Patent application title: METHOD FOR THE ALTERATION OF PLANTS USING CLE POLYPEPTIDES/PEPTIDES
Inventors:
Simon Turner (Manchester, GB)
Peter Etchells (Manchester, GB)
Assignees:
THE UNIVERSITY OF MANCHESTER
IPC8 Class: AA01H106FI
USPC Class:
800290
Class name: Multicellular living organisms and unmodified parts thereof and related processes method of introducing a polynucleotide molecule into or rearrangement of genetic material within a plant or plant part the polynucleotide alters plant part growth (e.g., stem or tuber length, etc.)
Publication date: 2011-08-04
Patent application number: 20110191911
Abstract:
The present invention relates to altering the biomass and/or structure of
a plant, in order to maximise its potential as a source of feedstock or
increase its potential as a feedstock for the paper industry. CLE41
and/or CLE42 are used to manipulate growth and structure of the vascular
tissue of the plant. The present invention also provides plants in which
the levels of CLE41 and/or CLE42 are increased compared to those of a
native plant grown under identical conditions, and parts of such plants.
Also provided are methods for using such plants or plant parts in the
production of plant derived products such as paper or biofuels.Claims:
1. A method of manipulating plant growth and/or structure, comprising:
providing polypeptide selected from the group consisting of: i) a CLE41
polypeptide; ii) a CLE42 polypeptide; iii) a polypeptide comprising an
amino acid sequence that is at least 70% identical to the amino acid
sequence of amino acids 124 to 137 of the consensus sequence of FIG. 10;
iv) a polypeptide comprising an amino acid sequence that is at least 70%
identical to the amino acid sequence of CLE41 of FIG. 13A or CLE42 of
FIG. 14A; and v) a polypeptide encoded by a nucleic acid molecule that is
at least 70% identical to the nucleotide sequence of CLE41 of FIG. 13B or
CLE42 of FIG. 14B; and using the polypeptide in the manipulation of plant
growth and/or structure.
2. A method of manipulating plant growth and/or structure, comprising: providing a nucleic acid molecule selected from the group consisting of: i) a nucleic acid molecule that encodes a CLE41 polypeptide; ii) a nucleic acid molecule that encodes a CLE42 polypeptide; iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of amino acids 124 to 137 of the consensus sequence of FIG. 10; iv) a nucleic acid molecule which is at least 70% identical to the nucleotide sequence of CLE41 of FIG. 13B or CLE42 of FIG. 14B; and v) a nucleic acid molecule that is hybridizes under stringent conditions to the nucleotide sequence i) or ii); and using the nucleic acid molecule in the manipulation of plant growth and/or structure.
3. The method according to claim 1 further comprising using the polypeptide in combination with a nucleic acid molecule selected from the group consisting of: i) a nucleic acid molecule that encodes a CLE41 receptor; ii) a nucleic acid molecule that encodes a CLE42 receptor; iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof; iv) a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of FIG. 15B or 15C; v) a nucleic acid molecule that is at least 70% identical to a nucleic acid molecule of i) or ii); and vi) a nucleic acid molecule that is hybridizes under stringent conditions to the nucleotide sequence i) or ii).
4. The method according to claim 1 further comprising using the polypeptide in combination with a second polypeptide selected from the group consisting of: i) a CLE41 receptor; ii) a CLE42 receptor; iii) a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof; iv) a polypeptide comprising an amino acid sequence that is at least 70% identical to the PXY sequence of FIG. 15A; v) a polypeptide sequence comprising an amino acid sequence which is at least 70% identical to a sequence encoding i) or ii); and vi) a polypeptide comprising an amino acid sequence encoded by a nucleic acid molecule that is at least 70% identical to the PXY nucleotide sequence of FIG. 15B or 15C.
5. The method according to claim 3 wherein said CLE41 or CLE42 receptor is PXY or a functional equivalent thereof.
6. A method of manipulating the growth and/or structure of a plant, comprising modulating the level of CLE41 and/or CLE42 or a functional equivalent thereof, in the plant.
7. A method according to claim 6 wherein the levels of CLE41 and/or CLE42 are modulated by introducing into a cell of the plant: i) a CLE41 polypeptide; ii) a CLE42 polypeptide; iii) a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of amino acids 124 to 137 of the consensus sequence of FIG. 10; iv) a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of CLE41 of FIG. 13A or CLE42 of FIG. 14A; or v) a polypeptide encoded by a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of CLE41 of FIG. 13B or CLE42 of FIG. 14B.
8. A method according to claim 6 wherein the levels of CLE41 and/or CLE42 are modulated by introducing into a cell of the plant: i) a nucleic acid molecule that encodes a CLE41 polypeptide; ii) a nucleic acid molecule that encodes a CLE42 polypeptide; iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of amino acids 124 to 137 of the consensus sequence of FIG. 10; iv) a nucleic acid molecule which is at least 70% identical to the nucleotide sequence of CLE41 of FIG. 13B or CLE42 of FIG. 14B; or v) a nucleic acid molecule that is hybridizes under stringent conditions to the nucleotide sequence i) or ii).
9. A method according to claim 6 wherein the levels of levels of CLE41 and/or CLE42 or a functional equivalent thereof are upregulated.
10. A method according to claim 6 further comprising introducing into a cell of the plant: i) a nucleic acid molecule that encodes a CLE41 receptor; ii) a nucleic acid molecule that encodes a CLE42 receptor; iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof; iv) a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of FIG. 15B or 15C; v) a nucleic acid molecule that is at least 70% identical to a nucleic acid molecule of i) or ii); or vi) a nucleic acid molecule that is hybridizes under stringent conditions to the nucleotide sequence i) or ii).
11. A method according to claim 6 further comprising introducing into a cell of the plant: i) a CLE41 receptor; ii) a CLE42 receptor; iii) a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof; iv) a polypeptide comprising an amino acid sequence that is at least 70% identical to the PXY sequence of FIG. 15A; v) a polypeptide sequence comprising an amino acid sequence which is at least 70% identical to a sequence encoding i) or ii); or vi) a polypeptide comprising an amino acid sequence encoded by a nucleic acid molecule that is at least 70% identical to the PXY nucleotide sequence of FIG. 15B or 15C.
12. A method according to claim 11 wherein said CLE41 and/or CLE42 receptor is PXY or a functional equivalent thereof.
13. A plant cell manipulated to express: i) a CLE41 polypeptide; ii) a CLE42 polypeptide; iii) a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of amino acids 124 to 137 of the consensus sequence of FIG. 10; iv) a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of CLE41 of FIG. 13A or CLE42 of FIG. 14A; or v) a polypeptide encoded by a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of CLE41 of FIG. 13B or CLE42 of FIG. 14B; optionally in combination with expression of a receptor for CLE41 and/or CLE42.
14. A plant cell manipulated to express i) a nucleic acid molecule that encodes a CLE41 polypeptide; ii) a nucleic acid molecule that encodes a CLE42 polypeptide; iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of amino acids 124 to 137 of the consensus sequence of FIG. 10; iv) a nucleic acid molecule which is at least 70% identical to the nucleotide sequence of CLE41 of FIG. 13B or CLE42 of FIG. 14B; or v) a nucleic acid molecule that is hybridizes under stringent conditions to the nucleotide sequence i) or ii).
15. A plant cell according to claim 13 further manipulated to express a nucleic acid molecule selected from the group consisting of: i) a nucleic acid molecule that encodes a CLE41 receptor; ii) a nucleic acid molecule that encodes a CLE42 receptor; iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof; iv) a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of FIG. 15B or 15C; v) a nucleic acid molecule that is at least 70% identical to a nucleic acid molecule of i) or ii); and vi) a nucleic acid molecule that is hybridizes under stringent conditions to the nucleotide sequence i) or ii).
16. A plant cell according to claim 13 further manipulated to express a polypeptide selected from the group consisting of: i) a CLE41 receptor; ii) a CLE42 receptor; iii) a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof; iv) a polypeptide comprising an amino acid sequence that is at least 70% identical to the PXY sequence of FIG. 15A; v) a polypeptide sequence comprising an amino acid sequence which is at least 70% identical to a sequence encoding i) or ii); and vi) a polypeptide comprising an amino acid sequence encoded by a nucleic acid molecule that is at least 70% identical to the PXY nucleotide sequence of FIG. 15B or 15C.
17. A plant comprising a cell according to claim 13.
18. A method of producing a plant-derived product, comprising using a cell or plant according to claim 13 in the production of the plant-derived product.
19. A method of manipulating the growth and/or structure of a plant, comprising the steps of: i) providing a cell/seed according to claim 13; ii) regenerating said cell/seed into a plant; and optionally iii) monitoring the levels of CLE41 and/or CLE42 or a receptor thereof, and and/or PXY or functional equivalents thereof in said regenerated plant.
20. An expression construct comprising a first nucleic acid sequence selected from the group consisting of: i) a nucleic acid molecule that encodes a CLE41 polypeptide; ii) a nucleic acid molecule that encodes a CLE42 polypeptide; iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of amino acids 124 to 137 of the consensus sequence of FIG. 10; iv) a nucleic acid molecule which is at least 70% identical to the nucleotide sequence of CLE41 of FIG. 13B or CLE42 of FIG. 14B; and v) a nucleic acid molecule that is hybridizes under stringent conditions to the nucleotide sequence i) or ii); and optionally a second nucleotide sequence encoding a regulatory sequence capable of expressing the first nucleotide sequence specifically in or adjacent to the vascular tissue of a plant.
21. An expression cassette according to claim 20 further comprising: i) a nucleic acid molecule that encodes a CLE41 receptor; ii) a nucleic acid molecule that encodes a CLE42 receptor; iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof; iv) a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of FIG. 15B or 15C; v) a nucleic acid molecule that is at least 70% identical to a nucleic acid molecule of i) or ii); or vi) a nucleic acid molecule that is hybridizes under stringent conditions to the nucleotide sequence i) or ii)
22. A host cell or organism comprising an expression construct of claim 20.
23. A transgenic plant seed comprising a cell according to claim 13.
24. A plant derived product produced by a method according to claim 1.
25. A method of producing a plant-derived product comprising: a) manipulating the growth and/or structure of a plant produced according to claim 1; b) growing the plant until it reaches a pre-determined lateral size; and optionally c) harvesting the plant derived product of the plant.
26. A method of altering the mechanical properties of a plant or plant derived product comprising: a) manipulating the growth and/or structure of a plant according to claim 1; b) growing the plant until it reaches a pre-determined size; and optionally c) harvesting a plant derived product of the plant.
27. The method according to claim 2 further comprising using the nucleic acid molecule in combination with a second nucleic acid molecule selected from the group consisting of: i) a nucleic acid molecule that encodes a CLE41 receptor; ii) a nucleic acid molecule that encodes a CLE42 receptor; iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof; iv) a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of FIG. 15B or 15C; v) a nucleic acid molecule that is at least 70% identical to a nucleic acid molecule of i) or ii); and vi) a nucleic acid molecule that is hybridizes under stringent conditions to the nucleotide sequence i) or ii).
28. The method according to claim 2 further comprising using the nucleic acid molecule in combination with a polypeptide selected from the group consisting of: i) a CLE41 receptor; ii) a CLE42 receptor; iii) a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof; iv) a polypeptide comprising an amino acid sequence that is at least 70% identical to the PXY sequence of FIG. 15A; v) a polypeptide sequence comprising an amino acid sequence which is at least 70% identical to a sequence encoding i) or ii); and vi) a polypeptide comprising an amino acid sequence encoded by a nucleic acid molecule that is at least 70% identical to the PXY nucleotide sequence of FIG. 15B or 15C.
29. The method according to claim 4 wherein said CLE41 or CLE42 receptor is PXY or a functional equivalent thereof.
30. A plant cell according to claim 14 further manipulated to express a nucleic acid molecule selected from the group consisting of: i) a nucleic acid molecule that encodes a CLE41 receptor; ii) a nucleic acid molecule that encodes a CLE42 receptor; iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof; iv) a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of FIG. 15B or 15C; v) a nucleic acid molecule that is at least 70% identical to a nucleic acid molecule of i) or ii); and vi) a nucleic acid molecule that is hybridizes under stringent conditions to the nucleotide sequence i) or ii).
31. A plant cell according to claim 14 further manipulated to express a polypeptide selected from the group consisting of: i) a CLE41 receptor; ii) a CLE42 receptor; iii) a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof iv) a polypeptide comprising an amino acid sequence that is at least 70% identical to the PXY sequence of FIG. 15A; v) a polypeptide sequence comprising an amino acid sequence which is at least 70% identical to a sequence encoding i) or ii); and vi) a polypeptide comprising an amino acid sequence encoded by a nucleic acid molecule that is at least 70% identical to the PXY nucleotide sequence of FIG. 15B or 15C.
32. A plant comprising a cell according to claim 14.
33. A method of producing a plant-derived product, comprising using a cell or plant according to claim 14 in the production of the plant-derived product.
34. The method of claim 33, wherein the plant-derived product is selected from the group consisting of biomass, fibres, forage, biocomposites, biopolymers, wood, biofuel and paper.
35. A method of manipulating the growth and/or structure of a plant, comprising the steps of: i) providing a cell/seed according to claim 14; ii) regenerating said cell/seed into a plant; and optionally iii) monitoring the levels of CLE41 and/or CLE42 or a receptor thereof, and/or PXY or functional equivalents thereof in said regenerated plant.
36. A transgenic plant seed comprising a cell according to claim 14.
37. A plant derived product produced by a method according to claim 2.
38. A method of producing a plant-derived product comprising: a) manipulating the growth and/or structure of a plant produced according to claim 2; b) growing the plant until it reaches a pre-determined lateral size; and optionally c) harvesting the plant derived product of the plant.
39. A method of altering the mechanical properties of a plant or plant derived product comprising: a) manipulating the growth and/or structure of a plant according to claim 2; b) growing the plant until it reaches a pre-determined size; and optionally c) harvesting a plant derived product of the plant.
40. A plant derived product produced by a method according to claim 6.
41. A method of producing a plant-derived product comprising: a) manipulating the growth and/or structure of a plant produced according to claim 6; b) growing the plant until it reaches a pre-determined lateral size; and optionally c) harvesting the plant derived product of the plant.
42. A method of altering the mechanical properties of a plant or plant derived product comprising: a) manipulating the growth and/or structure of a plant according to claim 6; b) growing the plant until it reaches a pre-determined size; and optionally c) harvesting a plant derived product of the plant.
43. The method of claim 18, wherein the plant-derived product is selected from the group consisting of biomass, fibres, forage, biocomposites, biopolymers, wood, biofuel and paper.
Description:
FIELD OF THE INVENTION
[0001] The present invention relates to manipulating the growth and/or structure of a plant through modulation of the amount of CLE41 and/or CLE42 expressed in the plant, and additionally or alternatively, modulating the amount of PXY in the plant. Manipulating the growth and/or structure of a plant can be used to alter the mechanical properties of a plant or plant derived product, or to maximise its potential for the production of plant derived products such as biofuels and paper.
BACKGROUND
[0002] In multi-cellular organisms, cells must communicate with each other in order for growth and development to occur in an ordered manner. In animals, it has long been known that polypeptides act as signalling molecules in mediating communication between cells, a common example being insulin in humans. These signalling molecules are responsible for initiating many cellular processes, typically by binding to a receptor at the cell surface, which in turn transmits a message to inside the cell via downstream signalling proteins such as membrane associated protein kinases (MAPK), tyrosine phosphatases and Ras proteins. In the cell, the cell signalling pathway end-point is usually a transcription factor target, which mediates a change in gene expression in the cell, thus causing a change in the growth and/or development of the cell in response to the initial extracellular signal.
[0003] In plants, it is also known that cell signalling occurs, and this was thought to be mediated by plant hormones such as auxin and cytokinin. More recently, the discovery of systemin has shown that polypeptides also play a role in cell-signalling in plants. One of the largest families of signalling polypeptides identified in plants is the Clavata3 (clv3)/Endosperm Surrounding Region (ESR)-related (CLE) family. These proteins are the most highly characterised family of small polypeptides in plants. The Arabidopsis thaliana genome contains 32 CLE genes. Clv3 is the best characterised CLE family member which acts together with a receptor kinase (CLAVATA 1) to play a role in regulating the proliferation of cells in the shoot (apical) meristem. At present, however, most of the CLE family remain functionally undefined.
[0004] The CLE gene family has been shown to be present in a variety of other plant species (Jun et al Cell. Mol. Life. Sci. 65 743-755 (2008) and Frickey et al BMC Plant Biology 2008, 8:1 10.1186/1471-2229-8-1) including rice, maize, tomato and alfalfa.
[0005] The polypeptides encoded by the CLE genes share common characteristics. They are less than 15 kDa in mass and comprise a short stretch of hydrophobic amino acids at the amino terminus which serves to target the polypeptide to the secretory pathway. This conserved stretch of 14 amino acids is known as the CLE domain (Jun et al supra).
[0006] Higher plants show post-embyronic development at shoot and root tips, which are known as the apical meristems. Stem cells at these meristems produce cells which differentiate to become flower, leaf, stem or root cells. A loss-of-function mutant resulting in an excess of stem cells at the apical meristem suggests that Clv3 plays a role in regulation of growth and/or differentiation at the growing tip. Over expression of CLV3 results in loss of apical stem cells, thus post-embryonic above ground parts of the plant are lost. The signalling pathway which CLV3 regulates has been elucidated and is described in Jun et al (supra). This pathway is thought to be conserved amongst other plants species.
[0007] Shiu and Bleecker suggest that the CLE family is likely to coordinate with a group of plant receptors known as the leucine-rich-repeat receptor-like (LLR-RLK) kinases (PNAS 98 10763-10768 (2001)).
[0008] U.S. Pat. No. 7,179,963 describes a maize clv3-like nucleotide sequence, and its use in modulating plant development and differentiation. U.S. Pat. No. 7,335,760 discloses nucleic acid sequences for use in genetically modifying a plant to increase plant yield and the mass of the plant, for example for biofuel production.
[0009] Other CLE family members have been shown to inhibit cell differentiation. For example, Frickey et al (supra) have looked at the CLE family and suggested that CLE family members CLE41 and/or 42 may play a role in vascular development. Ito et al (Science Vol 313 842-845 (2006)) show that dodecapeptides are important in preventing vascular cell differentiation.
[0010] In contrast, however, Strabala et al (Plant Physiology vol. 140 1331-1344 (2006)) show that CLE41 and/or 42 are genuine expressed members of the CLE family. Although general over-expression of CLE42 throughout the plant results in a dwarf phenotype, Strabala et al report that CLE42 is likely to be a functionally redundant molecule.
[0011] The source of biomass in plants is their woody tissue, derived from the vascular meristems of the plant such as the cambium and procambium, which divide to form the phloem and xylem cells of the vascular tissue within the plant stems and roots. The cambium and procambium (collectively known as the vascular meristems) are growth zones which enable the plant to grow laterally, thus generating the majority of biomass. Enhancing lateral growth by genetically altering the rates of procambial or cambial cell division may lead to an increase in the plant biomass. This would provide an additional source of biomass for various industries dependent upon plant derived products, such as the biofuel or paper industries.
[0012] Increasing the yield of biomass of plants, for example for paper and fuel production has previously been done by breeding programs, but in recent years there is interest in the use of genetic manipulation or plant modification for such purposes.
[0013] The division of cells to form the vascular tissue is a highly ordered process. Prominent polarity of cells destined to become either phloem cells or xylem cells is observed, the latter eventually forming the woody tissue of the plant. Xylem is principally water transporting tissue of the plant, and together with phloem, forms a vascular network for the plant. The cells of the xylem which are principally responsible for carrying water are the tracheary elements, of which there are two types--tracheids and vessels.
[0014] However, whilst there has been much investigation into the regulation of growth at the apical meristems, there is less understanding of the growth of the vascular tissue. Fisher et al (Current Biology 17 1061-1066 (2007)) report a loss of function mutant in which the spatial organisation of the vascular tissue is lost and the xylem and phloem cells are interspersed. The mutant is in a gene named PXY, which encodes a receptor-like kinase.
[0015] Tracheary elements (TEs) are cells in the xylem that are highly specialized for transporting water and solutes up the plant. They are produced from xylem cells by a process which involves specification, enlargement, patterned cell wall deposition, programmed cell death and cell wall removal. This results in adjacent TEs being joined together to form a continuous network for water transport.
[0016] Jun et al (supra) disclose that the CLE domain of CLE41 is identical to Tracheal Element Differentiation Inhibitory Factor (TDIF), which has been shown to inhibit cell differentiation, and CLE42 differs by only one amino acid from the TDIF sequence. When exogenously applied to cell cultures, synthetic CLE41 and CLE42 suppressed the formation of tracheary element cells from the xylem (Ito et al, supra).
[0017] There remains a need for identification of genetic elements, the manipulation of which can be used to alter the growth and/or structure of the plant.
BRIEF SUMMARY OF THE INVENTION
[0018] In a first aspect the invention provides the use of a polypeptide selected from the group consisting of: [0019] i) a CLE41 polypeptide; [0020] ii) a CLE42 polypeptide; [0021] iii) a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of amino acids 124 to 137 of the consensus sequence of FIG. 10; [0022] iv) a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of CLE41 of FIG. 13A or CLE42 of FIG. 14A; [0023] v) a polypeptide encoded by a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of CLE41 of FIG. 13B or CLE42 of FIG. 14B; in the manipulation of plant growth and/or structure.
[0024] In a second aspect the invention provides the use of a nucleic acid molecule selected from the group consisting of: [0025] i) a nucleic acid molecule that encodes a CLE41 polypeptide; [0026] ii) a nucleic acid molecule that encodes a CLE42 polypeptide; [0027] iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of amino acids 124 to 137 of the consensus sequence of FIG. 10; [0028] iv) a nucleic acid molecule which is at least 70% identical to the nucleotide sequence of CLE41 of FIG. 13B or CLE42 of FIG. 14B; [0029] v) a nucleic acid molecule that hybridizes under stringent conditions to the nucleotide sequence i) or ii) in the manipulation of plant growth and/or structure.
[0030] Preferably, the use of the first or second aspect is use of the polypeptide or nucleic acid in combination with a nucleic acid molecule selected from the group consisting of: [0031] i) a nucleic acid molecule that encodes a CLE41 receptor; [0032] ii) a nucleic acid molecule that encodes a CLE42 receptor; [0033] iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof; [0034] iv) a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of FIG. 15B or 15C; [0035] v) a nucleic acid molecule that is at least 70% identical to a nucleic acid molecule of i) or ii); [0036] vi) a nucleic acid molecule that hybridizes under stringent conditions to the nucleotide sequence i) or ii).
[0037] Preferably, the use of the first or second aspect is use of the polypeptide or nucleic acid in combination with a polypeptide selected from the group consisting of: [0038] i) a CLE41 receptor; [0039] ii) a CLE42 receptor; [0040] iii) a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof [0041] iv) a polypeptide comprising an amino acid sequence that is at least 70% identical to the PXY sequence of FIG. 15A; [0042] v) a polypeptide sequence comprising an amino acid sequence which is at least 70% identical to a sequence encoding i) or ii); [0043] vi) a polypeptide comprising an amino acid sequence encoded by a nucleic acid molecule that is at least 70% identical to the PXY nucleotide sequence of FIG. 15B or 15C.
[0044] Preferably said CLE41 or CLE42 receptor is PXY or a functional equivalent thereof.
[0045] Preferably said manipulation of the plant growth and/or structure is an increase or decrease in the amount of growth and/or division of the procambial and/or cambial cells in a plant, specifically the number of cells generated. More specifically, it is an increase or decrease in the rate of division of such cells. Thus, the manipulation of growth and/or structure can be said to be an increase or decrease in the secondary growth of the plant, and/or an increase or decrease in the degree of organisation of the secondary structure, at the cellular level. By secondary growth is preferably meant the woody tissue of a plant, or the vascular or interfasicular tissue. Preferably, where there is an increase in the number of procambial and/or cambial cells, these cells differentiate into xylem and/or phloem cells, preferably the former.
[0046] In a further aspect, the present invention provides a method of manipulating the growth and/or structure of a plant, comprising modulating the level of CLE41 and/or CLE42 or a functional equivalent thereof, in the plant.
[0047] Preferably the levels of CLE41 and/or CLE42 are modulated by introducing into a cell of the plant: [0048] i) a CLE41 polypeptide; [0049] ii) a CLE42 polypeptide; [0050] iii) a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of amino acids 124 to 137 of the consensus sequence of FIG. 10; [0051] iv) a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of CLE41 of FIG. 13A or CLE42 of FIG. 14A; [0052] v) a polypeptide encoded by a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of CLE41 of FIG. 13B or CLE42 of FIG. 14B.
[0053] Alternatively the levels of CLE41 and/or CLE42 are modulated by introducing into a cell of the plant: [0054] i) a nucleic acid molecule that encodes a CLE41 polypeptide; [0055] ii) a nucleic acid molecule that encodes a CLE42 polypeptide; [0056] iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of amino acids 124 to 137 of the consensus sequence of FIG. 10; [0057] iv) a nucleic acid molecule which is at least 70% identical to the nucleotide sequence of CLE41 of FIG. 13B or CLE42 of FIG. 14B; [0058] v) a nucleic acid molecule that hybridizes under stringent conditions to the nucleotide sequence i) or ii).
[0059] Preferably, the levels of levels of CLE41 and/or CLE42 or a functional equivalent thereof are upregulated.
[0060] Optionally, the method further comprises introducing into a cell of the plant: [0061] i) a nucleic acid molecule that encodes a CLE41 receptor; [0062] ii) a nucleic acid molecule that encodes a CLE42 receptor; [0063] iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof; [0064] iv) a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of FIG. 15B or 15C; [0065] v) a nucleic acid molecule that is at least 70% identical to a nucleic acid molecule of i) or ii); [0066] vi) a nucleic acid molecule that hybridizes under stringent conditions to the nucleotide sequence i) or ii).
[0067] Alternatively, the method further comprises introducing into cell of a plant: [0068] i) a CLE41 receptor; [0069] ii) a CLE42 receptor; [0070] iii) a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof [0071] iv) a polypeptide comprising an amino acid sequence that is at least 70% identical to the PXY sequence of FIG. 15A; [0072] v) a polypeptide sequence comprising an amino acid sequence which is at least 70% identical to a sequence encoding i) or ii); [0073] vi) a polypeptide comprising an amino acid sequence encoded by a nucleic acid molecule that is at least 70% identical to the PXY nucleotide sequence of FIG. 15B or 15C.
[0074] Preferably said CLE41 and/or CLE42 receptor is PXY or a functional equivalent thereof.
[0075] In aspects where the levels of two or more of CLE41, CLE42 and PXY are to be manipulated in a plant, this may be achieved by: [0076] (i) manipulating the levels of CLE41 and/or CLE42 as hereinbefore described, in a first plant; [0077] (ii) manipulating the levels of a CLE41 and/or CLE42 receptor as hereinbefore described, in a second plant; [0078] (iii) crossing said first and second plants to obtain a plant in which the levels of CLE41 and/or CLE42 and said receptor are manipulated. Also provided in the present invention is the plant produced by the crossing of the first and second plants, and progeny thereof which express the non-native nucleotide and/or polypeptide sequences.
[0079] In a further aspect, the present invention provides a plant cell manipulated to express: [0080] i) a CLE41 polypeptide; [0081] ii) a CLE42 polypeptide; [0082] iii) a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of amino acids 124 to 137 of the consensus sequence of FIG. 10; [0083] iv) a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of CLE41 of FIG. 13A or CLE42 of FIG. 14A; [0084] v) a polypeptide encoded by a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of CLE41 of FIG. 13B or CLE42 of FIG. 14B; optionally in combination with expression of a receptor for CLE41 and/or CLE42.
[0085] In a further aspect, the present invention provides a plant cell manipulated to express [0086] i) a nucleic acid molecule that encodes a CLE41 polypeptide; [0087] ii) a nucleic acid molecule that encodes a CLE42 polypeptide; [0088] iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of amino acids 124 to 137 of the consensus sequence of FIG. 10; [0089] iv) a nucleic acid molecule which is at least 70% identical to the nucleotide sequence of CLE41 of FIG. 13B or CLE42 of FIG. 14B; [0090] v) a nucleic acid molecule that hybridizes under stringent conditions to the nucleotide sequence i) or ii).
[0091] Preferably, said plant cell is further manipulated to express a nucleic acid molecule selected from the group consisting of: [0092] i) a nucleic acid molecule that encodes a CLE41 receptor; [0093] ii) a nucleic acid molecule that encodes a CLE42 receptor; [0094] iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof; [0095] iv) a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of FIG. 15B or 15C; [0096] v) a nucleic acid molecule that is at least 70% identical to a nucleic acid molecule of i) or ii); [0097] vi) a nucleic acid molecule that hybridizes under stringent conditions to the nucleotide sequence i) or ii).
[0098] Alternatively, said plant cell is further manipulated to express a polypeptide selected from the group consisting of: [0099] i) a CLE41 receptor; [0100] ii) a CLE42 receptor; [0101] iii) a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof [0102] iv) a polypeptide comprising an amino acid sequence that is at least 70% identical to the PXY sequence of FIG. 15A; [0103] v) a polypeptide sequence comprising an amino acid sequence which is at least 70% identical to a sequence encoding i) or ii); [0104] vi) a polypeptide comprising an amino acid sequence encoded by a nucleic acid molecule that is at least 70% identical to the PXY nucleotide sequence of FIG. 15B or 15C.
[0105] Preferably said CLE41 and/or CLE42 receptor is PXY or a functional equivalent thereof.
[0106] In a further aspect, there is provided a nucleic acid molecule encoding a functional equivalent of PXY, preferably derived from Arabidopsis thaliana, poplar or rice, and more preferably encoding the amino acid sequence of the consensus sequence of FIG. 12. Also provided is a polypeptide sequence encoding a functional equivalent of PXY, preferably derived from Arabidopsis thaliana, poplar or rice, and preferably comprising an amino acid sequence of the consensus sequence of FIG. 12. Preferably, the amino acid sequence comprises the sequence of pttPXY, PXYL-1, PXYL-2 or Os02g02140.1, or Os03g05140.1 of FIG. 12. Also included are sequences having 70% sequence identity or sequence homology thereto.
[0107] It is apparent that the levels of CLE41, CLE42 and or a receptor thereof, such as PXY, in each of the aspects of the present invention may be manipulated by altering the expression of native CLE41, CLE42 and or a receptor thereof within the plant cell. This may be achieved by placing the native nucleotide sequence under the control of a nucleotide sequence which modifies expression of a native gene to allow modify expression thereof. The nucleotide sequence may be a regulatory sequence, as defined herein, or may encode a regulatory protein, such as a transcription factor, or may encode a DNA or RNA antisense sequence. As such, the nucleotide sequence or its expression product can modify expression, amount and/or activity of a native gene/polypeptide. Methods of function of such regulatory proteins, expression products and antisense will be known to persons skilled in the art.
[0108] In a yet further aspect, the present invention provides a plant comprising a cell according to the invention. Also provided are progeny of the plants of the invention.
[0109] In a further aspect, there is provided the use of a cell or plant of the invention in the production of a plant-derived product. A plant-derived product may include biomass, fibres, forage, biocomposites, biopolymers, wood, biofuel or paper. In addition, the invention provides the use of a cell or a plant of the invention in altering the mechanical properties of a plant or a plant derived product.
[0110] In a further aspect, the present invention provides a method of manipulating the growth and/or structure of a plant, comprising the steps of: [0111] i) providing a cell/seed according to the invention; [0112] ii) regenerating said cell/seed into a plant; and optionally [0113] iii) monitoring the levels of CLE41 and/or CLE42 or a receptor thereof, and or PXY or functional equivalents thereof in said regenerated plant.
[0114] In a further aspect, there is provided an expression construct comprising a first nucleic acid sequence selected from the group consisting of: [0115] i) a nucleic acid molecule that encodes a CLE41 polypeptide; [0116] ii) a nucleic acid molecule that encodes a CLE42 polypeptide; [0117] iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the amino acid sequence of amino acids 124 to 137 of the consensus sequence of FIG. 10; [0118] iv) a nucleic acid molecule which is at least 70% identical to the nucleotide sequence of CLE41 of FIG. 13B or CLE42 of FIG. 14B; [0119] v) a nucleic acid molecule that hybridizes under stringent conditions to the nucleotide sequence i) or ii); and optionally a second nucleic sequence encoding a regulatory sequence capable of expressing the first nucleic sequence specifically in or adjacent to the vascular tissue of a plant.
[0120] Preferably, the regulatory sequence will be capable of directing expression of a nucleotide sequence specifically to the vascular tissue, preferably to the cambial/procambial cells and more preferably to tissue adjacent to the cambial/procambial cells i.e. the phloem and/or xylem tissue. Most preferably, a regulatory sequence used in the present invention will be capable of directing expression specifically to the phloem cells. Examples of suitable phloem specific regulatory sequences are SUC2 and APL, KAN1, KAN2, At4g33660, At3g61380, At1g79380. Xylem specific regulatory sequences may also be used in the present invention. Examples include REV, IRX1 COBL4, KOR, At2g38080, and At1g27440, the promoter sequence for the irregular xylem3 (irx3) (AtCESA7) gene, the promoter sequence for the irregular xylem? (FRAGILE FIBER 8) gene, and the promoter sequence for the irregular xylem12 (ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4) gene (Brown et al. The Plant Cell, Vol. 17, 2281-2295).
[0121] Optionally, the expression cassette may further comprise a third nucleic acid sequence selected from the group consisting of: [0122] i) a nucleic acid molecule that encodes a CLE41 receptor; [0123] ii) a nucleic acid molecule that encodes a CLE42 receptor; [0124] iii) a nucleic acid molecule that encodes a polypeptide comprising an amino acid sequence that is at least 70% identical to the consensus sequence of FIG. 12, or a functional equivalent thereof; [0125] iv) a nucleic acid molecule that is at least 70% identical to the nucleotide sequence of FIG. 15B or 15C; [0126] v) a nucleic acid molecule that is at least 70% identical to a nucleic acid molecule of i) or ii); [0127] vi) a nucleic acid molecule that hybridizes under stringent conditions to the nucleotide sequence i) or ii)
[0128] Preferably, the expression cassette comprises a nucleic acid encoding PXY or a functional equivalent thereof. The third nucleic acid sequence may be provided on the same expression cassette as the first and/or second nucleic acid sequence, or on a separate expression cassette to the first nucleic acid sequence. The third nucleic acid sequence may be under the control of fourth nucleic acid sequence encoding a regulatory sequence capable of expressing the third nucleic sequence specifically in or adjacent to the vascular tissue of a plant.
[0129] The second nucleic acid sequence may be the same or different to the fourth nucleic acid sequence.
[0130] In a further aspect, there is provided a host cell or organism comprising an expression construct of the invention.
[0131] According to a further aspect of the invention there is provided a transgenic plant seed comprising a cell according to the invention.
[0132] The present invention also provides a plant derived product produced by a method of the invention.
[0133] The present invention also provides a host cell or organism comprising an expression construct of the invention. A host cell or organism may be a plant cell, plant seed, plant, or other plant material.
[0134] The present a method of producing a plant-derived product comprising: [0135] a) manipulating the growth and/or structure of a plant using the methods of the invention; [0136] b) growing the plant until it reaches a pre-determined lateral size; optionally [0137] c) harvesting the plant derived product of the plant.
[0138] A plant-derived product may include biomass, fibres, forage, biocomposites, biopolymers, wood, biofuel or paper.
[0139] The present invention also provides a method of altering the mechanical properties of a plant or plant derived product comprising: [0140] a) manipulating the growth and/or structure of a plant using the methods of the invention; [0141] b) growing the plant until it reaches a pre-determined size; and optionally [0142] c) harvesting a plant derived product of the plant.
BRIEF DESCRIPTION OF THE DRAWINGS
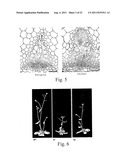
[0143] FIG. 1 shows the cambial meristem of birch (A) and Arabidopsis (B). (A) Transverse section of the growing cambium in Birch. (B) Cross section of a vascular bundle from an Arabidopsis stem. Phloem (ph), xylem (xy), procambium (pc) and cambium (c) are indicated.
[0144] FIG. 2 is a diagrammatic representation of vascular development by the procambium (in, for example, Aribidopsis) or by the cambium (for example in trees such as birch). New cells arise from division of procambial cells that subsequently differentiate into the phloem of xylem. In this model a ligand expressed in the xylem signals cell division in the procambium.
[0145] FIG. 3 shows the effects of over-expressing CLE41 (B,C) and CLE42 (D) on Arabidopsis vascular development compared to the wild type (A) in 35 day old plants. See also Table 1.
[0146] FIG. 4 is a graph showing the average number of cells in the vascular bundles of wild type compared to 35S::CLE41 and 35S::CLE42 Arabidopsis plants at 35 days.
[0147] FIG. 5 shows the effect of over-expressing CLE41 on the stem vascular bundle of an Arabidopsis compared to the wild-type. Sections through stem vascular bundles of Wild type, 35S::CLE41 and 35S::CLE42 from 50 day old plants. A large number of the extra cells in 35S::CLE41 plants (c.f. FIGS. 3 and 4) have differentiated into xylem cells. 35S::CLE41 therefore has more xylem cells than wild type.
[0148] FIG. 6 shows the effects on plant stature of wild type compared to 35S::CLE41 and 35S::CLE42 in Arabidopsis.
[0149] FIG. 7 shows the disrupted hypocotyl structure of a Arabidopsis plant where either CLE41 (M) or CLE42 (N) is over-expressed and compared with the wild type (L). In the transgenic lines (M, N), hypocotyls are much larger.
[0150] FIG. 8 shows the effect of over expressing both CLE41 and/or CLE42 and PXY on the structure and amount of cells in the vascular bundle and interfascicular region of stems from an Arabidopsis thaliana plant. Simultaneous over-expression of PXY and CLE41/42 gives increased in vascular cell number compared to plants over expressing CLE41/42 alone (c.f. FIG. 3).
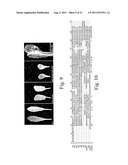
[0151] FIG. 9 shows the effect of over expression of both CLE41 and/or CLE42 and PXY on the leaf structure of an Arabidopsis thaliana plant. Multiple midveins in 35S::CLE42 35S::PXY plants demonstrate that expression of CLE42 and PXY can initiate vascular tissue.
[0152] FIG. 10 is an alignment of rice, poplar and Arabidopsis thaliana putative PXY ligands.
[0153] FIG. 11 shows the conservation of residues in the CLE signalling domain--the dashed line indicates the group 5 that contains all the putative PXY ligands.
[0154] FIG. 12 is a comparison of the kinase domain of PXY (Arabidopsis thaliana) from proteins in rice (Os02g02140.1), poplar (PttPXY) and Arabidopsis thaliana (PXL1 and PXL2).
[0155] FIG. 13 shows the amino acid sequence of the CLE41 proteins (A) and nucleotide sequence of the CLE41 gene (B).
[0156] FIG. 14 shows the amino acid sequence of the CLE42 proteins (A) and nucleotide sequence of the CLE42 gene (B).
[0157] FIG. 15 shows the amino acid sequence of the PXY proteins (A) and nucleotide sequence of the PXY gene without (B) or with (C) the intron.
[0158] FIG. 16 shows preferred promoter and terminator sequences for use in the invention.
[0159] FIG. 17 shows the diagram of the multisite gateway kit for cloning.
[0160] FIG. 18 is an alignment of full length PXY and related PXY proteins from poplar (PttPXY) and Arabidopsis thaliana (PXYL1 and 2)
[0161] FIG. 19 the heights of Nicotiana plants which over express CLE41, PXY or both are shown. A height defect is associated with plants carrying the 35S::CLE41 construct. Normal plant height is restored when plants harbour both 35S::CLE41 and 35S::PXY cassettes.
[0162] FIG. 20 shows the cross section of the Nicotiana plants showing tissue structure and size. 35S::CLE41 35S::PXY plants have hypocotyls larger than wild type
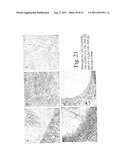
[0163] FIG. 21 shows cell organisation in Nicotiana plants in transverse section. 35S::CLE41 and 35S::CLE42 plants have more vascular tissue than wild type, but it is not ordered. 35S::CLE41 35S::PXY plants have ordered vasculature. Given that these plants have larger hypocotyls than wild type (see FIG. 20), and are of normal height (see FIG. 19), these plants clearly have more vascular tissue than wild type.
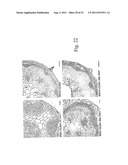
[0164] FIG. 22 shows the effect of phloem specific promoter SUC2 and xylem specific promoter IRX3 on cell organisation in Arabidopsis. IRX3::CLE41 plants have large vascular bundles. IRX3::CLE41 35S::PXY plants have large vascular bundles with large amounts of secondary growth (c.f. FIG. 8 wild type). SUC2::CLE41 and SUC2::CLE41 35S::PXY plants have vascular tissue that is highly ordered with many more vascular cells than wild type.
[0165] FIG. 23 is a graph showing the mean number of cells per vascular bundle. Expression of CLE41 in phloem cells under SUC2 gives more cells per vascular bundle in plants 6 week old Arabidopsis plants.
[0166] FIG. 24 is a graph showing the effect of over expression of CLE41and CLE42 on hypocotyl diameter in Arabidopsis.
[0167] FIG. 25 shows the effect of over expression of CLE41 in poplar trees using the SUC2 or 35S promoters. The bracket denotes xylem cells, of which there are more in 35S::CLE41 and SUC2::CLE41 than wild type.
DETAILED DESCRIPTION OF THE INVENTION
[0168] The present invention is based upon the finding that CLE41 and CLE42 function as ligands for the transmembrane receptor kinase PXY in plants, and modify and/or initiate the regulatory pathway which controls cell division and differentiation in the vascular tissue of a plant. Thus, by modulating the levels of CLE41 and/or CLE42 in a plant, optionally in combination with PXY, the growth and/or structure of the plant can be manipulated, as hereinbefore described.
[0169] In particular, the present invention is based upon the finding that individual over-expression of CLE41 and/or CLE42 leads to an excess of undifferentiated cells in the vascular meristem and a subsequent increase in the radial thickness of the xylem. Further, over-expression of PXY or a functional equivalent thereof, together with a PXY ligand such as CLE41 and/or CLE42, results in an excess of undifferentiated cells in the vascular meristem of the plant, which show a highly ordered structure. This excess of cells in the vascular meristem have been shown to then differentiate into xylem cells, thus increasing the radial thickness of the xylem and the biomass of the plant.
[0170] A "non-essential" amino acid residue is a residue that can be altered from the wild-type sequence of without abolishing or, more preferably, without substantially altering a biological activity, whereas an "essential" amino acid residue results in such a change. For example, amino acid residues that are conserved among the polypeptides of the present invention, e.g., those present in the conserved potassium channel domain are predicted to be particularly non-amenable to alteration, except that amino acid residues in transmembrane domains can generally be replaced by other residues having approximately equivalent hydrophobicity without significantly altering activity.
[0171] A "conservative amino acid substitution" is one in which the amino acid residue is replaced with an amino acid residue having a similar side chain. Families of amino acid residues having similar side chains have been defined in the art. These families include amino acids with basic side chains (e.g., lysine, arginine, histidine), acidic side chains (e.g., aspartic acid, glutamic acid), uncharged polar side chains (e.g., glycine, asparagine, glutamine, serine, threonine, tyrosine, cysteine), non-polar side chains (e.g., alanine, valine, leucine, isoleucine, proline, phenylalanine, methionine, tryptophan), beta-branched side chains (e.g., threonine, valine, isoleucine) and aromatic side chains (e.g., tyrosine, phenylalanine, tryptophan, histidine). Thus, a nonessential amino acid residue in protein is preferably replaced with another amino acid residue from the same side chain family. Alternatively, in another embodiment, mutations can be introduced randomly along all or part of coding sequences, such as by saturation mutagenesis, and the resultant mutants can be screened for biological activity to identify mutants that retain activity. Following mutagenesis of a polypeptide, the encoded proteins can be expressed recombinantly and the activity of the protein can be determined.
[0172] As used herein, a "biologically active fragment" of protein includes fragment of protein that participate in an interaction between molecules and non-molecules. Biologically active portions of protein include peptides comprising amino acid sequences sufficiently homologous to or derived from the amino acid sequences of the protein, which include fewer amino acids than the full length protein, and exhibit at least one activity of protein. Typically, biologically active portions comprise a domain or motif with at least one activity of the protein, e.g., the ability to modulate membrane excitability, intracellular ion concentration, membrane polarization, and action potential.
[0173] As used herein, the term "nucleic acid molecule" includes DNA molecules (e.g., a cDNA or genomic DNA) and RNA molecules (e.g., a mRNA) and analogs of the DNA or RNA generated, e.g., by the use of nucleotide analogs. The nucleic acid molecule can be single-stranded or double-stranded, but preferably is double-stranded DNA. The nucleotide sequence may be RNA or DNA, including cDNA.
[0174] With regards to genomic DNA, the term "isolated" includes nucleic acid molecules that are separated from the chromosome with which the genomic DNA is naturally associated. Preferably, an "isolated" nucleic acid is free of sequences that naturally flank the nucleic acid (i.e., sequences located at the 5'- and/or 3'-ends of the nucleic acid) in the genomic DNA of the organism from which the nucleic acid is derived. Moreover, an "isolated" nucleic acid molecule, such as a cDNA molecule, can be substantially free of other cellular material, or culture medium when produced by recombinant techniques, or substantially free of chemical precursors or other chemicals when chemically synthesized.
[0175] As used herein, a "naturally-occurring" nucleic acid molecule refers to an RNA or DNA molecule having a nucleotide sequence that occurs in nature (e.g., encodes a natural protein).
[0176] As used herein, the terms "gene" and "recombinant gene" refer to nucleic acid molecules which include an open reading frame encoding protein, and can further include non-coding regulatory sequences and introns.
[0177] In the present invention, CLE41 and CLE42 are polypeptides comprising the amino acid sequence of FIG. 13A or 14A, respectively. CLE41 and CLE42 are ligands which are able to activate a kinase receptor, and result in phosphorylation of itself or its target. References to CLE41 and/or CLE42 include functional equivalents thereof. By a functional equivalent of CLE41 or CLE42 is meant a polypeptide which is derived from the consensus sequence of FIG. 10 by addition, deletion or substitution of one or more amino acids, preferably non-essential amino acids. Preferably, a substitution is a conservative substitution. A functional equivalent of CLE41 and/or CLE42 for use in the present invention will be biologically active, and preferably have some or all of the desired biological activity of the native polypeptide, preferably the ability to bind to PXY or a functional equivalent thereof and regulate growth and/or differentiation of the vascular tissue. Preferably, the equivalent is a signalling protein, preferably of less than 15 kDa in mass, and preferably comprising a hydrophobic region at the amino terminus. Functional equivalents may exhibit altered binding characteristics to PXY compared to a native CLE41 and/or CLE42 protein, but will mediate the same downstream signalling pathway. Preferred functional equivalents may show reduced non-desirable biological activity compared to the native protein Preferably, the equivalent comprises a conserved region of 14 amino acids having a sequence which is at least 70% more preferably 80%, 85%, 90%, 95%, 96%, 97%, 98%, 99% or 100% identical to the conserved region of amino acids 124 to 137 of the consensus of FIG. 10, and more preferably across the full length of the consensus sequence. A functional equivalent of CLE41 and/or CLE42 preferably also shares at least 50%, even more preferably at least 60%, and even more preferably at least 70%, 75%, 80%, 82%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence homology with CLE41 or CLE42. Preferably, a functional equivalent may also share sequence identity with the CLE41 and/or CLE42 sequence of FIGS. 13A and 14A, respectively. Preferably, functional equivalents have at least preferably at least 50%, even more preferably at least 60%, and even more preferably at least 70%, 75%, 80%, 82%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity with the amino acid sequence of FIGS. 13A and 14A respectively.
[0178] References to CLE41 and/or CLE42 also include fragments of the CLE41 and/or CLE42 polypeptides or their functional equivalents. A fragment is a portion of a polypeptide sequence, preferably which retains some or all of the biological activity of the full length sequence. Preferably, fragments of CLE41 and/or CLE42 retain the ability to bind PXY and regulate the growth and/or differentiation of the vascular tissue of a plant. Preferably, a fragment may be at least 7 amino acids in length, preferably at least 8, 9, or 10, 15, 20, 25, 30, 40, 50, 60, 70, 80, 90 or 100 amino acids in length, up to the full length CLE polypeptide. Most preferably, a fragment will comprise the conserved region consisting of amino acids 124 to 137 of the consensus sequence of FIG. 10.
[0179] Nucleic acid molecules encoding CLE41 and CLE42 are preferably those which encode an amino acid sequence as defined by the consensus sequence of FIG. 10, and preferably having the sequences as shown in FIG. 13B or 14B. References to nucleic acid molecules encoding CLE41 and CLE42 also include variants of the sequences of FIGS. 13B or 14B. A variant sequence is derived from the sequence of FIG. 13B or 14B by the addition, deletion or substitution of one or more nucleotide residues. The variant preferably encodes a polypeptide having CLE41 or CLE42 or a functional equivalent thereof, as defined herein. Preferably, a variant of a nucleotide sequence of FIG. 13B or 14B will have at least 50%, even more preferably at least 60%, and even more preferably at least 70%, 75%, 80%, 82%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity with a sequence of FIG. 13B or 14B. Alternatively, a variant sequence which is substantially identical to a sequence of FIG. 13B or 14B may also be defined as one which hybridises under stringent conditions to the complement of a nucleotide sequence of FIG. 13B or 14B.
[0180] Nucleic acid molecules encoding CLE41 and/or CLE42 may be derived from Arabidopsis, or may be derived from any other plant and will preferably share preferably at least 50%, even more preferably at least 60%, and even more preferably at least 70%, 75%, 80%, 82%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity with a CLE41 and/or CLE42 gene from Arabidopsis thaliana, as shown in FIG. 13B or 14B.
[0181] Also encompassed by the present invention are fragments of the nucleic acid molecules encoding CLE41 and/or or CLE42. Preferably, such fragments encode a fragment of a CLE41 or CLE42 polypeptide as defined herein. A fragment of a nucleic acid molecules encoding CLE41 or CLE42 will preferably comprise at least 21 nucleotides in length, more preferably at least 24, 27, 30 or 33 nucleotides, up to the total number of nucleotide residues in a full length sequence of FIG. 13B or 14B.
[0182] In the present invention, PXY is a polypeptide having the amino acid sequences shown in FIG. 15A. References to PXY include functional equivalents thereof. By a functional equivalent of PXY is meant a polypeptide which is derived from the native PXY polypeptide sequence of FIG. 15 by addition, deletion or substitution of one or more amino acids. A functional equivalent of PXY for use in the present invention will be biologically active, and preferably have some or all of the desired biological activity of the native polypeptide, preferably the ability to bind to CLE41 and/or CLE42 and regulate growth and/or differentiation of the vascular tissue. Functional equivalents may exhibit altered binding characteristics to CLE41 and/or CLE42 compared to a native PXY protein. Preferred functional equivalents may show reduced non-desirable biological activity compared to the native protein.
[0183] In the present invention, PXY and functional equivalents thereof are proteins found in undifferentiated procambial cells, which mediate activation of the a signalling pathway when bound by CLE41 and/or CLE42, resulting in division of the cambial cells. Preferably, PXY or its functional equivalents is a protein kinase, preferably comprising a leucine rich domain. More preferably, it comprises a LLR-RLK (Leucine Rich Repeat-Receptor-Like-Kinase) protein. Preferably, PXY or its functional equivalents are members of the XI family of Arabidopsis thaliana RLK proteins, and preferably comprise a conserved region in the kinase domain having the sequence comprising the consensus sequence of FIG. 12 or a biologically active portion thereof, or a sequence having at least 30%, 40%, 50%, 55%, 60, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, 99% or 100% sequence identity to the consensus sequence of FIG. 12. Most preferably, a functional equivalent thereof will preferably comprise an amino acid having at least 70% sequence identity to the consensus sequence of FIG. 12 and preferably will bind a CLE 41 and/or CLE 42 polypeptide or fragment thereof. A functional equivalent of PXY preferably also shares preferably at least 50%, even more preferably at least 60%, and even more preferably at least 70%, 75%, 80%, 82%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence homology with native PXY. Preferably, a functional equivalent may also share sequence identity with the PXY sequence of FIG. 15A, respectively
[0184] References to PXY also include fragments of the PXY polypeptides or its functional equivalents. A fragment is a portion of a polypeptide sequence, preferably which retains some or all of the biological activity of the full length sequence. Preferably, fragments of PXY retain the ability to bind a ligand and regulate the growth and/or differentiation of the vascular tissue of a plant. Preferably, a fragment will comprise at least a portion of the kinase domain, preferably a biologically active portion thereof, up to the full length kinase domain. Most preferably, a fragment will further comprise at least a portion of the extracellular domain, and will preferably comprise at least a portion of the LLR region.
[0185] Nucleic acid molecules encoding PXY are preferably those which encode an amino acid sequence as defined in FIG. 15A, and preferably having the sequences as shown in FIG. 15B or C. References to nucleic acid molecules encoding PXY also include variants of the sequences of FIG. 15B or C. A variant sequence is a nucleic acid molecules which is derived from the sequence of FIG. 15B or C by the addition, deletion or substitution of one or more nucleotide residues. The variant preferably encodes a polypeptide having PXY activity, as defined herein. Preferably, a variant of a nucleotide sequence of FIG. 15B or C will have at least 50%, even more preferably at least 60%, and even more preferably at least 70%, 75%, 80%, 82%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity with a sequence of FIG. 15B or C. Alternatively, a variant sequence which is substantially identical to a sequence of FIG. 15B or C may also be defined as one which hybridises under stringent conditions to the complement of a nucleotide sequence of FIG. 15B or C.
[0186] Nucleotide sequences encoding PXY may be derived from Arabidopsis, or may be derived from any other plant and will preferably share preferably at least 50%, even more preferably at least 60%, and even more preferably at least 70%, 75%, 80%, 82%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93% , 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity with a PXY gene from Arabidopsis thaliana, as shown in FIG. 15B or C. Genbank references are PXY=At5g61480 (TAIR), PXL1=At1g08590 (TAIR), PXL2=At4g28650 (TAIR).
[0187] Also encompassed by the present invention are fragments of the nucleic acid molecule encoding PXY. Preferably, such fragments encode a fragment of a PXY polypeptide as defined herein. A fragment of a nucleic acid molecule encoding PXY will preferably comprise at least 10, 20, 30, 40, 50, 60, 70, 80 or 90 or 100, 200 or 300 or more nucleotides in length, up to the total number of nucleotide residues in a full length sequence of FIG. 15A.
[0188] Also provided in the present invention are antisense sequences of the above mentioned nucleic acid molecules, which hybridise under stringent conditions to the nucleotide sequences encoding CLE41 and/or CLE42 or PXY, or a functional equivalents thereof, as defined above. Such sequences are useful in down regulating expression of the CLE41 and/or CLE42 and/or PXY or functional equivalents thereof. Whilst in a preferred embodiment, both receptor and ligand will be either up-regulated (over-expressed) or down-regulated in a cell of a plant, it is envisaged that it may in certain circumstances be desirable to up-regulate either the receptor whilst down-regulating the ligand, or vice versa.
[0189] As used herein, the term "hybridizes under stringent conditions" describes conditions for hybridization and washing. Stringent conditions are known to those skilled in the art and can be found in available references (e.g., Current Protocols in Molecular Biology, John Wiley & Sons, N.Y., 1989, 6.3.1-6.3.6). Aqueous and non-aqueous methods are described in that reference and either can be used. A preferred example of stringent hybridization conditions are hybridization in 6× sodium chloride/sodium citrate (SSC) at about 45° C., followed by one or more washes in 0.2×SSC, 0.1% (w/v) SDS at 50° C. Another example of stringent hybridization conditions are hybridization in 6×SSC at about 45° C., followed by one or more washes in 0.2×SSC, 0.1% (w/v) SDS at 55° C. A further example of stringent hybridization conditions are hybridization in 6×SSC at about 45° C., followed by one or more washes in 0.2×SSC, 0.1% (w/v) SDS at 60° C. Preferably, stringent hybridization conditions are hybridization in 6×SSC at about 45° C., followed by one or more washes in 0.2×SSC, 0.1% (w/v) SDS at 65° C. Particularly preferred stringency conditions (and the conditions that should be used if the practitioner is uncertain about what conditions should be applied to determine if a molecule is within a hybridization limitation of the invention) are 0.5 molar sodium phosphate, 7% (w/v) SDS at 65° C., followed by one or more washes at 0.2×SSC, 1% (w/v) SDS at 65° C. Preferably, an isolated nucleic acid molecule of the invention that hybridizes under stringent conditions to the sequence of FIG. 15B or C corresponds to a naturally-occurring nucleic acid molecule.
[0190] Sequence identity is determined by comparing the two aligned sequences over a pre-determined comparison window, and determining the number of positions at which identical residues occur. Typically, this is expressed as a percentage. The measurement of sequence identity of a nucleotide sequences is a method well known to those skilled in the art, using computer implemented mathematical algorithms such as ALIGN (Version 2.0), GAP, BESTFIT, BLAST (Altschul et al J. Mol. Biol. 215: 403 (1990)), FASTA and TFASTA (Wisconsin Genetic Software Package Version 8, available from Genetics Computer Group, Accelrys Inc. San Diego, Calif.), and CLUSTAL (Higgins et al, Gene 73: 237-244 (1998)), using default parameters.
[0191] Calculations of sequence homology or identity between sequences are performed as follows:
[0192] To determine the percent identity of two amino acid sequences, or of two nucleic acid sequences, the sequences are aligned for optimal comparison purposes (e.g., gaps can be introduced in one or both of a first and a second amino acid or nucleic acid sequence for optimal alignment and non-homologous sequences can be disregarded for comparison purposes). In a preferred embodiment, the length of a reference sequence aligned for comparison purposes is at least 30%, preferably at least 40%, more preferably at least 50%, even more preferably at least 60%, and even more preferably at least 70%, 75%, 80%, 82%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% of the length of the reference sequence. The amino acid residues or nucleotides at corresponding amino acid positions or nucleotide positions are then compared. When a position in the first sequence is occupied by the same amino acid residue or nucleotide as the corresponding position in the second sequence, then the molecules are identical at that position. The percent identity between the two sequences is a function of the number of identical positions shared by the sequences, taking into account the number of gaps, and the length of each gap, which need to be introduced for optimal alignment of the two sequences.
[0193] The comparison of sequences and determination of percent identity between two sequences can be accomplished using a mathematical algorithm. In a preferred embodiment, the percent identity between two amino acid sequences is determined using the Needleman et al. (1970) J. Mol. Biol. 48:444-453) algorithm which has been incorporated into the GAP program in the GCG software package (available at http://www.gcg.com), using either a BLOSUM 62 matrix or a PAM250 matrix, and a gap weight of 16, 14, 12, 10, 8, 6, or 4 and a length weight of 1, 2, 3, 4, 5, or 6. In yet another preferred embodiment, the percent identity between two nucleotide sequences is determined using the GAP program in the GCG software package (available at http://www.gcg.com), using a NWSgapdna.CMP matrix and a gap weight of 40, 50, 60, 70, or 80 and a length weight of 1, 2, 3, 4, 5, or 6. A particularly preferred set of parameters (and the one that should be used if the practitioner is uncertain about what parameters should be applied to determine if a molecule is within a sequence identity or homology limitation of the invention) are a BLOSUM 62 scoring matrix with a gap penalty of 12, a gap extend penalty of 4, and a frameshift gap penalty of 5.
[0194] The percent identity between two amino acid or nucleotide sequences can be determined using the algorithm of Meyers et al. (1989) CABIOS 4:11-17) which has been incorporated into the ALIGN program (version 2.0), using a PAM120 weight residue table, a gap length penalty of 12 and a gap penalty of 4.
[0195] The nucleic acid and protein sequences described herein can be used as a "query sequence" to perform a search against public databases to, for example, identify other family members or related sequences. Such searches can be performed using the NBLAST and XBLAST programs (version 2.0) of Altschul, et al. (1990) J. Mol. Biol. 215:403-410). BLAST nucleotide searches can be performed with the NBLAST program, score=100, wordlength=12 to obtain nucleotide sequences homologous to nucleic acid molecules of the invention. BLAST protein searches can be performed with the XBLAST program, score=50, wordlength=3 to obtain amino acid sequences homologous to protein molecules of the invention. To obtain gapped alignments for comparison purposes, gapped BLAST can be utilized as described in Altschul et al. (1997, Nucl. Acids Res. 25:3389-3402). When using BLAST and gapped BLAST programs, the default parameters of the respective programs (e.g., XBLAST and NBLAST) can be used. See <http://www.ncbi.nlm.nih.gov>.
[0196] Sequence comparisons are preferably made over the full-length of the relevant sequence described herein.
[0197] The polypeptide sequences and nucleic acid molecules used in the present invention may be isolated or purified. By "purified" is meant that they are substantially free from other cellular components or material, or culture medium. "Isolated" means that they may also be free of naturally occurring sequences which flank the native sequence, for example in the case of nucleic acid molecule, isolated may mean that it is free of 5' and 3' regulatory sequences.
[0198] The polypeptide and nucleic acid molecule used in the invention may be naturally occurring or may be synthetic. The nucleic acid molecule may be recombinant.
[0199] The present invention is based upon using either CLE41 and/or CLE42 and/or a PXY, or functional equivalents thereof, to manipulate the growth and/or structure of a plant. By "manipulate" is meant altering the native growth pattern of a plant, compared to that of a non-manipulated plant of the same species, grown under identical conditions. The manipulation is preferably effected by altering the levels of said receptor and ligand in a cell of the plant.
[0200] A plant having increased levels of said CLE41 and/or CLE42 and/or a PXY, or functional equivalents thereof in a particular tissue and at a pre-selected developmental stage, compared to the native levels in the same tissue of a native plant of the same species, at the same developmental stage and grown in identical conditions.
[0201] Herein, the growth of a plant refers to the size of a plant, preferably the secondary growth, and preferably the amount of vascular and/or interfasicular tissue, more preferably the amount of xylem cells, also referred to as the woody tissue or biomass of a plant.
[0202] By identical conditions is meant conditions which are the substantially the same in terms of temperature, light, and availability of nutrients and water. By substantially is meant that the conditions may vary slightly, but not to an extent to which is known to affect the growth of a plant.
[0203] The structure of a plant refers to the organisation of tissue in a plant, preferably the vascular tissue, most preferably the polarity of the phloem and xylem cells.
[0204] The use of said CLE41 and/or CLE42 and/or a PXY, or functional equivalents thereof as defined herein to manipulate the growth and/or structure of a plant may be achieved in any manner which alters the regulation of the signalling pathway mediated by CLE41 and/or CLE42 binding to PXY. Preferably, the invention may be achieved in any manner which up-regulates the signalling pathway. Preferably, the manipulation is mediated via a PXY ligand as defined herein, preferably CLE41 and/or CLE42, or via a CLE41 and/or CLE42 receptor, preferably PXY. For example, manipulation may comprise altering their expression pattern within the plant, altering the amount of said receptor and/or ligand within the plant, or altering the binding pattern thereof.
[0205] By modulation of the levels of the CLE41 and/or CLE42 and/or a PXY, or functional equivalents thereof, is meant an increase or decrease in the levels of in the plant, preferably the levels localised in the vascular tissue, and preferably in the cambium or procambium of a plant, as compared to the levels in the same tissue in a native plant of the same species at the same stage if developed and grown under identical conditions, and in which no modulation has been made. Preferably, the levels of CLE41 and/or CLE42 and/or a PXY, or functional equivalents thereof, are increased. Preferred levels of PXY ligand are at least 5, 10, 20, 30, 40, 50, 60, 70, 80, 90% more or less relative to said native plant. Preferred levels of CLE41 and/or CLE42 receptor are 5, 10, 20, 30, 40, 50, 60, 70, 80, 90% more or less relative to said native plant.
[0206] The alteration in levels of CLE41 and/or CLE42 and/or a PXY, or functional equivalents thereof, as defined above preferably increases or decreases the activity by at least about 2-fold compared to a basal level of activity. More preferably said activity is increased or decreased by at least about 5 fold; 10 fold; 20 fold, 30 fold, 40 fold, 50 fold. Preferably said activity is increased or decreased by between at least 50 fold and 100 fold. Preferably said increase or decrease is greater than 100-fold.
[0207] It will be apparent that means to modulate the activity of a polypeptide encoded by a nucleic acid molecule are known to the skilled artisan. For example, and not by limitation, altering the gene dosage by providing a cell with multiple copies of said gene or its complement. Alternatively, or in addition, a gene(s) may be placed under the control of a powerful promoter sequence or an inducible promoter sequence to elevate expression of mRNA encoded by said gene. The modulation of mRNA stability is also a mechanism used to alter the steady state levels of an mRNA molecule, typically via alteration to the 5' or 3' untranslated regions of the mRNA.
[0208] It is envisaged that where a plant naturally expresses said CLE41 and/or CLE42 receptor and/or PXY ligand, their modulation may be achieved by altering the expression pattern of the native gene(s) and/or production of the polypeptide. This may be achieved by any suitable method, including altering transcription of the gene, and/or translation of the mRNA into polypeptide, and post-translational modification of the polypeptide.
[0209] Altering the expression pattern of a native gene may be achieved by placing it under control of a heterologous regulatory sequence, which is capable of directing the desired expression pattern of the gene. Suitable regulatory sequences are described herein. Alternatively, regulation of expression of the native gene may be altered through changing the pattern of transcription factors which mediate expression of the gene. This may require the use of modified transcription factors, whose binding pattern is altered to obtain a desired expression pattern of the gene. Alternatively, the copy number of the native gene may be increased or decreased, in order to change the amount of expression of the gene. Suitable methods for carrying out these embodiments of the invention are known to persons skilled in the art, and may employ the use of an expression construct according to the invention.
[0210] Plants transformed with a nucleic acid molecule or expression construct of the invention may be produced by standard techniques known in the art for the genetic manipulation of plants. DNA can be introduced into plant cells using any suitable technology, such as a disarmed Ti-plasmid vector carried by Agrobacterium exploiting its natural gene transferability (EP-A-270355, EP-A-0116718, NAR 12(22):8711-87215 (1984), Townsend et al., U.S. Pat. No. 5,563,055); particle or microprojectile bombardment (U.S. Pat. No. 5,100,792, EP-A-444882, EP-A-434616; Sanford et al, U.S. Pat. No. 4,945,050; Tomes et al. (1995) "Direct DNA Transfer into Intact Plant Cells via Microprojectile Bombardment", in Plant Cell, Tissue and Organ Culture: Fundamental Methods, ed. Gamborg and Phillips (Springer-Verlag, Berlin); and McCabe et al. (1988) Biotechnology 6: 923-926); microinjection (WO 92/09696, WO 94/00583, EP 331083, EP 175966, Green et al. 91987) Plant Tissue and Cell Culture, Academic Press, Crossway et al. (1986) Biotechniques 4:320-334); electroporation (EP 290395, WO 8706614, Riggs et al. (1986) Proc. Natl. Acad. Sci. USA 83:5602-5606; D'Halluin et al. 91992). Plant Cell 4:1495-1505) other forms of direct DNA uptake (DE 4005152, WO 9012096, U.S. Pat. No. 4,684,611, Paszkowski et al. (1984) EMBO J. 3:2717-2722); liposome-mediated DNA uptake (e.g. Freeman et al (1984) Plant Cell Physiol, 29:1353); or the vortexing method (e.g. Kindle (1990) Proc. Nat. Acad. Sci. USA 87:1228). Physical methods for the transformation of plant cells are reviewed in Oard (1991) Biotech. Adv. 9:1-11. See generally, Weissinger et al. (1988) Ann. Rev. Genet. 22:421-477; Sanford et al. (1987) Particulate Sciences and Technology 5:27-37; Christou et al. (1988) Plant Physiol. 87:671-674; McCabe et al. (1988) Bio/Technology 6:923-926; Finer and McMullen (1991) In Vitro Cell Dev. Biol. 27P:175-182; Singh et al. (1988) Theor. Appl. Genet. 96:319-324; Datta et al. (1990) Biotechnology 8:736-740; Klein et al. (1988) Proc. Natl. Acad. Sci. USA 85: 4305-4309; Klein et al. (1988) Biotechnology 6:559-563; Tomes, U.S. Pat. No. 5,240,855; Buising et al. U.S. Pat. Nos. 5,322,783 and 5,324,646; Klein et al. (1988) Plant Physiol 91: 440-444; Fromm et al (1990) Biotechnology 8:833-839; Hooykaas-Von Slogteren et al. 91984). Nature (London) 311:763-764; Bytebier et al. (1987) Proc. Natl. Acad. Sci. USA 84:5345-5349; De Wet et al. (1985) in The Experimental Manipulation of Ovule Tissues ed. Chapman et al. (Longman, New York), pp. 197-209; Kaeppler et al. (1990) Plant Cell Reports 9:415-418 and Kaeppler et al. (1992) Theor. Appl. Genet. 84:560-566; Li et al. (1993) Plant Cell Reports 12: 250-255 and Christou and Ford (1995) Annals of Botany 75: 407-413; Osjoda et al. (1996) Nature Biotechnology 14:745-750, all of which are herein incorporated by reference.
[0211] Agrobacterium transformation is widely used by those skilled in the art to transform dicotyledonous species. Recently, there has been substantial progress towards the routine production of stable, fertile transgenic plants in almost all economically relevant monocot plants (Toriyama et al. (1988) Bio/Technology 6: 1072-1074; Zhang et al. (1988) Plant Cell rep. 7379-384; Zhang et al. (1988) Theor. Appl. Genet. 76:835-840; Shimamoto et al. (1989) Nature 338:274-276; Datta et al. (1990) Bio/Technology 8: 736-740; Christou et al. (1991) Bio/Technology 9:957-962; Peng et al (1991) International Rice Research Institute, Manila, Philippines, pp. 563-574; Cao et al. (1992) Plant Cell Rep. 11: 585-591; Li et al. (1993) Plant Cell Rep. 12: 250-255; Rathore et al. (1993) Plant Mol. Biol. 21:871-884; Fromm et al (1990) Bio/Technology 8:833-839; Gordon Kamm et al. (1990) Plant Cell 2:603-618; D'Halluin et al. (1992) Plant Cell 4:1495-1505; Walters et al. (1992) Plant Mol. Biol. 18:189-200; Koziel et al. (1993). Biotechnology 11194-200; Vasil, I. K. (1994) Plant Mol. Biol. 25:925-937; Weeks et al (1993) Plant Physiol. 102:1077-1084; Somers et al. (1992) Bio/Technology 10:1589-1594; WO 92/14828. In particular, Agrobacterium mediated transformation is now emerging also as an highly efficient transformation method in monocots. (Hiei, et al. (1994) The Plant Journal 6:271-282). See also, Shimamoto, K. (1994) Current Opinion in Biotechnology 5:158-162; Vasil, et al. (1992) Bio/Technology 10:667-674; Vain, et al. (1995) Biotechnology Advances 13(4):653-671; Vasil, et al. (1996) Nature Biotechnology 14: 702).
[0212] Microprojectile bombardment, electroporation and direct DNA uptake are preferred where Agrobacterium is inefficient or ineffective. Alternatively, a combination of different techniques may be employed to enhance the efficiency of the transformation process, e.g. bombardment with Agrobacterium-coated microparticles (EP-A-486234) or microprojectile bombardment to induce wounding followed by co-cultivation with Agrobacterium (EP-A-486233).
[0213] Altering the production of a polypeptide may be achieved by increasing the amount of mRNA produced, increasing the stability of protein, altering the rate of post translational modification for example altering rates of proteolytic cleavage.
[0214] Altering the post-translational modification of a polypeptide may also affect its structure and function, and may be used to alter the expression of the native polypeptide. For example, the ligand is likely to be only a portion of the full length proteins and the active ligand is probably released by proteolysis.
[0215] Alternatively, a polypeptide or nucleic acid molecule as defined herein may be introduced into the plant, by any suitable means such as spraying, uptake by the roots, or injection into phloem. To down-regulate said receptor or ligand in a plant, an enzyme may be introduced which inhibits or digests one or both of the receptor or ligand.
[0216] In addition, modulating the activity mediated by CLE41 and/or CLE42 and/or a PXY, or functional equivalents thereof, by altering their binding pattern, in order to up-or-down-regulate the downstream signalling pathway. The binding pattern may be altered in any suitable way, for example by altering the structure, binding affinity, temporal binding pattern, selectivity and amount available for binding on the cell surface of CLE41 and/or CLE42 and/or a PXY, or functional equivalents thereof.
[0217] The binding pattern may be altered by making appropriate variations to the ligand polypeptide, for example to change its binding site to the receptor, using known methods for mutagenesis. Alternatively, non-protein analogues may be used. Methods for manipulating a polypeptide used in the present invention are known in the art, and include for example altering the nucleic acid sequence encoding the polypeptide. Methods for mutagenesis are well known. Preferably, where variants are produced using mutagenesis of the nucleic acid coding sequence, this is done in a manner which does not affect the reading frame of the sequence and which does not affect the polypeptide in a manner which affects the desired biological activity.
[0218] In selecting suitable variants for use in the present invention, routine assays may be used to screen for those which have the desired properties. This may be done by visual observation of plants and plant material, or measuring the biomass of the plant or plant material.
[0219] Thus, for use in altering the expression of the CLE41 and/or CLE42 and/or a PXY, or functional equivalents thereof, in a cell of a plant, there is provided an expression cassette comprising a regulatory sequence to modulate the expression of the native CLE41 and/or CLE42 or PXY genes in a plant. Preferably, the regulatory sequences are designed to be operably linked to the native gene, in order to direct expression in a manner according to the present invention.
[0220] The nucleic acid molecules as described herein, and/or a regulatory sequence are preferably provided as part of an expression cassette, for expression of the sequence in a cell of interest. Suitable expression cassettes may also comprise 5' and 3' regulatory sequences operably linked to the sequences of interest. In addition, genes encoding, for example, selectable markers and reporter genes may be included. The expression cassette will preferably also contain one or more restriction sites, to enable insertion of the nucleotide sequence and/or a regulatory sequence into the plant genome, at a pre-selected position. Also provided on the expression cassette may be transcription and translation initiation regions, to enable expression of the incoming genes, transcription and translational termination regions, and regulatory sequences. These sequences may be native to the plant being transformed, or may be heterologous and/or foreign.
[0221] Heterologous sequences are sequences which in nature are not operably linked to each other and/or are not found next to each other in a native sequence. In contrast, homologous sequences refer to sequences which share sequence similarity, which may be described as sequence homology. Homology is usually in a fragment of the sequence, typically in a functional domain of the sequence.
[0222] A foreign sequence is one which is not found in the native genome of the plant being transformed.
[0223] A regulatory sequence is a nucleotide sequence which is capable of influencing transcription or translation of a gene or gene product, for example in terms of initiation, rate, stability, downstream processing, and mobility. Examples of regulatory sequences include promoters, 5' and 3' UTR's, enhancers, transcription factor or protein binding sequences, start sites and termination sequences, ribozyme binding sites, recombination sites, polyadenylation sequences, sense or antisense sequences. They may be DNA, RNA or protein. The regulatory sequences may be plant- or virus derived, and preferably may be derived from the same species of plant as the plant being modulated.
[0224] By "promoter" is meant a nucleotide sequence upstream from the transcriptional initiation site and which contains all the regulatory regions required for transcription. Suitable promoters include constitutive, tissue-specific, inducible, developmental or other promoters for expression in plant cells comprised in plants depending on design. Such promoters include viral, fungal, bacterial, animal and plant-derived promoters capable of functioning in plant cells.
[0225] Constitutive promoters include, for example CaMV 35S promoter (Odell et al. (1985) Nature 313, 9810-812); rice actin (McElroy et al. (1990) Plant Cell 2: 163-171); ubiquitin (Christian et al. (1989) Plant Mol. Biol. 18 (675-689); pEMU (Last et al. (1991) Theor Appl. Genet. 81: 581-588); MAS (Velten et al. (1984) EMBO J. 3. 2723-2730); ALS promoter (U.S. application Ser. No. 08/409,297), and the like. Other constitutive promoters include those in U.S. Pat. Nos. 5,608,149; 5,608,144; 5,604,121; 5,569,597; 5,466,785; 5,399,680, 5,268,463; and 5,608,142.
[0226] Chemical-regulated promoters can be used to modulate the expression of a gene in a plant through the application of an exogenous chemical regulator. Depending upon the objective, the promoter may be a chemical-inducible promoter, where application of the chemical induced gene expression, or a chemical-repressible promoter, where application of the chemical represses gene expression. Chemical-inducible promoters are known in the art and include, but are not limited to, the maize In2-2 promoter, which is activated by benzenesulfonamide herbicide safeners, the maize GST promoter, which is activated by hydrophobic electrophilic compounds that are used as pre-emergent herbicides, and the tobacco PR-1 a promoter, which is activated by salicylic acid. Other chemical-regulated promoters of interest include steroid-responsive promoters (see, for example, the glucocorticoid-inducible promoter in Schena et al. (1991) Proc. Natl. Acad. Sci. USA 88: 10421-10425 and McNellis et al. (1998) Plant J. 14(2): 247-257) and tetracycline-inducible and tetracycline-repressible promoters (see, for example, Gatz et al. (1991) Mol. Gen. Genet. 227: 229-237, and U.S. Pat. Nos. 5,814,618 and 5,789,156, herein incorporated by reference.
[0227] Where enhanced expression in particular tissues is desired, tissue-specific promoters can be utilised. Tissue-specific promoters include those described by Yamamoto et al. (1997) Plant J. 12(2): 255-265; Kawamata et al. (1997) Plant Cell Physiol. 38(7): 792-803; Hansen et al. (1997) Mol. Gen. Genet. 254(3): 337-343; Russell et al. (1997) Transgenic Res. 6(2): 157-168; Rinehart et al. (1996) Plant Physiol. 112(3): 1331-1341; Van Camp et al. (1996) Plant Physiol. 112(2): 525-535; Canevascni et al. (1996) Plant Physiol. 112(2): 513-524; Yamamoto et al. (1994) Plant Cell Physiol. 35(5): 773-778; Lam (1994) Results Probl. Cell Differ. 20: 181-196; Orozco et al. (1993) Plant Mol. Biol. 23(6): 1129-1138; Mutsuoka et al. (1993) Proc. Natl. Acad. Sci. USA 90 (20): 9586-9590; and Guevara-Garcia et al (1993) Plant J. 4(3): 495-50.
[0228] "Operably linked" means joined as part of the same nucleic acid molecule, suitably positioned and oriented for transcription to be initiated from the promoter. DNA operably linked to a promoter is "under transcriptional initiation regulation" of the promoter. In a preferred aspect, the promoter is an inducible promoter or a developmentally regulated promoter.
[0229] The promoters which control the expression of CLE41 and/or CLE42 are preferably tissue or organ specific, such that expression of CLE41 and/or CLE42 can be directed to a particular organ or tissue, such as the vascular tissue, preferably the cambium or procambium, and most preferably phloem or xylem tissue. The promoters may be constitutive, whereby they direct expression under most environmental or developmental conditions. More preferably, the promoter is inducible, and will direct expression in response to environmental or developmental cues, such as temperature, chemicals, drought, and others. The promoter may also be developmental stage specific.
[0230] Examples of suitable promoter sequences include those of the T-DNA of A. tumefaciens, including mannopine synthase, nopaline synthase, and octopine synthase; alcohol dehydrogenase promoter from corn; light inducible promoters such as ribulose-biphosphate-carboxylase small subunit gene from various species and the major chlorophyll a/b binding protein gene promoter; histone promoters (EP 507 698), actin promoters; maize ubiquitin 1 promoter (Christensen et al. (1996) Transgenic Res. 5:213); 35S and 19S promoters of cauliflower mosaic virus; developmentally regulated promoters such as the waxy, zein, or bronze promoters from maize; as well as synthetic or other natural promoters including those promoters exhibiting organ specific expression or expression at specific development stage(s) of the plant, like the alpha-tubulin promoter disclosed in U.S. Pat. No. 5,635,618. Preferred phloem specific promoters include SUC2, APL, KAN1, KAN2, At4g33660, At3g61380, and At1g79380. Preferred xylem specific promoters include REV, IRX1 COBL4, KOR, At2g38080, and At1g2744.
[0231] Suitable expression cassettes for use in the present invention can be constructed, containing appropriate regulatory sequences, including promoter sequences, terminator fragments, polyadenylation sequences, enhancer sequences, marker genes and other sequences as appropriate. For further details see, for example, Molecular Cloning: Laboratory Manual: 2nd edition, Sambrook et al. 1989, Cold Spring Harbor Laboratory Press or Current Protocols in Molecular Biology, Second Edition, Ausubel et al. Eds., John Wiley & Sons, 1992. The expression cassettes may be a bi-functional expression cassette which functions in multiple hosts. In the case of GTase genomic DNA this may contain its own promoter or other regulatory elements and in the case of cDNA this may be under the control of an appropriate promoter or other regulatory elements for expression in the host cell.
[0232] An expression cassette including a nucleic acid molecule according to the invention need not include a promoter or other regulatory sequence, particularly if the vector is to be used to introduce the nucleic acid into cells for recombination into the gene.
[0233] Suitable selectable marker or reporter genes may be used to facilitate identification and selection of transformed cells. These will confer a selective phenotype on the plant or plant cell to enable selection of those cells which comprise the expression cassette. Preferred genes include the chloramphenicol acetyl transferase (cat) gene from Tn9 of E. coli, the beta-gluronidase (gus) gene of the uidA locus of E. coli, the green fluorescence protein (GFP) gene from Aequoria victoria, and the luciferase (luc) gene from the firefly Photinus pyralis. If desired, selectable genetic markers may be included in the construct, such as those that confer selectable phenotypes such as resistance to antibodies or herbicides (e.g. kanamycin, hygromycin, phosphinotricin, chlorsulfuron, methotrexate, gentamycin, spectinomycin, imidazolinones and glyphosate).
[0234] Reporter genes which encode easily assayable marker proteins are well known in the art. In general, a reporter gene is a gene which is not present or expressed by the recipient organism or tissue and which encodes a protein whose expression is manifested by some easily detectable property, e.g. phenotypic change or enzymatic activity.
[0235] The selectable marker or reporter gene may be carried on a separate expression cassette and co-transformed with the expression cassette of the invention. The selectable markers and/or reporter genes may be flanked with appropriate regulatory sequences to enable their expression in plants.
[0236] The expression cassette may also comprise elements such as introns, enhancers, and polyadenylation sequences. These elements must be compatible with the remainder of the expression cassette. These elements may not be necessary for the expression or function of the gene but may serve to improve expression or functioning of the gene by affecting transcription, stability of the mRNA, or the like. Therefore, such elements may be included in the expression construct to obtain the optimal expression and function of CLE41 and/or CLE42 and/or PXY in the plant.
[0237] The expression cassette comprising the heterologous nucleic acid may also comprise sequences coding for a transit peptide, to drive the protein coded by the heterologous gene into a desired part of the cell, for example the chloroplasts. Such transit peptides are well known to those of ordinary skill in the art, and may include single transit peptides, as well as multiple transit peptides obtained by the combination of sequences coding for at least two transit peptides. One preferred transit peptide is the Optimized Transit Peptide disclosed in U.S. Pat. No. 5,635,618, comprising in the direction of transcription a first DNA sequence encoding a first chloroplast transit peptide, a second DNA sequence encoding an N-terminal domain of a mature protein naturally driven into the chloroplasts, and a third DNA sequence encoding a second chloroplast transit peptide.
[0238] In the present invention, any plant species may be used, including both monocots and dicots. Preferred plants for use in the present invention are those which are targets for biomass, and/or are readily grown, exhibit high growth rates, are easily harvested, and can be readily converted to a biofuel. Preferred plants include grasses, trees, crops, and shrubs.
[0239] Suitable plants for use in the present invention are those which in their native form produce a high yield of feedstock, for paper or fuel production. Examples of suitable plant types include perennial fast growing herbaceous and woody plants, for example trees, shrubs and grasses. Preferred trees for use in the invention include poplar, hybrid poplar, willow, silver maple, black locust, sycamore, sweetgum and eucalyptus. Preferred shrubs include tobacco. Perennial grasses include switchgrass, reed canary grass, prairie cordgrass, tropical grasses, Brachypodiumdistachyon, and Miscanthes. Crops include wheat, soybean, alphalpha, corn, rice, maize, and sugar beet.
[0240] In yet still a further preferred embodiment of the invention said plant is a woody plant selected from: poplar; eucalyptus; Douglas fir; pine; walnut; ash; birch; oak; teak; spruce. Preferably said woody plant is a plant used typically in the paper industry, for example poplar.
[0241] Methods to transform woody species of plant are well known in the art. For example the transformation of poplar is disclosed in U.S. Pat. No. 4,795,855 and WO9118094. The transformation of eucalyptus is disclosed in EP1050209 and WO9725434. Each of these patents is incorporated in their entirety by reference.
[0242] In a still further preferred embodiment of the invention said plant is selected from: corn (Zea mays), canola (Brassica napus, Brassica rapa ssp.), alfalfa (Medicago sativa), rice (Oryza sativa), rye (Secale cerale), sorghum (Sorghum bicolor, Sorghum vulgare), sunflower (helianthus annuas), wheat (Tritium aestivum), soybean (Glycine max), tobacco (Nicotiana tabacum), potato (Solanum tuberosum), peanuts (Arachis hypogaea), cotton (Gossypium hirsutum), sweet potato (Iopmoea batatus), cassava (Manihot esculenta), coffee (Cofea spp.), coconut (Cocos nucifera), pineapple (Anana comosus), citrus tree (Citrus spp.) cocoa (Theobroma cacao), tea (Camellia senensis), banana (Musa spp.), avocado (Persea americana), fig (Ficus casica), guava (Psidium guajava), mango (Mangifer indica), olive (Olea europaea), papaya (Carica papaya), cashew (Anacardium occidentale), macadamia (Macadamia intergrifolia), almond (Prunus amygdalus), sugar beets (Beta vulgaris), oats, barley, vegetables and ornamentals.
[0243] Preferably, plants of the present invention are crop plants (for example, cereals and pulses, maize, wheat, potatoes, tapioca, rice, sorghum, millet, cassava, barley, pea, and other root, tuber or seed crops. Important seed crops are oil-seed rape, sugar beet, maize, sunflower, soybean, and sorghum. Horticultural plants to which the present invention may be applied may include lettuce, endive, and vegetable brassicas including cabbage, broccoli, and cauliflower, and carnations and geraniums. The present invention may be applied in tobacco, cucurbits, carrot, strawberry, sunflower, tomato, pepper, chrysanthemum.
[0244] Grain plants that provide seeds of interest include oil-seed plants and leguminous plants. Seeds of interest include grain seeds, such as corn, wheat, barley, rice, sorghum, rye, etc. Oil-seed plants include cotton, soybean, safflower, sunflower, Brassica, maize, alfalfa, palm, coconut, etc. Leguminous plants include beans and peas. Beans include guar, locust bean, fenugreek, soybean, garden beans, cowpea, mungbean, lima bean, fava been, lentils, chickpea, etc.
[0245] In a preferred embodiment of the invention said seed is produced from a plant selected from the group consisting of: corn (Zea mays), canola (Brassica napus, Brassica rapa ssp.), flax (Linum usitatissimum), alfalfa (Medicago sativa), rice (Oryza sativa), rye (Secale cerale), sorghum (Sorghum bicolor, Sorghum vulgare), sunflower (Helianthus annus), wheat (Tritium aestivum), soybean (Glycine max), tobacco (Nicotiana tabacum), potato (Solanum tuberosum), peanuts (Arachis hypogaea), cotton (Gossypium hirsutum), sweet potato (Iopmoea batatus), cassava (Manihot esculenta), coffee (Cofea spp.), coconut (Cocos nucifera), pineapple (Anana comosus), citrus tree (Citrus spp.) cocoa (Theobroma cacao), tea (Camellia senensis), banana (Musa spp.), avocado (Persea americana), fig (Ficus casica), guava (Psidium guajava), mango (Mangifer indica), olive (Olea europaea), papaya (Carica papaya), cashew (Anacardium occidentale), macadamia (Macadamia intergrifolia), almond (Prunus amygdalus), sugar beets (Beta vulgaris), oats, barley, vegetables.
[0246] The present invention has uses in methods which require increased biomass in plants, for example where plant biomass is used in the manufacture of products such as biofuels and paper. The invention is not limited to methods of making these particular products, and it is envisaged that the invention will be applicable to the manufacture of a variety of plant based products. In addition, the invention is also useful in altering the characteristics of plant material, such that the plant material can be adapted for particular purposes. In one such embodiment, over expression of the ligand and/or receptor as defined herein may be used to increase the number of cells in the vascular tissue of a plant, but without increasing the actual biomass of the plant (i.e. the number of cells may be increased, but the size of these cells is smaller). This has the effect of increasing the density of the vascular tissue, and therefore producing a harder wood. Thus, the invention includes methods for the production of a wood product having a particular density. In addition, it is envisaged that by manipulating plant cells to differentiate their vascular tissue, and therefore grow, environmental growth signals may be bypassed and the present invention may be used to extend the growth season of plants, beyond that which would be possible in a native plant.
[0247] The embodiments described in relation to the each aspect apply to the other aspects of the invention, mutatis mutandis.
[0248] Throughout the description and claims of this specification, the words "comprise" and "contain" and variations of the words, for example "comprising" and "comprises", means "including but not limited to", and is not intended to (and does not) exclude other moieties, additives, components, integers or steps.
[0249] Throughout the description and claims of this specification, the singular encompasses the plural unless the context otherwise requires. In particular, where the indefinite article is used, the specification is to be understood as contemplating plurality as well as singularity, unless the context requires otherwise.
[0250] Features, integers, characteristics, compounds, chemical moieties or groups described in conjunction with a particular aspect, embodiment or example of the invention are to be understood to be applicable to any other aspect, embodiment or example described herein unless incompatible therewith.
[0251] The present invention will now be described with reference to the following non-limiting examples:
EXAMPLES
[0252] DNA manipulation was carried out using standard methods. Over expression (35S) constructs for plant transformation were generated by cloning CLE41, CLE42, and PXY genomic DNA sequences into pK2GW7,0 (M. Karimi, D. Inze, A. Depicker, Trends in Plant Science 7, 193 (2002)) using gateway technology (invitrogen) with primers listed in the table. Sequences were amplified by PCR and cloned into pENTR-D-TOPO. Subsequently, reactions containing LR clonase II and pK2GW7,0 and the relevant TOPO vector, sequences were used to transfer sequences in pENTR-D-TOPO to the binary plasmid. pIRX3::CLE41/42 were constructed using the p3HSC Gateway destination vector (Atanassov et al. 2008) derived from pCB1300 by insertion of the 1.7 kb promoter sequences of irx3, the frame A (attR1/CmR/ccdB/attR2) cassette (Invitrogen) and the NOS terminator region from pGPTV-BAR. For SUC2::CLE41 we used overlapping PCR. The SUC2 promoter and CLE41 coding sequence were amplified separately with overlapping ends. These products were mixed, annealed and elongated prior to amplification with SUC2 and CLE41 forward and reverse oligos respectively. The resulting PCR product was cloned into pTF101.gw1 (Paz et al. 2004) via pENTR-D-TOPO. For tissue specific expression, promoters known to give xylem (IRX3; (Gardiner et al. 2003) or phloem (SUC2; (Truernit and Sauer 1995) specific expression were used. Plasmids were sequenced and transformed into Arabidopsis using the method of Clough and Bent (S. J. Clough, A. F. Bent, Plant Journal 16, 735 (December 1998)),
[0253] In order to understand if over expression of PXY and CLE41 had the same effect in poplar, plants were transformed in tissue culture using Agrobacterium to transfer the constructs into poplar tissue using the method of Meilan and Ma (R. Meilan, C. Ma, Methods in Molecular Biology 344, 143 (2006). 35S::CLE41, SUC::CLE41 and wild type were grown in magenta boxes in the growth cabinet under the same conditions. Similarly, plasmids were transformed into Nicotiana using the method of Horsch et al (R. B. Horsch et al., Science 227, 1229 (Mar. 8, 1985). Maize was transformed with the plasmids by contracting out transformation services. Similar phenotypes were confirmed in 10 independent transgenic lines for 35S::CLE constructs in Arabidopsis. Increases in expression were confirmed in 5 lines per construct by RT-PCR. RT-PCR analysis was carried out using the gene-specific primers listed in the table. RNA was isolated using Trizol reagent (Invitrogen). cDNA synthesis, following DNase treatment, was performed using Superscript III reverse transcriptase (Invitrogen). Expression levels of CLE41 in wild type were compared to that of 35S::CLE41 by qRT-PCR. All samples were measured in technical triplicates on biological triplicates. The qRT-PCR reaction was performed using SYBR Green JumpStart Taq ReadyMix (Sigma) using an ABI Prism 7000 machine (Applied Biosystems). PCR conditions were as follows: 50° C. for 2 min, 95° C. for 10 min, and 40 cycles of 95° C. for 15 s and 60° C. for 60 s. A melting curve was produced at the end of every experiment to ensure that only single products were formed. Gene expression was determined using a version of the comparative threshold cycle (Ct) method. The average amplification efficiency of each target was determined using LinReg (M. Hardstedt et al., Xenotransplantation 12, 293 (2005)).
[0254] Arabidopsis lines which carried 35S::CLE41 35S::PXY and 35S::CLE42 35S::PXY were generated by crossing and identified in the F2 population. IRX3::CLE41 35S::PXY and SUC2::CLE41 35S::PXY lines were generated by directly transforming plants carrying the 35S::PXY construct with pIRX3::CLE41 or pSUC2::CLE41. SUC2::CLE41 and 35S::CLE41 cell counts were carried out on 10 independent T2's (2 bundles/plant) and 6 independent T1's (3 bundles/plant) respectively. 5 week plants were used. Nicotiana lines carrying 35S::CLE41 35S::PXY were also generated by crossing.
TABLE-US-00001 TABLE 1 Oligonucleotides used in Invention. Oligo Name Sequence (5'-3') Used for CLE41F CACCATGGCAACATCAAATGAC 35S::CLE41 construct CLE41R AAACCAGATGTGCCAACTCA 35S::CLE41 construct and genotyping CLE42F CACCATGAGATCTCCTCACATC 35S::CLE42 construct CLE42R TGAATCAAACAAGCAACATAACAA 35S::CLE42 construct and genotyping PXY_ORF_f CACCTTAAATCCACCATTGTCA 35S::PXY construct PXY_ORF_r CCAAGATAATGGACGCCAAC 35S::PXY construct SUC2promFtopo caccaacacatgttgccgagtca SUC2::CLE41 overlap PCR entry clone SUC2pro/CLE41(1) GTCATTTGATGTTGCCAT SUC2::CLE41 overlap PCR entry clone gaaatttctttgagagggtttttg SUC2pro/CLE41(2) caaaaaccctctcaaagaaatttc SUC2::CLE41 overlap PCR entry clone ATGGCAACATCAAATGAC CLE41_RTF CCATGACTCGTCATCAGTCC RT-PCR CLE41_RTR TTTGGACCACTAGGAACCTCA RT-PCR CLE42_RTF TCCAAACCCATCAAAGAACC RT-PCR CLE42_RTR ATTGGCACCGATCATCTTTC RT-PCR PXY1_RTF AACCTAGCAATATCCTCCTCGAC RT-PCR PXY1_RTR GGTTCCACCGATCTTTTTCC RT-PCR ACTS-1 ATGAAGATTAAGGTCGTGGCA RT-PCR control ACTS-2 CCGAGTTTGAAGAGGCTAC RT-PCR control qCLE41f TCAAGAGGGTTCTCCTCGAA qRT-PCR qCLE41r TGTGCTAGCCTTTGGACGTA qRT-PCR 18s rRNA F CATCAGCTCGCGTTGACTAC qRT-PCR control 18s rRNA R GATCCTTCCGCAGGTTCAC qRT-PCR control 35S promoter F CGCACAATCCCACTATCCTT Genotyping pxy-3-r TTACCGTTTGATCCAAGCTTG Genotyping
Histology
[0255] Analysis of tobacco, poplar and Arabidopsis vasculature was carried out using thin transverse sections cut from JB4 resin embedded material as described previously (Pinon et al. 2008). Tissue was fixed in 3% glutaraldehyde or FAA, dehydrated through an ethanol series to 100% ethanol and embedded in JB4 resin (Agar Scientific). Embedded tissue was sectioned at 3 μm and subsequently stained with 0.02% Toluidine Blue. For hand cut sections, tissue was stained with either aqueous 0.02% Toluidine Blue or 0.05M Anniline blue in 100 mM Phosphate buffer, pH7.2.
[0256] Stems were analyzed at 8 weeks for Arabidopsis, 50 days for Nicotiana and four weeks after transfer to rooting medium for poplar.
[0257] Comparison of Cell Numbers in 35S::CLE41/42 Lines in Arabidopsis.
[0258] At the base of 6 week old inflorescence stems, lines over-expressing either CLE41 or CLE42 had, on average, more undifferentiated cells in vascular tissue (105.7 and 89.1, respectively) than those of wild type (58.6). When assaying cell numbers in vascular bundles from multiple insertion lines, both 35S::CLE41 and 35S::CLE42 plants had more vascular cells, although only in the case of 35S::CLE41 plants was this result statistically significant. There was no difference in the number of differentiated vascular cells in either 35S::CLE41 or 35S::CLE42 plants compared to the wild type (Table 1). We analysed progeny from two of the stronger transformed lines which were also used in subsequent genetic analysis. Stems from these lines had significantly more cells per vascular bundle (318.7 and 373.7 for 35S::CLE41 and 35S::CLE42, respectively) than wild type (273.7) clearly demonstrating that these genes are capable of increasing procambial cell divisions. In the case of 35S::CLE42 lines there was also a statistically significant increase in the number of differentiated cells.
[0259] In order to determine whether these extra procambial cells would remain undifferentiated or would differentiate into xylem and phloem, we looked at the base of plant stems at senescence. In all genotypes the vast majority of vascular cells in the stem were fully differentiated (FIG. 5), including areas in 35S::CLE41/42 where large numbers of undifferentiated cells were present at earlier stages of development. Therefore, early on 35S::CLE41/42 plants have more undifferentiated cells but these ultimately become differentiated in inflorescence stems.
TABLE-US-00002 Mean vascular cell number from 19 independent transgenic lines. Col (n = 10) 35S::CLE41 (n = 9) 35S::CLE42 (n = 10) Total Cells 311.6 ± 15.6 373.2.sup.φ ± 24.3 341.8 ± 19.1 Undifferentiated Cells 58.6 ± 4.4 105.7* ± 9.8 89.0* ± 7.2 Differentiated Cells (Xylem and 253 ± 12.6 267.5 ± 15.2 252.8 ± 14.4 Phloem) *Significantly different from Col P < 0.001. .sup.φSignificantly different from Col p < 0.05. ± Standard error.
[0260] Over-Expression of CLE41 and CLE42 in Conjunction with PXY Further Enhances Effects on Secondary Growth
[0261] We addressed the consequences of expressing PXY and CLE41 by using a 35S::PXY construct in a 35S::CLE41/42 background. The stems of 35S::CLE41/42 35S::PXY plants were characterised by dramatic increases in cell number in both the vascular bundle and in the interfascicular region such that a continuous ring of additional tissue within the stem. New cells were generated between the xylem and phloem in vascular bundles and also outside the interfascicular cells making the phenotype characteristic of dramatically increased secondary growth (FIG. 8). These results provide strong genetic evidence that CLE41/42 and PXY are sufficient for induction of vascular cell division within the procambium and elsewhere.
[0262] Interestingly, the majority of increased cell divisions occurring when both CLE41/42 and PXY are over-expressed were relatively ordered, although aberrant cells divisions are still present. We made lines harbouring both IRX3::CLE41 and 35S::PXY constructs. We found that vascular organisation was disrupted in 35S::PXY IRX3::CLE41 plants (FIG. 25?), but increased secondary growth was also observed. 35S::PXY SUC2::CLE41 plants also demonstrated enhanced secondary growth (FIG. 22), but in contrast to 35S::PXY IRX3::CLE41, vascular tissue was highly ordered.
[0263] An additional phenotype was observed in the leaves of 35S::CLE41/42 35S::PXY plants. In Col, 35S::CLE41/42 (FIG. 9) and 35S::PXY, leaves have a single midvein, however, in a minority of 35S::CLE42 35S::PXY plants the leaves appeared to exhibit increased vascular development. This additional vascular tissue develops together with the associated lamina suggesting development of ectopic vascular tissue.
[0264] Identification of CLE and PXY Homologues
[0265] Identification of Populus Trichocarpa CLE family was carried out by subjecting CLE41/42 to a WU-BLAST search against green plant GB genomic (DNA) datatsets using TBLASTN: AA query to NTdb parameters on the TAIR website (www.arabidopsis.orq). All Popolus trichocarpa hits (genomic region) with probability value (P) less than 1 were selected. These hits were subsequently on the Populus gene map (http://www.ncbi.nlm.nih.gov/projects/mapview/map search.cgi?taxid=3694). ±1 kb from the WU-BLAST hit region was then analyzed with the NCBI ORF finder (http://www.ncbi.nlm.nih.gov/projects/gorf/) and all coding regions containing similar 12 AA sequences to the output CLE sequence were examined. All putative proteins were aligned using the ClustalW algorithm using default settings.
[0266] The PXY homolog in Oryza Sativa was identified by locating PXY (At5g61480) in the homology tree from Shiu et al.(S.-H. Shiu et al., Plant Cell 16, 1220 (May 1, 2004, 2004)). The putative homolog was Osi056321.1 (Oryza Sativa Indica). This sequence was then subjected to a BLASTP protein search against O. sativa (japonica cultivar-group) Non-RefSeq protein. The top hit was EAZ41508.1: hypothetical protein OsJ--024991 and was confirmed as being the PXY orthologue by performing a BLASTP OsJ--024991 against Non-RefSeq protein database, Arabidopsis Thaliana, NCBI.
TABLE-US-00003 TABLE 2 ID CLE Representative Synonyms Organism (representative seq) Protein/Gene Annotation 41 AT_GEN_At3g24770.1 CLE41 Arabidopsis AT_GEN_At3g24770.1 At3g24770.1 thaliana 68416.m03109 CLE41, putative CLAVATA/ESR- Related 41 (CLE41) ATEST_TC255991_+1 TC255991 AT- TA13011 3702 Putative TA_TA13011_3702_+1 CLE41 protein related cluster 42 ATEST_NP1098871_+1 CLE42 Arabidopsis ATEST_NP1098871_+1 NP1098871 putative thaliana CLAVATA3/ESR-related 42 precursor [Arabidopsis thaliana] 44 AT_GEN_At4g13195.1 CLE44 Arabidopsis AT_GEN_At4g13195.1 At4g13195.1 thaliana 68417.m02052 expressed protein ATEST_TC275167_+3 TC275167 GB|AAO11557.1 |27363276| BT002641 At4g13194/At4g13194 {Arabidopsis thaliana;}, complete 51 GMEST_TC171126_+3 Glycine max GMEST_TC171126_+3 TC171126 similar to PIR|S61040|S61040 probable membrane protein YDL172c - yeast (Saccharomyces cerevisiae), partial (11%) GM- TA36215 3847 TA_TA36215_3847_-3 53 GMEST_TC162846_+3 Glycine max GMEST_TC162846_+3 TC162846 homologue to GP|21618281|gb| AAM67331.1 unknown {Arabidopsis thaliana}, partial (27%) GMEST_TC162847_+1 TC162847 homologue to GP|21618281|gb| AAM67331.1 unknown {Arabidopsis thaliana}, partial (19%) GM-TA_BQ627547_-1 BQ627547 Hypothetical protein CBG22664 related cluster GM- TA14900 3847 TA_TA14900_3847_+1 GM- TA8421 3847 SPBC215.13 TA_TA8421_3847_+3 protein related cluster 60 GMEST_BE658554_-3 Glycine max GMEST_BE658554_-3 BE658554 homologue to PIR|H72173|H7217 D5L protein - variola minor virus (strain Garcia- 1966), partial (23%) GM-TA_BE658554_-3 BE658554 61 GMEST_BM085374_+2 Glycine max GMEST_BM085374_+2 BM085374 homologue to GP|16945432|emb related to GLUCAN 1 3-BETA- GLUCOSIDASE PRECURSOR protein {Neurospora {Neurospora crassa}, partial (1%) GM- TA39380 3847 TA_TA39380_3847_+1 62 GMEST_TC171467_+2 Glycine max GMEST_TC171467_+2 TC171467 GM- TA31733 3847 TA_TA31733_3847_+2 63 GMEST_BU763224_+1 Glycine max GMEST_BU763224_+1 BU763224 similar to GP|21592472|gb|CLE gene family putative {Arabidopsis thaliana}, partial (15%) GM-TA_BU763224_+1 BU763224 64 MT_GEN_IMGA|AC137080_19.1 Medicago MT_GEN_IMGA|AC137080_19.1 IMGA|AC137080_19.1 truncatula AC137080.13 104569- 102347 E EGN_Mt041209 20041210 hypothetical protein MT_GEN_IMGA|AC147499_5.1 IMGA|AC147499_5.1 AC147499.5 26650-24428 E EGN_Mt041209 20041210 hypothetical protein MTEST_BI311733_+1 BI311733 MT-TA_BI311733_+1 BI311733 90 OS_GEN_Os02g56490.1 Oryza sativa OS_GEN_Os02g56490.1 Os02g56490.1|11972.m333 18|protein expressed protein OSEST_TC278386_+3 TC278386 Oryza sativa (japonica cultivar- group) cDNA clone: J033127D10, full insert sequence OS- TA21276 4530 TA_TA21276_4530_+2 Hypothetical protein OJ1520 C09.33 related cluster 116 PT_GEN_63277 Populus PT_GEN_63277 jgi|Poptr1|63277|fgenes trichocarpa h1_pg.C_LG_I000629 119 PT_GEN_569594 Populus PT_GEN_569594 jgi|Poptr1|569594|eugene3. trichocarpa 00120247 148 ZMEST_DR801316_+1 Zea mays ZMEST_DR801316_+1 DR801316 ZM-TA_DR801316_-1 DR801316 Hypothetical protein P0617C02.125 related cluster 149 ZMEST_BM350390_-2 Zea mays ZMEST_BM350390_-2 BM350390 similar to UP|Q4NZF2 9DELT (Q4NZF2) PE-PGRS family protein, partial (4%) ZM-TA_BM350390_+2 BM350390 PE-PGRS family protein related cluster 166 STEST_TC114822_+3 Solanum STEST_TC114822_+3 TC114822 TIGR tuberosum Ath1|At4g13195.1 68417.m02052 expressed protein, partial (12%) ST- TA8910 4113 TA_TA8910_4113_+3 Hypothetical protein MTH423 related cluster 167 STEST_BF187584_+1 Solanum STEST_BF187584_+1 BF187584 tuberosum ST- TA17128 4113 Cluster TA_TA17128_4113_+2 related to UPI0000517AA3 168 BN- Brassica BN-TA_CX187708_+3 CX187708 F20P5.29 TA_CX187708_+3 napus protein related cluster 169 PV- Phaseolus PV-TA_CV532906_+1 CV532906 Hypothetical TA_CV532906_+1 vulagaris protein related cluster 172 STEST_TC129811_+3 Solanum STEST_TC129811_+3 TC129811 similar to tuberosum TIGR Ath1|At4g13195.1 68417.m02052 expressed protein, partial (12%) ST- TA11604 4113 TA_TA11604_4113_+3 Hypothetical protein related cluster 173 STEST_CV500295_+3 Solanum STEST_CV500295_+3 CV500295 tuberosum ST- TA19709 4113 TA_TA19709_4113_+3 Hypothetical protein related cluster
Nicotiana Over Expressing CLE41/42 and PXY
[0267] In order to observe the phenotypic differences between the transgenic plants and wild type in Nicotiana, 35S::PXY, 35S::CLE41, 35S::CLE42 and wild type plants were grown in individual pots and places in the growth cabinet under same conditions. The height (from soil surface to the plant top in cM), hypocotyl width and stem width (diameter in mm) were measured when plants were 50 days old. The results of mean, standard error (SE), standard deviation (STD), minimum (Min) and maximum (Max) value were summarised in tables 3, 4, and 5. Single ANOVA between transgenic lines and wild type have been analysed and the P-value were given in the tables as well.
[0268] In table 3, the results show that the height of transgenic lines are highly significant difference between wild type (P <0.001****), the means of 35S::CLE41 and 35S::CLE42 are similar. 35S::CLE41 and 35S::CLE42 are also 20 cM and 12 cM shorter than wild type and 35S::PXY respectively. The results are consistent with the phenotypes being induced by over expression of CLE41 and CLE42.
TABLE-US-00004 TABLE 3 Height of Nicotiana at 50 days (cM) Name of plant N Mean ±SE STD Min Max P-value 35S::PXY 10 26.70 1.10 3.49 18 30.5 <0.001**** 35S::CLE41 10 14.40 1.37 4.34 5.5 20 <0.001**** 35S::CLE42 8 15.98 3.32 9.38 6 29 <0.001**** Wild type 10 35.35 1.20 3.79 27 39
[0269] Table 4, shows that Hypocotyl width is not significantly different between 35S::PXY and wild type, however, there is a highly significant difference between 35S::CLE41 or 35S::CLE42 and Wild type (P<0.0001***). The means of 35S::CLE41 and 35S::CLE42 are about 2.3 mm thicker than wild type. The maximum hypocotyls width is 10.29 mm in 35S::CLE41 compared to 7.1 mm in wild type, there is a 3.19 mm difference, demonstrating that overexpression of CLE41 and CLE42 increases hypocotyl width.
TABLE-US-00005 TABLE 4 Hypocotyl width at 50 days (mm) (Nicotiana) Name of plant N Mean ±SE STD Min Max P-value 35S::PXY 10 6.48 0.20 0.63 5.2 7.41 >0.05 35S::CLE41 10 8.71 0.30 0.96 6.87 10.29 <0.0001*** 35S::CLE42 8 8.93 0.35 0.99 7.1 10.06 <0.0001*** Wild type 10 6.34 0.14 0.45 5.3 7.1
[0270] In table 5, the results show that there is no significant difference between 35S::PXY and wild type stem width, however, there is highly significant difference between 35S::CLE41 or 35S::CLE42 and Wild type (P<0.0001****). The maximum stem width is 8.62 mm in 35S::CLE41 compared with 5.92 mm in wild type, there is 2.7 mm different. The results shown that 35S::PXY did not affect the stem width, while the overexpression of CLE41 and CLE42 made the stem thicker than wild type.
TABLE-US-00006 TABLE 5 Stem width at 50 days (mm) (Nicotiana) Name of plant N Mean ±SE STD Min Max P-Value 35S::PXY 10 5.23 0.22 0.68 4.3 6.67 >0.05 35S::CLE41 10 7.06 0.33 1.03 5.24 8.62 <0.001**** 35S::CLE42 8 6.89 0.40 1.13 4.96 8.06 <0.001**** Wild type 10 5.49 0.11 0.36 4.8 5.92
[0271] In summary, over expression of PXY results in a significant change to the plant height compared to the wild type. However, over expression of CLE41 and CLE42 significantly alter the plants phenotype in terms of height, hypocotyl width and stem width.
2. Nicotiana Images
[0272] In order to observe the phenotypic changes between the transgenic lines and wild type, the hypocotyl sections of transgenic lines 35S::PXY, 35S::CLE41, 35S::CLE42, 35S::CLE41 35S::PXY and wild type were cut when the plants were 50 days old. The images of whole plants and hypocotyl cross sections in FIGS. 19 and 20 illustrate the phenotypic differences between the lines. FIG. 19, shows that the over expression CLE41 results in a dwarf phenotype as documented in the table 4. The photographs demonstrate that this defect is much less when both CLE41 and PXY are over expressed.
[0273] In FIG. 20, cross sections of hypocotyls of plants demonstrates that plants over expressing PXY and CLE41 have thicker hypocotyls.
3. Histological Analysis of Nicotiana Hypocotyl Sections.
[0274] FIG. 21, wild type (A), 35S::CLE41 (B), 35S::CLE42 (C), 35S::PXY (D), 35S::CLE41 35S::PXY(E) Organisation is lost in plants over expressing CLE41/CLE42. Organisation is restored in plants over expressing both CLE41 and PXY.
[0275] A further experiment illustrates differences between single over expression of PXY, CLE41 and both PXY and CLE41. 10 plants of genotypes 35S::PXY, 35S::CLE41, 35S::CLE41 35S::PXY and wild type were planted in an individual pots and grown in a growth cabinet in identical conditions. The height (from soil surface to the plant top in cm), hypocotyl width and stem width (diameter in mm) were measured when plants were 42 days old. The results of mean, standard error (SE), standard deviation (STD), minimum (Min) and maximum (Max) value are presented in tables 6, 7, and 8. Single ANOVA between transgenic lines and wild type have been analysed and the P-value were given in the tables.
TABLE-US-00007 TABLE 6 The height of 35S::CLE41 plants are significantly smaller than wild type (P < 0.001****), however, there is no difference between 35S::CLE41 35S::PXY and wild type in height. Table 6 Height at 42 days (cM) Name N Mean SE STD Min Max P-value 35S::CLE41 10 9.6 0.42 1.34 7 11.5 P < 0.0001**** 35S::PXY 10 23.8 1.34 4.26 16 31.5 P < 0.01** 35S::CLE41 10 16.95 1.45 4.58 12 23 P > 0.05 35S::PXY Wild type 10 19.12 0.62 1.98 16.5 22
TABLE-US-00008 TABLE 7 Hypocotyl width was significantly larger in 35S::CLE41 and 35S::CLE41 35S::PXY compared to Wild type (P < 0.0001***). The mean of 35S::CLE41 is about 2.4 mm thicker than wild type. The mean of 35S::CLE41 35S::PXY is 2.7 mm thicker than wild type. Table 7 Nicotiana Hypocotyls at 42 days (mm) Name N Mean SE STD Min Max P-value 35S::CLE41 10 9.81 0.35 1.11 8.27 11.58 P < 0.0001**** 35S::PXY 10 8.31 0.39 1.25 6.47 9.86 P > 0.05 35S::CLE41 10 10.22 0.39 1.24 8.19 11.58 P < 0.0001**** 35S::PXY Wild type 10 7.45 0.27 0.87 5.99 8.87
TABLE-US-00009 TABLE 8 There is highly significant difference between 35S::CLE41 and 35S::CLE41 35S::PXY compared to wild type (P < 0.0001****). There is also a significant difference between 35S:PXY and Wild type. The maximum stem width is 9.43 mm in 35S::CLE41 compare 6.74 mm in wild type, a difference of 2.7 mm. The results demonstrate that overexpression of both CLE41 and PXY increase stem thickness compared to wild type. Table 8 Stem width at 42 days (mm) Name N Mean SE STD Min Max P-value 35S::CLE41 10 8.49 0.22 0.70 7.33 9.43 P < 0.0001**** 35S::PXY 10 6.81 0.37 1.19 4.58 8.27 P < 0.05* 35S::CLE41 10 8.18 0.36 1.14 6.86 9.69 P < 0.0001**** 35S::PXY Wild type 10 5.88 0.16 0.53 5.14 6.74
Poplar Harbouring 35S::CLE41 or SUC2::CLE41 Constructs Generate More Vascular Tissue than Wild Type.
[0276] Poplar transformed with 35S::CLE41 or SUC2::CLE41 were in JB4 sections were found to have more xylem tissue (see brackets in FIG. 25) demonstrating increases in vascular tissue.
Sequence CWU
1
631131PRTPopulus tremula x Populus tremuloides 1Met Asp Ile Glu Pro Leu
Trp Ala Leu Gly Gly Trp Phe Leu Phe Ser1 5
10 15Ile Thr Cys Met Ala Thr Pro Lys Ser Gln Ser Thr
Ile Ser Glu Thr 20 25 30Phe
Lys Arg Ser His His Phe Phe Leu Phe Leu Ala Leu Leu Phe Val 35
40 45Phe Ile Leu Leu Thr Ser Pro Ser Lys
Pro Ile Asn Pro Thr Asn Thr 50 55
60Val Ala Ser Ile Ser Ile Lys Arg Leu Leu Leu Glu Ser Ser Glu Pro65
70 75 80Ala Ser Thr Thr Met
Asn Leu His Pro Lys His Thr Gln Gly Thr Arg 85
90 95Thr Ser Ser Ser Ser Ser Ser Pro Pro Ser Ser
Lys Ser Thr Arg Lys 100 105
110Lys Phe Gly Ala Gln Ala His Glu Val Pro Ser Gly Pro Asn Pro Ile
115 120 125Ser Asn Arg
1302113PRTPopulus tremula x Populus tremuloides 2Met Ala Thr Pro Lys Thr
Gln Ser Thr Thr Ile Ser Asp His Gln Thr1 5
10 15Cys Thr Lys Ala His His Phe Leu Ser Leu Leu Ala
Leu Leu Phe Ile 20 25 30Phe
Ile Leu Leu Thr Thr Ser Thr Lys Pro Ile Asn Pro Thr Asn Met 35
40 45Ala Ala Ser Ile Ser Ile Lys Arg Leu
Leu Leu Glu Ser Ser Glu Pro 50 55
60Ala Ser Thr Thr Met Asn Leu His Pro Lys Gln Thr Gln Asp Ala Arg65
70 75 80Thr Ser Ser Ser Ser
Thr Ser Ser Ser Lys Ser Thr Arg Thr Lys Phe 85
90 95Gly Ala Ala Ala His Glu Val Pro Ser Gly Pro
Asn Pro Ile Ser Asn 100 105
110Arg399PRTArabidopsis thaliana 3Met Ala Thr Ser Asn Asp Gln Thr Asn Thr
Lys Ser Ser His Ser Arg1 5 10
15Thr Leu Leu Leu Leu Phe Ile Phe Leu Ser Leu Leu Leu Phe Ser Ser
20 25 30Leu Thr Ile Pro Met Thr
Arg His Gln Ser Thr Ser Met Val Ala Pro 35 40
45Phe Lys Arg Val Leu Leu Glu Ser Ser Val Pro Ala Ser Ser
Thr Met 50 55 60Asp Leu Arg Pro Lys
Ala Ser Thr Arg Arg Ser Arg Thr Ser Arg Arg65 70
75 80Arg Glu Phe Gly Asn Asp Ala His Glu Val
Pro Ser Gly Pro Asn Pro 85 90
95Ile Ser Asn4110PRTArabidopsis thaliana 4Met Ile Thr Ile Asp Gln
Thr Ser Ile Lys Ser Leu His Phe His Gln1 5
10 15Val Ile Arg Leu Ile Ile Thr Ile Ile Phe Leu Ala
Phe Leu Phe Leu 20 25 30Ile
Gly Pro Thr Ser Ser Met Asn His Leu His Glu Ser Ser Ser Lys 35
40 45Asn Thr Met Ala Pro Ser Lys Arg Phe
Leu Leu Gln Pro Ser Thr Pro 50 55
60Ser Ser Ser Thr Met Lys Met Arg Pro Thr Ala His Pro Arg Arg Ser65
70 75 80Gly Thr Ser Ser Ser
Ser Ala Arg Lys Arg Arg Arg Glu Phe Arg Ala 85
90 95Glu Ala His Glu Val Pro Ser Gly Pro Asn Pro
Ile Ser Asn 100 105
1105115PRTPopulus tremula x Populus tremuloides 5Met Ala Ser Asp Val Gly
Ser Pro Tyr Pro Thr Ser Leu Thr Ile Leu1 5
10 15Phe Phe Leu Leu Ile Met Ser His Thr Thr Met Ala
Thr Lys Glu His 20 25 30Arg
Phe Leu Leu Gly Thr Ser Arg Asp Gly Glu Ile Lys Lys Asn Asp 35
40 45Met Glu Tyr Phe Ala Asn Arg Arg His
Asp Met Gly Asn Ala Lys Thr 50 55
60Val Ser Lys Ala Asn Ile Ile His Ile Pro Pro Pro Ser Ser Arg Arg65
70 75 80Arg Gly Arg Phe Arg
Ala His Arg Ser Pro Leu Pro Trp Gln Glu Gly 85
90 95Val Glu Asn Asp Ser Ala His Glu Val Pro Ser
Gly Pro Asn Pro Ile 100 105
110Ser Asn Arg 1156115PRTPopulus tremula x Populus tremuloides
6Met Ala Ser Asp Val Gly Ser Pro Asn Leu Thr Ser Leu Thr Ile Leu1
5 10 15Phe Phe Leu Leu Ile Met
Phe His Thr Thr Met Ala Asn Lys Asp His 20 25
30Arg Phe Leu Leu Ser Thr Thr Arg Asp Gly Gly Tyr Phe
Lys Lys Ser 35 40 45Leu Met Glu
Phe Ser Thr Thr Arg Pro Asp Met Gly Asn Ala Lys Thr 50
55 60Val Ser Lys Ala Asn Val Ile His Ile Pro Pro Gln
Ser Ser Arg Arg65 70 75
80Arg Gly Arg Phe Arg Ala His Arg Ser Pro Leu Pro Trp Gln Glu Gly
85 90 95Ile Phe Ser Ala Ser Ala
His Glu Val Pro Ser Gly Pro Asn Pro Ile 100
105 110Ser Asn Arg 115745PRTPopulus tremula x
Populus tremuloides 7Met Gln Met Ile Asp Ala Phe Thr Leu Leu Val Leu Ser
Phe Met Leu1 5 10 15Arg
His Lys Gln Val Ala Glu Lys Arg Ile His Lys Ser Pro Ser Gly 20
25 30Pro Asn Pro Val Gly Asn His Asn
Pro Pro Ser Lys Gln 35 40
45876PRTArabidopsis thaliana 8Met Arg Arg His Asp Ile Ile Ile Lys Leu Leu
Leu Leu Met Cys Leu1 5 10
15Leu Leu Ser Arg Phe Val Thr Arg Glu Cys Gln Glu Val His Phe Lys
20 25 30Ile Gly Pro Ala Lys Ile Ile
Ala Lys Pro Asn Asn Ala Arg Val Asn 35 40
45Pro Thr Trp Gly Glu Glu Lys Lys Trp His Lys His Pro Ser Gly
Pro 50 55 60Asn Pro Thr Gly Asn Arg
His Pro Pro Val Lys His65 70
759104PRTOryza sativa 9Met Ala Arg Ala Arg Trp Ile Gly Asp Gly Arg Arg
Pro Ala Ala Ala1 5 10
15Leu Pro Leu Leu Gly Leu Cys Ala Phe Leu Cys Ala Val Met Leu Val
20 25 30Val Ser Leu Ala Pro Pro Pro
Gly Glu Glu Glu Glu Glu Ala Lys Val 35 40
45Arg Ser Ser Ser Leu Pro Ala Ala Ala Thr Ser Val Pro Ala Gly
Gly 50 55 60Arg Arg Leu Leu Leu Pro
Ala Ala Arg Thr Arg Arg Phe Arg Pro Arg65 70
75 80Arg Trp Asn Ser Ala Gly Ile Asp Asp Ser Lys
His Glu Val Pro Ser 85 90
95Gly Pro Asn Pro Asp Ser Asn Arg 1001088PRTArabidopsis
thaliana 10Met Arg Ser Pro His Ile Thr Ile Ser Leu Val Phe Leu Phe Phe
Leu1 5 10 15Phe Leu Ile
Ile Gln Thr His Gln Arg Thr Ile Asp Gln Thr His Gln 20
25 30Ile Gly Ser Asn Val Gln His Val Ser Asp
Met Ala Val Thr Ser Pro 35 40
45Glu Gly Lys Arg Arg Glu Arg Phe Arg Val Arg Arg Pro Met Thr Thr 50
55 60Trp Leu Lys Gly Lys Met Ile Gly Ala
Asn Glu His Gly Val Pro Ser65 70 75
80Gly Pro Asn Pro Ile Ser Asn Arg
8511120PRTOryza sativa 11Met Asp Thr Ala Arg Pro Val His Pro Leu Arg Val
His Gly Glu Ser1 5 10
15Ile Arg Gly Leu Leu Leu Leu Leu Leu Leu Phe Val Val Gln Cys Ser
20 25 30Leu Leu Ser Cys Cys Leu Ala
His Ala Ala Ala Ala Ala Asp Ala Val 35 40
45Asp Arg Asp Asp Pro Val Val Thr Ala Thr Ala Gly Arg Gly Arg
Arg 50 55 60Phe Leu Pro Ser Pro Ala
Leu Gln Leu His Ser Val Gln Val Asn Val65 70
75 80Ala Ala His Pro Trp Ser Lys Glu Arg Arg Arg
Ser Arg Arg Arg Arg 85 90
95Arg Arg Arg Ala Ala Thr Leu Met Ala Val Ser Lys His Gln Val Pro
100 105 110Thr Gly Ala Asn Pro Asp
Ser Asn 115 12012144PRTArtificial
SequenceConsensus gene for CLE41 protein derived from Arabidopsis
thaliana. 12Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa1 5 10 15Xaa Xaa Xaa
Met Ala Thr Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Ser Xaa 20
25 30Xaa Xaa Xaa Xaa Xaa Leu Leu Xaa Leu Phe
Xaa Xaa Xaa Leu Ala Leu 35 40
45Leu Xaa Xaa Xaa Xaa Leu Leu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 50
55 60Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa65 70 75
80Xaa Xaa Ser Ser Val Xaa Xaa Ser Xaa Thr Met Xaa Leu Xaa
Pro Xaa 85 90 95Ala Ser
Xaa Arg Arg Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 100
105 110Xaa Xaa Xaa Xaa Xaa Xaa Xaa Phe Xaa
Xaa Xaa Ala His Glu Val Pro 115 120
125Ser Gly Pro Asn Pro Ile Ser Asn Arg Xaa Xaa Xaa Xaa Xaa Xaa Xaa
130 135 14013313PRTOryza sativa 13Val Lys
Glu Ala Asn Val Val Gly Met Gly Ala Thr Gly Val Val Tyr1 5
10 15Lys Ala Glu Leu Pro Arg Ala Arg
Ala Val Ile Ala Val Lys Lys Leu 20 25
30Trp Arg Pro Ala Ala Ala Ala Glu Ala Ala Ala Ala Ala Pro Glu
Leu 35 40 45Thr Ala Glu Val Leu
Lys Glu Val Gly Leu Leu Gly Arg Leu Arg His 50 55
60Arg Asn Ile Val Arg Leu Leu Gly Tyr Met His Asn Glu Ala
Asp Ala65 70 75 80Met
Met Leu Tyr Glu Phe Met Pro Asn Gly Ser Leu Trp Glu Ala Leu
85 90 95His Gly Pro Pro Glu Arg Arg
Thr Leu Val Asp Trp Val Ser Arg Tyr 100 105
110Asp Val Ala Ala Gly Val Ala Gln Gly Leu Ala Tyr Leu His
His Asp 115 120 125Cys His Pro Pro
Val Ile His Arg Asp Ile Lys Ser Asn Asn Ile Leu 130
135 140Leu Asp Ala Asn Met Glu Ala Arg Ile Ala Asp Phe
Gly Leu Ala Arg145 150 155
160Ala Leu Gly Arg Ala Gly Glu Ser Val Ser Val Val Ala Gly Ser Tyr
165 170 175Gly Tyr Ile Ala Pro
Glu Tyr Gly Tyr Thr Met Lys Val Asp Gln Lys 180
185 190Ser Asp Thr Tyr Ser Tyr Gly Val Val Leu Met Glu
Leu Ile Thr Gly 195 200 205Arg Arg
Ala Val Glu Ala Ala Phe Gly Glu Gly Gln Asp Ile Val Gly 210
215 220Trp Val Arg Asn Lys Ile Arg Ser Asn Thr Val
Glu Asp His Leu Asp225 230 235
240Gly Gln Leu Val Gly Ala Gly Cys Pro His Val Arg Glu Glu Met Leu
245 250 255Leu Val Leu Arg
Thr Ala Val Leu Cys Thr Ala Arg Leu Pro Arg Asp 260
265 270Arg Pro Ser Met Arg Asp Val Ile Thr Met Leu
Gly Glu Ala Lys Pro 275 280 285Arg
Arg Lys Ser Gly Ser Ser Thr Gly Ser Ala Ser Ala Lys Ala Pro 290
295 300Thr Pro Ala Pro Pro Ala Val Ala Ala305
31014305PRTOryza sativa 14Ile Lys Glu Ala Asn Ile Val Gly
Met Gly Gly Thr Gly Val Val Tyr1 5 10
15Arg Ala Asp Met Pro Arg His His Ala Val Val Ala Val Lys
Lys Leu 20 25 30Trp Arg Ala
Ala Gly Cys Pro Glu Glu Ala Thr Thr Val Asp Gly Arg 35
40 45Thr Asp Val Glu Ala Gly Gly Glu Phe Ala Ala
Glu Val Lys Leu Leu 50 55 60Gly Arg
Leu Arg His Arg Asn Val Val Arg Met Leu Gly Tyr Val Ser65
70 75 80Asn Asn Leu Asp Thr Met Val
Ile Tyr Glu Tyr Met Val Asn Gly Ser 85 90
95Leu Trp Asp Ala Leu His Gly Gln Arg Lys Gly Lys Met
Leu Met Asp 100 105 110Trp Val
Ser Arg Tyr Asn Val Ala Ala Gly Val Ala Ala Gly Leu Ala 115
120 125Tyr Leu His His Asp Cys Arg Pro Pro Val
Ile His Arg Asp Val Lys 130 135 140Ser
Ser Asn Val Leu Leu Asp Asp Asn Met Asp Ala Lys Ile Ala Asp145
150 155 160Phe Gly Leu Ala Arg Val
Met Ala Arg Ala His Glu Thr Val Ser Val 165
170 175Val Ala Gly Ser Tyr Gly Tyr Ile Ala Pro Glu Tyr
Gly Tyr Thr Leu 180 185 190Lys
Val Asp Gln Lys Ser Asp Ile Tyr Ser Phe Gly Val Val Leu Met 195
200 205Glu Leu Leu Thr Gly Arg Arg Pro Ile
Glu Pro Glu Tyr Gly Glu Ser 210 215
220Gln Asp Ile Val Gly Trp Ile Arg Glu Arg Leu Arg Ser Asn Thr Gly225
230 235 240Val Glu Glu Leu
Leu Asp Ala Ser Val Gly Gly Arg Val Asp His Val 245
250 255Arg Glu Glu Met Leu Leu Val Leu Arg Val
Ala Val Leu Cys Thr Ala 260 265
270Lys Ser Pro Lys Asp Arg Pro Thr Met Arg Asp Val Val Thr Met Leu
275 280 285Gly Glu Ala Lys Pro Arg Arg
Lys Ser Ser Ser Ala Thr Val Ala Ala 290 295
300Thr30515309PRTOryza sativa 15Gly Ser Asp Gly Ile Val Gly Ala Gly
Ser Ser Gly Thr Val Tyr Arg1 5 10
15Ala Lys Met Pro Asn Gly Glu Val Ile Ala Val Lys Lys Leu Trp
Gln 20 25 30Ala Pro Ala Ala
Gln Lys Glu Ala Ala Ala Pro Thr Glu Gln Asn Gln 35
40 45Lys Leu Arg Gln Asp Ser Asp Gly Gly Gly Gly Gly
Lys Arg Thr Val 50 55 60Ala Glu Val
Glu Val Leu Gly His Leu Arg His Arg Asn Ile Val Arg65 70
75 80Leu Leu Gly Trp Cys Thr Asn Gly
Glu Ser Thr Met Leu Leu Tyr Glu 85 90
95Tyr Met Pro Asn Gly Ser Leu Asp Glu Leu Leu His Gly Ala
Ala Ala 100 105 110Lys Ala Arg
Pro Gly Trp Asp Ala Arg Tyr Lys Ile Ala Val Gly Val 115
120 125Ala Gln Gly Val Ser Tyr Leu His His Asp Cys
Leu Pro Ala Ile Ala 130 135 140His Arg
Asp Ile Lys Pro Ser Asn Ile Leu Leu Asp Asp Asp Met Glu145
150 155 160Ala Arg Val Ala Asp Phe Gly
Val Ala Lys Ala Leu Gln Ser Ala Ala 165
170 175Pro Met Ser Val Val Ala Gly Ser Cys Gly Tyr Ile
Ala Pro Glu Tyr 180 185 190Thr
Tyr Thr Leu Lys Val Asn Glu Lys Ser Asp Val Tyr Ser Pro Gly 195
200 205Val Val Leu Leu Glu Ile Leu Thr Gly
Arg Arg Ser Val Glu Ala Glu 210 215
220Tyr Gly Glu Gly Asn Asn Ile Val Asp Trp Val Arg Arg Lys Val Ala225
230 235 240Gly Gly Gly Val
Gly Asp Val Ile Asp Ala Ala Ala Trp Ala Asp Asn 245
250 255Asp Val Gly Gly Thr Arg Asp Glu Met Ala
Leu Ala Leu Arg Val Ala 260 265
270Leu Leu Cys Thr Ser Arg Cys Pro Gln Glu Arg Pro Ser Met Arg Glu
275 280 285Val Leu Ser Met Leu Gln Glu
Ala Arg Pro Lys Arg Lys Asn Ser Ala 290 295
300Lys Lys Gln Val Lys30516305PRTPopulus tremula x Populus
tremuloides 16Met Ser Asp Lys Ile Leu Gly Met Gly Ser Thr Gly Thr Val Tyr
Lys1 5 10 15Ala Glu Met
Pro Gly Gly Glu Ile Ile Ala Val Lys Lys Leu Trp Gly 20
25 30Lys His Lys Glu Asn Ile Arg Arg Arg Arg
Gly Val Leu Ala Glu Val 35 40
45Asp Val Leu Gly Asn Val Arg His Arg Asn Ile Val Arg Leu Leu Gly 50
55 60Cys Cys Ser Asn Arg Glu Cys Thr Met
Leu Leu Tyr Glu Tyr Met Pro65 70 75
80Asn Gly Asn Leu His Asp Leu Leu His Gly Lys Asn Lys Gly
Asp Asn 85 90 95Leu Val
Gly Asp Trp Leu Thr Arg Tyr Lys Ile Ala Leu Gly Val Ala 100
105 110Gln Gly Ile Cys Tyr Leu His His Asp
Cys Asp Pro Val Ile Val His 115 120
125Arg Asp Leu Lys Pro Ser Asn Ile Leu Leu Asp Gly Glu Met Glu Ala
130 135 140Arg Val Arg Asp Phe Gly Val
Ala Lys Leu Ile Gln Ser Asp Glu Ser145 150
155 160Met Ser Val Ile Ala Gly Ser Tyr Gly Tyr Ile Ala
Pro Glu Tyr Ala 165 170
175Tyr Thr Leu Gln Val Asp Glu Lys Ser Asp Ile Tyr Ser Tyr Gly Val
180 185 190Val Leu Met Glu Ile Ile
Ser Gly Lys Arg Ser Val Asp Ala Glu Phe 195 200
205Gly Asp Gly Asn Ser Ile Val Asp Trp Val Arg Ser Lys Ile
Lys Ala 210 215 220Lys Asp Gly Val Asn
Asp Ile Leu Asp Lys Asp Ala Gly Ala Ser Ile225 230
235 240Ala Ser Val Arg Glu Glu Met Met Gln Met
Leu Arg Ile Ala Leu Leu 245 250
255Cys Thr Ser Arg Asn Pro Ala Asp Arg Pro Ser Met Arg Asp Val Val
260 265 270Leu Met Leu Gln Glu
Ala Lys Pro Lys Arg Lys Leu Pro Gly Ser Ile 275
280 285Val Ser Val Gly Ser Gly Asp His Ile Val Thr Val
Asp Gly Ala Ile 290 295
300Ala30517251PRTPopulus tremula x Populus tremuloides 17Lys Thr Asp Asn
Ile Leu Gly Met Gly Ser Thr Gly Thr Val Tyr Lys1 5
10 15Ala Glu Met Pro Asn Gly Glu Ile Ile Ala
Val Lys Lys Leu Trp Gly 20 25
30Lys Asn Lys Glu Asn Gly Lys Ile Arg Arg Arg Lys Ser Gly Val Leu
35 40 45Ala Glu Val Asp Val Leu Gly Asn
Val Arg His Arg Asn Ile Val Arg 50 55
60Leu Leu Gly Cys Cys Thr Asn Arg Asp Cys Thr Met Leu Leu Tyr Glu65
70 75 80Tyr Met Pro Asn Gly
Ser Leu Asp Asp Leu Leu His Gly Gly Asp Lys 85
90 95Thr Met Thr Ala Ala Ala Glu Trp Thr Ala Leu
Tyr Gln Ile Ala Ile 100 105
110Gly Val Ala Gln Gly Ile Cys Tyr Leu His His Asp Cys Asp Pro Val
115 120 125Ile Val His Arg Asp Leu Lys
Pro Ser Asn Ile Leu Leu Asp Ala Asp 130 135
140Phe Glu Ala Arg Val Ala Asp Phe Gly Val Ala Lys Leu Ile Gln
Thr145 150 155 160Asp Glu
Ser Met Ser Val Val Ala Gly Ser Tyr Gly Tyr Ile Ala Pro
165 170 175Glu Tyr Ala Tyr Thr Leu Gln
Val Asp Lys Lys Ser Asp Ile Tyr Ser 180 185
190Tyr Gly Val Ile Leu Leu Glu Ile Ile Thr Gly Lys Arg Ser
Val Glu 195 200 205Pro Glu Phe Gly
Glu Gly Asn Ser Ile Val Asp Trp Val Arg Ser Lys 210
215 220Leu Lys Thr Lys Glu Asp Val Glu Glu Val Leu Asp
Lys Ser Met Gly225 230 235
240Arg Ser Cys Ser Leu Ile Arg Glu Glu Met Lys 245
25018287PRTPopulus tremula x Populus tremuloides 18Ile Lys Glu
Ser Asn Ile Ile Gly Met Gly Ala Ile Gly Ile Val Tyr1 5
10 15Lys Ala Glu Val Met Arg Arg Pro Leu
Leu Thr Val Ala Val Lys Lys 20 25
30Leu Trp Arg Ser Pro Ser Pro Gln Asn Asp Ile Glu Asp His His Gln
35 40 45Glu Glu Asp Glu Glu Asp Asp
Ile Leu Arg Glu Val Asn Leu Leu Gly 50 55
60Gly Leu Arg His Arg Asn Ile Val Lys Ile Leu Gly Tyr Val His Asn65
70 75 80Glu Arg Glu Val
Met Met Val Tyr Glu Tyr Met Pro Asn Gly Asn Leu 85
90 95Gly Thr Ala Leu His Ser Lys Asp Glu Lys
Phe Leu Leu Arg Asp Trp 100 105
110Leu Ser Arg Tyr Asn Val Ala Val Gly Val Val Gln Gly Leu Asn Tyr
115 120 125Leu His Asn Asp Cys Tyr Pro
Pro Ile Ile His Arg Asp Ile Lys Ser 130 135
140Asn Asn Ile Leu Leu Asp Ser Asn Leu Glu Ala Arg Ile Ala Asp
Phe145 150 155 160Gly Leu
Ala Lys Met Met Leu His Lys Asn Glu Thr Val Ser Met Val
165 170 175Ala Gly Ser Tyr Gly Tyr Ile
Ala Pro Glu Tyr Gly Tyr Thr Leu Lys 180 185
190Ile Asp Glu Lys Ser Asp Ile Tyr Ser Leu Gly Val Val Leu
Leu Glu 195 200 205Leu Val Thr Gly
Lys Met Pro Ile Asp Pro Ser Phe Glu Asp Ser Ile 210
215 220Asp Val Val Glu Trp Ile Arg Arg Lys Val Lys Lys
Asn Glu Ser Leu225 230 235
240Glu Glu Val Ile Asp Ala Ser Ile Ala Gly Asp Cys Lys His Val Ile
245 250 255Glu Glu Met Leu Leu
Ala Leu Arg Ile Ala Leu Leu Cys Thr Ala Lys 260
265 270Leu Pro Lys Asp Arg Pro Ser Ile Arg Asp Val Ile
Thr Met Leu 275 280
28519276PRTPopulus tremula x Populus tremuloides 19Lys Glu Ser Asn Met
Ile Gly Met Gly Ala Thr Gly Ile Val Tyr Lys1 5
10 15Ala Glu Met Ser Arg Ser Ser Thr Val Leu Ala
Val Lys Lys Leu Trp 20 25
30Arg Ser Ala Ala Asp Ile Glu Asp Gly Thr Thr Gly Asp Phe Val Gly
35 40 45Glu Val Asn Leu Leu Gly Lys Leu
Arg His Arg Asn Ile Val Arg Leu 50 55
60Leu Gly Phe Leu Tyr Asn Asp Lys Asn Met Met Ile Val Tyr Glu Phe65
70 75 80Met Leu Asn Gly Asn
Leu Gly Asp Ala Ile His Gly Lys Asn Ala Ala 85
90 95Gly Arg Leu Leu Val Asp Trp Val Ser Arg Tyr
Asn Ile Ala Leu Gly 100 105
110Val Ala His Gly Leu Tyr Leu His His Asp Cys His Pro Pro Val Ile
115 120 125His Arg Asp Ile Lys Ser Asn
Asn Ile Leu Leu Asp Ala Asn Leu Asp 130 135
140Ala Arg Ile Ala Asp Phe Gly Leu Ala Arg Met Met Ala Arg Lys
Lys145 150 155 160Glu Thr
Val Ser Met Val Ala Gly Ser Tyr Gly Tyr Ile Ala Pro Glu
165 170 175Tyr Gly Tyr Thr Leu Lys Val
Asp Glu Lys Ile Asp Ile Tyr Ser Tyr 180 185
190Gly Val Val Leu Leu Glu Leu Leu Thr Gly Arg Arg Leu Glu
Pro Glu 195 200 205Phe Gly Glu Ser
Val Asp Ile Val Glu Trp Val Arg Arg Lys Ile Arg 210
215 220Asp Asn Ile Ser Leu Glu Glu Ala Leu Asp Pro Asn
Val Gly Asn Cys225 230 235
240Arg Tyr Val Gln Glu Glu Met Leu Leu Val Leu Gln Ile Ala Leu Leu
245 250 255Cys Thr Thr Lys Leu
Pro Lys Asp Arg Pro Ser Met Arg Asp Val Ile 260
265 270Ser Met Leu Gly 27520308PRTArtificial
SequenceConsensus artificial amino acid sequence for PXY protein
dervied from Arabidopsis thaliana. 20Xaa Lys Glu Ala Asn Ile Val Gly Met
Gly Ala Thr Gly Ile Val Tyr1 5 10
15Lys Ala Glu Met Pro Arg Xaa Xaa Xaa Xaa Val Ile Ala Val Lys
Lys 20 25 30Leu Trp Arg Ala
Xaa Ala Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 35
40 45Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 50 55 60Asp Val Leu
Ala Glu Val Xaa Leu Leu Gly Xaa Leu Arg His Arg Asn65 70
75 80Ile Val Arg Leu Leu Gly Tyr Val
Ser Asn Xaa Xaa Xaa Thr Met Leu 85 90
95Leu Tyr Glu Tyr Met Pro Asn Gly Ser Leu Xaa Asp Ala Leu
His Gly 100 105 110Xaa Xaa Xaa
Ala Xaa Lys Leu Leu Xaa Asp Trp Val Ser Arg Tyr Asn 115
120 125Ile Ala Leu Gly Val Ala Gln Gly Leu Ala Tyr
Leu His His Asp Cys 130 135 140Xaa Pro
Pro Ile Ile His Arg Asp Ile Lys Ser Ser Asn Ile Leu Leu145
150 155 160Asp Ala Asn Met Glu Ala Arg
Ile Ala Asp Phe Gly Leu Ala Lys Leu 165
170 175Met Xaa Xaa Xaa Xaa Glu Ser Val Ser Val Val Ala
Gly Ser Tyr Gly 180 185 190Tyr
Ile Ala Pro Glu Tyr Gly Tyr Thr Leu Lys Val Asp Glu Lys Ser 195
200 205Asp Ile Tyr Ser Tyr Gly Val Val Leu
Leu Glu Leu Ile Thr Gly Arg 210 215
220Arg Ser Val Glu Pro Glu Phe Gly Glu Gly Asn Asp Ile Val Asp Trp225
230 235 240Val Arg Xaa Lys
Ile Lys Xaa Xaa Asn Xaa Val Xaa Glu Xaa Leu Asp 245
250 255Xaa Ala Ala Val Ala Xaa Xaa Xaa Cys Xaa
His Val Arg Glu Glu Met 260 265
270Leu Leu Val Leu Arg Ile Ala Leu Leu Cys Thr Ala Lys Xaa Pro Lys
275 280 285Asp Arg Pro Ser Met Arg Asp
Val Ile Thr Met Leu Xaa Glu Ala Lys 290 295
300Pro Lys Arg Lys3052199PRTArabidopsis thaliana 21Met Ala Thr Ser
Asn Asp Gln Thr Asn Thr Lys Ser Ser His Ser Arg1 5
10 15Thr Leu Leu Leu Leu Phe Ile Phe Leu Ser
Leu Leu Leu Phe Ser Ser 20 25
30Leu Thr Ile Pro Met Thr Arg His Gln Ser Thr Ser Met Val Ala Pro
35 40 45Phe Lys Arg Val Leu Leu Glu Ser
Ser Val Pro Ala Ser Ser Thr Met 50 55
60Asp Leu Arg Pro Lys Ala Ser Thr Arg Arg Ser Arg Thr Ser Arg Arg65
70 75 80Arg Glu Phe Gly Asn
Asp Ala His Glu Val Pro Ser Gly Pro Asn Pro 85
90 95Ile Ser Asn22300DNAArabidopsis thaliana
22atggcaacat caaatgacca aaccaatact aaatcatcac attctcgtac tcttctcctt
60ctcttcatct tcttatccct ccttctcttc agtagcctta caatccccat gactcgtcat
120cagtccacat ctatggttgc tcccttcaag agggttctcc tcgaatcttc agttccagct
180tcatcaacaa tggatctacg tccaaaggct agcacacgac gcagccgcac ttctagaagg
240agagagtttg gaaatgatgc tcatgaggtt cctagtggtc caaaccctat ttccaactag
3002388PRTArabidopsis thaliana 23Met Arg Ser Pro His Ile Thr Ile Ser Leu
Val Phe Leu Phe Phe Leu1 5 10
15Phe Leu Ile Ile Gln Thr His Gln Arg Thr Ile Asp Gln Thr His Gln
20 25 30Ile Gly Ser Asn Val Gln
His Val Ser Asp Met Ala Val Thr Ser Pro 35 40
45Glu Gly Lys Arg Arg Glu Arg Phe Arg Val Arg Arg Pro Met
Thr Thr 50 55 60Trp Leu Lys Gly Lys
Met Ile Gly Ala Asn Glu His Gly Val Pro Ser65 70
75 80Gly Pro Asn Pro Ile Ser Asn Arg
8524267DNAArabidopsis thaliana 24atgagatctc ctcacatcac catttcactt
gttttcttgt tctttctttt tctaatcatc 60caaacccatc aaagaaccat tgatcaaact
caccagattg gctccaatgt tcaacatgtc 120agtgacatgg cggtgacttc gcctgaaggg
aaaagaagag agaggtttag agttcggcgg 180ccgatgacga catggctgaa gggaaagatg
atcggtgcca atgaacatgg agtcccaagt 240ggtccaaatc ccatctccaa taggtag
267251041PRTArabidopsis thaliana 25Met
Lys Lys Lys Asn Ile Ser Pro Ser Leu Val Leu His Pro Leu Leu1
5 10 15Leu Leu Leu Leu Pro Phe Phe
Ala Phe Asn Ser Leu Ala Leu Lys Phe 20 25
30Ser Pro Gln Leu Leu Ser Leu Leu Ser Leu Lys Thr Ser Leu
Ser Gly 35 40 45Pro Pro Ser Ala
Phe Gln Asp Trp Lys Val Pro Val Asn Gly Gln Asn 50 55
60Asp Ala Val Trp Cys Ser Trp Ser Gly Val Val Cys Asp
Asn Val Thr65 70 75
80Ala Gln Val Ile Ser Leu Asp Leu Ser His Arg Asn Leu Ser Gly Arg
85 90 95Ile Pro Ile Gln Ile Arg
Tyr Leu Ser Ser Leu Leu Tyr Leu Asn Leu 100
105 110Ser Gly Asn Ser Leu Glu Gly Ser Phe Pro Thr Ser
Ile Phe Asp Leu 115 120 125Thr Lys
Leu Thr Thr Leu Asp Ile Ser Arg Asn Ser Phe Asp Ser Ser 130
135 140Phe Pro Pro Gly Ile Ser Lys Leu Lys Phe Leu
Lys Val Phe Asn Ala145 150 155
160Phe Ser Asn Asn Phe Glu Gly Leu Leu Pro Ser Asp Val Ser Arg Leu
165 170 175Arg Phe Leu Glu
Glu Leu Asn Phe Gly Gly Ser Tyr Phe Glu Gly Glu 180
185 190Ile Pro Ala Ala Tyr Gly Gly Leu Gln Arg Leu
Lys Phe Ile His Leu 195 200 205Ala
Gly Asn Val Leu Gly Gly Lys Leu Pro Pro Arg Leu Gly Leu Leu 210
215 220Thr Glu Leu Gln His Met Glu Ile Gly Tyr
Asn His Phe Asn Gly Asn225 230 235
240Ile Pro Ser Glu Phe Ala Leu Leu Ser Asn Leu Lys Tyr Phe Asp
Val 245 250 255Ser Asn Cys
Ser Leu Ser Gly Ser Leu Pro Gln Glu Leu Gly Asn Leu 260
265 270Ser Asn Leu Glu Thr Leu Phe Leu Phe Gln
Asn Gly Phe Thr Gly Glu 275 280
285Ile Pro Glu Ser Tyr Ser Asn Leu Lys Ser Leu Lys Leu Leu Asp Phe 290
295 300Ser Ser Asn Gln Leu Ser Gly Ser
Ile Pro Ser Gly Phe Ser Thr Leu305 310
315 320Lys Asn Leu Thr Trp Leu Ser Leu Ile Ser Asn Asn
Leu Ser Gly Glu 325 330
335Val Pro Glu Gly Ile Gly Glu Leu Pro Glu Leu Thr Thr Leu Phe Leu
340 345 350Trp Asn Asn Asn Phe Thr
Gly Val Leu Pro His Lys Leu Gly Ser Asn 355 360
365Gly Lys Leu Glu Thr Met Asp Val Ser Asn Asn Ser Phe Thr
Gly Thr 370 375 380Ile Pro Ser Ser Leu
Cys His Gly Asn Lys Leu Tyr Lys Leu Ile Leu385 390
395 400Phe Ser Asn Met Phe Glu Gly Glu Leu Pro
Lys Ser Leu Thr Arg Cys 405 410
415Glu Ser Leu Trp Arg Phe Arg Ser Gln Asn Asn Arg Leu Asn Gly Thr
420 425 430Ile Pro Ile Gly Phe
Gly Ser Leu Arg Asn Leu Thr Phe Val Asp Leu 435
440 445Ser Asn Asn Arg Phe Thr Asp Gln Ile Pro Ala Asp
Phe Ala Thr Ala 450 455 460Pro Val Leu
Gln Tyr Leu Asn Leu Ser Thr Asn Phe Phe His Arg Lys465
470 475 480Leu Pro Glu Asn Ile Trp Lys
Ala Pro Asn Leu Gln Ile Phe Ser Ala 485
490 495Ser Phe Ser Asn Leu Ile Gly Glu Ile Pro Asn Tyr
Val Gly Cys Lys 500 505 510Ser
Phe Tyr Arg Ile Glu Leu Gln Gly Asn Ser Leu Asn Gly Thr Ile 515
520 525Pro Trp Asp Ile Gly His Cys Glu Lys
Leu Leu Cys Leu Asn Leu Ser 530 535
540Gln Asn His Leu Asn Gly Ile Ile Pro Trp Glu Ile Ser Thr Leu Pro545
550 555 560Ser Ile Ala Asp
Val Asp Leu Ser His Asn Leu Leu Thr Gly Thr Ile 565
570 575Pro Ser Asp Phe Gly Ser Ser Lys Thr Ile
Thr Thr Phe Asn Val Ser 580 585
590Tyr Asn Gln Leu Ile Gly Pro Ile Pro Ser Gly Ser Phe Ala His Leu
595 600 605Asn Pro Ser Phe Phe Ser Ser
Asn Glu Gly Leu Cys Gly Asp Leu Val 610 615
620Gly Lys Pro Cys Asn Ser Asp Arg Phe Asn Ala Gly Asn Ala Asp
Ile625 630 635 640Asp Gly
His His Lys Glu Glu Arg Pro Lys Lys Thr Ala Gly Ala Ile
645 650 655Val Trp Ile Leu Ala Ala Ala
Ile Gly Val Gly Phe Phe Val Leu Val 660 665
670Ala Ala Thr Arg Cys Phe Gln Lys Ser Tyr Gly Asn Arg Val
Asp Gly 675 680 685Gly Gly Arg Asn
Gly Gly Asp Ile Gly Pro Trp Lys Leu Thr Ala Phe 690
695 700Gln Arg Leu Asn Phe Thr Ala Asp Asp Val Val Glu
Cys Leu Ser Lys705 710 715
720Thr Asp Asn Ile Leu Gly Met Gly Ser Thr Gly Thr Val Tyr Lys Ala
725 730 735Glu Met Pro Asn Gly
Glu Ile Ile Ala Val Lys Lys Leu Trp Gly Lys 740
745 750Asn Lys Glu Asn Gly Lys Ile Arg Arg Arg Lys Ser
Gly Val Leu Ala 755 760 765Glu Val
Asp Val Leu Gly Asn Val Arg His Arg Asn Ile Val Arg Leu 770
775 780Leu Gly Cys Cys Thr Asn Arg Asp Cys Thr Met
Leu Leu Tyr Glu Tyr785 790 795
800Met Pro Asn Gly Ser Leu Asp Asp Leu Leu His Gly Gly Asp Lys Thr
805 810 815Met Thr Ala Ala
Ala Glu Trp Thr Ala Leu Tyr Gln Ile Ala Ile Gly 820
825 830Val Ala Gln Gly Ile Cys Tyr Leu His His Asp
Cys Asp Pro Val Ile 835 840 845Val
His Arg Asp Leu Lys Pro Ser Asn Ile Leu Leu Asp Ala Asp Phe 850
855 860Glu Ala Arg Val Ala Asp Phe Gly Val Ala
Lys Leu Ile Gln Thr Asp865 870 875
880Glu Ser Met Ser Val Val Ala Gly Ser Tyr Gly Tyr Ile Ala Pro
Glu 885 890 895Tyr Ala Tyr
Thr Leu Gln Val Asp Lys Lys Ser Asp Ile Tyr Ser Tyr 900
905 910Gly Val Ile Leu Leu Glu Ile Ile Thr Gly
Lys Arg Ser Val Glu Pro 915 920
925Glu Phe Gly Glu Gly Asn Ser Ile Val Asp Trp Val Arg Ser Lys Leu 930
935 940Lys Thr Lys Glu Asp Val Glu Glu
Val Leu Asp Lys Ser Met Gly Arg945 950
955 960Ser Cys Ser Leu Ile Arg Glu Glu Met Lys Gln Met
Leu Arg Ile Ala 965 970
975Leu Leu Cys Thr Ser Arg Ser Pro Thr Asp Arg Pro Pro Met Arg Asp
980 985 990Val Leu Leu Ile Leu Gln
Glu Ala Lys Pro Lys Arg Lys Thr Val Gly 995 1000
1005Asp Asn Val Ile Val Val Gly Asp Val Asn Asp Val
Asn Phe Glu 1010 1015 1020Asp Val Cys
Ser Val Asp Val Gly His Asp Val Lys Cys Gln Arg 1025
1030 1035Ile Gly Val 1040263126DNAArabidopsis
thaliana 26atgaaaaaga agaacatttc tccttctctt gttcttcatc cccttctcct
tcttctactt 60cctttctttg ctttcaattc cttagctctc aagttttcac ctcaactctt
gtctctcctt 120tcccttaaaa catctctctc tggccctccc tctgcctttc aagactggaa
agtccccgtt 180aacggtcaaa acgacgccgt ttggtgttct tggtccggtg tagtctgtga
taatgtaacg 240gctcaagtca tttccctcga cctctctcac cggaacctct ctggtcgtat
tcctatacag 300attcgttact tgtcgagctt actctactta aatctcagtg ggaattcttt
ggaaggttcg 360tttccaactt ctatctttga tctcaccaag ctcactaccc tcgacatcag
ccgtaactcg 420ttcgactcga gttttcctcc cggaatctcc aagcttaagt tcttaaaagt
cttcaatgcg 480ttcagcaaca acttcgaagg tctattacct agtgacgtgt ctcgtcttcg
tttcttggaa 540gagcttaact ttggtggaag ttactttgaa ggagagattc cagcagctta
cggtggttta 600cagagattga agtttattca tttagctgga aatgtcctcg gaggtaaact
acctcctaga 660ttaggactct taacagagct ccaacacatg gaaatcggtt ataatcactt
caacggaaac 720ataccttcgg agtttgcctt actctcaaat ctcaagtact ttgacgtttc
caattgcagc 780ctctctggtt ctctgcctca agaactcggg aatctctcaa acctagagac
tttatttcta 840ttccaaaacg gtttcaccgg tgaaatccca gagagttata gcaacttgaa
atccctcaag 900cttctcgatt tttcgagtaa tcagctttct ggtagtatcc catcaggctt
ctcgaccttg 960aagaacctca catggctaag cttaatcagc aataacctct caggtgaagt
acctgaagga 1020atcggtgaac tccctgagct tactacattg tttctatgga acaataactt
caccggagtt 1080ttgccacaca agcttggatc aaacggtaaa cttgagacaa tggacgtctc
taacaattca 1140ttcaccggaa caatcccttc ttctctctgc catggaaaca agctatacaa
actcatcctc 1200ttctccaaca tgtttgaagg tgagctacca aagagcttga ctcgttgcga
atctctatgg 1260cggtttcgga gtcaaaacaa tcgattaaac ggcacaattc cgatcggatt
cggctctcta 1320cgtaacctca ctttcgttga tttaagcaac aacagattca ccgatcaaat
tccggcggat 1380ttcgccaccg ctcctgttct tcagtacttg aatctctcaa ccaatttctt
ccacaggaaa 1440ctaccggaaa acatatggaa agctccgaat ctacagatct tctcagcgag
tttcagcaat 1500ttgatcggtg aaatcccaaa ttacgttgga tgcaaaagct tctacaggat
tgaactacaa 1560ggaaactcac tcaacggaac gattccatgg gacatcggac attgcgagaa
gcttctctgt 1620ttgaatctca gccaaaatca tctcaacgga atcattccat gggagatttc
aactcttccg 1680tcaatcgccg acgtagatct ttctcataat ctcttaaccg gaacaatccc
ttccgatttc 1740ggaagctcta agacgatcac aaccttcaac gtttcgtata atcagctaat
cggtccgatt 1800ccaagtggtt ctttcgctca tctgaatccg tcgttcttct cctcaaacga
aggactctgt 1860ggagatctcg tcggaaaacc ttgcaattct gataggttta acgccggaaa
tgcagatata 1920gacggtcatc ataaagagga acgacctaag aaaacagccg gagctattgt
ttggatattg 1980gcggcggcga ttggggttgg attcttcgtc cttgtagccg ccactagatg
cttccagaaa 2040agctacggaa acagagtcga cggtggtgga agaaacggcg gagatatagg
accgtggaag 2100ctaacggctt ttcagagact aaacttcacg gcggatgatg tggttgagtg
tctctcaaag 2160actgataaca tcctcggaat gggatcaaca ggaacagtgt acaaagcaga
gatgcctaat 2220ggagaaataa tcgccgtgaa aaaactttgg ggaaaaaaca aagagaacgg
caaaatccgg 2280cggcggaaga gcggcgtatt ggcggaggtt gatgttctag ggaacgtacg
tcaccggaac 2340atcgttcgtc tccttggatg ttgcacgaat cgagattgca cgatgctttt
atacgaatac 2400atgcctaatg gaagcttaga cgatcttctt cacggtgggg ataagacgat
gaccgcggcg 2460gcggaatgga cggctttgta tcagatcgcg attggagtgg ctcaagggat
ctgttatctc 2520caccatgatt gtgatccggt gattgtacac cgtgacctga aacctagcaa
tatcctcctc 2580gacgccgatt tcgaggcgcg tgtggcggac ttcggcgtcg cgaagcttat
tcaaaccgac 2640gaatccatgt ccgtcgtcgc cggttcgtac ggttacattg caccagaata
tgcttacact 2700ttacaagtgg ataaaaagag tgatatctat agctatggag tgattttatt
agagataatc 2760accggaaaaa gatcggtgga accggaattt ggagaaggta acagtatcgt
ggattgggtt 2820agatcaaagt tgaagacgaa agaagatgta gaagaagttc tagacaaaag
catgggtagg 2880tcgtgtagtc ttataagaga agagatgaaa caaatgttga gaattgcgtt
gttgtgtaca 2940agccggagtc cgacagacag accgccgatg agagatgtgt tgttgattct
tcaagaggca 3000aagccaaaga ggaagacagt aggggataat gtgatcgtcg ttggtgatgt
taatgatgtc 3060aatttcgaag atgtttgtag tgttgatgtt ggtcatgatg ttaaatgtca
aaggattggg 3120gtgtga
3126273302DNAArabidopsis thaliana 27atgaaaaaga agaacatttc
tccttctctt gttcttcatc cccttctcct tcttctactt 60cctttctttg ctttcaattc
cttagctctc aagttttcac ctcaactctt gtctctcctt 120tcccttaaaa catctctctc
tggccctccc tctgcctttc aagactggaa agtccccgtt 180aacggtcaaa acgacgccgt
ttggtgttct tggtccggtg tagtctgtga taatgtaacg 240gctcaagtca tttccctcga
cctctctcac cggaacctct ctggtcgtat tcctatacag 300attcgttact tgtcgagctt
actctactta aatctcagtg ggaattcttt ggaaggttcg 360tttccaactt ctatctttga
tctcaccaag ctcactaccc tcgacatcag ccgtaactcg 420ttcgactcga gttttcctcc
cggaatctcc aagcttaagt tcttaaaagt cttcaatgcg 480ttcagcaaca acttcgaagg
tctattacct agtgacgtgt ctcgtcttcg tttcttggaa 540gagcttaact ttggtggaag
ttactttgaa ggagagattc cagcagctta cggtggttta 600cagagattga agtttattca
tttagctgga aatgtcctcg gaggtaaact acctcctaga 660ttaggactct taacagagct
ccaacacatg gaaatcggtt ataatcactt caacggaaac 720ataccttcgg agtttgcctt
actctcaaat ctcaagtact ttgacgtttc caattgcagc 780ctctctggtt ctctgcctca
agaactcggg aatctctcaa acctagagac tttatttcta 840ttccaaaacg gtttcaccgg
tgaaatccca gagagttata gcaacttgaa atccctcaag 900cttctcgatt tttcgagtaa
tcagctttct ggtagtatcc catcaggctt ctcgaccttg 960aagaacctca catggctaag
cttaatcagc aataacctct caggtgaagt acctgaagga 1020atcggtgaac tccctgagct
tactacattg tttctatgga acaataactt caccggagtt 1080ttgccacaca agcttggatc
aaacggtaaa cttgagacaa tggacgtctc taacaattca 1140ttcaccggaa caatcccttc
ttctctctgc catggaaaca agctatacaa actcatcctc 1200ttctccaaca tgtttgaagg
tgagctacca aagagcttga ctcgttgcga atctctatgg 1260cggtttcgga gtcaaaacaa
tcgattaaac ggcacaattc cgatcggatt cggctctcta 1320cgtaacctca ctttcgttga
tttaagcaac aacagattca ccgatcaaat tccggcggat 1380ttcgccaccg ctcctgttct
tcagtacttg aatctctcaa ccaatttctt ccacaggaaa 1440ctaccggaaa acatatggaa
agctccgaat ctacagatct tctcagcgag tttcagcaat 1500ttgatcggtg aaatcccaaa
ttacgttgga tgcaaaagct tctacaggat tgaactacaa 1560ggaaactcac tcaacggaac
gattccatgg gacatcggac attgcgagaa gcttctctgt 1620ttgaatctca gccaaaatca
tctcaacgga atcattccat gggagatttc aactcttccg 1680tcaatcgccg acgtagatct
ttctcataat ctcttaaccg gaacaatccc ttccgatttc 1740ggaagctcta agacgatcac
aaccttcaac gtttcgtata atcagctaat cggtccgatt 1800ccaagtggtt ctttcgctca
tctgaatccg tcgttcttct cctcaaacga aggactctgt 1860ggagatctcg tcggaaaacc
ttgcaattct gataggttta acgccggaaa tgcagatata 1920gacggtcatc ataaagagga
acgacctaag aaaacagccg gagctattgt ttggatattg 1980gcggcggcga ttggggttgg
attcttcgtc cttgtagccg ccactagatg cttccagaaa 2040agctacggaa acagagtcga
cggtggtgga agaaacggcg gagatatagg accgtggaag 2100ctaacggctt ttcagagact
aaacttcacg gcggatgatg tggttgagtg tctctcaaag 2160actgataaca tcctcggaat
gggatcaaca ggaacagtgt acaaagcaga gatgcctaat 2220ggagaaataa tcgccgtgaa
aaaactttgg ggaaaaaaca aagagaacgg caaaatccgg 2280cggcggaaga gcggcgtatt
ggcggaggtt gatgttctag ggaacgtacg tcaccggaac 2340atcgttcgtc tccttggatg
ttgcacgaat cgagattgca cgatgctttt atacgaatac 2400atgcctaatg gaagcttaga
cgatcttctt cacggtgggg ataagacgat gaccgcggcg 2460gcggaatgga cggctttgta
tcagatcgcg attggagtgg ctcaagggat ctgttatctc 2520caccatgatt gtgatccggt
gattgtacac cgtgacctga aacctagcaa tatcctcctc 2580gacgccgatt tcgaggcgcg
tgtggcggac ttcggcgtcg cgaagcttat tcaaaccgac 2640gaatccatgt ccgtcgtcgc
cggttcgtac ggttacattg caccaggtac ccttaacttt 2700ttttgattat tctttacttt
ccccaaattt taaattttgt acttttttgt ccctttgttt 2760ttattattcg aattttgtcc
gtttgttaaa cattcttttt gttgggatga caacatctga 2820caaatatgac taaaatttta
attttgtttg ttttggttac agaatatgct tacactttac 2880aagtggataa aaagagtgat
atctatagct atggagtgat tttattagag ataatcaccg 2940gaaaaagatc ggtggaaccg
gaatttggag aaggtaacag tatcgtggat tgggttagat 3000caaagttgaa gacgaaagaa
gatgtagaag aagttctaga caaaagcatg ggtaggtcgt 3060gtagtcttat aagagaagag
atgaaacaaa tgttgagaat tgcgttgttg tgtacaagcc 3120ggagtccgac agacagaccg
ccgatgagag atgtgttgtt gattcttcaa gaggcaaagc 3180caaagaggaa gacagtaggg
gataatgtga tcgtcgttgg tgatgttaat gatgtcaatt 3240tcgaagatgt ttgtagtgtt
gatgttggtc atgatgttaa atgtcaaagg attggggtgt 3300ga
330228345DNACauliflower
mosaic virus 28tgagactttt caacaaaggg taatatccgg aaacctcctc ggattccatt
gcccagctat 60ctgtcacttt attgtgaaga tagtggaaaa ggaaggtggc tcctacaaat
gccatcattg 120cgataaagga aaggccatcg ttgaagatgc ctctgccgac agtggtccca
aagatggacc 180cccaccccac gaggagcatc gtggaaaaag aagacgttcc aaccacgtct
tcaaagcaag 240tggattgatg tgatatctcc actgacgtaa gggatgacgc acaatcccac
tatccttcgc 300aagacccttc ctctatataa ggaagttcat ttcatttgga gagga
345291122DNAArabidopsis thaliana 29aaaaataagt aaaagatctt
ttagttgttt gctttgtatg ttgcgaacag tttgattctg 60tttttctttt tccttttttt
gggtaatttt cttataactt ttttcatagt ttcgattatt 120tggataaaat tttcagattg
aggatcattt tatttattta ttagtgtagt ctaatttagt 180tgtataacta taaaattgtt
gtttgtttcc gaatcataag tttttttttt ttttggtttt 240gtattgatag gtgcaagaga
ctcaaaattc tggtttcgat gttaacagaa ttcaagtagc 300tgcccacttg attcgatttg
ttttgtattt ggaaacaacc atggctggtc aaggcccagc 360ccgttgtgct tctgaacctg
cctagtccca tggactagat ctttatccgc agactccaaa 420agaaaaagga ttggcgcaga
ggaattgtca tggaaacaga atgaacaaga aagggtgaag 480aagatcaaag gcatatatga
tctttacatt ctctttagct tatgtatgca gaaaattcac 540ctaattaagg acagggaacg
taacttggct tgcactcctc tcaccaaacc ttacccccta 600actaatttta attcaaaatt
actagtattt tggccgatca ctttatataa taagatacca 660gatttattat atttacgaat
tatcagcatg catatactgt atatagtttt ttttttgtta 720aagggtaaaa taataggatc
cttttgaata aaatgaacat atataattag tataatgaaa 780acagaaggaa atgagattag
gacagtaagt aaaatgagag agacctgcaa aggataaaaa 840agagaagctt aaggaaaccg
cgacgatgaa agaaagacat gtcatcagct gatggatgtg 900agtgatgagt ttgttgcagt
tgtgtagaaa tttttactaa aacagttgtt tttacaaaaa 960agaaataata taaaacgaaa
gcttagcttg aaggcaatgg agactctaca acaaactatg 1020taccatacag agagagaaac
taaaagcttt tcacacataa aaaccaaact tattcgtctc 1080tcattgatca ccgttttgtt
ctctcaagat cgctgctaat ct 1122301569DNAArabidopsis
thaliana 30gtgtttttgg acataatgat ttgaatgata ttaaacaaaa aaaaggcatc
ttattgtaat 60taaattaata aagtaatgga cgtttttgtt agtgagacgc ctgaacttgt
catgacatac 120agacatacca attccattca tgcaggagaa tgttgaataa tgtgagggaa
cgtgaataga 180ggaactattt ttgggtaaat gcaatcacat cctctcattc tcaaacctcc
aaaccaaaat 240gtgtagtttt cttcctctcc aagaaacaat actcttatga ctgcaatacc
acactttagt 300ctgtgtgtgt tttttaatag aatggtataa agaagcatac tgaaatggtg
tttttaaatg 360aaagatcaaa tcacaacaat atccaaaacc taaaataaaa agattgtggg
cttatttatt 420aggcccatag tctttattct ttaagggcaa actttcagag atgcgggtct
acattatttg 480gcccgaacca gtaaaacctg caggcttcta gagaaaccca tacgaatggc
gagttaccgg 540tcagaagcgt aatcaccggt ccttaaattt catttccagg cgaacgattt
ggaaaagtcg 600gtgtcgttaa gaaaagacaa ttcctaccct ttttgtcgtc atttgtttgc
taattgctac 660ctttataatg taagggagga gtggagttgc tattattatt ccgacattta
agtgttatgt 720gtttgtcgga tcggattcta tcttaagaat cttatcttat tccccccgaa
tatcattgac 780tttgtcttaa atcttaattt gttatgtttg cacttaaact ctctcttttt
cgttagtgca 840cacaactcgt attccatcaa aatcaatgat aaaataattc ttaccaaatt
aacaaaaagt 900gaataaatat cacattattc atactaatct tcatgatatc atcacaagaa
taatgtgtgc 960acacaaaaaa ccactgtttg tttccgcgtg aagtgaactc tcaagagtct
ccaatgttga 1020ccaaatcaaa caacctctta acttctttga ccaacaattt ttaaaaccat
gaaataagtt 1080acatacgaag acttgacttg tttctttctc ttaaaaatca aagtttaact
gcttcagagt 1140ttaattttca atgtgtccat atacaattca ttttaaatct aaagcaaatt
cctcttcttt 1200tttttccttt aatttatttt attttttatg gagtgagttg agttctgtat
acattctttg 1260taaatggaaa aaaataaaaa aacagcttga ttaaaaaaaa taaagaaatt
gagaaaaaga 1320caagaattta aataataata aaataatgta aaaagaaagt gaacaacaaa
aaaagacaca 1380aaaaagtaaa actgaaaagg agtatttctc tgtcatttcc cacaccaatc
gcataatcga 1440tttcttccaa cttcaataaa ggggaaccaa cgtaacccta attttgcttt
ctcctctttg 1500ttcagaaaat tttcccttta ctctcaaatt ccttttcgat ttccctctct
taaacctccg 1560aaagctcac
1569311000DNAArtificial SequenceSynthetic Vector promoter DNA
derived from synthetic construct. 31gagggttgtc atgtcagcta aacagcagaa
aaataaaaga atgtgggaga caggctttgc 60ccttttacac atttttaagc tctcttcttt
attttctaaa aacactatag tacacagtta 120aaaatttgcg ggccctttgt ttaatcgatt
tagatttaac caacagcttc atagtcgttt 180cttcttgtta gaaatcaata aatgaaaagt
tggttgtttt gaaagcatat aattaactaa 240caatgtatca tgttgtgaaa caaaagctga
tgataaatgt taaatatagt gtccaaaatt 300ttgggaatgt cttcaaattt tgattttaaa
acatatgaga tgtaccaaca tccacaaaat 360gtttgtgaag ttgtgactta gtgagacgtt
gtcactttat tatctcgttt tggtaacggt 420aagtgcatgt gaacgtgttt gattataagt
ttaacccgac tttgtttacg tggtcatagt 480ttccaataag gctaagtaga atagtgtatg
atagttttca attcaagagt cttttttttc 540caagaaaatc cgaaaacaca atcgtttatt
ttaatatctc aaaagaaatc atatttctta 600ggtaaaacta tcatacgttg agtttttctt
tctccttttg cgattttccc tgaaatatat 660ttatgttgtg tgtgtgcttt gtaaacaaaa
taataatgaa cgtaataata gtaaaacaaa 720aagtaaccta tattattatc atacttgata
taacccaata gagaaataat aatataaatt 780aaaattttat ttcccaattc aaaaatcata
atcaaggagt gaatgctgcc agtagaccaa 840agtaacaata tttggtgtgt acatcaaatg
ataggaaaca acaaaaactc attaatatac 900ctaaatccat atatatatat atatatatat
atttcaatat ttcacattgt tataattagg 960tttaatagat accatattag aaatctcagt
atggtggttc 100032178DNAArtificial
SequenceSynthetic Vector transcriptional terminator DNA sequence
derived from synthetic construct. 32ggaattagaa attttattga tagaagtatt
ttacaaatac aaatacatac taagggtttc 60ttatatgctc aacacatgag cgaaacccta
taagaaccct aattccctta tctgggaact 120actcacacat tattatagag agagatagat
ttgtagagag agactggtga tttcagcg 17833255DNAArtificial
SequenceSynthetic Vector transcriptional terminator DNA sequence
derived from synthetic construct. 33gatcgttcaa acatttggca ataaagtttc
ttaagattga atcctgttgc cggtcttgcg 60atgattatca tataatttct gttgaattac
gttaagcatg taataattaa catgtaatgc 120atgacgttat ttatgagatg ggtttttatg
attagagtcc cgcaattata catttaatac 180gcgatagaaa acaaaatata gcgcgcaaac
taggataaat tatcgcgcgc ggtgtcatct 240atgttactag atcgg
25534931PRTArabidopsis thaliana 34Met
Ala Met Arg Leu Leu Lys Thr His Leu Leu Phe Leu His Leu Tyr1
5 10 15Leu Phe Phe Ser Pro Cys Phe
Ala Tyr Thr Asp Met Glu Val Leu Leu 20 25
30Asn Leu Lys Ser Ser Met Ile Gly Pro Lys Gly His Gly Leu
His Asp 35 40 45Trp Ile His Ser
Ser Ser Pro Asp Ala His Cys Ser Phe Ser Gly Val 50 55
60Ser Cys Asp Asp Asp Ala Arg Val Ile Ser Leu Asn Val
Ser Phe Thr65 70 75
80Pro Leu Phe Gly Thr Ile Ser Pro Glu Ile Gly Met Leu Thr His Leu
85 90 95Val Asn Leu Thr Leu Ala
Ala Asn Asn Phe Thr Gly Glu Leu Pro Leu 100
105 110Glu Met Lys Ser Leu Thr Ser Leu Lys Val Leu Asn
Ile Ser Asn Asn 115 120 125Gly Asn
Leu Thr Gly Thr Phe Pro Gly Glu Ile Leu Lys Ala Met Val 130
135 140Asp Leu Glu Val Leu Asp Thr Tyr Asn Asn Asn
Phe Asn Gly Lys Leu145 150 155
160Pro Pro Glu Met Ser Glu Leu Lys Lys Leu Lys Tyr Ser Phe Gly Gly
165 170 175Asn Phe Phe Ser
Gly Glu Ile Pro Glu Ser Tyr Gly Asp Ile Gln Ser 180
185 190Leu Glu Tyr Leu Gly Leu Asn Gly Ala Gly Leu
Ser Gly Lys Ser Pro 195 200 205Ala
Phe Leu Ser Arg Leu Lys Asn Leu Glu Arg Glu Met Tyr Ile Gly 210
215 220Tyr Tyr Asn Ser Tyr Thr Gly Gly Val Pro
Pro Glu Phe Gly Gly Leu225 230 235
240Thr Lys Leu Glu Ile Leu Asp Met Ala Ser Cys Thr Leu Thr Gly
Glu 245 250 255Ile Pro Thr
Ser Leu Ser Asn Leu Lys His Leu His Thr Leu Phe Leu 260
265 270His Ile Asn Asn Leu Thr Gly His Ile Pro
Pro Glu Leu Ser Gly Leu 275 280
285Val Ser Leu Lys Ser Leu Asp Leu Ser Ile Asn Leu Thr Gly Glu Ile 290
295 300Pro Gln Ser Phe Ile Asn Leu Gly
Asn Ile Thr Leu Ile Asn Leu Phe305 310
315 320Arg Asn Asn Leu Tyr Gly Gln Ile Pro Glu Ala Ile
Gly Glu Leu Pro 325 330
335Lys Leu Glu Val Phe Glu Val Trp Glu Asn Asn Phe Thr Leu Gln Leu
340 345 350Pro Ala Asn Leu Gly Arg
Asn Gly Asn Leu Ile Lys Leu Asp Val Ser 355 360
365Asp Asn His Leu Thr Gly Leu Ile Pro Lys Asp Leu Cys Arg
Gly Glu 370 375 380Lys Leu Glu Met Leu
Ile Leu Ser Asn Asn Phe Phe Phe Gly Pro Ile385 390
395 400Pro Glu Glu Leu Gly Lys Cys Lys Ser Leu
Thr Lys Ile Lys Arg Ile 405 410
415Val Lys Asn Leu Leu Asn Gly Thr Val Pro Ala Gly Leu Phe Asn Leu
420 425 430Pro Leu Val Thr Ile
Ile Glu Leu Thr Asp Asn Phe Phe Ser Gly Glu 435
440 445Leu Pro Val Thr Met Ser Gly Asp Val Leu Asp Gln
Ile Tyr Leu Ser 450 455 460Asn Asn Trp
Phe Ser Gly Glu Ile Pro Pro Ala Ile Gly Asn Phe Pro465
470 475 480Asn Leu Gln Thr Leu Phe Leu
Asp Arg Asn Arg Phe Arg Gly Asn Ile 485
490 495Pro Arg Glu Ile Phe Glu Leu Lys His Leu Ser Arg
Ile Asn Thr Ser 500 505 510Ala
Asn Asn Ile Thr Gly Gly Ile Pro Asp Ser Ile Ser Arg Cys Ser 515
520 525Thr Leu Ile Ser Val Asp Leu Ser Arg
Asn Ile Asn Gly Glu Ile Pro 530 535
540Lys Gly Ile Asn Asn Val Lys Asn Leu Gly Thr Leu Asn Ile Ser Gly545
550 555 560Asn Gln Leu Thr
Gly Ser Ile Pro Thr Gly Ile Gly Asn Met Thr Ser 565
570 575Leu Thr Thr Leu Asp Leu Ser Phe Asn Asp
Leu Ser Gly Arg Val Pro 580 585
590Leu Gly Gly Gln Phe Leu Val Phe Asn Glu Thr Ser Phe Ala Gly Asn
595 600 605Thr Tyr Leu Cys Leu Pro His
Arg Val Ser Cys Pro Thr Arg Pro Gly 610 615
620Gln Thr Ser Asp His Asn His Thr Ala Leu Phe Ser Pro Ser Arg
Ile625 630 635 640Val Ile
Thr Val Ile Ala Ala Ile Thr Gly Leu Ile Leu Ile Ser Val
645 650 655Ala Ile Arg Gln Met Asn Lys
Lys Lys Asn Gln Lys Ser Leu Ala Trp 660 665
670Lys Leu Thr Ala Phe Gln Lys Leu Asp Phe Lys Ser Glu Asp
Val Leu 675 680 685Glu Cys Leu Lys
Glu Glu Asn Ile Ile Gly Lys Gly Gly Ala Gly Ile 690
695 700Val Tyr Arg Gly Ser Met Pro Asn Asn Val Asp Val
Ala Ile Lys Arg705 710 715
720Leu Val Gly Arg Gly Thr Gly Arg Ser Asp His Gly Phe Thr Ala Glu
725 730 735Ile Gln Thr Leu Gly
Arg Ile Arg His Arg His Ile Val Arg Leu Leu 740
745 750Gly Tyr Val Ala Asn Lys Asp Thr Asn Leu Leu Leu
Tyr Glu Tyr Met 755 760 765Pro Asn
Gly Ser Leu Gly Glu Leu Leu His Gly Ser Lys Gly Gly His 770
775 780Leu Gln Trp Glu Thr Arg His Arg Val Ala Val
Glu Ala Ala Lys Gly785 790 795
800Leu Cys Tyr Leu His His Asp Cys Ser Pro Leu Ile Leu His Arg Asp
805 810 815Val Lys Ser Asn
Asn Ile Leu Leu Asp Ser Phe Glu Ala His Val Ala 820
825 830Asp Phe Gly Leu Ala Lys Phe Leu Val Asp Gly
Ala Ala Ser Glu Cys 835 840 845Met
Ser Ser Ile Ala Gly Ser Tyr Gly Tyr Ile Ala Pro Glu Tyr Ala 850
855 860Tyr Thr Leu Lys Val Asp Glu Lys Ser Asp
Val Tyr Ser Phe Gly Val865 870 875
880Val Leu Leu Glu Leu Ile Ala Gly Lys Lys Pro Val Gly Glu Phe
Gly 885 890 895Glu Gly Val
Asp Ile Val Arg Trp Val Arg Asn Thr Glu Glu Glu Ile 900
905 910Thr Gln Pro Ser Asp Ala Ala Ile Val Val
Ala Ile Val Asp Pro Arg 915 920
925Leu Thr Gly 93035961PRTPopulus tremula x Populus tremuloides 35Met
Lys Lys Lys Asn Ile Ser Pro Ser Leu Val Leu His Pro Leu Leu1
5 10 15Leu Leu Leu Leu Pro Phe Phe
Ala Phe Asn Ser Leu Ala Leu Lys Phe 20 25
30Ser Pro Gln Leu Leu Ser Leu Leu Ser Leu Lys Thr Ser Leu
Ser Gly 35 40 45Pro Pro Ser Ala
Phe Gln Asp Trp Lys Val Pro Val Asn Gly Gln Asn 50 55
60Asp Ala Val Trp Cys Ser Trp Ser Gly Val Val Cys Asp
Asn Val Thr65 70 75
80Ala Gln Val Ile Ser Leu Asp Leu Ser His Arg Asn Leu Ser Gly Arg
85 90 95Ile Pro Ile Gln Ile Arg
Tyr Leu Ser Ser Leu Leu Tyr Leu Asn Leu 100
105 110Ser Gly Asn Ser Leu Glu Gly Ser Phe Pro Thr Ser
Ile Phe Asp Leu 115 120 125Thr Lys
Leu Thr Thr Leu Asp Ile Ser Arg Asn Ser Phe Asp Ser Ser 130
135 140Phe Pro Pro Gly Ile Ser Lys Leu Lys Phe Leu
Lys Val Phe Asn Ala145 150 155
160Phe Ser Asn Asn Phe Glu Gly Leu Leu Pro Ser Asp Val Ser Arg Leu
165 170 175Arg Phe Leu Glu
Glu Leu Asn Phe Gly Gly Ser Tyr Phe Glu Gly Ile 180
185 190Pro Ala Ala Tyr Gly Gly Leu Gln Arg Leu Lys
Phe Ile His Leu Ala 195 200 205Gly
Asn Val Leu Gly Gly Lys Leu Pro Pro Arg Leu Gly Leu Leu Thr 210
215 220Glu Leu Gln His Met Glu Ile Gly Tyr Asn
His Phe Asn Gly Asn Ile225 230 235
240Pro Ser Glu Phe Ala Leu Leu Ser Asn Leu Lys Tyr Phe Asp Val
Ser 245 250 255Asn Cys Ser
Leu Ser Gly Ser Leu Pro Gln Glu Leu Gly Asn Leu Ser 260
265 270Asn Leu Glu Thr Leu Phe Leu Phe Gln Asn
Gly Phe Thr Gly Glu Ile 275 280
285Pro Glu Ser Tyr Ser Asn Leu Lys Ser Leu Lys Leu Leu Asp Phe Ser 290
295 300Ser Asn Gln Leu Ser Gly Ser Ile
Pro Ser Gly Phe Ser Thr Leu Lys305 310
315 320Asn Leu Thr Trp Leu Ser Leu Ile Ser Asn Asn Leu
Ser Gly Glu Val 325 330
335Pro Glu Gly Ile Gly Glu Leu Pro Glu Leu Thr Thr Leu Phe Leu Trp
340 345 350Asn Asn Asn Phe Thr Gly
Val Leu Pro His Lys Leu Gly Ser Asn Gly 355 360
365Lys Leu Glu Thr Met Asp Val Ser Asn Asn Ser Phe Thr Gly
Thr Ile 370 375 380Pro Ser Ser Leu Cys
His Gly Asn Lys Leu Tyr Lys Leu Ile Leu Phe385 390
395 400Ser Asn Met Phe Glu Gly Glu Leu Pro Lys
Ser Leu Thr Arg Cys Glu 405 410
415Ser Leu Trp Arg Phe Arg Ser Gln Asn Asn Arg Leu Asn Gly Thr Ile
420 425 430Pro Ile Gly Phe Gly
Ser Leu Arg Asn Leu Thr Phe Val Asp Leu Ser 435
440 445Asn Asn Arg Phe Thr Asp Gln Ile Pro Ala Asp Phe
Ala Thr Ala Pro 450 455 460Val Leu Gln
Tyr Leu Asn Leu Ser Thr Asn Phe Phe His Arg Lys Leu465
470 475 480Pro Glu Asn Ile Trp Lys Ala
Pro Asn Leu Gln Ile Phe Ser Ala Ser 485
490 495Phe Ser Asn Leu Ile Gly Glu Ile Pro Asn Tyr Val
Gly Cys Lys Ser 500 505 510Phe
Tyr Arg Ile Glu Leu Gln Gly Asn Ser Leu Asn Gly Thr Ile Pro 515
520 525Trp Asp Ile Gly His Cys Glu Lys Leu
Leu Cys Leu Asn Leu Ser Gln 530 535
540Asn His Leu Asn Gly Ile Ile Pro Trp Glu Ile Ser Thr Leu Pro Ser545
550 555 560Ile Ala Asp Val
Asp Leu Ser His Asn Leu Leu Thr Gly Ile Pro Ser 565
570 575Asp Phe Gly Ser Ser Lys Thr Ile Thr Thr
Phe Asn Val Ser Tyr Asn 580 585
590Gln Leu Ile Gly Pro Ile Pro Ser Gly Ser Phe Ala His Leu Asn Pro
595 600 605Ser Phe Phe Ser Ser Asn Glu
Gly Leu Cys Gly Asp Leu Val Gly Lys 610 615
620Pro Cys Asn Ser Asp Arg Phe Asn Ala Gly Asn Ala Asp Ile Asp
Gly625 630 635 640His His
Lys Glu Glu Arg Pro Lys Lys Thr Ala Gly Ala Ile Val Trp
645 650 655Ile Leu Ala Ala Ala Ile Gly
Val Gly Phe Phe Val Leu Val Ala Ala 660 665
670Thr Arg Cys Phe Gln Lys Ser Tyr Gly Asn Arg Val Asp Gly
Gly Gly 675 680 685Arg Asn Gly Gly
Asp Ile Gly Pro Trp Lys Leu Thr Ala Phe Gln Arg 690
695 700Leu Asn Phe Thr Ala Asp Asp Val Val Glu Cys Leu
Ser Lys Thr Asp705 710 715
720Asn Ile Leu Gly Met Gly Ser Thr Gly Thr Val Tyr Lys Ala Glu Met
725 730 735Pro Asn Gly Glu Ile
Ile Ala Val Lys Lys Leu Trp Gly Lys Asn Lys 740
745 750Glu Asn Gly Lys Ile Arg Arg Arg Lys Ser Gly Val
Leu Ala Glu Val 755 760 765Asp Val
Leu Gly Asn Val Arg His Arg Asn Ile Val Arg Leu Leu Gly 770
775 780Cys Cys Thr Asn Arg Asp Cys Thr Met Leu Leu
Tyr Glu Tyr Met Pro785 790 795
800Asn Gly Ser Leu Asp Asp Leu Leu His Gly Gly Asp Lys Thr Met Thr
805 810 815Ala Ala Ala Glu
Trp Thr Ala Leu Tyr Gln Ile Ala Ile Gly Val Ala 820
825 830Gln Gly Ile Cys Tyr Leu His His Asp Cys Asp
Pro Val Ile Val His 835 840 845Arg
Asp Leu Lys Pro Ser Asn Ile Leu Leu Asp Ala Asp Phe Glu Ala 850
855 860Arg Val Ala Asp Phe Gly Val Ala Lys Leu
Ile Gln Thr Asp Glu Ser865 870 875
880Met Ser Val Val Ala Gly Ser Tyr Gly Tyr Ile Ala Pro Glu Tyr
Ala 885 890 895Tyr Thr Leu
Gln Val Asp Lys Lys Ser Asp Ile Tyr Ser Tyr Gly Val 900
905 910Ile Leu Leu Glu Ile Ile Thr Gly Lys Arg
Ser Val Glu Pro Glu Phe 915 920
925Gly Glu Gly Asn Ser Ile Val Asp Trp Val Arg Ser Lys Leu Lys Thr 930
935 940Lys Glu Asp Val Glu Glu Val Leu
Asp Lys Ser Met Gly Arg Ser Cys945 950
955 960Ser36878PRTPopulus tremula x Populus tremuloides
36Met Ala Ile Pro Arg Leu Phe Phe Leu Phe Tyr Tyr Ile Gly Phe Ala1
5 10 15Leu Phe Pro Phe Val Ser
Ser Glu Thr Phe Gln Asn Ser Glu Gln Glu 20 25
30Ile Leu Leu Ala Phe Lys Ser Asp Leu Phe Asp Pro Ser
Asn Asn Leu 35 40 45Gln Asp Trp
Lys Arg Pro Glu Asn Ala Thr Thr Phe Ser Glu Leu Val 50
55 60His Cys His Trp Thr Gly Val His Cys Asp Ala Asn
Gly Tyr Val Ala65 70 75
80Lys Leu Leu Leu Ser Asn Met Asn Leu Ser Gly Asn Val Ser Asp Gln
85 90 95Ile Gln Ser Phe Pro Ser
Leu Gln Ala Leu Asp Leu Ser Asn Asn Ala 100
105 110Phe Glu Ser Ser Leu Pro Lys Ser Leu Ser Asn Leu
Thr Ser Leu Lys 115 120 125Val Ile
Asp Val Ser Val Asn Ser Phe Phe Gly Thr Phe Pro Tyr Gly 130
135 140Leu Gly Met Ala Thr Gly Leu Thr His Val Asn
Ala Ser Ser Asn Asn145 150 155
160Phe Ser Gly Phe Leu Pro Glu Asp Leu Gly Asn Ala Thr Thr Leu Glu
165 170 175Val Leu Asp Phe
Arg Gly Gly Tyr Phe Glu Gly Ser Val Pro Ser Ser 180
185 190Phe Lys Asn Leu Lys Asn Leu Lys Phe Leu Gly
Leu Ser Gly Asn Asn 195 200 205Phe
Gly Gly Lys Val Pro Lys Val Ile Gly Glu Leu Ser Ser Leu Glu 210
215 220Thr Ile Ile Leu Gly Tyr Asn Gly Phe Met
Gly Glu Ile Pro Glu Glu225 230 235
240Phe Gly Lys Leu Thr Arg Leu Gln Tyr Leu Asp Leu Ala Val Gly
Asn 245 250 255Leu Thr Gly
Gln Ile Pro Ser Ser Leu Gly Gln Leu Lys Gln Leu Thr 260
265 270Thr Val Tyr Leu Tyr Gln Asn Arg Leu Thr
Gly Lys Leu Pro Arg Glu 275 280
285Leu Gly Gly Met Thr Ser Leu Val Phe Leu Asp Leu Ser Asp Asn Gln 290
295 300Ile Thr Gly Glu Ile Pro Met Glu
Val Gly Glu Leu Lys Asn Leu Gln305 310
315 320Leu Leu Asn Leu Met Arg Asn Gln Leu Thr Gly Ile
Ile Pro Ser Lys 325 330
335Ile Ala Glu Leu Pro Asn Leu Glu Val Leu Glu Leu Trp Gln Asn Ser
340 345 350Leu Met Gly Ser Leu Pro
Val His Leu Gly Lys Asn Ser Pro Leu Lys 355 360
365Trp Leu Asp Val Ser Ser Asn Lys Leu Ser Gly Asp Ile Pro
Ser Gly 370 375 380Leu Cys Tyr Ser Arg
Asn Leu Thr Lys Leu Ile Leu Phe Asn Asn Ser385 390
395 400Phe Ser Gly Gln Ile Pro Glu Glu Ile Phe
Ser Cys Pro Thr Leu Val 405 410
415Arg Val Arg Ile Gln Lys Asn His Ile Ser Gly Ser Ile Pro Ala Gly
420 425 430Ser Gly Asp Leu Pro
Met Leu Gln His Leu Glu Leu Ala Lys Asn Asn 435
440 445Leu Thr Gly Lys Ile Pro Asp Asp Ile Ala Leu Ser
Thr Ser Leu Ser 450 455 460Phe Ile Asp
Ile Ser Phe His Asn Leu Ser Ser Leu Ser Ser Ser Ile465
470 475 480Phe Ser Ser Pro Asn Leu Gln
Thr Phe Ile Ala Ser His Asn Asn Phe 485
490 495Ala Gly Lys Ile Pro Asn Gln Ile Gln Asp Arg Pro
Ser Leu Ser Val 500 505 510Leu
Asp Leu Ser Phe Asn His Phe Ser Gly Gly Ile Pro Glu Arg Ile 515
520 525Ala Ser Phe Glu Lys Leu Val Ser Leu
Asn Leu Lys Ser Asn Gln Leu 530 535
540Val Gly Glu Ile Pro Lys Ala Leu Ala Gly Met His Met Leu Ala Val545
550 555 560Leu Asp Leu Ser
Asn Asn Ser Leu Thr Gly Asn Ile Pro Ala Asp Leu 565
570 575Gly Ala Ser Pro Thr Leu Glu Met Leu Asn
Val Ser Phe Asn Lys Leu 580 585
590Asp Gly Pro Ile Pro Ser Asn Met Leu Phe Ala Ala Ile Asp Pro Lys
595 600 605Asp Leu Val Gly Asn Asn Gly
Leu Cys Phe Thr Ala Gly Asp Ile Leu 610 615
620Ser His Ile Lys Glu Ser Asn Ile Ile Gly Met Gly Ala Ile Gly
Ile625 630 635 640Val Tyr
Lys Ala Glu Val Met Arg Arg Arg Leu Leu Thr Val Ala Val
645 650 655Lys Lys Leu Trp Arg Ser Pro
Ser Pro Gln Asn Asp Ile Glu Asp His 660 665
670His Gln Glu Glu Asp Glu Glu Asp Asp Ile Leu Arg Glu Val
Asn Leu 675 680 685Leu Gly Gly Leu
Arg His Arg Asn Ile Val Lys Ile Leu Gly Tyr Val 690
695 700His Asn Glu Arg Glu Val Met Met Val Tyr Glu Tyr
Met Pro Asn Gly705 710 715
720Asn Leu Gly Thr Ala Leu His Ser Lys Asp Glu Lys Phe Leu Leu Arg
725 730 735Asp Trp Leu Ser Arg
Tyr Asn Val Ala Val Gly Val Val Gln Gly Leu 740
745 750Asn Tyr Leu His Asn Asp Cys Tyr Pro Pro Ile Ile
His Arg Asp Ile 755 760 765Lys Ser
Asn Ile Leu Leu Asp Ser Asn Leu Glu Ala Arg Ile Ala Asp 770
775 780Phe Gly Leu Ala Lys Met Met Leu His Lys Asn
Glu Thr Val Ser Met785 790 795
800Val Ala Gly Ser Tyr Gly Tyr Ile Ala Pro Glu Tyr Gly Tyr Thr Leu
805 810 815Lys Ile Asp Glu
Lys Ser Asp Ile Tyr Ser Leu Gly Val Val Leu Leu 820
825 830Glu Leu Val Thr Gly Lys Met Pro Ile Asp Pro
Ser Phe Glu Asp Ser 835 840 845Ile
Asp Val Val Glu Trp Ile Arg Arg Lys Val Lys Lys Asn Glu Ser 850
855 860Leu Glu Glu Val Ile Asp Ala Ser Ile Ala
Gly Asp Cys Lys865 870 87537941PRTPopulus
tremula x Populus tremuloides 37Met Lys Met Lys Ile Ile Val Leu Phe Leu
Tyr Tyr Cys Tyr Ile Gly1 5 10
15Ser Thr Ser Ser Val Leu Ala Ser Ile Asp Asn Val Asn Glu Leu Ser
20 25 30Val Leu Leu Ser Val Lys
Ser Thr Leu Val Asp Pro Leu Asn Phe Leu 35 40
45Lys Asp Trp Lys Leu Ser Asp Thr Ser Asp His Cys Asn Trp
Thr Gly 50 55 60Val Arg Cys Asn Ser
Asn Gly Asn Val Glu Lys Leu Asp Leu Ala Gly65 70
75 80Met Asn Leu Thr Gly Lys Ile Ser Asp Ser
Ile Ser Gln Leu Ser Ser 85 90
95Leu Val Ser Phe Asn Ile Ser Cys Asn Gly Phe Glu Ser Leu Leu Pro
100 105 110Lys Ser Ile Pro Leu
Pro Lys Ser Ile Asp Ile Ser Gln Asn Ser Phe 115
120 125Ser Gly Ser Leu Phe Leu Phe Ser Asn Glu Ser Leu
Gly Leu Val His 130 135 140Leu Asn Ala
Ser Gly Asn Asn Leu Ser Gly Asn Leu Thr Glu Asp Leu145
150 155 160Gly Asn Leu Val Ser Leu Glu
Val Leu Asp Leu Arg Gly Asn Phe Phe 165
170 175Gln Gly Ser Leu Pro Ser Ser Phe Lys Asn Leu Gln
Lys Leu Arg Phe 180 185 190Leu
Gly Leu Ser Gly Asn Asn Leu Thr Gly Glu Leu Pro Ser Val Leu 195
200 205His Gln Leu Pro Ser Leu Glu Thr Ala
Ile Leu Gly Tyr Asn Glu Phe 210 215
220Lys Gly Pro Ile Pro Pro Glu Phe Gly Asn Ile Asn Ser Leu Lys Tyr225
230 235 240Leu Asp Leu Ala
Ile Gly Lys Leu Ser Gly Glu Ile Pro Ser Glu Leu 245
250 255Gly Lys Leu Lys Ser Leu Glu Thr Leu Leu
Leu Tyr Glu Asn Asn Phe 260 265
270Thr Gly Thr Ile Pro Arg Glu Ile Gly Ser Ile Thr Thr Leu Lys Val
275 280 285Leu Asp Phe Ser Asp Asn Ala
Leu Thr Gly Glu Ile Pro Met Glu Ile 290 295
300Thr Lys Leu Lys Asn Leu Gln Leu Leu Asn Leu Met Glu Asn Lys
Leu305 310 315 320Ser Gly
Ser Ile Pro Pro Ala Ile Ser Ser Leu Ala Gln Leu Val Leu
325 330 335Glu Leu Trp Asn Asn Thr Leu
Ser Gly Glu Leu Pro Ser Asp Leu Gly 340 345
350Lys Asn Ser Pro Leu Gln Trp Leu Asp Val Ser Ser Asn Ser
Phe Ser 355 360 365Gly Glu Ile Pro
Ser Thr Leu Cys Asn Lys Gly Asn Leu Thr Lys Leu 370
375 380Ile Leu Phe Asn Asn Thr Phe Thr Gly Gln Ile Pro
Ala Thr Leu Ser385 390 395
400Thr Cys Gln Ser Leu Val Arg Val Arg Met Gln Asn Asn Leu Leu Asn
405 410 415Gly Ser Ile Pro Ile
Gly Phe Gly Lys Leu Glu Lys Leu Gln Arg Leu 420
425 430Glu Leu Ala Gly Asn Arg Leu Ser Gly Gly Ile Pro
Gly Asp Ile Ser 435 440 445Asp Ser
Val Ser Leu Ser Phe Ile Asp Phe Ser Arg Asn Gln Ile Arg 450
455 460Ser Ser Leu Pro Ser Thr Ile Leu Ser Ile His
Asn Leu Gln Ala Phe465 470 475
480Leu Val Ala Asp Asn Phe Ile Ser Gly Glu Val Pro Asp Gln Phe Gln
485 490 495Asp Cys Pro Ser
Leu Ser Asn Leu Asp Leu Ser Ser Asn Thr Leu Thr 500
505 510Gly Thr Ile Pro Ser Ser Ile Ala Ser Cys Glu
Lys Leu Val Ser Leu 515 520 525Asn
Leu Arg Asn Asn Asn Leu Thr Gly Glu Ile Pro Arg Gln Ile Thr 530
535 540Thr Met Ser Ala Leu Ala Val Leu Asp Leu
Ser Asn Asn Ser Leu Thr545 550 555
560Gly Val Leu Pro Glu Ser Ile Gly Thr Ser Pro Ala Leu Glu Leu
Leu 565 570 575Asn Val Ser
Tyr Asn Lys Leu Thr Gly Pro Val Pro Ile Asn Gly Phe 580
585 590Leu Lys Thr Ile Asn Pro Asp Asp Leu Arg
Gly Asn Ser Gly Leu Cys 595 600
605Gly Gly Val Leu Pro Pro Cys Ser Lys Phe Gln Arg Ala Thr Ser Ser 610
615 620His Ser Ser Leu His Gly Lys Arg
Ile Val Ala Gly Trp Leu Ile Gly625 630
635 640Ile Ala Ser Val Leu Ala Leu Gly Ile Leu Thr Ile
Val Thr Arg Thr 645 650
655Leu Tyr Lys Lys Trp Tyr Ser Asn Gly Phe Cys Gly Asp Glu Thr Ala
660 665 670Ser Lys Gly Glu Trp Pro
Trp Arg Leu Met Ala Phe His Arg Leu Gly 675 680
685Phe Thr Ala Ser Asp Ile Leu Ala Cys Ile Lys Glu Ser Asn
Met Ile 690 695 700Gly Met Gly Ala Thr
Gly Ile Val Tyr Lys Ala Glu Met Ser Arg Ser705 710
715 720Ser Thr Val Leu Ala Val Lys Lys Leu Trp
Arg Ser Ala Ala Asp Ile 725 730
735Glu Asp Gly Thr Thr Gly Asp Phe Val Gly Glu Val Asn Leu Leu Gly
740 745 750Lys Leu Arg His Arg
Asn Ile Val Arg Leu Leu Gly Phe Leu Tyr Asn 755
760 765Asp Lys Asn Met Met Ile Val Tyr Glu Phe Met Leu
Asn Gly Asn Leu 770 775 780Gly Asp Ala
Ile His Gly Lys Asn Ala Ala Gly Arg Leu Leu Val Asp785
790 795 800Trp Val Ser Arg Tyr Asn Ile
Ala Leu Gly Val Ala His Gly Leu Ala 805
810 815Tyr Leu His His Asp Cys His Pro Pro Val Ile His
Arg Asp Ile Lys 820 825 830Ser
Asn Asn Ile Leu Leu Asp Ala Asn Leu Asp Ala Arg Ile Ala Asp 835
840 845Phe Gly Leu Ala Arg Met Met Ala Arg
Lys Lys Glu Thr Val Ser Met 850 855
860Val Ala Gly Ser Tyr Gly Tyr Ile Ala Pro Glu Tyr Gly Tyr Thr Leu865
870 875 880Lys Val Asp Glu
Lys Ile Asp Ile Tyr Ser Tyr Gly Val Val Leu Leu 885
890 895Glu Leu Leu Thr Gly Arg Arg Pro Leu Glu
Pro Glu Phe Gly Glu Ser 900 905
910Val Asp Ile Val Glu Trp Val Arg Ile Lys Ile Arg Asp Asn Ile Ser
915 920 925Leu Glu Glu Ala Leu Asp Pro
Asn Val Gly Asn Cys Arg 930 935
94038958PRTOryza sativa 38Met Glu Ala Thr Val Pro Val Leu Leu Leu Val Thr
Val Leu Ser Leu1 5 10
15Ile Leu Pro Ser Gly Ile Gly Ala Ala Ala Ala Gly Asp Glu Arg Ser
20 25 30Ala Leu Leu Ala Lys Ala Gly
Phe Val Asp Thr Val Ser Ala Leu Ala 35 40
45Asp Trp Thr Asp Gly Gly Lys Ala Ser Pro His Cys Lys Trp Thr
Gly 50 55 60Val Gly Cys Asn Ala Ala
Gly Leu Val Asp Arg Leu Arg Leu Ser Gly65 70
75 80Lys Asn Leu Ser Gly Lys Val Ala Asp Asp Val
Phe Arg Leu Pro Ala 85 90
95Leu Ala Val Leu Asn Ile Ser Asn Asn Ala Phe Ala Thr Thr Leu Pro
100 105 110Lys Ser Leu Pro Ser Leu
Pro Ser Leu Lys Val Phe Asp Val Ser Gln 115 120
125Asn Ser Phe Glu Gly Gly Phe Pro Ala Gly Leu Gly Gly Cys
Ala Asp 130 135 140Leu Val Ala Val Asn
Ala Ser Gly Asn Asn Phe Ala Gly Pro Leu Pro145 150
155 160Glu Asp Leu Ala Asn Ala Thr Ser Leu Glu
Thr Ile Asp Met Arg Gly 165 170
175Ser Phe Phe Gly Gly Ala Ile Pro Ala Ala Tyr Arg Arg Leu Tyr Lys
180 185 190Leu Lys Phe Leu Gly
Leu Ser Gly Asn Asn Ile Thr Gly Lys Ile Pro 195
200 205Pro Glu Ile Gly Glu Met Glu Ser Leu Glu Ser Leu
Ile Ile Gly Tyr 210 215 220Asn Glu Leu
Glu Gly Gly Ile Pro Pro Glu Leu Gly Asn Leu Ala Asn225
230 235 240Leu Gln Tyr Leu Asp Leu Ala
Val Gly Asn Leu Asp Gly Pro Ile Pro 245
250 255Pro Glu Leu Gly Lys Leu Pro Ala Leu Thr Ser Leu
Tyr Leu Tyr Lys 260 265 270Asn
Asn Leu Glu Gly Lys Ile Pro Pro Glu Leu Gly Asn Ile Ser Thr 275
280 285Leu Val Phe Leu Asp Leu Ser Asp Asn
Ala Phe Thr Gly Ala Ile Pro 290 295
300Asp Glu Val Ala Gln Leu Ser His Leu Arg Leu Leu Asn Leu Met Cys305
310 315 320Asn His Leu Asp
Gly Val Val Pro Ala Ala Ile Gly Asp Met Pro Lys 325
330 335Leu Glu Val Leu Glu Leu Trp Asn Asn Ser
Leu Thr Gly Ser Leu Pro 340 345
350Ala Ser Leu Gly Arg Ser Ser Pro Leu Gln Trp Val Asp Val Ser Ser
355 360 365Asn Gly Phe Thr Gly Gly Ile
Pro Ala Gly Ile Cys Asp Gly Lys Ala 370 375
380Leu Ile Lys Leu Ile Met Phe Asn Asn Gly Phe Thr Gly Gly Ile
Pro385 390 395 400Ala Gly
Leu Ala Ser Cys Ala Ser Leu Val Arg Met Arg Val His Gly
405 410 415Asn Arg Leu Asn Gly Thr Ile
Pro Val Gly Phe Gly Lys Leu Pro Leu 420 425
430Leu Gln Arg Leu Glu Leu Ala Gly Asn Asp Leu Ser Gly Glu
Ile Pro 435 440 445Gly Asp Leu Ala
Ser Ser Ala Ser Leu Ser Phe Ile Asp Val Ser Arg 450
455 460Asn His Leu Gln Tyr Ser Ile Pro Ser Ser Leu Phe
Thr Ile Pro Thr465 470 475
480Leu Gln Ser Phe Leu Ala Ser Asp Asn Met Ile Ser Gly Glu Leu Pro
485 490 495Asp Gln Phe Gln Asp
Cys Pro Ala Leu Ala Ala Leu Asp Leu Ser Asn 500
505 510Asn Arg Leu Ala Gly Ala Ile Pro Ser Ser Leu Ala
Ser Cys Gln Arg 515 520 525Leu Val
Lys Leu Asn Leu Arg Arg Asn Lys Leu Ala Gly Glu Ile Pro 530
535 540Arg Ser Leu Ala Asn Met Pro Ala Leu Ala Ile
Leu Asp Leu Ser Ser545 550 555
560Asn Val Leu Thr Gly Gly Ile Pro Glu Asn Phe Gly Ser Ser Pro Ala
565 570 575Leu Glu Thr Leu
Asn Leu Ala Tyr Asn Asn Leu Thr Gly Pro Val Pro 580
585 590Gly Asn Gly Val Leu Arg Ser Ile Asn Pro Asp
Glu Leu Ala Gly Asn 595 600 605Ala
Gly Leu Cys Gly Gly Val Leu Pro Pro Cys Ser Gly Ser Arg Ser 610
615 620Thr Ala Ala Gly Pro Arg Ser Arg Gly Ser
Ala Arg Leu Arg His Ile625 630 635
640Ala Val Gly Trp Leu Val Gly Met Val Ala Val Val Ala Ala Phe
Ala 645 650 655Ala Leu Phe
Gly Gly His Tyr Ala Tyr Arg Arg Trp Tyr Val Asp Gly 660
665 670Ala Gly Cys Cys Asp Asp Glu Asn Leu Gly
Gly Glu Ser Gly Ala Trp 675 680
685Pro Trp Arg Leu Thr Ala Phe Gln Arg Leu Gly Phe Thr Cys Ala Glu 690
695 700Val Leu Ala Cys Val Lys Glu Ala
Asn Val Val Gly Met Gly Ala Thr705 710
715 720Gly Val Val Tyr Lys Ala Glu Leu Pro Arg Ala Arg
Ala Val Ile Ala 725 730
735Val Lys Lys Leu Trp Arg Pro Ala Ala Ala Ala Glu Ala Ala Ala Ala
740 745 750Ala Pro Glu Leu Thr Ala
Glu Val Leu Lys Glu Val Gly Leu Leu Gly 755 760
765Arg Leu Arg His Arg Asn Ile Val Arg Leu Leu Gly Tyr Met
His Asn 770 775 780Glu Ala Asp Ala Met
Met Leu Tyr Glu Phe Met Pro Asn Gly Ser Leu785 790
795 800Trp Glu Ala Leu His Gly Pro Pro Glu Arg
Arg Thr Tyr Leu Val Asp 805 810
815Trp Val Ser Arg Tyr Asp Val Ala Ala Gly Val Ala Gln Gly Leu Ala
820 825 830Tyr Leu His His Asp
Cys His Pro Pro Val Ile His Arg Asp Ile Lys 835
840 845Ser Asn Asn Ile Leu Leu Asp Ala Asn Met Glu Ala
Arg Ile Ala Asp 850 855 860Phe Gly Leu
Ala Arg Ala Leu Gly Arg Ala Gly Glu Ser Val Ser Val865
870 875 880Val Ala Gly Ser Tyr Gly Tyr
Ile Ala Pro Glu Tyr Gly Tyr Thr Met 885
890 895Lys Val Asp Gln Lys Ser Asp Thr Tyr Ser Tyr Gly
Val Val Leu Met 900 905 910Glu
Leu Ile Thr Gly Arg Arg Ala Val Glu Ala Ala Phe Gly Glu Gly 915
920 925Gln Asp Ile Val Gly Trp Val Arg Asn
Lys Ile Arg Ser Asn Thr Val 930 935
940Glu Asp His Leu Asp Gly Gln Leu Val Gly Ala Gly Cys Pro945
950 95539794PRTPopulus tremula x Populus tremuloides
39Met Met Ser Ser Leu Gln Lys Pro Phe Ser Met Phe Leu Arg Val Leu1
5 10 15Phe Phe Leu Leu Leu Met
Cys Ile Ile Pro Ser Phe Phe Ala Phe Pro 20 25
30Ser Asn Ser Ser Ala Thr Ser Phe Gly Ala Ala Lys Tyr
Glu Ala Ala 35 40 45Glu Gly Asn
Glu Glu Ala Glu Ala Leu Leu Lys Trp Arg Ala Ser Leu 50
55 60Asp Asp Ser His Ser Gln Ser Val Leu Ser Ser Trp
Val Gly Ser Ser65 70 75
80Pro Cys Lys Trp Leu Gly Ile Thr Cys Asp Asn Ser Gly Ser Val Ala
85 90 95Asn Phe Ser Leu Pro His
Phe Gly Leu Arg Gly Thr Leu His Ser Phe 100
105 110Asn Phe Ser Ser Phe Pro Asn Leu Leu Thr Pro Asn
Leu Arg Asn Asn 115 120 125Ser Leu
Tyr Gly Thr Ile Pro Ser His Ile Ser Asn Leu Thr Lys Ile 130
135 140Thr Asn Leu Asn Leu Cys His Asn His Phe Asn
Gly Ser Leu Pro Pro145 150 155
160Glu Met Asn Asn Leu Thr His Leu Met Val Leu His Leu Phe Ser Asn
165 170 175Asn Phe Thr Gly
His Leu Pro Arg Asp Leu Cys Leu Gly Gly Leu Leu 180
185 190Val Asn Phe Thr Ala Ser Tyr Asn His Phe Ser
Gly Pro Ile Pro Lys 195 200 205Ser
Leu Arg Asn Cys Thr Ser Leu Phe Arg Val Arg Leu Asp Trp Asn 210
215 220Gln Leu Thr Gly Asn Ile Ser Glu Asp Phe
Gly Leu Tyr Pro Asn Leu225 230 235
240Asn Tyr Val Asp Leu Ser His Asn Asn Leu Tyr Gly Glu Leu Thr
Trp 245 250 255Lys Trp Gly
Gly Phe Asn Asn Leu Thr Ser Leu Lys Leu Ser Asn Asn 260
265 270Asn Ile Thr Gly Glu Ile Pro Ser Glu Ile
Ala Lys Ala Thr Gly Leu 275 280
285Gln Met Ile Asp Leu Ser Ser Asn Leu Leu Lys Gly Thr Ile Pro Lys 290
295 300Glu Leu Gly Lys Leu Lys Ala Leu
Tyr Asn Leu Thr Leu His Asn Asn305 310
315 320His Leu Phe Gly Val Val Pro Phe Glu Ile Gln Met
Leu Ser Gln Leu 325 330
335Arg Ala Leu Asn Leu Ala Ser Asn Asn Leu Gly Gly Ser Ile Pro Lys
340 345 350Gln Leu Gly Glu Cys Ser
Asn Leu Leu Gln Leu Asn Leu Ser His Asn 355 360
365Lys Phe Ile Gly Ser Ile Pro Ser Glu Ile Gly Phe Leu His
Phe Leu 370 375 380Gly Asp Leu Asp Leu
Ser Gly Asn Leu Leu Ala Gly Ile Glu Pro Ser385 390
395 400Glu Ile Gly Gln Leu Lys Gln Leu Glu Thr
Met Asn Leu Ser His Asn 405 410
415Lys Leu Ser Gly Leu Ile Pro Thr Ala Phe Val Asp Leu Val Ser Leu
420 425 430Thr Thr Val Asp Ile
Ser Tyr Asn Glu Leu Glu Gly Pro Ile Pro Lys 435
440 445Ile Lys Gly Phe Ile Glu Ala Pro Leu Glu Ala Phe
Met Asn Asn Ser 450 455 460Gly Leu Cys
Gly Asn Ala Asn Gly Leu Lys Pro Cys Thr Leu Leu Thr465
470 475 480Ser Arg Lys Lys Ser Asn Lys
Ile Val Ile Leu Ile Leu Phe Pro Leu 485
490 495Leu Gly Ser Leu Leu Leu Leu Leu Ile Met Val Gly
Cys Leu Tyr Phe 500 505 510His
His Gln Thr Ser Arg Glu Arg Ile Ser Cys Leu Gly Glu Arg Gln 515
520 525Ser Pro Leu Ser Phe Val Val Trp Gly
His Glu Glu Glu Ile His Glu 530 535
540Thr Ile Ile Gln Ala Ala Asn Asn Phe Asn Phe Asn Asn Cys Ile Gly545
550 555 560Lys Gly Gly Tyr
Gly Ile Val Tyr Arg Ala Met Leu Pro Thr Gly Gln 565
570 575Val Val Ala Val Lys Lys Phe His Pro Ser
Arg Asp Gly Glu Leu Met 580 585
590Asn Leu Arg Thr Phe Arg Asn Glu Ile Arg Met Leu Ile Asp Ile Arg
595 600 605His Arg Asn Ile Val Lys Leu
His Gly Phe Cys Ser Leu Ile Glu His 610 615
620Ser Phe Leu Val Tyr Glu Phe Ile Glu Arg Gly Ser Leu Lys Met
Asn625 630 635 640Leu Ser
Ser Glu Glu Gln Val Met Asp Leu Asp Trp Asn Arg Arg Leu
645 650 655Asn Val Val Lys Gly Val Ala
Ser Ala Leu Ser Tyr Leu His His Asp 660 665
670Cys Ser Pro Pro Ile Ile His Arg Asp Ile Ser Ser Ser Asn
Val Leu 675 680 685Leu Asp Ser Glu
Tyr Glu Ala His Val Ser Asp Phe Gly Thr Ala Glu 690
695 700Leu Leu Met Pro Asp Ser Thr Asn Trp Thr Ser Phe
Ala Gly Thr Leu705 710 715
720Gly Tyr Thr Ala Pro Glu Leu Ala Tyr Thr Met Arg Val Asn Glu Lys
725 730 735Cys Asp Val Tyr Ser
Phe Gly Val Val Thr Met Glu Val Ile Met Gly 740
745 750Met His Pro Gly Asp Leu Ile Ser Phe Leu Tyr Ala
Ser Ala Phe Ser 755 760 765Ser Ser
Ser Cys Ser Gln Ile Asn Gln His Ala Leu Leu Lys Asp Val 770
775 780Ile Asp Gln Arg Ile Pro Leu Pro Glu Asn785
79040992PRTArtificial SequenceSynthetic consensus protein
sequence for PXY derived from Arabidopsis thaliana. 40Xaa Xaa Xaa
Xaa Xaa Met Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa1 5
10 15Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa 20 25
30Xaa Xaa Xaa Xaa Xaa Xaa Leu Leu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
35 40 45Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 50 55
60Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa65
70 75 80Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 85
90 95Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa 100 105
110Xaa Xaa Xaa Xaa Asn Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
115 120 125Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Ile Xaa Xaa Xaa Xaa Xaa 130 135
140Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa145 150 155 160Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Xaa Xaa Xaa Xaa
165 170 175Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 180 185
190Xaa Xaa Xaa Xaa Xaa Ile Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa 195 200 205Xaa Tyr Xaa Xaa
Leu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 210
215 220Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa225 230 235
240Asn Xaa Xaa Xaa Gly Xaa Val Pro Xaa Xaa Xaa Xaa Xaa Leu Xaa Xaa
245 250 255Leu Xaa Xaa Xaa Xaa
Met Xaa Xaa Xaa Xaa Xaa Xaa Gly Xaa Ile Pro 260
265 270Xaa Xaa Leu Xaa Xaa Leu Xaa Xaa Leu Xaa Xaa Leu
Xaa Leu Xaa Xaa 275 280 285Asn Xaa
Xaa Xaa Gly Xaa Ile Pro Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 290
295 300Leu Xaa Xaa Xaa Xaa Xaa Ser Xaa Asn Xaa Xaa
Thr Gly Xaa Ile Pro305 310 315
320Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Ile Xaa Xaa Ile Xaa Leu Xaa Xaa
325 330 335Asn Xaa Leu Xaa
Gly Xaa Ile Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 340
345 350Leu Xaa Xaa Xaa Xaa Val Xaa Xaa Asn Xaa Xaa
Xaa Xaa Xaa Leu Xaa 355 360 365Xaa
Xaa Xaa Gly Xaa Xaa Xaa Xaa Leu Xaa Xaa Leu Xaa Val Ser Xaa 370
375 380Asn Xaa Xaa Thr Gly Xaa Ile Pro Xaa Xaa
Leu Xaa Xaa Xaa Xaa Xaa385 390 395
400Leu Xaa Xaa Leu Xaa Leu Xaa Xaa Asn Xaa Xaa Xaa Gly Xaa Ile
Pro 405 410 415Xaa Xaa Leu
Xaa Xaa Xaa Xaa Xaa Leu Xaa Xaa Xaa Xaa Xaa Xaa Xaa 420
425 430Asn Xaa Leu Xaa Gly Xaa Val Pro Xaa Xaa
Xaa Xaa Xaa Leu Xaa Xaa 435 440
445Val Xaa Xaa Ile Xaa Leu Xaa Xaa Asn Xaa Xaa Xaa Xaa Xaa Xaa Xaa 450
455 460Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa465 470
475 480Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 485 490
495Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Xaa Ile Pro
500 505 510Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Ile Xaa Xaa Xaa Xaa 515 520
525Asn Xaa Xaa Xaa Gly Xaa Ile Pro Xaa Xaa Ile Xaa Xaa Xaa
Xaa Xaa 530 535 540Leu Xaa Xaa Val Xaa
Leu Xaa Xaa Asn Xaa Ile Xaa Gly Xaa Ile Pro545 550
555 560Xaa Xaa Ile Xaa Xaa Val Xaa Xaa Leu Xaa
Xaa Leu Xaa Ile Ser Xaa 565 570
575Asn Xaa Leu Thr Gly Xaa Ile Pro Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
580 585 590Leu Xaa Xaa Xaa Xaa
Leu Xaa Phe Asn Xaa Leu Xaa Gly Xaa Val Pro 595
600 605Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Asn 610 615 620Xaa Xaa Leu
Cys Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa625
630 635 640Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 645
650 655Xaa Xaa Xaa Xaa Leu Ile Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa 660 665 670Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 675
680 685Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa 690 695
700Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Phe Xaa Lys Xaa Xaa Xaa705
710 715 720Xaa Xaa Xaa Asp
Xaa Leu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Asn 725
730 735Xaa Ile Gly Xaa Gly Xaa Xaa Gly Xaa Val
Tyr Arg Gly Xaa Met Xaa 740 745
750Xaa Xaa Xaa Xaa Xaa Val Ala Ile Lys Arg Xaa Xaa Xaa Xaa Xaa Xaa
755 760 765Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 770 775
780Xaa Xaa Xaa Xaa Xaa Glu Ile Xaa Xaa Leu Xaa Xaa Ile Arg His
Arg785 790 795 800Xaa Ile
Val Arg Leu Xaa Gly Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
805 810 815Leu Leu Tyr Glu Tyr Met Xaa
Xaa Gly Leu Xaa Xaa Xaa Leu Xaa Xaa 820 825
830Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Trp Xaa Xaa Xaa
Xaa Xaa 835 840 845Val Xaa Xaa Xaa
Xaa Xaa Xaa Gly Leu Xaa Tyr Leu His Xaa Asp Cys 850
855 860Xaa Pro Xaa Ile Leu His Arg Asp Val Xaa Xaa Xaa
Asn Ile Leu Leu865 870 875
880Asp Xaa Xaa Xaa Glu Ala His Val Xaa Asp Phe Gly Xaa Ala Lys Xaa
885 890 895Leu Xaa Leu Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Ser Xaa Xaa Ala 900
905 910Gly Ser Xaa Gly Tyr Xaa Ala Pro Glu Xaa Ala Tyr
Thr Leu Xaa Val 915 920 925Xaa Xaa
Lys Xaa Asp Xaa Tyr Ser Xaa Gly Val Val Xaa Leu Glu Leu 930
935 940Ile Xaa Gly Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa945 950 955
960Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Ile Xaa Xaa Xaa Xaa
965 970 975Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa 980
985 9904122DNAArtificial SequenceSynthetic forward
primer for CLE41 derived from Arabidopsis thaliana. 41caccatggca
acatcaaatg ac
224220DNAArtificial SequenceSynthetic reverse primer for CLE41 derived
from Arabidopsis thaliana. 42aaaccagatg tgccaactca
204322DNAArtificial SequenceSynthetic
forward primer for CLE42 derived from Arabidopsis thaliana.
43caccatgaga tctcctcaca tc
224424DNAArtificial SequenceSynthetic reverse primer for CLE42 derived
from Arabidopsis thaliana. 44tgaatcaaac aagcaacata acaa
244522DNAArtificial SequenceSynthetic
forward primer for PXY construct derived from Arabidopsis thaliana.
45caccttaaat ccaccattgt ca
224620DNAArtificial SequenceSynthetic reverse primer for PXY construct
derived from Arabidopsis thaliana. 46ccaagataat ggacgccaac
204723DNAArtificial SequenceSynthetic
forward primer for SUC2::CLE41 overlap PCR entry clone derived from
Arabidopsis thaliana. 47caccaacaca tgttgccgag tca
234842DNAArtificial SequenceSynthetic forward primer
for SUC2::CLE41 overlap PCR entry clone derived from Arabidopsis
thaliana. 48gtcatttgat gttgccatga aatttctttg agagggtttt tg
424942DNAArtificial SequenceSynthetic reverse primer for CLE41
overlap PCR entry clone derived from Arabidopsis thaliana.
49caaaaaccct ctcaaagaaa tttcatggca acatcaaatg ac
425020DNAArtificial SequenceSynthetic forward primer for CLE41 RT-PCR
derived from Arabidopsis thaliana. 50ccatgactcg tcatcagtcc
205121DNAArtificial SequenceSynthetic
reverse primer for CLE41 RT-PCR derived from Arabidopsis thaliana.
51tttggaccac taggaacctc a
215220DNAArtificial SequenceSynthetic forward primer for CLE42 RT-PCR
derived from Arabidopsis thaliana. 52tccaaaccca tcaaagaacc
205320DNAArtificial SequenceSynthetic
reverse primer for CLE42 RT-PCR derived from Arabidopsis thaliana.
53attggcaccg atcatctttc
205423DNAArtificial SequenceSynthetic primer for CLE42 RT-PCR derived
from Arabidopsis thaliana. 54aacctagcaa tatcctcctc gac
235520DNAArtificial SequenceSynthetic
reverse primer for PXY RT-PCR derived from Arabidopsis thaliana.
55ggttccaccg atctttttcc
205621DNAArtificial SequenceSynthetic forward primer for RT-PCR control
derived from Arabidopsis thaliana. 56atgaagatta aggtcgtggc a
215719DNAArtificial SequenceSynthetic
reverse primer for RT-PCR control derived from Arabidopsis thaliana.
57ccgagtttga agaggctac
195820DNAArtificial SequenceSynthetic forward primer for CLE41 qRT-PCR
derived from Arabidopsis thaliana. 58tcaagagggt tctcctcgaa
205920DNAArtificial SequenceSynthetic
reverse primer for CLE41 qRT-PCR derived from Arabidopsis thaliana.
59tgtgctagcc tttggacgta
206020DNAArtificial SequenceSynthetic forward primer to 18s rRNA qRT-PCR
control derived from Arabidopsis thaliana. 60catcagctcg cgttgactac
206119DNAArtificial
SequenceSynthetic reverse primer to 18s rRNA qRT-PCR control derived
from Arabidopsis thaliana. 61gatccttccg caggttcac
196220DNAArtificial SequenceSynthetic forward
primer to 35S for Genotyping derived from Arabidopsis thaliana.
62cgcacaatcc cactatcctt
206321DNAArtificial SequenceSynthetic reverse primer to pxy-3 for
Genotyping derived from Arabidopsis thaliana. 63ttaccgtttg atccaagctt g
21
User Contributions:
Comment about this patent or add new information about this topic:
| People who visited this patent also read: | |
| Patent application number | Title |
|---|---|
| 20120182659 | Dual-directional electrostatic discharge protection method |
| 20120182658 | Low-Power Magnetic Slope Detecting Circuit |
| 20120182657 | RATE OF CHANGE DIFFERENTIAL PROTECTION |
| 20120182656 | ACTIVE TRANSIENT CURRENT CONTROL IN ELECTRONIC CIRCUIT BREAKERS |
| 20120182655 | Methods and Systems Involving Monitoring Circuit Connectivity |