Patent application title: Polysaccharide Synthases
Inventors:
Rachel Anita Burton (Panorama, AU)
Geoffrey Bruce Fincher (Hazelwood Park, AU)
Antony Bacic (Victoria, AU)
Assignees:
ADELAIDE RESEARCH & INNOVATION PTY LTD
GRAINS RESEARCH & DEVELOPMENT CORPORATION
The University of Melboume
IPC8 Class: AA01H500FI
USPC Class:
800298
Class name: Multicellular living organisms and unmodified parts thereof and related processes plant, seedling, plant seed, or plant part, per se higher plant, seedling, plant seed, or plant part (i.e., angiosperms or gymnosperms)
Publication date: 2008-12-11
Patent application number: 20080307549
Claims:
1-46. (canceled)
47. A method for modulating the level and/or activity of a (1,3;1,4)-.beta.-D-glucan synthase in a cell, the method comprising modulating the expression of a (1,3;1,4)-.beta.-D-glucan synthase encoding nucleic acid in the cell.
48. A method according to claim 47 wherein the (1,3;1,4)-.beta.-D-glucan synthase encoding nucleic acid is a CslF gene or functional homolog thereof.
49. A method according to claim 47 wherein modulating the expression of a (1,3;1,4)-.beta.-D-glucan synthase encoding nucleic acid in the cell modulates the level of (1,3;1,4)-.beta.-D-glucan produced by the cell.
50. A method according to claim 49, wherein the method is a method for producing (1,3;1,4)-.beta.-D-glucan and modulating the expression of a (1,3;1,4)-.beta.-D-glucan synthase encoding nucleic acid in the cell comprises transforming the cell with an isolated (1,3;1,4)-.beta.-D-glucan synthase encoding nucleic acid and expression of the (1,3;1,4)-.beta.-D-glucan synthase encoding nucleic acid in the cell effects and/or increases production of (1,3;1,4)-.beta.-D-glucan by the cell.
51. (1,3;1,4)-.beta.-D-glucan produced according to the method of claim 50.
52. A cell comprising any one or more of:(i) a modulated level of (1,3;1,4)-.beta.-D-glucan relative to a wild type cell of the same taxon;(ii) a modulated level and/or activity of (1,3;1,4)-.beta.-D-glucan synthase relative to a wild type cell of the same taxon; and/or(iii) modulated expression of a (1,3;1,4)-.beta.-D-glucan synthase encoding nucleic acid relative to a wild type cell of the same taxon.
53. A multicellular structure comprising one or more cells according to claim 52.
54. A multicellular structure according to claim 53 wherein the multicellular structure is selected from the list consisting of: a whole plant, a plant tissue, a plant organ, a plant part, plant reproductive material and/or cultured plant tissue.
55. A multicellular structure according to claim 54, wherein the multicellular structure comprises a cereal grain comprising an altered level of (1,3;1,4)-.beta.-D-glucan, wherein the grain comprises one or more cells comprising an altered level and/or activity of (1,3;1,4)-.beta.-D-glucan synthase and/or altered expression of a (1,3;1,4)-.beta.-D-glucan synthase encoding nucleic acid molecule.
56. Flour comprising flour produced by the milling of a grain according to claim 55 and, optionally, flour produced by the milling of one or more other grains.
57. An isolated nucleic acid molecule selected from the list consisting of:(i) an isolated nucleic acid molecule that encodes a (1,3;1,4)-.beta.-D-glucan synthase;(ii) an isolated nucleic acid molecule comprising the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9 and SEQ ID NO: 11;(iii) an isolated nucleic acid molecule comprising a nucleotide sequence which is at least 50% identical to the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9 and SEQ ID NO: 11;(iv) an isolated nucleic acid molecule comprising a nucleotide sequence which encodes the amino acid sequence set forth in any of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8, SEQ ID NO: 10 and SEQ ID NO: 12;(v) an isolated nucleic acid molecule comprising a nucleotide sequence which hybridises to any of (i) to (iv) under stringent conditions;(vi) an isolated nucleic acid molecule comprising a nucleotide sequence which is the complement or reverse complement of the nucleotide sequence of the isolated nucleic acid molecule of any of (i) to (v); and/or(vii) a fragment of any one of the isolated nucleic acid molecule of any of (i) to (vi).
58. A genetic construct or vector comprising an isolated nucleic acid molecule according to claim 57.
59. A cell comprising a genetic construct according to claim 58.
60. A multicellular structure which comprises one or more of cells according to claim 59.
61. The multicellular structure according to claim 60 wherein the multicellular structure is selected from the list consisting of a whole plant, a plant tissue, a plant organ, a plant part, plant reproductive material or cultured plant tissue.
62. An isolated polypeptide selected from the list consisting of:(i) an isolated polypeptide comprising an amino acid sequence encoding a (1,3;1,4)-.beta.-D-glucan synthase;(ii) an isolated polypeptide comprising the amino acid sequence set forth in any of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8, SEQ ID NO: 10 and SEQ ID NO: 12;(iii) an isolated polypeptide comprising an amino acid sequence comprising at least 50% identity to the amino acid sequence set forth in any of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8, SEQ ID NO: 10 and SEQ ID NO: 12;(iv) an isolated polypeptide comprising an amino acid sequence encoded by the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9 and SEQ ID NO: 11; and/or(v) a fragment of the isolated polypeptide of any one of (i) to (iv).
63. An antibody, or an epitope binding fragment thereof, raised against the isolated polypeptide or fragment thereof according to claim 62.
Description:
FIELD OF THE INVENTION
[0001]The present invention relates generally to polysaccharide synthases. More particularly, the present invention relates to (1,3;1,4)-β-D-glucan synthases.
BACKGROUND OF THE INVENTION
[0002]The various tissues of cereal grains have diverse functions during grain development, dormancy and after germination.
[0003]For example, the pericarp and seed coat tissues are concerned with the protection of the seed during development and during dormancy. However, by grain maturity, these outer grain tissues have died and the tissue residues consist almost entirely of cell wall residues. The nucellar tissue between the seed coat and the aleurone surface is involved in transfer of nutrients to the developing grain, however, at maturity, this tissue has also collapsed to leave cell wall remnants. The thin walled cells of the starchy endosperm of mature grain are dead, but are packed with starch and storage protein. In contrast, the thick-walled, nucleated, aleurone cells are alive at grain maturity, and are packed with protein bodies and lipid droplets. At the interface of the starchy endosperm lies the scutellum, which functions in delivering nutrients to the developing endosperm and, during germination, transfers digestion products of the endosperm reserves to the developing embryo.
[0004]The different structure and function of each tissue type in the grain are determined, at least in part, by the cell wall composition of each of these cell types.
[0005]Non-cellulosic polysaccharides are key components in the cell walls of cereal grain tissues and include, for example, (1,3;1,4)-β-D-glucans, heteroxylans (mainly arabinoxylans), glucomannans, xyloglucans, pectic polysaccharides and callose. These non-cellulosic polysaccharides usually constitute less than 10% of the overall weight of the grain, but nevertheless are key determinants of grain quality.
[0006]Although the precise physical relationships between individual non-cellulosic polysaccharides and other wall components have not been described, it is generally considered that in the wall, microfibrils of cellulose are embedded in a matrix phase of non-cellulosic polysaccharides and protein. Wall integrity is maintained predominantly through extensive non-covalent interactions, especially hydrogen bonding, between the matrix phase and microfibrillar constituents. In the walls of some grain tissues covalent associations between heteroxylans, lignin and proteins are present. The extent of covalent associations between components also varies with the wall type and genotype.
[0007]Non-cellulosic polysaccharides, especially heteroxylans and (1,3;1,4)-β-D-glucans, constitute a relatively high proportion of the walls of the aleurone and starchy endosperm, and probably also of the scutellum. In these tissues, cellulose contents are correspondingly lower. The generally low cellulose content of these walls, together with the fact that they contain no lignin, are thought to be related to a limited requirement for structural rigidity of walls in central regions of the grain, and to a requirement to rapidly depolymerize wall components following germination of the grain.
[0008]In contrast, in the cell walls of the pericarp-seed coat, which provides a protective coat for the embryo and endosperm and which is not mobilized during germination, cellulose and lignin contents are much higher and the concentrations of non-cellulosic polysaccharides are correspondingly lower.
[0009](1,3;1,4)-β-D-glucans, also referred to as mixed-linkage or cereal β-glucans, are non-cellulosic polysaccharides which naturally occur in plants of the monocotyledon family Poaceae, to which the cereals and grasses belong, and in related families of the order Poales.
[0010]These non-cellulosic polysaccharides are important constituents of the walls of the starchy endosperm and aleurone cells of most cereal grains, where they can account for up to 70%-90% by weight of the walls.
[0011]Barley, oat and rye grains are rich sources of (1,3;1,4)-β-D-glucan, whereas wheat, rice and maize have lower concentrations of this polysaccharide. The (1,3;1,4)-β-D-glucans are also relatively minor components of walls in vegetative tissues of cereals and grasses. Although present as a relatively minor component in vegetative tissues (1,3;1,4)-β-D-glucan) is still important in terms of, for example, the digestibility of vegetative tissue by animals and in the use of crop residues for bioethanol production.
[0012](1,3;1,4)-β-D-glucans are important in large-scale food processing activities that include brewing and stockfeed manufacture. Moreover, the non-starchy polysaccharides of cereals, such as (1,3;1,4)-β-D-glucans, have attracted renewed interest in recent years because of their potentially beneficial effects in human nutrition.
[0013]However, despite this interest, major gaps remain in our knowledge of the genes and enzymes that control non-cellulosic polysaccharide biosynthesis, including (1,3;1,4)-β-D-glucan biosynthesis, in cereal grain.
[0014](1,3;1,4)-β-D-glucan concentrations in grain are thought to be influenced by both genotype and environment. For example, the concentration of (1,3;1,4)-β-D-glucan in cereal grains depends on the genotype, the position of the grain on the spike and environmental factors such as planting location, climatic conditions during development and soil nitrogen.
[0015]However, the genes that contribute to (1,3;1,4)-β-D-glucan content in grain have not yet been identified.
[0016]The identification of genes encoding (1,3;1,4)-β-D-glucan synthases through traditional biochemical approaches has been seriously hampered by an inability to purify the enzymes to homogeneity. (1,3;1,4)-β-D-glucan synthases are membrane-bound and, therefore, are difficult to solubilise in an active form. In addition, (1,3;1,4)-β-D-glucan synthases rapidly lose activity following disruption of cells, and are likely to be present at very low abundance in the cell. Despite numerous attempts, purification of (1,3;1,4)-β-D-glucan synthases to homogeneity has not been achieved and, as a result, there are no reports of amino acid sequences obtained from the enzymes themselves. The inability to obtain even partial amino acid sequences from the purified (1,3;1,4)-β-D-glucan synthase enzyme has also prevented the identification and isolation of genes encoding (1,3;1,4)-β-D-glucan synthases.
[0017]However, identification of the genes encoding (1,3;1,4)-β-D-glucan synthases would be desirable, as this would facilitate modulation of the level of (1,3;1,4)-β-D-glucan produced by a cell, and therefore, allow the qualities of grain or vegetative tissue to be altered. Therefore, in order to enable the modulation of the level of (1,3;1,4)-β-D-glucan in a cell and associated changes in grain or vegetative tissue quality, there is a clear need to identify genes that encode (1,3;1,4)-β-D-glucan synthases.
[0018]Reference to any prior art in this specification is not, and should not be taken as, an acknowledgment or any form of suggestion that this prior art forms part of the common general knowledge in any country.
SUMMARY OF THE INVENTION
[0019]The present invention is predicated, in part, on the identification of genes which encode the biosynthetic enzyme for (1,3;1,4)-β-D-glucans, referred to herein as "(1,3;1,4)-β-D-glucan syntheses".
[0020]In accordance with the present invention, it has been revealed that (1,3;1,4)-β-D-glucan synthases are encoded by members of the CslF gene family.
[0021]As a result of the identification of the nucleotide sequences, and corresponding amino acid sequences that encode (1,3;1,4)-β-D-glucan synthases, the present invention provides, inter alia, methods and compositions for influencing the level and/or activity of (1,3;1,4)-β-D-glucan synthase in a cell and thereby the level of (1,3;1,4)-β-D-glucan produced by the cell.
[0022]Therefore, in a first aspect, the present invention provides a method for influencing the level of (1,3;1,4)-β-D-glucan produced by a cell, the method comprising modulating the level and/or activity of a (1,3;1,4)-β-D-glucan synthase in the cell.
[0023]In one particularly preferred embodiment, the cell is a plant cell, more preferably a monocot plant cell and most preferably a cereal crop plant cell.
[0024]In a second aspect, the present invention provides a method for modulating the level and/or activity of a (1,3;1,4)-β-D-glucan synthase in a cell, the method comprising modulating the expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in the cell.
[0025]In a third aspect, the present invention provides a method for modulating the level and/or activity of a (1,3;1,4)-β-D-glucan synthase in a cell, the method comprising modulating the expression of a CslF gene or functional homolog thereof in the cell.
[0026]In a fourth aspect, the present invention provides a method for producing (1,3;1,4)-β-D-glucan, the method comprising expressing a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in a cell.
[0027]In a fifth aspect, the present invention also provides (1,3;1,4)-β-D-glucan produced according to the method of the fourth aspect of the invention.
[0028]In a sixth aspect, the present invention provides a cell comprising any one or more of: [0029](i) a modulated level of (1,3;1,4)-β-D-glucan relative to a wild type cell of the same taxon; [0030](ii) a modulated level and/or activity of (1,3;1,4)-β-D-glucan synthase relative to a wild type cell of the same taxon; [0031](iii) modulated expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid relative to a wild type cell of the same taxon.
[0032]Furthermore, in a seventh aspect, the present invention provides a multicellular structure comprising one or more cells according to the sixth aspect of the invention.
[0033]As mentioned above, in one preferred embodiment of the invention, the cell is a plant cell and as such, the present invention includes a whole plant, plant tissue, plant organ, plant part, plant reproductive material or cultured plant tissue, comprising one or more plant cells according to the sixth aspect of the invention. In a more preferred embodiment, the present invention provides a cereal plant comprising one or more cells according to the sixth aspect of the invention. In a particularly preferred embodiment, the present invention provides cereal grain comprising one or more cells according to the sixth aspect of the invention.
[0034]Therefore, in an eighth aspect, the present invention provides a cereal grain comprising an altered level of (1,3;1,4)-β-D-glucan, wherein the grain comprises one or more cells comprising an altered level and/or activity of (1,3;1,4)-β-D-glucan synthase and/or altered expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid molecule.
[0035]In a ninth aspect, the present invention also provides flour comprising: [0036](i) flour produced by the milling of the grain of the eighth aspect of the invention; and [0037](ii) optionally, flour produced by the milling of one or more other grains.
[0038]As set out above, the present invention is predicated, in part, on the identification and isolation of nucleotide and amino acid sequences that encode (1,3;1,4)-β-D-glucan synthases.
[0039]Therefore, in a tenth aspect, the present invention provides an isolated nucleic acid molecule that encodes a (1,3;1,4)-β-D-glucan synthase.
[0040]In an eleventh aspect, the present invention also provides an isolated nucleic acid molecule comprising one or more of: [0041](i) the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9 and SEQ ID NO: 11; [0042](ii) a nucleotide sequence which is at least 50% identical to the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9 and SEQ ID NO: 11; [0043](iii) a nucleotide sequence which hybridises to a nucleic acid molecule comprising the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9 and SEQ ID NO: 11 under low stringency, more preferably medium stringency and most preferably high stringency conditions; [0044](iv) a nucleotide sequence which encodes the amino acid sequence set forth in any of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8, SEQ ID NO: 10 and SEQ ID NO: 12. [0045](v) a nucleotide sequence which is the complement of any one of (i) to (iv); [0046](vi) a nucleotide sequence which is the reverse complement of any one of (i) to (iv); [0047](vii) a fragment of any one of (i) to (vi).
[0048]In a twelfth aspect, the present invention provides a genetic construct or vector comprising an isolated nucleic acid molecule of the eleventh aspect of the invention.
[0049]In a thirteenth aspect, the present invention extends to a cell comprising the isolated nucleic acid molecule of the tenth or eleventh aspects of the invention or genetic construct of the twelfth aspect of the invention.
[0050]In a fourteenth aspect, the present invention provides a multicellular structure which comprises one or more of the cells of the thirteenth aspect of the invention.
[0051]As set out above, the present invention also provides amino acid sequences for (1,3;1,4)-β-D-glucan syntheses.
[0052]Accordingly, in a fifteenth aspect, the present invention provides an isolated polypeptide comprising an amino acid sequence encoding a (1,3;1,4)-β-D-glucan synthase protein.
[0053]In a sixteenth aspect, the present invention provides an isolated polypeptide comprising one or more of: [0054](i) the amino acid sequence set forth in any of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8, SEQ ID NO: 10 and SEQ ID NO: 12; [0055](ii) an amino acid sequence comprising at least 50% identity to the amino acid sequence set forth in any of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8, SEQ ID NO: 10 and SEQ ID NO: 12; [0056](iii) an amino acid sequence encoded by the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9 and SEQ ID NO: 11; and/or [0057](iv) a fragment of any one of (i), (ii) or (iii).
[0058]In a preferred embodiment, the isolated polypeptide of the present invention comprises an amino acid sequence defining a "(1,3;1,4)-β-D-glucan synthase" as hereinbefore defined.
[0059]As set out above, the sixteenth aspect of the invention also provides fragments of isolated polypeptides including (1,3;1,4)-β-D-glucan synthase epitopes.
[0060]The isolated polypeptides and (1,3;1,4)-β-D-glucan synthase epitope-bearing polypeptides of the sixteenth aspect of the invention are useful, for example, in the generation of antibodies that bind to the isolated (1,3;1,4)-β-D-glucan synthase proteins
[0061]Accordingly, in a seventeenth aspect, the present invention provides an antibody or an epitope binding fragment thereof, raised against an isolated (1,3;1,4)-β-D-glucan synthase protein as hereinbefore defined or an epitope thereof.
[0062]Throughout this specification, unless the context requires otherwise, the word "comprise", or variations such as "comprises" or "comprising", will be understood to imply the inclusion of a stated element or integer or group of elements or integers but not the exclusion of any other element or integer or group of elements or integers.
[0063]Nucleotide and amino acid sequences are referred to herein by a sequence identifier number (SEQ ID NO:). The SEQ ID NOs: correspond numerically to the sequence identifiers <400> 1 (SEQ ID NO: 1), <400> 2 (SEQ ID NO: 2), etc. A summary of the sequence identifiers is provided in Table 1. A sequence listing is provided at the end of the specification.
TABLE-US-00001 TABLE 1 Summary of Sequence Identifiers Sequence Identifier Sequence SEQ ID NO: 1 HvCslF1 coding region nucleotide sequence SEQ ID NO: 2 HvCslF1 amino acid sequence SEQ ID NO: 3 HvCslF2 coding region nucleotide sequence SEQ ID NO: 4 HvCslF2 amino acid sequence SEQ ID NO: 5 HvCslF3 coding region nucleotide sequence SEQ ID NO: 6 HvCslF3 amino acid sequence SEQ ID NO: 7 HvCslF4 coding region nucleotide sequence SEQ ID NO: 8 HvCslF4 amino acid sequence SEQ ID NO: 9 HvCslF5 coding region nucleotide sequence SEQ ID NO: 10 HvCslF5 amino acid sequence SEQ ID NO: 11 HvCslF6 coding region nucleotide sequence SEQ ID NO: 12 HvCslF6 amino acid sequence SEQ ID NO: 13 HvCslF1 genomic nucleotide sequence SEQ ID NO: 14 HvCslF2 genomic nucleotide sequence SEQ ID NO: 15 HvCslF3 genomic nucleotide sequence SEQ ID NO: 16 HvCslF4 genomic nucleotide sequence SEQ ID NO: 17 HvCslF5 genomic nucleotide sequence SEQ ID NO: 18 HvCslF6 genomic nucleotide sequence SEQ ID NO: 19 OsCslF1 nucleotide sequence SEQ ID NO: 20 OsCslF1 amino acid sequence SEQ ID NO: 21 OsCslF2 nucleotide sequence SEQ ID NO: 22 OsCslF2 amino acid sequence SEQ ID NO: 23 OsCslF3 nucleotide sequence SEQ ID NO: 24 OsCslF3 amino acid sequence SEQ ID NO: 25 OsCslF4 nucleotide sequence SEQ ID NO: 26 OsCslF4 amino acid sequence SEQ ID NO: 27 OsCslF5 nucleotide sequence SEQ ID NO: 28 OsCslF5 amino acid sequence SEQ ID NO: 29 OsCslF7 nucleotide sequence SEQ ID NO: 30 OsCslF7 amino acid sequence SEQ ID NO: 31 OsCslF8 nucleotide sequence SEQ ID NO: 32 OsCslF8 amino acid sequence SEQ ID NO: 33 OsCslF9 nucleotide sequence SEQ ID NO: 34 OsCslF9 amino acid sequence SEQ ID NO: 35 OsF2Bll5 oligonucleotide primer SEQ ID NO: 36 OsF2ML3 oligonucleotide primer SEQ ID NO: 37 OsF3Bll5 oligonucleotide primer SEQ ID NO: 38 OsF3ML3 oligonucleotide primer SEQ ID NO: 39 OsF4H5 oligonucleotide primer SEQ ID NO: 40 OsF4S3 oligonucleotide primer SEQ ID NO: 41 OsF8H5 oligonucleotide primer SEQ ID NO: 42 OsF8S3 oligonucleotide primer SEQ ID NO: 43 GAPDH At oligonucleotide primer (forward) SEQ ID NO: 44 GAPDH At oligonucleotide primer (reverse) SEQ ID NO: 45 Tubulin At oligonucleotide primer (forward) SEQ ID NO: 46 Tubulin At oligonucleotide primer (reverse) SEQ ID NO: 47 Actin At oligonucleotide primer (forward) SEQ ID NO: 48 Actin At oligonucleotide primer (reverse) SEQ ID NO: 49 Cyclophilin At oligonucleotide primer (forward) SEQ ID NO: 50 Cyclophilin At oligonucleotide primer (reverse) SEQ ID NO: 51 OsCslF2 oligonucleotide primer (forward) SEQ ID NO: 52 OsCslF2 oligonucleotide primer (reverse) SEQ ID NO: 53 OsCslF3 oligonucleotide primer (forward) SEQ ID NO: 54 OsCslF3 oligonucleotide primer (reverse) SEQ ID NO: 55 OsCslF4 oligonucleotide primer (forward) SEQ ID NO: 56 OsCslF4 oligonucleotide primer (reverse) SEQ ID NO: 57 OsCslF5 oligonucleotide primer (forward) SEQ ID NO: 58 OsCslF5 oligonucleotide primer (reverse) SEQ ID NO: 59 HvFD5END oligonucleotide primer SEQ ID NO: 60 HvFDRQ oligonucleotide primer SEQ ID NO: 61 HvFC5N oligonucleotide primer SEQ ID NO: 62 HvFC3N oligonucleotide primer SEQ ID NO: 63 HvFH5 oligonucleotide primer SEQ ID NO: 64 HvFF3N oligonucleotide primer SEQ ID NO: 65 Hyg oligonucleotide primer (forward) SEQ ID NO: 66 Hyg oligonucleotide primer (reverse) SEQ ID NO: 67 HvCslF1 oligonucleotide primer (forward) SEQ ID NO: 68 HvCslF1 oligonucleotide primer (reverse) SEQ ID NO: 69 HvCslF4 oligonucleotide primer (forward) SEQ ID NO: 70 HvCslF4 oligonucleotide primer (reverse) SEQ ID NO: 71 HvCslF6 oligonucleotide primer (forward) SEQ ID NO: 72 HvCslF6 oligonucleotide primer (reverse)
BRIEF DESCRIPTION OF THE FIGURES
[0064]FIG. 1 shows a region on chromosome 7 of rice which is syntenous to a region of barley chromosome 2H, where a cluster of six cellulose synthase-like (Csl) genes was detected within an interval of 119 Kb, corresponding to the 21.59-21.72 Mb region of the chromosome.
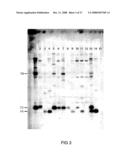
[0065]FIG. 2 shows a vector map of the pAJ22 vector used to express CslF genes in Arabidopsis.
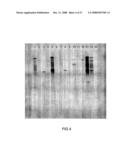
[0066]FIG. 3 is a Southern Blot showing XbaI and ScaI digested DNA derived from transformed Arabidopsis plants, which has been probed with fragments from OsCslF2, OsCslF4 and OsCslF8. The hybridizing fragments for these are marked on the figure as F2, F4 and F8. Track Numbers 1 to 14 are plant lines A2, A3, A7, A12, A16, A18, A21, A23, A28, A29, A31, A33, A41 and A42, respectively, while track 15 shows DNA derived from a wild-type Columbia plant.
[0067]FIG. 4 is a Southern Blot showing XbaI digested DNA derived from transformed Arabidopsis plants, which has been probed with a fragment of the BAR gene. Track Numbers 1 to 14 are plant lines A2, A3, A7, A12, A16, A18, A21, A23, A28, A29, A31, A33, A41 and A42, respectively, while track 15 shows DNA derived from a wild-type Columbia plant.
[0068]FIG. 5 shows normalized mRNA levels, as determined by Q-PCR, in the leaves of 14-day old transgenic Arabidopsis plant which express one or more of OsCslF2, OsCslF4 or OsCslF8.
[0069]FIG. 6 shows a ClustalW multiple sequence alignment of CslF amino acid sequences derived from Barley (Hordeum vulgare) and Rice (Oryza sativa).
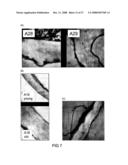
[0070]FIG. 7 shows transmission electron micrographs illustrating the detection of (1,3;1,4)-β-D-glucan in cell walls of several transgenic Arabidopsis plants with specific monoclonal antibodies. Panel A shows the detection of (1,3;1,4)-β-D-glucan in transformed Arabidopsis lines A28 and A29. In Panel B, walls from the epidermal layers of leaves from transgenic Arabidopsis line A18 are shown to accumulate (1,3;1,4)-β-D-glucan over a period of about fourteen days. Finally, Panel C shows a representative section of WT Arabidopsis leaf epidermal cell wall where minimal or no background labelling is commonly observed.
[0071]FIG. 8 shows the nucleotide sequence identity, protein sequence identity and protein sequence similarity between CslF sequences derived from Rice (Oryza sativa) and Barley (Hordeum vulgare)
[0072]FIG. 9 is a phylogenetic tree showing the relationship of complete and partial CslF amino acid sequences derived from Barley (Hordeum vulgare) and Rice (Oryza sativa).
[0073]FIG. 10 shows the location of HvCslF2, 4, 5 and 6 genes on chromosome 2H of the Steptoe×Morex (S×M 2H) Bin map. Key markers (as FIG. 1) are shown on the right-hand side and distances from the top of the chromosome in centimorgans are indicated on the left-hand side.
[0074]FIG. 11 shows a map of the pMDC32 vector
[0075]FIG. 12 shows the results of the QPCR analysis of hygromycin transcript levels in control and transgenic barley plants
[0076]FIG. 13 shows the results of the QPCR analysis of HvCslF1 transcript levels in control and transgenic barley plants.
[0077]FIG. 14 shows the results of the QPCR analysis of HvCslF4 transcript levels in control and transgenic barley plants
[0078]FIG. 15 shows the results of the QPCR analysis of HvCslF6 transcript levels in control and transgenic barley plants
[0079]FIG. 16 shows leaf autofluorescence under UV to demonstrate cell morphology. ab=abaxial surface, ad=adaxial surface, bs=bundle sheath cell, bul=bulliform cell, e=epidermal cell, m=mesophyll cell, p=phloem, scl=sclerenchyma fibre, st=stomate, x=xylem.
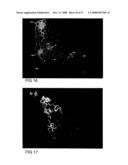
[0080]FIG. 17 shows G98-10 with both primary and secondary antibodies omitted from the labeling procedure, photographed at 7 seconds exposure under the 13 filter.
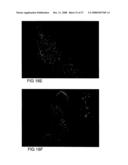
[0081]FIG. 18 shows transgenic plants (A,B,C,D) compared with control plants (E and F) all photographed at 7 seconds exposure under the I3 filter: A) G98-10 and B) G98-24, both showing increased fluorescence in the epidermal cells and the sclerenchyma fibre cells on the leaf tip, when compared with control sections; C) G103-5 showing increased fluorescence in all cell types when compared with the control sections; D) G99-12 showing increased fluorescence in stomata and vascular tissue when compared with the control sections; E) WT control showing fluorescence signal from endogenous (1,3;1,4)-β-D-glucans; F) transgene control G89-1 showing fluorescence from endogenous (1,3;1,4)-β-D-glucans.
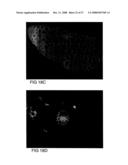
[0082]FIG. 19 shows transmission electron micrographs. Panel A shows a representative epidermal cell wall of the transgenic control G89-1, showing labeling of endogenous levels of (1,3;1,4)-β-D-glucan. Panel B shows a representative epidermal cell wall of the transgenic G98-10 showing significantly heavier labeling of (1,3;1,4)-β-D-glucan in the walls of these plants. Panel C shows a representative epidermal cell wall of transgenic G103-5 showing significantly heavier labeling of (1,3;1,4)-β-D-glucan in the walls of these plants.
[0083]FIG. 20 shows transmission electron micrographs. Panel A is a representative sclerenchyma fibre cell wall from the transgenic control G89-1 showing labeling of endogenous levels of (1,3;1,4)-β-D-glucan. Panel B shows a representative sclerenchyma fibre cell wall from the transgenic G98-10 showing heavier labeling of the (1,3;1,4)-β-D-glucan. Panel C shows a representative sclerenchyma fibre cell wall from the transgenic G103-5 showing heavier labeling of the (1,3;1,4)-β-D-glucan.
DESCRIPTION OF PREFERRED EMBODIMENTS
[0084]It is to be understood that following description is for the purpose of describing particular embodiments only and is not intended to be limiting with respect to the above description.
[0085]The present invention is predicated, in part, on the identification of genes which encode the biosynthetic enzyme for (1,3;1,4)-β-D-glucans, referred to herein as "(1,3;1,4)-β-D-glucan synthases".
[0086](1,3;1,4)-β-D-glucans" should be understood to include linear, unbranched polysaccharides in which β-D-glucopyranosyl monomers are polymerized through both (1→4)- and (1→3)-linkages.
[0087]The ratio of (1→4)- to (1→3)-linkages, in naturally occurring (1,3;1,4)-β-D-glucans, is generally in the range 2.2-2.6:1, although the ratio may also be outside of this range. For example, in the (1,3;1,4)-β-D-glucan from sorghum endosperm the ratio is 1.15:1. The two types of linkages are not arranged in regular, repeating sequences. Single (1→3)-linkages are separated by two or more (1→4)-linkages. Regions of two or three adjacent (1→4)-linkages predominate, but again there is no regularity in the arrangement of these units. The linkage sequence does not depend on preceding linkages further away than two glucose units and follows a second order Markov chain distribution. Moreover, up to 10% of the chain may consist of longer stretches of 5 to 20 adjacent (1→4)-linkages. Thus, cereal (1,3;1,4)-β-D-glucans may be considered as (1→3)-β-linked copolymers of cellotriosyl (G4G4GRed), cellotetraosyl (G4G4G4GRed) units and longer (1→4)-β-D-oligoglucosyl units.
[0088]The ratio of tri- to tetra-saccharide units in endogenous (1,3;1,4)-β-D-glucans varies between cereal species. For example, in wheat the ratio is 3.0-4.5:1, in barley 2.9-3.4:1, in rye 2.7:1 and in oats 1.8-2.3:1. Furthermore, the observed ratios may also vary according to the temperature and conditions of (1,3;1,4)-β-D-glucan extraction.
[0089]The average molecular masses reported for cereal (1,3;1,4)-β-D-glucans range from 48,000 (DP ˜300) to 3,000,000 (DP ˜1850), depending on the cereal species, cell wall type, extraction procedure and the method used for molecular mass determination. They are invariably polydisperse with respect to molecular mass and this is illustrated by a weight average to number average molecular mass ratio (Mw/Mn) of 1.18 for barley (1,3;1,4)-β-D-glucan. Certain barley (1,3;1,4)-β-D-glucans are also covalently-associated with small amounts of protein and have estimated molecular masses of up to 40,000,000.
[0090]The extractability of (1,3;1,4)-β-D-glucans from walls of cereal grains is a function of their degree of self-association and their association with other wall polysaccharides and proteins. In particular, extractability depends on the molecular mass and linkage distribution in the (1,3;1,4)-β-D-glucan chains. Extensive association with other polymers and very high molecular masses render the (1,3;1,4)-β-D-glucans more difficult to extract from grain.
[0091]For example, a portion of the (1,3;1,4)-β-D-glucan from barley, oat and rye flours may be extracted by water at pH 7.0 and 40° C. Further fractions can be solubilized at higher temperatures. The proportion of total (1,3;1,4)-β-D-glucan that is water-soluble at 40° C. varies within and between species. For example, waxy (high amylose) barleys have a higher proportion of water-soluble (1,3;1,4)-β-D-glucan than normal barleys. (1,3;1,4)-β-D-glucans extracted from barley at 40° C. have a slightly lower tri-/tetrasaccharide ratio (1.7:1) than those extracted at 65° C. (2.0:1). Complete extraction of cereal (1,3;1,4)-β-D-glucans from grain requires the use of alkaline extractants such as 4 M NaOH or aqueous Ba(OH)2, containing NaBH4 to prevent alkali-induced degradation from the reducing terminus. Alkali-extracted barley (1,3;1,4)-β-D-glucan fractions have higher molecular masses, higher ratios of (1→4):(1→3) linkages, more contiguously linked (1→4)-linked segments and higher tri-: tetra-saccharide ratios than their water-extractable counterparts. Other extractants, such as dimethylsulphoxide, hot perchloric acid, trichloroacetic acid, N-methylmorpholino-N-oxide and dimethylacetamide-LiCl, may also be used to solubilize (1,3;1,4)-β-D-glucans, but these extractants may cause some depolymerisation or degradation of the polymer. Once extracted with hot water or alkali, the (1,3;1,4)-β-D-glucans are often soluble at neutral pH and room temperature. However, upon cooling, (1,3;1,4)-β-D-glucans can aggregate and precipitate.
[0092]As mentioned above, the present invention is predicated, in part, on the identification of the biosynthetic enzyme, and encoding gene, that catalyses the synthesis of (1,3;1,4)-β-D-glucan. As used herein, this enzyme is referred to herein as "(1,3;1,4)-β-D-glucan synthase".
[0093]The present invention arises, in part, from an analysis of expressed sequence tag libraries and other sequence databases including cellulose synthase (CesA) genes. More particularly, it was noted in these analyses that the CesA genes were in fact members of a much larger super-family of genes, which included both the CesA genes and the cellulose synthase-like (Csl) gene family.
[0094]However, despite significant research effort, the particular functions of individual Csl genes are largely unknown. The Csl genes have been sub-divided into eight groups, designated CslA-CslH. However, the only Csl gene for which a specific biochemical function has been defined are CslA genes from guar and Arabidopsis, which encodes (1→4)-β-D-mannan synthases.
[0095]Given the similarities in structures of cellulose and (1,3;1,4)-β-D-glucan, the present inventors postulated that genes encoding (1,3;1,4)-β-D-glucan synthases might be members of the Csl gene family.
[0096]However, the Csl gene families in most vascular plants are very large and have been divided into several groups, designated CslA to CslH. In Arabidopsis thaliana there are 29 known Csl genes and in rice about 37. Overall, the Arabidopsis genome is believed to contain more than 700 genes involved in cell wall metabolism. However, in general, the specific functions of these genes are poorly understood. For example, the specific functions of only two of more than 170 genes involved in pectin biosynthesis have been defined. Furthermore, in contrast to the CesA genes, it has proved difficult to define the functions of the Csl genes. In fact, of the multiple Csl genes in higher plants, only the CslA group has been assigned a function.
[0097]The present invention used a genetic approach to identify the nucleotide sequences, and corresponding amino acid sequences, that encode (1,3;1,4)-β-D-glucan synthase. In accordance with the present invention, it has been revealed that (1,3;1,4)-β-D-glucan synthases are encoded by members of the CslF gene family.
[0098]As a result of the identification of the nucleotide sequences, and corresponding amino acid sequences that encode (1,3;1,4)-β-D-glucan synthases, the present invention provides, inter alia, methods and compositions for influencing the level and/or activity of (1,3;1,4)-β-D-glucan synthase in a cell and thereby the level of (1,3;1,4)-β-D-glucan produced by the cell.
[0099]Therefore, in a first aspect, the present invention provides a method for influencing the level of (1,3;1,4)-β-D-glucan produced by a cell, the method comprising modulating the level and/or activity of a (1,3;1,4)-β-D-glucan synthase in the cell.
[0100]The "cell" may be any suitable eukaryotic or prokaryotic cell. As such, a "cell" as referred to herein may be a eukaryotic cell including a fungal cell such as a yeast cell or mycelial fungus cell; an animal cell such as a mammalian cell or an insect cell; or a plant cell. Alternatively, the cell may also be a prokaryotic cell such as a bacterial cell including an E. coli cell, or an archaea cell.
[0101]Preferably, the cell is a plant cell, more preferably a vascular plant cell, including a monocotyledonous or dicotyledonous angiosperm plant cell or a gymnosperm plant cell. In an even more preferred embodiment, the plant is a monocotyledonous plant cell.
[0102]In one particularly preferred embodiment, the monocotyledonous plant cell is a cereal crop plant cell.
[0103]As used herein, the term "cereal crop plant" includes members of the Poales (grass family) that produce edible grain for human or animal food. Examples of Poales cereal crop plants which in no way limit the present invention include wheat, rice, maize, millets, sorghum, rye, triticale, oats, barley, teff, wild rice, spelt and the like. However, the term cereal crop plant should also be understood to include a number of non-Poales species that also produce edible grain and are known as the pseudocereals, such as amaranth, buckwheat and quinoa.
[0104]Although cereal crop plants are particularly preferred monocotyledonous plants, the other monocotyledonous plants are also preferred, such as other non-cereal plants of the Poales, specifically including pasture grasses such as Lolium spp.
[0105]As set out above, the present invention is predicated, in part, on modulating the level and/or activity of (1,3;1,4)-β-D-glucan synthase in a cell.
[0106](1,3;1,4)-β-D-glucan synthase" should be regarded as any protein which catalyses the synthesis of (1,3;1,4)-β-D-glucan and, optionally, catalyses the polymerisation of glucopyranosyl monomers.
[0107]Preferably, the (1,3;1,4)-β-D-glucan synthase comprises the amino acid sequence set forth in any of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 20, SEQ ID NO: 22, SEQ ID NO: 24, SEQ ID NO: 26, SEQ ID NO: 28, SEQ ID NO: 30, SEQ ID NO: 32 and SEQ ID NO: 34, or an amino acid sequence which is at least 40% identical thereto.
[0108]More preferably, the (1,3;1,4)-β-D-glucan synthase comprises at least 50% amino acid sequence identity, yet more preferably at least 60% amino acid sequence identity, even more preferably at least 70% amino acid sequence identity, and even more preferably at least 80% amino acid sequence identity and most preferably at least 90% amino acid sequence identity to any SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 20, SEQ ID NO: 22, SEQ ID NO: 24, SEQ ID NO: 26, SEQ ID NO: 28, SEQ ID NO: 30, SEQ ID NO: 32 and SEQ ID NO: 34. In a particularly preferred embodiment, the (1,3;1,4)-β-D-glucan synthase comprises the amino acid sequence set forth in any of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 20, SEQ ID NO: 22, SEQ ID NO: 24, SEQ ID NO: 26, SEQ ID NO: 28, SEQ ID NO: 30, SEQ ID NO: 32 and SEQ ID NO: 34.
[0109]When comparing amino acid sequences, the compared sequences should be compared over a comparison window of at least 100 amino acid residues, more preferably at least 200 amino acid residues, yet more preferably at least 400 amino acid residues, even more preferably at least 800 amino acid residues and most preferably over the full length of any of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 20, SEQ ID NO: 22, SEQ ID NO: 24, SEQ ID NO: 26, SEQ ID NO: 28, SEQ ID NO: 30, SEQ ID NO: 32 and SEQ ID NO: 34. The comparison window may comprise additions or deletions (i.e. gaps) of about 20% or less as compared to the reference sequence (which does not comprise additions or deletions) for optimal alignment of the two sequences. Optimal alignment of sequences for aligning a comparison window may be conducted by computerized implementations of algorithms such the BLAST family of programs as, for example, disclosed by Altschul et al., (Nucl. Acids Res. 25: 3389-3402, 1997). A detailed discussion of sequence analysis can be found in Unit 19.3 of Ausubel et al., ("Current Protocols in Molecular Biology" John Wiley & Sons Inc, 1994-1998, Chapter 15, 1998).
[0110]In a more preferred embodiment, the (1,3;1,4)-β-D-glucan synthase is encoded by a CslF gene or a functional homolog thereof (as defined later).
[0111]As referred to herein, the modulation of the "level" of the (1,3;1,4)-β-D-glucan synthase should be understood to include modulation of the level of (1,3;1,4)-β-D-glucan synthase transcripts and/or polypeptides in the cell. Modulation of the "activity" of the (1,3;1,4)-β-D-glucan synthase should be understood to include modulation of the total activity, specific activity, half-life and/or stability of the (1,3;1,4)-β-D-glucan synthase in the cell.
[0112]By "modulating" with regard to the level and/or activity of the (1,3;1,4)-β-D-glucan synthase is intended decreasing or increasing the level and/or activity of (1,3;1,4)-β-D-glucan synthase in the cell. By "decreasing" is intended, for example, a 1%, 5%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 100% reduction in the level or activity of (1,3;1,4)-β-D-glucan synthase in the cell. By "increasing" is intended, for example, a 1%, 5%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 100%, 2-fold, 5-fold, 10-fold, 20 fold, 50-fold, 100-fold increase in the level of activity of (1,3;1,4)-β-D-glucan synthase in the cell. "Modulating" also includes introducing a (1,3;1,4)-β-D-glucan synthase into a cell which does not normally express the introduced enzyme, or the substantially complete inhibition of (1,3;1,4)-β-D-glucan synthase activity in a cell that normally has such activity.
[0113]In one preferred embodiment, the level of (1,3;1,4)-β-D-glucan produced by a cell is increased by increasing the level and/or activity of (1,3;1,4)-β-D-glucan synthase in the cell. In another preferred embodiment, the level of (1,3;1,4)-β-D-glucan produced by a cell is decreased by decreasing the level and/or activity of (1,3;1,4)-β-D-glucan synthase in the cell.
[0114]The methods of the present invention contemplates any means known in the art by which the level and/or activity of (1,3;1,4)-β-D-glucan synthase in a cell may be modulated. This includes methods such as the application of agents which modulate (1,3;1,4)-β-D-glucan synthase activity in a cell, such as the application of a (1,3;1,4)-β-D-glucan synthase agonist or antagonist; the application of agents which mimic (1,3;1,4)-β-D-glucan synthase activity in a cell; modulating the expression of a nucleic acid which encodes (1,3;1,4)-β-D-glucan synthase in the cell; or effecting the expression of an altered or mutated (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in a cell such that a (1,3;1,4)-β-D-glucan synthase with increased or decreased specific activity, half-life and/or stability is expressed by the cell.
[0115]In a preferred embodiment, the level and/or activity of the (1,3;1,4)-β-D-glucan synthase is modulated by modulating the expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in the cell.
[0116]Therefore, in a second aspect, the present invention provides a method for modulating the level and/or activity of a (1,3;1,4)-β-D-glucan synthase in a cell, the method comprising modulating the expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in the cell.
[0117]As described herein, it has been identified that (1,3;1,4)-β-D-glucan synthase is encoded by members of the CslF gene family. Therefore, In a preferred embodiment, the (1,3;1,4)-β-D-glucan synthase encoding nucleic acid is a CslF gene or a functional homolog thereof.
[0118]Accordingly, in a third aspect, the present invention provides a method for modulating the level and/or activity of a (1,3;1,4)-β-D-glucan synthase in a cell, the method comprising modulating the expression of a CslF gene or functional homolog thereof in the cell.
[0119]As used herein, the term "CslF gene or functional homolog thereof" should be understood to include to a nucleic acid molecule which: [0120](i) encodes a (1,3;1,4)-β-D-glucan synthase as defined herein; and [0121](ii) preferably, comprises at least 50% nucleotide sequence identity to the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 19, SEQ ID NO: 21, SEQ ID NO: 23, SEQ ID NO: 25, SEQ ID NO: 27, SEQ ID NO: 29, SEQ ID NO: 31 and SEQ ID NO: 33; and/or [0122](iii) preferably, hybridises to a nucleic acid molecule comprising the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 19, SEQ ID NO: 21, SEQ ID NO: 23, SEQ ID NO: 25, SEQ ID NO: 27, SEQ ID NO: 29, SEQ ID NO: 31 and SEQ ID NO: 33 under stringent conditions.
[0123]More preferably, the CslF gene functional homolog thereof comprises a nucleotide sequence which is at least 54% identical to any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 19, SEQ ID NO: 21, SEQ ID NO: 23, SEQ ID NO: 25, SEQ ID NO: 27, SEQ ID NO: 29, SEQ ID NO: 31 and SEQ ID NO: 33, more preferably the CslF gene or functional homolog thereof comprises a nucleotide sequence which is at least 70% identical to any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 19, SEQ ID NO: 21, SEQ ID NO: 23, SEQ ID NO: 25, SEQ ID NO: 27, SEQ ID NO: 29, SEQ ID NO: 31 and SEQ ID NO: 33 and most preferably the CslF gene or functional homolog thereof comprises a nucleotide sequence which is at least 85% identical to any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 19, SEQ ID NO: 21, SEQ ID NO: 23, SEQ ID NO: 25, SEQ ID NO: 27, SEQ ID NO: 29, SEQ ID NO: 31 and SEQ ID NO: 33.
[0124]In a particularly preferred embodiment, the CslF gene or functional homolog thereof comprises the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 19, SEQ ID NO: 21, SEQ ID NO: 23, SEQ ID NO: 25, SEQ ID NO: 27, SEQ ID NO: 29, SEQ ID NO: 31 and SEQ ID NO: 33.
[0125]When comparing nucleic acid sequences to any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 19, SEQ ID NO: 21, SEQ ID NO: 23, SEQ ID NO: 25, SEQ ID NO: 27, SEQ ID NO: 29, SEQ ID NO: 31 and SEQ ID NO: 33 to calculate a percentage identity, the compared nucleotide sequences should be compared over a comparison window of at least 300 nucleotide residues, more preferably at least 600 nucleotide residues, yet more preferably at least 1200 nucleotide residues, even more preferably at least 2400 nucleotide residues and most preferably over the full length of SEQ ID NO: 1. The comparison window may comprise additions or deletions (ie. gaps) of about 20% or less as compared to the reference sequence (which does not comprise additions or deletions) for optimal alignment of the two sequences. Optimal alignment of sequences for aligning a comparison window may be conducted by computerized implementations of algorithms such the BLAST family of programs as, for example, disclosed by Altschul et al. (Nucl. Acids Res. 25: 3389-3402, 1997). A detailed discussion of sequence analysis can be found in Unit 19.3 of Ausubel et al., ("Current Protocols in Molecular Biology" John Wiley & Sons Inc, 1994-1998, Chapter 15, 1998).
[0126]As set out above, the CslF gene or functional homolog thereof may also comprise a nucleic acid, which hybridises to a nucleic acid molecule comprising the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 19, SEQ ID NO: 21, SEQ ID NO: 23, SEQ ID NO: 25, SEQ ID NO: 27, SEQ ID NO: 29, SEQ ID NO: 31 and SEQ ID NO: 33, under stringent conditions. As used herein, "stringent" hybridisation conditions will be those in which the salt concentration is less than about 1.5 M Na ion, typically about 0.01 to 1.0 M Na ion concentration (or other salts) at pH 7.0 to 8.3 and the temperature is at least 30° C. Stringent conditions may also be achieved with the addition of destabilizing agents such as formamide. Stringent hybridisation conditions may be low stringency conditions, more preferably medium stringency conditions and most preferably high stringency conditions. Exemplary low stringency conditions include hybridisation with a buffer solution of 30 to 35% formamide, 1 M NaCl, 1% SDS (sodium dodecyl sulphate) at 37° C., and a wash in 1× to 2×SSC (20×SSC=3.0 M NaCl/0.3 M trisodium citrate) at 50 to 55° C. Exemplary moderate stringency conditions include hybridisation in 40 to 45% formamide, 1.0 M NaCl, 1% SDS at 37° C., and a wash in 0.5× to 1×SSC at 55 to 60° C. Exemplary high stringency conditions include hybridisation in 50% formamide, 1 M NaCl, 1% SDS at 37° C., and a wash in 0.1×SSC at 60 to 65° C. Optionally, wash buffers may comprise about 0.1% to about 1% SDS. Duration of hybridization is generally less than about 24 hours, usually about 4 to about 12 hours.
[0127]Specificity of hybridisation is typically the function of post-hybridization washes, the critical factors being the ionic strength and temperature of the final wash solution. For DNA-DNA hybrids, the Tm can be approximated from the equation of Meinkoth and Wahl (Anal. Biochem. 138: 267-284, 1984), ie. Tm=81.5° C.+16.6 (log M)+0.41 (% GC)-0.61 (% form)-500/L; where M is the molarity of monovalent cations, % GC is the percentage of guanosine and cytosine nucleotides in the DNA, % form is the percentage of formamide in the hybridization solution, and L is the length of the hybrid in base pairs. The Tm is the temperature (under defined ionic strength and pH) at which 50% of a complementary target sequence hybridizes to a perfectly matched probe. Tm is reduced by about 1° C. for each 1% of mismatching; thus, Tm, hybridization, and/or wash conditions can be adjusted to hybridize to sequences of different degrees of complementarity. For example, sequences with >90% identity can be hybridised by decreasing the Tm by about 10° C. Generally, stringent conditions are selected to be about 5° C. lower than the thermal melting point (Tm) for the specific sequence and its complement at a defined ionic strength and pH. However, high stringency conditions can utilize a hybridization and/or wash at, for example, 1, 2, 3, or 4° C. lower than the thermal melting point (Tm); medium stringency conditions can utilize a hybridization and/or wash at, for example, 6, 7, 8, 9, or 10° C. lower than the thermal melting point (Tm); low stringency conditions can utilize a hybridization and/or wash at, for example, 11, 12, 13, 14, 15, or 20° C. lower than the thermal melting point (Tm). Using the equation, hybridization and wash compositions, and desired Tm, those of ordinary skill will understand that variations in the stringency of hybridization and/or wash solutions are inherently described. If the desired degree of mismatching results in a Tm of less than 45° C. (aqueous solution) or 32° C. (formamide solution), it is preferred to increase the SSC concentration so that a higher temperature can be used. An extensive guide to the hybridization of nucleic acids is found in Tijssen (Laboratory Techniques in Biochemistry and Molecular Biology-Hybridization with Nucleic Acid Probes, Pt I, Chapter 2, Elsevier, N.Y., 1993), Ausubel et al., eds. (Current Protocols in Molecular Biology, Chapter 2, Greene Publishing and Wiley-Interscience, New York, 1995) and Sambrook et al. (Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory Press, Plainview, N.Y., 1989).
[0128]The CslF gene or functional homolog thereof may also comprise a genomic nucleotide sequence from an organism which may include one or more non-protein-coding regions or one or more intronic regions. Exemplary genomic nucleotide sequences which comprise a CslF gene including the Hordeum vulgare genomic nucleotide sequences set forth in any of SEQ ID NO: 13, SEQ ID NO: 14, SEQ ID NO: 15, SEQ ID NO: 16, SEQ ID NO: 17 and SEQ ID NO: 18.
[0129]As mentioned above, the present invention provides methods for modulating the expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in a cell. The present invention contemplates any method by which the expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid molecule in a cell may be modulated.
[0130]Preferably, the term "modulating" with regard to the expression of the (1,3;1,4)-β-D-glucan synthase encoding nucleic acid is intended decreasing or increasing the transcription and/or translation of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid. By "decreasing" is intended, for example, a 1%, 5%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 100% reduction in the transcription and/or translation of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid. By "increasing" is intended, for example a 1%, 5%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 100%, 2-fold, 5-fold, 10-fold, 20-fold, 50-fold, 100-fold or greater increase in the transcription and/or translation of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid. Modulating also comprises introducing expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid not normally found in a particular cell; or the substantially complete inhibition (eg. knockout) of expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in a cell that normally has such activity.
[0131]Methods for modulating the expression of a particular nucleic acid molecule in a cell are known in the art and the present invention contemplates any such method. Exemplary methods for modulating the expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid include: genetic modification of the cell to upregulate or downregulate endogenous (1,3;1,4)-β-D-glucan synthase expression; genetic modification by transformation with a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid; administration of a nucleic acid molecule to the cell which modulates expression of an endogenous (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in the cell; and the like.
[0132]In one preferred embodiment, the expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid is modulated by genetic modification of the cell. The term "genetically modified", as used herein, should be understood to include any genetic modification that effects an alteration in the expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in the genetically modified cell relative to a non-genetically modified form of the cell. Exemplary types of genetic modification contemplated herein include: random mutagenesis such as transposon, chemical, UV and phage mutagenesis together with selection of mutants which overexpress or underexpress an endogenous (1,3;1,4)-β-D-glucan synthase encoding nucleic acid; transient or stable introduction of one or more nucleic acid molecules into a cell which direct the expression and/or overexpression of (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in the cell; inhibition of an endogenous (1,3;1,4)-β-D-glucan synthase by site-directed mutagenesis of an endogenous (1,3;1,4)-β-D-glucan synthase encoding nucleic acid; introduction of one or more nucleic acid molecules which inhibit the expression of an endogenous (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in the cell, eg. a cosuppression construct or an RNAi construct; and the like.
[0133]In one particularly preferred embodiment, the genetic modification comprises the introduction of a nucleic acid into a cell of interest.
[0134]The nucleic acid may be introduced using any method known in the art which is suitable for the cell type being used, for example, those described in Sambrook and Russell (Molecular Cloning-A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, 2000).
[0135]In preferred embodiments of the invention, wherein the cell is a plant cell, suitable methods for introduction of a nucleic acid molecule may include: Agrobacterium-mediated transformation, microprojectile bombardment based transformation methods and direct DNA uptake based methods. Roa-Rodriguez et al. (Agrobacterium-mediated transformation of plants, 3rd Ed. CAMBIA Intellectual Property Resource, Canberra, Australia, 2003) review a wide array of suitable Agrobacterium-mediated plant transformation methods for a wide range of plant species. Microprojectile bombardment may also be used to transform plant tissue and methods for the transformation of plants, particularly cereal plants, and such methods are reviewed by Casas et al. (Plant Breeding Rev. 13: 235-264, 1995). Direct DNA uptake transformation protocols such as protoplast transformation and electroporation are described in detail in Galbraith et al. (eds.), Methods in Cell Biology Vol. 50, Academic Press, San Diego, 1995). In addition to the methods mentioned above, a range of other transformation protocols may also be used. These include infiltration, electroporation of cells and tissues, electroporation of embryos, microinjection, pollen-tube pathway, silicon carbide- and liposome mediated transformation. Methods such as these are reviewed by Rakoczy-Trojanowska (Cell. Mol. Biol. Lett. 7: 849-858, 2002). A range of other plant transformation methods may also be evident to those of skill in the art.
[0136]The introduced nucleic acid may be single stranded or double stranded. The nucleic acid may be transcribed into mRNA and translated into (1,3;1,4)-β-D-glucan synthase or another protein; may encode for a non-translated RNA such as an RNAi construct, cosuppression construct, antisense RNA, tRNA, miRNA, siRNA, ntRNA and the like; or may act directly in the cell. The introduced nucleic acid may be an unmodified DNA or RNA or a modified DNA or RNA which may include modifications to the nucleotide bases, sugar or phosphate backbones but which retain functional equivalency to a nucleic acid. The introduced nucleic acid may optionally be replicated in the cell; integrated into a chromosome or any extrachromosomal elements of the cell; and/or transcribed by the cell. Also, the introduced nucleic acid may be either homologous or heterologous with respect to the host cell. That is, the introduced nucleic acid may be derived from a cell of the same species as the genetically modified cell (ie. homologous) or the introduced nucleic may be derived from a different species (ie. heterologous). The transgene may also be a synthetic transgene.
[0137]In one particularly preferred embodiment, the present invention contemplates increasing the level of (1,3;1,4)-β-D-glucan produced by a cell, by introducing a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid into the cell. More preferably, the (1,3;1,4)-β-D-glucan synthase encoding nucleic acid comprises a CslF gene or functional homolog thereof.
[0138]By identifying the nucleotide sequences which encode (1,3;1,4)-β-D-glucan synthases, in further embodiments the present invention also provides methods for down-regulating expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in a cell.
[0139]For example, the identification of (1,3;1,4)-β-D-glucan synthase encoding nucleic acid sequences, in accordance with the present invention, facilitates methods such as knockout or knockdown of an endogenous (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in a cell using methods such as: [0140](i) insertional mutagenesis of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in a cell including knockout or knockdown of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in a cell by homologous recombination with a knockout construct (for an example of targeted gene disruption in plants see Terada et al., Nat. Biotechnol. 20: 1030-1034, 2002); [0141](ii) post-transcriptional gene silencing (PTGS) or RNAi of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in a cell (for review of PTGS and RNAi see Sharp, Genes Dev. 15(5): 485-490, 2001; and Hannon, Nature 418: 244-51, 2002); [0142](iii) transformation of a cell with an antisense construct directed against a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid (for examples of antisense suppression in plants see van der Krol et al., Nature 333: 866-869; van der Krol et al., BioTechniques 6: 958-967; and van der Krol et al., Gen. Genet. 220: 204-212); [0143](iv) transformation of a cell with a co-suppression construct directed against a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid (for an example of co-suppression in plants see van der Krol et al., Plant Cell 2(4): 291-299); [0144](v) transformation of a cell with a construct encoding a double stranded RNA directed against a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid (for an example of dsRNA mediated gene silencing see Waterhouse et al., Proc. Natl. Acad. Sci. USA 95: 13959-13964, 1998); and [0145](vi) transformation of a cell with a construct encoding an siRNA or hairpin RNA directed against a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid (for an example of siRNA or hairpin RNA mediated gene silencing in plants see Lu et al., Nucl. Acids Res. 32 (21): e171; doi:10.1093/nar/gnh170, 2004).
[0146]The present invention also facilitates the downregulation of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in a cell via the use of synthetic oligonucleotides such as siRNAs or microRNAs directed against a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid which are administered to a cell (for examples of synthetic siRNA mediated silencing see Caplen et al., Proc. Natl. Acad. Sci. USA 98: 9742-9747, 2001; Elbashir et al., Genes Dev. 15: 188-200, 2001; Elbashir et al., Nature 411: 494-498, 2001; Elbashir et al., EMBO J. 20: 6877-6888, 2001; and Elbashir et al., Methods 26: 199-213, 2002).
[0147]In addition to the examples above, the introduced nucleic acid may also comprise a nucleotide sequence which is not directly related to a (1,3;1,4)-β-D-glucan synthase sequence but, nonetheless, may directly or indirectly modulate the expression of (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in a cell. Examples include nucleic acid molecules that encode transcription factors or other proteins which promote or suppress the expression of an endogenous (1,3;1,4)-β-D-glucan synthase encoding nucleic acid molecule in a cell; and other non-translated RNAs which directly or indirectly promote or suppress endogenous (1,3;1,4)-β-D-glucan synthase expression and the like.
[0148]In order to effect expression of an introduced nucleic acid in a genetically modified cell, where appropriate, the introduced nucleic acid may be operably connected to one or more control sequences. The term "control sequences" should be understood to include all components known in the art, which are necessary or advantageous for the transcription, translation and or post-translational modification of the operably connected nucleic acid or the transcript or protein encoded thereby. Each control sequence may be native or foreign to the operably connected nucleic acid. The control sequences may include, but are not limited to, a leader, polyadenylation sequence, propeptide sequence, promoter, enhancer or upstream activating sequence, signal peptide sequence, and transcription terminator. Typically, a control sequence at least includes a promoter.
[0149]The term "promoter" as used herein, describes any nucleic acid which confers, activates or enhances expression of a nucleic acid molecule in a cell. Promoters are generally positioned 5' (upstream) to the genes that they control. In the construction of heterologous promoter/structural gene combinations, it is generally preferred to position the promoter at a distance from the gene transcription start site that is approximately the same as the distance between that promoter and the gene it controls in its natural setting, ie. the gene from which the promoter is derived. As is known in the art, some variation in this distance can be accommodated without loss of promoter function. Similarly, the preferred positioning of a regulatory sequence element with respect to a heterologous gene to be placed under its control is defined by the positioning of the element in its natural setting, ie. the genes from which it is derived. Again, as is known in the art, some variation in this distance can also occur.
[0150]A promoter may regulate the expression of an operably connected nucleotide sequence constitutively, or differentially with respect to the cell, tissue, organ or developmental stage at which expression occurs, in response to external stimuli such as physiological stresses, pathogens, or metal ions, amongst others, or in response to one or more transcriptional activators. As such, the promoter used in accordance with the methods of the present invention may include a constitutive promoter, an inducible promoter, a tissue-specific promoter or an activatable promoter.
[0151]The present invention contemplates the use of any promoter which is active in a cell of interest. As such, a wide array of promoters which are active in any of bacteria, fungi, animal cells or plant cells would be readily ascertained by one of ordinary skill in the art. However, in particularly preferred embodiments of the invention, plant cells are used. Therefore, plant-active constitutive, inducible, tissue-specific or activatable promoters are particularly preferred.
[0152]Plant constitutive promoters typically direct expression in nearly all tissues of a plant and are largely independent of environmental and developmental factors. Examples of constitutive promoters that may be used in accordance with the present invention include plant viral derived promoters such as the Cauliflower Mosaic Virus 35S and 19S (CaMV 35S and CaMV 19S) promoters; bacterial plant pathogen derived promoters such as opine promoters derived from Agrobacterium spp., eg. the Agrobacterium-derived nopaline synthase (nos) promoter; and plant-derived promoters such as the rubisco small subunit gene (rbcS) promoter, the plant ubiquitin promoter (Pubi) and the rice actin promoter (Pact).
[0153]Inducible" promoters include, but are not limited to, chemically inducible promoters and physically inducible promoters. Chemically inducible promoters include promoters which have activity that is regulated by chemical compounds such as alcohols, antibiotics, steroids, metal ions or other compounds. Examples of chemically inducible promoters include: alcohol regulated promoters (eg. see European Patent 637 339); tetracycline regulated promoters (eg. see U.S. Pat. No. 5,851,796 and U.S. Pat. No. 5,464,758); steroid responsive promoters such as glucocorticoid receptor promoters (eg. see U.S. Pat. No. 5,512,483), estrogen receptor promoters (eg. see European Patent Application 1 232 273), ecdysone receptor promoters (eg. see U.S. Pat. No. 6,379,945) and the like; metal-responsive promoters such as metallothionein promoters (eg. see U.S. Pat. No. 4,940,661, U.S. Pat. No. 4,579,821 and U.S. Pat. No. 4,601,978); and pathogenesis related promoters such as chitinase or lysozyme promoters (eg. see U.S. Pat. No. 5,654,414) or PR protein promoters (eg. see U.S. Pat. No. 5,689,044, U.S. Pat. No. 5,789,214, Australian Patent 708850, U.S. Pat. No. 6,429,362).
[0154]The inducible promoter may also be a physically regulated promoter which is regulated by non-chemical environmental factors such as temperature (both heat and cold), light and the like. Examples of physically regulated promoters include heat shock promoters (eg. see U.S. Pat. No. 5,447,858, Australian Patent 732872, Canadian Patent Application 1324097); cold inducible promoters (eg. see U.S. Pat. No. 6,479,260, U.S. Pat. No. 6,084,08, U.S. Pat. No. 6,184,443 and U.S. Pat. No. 5,847,102); light inducible promoters (eg. see U.S. Pat. No. 5,750,385 and Canadian Patent 132 1563); light repressible promoters (eg. see New Zealand Patent 508103 and U.S. Pat. No. 5,639,952).
[0155]Tissue specific promoters" include promoters which are preferentially or specifically expressed in one or more specific cells, tissues or organs in an organism and/or one or more developmental stages of the organism. It should be understood that a tissue specific promoter may be either constitutive or inducible.
[0156]Examples of plant tissue specific promoters include: root specific promoters such as those described in US Patent Application 2001047525; fruit specific promoters including ovary specific and receptacle tissue specific promoters such as those described in European Patent 316 441, U.S. Pat. No. 5,753,475 and European Patent Application 973 922; and seed specific promoters such as those described in Australian Patent 612326 and European Patent application 0 781 849 and Australian Patent 746032.
[0157]In one preferred embodiment, the tissue specific promoter is a seed and/or grain specific promoter. Exemplary seed or grain specific promoters include puroindoline-b gene promoters (for example see Digeon et al., Plant Mol. Biol. 39: 1101-1112, 1999); Pbf gene promoters (for example see Mena et al., Plant J. 16: 53-62, 1998); GS1-2 gene promoters (for example see Muhitch et al., Plant Sci. 163: 865-872, 2002); glutelin Gt1 gene promoters (for example see Okita et al., J. Biol. Chem. 264: 12573-12581, 1989; Zheng et al., Plant J. 4: 357-366, 1993; Sindhu et al., Plant Sci. 130: 189-196, 1997; Nandi et al., Plant Sci. 163: 713-722, 2002); Hor2-4 gene promoters (for example see Knudsen and Muller, Planta 195: 330-336, 1991; Patel et al., Mol. Breeding 6: 113-123, 2000; Wong et al., Proc. Natl. Acad. Sci. USA 99: 16325-16330, 2002); lipoxygenase 1 gene promoters (for example see Rouster et al., Plant J. 15: 435-440, 1998); Chi26 gene promoters (for example see Leah et al., Plant J. 6: 579-589, 1994); Glu-D1-1 gene promoters (for example see Lamacchia et al., J. Exp. Bot. 52: 243-250, 2001; Zhang et al., Theor. Appl. Genet. 106: 1139-1146, 2003); Hor3-1 gene promoters (for example see Sorensen et al., Mol. Gen. Genet. 250: 750-760, 1996; Horvath et al., Proc. Nat. Acad. Sci. USA 97: 1914-1919, 2000) and Waxy (Wx) gene promoters (for example see Yao et al., Acta Phytophysiol. Sin. 22: 431-436, 1996; Terada et al., Plant Cell Physiol. 41: 881-888, 2000; Liu et al., Transgenic Res. 12: 71-82, 2003). In a particularly preferred embodiment, the seed specific promoter is an endosperm specific promoter.
[0158]The promoter may also be a promoter that is activatable by one or more transcriptional activators, referred to herein as an "activatable promoter". For example, the activatable promoter may comprise a minimal promoter operably connected to an Upstream Activating Sequence (UAS), which comprises, inter alia, a DNA binding site for one or more transcriptional activators.
[0159]As referred to herein the term "minimal promoter" should be understood to include any promoter that incorporates at least an RNA polymerase binding site and, preferably a TATA box and transcription initiation site and/or one or more CAAT boxes. More preferably, when the cell is a plant cell, the minimal promoter may be derived from the Cauliflower Mosaic Virus 35S (CaMV 35S) promoter. Preferably, the CaMV 35S derived minimal promoter may comprise a sequence that corresponds to positions -90 to +1 (the transcription initiation site) of the CaMV 35S promoter (also referred to as a -90 CaMV 35S minimal promoter), -60 to +1 of the CaMV 35S promoter (also referred to as a -60 CaMV 35S minimal promoter) or -45 to +1 of the CaMV 35S promoter (also referred to as a -45 CaMV 35S minimal promoter).
[0160]As set out above, the activatable promoter may comprise a minimal promoter fused to an Upstream Activating Sequence (UAS). The UAS may be any sequence that can bind a transcriptional activator to activate the minimal promoter. Exemplary transcriptional activators include, for example: yeast derived transcription activators such as Gal4, Pdr1, Gcn4 and Ace1; the viral derived transcription activator, VP16; Hap1 (Hach et al., J Biol Chem 278: 248-254, 2000); Gaf1 (Hoe et al., Gene 215(2): 319-328, 1998); E2F (Albani et al., J Biol Chem 275: 19258-19267, 2000); HAND2 (Dai and Cserjesi, J Biol Chem 277: 12604-12612, 2002); NRF-1 and EWG (Herzig et al., J Cell Sci 113: 4263-4273, 2000); P/CAF (Itoh et al., Nucl Acids Res 28: 4291-4298, 2000); MafA (Kataoka et al., J Biol Chem 277: 49903-49910, 2002); human activating transcription factor 4 (Liang and Hai, J Biol Chem 272: 24088-24095, 1997); Bcl10 (Liu et al., Biochem Biophys Res Comm 320(1): 1-6, 2004); CREB-H (Omori et al., Nucl Acids Res 29: 2154-2162, 2001); ARR1 and ARR2 (Sakai et al., Plant J 24(6): 703-711, 2000); Fos (Szuts and Bienz, Proc Natl Acad Sci USA 97: 5351-5356, 2000); HSF4 (Tanabe et al., J Biol Chem 274: 27845-27856, 1999); MAML1 (Wu et al., Nat Genet. 26: 484-489, 2000).
[0161]In one preferred embodiment, the UAS comprises a nucleotide sequence that is able to bind to at least the DNA-binding domain of the GAL4 transcriptional activator. UAS sequences, which can bind transcriptional activators that comprise at least the GAL4 DNA binding domain, are referred to herein as UASG. In a particularly preferred embodiment, the UASG comprises the sequence 5'-CGGAGTACTGTCCTCCGAG-3' or a functional homolog thereof.
[0162]As referred to herein, a "functional homolog" of the UASG sequence should be understood to refer to any nucleotide sequence which can bind at least the GAL4 DNA binding domain and which preferably comprises a nucleotide sequence having at least 50% identity, more preferably at least 65% identity, even more preferably at least 80% identity and most preferably at least 90% identity with the UASG nucleotide sequence.
[0163]The UAS sequence in the activatable promoter may comprise a plurality of tandem repeats of a DNA binding domain target sequence. For example, in its native state, UASG comprises four tandem repeats of the DNA binding domain target sequence. As such, the term "plurality" as used herein with regard to the number of tandem repeats of a DNA binding domain target sequence should be understood to include at least 2 tandem repeats, more preferably at least 3 tandem repeats and even more preferably at least 4 tandem repeats.
[0164]As mentioned above, the control sequences may also include a terminator. The term "terminator" refers to a DNA sequence at the end of a transcriptional unit which signals termination of transcription. Terminators are 3'-non-translated DNA sequences generally containing a polyadenylation signal, which facilitates the addition of polyadenylate sequences to the 3'-end of a primary transcript. As with promoter sequences, the terminator may be any terminator sequence which is operable in the cells, tissues or organs in which it is intended to be used. Examples of suitable terminator sequences which may be useful in plant cells include: the nopaline synthase (nos) terminator, the CaMV 35S terminator, the octopine synthase (ocs) terminator, potato proteinase inhibitor gene (pin) terminators, such as the pinII and pinIII terminators and the like.
[0165]As would be appreciated by one of skill in the art, the method of the present invention for modulating the level of (1,3;1,4)-β-D-glucan in a cell, by modulating the level and/or activity of (1,3;1,4)-β-D-glucan synthase in the cell, has several industrial applications.
[0166]For example, (1,3;1,4)-β-D-glucans are known to form viscous solutions. The viscosity-generating properties of soluble cereal (1,3;1,4)-β-D-glucans are critical determinants in many aspects of cereal processing. For example, incompletely degraded (1,3;1,4)-β-D-glucans from malted barley and cereal adjuncts can contribute to wort and beer viscosity and are associated with problems in wort separation and beer filtration (eg. see Bamforth, Brew. Dig. 69 (5): 12-16, 1994) Therefore, for example, in one embodiment, the present invention may be applied to reduce the level of (1,3;1,4)-β-D-glucan in barley grain, by reducing the level and/or activity of (1,3;1,4)-β-D-glucan synthase in one or more cells of the barley grain, to increase its suitability for beer production.
[0167]Soluble cereal (1,3;1,4)-β-D-glucans are also considered to have antinutritive effects in monogastric animals such as pigs and poultry. The "antinutritive" effects have been attributed to the increased viscosity of gut contents, which slows both the diffusion of digestive enzymes and the absorption of degradative products of enzyme action. This, in turn, leads to slower growth rates. Moreover, in dietary formulations for poultry, high (1,3;1,4)-β-D-glucan concentrations are associated with `sticky` faeces, which are indicative of the poor digestibility of the (1,3;1,4)-β-D-glucans and which may present major handling and hygiene problems for producers. Therefore, in another embodiment, the present invention may be applied to reducing the level of (1,3;1,4)-β-D-glucan in one or more cells of a plant used for animal feed, to improve the suitability of the plant as animal feed.
[0168]However, cereal (1,3;1,4)-β-D-glucans are important components of dietary fibre in human and animal diets. As used herein, the term "dietary fibre" should be understood to include the edible parts of plants or analogous carbohydrates that are resistant to digestion and absorption in the human small intestine with complete or partial fermentation in the large intestine. "Dietary fibre" includes polysaccharides (specifically including (1,3;1,4)-β-D-glucans), oligosaccharides, lignin and associated plant substances. In at least human diets, dietary fibres promote beneficial physiological effects including general bowel health, laxation, blood cholesterol attenuation, and/or blood glucose attenuation.
[0169]Humans and monogastric animals produce no enzymes that degrade (1,3;1,4)-β-D-glucans, although there are indications that some depolymerization occurs in the stomach and small intestine, presumably due to the activity of commensal microorganisms. By comparison the soluble (1,3;1,4)-β-D-glucans and other non-starchy polysaccharides are readily fermented by colonic micro-organisms and make a small contribution to digestible energy. In contrast to their antinutritive effects in monogastric animals, oat and barley (1,3;1,4)-β-D-glucans at high concentrations in human foods have beneficial effects, especially for non-insulin-dependent diabetics, by flattening glucose and insulin responses that follow a meal. High concentrations of (1,3;1,4)-β-D-glucans (20% w/v) in food have also been implicated in the reduction of serum cholesterol concentrations, by lowering the uptake of dietary cholesterol or resorption of bile acids from the intestine.
[0170]Therefore, in another embodiment, the present invention may be applied to increasing the dietary fibre content of an edible plant or edible plant part, by increasing the level of (1,3;1,4)-β-D-glucan in the plant, or part thereof. In a particularly preferred embodiment, the edible plant or edible part of a plant is a cereal crop plant or part thereof.
[0171](1,3;1,4)-β-D-glucans, in common with a number of other polysaccharides, in particular (1→3)-β-D-glucans, are also thought to modify immunological responses in humans by a process that is mediated through binding to receptors on cells of the reticuloendothelial system (leucocytes and macrophages). In addition, they may have the capacity to activate the proteins of the human complement pathway, a system that is invoked as a first line of defense before circulating antibodies are produced.
[0172]The method of the first aspect of the present invention also facilitates the production of (1,3;1,4)-β-D-glucan in a recombinant expression system. For example, a (1,3;1,4)-β-D-glucan may be recombinantly produced by introducing a (1,3;1,4)-β-D-glucan synthase encoding nucleotide sequence as described herein, under the control of a promoter, into a cell, wherein the cell subsequently expresses the (1,3;1,4)-β-D-glucan synthase and produces (1,3;1,4)-β-D-glucan.
[0173]A vast array of recombinant expression systems that may be used to express a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid are known in the art. Exemplary recombinant expression systems include: bacterial expression systems such as E. coli expression systems (reviewed in Baneyx, Curr. Opin. Biotechnol. 10: 411-421, 1999; eg. see also Gene expression in recombinant microorganisms, Smith (Ed.), Marcel Dekker, Inc. New York, 1994; and Protein Expression Technologies Current Status and Future Trends, Baneyx (Ed.), Chapters 2 and 3, Horizon Bioscience, Norwich, UK, 2004), Bacillus spp. expression systems (eg. see Protein Expression Technologies: Current Status and Future Trends, supra, chapter 4) and Streptomyces spp. expression systems (eg. see Practical Streptomyces Genetics, Kieser et al., (Eds.), Chapter 17, John Innes Foundation, Norwich, UK, 2000); fungal expression systems including yeast expression systems such as Saccharomyces spp., Schizosaccharomyces pombe, Hansenula polymorpha and Pichia spp. expression systems and filamentous fungi expression systems (eg. see Protein Expression Technologies Current Status and Future Trends, supra, chapters 5, 6 and 7; Buckholz and Gleeson, Bio/Technology 9(11): 1067-1072, 1991; Cregg et al., Mol. Biotechnol. 16(1): 23-52, 2000; Cereghino and Cregg, FEMS Microbiology Reviews 24: 45-66, 2000; Cregg et al., Bio/Technology 11: 905-910, 1993); mammalian cell expression systems including Chinese Hamster Ovary (CHO) cell based expression systems (eg. see Protein Expression Technologies: Current Status and Future Trends, supra, chapter 9); insect cell cultures including baculovirus expression systems (eg. see Protein Expression Technologies: Current Status and Future Trends, supra, chapter 8; Kost and Condreay, Curr. Opin. Biotechnol. 10: 428-433, 1999; Baculovirus Expression Vectors: A Laboratory Manual WH Freeman & Co., New York, 1992; and The Baculovirus Expression System: A Laboratory Manual, Chapman & Hall, London, 1992); Plant cell expression systems such as tobacco, soybean, rice and tomato cell expression systems (eg. see review of Hellwig et al., Nat Biotechnol 22: 1415-1422, 2004); and the like.
[0174]Therefore, in a fourth aspect, the present invention provides a method for producing (1,3;1,4)-β-D-glucan, the method comprising expressing a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid in a cell.
[0175]In one preferred embodiment, the cell is a cell from a recombinant expression system as hereinbefore defined.
[0176]In another preferred embodiment, the (1,3;1,4)-β-D-glucan synthase encoding nucleic acid is a CslF gene or functional homolog thereof.
[0177]In a fifth aspect, the present invention also provides (1,3;1,4)-β-D-glucan produced according to the method of the fourth aspect of the invention.
[0178]In a sixth aspect, the present invention also provides a cell comprising any one or more of: [0179](i) a modulated level of (1,3;1,4)-β-D-glucan relative to a wild type cell of the same taxon; [0180](ii) a modulated level and/or activity of (1,3;1,4)-β-D-glucan synthase relative to a wild type cell of the same taxon; [0181](iii) modulated expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid relative to a wild type cell of the same taxon.
[0182]In one preferred embodiment, the cell of the sixth aspect of the invention is produced according to the methods of the first, second or third aspects of the present invention as described herein. In another preferred embodiment, the cell is a plant cell, more preferably a monocot plant cell and most preferably a cereal crop plant cell.
[0183]Furthermore, in a seventh aspect, the present invention provides a multicellular structure comprising one or more cells according to the sixth aspect of the invention.
[0184]As referred to herein, a "multicellular structure" includes any aggregation of one or more cells. As such, a multicellular structure specifically encompasses tissues, organs, whole organisms and parts thereof. Furthermore, a multicellular structure should also be understood to encompass multicellular aggregations of cultured cells such as colonies, plant calli, suspension cultures and the like.
[0185]As mentioned above, in one preferred embodiment of the invention, the cell is a plant cell and as such, the present invention includes a whole plant, plant tissue, plant organ, plant part, plant reproductive material or cultured plant tissue, comprising one or more plant cells according to the sixth aspect of the invention.
[0186]In a more preferred embodiment, the present invention provides a cereal plant comprising one or more cells according to the sixth aspect of the invention.
[0187]In a particularly preferred embodiment, the present invention provides cereal grain comprising one or more cells according to the sixth aspect of the invention.
[0188]Therefore, in an eighth aspect, the present invention provides a cereal grain comprising an altered level of (1,3;1,4)-β-D-glucan, wherein the grain comprises one or more cells comprising an altered level and/or activity of (1,3;1,4)-β-D-glucan synthase and/or altered expression of a (1,3;1,4)-β-D-glucan synthase encoding nucleic acid molecule.
[0189]In one embodiment, the grain of the eighth aspect of the invention may have an increased level of (1,3;1,4)-β-D-glucan compared to wild type grain from the same species. In an alternate embodiment, the grain may have a decreased level of (1,3;1,4)-β-D-glucan compared to wild type grain from the same species.
[0190]In a ninth aspect, the present invention also provides flour comprising: [0191](i) flour produced by the milling of the grain of the eighth aspect of the invention; and [0192](ii) optionally, flour produced by the milling of one or more other grains.
[0193]As such, the flour produced by the milling of the grain of the eighth aspect of the invention may comprise, for example approximately 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90% or 100% by weight of the flour of the ninth aspect of the invention.
[0194]As referred to herein "milling" contemplates any method known in the art for milling grain, such as those described by Brennan et al. (Manual of Flour and Husk Milling, Brennan et al. (Eds.), AgriMedia, ISBN: 3-86037-277-7).
[0195]Preferably the flour produced by the milling of the grain of the eighth aspect of the invention used in the flour of the ninth aspect of the invention comprises an increased level of (1,3;1,4)-β-D-glucan compared to wild type flour.
[0196]The "flour produced by the milling of one or more other grains" may be flour produced by milling grain derived from any cereal plant, as hereinbefore defined. This component of the flour of the eighth aspect of the invention may, for example, comprise 0%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80% or 90% by weight.
[0197]In a preferred embodiment, the flour produced by the milling of one or more other grains is a wheat flour and, therefore, the flour of the ninth aspect of the invention may be suitable for producing bread, cakes, biscuits and the like.
[0198]As set out above, the present invention is predicated, in part, on the identification and isolation of nucleotide and amino acid sequences that encode (1,3;1,4)-β-D-glucan synthases.
[0199]Therefore, in a tenth aspect, the present invention provides an isolated nucleic acid molecule that encodes a (1,3;1,4)-β-D-glucan synthase.
[0200]In the present invention, "isolated" refers to material removed from its original environment (e.g., the natural environment if it is naturally occurring), and thus is altered "by the hand of man" from its natural state. For example, an isolated polynucleotide could be part of a vector or a composition of matter, or could be contained within a cell, and still be "isolated" because that vector, composition of matter, or particular cell is not the original environment of the polynucleotide. An "isolated" nucleic acid molecule should also be understood to include a synthetic nucleic acid molecule, including those produced by chemical synthesis using known methods in the art or by in-vitro amplification (eg. polymerase chain reaction and the like).
[0201]The isolated nucleic acid molecules of the present invention may be composed of any polyribonucleotide or polydeoxyribonucleotide, which may be unmodified RNA or DNA or modified RNA or DNA. For example, the isolated nucleic acid molecules of the invention can be composed of single- and double-stranded DNA, DNA that is a mixture of single- and double-stranded regions, single- and double-stranded RNA, and RNA that is mixture of single- and double-stranded regions, hybrid molecules comprising DNA and RNA that may be single-stranded or, more typically, double-stranded or a mixture of single- and double-stranded regions. In addition, the isolated nucleic acid molecules can be composed of triple-stranded regions comprising RNA or DNA or both RNA and DNA. The isolated nucleic acid molecules may also contain one or more modified bases or DNA or RNA backbones modified for stability or for other reasons. "Modified" bases include, for example, tritylated bases and unusual bases such as inosine. A variety of modifications can be made to DNA and RNA; thus, "polynucleotide" embraces chemically, enzymatically, or metabolically modified forms.
[0202]In an eleventh aspect, the present invention also provides an isolated nucleic acid molecule comprising one or more of: [0203](i) the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9 and SEQ ID NO: 11; [0204](ii) a nucleotide sequence which is at least 50% identical to the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9 and SEQ ID NO: 11; [0205](iii) a nucleotide sequence which hybridises to a nucleic acid molecule comprising the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9 and SEQ ID NO: 11 under low stringency, more preferably medium stringency and most preferably high stringency conditions; [0206](iv) a nucleotide sequence which encodes the amino acid sequence set forth in any of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8, SEQ ID NO: 10 and SEQ ID NO: 12. [0207](v) a nucleotide sequence which is the complement of any one of (i) to (iv); [0208](vi) a nucleotide sequence which is the reverse complement of any one of (i) to (iv); [0209](vii) a fragment of any one of (i) to (vi).
[0210]As referred to in this eleventh aspect of the invention, the term "at least 50% identical" should be understood to also include nucleotide sequence percentage identities greater than 50%. For example, the term "at least 50% identical" preferably encompasses at least 60% identity, at least 70% identity, at least 80% identity, at least 90% identity and at least 95% identity.
[0211]In a preferred embodiment, the isolated nucleic acid molecule or fragment thereof comprises a nucleotide sequence encoding a (1,3;1,4)-β-D-glucan synthase, as herein before defined. In a more preferred embodiment, the isolated nucleic acid molecule comprises a nucleotide sequence defining a CslF gene, or functional homolog thereof, as hereinbefore defined.
[0212]As set out above, the eleventh aspect of the invention provides fragments of a nucleotide sequence. "Fragments" of a nucleotide sequence should be at least 15 nucleotides (nt), and more preferably at least 20 nt, still more preferably at least 30 nt, and even more preferably, at least 40, 50, 100, 150, 200, 250, 300, 325, 350, 375, 400, 450, 500, 550, or 600 nt in length. These fragments have numerous uses that include, but are not limited to, diagnostic probes and primers. Of course, larger fragments, such as those of 601-3000 nt in length are also useful according to the present invention as are fragments corresponding to most, if not all, of the nucleotide sequences SEQ ID NO: 1. By a fragment at least 20 nt in length, for example, is intended fragments which include 20 or more contiguous bases from, for example, the nucleotide sequence of SEQ ID NO: 1.
[0213]Preferably, the polynucleotide fragments of the invention encode a polypeptide, having (1,3;1,4)-β-D-glucan synthase functional activity as defined herein.
[0214]Polypeptides or proteins encoded by these polynucleotides are also encompassed by the invention.
[0215]In a twelfth aspect, the present invention provides a genetic construct or vector comprising an isolated nucleic acid molecule of the eleventh aspect of the invention.
[0216]In addition to the nucleic acid of the eleventh aspect of the invention, the vector or construct of the twelfth aspect of the invention preferably further comprises one or more of: an origin of replication for one or more hosts; a selectable marker gene which is active in one or more hosts; or one or more control sequences which enable transcription of the isolated nucleic acid molecule in a cell.
[0217]As used herein, the term "selectable marker gene" includes any gene that confers a phenotype on a cell, in which it is expressed, to facilitate the identification and/or selection of cells which are transfected or transformed with a genetic construct of the invention.
[0218]Selectable marker genes" include any nucleotide sequences which, when expressed by a cell, confer a phenotype on the cell that facilitates the identification and/or selection of these transformed cells. A range of nucleotide sequences encoding suitable selectable markers are known in the art. Exemplary nucleotide sequences that encode selectable markers include: antibiotic resistance genes such as ampicillin-resistance genes, tetracycline-resistance genes, kanamycin-resistance genes, the AURI-C gene which confers resistance to the antibiotic aureobasidin A, neomycin phosphotransferase genes (eg. nptI and nptII) and hygromycin phosphotransferase genes (eg. hpt); herbicide resistance genes including glufosinate, phosphinothricin or bialaphos resistance genes such as phosphinothricin acetyl transferase encoding genes (eg. bar), glyphosate resistance genes including 3-enoyl pyruvyl shikimate 5-phosphate synthase encoding genes (eg. aroA), bromyxnil resistance genes including bromyxnil nitrilase encoding genes, sulfonamide resistance genes including dihydropterate synthase encoding genes (eg. sul) and sulfonylurea resistance genes including acetolactate synthase encoding genes; enzyme-encoding reporter genes such as GUS and chloramphenicolacetyltransferase (CAT) encoding genes; fluorescent reporter genes such as the green fluorescent protein-encoding gene; and luminescence-based reporter genes such as the luciferase gene, amongst others.
[0219]Furthermore, it should be noted that the selectable marker gene may be a distinct open reading frame in the construct or may be expressed as a fusion protein with the (1,3;1,4)-β-D-glucan synthase protein.
[0220]The twelfth aspect of the invention extends to all genetic constructs essentially as described herein, which include further nucleotide sequences intended for the maintenance and/or replication of the genetic construct in prokaryotes or eukaryotes and/or the integration of the genetic construct or a part thereof into the genome of a eukaryotic or prokaryotic cell.
[0221]In one preferred embodiment, the construct of the twelfth aspect of the invention is adapted to be at least partially transferred into a plant cell via Agrobacterium-mediated transformation. Accordingly, in a particularly preferred embodiment, the construct according to the twelfth aspect of the invention comprises left and/or right T-DNA border sequences.
[0222]Suitable T-DNA border sequences would be readily ascertained by one of skill in the art. However, the term "T-DNA border sequences" should be understood to encompass any substantially homologous and substantially directly repeated nucleotide sequences that delimit a nucleic acid molecule that is transferred from an Agrobacterium sp. cell into a plant cell susceptible to Agrobacterium-mediated transformation. By way of example, reference is made to the paper of Peralta and Ream (Proc. Natl. Acad. Sci. USA, 82(15): 5112-5116, 1985) and the review of Gelvin (Microbiology and Molecular Biology Reviews, 67(1): 16-37, 2003).
[0223]Although in one preferred embodiment, the construct of the twelfth aspect of the invention is adapted to be transferred into a plant via Agrobacterium-mediated transformation, the present invention also contemplates any suitable modifications to the genetic construct which facilitate bacterial mediated insertion into a plant cell via bacteria other than Agrobacterium sp., as described in Broothaerts et al. (Nature 433: 629-633, 2005).
[0224]Those skilled in the art will be aware of how to produce the constructs described herein and of the requirements for obtaining the expression thereof, when so desired, in a specific cell or cell-type under the conditions desired. In particular, it will be known to those skilled in the art that the genetic manipulations required to perform the present invention may require the propagation of a genetic construct described herein or a derivative thereof in a prokaryotic cell such as an E. coli cell or a plant cell or an animal cell. Exemplary methods for cloning nucleic acid molecules are described in Sambrook et al. (2000, supra)
[0225]In a thirteenth aspect, the present invention provides a cell comprising the isolated nucleic acid molecule of the tenth or eleventh aspects of the invention or genetic construct of the twelfth aspect of the invention.
[0226]The isolated nucleic acid molecule of the tenth or eleventh aspects of the invention or genetic construct of the twelfth aspect of the invention may be introduced into a cell via any means known in the art.
[0227]The isolated nucleic acid molecule or construct referred to above may be maintained in the cell as a DNA molecule, as part of an episome (eg. a plasmid, cosmid, artificial chromosome or the like) or it may be integrated into the genomic DNA of the cell.
[0228]As used herein, the term "genomic DNA" should be understood in it's broadest context to include any and all DNA that makes up the genetic complement of a cell. As such, the genomic DNA of a cell should be understood to include chromosomes, mitochondrial DNA, plastid DNA, chloroplast DNA, endogenous plasmid DNA and the like. As such, the term "genomically integrated" contemplates chromosomal integration, mitochondrial DNA integration, plastid DNA integration, chloroplast DNA integration, endogenous plasmid integration, and the like.
[0229]Preferably, the isolated nucleic acid molecule is operably connected to, inter alia, a promoter such that the cell may express the isolated nucleic acid molecule.
[0230]The cell of the thirteenth aspect of the invention may be any prokaryotic or eukaryotic cell. As such, the cell may be a prokaryotic cell such as a bacterial cell including an E. coli cell or an Agrobacterium spp. cell, or an archaea cell. The cell may also be a eukaryotic cell including a fungal cell such as a yeast cell or mycelial fungus cell; an animal cell such as a mammalian cell or an insect cell; or a plant cell. In a preferred embodiment, the cell is a plant cell. In a more preferred embodiment, the plant cell is a monocot plant cell. In a most preferred embodiment, the plant cell is a cereal plant cell.
[0231]In a fourteenth aspect, the present invention provides a multicellular structure, as hereinbefore defined, comprising one or more of the cells of the thirteenth aspect of the invention.
[0232]As mentioned above, in one preferred embodiment, the cell is a plant cell and as such, the present invention should be understood to specifically include a whole plant, plant tissue, plant organ, plant part, plant reproductive material, or cultured plant tissue, comprising one or more cells of the thirteenth aspect of the invention.
[0233]In a more preferred embodiment, the present invention provides a cereal plant or part thereof, comprising one or more cells of the thirteenth aspect of the invention.
[0234]In a particularly preferred embodiment, the fourteenth aspect of the invention provides cereal grain comprising one or more cells of the thirteenth aspect of the invention.
[0235]As set out above, the present invention also provides amino acid sequences for (1,3;1,4)-β-D-glucan synthases.
[0236]Accordingly, in a fifteenth aspect, the present invention provides an isolated polypeptide comprising an amino acid sequence encoding a (1,3;1,4)-β-D-glucan synthase protein. Accordingly, the present invention provides an isolated (1,3;1,4)-β-D-glucan synthase protein.
[0237]As used herein, the term "polypeptide" should be understood to include any length polymer of amino acids. As such the term "polypeptide" should be understood to encompass peptides, polypeptides and proteins.
[0238]In a sixteenth aspect, the present invention provides an isolated polypeptide comprising one or more of: [0239](i) the amino acid sequence set forth in any of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8, SEQ ID NO: 10 and SEQ ID NO: 12; [0240](ii) an amino acid sequence comprising at least 50% identity to the amino acid sequence set forth in any of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8, SEQ ID NO: 10 and SEQ ID NO: 12; [0241](iii) an amino acid sequence encoded by the nucleotide sequence set forth in any of SEQ ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7, SEQ ID NO: 9 and SEQ ID NO: 11; and/or [0242](iv) a fragment of any one of (i), (ii) or (iii).
[0243]As referred to in this sixteenth aspect of the invention, the term "at least 50% identical" should be understood to also include percentage amino acid sequence identities greater than 50%. For example, the term "at least 50% identical" preferably encompasses at least 60% identity, at least 70% identity, at least 80% identity, at least 90% identity and at least 96% identity.
[0244]In a preferred embodiment, the isolated polypeptide of the present invention comprises an amino acid sequence defining a "(1,3;1,4)-β-D-glucan synthase" as hereinbefore defined.
[0245]The isolated polypeptides of the sixteenth aspect may be composed of amino acids joined to each other by peptide bonds or modified peptide bonds, ie., peptide isosteres, and may contain amino acids other than the 20 gene-encoded amino acids. The isolated polypeptides of the present invention may be modified by either natural processes, such as post-translational processing, or by chemical modification techniques which are well known in the art. Such modifications are well described in basic texts and in more detailed monographs, as well as in the literature.
[0246]Modifications can occur anywhere in the isolated polypeptide, including the peptide backbone, the amino acid side-chains and/or the termini. It will be appreciated that the same type of modification may be present in the same or varying degrees at several sites In a given isolated polypeptide. Also, an isolated polypeptide of the present invention may contain many types of modifications.
[0247]The proteins may be branched, for example, as a result of ubiquitination, and/or they may be cyclic, with or without branching. Cyclic, branched, and branched cyclic polypeptides may result from post-translation natural processes or may be made by synthetic methods.
[0248]Modifications include acetylation, acylation, ADP-ribosylation, amidation, covalent attachment of flavin, covalent attachment of a heme moiety, covalent attachment of a nucleotide or nucleotide derivative, covalent attachment of a lipid or lipid derivative, covalent attachment of phosphatidylinositol, cross-linking, cyclization, disulfide bond formation, demethylation, formation of covalent cross-links, formation of cysteine, formation of pyroglutamate, formylation, gamma-carboxylation, glycosylation, GPI anchor formation, hydroxylation, iodination, methylation, myristoylation, oxidation, PEGylation, proteolytic processing, phosphorylation, prenylation, racemization, selenoylation, sulfation transfer-RNA mediated addition of amino acids to proteins such as arginylation, and ubiquitination. (See, for instance, Proteins-Structure And Molecular Properties 2nd Ed., Creighton (ed.), W.H. Freeman and Company, New York, 1993); Posttranslational Covalent Modification Of Proteins, Johnson (Ed.), Academic Press, New York, 1983; Seifter et al., Meth Enzymol 182: 620-646, 1990); Rattan et al., Ann NY Acad Sci 663: 48-62, 1992.).
[0249]As set out above, the sixteenth aspect of the invention also provides fragments of isolated polypeptides. Polypeptide fragments may be "free-standing," or comprised within a larger polypeptide of which the fragment forms a part or region, most preferably as a single continuous region.
[0250]The protein fragments can be at least 3, 4, 5, 6, 8, 9, 10, 11, 12, 13, 15, 20, 25, 30, 35, 40, 45, 50, 60, 70, 80, 90, 100, 110, 120, 130, 140, or 150 amino acids in length. In one preferred embodiment, the fragment comprises an amino acid sequence which is a part of the sequence set forth in SEQ ID NO: 2.
[0251]In one preferred embodiment, the fragment comprises (1,3;1,4)-β-D-glucan synthase functional activity. However, even if the fragment does not retain one or more biological functions of the (1,3;1,4)-β-D-glucan synthase protein, other functional activities may still be retained. For example, the fragments may lack (1,3;1,4)-β-D-glucan synthase functional activity but retain the ability to induce and/or bind to antibodies which recognize the complete or mature forms of an isolated (1,3;1,4)-β-D-glucan synthase protein. A peptide, polypeptide or protein fragment which has the ability to induce and/or bind to antibodies which recognize the complete or mature forms of the isolated (1,3;1,4)-β-D-glucan synthase protein is referred to herein as a "(1,3;1,4)-β-D-glucan synthase epitope".
[0252]A (1,3;1,4)-β-D-glucan synthase epitope may comprise as few as three or four amino acid residues, preferably at least 5 amino acids and more preferably at least 10 amino acid residues. Whether a particular epitope of an isolated (1,3;1,4)-β-D-glucan synthase protein retains such immunologic activities can readily be determined by methods known in the art. As such, one preferred (1,3;1,4)-β-D-glucan synthase protein fragment is a polypeptide comprising one or more (1,3;1,4)-β-D-glucan synthase epitopes.
[0253]A polypeptide comprising one or more (1,3;1,4)-β-D-glucan synthase epitopes may be produced by any conventional means for making polypeptides including synthetic and recombinant methods known in the art. In one embodiment, (1,3;1,4)-β-D-glucan synthase epitope-bearing polypeptides may be synthesized using known methods of chemical synthesis. For instance, Houghten has described a simple method for the synthesis of large numbers of peptides (Houghten, Proc. Natl. Acad. Sci. USA 82: 5131-5135, 1985).
[0254]The isolated polypeptides and (1,3;1,4)-β-D-glucan synthase epitope-bearing polypeptides of the sixteenth aspect of the invention are useful, for example, in the generation of antibodies that bind to the isolated (1,3;1,4)-β-D-glucan synthase proteins of the invention.
[0255]Such antibodies are useful, inter alia, in the detection and focalization of (1,3;1,4)-β-D-glucan synthase proteins and in affinity purification of (1,3;1,4)-β-D-glucan synthase proteins. The antibodies may also routinely be used in a variety of qualitative or quantitative immunoassay using methods known in the art. For example see Harlow et al., Antibodies: A Laboratory Manual, (Cold Spring Harbor Laboratory Press 2nd Ed., 1988).
[0256]Accordingly, in a seventeenth aspect, the present invention provides an antibody or an epitope binding fragment thereof, raised against an isolated (1,3;1,4)-β-D-glucan synthase protein as hereinbefore defined or an epitope thereof.
[0257]The antibodies of the present invention include, but are not limited to, polyclonal, monoclonal, multispecific, chimeric antibodies, single chain antibodies, Fab fragments, F(ab') fragments, fragments produced by a Fab expression library and epitope-binding fragments of any of the above.
[0258]The term "antibody", as used herein, refers to immunoglobulin molecules and immunologically active portions of immunoglobulin molecules, i.e., molecules that contain an antigen-binding site that immunospecifically binds an antigen. The immunoglobulin molecules of the invention can be of any type (e.g., IgG, IgE, IgM, IgD, IgA and IgY), class (e.g., IgG1, IgG2, IgG3, IgG4, IgA1 and IgA2) or subclass of immunoglobulin molecule.
[0259]The antibodies of the present invention may be monospecific, bispecific, trispecific, or of greater multispecificity. Multispecific antibodies may be specific for different epitopes of a polypeptide of the present invention or may be specific for both a polypeptide of the present invention as well as for a heterologous epitope, such as a heterologous polypeptide or solid support material, For example, see PCT publications WO 93/1715; WO 92/08802; WO 91/00360; WO 92/05793; Tutt et al., J. Immunol. 147: 60-69, 1991; U.S. Pat. Nos. 4,474,893; 4,714,681; 4,926,648; 5,573,020; 5,601,819; and Kostelny et al, J. Immunol, 148: 1547-1553, 1992).
[0260]In one embodiment, the antibodies of the present invention may act as agonists or antagonists of (1,3;1,4)-β-D-glucan synthase. In further embodiments, the antibodies of the present invention may be used, for example, to purify, detect, and target the polypeptides of the present invention, including both in vitro and in vivo diagnostic and therapeutic methods. For example, the antibodies have use in immunoassays for qualitatively and quantitatively measuring levels of (1,3;1,4)-β-D-glucan synthase in biological samples. See, e.g., Harlow et al., Antibodies: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2nd ed. 1988).
[0261]The term "antibody", as used herein, should be understood to encompass derivatives that are modified, eg. by the covalent attachment of any type of molecule to the antibody such that covalent attachment does not prevent the antibody from binding to (1,3;1,4)-β-D-glucan synthase or an epitope thereof. For example, the antibody derivatives include antibodies that have been modified, eg., by glycosylation, acetylation, pegylation, phosphorylation, amidation, derivatization by known protecting/blocking groups, proteolytic cleavage, linkage to a cellular ligand or other protein, etc. Furthermore, any of numerous chemical modifications may also be made using known techniques. These include specific chemical cleavage, acetylation, formylation, metabolic synthesis of tunicamycin, etc. Additionally, the derivative may contain one or more non-classical amino acids.
[0262]Antibodies may be generated using methods known in the art, such as in vivo immunization, in vitro immunization, and phage display methods. For example, see Bittle et al. (J. Gen. Virol. 66: 2347-2354, 1985).
[0263]If in vivo immunization is used, animals may be immunized with free peptide; however, anti-peptide antibody titer may be boosted by coupling of the peptide to a macromolecular carrier, such as keyhole limpet hemacyanin (KLH) or tetanus toxoid. For example, peptides containing cysteine residues may be coupled to a carrier using a linker such as maleimidobenzoyl-N-hydroxysuccinimide ester (MBS), while other peptides may be coupled to carriers using a more general linking agent such as glutaraldehyde.
[0264]Animals such as rabbits, rats and mice are immunized with either free or carrier-coupled peptides, for instance, by intraperitoneal and/or intradermal. Injection of emulsions containing about 100 micrograms of peptide or carrier protein and Freund's adjuvant. Several booster injections may be needed, for example, at intervals of about two weeks, to provide a useful titer of anti-peptide antibody which can be detected, for example, by ELISA assay using free peptide adsorbed to a solid surface. The titer of anti-peptide antibodies in serum from an immunized animal may be increased by selection of anti-peptide antibodies, for instance, by adsorption to the peptide on a solid support and elution of the selected antibodies according to methods well known in the art.
[0265]For example, polygonal antibodies to a (1,3;1,4)-β-D-glucan synthase protein or a polypeptide comprising one or more (1,3;1,4)-β-D-glucan synthase epitopes can be produced by various procedures well known in the art. For example, a polypeptide of the invention can be administered to various host animals including, but not limited to, rabbits, mice, rats, etc to induce the production of sera containing polyclonal antibodies specific for the antigen Various adjuvants may be used to increase the immunological response, depending on the host species, for example, Freund's (complete and incomplete), mineral gels such as aluminum hydroxide, surface active substances such as lysolecithin, pluronic polyols, polyanions, peptides, oil emulsions, keyhole limpet hemocyanins, dinitrophenol, and potentially useful human adjuvants such as BCG (bacille Calmette-Guerin) and Corynebacterium parvum. Such adjuvants are also well known in the art.
[0266]As another example, monoclonal antibodies can be prepared using a wide variety of techniques known in the art including the use of hybridoma, recombinant, and phage display technologies, or a combination thereof. For example, monoclonal antibodies can be produced using hybridoma techniques including those known in the art and taught, for example, in Harlow et al., Antibodies: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2nd ed., 1988) and Hammerling et al., in: Monoclonal Antibodies and T-Cell Hybridomas (Elsevier, N.Y., 1981). The term "monoclonal antibody" as used herein is not limited to antibodies produced through hybridoma technology. The term "monoclonal antibody" refers to an antibody that is derived from a single clone, including any eukaryotic, prokaryotic, or phase clone, and not the method by which it is produced.
[0267]Methods for producing and screening for specific antibodies using hybridoma technology are routine and well known in the art, For example, mice can be immunized with a polypeptide of the invention or a cell expressing such peptide. Once an immune response is detected, e.g., antibodies specific for the antigen are detected in the mouse serum, the mouse spleen is harvested and splenocytes isolated. The splenocytes are then fused by well-known techniques to any suitable myeloma cells, for example cells from cell line SP20 available from the ATCC. Hybridomas are selected and cloned by limited dilution. The hybridoma clones are then assayed by methods known in the art for cells that secrete antibodies capable of binding a polypeptide of the invention. Ascites fluid, which generally contains high levels of antibodies, can be generated by immunizing mice with positive hybridoma clones.
[0268]Antibody fragments which recognize one or more (1,3;1,4)-β-D-glucan synthase epitopes may also be generated by known techniques. For example, Fab and F(ab')2 fragments may be produced by proteolytic cleavage of immunoglobulin molecules, using enzymes such as papain (to produce Fab fragments) or pepsin (to produce F(ab')2 fragments). F(ab')2 fragments contain the variable region, the light chain constant region and the CH1 domain of the heavy chain.
[0269]The antibodies of the present invention can also be generated using various phage display methods known in the art. In phage display methods, functional antibody domains are displayed on the surface of phage particles which carry the polynucleotide sequences encoding them. In a particular embodiment, such phage can be utilized to display antigen-binding domains expressed from a repertoire or combinatorial antibody library (e.g., human or murine). Phage expressing an antigen binding domain that binds the antigen of interest can be selected or identified with antigen, e.g., using labeled antigen or antigen bound or captured to a solid surface or bead. Phages used in these methods are typically filamentous phage including fd and M13 binding domains expressed from phage with Fab, Fv or disulfide stabilized Fv antibody domains recombinantly fused to either the phage gene III or gene VIII protein.
[0270]Examples of phage display methods that can be used to make the antibodies of the present invention include those disclosed by Brinkman et al. (J. Immunol. Methods 182: 41-50, 1995), Ames et al. (J. Immunol. Methods 184: 177-186, 1995), Kettleborough et al. (Eur. J. Immunol. 24: 952-958, 1994), Persic et al. (Gene 187: 9-18, 1997), Burton et al. (Advances in Immunology 57: 191-280, 1994); PCT publications WO 90/02809; WO 91/10737; WO 92/01047; WO 92/18619; WO 93/11236; WO 95/15982; WO 95/20401; and U.S. Pat. Nos. 5,698,426; 5,223,409; 5,403,484; 5,580,717; 5,427,908; 6,750,753; 5,821,047; 5,571,698; 5,427,908; 5,516,637; 5,780,225; 5,658,727; 5,733,743 and 5,969,108.
[0271]After phage selection, the antibody coding regions from the phage can be isolated and used to generate whole antibodies or any other desired antigen binding fragment, and expressed in any desired host, including mammalian cells, insect cells, plant cells, yeast, and bacteria. For example, techniques to recombinantly produce Fab, Fab' and F(fab')2 fragments can also be employed using methods known in the art such as those disclosed in PCT publication WO 92/22324; Mullinax et al. (Bio Techniques 12 (6): 864-869, 1992); and Sawal et al. (AJRI 34:26-34, 1995); and Better at al. (Science 240: 1041-1043, 1988).
[0272]Examples of techniques which can be used to produce single-chain Fvs and antibodies include those described in U.S. Pat. Nos. 4,946,778 and 5,258,498; Huston et al. (Methods in Enzymology 203; 46-88, 1991); Shu et al. (Proc. Natl. Acad. Sci. USA 90: 7995-7999, 1993); and Skerra et al. (Science 240: 1038-1040, 1988).
[0273]The present invention is further described by the following non-limiting examples.
Example 1
Identification of Candidate Genes Through Natural Variation of (1,3;1,4)-β-D-Glucan Content in Barley Grain
[0274]Comparative mapping studies have revealed that there is a high level of conservation of gene order along chromosomes of species of the Poaceae, although macro-colinearity at this level does not always predict gene presence or order at the micro level, Nevertheless, co-linearity at the megabase level is essential for the use of model species for positional cloning of genes, for development of molecular markers and for identifying candidate genes that affect a trait of interest in one species through reference to the syntenous region of a model species. Therefore, this approach was adopted to identify candidate genes for (1,3;1,4)-β-D-glucan synthases in cereals.
[0275]Quantitative trait loci (QTL) mapping and comparative genomic has been used to identify genes involved in cell wall biosynthesis in maize (Zea mays). Because of the central role played by (1,3;1,4)-β-D-glucans in malting and brewing quality, QTL analyses of grain (1,3;1,4)-β-D-glucan content are available. As shown in FIG. 1, the QTL that has the largest effect on grain (1,3;1,4)-β-D-glucan content was located on barley chromosome 2H, between the Adh8 and ABG019 markers.
[0276]Using the sequences of the two DNA markers that flank the barley QTL on chromosome 2H, a syntenous region was located on chromosome 7 of rice, where a cluster of six cellulose synthase-like (Csl) genes was detected within an interval of 119 Kb, corresponding to the 21.59-21.72 Mb region of the chromosome (FIG. 1).
[0277]Each of these genes was classified in the CslF group of rice and they were designated OsCslF1 (SEQ ID NO: 19), OsCslF2 (SEQ ID NO: 21), OsCslF3 (SEQ ID NO: 23), OsCslF4 (SEQ ID NO: 25), OsCslF8 (SEQ ID NO: 31) and OsCslF9 (SEQ ID NO: 33). Other known genes in this interval of rice chromosome 7 include truncated OsCslF genes that might represent pseudogenes.
[0278]The OsCSslF5 (SEQ ID NO: 27) and OsCslF7 (SEQ ID NO: 29) genes are located elsewhere on the rice genome (data not shown).
[0279]On this basis, the comparative genomics approach enabled the identification of the CslF group of genes as potential candidate genes for (1,3;1,4)-β-D-glucan synthases in cereals. It is noteworthy that the CslF group of CSl genes is only found in monocotyledons, which is consistent with the exclusive occurrence of (1,3;1,4)-β-D-glucans in cell walls of the Poales.
Materials and Methods
(i) Plant Tissues
[0280]Tissues were collected from mature rice plants (Oryzae sativa cv Nippon Bare) grown at 28° C. day and 22° C. night temperatures under high humidity, a photointensity of 300 umol/m/s and an 11/13 hour day/night regime. Material was also collected from five day-old seedlings germinated at 28° C. in the dark on damp filter paper in Petri dishes.
(ii) Synteny Analysis
[0281]DNA sequences for the markers under the QTLs for barley (1,3;1,4)-β-D-glucan were obtained from the GrainGenes database (http://wheat.pw.usda.gov). Additional markers from within the corresponding chromosomal locations on the Barley-Consensus2 (Qi et al., Genome 39: 379-394, 1996) and Barley-Consensus2003 (Karakousis et al., Australian Journal of Agricultural Research 54: 1173-1185, 2003) maps were also included in the investigation. The syntenic chromosomal location(s) for the markers on the rice genome were determined by BLASTN analyses at the GRAMENE website (http://www.gramene.org). Syntenic regions were examined for gene annotations of enzymes encoding for synthesis of cell wall polysaccharides. A thorough analysis of the region on rice chromosome 7 that corresponded to the QTL peak for (1,3;1,4)-β-D-glucan an barley chromosome 2H was carried out and six co-located CslF genes were identified for further analyses.
Example 2
Transformation of Arabidopsis thaliana with Rice CslF Genes
[0282]The possible role of the rice OsCslF genes in (1,3;1,4)-β-D-glucan synthesis was tested by gain-of-function in transgenic Arabidopsis plants. Arabidopsis walls contain no (1,3;1,4)-β-D-glucan and the Arabidopsis genome does not contain any known CslF genes. Therefore, the deposition of (1,3;1,4)-β-D-glucan into walls of transgenic Arabidopsis plants carrying rice OsCslF genes would indicate that the introduced gene(s) encoded (1,3;1,4)-β-D-glucan synthases. This approach assumed and depended upon the availability in Arabidopsis of any precursors, intermediates, cofactors or ancillary enzymes needed for (1,3;1,4)-β-D-glucan synthesis.
[0283]Accordingly, the rice CslF1, 2, 3, 4, 8 genes were successfully amplified from cDNA by PCR and cloned into the pAJ22 binary, vector, behind the 35S promoter, as shown in FIG. 2.
[0284]The plasmid vectors were subsequently inserted into Agrobacterium tumefaciens, which was used to transform Arabidopsis by standard floral dip procedures (Clough and Bent, Plant J. 16: 735, 1998). In case multiple OsCslF genes might be required for (1,3;1,4)-β-D-glucan synthesis, transformation was performed not only with single gene constructs, but also with various combinations of the OsCslF genes.
[0285]Following selection with the herbicide BASTA, DNA and RNA were isolated from selected transgenic plants to check for the presence of the transgene(s) and to monitor transcription of the transgenes by real-time, quantitative PCR (Q-PCR; as described by Burton et al. (2004, supra).
[0286]Southern hybridization analyses confirmed the presence of the transgenes (FIG. 3). As shown in FIG. 4, at least some lines were found to contain single copies of the various OsCslF genes. Where the Arabidopsis was transformed with multiple OsCslF genes, all of those genes could be detected (FIG. 3).
[0287]Transcription of the OsCslF genes in 14-day old leaves of the transgenic lines was also confirmed. Normalized mRNA levels are shown for selected transgenic plants in FIG. 5, where large differences in transcriptional activity of the transgenes are evident between plant lines and where similarly large differences are observed between individual OsCslF genes in lines carrying more than one transgene. For example, in lines transformed with the OsCslF2, OsCslF4 and OsCslF8 genes, OsCslF4 transcripts were usually the highest n abundance. The results showed that the 35S promoter was clearly driving high level transcription in many lines.
Materials and Methods
(i) Plants
[0288]Arabidopsis plants were grown in Arabidopsis soil mix at 23° C. in a growth chamber under either long 12/12 hr day/night or short 8/16 hr day/night conditions. Seed collected from transgenic plants was dried, cleaned, vernalised for 2 days at 4° C. and sown onto solid MS media containing 25 mg/l Bialophos for selection. Survivors were transplanted into soil at the five leaf stage and grown in growth rooms under the conditions described above.
(ii) Binary Vector Construction
[0289]The binary vector pAJ22 (FIG. 2) was kindly supplied by Dr. Andrew Jacobs (University of Adelaide) and is based on the pAMPAT-MCS backbone (accession no. AY436765). It contains a double 35S promoter with a pNOS terminator region, separated by a modified multiple cloning site that incorporates a triple HA epitope, which was not used in this instance. Full-length PCR products corresponding to the rice OsCslF cDNAs amplified as described above were cloned into the TEASY vector (Promega). Clones carrying an insert of the correct size were digested with the appropriate restriction enzymes (Table 2) and an enzyme to cut the TEASY backbone into two segments. The reactions were separated by agarose gel electrophoresis, the CslF fragments were excised and purified using the QIAquick (QIAGEN) gel extraction kit according to the manufacturer's instructions. The binary vector pAJ22 was digested with the corresponding pair of restriction enzymes and the CslF fragment was ligated into the pAJ22 vector. Plasmid DNA was extracted from positive clones using the QIAquick miniprep kit and inserts were sequenced using BigDye 3.1 chemistry (ABI) on an Applied Biosystems ABI3700 capillary sequencer. Plasmid DNA preparations containing verified inserts were transformed into Agrobacterium tumefaciens cv GV3101 via electroporation using the method of Mersereau et al. (Gene 90: 149, 1990) and positive colonies were selected on media containing 25 mg/l rifampicin, 48 mg/l carbenicillin and 50 mg/l kanamycin.
(iii) Arabidopsis Transformation
[0290]Arabidopsis transformants were generated by the floral dip method of Clough and Bent (Plant J. 16: 735, 1998).
(iv) DNA Extraction and Southern Hybridisation Analyses
[0291]Genomic DNA was extracted from young leaves and flower buds using the Qiagen miniprep plant kit. Approximately 5 μg genomic DNA per plant was digested with the relevant restriction enzymes and separated on a 1% agarose TAE gel. DNA was transferred to Highbond+membranes. Membranes were pre-hybridised and hybridised and probe fragments were labelled using the Rediprime labelling kit (Amersham, High Wycome, UK) following the manufacturer's instructions.
(v) RNA extraction, cDNA synthesis and RT-PCR
[0292]All RNA extractions and cDNA syntheses were carried out as described in Burton et al. (Plant Physiol. 134: 224-236, 2004). Samples of cDNA from appropriate tissues were used as templates to amplify full-length CslF sequences, using Elongase Taq polymerase (Invitrogen) by PCR. Primer pairs, as listed in Table 2, were used in the PCR, following a standard recipe suggested by the manufacturer. Dimethylsulphoxide (DMSO, 5% v/v; Sigma, St Louis, Mo., USA) was added and PCR was performed for 40 cycles as follows; 94° C. for 30 sec, 50° C. to 58° C. (depending on the Tm of individual primers) for 30 sec and 68° C. for 3 min. The primers contained restriction sites at each end, as indicated in Table 2, to facilitate cloning of the amplified fragment into the binary vector.
TABLE-US-00002 TABLE 2 Oligonucleotides used for amplification of rice CsIF cDNAs. Oligo- nucleotide RE. Sequence cDNA Oligo sequence Site Identifier OsCsIF2 OsF2BII5 AGTCAGATCT BgIII SEQ ID NO: 35 GTTCCGTGCA TGGCGGCCAC CG OsCsLF2 OsF2ML3 CAGTACGCGT MIul SEQ ID NO: 36 CGCGATCGAA CTGTCCCTAC CC OsCsIF3 OsF3BII5 AGTCAGATCT BgIII SEQ ID NO: 37 ATAGAGTGCT CGTCATGGC OsCsIF3 OsF3ML3 CAGTACGCGT MIuI SEQ ID NO: 38 TTTATCTATG CACCTAGAAT GG OsCsIF4 OsF4H5 AGTCAAGCTT HindIII SEQ ID NO: 39 GCTACGGCCT CCACGATGTC CG OsCsIF4 OsF4S3 CAGTACTAGT SpeI SEQ ID NO: 40 CATGTCGTCC CTACCCAGAT GG OsCsIF8 OsF8H5 AGTCAAGCTT HindIII SEQ ID NO: 41 GCGACGATCG ATGGCGCTTT CG OsCsIF8 OsF8S3 CAGTACTACT SpeI SEQ ID NO: 42 CAATCAGAAA CCCCGC
(vi) Quantitative Real Time PCR (Q-PCR) Analysis
[0293]The primer pairs for control genes and specific CslF genes were used as indicated in Table 3. Stock solutions of PCR products for the preparation of dilution series were prepared by PCR from a cDNA derived from either a composite of rice or Arabidopsis tissue cDNAs, and was subsequently purified and quantified by HPLC, as described by Burton et al. (2004, supra). A dilution series covering seven orders of magnitude was prepared from the 109 copies/μl stock solution as follows; one microlitre of the stock solution was added to 99 μl of water, and six 1:10 serial dilutions were prepared to produce a total of seven solutions covering 107 copies/μl to 101 copies/μl. Three replicates of each of the seven standard solutions were included with every Q-PCR experiment, together with a minimum of three no-template controls. For all genes, a 1:20 dilution of the cDNA was sufficient to produce expression data with an acceptable standard deviation. Three replicate PCRs for each of the cDNAs were included in every run. All Q-PCR reaction mixes were prepared on a CAS-1200 robot (Corbett Robotics, Brisbane, Australia).
[0294]Two microlitres of the diluted cDNA solution were used in a reaction containing 5 μl QuantiTect SYBR Green PCR reagent, 1 μl each of the forward and reverse primers at 4 μM, 0.3 μl 10× SYBR Green in water (10,000× in DMSO, BioWhittaker Molecular Applications, Rockland, USA, 0.5 μl in 500 μl of water, prepared daily) and 0.7 μl water. The total volume of each Q-PCR reaction mixture was 10 μl. Reactions were performed in a RG 3000 Rotor-Gene Real Time Thermal Cycler (Corbett Research, Sydney, Australia) as follows; 15 min at 95° followed by 45 cycles of 20 sec at 95°, 30 sec at 55°, 30 sec at 72° and 15 sec at the optimal acquisition temperature (AT) described in Table 3. A melt curve was obtained from the product at the end of the amplification by heating from 70° to 99°. After the experiment, the optimal cycle threshold (CT) was determined from the dilution series and the raw expression data was derived. The mean expression level and standard deviation for each set of three replicates for each cDNA was calculated.
[0295]The raw expression data for the exogenous CslF genes was scaled using the approach of Vandesompele et al. (Genome Biol. 3: 1-11, 2002). The normalisation factor derived from the best three of four Arabidopsis control genes was generated using the Genorm software (Vandesompele et al., supra, 2002). The raw expression data for the exogenous CslF genes in each cDNA was scaled by dividing the raw expression value by the normalisation factor for the particular cDNA.
TABLE-US-00003 TABLE 3 Primers used for Q-PCR analysis. amplicon Gene Forward Primer Reverse Primer (bp) A T (° C.) GAPDH At TGGTTGATCTCGTTG GTCAGCCAAGTCAAC 262 77 TGCAGGTCTC AACTCTCTG (SEQ ID NO: 43) (SEQ ID NO: 44) Tubulin At ATGTGGGTGAGGGTA CCGACAACCTTCTTA 143 78 TGGAA GTxCTCCTCT (SEQ ID NO: 45) (SEQ ID NO: 46) Actin At GAGTTCTTCACGCGA GACCACCTTTATTAA 180 76 TACCTCCA CCCCATTTACCA (SEQ ID NO: 47) (SEQ ID NO: 48) Cyclophilin TGGCGAACGCTGGTC CAAAAACTCCTCTGC 223 79 At CTAATACA CCCAATCAA (SEQ ID NO: 49) (SEQ ID NO: 50) OsCSLF2 GTGCGCATACGAGGA AGAACATCTCCAGCG 220 83 TGGGACG AGCCGCC (SEQ ID NO: 51) (SEQ ID NO: 52) OsCSLF3 CCGATTGGGGCAAGG GACACGCTGGAGAGG 256 79 GTGTTGG TTGGAGC (SEQ ID NO: 53) (SEQ ID NO: 54) OsCSLF4 CTCCGTGTACACCTC CTCGGAGATGAGCCA 255 82 CATGGAG CATCACC (SEQ ID NO: 55) (SEQ ID NO: 56) OsCSLF8 TACGACATCGCGACG GTCATGTTGGCGTAC 244 83 GAGGACG GCGACGC (SEQ ID NO: 57) (SEQ ID NO: 58)
Example 3
Immunological Characterization of Transgenic Arabidopsis Lines
[0296]Transgenic Arabidopsis lines in which OsCslF transcript levels were highest were chosen for further analysis, in particular with respect to the deposition of (1,3;1,4)-β-D-glucan in cell walls. In the first instance, immunocytochemical methods involving monoclonal antibodies specific for (1,3;1,4)-β-D-glucans and electron microscopy were used to screen transgenic lines for the presence of the polysaccharide in the Arabidopsis lines. The antibody used does not bind with cellooligosaccharides or the (1→3)-β-D-glucan, callose. Inhibition studies showed that it binds relatively weakly to (1,3;1,4)-β-D-oligoglucosides.
[0297]Pieces of 14- and 28-day old leaves were sectioned for monoclonal antibody probing, and the antibody was routinely checked by pre-incubating tissue sections with commercially available barley (1,3;1,4)-β-D-glucan. Pre-incubation with the polysaccharide blocks the binding of gold-labelled secondary antibody. (1,3;1,4)-β-D-glucan was detected in cell walls of several transgenic Arabidopsis plants with the specific monoclonal antibodies, such as Arabidopsis lines A28, A29 and A18, as shown in FIG. 7. In FIG. 7, panel B, walls from the epidermal layers of leaves from transgenic Arabidopsis line A18 are shown to accumulate (1,3;1,4)-β-D-glucan over a period of about fourteen days. The polysaccharide was not detected in other tissues of this line. Finally, Panel C shows a representative control panel of a section of WT Arabidopsis leaf epidermal cell wall where minimal or no background labelling is commonly observed.
Materials and Methods
(i) Preparation of Transformed Arabidopsis Leaves for Electron Microscopy
[0298]Arabidopsis leaves were fixed in 4% (v/v) glutaraldehyde (EM Grade) in phosphate-buffered saline (PBS), pH 7.2, and stored at 4° C. Samples were washed three times in PBS and post-fixed in a 2% osmium tetroxide solution in PBS for 1 h at room temperature. After three rinses in MilliQ water the samples were dehydrated in a graded ethanol series and slowly infiltrated with LR White resin over several days. Individual leaves were placed in gelatin capsules, which were filled with fresh resin and polymerized overnight at 65° C.
(ii) Immunolocation for Transmission Electron Microscopy
[0299]Sections (80 nm) of Arabidopsis leaves were prepared on a Leica Ultracut R microtome using a diamond knife and collected on 100 and 200 mesh, Formvar coated gold grids. The ultrathin sections were blocked for 30 min in 1% bovine serum albumin in PBS before incubation in murine monoclonal antibodies raised against barley (1,3;1,4)-β-D-glucan (diluted 1:500; Biosupplies Australia, Parkville, VIC 3052, Australia) for 1 hr at room temperature and overnight at 4° C. The grids were washed twice in PBS and three times in blocking buffer before a 1 h incubation in 18 nm Colloidal Gold-AffiniPure Goat-Anti Mouse IgG+IgM (H+L) (Jackson ImmunoResearch Laboratories, Inc., PA, USA). All grids were washed twice in PBS and several times in MilliQ water before staining in 2% aqueous uranyl acetate followed by triple lead citrate stain. The sections were viewed on a Philips BioTwin Transmission Electron Microscope and images captured on a Gatan Multiscan CCD Camera.
[0300]In some experiments the primary antibody was omitted to control for non-specific secondary antibody binding. Other control experiments involved pre-absorbing the primary antibodies to their respective polysaccharides to ensure the specificity of the antibody. Solutions (1 mg/ml) of (1,3;1,4)-β-D-GLUCAN from barley (Biosupplies Australia) were mixed in equal volumes with their respective diluted primary antibodies. No labelling was observed in any of these negative control experiments.
[0301]Supplementary data relating to this experiment can be found in Burton et al., Science 311: 1940-1942, 2006.
Example 4
Identification of CslF Sequences from Barley
[0302]Where available, partial EST barley sequences were assembled into complete CslF sequences, using the rice CslF sequences as a guide.
[0303]Where no barley EST sequences were available, putative wheat CslF EST sequences were identified, which were potentially highly homologous to the equivalent barley sequences. Primers were then designed on the basis of the wheat EST sequences and were then used on barley cDNA populations to amplify the equivalent barley sequence. 3' and 5' RACE approaches were then used to extend the barley sequences.
[0304]In a few cases additional parts of closely related barley genes for which there were no wheat ESTs were amplified, and these were also extended using RACE. In total, 6 different barley CslF sequences were identified, which were designated HvCslF1, HvCslF2, HvCslF3, HvCslF4, HvCslF5 and HvCslF6.
Example 5
Alignment of CslF DNA and Amino Acid Sequences from Rice and Barley
[0305]An alignment of the DNA and amino acid sequences for the CslF sequences in both rice and barley was performed, the results of which are shown in FIG. 8.
[0306]The protein sequences were aligned and compared using the default parameters for the bl2seq pairwise alignment program at NCBI http://www.ncbi.nim.nih.gov/blast/bl2seq/wblast2.cgi. For the DNA alignments, the EMBOSS pairwise alignment algorithms http://www.ebi.ac.uk/emboss/align/ with the water (local) method was used.
[0307]Multiple sequence alignments and phylogenetic tree generation was performed using the ClustaIX program as described by Thompson et al. (Nucl Acids Res 25: 4876-4882, 1997). The resultant phylogenetic tree is shown in FIG. 9.
Example 6
Mapping of the Barley CslF Genes
[0308]The QTL that has the largest effect on grain (1,3;1,4)-β-D-glucan content is located on barley chromosome 2H, between the Adh8 and ABG019 markers, markers, as mapped in the Steptoe×Morex doubled haploid (DH) population by Han et al. (Theor. Appl. Genet. 91: 921, 1995).
[0309]Specific gene fragments of the six barley CslF cDNAs generated by PCR were radiolabelled and used as probes on DNA from a set of wheat barley addition lines (Islam and the Clipper X Sahara barley DH populations to establish firstly their chromosomal location and then to fine map the genes.
[0310]Use of the wheat-barley addition lines showed that HvCslF2, 4, 5 and 6 are found on chromosome 2H, HvCslF1 is found on 7H and HvCslF3 is found on 1HS. Identification of polymorphisms between the Clipper and Sahara barley cultivars (parent lines) for HvCsl2, 4, 5 and 6 and the subsequent screening of the DH mapping population created from these parents allowed the accurate map location of these genes to be defined (FIG. 10). These four barley CslF genes are therefore found to be coincident with the major QTL for grain (1,3;1,4)-β-D-glucan content on barley chromosome 2H. This implies that one or more of these genes is likely to directly influence barley grain (1,3;1,4)-β-D-glucan content.
Materials and Methods
[0311]Filters of digested genomic DNA of the wheat barley addition lines (Islam et al., Heredity 46: 161-174 1981) were used to map the genes to the chromosome level. The barley DH mapping population Clipper×Sahara was used to fine map the HvCslF genes (Karakousis et al., Aust. J. Ag. Res. 54: 1137-1140, 2003). Professor Peter Langridge (Australian Centre for Plant Functional Genomics, University of Adelaide) kindly supplied both sets of filters of digested genomic DNA for Southern Hybridization analyses using standard methods. Loci were positioned using the Map Manager QTX software (Manly et al., Mammalian Genome 12: 930-932, 2001).
Example 7
Overexpression of the Barley CslF Genes in Transgenic Barley Plants
[0312]The role of individual members of the barley CslF gene family in (1,3;1,4)-β-D-glucan synthesis was tested by inserting the genes under the control of the strong constitutive promoter, CaMV 35S, into the genome of Golden Promise barley plants. The complete cDNAs for the barley genes CslF1, CslF4 and CslF6 were amplified by PCR using the primers shown in Table 4 and a high fidelity polymerase and sequenced to ensure that they contained no PCR-induced errors. The PCR fragments were cloned into the binary vector pMDC32, shown in FIG. 11.
[0313]The plasmid vectors were subsequently inserted into Agrobacterium tumefaciens, which was used to transform the barley cultivar Golden Promise by standard transformation procedures (Tingay et al. Plant J. 11: 1369-1376, 1997; Matthews et al. Mol. Breeding 7: 195-202, 2001)
[0314]Following selection with hygromycin, total RNA was extracted from the leaves of transgenic plantlets to monitor transcription of the endogenous and the integrated transgenes by Q-PCR (Burton et al. Plant Phys. 134: 224-236, 2004). The QPCR results for transcription of the selectable marker gene, hygromycin, using the primers shown in Table 5, are shown in FIG. 12. There is no PCR product for hygromycin in the cDNA of the wild type, non-transformed control plants (lines WT1-3) whilst all plants transgenic for the hygromycin transgene, including the transformed controls (G89 lines) which contain a gene unrelated to the CslF family, show positive levels of hygromycin QPCR product (FIG. 2)
[0315]Transcription of the three HvCslF genes was also examined in these plants using the pimers given in Table 5. Normalized mRNA levels are shown for HvCslF1, HvCslF4 and HvCslF6 across the whole transgenic population and the control plants. Plants transformed with 35S:HvCslF1 are designated G98 and overexpression of the HvCslF1 gene in these lines is evident at significant levels above that of the endogenous gene in the WTC, G89, G99 and G103 groups (FIG. 13). Particularly high levels of transcripts are seen in plants 98-10, 98-11 and 98-24 (FIG. 13). Plants transformed with 35S:HvCslF4 are designated G103 and overexpression of the HvCslF4 gene in these lines is evident at significant levels above that of the endogenous gene in the WTC, G89, G98 and G99 groups (FIG. 14). The highest level of transcript is seen in plant 103-5 (FIG. 14). Finally, plants transformed with 35S:HvCslF6 are designated G99 and overexpression of the HvCslF6 gene in these lines is evident at significant levels above that of the endogenous gene in the WTC, G89, G98 and G103 groups (FIG. 15), with the highest number of transcripts seen in lines 99-6 and 99-11 (FIG. 15).
Materials and Methods.
(i) Binary Vector Construction
[0316]The binary vector pMDC32 was obtained from Dr Mark Curtis, University of Zurich (http://www.unizh.ch/botinst/Devo Website/curtisvector/index 2.html) and is a Gateway-enabled (Invitrogen) binary vector carrying the hygromycin resistance gene and the CaMV 35S promoter suitable for use in barley transformation experiments where gene over-expression is desired (Curtis and Gossniklaus, Plant Phys 133: 462-469, 2003) Full-length PCR products corresponding to the barley HvCslF cDNAs amplified with the primers given in Table 4 were sequenced using BigDye 3.1 chemistry (ABI) on an Applied Biosystems ABI3700 capillary sequencer. Correct cDNAs were recombined into the Gateway entry vector pDENTR-Topo (Invitrogen). The orientation of the cDNA was verified by restriction enzyme digestion and then the entry clone was used in an LR recombination reaction (Invitrogen) with pMDC32 as the destination vector. Successful insertion into pMDC32 was confirmed by restriction enzyme digestion and plasmid DNA preparations containing verified inserts were transformed into Agrobacterium tumefaciens cv AGL0 via electroporation using the method of Mersereau at al. (Gene 90: 149, 1990) and positive colonies were selected on media containing 25 mg/l rifampicin, and 25 mg/l kanamycin.
(ii) Barley Transformation
[0317]Agrobacterium tumefaciens-mediated transformation experiments were performed using the procedure developed by Tingay et al. (1997, supra) and modified by Matthews et al. (2001, supra). The developing spikes were harvested from donor plants (cv. Golden Promise) grown in the glasshouse when the immature embryos were approximately 1-2 mm in diameter. The immature embryos were aseptically excised from the surface-sterilised grain, and the scutella were isolated by removing the embryonic axis. Twenty five freshly isolated scutella were cultured cut side-up in the centre of a 90 mm×10 mm Petri dish that contained callus induction medium, based on the recipe of Wan and Lemaux (Plant Phys. 104: 37-48, 1994). This medium was composed of MS macro-nutrients (Murashige and Skoog, Physiologia Plant. 15: 473-497, 1962), FHG micro-nutrients supplemented with 30 g/L maltose, 1 mg/L thiamine-HCl, 0.25 g/L myo-inositol, 1 g/L casein hydrolysate, 0.69 g/L L-proline, 10 μM CuSO4, 2.5 mg/L Dicamba (3,6-dichloro-o-anisic acid), and was solidified with 3.5 g/L Phytagel® (Sigma Chemicals, St. Louis, Mo., USA). Agrobacterium suspension (50 μl) was aliquotted onto the scutella, and the Petri dish was held at a 45° angle to drain away excess bacterial suspension. The explants were turned over and dragged across the surface of the medium to the edge of the Petri dish. The scutella were transferred to a fresh plate of callus induction medium and cultured cut side-up for three days in the dark at 22-24° C. Following co-cultivation, the scutella were removed to fresh callus induction medium containing 95 μM hygromycin B (Becton Dickinson Biosciences, Palo Alto, Calif., USA) and cultured in the dark. The entire callus of an individual scutellum was transferred to fresh selection medium every fortnight for a further six weeks. At the end of the callus selection period, the callus derived from each treated scutellum was transferred to shoot regeneration medium. This medium was based on the FHG recipe of Wan and Lemaux (1994, supra). It contained FHG macro- and micro-nutrients, 1 mg/L thiamine-HCl, 1 mg/L benzylaminopurine, 0.25 g/L myo-inositol, 0.73 g/L L-glutamine, 62 g/L maltose, 10 μM CuSO4, 38 μM hygromycin B, and was solidified with 3.5 g/L Phytagel®. The cultures were exposed to light (16 h day/8 h night photo-period) for three to four weeks at 22-24° C. The regenerated shoots were excised from the callus and transferred to culture boxes (Magenta Corporation, Chicago, Ill., USA) that contained hormone-free callus induction medium, supplemented with 95 μM hygromycin B to induce root formation. The tissue culture-derived plants that grew vigorously were established in soil and grown to maturity (Singh et al. Plant Cell, Tissue and Organ Culture 49:121-127 1997). All the media contained 150 mg/L Timentin® (SmithKline Beecham, Pty. Ltd., Melbourne, Australia) to inhibit the growth of Agrobacterium tumefaciens following co-cultivation.
(iii) RNA Extraction and cDNA Synthesis.
[0318]Total RNA was extracted from the leaves of plantlets growing in Magenta boxes, as described above, using TRIZOL, and cDNA was synthesised using the reverse transcriptase Superscript III (Invitrogen) as described in Burton et al. (Plant Phys 134: 224-236, 2004).
(iv) Quantitative Real Time PCR (Q-PCR) Analysis
[0319]The primer pairs for control genes (Burton et al., 2004, supra) and specific CslF genes were used as indicated in Table 5. Stock solutions of PCR products for the preparation of dilution series were prepared by PCR from a cDNA derived from either a composite of barley tissue cDNAs, and was subsequently purified and quantified by HPLC, as described by Burton et al. (2004, supra). A dilution series covering seven orders of magnitude was prepared from the 109 copies/μl stock solution as follows; one microlitre of the stock solution was added to 99 μl of water, and six 1:10 serial dilutions were prepared to produce a total of seven solutions covering 107 copies/μl to 101 copies/μl. Three replicates of each of the seven standard solutions were included with every Q-PCR experiment, together with a minimum of three no-template controls. For all genes, a 1:20 dilution of the cDNA was sufficient to produce expression data with an acceptable standard deviation. Three replicate PCRs for each of the cDNAs were included in every run. All Q-PCR reaction mixes were prepared on a CAS-1200 robot (Corbett Robotics, Brisbane, Australia).
[0320]Two microlitres of the diluted cDNA solution were used in a reaction containing 5 μl QuantiTect SYBR Green PCR reagent, 1 μl each of the forward and reverse primers at 4 μM, 0.3 μl 10× SYBR Green in water (10,000× in DMSO, BioWhittaker Molecular Applications, Rockland, USA, 0.5 μl in 500 μl of water, prepared daily) and 0.7 μl water. The total volume of each Q-PCR reaction mixture was 10 μl. Reactions were performed in a RG 3000 Rotor-Gene Real Time Thermal Cycler (Corbett Research, Sydney, Australia) as follows; 15 min at 95° followed by 45 cycles of 20 sec at 95°, 30 sec at 55°, 30 sec at 72° and 15 sec at the optimal acquisition temperature (AT) described in Table 5. A melt curve was obtained from the product at the end of the amplification by heating from 70° to 99°. After the experiment, the optimal cycle threshold (CT) was determined from the dilution series and the raw expression data was derived. The mean expression level and standard deviation for each set of three replicates for each cDNA was calculated.
[0321]The raw expression data for the HvCslF genes was scaled using the approach of Vandesompele et al. (Genome Biol. 3: 1-11, 2002). The normalisation factor derived from the best three of four barley control genes (Burton et al., 2004, supra) was generated using the Genorm software. (Vandesompele et al., 2002, supra). The raw expression data for the exogenous CslF genes in each cDNA was scaled by dividing the raw expression value by the normalisation factor for the particular cDNA.
TABLE-US-00004 TABLE 4 Primers used for amplification of barley Cs/F cDNAs Hv CsIF1 HvFD5END GGAGAGCGCGTGCATTGAG SEQ ID NO: 59 GACG Hv CsIF1 HvFDRQ TGTCCGGGCAAAGTCAT SEQ ID NO: 60 CAA Hv CsIF4 HvFC5N GCACGGTAGCCACTTACAC SEQ ID NO: 61 TATGG Hv CsIF4 HvFC3N TTGCAGTGACTCTGGCTGT SEQ ID NO: 62 ACTTG Hv CsI F6 HvFH5 GTAGCTGGCTACTGTGCAT SEQ ID NO: 63 AGC HvCSLF6 HvFF3N GAACTTACAAACCCCAGCT SEQ ID NO: 64 TGTGG Hv CsIF1 HvFD5END GGAGAGCGCGTGCATTGAG SEQ ID NO: 59 GACG
TABLE-US-00005 TABLE 5 Primers used for Q-PCR analysis. amplicon Gene Forward Primer Reverse Primer (bp) A T (° C.) Hyg GTCGATCGACAGATC GGCAGTTTAGCGAGA 291 82 CGGTC GCCTG (SEQ ID NO: 65) (SEQ ID NO: 66) Hv CsIF1 TGGGCATTCACCTTC TGTCCGGGCAAACTC 157 81 GTCAT ATCAA (SEQ ID NO: 67) (SEQ ID NO: 68) Hv CsIF4 CCGTCGGGCTCGTCT TTGCAGTGACTCTGG 144 79 ATGTC CTGTACTTG (SEQ ID NO: 69) (SEQ ID NO: 70) Hv CsIF6 GGGATTGTTCGGTTC GCTGTTGCTTTGCCA 250 77 CACTTT CATCTC (SEQ ID NO: 71) (SEQ ID NO: 72)
Example 8
Immunological Detection of (1,3;1,4)-β-D-Glucan in Transgenic Barley Lines at the Light Microscope Level
[0322]Transgenic barley lines as described in Example 7, in which the HvCslF1, HvCslF4 or HvCslF6 transcript levels driven by the 35S promoter were highest, were chosen for further analysis, with respect to the deposition of (1,3;1,4)-beta-D-glucan in the cell walls. Leaf sections were screened for the presence of the polysaccharide in the barley lines using an immunocytochemical method in which a monoclonal antibody specific for (1,3;1,4)-β-D-glucan (as described in Example 3) is detected by a fluorophore-conjugated secondary antibody and observed by light microscopy. Due to expression of endogenous CslF genes (1,3;1,4)-β-D-glucans are normally deposited in the cell walls of vegetative tissues such as leaf, and their occurrence and distribution in the emergent tissues of the barley seedling has been documented at the TEM level by Trethewey and Harris (New Phytologist 154: 347-358, 2002). Here we contrast the distribution pattern of endogenous (1,3;1,4)-β-D-glucans in control leaf sections with patterns displayed by transgenic leaf samples over-expressing barley CslF genes.
[0323]Leaf pieces representative of the plant groups described in example 7 were harvested, fixed and embedded in paraffin. Slide-mounted sections were treated with the specific monoclonal antibody which binds to (1,3;1,4)-beta-D-glucan (primary antibody) and, after washing, fluorophore-conjugated Alexa 488 (secondary antibody) was added. Sections were rinsed with buffer, mountant was added, and images were captured using a microscope with appropriate fluorescence filters. All images were taken at a standard exposure time of seven seconds. Overall morphology and the position of the various cell types were identified using the UV filter, which causes all cell wall material to fluoresce non-specifically (FIG. 16). Specific antibody signal was viewed using the 13 filter (FIGS. 17 and 18).
[0324]All control sections taken from any sample treated without either primary, secondary or both antibodies showed very low levels of fluorescence at 400× magnification (FIG. 17). Labeling with both primary and secondary antibodies appears as a green signal when using the 13 filter (FIG. 18). As expected with both antibodies, a level of endogenous (1,3;1,4)-β-D-glucans was evident on tissue sections taken from wild type (WT, FIG. 18E) and transgenic control (G89, FIG. 18F), where signal was mainly concentrated in the mesophyll cells and vascular bundles in the mid regions of the sections. Under UV it was evident that all cells in all sections were present and intact. In contrast, antibody-labeled sections taken from the transgenic G98 and G103 plants, such as G98-10 (FIG. 18A), G98-24 (FIG. 18B) and G103-5 (FIG. 18c), display an increased intensity of signal, where the walls of the epidermal cells and sclerenchyma fibre cells are much more heavily labelled. The sclerenchyma cells have thickened secondary cell walls and Trethewey and Harris (2002, supra) detected only sparse labelling at the TEM level in these cells from wild type seedlings. The labelling pattern displayed by G99 plants, such as G99-12 (FIG. 18D), varies from that shown by the wild type and transgenic controls and from the G98 and G103 plants. In this case fluorescence is seen only in the stomatal cells and parts of the vascular bundle.
[0325]These results indicate that over-expression of individual barley CslF genes may lead to elevated protein levels of the glucan synthase enzymes, which in turn leads to increased deposition of (1,3;1,4)-beta-D-glucans in the cell walls of these transgenic plants.
Materials and Methods
(i) Preparation of Transformed Barley Leaves for Light Microscopy
[0326]Barley leaf pieces were fixed in 0.25% (v/v) glutaraldehyde, 4% (v/v) paraformaldehyde (EM Grade), and 4% sucrose in phosphate-buffered saline (PBS), pH 7.2, and stored at 4° C. overnight. Samples were washed three times in PBS, dehydrated in a graded ethanol series and slowly infiltrated with paraffin over several days. Blocks were trimmed and sectioned at 6 μm.
(ii) Immunolabeling for Light Microscopy with a Fluorophore-Conjugated Antibody
[0327]Sections were dewaxed in xylene and rehydrated progressively through 100%, 90% and 70% ethanol solutions. After two rinses with 1× phosphate buffered saline (PBS) the sections were incubated with 0.05M glycine for 20 mins to inactivate residual aldehyde groups. Sections were blocked in incubation buffer (1% bovine serum albumin (BSA) in 1×PBS) for 2×10 mins to prevent non-specific binding. Slides were drained and the specific primary antibody, BG1 (BioSupplies, Melbourne, Australia) was added at a dilution of 1:50 and left to incubate for one hour in a humidity chamber. Unbound primary antibody was removed by rinsing with incubation buffer for 3×10 mins and the secondary antibody, Alexa Fluor® 488 goat anti-mouse IgG (Molecular Probes, Eugene, USA), was added. Slides were wrapped in aluminium foil to exclude light and incubated for 2 hours at room temperature. Unbound secondary antibody was removed by rinsing 3×10 mins with incubation buffer before a few drops of mountant (90% glycerol: 10% water) were applied and sections cover-slipped. Controls were included which omitted either the primary antibody, the secondary antibody or both antibodies. Images were captured on a Leica AS LMD microscope under filter D (UV, excitation 355-425 nm) or filter I3 (blue, excitation 450-490 nm) using a DFC480 CCD camera. All images shown were taken at 400× magnification with a 7 second exposure time to standardise fluorescence intensity.
Example 9
Immunological Detection of (1,3;1,4)-β-D-Glucans in Transgenic Barley Using Transmission Electron Microscopy
[0328]Transgenic barley lines, as described in Examples 7 and 8, in which the HvCslF1, 4 or 6 transcript levels driven by the 35S promoter were highest, were chosen for further analysis, with respect to the deposition of (1,3;1,4)-β-D-glucan in the cell walls. An immunocytochemical method using the monoclonal antibody specific for (1,3;1,4)-β-D-glucan, as described in Example 3, and electron microscopy, was employed to screen leaf sections for the presence of the polysaccharide in the barley lines. Due to expression of the endogenous CslF genes (1,3;1,4)-β-D-glucans are normally deposited in the cell walls of vegetative tissues, such as leaf, and their occurrence and distribution in the emergent tissues of the barley seedling has been documented at the TEM level by Trethewey and Harris (New Phytologist 154: 347-358, 2002). In Example 7 we contrasted the distribution of endogenous (1,3;1,4)-β-D-glucans at the light microscope level in control material with that displayed by leaf sections of plantlets over-expressing barley CslF genes. A repeat analysis of a sub-set of the same lines using the specific monoclonal antibody and TEM, which allows a much closer examination of individual cell walls, is described here.
[0329]Leaf pieces representative of the plant groups described in Example 7 were harvested, fixed and embedded in LR white resin. Mounted sections were treated with the specific monoclonal antibody which binds to (1,3;1,4)-quadrature-D-glucan and colloidal gold and examined on the transmission electron microscope.
[0330]The presence of endogenous (1,3;1,4)-quadrature-D-glucans was clearly evident, as expected, on sections taken from wild type (WT) and transgenic control (G89) leaves. In contrast, sections taken from the transgenic G98 and G103 plants, such as G98-10 and G103-5, display much heavier labeling. A labeled epidermal cell wall of the control G89 is shown in FIG. 19A. Equivalent epidermal cell walls of the transgenics G98-10 and G103-5 are shown in FIG. 19B (G98-10) and 19C (G103-5), where the increased amount of labeling is clearly evident. FIG. 20A shows the wall of a sclerenchyma fibre cell in the G89 control which is lightly labeled. Such cells have thickened secondary cell walls and Trethewey and Harris (2002, supra) also detected sparse labeling at the TEM level in these fibres from wild type seedlings. In comparison the fibre cell walls from the transgenic G98-10 and G103-5 plants, shown in FIGS. 20B and 20C respectively, are more heavily labeled. These results indicate that over-expression of individual CslF genes has the potential to increase deposition of (1,3;1,4)-quadrature-D-glucans in the cell walls of transgenic plants. This has been demonstrated using both a fluorophore for glucan detection at the light microscope level (Example 7) and, as demonstrated here, by employing immunogold labeling at the TEM level.
Materials and Methods
(i) Preparation of Transformed Barley Leaves for Electron Microscopy
[0331]Pieces of barley plantlet leaves were fixed in 0.25% glutaraldehyde, 4% (v/v) paraformaldehyde (EM Grade) and 4% sucrose in phosphate-buffered saline (PBS), pH 7.2, and stored at 4° C. overnight. After three rinses in MilliQ water the samples were dehydrated in a graded ethanol series and slowly infiltrated with LR White resin over several days. Individual leaf pieces were placed in gelatin capsules, which were filled with fresh resin and polymerized overnight at 65° C.
(ii) Immunolocation for Transmission Electron Microscopy
[0332]Sections (80 nm) of barley leaves were prepared on a Leica Ultracut R microtome using a diamond knife and collected on 100 and 200 mesh, Formvar coated gold grids. The ultrathin sections were blocked for 30 min in 1% bovine serum albumin in PBS before incubation in murine monoclonal antibodies raised against barley (1,3;1,4)-β-D-glucan (diluted 1:500; Biosupplies Australia, Parkville, VIC 3052, Australia) for 1 hr at room temperature and overnight at 4° C. The grids were washed twice in PBS and three times in blocking buffer before a 1 h incubation in 18 nm Colloidal Gold-AffiniPure Goat-Anti Mouse IgG+IgM (H+L) (Jackson ImmunoResearch Laboratories, Inc., PA, USA). All grids were washed twice in PBS and several times in MilliQ water before viewing on a Philips BioTwin Transmission Electron Microscope and images captured on a Gatan Multiscan CCD Camera.
[0333]Those skilled in the art will appreciate that the invention described herein is susceptible to variations and modifications other than those specifically described. It is to be understood that the invention includes all such variations and modifications. The invention also includes all of the steps, features, compositions and compounds referred to, or indicated in this specification, individually or collectively, and any and all combinations of any two or more of the steps or features.
[0334]Also, it must be noted that, as used herein, the singular forms "a", "an" and "the" include plural aspects unless the context already dictates otherwise. Thus, for example, reference to "a transgene" includes a single transgene as well as two or more transgenes; "a plant cell" includes a single cell as well as two or more cells; and so forth.
Sequence CWU
1
7212841DNAHordeum vulgare 1atggcgccag cggtggccgg agggggccgc gtgcggagca
atgagccggt tgctgctgct 60gccgccgcgc cggcggccag cggcaagccc tgcgtgtgcg
gcttccaggt ttgcgcctgc 120acggggtcgg ccgcggtggc ctccgccgcc tcgtcgctgg
acatggacat cgtggccatg 180gggcagatcg gcgccgtcaa cgacgagagc tgggtgggcg
tggagctcgg cgaagatggc 240gagaccgacg aaagcggtgc cgccgttgac gaccgccccg
tattccgcac cgagaagatc 300aagggtgtcc tcctccaccc ctaccgggtg ctgattttcg
ttcgtctgat cgccttcacg 360ctgttcgtga tctggcgtat ctcccacaag aacccagacg
cgatgtggct gtgggtgaca 420tccatctgcg gcgagttctg gttcggtttc tcgtggctgc
tagatcagct gcccaagctg 480aaccccatca accgcgtgcc ggacctggcg gtgctgcggc
agcgcttcga ccgccccgac 540ggcacctcca cgctcccggg gctggacatc ttcgtcacca
cggccgaccc catcaaggag 600cccatcctct ccaccgccaa ctcggtgctc tccatcctgg
ccgccgacta ccccgtggac 660cgcaacacat gctacgtctc cgacgacagt ggcatgctgc
tcacctacga ggccctggca 720gagtcctcca agttcgccac gctctgggtg cccttctgcc
gcaagcacgg gatcgagccc 780aggggtccgg agagctactt cgagctcaag tcacaccctt
acatggggag agcccaggac 840gagttcgtca acgaccgccg ccgcgttcgc aaggagtacg
acgagttcaa ggccaggatc 900aacagcctgg agcatgacat caagcagcgc aacgacgggt
acaacgccgc cattgcccac 960agccaaggcg tgccccggcc cacctggatg gcggacggca
cccagtggga gggcacatgg 1020gtcgacgcct ccgagaacca ccgcaggggc gaccacgccg
gcatcgtact ggtgctgctg 1080aaccacccga gccaccgccg gcagacgggc ccgccggcga
gcgctgacaa cccactggac 1140ttgagcggcg tggatgtgcg tctccccatg ctggtgtacg
tgtcccgtga gaagcgcccc 1200gggcacgacc accagaagaa ggccggtgcc atgaacgcgc
ttacccgcgc ctcggcgctg 1260ctctccaact cccccttcat cctcaacctc gactgcgatc
attacatcaa caactcccag 1320gcccttcgcg ccggcatctg cttcatggtg ggacgggaca
gcgacacggt tgccttcgtc 1380cagttcccgc agcgcttcga gggcgtcgac cccaccgacc
tctacgccaa ccacaaccgc 1440atcttcttcg acggcaccct ccgtgccctg gacggcatgc
agggccccat ctacgtcggc 1500actgggtgtc tcttccgccg catcaccgtc tacggcttcg
acccgccgag gatcaacgtc 1560ggcggtccct gcttccccag gctcgccggg ctcttcgcca
agaccaagta cgagaagccc 1620gggctcgaga tgaccacggc caaggccaag gccgcgcccg
tgcccgccaa gggtaagcac 1680ggcttcttgc cactgcccaa gaagacgtac ggcaagtcgg
acgccttcgt ggacaccatc 1740ccgcgcgcgt cgcacccgtc gccctacgcc gcggcggctg
aggggatcgt ggccgacgag 1800gcgaccatcg tcgaggcggt gaacgtgacg gccgccgcgt
tcgagaagaa gaccggctgg 1860ggcaaagaga tcggctgggt gtacgacacc gtcacggagg
acgtggtcac cggctaccgg 1920atgcatatca aggggtggcg gtcacgctac tgctccatct
acccacacgc cttcatcggc 1980accgccccca tcaacctcac ggagaggctc ttccaggtgc
tccgctggtc cacgggatcc 2040ctcgagatct tcttctccaa gaacaacccg ctcttcggca
gcacatacct ccacccgctg 2100cagcgcgtcg cctacatcaa catcaccact taccccttca
ccgccatctt cctcatcttc 2160tacaccaccg tgccggcgct atccttcgtc accggccact
tcatcgtgca gcgcccgacc 2220accatgttct acgtctacct gggcatcgtg ctatccacgc
tgctcgtcat cgccgtgctg 2280gaggtcaagt gggccggggt cacagtcttc gagtggttca
ggaacggcca gttctggatg 2340acagcaagtt gctccgccta cctcgccgcc gtctgccagg
tgctgaccaa ggtgatattc 2400cggcgggaca tctccttcaa gctcacatcc aagctaccct
cgggagacga gaagaaggac 2460ccctacgccg acctctacgt ggtgcgctgg acgccgctca
tgattacacc catcatcatc 2520atcttcgtca acatcatcgg atccgccgtg gccttcgcca
aggttctcga cggcgagtgg 2580acgcactggc tcaaggtcgc cggcggcgtc ttcttcaact
tctgggtgct cttccacctc 2640taccccttcg ccaagggcat cctggggaag cacggaaaga
cgccagtcgt ggtgctcgtc 2700tggtgggcat tcaccttcgt catcaccgcc gtgctctaca
tcaacatccc ccacatgcat 2760acctcgggag gcaagcacac aacggtgcat ggtcaccatg
gcaagaagtt ggtcgacaca 2820gggctctatg gctggctcca t
28412947PRTHordeum vulgare 2Met Ala Pro Ala Val Ala
Gly Gly Gly Arg Val Arg Ser Asn Glu Pro1 5
10 15Val Ala Ala Ala Ala Ala Ala Pro Ala Ala Ser Gly
Lys Pro Cys Val 20 25 30Cys
Gly Phe Gln Val Cys Ala Cys Thr Gly Ser Ala Ala Val Ala Ser 35
40 45Ala Ala Ser Ser Leu Asp Met Asp Ile
Val Ala Met Gly Gln Ile Gly 50 55
60Ala Val Asn Asp Glu Ser Trp Val Gly Val Glu Leu Gly Glu Asp Gly65
70 75 80Glu Thr Asp Glu Ser
Gly Ala Ala Val Asp Asp Arg Pro Val Phe Arg 85
90 95Thr Glu Lys Ile Lys Gly Val Leu Leu His Pro
Tyr Arg Val Leu Ile 100 105
110Phe Val Arg Leu Ile Ala Phe Thr Leu Phe Val Ile Trp Arg Ile Ser
115 120 125His Lys Asn Pro Asp Ala Met
Trp Leu Trp Val Thr Ser Ile Cys Gly 130 135
140Glu Phe Trp Phe Gly Phe Ser Trp Leu Leu Asp Gln Leu Pro Lys
Leu145 150 155 160Asn Pro
Ile Asn Arg Val Pro Asp Leu Ala Val Leu Arg Gln Arg Phe
165 170 175Asp Arg Pro Asp Gly Thr Ser
Thr Leu Pro Gly Leu Asp Ile Phe Val 180 185
190Thr Thr Ala Asp Pro Ile Lys Glu Pro Ile Leu Ser Thr Ala
Asn Ser 195 200 205Val Leu Ser Ile
Leu Ala Ala Asp Tyr Pro Val Asp Arg Asn Thr Cys 210
215 220Tyr Val Ser Asp Asp Ser Gly Met Leu Leu Thr Tyr
Glu Ala Leu Ala225 230 235
240Glu Ser Ser Lys Phe Ala Thr Leu Trp Val Pro Phe Cys Arg Lys His
245 250 255Gly Ile Glu Pro Arg
Gly Pro Glu Ser Tyr Phe Glu Leu Lys Ser His 260
265 270Pro Tyr Met Gly Arg Ala Gln Asp Glu Phe Val Asn
Asp Arg Arg Arg 275 280 285Val Arg
Lys Glu Tyr Asp Glu Phe Lys Ala Arg Ile Asn Ser Leu Glu 290
295 300His Asp Ile Lys Gln Arg Asn Asp Gly Tyr Asn
Ala Ala Ile Ala His305 310 315
320Ser Gln Gly Val Pro Arg Pro Thr Trp Met Ala Asp Gly Thr Gln Trp
325 330 335Glu Gly Thr Trp
Val Asp Ala Ser Glu Asn His Arg Arg Gly Asp His 340
345 350Ala Gly Ile Val Leu Val Leu Leu Asn His Pro
Ser His Arg Arg Gln 355 360 365Thr
Gly Pro Pro Ala Ser Ala Asp Asn Pro Leu Asp Leu Ser Gly Val 370
375 380Asp Val Arg Leu Pro Met Leu Val Tyr Val
Ser Arg Glu Lys Arg Pro385 390 395
400Gly His Asp His Gln Lys Lys Ala Gly Ala Met Asn Ala Leu Thr
Arg 405 410 415Ala Ser Ala
Leu Leu Ser Asn Ser Pro Phe Ile Leu Asn Leu Asp Cys 420
425 430Asp His Tyr Ile Asn Asn Ser Gln Ala Leu
Arg Ala Gly Ile Cys Phe 435 440
445Met Val Gly Arg Asp Ser Asp Thr Val Ala Phe Val Gln Phe Pro Gln 450
455 460Arg Phe Glu Gly Val Asp Pro Thr
Asp Leu Tyr Ala Asn His Asn Arg465 470
475 480Ile Phe Phe Asp Gly Thr Leu Arg Ala Leu Asp Gly
Met Gln Gly Pro 485 490
495Ile Tyr Val Gly Thr Gly Cys Leu Phe Arg Arg Ile Thr Val Tyr Gly
500 505 510Phe Asp Pro Pro Arg Ile
Asn Val Gly Gly Pro Cys Phe Pro Arg Leu 515 520
525Ala Gly Leu Phe Ala Lys Thr Lys Tyr Glu Lys Pro Gly Leu
Glu Met 530 535 540Thr Thr Ala Lys Ala
Lys Ala Ala Pro Val Pro Ala Lys Gly Lys His545 550
555 560Gly Phe Leu Pro Leu Pro Lys Lys Thr Tyr
Gly Lys Ser Asp Ala Phe 565 570
575Val Asp Thr Ile Pro Arg Ala Ser His Pro Ser Pro Tyr Ala Ala Ala
580 585 590Ala Glu Gly Ile Val
Ala Asp Glu Ala Thr Ile Val Glu Ala Val Asn 595
600 605Val Thr Ala Ala Ala Phe Glu Lys Lys Thr Gly Trp
Gly Lys Glu Ile 610 615 620Gly Trp Val
Tyr Asp Thr Val Thr Glu Asp Val Val Thr Gly Tyr Arg625
630 635 640Met His Ile Lys Gly Trp Arg
Ser Arg Tyr Cys Ser Ile Tyr Pro His 645
650 655Ala Phe Ile Gly Thr Ala Pro Ile Asn Leu Thr Glu
Arg Leu Phe Gln 660 665 670Val
Leu Arg Trp Ser Thr Gly Ser Leu Glu Ile Phe Phe Ser Lys Asn 675
680 685Asn Pro Leu Phe Gly Ser Thr Tyr Leu
His Pro Leu Gln Arg Val Ala 690 695
700Tyr Ile Asn Ile Thr Thr Tyr Pro Phe Thr Ala Ile Phe Leu Ile Phe705
710 715 720Tyr Thr Thr Val
Pro Ala Leu Ser Phe Val Thr Gly His Phe Ile Val 725
730 735Gln Arg Pro Thr Thr Met Phe Tyr Val Tyr
Leu Gly Ile Val Leu Ser 740 745
750Thr Leu Leu Val Ile Ala Val Leu Glu Val Lys Trp Ala Gly Val Thr
755 760 765Val Phe Glu Trp Phe Arg Asn
Gly Gln Phe Trp Met Thr Ala Ser Cys 770 775
780Ser Ala Tyr Leu Ala Ala Val Cys Gln Val Leu Thr Lys Val Ile
Phe785 790 795 800Arg Arg
Asp Ile Ser Phe Lys Leu Thr Ser Lys Leu Pro Ser Gly Asp
805 810 815Glu Lys Lys Asp Pro Tyr Ala
Asp Leu Tyr Val Val Arg Trp Thr Pro 820 825
830Leu Met Ile Thr Pro Ile Ile Ile Ile Phe Val Asn Ile Ile
Gly Ser 835 840 845Ala Val Ala Phe
Ala Lys Val Leu Asp Gly Glu Trp Thr His Trp Leu 850
855 860Lys Val Ala Gly Gly Val Phe Phe Asn Phe Trp Val
Leu Phe His Leu865 870 875
880Tyr Pro Phe Ala Lys Gly Ile Leu Gly Lys His Gly Lys Thr Pro Val
885 890 895Val Val Leu Val Trp
Trp Ala Phe Thr Phe Val Ile Thr Ala Val Leu 900
905 910Tyr Ile Asn Ile Pro His Met His Thr Ser Gly Gly
Lys His Thr Thr 915 920 925Val His
Gly His His Gly Lys Lys Leu Val Asp Thr Gly Leu Tyr Gly 930
935 940Trp Leu His94532553DNAHordeum vulgare
3atggcgtcgg cggccggtgc tgctgggtca aatgccagcc tcgccgcccc gctgctggcg
60agccgcgagg gaggagccaa gaagccggtc ggtgccaagg gcaagcactg ggaggccgcc
120gacaaggacg agcggcgggc cgccaaggag agcggcggcg aggacggcag gccgctgctg
180ttccggacgt acaaggtcaa aggcaccctc ctgcacccat acagggcgct aatcttcatt
240cgcttaattg cggtccttct attcttcgta tggcgcatca agcacaacaa atccgacatc
300atgtggtttt ggacaatatc agtcgtcggg gacgtatggt tcgggttctc gtggctgctc
360aaccaactcc caaagttcaa ccctatcaaa accatacctg atatggtcgc ccttaggcga
420caatacgatc tttcagatgg gacatctaca ctcccgggca tagatgtctt tgtcaccacc
480gctgacccaa tcgatgagcc gatactatac accatgaatt gtgtcctttc tatccttgct
540tctgactatc ctgtcgatag gtgtgcctgc tatctcccag atgatagtgg agcattgatt
600caatacgagg ccttagttga gaccgcaaag tttgctactt tgtgggtccc attttgtcgg
660aagcattgca ttgagccaag agccccagaa agctactttg aaatagaggc accgttgtac
720actggaactg caccagagga gttcaagaat gattatagta gtgtacataa agagtatgat
780gagttcaaag agcgcttgga ctcactatcc gatgctattt ccaagcgttc tgatgcttac
840aacagcatga agactgagga aggagatgca aaggccacgt ggatggcaaa tgggacacaa
900tggccaggat catggattga cacaacggaa atccatagaa aaggacatca tgccggaatt
960gttaaggttg tgttggacca ttcgatccgt gggcataatc ttggttcaca agaaagcacc
1020cacaacctca gtttcgccaa caccgatgag cgcctcccga tgcttgtgta tatctctcgt
1080ggaaagaacc caagctatga ccacaacaag aaagctggtg ccttgaatgc gcaattgcgt
1140gcctctgcac tactctccaa cgcacaattc atcatcaact ttgactgcga ccactacatc
1200aacaactctc aagccctacg tgcagctatg tgcttcatgc ttgatcaaag gcaaggtgat
1260aacactgcct ttgttcaatt ccctcaacgc ttcgacaatg ttgatccatc agaccgatat
1320ggaaaccaca accgtgtctt ctttgacggc acaatgctcg ccctcaatgg cctccaaggg
1380ccatcttacc ttggcactgg ttgcatgttc cgccgcatag cactttatgg cattgaccca
1440cctgactgga gacatgacaa catcatagtt gatgataaaa agtttggtag ctccataccc
1500ttcctagatt ccgtatcaaa agccataaac caagaaaggt ctaccatacc tccacccatt
1560agtgaaacat tggtggctga gatggaaagg gttgtgtcgg cttcacacga taaagccact
1620ggttggggca agggtgttgg gtacatatat gacatagcca cagaggatat cgtgactggt
1680ttccgcatcc atgggcaagg ttggcgttcc atgtattgta caatggagcg tgacgccttc
1740tgtggcattg caccaatcaa cctaaccgag cgcctccacc aaattgtgcg ctggtccggt
1800ggatctttag agatgttctt ctcactaaat aacccactca taggtggtcg ccggatccac
1860gcccttcagc gtgtctccta cctcaacatg acagtctacc cagtcacatc actctttatc
1920ctactctatg ctctcagccc agtgatgtgg cttatccctg atgaagtata catccagagg
1980ccattcacca aatatgtcgt gttccttctc gtgatcattc tgatgatcca tataattggg
2040tggctcgaga taaaatgggc gggggtcaca tggttggatt actggaggaa tgaacagttc
2100tttatgatcg ggtcgacgag tgcataccca gcagccgtgc tgcacatggt ggtgaatctc
2160cttacaaaga agggtataca cttcagagtt acttcgaagc aaacaacggc agacaccaat
2220gacaagtttg ctgacttgta tgacatgcga tgggtgccaa tgttaatccc tacaacagtg
2280gtgctgattg ccaatgttgg tgcaatcggt gtagccatgg gtaaaacgat agtatacatg
2340ggagcatgga caattgcaca gaagacacat gccgcattgg gtctgctctt caacgtgtgg
2400atcatggtcc tgctctatcc gtttgcattg gcgatcatgg gacggtgggc aaagaggcca
2460gtcatcctgg tggtcttgtt gccggttgcc tttacaatag tttgccttgt atatgtttct
2520gttcatatat tacttcttag ttttcttcca ttt
25534851PRTHordeum vulgare 4Met Ala Ser Ala Ala Gly Ala Ala Gly Ser Asn
Ala Ser Leu Ala Ala1 5 10
15Pro Leu Leu Ala Ser Arg Glu Gly Gly Ala Lys Lys Pro Val Gly Ala
20 25 30Lys Gly Lys His Trp Glu Ala
Ala Asp Lys Asp Glu Arg Arg Ala Ala 35 40
45Lys Glu Ser Gly Gly Glu Asp Gly Arg Pro Leu Leu Phe Arg Thr
Tyr 50 55 60Lys Val Lys Gly Thr Leu
Leu His Pro Tyr Arg Ala Leu Ile Phe Ile65 70
75 80Arg Leu Ile Ala Val Leu Leu Phe Phe Val Trp
Arg Ile Lys His Asn 85 90
95Lys Ser Asp Ile Met Trp Phe Trp Thr Ile Ser Val Val Gly Asp Val
100 105 110Trp Phe Gly Phe Ser Trp
Leu Leu Asn Gln Leu Pro Lys Phe Asn Pro 115 120
125Ile Lys Thr Ile Pro Asp Met Val Ala Leu Arg Arg Gln Tyr
Asp Leu 130 135 140Ser Asp Gly Thr Ser
Thr Leu Pro Gly Ile Asp Val Phe Val Thr Thr145 150
155 160Ala Asp Pro Ile Asp Glu Pro Ile Leu Tyr
Thr Met Asn Cys Val Leu 165 170
175Ser Ile Leu Ala Ser Asp Tyr Pro Val Asp Arg Cys Ala Cys Tyr Leu
180 185 190Pro Asp Asp Ser Gly
Ala Leu Ile Gln Tyr Glu Ala Leu Val Glu Thr 195
200 205Ala Lys Phe Ala Thr Leu Trp Val Pro Phe Cys Arg
Lys His Cys Ile 210 215 220Glu Pro Arg
Ala Pro Glu Ser Tyr Phe Glu Ile Glu Ala Pro Leu Tyr225
230 235 240Thr Gly Thr Ala Pro Glu Glu
Phe Lys Asn Asp Tyr Ser Ser Val His 245
250 255Lys Glu Tyr Asp Glu Phe Lys Glu Arg Leu Asp Ser
Leu Ser Asp Ala 260 265 270Ile
Ser Lys Arg Ser Asp Ala Tyr Asn Ser Met Lys Thr Glu Glu Gly 275
280 285Asp Ala Lys Ala Thr Trp Met Ala Asn
Gly Thr Gln Trp Pro Gly Ser 290 295
300Trp Ile Asp Thr Thr Glu Ile His Arg Lys Gly His His Ala Gly Ile305
310 315 320Val Lys Val Val
Leu Asp His Ser Ile Arg Gly His Asn Leu Gly Ser 325
330 335Gln Glu Ser Thr His Asn Leu Ser Phe Ala
Asn Thr Asp Glu Arg Leu 340 345
350Pro Met Leu Val Tyr Ile Ser Arg Gly Lys Asn Pro Ser Tyr Asp His
355 360 365Asn Lys Lys Ala Gly Ala Leu
Asn Ala Gln Leu Arg Ala Ser Ala Leu 370 375
380Leu Ser Asn Ala Gln Phe Ile Ile Asn Phe Asp Cys Asp His Tyr
Ile385 390 395 400Asn Asn
Ser Gln Ala Leu Arg Ala Ala Met Cys Phe Met Leu Asp Gln
405 410 415Arg Gln Gly Asp Asn Thr Ala
Phe Val Gln Phe Pro Gln Arg Phe Asp 420 425
430Asn Val Asp Pro Ser Asp Arg Tyr Gly Asn His Asn Arg Val
Phe Phe 435 440 445Asp Gly Thr Met
Leu Ala Leu Asn Gly Leu Gln Gly Pro Ser Tyr Leu 450
455 460Gly Thr Gly Cys Met Phe Arg Arg Ile Ala Leu Tyr
Gly Ile Asp Pro465 470 475
480Pro Asp Trp Arg His Asp Asn Ile Ile Val Asp Asp Lys Lys Phe Gly
485 490 495Ser Ser Ile Pro Phe
Leu Asp Ser Val Ser Lys Ala Ile Asn Gln Glu 500
505 510Arg Ser Thr Ile Pro Pro Pro Ile Ser Glu Thr Leu
Val Ala Glu Met 515 520 525Glu Arg
Val Val Ser Ala Ser His Asp Lys Ala Thr Gly Trp Gly Lys 530
535 540Gly Val Gly Tyr Ile Tyr Asp Ile Ala Thr Glu
Asp Ile Val Thr Gly545 550 555
560Phe Arg Ile His Gly Gln Gly Trp Arg Ser Met Tyr Cys Thr Met Glu
565 570 575Arg Asp Ala Phe
Cys Gly Ile Ala Pro Ile Asn Leu Thr Glu Arg Leu 580
585 590His Gln Ile Val Arg Trp Ser Gly Gly Ser Leu
Glu Met Phe Phe Ser 595 600 605Leu
Asn Asn Pro Leu Ile Gly Gly Arg Arg Ile His Ala Leu Gln Arg 610
615 620Val Ser Tyr Leu Asn Met Thr Val Tyr Pro
Val Thr Ser Leu Phe Ile625 630 635
640Leu Leu Tyr Ala Leu Ser Pro Val Met Trp Leu Ile Pro Asp Glu
Val 645 650 655Tyr Ile Gln
Arg Pro Phe Thr Lys Tyr Val Val Phe Leu Leu Val Ile 660
665 670Ile Leu Met Ile His Ile Ile Gly Trp Leu
Glu Ile Lys Trp Ala Gly 675 680
685Val Thr Trp Leu Asp Tyr Trp Arg Asn Glu Gln Phe Phe Met Ile Gly 690
695 700Ser Thr Ser Ala Tyr Pro Ala Ala
Val Leu His Met Val Val Asn Leu705 710
715 720Leu Thr Lys Lys Gly Ile His Phe Arg Val Thr Ser
Lys Gln Thr Thr 725 730
735Ala Asp Thr Asn Asp Lys Phe Ala Asp Leu Tyr Asp Met Arg Trp Val
740 745 750Pro Met Leu Ile Pro Thr
Thr Val Val Leu Ile Ala Asn Val Gly Ala 755 760
765Ile Gly Val Ala Met Gly Lys Thr Ile Val Tyr Met Gly Ala
Trp Thr 770 775 780Ile Ala Gln Lys Thr
His Ala Ala Leu Gly Leu Leu Phe Asn Val Trp785 790
795 800Ile Met Val Leu Leu Tyr Pro Phe Ala Leu
Ala Ile Met Gly Arg Trp 805 810
815Ala Lys Arg Pro Val Ile Leu Val Val Leu Leu Pro Val Ala Phe Thr
820 825 830Ile Val Cys Leu Val
Tyr Val Ser Val His Ile Leu Leu Leu Ser Phe 835
840 845Leu Pro Phe 85052571DNAHordeum vulgare
5atggcttctc cggcggccgt cggcgggggt cgtctagccg acccactgct ggccgccgac
60gtcgtcgtcg tcggcgccaa agacaagtac tgggtgcccg ccgacgagag agagatcctg
120gcgtcgcaga gcagcggcgg cggtgaacag gacggccggg caccgctgct ataccgcacg
180ttcagggtca agggcttctt catcaacctt tacaggttat tgactctggt cagagttatc
240gtggttattc tattcttcac gtggcgcatg aggcaccggg actcggacgc gatgtggctg
300tggtggatct cggtcgtggg cgacctctgg ttcggagtca cctggctgct caaccagatc
360accaagctca agcccaggaa atgcgtcccc agcatctccg tcctgagaga gcagctcgac
420cagcccgacg gcggctccga cctgcccctt ctcgacgtgt tcatcaacac cgtcgacccg
480gtggacgagc cgatgctcta caccatgaac tccatcctct ccatcctggc caccgactac
540cccgtccaga agtacgccac ctatttctcc gatgacggcg ggtcgctggt gcactacgag
600gggctgctgc tgacggcgga gttcgccgcg tcgtgggtcc cgttctgccg gaagcattgc
660gtcgagcctc gcgccccgga gagctacttc tgggccaaga tgcgcgggga gtacgccggc
720agcgcggcca aggagttcct tgacgaccat cggaggatgc gcgcggcgta tgaggagttc
780aaggcgaggc tggacgggct ttctgccgtc atcgagcagc ggtccgaggc gtgcaaccgc
840gctgcaaacg agaaagaagg gtgtgggaac gcgacttgga tggccgatgg gtcgacgcaa
900tggcagggga cgtggatcaa gccggccaag ggccaccgga aaggacacca tcctgcaatt
960cttcaggtta tgctggatca acctagcaag gatcctgagc tgggaatggc ggcgagctcc
1020gaccaccctc tggatttcag cgccgtggac gtgcgcctcc cgatgctggt ctacattgcc
1080cgggagaagc ggcctgggta tgaccaccag aagaaggcgg gcgccatgaa cgtgcagctg
1140cgcgtgtccg cgctgctctc caacgcgccc ttcatcatca acttcgacgg cgaccactac
1200atcaacaact cgcaggcctt ccgcgccgcc atgtgcttca tgctcgaccc gcgcgacggc
1260gccgacaccg ccttcgtcca gttcccgcag cgcttcgacg acgtcgaccc caccgaccgc
1320tactgcaacc acaaccgcat gttcttcgac gccaccctcc tcggcctcaa cggcatccag
1380ggcccctcct tcgtcggcac cggatgcatg ttccgccgcg tcgctctcta cagcgccgac
1440cctccacggt ggcggtccga cgacgccaag gaggccaagg cctcgcacag gcccaacatg
1500tttggcaagt ctacgtcctt catcaactca atgccggcgg ccgccaacca agaacggtcc
1560gtcccgtcac cggcgacagt cggcgaggcg gagctcgcag acgcgatgac ttgcgcgtac
1620gaggacggca ccgagtgggg caacgacgtt gggtgggtgt acaacatcgc gacggaggac
1680gtggtgaccg gcttccggct gcaccggacg gggtggcgct ccacgtactg cgccatggag
1740cccgacgcgt tccgcggcac ggcgcccatc aacctcaccg agcgcctgta ccagatcctg
1800cgttggtcgg ggggatccct cgagatgttc ttctcccgct tctgcccgct cctggccggc
1860cgccgcctcc accccatgca gcgcgtcgcc tacatcaaca tgaccaccta cccggtctcc
1920accttcttca tcctcatgta ttacttctac ccggtcatgt ggctcttcca gggggagttc
1980tacatccaga ggccgttcca gacgttcgcg ctcttcgtcg tcgtcgtcat cgccacggtg
2040gagctcatcg gcatggtgga gatcaggtgg gcaggcctca cgctgctcga ctgggtccgc
2100aacgagcagt tctacatcat cggcaccacc ggcgtgtacc cgatggccat gctgcacatc
2160ctcctcaggt ccctcggcat aaagggggtg tccttcaagc tgacggccaa gaagctcacg
2220gggggcgcca gggagaggct cgcggagctg tacgacgtgc agtgggtgcc gttgctggtg
2280cccaccgtgg tggtcatggc cgtgaacgtg gccgccatcg gcgcggcggc gggcaaggcg
2340atcgttgggc ggtggtcggc agcgcaggtc gcgggggcgg cgagcgggct tgttttcaac
2400gtgtggatgc tgctgctgct ctacccgttc gcgctcggga taatggggca ctggagcaag
2460aggccctaca tcctgttcct tgtgctggtg accgcggtcg ctgccaccgc gtccgtgtac
2520gtcgcactcg cggggtcctt gctgtacttg cattcgggga taaaactagt t
25716857PRTHordeum vulgare 6Met Ala Ser Pro Ala Ala Val Gly Gly Gly Arg
Leu Ala Asp Pro Leu1 5 10
15Leu Ala Ala Asp Val Val Val Val Gly Ala Lys Asp Lys Tyr Trp Val
20 25 30Pro Ala Asp Glu Arg Glu Ile
Leu Ala Ser Gln Ser Ser Gly Gly Gly 35 40
45Glu Gln Asp Gly Arg Ala Pro Leu Leu Tyr Arg Thr Phe Arg Val
Lys 50 55 60Gly Phe Phe Ile Asn Leu
Tyr Arg Leu Leu Thr Leu Val Arg Val Ile65 70
75 80Val Val Ile Leu Phe Phe Thr Trp Arg Met Arg
His Arg Asp Ser Asp 85 90
95Ala Met Trp Leu Trp Trp Ile Ser Val Val Gly Asp Leu Trp Phe Gly
100 105 110Val Thr Trp Leu Leu Asn
Gln Ile Thr Lys Leu Lys Pro Arg Lys Cys 115 120
125Val Pro Ser Ile Ser Val Leu Arg Glu Gln Leu Asp Gln Pro
Asp Gly 130 135 140Gly Ser Asp Leu Pro
Leu Leu Asp Val Phe Ile Asn Thr Val Asp Pro145 150
155 160Val Asp Glu Pro Met Leu Tyr Thr Met Asn
Ser Ile Leu Ser Ile Leu 165 170
175Ala Thr Asp Tyr Pro Val Gln Lys Tyr Ala Thr Tyr Phe Ser Asp Asp
180 185 190Gly Gly Ser Leu Val
His Tyr Glu Gly Leu Leu Leu Thr Ala Glu Phe 195
200 205Ala Ala Ser Trp Val Pro Phe Cys Arg Lys His Cys
Val Glu Pro Arg 210 215 220Ala Pro Glu
Ser Tyr Phe Trp Ala Lys Met Arg Gly Glu Tyr Ala Gly225
230 235 240Ser Ala Ala Lys Glu Phe Leu
Asp Asp His Arg Arg Met Arg Ala Ala 245
250 255Tyr Glu Glu Phe Lys Ala Arg Leu Asp Gly Leu Ser
Ala Val Ile Glu 260 265 270Gln
Arg Ser Glu Ala Cys Asn Arg Ala Ala Asn Glu Lys Glu Gly Cys 275
280 285Gly Asn Ala Thr Trp Met Ala Asp Gly
Ser Thr Gln Trp Gln Gly Thr 290 295
300Trp Ile Lys Pro Ala Lys Gly His Arg Lys Gly His His Pro Ala Ile305
310 315 320Leu Gln Val Met
Leu Asp Gln Pro Ser Lys Asp Pro Glu Leu Gly Met 325
330 335Ala Ala Ser Ser Asp His Pro Leu Asp Phe
Ser Ala Val Asp Val Arg 340 345
350Leu Pro Met Leu Val Tyr Ile Ala Arg Glu Lys Arg Pro Gly Tyr Asp
355 360 365His Gln Lys Lys Ala Gly Ala
Met Asn Val Gln Leu Arg Val Ser Ala 370 375
380Leu Leu Ser Asn Ala Pro Phe Ile Ile Asn Phe Asp Gly Asp His
Tyr385 390 395 400Ile Asn
Asn Ser Gln Ala Phe Arg Ala Ala Met Cys Phe Met Leu Asp
405 410 415Pro Arg Asp Gly Ala Asp Thr
Ala Phe Val Gln Phe Pro Gln Arg Phe 420 425
430Asp Asp Val Asp Pro Thr Asp Arg Tyr Cys Asn His Asn Arg
Met Phe 435 440 445Phe Asp Ala Thr
Leu Leu Gly Leu Asn Gly Ile Gln Gly Pro Ser Phe 450
455 460Val Gly Thr Gly Cys Met Phe Arg Arg Val Ala Leu
Tyr Ser Ala Asp465 470 475
480Pro Pro Arg Trp Arg Ser Asp Asp Ala Lys Glu Ala Lys Ala Ser His
485 490 495Arg Pro Asn Met Phe
Gly Lys Ser Thr Ser Phe Ile Asn Ser Met Pro 500
505 510Ala Ala Ala Asn Gln Glu Arg Ser Val Pro Ser Pro
Ala Thr Val Gly 515 520 525Glu Ala
Glu Leu Ala Asp Ala Met Thr Cys Ala Tyr Glu Asp Gly Thr 530
535 540Glu Trp Gly Asn Asp Val Gly Trp Val Tyr Asn
Ile Ala Thr Glu Asp545 550 555
560Val Val Thr Gly Phe Arg Leu His Arg Thr Gly Trp Arg Ser Thr Tyr
565 570 575Cys Ala Met Glu
Pro Asp Ala Phe Arg Gly Thr Ala Pro Ile Asn Leu 580
585 590Thr Glu Arg Leu Tyr Gln Ile Leu Arg Trp Ser
Gly Gly Ser Leu Glu 595 600 605Met
Phe Phe Ser Arg Phe Cys Pro Leu Leu Ala Gly Arg Arg Leu His 610
615 620Pro Met Gln Arg Val Ala Tyr Ile Asn Met
Thr Thr Tyr Pro Val Ser625 630 635
640Thr Phe Phe Ile Leu Met Tyr Tyr Phe Tyr Pro Val Met Trp Leu
Phe 645 650 655Gln Gly Glu
Phe Tyr Ile Gln Arg Pro Phe Gln Thr Phe Ala Leu Phe 660
665 670Val Val Val Val Ile Ala Thr Val Glu Leu
Ile Gly Met Val Glu Ile 675 680
685Arg Trp Ala Gly Leu Thr Leu Leu Asp Trp Val Arg Asn Glu Gln Phe 690
695 700Tyr Ile Ile Gly Thr Thr Gly Val
Tyr Pro Met Ala Met Leu His Ile705 710
715 720Leu Leu Arg Ser Leu Gly Ile Lys Gly Val Ser Phe
Lys Leu Thr Ala 725 730
735Lys Lys Leu Thr Gly Gly Ala Arg Glu Arg Leu Ala Glu Leu Tyr Asp
740 745 750Val Gln Trp Val Pro Leu
Leu Val Pro Thr Val Val Val Met Ala Val 755 760
765Asn Val Ala Ala Ile Gly Ala Ala Ala Gly Lys Ala Ile Val
Gly Arg 770 775 780Trp Ser Ala Ala Gln
Val Ala Gly Ala Ala Ser Gly Leu Val Phe Asn785 790
795 800Val Trp Met Leu Leu Leu Leu Tyr Pro Phe
Ala Leu Gly Ile Met Gly 805 810
815His Trp Ser Lys Arg Pro Tyr Ile Leu Phe Leu Val Leu Val Thr Ala
820 825 830Val Ala Ala Thr Ala
Ser Val Tyr Val Ala Leu Ala Gly Ser Leu Leu 835
840 845Tyr Leu His Ser Gly Ile Lys Leu Val 850
85572616DNAHordeum vulgare 7atggccccgg cagtcactcg ccgagccaac
gctctccgcg tcgaggcccc ggacggcaat 60gccgagagcg ggcgcgccag cctagcagca
gactcccccg cggccaagcg ggccatcgat 120gccaaggacg atgtgtgggt ggccgcggct
gagggagacg cgtctggagc cagcgccggc 180aacggcgacc ggccgccgct gttccggacc
atgaaggtca agggaagcat cctccatcct 240tacaggttca tgatcctcgt gcgcttggtc
gccgtcgtcg cgttcttcgc gtggcgcctg 300aagcacaaga accacgacgg catgtggctc
tgggccacgt ccatggtcgc cgacgtctgg 360ttcggcttct catggctcct caaccagctg
cccaagctca accccatcaa gcgcgtcccc 420gacctggccg ccctcgccga ccagtgcggc
tcctccggcg acgccaacct gccaggcatc 480gacatctttg tcaccaccgt ggaccccgtg
gacgaaccca tcttgtacac cgtgaacacc 540atactctcca tcctcgccac cgactaccct
gtcgataagt acgcctgcta cctctcagac 600gacggcggca cgttggtgca ctacgaggcc
atgatcgaag tggccaattt cgcggtgatg 660tgggtccctt tttgccggaa gcactgtgtc
gagccaaggt cccccgagaa ctactttggg 720atgaaaacgc agccgtacgt cgggagtatg
gctggagaat tcatgaggga gcataggcgt 780gtgcgcagag agtatgatga gttcaaggtg
aggatagact ccctgtccac caccatccgc 840caaagatctg atgcgtacaa ctcgagcaac
aaaggagatg gtgtgcgtgc aacctggatg 900gctgatggga cacaatggcc tggtacgtgg
attgagcagg ttgagaacca ccggagagga 960caacatgctg gaattgttca ggtcatacta
agccatccta gttgcaaacc gcaactgggg 1020tctccggcga gcactgacaa tccacttgac
ttcagcaacg ttgacacgag gctgcccatg 1080ctcgtctaca tgtcccggga gaagcgcccc
ggttataacc accaaaagaa ggcaggcgcc 1140atgaacgtga tgctccgtgt ctcggcgttg
ctctccaacg cgccattcgt cgtcaatttt 1200gactgcgacc actacatcaa caacacgcaa
gctctccgcg cccctatgtg cttcatgctc 1260gaccctcgcg acggtcagaa cacggccttc
gtccagtttc cgcagcgctt cgacgacgtc 1320gacccgacgg accgctacgc caaccacaac
cgtgtcttct tcgacggtac catgctctcc 1380ctcaacggcc ttcaagggcc ttcctacctc
ggcactggca ccatgttccg tcgtgtcacg 1440ctctatggca tggagccacc acgttacaga
gcggagaaca tcaagcttgt aggtaagacc 1500tatgagttcg gtagctcgac gtctttcatc
aattccatgc cggacggcgc aatccaagag 1560cggtctatca cgccggtgtt ggtcgacgag
gcactcagca atgacctggc taccctgatg 1620acgtgtgctt acgaggacgg gacctcatgg
gggagagacg ttgggtgggt gtacaacatc 1680gcgacggagg acgtggtgac cggattccgc
atgcaccggc aggggtggcg ctccatgtat 1740tgctccatgg agccggccgc cttccgcgga
acagcgccga tcaacctcac cgagcgcctt 1800taccaggtgc tccggtggtc gggcggctct
ctcgagatgt tcttctccca cagcaacgct 1860ctcatggccg gccgccgtat ccaccctctg
cagcgtgtcg cgtacctcaa catgtcgacc 1920tacccgatcg tcacggtgtt catcctggcc
tacaacctct tccccgtcat gtggctcttc 1980tccgagcagt tctacatcca gaggccgttc
ggcacgtaca tcatgtacct cgtcggcgtc 2040atagcgatga ttcacgtgat cggcatgttc
gaggtgaaat gggcggggat cacgctgctc 2100gactggtgcc gcaacgagca gttctacatg
atcggggcga cgggcgtgta cccgacggcg 2160gtgctttaca tggcgctcaa gcttgtcacg
gggaagggga tatacttcag gctcacatcc 2220aagcagacgg acgcttgctc caacgacaag
ttcgccgacc tgtacacggt gcggtgggtg 2280ccgctgctgt tcccgacggt cgcagtgctc
atcgtgaacg tcgcggctgt cggggcagcg 2340ataggcaagg cagcagcgtg gggcttcttc
acggaccagg cgcggcacgt gctgctcggg 2400atggtgttca acgtgtggat cctcgtgctc
ctctacccgt ttgcgctcgg gatcatgggg 2460aaatggggga agagacccat catcctgttc
gtcatgttga tcatggccat tggcgccgtc 2520gggctcgtgt atgtcgcctt ccatgatccc
tacccaactg atttttcaga agttgcagct 2580tctcttggtg aagcatcgct gaccgggcca
tctggg 26168872PRTHordeum vulgare 8Met Ala
Pro Ala Val Thr Arg Arg Ala Asn Ala Leu Arg Val Glu Ala1 5
10 15Pro Asp Gly Asn Ala Glu Ser Gly
Arg Ala Ser Leu Ala Ala Asp Ser 20 25
30Pro Ala Ala Lys Arg Ala Ile Asp Ala Lys Asp Asp Val Trp Val
Ala 35 40 45Ala Ala Glu Gly Asp
Ala Ser Gly Ala Ser Ala Gly Asn Gly Asp Arg 50 55
60Pro Pro Leu Phe Arg Thr Met Lys Val Lys Gly Ser Ile Leu
His Pro65 70 75 80Tyr
Arg Phe Met Ile Leu Val Arg Leu Val Ala Val Val Ala Phe Phe
85 90 95Ala Trp Arg Leu Lys His Lys
Asn His Asp Gly Met Trp Leu Trp Ala 100 105
110Thr Ser Met Val Ala Asp Val Trp Phe Gly Phe Ser Trp Leu
Leu Asn 115 120 125Gln Leu Pro Lys
Leu Asn Pro Ile Lys Arg Val Pro Asp Leu Ala Ala 130
135 140Leu Ala Asp Gln Cys Gly Ser Ser Gly Asp Ala Asn
Leu Pro Gly Ile145 150 155
160Asp Ile Phe Val Thr Thr Val Asp Pro Val Asp Glu Pro Ile Leu Tyr
165 170 175Thr Val Asn Thr Ile
Leu Ser Ile Leu Ala Thr Asp Tyr Pro Val Asp 180
185 190Lys Tyr Ala Cys Tyr Leu Ser Asp Asp Gly Gly Thr
Leu Val His Tyr 195 200 205Glu Ala
Met Ile Glu Val Ala Asn Phe Ala Val Met Trp Val Pro Phe 210
215 220Cys Arg Lys His Cys Val Glu Pro Arg Ser Pro
Glu Asn Tyr Phe Gly225 230 235
240Met Lys Thr Gln Pro Tyr Val Gly Ser Met Ala Gly Glu Phe Met Arg
245 250 255Glu His Arg Arg
Val Arg Arg Glu Tyr Asp Glu Phe Lys Val Arg Ile 260
265 270Asp Ser Leu Ser Thr Thr Ile Arg Gln Arg Ser
Asp Ala Tyr Asn Ser 275 280 285Ser
Asn Lys Gly Asp Gly Val Arg Ala Thr Trp Met Ala Asp Gly Thr 290
295 300Gln Trp Pro Gly Thr Trp Ile Glu Gln Val
Glu Asn His Arg Arg Gly305 310 315
320Gln His Ala Gly Ile Val Gln Val Ile Leu Ser His Pro Ser Cys
Lys 325 330 335Pro Gln Leu
Gly Ser Pro Ala Ser Thr Asp Asn Pro Leu Asp Phe Ser 340
345 350Asn Val Asp Thr Arg Leu Pro Met Leu Val
Tyr Met Ser Arg Glu Lys 355 360
365Arg Pro Gly Tyr Asn His Gln Lys Lys Ala Gly Ala Met Asn Val Met 370
375 380Leu Arg Val Ser Ala Leu Leu Ser
Asn Ala Pro Phe Val Val Asn Phe385 390
395 400Asp Cys Asp His Tyr Ile Asn Asn Thr Gln Ala Leu
Arg Ala Pro Met 405 410
415Cys Phe Met Leu Asp Pro Arg Asp Gly Gln Asn Thr Ala Phe Val Gln
420 425 430Phe Pro Gln Arg Phe Asp
Asp Val Asp Pro Thr Asp Arg Tyr Ala Asn 435 440
445His Asn Arg Val Phe Phe Asp Gly Thr Met Leu Ser Leu Asn
Gly Leu 450 455 460Gln Gly Pro Ser Tyr
Leu Gly Thr Gly Thr Met Phe Arg Arg Val Thr465 470
475 480Leu Tyr Gly Met Glu Pro Pro Arg Tyr Arg
Ala Glu Asn Ile Lys Leu 485 490
495Val Gly Lys Thr Tyr Glu Phe Gly Ser Ser Thr Ser Phe Ile Asn Ser
500 505 510Met Pro Asp Gly Ala
Ile Gln Glu Arg Ser Ile Thr Pro Val Leu Val 515
520 525Asp Glu Ala Leu Ser Asn Asp Leu Ala Thr Leu Met
Thr Cys Ala Tyr 530 535 540Glu Asp Gly
Thr Ser Trp Gly Arg Asp Val Gly Trp Val Tyr Asn Ile545
550 555 560Ala Thr Glu Asp Val Val Thr
Gly Phe Arg Met His Arg Gln Gly Trp 565
570 575Arg Ser Met Tyr Cys Ser Met Glu Pro Ala Ala Phe
Arg Gly Thr Ala 580 585 590Pro
Ile Asn Leu Thr Glu Arg Leu Tyr Gln Val Leu Arg Trp Ser Gly 595
600 605Gly Ser Leu Glu Met Phe Phe Ser His
Ser Asn Ala Leu Met Ala Gly 610 615
620Arg Arg Ile His Pro Leu Gln Arg Val Ala Tyr Leu Asn Met Ser Thr625
630 635 640Tyr Pro Ile Val
Thr Val Phe Ile Leu Ala Tyr Asn Leu Phe Pro Val 645
650 655Met Trp Leu Phe Ser Glu Gln Phe Tyr Ile
Gln Arg Pro Phe Gly Thr 660 665
670Tyr Ile Met Tyr Leu Val Gly Val Ile Ala Met Ile His Val Ile Gly
675 680 685Met Phe Glu Val Lys Trp Ala
Gly Ile Thr Leu Leu Asp Trp Cys Arg 690 695
700Asn Glu Gln Phe Tyr Met Ile Gly Ala Thr Gly Val Tyr Pro Thr
Ala705 710 715 720Val Leu
Tyr Met Ala Leu Lys Leu Val Thr Gly Lys Gly Ile Tyr Phe
725 730 735Arg Leu Thr Ser Lys Gln Thr
Asp Ala Cys Ser Asn Asp Lys Phe Ala 740 745
750Asp Leu Tyr Thr Val Arg Trp Val Pro Leu Leu Phe Pro Thr
Val Ala 755 760 765Val Leu Ile Val
Asn Val Ala Ala Val Gly Ala Ala Ile Gly Lys Ala 770
775 780Ala Ala Trp Gly Phe Phe Thr Asp Gln Ala Arg His
Val Leu Leu Gly785 790 795
800Met Val Phe Asn Val Trp Ile Leu Val Leu Leu Tyr Pro Phe Ala Leu
805 810 815Gly Ile Met Gly Lys
Trp Gly Lys Arg Pro Ile Ile Leu Phe Val Met 820
825 830Leu Ile Met Ala Ile Gly Ala Val Gly Leu Val Tyr
Val Ala Phe His 835 840 845Asp Pro
Tyr Pro Thr Asp Phe Ser Glu Val Ala Ala Ser Leu Gly Glu 850
855 860Ala Ser Leu Thr Gly Pro Ser Gly865
87092637DNAHordeum vulgare 9atgttgtcgc cccggacaga cgccggcgcc
ggcgccgcca ccgacctcag ccagccactt 60ctctggaacc gcaatggcgt tcacgcagga
gcattggtcg tcatgccagt cgtggccaat 120ggtcacggcg gcggcgacaa gcttaagggc
gccccgaaag ccaaggacaa gtactggaaa 180gacgtcgacc agccggacga catggcggca
gcgccagacc tggacaacgg cggcggccgg 240ccgctgctgt tctcgaactt gagagtccag
aatatcatcc tgtaccccgg cgaggtattg 300atcctgatac gagtaatcgc cgtaatctta
tttgttggat ggcgcatcaa gcataacaat 360tcagatgtca tgtggttttg gatgatgtcc
gtcgtcgcag acgtgtggtt tagcttatca 420tggctaagct accaactgcc aaagtataat
cccgttaaaa ggatacccga ccttgctaca 480ctcaggaaac aatatgacac accagggagg
agctcccagc tgccaagcat tgacgtcatc 540gtcaccactg ccagtgctac cgatgagccc
atattgtaca ccatgaactg tgttctctct 600atacttgcag ctgactatca tattggcagg
tgcaactgct acctatcaga tgatagcggc 660tcattggtcc tttatgaggc attggttgag
actgcaaagt ttgctgcttt atgggttcct 720ttctgtagaa agcatcagat tgagccaaga
gcaccggaaa gctattttga actaaagggc 780ccgttgtatg gagggacgcc acataaggag
ttctttcagg attataagca tgtacgtaca 840caatatgaag agttcaagaa gaatttagat
atgcttccta acaccatcca tcaaaggtcg 900ggaacttaca gtaaaacagg aacggaggat
gaagatgcaa aagtgacttg gatggctgac 960ggaacacaat ggccaggcac atggcttgac
ccagcagaaa aacatagggc cgggcatcat 1020gcaggaattg ttaagattgt gcagagccat
ccagaacatg tggttcaacc aggcgtacaa 1080gagagccttg acaacccact cagctttgac
gatgttgatg tgcgcctgcc catgttggta 1140tatgtggctc gtgaaaagag tccaggtatc
gagcataaca aaaaggcagg cgctttgaat 1200gcagagctac gtatctcagc tctactctct
aatgcacctt tcttcattaa ctttgactgc 1260gaccactaca tcaacaattc agaagcccta
cgtgcagctg tttgcttcat gctagaccca 1320cgtgaagggg ataatactgg atttgttcag
ttcccgcaaa gatttgataa tgtcgaccca 1380actgaccggt atggaaacca taatcgagtc
ttttttgatg gtgccatgta tggcctcaat 1440ggtcaacaag ggcctactta ccttggcaca
ggttgcatgt tccgtcccct tgcactctat 1500ggaattgatc caccttgctg gagagccgag
gacatcatag tcgacagtaa caggtttggc 1560aactcattac ccttcctcaa ctcagtacta
gcagccataa agcaagagga aggtgtcaca 1620ctaccaccac cactagatga ttcatttctt
gaagagatga caaaagttgt gtcatgttcc 1680tatgatgatt ccactgattg gggtaggggc
attggctaca tatacaatat ggcaacagaa 1740gacatagtaa caggatttcg tatccatggg
caagggtggt gctccatgta tgttaccatg 1800gaacgtgaag cgttccgtgg cactgcaccg
atcaatctaa cagagcgcct ccgccaaata 1860gtgcgatggt ctggtggttc cctagagatg
ttcttctcgc acatcagccc actattcgct 1920ggtcgtcgac tcagtttggt gcagcgactc
tcgtacatca atttcactat atacccattg 1980acatcactct ttatcctaat gtatgccttc
tgtccagtga tgtggcttct tccaacagaa 2040atacttatac aaaggccata taccaggtac
attgtgtacc ttatcattgt cgtcgcgatg 2100atccatgtga ttggcatgtt tgagataatg
tgggcaggaa tcacatggtt ggattggtgg 2160cgcaacgagc aatttttcat gatcggctcg
gtaactgcat atccaacggc ggtgttgcac 2220atggtggtga atatccttac aaaaaagggt
atacacttca gagtaaccac aaagcaacca 2280gtggctgata cagatgacaa gtatgctgag
atgtatgaag tgcattgggt acccatgatg 2340gtccccgcgg ttgtggtatt gttttccaac
atcttggcta ttggtgtagc aattggtaaa 2400tcagtcttat acatggggac atggtctgta
gcacagaaaa ggcatggtgc actagggcta 2460ttgttcaacc tgtggattat ggtgctcctt
tacccatttg cattggcgat tattggaaga 2520tgggccaaga gaaccggaat cctattcatc
ttactaccca ttgctttctt ggccaccgca 2580ttgatgtaca ttggcatcca tacattcctt
ttacatttct ttccatccat gttggta 263710879PRTHordeum vulgare 10Met Leu
Ser Pro Arg Thr Asp Ala Gly Ala Gly Ala Ala Thr Asp Leu1 5
10 15Ser Gln Pro Leu Leu Trp Asn Arg
Asn Gly Val His Ala Gly Ala Leu 20 25
30Val Val Met Pro Val Val Ala Asn Gly His Gly Gly Gly Asp Lys
Leu 35 40 45Lys Gly Ala Pro Lys
Ala Lys Asp Lys Tyr Trp Lys Asp Val Asp Gln 50 55
60Pro Asp Asp Met Ala Ala Ala Pro Asp Leu Asp Asn Gly Gly
Gly Arg65 70 75 80Pro
Leu Leu Phe Ser Asn Leu Arg Val Gln Asn Ile Ile Leu Tyr Pro
85 90 95Gly Glu Val Leu Ile Leu Ile
Arg Val Ile Ala Val Ile Leu Phe Val 100 105
110Gly Trp Arg Ile Lys His Asn Asn Ser Asp Val Met Trp Phe
Trp Met 115 120 125Met Ser Val Val
Ala Asp Val Trp Phe Ser Leu Ser Trp Leu Ser Tyr 130
135 140Gln Leu Pro Lys Tyr Asn Pro Val Lys Arg Ile Pro
Asp Leu Ala Thr145 150 155
160Leu Arg Lys Gln Tyr Asp Thr Pro Gly Arg Ser Ser Gln Leu Pro Ser
165 170 175Ile Asp Val Ile Val
Thr Thr Ala Ser Ala Thr Asp Glu Pro Ile Leu 180
185 190Tyr Thr Met Asn Cys Val Leu Ser Ile Leu Ala Ala
Asp Tyr His Ile 195 200 205Gly Arg
Cys Asn Cys Tyr Leu Ser Asp Asp Ser Gly Ser Leu Val Leu 210
215 220Tyr Glu Ala Leu Val Glu Thr Ala Lys Phe Ala
Ala Leu Trp Val Pro225 230 235
240Phe Cys Arg Lys His Gln Ile Glu Pro Arg Ala Pro Glu Ser Tyr Phe
245 250 255Glu Leu Lys Gly
Pro Leu Tyr Gly Gly Thr Pro His Lys Glu Phe Phe 260
265 270Gln Asp Tyr Lys His Val Arg Thr Gln Tyr Glu
Glu Phe Lys Lys Asn 275 280 285Leu
Asp Met Leu Pro Asn Thr Ile His Gln Arg Ser Gly Thr Tyr Ser 290
295 300Lys Thr Gly Thr Glu Asp Glu Asp Ala Lys
Val Thr Trp Met Ala Asp305 310 315
320Gly Thr Gln Trp Pro Gly Thr Trp Leu Asp Pro Ala Glu Lys His
Arg 325 330 335Ala Gly His
His Ala Gly Ile Val Lys Ile Val Gln Ser His Pro Glu 340
345 350His Val Val Gln Pro Gly Val Gln Glu Ser
Leu Asp Asn Pro Leu Ser 355 360
365Phe Asp Asp Val Asp Val Arg Leu Pro Met Leu Val Tyr Val Ala Arg 370
375 380Glu Lys Ser Pro Gly Ile Glu His
Asn Lys Lys Ala Gly Ala Leu Asn385 390
395 400Ala Glu Leu Arg Ile Ser Ala Leu Leu Ser Asn Ala
Pro Phe Phe Ile 405 410
415Asn Phe Asp Cys Asp His Tyr Ile Asn Asn Ser Glu Ala Leu Arg Ala
420 425 430Ala Val Cys Phe Met Leu
Asp Pro Arg Glu Gly Asp Asn Thr Gly Phe 435 440
445Val Gln Phe Pro Gln Arg Phe Asp Asn Val Asp Pro Thr Asp
Arg Tyr 450 455 460Gly Asn His Asn Arg
Val Phe Phe Asp Gly Ala Met Tyr Gly Leu Asn465 470
475 480Gly Gln Gln Gly Pro Thr Tyr Leu Gly Thr
Gly Cys Met Phe Arg Pro 485 490
495Leu Ala Leu Tyr Gly Ile Asp Pro Pro Cys Trp Arg Ala Glu Asp Ile
500 505 510Ile Val Asp Ser Asn
Arg Phe Gly Asn Ser Leu Pro Phe Leu Asn Ser 515
520 525Val Leu Ala Ala Ile Lys Gln Glu Glu Gly Val Thr
Leu Pro Pro Pro 530 535 540Leu Asp Asp
Ser Phe Leu Glu Glu Met Thr Lys Val Val Ser Cys Ser545
550 555 560Tyr Asp Asp Ser Thr Asp Trp
Gly Arg Gly Ile Gly Tyr Ile Tyr Asn 565
570 575Met Ala Thr Glu Asp Ile Val Thr Gly Phe Arg Ile
His Gly Gln Gly 580 585 590Trp
Cys Ser Met Tyr Val Thr Met Glu Arg Glu Ala Phe Arg Gly Thr 595
600 605Ala Pro Ile Asn Leu Thr Glu Arg Leu
Arg Gln Ile Val Arg Trp Ser 610 615
620Gly Gly Ser Leu Glu Met Phe Phe Ser His Ile Ser Pro Leu Phe Ala625
630 635 640Gly Arg Arg Leu
Ser Leu Val Gln Arg Leu Ser Tyr Ile Asn Phe Thr 645
650 655Ile Tyr Pro Leu Thr Ser Leu Phe Ile Leu
Met Tyr Ala Phe Cys Pro 660 665
670Val Met Trp Leu Leu Pro Thr Glu Ile Leu Ile Gln Arg Pro Tyr Thr
675 680 685Arg Tyr Ile Val Tyr Leu Ile
Ile Val Val Ala Met Ile His Val Ile 690 695
700Gly Met Phe Glu Ile Met Trp Ala Gly Ile Thr Trp Leu Asp Trp
Trp705 710 715 720Arg Asn
Glu Gln Phe Phe Met Ile Gly Ser Val Thr Ala Tyr Pro Thr
725 730 735Ala Val Leu His Met Val Val
Asn Ile Leu Thr Lys Lys Gly Ile His 740 745
750Phe Arg Val Thr Thr Lys Gln Pro Val Ala Asp Thr Asp Asp
Lys Tyr 755 760 765Ala Glu Met Tyr
Glu Val His Trp Val Pro Met Met Val Pro Ala Val 770
775 780Val Val Leu Phe Ser Asn Ile Leu Ala Ile Gly Val
Ala Ile Gly Lys785 790 795
800Ser Val Leu Tyr Met Gly Thr Trp Ser Val Ala Gln Lys Arg His Gly
805 810 815Ala Leu Gly Leu Leu
Phe Asn Leu Trp Ile Met Val Leu Leu Tyr Pro 820
825 830Phe Ala Leu Ala Ile Ile Gly Arg Trp Ala Lys Arg
Thr Gly Ile Leu 835 840 845Phe Ile
Leu Leu Pro Ile Ala Phe Leu Ala Thr Ala Leu Met Tyr Ile 850
855 860Gly Ile His Thr Phe Leu Leu His Phe Phe Pro
Ser Met Leu Val865 870
875112691DNAHordeum vulgare 11atgggttctt tggcggcagc caacggggcc ggtcatgcga
gcaatggcgc cggcgtcgcg 60gaccaggcgc tggcactgga gaacggcacc ggcaatgggc
acaaggccag cgacgccaac 120cgagcgacgc cggtacagca ggcaaacggc agcagcaagg
ccgcggggaa ggttagcccg 180aaggacaagt actgggtggc cgtcgatgag ggagagatgg
cggccgccat agcggacggc 240ggcgaggacg gccggcgacc gctgctgtac cggacgttca
aggtcaaggg catcctcctg 300catccctaca ggttgctgag cttgatcaga ttggttgcta
tcgtcctatt tttcgtatgg 360cgtgtcaggc acccatacgc tgatggcatg tggctctggt
ggatatcgat ggttggggat 420ctttggtttg gcgtcacttg gttgctaaac caagttgcaa
agctcaaccc tgtcaagcgt 480gtccccaacc ttgcgctttt gcaacagcag tttgatctcc
ctgacggcaa ctccaacctt 540ccttgtcttg atgtcttcat caacaccgtt gatcccatta
atgaacctat gatatacact 600atgaactcca tcatatccat ccttgctgca gactatccgg
ttgacaagca tgcttgctac 660ctttcagatg atggtgggtc aataatccat tatgatggtt
tgcttgagac tgcaaaattt 720gctgcattat gggttccctt ttgcagaaaa cattccattg
agccaagagc ccctgagagc 780tatttttctt tgaatacacg cccatacact ggaaatgcac
cacaagactt tgtcaatgac 840cgcagacaca tgtgtagaga gtatgatgag ttcaaggagc
gcttagatgc actttttacc 900ctcattccca aacggtcaga tgtgtacaat catgctgctg
gcaaagaagg tgcaaaggca 960acttggatgg cagatgggac acagtggcca ggcacatgga
ttgacccagc tgaaaaccat 1020aagaaaggac aacatgctgg gatcgttaag gttttgttga
aacatccaag ttatgaacca 1080gaacttggtc taggagcaag caccaacagt cctctagact
tcagtgcagt tgatgtgcgc 1140ctcccaatgc tcgtttacat ctcccgtgag aagagtccaa
gctgtgatca tcaaaagaag 1200gcaggtgcca tgaacgtaca gttgcgagtc tctgccctcc
tgaccaatgc gccctttatc 1260atcaactttg atggtgacca ctacgtcaac aactcgaaag
ccttccgtgc tggcatatgt 1320ttcatgctcg atcgccgtga aggtgacaat actgcctttg
tccagtttcc ccaacgcttc 1380gatgatgttg atcccacaga taggtactgc aatcacaatc
gtgtcttctt tgacgccacc 1440ttgctcggct ccaatggcat ccaagggccg tcttatgttg
gcactggttg catgttccgc 1500cgtgtcgcac tttacggtgt tgacccacct cgctggagac
ctgatgacgt gaagatcgtg 1560gacagctcca gcaagtttgg cagttcagag tcattcatca
gctcaatact gccagcagca 1620gaccaagaac gctccatcat gtcgccaccg gcacttgaag
agtctgtcat ggctgactta 1680gctcatgtca tgacttgtgc atatgaggac gggactgaat
ggggcagaga agttggttgg 1740gtgtacaaca ttgcaactga ggatgtggtg accggcttcc
ggctgcaccg gaatgggtgg 1800cgatccatgt actgccgcat ggaaccagat gcattcgccg
gcaccgcgcc aatcaacctc 1860actgagcggc tctaccagat cctgcgctgg tcggggggct
cccttgagat gttcttctcg 1920cacaactgcc cactcctggc tggccgccgc ctccacccaa
tgcaaagaat tgcctatgcc 1980aacatgacag cctacccagt ttcatctgtc tttcttgtgt
tctatctcct cttcccggtg 2040atatggatct tccgtgggca attctacata cagaagccat
tccccacgta tgtgttgtac 2100ctcgtcatcg tcatagccct gaccgagtta atcggtatgg
ttgagatcaa gtgggctggg 2160ctcacgctgc tggactggat ccgcaacgag cagttctaca
ttattggtgc aacagccgtg 2220taccctacag cagtatttca catagtgctg aagctgtttg
gcctgaaggg tgtttcattc 2280aagctgacgg caaaacaggt agcaagcagt accagcgata
agtttgctga actgtatgcc 2340gtgcagtggg ctccgatgct gatccctacc atggtggtta
tagcggtgaa tgtttgtgcc 2400attggcgcgt cgataggcaa ggcggtagtg ggaggatggt
cactgatgca gatggccgat 2460gcaggacttg ggctggtgtt caacgcgtgg attctggtgc
tgatctaccc gtttgcactg 2520ggcatgattg gacggtggag caagaggccc tacatcctgt
tcattctgtt tgtcattgcg 2580tttattttga tcgcattggt ggatatcgcc atccaggcca
tgcggtctgg gattgttcgg 2640ttccacttta aaagctcagg tggcgccact tttcccacaa
gctggggttt g 269112897PRTHordeum vulgare 12Met Gly Ser Leu Ala
Ala Ala Asn Gly Ala Gly His Ala Ser Asn Gly1 5
10 15Ala Gly Val Ala Asp Gln Ala Leu Ala Leu Glu
Asn Gly Thr Gly Asn 20 25
30Gly His Lys Ala Ser Asp Ala Asn Arg Ala Thr Pro Val Gln Gln Ala
35 40 45Asn Gly Ser Ser Lys Ala Ala Gly
Lys Val Ser Pro Lys Asp Lys Tyr 50 55
60Trp Val Ala Val Asp Glu Gly Glu Met Ala Ala Ala Ile Ala Asp Gly65
70 75 80Gly Glu Asp Gly Arg
Arg Pro Leu Leu Tyr Arg Thr Phe Lys Val Lys 85
90 95Gly Ile Leu Leu His Pro Tyr Arg Leu Leu Ser
Leu Ile Arg Leu Val 100 105
110Ala Ile Val Leu Phe Phe Val Trp Arg Val Arg His Pro Tyr Ala Asp
115 120 125Gly Met Trp Leu Trp Trp Ile
Ser Met Val Gly Asp Leu Trp Phe Gly 130 135
140Val Thr Trp Leu Leu Asn Gln Val Ala Lys Leu Asn Pro Val Lys
Arg145 150 155 160Val Pro
Asn Leu Ala Leu Leu Gln Gln Gln Phe Asp Leu Pro Asp Gly
165 170 175Asn Ser Asn Leu Pro Cys Leu
Asp Val Phe Ile Asn Thr Val Asp Pro 180 185
190Ile Asn Glu Pro Met Ile Tyr Thr Met Asn Ser Ile Ile Ser
Ile Leu 195 200 205Ala Ala Asp Tyr
Pro Val Asp Lys His Ala Cys Tyr Leu Ser Asp Asp 210
215 220Gly Gly Ser Ile Ile His Tyr Asp Gly Leu Leu Glu
Thr Ala Lys Phe225 230 235
240Ala Ala Leu Trp Val Pro Phe Cys Arg Lys His Ser Ile Glu Pro Arg
245 250 255Ala Pro Glu Ser Tyr
Phe Ser Leu Asn Thr Arg Pro Tyr Thr Gly Asn 260
265 270Ala Pro Gln Asp Phe Val Asn Asp Arg Arg His Met
Cys Arg Glu Tyr 275 280 285Asp Glu
Phe Lys Glu Arg Leu Asp Ala Leu Phe Thr Leu Ile Pro Lys 290
295 300Arg Ser Asp Val Tyr Asn His Ala Ala Gly Lys
Glu Gly Ala Lys Ala305 310 315
320Thr Trp Met Ala Asp Gly Thr Gln Trp Pro Gly Thr Trp Ile Asp Pro
325 330 335Ala Glu Asn His
Lys Lys Gly Gln His Ala Gly Ile Val Lys Val Leu 340
345 350Leu Lys His Pro Ser Tyr Glu Pro Glu Leu Gly
Leu Gly Ala Ser Thr 355 360 365Asn
Ser Pro Leu Asp Phe Ser Ala Val Asp Val Arg Leu Pro Met Leu 370
375 380Val Tyr Ile Ser Arg Glu Lys Ser Pro Ser
Cys Asp His Gln Lys Lys385 390 395
400Ala Gly Ala Met Asn Val Gln Leu Arg Val Ser Ala Leu Leu Thr
Asn 405 410 415Ala Pro Phe
Ile Ile Asn Phe Asp Gly Asp His Tyr Val Asn Asn Ser 420
425 430Lys Ala Phe Arg Ala Gly Ile Cys Phe Met
Leu Asp Arg Arg Glu Gly 435 440
445Asp Asn Thr Ala Phe Val Gln Phe Pro Gln Arg Phe Asp Asp Val Asp 450
455 460Pro Thr Asp Arg Tyr Cys Asn His
Asn Arg Val Phe Phe Asp Ala Thr465 470
475 480Leu Leu Gly Ser Asn Gly Ile Gln Gly Pro Ser Tyr
Val Gly Thr Gly 485 490
495Cys Met Phe Arg Arg Val Ala Leu Tyr Gly Val Asp Pro Pro Arg Trp
500 505 510Arg Pro Asp Asp Val Lys
Ile Val Asp Ser Ser Ser Lys Phe Gly Ser 515 520
525Ser Glu Ser Phe Ile Ser Ser Ile Leu Pro Ala Ala Asp Gln
Glu Arg 530 535 540Ser Ile Met Ser Pro
Pro Ala Leu Glu Glu Ser Val Met Ala Asp Leu545 550
555 560Ala His Val Met Thr Cys Ala Tyr Glu Asp
Gly Thr Glu Trp Gly Arg 565 570
575Glu Val Gly Trp Val Tyr Asn Ile Ala Thr Glu Asp Val Val Thr Gly
580 585 590Phe Arg Leu His Arg
Asn Gly Trp Arg Ser Met Tyr Cys Arg Met Glu 595
600 605Pro Asp Ala Phe Ala Gly Thr Ala Pro Ile Asn Leu
Thr Glu Arg Leu 610 615 620Tyr Gln Ile
Leu Arg Trp Ser Gly Gly Ser Leu Glu Met Phe Phe Ser625
630 635 640His Asn Cys Pro Leu Leu Ala
Gly Arg Arg Leu His Pro Met Gln Arg 645
650 655Ile Ala Tyr Ala Asn Met Thr Ala Tyr Pro Val Ser
Ser Val Phe Leu 660 665 670Val
Phe Tyr Leu Leu Phe Pro Val Ile Trp Ile Phe Arg Gly Gln Phe 675
680 685Tyr Ile Gln Lys Pro Phe Pro Thr Tyr
Val Leu Tyr Leu Val Ile Val 690 695
700Ile Ala Leu Thr Glu Leu Ile Gly Met Val Glu Ile Lys Trp Ala Gly705
710 715 720Leu Thr Leu Leu
Asp Trp Ile Arg Asn Glu Gln Phe Tyr Ile Ile Gly 725
730 735Ala Thr Ala Val Tyr Pro Thr Ala Val Phe
His Ile Val Leu Lys Leu 740 745
750Phe Gly Leu Lys Gly Val Ser Phe Lys Leu Thr Ala Lys Gln Val Ala
755 760 765Ser Ser Thr Ser Asp Lys Phe
Ala Glu Leu Tyr Ala Val Gln Trp Ala 770 775
780Pro Met Leu Ile Pro Thr Met Val Val Ile Ala Val Asn Val Cys
Ala785 790 795 800Ile Gly
Ala Ser Ile Gly Lys Ala Val Val Gly Gly Trp Ser Leu Met
805 810 815Gln Met Ala Asp Ala Gly Leu
Gly Leu Val Phe Asn Ala Trp Ile Leu 820 825
830Val Leu Ile Tyr Pro Phe Ala Leu Gly Met Ile Gly Arg Trp
Ser Lys 835 840 845Arg Pro Tyr Ile
Leu Phe Ile Leu Phe Val Ile Ala Phe Ile Leu Ile 850
855 860Ala Leu Val Asp Ile Ala Ile Gln Ala Met Arg Ser
Gly Ile Val Arg865 870 875
880Phe His Phe Lys Ser Ser Gly Gly Ala Thr Phe Pro Thr Ser Trp Gly
885 890 895Leu133489DNAHordeum
vulgare 13tgcatcccag cccataaatc ccccacgtac tttactcgtc atttctcgca
cctcctcctc 60cccctccccc tcgcctacat tcattcattg cttccttttc tctctccctg
cccatgttaa 120agcctgcctc ggccattgcc tgagcctgcc attgttggac cttggacctg
ttctccttgt 180cgtaaaggca aaggagagcg cgtgcattga ggacgacggc catggcgcca
gcggtggccg 240gagggggccg cgtgcggagc aatgagccgg ttgctgctgc tgccgccgcg
ccggcggcca 300gcggcaagcc ctgcgtgtgc ggcttccagg tttgcgcctg cacggggtcg
gccgcggtgg 360cctccgccgc ctcgtcgctg gacatggaca tcgtggccat ggggcagatc
ggcgccgtca 420acgacgagag ctgggtgggc gtggagctcg gcgaagatgg cgagaccgac
gaaagcggtg 480ccgccgttga cgaccgcccc gtattccgca ccgagaagat caagggtgtc
ctcctccacc 540cctaccgggt gctgattttc gttcgtctga tcgccttcac gctgttcgtg
atctggcgta 600tctcccacaa gaacccagac gcgatgtggc tgtgggtgac atccatctgc
ggcgagttct 660ggttcggttt ctcgtggctg ctagatcagc tgcccaagct gaaccccatc
aaccgcgtgc 720cggacctggc ggtgctgcgg cagcgcttcg accgccccga cggcacctcc
acgctcccgg 780ggctggacat cttcgtcacc acggccgacc ccatcaagga gcccatcctc
tccaccgcca 840actcggtgct ctccatcctg gccgccgact accccgtgga ccgcaacaca
tgctacgtct 900ccgacgacag tggcatgctg ctcacctacg aggccctggc agagtcctcc
aagttcgcca 960cgctctgggt gcccttctgc cgcaagcacg ggatcgagcc caggggtccg
gagagctact 1020tcgagctcaa gtcacaccct tacatgggga gagcccagga cgagttcgtc
aacgaccgcc 1080gccgcgttcg caaggagtac gacgagttca aggccaggat caacagcctg
gagcatgaca 1140tcaagcagcg caacgacggg tacaacgccg ccattgccca cagccaaggc
gtgccccggc 1200ccacctggat ggcggacggc acccagtggg agggcacatg ggtcgacgcc
tccgagaacc 1260accgcagggg cgaccacgcc ggcatcgtac tggtgctgct gaaccacccg
agccaccgcc 1320ggcagacggg cccgccggcg agcgctgaca acccactgga cttgagcggc
gtggatgtgc 1380gtctccccat gctggtgtac gtgtcccgtg agaagcgccc cgggcacgac
caccagaaga 1440aggccggtgc catgaacgcg cttacccgcg cctcggcgct gctctccaac
tcccccttca 1500tcctcaacct cgactgcgat cattacatca acaactccca ggcccttcgc
gccggcatct 1560gcttcatggt gggacgggac agcgacacgg ttgccttcgt ccagttcccg
cagcgcttcg 1620agggcgtcga ccccaccgac ctctacgcca accacaaccg catcttcttc
gacggcaccc 1680tccgtgccct ggacggcatg cagggcccca tctacgtcgg cactgggtgt
ctcttccgcc 1740gcatcaccgt ctacggcttc gacccgccga ggatcaacgt cggcggtccc
tgcttcccca 1800ggctcgccgg gctcttcgcc aagaccaagt acgagaagcc cgggctcgag
atgaccacgg 1860ccaaggccaa ggccgcgccc gtgcccgcca agggtaagca cggcttcttg
ccactgccca 1920agaagacgta cggcaagtcg gacgccttcg tggacaccat cccgcgcgcg
tcgcacccgt 1980cgccctacgc cgcggcggct gaggggatcg tggccgacga ggcgaccatc
gtcgaggcgg 2040tgaacgtgac ggccgccgcg ttcgagaaga agaccggctg gggcaaagag
atcggctggg 2100tgtacgacac cgtcacggag gacgtggtca ccggctaccg gatgcatatc
aaggggtggc 2160ggtcacgcta ctgctccatc tacccacacg ccttcatcgg caccgccccc
atcaacctca 2220cggagaggct cttccaggtg ctccgctggt ccacgggatc cctcgagatc
ttcttctcca 2280agaacaaccc gctcttcggc agcacatacc tccacccgct gcagcgcgtc
gcctacatca 2340acatcaccac ttaccccttc accgccatct tcctcatctt ctacaccacc
gtgccggcgc 2400tatccttcgt caccggccac ttcatcgtgc agcgcccgac caccatgttc
tacgtctacc 2460tgggcatcgt gctatccacg ctgctcgtca tcgccgtgct ggaggtcaag
tgggccgggg 2520tcacagtctt cgagtggttc aggaacggcc agttctggat gacagcaagt
tgctccgcct 2580acctcgccgc cgtctgccag gtgctgacca aggtgatatt ccggcgggac
atctccttca 2640agctcacatc caagctaccc tcgggagacg agaagaagga cccctacgcc
gacctctacg 2700tggtgcgctg gacgccgctc atgattacac ccatcatcat catcttcgtc
aacatcatcg 2760gatccgccgt ggccttcgcc aaggttctcg acggcgagtg gacgcactgg
ctcaaggtcg 2820ccggcggcgt cttcttcaac ttctgggtgc tcttccacct ctaccccttc
gccaagggca 2880tcctggggaa gcacggaaag acgccagtcg tggtgctcgt ctggtgggca
ttcaccttcg 2940tcatcaccgc cgtgctctac atcaacatcc cccacatgca tacctcggga
ggcaagcaca 3000caacggtgca tggtcaccat ggcaagaagt tggtcgacac agggctctat
ggctggctcc 3060attgatgact ttgcccggac aagacgacct gagacaagaa acaactcatc
cactcaacag 3120tcagtgcatg catccatctc atcgagaagc agagcccgcc aaagtttgaa
ttttttaatt 3180ttttttcttc acttttttgc ccgtttcttt ttagttttgt ccagaaaaaa
gatggtgttg 3240atttgattta gtttataatt acctgtggta attaattatg tacttaatta
tacattccgc 3300gaacaacaag ggagacagac gacttacggg gtactggctc gggtggtaag
agcttgcact 3360gtactgtaca tgctcgacga tgtatagaga tgcacagagg agaggatggg
agtgctggga 3420ccgtggggtg gacggcggta ttcttttagt attatatatg gaaacaataa
atttaatttc 3480attaaaaaa
3489142799DNAHordeum vulgare 14gtgcaccacc ggcgccgctg
gctcacatcc tagcttctcc tcccttcctg ctccggtcgg 60gaaacctggg agtgggagac
atggcgtcgg cggccggtgc tgctgggtca aatgccagcc 120tcgccgcccc gctgctggcg
agccgcgagg gaggagccaa gaagccggtc ggtgccaagg 180gcaagcactg ggaggccgcc
gacaaggacg agcggcgggc cgccaaggag agcggcggcg 240aggacggcag gccgctgctg
ttccggacgt acaaggtcaa aggcaccctc ctgcacccat 300acagggcgct aatcttcatt
cgcttaattg cggtccttct attcttcgta tggcgcatca 360agcacaacaa atccgacatc
atgtggtttt ggacaatatc agtcgtcggg gacgtatggt 420tcgggttctc gtggctgctc
aaccaactcc caaagttcaa ccctatcaaa accatacctg 480atatggtcgc ccttaggcga
caatacgatc tttcagatgg gacatctaca ctcccgggca 540tagatgtctt tgtcaccacc
gctgacccaa tcgatgagcc gatactatac accatgaatt 600gtgtcctttc tatccttgct
tctgactatc ctgtcgatag gtgtgcctgc tatctcccag 660atgatagtgg agcattgatt
caatacgagg ccttagttga gaccgcaaag tttgctactt 720tgtgggtccc attttgtcgg
aagcattgca ttgagccaag agccccagaa agctactttg 780aaatagaggc accgttgtac
actggaactg caccagagga gttcaagaat gattatagta 840gtgtacataa agagtatgat
gagttcaaag agcgcttgga ctcactatcc gatgctattt 900ccaagcgttc tgatgcttac
aacagcatga agactgagga aggagatgca aaggccacgt 960ggatggcaaa tgggacacaa
tggccaggat catggattga cacaacggaa atccatagaa 1020aaggacatca tgccggaatt
gttaaggttg tgttggacca ttcgatccgt gggcataatc 1080ttggttcaca agaaagcacc
cacaacctca gtttcgccaa caccgatgag cgcctcccga 1140tgcttgtgta tatctctcgt
ggaaagaacc caagctatga ccacaacaag aaagctggtg 1200ccttgaatgc gcaattgcgt
gcctctgcac tactctccaa cgcacaattc atcatcaact 1260ttgactgcga ccactacatc
aacaactctc aagccctacg tgcagctatg tgcttcatgc 1320ttgatcaaag gcaaggtgat
aacactgcct ttgttcaatt ccctcaacgc ttcgacaatg 1380ttgatccatc agaccgatat
ggaaaccaca accgtgtctt ctttgacggc acaatgctcg 1440ccctcaatgg cctccaaggg
ccatcttacc ttggcactgg ttgcatgttc cgccgcatag 1500cactttatgg cattgaccca
cctgactgga gacatgacaa catcatagtt gatgataaaa 1560agtttggtag ctccataccc
ttcctagatt ccgtatcaaa agccataaac caagaaaggt 1620ctaccatacc tccacccatt
agtgaaacat tggtggctga gatggaaagg gttgtgtcgg 1680cttcacacga taaagccact
ggttggggca agggtgttgg gtacatatat gacatagcca 1740cagaggatat cgtgactggt
ttccgcatcc atgggcaagg ttggcgttcc atgtattgta 1800caatggagcg tgacgccttc
tgtggcattg caccaatcaa cctaaccgag cgcctccacc 1860aaattgtgcg ctggtccggt
ggatctttag agatgttctt ctcactaaat aacccactca 1920taggtggtcg ccggatccac
gcccttcagc gtgtctccta cctcaacatg acagtctacc 1980cagtcacatc actctttatc
ctactctatg ctctcagccc agtgatgtgg cttatccctg 2040atgaagtata catccagagg
ccattcacca aatatgtcgt gttccttctc gtgatcattc 2100tgatgatcca tataattggg
tggctcgaga taaaatgggc gggggtcaca tggttggatt 2160actggaggaa tgaacagttc
tttatgatcg ggtcgacgag tgcataccca gcagccgtgc 2220tgcacatggt ggtgaatctc
cttacaaaga agggtataca cttcagagtt acttcgaagc 2280aaacaacggc agacaccaat
gacaagtttg ctgacttgta tgacatgcga tgggtgccaa 2340tgttaatccc tacaacagtg
gtgctgattg ccaatgttgg tgcaatcggt gtagccatgg 2400gtaaaacgat agtatacatg
ggagcatgga caattgcaca gaagacacat gccgcattgg 2460gtctgctctt caacgtgtgg
atcatggtcc tgctctatcc gtttgcattg gcgatcatgg 2520gacggtgggc aaagaggcca
gtcatcctgg tggtcttgtt gccggttgcc tttacaatag 2580tttgccttgt atatgtttct
gttcatatat tacttcttag ttttcttcca ttttagccaa 2640ttgatatagt acaaaaccat
tgtattgttt aggttctgta gtaagctggg agccaaatat 2700gctatgtgta tccaataaac
cattgtagta cttgtacttt gtaccaatta atagcggatg 2760gttataattt aggccttgat
ggctaatgtt acaacttac 2799152785DNAHordeum
vulgare 15tcagttcact gcttggctgc ttgctcccct acatcactct ctcactctca
ctcactcagg 60gtgcaagaac aggctctgct acttggtagg cggaccaaag gtggcatggc
ttctccggcg 120gccgtcggcg ggggtcgtct agccgaccca ctgctggccg ccgacgtcgt
cgtcgtcggc 180gccaaagaca agtactgggt gcccgccgac gagagagaga tcctggcgtc
gcagagcagc 240ggcggcggtg aacaggacgg ccgggcaccg ctgctatacc gcacgttcag
ggtcaagggc 300ttcttcatca acctttacag gttattgact ctggtcagag ttatcgtggt
tattctattc 360ttcacgtggc gcatgaggca ccgggactcg gacgcgatgt ggctgtggtg
gatctcggtc 420gtgggcgacc tctggttcgg agtcacctgg ctgctcaacc agatcaccaa
gctcaagccc 480aggaaatgcg tccccagcat ctccgtcctg agagagcagc tcgaccagcc
cgacggcggc 540tccgacctgc cccttctcga cgtgttcatc aacaccgtcg acccggtgga
cgagccgatg 600ctctacacca tgaactccat cctctccatc ctggccaccg actaccccgt
ccagaagtac 660gccacctatt tctccgatga cggcgggtcg ctggtgcact acgaggggct
gctgctgacg 720gcggagttcg ccgcgtcgtg ggtcccgttc tgccggaagc attgcgtcga
gcctcgcgcc 780ccggagagct acttctgggc caagatgcgc ggggagtacg ccggcagcgc
ggccaaggag 840ttccttgacg accatcggag gatgcgcgcg gcgtatgagg agttcaaggc
gaggctggac 900gggctttctg ccgtcatcga gcagcggtcc gaggcgtgca accgcgctgc
aaacgagaaa 960gaagggtgtg ggaacgcgac ttggatggcc gatgggtcga cgcaatggca
ggggacgtgg 1020atcaagccgg ccaagggcca ccggaaagga caccatcctg caattcttca
ggttatgctg 1080gatcaaccta gcaaggatcc tgagctggga atggcggcga gctccgacca
ccctctggat 1140ttcagcgccg tggacgtgcg cctcccgatg ctggtctaca ttgcccggga
gaagcggcct 1200gggtatgacc accagaagaa ggcgggcgcc atgaacgtgc agctgcgcgt
gtccgcgctg 1260ctctccaacg cgcccttcat catcaacttc gacggcgacc actacatcaa
caactcgcag 1320gccttccgcg ccgccatgtg cttcatgctc gacccgcgcg acggcgccga
caccgccttc 1380gtccagttcc cgcagcgctt cgacgacgtc gaccccaccg accgctactg
caaccacaac 1440cgcatgttct tcgacgccac cctcctcggc ctcaacggca tccagggccc
ctccttcgtc 1500ggcaccggat gcatgttccg ccgcgtcgct ctctacagcg ccgaccctcc
acggtggcgg 1560tccgacgacg ccaaggaggc caaggcctcg cacaggccca acatgtttgg
caagtctacg 1620tccttcatca actcaatgcc ggcggccgcc aaccaagaac ggtccgtccc
gtcaccggcg 1680acagtcggcg aggcggagct cgcagacgcg atgacttgcg cgtacgagga
cggcaccgag 1740tggggcaacg acgttgggtg ggtgtacaac atcgcgacgg aggacgtggt
gaccggcttc 1800cggctgcacc ggacggggtg gcgctccacg tactgcgcca tggagcccga
cgcgttccgc 1860ggcacggcgc ccatcaacct caccgagcgc ctgtaccaga tcctgcgttg
gtcgggggga 1920tccctcgaga tgttcttctc ccgcttctgc ccgctcctgg ccggccgccg
cctccacccc 1980atgcagcgcg tcgcctacat caacatgacc acctacccgg tctccacctt
cttcatcctc 2040atgtattact tctacccggt catgtggctc ttccaggggg agttctacat
ccagaggccg 2100ttccagacgt tcgcgctctt cgtcgtcgtc gtcatcgcca cggtggagct
catcggcatg 2160gtggagatca ggtgggcagg cctcacgctg ctcgactggg tccgcaacga
gcagttctac 2220atcatcggca ccaccggcgt gtacccgatg gccatgctgc acatcctcct
caggtccctc 2280ggcataaagg gggtgtcctt caagctgacg gccaagaagc tcacgggggg
cgccagggag 2340aggctcgcgg agctgtacga cgtgcagtgg gtgccgttgc tggtgcccac
cgtggtggtc 2400atggccgtga acgtggccgc catcggcgcg gcggcgggca aggcgatcgt
tgggcggtgg 2460tcggcagcgc aggtcgcggg ggcggcgagc gggcttgttt tcaacgtgtg
gatgctgctg 2520ctgctctacc cgttcgcgct cgggataatg gggcactgga gcaagaggcc
ctacatcctg 2580ttccttgtgc tggtgaccgc ggtcgctgcc accgcgtccg tgtacgtcgc
actcgcgggg 2640tccttgctgt acttgcattc ggggataaaa ctagtttaat ttttgtcaag
taatgctgca 2700waacctgtaa aagctgtgaa acaaaacagc tttaattgca gcatataata
aagtttacct 2760tgaaaaaaaa aaaaaaaaaa aaaaa
2785162864DNAHordeum vulgare 16ggcagggaaa tcagctgtct
agctagtcgc acgtaggcac ttacactatg gccccggcag 60tcactcgccg agccaacgct
ctccgcgtcg aggccccgga cggcaatgcc gagagcgggc 120gcgccagcct agcagcagac
tcccccgcgg ccaagcgggc catcgatgcc aaggacgaag 180accgggtggc cgcggctgag
ggagacgcgt ctggagccag cgccggcaac ggcgaccggc 240cgccgctgtt ccggaccatg
aaggtcaagg gaagcatcct ccatccttac aggttcatga 300tcctcgtgcg cttggtcgcc
gtcgtcgcgt tcttcgcgtg gcgcctgaag cacaagaacc 360acgacggcat gtggctctgg
gccacgtcca tggtcgccga cgtctggttc ggcttctcat 420ggctcctcaa ccagctgccc
aagctcaacc ccatcaagcg cgtccccgac ctggccgccc 480tcgccgacca gtgcggctcc
tccggcgacg ccaacctgcc aggcatcgac atctttgtca 540ccaccgtgga ccccgtggac
gaacccatct tgtacaccgt gaacaccata ctctccatcc 600tcgccaccga ctaccctgtc
gataagtacg cctgctacct ctcagacgac ggcggcacgt 660tggtgcacta cgaggccatg
atcgaagtgg ccaatttcgc ggtgatgtgg gtcccttttt 720gccggaagca ctgtgtcgag
ccaaggtccc ccgagaacta ctttgggatg aaaacgcagc 780cgtacgtcgg gagtatggct
ggagaattca tgagggagca taggcgtgtg cgcagagagt 840atgatgagtt caaggtgagg
atagactccc tgtccaccac catccgccaa agatctgatg 900cgtacaactc gagcatctca
ggagatggtg tgcgtgcaac ctggatggct gatggggcac 960aatggcctgg tacgtggatt
gagcagtttg agaaccaccg gagaggacaa catgctggaa 1020ttgttcaggt catactaagc
catcctagtt gcaaaccgca actggggtct ccggcgagca 1080ctgacaatcc acttgacttc
agcaacgttg acacgaggct gcccatgctc gtctacatgt 1140cccgggagaa gcgccccggt
tataaccacc aaaagaaggc aggcgccatg aacgtgatgc 1200tccgtgtctc ggcgttgctc
tccaacgcgc cattcgtcgt caattttgac tgcgaccact 1260acatcaacaa cacgcaagct
ctccgcgccc ctatgtgctt catgctcgac cctcgcgacg 1320gtcagaacac ggccttcgtc
cagtttccgc agcgcttcga cgacgtcgac ccgacggacc 1380gctacgccaa ccacaaccgt
gtcttcttcg acggtaccat gctctccctc aacggccttc 1440aagggccttc ctacctcggc
actggcacca tgttccgtcg tgtcacgctc tatggcatgg 1500agccaccacg ttacagagcg
gagaacatca agcttgtagg taagacctat gagttcggta 1560gctcgacgtc tttcatcaat
tccatgccgg acggcgcaat ccaagagcgg tctatcacgc 1620cggtgttggt cgacgaggca
ctcagcaatg acctggctac cctgatgacg tgtgcttacg 1680aggacgggac ctcatggggg
agagacgttg ggtgggtgta caacatcgcg acggaggacg 1740tggtgaccgg attccgcatg
caccggcagg ggtggcgctc catgtattgc tccatggagc 1800cggccgcctt ccgcggaaca
gcgccgatca acctcaccga gcgcctttac caggtgctcc 1860ggtggtcggg cggctctctc
gagatgttct tctcccacag caacgctctc atggccggcc 1920gccgtatcca ccctctgcag
cgtgtcgcgt acctcaacat gtcgacctac ccgatcgtca 1980cggtgttcat cctggcctac
aacctcttcc ccgtcatgtg gctcttctcc gagcagttct 2040acatccagag gccgttcggc
acgtacatca tgtacctcgt cggcgtcata gcgatgattc 2100acgtgatcgg catgttcgag
gtgaaatggg cggggatcac gctgctcgac tggtgccgca 2160acgagcagtt ctacatgatc
ggggcgacgg gcgtgtaccc gacggcggtg ctttacatgg 2220cgctcaagct tgtcacgggg
aaggggatat acttcaggct cacatccaag cagacggacg 2280cttgctccaa cgacaagttc
gccgacctgt acacggtgcg gtgggtgccg ctgctgttcc 2340cgacggtcgc agtgctcatc
gtgaacgtcg cggctgtcgg ggcagcgata ggcaaggcag 2400cagcgtgggg cttcttcacg
gaccaggcgc ggcacgtgct gctcgggatg gtgttcaacg 2460tgtggatcct cgtgctcctc
tacccgtttg cgctcgggat catggggaaa tgggggaaga 2520gacccatcat cctgttcgtc
atgttgatca tggtcattgg cgccgtcggg ctcgtgtatg 2580tcgccttcca tgatccctac
ccactgattt tcagagttgc agctcttggt gaagcatcgc 2640tgaccgggcc atctgggtag
acacgtacgg ctcttttttt tacaagtaca gccagagtca 2700ctgcaataat ttgagtgtgt
gtattcatgt ctacttatat agcatgagaa ctggtcattg 2760tgtgccactc ctctactcta
gtagagtata aatgtaccta tcttttcttt ggaaaaaaac 2820tgaagtgcgg cttgtgctct
tttggtgaga actaaaaaaa aaaa 2864172910DNAHordeum
vulgare 17tacaactaag cggactctag tatcgtggtt gatgattgca atccggcaat
gttgtcgccc 60cggacagacg ccggcgccgg cgccgccacc gacctcagcc agccacttct
ctggaaccgc 120aatggcgttc acgcaggagc attggtcgtc atgccagtcg tggccaatgg
tcacggcggc 180ggcgacaagc ttaagggcgc cccgaaagcc aaggacaagt actggaaaga
cgtcgaccag 240ccggacgaca tggcggcagc gccagacctg gacaacggcg gcggccggcc
gctgctgttc 300tcgaacttga gagtccagaa tatcatcctg taccccggcg aggtattgat
cctgatacga 360gtaatcgccg taatcttatt tgttggatgg cgcatcaagc ataacaattc
agatgtcatg 420tggttttgga tgatgtccgt cgtcgcagac gtgtggttta gcttatcatg
gctaagctac 480caactgccaa agtataatcc cgttaaaagg atacccgacc ttgctacact
caggaaacaa 540tatgacacac cagggaggag ctcccagctg ccaagcattg acgtcatcgt
caccactgcc 600agtgctaccg atgagcccat attgtacacc atgaactgtg ttctctctat
acttgcagct 660gactatcata ttggcaggtg caactgctac ctatcagatg atagcggctc
attggtcctt 720tatgaggcat tggttgagac tgcaaagttt gctgctttat gggttccttt
ctgtagaaag 780catcagattg agccaagagc accggaaagc tattttgaac taaagggccc
gttgtatgga 840gggacgccac ataaggagtt ctttcaggat tataagcatt taggcacaca
atatgaagag 900ttcaagaaga atttagatat gcttcctaac accatccatc aaaggtcggg
aacttacagt 960aaaacaggaa cggaggatga agatgcaaaa gtgacttgga tggctgacgg
aacacaatgg 1020ccaggcacat ggcttgaccc agcagaaaaa catagggccg ggcatcatgc
aggaattgtt 1080aagattgtgc agagccatcc agaacatgtg gttcaaccag gcgtacaaga
gagccttgac 1140aacccactca gctttgacga tgttgatgtg cgcctgccca tgttggtata
tgtggctcgt 1200gaaaagagtc caggtatcga gcataacaaa aaggcaggcg ctttgaatgc
agagctacgt 1260atctcagctc tactctctaa tgcacctttc ttcattaact ttgactgcga
ccactacatc 1320aacaattcag aagccctacg tgcagctgtt tgcttcatgc tagacccacg
tgaaggggat 1380aatactggat ttgttcagtt cccgcaaaga tttgataatg tcgacccaac
tgaccggtat 1440ggaaaccata atcgagtctt ttttgatggt gccatgtatg gcctcaatgg
tcaacaaggg 1500cctacttacc ttggcacagg ttgcatgttc cgtccccttg cactctatgg
aattgatcca 1560ccttgctgga gagccgagga catcatagtc gacagtaaca ggtttggcaa
ctcattaccc 1620ttcctcaact cagtactagc agccataaag caagaggaag gtgtcacact
accaccacca 1680ctagatgatt catttcttga agagatgaca aaagttgtgt catgttccta
tgatgattcc 1740actgattggg gtaggggcat tggctacata tacaatatgg caacagaaga
catagtaaca 1800ggatttcgta tccatgggca agggtggtgc tccatgtatg ttaccatgga
acgtgaagcg 1860ttccgtggca ctgcaccgat caatctaaca gagcgcctcc gccaaatagt
gcgatggtct 1920ggtggttccc tagagatgtt cttctcgcac atcagcccac tattcgctgg
tcgtcgactc 1980agtttggtgc agcgactctc gtacatcaat ttcactatat acccattgac
atcactcttt 2040atcctaatgt atgccttctg tccagtgatg tggcttcttc caacagaaat
acttatacaa 2100aggccatata ccaggtacat tgtgtacctt atcattgtcg tcgcgatgat
ccatgtgatt 2160ggcatgtttg agataatgtg ggcaggaatc acatggttgg attggtggcg
caacgagcaa 2220tttttcatga tcggctcggt aactgcatat ccaacggcgg tgttgcacat
ggtggtgaat 2280atccttacaa aaaagggtat acacttcaga gtaaccacaa agcaaccagt
ggctgataca 2340gatgacaagt atgctgagat gtatgaagtg cattgggtac ccatgatggt
ccccgcggtt 2400gtggtattgt tttccaacat cttggctatt ggtgtagcaa ttggtaaatc
agtcttatac 2460atggggacat ggtctgtagc acagaaaagg catggtgcac tagggctatt
gttcaacctg 2520tggattatgg tgctccttta cccatttgca ttggcgatta ttggaagatg
ggccaagaga 2580accggaatcc tattcatctt actacccatt gctttcttgg ccaccgcatt
gatgtacatt 2640ggcatccata cattcctttt acatttcttt ccatccatgt tggtatagaa
aatacctagc 2700acacttttct atggatcagt atgacaaatg aagatatctc cttcaggaag
ggacttatgt 2760agttgaacgt gtactttgtg atatagtaat atactagaag ccaacgtgtg
cttcgccaca 2820ccgttcttaa aaaaaaaaaa gaaacacggg gcgttacgga ttccaggaat
agatgagtgg 2880agccgatctc gaaaggaaaa gtaatgggcc
2910182943DNAHordeum vulgare 18aatcgtggga catgggttct
ttggcggcag ccaacggggc cggtcatgcg agcaatggcg 60ccggcgtcgc ggaccaggcg
ctggcactgg agaacggcac cggcaatggg cacaaggcca 120gcgacgccaa ccgagcgacg
ccggtacagc aggcaaacgg cagcagcaag gccgcgggga 180aggttagccc gaaggacaag
tactgggtgg ccgtcgatga gggagagatg gcggccgcca 240tagcggacgg cggcgaggac
ggccggcgac cgctgctgta ccggacgttc aaggtcaagg 300gcatcctcct gcatccctac
aggttgctga gcttgatcag attggttgct atcgtcctat 360ttttcgtatg gcgtgtcagg
cacccatacg ctgatggcat gtggctctgg tggatatcga 420tggttgggga tctttggttt
ggcgtcactt ggttgctaaa ccaagttgca aagctcaacc 480ctgtcaagcg tgtccccaac
cttgcgcttt tgcaacagca gtttgatctc cctgacggca 540actccaacct tccttgtctt
gatgtcttca tcaacaccgt tgatcccatt aatgaaccta 600tgatatacac tatgaactcc
atcatatcca tccttgctgc agactatccg gttgacaagc 660atgcttgcta cctttcagat
gatggtgggt caataatcca ttatgatggt ttgcttgaga 720ctgcaaaatt tgctgcatta
tgggttccct tttgcagaaa acattccatt gagccaagag 780cccctgagag ctatttttct
ttgaatacac gcccatacac tggaaatgca ccacaagact 840ttgtcaatga ccgcagacac
atgtgtagag agtatgatga gttcaaggag cgcttagatg 900cactttttac cctcattccc
aaacggtcag atgtgtacaa tcatgctgct ggcaaagaag 960gtgcaaaggc aacttggatg
gcagatggga cacagtggcc aggcacatgg attgacccag 1020ctgaaaacca taagaaagga
caacatgctg ggatcgttaa ggttttgttg aaacatccaa 1080gttatgaacc agaacttggt
ctaggagcaa gcaccaacag tcctctagac ttcagtgcag 1140ttgatgtgcg cctcccaatg
ctcgtttaca tctcccgtga gaagagtcca agctgtgatc 1200atcaaaagaa ggcaggtgcc
atgaacgtac agttgcgagt ctctgccctc ctgaccaatg 1260cgccctttat catcaacttt
gatggtgacc actacgtcaa caactcgaaa gccttccgtg 1320ctggcatatg tttcatgctc
gatcgccgtg aaggtgacaa tactgccttt gtccagtttc 1380cccaacgctt cgatgatgtt
gatcccacag ataggtactg caatcacaat cgtgtcttct 1440ttgacgccac cttgctcggc
tccaatggca tccaagggcc gtcttatgtt ggcactggtt 1500gcatgttccg ccgtgtcgca
ctttacggtg ttgacccacc tcgctggaga cctgatgacg 1560tgaagatcgt ggacagctcc
agcaagtttg gcagttcaga gtcattcatc agctcaatac 1620tgccagcagc agaccaagaa
cgctccatca tgtcgccacc ggcacttgaa gagtctgtca 1680tggctgactt agctcatgtc
atgacttgtg catatgagga cgggactgaa tggggcagag 1740aagttggttg ggtgtacaac
attgcaactg aggatgtggt gaccggcttc cggctgcacc 1800ggaatgggtg gcgatccatg
tactgccgca tggaaccaga tgcattcgcc ggcaccgcgc 1860caatcaacct cactgagcgg
ctctaccaga tcctgcgctg gtcggggggc tcccttgaga 1920tgttcttctc gcacaactgc
ccactcctgg ctggccgccg cctccaccca atgcaaagaa 1980ttgcctatgc caacatgaca
gcctacccag tttcatctgt ctttcttgtg ttctatctcc 2040tcttcccggt gatatggatc
ttccgtgggc aattctacat acagaagcca ttccccacgt 2100atgtgttgta cctcgtcatc
gtcatagccc tgaccgagtt aatcggtatg gttgagatca 2160agtgggctgg gctcacgctg
ctggactgga tccgcaacga gcagttctac attattggtg 2220caacagccgt gtaccctaca
gcagtatttc acatagtgct gaagctgttt ggcctgaagg 2280gtgtttcatt caagctgacg
gcaaaacagg tagcaagcag taccagcgat aagtttgctg 2340aactgtatgc cgtgcagtgg
gctccgatgc tgatccctac catggtggtt atagcggtga 2400atgtttgtgc cattggcgcg
tcgataggca aggcggtagt gggaggatgg tcactgatgc 2460agatggccga tgcaggactt
gggctggtgt tcaacgcgtg gattctggtg ctgatctacc 2520cgtttgcact gggcatgatt
ggacggtgga gcaagaggcc ctacatcctg ttcattctgt 2580ttgtcattgc gtttattttg
atcgcattgg tggatatcgc catccaggcc atgcggtctg 2640ggattgttcg gttccacttt
aaaagctcag gtggcgccac ttttcccaca agctggggtt 2700tgtaagttcc tccctttgct
gggaagttcc ttttcctttg tggtgtgggt tttttctttt 2760ttccttttct tgttattgca
tatatagacc atttgtggtg tgaatcatat ataggctgta 2820caagtttgtg gaaataatgt
aaaattggcc atttgcctgg aagaaaaaga gatgtggcaa 2880agcaacagct gtagtatatt
ggatataatg ttgaggatgc ctggaagtat atctattcct 2940gta
2943192583DNAOryza sativa
19atgtccgcgg cggcagcggt gacaagctgg actaacggat gctggtcgcc cgcggctacg
60cgggtgaacg acggcggcaa ggacgatgtg tgggtggccg tcgacgaagc ggacgtgtcg
120ggggcccgcg gcagcgacgg cggcggccgg ccgccgctgt tccagacgta caaggtcaag
180ggcagcatcc ttcatcctta caggttcttg atcctggcgc gactgatcgc catcgtcgcc
240ttcttcgcgt ggcgcatacg tcacaagaac cgcgacggcg cgtggctgtg gacaatgtcc
300atggtcggcg acgtctggtt cggcttctcg tgggtgctca accagctacc gaagcagagc
360cccatcaagc gcgtcccgga catcgccgcc ctcgccgacc ggcactccgg cgacctaccc
420ggcgtcgacg tcttcgtcac caccgtcgac cctgtcgacg agccgatact ctacaccgtg
480aacaccatcc tctccattct cgccgccgac tacccggtgg acaggtacgc ttgctacctc
540tccgacgacg gcgggacgct ggtccactac gaggctatgg tggaggtcgc caagttcgcc
600gagctgtggg tgcccttctg ccggaagcac tgcgtcgagc cgaggtcgcc ggagaactac
660ttcgcgatga agacgcaggc gtacaaaggc ggcgtccccg gcgagctgat gagcgatcac
720cggcgtgtgc ggcgagagta cgaggagttc aaggtcagga tcgactccct ctcgagcacc
780attcgccagc gatctgatgt gtacaacgcc aaacatgccg gcgaaaatgc gacatggatg
840gctgatggca cacattggcc cggcacatgg tttgagccgg ctgacaacca ccagagaggg
900aaacatgctg gaattgttca ggttttactg aaccatccaa gctgtaaacc gaggcttgga
960ttggcggcga gtgctgagaa tccggttgat ttcagcggtg tcgacgtgcg gctccccatg
1020ctggtgtaca tctcgcgcga gaagcggccc ggatacaacc accagaagaa ggccggcgcc
1080atgaacgtga tgctccgcgt gtccgcgctg ctgtcgaacg cgccgttcgt catcaacttc
1140gacggcgacc actacgtcaa caactcgcag gcgttcaggg cgccgatgtg cttcatgctc
1200gacggccgcg gccgcggcgg cgagaacacg gcgttcgtcc agttcccgca gcggttcgac
1260gacgttgacc cgacggaccg gtacgcgaac cataaccgcg tcttcttcga cggcaccatg
1320ctctccctca acggcctcca ggggccctcc tacctcggca ccggcaccat gttccgccgc
1380gtcgcgctct acggcgtgga gccgccgcgc tggggagcgg cggcgagcca gatcaaggct
1440atggacatcg ccaacaagtt cggcagctcg acgtcgttcg tcggcacgat gctggacggc
1500gccaaccaag aacggtcgat cacgccgctg gcggtgctcg acgagtcggt cgccggcgac
1560ctcgccgccc tgacggcgtg cgcgtatgag gacgggacgt catgggggag agacgtcggg
1620tgggtgtaca acatcgcgac ggaggacgtg gtgaccgggt tccgcatgca ccggcagggg
1680tggcgctccg tatacgcctc agtggagccc gccgcgttcc gcggcacggc gccgatcaac
1740ctcaccgagc gcctctacca gatcctccgg tggtcgggcg gctcgctgga gatgttcttc
1800tcccacagca acgcgctcct cgccggccgc cgcctccacc cgctgcagcg cgtcgcctac
1860ctcaatatgt ccacctaccc gatcgtgacc gtgttcatct tcttctacaa cctcttcccg
1920gtgatgtggc tcatctccga gcagtattac atccagcggc cgttcggcga gtacctcctc
1980tacctcgtcg ccgtcatcgc catgatccac gtgatcggca tgttcgaggt gaagtgggct
2040ggcatcacgc tgctggactg gtgccgcaac gagcagttct acatgatcgg atccacgggg
2100gtgtacccga cggcggtgct gtacatggcg ctcaagctcg tcaccgggaa gggcatctac
2160ttccgcctca cgtcgaagca gacggcagcc agctccggcg acaagttcgc cgacctgtac
2220accgtgcggt gggtgcctct gctgatcccg accatcgtca tcatggtcgt gaacgtcgcc
2280gccgtcgggg tggcggtcgg caaggcggcg gcgtgggggc cgctcaccga gccggggtgg
2340ctcgccgtgc tcgggatggt gttcaacgtg tggatcctgg tgctcctcta cccgttcgcg
2400ctcggggtca tgggtcaatg ggggaagcgg ccggccgtgc tgttcgtggc gatggcgatg
2460gccgtcgccg ccgtggcggc catgtacgtc gccttcggtg caccgtacca agctgagttg
2520tcaggtgttg ctgcttctct cggtaaagtg gcggcggcat cgctgactgg gccatctggg
2580tag
258320860PRTOryza sativa 20Met Ser Ala Ala Ala Ala Val Thr Ser Trp Thr
Asn Gly Cys Trp Ser1 5 10
15Pro Ala Ala Thr Arg Val Asn Asp Gly Gly Lys Asp Asp Val Trp Val
20 25 30Ala Val Asp Glu Ala Asp Val
Ser Gly Ala Arg Gly Ser Asp Gly Gly 35 40
45Gly Arg Pro Pro Leu Phe Gln Thr Tyr Lys Val Lys Gly Ser Ile
Leu 50 55 60His Pro Tyr Arg Phe Leu
Ile Leu Ala Arg Leu Ile Ala Ile Val Ala65 70
75 80Phe Phe Ala Trp Arg Ile Arg His Lys Asn Arg
Asp Gly Ala Trp Leu 85 90
95Trp Thr Met Ser Met Val Gly Asp Val Trp Phe Gly Phe Ser Trp Val
100 105 110Leu Asn Gln Leu Pro Lys
Gln Ser Pro Ile Lys Arg Val Pro Asp Ile 115 120
125Ala Ala Leu Ala Asp Arg His Ser Gly Asp Leu Pro Gly Val
Asp Val 130 135 140Phe Val Thr Thr Val
Asp Pro Val Asp Glu Pro Ile Leu Tyr Thr Val145 150
155 160Asn Thr Ile Leu Ser Ile Leu Ala Ala Asp
Tyr Pro Val Asp Arg Tyr 165 170
175Ala Cys Tyr Leu Ser Asp Asp Gly Gly Thr Leu Val His Tyr Glu Ala
180 185 190Met Val Glu Val Ala
Lys Phe Ala Glu Leu Trp Val Pro Phe Cys Arg 195
200 205Lys His Cys Val Glu Pro Arg Ser Pro Glu Asn Tyr
Phe Ala Met Lys 210 215 220Thr Gln Ala
Tyr Lys Gly Gly Val Pro Gly Glu Leu Met Ser Asp His225
230 235 240Arg Arg Val Arg Arg Glu Tyr
Glu Glu Phe Lys Val Arg Ile Asp Ser 245
250 255Leu Ser Ser Thr Ile Arg Gln Arg Ser Asp Val Tyr
Asn Ala Lys His 260 265 270Ala
Gly Glu Asn Ala Thr Trp Met Ala Asp Gly Thr His Trp Pro Gly 275
280 285Thr Trp Phe Glu Pro Ala Asp Asn His
Gln Arg Gly Lys His Ala Gly 290 295
300Ile Val Gln Val Leu Leu Asn His Pro Ser Cys Lys Pro Arg Leu Gly305
310 315 320Leu Ala Ala Ser
Ala Glu Asn Pro Val Asp Phe Ser Gly Val Asp Val 325
330 335Arg Leu Pro Met Leu Val Tyr Ile Ser Arg
Glu Lys Arg Pro Gly Tyr 340 345
350Asn His Gln Lys Lys Ala Gly Ala Met Asn Val Met Leu Arg Val Ser
355 360 365Ala Leu Leu Ser Asn Ala Pro
Phe Val Ile Asn Phe Asp Gly Asp His 370 375
380Tyr Val Asn Asn Ser Gln Ala Phe Arg Ala Pro Met Cys Phe Met
Leu385 390 395 400Asp Gly
Arg Gly Arg Gly Gly Glu Asn Thr Ala Phe Val Gln Phe Pro
405 410 415Gln Arg Phe Asp Asp Val Asp
Pro Thr Asp Arg Tyr Ala Asn His Asn 420 425
430Arg Val Phe Phe Asp Gly Thr Met Leu Ser Leu Asn Gly Leu
Gln Gly 435 440 445Pro Ser Tyr Leu
Gly Thr Gly Thr Met Phe Arg Arg Val Ala Leu Tyr 450
455 460Gly Val Glu Pro Pro Arg Trp Gly Ala Ala Ala Ser
Gln Ile Lys Ala465 470 475
480Met Asp Ile Ala Asn Lys Phe Gly Ser Ser Thr Ser Phe Val Gly Thr
485 490 495Met Leu Asp Gly Ala
Asn Gln Glu Arg Ser Ile Thr Pro Leu Ala Val 500
505 510Leu Asp Glu Ser Val Ala Gly Asp Leu Ala Ala Leu
Thr Ala Cys Ala 515 520 525Tyr Glu
Asp Gly Thr Ser Trp Gly Arg Asp Val Gly Trp Val Tyr Asn 530
535 540Ile Ala Thr Glu Asp Val Val Thr Gly Phe Arg
Met His Arg Gln Gly545 550 555
560Trp Arg Ser Val Tyr Ala Ser Val Glu Pro Ala Ala Phe Arg Gly Thr
565 570 575Ala Pro Ile Asn
Leu Thr Glu Arg Leu Tyr Gln Ile Leu Arg Trp Ser 580
585 590Gly Gly Ser Leu Glu Met Phe Phe Ser His Ser
Asn Ala Leu Leu Ala 595 600 605Gly
Arg Arg Leu His Pro Leu Gln Arg Val Ala Tyr Leu Asn Met Ser 610
615 620Thr Tyr Pro Ile Val Thr Val Phe Ile Phe
Phe Tyr Asn Leu Phe Pro625 630 635
640Val Met Trp Leu Ile Ser Glu Gln Tyr Tyr Ile Gln Arg Pro Phe
Gly 645 650 655Glu Tyr Leu
Leu Tyr Leu Val Ala Val Ile Ala Met Ile His Val Ile 660
665 670Gly Met Phe Glu Val Lys Trp Ala Gly Ile
Thr Leu Leu Asp Trp Cys 675 680
685Arg Asn Glu Gln Phe Tyr Met Ile Gly Ser Thr Gly Val Tyr Pro Thr 690
695 700Ala Val Leu Tyr Met Ala Leu Lys
Leu Val Thr Gly Lys Gly Ile Tyr705 710
715 720Phe Arg Leu Thr Ser Lys Gln Thr Ala Ala Ser Ser
Gly Asp Lys Phe 725 730
735Ala Asp Leu Tyr Thr Val Arg Trp Val Pro Leu Leu Ile Pro Thr Ile
740 745 750Val Ile Met Val Val Asn
Val Ala Ala Val Gly Val Ala Val Gly Lys 755 760
765Ala Ala Ala Trp Gly Pro Leu Thr Glu Pro Gly Trp Leu Ala
Val Leu 770 775 780Gly Met Val Phe Asn
Val Trp Ile Leu Val Leu Leu Tyr Pro Phe Ala785 790
795 800Leu Gly Val Met Gly Gln Trp Gly Lys Arg
Pro Ala Val Leu Phe Val 805 810
815Ala Met Ala Met Ala Val Ala Ala Val Ala Ala Met Tyr Val Ala Phe
820 825 830Gly Ala Pro Tyr Gln
Ala Glu Leu Ser Gly Val Ala Ala Ser Leu Gly 835
840 845Lys Val Ala Ala Ala Ser Leu Thr Gly Pro Ser Gly
850 855 860212670DNAOryza sativa
21atggcggcca ccgcggcttc cacgatgtcc gcagcggcgg cagtgactcg ccggatcaac
60gctgccctcc gcgtggacgc caccagcggt gacgtcgcgg ccggcgccga cgggcagaac
120gggcgccggt cgcccgtggc caagcgggtg aacgacggcg gtggcggcaa ggatgacgtg
180tgggtggccg tcgacgaaaa ggacgtgtgc ggggcccgcg gcggcgatgg cgccgcccgg
240ccgccgctgt tccggacgta caaggtcaag ggcagcatcc ttcatcctta caggttcctg
300atccttcttc gactgatcgc catcgtcgcc ttcttcgcgt ggcgcgtacg tcacaagaac
360cgcgacggcg tgtggctgtg gacaatgtcc atggtcggcg acgtctggtt cggcttctcg
420tgggtgctca accagctccc gaagctgagc cccatcaagc gcgtcccgga cctcgccgcc
480ctcgccgacc ggcactccgg cgacctaccc ggcgtcgacg tcttcgtcac caccgtcgac
540cccgtcgacg agccgatact ctacaccgtg aacaccatcc tctccatcct cgccgccgac
600tacccggtgg acaggtacgc ctgctacctg tccgacgacg gcgggacgct ggtccactac
660gaggccatgg tggaggtcgc caagttcgcc gagctgtggg tgcccttctg ccggaagcac
720tgcgtcgagc cgaggtcgcc ggagaactac ttcgcgatga agacgcaggc gtacaaaggc
780ggcgtccccg gcgagctgat gagcgatcac cggcgtgtgc ggcgagagta cgaggagttc
840aaggtcagga tcgactccct ctcgagcacc attcgccagc gatcagatgt gtacaacgcc
900aaacatgccg gcgaaaatgc gacatggatg gctgatggca cacattggcc cggcacatgg
960tttgagccgg ctgacaacca ccagagaggg aaacatgctg gaattgttca ggttttactg
1020aaccatccaa gctgtaaacc gaggcttgga ttggcggcga gtgctgagaa tccggttgat
1080ttcagcggtg tcgacgtgcg gctccccatg ctggtgtaca tctcgcgcga gaagaggccc
1140ggatacaacc accagaagaa ggccggcgcc atgaacgtga tgctccgcgt gtccgcgctg
1200ctgtcgaacg cgccgttcgt catcaacttc gacggcgacc actacgtcaa caactcgcag
1260gcgttcaggg cgccgatgtg cttcatgctc gacggccgcg gccgcggcgg cgagaacacg
1320gcgttcgtcc agttcccgca gcggttcgac gacgttgacc cgacggaccg gtacgcgaac
1380cataaccgcg tcttcttcga cggcaccatg ctctccctca acggcctcca ggggccctcc
1440tacctcggca ccggcaccat gttccgccgc gtcgcgctct acggcgtgga gccgccgcgc
1500tggggagcgg cggcgagcca gatcaaggct atggacatcg ccaacaagtt cggcagctcg
1560acgtcgttcg tcggcacgat gctggacggc gccaaccaag aacggtcgat cacgccgctg
1620gcggtgctcg acgagtcggt cgccggcgac ctcgccgccc tgacggcgtg cgcgtatgag
1680gacgggacgt catgggggag agacgtcggg tgggtgtaca acatcgcgac ggaggacgtg
1740gtgaccgggt tccgcatgca ccggcagggg tggcgctccg tatacgcctc agtggagccc
1800gccgcgttcc gcggcacggc gccgatcaac ctcaccgagc gcctctacca gatcctccgg
1860tggtcgggcg gctcgctgga gatgttcttc tcccacagca acgcgctcct cgccggccgc
1920cgcctccacc cgctgcagcg cgtcgcctac ctcaatatgt ccacctaccc gatcgtgacc
1980gtgttcatct tcttctacaa cctcttcccg gtgatgtggc tcatctccga gcagtattac
2040atccagcggc cgttcggcga gtacctcctc tacctcgtcg ccgtcatcgc catgatccac
2100gtgatcggca tgttcgaggt gaagtgggct ggcatcacgc tgctggactg gtgccgcaac
2160gagcagttct acatgatcgg atccacgggg gtgtacccga cggcggtgct gtacatggcg
2220ctcaagctcg tcaccgggaa gggcatctac ttccggctca cgtcgaagca gacgacggcc
2280agctccggcg acaagttcgc cgacctgtac accgtgcggt gggtgccgct gctgataccg
2340accatcgtca tcattgtcgt gaacgtcgcc gccgtcgggg tggcggtcgg caaggcggcg
2400gcgtgggggc cgctcaccga gccggggtgg ctcgccgtgc tcgggatggt gttcaacgtg
2460tggatcctgg tgctcctcta cccgttcgcg ctcggggtca tgggtcaatg ggggaagcgg
2520ccggccgtgc tgttcgtggc gatggcgatg gccgtcgccg ccgtggcggc catgtacgtc
2580gccttcggtg caccgtacca agctgagttg tcaggtggtg ctgcttctct cggaaaagcg
2640gcggcgtcgc tgaccgggcc atccgggtag
267022889PRTOryza sativa 22Met Ala Ala Thr Ala Ala Ser Thr Met Ser Ala
Ala Ala Ala Val Thr1 5 10
15Arg Arg Ile Asn Ala Ala Leu Arg Val Asp Ala Thr Ser Gly Asp Val
20 25 30Ala Ala Gly Ala Asp Gly Gln
Asn Gly Arg Arg Ser Pro Val Ala Lys 35 40
45Arg Val Asn Asp Gly Gly Gly Gly Lys Asp Asp Val Trp Val Ala
Val 50 55 60Asp Glu Lys Asp Val Cys
Gly Ala Arg Gly Gly Asp Gly Ala Ala Arg65 70
75 80Pro Pro Leu Phe Arg Thr Tyr Lys Val Lys Gly
Ser Ile Leu His Pro 85 90
95Tyr Arg Phe Leu Ile Leu Leu Arg Leu Ile Ala Ile Val Ala Phe Phe
100 105 110Ala Trp Arg Val Arg His
Lys Asn Arg Asp Gly Val Trp Leu Trp Thr 115 120
125Met Ser Met Val Gly Asp Val Trp Phe Gly Phe Ser Trp Val
Leu Asn 130 135 140Gln Leu Pro Lys Leu
Ser Pro Ile Lys Arg Val Pro Asp Leu Ala Ala145 150
155 160Leu Ala Asp Arg His Ser Gly Asp Leu Pro
Gly Val Asp Val Phe Val 165 170
175Thr Thr Val Asp Pro Val Asp Glu Pro Ile Leu Tyr Thr Val Asn Thr
180 185 190Ile Leu Ser Ile Leu
Ala Ala Asp Tyr Pro Val Asp Arg Tyr Ala Cys 195
200 205Tyr Leu Ser Asp Asp Gly Gly Thr Leu Val His Tyr
Glu Ala Met Val 210 215 220Glu Val Ala
Lys Phe Ala Glu Leu Trp Val Pro Phe Cys Arg Lys His225
230 235 240Cys Val Glu Pro Arg Ser Pro
Glu Asn Tyr Phe Ala Met Lys Thr Gln 245
250 255Ala Tyr Lys Gly Gly Val Pro Gly Glu Leu Met Ser
Asp His Arg Arg 260 265 270Val
Arg Arg Glu Tyr Glu Glu Phe Lys Val Arg Ile Asp Ser Leu Ser 275
280 285Ser Thr Ile Arg Gln Arg Ser Asp Val
Tyr Asn Ala Lys His Ala Gly 290 295
300Glu Asn Ala Thr Trp Met Ala Asp Gly Thr His Trp Pro Gly Thr Trp305
310 315 320Phe Glu Pro Ala
Asp Asn His Gln Arg Gly Lys His Ala Gly Ile Val 325
330 335Gln Val Leu Leu Asn His Pro Ser Cys Lys
Pro Arg Leu Gly Leu Ala 340 345
350Ala Ser Ala Glu Asn Pro Val Asp Phe Ser Gly Val Asp Val Arg Leu
355 360 365Pro Met Leu Val Tyr Ile Ser
Arg Glu Lys Arg Pro Gly Tyr Asn His 370 375
380Gln Lys Lys Ala Gly Ala Met Asn Val Met Leu Arg Val Ser Ala
Leu385 390 395 400Leu Ser
Asn Ala Pro Phe Val Ile Asn Phe Asp Gly Asp His Tyr Val
405 410 415Asn Asn Ser Gln Ala Phe Arg
Ala Pro Met Cys Phe Met Leu Asp Gly 420 425
430Arg Gly Arg Gly Gly Glu Asn Thr Ala Phe Val Gln Phe Pro
Gln Arg 435 440 445Phe Asp Asp Val
Asp Pro Thr Asp Arg Tyr Ala Asn His Asn Arg Val 450
455 460Phe Phe Asp Gly Thr Met Leu Ser Leu Asn Gly Leu
Gln Gly Pro Ser465 470 475
480Tyr Leu Gly Thr Gly Thr Met Phe Arg Arg Val Ala Leu Tyr Gly Val
485 490 495Glu Pro Pro Arg Trp
Gly Ala Ala Ala Ser Gln Ile Lys Ala Met Asp 500
505 510Ile Ala Asn Lys Phe Gly Ser Ser Thr Ser Phe Val
Gly Thr Met Leu 515 520 525Asp Gly
Ala Asn Gln Glu Arg Ser Ile Thr Pro Leu Ala Val Leu Asp 530
535 540Glu Ser Val Ala Gly Asp Leu Ala Ala Leu Thr
Ala Cys Ala Tyr Glu545 550 555
560Asp Gly Thr Ser Trp Gly Arg Asp Val Gly Trp Val Tyr Asn Ile Ala
565 570 575Thr Glu Asp Val
Val Thr Gly Phe Arg Met His Arg Gln Gly Trp Arg 580
585 590Ser Val Tyr Ala Ser Val Glu Pro Ala Ala Phe
Arg Gly Thr Ala Pro 595 600 605Ile
Asn Leu Thr Glu Arg Leu Tyr Gln Ile Leu Arg Trp Ser Gly Gly 610
615 620Ser Leu Glu Met Phe Phe Ser His Ser Asn
Ala Leu Leu Ala Gly Arg625 630 635
640Arg Leu His Pro Leu Gln Arg Val Ala Tyr Leu Asn Met Ser Thr
Tyr 645 650 655Pro Ile Val
Thr Val Phe Ile Phe Phe Tyr Asn Leu Phe Pro Val Met 660
665 670Trp Leu Ile Ser Glu Gln Tyr Tyr Ile Gln
Arg Pro Phe Gly Glu Tyr 675 680
685Leu Leu Tyr Leu Val Ala Val Ile Ala Met Ile His Val Ile Gly Met 690
695 700Phe Glu Val Lys Trp Ala Gly Ile
Thr Leu Leu Asp Trp Cys Arg Asn705 710
715 720Glu Gln Phe Tyr Met Ile Gly Ser Thr Gly Val Tyr
Pro Thr Ala Val 725 730
735Leu Tyr Met Ala Leu Lys Leu Val Thr Gly Lys Gly Ile Tyr Phe Arg
740 745 750Leu Thr Ser Lys Gln Thr
Thr Ala Ser Ser Gly Asp Lys Phe Ala Asp 755 760
765Leu Tyr Thr Val Arg Trp Val Pro Leu Leu Ile Pro Thr Ile
Val Ile 770 775 780Ile Val Val Asn Val
Ala Ala Val Gly Val Ala Val Gly Lys Ala Ala785 790
795 800Ala Trp Gly Pro Leu Thr Glu Pro Gly Trp
Leu Ala Val Leu Gly Met 805 810
815Val Phe Asn Val Trp Ile Leu Val Leu Leu Tyr Pro Phe Ala Leu Gly
820 825 830Val Met Gly Gln Trp
Gly Lys Arg Pro Ala Val Leu Phe Val Ala Met 835
840 845Ala Met Ala Val Ala Ala Val Ala Ala Met Tyr Val
Ala Phe Gly Ala 850 855 860Pro Tyr Gln
Ala Glu Leu Ser Gly Gly Ala Ala Ser Leu Gly Lys Ala865
870 875 880Ala Ala Ser Leu Thr Gly Pro
Ser Gly 885232607DNAOryza sativa 23atggcttcgc cggcgtcggt
cgccggcggt ggtgaggata gcaatggctg cagcagcctc 60atcgacccgc tgctagtgag
ccgcacgagc agcatcggcg gcgcggagag gaaggcggcc 120ggcggcggcg gcggcggcgc
caaggggaag cactgggccg ccgccgataa gggggagcgg 180cgcgcggcga aggagtgcgg
cggcgaggac ggccgccggc cgctgctgtt caggtcgtac 240agggtcaagg gctccctcct
gcacccgtac agggctctga tctttgcacg cttgattgcc 300gttctcctct tcttcggatg
gcggatcagg cacaataatt ctgacataat gtggttctgg 360acaatgtcag tcgccggtga
tgtttggttt ggtttttcat ggctacttaa ccaactccca 420aagttcaatc cggtcaaaac
catacctgac cttactgccc taaggcagta ctgtgatctc 480gccgacggaa gctacagact
tcccggcatc gatgttttcg tcaccaccgc tgatccaatc 540gacgaaccgg ttctatacac
catgaattgt gttctctcta ttcttgcggc tgactaccct 600gttgataggt cagcctgcta
tctctctgat gatagtggag cattgatcct atatgaagca 660ttggttgaga cagccaaatt
tgctactcta tgggttccat tttgccggaa gcattgcatt 720gagcctagat ccccagagag
ctactttgag cttgaggcac catcgtatac tggaagtgca 780ccagaggagt tcaagaatga
ctctaggatt gtgcatcttg agtatgatga gttcaaggtg 840cgattggaag cacttcctga
gactattcgt aaacgatcag atgtttacaa tagtatgaaa 900actgatcaag gagcaccaaa
tgcgacttgg atggctaatg ggacccaatg gccaggcacg 960tggattgagc caatagaaaa
tcacaggaaa ggacaccatg ctggaattgt taaggttgtg 1020ttggaccatc ccatccgtgg
ccacaatctt agcctgaagg atagcacggg caacaatctt 1080aattttaatg ccactgatgt
gcgcatcccg atgcttgtct atgtgtctcg tggaaagaac 1140ccaaattatg atcataataa
gaaggcgggt gcattaaatg cgcaacttcg tgcctctgct 1200ctactctcca atgcacaatt
catcatcaac tttgattgtg atcactacat caacaattct 1260caagccttcc gtgcagcaat
ttgtttcatg cttgaccaaa gagaaggtga taatactgcc 1320tttgttcagt tcccacaacg
ctttgacaat gttgacccaa aagaccgata tggcaatcat 1380aatcgtgtat tctttgatgg
cacaatgctt gccctaaatg gtctccaagg accttcatac 1440cttggtactg gttgcatgtt
ccgtcgctta gctctctatg gtattgatcc tcctcattgg 1500agacaagaca acatcacacc
tgaagctagc aagtttggta actccatact cttattagag 1560tcagtgttag aagccctaaa
ccaagaccga tttgctacac catcaccggt caatgacata 1620tttgtcaatg agctggagat
ggttgtgtca gcttcattcg acaaagaaac cgattggggc 1680aagggtgttg gatacatata
tgacatagcc acagaagata tagtcacggg ttttcgcatc 1740catgggcaag gttggcgatc
catgtattgc accatggagc atgatgcatt ctgtggcact 1800gcacctataa atctaacaga
acgtcttcac caaattgtac gttggtctgg tggatcccta 1860gagatgttct tctcccacaa
taacccactt attggaggtc gtcggctcca acctctccag 1920cgtgtctcat acctcaatat
gacaatctac ccggtgacat cactctttat tttactctat 1980gctatcagcc ctgtgatgtg
gcttatcccc gatgaagtat atattcagag gccattcact 2040aggtatgtgg tgtaccttct
cgtgatcatt ttgatgattc atatgatcgg atggctcgag 2100ataaagtggg cagggatcac
atggttagat tattggcgca atgagcagtt cttcatgatc 2160ggctcaacga gtgcttaccc
aacagctgtg cttcatatgg tggtcaatct tctcacaaag 2220aagggtatac attttagagt
cacttcaaag caaacaaccg ctgacaccaa cgataaattt 2280gctgacttat atgagatgag
atgggttccc atgttaatcc caacaatggt agttttagtt 2340gctaatatcg gtgccattgg
tgtagctatt ggaaagacgg cagtatatat gggagtatgg 2400acgatagcac agaagagaca
tgctgcaatg ggactcctat tcaacatgtg ggttatgttt 2460ctcctttacc catttgcact
agcaatcatg gggagatggg caaagaggtc aatcattctt 2520gttgttttgt tgccaattat
ctttgtaatt gttgcccttg tatatgttgc tacccatatc 2580ttactagcaa acattattcc
attctag 260724868PRTOryza sativa
24Met Ala Ser Pro Ala Ser Val Ala Gly Gly Gly Glu Asp Ser Asn Gly1
5 10 15Cys Ser Ser Leu Ile Asp
Pro Leu Leu Val Ser Arg Thr Ser Ser Ile 20 25
30Gly Gly Ala Glu Arg Lys Ala Ala Gly Gly Gly Gly Gly
Gly Ala Lys 35 40 45Gly Lys His
Trp Ala Ala Ala Asp Lys Gly Glu Arg Arg Ala Ala Lys 50
55 60Glu Cys Gly Gly Glu Asp Gly Arg Arg Pro Leu Leu
Phe Arg Ser Tyr65 70 75
80Arg Val Lys Gly Ser Leu Leu His Pro Tyr Arg Ala Leu Ile Phe Ala
85 90 95Arg Leu Ile Ala Val Leu
Leu Phe Phe Gly Trp Arg Ile Arg His Asn 100
105 110Asn Ser Asp Ile Met Trp Phe Trp Thr Met Ser Val
Ala Gly Asp Val 115 120 125Trp Phe
Gly Phe Ser Trp Leu Leu Asn Gln Leu Pro Lys Phe Asn Pro 130
135 140Val Lys Thr Ile Pro Asp Leu Thr Ala Leu Arg
Gln Tyr Cys Asp Leu145 150 155
160Ala Asp Gly Ser Tyr Arg Leu Pro Gly Ile Asp Val Phe Val Thr Thr
165 170 175Ala Asp Pro Ile
Asp Glu Pro Val Leu Tyr Thr Met Asn Cys Val Leu 180
185 190Ser Ile Leu Ala Ala Asp Tyr Pro Val Asp Arg
Ser Ala Cys Tyr Leu 195 200 205Ser
Asp Asp Ser Gly Ala Leu Ile Leu Tyr Glu Ala Leu Val Glu Thr 210
215 220Ala Lys Phe Ala Thr Leu Trp Val Pro Phe
Cys Arg Lys His Cys Ile225 230 235
240Glu Pro Arg Ser Pro Glu Ser Tyr Phe Glu Leu Glu Ala Pro Ser
Tyr 245 250 255Thr Gly Ser
Ala Pro Glu Glu Phe Lys Asn Asp Ser Arg Ile Val His 260
265 270Leu Glu Tyr Asp Glu Phe Lys Val Arg Leu
Glu Ala Leu Pro Glu Thr 275 280
285Ile Arg Lys Arg Ser Asp Val Tyr Asn Ser Met Lys Thr Asp Gln Gly 290
295 300Ala Pro Asn Ala Thr Trp Met Ala
Asn Gly Thr Gln Trp Pro Gly Thr305 310
315 320Trp Ile Glu Pro Ile Glu Asn His Arg Lys Gly His
His Ala Gly Ile 325 330
335Val Lys Val Val Leu Asp His Pro Ile Arg Gly His Asn Leu Ser Leu
340 345 350Lys Asp Ser Thr Gly Asn
Asn Leu Asn Phe Asn Ala Thr Asp Val Arg 355 360
365Ile Pro Met Leu Val Tyr Val Ser Arg Gly Lys Asn Pro Asn
Tyr Asp 370 375 380His Asn Lys Lys Ala
Gly Ala Leu Asn Ala Gln Leu Arg Ala Ser Ala385 390
395 400Leu Leu Ser Asn Ala Gln Phe Ile Ile Asn
Phe Asp Cys Asp His Tyr 405 410
415Ile Asn Asn Ser Gln Ala Phe Arg Ala Ala Ile Cys Phe Met Leu Asp
420 425 430Gln Arg Glu Gly Asp
Asn Thr Ala Phe Val Gln Phe Pro Gln Arg Phe 435
440 445Asp Asn Val Asp Pro Lys Asp Arg Tyr Gly Asn His
Asn Arg Val Phe 450 455 460Phe Asp Gly
Thr Met Leu Ala Leu Asn Gly Leu Gln Gly Pro Ser Tyr465
470 475 480Leu Gly Thr Gly Cys Met Phe
Arg Arg Leu Ala Leu Tyr Gly Ile Asp 485
490 495Pro Pro His Trp Arg Gln Asp Asn Ile Thr Pro Glu
Ala Ser Lys Phe 500 505 510Gly
Asn Ser Ile Leu Leu Leu Glu Ser Val Leu Glu Ala Leu Asn Gln 515
520 525Asp Arg Phe Ala Thr Pro Ser Pro Val
Asn Asp Ile Phe Val Asn Glu 530 535
540Leu Glu Met Val Val Ser Ala Ser Phe Asp Lys Glu Thr Asp Trp Gly545
550 555 560Lys Gly Val Gly
Tyr Ile Tyr Asp Ile Ala Thr Glu Asp Ile Val Thr 565
570 575Gly Phe Arg Ile His Gly Gln Gly Trp Arg
Ser Met Tyr Cys Thr Met 580 585
590Glu His Asp Ala Phe Cys Gly Thr Ala Pro Ile Asn Leu Thr Glu Arg
595 600 605Leu His Gln Ile Val Arg Trp
Ser Gly Gly Ser Leu Glu Met Phe Phe 610 615
620Ser His Asn Asn Pro Leu Ile Gly Gly Arg Arg Leu Gln Pro Leu
Gln625 630 635 640Arg Val
Ser Tyr Leu Asn Met Thr Ile Tyr Pro Val Thr Ser Leu Phe
645 650 655Ile Leu Leu Tyr Ala Ile Ser
Pro Val Met Trp Leu Ile Pro Asp Glu 660 665
670Val Tyr Ile Gln Arg Pro Phe Thr Arg Tyr Val Val Tyr Leu
Leu Val 675 680 685Ile Ile Leu Met
Ile His Met Ile Gly Trp Leu Glu Ile Lys Trp Ala 690
695 700Gly Ile Thr Trp Leu Asp Tyr Trp Arg Asn Glu Gln
Phe Phe Met Ile705 710 715
720Gly Ser Thr Ser Ala Tyr Pro Thr Ala Val Leu His Met Val Val Asn
725 730 735Leu Leu Thr Lys Lys
Gly Ile His Phe Arg Val Thr Ser Lys Gln Thr 740
745 750Thr Ala Asp Thr Asn Asp Lys Phe Ala Asp Leu Tyr
Glu Met Arg Trp 755 760 765Val Pro
Met Leu Ile Pro Thr Met Val Val Leu Val Ala Asn Ile Gly 770
775 780Ala Ile Gly Val Ala Ile Gly Lys Thr Ala Val
Tyr Met Gly Val Trp785 790 795
800Thr Ile Ala Gln Lys Arg His Ala Ala Met Gly Leu Leu Phe Asn Met
805 810 815Trp Val Met Phe
Leu Leu Tyr Pro Phe Ala Leu Ala Ile Met Gly Arg 820
825 830Trp Ala Lys Arg Ser Ile Ile Leu Val Val Leu
Leu Pro Ile Ile Phe 835 840 845Val
Ile Val Ala Leu Val Tyr Val Ala Thr His Ile Leu Leu Ala Asn 850
855 860Ile Ile Pro Phe865252670DNAOryza sativa
25atgtccgcgg cggccgtgac tcgccggatc aacgcgggcg gcctccgcgt cgaggtcacc
60aacggcaatg gcgcggccgg cgtctacgtg gcggcggcgg cggcaccgtg ctcgccggcg
120gccaagcggg tgaacgacgg cggcggcaag gatgacgtgt gggtggccgt cgacgaggcg
180gacgtgtcgg ggcccagcgg cggcgatggc gttcggccga cgctgttccg gacgtacaag
240gtcaagggca gcatcctgca tccttacagg ttcttgatcc tagttcgact gatcgccatc
300gtcgccttct tcgcgtggcg cgtacgccac aagaaccgcg acggcgcgtg gctgtggaca
360atgtccatgg ccggcgacgt ctggttcggc ttctcgtggg cgctcaacca gctcccgaag
420ctgaacccca tcaagcgcgt cgcggacctc gccgccctcg ccgaccggca gcagcacggc
480acctccggcg gcggcgagct ccccggcgtc gacgtcttcg tcaccaccgt cgaccccgtc
540gacgagccga tcctctacac cgtgaactcc atcctctcca tcctcgccgc cgactacccg
600gtggacaggt acgcctgcta cctgtccgac gacggcggga cgctggtcca ctacgaggcc
660atggtggagg tcgccaagtt cgctgagctg tgggtgccct tctgccggaa gcactgcgtc
720gagccgaggg cgccggagag ctacttcgcg atgaagacgc aggcgtacag gggcggcgtc
780gccggcgagc tgatgagcga tcgccgccgc gtgcggcgag agtacgagga gttcaaggtc
840aggatcgact cgctgttcag caccattcgc aagcgatctg acgcgtacaa cagagcgaag
900gatggcaaag atgacggtga aaacgcgaca tggatggctg atgggacgca ttggcccggc
960acatggtttg agccggcgga gaatcaccgg aaagggcaac acgctgggat tgttcaggtt
1020ttactgaacc atcccaccag taagccacgg tttggagtgg cggcgagtgt tgacaacccg
1080ttggacttca gcggcgtgga cgtgcggctc cccatgctgg tgtacatctc gcgcgagaag
1140cgccccgggt acaaccacca gaagaaggcc ggcgccatga acgcgctgct ccgcgtgtcc
1200gcgctgctgt cgaacgcgcc cttcatcatc aacttcgact gcgaccacta cgtcaacaac
1260tcgcaggcgt tccgtgcgcc gatgtgcttc atgctcgacc ggcgcggcgg cggcgacgac
1320gtggcgttcg tccagttccc gcagcggttc gacgacgtcg acccgacgga ccggtacgcg
1380aaccacaacc gcgtcttctt cgacggcacc acgctctccc tcaacggcct ccagggcccc
1440tcctacctcg gcaccggcac catgttccgc cgcgccgcgc tctacggcct ggagccgccg
1500cggtgggggg cggcggggag ccagatcaag gccatggaca atgccaacaa gttcggcgcc
1560tcgtcgacgc tagtcagctc gatgctggac ggcgccaacc aagaacggtc gatcacgccg
1620cccgtggcga tcgacgggtc ggtcgcccgt gacctcgccg ccgtgacggc gtgcggctac
1680gacctcggga cgtcgtgggg gagagacgcc gggtgggtgt acgacatcgc gacggaggac
1740gtggcgaccg ggttccgcat gcaccagcag ggatggcgct ccgtgtacac ctccatggag
1800cccgccgcgt tccgcggcac ggcgccgatc aacctcaccg agcgcctcta ccagatccta
1860aggtggtcgg gcggctcgct cgagatgttc ttctcccaca gcaacgcgct cctcgccggc
1920cgccgcctcc acccgctgca gcgcatcgcc tacctcaaca tgtcgaccta cccgatcgtc
1980accgtgttca tcttcttcta caacctcttc ccggtgatgt ggctcatctc cgagcagtac
2040tacatccagc agccattcgg cgagtacctc ctctacctcg tcgccatcat cgccatgatc
2100cacgtgatcg gcatgttcga ggtgaagtgg tcgggcatca cggtgctgga ctggtgccgc
2160aacgagcagt tctacatgat cggctccacg ggggtgtacc cgacggcggt gctgtacatg
2220gcgctcaagc tcttcaccgg gaagggcatc cacttcaggc tcacgtcgaa gcagacgacg
2280gccagctccg gcgacaagtt cgccgacctg tacaccgtgc ggtgggtgcc tctgctgatc
2340ccgaccatcg tcgtcctggc cgtgaacgtc ggcgccgtcg gggtggcggt cggcaaggca
2400gcggcgtggg ggttgctcac cgagcagggg cggttcgcgg tgctcgggat ggtgttcaac
2460gtgtggatcc tggcgctcct ctacccgttc gcgctgggga tcatggggca gcgggggaag
2520cggccggcgg tgctgttcgt ggcgacggtg atggccgtcg ccgccgtggc gatcatgtac
2580gccgccttcg gtgcgccgta ccaagctggg ttgtcaggtg tcgcggcttc tctcggtaaa
2640gcggcgtcgc tgaccgggcc atctgggtag
267026889PRTOryza sativa 26Met Ser Ala Ala Ala Val Thr Arg Arg Ile Asn
Ala Gly Gly Leu Arg1 5 10
15Val Glu Val Thr Asn Gly Asn Gly Ala Ala Gly Val Tyr Val Ala Ala
20 25 30Ala Ala Ala Pro Cys Ser Pro
Ala Ala Lys Arg Val Asn Asp Gly Gly 35 40
45Gly Lys Asp Asp Val Trp Val Ala Val Asp Glu Ala Asp Val Ser
Gly 50 55 60Pro Ser Gly Gly Asp Gly
Val Arg Pro Thr Leu Phe Arg Thr Tyr Lys65 70
75 80Val Lys Gly Ser Ile Leu His Pro Tyr Arg Phe
Leu Ile Leu Val Arg 85 90
95Leu Ile Ala Ile Val Ala Phe Phe Ala Trp Arg Val Arg His Lys Asn
100 105 110Arg Asp Gly Ala Trp Leu
Trp Thr Met Ser Met Ala Gly Asp Val Trp 115 120
125Phe Gly Phe Ser Trp Ala Leu Asn Gln Leu Pro Lys Leu Asn
Pro Ile 130 135 140Lys Arg Val Ala Asp
Leu Ala Ala Leu Ala Asp Arg Gln Gln His Gly145 150
155 160Thr Ser Gly Gly Gly Glu Leu Pro Gly Val
Asp Val Phe Val Thr Thr 165 170
175Val Asp Pro Val Asp Glu Pro Ile Leu Tyr Thr Val Asn Ser Ile Leu
180 185 190Ser Ile Leu Ala Ala
Asp Tyr Pro Val Asp Arg Tyr Ala Cys Tyr Leu 195
200 205Ser Asp Asp Gly Gly Thr Leu Val His Tyr Glu Ala
Met Val Glu Val 210 215 220Ala Lys Phe
Ala Glu Leu Trp Val Pro Phe Cys Arg Lys His Cys Val225
230 235 240Glu Pro Arg Ala Pro Glu Ser
Tyr Phe Ala Met Lys Thr Gln Ala Tyr 245
250 255Arg Gly Gly Val Ala Gly Glu Leu Met Ser Asp Arg
Arg Arg Val Arg 260 265 270Arg
Glu Tyr Glu Glu Phe Lys Val Arg Ile Asp Ser Leu Phe Ser Thr 275
280 285Ile Arg Lys Arg Ser Asp Ala Tyr Asn
Arg Ala Lys Asp Gly Lys Asp 290 295
300Asp Gly Glu Asn Ala Thr Trp Met Ala Asp Gly Thr His Trp Pro Gly305
310 315 320Thr Trp Phe Glu
Pro Ala Glu Asn His Arg Lys Gly Gln His Ala Gly 325
330 335Ile Val Gln Val Leu Leu Asn His Pro Thr
Ser Lys Pro Arg Phe Gly 340 345
350Val Ala Ala Ser Val Asp Asn Pro Leu Asp Phe Ser Gly Val Asp Val
355 360 365Arg Leu Pro Met Leu Val Tyr
Ile Ser Arg Glu Lys Arg Pro Gly Tyr 370 375
380Asn His Gln Lys Lys Ala Gly Ala Met Asn Ala Leu Leu Arg Val
Ser385 390 395 400Ala Leu
Leu Ser Asn Ala Pro Phe Ile Ile Asn Phe Asp Cys Asp His
405 410 415Tyr Val Asn Asn Ser Gln Ala
Phe Arg Ala Pro Met Cys Phe Met Leu 420 425
430Asp Arg Arg Gly Gly Gly Asp Asp Val Ala Phe Val Gln Phe
Pro Gln 435 440 445Arg Phe Asp Asp
Val Asp Pro Thr Asp Arg Tyr Ala Asn His Asn Arg 450
455 460Val Phe Phe Asp Gly Thr Thr Leu Ser Leu Asn Gly
Leu Gln Gly Pro465 470 475
480Ser Tyr Leu Gly Thr Gly Thr Met Phe Arg Arg Ala Ala Leu Tyr Gly
485 490 495Leu Glu Pro Pro Arg
Trp Gly Ala Ala Gly Ser Gln Ile Lys Ala Met 500
505 510Asp Asn Ala Asn Lys Phe Gly Ala Ser Ser Thr Leu
Val Ser Ser Met 515 520 525Leu Asp
Gly Ala Asn Gln Glu Arg Ser Ile Thr Pro Pro Val Ala Ile 530
535 540Asp Gly Ser Val Ala Arg Asp Leu Ala Ala Val
Thr Ala Cys Gly Tyr545 550 555
560Asp Leu Gly Thr Ser Trp Gly Arg Asp Ala Gly Trp Val Tyr Asp Ile
565 570 575Ala Thr Glu Asp
Val Ala Thr Gly Phe Arg Met His Gln Gln Gly Trp 580
585 590Arg Ser Val Tyr Thr Ser Met Glu Pro Ala Ala
Phe Arg Gly Thr Ala 595 600 605Pro
Ile Asn Leu Thr Glu Arg Leu Tyr Gln Ile Leu Arg Trp Ser Gly 610
615 620Gly Ser Leu Glu Met Phe Phe Ser His Ser
Asn Ala Leu Leu Ala Gly625 630 635
640Arg Arg Leu His Pro Leu Gln Arg Ile Ala Tyr Leu Asn Met Ser
Thr 645 650 655Tyr Pro Ile
Val Thr Val Phe Ile Phe Phe Tyr Asn Leu Phe Pro Val 660
665 670Met Trp Leu Ile Ser Glu Gln Tyr Tyr Ile
Gln Gln Pro Phe Gly Glu 675 680
685Tyr Leu Leu Tyr Leu Val Ala Ile Ile Ala Met Ile His Val Ile Gly 690
695 700Met Phe Glu Val Lys Trp Ser Gly
Ile Thr Val Leu Asp Trp Cys Arg705 710
715 720Asn Glu Gln Phe Tyr Met Ile Gly Ser Thr Gly Val
Tyr Pro Thr Ala 725 730
735Val Leu Tyr Met Ala Leu Lys Leu Phe Thr Gly Lys Gly Ile His Phe
740 745 750Arg Leu Thr Ser Lys Gln
Thr Thr Ala Ser Ser Gly Asp Lys Phe Ala 755 760
765Asp Leu Tyr Thr Val Arg Trp Val Pro Leu Leu Ile Pro Thr
Ile Val 770 775 780Val Leu Ala Val Asn
Val Gly Ala Val Gly Val Ala Val Gly Lys Ala785 790
795 800Ala Ala Trp Gly Leu Leu Thr Glu Gln Gly
Arg Phe Ala Val Leu Gly 805 810
815Met Val Phe Asn Val Trp Ile Leu Ala Leu Leu Tyr Pro Phe Ala Leu
820 825 830Gly Ile Met Gly Gln
Arg Gly Lys Arg Pro Ala Val Leu Phe Val Ala 835
840 845Thr Val Met Ala Val Ala Ala Val Ala Ile Met Tyr
Ala Ala Phe Gly 850 855 860Ala Pro Tyr
Gln Ala Gly Leu Ser Gly Val Ala Ala Ser Leu Gly Lys865
870 875 880Ala Ala Ser Leu Thr Gly Pro
Ser Gly 885272859DNAOryza sativa 27atggcgccag cggtggccgg
cggcggaggg aggaggaaca atgagggggt gaacgggaac 60gcggcggcgc cggcgtgcgt
gtgcgggttc ccggtgtgcg cgtgcgcggg ggcggcggcg 120gtggcgtcgg cggcgtcgtc
ggcggacatg gacatcgtgg cggcggggca gatcggcgcc 180gtcaacgacg agagctgggt
cgccgtcgac ctcagcgaca gcgacgacgc ccccgccgcc 240ggcgacgtcc agggcgccct
cgacgaccgc cccgtcttcc gtaccgagaa gatcaagggc 300gtcctcctcc acccctaccg
ggtgctgatc tttgtgaggc tgatcgcgtt cacactgttc 360gtgatatggc gtatcgagca
caagaacccg gacgcgatgt ggctgtgggt gacgtcgatc 420gccggcgagt tctggttcgg
gttctcgtgg ctgctcgacc agctccccaa gctgaacccg 480atcaaccgcg tccccgacct
cgccgtcctc cgccgccgct tcgaccacgc cgacgggacc 540tcctccctcc cggggctgga
catcttcgtc accaccgccg acccgatcaa ggagcccatc 600ctgtcgacgg cgaactccat
cctctccatc ctcgccgccg actaccccgt cgaccgcaac 660acctgctacc tctccgacga
ctctgggatg ctcctcacct acgaggccat ggcggaggcg 720gccaagttcg cgacgctgtg
ggtgcccttc tgccggaagc acgccatcga gccgcgcggg 780cctgagagct acttcgagct
caagtcccac ccctacatgg ggagggcgca ggaggagttc 840gtcaacgacc gccgccgcgt
ccgcaaggag tacgacgact tcaaggccag gatcaacggc 900ctcgagcacg acatcaagca
gaggtccgac tcctacaacg ccgccgccgg cgtcaaggac 960ggcgagcccc gcgccacctg
gatggccgac gggtcgcagt gggagggcac ctggatcgag 1020cagtcggaga accaccgcaa
gggcgaccac gccggcatcg tcctggtgtt gctgaaccac 1080ccgagccacg cacggcagct
ggggccgccg gcgagcgccg acaacccgct ggacttcagc 1140ggcgtggacg tgcggctgcc
gatgctggtg tacgtcgcac gtgagaagcg ccccgggtgc 1200aaccaccaga agaaggccgg
cgccatgaac gcgctgaccc gcgcctccgc cgtgctctcc 1260aactccccct tcatcctcaa
cctcgactgc gaccactaca tcaacaactc ccaggcgctc 1320cgcgccggca tctgcttcat
gctcggccgc gacagcgaca ccgtcgcgtt cgtccagttc 1380ccgcagcgct tcgagggcgt
cgaccccacc gacctctatg ctaaccacaa ccgtatcttc 1440ttcgacggca cgctccgtgc
cctcgacggg ctgcaggggc ctatctacgt cggcaccggg 1500tgtctcttcc gccgcatcac
gctgtacggg ttcgagccgc cgaggatcaa cgtcggcgga 1560ccgtgcttcc cgaggctcgg
tgggatgttc gccaagaaca ggtaccagaa gcctgggttc 1620gagatgacca agcctggtgc
caagccggtg gcgccgccgc cggcggcgac ggtggcgaag 1680gggaagcacg ggttcctgcc
gatgcccaag aaggcgtacg gcaagtcgga cgcgttcgcc 1740gacaccatcc cgcgcgcgtc
gcacccgtcg ccgtacgcgg cggaggcggc ggtggcggcc 1800gacgaggcgg cgatcgcgga
ggccgtgatg gtgacggcgg cggcgtacga gaagaagacc 1860gggtggggga gcgacatcgg
gtgggtgtac ggcacggtga cggaggacgt ggtgaccggc 1920taccggatgc acatcaaggg
gtggaggtcg cgctactgct ccatctaccc gcacgcgttc 1980atcgggacgg cgccgatcaa
cctgacggag aggctgttcc aggtgctccg gtggtcgacg 2040ggttcgctgg agatcttctt
ctcgaggaac aacccgctgt tcgggagcac gttcctgcac 2100ccgctgcagc gcgtggcgta
catcaacatc accacctacc cgttcacggc gctgttcctc 2160atcttctaca ccaccgtgcc
ggcgctgtcg ttcgtgacgg ggcacttcat cgtgcagagg 2220ccgaccacca tgttctacgt
ctacctcgcc atcgtgctcg ggacgctgct catcctcgcc 2280gtgctggagg tgaagtgggc
gggggtcacc gtgttcgagt ggttcaggaa cgggcagttc 2340tggatgacgg ccagctgctc
cgcctacctc gccgccgtgc tgcaggtggt caccaaggtg 2400gtgttccggc gggacatctc
gttcaagctc acctccaagc tccccgccgg cgacgagaag 2460aaggacccct acgccgacct
gtacgtggtg cggtggacgt ggctcatgat cacccccatc 2520atcatcatcc tcgtcaacat
catcggctcc gccgtcgcct tcgccaaggt gctcgacggc 2580gagtggacgc actggctcaa
ggtcgccggc ggcgtgttct tcaacttctg ggtcctcttc 2640cacctctacc ccttcgccaa
gggcatcctc gggaagcacg gcaagacgcc ggtggtggtg 2700ctcgtctggt gggccttcac
cttcgtcatc accgccgtgc tctacatcaa catcccccac 2760atccatggcc ccggccgcca
cggcgccgcc tcaccatccc acggccacca cagcgcccat 2820ggcaccaaga agtacgactt
cacctacgcc tggccatga 285928952PRTOryza sativa
28Met Ala Pro Ala Val Ala Gly Gly Gly Gly Arg Arg Asn Asn Glu Gly1
5 10 15Val Asn Gly Asn Ala Ala
Ala Pro Ala Cys Val Cys Gly Phe Pro Val 20 25
30Cys Ala Cys Ala Gly Ala Ala Ala Val Ala Ser Ala Ala
Ser Ser Ala 35 40 45Asp Met Asp
Ile Val Ala Ala Gly Gln Ile Gly Ala Val Asn Asp Glu 50
55 60Ser Trp Val Ala Val Asp Leu Ser Asp Ser Asp Asp
Ala Pro Ala Ala65 70 75
80Gly Asp Val Gln Gly Ala Leu Asp Asp Arg Pro Val Phe Arg Thr Glu
85 90 95Lys Ile Lys Gly Val Leu
Leu His Pro Tyr Arg Val Leu Ile Phe Val 100
105 110Arg Leu Ile Ala Phe Thr Leu Phe Val Ile Trp Arg
Ile Glu His Lys 115 120 125Asn Pro
Asp Ala Met Trp Leu Trp Val Thr Ser Ile Ala Gly Glu Phe 130
135 140Trp Phe Gly Phe Ser Trp Leu Leu Asp Gln Leu
Pro Lys Leu Asn Pro145 150 155
160Ile Asn Arg Val Pro Asp Leu Ala Val Leu Arg Arg Arg Phe Asp His
165 170 175Ala Asp Gly Thr
Ser Ser Leu Pro Gly Leu Asp Ile Phe Val Thr Thr 180
185 190Ala Asp Pro Ile Lys Glu Pro Ile Leu Ser Thr
Ala Asn Ser Ile Leu 195 200 205Ser
Ile Leu Ala Ala Asp Tyr Pro Val Asp Arg Asn Thr Cys Tyr Leu 210
215 220Ser Asp Asp Ser Gly Met Leu Leu Thr Tyr
Glu Ala Met Ala Glu Ala225 230 235
240Ala Lys Phe Ala Thr Leu Trp Val Pro Phe Cys Arg Lys His Ala
Ile 245 250 255Glu Pro Arg
Gly Pro Glu Ser Tyr Phe Glu Leu Lys Ser His Pro Tyr 260
265 270Met Gly Arg Ala Gln Glu Glu Phe Val Asn
Asp Arg Arg Arg Val Arg 275 280
285Lys Glu Tyr Asp Asp Phe Lys Ala Arg Ile Asn Gly Leu Glu His Asp 290
295 300Ile Lys Gln Arg Ser Asp Ser Tyr
Asn Ala Ala Ala Gly Val Lys Asp305 310
315 320Gly Glu Pro Arg Ala Thr Trp Met Ala Asp Gly Ser
Gln Trp Glu Gly 325 330
335Thr Trp Ile Glu Gln Ser Glu Asn His Arg Lys Gly Asp His Ala Gly
340 345 350Ile Val Leu Val Leu Leu
Asn His Pro Ser His Ala Arg Gln Leu Gly 355 360
365Pro Pro Ala Ser Ala Asp Asn Pro Leu Asp Phe Ser Gly Val
Asp Val 370 375 380Arg Leu Pro Met Leu
Val Tyr Val Ala Arg Glu Lys Arg Pro Gly Cys385 390
395 400Asn His Gln Lys Lys Ala Gly Ala Met Asn
Ala Leu Thr Arg Ala Ser 405 410
415Ala Val Leu Ser Asn Ser Pro Phe Ile Leu Asn Leu Asp Cys Asp His
420 425 430Tyr Ile Asn Asn Ser
Gln Ala Leu Arg Ala Gly Ile Cys Phe Met Leu 435
440 445Gly Arg Asp Ser Asp Thr Val Ala Phe Val Gln Phe
Pro Gln Arg Phe 450 455 460Glu Gly Val
Asp Pro Thr Asp Leu Tyr Ala Asn His Asn Arg Ile Phe465
470 475 480Phe Asp Gly Thr Leu Arg Ala
Leu Asp Gly Leu Gln Gly Pro Ile Tyr 485
490 495Val Gly Thr Gly Cys Leu Phe Arg Arg Ile Thr Leu
Tyr Gly Phe Glu 500 505 510Pro
Pro Arg Ile Asn Val Gly Gly Pro Cys Phe Pro Arg Leu Gly Gly 515
520 525Met Phe Ala Lys Asn Arg Tyr Gln Lys
Pro Gly Phe Glu Met Thr Lys 530 535
540Pro Gly Ala Lys Pro Val Ala Pro Pro Pro Ala Ala Thr Val Ala Lys545
550 555 560Gly Lys His Gly
Phe Leu Pro Met Pro Lys Lys Ala Tyr Gly Lys Ser 565
570 575Asp Ala Phe Ala Asp Thr Ile Pro Arg Ala
Ser His Pro Ser Pro Tyr 580 585
590Ala Ala Glu Ala Ala Val Ala Ala Asp Glu Ala Ala Ile Ala Glu Ala
595 600 605Val Met Val Thr Ala Ala Ala
Tyr Glu Lys Lys Thr Gly Trp Gly Ser 610 615
620Asp Ile Gly Trp Val Tyr Gly Thr Val Thr Glu Asp Val Val Thr
Gly625 630 635 640Tyr Arg
Met His Ile Lys Gly Trp Arg Ser Arg Tyr Cys Ser Ile Tyr
645 650 655Pro His Ala Phe Ile Gly Thr
Ala Pro Ile Asn Leu Thr Glu Arg Leu 660 665
670Phe Gln Val Leu Arg Trp Ser Thr Gly Ser Leu Glu Ile Phe
Phe Ser 675 680 685Arg Asn Asn Pro
Leu Phe Gly Ser Thr Phe Leu His Pro Leu Gln Arg 690
695 700Val Ala Tyr Ile Asn Ile Thr Thr Tyr Pro Phe Thr
Ala Leu Phe Leu705 710 715
720Ile Phe Tyr Thr Thr Val Pro Ala Leu Ser Phe Val Thr Gly His Phe
725 730 735Ile Val Gln Arg Pro
Thr Thr Met Phe Tyr Val Tyr Leu Ala Ile Val 740
745 750Leu Gly Thr Leu Leu Ile Leu Ala Val Leu Glu Val
Lys Trp Ala Gly 755 760 765Val Thr
Val Phe Glu Trp Phe Arg Asn Gly Gln Phe Trp Met Thr Ala 770
775 780Ser Cys Ser Ala Tyr Leu Ala Ala Val Leu Gln
Val Val Thr Lys Val785 790 795
800Val Phe Arg Arg Asp Ile Ser Phe Lys Leu Thr Ser Lys Leu Pro Ala
805 810 815Gly Asp Glu Lys
Lys Asp Pro Tyr Ala Asp Leu Tyr Val Val Arg Trp 820
825 830Thr Trp Leu Met Ile Thr Pro Ile Ile Ile Ile
Leu Val Asn Ile Ile 835 840 845Gly
Ser Ala Val Ala Phe Ala Lys Val Leu Asp Gly Glu Trp Thr His 850
855 860Trp Leu Lys Val Ala Gly Gly Val Phe Phe
Asn Phe Trp Val Leu Phe865 870 875
880His Leu Tyr Pro Phe Ala Lys Gly Ile Leu Gly Lys His Gly Lys
Thr 885 890 895Pro Val Val
Val Leu Val Trp Trp Ala Phe Thr Phe Val Ile Thr Ala 900
905 910Val Leu Tyr Ile Asn Ile Pro His Ile His
Gly Pro Gly Arg His Gly 915 920
925Ala Ala Ser Pro Ser His Gly His His Ser Ala His Gly Thr Lys Lys 930
935 940Tyr Asp Phe Thr Tyr Ala Trp Pro945
950292493DNAOryza sativa 29atgccgccgt cagcaggctt
ggccactgag agcttgccgg cggcgacatg cccggccaag 60aaggatgcct atgccgcggc
ggcgtcgccg gagtccgaga cgaagctggc cgccggcgac 120gagagggcgc cgctcgtccg
gacgactcgc atctcgacaa ctaccatcaa gttatacagg 180ctcaccatct ttgttcgcat
cgccatcttc gtgctcttct tcaagtggag aatcacctac 240gctgctcgcg ccatcagctc
caccgacgcc ggcggcatcg gcatgagcaa ggcggcgaca 300ttttggacgg cgtccatcgc
cggcgagctc tggttcgcgt tcatgtgggt gctcgaccag 360ctgcccaaga cgatgcccgt
ccggcgcgcc gtcgacgtca cggcgctgaa cgacgacacg 420ctgctcccgg cgatggacgt
gttcgtcacc accgccgacc ccgacaagga gccgccgctc 480gccacggcga acaccgtgct
gtccatcctc gccgcgggct accccgccgg caaggtgacg 540tgctacgtct ccgacgacgc
cggcgcggag gtgacacgcg gggcggtcgt ggaggcggcc 600cggttcgcgg cactgtgggt
gcccttctgc cggaagcacg gcgtcgagcc gaggaacccg 660gaggcgtact tcaacggcgg
cgagggtggc ggtggtggcg gcaaggcgag ggtggtggcg 720agggggagct acaaggggag
ggcgtggccg gagctggtgc gcgacaggag gcgggtgcgc 780cgcgagtacg aggagatgcg
gctgcggatc gacgcgctgc aggccgccga cgcgcgccgc 840cggcgctgcg gcgcggccga
tgaccacgcc ggagttgtgc aggtactgat cgattctgct 900gggagcgcgc cacagctcgg
cgtcgcggac gggagcaagc tcatcgacct cgcctccgtc 960gacgtgcgcc tcccggcgct
tgtgtacgtg tgccgcgaga agcgccgcgg ccgcgcacac 1020caccggaagg ccggcgccat
gaacgcgctg ctgcgcgcat ccgccgtgct ctcgaacgcg 1080cccttcatcc tcaacctcga
ctgcgaccac tatgtcaaca actcgcaggc cctccgcgcc 1140ggcatttgct tcatgatcga
acgccgcggc ggcggcgccg aagacgccgg cgatgtcgcg 1200ttcgtccagt tcccgcagcg
gttcgacggc gtcgatcccg gcgaccgcta cgccaaccac 1260aaccgcgtct tcttcgactg
taccgagctt ggcctcgacg gcctccaggg ccccatctac 1320gtcggcaccg gctgcttgtt
ccgccgcgtc gcgctctacg gcgtcgaccc accgcgctgg 1380agatcgcccg gcggcggtgt
cgccgcggac cctgccaagt tcggcgagtc ggcgccgttc 1440ctagcctccg ttcgggcgga
gcagagtcac agtcgcgacg acggcgacgc cattgccgag 1500gcgagtgcgc tcgtgtcgtg
cgcgtacgag gacgggacgg cgtggggcag ggacgtcggc 1560tgggtgtacg gcaccgtgac
ggaggacgtg gccacgggct tctgcatgca ccggcgaggg 1620tggcgctccg cctactacgc
cgccgcgccc gacgcgttcc gcggcacggc gccgatcaac 1680ctcgccgacc gcctccacca
ggtgctccgc tgggcggcgg gctccctcga gatcttcttc 1740tcccgcaaca acgcactcct
cgccggcggc cgccgccgcc tccacccgct gcagcgcgcc 1800gcctacctca acacgacggt
gtacccgttc acgtcgctct tcctcatggc ctactgcctc 1860ttcccggcga tcccgctcat
cgccggcggc ggcggctgga acgccgcgcc gacaccgacg 1920tacgtcgcgt tcctggcggc
gctgatggtg acgctcgcgg cggtggccgt gctggagacg 1980aggtggtcgg ggatcgcgct
gggtgagtgg tggcggaacg agcagttctg gatggtgtcc 2040gcgacgagcg cgtacctcgc
cgcggtggcg caggtggcgc tcaaggtcgc gacggggaag 2100gaaatatcgt tcaagctgac
ctcgaagcat ctcgcgtcgt cggcgacgcc ggtcgccggt 2160aaggataggc agtacgcgga
gctgtacgcc gtgaggtgga cggcgctgat ggcgccgacg 2220gcggcggctc tggcagtgaa
cgtggcgtcg atggcggcgg cgggtggtgg tggccggtgg 2280tggtggtggg acgctccgtc
ggcggcggcg gcggcggcgg cggcactccc ggtggcgttc 2340aacgtgtggg tggtggtgca
tctctacccg ttcgcgctcg ggctgatggg tcgccggagc 2400aaggcggtgc gccccattct
gttcctgttc gccgtcgtcg cctacctcgc cgtccgcttc 2460ctttgtctct tgttacaatt
ccatacggct taa 249330830PRTOryza sativa
30Met Pro Pro Ser Ala Gly Leu Ala Thr Glu Ser Leu Pro Ala Ala Thr1
5 10 15Cys Pro Ala Lys Lys Asp
Ala Tyr Ala Ala Ala Ala Ser Pro Glu Ser 20 25
30Glu Thr Lys Leu Ala Ala Gly Asp Glu Arg Ala Pro Leu
Val Arg Thr 35 40 45Thr Arg Ile
Ser Thr Thr Thr Ile Lys Leu Tyr Arg Leu Thr Ile Phe 50
55 60Val Arg Ile Ala Ile Phe Val Leu Phe Phe Lys Trp
Arg Ile Thr Tyr65 70 75
80Ala Ala Arg Ala Ile Ser Ser Thr Asp Ala Gly Gly Ile Gly Met Ser
85 90 95Lys Ala Ala Thr Phe Trp
Thr Ala Ser Ile Ala Gly Glu Leu Trp Phe 100
105 110Ala Phe Met Trp Val Leu Asp Gln Leu Pro Lys Thr
Met Pro Val Arg 115 120 125Arg Ala
Val Asp Val Thr Ala Leu Asn Asp Asp Thr Leu Leu Pro Ala 130
135 140Met Asp Val Phe Val Thr Thr Ala Asp Pro Asp
Lys Glu Pro Pro Leu145 150 155
160Ala Thr Ala Asn Thr Val Leu Ser Ile Leu Ala Ala Gly Tyr Pro Ala
165 170 175Gly Lys Val Thr
Cys Tyr Val Ser Asp Asp Ala Gly Ala Glu Val Thr 180
185 190Arg Gly Ala Val Val Glu Ala Ala Arg Phe Ala
Ala Leu Trp Val Pro 195 200 205Phe
Cys Arg Lys His Gly Val Glu Pro Arg Asn Pro Glu Ala Tyr Phe 210
215 220Asn Gly Gly Glu Gly Gly Gly Gly Gly Gly
Lys Ala Arg Val Val Ala225 230 235
240Arg Gly Ser Tyr Lys Gly Arg Ala Trp Pro Glu Leu Val Arg Asp
Arg 245 250 255Arg Arg Val
Arg Arg Glu Tyr Glu Glu Met Arg Leu Arg Ile Asp Ala 260
265 270Leu Gln Ala Ala Asp Ala Arg Arg Arg Arg
Cys Gly Ala Ala Asp Asp 275 280
285His Ala Gly Val Val Gln Val Leu Ile Asp Ser Ala Gly Ser Ala Pro 290
295 300Gln Leu Gly Val Ala Asp Gly Ser
Lys Leu Ile Asp Leu Ala Ser Val305 310
315 320Asp Val Arg Leu Pro Ala Leu Val Tyr Val Cys Arg
Glu Lys Arg Arg 325 330
335Gly Arg Ala His His Arg Lys Ala Gly Ala Met Asn Ala Leu Leu Arg
340 345 350Ala Ser Ala Val Leu Ser
Asn Ala Pro Phe Ile Leu Asn Leu Asp Cys 355 360
365Asp His Tyr Val Asn Asn Ser Gln Ala Leu Arg Ala Gly Ile
Cys Phe 370 375 380Met Ile Glu Arg Arg
Gly Gly Gly Ala Glu Asp Ala Gly Asp Val Ala385 390
395 400Phe Val Gln Phe Pro Gln Arg Phe Asp Gly
Val Asp Pro Gly Asp Arg 405 410
415Tyr Ala Asn His Asn Arg Val Phe Phe Asp Cys Thr Glu Leu Gly Leu
420 425 430Asp Gly Leu Gln Gly
Pro Ile Tyr Val Gly Thr Gly Cys Leu Phe Arg 435
440 445Arg Val Ala Leu Tyr Gly Val Asp Pro Pro Arg Trp
Arg Ser Pro Gly 450 455 460Gly Gly Val
Ala Ala Asp Pro Ala Lys Phe Gly Glu Ser Ala Pro Phe465
470 475 480Leu Ala Ser Val Arg Ala Glu
Gln Ser His Ser Arg Asp Asp Gly Asp 485
490 495Ala Ile Ala Glu Ala Ser Ala Leu Val Ser Cys Ala
Tyr Glu Asp Gly 500 505 510Thr
Ala Trp Gly Arg Asp Val Gly Trp Val Tyr Gly Thr Val Thr Glu 515
520 525Asp Val Ala Thr Gly Phe Cys Met His
Arg Arg Gly Trp Arg Ser Ala 530 535
540Tyr Tyr Ala Ala Ala Pro Asp Ala Phe Arg Gly Thr Ala Pro Ile Asn545
550 555 560Leu Ala Asp Arg
Leu His Gln Val Leu Arg Trp Ala Ala Gly Ser Leu 565
570 575Glu Ile Phe Phe Ser Arg Asn Asn Ala Leu
Leu Ala Gly Gly Arg Arg 580 585
590Arg Leu His Pro Leu Gln Arg Ala Ala Tyr Leu Asn Thr Thr Val Tyr
595 600 605Pro Phe Thr Ser Leu Phe Leu
Met Ala Tyr Cys Leu Phe Pro Ala Ile 610 615
620Pro Leu Ile Ala Gly Gly Gly Gly Trp Asn Ala Ala Pro Thr Pro
Thr625 630 635 640Tyr Val
Ala Phe Leu Ala Ala Leu Met Val Thr Leu Ala Ala Val Ala
645 650 655Val Leu Glu Thr Arg Trp Ser
Gly Ile Ala Leu Gly Glu Trp Trp Arg 660 665
670Asn Glu Gln Phe Trp Met Val Ser Ala Thr Ser Ala Tyr Leu
Ala Ala 675 680 685Val Ala Gln Val
Ala Leu Lys Val Ala Thr Gly Lys Glu Ile Ser Phe 690
695 700Lys Leu Thr Ser Lys His Leu Ala Ser Ser Ala Thr
Pro Val Ala Gly705 710 715
720Lys Asp Arg Gln Tyr Ala Glu Leu Tyr Ala Val Arg Trp Thr Ala Leu
725 730 735Met Ala Pro Thr Ala
Ala Ala Leu Ala Val Asn Val Ala Ser Met Ala 740
745 750Ala Ala Gly Gly Gly Gly Arg Trp Trp Trp Trp Asp
Ala Pro Ser Ala 755 760 765Ala Ala
Ala Ala Ala Ala Ala Leu Pro Val Ala Phe Asn Val Trp Val 770
775 780Val Val His Leu Tyr Pro Phe Ala Leu Gly Leu
Met Gly Arg Arg Ser785 790 795
800Lys Ala Val Arg Pro Ile Leu Phe Leu Phe Ala Val Val Ala Tyr Leu
805 810 815Ala Val Arg Phe
Leu Cys Leu Leu Leu Gln Phe His Thr Ala 820
825 830312655DNAOryza sativa 31atggcgcttt cgccggcggc
ggctggccgt accggccgaa acaacaataa cgacgccggc 60ctcgcggacc ctctgctgcc
ggctggcggc ggcggcggcg gcggtaagga caagtactgg 120gtgcccgccg acgaggagga
agagatttgc cgcggcgagg acggcggccg cccgccggcg 180ccgccgctgc tgtaccggac
gttcaaggtc agcggcgtcc tcctccatcc ctacaggtta 240ctgaccttag tcaggctgat
cgccgtcgtc ctcttcctcg catggcgcct gaagcaccgg 300gactccgacg ccatgtggct
ctggtggatc tcgatcgccg gcgacttctg gttcggcgtc 360acctggctgc tcaaccaggc
ctccaagctc aaccccgtca agcgcgtccc cgacctctcc 420ctcctcaggc ggcgcttcga
cgacggcggc ctccccggca tcgacgtgtt catcaacacc 480gtcgaccccg tcgacgagcc
gatgctctac accatgaact ccatcctgtc catcctcgcc 540accgactacc ccgccgaccg
gcacgccgcc tacctctccg acgacggcgc gtcgctggcc 600cactacgagg ggctgatcga
gacggcgagg ttcgccgcgc tgtgggtccc gttctgccgg 660aagcaccgcg tcgagccgag
ggcgcccgag agctacttcg cggcgaaggc ggcgccgtac 720gccgggccgg cgctgccgga
ggagttcttc ggcgaccgca ggctcgtgcg ccgggagtac 780gaggagttca aggcgcggct
cgatgcgctg ttcactgaca ttccgcaacg atcggaagcg 840agtgttggca atgcaaacac
caaaggcgcg aaggccactc tgatggcaga tgggacccct 900tggccaggga catggaccga
gccagcagag aatcacaaaa aaggacagca cgccggaatc 960gttaaggtaa tgttgagcca
tccgggtgaa gagcctcagc tcggcatgcc ggcgagctcc 1020ggccaccctc tggacttcag
cgccgtcgac gtgcgcctcc cgatactggt ctacatcgcc 1080cgggagaagc ggccgggata
cgaccaccag aagaaggccg gcgccatgaa cgcgcagctg 1140cgcgtatccg cgctgctgtc
gaacgcgccc ttcatcttca acttcgacgg cgaccactac 1200atcaacaact cccaggcgtt
ccgcgccgcc ctgtgcttca tgctcgactg ccgccacggc 1260gacgacaccg cattcgtcca
gttcccgcag cgcttcgacg acgtcgaccc gaccgaccgg 1320tactgcaacc acaaccgcgt
cttcttcgac gccacgctgc tcggcctcaa cggcgtccag 1380ggcccgtcct acgtcggcac
cggctgcatg ttccgccgcg tcgcgctcta cggcgccgac 1440ccgccgcggt ggaggccgga
ggacgacgac gccaaggcgc tgggctgccc cggcaggtat 1500ggaaactcga tgccgttcat
caacacgata ccggcggcgg cgagccaaga acggtccatc 1560gcgtcgccgg cggcggcgtc
gctcgacgag acggcggcca tggcggaggt ggaggaggtg 1620atgacgtgcg cgtacgagga
cgggacggaa tggggcgacg gcgtcgggtg ggtgtacgac 1680atcgcgacgg aggacgtggt
gaccggcttc cggctgcacc ggaaggggtg gcggtccatg 1740tactgcgcca tggagcccga
cgcgttccgc ggcacggcgc cgatcaacct cacggagcgc 1800ctctaccaga tcctgcggtg
gtcgggcggc tcgctcgaga tgttcttctc ccgcaactgc 1860ccgctcctcg ccggctgccg
gctgcgcccc atgcagcgcg tcgcgtacgc caacatgacg 1920gcgtacccgg tctcggcgct
gttcatggtc gtctacgacc tcctcccggt gatctggctc 1980tcccaccacg gcgagttcca
catccagaag ccgttctcga cgtacgtcgc ctacctcgtc 2040gccgtcatcg ccatgatcga
ggtgatcggc ctggtcgaga tcaagtgggc ggggctcacc 2100ctgctcgact ggtggcgcaa
cgagcagttc tacatgatcg gcgccacggg ggtgtacctg 2160gcggcggtgc tgcacatcgt
gctcaagagg ctcctcggac tgaagggcgt gcggttcaag 2220ctgacggcga agcagctggc
cggcggcgcg agggagaggt tcgcggagct gtacgacgtg 2280cactggtcgc cgctgctggc
gccgacggtg gtggtgatgg cggtgaacgt gactgccatc 2340ggcgcggcgg cggggaaggc
ggtcgtcggg gggtggacgc cggcgcaggt cgccggcgcg 2400tcggcggggc tggtgttcaa
cgtgtgggtc ctggtgctgc tctacccgtt cgcgctcggg 2460atcatgggga ggtggagcaa
gaggccgtgc gcgctcttcg cgctgctcgt ggccgcgtgc 2520gcggcggtcg cggcggggtt
cgtcgccgtc catgccgtgc tcgccgccgg ctccgccgcg 2580ccgtcctggt tgggatggtc
tcgtggcgcc actgccattt tgccgtcaag ctggcgactt 2640aagcggggtt tctga
265532884PRTOryza sativa
32Met Ala Leu Ser Pro Ala Ala Ala Gly Arg Thr Gly Arg Asn Asn Asn1
5 10 15Asn Asp Ala Gly Leu Ala
Asp Pro Leu Leu Pro Ala Gly Gly Gly Gly 20 25
30Gly Gly Gly Lys Asp Lys Tyr Trp Val Pro Ala Asp Glu
Glu Glu Glu 35 40 45Ile Cys Arg
Gly Glu Asp Gly Gly Arg Pro Pro Ala Pro Pro Leu Leu 50
55 60Tyr Arg Thr Phe Lys Val Ser Gly Val Leu Leu His
Pro Tyr Arg Leu65 70 75
80Leu Thr Leu Val Arg Leu Ile Ala Val Val Leu Phe Leu Ala Trp Arg
85 90 95Leu Lys His Arg Asp Ser
Asp Ala Met Trp Leu Trp Trp Ile Ser Ile 100
105 110Ala Gly Asp Phe Trp Phe Gly Val Thr Trp Leu Leu
Asn Gln Ala Ser 115 120 125Lys Leu
Asn Pro Val Lys Arg Val Pro Asp Leu Ser Leu Leu Arg Arg 130
135 140Arg Phe Asp Asp Gly Gly Leu Pro Gly Ile Asp
Val Phe Ile Asn Thr145 150 155
160Val Asp Pro Val Asp Glu Pro Met Leu Tyr Thr Met Asn Ser Ile Leu
165 170 175Ser Ile Leu Ala
Thr Asp Tyr Pro Ala Asp Arg His Ala Ala Tyr Leu 180
185 190Ser Asp Asp Gly Ala Ser Leu Ala His Tyr Glu
Gly Leu Ile Glu Thr 195 200 205Ala
Arg Phe Ala Ala Leu Trp Val Pro Phe Cys Arg Lys His Arg Val 210
215 220Glu Pro Arg Ala Pro Glu Ser Tyr Phe Ala
Ala Lys Ala Ala Pro Tyr225 230 235
240Ala Gly Pro Ala Leu Pro Glu Glu Phe Phe Gly Asp Arg Arg Leu
Val 245 250 255Arg Arg Glu
Tyr Glu Glu Phe Lys Ala Arg Leu Asp Ala Leu Phe Thr 260
265 270Asp Ile Pro Gln Arg Ser Glu Ala Ser Val
Gly Asn Ala Asn Thr Lys 275 280
285Gly Ala Lys Ala Thr Leu Met Ala Asp Gly Thr Pro Trp Pro Gly Thr 290
295 300Trp Thr Glu Pro Ala Glu Asn His
Lys Lys Gly Gln His Ala Gly Ile305 310
315 320Val Lys Val Met Leu Ser His Pro Gly Glu Glu Pro
Gln Leu Gly Met 325 330
335Pro Ala Ser Ser Gly His Pro Leu Asp Phe Ser Ala Val Asp Val Arg
340 345 350Leu Pro Ile Leu Val Tyr
Ile Ala Arg Glu Lys Arg Pro Gly Tyr Asp 355 360
365His Gln Lys Lys Ala Gly Ala Met Asn Ala Gln Leu Arg Val
Ser Ala 370 375 380Leu Leu Ser Asn Ala
Pro Phe Ile Phe Asn Phe Asp Gly Asp His Tyr385 390
395 400Ile Asn Asn Ser Gln Ala Phe Arg Ala Ala
Leu Cys Phe Met Leu Asp 405 410
415Cys Arg His Gly Asp Asp Thr Ala Phe Val Gln Phe Pro Gln Arg Phe
420 425 430Asp Asp Val Asp Pro
Thr Asp Arg Tyr Cys Asn His Asn Arg Val Phe 435
440 445Phe Asp Ala Thr Leu Leu Gly Leu Asn Gly Val Gln
Gly Pro Ser Tyr 450 455 460Val Gly Thr
Gly Cys Met Phe Arg Arg Val Ala Leu Tyr Gly Ala Asp465
470 475 480Pro Pro Arg Trp Arg Pro Glu
Asp Asp Asp Ala Lys Ala Leu Gly Cys 485
490 495Pro Gly Arg Tyr Gly Asn Ser Met Pro Phe Ile Asn
Thr Ile Pro Ala 500 505 510Ala
Ala Ser Gln Glu Arg Ser Ile Ala Ser Pro Ala Ala Ala Ser Leu 515
520 525Asp Glu Thr Ala Ala Met Ala Glu Val
Glu Glu Val Met Thr Cys Ala 530 535
540Tyr Glu Asp Gly Thr Glu Trp Gly Asp Gly Val Gly Trp Val Tyr Asp545
550 555 560Ile Ala Thr Glu
Asp Val Val Thr Gly Phe Arg Leu His Arg Lys Gly 565
570 575Trp Arg Ser Met Tyr Cys Ala Met Glu Pro
Asp Ala Phe Arg Gly Thr 580 585
590Ala Pro Ile Asn Leu Thr Glu Arg Leu Tyr Gln Ile Leu Arg Trp Ser
595 600 605Gly Gly Ser Leu Glu Met Phe
Phe Ser Arg Asn Cys Pro Leu Leu Ala 610 615
620Gly Cys Arg Leu Arg Pro Met Gln Arg Val Ala Tyr Ala Asn Met
Thr625 630 635 640Ala Tyr
Pro Val Ser Ala Leu Phe Met Val Val Tyr Asp Leu Leu Pro
645 650 655Val Ile Trp Leu Ser His His
Gly Glu Phe His Ile Gln Lys Pro Phe 660 665
670Ser Thr Tyr Val Ala Tyr Leu Val Ala Val Ile Ala Met Ile
Glu Val 675 680 685Ile Gly Leu Val
Glu Ile Lys Trp Ala Gly Leu Thr Leu Leu Asp Trp 690
695 700Trp Arg Asn Glu Gln Phe Tyr Met Ile Gly Ala Thr
Gly Val Tyr Leu705 710 715
720Ala Ala Val Leu His Ile Val Leu Lys Arg Leu Leu Gly Leu Lys Gly
725 730 735Val Arg Phe Lys Leu
Thr Ala Lys Gln Leu Ala Gly Gly Ala Arg Glu 740
745 750Arg Phe Ala Glu Leu Tyr Asp Val His Trp Ser Pro
Leu Leu Ala Pro 755 760 765Thr Val
Val Val Met Ala Val Asn Val Thr Ala Ile Gly Ala Ala Ala 770
775 780Gly Lys Ala Val Val Gly Gly Trp Thr Pro Ala
Gln Val Ala Gly Ala785 790 795
800Ser Ala Gly Leu Val Phe Asn Val Trp Val Leu Val Leu Leu Tyr Pro
805 810 815Phe Ala Leu Gly
Ile Met Gly Arg Trp Ser Lys Arg Pro Cys Ala Leu 820
825 830Phe Ala Leu Leu Val Ala Ala Cys Ala Ala Val
Ala Ala Gly Phe Val 835 840 845Ala
Val His Ala Val Leu Ala Ala Gly Ser Ala Ala Pro Ser Trp Leu 850
855 860Gly Trp Ser Arg Gly Ala Thr Ala Ile Leu
Pro Ser Ser Trp Arg Leu865 870 875
880Lys Arg Gly Phe332661DNAOryza sativa 33atggcggcca acggcggcgg
cggcggcgcc ggtggctgca gcaatggtgg cggtggcggc 60gccgtgaacg gcgcggcggc
gaatggcggc ggcggcggag gaggcggcag taagggcgcg 120acgacgagga gggcgaaggt
cagcccgatg gacaggtact gggtgcccac cgacgagaag 180gagatggcgg cggcggtcgc
cgacggcggc gaggacggcc ggcggccgct gctgttccgg 240acgttcacgg tcaggggcat
cctcctccac ccctacaggt tattgacatt ggtcagattg 300gttgctatcg tcctattttt
catatggcgt atcaggcacc cgtacgccga tggcatgttt 360ttctggtgga tatctgtgat
tggagatttt tggtttggtg ttagttggct gctaaaccaa 420gtggcaaagc tgaaaccgat
caggcgtgtt cccgatctta acctcttaca acaacagttt 480gatcttcctg atggaaactc
caacctccct ggccttgatg tattcatcaa caccgtcgat 540cccataaatg agcctatgat
atacactatg aacgccattt tatccattct tgcagcagac 600tacccagttg acaagcatgc
ttgctatctt tcggatgatg gtggatcgat catccattat 660gatggtttac ttgagactgc
aaagtttgct gcgttatggg ttcccttttg ccgaaaacat 720tccattgagc ctagggcccc
tgagagctat tttgcagtga agtcacgtcc atacgctgga 780agtgcaccag aggattttct
cagtgaccac agatacatgc gtagggagta tgatgagttc 840aaggtacgct tagatgcgct
ttttactgtc attcccaaac ggtcagatgc atacaaccag 900gcacatgccg aagaaggtgt
gaaggcaacc tggatggcag atgggacaga gtggcctggt 960acatggattg atccatctga
gaaccataag aaaggaaatc acgctggaat tgttcaggtt 1020atgttgaacc atccgagcaa
tcaacctcaa cttggtctac cagcaagcac tgacagccct 1080gtggacttca gcaatgttga
tgtgcgcctt cctatgcttg tatacatagc ccgcgagaag 1140cgcccaggct atgaccacca
aaagaaggca ggtgccatga acgtgcagct gcgagtatct 1200gccctcctca ccaatgcacc
attcatcatc aactttgatg gtgaccacta tgtcaacaac 1260tcgaaggcct tccgtgctgg
tatatgtttc atgctcgatc gacgtgaagg tgacaacact 1320gcctttgtcc aattccccca
acgttttgat gacgttgacc caactgatag gtattgcaat 1380cacaaccgag tcttctttga
tgctactttg ctcggcctca atggtatcca gggcccctct 1440tatgttggca ccggttgcat
gttccgccga gtcgcactgt atggtgttga cccacctcgt 1500tggagacccg atgatggcaa
tattgtggat agctccaaaa agttcggcaa cttggactcc 1560ttcatcagct caatacctat
agcagcaaac caagaacgct caatcatatc accacctgcc 1620cttgaagagt ctatcctgca
ggagttgagt gatgccatgg catgtgcata tgaggatggg 1680actgactggg gcaaggatgt
tggttgggtt tacaatattg caaccgagga tgtggtgact 1740ggtttccgat tgcatcggac
agggtggcgc tcaatgtact gccgcatgga gcctgatgca 1800ttccgcggca ctgcaccaat
caacctcact gagcgcctct accagattct gcgctggtca 1860ggtggctccc ttgagatgtt
cttctcacat aactgcccac tccttgctgg ccgccgactc 1920aactttatgc aacgaattgc
ttacattaac atgacaggct acccagttac atcagtcttc 1980cttttgttct atctcctctt
ccctgtcata tggatctttc gcggcatatt ctacatacag 2040aagccatttc ctacatatgt
attgtacctt gtgatcgtca tatttatgtc agaaatgatc 2100ggtatggttg agatcaagtg
ggcggggcta acactactgg actggatccg caatgaacag 2160ttctacatta ttggagcaac
agctgtttac ccgcttgcag tcctgcacat agtgctgaag 2220tgttttggtt tgaagggtgt
ctcattcaag ctgacagcaa aacaagtagc aagcagcacc 2280agcgagaagt ttgcagaact
gtatgatgtt caatgggcac cattgttgtt cccgacgata 2340gtggtgatag cagtgaatat
ctgtgccatt ggcgcggcaa taggcaaggc tctctttgga 2400ggatggtcac tgatgcagat
gggagatgcg tcgctcgggc tggtattcaa cgtgtggatc 2460ctgctgctga tatatccatt
tgcactgggt atcatgggaa gatggagcaa gagaccctat 2520atcctgttcg tcttgattgt
gatttcattt gttataatcg cattggccga tattgccatc 2580caggcaatgc gttctggatc
cgttcggctc cactttagac ggtcaggtgg agccaacttc 2640cctacaagct gggggtttta g
266134886PRTOryza sativa
34Met Ala Ala Asn Gly Gly Gly Gly Gly Ala Gly Gly Cys Ser Asn Gly1
5 10 15Gly Gly Gly Gly Ala Val
Asn Gly Ala Ala Ala Asn Gly Gly Gly Gly 20 25
30Gly Gly Gly Gly Ser Lys Gly Ala Thr Thr Arg Arg Ala
Lys Val Ser 35 40 45Pro Met Asp
Arg Tyr Trp Val Pro Thr Asp Glu Lys Glu Met Ala Ala 50
55 60Ala Val Ala Asp Gly Gly Glu Asp Gly Arg Arg Pro
Leu Leu Phe Arg65 70 75
80Thr Phe Thr Val Arg Gly Ile Leu Leu His Pro Tyr Arg Leu Leu Thr
85 90 95Leu Val Arg Leu Val Ala
Ile Val Leu Phe Phe Ile Trp Arg Ile Arg 100
105 110His Pro Tyr Ala Asp Gly Met Phe Phe Trp Trp Ile
Ser Val Ile Gly 115 120 125Asp Phe
Trp Phe Gly Val Ser Trp Leu Leu Asn Gln Val Ala Lys Leu 130
135 140Lys Pro Ile Arg Arg Val Pro Asp Leu Asn Leu
Leu Gln Gln Gln Phe145 150 155
160Asp Leu Pro Asp Gly Asn Ser Asn Leu Pro Gly Leu Asp Val Phe Ile
165 170 175Asn Thr Val Asp
Pro Ile Asn Glu Pro Met Ile Tyr Thr Met Asn Ala 180
185 190Ile Leu Ser Ile Leu Ala Ala Asp Tyr Pro Val
Asp Lys His Ala Cys 195 200 205Tyr
Leu Ser Asp Asp Gly Gly Ser Ile Ile His Tyr Asp Gly Leu Leu 210
215 220Glu Thr Ala Lys Phe Ala Ala Leu Trp Val
Pro Phe Cys Arg Lys His225 230 235
240Ser Ile Glu Pro Arg Ala Pro Glu Ser Tyr Phe Ala Val Lys Ser
Arg 245 250 255Pro Tyr Ala
Gly Ser Ala Pro Glu Asp Phe Leu Ser Asp His Arg Tyr 260
265 270Met Arg Arg Glu Tyr Asp Glu Phe Lys Val
Arg Leu Asp Ala Leu Phe 275 280
285Thr Val Ile Pro Lys Arg Ser Asp Ala Tyr Asn Gln Ala His Ala Glu 290
295 300Glu Gly Val Lys Ala Thr Trp Met
Ala Asp Gly Thr Glu Trp Pro Gly305 310
315 320Thr Trp Ile Asp Pro Ser Glu Asn His Lys Lys Gly
Asn His Ala Gly 325 330
335Ile Val Gln Val Met Leu Asn His Pro Ser Asn Gln Pro Gln Leu Gly
340 345 350Leu Pro Ala Ser Thr Asp
Ser Pro Val Asp Phe Ser Asn Val Asp Val 355 360
365Arg Leu Pro Met Leu Val Tyr Ile Ala Arg Glu Lys Arg Pro
Gly Tyr 370 375 380Asp His Gln Lys Lys
Ala Gly Ala Met Asn Val Gln Leu Arg Val Ser385 390
395 400Ala Leu Leu Thr Asn Ala Pro Phe Ile Ile
Asn Phe Asp Gly Asp His 405 410
415Tyr Val Asn Asn Ser Lys Ala Phe Arg Ala Gly Ile Cys Phe Met Leu
420 425 430Asp Arg Arg Glu Gly
Asp Asn Thr Ala Phe Val Gln Phe Pro Gln Arg 435
440 445Phe Asp Asp Val Asp Pro Thr Asp Arg Tyr Cys Asn
His Asn Arg Val 450 455 460Phe Phe Asp
Ala Thr Leu Leu Gly Leu Asn Gly Ile Gln Gly Pro Ser465
470 475 480Tyr Val Gly Thr Gly Cys Met
Phe Arg Arg Val Ala Leu Tyr Gly Val 485
490 495Asp Pro Pro Arg Trp Arg Pro Asp Asp Gly Asn Ile
Val Asp Ser Ser 500 505 510Lys
Lys Phe Gly Asn Leu Asp Ser Phe Ile Ser Ser Ile Pro Ile Ala 515
520 525Ala Asn Gln Glu Arg Ser Ile Ile Ser
Pro Pro Ala Leu Glu Glu Ser 530 535
540Ile Leu Gln Glu Leu Ser Asp Ala Met Ala Cys Ala Tyr Glu Asp Gly545
550 555 560Thr Asp Trp Gly
Lys Asp Val Gly Trp Val Tyr Asn Ile Ala Thr Glu 565
570 575Asp Val Val Thr Gly Phe Arg Leu His Arg
Thr Gly Trp Arg Ser Met 580 585
590Tyr Cys Arg Met Glu Pro Asp Ala Phe Arg Gly Thr Ala Pro Ile Asn
595 600 605Leu Thr Glu Arg Leu Tyr Gln
Ile Leu Arg Trp Ser Gly Gly Ser Leu 610 615
620Glu Met Phe Phe Ser His Asn Cys Pro Leu Leu Ala Gly Arg Arg
Leu625 630 635 640Asn Phe
Met Gln Arg Ile Ala Tyr Ile Asn Met Thr Gly Tyr Pro Val
645 650 655Thr Ser Val Phe Leu Leu Phe
Tyr Leu Leu Phe Pro Val Ile Trp Ile 660 665
670Phe Arg Gly Ile Phe Tyr Ile Gln Lys Pro Phe Pro Thr Tyr
Val Leu 675 680 685Tyr Leu Val Ile
Val Ile Phe Met Ser Glu Met Ile Gly Met Val Glu 690
695 700Ile Lys Trp Ala Gly Leu Thr Leu Leu Asp Trp Ile
Arg Asn Glu Gln705 710 715
720Phe Tyr Ile Ile Gly Ala Thr Ala Val Tyr Pro Leu Ala Val Leu His
725 730 735Ile Val Leu Lys Cys
Phe Gly Leu Lys Gly Val Ser Phe Lys Leu Thr 740
745 750Ala Lys Gln Val Ala Ser Ser Thr Ser Glu Lys Phe
Ala Glu Leu Tyr 755 760 765Asp Val
Gln Trp Ala Pro Leu Leu Phe Pro Thr Ile Val Val Ile Ala 770
775 780Val Asn Ile Cys Ala Ile Gly Ala Ala Ile Gly
Lys Ala Leu Phe Gly785 790 795
800Gly Trp Ser Leu Met Gln Met Gly Asp Ala Ser Leu Gly Leu Val Phe
805 810 815Asn Val Trp Ile
Leu Leu Leu Ile Tyr Pro Phe Ala Leu Gly Ile Met 820
825 830Gly Arg Trp Ser Lys Arg Pro Tyr Ile Leu Phe
Val Leu Ile Val Ile 835 840 845Ser
Phe Val Ile Ile Ala Leu Ala Asp Ile Ala Ile Gln Ala Met Arg 850
855 860Ser Gly Ser Val Arg Leu His Phe Arg Arg
Ser Gly Gly Ala Asn Phe865 870 875
880Pro Thr Ser Trp Gly Phe
8853532DNAArtificialOligonucleotide Primer 35agtcagatct gttccgtgca
tggcggccac cg
323632DNAArtificialOligonucleotide Primer 36cagtacgcgt cgcgatcgaa
ctgtccctac cc
323729DNAArtificialOligonucleotide Primer 37agtcagatct atagagtgct
cgtcatggc
293832DNAArtificialOligonucleotide Primer 38cagtacgcgt tttatctatg
cacctagaat gg
323932DNAArtificialOligonucleotide Primer 39agtcaagctt gctacggcct
ccacgatgtc cg
324032DNAArtificialOligonucleotide Primer 40cagtactagt catgtcgtcc
ctacccagat gg
324132DNAArtificialOligonucleotide Primer 41agtcaagctt gcgacgatcg
atggcgcttt cg
324231DNAArtificialOligonucleotide Primer 42cagtactact tgcatcaatc
agaaaccccg c
314325DNAArtificialOligonucleotide Primer 43tggttgatct cgttgtgcag gtctc
254424DNAArtificialOligonucleotide Primer 44gtcagccaag tcaacaactc tctg
244520DNAArtificialOligonucleotide Primer 45atgtgggtga gggtatggaa
204625DNAArtificialOligonucleotide Primer 46ccgacaacct tcttagtnct cctct
254723DNAArtificialOligonucleotide Primer 47gagttcttca cgcgatacct cca
234827DNAArtificialOligonucleotide Primer 48gaccaccttt attaacccca tttacca
274923DNAArtificialOligonucleotide Primer 49tggcgaacgc tggtcctaat aca
235024DNAArtificialOligonucleotide Primer 50caaaaactcc tctgccccaa tcaa
245122DNAArtificialOligonucleotide Primer 51gtgcgcatac gaggatggga cg
225222DNAArtificialOligonucleotide Primer 52agaacatctc cagcgagccg cc
225322DNAArtificialOligonucleotide Primer 53ccgattgggg caagggtgtt gg
225422DNAArtificialOligonucleotide Primer 54gacacgctgg agaggttgga gc
225522DNAArtificialOligonucleotide Primer 55ctccgtgtac acctccatgg ag
225622DNAArtificialOligonucleotide Primer 56ctcggagatg agccacatca cc
225722DNAArtificialOligonucleotide Primer 57tacgacatcg cgacggagga cg
225822DNAArtificialOligonucleotide Primer 58gtcatgttgg cgtacgcgac gc
225923DNAArtificialoligonucleotide primer 59ggagagcgcg tgcattgagg acg
236020DNAArtificialoligonucleotide primer 60tgtccgggca aagtcatcaa
206122DNAArtificialoligonucleotide primer 61cggtcgggaa acctgggagt gg
226222DNAArtificialoligonucleotide primer 62gctcccagct tactacagaa cc
226322DNAArtificialoligonucleotide primer 63gtagctggct actgtgcata gc
226424DNAArtificialoligonucleotide primer 64gaacttacaa accccagctt gtgg
246520DNAArtificialoligonucleotide primer 65gtcgatcgac agatccggtc
206620DNAArtificialoligonucleotide primer 66gggagtttag cgagagcctg
206720DNAArtificialoligonucleotide primer 67tgggcattca ccttcgtcat
206820DNAArtificialoligonucleotide primer 68tgtccgggca aactcatcaa
206920DNAArtificialoligonucleotide primer 69ccgtcgggct cgtgtatgtc
207024DNAArtificialoligonucleotide primer 70ttgcagtgac tctggctgta cttg
247121DNAArtificialoligonucleotide primer 71gggattgttc ggttccactt t
217221DNAArtificialoligonucleotide primer 72gctgttgctt tgccacatct c
21
User Contributions:
Comment about this patent or add new information about this topic: