro Asp Pro Ala Ala Ala Phe Val Asn Gln His Leu Cys Gly
20 25 30Ser His Leu Val Glu Ala Leu
Tyr Leu Val Cys Gly Glu Arg Gly Phe 35 40
45Phe Tyr Thr Pro Lys Thr Arg Arg Glu Ala Glu Asp Leu Gln Val
Gly 50 55 60Gln Val Glu Leu Gly Gly
Gly Pro Gly Ala Gly Ser Leu Gln Pro Leu65 70
75 80Ala Leu Glu Gly Ser Leu Gln Lys Arg Gly Ile
Val Glu Gln Cys Cys 85 90
95Thr Ser Ile Cys Ser Leu Tyr Gln Leu Glu Asn Tyr Cys Asn 100
105 11031585PRTHomo sapiens 31Met Ala Ser
Pro Gly Ser Gly Phe Trp Ser Phe Gly Ser Glu Asp Gly1 5
10 15Ser Gly Asp Ser Glu Asn Pro Gly Thr
Ala Arg Ala Trp Cys Gln Val 20 25
30Ala Gln Lys Phe Thr Gly Gly Ile Gly Asn Lys Leu Cys Ala Leu Leu
35 40 45Tyr Gly Asp Ala Glu Lys Pro
Ala Glu Ser Gly Gly Ser Gln Pro Pro 50 55
60Arg Ala Ala Ala Arg Lys Ala Ala Cys Ala Cys Asp Gln Lys Pro Cys65
70 75 80Ser Cys Ser Lys
Val Asp Val Asn Tyr Ala Phe Leu His Ala Thr Asp 85
90 95Leu Leu Pro Ala Cys Asp Gly Glu Arg Pro
Thr Leu Ala Phe Leu Gln 100 105
110Asp Val Met Asn Ile Leu Leu Gln Tyr Val Val Lys Ser Phe Asp Arg
115 120 125Ser Thr Lys Val Ile Asp Phe
His Tyr Pro Asn Glu Leu Leu Gln Glu 130 135
140Tyr Asn Trp Glu Leu Ala Asp Gln Pro Gln Asn Leu Glu Glu Ile
Leu145 150 155 160Met His
Cys Gln Thr Thr Leu Lys Tyr Ala Ile Lys Thr Gly His Pro
165 170 175Arg Tyr Phe Asn Gln Leu Ser
Thr Gly Leu Asp Met Val Gly Leu Ala 180 185
190Ala Asp Trp Leu Thr Ser Thr Ala Asn Thr Asn Met Phe Thr
Tyr Glu 195 200 205Ile Ala Pro Val
Phe Val Leu Leu Glu Tyr Val Thr Leu Lys Lys Met 210
215 220Arg Glu Ile Ile Gly Trp Pro Gly Gly Ser Gly Asp
Gly Ile Phe Ser225 230 235
240Pro Gly Gly Ala Ile Ser Asn Met Tyr Ala Met Met Ile Ala Arg Phe
245 250 255Lys Met Phe Pro Glu
Val Lys Glu Lys Gly Met Ala Ala Leu Pro Arg 260
265 270Leu Ile Ala Phe Thr Ser Glu His Ser His Phe Ser
Leu Lys Lys Gly 275 280 285Ala Ala
Ala Leu Gly Ile Gly Thr Asp Ser Val Ile Leu Ile Lys Cys 290
295 300Asp Glu Arg Gly Lys Met Ile Pro Ser Asp Leu
Glu Arg Arg Ile Leu305 310 315
320Glu Ala Lys Gln Lys Gly Phe Val Pro Phe Leu Val Ser Ala Thr Ala
325 330 335Gly Thr Thr Val
Tyr Gly Ala Phe Asp Pro Leu Leu Ala Val Ala Asp 340
345 350Ile Cys Lys Lys Tyr Lys Ile Trp Met His Val
Asp Ala Ala Trp Gly 355 360 365Gly
Gly Leu Leu Met Ser Arg Lys His Lys Trp Lys Leu Ser Gly Val 370
375 380Glu Arg Ala Asn Ser Val Thr Trp Asn Pro
His Lys Met Met Gly Val385 390 395
400Pro Leu Gln Cys Ser Ala Leu Leu Val Arg Glu Glu Gly Leu Met
Gln 405 410 415Asn Cys Asn
Gln Met His Ala Ser Tyr Leu Phe Gln Gln Asp Lys His 420
425 430Tyr Asp Leu Ser Tyr Asp Thr Gly Asp Lys
Ala Leu Gln Cys Gly Arg 435 440
445His Val Asp Val Phe Lys Leu Trp Leu Met Trp Arg Ala Lys Gly Thr 450
455 460Thr Gly Phe Glu Ala His Val Asp
Lys Cys Leu Glu Leu Ala Glu Tyr465 470
475 480Leu Tyr Asn Ile Ile Lys Asn Arg Glu Gly Tyr Glu
Met Val Phe Asp 485 490
495Gly Lys Pro Gln His Thr Asn Val Cys Phe Trp Tyr Ile Pro Pro Ser
500 505 510Leu Arg Thr Leu Glu Asp
Asn Glu Glu Arg Met Ser Arg Leu Ser Lys 515 520
525Val Ala Pro Val Ile Lys Ala Arg Met Met Glu Tyr Gly Thr
Thr Met 530 535 540Val Ser Tyr Gln Pro
Leu Gly Asp Lys Val Asn Phe Phe Arg Met Val545 550
555 560Ile Ser Asn Pro Ala Ala Thr His Gln Asp
Ile Asp Phe Leu Ile Glu 565 570
575Glu Ile Glu Arg Leu Gly Gln Asp Leu 580
5853274PRTUnknownDescription of Unknown Composite self epitope
Insulin-GAD peptide 32Cys Gly Ser His Leu Val Glu Ala Leu Tyr Leu Val Cys
Gly Glu Arg1 5 10 15Gly
Xaa Xaa Xaa Xaa Pro Arg Leu Ile Ala Phe Thr Ser Glu His Ser 20
25 30His Phe Ser Leu Xaa Xaa Xaa Xaa
Leu Tyr Asn Ile Ile Lys Asn Arg 35 40
45Glu Gly Tyr Glu Met Val Phe Xaa Xaa Xaa Xaa Pro Ser Leu Arg Thr
50 55 60Leu Glu Asp Asn Glu Glu Arg Met
Ser Arg65 703354PRTUnknownDescription of Unknown
Composite non-self epitope Tetanus-gp100, MART-1, TRP-2 peptide
33Gln Tyr Ile Lys Ala Asn Ser Lys Phe Ile Gly Ile Thr Glu Leu Xaa1
5 10 15Xaa Xaa Xaa Phe Leu Asp
Gln Val Ala Phe Ser Val Xaa Xaa Xaa Xaa 20 25
30Ala Ala Gly Ile Gly Ile Leu Thr Val Xaa Xaa Xaa Xaa
Ser Val Arg 35 40 45Asp Thr Leu
Leu Gly Gly 50348PRTUnknownDescription of Unknown MHC class
I-restricted peptide 34Ser Ile Ile Asn Phe Glu Lys Leu1
53524PRTArtificial SequenceDescription of Artificial Sequence Synthetic
peptide 35Met Lys His Leu Trp Phe Phe Leu Leu Leu Val Ala Ala Pro Arg
Trp1 5 10 15Val Leu Ser
Gln Val Gln Leu Gln 20369PRTHomo sapiens 36Phe Leu Asp Gln Val
Ala Phe Ser Val1 5379PRTHomo sapiens 37Phe Leu Asp Gln Arg
Val Phe Val Val1 5389PRTHomo sapiens 38Phe Leu Phe Leu Trp
Phe Phe Glu Val1 539661PRTHomo sapiens 39Met Asp Leu Val
Leu Lys Arg Cys Leu Leu His Leu Ala Val Ile Gly1 5
10 15Ala Leu Leu Ala Val Gly Ala Thr Lys Val
Pro Arg Asn Gln Asp Trp 20 25
30Leu Gly Val Ser Arg Gln Leu Arg Thr Lys Ala Trp Asn Arg Gln Leu
35 40 45Tyr Pro Glu Trp Thr Glu Ala Gln
Arg Leu Asp Cys Trp Arg Gly Gly 50 55
60Gln Val Ser Leu Lys Val Ser Asn Asp Gly Pro Thr Leu Ile Gly Ala65
70 75 80Asn Ala Ser Phe Ser
Ile Ala Leu Asn Phe Pro Gly Ser Gln Lys Val 85
90 95Leu Pro Asp Gly Gln Val Ile Trp Val Asn Asn
Thr Ile Ile Asn Gly 100 105
110Ser Gln Val Trp Gly Gly Gln Pro Val Tyr Pro Gln Glu Thr Asp Asp
115 120 125Ala Cys Ile Phe Pro Asp Gly
Gly Pro Cys Pro Ser Gly Ser Trp Ser 130 135
140Gln Lys Arg Ser Phe Val Tyr Val Trp Lys Thr Trp Gly Gln Tyr
Trp145 150 155 160Gln Val
Leu Gly Gly Pro Val Ser Gly Leu Ser Ile Gly Thr Gly Arg
165 170 175Ala Met Leu Gly Thr His Thr
Met Glu Val Thr Val Tyr His Arg Arg 180 185
190Gly Ser Arg Ser Tyr Val Pro Leu Ala His Ser Ser Ser Ala
Phe Thr 195 200 205Phe Leu Asp Gln
Val Ala Phe Ser Val Ser Val Ser Gln Leu Arg Ala 210
215 220Leu Asp Gly Gly Asn Lys His Phe Leu Arg Asn Gln
Pro Leu Thr Phe225 230 235
240Ala Leu Gln Leu His Asp Pro Ser Gly Tyr Leu Ala Glu Ala Asp Leu
245 250 255Ser Tyr Thr Trp Asp
Phe Gly Asp Ser Ser Gly Thr Leu Ile Ser Arg 260
265 270Ala Leu Val Val Thr His Thr Tyr Leu Glu Pro Gly
Pro Val Thr Ala 275 280 285Gln Val
Val Leu Gln Ala Ala Ile Pro Leu Thr Ser Cys Gly Ser Ser 290
295 300Pro Val Pro Gly Thr Thr Asp Gly His Arg Pro
Thr Ala Glu Ala Pro305 310 315
320Asn Thr Thr Ala Gly Gln Val Pro Thr Thr Glu Val Val Gly Thr Thr
325 330 335Pro Gly Gln Ala
Pro Thr Ala Glu Pro Ser Gly Thr Thr Ser Val Gln 340
345 350Val Pro Thr Thr Glu Val Ile Ser Thr Ala Pro
Val Gln Met Pro Thr 355 360 365Ala
Glu Ser Thr Gly Met Thr Pro Glu Lys Val Pro Val Ser Glu Val 370
375 380Met Gly Thr Thr Leu Ala Glu Met Ser Thr
Pro Glu Ala Thr Gly Met385 390 395
400Thr Pro Ala Glu Val Ser Ile Val Val Leu Ser Gly Thr Thr Ala
Ala 405 410 415Gln Val Thr
Thr Thr Glu Trp Val Glu Thr Thr Ala Arg Glu Leu Pro 420
425 430Ile Pro Glu Pro Glu Gly Pro Asp Ala Ser
Ser Ile Met Ser Thr Glu 435 440
445Ser Ile Thr Gly Ser Leu Gly Pro Leu Leu Asp Gly Thr Ala Thr Leu 450
455 460Arg Leu Val Lys Arg Gln Val Pro
Leu Asp Cys Val Leu Tyr Arg Tyr465 470
475 480Gly Ser Phe Ser Val Thr Leu Asp Ile Val Gln Gly
Ile Glu Ser Ala 485 490
495Glu Ile Leu Gln Ala Val Pro Ser Gly Glu Gly Asp Ala Phe Glu Leu
500 505 510Thr Val Ser Cys Gln Gly
Gly Leu Pro Lys Glu Ala Cys Met Glu Ile 515 520
525Ser Ser Pro Gly Cys Gln Pro Pro Ala Gln Arg Leu Cys Gln
Pro Val 530 535 540Leu Pro Ser Pro Ala
Cys Gln Leu Val Leu His Gln Ile Leu Lys Gly545 550
555 560Gly Ser Gly Thr Tyr Cys Leu Asn Val Ser
Leu Ala Asp Thr Asn Ser 565 570
575Leu Ala Val Val Ser Thr Gln Leu Ile Met Pro Gly Gln Glu Ala Gly
580 585 590Leu Gly Gln Val Pro
Leu Ile Val Gly Ile Leu Leu Val Leu Met Ala 595
600 605Val Val Leu Ala Ser Leu Ile Tyr Arg Arg Arg Leu
Met Lys Gln Asp 610 615 620Phe Ser Val
Pro Gln Leu Pro His Ser Ser Ser His Trp Leu Arg Leu625
630 635 640Pro Arg Ile Phe Cys Ser Cys
Pro Ile Gly Glu Asn Ser Pro Leu Leu 645
650 655Ser Gly Gln Gln Val 66040661PRTHomo
sapiens 40Met Asp Leu Val Leu Lys Arg Cys Leu Leu His Leu Ala Val Ile
Gly1 5 10 15Ala Leu Leu
Ala Val Gly Ala Thr Lys Val Pro Arg Asn Gln Asp Trp 20
25 30Leu Gly Val Ser Arg Gln Leu Arg Thr Lys
Ala Trp Asn Arg Gln Leu 35 40
45Tyr Pro Glu Trp Thr Glu Ala Gln Arg Leu Asp Cys Trp Arg Gly Gly 50
55 60Gln Val Ser Leu Lys Val Ser Asn Asp
Gly Pro Thr Leu Ile Gly Ala65 70 75
80Asn Ala Ser Phe Ser Ile Ala Leu Asn Phe Pro Gly Ser Gln
Lys Val 85 90 95Leu Pro
Asp Gly Gln Val Ile Trp Val Asn Asn Thr Ile Ile Asn Gly 100
105 110Ser Gln Val Trp Gly Gly Gln Pro Val
Tyr Pro Gln Glu Thr Asp Asp 115 120
125Ala Cys Ile Phe Pro Asp Gly Gly Pro Cys Pro Ser Gly Ser Trp Ser
130 135 140Gln Lys Arg Ser Phe Val Tyr
Val Trp Lys Thr Trp Gly Gln Tyr Trp145 150
155 160Gln Val Leu Gly Gly Pro Val Ser Gly Leu Ser Ile
Gly Thr Gly Arg 165 170
175Ala Met Leu Gly Thr His Thr Met Glu Val Thr Val Tyr His Arg Arg
180 185 190Gly Ser Arg Ser Tyr Val
Pro Leu Ala His Ser Ser Ser Ala Phe Thr 195 200
205Phe Leu Asp Gln Arg Val Phe Val Val Ser Val Ser Gln Leu
Arg Ala 210 215 220Leu Asp Gly Gly Asn
Lys His Phe Leu Arg Asn Gln Pro Leu Thr Phe225 230
235 240Ala Leu Gln Leu His Asp Pro Ser Gly Tyr
Leu Ala Glu Ala Asp Leu 245 250
255Ser Tyr Thr Trp Asp Phe Gly Asp Ser Ser Gly Thr Leu Ile Ser Arg
260 265 270Ala Leu Val Val Thr
His Thr Tyr Leu Glu Pro Gly Pro Val Thr Ala 275
280 285Gln Val Val Leu Gln Ala Ala Ile Pro Leu Thr Ser
Cys Gly Ser Ser 290 295 300Pro Val Pro
Gly Thr Thr Asp Gly His Arg Pro Thr Ala Glu Ala Pro305
310 315 320Asn Thr Thr Ala Gly Gln Val
Pro Thr Thr Glu Val Val Gly Thr Thr 325
330 335Pro Gly Gln Ala Pro Thr Ala Glu Pro Ser Gly Thr
Thr Ser Val Gln 340 345 350Val
Pro Thr Thr Glu Val Ile Ser Thr Ala Pro Val Gln Met Pro Thr 355
360 365Ala Glu Ser Thr Gly Met Thr Pro Glu
Lys Val Pro Val Ser Glu Val 370 375
380Met Gly Thr Thr Leu Ala Glu Met Ser Thr Pro Glu Ala Thr Gly Met385
390 395 400Thr Pro Ala Glu
Val Ser Ile Val Val Leu Ser Gly Thr Thr Ala Ala 405
410 415Gln Val Thr Thr Thr Glu Trp Val Glu Thr
Thr Ala Arg Glu Leu Pro 420 425
430Ile Pro Glu Pro Glu Gly Pro Asp Ala Ser Ser Ile Met Ser Thr Glu
435 440 445Ser Ile Thr Gly Ser Leu Gly
Pro Leu Leu Asp Gly Thr Ala Thr Leu 450 455
460Arg Leu Val Lys Arg Gln Val Pro Leu Asp Cys Val Leu Tyr Arg
Tyr465 470 475 480Gly Ser
Phe Ser Val Thr Leu Asp Ile Val Gln Gly Ile Glu Ser Ala
485 490 495Glu Ile Leu Gln Ala Val Pro
Ser Gly Glu Gly Asp Ala Phe Glu Leu 500 505
510Thr Val Ser Cys Gln Gly Gly Leu Pro Lys Glu Ala Cys Met
Glu Ile 515 520 525Ser Ser Pro Gly
Cys Gln Pro Pro Ala Gln Arg Leu Cys Gln Pro Val 530
535 540Leu Pro Ser Pro Ala Cys Gln Leu Val Leu His Gln
Ile Leu Lys Gly545 550 555
560Gly Ser Gly Thr Tyr Cys Leu Asn Val Ser Leu Ala Asp Thr Asn Ser
565 570 575Leu Ala Val Val Ser
Thr Gln Leu Ile Met Pro Gly Gln Glu Ala Gly 580
585 590Leu Gly Gln Val Pro Leu Ile Val Gly Ile Leu Leu
Val Leu Met Ala 595 600 605Val Val
Leu Ala Ser Leu Ile Tyr Arg Arg Arg Leu Met Lys Gln Asp 610
615 620Phe Ser Val Pro Gln Leu Pro His Ser Ser Ser
His Trp Leu Arg Leu625 630 635
640Pro Arg Ile Phe Cys Ser Cys Pro Ile Gly Glu Asn Ser Pro Leu Leu
645 650 655Ser Gly Gln Gln
Val 66041661PRTHomo sapiens 41Met Asp Leu Val Leu Lys Arg Cys
Leu Leu His Leu Ala Val Ile Gly1 5 10
15Ala Leu Leu Ala Val Gly Ala Thr Lys Val Pro Arg Asn Gln
Asp Trp 20 25 30Leu Gly Val
Ser Arg Gln Leu Arg Thr Lys Ala Trp Asn Arg Gln Leu 35
40 45Tyr Pro Glu Trp Thr Glu Ala Gln Arg Leu Asp
Cys Trp Arg Gly Gly 50 55 60Gln Val
Ser Leu Lys Val Ser Asn Asp Gly Pro Thr Leu Ile Gly Ala65
70 75 80Asn Ala Ser Phe Ser Ile Ala
Leu Asn Phe Pro Gly Ser Gln Lys Val 85 90
95Leu Pro Asp Gly Gln Val Ile Trp Val Asn Asn Thr Ile
Ile Asn Gly 100 105 110Ser Gln
Val Trp Gly Gly Gln Pro Val Tyr Pro Gln Glu Thr Asp Asp 115
120 125Ala Cys Ile Phe Pro Asp Gly Gly Pro Cys
Pro Ser Gly Ser Trp Ser 130 135 140Gln
Lys Arg Ser Phe Val Tyr Val Trp Lys Thr Trp Gly Gln Tyr Trp145
150 155 160Gln Val Leu Gly Gly Pro
Val Ser Gly Leu Ser Ile Gly Thr Gly Arg 165
170 175Ala Met Leu Gly Thr His Thr Met Glu Val Thr Val
Tyr His Arg Arg 180 185 190Gly
Ser Arg Ser Tyr Val Pro Leu Ala His Ser Ser Ser Ala Phe Thr 195
200 205Phe Leu Phe Leu Trp Phe Phe Glu Val
Ser Val Ser Gln Leu Arg Ala 210 215
220Leu Asp Gly Gly Asn Lys His Phe Leu Arg Asn Gln Pro Leu Thr Phe225
230 235 240Ala Leu Gln Leu
His Asp Pro Ser Gly Tyr Leu Ala Glu Ala Asp Leu 245
250 255Ser Tyr Thr Trp Asp Phe Gly Asp Ser Ser
Gly Thr Leu Ile Ser Arg 260 265
270Ala Leu Val Val Thr His Thr Tyr Leu Glu Pro Gly Pro Val Thr Ala
275 280 285Gln Val Val Leu Gln Ala Ala
Ile Pro Leu Thr Ser Cys Gly Ser Ser 290 295
300Pro Val Pro Gly Thr Thr Asp Gly His Arg Pro Thr Ala Glu Ala
Pro305 310 315 320Asn Thr
Thr Ala Gly Gln Val Pro Thr Thr Glu Val Val Gly Thr Thr
325 330 335Pro Gly Gln Ala Pro Thr Ala
Glu Pro Ser Gly Thr Thr Ser Val Gln 340 345
350Val Pro Thr Thr Glu Val Ile Ser Thr Ala Pro Val Gln Met
Pro Thr 355 360 365Ala Glu Ser Thr
Gly Met Thr Pro Glu Lys Val Pro Val Ser Glu Val 370
375 380Met Gly Thr Thr Leu Ala Glu Met Ser Thr Pro Glu
Ala Thr Gly Met385 390 395
400Thr Pro Ala Glu Val Ser Ile Val Val Leu Ser Gly Thr Thr Ala Ala
405 410 415Gln Val Thr Thr Thr
Glu Trp Val Glu Thr Thr Ala Arg Glu Leu Pro 420
425 430Ile Pro Glu Pro Glu Gly Pro Asp Ala Ser Ser Ile
Met Ser Thr Glu 435 440 445Ser Ile
Thr Gly Ser Leu Gly Pro Leu Leu Asp Gly Thr Ala Thr Leu 450
455 460Arg Leu Val Lys Arg Gln Val Pro Leu Asp Cys
Val Leu Tyr Arg Tyr465 470 475
480Gly Ser Phe Ser Val Thr Leu Asp Ile Val Gln Gly Ile Glu Ser Ala
485 490 495Glu Ile Leu Gln
Ala Val Pro Ser Gly Glu Gly Asp Ala Phe Glu Leu 500
505 510Thr Val Ser Cys Gln Gly Gly Leu Pro Lys Glu
Ala Cys Met Glu Ile 515 520 525Ser
Ser Pro Gly Cys Gln Pro Pro Ala Gln Arg Leu Cys Gln Pro Val 530
535 540Leu Pro Ser Pro Ala Cys Gln Leu Val Leu
His Gln Ile Leu Lys Gly545 550 555
560Gly Ser Gly Thr Tyr Cys Leu Asn Val Ser Leu Ala Asp Thr Asn
Ser 565 570 575Leu Ala Val
Val Ser Thr Gln Leu Ile Met Pro Gly Gln Glu Ala Gly 580
585 590Leu Gly Gln Val Pro Leu Ile Val Gly Ile
Leu Leu Val Leu Met Ala 595 600
605Val Val Leu Ala Ser Leu Ile Tyr Arg Arg Arg Leu Met Lys Gln Asp 610
615 620Phe Ser Val Pro Gln Leu Pro His
Ser Ser Ser His Trp Leu Arg Leu625 630
635 640Pro Arg Ile Phe Cys Ser Cys Pro Ile Gly Glu Asn
Ser Pro Leu Leu 645 650
655Ser Gly Gln Gln Val 660429PRTHomo sapiens 42Leu Leu Pro Gly
Gly Arg Pro Tyr Arg1 5438PRTHomo sapiens 43Ser Val Tyr Asp
Phe Phe Val Trp1 544235PRTHomo sapiens 44Met Ser Pro Leu
Trp Trp Gly Phe Leu Leu Ser Cys Leu Gly Cys Lys1 5
10 15Ile Leu Pro Gly Ala Gln Gly Gln Phe Pro
Arg Val Cys Met Thr Val 20 25
30Asp Ser Leu Val Asn Lys Glu Cys Cys Pro Arg Leu Gly Ala Glu Ser
35 40 45Ala Asn Val Cys Gly Ser Gln Gln
Gly Arg Gly Gln Cys Thr Glu Val 50 55
60Arg Ala Asp Thr Arg Pro Trp Ser Gly Pro Tyr Ile Leu Arg Asn Gln65
70 75 80Asp Asp Arg Glu Leu
Trp Pro Arg Lys Phe Phe His Arg Thr Cys Lys 85
90 95Cys Thr Gly Asn Phe Ala Gly Tyr Asn Cys Gly
Asp Cys Lys Phe Gly 100 105
110Trp Thr Gly Pro Asn Cys Glu Arg Lys Lys Pro Pro Val Ile Arg Gln
115 120 125Asn Ile His Ser Leu Leu Pro
Gly Gly Arg Pro Tyr Arg Leu Gly Ala 130 135
140Leu Asp Leu Ala Lys Lys Arg Val His Pro Asp Tyr Val Ile Thr
Thr145 150 155 160Gln His
Trp Val Gly Leu Leu Gly Pro Asn Gly Thr Gln Pro Gln Phe
165 170 175Ala Asn Cys Ser Val Tyr Asp
Phe Phe Val Trp Leu His Tyr Ser Val 180 185
190Tyr Asp Phe Phe Val Trp Phe Phe Pro Trp Leu Lys Val Tyr
Tyr Tyr 195 200 205Arg Phe Val Ile
Gly Leu Arg Val Trp Gln Trp Glu Val Ile Ser Cys 210
215 220Lys Leu Ile Lys Arg Ala Thr Thr Arg Gln Pro225
230 23545146PRTHomo sapiens 45Met Gly Phe Leu
Arg Arg Leu Ile Tyr Arg Arg Arg Pro Met Ile Tyr1 5
10 15Val Glu Ser Ser Glu Glu Ser Ser Asp Glu
Gln Pro Asp Glu Val Glu 20 25
30Ser Pro Thr Gln Ser Gln Asp Ser Thr Pro Ala Glu Glu Arg Glu Asp
35 40 45Glu Gly Ala Ser Ala Ala Gln Gly
Gln Glu Pro Glu Ala Asp Ser Gln 50 55
60Glu Leu Val Gln Pro Lys Thr Gly Cys Glu Pro Gly Asp Gly Pro Asp65
70 75 80Thr Lys Arg Val Cys
Leu Arg Asn Glu Glu Gln Met Lys Leu Pro Ala 85
90 95Glu Gly Pro Glu Pro Glu Ala Asp Ser Gln Glu
Gln Val His Pro Lys 100 105
110Thr Gly Cys Glu Arg Gly Asp Gly Pro Asp Val Gln Glu Leu Gly Leu
115 120 125Pro Asn Pro Glu Glu Val Lys
Thr Pro Glu Glu Asp Glu Gly Gln Ser 130 135
140Gln Pro14546339PRTHomo sapiens 46Met Glu Ser Arg Lys Asp Ile Thr
Asn Gln Glu Glu Leu Trp Lys Met1 5 10
15Lys Pro Arg Arg Asn Leu Glu Glu Asp Asp Tyr Leu His Lys
Asp Thr 20 25 30Gly Glu Thr
Ser Met Leu Lys Arg Pro Val Leu Leu His Leu His Gln 35
40 45Thr Ala His Ala Asp Glu Phe Asp Cys Pro Ser
Glu Leu Gln His Thr 50 55 60Gln Glu
Leu Phe Pro Gln Trp His Leu Pro Ile Lys Ile Ala Ala Ile65
70 75 80Ile Ala Ser Leu Thr Phe Leu
Tyr Thr Leu Leu Arg Glu Val Ile His 85 90
95Pro Leu Ala Thr Ser His Gln Gln Tyr Phe Tyr Lys Ile
Pro Ile Leu 100 105 110Val Ile
Asn Lys Val Leu Pro Met Val Ser Ile Thr Leu Leu Ala Leu 115
120 125Val Tyr Leu Pro Gly Val Ile Ala Ala Ile
Val Gln Leu His Asn Gly 130 135 140Thr
Lys Tyr Lys Lys Phe Pro His Trp Leu Asp Lys Trp Met Leu Thr145
150 155 160Arg Lys Gln Phe Gly Leu
Leu Ser Phe Phe Phe Ala Val Leu His Ala 165
170 175Ile Tyr Ser Leu Ser Tyr Pro Met Arg Arg Ser Tyr
Arg Tyr Lys Leu 180 185 190Leu
Asn Trp Ala Tyr Gln Gln Val Gln Gln Asn Lys Glu Asp Ala Trp 195
200 205Ile Glu His Asp Val Trp Arg Met Glu
Ile Tyr Val Ser Leu Gly Ile 210 215
220Val Gly Leu Ala Ile Leu Ala Leu Leu Ala Val Thr Ser Ile Pro Ser225
230 235 240Val Ser Asp Ser
Leu Thr Trp Arg Glu Phe His Tyr Ile Gln Ser Lys 245
250 255Leu Gly Ile Val Ser Leu Leu Leu Gly Thr
Ile His Ala Leu Ile Phe 260 265
270Ala Trp Asn Lys Trp Ile Asp Ile Lys Gln Phe Val Trp Tyr Thr Pro
275 280 285Pro Thr Phe Met Ile Ala Val
Phe Leu Pro Ile Val Val Leu Ile Phe 290 295
300Lys Ser Ile Leu Phe Leu Pro Cys Leu Arg Lys Lys Ile Leu Lys
Ile305 310 315 320Arg His
Gly Trp Glu Asp Val Thr Lys Ile Asn Lys Thr Glu Ile Cys
325 330 335Ser Gln Leu47102PRTHomo
sapiens 47Met Ser Ala Arg Val Arg Ser Arg Ser Arg Gly Arg Gly Asp Gly
Gln1 5 10 15Glu Ala Pro
Asp Val Val Ala Phe Val Ala Pro Gly Glu Ser Gln Gln 20
25 30Glu Glu Pro Pro Thr Asp Asn Gln Asp Ile
Glu Pro Gly Gln Glu Arg 35 40
45Glu Gly Thr Pro Pro Ile Glu Glu Arg Lys Val Glu Gly Asp Cys Gln 50
55 60Glu Met Asp Leu Glu Lys Thr Arg Ser
Glu Arg Gly Asp Gly Ser Asp65 70 75
80Val Lys Glu Lys Thr Pro Pro Asn Pro Lys His Ala Lys Thr
Lys Glu 85 90 95Ala Gly
Asp Gly Gln Pro 100481255PRTHomo sapiens 48Met Thr Pro Gly Thr
Gln Ser Pro Phe Phe Leu Leu Leu Leu Leu Thr1 5
10 15Val Leu Thr Val Val Thr Gly Ser Gly His Ala
Ser Ser Thr Pro Gly 20 25
30Gly Glu Lys Glu Thr Ser Ala Thr Gln Arg Ser Ser Val Pro Ser Ser
35 40 45Thr Glu Lys Asn Ala Val Ser Met
Thr Ser Ser Val Leu Ser Ser His 50 55
60Ser Pro Gly Ser Gly Ser Ser Thr Thr Gln Gly Gln Asp Val Thr Leu65
70 75 80Ala Pro Ala Thr Glu
Pro Ala Ser Gly Ser Ala Ala Thr Trp Gly Gln 85
90 95Asp Val Thr Ser Val Pro Val Thr Arg Pro Ala
Leu Gly Ser Thr Thr 100 105
110Pro Pro Ala His Asp Val Thr Ser Ala Pro Asp Asn Lys Pro Ala Pro
115 120 125Gly Ser Thr Ala Pro Pro Ala
His Gly Val Thr Ser Ala Pro Asp Thr 130 135
140Arg Pro Ala Pro Gly Ser Thr Ala Pro Pro Ala His Gly Val Thr
Ser145 150 155 160Ala Pro
Asp Thr Arg Pro Ala Pro Gly Ser Thr Ala Pro Pro Ala His
165 170 175Gly Val Thr Ser Ala Pro Asp
Thr Arg Pro Ala Pro Gly Ser Thr Ala 180 185
190Pro Pro Ala His Gly Val Thr Ser Ala Pro Asp Thr Arg Pro
Ala Pro 195 200 205Gly Ser Thr Ala
Pro Pro Ala His Gly Val Thr Ser Ala Pro Asp Thr 210
215 220Arg Pro Ala Pro Gly Ser Thr Ala Pro Pro Ala His
Gly Val Thr Ser225 230 235
240Ala Pro Asp Thr Arg Pro Ala Pro Gly Ser Thr Ala Pro Pro Ala His
245 250 255Gly Val Thr Ser Ala
Pro Asp Thr Arg Pro Ala Pro Gly Ser Thr Ala 260
265 270Pro Pro Ala His Gly Val Thr Ser Ala Pro Asp Thr
Arg Pro Ala Pro 275 280 285Gly Ser
Thr Ala Pro Pro Ala His Gly Val Thr Ser Ala Pro Asp Thr 290
295 300Arg Pro Ala Pro Gly Ser Thr Ala Pro Pro Ala
His Gly Val Thr Ser305 310 315
320Ala Pro Asp Thr Arg Pro Ala Pro Gly Ser Thr Ala Pro Pro Ala His
325 330 335Gly Val Thr Ser
Ala Pro Asp Thr Arg Pro Ala Pro Gly Ser Thr Ala 340
345 350Pro Pro Ala His Gly Val Thr Ser Ala Pro Asp
Thr Arg Pro Ala Pro 355 360 365Gly
Ser Thr Ala Pro Pro Ala His Gly Val Thr Ser Ala Pro Asp Thr 370
375 380Arg Pro Ala Pro Gly Ser Thr Ala Pro Pro
Ala His Gly Val Thr Ser385 390 395
400Ala Pro Asp Thr Arg Pro Ala Pro Gly Ser Thr Ala Pro Pro Ala
His 405 410 415Gly Val Thr
Ser Ala Pro Asp Thr Arg Pro Ala Pro Gly Ser Thr Ala 420
425 430Pro Pro Ala His Gly Val Thr Ser Ala Pro
Asp Thr Arg Pro Ala Pro 435 440
445Gly Ser Thr Ala Pro Pro Ala His Gly Val Thr Ser Ala Pro Asp Thr 450
455 460Arg Pro Ala Pro Gly Ser Thr Ala
Pro Pro Ala His Gly Val Thr Ser465 470
475 480Ala Pro Asp Thr Arg Pro Ala Pro Gly Ser Thr Ala
Pro Pro Ala His 485 490
495Gly Val Thr Ser Ala Pro Asp Thr Arg Pro Ala Pro Gly Ser Thr Ala
500 505 510Pro Pro Ala His Gly Val
Thr Ser Ala Pro Asp Thr Arg Pro Ala Pro 515 520
525Gly Ser Thr Ala Pro Pro Ala His Gly Val Thr Ser Ala Pro
Asp Thr 530 535 540Arg Pro Ala Pro Gly
Ser Thr Ala Pro Pro Ala His Gly Val Thr Ser545 550
555 560Ala Pro Asp Thr Arg Pro Ala Pro Gly Ser
Thr Ala Pro Pro Ala His 565 570
575Gly Val Thr Ser Ala Pro Asp Thr Arg Pro Ala Pro Gly Ser Thr Ala
580 585 590Pro Pro Ala His Gly
Val Thr Ser Ala Pro Asp Thr Arg Pro Ala Pro 595
600 605Gly Ser Thr Ala Pro Pro Ala His Gly Val Thr Ser
Ala Pro Asp Thr 610 615 620Arg Pro Ala
Pro Gly Ser Thr Ala Pro Pro Ala His Gly Val Thr Ser625
630 635 640Ala Pro Asp Thr Arg Pro Ala
Pro Gly Ser Thr Ala Pro Pro Ala His 645
650 655Gly Val Thr Ser Ala Pro Asp Thr Arg Pro Ala Pro
Gly Ser Thr Ala 660 665 670Pro
Pro Ala His Gly Val Thr Ser Ala Pro Asp Thr Arg Pro Ala Pro 675
680 685Gly Ser Thr Ala Pro Pro Ala His Gly
Val Thr Ser Ala Pro Asp Thr 690 695
700Arg Pro Ala Pro Gly Ser Thr Ala Pro Pro Ala His Gly Val Thr Ser705
710 715 720Ala Pro Asp Thr
Arg Pro Ala Pro Gly Ser Thr Ala Pro Pro Ala His 725
730 735Gly Val Thr Ser Ala Pro Asp Thr Arg Pro
Ala Pro Gly Ser Thr Ala 740 745
750Pro Pro Ala His Gly Val Thr Ser Ala Pro Asp Thr Arg Pro Ala Pro
755 760 765Gly Ser Thr Ala Pro Pro Ala
His Gly Val Thr Ser Ala Pro Asp Thr 770 775
780Arg Pro Ala Pro Gly Ser Thr Ala Pro Pro Ala His Gly Val Thr
Ser785 790 795 800Ala Pro
Asp Thr Arg Pro Ala Pro Gly Ser Thr Ala Pro Pro Ala His
805 810 815Gly Val Thr Ser Ala Pro Asp
Thr Arg Pro Ala Pro Gly Ser Thr Ala 820 825
830Pro Pro Ala His Gly Val Thr Ser Ala Pro Asp Thr Arg Pro
Ala Pro 835 840 845Gly Ser Thr Ala
Pro Pro Ala His Gly Val Thr Ser Ala Pro Asp Thr 850
855 860Arg Pro Ala Pro Gly Ser Thr Ala Pro Pro Ala His
Gly Val Thr Ser865 870 875
880Ala Pro Asp Thr Arg Pro Ala Pro Gly Ser Thr Ala Pro Pro Ala His
885 890 895Gly Val Thr Ser Ala
Pro Asp Thr Arg Pro Ala Pro Gly Ser Thr Ala 900
905 910Pro Pro Ala His Gly Val Thr Ser Ala Pro Asp Thr
Arg Pro Ala Pro 915 920 925Gly Ser
Thr Ala Pro Pro Ala His Gly Val Thr Ser Ala Pro Asp Asn 930
935 940Arg Pro Ala Leu Gly Ser Thr Ala Pro Pro Val
His Asn Val Thr Ser945 950 955
960Ala Ser Gly Ser Ala Ser Gly Ser Ala Ser Thr Leu Val His Asn Gly
965 970 975Thr Ser Ala Arg
Ala Thr Thr Thr Pro Ala Ser Lys Ser Thr Pro Phe 980
985 990Ser Ile Pro Ser His His Ser Asp Thr Pro Thr
Thr Leu Ala Ser His 995 1000
1005Ser Thr Lys Thr Asp Ala Ser Ser Thr His His Ser Ser Val Pro
1010 1015 1020Pro Leu Thr Ser Ser Asn
His Ser Thr Ser Pro Gln Leu Ser Thr 1025 1030
1035Gly Val Ser Phe Phe Phe Leu Ser Phe His Ile Ser Asn Leu
Gln 1040 1045 1050Phe Asn Ser Ser Leu
Glu Asp Pro Ser Thr Asp Tyr Tyr Gln Glu 1055 1060
1065Leu Gln Arg Asp Ile Ser Glu Met Phe Leu Gln Ile Tyr
Lys Gln 1070 1075 1080Gly Gly Phe Leu
Gly Leu Ser Asn Ile Lys Phe Arg Pro Gly Ser 1085
1090 1095Val Val Val Gln Leu Thr Leu Ala Phe Arg Glu
Gly Thr Ile Asn 1100 1105 1110Val His
Asp Val Glu Thr Gln Phe Asn Gln Tyr Lys Thr Glu Ala 1115
1120 1125Ala Ser Arg Tyr Asn Leu Thr Ile Ser Asp
Val Ser Val Ser Asp 1130 1135 1140Val
Pro Phe Pro Phe Ser Ala Gln Ser Gly Ala Gly Val Pro Gly 1145
1150 1155Trp Gly Ile Ala Leu Leu Val Leu Val
Cys Val Leu Val Ala Leu 1160 1165
1170Ala Ile Val Tyr Leu Ile Ala Leu Ala Val Cys Gln Cys Arg Arg
1175 1180 1185Lys Asn Tyr Gly Gln Leu
Asp Ile Phe Pro Ala Arg Asp Thr Tyr 1190 1195
1200His Pro Met Ser Glu Tyr Pro Thr Tyr His Thr His Gly Arg
Tyr 1205 1210 1215Val Pro Pro Ser Ser
Thr Asp Arg Ser Pro Tyr Glu Lys Val Ser 1220 1225
1230Ala Gly Asn Gly Gly Ser Ser Leu Ser Tyr Thr Asn Pro
Ala Val 1235 1240 1245Ala Ala Ala Ser
Ala Asn Leu 1250 125549263PRTOvine respiratory
syncytial virus 49Met Ser Asn His Thr His His Phe Glu Phe Lys Thr Leu Lys
Lys Ala1 5 10 15Trp Lys
Ala Ser Lys Tyr Phe Ile Val Gly Leu Ser Cys Leu Tyr Lys 20
25 30Leu Asn Leu Lys Ser Leu Val Gln Met
Ala Leu Ser Ala Leu Ala Met 35 40
45Ile Thr Leu Val Ser Leu Thr Ile Thr Ala Ile Ile Tyr Ile Ser Thr 50
55 60Gly Asn Thr Lys Ala Lys Pro Met Pro
Thr Pro Thr Ile Gln Ile Thr65 70 75
80Gln Gln Phe Gln Asn His Thr Ser Leu Pro Pro Thr Glu His
Asn His 85 90 95Asn Ser
Thr His Ser Pro Thr Gln Gly Thr Thr Ser Pro His Thr Phe 100
105 110Ala Val Asp Val Thr Glu Gly Thr Arg
Tyr Tyr His Leu Thr Leu Lys 115 120
125Thr Gln Gly Gly Lys Thr Lys Gly Pro Pro Thr Pro His Ala Thr Arg
130 135 140Lys Pro Pro Ile Ser Ser Gln
Lys Ser Asn Pro Ser Glu Ile Gln Gln145 150
155 160Asp Tyr Ser Asp Phe Gln Ile Leu Pro Tyr Val Pro
Cys Asn Ile Cys 165 170
175Glu Gly Asp Ser Ala Cys Leu Ser Leu Cys Gln Asp Arg Ser Glu Ser
180 185 190Ile Leu Asp Lys Ala Leu
Thr Thr Thr Pro Lys Lys Thr Pro Lys Pro 195 200
205Met Thr Thr Lys Lys Pro Thr Lys Thr Ser Thr His His Arg
Thr Ser 210 215 220Leu Arg Asn Lys Leu
Tyr Ile Lys Thr Asn Met Thr Thr Pro Pro His225 230
235 240Gly Leu Ile Ser Thr Ala Lys His Asn Lys
Asn Gln Ser Thr Val Gln 245 250
255Asn Pro Arg His Thr Leu Ala 26050192PRTHuman
respiratory syncytial virus 50Ala Thr Asp Gln Ile Lys Asn Thr Thr Pro Thr
Tyr Leu Thr Gln Asn1 5 10
15Pro Gln Leu Gly Ile Ser Phe Ser Asn Leu Ser Glu Thr Thr Ser Gln
20 25 30Pro Thr Thr Ile Leu Ala Ser
Thr Thr Pro Ser Ala Glu Ser Thr Pro 35 40
45Gln Ser Thr Thr Val Lys Ile Lys Asn Thr Thr Thr Thr Gln Ile
Gln 50 55 60Pro Ser Lys Pro Thr Thr
Lys Gln Arg Gln Asn Lys Pro Gln Asn Lys65 70
75 80Pro Asn Asn Asp Phe His Phe Glu Val Phe Asn
Phe Val Pro Cys Ser 85 90
95Ile Cys Ser Asn Asn Pro Thr Cys Trp Ala Ile Cys Lys Arg Ile Pro
100 105 110Asn Lys Lys Pro Gly Lys
Lys Thr Thr Thr Lys Pro Thr Lys Lys Pro 115 120
125Thr Ile Lys Thr Thr Lys Lys Asp Pro Lys Pro Gln Thr Thr
Lys Pro 130 135 140Lys Glu Val Leu Thr
Thr Lys Pro Thr Glu Lys Pro Thr Ile Ser Thr145 150
155 160Thr Lys Thr Asn Ile Arg Thr Thr Leu Leu
Thr Ser Asn Thr Thr Gly 165 170
175Asn Pro Glu His Thr Ser Gln Lys Gly Asn Pro Pro Leu Asn His Leu
180 185 1905194PRTHuman herpes
virus 1 51Thr Pro Pro Met Pro Ser Ile Gly Leu Glu Glu Glu Glu Glu Glu
Glu1 5 10 15Gly Ala Gly
Asp Gly Glu His Leu Glu Gly Gly Asp Gly Thr Arg Asp 20
25 30Thr Leu Pro Gln Ser Pro Gly Pro Ala Phe
Pro Leu Ala Glu Asp Val 35 40
45Glu Lys Asp Lys Pro Asn Arg Pro Val Val Pro Ser Pro Asp Pro Asn 50
55 60Asn Ser Pro Ala Arg Pro Glu Thr Ser
Arg Pro Lys Thr Pro Pro Thr65 70 75
80Ile Ile Gly Pro Leu Ala Thr Arg Pro Thr Thr Arg Leu Thr
85 905214PRTHuman herpes virus 1 52Met Ser
Trp Ala Leu Glu Met Ala Asp Thr Phe Leu Asp Thr1 5
1053261PRTHuman herpes virus 2 53Met Ser Arg Arg Arg Gly Pro Arg
Arg Arg Gly Pro Arg Arg Arg Pro1 5 10
15Arg Pro Gly Ala Pro Ala Val Pro Arg Pro Gly Ala Pro Ala
Val Pro 20 25 30Arg Pro Gly
Ala Leu Pro Thr Ala Asp Ser Gln Met Val Pro Ala Tyr 35
40 45Asp Ser Gly Thr Ala Val Glu Ser Ala Pro Ala
Ala Ser Ser Leu Leu 50 55 60Arg Arg
Trp Leu Leu Val Pro Gln Ala Asp Asp Ser Asp Asp Ala Asp65
70 75 80Tyr Ala Gly Asn Asp Asp Ala
Glu Trp Ala Asn Ser Pro Pro Ser Glu 85 90
95Gly Gly Gly Lys Ala Pro Glu Ala Pro His Ala Ala Pro
Ala Ala Ala 100 105 110Cys Pro
Pro Pro Pro Pro Arg Lys Glu Arg Gly Pro Gln Arg Pro Leu 115
120 125Pro Pro His Leu Ala Leu Arg Leu Arg Thr
Thr Thr Glu Tyr Leu Ala 130 135 140Arg
Leu Ser Leu Arg Arg Arg Arg Pro Pro Ala Ser Pro Pro Ala Asp145
150 155 160Ala Pro Arg Gly Lys Val
Cys Phe Ser Pro Arg Val Gln Val Arg His 165
170 175Leu Val Ala Trp Glu Thr Ala Ala Arg Leu Ala Arg
Arg Gly Ser Trp 180 185 190Ala
Arg Glu Arg Ala Asp Arg Asp Arg Phe Arg Arg Arg Val Ala Ala 195
200 205Ala Glu Ala Val Ile Gly Pro Cys Leu
Glu Pro Glu Ala Arg Ala Arg 210 215
220Ala Arg Ala Arg Ala Arg Ala His Glu Asp Gly Gly Pro Ala Glu Glu225
230 235 240Glu Glu Ala Ala
Ala Ala Ala Arg Gly Ser Ser Ala Ala Ala Gly Pro 245
250 255Gly Arg Arg Ala Val 260
User Contributions:
Comment about this patent or add new information about this topic:
| People who visited this patent also read: | |
| Patent application number | Title |
|---|---|
| 20180000927 | CIRCOVIRUS SEQUENCES ASSOCIATED WITH PIGLET WEIGHT LOSS DISEASE (PWD) |
| 20180000926 | METHODS OF INDUCING AN IMMUNE RESPONSE TO HEPATITIS C VIRUS |
| 20180000925 | VIRAL RNA SEGMENTS AS IMMUNOMODULATORY AGENTS AND VACCINE COMPONENTS |
| 20180000924 | A VACCINE FOR USE AGAINST SUBCLINICAL LAWSONIA INFECTION IN A PIG |
| 20180000923 | NEISSERIA MENINGITIDIS COMPOSITIONS AND METHODS THEREOF |