Patent application title: Inhibition of HIV-1 Replication by Disruption of the Processing of the Viral Capsid-Spacer Peptide 1 Protein
Inventors:
Karl Salzwedel (Olney, MD, US)
Feng Li (Gaithersburg, MD, US)
Carl T. Wild (Gaithersburg, MD, US)
Graham P. Allaway (Darnestown, MD, US)
Eric O. Freed (Frederick, MD, US)
Assignees:
Panacos Pharmaceuticals, Inc.
The Government of the USA as Represented by the Secretary, Department of Health & Human Services
IPC8 Class: AC12Q170FI
USPC Class:
435 5
Class name: Chemistry: molecular biology and microbiology measuring or testing process involving enzymes or micro-organisms; composition or test strip therefore; processes of forming such composition or test strip involving virus or bacteriophage
Publication date: 2008-09-25
Patent application number: 20080233559
Claims:
1. A method of determining if an individual is infected with HIV-1 that is
susceptible to inhibition by a compound that inhibits p25 (CA-SP1)
processing to p24 (CA), comprising taking a sample of blood from the
patient, and determining whether HIV-1 gag in said sample is susceptible
to inhibition of p25 (CA-SP1) processing to p24 (CA) by the compound.
2. The method of claim 1, which comprises determining whether said HIV-1 gag contains a mutation that results in a decrease in inhibition of processing by said compound.
3. The method of claim 2, which comprises genotyping said HIV-1 gag.
4. The method of claim 3, which comprises genotyping the region of the CA-SP1 cleavage site.
5. The method of claim 3, which comprises determining whether said HIV-1 gag encodes a substitution of Ala to Val at a position corresponding to residue 364 of HIV-1 Gag (residue 1 of SP1).
6. The method of claim 3, which comprises determining whether said HIV-1 gag encodes a substitution of Ala to Val at a position corresponding to residue 366 of HIV-1 Gag (residue 3 of SP1).
7. The method of claim 3, which comprises determining whether said HIV-1 gag encodes a substitution of H is to Tyr at a position corresponding to residue 358 of HIV-1 Gag.
8. The method of claim 3, which comprises determining whether said HIV-1 gag encodes a substitution of Leu to Phe at a position corresponding to residue 363 of HIV-1 Gag (the C-terminal residue of CA).
9. The method of claim 3, which comprises determining whether said HIV-1 gag encodes a substitution of Leu to Met at a position corresponding to residue 363 of HIV-1 Gag (the C-terminal residue of CA).
10. The method of claim 3, which comprises determining whether said HIV-1 gag encodes a deletion of the residue at a position corresponding to residue 370 of HIV-1 Gag (position 7 of SP1).
11. The method of claim 3, which comprises determining whether said HIV-1 gag encodes a substitution of Ala to Val at a position corresponding to residue 366 of HIV-1 Gag (position 3 of SP1) and a Ser residue at a position corresponding to residue 357 of HIV-1 Gag.
12. The method of claim 3, which comprises determining whether said HIV-1 gag encodes a substitution of Ala to Val at a position corresponding to residue 366 of HIV-1 Gag (position 3 of SP1) and a substitution of Gly to Ser at a position corresponding to residue 357 of HIV-1 Gag.
Description:
RELATED U.S. APPLICATION DATA
[0001]This application is a divisional of U.S. application Ser. No. 10/851,637, filed May 24, 2004, which is a continuation-in-part of U.S. application Ser. No. 10/706,528, filed Jan. 29, 2004, which claims the benefit of U.S. Provisional Application Nos. 60/496,660, filed Aug. 21, 2003, and 60/443,180, filed Jan. 29, 2003, all of which are entirely incorporated by reference herein.
BACKGROUND OF THE INVENTION
[0003]1. Field of the Invention
[0004]The invention includes methods of inhibiting, inhibitors and methods of discovery of inhibitors of HIV infection.
[0005]2. Background
[0006]Human Immunodeficiency Virus (HIV) is a member of the lentiviruses, a subfamily of retroviruses. The viral genome contains many regulatory elements which allow the virus to control its rate of replication in both resting and dividing cells. Most importantly, HIV infects and invades cells of the immune system; it breaks down the body's immune system and renders the patient susceptible to opportunistic infections and neoplasms. The immune defect appears to be progressive and irreversible, with a high mortality rate that approaches 100% over several years.
[0007]HIV-1 is trophic and cytopathic for T4 lymphocytes, cells of the immune system which express the cell surface differentiation antigen CD4, also known as OKT4, T4 and leu3. The viral tropism is due to the interactions between the viral envelope glycoprotein, gp120, and the cell-surface CD4 molecules (Dalgleish et al., Nature 312:763-767 (1984)). These interactions not only mediate the infection of susceptible cells by HIV, but are also responsible for the virus-induced fusion of infected and uninfected T cells. This cell fusion results in the formation of giant multinucleated syncytia, cell death, and progressive depletion of CD4 cells in HIV-infected patients. These events result in HIV-induced immunosuppression and its subsequent sequelae, opportunistic infections and neoplasms.
[0008]In addition to CD4.sup.+ T cells, the host range of HIV includes cells of the mononuclear phagocytic lineage (Dalgleish et al., supra), including blood monocytes, tissue macrophages, Langerhans cells of the skin and dendritic reticulum cells within lymph nodes. HIV is also neurotropic, capable of infecting monocytes and macrophages in the central nervous system causing severe neurologic damage. Macrophage and monocytes are major reservoirs of HIV. They can interact and fuse with CD4-bearing T cells, causing T cell depletion and thus contributing to the pathogenesis of AIDS.
[0009]Considerable progress has been made in the development of drugs for HIV-1 therapy. Therapeutic agents for HIV can include, but not are not limited to, at least one of AZT, 3TC, ddC, d4T, ddI, tenofovir, abacavir, nevirapine, delavirdine, emtricitabine, efavirenz, saquinavir, ritonavir, indinavir, nelfinavir, lopinavir, amprenavir, atazanavir and fosamprenavir, or any other antiretroviral drugs or antibodies in combination with each other, or associated with a biologically based therapeutic, such as, for example, gp41-derived peptides enfuvirtide (Fuzeon; Timeris-Roche) and T-1249 (Trimeris), or soluble CD4, antibodies to CD4, and conjugates of CD4 or anti-CD4, or as additionally presented herein. Combinations of these drugs are particularly effective and can reduce levels of viral RNA to undetectable levels in the plasma and slow the development of viral resistance, with resulting improvements in patient health and life span.
[0010]Despite these advances, there are still problems with the currently available drug regimens. Many of the drugs exhibit severe toxicities, have other side-effects (e.g., fat redistribution) or require complicated dosing schedules that reduce compliance and thereby limit efficacy. Resistant strains of HIV often appear over extended periods of time even on combination therapy. The high cost of these drugs is also a limitation to their widespread use, especially outside of developed countries.
[0011]There is still a major need for the development of additional drugs to circumvent these issues. Ideally these would target different stages in the viral life cycle, adding to the armamentarium for combination therapy, and exhibit minimal toxicity, yet have lower manufacturing costs.
[0012]HIV virion assembly takes place at the surface membrane of the infected cell where the viral Gag polyprotein accumulates, leading to the assembly of immature virions that bud from the cell surface. Within the virion, Gag is cleaved by the viral proteinase (PR) into the matrix (MA), capsid (CA), nucleocapsid (NC), and C-terminal p6 structural proteins (Wiegers K. et al., J. Virol. 72:2846-2854 (1998)). Gag processing induces a reorganization of the internal virion structure, a process termed "maturation." In mature HIV particles, MA lines the inner surface of the membrane, while CA forms the conical core which encases the genomic RNA that is complexed with NC. Cleavage and maturation are not required for particle formation but are essential for infectivity (Kohl, N. et al, Proc. Natl. Acad. Sci. USA 85:4686-4690, (1998)).
[0013]CA and NC as well as NC and p6 are separated on the Gag polyprotein by short spacer peptides of 14 and 10 amino acids (p2), respectively (spacer peptide 1 (SP1) and SP2, respectively) (Wiegers K. et al., J. Virol. 72:2846-2854 (1998), Pettit, S. C. et al., J. Virol. 68:8017-8027 (1994), Liang et al. J. Virol. 76:11729-11737 (2002)). These spacer peptides are released by PR-mediated cleavages at their N and C termini during particle maturation. The individual cleavage sites on the HIV Gag and Gag-Pol polyproteins are processed at different rates and this sequential processing results in Gag intermediates appearing transiently before the final products. Such intermediates may be important for virion morphogenesis or maturation but do not contribute to the structure of the mature viral particle (Weigers et al. and Pettit, et al., supra). The initial Gag cleavage event occurs at the C terminus of SP1 and separates an N-terminal MA-CA-SP1 intermediate from a C-terminal NC-SP2-p6 intermediate. Subsequent cleavages separating MA from CA-SP1 and NC-SP2 from p6 occur at an approximately 10-fold-lower rate. Cleavage of SP1 from the C terminus of CA is a late event and occurs at a 400-fold-lower rate than cleavage at the SP1-NC site (Weigers et al. and Pettit, et al., supra). The uncleaved CA-SP1 intermediate protein is alternatively termed "p25," whereas the cleaved CA protein is alternatively termed "p24" and the cleaved SP1 peptide is alternatively termed "p2".
[0014]Cleavage of SP1 from the C terminus of CA appears to be one of the last events in the Gag processing cascade and is required for final capsid condensation and formation of mature, infectious viral particles. Electron micrographs of mature virions reveal particles having electron dense conical cores. On the other hand, electron microscopy studies of viral particles defective for CA-SP1 cleavage show particles having a spherical electron-dense ribonucleoprotein core and a crescent-shaped, electron-dense layer located just inside the viral membrane (Weigers et al., supra). Mutations at or near the CA-SP1 cleavage site have been shown to inhibit Gag processing and disrupt the normal maturation process, thereby resulting in the production of non-infectious viral particles (Weigers et al., supra). Phenotypically, these particles exhibit a defect in Gag processing (which manifests itself in the presence of a p25 (CA-SP1) band in Western blot analysis) and the aberrant particle morphology described above which results from defective capsid condensation.
[0015]Previously, betulinic acid and platanic acid were isolated from Syzigium claviflorum and were determined to have anti-HIV activity. Betulinic acid and platanic acid exhibited inhibitory activity against HIV-1 replication in H9 lymphocyte cells with EC50 values of 1.4 μM and 6.5 μM, respectively, and therapeutic index (T.I.) values of 9.3 and 14, respectively. Hydrogenation of betulinic acid yielded dihydrobetulinic acid, which showed slightly more potent anti-HIV activity with an EC50 value of 0.9 and a T.I. value of 14 (Fujioka, T., et al., J. Nat. Prod. 57:243-247 (1994)). Esterification of betulinic acid with certain substituted acyl groups, such as 3',3'-dimethylglutaryl and 3',3'-dimethylsuccinyl groups produced derivatives having enhanced activity (Kashiwada, Y., et al., J. Med. Chem. 39:1016-1017 (1996)). Acylated betulinic acid and dihydrobetulinic acid derivatives that are potent anti-HIV agents are also described in U.S. Pat. No. 5,679,828. Anti-HIV assays indicated that 3-O-(3',3'-dimethylsuccinyl)-betulinic acid (DSB) and the dihydrobetulinic acid analog both demonstrated extremely potent anti-HIV activity in acutely infected H9 lymphocytes with EC50 values of less than 1.7×10-5 μM, respectively. These compounds exhibited remarkable T.I. values of more than 970,000 and more than 400,000, respectively.
[0016]U.S. Pat. No. 5,468,888 discloses 28-amido derivatives of lupanes that are described as having a cytoprotecting effect for HIV-infected cells.
[0017]Japanese Patent Application No. JP 01 143,832 discloses that betulin and 3,28-diesters thereof are useful in the anti-cancer field.
[0018]U.S. Pat. No. 6,172,110 discloses betulinic acid and dihydrobetulin derivatives which have the following formulae or pharmaceutically acceptable salts thereof,
Betulin and Dihydrobetulin Derivatives
[0019]wherein R1 is a C2-C20 substituted or unsubstituted carboxyacyl, R2 is a C2-C20 substituted or unsubstituted carboxyacyl; and R3 is hydrogen, halogen, amino, optionally substituted mono- or di-alkylamino, or --OR4, where R4 is hydrogen, C1-4 alkanoyl, benzoyl, or C2-C20 substituted or unsubstituted carboxyacyl; wherein the dashed line represents an optional double bond between C20 and C29.
[0020]U.S. Patent Application No. 60/413,451 discloses 3,3-dimethylsuccinyl betulin and is herein incorporated by reference. Zhu, Y-M. et al., Bioorg. Chem. Lett. 11:3115-3118 (2001); Kashiwada Y. et al., J. Nat. Prod. 61:1090-1095 (1998); Kashiwada Y. et al., J. Nat. Prod. 63:1619-1622 (2000); and Kashiwada Y. et al., Chem. Pharm. Bull. 48:1387-1390 (2000) disclose dimethylsuccinyl betulinic acid and dimethylsuccinyl oleanolic acid. Esterification of the 3' carbon of betulin with succinic acid produced a compound capable of inhibiting HIV-1 activity (Pokrovskii, A. G. et al., Gos. Nauchnyi Tsentr Virusol. Biotekhnol. "Vector," 9:485-491 (2001)).
[0021]Published International Application No. WO 02/26761 discloses the use of betulin and analogs thereof for treating fungal infections.
[0022]There exists a need for new HIV inhibition methods that are effective against drug resistant strains of the virus. The strategy of this invention is to provide therapeutic methods and compounds that inhibit the virus in different ways from approved therapies.
[0023]The compound and methods of the present invention have a novel mechanism of action and therefore are active against HIV strains that are resistant to current reverse transcriptase and protease inhibitors. As such, this invention offers a completely new approach for treating HIV/AIDS.
BRIEF SUMMARY OF THE INVENTION
[0024]Generally, the invention provides methods of inhibiting, inhibitory compounds and methods of identifying inhibitory compounds that target proteolytic processing of the HIV-1 Gag protein. In one embodiment, such compounds may directly or indirectly inhibit the interaction of a protease enzyme with HIV-1 Gag protein. In another embodiment, such inhibition of interaction occurs via the binding of a compound to Gag. The inhibition of protease cleavage of the CA-SP1 protein of HIV-1 Gag by 3-O-(3',3'-dimethylsuccinyl)betulinic acid (DSB) is one example, but other proteolytic cleavage sites can be targeted by a similar approach using inhibitory compounds that interact with the substrate in a manner similar to that in which DSB interacts with Gag.
[0025]Another aspect of the invention is directed to a method of inhibiting the processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), but having no effect on other Gag processing steps.
[0026]A further aspect of the invention is directed to a method for identifying compounds that inhibit processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), but have no effect on other Gag processing steps.
[0027]In one aspect, the invention is drawn to a compound or pharmaceutical composition identified by the method for identifying compounds that inhibit HIV-1 replication disclosed herein.
[0028]In another aspect, the present invention is directed to a polynucleotide comprising a sequence which encodes an amino acid sequence containing a mutation in the Gag p25 protein, said mutation resulting in a decrease in the inhibition of processing of p25 to p24 by 3-O-(3',3'-dimethylsuccinyl)betulinic acid. This aspect of the invention is also directed to a vector, virus and host cell comprising said polynucleotide, and a method of making said protein.
[0029]A further aspect of the present invention is directed to an amino acid sequence containing a mutation in the Gag p25 protein, said mutation resulting in a decrease in the inhibition of processing of p25 to p24 by 3-O-(3',3'-dimethylsuccinyl)betulinic acid.
[0030]An additional aspect of the invention is directed to an antibody which selectively binds an amino acid sequence containing a mutation in the Gag p25 protein, said mutation resulting in a decrease in the inhibition of processing of p25 to p24 by 3-O-(3',3'-dimethylsuccinyl)betulinic acid. Also included in this aspect of the invention are a method of making said antibody, a hybridoma producing said antibody and a method of making said hybridoma.
[0031]In a further embodiment, the invention is directed to a kit comprising a polynucleotide, polypeptide or antibody disclosed herein.
[0032]The invention further relates to a method of inhibiting HIV-1 infection in cells of an animal by contacting said cells with a compound that blocks the maturation of virus particles released from treated infected cells. In one embodiment, the released virus particles exhibit non-condensed cores and a distinctive thin electron-dense layer near the viral membrane and have reduced infectivity. A method is included of contacting animal cells with a compound that both inhibits processing of the viral Gag p25 protein and that disrupts the maturation of virus particles. Also, included is a method of treating HIV-infected cells, wherein the HIV infecting said cells does not respond to other HIV therapies.
[0033]This invention further includes a method for identifying compounds that inhibit processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), but have no significant effect on other Gag processing steps. The method involves contacting HIV-1 infected cells with a test compound, and thereafter analyzing virus particles that are released to detect the presence of p25. Methods to detect p25 include western blotting of viral proteins and detecting using an antibody to p25, gel electrophoresis, and imaging of metabolically labeled proteins. Methods to detect p25 also include immunoassays using an antibody to p25 or SP1 (p2) or to an epitope tag inserted into the SP1 sequence.
[0034]The invention is further directed to a method for identifying compounds involving contacting HIV-1 infected cells with a compound, and thereafter analyzing virus particles released by the contacted cells, by thin-sectioning and transmission electron microscopy, and determining whether viral particles with non-condensed cores and a distinctive thin electron-dense layer near the viral membrane are present.
[0035]The invention is also directed to compounds identified by the aforementioned screening methods. In additional embodiments, the invention is drawn to a method of treating HIV-1 infection in a patient by administering a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), but does not significantly affect other Gag processing steps. In related embodiments, such inhibition may be accompanied by a different observable phenotypes. For example, inhibition may not necessarily significantly reduce the quantity of virions released from treated infected cells; and/or said inhibition may have little or no significant effect on the amount of RNA incorporation into the released virions; and/or said inhibition disrupts the maturation of virions released from infected cells treated with said compound. In related embodiments, the virion structure may be affected, and a majority of virions released from treated infected cells exhibit spherical, electron-dense cores that are acentric with respect to the viral particle; and/or possess crescent-shaped electron-dense layers lying just inside the viral membrane; and/or and have reduced or no infectivity.
[0036]In additional embodiments, the invention is drawn to a method of treating HIV-1 infection in a patient by administering a compound that inhibits the interaction of HIV protease with CA-SP1, which results in the inhibition of the processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), but has no significant effect on other Gag processing steps. Such inhibition may be direct or, alternatively, indirect; and/or may involve said compound binding to the viral Gag protein such that interaction of HIV protease with CA-SP1 is inhibited. The invention is also drawn to a method of treating HIV in a patient with a compound that binds at or near the site of cleavage of the viral Gag p25 protein (CA-SP1) to p24 (CA), thereby inhibiting the interaction of HIV protease with the CA-SP1 cleavage site and resulting in the inhibition of processing of p25 to p24.
[0037]In other embodiments, the invention is drawn to a method of treating HIV-1-infection in a patient by administering a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), wherein said compound binds to a polypeptide with an amino acid sequence having at least about 40%, 50%, 60%, 70%, 80%, 90% identity, or which is identical to a sequence selected from the group consisting of:
TABLE-US-00001 (SEQ ID NO: 21) (a) KNWMTETFLVQNANPDCKTILKALGPAATLEEMMTACQGVGGPHKA RILAEAMSQVTNSATIM; (SEQ ID NO: 22) (b) KNWMTETLLVQNANPDCKTILKALGPGATLEEMMTACQGVGGPGHK ARVLAEAMSQVTNPATIM; (SEQ ID NO: 23) (c) TACQGVGGPSHKARILAEAMSQVTNSATIM; (SEQ ID NO: 24) (d) MTACQGVGGPGHKARVLAEAMSQVTKPATIM; (SEQ ID NO: 25) (e) SHKARILAEAMSQV and (SEQ ID NO: 26) (f) GHKARVLAEAMSQV.
[0038]In other embodiments, the invention is drawn to a method of treating HIV-1-infection in a patient by administering a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), wherein said compound binds to a polypeptide encoded by a polynucleotide sequence having at least about 40%, 50%, 60%, 70%, 80%, 90% identity, or which is identical to a polynucleotide selected from the group consisting of: (a) about nucleotides 1243-1435 of; (b) about nucleotides 1729-1920 of SEQ ID NO: 19; (c) about nucleotides 1344-1435 of SEQ ID NO: 18; (d) about nucleotides 1828-1920 of SEQ ID NO: 19; (e) about nucleotides 1370-1413 of SEQ ID NO: 18; and (f) about nucleotides 1857-1899 of SEQ ID NO: 19.
[0039]In another aspect, the invention is drawn to a method of inhibiting processing of the viral Gag p25 protein (CA-SP1) by administration of a compound. In related embodiments, such a compound binds to a polypeptide with an amino acid sequence having at least about 40%, 50%, 60%, 70%, 80%, 90% identity, or which is identical to a sequence selected from the group consisting of:
TABLE-US-00002 (SEQ ID NO: 21) (a) KNWMTETFLVQNANPDCKTILKALGPAATLEEMMTACQGVGGPHKA RILAEAMSQVTNSATIM; (SEQ ID NO: 22) (b) KNWMTETLLVQNANPDCKTILKALGPGATLEEMMTACQGVGGPGHK ARVLAEAMSQVTNPATIM; (SEQ ID NO: 23) (c) TACQGVGGPSHKARILAEAMSQVTNSATIM; (SEQ ID NO: 24) (d) MTACQGVGGPGHKARVLAEAMSQVTKPATIM; (SEQ ID NO: 25) (e) SHKARILAEAMSQV; and (SEQ ID NO: 26) (f) GHKARVLAEAMSQV.
[0040]In related embodiments; the invention is drawns to a method of inhibiting processing of the viral Gag p25 protein (CA-SP1) by administration of a compound wherein said compound binds to a polypeptide encoded by a polynucleotide sequence having at least about 40%, 50%, 60%, 70%, 80%, 90% identity, or which is identical to a polynucleotide selected from the group consisting of:
[0041](a) about nucleotides 1243-1435 of SEQ ID NO: 18; (b) about nucleotides 1729-1920 of SEQ ID NO: 19; (c) about nucleotides 1344-1435 of SEQ ID NO: 18; (d) about nucleotides 1828-1920 of SEQ ID NO: 19; (e) about nucleotides 1370-1413 of SEQ ID NO: 18; and (f) about nucleotides 1857-1899 of SEQ ID NO: 19.
[0042]The invention may be useful in the treatment of HIV in patients who are not adequately treated by other HIV-1 therapies. Accordingly, the invention is also drawn to a method of treating a patient in need of therapy, wherein the HIV-1 infecting said cells does not respond to other HIV-1 therapies. In another embodiment, methods of the invention are practiced on a subject infected with an HIV that is resistant to a drug used to treat HIV infection. In one application, the HIV is resistant to a protease inhibitor, a polymerase inhibitor, a nucleoside analog, a vaccine, a binding inhibitor, an immunomodulator, or any other inhibitor. In another embodiment, methods of the invention are practiced on a subject infected with an HIV that is resistant to a drug used to treat HIV infection is selected from the group consisting of zidovudine, lamivudine, didanosine, zalcitabine, stavudine, abacavir, nevirapine, delavirdine, emtricitabine, efavirenz, saquinavir, ritonavir, indinavir, nelfinavir, tenofovir, amprenavir, adefovir, atazanavir, fosamprenavir, hydroxyurea, AL-721, ampligen, butylated hydroxytoluene; polymannoacetate, castanospermine; contracan; creme pharmatex, CS-87, penciclovir, famciclovir, acyclovir, cytofovir, ganciclovir, dextran sulfate, D-penicillamine trisodium phosphonoformate, fusidic acid, HPA-23, eflornithine, nonoxynol, pentamidine isethionate, peptide T, phenyloin, isoniazid, ribavirin, rifabutin, ansamycin, trimetrexate, SK-818, suramin, UA001, and combinations thereof.
[0043]Compounds of the invention are also useful as part of combination of therapies. Accordingly, in one aspect the invention is drawn to a method of treating HIV in a patient, wherein said patient is administered said compound in combination with at least one anti-viral agent. Anti-viral agents suitable include, but are not limited to: zidovudine, lamivudine, didanosine, zalcitabine, stavudine, abacavir, nevirapine, delavirdine, emtricitabine, efavirenz, saquinavir, ritonavir, indinavir, nelfinavir, amprenavir, tenofovir, adefovir, atazanavir, fosamprenavir, hydroxyurea, AL-721, ampligen, butylated hydroxytoluene; polymannoacetate, castanospermine; contracan; creme pharmatex, CS-87, penciclovir, famciclovir, acyclovir, cytofovir, ganciclovir, dextran sulfate, D-penicillamine trisodium phosphonoformate, fusidic acid, HPA-23, eflornithine, nonoxynol, pentamidine isethionate, peptide T, phenyloin, isoniazid, ribavirin, rifabutin, ansamycin, trimetrexate, SK-818, suramin, UA001, enfuvirtide, gp41-derived peptides, antibodies to CD4, soluble CD4, CD4-containing molecules, CD4-IgG2, and combinations thereof. In another embodiment, the patient is administered said compound in combination with an immunomodulating agent, anticancer agent, antibacterial agent, antifungal agent, or a combination thereof.
[0044]The invention is also directed to compounds. Such compounds are useful in a method of treating patients infected with HIV; in a method for inhibiting processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), or in a method for treating human blood and human blood products. Such compounds useful in the present invention include, but are not limited to derivatives of dimethylsuccinyl betulinic acid or dimethylsuccinyl betulin, or is selected from the group consisting of 3-O-(3',3'-dimethylsuccinyl)betulinic acid, 3-O-(3',3'-dimethylsuccinyl)betulin, 3-O-(3',3'-dimethylglutaryl)betulin, 3-O-(3',3'-dimethylsuccinyl)dihydrobetulinic acid; 3-O-(3',3'-dimethylglutaryl)betulinic acid, (3',3'-dimethylglutaryl)dihydrobetulinic acid, 3-O-diglycolyl-betulinic acid, 3-O-diglycolyl-dihydrobetulinic acid and combinations thereof.
[0045]Compounds of the invention may be used alone, or administered with additional compounds, including zidovudine, lamivudine, didanosine, zalcitabine, stavudine, abacavir, nevirapine, delavirdine, emtricitabine, efavirenz, saquinavir, ritonavir, indinavir, nelfinavir, amprenavir, tenofovir, adefovir, atazanavir, fosamprenavir, hydroxyurea, AL-721, ampligen, butylated hydroxytoluene; polymannoacetate, castanospermine; contracan; creme pharmatex, CS-87, penciclovir, famciclovir, acyclovir, cytofovir, ganciclovir, dextran sulfate, D-penicillamine trisodium phosphonoformate, fusidic acid, HPA-23, eflornithine, nonoxynol, pentamidine isethionate, peptide T, phenyloin, isoniazid, ribavirin, rifabutin, ansamycin, trimetrexate, SK-818, suramin, UA001, enfuvirtide, gp41-derived peptides, antibodies to CD4, soluble CD4, CD4-containing molecules, CD4-IgG2, and combinations thereof; an antiviral, an immunomodulating agent, anti-cancer agent, antibacterial agent, an anti-fungal agent, or combinations thereof.
[0046]In further embodiments, the invention is directed to a method of treating human blood products comprising contacting said blood products with a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA). In one aspect, said compound does not significantly affect other Gag processing steps. In related embodiments of this method, said inhibition does not significantly reduce the quantity of virions released from treated infected cell; and/or has little or no significant effect on the amount of RNA incorporation into the released virions; and/or inhibits the maturation of virions released from infected cells treated with said compound; and/or affects viral morphology. Such effects on viral morphology include, but are not limited to: the virions released from treated infected cells to exhibit spherical, electron-dense cores that are acentric with respect to the viral particle; and/or possess crescent-shaped electron-dense layers lying just inside the viral membrane; and/or and have reduced or no infectivity. In related embodiments, the method involves the administration of the compound which inhibits the interaction of HIV protease with CA-SP1, which results in the inhibition of the processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) but has no significant effect on other Gag processing steps. This may be via direct, or indirect inhibition of the interaction of HIV protease with CA-SP 1; and/or may involve said compound binds to the viral Gag protein such that interaction of HIV protease with CA-SP1 is inhibited; and/or said compound binds at or near the site of cleavage of the viral Gag p25 protein (CA-SP1) to p24 (CA), thereby inhibiting the interaction of HIV protease with the CA-SP1 cleavage site and resulting in the inhibition of processing of p25 to p24.
[0047]In a further embodiment, the invention is drawn to a method of treating human blood products comprising contacting said blood products with a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), wherein said compound binds to a polypeptide with an amino acid sequence having at least about 40%, 50%, 60%, 70%, 80%, 90% identity, or which is identical to a sequence selected from the group consisting of:
TABLE-US-00003 (SEQ ID NO: 21) (a) KNWMTETFLVQNANPDCKTILKALGPAATLEEMMTACQGVGGPHKA RILAEAMSQVTNSATIM; (SEQ ID NO: 22) (b) KNWMTETLLVQNANPDCKTILKALGPGATLEEMMTACQGVGGPGHK ARVLAEAMSQVTNPATIM; (SEQ ID NO: 23) (c) TACQGVGGPSHKARILAEAMSQVTNSATIM; (SEQ ID NO: 24) (d) MTACQGVGGPGHKARVLAEAMSQVTKPATIM; (SEQ ID NO: 25) (e) SHKARILAEAMSQV; and (SEQ ID NO: 26) (f) GHKARVLAEAMSQV.
[0048]In a related embodiment, the invention is drawn to a method of treating human blood products comprising contacting said blood products with a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), wherein said compound binds to a polypeptide encoded by a polynucleotide sequence having at least about 40%, 50%, 60%, 70%, 80%, 90% identity, or which is identical a polynucleotide selected from the group consisting of:
[0049](a) about nucleotides 1243-1435 of SEQ ID NO: 18; (b) about nucleotides 1729-1920 of SEQ ID NO: 19; (c) about nucleotides 1344-1435 of SEQ ID NO: 18; (d) about nucleotides 1828-1920 of SEQ ID NO: 19; (e) about nucleotides 1370-1413 of SEQ ID NO: 18; and (f) about nucleotides 1857-1899 of SEQ ID NO: 19.
[0050]The invention also embodies methods for identifying compounds that inhibit HIV-1 replication. Accordingly, the invention also includes a method of identifying compounds that inhibit HIV-1 replication in cells of an animal, comprising: contacting a Gag protein comprising a CA-SP1 cleavage site with a test compound; adding a labeled substance that selectively binds near the CA-SP1 cleavage site; and measuring competition between the binding of the test compound and the labeled substance to the CA-SP1 cleavage site. In further embodiments of this method, the compounds inhibits the interaction of HIV-1 protease with a target site by binding to said target site.
[0051]These methods also include embodiments wherein the CA-SP1 cleavage site region is contained within a polypeptide fragment or recombinant peptide; and/or wherein the labeled substance is a labeled antibody specific for CA-SP1, and measuring the change in the amount of labeled antibody bound to the protein in the presence of test compound compared with a control. Labels include, but are not limited to, an enzyme, fluorescent substance, chemiluminescent substance, horseradish peroxidase, alkaline phosphatase, biotin, avidin, electron dense substance, radioisotope and a combination thereof.
[0052]The method of identifying compounds that inhibit HIV-1 replication in cells of an animal also comprises, in one embodiment, measuring the change in the amount of labeled 3-O-(3',3'-dimethylsuccinyl)betulinic acid bound to the protein in the presence of test compound, compared with a control, and wherein the labeled substance is 3-O-(3',3'-dimethylsuccinyl)betulinic acid.
[0053]In an alternative embodiment, the invention comprises a method for identifying compounds that inhibit HIV-1 replication in the cells of an animal which comprises: contacting a polypeptide comprising a CA-SP1 cleavage site, with a protease in the presence of a test compound. Preferably the protease is related to HIV-1 protease, or is HIV protease. In one embodiment, the method comprises; contacting a polypeptide comprising a wild type CA-SP1 cleavage site, with a protease in the presence of a test compound and also contacting a polypeptide comprising a mutant CA-SP1 cleavage site or a protein comprising an alternative protease cleavage site with HIV-1 protease in the presence of the test compound, detecting the cleavage, and comparing the amount of cleavage of the native wild-type polypeptide to the amount of cleavage of the mutant polypeptide or to amount of cleavage of the protein comprising an alternative protease cleavage site. In a related aspect of this method, the wild-type CA-SP1 or mutant CA-SP1 or alternative protease cleavage site region is contained within a polypeptide fragment or recombinant peptide. In a further related aspect, the polypeptide is labeled with a fluorescent moiety and a fluorescence quenching moiety, each bound to opposite sides of the CA-SP1 cleavage site, and wherein said detecting comprises measuring the signal from the fluorescent moiety. In another related embodiment, the polypeptide is labeled with two fluorescent moieties, each bound to opposite sides of the CA-SP1 cleavage site, and wherein said detecting comprises measuring the transfer of fluorescent energy from one moiety to the other in the presence of the test compound. In a further embodiment, the effect of the test compound on cleavage of the polypeptide is detected by measuring the amount of a labeled antibody that is bound to SP1 or p24 (CA). In a related aspect, the labeled antibody that binds CA, or the antibody that binds SP1 is labeled with a molecule selected from the group consisting of enzyme, fluorescent substance, chemiluminescent substance, horseradish peroxidase, alkaline phosphatase, biotin, avidin, electron dense substance, radioisotope, and combinations thereof.
[0054]The invention is also directed to a method for identifying compounds that inhibit HIV-1 replication in cells of an animal. In one embodiment, the method comprises: contacting a test compound with cells infected with wild-type virus isolates and with cells infected with virus isolates having significantly reduced sensitivity to 3-O-(3',3'-dimethylsuccinyl)betulinic acid; and selecting test compounds that are more active against the wild-type virus isolate compared with virus isolates that have reduced sensitivity to 3-O-(3',3'-dimethylsuccinyl)betulinic acid. In another embodiment, the method comprises contacting HIV-1 infected cells with a test compound; lysing the infected cells or the released viral particles to form a lysate, and analyzing the lysate to determine whether cleavage of the CA-SP1 protein has occurred. In this latter embodiment, said analyzing may comprise measuring the presence or absence of p25; and or performing a western blot of viral proteins and detecting p25 using an antibody to p25; and/or performing a gel electrophoresis of viral proteins and imaging of metabolically labeled proteins; and/or performing an immunoassay. Such an immunoassay may be performed by any methods known in the art, including, but not limited to:
[0055](a) capturing p25 and p24 on a substrate using an antibody that selectively binds p24; and
[0056](b) detecting the presence or absence of p25 on the substrate by using an antibody that selectively binds p25. The invention also includes such modifications of the above assay as would be obvious to one of ordinary skill in the art.
[0057]In a further embodiment, the method of identifying a compound according to the invention comprises the use of an epitope tag sequence inserted into SP1 and the selective detection of p25 is performed using an antibody to the epitope tag.
[0058]The invention is also directed to a method for identifying compounds that inhibit HIV-1 replication in the cells of an animal comprising: contacting HIV-1 infected cells with a test compound and thereafter analyzing the virus particles using transmission electron microscopy. Such analysis includes for example, looking for the presence of spherical cores that are acentric with respect to the viral particle; and/or having crescent-shaped, electron-dense layers lying just inside the viral membrane.
[0059]In additional aspects, the invention is drawn to an isolated polynucleotide comprising a sequence which encodes an amino acid sequence containing a mutation in an HIV Gag p25 protein (CA SP1), said mutation resulting in a decrease in inhibition of processing of p25 (CA-SP1) to p24 (CA) by 3-O-(3',3'-dimethylsuccinyl)betulinic acid (DSB). This inhibition of processing of p25 may be due to a decrease in inhibition of the interaction of HIV-1 protease with Gag; and/or a decrease in the binding of 3-O-(3',3'-dimethylsuccinyl)betulinic acid to Gag; and/or a decrease in the binding of DSB at or near the CA-SP1 cleavage site of Gag. Suitable polynucleotides also include those encoding a mutation at or near the CA-SP1 cleavage site or in the SP1 domain of CA-SP1; and/or those encoding a mutation at or near the amino acid sequence G/SHKARV/ILAEAMSQV (SEQ ID NO: 1); and/or those encoding the amino acid sequences GHKARVLVEAMSQV (SEQ ID NO: 2) or SHKARILAEAMSQV (SEQ ID NO: 3); and/or isolated polynucleotide which is selected from the group consisting of SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8 and SEQ ID NO: 9; and/or having at least about 95% identity to a polynucleotide selected from the group consisting of SEQ ID NO: 4, and SEQ ID NO: 6; and/or having at least about 80% identity to a polynucleotide selected from the group consisting of SEQ ID NO: 8 and SEQ ID NO: 9; and/or having at least about 95% identity to a polynucleotide selected from the group consisting of SEQ NO: 5 and SEQ ID NO: 7; and/or having at least about 80% identity to a polynucleotide of SEQ ID NO: 10. In additional embodiments, the polynucleotide having more than about 40%, 50%, 60%, 70%, 80%, 90%, 95%, 99% identity or which is identical to the polynucleotide sequences listed above.
[0060]The invention is also drawn to vectors comprising such polynucleotides as described above; to a host cell comprising such a vector; and to a method of producing a polypeptide comprising incubating the host cell containing such a vector in a medium and recovering the polypeptide from said medium.
[0061]In one embodiment, the invention is directed to an antibody. Such an antibody may bind to a polypeptide with an amino acid sequence having at least about 40%, 50%, 60%, 70%, 80%, 90% identity, or which is identical to a sequence selected from the group consisting of:
TABLE-US-00004 (SEQ ID NO: 21) (a) KNWMTETFLVQNANPDCKTILKALGPAATLEEMMTACQGVGGPHKA RILAEAMSQVTNSATIM; (SEQ ID NO: 22) (b) KNWMTETLLVQNANPDCKTILKALGPGATLEEMMTACQGVGGPGHK ARVLAEAMSQVTNPATIM; (SEQ ID NO: 23) (c) TACQGVGGPSHKARILAEAMSQVTNSATIM; (SEQ ID NO: 24) (d) MTACQGVGGPGHKARVLAEAMSQVTKPATIM; (SEQ ID NO: 25) (e) SHKARILAEAMSQV; and (SEQ ID NO: 26) (f) GHKARVLAEAMSQV.
[0062]In a further related embodiment, the invention is drawn to an antibody which binds to a polypeptide encoded by a polynucleotide sequence having at least about 40%, 50%, 60%, 70%, 80%, 90% identity, or which is identical to a polynucleotide with a sequence selected from the group consisting of: (a) about nucleotides 1243-1435 of SEQ ID NO: 18; (b) about nucleotides 1729-1920 of SEQ ID NO: 19; (c) about nucleotides 1344-1435 of SEQ ID NO: 18; (d) about nucleotides 1828-1920 of SEQ ID NO: 19; (e) about nucleotides 1370-1413 of SEQ ID NO: 18; and (f) about nucleotides 1857-1899 of SEQ ID NO: 19.
[0063]In one embodiment, the antibody binds to amino acids of the CA-SP1 region of the HIV-1 Gag polypeptide, wherein said amino acids comprise: SHKARILAEAMSQV (SEQ ID NO: 25) or GHKARVLAEAMSQV (SEQ ID NO: 26).
[0064]In one embodiment, the invention is drawn to an antibody that inhibits the binding of 3-O-(3',3'-dimethylsuccinyl)betulinic acid to the CA-SP1 region of the Gag polypeptide.
[0065]The invention is also drawn to mutant HIV-1 viruses. In one such embodiment, the invention is an isolated mutant recombinant HIV-1 virus, wherein the processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) in said virus is not significantly inhibited by 3-O-(3',3'-dimethylsuccinyl)betulinic acid. In related embodiments, this virus is not inhibited by 3-O-(3',3'-dimethylsuccinyl)betulinic acid. In another embodiment, 3-O-(3',3'-dimethylsuccinyl)betulinic acid does not inhibit the interaction of protease with the Gag polypeptide in this virus. In another, the virus does not bind to 3-O-(3',3'-dimethylsuccinyl)betulinic acid. In further embodiments the invention is drawn to viruses wherein the amino acids of the CA-SP1 region are replaced with alternative amino acids, or amino acids are added to the CA-SP1 region, or where amino acids are deleted. In one embodiment, one or more amino acids are deleted from the AEAMSQV (amino acid no. 8-14 of SEQ ID NO:26) amino acid sequence in the CA-SP1 region.
[0066]A mutant viruses may be used in the methods of the invention described elsewhere herein. For example, such viruses are useful in a method of identifying a compound which inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), the method comprising comparing the ability of said compound to inhibit HIV-1 replication compared with the replication of a the mutant virus outlined above. Such inhibition may be examined in a cell, or in an animal, or in vitro.
[0067]The invention is also drawn to non-HIV-1 retroviruses that are sensitive to 3-O-(3',3'-dimethylsuccinyl)betulinic acid. In some embodiment, said retrovirus encodes a CA-SP1 polypeptide with an amino acid sequence comprising the sequence AEAMSQV (amino acid no. 8-14 of SEQ ID NO: 26) at or near the CA-SP1 cleavage site. In another embodiment, the retrovirus encodes a CA-SP1 polypeptide with an amino acid sequence comprising the sequence VLAEAMSQV (amino acid no. 6-14 of SEQ ID NO: 26) at or near the CA-SP1 cleavage site. In another embodiment, the retrovirus encodes a CA-SP1 polypeptide with an amino acid sequence comprising the sequence GHKARVLAEAMSQV (SEQ ID NO: 26) at or near the CA-SP1 cleavage site; in another the retrovirus comprises the amino acid sequence having at least 60%, 70%, 80%, 90% identity or which is identical to the sequence encoded by the polynucleotide of SEQ ID NO:26, SEQ ID NO: 90; SEQ ID NO: 92; SEQ ID NO: 94; SEQ ID NO: 96; or SEQ ID NO: 98; in another embodiment the retrovirus comprises the amino acid sequence having at least 60%, 70%, 80%, 90% identity or which is identical to the sequence of SEQ ID NO: 91; SEQ ID NO: 93; SEQ ID NO: 95; SEQ ID NO: 97; or SEQ ID NO: 99. In another embodiment, the retrovirus comprises the nucleic acid sequence having at least 70%, 80%, 90% or which is identical to the sequence of SEQ ID NO: 90; SEQ ID NO: 92; SEQ ID NO: 94; SEQ ID NO: 96; or SEQ ID NO: 98.
[0068]Retroviruses of this embodiment of the invention include, but are not limited to HIV-2, HTLV-I, HTLV-II, SIV, avian leukosis virus (ALV), endogenous avian retrovirus (EAV), mouse mammary tumor virus (MMTV), feline immunodeficiency virus (FIV), Bovine immunodeficiency virus (BIV), caprine arthritis encephalitis virus (CAEV), Visna-maedi virus, or feline leukemia virus (FeLV).
[0069]In a related embodiment, the invention is drawn to a method of making a recombinant non-HIV-1 lentivirus sensitive to DSB. This method comprises: deleting from the genome of said lentivirus the nucleotides which corresponds nucleotides deleting from the genome of said lentivirus the nucleotides which correspond to nucleotides 1370-1413 from SEQ ID NO: 18, in HIV-1; and inserting nucleotides 1370-1413 from SEQ ID NO: 18 or nucleotides 1857-1899 of SEQ ID NO: 19 into said region of said non-HIV-1 lentivirus.
[0070]Examples of chimeric lentiviruses that were, are or may be constructed by this method are described in FIG. 10.
[0071]Such viruses may be used in the methods of the invention described elsewhere herein. For example, such recombinant non-HIV-1 lentiviruses may be used in a method of identifying a compound which inhibit processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), the method consisting of comparing of the ability of said compound to inhibit replication of a wild-type non-HIV-1 lentivirus with the DSB-sensitive recombinant variant thereof. Such inhibition may occur in a cell; in an animal; or in vitro.
[0072]The invention is also drawn to an animal model of lentivirus infection comprising a suitable non-human animal host infected with a lentivirus sensitive to 3-O-(3',3'-dimethylsuccinyl)betulinic acid. In such an embodiment, the lentivirus may include, but is not limited to SIV; FIV; EIAV; BIV; CAEV; and Visna-Maedi virus.
[0073]The invention is also drawn to isolated polypeptides. In one embodiment, the invention is drawn to a polypeptide containing a mutation in an HIV CA-SP1 protein, said mutation which results in a decrease in inhibition of processing of p25 by 3-O-(3',3'-dimethylsuccinyl)betulinic acid. In a related embodiment, this polypeptide is encoded by a polynucleotide that contains a mutation located at or near the CA-SP1 cleavage site or in the SP1 domain encoded by SEQ ID NO: 5, SEQ ID NO: 7, or SEQ ID NO: 10 and/or is encoded by a polynucleotide selected from the group consisting of SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8 and SEQ ID NO: 9; and/or comprises a sequence that is selected from the group consisting of GHKARVLVEAMSQV (SEQ ID NO: 2) or SHKARILAEVMSQV (SEQ ID NO: 3); and/or is encoded by an isolated polynucleotide which hybridizes under stringent conditions to a polynucleotide selected from the group consisting of SEQ ID NO: 5, SEQ ID NO: 7, and 10; and/or is part of a chimeric or fusion protein.
[0074]The invention is also drawn to antibodies which selectively bind to an amino acid sequence containing a mutation in an HIV CA-SP1 protein which results in a decrease in the inhibition of processing of p25 (CA-SP1) to p24 (CA) by 3-O-(3'3'-dimethylsuccinyl)betulinic acid. In one such embodiment, the antibody selectively binds to a mutation located at or near the CA-SP1 cleavage site or in the SP1 domain of CA-SP1; in another, the antibody selectively binds to a mutation comprising a sequence that is selected from the group consisting of GHKARVLVEAMSQV (SEQ ID NO: 2) or SHKARILAEVMSQV (SEQ ID NO: 3); in another embodiment, the antibody selectively binds an amino acid sequence selected from the group consisting of SEQ ID NO: 2 and SEQ ID NO: 3.
[0075]In another embodiment, the invention is drawn to an antibody that selectively binds SP1 but not CA-SP1; another that selectively binds CA-SP1 but not CA; another that selectively binds CA but not CA-SP1; and a further antibody that selectively binds at or near the CA-SP1 cleavage site.
[0076]The invention is also directed to a compound identified by any of the methods elucidated herein. In one embodiment, the compounds is not a compound selected from the group consisting of 3-O-(3',3'-dimethylsuccinyl)betulinic acid, 3-O-(3',3'-dimethylsuccinyl)betulin, 3-O-(3',3'-dimethylglutaryl)betulin, 3-O-(3',3'-dimethylsuccinyl)dihydrobetulinic acid, 3-O-(3',3'-dimethylglutaryl)betulinic acid, (3',3'-dimethylglutaryl)dihydrobetulinic acid, 3-O-diglycolyl-betulinic acid, 3-O-diglycolyl-dihydrobetulinic acid, and combinations thereof.
[0077]The invention is also drawn to a pharmaceutical composition. In one embodiment, the pharmaceutical composition comprises derivatives of dimethylsuccinyl betulinic acid or dimethylsuccinyl betulin; in another, the pharmaceutical composition comprises a compound selected from the group consisting of 3-O-(3',3'-dimethylsuccinyl)betulinic acid, 3-O-(3',3'-dimethylsuccinyl)betulin, 3-O-(3',3'-dimethylglutaryl)betulin, 3-O-(3',3'-dimethylsuccinyl)dihydrobetulinic acid, 3-O-(3',3'-dimethylglutaryl)betulinic acid, (3',3'-dimethylglutaryl)dihydrobetulinic acid, 3-O-diglycolyl-betulinic acid, 3-O-diglycolyl-dihydrobetulinic acid, and combinations thereof. In another embodiment, the pharmaceutical composition comprises one or more compounds identified according to the methods of the invention which are not otherwise listed; or any pharmaceutically acceptable salt, ester or prodrug thereof, and a pharmaceutically acceptable carrier. In another embodiment, the pharmaceutical composition further comprising an anti-viral agent which may include any one of zidovudine, lamivudine, didanosine, zalcitabine, stavudine, abacavir, nevirapine, delavirdine, emtricitabine, efavirenz, saquinavir, ritonavir, indinavir, nelfinavir, tenofovir, amprenavir, adefovir, atazanavir, fosamprenavir, hydroxyurea, AL-721, ampligen, butylated hydroxytoluene; polymannoacetate, castanospermine; contracan; creme pharmatex, CS-87, penciclovir, famciclovir, acyclovir, cytofovir, ganciclovir, dextran sulfate, D-penicillamine trisodium phosphonoformate, fusidic acid, HPA-23, eflornithine, nonoxynol, pentamidine isethionate, peptide T, phenyloin, isoniazid, ribavirin, rifabutin, ansamycin, trimetrexate, SK-818, suramin, UA001, combinations thereof, any other antiviral, immunomodulating agent, anti-cancer agent, anti-fungal agent, anti-bacterial agent, or combinations thereof.
[0078]The invention is also drawn to a method of determining if an individual is infected with HIV-1 that is susceptible to treatment by a compound that inhibits p25 processing. In one embodiment, the method involves taking blood from the patient, genotyping the viral RNA and determining whether the viral RNA contains mutations in the sequence encoding the region of the CA-SP1 cleavage site.
[0079]The invention is also drawn to a method of treating a disease in a patient in need thereof comprising:
[0080]identifying a compound which inhibits the processing of viral Gag p25 protein (CA-SP1) to p24 (CA), but has no significant effect on other Gag processing steps;
[0081]obtaining regulatory approval for the sale and use of said compound;
[0082]packaging the compound for sale and treatment of a disease in a patient in need thereof.
BRIEF DESCRIPTION OF THE DRAWINGS
[0083]FIG. 1. DSB does not disrupt the activity of HIV-1 protease at a concentration of 50 μg/mL. In DSB-containing samples recombinant Gag is processed correctly. In contrast, indinavir blocks protease activity at 5 μg/mL as evidenced by the absence of bands corresponding to p24 and the MA-CA precursor.
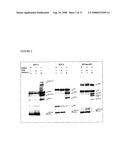
[0084]FIG. 2. Western blots of virion-associated Gag derived from chronically infected H9/HIV-1IIIB1, H9/HIV-2ROD, and H9/SIVmac251 in the presence of DSB (1 μg/mL), indinavir (1 μg/mL) or control (DMSO). Gag proteins were visualized using HIV-Ig (HIV-1) or monkey anti-SIVmac251 serum (HIV-2 and SIV; NIH AIDS Research and Reference Reagent Program).
[0085]FIG. 3. EM analysis of DSB-treated HIV-1 infected cells. The EM data show two primary differences between DSB-treated and untreated samples. Virions generated in the presence of DSB are characterized by an absence of conical, mature cores. In these samples the cores are uniformly spherical and often acentric. Secondly, many virions display an electron dense layer inside the lipid bilayer but outside the core (indicated with arrows in the DSB-treated sample panels). In the DSB-treated samples no mature viral particles were observed.
[0086]FIG. 4 depicts amino acid sequences in the region of the CA-SP1 cleavage site from DSB-sensitive HIV-1 isolates NL4-3 and RF (#1; SEQ ID NO: 1) and DSB-resistant HIV-1 isolates (#2; SEQ ID NO: 2 (NL4-3), and #3; SEQ ID NO: 3 (RF)). The differences between the native and DSB-resistant sequences involve an alanine to valine change at the first downstream residue (#2) and an alanine to valine change in the third downstream residue (#3) from the CA-SP1 cleavage site (-|-). These residues are underlined and bolded for ease of identification.
[0087]FIG. 5 depicts the + sense consensus sequence for the A364V DSB-resistant NL4-3 mutant (SEQ ID NO: 4) beginning with the start of gag and continuing into pol, including the entire protease coding region. Missense mutations not found in the wild-type NL4-3 GENBANK M19921 sequence are in bold and gray shadowing. The coding sequence for the consensus CA-SP1 cleavage site region is underlined. The shaded area including the cleavage site denotes the SP1 sequence. The first mutation is the A364V mutation.
[0088]The second amino acid change (in protease) was also found in the parental clone and has been confirmed to correspond to a sequencing error in the original GENBANK entry. Therefore, no mutations actually occurred in protease.
[0089]FIG. 6 depicts the + sense consensus sequence for the DSB-sensitive NL4-3 parental isolate (SEQ ID NO: 5) that was passaged in the absence of drug in parallel with the A364V mutant isolate.
[0090]FIG. 7 depicts the + sense consensus sequence for the A366V DSB-resistant HIV-1 RF mutant (SEQ ID NO: 6) beginning with the start of the gag and continuing into pol, including the entire protease coding region. Missense mutations not found in the wild-type HIV-1 RF GENBANK M17451 sequence are shadowed in gray. The region of the CA-SP1 cleavage site is underlined. The only missense mutation not also found in the identically passaged DSB-sensitive isolate is the A366V mutation in the CA-SP1 cleavage site.
[0091]FIG. 8 depicts the + sense consensus sequence for the DSB-sensitive HIV-1 RF parental isolate (SEQ ID NO: 7), that was passaged in the absence of drug in parallel with the A366V mutant isolate.
[0092]FIG. 9 depicts the polynucleotide sequences, SEQ ID NO: 8 and SEQ ID NO: 9, which encode the polypeptides designated herein as SEQ ID NO: 2 and SEQ ID NO: 3, respectively. SEQ ID NO: 10 and 12 depict the nucleotide sequences that encode the parental polypeptide sequences designated as SEQ ID NO: 1. SEQ ID NO: 1 is a consensus sequence based on the sequences of the region from NL4-3 and RF
[0093]FIG. 10:
[0094]10A. Amino acid sequences in the CA-SP1 region of lentiviruses
[0095]10B: Amino acid sequences of the CA-SP1 region in HIV-1 strains RF and NL4-3
[0096]10C-10D: Nucleotide sequences of gag gene chimeric SIVs. The 42 nucleotide sequence encoding the seven amino acids upstream and seven amino acids downstream of the CA-SP1 cleavage site is underlined and in bold.
[0097]10E-H Nucleotide sequences of gag gene of chimeric FIV, EIAV and BIV to be made according to the invention. The 42 nucleotide sequences encoding the seven amino acids upstream and seven amino acids downstream of the CA-SP1 cleavage sites are underlined and in bold.
[0098]FIG. 11: Replication kinetics of PA-457 (DSB)-resistant mutants
[0099]FIG. 12: Sequential SP1 point deletions in the context of NL4-3 used to identify residues necessary for DSB activity. The amino acid sequence of SP1 domain in NL4-3 is shown. "Δ" indicates the deletion and "-" means identical residues between point deletion mutants and NL4-3
[0100]FIG. 13. Summary of particle production and infectivity of point deletions mutants.
[0101]FIG. 14. Western blots for viruses containing point deletions in SP1, in the presence (+) and absence (-) of DSB.
[0102]FIG. 15. Substitution of HIV-1 CA-SP1 residues VL-AEAMSQV into SIVmac239 backbone renders SIVmac239 sensitive to DSB.
[0103](Top panel) Amino acid sequences near the CA-SP1 cleavage site (including entire SP1 region) are shown for SIVmac239, HIV-1 NL4-3 and a series of SIV mutants into which various NL4-3 residues (underlined) were inserted. Dashes ("-") indicates the residues are the same as those in SIVmac239.
[0104](Bottom panel) Western blots showing the CA and CA-SP1 proteins for this series of viruses in the presence (+) or absence (-) of DSB.
[0105]FIG. 16: Sequence conservation in the CA-SP1 region of Lentiviruses.
[0106]FIG. 16. Cloning Strategy: Substituting HIV-1 specific CA-SP1 residues into the corresponding Gag region of FIV, EIAV or BIV
[0107]FIG. 17. HIV-1 NL4-3 SP1 tagged with an epitope. Sequences of SP1 peptides with peptide tags inserted are shown. "Δ" indicates deleted residue and "-" indicates that the residue is identical to that in NL4-3 SP1
[0108]FIG. 18A-C: HIV-1 strain RF polynucleotide sequence. The nucleotide sequence of the Gag polyprotein is underlined and in bold. The 42 nucleotide sequence encoding the seven amino acids upstream and seven amino acids downstream of the CA-SP1 cleavage site is highlighted in green. An additional 129 nucleotides (43 amino acid residues) upstream of the cleavage site in CA and the remaining 21 nucleotides (seven amino acids residues) in SP1 are highlighted.
[0109]FIG. 19A-E: HIV-1 strain NL4-3 polynucleotide sequence. The nucleotide sequence of the Gag polyprotein is underlined and in bold. The 42 nucleotide sequence encoding the seven amino acids upstream and seven amino acids downstream of the CA-SP1 cleavage site is highlighted in green. An additional 129 nucleotides (43 amino acid residues) upstream of the cleavage site in CA and the remaining 21 nucleotides (seven amino acids residues) in SP1 are highlighted.
DETAILED DESCRIPTION OF THE INVENTION
[0110]The present invention is directed to methods of inhibiting HIV-1 replication in the cells of an animal. More specifically, the invention involves methods of inhibiting HIV-1 replication in the cells of a mammal by contacting infected cells with a compound that inhibits the processing of the viral Gag p25 protein (CA-SP1) to the p24 protein (CA). More specifically, such compounds inhibit the processing of the viral Gag p25 protein (CA-SP1) to the p24 protein (CA) without significantly affecting other Gag processing steps.
[0111]A compound that does not significantly affect other Gag processing steps" means that the compound in question predominantly inhibits processing of p25 to p24, but does not necessarily preclude the possibility of having additional minor effects on other Gag processing steps.
[0112]Significant" or "Significantly," where not otherwise defined herein, means an observable or measurable change compared to the process in the absence of a compound. However, not all observable or measureable changes may necessarily be significant.
[0113]A number of viral phenotypes may also be observed in practicing the method of the invention. One result of contacting an infected cell with the compounds of the invention may be the formation of noninfectious viral particles. Alternatively, or in addition, contacting infected cells with a compound that inhibits p25 to p24 processing, results in the formation of non-infectious viral particles, but where there is no significant effect on other Gag processing steps. This may not significantly reduce the quantity of virus released from treated cells and/or has no little or no significant effect on the amount of RNA incorporation into the released virions.
[0114]Accordingly, the invention is also drawn to a method of inhibiting HIV infection in cells of an animal comprising contacting said cells with a compound that inhibits p25 processing and also affects other viral phenotypes, described above.
[0115]Mutant viruses defective in CA-SP1 cleavage have been shown to be non-infectious (Wiegers K. et al., J. Virol. 72:2846-2854 (1998)). 3-O-(3',3'-dimethylsuccinyl)betulinic acid (DSB) is an example of a compound that disrupts p25 to p24 processing and potently inhibits HIV-1 replication. This compound's activity is specific for the p25 to p24 processing step, not other steps in Gag processing. Furthermore, DSB treatment results in the aberrant HIV particle morphology as described in FIG. 3.
Identification of HIV-1 Determinants Associated with Sensitivity to 3-O-(3',3'-dimethylsuccinyl)betulinic Acid
[0116]Generation and Selection of HIV-1 Viruses Resistant to DSB.
[0117]Mutant forms of HIV-1 have been generated in which the amino acid sequence in the region of the CA-SP1 cleavage site is modified, decreasing the sensitivity of these strains to compounds that disrupt CA-SP1 processing. Data on these mutant viruses have been used to identify the amino acid residues in wild-type Gag that are implicated in the antiviral activity of these compounds. In one embodiment, compounds that disrupt CA-SP1 processing directly or indirectly inhibit the interaction of HIV-1 protease with the region of the Gag protein containing these amino acid residues. In another embodiment, compounds that disrupt CA-SP1 processing bind to the region containing these amino acid residues. As used herein, the terms "bind," "bound" or "binding" refers to binding or attachment including, e.g., ionic interactions, electrostatic hydrophobic interactions, hydrogen bonds, etc; and also includes associations that may be covalent, e.g., by chemically coupling. Covalent bonds can be, for example, ester, ether, phosphoester, thioester, thioether, urethane, amide, amine, peptide, imide, hydrazone, hydrazide, carbon-sulfur bonds, carbon-phosphorus bonds, and the like. The term "bound" is broader than and includes terms such as "coupled," "conjugated" and "attached."
[0118]In another embodiment, compounds that disrupt CA-SP1 processing bind to another region of Gag and thereby inhibit the interaction of HIV-1 protease with the region of the CA-SP1 cleavage site. In another embodiment, viruses or recombinant proteins that contain mutations in the region of the CA-SP1 cleavage site can be used in screening assays to identify compounds that disrupt CA-SP1 processing.
[0119]In one set of experiments, amino acid residues in HIV-1 Gag that are involved in the disruption of CA-SP1 processing by 3-O-(3',3'-dimethylsuccinyl)betulinic acid (DSB) were identified by sequencing the gag-pol gene of virus isolates that had been selected for resistance to DSB. The amino acid sequences from these resistant viruses were compared with the gag-pol gene sequences from DSB-sensitive HIV-1 isolates. Two single amino acid changes were identified in the DSB-resistant viruses, an alanine (Ala) to valine (Val) substitution at residue 364 (SEQ ID NO: 4) and in a second isolate, at residue 366 (SEQ ID NO: 6), in the Gag polyprotein (see FIG. 4). These residues are located immediately downstream of the CA-SP1 cleavage site (at the N-terminus of SP1). Alanine is highly conserved at these positions throughout all HIV-1 subtypes listed in the Los Alamos National Laboratory database. The five amino acid residues upstream and downstream of the CA-SP1 cleavage site are also highly conserved among the various subtypes. However, isoleucine replaces valine at the position two residues upstream of the cleavage site in a number of clades (c.f., FIG. 4, SEQ ID NO. 1). ("HIV Sequence Compendium 2002," Kuiken et al. eds. Los Alamos National Laboratory, Los Alamos, N. Mex.)
[0120]In order to more extensively map the viral genetic determinants for DSB resistance, additional experiments were performed to select for viruses in vitro that are drug resistant. Multiple parallel cultures of Jurkat T cells (5×105 each) were transfected with the proviral DNA clone pNL4-3 in the presence or absence of 10-50 ng/ml DSB. The cells were passaged every two days, and fresh drug was added at each passage. Virus replication was monitored by measuring reverse transcriptase activity in culture supernatants. Virus was isolated from culture supernatants harvested at selected timepoints, and genomic DNA was amplified by RT-PCR using primers that spanned the coding region between the N-terminus of CA and the N-terminus of RT. The amplified product was then sequenced using the same set of primers.
[0121]In one experiment, an A366V mutation was identified in the SP1 region of NL4-3 virus cultured in the presence of DSB (note: numbering is relative to the Gag polyprotein). Upon further passaging, a double mutant was identified that contained a G357S mutation in CA as well as the A366V mutation in SP1. The A366V mutation was identified previously in experiments selecting for resistant variants of the RF isolate. Interestingly, the wild-type RF sequence also contains a serine residue at position 357 in CA (FIG. 4). Since serine is present at this position in isolates (such as RF) that are sensitive to DSB, the CA G357S mutation alone is not sufficient to confer resistance to DSB. To determine the contribution of each of these mutations to drug resistance, the A366V mutation and the A366V/G357S double mutation were re-engineered into the wild-type NL4-3 backbone by site-directed mutagenesis. The resulting constructs were transfected into Jurkat T cells and characterized in a virus replication assay as described above for the selection of resistance. SDS-PAGE analysis of transfected cell lysates and virus released into the media demonstrated that the A366V mutant Gag was processed and released from cells inefficiently (data not shown) and thus replicated very poorly even in the absence of drug (FIG. 11) However, the A366V/G357S double mutant replicated efficiently in the absence or presence of DSB. There data indicate that the resistant mutant, A366V, requires a serine at the 357 position in the CA region of Gag to compensate for a deleterious effect on virus replication (FIG. 11).
[0122]In a further experiment, ten different resistant isolates were generated. Sequencing of these isolates identified four additional mutations not previously seen in resistance selection experiments. These were H358Y, L363F and L363M in CA, and A402T in the NC region of Gag. None of these mutations are present in the consensus sequences for HIV-1 clades A-O, reflecting the breadth of activity of DSB against genetically diverse clades of HIV-1. The L363M substitution in CA was found in the consensus sequence for HIV-2, which may, in part, explain the specificity of DSB for HIV-1.
[0123]These results demonstrate the presence of specific genetic determinants for DSB activity in HIV-1, and that these determinants are centered around the CA-SP1 cleavage site.
[0124]HIV-1 NL4-3 Deletion and SIV Insertion Studies Used to Identify Viral Genetic Determinants of DSB Sensitivity
[0125]Results from in vitro resistance selection experiments indicated that the determinants of DSB HIV-1 inhibitory activity map to the region of Gag flanking the CA-SP1 cleavage site. In order to better define the viral genetic determinant for DSB, HIV-1 point-deletion mutagenesis and SIV insertion studies were undertaken to identify the specific amino acid residues associated with compound activity. The study was carried out as follows. Single residue deletions starting with residue E365 and continuing through residue M377 were engineered into the SP1 domain of the infectious HIV-1 molecular clone NL4-3 (FIG. 12). The effect of these point deletions on viral particle production, infectivity, Gag processing and sensitivity to DSB was determined. The results of these experiments were used to identify the Gag residues in the region of the CA-SP1 cleavage site that are associated with DSB activity. The residues associated with activity were inserted into the CA-SP1 cleavage site region of the DSB-resistant virus SIV (Mac 239 isolate) to generate a HIV-1, SIV chimeric virus (SHIV). Point substitution of HIV-1 residues from the N-terminus of the CA protein were made into this chimeric virus until the minimal sequence necessary to rescue DSB activity was identified. This minimal sequence necessary to gain DSB activity is considered a primary viral genetic determinant of DSB activity. It may suggest the molecular determinant of DSB activity.
Methods:
[0126]Construction of NL4-3 Single Point-Deletion Mutants.
[0127]Single point-deletion constructs were generated using the PCR-ligation-PCR (PLP) strategy as previously described. HIV-1 NL4-3 plasmid DNA was used as the template to perform all PCR reactions for generating point deletions spanning the complete Gag SP1 domain with the exception of the first residue of SP1.
[0128]E365 was generated using NL4-3 as the template with Vent DNA polymerase (NEB) by using deletion-specific downstream primer (Primer 1) with universal upstream primer (Primer 2) (Table 1). The fragment derived from this was termed as a first flanking PCR fragment. A second flanking fragment was amplified using deletion-specific upstream primer (Primer. 3) and universal downstream primer (Primer 4) (Table 1). To generate other deletion constructs (ΔA366, ΔM367, ΔS368, ΔQ369, ΔV370, ΔT371, ΔN372, ΔP373, ΔA374, ΔT375, ΔI376, and ΔM377). PCR procedures were similarly performed by varying deletion-specific downstream and upstream primers corresponding to each specific point deletion (Table 1).
[0129]Each of these parallel two adjacent PCR fragments was gel purified, phosphorylated using T4 polynucleotide kinase (NEB), and ligated by using T4 DNA ligase (NEB). After inactivation at 65° C. for 15 minutes, the ligation reaction was used for a subsequent amplification with universal upstream primer (Primer. 2) and downstream primer (Primer. 4). This product was gel purified, digested with SpeI and ApaI, and then ligated into the SpeI and ApaI sites of NL4-3 proviral DNA clone.
[0130]Standard PCR conditions were used for the above-described reactions. These included, one cycle of denaturation at 95° C. for 1 minutes 30 seconds, followed by 30 cycles of denaturation at 95° C. for 30 seconds, 60° C. for 30 seconds and 72° C. for 30 seconds. The PCR reactions were set up using the following components: δ5 μL 10×NEB Thermophilic buffer
[0131]2 μL 10 mM dNTPs
[0132]1 μL 10 nM MgSO2
[0133]1 μL 50 μmol upstream primer
[0134]1 μL 50 μmol downstream primer
[0135]1 μL 50 ng/mL template DNA
[0136]0.5 μL Vent DNA polymerase
[0137]38.5 μL ddH2O
[0138]A 10 μL aliquot was run on a 1.0% agarose gel to make sure the correct size product was amplified. The PCR products were then gel isolated and purified with a Qiaex II gel extraction Kit (Qiagen). The gel-purified two adjacent PCR fragments were individually phosphorylated in the following reaction by using T4 polynucleotide kinase (NEB) prior to ligation. The phosphorylation reaction was set up as follows:
[0139]2 μL 10×T4 polynucleotide kinase buffer
[0140]2 μL 10 mM ATP
[0141]1 μL T4 polynucleotide kinase
[0142]15 μL gel purified DNA of each of these two adjacent PCR fragments The reaction was incubated at 37° C. for 1 hour. Following the inactivation at 65° C. for 10 minutes, the adjacent phosphorylated PCR fragments were then ligated together by using T4 DNA ligase (NEB) under following conditions:
[0143]3 μL 10×T4 DNA ligase buffer
[0144]13 μL of each of two adjacent PCR fragments
[0145]1 μL T4 DNA ligase
[0146]After overnight incubation at 16° C. the ligation reaction product was used in a second round PCR reaction to amplify the full-length PCR fragment spanning these two adjacent PCR products. The second round PCR reaction was performed as described above with the exception that only universal upstream primer (Primer. 2) and downstream primer (Primer. 4) were used. Again, a 10 μL aliquot was run on a agarose gel to make sure the correct product was amplified. The full-length PCR fragments were then gel isolated and purified using a Qiaex II kit. The purified full-length PCR fragment, together with NL4-3, were then cut with SpeI and ApaI under the following conditions:
[0147]2 μL 10×NE buffer 4 (NEB)
[0148]1 μL ApaI (NEB)
[0149]1 μL SpeI (NEB)
[0150]16 μL full length PCR product (1 μg) or NL4-3 (500 ng)
[0151]The above restriction enzyme digestion mixture was incubated at 37° C. for 2 hours. Digested DNA fragments for the full-length PCR product and the NL4-3 plasmid were individually gel isolated and purified using a Qiaex II kit. The digested vector NL4-3 and full length PCR fragment were ligated using T4 DNA ligase under the following procedure:
[0152]1 μL 10×T4 DNA ligase buffer
[0153]1 μL (25-50 ng) digested NL4-3 vector
[0154]7 μL digested (200 ng-400 ng) digested PCR fragment (700 bp)
[0155]1 μL T4 DNA ligase
[0156]The ligation reaction was incubated at 16° C. overnight and the ligated products were transformed into Escherichia coli Max Efficiency Stb12 (Invitrogen) by heat shock according to instruction (Invitrogen). The proviral DNA clones were then screened by automatically sequencing using a Taq Dye Deoxy Terminator cycle Sequencer Kit (Applied Biosystems) individually using internal primers (Primer 29 and 30) Following the verification the mutations the proviral DNA clones were used for various future studies.
Construction of SIV Chimeric Mutants
[0157]A panel of SIV chimeric constructs harboring various residues of NL4-3 CA-SP1 boundary region was generated using the SIVmac239 molecular clone by employing PCR and cloning procedures described above. These constructs and their amino acid sequences in the CA-SP1 boundary region are shown in FIG. 15. SIV mac239 was used to generate the SIV DD and DE constructs. The SIV DD construct was used to generate SIV DM. Different SIV chimeric constructs were produced in the PCR by varying respective mutagenic upstream and downstream primers corresponding to each chimera (Table 1). Each of these parallel two adjacent PCR fragments was gel purified and directly used without phosphohorylation treatment for a subsequent amplification with universal upstream primer (Primer. 31) and downstream primer (Primer 32). This product was gel purified, digested with BamHI and SbfI, and then ligated into the BamHI and SbfI sites of SIVmac239 proviral DNA clone. The proviral DNA clones were then screened by automatically sequencing using a Taq Dye Deoxy Terminator cycle Sequencer Kit (Applied Biosystems) individually using an internal primer (Primer. 39). Following the verification the mutations the proviral DNA clones were used for various future studies.
Cell Culture and DNA Transfection
[0158]HeLa cells were maintained in DMEM (Invitrogen) (10% FBS, 100 U/ml penicillin, and 100 μg/ml Streptomycin) and passaged upon confluence. Jurkat cells were cultured in RPMI 1640 (Invitrogen) (10% FBS, 100 U/ml penicillin, and 100 μg/ml Streptomycin) and passaged every two or three days.
[0159]To characterize the effect of deletion or substitution on viral particle production and Gag polyprotein processing, wild-type HIV-1 NL4-3 or SIVmac239 and respective mutant proviral DNAs were transfected into HeLa cells by employing FuGENE 6 transfection reagent (Roche). Briefly, cells were seeded into a 6-well plate (Corning) at a concentration of 0.5×105 per well the day before transfection to reach 60 to 80% confluence on the day of transfection. For each transfection, 3 μl of FuGENE 6 was diluted into 100 μl of serum-free DMEM followed by the addition of 1 μg of DNA. After gently mixing, the mixture of DNA-lipid complexes was gently added drop wise into the cells containing 2 ml of complete DMEM medium. Twenty-four hours post-transfection, medium containing DNA-FuGENE 6 complexes was removed, 2 ml of fresh DMEM was added into the transfected cells. At 48 h post-transfection, medium containing viral particles was collected and clarified by centrifugation at 2,000 rpm at 4° C. for 20 min in a Sorvall RT 6000B centrifuge. Virus particle-containing supernatants were then concentrated through a 20% sucrose cushion in a microcentrifuge at 13,000 rpm at 4° C. for 120 min and pellets were resuspended in a lysis buffer (150 mM Tris-HCl, 5% Triton X-100, 1% deoxycholate, pH 8.0). The level of viral particle production for wild type NL4-3 and point deletion mutants was determined by p24 antigen capture ELISA (ZeptoMetrix, Buffalo, N.Y.).
[0160]To examine the effect of deletion or substitution on Gag polyprotein processing (in the absence of DSB), SDS-PAGE and Western-Blot was performed. In brief, viral proteins were separated on a 12% NuPAGE Bis-Tris Gel (Invitrogen) and transferred to a nitrocellulose membrane (Invitrogen) followed by blocking in a PBS buffer containing 0.5% Tween and 5% dry milk. The membrane was incubated with immunoglobulin from HIV-1-infected patients (HIV-Ig) (NIH AIDS research and reference reagent program) and hybridized with goat anti-human horseradish peroxidase (Sigma). For the membrane containing SIV proteins, the membrane was incubated with a reference polyclonal immune serum from a SIV-infected monkey (NIH AIDS Research and Reference Reagent Program) and hybridized with goat-anti-monkey horseradish peroxidase (Sigma). The immune complex was visualized with an ECL system (Amersham Pharmacis Biotech) according to the instructions provided by the manufacturer.
[0161]To address the effect of deletion or substitution on the ability of DSB to inhibit CA-SP1 processing, HeLa cells were transfected with wild-type HIV-1 NL4-3 or SIVmac239 and respective mutant proviral DNAs by employing the procedure described above. DSB at a concentration of 1 μg/ml and DMSO control were maintained throughout the entire culture and SDS-PAGE/Western-Blot for analyzing viral proteins derived from these transfections were performed as described in the previous paragraph.
[0162]The 50%-Tissue Culture Infectious Dose (TCID50) per ml was used as a measure of the infectivity of each deletion mutant. Mutant viruses derived from transfections in HeLa cells were used to infect U87 CD4.CXCR4 cells. Each virus stock was tested in triplicate at a starting dilution of 1:10, followed by four-fold serial dilutions. Cells were plated the day before infection at a density of 3×103 cells/well. On the day of infection, culture media was removed from the cell plate and 90 μl of diluted virus was added. On days 1, 3, and 6 post infection, virus was removed from plate and 200 μl of culture media was added. On days 6 and 8 post infection, supernatant was collected for p24 ELISA analysis. The virus dilution that caused 50% of the culture to be infected (TCID50) was determined according to the method of Reed and Muench (Aldovini A. and B. Walker 1990; Dulbecco R. 1988).
Results
[0163]Viruses containing sequential point deletions within the Gag SP1 domain (FIG. 12) were characterized for particle production, infectivity, Gag processing and sensitivity to DSB. The results from these experiments were used to identify SP1 residues associated with DSB activity.
[0164]As expected, the effect of point deletions on viral particle production varied as a function of the proximity of the change from the proteolytic cleavage site. The results from these experiments are summarized in FIG. 13. Viruses with deletions at residues E366, A367 and M368 were most affected, generating <25% the number of particles normally observed in wild-type virus infection. In vitro infectivity assays were used to characterize the ability of the deletion mutants to support virus replication. These experiments indicated that deletion of single residues at any of the five positions E365 through Q369 resulted in a virus that was either non-infectious or significantly impaired for replication (FIG. 13). In contrast, starting with residue V370 and extending away from the CA-SP1 cleavage site, none of the characterized point deletions resulted in a decrease in virus infectivity (FIG. 13). With the exception of viruses with deletions at positions I376 and M377 all mutant viruses exhibited a normal or near normal Gag processing phenotype (FIG. 13). The results from these three sets of experiments permitted the design and interpretation of experiments to identify the genetic determinants of DSB activity.
[0165]Sensitivity to DSB was determined in experiments that characterized the effect of DSB on a late step in Gag processing, CA-SP1 cleavage. Specifically, these assays measured the ability of DSB to disrupt CA-SP1 processing. As seen, e.g. Example 8, the DSB-induced defect in Gag processing correlates with the ability of the compound to inhibit virus replication. Results from these experiments indicate that deletion of a single residue at any of the six positions E365 through V370 significantly reduces the affect of DSB on CA-SP1 processing (FIG. 14). In contrast, starting with residue T372 and extending away from the CA-SP1 cleavage site, all of the characterized point deletions are fully sensitive to DSB-induced disruption of CA-SP1 processing (FIG. 14).
[0166]The SP1 residues associated with DSB activity consist of the contiguous residues E365 through V370.
[0167]Residues A364 through V370 were inserted into the analogous position of the Gag SP1 domain in the DSB-resistant retrovirus SIV (Mac 239 isolate). Additionally, the N-terminus of the CA protein of this chimeric virus was modified by cumulative substitution of residues found in SIV with HIV-1-specific residues. This approach is summarized in FIG. 15. Next, the effect of DSB on the Gag processing phenotype of each of the chimeric viruses was determined. As shown in FIG. 15, the SIV.DM virus displays a Gag processing phenotype indicative of sensitivity to DSB. Thus, the minimum sequence of HIV-1 CA-SP1-specific residues that needs to be inserted to rescue DSB activity in the SHIVs extends from V362 to V370
TABLE-US-00005 TABLE 1 PCR Mutagenesis Primers Primer PCR ID Sequence (5' to 3') Construct application 1 agccaaaactcttgctttatggcc ΔE365 First PCR (SEQ ID NO: 37) fragment (with No. 2 primer) 2 agtcagtgtggaaaatctctagcagtgg All All first (SEQ ID NO: 38) deletion PCR constructs fragments of NL4-3 3 gcaatgagccaagtaacaaatcca ΔE365 Second PCR (SEQ ID NO: 39) fragment (with No. 4 primer) 4 aggtatggtaaatgcagtatacttcctgaag All All second (SEQ ID NO: 40) deletion PCR constructs fragments 5 ttcagccaaaactcttgctttatggcc ΔA366 First PCR (SEQ ID NO: 41) fragment with No. 2 primer) 6 atgagccaagtaacaaatccagc ΔA366 Second PCR (SEQ ID NO: 42) fragment (with No. 4 primer) 7 tgcttcagccaaaactcttgc ΔM367 First PCR (SEQ ID NO: 43) fragment (with No. 2 primer) 8 agccaagtaacaaatccagct ΔM367 Second PCR (SEQ ID NO: 44) fragment (with No. 4 primer) 9 cattgcttcagccaaaactcttgc ΔS368 First PCR (SEQ ID NO: 45) fragment (with No. 2 primer) 10 caagtaacaaatccagctacca ΔS368 Second PCR (SEQ ID NO: 46) fragment (with No. 4 primer) 11 gctcattgcttcagccaaaactctt ΔQ369 First PCR (SEQ ID NO: 47) fragment (with No. 2 primer) 12 gtaacaaatccagctaccataa ΔQ369 Second PCR (SEQ ID NO: 48) fragment (with No. 4 primer) 13 acaaatccagctaccataatgatac ΔV370 First PCR (SEQ ID NO: 49) fragment (with No. 2 primer) 14 ttggctcattgcttcagccaaaactc ΔV370 Second PCR (SEQ ID NO: 50) fragment (with No. 4 primer) 15 tacttggctcattgcttcagccaa ΔT371 First PCR (SEQ ID NO: 51) fragment (with No. 2 primer) 16 aatccagctaccataatgatacag ΔT371 Second PCR (SEQ ID NO: 52) fragment (with No. 4 primer) 17 tgttacttggctcattgcttc ΔN372 First PCR (SEQ ID NO: 53) fragment (with No. 2 primer) 18 ccagctaccataatgatacagaaa ΔN372 Second PCR (SEQ ID NO: 54) fragment (with No. 4 primer) 19 atttgttacttggctcattgcttc ΔP373 First PCR (SEQ ID NO: 55) fragment (with No. 2 primer) 20 gctaccataatgatacagaaaggcaa ΔP373 Second PCR (SEQ ID NO: 56) fragment (with No. 4 primer) 21 tggatttgttacttggctcattgc ΔA374 First PCR (SEQ ID NO: 57) fragment (with No. 2 primer) 22 accataatgatacagaaaggc ΔA374 Second PCR (SEQ ID NO: 58) fragment (with No. 4 primer) 23 agctggatttgttacttggctc ΔT375 First PCR (SEQ ID NO: 59) fragment (with No. 2 primer) 24 ataatgatacagaaaggcaattttagg ΔT375 Second PCR (SEQ ID NO: 60) fragment (with No. 4 primer) 25 ggtagctggatttgttacttg ΔI376 First PCR (SEQ ID NO: 61) fragment (with No. 2 primer) 26 atgatacagaaaggcaattttaggaacc ΔI376 Second PCR (SEQ ID NO: 62) fragment (with No. 4 primer) 27 tatggtagctggatttgttac ΔM377 First PCR (SEQ ID NO: 63) fragment (with No. 2 primer) 28 atacagaaaggcaattttagg ΔM377 Second PCR (SEQ ID NO: 64) fragment (with No. 4 primer) 29 ccacctatcccagtaggag Sequencing For NL4-3 (SEQ ID NO: 65) primer mutants 30 ggcacagcaagcagcagctg Sequencing For NL4-3 (SEQ ID NO: 66) primer mutants 31 gtagaccaacagcaccatctagcggcaga All For SIV (SEQ ID NO: 67) substitution mutants constructs of SIV 32 ggtaaagtaaaggcagtgtactgcctaa All For SIV (SEQ ID NO: 68) substitution mutants constructs of SIV 33 cactggtgcgaggacctgactcatggcttctgccatt SIV DD First PCR (SEQ ID NO: 69) fragment (with No. 31 primer) 34 aatggcagaagccatgagtcaggtcctcgcaccagtg SIV DD Second PCR (SEQ ID NO: 70) fragment (with No. 32 primer) 35 ggcttctgccagtactctagccttctgt SIV DE First PCR (SEQ ID NO: 71) fragment (with No. 31 primer) 36 acagaaggctagagtactggcagaagcc SIV DE Second PCR (SEQ ID NO: 72) fragment (with No. 32 primer) 37 ggcttctgccagtactctagccttctgt SIV DM First PCR (SEQ ID NO: 73) fragment (with No. 31 primer) 38 acagaaggctagagtactggcagaagcc SIV DM Second PCR (SEQ ID NO: 74) fragment (with No. 32 primer) 39 atccaactggggttgcaaaaatgtg Sequencing For SIV (SEQ ID NO: 75) primer mutant
[0168]The resistance and mutagenesis data presented above suggest that the GHKARVL-AEAMSQV amino acid sequence in the region of the HIV-1 Gag CA-SP1 cleavage site serves as a genetic determinant of viral sensitivity to DSB.
Extending the Determinants of PA-457DSB Sensitivity to Other Lentiviruses
CA-SP1 Chimeras as Animal Efficacy Models for Development of Maturation Inhibitors
[0169]The development of anti-HIV therapeutics has been hindered by the lack of an animal efficacy model. This lack of an animal model is primarily due to the inability of most HIV-1 strains to replicate and cause disease in non-human primates. In some instances this problem has been overcome through the use of chimeric viruses that incorporate the region(s) of interest from the HIV-1 viral target into an SIV viral backbone that will support replication in a non-human primate. The most notable example of this approach involves the HIV-1/SIV (SHIV) chimeric viruses in which the proteins making up the infectious virus are exclusively SIV in origin with the exception of Env (gp120/gp41) which is derived form HIV-1. These SHIV envelope chimeras have been used extensively in HIV-1 vaccine development.
[0170]HIV-1 maturation inhibitors disrupt Gag CA-SP1 processing, which results in the formation and release of non-infectious viral particles exhibiting aberrant core morphology. See e.g. Li et al. Proc Natl Acad Sci USA. 100:13555-60 (2003). The betulinic acid derivative PA-457 DSB is an example of this class of inhibitors. The viral genetic determinants critical that are associated with the activity of maturation inhibitors map to amino acid residues flanking the HIV-1 CA-SP1 cleavage site. When this determinant is introduced into the CA-SP1 cleavage sites of PA-457DSB-resistant non-HIV-1 viruses, maturation inhibitor sensitive chimeras result. These CA-SP1 chimeric viruses serve as the basis for an animal efficacy model for HIV-1 maturation inhibitors.
[0171]The region of HIV-1 CA-SP1 necessary for maturation inhibitor sensitivity is introduced into selected lentiviruses. Amino acid residues from HIV-1 CA-SP1 junction that are determinants of PA-457DSB sensitivity were used to replace the corresponding CA-SP1 amino acids in the genome of Simian Immunodeficiency (SIV). Similarly, the amino acid residues from HIV-1 CA-SP1 junction that are determinants of PA-457DSB can be replaced in Feline Immunodeficiency virus (FIV), Bovine Immunodeficiency Virus (BIV), Equine Infectious Anemia Virus (EIAV), Visna-Maedi, and Caprine Arthritis Encephalitis virus (CAEV). Table 2 depicts the Gag polypeptide sequence for HIV-1, SIV, FUV, EIAV and BIV in the region of the CA-SP1 cleavage site.
TABLE-US-00006 TABLE 2 Sequence comparison in the region of the CA-SP1 cleavage site region of HIV-1 with SIV, FIV, EIAV and BIV CA SP1 NC HIV-1 GHKARVL AEAMSQVTNPATIM IQKG (SEQ ID NO: 76) FIV GYKMQLL AEALTKVQ VVQS (SEQ ID NO: 77) EIAV KQKMMLL AKALQ TGLA (SEQ ID NO: 78) BIV KSKMQFL VAAMKEMGIQSPIPAVLPHTPEAYA SQTS (SEQ ID NO: 79)
[0172]The HIV-1 CA-SP1 sequence used for replacement is as follows:
TABLE-US-00007 CA SP1 GHKARVL AEAMSQV (SEQ ID NO: 80)
[0173]The method described above for generating the SHIV CA-SP1 chimeric provirus DNA clone is used to generate FIV, EIAV and BIV provirus clones containing selected residues or extended region from CA-SP1 region of HIV-1 replacing the corresponding wild-type sequence (FIG. 16).
[0174]The SHIV CA-SP1 chimeric-provirus DNA clone was generated by site-directed mutagenesis employing standard molecular biology techniques. Briefly, the unique restriction enzyme sites in the SIV Gag that surrounding the CA-SP1 region were identified i.e., BamHI (in matrix) and Sbf-I (in NC). Starting from the CA-SP1 region where the mutagenisis is intended two overlapping primers, a forward and a reverse primer incorporating the mutated sequence i.e., HIV CA-SP1 at their 5' ends were synthesized. Using the wild-type SIV provirus DNA as a template, two separate PCR reactions were set up to amplify SIV-Gag fragments in either direction from the site of mutagenisis (CA-SP1 region), i.e., yield two amplified fragments that overlapped in the mutated CA-SP1 region, a Bam HI-CA-SP1 fragment and a CA-SP1-Sbf-I fragment. In a third PCR reaction, the fragments, Bam HI-CA-SP1 and CA-SP1-Sbf-I were annealed at their common HIV-CA-SP1 sequence and amplified with a forward SIV Bam HI primer and a reverse SIV Sbf-I primer to generate a full-length chimeric SHIV CA-SP1 gag fragment. The chimeric SHIV CA-SP1 PCR fragment was cloned into BamHI-Sbf-I window of SIV provirus clone replacing the SIV-Gag wild-type sequence to yield the SHIV CA-SP1 provirus cDNA clone.
[0175]Similarly, unique restriction enzyme cloning sites surrounding the CA-SP1 regions in FIV (Genbank Accession # NC--001482), EIAV (GA# AF016316), and BIV (GA# M32690) genome have been identified (FIG. 16). Specific FIV/EIAV/BIV-HIV1CA-SP1 chimeric primers along with genome specific primers of FIV, EIAV or BIV incorporating the specific cloning site sequence are synthesized. These primers along with corresponding provirus DNA clone as template (FIV, EIAV or BIAV) in PCR reactions to generate the Chimeric HIV-1 CA-SP1 fragment. The chimeric HIV-1 CA-SP1 fragment is digested with the appropriate restriction enzyme and cloned into SacI-EcoRI window of FIV provirus; or (ii) KasI-EcoRV window of EIAV provirus; or (iii) BsrGI-ApaI window of BIV provirus replacing the corresponding wild type sequence (FIG. 16). The chimeric FIV/EIAV/BIV-HIV-1CA-SP1 provirus DNA clones are sequenced to confirm the presence of intended mutations. Based on observed results that indicate the transfer of PA-457DSB sensitivity, additional constructs are generated employing the above strategy in order to optimize the results.
[0176]In summary, a chimeric virus was generated in which the CA-SP1 determinant of HIV-1 maturation inhibitor sensitivity has replaced the analogous region of Gag in the maturation inhibitor-resistant simian immunodeficiency virus (SIV). Transfer of this region of HIV-1 into the genome of SIV results in a maturation inhibitor-sensitive phenotype. Infection of a non-human primate with this HIV-1/SIV chimeric virus should result in an animal efficacy model for therapeutic development of maturation inhibitors.
[0177]Analogous approaches are used to prepare and characterize HIV-1 CA-SP1 chimeras with FIV, BIV and EIAV. These additional DSB-sensitive chimeric viruses should enable the development of additional animal efficacy models for the study of HIV-1 maturation inhibitors.
Uses of Mutant and Chimeric Viruses
[0178]The mutant and chimeric viruses of the present invention, as described above, are useful in a variety of cell based as well as animal based assays.
[0179]by comparing the phenotypes associated with a virus that is resistant to DSB, with a virus that is sensitive DSB, one may identify compounds that act by a mechanism similar to that of DSB. Thus the invention includes a method of identifying a compound that inhibits cleavage of p25 to p24 in wild type HIV-1, but does not inhibit CA-SP1 processing in HIV-1 containing a deletion in the CA-SP1 region. Compounds obtained by such a method are also included in the present invention.
[0180]Chimeras of SIV and other lentiviruses that do not readily infect humans have additional advantages. Firstly, these viruses pose a lesser safety hazard to laboratory workers. As a result, cell based assays to identify novel compounds that inhibit CA-SP1 processing, for example, can be conducted with less risk. The lower risk may allow assays to be performed that cannot be performed readily or safely with HIV, and may also lower the cost of such assays.
[0181]Furthermore, such chimeric viruses are useful in animal models. For example chimeric SIV that is sensitive to DSB may be used to identify novel compounds that inhibit CA-SP1 processing, for example; to identify pharmaceutical compositions, routes of administration and dosage regimes for treatment of disease; and for studying the effect of combination therapies, such as DSB with protease inhibitors.
[0182]As SIV is generally limited to infection of monkeys, the generation of additional lentiviral chimeras allows animal studies to be performed in animals that are less expensive, easier to handle, have a faster disease progression or otherwise more appropriate for a particular aspect of human disease, for example.
[0183]Furthermore, animal models may be used to identify appropriate pharmaceutical compositions for the treatment of animal diseases, of interest in the treatment of companion animals and other high value animals, such as agricultural breeding stock and race horses.
[0184]Chimeric viruses may be derived from any retrovirus. For example, derived HIV-2, HTLV-I, HTLV-II, SIV, avian leukosis virus (ALV), endogenous avian retrovirus (EAV), mouse mammary tumor virus (MMTV), feline immunodeficiency virus (FIV), Bovine immunodeficiency virus (BIV), caprine arthritis encephalitis virus (CAEV), Equine infectious anemia virus (EIAV), Visna-maedi virus, or feline leukemia virus (FeLV).
[0185]Such chimeric viruses may be used in the methods of the invention described elsewhere herein. For example, such recombinant non-HIV-1 lentiviruses may be used in a method of identifying a compound which inhibit processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), the method consisting of comparing of the ability of said compound to inhibit replication of a wild-type non-HIV-1 lentivirus with the DSB-sensitive recombinant variant thereof. Such inhibition may occur in a cell; in an animal; or in vitro.
Construction and Use of Viruses or Polypeptides with Epitope Tags
[0186]The present invention is also drawn to recombinant retroviruses with epitope tags in the CA-SP1 region of Gag. Epitope tags may be inserted in the CA domain and/or in the SP1 domain. Suitable tags are well known to those of ordinary skill in the art, and include haemagglutinin epitope HA (YPYDVPDYA) (SEQ ID NO: 81), bluetongue virus epitope VP7 (QYPALT) (SEQ ID NO: 82), α-tubulin epitope (EEF), Flag (DYKDDDDK) (SEQ ID NO: 83), and VSV-G (YTDOEMNRLGK) (SEQ ID NO: 84). Examples of SP1 containing epitope tags are illustrated in FIG. 17.
[0187]Such epitope tagged viruses and fragments thereof are useful in identifying novel compounds that inhibit CA-SP1 processing in vitro, in cell based assays, and in vivo, including in animal models. Additional uses of such epitope tagged viruses and fragments thereof are described elsewhere herein.
Polynucleotides, Polypeptides and Antibodies of the Invention
[0188]The invention also includes isolated polypeptides and polynucleotides. In one embodiment, the invention includes polypeptides at least about 40%, 50%, 60%, 70%, 80%, 90%, 99% or 100% identical to an amino acid sequence selected from the group consisting of: (a) KNWMTETFLVQNANPDCKTILKALGPAATLEEMMTACQGVGGPH KARILAEAMSQVTNSATIM (SEQ ID NO: 21); (b) KNWMTETLLVQNANPDCKTILKALGPGATLEEMMTACQGVGGPG HKARVLAEAMSQVTNPATIM (SEQ ID NO: 22); (c) TACQGVGGPSHKARILAEAMSQVTNSATIM (SEQ ID NO: 23); (d) MTACQGVGGPGHKARVLAEAMSQVTNPATIM (SEQ ID NO: 24); (e) SHKARILAEAMSQV (SEQ ID NO: 25); and (f) GHKARVLAEAMSQV (SEQ ID NO: 26).
[0189]In another embodiment the invention includes polynucleotides encoding the above polypeptides. Polynucleotides of the invention include degenerate variants, such as those that differ in the third base of the codon but nevertheless encodes the same amino acid due to coding "degeneracy".
[0190]The term "about" as used herein refers to a value that is 10% more or less than the stated value, and preferably is 5% more or less.
[0191]The polypeptides and polynucleotides of the invention are useful in the methods of the invention. In one aspect, they may be used in an in vitro assay to identify compounds that bind to the CA-SP1 region of Gag. In another, they may be used in the production of antibodies useful in other methods described elsewhere herein. In another, a polynucleotide may be inserted into a vector and thereupon into a host cell for production of polypeptide. The above embodiments are exemplary and are not intended to be limiting.
[0192]The present invention comprises a polynucleotide comprising a sequence which encodes an amino acid sequence containing a mutation in the HIV Gag p25 protein (CA-SP1), said mutation resulting in a decrease in the inhibition of processing of p25 (CA-SP1) to p24 (CA) by DSB. The polynucleotide of the invention includes a mutation which is optionally located at or near the CA-SP1 cleavage site or located in the SP1 domain of CA-SP1. Said mutation can be present in an amino acid sequence that is selected from the group consisting of GHKARVLVEAMSQV (SEQ ID NO: 2) and SHKARILAEVMSQV (SEQ ID NO: 3). The polynucleotide of this invention is also drawn to sequences designated as SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8 and SEQ ID NO: 9. The invention also includes a vector comprising said polynucleotide, a host cell comprising said vector and a method of producing said polypeptides comprising incubating said host cell in a medium and recovering the polypeptide from the medium.
[0193]The invention further includes a polynucleotide that hybridizes under stringent conditions to a polynucleotide selected from the group consisting of SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8 and SEQ ID NO: 9. The invention also includes a polynucleotide which hybridizes to SEQ NO: 5, SEQ ID NO: 7 or SEQ ID NO: 10 or 12, which contains a mutation which results in the decrease in the inhibition of processing of p25 to p24 by 3-O-(3',3'-dimethylsuccinyl)betulinic acid, and also wherein said mutation is optionally located at or near the CA-SP1 cleavage site or in the SP1 domain of CA-SP1. The invention is also directed to a vector comprising said polynucleotides, a host cell comprising said vector and a method of producing said polypeptides, comprising incubating said host cell in a medium and recovering said polypeptide from the medium.
[0194]Near" or "adjacent," as used herein in reference to polypeptides is meant to include about 50, about 25, about 20, or about 15 residues from the point of reference. For example, near may encompass about 50, about 25, about 20 or about 15 residues on either side of the HIV-1 Gag CA-SP1 cleavage site; more preferably about ten residues on either side of the HIV-1 Gag CA-SP1 cleavage site; and most preferably about seven residues on either side of the HIV-1 Gag CA-SP1 cleavage site. In reference to polynucleotides, the terms "near" or "adjacent refer to about 150, about 75, about 60, about 45, or about 30 nucleotides from the point of reference.
[0195]Isolated" means altered "by the hand of man" from the natural state. If an "isolated" composition or substance occurs in nature, it has been changed or removed from its original environment, or both. Also, "isolated" nucleic acid molecule(s) of the invention is intended a nucleic acid molecule, DNA or RNA, which has been removed from its native environment For example, recombinant DNA molecules contained in a vector are considered isolated for the purposes of the present invention. Further examples of isolated DNA molecules include recombinant DNA molecules maintained in heterologous host cells or purified (partially or substantially) DNA molecules in solution. Isolated RNA molecules include in vivo or in vitro RNA transcripts of the DNA molecules of the present invention. Isolated nucleic acid molecules according to the present invention further include such molecules produced synthetically.
[0196]Polynucleotide" generally refers to any polyribonucleotide or polydeoxyribonucleotide, which may be unmodified RNA or DNA or modified RNA or DNA. "Polynucleotides" include, without limitation single- and double-stranded DNA, DNA that is a mixture of single- and double-stranded regions, single- and double-stranded RNA, and RNA that is mixture of single- and double-stranded regions, hybrid molecules comprising DNA and RNA that may be single-stranded or, more typically, double-stranded or a mixture of single- and double-stranded regions. In addition, "polynucleotide" refers to triple-stranded regions comprising RNA or DNA or both RNA and DNA. The term polynucleotide also includes DNAs or RNAs containing one or more modified bases and DNAs or RNAs with backbones modified for stability or for other reasons. "Modified" bases include, for example, tritiated bases and unusual bases such as inosine. A variety of modifications has been made to DNA and RNA; thus, "polynucleotide" embraces chemically, enzymatically or metabolically modified forms of polynucleotides as typically found in nature, as well as the chemical forms of DNA and RNA characteristic of viruses and cells. "Polynucleotide" also embraces relatively short polynucleotides, often referred to as oligonucleotides.
[0197]Polypeptide" refers to any peptide or protein comprising two or more amino acids joined to each other by peptide bonds or modified peptide bonds, i.e., peptide isosteres. "Polypeptide" refers to both short chains, commonly referred to as peptides, oligopeptides or oligomers, and to longer chains, generally referred to as proteins. Polypeptides may contain amino acids other than the 20 gene-encoded amino acids. "Polypeptides" include amino acid sequences modified either by natural processes, such as post-translational processing, or by chemical modification techniques which are well known in the art. Such modifications are well described in basic texts and in more detailed monographs, as well as in a voluminous research literature. Modifications can occur anywhere in a polypeptide, including the peptide backbone, the amino acid side-chains and the amino or carboxyl termini. It will be appreciated that the same type of modification may be present in the same or varying degrees at several sites in a given polypeptide. Also, a given polypeptide may contain many types of modifications. Polypeptides may be branched as a result of ubiquitination, and they may be cyclic, with or without branching. Cyclic, branched and branched cyclic polypeptides may result from post-translation natural processes or may be made by synthetic methods. Modifications include acetylation, acylation, ADP-ribosylation, amidation, covalent attachment of flavin, covalent attachment of a heme moiety, covalent attachment of a nucleotide or nucleotide derivative, covalent attachment of a lipid or lipid derivative, covalent attachment of phosphotidylinositol, cross-linking, cyclization, disulfide bond formation, demethylation, formation of covalent cross-links, formation of cystine, formation of pyroglutamate, formylation, gamma-carboxylation, glycosylation, GPI anchor formation, hydroxylation, iodination, methylation, myristoylation, oxidation, proteolytic processing, phosphorylation, prenylation, racemization, selenoylation, sulfation, transfer-RNA mediated addition of amino acids to proteins such as arginylation, and ubiquitination.
[0198]Mutant" as the term is used herein, is a polynucleotide or polypeptide that differs from a reference polynucleotide or polypeptide respectively. A typical mutant of a polynucleotide differs in nucleotide sequence from another, reference polynucleotide. Changes in the nucleotide sequence of the mutant may or may not alter the amino acid sequence of a polypeptide encoded by the reference polynucleotide. Nucleotide changes may result in amino acid substitutions, additions, deletions, fusions and truncations in the polypeptide encoded by the reference sequence, as discussed below. A typical mutant of a polypeptide differs in amino acid sequence from another, reference polypeptide. Generally, differences are limited so that the sequences of the reference polypeptide and the variant are closely similar overall and, in many regions, identical. A mutant and reference polypeptide may differ in amino acid sequence by one or more substitutions, additions, deletions in any combination. A substituted or inserted amino acid residue may or may not be one encoded by the genetic code. A mutant of a polynucleotide or polypeptide may be a naturally occurring such as an allelic variant, or it may be a mutant that is not known to occur naturally. Non-naturally occurring mutants of polynucleotides and polypeptides may be made by mutagenesis techniques or by direct synthesis.
[0199]Thus, the mutant, (or fragments, derivatives or analogs) of a polypeptide encoded by any one of the polynucleotides described herein may be (i) one in which at least one or more of the amino acid residues are substituted with a conserved or non-conserved amino acid residue (a conserved amino acid residue(s), or at least one but less than ten conserved amino acid residues) and such substituted amino acid residue may or may not be one encoded by the genetic code, (ii) one in which one or more of the amino acid residues includes a substituent group, (iii) one in which the mature polypeptide is fused with another compound, such as a compound to increase the half-life of the polypeptide (for example, polyethylene glycol), or (iv) one in which the additional amino acids are fused to the mature polypeptide, such as an IgG:Fc fusion region peptide or leader or secretory sequence or a sequence which is employed for purification of the mature polypeptide or a proprotein sequence. Such mutants are deemed to be within the scope of those skilled in the art from the teachings herein. Polynucleotides encoding these mutants are also encompassed by the invention. "Mutant" as used herein is equivalent to the term "variant."
[0200]Substitutions of charged amino acids with another charged amino acids and with neutral or negatively charged amino acids are included. Additionally, one or more of the amino acid residues of the polypeptides of the invention (e.g., arginine and lysine residues) may be deleted or substituted with another residue to eliminate undesired processing by proteases such as, for example, furins or kexins. The prevention of aggregation is highly desirable. Aggregation of proteins not only results in a loss of activity but can also be problematic when preparing pharmaceutical formulations, because they can be immunogenic. (Pinckard et al., Clin Exp. Immunol. 2:331-340 (1967); Robbins et al., Diabetes 36:838-845 (1987); Cleland et al Crit. Rev. Therapeutic Drug Carrier Systems 10:307-377 (1993)). Thus, the polypeptides of the present invention may include one or more amino acid substitutions, deletions or additions, either from natural mutations or human manipulation.
[0201]As indicated, changes are preferably of a minor nature, such as conservative amino acid substitutions that do not significantly affect the folding or activity of the protein (see Table 3).
TABLE-US-00008 TABLE 3 Conservative Amino Acid Substitutions Aromatic Phenylalanine Tryptophan Tyrosine Hydrophobic Leucine Isoleucine Valine Polar Glutamine Asparagine Basic Arginine Lysine Histidine Acidic Aspartic Acid Glutamic Acid Small Alanine Serine Threonine Methionine Glycine
[0202]However, in some embodiments, it is desirable to use nonconservative substitutions of amino acids. For example nonconservative substitution of amino acids is used to render a DSB sensitive virus resistant to DSB.
[0203]The polynucleotides encompassed by this invention may have at least about 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95% or 100% identity with a reference sequence, providing the reference polynucleotide encodes an amino acid sequence containing a mutation in the CA-SP1 protein, said mutation which results in the decrease in the inhibition of processing of p25 to p24 by a 3-O-(3',3'-dimethylsuccinyl)betulinic acid. The polynucleotides also encompassed by this invention include those mutations which are "silent," in which different codons encode the same amino acid (wobble).
[0204]Identity" is a measure of the identity of nucleotide sequences or amino acid sequences. The term "identity" is used interchangeably with the word "homology" herein. In general, the sequences are aligned so that the highest order match is obtained. "Identity" per se has an art-recognized meaning and can be calculated using published techniques. While there exist a number of methods to measure identity between two polynucleotide or polypeptide sequences, the term "identity" is well known to skilled artisans. Methods commonly employed to determine identity or similarity between two sequences include, but are not limited to, those disclosed in Baxevanis and Oullette, Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins, Second Edition, Wiley-Interscience, New York, (2001). Methods to determine identity and similarity are codified in computer programs. Preferred computer program methods to determine identity and similarity between two sequences include, but are not limited to, GCS program package (Devereux, J. et al., Nucleic Acids Research 12(1):387, (1984)), BLASTP, BLASTN, FASTA (Atschul, S. F. et al., J. Molec. Biol. 215:403, (1990)).
[0205]A polynucleotide having a nucleotide sequence having at least, for example, 95% "identity" to a reference nucleotide sequence is intended that the nucleotide sequence of the polynucleotide is identical to the reference sequence except that the polynucleotide sequence may include up to five point mutations per each 100 nucleotides of the reference nucleotide sequence, up to 5% of the nucleotides in the reference sequence may be deleted or substituted with another nucleotide, or a number of nucleotides up to 5% of the total nucleotides in the reference sequence may be inserted into the reference sequence. These mutations of the reference sequence may occur at the 5' or 3' terminal positions of the reference nucleotide sequence or anywhere between those terminal positions, interspersed either individually among nucleotides in the reference sequence or in one or more contiguous groups within the reference sequence.
[0206]Similarly, by a polypeptide having an amino acid sequence having at least, for example, 95% "identity" to a reference amino acid sequence, is intended that the amino acid sequence of the polypeptide is identical to the reference sequence except that the polypeptide sequence may include up to five amino acid alterations per each 100 amino acids of the reference amino acid. To obtain a polypeptide having an amino acid sequence at least 95% identical to a reference amino acid sequence, up to 5% of the amino acid residues in the reference sequence may be deleted or substituted with another amino acid, or a number of amino acids up to 5% of the total amino acid residues in the reference sequence may be inserted into the reference sequence. These alterations of the reference sequence may occur at the amino or carboxy terminal positions of the reference amino acid sequence or anywhere between those terminal positions, interspersed either individually among residues in the reference sequence or in one or more contiguous groups within the reference sequence. The reference (query) sequence may be the entire nucleotide sequence of any one of the nucleotide sequences of the invention or any polynucleotide fragment (e.g., a polynucleotide encoding the amino acid sequence of the invention and/or C terminal deletion).
[0207]Whether any particular nucleic acid molecule having at least 40%, 50%, 60%, 70%, 80%, 85%, 90%, 92%, 95%, 96%, 97%, 98%, 99% identity or which are identical to, for instance, the nucleotide sequences of the invention can be determined conventionally using known computer programs such as the BESTFIT program (Wisconsin Sequence Analysis Package, Version 8 for Unix, Genetics Computer Group, University Research Park, 575 Science Drive, Madison, Wis. 53711). BESTFIT uses the local homology algorithm of Smith and Waterman, (Advances in Applied Mathematics 2:482-489 (1981)), to find the best segment of homology between two sequences. When using BESTFIT or any other sequence alignment program to determine whether a particular sequence is, for instance, 95% identical to a reference sequence according to the present invention, the parameters are set, such that the percentage of identity is calculated over the full length of the reference nucleotide sequence and that gaps in homology of up to 5% of the total number of nucleotides in the reference sequence are allowed.
[0208]In a specific embodiment, the identity between a sequence of the present invention and a subject sequence, also referred to as a global sequence alignment, is determined using the FASTDB computer program based on the algorithm of Brutlag et al. (Comp. App. Biosci. 6:237-245 (1990)). Preferred parameters used in a FASTDB alignment of DNA sequences to calculate percent identity are: Matrix=Unitary, k-tuple=4, Mismatch Penalty=1, Joining Penalty=30, Randomization Group Length=0, Cutoff Score=1, Gap Penalty=5, Gap Size Penalty 0.05, Window Size=500 or the length of the subject nucleotide sequence, whichever is shorter. According to this embodiment, if the subject sequence is shorter than the reference sequence because of 5' or 3' deletions, not because of internal deletions, a manual correction is made to the results to take into consideration the fact that the FASTDB program does not account for 5' and 3' truncations of the subject sequence when calculating percent identity. For subject sequences truncated at the 5' or 3' ends, relative to the query sequence, the percent identity is corrected by calculating the number of bases of the query sequence that are 5' and 3' of the subject sequence, which are not matched/aligned, as a percent of the total bases of the query sequence. A determination of whether a nucleotide is matched/aligned is determined by results of the FASTDB sequence alignment. This percentage is then subtracted from the percent identity, calculated by the above FASTDB program using the specified parameters, to arrive at a final percent identity score. This corrected score is what is used for the purposes of this embodiment. Only bases outside the 5' and 3' bases of the subject sequence, as displayed by the FASTDB alignment, which are not matched/aligned with the query sequence, are calculated for the purposes of manually adjusting the percent identity score. For example, a 90 base subject sequence is aligned to a 100 base query sequence to determine percent identity. The deletions occur at the 5' end of the subject sequence and therefore, the FASTDB alignment does not show a matched/alignment of the first 10 bases at 5' end. The 10 unpaired bases represent 10% of the sequence (number of bases at the 5' and 3' ends not matched/total number of bases in the query sequence) so 10% is subtracted from the percent identity score calculated by the FASTDB program. If the remaining 90 bases were perfectly matched the final percent identity would be 90%. In another example, a 90 base subject sequence is compared with a 100 base query sequence. This time the deletions are internal deletions so that there are no bases on the 5' or 3' of the subject sequence, which are not matched/aligned with the query. In this case the percent identity calculated by FASTDB is not manually corrected. Only bases 5' and 3' of the subject sequence which are not matched/aligned with the query sequence are manually corrected. No other manual corrections are made for the purposes of this embodiment.
[0209]The present application is directed to nucleic acid molecules having at least about 40%, 50%, 60%, 70%, 80%, 85%, 90%, 92%, 95%, 96%, 97%, 98% or 99% identity or which is identical to the nucleic acid sequence disclosed herein, or fragments thereof, irrespective of whether they encode a polypeptide having the disclosed functional activity. This is because even where a particular nucleic acid molecule does not encode a polypeptide having the disclosed functional activity, one of skill in the art would still know how to use the nucleic acid molecule, for instance, as a hybridization probe or a polymerase chain reaction (PCR) primer. Uses of the nucleic acid molecules of the present invention that do not encode a polypeptide having the disclosed functional activity include, inter alia: (1) isolating the variants thereof in a cDNA library; (2) in situ hybridization (e.g., "FISH") to determine cellular location or presence of the disclosed sequences, and (3) Northern Blot analysis for detecting mRNA expression in specific tissues.
[0210]As used herein, the term "PCR" refers to the polymerase chain reaction that is the subject of U.S. Pat. Nos. 4,683,195 and 4,683,202 to Mullis et al., as well as improvements now known in the art. In accordance with the present invention there may be employed conventional molecular biology, microbiology, and recombinant DNA techniques within the skill of the art. Such techniques are explained fully in the literature. See, for example, Sambrook, J. and Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
[0211]The term "stringent conditions," as used herein refers to homology in hybridization, is based upon combined conditions of salt, temperature, organic solvents, and other parameters typically controlled in hybridization reactions, and well known in the art (Sambrook, et al. supra). The invention includes an isolated nucleic acid molecule comprising, a polynucleotide which hybridizes under stringent hybridization conditions to a portion of the polynucleotide in a nucleic acid molecule of the invention described above, for instance, the sequence complementary to the coding and/or noncoding (i.e., transcribed, untranslated) sequence of any polynucleotide or a polynucleotide fragment as described herein. By "stringent hybridization conditions" is intended overnight incubation at 42° C. in a solution comprising, or alternatively consisting of: 50% formamide, 5×SSC (750 mM NaCl, 75 mM trisodium citrate), 50 mM sodium phosphate (pH 7.6), 5×Denhardt's solution, 10% dextran sulfate, and 20 μg/ml denatured, sheared salmon sperm DNA, followed by washing in 0.1×SSC at about 65° C. Polypeptides encoded by these polynucleotides are also encompassed by the invention.
[0212]The invention also includes a virus comprising the polynucleotides of the invention, and wherein the virus includes a retrovirus comprising said polynucleotides, and wherein the retrovirus may be a member of the group consisting of HIV-1, HIV-2, HTLV-I, HTLV-II, SIV, avian leukosis virus (ALV), endogenous avian retrovirus (EAV), mouse mammary tumor virus (MMTV), feline immunodeficiency virus (FIV), or feline leukemia virus (FeLV).
[0213]The invention further includes a polypeptide containing a mutation in the CA-SP1 protein, said mutation which results in the decrease in inhibition of processing of p25 to p24 by 3-O-(3',3'-dimethylsuccinyl)betulinic acid, and also wherein said mutation is optionally located at or near the CA-SP1 cleavage site or located in the SP1 domain of SEQ ID NO: 5 or SEQ ID NO: 7 (parental polynucleotide sequences) encoding the CA-SP1 protein. Said polypeptide may be encoded by a polynucleotide selected from the group consisting of SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8 and SEQ ID NO: 9, or may comprise a sequence that is selected from the group consisting of GHKARVLVEAMSQV (SEQ ID NO: 2) and SHKARILAEVMSQV (SEQ ID NO: 3). The polypeptide of this invention may further be encoded by a polynucleotide which hybridizes to a polynucleotide selected from the group consisting of SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8 and SEQ ID NO: 9. The invention also includes a polypeptide encoded by a polynucleotide which hybridizes to SEQ NO: 5, SEQ ID NO: 7 or SEQ ID NO: 10 or 12, which contains a mutation that results in decrease in inhibition of processing of p25 to p24 by 3-O-(3',3'-dimethylsuccinyl)betulinic acid, and also wherein said mutation is optionally located at or near the CA-SP1 cleavage site or in the SP1 domain of CA-SP1. The polypeptide of this invention further includes polypeptides that are part of a chimeric or fusion protein. Said chimeric proteins may be derived from species which include, but are not limited to: primates, including simian and human; rodentia, including rat and mouse; feline; bovine; ovine; including goat and sheep; canine; or porcine. Fusion proteins may include synthetic peptide sequences, bifunctional antibodies, peptides linked with proteins from the above species, or with linker peptides. Polypeptides of the invention may be further linked with detectable labels; metal compounds; cofactors; chromatography separation tags, such as, but not limited to: histidine, protein A, or the like, or linkers; blood stabilization moieties such as, but not limited to: transferrin, or the like; therapeutic agents, and so forth.
[0214]The invention also includes an antibody which selectively binds an amino acid sequence containing a mutation in the CA-SP1 protein that results in a decrease in the inhibition of processing of p25 (CA-SP1) to p24 (CA) by 3-O-(3',3'-dimethylsuccinyl)betulinic acid and also wherein said mutation is optionally located at or near the CA-SP1 cleavage site or in the SP1 domain of CA-SP1. The invention also includes an antibody which selectively binds the polypeptide having a mutation which comprises a sequence that is one of GHKARVLVEAMSQV (SEQ ID NO: 2), SHKARILAEVMSQV (SEQ ID NO: 3). Said antibody can selectively bind the polypeptide encoded by a polynucleotide sequence selected from the group consisting of SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8 and SEQ ID NO: 9. Said antibody can also selectively bind the polypeptide encoded by a polynucleotide which hybridizes under highly stringent conditions to a polynucleotide selected from the group consisting of SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8 and SEQ ID NO: 9. The invention also includes an antibody that selectively binds SP1, which would enable one to distinguish SP1 from CA-SP1 (p25). The invention also includes an antibody that selectively binds CA (p24), which would enable one to distinguish CA from CA-SP1. The invention also includes an antibody that selectively binds CA-SP1, which would enable one to distinguish CA from CA-SP1. The invention additionally includes an antibody that selectively binds at or near the CA-SP1 cleavage site. The antibody of this invention may be a polyclonal antibody, a monoclonal antibody or said antibody may be chimeric or bifunctional, or part of a fusion protein. The invention further includes a portion of any antibody of this invention, including single chain, light chain, heavy chain, CDR, F(ab')2, Fab, Fab', Fv, sFv, or dsFv, or any combinations thereof.
[0215]As used herein, an antibody "selectively binds" a target peptide when it binds the target peptide and does not significantly bind to unrelated proteins. The term "selectively binds" also comprises determining whether the antibody selectively binds to the target mutant sequence relative to a native target sequence. An antibody which "selectively binds" a target peptide is equivalent to an antibody which is "specific" to a target peptide, as used herein. An antibody is still considered to selectively bind a peptide even if it also binds to other proteins that are not substantially homologous with the target peptide so long as such proteins share homology with a fragment or domain of the peptide target of the antibody. In this case, it would be understood that antibody binding to the peptide is still selective despite some degree of cross-reactivity. In another embodiment, the determination whether the antibody selectively binds to the mutant target sequence comprises: (a) determining the binding affinity of the antibody for the mutant target sequence and for the native target sequences; and (b) comparing the binding affinities so determined, the presence of a higher binding affinity for the mutant target sequence than for the native indicating that the antibody selectively binds to the mutant target sequence.
[0216]The invention is further drawn to an antibody immobilized on an insoluble carrier comprising any of the antibodies disclosed herein. The antibody immobilized on an insoluble carrier includes multiple well plates, culture plates, culture tubes, test tubes, beads, spheres, filters, electrophoresis material, microscope slides, membranes, or affinity chromatography medium.
[0217]The invention also includes labeled antibodies, comprising a detectable signal. The labeled antibodies of this invention are labeled with a detectable molecule, which includes an enzyme, a fluorescent substance, a chemiluminescent substance, horseradish peroxidase, alkaline phosphatase, biotin, avidin, an electron dense substance, and a radioisotope, or any combination thereof.
[0218]The invention further includes a method of producing a hybridoma comprising fusing a mammalian myeloma cell with a mammalian B cell that produces a monoclonal antibody which selectively binds an amino acid sequence containing a mutation in the CA-SP1 protein, said mutation resulting in a decrease in the inhibition of processing of p25 to p24 by 3-O-(3',3'-dimethylsuccinyl)betulinic acid and a hybridoma producing any of the monoclonal antibodies disclosed herein. The invention further includes a method of producing an antibody comprising growing a hybridoma producing the monoclonal antibodies disclosed herein in an appropriate medium and isolating the antibodies from the medium, as is well known in the art. The invention also includes the production of polyclonal antibodies comprising the injection, either one injection or multiple injections of any of the polypeptides of the inventions into any animal known in the art to be useful for the production of polyclonal antibodies, including, but not limited to mouse, rat, hamster, rabbit, goat, sheep, deer, guinea pig, or primate, and recovering the antibodies in sera produced therein. The invention includes high avidity or high affinity antibodies produced therein. The invention also includes B cells produced from the listed species to be further used in cell fusion procedures for the manufacture of monoclonal antibody-producing hybridomas as disclosed herein.
[0219]The invention is further drawn to a kit comprising the antibody or a portion thereof as disclosed herein, a container comprising said antibody and instructions for use, a kit comprising the polypeptides of this invention and instructions for use and a kit comprising the polynucleotide of the invention, a container comprising said polynucleotide and instructions for use, or any combinations thereof. These kits would include, but not be limited to nucleic acid detection kits, which may, or may not, utilize PCR and immunoassay kits. Such kits are useful for clinical diagnostic use and provide standardized reagents as required in current clinical practice. These kits could either provide information as to the presence or absence of mutations prior to treatment or monitor the progress of the patient during therapy. The kits of the invention may also be used to provide standardized reagents for use in research laboratory studies.
Compounds of the Invention
[0220]In one aspect, the invention is also directed to a compound, a method of using a compound, a method of identifying a compound and the like.
[0221]The term "a", "an" or "one", as used in the present invention may refer to either the singular or the plural. For example, "a compound" encompasses one or more compounds.
[0222]Compounds useful in the methods of the present invention include derivatives of betulinic acid and betulin that are presented in U.S. Pat. Nos. 5,679,828 and 6,172,110 respectively, and in U.S. application Nos. 60/443,180 and 10/670,797, which are herein incorporated by reference. Additional useful compounds include oleanolic acid derivatives disclosed by Zhu et al. (Bioorg. Chem. Lett. 11:3115-3118 (2001)); oleanolic acid and pomolic acid derivatives disclosed by Kashiwada et al. (J. Nat. Prod. 61:1090-1095 (1998)); 3-O-acyl ursolic acid derivatives described by Kashiwada et al. (J. Nat. Prod. 63:1619-1622 (2000)); and 3-alkylamido-3-deoxy-betulinic acid derivatives, disclosed by Kashiwada et al. (Chem. Pharm. Bull. 48:1387-1390 (2000)). (All references incorporated by reference).
[0223]Compounds useful in the present invention include, but are not limited to those betulinic acid derivatives having the general Formula I and dihydrobetulinic acid derivatives of Formula II:
[0224]or a pharmaceutically acceptable salt thereof, wherein,
[0225]R is a C2-C20 substituted or unsubstituted carboxyacyl,
[0226]R' is hydrogen or a C2-C10 substituted and unsubstituted alkyl or aryl group. Preferred compounds are those wherein R is one of the substituents in Table 4, below, and R' is hydrogen.
[0227]Additional useful compounds include derivatives of betulin and dihydrobetulin of Formula III:
[0228]or a pharmaceutically acceptable salt thereof, wherein,
[0229]R1 is a C2-C20 substituted or unsubstituted carboxyacyl,
[0230]R2 is hydrogen, C(C6H5)3, or a C2-C20 substituted or unsubstituted carboxyacyl; and
[0231]R3 is hydrogen, halogen, amino, optionally substituted mono- or di-alkylamino, or --OR4, where R4 is hydrogen, C1-4 alkanoyl, benzoyl, or C2-C20 substituted or unsubstituted carboxyacyl;
[0232]wherein the dashed line represents an optional double bond between C20 and C29.
[0233]Preferred compounds useful in the present invention are those where
[0234]R1 is one of the substituents in Table 4, R2 is hydrogen or one of the substituents in Table 4 and R3 is hydrogen.
TABLE-US-00009 TABLE 4 Preferred Substituents for R1, R2.
[0235]More preferred compounds are 3-O-(3',3'-dimethylsuccinyl)betulinic acid, 3-O-(3',3'-dimethylsuccinyl)dihydrobetulinic acid, 3-O-(3',3'-dimethylsuccinyl)betulin, and 3-O-(3',3'-dimethylsuccinylglutaryl)dihydrobetulin.
[0236]A particularly preferred compound is 3-O-(3',3'-dimethylsuccinyl)betulinic acid.
[0237]Additional compounds useful in the present invention are described by the Formulas IV, V, VI and VII.
[0238]R11=--OR14 or --NHR15; [0239]R12=COOR17, COO-A.sup.+, or CHOR17 [0240]R13=--H, halogen, amino, optionally substituted mono- or di-alkylamino, or --OR16; [0241]R14--H, C2-C20 substituted or unsubstituted carboxyacyl; [0242]R15=--H, C2-C20 substituted or unsubstituted carboxyacyl; [0243]R16=--H, C4-C7 alkanoyl, benzyloyl, or C2-C20 substituted or unsubstituted carboxyacyl; [0244]R17=--H, C(C6H5)3, or C2-C20 substituted or unsubstituted carboxyacyl;
[0245]wherein dashed line represents optional bond between C20 and C29, and
[0246]wherein A=Na+, K.sup.+, or other cation
TABLE-US-00010 VII R31 R32 R33 R34 R35 R36 R37 R38 R39 R40 R41 1 CH2NH2 H H H H H H H H H H 2 H H H H H H H H H 3 H H H H H H H H H 4 COOH H H H H H H H H H H 5 COOH H H H H H H H H H 6 COOH H H H H H H H H H 7 COOH H H H H H H H H H 8 COOH H H H H H H H H H 9 COOH H H H H H H H H H 10 COOH H H H H H H H H H 11 COOH H H H H H H H H H 12 COOH H H H H H H H H H 13 COOH H H H H H H H H H 14 COOH H H H H H H H H H 15 COOH H H H H H H H H H 16 COOH H H H H H H H H H H 17 COOH H H H H H OH H H OH H 18 COOH H H H H H OH H OH H H 19 COOH H H H H H H H H H OH 20 COOH H OH H H H H H H H H 21 COOH H H H H H H H H H H 22 COOH H H H H H OH H H H H 23 COOH H H H H H OH H OH H H 24 COOH H OH H H H OH H H H H 25 COOH H OH H H O H H H H 26 COOH H OH H OH O H H H H 27 COOH H OH H OH OH H H H H H 28 COOH H OH H OH H OH H H H H 29 COOH H OH H HH OH H H H H 30 COOH CH3 H H H H H H H H H 31 COOH CH3 H H H H H H H H 32 COOH H H CH3 H H H H H H H 33 COOH H H H H H H H H H
[0247]R38 moieties other than hydrogen are attached to R33 oxygen by a covalent bond to the carbonyl carbon.
[0248]Preferred compounds are those where R38 is not hydrogen.
[0249]Compounds useful in the methods of the invention also include those described in U.S. Provisional Application No. 60/559,358, which is entirely incorporated by reference. In one aspect, these compounds are described by reference to the following compounds VIII to XI:
[0250]Additional compounds useful in the present invention have the general Formula VIII:
or a pharmaceutically acceptable salt or ester thereof:
[0251]wherein A is a fused ring of formula
[0252]wherein the ring carbons designated x and y in the formulas of A are the same as the ring carbons designated x and y in Formula VIII;
[0253]R1 is a carboxyalkanoyl, where the alkanoyl chain can be optionally substituted by one or more hydroxy or halo, or can be interrupted by a nitrogen, sulfur or oxygen atom, or combinations thereof;
[0254]R2, R3 and R4 are independently hydrogen, methyl, halogen, or hydroxy;
[0255]R5 is carboxyalkoxycarbonyl, alkoxycarbonyl, alkanoyloxymethyl, carboxyalkanoyloxymethyl, alkoxymethyl or carboxyalkoxymethyl, any of which is optionally substituted by one or more hydroxy or halo, or R5 is a carboxyl or hydroxymethyl;
[0256]R6 is hydrogen, methyl, hydroxy or halogen;
[0257]R7 and R8 are independently hydrogen or C1-6 alkyl;
[0258]R9 is CH2 or CH3;
[0259]R10 is hydrogen, hydroxy or methyl;
[0260]R11 is methyl, methoxycarbonyl, carboxyalkoxycarbonyl, alkanoyloxymethyl, alkoxymethyl or carboxyalkoxymethyl, any of which is optionally substituted by one or more hydroxy or halo;
[0261]R12 is hydrogen or methyl;
[0262]R13 is hydrogen or methyl;
[0263]R14 is hydrogen or hydroxy;
[0264]R15 is hydrogen if C12 and C13 form a single bond, or R15 is absent if C12 and C13 form a double bond; and
[0265]wherein the straight dashed line represents an optional double bond between C12 and C13 or C20 and C29;
[0266]with the proviso that when A is
then R1 cannot be glutaryl or succinyl when a double bond exists between C12 and C13;
[0267]when A is (ii) and R11 is methyl, then R1 cannot be succinyl;
[0268]when A is (iii) and R2, R3 and R13 are each hydrogen, then R1 cannot be succinyl; and
[0269]with the proviso that A (i) cannot be
when R2 and R3 are both methyl and a double bond exists between C12 and C13.
[0270]In some embodiments, R1 is a carboxy(C2-6)alkylcarbonyl group or a carboxy(C2-6)alkoxy(C1-6)alkylcarbonyl group. Suitable groups are selected from the group consisting of:
[0271]According to the invention, in some embodiments the compounds have Formula IX:
wherein R1, R4, R5, R6, R7, R8 and R14 are as defined above for Formula VIII. In one embodiment, R6 is β-methyl, R8 is hydrogen, R5 is hydroxymethyl and R1 is 3',3'-dimethylglutaryl, 3',3'-dimethylsuccinyl, glutaryl or succinyl. In another embodiment, R6 is hydrogen, R7 and R8 are both methyl, R5 is carboxyl and R1 is 3',3'-dimethylglutaryl, 3',3'-dimethylsuccinyl, glutaryl or succinyl.
[0272]In some embodiments, R5 is carboxyalkoxycarbonyl, alkoxycarbonyl, alkanoyloxymethyl, carboxyalkanoyloxymethyl, alkoxymethyl or carboxyalkoxymethyl, any of which is optionally substituted by one or more hydroxy or halo, or R5 is a carboxyl or hydroxymethyl. In some embodiments, R5 is selected from a group consisting of carboxyl, hydroxymethyl, --CO2(CH2)nCOOH, --CO2(CH2)nCH3, --CH2OC(O)(CH2)nCH3, --CH2OC(O)(CH2)nCOOH, --CH2--O--(CH2)nCH3 and --CH2--O--(CH2)nCOOH. In some embodiments, R5 is selected from a group consisting of
[0273]In some embodiments, R5 is hydroxymethyl. In some embodiments, R5 is carboxyl. In some embodiments, n is from 0 to 20. In some embodiments, n is from 1 to 10. In some embodiments, n is from 2 to 8. In some embodiments, n is from 1 to 6. In some embodiments, n is from 2 to 6.
[0274]In some embodiments, compounds useful in the present invention have the Formula X:
wherein R1, R9, R10, and R11 are as defined above for Formula VIII. In one embodiment, R1 is 3',3'-dimethylglutaryl, 3',3'-dimethylsuccinyl, glutaryl or succinyl.
[0275]In some embodiments, R11 is methyl, methoxycarbonyl, carboxyalkoxycarbonyl, alkanoyloxymethyl, alkoxymethyl or carboxyalkoxymethyl, any of which is optionally substituted by one or more hydroxy or halo. In some embodiments, R11 is selected from the group consisting of methyl, --CO2(CH2)nCOOH, --CH2OC(O)(CH2)nCH3, --CH2--O--(CH2)nCH3 and --CH2--O--(CH2)nCOOH.
[0276]In some embodiments, n is from 0 to 20. In some embodiments, n is from 1 to 10. In some embodiments, n is from 2 to 8. In some embodiments, n is from 1 to 6. In some embodiments, n is from 2 to 6. In some embodiments, R11 is methyl. In some embodiments, R11 is methoxycarbonyl. In some embodiments, R11 is selected from the group consisting of methoxymethyl and ethoxymethyl. In some embodiments, methyl groups found in R11 can be substituted with a halogen or a hydroxy.
[0277]In some embodiments, the compounds useful in the present invention have Formula XI:
wherein R1, R2, R3, R4, and R13 are as defined above for Formula VIII. In one embodiment, R1 is 3',3'-dimethylglutaryl, 3',3'-dimethylsuccinyl, glutaryl or succinyl. In one embodiment, both R2 and R3 are methyl.
[0278]Any triterpene which falls within the scope of Formula VIII can be used. According to the invention, in some embodiments the compounds of Formula VIII are selected from the group consisting of derivatives of uvaol, ursolic acid, erythrodiol, echinocystic acid, oleanolic acid, sumaresinolic acid, lupeol, dihydrolupeol, betulinic acid methylester, dihydrobetulinic acid methylester, 17-α-methyl-androstanediol, androstanediol, and 4,4-dimethyl-androstanediol.
[0279]In some embodiments, the compounds of the present invention are defined as in Formula VIII, wherein R2 and R3 are both methyl. In some embodiments, the compounds of the present invention are defined as in Formula VIII, wherein R1 is 3',3'-dimethylsuccinyl. In some embodiments, the compounds of the present invention are defined as in Formula VIII, wherein R1 is succinyl, i.e.,
[0280]According to the invention, in some embodiments the stereochemistry of the sidechain substituents is important. In some embodiments, the compounds of the present invention are defined as in Formula VIII, wherein A is (i) and R5 is in the β position. In some embodiments, the compounds of the present invention are defined as in Formula VIII, wherein A is (i) and R6 is in the β position. In some embodiments, the compounds of the present invention are defined as in Formula VIII, wherein A is (i) and R14 is in the α position. In some embodiments, the compounds of the present invention are defined as in Formula VIII, wherein A is (i), R7 is α-methyl, and R8 is hydrogen. In some embodiments, the compounds of the present invention are defined as in Formula VIII, wherein A is (i), R8 is α-methyl, and R7 is hydrogen. In some embodiments, the compounds of the present invention are defined as in Formula VIII, wherein A is (i) and both R7 and R8 are methyl. In some embodiments, the compounds of the present invention are defined as in Formula VIII, wherein A is (ii) and R11 is in the β position.
[0281]In some embodiments, 3',3'-dimethylsuccinyl is at the C3 position. In some embodiments, the compounds of Formula IX are 3-O-(3',3'-dimethylsuccinyl)uvaol; 3-O-(3',3'-dimethylsuccinyl)erythrodiol; 3-O-(3',3'-dimethylsuccinyl)echinocystic acid or 3-O-(3',3'-dimethylsuccinyl)sumaresinolic acid. In some embodiments, the compounds of Formula X are 3-O-(3',3'-dimethylsuccinyl)lupeol; 3-O-(3',3'-dimethylsuccinyl)dihydrolupeol; 3-O-(3',3'-dimethylsuccinyl)17β-methylester-betulinic acid; or 3-O-(3',3'-dimethylsuccinyl) 17β-methylester-dihydrobetulinic acid. In some embodiments, the compounds of Formula XI are 3-O-(3',3'-dimethylsuccinyl) 4,4-dimethylandrostanediol; 3-O-(3',3'-dimethylsuccinyl) 17α-methylandrostanediol; 3-O-(3',3'-dimethylsuccinyl)androstanediol.
[0282]Alkyl groups and alkyl containing groups of the compounds of the present invention can be straight chain or branched alkyl groups, preferably having one to ten carbon atoms. In some embodiments, the alkyl groups or alkyl containing groups of the present invention can be substituted with a C3-7 cycloalkyl group. In some embodiments, the cycloalkyl group may include, but is not limited to, a cyclobutyl, cyclopentyl or cyclohexyl group.
[0283]Also, included within the scope of the present invention are the non-toxic pharmaceutically acceptable salts of the compounds of the present invention. These salts can be prepared in situ during the final isolation and purification of the compounds or by separately reacting the purified compound in its free acid form with a suitable organic or inorganic base and isolating the salt thus formed. These can include cations based on the alkali and alkaline earth metals, such as sodium, lithium, potassium, calcium, magnesium, and the like, as well as nontoxic ammonium, quaternary ammonium and amine cations including, but not limited to ammonium, tetramethylammonium, tetraethylammonium, methylamine, dimethylamine, trimethylamine, ethylamine, N-methyl glucamine and the like.
[0284]Also, included within the scope of the present invention are the non-toxic pharmaceutically acceptable esters of the compounds of the present invention. Ester groups are preferably of the type which are relatively readily hydrolyzed under physiological conditions. Examples of pharmaceutically acceptable esters of the compounds of the invention include C1-6 alkyl esters wherein the alkyl group is a straight or branched chain. Acceptable esters also include C5-7 cycloalkyl esters as well as arylalkyl esters, such as, but not limited to benzyl. C1-4 alkyl esters are preferred. In some embodiments, the esters are selected from the group consisting of alkylcarboxylic acid esters, such as acetic acid esters, and mono- or dialkylphosphate esters, such as methylphoshate ester or dimethylphosphate ester. Esters of the compounds of the present invention can be prepared according to conventional methods.
[0285]Certain compounds within the scope of Formulae VIII, IX, X and XI are derivatives referred to as "prodrugs". The expression "prodrug" refers to compounds that are rapidly transformed in vivo by an enzymatic or chemical process, to yield the parent compound of the above formulas, for example, by hydrolysis in blood. A thorough discussion is provided by Higuchi, T. and V. Stella in Pro-drugs as Novel Delivery Systems, Vol. 14, A.C.S. Symposium Series, and in Bioreversible Carriers in Drug Design, Ed. Edward B. Roche, American Pharmaceutical Association, Pergamon Press, 1987. Useful prodrugs can be esters of the compounds of Formulae VIII, IX, X, and XI. In some prodrug embodiments, a lower alkyl group is substituted with one or more hydroxy or halo groups by a suitable acid. Suitable acids include, e.g., carboxylic acids, sulfonic acids, phosphoric acid or lower alkyl esters thereof, and phosphonic acid or lower alkyl esters thereof. For example, suitable carboxylic acids include alkylcarboxylic acids, such as acetic acid, arylcarboxylic acids and arylalkylcarboxylic acids. Suitable sulfonic acids include alkylsulfonic acids, arylsulfonic acids and arylalkylsulfonic acids. Suitable phosphoric and phosphonic acid esters are methyl or ethyl esters. In some embodiments, the C3 acyl groups having dimethyl groups or oxygen at the C3' position can be the most active compounds. This observation suggests that these types of acyl groups might be important to the enhanced anti-HIV activity.
[0286]In an additional embodiment, the invention includes compounds and methods that use compounds of Formula XII:
where R72 is one of:
wherein
[0287]X is hydroxy or halogen; and
[0288]R73 is lower alkyl, such as methyl, ethyl or propyl.
[0289]In one embodiment, the invention is drawn to a method of treating HIV-1 infection in a patient by administering a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), but does not significantly affect other Gag processing steps, wherein said compound is a compound of Formula I through XII
[0290]In one embodiment, the invention is drawn to a method of treating HIV-1 infection in a patient by administering a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), but does not significantly affect other Gag processing steps, wherein said compound is a compound of Formula I through XII, with the exception of DSB.
[0291]In one embodiment, the invention is drawn to a method of treating HIV-1 infection in a patient by administering a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), but does not significantly affect other Gag processing steps, wherein said compound is other than a compound of Formula I through VII. In one embodiment, the invention is drawn to a method of treating HIV-1 infection in a patient by administering a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), but does not significantly affect other Gag processing steps, wherein said compound is other than a compound of Formula I through XI.
[0292]In another embodiment, the invention is drawn to a method of inhibiting processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) in a cell, but without significantly affect other Gag processing steps, wherein said compound is a compound of Formula Groups I through XII.
[0293]In another embodiment, the invention is drawn to a method of inhibiting processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) in a cell, but without significantly affect other Gag processing steps, wherein said compound is other than a compound of Formula I through VII.
[0294]In another embodiment, the invention is drawn to a method of inhibiting processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) in a cell, but without significantly affect other Gag processing steps, wherein said compound is other than a compound of Formula I through XI.
[0295]In another embodiment, the invention is drawn to a method of treating human blood or blood products by inhibiting processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) in a cell, but without significantly affect other Gag processing steps, wherein said compound is a compound of Formula I through XII.
[0296]In another embodiment, the invention is drawn to a method of treating human blood or blood products by inhibiting processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) in a cell, but without significantly affect other Gag processing steps, wherein said compound is other than a compound of Formula I through VII.
[0297]In another embodiment, the invention is drawn to a method of treating human blood or blood products by inhibiting processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) in a cell, but without significantly affect other Gag processing steps, wherein said compound is other than a compound of Formula I through XI.
Synthesis of DSB and Related Compounds
[0298]Reaction of betulinic acid and dihydrobetulinic acid with dimethylsuccinic anhydride produced a mixture of 3-O-(2',2'-dimethylsuccinyl) and 3-O-(3',3'-dimethylsuccinyl)-betulinic acid and dihydrobetulinic acid, respectively. The mixtures were successfully separated by preparative scale HPLC yielding pure samples. The structures of these isomers were assigned by long-range 1H-13C COSY examinations.
[0299]The derivatives of betulinic acid and dihydrobetulinic acid of the present invention were all synthesized by refluxing a solution of betulinic acid or dihydrobetulinic acid, dimethylaminopyridine (1 equivalent mol), and an appropriate anhydride (2.5-10 equivalent mol) in anhydrous pyridine (5-10 mL). The reaction mixture was then diluted with ice water and extracted with CHCl3. The organic layer was washed with water, dried over MgSO4, and concentrated under reduced pressure. The residue was chromatographed using silica gel column or semi-preparative-scale HPLC to yield the product.
[0300]Preparation of 3-O-(3',3'-dimethylsuccinyl)betulinic acid: yield 70% (starting with 542 mg of betulinic acid); crystallization from MeOH gave colorless needles; mp 274°-276° C.; [α]D19+23.5° (c=0.71), CHCl3-MeOH [1:1]); Positive FABMS m/z 585 (M+H).sup.+; Negative FABMS m/z 583 (M-H)-; HR-FABMS calcd for C36H57O6 585.4155, found m/z 585.4161; 1H NMR (pyridine-d5): 0.73, 0.92, 0.97, 1.01, 1.05 (each 3H, s; 4-(CH3)2, 8-CH3, 10-CH3, 14-CH3), 1.55 (6H, s, 3'-CH3×2), 1.80 (3H, s, 20-CH3), 2.89, 2.97 (each 1H, d, J=15.5 Hz, H-2'), 3.53 (1H, m, H-19), 4.76 (1H, dd, J=5.0, 11.5 Hz, H-3), 4.78, 4.95 (each 1H, br s, H-30).
[0301]3-O-(3',3'-dimethylsuccinyl)dihydrobetulinic acid: yield 24.5% (starting with 155.9 mg of dihydrobetulinic acid); crystallization from MeOH--H2O gave colorless needles; mp 291°-292° C.; [α]D20-13.4° (c=1.1, CHCl3-MeOH [1:1], 1H NMR (pyridine-d5): 0.85, 0.94 (each 3H, d, J=7.0 Hz; 20-(CH3)2), 0.75, 0.93, 0.97, 1.01, 1.03 (each 3H, s; 4-(CH3)2, 8-CH3, 10-CH3, 14-CH3), 1.55 (6H, s; 3'-CH3×2), 2.89, 2.97 (each 1H, d, J=15.5 Hz; H-2'), 4.77 (1H, dd, J=5.0, 11.0 Hz, H-3); Anal. Calcd for C36H58O6.5/2H2O: C, 68.43; H, 10.04; found C, 68.64; H 9.78.
[0302]The synthesis of 3-O-(3',3'-dimethylglutaryl)betulinic acid was disclosed U.S. Pat. No. 5,679,828, as COMPOUND NO. 4.
[0303]3-O-(3',3'-dimethylglutaryl)dihydrobetulinic acid: yield 93.3% (starting with 100.5 mg of dihydrobetulinic acid); crystallization from needles MeOH--H2O gave colorless needles; mp 287°-289° C.; [α]D20-17.9° (c=0.5, CHCl3-MeOH[1:1]); 1H-NMR (pyridine-d5): 0.86, 0.93 (each 3H, d, J=6.5 Hz; 20-(CH3)2), 0.78, 0.92, 0.96, 1.02, 1.05 (each 3H, s; 4-(CH3)2, 8-CH3, 10-CH3, 14-CH3), 1.38, 1.39 (each 3H, s; 3'-CH3×2), 2.78 (4H, m, H2-2' and 4'), 4.76 (1H, dd, J=4.5, 11.5 Hz; H-3). Anal. Calcd for C37H60O6: C, 73.96; H, 10.06; found C, 73.83; H 10.10.
[0304]The synthesis for 3-O-diglycolyl-betulinic acid was disclosed in U.S. Pat. No. 5,679,828, as COMPOUND NO. 5.
[0305]3-O-diglycolyl-dihydrobetulinic acid: yield 79.2% (starting with 103.5 mg of dihydrobetulinic acid); an off-white amorphous powder; [α]D20-9.8° (c=1.1, CHCl3-MeOH[1:1]); 1H-NMR (pyridine-d5): 0.79, 0.87 (each 3H, d, J=6.5 Hz; 20-(CH3)2), 0.87, 0.88, 0.91, 0.98, 1.01 (each 3H, s; 4-(CH3)2, 8-CH3, 10-CH3, 14-CH3), 4.21, 4.23 (each 2H, s, H2-2' and 4'), 4.57 (1H, dd, J=6.5, 10.0 Hz, H-3); Anal. Calcd for C34H54O7.2H2O: C, 66.85; H, 9.57; found C, 67.21; H, 9.33.
[0306]The syntheses of 3-O-(3',3'-dimethylsuccinyl)betulin and 3-O-(3',3'-dimethylglutaryl)betulin were disclosed in U.S. application Ser. No. 10/670,797.
Methods of Inhibiting HIV with a Compound
[0307]Methods of "inhibiting HIV" or "inhibition of HIV" as used herein, means any interference in, inhibition of, and/or prevention of HIV using the methods of the invention. As such, methods of inhibition are useful in inhibiting with the infectivity of HIV, inhibition of p25 processing, inhibition of viral maturation, formation of virions that exhibit altered phenotypes, and the like. Preferably, methods of the invention act upon p25 processing in the cells of an animal, but are not limited by that method of action.
[0308]A method of inhibiting HIV with a compound may be relevant to a method of treating HIV infection in a patient. Therefore, a method of inhibiting HIV with a compound may similarly be used to treat a patient.
[0309]The methods of inhibiting HIV-1 replication in cells of an animal includes contacting infected cells with a compound of Formula I through XII, above. Related embodiments include a method of treating a HIV-1 infectionin a patient comprising administration of a compound of Formula I through XII; a method of inhibiting p25 processing either in a cell, in vivo, and/or in vitro, by administration of a compound that inhibits said p25 processing; and a method of treating human blood or blood products by administering a compound of Formula I through XII. Also included are a method of identifying a compound that inhibits any one of p25 processing, HIV maturation, HIV infectivity, HIV virion phenotypes and the like.
[0310]In one embodiment, the compound is a derivative of betulinic acid, betulin, or dihydrobetulinic acid or dihydrobetulin and which includes the preferred substituents of Table 4. Preferred compounds include but are not limited to 3-O-(3',3'-dimethylsuccinyl)betulinic acid, 3-O-(3',3'-dimethylsuccinyl)betulin, 3-O-(3',3'-dimethylglutaryl)betulin, 3-O-(3',3'-di methylsuccinyl)dihydrobetulinic acid, 3-O-(3',3'-dimethylglutaryl)betulinic acid, (3',3'-dimethylglutaryl)dihydrobetulinic acid, 3-O-diglycolyl-betulinic acid, and 3-O-diglycolyl-dihydrobetulinic acid.
[0311]In one embodiment, the invention is drawn to a method inhibiting HIV-1 replication in cells of an animal by contacting infected cells with a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), but does not significantly affect other Gag processing steps, wherein said compound is a compound of Formulas I through XII above.
[0312]In one embodiment, the invention is drawn to a method of inhibiting HIV-1 replication in cells of an animal by contacting infected cells with a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), but does not significantly affect other Gag processing steps, wherein said compound is compound of Formulas I through XII, with the exception of DSB.
[0313]In one embodiment, the invention is drawn to a method of inhibiting HIV-1 replication in cells of an animal by contacting infected cells with a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), but does not significantly affect other Gag processing steps, wherein said compound is one that is excluded from those of Formulas I through VI. In one embodiment, the invention is drawn to a method of treating HIV-1 infection in a patient by administering a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), but does not significantly affect other Gag processing steps, wherein said compound is one that is other than those of Formulas I through XI.
[0314]In another embodiment, the invention is drawn to a method of inhibiting processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) in a cell, but without significantly affecting other Gag processing steps, wherein said compound is a compound of Formulas I through XII.
[0315]In another embodiment, the invention is drawn to a method of inhibiting processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) in a cell, but without significantly affecting other Gag processing steps, wherein said compound is a compound other than those of Formulas I through VI.
[0316]In another embodiment, the invention is drawn to a method of inhibiting processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) in a cell, but without significantly affecting other Gag processing steps, wherein said compound is a compound other than those of Formulas I through XI.
[0317]In another embodiment, the invention is drawn to a method of inhibiting processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) in a cell, but without significantly affecting other Gag processing steps, wherein said compound is a compound other than those of Formulas I through XI. In another embodiment, the invention is drawn to a method of treating human blood or blood products by inhibiting processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) in a cell, but without significantly affect other Gag processing steps, wherein said compound is a compound of Formulas I through XII.
[0318]In another embodiment, the invention is drawn to a method of treating human blood or blood products by inhibiting processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) in a cell, but without significantly affect other Gag processing steps, wherein said compound is a compound other than those of Formulas I through VI.
[0319]In another embodiment, the invention is drawn to a method of treating human blood or blood products by inhibiting processing of the viral Gag p25 protein (CA-SP1) to p24 (CA) in a cell, but without significantly affect other Gag processing steps, wherein said compound is a compound other than those of Formulas I through XI.
[0320]The method disclosed herein, further comprises contacting said cells with one or more drugs selected from the group consisting of anti-viral agents, anti-fungal agents, anti-bacterial agents, anti-cancer agents, immunostimulating agents, and combinations thereof. The method may include the treatment of human blood products.
[0321]The invention may also be used in conjunction with a method of treating cancer comprising the administration to an animal of one or more anti-neoplastic agents, exposing an animal to a cancer cell-killing amount of radiation, or a combination of both.
Methods of Identifying Compounds
[0322]The invention further includes a method for identifying compounds that inhibit HIV-1 replication in cells of an animal disclosed herein. In one embodiment, said method comprises:
[0323](a) contacting a Gag polypeptide comprising a CA-SP1 cleavage site with a test compound;
[0324](b) adding a labeled substance that selectively binds at or near the CA-SP1 cleavage site; and
[0325](c) measuring the binding of the test compound at or near the CA-SP1 cleavage site.
[0326]Labeled substances or molecules include labeled antibodies or labeled DSB and the label includes an enzyme, fluorescent substance, chemiluminescent substance, horseradish peroxidase, alkaline phosphatase, biotin, avidin, electron dense substance, such as gold, osmium tetroxide, lead or uranyl acetate, and radioisotope, antibodies labeled with such substances of molecules or a combination thereof. The assays could include, but are not limited to ELISA, single and double sandwich techniques, immunodiffusion or immunoprecipitation techniques, as known in the art ("Immunoassay Handbook, 2nd ed.," D. Wild, Nature Publishing Group, (2001)). Said methods of identifying also could include, but are not limited to Western blot assays, calorimetric assays, light and electron microscopic techniques, confocal microscopy, or other techniques known in the art.
[0327]A method of identifying compounds that inhibit HIV replication in cells of an animal further comprises:
[0328](a) contacting a Gag protein comprising a wild-type CA-SP1 cleavage site, with HIV-1 protease in the presence of a test compound;
[0329](b) separately, contacting a Gag protein comprising a mutant CA-SP1 cleavage site or a protein comprising an alternative protease cleavage site with HIV-1 protease in the presence of the test compound; and
[0330](c) comparing the cleavage of the native wild-type Gag protein to the amount of cleavage of the mutant Gag protein or to the amount of cleavage of the peptide comprising an alternative protease cleavage site.
[0331]Step (b) above is performed as a control in order to eliminate compounds that might bind directly to, and therefore inhibit, the protease enzyme. The above method also includes the method wherein the wild-type CA-SP1, mutant CA-SP1 or alternative protease cleavage site is contained within a polypeptide fragment or recombinant peptide.
[0332]The method for identifying compounds that inhibit HIV-1 disclosed herein, also includes a method wherein said peptide or protein is labeled with a fluorescent moiety and a fluorescence quenching moiety, each bound to opposite sides of the CA-SP1 cleavage site, and wherein said detecting comprises measuring the signal from the fluorescent moiety, or wherein said peptide or protein is labeled with two fluorescent moieties, each bound to opposite sides of the CA-SP1 cleavage site, and wherein said detecting comprises measuring the transfer of fluorescent energy from one moiety to the other in the presence of the test compound and HIV-1 protease and comparing said transfer of fluorescent energy to that observed when the same procedure is applied to a peptide that comprises a sequence containing a mutation in the CA-SP1 cleavage site or that comprises a sequence containing another cleavage site. Examples of fluorescence-based assays of protease activity are well known in the art. In one such example, a protease substrate is labeled with green fluorescent dye molecules, which fluoresce when the substrate is cleaved by the protease enzyme (Molecular Probes, Protease Assay Kit).
[0333]The method of comparing the cleavage, above, also includes using a labeled antibody that selectively binds CA or SP1 or CA-SP1 in order to measure the extent to which the test compound inhibits CA-SP1 cleavage. The antibody can be labeled with a molecule selected from the group consisting of enzyme, fluorescent substance, chemiluminescent substance, horseradish peroxidase, alkaline phosphatase, biotin, avidin, electron dense substance, and radioisotope, or combinations thereof. The method also includes the use of an antibody to a specific epitope tag sequence to selectively detect CA-SP1 (p25) or SP1, into which the amino acid sequence for that epitope tag has been engineered according to standard methods in the art. Suitable tags are well known to those of ordinary skill in the art, and include haemagglutinin epitope HA (YPYDVPDYA) (SEQ ID NO: 81), bluetongue virus epitope VP7 (QYPALT) (SEQ ID NO: 82), α-tubulin epitope (EEF), Flag (DYKDDDDK) (SEQ ID NO: 83), and VSV-G (YTDOEMNRLGK) (SEQ ID NO: 84). Examples of SP1 containing epitope tags are illustrated in FIG. 17.
[0334]As an example, the sequence of the FLAG epitope tag (Sigma-Aldrich) is inserted into the p2 (SP1) region of Gag by oligonucleotide-directed mutagenesis of a Gag expression plasmid. The presence of the SP1 domain in the cell-expressed protein is then be detected using commercially available anti-FLAG monoclonal antibodies (Sigma-Aldrich). (Hopp, T. P. Biotechnology 6: 1204-1210 (1988)).
[0335]The method of identifying compounds that disrupt CA-SP1 cleavage also includes the addition of a compound to cells infected with HIV-1 and the detection of CA-SP1 cleavage products by lysing and analyzing the cells or the released virions. The method included in the invention can be performed using a western blot analysis of viral proteins and detecting p25 using an antibody to p25 or wherein said mixture is analyzed by performing a gel electrophoresis of viral proteins and imaging of metabolically labeled proteins, or wherein the mixture is analyzed using immunoassays that use an antibody that selectively binds p25 or an antibody that selectively binds in order to distinguish p25 from p24. The invention includes the use of an antibody to a specific epitope tag sequence inserted into the C-terminal domain of SP1 to selectively detect p25 or SP1. For example, a sandwich ELISA assay can be performed where p25 and p24 in detergent-solubilized virus are captured using an antibody that selectively binds to the CA region of Gag, which antibody is bound to a multiple well plate. Following a washing step, bound p25 is detected using an antibody to an epitope tag inserted in SP1, which is conjugated to an appropriate detection reagent (e.g. alkaline phosphatase for an enzyme-linked immunosorbent assay). Virus released by cells treated with compounds that act via this mechanism will generally have increased levels of p25 compared with untreated virions.
[0336]The disclosed method is drawn to an antibody that selectively binds p25, or an antibody that selectively binds SP1, or an antibody to an epitope tag sequence inserted into SP1, which is labeled with a molecule selected from the group consisting of enzyme, fluorescent substance, chemiluminescent substance, horseradish peroxidase, alkaline phosphatase, biotin, avidin, electron dense substance, and radioisotope, or combinations thereof.
[0337]Infected cells," as used herein, includes cells infected naturally by membrane fusion and subsequent insertion of the viral genome into the cells, or transfection of the cells with viral genetic material through artificial means. These methods include, but are not limited to, calcium phosphate transfection, DEAE-dextran mediated transfection, microinjection, lipid-mediated transfection, electroporation or infection.
[0338]The invention may be practiced by infecting target cells in vitro with an infectious strain of HIV and in the presence of test compound, under appropriate culture conditions and for varying periods of time. Infected cells or supernatant fluid can be processed and loaded onto a polyacrylamide gel for the detection of virus levels, by methods that are well known in the art. Non-infected and non-treated cells can be used as negative and positive infection controls, respectively. Alternatively, the invention may be practiced by culturing the target cells in the presence of test compound prior to infecting the cells with an HIV strain.
[0339]The invention also includes a method for identifying compounds that inhibit HIV-1 replication in the cells of an animal, comprising:
[0340](a) contacting a test compound with wild-type virus isolates and separately with virus isolates having reduced sensitivity to 3-O-(3',3'-dimethylsuccinyl)betulinic acid; and
[0341](b) selecting test compounds that are more active against the wild-type virus isolate compared with virus isolates that have reduced sensitivity to 3-O-(3',3'-dimethylsuccinyl)betulinic acid.
[0342]This invention further includes a method for identifying compounds that act by any of the abovementioned mechanism, involving treating HIV-1 infected or transfected cells with a compound then analyzing the virus particles released by compound-treated cells by thin-sectioning and transmission electron microscopy, by standard methods well known in the art. A compound acts by the abovementioned mechanism if particles are detected that exhibit spherical condensed cores that are acentric with respect to the viral particle and/or a crescent-shaped electron-dense layer just inside the viral membrane.
[0343]For electron microscopic studies, infected cells or centrifuged virus pellets obtained from the supernatant fluid can be contacted with a fixative, such as glutaraldehyde or freshly-made paraformaldehyde, and/or osmium tetroxide or other electron microscopy compatible fixative that is known in the art. The virus from the supernatant fluid or the cells, is dehydrated and embedded in an electron-lucent polymer such as an epoxy resin or methacrylate, thin sectioned using an ultramicrotome, stained using electron dense stains such as uranyl acetate, and/or lead citrate, and viewed in a transmission electron microscope. Non-infected and non-treated cells can be used as negative and positive infection controls, respectively. Alternatively, the invention may be practiced by culturing the target cells in the presence of test compound prior to infecting the cells with an HIV strain. Maturation defects caused by the compounds of the present invention are determined by the presence of morphologically aberrant viral particles, compared with controls, as described herein.
[0344]For cell culture studies, the virus-infected cells may be observed for the formation of syncytia, or the supernatant may be tested for the presence of HIV particles. Virus present in the supernatant may be harvested to infect other naive cultures to determine infectivity.
[0345]Also included in the invention, is a method of determining if an individual is infected with HIV-1, is susceptible to treatment by a compound that inhibits p25 processing, the method involves taking blood from the patient, genotyping the viral RNA and determining whether the viral RNA contains mutations in the CA-SP1 cleavage site.
[0346]The invention also includes a method for identifying compounds that act by the abovementioned mechanisms, involving testing by a combination of the methods disclosed herein.
[0347]HIV Gag protein and fragments thereof for use in the aforementioned assays may be expressed or synthesized using a variety of methods familiar to those skilled in the art. Gag can be produced in an in vitro transcription and translation system using a rabbit reticulocyte lysate. Gag expressed in this system has been shown to be processed sequentially in a pattern similar to that observed in infected cells (Pettit, S. C. et al. J. Virol. 76:10226-10233 (2002)). Moreover, Gag expressed by this method is capable of assembling into immature viral particles when fused to a heterologous type D retroviral cytoplasmic self-assembly domain (Sakalian, M. et al., J. Virol. 76:10811-10820 (2002)). The plasmid pDAB72, available from the NIH AIDS Reagent Program can be used for this purpose (Erickson-Viitanen, S. et al., AIDS Res. Hum. Retroviruses. 5:577-91 (1989); Sidhu M. K. et al., Biotechniques, 18:20, 22, 24 (1995)). Other in vitro transcription/translation systems based on wheat germ or bacterial lysates can also be used for this purpose. HIV Gag may also be expressed in transfected cells using a variety of commercially available expression vectors. The plasmid p55-GAG/GFP, available from the NIH AIDS Reagent Program, may be used to express an HIV Gag-green fluorescent protein fusion protein in mammalian cells for drug interaction studies (Sandefur, S. et al., J. Virol. 72:2723-2732 (1998)). This construct would permit the capture and purification of Gag fusion protein using GFP-specific monoclonal antibodies. In addition, Gag may be expressed in cells using recombinant viral vectors, such as those used in the vaccinia virus, adenovirus, or baculovirus systems. Gag can also be expressed by infecting cells with HIV or by transfecting cells with proviral DNA. Finally, Gag may be expressed in yeast or bacterial cells transformed with the appropriate expression vectors.
[0348]In addition to Gag proteins expressed in cells or in vitro using cell lysates, peptides corresponding to various regions of Gag may be commercially synthesized from using standard peptide synthesis techniques.
[0349]The invention further encompasses compounds identified by the method of this invention and/or a compound which inhibits HIV-1 replication according to the methods of this invention and pharmaceutical compositions comprising one or more compounds as disclosed herein, or pharmaceutically acceptable salts, esters or prodrugs thereof, and pharmaceutically acceptable carriers.
Pharmaceutical Compositions
[0350]Compounds according to the present invention have been found to possess anti-retroviral, particularly anti-HIV, activity. The salts and other formulations of the present invention are expected to have improved water solubility, and enhanced oral bioavailability. Also, due to the improved water solubility, it will be easier to formulate the salts of the present invention into pharmaceutical preparations. Further, compounds according to the present invention are expected to have improved biodistribution properties.
[0351]In one embodiment, the compounds are those of Formula I through XII, in another they are compounds other than the compounds of Formula I through XI.
[0352]This invention also includes a pharmaceutical composition comprising a compound that inhibits processing of the viral Gag p25 protein (CA-SP1) to p24 (CA), but has no significant effect on other Gag processing steps, or that inhibits the maturation of virus particles released from treated infected cells, such as the compounds of Formula I through XII. The invention includes a pharmaceutical composition comprising one or more compounds disclosed herein, or pharmaceutically acceptable salts, esters or prodrugs thereof, and pharmaceutically acceptable carriers, wherein said compound is of Formulae I through XII above, or preferably, wherein said compound is selected from the group consisting of 3-O-(3',3'-dimethylsuccinyl)betulinic acid, 3-O-(3',3'-dimethylsuccinyl)betulin, 3-O-(3',3'-dimethylglutaryl)betulin, 3-O-(3',3'-dimethylsuccinyl)dihydrobetulinic acid, 3-O-(3',3'-dimethylglutaryl)betulinic acid, (3',3'-dimethylglutaryl)dihydrobetulinic acid, 3-O-diglycolyl-betulinic acid, and 3-O-diglycolyl-dihydrobetulinic acid. The pharmaceutical compositions according to the invention, further comprise one or more drugs selected from an anti-viral agent, anti-fungal agent, anti-cancer agent or an immunostimulating agent.
[0353]Pharmaceutical compositions of the present invention can comprise at least one of the compounds of Formulae I through XII disclosed herein. Pharmaceutical compositions according to the present invention can also further comprise other anti-viral agents such as, but not limited to, AZT (zidovudine, RETROVIR®, GlaxoSmithKline), 3TC (lamivudine, EPIVIR®, GlaxoSmithKline), AZT+3TC, (COMBIVIR®, GlaxoSmithKline), AZT+3TC+abacavir (TRIZIVIR®, GlaxoSmithKline), ddI (didanosine, VIDEX®, Bristol-Myers Squibb), ddC (zalcitabine, HIVID®, Hoffmann-La Roche), D4T (stavudine, ZERIT®, Bristol-Myers Squibb), abacavir (ZIAGEN®, GlaxoSmithKline), tenofovir (VIREAD®, Gilead Sciences), nevirapine (VIRAMUNE®, Boehringer Ingelheim), delavirdine (Pfizer), emtricitabine (EMTRIVA®, Gilead Sciences), efavirenz (SUSTIVA®, DuPont Pharmaceuticals), saquinavir (INVIRASE®, FORTOVASE®, Hoffmann-LaRoche), ritonavir (NORVIR®, Abbott Laboratories), indinavir (CRIXIVAN®, Merck and Company), nelfinavir (VIRACEPT®, Pfizer), amprenavir (AGENERASE®, GlaxoSmithKline), adefovir (PREVEON®, HEPSERA®, Gilead Sciences), atazanavir (Bristol-Myers Squibb), fosamprenavir (LEXIVA®, GlaxoSmithKline) and hydroxyurea (HYDREA®, Bristol-Meyers Squibb), or any other antiretroviral drugs or antibodies in combination with each other, or associated with a biologically based therapeutic, such as, for example, gp41-derived peptides enfuvirtide (FUZEON®, Roche and Trimeris) and T-1249, or soluble CD4, antibodies to CD4, and conjugates of CD4 or anti-CD4, or as additionally presented herein.
[0354]Additional suitable antiviral agents for optimal use with one of the compounds of Formulae I through XII of the present invention can include, but are not limited to, amphotericin B (FUNGIZONE®); Ampligen (mismatched RNA) developed by Hemispherx Biopharma; BETASERON® (β-interferon, Chiron); butylated hydroxytoluene; Carrosyn (polymannoacetate); Castanospermine; Contracan (stearic acid derivative); Creme Pharmatex (containing benzalkonium chloride); 5-unsubstituted derivative of zidovudine; penciclovir (DENAVIR® Novartis); famciclovir (FAMVIR® Novartis); acyclovir (ZOVIRAX® GlaxoSmithKline); cytofovir (VISTIDE® Gilead); ganciclovir (CYTOVENE®, Hoffman LaRoche); dextran sulfate; D-penicillamine (3-mercapto-D-valine); FOSCARNET® (trisodium phosphonoformate; AstraZeneca); fusidic acid; glycyrrhizin (a constituent of licorice root); HPA-23 (ammonium-21-tungsto-9-antimonate); ORNIDYL® (eflornithine; Aventis); nonoxynol; pentamidine isethionate (PENTAM-300); Peptide T (octapeptide sequence; Peninsula Laboratories); Phenyloin (Pfizer); INH or isoniazid; ribavirin (VIRAZOLE®, Valeant Pharmaceuticals); rifabutin, ansamycin (MYCOBUTIN® Pfizer); CD4-IgG2 (Progenics Pharmaceuticals) or other CD4-containing or CD4-based molecules; Trimetrexate (Medimmune); suramin and analogues thereof (Bayer); and WELLFERON® (α-interferon, GlaxoSmithKline).
[0355]Pharmaceutical compositions of the present invention can also further comprise immunomodulators. Suitable immunomodulators for optional use with a betulinic acid or betulin derivative of the present invention in accordance with the present invention can include, but are not limited to: ABPP (Bropririmine); Ampligen (mismatched RNA) Hemispherx Biopharma; anti-human interferon-α-antibody; ascorbic acid and derivatives thereof; interferon-β; Ciamexon; cyclosporin; cimetidine; CL-246,738; colony stimulating factors, including GM-CSF; dinitrochlorobenzene; HE2000 (Hollis-Eden Pharmaceuticals); inteferon-γ; glucan; hyperimmune gamma-globulin (Bayer); immuthiol (sodium diethylthiocarbamate); interleukin-1 (Hoffmann-LaRoche; Amgen), interleukin-2 (IL-2) (Chiron); isoprinosine (inosine pranobex); Krestin; LC-9018 (Yakult); lentinan (Yamanouchi); LF-1695; methionine-enkephalin; Minophagen C; muramyl tripeptide, MTP-PE; naltrexone (Barr Laboratories); RNA immunomodulator; REMUNE® (Immune Response Corporation); RETICULOSE® (Advanced Viral Research Corporation); shosaikoto; ginseng; thymic humoral factor; Thymopentin; thymosin factor 5; thymosin 1 (ZADAXIN®, SciClone), thymostimulin, TNF (tumor necrosis factor, Genentech), and vitamin preparations.
[0356]Pharmaceutical compositions of the present invention can also further comprise anti-cancer therapeutic agents. Suitable anti-cancer therapeutic agents for optional use include an anti-cancer composition effective to inhibit neoplasia comprising a compound, or a pharmaceutically acceptable salt or prodrug of said anti-cancer agent, which can be used for combination therapy include, but are not limited to alkylating agents, such as busulfan, cis-platin, mitomycin C, and carboplatin antimitotic agents, such as colchicine, vinblastine, taxols, such as paclitaxel (TAXOL®, Bristol-Meyers Squibb) docetaxel (TAXOTERE®, Aventis), topo I inhibitors, such as camptothecin, irinotecan and topotecan (HYCAMTIN®, GlaxoSmithKline), topo II inhibitors, such as doxorubicin, daunorubicin and etoposides such as VP16; RNA/DNA antimetabolites, such as 5-azacytidine, 5-fluorouracil and methotrexate, DNA antimetabolites, such as 5-fluoro-2'-deoxy-uridine, ara-C, hydroxyurea, thioguanine, and antibodies, such as trastuzumab (HERCEPTIN®, Genentech), and rituximab (RITUXAN®, Genentech and Biogen-Idec), melphalan, chlorambucil, cyclophosamide, ifosfamide, vincristine, mitoguazone, epirubicin, aclarubicin, bleomycin, mitoxantrone, elliptinium, fludarabine, octreotide, retinoic acid, tamoxifen, alanosine, and combinations thereof.
[0357]The invention further provides methods for providing anti-bacterial therapeutics, anti-parasitic therapeutics, and anti-fungal therapeutics, for use in combination with the compounds of the invention and pharmaceutically-acceptable salts thereof. Examples of anti-bacterial therapeutics include compounds such as penicillins, ampicillin, amoxicillin, cyclacillin, epicillin, methicillin, nafcillin, oxacillin, cloxacillin, dicloxacillin, flucloxacillin, carbenicillin, cephalexin, cepharadine, cefadoxil, cefaclor, cefoxitin, cefotaxime, ceftizoxime, cefinenoxine, ceftriaxone, moxalactam, imipenem, clavulanate, timentin, sulbactam, erythromycin, neomycin, gentamycin, streptomycin, metronidazole, chloramphenicol, clindamycin, lincomycin, quinolones, rifampin, sulfonamides, bacitracin, polymyxin B, vancomycin, doxycycline, methacycline, minocycline, tetracycline, amphotericin B, cycloserine, ciprofloxacin, norfloxacin, isoniazid, ethambutol, and nalidixic acid, as well as derivatives and altered forms of each of these compounds.
[0358]Examples of anti-parasitic therapeutics include bithionol, diethylcarbamazine citrate, mebendazole, metrifonate, niclosamine, niridazole, oxamniquine and other quinine derivatives, piperazine citrate, praziquantel, pyrantel pamoate and thiabendazole, as well as derivatives and altered forms of each of these compounds.
[0359]Examples of anti-fungal therapeutics include amphotericin B, clotrimazole, econazole nitrate, flucytosine, griseofulvin, ketoconazole and miconazole, as well as derivatives and altered forms of each of these compounds. Anti-fungal compounds also include aculeacin A and papulocandin B.
[0360]The preferred animal subject of the present invention is a mammal. By the term "mammal" is meant an individual belonging to the class Mammalia. The invention is particularly useful in the treatment of human patients.
[0361]The term "treating" means the administering to subjects a compound of Formulae I through XII or a compound identified by one or more assays within the present invention, for purposes which can include prevention, amelioration, or cure of a retroviral-related pathology. Said compounds for treating a subject that are identified by one or more assays within the present inventions are identified as compounds which have the ability to disrupt Gag processing, described herein.
[0362]The term "inhibits the interaction" as used herein, means preventing, or reducing the rate of, direct or indirect association of one or more molecules, peptides, proteins, enzymes, or receptors; or preventing or reducing the normal activity of one or more molecules, peptides, proteins, enzymes or receptors.
[0363]Medicaments are considered to be provided "in combination" with one another if they are provided to the patient concurrently or if the time between the administration of each medicament is such as to permit an overlap of biological activity.
[0364]In one preferred embodiment, at least one compound of Formulae I through XII above comprises a single pharmaceutical composition.
[0365]Pharmaceutical compositions for administration according to the present invention can comprise at least one compound of Formulae I through XII above or compounds identified by one or more assays within the present invention. Said compounds for treating a subject that are identified by one or more assays within the present inventions are identified as compounds which have the ability to disrupt Gag processing, described herein. The compounds according to the present invention are further included in a pharmaceutically acceptable form optionally combined with a pharmaceutically acceptable carrier. These compositions can be administered by any means that achieve their intended purposes. Amounts and regimens for the administration of a compound of Formulae I through XII according to the present invention can be determined readily by those with ordinary skill in the clinical art of treating a retroviral pathology.
[0366]For example, administration can be by parenteral, such as subcutaneous, intravenous, intramuscular, intraperitoneal, transdermal, transmucosal, ocular, rectal, intravaginal, or buccal routes. Alternatively, or concurrently, administration can be by the oral route. The administration may be as an oral or nasal spray, or topically, such as powders, ointments, drops or a patch. The dosage administered depends upon the age, health and weight of the recipient, type of previous or concurrent treatment, if any, frequency of treatment, and the nature of the effect desired.
[0367]Compounds and methods of the invention are useful in additional ways. For example, such compounds may be used prophylatically, to minimize the risk of infection. In another embodiment, a compound may be used to minimize spread of the disease from an infected person.
[0368]The invention is also directed to novel methods of treating HIV in an infected individual. In one embodiment, the invention is particularly useful in stimulating an immune response in a person infected with HIV. For example, by allowing noninfectious virus to be released from infected cells, such infected cells continue to expose antigens and may be effectively targeted by the immune system or other therapies directed against such antigens. In another example, by continuing to permit the release of noninfectious virus, an infected individual continues to develop an immune response to said virus without suffering the deleterious effects of such a virus.
[0369]The invention is also useful in expanding the scope of treatment, and offers novel means of treating disease in patients in need thereof. In another embodiment, the invention may be practiced in a patient who does not respond to other therapy for reasons other than viral resistance. For example, conventional methods of treating HIV, as known in the art, are associated with deleterious side effects. In one embodiment, the methods and compositions of the invention are useful in treating a patient without a reduction in one or more deleterious side effects. In one embodiment the invention includes a method of treating a patient with a compound that does not have a particular side effect or has less of a particular side effect.
[0370]The bioavailability of drugs is also relevant in treatment. In an embodiment, the invention may be practiced such that compounds are more effectively absorbed into infected cells. In one embodiment the invention encompasses improved methods of delivering a drug to a cell infected with HIV.
[0371]Compositions within the scope of this invention include all compositions comprising at least one compound of Formulae I through XII above according to the present invention in an amount effective to achieve its intended purpose. While individual needs vary, determination of optimal ranges of effective amounts of each component is within the skill of the art. For example, a dose may comprise 0.0001 mg to 10 g/kg of body weight. Typical dosages comprise about 0.1 to about 100 mg/kg body weight. The preferred dosages comprise about 1 to about 100 mg/kg body weight of the active ingredient. More preferred dosages comprise about 5 to about 50 mg/kg body weight.
[0372]Administration of a compound of the present invention can also optionally include previous, concurrent, subsequent or adjunctive therapy using immune system boosters or immunomodulators. In addition to the pharmacologically active compounds, a pharmaceutical composition of the present invention can also contain suitable pharmaceutically acceptable carriers comprising excipients and auxiliaries which facilitate processing of the active compounds into preparations which can be used pharmaceutically. Preferably, the preparations, particularly those preparations which can be administered orally and which can be used for the preferred type of administration, such as tablets, dragees, and capsules, and also preparations which can be administered rectally, such as suppositories, as well as suitable solutions for administration by injection or orally, contain from about 0.01 to 99 percent, preferably from about 20 to 75 percent of active compound(s), together with the excipient.
[0373]Pharmaceutical preparations of the present invention are manufactured in a manner which is itself known, for example, by means of conventional mixing, granulating, dragee-making, dissolving, or lyophilizing processes. Thus, pharmaceutical preparations for oral use can be obtained by combining the active compounds with solid excipients, optionally grinding the resulting mixture, and processing the mixture of granules, after adding suitable auxiliaries, if desired or necessary, to obtain tablets or dragee cores.
[0374]Suitable excipients are, e.g., fillers such as saccharide, for example, lactose or sucrose, mannitol or sorbitol; cellulose preparations and/or calcium phosphates, such as tricalcium phosphate or calcium hydrogen phosphate; as well as binders such as starch paste, using, for example, maize starch, wheat starch, rice starch, potato starch, gelatin, tragacanth, cellulose, methyl cellulose, hydroxypropylmethylcellulose, sodium carboxymethylcellulose, and/or polyvinyl pyrrolidone. If desired, disintegrating agents can be added such as the above-mentioned starches and also carboxymethyl starch, cross-linked polyvinyl pyrrolidone, agar, or alginic acid or a salt thereof, such as sodium alginate. Auxiliaries are, above all, flow-regulating agents and lubricants, for example, silica, talc, stearic acid or salts thereof, such as magnesium stearate or calcium stearate, and/or polyethylene glycol. Dragee cores are provided with suitable coatings which, if desired, are resistant to gastric juices. For this purpose, concentrated saccharide solutions can be used, which can optionally contain gum arabic, talc, polyvinyl pyrrolidone, polyethylene glycol and/or titanium dioxide, lacquer solutions and suitable organic solvents or solvent mixtures. In order to produce coatings resistant to gastric juices, solutions of suitable cellulose preparations such as acetylcellulose phthalate or hydroxypropylmethyl cellulose phthalate are used. Dyestuffs or pigments can be added to the tablets or dragee coatings, for example, for identification or in order to characterize combinations of active compound doses.
[0375]Other pharmaceutical preparations which an be used orally include push-fit capsules made of gelatin, as well as soft, sealed capsules made of gelatin and a plasticizer such as glycerol or sorbitol. The push-fit capsules can contain the active compounds in the form of granules which can be mixed with fillers such as lactose, binders such as starches, and/or lubricants such as talc or magnesium stearate and, optionally, stabilizers. In soft capsules, the active compounds are preferably dissolved or suspended in suitable liquids, such as fatty oils or liquid paraffin. In addition, stabilizers can be added.
[0376]Possible pharmaceutical preparations which can be used rectally include, for example, suppositories which consist of a combination of the active compounds with a suppository base. Suitable suppository bases are, for example, natural or synthetic triglycerides, or paraffin hydrocarbons. In addition, it is also possible to use gelatin rectal capsules which consist of a combination of the active compounds with a base. Possible base materials include, for example, liquid triglycerides, polyethylene glycols, or paraffin hydrocarbons.
[0377]Suitable formulations for parenteral administration include aqueous solutions of the active compounds in water-soluble form, for example, water-soluble salts. In addition, suspensions of the active compounds as appropriate oily injection suspensions can be administered. Suitable lipophilic solvents or vehicles include fatty oils, such as sesame oil, or synthetic fatty acid esters, such as ethyl oleate, triglycerides or glycol-400. Aqueous injection suspensions that can contain substances which increase the viscosity of the suspension include, for example, sodium carboxymethyl cellulose, sorbitol, and/or dextran. Optionally, the suspension can also contain stabilizers.
[0378]Liquid dosage forms for oral administration include pharmaceutically acceptable emulsions, solutions, suspensions, syrups, and elixirs. In addition to the active compounds, the liquid dosage forms may contain inert diluents commonly used in the art such as, for example, water or other solvents, solubilizing agents and emulsifiers such as, for example, water or other solvents, solubilizing agents and emulsifiers such as ethyl alcohol, isopropyl alcohol, ethyl carbonate, ethyl acetate, benzyl alcohol, benzyl benzoate, propylene glycol, 1,3-butylene glycol, dimethyl formamide, oils such as cottonseed, groundnut, corn, germ, olive, castor, and sesame oils, glycerol, tetrahydrofurfuryl alcohol, polyethylene glycols and fatty acid esters of sorbitan, and mixtures thereof.
[0379]Suspensions, in addition to the active compounds, may contain suspending agents as, for example, ethoxylated isostearyl alcohols, polyoxyethylene sorbitol and sorbitan esters, cellulose, microcrystalline cellulose, aluminum metahydroxide, bentonite, agar-agar, and tragacanth, and combinations thereof.
[0380]Pharmaceutical compositions for topical administration include formulations appropriate for administration to the skin, mucosa, surfaces of the lung or eye. Compositions may be prepared as a pressurized or non-pressurized dry powder, liquid or suspension. The active ingredients in non-pressurized powdered formulations may be admixed in a finely divided form in a pharmaceutically-acceptable inert carrier, including but not limited to mannitol, fructose, dextrose, sucrose, lactose, saccharin or other sugars or sweeteners.
[0381]The pressurized composition may contain a compressed gas, such as nitrogen, or a liquefied gas propellant. The propellant may also contain a surface-active ingredient, which may be a liquid or solid non-ionic or anionic agent. The anionic agent may be in the form of a sodium salt.
[0382]A formulation for use in the eye would comprise a pharmaceutically acceptable ophthalmic carrier, such as an ointment, oils, such as vegetable oils, or an encapsulating material. The regions of the eye to be treated include the corneal region, or internal regions such as the iris, lens, ciliary body, anterior chamber, posterior chamber, aqueous humor, vitreous humor, choroid or retina.
[0383]Compositions for rectal administration may be in the form of suppositories. Compositions for use in the vagina may be in the form of suppositories, creams, foams, or in-dwelling vaginal inserts.
[0384]The compositions may be administered in the form of liposomes. Liposomes may be made from phospholipids, phosphatidyl cholines (lecithins) or other lipoidal compounds, natural or synthetic, as known in the art. Any non-toxic, pharmacologically acceptable lipid capable of forming liposomes may be used. The liposomes may be multilamellar or mono-lamellar.
[0385]A pharmaceutical formulation for systemic administration according to the invention can be formulated for enteral, parenteral or topical administration. Indeed, all three types of formulation can be used simultaneously to achieve systemic administration of the active ingredient.
[0386]Suitable formulations for oral administration include hard or soft gelatin capsules, dragees, pills, tablets, including coated tablets, elixirs, suspensions, syrups or inhalations and controlled release forms thereof.
[0387]The compounds of Formula I or II above or compounds identified by one or more assays within the present invention and have the ability to disrupt Gag processing, can also be administered in the form of an implant when compounded with a biodegradable slow-release carrier. Alternatively, the compounds of the present invention can be formulated as a transdermal patch for continuous release of the active ingredient.
[0388]The following examples are illustrative only and are not intended to limit the scope of the invention as defined by the appended claims. It will be apparent to those skilled in the art that various modifications and variations can be made in the methods of the present invention without departing from the spirit and scope of the invention. Thus, it is intended that the present invention covers the modifications and variations of this invention provided they come within the scope of the appended claims and their equivalents.
EXAMPLES
Example 1
Anti-Viral Activity Against Primary HIV-1 Isolates
[0389]A robust virus inhibition assay was used to evaluate the anti-viral activity of DSB against primary HIV-1 isolates propagated in PMBC. Briefly, serial dilutions of DSB were made in medium into 96-well tissue culture plates. 25-250 TCID50 of virus and 5×105 PHA-stimulated PBMCs were added to each well. On days 1, 3 and 5 post-infection, media was removed from each well and replaced with fresh media containing DSB at the appropriate concentration. On day 7 post-infection, culture supernatant was removed from each well for p24 detection of virus replication and 50% inhibitory concentrations (IC50) were calculated by standard methods.
[0390]Table 5 shows the potent anti-viral activity of DSB against a panel of primary HIV-1 isolates. DSB exhibits levels of activity similar to approved drugs that were tested in parallel. Importantly, the activity of DSB was not restricted by co-receptor usage.
TABLE-US-00011 TABLE 5 Inhibitory activity (IC50) of DSB and two approved drugs against a panel of primary Clade B HIV-1 isolates. Clinical HIV-1 isolates denoted by * were isolated at Panacos. All other virus isolates were obtained from the NIH AIDS Reference Repository. IC50 (nM) Virus Isolate # Co-Receptor usage DSB AZT Nevirapine BZ167 X4 4.0 2.2 31.2 92HT599 X4 9.8 5.8 25.3 US1 R5 5.6 0.9 22.1 19101N* R5 3.8 2.4 59.4 3401N* R5/X4 12.0 17.5 32.1 92US723 R5/X4 4.6 1.2 26.8 22101N* R5/X4 2.6 0.9 4.9 Mean 6.1 4.4 28.8 Note: R5 and X4 refer to the chemokine receptors CCR5 and CXCR4 respectively.
[0391]Toxicity of DSB was analyzed by incubating with PHA-stimulated PBMC for 7 days at a range of concentrations, then determining cell viability using the XTT method. The 50% cytotoxic concentration was >30 μM, corresponding to an in vitro therapeutic index of approximately 5000.
Example 2
Anti-Viral Activity of DSB Against Drug Resistant HIV-1 Isolates
[0392]The activity of DSB was tested against a panel of HIV-1 isolates resistant to approved drugs. These viruses were obtained from the NIH AIDS Research and Reference Reagent Program. Assays were performed using virus propagated in PBMCs with a p24 endpoint (above), or using cell line targets (MT-2 cells) and a cell killing endpoint. The MT-2 assay format was as follows. Serial dilutions of DSB, or each approved drug, were prepared in 96 well plates. To each sample well was added media containing MT-2 cells at 3×105 cells/mL and virus inoculum at a concentration necessary to result in 80% killing of the cell targets at 5 days post-infection (PI). On day 5 post-infection, virus-induced cell killing was determined by the XTT method and the inhibitory activity of the compound was determined.
[0393]Table 6 shows the potent anti-viral activity of DSB against a panel of drug-resistant HIV-1 isolates. The results were not significantly different from those obtained with the panel of wild-type isolates (Table 5), demonstrating that DSB retains its activity against virus strains resistant to all of the major classes of approved drugs.
TABLE-US-00012 TABLE 6 Inhibitory activity (nM IC50) of DSB against a panel of drug resistant HIV-1 isolates. Assays were done in fresh PBMC with a p24 endpoint except for the NNRTI-resistant isolates that were performed in MT-2 cells with a cell viability (XTT) endpoint. Virus Co- Isolate Receptor IC50(nM) # Mutation(s) usage DSB AZT Nevirapine Indinavir NRTI-resistant 1 K70R R5/X4 4.4 86.4 (54X)* ND 9.8 T215Y/F 2 K70R R5/X4 4.2 63.4 (40X) ND 6.1 T215Y/F NNRTI-resistant 3 Y181C X4 1.0 5.1 >3800 2.5 (>177X) 4 K103N X4 1.3 2.0 2630 4.5 Y181C (122X) Protease-resistant 5 V82A X4 5.6 13.1 ND 39.7 (12X) 6 I84V X4 5.5 14.4 ND 32.7 (10X) 7 L10R/M46I/ X4 12.9 3.5 ND 72.5 L63P/V82T/I84V (23X) *Fold Resistance. Note: R5 and X4 refer to the chemokine receptors CCR5 and CXCR4 respectively.
Example 3
DSB Inhibits HIV-1 Replication at a Late Step in the Virus Life Cycle
[0394]To distinguish the inhibitory activity of DSB against early and late replication targets, a multinuclear activation of a galactosidase indicator (MAGI) assay was used. In this assay, the targets are HeLa cells stably expressing CD4, CXCR4, CCR5 and a reporter construct consisting of the -galactosidase gene (modified to localize to the nucleus) driven by a truncated HIV-1 LTR. Infection of these cells results in expression of Tat that drives activation of the β-galactosidase reporter gene. Expression of β-galactosidase in infected cells is detected using the chromogenic substrate X-gal. As shown in Table 7, the entry inhibitor T-20, the NRTI AZT and the NNRTI nevirapine caused significant reductions in β-galactosidase gene expression in HIV-1 infected MAGI cells due to their ability to disrupt early steps in viral replication that affect Tat protein expression. In contrast, the protease inhibitor indinavir targets a late step in virus replication (following Tat expression) and does not prevent β-galactosidase gene expression in this system. Similar results were obtained with DSB as with indinavir, indicating that DSB blocks virus replication at a time point following the completion of proviral DNA integration and synthesis of the viral transactivating protein (Table 7).
TABLE-US-00013 TABLE 7 Effect of DSB and inhibitors of entry (the gp41 peptide T-20), RT (AZT and Nevirapine) and protease (indinavir) on expression of b-galactosidase in HIV-1 infected MAGI cells. The DMSO control contained no drug. Inhibitor DMSO T-20 AZT Nevirapine Indinavir DSB % Decrease 0 98 82 85 10 12 (β- galactosidase expression)
[0395]Kanamoto et al. (Antimicrob. Agents Chemother., April; 45(4):1225-30, (2002)) have also reported that DSB acts at a late step in HIV replication. However, they reported that the compound inhibits release of virus from chronically-infected cells. In contrast, our data using a variety of experimental systems indicate that DSB does not have a significant effect on virus release (e.g. Example 6).
Example 4
DSB does not Inhibit HIV-1 Protease Activity
[0396]It was previously determined that DSB had no effect on HIV-1 protease function using a cell-free fluorometric assay that characterized enzyme activity by following the cleavage of a synthetic peptide substrate. The results of these experiments indicated that at concentrations up to 50 μg/mL that DSB had no effect on protease function. As a result of the observation that DSB blocks virus replication at a late step, studies were also performed using a recombinant form of the Gag protein, which is a more relevant system than the synthetic peptide substrate used in the initial assays. The use of the recombinant Gag protein as substrate resulted in a similar experimental outcome. In these experiments DSB did not disrupt protease-mediated Gag protein processing at concentrations as high as 50 μg/mL. In contrast, as expected, the protease inhibitor indinavir blocked Gag protein processing at 5 μg/mL (FIG. 1).
Example 5
DSB Causes a Defect in the Final Step of Gag Processing (CA-SP1 Cleavage) that has been Associated with Viral Maturation Defects
[0397]In order to better define DSB's mechanism of action, a detailed examination was undertaken of the virus produced from HIV-1-infected cell lines treated with DSB. Briefly, H9 cells chronically infected with the HIV-1IIIB isolate were treated with DSB at 1 μg/mL for a period of 48 hrs. Indinavir was used as a control. At the 48 hr time-point, spent media was removed and fresh media containing compound was added. At 24, 48 and 72 hrs post fresh compound addition, both cells and supernatant were recovered for analysis. The level of virus in the culture supernatant was determined and western blots were used to characterize viral protein production in both cell-associated and cell-free virus. As observed in previous experiments, DSB did not cause a significant reduction in the amount of virus produced by chronically infected H9 cells, however, there was a defect in Gag processing in both cell-associated and cell-free virus. This defect took the form of an additional band in the western blots corresponding to p25 (FIG. 2). This p25 band results from the incomplete processing of the capsid CA-SP1 precursor. DSB treatment of HIV-2 and SIV chronically infected cell lines exhibited normal Gag processing consistent with the observed lack of antiviral activity against these viruses. The Gag processing defect seen in the presence of DSB is completely distinct from that observed with the protease inhibitor indinavir (FIG. 2). As discussed above, mutations at the p25 to p24 cleavage site that prevent processing are associated with defects in viral maturation and infectivity (Wiegers K. et al., J. Virol. 72:2846-54 (1998)).
[0398]As previously discussed (C. T. Wild et al., XIV Int. AIDS Conf. Barcelona, Spain, Abstract MoPeA3030, (July 2002)), abnormal p25 to p24 processing is also seen in other maturation budding defects. These include mutations in the Gag late domain (PTAP) or defects in TSG-101 mediated viral assembly that disrupt budding (Garrus, J. E et al., Cell, 107:55-65, (2001); Demirov, D. G. et al., J. Virology 76:105-117, (2002)). However, these mutations cause inhibition of virus release, while DSB treatment does not have a significant effect on virus release. The morphology of these maturation/budding mutants is also quite distinct from that following DSB-treatment (see Example 6).
[0399]In addition, mutations that interfere with viral RNA dimerization and lead to the production of immature virus with defective core structures give a similar Gag processing phenotype (Liang, C. et al., J. Virology, 73:6147-6151, (1999)). However, in those cases RNA incorporation is inhibited and the morphology of particles released is distinct from those following DSB treatment (see Example 6).
Example 6
DSB Treatment Effects HIV-1 Maturation as Determined by Electron Microscopy (EM)
[0400]It has been demonstrated that mutations in HIV-1 Gag that disrupt p25 to p24 processing give rise to non-infectious viral particles characterized by an internal morphology distinct from normal virus (Wiegers K. et al., J. Virol. 72:2846-54 (1998)). To determine if virus generated in the presence of DSB exhibited this distinct morphology the following experiment was carried out.
[0401]HeLa cells were transfected with HIV-1 infectious molecular clone pNL4-3 and treated as described previously with DSB. Following treatment, DSB-treated infected cells were fixed in glutaraldehyde and analyzed by EM. The results of this analysis are shown in FIG. 3.
[0402]These results are consistent with a compound that disrupts p25 to p24 processing which generates non-infectious morphologically aberrant viral particles.
[0403]3-O-(3',3'-dimethylsuccinyl)betulinic acid (DSB) is an example of a compound that disrupts p25 to p24 processing and potently inhibits HIV-1 replication. However, this compound does not inhibit PR activity, and its action is specific for the p25 to p24 processing step, not other steps in Gag processing. Furthermore, DSB treatment results in the aberrant HIV particle morphology described above.
Example 7
[0404]In vitro selection for HIV-1 isolates resistant to compounds that disrupt the processing of the viral Gag capsid (CA) protein from the CA-spacer peptide 1 protein precursor.
[0405]A series of experiments were performed to select for viruses resistant to inhibition by 3-O-(3',3'-dimethylsuccinyl)betulinic acid (DSB), an inhibitor HIV-1 maturation. For each experiment, either NL4-3 or RF virus isolate was used to infect two cell cultures. Following infection, one culture was maintained in growth medium containing DSB, while the other culture was maintained in parallel in growth medium lacking DSB.
[0406]In one experiment, H9 cells that had been infected with RF virus were maintained in the presence or absence of increasing concentrations of DSB (0.05-1.6 μg/ml). The cells were passaged every 2-3 days with the addition of fresh drug. Virus replication was monitored by p24 ELISA every 7 days. At that time, DSB-treated cultures with high levels of p24 were passaged by co-cultivation with fresh uninfected H9 cells at a 1:1 ratio of cells in the presence of 1× or 2× the original concentration of DSB. After 8 weeks of co-cultivation, cell-free virus was collected from the culture containing DSB at a concentration of 1.6 μg/ml and used to infect fresh H9 cells. Every 7 days, virus from cultures containing high levels of p24 was passaged by cell-free infection in the presence of 1× or 2× the original concentration of DSB. After 5 weeks of cell-free passaging, virus from the culture containing 3.2 μg/ml DSB was collected and used to infect MT-2 cells. Virus replication in the MT-2 cells, was monitored by observing syncytia formation microscopically. Every 1-3 days, the cells were washed to remove input virus, and fresh drug was added to the culture under selection. Every 3-4 days, following the emergence of extensive syncytia in the culture under selection, supernatant from each culture was collected and passed through a 0.45 μm filter to remove cell debris. This filtered virus supernatant was then used to infect fresh MT-2 cells in the presence or absence of fresh drug. After 4 rounds of cell-free infection (approximately 2 weeks in culture), with the concentration of drug at 3.2 μg/ml, virus stocks were collected and frozen for further analysis.
[0407]In a second experiment, a stock of virus derived from the molecular clone pNL4-3 (5.7×104 TCID50) was used to infect MT-2 cells (6×106 cells) and cultures were maintained in the presence or absence of PA-457DSB at a concentration of 1.6 μg/ml. Every 1-3 days, the cells were washed to remove input virus, and fresh drug was added to the culture under selection. Virus replication was monitored by observing syncytia formation microscopically. Every 3-7 days, following the emergence of extensive syncytia in the culture under selection, supernatant from each culture was collected and passed through a 0.45 μm filter to remove cell debris. This filtered virus supernatant was then used to infect fresh MT-2 cells in the presence or absence of fresh drug. After 5 rounds of cell-free infection, and every other round thereafter, the concentration of drug was doubled. After 10 rounds of cell-free infection (approximately 7 weeks in culture), when the concentration of drug reached 12.8 μg/ml, virus stocks were collected and frozen for further analysis.
Example 8
[0408]Characterization of HIV-1 isolates selected for resistance to compounds that disrupt the processing of the viral Gag capsid (CA) protein from the CA-spacer peptide 1 protein precursor.
[0409]Virus stocks derived as described above were further analyzed both phenotypically and genotypically to characterize the nature of their drug-resistance. The resistance of the viruses to 3-O-(3',3'-dimethylsuccinyl)-betulinic acid (DSB) was determined in virus replication assays. Briefly, the virus stocks were first titered in H9 cells by quantitating the levels of p24 (by ELISA) in cultures 8 days after infection with serial 4-fold dilutions of virus. Virus input was then normalized for a second assay in which each virus is cultured for 8 days in the presence of serial 4-fold dilutions of drug. The IC50 for each virus was determined as the dilution of drug that reduced the p24 endpoint level by 50% as compared to the no-drug control. In these assays, the two independently derived virus stocks resulted in IC50 values greater than 2 μM for DSB, as compared to an IC50 of 0.02 μM for virus that had been cultured in parallel in the absence of drug. In a subsequent series of experiments, the A364V mutation was engineered into the HIV-1 NL4-3 proviral DNA, which was subsequently transfected into HeLa cells. Resulting virus was collected and used to test the activity of DSB in a viral replication assay, as described above. In these assays, the DSB-resistant virus resulted in an IC50 value of 0.1 μM whereas wild-type NL4-3 gave an IC50 value of 0.01 μM.
[0410]To determine if the resistant viruses were able to escape the CA-SP1 cleavage defect caused by DSB in wild-type virus, stocks of each virus grown in either the presence or absence of drug were analyzed by Western blot. Virus was pelleted through a 20% sucrose cushion from filtered culture supernatants that were collected 60 hr post-infection and 18 hr after the cells had been washed and fresh drug added. The viruses were lysed, and the amount of each virus was normalized by quantitating p24 levels in each sample. Western blot analysis of the viral proteins in each sample demonstrated that the drug-resistant viruses did not contain the CA-SP1 product in the presence of DSB, confirming that these viruses were resistant to the effects of the drug on this cleavage event.
[0411]Finally, to identify the genetic determinants of DSB resistance, the entire Gag and PR coding regions of the viral genomes were amplified by high-fidelity RT-PCR for sequencing. The viral RNA was purified from each virus lysate prepared as described above and digested with DNase to remove any contaminating DNA. The RT-PCR products were then gel-purified to remove any non-specific PCR products. Finally, both strands of the resulting DNA fragments were sequenced using overlapping a series of primers. Two amino acid mutations were identified that are independently capable of conferring resistance to DSB, an alanine to valine substitution in the Gag polyprotein at residue 364 in the NL4-3 isolate and at residue 366 in the RF isolate. These are the first and the third residues, respectively, downstream of the CA-SP1 cleavage site (the N-terminus of SP1). Alanine is highly conserved at each of these positions throughout all HIV-1 clades in the database.
[0412]Having now fully described this invention, it will be understood to those of ordinary skill in the art that the same can be performed within a wide and equivalent range of conditions, formulations and other parameters without affecting the scope of the invention or any embodiment thereof. All patents, applications and publications cited herein are fully incorporated by reference in their entirety.
Sequence CWU
1
SEQUENCE LISTING
<160> NUMBER OF SEQ ID NOS: 115
<210> SEQ ID NO 1
<211> LENGTH: 14
<212> TYPE: PRT
<213> ORGANISM: HIV-1
<220> FEATURE:
<221> NAME/KEY: MISC_FEATURE
<222> LOCATION: (1)..(1)
<223> OTHER INFORMATION: Xaa can be either Glycine or Serine
<220> FEATURE:
<221> NAME/KEY: MISC_FEATURE
<222> LOCATION: (6)..(6)
<223> OTHER INFORMATION: Xaa can be either Valine or Isoleucine
<400> SEQUENCE: 1
Xaa His Lys Ala Arg Xaa Leu Ala Glu Ala Met Ser Gln Val
1 5 10
<210> SEQ ID NO 2
<211> LENGTH: 14
<212> TYPE: PRT
<213> ORGANISM: HIV-1 NL4-3
<400> SEQUENCE: 2
Gly His Lys Ala Arg Val Leu Val Glu Ala Met Ser Gln Val
1 5 10
<210> SEQ ID NO 3
<211> LENGTH: 14
<212> TYPE: PRT
<213> ORGANISM: HIV-1 RF
<400> SEQUENCE: 3
Ser His Lys Ala Arg Ile Leu Ala Glu Val Met Ser Gln Val
1 5 10
<210> SEQ ID NO 4
<211> LENGTH: 1823
<212> TYPE: DNA
<213> ORGANISM: HIV-1 NL4-3
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (1365)..(1365)
<223> OTHER INFORMATION: N can be any nucleotide: a, t, g, c
<400> SEQUENCE: 4
atgggtgcga gagcgtcggt attaagcggg ggagaattag ataaatggga aaaaattcgg 60
ttaaggccag ggggaaagaa acaatataaa ctaaaacata tagtatgggc aagcagggag 120
ctagaacgat tcgcagttaa tcctggcctt ttagagacat cagaaggctg tagacaaata 180
ctgggacagc tacaaccatc ccttcagaca ggatcagaag aacttagatc attatataat 240
acaatagcag tcctctattg tgtgcatcaa aggatagatg taaaagacac caaggaagcc 300
ttagataaga tagaggaaga gcaaaacaaa agtaagaaaa aggcacagca agcagcagct 360
gacacaggaa acaacagcca ggtcagccaa aattacccta tagtgcagaa cctccagggg 420
caaatggtac atcaggccat atcacctaga actttaaatg catgggtaaa agtagtagaa 480
gagaaggctt tcagcccaga agtaataccc atgttttcag cattatcaga aggagccacc 540
ccacaagatt taaataccat gctaaacaca gtggggggac atcaagcagc catgcaaatg 600
ttaaaagaga ccatcaatga ggaagctgca gaatgggata gattgcatcc agtgcatgca 660
gggcctattg caccaggcca gatgagagaa ccaaggggaa gtgacatagc aggaactact 720
agtacccttc aggaacaaat aggatggatg acacataatc cacctatccc agtaggagaa 780
atctataaaa gatggataat cctgggatta aataaaatag taagaatgta tagccctacc 840
agcattctgg acataagaca aggaccaaag gaacccttta gagactatgt agaccgattc 900
tataaaactc taagagccga gcaagcttca caagaggtaa aaaattggat gacagaaacc 960
ttgttggtcc aaaatgcgaa cccagattgt aagactattt taaaagcatt gggaccagga 1020
gcgacactag aagaaatgat gacagcatgt cagggagtgg ggggacccgg ccataaagca 1080
agagttttgg ttgaagcaat gagccaagta acaaatccag ctaccataat gatacagaaa 1140
ggcaatttta ggaaccaaag aaagactgtt aagtgtttca attgtggcaa agaagggcac 1200
atagccaaaa attgcagggc ccctaggaaa aagggctgtt ggaaatgtgg aaaggaagga 1260
caccaaatga aagattgtac tgagagacag gctaattttt tagggaagat ctggccttcc 1320
cacaagggaa ggccagggaa ttttcttcag agcagaccag agccnacagc cccaccagaa 1380
gagagcttca ggtttgggga agagacaaca actccctctc agaagcagga gccgatagac 1440
aaggaactgt atcctttagc ttccctcaga tcactctttg gcagcgaccc ctcgtcacaa 1500
taaagatagg ggggcaatta aaggaagctc tattagatac aggagcagat gatacagtat 1560
tagaagaaat gaatttgcca ggaagatgga aaccaaaaat gataggggga attggaggtt 1620
ttatcaaagt aagacagtat gatcagatac tcatagaaat ctgcggacat aaagctatag 1680
gtacagtatt agtaggacct acacctgtca acataattgg aagaaatctg ttgactcaga 1740
ttggctgcac tttaaatttt cccattagtc ctattgagac tgtaccagta aaattaaagc 1800
caggaatgga tggcccaaaa gtt 1823
<210> SEQ ID NO 5
<211> LENGTH: 1820
<212> TYPE: DNA
<213> ORGANISM: HIV-1 NL4-3
<400> SEQUENCE: 5
atgggtgcga gagcgtcggt attaagcggg ggagaattag ataaatggga aaaaattcgg 60
ttaaggccag ggggaaagaa acaatataaa ctaaaacata tagtatgggc aagcagggag 120
ctagaacgat tcgcagttaa tcctggcctt ttagagacat cagaaggctg tagacaaata 180
ctgggacagc tacaaccatc ccttcagaca ggatcagaag aacttagatc attatataat 240
acaatagcag tcctctattg tgtgcatcaa aggatagatg taaaagacac caaggaagcc 300
ttagataaga tagaggaaga gcaaaacaaa agtaagaaaa aggcacagca agcagcagct 360
gacacaggaa acaacagcca ggtcagccaa aattacccta tagtgcagaa cctccagggg 420
caaatggtac atcaggccat atcacctaga actttaaatg catgggtaaa agtagtagaa 480
gagaaggctt tcagcccaga agtaataccc atgttttcag cattatcaga aggagccacc 540
ccacaagatt taaataccat gctaaacaca gtggggggac atcaagcagc catgcaaatg 600
ttaaaagaga ccatcaatga ggaagctgca gaatgggata gattgcatcc agtgcaggca 660
gggcctattg caccaggcca gatgagagaa ccaaggggaa gtgacatagc aggaactact 720
agtacccttc aggaacaaat aggatggatg acacataatc cacctatccc agtaggagaa 780
atctataaaa gatggataat cctgggatta aataaaatag taagaatgta tagccctacc 840
agcattctgg acataagaca aggaccaaag gaacccttta gagactatgt agaccgattc 900
tataaaactc taagagccga gcaagcttca caagaggtaa aaaattggat gacagaaacc 960
ttgttggtcc aaaatgcgaa cccagattgt aagactattt taaaagcatt gggaccagga 1020
gcgacactag aagaaatgat gacagcatgt cagggagtgg ggggacccgg ccataaagca 1080
agagttttgg ctgaagcaat gagccaagta acaaatccag ctaccataat gatacagaaa 1140
ggcaatttta ggaaccaaag aaagactgtt aagtgtttca attgtggcaa agaagggcac 1200
atagccaaaa attgcagggc ccctaggaaa aagggctgtt ggaaatgtgg aaaggaagga 1260
caccaaatga aagattgtac tgagagacag gctaattttt tagggaagat ctggccttcc 1320
cacaagggaa ggccagggaa ttttcttcag agcagaccag agccaacagc cccaccagaa 1380
gagagcttca ggtttgggga agagacaaca actccctctc agaagcagga gccgatagac 1440
aaggaactgt atcctttagc ttccctcaga tcactctttg gcagcgaccc ctcgtcacaa 1500
taaagatagg ggggcaatta aaggaagctc tattagatac aggagcagat gatacagtat 1560
tagaagaaat gaatttgcca ggaagatgga aaccaaaaat gataggggga attggaggtt 1620
ttatcaaagt aagacagtat gatcagatac tcatagaaat ctgcggacat aaagctatag 1680
gtacagtatt agtaggacct acacctgtca acataattgg aagaaatctg ttgactcaga 1740
ttggctgcac tttaaatttt cccattagtc ctattgagac tgtaccagta aaattaaagc 1800
caggaatgga tggcccaaag 1820
<210> SEQ ID NO 6
<211> LENGTH: 1982
<212> TYPE: DNA
<213> ORGANISM: HIV-1 RF
<220> FEATURE:
<221> NAME/KEY: misc_feature
<222> LOCATION: (1961)..(1961)
<223> OTHER INFORMATION: n can be any nucleotide: a, t, g, c
<400> SEQUENCE: 6
atgggtgcga gagcgtcagt attaagcggc ggaaaattag acaaatggga aaaaattcgg 60
ttaaggccag ggggaaagaa aagatataag ttaaaacata taatatgggc aagcagggag 120
ctagaacgat ttgctgtcaa tcctggcctt ttagagacag cagagggctg tagacaaata 180
ctgggacagc tacaaccagc ccttcagaca ggatcagaag aacttaaatc attatataat 240
gcagtagcaa ccctctattg tgtacatcaa aatatagagg taagagacac caaggaagct 300
ttagacaaga tagaggaaga gcaaaacaaa agtaagaaaa aagcacagca agcagcagct 360
gacacaggaa acggcagcca ggtcagccaa aattacccta tagtgcagaa ccttcagggg 420
caaatggtac atcaagccat atcacctaga actttaaatg catgggtaaa agtagtagaa 480
gagaaggctt ttagcccaga agtaataccc atgttttcag cattatcaga aggagccacc 540
ccacaagatt taaacaccat gctaaacaca gtggggggac atcaagcagc catgcaaatg 600
ttaaaagaga ctatcaatga ggaagctgca gaatgggata gattgcatcc agtgcaagca 660
gggcctattg caccaggcca gatgagagaa ccaaggggaa gtgacatagc aggaaccact 720
agtacccttc aggaacaaat aggatggatg acaaataatc cacctatccc agtaggagaa 780
atctataaaa ggtggataat tctgggatta aataaaatag taagaatgta tagccccatc 840
agcattctgg acataagaca aggacctaag gaacccttta gagactatgt agaccggttc 900
tataaaactc taagagccga gcaagcttca caggatgtaa aaaattggat gacagaaacc 960
ttgctggtcc aaaatgcgaa cccagattgt aaaactattt taaaagcatt gggaccagca 1020
gctacactag aagaaatgat gacagcatgt cagggagtag ggggacccag ccataaagca 1080
agaattttgg ctgaagtaat gagccaagta acaaattcag ctaccataat gctgcagaaa 1140
ggtaatttta gggaccaaag aaaaattgtt aagtgtttca actgtggcaa agtagggcac 1200
atagccaaaa attgcagggc ccctaggaaa aagggctgtt ggaaatgtgg aaaggaagga 1260
caccaaatga aagattgcac tactgaggga cgacaggcta attttttagg gaaaatctgg 1320
ccttcccaca agggaaggcc agggaacttt cttcagagca gaccagagcc aacagcccca 1380
ccagaagaga gcttcaggtt tggggaagag acaactccct ctcagaagca ggagaagata 1440
gacaaggaac tgtatccttt agcttccctc aaatcactct ttggcaacga cccatcgtca 1500
cagtaaagat aggggggcaa ttaaaggaag ctctattaga tacaggagca gatgatacag 1560
tattagaaga aatgaatttg ccaggaaaat ggaaaccaaa aatgataggg ggaattggag 1620
gttttatcaa agtaaggcag tatgatcaaa tactcataga aatctgtgga cataaagcta 1680
taggtacagt attagtagga cctacacctg tcaacataat tggaagaaat ctgttgactc 1740
agattggttg cactttaaat tttcccatta gtcctattga aactatacca gtaaaattaa 1800
agccaggaat ggatggccca aaagttaaac aatggccatt gacagaggaa aaaataaaag 1860
cattgataga aatttgtaca gaaatggaaa aggaaggaaa aatttcaaaa attgggcctg 1920
aaaatccata caatactcca gtatttgcca taaagaaaaa ngacagtact aaatggagaa 1980
aa 1982
<210> SEQ ID NO 7
<211> LENGTH: 1904
<212> TYPE: DNA
<213> ORGANISM: HIV-1 RF
<400> SEQUENCE: 7
atgggtgcga gagcgtcagt attaagcggc ggaaaattag acaaatggga aaaaattcgg 60
ttaaggccag ggggaaagaa aagatataag ttaaaacata taatatgggc aagcagggag 120
ctagaacgat ttgctgtcaa tcctggcctt ttagagacag cagagggctg tagacaaata 180
ctgggacagc tacaaccagc ccttcagaca ggatcagaag aacttaaatc attatataat 240
gcagtagcaa ccctctattg tgtacatcaa aatatagagg taagagacac caaggaagct 300
ttagacaaga tagaggaaga gcaaaacaaa agtaagaaaa aagcacagca agcagcagct 360
gacacaggaa acggcagcca ggtcagccaa aattacccta tagtgcagaa ccttcagggg 420
caaatggtac atcaagccat atcacctaga actttaaatg catgggtaaa agtagtagaa 480
gagaaggctt ttagcccaga agtaataccc atgttttcag cattatcaga aggagccacc 540
ccacaagatt taaacaccat gctaaacaca gtggggggac atcaagcagc catgcaaatg 600
ttaaaagaga ctatcaatga ggaagctgca gaatgggata gattgcatcc agtgcaagca 660
gggcctattg caccaggcca gatgagagaa ccaaggggaa gtgacatagc aggaaccact 720
agtacccttc aggaacaaat aggatggatg acaaataatc cacctatccc agtaggagaa 780
atctataaaa ggtggataat tctgggatta aataaaatag taagaatgta tagccccatc 840
agcattctgg acataagaca aggacctaag gaacccttta gagactatgt agaccggttc 900
tataaaactc taagagccga gcaagcttca caggatgtaa aaaattggat gacagaaacc 960
ttgctggtcc aaaatgcgaa cccagattgt aaaactattt taaaagcatt gggaccagca 1020
gctacactag aagaaatgat gacagcatgt cagggagtag ggggacccag ccataaagca 1080
agaattttgg ctgaagcaat gagccaagta acaaattcag ctaccataat gctgcagaaa 1140
ggtaatttta gggaccaaag aaaaattgtt aagtgtttca actgtggcaa agtagggcac 1200
atagccaaaa attgcagggc ccctaggaaa aagggctgtt ggaaatgtgg aaaggaagga 1260
caccaaatga aagattgcac tactgaggga cgacaggcta attttttagg gaaaatctgg 1320
ccttcccaca agggaaggcc agggaacttt cttcagagca gaccagagcc aacagcccca 1380
ccagaagaga gcttcaggtt tggggaagag acaactccct ctcagaagca ggagaagata 1440
gacaaggaac tgtatccttt agcttccctc aaatcactct ttggcaacga cccatcgtca 1500
cagtaaagat aggggggcaa ttaaaggaag ctctattaga tacaggagca gatgatacag 1560
tattagaaga aatgaatttg ccaggaaaat ggaaaccaaa aatgataggg ggaattggag 1620
gttttatcaa agtaaggcag tatgatcaaa tactcataga aatctgtgga cataaagcta 1680
taggtacagt attagtagga cctacacctg tcaacataat tggaagaaat ctgttgactc 1740
agattggttg cactttaaat tttcccatta gtcctattga aactatacca gtaaaattaa 1800
agccaggaat ggatggccca aaagttaaac aatggccatt gacagaggaa aaaataaaag 1860
cattgataga aatttgtaca gaaatggaaa aggaaggaaa aatt 1904
<210> SEQ ID NO 8
<211> LENGTH: 42
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Region of the CA-SP1 cleavage site from
DSB-sensitive HIV-1 isolates NL4-3
<400> SEQUENCE: 8
ggccataaag caagagtttt ggttgaagca atgagccaag ta 42
<210> SEQ ID NO 9
<211> LENGTH: 42
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Region of the CA-SP1 cleavage site from
DSB-sensitive HIV-1 isolates RF
<400> SEQUENCE: 9
agccataaag caagaatttt ggctgaagta atgagccaag ta 42
<210> SEQ ID NO 10
<211> LENGTH: 42
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Region of the CA-SP1 cleavage site from
DSB-sensitive HIV-1 isolates NL4-3
<400> SEQUENCE: 10
ggccataaag caagagtttt ggctgaagca atgagccaag ta 42
<210> SEQ ID NO 11
<211> LENGTH: 64
<212> TYPE: PRT
<213> ORGANISM: HIV-1 RF
<400> SEQUENCE: 11
Lys Asn Trp Met Thr Glu Thr Phe Leu Val Gln Asn Ala Asn Pro Asp
1 5 10 15
Cys Lys Thr Ile Leu Lys Ala Leu Gly Pro Ala Ala Thr Leu Glu Glu
20 25 30
Met Met Thr Ala Cys Gln Gly Val Gly Gly Pro Ser His Lys Ala Arg
35 40 45
Ile Leu Ala Glu Ala Met Ser Gln Val Thr Asn Ser Ala Thr Ile Met
50 55 60
<210> SEQ ID NO 12
<211> LENGTH: 42
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Region of the CA-SP1 cleavage site from
DSB-sensitive HIV-1 isolates RF
<400> SEQUENCE: 12
agccataaag caagaatttt ggctgaagca atgagccaag ta 42
<210> SEQ ID NO 13
<211> LENGTH: 64
<212> TYPE: PRT
<213> ORGANISM: HIV-1 NL4-3
<400> SEQUENCE: 13
Lys Asn Trp Met Thr Glu Thr Leu Leu Val Gln Asn Ala Asn Pro Asp
1 5 10 15
Cys Lys Thr Ile Leu Lys Ala Leu Gly Pro Gly Ala Thr Leu Glu Glu
20 25 30
Met Met Thr Ala Cys Gln Gly Val Gly Gly Pro Gly His Lys Ala Arg
35 40 45
Val Leu Ala Glu Ala Met Ser Gln Val Thr Asn Pro Ala Thr Ile Met
50 55 60
<210> SEQ ID NO 14
<211> LENGTH: 24
<212> TYPE: PRT
<213> ORGANISM: SIV mac239
<400> SEQUENCE: 14
Gly Gln Lys Ala Arg Leu Met Ala Glu Ala Leu Lys Glu Ala Leu Ala
1 5 10 15
Pro Val Pro Ile Pro Phe Ala Ala
20
<210> SEQ ID NO 15
<211> LENGTH: 24
<212> TYPE: PRT
<213> ORGANISM: SIV DD
<400> SEQUENCE: 15
Gly Gln Lys Ala Arg Leu Met Ala Glu Ala Met Ser Gln Val Leu Ala
1 5 10 15
Pro Val Pro Ile Pro Phe Ala Ala
20
<210> SEQ ID NO 16
<211> LENGTH: 42
<212> TYPE: DNA
<213> ORGANISM: HIV-1 NL4-3
<220> FEATURE:
<221> NAME/KEY: CDS
<222> LOCATION: (1)..(42)
<400> SEQUENCE: 16
ggc cat aaa gca aga gtt ttg gct gaa gca atg agc caa gta 42
Gly His Lys Ala Arg Val Leu Ala Glu Ala Met Ser Gln Val
1 5 10
<210> SEQ ID NO 17
<211> LENGTH: 14
<212> TYPE: PRT
<213> ORGANISM: HIV-1 NL4-3
<400> SEQUENCE: 17
Gly His Lys Ala Arg Val Leu Ala Glu Ala Met Ser Gln Val
1 5 10
<210> SEQ ID NO 18
<211> LENGTH: 9128
<212> TYPE: DNA
<213> ORGANISM: HIV-1 RF
<400> SEQUENCE: 18
gagctctctg gctagctagg gaacccactg cttaagcctc aataaagctt gccttgagtg 60
cttcaagtag tgtgtgcccg tctgttgtgt gactctggta actagagatc cctcagacct 120
ctttagtcag tgtggaaaat ctctagcagt ggcgcccgaa cagggaccag aaagcgaaag 180
tagaaccaga ggagatctct cgacgcagga ctcggcttgc tgaagcgcgc gcggcaagag 240
gcgaggggcg gcgaacggtg agtacgccga aaattttgac tagcggaggc tagaaggaga 300
gagatgggtg cgagagcgtc agtattaagc ggcggaaaat tagacaaatg ggaaaaaatt 360
cggttaaggc caaggggaaa gaaaagatat aagttaaaac atatagtatg ggcaagcagg 420
gagctagaac gatttgctgt caatcctagc cttttagaga cagcagaggg ctgtagacaa 480
atactgggac agctacaacc agcccttcag acaggatcag aagaacttaa atcattatat 540
aatgcagtag caaccctcta ttgtgtacat caaaatatag aggtaagaga caccaaggaa 600
gctttagaca agatagagga agagcaaaac aaaagtaaga aaaaagcaca gcaagcagca 660
gctgacacag gaaacggcag ccaggtcagc caaaattacc ctatagtgca gaaccttcag 720
gggcaaatgg tacatcaagc catatcacct agaactttaa atgcatgggt aaaagtagta 780
gaagagaagg cttttagccc agaagtaata cccatgtttt cagcattatc agaaggagcc 840
accccacaag atttaaacac catgctaaac acagtggggg gacatcaagc agccatgcaa 900
atgttaaaag agactatcaa tgaggaagct gcagaatggg atagattgca tccagtgcat 960
gcagggccta ttgcaccagg tcagatgaga gaaccaaggg gaagtgacat agcaggaacc 1020
actagtaccc ttcaggaaca aataggatgg atgacaaata atccacctat cccagtagga 1080
gaaatctata aaaggtggat aattctggga ttaaataaaa tagtaagaat gtatagcccc 1140
atcagcattc tggacataag acaaggacct aaggaaccct ttagagacta tgtagaccgg 1200
ttctataaaa ctctaagagc cgagcaagct tcacaggatg taaaaaattg gatgacagaa 1260
accttcctgg tccaaaatgc gaacccagat tgtaaaacta ttttaaaagc attgggacca 1320
gcagctacac tagaagaaat gatgacagca tgtcagggag tagggggacc cagccataaa 1380
gcaagaattt tggctgaagc aatgagccaa gtaacaaatt cagctaccat aatgctgcag 1440
aaaggtaatt ttagggacca aagaaaaatt gttaagtgtt tcaactgtgg caaagtaggg 1500
cacatagcca aaaattgcag ggcccctagg aaaaagggct gttggaaatg tggaaaggaa 1560
ggacaccaaa tgaaagattg cactaatgag ggacgacagg ctaatttttt agggaaaatc 1620
tggccttccc acaagggaag gccagggaac tttcttcaga gcagaccaga gccaacagcc 1680
ccaccagaag agagcttcag gtttggggaa gagacaactc cctctcagaa gcaggagaag 1740
atagacaagg aactgtatcc tttagcttcc ctcaaatcac tctttggcaa cgacccatcg 1800
tcacagtaaa gatagggggg caattaaagg aagctctatt agatacagga gcagatgata 1860
cagtattaga agaaatgaat ttgccaggaa aatggaaacc aaaaatgata gggggaattg 1920
gaggttttat caaagtgaga cagtatgatc aaatactcat agaaatctgt ggacataaag 1980
ctataggtac agtattagta ggacctacac ctgtcaacat aattggaaga aatctgttga 2040
ctcagattgg ttgcacttta aattttccca ttagtcctat tgaaactgta ccagtaaaat 2100
taaagccagg aatggatggc ccaaaagtta aacaatggcc attgacagag gaaaaaataa 2160
aagcattggt agaaatttgt acagaaatgg aaaaggaagg aaaaatttcc aaaattgggc 2220
ctgaaaatcc atacaatact ccagtatttg ccataaagaa aaaagacagt actaaatgga 2280
gaaaattagt agatttcaga gaacttaata agagaactca agacttctgg gaagttcagt 2340
taggaatacc acatcctgca gggttaaaaa agaagaaatc agtaacagta ttggatgtgg 2400
gtgatgcata tttttcagtt cccttagata aagagttcag gaagtatact gcatttacca 2460
tacctagtat aaacaatgaa acaccacgga ttagatatca gtacaatgtg cttccacaag 2520
ggtggaaagg atcaccagca atattccaaa gtagtatgac aaaaatctta gagcctttta 2580
aaaaacaaaa tccagaaata gttatctatc aatacatgga tgatttgtat gtaggatctg 2640
atttagaaat agggcagcat agaataaaaa tagaggaact gagagaacat ctgttaaagt 2700
gggggtttac cacaccggac aagaaacatc agaaagaacc tccatttctt tggatgggtt 2760
atgaactcca tcctgataaa tggacagtac agcctatagt gctgccagaa aaagacagct 2820
ggactgtcaa tgacatacag aagttagtgg gaaaattgaa ttgggcaagt cagatttatg 2880
cagggattaa agtaaagcaa ttatgtaaac tccttagggg aaccaaagca ctaacagaag 2940
tagtacaact aacaaaagaa gcagagctag aactggcaga aaatagggag attctaaaag 3000
aaccagtaca tggagtgtat tatgacccat caaaagactt aatagcagaa atacagaagc 3060
aggggcaagg ccaatggaca taccaaattt atcaagagcc atttaaaaac ctgaaaacag 3120
gaaagtatgc aagaatgagg ggtgcccaca ctaatgatgt aaaacaatta acagaggcag 3180
tacaaaaagt agccacagaa agcatagtaa tatggggaaa gactcctaaa tttaaactac 3240
ccatacaaaa agaaacatgg gaggcatggt ggacagagta ttggcaagcc acctggattc 3300
ctgagtggga gtttgtcaat acccctccct tagtaaaatt gtggtaccag ttagaaaaag 3360
aacccataat aggagcagaa actttctatg tagatggggc agctaataga gagactaaat 3420
taggaaaagc aggatatgtt actgacagag gaagacaaaa agttgtctcc ctaactgaca 3480
caacaaatca gaagactgag ttacaagcaa ttcatctagc tttgcaggat tcgggattag 3540
aagtaaacat agtaacagac tcacaatatg cattgggaat cattcaagca caaccagata 3600
aaagtgaatc agagttagtc agtcagataa tagagcagtt aataaaaaag gaaaaggtct 3660
acctggcatg ggtaccagca cacaaaggga ttggaggaaa tgaacaagta gatagattag 3720
tcagtactgg aatcaggaaa gtactatttt tggatggaat agataaggcc caagatgaac 3780
atgagaaata tcacagtaat tggagagcaa tggctagtga ttttaacctg ccacctgtag 3840
tagcaaaaga aatagtagcc agctgtgata aatgtcagct aaaaggagaa gccatgcatg 3900
gacaagtaga ttgtagtcca ggaatatggc aactagattg tacacatcta gaaggaaaaa 3960
ttatcctggt agcagttcat gtagccagtg gctatataga agcagaagtt attccagcag 4020
aaacaggaca ggaaacagca tactttatct taaaattagc aggaagatgg ccagtaaaag 4080
taatacatac agacaatggc agcaatttca ccagtactac agttaaggcc gcctgttggt 4140
gggcagggat caagcaggaa tttggcattc cctacaatcc ccaaagtcaa ggagtagtag 4200
aatctatgaa taaacaatta aagcaaatta taggacaggt aagagatcag gctgaacatc 4260
ttaagacagc agtacaaatg gcagtattca tccacaattt taaaagaaaa ggggggattg 4320
gggggtacag tgcaggggaa agaatagtag acataatagc aacagacata caaactaaag 4380
aactacaaaa acaaattaca aaaattcaaa attttcgggt ttattacagg gacagcagag 4440
atccactttg gaaaggacac gcaaagcttc tctggaaagg tgaaggggca gtagtaatac 4500
aagataatag tgacataaaa gtagtgccaa gaagaaaagc aaagatcatt agggattatg 4560
gaaaacagat ggcaggtgat gattgtgtgg caagtagaca ggatgaggat tagaacatgg 4620
aaaagtttag taaaacacca tatgtatatt tcaaggaaag ctaagggatg gttttataga 4680
catcactatg aaagcactca tccaagaata agttcagagg tacacatccc accaggggat 4740
gaaaggttgg taataacaac atattggggt ctgcatacag gagaaagaga ctggcatttg 4800
ggtcagggag tctccataga atggaggaaa aggagatata gcacacaagt agaccctgac 4860
ctagcagacc aactaattca cctgtactat tttgattgtt tttcagaatc tgctataaga 4920
aagccatcat taggacatat agttagtcct aggtgtgaat atcaagcagg acataacaag 4980
gtaggatctc tacagtacct ggcactagca gcattaacaa caccaaaaaa gataaagcca 5040
cctttgccta gtgttaagaa actgacagag gatagatgga acaagcccca gaagaccaag 5100
ggccacagag ggagccatac aatgaatgga cactagagct tttagaggag cttaagagtg 5160
aagctgtcag acattttcct aggctatggc tccatagctt aggacaacat atctatgaaa 5220
cttatgggga tacatgggca ggagtggaag ctataataag aattctgcaa caactgctgt 5280
ttattcattt cagaattggg tgtcaacata gcagaatagg cattactcga caaagaagag 5340
caagaaatgg agccagtaga tcctagacta gagccctgga agcatccagg aagtcagcct 5400
aagactgctt gtaacaattg ctattgtaaa aagtgttgct atcattgcca agtttgcttc 5460
ttaacaaaag gcttaggcat ctcctatggc aggaagaaga ggagacagcg acgaggacct 5520
cctcaaggca gtcagactca tcaagtctct ttatcaaagc agtaagtagt atatgtaatg 5580
caatctttag aaatattagc aatagtagca ttagtagtag cagcaatact agcaatagtt 5640
gtgtggacca tagttggcat agaaattagg aaaacattaa ggcaaaaaaa aaaatagaca 5700
ggttaattga tagaataaga gaaagagcag aagacagtgg caatgagagt gatggagatg 5760
aggaagaatt gtcagcactt gtggaaatgg ggcaccatgc tccttgggat gttgatgatc 5820
tgtagtgctg cagaggactt gtgggtcaca gtctattatg gggtacctgt gtggaaagaa 5880
gcaaccacca ctctattttg tgcatcagaa gctaaagcat ataaaacaga ggtacataat 5940
gtctgggcca aacatgcttg tgtacctaca gaccccaacc cacaagaagt actattggaa 6000
aatgtgacag aaaattttaa catgtggaaa aataacatgg tagaacagat gcatgaggat 6060
ataatcagtt tatgggatca aagcctaaag ccatgtgtaa aattaacccc actctgtgtt 6120
actttaaatt gcactgatgc taacttgaat ggtactaatg tcactagtag tagcggggga 6180
acaatgatgg agaacggaga aataaaaaac tgctctttcc aagttaccac aagtagaaga 6240
gataagacgc agaaaaaata tgcacttttt tataaacttg atgtggtacc aatagagaag 6300
ggtaatatta gccctaagaa taatactagc aataatacta gctatggtaa ctatacattg 6360
atacattgta attcctcagt cattacacag gcctgtccaa aggtatcctt tgagccaatt 6420
cccatacatt attgcacccc ggctggtttt gcgattctaa agtgtaatga taagaagttc 6480
aatggaacag gaccatgtaa aaatgtcagc acagtacaat gtacacatgg aattaggcca 6540
gtagtgtcaa ctcaactgct gttaaatggc agtctagcag aagaagaggt agtaattaga 6600
tctgaaaatt tcacggacaa tgttaaaacc ataatagtac agctgaatgc atctgtacaa 6660
attaattgta caagacccaa caacaataca agaaaaagta taactaaggg accagggaga 6720
gtaatttatg caacaggaca aataatagga gatataagaa aagcacattg taaccttagt 6780
agagcacaat ggaataacac tttaaaacag gtagttacaa aattaagaga acaatttgac 6840
aataaaacaa tagtctttac gtcatcctca ggaggggacc cagaaattgt acttcacagt 6900
tttaattgtg gaggggaatt tttctactgt aatacaacac aactgtttaa tagtacttgg 6960
aatagtactg aagggtcaaa taacactgga ggaaatgaca caatcacact cccatgcaga 7020
ataaaacaaa ttgtaaacat gtggcaggaa gtaggaaaag caatgtatgc ccctcccatc 7080
agtggacaaa ttaaatgtat atcaaatatt acagggctac tattaacaag agatgggggt 7140
gaagatacaa ctaatactac agagatcttc agacttggag gaggaaatat gagggacaat 7200
tggagaagtg aattatataa atataaagtg gtaagaattg agccattagg agtggcaccc 7260
actagggcaa agagaagagt ggtgcaaaga gaaaaaagag cagtgggaac aataggagct 7320
atgttccttg ggttcttggg agcagcagga agcactatgg gcgcaggctc aataacgcta 7380
acggtacagg ccagacactt attgtctggt atagtgcaac agcaaaacaa tttgctgagg 7440
gctattgagg cgcaacagca tctgttgcaa ctcacggtct ggggcatcaa acagctccag 7500
gcaagagtcc tggctgtgga aagataccta agggatcaac agctcctagg aatttgggga 7560
tgctctggaa aactcatttg caccactact gtgccttgga atgctagttg gagtaataaa 7620
tctctgaata tgatttggaa taacatgacc tggatgcagt gggaaagaga aattgacaat 7680
tacacaggca taatatacaa cttacttgaa gaatcgcaga accagcaaga aaagaatgaa 7740
caagaattat tggaattgga taaatgggca aatttgtgga attggtttga cataacacaa 7800
tggctgtggt atataagaat attcataatg atagtaggag gcttggtagg tctaaaaata 7860
gtttttgctg tgctttctat agtgaataga gttaggcagg gatactcacc attatcattt 7920
cagacccacc tcccagcccc gaggggaccc gacaggcccg aaggaatcga aggagaaggt 7980
ggagagagag acagagacag atccggcggt gcagtgaatg gattcttgac acttatctgg 8040
gacgatctgt ggaccctgtg cagcttcagc taccaccgct tgagagactt actcttgata 8100
gtagtgagga ttgtggaact tctgggacgc agggggtggg aagccctcaa gtattggtgg 8160
aatctcctgc agtattggag tcaggagcta aagaatagtg ctgttagctt gcttaatacc 8220
acagcaatag cagtagctga agggacagat aggattatag aagtagcaca aagaattctt 8280
agagcttttc ttcacatacc tagaagaata agacagggct tagaaagggc tttgctgtaa 8340
aatgggtggc aagtggtcaa aaagtaagat gggtggatgg cctgctgtaa gggaaagaat 8400
gcaaaaagct gagccagcag cagatggggt gggagcagca tctcgagacc tggagaaaca 8460
tggaacaatc acaagtagca atacagcagc taataatgct gcttgcacct ggctagaagc 8520
acaagaggat gaggatgagg aggtgggttt tccagtcaga cctcaggtac ctttaaggcc 8580
tatgactttc aaggcagctg tagatcttag ccacttttta aaagaaaagg ggggactgga 8640
tgggctagtg ttctcccaga aaagacaaga tatccttgat ctgtgggttt accacacaca 8700
aggctacttc cctgactggc agaactacac accagggcca gggaccagat atccactgac 8760
ctttggatgg tgcttcaagc tagtaccagt tgagccagat aaggtagaag aggccactga 8820
aggagagaac aacagcttgt tacaccctat atgcctgcat gggatggatg acccagagaa 8880
agaagtgtta gtgtggaagt ttgacagccg cctcgcattt catcacgtcg cccgagagaa 8940
gcatccggag tactacaaag actgctgaca tcgagttttc tacaagggac tttccgctgg 9000
ggactttcca gggaggtgtg gcctgggcgg gactggggag tggcgagccc tcagatgctg 9060
catataagca gctgcttttt gcctgtactg ggtctctctg gttagaccag atctgagctt 9120
gggagctc 9128
<210> SEQ ID NO 19
<211> LENGTH: 14824
<212> TYPE: DNA
<213> ORGANISM: HIV-1 NL4-3
<400> SEQUENCE: 19
tggaagggct aatttggtcc caaaaaagac aagagatcct tgatctgtgg atctaccaca 60
cacaaggcta cttccctgat tggcagaact acacaccagg gccagggatc agatatccac 120
tgacctttgg atggtgcttc aagttagtac cagttgaacc agagcaagta gaagaggcca 180
aataaggaga gaagaacagc ttgttacacc ctatgagcca gcatgggatg gaggacccgg 240
agggagaagt attagtgtgg aagtttgaca gcctcctagc atttcgtcac atggcccgag 300
agctgcatcc ggagtactac aaagactgct gacatcgagc tttctacaag ggactttccg 360
ctggggactt tccagggagg tgtggcctgg gcgggactgg ggagtggcga gccctcagat 420
gctacatata agcagctgct ttttgcctgt actgggtctc tctggttaga ccagatctga 480
gcctgggagc tctctggcta actagggaac ccactgctta agcctcaata aagcttgcct 540
tgagtgctca aagtagtgtg tgcccgtctg ttgtgtgact ctggtaacta gagatccctc 600
agaccctttt agtcagtgtg gaaaatctct agcagtggcg cccgaacagg gacttgaaag 660
cgaaagtaaa gccagaggag atctctcgac gcaggactcg gcttgctgaa gcgcgcacgg 720
caagaggcga ggggcggcga ctggtgagta cgccaaaaat tttgactagc ggaggctaga 780
aggagagaga tgggtgcgag agcgtcggta ttaagcgggg gagaattaga taaatgggaa 840
aaaattcggt taaggccagg gggaaagaaa caatataaac taaaacatat agtatgggca 900
agcagggagc tagaacgatt cgcagttaat cctggccttt tagagacatc agaaggctgt 960
agacaaatac tgggacagct acaaccatcc cttcagacag gatcagaaga acttagatca 1020
ttatataata caatagcagt cctctattgt gtgcatcaaa ggatagatgt aaaagacacc 1080
aaggaagcct tagataagat agaggaagag caaaacaaaa gtaagaaaaa ggcacagcaa 1140
gcagcagctg acacaggaaa caacagccag gtcagccaaa attaccctat agtgcagaac 1200
ctccaggggc aaatggtaca tcaggccata tcacctagaa ctttaaatgc atgggtaaaa 1260
gtagtagaag agaaggcttt cagcccagaa gtaataccca tgttttcagc attatcagaa 1320
ggagccaccc cacaagattt aaataccatg ctaaacacag tggggggaca tcaagcagcc 1380
atgcaaatgt taaaagagac catcaatgag gaagctgcag aatgggatag attgcatcca 1440
gtgcatgcag ggcctattgc accaggccag atgagagaac caaggggaag tgacatagca 1500
ggaactacta gtacccttca ggaacaaata ggatggatga cacataatcc acctatccca 1560
gtaggagaaa tctataaaag atggataatc ctgggattaa ataaaatagt aagaatgtat 1620
agccctacca gcattctgga cataagacaa ggaccaaagg aaccctttag agactatgta 1680
gaccgattct ataaaactct aagagccgag caagcttcac aagaggtaaa aaattggatg 1740
acagaaacct tgttggtcca aaatgcgaac ccagattgta agactatttt aaaagcattg 1800
ggaccaggag cgacactaga agaaatgatg acagcatgtc agggagtggg gggacccggc 1860
cataaagcaa gagttttggc tgaagcaatg agccaagtaa caaatccagc taccataatg 1920
atacagaaag gcaattttag gaaccaaaga aagactgtta agtgtttcaa ttgtggcaaa 1980
gaagggcaca tagccaaaaa ttgcagggcc cctaggaaaa agggctgttg gaaatgtgga 2040
aaggaaggac accaaatgaa agattgtact gagagacagg ctaatttttt agggaagatc 2100
tggccttccc acaagggaag gccagggaat tttcttcaga gcagaccaga gccaacagcc 2160
ccaccagaag agagcttcag gtttggggaa gagacaacaa ctccctctca gaagcaggag 2220
ccgatagaca aggaactgta tcctttagct tccctcagat cactctttgg cagcgacccc 2280
tcgtcacaat aaagataggg gggcaattaa aggaagctct attagataca ggagcagatg 2340
atacagtatt agaagaaatg aatttgccag gaagatggaa accaaaaatg atagggggaa 2400
ttggaggttt tatcaaagta agacagtatg atcagatact catagaaatc tgcggacata 2460
aagctatagg tacagtatta gtaggaccta cacctgtcaa cataattgga agaaatctgt 2520
tgactcagat tggctgcact ttaaattttc ccattagtcc tattgagact gtaccagtaa 2580
aattaaagcc aggaatggat ggcccaaaag ttaaacaatg gccattgaca gaagaaaaaa 2640
taaaagcatt agtagaaatt tgtacagaaa tggaaaagga aggaaaaatt tcaaaaattg 2700
ggcctgaaaa tccatacaat actccagtat ttgccataaa gaaaaaagac agtactaaat 2760
ggagaaaatt agtagatttc agagaactta ataagagaac tcaagatttc tgggaagttc 2820
aattaggaat accacatcct gcagggttaa aacagaaaaa atcagtaaca gtactggatg 2880
tgggcgatgc atatttttca gttcccttag ataaagactt caggaagtat actgcattta 2940
ccatacctag tataaacaat gagacaccag ggattagata tcagtacaat gtgcttccac 3000
agggatggaa aggatcacca gcaatattcc agtgtagcat gacaaaaatc ttagagcctt 3060
ttagaaaaca aaatccagac atagtcatct atcaatacat ggatgatttg tatgtaggat 3120
ctgacttaga aatagggcag catagaacaa aaatagagga actgagacaa catctgttga 3180
ggtggggatt taccacacca gacaaaaaac atcagaaaga acctccattc ctttggatgg 3240
gttatgaact ccatcctgat aaatggacag tacagcctat agtgctgcca gaaaaggaca 3300
gctggactgt caatgacata cagaaattag tgggaaaatt gaattgggca agtcagattt 3360
atgcagggat taaagtaagg caattatgta aacttcttag gggaaccaaa gcactaacag 3420
aagtagtacc actaacagaa gaagcagagc tagaactggc agaaaacagg gagattctaa 3480
aagaaccggt acatggagtg tattatgacc catcaaaaga cttaatagca gaaatacaga 3540
agcaggggca aggccaatgg acatatcaaa tttatcaaga gccatttaaa aatctgaaaa 3600
caggaaaata tgcaagaatg aagggtgccc acactaatga tgtgaaacaa ttaacagagg 3660
cagtacaaaa aatagccaca gaaagcatag taatatgggg aaagactcct aaatttaaat 3720
tacccataca aaaggaaaca tgggaagcat ggtggacaga gtattggcaa gccacctgga 3780
ttcctgagtg ggagtttgtc aatacccctc ccttagtgaa gttatggtac cagttagaga 3840
aagaacccat aataggagca gaaactttct atgtagatgg ggcagccaat agggaaacta 3900
aattaggaaa agcaggatat gtaactgaca gaggaagaca aaaagttgtc cccctaacgg 3960
acacaacaaa tcagaagact gagttacaag caattcatct agctttgcag gattcgggat 4020
tagaagtaaa catagtgaca gactcacaat atgcattggg aatcattcaa gcacaaccag 4080
ataagagtga atcagagtta gtcagtcaaa taatagagca gttaataaaa aaggaaaaag 4140
tctacctggc atgggtacca gcacacaaag gaattggagg aaatgaacaa gtagatgggt 4200
tggtcagtgc tggaatcagg aaagtactat ttttagatgg aatagataag gcccaagaag 4260
aacatgagaa atatcacagt aattggagag caatggctag tgattttaac ctaccacctg 4320
tagtagcaaa agaaatagta gccagctgtg ataaatgtca gctaaaaggg gaagccatgc 4380
atggacaagt agactgtagc ccaggaatat ggcagctaga ttgtacacat ttagaaggaa 4440
aagttatctt ggtagcagtt catgtagcca gtggatatat agaagcagaa gtaattccag 4500
cagagacagg gcaagaaaca gcatacttcc tcttaaaatt agcaggaaga tggccagtaa 4560
aaacagtaca tacagacaat ggcagcaatt tcaccagtac tacagttaag gccgcctgtt 4620
ggtgggcggg gatcaagcag gaatttggca ttccctacaa tccccaaagt caaggagtaa 4680
tagaatctat gaataaagaa ttaaagaaaa ttataggaca ggtaagagat caggctgaac 4740
atcttaagac agcagtacaa atggcagtat tcatccacaa ttttaaaaga aaagggggga 4800
ttggggggta cagtgcaggg gaaagaatag tagacataat agcaacagac atacaaacta 4860
aagaattaca aaaacaaatt acaaaaattc aaaattttcg ggtttattac agggacagca 4920
gagatccagt ttggaaagga ccagcaaagc tcctctggaa aggtgaaggg gcagtagtaa 4980
tacaagataa tagtgacata aaagtagtgc caagaagaaa agcaaagatc atcagggatt 5040
atggaaaaca gatggcaggt gatgattgtg tggcaagtag acaggatgag gattaacaca 5100
tggaaaagat tagtaaaaca ccatatgtat atttcaagga aagctaagga ctggttttat 5160
agacatcact atgaaagtac taatccaaaa ataagttcag aagtacacat cccactaggg 5220
gatgctaaat tagtaataac aacatattgg ggtctgcata caggagaaag agactggcat 5280
ttgggtcagg gagtctccat agaatggagg aaaaagagat atagcacaca agtagaccct 5340
gacctagcag accaactaat tcatctgcac tattttgatt gtttttcaga atctgctata 5400
agaaatacca tattaggacg tatagttagt cctaggtgtg aatatcaagc aggacataac 5460
aaggtaggat ctctacagta cttggcacta gcagcattaa taaaaccaaa acagataaag 5520
ccacctttgc ctagtgttag gaaactgaca gaggacagat ggaacaagcc ccagaagacc 5580
aagggccaca gagggagcca tacaatgaat ggacactaga gcttttagag gaacttaaga 5640
gtgaagctgt tagacatttt cctaggatat ggctccataa cttaggacaa catatctatg 5700
aaacttacgg ggatacttgg gcaggagtgg aagccataat aagaattctg caacaactgc 5760
tgtttatcca tttcagaatt gggtgtcgac atagcagaat aggcgttact cgacagagga 5820
gagcaagaaa tggagccagt agatcctaga ctagagccct ggaagcatcc aggaagtcag 5880
cctaaaactg cttgtaccaa ttgctattgt aaaaagtgtt gctttcattg ccaagtttgt 5940
ttcatgacaa aagccttagg catctcctat ggcaggaaga agcggagaca gcgacgaaga 6000
gctcatcaga acagtcagac tcatcaagct tctctatcaa agcagtaagt agtacatgta 6060
atgcaaccta taatagtagc aatagtagca ttagtagtag caataataat agcaatagtt 6120
gtgtggtcca tagtaatcat agaatatagg aaaatattaa gacaaagaaa aatagacagg 6180
ttaattgata gactaataga aagagcagaa gacagtggca atgagagtga aggagaagta 6240
tcagcacttg tggagatggg ggtggaaatg gggcaccatg ctccttggga tattgatgat 6300
ctgtagtgct acagaaaaat tgtgggtcac agtctattat ggggtacctg tgtggaagga 6360
agcaaccacc actctatttt gtgcatcaga tgctaaagca tatgatacag aggtacataa 6420
tgtttgggcc acacatgcct gtgtacccac agaccccaac ccacaagaag tagtattggt 6480
aaatgtgaca gaaaatttta acatgtggaa aaatgacatg gtagaacaga tgcatgagga 6540
tataatcagt ttatgggatc aaagcctaaa gccatgtgta aaattaaccc cactctgtgt 6600
tagtttaaag tgcactgatt tgaagaatga tactaatacc aatagtagta gcgggagaat 6660
gataatggag aaaggagaga taaaaaactg ctctttcaat atcagcacaa gcataagaga 6720
taaggtgcag aaagaatatg cattctttta taaacttgat atagtaccaa tagataatac 6780
cagctatagg ttgataagtt gtaacacctc agtcattaca caggcctgtc caaaggtatc 6840
ctttgagcca attcccatac attattgtgc cccggctggt tttgcgattc taaaatgtaa 6900
taataagacg ttcaatggaa caggaccatg tacaaatgtc agcacagtac aatgtacaca 6960
tggaatcagg ccagtagtat caactcaact gctgttaaat ggcagtctag cagaagaaga 7020
tgtagtaatt agatctgcca atttcacaga caatgctaaa accataatag tacagctgaa 7080
cacatctgta gaaattaatt gtacaagacc caacaacaat acaagaaaaa gtatccgtat 7140
ccagagggga ccagggagag catttgttac aataggaaaa ataggaaata tgagacaagc 7200
acattgtaac attagtagag caaaatggaa tgccacttta aaacagatag ctagcaaatt 7260
aagagaacaa tttggaaata ataaaacaat aatctttaag caatcctcag gaggggaccc 7320
agaaattgta acgcacagtt ttaattgtgg aggggaattt ttctactgta attcaacaca 7380
actgtttaat agtacttggt ttaatagtac ttggagtact gaagggtcaa ataacactga 7440
aggaagtgac acaatcacac tcccatgcag aataaaacaa tttataaaca tgtggcagga 7500
agtaggaaaa gcaatgtatg cccctcccat cagtggacaa attagatgtt catcaaatat 7560
tactgggctg ctattaacaa gagatggtgg taataacaac aatgggtccg agatcttcag 7620
acctggagga ggcgatatga gggacaattg gagaagtgaa ttatataaat ataaagtagt 7680
aaaaattgaa ccattaggag tagcacccac caaggcaaag agaagagtgg tgcagagaga 7740
aaaaagagca gtgggaatag gagctttgtt ccttgggttc ttgggagcag caggaagcac 7800
tatgggctgc acgtcaatga cgctgacggt acaggccaga caattattgt ctgatatagt 7860
gcagcagcag aacaatttgc tgagggctat tgaggcgcaa cagcatctgt tgcaactcac 7920
agtctggggc atcaaacagc tccaggcaag aatcctggct gtggaaagat acctaaagga 7980
tcaacagctc ctggggattt ggggttgctc tggaaaactc atttgcacca ctgctgtgcc 8040
ttggaatgct agttggagta ataaatctct ggaacagatt tggaataaca tgacctggat 8100
ggagtgggac agagaaatta acaattacac aagcttaata cactccttaa ttgaagaatc 8160
gcaaaaccag caagaaaaga atgaacaaga attattggaa ttagataaat gggcaagttt 8220
gtggaattgg tttaacataa caaattggct gtggtatata aaattattca taatgatagt 8280
aggaggcttg gtaggtttaa gaatagtttt tgctgtactt tctatagtga atagagttag 8340
gcagggatat tcaccattat cgtttcagac ccacctccca atcccgaggg gacccgacag 8400
gcccgaagga atagaagaag aaggtggaga gagagacaga gacagatcca ttcgattagt 8460
gaacggatcc ttagcactta tctgggacga tctgcggagc ctgtgcctct tcagctacca 8520
ccgcttgaga gacttactct tgattgtaac gaggattgtg gaacttctgg gacgcagggg 8580
gtgggaagcc ctcaaatatt ggtggaatct cctacagtat tggagtcagg aactaaagaa 8640
tagtgctgtt aacttgctca atgccacagc catagcagta gctgagggga cagatagggt 8700
tatagaagta ttacaagcag cttatagagc tattcgccac atacctagaa gaataagaca 8760
gggcttggaa aggattttgc tataagatgg gtggcaagtg gtcaaaaagt agtgtgattg 8820
gatggcctgc tgtaagggaa agaatgagac gagctgagcc agcagcagat ggggtgggag 8880
cagtatctcg agacctagaa aaacatggag caatcacaag tagcaataca gcagctaaca 8940
atgctgcttg tgcctggcta gaagcacaag aggaggaaga ggtgggtttt ccagtcacac 9000
ctcaggtacc tttaagacca atgacttaca aggcagctgt agatcttagc cactttttaa 9060
aagaaaaggg gggactggaa gggctaattc actcccaaag aagacaagat atccttgatc 9120
tgtggatcta ccacacacaa ggctacttcc ctgattggca gaactacaca ccagggccag 9180
gggtcagata tccactgacc tttggatggt gctacaagct agtaccagtt gagccagata 9240
aggtagaaga ggccaataaa ggagagaaca ccagcttgtt acaccctgtg agcctgcatg 9300
gaatggatga ccctgagaga gaagtgttag agtggaggtt tgacagccgc ctagcatttc 9360
atcacgtggc ccgagagctg catccggagt acttcaagaa ctgctgacat cgagcttgct 9420
acaagggact ttccgctggg gactttccag ggaggcgtgg cctgggcggg actggggagt 9480
ggcgagccct cagatgctgc atataagcag ctgctttttg cctgtactgg gtctctctgg 9540
ttagaccaga tctgagcctg ggagctctct ggctaactag ggaacccact gcttaagcct 9600
caataaagct tgccttgagt gcttcaagta gtgtgtgccc gtctgttgtg tgactctggt 9660
aactagagat ccctcagacc cttttagtca gtgtggaaaa tctctagcac ccaggaggta 9720
gaggttgcag tgagccaaga tcgcgccact gcattccagc ctgggcaaga aaacaagact 9780
gtctaaaata ataataataa gttaagggta ttaaatatat ttatacatgg aggtcataaa 9840
aatatatata tttgggctgg gcgcagtggc tcacacctgc gcccggccct ttgggaggcc 9900
gaggcaggtg gatcacctga gtttgggagt tccagaccag cctgaccaac atggagaaac 9960
cccttctctg tgtattttta gtagatttta ttttatgtgt attttattca caggtatttc 10020
tggaaaactg aaactgtttt tcctctactc tgataccaca agaatcatca gcacagagga 10080
agacttctgt gatcaaatgt ggtgggagag ggaggttttc accagcacat gagcagtcag 10140
ttctgccgca gactcggcgg gtgtccttcg gttcagttcc aacaccgcct gcctggagag 10200
aggtcagacc acagggtgag ggctcagtcc ccaagacata aacacccaag acataaacac 10260
ccaacaggtc caccccgcct gctgcccagg cagagccgat tcaccaagac gggaattagg 10320
atagagaaag agtaagtcac acagagccgg ctgtgcggga gaacggagtt ctattatgac 10380
tcaaatcagt ctccccaagc attcggggat cagagttttt aaggataact tagtgtgtag 10440
ggggccagtg agttggagat gaaagcgtag ggagtcgaag gtgtcctttt gcgccgagtc 10500
agttcctggg tgggggccac aagatcggat gagccagttt atcaatccgg gggtgccagc 10560
tgatccatgg agtgcagggt ctgcaaaata tctcaagcac tgattgatct taggttttac 10620
aatagtgatg ttaccccagg aacaatttgg ggaaggtcag aatcttgtag cctgtagctg 10680
catgactcct aaaccataat ttcttttttg tttttttttt tttatttttg agacagggtc 10740
tcactctgtc acctaggctg gagtgcagtg gtgcaatcac agctcactgc agcctcaacg 10800
tcgtaagctc aagcgatcct cccacctcag cctgcctggt agctgagact acaagcgacg 10860
ccccagttaa tttttgtatt tttggtagag gcagcgtttt gccgtgtggc cctggctggt 10920
ctcgaactcc tgggctcaag tgatccagcc tcagcctccc aaagtgctgg gacaaccggg 10980
cccagtcact gcacctggcc ctaaaccata atttctaatc ttttggctaa tttgttagtc 11040
ctacaaaggc agtctagtcc ccagcaaaaa gggggtttgt ttcgggaaag ggctgttact 11100
gtctttgttt caaactataa actaagttcc tcctaaactt agttcggcct acacccagga 11160
atgaacaagg agagcttgga ggttagaagc acgatggaat tggttaggtc agatctcttt 11220
cactgtctga gttataattt tgcaatggtg gttcaaagac tgcccgcttc tgacaccagt 11280
cgctgcatta atgaatcggc caacgcgcgg ggagaggcgg tttgcgtatt gggcgctctt 11340
ccgcttcctc gctcactgac tcgctgcgct cggtcgttcg gctgcggcga gcggtatcag 11400
ctcactcaaa ggcggtaata cggttatcca cagaatcagg ggataacgca ggaaagaaca 11460
tgtgagcaaa aggccagcaa aaggccagga accgtaaaaa ggccgcgttg ctggcgtttt 11520
tccataggct ccgcccccct gacgagcatc acaaaaatcg acgctcaagt cagaggtggc 11580
gaaacccgac aggactataa agataccagg cgtttccccc tggaagctcc ctcgtgcgct 11640
ctcctgttcc gaccctgccg cttaccggat acctgtccgc ctttctccct tcgggaagcg 11700
tggcgctttc tcatagctca cgctgtaggt atctcagttc ggtgtaggtc gttcgctcca 11760
agctgggctg tgtgcacgaa ccccccgttc agcccgaccg ctgcgcctta tccggtaact 11820
atcgtcttga gtccaacccg gtaagacacg acttatcgcc actggcagca gccactggta 11880
acaggattag cagagcgagg tatgtaggcg gtgctacaga gttcttgaag tggtggccta 11940
actacggcta cactagaagg acagtatttg gtatctgcgc tctgctgaag ccagttacct 12000
tcggaaaaag agttggtagc tcttgatccg gcaaacaaac caccgctggt agcggtggtt 12060
tttttgtttg caagcagcag attacgcgca gaaaaaaagg atctcaagaa gatcctttga 12120
tcttttctac ggggtctgac gctcagtgga acgaaaactc acgttaaggg attttggtca 12180
tgagattatc aaaaaggatc ttcacctaga tccttttaaa ttaaaaatga agttttaaat 12240
caatctaaag tatatatgag taaacttggt ctgacagtta ccaatgctta atcagtgagg 12300
cacctatctc agcgatctgt ctatttcgtt catccatagt tgcctgactc cccgtcgtgt 12360
agataactac gatacgggag ggcttaccat ctggccccag tgctgcaatg ataccgcgag 12420
acccacgctc accggctcca gatttatcag caataaacca gccagccgga agggccgagc 12480
gcagaagtgg tcctgcaact ttatccgcct ccatccagtc tattaattgt tgccgggaag 12540
ctagagtaag tagttcgcca gttaatagtt tgcgcaacgt tgttgccatt gctacaggca 12600
tcgtggtgtc acgctcgtcg tttggtatgg cttcattcag ctccggttcc caacgatcaa 12660
ggcgagttac atgatccccc atgttgtgca aaaaagcggt tagctccttc ggtcctccga 12720
tcgttgtcag aagtaagttg gccgcagtgt tatcactcat ggttatggca gcactgcata 12780
attctcttac tgtcatgcca tccgtaagat gcttttctgt gactggtgag tactcaacca 12840
agtcattctg agaatagtgt atgcggcgac cgagttgctc ttgcccggcg tcaatacggg 12900
ataataccgc gccacatagc agaactttaa aagtgctcat cattggaaaa cgttcttcgg 12960
ggcgaaaact ctcaaggatc ttaccgctgt tgagatccag ttcgatgtaa cccactcgtg 13020
cacccaactg atcttcagca tcttttactt tcaccagcgt ttctgggtga gcaaaaacag 13080
gaaggcaaaa tgccgcaaaa aagggaataa gggcgacacg gaaatgttga atactcatac 13140
tcttcctttt tcaatattat tgaagcattt atcagggtta ttgtctcatg agcggataca 13200
tatttgaatg tatttagaaa aataaacaaa taggggttcc gcgcacattt ccccgaaaag 13260
tgccacctga cgtctaagaa accattatta tcatgacatt aacctataaa aataggcgta 13320
tcacgaggcc ctttcgtctc gcgcgtttcg gtgatgacgg tgaaaacctc tgacacatgc 13380
agctcccgga gacggtcaca gcttgtctgt aagcggatgc cgggagcaga caagcccgtc 13440
agggcgcgtc agcgggtgtt ggcgggtgtc ggggctggct taactatgcg gcatcagagc 13500
agattgtact gagagtgcac catatgcggt gtgaaatacc gcacagatgc gtaaggagaa 13560
aataccgcat caggcgccat tcgccattca ggctgcgcaa ctgttgggaa gggcgatcgg 13620
tgcgggcctc ttcgctatta cgccagggga ggcagagatt gcagtaagct gagatcgcag 13680
cactgcactc cagcctgggc gacagagtaa gactctgtct caaaaataaa ataaataaat 13740
caatcagata ttccaatctt ttcctttatt tatttattta ttttctattt tggaaacaca 13800
gtccttcctt attccagaat tacacatata ttctattttt ctttatatgc tccagttttt 13860
tttagacctt cacctgaaat gtgtgtatac aaaatctagg ccagtccagc agagcctaaa 13920
ggtaaaaaat aaaataataa aaaataaata aaatctagct cactccttca catcaaaatg 13980
gagatacagc tgttagcatt aaataccaaa taacccatct tgtcctcaat aattttaagc 14040
gcctctctcc accacatcta actcctgtca aaggcatgtg ccccttccgg gcgctctgct 14100
gtgctgccaa ccaactggca tgtggactct gcagggtccc taactgccaa gccccacagt 14160
gtgccctgag gctgcccctt ccttctagcg gctgccccca ctcggctttg ctttccctag 14220
tttcagttac ttgcgttcag ccaaggtctg aaactaggtg cgcacagagc ggtaagactg 14280
cgagagaaag agaccagctt tacagggggt ttatcacagt gcaccctgac agtcgtcagc 14340
ctcacagggg gtttatcaca ttgcaccctg acagtcgtca gcctcacagg gggtttatca 14400
cagtgcaccc ttacaatcat tccatttgat tcacaatttt tttagtctct actgtgccta 14460
acttgtaagt taaatttgat cagaggtgtg ttcccagagg ggaaaacagt atatacaggg 14520
ttcagtacta tcgcatttca ggcctccacc tgggtcttgg aatgtgtccc ccgaggggtg 14580
atgactacct cagttggatc tccacaggtc acagtgacac aagataacca agacacctcc 14640
caaggctacc acaatgggcc gccctccacg tgcacatggc cggaggaact gccatgtcgg 14700
aggtgcaagc acacctgcgc atcagagtcc ttggtgtgga gggagggacc agcgcagctt 14760
ccagccatcc acctgatgaa cagaacctag ggaaagcccc agttctactt acaccaggaa 14820
aggc 14824
<210> SEQ ID NO 20
<211> LENGTH: 24
<212> TYPE: PRT
<213> ORGANISM: SIV DE
<400> SEQUENCE: 20
Gly Gln Lys Ala Arg Val Leu Ala Glu Ala Leu Lys Glu Ala Leu Ala
1 5 10 15
Pro Val Pro Ile Pro Phe Ala Ala
20
<210> SEQ ID NO 21
<211> LENGTH: 64
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Used to inhibit the viral Gag p25 protein
(CA-SP1)
<400> SEQUENCE: 21
Lys Asn Trp Met Thr Glu Thr Phe Leu Val Gln Asn Ala Asn Pro Asp
1 5 10 15
Cys Lys Thr Ile Leu Lys Ala Leu Gly Pro Ala Ala Thr Leu Glu Glu
20 25 30
Met Met Thr Ala Cys Gln Gly Val Gly Gly Pro Ser His Lys Ala Arg
35 40 45
Ile Leu Ala Glu Ala Met Ser Gln Val Thr Asn Ser Ala Thr Ile Met
50 55 60
<210> SEQ ID NO 22
<211> LENGTH: 64
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Used to inhibit the viral Gag p25 protein
(CA-SP1)
<400> SEQUENCE: 22
Lys Asn Trp Met Thr Glu Thr Leu Leu Val Gln Asn Ala Asn Pro Asp
1 5 10 15
Cys Lys Thr Ile Leu Lys Ala Leu Gly Pro Gly Ala Thr Leu Glu Glu
20 25 30
Met Met Thr Ala Cys Gln Gly Val Gly Gly Pro Gly His Lys Ala Arg
35 40 45
Val Leu Ala Glu Ala Met Ser Gln Val Thr Asn Pro Ala Thr Ile Met
50 55 60
<210> SEQ ID NO 23
<211> LENGTH: 30
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Used to inhibit the viral Gag p25 protein
(CA-SP1)
<400> SEQUENCE: 23
Thr Ala Cys Gln Gly Val Gly Gly Pro Ser His Lys Ala Arg Ile Leu
1 5 10 15
Ala Glu Ala Met Ser Gln Val Thr Asn Ser Ala Thr Ile Met
20 25 30
<210> SEQ ID NO 24
<211> LENGTH: 31
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Used to inhibit the viral Gag p25 protein
(CA-SP1)
<400> SEQUENCE: 24
Met Thr Ala Cys Gln Gly Val Gly Gly Pro Gly His Lys Ala Arg Val
1 5 10 15
Leu Ala Glu Ala Met Ser Gln Val Thr Asn Pro Ala Thr Ile Met
20 25 30
<210> SEQ ID NO 25
<211> LENGTH: 14
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Used to inhibit the viral Gag p25 protein
(CA-SP1)
<400> SEQUENCE: 25
Ser His Lys Ala Arg Ile Leu Ala Glu Ala Met Ser Gln Val
1 5 10
<210> SEQ ID NO 26
<211> LENGTH: 14
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Used to inhibit the viral Gag p25 protein
(CA-SP1)
<400> SEQUENCE: 26
Gly His Lys Ala Arg Val Leu Ala Glu Ala Met Ser Gln Val
1 5 10
<210> SEQ ID NO 27
<211> LENGTH: 24
<212> TYPE: PRT
<213> ORGANISM: SIV DM
<400> SEQUENCE: 27
Gly Gln Lys Ala Arg Val Leu Ala Glu Ala Met Ser Gln Val Leu Ala
1 5 10 15
Pro Val Pro Ile Pro Phe Ala Ala
20
<210> SEQ ID NO 28
<211> LENGTH: 24
<212> TYPE: PRT
<213> ORGANISM: SIV DJ
<400> SEQUENCE: 28
Gly Gln Lys Ala Arg Val Met Ala Glu Ala Met Ser Gln Val Leu Ala
1 5 10 15
Pro Val Pro Ile Pro Phe Ala Ala
20
<210> SEQ ID NO 29
<211> LENGTH: 15
<212> TYPE: PRT
<213> ORGANISM: FIV
<400> SEQUENCE: 29
Gly Tyr Lys Met Gln Leu Leu Ala Glu Ala Leu Thr Lys Val Gln
1 5 10 15
<210> SEQ ID NO 30
<211> LENGTH: 12
<212> TYPE: PRT
<213> ORGANISM: EIAV
<400> SEQUENCE: 30
Lys Gln Lys Met Met Leu Leu Ala Lys Ala Leu Gln
1 5 10
<210> SEQ ID NO 31
<211> LENGTH: 32
<212> TYPE: PRT
<213> ORGANISM: BIV
<400> SEQUENCE: 31
Lys Ser Lys Met Gln Phe Leu Val Ala Ala Met Lys Glu Met Gly Ile
1 5 10 15
Gln Ser Pro Ile Pro Ala Val Leu Pro His Thr Pro Glu Ala Tyr Ala
20 25 30
<210> SEQ ID NO 32
<211> LENGTH: 9
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: HIV-1 CA-SP1 residues to be inserted into
SIVmac239 backbone
<400> SEQUENCE: 32
Val Leu Ala Glu Ala Met Ser Gln Val
1 5
<210> SEQ ID NO 33
<211> LENGTH: 13
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Mutant SP1 region in HIV-1 NL4-3
<400> SEQUENCE: 33
Ala Ala Met Ser Gln Val Thr Asn Pro Ala Thr Ile Met
1 5 10
<210> SEQ ID NO 34
<211> LENGTH: 13
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Mutant SP1 region in HIV-1 NL4-3
<400> SEQUENCE: 34
Ala Glu Met Ser Gln Val Thr Asn Pro Ala Thr Ile Met
1 5 10
<210> SEQ ID NO 35
<211> LENGTH: 13
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Mutant SP1 region in HIV-1 NL4-3
<400> SEQUENCE: 35
Ala Glu Ala Ser Gln Val Thr Asn Pro Ala Thr Ile Met
1 5 10
<210> SEQ ID NO 36
<211> LENGTH: 13
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Mutant SP1 region in HIV-1 NL4-3
<400> SEQUENCE: 36
Ala Glu Ala Met Gln Val Thr Asn Pro Ala Thr Ile Met
1 5 10
<210> SEQ ID NO 37
<211> LENGTH: 24
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 37
agccaaaact cttgctttat ggcc 24
<210> SEQ ID NO 38
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 38
agtcagtgtg gaaaatctct agcagtgg 28
<210> SEQ ID NO 39
<211> LENGTH: 24
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 39
gcaatgagcc aagtaacaaa tcca 24
<210> SEQ ID NO 40
<211> LENGTH: 31
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 40
aggtatggta aatgcagtat acttcctgaa g 31
<210> SEQ ID NO 41
<211> LENGTH: 27
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 41
ttcagccaaa actcttgctt tatggcc 27
<210> SEQ ID NO 42
<211> LENGTH: 23
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 42
atgagccaag taacaaatcc agc 23
<210> SEQ ID NO 43
<211> LENGTH: 21
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 43
tgcttcagcc aaaactcttg c 21
<210> SEQ ID NO 44
<211> LENGTH: 21
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 44
agccaagtaa caaatccagc t 21
<210> SEQ ID NO 45
<211> LENGTH: 24
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 45
cattgcttca gccaaaactc ttgc 24
<210> SEQ ID NO 46
<211> LENGTH: 22
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 46
caagtaacaa atccagctac ca 22
<210> SEQ ID NO 47
<211> LENGTH: 25
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 47
gctcattgct tcagccaaaa ctctt 25
<210> SEQ ID NO 48
<211> LENGTH: 22
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 48
gtaacaaatc cagctaccat aa 22
<210> SEQ ID NO 49
<211> LENGTH: 25
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 49
acaaatccag ctaccataat gatac 25
<210> SEQ ID NO 50
<211> LENGTH: 26
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 50
ttggctcatt gcttcagcca aaactc 26
<210> SEQ ID NO 51
<211> LENGTH: 24
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 51
tacttggctc attgcttcag ccaa 24
<210> SEQ ID NO 52
<211> LENGTH: 24
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 52
aatccagcta ccataatgat acag 24
<210> SEQ ID NO 53
<211> LENGTH: 21
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 53
tgttacttgg ctcattgctt c 21
<210> SEQ ID NO 54
<211> LENGTH: 24
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 54
ccagctacca taatgataca gaaa 24
<210> SEQ ID NO 55
<211> LENGTH: 24
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 55
atttgttact tggctcattg cttc 24
<210> SEQ ID NO 56
<211> LENGTH: 26
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 56
gctaccataa tgatacagaa aggcaa 26
<210> SEQ ID NO 57
<211> LENGTH: 24
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 57
tggatttgtt acttggctca ttgc 24
<210> SEQ ID NO 58
<211> LENGTH: 21
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 58
accataatga tacagaaagg c 21
<210> SEQ ID NO 59
<211> LENGTH: 22
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 59
agctggattt gttacttggc tc 22
<210> SEQ ID NO 60
<211> LENGTH: 27
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 60
ataatgatac agaaaggcaa ttttagg 27
<210> SEQ ID NO 61
<211> LENGTH: 21
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 61
ggtagctgga tttgttactt g 21
<210> SEQ ID NO 62
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 62
atgatacaga aaggcaattt taggaacc 28
<210> SEQ ID NO 63
<211> LENGTH: 21
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 63
tatggtagct ggatttgtta c 21
<210> SEQ ID NO 64
<211> LENGTH: 21
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 64
atacagaaag gcaattttag g 21
<210> SEQ ID NO 65
<211> LENGTH: 19
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 65
ccacctatcc cagtaggag 19
<210> SEQ ID NO 66
<211> LENGTH: 20
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 66
ggcacagcaa gcagcagctg 20
<210> SEQ ID NO 67
<211> LENGTH: 29
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 67
gtagaccaac agcaccatct agcggcaga 29
<210> SEQ ID NO 68
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 68
ggtaaagtaa aggcagtgta ctgcctaa 28
<210> SEQ ID NO 69
<211> LENGTH: 37
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 69
cactggtgcg aggacctgac tcatggcttc tgccatt 37
<210> SEQ ID NO 70
<211> LENGTH: 37
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 70
aatggcagaa gccatgagtc aggtcctcgc accagtg 37
<210> SEQ ID NO 71
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 71
ggcttctgcc agtactctag ccttctgt 28
<210> SEQ ID NO 72
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 72
acagaaggct agagtactgg cagaagcc 28
<210> SEQ ID NO 73
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 73
ggcttctgcc agtactctag ccttctgt 28
<210> SEQ ID NO 74
<211> LENGTH: 28
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 74
acagaaggct agagtactgg cagaagcc 28
<210> SEQ ID NO 75
<211> LENGTH: 25
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Sequence inserted into Gag SP1 domain
<400> SEQUENCE: 75
atccaactgg ggttgcaaaa atgtg 25
<210> SEQ ID NO 76
<211> LENGTH: 25
<212> TYPE: PRT
<213> ORGANISM: HIV-1
<400> SEQUENCE: 76
Gly His Lys Ala Arg Val Leu Ala Glu Ala Met Ser Gln Val Thr Asn
1 5 10 15
Pro Ala Thr Ile Met Ile Gln Lys Gly
20 25
<210> SEQ ID NO 77
<211> LENGTH: 19
<212> TYPE: PRT
<213> ORGANISM: FIV
<400> SEQUENCE: 77
Gly Tyr Lys Met Gln Leu Leu Ala Glu Ala Leu Thr Lys Val Gln Val
1 5 10 15
Val Gln Ser
<210> SEQ ID NO 78
<211> LENGTH: 16
<212> TYPE: PRT
<213> ORGANISM: EIAV
<400> SEQUENCE: 78
Lys Gln Lys Met Met Leu Leu Ala Lys Ala Leu Gln Thr Gly Leu Ala
1 5 10 15
<210> SEQ ID NO 79
<211> LENGTH: 36
<212> TYPE: PRT
<213> ORGANISM: BIV
<400> SEQUENCE: 79
Lys Ser Lys Met Gln Phe Leu Val Ala Ala Met Lys Glu Met Gly Ile
1 5 10 15
Gln Ser Pro Ile Pro Ala Val Leu Pro His Thr Pro Glu Ala Tyr Ala
20 25 30
Ser Gln Thr Ser
35
<210> SEQ ID NO 80
<211> LENGTH: 14
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: HIV-1 CA-SP1 sequence used for replacement
<400> SEQUENCE: 80
Gly His Lys Ala Arg Val Leu Ala Glu Ala Met Ser Gln Val
1 5 10
<210> SEQ ID NO 81
<211> LENGTH: 9
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: haemagglutinin epitope HA
<400> SEQUENCE: 81
Tyr Pro Tyr Asp Val Pro Asp Tyr Ala
1 5
<210> SEQ ID NO 82
<211> LENGTH: 6
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Bluetongue virus epitope VP7
<400> SEQUENCE: 82
Gln Tyr Pro Ala Leu Thr
1 5
<210> SEQ ID NO 83
<211> LENGTH: 8
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Flag epitope
<400> SEQUENCE: 83
Asp Tyr Lys Asp Asp Asp Asp Lys
1 5
<210> SEQ ID NO 84
<211> LENGTH: 11
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: VSV-G epitope
<400> SEQUENCE: 84
Tyr Thr Asp Ile Glu Met Asn Arg Leu Gly Lys
1 5 10
<210> SEQ ID NO 85
<211> LENGTH: 13
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Mutant SP1 region in HIV-1 NL4-3
<400> SEQUENCE: 85
Ala Glu Ala Met Ser Val Thr Asn Pro Ala Thr Ile Met
1 5 10
<210> SEQ ID NO 86
<211> LENGTH: 13
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Mutant SP1 region in HIV-1 NL4-3
<400> SEQUENCE: 86
Ala Glu Ala Met Ser Gln Thr Asn Pro Ala Thr Ile Met
1 5 10
<210> SEQ ID NO 87
<211> LENGTH: 13
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Mutant SP1 region in HIV-1 NL4-3
<400> SEQUENCE: 87
Ala Glu Ala Met Ser Gln Val Asn Pro Ala Thr Ile Met
1 5 10
<210> SEQ ID NO 88
<211> LENGTH: 13
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Mutant SP1 region in HIV-1 NL4-3
<400> SEQUENCE: 88
Ala Glu Ala Met Ser Gln Val Thr Pro Ala Thr Ile Met
1 5 10
<210> SEQ ID NO 89
<211> LENGTH: 13
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Mutant SP1 region in HIV-1 NL4-3
<400> SEQUENCE: 89
Ala Glu Ala Met Ser Gln Val Thr Asn Ala Thr Ile Met
1 5 10
<210> SEQ ID NO 90
<211> LENGTH: 1530
<212> TYPE: DNA
<213> ORGANISM: SIV DM
<400> SEQUENCE: 90
atgggcgtga gaaactccgt cttgtcaggg aagaaagcag atgaattaga aaaaattagg 60
ctacgaccca acggaaagaa aaagtacatg ttgaagcatg tagtatgggc agcaaatgaa 120
ttagatagat ttggattagc agaaagcctg ttggagaaca aagaaggatg tcaaaaaata 180
ctttcggtct tagctccatt agtgccaaca ggctcagaaa atttaaaaag cctttataat 240
actgtctgcg tcatctggtg cattcacgca gaagagaaag tgaaacacac tgaggaagca 300
aaacagatag tgcagagaca cctagtggtg gaaacaggaa caacagaaac tatgccaaaa 360
acaagtagac caacagcacc atctagcggc agaggaggaa attacccagt acaacaaata 420
ggtggtaact atgtccacct gccattaagc ccgagaacat taaatgcctg ggtaaaattg 480
atagaggaaa agaaatttgg agcagaagta gtgccaggat ttcaggcact gtcagaaggt 540
tgcaccccct atgacattaa tcagatgtta aattgtgtgg gagaccatca agcggctatg 600
cagattatca gagatattat aaacgaggag gctgcagatt gggacttgca gcacccacaa 660
ccagctccac aacaaggaca acttagggag ccgtcaggat cagatattgc aggaacaact 720
agttcagtag atgaacaaat ccagtggatg tacagacaac agaaccccat accagtaggc 780
aacatttaca ggagatggat ccaactgggg ttgcaaaaat gtgtcagaat gtataaccca 840
acaaacattc tagatgtaaa acaagggcca aaagagccat ttcagagcta tgtagacagg 900
ttctacaaaa gtttaagagc agaacagaca gatgcagcag taaagaattg gatgactcaa 960
acactgctga ttcaaaatgc taacccagat tgcaagctag tgctgaaggg gctgggtgtg 1020
aatcccaccc tagaagaaat gctgacggct tgtcaaggag taggggggcc gggacagaag 1080
gctagagtat tggcagaagc catgagtcag gtcctcgcac cagtgccaat cccttttgca 1140
gcagcccaac agaggggacc aagaaagcca attaagtgtt ggaattgtgg gaaagaggga 1200
cactctgcaa ggcaatgcag agccccaaga agacagggat gctggaaatg tggaaaaatg 1260
gaccatgtta tggccaaatg cccagacaga caggcgggtt ttttaggcct tggtccatgg 1320
ggaaagaagc cccgcaattt ccccatggct caagtgcatc aggggctgat gccaactgct 1380
cccccagagg acccagctgt ggatctgcta aagaactaca tgcagttggg caagcagcag 1440
agagaaaagc agagagaaag cagagagaag ccttacaagg aggtgacaga ggatttgctg 1500
cacctcaatt ctctctttgg aggagaccag 1530
<210> SEQ ID NO 91
<211> LENGTH: 67
<212> TYPE: PRT
<213> ORGANISM: SIV DM
<400> SEQUENCE: 91
Lys Asn Trp Met Thr Gln Thr Leu Leu Ile Gln Asn Ala Asn Pro Asp
1 5 10 15
Cys Lys Leu Val Leu Lys Gly Leu Gly Val Asn Pro Thr Leu Glu Glu
20 25 30
Met Leu Thr Ala Cys Gln Gly Val Gly Gly Pro Gly Gln Lys Ala Arg
35 40 45
Val Leu Ala Glu Ala Met Ser Gln Val Leu Ala Pro Val Pro Ile Pro
50 55 60
Phe Ala Ala
65
<210> SEQ ID NO 92
<211> LENGTH: 1530
<212> TYPE: DNA
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: GAG gene of SIV Chimera with Q358 changed
to
H358
<400> SEQUENCE: 92
atgggcgtga gaaactccgt cttgtcaggg aagaaagcag atgaattaga aaaaattagg 60
ctacgaccca acggaaagaa aaagtacatg ttgaagcatg tagtatgggc agcaaatgaa 120
ttagatagat ttggattagc agaaagcctg ttggagaaca aagaaggatg tcaaaaaata 180
ctttcggtct tagctccatt agtgccaaca ggctcagaaa atttaaaaag cctttataat 240
actgtctgcg tcatctggtg cattcacgca gaagagaaag tgaaacacac tgaggaagca 300
aaacagatag tgcagagaca cctagtggtg gaaacaggaa caacagaaac tatgccaaaa 360
acaagtagac caacagcacc atctagcggc agaggaggaa attacccagt acaacaaata 420
ggtggtaact atgtccacct gccattaagc ccgagaacat taaatgcctg ggtaaaattg 480
atagaggaaa agaaatttgg agcagaagta gtgccaggat ttcaggcact gtcagaaggt 540
tgcaccccct atgacattaa tcagatgtta aattgtgtgg gagaccatca agcggctatg 600
cagattatca gagatattat aaacgaggag gctgcagatt gggacttgca gcacccacaa 660
ccagctccac aacaaggaca acttagggag ccgtcaggat cagatattgc aggaacaact 720
agttcagtag atgaacaaat ccagtggatg tacagacaac agaaccccat accagtaggc 780
aacatttaca ggagatggat ccaactgggg ttgcaaaaat gtgtcagaat gtataaccca 840
acaaacattc tagatgtaaa acaagggcca aaagagccat ttcagagcta tgtagacagg 900
ttctacaaaa gtttaagagc agaacagaca gatgcagcag taaagaattg gatgactcaa 960
acactgctga ttcaaaatgc taacccagat tgcaagctag tgctgaaggg gctgggtgtg 1020
aatcccaccc tagaagaaat gctgacggct tgtcaaggag taggggggcc gggacataag 1080
gctagagtat tggcagaagc catgagtcag gtcctcgcac cagtgccaat cccttttgca 1140
gcagcccaac agaggggacc aagaaagcca attaagtgtt ggaattgtgg gaaagaggga 1200
cactctgcaa ggcaatgcag agccccaaga agacagggat gctggaaatg tggaaaaatg 1260
gaccatgtta tggccaaatg cccagacaga caggcgggtt ttttaggcct tggtccatgg 1320
ggaaagaagc cccgcaattt ccccatggct caagtgcatc aggggctgat gccaactgct 1380
cccccagagg acccagctgt ggatctgcta aagaactaca tgcagttggg caagcagcag 1440
agagaaaagc agagagaaag cagagagaag ccttacaagg aggtgacaga ggatttgctg 1500
cacctcaatt ctctctttgg aggagaccag 1530
<210> SEQ ID NO 93
<211> LENGTH: 67
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: GAG gene of SIV Chimera with Q358 changed
to
H358
<400> SEQUENCE: 93
Lys Asn Trp Met Thr Gln Thr Leu Leu Ile Gln Asn Ala Asn Pro Asp
1 5 10 15
Cys Lys Leu Val Leu Lys Gly Leu Gly Val Asn Pro Thr Leu Glu Glu
20 25 30
Met Leu Thr Ala Cys Gln Gly Val Gly Gly Pro Gly His Lys Ala Arg
35 40 45
Val Leu Ala Glu Ala Met Ser Gln Val Leu Ala Pro Val Pro Ile Pro
50 55 60
Phe Ala Ala
65
<210> SEQ ID NO 94
<211> LENGTH: 1353
<212> TYPE: DNA
<213> ORGANISM: FIV
<400> SEQUENCE: 94
atggggaatg gacaggggcg agattggaaa atggccatta agagatgtag taatgttgct 60
gtaggagtag gggggaagag taaaaaattt ggagaaggga atttcagatg ggccattaga 120
atggctaatg tatctacagg acgagaacct ggtgatatac cagagacttt agatcaacta 180
aggttggtta tttgcgattt acaagaaaga agagaaaaat ttggatctag caaagaaatt 240
gatatggcaa ttgtgacatt aaaagtcttt gcggtagcag gacttttaaa tatgacggtg 300
tctactgctg ctgcagctga aaatatgtat tctcaaatgg gattagacac taggccatct 360
atgaaagaag caggtggaaa agaggaaggc cctccacagg catatcctat tcaaacagta 420
aatggagtac cacaatatgt agcacttgac ccaaaaatgg tgtccatttt tatggaaaag 480
gcaagagaag gactaggagg tgaggaagtt caactatggt ttactgcctt ctctgcaaat 540
ttaacaccta ctgacatggc cacattaata atggccgcac cagggtgcgc tgcagataaa 600
gaaatattgg atgaaagctt aaagcaactg acagcagaat atgatcgcac acatccccct 660
gatgctccca gaccattacc ctattttact gcagcagaaa ttatgggtat aggattaact 720
caagaacaac aagcagaagc aagatttgca ccagctagga tgcagtgtag agcatggtat 780
ctcgaggcat taggaaaatt ggctgccata aaagctaagt ctcctcgagc tgtgcagtta 840
agacaaggag ctaaggaaga ttattcatcc tttatagaca gattgtttgc ccaaatagat 900
caagaacaaa atacagctga agttaagtta tatttaaaac agtcattgag catagctaat 960
gctaatgcag actgtaaaaa ggcaatgagc caccttaagc cagaaagtac cctagaagaa 1020
aagttgagag cttgtcaaga aataggctca ccaggccata aagcaagagt tttggctgaa 1080
gcaatgagcc aagtacaagt agtgcaatca aaaggatcag gaccagtgtg ttttaattgt 1140
aaaaaaccag gacatctagc aagacaatgt agagaagtga aaaaatgtaa taaatgtgga 1200
aaacctggtc atgtagctgc caaatgttgg caaggaaata gaaagaattc gggaaactgg 1260
aaggcggggc gagctgcagc cccagtgaat caaatgcagc aagcagtaat gccatctgca 1320
cctccaatgg aggagaaact attggattta taa 1353
<210> SEQ ID NO 95
<400> SEQUENCE: 95
000
<210> SEQ ID NO 96
<211> LENGTH: 1467
<212> TYPE: DNA
<213> ORGANISM: EIAV
<400> SEQUENCE: 96
atgggagacc ctttgacatg gagcaaggcg ctcaagaagt tagagaaggt gacggtacaa 60
gggtctcaga aattaactac tggtaactgt aattgggcgc taagtctagt agacttattt 120
catgatacca actttgtaaa agaaaaggac tggcagctga gggatgtcat tccattgctg 180
gaagatgtaa ctcagacgct gtcaggacaa gaaagagagg cctttgaaag aacatggtgg 240
gcaatttctg ctgtaaagat gggcctccag attaataatg tagtagatgg aaaggcatca 300
ttccagctcc taagagcgaa atatgaaaag aagactgcta ataaaaagca gtctgagccc 360
tctgaagaat atccaatcat gatagatggg gctggaaaca gaaattttag acctctaaca 420
cctagaggat atactacttg ggtgaatacc atacagacaa atggtctatt aaatgaagct 480
agtcaaaact tatttgggat attatcagta gactgtactt ctgaagaaat gaatgcattt 540
ttggatgtgg tacctggcca ggcaggacaa aagcagatat tacttgatgc aattgataag 600
atagcagatg attgggataa tagacatcca ttaccgaatg ctccactggt ggcaccacca 660
caagggccta ttcccatgac agcaaggttt attagaggtt taggagtacc tagagaaaga 720
cagatggagc ctgcttttga tcagtttagg cagacatata gacaatggat aatagaagcc 780
atgtcagaag gcatcaaagt gatgattgga aaacctaaag ctcaaaatat taggcaagga 840
gctaaggaac cttacccaga atttgtagac agactattat cccaaataaa aagtgaggga 900
catccacaag agatttcaaa attcttgact gatacactga ctattcagaa cgcaaatgag 960
gaatgtagaa atgctatgag acatttaaga ccagaggata cattagaaga gaaaatgtat 1020
gcttgcagag acattggaac tacaggccat aaagcaagag ttttggctga agcaatgagc 1080
caagtaactg gtcttgcggg cccatttaaa ggtggagcct tgaaaggagg gccactaaag 1140
gcagcacaaa catgttataa ctgtgggaag ccaggacatt tatctagtca atgtagagca 1200
cctaaagtct gttttaaatg taaacagcct ggacatttct caaagcaatg cagaagtgtt 1260
ccaaaaaacg ggaagcaagg ggctcaaggg aggccccaga aacaaacttt cccgatacaa 1320
cagaagagtc agcacaacaa atctgttgta caagagactc ctcagactca aaatctgtac 1380
ccagatctga gcgaaataaa aaaggaatac aatgtcaagg agaaggatca agtagaggat 1440
ctcaacctgg acagtttgtg ggagtaa 1467
<210> SEQ ID NO 97
<400> SEQUENCE: 97
000
<210> SEQ ID NO 98
<211> LENGTH: 1431
<212> TYPE: DNA
<213> ORGANISM: BIV
<400> SEQUENCE: 98
atgaagagaa gggagttaga aaagaagctt cgtaaggtta gggtgacacc ccaacaggat 60
aaatattata ctatagggaa tcttcaatgg gccattagaa tgataaatct aatggggatc 120
aaatgtgtgt gtgacgagga gtgctcggca gcagaggtag cccttatcat aacccaattt 180
tcagctttag acttagaaaa ttctcctatc agaggtaagg aggaggtggc cataaaaaat 240
actctgaagg ttttctggtc cctgctggcg gggtacaaac cagagagtac agaaacggcc 300
ctaggatatt gggaggcctt tacatataga gaaagggagg ccagagctga taaggaaggc 360
gaaattaaga gtatttaccc ttccctaaca cagaacacac agaataagaa gcagacatcg 420
aatcagacaa acactcaatc attaccagct atcactactc aagatggtac tcctaggttt 480
gatcctgacc tcatgaagca gcttaagatc tggtcagacg ccactgaaag aaatggggtt 540
gaccttcatg cagtgaatat attaggggtc attacagcaa acctagtaca ggaagaaatt 600
aaactcctct tgaatagtac acccaagtgg agattagatg tacaacttat agaatcaaaa 660
gtaagagaga aagaaaatgc ccacagaacg tggaaacagc atcatccaga agccccaaaa 720
acagatgaaa tcatcggtaa ggggcttagt tctgctgaac aagccaccct gatctcagta 780
gaatgcagag aaactttcag acagtgggtg ctgcaggcag ctatggaggt ggcacaggca 840
aaacatgcta ccccaggtcc catcaacatt catcagggac ccaaggagcc gtacacagac 900
tttataaata gattagtggc agcccttgaa ggtatggcgg ctccagaaac cacaaaagaa 960
tacttactcc aacatctatc tattgatcat gccaatgaag actgccagtc tattctaaga 1020
cctttgggac ccaacacccc aatggagaaa aaattagaag catgtagggt agtgggatct 1080
cagggccata aagcaagagt tttggctgaa gcaatgagcc aagtagggat ccaatcacca 1140
attccagcag tcttgcctca cacaccagaa gcatatgcct cccaaacctc agggcccgag 1200
gatggtagga gatgttacgg atgtgggaag acaggacatt tgaagaggaa ttgtaaacag 1260
caaaaatgct accattgtgg caaacctggc caccaagcaa gaaactgcag gtcaaaaaac 1320
gggaagtgct cctctgcccc ttatgggcag aggagccaac cacagaacaa ttttcaccag 1380
agcaacatga gttctgtgac cccatctgca ccccctctta tattagatta g 1431
<210> SEQ ID NO 99
<211> LENGTH: 10
<212> TYPE: DNA
<213> ORGANISM: Abudefduf declivifrons
<400> SEQUENCE: 99
aaaaaaaaaa 10
<210> SEQ ID NO 100
<211> LENGTH: 13
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Mutant SP1 region in HIV-1 NL4-3
<400> SEQUENCE: 100
Ala Glu Ala Met Ser Gln Val Thr Asn Pro Thr Ile Met
1 5 10
<210> SEQ ID NO 101
<211> LENGTH: 13
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Mutant SP1 region in HIV-1 NL4-3
<400> SEQUENCE: 101
Ala Glu Ala Met Ser Gln Val Thr Asn Pro Ala Ile Met
1 5 10
<210> SEQ ID NO 102
<211> LENGTH: 13
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Mutant SP1 region in HIV-1 NL4-3
<400> SEQUENCE: 102
Ala Glu Ala Met Ser Gln Val Thr Asn Pro Ala Thr Met
1 5 10
<210> SEQ ID NO 103
<211> LENGTH: 13
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: Mutant SP1 region in HIV-1 NL4-3
<400> SEQUENCE: 103
Ala Glu Ala Met Ser Gln Val Thr Asn Pro Ala Thr Ile
1 5 10
<210> SEQ ID NO 104
<211> LENGTH: 20
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: CA-SP1 in HIV-1 NL4-3 with HA epitope
<400> SEQUENCE: 104
Ala Glu Ala Met Ser Gln Val Thr Tyr Pro Tyr Asp Val Pro Asp Tyr
1 5 10 15
Ala Thr Ile Met
20
<210> SEQ ID NO 105
<211> LENGTH: 19
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: CA-SP1 in HIV-1 NL4-3 with HA epitope
<400> SEQUENCE: 105
Ala Glu Ala Met Ser Gln Val Thr Tyr Pro Tyr Asp Val Pro Asp Tyr
1 5 10 15
Ala Ile Met
<210> SEQ ID NO 106
<211> LENGTH: 19
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: CA-SP1 in HIV-1 NL4-3 with HA epitope
<400> SEQUENCE: 106
Ala Glu Ala Met Ser Gln Val Tyr Pro Tyr Asp Val Pro Asp Tyr Ala
1 5 10 15
Thr Ile Met
<210> SEQ ID NO 107
<211> LENGTH: 18
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: CA-SP1 in HIV-1 NL4-3 with HA epitope
<400> SEQUENCE: 107
Ala Glu Ala Met Ser Gln Val Tyr Pro Tyr Asp Val Pro Asp Tyr Ala
1 5 10 15
Ile Met
<210> SEQ ID NO 108
<211> LENGTH: 15
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: CA-SP1 in HIV-1 NL4-3 with blue-tongue
virus
VP7 epitope
<400> SEQUENCE: 108
Ala Glu Ala Met Ser Gln Val Gln Tyr Pro Ala Leu Thr Ile Met
1 5 10 15
<210> SEQ ID NO 109
<211> LENGTH: 17
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: CA-SP1 in HIV-1 NL4-3 with blue-tongue
virus
VP7 epitope
<400> SEQUENCE: 109
Ala Glu Ala Met Ser Gln Val Gln Tyr Pro Ala Leu Thr Ala Thr Ile
1 5 10 15
Met
<210> SEQ ID NO 110
<211> LENGTH: 17
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: CA-SP1 in HIV-1 NL4-3 with alpha-tubulin
epitope
<400> SEQUENCE: 110
Ala Glu Ala Met Ser Gln Val Thr Asn Pro Glu Glu Phe Ala Thr Ile
1 5 10 15
Met
<210> SEQ ID NO 111
<211> LENGTH: 18
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: CA-SP1 in HIV-1 NL4-3 with alpha-tubulin
epitope
<400> SEQUENCE: 111
Ala Glu Ala Met Ser Gln Val Thr Glu Glu Phe Glu Glu Phe Ala Thr
1 5 10 15
Ile Met
<210> SEQ ID NO 112
<211> LENGTH: 20
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: CA-SP1 in HIV-1 NL4-3 with flag epitope
<400> SEQUENCE: 112
Ala Glu Ala Met Ser Gln Val Thr Asp Tyr Lys Asp Asp Asp Asp Lys
1 5 10 15
Ala Thr Ile Met
20
<210> SEQ ID NO 113
<211> LENGTH: 18
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: CA-SP1 in HIV-1 NL4-3 with flag epitope
<400> SEQUENCE: 113
Ala Glu Ala Met Ser Gln Val Thr Asp Tyr Lys Asp Asp Asp Asp Lys
1 5 10 15
Ile Met
<210> SEQ ID NO 114
<211> LENGTH: 22
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: CA-SP1 in HIV-1 NL4-3 with flag epitope
<400> SEQUENCE: 114
Ala Glu Ala Met Ser Gln Val Thr Asn Pro Ala Thr Ile Asp Tyr Lys
1 5 10 15
Asp Asp Asp Asp Ile Met
20
<210> SEQ ID NO 115
<211> LENGTH: 23
<212> TYPE: PRT
<213> ORGANISM: Artificial Sequence
<220> FEATURE:
<223> OTHER INFORMATION: CA-SP1 in HIV-1 NL4-3 with VSV-G-tag
construct
<400> SEQUENCE: 115
Ala Glu Ala Met Ser Gln Val Thr Tyr Thr Asp Ile Glu Met Asn Arg
1 5 10 15
Leu Gly Lys Ala Thr Ile Met
20
User Contributions:
Comment about this patent or add new information about this topic:
| People who visited this patent also read: | |
| Patent application number | Title |
|---|---|
| 20160046565 | XYLYLENE DICARBAMATE, METHOD FOR PRODUCING XYLYLENE DIISOCYANATE, XYLYLENE DIISOCYANATE, AND METHOD FOR RESERVING XYLYLENE DICARBAMATE |
| 20160046564 | AMIDINE COMPOUND AND USE THEREOF |
| 20160046563 | SYNTHETIC BUILDING BLOCKS FOR THE PRODUCTION OF MATERIALS FOR ORGANIC ELECTROLUMINESCENCE DEVICES |
| 20160046562 | Styrenated Phenol Compound and Method of Preparing the Same |
| 20160046561 | SUBSTITUTED TETRACYCLINE COMPOUNDS |