Patent application title: Modulation of plant cell number
Inventors:
Gerda Cnops (Gent, BE)
Delphine Fleury (Belair, AU)
Dirk G. Inze (Moorsel-Aalst, BE)
Dirk G. Inze (Moorsel-Aalst, BE)
Maria Van Lijsebettens (Merelbeke, BE)
Assignees:
VIB VZW
UNIVERSITEIT GENT
IPC8 Class: AC12N1582FI
USPC Class:
800290
Class name: Multicellular living organisms and unmodified parts thereof and related processes method of introducing a polynucleotide molecule into or rearrangement of genetic material within a plant or plant part the polynucleotide alters plant part growth (e.g., stem or tuber length, etc.)
Publication date: 2011-03-31
Patent application number: 20110078825
Claims:
1.-6. (canceled)
7. A method of modulating cell number in a plant organ or part thereof, said method comprising:transforming the plant organ or part thereof with a nucleic acid molecule encoding a protein comprising a variant of SEQ ID NO:2 that modulates cell number in a plant organ or part thereof, wherein the nucleic acid molecule is operably lined to a promoter; andexpression said protein so as to modulate cell number in the plant organ or part thereof.
8. The method according to claim 7, wherein the variant is selected from the group consisting of SEQ ID NOs: 49-54.
9. The method according to claim 7, wherein the protein consists of a variant of SEQ ID NO:2.
10. The method according to claim 7, wherein the nucleic acid molecule is overexpressed.
11. The method according to claim 7, further comprising modulate leaf morphology with the variant.
12. The method according to claim 7, wherein said plant organ comprises a leaf palisade cell.
13. The method according to claim 7, wherein said plant organ is a plant leaf or a plant root.
14. The method according to claim 13, wherein said plant organ comprises a leaf palisade cell.
15. A method of increasing plant biomass, said method comprising:transforming the plant with a nucleic acid molecule encoding a protein comprising a variant of SEQ ID NO:2, wherein said protein, when expressed, modulates plant biomass, wherein the nucleic acid molecule is operably lined to a promoter; andexpressing the protein so as to modulate plant biomass.
16. The method according to claim 15, wherein the nucleic acid is overexpressed.
17. The method according to claim 15, wherein the variant is selected from the group consisting of SEQ ID NOs: 49-54.
18. A method of modulating cell number in a plant organ or part thereof, said method comprising:providing the plant organ or part thereof;transforming the plant organ or part thereof with a means for modulating cell number in the plant organ or part thereof; andexpressing the means for modulating cell number in the plant organ or part thereof so as to modulate cell number in the plant organ or part thereof.
19. The method according to claim 18, wherein the means comprises a nucleic acid sequence selected from the group consisting of SEQ ID NOs: 49-54.
20. A method of increasing plant biomass, said method comprising:providing a plant;transforming the plant with a means for increasing plant biomass; andexpressing the means for increasing plant biomass so as increase the biomass of the plant.
21. The method according to claim 20, wherein the means comprises a nucleic acid sequence selected from the group consisting of SEQ ID NOs: 49-54.
Description:
CROSS-REFERENCE TO RELATED APPLICATION(S)
[0001]This application is a continuation of co-pending U.S. patent application Ser. No. 11/660,483, filed Apr. 22, 2008, now U.S. Pat. No. ______, which itself is the national stage of PCT International Application Number PCT/EP2005/054031, filed Aug. 16, 2005, which itself claims priority to EP 04103971.0, filed Aug. 18, 2004, the disclosure of each of which is hereby incorporated herein by this reference in their entirety.
TECHNICAL FIELD
[0002]The invention relates to the use of the ANG4 gene, or a variant thereof, to modulate the cell number of a plant organ. The modulation can be used to increase the plant biomass, or to adapt the plant architecture.
BACKGROUND
[0003]Plant architecture, especially leaf and root morphology, is an important factor in the determination of the plant productivity. Therefore, the study of genes involved in plant architecture and their regulation has drawn a lot of attention by several research groups.
[0004]The isolation, identification, characterization and manipulation of genes that are candidates for controlling leaf development is a key in understanding how plant leaves are constructed. Several methods have been used to study genes and their functions that regulate leaf development such as forward or reverse genetics. During leaf development processes, there are at least two factors that affect the leaf phenotype, at first cell division, that results in a given cell number, and second is cell expansion, which is required for the establishment of the cell size and shape. The length and width of leaves are regulated by cell division and cell expansion according to a gradient (Pyke et al., 1991; Van Lijsebettens and Clarke, 1998). In addition, the leaves are also modulated by environmental factors such as water, nutrients, light and CO2 concentration. Berna et al. (1999) gives an overview of mutations and phenotypic classes that influence leaf morphology in Arabidopsis. Some of those mutations were characterized on gene level. Genes that regulate cell number along the width axis are DRL1 and SWP1 genes that act mainly on lateral growth of the lamina (Nelissen et al., 2003, and Autran et al., 2002).
[0005]Although these genes might be used to modulate the plant biomass, there is still a further need for genes controlling plant architecture, especially for genes capable of controlling the cell number in specific plant organs.
[0006]In this invention, we studied a mutant with narrow leaves, angusta4, from the seed collection of Berna et al. (1999), and identified the causal gene, which we called ANG4. The mutant was originally created by EMS method (FIG. 1, Panel A). Molecular analysis surprisingly showed that the causal gene for the angusta4 mutation, which is located on chromosome 2, is a RING finger protein (Anami, 2004; Stone et al., 2005) with E3 ligase activity. This activity is related to protein degradation, but has never been linked to altered leaf morphology. The width and length of angusta4 laminas was compared to wild-type (Landsberg erecta) (FIG. 1, Panel A). The data showed that total length lamina in the angusta4 leaves is significantly reduced compared to Ler. angusta 4 had narrow first leaf and shorter petioles than Landsberg. The epidermal and palisade cell area in the angusta4 (11 mm2) is smaller than in wild-type (19 mm2) as well. Even more surprisingly, we found that that the phenotype of the leaf is due to a drastic reduction in the number of palisade cells. Moreover, we found that the same mutation has a dramatical effect on root growth too, making the gene an interesting tool for biomass modulation.
DISCLOSURE
[0007]A first aspect of the invention is the use of a gene encoding a protein comprising SEQ ID NO:2 (TAIR_At2g44950, FIG. 10), or a functional fragment or variant thereof, to modulate the cell number of a plant organ, or a part thereof. Preferably, the gene encodes a protein consisting of SEQ ID NO:2. Gene as used here, refers to the coding sequence, which may be linked to its own promoter, but is preferably operably linked to a promoter which is not its own. The promoter can be any promoter suitable for expression in plants. Preferably, the promoter is a strong promoter, such as, but not limited to the 35S promoter. "Gene" refers both to the genomic sequence (including possible introns) as well as to the cDNA derived from the spliced messenger, operably linked to a promoter sequence. Coding sequence is a nucleotide sequence, which is transcribed into mRNA and/or translated into a polypeptide when placed under the control of appropriate regulatory sequences. The boundaries of the coding sequence are determined by a translation start codon at the 5'-terminus and a translation stop codon at the 3'-terminus. A coding sequence can include, but is not limited to mRNA, cDNA, recombinant nucleotide sequences or genomic DNA, while introns may be present as well under certain circumstances.
[0008]Operably linked refers to a juxtaposition wherein the components so described are in a relationship permitting them to function in their intended manner. A promoter sequence "operably linked" to a coding sequence is ligated in such a way that expression of the coding sequence is achieved under conditions compatible with the promoter sequence.
[0009]A variant, as used herein, is a plant gene comprising a ring finger, with a homology with SEQ ID NO:2 of at least 25% identities and/or 45% positives, preferably at least 35% identifies and/or 55% positives, more preferably at least 45% identities and/or 65% positives, even more preferably at least 55% identities and/or 75% positives, most preferably at least 65% identities and/or 85% positives, as measured by a protein-protein Blast search. Preferably, the variant has E3 ubiquitin protein ligase activity. Preferred variants are the Oryza sativa ANG4 homologues CAD41603 and NP922769, as listed in FIG. 10. Plant organs, as used here, comprise roots, stem, leaves and flowers. Preferably, the plant organ is a plant leaf and/or a plant root. Parts of a plant organ are, as a non-limiting example, the palisade cells of the leaves, or the lateral roots. One preferred embodiment is the use according to the invention, whereby the modulation of the cell number is used to modulate the leaf morphology. A functional fragment, as used here, is any fragment that still has the E3 ubiquitin-protein ligase activity.
[0010]Still another aspect of the invention is the use of a gene encoding a protein comprising, preferably consisting of SEQ ID NO:2, or a functional fragment or variant thereof, to modulate the root length. Preferably, the gene, variant of functional fragment is overexpressed and the modulation is an increase in root length. Preferably, the gene comprises SEQ ID NO:1 (genbank NM--130060).
[0011]Another aspect of the invention is the use of a gene encoding a protein comprising, preferably consisting of SEQ ID NO:2, or a functional fragment or variant thereof, to increase biomass. Preferably, the increase of biomass is obtained by an overexpression of the gene, variant or functional fragment.
BRIEF DESCRIPTION OF THE FIGURES
[0012]FIG. 1: Leaf phenotype of angusta4 and wild-type. Panel A: In vivo condition, fully grown rosettes of Ler and angusta4. Panel B: Juvenile and adult fully expanded leaves of Ler. Panel C: Juvenile and adult fully expanded leaves of angusta4.
[0013]FIG. 2: In vitro leaf phenotype of wild-type and angusta4 mutant plant (26 days after germination)
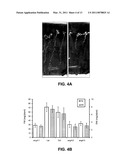
[0014]FIG. 3: Leaf phenotype of angusta4. Panel A: Transversal sections at the widest location of expanded lamina of first leaves of wild-type (Ler)-(top) and angusta4 (bottom). Panel B: Mean value of palisade cell number in first leaves of angusta4 and the wild-type (asterisk indicates statically significant difference). Panel C: Cross section of wild-type at the midvein (Adaxial surface is up). Panel D: Close up of wild-type vascular tissue. Panel E: Cross section of angusta4 at the midvein (Adaxial surface is up). Panel F: Close up of angusta4 vascular tissue (V: vascular bundle; P: palisade cell; x: xylem; ph: phloem; I: inter cellular space). Panels G, H, I: Morphological data of expanded leaves of ang4-1 mutant (Panels A, B, C). Bars represent mean values and standard deviation. *** means statistical difference at p<0.001 from the t test. Histological observations of expanded leaves of ang4 mutant and Ler.
[0015]FIG. 4A: In vitro root growth after 15 days of germination. FIG. 4B: Biomass of ang4-1, ang4-2, ang4-3 mutants, Ler and Col in soil conditions. Average of two assays, four blocks per assay and eight plants per block. The bars correspond to the standard deviation.
[0016]FIG. 5: Panel A: Root growth kinetics of angusta4 in comparison with wild-type (Ler). Panels B and C: Longitudinal sections by confocal microscopy through the root apical meristem in the root tip in wild-type and the angusta4 mutants.
[0017]FIG. 6: Fine mapping of the ANG4 gene: The figure indicates the map-based cloning strategy where a set of eight AFLP primer combinations was applied to 20 F2 individual mutants that indicated that the ANG4 mutation was located on chromosome 2 (blue and white pattern). Further application of AFLP marker SM33--202 and SM26--495 narrowed the ANG4 interval to 293 kb. Finally, InDel and SNP markers were used on a total of 1062 recombinants that delimited the ANG4 area to 27 kb at the bottom of chromosome 2 flanked by SNP markers CER458218 and CER458367. The 27 kb region contained four genes, one of which was the ANG4 candidate gene.
[0018]FIG. 7: Separation of PCR products of At2g44960 gene following amplification with two primer sets. Lane 1 contains a 1 kb molecular weight marker. Lanes 2-7 contains PCR products from Ler At2g44960 gene while lanes 8-13 contains PCR products from ANG4 At2g44960 gene. PCR products of lanes 2, 3, 4, 8, 9, and 10 were amplified with primer combinations Defle 12 and Defle 13 while PCR products of lanes 5, 6, 7, 11, 12 and 13 were amplified with primer combination: Defle 14 and Defle 15.
[0019]FIG. 8: Example of an alignment performed by CLUSTALW 1.8. This alignment is between 2652 bp and 3873 bp part of the At2g44950 gene that was amplified by 5'CTCGCCCATTGTTGTTTCAG3' (SEQ ID NO:3) and 5'AATTGCGGAAACCATGTTCC 3' (SEQ ID NO:4) primer combination. It clearly demonstrates the point mutation induced by EMS as a C was changed to a T generating a stop codon UAG. Aligned Ang sequence in FIG. 8 is represented in SEQ ID NO:47. Aligned Ler sequence in FIG. 8 is represented in SEQ ID NO:48.
[0020]FIG. 9: ANG4 gene structure. Shown are the ANG4 candidate genes covering a 27 kb region on chromosome 2 and linked by CER458218 and CER458367 SNP markers. The unspliced mRNA of ANG4 has 19 exons and 18 introns covering a region of 6298 bp while the full length cDNA covers a region of 2637 bp. EMS mutagenization caused a C to change to a T generating a stop codon at the end of exon 16 hence truncating the protein from 878 amino acids to 844 amino acids (Exons in blue boxes, and introns in orange boxes). Figure drawn to scale; for the candidate genes structure, 1 cm=2 kb and for the unspliced mRNA and the spliced mRNA, 1 cm=1 kb.
[0021]FIG. 10: An alignment of the ANG4 homologues in different species. The orange underlined sequence indicates the conserved RING finger motif. Conserved cystein and histidine residues are colored with red and blue colors respectively. At2g44950 is ANG4 sequence with 878 amino acid residues. At1g55250 is the ANG4 homologue on chromosome 1 in Arabidopsis with 899 amino acids. NP--55586 and AAK58539 are ANG4 homologues in human genome with 1001 and 975 amino acids respectively. CAD41603 and NP922769 are the ANG4 homologues in Oryza sativa with 883 and 789 amino acids respectively. TAIR_At2g44950 in FIG. 10 is represented in SEQ ID NO:2. AAP36593.1 in FIG. 10 is represented in SEQ ID NO:49. AAK58539_RFP--20 in FIG. 10 is represented in SEQ ID NO:50. CAD41603.3 in FIG. 10 is represented in SEQ ID NO:51. MIPS_At1g55250 in FIG. 10 is represented in SEQ ID NO:52. NP055586_RFP--40 in FIG. 10 is represented in SEQ ID NO:53. NP922769.1 in FIG. 10 is represented in SEQ ID NO:54.
[0022]FIG. 11: RT-PCR analysis of ANG4 gene expression in different Ler organs. The expression pattern was visualized on acrylamide gel. Four μl samples were loaded an acrylamide gel in 1× Tris-Boric acid-EDTA buffer and electrophoresed at 3000V. Primers Defle 44 and syana--01 were labeled with P33. Numbers on the gel indicate different Ler organs as follows: 1--Ler apex, 2--Ler shoot apex, 3--Ler roots, 4--Ler cotyledons, 5--Ler young leaves, 6--Ler Expanded leaves, 7--Ler flowers and 8--water as a control sample.
[0023]FIG. 12: Summary of Arabidopsis genes with altered mRNA expression in ang4 and two other leaf development mutants, elo2 and drl1-2. RNA was extracted from shoot apex of young plants and expression measured using ATH1 microarrays (Affymetrix) method in triplicates. Comparisons of expression level were done between each mutant and the wild-type Ler following the Bayesian test of linear model performed with Bioconductor programs. Values without parenthesis are the number of DE genes equally expressed in different mutants, and values in parenthesis, the number of DE genes up-regulated in one mutant and down-regulated in another.
[0024]FIG. 13: Kinematic analysis of leaf growth of the first leaf pair of the wild-type Ler and the ang4-1 mutant. (Panel A) leaf blade area, (Panel B) epidermal cell number on the abaxial side of the leaf, (Panel C) relative leaf expansion rate, (Panel D) average cell division rates of the epidermal cells on the abaxial side of the leaf, (Panel E) average epidermal cell size on the abaxial side of the leaf, (Panel F) stomatal index on the abaxial side of the leaf. Error bars correspond to the standard deviation (n=5).
[0025]FIG. 14A: Flow cytometry analysis of nuclear DNA content of the Ler. FIG. 14B: Flow cytometry analysis of nuclear DNA content of the ang4-1 mutant (B).
[0026]FIG. 15: Ler wild-type and OE-ANG4 (T1) plants two weeks after transfer to soil.
DETAILED DESCRIPTION OF THE INVENTION
Examples
Materials and Methods According to the Invention
Plant Material and Growth Conditions
[0027]Seeds of the Arabidopsis thaliana (L.) Heynh. Landsberg erecta (Ler) and the ang4-2 mutant (SALK--122512) were obtained from the Nottingham Arabidopsis Stock Centre. The ang4-1 homozygous mutant was provided by J. L. Micol (Universidad Miguel Hernandez, Alicante, Spain) (Berna et al., 1999). The T-DNA insertion line ang4-3 (GABI--276D08) was supplied from GABI-Kat.
[0028]Plants of the wild-type Landsberg erecta (Ler) and angusta4 (ang4) were grown in in vitro conditions with following conditions: 16/8 hours (d/n) with white light (Neon tubes, cool white), 100 μEm-2h-1 PAR and 20° C. The medium was 2.15 g/l MS salts (micro and macro elements), 1g/l sucrose, 0.5 g/l MES, pH 6.0, 6 g/l plant tissue culture agar. Seeds were sowed in 150×25 mm round dishes, sealed with Urgopore tape. Sixty seeds were sowed per plate. The vernalization period was three days after sowing.
[0029]For the root growth experiment, one lane of five plants was sowed in square plate in vertical position. The homozygous ang4-2 and ang4-3 lines were selected in in vitro medium containing kanamycin 25 mg/l for the ang4-2 or sulfadiazine 11.25 mg/l for the ang4-3 line. The phenotype of the T-DNA insertion lines was scored in soil growth conditions.
Standard Leaf Analysis
[0030]Eight to twelve expanded first and third leaves of 30-days-old and 40-days-old Ler and ang4 in vitro grown plants have been harvested, treated with 100% methanol 0/N, cleared with 90% lactic acid for two to three days O/N and put on a slide for image analysis. Petiole, lamina and leaf length, lamina width and area of first and third leaves have been measured with the Scion Image software (version β-3b; Scion Corp., Frederick, Md.) from digital pictures directly taken from binocular observations.
[0031]The statistical significance of the mean differences (p≦0.05) was analyzed by t-test using the SPSS (Statistical Package for the Social Sciences, version 10.0.5, SPSS, Inc.; Chicago, Ill.) software on normally distributed data.
Root Growth Kinetics
[0032]Fifteen seeds of each angusta4 line was sown out (made only or one row per plates) in the square plates with GM medium contain vitamin. The plates were oriented in a vertical position. By using scalpel, roots of these lines were marked every two days until 14 days.
Differential Interference Contrast (DIC) Optic Analysis
[0033]The cleared first and third Ler and ang4 leaves prepared for the imaging analysis have been used to perform DIC (Differential Interference Contrast) optics analysis. This technique allows counting the number of cells of a determinate histological tissue layer and most importantly measuring the cell area from the adaxial side using a Scion Image.
Leaf Histology: Determination of Palisade Cell Number (PCN)
[0034]Twenty-six-day-old fully expanded first and third leaves of Ler and angusta4 plants were harvested and immediately fixed in FAA (90% EtOH, 5% acetic acid, 5% formaldehyde) at 4° C. overnight. The process of dehydratation was done by increasing concentrations of EtOH as followed: 2×30 minutes EtOH 50%, 2 hours EtOH 50%, 2 hours EtOH 70%, 2 hours EtOH 80%, 0/N EtOH 80%, 2×2 hours EtOH 90% and ultimately 0/N EtOH 95%. Tissue infiltration was realized in a gradually permeation of Historesin and was achieved by first putting the leaves for 4 hours in a mix of 50% EtOH and 50% Historesin, followed by another mix of 30% EtOH and 70% Historesin for 4 hours and finally in 100% Historesin for 4 hours. During that time, the samples were always kept for 30 minutes in vacuum. During the last step, the leaves were shacking at room temperature for three days. The leaves were then immerged in a new basic resin solution containing a 1% temperature-sensitive activator and left shaking ON. Leaves were finally oriented in beds which were half-filled with the resin solution, covered with new resin and left polymerizing at 45° C. for at least 2 hours. The histology analysis has been performed on 5 μm sections collected on glass slides by using a Reichert Jung Ultracut Microtome using homemade glass knives. The Historesin leaf-containing blocks obtained after polymerization were oriented on a plastic cube and fixed with super-glue. The plastic cubes were holder by the micro tube climb.
[0035]Cytoplasm were stained in each sections by toluidin blue following the process below: The treated glass slides were stained for 8 minutes in 0.05% Teledyne blue and 0.1 M phosphate buffer, pH 6.8 for 10 minutes. After two washes (5 to 10 minutes each) in sterilize water, the slides were dried and mounted with DePex. Photographs were taken by using an Olympus CAMEDIA C-3040 digital camera zoom 3.3 mega pixel at the same magnification and pictures image were performed by Adobe Photoshop 6.0 program.
[0036]Five μm transversal sections of 28-day-old Ler and ang4 first and third full-expanded leaves have been made with a Reichert Jung Ultracut microtome in order to determine with the aid of a binocular microscope the Palisade Cell Number (PCN) present at the widest part of the lamina. This parameter is an indicator of leaf blade lateral growth (Tsuge et al., 1996). Several leaves have been entirely sectioned from tip to petiole: one section every ten has been collected and put on a glass slide. The glass slides were subsequently stained with toluidine blue and mounted with DePex.
Confocal Microscopy Observations of Root Meristem
[0037]Seven-day-old seedling of angusta4 were stained with 100 ng/ml propidium iodide solution for 3 minutes and washed three times by sterilized water. Stained root were observed under a MRC600 Biorad confocal microscope using 543 nm excitation 560 LB light.
Flow Cytometry
[0038]The flow cytometry analysis was performed as described by De Veylder et al. (2001). The first two leaves were chopped with a razor blade in 300 μl of buffer (45 mM MgCl2, 30 mM sodium citrate, 20 mM 3-[N-morpholino]propanesulphonic acid, pH 7, and 1% Triton X-100) (Galbraight et al., 1991). To the supernatant, which was filtered over a 30-μm mesh, 1 μl of 4,6-diamidino-2-phenylindole (DAPI) from a stock of 1 mg/mL was added. The nuclei were analyzed with the BRYTE HS flow cytometer, using Win-Bryte software (Bio-Rad, Hercules, Calif.). Of each time point, two biological and three technical replicates were taken.
Leaf Growth Kinematic Analysis
[0039]Leaf growth was analyzed kinematically from 5 to 28 days after sowing as described (De Veylder et al., 2001). The wild-type and ang4-1 plants were germinated and grown in in vitro conditions in GM+V medium. The following parameters were determined: total area of all cells in the drawing, total number of cells, and number of guard cells. From these data, we calculated the average cell area and estimated the total number of cells per leaf by dividing the leaf area by the average cell area (averaged between the apical and basal positions). Finally, average cell division rates for the whole leaf were determined as the slope of the log2-transformed number of cells per leaf, which was done using five-point differentiation formulas (Erickson, 1976).
Map-Based Cloning Procedure
[0040]The DNA extraction, AFLP, insertion/deletion (InDel) and single-nucleotide polymorphism (SNP) analysis were done according to (Peters et al., 2004) and (Cnops et al., 2004). A standard set of eight AFLP markers were analyzed on 20 F2 mutants and identified the mutation in a 493 kb interval on chromosome 2. The fine-mapping of the ANG4 locus was done using the InDel and SNP markers described in Table S1. Recombinants were used for fine-mapping and delineated the locus to 97 and 27 kb regions flanked by SNP markers. The last interval covered a 27 kb region between CER458218 and CER458367 SNP markers and contained four genes that were sequenced.
[0041]The candidate genes identified in the last mapping interval were amplified from DNA and cDNA, and fully sequenced in at least three replicates to identify the base exchange in the ang4-1 mutant compared to Ler.
Microarrays Experiment
[0042]The studied organs, shoot apex of plants (comprising shoot apex meristem, first and second rosette leaf primordia at petiole-less stage), were harvested removing the cotyledons and the hypocotyls. The harvesting was done in the laboratory conditions under additional light and 20° C. between 11 am and 6 pm. The age of the plants at the harvesting step were between 8 and 15 days after germination depending of the delay of mutant development. RNA were extracted with TRIzol Reagent (Life Technologies, Breda, The Netherlands). The experimental design comprised three replicates of Ler and ang4, one replicate corresponding to one RNA extraction and about 150 apexis.
[0043]Microarrays experiment was done by the VIB Microarrays Facility lab (Paul van Hummelen, Leuven, Belgium; worldwideweb.microarras.be/) using ATH1 Affymetrix chips of 23,800 probes sets for Arabidopsis thaliana. The raw data were normalized and summarized using Robust Multi-Array average method from affy package of Bioconductor statistical R programs (Wu and Irizarry, 2004). The genes were ranking in order of evidence for differential expression DE between mutant and wild-type using an empirical Bayes method performed with the limma package of Bioconductor. This method consists to combine at the gene level with means and standard deviation from the three replicates to form a statistic B which is a Bayes log posterior log-odds that each gene is DE (Lonnstedt and Speed, 2002; Smyth et al., 2003). The p value calculating from B data was corrected by Holm's method and the cut-off value of p was 0.01.
Alleles Characterization
[0044]The ang4-2 and ang4-3 mutants with T-DNA insertion respectively in the exon 6 and the exon 19 of ANG4 gene were studied (worldwideweb.arabidopsis.org). The T-DNA insertion was checked by PCR on F2 plants using primers designed before (P1) and after (P3) the putative position of the T-DNA and a primer specific of the left border of the T-DNA (P2). A positive amplification between P1 and P2 validates the position of the T-DNA insertion. A coincident positive or negative amplification using P1 and P3 shows that the line is respectively heterozygous or homozygous.
Over-Expression Construct and Plant Transformation
[0045]To obtain overexpression lines of ANG4 the open reading frame (including ATG and stop codon) of ANG4 (2637 bp) was amplified by Pfu polymerase and cloned into the pDONRT221 vector using the GATEWAY recombination strategy (Invitrogen) to obtain ENTRY clones. The ENTRY clone was recombined with the pK7WG2 vector (Karimi et al., 2002) to obtain a DESTINATION vector with the ORF under the control of a 35S promotor. This construct was introduced into Agrobacterium tumefaciens and subsequently Ler or ang4-1 plants were transformed with the Agrobacterium tumefaciens suspension through floral dip. The T0 seeds were grown in high density on growth medium containing Kanamycin (50 μg/ml), Nystatin (50 μg/ml) and Carbenicillin (250 μg/ml) to select the transformants. These T1 transformants were transferred to soil to obtain T2 seeds.
Example 1
Histological Analysis of the Ang4 Mutation
[0046]We performed an anatomical analysis in the first leaf by using light microscopy to identify phenotypic functions of the ANGUSTA 4 gene. Our interest was to focus on the number of palisade cells, structure of vascular tissue in the leaf as well as primary root development of mutant plants in comparison with that of wild-type.
[0047]We looked at the anatomy of angusta4 leaves to determine whether cell division or cell expansion was affected and to check polarity and studied root growth kinetic as a measure for root apical meristem activity in the mutants. In plant, cell expansion and cell division are key parameter in the determination of organ shape.
[0048]Major trait of the angusta class of mutants is narrow leaf lamina (Berna et al., 1999; FIG. 1). The reduced leaf size in ang4-1 mutant was confirmed by morphological measurements of expanded leaves. The measurements showed a significant decrease of lamina length and width, petiole length and total lamina and petiole length (FIG. 3, Panel G). The lamina area of ang4-1 first and second expanded leaves was 10.7±2.4 mm2, i.e., 55% of Ler lamina area which was 19.3±2.5 mm2 (FIG. 3, Panel H). The length/width ratio of the lamina was significantly increased in ang4-1 mutant showing a modification of the lamina proportions and to a narrower shape (FIG. 3, Panel I). The fresh weight of the rosette leaves at the flower emergence stage of development were significantly reduced in ang4 mutants: ang4-1 biomass was 40% of Ler biomass, and ang4-2 and ang4-3 fresh weight were respectively 51% and 55% compared to Col (FIG. 4B). The dry weight was also strongly affected by the mutation in ANG4 with 39% for the ang4-1 plants compared to Ler, and respectively 45% and 49% for the ang4-2 and ang4-3 plants compared to Col.
[0049]Serial sections through historesin embedded expanded first leaves (26-day-old seedlings) were taken (FIG. 3, Panel A). The number of palisade cells was counted in a number of serial sections at the widest width of the leaf to be used as a measure for lateral growth of the leaf lamina (Tsuge et al., 1996). The number of palisade cells of angusta4 was smaller than wild-type. The data showed that the number of palisade cells is 30 in the angusta4, and about 66 cells in Ler (FIG. 3, Panel B). Thus, palisade cells were reduced by about 50% in angusta4 compared to wild-type. The structure of palisade cell was larger and distributed more irregularly than in Landsberg (FIG. 3, Panels C and E).
[0050]The vascular tissue of Ler wild-type and angusta4 mutants was also visualized under the microscope. The polarity was correct in the mutant: xylem at the dorsal side and phloem at the ventral side. The midvein of wild-type and mutant are shown (FIG. 3, Panels D-F). In the angusta4 mutants, cells surrounding xylem and phloem were bigger than in Ler. The number of cells is also higher in the vascular bundle in the angusta4 midvein (FIG. 3, Panels E and F). These data show that the ANGUSTA4 gene is involved in the regulation of cell number during leaf growth; it has no function in leaf polarity.
[0051]To investigate in more detail the function of the ANGUSTA4 gene, primary root growth was analysed. Sixty seedlings of angusta4 and Ler were germinated in the square plates and kept in vertical position in the tissue culture room. The root tip was marked every two days with a scalpel blade. The mean value was calculated for each time point. A graphical representation of these mean values is shown in FIG. 5, Panel A. After 15 days, the length of angusta4 reached 1 cm, which is much shorter than the 5 cm of the Ler line. In addition, angusta4 roots started to form adventitious roots after four days germination; each angusta4 plant had two to three adventitious roots.
[0052]Moreover, apical sections from in planta Arabidopsis roots (seven-day-old seedling and n=20) were visualized under confocal microscope to investigate the structure of the root apical meristem. FIG. 5, Panels B and C showed the meristem zone of the primaty root in angusta4 and wild-type. Longitudinal section of root meristem region of angusta4 showed no difference in cell division and cell expansion. It indicates no defective root meristem activity.
[0053]The flower organization is also altered by ANG4 mutation. The floral diagrams of ang4-1 showed an asymmetric position of the petals and missing anther or carpel. The flower of ang4-2 and ang4-3 plants was not modified but the inflorescence stem appeared thinner as compared to Col. To verify if ang4 mutation only affected aerial organs, the root growth rate was analyzed and compared to Col alleles and wild-types. The root growth was strongly decreased in ang4-1 plants compared to the wild-type Ler. However, the root growth of ang4-2 and ang4-3 was similar to that of the wild-type Col suggesting that the mutation of ANG4 gene does not alter the root growth in the genetic background of Col.
[0054]Thus, the ANG4 gene has a function in leaf and flower development and root growth.
Example 2
Mapping of ANG4 Leaf Form Mutation
[0055]The mutant, ang4, was obtained from the collection of 255 mutant lines induced by EMS mutagenesis (Berna et al., 1999). The aim of this work was to verify the ANG4 region delimited by AFLP, InDel and SNPs markers and by recombinant analysis. The Ler mutant was crossed with Col-0 wild-type and the resulting F1s were allowed to self in order to produce F2 mapping populations (Robles and Micol., 2001). 320 F2 mutants together with their Ler and Col-0 parents were analyzed using a standard set of eight AFLP primer combinations shown in Table 1 in order to visualize 85 AFLP markers on the genome (Peters et al., 2004). After scoring the resulting 85 AFLP markers, linkage to chromosome 2 and non-linkage to other chromosomes was observed. Table 2 shows the genotypic scoring that was done using AFLP, InDel and SNP markers. Presence of the AFLP marker signifies that the marker behaves as the Col parent and is represented in Table 2 as number 1. For the F2 individuals this means that the marker is either homozygous or heterozygous. Absence of the AFLP marker indicates that the marker is homozygous Ler and it is indicated as number zero (0) in Table 2.
[0056]Initially, as shown in Table 2, F3 recombinants 670, 227, and 1389 were scored as homozygous mutants (100% ang4) while recombinants 635,1472, 1747 and 387, 1607,1716 were scored as heterozygote (1 ang4: 3 wild-type) and homozygote (100% Ler) respectively. During meiosis, for recombinant 1747, a cross-over event took place between markers CER458218 and CER442324. This recombinant was used to delimit ANG4 mutation from the top of chromosome 2 and hence marker CER442324 was taken as the top marker that limited the ANG4 interval. In contrast, a cross over event occurred between markers CER458218 and CER458219 for recombinant 670 and 227, markers CER442324 and CER458218 for recombinant 1389 and markers CER442323 and CER458367 for recombinants1472. All these markers delimited the ANG4 mutation from the bottom of chromosome 2. Delimiting the ANG4 region became rather difficult because the mutant was phenotypically very clear in the Ler background and less clear after crossing (i.e., it was difficult to score the phenotype in the F2 derived F3 populations).
[0057]In order to verify the ANG4 interval of 27 kb, and probably reduce this region to about 10 to 20 kb, phenotypic scores of the F3 of nine recombinants that were not very informative in the previous scoring were repeated. In vitro, thirty seeds of each recombinant were planted on GM medium in 150×25 mm Petri dishes in replicate. Two hundred seeds of each recombinant were planted in vivo on trays containing 52 wells in which one seed was planted in each well. Phenotypic scores were done at four time points over a period of 4 weeks to determine whether the F3 was homozygous mutant (100% ang4), heterozygous (1 ang4: 3 wild-type) or homozygous wild-type (100% wild-type Ler) and these scores are summarized in Table 3, and compared to the previous less extensive scoring. Recombinants 635, 670, and 1389 were scored differently compared to previous scoring.
[0058]Earlier phenotypic scores had shown that recombinant 635 was heterozygote while recombinants 670, 1385, and 1472 were homozygote mutant. Table 3, which indicates new phenotypic scoring, revised these earlier scores. Recombinant 227 was very difficult to score in the second round of phenotype scoring, as it was less clear in vitro. Scores from in vivo growth conditions indicated that it was homozygous mutant.
[0059]For instance, recombinant 670 was scored as a homozygous mutant before and from Table 3, it was scored as heterozygous (1 mutant: 3 wild-type). It was therefore decided that a number of recombinants that were not clearly scored and therefore not very informative, including recombinants 387,670,1389,1607 and 1716 would be ignored and that recombinants that were clearly scored as shown in Table 4 will be used to delimit the ANG4 mutation. As indicated in Table 4, the SNP marker that delimited ANG4 mutation from the top of chromosome 2 was CER458218 based on recombinant 227 while marker CER458367 delimited ANG4 mutation from the bottom of chromosome 2 based on recombinant 1472. These markers are within a 27 kb (26,647 mb) region. This region was the minimal region delimited by markers while the maximal ANG4 region was between CER458219 as the top marker based on recombinants 377 and 1775 and CER458367 as the bottom marker based on recombinant 1472. Recombinant lines that were most informative were those with Ler scoring because Col-0 is a recombinant inbreed line (RIL) and as such any cross over event in it does not necessarily indicate linkage to the mutation of interest as shown in Table 4 for recombinant 635.
[0060]Verification of the ANG4 mutation after the phenotypic scores showed that, indeed the ANG4 mutation was within the 27 kb region delimited by genotypic scoring as indicated in Table 2 and FIG. 6. Within this 27 kb region; there are four intact genes, one of which has to be ANG4 gene.
[0061]The ANG4 interval was determined at 27 kb and flanked by CER458218 and CER458367 markers. This was based on the recombinant analysis of 1062 F2 plants. We checked the phenotypic region of the remaining recombinants in the F3 generation both in vivo and in vitro at four time points over a period of four weeks. The ANG4 region was determined and allowed to deduce the F2 genotypes. This F2 genotypic information was integrated in Table 4 and the ANG4 interval delineated to a 27 kb region containing 4 intact genes one of which has to be ANG4 gene.
Example 3
Sequencing of Candidate ANG4 Genes
[0062]Four candidate genes are situated in the 27 kb interval delimited by the recombinant analysis and are listed in Table 5 with their respective functions. For each gene, the genomic DNA was amplified from the ang4 mutant and compared to the wild-type Ler in order to determine the single base change.
[0063]Total genomic DNA from ang4 mutant and Ler were extracted using the CTAB method and DNeasy Plant mini kit. Ler DNA acted as a control. For ang4 mutant, four candidate genes were amplified by performing three independent PCR for each of the primer combinations (Table 9). The same primer combinations were used to three independent PCR to amplify the four genes present in the genomic DNA of Ler. Primer pairs were designed for all the candidate genes that amplified overlapping segments of 800 bp-1200 bp spanning the entire 27 kb region (FIG. 6; Table 9). Three independent PCR reactions of these segments were sequenced. An example of this PCR amplification is shown in FIG. 7 where each band indicates DNA amplified with two primer sets. Sequence alignment was performed by CLUSTALW 1.8 software and compared with that of the wild-type plant Ler. An example of sequence alignment is shown in FIG. 8 with the gene At2g44950. Sequencing of these fragments and comparison with the wild-type Ler sequence identified a mis-sense change in the candidate gene At2g44950 generating a stop codon UAG instead of the CAG codon corresponding to amino acid glutamine in the predicted exon 16 (FIG. 9). Sequence alignment of other candidate genes, At2g44940, At2g44970 and At2g44980 genes did not show any mutation.
[0064]The At2g44950 gene is within the 27 kb region on chromosome 2 together with At2g44940, At2g44970 and At2g44980 genes flanked with CER458218 marker from the top of chromosome 2 and CER458367 marker from the bottom of chromosome 2 as shown in FIG. 9. Amongst these candidate genes, ANG4 is the largest covering a region of 6298 bp with an open reading frame (ORF) of 5245 bp; while the At2g44940, At2g44970 and At2g44980 genes covers 1157 bp with an ORF of 887 bp, 3337 bp with an ORF of 3020 bp and 4230 bp with the same number of base pairs as its ORF respectively. ANG4 gene has two untranslated regions, one at the 5' end covering a region of 344 bp and the other at the 3' end with 307 bp. It consists of 19 exons and 18 introns. Once the introns have been spliced, the exons form the full length cDNA that consists of 2637 bp and this is translated in a protein of 878 amino acids (worldwideweb.arabidopsis.org).
[0065]The mutation that was found in the At2g44950 gene (FIG. 8), truncated the protein from 878 to 844 amino acids. This was as a result of the stop codon, UAG that was created at position 5183 in the unspliced mRNA and at position 2134 in the spliced mRNA when cytosine nucleotide changed to a tyrosine nucleotide that was caused by an EMS mutagenesis.
[0066]The At2g44950 gene has a RING-finger motif that begins with the amino acid cystein at position 826 in the amino acid sequence and ends with amino acid cystein at position 864 (CKACNDR-PKEVVITKCYHLFCNPCVQK-LTGTRQKKCPTC) as shown in FIG. 10 (SEQ ID NO:2). Eighteen amino acids of the RING finger motif are part of the 844 amino acids that makes a protein after the mutation and 23 amino acids of the RING finger motif are lost (FIG. 9). This means that the RING finger motif that functions as part of the E3 ligase was inactivated in the ang4 mutant and that this might have lead to defect in the degradation of a number of proteins in the proteasome.
[0067]Molecular cloning of ANG4 demonstrates that map-based cloning using AFLP markers is a reliable strategy for accessing genes from the genome of Arabidopsis thaliana. Cloning of ANG4 will facilitate studies on its function for crop improvement.
Example 4
ANG4 Homologues and Functional Domain
[0068]Data base searches revealed the presence of At2g44950 homologues as uncharacterized cDNA or open reading frames obtained from genome projects in a number of organisms including Arabidopsis thaliana, humans, and rice. ANG4 has a close homologue in Arabidopsis thaliana located on chromosome 1 (At1g55250). Sequence comparison analysis indicates that NP--055586 is the human orthologue of the Arabidopsis ANG4. The human genome also contains a second ANG4 homologue, AAK58539 (RING finger protein 20), which is encoded by a gene that is distinct from the NP--055586 gene (RING finger protein 40). In Oryza sativa (japonica cultivar-group), there appears to be two ANG4 homologues with accession numbers CAD41603 and NP922769. FIG. 10 shows an alignment of the amino acids of ang4 mutant and its homologues in humans, Arabidopsis and rice which revealed a conserved Really Interesting New Gene motif (RING finger) at the end of the sequences indicating that ANG4 is an evolutionary conserved protein. The RING finger domain has been classified into 20 different subgroups in Arabidopsis thaliana (Stone et al., 2005). In this sub groups, ANG4 was classified as having an ATP binding domain. We searched for this ATP binding domain (the P-loop) using Prosite (worldwideweb.expasy.org/cgi-bin/prosite/S) and was not found though the ANG4 homology to ATPases involved in chromosome segregation and cell division was found. Search for other functional motifs was done but no other functional domain was found besides the RING finger.
Example 5
Alleles in ANG4
[0069]A number of alleles for At2g44950 gene with T-DNA insertions are available from Signal (worldwideweb.signal.salk.edu/cgi-bin/tdnaexpress?GENE=at2g44950&FUNCTION- =& TDNA=) and GABI (worldwideweb.mpiz-koeln.mpg.de/GABI-Kat/db/search.php?type=seq&term=60-K- 015154-022-276-D08-8409) collections. T-DNA insertion lines are also available for the ANG4 homologue in Arabidopsis (At1g55250) (Tables 6 A and 6B).
Example 6
ANG4 Expression Patterns in Different Arabidopsis Organs
[0070]To examine the expression pattern of ANG4 gene in Ler organs, we performed semi-quantitative reverse transcriptase (RT)-PCR analysis with different tissues including shoot apex, flower, young leaves, expanded leaves, cotyledon and roots. RNA was extracted from different frozen ground Ler organs using TRIZOL reagent (Life Technologies, Paisley, UK) according to manufacturer's protocol. The cDNA samples were standardized on actin transcript (At3g18780) amount using primers Defle 44 and Defle 45 with the following sequences: TGCTGGACGTGACCTTACTG (SEQ ID NO:5) as a forward primer and GGGCTGGAACAAGACTTCTG (SEQ ID NO:6) as a reverse primer. The melting temperature for these standard primers that acted as control in this experiment was 59° C. for both. For ANG4 gene, the following gene specific primers were used: syana--01 as a forward primer and syana--02 as a reverse primer with the following sequences: TGCTCGAATCAGATGGAAGA (SEQ ID NO:7) and AGCTAGCTGACCGCACAAAT (SEQ ID NO:8), respectively. The melting temperature for syana--01 was 59° C. while for syana--02 was 60° C. Actin is a fundamental component of the cytoskeleton in all eukaryotes and directs the spatial organization of many crucial sub cellular processes. Hightower and Meagher (1986) proposed that the six subclasses of actin have been conserved during vascular plant evolution and hence it can be used as a reference for expression analysis of other plant genes. FIG. 11 shows the result of a typical RT-PCR analysis of the expression pattern of ANG4 in different Ler organs. Primers Defle 44 and Defle 45 amplified a single 253 bp actin PCR product while primers syana--01 and syana--02 amplified a predicted single 164 bp ANG4 PCR product. This analysis shows that the ANG4 gene is expressed in all organs studied.
[0071]The expression pattern of ANG4 gene in all Ler organs studied could indicate that it may play a basic role in all these organs. The understanding of whether ANG4 gene may be involved in other possible roles, it would be important to investigate its expression levels in response to hormone and stress treatment. In addition, the expression analysis at the cellular level will be analyzed using the GFP marker line. Expression of At2g44950 gene in all organs means that it is required for fundamental or basic processes in all plant organs and throughout the life cycle. Cellular experimental analysis would also indicate whether ANG4 gene function is related to cell division processes.
Example 7
Genome-Wide Expression in ang4 Shoot Apex
[0072]A total of 1821 genes were differentially expressed (DE) in the apex of young plants of ang4 compared to Ler, that represents 8% of the Arabidopsis genome. Comparing these results with those obtained in other narrow leaf mutants (elo2 and drl1-2 involved in independent process to ang4), 1314 genes appeared differentially expressed specifically in ang4 (FIG. 12). Considering the level of expression, 494 genes are DE at a two-fold change expression threshold. The number of genes regulated by ANG4 is higher than those regulated by the DRL1 and ELP1 genes, respectively mutated in drl1-2 and elo2 mutants, showing the general function of ANG4 in the development.
[0073]Most of the genes regulated by ANG4 are involved in cytokinesis and cell cycle. A partial list of the DE genes in ang4 shows that 24 cell cycle genes and 27 microtubule and myosin related genes, are regulated in ang4 mutant (Table 7). Among these, one finds eight genes related to E2F-DP complex regulating the G1 to S transition in plants (De Veylder et al., 2003). Eight A- and B-type cyclins genes and three B-type cyclin-dependent kinase genes involved in G2 to M transition in cell cycle are down-regulated in ang4 genotype. Kinesins represent a super-family of microtubule motor proteins involved in the transport of vesicles and organelles, spindle formation and elongation, chromosome segregation, microtubule dynamics and morphogenesis (Reddy and Day, 2001). Among the 61 kinesin genes identified in Arabidopsis genome, 19 are down-regulated in ang4, that TETRASPORE involved in the formation of tetrad of microspores after meiosis (Yang et al., 2003). The HINKEL gene, another kinesin, plays a role in the reorganization of phragmoplast microtubules during cell plate formation (Strompen et al., 2002). Other cytokinesis related genes are also DE in ang4, as the cytoskeletal components actin 8, tubulins, myosin like proteins and microtubule-associated proteins. The PLEIADE gene that has a function in the stabilization of cytokinetic structures of cell plate during cytokinesis is also down-regulated in ang4 mutant (Muller et al., 2002). The KNOLLE gene, a cell-cycle-regulated syntaxin involved in membrane fusion in cytokinesis, is also repressed in ang4 (Muller et al., 2003). The SIAMESE gene, required for coordinating cell division and cell differentiation during the development of trichomes and may function as a repressor of mitosis in the endoreduplication cell cycle, is up-regulated in ang4. These results suggest an implication of ANG4 gene in cell cycle regulation.
[0074]Some genes related to plant development are also regulated by ANG4 gene expression (Table 8). The GLABRA1 gene is a MYB transcription factor that specify the primary cell fate during development of epidermal hairs in Arabidopsis (Schiefelbein, 2003). The homeobox genes KNAT2 and KNAT6 have a role in meristem initiation and maintenance (Tsiantis and Hay, 2003). The genes NAM and AINTEGUMENTA are known to be involved in organ initiation and separation (Traas and Vernoux, 2002). In Arabidopsis, SCARECROW (SCR) is essential for the asymmetric division of the cortex/endodermis progenitor cell in the root (Kamiya et al., 2003). Two genes related to auxins are DE in ang4: a putative ARF1 auxin responsive transcription factor and a putative AUX1-like permease, a regulator of root gravitropism (Liscum and Reed, 2002).
Example 8
Effect of ang4 Mutation on Endoreduplication and Cell Expansion
[0075]The effect of the ang4 mutation on leaf development and cell cycle duration was analyzed by a kinematic analysis on the first leaf pair of in vitro grown plants. Leaf blade area was similar in Ler and the ang4-1 mutant at the earliest observations. However, the increase in leaf area was slower in ang4-1 compared to Ler between 5 and 8 DAS (FIG. 13, Panel A). At maturity, the leaf blade area of ang4-1 was about 47% of those of Ler, with respectively 11 and 24 mm2. During the same period, the number of cells per leaf also increased quicker in Ler than in ang4-1 (FIG. 13, Panel B). So, at maturity (after 18 DAS), the ang4-1 leaves contained only 48% the number of epidermal cells of Ler. Differences in the rate of increase of leaf area and cell number must be due to effects on cell expansion and division respectively. Indeed, between 5 and 10 DAS, the cell division rate and the relative leaf expansion rate (RLE) were lower in ang4-1 compared to Ler, but they decreased more slowly in the mutant (FIG. 13, Panels C and D). Consequently, the cell division rate and the RLE rate became similar in ang4-1 and Ler from the 10 DAS and along the expansion phase until no cell was dividing anymore at the 15 DAS. The expansion continued in both Ler and ang4-1 until the 18 DAS when the leaf reached the maturity. So, the ANG4 mutation alters the cell division and the leaf expansion only during the early stage of leaf development.
[0076]At this stage, the cells of ang4-1 were bigger than in Ler with an average cell area respectively of 82 μm2 and 54 μm2 at day 5 (FIG. 13, Panel E). After 7 DAS, no difference of the cell area could be observed at the later stages between ang4-1 and Ler showing that the balance between division and expansion rates is the same in ang4-1 and in Ler.
[0077]Because the final divisions give rise to stomata, the stomata index (SI) indicates the exit from cell cycle and the end of proliferation activity, which starts from the tip to the base of the leaf in Arabidopsis (De Veylder, 2001). The SI also increased slower in ang4-1 compared to Ler between 5 and 8 DAS, resulting in the final SI in mature leaves being lower with 0.23 in average for ang4-1 and 0.35 for Ler (FIG. 13, Panel F). These data validate the previous data showing that the ANG4 mutation decreases the cell division activity at the early stage of leaf growth without modifying the duration of the proliferation and expansion phases. At 5 DAS, the average cell cycle duration, which is the inverse of cell division rate, was almost 50% longer in ang4-1 (20.6 hours) than in Ler (14.1 hours), and it was longer until 11 DAS where the cell cycle duration was the same in both genotypes (respectively 48.4 hours and 50.7 hours for Ler and ang4-1).
[0078]To investigate deeper the effect of ang4 mutation on cell cycle progression, we analyzed wild-type and mutant leaves by means of flow cytometry. The ploidy level of the first leaf pair was determined throughout the development of wild-type and mutant leaves to reveal the changes in relative duration of G1/G2 phase during mitotic cell division and timing and amount of endoreduplication in the ang4-1 mutant.
[0079]At 8 DAS when leaves could first be harvested, a shift is seen in the G1-to-G2 populations in the ang4-1 mutant compared to Ler (FIG. 14). In the mutant, the population of cells in 4C is similar to that in 2C (ang4-1; 2C=46.2%, 4C=44.0%), while in wild-type, the number of cells in 4C is only half of those in 2C (Ler; 2C=66.2%, 4C=33.8%), suggesting that increased cell cycle duration is associated with a block at the G2-to-M transition point of the cell cycle in ang4-1. The exit from mitosis coincided with the start of the endocycles and could be seen by the increase of 4C content and the appearance of higher ploidy levels (8C, 16C). Cell cycle activity ended as evidenced by a stable DNA distribution around 18 DAS, coinciding with the end of growth. The endocycle was enhanced in the ang4-1 mutant from earliest stage with already 10% of the cells in 8C at 8 DAS, while this level of 8C was only reached at 13 DAS in Ler. The consequence was a higher ploidy levels in ang4-1. In the mature leaves, more than 4% of the cells contained a ploidy level of 32C in the ang4-1 mutant, while the ploidy level in mature Ler leaves only reached 16C. The exit from the endocycle occurred at the same date for both ang4-1 and Ler, at 18 DAS. So, when ANG4 is mutated, cells arrest in the G2/M phase of the cell cycle and proceed into endocycles instead. We postulate that the ANG4 protein has a function in the degradation of a cell cycle regulator(s) working at the G2-M transition of the cell cycle during early organ growth.
[0080]To confirm that these effects were not specific for the leaves, flow cytometry was done on roots, hypocotyls and first leaves at one time point in development (12 DAS). The ploidy levels obtained for the root and hypocotyls were comparable to those of the first leaves, indicating that ANG4 affects the cell cycle throughout plant development.
[0081]The flow cytometry profile of the ang1 allele, GABI--634H04, differs from that of the Col control and is similar but weaker to that of ang4: more endopolyploidy (presence of 32C), slight shift in the G1-to-G2 cell populations (reduced 2C cell number and increased 4C cell number). The mutational analysis of the ang1 allele indicates that ANGL (At1g55250) is also functional and might have functional redundancy with the ANG4 gene (At2g44950).
Example 9
ANG4 Overexpression Increases Leaf Size
[0082]Photographical observations of ANG4 overexpression plants (T1) clearly indicate that the plants have improved growth performance compared to wild-type plants. For example, the rosette leaf size of the overexpression plants are considerably increased as can be seen in FIG. 15.
TABLES
TABLE-US-00001 [0083]TABLE 1 Standard set of eight AFLP primer combinations used to detect linkage between 85 Col/Ler AFLP markers and ANG4 locus. Table obtained from Peters et al., 2004. Selective Selective Selective Selective Marker nucleotide nucleotide nucleotide nucleotide Number of code SacI + 1 SacI + 2 MseI + 1 MseI + 2 AFLP markers SM8 A A C T 9 SM57 A T G A 13 SM61 A T T A 9 SM205 T A T A 12 SM229 T G C A 14 SM233 T G G A 10 SM236 T G G T 10 SM240 T G T T 8
TABLE-US-00002 TABLE 2 Genotypic scores of nine recombinants using AFLP, InDel and SNP markers and L - indicates co-dominant marker, 1 - dominant marker, 0 -3 No marker MARKER POSITION ON PARENTS F3 RECOMBINANTS NAME CONTINUOUS SEQ COL-0 Ler 227 387 635 670 1389 1472 1607 1716 1747 CER458222 18657105 C L H C C H H H H C L CER442328 18519888 C L H C C H H CER458219 18528422 C L H C H H H H C C L CER458218 18539154 C L L C H H H C C L CER442324 18549847 C L L C H L L H C ? H CER442323 18559130 C L L C H L L H C C H CER458367 18565840 C L L C H L L L C C H CER442612 C L L C H L L L C C/H H SM33_202.4 18745473 1 0 0 1 1 0 0 CER458362 18753831 C L L H H L L L C H H H--heterozygous. Numbers in top row indicate the F3 individual recombinants. Recombinants indicated in blue were scored as ang4 mutants, green as wild-type and turquoise as heterozygote. indicates data missing or illegible when filed
TABLE-US-00003 TABLE 3 Phenotypic scores of nine recombinants: The scores were done at four time points over a period of four weeks both in soil and in vitro. In both growth conditions the scores were the same. Heterozygous indicates that the wild-type and the mutants were observed while homozygous mutant implies only mutants were observed. Homozygous wild-type indicates no mutant was observed in those recombinants. F3 PROGENY ang4 NO. OF PREVIOUS RECOMBINANT WILD- NO. OF NEW PHENOTYPIC PHENOTYPIC LINE TYPE MUTANTS INTERPRETATION INTERPRETATION 227 0 66 Homozygous mutant Homozygous mutant 387 53 0 Homozygous wild-type Homozygous wild-type 635 52 0 Homozygous wild-type Heterozygous 670 118 25 Heterozygous Homozygous mutant 1389 28 5 Heterozygous Homozygous wild-type 1472 170 50 Heterozygous Heterozygous 1607 53 0 Homozygous wild-type Homozygous wild-type 1716 39 0 Homozygous wild-type Homozygous wild-type 1747 63 18 Heterozygous Heterozygous
TABLE-US-00004 TABLE 4 Recombinant used to delimit the ang4 mutation. Marker Position on F3 RECOMBINANTS Name Continuous Seq. Col Ler 227 377 635 1472 1607 1775 1747 CER458222 18657105 C L H L C H H H L CER442328 C L H L C CER458219 18670459 C L H H H H C H L CER458218 18681209 C L L H H H C C L CER442324 18691819 C L L H H H C ? ? CER442323 C L L H H H C C/H H CER458367 18707856 C L L H H L C C H CER442612 C L L H H L C C/H H SM33_202,4 18745473 1 0 0 1 1 CER458362 18753831 C L L H H L C C H Only seven recombinants shown on the top row of the Table from recombinant 227 through recombinant 1747 were selected because they were the most informative recombinants while the other recombinants were ignored. The SNP markers and their position on continuous sequence are indicated indicates heterozygozity after a cross over event during meiosis. L--Ler, and C--Col ecotypes. The (?) in the Table means that the scoring of the recombinants was not clear.
TABLE-US-00005 TABLE 5 ANG4 candidate genes: The four candidate genes in the 27 kb region and their functions based on TAIR annotation. GENE CODE FUNCTION At2g44940 Involved in DNA binding and transcription regulation by its AP2 domain At2g44950 It is a C3HC4 type zinc finger protein involved in zinc ion binding and as a E3ligase At2g44970 It is an expressed protein playing a role in lipid metabolism and catalytic activity At2g44980 Putative SNF2 transcription regulatory protein involved in ATP, DNA binding (Helicase activity) At--Arabidopsis thaliana, g--genomic.
TABLE-US-00006 TABLE 6 INSERTION SALK LINES GABI LINES SITE A - At2g44950 POSITION IN CHROMOSOME 2 SALK_122512 EXON 1 18549684 SALK_044415 INTRON 3 18550469 GABI_306H08 INTRON 2 18550191 GABI_276D08 INTRON 13 18553269 B - At1g55250 POSITION ON CHROMOSOME 1 SALK_071289 EXON 17 20615235 SALK_141948 EXON 19 20615962 GABI_634H04 EXON 13 20614348 GABI_529603 INTRON19 20616201 Alleles for At2g44950 and At1g55250 genes: A, ANG4 alleles. Two SALK lines, SALK_122512 and SALK_044415 from SIGnAL collections and two GABI line, GABI_276D08, and GABI_306H08. B, alleles for the ANG4 homologues in Arabidopsis (At1g55250); SALK_071289 and SAKL_141948 from SIGnAL collections and GABI_634H04 and GABI_529603 from GABI collections.
TABLE-US-00007 TABLE 7 Differentially expressed genes in ang4 mutant compared to Ler and related to cell cycle and cytokinesis. Data were performed on microarrays ATH1 experiment with RNA from shoot apex of young plants grown in in vitro conditions. The p values are calculated according a Bayesian test of linear model and corrected by Holm's method. probes name Sequence P. value expression fold change derived from gene descriptions process related 1.29E-05 248413_at 0.21 At5g51600 PLEIADE gene cytokinesis 5.78E-06 258098_at 0.23 At3g23670 hypothetical protein similar to kinesin like protein cytokinesis 6.79E-06 248057_at 0.26 At5g55520 putative myosin heavy chain protein cytokinesis 2.18E-06 261660_at 0.26 At1g18370 HINKEL, kinesin heavy chain isolog cytokinesis 3.23E-06 264802_at 0.26 At1g08560 KNOLLE, putative syntaxin-related protein cytokinesis 5.42E-07 261159_s_at 0.28 At1g34460 putative cyclin cell cycle 2.89E-05 252691_at 0.30 At3g44050 kinesin-like protein KLP2 protein cytokinesis 1.31E-04 257115_at 0.30 At3g20150 kinesin-like protein cytokinesis 1.74E-06 247039_at 0.31 At5g67270 putative microtubule-associated protein cytokinesis 2.97E-05 245607_at 0.32 At4g14330 kinesin like protein cytokinesis 3.40E-06 255265_at 0.32 At4g05190 kinesin like protein A cytokinesis 9.40E-05 253978_at 0.32 At4g26660 putative kinesin cytokinesis 5.48E-05 263441_at 0.33 At2g28620 putative kinesin-like spindle protein cytokinesis 1.12E-04 266009_at 0.33 At2g37420 putative kinesin heavy chain cytokinesis 4.74E-05 261780_at 0.36 At1g76310 CYCB2_4 (cyclin) cell cycle 2.53E-05 259151_at 0.36 At3g10310 kinesin-like protein similar to carboxy-terminal kinesin 2 cytokinesis 8.84E-06 265349_at 0.36 At2g22610 putative kinesin heavy chain cytokinesis 5.35E-06 257008_at 0.38 At3q14210 Myrosinase-associated protein cell cycle 5.45E-04 253148_at 0.38 At4g35620 CYCB2_2 (cyclin) cell cycle 2.41E-05 267618_at 0.38 At2g26760 CYCB1_4 (cyclin) cell cycle 3.51E-05 254400_at 0.40 At4g21270 kinesin-related protein katA cytokinesis 6.86E-05 245739_at 0.40 At1g44110 CYCA1_1 (cyclin) cell cycle 1.80E-06 258573_at 0.41 At3g04260 BC010 (E2Fb binding protein) cell cycle 2.96E-05 259851_at 0.42 At1g72250 putative kinesin cytokinesis 1.53E-06 259978_at 0.43 At1g76540 CDKB2_1 (Cyclin dependent kinase) cell cycle 2.71E-04 266401_s_at 0.43 At2g38620 CDKB1_2 (Cyclin-dependent kinase) cell cycle 3.12E-04 262802_at 0.45 At1g20930 CDKB2_2 (Cyclin-dependent kinase) cell cycle 2.30E-04 257267_at 0.46 At3g15030 TCP family (E2Fa-DPa induced Transcription factor) cell cycle 1.69E-05 257524_at 0.46 At3g01330 DEL3 (E2F-DP-like protein) cell cycle 3.53E-06 248150_at 0.46 At5g54670 kinesin-like protein cytokinesis 3.35E-04 262081_at 0.47 At1g59540 kinesin motor protein (kin2) cytokinesis 6.18E-05 245259_at 0.47 At4g14150 kinesin like protein cytokinesis 2.72E-04 261605_at 0.48 At1g49580 CDPK-related protein kinase cell cycle 1.59E-05 260329_at 0.49 At1g80370 CYCA2_4 (cyclin) cell cycle 5.13E-05 263017_at 0.49 At2g17620 CYCB2_1 (cyclin) cell cycle 4.54E-04 262752_at 0.49 At1g16330 CYCB3_1 (cyclin) cell cycle 2.60E-05 266295_at 0.49 At2g29550 tubulin beta-7 chain cytokinesis 3.78E-04 261765_at 0.51 At1g15570 CYCA2_3 (cyclin) cell cycle 4.48E-04 252736_at 0.52 At3g43210 TETRASPORE (TES), kinesin-like protein ZCF125 cytokinesis 7.37E-04 262494_at 0.54 At1g21810 myosin-like protein cytokinesis 8.43E-04 265464_at 0.54 At2g37080 putative myosin heavy chain cytokinesis 7.93E-04 250386_at 0.55 At5g11510 MYB3R4 (transcription factor) cell cycle 1.06E-03 264061_at 0.55 At2g27970 CKS2 (CDK binding protein) cell cycle 1.06E-03 246683_at 0.56 At5g33300 putative protein chromokinesin KIF4 cytokinesis 8.76E-03 250685_at 0.56 At5g06670 kinesin heavy chain-like protein cytokinesis 8.23E-04 261639_at 0.57 At1g50010 putative tubulin alpha-2/alpha-4 chain cytokinesis 3.16E-03 245576_at 0.57 At4g14770 CPP1-related transcription factor family (E2Fa-DPa cell cycle induced TF). 4.07E-03 249095_at 1.44 At5g43900 myosin heavy chain MYA2 cytokinesis 1.08E-03 251052_at 1.54 At5g02470 DPA transcription factor cell cycle 1.45E-04 250923_at 1.69 At5g03455 GTPV2 (putative CDC25 homolog) cell cycle 4.59E-04 260765_at 1.79 At1g49240 actin 8 cytokinesis 2.13E-04 250844_at 1.97 At5g04470 SIAMESE gene (SIM) cell cycle 4.89E-03 264006_at 2.02 At2g22430 homeodomain (ATHB-6) (E2Fa-DPa induced TF) cell cycle 1.93E-06 253217_at 2.58 At4g34970 actin depolymerizing factor-like protein cytokinesis 1.17E-03 250666_at 3.25 At5g07100 WRKY family (E2Fa-DPa induced Transcription factor) cell cycle 1.65E-09 253890_s_at 3.84 At5g54100 Putative protein contains similarity to stomatin like cell cycle protein
TABLE-US-00008 TABLE 8 Differentially expressed genes in ang4 mutant compared to Ler and related to plant development. Data were performed on microarrays ATH1 experiment with RNA from shoot apex of young plants grown in in vitro conditions. The p values are calculated according a Bayesian test of linear model and corrected by Holm's method. fold change Sequence P. value probes name expression derived from gene descriptions 3.28E-06 259686_at 0.26 At1g63100 transcription factor SCARECROW development 2.15E-03 257221_at 0.46 At3g27920 GLABRA1 (GL1), MYB family transcription factor development 1.26E-05 265454_at 0.50 At2g46530 putative ARF1 family auxin responsive transcription development factor 1.86E-03 263013_at 1.92 At1g23380 KNAT6, knotted-like homeobox protein development 9.50E-05 260334_at 1.96 At1g70510 KNAT2, homeotic protein (ATK1) development 7.91E-04 263194_at 2.58 At1g36060 AP2 domain transcription factor development 5.10E-07 259680_at 2.86 At1g77690 putative AUX1-like permease development 3.21E-08 265813_at 3.47 At2g18060 putative NAM (no apical meristem)-like protein development 3.23E-10 245173_at 8.79 At2g47520 putative AP2 domain transcription factor, development aintegumenta-like protein
TABLE-US-00009 TABLE 9 Primers used for ANG4 candidate genes amplification and sequencing GENE PRIMER MIPS FRAGMENT NAME DIRECTION POSITION PRIMER SEQUENCE SIZE At2g44940 F 67 TGTT AAGAGGTGAC GCACATG 943 (SEQ ID NO: 9) R 1010 CGGCGGCTTGAATGTCTTTA (SEQ ID NO: 10) At2g44940 F 800 T AGAGGAGTGA GGATGAGGAG TT 723 (SEQ ID NO: 11) R 1523 GCTAGGAAAAAAAAAAGAAATTGT (SEQ ID NO: 12) At2g44960 F 166 TTTTT GTTTCTGTGAGTGCTGTG 1119 (SEQ ID NO: 13) R 1285 TCTCAGTAGCACCAGTTTCAAG (SEQ ID NO: 14) At2g44960 F 625 CTTCT TCATCTCCCC CTTGTGC 1327 (SEQ ID NO: 15) R 1952 ATAAATACACAGGCGTGGAATTGG (SEQ ID NO: 16) At2g44970 F 86 TATGT GTCGCCCGTC TTCTTTCTT 854 (SEQ ID NO: 17) R 940 ATGCCAATGAACAACAAGTAAAGA (SEQ ID NO: 18) At2g44970 F 800 TTCCTAATGT TGTTTGCCGTTTCA 827 (SEQ ID NO: 19) R 1627 CATGGGGGTGGAAATAGTATCCT (SEQ ID NO: 20) At2g44970 F 1468 TAC TCAGTATGCA ATTCCACGTT CATAT 1114 (SEQ ID NO: 21) R 2582 TCTCTCTCGCATTTTTCTCAACCG (SEQ ID NO: 22) At2g44970 F 2443 AACGAAAT TCTCAAAGATGGGTTT 1238 (SEQ ID NO: 23) R 3681 AACGAAATTCTCAAAGATGGGTTT (SEQ ID NO: 24) At2g44980 F 73 CGTCACAC CATCCACACC ACTTG 1120 (SEQ ID NO: 25) R 1193 GACGGCAATACTTATCGCCAACATAT (SEQ ID NO: 26) At2g44980 F 927 AATT GGACCAGATG GGATTGGGAA AG 1032 (SEQ ID NO: 27) R 1959 TCCACACAAAAATGTCAGAGTGCTTAGC (SEQ ID NO: 28) At2g44980 F 1799 CTTGATTACTGG CACACCTATC CA 943 (SEQ ID NO: 29) R 2742 GGGAAAAGAGGAGGACACGATG (SEQ ID NO: 30) At2g44980 F 2480 T GTTTATCTCC CTATCTATTT CCTTG 927 (SEQ ID NO: 31) R 3404 CTTTCTCTCTGCCCTCCTCAA (SEQ ID NO: 32) At2g44980 F 3122 TGGAATACA TCGGCATAGA GAAAG 1053 (SEQ ID NO: 33) R 4175 TAAACTCGGATGCTCGGTGATAAG (SEQ ID NO: 34) At2g44980 F 3836 CCAGGAAAAAGGCA GAAGAGAAGA 980 (SEQ ID NO: 35) R 4816 CATTGTGTGATTCAGGGAGATCGA (SEQ ID NO: 36) At2g44950 F 11 GGGCGTTTTTCCCAGTGTTG 1014 (SEQ ID NO: 37) R 1005 TCAGCCCGCAGAGAATGAAT (SEQ ID NO: 38) At2g44950 F 845 TCCCACCCACACCTGTTTCA 1182 (SEQ ID NO: 39) R 2007 TTCCGCAGCAGCCAACATTT (SEQ ID NO: 40) At2g44950 F 1886 GAAGCCAAGGAACAGGAGTA 947 (SEQ ID NO: 41) R 2813 CATACGGGCACACACAGATA (SEQ ID NO: 42) At2g44950 F 2652 CTCGCCCATTGTTGTTTCAG 1241 (SEQ ID NO: 43) R 3873 AATTGCGGAAACCATGTTCC (SEQ ID NO: 44) At2g44950 F 3065 TGGGGCATTAGAACTGGAAC 1010 (SEQ ID NO: 45) R 4055 TCCCAAGGATCGAAGTCTTT (SEQ ID NO: 46)
REFERENCES
[0084]Anami S. (2004). Cloning and functional analysis of genes controlling organ growth and development in Arabidopsis thaliana. Masters thesis for International Post-graduate course on Molecular Biology (VUB, Brussels). [0085]Autran D., C. Jonak, K. Belcram, G. T. S. Beemster, J. Kronenberger, O. Grandjean, D. Inze, and J. Traas (2002). Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. The EMBO Journal 21:6036-6049. [0086]Berna G., P. Robles, and J. L. Micol (1999). A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152:729-742. [0087]De Veylder L., J. Joubes, and D. Inze (2003). Plant cell cycle transitions. Curr. Opin. Plant Biol. 6:536-543. [0088]Cnops G., S. Jover-Gil, J. Peters, P. Neyt, S. De Block, P. Robles, M. Ponce, T. Gerats, J. Micol, and M. Van Lijsebettens (2004). The rotunda2 mutants identify a role for the LEUNIG gene in vegetative leaf morphogenesis. Journal of Experimental Botany 55:1529-1539. [0089]De Veylder L., T. Beeckman, G. T. S. Beemster, L. Krols, F. Terras, I. Landrieu, E. Van Der Schueren, S. Maes, M. Naudts, and D. Inze (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13:1653-1667. [0090]Erickson R. O. (1976). Modeling of plant growth. Annu. Rev. Plant Physiol. 27:407-434. [0091]Galbraight D. W., K. R. Harkins, and S. Knapp (1991). Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol. 96:985-989. [0092]Hightower R. C. and R. B. Meagher (1986). The molecular evolution of actin. Genetics 114:315-332. [0093]Kamiya N., J. Itoh, A. Morikami, Y. Nagato, and M. Matsuoka (2003). The SCARECROW gene's role in asymmetric cell divisions in rice plants. Plant J. 36:45-54. [0094]Karimi M., D. Inze, and A. Depicker (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Sciences 7:193-195. [0095]Liscum E, and J. W. Reed (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49:387-400. [0096]Lonnstedt I. and T. Speed (2002). Replicated microarray data. Statistica Sinica 12:31-46. [0097]Muller S., E. Fuchs, M. Ovecka, J. Wysocka-Diller, P. N. Benfey, and M. T. Hauser (2002). Two new loci, PLEIADE and HYADE, implicate organ-specific regulation of cytokinesis in Arabidopsis. Plant Physiol. 130:312-324. [0098]Muller I., W. Wagner, A. Volker, S. Schellmann, P. Nacry, F. Kuttner, Z. Schwarz-Sommer, U. Mayer, and G. Jurgens (2003). Syntaxin specificity of cytokinesis in Arabidopsis. Nat. Cell. Biol. 5:531-534. [0099]Nelissen H., J. H. Clarke, M. De Block, S. De Block, R. Vanderhaeghen, R. E. Zielinski, T. Dyer, S. Lust, D. Inze, and M. Van Lijsebettens (2003). DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. The Plant Cell 15:639-654. [0100]Peters J. L., G. Cnops, P. Neyt, J. Zethof, K. Cornelis, M. Van Lijsebettens, and T. Gerats (2004). An AFLP-based genome-wide mapping strategy: a practical approach to positional cloning. Theoretical and Applied Genetics 108:321-327. [0101]Pyke K. A., J. L. Marrison, and R. M. Leech (1991). Temporal and spatial development of the cells of the expanding first leaf of Arabidopsis thaliana (L.) Heynh. Journal of Experimental Botany 42:1407-1416. [0102]Reddy A. S., and I. S. Day (2001). Kinesins in the Arabidopsis genome: a comparative analysis among eukaryotes. BMC Genomics. 2, Epub 25. [0103]Robles P. and J. L. Micol (2001). Genome-wide linkage analysis of Arabidopsis genes required for leaf development. Mol. Genet. Genomics 266:12-19. [0104]Schiefelbein J. (2003). Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 6:74-78. [0105]Smyth G. K., Y. H. Yang, and T. Speed (2003). Statistical issues in cDNA microarray data analysis. Meth. Mol. Biol. 224:111-136. [0106]Stone S., H. Hauksdottir, J. Herschleb, E. Kraft, and J. Callis (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137:13-30. [0107]Strompen G., F. El Kasmi, S. Richter, W. Lukowitz, F. F. Assaad, G. Jurgens, and U. Mayer (2002). The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr Biol. 12:153-158. [0108]Traas J., and T. Vernoux (2002). The shoot apical meristem: the dynamics of a stable structure. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 357:737-747. [0109]Tsiantis M., and A. Hay (2003). Comparative plant development: the time of the leaf? Nat. Rev. Genet. 4:169-180. [0110]Tsuge T., H. Tsukaya, and H. Uchimiya (1996). Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122:1589-1600. [0111]Van Lijsebettens M., and J. Clarke (1998). Leaf development in Arabidopsis. Plant Physiology and Biochemistry 36:47-60. [0112]Wu Z., and K. A. Irizarry (2004). Preprocessing of oligonucleotide array data. Nat. Biotechnol. 22:656-658. [0113]Yang C. Y., M. Spielman, J. P. Coles, Y. Li, S. Ghelani, V. Bourdon, R. C. Brown, B. E. Lemmon, R. J. Scott, and H. G. Dickinson (2003). TETRASPORE encodes a kinesin required for male meiotic cytokinesis in Arabidopsis. Plant J. 34:229-240.
Sequence CWU
1
5413289DNAArabidopsis thalianamisc_featuregenbank NM_130060 1tcactcttgt
ttttgtttct gtgagtgctg tgtgtgttct cggtttgtaa tatctaagtc 60gcacaaagat
cagacaaagt ctgtttgcgt ttgatagcat tagagtctga tcgaatttgg 120aagacctgtg
agattagggt ttctttttgt ttgttttttt ttattccgaa ggagagctgc 180ggagcaattg
gaattacctc ggttaaagca atcacgtagc tttggatcgt ttagggtttt 240ctcattgggt
ttgggggatt tgtattcgag cttcctcaaa ctcaatctga tggtgatgtg 300agacaaggct
taatcctgtt aattggcgat tctagggttt ttcaatggcg agcacaggcg 360agcctgaccg
taaaaggcgt cactttagct ccatatcacc ttctgaagct gcagccgccg 420taaagaaaca
gcctttcttt tggccctcct ccgaggacaa gcttgatact gcagttcttc 480agttccaaaa
tcttaagcta tcacaaaagc tagaggctca gcaggttgag tgttctattc 540ttgaggataa
actctctcag atcaaggaaa aacaattacc atacaactcc agtttgaaga 600ctgtccataa
gtcttgggaa aagcttacag cttcagtgga atcatgctct gttcgtgtga 660gtgattcaag
cagcggagct cataggtttg taaacaagga ggatgggtct tctccagccg 720tgaaaaacga
tttcatcaac cggctacttg aaactggtgc tactgagagc tcctcatcca 780atatctgctc
gaatcagatg gaagaaaatg gagtgaatac gtcaagccag atgacgcaaa 840ccttgtataa
tctagtagcc gcgacagagg atttgaggtg tctgaaggat gaattatatc 900ccacagttct
cagaaccaat cttggtaaag atttgtgcgg tcagctagct ctgagtgagt 960tggaatcaga
aattaaaagt ttcagagggg atctagatga tgtacttgtg aagttcaaat 1020cactttctag
agaattgcag agtcatcgcg atgctgatgc taaagttaga gtagacctca 1080aacgaataag
aggggagcta gaggatgagg ttgtggagct tcagcagtgt aatggtgact 1140tgtcagcatt
gagagcagaa agggatgcaa cagctggggc gtttttccca gtgttgagtc 1200ttggaaataa
gcttgctacc agtgatcggg agagggataa acaaagggat ctgcaagaca 1260tggaaacagt
tctgaaagag ttaacggtcc tggcttcagg caggctacaa cagctaaaaa 1320atcttcatga
ggagaggaca aagatgcttg gaaaaatgag taatttacag aacaagtcaa 1380agtctgtgag
gtgcatctca tcttctcaag cctgcctttc tttgaaagac cagctagaaa 1440aatccaaaga
agcagttttc cagtatatgg ctttacttga gaaactgcag gttgaaaaag 1500atagtatagt
ctggaaggaa agggagataa atataaaaaa tgaactaggt gatgtttctc 1560gaaagacgtc
tgctgttact gattctagaa tggcttcttt ggattcggag atacagaaac 1620aactggatga
aaaaatgcga atcaagacta ggctgggaaa tatatcaaga gagcgaggta 1680gaaaagaaat
ctttgcagat atgaaggcat taatttcttc gttccccgag gaaatgagtt 1740ccatgcgtag
tcaattaaac aattataaag agactgctgg aggtattcat tctctgcggg 1800ctgatgtcca
gtccctctct ggggttctat gtaggaagac aaaagagtat gaagcattgc 1860aattgagatc
agctgattat gcttctcagt taggtgacct gaatgctacg gtttgtgatt 1920tgaagaacag
tcatgaggag ttaaagttgt ttctggacat gtataaacgt gagtccactg 1980atgcgaggga
catagctgaa gccaaggaac aggagtacag ggcttgggct catgttcaga 2040gtttgaaatc
atcccttgat gagcaaaatc tggagttgcg cgttaaggca gcaaatgaag 2100ctgaagccgt
ttcccaacaa atgttggctg ctgcggaagc agagattgct gatttaaggc 2160agaaaatgga
tgattgtaaa agggatgtcg ccaagcattc tgatatcttg aaatctaaac 2220atgaagaaca
tggaacatat ctttctgaaa tacagacaat tggaagtgcc tatgaggaca 2280ttgtaccgca
aaaccaacag cttttgcttc aagttacaga gagggatgac tataacatca 2340agcttttctt
ggaaggcata acttctaggc agatgcaaga tactctgctt atcgataaat 2400acatcatgga
taaggatata cagcaaggca gtgcatatgc cagtttccta tccaagaaat 2460catcaagaat
tgaagatcag ttgaggttct gcacagatca gtttcagaaa ctagcggaag 2520ataaatatca
aaagtctgtt tctcttgaaa atctgcaaaa gaaacgtgca gacatcggga 2580atggcttgga
acaagctagg tcaaggctgg aggagtccca ttctaaagtt gagcaaagtc 2640ggctggatta
tggggcatta gaactggaac tggagattga aaggttcaat aggagaagga 2700tagaggagga
aatggaaata gccaaaaaga aagtttctcg tcttcggtct ctcatagaag 2760gatcatcggc
cattcaaaag ctccgacaag aactcagtga atttaaagaa attctgaagt 2820gtaaggcctg
caacgatcgc ccaaaagagg tggtgattac gaagtgctac catttgttct 2880gcaacccatg
tgtgcaaaag ctcacaggaa ctcgacaaaa gaagtgtcca acatgctcag 2940caagttttgg
accaaatgat attaaaccta tctacatatg accgcaccaa aacactctga 3000gcatgatgat
gtatgatgaa actgtgaaac acacacaggt actttttctc atataggata 3060aacattagat
ctctctgtaa ttattactct cttttattgg gaaaggtcat gaagaataat 3120tggatatggc
aaatcagagt ttttgaggaa ccattttggg attttgataa tgtgatagag 3180aataggtaac
actgttaggg tttatgtctt tggtgatact tcctttttgt ttgtaatttg 3240gaacatggtt
tccgcaattg atttttcaat taaatcttct tttttgttc
32892878PRTArabidopsis thalianaMISC_FEATURE(826)..(864)RING finger motif
2Met Ala Ser Thr Gly Glu Pro Asp Arg Lys Arg Arg His Phe Ser Ser1
5 10 15Ile Ser Pro Ser Glu Ala
Ala Ala Ala Val Lys Lys Gln Pro Phe Phe 20 25
30Trp Pro Ser Ser Glu Asp Lys Leu Asp Thr Ala Val Leu
Gln Phe Gln 35 40 45Asn Leu Lys
Leu Ser Gln Lys Leu Glu Ala Gln Gln Val Glu Cys Ser 50
55 60Ile Leu Glu Asp Lys Leu Ser Gln Ile Lys Glu Lys
Gln Leu Pro Tyr65 70 75
80Asn Ser Ser Leu Lys Thr Val His Lys Ser Trp Glu Lys Leu Thr Ala
85 90 95Ser Val Glu Ser Cys Ser
Val Arg Val Ser Asp Ser Ser Ser Gly Ala 100
105 110His Arg Phe Val Asn Lys Glu Asp Gly Ser Ser Pro
Ala Val Lys Asn 115 120 125Asp Phe
Ile Asn Arg Leu Leu Glu Thr Gly Ala Thr Glu Ser Ser Ser 130
135 140Ser Asn Ile Cys Ser Asn Gln Met Glu Glu Asn
Gly Val Asn Thr Ser145 150 155
160Ser Gln Met Thr Gln Thr Leu Tyr Asn Leu Val Ala Ala Thr Glu Asp
165 170 175Leu Arg Cys Leu
Lys Asp Glu Leu Tyr Pro Thr Val Leu Arg Thr Asn 180
185 190Leu Gly Lys Asp Leu Cys Gly Gln Leu Ala Leu
Ser Glu Leu Glu Ser 195 200 205Glu
Ile Lys Ser Phe Arg Gly Asp Leu Asp Asp Val Leu Val Lys Phe 210
215 220Lys Ser Leu Ser Arg Glu Leu Gln Ser His
Arg Asp Ala Asp Ala Lys225 230 235
240Val Arg Val Asp Leu Lys Arg Ile Arg Gly Glu Leu Glu Asp Glu
Val 245 250 255Val Glu Leu
Gln Gln Cys Asn Gly Asp Leu Ser Ala Leu Arg Ala Glu 260
265 270Arg Asp Ala Thr Ala Gly Ala Phe Phe Pro
Val Leu Ser Leu Gly Asn 275 280
285Lys Leu Ala Thr Ser Asp Arg Glu Arg Asp Lys Gln Arg Asp Leu Gln 290
295 300Asp Met Glu Thr Val Leu Lys Glu
Leu Thr Val Leu Ala Ser Gly Arg305 310
315 320Leu Gln Gln Leu Lys Asn Leu His Glu Glu Arg Thr
Lys Met Leu Gly 325 330
335Lys Met Ser Asn Leu Gln Asn Lys Ser Lys Ser Val Arg Cys Ile Ser
340 345 350Ser Ser Gln Ala Cys Leu
Ser Leu Lys Asp Gln Leu Glu Lys Ser Lys 355 360
365Glu Ala Val Phe Gln Tyr Met Ala Leu Leu Glu Lys Leu Gln
Val Glu 370 375 380Lys Asp Ser Ile Val
Trp Lys Glu Arg Glu Ile Asn Ile Lys Asn Glu385 390
395 400Leu Gly Asp Val Ser Arg Lys Thr Ser Ala
Val Thr Asp Ser Arg Met 405 410
415Ala Ser Leu Asp Ser Glu Ile Gln Lys Gln Leu Asp Glu Lys Met Arg
420 425 430Ile Lys Thr Arg Leu
Gly Asn Ile Ser Arg Glu Arg Gly Arg Lys Glu 435
440 445Ile Phe Ala Asp Met Lys Ala Leu Ile Ser Ser Phe
Pro Glu Glu Met 450 455 460Ser Ser Met
Arg Ser Gln Leu Asn Asn Tyr Lys Glu Thr Ala Gly Gly465
470 475 480Ile His Ser Leu Arg Ala Asp
Val Gln Ser Leu Ser Gly Val Leu Cys 485
490 495Arg Lys Thr Lys Glu Tyr Glu Ala Leu Gln Leu Arg
Ser Ala Asp Tyr 500 505 510Ala
Ser Gln Leu Gly Asp Leu Asn Ala Thr Val Cys Asp Leu Lys Asn 515
520 525Ser His Glu Glu Leu Lys Leu Phe Leu
Asp Met Tyr Lys Arg Glu Ser 530 535
540Thr Asp Ala Arg Asp Ile Ala Glu Ala Lys Glu Gln Glu Tyr Arg Ala545
550 555 560Trp Ala His Val
Gln Ser Leu Lys Ser Ser Leu Asp Glu Gln Asn Leu 565
570 575Glu Leu Arg Val Lys Ala Ala Asn Glu Ala
Glu Ala Val Ser Gln Gln 580 585
590Met Leu Ala Ala Ala Glu Ala Glu Ile Ala Asp Leu Arg Gln Lys Met
595 600 605Asp Asp Cys Lys Arg Asp Val
Ala Lys His Ser Asp Ile Leu Lys Ser 610 615
620Lys His Glu Glu His Gly Thr Tyr Leu Ser Glu Ile Gln Thr Ile
Gly625 630 635 640Ser Ala
Tyr Glu Asp Ile Val Pro Gln Asn Gln Gln Leu Leu Leu Gln
645 650 655Val Thr Glu Arg Asp Asp Tyr
Asn Ile Lys Leu Phe Leu Glu Gly Ile 660 665
670Thr Ser Arg Gln Met Gln Asp Thr Leu Leu Ile Asp Lys Tyr
Ile Met 675 680 685Asp Lys Asp Ile
Gln Gln Gly Ser Ala Tyr Ala Ser Phe Leu Ser Lys 690
695 700Lys Ser Ser Arg Ile Glu Asp Gln Leu Arg Phe Cys
Thr Asp Gln Phe705 710 715
720Gln Lys Leu Ala Glu Asp Lys Tyr Gln Lys Ser Val Ser Leu Glu Asn
725 730 735Leu Gln Lys Lys Arg
Ala Asp Ile Gly Asn Gly Leu Glu Gln Ala Arg 740
745 750Ser Arg Leu Glu Glu Ser His Ser Lys Val Glu Gln
Ser Arg Leu Asp 755 760 765Tyr Gly
Ala Leu Glu Leu Glu Leu Glu Ile Glu Arg Phe Asn Arg Arg 770
775 780Arg Ile Glu Glu Glu Met Glu Ile Ala Lys Lys
Lys Val Ser Arg Leu785 790 795
800Arg Ser Leu Ile Glu Gly Ser Ser Ala Ile Gln Lys Leu Arg Gln Glu
805 810 815Leu Ser Glu Phe
Lys Glu Ile Leu Lys Cys Lys Ala Cys Asn Asp Arg 820
825 830Pro Lys Glu Val Val Ile Thr Lys Cys Tyr His
Leu Phe Cys Asn Pro 835 840 845Cys
Val Gln Lys Leu Thr Gly Thr Arg Gln Lys Lys Cys Pro Thr Cys 850
855 860Ser Ala Ser Phe Gly Pro Asn Asp Ile Lys
Pro Ile Tyr Ile865 870
875320DNAArtificial Sequenceprimer 3ctcgcccatt gttgtttcag
20420DNAArtificial Sequenceprimer
4aattgcggaa accatgttcc
20520DNAArtificial Sequenceprimer Defle44 5tgctggacgt gaccttactg
20620DNAArtificial Sequenceprimer
Defle45 6gggctggaac aagacttctg
20720DNAArtificial Sequenceprimer syana_01 7tgctcgaatc agatggaaga
20820DNAArtificial
Sequenceprimer syana_02 8agctagctga ccgcacaaat
20921DNAArtificial Sequenceprimer used for ANG4
candidate genes amplification and sequencing (table 9) 9tgttaagagg
tgacgcacat g
211020DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 10cggcggcttg aatgtcttta
201123DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
11tagaggagtg aggatgagga gtt
231224DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 12gctaggaaaa aaaaaagaaa ttgt
241323DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
13tttttgtttc tgtgagtgct gtg
231422DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 14tctcagtagc accagtttca ag
221522DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
15cttcttcatc tcccccttgt gc
221624DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 16ataaatacac aggcgtggaa ttgg
241724DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
17tatgtgtcgc ccgtcttctt tctt
241824DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 18atgccaatga acaacaagta aaga
241924DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
19ttcctaatgt tgtttgccgt ttca
242023DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 20catgggggtg gaaatagtat cct
232128DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
21tactcagtat gcaattccac gttcatat
282224DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 22tctctctcgc atttttctca accg
242324DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
23aacgaaattc tcaaagatgg gttt
242424DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 24aacgaaattc tcaaagatgg gttt
242523DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
25cgtcacacca tccacaccac ttg
232626DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 26gacggcaata cttatcgcca acatat
262726DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
27aattggacca gatgggattg ggaaag
262828DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 28tccacacaaa aatgtcagag tgcttagc
282924DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
29cttgattact ggcacaccta tcca
243022DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 30gggaaaagag gaggacacga tg
223126DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
31tgtttatctc cctatctatt tccttg
263221DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 32ctttctctct gccctcctca a
213324DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
33tggaatacat cggcatagag aaag
243424DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 34taaactcgga tgctcggtga taag
243524DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
35ccaggaaaaa ggcagaagag aaga
243624DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 36cattgtgtga ttcagggaga tcga
243720DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
37gggcgttttt cccagtgttg
203820DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 38tcagcccgca gagaatgaat
203920DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
39tcccacccac acctgtttca
204020DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 40ttccgcagca gccaacattt
204120DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
41gaagccaagg aacaggagta
204220DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 42catacgggca cacacagata
204320DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
43ctcgcccatt gttgtttcag
204420DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 44aattgcggaa accatgttcc
204520DNAArtificial Sequenceprimer
used for ANG4 candidate genes amplification and sequencing (table 9)
45tggggcatta gaactggaac
204620DNAArtificial Sequenceprimer used for ANG4 candidate genes
amplification and sequencing (table 9) 46tcccaaggat cgaagtcttt
204752DNAArabidopsis
thalianamisc_featureAligned Ang sequence in fig 8 47aatcatcaag aattgaagat
taggtatatc tgtgtgtgcc cgtatgctca aa 524852DNAArabidopsis
thalianamisc_featureAligned Ler sequence in fig 8 48aatcatcaag aattgaagat
caggtatatc tgtgtgtgcc cgtatgctca aa 52491002PRTHomo
sapiensMISC_FEATUREAAP36593.1 in Fig. 10 49Met Ser Gly Pro Gly Asn Lys
Arg Ala Ala Gly Asp Gly Gly Ser Gly1 5 10
15Pro Pro Glu Lys Lys Leu Ser Arg Glu Glu Lys Thr Thr
Thr Thr Leu 20 25 30Ile Glu
Pro Ile Arg Leu Gly Gly Ile Ser Ser Thr Glu Glu Met Asp 35
40 45Leu Lys Val Leu Gln Phe Lys Asn Lys Lys
Leu Ala Glu Arg Leu Glu 50 55 60Gln
Arg Gln Ala Cys Glu Asp Glu Leu Arg Glu Arg Ile Glu Lys Leu65
70 75 80Glu Lys Arg Gln Ala Thr
Asp Asp Ala Thr Leu Leu Ile Val Asn Arg 85
90 95Tyr Trp Ala Gln Leu Asp Glu Thr Val Glu Ala Leu
Leu Arg Cys His 100 105 110Glu
Ser Gln Gly Glu Leu Ser Ser Ala Pro Glu Ala Pro Gly Thr Gln 115
120 125Glu Gly Pro Thr Cys Asp Gly Thr Pro
Leu Pro Glu Pro Gly Thr Ser 130 135
140Glu Leu Arg Asp Pro Leu Leu Met Gln Leu Arg Pro Pro Leu Ser Glu145
150 155 160Pro Ala Leu Ala
Phe Val Val Ala Leu Gly Ala Ser Ser Ser Glu Glu 165
170 175Val Glu Leu Glu Leu Gln Gly Arg Met Glu
Phe Ser Lys Ala Ala Val 180 185
190Ser Arg Val Val Glu Ala Ser Asp Arg Leu Gln Arg Arg Val Glu Glu
195 200 205Leu Cys Gln Arg Val Tyr Ser
Arg Gly Asp Ser Glu Pro Leu Ser Glu 210 215
220Ala Ala Gln Ala His Thr Arg Glu Leu Gly Arg Glu Asn Arg Arg
Leu225 230 235 240Gln Asp
Leu Ala Thr Gln Leu Gln Glu Lys His His Arg Ile Ser Leu
245 250 255Glu Tyr Ser Glu Leu Gln Asp
Lys Val Thr Ser Ala Glu Thr Lys Val 260 265
270Leu Glu Met Glu Thr Thr Val Glu Asp Leu Gln Trp Asp Ile
Glu Lys 275 280 285Leu Arg Lys Arg
Glu Gln Lys Leu Asn Lys His Leu Ala Glu Ala Leu 290
295 300Glu Gln Leu Asn Ser Gly Tyr Tyr Val Ser Gly Ser
Ser Ser Gly Phe305 310 315
320Gln Gly Gly Gln Ile Thr Leu Ser Met Gln Lys Phe Glu Met Leu Asn
325 330 335Ala Glu Leu Glu Glu
Asn Gln Glu Leu Ala Asn Ser Arg Met Ala Glu 340
345 350Leu Glu Lys Leu Gln Ala Glu Leu Gln Gly Ala Val
Arg Thr Asn Glu 355 360 365Arg Leu
Lys Val Ala Leu Arg Ser Leu Pro Glu Glu Val Val Arg Glu 370
375 380Thr Gly Glu Tyr Arg Met Leu Gln Ala Gln Phe
Ser Leu Leu Tyr Asn385 390 395
400Glu Ser Leu Gln Val Lys Thr Gln Leu Asp Glu Ala Arg Gly Leu Leu
405 410 415Leu Ala Thr Lys
Asn Ser His Leu Arg His Ile Glu His Met Glu Ser 420
425 430Asp Glu Leu Gly Leu Gln Lys Lys Leu Arg Thr
Glu Val Ile Gln Leu 435 440 445Glu
Asp Thr Leu Ala Gln Val Arg Lys Glu Tyr Glu Met Leu Arg Ile 450
455 460Glu Phe Glu Gln Asn Leu Ala Ala Asn Glu
Gln Ala Gly Pro Ile Asn465 470 475
480Arg Glu Met Arg His Leu Ile Ser Ser Leu Gln Asn His Asn His
Gln 485 490 495Leu Lys Gly
Asp Ala Gln Arg Tyr Lys Arg Lys Leu Arg Glu Val Gln 500
505 510Ala Glu Ile Gly Lys Leu Arg Ala Gln Ala
Ser Gly Ser Ala His Ser 515 520
525Thr Pro Asn Leu Gly His Pro Glu Asp Ser Gly Val Ser Ala Pro Ala 530
535 540Pro Gly Lys Glu Glu Gly Gly Pro
Gly Pro Val Ser Thr Pro Asp Asn545 550
555 560Arg Lys Glu Met Ala Pro Val Pro Gly Thr Thr Thr
Thr Thr Thr Ser 565 570
575Val Lys Lys Glu Glu Leu Val Pro Ser Glu Glu Asp Phe Gln Gly Ile
580 585 590Thr Pro Gly Ala Gln Gly
Pro Ser Ser Arg Gly Arg Glu Pro Glu Ala 595 600
605Arg Pro Lys Arg Glu Leu Arg Glu Arg Glu Gly Pro Ser Leu
Gly Pro 610 615 620Pro Pro Val Ala Ser
Ala Leu Ser Arg Ala Asp Arg Glu Lys Ala Lys625 630
635 640Val Glu Glu Thr Lys Arg Lys Glu Ser Glu
Leu Leu Lys Gly Leu Arg 645 650
655Ala Glu Leu Lys Lys Ala Gln Glu Ser Gln Lys Glu Met Lys Leu Leu
660 665 670Leu Asp Met Tyr Lys
Ser Ala Pro Lys Glu Gln Arg Asp Lys Val Gln 675
680 685Leu Met Ala Ala Glu Arg Lys Ala Lys Ala Glu Val
Asp Glu Leu Arg 690 695 700Ser Arg Ile
Arg Glu Leu Glu Glu Arg Asp Arg Arg Glu Ser Lys Lys705
710 715 720Ile Ala Asp Glu Asp Ala Leu
Arg Arg Ile Arg Gln Ala Glu Glu Gln 725
730 735Ile Glu His Leu Gln Arg Lys Leu Gly Ala Thr Lys
Gln Glu Glu Glu 740 745 750Ala
Leu Leu Ser Glu Met Asp Val Thr Gly Gln Ala Phe Glu Asp Met 755
760 765Gln Glu Gln Asn Gly Arg Leu Leu Gln
Gln Leu Arg Glu Lys Asp Asp 770 775
780Ala Asn Phe Lys Leu Met Ser Glu Arg Ile Lys Ala Asn Gln Ile His785
790 795 800Lys Leu Leu Arg
Glu Glu Lys Asp Glu Leu Gly Glu Gln Val Leu Gly 805
810 815Leu Lys Ser Gln Val Asp Ala Gln Leu Leu
Thr Val Gln Lys Leu Glu 820 825
830Glu Lys Glu Arg Ala Leu Gln Gly Ser Leu Gly Gly Val Glu Lys Glu
835 840 845Leu Thr Leu Arg Ser Gln Ala
Leu Glu Leu Asn Lys Arg Lys Ala Val 850 855
860Glu Ala Ala Gln Leu Ala Glu Asp Leu Lys Val Gln Leu Glu His
Val865 870 875 880Gln Thr
Arg Leu Arg Glu Ile Gln Pro Cys Leu Ala Glu Ser Arg Ala
885 890 895Ala Arg Glu Lys Glu Ser Phe
Asn Leu Lys Arg Ala Gln Glu Asp Ile 900 905
910Ser Arg Leu Arg Arg Lys Leu Glu Lys Gln Arg Lys Val Glu
Val Tyr 915 920 925Ala Asp Ala Asp
Glu Ile Leu Gln Glu Glu Ile Lys Glu Tyr Lys Ala 930
935 940Arg Leu Thr Cys Pro Cys Cys Asn Thr Arg Lys Lys
Asp Ala Val Leu945 950 955
960Thr Lys Cys Phe His Val Phe Cys Phe Glu Cys Val Arg Gly Arg Tyr
965 970 975Glu Ala Arg Gln Arg
Lys Cys Pro Lys Cys Asn Ala Ala Phe Gly Ala 980
985 990His Asp Phe His Arg Ile Tyr Ile Ser Leu
995 100050975PRTHomo sapiensMISC_FEATUREAAK58539_RFP_20
in Fig. 10 50Met Ser Gly Ile Gly Asn Lys Arg Ala Ala Gly Glu Pro Gly Thr
Ser1 5 10 15Met Pro Pro
Glu Lys Lys Ala Ala Val Glu Asp Ser Gly Thr Thr Val 20
25 30Glu Thr Ile Lys Leu Gly Gly Val Ser Ser
Thr Glu Glu Leu Asp Ile 35 40
45Arg Thr Leu Gln Thr Lys Asn Arg Lys Leu Ala Glu Met Leu Asp Gln 50
55 60Arg Gln Ala Ile Glu Asp Glu Leu Arg
Glu His Ile Glu Lys Leu Glu65 70 75
80Arg Arg Gln Ala Thr Asp Asp Ala Ser Leu Leu Ile Val Asn
Arg Tyr 85 90 95Trp Ser
Gln Phe Asp Glu Asn Ile Arg Ile Ile Leu Lys Arg Tyr Asp 100
105 110Leu Glu Gln Gly Leu Gly Asp Leu Leu
Thr Glu Arg Lys Ala Leu Val 115 120
125Val Pro Glu Pro Glu Pro Asp Ser Asp Ser Asn Gln Glu Arg Lys Asp
130 135 140Asp Arg Glu Arg Gly Glu Gly
Gln Glu Pro Ala Phe Ser Phe Leu Ala145 150
155 160Thr Leu Ala Ser Ser Ser Ser Glu Glu Met Glu Ser
Gln Leu Gln Glu 165 170
175Arg Val Glu Ser Ser Arg Arg Ala Val Ser Gln Ile Val Thr Val Tyr
180 185 190Asp Lys Leu Gln Glu Lys
Val Glu Leu Leu Ser Arg Lys Leu Asn Ser 195 200
205Gly Asp Asn Leu Ile Val Glu Glu Ala Val Gln Glu Leu Asn
Ser Phe 210 215 220Leu Ala Gln Glu Asn
Met Arg Leu Gln Glu Leu Thr Asp Leu Leu Gln225 230
235 240Glu Lys His Arg Thr Met Ser Gln Glu Phe
Ser Lys Leu Gln Ser Lys 245 250
255Val Glu Thr Ala Glu Ser Arg Val Ser Val Leu Glu Ser Met Ile Asp
260 265 270Asp Leu Gln Trp Asp
Ile Asp Lys Ile Arg Lys Arg Glu Gln Arg Leu 275
280 285Asp Arg His Leu Ala Glu Val Leu Glu Arg Val Asn
Ser Lys Gly Tyr 290 295 300Lys Val Tyr
Gly Ala Gly Ser Ser Leu Tyr Gly Gly Thr Ile Thr Ile305
310 315 320Asn Ala Arg Lys Phe Glu Glu
Met Asn Ala Glu Leu Glu Glu Asn Lys 325
330 335Glu Leu Ala Gln Asn Arg Leu Cys Glu Leu Glu Lys
Leu Arg Gln Asp 340 345 350Phe
Glu Glu Val Thr Thr Gln Asn Glu Lys Leu Lys Val Glu Leu Arg 355
360 365Ser Ala Val Glu Gln Val Val Lys Glu
Thr Pro Glu Tyr Arg Cys Met 370 375
380Gln Ser Gln Phe Ser Val Leu Tyr Asn Glu Ser Leu Gln Leu Lys Ala385
390 395 400His Leu Asp Glu
Ala Arg Thr Leu Leu His Gly Thr Arg Gly Thr His 405
410 415Gln His Gln Val Glu Leu Ile Glu Arg Asp
Glu Val Ser Leu His Lys 420 425
430Lys Leu Arg Thr Glu Val Ile Gln Leu Glu Asp Thr Leu Ala Gln Val
435 440 445Arg Lys Glu Tyr Glu Met Leu
Arg Ile Glu Phe Glu Gln Thr Leu Ala 450 455
460Ala Asn Glu Gln Ala Gly Pro Ile Asn Arg Glu Met Arg His Leu
Ile465 470 475 480Ser Ser
Leu Gln Asn His Asn His Gln Leu Lys Gly Glu Val Leu Arg
485 490 495Tyr Lys Arg Lys Leu Arg Glu
Ala Gln Ser Asp Leu Asn Lys Thr Arg 500 505
510Leu Arg Ser Gly Ser Ala Leu Leu Gln Ser Gln Ser Ser Thr
Glu Asp 515 520 525Pro Lys Asp Glu
Pro Ala Glu Leu Lys Pro Asp Ser Glu Asp Leu Ser 530
535 540Ser Gln Ser Ser Ala Ser Lys Ala Ser Gln Glu Asp
Ala Asn Glu Ile545 550 555
560Lys Ser Lys Arg Asp Glu Glu Glu Arg Glu Arg Glu Arg Arg Glu Lys
565 570 575Glu Arg Glu Arg Glu
Arg Glu Arg Glu Lys Glu Lys Glu Arg Glu Arg 580
585 590Glu Lys Gln Lys Leu Lys Glu Ser Glu Lys Glu Arg
Asp Ser Ala Lys 595 600 605Asp Lys
Glu Lys Gly Lys His Asp Asp Gly Arg Lys Lys Glu Ala Glu 610
615 620Ile Ile Lys Gln Leu Lys Ile Glu Leu Lys Lys
Ala Gln Glu Ser Gln625 630 635
640Lys Glu Met Lys Leu Leu Leu Asp Met Tyr Arg Ser Ala Pro Lys Glu
645 650 655Gln Arg Asp Lys
Val Gln Leu Met Ala Ala Glu Lys Lys Ser Lys Ala 660
665 670Glu Leu Glu Asp Leu Arg Gln Arg Leu Lys Asp
Leu Glu Asp Lys Glu 675 680 685Lys
Lys Glu Asn Thr Lys Met Ala Asp Glu Asp Ala Leu Arg Lys Ile 690
695 700Arg Ala Val Glu Glu Gln Ile Glu Tyr Leu
Gln Lys Lys Leu Ala Met705 710 715
720Ala Lys Gln Glu Glu Glu Ala Leu Leu Ser Glu Met Asp Val Thr
Gly 725 730 735Gln Ala Phe
Glu Asp Met Gln Glu Gln Asn Ile Arg Leu Met Gln Gln 740
745 750Leu Arg Glu Lys Asp Asp Ala Asn Phe Lys
Leu Met Ser Glu Arg Ile 755 760
765Lys Ser Asn Gln Ile His Lys Leu Leu Lys Glu Glu Lys Glu Glu Leu 770
775 780Ala Asp Gln Val Leu Thr Leu Lys
Thr Gln Val Asp Ala Gln Leu Gln785 790
795 800Val Val Arg Lys Leu Glu Glu Lys Glu His Leu Leu
Gln Ser Asn Ile 805 810
815Gly Thr Gly Glu Lys Glu Leu Gly Leu Arg Thr Gln Ala Leu Glu Met
820 825 830Asn Lys Arg Lys Ala Met
Glu Ala Ala Gln Leu Ala Asp Asp Leu Lys 835 840
845Ala Gln Leu Glu Leu Ala Gln Lys Lys Leu His Asp Phe Gln
Asp Glu 850 855 860Ile Val Glu Asn Ser
Val Thr Lys Glu Lys Asp Met Phe Asn Phe Lys865 870
875 880Arg Ala Gln Glu Asp Ile Ser Arg Leu Arg
Arg Lys Leu Glu Thr Thr 885 890
895Lys Lys Pro Asp Asn Val Pro Lys Cys Asp Glu Ile Leu Met Glu Glu
900 905 910Ile Lys Asp Tyr Lys
Ala Arg Leu Thr Cys Pro Cys Cys Asn Met Arg 915
920 925Lys Lys Asp Ala Val Leu Thr Lys Cys Phe His Val
Phe Cys Phe Glu 930 935 940Cys Val Lys
Thr Arg Tyr Asp Thr Arg Gln Arg Lys Cys Pro Lys Cys945
950 955 960Asn Ala Ala Phe Gly Ala Asn
Asp Phe His Arg Ile Tyr Ile Gly 965 970
97551883PRTOryza sativaMISC_FEATURECAD41603.3 in Fig. 10
51Met Gly Ser Thr Gly Glu Pro Asp Arg Lys Arg Arg Leu Ser Ser Ser1
5 10 15Val Ala Pro Gly Gly Gly
Ala Pro Val Ser Pro Ala Lys Arg Leu Ala 20 25
30Val Ala Pro Thr Ser Glu Asp Lys Lys Leu Asp Phe Thr
Val Leu Lys 35 40 45Tyr Lys Asn
Gln Lys Leu Ser Glu Gln Leu Glu Ala His Lys Phe Glu 50
55 60Tyr Arg Ala Leu Glu Asn Lys Phe Ala Gly Leu Lys
Glu Lys Gln Arg65 70 75
80Thr His Asn Glu Thr Leu Ser Leu Val Asn Ser Ser Trp Glu Gln Leu
85 90 95Val Ala Asp Leu Lys Ser
Arg Ser Phe Cys Lys Ser Gly Ser Pro Asn 100
105 110Ser Ser Pro Gly Ser Gly His Asn Asn Val Gln Lys
Asp Gly Thr Cys 115 120 125Ala Pro
Ile Glu Arg Asp Thr Leu Arg Ser Leu Val Glu Ser Gly Ala 130
135 140Thr Glu Ser Ser Gly Cys Leu Pro Gly Cys His
Leu Gly Ser Asp Ala145 150 155
160Pro Pro Leu His Leu Ser Thr Ala Asn Ala Leu Gly Asp Ile Phe Phe
165 170 175Pro Ser Ser Asp
Leu Leu Gln Ala Asn Glu Glu Cys Ala Leu Ala Ala 180
185 190Leu Thr Lys Leu Pro Glu Asn Asp Arg Ser Lys
Gln Leu Gln Ser Thr 195 200 205Ser
Ser Asn Leu Leu Ser Ser Leu Asn Asn Val Val Gln Ala Leu Ser 210
215 220Asn Leu Gln Leu Lys His Lys Gln Leu Ala
Glu Asp Tyr Gln Asn Gln225 230 235
240Arg Asp Ser Ser Ala Arg Lys Arg Ala Glu His Arg Arg Leu Lys
Glu 245 250 255Glu Leu Ala
Ser Ala Ala Ser Glu Leu Glu Glu Thr Asn Tyr Lys Leu 260
265 270Ala Ala Leu Lys Ala Gln Arg Asp Asn Thr
Gln Gly Ala Arg Ile Pro 275 280
285Tyr Pro Thr Leu Gly Asn Lys Asn Met Pro Glu Asp Lys Glu Leu Ile 290
295 300Ser Lys Arg Leu Val Glu Ile Lys
Arg Leu His Glu Glu Arg Ile Glu305 310
315 320Ile Leu Asn Lys Ile Ala Thr Phe Gln Asn Ile Leu
Met Asp Phe Lys 325 330
335Ser Ile Arg Ser Ser Lys Ala Phe Gln Leu Val Asn Asp Arg Leu Gln
340 345 350Lys Ser Gln Ala Glu Leu
Asp His Tyr Gln Thr Leu Leu Glu Lys Leu 355 360
365Gln Val Asp Lys Asp Lys Phe Val Trp Gln Glu Arg Gln Phe
Asn Leu 370 375 380Lys Val Asp Leu Ala
Glu Ile Pro Glu Arg Val Ser Thr Tyr Cys Arg385 390
395 400Asn Gln Val Ile Thr Lys Phe Lys Ala Leu
Val Ser Ser Ile Pro Arg 405 410
415Glu Met Gly Ala Met Gln Ser Glu Met Thr Lys His Lys Glu Ala Ser
420 425 430Leu Glu Leu Asn Ser
Leu Arg Ala Glu Val His Ser Leu Ser Arg Ile 435
440 445Leu Ser Arg Lys Glu Arg Asp Asn Glu Glu Ala Ser
Cys Arg Ser Ala 450 455 460Arg Ala Gly
Ser Asp Ile Thr Gln Leu Gln Ser Val Ile Ser Asp Leu465
470 475 480Lys Gln Thr Asn Lys Glu Leu
Lys Leu Phe Ala Asp Met Tyr Lys Arg 485
490 495Glu Ser Thr Asp Ser Arg Glu Ile Met Glu Ser Arg
Asp Arg Glu Phe 500 505 510Leu
Glu Trp Ala His Val His Ala Leu Lys Ser Ser Leu Asp Glu Ser 515
520 525Lys Leu Glu Gln Arg Val Lys Ala Ala
Asn Glu Ala Glu Ala Ile Thr 530 535
540Gln Gln Arg Leu Ala Thr Ala Glu Ala Glu Ile Ala Glu Ser Gly Gln545
550 555 560Lys Leu Gly Thr
Ser Arg Lys Tyr Arg Ile Met Leu Leu Asn Ile Val 565
570 575Ser Leu Arg Thr Val Glu Val Gly Val Thr
Ser Leu Leu Gly Asp Leu 580 585
590Val Ser Leu Ser His Met Leu Lys Ser Lys Gln Glu Glu Cys Glu Ala
595 600 605Tyr Arg Val Glu Val Glu Cys
Ile Gly Gln Ala Tyr Glu Asp Ile Gln 610 615
620Ala Gln Asn Gln Gln Leu Leu Gln Gln Ile Ile Glu Arg Asp Asp
Asp625 630 635 640Asn Thr
Lys Asp Val Arg Phe Gly Tyr Ile Val Asn Leu Ile Val Pro
645 650 655Glu Thr Gln Tyr Phe Ile Glu
Lys Leu Phe Thr Cys Val Lys Leu Ile 660 665
670Phe Met Glu Gly Val Lys Ala Lys Gln Thr Gln Asp Ala Leu
His Leu 675 680 685Glu Thr Tyr Ser
Leu Arg Arg Asn Leu Gln Gln Glu Ser Ser Leu Met 690
695 700Asp Leu Tyr Asn Gln Lys Ile Val Ser Leu Glu Asp
Gln Leu Lys Met705 710 715
720Trp Ser Asp Arg Val Gly Lys Leu Gln Glu Asp Gly Trp Gln Gln Ser
725 730 735Val Ser Leu Ser Asn
Tyr Gln Arg Lys Leu Val Asp Val His Arg Asp 740
745 750Ala Gln Lys Leu Met Gln Ser Leu Asp Gly Ile Gln
Ala Asn Val Gly 755 760 765Ser Ser
Arg Leu Glu Val Ala Asp Leu Leu Ile Glu Leu Glu Lys Glu 770
775 780Arg Phe Ser Lys Lys Arg Ile Glu Asp Asp Leu
Glu Val Met Ser Arg785 790 795
800Lys Ala Ser Ser Leu Arg Ala Lys Ala Arg Glu Ser Ala Val Leu Glu
805 810 815Lys Leu Arg His
Glu Val Lys Glu Tyr Arg Gly Ile Leu Lys Cys Gly 820
825 830Ile Cys His Asp Arg Gln Lys Glu Val Val Ile
Thr Lys Cys Tyr His 835 840 845Leu
Phe Cys Asn Gln Cys Ile Gln Lys Ser Leu Gly Asn Arg Gln Arg 850
855 860Arg Cys Pro Ser Cys Ser Leu Ser Phe Gly
Ala Asn Asp Val Lys Pro865 870 875
880Ile Tyr Ile52899PRTArabidopsis
thalianaMISC_FEATUREMIPS_At1g55250 in Fig. 10 52Met Glu Asn Gln Glu Ser
Asp Glu Pro Met Gln Lys Lys Pro His Leu1 5
10 15Leu Asp Ser Val Ser Pro Asn Ser Met Ala Arg Asn
Ser Ser Pro Ser 20 25 30His
Pro Ile Ala Lys Ser Val Ser Phe Phe Asp Cys Asp Phe Ser Leu 35
40 45Leu Cys Leu Arg Leu Val Asp Tyr Glu
Ile Asp Val Asp Ala Thr Val 50 55
60Leu Gln Leu Gln Asn Gln Lys Leu Val Gln Gln Leu Asp Leu Gln Lys65
70 75 80Lys Gln Leu Tyr Asp
Val Glu Ser Lys Ile Gln Glu Leu Gln Leu Asn 85
90 95Gln Thr Ser Tyr Asp Asp Glu Leu Ile Ser Val
Asn Gln Leu Trp Asn 100 105
110Gln Leu Val Asp Asp Leu Ile Leu Leu Gly Val Arg Ala Gly Ala Asn
115 120 125Gln Glu Ala Leu Asn Tyr Leu
Asp Ile Val Asp Lys Lys Arg Val Pro 130 135
140Pro Cys Ala Ala Asp Glu Thr Phe Leu Cys Arg Leu Leu Gln Val
Asp145 150 155 160Ser Leu
Asp Thr Ser Lys Ser Asp Glu Val Val Arg Lys Val Glu Glu
165 170 175Ala Leu Ala Leu Arg His Ser
Ser Thr Met Glu Leu Met Gly Leu Phe 180 185
190Glu Asn Thr Ile Asp Thr Gln Lys Thr Lys Ala Glu Ser Ile
Ser Gln 195 200 205Ser Leu His Ala
Val Lys Ser Thr Glu Asp Ala Thr Ile Gln Leu Ser 210
215 220Ser Ile Asn Asp Leu Met Lys Glu Glu Ser Lys Asn
Leu Arg Glu Met225 230 235
240Ile Asp Ala Leu His Val Arg His Lys Glu His Ser Glu Gln Ile Gln
245 250 255Ala Tyr Ile Ser Ser
His Ser Thr Asp Gln Ser Glu Leu Lys His Leu 260
265 270Lys Gly Gln Leu Glu Glu Ile Lys Ala Glu Leu Glu
Glu Asn Arg Arg 275 280 285Lys Leu
Ile Thr Leu Lys Met Gln Lys Asp Ala Ala Cys Glu Gly His 290
295 300Val Thr Ser Pro Ala Ile Ala Asn Gly Ser Leu
Ser Pro Glu Lys Pro305 310 315
320Val Asp Lys Thr Lys Leu Arg Glu Leu Lys Asp Ser Ile Asp Glu Ile
325 330 335Lys Ile Met Ala
Glu Gly Arg Leu Ser Glu Leu Gln Ala Ser Gln Glu 340
345 350Tyr Asn Leu Ser Leu Ser Arg Gln Cys Gln Asp
Ile Glu Asn Glu Leu 355 360 365Lys
Asp Asp Gln Tyr Ile Tyr Ser Ser Arg Leu Tyr Ser Leu Ile Asn 370
375 380Asp Arg Ile His His Trp Asn Ala Glu Leu
Asp Arg Tyr Lys Ile Leu385 390 395
400Thr Glu Ala Ile Gln Ala Glu Arg Ser Phe Val Met Arg Arg Asp
Lys 405 410 415Glu Leu Asn
Leu Arg Ala Glu Ser Leu Glu Ala Ala Asn His Lys Thr 420
425 430Thr Thr Val Gly Ser Arg Ile Glu Val Leu
Glu Lys Lys Leu Gln Ser 435 440
445Cys Ile Ile Glu Lys Asn Gly Leu Glu Leu Glu Thr Glu Glu Ala Ile 450
455 460Gln Asp Ser Glu Arg Gln Asp Ile
Lys Ser Glu Phe Ile Ala Met Ala465 470
475 480Ser Thr Leu Ser Lys Glu Met Glu Met Met Glu Ala
Gln Leu Lys Arg 485 490
495Trp Lys Asp Thr Ala Gln Asp Ala Leu Tyr Leu Arg Glu Gln Ala Gln
500 505 510Ser Leu Arg Val Ser Leu
Ser Asn Lys Ala Asp Glu Gln Lys Gly Leu 515 520
525Glu Asp Lys Cys Ala Lys Gln Met Ala Glu Ile Lys Ser Leu
Lys Ala 530 535 540Leu Ile Glu Lys Leu
Leu Lys Glu Lys Leu Gln Leu Gln Asn Leu Ala545 550
555 560Ser Ile Cys Thr Arg Glu Cys Asn Asp Asp
Arg Gly Leu Ala Glu Ile 565 570
575Lys Asp Ser Gln Arg Lys Ala Gln Ala Gln Ala Glu Glu Leu Lys Asn
580 585 590Val Leu Asp Glu His
Phe Leu Glu Leu Arg Val Lys Ala Ala His Glu 595
600 605Thr Glu Ser Ala Cys Gln Glu Arg Leu Ala Thr Ala
Lys Ala Glu Ile 610 615 620Ala Glu Leu
Arg Thr Gln Leu Asp Leu Ser Glu Arg Glu Val Leu Glu625
630 635 640Leu Lys Glu Gly Ile Lys Val
Lys Glu Gln Glu Ala Glu Ala Ser Ile 645
650 655Ala Glu Met Glu Thr Ile Gly Gln Ala Tyr Glu Asp
Met Gln Thr Gln 660 665 670Asn
Gln His Leu Leu Gln Gln Val Ala Glu Arg Asp Asp Tyr Asn Ile 675
680 685Lys Leu Val Ser Glu Ser Val Lys Thr
Lys His Ala Tyr Asn Thr His 690 695
700Leu Ser Glu Lys Gln Val Met Glu Lys Gln Leu His Gln Val Asn Ala705
710 715 720Ser Val Glu Asn
Phe Lys Ala Arg Ile Ala His Asn Glu Glu Gln Met 725
730 735Lys Gly Cys Phe Ser Glu Ala Tyr Lys Leu
Ile Gln Glu Asp Arg His 740 745
750Leu Val Ile Ser Leu Glu Thr Thr Lys Trp Glu Val Ala Asp Ala Asp
755 760 765Lys Glu Phe Arg Trp Leu Lys
Ser Ala Val Ser Ser Ser Glu Lys Glu 770 775
780Tyr Glu Gln Ile Ser Arg Arg Thr Asp Asp Ile Lys Leu Glu Leu
Asp785 790 795 800Asp Glu
Arg Glu Lys Lys Lys Leu Glu Glu Glu Leu Met Glu Leu Asn
805 810 815Lys Glu Leu Glu Glu Leu Gly
Ser Glu Ser Val Glu Ala Ala Ile Val 820 825
830Arg Leu Gln Glu Glu Val Lys Asn Cys Lys Asn Ile Leu Lys
Cys Gly 835 840 845Val Cys Phe Asp
Arg Pro Lys Glu Val Val Ile Val Lys Cys Tyr His 850
855 860Leu Phe Cys Gln Gln Cys Ile Gln Arg Ser Leu Glu
Ile Arg His Arg865 870 875
880Lys Cys Pro Gly Cys Gly Thr Ala Phe Gly Gln Asn Asp Val Arg Leu
885 890 895Val Lys
Met531001PRTHomo sapiensMISC_FEATURENP055586_RFP_40 in Fig. 10 53Met Ser
Gly Pro Gly Asn Lys Arg Ala Ala Gly Asp Gly Gly Ser Gly1 5
10 15Pro Pro Glu Lys Lys Leu Ser Arg
Glu Glu Lys Thr Thr Thr Thr Leu 20 25
30Ile Glu Pro Ile Arg Leu Gly Gly Ile Ser Ser Thr Glu Glu Met
Asp 35 40 45Leu Lys Val Leu Gln
Phe Lys Asn Lys Lys Leu Ala Glu Arg Leu Glu 50 55
60Gln Arg Gln Ala Cys Glu Asp Glu Leu Arg Glu Arg Ile Glu
Lys Leu65 70 75 80Glu
Lys Arg Gln Ala Thr Asp Asp Ala Thr Leu Leu Ile Val Asn Arg
85 90 95Tyr Trp Ala Gln Leu Asp Glu
Thr Val Glu Ala Leu Leu Arg Cys His 100 105
110Glu Ser Gln Gly Glu Leu Ser Ser Ala Pro Glu Ala Pro Gly
Thr Gln 115 120 125Glu Gly Pro Thr
Cys Asp Gly Thr Pro Leu Pro Glu Pro Gly Thr Ser 130
135 140Glu Leu Arg Asp Pro Leu Leu Met Gln Leu Arg Pro
Pro Leu Ser Glu145 150 155
160Pro Ala Leu Ala Phe Val Val Ala Leu Gly Ala Ser Ser Ser Glu Glu
165 170 175Val Glu Leu Glu Leu
Gln Gly Arg Met Glu Phe Ser Lys Ala Ala Val 180
185 190Ser Arg Val Val Glu Ala Ser Asp Arg Leu Gln Arg
Arg Val Glu Glu 195 200 205Leu Cys
Gln Arg Val Tyr Ser Arg Gly Asp Ser Glu Pro Leu Ser Glu 210
215 220Ala Ala Gln Ala His Thr Arg Glu Leu Gly Arg
Glu Asn Arg Arg Leu225 230 235
240Gln Asp Leu Ala Thr Gln Leu Gln Glu Lys His His Arg Ile Ser Leu
245 250 255Glu Tyr Ser Glu
Leu Gln Asp Lys Val Thr Ser Ala Glu Thr Lys Val 260
265 270Leu Glu Met Glu Thr Thr Val Glu Asp Leu Gln
Trp Asp Ile Glu Lys 275 280 285Leu
Arg Lys Arg Glu Gln Lys Leu Asn Lys His Leu Ala Glu Ala Leu 290
295 300Glu Gln Leu Asn Ser Gly Tyr Tyr Val Ser
Gly Ser Ser Ser Gly Phe305 310 315
320Gln Gly Gly Gln Ile Thr Leu Ser Met Gln Lys Phe Glu Met Leu
Asn 325 330 335Ala Glu Leu
Glu Glu Asn Gln Glu Leu Ala Asn Ser Arg Met Ala Glu 340
345 350Leu Glu Lys Leu Gln Ala Glu Leu Gln Gly
Ala Val Arg Thr Asn Glu 355 360
365Arg Leu Lys Val Ala Leu Arg Ser Leu Pro Glu Glu Val Val Arg Glu 370
375 380Thr Gly Glu Tyr Arg Met Leu Gln
Ala Gln Phe Ser Leu Leu Tyr Asn385 390
395 400Glu Ser Leu Gln Val Lys Thr Gln Leu Asp Glu Ala
Arg Gly Leu Leu 405 410
415Leu Ala Thr Lys Asn Ser His Leu Arg His Ile Glu His Met Glu Ser
420 425 430Asp Glu Leu Gly Leu Gln
Lys Lys Leu Arg Thr Glu Val Ile Gln Leu 435 440
445Glu Asp Thr Leu Ala Gln Val Arg Lys Glu Tyr Glu Met Leu
Arg Ile 450 455 460Glu Phe Glu Gln Asn
Leu Ala Ala Asn Glu Gln Ala Gly Pro Ile Asn465 470
475 480Arg Glu Met Arg His Leu Ile Ser Ser Leu
Gln Asn His Asn His Gln 485 490
495Leu Lys Gly Asp Ala Gln Arg Tyr Lys Arg Lys Leu Arg Glu Val Gln
500 505 510Ala Glu Ile Gly Lys
Leu Arg Ala Gln Ala Ser Gly Ser Ala His Ser 515
520 525Thr Pro Asn Leu Gly His Pro Glu Asp Ser Gly Val
Ser Ala Pro Ala 530 535 540Pro Gly Lys
Glu Glu Gly Gly Pro Gly Pro Val Ser Thr Pro Asp Asn545
550 555 560Arg Lys Glu Met Ala Pro Val
Pro Gly Thr Thr Thr Thr Thr Thr Ser 565
570 575Val Lys Lys Glu Glu Leu Val Pro Ser Glu Glu Asp
Phe Gln Gly Ile 580 585 590Thr
Pro Gly Ala Gln Gly Pro Ser Ser Arg Gly Arg Glu Pro Glu Ala 595
600 605Arg Pro Lys Arg Glu Leu Arg Glu Arg
Glu Gly Pro Ser Leu Gly Pro 610 615
620Pro Pro Val Ala Ser Ala Leu Ser Arg Ala Asp Arg Glu Lys Ala Lys625
630 635 640Val Glu Glu Thr
Lys Arg Lys Glu Ser Glu Leu Leu Lys Gly Leu Arg 645
650 655Ala Glu Leu Lys Lys Ala Gln Glu Ser Gln
Lys Glu Met Lys Leu Leu 660 665
670Leu Asp Met Tyr Lys Ser Ala Pro Lys Glu Gln Arg Asp Lys Val Gln
675 680 685Leu Met Ala Ala Glu Arg Lys
Ala Lys Ala Glu Val Asp Glu Leu Arg 690 695
700Ser Arg Ile Arg Glu Leu Glu Glu Arg Asp Arg Arg Glu Ser Lys
Lys705 710 715 720Ile Ala
Asp Glu Asp Ala Leu Arg Arg Ile Arg Gln Ala Glu Glu Gln
725 730 735Ile Glu His Leu Gln Arg Lys
Leu Gly Ala Thr Lys Gln Glu Glu Glu 740 745
750Ala Leu Leu Ser Glu Met Asp Val Thr Gly Gln Ala Phe Glu
Asp Met 755 760 765Gln Glu Gln Asn
Gly Arg Leu Leu Gln Gln Leu Arg Glu Lys Asp Asp 770
775 780Ala Asn Phe Lys Leu Met Ser Glu Arg Ile Lys Ala
Asn Gln Ile His785 790 795
800Lys Leu Leu Arg Glu Glu Lys Asp Glu Leu Gly Glu Gln Val Leu Gly
805 810 815Leu Lys Ser Gln Val
Asp Ala Gln Leu Leu Thr Val Gln Lys Leu Glu 820
825 830Glu Lys Glu Arg Ala Leu Gln Gly Ser Leu Gly Gly
Val Glu Lys Glu 835 840 845Leu Thr
Leu Arg Ser Gln Ala Leu Glu Leu Asn Lys Arg Lys Ala Val 850
855 860Glu Ala Ala Gln Leu Ala Glu Asp Leu Lys Val
Gln Leu Glu His Val865 870 875
880Gln Thr Arg Leu Arg Glu Ile Gln Pro Cys Leu Ala Glu Ser Arg Ala
885 890 895Ala Arg Glu Lys
Glu Ser Phe Asn Leu Lys Arg Ala Gln Glu Asp Ile 900
905 910Ser Arg Leu Arg Arg Lys Leu Glu Lys Gln Arg
Lys Val Glu Val Tyr 915 920 925Ala
Asp Ala Asp Glu Ile Leu Gln Glu Glu Ile Lys Glu Tyr Lys Ala 930
935 940Arg Leu Thr Cys Pro Cys Cys Asn Thr Arg
Lys Lys Asp Ala Val Leu945 950 955
960Thr Lys Cys Phe His Val Phe Cys Phe Glu Cys Val Arg Gly Arg
Tyr 965 970 975Glu Ala Arg
Gln Arg Lys Cys Pro Lys Cys Asn Ala Ala Phe Gly Ala 980
985 990His Asp Phe His Arg Ile Tyr Ile Ser
995 100054789PRTOryza sativaMISC_FEATURENP922769.1 in
Fig. 10 54Met Asp Ala Ala Ala Leu Gln Tyr Glu Asn Gln Lys Leu Val Gln
Gln1 5 10 15Leu Glu Ala
Gln Lys Ser Lys Met Arg Ala Leu Glu Gly Lys Phe Lys 20
25 30Glu Leu Arg Asp Glu Gln Cys Ser Tyr Asp
Asn Thr Leu Ile Cys Leu 35 40
45Asn Lys Met Trp Asn Gln Leu Ile Asp Asp Leu Val Leu Leu Gly Val 50
55 60Arg Ala Gly Gly Asp Leu Asn Gly Leu
Gln Ala Leu Asp His Glu Glu65 70 75
80Met Ser Glu Glu Ser Leu Glu Ser Cys Pro Ser Glu Glu Ile
Phe Leu 85 90 95Phe Arg
Leu Leu Asn Ser Arg Asn Phe Arg Asn Asn Asp Asp Ser Ser 100
105 110Leu Ser Lys Leu Val Glu Glu Ala Leu
Ala Leu Arg Tyr Ser Thr Thr 115 120
125Val Thr Leu Met Lys Ser Leu Gln Glu Ala Phe Ala Val Gln Gln Ala
130 135 140Arg Ser Glu Ser Leu Ser Leu
Ala Leu Asn Gly Gln Asn Ser Ser Glu145 150
155 160Asp Val Ile Val Ala Leu Glu Asn His Asn Asp Tyr
Leu Lys Glu Val 165 170
175Val Asp Asn Leu Arg Gln Ala Val Ser Ile Ile Asn Arg Lys His Glu
180 185 190Lys Tyr Leu Asp Glu Ile
Glu Ala Phe Lys Asn Asn Gln Ser Arg Glu 195 200
205Leu His Glu Val Lys Cys Leu Ser Gly Glu Leu Glu Glu Ser
Met Ala 210 215 220Glu Leu Glu Glu Ser
Arg Arg Lys Leu Ala Val Leu Gln Leu Gln Thr225 230
235 240Gly Gly Gly Ser Leu Met Asn Thr Ser Ala
Pro Asn Gly Val Asn Gly 245 250
255Ser Val Ser Thr Asp Lys Ser Ser Asp Lys Gly Met Gly Trp Arg Asp
260 265 270Leu Lys Asp Ala Val
Glu Glu Ala Lys Thr Leu Ala Ala Asn Arg Leu 275
280 285Phe Glu Leu His Glu Thr Gln Glu Asp Asn Leu Ile
Leu Ser Lys Gln 290 295 300Leu Glu Asp
Ile Gln Asp Gln Leu Lys Asp Glu Asn Tyr Ile Val Thr305
310 315 320Ser Lys Pro Tyr Thr Ile Leu
Ser Asp Gln Leu His His Leu Asn Ala 325
330 335Glu Ile Glu Arg Tyr Arg Gly Leu Val Glu Val Leu
Gln Ala Lys Ile 340 345 350Glu
Asp Leu Glu His Glu Ile Gln Lys Leu Met Ala Glu Lys Asn Asp 355
360 365Leu Glu Ile Lys Ala Glu Glu Ala Leu
Gln Asp Ser Gly Lys Lys Asp 370 375
380Phe Lys Asp Glu Ile His Val Met Ala Ala Ser Leu Ser Lys Glu Met385
390 395 400Glu Leu Leu Asp
Asn Gln Met Asn Arg Ser Lys Asp Ala Ala Ser Glu 405
410 415Ala Leu Ala Leu Arg Glu Glu Ala Asp Tyr
Leu Arg Thr Leu Leu Ala 420 425
430Lys Lys Ile Glu Thr Leu Asp Gln Glu Lys Gln Glu Leu Gln Phe Ile
435 440 445Val Asp Met Leu Gly Lys Glu
Cys Ser Glu Ser Arg Ala Ile Ser Glu 450 455
460Ile Glu Glu Ser Glu Asn Arg Ala Arg Lys Gln Ala Glu Tyr Leu
Arg465 470 475 480Lys Cys
Leu Glu Glu His Asn Leu Glu Leu Arg Val Lys Ala Ala Asn
485 490 495Glu Ala Glu Thr Ala Cys Gln
Gln Arg Leu Ser Ile Ala Glu Ala Glu 500 505
510Leu Glu Asp Leu Arg Ala Lys Val Asp Ala Ser Glu Arg Asp
Val Met 515 520 525Lys Leu Lys Glu
Ser Ile Arg Ile Lys Glu Ala Glu Val Asp Gly His 530
535 540Ile Ser Glu Ile Glu Thr Ile Gly Gln Ala Tyr Glu
Asp Met Gln Thr545 550 555
560Gln Asn Gln His Leu Leu Gln Gln Val Ala Asp Arg Asp Asp Phe Asn
565 570 575Ile Lys Leu Val Ser
Asp Ser Val Lys Met Lys Gln Ala Tyr Gly Ser 580
585 590Leu Leu Ala Glu Lys Asn Met Leu Gln Lys Gln Leu
Gln His Val Asn 595 600 605Ser Ser
Leu Glu Ser Ser Lys Leu Lys Ile Thr Ser Gly Glu Glu Gln 610
615 620Met Lys Thr Tyr Val Ala Gln Ala Met Lys Ser
Ser Ser Glu Asn Arg625 630 635
640His Leu Ala Ile Ser Leu Glu Arg Thr Met Leu Glu Val Ser Asp Ala
645 650 655Glu Lys Glu Leu
Lys Trp Leu Arg Ser Ala Thr Gly Ser Ala Glu Lys 660
665 670Glu Tyr Glu Ile Asn Gln Lys Lys Ile Ala Glu
Leu Lys Met Glu Leu 675 680 685Glu
Arg Glu Arg Asn Glu Arg Ile Lys Leu Glu Glu Glu Tyr Glu Glu 690
695 700Val Lys Asn Glu Val Ser Glu Leu Thr Ser
Glu Thr Glu Glu Thr Thr705 710 715
720Ile Gln Lys Leu Gln Asp Glu Ile Lys Glu Cys Lys Ala Ile Leu
Lys 725 730 735Cys Gly Val
Cys Phe Asp Arg Pro Lys Glu Val Val Ile Thr Lys Cys 740
745 750Phe His Leu Phe Cys Ser Pro Cys Ile Gln
Arg Asn Leu Glu Ile Arg 755 760
765His Arg Lys Cys Pro Gly Cys Gly Thr Pro Phe Gly Gln Ser Asp Val 770
775 780Arg Glu Val Lys Ile785
User Contributions:
Comment about this patent or add new information about this topic: